Abstract
The internal representation of an experience is thought to be encoded by long-lasting physical changes to the brain (“engrams”) . Previously, we and others showed within the lateral amygdala (LA), a region critical for auditory conditioned fear, eligible neurons compete against one other for allocation to an engram. Neurons with relatively higher function of the transcription factor CREB were more likely to be allocated to the engram. In these studies, though, CREB function was artificially increased for several days before training. Precisely when increased CREB function is important for allocation remains an unanswered question. Here, we took advantage of a novel optogenetic tool (opto-DN-CREB) to gain spatial and temporal control of CREB function in freely behaving mice. We found increasing CREB function in a small, random population of LA principal neurons in the minutes, but not 24 h, before training was sufficient to enhance memory, likely because these neurons were preferentially allocated to the underlying engram. However, similarly increasing CREB activity in a small population of random LA neurons immediately after training disrupted subsequent memory retrieval, likely by disrupting the precise spatial and temporal patterns of offline post-training neuronal activity and/or function required for consolidation. These findings reveal the importance of the timing of CREB activity in regulating allocation and subsequent memory retrieval, and further, highlight the potential of optogenetic approaches to control protein function with temporal specificity in behaving animals.
Subject terms: Long-term memory, Fear conditioning
Introduction
Memory for an experience is thought to be represented in the brain by a sparsely distributed cell ensemble, or engram [1–6]. Neurons active at the time of an experience are hypothesized to be recruited to an engram, with subsequent reactivation of these neurons supporting memory retrieval. Although the engram is a hypothetical construct dating back to the turn of the last century [1–4, 7, 8], recently, progress has been made in identifying and manipulating key components of an engram supporting a specific memory. Largely, this progress has been enabled by an array of new tools to identify and control the activity of individual neurons hypothesized to be part of an engram (“engram neurons”).
Two complementary strategies have been used to identify and manipulate engram neurons. First, neurons active at the time of a training experience can be genetically “tagged” [9–12], or, second, the excitability of random neurons can be artificially increased around the time of training to bias their recruitment or allocation into an engram [13–15]. Inactivation of tagged or allocated neurons disrupts subsequent memory retrieval [9, 16–19] while artificial reactivation of these neurons induces memory expression in the absence of an external sensory retrieval cue [11, 18, 20–27]. Although these findings suggest that neurons allocated to an engram are indispensable for subsequent memory retrieval and therefore, are key components of the underlying engram, the precise mechanisms underlying allocation are not fully understood.
Previous research shows neuronal allocation to an engram within a given brain region is a competitive process between eligible neurons [13–15, 23, 24, 28–37]. Although the outcome of this neuronal competition may be mediated by many things, converging results indicate the transcription factor CREB (cAMP-response element binding protein) plays a vital role in this process [13, 14, 36]. First, endogenous (non-manipulated) memory formation is correlated with an increase in CREB function in a small, seemingly random population of neurons [19, 29], consistent with the notion that these neurons were key components of an engram. Second, artificially increasing CREB function in a similar small portion (roughly 10%) of random excitatory neurons in the lateral amygdala (LA) enhanced auditory fear conditioned memory [14, 24, 29]. Third, neurons with relatively higher CREB function were more likely to be allocated to an engram than their neighbors [14, 24, 29]. Although these results are compelling, in these manipulation experiments, CREB function was increased for 2–3 d before training. This leaves open the crucial issue of when, relative to training, increased CREB function is important for neuronal allocation and memory enhancement.
To address this question, we took advantage of a novel tool to optically control CREB function in vivo. Recently, we engineered a light-sensitive CREB protein (opto-DN-CREB) by fusing photoactive yellow protein (PYP, a blue-light photoreceptor) to a dominant-negative inhibitor of CREB (A-CREB, DN) [38]. A-CREB is a specific and strong inhibitor of CREB transcription [39], while PYP is a photo-switchable protein that changes conformation in response to blue light (BL). Therefore, in the absence of BL, the opto-DN-CREB construct would repress CREB function. However, once activated by BL, the structure of PYP changes, sequestering A-CREB, and thereby liberating endogenous CREB function [38]. In this way, it is possible to temporally control CREB function with BL.
We found acutely increasing CREB function (photoactivation of opto-DN-CREB) 1 h, but not 24 h, before training enhanced the formation of an auditory fear memory, similar to overexpressing wild-type CREB for 2–3 d before training [23]. This finding indicates that high CREB function in the minutes-to-hours before the training event is sufficient to govern allocation and enhance memory. In contrast, increasing CREB in a random portion of neurons immediately after training impaired subsequent fear memory retrieval, perhaps by interrupting the precise timing of post-training neuronal reactivation that may be necessary for memory consolidation. Together, these results show the importance of the timing of increased CREB function in governing neuronal allocation and memory.
Materials and methods
Mice
All experiments were performed according to the guidelines provided by The Hospital for Sick Children Animal Care and Use Committee in addition to the Canadian Council on Animal Care (CCAC) and the NIH Guidelines on the Care and Use of Laboratory Animals. Adult mice between the ages of 8 and 12 weeks of age were used for all experiments. Experiments were conducted with male and female hybrid (C57BL/6Ntac × 129S6/SvEvTac) mice. As we observed no differences in our dependent variable between sexes, we pooled the data. Mice were bred and housed at The Hospital for Sick Children on a 12-h light–dark cycle, with food and water available ad libitum. Behavioral experiments were performed during the light cycle.
opto-DN-CREB
The opto-DN-CREB construct contains two important components: (1) a dominant-negative inhibitor of CREB (A-CREB) and (2) a PYP light-sensitive domain. The dominant-negative inhibitor of CREB, originally published by Ahn and colleagues [39], was constructed by adding an acidic extension to the N-terminus of the CREB basic leucine zipper domain. This acidic extension interacts with the basic region of CREB to form a coiled-coil extension, preventing wild-type CREB from binding to DNA. A-CREB heterodimerizes with endogenous CREB and prevents their interaction with CRE sites [40]. Many labs have shown that A-CREB functions as a specific inhibitor of CREB function [41, 42].
PYP is a small soluble protein found in purple sulfur bacteria, where it senses BL [43]. BL bleaches PYP; however, its color recovers in the dark within roughly 1 s [43]. This light-induced change in the color, and quick reversal in the dark, likely parallels the conformational changes of PYP. We fused PYP to A-CREB to confer light responsivity to this dominant-negative CREB inhibitor. In the dark, opto-DN-CREB functions as a CREB inhibitor. However, upon BL stimulation, PYP undergoes a conformational change and sequesters A-CREB, thereby liberating endogenous CREB [38] (see Fig. 1a). That is, upon BL stimulation, opto-DN-CREB increases CREB function.
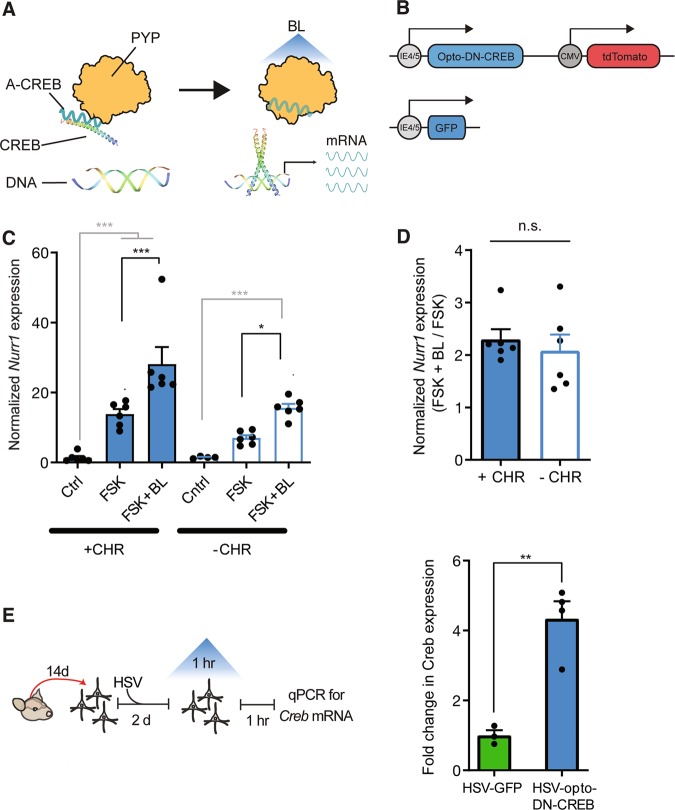
Fig. 1. Photostimulation of opto-DN-CREB, even without the exogenous addition of a chromophore, increases expression of CREB and CREB target genes.
a Schematic of opto-DN-CREB construct. The photoreceptor PYP (photoactive yellow protein) is bound to a dominant-negative inhibitor of CREB (A-CREB). In the lights-off condition, A-CREB (fused with PYP) binds to endogenous CREB, thereby blocking CREB function. Upon blue light (BL) stimulation, PYP undergoes a conformational change, sequestering A-CREB, thereby liberating endogenous CREB and increasing CREB function. b (Top) Schematic of HSV-opto-DN-CREB construct. (Bottom) HSV-GFP control construct. c Exogenous addition of the PYP chromophore (CHR) is not required for opto-DN-CREB to increase CREB function in HEK cells. As expected, in HEK cells transfected with opto-DN-CREB, addition of the adenylyl cyclase activator forskolin (FSK) increased expression of the CREB target gene Nurr1 compared to control cells (Cntrl, transfected with opto-DN-CREB, no FSK). Nurr1 expression was further increased with BL, whether exogenous chromophore (CHR) was (Left) added (+CHR) [Cntrl (n = 4), FSK (n = 7), FSK + BL (n = 6)] (Right) or not (−CHR) [Cntrl (n = 4), FSK (n = 6), FSK + BL (n = 6)]. d Normalization of FSK + BL groups from c with, and without, exogenous CHR show similar increases in Nurr1 expression, indicating exogenous chromophore is not required for opto-DN-CREB function. e (Left) Experimental design for experiment showing opto-DN-CREB enhances CREB function. (Right) In cultured hippocampal neurons expressing opto-DN-CREB, 1 h of BL increased Creb expression [HSV-GFP (n = 3), HSV-opto-DN-CREB (n = 4)]. All error bars represent SEM; n.s. represents p > 0.05, * represents p < 0.05, ** represents p < 0.001.
Viral vectors
We used three replication-defective herpes simplex viral vectors (HSV) (see Fig. 1b): (1) HSV-CREB, originally described in ref. [29], contains a fusion protein of wild-type (WT) CREB with GFP expressed under the control of the HSV IE 4/5 promoter, (2) control vectors HSV-GFP or HSV-tdTomato (tdT) that express GFP or tdT after the HSV IE4/5 promoter, and (3) HSV-opto-DN-CREB, in which the light-inducible dominant-negative form of CREB is expressed after the IE 4/5 promoter. To identify neurons infected with HSV-opto-DN-CREB, tdT was also expressed (driven by the CMV promoter in the same vector).
Replication-deficient HSV-derived particles were packaged in-house as previously described [29] to titers > 1 × 108 infectious units/mL. HSV randomly infects ~10–20% of principal (excitatory) neurons when injected into the LA [23] and reaches maximal expression levels between 2 and 4 d post-injection [44–47].
In vitro analysis of opto-DN-CREB function
We previously characterized opto-DN-CREB in a series of biochemical studies and showed this construct functions as a light-responsive dominant-negative inhibitor of CREB [38]. Here we examined whether functional opto-DN-CREB requires addition of an exogenous chromophore and whether blue-light activation of opto-DN-CREB increases Creb expression, thereby increasing CREB function.
HEK cells
To examine the necessity of exogenously adding the chromophore (CHR) for opto-DN-CREB function, we treated opto-DN-CREB-expressing HEK cells with forskolin (FSK) (an activator of adenylyl cyclase) and BL, in the presence or absence of CHR. As our downstream readout of CREB function, we examined levels of the CREB-responsive gene Nurr1. In HEK cells, Nurr1 is one of the most highly CREB-responsive genes [48]. Because there is no robust antibody to detect Nurr1, we quantified Nurr1 levels using quantitative real-time PCR (RT-qPCR).
HEK293T cells were grown in complete feeding medium (Dulbecco’s modified eagle medium (DMEM, Life Technologies, Inc.) with 10% fetal bovine serum (FBS) at 37 °C in the dark, in 6-well plates and transfected (Lipofectamine 2000, Thermo Fisher) with opto-DN-CREB or control plasmid. Approximately 48 h later, cells were washed and chromophore (activated p-coumaric acid, S-thiophenylester [38], 0.1 mg/mL, referred to as p-coumaric acid, CHR) or vehicle added. Cells were treated with FSK (50 μM) for 2 h in serum-free medium before being moved to an incubator with or without BL illumination (0.2 mW/cm2) for 1 h.
Primary hippocampal neurons
To examine whether BL activation of opto-DN-CREB enhanced Creb expression, we used primary hippocampal neurons as described in [49, 50]. After 14 d in culture, 2 µL of virus (HSV-opto-DN-CREB or HSV-GFP) was added for 2 d. Neurons were then illuminated with BL (0.2 mW/cm2) for 1 h, and 1 h following this, neurons were processed for RT-qPCR to examine changes in Creb expression.
RT-qPCR
Cell culture medium was aspirated, and cells were flash frozen before RNA isolation (EZ-10 RNA isolation kit, Bio-Basic). Total RNA was quantified, and 500–1000 ng of RNA was made into cDNA using a high capacity cDNA reverse transcriptase kit (Life Technologies Inc). RT-qPCR was performed on 2 ng of cDNA using Evagreen mastermix (ABM) in a Biorad CFX96 real-time detection system. Ct thresholds were determined automatically and compared using the ∆∆Ct method. The following primers were added to each reaction (final concentration 500 nM):
Nurr1 F: 5′ CAACTACAGCACAGGCTACGA 3′;
Nurr1 R: 5′ GCATCTGAATGTCTTCTACCTTAATG 3′;
Creb F: 5′ GAACGAAAGCAGTGACGGA 3′;
Creb R: 5′ CTAAGGTTACAGTGGGAGCAG 3′.
Between 4 and 6 replicates were measured for each condition and levels of target transcript were normalized to levels of the housekeeping gene, Hprt (primers: 5′ GGAGTCCTGTTGATGTTGCCAGTA 3′; 5′ GGGACGCAGCAACTGACATTTCTA 3′) within each sample. Data were normalized to transfected/infected cells in the control condition.
Surgery
Mice were pre-treated with atropine sulfate (0.1 mg/kg, i.p.), anesthetized with chloral hydrate (400 mg/kg, i.p.) and placed in a stereotaxic frame. Topical lidocaine was applied to the incision site. The skull was exposed, and holes drilled bilaterally above the LA (AP −1.2, ML ±3.4, DV −5.0) [51]. Glass micropipettes filled with virus of interest were slowly lowered into the brain and virus (1.5 μL) was injected over 20 min. Following microinfusion, the micropipette was left in place for an additional 15 min to ensure viral diffusion. Following surgery, mice were given the analgesic Metacam (2 mg/kg), and recovered for 4 h before being returned to normal housing conditions.
For in vivo optogenetic experiments requiring BL stimulation, we also implanted bilateral optical fibers (fibers consisted of a 5-mm-long, 200 µm diameter, 0.39 numerical aperture fiber attached to a 1.25-mm zirconia ferrule) 0.5 mm above virus injection site, as previously described [19]. Optical fibers were constructed in-house by cutting a 10-mm-long fiber, polishing the fiber, and fixing it into a ferrule using epoxy resin. Optical fibers were secured in place using screws and dental cement.
Optogenetic stimulation
Two days after surgery, mice received BL stimulation (or no light stimulation, as a control) at different times relative to auditory fear conditioning.
Pre-training BL stimulation
Mice were individually placed in cages and received either BL stimulation (473 nm, 20 Hz, 10% duty cycle, 1–1.5 mW) or no stimulation for 1 h (Fig. 3b, e, f). Mice were then returned to homecage for 1 h before being placed in the fear conditioning apparatus and trained. We chose the parameters for photostimulation based on a similar system [52] and because our pilot data showed that BL stimulation for 5 min immediately before training was not sufficient to induce changes in memory (data not shown, F1,8 = 0.01, p > 0.05). In a separate group of mice (Fig. 3e), the same procedure was followed except that mice received BL stimulation 24 h before training.
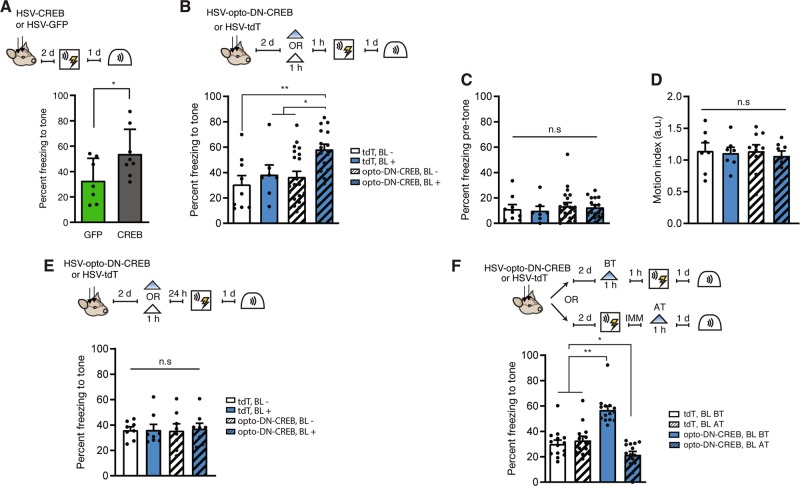
Fig. 3. Photostimulation of opto-DN-CREB in the minutes (but not 24 h) before (but not immediately after) conditioning enhances formation of an auditory fear memory.
a (Left) Outline of experimental design. (Right) Similar to previous studies, virally overexpressing wild-type CREB for 2–3 d before training enhanced auditory fear memory (HSV-CREB, (n = 8); HSV-GFP, (n = 7)). b (Top) Outline of experimental design. (Bottom) Photostimulating opto-DN-CREB-expressing LA neurons for 1 h, 1 h before training, enhanced auditory fear memory (percent time freezing to the tone) (tdT BL− (n = 9), tdT BL+ (n = 7), opto-DN-CREB BL− (n = 21), opto-DN-CREB BL+ (n = 16)). c Photostimulating opto-DN-CREB-expressing LA neurons did not enhance freezing or d impact general motion before tone presentation in these mice (for c, tdT BL− (n = 9), tdT BL+ (n = 7), opto-DN-CREB BL− (n = 21), opto-DN-CREB BL+ (n = 16); for d, tdT BL− (n = 7), tdT BL+ (n = 7), opto-DN-CREB BL− (n = 10), opto-DN-CREB BL+ (n = 8)). e Photostimulating opto-DN-CREB-expressing LA neurons for 1 h, 24 h before training, failed to enhance auditory fear memory (for all groups, n = 8). f Photostimulating opto-DN-CREB-expressing LA neurons for 1 h, immediately after training, impaired auditory fear memory (tdT + BL before training (BT) (n = 14), tdT + BL after training (AT) (n = 14), opto-DN-CREB + BL BT (n = 14), opto-DN-CREB + BL AT (n = 14)]. Error bars = SEM; n.s. p > 0.05, * p < 0.05, ** p < 0.001.
Post-training BL stimulation
Immediately after fear conditioning, mice were individually placed in a cage and either BL stimulation or no stimulation was applied for 1 h, as above (Fig. 3f). Mice were then returned to homecage.
Auditory fear training and testing
Training
Mice were placed in a conditioning chamber (31 × 24 × 21 cm) with a stainless-steel grid floor through which shocks could be delivered (Med Associates, St Albans, VT). After 2 min, a 30-s tone (93 dB, 2800 Hz) conditioned stimulus (CS) that co-terminated with a 2-s footshock (0.4 mA), the unconditioned stimulus (US), was played. Mice remained in the chambers for an additional 30 s before being returned to the homecage.
Testing
Memory was tested 24 h later, by placing mice in novel context (a chamber with a white plastic floor and semi-circular white plastic walls). Following 2 min, the tone CS was played for 1 min. Our index of memory was the amount of time spent freezing during the tone. Freezing is an active defensive response defined as cessation of movement, except for breathing [53, 54]. Video Freeze software (version 2.7.1.107, Med Associates, Inc.) was used to assess freezing both before and during the tone CS, as previously described [13, 23, 29, 30]. The freezing scores generated by this software correlate well with human scored freezing levels.
To more thoroughly investigate potential differences in movement between groups before tone onset in the memory test, we also used DeepLabCut software as a means of developing a motion index [55, 56]. DeepLabCut uses deep learning to estimate body part positions from unlabeled videos. We trained the DeepLabCut neural network by manually labeling the positions of the snout, ear, and base of tail in randomly selected frames from a subset of our behavioral videos. Our trained model was then able to extract the positions of these body parts across videos from a randomly selected subset of mice in each of the four experimental groups (tdT BL− (n = 7), tdT BL+ (n = 7); opto-DN-CREB BL− (n = 10), opto-DN-CREB BL+ (n = 8)). To calculate motion index, we computed the change in position for each of the three body parts from one frame to the next and averaged across the pre-tone period.
Verification of virus and fiber placement
Following behavioral experiments, mice were perfused, and their brains fixed with 4% paraformaldehyde (PFA) for 1 d, then transferred to a 30% sucrose solution for 2 d. Brains were sliced coronally (40 μm) using a cryostat. Slices were mounted onto a gelatin-coated slides, and cover-slipped with a Vectashield fluorescence mounting medium containing DAPI (to label nuclei). The LA was imaged on a laser confocal microscope (LSM 710; Zeiss). Images were analyzed using ImageJ software. To estimate the percentage of infection, we counted the number of tdT- or GFP-positive neurons within LA fields of view and divided by the total number of neurons (estimated by number of DAPI-labeled cells, excluding DAPI signal that was brightly colored and concentrated nuclei likely representing glial cells [57]).
Only data corresponding to mice with robust bilateral transgene expression limited to the LA were included in subsequent analysis (“hits”). A “hit” was defined as robust tdT or GFP fluorescence (depending on vector used) bilaterally in each LA across four or more consecutive LA slices. Similar to several previous reports [13, 23, 29, 45, 49, 50, 58], HSV microinjection resulted in expression of the transgene in a small proportion of LA neurons (20.5% ± 1.74, n = 3 for HSV-tdT and 19.9% ± 2.24, n = 3 for HSV-opto-DN-CREB) with minimal damage to surrounding tissue (Fig. 2b). Similar criteria were used to verify fiber placement, with the defining criteria being placement ~0.5 mm dorsal to the LA.
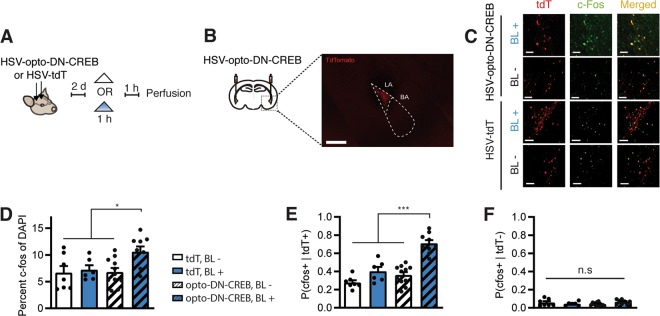
Fig. 2. Photostimulation of LA neurons expressing opto-DN-CREB increases expression of the CREB-dependent immediate early gene c-Fos.
a Outline of experimental design. b Microinjection of HSV-opto-DN-CREB (or HSV-tdTomato, HSV-tdT) into LA produced robust expression in small, random proportion of neurons. Scale bar = 100 μm. Dashed box outline shows damage from optic fiber, dashed triangle outlines LA. c Representative images showing increase in c-Fos (green) after blue light (BL+) stimulation, specifically in neurons expressing opto-DN-CREB (red). Scale bars = 50 μm. Images quantified in d–f. d In mice expressing opto-DN-CREB, BL stimulation increased overall number of c-Fos+ cells in the LA (as a function of DAPI). e, f Quantification of c-Fos expression in infected (tdT+) (e) and non-infected (tdT−) (f) LA neurons in mice microinjected with HSV-opto-DN-CREB or HSV-tdT. e Increase in number of c-Fos+ cells in infected neurons in mice expressing opto-DN-CREB following blue-light stimulation (and not in mice expressing tdT alone). f No increase in the number of c-Fos+ cells in non-infected neurons following BL stimulation (or no stimulation, BL−) in mice microinjected with HSV-tdT or HSV-opto-DN-CREB (for d–f, tdT BL+ (n = 6 slices from two mice); tdT BL− (n = 7 slices from three mice); opto-DN-CREB BL+ (n = 9 slices from two mice); opto-DN-CREB BL− (n = 11 slices from three mice)). Error bars = SEM; n.s. p > 0.05, *p < 0.05, **p < 0.001.
Ex vivo analysis of opto-DN-CREB
To assess the effectiveness of BL activation of opto-DN-CREB in vivo, we examined c-Fos protein levels as a measure of downstream CREB function. Two days after surgery in which mice were microinfused with HSV-opto-DN-CREB or control virus, HSV-tdT, mice received BL stimulation (473 nm, 20 Hz, 1–1.5 mW, the same stimulation protocol used in behavioral experiments) or no BL stimulation for 1 h. One hour later, mice were perfused and brains removed. Brains were post-fixed with 4% PFA for 1 d then transferred to a 30% sucrose solution for 2 d.
Brains were sliced coronally (40 μm) using a cryostat. Slices were washed and incubated with rabbit anti-c-Fos polyclonal antibody (1:500, Santa Cruz) at −4 °C. Staining was visualized with a fluorescent green secondary goat anti-rabbit Alexa 488 (1:500, Invitrogen). Slices were mounted onto a gelatin-coated slides, and cover-slipped with a Vectashield fluorescence mounting medium containing DAPI (to label nuclei).
Data analysis and statistics
Data were analyzed using ANOVAs to test differences of means between experimental groups. All statistical tests were considered statistically reliable at a p-value of <0.05. Significant main effects were further analyzed using Newman–Keuls post-hoc comparisons. All analyses were conducted in Statistica software package (Dell Inc. 2016, version 13).
Results
Photostimulation of opto-DN-CREB, even without exogenous chromophore, promotes transcription of CREB target genes
Conformational changes in PYP structure are initiated by the absorption of light by the covalently attached 4-hydroxycinnamic acid (p-coumaric acid) chromophore [59]. In the total absence of this p-coumaric acid chromophore, PYP is not thought to undergo conformational change, and we would expect our PYP-based opto-DN-CREB construct to be insensitive to BL stimulation (Fig. 1a). Previous data from Escherichia coli and yeast show that addition of exogenous chromophore is required to produce BL-activated PYP [60]. Therefore, in our previous in vitro experiments, we added p-coumaric acid directly to the medium. Exogenously adding this chromophore, though, complicates in vivo experiments. Here, we assessed whether exogenous application of p-coumaric acid is essential for PYP function under all experimental conditions. To examine the necessity of adding exogenous p-coumaric acid for PYP function in opto-DN-CREB in HEK cells, we compared the expression levels of the CREB target gene Nurr1 in cells expressing opto-DN-CREB both in the presence and absence of exogenous p-coumaric acid. To increase CREB function in these cells, we first stimulated with FSK (to activate CREB) and then applied BL (to photoactivate the PYP in our opto-DN-CREB construct) or not.
As expected, FSK increased the expression of the CREB target gene Nurr1 in HEK cells. This increase in Nurr1 expression was further enhanced with BL stimulation of opto-DN-CREB, regardless of whether the exogenous chromophore (CHR) was added or not (Fig. 1c, two-way ANOVA with between-group factors, p-coumaric acid (CHR+ or CHR−) and Treatment (Ctrl, FSK or FSK + BL) revealed a significant CHR × Treatment interaction F2,29 = 7.49, p < 0.01. Post-hoc Newman–Keuls tests showed that in both the CHR+ and CHR− conditions, BL application further increased the levels of FSK-induced Nurr1 expression. Furthermore, the relative magnitude of the increase in Nurr1 expression (FSK + BL normalized to FSK) did not differ between the two conditions (with and without exogenous application of p-coumaric acid, Fig. 1d, F1,10 = 0.06, p > 0.05). These data indicate that exogenous application of the chromophore is not required for opto-DN-CREB to activate CREB transcription under these conditions. Thus, the necessity for exogenous chromophore in PYP function may depend on cell type or culture conditions, as was the case for channelrhodopsin and the chromophore retinal [61].
Photostimulation of opto-DN-CREB increased CREB transcription in cultured hippocampal neurons
Evidence suggests CREB promotes its own expression [62, 63]. Therefore, we assessed whether optically relieving CREB inhibition (via BL activation of opto-DN-CREB) enhances Creb expression in cultured neurons. Isolated hippocampal neurons (14 days in vitro, (DIV)) were infected with opto-DN-CREB or GFP control virus. Two days later, BL was applied for 1 h followed by 1 h of rest. Neurons were then processed for RNA, and Creb expression assayed. Importantly, exogenous chromophore was not added. A robust increase in Creb expression was observed following BL stimulation in neurons expressing opto-DN-CREB compared to GFP control virus (Fig. 1e, F1,5 = 30.98, p < 0.01). Therefore, BL activation of opto-DN-CREB increased CREB function in cultured neurons (without addition of the exogenous chromophore).
Photostimulation of opto-DN-CREB in vivo increased the levels of a CREB-dependent protein
To verify that BL stimulation of opto-DN-CREB increased CREB function in vivo, we examined c-Fos expression in mice microinjected with opto-DN-CREB virus (or control virus) into the LA and treated with BL stimulation (or not) (Fig. 2a). We examined c-Fos expression because this immediate early gene contains consensus CRE sites in its promoter region and its transcription is CREB-dependent [62, 63].
Consistent with previous results using similar HSV viruses in other brain regions [23, 58], we observed robust transgene expression in a small portion of principal neurons in the LA (Fig. 2b). Mice expressing opto-DN-CREB and stimulated with BL showed higher overall c-Fos+ cells in the LA than control groups (not stimulated with BL or not expressing opto-DN-CREB) (Fig. 2d, two-way ANOVA with between-groups factors Virus (opto-DN-CREB or Control) and Light (BL or BL−) showed a significant interaction, F1,29 = 4.86, p < 0.05, post-hoc Newman–Keuls revealed mice with opto-DN-CREB + BL showed higher levels of c-Fos than all other groups (p < 0.05), which did not differ from each other). This overall increase in c-Fos expression was cell-autonomous. That is, there was an increase in c-Fos in photostimulated opto-DN-CREB-expressing neurons only and not in surrounding neurons (Fig. 2e; two-way ANOVA with between-group factors Light (BL+ or BL−) and Virus (opto-DN-CREB or TdTomato) showed a significant interaction F1,29 = 9.52, p < 0.05. Post-hoc Newman–Keuls tests revealed that c-Fos+ was higher in opto-DN-CREB + BL group than all other groups (p < 0.001); Fig. 2f, two-way ANOVA, Light × Virus interaction, F1,29 = 3.95, p > 0.05]. These in vivo results are consistent with our in vitro data and indicate photostimulation of opto-DN-CREB promotes expression of CREB target genes specifically in neurons expressing opto-DN-CREB, even in the absence of exogenous chromophore.
In vivo photostimulation of opto-DN-CREB in a small proportion of LA neurons 1 h before auditory fear conditioning enhanced memory formation
Next, we used opto-DN-CREB to manipulate CREB function at different time points relative to auditory fear conditioning. Previously, we [13, 23, 29] and others [14, 64] showed that prolonged overexpression of CREB (2–3 d) in a small, but random, portion of LA principal neurons enhanced auditory fear conditioned memory, as neurons with relatively increased CREB function were preferentially allocated to the engram supporting this memory. First, we replicated these results. Mice microinjected with HSV-CREB (a virus in which wild-type CREB is constitutively expressed) 2 d before auditory fear conditioning showed enhanced memory compared to mice microinjected with control vector (Fig. 3a, F1,13 = 4.79, p < 0.05).
To examine whether restricting the increase in CREB function to the minutes (rather than days) before fear conditioning, we microinjected mice with HSV-opto-DN-CREB or control virus. Two days later, mice received BL stimulation (for 1 h, or no BL stimulation) 1 h before fear conditioning. Mice microinjected with opto-DN-CREB and stimulated with BL showed enhanced freezing to the tone compared to all other groups (Fig. 3b; two-way ANOVA with between-group factors Light (BL+ or BL−) and Virus (opto-DN-CREB or TdTomato): there was a significant main effect of light F1,49 = 6.54, p < 0.05, and a significant main effect of virus F1,49 = 5.13, p < 0.05. Post-hoc Newman–Keuls tests revealed the opto-DN-CREB group froze significantly more than all other groups (p < 0.05)). Interestingly, the size of the memory enhancement was strikingly similar to that produced by increasing CREB via microinjection of HSV-CREB 2 d before training (Fig. 3a). This increase in freezing behavior was specific to the tone, as all groups showed similarly low freezing in the test session before tone onset (Fig. 3c; two-way ANOVA with between-group factors Light and Virus: Light × Virus interaction F1,49 = 0.01, p > 0.05]. Furthermore, there was no difference in activity between the groups before tone onset (Fig. 3d; two-way ANOVA with between-group factors Light and Virus: Light × Virus interaction F1,28 = 0.85, p > 0.05). These results converge to show the specificity of the effects of increasing CREB function on memory.
Importantly, similarly photostimulating opto-DN-CREB for 1 h, 24 h before fear conditioning did not enhance freezing showing the reversibility of increased CREB function with opto-DN-CREB (Fig. 3e; two-way ANOVA with between-group factors Light and Virus: Light × Virus interaction F1,28 = 0.03, p > 0.05). Together, these behavioral data indicate that multi-day overexpression is not required for the memory-enhancing effects of CREB, as a relatively short increase in CREB function in the minutes (but not 24 h) before training is sufficient to enhance memory.
In vivo photostimulation of opto-DN-CREB in a small proportion of LA neurons after auditory fear conditioning disrupts memory
Finally, opto-DN-CREB allowed us, for the first time, to examine the mnemonic effects of increasing CREB function in a small population of random neurons immediately after training. In this experiment, BL was applied immediately after, rather than before, training. Neurons expressing opto-DN-CREB would likely not be allocated to the engram supporting the conditioned fear memory as training was conducted in the absence of BL at a time when these neurons likely had decreased CREB function. We hypothesized that increasing CREB function in these random (non-engram) neurons might impair subsequent memory expression of this particular memory by adding post-training transcriptional or population activity “noise”.
We microinjected mice with opto-DN-CREB or control virus and photostimulated either before (as a control) or immediately after training. As before (Fig. 3b), photostimulating opto-DN-CREB-expressing neurons 1 h before training enhanced subsequent memory retrieval (Fig. 3f; two-way ANOVA with between-group factors Virus (opto-DN-CREB or TdTomato) and Time (stimulation before training or after training), Virus × Time interaction, F1,52 = 40.54, p < 0.0001. Post-hoc Newman–Keuls tests revealed the opto-DN-CREB group froze significantly more than all other groups (p < 0.001). However, photostimulating opto-DN-CREB expressing neurons immediately after training disrupted subsequent memory retrieval (Fig. 3f, post-hoc Newman–Keuls tests revealed that post-training opto-DN-CREB stimulation significantly reduced freezing compared to TdTomato control groups (p < 0.05)). Therefore, artificially increasing CREB function in a small subset of random (non-engram) neurons after training inhibited subsequent memory retrieval. Together, these data again show the critical role of CREB in memory allocation and, furthermore, highlight the importance of the timing of increased CREB function in memory processes.
Discussion
Here we used a novel light-activated protein system to examine the effects of acutely increasing CREB levels in a small, random portion of LA neurons on neuronal allocation to an engram supporting auditory conditioned fear, and the subsequent effects on memory consolidation. Previous experiments show that overexpressing CREB in LA neurons for 2–3 d before training enhanced memory as neurons overexpressing CREB were preferentially allocated to an engram supporting that memory. Here we showed that enhancing CREB in the minutes, but not 24 hours, before training was sufficient to enhance memory formation. In stark contrast, similarly enhancing CREB function in random neurons immediately after training disrupted subsequent retrieval of that memory. These results not only show the importance of the timing of increased CREB activity in relation to the training event on subsequent memory expression but also highlight the potential of optogenetic approaches to control protein function with temporal specificity.
In vivo use of PYP-based vectors
In developing and initially characterizing opto-DN-CREB, we focused on finding optimal conditions for controlling the PYP protein. To this end, we supplemented the feeding medium with activated p-coumaric acid, a chromophore necessary for conferring photosensitivity to PYP [38]. However, the in vivo utility of using opto-DN-CREB, or similar PYP-based protein systems, would be enhanced greatly if exogenous administration of this chromophore was not required. In HEK cells, isolated mouse neurons, and in behaving mice, we found that photostimulation of opto-DN-CREB increased CREB function, even in the absence of exogenously delivered chromophore. Therefore, under some conditions, sufficient levels of the chromophore may be available endogenously to enable PYP to undergo conformational change. p-Coumaric acid, for instance, is found in many plants [65] and is absorbed through the intestine [66]. Indeed, p-coumaric acid has been detected in several human metabolic studies [67, 68]. Therefore, mice may acquire sufficient p-coumaric through their diet to allow photostimulation to induce conformational changes in PYP. It remains to be determined, however, whether p-coumaric acid levels via diet are sufficient for producing optimal functioning of PYP proteins in cells and tissues other than those studied here.
Using opto-DN-CREB to enhance CREB function
We hypothesized that relieving CREB inhibition (by photostimulating opto-DN-CREB-expressing neurons) produces an abrupt increase in CREB function. Supporting this idea, photostimulating cultured neurons expressing opto-DN-CREB increased Creb transcription fourfold above control conditions. Furthermore, photostimulating LA neurons expressing opto-DN-CREB increased levels of c-Fos. Together, these findings provide compelling convergent evidence the opto-DN-CREB construct can be used to acutely increase CREB function. The current method offers advantages in terms of speed and molecular specificity over other common inducible expression systems to manipulate CREB (or indeed other protein) function.
Enhancing CREB function in the minutes-to-hours before training enhances memory
CREB is a transcription factor involved in multiple processes [69]. A number of these processes have been proposed to mediate neuronal allocation to an engram (Josselyn et al. 2018; [24, 28, 70, 71]). For instance, increasing CREB function increases neuronal excitability [14, 23, 72–76] and artificially increasing neuronal excitability without directly targeting CREB produces a similar effect (enhancing memory and biasing neuronal allocation) [23]. In addition, increasing CREB function increases dendritic spine density [73, 77], providing a complementary potential mechanism for the allocation effect.
Here we replicated this basic CREB-allocation effect, and, furthermore, found that increasing CREB function in the minutes before auditory fear conditioning was sufficient to enhance memory. The magnitude of this memory enhancement was similar to that observed when CREB was overexpressed for 2–3 d before conditioning suggesting that it is the relative level of CREB function in the minutes before training that plays a critical role in neuronal allocation.
Enhancing CREB function in random neurons immediately after training disrupts memory
This novel optogenetic tools allowed us to examine the effects of increasing CREB function immediately after training. In contrast to the effects of increasing CREB function before training, increasing CREB function after training impaired subsequent memory retrieval. We postulate that at least two processes may contribute to the observed memory weakening. First, tightly controlled “waves of transcription” [78, 79] have been described after memory formation, and inappropriately activating an important transcription factor such as CREB may impair the endogenous training-initiated genetic program and negatively impact memory formation. Second, in vivo electrophysiological data suggest that memory formation requires offline post-encoding reactivation of the neuronal activity patterns present during the initial learning experience. This reactivation or “replay” occurs with precise spatial and temporal fidelity [80–83]. Although most studied in the hippocampus, some types of memory involve offline reactivation of amygdala neurons [15, 84–86]. Therefore, non-discriminately increasing CREB in a random population of non-engram neurons may interfere with the precise reactivation required for memory consolidation. Consistent with this, we observed a similar disruption in subsequent memory expression using chemogenetics to artificially increase neuronal excitability in the LA 18 h after training in a rewarding task (cocaine-induced conditioned place preference) [15].
Use of PYP to modify the function of other proteins
Since the term “optogenetics” was coined [87, 88], there has been an explosion of tools that make use of light to either activate or repress cells, enzymes, or signaling pathways [87–89]. Some optogenetic tools, such as the light-gated ion channels (including channelrhodopsins), have gained popularity, due to their ability to modulate the electrical properties of cells. Although the widespread use of optogenetics and light to modify the function of proteins and other molecules is relatively slower, this field is also showing remarkable progress [90].
The PYP protein used in the present study to allow light-induced modification of CREB function is part of large protein domain family the Per-ARNT-Sim (PAS) family. This family also includes the light–oxygen–voltage domain (LOV) proteins. LOV-based optogenetic tools use endogenous flavin chromophores [90]. A number of LOV-based optogenetic tools have been used to modulate the activity of the protein RAC1 within spines [91], dimerization of proteins, and affinity of proteins to their binding partners (reviewed in [90, 92]). Although PYP domains are not as commonly used as LOV domains for designing optogenetic tools, this may derive from the expectation in vivo experiments would require exogenous addition of the chromophore [90]. The present findings show exogenous chromophore is not always required for PYP, thus increasing the potential applicability of PYP-based optogenetic tools for in vivo applications requiring temporally restricted, rapid, and reversible control of proteins.
Funding and disclosure
The authors declare no competing interests.
Acknowledgements
This work was supported by grants from the Canadian Institutes of Health Research (CIHR, grant numbers FDN-388455 to SAJ, FDN143227 to PWF), Natural Science and Engineering Council of Canada (NSERCs to SAJ and PWF), CIFAR catalyst award (SAJ, PWF) and an NIH (NIMH, 1 R01 MH119421-01) (SAJ, PWF). BJW was partially supported by a SickKids Research Training Centre Restracomp Fellowship and ADJ by a Canadian Open Neuroscience Platform Student Scholar Award (in partnership with Brain Canada).
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Albert Park, Alexander D. Jacob, Brandon J. Walters, Sungmo Park
Reference
- 1.Josselyn SA, Kohler S, Frankland PW. Finding the engram. Nat Rev Neurosci. 2015;16:521–34. doi: 10.1038/nrn4000. [DOI] [PubMed] [Google Scholar]
- 2.Josselyn SA, Kohler S, Frankland PW. Heroes of the Engram. J Neurosci. 2017;37:4647–57. doi: 10.1523/JNEUROSCI.0056-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tonegawa S, Liu X, Ramirez S, Redondo R. Memory engram cells have come of age. Neuron. 2015;87:918–31. doi: 10.1016/j.neuron.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 4.Schacter DL. Forgotten ideas, neglected pioneers: Richard Semon and the story of memory. Philadelphia: Psychology Press; 2001.
- 5.Semon R. Die Mneme als erhaltendes Prinzip im Wechsel des organischen Geschehens. Leipzig;1904.
- 6.Schacter DL, Eich JE, Tulving E. Richard Semon's theory of memory. J Verbal Learn Verbal Behav. 1978;17:721–43. [Google Scholar]
- 7.Ryan TJ, Tonegawa S. Rehebbilitating memory. Neuropsychopharmacology. 2016;41:1437. doi: 10.1038/npp.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frankland PW, Josselyn SA, Kohler S. The neurobiological foundation of memory retrieval. Nat Neurosci. 2019;22:1576–85. doi: 10.1038/s41593-019-0493-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denny CA, Kheirbek MA, Alba EL, Tanaka KF, Brachman RA, Laughman KB, et al. Hippocampal memory traces are differentially modulated by experience, time, and adult neurogenesis. Neuron. 2014;83:189–201. doi: 10.1016/j.neuron.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reijmers LG, Perkins BL, Matsuo N, Mayford M. Localization of a stable neural correlate of associative memory. Science. 2007;317:1230–3. doi: 10.1126/science.1143839. [DOI] [PubMed] [Google Scholar]
- 11.DeNardo LA, Liu CD, Allen WE, Adams EL, Friedmann D, Fu L, et al. Temporal evolution of cortical ensembles promoting remote memory retrieval. Nat Neurosci. 2019;22:460–9. doi: 10.1038/s41593-018-0318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sørensen AT, Cooper YA, Baratta MV, Weng F-J, Zhang Y, Ramamoorthi K, et al. A robust activity marking system for exploring active neuronal ensembles. eLife. 2016;5:e13918. doi: 10.7554/eLife.13918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han JH, Kushner SA, Yiu AP, Hsiang HL, Buch T, Waisman A, et al. Selective erasure of a fear memory. Science. 2009;323:1492–6. doi: 10.1126/science.1164139. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Y, Won J, Karlsson MG, Zhou M, Rogerson T, Balaji J, et al. CREB regulates excitability and the allocation of memory to subsets of neurons in the amygdala. Nat Neurosci. 2009;12:1438–43. doi: 10.1038/nn.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsiang HL, Epp JR, van den Oever MC, Yan C, Rashid AJ, Insel N, et al. Manipulating a "cocaine engram" in mice. J Neurosci. 2014;34:14115–27. doi: 10.1523/JNEUROSCI.3327-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koya E, Golden SA, Harvey BK, Guez-Barber DH, Berkow A, Simmons DE, et al. Targeted disruption of cocaine-activated nucleus accumbens neurons prevents context-specific sensitization. Nat Neurosci. 2009;12:1069–73. doi: 10.1038/nn.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanaka KZ, Pevzner A, Hamidi AB, Nakazawa Y, Graham J, Wiltgen BJ. Cortical representations are reinstated by the hippocampus during memory retrieval. Neuron. 2014;84:347–54. doi: 10.1016/j.neuron.2014.09.037. [DOI] [PubMed] [Google Scholar]
- 18.Lacagnina AF, Brockway ET, Crovetti CR, Shue F, McCarty MJ, Sattler KP, et al. Distinct hippocampal engrams control extinction and relapse of fear memory. Nat Neurosci. 2019;22:753–61. doi: 10.1038/s41593-019-0361-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rashid AJ, Yan C, Mercaldo V, Hsiang HL, Park S, Cole CJ, et al. Competition between engrams influences fear memory formation and recall. Science. 2016;353:383–7. doi: 10.1126/science.aaf0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X, Ramirez S, Pang PT, Puryear CB, Govindarajan A, Deisseroth K, et al. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature. 2012;484:381–5. doi: 10.1038/nature11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cowansage KK, Shuman T, Dillingham BC, Chang A, Golshani P, Mayford M. Direct reactivation of a coherent neocortical memory of context. Neuron. 2014;84:432–41. doi: 10.1016/j.neuron.2014.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Redondo RL, Kim J, Arons AL, Ramirez S, Liu X, Tonegawa S. Bidirectional switch of the valence associated with a hippocampal contextual memory engram. Nature. 2014;513:426–30. doi: 10.1038/nature13725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yiu AP, Mercaldo V, Yan C, Richards B, Rashid AJ, Hsiang HL, et al. Neurons are recruited to a memory trace based on relative neuronal excitability immediately before training. Neuron. 2014;83:722–35. doi: 10.1016/j.neuron.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 24.Rogerson T, Cai DJ, Frank A, Sano Y, Shobe J, Lopez-Aranda MF, et al. Synaptic tagging during memory allocation. Nat Rev Neurosci. 2014;15:157–69. doi: 10.1038/nrn3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J, Kwon JT, Kim HS, Josselyn SA, Han JH. Memory recall and modifications by activating neurons with elevated CREB. Nat Neurosci. 2014;17:65–72. doi: 10.1038/nn.3592. [DOI] [PubMed] [Google Scholar]
- 26.Abdou K, Shehata M, Choko K, Nishizono H, Matsuo M, Muramatsu SI, et al. Synapse-specific representation of the identity of overlapping memory engrams. Science. 2018;360:1227–31. doi: 10.1126/science.aat3810. [DOI] [PubMed] [Google Scholar]
- 27.Guskjolen A, Kenney JW, de la Parra J, Yeung BA, Josselyn SA, Frankland PW. Recovery of "lost" infant memories in mice. Curr Biol. 2018;28:2283–90.e3. doi: 10.1016/j.cub.2018.05.059. [DOI] [PubMed] [Google Scholar]
- 28.Josselyn SA, Frankland PW. Memory allocation: mechanisms and function. Annu Rev Neurosci. 2018;41:389–13. doi: 10.1146/annurev-neuro-080317-061956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han JH, Kushner SA, Yiu AP, Cole CJ, Matynia A, Brown RA, et al. Neuronal competition and selection during memory formation. Science. 2007;316:457–60. doi: 10.1126/science.1139438. [DOI] [PubMed] [Google Scholar]
- 30.Han JH, Yiu AP, Cole CJ, Hsiang HL, Neve RL, Josselyn SA. Increasing CREB in the auditory thalamus enhances memory and generalization of auditory conditioned fear. Learn Mem. 2008;15:443–53. doi: 10.1101/lm.993608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Josselyn SA. Continuing the search for the engram: examining the mechanism of fear memories. J Psychiatry Neurosci. 2010;35:221–8. doi: 10.1503/jpn.100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silva AJ, Zhou Y, Rogerson T, Shobe J, Balaji J. Molecular and cellular approaches to memory allocation in neural circuits. Science. 2009;326:391–5. doi: 10.1126/science.1174519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park S, Kramer EE, Mercaldo V, Rashid AJ, Insel N, Frankland PW, et al. Neuronal allocation to a hippocampal engram. Neuropsychopharmacology. 2016;41:2987–93. doi: 10.1038/npp.2016.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sekeres MJ, Neve RL, Frankland PW, Josselyn SA. Dorsal hippocampal CREB is both necessary and sufficient for spatial memory. Learn Mem. 2010;17:280–3. doi: 10.1101/lm.1785510. [DOI] [PubMed] [Google Scholar]
- 35.Sekeres MJ, Mercaldo V, Richards B, Sargin D, Mahadevan V, Woodin MA, et al. Increasing CRTC1 function in the dentate gyrus during memory formation or reactivation increases memory strength without compromising memory quality. J Neurosci. 2012;32:17857–68. doi: 10.1523/JNEUROSCI.1419-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sano Y, Shobe JL, Zhou M, Huang S, Shuman T, Cai DJ, et al. CREB regulates memory allocation in the insular cortex. Curr Biol. 2014;24:2833–7. doi: 10.1016/j.cub.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Czajkowski R, Jayaprakash B, Wiltgen B, Rogerson T, Guzman-Karlsson MC, Barth AL, et al. Encoding and storage of spatial information in the retrosplenial cortex. Proc Natl Acad Sci USA. 2014;111:8661–6. doi: 10.1073/pnas.1313222111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ali AM, Reis JM, Xia Y, Rashid AJ, Mercaldo V, Walters BJ, et al. Optogenetic inhibitor of the transcription factor CREB. Chem Biol. 2015;22:1531–9. doi: 10.1016/j.chembiol.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahn S, Olive M, Aggarwal S, Krylov D, Ginty DD, Vinson C. A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol Cell Biol. 1998;18:967–77. doi: 10.1128/mcb.18.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–61. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- 41.Landeira BS, Santana T, Araujo JAM, Tabet EI, Tannous BA, Schroeder T, et al. Activity-independent effects of CREB on neuronal survival and differentiation during mouse cerebral cortex development. Cereb Cortex. 2018;28:538–48. doi: 10.1093/cercor/bhw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jancic D, Lopez, de Armentia M, Valor LM, Olivares R, Barco A. Inhibition of cAMP response element-binding protein reduces neuronal excitability and plasticity, and triggers neurodegeneration. Cereb Cortex. 2009;19:2535–47. doi: 10.1093/cercor/bhp004. [DOI] [PubMed] [Google Scholar]
- 43.Imamoto Y, Kataoka M. Structure and photoreaction of photoactive yellow protein, a structural prototype of the PAS domain superfamily. Photochemistry Photobiol. 2007;83:40–49. doi: 10.1562/2006-02-28-IR-827. [DOI] [PubMed] [Google Scholar]
- 44.Carlezon WA, Jr., Neve RL. Viral-mediated gene transfer to study the behavioral correlates of CREB function in the nucleus accumbens of rats. Methods Mol Med. 2003;79:331–50. doi: 10.1385/1-59259-358-5:331. [DOI] [PubMed] [Google Scholar]
- 45.Carlezon WA, Jr., Nestler EJ, Neve RL. Herpes simplex virus-mediated gene transfer as a tool for neuropsychiatric research. Crit Rev Neurobiol. 2000;14:47–67. doi: 10.1080/08913810008443546. [DOI] [PubMed] [Google Scholar]
- 46.Neve RL, Neve KA, Nestler EJ, Carlezon WA., Jr Use of herpes virus amplicon vectors to study brain disorders. Biotechniques. 2005;39:381–91. doi: 10.2144/05393PS01. [DOI] [PubMed] [Google Scholar]
- 47.Barrot M, Olivier JD, Perrotti LI, DiLeone RJ, Berton O, Eisch AJ, et al. CREB activity in the nucleus accumbens shell controls gating of behavioral responses to emotional stimuli. Proc Natl Acad Sci USA. 2002;99:11435–40. doi: 10.1073/pnas.172091899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci USA. 2005;102:4459–64. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walters BJ, Mercaldo V, Gillon CJ, Yip M, Neve RL, Boyce FM, et al. The role of the RNA demethylase FTO (fat mass and obesity-associated) and mRNA methylation in hippocampal memory formation. Neuropsychopharmacology. 2017;42:1502–10. doi: 10.1038/npp.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cole CJ, Mercaldo V, Restivo L, Yiu AP, Sekeres MJ, Han JH, et al. MEF2 negatively regulates learning-induced structural plasticity and memory formation. Nat Neurosci. 2012;15:1255–64. doi: 10.1038/nn.3189. [DOI] [PubMed] [Google Scholar]
- 51.Paxinos G, Franklin KBJ. The mouse brain in stereotaxic coordinates. 2nd ed. San Diego: Academic Press; 2001. [Google Scholar]
- 52.Murakoshi H, Shin ME, Parra-Bueno P, Szatmari EM, Shibata ACE, Yasuda R. Kinetics of endogenous CaMKII required for synaptic plasticity revealed by optogenetic kinase inhibitor. Neuron. 2017;94:690. doi: 10.1016/j.neuron.2017.04.027. [DOI] [PubMed] [Google Scholar]
- 53.Blanchard RJ, Blanchard DC. Crouching as an index of fear. J Comp Physiol. Psychol. 1969;67:370–5. doi: 10.1037/h0026779. [DOI] [PubMed] [Google Scholar]
- 54.Fanselow MS, Lester LS. A functional behavioristic approach to aversively motivated behavior: predatory imminence as a determinant of the topography of defensive behavior. In: Bolles, RC, Beecher, MD. editors. Evolution and learning. Hillsdale, NJ, US: Lawrence Erlbaum Associates, Inc; 1988. p. 185–212.
- 55.Mathis A, Mamidanna P, Cury KM, Abe T, Murthy VN, Mathis MW, et al. DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nat Neurosci. 2018;21:1281–9. doi: 10.1038/s41593-018-0209-y. [DOI] [PubMed] [Google Scholar]
- 56.Nath T, Mathis A, Chen AC, Patel A, Bethge M, Mathis MW. Using DeepLabCut for 3D markerless pose estimation across species and behaviors. Nat Protoc. 2019;14:2152–76. doi: 10.1038/s41596-019-0176-0. [DOI] [PubMed] [Google Scholar]
- 57.Chawla MK, Lin G, Olson K, Vazdarjanova A, Burke SN, McNaughton BL, et al. 3D-catFISH: a system for automated quantitative three-dimensional compartmental analysis of temporal gene transcription activity imaged by fluorescence in situ hybridization. J Neurosci Methods. 2004;139:13–24. doi: 10.1016/j.jneumeth.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 58.Calipari ES, Godino A, Salery M, Damez-Werno DM, Cahill ME, Werner CT, et al. Synaptic microtubule-associated protein EB3 and SRC phosphorylation mediate structural and behavioral adaptations during withdrawal from cocaine self-administration. J Neurosci. 2019;39:5634–46. [DOI] [PMC free article] [PubMed]
- 59.Brechun KE, Zhen D, Jaikaran A, Borisenko V, Kumauchi M, Hoff WD, et al. Detection of incorporation of p-coumaric acid into photoactive yellow protein variants in vivo. Biochemistry. 2019;58:2682–94. doi: 10.1021/acs.biochem.9b00279. [DOI] [PubMed] [Google Scholar]
- 60.Reis JM, Xu X, McDonald S, Woloschuk RM, Jaikaran ASI, Vizeacoumar FS, et al. Discovering selective binders for photoswitchable proteins using phage display. ACS Synth Biol. 2018;7:2355–64. doi: 10.1021/acssynbio.8b00123. [DOI] [PubMed] [Google Scholar]
- 61.Fang-Yen C, Alkema MJ, Samuel AD. Illuminating neural circuits and behaviour in Caenorhabditis elegans with optogenetics. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140212. doi: 10.1098/rstb.2014.0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lesiak A, Pelz C, Ando H, Zhu M, Davare M, Lambert TJ, et al. A genome-wide screen of CREB occupancy identifies the RhoA inhibitors Par6C and Rnd3 as regulators of BDNF-induced synaptogenesis. PLoS ONE. 2013;8:e64658. doi: 10.1371/journal.pone.0064658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walker WH, Daniel PB, Habener JF. Inducible cAMP early repressor ICER down-regulation of CREB gene expression in Sertoli cells. Mol Cell Endocrinol. 1998;143:167–78. doi: 10.1016/s0303-7207(98)00082-3. [DOI] [PubMed] [Google Scholar]
- 64.Rogerson T, Jayaprakash B, Cai DJ, Sano Y, Lee YS, Zhou Y, et al. Molecular and cellular mechanisms for trapping and activating emotional memories. PLoS ONE. 2016;11:e0161655. doi: 10.1371/journal.pone.0161655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pei K, Ou J, Huang J, Ou S. p-Coumaric acid and its conjugates: dietary sources, pharmacokinetic properties and biological activities. J Sci food agriculture. 2016;96:2952–62. doi: 10.1002/jsfa.7578. [DOI] [PubMed] [Google Scholar]
- 66.Konishi Y, Hitomi Y, Yoshioka E. Intestinal absorption of p-coumaric and gallic acids in rats after oral administration. J Agric food Chem. 2004;52:2527–32. doi: 10.1021/jf035366k. [DOI] [PubMed] [Google Scholar]
- 67.Loke WM, Jenner AM, Proudfoot JM, McKinley AJ, Hodgson JM, Halliwell B, et al. A metabolite profiling approach to identify biomarkers of flavonoid intake in humans. J Nutr. 2009;139:2309–14. doi: 10.3945/jn.109.113613. [DOI] [PubMed] [Google Scholar]
- 68.Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vazquez-Fresno R, et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46:D608–17. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–23. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 70.Rao-Ruiz P, Yu J, Kushner SA, Josselyn SA. Neuronal competition: microcircuit mechanisms define the sparsity of the engram. Curr Opin Neurobiol. 2019;54:163–70. doi: 10.1016/j.conb.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lisman J, Cooper K, Sehgal M, Silva AJ. Memory formation depends on both synapse-specific modifications of synaptic strength and cell-specific increases in excitability. Nat Neurosci. 2018;21:309–14. doi: 10.1038/s41593-018-0076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dong Y, Green T, Saal D, Marie H, Neve R, Nestler EJ, et al. CREB modulates excitability of nucleus accumbens neurons. Nat Neurosci. 2006;9:475–7. doi: 10.1038/nn1661. [DOI] [PubMed] [Google Scholar]
- 73.Marie H, Morishita W, Yu X, Calakos N, Malenka RC. Generation of silent synapses by acute in vivo expression of CaMKIV and CREB. Neuron. 2005;45:741–52. doi: 10.1016/j.neuron.2005.01.039. [DOI] [PubMed] [Google Scholar]
- 74.Han MH, Bolanos CA, Green TA, Olson VG, Neve RL, Liu RJ, et al. Role of cAMP response element-binding protein in the rat locus ceruleus: regulation of neuronal activity and opiate withdrawal behaviors. J Neurosci. 2006;26:4624–9. doi: 10.1523/JNEUROSCI.4701-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lopez de Armentia M, Jancic D, Lugan R, Kandel ER, Barco A. The survival of CA1 hippocampal neurons depend on normal levels of activity in the CREB pathway. Society for Neuroscience Abstract MM100. 2006.
- 76.Benito E, Barco A. CREB's control of intrinsic and synaptic plasticity: implications for CREB-dependent memory models. Trends Neurosci. 2010;33:230–40. doi: 10.1016/j.tins.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 77.Sargin D, Mercaldo V, Yiu AP, Higgs G, Han JH, Frankland PW, et al. CREB regulates spine density of lateral amygdala neurons: implications for memory allocation. Front Behav Neurosci. 2013;7:209. doi: 10.3389/fnbeh.2013.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Igaz LM, Vianna MR, Medina JH, Izquierdo I. Two time periods of hippocampal mRNA synthesis are required for memory consolidation of fear-motivated learning. J Neurosci. 2002;22:6781–9. doi: 10.1523/JNEUROSCI.22-15-06781.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rodriguez-Blanco LA, Rivera-Olvera A, Escobar ML. Consolidation of an aversive taste memory requires two rounds of transcriptional and epigenetic regulation in the insular cortex. Behav Brain Res. 2019;356:371–4. doi: 10.1016/j.bbr.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 80.Joo HR, Frank LM. The hippocampal sharp wave-ripple in memory retrieval for immediate use and consolidation. Nat Rev Neurosci. 2018;19:744–57. doi: 10.1038/s41583-018-0077-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Buzsaki G. Hippocampal sharp wave-ripple: a cognitive biomarker for episodic memory and planning. Hippocampus. 2015;25:1073–188. doi: 10.1002/hipo.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Carr MF, Jadhav SP, Frank LM. Hippocampal replay in the awake state: a potential substrate for memory consolidation and retrieval. Nat Neurosci. 2011;14:147–53. doi: 10.1038/nn.2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kudrimoti HS, Barnes CA, McNaughton BL. Reactivation of hippocampal cell assemblies: effects of behavioral state, experience, and EEG dynamics. J Neurosci. 1999;19:4090–101. doi: 10.1523/JNEUROSCI.19-10-04090.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Girardeau G, Inema I, Buzsáki G. Reactivations of emotional memory in the hippocampus–amygdala system during sleep. Nat Neurosci. 2017;20:1634. doi: 10.1038/nn.4637. [DOI] [PubMed] [Google Scholar]
- 85.Kanta V, Pare D, Headley DB. Closed-loop control of gamma oscillations in the amygdala demonstrates their role in spatial memory consolidation. Nat Commun. 2019;10:3970. doi: 10.1038/s41467-019-11938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Feng F, Headley DB, Amir A, Kanta V, Chen Z, Pare D, et al. Gamma oscillations in the basolateral amygdala: biophysical mechanisms and computational consequences. eNeuro. 2019;6:ENEURO.0388-18.2018. doi: 10.1523/ENEURO.0388-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Deisseroth K, Feng G, Majewska AK, Miesenbock G, Ting A, Schnitzer MJ. Next-generation optical technologies for illuminating genetically targeted brain circuits. J Neurosci. 2006;26:10380–6. doi: 10.1523/JNEUROSCI.3863-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Deisseroth K. Optogenetics: 10 years of microbial opsins in neuroscience. Nat Neurosci. 2015;18:1213–25. doi: 10.1038/nn.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Josselyn SA. The past, present and future of light-gated ion channels and optogenetics. eLife. 2018;7: e42367. doi: 10.7554/eLife.42367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Losi A, Gardner KH, Moglich A. Blue-light receptors for optogenetics. Chem Rev. 2018;118:10659–709. doi: 10.1021/acs.chemrev.8b00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hayashi-Takagi A, Yagishita S, Nakamura M, Shirai F, Wu YI, Loshbaugh AL, et al. Labelling and optical erasure of synaptic memory traces in the motor cortex. Nature. 2015;525:333–8. doi: 10.1038/nature15257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rost BR, Schneider-Warme F, Schmitz D, Hegemann P. Optogenetic tools for subcellular applications in neuroscience. Neuron. 2017;96:572–603. doi: 10.1016/j.neuron.2017.09.047. [DOI] [PubMed] [Google Scholar]