Animal models of viral pathogenesis are essential tools in human disease research. Human papillomaviruses (HPVs) are a significant public health issue due to their widespread sexual transmission and oncogenic potential. Infection-based models of papillomavirus pathogenesis have been complicated by their strict species and tissue specificity. In this Gem, we discuss the discovery of a murine papillomavirus, Mus musculus papillomavirus 1 (MmuPV1), and how its experimental use represents a major advancement in models of papillomavirus-induced pathogenesis/carcinogenesis, and their transmission.
KEYWORDS: animal models, human papillomavirus, murine papillomavirus, papillomavirus, viral oncology, viral pathogenesis, MmuPV1
ABSTRACT
Animal models of viral pathogenesis are essential tools in human disease research. Human papillomaviruses (HPVs) are a significant public health issue due to their widespread sexual transmission and oncogenic potential. Infection-based models of papillomavirus pathogenesis have been complicated by their strict species and tissue specificity. In this Gem, we discuss the discovery of a murine papillomavirus, Mus musculus papillomavirus 1 (MmuPV1), and how its experimental use represents a major advancement in models of papillomavirus-induced pathogenesis/carcinogenesis, and their transmission.
INTRODUCTION
Viruses significantly affect human health, and the prevention, control, and treatment of viral infections require a fundamental understanding of their pathogenesis. Such knowledge demands investigation not only at the molecular and cellular levels but also in the organisms they infect. Animal viruses are vital to our understanding of human viruses, including infection, transmission, host responses, and pathogenesis. Moreover, they provide critical preclinical models in which to identify preventative and therapeutic approaches to human viral infections and disease. Such models have provided pivotal insight into viruses such as influenza virus, herpesviruses, Zika virus, and emerging viruses (1–5). Studying human viruses using genetically tractable and cost-effective animal models is often complicated by their strict species tropism. This is true for human papillomaviruses (HPVs), thus limiting the use of infection models to study these common and important human pathogens.
Papillomaviridae is a large and diverse family of nonenveloped, double-stranded circular DNA viruses that by and large exhibit rigid species and tissue tropism. There are more than 220 formally accepted types that infect humans (6–8). Human papillomaviruses infect stratified squamous epithelia in the oral cavity and upper respiratory tract, the anogenital tract, and the skin and cause a range of pathologies, from warts (papillomas) to dysplasia and cancer. HPV genotypes are classified by their oncogenic potential. Low-risk HPV types cause benign skin, oral/respiratory, and genital papillomas, whereas high-risk HPVs cause cancer (9). High-risk mucosal HPVs are the etiological factors of nearly all cervical cancers, a large number of vaginal, penile, and anal cancers, and a subset of head and neck cancers, particularly of the oropharynx (10). Certain high-risk HPVs, such as HPV16 and HPV18, cause the majority of HPV-associated cancers (11–13). Cutaneous HPVs are also linked to certain types of skin malignancies (14). Given their broad diversity, prevalence, and oncogenic potential, HPVs are one of the top infectious causes of human cancer, causing approximately 5% of cancers worldwide (10, 15).
The public health threat of mucosotropic HPVs is exacerbated by their being the most common sexually transmitted infection in the United States (16). While most infections are cleared (17), persistent HPV infections can be established and are a major risk factor for progression to cancer (18, 19). Particular high-risk HPVs such as HPV16 are more likely to establish persistent infections, contributing to their oncogenic potential (20). HPV persistence is often accompanied by viral genome integration into host DNA, which occurs at random sites but with a preference for chromosomal fragile sites, genes, and enhancers (21–24). Integration is thought to potentiate HPV-mediated oncogenic progression by increasing the amount and stability of transcripts of the viral oncogenes E6 and E7 (25, 26), providing a selective growth advantage to cells (27). High-risk HPV E6 and E7 are well-validated, potent oncogenes. The highly multifunctional proteins they encode contribute to carcinogenesis at least in part by inactivating the major tumor suppressors p53 and pRb, both common targets of DNA tumor viruses (28, 29).
Prophylactic HPV vaccines are a significant milestone in the effort to control HPV-mediated cancers (30). In Australia, where HPV vaccination is mandatory for young girls and boys, cervical cancer is predicted to be eliminated as a public health problem within the next 20 years (31). However, inadequate vaccine availability and vaccination coverage have allowed HPV to remain a significant public health issue elsewhere (32, 33). Vaccination is also ineffective against preexisting HPV infections. For these reasons, there remains a pressing need to study HPV infection and persistence and the contribution of host and environmental factors to HPV transmission and subsequent disease. Many of the underlying mechanisms that govern these aspects of HPV pathogenesis have not been fully elucidated. Tractable animal models of papillomaviral pathogenesis are essential to advance our understanding of these viruses. In this Gem, we discuss existing comparative models of HPV pathogenesis and disease and focus on new and emerging models utilizing a murine papillomavirus, Mus musculus papillomavirus 1 (MmuPV1). These MmuPV1-based models have the potential to transform our ability to study the molecular basis of PV infection and pathogenesis and provide an opportunity to identify therapeutic interventions to control HPV transmission and disease.
EXISTING ANIMAL MODELS OF PAPILLOMAVIRUS PATHOGENESIS
As a result of the stringent host species specificity of PVs, animals do not support productive HPV infections. Researchers therefore rely on the use of animal PVs to establish infection models in their respective hosts or use genetically engineered mouse models, such as transgenic mice, to study the role of specific HPV genes in neoplastic disease.
Animal models of papillomavirus pathogenesis.
Modern sequencing techniques and increased sampling have started to reveal the broad diversity of animal papillomaviruses (8, 34–36). Animal models have contributed significantly to our understanding of papillomaviral pathogenesis, tissue tropism, and disease (for reviews, see references 37 to 40). The first animal papillomavirus studied was cottontail rabbit papillomavirus (CRPV), described in the 1930s as causing papillomas in rabbits (41). CRPV was subsequently found to promote cutaneous malignancies (42) and provided insights into a variety of virus-host interactions for PVs with cutaneous tropism (43). A mucosotropic PV, rabbit oral papillomavirus (ROPV), was later isolated from domestic rabbits and paved the way for studies in the oral mucosa as well as male and female genital tissues (44, 45). Given their divergent tropisms, CRPV and ROPV were useful models to study the underlying molecular mechanisms of PV tissue tropism. These models, along with canine oral PV (COPV) (46), were heavily utilized in testing vaccines, leading to the current HPV prophylactic vaccines (for reviews, see references 39, 47, and 48). Papillomaviruses that infect the multimammate rat species Mastomys natalensis (Mastomys natalensis papillomavirus 1 [MnPV1]) and Mastomys coucha (Mastomys coucha papillomavirus 2 [McPV2]) can establish persistent infections and promote tumorigenesis in the skin and anogenital tissues, respectively (49, 50). The Rosl group has extensively studied various aspects of PV biology using these Mastomys models (for a review, see reference 51), contributing novel observations related to viral mRNA splicing patterns in vivo (52) and the role of environmental factors such as UV radiation in cutaneous PV-associated carcinogenesis (53).
Comparative models of PV pathogenesis have also been established in cattle and nonhuman primates. Bovine papillomaviruses (BPVs) cause various pathologies in cattle (54) and horses (55). BPV-1 was the first fully sequenced PV genome (56) and was the subject of many in vitro transformation studies and the first papillomavirus transgenic mouse model (57). BPV-1 studies facilitated the discovery of the viral E5 oncoprotein (58, 59), which is also expressed by high-risk mucosal HPVs (for a review, see reference 60). The evolutionary proximity of nonhuman primates to humans made these species attractive models for studying HPV pathogenesis. Papillomaviruses have been associated with cutaneous and mucosal disease in colobus monkeys, rhesus macaques, and chimpanzees (35, 61–66). Some of these PVs show close sequence similarity to high-risk HPVs and are associated with neoplastic disease, including precancerous lesions and carcinomas (65, 67, 68). The first preclinical models of PV sexual transmission also were in nonhuman primates (67, 69). While the contribution of these animal models to our understanding of PV infection and pathogenesis cannot be overstated, they present challenges related to cost, availability of technical reagents, and genetic tractability.
HPV transgenic mouse models.
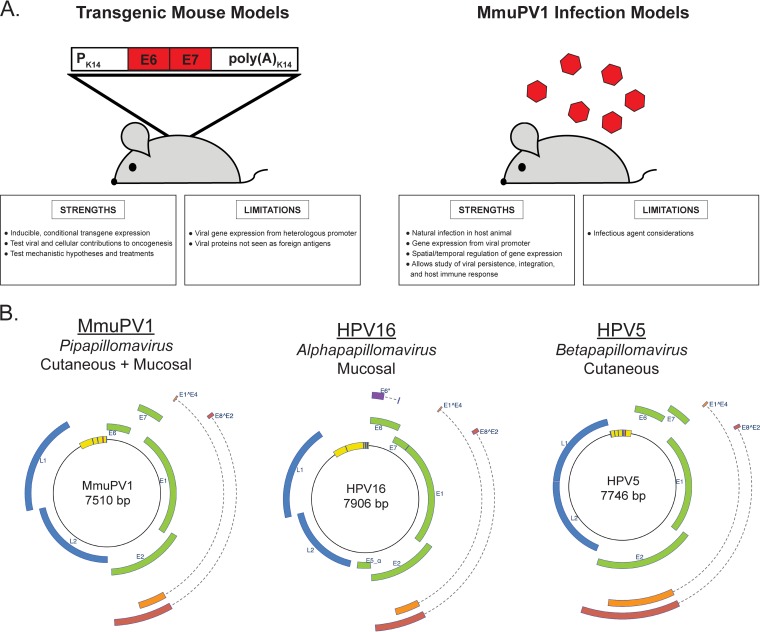
The most widely used, well-characterized, and technically well-supported animal used as a model system remains the laboratory mouse, Mus musculus (70). Until the discovery of a murine papillomavirus, the PV field lacked an infection-based system to model HPV-mediated carcinogenesis in laboratory mice. Instead, researchers employed genetically engineered, transgenic mice (for a review, see reference 71). The first such model involved insertion of 1.69 tandem copies of the BPV-1 genome into the mouse genome (57). These transgenic mice developed cutaneous fibropapillomas and fibrosarcomas, both of which contained replicating extrachromosomal BPV-1; however, the virus remained transcriptionally inactive in asymptomatic skin (72). This model established the feasibility of using transgenic mice to study PV-associated diseases. The high-risk mucosotropic HPV16 E6 and E7 oncogenes were then studied using transgenic mice (73–76), providing key insights into their contributions to tumorigenesis, including the abilities of E6 to inhibit apoptosis through p53-dependent and -independent means (74, 77, 78) and E7 to induce hyperplasia through its inactivation of the tumor suppressor pRb and dysregulation of E2F-dependent gene expression (77, 79). Epidermis-specific expression of E6 and E7 also induced skin tumors (76). A subsequent generation of transgenic mice directed HPV16 protein expression to the natural site of infection, the basal cells of the stratified squamous epithelia, using the keratin 14 (K14) promoter. These models expressed either the entire early region of the HPV16 genome (80) or the individual HPV16 viral oncogene E5 (81), E6 (82), or E7 (83). These transgenic mice have been instrumental in modeling HPV-induced progressive neoplastic disease and cancer development in the cervix (84–88), head and neck (89, 90), and anus (91). HPV-transgenic mice have allowed researchers to establish the relative potencies of individual HPV oncogenes in neoplastic disease (88, 89, 92–95), the role of host factors in HPV-associated disease (84–86, 90, 96–100), and therapeutic treatment efficacy (99, 101–103). Transgenic mice have also been developed to study cutaneotropic HPVs, such as HPV8 (104) and HPV38 (105), and their role together with UV radiation in promoting cutaneous disease and carcinogenesis (106–109). Clearly, HPV-transgenic models have provided a vital platform to study various aspects of papillomavirus-induced disease in vivo. However, their ability to model other key events during PV infection and pathogenesis is limited (Fig. 1A).
FIG 1.
Comparison of HPV transgenic models and MmuPV1 infection-based models. (A) Strengths and limitations of HPV transgenic mouse models and MmuPV1 infection-based models. (B) MmuPV1 genomic organization, classification, and tissue tropism compared to an Alphapapillomavirus, HPV16, and a Betapapillomavirus, HPV5. All viral genomes were generated and images adapted from the PAVE database (36; http://pave.niaid.nih.gov).
MmuPV1 DISCOVERY AND MOLECULAR VIROLOGY
Researchers have long sought an infection-based model of papillomaviral pathogenesis in laboratory mice to study aspects of viral pathogenesis not possible in transgenic mice, such as virus replication, persistence, transmission, and infection-mediated carcinogenesis (Fig. 1A). Until recently, no murine papillomavirus had been discovered. In 2011, Ingle and colleagues reported the isolation of a murine papillomavirus (MmuPV1) from cutaneous papillomas present on the skin of immunodeficient NMR1-FoxN1nu/nu mice (110). A highly similar MmuPV1 variant and a novel PV were subsequently isolated from normal skin of a house mouse (Mus musculus) and a wood mouse (Apodemus sylvaticus), respectively (111). Phylogenetically, MmuPV1 is classified in the genus Pipapillomavirus and is most closely related to other rodent papillomaviruses (40, 112). The genomic organization of MmuPV1 is comparable to that of other PV genomes, including common HPV genotypes (Fig. 1B). The MmuPV1 double-stranded DNA circular genome is composed of a noncoding upstream regulatory region (also known as the long control region) and early and late regions containing 7 translational open reading frames (ORFs). The early region ORFs encode the early (E) proteins E1, E2, E4, E6, and E7. Two late (L)-region ORFs encode the major and minor capsid proteins L1 and L2, respectively. MmuPV1 also expresses two spliced gene products, E1̂E4 and E8̂E2. There is no E5 ORF in the MmuPV1 genome, a trait that is shared with cutaneotropic beta HPVs and that differs from mucotropic alpha HPVs (Fig. 1B).
Despite their similarities, there are molecular and biochemical differences between MmuPV1 and HPVs. MmuPV1 shares little nucleotide sequence similarity (<70%) with other PVs (112), with MmuPV1 and HPV16, a prototypic oncogenic alpha PV, being 49.8% identical in sequence. In high-risk alpha HPVs, the major viral oncogenes E6 and E7 are transcribed from a single early promoter. In MmuPV1, E6 and E7 are transcribed from two separate early viral promoters (113). HPV16 E6 and E7 proteins share approximately 45% and 40% sequence identity with their MmuPV1 counterparts, respectively (112). At first glance, these differences call into question the use of MmuPV1 as a model for HPV-associated carcinogenesis, but experimental studies demonstrate that activities associated with transformation by high-risk alpha HPV oncoproteins are retained in MmuPV1 E6 and E7. High-risk alpha HPV E7 proteins contain an LXCXE motif, which binds pocket proteins, including the RB1 tumor suppressor. In MmuPV1, the LXCXE motif is present in E6 but not E7 (111, 112). However, MmuPV1 E7 is similar to the gamma PV HPV197 E7 protein in that it binds RB1 through LXCXE-independent mechanisms (114). Another possibility is that MmuPV1 E7 has RB1-independent oncogenic potential. White and colleagues recently reported that MmuPV1 E7 binds to the cellular nonreceptor protein tyrosine phosphatase PTPN14 (115), a protein targeted for HPV16 E7-mediated degradation to impair keratinocyte differentiation in a process independent of RB1 binding (116). The MmuPV1 E6 protein inhibits Notch and transforming growth factor β (TGF-β) signaling, both tumor suppressor pathways, to delay differentiation and promote proliferation, functions shared with high-risk beta HPV E6 proteins (117, 118). Like high-risk beta HPV E6 proteins, MmuPV1 E6 does not bind directly to p53. Schulz et al. noted that the C terminus of the MmuPV1 E7 protein contains a putative PDZ-binding motif, a feature present in alpha HPV E6 that interacts with cell polarity and motility proteins (111). Therefore, MmuPV1 E6 and E7 appear to retain multiple, potentially tumorigenic properties. Most importantly, MmuPV1 clearly exhibits oncogenic potential, as discussed below. Additional studies are required to further characterize binding partners of the MmuPV1 E6 and E7 proteins and to determine whether and to what extent these interactions contribute to pathogenesis.
MmuPV1 EXHIBITS EXPANDED TROPISM IN MICE
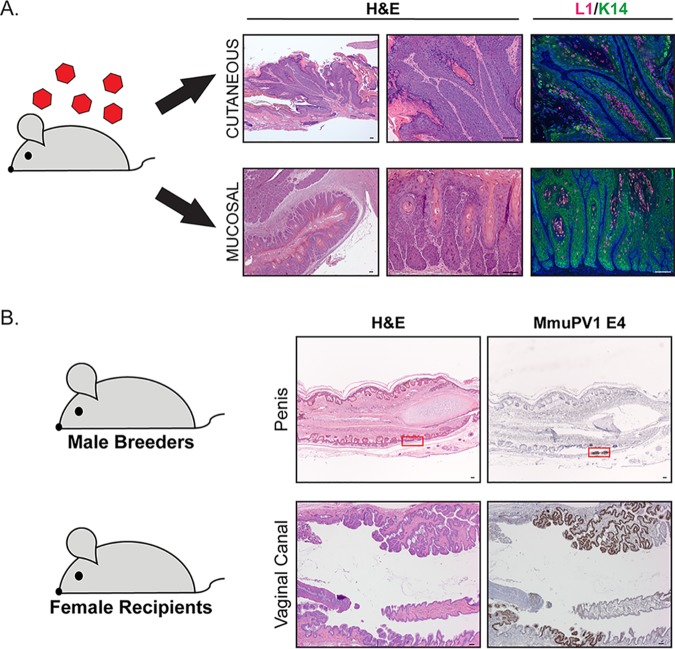
Given its isolation from cutaneous papillomas and its genomic similarities with cutaneous beta HPVs, MmuPV1 was initially considered a cutaneotropic virus (110). MmuPV1 infects and causes disease at cutaneous sites, including the tail, muzzle, back, and ears (110, 119–125). There are conflicting reports on whether dorsal skin supports MmuPV1 infection, suggesting strain- and skin-specific variability in susceptibility to MmuPV1 infection (120, 123, 126, 127). Importantly, mucosal epithelia also support experimental MmuPV1 infection in immunodeficient mice (119, 123, 127–130), and studies in our laboratory have verified this expanded tissue tropism in immunocompetent FVB/N mice (Fig. 2A) (131–133). Lateral transmission in FoxN1nu/nu immunodeficient mice between experimentally infected cutaneous sites and oral and vaginal mucosae (134) provides further evidence for dual tropism. Like MmuPV1, certain beta HPVs also exhibit dual tropism (135, 136). The expanded tissue tropism of MmuPV1 has facilitated the development of novel models of PV pathogenesis in both cutaneous and mucosal epithelia, described below.
FIG 2.
MmuPV1 exhibits expanded tissue tropism in mice and is a sexually transmitted virus. (A) MmuPV1 infects cutaneous and mucosal epithelia in immunocompetent mice, causing papillomas in skin (top row) and neoplastic disease in the female cervicovaginal mucosal epithelium (bottom row). Representative images of H&E (hematoxylin and eosin)-stained tissue show pathology. Productive MmuPV1 infection is indicated by immunofluorescence for MmuPV1 L1 protein (green), and keratin staining (red) highlights the epithelium. Bars = 100 μM. (B) MmuPV1 is a sexually transmitted virus. Shown are male (penile epithelium; top row) and female (vaginal canal; bottom row) reproductive organs of FVB/N mice that acquired MmuPV1 through sexual transmission. Representative images of H&E-stained tissue are shown, and RNA in situ hybridization for the E4 transcript (brown) indicates infected regions of epithelia (the red box highlights the region in male penile epithelium). Bars = 100 μM.
CURRENT MmuPV1 MODELS OF PAPILLOMAVIRUS INFECTION AND PATHOGENESIS
Virus entry and species and tissue tropism.
MmuPV1 provides an opportunity to explore mechanisms that govern PV species and tissue tropism. Much of the research comparing MmuPV1 and HPV entry and tropism has been performed using in vivo cervicovaginal and/or cutaneous infections with pseudoviruses (PsV), which do not contain the viral genome but rather a reporter gene (e.g., that encoding luciferase or green fluorescent protein) encapsidated into virus-like particles composed of papillomaviral capsid proteins L1 and L2. Initial studies using MmuPV1 and HPV PsVs revealed similar mechanisms for virus entry (137); however, some differences have emerged. Day and colleagues found that, while MmuPV1 PsV initiates infection at the basement membrane, it does so, at least initially, in a heparan sulfate proteoglycan (HSPG)-independent manner (138), unlike HPV16 and other mucosotropic alpha PVs, which associate with the basement membrane using HSPG-dependent mechanisms. Interestingly, cutaneotropic beta PVs, like HPV5, interact with heparin moieties in a manner that is also distinct from that of alpha PVs (139). Nevertheless, postentry intracellular trafficking of MmuPV1 PsV is similar to that of HPV PsV (138). These findings illuminate key differences and similarities in papillomavirus entry processes that are consistent with a previous report finding variations in how accurately animal PV replication models the HPV life cycle (140). Such limitations are inherent to animal models and warrant caution in extrapolating findings made in the use of MmuPV1 as a model for HPVs.
Studies of host immune response.
A major strength of infection-based models is the opportunity they provide to study the host immune response during a natural infection. Initially, several laboratories reported that MmuPV1 fails to infect and/or efficiently promote disease in common strains of immunocompetent mice (110, 120, 121, 123–125, 141). However, subsequent studies have revealed a more nuanced relationship. Immunocompetent SENCAR and S/RV/Cri-ba/ba bare mice are susceptible to MmuPV1 infection and disease when high virus titers are used (110, 121, 123), an observation confirmed in FVB/N mice in our laboratory (133, 142). Outbred immunocompetent SKH1 mice are also susceptible (122, 125). Mice of the common C57BL/6 strain seem particularly resistant to MmuPV1-induced disease (121, 123–125, 132), while they develop antibody responses and biomarkers for MmuPV1 infection in asymptomatically infected skin (121, 123, 125, 141), demonstrating that the genetic background of mice affects susceptibility to MmuPV1 infection.
The different sensitivities of various murine strains to MmuPV1 have helped define a role for the immune system, and more specifically, adaptive immunity, in regulating MmuPV1 infection. MmuPV1 was originally isolated from FoxN1nu/nu mice, which are homozygously null for the FoxN1 gene, resulting in athymic mice that are T-cell deficient (143). Using a combination of approaches, researchers discovered that T-cell deficiency renders mice susceptible to MmuPV1 infection and disease (110, 120, 121, 123–125, 127, 129, 134, 141). For instance, several other strains of mice carrying the FoxN1 genetic mutation are also susceptible to MmuPV1 infection (for a review, see reference 40). Various regimens of T-cell depletion revealed that complete T-cell deficiency correlates with susceptibility to MmuPV1 infection (121, 125). Systemic immunosuppression, induced with continuous cyclosporine treatment (121) or UV radiation (124), also can make resistant strains of mice more susceptible to MmuPV1 infection.
Several labs are using MmuPV1-based models to explore PV immunity. Pretreatment with an MmuPV1 L1 rabbit antiserum prevented MmuPV1-induced papillomas (120). Likewise, hyperimmune serum from mice immunized with MmuPV1 virus-like particles (VLPs) prevented MmuPV1 infection in highly susceptible T-cell-deficient strains of mice (127). Many studies continue to define the role of T-cell antitumor immunity to MmuPV1-based disease. The Roden lab revealed that MmuPV1 E6- and E7-specific CD8+ T-cell responses promote papilloma clearance and regression and that adoptively transferred E6-specific CD8+ T cells alone prevent MmuPV1-dependent tumor growth in nude mice (122, 125). In another study, MmuPV1-induced cutaneous tumors in T-cell-deficient mice regressed following adoptive transfer of hyperimmune splenocytes from congenic mice (144). Our laboratory discovered that a host stress keratin, keratin 17, is upregulated following MmuPV1 infection and prevents T cell recruitment to confer protection against cutaneous disease (142), highlighting a critical virus-host interaction involved in PV pathogenesis. Recently, a provocative study used an MmuPV1 cutaneous infection model to argue that immunity to cutaneous PVs protects against UV- and chemical-induced skin cancer (145). However, data from our laboratory contradict these findings, in that UV B radiation (UVB) treatment and MmuPV1 infection led to the development of cutaneous squamous cell carcinomas (SCCs) (124). Ongoing studies continue to evaluate other aspects of immunity, including the role of interferon signaling and neutrophil infiltration in MmuPV1 infection and disease (125, 126). More comprehensive reviews on MmuPV1 models and host immune response have been written (40, 146).
Studies of MmuPV1 viral gene products and host factors.
The alpha HPV E6 and E7 proteins, in addition to their oncogenic properties (28), function during the maintenance and productive phases of the HPV life cycle (147, 148). We found that MmuPV1-induced papilloma formation requires the E6 protein, as an E6-null (E6STOP) mutant MmuPV1 mutant failed to induce papillomas in nude mice (118). Likewise, an MmuPV1 E6 mutant that cannot bind MAML1 (E6R130A) also failed to induce papillomas. These findings suggest that E6 protein functions, including its inhibition of Notch signaling, are critical to MmuPV1-induced pathogenesis. Using antisera against MmuPV1 L1 and L2, Handisurya and colleagues found that during MmuPV1 cutaneous infections, L1 is expressed throughout all epithelial layers in papillomas instead of just suprabasal layers and is localized to the cytoplasm in the absence of L2, suggesting that a unique pattern of late gene expression and virion assembly may occur during the MmuPV1 life cycle (120). Additional studies are necessary to characterize the role of other MmuPV1 proteins in the viral life cycle, pathogenesis, and carcinogenesis.
The lack of an E5 protein is a key difference between MmuPV1 and high-risk alpha HPV genomes (Fig. 1B). We infected the skin and female reproductive tracts of K14E5 HPV16 transgenic mice with MmuPV1 to determine the effect of epithelial E5 expression on MmuPV1-associated pathogenesis (133). In MmuPV1-infected K14E5 mice, skin lesions exhibited earlier onset, higher incidence, and reduced frequency of spontaneous regression compared to nontransgenic mice. Moreover, estrogen-treated K14E5 mice were more likely to develop cervicovaginal cancers than their nontransgenic counterparts. Therefore, HPV16 E5 potentiates MmuPV1 pathogenesis. Further studies are necessary to determine mechanisms of E5 function in this context, which could involve the role of HPV16 E5 in suppressing immune responses (149). Complementation studies combining HPV16 transgenic mice and MmuPV1 infection provide a unique platform to study the role of high-risk alpha HPV proteins in pathogenesis.
New MmuPV1-based infection models provide important opportunities to study host factors that promote PV persistence, a key risk factor for subsequent malignant progression (18, 19). In the MmuPV1 cervicovaginal model, persistent infections are established in the mucosal epithelia of immunocompetent FVB/N mice that persist for at least 10 months (131, 132). Estrogen increases the severity of disease, and this correlates with the establishment of persistent infections (132). Notably, MmuPV1 viral copy numbers are highest during the estrus phase in immunodeficient mice (130). That estrogen is being revealed as a potential persistence factor is just one example of how MmuPV1 has the potential to illuminate roles of host factors in PV pathogenesis and disease.
MmuPV1 cancer models.
MmuPV1 E6 delays differentiation and promotes proliferation in keratinocytes in vitro and is necessary for papilloma development in vivo (118). However, there is relatively little published biochemical evidence for the transforming activity of the MmuPV1 viral proteins. That being said, there are now multiple studies demonstrating that MmuPV1 displays oncogenic potential in vivo. In cutaneous sites, immunodeficient nude mice experimentally infected with MmuPV1 developed poorly differentiated, locally invasive tumors that histologically resembled human trichoblastomas (123). Squamous cell carcinomas were also observed at cutaneous sites of secondary MmuPV1 infections in nude mice at 9.5 months postinfection (134). FVB/NJ mice infected with MmuPV1 and treated with UVB were found to develop SCC of the skin, demonstrating that MmuPV1 can cause cancer in immunocompetent mice (124).
MmuPV1 also displays oncogenic potential in mucosal tissues. Cladel et al. first reported that heterozygous nude mice (FoxN1nu/+) infected for 7.5 months with MmuPV1 develop carcinoma in situ in the female reproductive tract (126). Later, it was found that high-grade precancerous lesions and SCC developed in the female reproductive tract of immunocompetent FVB/N mice at 6 months postinfection (132). Similar to its role as a cocarcinogen in HPV16 transgenic mice (84–86, 88), estrogen significantly increased the incidence and severity of high-grade disease and cancer in MmuPV1-infected FVB/N mice (132). Further exploration of the role of estrogen and of whether anti-estrogen drugs are efficacious in preventing and treating disease, as is the case in HPV16-transgenic mice (101, 102), is warranted in MmuPV1-based infection models of cervicovaginal disease. These established and emerging MmuPV1 models show great potential for studying all stages of papillomavirus-mediated carcinogenesis, the role of host cofactors, and therapeutic treatments.
Models of PV transmission.
MmuPV1 models have the potential to identify unexplored facets of PV transmission relevant to public health. Mucosal HPV transmission occurs most frequently through sexual contact, and HPV infections are the most common sexually transmitted infections in the United States (16). Epidemiological data are used to understand HPV sexual transmission in humans (150); however, laboratory models to study the underlying molecular mechanisms involved in papillomavirus sexual transmission are lacking. While rhesus macaque PV 1 (RhPV-1) is sexually transmitted (67, 69), the cost, scalability and ethical considerations in the use of nonhuman primates limit its application to the study of sexual transmission of papillomaviruses. MmuPV1 was recently described as being sexually transmitted (131). Female FVB/N donor mice, experimentally infected in their cervicovaginal tracts with MmuPV1, transmitted MmuPV1 to untreated FVB/N male breeders through sexual transmission, indicating that the penile epithelium supports MmuPV1 infection (Fig. 2B). The infected male breeders subsequently transmitted MmuPV1 to untreated naive FVB/N recipient female mice (Fig. 2B). Approximately one-third of these recipient female mice acquired MmuPV1 infections, some transient and some prolonged, through natural sexual transmission. Therefore, MmuPV1 can be sexually transmitted in wild-type laboratory mice in the absence of any environmental or genetic manipulation. This powerful new animal model of natural PV sexual transmission promises to provide new insights into the molecular dynamics of PV sexual transmission in both male and female reproductive organs.
An MmuPV1 model has also provided potential evidence for blood-borne PV transmission (151). MmuPV1 was introduced into immunodeficient FoxN1nu/nu mice via tail vein injection; infections developed at prewounded cutaneous and mucosal sites, and virus was detected in the stomach. Furthermore, naive mice receiving blood from MmuPV1-infected animals developed disease in both cutaneous and mucosal epithelia. While these results are intriguing, they should be carefully interpreted. Experimentally wounded immunodeficient mice used in this study are highly susceptible to lateral transmission and infection by MmuPV1, which could be inadvertently introduced through a variety of environmental routes (animal bedding, handling, etc.). Therefore, the incorporation of appropriate mock-infected and unwounded controls into such experiments is critical for conclusive interpretation. Nevertheless, MmuPV1 promises to uncover new molecular insights into PV transmission.
CONCLUSIONS AND FUTURE PERSPECTIVES
The discovery of MmuPV1 has ultimately provided a practical and genetically tractable infection-based system to model HPV infection, transmission, and pathogenesis. Preclinical animal models of viral pathogenesis are inherently limited in their ability to unequivocally recapitulate every aspect of human infection and disease. However, as discussed in this Gem, MmuPV1-based infection models hold incredible promise for providing insight into facets of HPV pathogenesis that were difficult to study before their development. For these reasons, established and emerging MmuPV1 infection models represent a new frontier in animal models of PV pathogenesis and promise to transform our understanding of HPV-associated human disease.
ACKNOWLEDGMENTS
We express regret to those colleagues whose work could not be referenced due to space limitations.
Megan E. Spurgeon is supported by a grant from the National Institutes of Health (CA211246). Paul F. Lambert is supported by grants from the National Institutes of Health (CA022443, CA210807, CA228543, and DE026787).
REFERENCES
- 1.Adachi A, Miura T. 2014. Animal model studies on viral infections. Front Microbiol 5:672. doi: 10.3389/fmicb.2014.00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouvier NM, Lowen AC. 2010. Animal models for influenza virus pathogenesis and transmission. Viruses 2:1530–1563. doi: 10.3390/v20801530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dudley DM, Aliota MT, Mohr EL, Newman CM, Golos TG, Friedrich TC, O'Connor DH. 2019. Using macaques to address critical questions in Zika virus research. Annu Rev Virol 6:481–500. doi: 10.1146/annurev-virology-092818-015732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morrison TE, Diamond MS. 2017. Animal models of Zika virus infection, pathogenesis, and immunity. J Virol 91:e00009-17. doi: 10.1128/JVI.00009-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Safronetz D, Geisbert TW, Feldmann H. 2013. Animal models for highly pathogenic emerging viruses. Curr Opin Virol 3:205–209. doi: 10.1016/j.coviro.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernard HU, Burk RD, Chen Z, van Doorslaer K, Zur Hausen H, de Villiers EM. 2010. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 401:70–79. doi: 10.1016/j.virol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Villiers EM, Fauquet C, Broker TR, Bernard HU, Zur Hausen H. 2004. Classification of papillomaviruses. Virology 324:17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 8.Van Doorslaer K, Chen Z, Bernard HU, Chan PKS, DeSalle R, Dillner J, Forslund O, Haga T, McBride AA, Villa LL, Burk RD, Ictv Report C. 2018. ICTV virus taxonomy profile: Papillomaviridae. J Gen Virol 99:989–990. doi: 10.1099/jgv.0.001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cogliano V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, WHO International Agency for Research on Cancer. 2005. Carcinogenicity of human papillomaviruses. Lancet Oncol 6:204. doi: 10.1016/s1470-2045(05)70086-3. [DOI] [PubMed] [Google Scholar]
- 10.zur Hausen H. 2009. Papillomaviruses in the causation of human cancers—a brief historical account. Virology 384:260–265. doi: 10.1016/j.virol.2008.11.046. [DOI] [PubMed] [Google Scholar]
- 11.D'Souza G, Kreimer AR, Viscidi R, Pawlita M, Fakhry C, Koch WM, Westra WH, Gillison ML. 2007. Case-control study of human papillomavirus and oropharyngeal cancer. N Engl J Med 356:1944–1956. doi: 10.1056/NEJMoa065497. [DOI] [PubMed] [Google Scholar]
- 12.Hoots BE, Palefsky JM, Pimenta JM, Smith JS. 2009. Human papillomavirus type distribution in anal cancer and anal intraepithelial lesions. Int J Cancer 124:2375–2383. doi: 10.1002/ijc.24215. [DOI] [PubMed] [Google Scholar]
- 13.Munoz N, Bosch FX, Castellsague X, Diaz M, de Sanjose S, Hammouda D, Shah KV, Meijer CJ. 2004. Against which human papillomavirus types shall we vaccinate and screen? The international perspective. Int J Cancer 111:278–285. doi: 10.1002/ijc.20244. [DOI] [PubMed] [Google Scholar]
- 14.Rollison DE, Viarisio D, Amorrortu RP, Gheit T, Tommasino M. 2019. An emerging issue in oncogenic virology: the role of beta human papillomavirus types in the development of cutaneous squamous cell carcinoma. J Virol 93:e01003-18. doi: 10.1128/JVI.01003-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, Plummer M. 2012. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol 13:607–615. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- 16.Satterwhite CL, Torrone E, Meites E, Dunne EF, Mahajan R, Ocfemia MC, Su J, Xu F, Weinstock H. 2013. Sexually transmitted infections among US women and men: prevalence and incidence estimates, 2008. Sex Transm Dis 40:187–193. doi: 10.1097/OLQ.0b013e318286bb53. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez AC, Schiffman M, Herrero R, Hildesheim A, Bratti C, Sherman ME, Solomon D, Guillen D, Alfaro M, Morales J, Hutchinson M, Katki H, Cheung L, Wacholder S, Burk RD. 2010. Longitudinal study of human papillomavirus persistence and cervical intraepithelial neoplasia grade 2/3: critical role of duration of infection. J Natl Cancer Inst 102:315–324. doi: 10.1093/jnci/djq001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bodily J, Laimins LA. 2011. Persistence of human papillomavirus infection: keys to malignant progression. Trends Microbiol 19:33–39. doi: 10.1016/j.tim.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moscicki AB, Schiffman M, Burchell A, Albero G, Giuliano AR, Goodman MT, Kjaer SK, Palefsky J. 2012. Updating the natural history of human papillomavirus and anogenital cancers. Vaccine 30 Suppl 5:F24–33. doi: 10.1016/j.vaccine.2012.05.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schiffman M, Rodriguez AC, Chen Z, Wacholder S, Herrero R, Hildesheim A, Desalle R, Befano B, Yu K, Safaeian M, Sherman ME, Morales J, Guillen D, Alfaro M, Hutchinson M, Solomon D, Castle PE, Burk RD. 2010. A population-based prospective study of carcinogenic human papillomavirus variant lineages, viral persistence, and cervical neoplasia. Cancer Res 70:3159–3169. doi: 10.1158/0008-5472.CAN-09-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jang MK, Shen K, McBride AA. 2014. Papillomavirus genomes associate with BRD4 to replicate at fragile sites in the host genome. PLoS Pathog 10:e1004117. doi: 10.1371/journal.ppat.1004117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luft F, Klaes R, Nees M, Durst M, Heilmann V, Melsheimer P, von Knebel Doeberitz M. 2001. Detection of integrated papillomavirus sequences by ligation-mediated PCR (DIPS-PCR) and molecular characterization in cervical cancer cells. Int J Cancer 92:9–17. doi:. [DOI] [PubMed] [Google Scholar]
- 23.McBride AA, Warburton A. 2017. The role of integration in oncogenic progression of HPV-associated cancers. PLoS Pathog 13:e1006211. doi: 10.1371/journal.ppat.1006211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wentzensen N, Ridder R, Klaes R, Vinokurova S, Schaefer U, Doeberitz M. 2002. Characterization of viral-cellular fusion transcripts in a large series of HPV16 and 18 positive anogenital lesions. Oncogene 21:419–426. doi: 10.1038/sj.onc.1205104. [DOI] [PubMed] [Google Scholar]
- 25.Jeon S, Lambert PF. 1995. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: implications for cervical carcinogenesis. Proc Natl Acad Sci U S A 92:1654–1658. doi: 10.1073/pnas.92.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warburton A, Redmond CJ, Dooley KE, Fu H, Gillison ML, Akagi K, Symer DE, Aladjem MI, McBride AA. 2018. HPV integration hijacks and multimerizes a cellular enhancer to generate a viral-cellular super-enhancer that drives high viral oncogene expression. PLoS Genet 14:e1007179. doi: 10.1371/journal.pgen.1007179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeon S, Allen-Hoffmann BL, Lambert PF. 1995. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J Virol 69:2989–2997. doi: 10.1128/JVI.69.5.2989-2997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munger K, Baldwin A, Edwards KM, Hayakawa H, Nguyen CL, Owens M, Grace M, Huh K. 2004. Mechanisms of human papillomavirus-induced oncogenesis. J Virol 78:11451–11460. doi: 10.1128/JVI.78.21.11451-11460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.zur Hausen H. 2000. Papillomaviruses causing cancer: evasion from host-cell control in early events in carcinogenesis. J Natl Cancer Inst 92:690–698. doi: 10.1093/jnci/92.9.690. [DOI] [PubMed] [Google Scholar]
- 30.Frazer IH, Leggatt GR, Mattarollo SR. 2011. Prevention and treatment of papillomavirus-related cancers through immunization. Annu Rev Immunol 29:111–138. doi: 10.1146/annurev-immunol-031210-101308. [DOI] [PubMed] [Google Scholar]
- 31.Hall MT, Simms KT, Lew JB, Smith MA, Brotherton JM, Saville M, Frazer IH, Canfell K. 2019. The projected timeframe until cervical cancer elimination in Australia: a modelling study. Lancet Public Health 4:e19–e27. doi: 10.1016/S2468-2667(18)30183-X. [DOI] [PubMed] [Google Scholar]
- 32.Frazer IH. 2018. Eradicating HPV-associated cancer through immunization: a glass half full. Viral Immunol 31:80–85. doi: 10.1089/vim.2017.0119. [DOI] [PubMed] [Google Scholar]
- 33.Jemal A, Simard EP, Dorell C, Noone AM, Markowitz LE, Kohler B, Eheman C, Saraiya M, Bandi P, Saslow D, Cronin KA, Watson M, Schiffman M, Henley SJ, Schymura MJ, Anderson RN, Yankey D, Edwards BK. 2013. Annual report to the nation on the status of cancer, 1975-2009, featuring the burden and trends in human papillomavirus (HPV)-associated cancers and HPV vaccination coverage levels. J Natl Cancer Inst 105:175–201. doi: 10.1093/jnci/djs491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rector A, Van Ranst M. 2013. Animal papillomaviruses. Virology 445:213–223. doi: 10.1016/j.virol.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 35.Van Doorslaer K, Dillner J. 2019. The launch of an international animal papillomavirus reference center. Viruses 11:55. doi: 10.3390/v11010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Doorslaer K, Li Z, Xirasagar S, Maes P, Kaminsky D, Liou D, Sun Q, Kaur R, Huyen Y, McBride AA. 2017. The Papillomavirus Episteme: a major update to the papillomavirus sequence database. Nucleic Acids Res 45:D499–D506. doi: 10.1093/nar/gkw879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campo MS. 2002. Animal models of papillomavirus pathogenesis. Virus Res 89:249–261. doi: 10.1016/s0168-1702(02)00193-4. [DOI] [PubMed] [Google Scholar]
- 38.Christensen ND, Budgeon LR, Cladel NM, Hu J. 2017. Recent advances in preclinical model systems for papillomaviruses. Virus Res 231:108–118. doi: 10.1016/j.virusres.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doorbar J. 2016. Model systems of human papillomavirus-associated disease. J Pathol 238:166–179. doi: 10.1002/path.4656. [DOI] [PubMed] [Google Scholar]
- 40.Uberoi A, Lambert PF. 2017. Rodent papillomaviruses. Viruses 9 9:362. doi: 10.3390/v9120362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shope RE, Hurst EW. 1933. Infectious papillomatosis of rabbits: with a note on the histopathology. J Exp Med 58:607–624. doi: 10.1084/jem.58.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rous P, Beard JW. 1935. The progression to carcinoma of virus-induced rabbit papillomas (Shope). J Exp Med 62:523–548. doi: 10.1084/jem.62.4.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brandsma JL. 2005. The cottontail rabbit papillomavirus model of high-risk HPV-induced disease. Methods Mol Med 119:217–235. doi: 10.1385/1-59259-982-6:217. [DOI] [PubMed] [Google Scholar]
- 44.Harvey SB, Cladel NM, Budgeon LR, Welsh PA, Griffith JW, Lang CM, Christensen ND. 1998. Rabbit genital tissue is susceptible to infection by rabbit oral papillomavirus: an animal model for a genital tissue-targeting papillomavirus. J Virol 72:5239–5244. doi: 10.1128/JVI.72.6.5239-5244.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sundberg JP, Junge RE, el Shazly MO. 1985. Oral papillomatosis in New Zealand white rabbits. Am J Vet Res 46:664–668. [PubMed] [Google Scholar]
- 46.Watrach AM, Small E, Case MT. 1970. Canine papilloma: progression of oral papilloma to carcinoma. J Natl Cancer Inst 45:915–920. [PubMed] [Google Scholar]
- 47.Kirnbauer R. 1996. Papillomavirus-like particles for serology and vaccine development. Intervirology 39:54–61. doi: 10.1159/000150475. [DOI] [PubMed] [Google Scholar]
- 48.Christensen ND. 2005. Cottontail rabbit papillomavirus (CRPV) model system to test antiviral and immunotherapeutic strategies. Antivir Chem Chemother 16:355–362. doi: 10.1177/095632020501600602. [DOI] [PubMed] [Google Scholar]
- 49.Amtmann E, Volm M, Wayss K. 1984. Tumour induction in the rodent Mastomys natalensis by activation of endogenous papilloma virus genomes. Nature 308:291–292. doi: 10.1038/308291a0. [DOI] [PubMed] [Google Scholar]
- 50.Nafz J, Schafer K, Chen SF, Bravo IG, Ibberson M, Nindl I, Stockfleth E, Rosl F. 2008. A novel rodent papillomavirus isolated from anogenital lesions in its natural host. Virology 374:186–197. doi: 10.1016/j.virol.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 51.Hasche D, Rosl F. 2019. Mastomys species as model systems for infectious diseases. Viruses 11:182. doi: 10.3390/v11020182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salvermoser M, Chotewutmontri S, Braspenning-Wesch I, Hasche D, Rösl F, Vinzón SE. 2016. Transcriptome analysis of Mastomys natalensis papillomavirus in productive lesions after natural infection. J Gen Virol 97:1658–1669. doi: 10.1099/jgv.0.000471. [DOI] [PubMed] [Google Scholar]
- 53.Hasche D, Stephan S, Braspenning-Wesch I, Mikulec J, Niebler M, Gröne H-J, Flechtenmacher C, Akgül B, Rösl F, Vinzón SE. 2017. The interplay of UV and cutaneous papillomavirus infection in skin cancer development. PLoS Pathog 13:e1006723. doi: 10.1371/journal.ppat.1006723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Campo MS. 1987. Papillomas and cancer in cattle. Cancer Surv 6:39–54. [PubMed] [Google Scholar]
- 55.Chambers G, Ellsmore VA, O'Brien PM, Reid SWJ, Love S, Campo MS, Nasir L. 2003. Association of bovine papillomavirus with the equine sarcoid. J Gen Virol 84:1055–1062. doi: 10.1099/vir.0.18947-0. [DOI] [PubMed] [Google Scholar]
- 56.Chen EY, Howley PM, Levinson AD, Seeburg PH. 1982. The primary structure and genetic organization of the bovine papillomavirus type 1 genome. Nature 299:529–534. doi: 10.1038/299529a0. [DOI] [PubMed] [Google Scholar]
- 57.Lacey M, Alpert S, Hanahan D. 1986. Bovine papillomavirus genome elicits skin tumours in transgenic mice. Nature 322:609–612. doi: 10.1038/322609a0. [DOI] [PubMed] [Google Scholar]
- 58.DiMaio D, Guralski D, Schiller JT. 1986. Translation of open reading frame E5 of bovine papillomavirus is required for its transforming activity. Proc Natl Acad Sci U S A 83:1797–1801. doi: 10.1073/pnas.83.6.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schiller JT, Vass WC, Vousden KH, Lowy DR. 1986. E5 open reading frame of bovine papillomavirus type 1 encodes a transforming gene. J Virol 57:1–6. doi: 10.1128/JVI.57.1.1-6.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DiMaio D, Petti LM. 2013. The E5 proteins. Virology 445:99–114. doi: 10.1016/j.virol.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bergin IL, Bell JD, Chen Z, Zochowski MK, Chai D, Schmidt K, Culmer DL, Aronoff DM, Patton DL, Mwenda JM, Wood CE, Burk RD. 2013. Novel genital alphapapillomaviruses in baboons (Papio hamadryas anubis) with cervical dysplasia. Vet Pathol 50:200–208. doi: 10.1177/0300985812439725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harari A, Wood CE, Van Doorslaer K, Chen Z, Domaingue MC, Elmore D, Koenig P, Wagner JD, Jennings RN, Burk RD. 2013. Condylomatous genital lesions in cynomolgus macaques from Mauritius. Toxicol Pathol 41:893–901. doi: 10.1177/0192623312467521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kloster BE, Manias DA, Ostrow RS, Shaver MK, McPherson SW, Rangen SR, Uno H, Faras AJ. 1988. Molecular cloning and characterization of the DNA of two papillomaviruses from monkeys. Virology 166:30–40. doi: 10.1016/0042-6822(88)90143-2. [DOI] [PubMed] [Google Scholar]
- 64.O'Banion MK, Sundberg JP, Shima AL, Reichmann ME. 1987. Venereal papilloma and papillomavirus in a colobus monkey (Colobus guereza). Intervirology 28:232–237. doi: 10.1159/000150020. [DOI] [PubMed] [Google Scholar]
- 65.Ostrow RS, LaBresh KV, Faras AJ. 1991. Characterization of the complete RhPV 1 genomic sequence and an integration locus from a metastatic tumor. Virology 181:424–429. doi: 10.1016/0042-6822(91)90519-h. [DOI] [PubMed] [Google Scholar]
- 66.Van Ranst M, Fuse A, Sobis H, De Meurichy W, Syrjanen SM, Billiau A, Opdenakker G. 1991. A papillomavirus related to HPV type 13 in oral focal epithelial hyperplasia in the pygmy chimpanzee. J Oral Pathol Med 20:325–331. doi: 10.1111/j.1600-0714.1991.tb00939.x. [DOI] [PubMed] [Google Scholar]
- 67.Ostrow RS, McGlennen RC, Shaver MK, Kloster BE, Houser D, Faras AJ. 1990. A rhesus monkey model for sexual transmission of a papillomavirus isolated from a squamous cell carcinoma. Proc Natl Acad Sci U S A 87:8170–8174. doi: 10.1073/pnas.87.20.8170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wood CE, Borgerink H, Register TC, Scott L, Cline JM. 2004. Cervical and vaginal epithelial neoplasms in cynomolgus monkeys. Vet Pathol 41:108–115. doi: 10.1354/vp.41-2-108. [DOI] [PubMed] [Google Scholar]
- 69.Wood CE, Chen Z, Cline JM, Miller BE, Burk RD. 2007. Characterization and experimental transmission of an oncogenic papillomavirus in female macaques. J Virol 81:6339–6345. doi: 10.1128/JVI.00233-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Justice MJ, Dhillon P. 2016. Using the mouse to model human disease: increasing validity and reproducibility. Dis Model Mech 9:101–103. doi: 10.1242/dmm.024547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lambert PF. 2016. Transgenic mouse models of tumor virus action. Annu Rev Virol 3:473–489. doi: 10.1146/annurev-virology-100114-054908. [DOI] [PubMed] [Google Scholar]
- 72.Sippola-Thiele M, Hanahan D, Howley PM. 1989. Cell-heritable stages of tumor progression in transgenic mice harboring the bovine papillomavirus type 1 genome. Mol Cell Biol 9:925–934. doi: 10.1128/mcb.9.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arbeit JM, Munger K, Howley PM, Hanahan D. 1993. Neuroepithelial carcinomas in mice transgenic with human papillomavirus type 16 E6/E7 ORFs. Am J Pathol 142:1187–1197. [PMC free article] [PubMed] [Google Scholar]
- 74.Griep AE, Herber R, Jeon S, Lohse JK, Dubielzig RR, Lambert PF. 1993. Tumorigenicity by human papillomavirus type 16 E6 and E7 in transgenic mice correlates with alterations in epithelial cell growth and differentiation. J Virol 67:1373–1384. doi: 10.1128/JVI.67.3.1373-1384.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kondoh G, Murata Y, Aozasa K, Yutsudo M, Hakura A. 1991. Very high incidence of germ cell tumorigenesis (seminomagenesis) in human papillomavirus type 16 transgenic mice. J Virol 65:3335–3339. doi: 10.1128/JVI.65.6.3335-3339.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lambert PF, Pan H, Pitot HC, Liem A, Jackson M, Griep AE. 1993. Epidermal cancer associated with expression of human papillomavirus type 16 E6 and E7 oncogenes in the skin of transgenic mice. Proc Natl Acad Sci U S A 90:5583–5587. doi: 10.1073/pnas.90.12.5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pan H, Griep AE. 1994. Altered cell cycle regulation in the lens of HPV-16 E6 or E7 transgenic mice: implications for tumor suppressor gene function in development. Genes Dev 8:1285–1299. doi: 10.1101/gad.8.11.1285. [DOI] [PubMed] [Google Scholar]
- 78.Pan H, Griep AE. 1995. Temporally distinct patterns of p53-dependent and p53-independent apoptosis during mouse lens development. Genes Dev 9:2157–2169. doi: 10.1101/gad.9.17.2157. [DOI] [PubMed] [Google Scholar]
- 79.McCaffrey J, Yamasaki L, Dyson NJ, Harlow E, Griep AE. 1999. Disruption of retinoblastoma protein family function by human papillomavirus type 16 E7 oncoprotein inhibits lens development in part through E2F-1. Mol Cell Biol 19:6458–6468. doi: 10.1128/mcb.19.9.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arbeit JM, Munger K, Howley PM, Hanahan D. 1994. Progressive squamous epithelial neoplasia in K14-human papillomavirus type 16 transgenic mice. J Virol 68:4358–4368. doi: 10.1128/JVI.68.7.4358-4368.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Genther Williams SM, Disbrow GL, Schlegel R, Lee D, Threadgill DW, Lambert PF. 2005. Requirement of epidermal growth factor receptor for hyperplasia induced by E5, a high-risk human papillomavirus oncogene. Cancer Res 65:6534–6542. doi: 10.1158/0008-5472.CAN-05-0083. [DOI] [PubMed] [Google Scholar]
- 82.Song S, Pitot HC, Lambert PF. 1999. The human papillomavirus type 16 E6 gene alone is sufficient to induce carcinomas in transgenic animals. J Virol 73:5887–5893. doi: 10.1128/JVI.73.7.5887-5893.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Herber R, Liem A, Pitot H, Lambert PF. 1996. Squamous epithelial hyperplasia and carcinoma in mice transgenic for the human papillomavirus type 16 E7 oncogene. J Virol 70:1873–1881. doi: 10.1128/JVI.70.3.1873-1881.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Arbeit JM, Howley PM, Hanahan D. 1996. Chronic estrogen-induced cervical and vaginal squamous carcinogenesis in human papillomavirus type 16 transgenic mice. Proc Natl Acad Sci U S A 93:2930–2935. doi: 10.1073/pnas.93.7.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chung SH, Shin MK, Korach KS, Lambert PF. 2013. Requirement for stromal estrogen receptor alpha in cervical neoplasia. Horm Cancer 4:50–59. doi: 10.1007/s12672-012-0125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chung SH, Wiedmeyer K, Shai A, Korach KS, Lambert PF. 2008. Requirement for estrogen receptor alpha in a mouse model for human papillomavirus-associated cervical cancer. Cancer Res 68:9928–9934. doi: 10.1158/0008-5472.CAN-08-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Elson DA, Riley RR, Lacey A, Thordarson G, Talamantes FJ, Arbeit JM. 2000. Sensitivity of the cervical transformation zone to estrogen-induced squamous carcinogenesis. Cancer Res 60:1267–1275. [PubMed] [Google Scholar]
- 88.Riley RR, Duensing S, Brake T, Munger K, Lambert PF, Arbeit JM. 2003. Dissection of human papillomavirus E6 and E7 function in transgenic mouse models of cervical carcinogenesis. Cancer Res 63:4862–4871. [PubMed] [Google Scholar]
- 89.Jabbar S, Strati K, Shin MK, Pitot HC, Lambert PF. 2010. Human papillomavirus type 16 E6 and E7 oncoproteins act synergistically to cause head and neck cancer in mice. Virology 407:60–67. doi: 10.1016/j.virol.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Strati K, Pitot HC, Lambert PF. 2006. Identification of biomarkers that distinguish human papillomavirus (HPV)-positive versus HPV-negative head and neck cancers in a mouse model. Proc Natl Acad Sci U S A 103:14152–14157. doi: 10.1073/pnas.0606698103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stelzer MK, Pitot HC, Liem A, Schweizer J, Mahoney C, Lambert PF. 2010. A mouse model for human anal cancer. Cancer Prev Res (Phila) 3:1534–1541. doi: 10.1158/1940-6207.CAPR-10-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brake T, Lambert PF. 2005. Estrogen contributes to the onset, persistence, and malignant progression of cervical cancer in a human papillomavirus-transgenic mouse model. Proc Natl Acad Sci U S A 102:2490–2495. doi: 10.1073/pnas.0409883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Maufort JP, Shai A, Pitot HC, Lambert PF. 2010. A role for HPV16 E5 in cervical carcinogenesis. Cancer Res 70:2924–2931. doi: 10.1158/0008-5472.CAN-09-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Song S, Liem A, Miller JA, Lambert PF. 2000. Human papillomavirus types 16 E6 and E7 contribute differently to carcinogenesis. Virology 267:141–150. doi: 10.1006/viro.1999.0106. [DOI] [PubMed] [Google Scholar]
- 95.Thomas MK, Pitot HC, Liem A, Lambert PF. 2011. Dominant role of HPV16 E7 in anal carcinogenesis. Virology 421:114–118. doi: 10.1016/j.virol.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Carchman EH, Matkowskyj KA, Meske L, Lambert PF. 2016. Dysregulation of autophagy contributes to anal carcinogenesis. PLoS One 11:e0164273. doi: 10.1371/journal.pone.0164273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nyman PE, Buehler D, Lambert PF. 2018. Loss of function of canonical Notch signaling drives head and neck carcinogenesis. Clin Cancer Res 24:6308–6318. doi: 10.1158/1078-0432.CCR-17-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shai A, Brake T, Somoza C, Lambert PF. 2007. The human papillomavirus E6 oncogene dysregulates the cell cycle and contributes to cervical carcinogenesis through two independent activities. Cancer Res 67:1626–1635. doi: 10.1158/0008-5472.CAN-06-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shin MK, Payne S, Bilger A, Matkowskyj KA, Carchman E, Meyer DS, Bentires-Alj M, Deming DA, Lambert PF. 2019. Activating mutations in Pik3ca contribute to anal carcinogenesis in the presence or absence of HPV-16 oncogenes. Clin Cancer Res 25:1889–1900. doi: 10.1158/1078-0432.CCR-18-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shin MK, Sage J, Lambert PF. 2012. Inactivating all three Rb family pocket proteins is insufficient to initiate cervical cancer. Cancer Res 72:5418–5427. doi: 10.1158/0008-5472.CAN-12-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chung SH, Lambert PF. 2009. Prevention and treatment of cervical cancer in mice using estrogen receptor antagonists. Proc Natl Acad Sci U S A 106:19467–19472. doi: 10.1073/pnas.0911436106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Spurgeon ME, Chung SH, Lambert PF. 2014. Recurrence of cervical cancer in mice after selective estrogen receptor modulator therapy. Am J Pathol 184:530–540. doi: 10.1016/j.ajpath.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stelzer MK, Pitot HC, Liem A, Lee D, Kennedy GD, Lambert PF. 2010. Rapamycin inhibits anal carcinogenesis in two preclinical animal models. Cancer Prev Res (Phila) 3:1542–1551. doi: 10.1158/1940-6207.CAPR-10-0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schaper ID, Marcuzzi GP, Weissenborn SJ, Kasper HU, Dries V, Smyth N, Fuchs P, Pfister H. 2005. Development of skin tumors in mice transgenic for early genes of human papillomavirus type 8. Cancer Res 65:1394–1400. doi: 10.1158/0008-5472.CAN-04-3263. [DOI] [PubMed] [Google Scholar]
- 105.Dong W, Kloz U, Accardi R, Caldeira S, Tong WM, Wang ZQ, Jansen L, Durst M, Sylla BS, Gissmann L, Tommasino M. 2005. Skin hyperproliferation and susceptibility to chemical carcinogenesis in transgenic mice expressing E6 and E7 of human papillomavirus type 38. J Virol 79:14899–14908. doi: 10.1128/JVI.79.23.14899-14908.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hufbauer M, Lazic D, Reinartz M, Akgul B, Pfister H, Weissenborn SJ. 2011. Skin tumor formation in human papillomavirus 8 transgenic mice is associated with a deregulation of oncogenic miRNAs and their tumor suppressive targets. J Dermatol Sci 64:7–15. doi: 10.1016/j.jdermsci.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 107.Viarisio D, Decker KM, Aengeneyndt B, Flechtenmacher C, Gissmann L, Tommasino M. 2013. Human papillomavirus type 38 E6 and E7 act as tumour promoters during chemically induced skin carcinogenesis. J Gen Virol 94:749–752. doi: 10.1099/vir.0.048991-0. [DOI] [PubMed] [Google Scholar]
- 108.Viarisio D, Mueller-Decker K, Kloz U, Aengeneyndt B, Kopp-Schneider A, Grone HJ, Gheit T, Flechtenmacher C, Gissmann L, Tommasino M. 2011. E6 and E7 from beta HPV38 cooperate with ultraviolet light in the development of actinic keratosis-like lesions and squamous cell carcinoma in mice. PLoS Pathog 7:e1002125. doi: 10.1371/journal.ppat.1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Viarisio D, Muller-Decker K, Accardi R, Robitaille A, Durst M, Beer K, Jansen L, Flechtenmacher C, Bozza M, Harbottle R, Voegele C, Ardin M, Zavadil J, Caldeira S, Gissmann L, Tommasino M. 2018. Beta HPV38 oncoproteins act with a hit-and-run mechanism in ultraviolet radiation-induced skin carcinogenesis in mice. PLoS Pathog 14:e1006783. doi: 10.1371/journal.ppat.1006783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ingle A, Ghim S, Joh J, Chepkoech I, Bennett Jenson A, Sundberg JP. 2011. Novel laboratory mouse papillomavirus (MusPV) infection. Vet Pathol 48:500–505. doi: 10.1177/0300985810377186. [DOI] [PubMed] [Google Scholar]
- 111.Schulz E, Gottschling M, Ulrich RG, Richter D, Stockfleth E, Nindl I. 2012. Isolation of three novel rat and mouse papillomaviruses and their genomic characterization. PLoS One 7:e47164. doi: 10.1371/journal.pone.0047164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Joh J, Jenson AB, King W, Proctor M, Ingle A, Sundberg JP, Ghim SJ. 2011. Genomic analysis of the first laboratory-mouse papillomavirus. J Gen Virol 92:692–698. doi: 10.1099/vir.0.026138-0. [DOI] [PubMed] [Google Scholar]
- 113.Xue XY, Majerciak V, Uberoi A, Kim BH, Gotte D, Chen X, Cam M, Lambert PF, Zheng ZM. 2017. The full transcription map of mouse papillomavirus type 1 (MmuPV1) in mouse wart tissues. PLoS Pathog 13:e1006715. doi: 10.1371/journal.ppat.1006715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Grace M, Munger K. 2017. Proteomic analysis of the gamma human papillomavirus type 197 E6 and E7 associated cellular proteins. Virology 500:71–81. doi: 10.1016/j.virol.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.White EA, Munger K, Howley PM. 2016. High-risk human papillomavirus E7 proteins target PTPN14 for degradation. mBio 7:e01530-16. doi: 10.1128/mBio.01530-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hatterschide J, Bohidar AE, Grace M, Nulton TJ, Kim HW, Windle B, Morgan IM, Munger K, White EA. 2019. PTPN14 degradation by high-risk human papillomavirus E7 limits keratinocyte differentiation and contributes to HPV-mediated oncogenesis. Proc Natl Acad Sci U S A 116:7033–7042. doi: 10.1073/pnas.1819534116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Meyers JM, Grace M, Uberoi A, Lambert PF, Munger K. 2018. Inhibition of TGF-β and NOTCH signaling by cutaneous papillomaviruses. Front Microbiol 9:389. doi: 10.3389/fmicb.2018.00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Meyers JM, Uberoi A, Grace M, Lambert PF, Munger K. 2017. Cutaneous HPV8 and MmuPV1 E6 proteins target the NOTCH and TGF-beta tumor suppressors to inhibit differentiation and sustain keratinocyte proliferation. PLoS Pathog 13:e1006171. doi: 10.1371/journal.ppat.1006171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cladel NM, Budgeon LR, Cooper TK, Balogh KK, Hu J, Christensen ND. 2013. Secondary infections, expanded tissue tropism, and evidence for malignant potential in immunocompromised mice infected with Mus musculus papillomavirus 1 DNA and virus. J Virol 87:9391–9395. doi: 10.1128/JVI.00777-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Handisurya A, Day PM, Thompson CD, Buck CB, Pang YY, Lowy DR, Schiller JT. 2013. Characterization of Mus musculus papillomavirus 1 infection in situ reveals an unusual pattern of late gene expression and capsid protein localization. J Virol 87:13214–13225. doi: 10.1128/JVI.02162-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Handisurya A, Day PM, Thompson CD, Bonelli M, Lowy DR, Schiller JT. 2014. Strain-specific properties and T cells regulate the susceptibility to papilloma induction by Mus musculus papillomavirus 1. PLoS Pathog 10:e1004314. doi: 10.1371/journal.ppat.1004314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jiang RT, Wang JW, Peng S, Huang TC, Wang C, Cannella F, Chang YN, Viscidi RP, Best SRA, Hung CF, Roden R. 2017. Spontaneous and vaccine-induced clearance of Mus musculus papillomavirus 1 infection. J Virol 91:e00699-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sundberg JP, Stearns TM, Joh J, Proctor M, Ingle A, Silva KA, Dadras SS, Jenson AB, Ghim SJ. 2014. Immune status, strain background, and anatomic site of inoculation affect mouse papillomavirus (MmuPV1) induction of exophytic papillomas or endophytic trichoblastomas. PLoS One 9:e113582. doi: 10.1371/journal.pone.0113582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Uberoi A, Yoshida S, Frazer IH, Pitot HC, Lambert PF. 2016. Role of ultraviolet radiation in papillomavirus-induced disease. PLoS Pathog 12:e1005664. doi: 10.1371/journal.ppat.1005664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang JW, Jiang R, Peng S, Chang YN, Hung CF, Roden RB. 2015. Immunologic control of Mus musculus papillomavirus type 1. PLoS Pathog 11:e1005243. doi: 10.1371/journal.ppat.1005243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cladel NM, Budgeon LR, Balogh KK, Cooper TK, Brendle SA, Christensen ND, Schell TD, Hu J. 2017. Mouse papillomavirus infection persists in mucosal tissues of an immunocompetent mouse strain and progresses to cancer. Sci Rep 7:16932. doi: 10.1038/s41598-017-17089-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Joh J, Ghim SJ, Chilton PM, Sundberg JP, Park J, Wilcher SA, Proctor ML, Bennett Jenson A. 2016. MmuPV1 infection and tumor development of T cell-deficient mice is prevented by passively transferred hyperimmune sera from normal congenic mice immunized with MmuPV1 virus-like particles (VLPs). Exp Mol Pathol 100:212–219. doi: 10.1016/j.yexmp.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 128.Cladel NM, Budgeon LR, Balogh KK, Cooper TK, Hu J, Christensen ND. 2015. A novel pre-clinical murine model to study the life cycle and progression of cervical and anal papillomavirus infections. PLoS One 10:e0120128. doi: 10.1371/journal.pone.0120128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cladel NM, Budgeon LR, Balogh KK, Cooper TK, Hu J, Christensen ND. 2016. Mouse papillomavirus MmuPV1 infects oral mucosa and preferentially targets the base of the tongue. Virology 488:73–80. doi: 10.1016/j.virol.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hu J, Budgeon LR, Cladel NM, Balogh K, Myers R, Cooper TK, Christensen ND. 2015. Tracking vaginal, anal and oral infection in a mouse papillomavirus infection model. J Gen Virol 96:3554–3565. doi: 10.1099/jgv.0.000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Spurgeon ME, Lambert PF. 2019. Sexual transmission of murine papillomavirus (MmuPV1) in Mus musculus. Elife 8:e50056. doi: 10.7554/eLife.50056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Spurgeon ME, Uberoi A, McGregor SM, Wei T, Ward-Shaw E, Lambert PF. 2019. A novel in vivo infection model to study papillomavirus-mediated disease of the female reproductive tract. mBio 10:e00180-19. doi: 10.1128/mBio.00180-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Torres AD, Spurgeon ME, Bilger A, Blaine-Sauer S, Uberoi A, Buehler D, McGregor SM, Ward-Shaw E, Lambert PF. 2019. The human papillomavirus 16 E5 gene potentiates MmuPV1-dependent pathogenesis. Virology 541:1–12. doi: 10.1016/j.virol.2019.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cladel NM, Budgeon LR, Cooper TK, Balogh KK, Christensen ND, Myers R, Majerciak V, Gotte D, Zheng ZM, Hu J. 2017. Mouse papillomavirus infections spread to cutaneous sites with progression to malignancy. J Gen Virol 98:2520–2529. doi: 10.1099/jgv.0.000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Forslund O, Johansson H, Madsen KG, Kofoed K. 2013. The nasal mucosa contains a large spectrum of human papillomavirus types from the Betapapillomavirus and Gammapapillomavirus genera. J Infect Dis 208:1335–1341. doi: 10.1093/infdis/jit326. [DOI] [PubMed] [Google Scholar]
- 136.Hampras SS, Rollison DE, Giuliano AR, McKay-Chopin S, Minoni L, Sereday K, Gheit T, Tommasino M. 2017. Prevalence and concordance of cutaneous beta human papillomavirus infection at mucosal and cutaneous sites. J Infect Dis 216:92–96. doi: 10.1093/infdis/jix245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Handisurya A, Day PM, Thompson CD, Buck CB, Kwak K, Roden RB, Lowy DR, Schiller JT. 2012. Murine skin and vaginal mucosa are similarly susceptible to infection by pseudovirions of different papillomavirus classifications and species. Virology 433:385–394. doi: 10.1016/j.virol.2012.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Day PM, Thompson CD, Lowy DR, Schiller JT. 2015. The HPV16 and MusPV1 papillomaviruses initially interact with distinct host components on the basement membrane. Virology 481:79–94. doi: 10.1016/j.virol.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Johnson KM, Kines RC, Roberts JN, Lowy DR, Schiller JT, Day PM. 2009. Role of heparan sulfate in attachment to and infection of the murine female genital tract by human papillomavirus. J Virol 83:2067–2074. doi: 10.1128/JVI.02190-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Peh WL, Middleton K, Christensen N, Nicholls P, Egawa K, Sotlar K, Brandsma J, Percival A, Lewis J, Liu WJ, Doorbar J. 2002. Life cycle heterogeneity in animal models of human papillomavirus-associated disease. J Virol 76:10401–10416. doi: 10.1128/jvi.76.20.10401-10416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Joh J, Jenson AB, Proctor M, Ingle A, Silva KA, Potter CS, Sundberg JP, Ghim SJ. 2012. Molecular diagnosis of a laboratory mouse papillomavirus (MusPV). Exp Mol Pathol 93:416–421. doi: 10.1016/j.yexmp.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 142.Wang W, Uberoi A, Spurgeon M, Gronski E, Majerciak V, Lobanov A, Hayes M, Loke A, Zheng ZM, Lambert PF. 2020. Stress keratin 17 enhances papillomavirus infection-induced disease by downregulating T cell recruitment. PLoS Pathog 16:e1008206. doi: 10.1371/journal.ppat.1008206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pantelouris EM. 1968. Absence of thymus in a mouse mutant. Nature 217:370–371. doi: 10.1038/217370a0. [DOI] [PubMed] [Google Scholar]
- 144.Joh J, Chilton PM, Wilcher SA, Zahin M, Park J, Proctor ML, Ghim SJ, Jenson AB. 2017. T cell-mediated antitumor immune response eliminates skin tumors induced by mouse papillomavirus, MmuPV1. Exp Mol Pathol 103:181–190. doi: 10.1016/j.yexmp.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 145.Strickley JD, Messerschmidt JL, Awad ME, Li T, Hasegawa T, Ha DT, Nabeta HW, Bevins PA, Ngo KH, Asgari MM, Nazarian RM, Neel VA, Jenson AB, Joh J, Demehri S. 2019. Immunity to commensal papillomaviruses protects against skin cancer. Nature 575:519–522. doi: 10.1038/s41586-019-1719-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hu J, Cladel NM, Budgeon LR, Balogh KK, Christensen ND. 2017. The mouse papillomavirus infection model. Viruses 9:246. doi: 10.3390/v9090246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Flores ER, Allen-Hoffmann BL, Lee D, Lambert PF. 2000. The human papillomavirus type 16 E7 oncogene is required for the productive stage of the viral life cycle. J Virol 74:6622–6631. doi: 10.1128/jvi.74.14.6622-6631.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Thomas JT, Hubert WG, Ruesch MN, Laimins LA. 1999. Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc Natl Acad Sci U S A 96:8449–8454. doi: 10.1073/pnas.96.15.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Miyauchi S, Sanders PD, Guram K, Kim SS, Paolini F, Venuti A, Cohen EEW, Gutkind JS, Califano JA, Sharabi AB. 2019. HPV16 E5 mediates resistance to PD-L1 blockade and can be targeted with rimantadine in head and neck cancer. Cancer Res 80(4):732–746. doi: 10.1158/0008-5472.CAN-19-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ault KA. 2006. Epidemiology and natural history of human papillomavirus infections in the female genital tract. Infect Dis Obstet Gynecol 2006(Suppl):40470. doi: 10.1155/IDOG/2006/40470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cladel NM, Jiang P, Li JJ, Peng X, Cooper TK, Majerciak V, Balogh KK, Meyer TJ, Brendle SA, Budgeon LR, Shearer DA, Munden R, Cam M, Vallur R, Christensen ND, Zheng ZM, Hu J. 2019. Papillomavirus can be transmitted through the blood and produce infections in blood recipients: evidence from two animal models. Emerg Microbes Infect 8:1108–1121. doi: 10.1080/22221751.2019.1637072. [DOI] [PMC free article] [PubMed] [Google Scholar]