Abstract
Conformational changes of proteins are essential to their functions. Yet it remains challenging to measure the amplitudes and timescales of protein motions. Here we show that the cytolysin A (ClyA) nanopore was used as a molecular tweezer to trap a single maltose-binding protein (MBP) within its lumen, which allows conformation changes to be monitored as electrical current fluctuations in real time. In contrast to the current two state binding model, the current measurements revealed three distinct ligand-bound states for MBP in the presence of reducing saccharides. Our analysis reveal that these three states represented MBP bound to different isomers of reducing sugars. These findings contribute to on the understanding of the mechanism of substrate recognition by MBP and illustrate that the nanopore tweezer is a powerful, label-free, single-molecule approach for studying protein conformational dynamics under functional conditions.
Keywords: nanopores, ionic current, structural dynamics, protein conformation, protein-ligand interactions, single-molecular detection
Proteins are dynamic entities that sample multiple conformations over a range of timescales.1,2 Understanding how proteins work requires a full description of their conformational dynamics over time. Structural approaches such as X-ray crystallography and electron microscopy, while yielding an abundance of information, are largely precluded from detailing time-dependent and transient protein dynamics. Nuclear magnetic resonance (NMR) spectroscopy can measure protein dynamics at multiple time scales,2,3 however, application is laborious and limited to relatively small proteins that can be stably prepared at high concentrations. Conformational heterogeneity, such as multiple weakly populated conformational states, is challenging to resolve from ensemble methods such as NMR due to averaging effects.4 Integrative approaches5,6 and single-molecule techniques7,8 can be advantageous for revealing transient protein dynamics and functional heterogeneities. For example, single-molecule fluorescence has revealed previously unknown enzyme conformational dynamics,9 transient intermediates,10 heterogeneity11 and stochasticity.12 Fluorophore labels, however, have limited photon flux, and may blink or photobleach over time, making very short-lived states or long duration monitoring more difficult.13 Electrical recording of single-molecule transient motions and intermediate states has also been achieved using field-effect transistors (FET’s),14 although the measurement still requires chemical labeling.
Nanopores have emerged as a powerful, single-molecule analytical tool to address fundamental problems in chemistry and biology, such as single molecular chemistry,15 DNA sequencing,16 protein sequencing,17 and protein detection and characterization.18–21 A nanopore detection platform is composed of a single hole in an insulating membrane that separates two electrolyte chambers. An analyte molecule interacting with the nanopore occludes the pore and causes a transient change in ionic current. Due to its sensitivity to the structure of biological molecules, nanopore analysis has been used to probe the structural flexibility and conformational change of proteins in solution.22–24 Taking advantage of the high temporal resolution (μs-ms) of the nanopore measurement, a biological nanopore Cytolysin A (ClyA) from Salmonella typhi (S. typhi) has been applied to confine a single protein within its lumen to continuously monitor fast action of substrate binding, or enzyme activities in real time.25–27 Current measurements from a ClyA nanopore from Escherichia coli (E.coli) have been shown to distinguish apo avidin from biotin-bound avidin,28 even though the root mean square deviation (RMSD) between the Cα atoms of the unbound and bound states is only 0.42 Å;29 this demonstrates that the nanopore approach has high spatial sensitivity and the potential to detect subtle differences between protein conformers.
Maltose-binding protein (MBP) is a periplasmic binding protein responsible for the uptake and efficient catabolism of maltodextrins in E. coli. MBP is ellipsoidal, comprised of two distinct globular domains with a ligand binding site located at the hinge region in-between two domains (Fig. 1a).30 Apo MBP adopts an open conformation where the groove between the domains is readily accessible to solvents and ligands.31 In the ligand bound state, MBP becomes more compact with the two domains moving towards each other by ~1 nm (Fig. 1a).31 The large-scale domain motion triggered by ligand-binding makes MBP a suitable model for studying the conformation dynamics by both ensemble and single molecule approaches, such as NMR and smFRET.13,32–34
Figure 1.
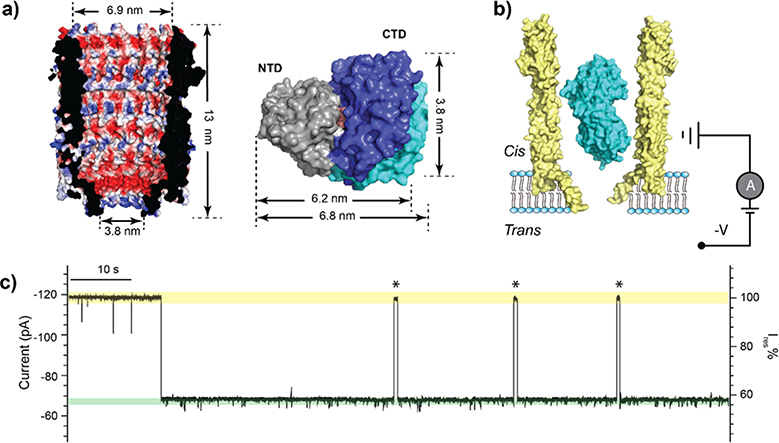
Trapping of MBP within the ClyA nanopore. (a) A cross-section view of the structure of the ClyA nanopore (PDB:2WCD) showing the electrostatic potential of its inner surface (left). Surface representation of MBP structures in the ligand-free (PDB:1JW4) and bound (PDB:1ANF) states (right). The two structures are aligned by the N-terminal domain (NTD, grey), with the C-terminal domains (CTD) shown in cyan and blue for the apo and holo states, respectively. The bound maltose is shown in orange. (b) Schematic representation of an MBP trapped in ClyA with negative potential applied on the trans side. (c) Representative current trace of ClyA in the presence of MBP. The open pore current is highlighted in green, while the blockades caused by MBP in yellow. Asterisks represent MBP escape events from the nanopore. Ires% represents the ratio of residual current to open pore current. The current traces were collected at −80 mV in the buffer 150 mM NaCl, 15 mM Tris-HCl, pH 7.5 in the presence of 56 nM MBP.
In this work, we apply the E. coli ClyA nanopore as a molecular tweezer to trap a single MBP and monitor its conformational changes during ligand binding. Our single-molecule recordings from the ClyA nanopore tweezer revealed the existence of three distinct ligand-bound states that has not been previously recognized. Combining molecule dynamic simulations and current measurements, we determine that these bound states are induced by the α- and β-isomers of maltooligosaccharides and quantify the binding affinities and kinetic rates of all complexes. Our findings contribute to the understanding of the mechanism of substrate recognition by MBP and provide a label-free framework for deploying the ClyA trapping approach to study the single-molecule protein dynamics in various biological processes.
Results
Confinement of a single MBP inside the ClyA nanopore
For single channel recording studies, ClyA pores were obtained by incubating ClyA monomers with n-dodecyl-β-maltoside.35 The pores were then added to the cis chamber of a recording apparatus. The resulting ClyA pores varied widely in conductance, which suggests that the pores are formed from oligomers of different stoichiometries and sizes.35,36 In this work, we chose to work with ClyA pores with a conductance of approximately 1.5 nS, which was the major population and appeared to be the most suitable for trapping MBP. The structure of the E. coli ClyA pore can be approximated as a conical frustum with an opening of 6.9 nm in diameter at the top and 3.8 nm at the bottom (Fig. 1a). Therefore, ClyA nanopores can accommodate a single MBP protein (~3.0 × 4.0 × 6.5 nm;3 see Fig. 1a) without it being translocated through. ClyA nanopores insert into lipid bilayers in a unidirectional manner due to the large soluble domain, with the nanopore lumen facing the cis chamber where both ClyA and MBP were added. When a negative voltage was applied to the trans side, current blockades were observed (Fig. 1b), indicating that an MBP was trapped in the pore. When a positive voltage was applied to the trans side, no current blockage was observed indicating that ClyA did not capture MBP. The inner surface of ClyA is predominantly negatively charged (Fig. 1a), and ClyA was previously determined to exhibit high cation selectivity.37 At pH 7.5, MBP is slightly negatively charged, which suggests that the electroosmotic force is dominant to drive MBP against the electrophoretic force. Our observation is in line with the previous trapping study with S. typhi ClyA that also showed the electroosmosis as the driving force for trapping proteinanalytes.25 The duration of MBP trapping is voltage dependent, with higher voltages leading to longer MBP trapping times (Supplementary Fig. 1). Appling voltages higher than −80 mV caused self-closure or gating of the ClyA nanopore; therefore, we applied a voltage of −80 mV throughout this study. The MBP trapping population induced current blockades with a residual current (Ires %) of 57.1 ± 1.3 % (n = 45, Fig. 1c). By measuring the dwell time (τoff = 178.0 ± 27.1 s, Supplementary Fig. 1) and inter-event interval (τon = 1.0 ± 0.1 s, Supplementary Fig. 1), we obtained a capture rate of (17.9 ± 1.8) μM−1 s−1 and an apparent affinity constant of (3.1 ± 0.6) x 10−10 M for the trapping of MBP within the ClyA nanopore.
Detection of MBP-ligand interaction within a nanopore
ClyA nanopores with trapped MBPs exhibited two types of current signals: one quiet (~60 % of the time) and one noisier with a smaller residual current (~40% of the time; Supplementary Fig. 2). We surmised that these two types of signals were induced by MBP that was trapped in ClyA in opposite orientations (Supplementary Fig. 2). Previous research has shown that proteins can adopt different orientations in nanopores.26 Relative to the ClyA lumen, MBP’s large diameter and ellipsoid geometry makes it unlikely change orientation once it is inside the nanopore (Fig. 1b). Calculated by using standard CHARMM charges, both MBP and ClyA appear to have large net dipole moments (~694.3 D for holo MBP, 773.7 D for apo MBP, and 7580.7 D for ClyA, shown in Supplementary Fig. 5a). The dipole-dipole interaction favors the N-terminal domain (NTD)–up orientation by ~0.3 kcal/mol under the experimental conditions, which is consistent with the observed percentages of quiet and noisy traces (Supplementary Fig. 2). We focused our analyses on MBPs trapped in the quiet, NTD–up orientation, because the resulting signals allowed more reliable data analysis.
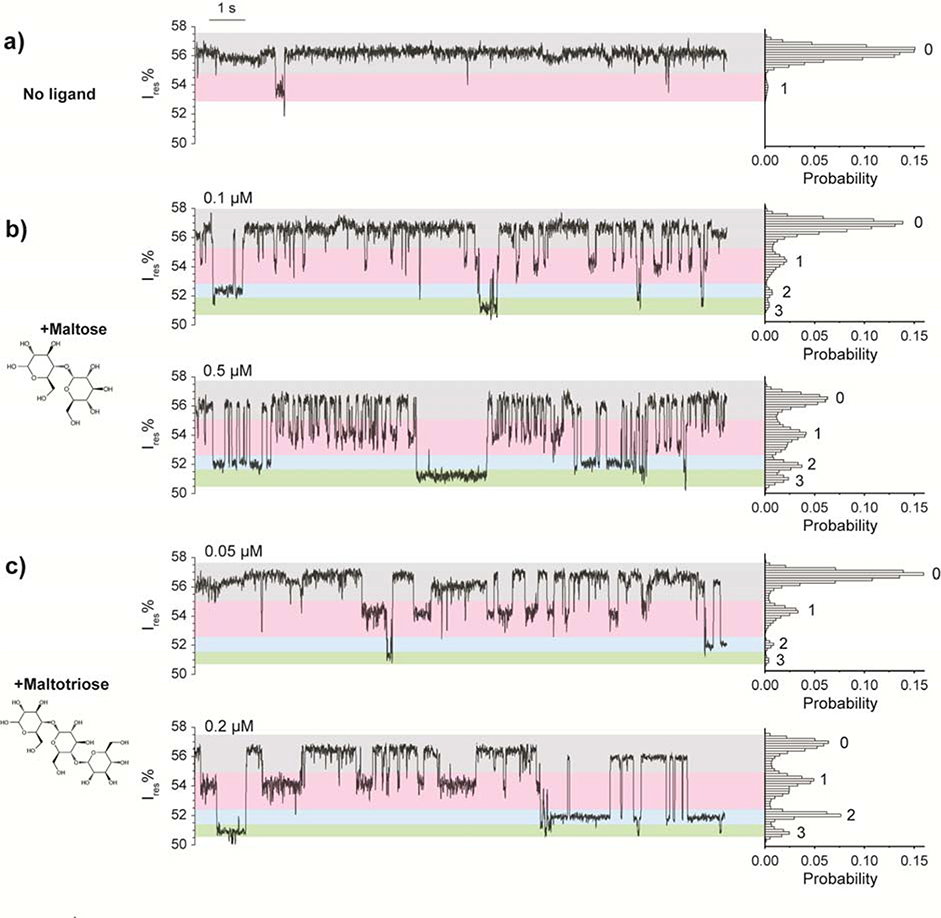
We observed that the apo MBP populated two states: state 0, a lower blockage state (57.1 ± 1.3 %, n= 45 MBP events) and state 1, a higher blockage state (54.9 ± 1.5 %, n= 39 MBP events) (Fig. 2a). State 0 occupied the majority of the recording time: 98.2 ± 2.4 %. These results may indicate spontaneous conformational fluctuations of MBP in the apo state and are consistent with NMR relaxation analyses showing that apo MBP can sample a “hidden” excited state with a population of ~5%.31 These results suggest that the nanopore tweezer method can resolve relatively subtle spontaneous protein conformational transitions.
Figure 2.
ClyA nanopore current responses induced by ligand interactions with trapped MBP. (a-c) Typical traces and corresponding histograms of ClyA pores containing a trapped MBP in the absence (a) and presence of ligand (b, c). Maltose or maltotriose was added to the cis chamber separately. The concentrations of maltose (b) and maltotriose (c) are indicated in the figure. All the measurements were performed in 150 mM NaCl, 15 mM Tris-HCl, pH 7.5 at −80 mV by applying a Bessel low-pass filter with a 2 kHz cutoff and sampled at 5 kHz.
MBP can bind specifically with maltose, maltodextrins, and cyclodextrins.33,34 To investigate if the nanopore can detect MBP/ligand binding, we added maltose or maltotriose to the recording chamber in various concentrations. In the presence of maltose, four major current states were observed: state 0, with a residual current of 56.5 ± 0.4 %; state 1, with a residual current of 54.0 ± 0.4 %; state 2, with a residual current of 52.0 ± 0.4 %; and state 3, with a residual current of 51.1 ± 0.4 % (Fig. 2b). Maltotriose induced four states that were almost identical to the states seen with maltose (Fig. 2c). Raising the maltose or maltotriose concentration increased the frequencies of states 1, 2, and 3 (Fig. 2b&c, Supplementary Fig. 3); this suggests that states 1, 2, and 3 represent the bound states of MBP. To exclude the possibility that the signal was caused by nonspecific binding of sugar molecules to the ClyA nanopore itself, we repeated the experiment without MBP and detected no changes in the current recording (data not shown). To further confirm that the observed signals were caused by specific ligand binding to the trapped MBP, we added sucrose, a non-substrate disaccharide, into the cis chamber and observed no signal change (Supplementary Fig. 4). Together, these results demonstrate that the ligand binding of MBP can be monitored by ClyA nanopore current recording at the single-molecule level in real time.
Molecular Dynamic Modeling and Simulation
X-ray crystallography shows that ligand-free MBP adopts a more open conformation than the ligand-bound MBP.30 Therefore, it would be reasonable to expect that apo MBP would cause a greater blockage and thus a lower residual current than bound MBP. However, the nanopore measurements showed that MBP bound to maltose or maltotriose blocked the nanopore more than the apo MBP did. We explored this unexpected result by performing molecular dynamics simulations to characterize how MBP interacts with ClyA.
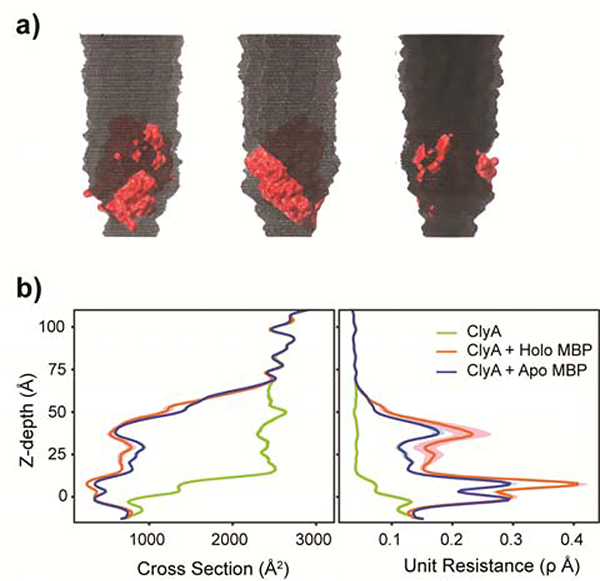
As expected, the modeling results showed that the membrane-embedded region of ClyA (from −15 to 15 Å in z-depth with 0 defined as the center of membrane) has the smallest solvated ion–accessible cross-section and gives rise to the greatest unit resistance, with or without trapped MBP (Fig. 3). However, trapped MBP forms additional constrictions above the membrane-embedded region (z-depth > 15 Å) (Fig. 3). These constricted regions have cross-sections comparable to the narrowest portion of the membrane-embedded region and contribute significantly to the net resistance, in contrast to the empty nanopore where the resistance is mainly attributed to the membrane-embedded region.38,39 This likely explains why the measured nanopore current depends sensitively on the conformational states of the trapped protein. The simulation further revealed that the holo MBP conformer, which is more compact, is trapped more deeply within the ClyA lumen than the apo conformer (Table 1), which leads to greater blockage of the current. The more elongated apo MBP is trapped in the higher, larger-diameter part of the lumen, thus leaving more solvated-ion accessible regions and gives rise to smaller unit resistance than bound MBP.
Figure 3.
Molecular basis of ClyA nanopore current blockades (a) Multiple views of the solvated ions accessible volume for ClyA nanopore with a trapped apo MBP. The color code for ClyA and MBP are grey and red respectively. Accessible volume of solvated ions inside the pore is shown in black. (b) Nanopore profile of solvated-ions accessible cross section and unit resistance. Results from holo and apo MBP simulations are shown in red and blue. The green line represents estimation for empty ClyA nanopore without trapped MBP. The colored shades are uncertainty estimated from the snapshots chosen periodically in the last 500 ns of the simulations. The unit of resistance is using unit resistance which is 100ρ. Here ρ is the physical parameter of charge density and assumed to be 1 in the estimation.
Table 1|.
Simulation
| Force | Conformation | Length | Depth | Net Unit Resistance* | |
|---|---|---|---|---|---|
| (kcal/mol/Å) | (μs) | (Å) | ClyA | ClyA + MBP | |
| 0.1 | apo | 2.0 | 29.3 ± 0.3 | 6.6 ± 0.0 | 15.7 ± 0.7 |
| 0.1 | holo | 2.0 | 30.5 ± 0.7 | 6.6 ± 0.0 | 13.3 ± 0.3 |
| 0.5 | apo | 2.0 | 26.5 ± 0.4 | 6.6 ± 0.0 | 22.1 ± 0.4 |
| 0.5 | holo | 1.0 | 27.4 ± 0.5 | 6.6 ± 0.0 | 17.0 ± 0.3 |
| 1.0 | apo | 1.0 | 26.3 ± 0.2 | 6.6 ± 0.0 | 22.6 ± 0.6 |
| 1.0 | holo | 1.0 | 27.2 ± 0.3 | 6.6 ± 0.0 | 17.2 ± 0.2 |
The last 500 ns of the simulation were used to calculate the z-depth and the net unit resistance. The net unit resistance is integrated the unit resistance through the whole pore. The uncertainty is due to the calculation using periodical snapshots in the last 500 ns.
Once net unit resistance is converted into current blockade, models predict that ClyA with trapped holo MBP will cause a 6 % to 10 % greater current blockade than ClyA with apo MBP, depending on the net drifting force constants used (which mimic the electroosmotic force under constant probing voltage; see Methods) (Table 1). The magnitudes of current reduction upon MBP trapping as well as ligand binding predicted by the simulation are in quantitative agreement with the actual measurements (Fig. 2). The model also indicated that when MBPs are trapped more deeply with a stronger force constant, the tilting angle of holo MBP is larger than that of apo MBP (Supplementary Fig. 5). This suggests that MBP adjusts its orientation as a result of nonspecific ClyA-MBP interactions and topology. These results suggest that it may be possible to resolve additional details of the conformational changes of a trapped protein by adjusting the applied voltage.
Anomer maltose-specific binding states
The current model of maltose-MBP binding has two states—bound and unbound. Therefore, we were surprised to find that the ClyA nanopore resistance recordings showed four states (Fig. 2b&c). One possible explanation for this result is that states 1, 2, and 3 represent a single holo MBP conformer that can become trapped within the lumen of the ClyA nanopore in three different configurations. If this was the case, the relative abundance of current states 1, 2, and 3 would be determined by the affinity constants of the MBP/ClyA complex in these alternative configurations and independent of the ligand concentration (Supplementary Fig. 6). However, our data clearly show that the relative abundance of the three states varies with ligand concentration (Fig. 2b&c).
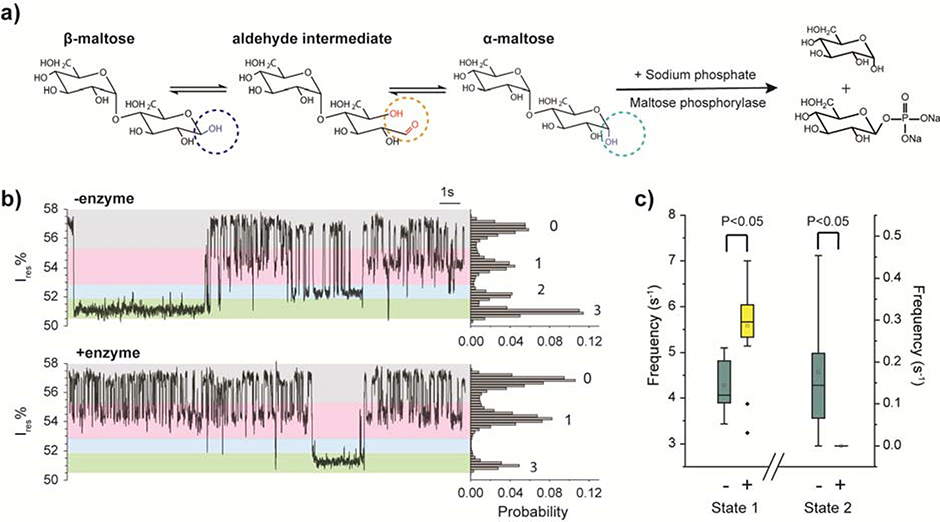
Another explanation for the additional states is that they result from an anomeric effect. Maltose is a reducing sugar with an α-anomer and a β-anomer that coexist in solution and mutarotate via an aldehyde intermediate (Fig. 4a). A previous NMR study showed the tritium-labeled α-maltose and β-maltose bound to MBP with different affinities.40 Inspired by this observation, we hypothesized that the multiple ligand-bound states represent MBP bound with different maltose anomers. We were not able to find a source of anomerically pure maltose to directly test the hypothesis. Instead, we introduced maltose phosphorylase into the buffer to deplete α-maltose in situ (Fig. 4a). Maltose phosphorylase is a 180 kDa enzyme (too large to be captured in ClyA) that specifically hydrolyzes α-maltose and generates glucose and β-D-glucose-1-phosphate (Fig. 4a).41 The spontaneous interconversion between α-maltose and β-maltose is a slow process with a rate constant of ~0.009 min−1 and a half-life of ~ 60 min.42 The turnover rate of 0.25 unit of maltose phosphorylase is 1.5 × 1017 min-1.43 Thus, the addition of maltose phosphorylase will both deplete the existing < 1 μM α-maltose in the recording chamber within one minute and prevent α-maltose from accumulating in a significant quantity. This results in a solution of primarily β-maltose. Control experiments showed that neither maltose phosphorylase nor its reaction products interfered with the current recording of MBP (Supplementary Fig. 4).
Figure 4.
Identification of anomeric binding to MBP by depletion of α-maltose with ClyA. (a) Reaction schemes of maltose mutarotation and maltose phosphorylase-catalyzed phosphorolysis of α-maltose. (b) Representative current traces and the corresponding all points histograms of a ClyA pore with a trapped MBP before and after removal of α-maltose by maltose phosphorylase. (c) Frequency of state 1and 2 before and after depletion of α-maltose. The two-tailed t-test was performed and the confidence interval is 95 %. Data were collected from 9 individual events before the depletion of α-maltose and 20 trapped MBP events after the depletion. Maltose and maltose phosphorylase were sequentially added to the cis chamber. All the measurements were performed in 150 mM NaCl, 15 mM Tris-HCl, pH 7.5 at −80 mV by applying a Bessel low-pass filter with a 2 kHz cutoff and sampled at 5 kHz.
After a single MBP protein was trapped inside the ClyA nanopore, we added 1 μM maltose and 10 μM sodium phosphate into the cis chamber and observed the typical trace with four current levels (Fig. 4b). Then 0.25 unit of maltose phosphorylase was added to the system, the current signal showed all three bound states for the first minute. After 1 min incubation the state 2 signal disappeared but state 1 and some state 3 events were still present (Fig. 4b bottom). The frequencies of state 1 and state 2 before and after the enzyme addition are summarized in Fig. 4c, with outliers removed on the basis of Grubbs’ test. Depletion of α-maltose led to the frequency of state 2 dropping from 0.18 ± 0.17 s−1 to 0 (n=4 pores). Because depletion of α-maltose led to a reduction in the state 2 signal, the data strongly suggest that state 2 represents MBP bound to the α-maltose anomer. In addition, we would expect β-maltose to bind to MBP more frequently after the α-maltose was depleted by the enzyme, because of the reduced competition from α-maltose, and indeed the frequency of state 1 increased from 4.28 ± 0.57 s−1 to 5.59 ± 0.89 s−1 after the α-maltose was depleted. For these reasons, we assigned state 1 as the β-maltose-bound state of MBP.
State 3 events were rare, with a frequency of 0.05 ± 0.04 s−1 before the depletion of α-maltose and 0.01 ± 0.02 s−1 after the depletion of α-maltose. Because of the relatively large uncertainty in these measurements, we cannot accurately assess the change in the abundance of state 3 before and after enzyme addition. Two possibilities were considered for the identities of state 3. One explanation is that state 3 results from MBP bound to an aldehyde intermediate. An equilibrated maltose solution contains 42 % α-maltose and 58 % β-maltose,44 as well as a very small fraction (0.0026 %) of aldehyde intermediate.45 The frequency of state 3 is significantly lower than that of the other two states, and thus it is possible that state 3 represents MBP bound to the aldehyde intermediate. Another possibility is that state 3 represents a second conformation of β-maltose bound to MBP. An NMR study of tritium-labeled maltose showed two separate chemical shifts for β-maltose upon interaction with MBP, which suggests that β-maltose can bind to MBP in two different modes.40 We investigated these two possibilities through the analysis of thermodynamic and kinetic parameters as described in the next section.
To further confirm that the three bound states were related to the anomeric forms of the ligand, we tested MBP bound with β-cyclodextrin, a ligand that does not undergo anomeric mutarotation because of its cyclic-ring structure.46 In the presence of β-cyclodextrin, we observed one major current level similar to state 1 of maltose. This indicates that β-cyclodextrin has only one binding mode with MBP (Supplementary Fig. 4). In addition, we observed that MBP bound to maltotetraose, another reducing ligand, exhibited three ligand-bound current levels similar to those of maltose and maltotriose (Supplementary Fig. 4). Taken together, these data strongly indicate that the multiple current levels of ligand-bound MBP are derived from its interaction with the α- and β-anomers of the reducing maltooligosaccharides.
Kinetic and thermodynamic analysis of MBP-ligand interactions
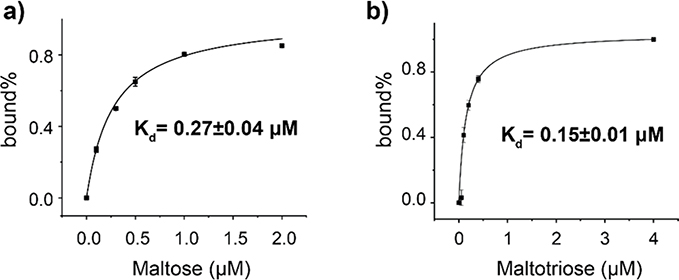
The ability of the nanopore tweezer to monitor the electrical current fluctuation for extended periods has allowed us to derive precise kinetic and thermodynamic parameters of MBP-ligand interactions. We first titrated trapped MBP with increasing concentrations of maltose or maltotriose (Table S1) and calculated the dissociation constant (Kd) of the ligand-MBP bound by using accumulative abundance of the three bound states (Table 2). The dissociation constants calculated by this method were 0.27 ± 0.04 μM (Fig. 5a) for maltose, which is similar to the 0.75 ± 0.13 μM measured by isothermal titration calorimetry in our lab (Supplementary Fig. 7) as well as reported values of 0.35 to 1.2 μM measured by fluorescence titration and isothermal titration calorimetry.30,47 The calculated Kd, 0.15 ± 0.01 μM (Fig. 5b), for maltotriose is comparable to the previously determined value of 0.16 to 0.7 μM.47 These nanopore-derived values are consistent with previous findings47 showing that maltotriose has a higher affinity for MBP than maltose.
Table 2|.
Kinetics and thermodynamics of MBP bound to different ligands
| Ligand | State | Anomer | Kd (μM) | koff (s−1) | kon (μM−1s−1) |
|---|---|---|---|---|---|
| Maltose | 1 | β | 0.31 ± 0.01 | 13.3 ± 1.6 | 43.6 ± 0.1 |
| 2 | α | 0.54 ± 0.05 | 3.5 ± 0.2 | 6.5 ± 0.1 | |
| 3 | β | 0.42 ± 0.02 | 1.3 ± 0.5 | 3.1 ± 0.4 | |
| Maltotriose | 1 | β | 0.12 ± 0.00 | 4.8 ± 0.3 | 40.8 ± 0.1 |
| 2 | α | 0.24 ± 0.01 | 1.6 ± 0.1 | 6.6 ± 0.1 | |
| 3 | β | 0.38 ± 0.02 | 1.9 ± 0.4 | 5.0 ± 0.2 | |
The errors represent the standard deviations of 5 independent pores for each ligand condition and 51 trapped MBPs.
Figure 5.
Determination of the binding affinities of MBP for maltose (a) and maltotriose (b). The MBP trapping experiments were performed in the presence of increasing concentration of maltose or maltotriose. All-point histograms of the traces, such as figure 2, were used to derive the percentage of the bound states calculates as the sum of state 1, 2 and 3 divided by the state 0. The dissociation constant (Kd) was derived by fitting the data with the single-ligand binding equation.
Because the three bound states were well resolved, we were also able to calculate the Kd for each anomeric species (Supplementary Fig. 8). The high temporal resolution of the nanopore-based current recording technique made it possible to capture transitions between states to derive the kinetic parameters. The dissociation rate constant, koff, was derived from the dwell time (τoff) of each binding model (Supplementary Fig. 9), which is independent of ligand concentration (Supplementary Fig. 10).
Quantification of kinetic parameters have also shed light on the identity of state 3. For this, we compared kinetic parameters calculated with two different assumptions, first, that state 3 represents MBP bound to an aldehyde intermediate of the sugar molecule, and second, that state 3 represents an alternate conformation of MBP bound to the β-anomer of the sugar molecule. Under the first assumption, we derived a kon of 6.9 ± 2.8 × 1010 M−1s−1, koff of 1.3 ± 0.5 s−1, and Kd of 1.9 ± 0.1 × 10−5 μM. The association rate calculated with this assumption was four orders of magnitude higher than that of the other two isomers and exceeds the known diffusion limit for any enzyme.48–51 This suggests that state 3 is highly unlikely to represent MBP bound to the aldehyde species. As such, we believe that state 3 may represent a second conformation of MBP bound to the β-anomer of maltose. Based on this assignment, the thermodynamic and kinetic parameters of all three states, are summarized in Table 2. These data show that the β-anomer exhibits the highest affinity for MBP when it adopts binding mode 1. Maltose has a higher Kd than maltotriose at each binding mode. Maltose and maltotriose have similar association rates but different dissociation rates, which results in their different binding affinities.
Conclusions
We applied the ClyA nanopore to capture a single MBP molecule and study its interaction with ligands over multiple cycles of association and dissociation. We observed that MBP could adopt three distinct binding modes upon interacting with ligands. Combining biochemical experiments and kinetic and thermodynamic analyses, we show that these three ligand-bound modes are induced by MBP binding to different anomeric forms of maltooligosaccharides. So far, X-ray crystallography obtained MBP structures with the preferential binding of only the α-form of linear maltooligosaccharides,30,52 despite 3H NMR studies with tritium-labeled ligands showed that MBP could bind to both α- and β-anomers.40 In this study, MBP has been observed to adopt different conformations upon binding to anomeric sugars. Nevertheless, the results can be explained by the structural arrangement of the substrate binding site within MBP. Regardless of the length of the oligosaccharides, the reducing glucose units of all oligosaccharides are buried in identical binding pockets that feature two amino acids, Asp 14 and Lys 15, which form hydrogen bonds with the α-anomeric hydroxyl (Supplementary Figure 11).30 The interactions between a β-maltooligosaccharide and Asp 14/Lys 15 would be disrupted because the β-anomic hydroxyl flips from the axial to the equatorial position. The loss of the two hydrogen bonds may cause the β-anomer–bound MBP to adopt a slightly different conformation than the α-anomer–bound MBP. The resulting difference in conformation of the sugar-MBP complex could be too small to be resolved by NMR;53 yet it generated distinct current levels measurable by the ClyA nanopore tweezer. Interestingly, the technique even detected two types of β-maltooligosaccharide bound states, supporting the previous observation by 3H NMR spectroscopic studies.33
Computer modeling and simulations have provided the molecular basis for the sensitivity of the ClyA trapping measurement to resolve conformations of the trapped MBP.54 The trapped protein in the lumen gives rise to additional constrictions above the membrane-embedded region, which can depend sensitively to the protein conformation (and configuration of entrapment). Specifically, the holo MBP, which is more compact than apo MBP, becomes trapped at a deeper location within the lumen of the ClyA nanopore and therefore causes a greater current blockade than apo MBP does. However, the present coarse-gained model did not reveal details about conformational differences between MBP bound with α-anomers and β-anomers. In the future, atomistic simulations would be useful to interpret the distinct current levels triggered by MBP bound to different anomers of the ligand. Particularly, the MD simulation of MBP may provide evidence for the two possible binding modes of β-anomers observed here and in the previous NMR study.
A major advantage of nanopore analysis of protein functional motion is that the time-resolved current fluctuations can be used to quantify thermodynamic and kinetic properties of the conformational transitions. Binding affinities of maltose and maltotriose for MBP calculated from nanopore measurements are highly consistent with reported values from multiple bulk assays including our isothermal titration calorimetry experiments. The kinetic rate constant kon and koff calculated from the ClyA experiments approximate the results obtained from the rapid-mixing, stopped flow technique.55 Our calculated values indicate that maltotriose has a higher affinity for MBP than maltose does, which is consistent with previous structural and biochemical analyses.30,47 Our data also show that within the similar binding mode, the kon values of maltose and maltotriose are very similar, while the koff of maltose is three times higher than that of maltotriose. These data could be readily explained by the structures of MBP because the additional glucose unit of maltotriose could make extra contact with the aromatic residues within the binding pocket of MBP.
The single-molecule ClyA nanopore tweezer study has further elucidated the anomic specificity of reduced maltooligosaccharides for MBP, a phenomenon which would be difficult to evaluate with other techniques because mutarotation occurs spontaneously in solution. Three anomeric maltooligosaccaride bound states exhibited current blockages at different levels with the major β-anomer state (state 1) showing the least blockage followed by the α-anomer bound state (state 2) and the minor β-anomer state (state 3) sequentially. Our simulation results revealed that the higher current state is associated with a more compact structure. Thus, the different current levels of the three ligand bound states indicate that the major β-anomer state has a more open structure than the α-anomer bound state. This result can be explained by the structure of ligand binding pocket that is consist of mostly aromatic residues (W230, Y155, W340, Y341) from the C-terminal domain (CTD) that stack with the hydrophobic glucose rings and polar residues (N12, D14, K15, E65) from the NTD that form H-bonding with the hydroxyl groups (Supplementary Fig. 11).52 In order for the D14 and K15 at the NTD to be within close proximity to the α-hydroxyl, the NTP needs to move close to the CTD to form a closed state. Thus the major β-anomer state probably forms at a less compacted “semi-closed” state that does not involve the hydrogen bonding with D14 and K15. This will explain our results that α-maltose and α-maltotriose had a slower koff values for MBP than the major binding state of β-maltose and β-maltotriose. Since the minor β-anomer bound state has the most compact form and the slowest koff, we speculate that NTD move even closer to the CTD to form a very tight binding pocket that even allows D14 and K15 to form H-bond with the β-anomeric hydroxyl. Interestingly, the association rate of the three states decrease with the increasing level of the blockages for both maltose and maltotriose. For example, the major β-anomer binding state, exhibited faster association rates by more than 3 times than the α-anomers for both maltose and maltotriose in our measurements. In nature, maltoporins have aromatic residues spiraling along its lumen to form the so-called “greasy ladder” which facilitates the maltose translocation.56 Here, we speculate that binding of the maltooligosacchride is initiated by the sugar sliding into the “greasy” aromatic pocket of the CTD which is followed by the two domains undergoing a large conformational change to move towards each other to allow the H-bonding formation of the NTD residues with the hydroxyl edge of the saccharide. To achieve the α-anomer bound state requires the conformational change to a larger scale than that to form the major β-anomer state, thus making α-anomer interaction a slower process. Similarly, the minor β-anomer bound state requires the conformational change to the largest scale, which causes this process to occur with the slowest rate. As a result, the major β-anomer binding mode shows a slightly higher affinity than α-anomer binding (Table 2). This observation is contradictory to results obtained from tritium NMR spectroscopy that show that α-maltose has a ~2.5-fold higher affinity for MBP than β-maltose.40 Here we observe that both the apo state 1 and the β-maltose bound state 1 has the same current level, which suggests that the two states have similar structures. Thus, we cannot exclude the possibility that the higher affinity of the β-anomer observed here may be because the trapped apo-MBP adopts conformational states that favor β-anomer association.
The ClyA tweezer platform deviates from classic nanopore sensing techniques in which analytes interact transiently with or translocate through nanopores. In this work, the funnel-shaped ClyA was used as a molecular tweezer to trap the analyte MBP for a duration (average ~200 s) long enough to observe the action of MBP over multiple cycles of ligand interactions. The label-free, single-molecule nanopore tweezer allows continuous monitoring of the protein’s motion at 10 μs to ms resolution. The approach could be modified to manipulate the trapping time by adjusting the applied potential. In addition, multiple analyte molecules could be probed separately by simply reversing the voltage to eject the trapped molecule and capture a new one. When combined with state-of-art multiple channel recording systems,57 the method is highly scalable and has the potential to become a high-throughput approach for monitoring protein-ligand interactions. Despite the advantages, the nanopore sensing technique described here has some limitations. Unlike the FRET and NMR, current signals do not contain distance information of the states that can be used for structure interpretation. Therefore, assigning the current states to biologically relevant conformational states can be difficult without previous knowledge of the conformers. To overcome the limitation, computational simulations will be essential to interpret the time-series of state transitions revealed by nanopore trapping measurements. It is noteworthy that the nanopore tweezer was previously used to describe an αHL nanopore detection platform in which an analyte molecule with a strong dipole can be trapped within the αHL lumen.58,59 To apply this approach, biological molecules would require further modification, such as by adding a positive and a negative tail at the two ends to introduce the dipole.58,59 Since the ClyA nanopore tweezer can trap an intact protein, this approach has the advantage of avoiding the interference of the modification on the functional dynamics of the protein analyte.
In summary, this study establishes the viability of applying the ClyA nanopore tweezer to investigate the conformational dynamics of MBP by continuously monitoring its motion with high temporal and spatial resolution. We believe that this approach will complement conventional smFRET and NMR methods and provide a valuable tool for comprehensive analysis of protein folding, enzymatic catalysis, and biomolecule recognition, as well as drug screening.
Methods
Protein preparation
The ClyA gene with C-terminus His6 was cloned into a pT7 plasmid vector using HindIII and Xhol restriction digestion sites. The MBP gene with N-terminus His8 was cloned into a pT7 plasmid vector using HindIII and NdeI restriction digestion sites. ClyA monomer and MBP were expressed and purified with the similar protocol according the previous study.35 Both plasmids were transformed into BL21(DE3) Competent E. coli cells to express proteins. Proteins were purified by using Ni-NTA affinity chromatography. The purified proteins were further purified by FPLC to remove the potential aggregates. Protein concentration was then determined by BCA assay and stored in −80 °C freezer for future use. The ClyA monomer was assembled into oligomer with 0.2 % DDM at room temperature for 20 min and then stored in 4 °C fridge for electrophysiology experiments.
Single channel planar bilayer recording experiments
Single channel recording is similar as described before.60 Briefly, a ClyA nanopore was inserted into a DPhPC lipid bilayer separating two chambers. Both chambers were filled with 900 μL buffer (150mM NaCl, 15 mM Tris-HCl, pH 7.5). ClyA was added from the cis chamber and inserted into the bilayer spontaneously. MBP was also added from the same chamber to be trapped in nanopore. −80 mV potential was applied across the bilayer and ionic current through nanopore was monitored in voltage-clamp mode by integrated patch clamp amplifier (Axopatch 200B, Molecular Devices). The signal was acquired by an analog-to-digital converter (Digidata 1440A, Molecular Devices) at a sampling rate of 5 kHz after processing with a 4-pole lowpass Bessel filter at 2 kHz. Data was recorded by Clampex software (Molecular Devices). Experiments were conducted at 23 °C.
For data analysis, all traces were filtered with a 50 Hz Gaussian filter. An in-house R script was developed to capture the state transitions of the current traces. Multi Gaussian fitting (mixtools in R) was then applied to fit the all-points histogram, and the peak value of current distribution was then determined. Different bound/unbound states were defined based on boundary of each peak, and assigned back to the original current value. Some current values overlapped between peak boundaries, and the treatment was to reassign the state to the previous state. The transition time of each state was then collected. Cumulated histograms of the transition time of the three bound states to the unbound state were used to calculate the mean first passage time as the average dwell time for each state.
Simulation System setups
Due to the large size, the system was represented using the MARTINI coarse-grained (CG) force field to allow sufficient sampling of potentially heterogeneous and nonspecific interactions between MBP and ClyA. MARTINI represents an approximate 4 to 1 mapping of heavy atoms to CG beads.61–63 Each CG bead is assigned a specific particle type based on the chemical nature of the underlying structure. The non-bonded interactions have been parameterized to capture to experimental thermodynamic data.61,62 Importantly, despite the CG nature, the MARTINI force field has been extensively parameterized provide a quantitative description of the thermodynamic balance of protein-water, protein-protein and water-water interactions. It has been successfully used to describe protein-protein interactions in both aqueous and membrane environments.64,65 Therefore, the MARTINI force field is expected to be appropriate for probing the interactions between MBP and ClyA.
Unbound MBP has a wider angle between two domains. The Cα RMSD between ligand bound and unbound MBP structures is 3.9 Å. The ClyA nanopore has 12 units of protomer, and each of them has 285 amino acid residues. The atomistic structures were mapped onto the MARTINI CG representation separately using CHARMM-GUI.66 MARTINI requires additional structural restrains to remain the protein structure. This was achieved by elastic network restraints with a force constant of 1.5 kcal/mol/Å2. The force constant was assigned by matching the flexibility of CG simulation to results from atomistic runs. For this, pioneer atomistic and CG simulations were performed to validate the force constant of restraining secondary structure in MARTINI CG model. In atomistic simulation, MBP was solvated in a cubic water box of 84.76 Å with TIP3P water.67 CHARMM was used to perform Langevin dynamics using CAHRMM 36m force field in constant temperature 298 K and constant pressure 1.0 atm.68,69 The SHAKE algorithm was applied to all hydrogen-containing bonds to allow a 2.0 fs timestep.70 Particle mesh Ewald was utilized for electrostatics with a real-space cutoff of 13 Å.71 Van der Waals interactions were cut off at 13 Å with a switching function between 12 and 13 Å. In CG simulation, MBP was solvated with MARTINI water box of 126.40 Å by 140.64 Å by 152.66 Å. NAMD 2.12 was used to perform the CG simulation with elastic network restraint (1.5 kcal/mol/Å2) applied to MBP. Parameters used are 5 fs timestep, switch distance cutoff 9.0 Å, real-space cutoff 12.0 Å, dielectric constant 15, and temperature 300 K. The martiniSwitching was on to enable the MARTINI Lennard-Jones switching function. The root-mean-square fluctuation (RMSF) profiles of MBP calculated from the last half of 50 ns atomistic and CG simulations is shown in Supplementary Fig. 5b. The RMSF comparison validates that the coarse-grained model is much stable than atomistic model for nonstructural region, but for the structural region, the CG model remains the same properties as atomistic simulation. That indicate the elastic network restraints in CG model do remain the protein structure property as atomistic model. The geometry of ClyA nanopore is well-characterized, and thus cylinder boundary condition with radius 62 Å and length 160 Å was used to reduce the computational cost for further production simulations. During the whole course of simulation, the MD parameters were set as described in the pioneer CG simulation. Additionally, position restraint with force constant 5 kcal/mol/Å2 was applied to MARTINI backbone atoms of ClyA nanopore to restraint the pore shape.
Generation of initial configurations and production simulations
To generate initial conformations with MBP captured inside the nanopore, MBP was first placed 30 Å above the pore entrance, and then 10 ns constant velocity pulling simulations were performed to guide MBP enter the pore. The pulling velocity was 0.00005 Å/timestep (i.e., 10 Å/ns) applied on the center of mass of MBP with direction to the bottom of the pore. Given the elongated geometry of MBP and the pore size, MBP can enter the ClyA pore in only two possible orientations, the N-terminus of MBP facing either up or down with the alignment to its the principal axis (Supplementary Fig. 5a shows the N-terminus up). The dipole moments of proteins were calculated using the PDB structures with standard CHARMM 36m atomic charges. The dipole-dipole interaction potentially provides a bias favoring the N-terminus up orientation by ~0.26 kcal/mol in aqueous solution. As such, the simulations have focused exclusively on cases where MBP is captured in the N-terminus up orientation. After the MBP entered the nanopore, 60 ns constant force pulling simulations (50 kcal/mol/Å) were performed to allow MBP travel deeper in the pore. The final coordinate of MBP inside the pore was then used as initial structure for production simulations. The initial z-depths of bound and unbound MBP are 26.6 Å and 25.9 Å, respectively. In production simulations, weak constant force pulling simulations (0.1, 0.5 and 1.0 kcal/mol/Å) were constantly performed to mimic the constant flow in the experiments. As MBP trapping free energy is about −12.6 kcal/mol from experiment, the proper force constant is arguable to be in the range of 0.1~0.2 kcal/mol/Å.
Data analyses
VMD was used for the structural visualizations and data analyses.72 The z-depth is measuring the center of mass of MBP, and the origin is at the center of member. Euler angles (φ, θ, and ψ) of MBP is using geometrical definition to identify the orientation in space. Due the pore symmetric, only the Euler angles θ needs to be considered and that reflects the tilting angle between MBP and nanopore. Both z-depth and Euler angles θ were then used as reaction coordinate for visualizing the free energy surface of sampling (Supplementary Fig. 5c). To estimate the solvated-ion cross section and unit resistance, the in-house python scripts were used. A box of gird points with 1 Å spacing was first built to cover the whole pore. The overlapped grids to the ClyA and MBP were then removed based on the radii of CG beads. We further considered chloride ion with an additional water probe as the solvated- ion with a radius of 3.2 Å to filter out the non-accessible grids. Finally, a filter was used to ensure the continuity of girds that contribute to conductance. The pore profiles of the solvated-ion cross section were then estimated based on the remaining grids (i.e., one grid ~ 1 Å2 cross section). Followed by the resistance approximation, the solvated-ion cross section can be converted to unit resistance, and the net resistance can be integrated through the whole pore:73
Here, length is the Z-depth of nanopore and for our estimation can be treated as 1 because the nanopore is the same. One assumption made here is charge density (ρ) doesn’t vary for apo-MBP and holo-MBP when they were trapped in nanopore. Thus, the unit resistance (ρÅ) has the reduced unit with ignoring the effect of the charge density. We equally chose 20 frames from the last 500 ns to get the statistic error of unit resistance estimation. Other analyses and plots were done with in-house scripts in R.74
Supplementary Material
Figure S1-11: supplement to computational methods, dwell time analysis of MBP trapped in ClyA, control experiments with no-substrate and non-reducing substrate, determination of the binding affinity and kinetic rates, structures of the ligand binding site of MBP.
Table S1: summary of the event numbers of all MBP trapped events.
Acknowledgement
This research was supported by the US National Institutes of Health grant R01-GM115442 (M Chen) and the US National Science Foundation grant No.1253565 (M Chen) and the US National Institutes of Health grant R01-GM114300 (J Chen).
Footnotes
Supporting Information
The supporting information is available free of charge on the ACS Publications website.
Associated Content
This work is associated with a preprint posted at biorxiv: https://www.biorxiv.org/content/10.1101/727503v1.75
References
- (1).Karplus M; Kuriyan J Molecular Dynamics and Protein Function. Proc. Natl. Acad. Sci. 2005, 102, 6679–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Henzler-Wildman K; Kern D Dynamic Personalities of Proteins. Nature 2007, 450, 964–972. [DOI] [PubMed] [Google Scholar]
- (3).Eisenmesser EZ; Bosco DA; Akke M; Kern D Enzyme Dynamics during Catalysis. Science 2002, 295, 1520–1523. [DOI] [PubMed] [Google Scholar]
- (4).Emwas AM The Strengths and Weaknesses of NMR Spectroscopy and Mass Spectrometry with Particular Focus on Metabolomics Research In Methods in Molecular Biology; Bjerrum JT, Ed.; Humana Press: New York, 2015; pp 161–193. [DOI] [PubMed] [Google Scholar]
- (5).van den Bedem H; Fraser JS Integrative, Dynamic Structural Biology at Atomic Resolution— It’s About Time. Nat Methods 2015, 12, 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Cerofolini L; Fragai M; Ravera E; Diebolder CA; Renault L; Calderone V Integrative Approaches in Structural Biology: A More Complete Picture from the Combination of Individual Techniques. Biomolecules 2019, 9, 0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Lerner E; Cordes T; Ingargiola A; Alhadid Y; Chung SY; Michalet X; Weiss S Toward Dynamic Structural Biology: Two Decades of Single-Molecule Förster Resonance Energy Transfer. Science 2018, 359, eaan1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Willems K; Van Meervelt V; Wloka C; Maglia G Single-Molecule Nanopore Enzymology. Philos. Trans. R. Soc. B 2017, 372, 20160230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Kapanidis AN; Margeat E; Ho SO; Kortkhonjia E; Weiss S; Ebright RH Initial Transcription by RNA Polymerase Proceeds through a DNA-Scrunching Mechanism. Science 2006, 314, 1144–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Ha T; Ting AY; Liang J; Caldwell WB; Deniz AA; Chemla DS; Schultz PG; Weiss S Single-Molecule Fluorescence Spectroscopy of Enzyme Conformational Dynamics and Cleavage Mechanism. Proc. Natl. Acad. Sci. U. S. A. 1999, 96, 893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Lu HP; Xun L; Xie XS Single-Molecule Enzymatic Dynamics. Science 1998, 282, 1877–1882. [DOI] [PubMed] [Google Scholar]
- (12).Cai L; Friedman N; Xie XS Stochastic Protein Expression in Individual Cells at the Single Molecule Level. Nature 2006, 440, 358–362. [DOI] [PubMed] [Google Scholar]
- (13).Kim E; Lee S; Jeon A; Choi JM; Lee HS; Hohng S; Kim HS A Single-Molecule Dissection of Ligand Binding to a Protein with Intrinsic Dynamics. Nat. Chem. Biol. 2013, 9, 313–318. [DOI] [PubMed] [Google Scholar]
- (14).Choi Y; Moody IS; Sims PC; Hunt SR; Corso BL; Perez I; Weiss GA; Collins PG Single-Molecule Lysozyme Dynamics Monitored by an Electronic Circuit. Science 2012, 335, 319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Cao J; Jia W; Zhang J; Xu X; Yan S; Wang Y; Zhang P; Chen HY; Huang S Giant Single Molecule Chemistry Events Observed from a Tetrachloroaurate(III) Embedded Mycobacterium Smegmatis Porin A Nanopore. Nat. Commun. 2019, 10, 5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Deamer D; Akeson M; Branton D Three Decades of Nanopore Sequencing. Nat. Biotechnol. 2016, 34, 518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Ouldali H; Sarthak K; Ensslen T; Piguet F; Manivet P; Pelta J; Behrends JC; Aksimentiev A; Oukhaled A Electrical Recognition of the Twenty Proteinogenic Amino Acids Using an Aerolysin Nanopore. Nat. Biotechnol. 2019, 019, 0345–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Oukhaled G; Mathé J; Biance a.-L. L.; Bacri L; Betton J-MM; Lairez D; Pelta J; Auvray L Unfolding of Proteins and Long Transient Conformations Detected by Single Nanopore Recording. Phys. Rev. Lett. 2007, 98, 98–101. [DOI] [PubMed] [Google Scholar]
- (19).Fahie MA; Yang B; Mullis M; Holden MA; Chen M Selective Detection of Protein Homologues in Serum Using an OmpG Nanopore. Anal. Chem. 2015, 87, 11143–11149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Thakur AK; Movileanu L Real-Time Measurement of Protein–Protein Interactions at Single-Molecule Resolution Using a Biological Nanopore. Nat. Biotechnol. 2018, 37, 96–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Harrington L; Alexander LT; Knapp S; Bayley H Single-Molecule Protein Phosphorylation and Dephosphorylation by Nanopore Enzymology. ACS Nano 2019, 13, 633–641. [DOI] [PubMed] [Google Scholar]
- (22).Asandei A; Schiopu I; Iftemi S; Mereuta L; Luchian T Investigation of Cu2+ Binding to Human and Rat Amyloid Fragments Aβ (1–16) with a Protein Nanopore. Langmuir 2013, 29, 15634–15642. [DOI] [PubMed] [Google Scholar]
- (23).Waduge P; Hu R; Bandarkar P; Yamazaki H; Cressiot B; Zhao Q; Whitford PC; Wanunu M Nanopore-Based Measurements of Protein Size, Fluctuations, and Conformational Changes. ACS Nano 2017, 11, 5706–5716. [DOI] [PubMed] [Google Scholar]
- (24).Hu R; Rodrigues JV; Waduge P; Yamazaki H; Cressiot B; Chishti Y; Makowski L; Yu D; Shakhnovich E; Zhao Q; Wanunu M Differential Enzyme Flexibility Probed Using Solid-State Nanopores. ACS Nano 2018, 12, 4494–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Soskine M; Biesemans A; Maglia G Single-Molecule Analyte Recognition with ClyA Nanopores Equipped with Internal Protein Adaptors. J. Am. Chem. Soc. 2015, 137, 5793–5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Van Meervelt V; Soskine M; Singh S; Schuurman-Wolters GK; Wijma HJ; Poolman B; Maglia G Real-Time Conformational Changes and Controlled Orientation of Native Proteins inside a Protein Nanoreactor. J. Am. Chem. Soc. 2017, 139, 18640–18646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Galenkamp NS; Soskine M; Hermans J; Wloka C; Maglia G Direct Electrical Quantification of Glucose and Asparagine from Bodily Fluids Using Nanopores. Nat. Commun. 2018, 9, 4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Lu B; Stokes C; Fahie M; Chen M; Golovchenko JA; Hau LV Protein Motion and Configurations in a Form-Fitting Nanopore: Avidin in ClyA. Biophys. J. 2018, 115, 801–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Livnah O; Bayer EA; Wilchek M; Sussman JL Three-Dimensional Structures of Avidin and the Avidin-Biotin Complex. Proc. Natl. Acad. Sci. U. S. A. 1993, 90, 5076–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Quiocho FA; Spurlino JC; Rodseth LE Extensive Features of Tight Oligosaccharide Binding Revealed in High-Resolution Structures of the Maltodextrin Transport/Chemosensory Receptor. Structure 1997, 5, 997–1015. [DOI] [PubMed] [Google Scholar]
- (31).Tang C; Schwieters CD; Clore GM Open-to-Closed Transition in Apo Maltose-Binding Protein Observed by Paramagnetic NMR. Nature 2007, 449, 1078–1082. [DOI] [PubMed] [Google Scholar]
- (32).Evenäs J; Tugarinov V; Skrynnikov NR; Goto NK; Muhandiram R; Kay LE Ligand-Induced Structural Changes to Maltodextrin-Binding Protein as Studied by Solution NMR Spectroscopy. J. Mol. Biol. 2001, 309, 961–974. [DOI] [PubMed] [Google Scholar]
- (33).Hall JA; Ganesan AK; Chen J; Nikaido H Two Modes of Ligand Binding in Maltose-Binding Protein of Escherichia Coli: Functional Significance in Active Transport. J. Biol. Chem. 1997, 272, 17615–17622. [DOI] [PubMed] [Google Scholar]
- (34).Staii C; Wood DW; Scoles G Ligand-Induced Structural Changes in Maltose Binding Proteins Measured by Atomic Force Microscopy. Nano Lett. 2008, 8, 2503–2509. [DOI] [PubMed] [Google Scholar]
- (35).Fahie M; Romano FB; Chisholm C; Heuck AP; Zbinden M; Chen M A Non-Classical Assembly Pathway of Escherichia Coli Pore-Forming Toxin Cytolysin A. J. Biol. Chem. 2013, 288, 31042–31051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Peng W; de Souza Santos M; Li Y; Tomchick DR; Orth K High-Resolution Cryo-EM Structures of the E. Coli Hemolysin ClyA Oligomers. PLoS One 2019, 14, e0213423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Ludwig A; Bauer S; Benz R; Bergmann B; Goebel W Analysis of the SlyA-Controlled Expression, Subcellular Localization and Pore-Forming Activity of a 34 KDa Haemolysin (ClyA) from Escherichia Coli K-12. Mol. Microbiol. 1999, 31, 557–567. [DOI] [PubMed] [Google Scholar]
- (38).Ficici E; Andricioaei I; Howorka S Dendrimers in Nanoscale Confinement: The Interplay between Conformational Change and Nanopore Entrance. Nano Lett. 2015, 15, 4822–4828. [DOI] [PubMed] [Google Scholar]
- (39).Asandei A; Ciuca A; Apetrei A; Schiopu I; Mereuta L; Seo CH; Park Y; Luchian T Nanoscale Investigation of Generation 1 PAMAM Dendrimers Interaction with a Protein Nanopore. Sci. Rep. 2017, 7, 6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Gehring K; Williams PG; Pelton JG; Morimoto H; Wemmer DE Tritium NMR Spectroscopy of Ligand Binding to Maltose-Binding Protein. Biochemistry 1991, 30, 5524–5531. [DOI] [PubMed] [Google Scholar]
- (41).Shirokane Y; Ichikawa K; Suzuki M A Novel Enzymic Determination of Maltose. Carbohydr. Res. 2000, 329, 699–702. [DOI] [PubMed] [Google Scholar]
- (42).Shirokane Y; Suzuki M A Novel Enzyme, Maltose 1-Epimerase from Lactobacillus Brevis IFO 3345. FEBS Lett. 1995, 367, 177–179. [DOI] [PubMed] [Google Scholar]
- (43).Miah F; Koliwer-Brandl H; Rejzek M; Field RA; Kalscheuer R; Bornemann S Flux through Trehalose Synthase Flows from Trehalose to the Alpha Anomer of Maltose in Mycobacteria. Chem. Biol. 2013, 20, 487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Hodge JE; Rendleman JA; Nelson EC Useful Properties of Maltose. Cereal Sci. Today 1972, 17, 180–188. [Google Scholar]
- (45).Los JM; Simpson LB; Wiesner K The Kinetics of Mutarotation of D-Glucose with Consideration of an Intermediate Free-Aldehyde Form. J. Am. Chem. Soc. 1956, 78, 1564–1568. [Google Scholar]
- (46).Bekiroglu S; Kenne L; Sandström C 1H NMR Studies of Maltose, Maltoheptaose, α-, β-, and γ-Cyclodextrins, and Complexes in Aqueous Solutions with Hydroxy Protons as Structural Probes. J. Org. Chem. 2003, 68, 1671–1678. [DOI] [PubMed] [Google Scholar]
- (47).Telmer PG; Shilton BH Insights into the Conformational Equilibria of Maltose-Binding Protein by Analysis of High Affinity Mutants. J. Biol. Chem. 2003, 278, 34555–34567. [DOI] [PubMed] [Google Scholar]
- (48).Li TT; Chou KC The Quantitative Relations between Diffusion-Controlled Reaction Rate and Characteristic Parameters in Enzyme-Substrate Reaction Systems. I. Neutral Substrates. Sci. Sin. 1976, 19, 117–136. [PubMed] [Google Scholar]
- (49).Kuo-Chen C The Kinetics of the Combination Reaction between Enzyme and Substrate. Sci. Sin. 1976, 19, 505–528. [PubMed] [Google Scholar]
- (50).Chou KC; Zhou GP Role of the Protein Outside Active Site on the Diffusion-Controlled Reaction of Enzyme. J. Am. Chem. Soc. 1982, 104, 1409–1413. [Google Scholar]
- (51).Koenig SH; Brown RD; Brewer CF Solvent Proton Magnetic Relaxation Dispersion in Solutions of Concanavalin A. Proc. Natl. Acad. Sci. 1972, 70, 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Duan X; Hall JA; Nikaido H; Quiocho FA Crystal Structures of the Maltodextrin/Maltose-Binding Protein Complexed with Reduced Oligosaccharides: Flexibility of Tertiary Structure and Ligand Binding. J. Mol. Biol. 2001, 306, 1115–1126. [DOI] [PubMed] [Google Scholar]
- (53).Gehring K; Zhang X; Hall J; Nikaido H; Wemmer DE An NMR Study of Ligand Binding by Maltodextrin Binding Protein. Biochem. Cell Biol. 1998, 76, 189–197. [DOI] [PubMed] [Google Scholar]
- (54).Bucher D; Grant BJ; McCammon JA Induced Fit or Conformational Selection? The Role of the Semi-Closed State in the Maltose Binding Protein. Biochemistry 2011, 50, 10530–10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Miller DM; Olson JS; Pflugrath JW; Quiocho FA Rates of Ligand Binding to Periplasmic Proteins Involved in Bacterial Transport and Chemotaxis. J. Biol. Chem. 1983, 258, 13665–13672. [PubMed] [Google Scholar]
- (56).Pothula KR; Solano CJF; Kleinekathöfer U Simulations of Outer Membrane Channels and Their Permeability. Biochim. Biophys. Acta, Biomembr. 2016, 1858, 1760–1771. [DOI] [PubMed] [Google Scholar]
- (57).Lu H; Giordano F; Ning Z Oxford Nanopore MinION Sequencing and Genome Assembly. Genomics, Proteomics Bioinf. 2016, 14, 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Asandei A; Chinappi M; Lee JK; Seo CH; Mereuta L; Park Y; Luchian T Placement of Oppositely Charged Amino Acids at a Polypeptide Termini Determines the Voltage-Controlled Braking of Polymer Transport through Nanometer-Scale Pores. Sci. Rep. 2015, 5, 10419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Chinappi M; Luchian T; Cecconi F Nanopore Tweezers: Voltage-Controlled Trapping and Releasing of Analytes. Phys. Rev. E. 2015, 92, 032714. [DOI] [PubMed] [Google Scholar]
- (60).Fahie M; Chisholm C; Chen M Resolved Single-Molecule Detection of Individual Species within a Mixture of Anti-Biotin Antibodies Using an Engineered Monomeric Nanopore. ACS Nano 2015, 9, 1089–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Marrink SJ; Risselada HJ; Yefimov S; Tieleman DP; De Vries AH The MARTINI Force Field: Coarse Grained Model for Biomolecular Simulations. J. Phys. Chem. B 2007, 111, 7812–7824. [DOI] [PubMed] [Google Scholar]
- (62).Marrink SJ; Tieleman DP Perspective on the Martini Model. Chem. Soc. Rev 2013, 42, 6801. [DOI] [PubMed] [Google Scholar]
- (63).Monticelli L; Kandasamy SK; Periole X; Larson RG; Tieleman DP; Marrink SJ The MARTINI Coarse-Grained Force Field: Extension to Proteins. J. Chem. Theory Comput. 2008, 4, 819–834. [DOI] [PubMed] [Google Scholar]
- (64).Marzinek JK; Holdbrook DA; Huber RG; Verma C; Bond PJ Pushing the Envelope: Dengue Viral Membrane Coaxed into Shape by Molecular Simulations. Structure 2016, 24, 1410–1420. [DOI] [PubMed] [Google Scholar]
- (65).Marrink SJ; de Vries AH; Mark AE Coarse Grained Model for Semiquantitative Lipid Simulations. J. Phys. Chem. B 2004, 108, 750–760. [Google Scholar]
- (66).Qi Y; Ingólfsson HI; Cheng X; Lee J; Marrink SJ; Im W CHARMM-GUI Martini Maker for Coarse-Grained Simulations with the Martini Force Field. J. Chem. Theory Comput. 2015, 11, 4486–4494. [DOI] [PubMed] [Google Scholar]
- (67).Jorgensen WL; Chandrasekhar J; Madura JD; Impey RW; Klein ML Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 174501. [Google Scholar]
- (68).Huang J; Rauscher S; Nawrocki G; Ran T; Feig M; De Groot BL; Grubmüller H; MacKerell AD CHARMM36m: An Improved Force Field for Folded and Intrinsically Disordered Proteins. Nat. Methods 2016, 14, 71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Van Gunsteren WF; Berendsen HJC; Rullmann JAC Stochastic Dynamics for Molecules with Constraints Brownian Dynamics of n-Alkanes. Mol. Phys. 1981, 44, 69–95. [Google Scholar]
- (70).Ryckaert JP; Ciccotti G; Berendsen HJC Numerical Integration of the Cartesian Equations of Motion of a System with Constraints: Molecular Dynamics of n-Alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar]
- (71).Darden T; York D; Pedersen L Particle Mesh Ewald: An N·log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar]
- (72).Humphrey W; Dalke A; Schulten K VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [DOI] [PubMed] [Google Scholar]
- (73).Smart OS; Neduvelil JG; Wang X; Wallace BA; Sansom MSP HOLE: A Program for the Analysis of the Pore Dimensions of Ion Channel Structural Models. J. Mol. Graph. 1996, 14, 354–360. [DOI] [PubMed] [Google Scholar]
- (74).R Core Team (2018). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austrailia: Available online at https://www.R-project.org/. [Google Scholar]
- (75).Li X; Lee K; Chen J; Chen M A ClyA Nanopore Tweezer for Analysis of Functional States of Protein-Ligand Interactions. bioRxiv 2019. https://www.biorxiv.org/content/10.1101/727503v1, (accessed August 6th 2019)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1-11: supplement to computational methods, dwell time analysis of MBP trapped in ClyA, control experiments with no-substrate and non-reducing substrate, determination of the binding affinity and kinetic rates, structures of the ligand binding site of MBP.
Table S1: summary of the event numbers of all MBP trapped events.