Sialic acids (Sia) are involved in numerous different cellular functions and are receptors for many pathogens. Sia come in chemically modified forms, but we lack a clear understanding of how they alter interactions with microbes. Here, we examine the expression of modified Sia in mouse tissues, on secreted mucus in saliva, and on erythrocytes, including those from IAV host species and animals used in IAV research. These Sia forms varied considerably among different animals, and their inhibitory effects on IAV NA and HA activities and on bacterial sialidases (neuraminidases) suggest a host-variable protective role in secreted mucus.
KEYWORDS: influenza, mucus, sialic acids
ABSTRACT
Sialic acids (Sia) are the primary receptors for influenza viruses and are widely displayed on cell surfaces and in secreted mucus. Sia may be present in variant forms that include O-acetyl modifications at C-4, C-7, C-8, and C-9 positions and N-acetyl or N-glycolyl at C-5. They can also vary in their linkages, including α2-3 or α2-6 linkages. Here, we analyze the distribution of modified Sia in cells and tissues of wild-type mice or in mice lacking CMP-N-acetylneuraminic acid hydroxylase (CMAH) enzyme, which synthesizes N-glycolyl (Neu5Gc) modifications. We also examined the variation of Sia forms on erythrocytes and in saliva from different animals. To determine the effect of Sia modifications on influenza A virus (IAV) infection, we tested for effects on hemagglutinin (HA) binding and neuraminidase (NA) cleavage. We confirmed that 9-O-acetyl, 7,9-O-acetyl, 4-O-acetyl, and Neu5Gc modifications are widely but variably expressed in mouse tissues, with the highest levels detected in the respiratory and gastrointestinal (GI) tracts. Secreted mucins in saliva and surface proteins of erythrocytes showed a high degree of variability in display of modified Sia between different species. IAV HAs from different virus strains showed consistently reduced binding to both Neu5Gc- and O-acetyl-modified Sia; however, while IAV NAs were inhibited by Neu5Gc and O-acetyl modifications, there was significant variability between NA types. The modifications of Sia in mucus may therefore have potent effects on the functions of IAV and may affect both pathogens and the normal flora of different mucosal sites.
IMPORTANCE Sialic acids (Sia) are involved in numerous different cellular functions and are receptors for many pathogens. Sia come in chemically modified forms, but we lack a clear understanding of how they alter interactions with microbes. Here, we examine the expression of modified Sia in mouse tissues, on secreted mucus in saliva, and on erythrocytes, including those from IAV host species and animals used in IAV research. These Sia forms varied considerably among different animals, and their inhibitory effects on IAV NA and HA activities and on bacterial sialidases (neuraminidases) suggest a host-variable protective role in secreted mucus.
INTRODUCTION
Sialic acids (Sia) are a family of nine-carbon monosaccharides that often serve as terminal residues of carbohydrate chains. They are present at high levels on cell membrane glycoproteins and glycolipids, as well as on secreted glycoproteins and mucus at all mucosal surfaces (Fig. 1A) (1, 2). Sia are key mediators of many normal cell and tissue functions through a wide variety of highly regulated cell-cell interactions during both development and homeostatic processes, where they may be bound by cellular receptors and members of the selectin family (3, 4). Their ubiquitous presence on cells, tissues, and mucosal surfaces also make Sia a key point of contact for commensal microbes and for invading pathogens, including viruses, bacteria, and parasites (3, 5, 6).
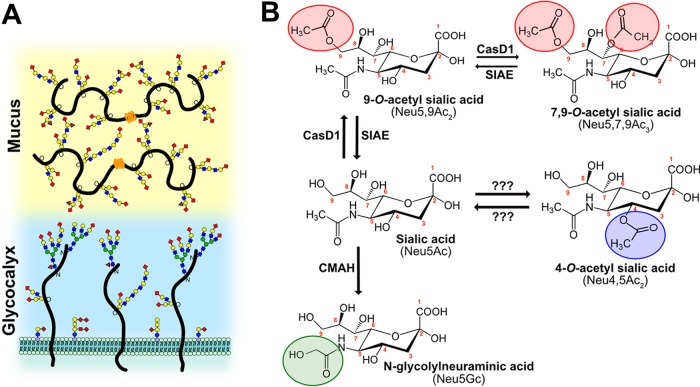
FIG 1.
(A) Sialic acids (red diamonds) terminate glycan chains on glycolipids and glycoproteins as part of the glycocalyx on the surfaces of cells. They can also terminate glycans on secreted glycoproteins, like mucins, which are an important component of the protective mucosal barrier in gastrointestinal and respiratory tissue. (B) Sialic acid (Neu5Ac) can be modified by the addition of O-acetyl modifications at the C-4, -7, and -9 positions or by the hydroxylation of the N-acetyl group at C-5 to form Neu5Gc by the enzyme CMAH. The sialate O-acetyltransferase CasD1 adds acetyl groups at C-7, from which it migrates to the C-9 position (Neu5,9Ac2) under physiological conditions. This can allow an additional acetyl group to be added by CasD1 to C-7 (Neu5,7,9Ac3). The SIAE can remove these acetyl modifications, restoring the unmodified Neu5Ac form of sialic acid. O-Acetyl modifications can also be added at the C-4 position by a specific 4-O-acetyltransferase (Neu4,5Ac2) and removed by a 4-O-acetylesterase. However, the genes for these enzymes have not yet been identified.
Sia are a highly diverse family of molecules that may be present as more than 50 structurally and chemically distinct modified variants. They are formed from the basic structure of N-acetylneuraminic acid (Neu5Ac) by the addition of chemical groups at various positions on the pyranose ring or the glycerol side chain. The modifications may include N-glycolyl and/or O-linked acetyl, sulfo, methyl, and lactyl groups, among others (1, 2, 7, 8). Many different enzymes and pathways introduce these chemical modifications, and some can be removed by regulatory enzymes. The different modified Sia are often themselves substrates for other modifying enzymes and transferases, so that each modification may alter the synthesis of other modified forms. This therefore leads to complex patterns and mixtures of modified Sia forms, with significant variation in both the levels and specific combinations of modifications in different hosts and tissues and under different physiological conditions (4, 9–12). The roles of these modified Sia forms in cellular functions and interactions with microbes have been understudied, largely due to a relative lack of reagents or experimental approaches. The recent development of specific probes for some Sia variants (13, 14) combined with existing methods for studying glycans has allowed us to determine the expression of modified Sia between species and tissues, as well as to begin to unravel their effects on host-pathogen interactions.
Sialic acid modifications.
While over 50 different chemical forms of Sia exist in nature, the commonest chemical additions seen in vertebrates include ester-bonded O-acetyl (O-Ac) modifications to C-4, -7, -8, and/or -9 positions, resulting in a variety of combinations of Sia forms, such as Neu4,5Ac2, Neu5,9Ac2, and Neu5,7,9Ac3, as well as their N-glycolylneuraminic acid (Neu5Gc) analogs with the same O-acetyl modifications (Fig. 1B). Neu5Gc is produced from Neu5Ac by the activity of CMP-N-acetylneuraminic acid hydroxylase (CMAH) in the cytoplasm of cells, and the enzyme is missing or inactive in some animals, including humans (10). The addition of O-Ac to the C-7 and/or C-9 position is mediated by the sialylate O-acetyltransferase Cas1 domain-containing 1 (CasD1), which has been suggested to add an O-acetyl group to the C-7 position, from which it would migrate to the C-8 and C-9 position under physiological conditions, allowing the possibility of the addition of another O-acetyl group to C-7 (7, 15, 16). The regulatory processes that control the number or positions of acetyl groups have not been well defined, although distinct differences in expression of 7,9-O-Ac and 9-O-Ac Sia have been reported in mouse and human tissues, in chicken embryos, and in some other animals (7). CasD1 modifies Sia in the activated CMP-Sia form before it is added to the glycan chain, and it likely has a preference for CMP-Neu5Ac as a substrate and is less active on CMP-Neu5Gc (16). The sialate O-acetylesterase (SIAE) can remove the 7,9-O- and 9-O-Ac modifications, although its activities and roles are not well understood (17–20). The 4-O-Ac Sia is produced in some tissues of many animals by a distinct sialate 4-O-acetyltransferase that is also likely expressed in the Golgi compartment; however, the gene for the enzyme has also not yet been identified (21–23).
The 7-, 8-, and/or 9-O-Ac Sia appear to be present at low levels—a few percent or less—in the cell-associated Sia on many cultured cells but may be present at higher levels (10% to 50%) in Sia on the secreted mucus of various animals and on erythrocyte-associated glycans (14, 24, 25). However, the expression, distribution, and regulation of these modified Sia are not well documented, nor do we understand their impact on pathogens, host homeostasis, and normal microbiota at different mucosal sites (10, 13, 14, 26).
While O-acetyl Sia and Neu5Gc are the most common chemically modified Sia forms and the focus of this study, a greater diversity of Sia exist in nature. These other chemically modified forms, including O-sulfo, O-lactyl, and O-methyl modifications, likely play important roles in development and cell-cell signaling (1). Both O-sulfo and O-methyl modifications have been detected in invertebrate species, such as sea stars and sea urchins, as well as some mouse tissues (27–29). O-Lactyl Sia has been detected in human gastric aspirate, along with O-acetyl-modified Sia (30). However, similar to O-acetyl and Neu5Gc modifications, these other Sia variants have also been understudied due to a lack of reagents and technology; thus, little is known about their specific regulation, expression, and functions in different animal species and tissues.
Modified sialic acid and pathogen interactions.
Many pathogens interact with Sia on host cells at various stages in their infection cycles, including various viruses, bacteria, and parasites (3, 5). The densely expressed Sia within various mucus layers on mucosal surfaces also act to bind incoming pathogens and likely regulate both the release and transmission of the pathogens (31, 32). Many pathogens therefore express proteins that attach to Sia, as well as expressing receptor-modifying enzymes, such as sialidases (neuraminidases), that remove the Sia from the underlying glycan. Bacterial adhesins and toxins may recognize Sia on the surfaces of cells, and many bacteria can also use Sia as a metabolic carbon source after release through the activities of neuraminidases and uptake into the cell by Sia transporters (3, 33–36). These bacterial interactions with Sia are potentially affected by chemical modifications (35, 37). Both enveloped and nonenveloped viruses may also bind Sia as primary receptors or coreceptors for cell recognition and infection, although only the enveloped viruses appear to express neuraminidases or sialate O-acetyl esterases, possibly to reduce aggregation of viral particles during budding (5, 38). For some viruses, Sia modifications are required for infection, as viral proteins specifically bind to modified Sia—examples include human coronavirus OC43 and HKU1 and influenza C and D viruses, which all require 9-O-Ac Sia for cell infection (39, 40).
Significant effects of different Sia modifications on the binding of pathogens or the activities of their sialidases (neuraminidases) have been suggested, but in general, they are still not well understood. Influenza A viruses (IAV) use Sia as primary receptors for host recognition and cell entry through the activities of two surface glycoproteins that interact with Sia, hemagglutinin (HA) and neuraminidase (NA). HA is a trimeric protein that binds Sia to initiate the endocytic uptake of the virus by the cell, leading to fusion between the viral envelope and the endosomal membrane after exposure of the virus to low pH (41). NA is a sialidase that cleaves Sia from the mucus and cell surface and from viral glycoproteins, allowing the virus to penetrate through the mucus to the epithelial cells and reducing the aggregation of virions after budding from the surfaces of cells (42). Previous studies have shown that modifications, such as 7,9-O- and 9-O-Ac Sia, are expressed on cells or in tissues of many IAV host species, and there is some evidence that these modifications may be inhibitory for NA activity and HA binding (25, 43, 44). However, the specific effects of 7,9-O- and 9-O-Ac on the binding of Sia by HA or the cleavage of Sia by IAV NA have not been examined in detail, and it is unclear whether these changes influence infection efficiency or viral shedding. For example, modification of HA binding might influence the attachment of virus to cells or to mucus, while inhibition of NA cleavage of O-acetyl-modified Sia may lead to virus being trapped in the mucus and cleared, reducing the efficiency of infection.
The difference between Neu5Gc and Neu5Ac has been found to influence the tropism of several different viruses, as well as some bacterial toxins (3, 5). Indeed, it has been proposed that the loss of the CMAH gene in humans was an adaptive response to pathogen pressures (45–47). Neu5Gc is highly expressed in some tissues of IAV natural host species, including pigs and horses, and is also present in the tissues of mice and guinea pigs, which are frequently used as animal models (10, 48). Neu5Gc has been seen to prevent binding of the HAs of some IAV, particularly in human-adapted strains (44, 49), but the effects on NA have not been well characterized. Nevertheless, examination of swine IAV isolates found distinct strain differences in the ability to cleave Neu5Gc by sialidase activity, which was generally lower than that against Neu5Ac (50).
In this study, we define the expression of 7,9-O-Ac-, 9-O-Ac-, and Neu5Gc-modified Sia in the mucus and saliva and on erythrocytes of different IAV host animals as an example to highlight the variability in expression of different Sia modifications between species. We also examine the display of modified Sia on the tissues and secreted mucus of mice, an important model species, not only for IAV research, but for many viruses and bacterial pathogens. Finally, we test the effects of these modifications on HA binding and NA activities of different strains of IAV, as well as their potential to alter virus infection. While we have focused on IAV, comparing the expression of modified Sia between different hosts and in mice has importance for many viruses and other organisms that infect or colonize mucosal tissues. Therefore, the findings here are broadly relevant and underline the need for understanding the expression of modified Sia between species and their potential roles in virus tropism and infection.
RESULTS
Distribution of modified Sia in mouse tissues.
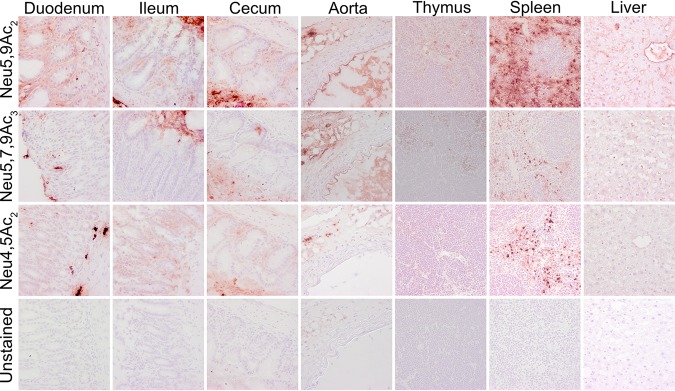
Previous studies on display of modified Sia in animal tissues and cultured cells have shown varying distributions of 7,9-O-Ac, 9-O-Ac, and 4-O-Ac Sia depending on the animal and the tissue examined (13, 14). Mice are an important model species for biomedical research, and some tissues have previously been screened for O-Ac display using probes derived from viral glycoproteins (virolectins) and by other methods (13, 14). We examined the distribution of modified Sia in a variety of tissues of wild-type (WT) C57/BL6 mice. Both 9-O- and 7,9-O-Ac were found throughout the lung and trachea, as well as in the tracheal submucosal glands that produce most mucus (Fig. 2A). These modified Sia were also found throughout the GI tract, with staining associated with epithelial cells, goblet cells, and associated mucus layers of the gastrointestinal tissues, including the stomach, small intestine, and colon (Fig. 2B). Interestingly, 9-O-Ac appeared to be present in larger amounts in most tissues, including salivary gland and esophagus, while the 7,9-O-Ac staining was minimal. However, 7,9-O-Ac did stain more strongly than 9-O-Ac in stomach-associated mucus and on tracheal epithelial cells. This seems to indicate that, while the same enzymes (CasD1 and SIAE) are considered to control the presence of these modifications, the expression of 9-O- and 7,9-O-Ac are differentially regulated in individual tissues. The 4-O-Ac Sia showed high levels of probe binding in the colon, primarily in mucus, and there was also some expression on the mucosal surfaces in the stomach, small intestine (jejunum), and trachea, as well as on cells within the red pulp of the spleen (Fig. 2A and B and Fig. 3).
FIG 2.
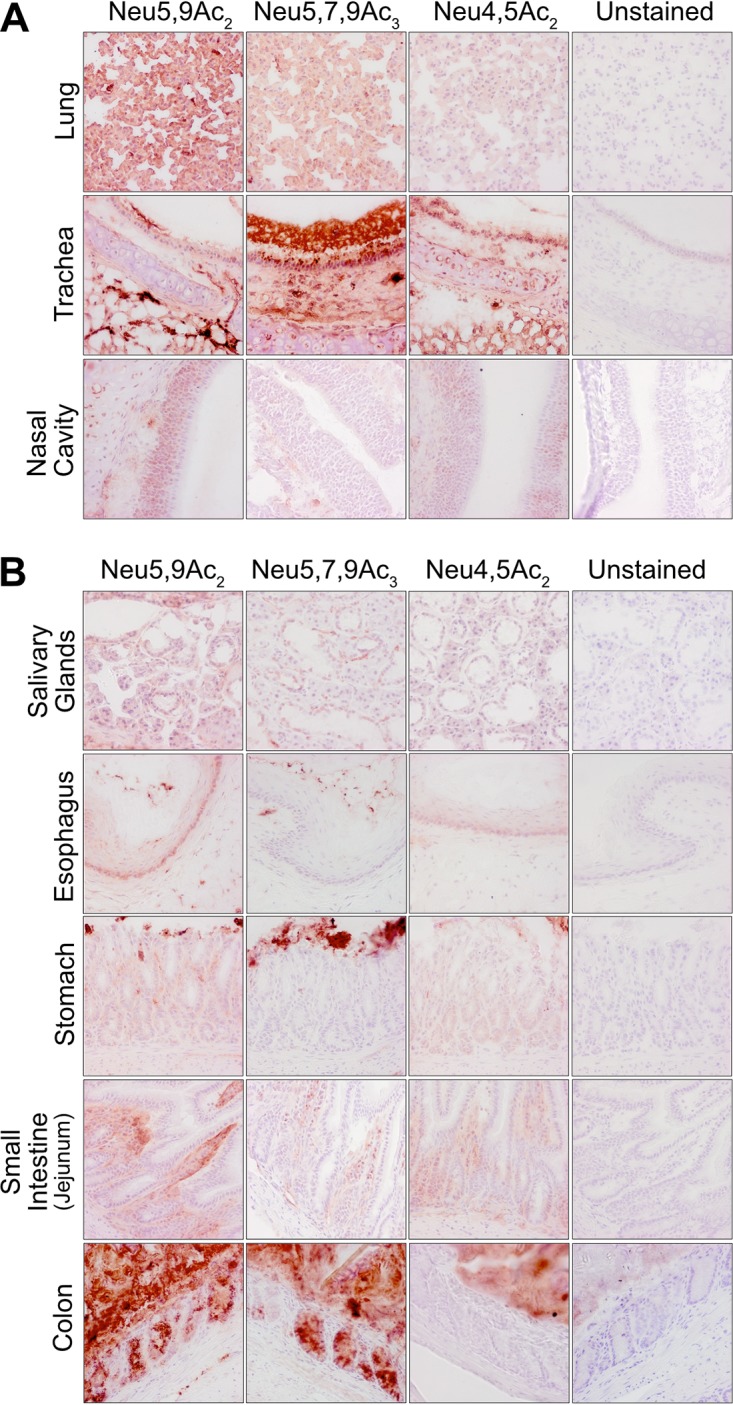
Expression of O-acetylated Sia varies between tissues in wild-type C57BL/6 mice. Frozen tissue sections from respiratory tissues (A) and gastrointestinal tissues (B) were stained using virolectins derived from the hemagglutinin esterases (HE-Fcs) of various nidoviruses with high specificity for the different O-acetyl-modified Sia forms. Sections were counterstained with hematoxylin and imaged at ×40 magnification.
FIG 3.
Expression of O-acetylated Sia varies between tissues in wild-type C57BL/6 mice. Frozen tissue sections were stained using virolectins derived from the hemagglutinin esterases (HE-Fcs) of various nidoviruses with high specificity for the different O-acetyl-modified Sia forms. Sections were counterstained with hematoxylin and imaged at ×40 magnification.
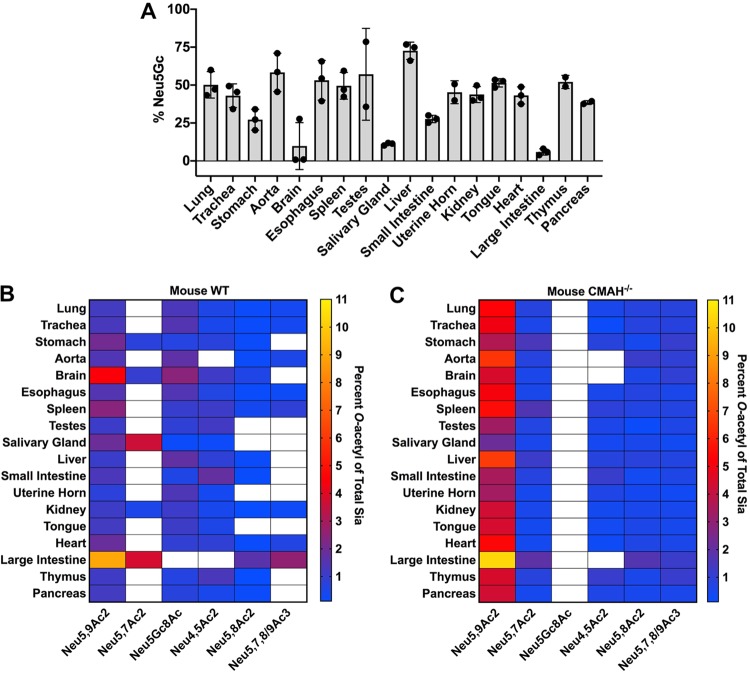
The virolectin probes used were sensitive but did not reveal the quantity of each of the modified Sia forms present. To determine the relative amounts of different Sia forms, we tested tissues from mice using 1,2-diamino-4,5-methylenedioxybenzene (DMB) labeling of the Sia and analysis with high-performance liquid chromatography (HPLC) under conditions that revealed the amounts of Neu5Ac and Neu5Gc and best preserved the O-acetylation of the Sia, although there may still be some loss and migration of O-acetyl modifications (51). Therefore, while this method gives us the best estimate of the amounts of O-acetyl modifications present, it likely still underestimates the true total. The WT mice showed varying levels of Neu5Gc (Fig. 4A and Table 1), while as expected, the CMAH knockout (CMAH−/−) mice showed only Neu5Ac in all tissues, similar to amounts reported previously for some of the tissues (52) (Table 2). The lack of Neu5Gc in CMAH−/− mice was also seen in GI tissues, indicating that Neu5Gc from dietary sources was not detectably being taken up by the mice, as is seen in humans who eat a diet containing Neu5Gc (53). In the WT mice, tissues showed a great deal of variability in Neu5Gc expression. Most tissues had around 50 to 60% Neu5Gc; however, some tissues, including the liver, had higher levels of over 70%, while the brain and salivary glands had only 10%. All O-acetyl Sia variants combined comprised between 2 and 16% of the Sia in most tissues, with the majority being 9-O-Ac (1 to 9% of total Sia) (Fig. 4B and Table 1). There were about 1.5- to 3-fold higher levels of O-acetyl Sia in most tissues of the CMAH knockout mice (Fig. 4C and Table 2), as has been reported previously (52). The levels of 4-O-Ac Sia were generally low, making up ∼2% of the Sia in the small intestine (duodenum) and ∼1% in the spleen, testes, and esophagus. Mouse colon samples showed the highest levels of total O-acetylation, with ∼17% of Sia having one or more O-acetyl modifications, again primarily 9-O-Ac. Given the patterns seen using hemagglutinin esterase (HE-Fc) virolectin staining, the 7,9-O- and 9-O-Ac forms must be present at high levels within or on certain cell subpopulations, as well as within mucus or mucus-secreting cells. For example, the high levels of 7,9-O- and 9-O-Ac found in the mouse colon were most likely associated with secreted mucus, as most of those modified Sia were present in goblet cells (Fig. 2B). However, the differences seen between probe binding and the ratios of the different modified Sia within the total Sia underscore the importance of quantifying the different forms.
FIG 4.
Neu5Gc and O-acetyl Sia modifications vary by tissue in wild-type C57BL/6 mice, and absence of Neu5Gc in CMAH−/− mice leads to an increase in O-acetylation across tissues. Total Sia was measured from tissue samples using HPLC analysis to determine relative Sia quantities. (A) Neu5Gc levels were measured in tissues from WT mice showing variable levels of expression between different tissues. CMAH−/− mice showed undetectable levels of Neu5Gc. Filled cirlces show the individual data points. The error bars indicate standard deviations. (B and C) Percentages of O-acetyl-modified Sia in different tissues from WT (B) and CMAH−/− (C) mice are given as a heat map showing variation across tissues. The white squares indicate that the Sia form was below the limit of detection. Values are given as the percentage of total Sia collected from tissue samples. The sample size for each tissue was three individual mice (n = 3) of each mouse strain, with average values for total sialic acid content given in Tables 1 and 2.
TABLE 1.
Average relative Sia quantities as determined by HPLC analysis for each tissue tested in WT C57BL/6 mice (n = 3)
| Tissue | % Siaa
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Neu5Gc | Neu5Ac | Neu5,9Ac2 | Neu5,7Ac2 | Neu5Gc8Ac | Neu4,5Ac2 | Neu5,8Ac2 | Neu5,7,8/9Ac3 | O-Ac sum | |
| Lung | 50.2 | 47.6 | 1.2 | BD | 1.3 | 0.5 | 0.2 | 0.4 | 3.6 |
| Trachea | 43.0 | 53.5 | 1.3 | BD | 1.5 | 0.4 | 0.2 | 0.4 | 3.7 |
| Stomach | 27.2 | 68.8 | 2.0 | 0.7 | 0.6 | 0.7 | 0.2 | BD | 4.3 |
| Aorta | 58.4 | 38.2 | 1.4 | BD | 1.7 | BD | 0.2 | BD | 3.3 |
| Brain | 9.8 | 83.1 | 4.5 | 1.0 | 2.5 | 1.1 | 0.5 | BD | 9.5 |
| Esophagus | 53.1 | 43.2 | 1.5 | BD | 1.4 | 0.6 | 0.2 | 0.2 | 3.9 |
| Spleen | 49.6 | 45.2 | 2.4 | BD | 1.0 | 1.1 | 0.3 | 0.8 | 5.6 |
| Testes | 57.1 | 40.4 | 1.1 | BD | 0.8 | 1.2 | BD | BD | 3.0 |
| Salivary gland | 11.1 | 85.5 | 1.9 | 3.6 | 0.2 | 0.2 | 0.1 | BD | 6.0 |
| Liver | 72.6 | 23.8 | 1.0 | BD | 1.7 | 0.8 | 0.2 | BD | 3.7 |
| Small intestine (duodenum) | 27.6 | 68.7 | 1.3 | BD | 0.6 | 1.8 | 0.1 | BD | 3.8 |
| Uterine horn | 45.3 | 52.4 | 0.8 | BD | 1.4 | 0.4 | 0.1 | BD | 2.6 |
| Kidney | 43.8 | 53.5 | 1.1 | 0.5 | 1.0 | 0.4 | 0.1 | 0.2 | 3.4 |
| Tongue | 51.5 | 45.8 | 1.2 | BD | 1.1 | 0.5 | n/d | BD | 2.8 |
| Heart | 43.2 | 53.3 | 1.8 | BD | 0.9 | 0.8 | 0.2 | 0.6 | 4.3 |
| Large intestine (colon) | 5.9 | 77.2 | 9.0 | 3.8 | BD | BD | 1.6 | 2.6 | 16.9 |
| Thymus | 52.1 | 44.8 | 1.1 | BD | 0.6 | 1.3 | 0.1 | BD | 3.1 |
| Pancreas | 38.5 | 59.1 | 1.2 | BD | 0.7 | 0.4 | 0.2 | BD | 2.5 |
Proportion of total Sia for each variant shown as a percentage, with the sum of all O-acetyl forms combined given in the far-right column (O-Ac sum). BD, the Sia form was below the limit of detection.
TABLE 2.
Average relative Sia quantities as determined by HPLC analysis for each tissue tested in CMAH−/− C57BL/6 mice (n = 3)
| Tissue | % Siaa
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Neu5Gc | Neu5Ac | Neu5,9Ac2 | Neu5,7Ac2 | Neu5Gc8Ac | Neu4,5Ac2 | Neu5,8Ac2 | Neu5,7,8/9Ac3 | O-Ac sum | |
| Lung | BD | 92.7 | 5.6 | 1.5 | BD | 0.5 | 0.8 | 1.0 | 9.4 |
| Trachea | BD | 93.4 | 5.8 | 1.1 | BD | 0.2 | 0.7 | 0.7 | 8.5 |
| Stomach | BD | 92.6 | 3.4 | 4.5 | BD | 0.8 | 0.4 | 0.9 | 10.0 |
| Aorta | BD | 90.5 | 5.4 | 2.3 | BD | BD | 1.2 | 1.1 | 10.0 |
| Brain | BD | 94.6 | 5.1 | 1.1 | BD | BD | 0.6 | BD | 6.7 |
| Esophagus | BD | 92.8 | 4.4 | 1.5 | BD | 0.5 | 0.7 | 0.6 | 7.6 |
| Spleen | BD | 90.4 | 4.9 | 4.9 | BD | 0.9 | 0.7 | 0.9 | 12.2 |
| Testes | BD | 95.7 | 5.2 | 0.7 | BD | 0.6 | 0.2 | BD | 6.7 |
| Salivary gland | BD | 96.5 | 2.0 | 1.5 | BD | 0.4 | 0.5 | 0.3 | 4.7 |
| Liver | BD | 90.1 | 5.3 | 3.6 | BD | 0.8 | 0.8 | 0.7 | 11.2 |
| Small intestine (duodenum) | BD | 94.3 | 3.8 | 2.2 | BD | 1.2 | 0.3 | 0.4 | 8.0 |
| Uterine horn | BD | 94.9 | 4.0 | 0.9 | BD | 0.6 | 0.4 | 0.3 | 6.2 |
| Kidney | BD | 93.8 | 5.0 | 1.5 | BD | 0.4 | 0.5 | 0.5 | 7.9 |
| Tongue | BD | 94.2 | 4.3 | 1.1 | BD | 0.4 | 0.6 | 0.4 | 6.7 |
| Heart | BD | 92.7 | 4.7 | 1.1 | BD | 0.3 | 0.6 | 0.6 | 7.3 |
| Large intestine (colon) | BD | 84.2 | 10.5 | 5.7 | BD | 0.1 | 1.7 | 1.8 | 19.8 |
| Thymus | BD | 92.6 | 4.3 | 2.4 | BD | 1.2 | 0.5 | 0.4 | 8.8 |
| Pancreas | BD | 94.1 | 3.9 | 1.3 | BD | 0.6 | 0.5 | 0.5 | 6.8 |
Proportion of total Sia for each variant shown as a percentage, with the sum of all O-acetyl forms combined given in the far-right column (O-Ac sum). BD, the Sia form was below the limit of detection.
Analysis of modified Sia in saliva and mucus and on erythrocytes.
It has been previously reported that human colonic mucin is highly enriched in 9-O-Ac Sia, which may regulate the activities of some sialidases and Sia transporters of the gut microflora (12, 37, 54, 55). Strong staining for 9-O-Ac in human respiratory tissues, and also within the submucosal glands of human respiratory tissue, have also been reported, indicating that mucus from these glands could be enriched in O-acetylated Sia (54, 55). To determine if human respiratory mucus was enriched in 7,9-O- and 9-O-Ac, secreted mucus from primary normal human bronchial epithelial (NHBE) cells, as well as conditioned medium from human alveolar basal epithelial adenocarcinoma A549 cells, was analyzed by HPLC to determine the Sia composition. We found that the secreted proteins in mucus from NHBE cell and A549 cell conditioned media contained primarily unmodified Neu5Ac, with ∼1 to 2% 9-O-Ac and no detectable levels of 7,9-O-Ac (Table 3). This indicates that secreted mucus from these respiratory cells in culture is not enriched for O-acetyl modifications, which differs from previous reports for colonic mucin (12, 54).
TABLE 3.
Analysis of A549 conditioned media and mucus collected from NHBE cell cultures for total sialic acid content using HPLC
| Cell type | % Siaa
|
||||||
|---|---|---|---|---|---|---|---|
| Source | Neu5Gc | Neu5Ac | Neu5,9Ac2 | Neu5,7Ac2 | Neu5,8Ac2 | Neu5,7,8/9Ac3 | |
| A549 | Conditioned mediab | BD | 98.16 | 1.84 | BD | BD | BD |
| NHBE | Mucus | BD | 98.6 | 1.40 | BD | BD | BD |
Proportion of total Sia for each variant shown as a percentage. BD, the Sia form was below the limit of detection. The percentages are averages of multiple conditioned-medium collections from A549 cells (n = 4) and multiple NHBE cell donors (n = 4).
Collected from A549 bronchial epithelial cell conditioned media; contained mucins.
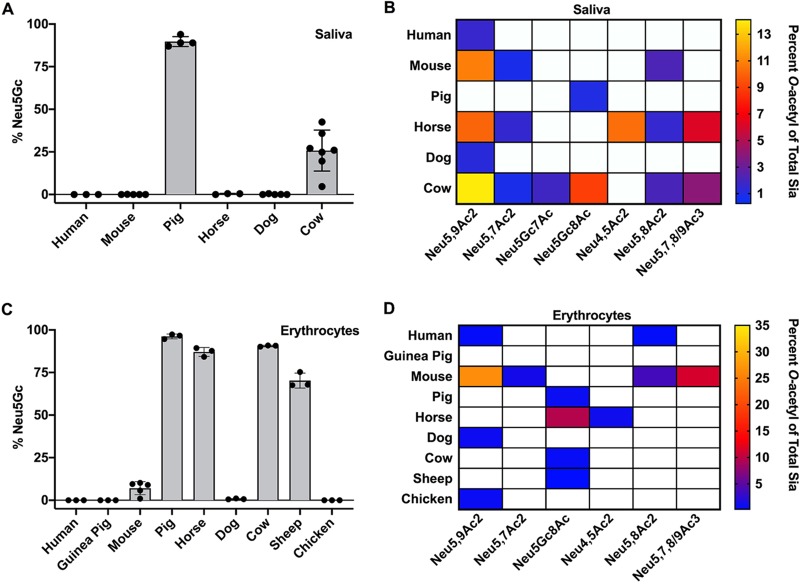
To look more broadly at the possible range of modified Sia present in secreted mucus in different animals, we examined saliva from a number of influenza virus host species, including human, pig, horse, and dog (Fig. 5A and B). While the proteins in saliva differ from those found in respiratory mucus, they contain some of the same heavily glycosylated proteins, including mucins like MUC5B (56–58). Human saliva was similar to the secreted mucus from NHBE and A549 cells in containing primarily Neu5Ac Sia with little 9-O-Ac Sia, and the composition of dog saliva showed a similar profile. However, most other animals showed far more diversity in their Sia profiles, with both mice and horses having enrichment for several different O-acetyl modifications. Laboratory mouse saliva contained a combined ∼17% O-acetylated Sia in the forms of 7-O-, 8-O-, and 9-O-Ac, while horse saliva contained ∼10% 4-O-Ac, as well as ∼19% other O-acetyl Sia variants combined. Pig saliva was unique among the IAV hosts examined in having ∼90% of the total Sia belonging to the Neu5Gc form. The diversity of modifications seen in mouse, horse, and pig salivas may have a strong influence on any Sia-binding pathogens, including influenza viruses, as well as on commensal bacterial communities in different species.
FIG 5.
Total Sia of saliva (A and B) and erythrocytes (C and D) were collected via acid hydrolysis and analyzed using HPLC. O-Acetyl Sia percentages are given as heat maps, with white squares indicating that a Sia form was below the limit of detection. Values are given as the percentage of total Sia collected from tissue samples. Saliva samples were as follows (n represents the number of individuals of each species): human (n = 3), mouse (n = 5), pig (n = 4), horse (n = 3), dog (n = 5), and cow (n = 7). Erythrocyte samples were as follows (n represents the number of individuals of each species): human (n = 3), guinea pig (n = 3), mouse (n = 5), pig (n = 3), horse (n = 3), cow (n = 3), sheep (n = 3), chicken (n = 3), and dog (n = 3). Filled circles show individual data points.
Erythrocytes (red blood cells [RBCs]) express high levels of sialylated surface molecules, primarily on glycophorins, and are used in IAV research to study the interactions of HA binding specificity, determining viral titers through the hemagglutination assay and inhibition of hemagglutination by antibodies (HAI assay) (59, 60). It has long been known that IAV varies in hemagglutination of RBCs from different species, at least in part due to differences in the Sia linkages present. The structures of HAs with Sia bound often suggest that modification of the C-4, -5, -7, and/or -9 position would influence IAV interactions with Sia. We found that chicken and guinea pig RBCs, which are often used to determine the titer of IAV and as the standard substrate for HAI assays, contained almost exclusively unmodified Neu5Ac, as did those from humans and dogs (Fig. 5C and D). In contrast, pig, horse, cow, and sheep RBCs contain high proportions of Neu5Gc, along with varying amounts of O-acetyl modifications. The high levels of Neu5Gc Sia present on the RBCs of these species had been previously reported, although not directly quantified as presented here (24, 61). The mouse erythrocytes tested here (from C57BL/6 mice) had the greatest diversity of modifications, with little Neu5Gc, around 50% Neu5Ac, and ∼43% of the Sia modified by 7-, 8-, and/or 9-O-acetylation, as previously reported (45).
Effects of modified Sia on NA cleavage.
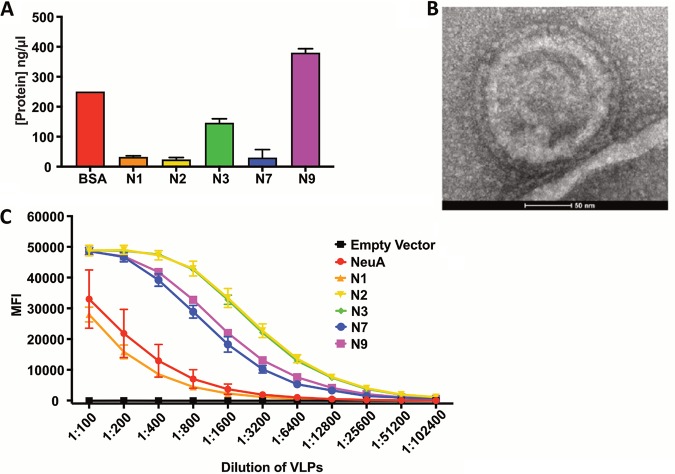
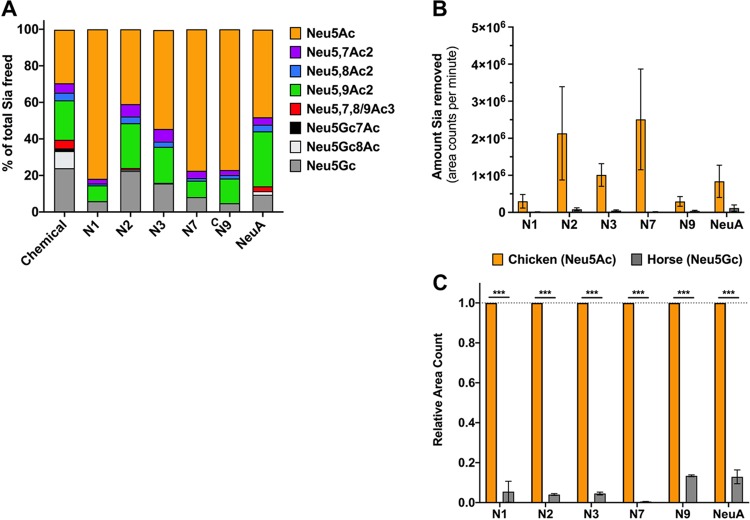
Neuraminidases (sialidases) expressed by bacteria and viruses cleave Sia from oligosaccharides and glycoconjugates, and some have been shown to be affected by various Sia modifications (43, 49, 61–63). However, little is known about the effects of modified Sia on IAV NAs from different strains. We examined the effects of Sia modifications on cleavage by several different IAV NAs, as well as on the activity of Arthrobacter ureafaciens neuraminidase (NeuA), which was included as a positive control for sialidase activity. The substrates used had high levels of 7,9-O- and 9-O-Ac Neu5Ac (bovine submaxillary mucin [BSM]), Neu5Gc (horse RBCs), or unmodified Neu5Ac (chicken RBCs). IAV NAs from a variety of different IAV strains (N1, N2, N3, N7, and N9) were expressed alone in cells (Fig. 6A) and recovered as purified virus-like particles (VLPs) (Fig. 6B) (64). These VLPs are composed primarily of cell membranes and NA glycoproteins, along with very few host membrane-derived proteins. The sources for the NA proteins included both human isolates (N1, N7, and N9) and avian isolates (N2 and N3). These NA VLPs were first tested for enzymatic activity using a standard NA cleavage assay with methylumbelliferyl N-acetylneuraminide (MuNANA) as the substrate, and a 1:100 dilution was chosen for further cleavage analysis, as this gave a high level of enzymatic activity for all VLPs (Fig. 6C) (49). The NA-expressing VLPs were incubated with BSM or RBCs, and the released Sia were collected and analyzed by HPLC. For BSM, HPLC profiles of total Sia were created to compare NA cleavage preferences (Fig. 7A). These profiles showed the Sia forms that were susceptible to NA cleavage and release, while the nonreleased forms were considered to be resistant to NA. The HPLC profiles were then compared to the total Sia released chemically by acid hydrolysis, an unbiased method that removes all Sia forms present in the original sample (51). All of the viral NA and the bacterial NeuA showed the highest level of cleavage for unmodified Neu5Ac compared to any of the modified forms, as more Neu5Ac was present in the released profiles than with chemical release. There was substantial variation in the cleavage activity against the modified Sia by the different viral NAs. N1 and N7 showed the lowest activities against any modified Sia, and N3 and N9 had intermediate activities, while N2 and NeuA were active against the greatest number of modified forms and most closely matched the chemical release profile. There was lower activity for mono-O-acetylated Sia (7-O-, 8-O-, and 9-O-Ac) by N1, N3, N7, and N9, while all NAs tested had lower activity against the di-O-acetylated Sia (7,8- and 9-O-Ac2) and mono-O-acetylated Neu5Gc forms. All the viral and bacterial NAs, apart from N2, had severalfold lower activities on Neu5Gc alone, as seen in the smaller proportion of that Sia form released compared to the chemical release profile.
FIG 6.
NA VLPs produced in HEK-293 cells are enzymatically functional. (A) NA protein levels by Coomassie blue staining of NA protein present in VLPs after control background subtraction. The data represent three replicates. (B) TEM micrograph of a VLP expressing N2. (C) Comparison of NA enzymatic activities, using a MuNANA assay, between different NA serotypes. The data represent three experimental replicates. The error bars indicate standard deviations.
FIG 7.
NA VLPs preferentially cleave unmodified Neu5Ac Sia, and NA activity is inhibited by O-acetyl and Neu5Gc modifications. (A) Bovine submaxillary mucin was treated with 1:100 NA VLPs or A. ureafaciens NA (NeuA) for 4 h at 37°C, and freed Sia was collected and analyzed using HPLC. The profiles of freed Sia were then compared to the profile of Sia removed chemically, a more unbiased approach. The profiles shown are the averages of the results of two independent experiments. (B and C) Chicken erythrocytes (Neu5Ac) or horse erythrocytes (Neu5Gc) were treated with 1:100 NA VLPs for 4 h at 37°C, freed Sia was collected, and the total Sia removed was determined using HPLC. (B) Average area counts per minute (area under the curve of the HLC chromatogram) were used as a measure of the absolute amount of Sia removal to compare the amounts of Sia released between chicken and horse erythrocytes. (C) Relative area counts compared between chicken and horse erythrocytes. The data shown are averages of the results of two independent experiments. The data were analyzed by t test using PRISM software. ***, P ≤ 0.001. The error bars indicate standard deviations.
To further test the abilities of these NA VLPs to cleave Neu5Gc compared to Neu5Ac, the amounts of Sia released from either horse RBCs (84% Neu5Gc) or chicken RBCs (99% Neu5Ac) were compared. All NAs showed levels of Neu5Gc Sia released from horse RBCs that were significantly lower than the levels of Neu5Ac released from chicken RBCs (Fig. 7B). When compared directly, NA VLPs removed 5 to 12% of the Neu5Gc relative to their activities on Neu5Ac (Fig. 7C). Again, variability was seen between NA VLPs here, with N7 having the lowest activity against Neu5Gc compared to Neu5Ac and N9 having the highest. There was also variability in cleavage activity between NAs from different strains, as seen in the variable amounts of Neu5Ac removed by the NA VLPs (Fig. 7B), but it is unclear whether this difference was due to the intrinsic activities of the NAs when expressed as VLPs or to innate differences in the specific activities of each NA enzyme, or both. It is clear, however, that O-acetyl and Neu5Gc modifications inhibit the activities of many different IAV NAs and of NeuA and that multiple modifications, such as di-O-acetyl modifications or Neu5Gc that is also O-acetylated, were even more inhibitory, showing high resistance to most of the viral and bacterial NAs tested here.
Effects of modified Sia on HA binding.
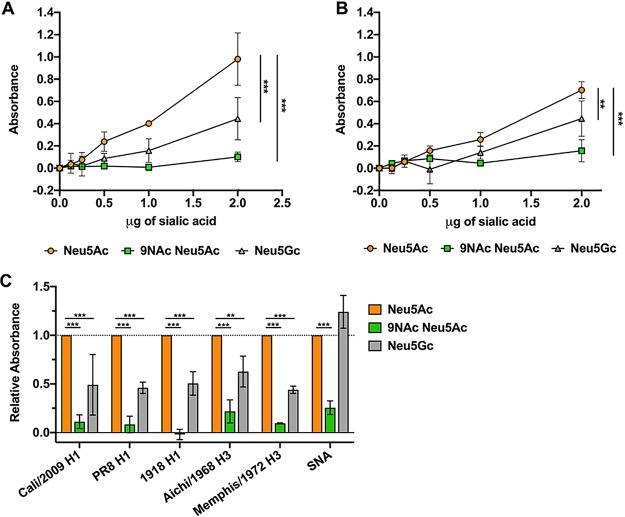
The initiation of IAV infection requires HA glycoprotein binding to Sia to allow the virus to be taken up into the cell, and there appears to be a direct relationship between Sia binding affinity and infection (65). To clearly determine the effect of Sia modifications on HA binding, we examined the binding of soluble HA fused to a human IgG1 Fc (HA-Fc) to synthetic, biotinylated α2-6-linked sialosides of Neu5Ac, Neu5Ac9NAc, and Neu5Gc (66). Neu5Ac9NAc was used instead of 9-O-acetyl Neu5Ac (Neu5,9Ac2) due to the increased stability of the 9-N-Ac group (67, 68). Briefly, enzyme-linked immunosorbent assay (ELISA) grade 96-well plates were coated with HA-Fc derived from California/04/2009 H1N1 and Aichi/2/1968 H3N2 strains and then incubated with the synthetic biotinylated sialosides. Binding of the biotinylated sialosides to HA-Fc was detected using a streptavidin-linked horseradish peroxidase (HRP) probe, as previously described (69, 70). Compared to Neu5Ac sialosides, both California/04/2009 H1 and Aichi/1968 H3 HA-Fcs had decreased binding to Neu5Gc and Neu5Ac9NAc (Fig. 8A). The addition of the N-acetyl group at C-9 blocked most binding, while Neu5Gc showed only a low level of binding. We saw the same binding dynamics for other H1 and H3 HA-Fcs, but the SNA lectin, which recognizes α2-6-linked Sia, bound equally well to Neu5Ac and Neu5Gc, but not to Neu5Ac9NAc (Fig. 8B). This shows that the presence of O-acetyl (and in this case N-acetyl) modifications can inhibit many Sia-binding proteins.
FIG 8.
Soluble HA-Fc binding to synthetic sialosides showed decreased binding to modified Sia in an ELLA. (A and B) Soluble HA constructs were developed by expressing HA proteins from different IAV strains fused to a human IgG1 Fc (HA-Fc). HA-Fc binding to synthetic α2-6-linked sialosides was assessed using an ELLA. Titration curves of sialoside binding by HA-Fc for A/California/04/2009 H1N1 (A) and A/Aichi/2/1968 H3N2 (B) were measured via colorimetric measurement. The error bars indicate standard deviations. (C) Sialoside binding for different H1 and H3 HA-Fcs were determined using 2 μg of sialic acid. Lectin from Sambucus nigra (SNA), which specifically binds α2-6-linked Sia, was also included as a control. The data are shown relative to HA-Fc binding to unmodified Neu5Ac. The data were analyzed by 2-way analysis of variance (ANOVA) using PRISM software and are averages of the results of three independent experiments. **, P ≤ 0.01; ***, P ≤ 0.001.
Effects of modified Sia on influenza A virus infection.
While the low surface expression of 9-O- and 7,9-O-Ac on cells does not reduce IAV infection, viruses also encounter modified Sia in mucus, which in many hosts and tissues has larger amounts of these modifications. To determine how the Sia modifications can affect IAV infection, untreated BSM or BSM treated with esterase to remove 9-O-Ac and 7,9-O-Ac was incubated with A/California/04/2009 (pH1N1), A/Puerto Rico/8/1934 (PR8 H1N1), and A/Victoria/361/2011 (Victoria H3N2) prior to infection of cells (Fig. 9A). Both untreated BSM and esterase-treated BSM were inhibitory toward all three IAV strains, with a trend toward higher inhibition by esterase-treated BSM and significantly more inhibition for the PR8 H1N1 strain. This suggests that removing the 7,9-O- and 9-O-Ac from the Sia may have increased virus binding to mucin and inhibition of viral infection.
FIG 9.
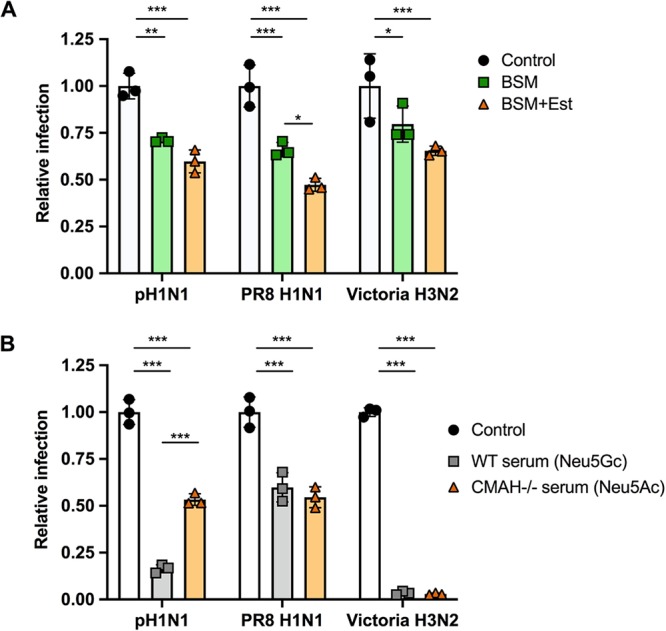
Virus infection is inhibited by mucin, with greater inhibition when O-acetyl groups are removed. Virus infection is also inhibited by serum, with no clear difference between Neu5Ac and Neu5Gc presence. (A) A/California/04/2009 (pH1N1), A/Puerto Rico/8/1934 (PR8, H1N1), and A/Victoria/361/2011 (Victoria H3N2) were mixed with 20 μg of BSM or BSM pretreated with esterase-active bovine coronavirus (BCoV HE-Fc) to remove O-acetyl modifications (BSM+Est). The mixture was then used to infect cells at an MOI of 0.5 for 10 h. Infectivity was determined by flow cytometry analysis for NP-positive cells. (B) A/California/04/2009 (pH1N1), A/Puerto Rico/8/1934 (PR8 H1N1), and A/Victoria/361/2011 (Victoria H3N2) were mixed with sera from either WT mice (Neu5Gc) or CMAH−/− mice (Neu5Ac). The mixture was then used to infect cells at an MOI of 0.5 for 10 h. Infectivity was determined by flow cytometry analysis for NP-positive cells. The data were analyzed by 2-way ANOVA using PRISM software. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Sia in sera have long been known to bind to influenza viruses, so sera are often treated with neuraminidase as a “receptor-destroying enzyme” prior to use in serological tests (71, 72). To specifically compare the effects of added Neu5Gc or Neu5Ac on the efficiency of infection, the same three IAV strains were incubated with mouse serum from either wild-type mice (>80% Neu5Gc) or CMAH−/− mice (100% Neu5Ac) prior to inoculation of cells (Fig. 9B). In this case, no specific trend was detected for the three viruses tested. Victoria H3N2 showed almost complete inhibition of infection by both sera, while PR8 H1N1 had a lower level of inhibition. Only pH1N1 showed a significant difference in responses to the sera, with the WT mouse serum having a much stronger inhibitory effect than the CMAH−/− serum. This suggests that Neu5Gc versus Neu5Ac inhibition may vary by virus strain and that it likely depends on some combination of HA binding specificity and NA activity.
DISCUSSION
Modified Sia are widely expressed within tissues and on mucosal surfaces of many animals, but with significant variation in the amounts of each modified Sia present (13, 14). Modified Sia are present at high levels on mucosal surfaces, including in GI and respiratory tissues, indicating that they are likely involved in tissue- and host-specific interactions with both pathogens and normal flora. While there have been suggestions that 9-O-Ac, 7,9-O-Ac, and Neu5Gc Sia might influence IAV infection by interfering with HA binding or NA activities, there is not a lot of direct evidence for their effects. The goal of this study was to reexamine and provide additional details on the tissue-specific expression of the 9-O-Ac, 7,9-O-Ac, and Neu5Gc Sia in mice, as well as in secreted mucus from different IAV host species. We also defined the effects of these Sia modifications on HA binding, NA activity, and infection for different IAV strains. While we focused on IAV here, the findings should be broadly relevant and underline the need to understand differences in the expression of modified Sia between animal species and their potential roles in tropism of and infection by the many viruses that interact with or utilize Sia during infection.
Mouse tissue distribution.
In previous studies, we have shown that there is variation in the expression of 9-O-, 7,9-O-, and 4-O-Ac Sia (14). To better understand the type of variation seen in other tissues, we examined the distribution of these forms, as well as Neu5Gc, in the different tissues of wild-type C57BL/6 mice, as well as for CMAH−/− mice. Using the HE-Fc virolectin probes, O-acetyl-modified Sia were found only in specific tissues and on certain cell subpopulations. Higher staining was seen for cells and mucus in the respiratory and GI tracts, with trachea and colon having particularly strong staining for O-acetyl Sia on epithelial cells and associated mucus layers. The percentages of O-acetyl-modified Sia in the tissues of mice varied widely, with high levels of 7,9-O-Ac2 and 9-O-Ac being found on erythrocytes (∼43%) and in the colon (∼19%). The levels in most other tissues varied between 2 and 9%, although this would likely be associated with higher levels on the smaller subsets of cells that were positive for staining with the probes, as processing the tissue in bulk for HPLC analysis would dilute higher levels of Sia on cell subpopulations. Neu5Gc was also present in many wild-type mouse tissues, with expression varying widely (between 10 and 80%). This is consistent with staining for Neu5Gc previously reported in some mouse tissues (52). 4-O-Ac Sia was found in small quantities in only a few tissues, with the highest levels found in the small intestine (∼2%). This is consistent with previous findings of 4-O-Ac levels in mouse brain and liver quantified by gas chromatography-mass spectrometry (GC-MS), although gut-associated levels of 4-O-Ac were much higher in that study (73). This discrepancy is likely due to the acetic acid hydrolysis used to prepare samples for HPLC analysis, which can lead to loss of Sia modifications, particularly 4-O-Ac modifications. This is a major challenge in studying Sia modifications, as they are often labile and easily lost by processing methods. The virolectins are advantageous, as they allow us to examine O-acetyl Sia in situ, but better techniques are still needed to study many of the Sia modifications with accuracy.
It is interesting that the levels of O-acetyl modifications were more prevalent on Neu5Ac Sia than on Neu5Gc and that the levels of O-acetyl-modified Sia were higher across most tissues tested in the CMAH−/− mice. This is consistent with previous analysis of some CMAH−/− tissues by immunohistochemical staining (52). However, Neu5Gc also had O-acetyl variants (Neu5Gc7Ac and Neu5Gc8Ac) that were present at low levels in mouse tissues, as well as in bovine mucin, which is consistent with previous reports (8, 29, 74). It has been previously shown that the O-acetyltransferase CasD1 prefers CMP-Neu5Ac as a substrate over CMP-Neu5Gc (16). However, the regulation of the O-acetyl modification placement on both Neu5Ac and Neu5Gc, and possible differences between the two Sia forms, remain to be fully elucidated.
Variation on erythrocytes, in saliva, and on other mucins.
Erythrocytes interact with viruses in the blood, and are an important tool in virology for analyzing virus binding to Sia in hemagglutination assays, as well as for determining virus titers and for antibody inhibition (HAI) assays (59, 60). It has long been known that many IAV strains vary in hemagglutination of RBCs from different species, at least in part due to differences in the Sia linkages present. The structures of HAs with Sia bound often suggest that modification of the C-4, -5, -7, and/or -9 position would influence IAV interactions with Sia (75, 76). We found that the Sia modifications on erythrocytes varied greatly between species, with mice having a high proportion of O-acetyl-modified Sia while horses, cows, sheep, and pigs have primarily Neu5Gc Sia. The reasons for such variation in Sia modifications are likely complex but may involve regulation of cell-cell signaling between erythrocytes and endothelial and immune cells, as well as of interactions with pathogens. It has recently been suggested that loss of Neu5Gc expression in humans could be related to pressure from an ancestral strain of Plasmodium falciparum (45, 47).
To define the variation of modified Sia present in secreted mucus across different animals, we examined saliva, as it contains many of the same heavily glycosylated proteins and mucins, including MUC5B, that are present in respiratory mucus (56–58). We found a great deal of variability in both the amounts and types of modified Sia present. Some animals (horses, mice, and cows) had larger amounts of O-acetyl Sia in their saliva, while humans and dogs had primarily unmodified Neu5Ac forms. Pigs are a natural IAV host, and their saliva contained primarily Neu5Gc, so pig saliva might inhibit IAV infection. Horses had around 10% 4-O-Ac Sia present in their saliva. The 4-O-Ac modification has been proposed to be a potent inhibitor of many types of viral and bacterial neuraminidases and to be the γ-inhibitor of horse serum, where it may be present at high levels on the α-2 macroglobulin protein (62, 71, 77). However, as the gene for the 4-O-sialyl acetyltransferase has not yet been identified, little is known about its synthesis, expression, or regulation (22, 23).
It has previously been reported that human colonic mucus is enriched for 7,9-O- and 9-O-Ac (12, 54). However, human saliva and secreted mucus from respiratory cells contained mostly unmodified Neu5Ac, suggesting different expression levels of modified Sia in the mucus of the respiratory and gastrointestinal tissues. This may be due to the particular functional characteristics of the microbiome in the GI tract compared to other mucosal sites, where 7,9-O- and 9-O-Ac on colonic mucus may decrease bacterial degradation of sialylated glycans, perhaps improving mucus integrity (78, 79). Microbiome interactions may also explain the increased O-acetyl modifications found in both cow and mouse saliva. Cows are ruminants and, as part of their digestion, regurgitate partially digested food from their microbe-rich rumen into the mouth to continue chewing as a cud to extract more nutrients. Somewhat similarly, mice are coprophagic and consume feces, along with associated microbes, to redigest to improve absorption of nutrients. This would give both cows and mice the potential for more complex mucus and microbe interactions in both the oral cavity and the gut, and thus, having O-acetyl modifications on sialylated glycans on saliva mucus proteins could be advantageous by preventing degradation. However, further research is needed on the roles of these modifications in the interactions of oral and colonic mucus with the microbiome.
Effects of modified Sia on influenza viruses.
IAV use Sia as their primary receptor for host infection, and the specific linkages of Sia to the underlying glycan chain have long been known to influence host tropism (41). We examined the effects of O-acetyl and Neu5Gc modifications on IAV HA binding and NA cleavage and on infection and found differences among the IAV strains examined. All the different IAV NAs tested showed preferential removal of Neu5Ac over any modified Sia form. Cleavage by N1, N7, and N9 were strongly inhibited by monoacetylated Sia, while N2 and N3 were less affected. All NAs had much lower activity against Neu5Gc and diacetylated Sia, and particularly against Neu5Gc forms with additional O-acetylations. This confirms previous reports showing or suggesting inhibition of NA cleavage in H1N1 and H3N2 strains (43, 49) but revealing that there is wide variation of effects on different NA types. These differences did not appear to follow broad structural groupings between group 1 NA proteins (N1, N4, N5, and N8) and group 2 NA proteins (N2, N3, N6, N7, and N9) (80). However, one possible source of variation could be the host tropism of the parent viruses for the NA proteins used in the VLPs. Both the N2 and N3 genes originated from avian isolates, while N1, N7, and N9 were from human isolates. Differences in NA cleavage abilities against Neu5Gc had previously been reported for swine IAV isolates (50), but comparisons between NA proteins against O-acetylated Sia forms have been restricted to human isolates (43). The structures of NA inhibitors, such as oseltamivir and zanamivir, resemble that of Sia, with side groups added along the glycerol side chain, analogous to C-7 through C-9, and at the C-5 and C-4 positions (81). The locations of these inhibitory side groups resemble the chemical modifications of NeuGc and 7,9-O- and 9-O-Ac Sia variants, which explains in part why these natural Sia forms are NA inhibitors. It will be interesting to examine NA cleavage efficiency against these natural modified Sia forms from IAV strains that were adapted to other hosts with Neu5Gc expression, such as horses and pigs, as well as NA proteins with resistance to synthetic NA inhibitors.
Acetylation and Neu5Gc modifications were also inhibitory for HA binding, with soluble HA-Fcs sourced from different H1N1 and H3N2 IAV strains showing significantly lower binding to these forms than to unmodified Neu5Ac in an enzyme-linked lectin assay (ELLA). Previous reports indicated that most natural IAV isolates do not bind to Neu5Gc, with the exception of an equine H7N7 strain (49). When the equine H7N7 strain and a laboratory-generated H5 Y161A mutant that bound Neu5Gc were compared, it was found that the structural changes in the 130 loop caused by the loss of hydrophobic interactions of the H1 Y161A mutation were also shared in the H7 structure (49). Thus, it appears that the receptor-binding pocket of most IAV HA proteins cannot accommodate the Neu5Gc modification due to steric hindrance of the HA1 130 loop. Structural analysis of HA proteins with O-acetyl variants have not been performed to the same level of detail; however, structures of HA bound to Neu5Ac show important hydrogen bond interactions between amino acids at the C-9 of Sia and positions 98, 190, and 228 in the receptor-binding pocket (82). It is likely that the O-acetyl modification at C-9 would block Sia from binding correctly in the receptor-binding pocket.
IAV infects cells at mucosal surfaces, including those of the gastrointestinal tract of birds and the respiratory tract of mammalian hosts, so it interacts directly with mucus both to initiate infection and during viral release and shedding. Using untreated or esterase-treated BSM (to remove 7,9-O- and 9-O-Ac), it was seen that the unmodified Neu5Ac was most inhibitory, suggesting that O-acetyl Sia reduces HA binding to the mucin, allowing binding to the unmodified Sia on the cell surface. Esterase treatment, therefore, allows more efficient binding to the BSM and greater inhibition of IAV infection. For some viruses, there is likely complementarity between the inhibitory effects of 7,9-O- and 9-O-Ac on HA and NA, where lower HA binding allows the viruses to avoid binding to Sia forms that NA cannot remove efficiently. This effect has been reported for virus grown in the presence of other NA inhibitors (65, 83) and is seen in the balance between the activities of HA and NA for the α2-3- and α2-6-linked Sia (83, 84). Incubation of virus with wild-type mouse serum (>80% Neu5Gc) compared to the sera of CMAH−/− mice (100% Neu5Ac) also showed varying effects. Inhibition by WT serum was seen only for pH1N1 virus, while Victoria H3N2 and PR8 H1N1 were inhibited by both sera. This could confirm previous findings that the density of Sia on these serum proteins is the strongest inhibitory factor, rather than the type of modified Sia present, as shown for incubation with horse serum (85). However, the variable results for different viruses may indicate variation in sensitivity to Neu5Gc between strains that requires further investigation. A recent paper showed that repeat passaging of human and canine strains of IAV in wild-type mice compared to CMAH−/− mice saw no mutations in HA or NA nor any differences in disease severity between mouse strains (86). It is possible that the amount of unmodified Sia present in mouse respiratory tissue is sufficient for infection, with virus avoiding binding to Neu5Gc. However, a natural isolate of equine H7N7 has previously been reported to preferentially bind Neu5Gc (49). It would therefore be worth comparing the interactions of IAV strains adapted to host species with Neu5Gc, particularly species with higher levels of modified Sia present in their respiratory tracts, such as horses and pigs.
In summary we have shown that both the O-acetyl and Neu5Gc modifications present on secreted glycoproteins in mucus and saliva, as well as on erythrocytes, vary greatly between different species. Some of these modifications inhibit HA binding and NA cleavage, but with significant variability between IAV strains. While the presence of these modifications can inhibit infection, how they affect virus host tropism and evolution is likely complex and is still not fully understood.
MATERIALS AND METHODS
Cells and virus.
Canine MDCK-NBL2 (ATCC) and A549 (ATCC) cells were grown in Dulbecco’s modified Eagle medium (DMEM) with 5% fetal bovine serum and 50 μg/ml gentamicin. Influenza A virus strains pH1N1 (A/California/04/2009), Victoria H3N2 (A/Victoria/361/2011), and PR8 (A/Puerto Rico/8/1934) were rescued from reverse-genetics plasmids using a previously established protocol (87). The rescued viruses were grown to low passage on MDCK-NBL2 cells using infection medium containing DMEM, 0.03% bovine serum albumin (BSA), and 1 μg/ml tosyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin.
Erythrocytes, mucus, and saliva.
Chicken, cow, sheep, guinea pig, and pig erythrocytes were sourced from Lampire Biological Laboratories (Pipersville, PA). Horse blood and mouse blood were sourced from the Baker Institute for Animal Health (Cornell University, Ithaca, NY). Dog blood was sourced from the Cornell Veterinary Hospital Diagnostic Center (Ithaca, NY). All the erythrocytes were washed in phosphate-buffered saline (PBS) three times and diluted to 5% (vol/vol) in PBS. Bovine submaxillary mucin was purchased from Sigma-Aldrich (St. Louis, MO). Animal studies were all subject to approved protocols from the Cornell Institutional Animal Care and Use Committee.
Saliva from humans was collected by passive drooling, following the protocol approved by the University at Buffalo Human Subjects Institutional Review Board (IRB) (study no. 030–505616). Informed consent was obtained from all human participants. Saliva from animals was collected by suction using commercially available devices containing absorbent sponges in a syringe-like receptacle (Super-SAL and Micro-SAL; Oasis Diagnostics, Vancouver, WA). Saliva from mice (laboratory strain C57BL/10SnJ) was kindly provided by Jill Kramer (University at Buffalo) using a collection procedure described previously (88). Saliva from dogs, cows, horses, and pigs was provided by Erin Daugherty and Luce E. Guanzini (Cornell University). The animals were not allowed to eat or drink prior to collection to ensure the oral cavity was free of food and other debris. The collection was performed using a commercially available device (Micro-SAL). Large animals were gently restrained, and a larger collection device (Super-SAL) was placed under the tongue for up to 3 min or until fully soaked. Saliva from castrated domestic pigs was also provided by Anja Globig (Friedrich-Loeffler-Institut, Insel Riems-Greifswald, Germany). Saliva from dogs was also kindly provided by Barbara McCabe (Buffalo, NY).
Normal human bronchial epithelial cells (Lonza; catalog no. CC-2540S) were seeded onto human placental collagen-coated permeable transwell inserts at a density of 2.5 × 104 cells per well and cultured in bronchial epithelial cell growth basal medium (BEBM) supplemented with bronchial epithelial cell growth SingleQuots (BEGM) in both the apical and basal compartments. After reaching confluence at approximately 7 days postseeding, the apical medium was aspirated, and the basal medium was replaced with air-liquid interface medium containing half DMEM and half BEBM, plus the BEGM SingleQuots to complete differentiation. Apical surfaces of NHBE cells were washed twice with PBS to collect mucins for HPLC analysis.
Conditioned medium from A549 cells was prepared by washing a fully confluent flask of cells to remove any serum and allowing the cells to grow in serum-free medium for 5 to 7 days. Conditioned medium was collected, dialyzed with three volumes of PBS, and concentrated using a 30-kDa centrifugal filter (Pall Corporation). The protein concentration was determined using a Qubit 4 fluorometer (Invitrogen).
Immunohistochemistry of mouse tissues.
Expression of O-acetyl-modified Sia in various tissues of mice was examined by preparing frozen sections of optimal-cutting-temperature compound (OCT)-embedded tissue. After a 30-min fixation in 10% buffered formalin, sections were incubated with recombinant virolectins made by expressing nidovirus HE glycoprotein fused to the Fc region of human IgG1, as described previously (13). Nidovirus HEs are specific for O-acetyl Sia modifications: mouse hepatitis virus (MHV) S for 4-O-Ac, bovine coronavirus (BCoV)-Mebus for 7,9-O-Ac (and low recognition of 9-O-Ac), and porcine torovirus (PToV) P4 for 9-O-Ac. Virolectins were then detected using a biotin-conjugated αFc region secondary antibody followed by incubation with the Vectastain ABC reagent and NovaRed substrate (Vector). Sections were counterstained with hematoxylin.
Quantification of sialic acid by HPLC.
The sialic acid compositions of tissues, mucin, and erythrocytes were analyzed by HPLC as previously described (51, 89). In brief, Sia from 20 to 30 mg of tissue, 50 μg of mucin or saliva, or 100 μl of washed 5% (vol/vol) erythrocytes were released using 2 M acetic acid at 80°C for 3 h, followed by filtration through a 10-kDa centrifugal filter (Microcon) and dried using a vacuum concentrator (SpeedVac). The released Sia were labeled with DMB (Sigma-Aldrich) for 2.5 h at 50°C. HPLC analysis was performed using a Dionex UltiMate 3000 system with an Acclaim C18 column (ThermoFisher) under isocratic elution in 7% methanol, 7% acetonitrile, and 86% water. The Sia standards were bovine submaxillary mucin, normal horse serum, and commercial standards for Neu5Ac and Neu5Gc (Sigma-Aldrich). Pretreatment of samples with 30 μg/ml esterase-active BCoV HE-Fc overnight at 37°C removed 7,9-O- and 9-O-acetyl modifications. Final data analysis was completed using PRISM software (GraphPad; version 8).
Biotinylated α2-6-linked sialosides.
Biotinylated α2-6-linked sialosides, Sia α2-6LacNAc-biotin containing Neu5Ac, Neu5Gc, or Neu5Ac9NAc as the sialic acid form, were synthesized from LacNAc-biotin (90) as the acceptor substrate and Neu5Ac, ManNGc (91), or ManNAc6NAc (68) as the donor precursor using a one-pot multienzyme sialylation system similar to one described previously (92).
IAV HA affinity for 9-O-Ac-modified Sia.
HA-Fc constructs were produced as previously described (93). HA-Fc binding to sialosides was performed as previously described (69, 70). In brief, ELISA grade 96-well plates (ThermoFisher Scientific) were coated with 5 μg of HA-Fc overnight at 4°C. The plates were then washed 3 times with PBS and blocked using 1× Carbo Free blocking buffer (Vector Laboratories, Burlingame, CA) for 1 h. After blocking, the plates were washed once with PBS and treated with sialosides diluted in PBS for 3 h at room temperature and then washed 3 times with PBS. The plates were then incubated with HRP-streptavidin complex (Vector Laboratories) for 45 min, washed 3 times with PBS, and then incubated with 3,3′,5,5′-tetramethyl benzidine (TMB) (Thermo Fisher Scientific). TMB development was stopped with 2 M sulfuric acid and then analyzed using a colorimetric plate reader (Multiskan EX; ThermoFisher Scientific). Data analysis was completed with PRISM software (GraphPad; version 8).
Generation of NA VLPs.
NA sequences were obtained from GenBank (A/Ohio/07/2009 N1, ACP44181; A/mallard/Ohio/11OS2045/2011 N2, AGC70842; A/chicken/Murree/NARC-01/1995 N3, ACL11962; A/Netherlands/219/2003 N7, AAR11367; A/Yunnan/0129/2017 N9, ARG43209). The sequences were codon optimized, tagged, and ordered through Biomatik in the pcDNA3.1(+) vector. To produce VLPs, HEK-293T cells were seeded in 15-cm plates and transfected when 80% confluent. Cells were transfected with 4 μl of polyethylenimine (PEI) (Polysciences; catalog no. 23966-2) at 1-mg/ml concentration for every 1 μg plasmid DNA stock in 9 ml of Opti-MEM. Eight hours posttransfection, 6 ml of prewarmed Opti-MEM was added. Supernatant was collected 72 h posttransfection and purified using ultracentrifugation (110,000 × g; 1.5 h; 4°C) through a 20% sucrose cushion, and then the pellet was resuspended in PBS and stored at 4°C.
NA cleavage assay with NA VLPs.
BSM or erythrocytes from horses and chickens were used as a substrate to determine the NA activities of the different NA VLPs. Briefly, 50 μg of BSM or 5% (vol/vol) washed erythrocytes in PBS were treated with 1:100 NA VLPs or 1:100 A. ureafaciens NA (NeuA; New England BioLabs) for 4 h at 37°C. These dilutions of VLPs were chosen based on the enzymatic activity as determined by a standard NA cleavage assay using MuNANA as the substrate (49). The conditions for NA cleavage were chosen to allow enough Sia to be released for HPLC analysis based on the enzymatic activity of the NA VLPs and NeuA control. Free Sia was collected and prepared for HPLC analysis as described above.
Mucin and serum inhibition of infection.
MDCK cells were seeded to ∼80% confluence in 12-well plates, with cells allowed to settle for 6 h. Virus was diluted in PBS to give a multiplicity of infection (MOI) of 0.5 and then mixed with 20 μg untreated BSM or esterase-treated BSM and incubated for 45 min at room temperature. For mouse serum inhibition, virus was mixed with 200 μg wild-type or CMAH−/− serum instead. Serum- or mucin-treated virus was then added to the washed MDCK cells and incubated for 1 h with tilting to prevent cell drying. The inoculum was then removed, medium was added, and the cells were incubated for 10 h. Cells were then harvested, stained with an anti-influenza A virus NP antibody, and analyzed for infection using a Millipore Guava EasyCyte Plus flow cytometer (EMD Millipore, Billerica, MA) with analysis using FlowJo software (TreeStar, Ashland, OR). Statistical analyses were performed in PRISM software (GraphPad; version 8).
ACKNOWLEDGMENTS
We thank Wendy Weichert for expert technical support and help with erythrocyte samples. We also thank Lubov Neznanova and Jill M. Kramer (University of Buffalo), along with Anja Globig (Friedrich Loeffler Institute, Germany), for sharing samples of saliva. Special thanks are due to Jessica Waltemyer (Cornell University) for additional erythrocyte samples.
The study was supported in part by CRIP (Center of Research in Influenza Pathogenesis), an NIAID-funded Center of Excellence in Influenza Research and Surveillance (CEIRS) contract (HHSN272201400008C to Colin R. Parrish), NIH grants R01 GM080533 and R21 AI142297 to Colin R. Parrish and R01AI130684 to Xi Chen and Ajit Varki, NIH Common Fund Grant U01CA199792 to Ajit Varki, and NIDCR R01DE019807 and NIH Common Fund Grant U01CA221244 to Stefan Ruhl.
REFERENCES
- 1.Varki A, Schauer R. 2009. Sialic acids, Chapter 14. In Essentials of glycobiology, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 2.Varki NM, Varki A. 2007. Diversity in cell surface sialic acid presentations: implications for biology and disease. Lab Invest 87:851–857. doi: 10.1038/labinvest.3700656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lehmann F, Tiralongo E, Tiralongo J. 2006. Sialic acid-specific lectins: occurrence, specificity and function. Cell Mol Life Sci 63:1331–1354. doi: 10.1007/s00018-005-5589-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varki A. 2007. Glycan-based interactions involving vertebrate sialic-acid-recognizing proteins. Nature 446:1023–1029. doi: 10.1038/nature05816. [DOI] [PubMed] [Google Scholar]
- 5.Wasik BR, Barnard KN, Parrish CR. 2016. Effects of sialic acid modifications on virus binding and infection. Trends Microbiol 20:1–11. doi: 10.1016/j.tim.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Breedam W, Pöhlmann S, Favoreel HW, de Groot RJ, Nauwynck HJ. 2014. Bitter-sweet symphony: glycan-lectin interactions in virus biology. FEMS Microbiol Rev 38:598–632. doi: 10.1111/1574-6976.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mandal C, Schwartz-Albiez R, Vlasak R. 2012. Functions and biosynthesis of O-acetylated sialic acids. Top Curr Chem 366:1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schauer R, Kamerling JP. 2018. Exploration of the sialic acid world, p 1–213. In Advances in carbohydrate chemistry and biochemistry. Elsevier, Philadelphia, PA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirose K, Amano M, Hashimoto R, Lee YC, Nishimura SI. 2011. Insight into glycan diversity and evolutionary lineage based on comparative avio-N-glycomics and sialic acid analysis of 88 egg whites of galloanserae. Biochemistry 50:4757–4774. doi: 10.1021/bi101940x. [DOI] [PubMed] [Google Scholar]
- 10.Peri S, Kulkarni A, Feyertag F, Berninsone PM, Alvarez-Ponce D. 2018. Phylogenetic distribution of CMP-Neu5Ac hydroxylase (CMAH), the enzyme synthetizing the proinflammatory human xenoantigen Neu5Gc. Genome Biol Evol 10:207–219. doi: 10.1093/gbe/evx251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aamelfot M, Dale OB, Weli SC, Koppang EO, Falk K. 2014. The in situ distribution of glycoprotein-bound {4-O-acetylated} sialic acids in vertebrates. Glycoconj J 31:327–335. doi: 10.1007/s10719-014-9529-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muchmore EA, Varki NM, Fukuda M, Varki A. 1987. Developmental regulation of sialic acid modifications in rat and human colon. FASEB J 1:229–235. doi: 10.1096/fasebj.1.3.3623000. [DOI] [PubMed] [Google Scholar]
- 13.Langereis MA, Bakkers MJ, Deng L, Vered P-K, Vervoort SJ, Hulswit RJ, van Vliet AL, Gerwig GJ, de Poot SA, Boot W, van Ederen AM, Heesters BA, van der Loos CM, van Kuppeveld FJ, Yu H, Huizinga EG, Chen X, Varki A, Kamerling JP, de Groot RJ. 2015. Complexity and diversity of the mammalian sialome revealed by nidovirus virolectins. Cell Rep 11:1966–1978. doi: 10.1016/j.celrep.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wasik BR, Barnard KN, Ossiboff RJ, Khedri Z, Feng KH, Yu H, Chen X, Perez DR, Varki A, Parrish CR. 2017. Distribution of O-acetylated sialic acids among the tissues of influenza hosts. mSphere 2:e00379-16. doi: 10.1128/mSphere.00379-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arming S, Wipfler D, Mayr J, Merling A, Vilas U, Schauer R, Schwartz-Albiez R, Vlasak R. 2011. The human Cas1 protein: a sialic acid-specific O-acetyltransferase? Glycobiology 21:553–564. doi: 10.1093/glycob/cwq153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baumann A-MT, Bakkers MJG, Buettner FFR, Hartmann M, Grove M, Langereis MA, de Groot RJ, Mühlenhoff M. 2015. 9-O-Acetylation of sialic acids is catalysed by CASD1 via a covalent acetyl-enzyme intermediate. Nat Commun 6:7673. doi: 10.1038/ncomms8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ravasio V, Damiati E, Zizioli D, Orizio F, Giacopuzzi E, Manzoni M, Bresciani R, Borsani G, Monti E. 2017. Genomic and biochemical characterization of sialic acid acetylesterase (siae) in zebrafish. Glycobiology 27:938–946. doi: 10.1093/glycob/cwx068. [DOI] [PubMed] [Google Scholar]
- 18.Orizio F, Damiati E, Giacopuzzi E, Benaglia G, Pianta S, Schauer R, Schwartz-Albiez R, Borsani G, Bresciani R, Monti E. 2015. Human sialic acid acetyl esterase: towards a better understanding of a puzzling enzyme. Glycobiology 25:992–1006. doi: 10.1093/glycob/cwv034. [DOI] [PubMed] [Google Scholar]
- 19.Takematsu H, Diaz S, Stoddart A, Zhang Y, Varki A. 1999. Lysosomal and cytosolic sialic acid 9-O-acetylesterase activities can be encoded by one gene via differential usage of a signal peptide-encoding exon at the N terminus. J Biol Chem 274:25623–25631. doi: 10.1074/jbc.274.36.25623. [DOI] [PubMed] [Google Scholar]
- 20.Zhu H, Chan HC, Zhou Z, Li J, Zhu H, Yin L, Xu M, Cheng L, Sha J. 2004. A gene encoding sialic-acid-specific 9-O-acetylesterase found in human adult testis. J Biomed Biotechnol 2004:130–136. doi: 10.1155/S1110724304307084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schauer R, Shukla AK. 2008. Isolation and properties of two sialate-O-acetylesterases from horse liver with 4- and 9-O-acetyl specificities. Glycoconj J 25:625–632. doi: 10.1007/s10719-008-9109-9. [DOI] [PubMed] [Google Scholar]
- 22.Iwersen M, Dora H, Kohla G, Gasa S, Schauer R. 2003. Solubilisation and properties of the {sialate-4-O-acetyltransferase} from guinea pig liver. Biol Chem 384:1035–1047. doi: 10.1515/BC.2003.116. [DOI] [PubMed] [Google Scholar]
- 23.Iwersen M, Vandamme-Feldhaus V, Schauer R, Institut B, Kiel D. 1998. Enzymatic 4-O-acetylation of N-acetylneuraminic acid in guinea-pig liver. Glycoconj J 904:895–904. [DOI] [PubMed] [Google Scholar]
- 24.Gentsch JR, Pacitti AF. 1987. Differential interaction of reovirus type 3 with sialylated receptor components on animal cells. Virology 161:245–248. doi: 10.1016/0042-6822(87)90192-9. [DOI] [PubMed] [Google Scholar]
- 25.Barnard KN, Wasik BR, LaClair JR, Buchholz DW, Weichert WS, Alford-Lawrence BK, Aguilar HC, Parrish CR. 2019. Expression of 9-O- and 7,9-O-acetyl modified sialic acid in cells and their effects on influenza viruses. mBio 10:e02490-19. doi: 10.1128/mBio.02490-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein A, Krishna M, Varki NM, Varki A. 1994. 9-O-Acetylated sialic acids have widespread but selective expression: analysis using a chimeric dual-function probe derived from influenza C hemagglutinin-esterase. Proc Natl Acad Sci U S A 91:7782–7786. doi: 10.1073/pnas.91.16.7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu H, Cao H, Tiwari VK, Li Y, Chen X. 2011. Chemoenzymatic synthesis of C8-modified sialic acids and related α2-3- and α2-6-linked sialosides. Bioorg Med Chem Lett 21:5037–5040. doi: 10.1016/j.bmcl.2011.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ravindranath RMH, Basilrose S. 2005. Localization of sulfated sialic acids in the dentinal tubules during tooth formation in mice. Acta Histochem 107:43–56. doi: 10.1016/j.acthis.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 29.Morimoto N, Nakano M, Kinoshita M, Kawabata A, Morita M, Oda Y, Kuroda R, Kakehi K. 2001. Specific distribution of sialic acids in animal tissues as examined by LC−ESI-MS after derivatization with 1,2-diamino-4,5-methylenedioxybenzene. Anal Chem 73:5422–5428. doi: 10.1021/ac0104328. [DOI] [PubMed] [Google Scholar]
- 30.Corfield AP, Wagner SA, Safe A, Mountford RA, Clamp JR, Kamerling JP, Vliegenthart JFG, Schauer R. 1993. Sialic acids in human gastric aspirates: detection of 9-O-lactyl- and 9-O-acetyl-N-acetylneuraminic acids and a decrease in total sialic acid concentration with age. Clin Sci 84:573–579. doi: 10.1042/cs0840573. [DOI] [PubMed] [Google Scholar]
- 31.Deplancke B, Gaskins HR. 2001. Microbial modulation of innate defense: goblet cells and the intestinal mucus layer. Am J Clin Nutr 73:1131S–1141S. doi: 10.1093/ajcn/73.6.1131S. [DOI] [PubMed] [Google Scholar]
- 32.Knowles MR, Boucher RC. 2002. Mucus clearance as a primary innate defense mechanism for mammalian airways. J Clin Invest 109:571–577. doi: 10.1172/JCI15217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Severi E, Hood DW, Thomas GH. 2007. Sialic acid utilization by bacterial pathogens. Microbiology 153:2817–2822. doi: 10.1099/mic.0.2007/009480-0. [DOI] [PubMed] [Google Scholar]
- 34.Thomas GH. 2016. Sialic acid acquisition in bacteria—one substrate, many transporters. Biochem Soc Trans 44:760–765. doi: 10.1042/BST20160056. [DOI] [PubMed] [Google Scholar]
- 35.Phansopa C, Kozak RP, Liew LP, Frey AM, Farmilo T, Parker JL, Kelly DJ, Emery RJ, Thomson RI, Royle L, Gardner RA, Spencer DIR, Stafford GP. 2015. Characterization of a sialate-O-acetylesterase (NanS) from the oral pathogen Tannerella forsythia that enhances sialic acid release by NanH, its cognate sialidase. Biochem J 472:157–167. doi: 10.1042/BJ20150388. [DOI] [PubMed] [Google Scholar]
- 36.Lewis AL, Lewis WG. 2012. Host sialoglycans and bacterial sialidases: a mucosal perspective. Cell Microbiol 14:1174–1182. doi: 10.1111/j.1462-5822.2012.01807.x. [DOI] [PubMed] [Google Scholar]
- 37.Robinson LS, Lewis WG, Lewis AL. 2017. The sialate O-acetylesterase EstA from gut Bacteroidetes species enables sialidase-mediated cross-species foraging of 9-O-acetylated sialoglycans. J Biol Chem 292:11861–11872. doi: 10.1074/jbc.M116.769232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matrosovich M, Herrler G, Klenk HD. 2015. Sialic acid receptors of viruses. Top Curr Chem 367:1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hulswit RJG, Lang Y, Bakkers MJG, Li W, Li Z, Schouten A, Ophorst B, van Kuppeveld FJM, Boons G-J, Bosch B-J, Huizinga EG, de Groot RJ. 2019. Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc Natl Acad Sci U S A 116:2681–2690. doi: 10.1073/pnas.1809667116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang X, Dong W, Milewska A, Golda A, Qi Y, Zhu QK, Marasco WA, Baric RS, Sims AC, Pyrc K, Li W, Sui J. 2015. Human coronavirus HKU1 spike protein uses O-acetylated sialic acid as an attachment receptor determinant and employs hemagglutinin-esterase protein as a receptor-destroying enzyme. J Virol 89:7202–7213. doi: 10.1128/JVI.00854-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Graaf M, Fouchier RAM. 2014. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J 33:823–841. doi: 10.1002/embj.201387442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohen M, Zhang X-Q, Senaati HP, Chen H-W, Varki NM, Schooley RT, Gagneux P. 2013. Influenza A penetrates host mucus by cleaving sialic acids with neuraminidase. Virol J 10:321. doi: 10.1186/1743-422X-10-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muñoz-Barroso I, García-Sastre A, Villar E, Manuguerra JC, Hannoun C, Cabezas JA. 1992. Increased influenza A virus sialidase activity with N-acetyl-9-O-acetylneuraminic acid-containing substrates resulting from influenza C virus O-acetylesterase action. Virus Res 25:145–153. doi: 10.1016/0168-1702(92)90106-j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Higa H, Rogers G, Paulson C. 1985. Influenza virus hemagglutinins differentiate between receptor determinants bearing N-acetyl-, N-glycollyl-, and N,O-diacetylneuraminic acids. Virology 144:279–282. doi: 10.1016/0042-6822(85)90325-3. [DOI] [PubMed] [Google Scholar]
- 45.Klotz FW, Orlandi PA, Reuter G, Cohen SJ, Haynes JD, Schauer R, Howard RJ, Palese P, Miller LH. 1992. Binding of Plasmodium falciparum 175-kilodalton erythrocyte binding antigen and invasion of murine erythrocytes requires N-acetylneuraminic acid but not its O-acetylated form. Mol Biochem Parasitol 51:49–54. doi: 10.1016/0166-6851(92)90199-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Varki A. 2001. Loss of N-glycolylneuraminic acid in humans: mechanisms, consequences, and implications for hominid evolution. Am J Phys Anthropol Suppl 33:54–69. doi: 10.1002/ajpa.10018.abs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Proto WR, Siegel SV, Dankwa S, Liu W, Kemp A, Marsden S, Zenonos ZA, Unwin S, Sharp PM, Wright GJ, Hahn BH, Duraisingh MT, Rayner JC. 2019. Adaptation of Plasmodium falciparum to humans involved the loss of an ape-specific erythrocyte invasion ligand. Nat Commun 10:4512. doi: 10.1038/s41467-019-12294-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Malykh YN, Shaw L, Schauer R. 1998. The role of CMP-N-acetylneuraminic acid hydroxylase in determining the level of N-glycolylneuraminic acid in porcine tissues. Glycoconj J 15:885–893. doi: 10.1023/a:1006959016011. [DOI] [PubMed] [Google Scholar]
- 49.Broszeit F, Tzarum N, Zhu X, Nemanichvili N, Eggink D, Leenders T, Li Z, Liu L, Wolfert MA, Papanikolaou A, Martínez-Romero C, Gagarinov IA, Yu W, García-Sastre A, Wennekes T, Okamatsu M, Verheije MH, Wilson IA, Boons G-J, de Vries RP. 2019. N-Glycolylneuraminic acid as a receptor for influenza A viruses. Cell Rep 27:3284–3294.e6. doi: 10.1016/j.celrep.2019.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu G, Suzuki T, Maejima Y, Mizoguchi T, Tsuchiya M, Kiso M, Hasegawa A, Suzuki Y. 1995. Sialidase of swine influenza A viruses: variation of the recognition specificities for sialyl linkages and for the molecular species of sialic acid with the year of isolation. Glycoconj J 12:156–161. doi: 10.1007/bf00731360. [DOI] [PubMed] [Google Scholar]
- 51.Varki A, Diaz S. 1984. The release and purification of sialic acids from glycoconjugates: methods to minimize the loss and migration of O-acetyl groups. Anal Biochem 137:236–247. doi: 10.1016/0003-2697(84)90377-4. [DOI] [PubMed] [Google Scholar]
- 52.Hedlund M, Tangvoranuntakul P, Takematsu H, Long JM, Housley GD, Kozutsumi Y, Suzuki A, Wynshaw-Boris A, Ryan AF, Gallo RL, Varki N, Varki A. 2007. N-Glycolylneuraminic acid deficiency in mice: implications for human biology and evolution. Mol Cell Biol 27:4340–4346. doi: 10.1128/MCB.00379-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu Q, Padler-Karavani V, Yu H, Chen X, Wu S-L, Varki A, Hancock WS. 2012. LC-MS analysis of polyclonal human anti-Neu5Gc xeno-autoantibodies immunoglobulin G subclass and partial sequence using multistep intravenous immunoglobulin affinity purification and multienzymatic digestion. Anal Chem 84:2761–2768. doi: 10.1021/ac2030893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen Y, Tiralongo J, Kohla G, Schauer R. 2004. Regulation of sialic acid O-acetylation in human colon mucosa. Biol Chem 385:145–152. doi: 10.1515/BC.2004.033. [DOI] [PubMed] [Google Scholar]
- 55.Campbell F, Appleton MAC, Fuller CE, Greeff MP, Hallgrimsson J, Katoh R, Ng OLI, Satir A, Williams GT, Williams ED. 1994. Racial variation in the O-acetylation phenotype of human colonic mucosa. J Pathol 174:169–174. doi: 10.1002/path.1711740305. [DOI] [PubMed] [Google Scholar]
- 56.Cross BW, Ruhl S. 2018. Glycan recognition at the saliva-oral microbiome interface. Cell Immunol 333:19–33. doi: 10.1016/j.cellimm.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holmén JM, Karlsson NG, Abdullah LH, Randell SH, Sheehan JK, Hansson GC, Davis CW. 2004. Mucins and their O-glycans from human bronchial epithelial cell cultures. Am J Physiol Lung Cell Mol Physiol 287:L824–L834. doi: 10.1152/ajplung.00108.2004. [DOI] [PubMed] [Google Scholar]
- 58.Joo NS, Evans IAT, Cho H-J, Park I-H, Engelhardt JF, Wine JJ. 2015. Proteomic analysis of pure human airway gland mucus reveals a large component of protective proteins. PLoS One 10:e0116756. doi: 10.1371/journal.pone.0116756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pretini V, Koenen MH, Kaestner L, Fens M, Schiffelers RM, Bartels M, Van Wijk R. 2019. Red blood cells: chasing interactions. Front Physiol 10:945. doi: 10.3389/fphys.2019.00945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ustinov NB, Zavyalova EG, Smirnova IG, Kopylov AM. 2017. The power and limitations of influenza virus hemagglutinin assays. Biochemistry 82:1234–1248. doi: 10.1134/S0006297917110025. [DOI] [PubMed] [Google Scholar]
- 61.Corfield AP, Veh RW, Wember M, Michalski JC, Schauer R. 1981. The release of N-acetyl- and N-glycolloyl-neuraminic acid from soluble complex carbohydrates and erythrocytes by bacterial, viral and mammalian sialidases. Biochem J 197:293–299. doi: 10.1042/bj1970293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hanaoka K, Pritchett TJ, Takasaki S, Kochibe N, Sabesan S, Paulson JC, Kobata A. 1989. 4-O-Acetyl-N-acetylneuraminic acid in the N-linked carbohydrate structures of equine and guinea pig a2-macroglobulins, potent inhibitors of influenza virus infection. J Biol Chem 264:9842–9849. [PubMed] [Google Scholar]
- 63.Hunter CD, Khanna N, Richards MR, Rezaei Darestani R, Zou C, Klassen JS, Cairo CW. 2018. Human neuraminidase isoenzymes show variable activities for 9-O-acetyl-sialoside substrates. ACS Chem Biol 13:922–932. doi: 10.1021/acschembio.7b00952. [DOI] [PubMed] [Google Scholar]
- 64.Thompson CM, Petiot E, Mullick A, Aucoin MG, Henry O, Kamen AA. 2015. Critical assessment of influenza VLP production in Sf9 and HEK293 expression systems. BMC Biotechnol 15:31. doi: 10.1186/s12896-015-0152-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mitnaul LJ, Matrosovich MN, Castrucci MR, Tuzikov AB, Bovin NV, Kobasa D, Kawaoka Y. 2000. Balanced hemagglutinin and neuraminidase activities are critical for efficient replication of influenza A virus. J Virol 74:6015–6020. doi: 10.1128/jvi.74.13.6015-6020.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chokhawala HA, Yu H, Chen X. 2007. High-throughput substrate specificity studies of sialidases by using chemoenzymatically synthesized sialoside libraries. Chembiochem 8:194–201. doi: 10.1002/cbic.200600410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li W, Xiao A, Li Y, Yu H, Chen X. 2017. Chemoenzymatic synthesis of Neu5Ac9NAc-containing α2-3- and α2-6-linked sialosides and their use for sialidase substrate specificity studies. Carbohydr Res 451:51–58. doi: 10.1016/j.carres.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khedri Z, Xiao A, Yu H, Landig CS, Li W, Diaz S, Wasik BR, Parrish CR, Wang L-P, Varki A, Chen X. 2016. A chemical biology solution to problems with studying biologically important but unstable 9-O-acetyl sialic acids. ACS Chem Biol 12:214–224. doi: 10.1021/acschembio.6b00928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kumari K, Gulati S, Smith DF, Gulati U, Cummings RD, Air GM. 2007. Receptor binding specificity of recent human H3N2 influenza viruses. Virol J 4:42. doi: 10.1186/1743-422X-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu W, Air GM. 2004. Binding of influenza viruses to sialic acids: reassortant viruses with A/NWS/33 hemagglutinin bind to α2,8-linked sialic acid. Virology 325:340–350. doi: 10.1016/j.virol.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 71.Matrosovich MN, Gambaryan AS, Chumakov MP. 1992. Influenza viruses differ in recognition of 4-O-acetyl substitution of sialic acid receptor determinant. J Virol 858:854–858. doi: 10.1016/0042-6822(92)90541-V. [DOI] [PubMed] [Google Scholar]
- 72.Matrosovich M, Gao P, Kawaoka Y. 1998. Molecular mechanisms of serum resistance of human influenza H3N2 virus and their involvement in virus adaptation in a new host. J Virol 72:6373–6380. doi: 10.1128/JVI.72.8.6373-6380.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rinninger A, Richet C, Pons A, Kohla G, Schauer R, Bauer H-C, Zanetta J-P, Vlasak R. 2006. Localisation and distribution of O-acetylated N-acetylneuraminic acids, the endogenous substrates of the hemagglutinin-esterases of murine coronaviruses, in mouse tissue. Glycoconj J 23:73–84. doi: 10.1007/s10719-006-5439-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reuter G, Pfeil R, Stoll S, Schauer R, Kamerling JP, Versluis C, Vliegenthart JF. 1983. Identification of new sialic acids derived from glycoprotein of bovine submandibular gland. Eur J Biochem 134:139–143. doi: 10.1111/j.1432-1033.1983.tb07542.x. [DOI] [PubMed] [Google Scholar]
- 75.Eisen MB, Sabesan S, Skehel JJ, Wiley DC. 1997. Binding of the influenza A virus to cell-surface receptors: structures of five hemagglutinin-sialyloligosaccharide complexes determined by X-ray crystallography. Virology 232:19–31. doi: 10.1006/viro.1997.8526. [DOI] [PubMed] [Google Scholar]
- 76.Lazniewski M, Dawson WK, Szczepinska T, Plewczynski D. 2018. The structural variability of the influenza A hemagglutinin receptor-binding site. Brief Funct Genomics 17:415–427. doi: 10.1093/bfgp/elx042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pepper DS. 1968. The sialic acids of horse serum with special reference to their virus inhibitory properties. Biochim Biophys Acta 156:317–325. doi: 10.1016/0304-4165(68)90261-4. [DOI] [PubMed] [Google Scholar]
- 78.Dickson RP, Erb-Downward JR, Martinez FJ, Huffnagle GB. 2016. The microbiome and the respiratory tract. Annu Rev Physiol 78:481–504. doi: 10.1146/annurev-physiol-021115-105238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sicard J-F, Le Bihan G, Vogeleer P, Jacques M, Harel J. 2017. Interactions of intestinal bacteria with components of the intestinal mucus. Front Cell Infect Microbiol 7:387. doi: 10.3389/fcimb.2017.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McAuley JL, Gilbertson BP, Trifkovic S, Brown LE, McKimm-Breschkin JL. 2019. Influenza virus neuraminidase structure and functions. Front Microbiol 10:39. doi: 10.3389/fmicb.2019.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Laborda P, Wang S-Y, Voglmeir J. 2016. Influenza neuraminidase inhibitors: synthetic approaches, derivatives and biological activity. Molecules 21:1513. doi: 10.3390/molecules21111513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lin YP, Xionga X, Whartona SA, Martinc SR, Coombsa PJ, Vachieria SG, Christodouloub E, Walkerb PA, Liua J, Skehela JJ, Gamblinb SJ, Haya AJ, Danielsa RS, McCauleya JW. 2013. Evolution of the receptor binding properties of the influenza A (H3N2) hemagglutinin. Proc Natl Acad Sci U S A 110:2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wagner R, Matrosovich M, Klenk HD. 2002. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev Med Virol 12:159–166. doi: 10.1002/rmv.352. [DOI] [PubMed] [Google Scholar]
- 84.Guo H, Rabouw H, Slomp A, Dai M, van der Vegt F, van Lent JWM, McBride R, Paulson JC, de Groot RJ, van Kuppeveld FJM, de Vries E, de Haan C. 2018. Kinetic analysis of the influenza A virus HA/NA balance reveals contribution of NA to virus-receptor binding and NA-dependent rolling on receptor-containing surfaces. PLoS Pathog 14:e1007233. doi: 10.1371/journal.ppat.1007233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pritchett TJ, Paulson JC. 1989. Basis for the potent inhibition of influenza virus infection by equine and guinea pig alpha 2-macroglobulin. J Biol Chem 264:9850–9858. [PubMed] [Google Scholar]
- 86.Wasik BR, Voorhees IEH, Barnard KN, Alford-Lawrence BK, Weichert WS, Hood G, Nogales A, Martínez-Sobrido L, Holmes EC, Parrish CR. 2019. Influenza viruses in mice: deep sequencing analysis of serial passage and effects of sialic acid structural variation. J Virol 93:e01039-19. doi: 10.1128/JVI.01039-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hoffmann E, Krauss S, Perez D, Webby R, Webster RG. 2002. Eight-plasmid system for rapid generation of influenza virus vaccines. Vaccine 20:3165–3170. doi: 10.1016/s0264-410x(02)00268-2. [DOI] [PubMed] [Google Scholar]
- 88.Kiripolsky J, McCabe LG, Gaile DP, Kramer JM. 2017. Myd88 is required for disease development in a primary Sjögren’s syndrome mouse model. J Leukoc Biol 102:1411–1420. doi: 10.1189/jlb.3A0717-311R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cao C, Wang WJ, Huang YY, Yao HL, Conway LP, Liu L, Voglmeir J. 2017. Determination of sialic acids in liver and milk samples of wild-type and CMAH knock-out mice. J Vis Exp 14:125. doi: 10.3791/56030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chokhawala HA, Huang S, Lau K, Yu H, Cheng J, Thon V, Hurtado-Ziola N, Guerrero JA, Varki A, Chen X. 2008. Combinatorial chemoenzymatic synthesis and high-throughput screening of sialosides. ACS Chem Biol 3:567–576. doi: 10.1021/cb800127n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yu H, Yu H, Karpel R, Chen X. 2004. Chemoenzymatic synthesis of CMP-sialic acid derivatives by a one-pot two-enzyme system: comparison of substrate flexibility of three microbial CMP-sialic acid synthetases. Bioorg Med Chem 12:6427–6435. doi: 10.1016/j.bmc.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 92.Yu H, Chokhawala HA, Huang S, Chen X. 2006. One-pot three-enzyme chemoenzymatic approach to the synthesis of sialosides containing natural and non-natural functionalities. Nat Protoc 1:2485–2492. doi: 10.1038/nprot.2006.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Feng KH, Gonzalez G, Deng L, Yu H, Tse VL, Huang L, Huang K, Wasik BR, Zhou B, Wentworth DE, Holmes EC, Chen X, Varki A, Murcia PR, Parrish CR. 2015. Equine and canine influenza H3N8 viruses show minimal biological differences despite phylogenetic divergence. J Virol 89:e00521-15-6873. doi: 10.1128/JVI.00521-15. [DOI] [PMC free article] [PubMed] [Google Scholar]