Abstract
Attention-Deficit/Hyperactivity Disorder (ADHD) is a common neurodevelopmental disorder with only symptomatic care available. Genome-wide association (GWA) studies can provide a starting point in the search for novel drug targets and possibilities of drug repurposing. Here, we explored the druggable genome in ADHD by utilising GWA studies on ADHD and its co-morbid conditions. First, we explored whether the genes targeted by current ADHD drugs show association with the disorder and/or its co-morbidities. Second, we aimed to identify genes and pathways involved in the biological processes underlying ADHD that can be targeted by pharmacological agents. These ADHD-associated druggable genes and pathways were also examined in co-morbidities of ADHD, as commonalities in their aetiology and management may lead to novel pharmacological insights. Strikingly, none of the genes encoding targets of first-line pharmacotherapeutics for ADHD were significantly associated with the disorder, suggesting that FDA-approved ADHD drugs may act through different mechanisms than those underlying ADHD. In the examined druggable genome, three loci on chromosomes 1, 4 and 12 revealed significant association with ADHD and contained nine druggable genes, five of which encode established drug targets for malignancies, autoimmune and neurodevelopmental disorders. To conclude, we present a framework to assess the druggable genome in a disorder, exemplified by ADHD. We highlight signal transduction and cell adhesion as potential novel avenues for ADHD treatment. Our findings add to knowledge on known ADHD drugs and present the exploration of druggable genome associated with ADHD, which may offer interventions at the aetiological level of the disorder.
Subject terms: ADHD, Psychiatric disorders
Introduction
Attention deficit/hyperactivity disorder (ADHD) is a common and highly heritable childhood-onset neurodevelopmental disorder that often persists into adulthood [1, 2]. The prevalence of the disorder in children is 6.5%, while in adults the estimates vary between 2.5 and 3.4% [3]. ADHD patients are at high risk of experiencing difficulties in their education and social integration [4], elevated rates of incarceration, unemployment and accidental deaths, all resulting in high societal and economic burden [1, 5–7]. To date, no treatments cure ADHD, although available therapies offer symptomatic relief.
Current management of ADHD is based on either non-pharmacologic or pharmacologic treatments as well as the combination of the two. The non-pharmacologic treatments usually involve psychological and/or behavioural therapies, while the pharmacologic interventions include stimulant and/or non-stimulant drugs [1, 8]. For ADHD treatment, the U.S. Food and Drug Administration (FDA) has approved the stimulants methylphenidate (MPH) and amphetamine (AMP) along with three non-stimulants: atomoxetine, clonidine and guanfacine [9]. In many guidelines, MPH and AMP are the first-line agents for ADHD pharmacotherapy, exerting their primary effect by increasing dopamine and norepinephrine activity [10, 11]. Atomoxetine is a selective norepinephrine re-uptake inhibitor, while both clonidine and guanfacine are alpha-2 adrenoreceptor agonists [11]. Although all the pharmaceuticals used to manage ADHD are believed to act on biological pathways underlying the disorder, their complete mechanisms of action remain unknown, as are the causal biological mechanisms of ADHD.
An important feature of ADHD relevant to the search of new medications is the existence of co-morbid conditions [12, 13]. As it has been postulated that biological processes underlying ADHD may also be involved in the development of its co-morbidities [14], it is prudent to examine the relationship between ADHD-associated druggable loci and those conditions. The exploration of associations between a gene encoding or being the target of a drug and a number of phenotypes has been proposed to aid pharmacotherapeutics by capturing a broader spectrum of relevant biological information and offering alternatives to existing pharmaceuticals to treat a disorder (drug repurposing) [15].
Randomised controlled studies have shown that pharmacotherapy reduces the ADHD symptom burden [16] and observational studies have reported that it improves important life outcomes, such as academic performance [17], social functioning [18, 19] and the rate of motor vehicle accidents [20]. Nonetheless, the current pharmacological treatment of ADHD is not curative and, although many patients improve markedly, optimal outcomes are difficult to achieve, especially with regards to signs of executive dysfunction and emotional dysregulation [21, 22]. There are also lingering concerns about long-term effects of stimulants on growth and weight [23–25]. Thus, there is a need for more efficient and safe pharmacological agents to treat and, eventually, cure ADHD.
Improvements in the pharmacotherapeutic options for ADHD may require a fuller understanding of its underlying biological processes [26]. As knowledge on the genetics of common disorders evolves, novel strategies for the development of new and improved pharmacotherapeutics are emerging. For complex disorders, such as ADHD, genome-wide association (GWA) studies can uncover genes and pathways involved in the disease aetiology, yielding innovative avenues for future drug development and repurposing [27, 28].
In this study, we explored the druggable genome in ADHD by utilising the summary statistics from GWA studies on ADHD and its major co-morbid conditions. We aimed to (1) explore whether the genes targeted by current FDA-approved ADHD drugs show association with the disorder and/or its co-morbidities, (2) identify genes and pathways involved in the biological processes underlying ADHD, its co-morbidities and quality of life phenotypes that can be targeted by pharmacological compounds and (3) examine the identified druggable genes and pathways as potential options for novel drug development and repurposing.
Materials and methods
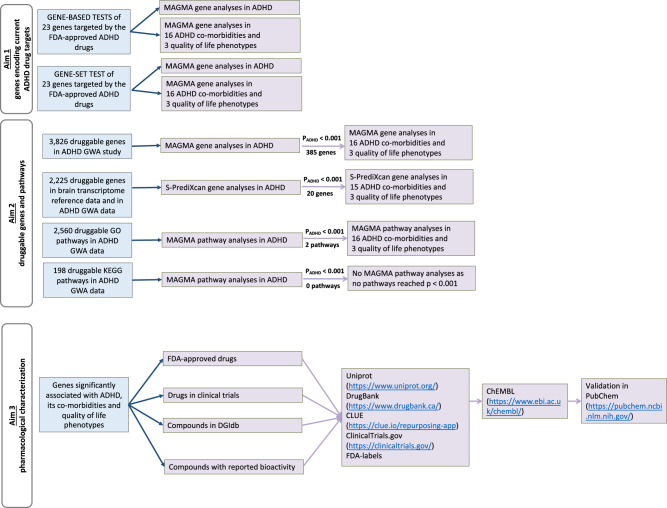
Figure 1 shows a flowchart summarising the steps of our study.
Fig. 1.
Study design flowchart. ADHD; attention deficit/hyperactivity disorder, BMI; body mass index, CHD; coronary heart disease, T2DM; type 2 diabetes mellitus, SWB; subjective well-being
Definition of the druggable genome
The druggable genome was defined as described in Finan et al. [29] as a selection of genes that are potential targets for pharmacological intervention.
The identification of these druggable genes was based on the protein targets of known and experimental drugs, sequence similarities to those targets (potential druggability), drug–gene interactions, biotherapeutics and a number of databases documenting pharmacological molecules and their therapeutic targets.
Definition of ADHD co-morbidities and quality of life phenotypes
For the purpose of this study, we focused only on common conditions with well-documented evidence for association with ADHD based on large-scale genetic [30] and epidemiological [31] studies, together with a systematic literature review [32] complemented by a PubMed search using the following criteria “((ADHD co-morbidity) AND English [Language]) AND (“2015” [Date-Publication]: “3000” [Date-Publication])”.
Additional criteria were the availability of large (≥20,000 individuals) GWA studies and their summary statistics. Thus, where such data were not available, we used proxy phenotypes (e.g. instead of insomnia disorder, we examined insomnia symptoms [33]).
We characterised the co-morbidities into the three main groups: (1) cardiometabolic, (2) immune-inflammatory-autoimmune (referred to as immune) and (3) neuropsychiatric.
In addition to well-defined clinical diagnoses, ADHD has been reported to be associated with reduced quality of life, reduced educational attainment and sleep disturbances [1]. Therefore, we also examined the druggable genome overlapping between ADHD and educational attainment, sleep duration and subjective well-being as proxies for quality of life and functional outcomes associated with ADHD.
Genetic data
We relied on summary statistics derived from large-scale GWA studies. Where possible, we restricted our analyses to individuals of European descent only, meta-analysed sample size equal to or larger than 70% of the total sample, variants with minor allele frequency above or equal to 1% and of good imputation quality (INFO ≥ 0.8). For ADHD, summary statistics were acquired from the large-scale meta-analysis of 19,099 cases and 34,194 controls [30]. For the co-morbidities, we curated data from openly available resources or through correspondence with the authors of the GWA studies of interest (Table 1).
Table 1.
Overview of the examined data of ADHD, its co-morbidities and quality of life phenotypes
| Condition | Sample sizeb | Data source and reference | Reason for inclusion (of co-morbidities and quality of life phenotypes) |
|---|---|---|---|
| ADHD | 53,293 | Psychiatric genetics consortium [30] http://www.med.unc.edu/pgc/results-and-downloads | |
| Cardiometabolic co-morbidities | |||
| Body mass indexc | 322,154 | Genetic Investigation of Anthropometric Traits consortium [58] https://portals.broadinstitute.org/collaboration/giant/index.php/GIANT_consortium_data_files | [30, 32]a |
| Coronary heart disease | 84,264 | Coronary Artery Disease Genome wide Replication and Meta-analysis plus The Coronary Artery Disease Genetics consortium [59] http://www.cardiogramplusc4d.org/data-downloads/ | [30] |
| Type 2 diabetes mellitus | 44,414 | Diabetes Genetics Replication And Meta-analysis [60] http://diagram-consortium.org/downloads.html | [30] |
| Immune co-morbidities | |||
| Asthma | 26,475 | GABRIEL Consortium [61] http://www.cng.fr/gabriel/results.html | [31, 32]a |
| Crohn’s disease | 49,970 | International Inflammatory Bowel Disease Genetics Consortium [62] https://www.ibdgenetics.org/downloads.html | [63] |
| Dermatitis | 40,835 | Early Genetics and Lifecourse Epidemiology eczema consortium [64] https://data.bris.ac.uk/data/dataset/28uchsdpmub118uex26ylacqm | [31] |
| Rheumatoid arthritis | 58,284 | Meta-analysis of GWA studies on rheumatoid arthritis [65] http://plaza.umin.ac.jp/yokada/datasource/software.htm | [30] |
| Ulcerative colitis | 43,823 | International Inflammatory Bowel Disease Genetics Consortium [62] https://www.ibdgenetics.org/downloads.html | [63, 66] |
| Neuropsychiatric co-morbidities | |||
| Alcohol intake frequencyd | 336,965 | UK biobank [67] https://docs.google.com/spreadsheets/d/1b3oGI2lUt57BcuHttWaZotQcI0-mBRPyZihz87Ms_No/edit#gid=1209628142t | [31] |
| Autism spectrum disorder | 46,351 | Psychiatric genetics consortium [68] http://www.med.unc.edu/pgc/results-and-downloads | [69, 70] |
| Number of Cigarettes smoked in a dayd | 38,181 | Tobacco and Genetics Consortium [71] https://www.med.unc.edu/pgc/results-and-downloads | [30, 31] |
| Epilepsy | 34,852 | International League Against Epilepsy Consortium on Complex Epilepsies [72] http://www.epigad.org/page/show/gwas_index | [73] |
| Insomnia symptomse | 113,006 | UK Biobank / Hammerchlag et al. [33] http://ctg.cncr.nl/software/summary_statistics | [32]a |
| Major depressive disorder | 260,929 | Psychiatric genetics consortium http://www.med.unc.edu/pgc/results-and-downloads [74] | [30, 31, 74] |
| Migraine | 205,094 | International Headache Genetics Consortium [75] | [31, 32]a |
| Schizophrenia | 77,096 | Psychiatric genetics consortium [76] http://www.med.unc.edu/pgc/results-and-downloads | [31] |
| Quality of life phenotypes | |||
| Educational attainment | 328,917 | Social Science Genetic Association Consortium [77] https://www.thessgac.org/data | [30] |
| Sleep duration | 111,980 | International Sleep Genetic Epidemiology Consortium [78] https://sleepgenetics.org/downloads/ | [32]a |
| Subjective Well Being | 298,420 | Social Science Genetic Association Consortium [79] https://www.thessgac.org/data | [30] |
ADHD attention deficit hyperactivity disorder
aOnly those co-morbidities were included in this study that revealed well-established evidence for association [32]
bMaximum total sample size
cProxy for obesity
dProxy for substance abuse
eProxy for insomnia disorder
Statistical analyses
Statistical analyses were divided into two main steps to address our first two aims: (1) examination of genes targeted by current FDA-approved ADHD drugs and (2) examination of the genes within the druggable genome and their pathways defined as known biological pathways containing at least one gene from the druggable genome. In step 1, we examined all genes in all curated GWA data. In step 2, we first examined ADHD and only genes and pathways that revealed suggestive association with it (p ≤ 0.001) were further analysed in the GWA data of its co-morbidities and quality of life phenotypes. We applied Bonferroni correction to account for multiple testing.
Step one: analyses of the genes targeted by current ADHD drugs
The genes targeted by the current FDA-approved ADHD medications were defined by Gaspar and Breen [34]. We examined these genes individually (gene-based tests) and altogether (gene-set analyses) in MAGMA software [35]. Each gene’s degree of association with a phenotype was calculated based on the individual single nucleotide polymorphisms’ (SNP) association p-values from their respective GWA studies. SNPs with chromosomal positions within the boundaries of a gene (start and end of a primary transcript) were assigned to that gene (i.e. the default settings of MAGMA). The 1000 Genomes CEU population was used as the reference panel to correct for linkage disequilibrium (LD). We conducted gene-based analyses of all genes on autosomal chromosomes. Genes represented by a single parameter (i.e. only one association signal) in MAGMA were excluded. To evaluate each gene’s contribution to the examined gene-set, the association p-value of each gene was converted to a Z-value and used as an outcome variable for a regression model with gene-set membership as a predictor. Gene size, gene-sets’ gene density and LD were taken into account to adjust for possible confounding effects and prevent spurious association.
Step two: analyses of genes and pathways within the druggable genome
The gene associations with ADHD, its co-morbidities and quality of life phenotypes were tested on two levels: (a) DNA variation and (b) gene expression. The first was tested in MAGMA as described above. The latter was tested in S-PrediXcan [36]. In short, S-PrediXcan first predicts tissue-specific gene expression level of each gene based on the reference transcriptome data [37] and then estimates the correlation between that level and a phenotype using GWA summary statistics. Given that ADHD is believed to be a disorder of the central nervous system, we restricted our S-PrediXcan analyses to its tissues. S-PrediXcan analyses were performed using the default settings of the software. Type 2 diabetes was excluded from these analyses as the available summary statistics did not contain the necessary data.
The biological pathways were defined as determined by Gene Ontology (GO) [38] and the Kyoto encyclopaedia of genes and genomes (KEGG) [39]. We restricted our analyses to pathways represented by more than 10 and less than 1000 genes. The analyses were conducted in MAGMA [35] as described above.
Characterisation of the druggable genome loci associated with ADHD and/or its co-morbidities and quality of life phenotypes
To address our third aim, we explored the pharmacology of genes (or their encoded proteins) pinpointed in our statistical analyses.
To identify the pharmacological agents, we developed a systematic pipeline utilising publically available databases, where the agents were assessed in four stages: (1) FDA-approved drugs, (2) drugs in clinical trials, (3) compounds reported in the Drug–Gene Interaction Database (DGIdb, http://dgidb.org/), and (4) small molecule compounds with reported molarity measurement for bioactivity.
First, we evaluated each gene of interest in Uniprot (https://www.uniprot.org/) and characterised the identified FDA-approved drugs and compounds in clinical trials using Drugbank (https://www.drugbank.ca/), CLUE Repurposing Hub (https://clue.io/repurposing-app), and DGIdb databases. The FDA approval and clinical trial status of the compounds were crosschecked using the publically available FDA labels and ClinicalTrials.gov (https://clinicaltrials.gov/) database. The paediatric approval status was investigated on Medscape (https://reference.medscape.com/) and in FDA labels. For FDA-approved compounds, the approved indication reported in FDA label was noted. For compounds in clinical trials, the indication was researched in ClinicalTrials.gov, applying the following filters: “not yet recruiting”, “recruiting”, “enroling by invitation”, “active, not recruiting” and “completed”, in order to select trials that are currently active. In addition, the mechanism of action on the specific gene of interest was noted from Drugbank.
Next, the genes were researched as targets in ChEMBL (https://www.ebi.ac.uk/chembl/), downloading all compounds reported to interact with a gene of interest along with their reported “Target Associated Bioactivity” as their affinity or potency to the human gene product stated in molarity (referred to as “Bioactivity”). The compounds from ChEMBL were then prioritised from the lowest “Bioactivity” value to the highest and, in order to investigate the most relevant compounds, the top 50 were characterised further by being individually investigated in PubChem (https://pubchem.ncbi.nlm.nih.gov/) to confirm their target-associated bioactivity.
Results
Definition of druggable genome and ADHD co-morbidities
The druggable human genome has been estimated to comprise 4479 genes [29], 3826 of which were represented by more than one association signal in the ADHD GWA data. These genes were present in 2758 pathways (2560 GO and 198 KEGG).
Genetic data
We obtained summary statistics from GWA studies for ADHD, eight neuropsychiatric disorders, three cardiometabolic diseases, five immune diseases and three quality of life phenotypes (Table 1).
Statistical analyses
In ADHD, we examined 3826 genes and 2759 gene sets in the druggable genome (2560 GO, 198 KEGG and one set of genes targeted by FDA-approved ADHD drugs), bringing the Bonferroni-corrected significance threshold to p = 7.59E−06.
In the co-morbidities and quality of life phenotypes, we examined 385 genes and three gene sets (two GO pathways and one set of FDA-approved ADHD genes) in the druggable genome. The Bonferroni-adjusted significance thresholds for these analyses was set to p = 1.29E−04.
Associations stronger than the determined Bonferroni thresholds were considered significant.
Step one: analyses of the genes targeted by current ADHD drugs
We identified 23 genes targeted by the FDA-approved ADHD drugs (and revealing more than one independent association signal in ADHD GWA summary statistics). Individually, none of these genes showed significant association with ADHD (Table 2). For co-morbid conditions, several significant associations were noted (Table S1). The strongest one was observed between DRD2 and the frequency of alcohol consumption (p = 2.88E−08), followed by associations between the same gene (DRD2) and SCZ (p = 1.55E−07), CYP2D6 and SCZ (p = 1.81E−06), CHRM2 and major depressive disorder (p = 2.56E−06). In addition, SLC6A3 revealed significant association with sleep duration (p = 2.41E−05). All of these genes encode protein targets of atomoxetine. Furthermore, SLC6A3 is also targeted by MPH and AMP, while DRD2 is a secondary target of MPH and AMP.
Table 2.
Association between ADHD and genes targeted by FDA-approved ADHD drugs
| Target gene | FDA approved ADHD drug | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Stimulants | Non-stimulants | |||||||||
| Gene | Chr | Start | End | Number of SNPs | p-value | Methylphenidate and derivatives | Amphetamine and derivatives | Atomoxetine | Clonidine | Guanfacine |
| ADRA1A | 8 | 26605667 | 26724790 | 469 | 0.668 | N | Y | N | Y | N |
| ADRA1B | 5 | 159343790 | 159399551 | 118 | 0.82 | N | Y | N | Y | N |
| ADRA1D | 20 | 4201329 | 4229721 | 89 | 0.499 | N | N | N | Y | N |
| ADRA2A | 10 | 112836790 | 112840658 | 5 | 0.43 | Y | Y | Y | Y | Y |
| ADRA2B | 2 | 96778707 | 96781984 | 5 | 0.399 | Y | N | N | Y | Y |
| CARTPT | 5 | 71014990 | 71016875 | 2 | 0.376 | N | Y | N | N | N |
| CHRM1 | 11 | 62676151 | 62689279 | 25 | 0.254 | N | N | Y | N | N |
| CHRM2 | 7 | 136553416 | 136705002 | 399 | 0.049 | N | N | Y | N | N |
| CYP2D6 | 22 | 42522501 | 42526908 | 29 | 0.689 | N | N | Y | N | N |
| DRD2 | 11 | 113280318 | 113346413 | 167 | 0.237 | N | N | Y | N | N |
| HRH1 | 3 | 11178779 | 11305243 | 308 | 0.505 | N | N | Y | N | N |
| HTR1B | 6 | 78171948 | 78173490 | 5 | 0.053 | N | N | Y | N | N |
| HTR1D | 1 | 23516993 | 23521222 | 9 | 0.301 | N | N | Y | N | N |
| HTR2A | 13 | 47405685 | 47471169 | 207 | 0.775 | N | N | Y | N | N |
| HTR6 | 1 | 19991780 | 20006055 | 27 | 0.026 | N | N | Y | N | N |
| HTR7 | 10 | 92500578 | 92617671 | 305 | 0.382 | N | N | Y | N | N |
| NISCH | 3 | 52489134 | 52527087 | 68 | 0.145 | N | N | N | Y | N |
| NPY1R | 4 | 164245113 | 164265984 | 48 | 0.032 | N | N | Y | N | N |
| OPRM1 | 6 | 154331631 | 154568001 | 609 | 0.217 | N | Y | N | N | N |
| SLC18A2 | 10 | 119000604 | 119038941 | 92 | 0.347 | N | Y | N | N | N |
| SLC6A2 | 16 | 55689516 | 55740104 | 154 | 0.223 | Y | Y | Y | N | Y |
| SLC6A3 | 5 | 1392909 | 1445545 | 100 | 0.836 | Y | Y | Y | N | Y |
| SLC6A4 | 17 | 28521337 | 28563020 | 65 | 0.483 | Y | N | Y | N | Y |
| TAAR1 | 6 | 132966123 | 132967142 | 3 | 0.767 | N | Y | N | N | N |
Genes reaching nominal association p-value below 0.05 are highlighted in bold.
“Y” (meaning “yes”) indicates that a gene is targeted by the drug, while “N” (meaning “no”) indicates that a gene is not targeted by the drug. “Chr” refers to the number of a chromosome where the gene of interest is located. “Start” and “End” refer to base pair location of genes of interest.
Examining all the genes as a set revealed no significant association with neither ADHD nor its co-morbidities (Table S2).
Step two: analyses of genes and pathways within the druggable genome
Analyses of genetic variation (MAGMA)
For ADHD, four loci on chromosomes one, three, four and twelve showed significant association (Table 3). The locus on chromosome one contains seven druggable genes, while the other three loci contain one druggable gene each (Figs. S1–S4). The most significant association was observed on chromosome one (ST3GAL3 gene, p = 3.10E−12, Table 3 and Fig. S1). While the loci on chromosomes one, four and twelve revealed strong association signals at the individual SNP level (Figs. S1–S3), the locus on chromosome three did not (Fig. S4) and, thus, was excluded from further analyses.
Table 3.
The druggable genes located within the three loci significantly associated with ADHD
| Locus | Gene | Number of SNPs per gene | p-value | Small molecule compound | Biotherapeutic | ADME | Drug in clinical trial | FDA approved drug |
|---|---|---|---|---|---|---|---|---|
| Chr1 | ST3GAL3 | 483 | 3.06E−12 | Yes | No | No | No | No |
| KDM4A | 71 | 2.11E−11 | Yes | No | No | No | No | |
| PTPRF | 225 | 5.06E−10 | Yes | No | No | Yes | Yes | |
| TIE1 | 30 | 2.01E−08 | Yes | No | No | Yes | Yes | |
| MPL | 12 | 4.69E−08 | Yes | Yes | No | Yes | Yes | |
| SLC6A9 | 67 | 2.39E−07 | Yes | No | No | Yes | Yes | |
| ARTN | 10 | 3.27E−06 | No | No | No | No | No | |
| Chr4 | MANBA | 203 | 5.99E−08 | Yes | No | No | No | No |
| Chr12 | KCNH3 | 40 | 4.23E−06 | Yes | No | No | Yes | Yes |
“p-value” column indicates the strength of association between a gene and ADHD
ADME: genes involved in the absorption, distribution, metabolism, and excretion of drugs; FDA: United States of America Food and Drug Administration; SNP: single nucleotide polymorphism
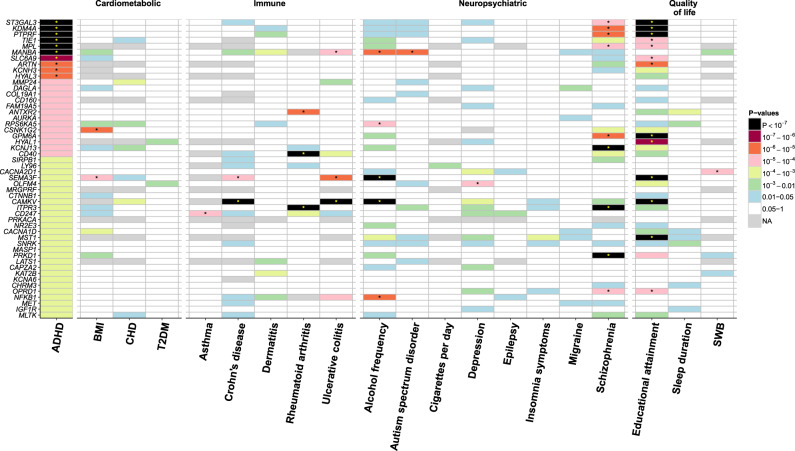
For the co-morbidities, the most significant association was noted between ITPR3 and rheumatoid arthritis (p = 4.88E−38, Fig. 2 and Table S3). This gene was also significantly associated with SCZ (p = 1.13E−09). Among the three loci associated with ADHD, those on chromosomes one and four revealed druggable genes also significantly associated with SCZ, ulcerative colitis, autism spectrum disorder and the frequency of alcohol consumption (Fig. 2 and Table S3). In addition, educational attainment showed the highest number of significantly associated genes (Table S3).
Fig. 2.
Tile plot of the association between druggable genes and ADHD (p < 0.001), its co-morbid conditions of ADHD and quality of life phenotypes. ADHD; attention deficit/hyperactivity disorder, BMI; body mass index, CHD; coronary heart disease, T2DM; type 2 diabetes mellitus, SWB; subjective well-being
Analyses of gene expression (S-PrediXcan)
In total, we examined 13 tissues of the central nervous system (Fig. S5). Transcriptome data for 2225 druggable genes (also examined in MAGMA) were present in at least one tissue of central nervous system.
For ADHD, the expression levels of two genes—MANBA (p = 1.63E-07 in”cerebellar hemisphere”) and LEPRE1 (p = 5.05E−09 in “frontal cortex”)—showed significant association and 18 additional genes showed signs of suggestive association (p < 0.001) (Table S4).
For co-morbidities, the expression of 13 genes revealed significant associations with a number of examined phenotypes (Fig. S5 B-S, Table S4). The most significant association was observed between the expression of HLA-DPB1 and rheumatoid arthritis (p = 2.96E-45 in “cerebellum”, Table S4). The expression levels of the two genes that showed significant association with ADHD (MANBA and LEPRE1) were also significantly associated with body mass index, rheumatoid arthritis and SCZ (Table S4).
Overall, gene expression analyses highlighted five druggable genes significantly associated with ADHD and/or its comorbidities and quality of life phenotypes in addition to those prioritised in analyses of genetic variation.
Pathway analyses
For ADHD, no significant association was noted among either GO or KEGG pathways, with the strongest signal observed for negative regulation of protein binding (GO:0032091, p = 1.5E−04). Two GO pathways revealed nominal associations with ADHD (p < 0.001, Table S5) and were analysed for association with its co-morbidities and quality of life phenotypes, showing no significant associations (Table S5).
The results of KEGG pathways are summarised in Table S6.
Characterisation of the druggable genome loci associated with ADHD and/or its co-morbidities and quality of life phenotypes
Genetic variation loci
Out of the nine druggable genes located within the three loci significantly associated with ADHD, the proteins encoded by five of them are interacting with pharmaceuticals that are FDA-approved or in clinical trials: PTPRF, TIE1, MPL, SLC6A9 and KCNH3 (Table 3). Among their indications we noted malignancies, autoimmune diseases, neuropsychiatric disorders (including ADHD, Parkinson’s and Alzheimer’s diseases), metabolic disorder, haematopoietic processes, inflammation, atrial fibrillation and spinal cord injury (Table S7). No FDA-approved drugs or drugs in clinical trials interacted with the druggable locus on chromosome four.
For co-morbid conditions, we examined 14 loci within the druggable genome that all showed suggestive association with ADHD (p < 0.001) and significant association with any of the examined co-morbidities. Among the 17 druggable genes within those 14 loci, 13 interact with drugs that are in clinical trials or are FDA-approved, with the majority of indications being autoimmune disorders and/or malignancies (Table S8).
Gene expression loci
Among the five genes pinpointed by S-PrediXcan (and not overlapping with those identified in MAGMA), three are targeted by compounds in clinical trials and two of them are also FDA-approved. All of these compounds are nutraceuticals, with malignancies and immune dysfunctions among their indications (Tables S9 and S10).
Discussion
Despite ADHD being a highly heritable disorder, it has been challenging to utilise genetic information in its treatment. Nonetheless, the more insight we gain into the molecular genetics of ADHD, the more options for its treatment may become available [40]. In this study, we explored the druggable genome in ADHD, its co-morbid conditions and quality of life phenotypes utilising large-scale GWA studies. We aimed to address three questions: (1) do any of the genes encoding targets of FDA-approved ADHD drugs show association with ADHD and/or its co-morbidities, (2) are ADHD and/or its co-morbidities and quality of life phenotypes associated with genetic variation and expression within the known druggable genome and if so, (3) can we use those association signals to identify gene targets for novel drug development and/or repurposing to treat ADHD.
To answer the first question, we examined the association between the genes encoding the immediate targets of the first-line ADHD pharmacotherapeutics and ADHD as well as its co-morbidities. We observed no significant association between these genes and ADHD, suggesting that these drugs may act through mechanisms different to those underlying ADHD. However, as the current GWA study on ADHD reveals only a small fraction of the biological processes underlying this condition [30], larger studies are needed to draw any definitive conclusions.
Overall, drugs that are FDA-approved or currently undergoing clinical trials to treat ADHD (e.g. dasotraline) target only a limited number of known pharmacological targets, essentially enhancing catecholamine signalling. This illustrates that all active ADHD drugs belong to a small pharmacological niche and we should aim to move beyond it. Hence, we examined whether any known druggable genes and pathways are associated with ADHD and/or its co-morbidities and quality of life phenotypes (second question), following by pharmacological characterisation of identified associations (third question). These analyses aimed to pinpoint novel avenues for ADHD drug development as well as repurposing. Because the de novo discovery and development of entirely new drugs targeting unique biology of a disorder is a tedious and expensive process with a low success rate, the possibility of repurposing already existing drugs towards new indications may be more effective [40]. Here, we highlight some of these potential new targets, although this list is not comprehensive.
Within the loci associated with ADHD, 5 druggable genes encode proteins interacting with drugs that are FDA-approved or are in clinical trials. The common indications of those pharmaceuticals are autoimmune disorders and malignancies, with some also being tested in clinical trials for treatment of neurodevelopmental disorders. Interestingly, autoimmune disorders and malignancies are also common indications for drugs interacting with genes associated with co-morbidities of ADHD, suggesting that these two fields of research could present novel paths for ADHD treatment.
The locus on chromosome one shows the strongest association with ADHD and also contains the most genes interacting with drugs that are FDA-approved or in clinical trials. Among them, PTPRF is the gene with the most prominent association signal. This gene encodes a tyrosine phosphatase, a signalling molecule involved in a myriad of cellular processes, including cell adhesion, neuronal development and functioning [41, 42]. PTPRF has mainly been studied in the context of cancer. However, its involvement in hyperactivity [42] and axonal growth [43] has also been reported. Another ADHD-associated gene interacting with drugs that are FDA-approved and/or are in clinical trials is SLC6A9, a gene encoding a glycine transporter that is targeted by such compounds as bitopertin, sarcosine and glycine [44]. In ADHD, glycine supplementation is currently under investigation as a potential treatment [45]. Similarly, sarcosine has also been tested as a possible ADHD drug, although the preliminary analyses indicate that its effect may be limited to oppositional symptoms only [46].
Outside the chromosome one locus, the KCNH3 gene is also interacting with drugs that are FDA-approved or are in clinical trials. This gene encodes a voltage-dependent potassium channel, a selective inhibitor of which was recently described [47]. It is also a non-specific target of blood–brain barrier penetrating drug dalfampridine [48] used to relieve the symptoms of multiple sclerosis and related neurologic disorders [44, 49]. Knocking out KCNH3 in mice has been reported to enhance cognitive skills, including attention, further supporting a potential role of dalfampridine-like drugs in the treatment of ADHD [50].
The aforementioned druggable genes also showed significant association with educational attainment, suggesting that drugs targeting them may have a possible impact on quality of life of ADHD patients.
The analyses of correlation between ADHD and gene expression levels in brain pinpointed druggable genes MANBA and LEPRE1, among which only LEPRE1 interacts with a number of compounds in clinical trials, such as nutraceutical ascorbate, succinic acid and L-proline. This gene encodes an enzyme needed for collagen synthesis and assembly, which has recently been proposed as a novel therapeutic vista for protection and regeneration of neurons [51]. Moreover, two additional genes, the expression of which correlated with the examined co-morbidities and quality of life phenotypes are also targeted by nutraceuticals.
In pathway analyses, the GO pathway of negative regulation of protein binding (GO:0032091) showed the strongest, albeit non-significant, association with ADHD. This pathway encompasses any process that negatively affects any protein binding, such as actin binding (e.g. synaptic plasticity), microtubule binding, receptor binding and homodimerization activity of a protein. The latter processes affect a G-protein-coupled receptor signalling, tapping into the largest class of targets in current drug development [52, 53] and presenting a myriad of potential opportunities for new drug discoveries in ADHD. Indeed, one of the novel approaches to pharmacotherapy of ADHD is the use of fasoracetam that acts on G-protein coupled glutamate receptor [40].
Among the examined co-morbidities, the neuropsychiatric (mostly SCZ) and immune groups revealed significant associations, with 12 genes interacting with compounds that are FDA-approved or in clinical trials. Interestingly, one of these genes is KCNJ13, encoding a druggable potassium channel targeted by dalfampridine, the same compound that also targets the KCNH3-protein discussed above.
The gene that revealed significant associations with the largest number of co-morbidities is SEMA3F. This gene also showed significant association with educational attainment. SEMA3F encodes semaphoring-3F protein involved in cell signalling, affecting cell adhesion and migration and being explored mostly in cancer therapies [54, 55]. Nonetheless, the range of therapeutic potential of semaphorins is large [56].
Our study has some limitations. As we examined associations observed in GWA studies, where it is difficult to obtain adequate sample sizes to detect associations of small effects, our findings are limited by their statistical power. Moreover, as we imposed a sample size limit of 20,000 individuals, some of the co-morbid conditions, where GWA studies of such size were not available, were replaced by proxy phenotypes.
The current statistical methods allow us to identify chromosomal loci only. Further studies on the genes of interest as well as fine mapping are needed to unambiguously establish which gene(s) lies on the causal pathway to developing ADHD. This knowledge would allow for a higher resolution search for therapeutic targets, especially on chromosome one locus where the LD structure is particularly complicated.
The gene expression analyses have several limitations [57], including the confounding by genetic associations due to LD, implying a possible substantial bias towards genes located in the loci revealing genome-wide association with the examined trait. In addition, the available transcriptome data are limited and are not available in many relevant tissues (e.g. lack of expression data for KCNH3 in brain tissues in reference transcriptome), preventing a comprehensive investigation of the transcriptome.
As we used publicly available databases, it was not possible to control their quality. Furthermore, the information provided in the utilised drug target databases may be incomplete.
To conclude, we present a framework for assessment of the druggable genome in a disorder, exemplified by ADHD. We present possibilities for drug repurposing (e.g. dalfampridine) and highlight processes of signal transduction and cell adhesion (negative regulation of protein binding, PTPRF, SEMA3F, KCNH3, KCNJ13) as potential novel avenues for ADHD treatment. Our findings add to the knowledge on known ADHD drugs and present an exploration of druggable genome associated with ADHD, which may offer intervention at the aetiological level of the disorder.
Supplementary information
Acknowledgements
We would like to express our profound gratitude to all individuals who have contributed DNA to all GWA studies. We also thank all individuals who have participated in collecting and analysing the genetic data, in addition to making the data available for the research community. We would especially like to thank Professor David A. van Heel and Risto Kajanne. We would also like to thank Benjamin Neale for his invaluable suggestions and discussions on this work. Our study was financed by Stiftelsen Kristian Gerhard Jebsen (SKGJ-MED02), the Western Norway Regional Health, Authorities (Helse Vest), the University of Bergen, The Norwegian national research network for ADHD, the European Union’s Horizon 2020 research and innovation programme under Grant Agreement No. 667302 (CoCA), Marie Sklodowska-Curie Grant Agreement No. 643051 (MiND) and the U.S. Department of Health & Human Services, National Institute of Mental Health, grant number 5U01MH109539-03 (Psychiatric Genomics Consortium (PGC)).
Disclaimer
This manuscript reflects only the authors’ view. The European Commission is not responsible for any use that may be made of the information it contain.
Compliance with ethical standards
Conflict of interest
During the past three years Jan Haavik has served as a speaker at meetings organised by Eli-Lilly, HB Pharma and Shire.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41380-019-0540-z) contains supplementary material, which is available to authorised users.
References
- 1.Faraone SV, Asherson P, Banaschewski T, Biederman J, Buitelaar JK, Ramos-Quiroga JA, et al. Attention-deficit/hyperactivity disorder. Nat Rev Dis Prim. 2015;1:15020. doi: 10.1038/nrdp.2015.20. [DOI] [PubMed] [Google Scholar]
- 2.Faraone SV, Larsson H. Genetics of attention deficit hyperactivity disorder. Mol Psychiatry. 2018. [DOI] [PMC free article] [PubMed]
- 3.Franke B, Michelini G, Asherson P, Banaschewski T, Bilbow A, Buitelaar JK, et al. Live fast, die young? A review on the developmental trajectories of ADHD across the lifespan. Eur Neuropsychopharmacol. 2018;28:1059–88. doi: 10.1016/j.euroneuro.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harpin VA. The effect of ADHD on the life of an individual, their family, and community from preschool to adult life. Arch Dis Child. 2005;90(Suppl 1):i2–7. doi: 10.1136/adc.2004.059006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohr-Jensen C, Steinhausen HC. A meta-analysis and systematic review of the risks associated with childhood attention-deficit hyperactivity disorder on long-term outcome of arrests, convictions, and incarcerations. Clin Psychol Rev. 2016;48:32–42. doi: 10.1016/j.cpr.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Lichtenstein P, Halldner L, Zetterqvist J, Sjolander A, Serlachius E, Fazel S, et al. Medication for attention deficit-hyperactivity disorder and criminality. New Engl J Med. 2012;367:2006–14. doi: 10.1056/NEJMoa1203241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalsgaard S, Ostergaard SD, Leckman JF, Mortensen PB, Pedersen MG. Mortality in children, adolescents, and adults with attention deficit hyperactivity disorder: a nationwide cohort study. Lancet. 2015;385:2190–6. doi: 10.1016/S0140-6736(14)61684-6. [DOI] [PubMed] [Google Scholar]
- 8.Wesemann D, Van Cleve SN. ADHD: From childhood to young adulthood. Nurse Pract. 2018;43:8–15. doi: 10.1097/01.NPR.0000530307.76316.cf. [DOI] [PubMed] [Google Scholar]
- 9.FDA-approved drugs to treat ADHD. J Psychosoc Nurs Ment Health Serv. 2018;56:13–4. https://www.ncbi.nlm.nih.gov/pubmed/29505085. [DOI] [PubMed]
- 10.Faraone SV. The pharmacology of amphetamine and methylphenidate: relevance to the neurobiology of attention-deficit/hyperactivity disorder and other psychiatric comorbidities. Neurosci Biobehav Rev. 2018;87:255–70. doi: 10.1016/j.neubiorev.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown KA, Samuel S, Patel DR. Pharmacologic management of attention deficit hyperactivity disorder in children and adolescents: a review for practitioners. Transl Pediatr. 2018;7:36–47. doi: 10.21037/tp.2017.08.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pauly V, Frauger E, Lepelley M, Mallaret M, Boucherie Q, Micallef J. Patterns and profiles of methylphenidate use both in children and adults. Br J Clin Pharm. 2018;84:1215–27. doi: 10.1111/bcp.13544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heal DJ, Smith SL, Gosden J, Nutt DJ. Amphetamine, past and present—a pharmacological and clinical perspective. J Psychopharmacol. 2013;27:479–96. doi: 10.1177/0269881113482532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sternat T, Katzman MA. Neurobiology of hedonic tone: the relationship between treatment-resistant depression, attention-deficit hyperactivity disorder, and substance abuse. Neuropsychiatr Dis Treat. 2016;12:2149–64. doi: 10.2147/NDT.S111818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diogo D, Tian C, Franklin CS, Alanne-Kinnunen M, March M, Spencer CCA, et al. Phenome-wide association studies across large population cohorts support drug target validation. Nat Commun. 2018;9:4285. doi: 10.1038/s41467-018-06540-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cortese S, Adamo N, Del Giovane C, Mohr-Jensen C, Hayes AJ, Carucci S, et al. Comparative efficacy and tolerability of medications for attention-deficit hyperactivity disorder in children, adolescents, and adults: a systematic review and network meta-analysis. Lancet Psychiatry. 2018;5:727–38. doi: 10.1016/S2215-0366(18)30269-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu Y, Sjolander A, Cederlof M, D’Onofrio BM, Almqvist C, Larsson H, et al. Association between medication use and performance on higher education entrance tests in individuals with attention-deficit/hyperactivity disorder. JAMA Psychiatry. 2017;74:815–22. doi: 10.1001/jamapsychiatry.2017.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arnold LE, Hodgkins P, Caci H, Kahle J, Young S. Effect of treatment modality on long-term outcomes in attention-deficit/hyperactivity disorder: a systematic review. PloS One. 2015;10:e0116407. doi: 10.1371/journal.pone.0116407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fredriksen M, Halmoy A, Faraone SV, Haavik J. Long-term efficacy and safety of treatment with stimulants and atomoxetine in adult ADHD: a review of controlled and naturalistic studies. Eur Neuropsychopharmacol. 2013;23:508–27. doi: 10.1016/j.euroneuro.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 20.Chang Z, Quinn PD, Hur K, Gibbons RD, Sjolander A, Larsson H, et al. Association between medication use for attention-deficit/hyperactivity disorder and risk of motor vehicle crashes. JAMA Psychiatry. 2017;74:597–603. doi: 10.1001/jamapsychiatry.2017.0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rostain A, Jensen PS, Connor DF, Miesle LM, Faraone SV. Toward quality care in ADHD: defining the goals of treatment. J Atten Disord. 2015;19:99–117. doi: 10.1177/1087054712473835. [DOI] [PubMed] [Google Scholar]
- 22.Faraone SV, Rostain AL, Blader J, Busch B, Childress AC, Connor DF, et al. Practitioner review: Emotional dysregulation in attention-deficit/hyperactivity disorder—implications for clinical recognition and intervention. J Child Psychol Psychiatry Allied Discip. 2018;60:133–50. doi: 10.1111/jcpp.12899. [DOI] [PubMed] [Google Scholar]
- 23.Charach A, Figueroa M, Chen S, Ickowicz A, Schachar R. Stimulant treatment over 5 years: effects on growth. J Am Acad Child Adolesc Psychiatry. 2006;45:415–21. doi: 10.1097/01.chi.0000199026.91699.20. [DOI] [PubMed] [Google Scholar]
- 24.Swanson JM, Elliott GR, Greenhill LL, Wigal T, Arnold LE, Vitiello B, et al. Effects of stimulant medication on growth rates across 3 years in the MTA follow-up. J Am Acad Child Adolesc Psychiatry. 2007;46:1015–27. doi: 10.1097/chi.0b013e3180686d7e. [DOI] [PubMed] [Google Scholar]
- 25.Faraone SV, Biederman J, Morley CP, Spencer TJ. Effect of stimulants on height and weight: a review of the literature. J Am Acad Child Adolesc Psychiatry. 2008;47:994–1009. doi: 10.1097/CHI.ObO13e31817eOea7. [DOI] [PubMed] [Google Scholar]
- 26.Gintant GA, George CH. Introduction to biological complexity as a missing link in drug discovery. Exp Opin Drug Discov. 2018;13:753–63. doi: 10.1080/17460441.2018.1480608. [DOI] [PubMed] [Google Scholar]
- 27.Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, et al. 10 years of GWAS discovery: biology, function, and translation. Am J Hum Genet. 2017;101:5–22. doi: 10.1016/j.ajhg.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lencz T, Malhotra AK. Targeting the schizophrenia genome: a fast track strategy from GWAS to clinic. Mol Psychiatry. 2015;20:820–6. doi: 10.1038/mp.2015.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finan C, Gaulton A, Kruger FA, Lumbers RT, Shah T, Engmann J et al. The druggable genome and support for target identification and validation in drug development. Sci Transl Med. 2017;9. pii: eaag1166. 10.1126/scitranslmed.aag1166. [DOI] [PMC free article] [PubMed]
- 30.Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, et al. Discovery of the first genome-wide significant risk loci for ADHD. Nat Genet. 2018;2017:14551–14543. doi: 10.1038/s41588-018-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang K, Gaitsch H, Poon H, Cox NJ, Rzhetsky A. Classification of common human diseases derived from shared genetic and environmental determinants. Nat Genet. 2017;49:1319–25. doi: 10.1038/ng.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Instanes JT, Klungsoyr K, Halmoy A, Fasmer OB, Haavik J. Adult ADHD and comorbid somatic disease: a systematic literature review. J Atten Disord. 2018;22:203–28. doi: 10.1177/1087054716669589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammerschlag AR, Stringer S, de Leeuw CA, Sniekers S, Taskesen E, Watanabe K, et al. Genome-wide association analysis of insomnia complaints identifies risk genes and genetic overlap with psychiatric and metabolic traits. Nat Genet. 2017;49:1584–92. doi: 10.1038/ng.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaspar HA, Breen G. Drug enrichment and discovery from schizophrenia genome-wide association results: an analysis and visualisation approach. Sci Rep. 2017;7:12460. doi: 10.1038/s41598-017-12325-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barbeira AN, Dickinson SP, Bonazzola R, Zheng J, Wheeler HE, Torres JM, et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun. 2018;9:1825. doi: 10.1038/s41467-018-03621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carithers LJ, Moore HM. The Genotype-Tissue Expression (GTEx) project. Biopreserv Biobank. 2015;13:307–8. doi: 10.1089/bio.2015.29031.hmm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic acids Res. 2015;43(Database issue):D1049–1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elia J, Ungal G, Kao C, Ambrosini A, De Jesus-Rosario N, Larsen L, et al. Fasoracetam in adolescents with ADHD and glutamatergic gene network variants disrupting mGluR neurotransmitter signaling. Nat Commun. 2018;9:4. doi: 10.1038/s41467-017-02244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Um JW, Ko J. LAR-RPTPs: synaptic adhesion molecules that shape synapse development. Trends Cell Biol. 2013;23:465–75. doi: 10.1016/j.tcb.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 42.Kolkman MJ, Streijger F, Linkels M, Bloemen M, Heeren DJ, Hendriks WJ, et al. Mice lacking leukocyte common antigen-related (LAR) protein tyrosine phosphatase domains demonstrate spatial learning impairment in the two-trial water maze and hyperactivity in multiple behavioural tests. Behav Brain Res. 2004;154:171–82. doi: 10.1016/j.bbr.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 43.Fisher D, Xing B, Dill J, Li H, Hoang HH, Zhao Z, et al. Leukocyte common antigen-related phosphatase is a functional receptor for chondroitin sulfate proteoglycan axon growth inhibitors. J Neurosci: Off J Soc Neurosci. 2011;31:14051–66. doi: 10.1523/JNEUROSCI.1737-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cotto KC, Wagner AH, Feng YY, Kiwala S, Coffman AC, Spies G, et al. DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic Acids Res. 2017;46:D1068–D1073. doi: 10.1093/nar/gkx1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sublingual glycine vs. placebo on attentional difficulties and hyperactivity in prepuberal children. https://clinicaltrials.gov/ct2/show/NCT02655276.
- 46.Tzang RF, Chang YC, Tsai GE, Lane HY. Sarcosine treatment for oppositional defiant disorder symptoms of attention deficit hyperactivity disorder children. J Psychopharmacol. 2016;30:976–82. doi: 10.1177/0269881116658986. [DOI] [PubMed] [Google Scholar]
- 47.Takahashi S, Inamura K, Yarimizu J, Yamazaki M, Murai N, Ni K. Neurochemical and neuropharmacological characterization of ASP2905, a novel potent selective inhibitor of the potassium channel KCNH3. Eur J Pharm. 2017;810:26–35. doi: 10.1016/j.ejphar.2017.05.045. [DOI] [PubMed] [Google Scholar]
- 48.Bever CT, Judge SI. Sustained-release fampridine for multiple sclerosis. Exp Opin Investig Drugs. 2009;18:1013–24. doi: 10.1517/13543780903002082. [DOI] [PubMed] [Google Scholar]
- 49.Blight AR, Henney HR, 3rd, Cohen R. Development of dalfampridine, a novel pharmacologic approach for treating walking impairment in multiple sclerosis. Ann N Y Acad Sci. 2014;1329:33–44. doi: 10.1111/nyas.12512. [DOI] [PubMed] [Google Scholar]
- 50.Miyake A, Takahashi S, Nakamura Y, Inamura K, Matsumoto S, Mochizuki S, et al. Disruption of the ether-a-go-go K+ channel gene BEC1/KCNH3 enhances cognitive function. J Neurosci. 2009;29:14637–45. doi: 10.1523/JNEUROSCI.0901-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ucar B, Humpel C. Collagen for brain repair: therapeutic perspectives. Neural Regen Res. 2018;13:595–8. doi: 10.4103/1673-5374.230273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.George SR, O’Dowd BF, Lee SP. G-protein-coupled receptor oligomerization and its potential for drug discovery. Nat Rev Drug Discov. 2002;1:808–20. doi: 10.1038/nrd913. [DOI] [PubMed] [Google Scholar]
- 53.Milligan G. G protein-coupled receptor hetero-dimerization: contribution to pharmacology and function. Br J Pharm. 2009;158:5–14. doi: 10.1111/j.1476-5381.2009.00169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neufeld G, Sabag AD, Rabinovicz N, Kessler O. Semaphorins in angiogenesis and tumor progression. Cold Spring Harb Perspect Med. 2012;2:a006718. doi: 10.1101/cshperspect.a006718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dupain C, Harttrampf AC, Boursin Y, Lebeurrier M, Rondof W, Robert-Siegwald G, et al. Discovery of new fusion transcripts in a cohort of pediatric solid cancers at relapse and relevance for personalized medicine. Mol Ther. 2019;27:200–18. doi: 10.1016/j.ymthe.2018.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Worzfeld T, Offermanns S. Semaphorins and plexins as therapeutic targets. Nat Rev Drug Discov. 2014;13:603–21. doi: 10.1038/nrd4337. [DOI] [PubMed] [Google Scholar]
- 57.Wainberg M, Sinnott-Armstrong N, Mancuso N, Barbeira AN, Knowles DA, Golan D, et al. Opportunities and challenges for transcriptome-wide association studies. Nat Genet. 2019;51:592–9. doi: 10.1038/s41588-019-0385-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–8. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gaulton KJ, Ferreira T, Lee Y, Raimondo A, Magi R, Reschen ME, et al. Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat Genet. 2015;47:1415–25. doi: 10.1038/ng.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. New Engl J Med. 2010;363:1211–21. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–86. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hegvik TA, Instanes JT, Haavik J, Klungsoyr K, Engeland A. Associations between attention-deficit/hyperactivity disorder and autoimmune diseases are modified by sex: a population-based cross-sectional study. Eur Child Adolesc Psychiatry. 2017;27:677. doi: 10.1007/s00787-017-1087-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paternoster L, Standl M, Waage J, Baurecht H, Hotze M, Strachan DP, et al. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat Genet. 2015;47:1449–56. doi: 10.1038/ng.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–81. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen MH, Su TP, Chen YS, Hsu JW, Huang KL, Chang WH, et al. Comorbidity of allergic and autoimmune diseases among patients With ADHD. J Atten Disord. 2017;21:219–27. doi: 10.1177/1087054712474686. [DOI] [PubMed] [Google Scholar]
- 67.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H, et al. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet. 2019;51:431–44. doi: 10.1038/s41588-019-0344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Diallo FB, Fombonne E, Kisely S, Rochette L, Vasiliadis HM, Vanasse A, et al. Prevalence and Correlates of Autism Spectrum Disorders in Quebec: prevalence et correlats des troubles du spectre de l’autisme au Quebec. Can J Psychiatry. 2018;63:231–9. doi: 10.1177/0706743717737031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Supekar K, Iyer T, Menon V. The influence of sex and age on prevalence rates of comorbid conditions in autism. Autism Res. 2017;10:778–89. doi: 10.1002/aur.1741. [DOI] [PubMed] [Google Scholar]
- 71.Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–7. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.International League Against Epilepsy Consortium on Complex Epilepsies. Genetic determinants of common epilepsies: a meta-analysis of genome-wide association studies. Lancet Neurol. 2014;13:893–903. doi: 10.1016/S1474-4422(14)70171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brikell I, Ghirardi L, D’Onofrio BM, Dunn DW, Almqvist C, Dalsgaard S, et al. Familial liability to epilepsy and attention-deficit/hyperactivity disorder: a nationwide cohort study. Biol Psychiatry. 2018;83:173–80. doi: 10.1016/j.biopsych.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50:668–81. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gormley P, Anttila V, Winsvold BS, Palta P, Esko T, Pers TH, et al. Meta-analysis of 375,000 individuals identifies 38 susceptibility loci for migraine. Nat Genet. 2016;48:856–66. doi: 10.1038/ng.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Okbay A, Beauchamp JP, Fontana MA, Lee JJ, Pers TH, Rietveld CA, et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016;533:539–42. doi: 10.1038/nature17671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lane JM, Liang J, Vlasac I, Anderson SG, Bechtold DA, Bowden J, et al. Genome-wide association analyses of sleep disturbance traits identify new loci and highlight shared genetics with neuropsychiatric and metabolic traits. Nat Genet. 2017;49:274–81. doi: 10.1038/ng.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Okbay A, Baselmans BM, De Neve JE, Turley P, Nivard MG, Fontana MA, et al. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet. 2016;48:624–33. doi: 10.1038/ng.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.