Abstract
Advances in next generation sequencing have transformed our ability to identify genetic variants associated with clinical disorders of the musculoskeletal system. However, the means to functionally validate and analyze the physiological repercussions of genetic variation have lagged behind the rate of genetic discovery. The zebrafish provides an efficient model to leverage genetic analysis in an in vivo context. Its utility for orthopaedic research is becoming evident in regard to both candidate gene validation as well as therapeutic discovery in tissues such as bone, tendon, muscle, and cartilage. With the development of new genetic and analytical tools to better assay aspects of skeletal tissue morphology, mineralization, composition, and biomechanics, researchers are emboldened to systematically approach how the skeleton develops and to identify the root causes, and potential treatments, of skeletal disease.
Keywords: zebrafish, bone, tendon, muscle, cartilage
1. INTRODUCTION
Small animal models amenable to rapid-throughput biology are needed to accelerate the discovery of new treatments for clinical disorders of the musculoskeletal system. Complex, multi-cellular interactions are difficult to recapitulate in a dish. While such processes can be studied in animal models, ready-made mutant lines often do not exist (e.g., see section 5.3). The zebrafish (Danio rerio) is a small, tropical freshwater fish that, by virtue of its unique experimental attributes (e.g., small size, low cost, genetic tractability, and optical transparency), has opened powerful avenues for biomedical research (including studies of development 1; 2, neuroscience 3, regeneration 4, and disease 5) that are difficult in other vertebrate models. Such avenues include in vivo imaging of cell dynamics, and genetic and chemical screens. Moreover, zebrafish can be used as a pre-screening tool to prioritize more labor and cost-intensive studies that require de novo mutant mouse generation. The potential benefits of incorporating zebrafish into a research program must be weighed with limitations, including infrastructure costs (which vary depending on institution), differences in zebrafish and human genetics and physiology, and the fact that many experimental approaches are still in their infancy, and thus remain to be rigorously validated. Indeed, the use of zebrafish to understand clinical disorders of the musculoskeletal system has only begun to be established. Here, we introduce experimental advantages of the zebrafish, discuss its genetic and physiological similarities and differences to humans, and survey recent applications to musculoskeletal development and disease. This review elaborates on a workshop of the same name at the 2019 Orthopaedic Research Society Meeting, conducted by the authors, the purpose of which was to introduce emerging orthopaedic research in zebrafish to facilitate cross-talk, establish foundations, and develop new models of clinical disorders.
2. GENETICS
2.1. Genetic similarity to humans
A key criterion in the selection of an appropriate disease model is its genetic similarity to humans. Approximately 71% of human protein coding genes possess at least one zebrafish ortholog 6. This is comparable to mouse, as ~80–90% of human protein coding genes possess at least one ortholog in mouse (http://www.informatics.jax.org/homology.shtml). Because zebrafish arose from a common ancestor that underwent an additional round of whole-genome duplication relative to mice and humans, zebrafish can have multiple co-orthologs for human genes (e.g., human RUNX2 has two zebrafish co-orthologs, runx2a and runx2b). While this can complicate testing of gene function due to issues such as functional redundancy, such difficulties can be alleviated through simultaneous knockdown or knockout of co-orthologs 1; 7. In other cases, maintenance of two copies of a gene in the zebrafish often is balanced by partitioning function of a gene, or subfunctionalization. As such, the retention of co-orthologs in the zebrafish often permits nuanced analysis of gene function in genes that might be lethal in mice. In mouse, analysis of many genes often requires combinatorial genetic techniques to provide conditional spatial or temporal regulation of gene function, whereas in the zebrafish simple genetic alterations can be studied (e.g. Fgfr1 8). The often partitioned function of paralogues in the zebrafish also permits loss-of-function analysis to model effect of more nuanced alleles such as regulatory shifts in gene function underlying many common skeletal pathologies. Such pathologies cannot easily be modeled with knock out strategies in the mouse and due to the complex anatomical nature of many skeletal disorders, often cannot be modeled in vitro even when allele-specific cell lines are constructed. A large number of skeletal disease models have been identified in zebrafish (recently reviewed in 9–11), and this number is steadily increasing.
2.2. Techniques
The ease of forward genetics in zebrafish gives this model an advantage for unbiased discovery of mutant phenotypes. N-ethyl-N-nitrosourea (ENU) mutagenesis screens in zebrafish have uncovered a large number of variants relevant to fundamental aspects of skeletogenesis 12–14, morphological evolution 15, and human skeletal diseases 16–20. Early examples involved the identification of a large collection of mutants with defects in formation of the jaw and branchial arches 12–14. The same screen also identified mutations specifically affecting the adult form 21. The mutants identified in these early large-scale screens served as foundation for experimental analysis and were proof that not only could specific mechanisms of skeletal development be identified, but that mutations could be in genes homologous to human genes associated with skeletal diseases. Such screens often yield specific defined point mutations which provide nuanced changes in gene function that simple loss-of-function mutations or frameshifts cannot. Screens for dominant mutations affecting skeletogenesis in which mutations often lead to dominant negative properties as well as alleles with increased functions (hyper- or neomorphic alleles), are proving to be informative and useful in disease modeling. For example, recent screens have identified dominant mutants closely mirroring collagenopathies and Osteogenesis Imperfecta (OI) (col1a1a/b, col1a2), Adams-Oliver Syndrome (dll4), and Hyperhidrotic Ectodermal dysplasia (edar) 16. Finally, the Zebrafish Mutation Project22, which has phenotyped a large number of zebrafish mutant alleles and made them available to the community, demonstrates the feasibility of systematic genome-wide analysis.
In addition to forward genetics, zebrafish are also readily amenable to reverse genetics, i.e., testing for phenotypic consequences following the targeted interference of gene function. The advent of TALEN-23 and CRISPR-based gene editing has substantially expanded the means by which the scientific community can approach reverse genetics in zebrafish. For gene editing using CRISPR, administration of Cas9:gRNA ribonucleoproteins (RNPs) generates double stranded breaks at defined loci. Errors in the non-homologous end joining (NHEJ) repair mechanism lead to insertions and deletions (indels) at the cut site, often leading to loss of function (e.g., due to non-sense mediated decay triggered by a premature stop codon). Alternatively, multiple RNPs can be used to induce site-spanning deletions that delete promoter regions or entire gene loci, which may help reduce activation compensatory pathways triggered by mRNA degradation 24. Moreover, Cas9:gRNAs can be co-injected with a donor template which, following homology directed repair (HDR), can result in precise gene edits. Because zebrafish develop externally, hundreds of embryos can be injected by a single user in one morning. This allows for efficient creation of induced mutations or replacements at specific genetic loci. Screening phenotypes in injected G0 founder “crispant” animals can further enable rapid and cost-effective assessment of gene function 1. In addition to alleviating the time and resources needed to breed alleles to homozygosity, G0 screens are also amenable to multiplexing strategies in which multiple genes are targeted in the same animal. The ability to detect adult skeletal phenotypes in G0 zebrafish for genes associated with recessive forms of OI (bmp1a and plod2) and genetic risk for osteoporosis (wnt16) was recently demonstrated 25.
Zebrafish also provide a versatile system to test gene function through use of transgenesis. This allows for stable or inducible (e.g., heat shock-induced) protein expression, or conditional gene targeting (e.g., Cre-mediated recombination). The Tol2 transposon system is commonly used for introducing transgenes. There exists a large, growing panel of zebrafish fluorescent reporter lines for cell types within the musculoskeletal system (Table 1). As zebrafish are also relatively transparent and develop externally, development can be easily observed in real time. With these two attributes, use of transgenic reporters for particular cell types and proteins have provided an unmatched ability to visualize the dynamics of skeletal patterning and regeneration. These advantages also permit the visualization of cell behaviors in specific genetic contexts to gain mechanistic understanding of disease etiology.
Table 1:
Transgenic lines for visualizing cells relevant to the musculoskeletal system.
| Tissue/cell type | Transgene | Reference | Notes |
|---|---|---|---|
| Early neural crest and CNC-derived cartilages | Tg(sox10:GFP) | 119 | |
| Early neural crest and CNC-derived cartilages | Tg(sox10-CreERt2) | 120 | Tamoxifen-inducible Cre |
| Cartilage/chondrocytes | Tg(col2a1:eGFP) | 121; 122 | |
| Cartilage/chondrocytes | Tg(Col2a1aBAC:mcherry) | 123 | |
| Cartilage/chondrocytes | Tg(1.7c2a1a:mEGFP) | 121 | Membrane-tagged EGFP |
| Tendon cells | Tg(scx:mCherry) | 49 | |
| Muscle cells | Tg(−0.5unc45b:mCherry) | 124 | |
| Bone/Pre-osteoblasts | Tg(runx2:GFP) | 125 | |
| Bone/Osteoblasts | TgBAC(col10a1a:Citrine) | 126 | |
| Bone/Osteoblasts | Tg(sp7:EGFP) | 127 | |
| Bone/Osteoblasts | Tg(osx:GFP) | 127 | |
| Bone/Osteoblasts | Tg(osx:CreERt2) | 74 | Tamoxifen-inducible Cre |
| Bone/Osteoblasts | Tg(Ola.Sp7:NLS-GFP) | 128 | Nuclear localization signal-tagged GFP |
| Bone/Osteoblasts | TgBAC(entpd5a:Citrine) | 129 | |
| Bone/Mature osteoblasts | Tg(ocn:GFP) | 74 | |
| Bone/Osteoclasts | Tg(ctsk:YFP) | 130 | |
| Bone/Osteoclasts | Tg(ctsk:DsRed) | Personal communication |
Because of their small size and low cost, zebrafish are also amenable to drug discovery via chemical screens. In such screens, large libraries of small molecules are tested to identify specific compounds that affect gene function or developmental processes. In a typical screen, zebrafish embryos/larvae are dispensed into 48 or 96 well plates, drugs are administered by adding them to the water, and phenotypes are assessed (e.g., via morphological, fluorescent, or behavioral readouts). This strategy can be adapted to adults 26; 27. The identification of dorsomorphin as a selective inhibitor of BMP type I receptors was discovered in a large zebrafish chemical screen and led to the development of analogs for treatment of heterotopic ossification 28. In another screen, phosphodiesterase (PDE) inhibitors were found to alter phenotypes in a zebrafish model of Duchenne muscular dystrophy 29. See 30 for a recent review of chemical screens in zebrafish.
3. FORMATION AND INTEGRATION OF THE ZEBRAFISH MUSCULOSKELETAL SYSTEM
3.1. Development and patterning
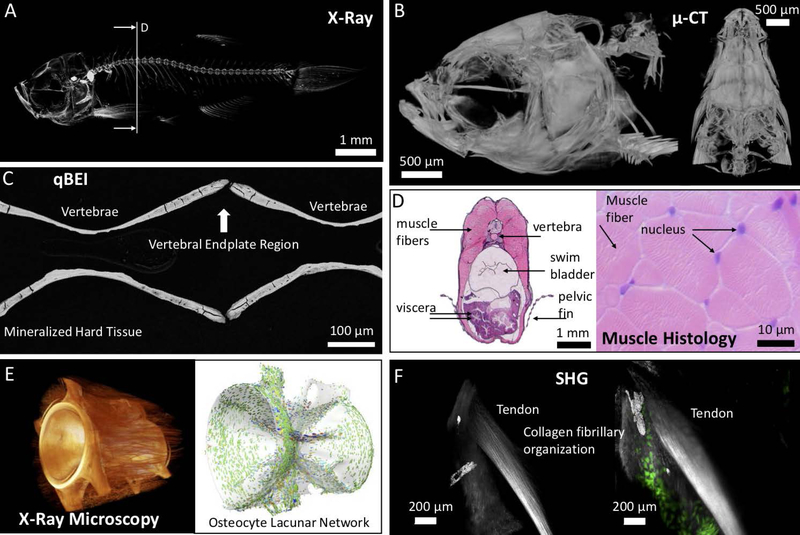
Fully developed, the zebrafish skeleton comprises several functional groups including the cranial skeleton, axial skeleton, caudal skeleton, unpaired fins (dorsal, anal, caudal fins), paired fins (pectoral, pelvic fins), and elasmoid scales (Fig. 1A–D). As in all vertebrates, the zebrafish cranial skeleton and its associated connective tissues, tendons and ligaments, arise from the cranial neural crest; the fin skeletal elements arise from the lateral plate mesoderm, and the myosepta and axial skeleton from somitic paraxial mesoderm 31–33. The cranial musculoskeletal system forms rapidly and can function by 5 days post fertilization (dpf). The pectoral fin cartilage and muscles are also developing at this time. Although the axial skeleton does not form cartilage and bone until later stages, it has the same somitic compartments, sclerotome, syndetome 34, and myotome, fated to become skeletal, tendon, and muscle tissues as in higher vertebrates. Prior to 5 dpf, the axial musculoskeletal structures primarily are composed of muscle and myosepta, a scleraxis-expressing myotendinous tissue that links the myomeres 35; 36. Bony elements form through direct/intramembranous ossification, or via a cartilage or cartilage-like template (e.g., via perichondral or endochondral ossification) 37–40. Modes of ossification can differ in zebrafish and mammals in similar bones. For example, in mouse, the vertebrae form by endochondral ossification 41; in zebrafish, vertebrae form by direct mineralization of the notochord sheath (perichordal ossification), without passing through a cartilaginous stage 42. In some bones, osteoblasts and osteoclasts act in concert to model bone shape into adulthood 43. Although uncommon, osteon-like structures in zebrafish have been reported for lateral ethmoid bone 40. Notably, these structures contained solely one lamella and no osteocytes. Indeed, most skeletal elements in adult zebrafish skeletons are osteocytic and do not show osteons or hemi-osteons indicative of human-like secondary remodeling. In vertebrae of adult zebrafish, osteocyte lacunar orientation shows a preferred orientation (Fig. 1E) 44. While the mechanosensing and remodeling characteristics of osteocytic bone in zebrafish remain to be fully understood, lacunae in zebrafish indicate smaller volumes with less numerous canaliculi compared to mice and humans.
Figure 1: Imaging of tissue structure, composition, and quality.
(A) Contact X-Ray of a juvenile zebrafish. The vertical line shows the histological plane for the image in (D). (B) Microcomputed tomography (2μm isotropic voxel size) of an adult zebrafish skull. (C) Quantitative backscattered scanning electron imaging (qBEI) in the spine of an adult zebrafish. Bone growth occurs at the vertebral endplates. (D) H&E stained section of the zebrafish trunk. Muscle fiber density and cross-sectional muscle fiber area are readily assessed. E) High-resolution imaging of a vertebral body via X-Ray Microscopy highlighting the osteocyte lacunar network. The osteocyte-lacunar orientation may reflect orientation of collagen fibers, and loading patterns in zebrafish vertebrae. The lacunar orientation follows a specific pattern, i.e. longitudinal orientation in the center of the vertebrae and circumferential orientation near the endplate regions. F) An adult tendon attached to the maxilla in zebrafish is imaged using in vivo Second Harmonic Generation (SHG) imaging, an indicator of type I collagen organization and density. The right panel shows dual imaging of tendon in concert with osteoblasts (green: osteocalcin+ cells expressing the ocn:GFP transgene).
Tendons are the tissue interface between muscle and bone 45. Concurrent to skeletal development, transcripts of scleraxis a (scxa) are found in the forming tendon cells adjacent to the developing cartilage and muscle by 2 dpf. These cells aggregate and differentiate, turning on expression of tendon matrix genes, tenomodulin (tnmd), thrombospndin-4 (tsp4b), and type I collagen (col1a1a/b, col1a2) 31; 36. As in mammals, initiation of the axial tendon program depends on signals from the muscle. Cranial and fin tendons form in the absence of muscle, but require muscle for tendon maintenance 31; 46. FGF and TGFβ are also important for proper tendon formation 31, and cyp26b1 loss of function studies suggest that retinoic acid is required for tendon cell condensation 47; 48. Recent studies have shown that mechanical force, through release of TGFβ, regulates the formation of tendon cell projections, which are thought to be involved in ECM production 49. In the adult, the cranial tendons have similar ultrastructure to mammalian tendons with highly ordered type I collagen fibrils observed by TEM 31. In addition, they can be readily visualized using second harmonic generation (SHG) imaging (Fig. 1F).
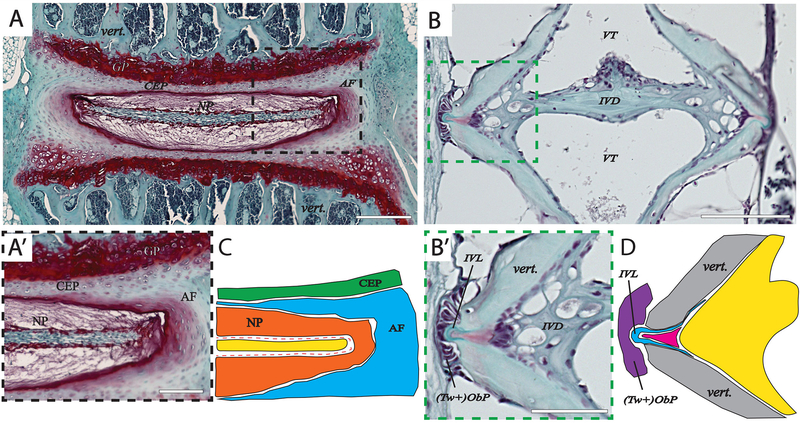
Analogous to higher vertebrates, striated muscle of zebrafish contain three main components: contractile proteins, lipids, and connective tissue 50. Vertebrae are connected by intervertebral ligaments 51. Zebrafish possess both slow- and fast-twitch muscle fibers, which are topographically separated 52. Together with cellular mineralized bone tissue, muscles, tendon, and other soft tissues, the zebrafish skeleton facilitates locomotion, provides mechanical support, and protects internal organs. In Fig. 2, we compare the inter-vertebral space in the zebrafish and mouse as a case study of how skeletal structures in each species typically exhibit both morphophysiological similarities and differences.
Figure 2: Comparison of the intervertebral disc (IVD) in mouse and zebrafish.
(A-A’) and (B-B’): Midline section of a Safranin-O/Fast green staining of an intervertebral disc region in mouse (6-months) (A-A’) and zebrafish (1-year) (B-B’). (C) and (D): Cartoon schematic of insets for mouse (C) and zebrafish (D). In mouse, the IVD is composed of a proteoglycan-rich lamellar fibrocartilaginous cartilage called the annulus fibrosus (AF) which surrounds the nucleus pulposus (NP) joins adjacent bony vertebrae at the level of the cartilaginous end plate (CEP). Zebrafish IVD retains notochord-derived vacuolated cells embedded in a fibrocartilaginous matrix, however, there is no NP-like structure observed in zebrafish. An analogous structure to the outer AF layer in is observed as a small acellular intervertebral ligament (IVL). Histologically, the NP in mouse appears to be composed of: an outer tissue layer which stains for Safranin-O (orange in (C)); an inner cell layer (dotted red line in (C)); and an inner tissue layer that does not stain well for Safranin-O (yellow in (C)). In contrast, the zebrafish IVD has only weak Safranin-O staining (magenta in (B’, D)) in an interior region adjacent to the intervertebral ligament (IVL) (B’, blue in (D)). The zebrafish does not display a true cartilaginous NP tissue, rather the IVD is composed of vacuolated cells and fibrocartilaginous matrix. In contrast to mouse vertebrae which contains bone marrow and trabecular bone filling the vertebrae, zebrafish vertebrae contain bone-shaped vacuolated tissue (VT). Twist positive osteoblast progenitor cells ((Tw+)ObP) are observed adjacent to the IVL. GP-Growth plate; CEP-Cartilaginous endplate; NP- Nucleus pulposus; AF- Annulus fibrosis; IVL-intervertebral ligament; (Tw+)ObP - Twist positive osteoblast progenitors; vert.- vertebrae.
3.2. Conservation of developmental programs
The molecules that govern zebrafish skeletal development are highly conserved with mammals. Sox9, a transcription factor necessary for chondrogenesis and skeletal development 53, has two co-orthologs in the zebrafish, sox9a and sox9b. They are expressed in overlapping and complementary patterns during development with sox9a in the pharyngeal arches and later restricted to the pre-chondrogenic mesenchyme that will form the jaw cartilage and fin scapulocoracoid, and with sox9b in the premigratory neural crest and fin endochondral disc 7. The sox9a expressing chondrocytes also express col2a1 and are Alcian Blue positive before 3 dpf. These skeletal elements will undergo perichondral or endochondral ossification and later become Alizarin red positive cranial bones. In perichondral ossification, perichondral cells become runx2a/b, osterix (sp7/osx), and collagen 10 positive osteoblasts and initiate ossification 54. Similar to other vertebrates, indian hedgehog co-orthologs (ihha/b) are expressed by chondrocytes and are thought to signal to patched, Hh receptors (ptc1/2) in the perichondrium and mediate bone formation 55; 56. Other cranial elements, such as the maxilla undergo direct intramembranous ossification via osx-expressing osteoblasts 40; 57. For many of these genes and cell types reporter and lineage tracing transgenic zebrafish lines have been generated, which, along with the optical access provided by zebrafish, allow unprecedented ability to visualize skeletogenesis (Fig. 3).
Figure 3: Live imaging of cell dynamics.
(A) Whole-body images of a larval zebrafish (top: lateral view; bottom: ventral view), showing osteoblasts expressing osterix in the cranial skeleton, spine, and fins. (image source: 25; use permitted under the Creative Commons Attribution 4.0 International License; image adapted from original). (B) Ventral view of 3 dpf zebrafish lower jaw showing scxa:mCherry expression in the forming tendon cells and col2a1:eGFP expression in chondrocytes (C) Tol2-clonal notochordal expressing twhh-mCherry-CAAX cells (magenta) and spine marked by calcein (green) in 16dpf zebrafish. (D) Representative imaging of caudal fin development demonstrating morphogenesis of the connective tissues (Tol2-EGFPj1184bGt; green), muscle (−503unc:mCherry; red), and bone (Alizarin Red - isolated with a far-red band filter; pseudocolored blue). (E) Osteoclast expressing ctsk:dsRed within resorption pit of an adult zebrafish scale stained with calcein to show mineralized matrix.
3.3. Physiology during development and in homeostasis
The skeleton serves as a key organ which mediates systemic signaling affecting physiology. Although many of these non-structural functions of the skeleton are just being identified, it is clear that many have conservation between humans and zebrafish. One key function of the skeleton is to facilitate mineral homeostasis. The skeleton participates in part by regulating phosphate homeostasis in the kidney through bone-kidney crosstalk. There is evidence that Fgf23, which in humans and mice, is synthesized in osteocytes and regulates kidney phosphate reabsorption, also regulates phosphate homeostasis in zebrafish 58. In mammals, the skeleton also serves as a calcium and phosphorus reservoir. Because calcium regulation can occur through the gills in fish, compared to humans, the physiological role of the skeleton in calcium homeostasis in fish may differ 39. Another function of the mammalian skeleton is acting as a site of hematopoiesis, as well as fat storage, in the marrow cavities. Zebrafish possess bone marrow spaces 39, which is evident in endochondral bones, which are filled with fatty tissue 40. However, unlike in humans, this is never colonized by hematopoietic stem cells (HSC). Thus, zebrafish bone marrow spaces lack hematopoietic tissue 39. A number of zebrafish bones possess adipocytes within their marrow spaces 40, however it is unknown whether this adiposity responds to metabolic demands, as it does in mice 59. Finally, the skeleton can regulate metabolic processes independent of mineral metabolism. For instance, the bone-derived hormone osteocalcin has been implicated in glucose homeostasis, cognition, and male fertility 60. Whether the zebrafish skeleton functions as an endocrine organ through osteocalcin secretion requires further investigation.
3.4. Aging
Compared to early development, processes such as homeostasis and aging have not been studied in depth in the zebrafish. As certain debilitating conditions arise in the skeleton as a function of age, such as osteopenia and osteoporosis, the ability of the zebrafish to model components of these processes would be important. Zebrafish typically have a lifespan of approximately 2–3 years (though 5 years or more is possible)61, and exhibit growth throughout life. There is evidence that zebrafish skeletal function declines with age. For instance, tendon mechanical properties diminish with age 62. Moreover, alterations of vertebral bone and disc are observed in aged zebrafish 63. Bone dependence on estrogen has been modeled in another small teleost, medaka, and thus basic properties of the etiology are likely present in zebrafish 64. With more analysis of late developmental stages, it is likely more insight will emerge from the zebrafish into how the skeletal system ages and its consequences.
3.5. Regeneration and repair
Zebrafish have not been used as a common model for understanding human fracture repair. This is in part due to lack of accessible long bones, as well as its high regenerative capacity which may utilize different repair mechanisms than in mammals. Previous studies have examined the repair properties of damaged membranous bones of the skull roof 65 as well as mandible 66. Although these are not directly comparable to analysis of long bone fractures studied in mouse and most commonly seen in patients, there were some similarities in terms of the genes and cell types involved. For example, runx2+ cells in the periosteum were likely involved in new bone formation and proper formation of the cartilage callus relied upon Indian hedgehog a (ihha) 66. In addition to examination of intrinsic regenerative mechanisms, the transparency of the zebrafish permits the analysis of extrinsic cell populations in the healing process. Studies have shown that the immune system plays an important role in mediating tissue regeneration 67; 68. Visualization of immune infiltration after injury can be accomplished through the use of transgenic reporter lines that either label all leukocytes (cd45:DsRed) 69 or are specific for neutrophils (mpx:GFPi114Tg) 70 or macrophages (mpeg:eGFP) 71. There are also several methods to functionally deplete immune cell populations (reviewed in 67), which can permit temporal control over cell type specific cell ablation to assess the role of immune cell populations at different stages of the regenerative process, as has been performed for tail fin regeneration 72.
Zebrafish have a significant capacity for epimorphic regeneration [3,14]. One example is the caudal fin, which regenerates following amputation 73. Similar to salamander limb regeneration, fin regeneration involves a heterogeneous pool of progenitors called the blastema, which is comprised, at least in part, of mature cells at the amputation stump that dedifferentiated, including osteoblasts 74. A variety of pathways known to be important for skeletogenesis in mammals are recapitulated during fin redevelopment, as reviewed in 73. An intact musculoskeletal system is required for normal regeneration as zebrafish subjected to injection of Botulinim toxin, which inhibits synaptic release at cholinergic nerves, exhibit impaired regeneration 75. This model has also revealed the existence of mesenchymal progenitor populations within specific regions that robustly respond to injury and generate new osx+ osteoblasts 76; 77. Dedifferentiation of mature osteoblasts also occurs during repair of zebrafish fin fractures and skull injuries 65. While osteoblast dedifferentiation is more limited in mammals, fin repair after fracture exhibits some similarities to mammalian long bone fracture, including formation of a remodeling callus 78, and recruitment of osteoclasts 79. Recently, it was shown that neutrophils dynamically colonize the fracture site. When infected with Staphylococcus aureus, neutrophils were retained in the fracture site, and repair was reduced 79. Further studies examining the utility of the fin fracture model to study aspects of fracture biology are warranted.
3.6. Musculoskeletal loading
While the zebrafish skeleton has a reduced role in resisting gravitational loads relative to humans, there is evidence that the zebrafish skeleton can respond to exercise, as well as disuse. Swim training routines have been established to force exercise and stimulate natural modes of skeletal loading in zebrafish. In this way, the complex interplay of cellular, structural, and compositional bone characteristics can be assessed using multiscale approaches in zebrafish to study the effects of genetic and environmental interactions on the skeletal system in vivo. During early development in zebrafish, swim training alters timing of skeletogenesis 80. In adult zebrafish, swim training increases vertebral bone formation and alters quality 44. Moreover, this type of forced exercise also induces muscle adaptations in adult zebrafish 81. This paradigm opens up avenues for genetic and small molecule screens to identify signaling pathways critical for musculoskeletal adaptation to loading and exercise.
4. PHENOTYPING
4.1. MicroCT
3D-high-resolution micro-computed tomography has become established as a powerful method to assess bone morphology and microstructure in zebrafish 18; 44; 82; 83. Using a 5μm voxel size, bone structure indices as vertebral bone volume, thickness, and eccentricity can be characterized 18; 44. Neural arch area, which reflects modeling arising from osteoblast and osteoclast activity, can also be captured 82. Because of their small size, whole body, high-resolution scans are readily acquired 83. Software for semi-automated segmentation enables in-depth phenotyping at a large number of skeletal sites. By quantifying hundreds of measures, this was shown to increase sensitivity in discriminating mutant populations 83. Moreover, the osteocyte lacunar network in the vertebral tissue can be imaged at high resolution with lab-based nano-CTs and 3D X-ray Microscopy (3DXRM). Orientation of the osteocyte lacunae in relation to the long and short axis of the vertebral bodies, sphericity, mean lacunar volume and lacunar density can be quantified 18; 44. Finally, synchrotron-based X-ray microCT, when combined with tissue contrast stains, can yield whole-organism images suitable for cell-level quantitative histological phenotyping in zebrafish 84.
4.2. Histomorphometry
In zebrafish, histologic sections stained using von Kossa/van Gieson, Goldner’s modified Masson-trichrome and toluidine blue enable static bone histomorphometry, and performed in accordance with standardized nomenclature set forth by the ASBMR nomenclature committee for practitioners of bone histomorphometry 85. Calcein labeling or double labeling with calcein and Alizarin Red S can be performed and double labels can be evaluated for dynamic bone histomorphometry 18; 44. Such an approach was used to quantify increases in mineral apposition rate (MAR), mineralizing surface per bone surface (MS/BS), and bone formation rate (BFR) at the vertebral endplates in zebrafish subjected to swimming exercise 44.
4.3. Assessment of bone composition, mineral density distribution, and mechanical properties
Recently, quantitative backscattered electron imaging (qBEI) has been established as an effective means to measure bone mineral density distribution in zebrafish 18; 44. Gray value histograms were used to assess the mean calcium content in the mineralized bone tissue, as well as the homogeneity of mineralization 18; 44. Vibrational spectroscopy methods (e.g. Fourier transform infrared spectroscopy (FTIR) and Raman spectroscopy) have also been adapted to zebrafish bone 18; 86. Parameters such as the mineral-to-matrix-ratio, carbonate-to-phosphate ratio, cross-link-ratio (collagen maturity), and crystallinity (purity, size of mineral crystals) of the bone were shown to provide information about the molecular and compositional bone characteristics 18. Finally, nanoindentation of vertebrae can be performed in zebrafish to assess local mechanical properties material properties such as Young’s Modulus (Elastic modulus), hardness, and fracture toughness 18. Biomechanical properties of zebrafish cranial tendons can also be measured. A maxillary tendon was found to have stress-strain nonlinearity and a linear modulus similar to mammalian tendon data 62.
5. DISEASE APPLICATIONS
5.1. Collagenopathies
OI is a disease of the collagen matrix which results in brittle bones and skeletal deformities. In humans, collagen type I is a heterotrimer composed by two alpha chains, α1(I) and α2(I), which trimerize in a 2:1 ratio, respectively, to form a fibril with a triple helix structure. In zebrafish, the collagen type I triple helix is composed of three α chains, α1(I), α2(I), and α3(I), which are encoded for by the genes col1a1a, col1a2, and col1a1b, respectively 87. Most human patients with OI are attributed to mutations in type I collagens, with the majority of mutations disrupting the conserved Gly-X-Y motifs responsible for fibrillar assembly of the collagen heterotrimers 88. In zebrafish, several dominant mutants have been identified carrying heterozygous glycine substitution in the α1 chain of collagen type I, and which exhibit severe, pathological features of classical OI. This was demonstrated in the chihuahua mutant, which exhibited changes in vertebral tissue composition 18. A large panel of zebrafish mutants of col1a1 genes with qualitative and quantitative defects in collagen type I have been characterized, and found to mirror genotype-phenotype relationships of the range of OI subtypes found in humans 16–20. Disease models have also been developed for mutations affecting COL1A2 17 as well as rare recessive forms of OI affecting non-collagenous proteins (e.g., PLOD2 83; 89 and BMP1 83; 90).
5.2. Spinal curvature
Adolescent idiopathic scoliosis (AIS) is defined as scoliosis without underlying vertebral malformations 91. While the pathogenesis of this disease is still controversial, the zebrafish has made significant advances in our understanding of this disorder. AIS-like scoliosis was demonstrated in cc2d2a mutant zebrafish, which has a role in vesicle trafficking and fusion at the transition zone of photoreceptor connecting cilium in the eye 92. Recently, mutant zebrafish displaying larval or late-onset scoliosis without vertebral malformations, analogous to AIS, have been described for c21orf59, ccdc40, ccdc151, dyx1c1, kif6, and ptk7 93; 94. A common mechanism has emerged from these studies, where loss of ependymal cell cilia function lining the ventricles of the brain, leading to reduced cerebrospinal fluid flow can generate AIS in zebrafish 93; 95. Interestingly, maternal-zygotic ptk7 mutant zebrafish display scoliosis with vertebral malformations, while strictly zygotic ptk7 mutant fish display late-onset AIS without vertebral malformations 96. This suggests that severity of scoliosis can be on a spectrum based on temporal requirements for gene function, which may explain the strong association of AIS in families of children with CS 97.
The cellular mechanism of AIS in ptk7 was demonstrated by a foxj1:ptk7 transgenic zebrafish, which can completely rescue the onset of scoliosis phenotypes observed in ptk7 mutant zebrafish 93. Foxj1 is a master transcriptional regulator of motile cilia 98, which labels motile cilia of the ventricles of the brain and in the pronephros but also labels subset of central canal lining ciliated CSF-fluid contacting neurons. Indeed, disruption of a major signaling receptor, pdk2l1, of CSF-contacting neurons led to mild alterations in spine curvature in zebrafish 99. While it is still unclear how these disruptions of CSF physiology cause scoliosis, recent studies demonstrated that CSF flow (i) helps to stimulate the proper formation of the extracellular Reissner’s fiber which can directly contribute to body straightness during embryonic development, via an unknown mechanisms 100; and (ii) that CSF flow transports adrenergic signals which stimulate the expression of urotensin neuropeptides from CSF-contacting neurons along the spinal cord and mutant zebrafish of the urotensin receptor uts2ra display AIS 101. How these studies will translate to mammalian physiology is still unclear. However, at least one candidate gene for AIS uncovered in zebrafish kif6 94 does not recapitulate AIS phenotypes when mutated in mouse or human 95.
Although having early differences in vertebral specification compared to mammals, the zebrafish may also serve as a model for aspects of congenital scoliosis (CS). Disruption of the extracellular sheath through chemical disruption of lysyl oxidases 102 or specific genetic disruptions of the sheath extracellular matrix components of the notochord sheath such as: col8a1a, col27a1a/b, or calymmin 16; 103; 104 can generate CS-like scoliosis with vertebral malformations in zebrafish suggesting potential underlying components of zebrafish development that can be used to assess gene function in CS etiology.
5.3. Disease loci
Human genome-wide association studies (GWAS) are a powerful means to understand genetic risk factors for chronic diseases such as osteoarthritis105 and osteoporosis 106. These loci may harbor novel drug targets for orthopedic diseases, as evidenced by the fact that OPG/RANK/RANKL and LRP5/SOST, all genes at BMD loci, are members of pathways targeted by osteoporosis drugs (Denosumab and Romosozumab, respectively). A recent analysis of UK-Biobank data identified 515 loci associated with eBMD 106. The causal genes responsible for most associations have yet to be assigned, and thus gene discovery in animal models is needed to complement GWAS in human populations 107. One limitation with knockout mice is the lack of coverage of genes at BMD loci. For instance, of the >23,000 protein coding genes in the mouse genome, <5% have been rigorously analyzed for bone phenotypes in knockout mice within ancillary bone phenotyping projects of the International Mouse Phenotyping Consortium 107. A recent study demonstrated the potential to rapidly generate mutations in CRISPR-edited G0 zebrafish to attribute functional contributions of candidate genes at BMD loci 25. This study also identified a robust, low bone mass phenotype in zebrafish with mutations in wnt16 25, a candidate gene with strong ties to osteoporosis-related traits in humans. Skeletal abnormalities have been observed in fish with altered function in other genes at BMD loci, including LRP5, OSX/SP7, and RANKL 108–110, further evidence that the identification of genes that contribute to human osteoporosis-related traits in zebrafish is feasible. The utility of zebrafish for human skeletal genomics is reviewed in 107.
5.4. Osteoarthritis
Recent data has shown zebrafish can be used to study the development of synovial joints and osteoarthritis. Mutations in col11a2 in zebrafish leads to joint pathologies reminiscent of early-onset osteoarthritis in humans 111. Further, as in humans and mice, zebrafish lubricin or proteoglycan-4b (prg4b) expression was found within some joint regions, such as articular chondrocytes in the jaw 112. These joint regions also expressed col10a1, acana, and matrilin1, which together with prg4b, show similarities to genes expressed in mammalian synovial joints. Loss of both prg4 orthologs resulted in synovial hyperplasia and deterioration of the joint surface by 6–12 months of age. This phenotype recapitulates that found in mouse where loss of Prg4 results in joint disease by 2 months 113. Although it is unknown why there is a significant delay in timing of osteoarthritis onset in the zebrafish compared with mouse genetic models, possible explanations are the zebrafish’s robust regenerative abilities combined with different mechanical environments. Even with these differences, this work establishes the conservation in synovial joint gene expression and function in the zebrafish.
6. SUMMARY/FUTURE DIRECTIONS
The genetic causes of skeletal disorders are rapidly becoming identified, and it is now clear that many common musculoskeletal disorders are fundamentally complex in their causes 106; 114. The field is faced with the need to more fully interrogate the functional consequences of genetic and environmental stresses in how the skeleton and its connecting tissues are formed, how it integrates with broader physiology, and how it repairs damage. There is now a strong rationale to validate newly discovered disease candidate genes in the zebrafish prior to extensive mouse analyses. The refinement of clonal analyses of gene function will expedite combinatorial analysis of gene function and permit a systematic testing platform for genetic association studies and analysis of genetic modifiers. With the ability to visualize cellular dynamics during formation of the skeleton as well as during repair, in different genetic contexts, the zebrafish provides a powerful system to bring functional characterization in line with the rate of genetic discoveries.
In addition to validation, the zebrafish is also a valuable platform for discovery. Due to its small size, low cost, and genetic malleability, the zebrafish has opened new screening methods that have already discovered small molecules efficacious in regulating skeletal phenotypes. Similarly, through unbiased mutational screening, new genes–and new functions for existing genes–have been discovered. These mutations have shown to be predictive of causes of skeletal disorders in humans 115; 116, and opened new areas of musculoskeletal development previously uncharacterized or neglected 101; 117.
As the use of zebrafish for orthopaedic research is still in its relative infancy, there are a number of open questions regarding developmental stages, bones, and phenotypic traits in zebrafish that best serve as a model for human skeletal biology 107. Morphophysiological differences can make one-to-one modeling of human skeletal phenotypes in zebrafish challenging. Origins of mammalian bones and their connections to fish bones (e.g., the mammalian middle ear bones, which derive from bones that form the jaw in fish 118) can sometimes be revealed through evolutionary analyses, however, such relationships cannot always be made. In this context, a community effort to phenotype zebrafish mutants for orthologs of genes examined in mutant mouse phenotyping consortiums may aid in identifying zebrafish phenotypes that are most consistently associated with phenotypic changes in the orthologous mutant mouse 107.
With the extension of zebrafish work into phenotypes of the skeleton beyond early development, the broad utility of this model has emerged. While there are differences stemming from the use of a non-mammalian vertebrate for modeling human disorders, the zebrafish model provides both genetic and anatomical foundations in which informed analyses can be made concerning the etiology of disorders and a tool in which to refine potential therapeutic strategies.
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health under award numbers AR071554 (JLG), AR074541 (JLG), AR072009 (RSG), (DE024434 (MPH), and AR066061 (RYK). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. BB would like to thank Hrishikesh A. Bale, Imke A. K. Fiedler and Tessa Krappa for imaging support. JLG would like to thank Marie Noedl and Xubo Niu for providing the tendon images.
REFERENCES
- 1.Shah AN, Davey CF, Whitebirch AC, et al. 2015. Rapid reverse genetic screening using CRISPR in zebrafish. Nat Methods 12:535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keller PJ, Schmidt AD, Wittbrodt J, et al. 2008. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 322:1065–1069. [DOI] [PubMed] [Google Scholar]
- 3.Ahrens MB, Li JM, Orger MB, et al. 2012. Brain-wide neuronal dynamics during motor adaptation in zebrafish. Nature 485:471–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang J, Hu J, Karra R, et al. 2016. Modulation of tissue repair by regeneration enhancer elements. Nature 532:201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan C, Brunson DC, Tang Q, et al. 2019. Visualizing Engrafted Human Cancer and Therapy Responses in Immunodeficient Zebrafish. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howe K, Clark MD, Torroja CF, et al. 2013. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496:498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan YL, Willoughby J, Liu D, et al. 2005. A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development 132:1069–1083. [DOI] [PubMed] [Google Scholar]
- 8.Rohner N, Bercsenyi M, Orban L, et al. 2009. Duplication of fgfr1 permits Fgf signaling to serve as a target for selection during domestication. Curr Biol 19:1642–1647. [DOI] [PubMed] [Google Scholar]
- 9.Mork L, Crump G. 2015. Zebrafish Craniofacial Development: A Window into Early Patterning. Curr Top Dev Biol 115:235–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergen DJM, Kague E, Hammond CL. 2019. Zebrafish as an Emerging Model for Osteoporosis: A Primary Testing Platform for Screening New Osteo-Active Compounds. Front Endocrinol (Lausanne) 10:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwon RY, Watson CJ, Karasik D. 2019. Using zebrafish to study skeletal genomics. Bone. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schilling TF, Piotrowski T, Grandel H, et al. 1996. Jaw and branchial arch mutants in zebrafish I: branchial arches. Development 123:329–344. [DOI] [PubMed] [Google Scholar]
- 13.Piotrowski T, Schilling TF, Brand M, et al. 1996. Jaw and branchial arch mutants in zebrafish II: anterior arches and cartilage differentiation. Development 123:345–356. [DOI] [PubMed] [Google Scholar]
- 14.Neuhauss SC, Solnica-Krezel L, Schier AF, et al. 1996. Mutations affecting craniofacial development in zebrafish. Development 123:357–367. [DOI] [PubMed] [Google Scholar]
- 15.Harris MP, Rohner N, Schwarz H, et al. 2008. Zebrafish eda and edar mutants reveal conserved and ancestral roles of ectodysplasin signaling in vertebrates. PLoS Genet 4:e1000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henke K, Daane JM, Hawkins MB, et al. 2017. Genetic Screen for Postembryonic Development in the Zebrafish (Danio rerio): Dominant Mutations Affecting Adult Form. Genetics 207:609–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gistelinck C, Kwon RY, Malfait F, et al. 2018. Zebrafish type I collagen mutants faithfully recapitulate human type I collagenopathies. Proc Natl Acad Sci U S A 115:E8037–E8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiedler IAK, Schmidt FN, Wolfel EM, et al. 2018. Severely Impaired Bone Material Quality in Chihuahua Zebrafish Resembles Classical Dominant Human Osteogenesis Imperfecta. J Bone Miner Res 33:1489–1499. [DOI] [PubMed] [Google Scholar]
- 19.Gioia R, Tonelli F, Ceppi I, et al. 2017. The chaperone activity of 4PBA ameliorates the skeletal phenotype of Chihuahua, a zebrafish model for dominant osteogenesis imperfecta. Hum Mol Genet 26:2897–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher S, Jagadeeswaran P, Halpern ME. 2003. Radiographic analysis of zebrafish skeletal defects. Dev Biol 264:64–76. [DOI] [PubMed] [Google Scholar]
- 21.Haffter P, Odenthal J, Mullins MC, et al. 1996. Mutations affecting pigmentation and shape of the adult zebrafish. Dev Genes Evol 206:260–276. [DOI] [PubMed] [Google Scholar]
- 22.Kettleborough RN, Busch-Nentwich EM, Harvey SA, et al. 2013. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature 496:494–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bedell VM, Wang Y, Campbell JM, et al. 2012. In vivo genome editing using a high-efficiency TALEN system. Nature 491:114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El-Brolosy MA, Kontarakis Z, Rossi A, et al. 2019. Genetic compensation triggered by mutant mRNA degradation. Nature 568:193–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watson CJ, Monstad-Rios AT, Bhimani RM, et al. 2018. Decoding G0 somatic mutants through deep phenotyping and mosaic pattern analysis in the zebrafish skeleton. bioRxiv doi: 10.1101/466185. [DOI] [Google Scholar]
- 26.Oppedal D, Goldsmith MI. 2010. A chemical screen to identify novel inhibitors of fin regeneration in zebrafish. Zebrafish 7:53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monstad-Rios AT, Watson CJ, Kwon RY. 2018. ScreenCube: A 3D Printed System for Rapid and Cost-Effective Chemical Screening in Adult Zebrafish. Zebrafish 15:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu PB, Hong CC, Sachidanandan C, et al. 2008. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat Chem Biol 4:33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawahara G, Karpf JA, Myers JA, et al. 2011. Drug screening in a zebrafish model of Duchenne muscular dystrophy. Proc Natl Acad Sci U S A 108:5331–5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiley DS, Redfield SE, Zon LI. 2017. Chemical screening in zebrafish for novel biological and therapeutic discovery. Methods Cell Biol 138:651–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen JW, Galloway JL. 2014. The development of zebrafish tendon and ligament progenitors. Development 141:2035–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma RC, Jacobs CT, Sharma P, et al. 2018. Stereotypic generation of axial tenocytes from bipartite sclerotome domains in zebrafish. PLoS Genet 14:e1007775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schilling TF, Kimmel CB. 1997. Musculoskeletal patterning in the pharyngeal segments of the zebrafish embryo. Development 124:2945–2960. [DOI] [PubMed] [Google Scholar]
- 34.Dubrulle J, Pourquie O. 2003. Welcome to syndetome: a new somitic compartment. Dev Cell 4:611–612. [DOI] [PubMed] [Google Scholar]
- 35.Charvet B, Malbouyres M, Pagnon-Minot A, et al. 2011. Development of the zebrafish myoseptum with emphasis on the myotendinous junction. Cell Tissue Res 346:439–449. [DOI] [PubMed] [Google Scholar]
- 36.Subramanian A, Schilling TF. 2014. Thrombospondin-4 controls matrix assembly during development and repair of myotendinous junctions. Elife 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bird NC, Mabee PM. 2003. Developmental morphology of the axial skeleton of the zebrafish, Danio rerio (Ostariophysi: Cyprinidae). Dev Dyn 228:337–357. [DOI] [PubMed] [Google Scholar]
- 38.Cubbage CC, Mabee PM. 1996. Development of the cranium and paired fins in the zebrafish Danio rerio (Ostariophysi, Cyprinidae). J Morphol 229:121–160. [DOI] [PubMed] [Google Scholar]
- 39.Apschner A, Schulte-Merker S, Witten PE. 2011. Not all bones are created equal - using zebrafish and other teleost species in osteogenesis research. Methods Cell Biol 105:239–255. [DOI] [PubMed] [Google Scholar]
- 40.Weigele J, Franz-Odendaal TA. 2016. Functional bone histology of zebrafish reveals two types of endochondral ossification, different types of osteoblast clusters and a new bone type. J Anat 229:92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lefebvre V, Bhattaram P. 2010. Vertebrate skeletogenesis. Curr Top Dev Biol 90:291–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fleming A, Keynes R, Tannahill D. 2004. A central role for the notochord in vertebral patterning. Development 131:873–880. [DOI] [PubMed] [Google Scholar]
- 43.Chatani M, Takano Y, Kudo A. 2011. Osteoclasts in bone modeling, as revealed by in vivo imaging, are essential for organogenesis in fish. Dev Biol 360:96–109. [DOI] [PubMed] [Google Scholar]
- 44.Suniaga S, Rolvien T, Vom Scheidt A, et al. 2018. Increased mechanical loading through controlled swimming exercise induces bone formation and mineralization in adult zebrafish. Sci Rep 8:3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen JW, Galloway JL. 2017. Using the zebrafish to understand tendon development and repair. Methods Cell Biol 138:299–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brent AE, Tabin CJ. 2004. FGF acts directly on the somitic tendon progenitors through the Ets transcription factors Pea3 and Erm to regulate scleraxis expression. Development 131:3885–3896. [DOI] [PubMed] [Google Scholar]
- 47.Pryce BA, Watson SS, Murchison ND, et al. 2009. Recruitment and maintenance of tendon progenitors by TGFbeta signaling are essential for tendon formation. Development 136:1351–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McGurk PD, Swartz ME, Chen JW, et al. 2017. In vivo zebrafish morphogenesis shows Cyp26b1 promotes tendon condensation and musculoskeletal patterning in the embryonic jaw. PLoS Genet 13:e1007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subramanian A, Kanzaki LF, Galloway JL, et al. 2018. Mechanical force regulates tendon extracellular matrix organization and tenocyte morphogenesis through TGFbeta signaling. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kiessling A, Ruohonen K, Bjørnevik M. 2006. Muscle fibre growth and quality in fish. Arch Tierz, Dummerstorf 49:137–146. [Google Scholar]
- 51.Inohaya K, Takano Y, Kudo A. 2007. The teleost intervertebral region acts as a growth center of the centrum: in vivo visualization of osteoblasts and their progenitors in transgenic fish. Dev Dyn 236:3031–3046. [DOI] [PubMed] [Google Scholar]
- 52.Lieschke GJ, Currie PD. 2007. Animal models of human disease: zebrafish swim into view. Nat Rev Genet 8:353–367. [DOI] [PubMed] [Google Scholar]
- 53.Akiyama H, Chaboissier MC, Martin JF, et al. 2002. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev 16:2813–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Felber K, Croucher P, Roehl HH. 2011. Hedgehog signalling is required for perichondral osteoblast differentiation in zebrafish. Mech Dev 128:141–152. [DOI] [PubMed] [Google Scholar]
- 55.Avaron F, Hoffman L, Guay D, et al. 2006. Characterization of two new zebrafish members of the hedgehog family: atypical expression of a zebrafish indian hedgehog gene in skeletal elements of both endochondral and dermal origins. Dev Dyn 235:478–489. [DOI] [PubMed] [Google Scholar]
- 56.Eames BF, Singer A, Smith GA, et al. 2010. UDP xylose synthase 1 is required for morphogenesis and histogenesis of the craniofacial skeleton. Dev Biol 341:400–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li N, Felber K, Elks P, et al. 2009. Tracking gene expression during zebrafish osteoblast differentiation. Dev Dyn 238:459–466. [DOI] [PubMed] [Google Scholar]
- 58.Elizondo MR, Arduini BL, Paulsen J, et al. 2005. Defective skeletogenesis with kidney stone formation in dwarf zebrafish mutant for trpm7. Curr Biol 15:667–671. [DOI] [PubMed] [Google Scholar]
- 59.de Paula FJ, Rosen CJ. 2013. Bone Remodeling and Energy Metabolism: New Perspectives. Bone Res 1:72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moser SC, van der Eerden BCJ. 2018. Osteocalcin-A Versatile Bone-Derived Hormone. Front Endocrinol (Lausanne) 9:794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gerhard GS, Kauffman EJ, Wang X, et al. 2002. Life spans and senescent phenotypes in two strains of Zebrafish (Danio rerio). Exp Gerontol 37:1055–1068. [DOI] [PubMed] [Google Scholar]
- 62.Shah RR, Nerurkar NL, Wang CC, et al. 2015. Tensile properties of craniofacial tendons in the mature and aged zebrafish. J Orthop Res 33:867–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hayes AJ, Reynolds S, Nowell MA, et al. 2013. Spinal deformity in aged zebrafish is accompanied by degenerative changes to their vertebrae that resemble osteoarthritis. Plos One 8:e75787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shanthanagouda AH, Guo BS, Ye RR, et al. 2014. Japanese medaka: a non-mammalian vertebrate model for studying sex and age-related bone metabolism in vivo. Plos One 9:e88165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Geurtzen K, Knopf F, Wehner D, et al. 2014. Mature osteoblasts dedifferentiate in response to traumatic bone injury in the zebrafish fin and skull. Development 141:2225–2234. [DOI] [PubMed] [Google Scholar]
- 66.Paul S, Schindler S, Giovannone D, et al. 2016. Ihha induces hybrid cartilage-bone cells during zebrafish jawbone regeneration. Development 143:2066–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Keightley MC, Wang CH, Pazhakh V, et al. 2014. Delineating the roles of neutrophils and macrophages in zebrafish regeneration models. Int J Biochem Cell Biol 56:92–106. [DOI] [PubMed] [Google Scholar]
- 68.Eming SA, Wynn TA, Martin P. 2017. Inflammation and metabolism in tissue repair and regeneration. Science 356:1026–1030. [DOI] [PubMed] [Google Scholar]
- 69.Richardson R, Slanchev K, Kraus C, et al. 2013. Adult zebrafish as a model system for cutaneous wound-healing research. J Invest Dermatol 133:1655–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Renshaw SA, Loynes CA, Elworthy S, et al. 2007. Modeling inflammation in the zebrafish: how a fish can help us understand lung disease. Exp Lung Res 33:549–554. [DOI] [PubMed] [Google Scholar]
- 71.Ellett F, Pase L, Hayman JW, et al. 2011. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 117:e49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Petrie TA, Strand NS, Yang CT, et al. 2014. Macrophages modulate adult zebrafish tail fin regeneration. Development 141:2581–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Watson CJ, Kwon RY. 2015. Osteogenic programs during zebrafish fin regeneration. Bonekey Rep 4:745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Knopf F, Hammond C, Chekuru A, et al. 2011. Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev Cell 20:713–724. [DOI] [PubMed] [Google Scholar]
- 75.Recidoro AM, Roof AC, Schmitt M, et al. 2014. Botulinum toxin induces muscle paralysis and inhibits bone regeneration in zebrafish. J Bone Miner Res 29:2346–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ando K, Shibata E, Hans S, et al. 2017. Osteoblast Production by Reserved Progenitor Cells in Zebrafish Bone Regeneration and Maintenance. Dev Cell 43:643–650 e643. [DOI] [PubMed] [Google Scholar]
- 77.Singh SP, Holdway JE, Poss KD. 2012. Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev Cell 22:879–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sousa S, Valerio F, Jacinto A. 2012. A new zebrafish bone crush injury model. Biol Open 1:915–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tomecka MJ, Ethiraj LP, Sanchez LM, et al. 2019. Clinical pathologies of bone fracture modelled in zebrafish. Dis Model Mech 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fiaz AW, Leon-Kloosterziel KM, Gort G, et al. 2012. Swim-training changes the spatio-temporal dynamics of skeletogenesis in zebrafish larvae (Danio rerio). Plos One 7:e34072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Palstra AP, Rovira M, Rizo-Roca D, et al. 2014. Swimming-induced exercise promotes hypertrophy and vascularization of fast skeletal muscle fibres and activation of myogenic and angiogenic transcriptional programs in adult zebrafish. Bmc Genomics 15:1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Charles JF, Sury M, Tsang K, et al. 2017. Utility of quantitative micro-computed tomographic analysis in zebrafish to define gene function during skeletogenesis. Bone 101:162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hur M, Gistelinck CA, Huber P, et al. 2017. MicroCT-based phenomics in the zebrafish skeleton reveals virtues of deep phenotyping in a distributed organ system. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ding Y, Vanselow DJ, Yakovlev MA, et al. 2019. Computational 3D histological phenotyping of whole zebrafish by X-ray histotomography. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dempster DW, Compston JE, Drezner MK, et al. 2013. Standardized nomenclature, symbols, and units for bone histomorphometry: a 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res 28:2–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fiedler IAK, Casanova M, Keplinger T, et al. 2018. Effect of short-term formaldehyde fixation on Raman spectral parameters of bone quality. J Biomed Opt 23:1–6. [DOI] [PubMed] [Google Scholar]
- 87.Gistelinck C, Gioia R, Gagliardi A, et al. 2016. Zebrafish Collagen Type I: Molecular and Biochemical Characterization of the Major Structural Protein in Bone and Skin. Scientific Reports 6:21540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ricard-Blum S 2011. The collagen family. Cold Spring Harb Perspect Biol 3:a004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gistelinck C, Witten PE, Huysseune A, et al. 2016. Loss of Type I Collagen Telopeptide Lysyl Hydroxylation Causes Musculoskeletal Abnormalities in a Zebrafish Model of Bruck Syndrome. J Bone Miner Res 31:1930–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Asharani PV, Keupp K, Semler O, et al. 2012. Attenuated BMP1 function compromises osteogenesis, leading to bone fragility in humans and zebrafish. Am J Hum Genet 90:661–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cheng JC, Castelein RM, Chu WC, et al. 2015. Adolescent idiopathic scoliosis. Nat Rev Dis Primers 1:15030. [DOI] [PubMed] [Google Scholar]
- 92.Bachmann-Gagescu R, Phelps IG, Stearns G, et al. 2011. The ciliopathy gene cc2d2a controls zebrafish photoreceptor outer segment development through a role in Rab8-dependent vesicle trafficking. Hum Mol Genet 20:4041–4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Grimes DT, Boswell CW, Morante NF, et al. 2016. Zebrafish models of idiopathic scoliosis link cerebrospinal fluid flow defects to spine curvature. Science 352:1341–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Buchan JG, Gray RS, Gansner JM, et al. 2014. Kinesin family member 6 (kif6) is necessary for spine development in zebrafish. Dev Dyn 243:1646–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Konjikusic MJ, Yeetong P, Boswell CW, et al. 2018. Mutations in Kinesin family member 6 reveal specific role in ependymal cell ciliogenesis and human neurological development. PLoS Genet 14:e1007817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hayes M, Gao X, Yu LX, et al. 2014. ptk7 mutant zebrafish models of congenital and idiopathic scoliosis implicate dysregulated Wnt signalling in disease. Nat Commun 5:4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Purkiss SB, Driscoll B, Cole WG, et al. 2002. Idiopathic scoliosis in families of children with congenital scoliosis. Clin Orthop Relat Res:27–31. [DOI] [PubMed] [Google Scholar]
- 98.Yu X, Ng CP, Habacher H, et al. 2008. Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nat Genet 40:1445–1453. [DOI] [PubMed] [Google Scholar]
- 99.Sternberg JR, Prendergast AE, Brosse L, et al. 2018. Pkd2l1 is required for mechanoception in cerebrospinal fluid-contacting neurons and maintenance of spine curvature. Nat Commun 9:3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cantaut-Belarif Y, Sternberg JR, Thouvenin O, et al. 2018. The Reissner Fiber in the Cerebrospinal Fluid Controls Morphogenesis of the Body Axis. Curr Biol 28:2479–2486 e2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang X, Jia S, Chen Z, et al. 2018. Cilia-driven cerebrospinal fluid flow directs expression of urotensin neuropeptides to straighten the vertebrate body axis. Nat Genet 50:1666–1673. [DOI] [PubMed] [Google Scholar]
- 102.Gansner JM, Mendelsohn BA, Hultman KA, et al. 2007. Essential role of lysyl oxidases in notochord development. Dev Biol 307:202–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gray RS, Wilm TP, Smith J, et al. 2014. Loss of col8a1a function during zebrafish embryogenesis results in congenital vertebral malformations. Dev Biol 386:72–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Christiansen HE, Lang MR, Pace JM, et al. 2009. Critical early roles for col27a1a and col27a1b in zebrafish notochord morphogenesis, vertebral mineralization and post-embryonic axial growth. Plos One 4:e8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tachmazidou I, Hatzikotoulas K, Southam L, et al. 2019. Identification of new therapeutic targets for osteoarthritis through genome-wide analyses of UK Biobank data. Nat Genet 51:230–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Morris JA, Kemp JP, Youlten SE, et al. 2019. An atlas of genetic influences on osteoporosis in humans and mice. Nat Genet 51:258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kwon RY, Watson CJ, Karasik D. 2019. Using zebrafish to study skeletal genomics. Bone 126:37–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Willems B, Tao S, Yu T, et al. 2015. The Wnt Co-Receptor Lrp5 Is Required for Cranial Neural Crest Cell Migration in Zebrafish. Plos One 10:e0131768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kague E, Roy P, Asselin G, et al. 2016. Osterix/Sp7 limits cranial bone initiation sites and is required for formation of sutures. Dev Biol 413:160–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.To TT, Witten PE, Renn J, et al. 2012. Rankl-induced osteoclastogenesis leads to loss of mineralization in a medaka osteoporosis model. Development 139:141–150. [DOI] [PubMed] [Google Scholar]
- 111.Lawrence EA, Kague E, Aggleton JA, et al. 2018. The mechanical impact of col11a2 loss on joints; col11a2 mutant zebrafish show changes to joint development and function, which leads to early-onset osteoarthritis. Philos Trans R Soc Lond B Biol Sci 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Askary A, Smeeton J, Paul S, et al. 2016. Ancient origin of lubricated joints in bony vertebrates. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hill A, Duran J, Purcell P. 2014. Lubricin protects the temporomandibular joint surfaces from degeneration. Plos One 9:e106497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Estrada K, Styrkarsdottir U, Evangelou E, et al. 2012. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet 44:491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Clement A, Wiweger M, von der Hardt S, et al. 2008. Regulation of zebrafish skeletogenesis by ext2/dackel and papst1/pinscher. PLoS Genet 4:e1000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wiweger MI, Zhao Z, van Merkesteyn RJ, et al. 2012. HSPG-deficient zebrafish uncovers dental aspect of multiple osteochondromas. Plos One 7:e29734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Daane JM, Lanni J, Rothenberg I, et al. 2018. Bioelectric-calcineurin signaling module regulates allometric growth and size of the zebrafish fin. Sci Rep 8:10391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Anthwal N, Joshi L, Tucker AS. 2013. Evolution of the mammalian middle ear and jaw: adaptations and novel structures. J Anat 222:147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kwak J, Park OK, Jung YJ, et al. 2013. Live image profiling of neural crest lineages in zebrafish transgenic lines. Mol Cells 35:255–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mongera A, Singh AP, Levesque MP, et al. 2013. Genetic lineage labeling in zebrafish uncovers novel neural crest contributions to the head, including gill pillar cells. Development 140:916–925. [DOI] [PubMed] [Google Scholar]
- 121.Dale RM, Topczewski J. 2011. Identification of an evolutionarily conserved regulatory element of the zebrafish col2a1a gene. Dev Biol 357:518–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Askary A, Mork L, Paul S, et al. 2015. Iroquois Proteins Promote Skeletal Joint Formation by Maintaining Chondrocytes in an Immature State. Dev Cell 35:358–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hammond CL, Schulte-Merker S. 2009. Two populations of endochondral osteoblasts with differential sensitivity to Hedgehog signalling. Development 136:3991–4000. [DOI] [PubMed] [Google Scholar]
- 124.Berger J, Currie PD. 2013. 503unc, a small and muscle-specific zebrafish promoter. Genesis 51:443–447. [DOI] [PubMed] [Google Scholar]
- 125.Kague E, Gallagher M, Burke S, et al. 2012. Skeletogenic fate of zebrafish cranial and trunk neural crest. Plos One 7:e47394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mitchell RE, Huitema LF, Skinner RE, et al. 2013. New tools for studying osteoarthritis genetics in zebrafish. Osteoarthritis Cartilage 21:269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.DeLaurier A, Eames BF, Blanco-Sanchez B, et al. 2010. Zebrafish sp7:EGFP: a transgenic for studying otic vesicle formation, skeletogenesis, and bone regeneration. Genesis 48:505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Spoorendonk KM, Peterson-Maduro J, Renn J, et al. 2008. Retinoic acid and Cyp26b1 are critical regulators of osteogenesis in the axial skeleton. Development 135:3765–3774. [DOI] [PubMed] [Google Scholar]
- 129.Bussmann J, Schulte-Merker S. 2011. Rapid BAC selection for tol2-mediated transgenesis in zebrafish. Development 138:4327–4332. [DOI] [PubMed] [Google Scholar]
- 130.Sharif F, de Bakker MA, Richardson MK. 2014. Osteoclast-like Cells in Early Zebrafish Embryos. Cell J 16:211–224. [PMC free article] [PubMed] [Google Scholar]