Abstract
Background
Recent evidence has linked the gut microbiome to host behavior via the gut–brain axis [1–3]; however, the underlying mechanisms remain unexplored. Here, we determined the links between host genetics, the gut microbiome and memory using the genetically defined Collaborative Cross (CC) mouse cohort, complemented with microbiome and metabolomic analyses in conventional and germ-free (GF) mice.
Results
A genome-wide association analysis (GWAS) identified 715 of 76,080 single-nucleotide polymorphisms (SNPs) that were significantly associated with short-term memory using the passive avoidance model. The identified SNPs were enriched in genes known to be involved in learning and memory functions. By 16S rRNA gene sequencing of the gut microbial community in the same CC cohort, we identified specific microorganisms that were significantly correlated with longer latencies in our retention test, including a positive correlation with Lactobacillus. Inoculation of GF mice with individual species of Lactobacillus (L. reuteri F275, L. plantarum BDGP2 or L. brevis BDGP6) resulted in significantly improved memory compared to uninoculated or E. coli DH10B inoculated controls. Untargeted metabolomics analysis revealed significantly higher levels of several metabolites, including lactate, in the stools of Lactobacillus-colonized mice, when compared to GF control mice. Moreover, we demonstrate that dietary lactate treatment alone boosted memory in conventional mice. Mechanistically, we show that both inoculation with Lactobacillus or lactate treatment significantly increased the levels of the neurotransmitter, gamma-aminobutyric acid (GABA), in the hippocampus of the mice.
Conclusion
Together, this study provides new evidence for a link between Lactobacillus and memory and our results open possible new avenues for treating memory impairment disorders using specific gut microbial inoculants and/or metabolites.
Video Abstract
Keywords: Collaborative Cross mouse model, Memory, Gut–brain axis, Lactobacillus, Germ-free, Metabolites, Lactate, GABA
Background
Specific members of the gut microbiome have been linked to host health and behavior [4]. Intriguingly, probiotics comprised of different Lactobacillus and/or Bifidobacterium strains have been shown to impact behavior in mice, including reduction of symptoms linked to anxiety [5–7] and improvement of memory [8, 9]. Administration of probiotic strains specifically results in an improvement in memory of objects and object location [8–11], but not object temporal order memory [8].
Metabolic clues to memory enhancement have been found by analyzing metabolic signatures in the brains of mice following administration with specific Lactobacillus strains. Increased levels of GABA in the brain is linked to improved working memory and novel object recognition [12, 13]. Mice fed with L. rhamnosus JB-1 had increased mRNA expression of the GABA receptor [5], and increased metabolic levels of GABA in the hippocampus [14]. Increased levels of GABA in the brain could also be due to increased production of GABA by gut bacteria [15, 16]. However, the metabolic mediator(s), if any, between the gut and the brain remain unknown. Recently, O'Hagan et al. (2017) found increased levels of lactate in the brains of mice that were fed supplements containing a mixture of lactobacilli and bifidobacteria: L. acidophilus CUL60, L. acidophilus CUL21, B. bifidum CUL20 and B. lactis CUL34) [8]. Together, these studies suggest a link between specific metabolites produced by lactobacilli and memory of the host via the gut–brain axis that remains to be further explored and validated.
The complex interplay between host genetics, environment, and lifestyle factors and the gut microbiome make studying the role of the microbiome on memory potential difficult in human populations. Model systems can help overcome this barrier and offer many advantages for the study of the genetic basis of complex phenotypes. The “Collaborative Cross” (CC) is a population-based mouse model system with genetic and phenotypic diversity on par with the human population [17]. The CC, which captures nearly 90% of the known variation present in laboratory mice, was established by combining the genomes of eight diverse founder strains (A/J, C57BL/6J, 129S1/SvImJ, NOD/LtJ, NZO/HlLtJ, CAST/EiJ, PWK/PhJ, and WSB/EiJ). The advantage of the CC is that genetic and environmental components of risk can be specified and tightly controlled allowing for a comprehensive analysis of the role of host genetics and the microbiome on memory. In this study for the first time, we performed an unbiased genetic screen using CC mice to identify host genetic and microbiome components that are associated with memory potential. Subsequently, we used this information to focus on specific strains that were correlated with memory in the CC mouse cohort and to evaluate their metabolic profiles in a gnotobiotic mouse system in order to better understand the metabolic mechanisms underlying memory improvement.
Results
We assessed memory using passive avoidance, a fear-motivated test to assess memory-dependent hippocampal function [18, 19], in 535 mice from 29 Collaborative Cross (CC) strains (Table S1). The passive avoidance memory test is based on latency of entry into a compartment where three days earlier, a mild foot shock (0.3 mA for 5 s) was experienced. Mice with good memory avoided entering the chamber where they had previously been exposed to the shock, whereas mice with poor memory entered the chamber. There were significant and reproducible variations in memory potentials across the different CC strains (Fig. 1a). The latency in entry time on the testing day ranged from 87.9 to 600 s (Fig. 1a). Mice from two strains (CC036 and CC010) never entered the chamber within the 600 s assay time. We observed sex differences in memory potential in two of the strains (CC019 and CC032) where memory potential was higher in male mice, whereas no significant sex difference was observed for any of the other strains (Figure S1).
Fig. 1.
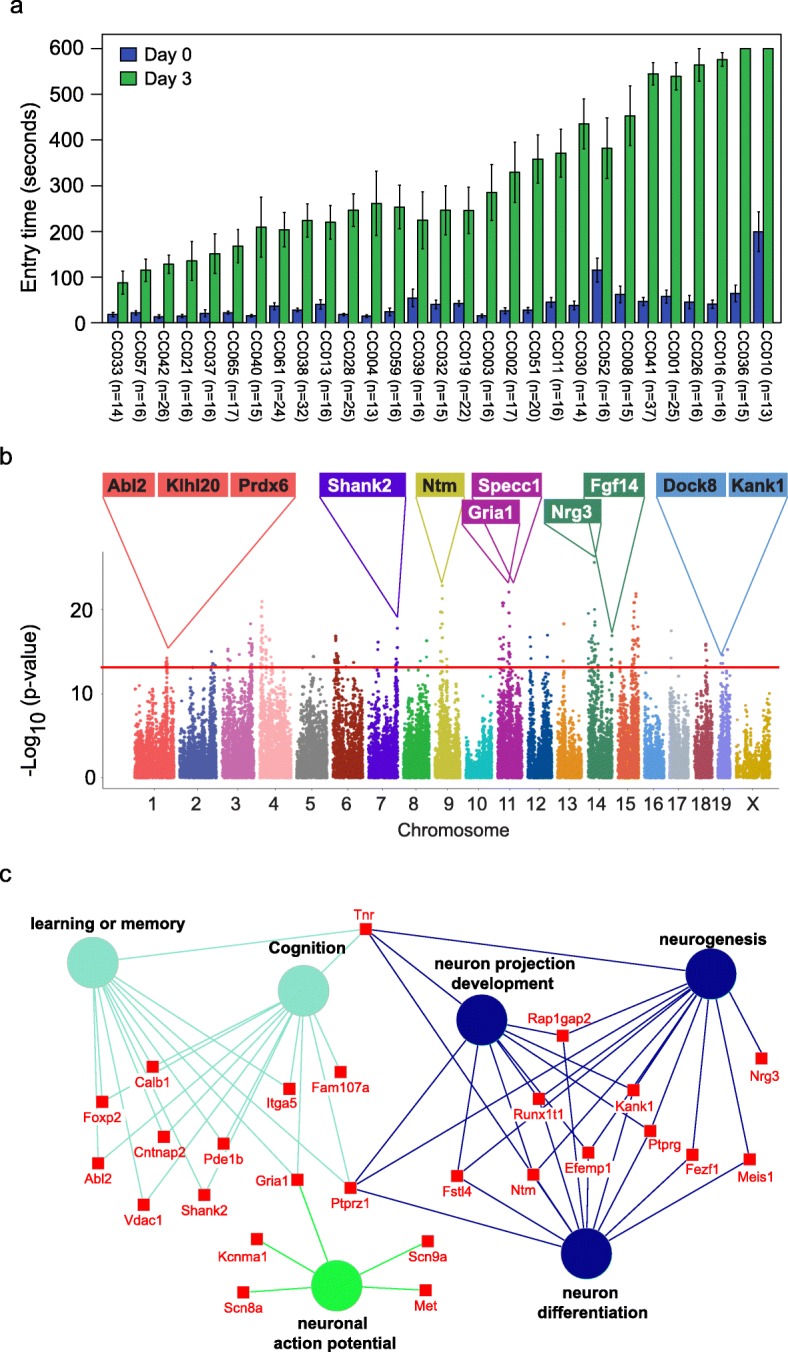
Identification of genetic variations and candidate genes associated with memory in CC mice. a Variations in memory across CC strains. Memory was assessed using passive avoidance, a fear-motivated test. The memory test is based on latency of entry into a compartment where 3 days earlier a mild foot shock (0.3 mA for 5 s) was experienced. Entry into the shock compartment on Day 0 is shown in blue, whereas entry 3 days after the foot shock is shown in green (Day 3). Mice with good memory avoided entering the chamber on Day 3, whereas mice with poor memory entered the chamber. Error bars indicate mean ± SEM. b Manhattan plot of the GWAS analysis for memory in CC mice (n = 535 mice). The – log10(P value) is shown for 76,080 SNPs ordered based on genomic position. The horizontal red line indicates the QTL significance threshold at − log10(P value) = 12. Candidate genes previously associated with memory, cognition, or other neurodevelopmental processes located in representative QTL are listed above the plot. c Gene ontology (GO) analysis of genes identified in QTL associated with memory potential in Fig. 1b (n = 535 mice). Genetic loci are significantly enriched for genes implicated in learning or memory, cognition, neuron projection development, neurogenesis, neuron differentiation, and neuronal action potential
The reproducible variation in memory potential across the CC cohort suggests that host genetics plays an important role in memory. To identify the potential genetic variations that contribute to memory, we performed GWAS with 76080 SNPs across 29 CC strains. We identified 715 SNPs significantly associated with memory (p value < 10−12), corresponding to 222 annotated genes (Fig. 1b, Table S2). Gene set enrichment analysis revealed that the 222 genes were significantly enriched in biological processes related to learning or memory (p = 1.87E–5), neuron cellular component (p = 3.97E–9), and abnormal learning/memory/conditioning phenotypes (p = 1.03E–4; Fig. 1c, Table S3) [20]. In addition to 71 genes known to be associated with memory and learning, our screen also identified 135 genes not previously associated with memory including 65 genes that show expression in the brain based on in situ hybridization data from the Mouse Brain Atlas (Allen Brain Atlas; Table S4). The spatial gene expression data suggest that these 65 genes may play a role in memory.
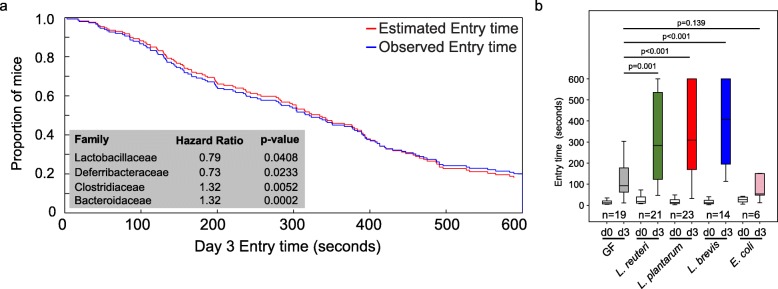
Previously, we demonstrated natural host variation in the gut microbiome composition across CC mice [21]. To determine links between specific members of the gut microbiome and memory, we correlated 16S rRNA gene sequence data to memory of individual CC strains. Sequence reads were mapped to 5761 OTUs corresponding to 72 bacterial families (Table S5). After filtering OTUs to those with > 100 reads 41 families remained. Four families (Lactobacillaceae, Deferribacteraceae, Bacteroidaceae, and Clostridiaceae) were significantly correlated with memory based on multivariate Cox regression analysis (p < 0.05; Fig. 2a). The hazard ratio (HR) indicated that higher relative abundances of Lactobacillaceae (specifically L. reuteri; HR = 0.79) and Deferribacteraceae (HR = 0.73) and lower relative abundances of Bacteroidaceae (HR = 1.32) and Clostridaceae (HR = 1.32) predicted improved memory potential (Fig. 2a). Because other Lactobacillus strains have previously been implicated in improving memory in mice, humans, and rats: e.g., L. rhamnosus JB-1 in mice [5], L. casei LC122 in aged mice [22], L. helveticus ROO52 in humans [6], and L. acidophilus strains CUL60 and CUL21 (in combination with bifidobacteria) in rats [8]. We focused our remaining studies on Lactobacillus, in particular L. reuteri that was identified in this study.
Fig. 2.
The impact of Lactobacillus or lactate treatment on memory. a Identification of microbes associated with memory in CC mice by multivariate Cox regression analysis. b Inoculation of GF mice with individual species of Lactobacillus (L. reuteri F275, L. brevis BDGP6 or L. plantarum BDGP2) resulted in significantly improved memory compared to uninoculated or E. coli inoculated controls. Error bars indicate mean ± SEM
To investigate the impact of Lactobacillus on memory, we colonized separate cohorts of germ-free (GF) mice by oral gavage with L. reuteri F275. This species has an exact 16S rRNA gene sequence match to the OTU that was significantly correlated with improved memory. In addition, we included two other Lactobacillus species for comparison (L. plantarum BDGP2 and L. brevis BDGP6). Within our three inoculated groups, we validated the presence of the individual Lactobacillus OTUs, each of which was dominant in a single treatment group and could be traced to our inoculated species (Figure S2). Memory was assessed using the same passive avoidance test in the Lactobacillus inoculated mice and compared to GF mice of the same genetic background. Eschericia coli DH10B inoculated mice were included as a negative control. Our data showed that all of the Lactobacillus mono-associated mice showed a significant improvement in memory compared to GF mice (p < 0.001; Fig. 2b). In contrast, we observed no memory improvement following application of Escherichia coli DH10B (Fig. 2b).
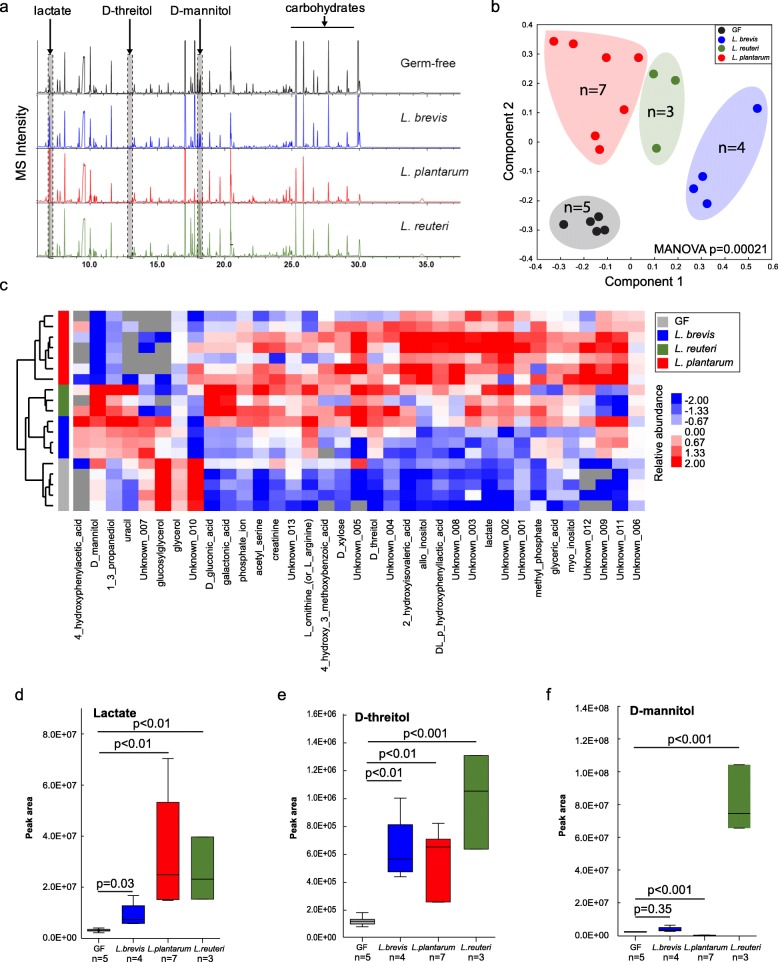
To identify metabolites produced by lactobacilli that are candidates for microbiome mediated memory enhancement, we assessed the metabolome in fecal samples collected from GF and mice mono-associated with one of each of the three Lactobacillus species: L. reuteri F275, L. plantarum BDGP2 or L. brevis BDGP6 (Fig. 3a, Figure S3, Table S6). The LLE (local-linear embedding analysis) plot shows that the metabolite composition in fecal samples from the Lactobacillus-colonized mice and GF mice were distinct (p = 0.00021; Fig. 3b,c), demonstrating that Lactobacillus inoculation significantly affected the gut metabolome. Based on gas chromatography–mass spectrometry (GC–MS) peak intensities, lactate and threitol were consistently higher in stool samples collected from mice that were colonized with any of the three strains (p < 0.01; Fig. 3d,e; Fig. S3), whereas some strain-specific differences in gut metabolites were seen, such as D-mannitol that was only higher in L. reuteri inoculated samples (p < 0.001; Fig. 3f; Fig. S3). Subsequent injection of mice with mannitol did not enhance memory (data not shown). Other examples of metabolites showing highed levels in fecal samples from Lactobacillus inoculated mice include: galactonic acid (L. reuteri); D-xylose, glyceric acid, and methyl phosphate (L. plantarum); and uracil (L. brevis; Fig. S3A). Also, many of the GC–MS peaks corresponding to “carbohydrates”, presumably from the mouse chow, were lower in intensity in stool samples colonized with the Lactobacillus species suggesting that the components were being degraded by the inoculants (Fig. 3a).
Fig. 3.
Metabolomics analysis of fecal samples from Lactobacillus-colonized and germ-free mice. a Representative GC–MS chromatograms of metabolite profiles in germ-free and Lactobacillus-colonized mouse fecal samples. Each chromatogram is a representative mass spectrometry profile from one cage of mice (4 mice/cage). b PCoA of metabolite profiles were measured in fecal samples. c Heatmap of metabolites differentiated between Lactobacillus-colonized and germ-free mice. d–f Relative abundance of select metabolites in fecal samples from individual mice. Error bars indicate mean ± SEM
To determine which metabolites could be possible mediators of the memory response, we also identified specific metabolites that were significantly higher in plasma and brain homogenates from the colonized mice compared to GF mice (Fig. S3). Fewer metabolite differences were found between species in plasma and brain, with the exception of higher plasma levels of arabitol, citric acid, glucose and L-tryptophan (L. plantarum; Fig. S3B). In the brain homogenates, species-specific differences included significantly higher levels of D-malic acid, dehydroascorbic acid, GABA, lactate, methyl phosphate, myo-inositol, and scyllo-inositol (L. plantarum); and glycine (L. reuteri; Fig. S3C). Examples of brain metabolites that were significantly higher in L. reuteri or L. plantarum inoculated mice compared to GF controls include glycerol, L-glutamic acid, L-serine, and N-acetyl-L-aspartic acid (Fig. S3C). Also, for both L. plantarum and L. brevis inoculated mice, 2,5-dihydroxypyrazine and citric acid were significantly higher in brain homogenates compared to GF controls (Fig. S3C).
Because we found an improved memory response with each of the three Lactobacillus species, we focused on fecal metabolites that were consistently higher across all three species compared to GF controls (Fig. S3), these included statistically (p < 0.05) higher levels of lactate (Fig. 3d), D-threitol (Fig. 3e), 2-hydoxyisovaleric acid, and acetyl-serine (Figure S3A). Interestingly, glycerol was significantly lower in fecal samples from mice inoculated with the three Lactobacillus species compared to GF controls (Fig. S3A). For the plasma samples, 1,5-anhydrohexitol, carbonate ion, pyruvic acid, and xylitol were significantly higher in all of the Lactobacillus inoculated mice compared to the GF controls (Fig. S3B). However, none of the identified metabolites were significantly higher in brain homogenates of Lactobacillus inoculated mice when compared to GF controls (Fig. S3C). Lactate did, however, have a trend towards higher levels in both plasma and brain, and was only significantly higher than GF controls (p < 0.05) in the brain samples from L. plantarum inoculated mice (Fig. S3C).
Based on these metabolite data and other studies of the role of lactate in memory formation [23], we hypothesized that lactate could be a mediator of the improved memory response in our trials because lactate is commonly produced by all lactobacilli. Also, lactate was recently shown to be higher in brain samples from rats that consumed a dietary supplement containing Lactobacillus and Bifidobacterium strains, and the rats had an improved memory as a result [8]. Therefore, we conducted an experiment to determine whether mice treated with dietary lactate had improved memory. CC042 mice, which have a relatively poor memory in the passive avoidance memory test (Fig. 1a), were treated with lactate through drinking water (0.5 g lactate/100 ml water) for 5 weeks. We found that dietary lactate treatment improved the average retention latency period of the CC042 mice from 92 s to 210 s (p = 0.01; Fig. 4).
Fig. 4.
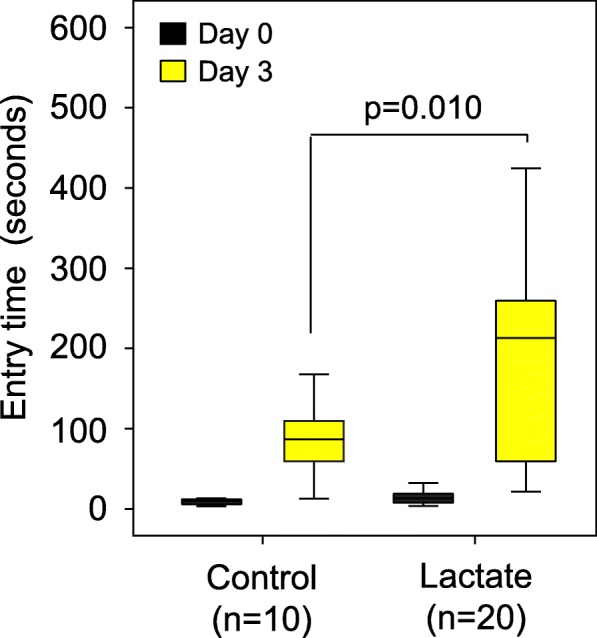
Dietary lactate treatment alone significantly boosted memory in CC042 mice. CC042 mice were treated with lactate through drinking water (n = 20; 0.5 g lactate/100 ml water) for 5 weeks or control (n = 20). Memory was assessed using passive avoidance. Dietary lactate treatment significantly improved the average retention latency period of the CC042 mice from 92 s to 210 s. Error bars indicate mean ± SEM
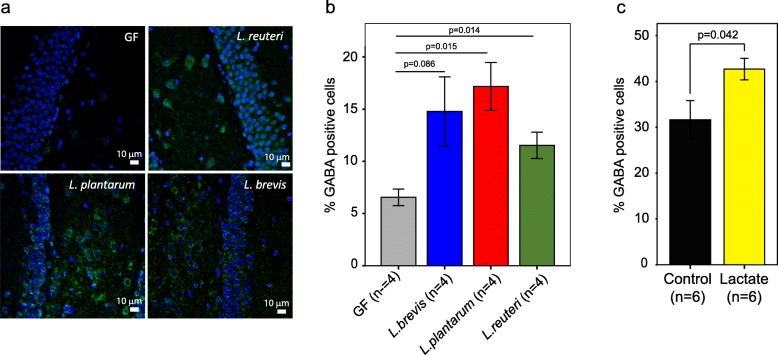
To further define the mechanism(s) underlying the improved memory response in mice following supplementation with Lactobacillus spp. or lactate, we quantified levels of GABA in the hippocampus proper, including the four Cornu Ammonis (CA) regions and the dentate gyrus (DG), using brain coronal sections taken from the treated mice. We chose to focus on the hippocampus as it is an essential area for the acquisition and formation of new memories, where GABA, the main inhibitory neurotransmitter in the brain, plays a critical role [24, 25]. As mentioned above, using metabolomic analysis we found that GABA levels were significantly higher in brain homogenates of mice inoculated with L. plantarum, but not with the other two species; although at a lower significance threshold GABA levels were also higher in the L. reuteri samples (p = 0.065; Fig. S3C). Using immunofluorescence, we quantified GABA expression in the hippocampus (Fig. S4) and GABA expression was compared between the GF controls and the three Lactobacillus-colonized mice groups (Fig. 5a, Fig. S4). All of the Lactobacillus-colonized mice showed higher fractions of cell bodies that expressed GABA in the hippocampus, compared to the germ-free control mice (Fig. 5b). However, our metabolite data showed no significant increases in GABA in stool or plasma samples of the inoculated mice compared to controls. Interestingly, we found that lactate treatment alone also increased the fractions of cell bodies that expressed GABA compared with control mice from 32% to 43% (p = 0.042; Fig. 5c), supporting the hypothesis that lactate could serve as the metabolic conduit between the gut and the brain.
Fig. 5.
Influence of Lactobacillus inoculation or lactate treatment on the levels of the neurotransmitter, gamma-aminobutyric acid (GABA), in the hippocampus of the mice. a Representative images of immunostaining for GABA, taken from the cell body layers in the dentate gyrus of a germ-free mouse and mice treated with Lactobacillus reuteri, brevis, or plantarum. Nuclei are shown in blue, GABA is shown in green. Using images covering the CA fields and the dentage gyrus, the percent GABA-positive cells was calculated by the fraction of cell bodies showing GABA (green color) from the total number of cells (identified by the blue nuclei). b and cLactobacillus inoculation (n = 4, 2 males and 2 females for each treatment) (b) or lactate treatment (n = 6, 3 males and 3 females for each treatment) (c) significantly increased the levels of GABA in the hippocampus of mice. Error bars indicate mean ± SEM
A possible mechanism underlying the observed increase in GABA expression in the hippocampus could be an increase in the expression level of glutamate decarboxylase (GAD), which converts glutamate to GABA, in this brain area. To test this possibility, we quantified GAD67 gene expression levels in the dentate gyrus using single-molecule-based fluorescence in situ hybridization approach (fliFISH) [26]. However, we found no significant difference between GAD67 gene expression levels in germ-free mice and mice colonized with Lactobacillus strains (Fig. S5). Sequence analysis of the Lactobacillus isolates used in our study showed that L. brevis has two genes encoding glutamate decarboxylase and L. plantarum and L. reuteri each have one, suggesting that Lactobacillus may be the source of the increased GABA levels observed in the hippocampus. Another possible mechanism for the observed increase in GABA in the hippocampus of lactobacilli-inoculated mice is the non-oxidative metabolism and conversion of lactate to GABA via α-ketoglutarate transamination [27]. These hypotheses remain to be validated in future studies.
To further assess the translatable nature of our findings to the human host, co-culture experiments were performed with L. reuteri and human epithelial cells (Caco-2) using the HuMiX model (Fig. S6A). HuMiX is a microfluidics-based human-microbial co-culture system which was designed to simulate the environment of the human gastrointestinal tract where microbial and mammalian epithelial cells interact via soluble molecular factors [28]. The bacterial cells were inoculated after the epithelial cells had fully differentiated into polarized monolayers (after 7 days). Metabolites were measured in collected fluid samples from the bacterial and epithelial cell perfusion chambers for two time points (6 h and 24 h after incubation in addition to just before inoculation as baseline). Similar to the in vivo studies, we observed that lactate and mannitol increased over the incubation time (Figs. S6B-C and Table S7). We focused on lactate and found that it was present at significantly higher levels in both bacterial and perfusion chambers inoculated with L. reuteri compared to uninoculated controls (Fig. S6B). These results demonstrate that lactate produced by L. reuteri could diffuse to the opposite side of the chamber, indicating that it likely diffuses through the gut epithelial cell lining in humans and is transported to the brain via the bloodstream.
Discussion
We conducted systematic genetic and microbial community profiling analyses to determine potential links between host genetics and the gut microbiome in memory. We demonstrated that members of the gut microbiome, specifically different Lactobacillus species, play a role in improving memory in mice. Also, we identified two sets of host genes, one previously associated with memory, cognition or other neurodevelopmental processes and the other set identified here as candidates for roles in cognition (Table S4). Examples of known human genes associated with memory also identified in our study include: NTM and KLHl20 that are associated with Alzheimer’s disease and DOCK8 and KANK1 that are associated with neurodevelopmental disorders including memory potential [29–31]. Examples of known mouse genes associated with memory include Specc1 that was identified in a mouse genetic screen for avoidance learning [32], Prdx6 that was associated with neurogenesis and Alzheimer’s disease in mice [33, 34] and knock-out mice for Abl2, Nrg3, Shank2, Gria1, and Fgf14 that caused neurodevelopmental defects [35–39]. The other set contains 135 genes not previously associated with memory of which 65 have brain expression [40] (http://connectivity.brain-map.org/). Additional experiments are required to assess their function in memory and to determine if and how they are linked to specific members of the gut microbiome.
We also provide metabolic evidence of links between the host, the gut microbiome and memory responses. Our untargeted approach in a complex microbial community-based system was able to identify significant correlations between memory and Lactobacillus within a background of the entire gut microbiome. Because we specifically identified L. reuteri in screening of the CC mouse cohort, we focused on that species, and included two other Lactobacillus species for comparison (L. plantarum and L. brevis). Subsequently, we confirmed that mono-association of gnotobiotic mice with any of the three Lactobacillus species was able to improve memory. This impact was not seen using E. coli as a negative control. This led us to hypothesize that lactobacilli produce specific metabolites that could cross the blood–brain barrier and influence memory of the host.
Because of recent reports which link specific Lactobacillus species and memory in mice (e.g., L. rhamnosus JB-1 [5], L. helveticus R0052 [6], L. helveticus NS8 [10]) and rats (e.g., L. acidophilus CUL60 and L. acidophilus CUL21 [8]), we focused on understanding the metabolic mechanism(s) underlying the memory improvement. Because we were interested in potential flow of metabolites from the gut, through plasma, to the brain, we examined samples from all three body locations for their metabolic compositions. Several metabolites were significantly higher in fecal samples of Lactobacillus-colonized mice, and less so in plasma and brain samples. In particular, in stool samples, there were specific metabolites that were significantly higher than GF controls for mice mono-associated with individual Lactobacillus strains, such as mannitol that was only observed for L. reuteri F275. In this case, mannitol injection did not improve memory over that of control mice. On the other hand, lactate was significantly higher in stool samples of mice inoculated with any one of the three lactobacilli and in brain samples with L. plantarum BDGP2 and trended towards higher levels in brain and plasma samples of mice inoculated with any one of the three species. Because lactate is produced by all lactobacilli and because it has previously been recognized as crucial for learning and long-term memory [41] we focused on lactate in subsequent targeted feeding experiments. Our results indicated that indeed lactate supplementation resulted in improvement in memory of CC042 mice that otherwise have a poor memory.
There are several clues in the literature towards the possible mechanism(s) underlying memory improvement with lactate. Suzuki et al. (2011) demonstrated that lactate derived from astrocytes via glycogenolysis was critical for long-term memory formation in rats [42]. Memory is an energy requiring process and the brain needs fuel to make and store memories. It is known that lactate is produced in astrocytes in the brain via glycolysis to provide energy to neurons [41]. Lactate administration by injection has previously been demonstrated to restore memory in chicks [43] and rats [42]. Lactate administration has also been found to enhance neuronal activity in vivo [44]. A recent study has shown that during exercise, lactate is released by the muscles, and crosses the blood–brain barrier to induce Bdnf expression and TRKB signaling in the hippocampus [45]. This enforces the hypothesis that a direct link from lactate to neurons exists that bypasses the need for glycogen energy storage in astrocytes [41]. While lactate has been implicated in several aspects of brain signaling, one of its roles might be metabolic [44]. Lactate has also been shown to signal changes in the NADH/NAD ratio [46] and lactate application increases intracellular levels of NADH [47]. Recent reviews have emphasized that the understanding of the role of lactate in memory is still in its infancy and there is the possibility that it plays several roles in this process [41, 48]. Also, we should note that other metabolites that we identified that were significantly higher in mice that were mono-associated with the different Lactobacillus species are potential targets for future study.
We also found an increase in GABA in the hippocampus of gnotobiotic mice that were mono-associated with either L. reuteri F275 L. plantarum BDGP2 or L. brevis BDGP6. These findings support previous findings of increased expression of GABA in the hippocampus of mice following treatment with Lactobacillus rhamnosus JB-1 [5, 12]. However, in older rats that were supplemented with a combined inoculum containing two strains of Lactobacillus acidophilus and two strains of Bifidobacterium, GABA increased in the frontal cortex, but not the hippocampus [8]. Recently, several gut microbes have been found to modulate GABA production pathways in the gut, suggesting a potential route for production of GABA that could serve to increase GABA levels in the brain [15]. Intriguingly, our finding that lactate treatment alone increased the fractions of cell bodies that expressed GABA compared with control mice (Fig. 4) supports the hypothesis that lactate could serve as a metabolic conduit for increased GABA production in the brain.
Conclusion
Our study provides new evidence that links the complex genetic–microbiome-metabolome interplay that can contribute to memory. One potential outcome of this research is the support of use of probiotic Lactobacillus strains to promote memory through their production of lactate and through their promotion of GABA accumulation in the hippocampus, although it remains to translate these findings from mice to humans. In addition, our findings suggest that the metabolic mechanisms underlying the improved memory response by Lactobacillus in the diet could be at least partly due to the production of lactate in the colon that migrates through the blood to the brain.
Methods
Mice
All CC strains were purchased from the Systems Genetics Core Facility at the University of North Carolina (UNC). Passive avoidance memory test was assessed at 10–11 weeks of age. The study was carried out in strict accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The Animal Welfare and Research Committee at Lawrence Berkeley National Laboratory approved the animal use protocol. Mice were maintained on PicoLab Rodent Diet 20 (5053), housed in standard micro-isolator cages on corn cobb bedding with enrichment consisting of crinkle cut, naturalistic paper strands. To test the effect of dietary lactate on memory, drinking water of CC042 mice was supplemented with sodium L-lactate (SIGMA; 71718) for 5 weeks starting at 4 weeks of age.
Germ-free mice
Germ-free C57BL/6NTac mice were purchased from Taconic and were maintained within germ-free isolators. The status of our germ-free mice colony was tested every other week and after each opening of the transfer port. A sample was removed from the isolator consisting of fecal samples from multiple cages of animals housed in the isolator, water from the drinking bottles of all cages in the isolator and swabs of the inside of the cages, the floor, and entry port of the isolator. A portion of each sample was streaked onto a sheep blood agar plate and the swab was cultured in thioglycollate medium at 37 °C for 3 days, after which they were maintained at room temperature for 11 more days and observed for growth at 24-h intervals. In addition, every other month, samples were collected and tested for aerobic, anaerobic, and fungal growth at an independent commercial laboratory (IDEXX). All results for our animals were negative for bacterial and/or fungal growth.
Mono-association of GF mice
At 3 weeks of age, GF mice were inoculated with either Lactobacillus reuteri F275, L. plantarum BDGP2 or L. brevis BDGP6 in the different isolators, which was confirmed by PCR and sequencing of their respective 16S rRNA genes. Lactobacillus reuteri F275 was purchased from ATCC (23272). L. plantarum BDGP2 [61] and L. brevis BDGP6 (unpublished; accession number CP024635) were isolated from Drosophila gut samples and verified by genome sequencing. E.coli strain DH10B was purchased from Invitrogen. GF mice were inoculated with 100 μl overnight cultures of Lactobacillus or E.coli.
Passive avoidance memory test
Short-term memory of mice was assessed by passive avoidance using the Panlab passive avoidance box (Panlab: LE870/872). During the acquisition phase, mice were placed in the light compartment. When the mice innately crossed to the dark compartment, they received a mild foot shock (5 s; 0.3 mA). Duration of mice in the light compartment before entering to the dark compartment was recorded. Three days after the acquisition phase mice were again placed in the light compartment and the passive avoidance response was evaluated by measuring the latency to enter the dark compartment.
QTL analysis of memory
Latency of entry into the dark compartment 3 days after the acquisition phase for all CC mice was used for genetic mapping. Genotype data for 134,593 SNPs was obtained from the UNC Systems Genetics Core website (http://csbio.unc.edu/CCstatus/index.py), and filtered for minor allele frequency > 5 out of the 29 CC strains, leaving 76,080 SNPs. At each SNP, latency to enter on day 3 for all CC mice were assigned to their respective alleles. We then used Mann–Whitney U to test the significance of associations between memory and allele classes at each SNP.
Microbiome analyses
Genomic DNA was extracted from the homogenized fecal samples using the PowerSoil DNA Isolation Kit (http://www.mobio.com/) according to the manufacturer’s instructions. PCR amplification of the V4 region of the 16S rRNA gene was performed using the protocol developed by the Earth Microbiome Project (http://press.igsb.anl.gov/earthmicrobiome/empstandard-protocols/16s/) and modern primers [49]. Amplicons were sequenced on an Illumina MiSeq using paired, 250 base-pair reads, according to the manufacturer’s instructions and are available on OSF (https://osf.io/jbt5g/). The Hundo amplicon processing protocol was used to process 16S and ITS amplicons [50]. In brief, sequences were trimmed and filtered of adapters and contaminants using BBDuk2 of the BBTools package. VSEARCH [51] was used to merge, filter to an expected error rate of 1, dereplicate, and remove singletons before preclustering reads for de novo and reference-based chimera checking. Reads were clustered into OTUs at 97% similarity and an OTU table in the BIOM format [52] was constructed by mapping filtered reads back to these clusters. BLAST+ [53] is used to align OTU sequences to the database curated by CREST [54] (SILVA v128 for 16S) and taxonomy was assigned based on the CREST LCA method. Graphing was performed in R, making use of the Phyloseq package [55].
L. plantarum BDGP2 [56] and L. brevis BDGP6 (unpublished) were sequenced using the PacBio long read strategy. After assembly the genomes were annotated for predicted protein-coding open reading frames using Rapid Annotation of microbial genomes using Subsystems Technology tool [57] and the GenBank annotation pipeline. The RAST and GenBank produced gene models for L. plantarum, L. brevis, and L. reuteri predict: six L-lactate dehydrogenase (EC 1.1.1.27) encoding genes, one D-lactate dehydrogenase (EC 1.1.1.28) encoding gene and one Glutamate decarboxylase (EC 4.1.1.15) encoding gene; two L-lactate dehydrogenase encoding genes, two D-lactate dehydrogenase encoding genes, and two Glutamate decarboxylase encoding genes; and five L-lactate dehydrogenase encoding genes, one D-lactate dehydrogenase encoding gene, and one Glutamate decarboxylase encoding gene, respectively.
Metabolome analyses
Metabolites were extracted from mouse fecal, plasma, and whole brain homogenate samples. Fecal samples were extracted with methanol as reported previously [21], and plasma and whole brain homogenates were extracted using the MLPEx method [58]. Briefly, 50 μL of plasma was extracted with 200 μL of chloroform/methanol (2:1, v/v), and extracted molecules in both aqueous and organic layers were combined and dried in vacuo. Whole brains were weighed and extracted using MPLEx, but the volume of solvent was added proportionally to the amount of tissue. All the extracts were stored at − 80 °C, and they were analyzed by gas chromatography coupled to mass spectrometry as reported previously [21]. All the raw MS data files are available at the OSF public data depository (https://osf.io/jbt5g/).
Immunostaining of GABA in mouse hippocampus
Mice brains were fixed in 4% paraformaldehyde and embedded in paraffin. Two male and two female mice were analyzed for each treatment (n = 4). Coronal sections were generated and placed on class slides. The sections were rehydrated by first immersing the slides in xylene (mixed isomers) for 20 min. The slides where then incubated in 100% ethanol for 20 min, followed by incubations in ethanol at decreasing concentrations (100%, 95%, 70%, and 50%), for 5 min each. The slides where then rinsed using deionized water. Heat-induced epitope retrieval was done by 5 min incubation of the slides in 10 mM Tris, 1 mM EDTA heated to 95 °C, followed by rinsing in water. Immunostaining of the hippocampus proper, including the Cornu Ammonis (CA) fields and the dentate gyrus (DG), was done using anti-GABA antibody produced in rabbit (Sigma A2052) and applied to the sections at 1:200 dilution, followed by a fluorescent (Alexa488) Goat anti-rabbit IgG H+L (ThermoFisher A11008) applied at 1:400 dilution. Antibody dilutions were made in PBS containing 0.25% Triton X-100 and 5% goat serum. The sections were then stained with DAPI (1 μg/ml in PBS for 10 min) and washed with PBS. Vectashield drops were placed on the sections and glass coverslips were placed on top of them and sealed using nail polish. Fluorescence imaging was done using inverted confocal fluorescence microscope (Zeiss). Initially, 10 × objective and tiling over the whole section was used, followed by imaging the hippocampus, including the CA fields and DG, using 40 × water immersion objective to achieve the magnification needed for identifying and counting cell bodies (Fig. S4). GABA expression was quantified by calculating the number of cell bodies showing GABA expression (green) as the percent of all cell bodies, detected by the DAPI-stained nuclei (blue). Significant differences between germ-free and Lactobacillus-treated mice were determined using t tests.
Glutamate decarboxylase (GAD67) gene expression using fliFISH in mouse brain sections
Fluctuation localization imaging-based FISH (fliFISH) was developed and performed as described in Cui et al. [26]. In short, fliFISH utilizes photoswitchable dyes and super-resolution localization microscopy to accurately count and localize mRNA molecules with a small number of oligonucleotide probes. The single-molecule on-time fraction (Fsingle) for Alexa647 was found to be 0.2% under 0.5 kW/cm2 excitation. When using 20 probes (n), each tagged with one dye molecule, to target a transcript, the ensemble on-time fraction (Fensemble= 1−(1−Fsingle)n) of 4% should be detected from a successfully hybridized RNA transcript position. In contrast, stray or nonspecifically bound probes would generate roughly a single-molecule on-time fraction values, while strong autofluorescence and aggregated probes would generate higher ensemble on-time fraction values. In addition, fliFISH enables to resolve multiple transcripts in a diffraction-limited area as the centroid of each blinking event is registered with 15–25 nm resolution in the super-resolution reconstructed image.
The 24 primary FISH probes used in this study were designed to have two segments: a GAD67 transcript targeting domain, and a terminal overhang. The targeting domain was generally 20 nucleotide-long, with 45–55% CG content, no self-repeats and inner loop-stem structures. The secondary probe was labeled with two Alexa647 dye molecule, one in each end, and was designed to hybridize with the overhang sequence. All the probe sequences were subjected to BLAST searching to avoid nonspecific targeting and purchased from Integrated DNA Technologies.
The hybridization procedure followed previously established protocols [59, 60]. Primary probes were mixed and hybridized with the secondary probe to form fluorescent complexes before introducing to the tissue sections following a published protocol [60]. One microliter from each of the 24 oligonucleotide probes 100 μM solutions was mixed together and water was added to a total of 120 μl. Six microliters were then taken out and mixed with 1.5 μl 100 μM secondary probe, 2 μl 10 × NEB3 buffer (containing 1 M NaCl, 0.5 M Tris-HCl, 0.1 M MgCl2), and 10.5 μl water. The mixture was heated to 85 °C and gradually cooled down to room temperature. The microscope slides with paraffin-embedded mouse brain sections were dipped in 100% xylenes for 10 min, followed by100%, 95%, and 70% ethanol for 10 min each. The slides were then left in 70% ethanol overnight at 4 °C and washed with PBS before hybridization. Hybridization was done by first washing the slides with “wash buffer” (containing 2 × SSC and 10% formamide). 200 μl “hybridization buffer” (containing 10% dextran sulfate, 2 × SSC and 10% formamide) were mixed with 3.3 μl of the FISH probe solution. Drops of this mixture were placed on the tissue sections and the slides were kept overnight at 46 °C. The slides were then washed twice by incubating in “wash buffer” for 20 min at 46 °C, followed by a wash with 2 × SSC buffer. The slides were then incubated in 1 μg/ml DAPI solution for 10 min and were washed using PBS.
The fliFISH images were taken using a home-built, Zeiss Axioobserver based single-molecule imaging system. A 100 × oil immersion objective lens (NA 1.4, Plan Apo) and an EMCCD camera (Andor iXon Ultra 897) were used. DIC and DAPI fluorescence images were taken in addition to fluorescent Alexa 647 single molecules images. Over 10,000 image frames were taken (at 25 Hz frame rate) for post-processing. Gaussian musk fitting algorithm was applied to find the central location of each emission event and nearby events were grouped together (assigned to the same transcript) using the DBSCAN algorithm. For the grouped emission events, the center of mass was determined to represent the possible existence of a transcript. All image processing was performed with MATLAB and C scripts that are available upon request.
HuMiX-based analyses
HuMiX-based co-cultures involving L. reuteri and the human epithelial cell line Caco-2 were performed as previously described [28]. Caco-2 were allowed to fully differentiate for 7 days at which point the co-cultures with L. reuteri were established. Eluates were collected from the microbial and perfusion chambers just before inoculation as well as after 6 and 24 h of co-culture and immediately flash-frozen for subsequent metabolomic analyses.
Statistics
To evaluate the association between the memory latency to enter time and the microbiome features, we employed Cox Proportional-Hazards Regression analysis, where both forward and backward feature selection strategies were used to optimize the subset of microbes that significantly impact memory. During forward selection, each individual microbe at the OTU level was evaluated through the Cox Proportional-Hazards Regression analysis, and only those with significant impact on memory (round (P value, 2) ≤ 0.05, where round (X,N) rounds X to its nearest N decimal digits) were selected (6 OTUs: f__Anaeroplasmataceae, f__Bacteroidaceae, f__Bacteroidales_S24-7_group, f__Lactobacillaceae, f__Deferribacteraceae, f__Clostridiaceae_1). During backward selection, the combined subset of features (multivariates) were further evaluated through the Cox Proportional-Hazards Regression analysis, where only the features with significant impact (round (P value, 2) ≤_0.05) were retained. Different from forward selection, the backward selection was performed in an iterative manner until all the features in the refined subset were significantly associated with memory. In our study, the backward selection with 2 iterations led to the refined final subset of four microbiome features (i.e., f__Bacteroidaceae, f__Lactobacillaceae, f__Deferribacteraceae, and f__Clostridiaceae_1), where f__Bacteroidaceae and f__Clostridiaceae_1 are “prognostic” unfavorable (hazard ratio >_1), and f__Lactobacillaceae and f__Deferribacteraceae are “prognostic” favorable (hazard ratio <_1).
Differences in memory potential between germ-free and Lactobacillus inoculated mice or between lactate treated and control mice was assessed by non-parametric test (Mann–Whitney test). Difference in metabolite abundance and GABA expression between germ-free and Lactobacillus inoculated mice was assessed by Student’s t test. Significance was determined at P_<_0.05. Multivariate analysis of variance (MANOVA) was used to examine for statistical differences in fecal metabolite profiles across germ-free and Lactobacillus-inoculated mice.
Supplementary information
Acknowledgements
The authors would like to thank Gabriela Fuentes-Creollo and Frank Ponce for maintenance and breeding of germ-free and gnotobiotic mice, Kenneth Wan and Benjamin Booth for assistance in 16S rRNA gene sequencing, and Emma L. Schymanski for technical assistance with HuMiX experiments
Abbreviations
- CC
Collaborative Cross
- GF
Germ-free
- GWAS
Genome-wide Association Analysis
- SNP
Single-nucleotide polymorphisms
- GABA
Gamma-aminobutyric acid
- GAD
Glutamate decarboxylase
- fliFISH
Single molecule-based fluorescence in situ hybridization
- MANOVA
Multivariate analysis of variance
Authors’ contributions
AMS, J-H M, and JKJ conceived of and designed the study and co-wrote the manuscript. AMS, J-HM, Y-XZ, YW, performed the mouse experiments and acquired the mouse data. AMS, J-HM, CZ, and SAL performed mouse data analysis and interpreted results. TOM and YMK performed metabolome data analysis, interpreted results and co-wrote the manuscript. CJB and HC performed microbiome data analysis and interpreted results. Y-XZ prepared microbiome samples and carried out microbiome sequencing. SEC and DWT interpreted results and co-wrote the manuscript. GO and DH performed immunostaining and fliFISH, interpreted results and co-wrote the manuscript. PW and BYLP performed HuMiX experiments and interpreted the data. All authors read and approved the final manuscript.
Funding
This work was primarily supported by the Office of Naval Research under ONR contract N0001415IP00021 (J.J.). Additional support was provided by the Lawrence Berkeley National Laboratory Directed Research and Development (LDRD) program funding (J.H.M. and A.M.S.). Partial support was also provided under the Microbiomes in Transition (MinT) Initiative as part of the Laboratory Directed Research and Development Program at PNNL. Metabolomic measurements, fluorescence microscopy, and mRNA FISH analyses were performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the US DOE OBER and located at PNNL in Richland, Washington. PNNL and LBNL are multi-program national laboratories operated by Battelle for the DOE under contract DE-AC05-76RLO 1830 and the University of California for the DOE under contract DE AC02-05CH11231, respectively.
Availability of data and materials
Mouse gut microbiome 16S rRNA gene sequence data is available on OSF (https://osf.io/jbt5g/).
All the raw metabolomic data are available at the OSF public data depository (https://osf.io/jbt5g/).
Ethics approval and consent to participate
The study was carried out in strict accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The Animal Welfare and Research Committee at Lawrence Berkeley National Laboratory approved the animal use protocol.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
The original version of this article was revised: Following publication of the original article, the authors reported that Sarah Fansler had been left off the author list in error. The author group has been updated above and the original article has been corrected.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jian-Hua Mao, Young-Mo Kim and Yan-Xia Zhou contributed equally to this work.
Change history
5/28/2020
An amendment to this paper has been published and can be accessed via the original article.
Contributor Information
Sarah Fansler, Email: Sarah.fansler@pnnl.gov.
Janet K. Jansson, Email: Janet.Jansson@pnnl.gov
Antoine M. Snijders, Email: AMSnijders@lbl.gov
Supplementary information
Supplementary information accompanies this paper at 10.1186/s40168-020-00817-w.
References
- 1.Burokas A, Moloney RD, Dinan TG, Cryan JF. Microbiota regulation of the mammalian gut-brain axis. Adv Appl Microbiol. 2015;91:1–62. doi: 10.1016/bs.aambs.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Carabotti M, Scirocco A, Maselli MA, Severi C. The gut-brain axis: interactions between enteric microbiota, central and enteric nervous systems. Ann Gastroenterol. 2015;28(2):203–209. [PMC free article] [PubMed] [Google Scholar]
- 3.Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13(10):701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. Current understanding of the human microbiome. Nat Med. 2018;24(4):392–400. doi: 10.1038/nm.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, Bienenstock J, Cryan JF. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci U S A. 2011;108(38):16050–16055. doi: 10.1073/pnas.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Messaoudi M, Violle N, Bisson JF, Desor D, Javelot H, Rougeot C. Beneficial psychological effects of a probiotic formulation (Lactobacillus helveticus R0052 and Bifidobacterium longum R0175) in healthy human volunteers. Gut Microbes. 2011;2(4):256–261. doi: 10.4161/gmic.2.4.16108. [DOI] [PubMed] [Google Scholar]
- 7.Mohammadi AA, Jazayeri S, Khosravi-Darani K, Solati Z, Mohammadpour N, Asemi Z, Adab Z, Djalali M, Tehrani-Doost M, Hosseini M, et al. The effects of probiotics on mental health and hypothalamic-pituitary-adrenal axis: a randomized, double-blind, placebo-controlled trial in petrochemical workers. Nutr Neurosci. 2016;19(9):387–395. doi: 10.1179/1476830515Y.0000000023. [DOI] [PubMed] [Google Scholar]
- 8.O'Hagan C, Li JV, Marchesi JR, Plummer S, Garaiova I, Good MA. Long-term multi-species Lactobacillus and Bifidobacterium dietary supplement enhances memory and changes regional brain metabolites in middle-aged rats. Neurobiol Learn Mem. 2017;144:36–47. doi: 10.1016/j.nlm.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 9.Smith CJ, Emge JR, Berzins K, Lung L, Khamishon R, Shah P, Rodrigues DM, Sousa AJ, Reardon C, Sherman PM, et al. Probiotics normalize the gut-brain-microbiota axis in immunodeficient mice. Am J Physiol Gastrointest Liver Physiol. 2014;307(8):G793–G802. doi: 10.1152/ajpgi.00238.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang S, Wang T, Hu X, Luo J, Li W, Wu X, Duan Y, Jin F. Administration of Lactobacillus helveticus NS8 improves behavioral, cognitive, and biochemical aberrations caused by chronic restraint stress. Neuroscience. 2015;310:561–577. doi: 10.1016/j.neuroscience.2015.09.033. [DOI] [PubMed] [Google Scholar]
- 11.Warburton EC, Brown MW. Neural circuitry for rat recognition memory. Behav Brain Res. 2015;285:131–139. doi: 10.1016/j.bbr.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thanapreedawat P, Kobayashi H, Inui N, Sakamoto K, Kim M, Yoto A, Yokogoshi H. GABA affects novel object recognition memory and working memory in rats. J Nutr Sci Vitaminol (Tokyo) 2013;59(2):152–157. doi: 10.3177/jnsv.59.152. [DOI] [PubMed] [Google Scholar]
- 13.Yoon JH, Grandelis A, Maddock RJ. Dorsolateral prefrontal cortex GABA concentration in humans predicts working memory load processing capacity. J Neurosci. 2016;36(46):11788–11794. doi: 10.1523/JNEUROSCI.1970-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janik R, Thomason LAM, Stanisz AM, Forsythe P, Bienenstock J, Stanisz GJ. Magnetic resonance spectroscopy reveals oral Lactobacillus promotion of increases in brain GABA, N-acetyl aspartate and glutamate. Neuroimage. 2016;125:988–995. doi: 10.1016/j.neuroimage.2015.11.018. [DOI] [PubMed] [Google Scholar]
- 15.Strandwitz Philip. Neurotransmitter modulation by the gut microbiota. Brain Research. 2018;1693:128–133. doi: 10.1016/j.brainres.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strandwitz P, Kim KH, Terekhova D, Liu JK, Sharma A, Levering J, McDonald D, Dietrich D, Ramadhar TR, Lekbua A, et al. GABA-modulating bacteria of the human gut microbiota. Nat Microbiol. 2019;4(3):396–403. doi: 10.1038/s41564-018-0307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, Beavis WD, Belknap JK, Bennett B, Berrettini W, et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet. 2004;36(11):1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- 18.Impey S, Smith DM, Obrietan K, Donahue R, Wade C, Storm DR. Stimulation of cAMP response element (CRE)-mediated transcription during contextual learning. Nat Neurosci. 1998;1(7):595–601. doi: 10.1038/2830. [DOI] [PubMed] [Google Scholar]
- 19.Stubley-Weatherly L, Harding JW, Wright JW. Effects of discrete kainic acid-induced hippocampal lesions on spatial and contextual learning and memory in rats. Brain Res. 1996;716(1-2):29–38. doi: 10.1016/0006-8993(95)01589-2. [DOI] [PubMed] [Google Scholar]
- 20.Wang Jing, Duncan Dexter, Shi Zhiao, Zhang Bing. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Research. 2013;41(W1):W77–W83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snijders AM, Langley SA, Kim YM, Brislawn CJ, Noecker C, Zink EM, Fansler SJ, Casey CP, Miller DR, Huang Y, et al. Influence of early life exposure, host genetics and diet on the mouse gut microbiome and metabolome. Nat Microbiol. 2016;2:16221. doi: 10.1038/nmicrobiol.2016.221. [DOI] [PubMed] [Google Scholar]
- 22.Ni Y, Yang X, Zheng L, Wang Z, Wu L, Jiang J, Yang T, Ma L, Fu Z. Lactobacillus and Bifidobacterium improves physiological function and cognitive ability in aged mice by the regulation of gut microbiota. Mol Nutr Food Res. 2019;63(22):e1900603. doi: 10.1002/mnfr.201900603. [DOI] [PubMed] [Google Scholar]
- 23.Scavuzzo CJ, Rakotovao I, Dickson CT. Differential effects of L- and D-lactate on memory encoding and consolidation: potential role of HCAR1 signaling. Neurobiol Learn Mem. 2019;168:107151. doi: 10.1016/j.nlm.2019.107151. [DOI] [PubMed] [Google Scholar]
- 24.Sibbe M, Kulik A. GABAergic regulation of adult hippocampal neurogenesis. Mol Neurobiol. 2017;54(7):5497–5510. doi: 10.1007/s12035-016-0072-3. [DOI] [PubMed] [Google Scholar]
- 25.Wideman CE, Jardine KH, Winters BD. Involvement of classical neurotransmitter systems in memory reconsolidation: focus on destabilization. Neurobiol Learn Mem. 2018;156:68–79. doi: 10.1016/j.nlm.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 26.Cui Y, Hu D, Markillie LM, Chrisler WB, Gaffrey MJ, Ansong C, Sussel L, Orr G. Fluctuation localization imaging-based fluorescence in situ hybridization (fliFISH) for accurate detection and counting of RNA copies in single cells. Nucleic Acids Res. 2018;46(2):e7. doi: 10.1093/nar/gkx874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maddock RJ, Casazza GA, Fernandez DH, Maddock MI. Acute modulation of cortical glutamate and GABA content by physical activity. J Neurosci. 2016;36(8):2449–2457. doi: 10.1523/JNEUROSCI.3455-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shah P, Fritz JV, Glaab E, Desai MS, Greenhalgh K, Frachet A, Niegowska M, Estes M, Jager C, Seguin-Devaux C, et al. A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nat Commun. 2016;7:11535. doi: 10.1038/ncomms11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gomez Ravetti M, Rosso OA, Berretta R, Moscato P. Uncovering molecular biomarkers that correlate cognitive decline with the changes of hippocampus' gene expression profiles in Alzheimer's disease. PLoS One. 2010;5(4):e10153. doi: 10.1371/journal.pone.0010153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krgovic D, Kokalj Vokac N, Zagorac A, Gregoric Kumperscak H. Rare structural variants in the DOCK8 gene identified in a cohort of 439 patients with neurodevelopmental disorders. Sci Rep. 2018;8(1):9449. doi: 10.1038/s41598-018-27824-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu F, Arias-Vasquez A, Sleegers K, Aulchenko YS, Kayser M, Sanchez-Juan P, Feng BJ, Bertoli-Avella AM, van Swieten J, Axenovich TI, et al. A genomewide screen for late-onset Alzheimer disease in a genetically isolated Dutch population. Am J Hum Genet. 2007;81(1):17–31. doi: 10.1086/518720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maroteaux G, Loos M, van der Sluis S, Koopmans B, Aarts E, van Gassen K, Geurts A, Neuro BMPC, Largaespada DA, Spruijt BM, et al. High-throughput phenotyping of avoidance learning in mice discriminates different genotypes and identifies a novel gene. Genes Brain Behav. 2012;11(7):772–784. doi: 10.1111/j.1601-183X.2012.00820.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeo IJ, Park MH, Son DJ, Kim JY, Nam KT, Hyun BK, Kim SY, Jung MH, Song MJ, Chun HO, et al. PRDX6 inhibits neurogenesis through downregulation of WDFY1-mediated TLR4 signal. Mol Neurobiol. 2018. [DOI] [PMC free article] [PubMed]
- 34.Yun HM, Jin P, Han JY, Lee MS, Han SB, Oh KW, Hong SH, Jung EY, Hong JT. Acceleration of the development of Alzheimer’s disease in amyloid beta-infused peroxiredoxin 6 overexpression transgenic mice. Mol Neurobiol. 2013;48(3):941–951. doi: 10.1007/s12035-013-8479-6. [DOI] [PubMed] [Google Scholar]
- 35.Freudenberg F, Resnik E, Kolleker A, Celikel T, Sprengel R, Seeburg PH. Hippocampal GluA1 expression in Gria1(-/-) mice only partially restores spatial memory performance deficits. Neurobiol Learn Mem. 2016;135:83–90. doi: 10.1016/j.nlm.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 36.Hayes LN, Shevelkin A, Zeledon M, Steel G, Chen PL, Obie C, Pulver A, Avramopoulos D, Valle D, Sawa A, et al. Neuregulin 3 knockout mice exhibit behaviors consistent with psychotic disorders. Mol Neuropsychiatry. 2016;2(2):79–87. doi: 10.1159/000445836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kerrisk ME, Koleske AJ. Arg kinase signaling in dendrite and synapse stabilization pathways: memory, cocaine sensitivity, and stress. Int J Biochem Cell Biol. 2013;45(11):2496–2500. doi: 10.1016/j.biocel.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim Chae-Seok, Kim Hyopil, Yu Nam-Kyung, Kang Sukjae Joshua, Kim TaeHyun, Ko Hyoung-Gon, Lee Jaehyun, Yang Jung-eun, Ryu Hyun-Hee, Park Taesung, Gim Jungsoo, Nam Hye Jin, Baek Sung Hee, Wegener Stephanie, Schmitz Dietmar, Boeckers Tobias M., Lee Min Goo, Kim Eunjoon, Lee Jae-Hyung, Lee Yong-Seok, Kaang Bong-Kiun. Enhancing inhibitory synaptic function reverses spatial memory deficits in Shank2 mutant mice. Neuropharmacology. 2017;112:104–112. doi: 10.1016/j.neuropharm.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 39.Wozniak DF, Xiao M, Xu L, Yamada KA, Ornitz DM. Impaired spatial learning and defective theta burst induced LTP in mice lacking fibroblast growth factor 14. Neurobiol Dis. 2007;26(1):14–26. doi: 10.1016/j.nbd.2006.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445(7124):168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 41.Alberini CM, Cruz E, Descalzi G, Bessieres B, Gao V. Astrocyte glycogen and lactate: New insights into learning and memory mechanisms. Glia. 2018;66(6):1244–1262. doi: 10.1002/glia.23250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki A, Stern SA, Bozdagi O, Huntley GW, Walker RH, Magistretti PJ, Alberini CM. Astrocyte-neuron lactate transport is required for long-term memory formation. Cell. 2011;144(5):810–823. doi: 10.1016/j.cell.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibbs ME, Lloyd HG, Santa T, Hertz L. Glycogen is a preferred glutamate precursor during learning in 1-day-old chick: biochemical and behavioral evidence. J Neurosci Res. 2007;85(15):3326–3333. doi: 10.1002/jnr.21307. [DOI] [PubMed] [Google Scholar]
- 44.Wyss MT, Jolivet R, Buck A, Magistretti PJ, Weber B. In vivo evidence for lactate as a neuronal energy source. J Neurosci. 2011;31(20):7477–7485. doi: 10.1523/JNEUROSCI.0415-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.El Hayek L, Khalifeh M, Zibara V, Abi Assaad R, Emmanuel N, Karnib N, El-Ghandour R, Nasrallah P, Bilen M, Ibrahim P, et al. Lactate mediates the effects of exercise on learning and memory through SIRT1-dependent activation of hippocampal brain-derived neurotrophic factor (BDNF) J Neurosci. 2019;39(13):2369–2382. doi: 10.1523/JNEUROSCI.1661-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brooks GA. Cell-cell and intracellular lactate shuttles. J Physiol. 2009;587(Pt 23):5591–5600. doi: 10.1113/jphysiol.2009.178350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang J, Ruchti E, Petit JM, Jourdain P, Grenningloh G, Allaman I, Magistretti PJ. Lactate promotes plasticity gene expression by potentiating NMDA signaling in neurons. Proc Natl Acad Sci U S A. 2014;111(33):12228–12233. doi: 10.1073/pnas.1322912111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steinman MQ, Gao V, Alberini CM. The role of lactate-mediated metabolic coupling between astrocytes and neurons in long-term memory formation. Front Integr Neurosci. 2016;10:10. doi: 10.3389/fnint.2016.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walters W, Hyde ER, Berg-Lyons D, Ackermann G, Humphrey G, Parada A, Gilbert JA, Jansson JK, Caporaso JG, Fuhrman JA, et al. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems. 2016;1:1. doi: 10.1128/mSystems.00009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown J, Zavoshy N, Brislawn CJ, McCue LA: Hundo: a Snakemake workflow for microbial community sequence data. Peer J Preprints 2018, 6:e27272v1.
- 51.Rognes Torbjørn, Flouri Tomáš, Nichols Ben, Quince Christopher, Mahé Frédéric. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McDonald D, Clemente JC, Kuczynski J, Rideout JR, Stombaugh J, Wendel D, Wilke A, Huse S, Hufnagle J, Meyer F, et al. The Biological Observation Matrix (BIOM) format or: how I learned to stop worrying and love the ome-ome. Gigascience. 2012;1(1):7. doi: 10.1186/2047-217X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lanzen A, Jorgensen SL, Huson DH, Gorfer M, Grindhaug SH, Jonassen I, Ovreas L, Urich T. CREST--classification resources for environmental sequence tags. PLoS One. 2012;7(11):e49334. doi: 10.1371/journal.pone.0049334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4):e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wan KH, Yu C, Park S, Hammonds AS, Booth BW, Celniker SE. Complete genome sequence of Lactobacillus plantarum oregon-R-modENCODE strain BDGP2 isolated from Drosophila melanogaster gut. Genome Announc. 2017;5:41. doi: 10.1128/genomeA.01155-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42(Database issue):D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakayasu ES, Nicora CD, Sims AC, Burnum-Johnson KE, Kim YM, Kyle JE, Matzke MM, Shukla AK, Chu RK, Schepmoes AA, et al. MPLEx: a robust and universal protocol for single-sample integrative proteomic, metabolomic, and lipidomic analyses. mSystems. 2016;1:3. doi: 10.1128/mSystems.00043-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5(10):877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsanov N, Samacoits A, Chouaib R, Traboulsi AM, Gostan T, Weber C, Zimmer C, Zibara K, Walter T, Peter M, et al. smiFISH and FISH-quant - a flexible single RNA detection approach with super-resolution capability. Nucleic Acids Res. 2016;44(22):e165. doi: 10.1093/nar/gkw784. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mouse gut microbiome 16S rRNA gene sequence data is available on OSF (https://osf.io/jbt5g/).
All the raw metabolomic data are available at the OSF public data depository (https://osf.io/jbt5g/).