Abstract
Hereditary hemorrhagic telangiectasia (HHT) is an autosomal dominant vascular disorder characterized by development of high-flow arteriovenous malformations (AVMs) that can lead to stroke or high-output heart failure. HHT2 is caused by heterozygous mutations in ACVRL1, which encodes an endothelial cell bone morphogenetic protein (BMP) receptor, ALK1. BMP9 and BMP10 are established ALK1 ligands. However, the unique and overlapping roles of these ligands remain poorly understood. To define the physiologically relevant ALK1 ligand(s) required for vascular development and maintenance, we generated zebrafish harboring mutations in bmp9 and duplicate BMP10 paralogs, bmp10 and bmp10-like. bmp9 mutants survive to adulthood with no overt phenotype. In contrast, combined loss of bmp10 and bmp10-like results in embryonic lethal cranial AVMs indistinguishable from acvrl1 mutants. However, despite embryonic functional redundancy of bmp10 and bmp10-like, bmp10 encodes the only required Alk1 ligand in the juvenile-to-adult period. bmp10 mutants exhibit blood vessel abnormalities in anterior skin and liver, heart dysmorphology, and premature death, and vascular defects correlate with increased cardiac output. Together, our findings support a unique role for Bmp10 as a non-redundant Alk1 ligand required to maintain the post-embryonic vasculature and establish zebrafish bmp10 mutants as a model for AVM-associated high-output heart failure, which is an increasingly recognized complication of severe liver involvement in HHT2.
Keywords: hereditary hemorrhagic telangiectasia, bone morphogenetic protein, vascular development, zebrafish
Introduction
Hereditary hemorrhagic telangiectasia (HHT) is an autosomal dominant vascular disorder that presents with variable age of onset and expressivity. The disease phenotype is characterized by development of arteriovenous malformations (AVMs), which are direct connections between arteries and veins that bypass the capillary bed [1]. Because capillaries are required for gas exchange and to slow blood flow as it travels from arteries to veins, AVMs may lead to tissue hypoxia and hemorrhage.
HHT is associated with decreased quality of life and may be associated with decreased life expectancy [2–4]. In HHT patients, AVMs in small caliber vessels (telangiectasias) in the nose and gastrointestinal tract may lead to hemorrhage and anemia. Pulmonary AVMs may lead to dyspnea, brain abscess, or embolic stroke, whereas liver AVMs may lead to decreased systemic vascular resistance and high-output heart failure (HOHF). While cautery, resection, or embolization can be effective treatments for telangiectasias or pulmonary AVMs, the primary treatment option for HHT patients with severe liver involvement and HOHF is liver transplantation [5]. Intravenous administration of antiangiogenics has shown some therapeutic value in HHT patients [6–8], but these therapies globally depress angiogenesis. As such, there is an unmet need for therapeutics that directly target the molecular defect that underlies HHT-associated AVMs.
HHT is caused by defects in bone morphogenetic protein (BMP) signaling in endothelial cells. In BMP signaling [9], extracellular ligands bind with the aid of non-signaling type III receptors to a complex of type II and type I transmembrane receptor serine/threonine kinases. The type II receptors phosphorylate and activate the type I receptors, which phosphorylate SMAD proteins (SMAD1, SMAD5, SMAD8). Phosphorylated SMADs (pSMADs) complex with SMAD4, enter the nucleus, bind to DNA, and regulate gene expression. Mutations in endoglin (ENG; type III receptor), ACVRL1 (or ALK1; type I receptor), and SMAD4 cause HHT1, HHT2, and juvenile polyposis with HHT, respectively [10–12]. ENG and ACVRL1 mutations account for 85–96% of HHT [13].
BMP9 and BMP10 are high affinity ligands for ALK1 and ENG that similarly induce expression of downstream target genes [14–19]. In mice, Bmp9 is synthesized in the liver by embryonic day (E) 9.75–10, with expression localized to stellate cells in the adult [20–24]. Bmp10 predominates in the heart, with expression restricted to ventricular trabecular myocardium at E8.0, atrial myocardium by E16.5, and right atrial myocardium in adults [25, 26]. Both proteins are detected in mouse and human peripheral blood [21, 23, 27–29], suggesting that they might act redundantly as endocrine ligands for endothelial ALK1. Yet, whereas Bmp9 null mice have no overt embryonic blood vessel phenotype and live to adulthood [23, 27], Bmp10 nulls develop embryonic lethal AVMs, phenocopying Acvrl1 mutants [23], These observations suggest a unique embryonic requirement for Bmp10 as the critical Alk1 ligand. However, there is a short developmental window (E8.0-E9.75) during which only Bmp10 and not Bmp9 is expressed, which could mask functional redundancy. In fact, functional redundancy is supported by observations that embryonic AVMs in Bmp10 nulls are rescued by inserting Bmp9 into the Bmp10 locus [23], and that neonatal retinal AVMs develop upon simultaneous blockade of Bmp9 and Bmp10 [30–32]. However, post-embryonic ligand requirements outside of the retina have not been explored.
Because HHT is a heterozygous condition, ligand-based therapeutics that increase signaling through wild type receptors may ameliorate disease phenotype. The utility of this approach is predicated on understanding ALK1 ligand requirements in different vascular beds throughout life. To this end, we took advantage of the embryonic redundancy of zebrafish duplicate paralogs of BMP10—bmp10 and bmp10-like—to explore Alk1 ligand requirements through adulthood. Zebrafish acvrl1 mutants develop embryonic lethal AVMs, and simultaneous knockdown of bmp10 and bmp10-like, but not bmp9, phenocopies acvrl1 mutants [33–35]. In this work, we generated zebrafish bmp9, bmp10, and bmp10-like mutants, all of which are viable. Although bmp9 or bmp10-like mutants develop no phenotype, bmp10 mutants develop external vascular lesions at similar sites to HHT patients, with variable age of onset and expressivity. Moreover, bmp10 mutants display liver vascular malformations, ventricular enlargement, and functional evidence of high-output heart failure, reminiscent of HHT patients. Taken together, our results support a unique and unexpected requirement for Bmp10 in endocrine activation of endothelial Alk1 throughout life, a requirement that must be considered in development of ligand-based therapy for HHT.
Materials and Methods
Zebrafish maintenance
All zebrafish (Danio rerio) experiments conformed to NIH guidelines and were approved by the University of Pittsburgh Institutional Animal Care and Use Committee. Adults were maintained and bred and embryos reared according to standard protocols [36]. For experiments requiring analysis of adult phenotypes, we reared fish at a density of 40 fish/2.8 L tank from 5 days post-fertilization (dpf) until 5–6 weeks, then decreased density to 20 fish/2.8L tank, or ~7 fish/L. To monitor phenotype progression, we transferred phenotypic fish to a new tank upon onset of phenotype and supplemented with fluorescent age-matched “Starfire Red” fish to maintain density at 7 fish/L.
Zebrafish lines
Mutant lines acvrl1ft09e and acvrl1y6 [33] and transgenic lines Tg(kdrl:GFP)la116 (endothelial cells) [37]; Tg(acta2:mCherry)ca8 (smooth muscle cells) [38]; Tg(flt1:tdTomato)hu5333 (arterial endothelial cells) [39]; Tg(flt4:Citrine)hu7135 (venous endothelial cells) [40]; Tg(myl7:actn3b-EGFP)sd10 (α-actinin 3B-EGFP fusion; marks sarcomeric Z-discs) and Tg(myl7:Mkate-CAAX)sd11 (cardiomyocyte membranes) [41]; Tg(myl7:EGFP)twu34 (cardiomyocytes) [42]; and Tg(myl7:DsRed2-NLS)f2 (cardiomyocyte nuclei) [43] have been described. bmp10-likesa11654 was obtained from the Sanger Institute Zebrafish Mutation Project [44].
We used genome editing to target bmp9 [also known as gdf2; chromosome 12; ENSDARG00000059173; ENSDART00000082220.5/NM_01171586 (1170 bp); F1QWZ4/NP_001165057 (389 amino acids)], bmp10 [chromosome 5; ENSDARG00000061769; ENSDART00000088492.5/NM_001130600 (1458 bp); A2BGK2/NP_001124072 (485 amino acids)], and bmp10-like [chromosome 10; ENSDARG00000109233, ENSDART00000192123.1 (1221 bp), A0A2R8RLC3 (406 amino acids]). To generate bmp9/gdf2 (pt533, pt536) and bmp10 (pt527, pt543) mutants, we injected into 1-celll stage embryos 250 pg capped, polyadenylated synthetic mRNA (mMessage mMachine T7 Ultra, Thermo Fisher, Waltham, MA USA) synthesized from pairs of TALEN-encoding plasmids, following established protocols [45]. Plasmids were designed and generated by Dr. Keith Joung (Harvard, Cambridge, MA USA) and deposited at Addgene (Watertown, MA USA; Supplementary Table 1). To generate additional bmp10-like mutant alleles (pt544, pt545), we synthesized a CRISPR single guide RNA (MegashortScript, Thermo Fisher) from a synthetic DNA template (Supplementary Table 1) inserted into plasmid DR274 (Addgene) along with 600 ng Cas9 mRNA (mMessage mMachine T7 Ultra, Thermo Fisher) generated from plasmid MLM3613 (Addgene). In all cases, we identified P0 founders by outcrossing and genotyping offspring by PCR and restriction digest and/or sequencing. F1 generations were outcrossed to isolate single alleles. All genotypes were confirmed using assays provided in Supplementary Table 2. Mutation nomenclature is consistent with the recommendations of the Human Genome Variation Society [46].
For bmp9, we used TALENs targeting coding nucleotides 122–180 (exon 1) and propagated two mutant lines (Fig. 1a). Allele pt533 harbors a 9-bp deletion/1-bp insertion, generating a frameshift at amino acid 52 and premature termination codon within the prodomain at amino acid 84 (c.155_163delinsC p.Phe52Serfs*33). Allele pt536 harbors a 7-bp deletion, generating a premature termination codon within the prodomain at amino acid 54 (c.159_165del p.Gly54*). To confirm the predicted effects of these mutations and assess nonsense-mediated decay (NMD), we synthesized cDNA from pooled embryos from heterozygous incrosses, PCR amplified across the mutation, and cloned and sequenced the PCR product. Sequencing confirmed the presence of the predicted mutant transcripts in 4/6 (pt533) and 5/6 (pt536) cDNA clones, providing no evidence of NMD.
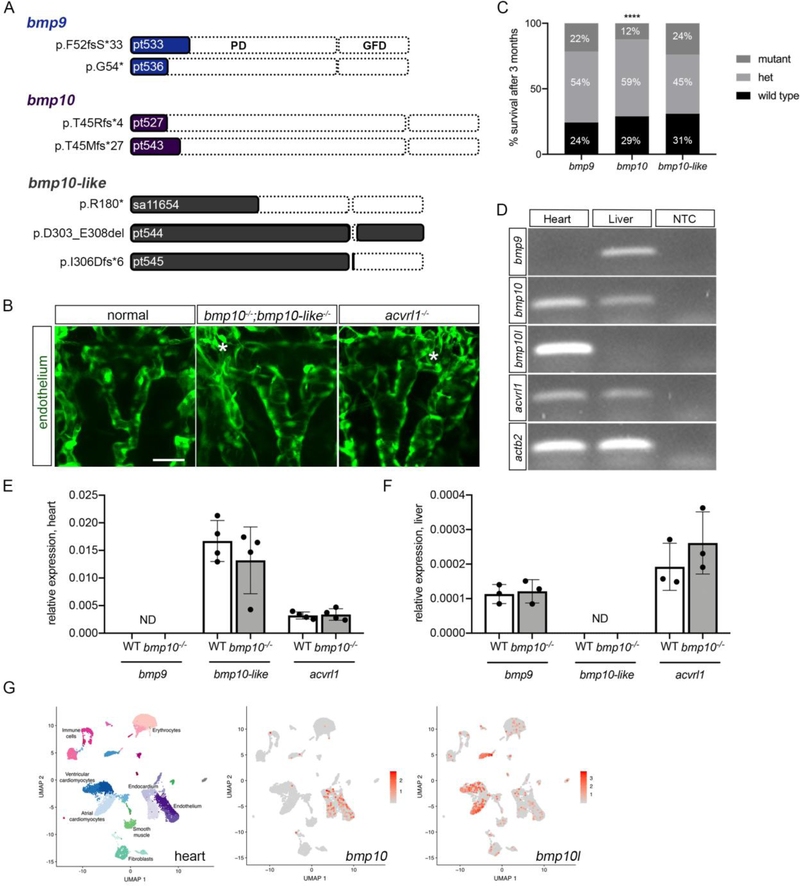
Fig. 1. Duplicate bmp10 paralogs are redundant during embryogenesis but not in adulthood.
a Schematics indicating zebrafish wild type Bmp9, Bmp10, and Bmp10-like protein structures and mutant alleles. For each schematic, mutation nomenclature is indicated to the left (following conventions of the Human Genome Variation Society), allele name is indicated within the shaded region, and truncation or deletion is indicated by the unshaded region. PD, prodomain; GFD, growth factor domain; fs, frame shift; *, premature termination codon; del, deletion. b Cranial vasculature in 48 hpf Tg(kdrl:GFP)la116 embryos from a bmp10pt527/+;bmp10-likesa11654/+ incross. “Normal” is bmp10pt527/+ and representative of all genotypes except double mutants. Asterisks denote AVMs in bmp10pt527/pt527;bmp10-likesa11654/;sa11654 double mutants and acvrl1y6/y6 mutants. Endothelial cells are green. 2D confocal projections, dorsal views, anterior up. Scale bar: 50 μm. Confocal images representative of 6 non-double mutants and 2 double mutants from a bmp10;bmp10-like double heterozygous incross. c Genotypes in adult offspring from bmp9, bmp10, and bmp10-like heterozygous incrosses. Data aggregated across all alleles within a single gene. bmp9: N = 4 clutches, 144 fish; bmp10: N = 25 clutches, 952 fish; bmp10-like: N = 6 clutches, 197 fish. Chi-squared test, ****P<0.0001. d RT-PCR from 6-month pooled wild type hearts and livers. NTC, no template control. e,f qRT-PCR, 3-month hearts (e) and livers (f), showing no transcriptional adaptation of bmp9, bmp10-like, or acvrl1 in bmp10 mutants. N = 3–4 pools of tissue from 4–10 fish. WT, wild type. Bars represent mean ± SD. Not significant by unpaired t-test. ND, not detected. g UMAP analysis plots of single cell transcriptome of three adult zebrafish hearts (n=14,472 cells), from published data (GSE106121). Left, cell types are annotated and subclusters within cell types are indicated by shades of the same color. Middle and right, feature plots demonstrating scaled expression values of bmp10 and bmp10-like projected on the UMAP plot. bmp10 predominates in endocardium/endothelium, and bmp10-like in cardiomyocytes.
For bmp10, we used TALENs targeting coding nucleotides 110–162 (exon 1) and propagated two mutant lines (Fig. 1a). Allele pt527 harbors an 8-bp deletion, generating a frameshift at amino acid 45 and premature stop codon within the prodomain at amino acid 48 (c.132_139del p.Thr45Argfs*4). Allele pt543 harbors a 7-bp deletion, generating a frameshift at amino acid 45 and a premature stop codon within the prodomain at amino acid 71 (c.131_137del p.Thr45Metfs*27). We confirmed the presence of the predicted mutant transcripts in 6/11 (pt527) and 7/14 (pt543) cDNA clones as described above, providing no evidence for NMD.
For bmp10-like, we obtained an ethylnitrosourea-induced mutant allele, sa11654 [44], which harbors a point mutation in exon 2 that generates a premature stop codon within the prodomain at amino acid 180 (c.538C>T p.R180*) (Fig. 1a). We generated additional bmp10-like alleles using CRISPR/Cas9 to target coding nucleotides 900–919 within exon 2 (Fig. 1a). Allele pt544 harbors an 18-bp in-frame deletion within the growth factor domain (c.907_924del p.Asp303-Glu308del) that included cysteine 304, which is required for intramolecular disulfide bonding. Allele pt545 harbors an 8-bp deletion, generating a frameshift at amino acid 306 and premature stop codon within the growth factor domain at amino acid 311 (c.916_923del p.Ile306Aspfs*6). Sequencing of RT-PCR products generated from heterozygous incrosses confirmed the presence of all predicted mutant transcripts. Because all mutations are in exon 2 of this two-exon gene, we did not evaluate NMD.
Histology and Imaging
Adult fish and whole heart imaging and measurements
We imaged adult fish and whole hearts using an MVX-10 MacroView microscope and DP71 camera (Olympus America, Center Valley, PA, USA). We measured fish length (nose to base of fin) using hand-held calipers. We measured ventricle cross-sectional area by tracing the maximal ventricle outline from images of ventricles (atrium removed) positioned in 5% methylcellulose with bulbus arteriosus to the right.
Histological staining and imaging
We stained 5 μm sections of Bouin’s fixed (4°C overnight), paraffin embedded hearts with Acid Fuchsin-Orange G (AFOG), Hart’s elastin, and Movat’s pentachrome stain, using standard protocols. We imaged stained sections using a BX51 upright compound microscope (Olympus America) and DP71 camera.
Confocal imaging, live embryos
To prevent pigmentation, we transferred zebrafish embryos to 30% Danieau [17 mmol/L NaCl, 2 mmol/L KCl, 0.12 mmol/L MgSO4, 1.8 mmol/L Ca(NO3)2, 1.5 mmol/L HEPES] containing 0.003% phenylthiourea (PTU; Sigma, St. Louis, MO, USA) at ~23 hours post-fertilization (hpf). Just prior to imaging, we anesthetized embryos in 30% Danieau/0.003% PTU containing 0.16 g/L tricaine methanesulfonate (Pentair, Cary, North Carolina, USA) (to arrest movement) or 0.8 g/ L tricaine (to stop heartbeat) and mounted them in 1% SeaPlaque low melting temperature agarose (Lonza, Walkersville, MD USA). We collected Z-series (3 μm steps) using a TCS SP5 or TCS SP8 upright confocal microscope (Leica Microsystems, Wetzlar, Germany) outfitted a multi-line argon laser (488 nm for EGFP; 514 nm for citrine) and 561 nm diode (for dsRed, dsRed2, mCherry, tdTomato; mKATE); an HCX IRAPO L 25x/0.95 water-dipping objective; and spectral detectors. We scanned embryos at 400 Hz with 3x line averaging or 8000 Hz with 16x line averaging.
Confocal imaging, adults
For skin vessel imaging, we fixed fish in 4% paraformaldehyde overnight at 4°C and imaged the skin without dissection, securing the tail in 1% low-melt agarose. For liver vessel imaging, we dissected the entire gut and liver as a single entity, fixed the tissue overnight at 4°C in 4% paraformaldehyde, then dissected the liver away from the gut. We mounted the right liver lobe in 1% low-melt agarose and imaged the lateral face. For all imaging, we collected Z-series (3 μm steps) as described above using a Leica TCS SP5.
Image Analysis
Using the “measure” tool in Fiji (version: 2.0.0-rc-65/1.51u; build: 961c5f1b7f, National Institutes of Health, Bethesda, MD USA), we measured basal communicating artery (BCA) diameters on 2D confocal projections at three sites, between and on either side of the paired middle mesencephalic central arteries [47]. Each data point represents an average value from a single embryo. For trabeculation measurements, we identified a single 3-μm optical section at the level of the AV canal, and used the “polygon selections” tool and “perimeter measurement” in Fiji to trace both the convoluted trabecular surface (T) and the minimal chamber delineated by the tips of the trabeculae (C). From these data, we calculated trabeculation as (T-C)/C, with higher values indicating more trabeculation. For cardiomyocyte cell counts, we used Imaris (x64 version 9.1.2, Bitplane, Berlin, Germany) to generate 3D projections and applied the spot tool, with manual correction.
RT-PCR
We isolated total RNA from pooled zebrafish hearts or livers (RNeasy mini kit, Qiagen, Germantown, MD, USA), generated cDNA (SuperScript IV, ThermoFisher), and performed PCR with gene-specific primers (Supplementary Table 3) using Amplitaq Gold (Thermo Fisher). For quantitative RT-PCR, we ran TaqMan assays (Thermo Fisher) on a QuantStudio 12k Flex Real Time PCR System (Thermo Fisher). We validated all TaqMan assays using a dilution series of cDNA, according to manufacturer recommendations (amplification efficiency, 1.8–2.2). Zebrafish assays included inventoried eef1a1l1 (Dr03432748_m1) and acvrl1 (Dr03144495_m1), and custom assays designed to target exon-exon boundaries in bmp9/gdf2 (ARKA3UA), bmp10 (ARMFXD9), bmp10-like (ARMFXD7), nppa (ARKA3UF), and nppb (APRWFMN). We analyzed data using the ΔΔCt method and present results as expression normalized to eef1a1l1.
In situ hybridization
We synthesized antisense riboprobes from T7-containing, PCR-generated templates using a DIG RNA labeling kit (Sigma-Aldrich, St. Louis, MO USA). Primers for ltbp3, nkx2.5, and tbx20 templates are listed in Supplementary Table 4. The cxcr4a riboprobe has been described [34].
Ultrasound imaging and analysis
We anesthetized zebrafish in 0.12 g/L tricaine for 2 minutes, then placed the fish in a supine position in a groove cut into a sponge saturated with 0.096 g/L tricaine and waited 2 minutes before scanning. We performed ultrasound echocardiography using a high-frequency ultrasound small animal imaging system (Vevo 2100, FujiFilm VisualSonics, Toronto, Canada) outfitted with a high frequency linear array transducer, MS 700 (bandwidth 30–70 MHz; centered at 50 MHz operating frequency). We acquired images in two planes: the longitudinal (long) axis view, with the transducer positioned parallel to the long axis of the fish, and the short axis view, with the transducer positioned across the gills and heart orthogonally to the long axis. In each view, we acquired B-mode, color Doppler and pulsed-wave Doppler images. Image acquisition time averaged approximately 5 minutes, after which we transferred the fish to fresh water to allow recovery. All fish maintained heart rate over the course of scanning.
We performed image analysis using the manufacturer-provided software package, Vevo LAB (FujiFilm VisualSonics). We used color Doppler images to locate the valve between the ventricle and bulbus arteriosus [ventriculobulbar (VB) valve], acquired long axis B-mode images at this location, and measured the diameter of the VB valve region. We used VB valve diameter to calculate the cross-sectional area of the VB valve region, assuming circularity. Along the long axis, we used pulsed-wave Doppler to record heart rate and the ventricular outflow signal, and color Doppler to aid in determining the location of maximal blood flow. We computed the velocity-time integral (VTI) at the VB valve and averaged VTI over 3 cardiac cycles. We then calculated stroke volume as the VTI at the VB valve multiplied by the cross-sectional area of the VB valve, and cardiac output as stroke volume multiplied by heart rate. We normalized stroke volume and cardiac output to weight and combined data from males and females [48].
Analysis of single cell RNAseq data from adult zebrafish heart
Cell Ranger outputs (matrix.tsv, barcodes.tsv and genes.tsv) from three adult zebrafish hearts [49] were downloaded from Gene Expression Omnibus (GSE106121). We used the R package Seurat (v3.1.1) for clustering and cell type identification [50–52] . We excluded genes found in fewer than three cells, and removed cells that had fewer than 200 or greater than 4000 genes or more than 25% mitochondrial content. The latter cut-off was not stringent because cardiomyocytes express a high percentage of mitochondrial genes. We followed the standard workflow for integration as recommended in the Seurat vignette (https://satijalab.org/seurat/v3.1/integration.html). We independently preprocessed each heart dataset using log-normalization, identified variable genes based on a variance stabilizing transformation (vst), and identified anchors. We integrated the data from the three hearts using 4000 anchor genes, producing an expression matrix for all cells (n=14,472). We then scaled the data and performed a principal component analysis using the first 30 components. We used UMAP dimensionality reduction [53] to cluster and visualize the data. Using differential gene expression results from a likelihood-ratio test (Seurat FindAllMarkers function), we identified broad cell types using gene expression data from the ZFIN database (https://zfin.org/) (Supplementary Fig. 1, Supplementary Dataset 1). We exported UMAP coordinates, expression values from the integrated assay “data” slot, and metadata from the Seurat data object, and used data.table [54] and ggplot2 [55] to create UMAP figures demonstrating cluster identities. We used Seurat DotPlot function to display markers used for cluster identification and FeaturePlot to display expression values of bmp10 and bmp10l on UMAP plots, with expressing cells in the foreground.
Statistical analysis
We analyzed all data and graphs using Prism (version 8.0.1, GraphPad, San Diego, CA USA). Sample sizes, experimental repeats, and statistical tests are indicated in figure legends. To avoid bias, all subjective analyses and image analyses were either performed prior to genotyping or by a blinded observer. All data presented are genotypic wild type versus genotypic mutant siblings unless otherwise stated. We generated all figures using Photoshop CC 2017.1.0 release (Adobe, San Jose, CA USA). We applied exposure correction to some images for presentation purposes only.
Results
Generation of zebrafish Alk1 ligand mutants
To explore the unique and overlapping roles of Alk1 ligands in development and disease, we used genome editing to generate mutations in zebrafish bmp9 (also known as gdf2), bmp10, and bmp10-like. These two-exon genes encode ligands that are generated as proproteins (Fig. 1a), with a furin cleavage site separating the N-terminal prodomain from the C-terminal growth factor domain [56]. The released disulphide-bonded growth factor domain dimers are the active ligands in BMP signaling [57]. For each ligand-encoding gene, we assessed two or three mutant alleles, most of which generated frame shifts and/or premature termination codons that truncated proteins within the prodomain or the N-terminus of the growth factor domain (Fig. 1a). Additional details on allele identity and verification are provided in Materials and Methods.
bmp10;bmp10-like mutants phenocopy acvrl1 mutants
Zebrafish acvrl1 mutants develop cranial AVMs connecting the basal communicating artery (BCA) or basilar artery to neighboring veins [33, 34, 58]. This larval lethal phenotype is detectable by 40–48 hpf and is 100% penetrant. Using morpholinos, we previously demonstrated that the Alk1 ligands Bmp10 and Bmp10-like are redundant with respect to Alk1 signaling in embryonic vascular development, with concomitant knockdown phenocopying acvrl1 mutants [35]. Analysis of blood flow in ligand mutants confirms these results: whereas bmp9, bmp10, and bmp10-like single mutants do not develop AVMs (data not shown), bmp10;bmp10-like double mutants develop high-flow shunts in cranial vessels (from bmp10pt527/+;bmp10-likesa11654/+ incrosses, shunts scored in 43/45 double mutants and 1/575 non-double mutants, over 2 experiments). Via confocal microscopy, we confirmed that these shunts represent AVMs between the BCA or basilar artery and flanking veins (primordial midbrain or hindbrain channel), which are indistinguishable from AVMs in acvrl1 mutants (Fig. 1b). As in acvrl1 mutants, this phenotype is fully penetrant and larval lethal. In contrast, we identified no shunts in embryos from bmp9pt533/+;bmp10pt527/+ incrosses (N = 255 embryos, 2 experiments), a bmp9pt536/+;bmp10-likesa11654/+ incross (N = 630 embryos, 1 experiment), or a bmp9pt536/pt536;bmp10-likesa11654/+ x bmp9pt536/+;bmp10-likesa11654/sa11654 cross (N = 54 embryos, 1 experiment). These data confirm that bmp10 and bmp10-like encode functionally redundant embryonic Alk1 ligands, with no embryonic requirement for bmp9.
bmp10 mutants die prematurely
To examine Alk1 ligand requirements post-embryonically, we monitored single-mutants through adulthood. bmp9 and bmp10-like mutants develop no overt phenotype and all alleles show expected genotype ratios as adults (Fig. 1c). In contrast, we recovered significantly fewer bmp10 mutant adults than expected (Fig. 1c). The lack of compensation for bmp10 loss by other Alk1 ligands is not caused by postembryonic silencing of these genes: at 6 months, we detected bmp9 in the liver, bmp10-like in the heart, and bmp10 and acvrl1 in both heart and liver (Fig. 1d). These expression patterns concur with those of mouse Bmp9 and Bmp10 [20–26]. Quantitative measurement of gene expression in adult heart and liver demonstrated no upregulation or ectopic expression of bmp9 or bmp10-like and no change in acvrl1 expression in bmp10 mutants (Fig. 1e,f), suggesting no transcriptional adaptation or genetic compensation [59]. Analysis of published single-cell RNAseq data from adult zebrafish hearts [49] revealed expression of bmp10 predominantly in endocardium/endothelium and bmp10-like predominantly in cardiomyocytes (Fig. 1g), similar to observations in embryonic zebrafish [35]. Together, these results demonstrate distinct cellular localization and subfunctionalization of zebrafish BMP10 paralogs in the post-embryonic period and a unique requirement only for bmp10. Because all bmp10 alleles showed similar adult phenotypes, hereafter, all mutant data represent bmp10pt527.
Embryonic cranial vessels are transiently enlarged in bmp10 mutants
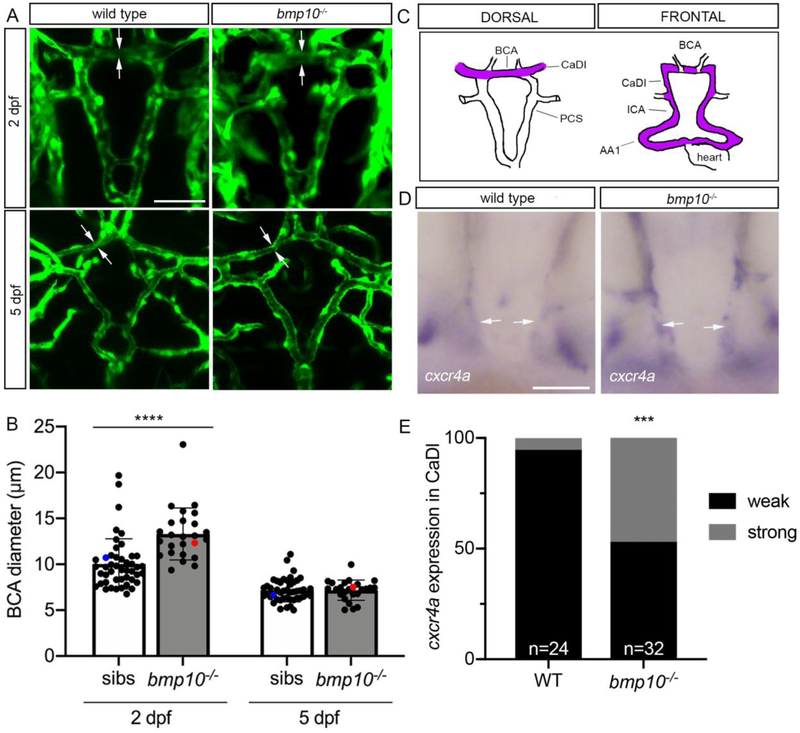
To understand the cause of premature death in bmp10 mutants, we first focused on the embryonic vasculature. Although we did not detect cranial shunts in bmp10 mutants, confocal imaging revealed enlargement of the acvrl1-positive BCA at 2 days post-fertilization (dpf) (Fig. 2a,b), a phenotype reminiscent of but less severe than bmp10/bmp10-like double mutants (Fig. 1b) and acvrl1 mutants [33, 34]. The 2 dpf BCA enlargement in bmp10 mutants correlated with increased cxcr4a staining in the caudal divisions of the internal carotid arteries (CaDI), which lead directly into the BCA (Fig. 2c–e). This increase in cxcr4a expression, characteristic of acvrl1 mutants [60], suggests decreased Alk1 activity. However, the bmp10 mutant cranial vascular phenotype is transient: longitudinal measurements revealed normal BCA diameter (Fig. 2a,b) and smooth muscle cell investment (Supplementary Fig. 2) of the BCA at 5 dpf. We speculate that the transient increase in arterial caliber stems from the later onset of bmp10-like expression [35], leaving a short window of time (~28–36 hpf) during which Alk1 signaling is inactive in bmp10 mutants. Based on our findings in acvrl1 mutants [58], this transient loss of Alk1 activity may allow limited migration of endothelial cells in the direction of blood flow and modest enlargement of distal arteries in bmp10 mutants.
Fig. 2. bmp10 mutant embryos develop transiently enlarged cranial vessels.
a Repeated imaging of cranial vessels in bmp10pt527;Tg(kdrl:GFP)la116 mutant and wild type siblings, 2 and 5 dpf. Endothelial cells are green. Arrows delineate width of basal communicating artery (BCA). 2D confocal projections, dorsal views, anterior up. Scale bar: 50 μm. b Quantification of BCA diameters. Blue dot, wild type embryo shown in a; red dot, mutant embryo shown in a. N = 46 non-mutant sibs, 25 mutants over 5 experiments. Bars represent mean ± SD. Repeated measures two-way ANOVA followed by Sidak’s multiple comparisons test, ****P<0.0001. c Wiring diagrams of cranial vessels, dorsal and frontal views, showing acvrl1-positive vessels in purple. AA1, first aortic arch; ICA, internal carotid artery; CaDI, caudal division of internal carotid artery; PCS, posterior communicating segments. d Whole-mount in situ hybridization for cxcr4a at 2 dpf, wild type and bmp10pt527 mutant. Arrows: CaDI. Frontal views, anterior top. Scale bar: 50 μm. e Qualitative evaluation of cxcr4a expression intensity in CaDI. WT, wild type. Fisher’s exact test, ***P<0.001.
Embryonic ventricular myocardium development is normal in bmp10 mutants
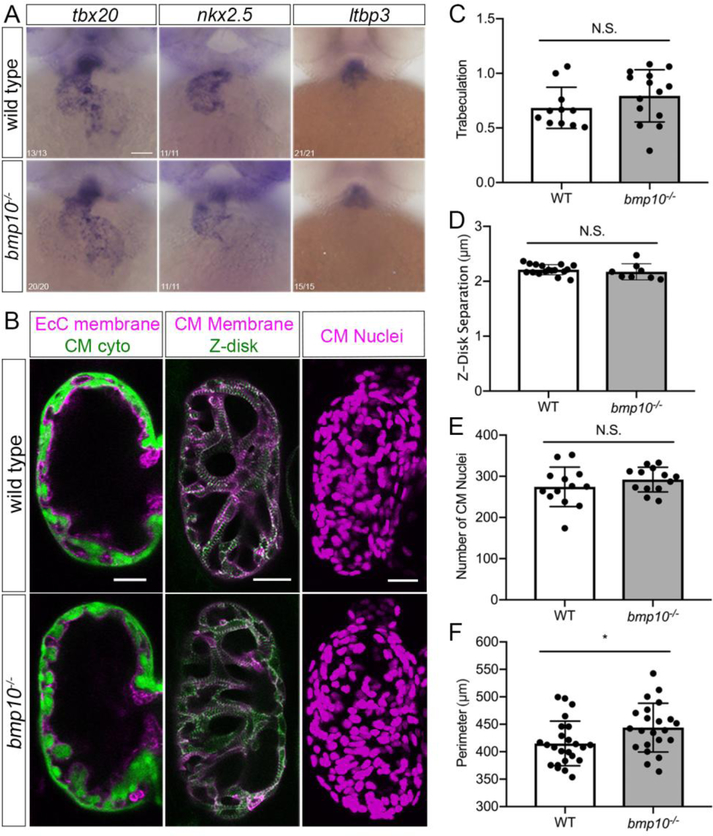
Because Bmp10 reportedly plays independent roles in embryonic heart and blood vessel development in mice [23], we next asked whether bmp10 mutants display embryonic heart defects that might contribute to post-embryonic lethality. In bmp10 mutants, early cardiac markers and putative downstream targets of Bmp10, tbx20 and nkx2.5 [23, 61, 62], are unaffected at 2 dpf (Fig. 3a). The second heart field marker, ltbp3 [63], is also unaffected (Fig. 3a). These observations, together with measurement of normal heart rate at 2 dpf (data not shown), suggest that the primitive heart forms and functions normally in bmp10 mutants.
Fig. 3. Embryonic ventricular myocardium development is unaffected bmp10 mutants.
a Whole-mount in situ hybridization for tbx20, nkx2.5, and ltbp3 at 2 dpf, wild type and bmp10pt527 mutant siblings. Ventral views, anterior top. Numbers in left-hand corner show number of embryos with shown phenotype over total number of embryos imaged over 3 experiments. Scale bar: 100 μm. b Ventricle morphology at 5 dpf in wild type and bmp10pt527 mutant siblings. Left panels, trabeculation: Tg(fli1a.ep:mRFP-CAAX)pt505;Tg(myl7:EGFP)twu34, endocardial cell (EcC) membranes magenta, cardiomyocyte (CM) cytoplasm green. Middle panels, sarcomeric architecture: Tg(myl7:mKATE-CAAX)sd11;Tg(myl7:actn3b-EGFP)sd10; CM membranes magenta, Actn3b (Z-disks) green. Right panels, CM number: Tg(myl7:dsRed2-NLS)f2; CM nuclei magenta. Confocal images, single planes (left, middle) or 2D projections (right). Scale bars: 25 μm. c Trabeculation [(trabecular trace-central chamber trace)/central chamber trace)] at 5 dpf, N = 11 wild types, 14 mutants over 4 experiments. d Z-disk separation, N = 17 wild types, 8 mutants over 5 experiments. e Number of CM nuclei, N = 13 wild types, 13 mutants over 4 experiments. f Ventricle outer perimeter, N = 24 wild type, 22 mutants over 8 experiments. Bars represent mean ± SD. Unpaired t-test, *P < 0.05, NS = not significant.
Trabeculation is the embryonic process by which the smooth heart wall generates folds that extend into the ventricle lumen and is required to generate a functionally competent, thickened ventricular wall [64]. Because mouse Bmp10 mutants develop hypoplastic ventricular myocardium that lacks trabeculae [25, 61, 65], we asked whether trabeculation defects might contribute to lethality in zebrafish bmp10 mutants. Zebrafish myocardium begins to trabeculate around 2.5 dpf at the outer curvature of the ventricle and by 5 dpf, a complex trabecular network is evident throughout the ventricle [66, 67]. However, we found no trabeculation defect in bmp10 mutant zebrafish at 5 dpf (Fig. 3b, c). Sarcomeric structure, as measured by distance between Z-disks (Fig. 3b, d), and cardiomyocyte number (Fig. 3b, e) were also unaffected, whereas ventricular outer perimeter (Fig. 3b, f) was slightly increased. Together, these data suggest that zebrafish bmp10 is not required for normal ventricular cardiomyocyte differentiation, proliferation, or trabeculation, possibly due to redundancy with bmp10-like.
bmp10 mutants develop external vascular phenotype with variable age of onset and expressivity
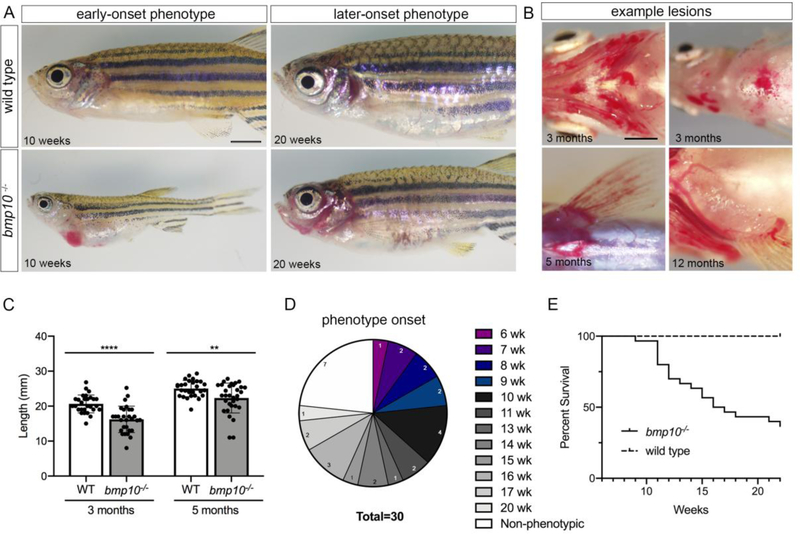
Next, we monitored bmp10 mutants for external phenotype and survival beginning at 6 weeks (juvenile period), prior to significant loss of bmp10 mutants. Earlier onset external phenotype (6–14 weeks) is characterized by abdominal edema and enlarged and/or hemorrhagic blood vessels in skin (Fig. 4a). Later onset external phenotype (15 weeks or later) is similar, but with less edema (Fig. 4a). The external vascular phenotype in bmp10 mutants predominates in anterior and ventral regions, including the face, gills, and pectoral fins (Fig. 4a,b). This phenotype is reminiscent of HHT patients, who present with telangiectasias in anterior skin and mucosal membranes and on the palmar surface of the hands, with variable age of onset and expressivity [68]. bmp10 mutants are smaller than their wild type siblings, particularly with earlier onset phenotype (Fig. 4a, c).
Fig. 4. bmp10 mutants develop external phenotype with variable age of onset and expressivity.
a Representative examples of early-onset (6–14 weeks; left) and later-onset (≥ 15 weeks; right) external phenotype in bmp10pt527 mutants, compared to wild type siblings. Lateral views, anterior left. Scale bars: 2 mm. b Dilated vessels and hemorrhages in pharyngeal arch arteries (top left), anterior abdomen (top right), and pectoral fins (bottom). Ventral views, anterior left. Scale bar: 1 mm. c Fish lengths at 3 months (N = 28 wild types, 29 mutants from 7 independent lines) and 5 months (N = 28 wild types, 32 mutants from 6 independent lines). Data include phenotypic and non-phenotypic mutants. Bars represent mean ± SD. Unpaired t-test,**P < 0.01; ****P < 0.0001. d-e Phenotype onset and survival in 1 clutch containing 30 bmp10pt527 mutants and 33 wild type siblings. d Age at onset of external phenotype. Numbers within chart indicate number of fish that became phenotypic during the indicated week, or remained non-phenotypic at 22 weeks. e Survival in bmp10 mutants and sibling wild types. Log-rank (Mantel-Cox) test, ****P < 0.0001.
To characterize variability in age of onset and expressivity, we tracked phenotype and survival in bmp10 mutant and wild type siblings from 6 until 22 weeks of age. We first observed external phenotype in mutants at 6 weeks and identified on average approximately 2 newly phenotypic fish per week, with 7/30 fish (23.3%) remaining non-phenotypic at 22 weeks (Fig. 4d). Only 37% of bmp10 mutants survived until 22 weeks compared to 100% of their wild type siblings (Fig. 4e). During the course of this study, we identified three acvrl1ft09e heterozygous adult zebrafish that phenocopied bmp10 mutants, with enlarged and hemorrhagic vasculature in anterior and ventral skin and gills (Supplementary Fig. 3). We speculate that the low penetrance of this acvrl1 heterozygous phenotype (ascertained anecdotally) may reflect the rare acquisition of second somatic hits that drive lesion development, as recently observed in HHT patient telangiectasias [69]. In sum, our data support the assertion that bmp10 encodes an Alk1 ligand that is required for vessel maintenance throughout life.
bmp10 mutants develop skin and liver vascular abnormalities
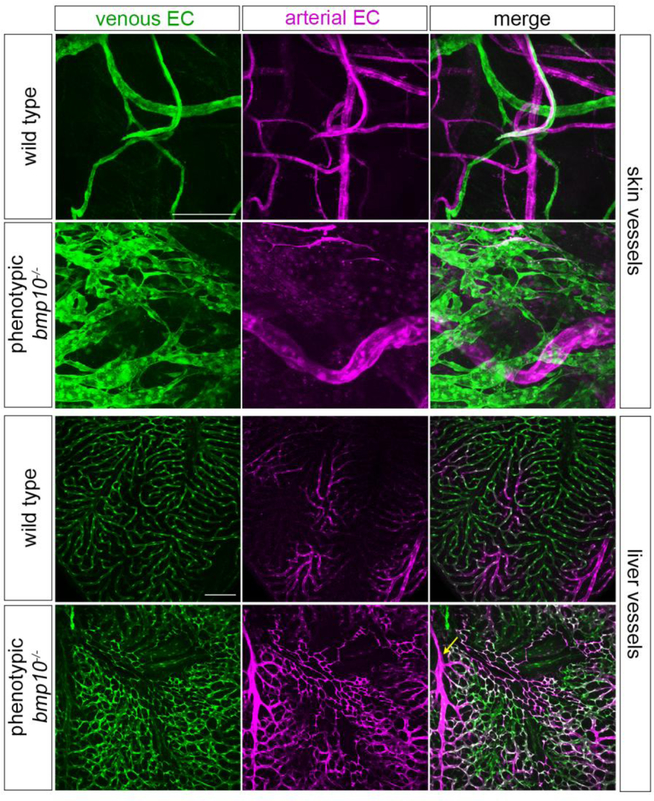
To further probe the architecture and location of vascular malformations in bmp10 mutants, we used confocal microscopy to image blood vessels in a double transgenic line that differentially marks arterial and venous endothelium. Although Tg(flt1:tdTomato)hu5333 has been used primarily to mark arterial endothelial cells and Tg(flt4:Citrine)hu7135 to mark venous and lymphatic endothelial cells in embryos [70–72], these transgenes also mark heart, skin, and fin arterial endothelium and fin venous endothelium, respectively, in adults [73–75]. In wild type adult skin, we found that both transgenes are expressed, revealing an interconnected network of thin, elongated arteries and veins that meander through the tissue in a somewhat stochastic pattern (Fig. 5). In contrast, in bmp10 mutants, blood-filled vascular malformations that were visible upon gross examination manifested as enlarged arteries and plexus-like veins (Fig. 5). Liver vasculature in phenotypic bmp10 mutants was also abnormal. In wild type adults, well separated clusters of small, branching arterial vessels connect to similarly sized straight sinusoids of venous character, which then drain into a large central vein (Fig. 5 and data not shown). In bmp10 mutants with external vascular phenotype, the liver vasculature was markedly less organized, with elongated, branching arteries and large swaths of plexus-like sinusoids (Fig. 5). Although we could not define liver AVMs per se, possibly because of the profoundly different, non-lobular architecture of the zebrafish liver [76, 77], this vascular phenotype is reminiscent of enlarged arteries and ectatic sinusoids that have been described in HHT patients [78, 79]. Based on a report of tortuous brain vessels in zebrafish adult eng mutants [80], we also examined the superficial brain vasculature in bmp10 mutants. However, we detected no brain vessel abnormalities using bright field (N = 11; 3 experiments) or confocal (N = 8; 2 experiments) microscopy. In sum, bmp10 mutants develop vascular malformations in skin and liver, two primary sites of AVM development in HHT. We hypothesize that the abdominal edema and ascites fluid in bmp10 mutants is related to liver vascular malformations and consequent liver dysfunction, as this is a common presentation of HHT with symptomatic liver involvement [81]. However, lymphatic defects may also cause edema, and we acknowledge reports of a role for Alk1 signaling in lymphatic vessel development and function [82–84]. Evaluation of effects of bmp10 mutation on the lymphatic vasculature will require further studies.
Fig. 5. bmp10 mutants develop skin and liver vascular malformations.
a Venous (green) and arterial (magenta) vasculature in anterior ventral skin, adult Tg(flt4:citrine)hu7135;Tg(flt1:tdTomato)hu5333 wild type and bmp10pt527 mutant siblings. Images are from 23-month fish and representative of N = 7 wild types and 8 phenotypic mutants from 2 independent lines, ages 3–23 months. Siblings were randomly paired to match imaging site in wild types to lesion site in mutants. 2D confocal projections. Scale bar: 50 μm. b Venous (green) and arterial (magenta) vasculature in distal tip of right liver lobe from adult Tg(flt4:citrine)hu7135;Tg(flt1:tdTomato)hu5333 wild type and bmp10pt527 mutant siblings. Images representative of N = 12 wild types and 9 phenotypic mutants from 2 independent lines, age 5 months. 2D confocal projections. Arrow, enlarged artery. Scale bar: 100 μm.
Vascular defects are associated with high output heart failure in bmp10 mutants
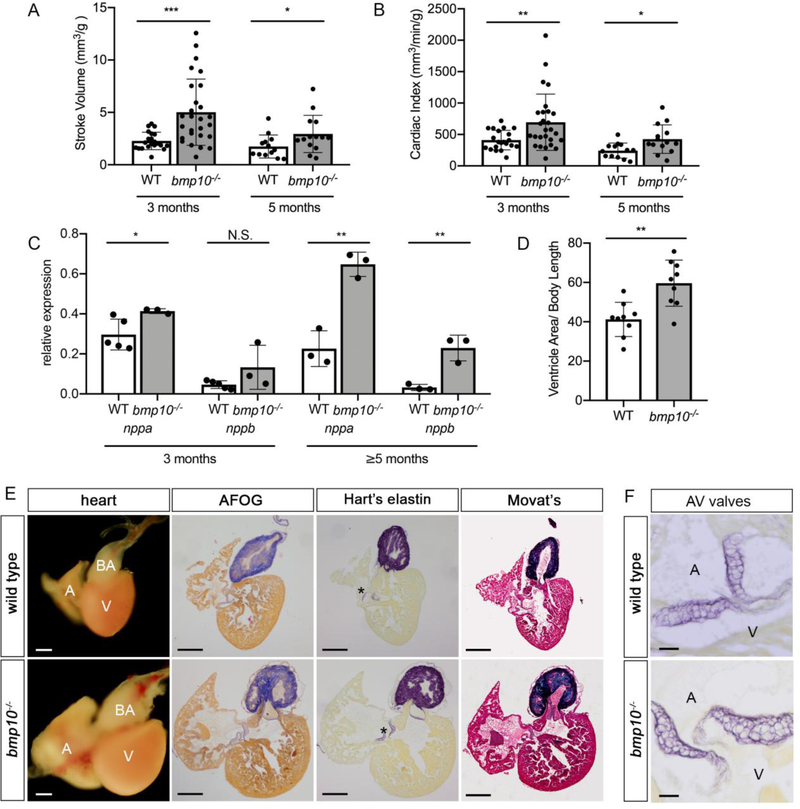
Because embryonic ventricular myocardium development is normal in bmp10 mutants and liver AVMs in HHT patients are associated with low vascular resistance, volume overload, and HOHF [85], we reasoned that early lethality in bmp10 mutants may be caused by HOHF. To test this hypothesis, we used echocardiography to assess heart function. At 3 and 5 months, externally phenotypic mutants exhibited significantly increased normalized stroke volume and cardiac index compared to wild type siblings (Fig. 6a,b), consistent with a high-output state. Additionally, mutant hearts expressed increased levels of nppa and nppb (Fig. 6c), both of which are elevated in high-output heart failure [86, 87]. Dissection of externally phenotypic bmp10 mutants at 3 months (Supplementary Fig. 4) and 5 months (Fig. 6d,e) revealed an enlarged heart with rounded ventricle. Qualitatively, the severity of the cardiac phenotype correlated with the severity of the external vascular phenotype. However, despite these obvious shape, size, and tissue integrity abnormalities, we failed to detect extracellular matrix remodeling or fibrosis, and elastin staining indicated normal development of the bulbus arteriosus (outflow tract) and atrioventricular valves (Fig. 6e–f; Supplementary Fig. 4). In sum, these data support the hypothesis that adult bmp10 mutants develop vascular malformations in skin and liver that lead to low systemic vascular resistance and volume overload, precipitating HOHF without fibrosis.
Fig. 6. bmp10 mutants develop high-output heart failure.
a,b Normalized stroke volume (a) and cardiac index (b) at 3 months [N = 21 wild types (16 males, 5 females), 28 bmp10pt527 mutants (25 males, 3 females) over 3 experiments] and 5 months [N = 13 wild types (11 males, 2 females), 16 bmp10pt527 mutants (13 males, 3 females) over 2 experiments]. c qRT-PCR quantification of nppa and nppb in the heart at 3 months (wild type, N= 4 pools, 6–10 hearts per pool; phenotypic bmp10pt527 mutants–, N = 3 pools, 4–10 hearts per pool) or 5 months (N= 3 pools, 10 hearts per pool, for both wild type and phenotypic bmp10pt527 mutants). d Relative ventricle size at 5 months, measured as maximal cross-sectional ventricle area/body length. N = 9 hearts (3 males, 6 females) per genotype, 1 experiment. Bars represent mean ± SD. Welch’s t-test (a,b) or unpaired Student’s t-test (c,d), *P<0.05,**P<0.01, ***P<0.001. NS = not significant. e Hearts from 5-month wild type and bmp10pt527 mutant siblings, whole mount or sections stained with acid fuchsin orange G (AFOG) for collagen (blue) and fibrin (red); Hart’s elastin stain (purple); or Movat’s pentachrome stain for muscle (red), collagen (yellow), elastin (blue-black), and ground substance (light blue). A, atrium; V, ventricle; BA, bulbus arteriosus; asterisks, atrioventricular valves. Scale bar: 200 μm. Images representative of N = 3 hearts per genotype, males from 2 independent lines. f Atrioventricular valves, 5-month wild type and bmp10pt527 mutant siblings, Hart’s elastin stain. A, atrium; V, ventricle; AV, atrioventricular. High magnification of different section of hearts shown in e. Scale bar: 20 μm.
In conclusion, our data demonstrate that Bmp10 is the only Alk1 ligand that is required for maintenance of the post-embryonic vasculature, a finding that should be taken into consideration in development of ligand-based therapies for HHT. In addition, we present zebrafish bmp10 mutants as a valuable model to investigate cardiac adaptation and the progression to heart failure in conditions of volume overload, especially in HHT2.
Discussion
The striking vascular phenotype in juvenile-to-adult bmp10 mutants and, rarely, acvrl1 heterozygotes, combined with the lack of phenotype in bmp9 and bmp10-like mutants, suggests that Bmp10 is the only required Alk1 ligand in post-embryonic zebrafish. The unique requirement for bmp10 was unexpected given the embryonic redundancy of bmp10 and bmp10-like [35] and the continued expression of bmp10-like in the adult heart. We speculate that these duplicate paralogs of mammalian BMP10 may have subfunctionalized in zebrafish, with endocardial- or liver-derived Bmp10 predominating over cardiomyocyte-derived Bmp10-like in circulation. We hypothesize that cardiomyocyte-derived Bmp10-like may act primarily at close range, with diminishing contribution to circulating Alk1 ligand concentration with increasing age. This explanation could account for the ability of bmp10-like to compensate for bmp10 during the embryonic period but not at later times. Further studies are required to confirm this hypothesis and to understand whether differential cell type of origin influences disposition and function of these Bmp10 paralogs.
The presence of post-embryonic phenotype in bmp10 mutants was also surprising given the continued expression of bmp9 in adult liver and the consensus that BMP9 and BMP10 are redundant ALK1 ligands. The notion of redundancy was initially derived from observations in mice showing that 1) Bmp9 nulls live to adulthood and do not develop AVMs; 2) Bmp10 nulls develop embryonic lethal AVMs at E9.5, similar to acvrl1 nulls; and 3) insertion of Bmp9 into the Bmp10 locus rescues embryonic AVMs in Bmp10 mutants [23, 27]. These data, combined with the fact that Bmp9 expression in liver begins at E9.75-E10, imply functional redundancy of BMP9 and BMP10, with respect to vascular requirements, at time points when both are expressed [23]. However, careful examination of this work reveals that rescued mice develop enlarged, hemorrhagic vessels by E17.5 [23], a phenotype that is remarkably similar to zebrafish bmp10 mutants, albeit at a much younger age. Additional support for redundancy of BMP9 and BMP10 derives from data showing that simultaneous depletion of these ligands in neonatal mice results in vascular dysplasia and/or AVMs [23, 27, 30–32]. However, these studies focused almost exclusively on the retinal vasculature, and the few studies that presented true retinal AVMs utilized combined but not independent BMP9/BMP10 antibody blockade [30–32]. In light of our zebrafish results, conditional knockout of Bmp10 at later developmental stages in mice might reveal unique requirements for Bmp10 in specific vascular beds.
The requirement for bmp10 but not bmp9 in vascular homeostasis in zebrafish is also surprising because heterozygous mutations in BMP9 are associated with an HHT-like syndrome: BMP9 patients do not develop AVMs but do develop telangiectasias, although the latter occur at sites that are not characteristic of HHT [88]. In contrast, there is no evidence linking mutations in BMP10 to HHT. Interestingly, the ExAC browser (exac.broadinstitute.org) calculates a probability of loss-of-function intolerance (pLI) of 0.72 for BMP10 and 0.09 for BMP9. The high pLI for BMP10 suggests that it is approaching intolerance to loss-of-function with a high likelihood of haploinsufficiency, whereas the low pLI for BMP9 suggests that the null state may be tolerated [89]. The association between heterozygous BMP9 mutations and pulmonary arterial hypertension (PAH) [90] contradicts the latter prediction, although the rarity and reduced penetrance of causal BMP9 mutations in PAH suggest that they are not a major contributor to disease.
Because HHT is an autosomal dominant disorder, ligand-based therapies may overcome haploinsufficiency. As proof of principle, administration of recombinant BMP9 in rodent models of PAH, which is primarily caused by BMP receptor 2 (BMPR2) heterozygous mutation or deficiency, enhances pSMAD1/5 and improves disease phenotype [91]. Our data suggest that BMP10, which is required for ALK1 signaling and, unlike BMP9, does not have strong osteogenic activity [92], may be the best choice for ligand-based therapy for HHT. Although we acknowledge a recent report identifying somatic second hits in some HHT telangiectasias [69], regardless of genetic mechanism of disease, this BMP10-based approach to therapy should remain a viable option for HHT1/ENG patients with intact ALK1.
The active form of TGF-β family ligands was originally described as disulfide-bonded homodimers, but developmental requirements for heterodimers (Bmp2/Bmp7; Ggf3/Nodal) have been reported [93, 94]. Notably, BMP9 and BMP10 can form biologically active heterodimers when co-expressed in vitro, and heterodimers, speculated to be produced by liver stellate cells, reportedly account for the large majority of ALK1-activating activity in mouse and human plasma [22]. However, BMP9/BMP10 heterodimers activate signaling with similar potency to homodimers [22], providing no clear biochemical advantage. Moreover, given the lack of a requirement for Bmp9 in AVM prevention in both mouse [27] and zebrafish, it is unlikely that the ALK1 ligand is an obligate BMP9/BMP10 heterodimer. Parsing out the differential functions of BMP10 homodimers versus BMP9/BMP10 heterodimers will require further investigation.
In mice, Bmp10 acts directly on embryonic ventricular cardiomyocytes to induce proliferation and trabeculation [23, 25, 61, 65, 95, 96]. However, we detected no changes in ventricular cardiomyocyte number or trabeculation in bmp10 mutant zebrafish embryos. It is possible that bmp10-like may compensate for the loss of bmp10 in zebrafish heart development. Alternatively, effects of Bmp10 loss on trabeculation in mice may be, at least in part, secondary to AVM development: AVMs disrupt flow patterns, and normal heartbeat and shear stress are required for proper trabeculation [66, 97]. In zebrafish, the progressive loss of blood flow subsequent to AVM development in acvrl1 [34] and bmp10;bmp10-like double mutants makes it difficult to distinguish between these two possibilities.
The zebrafish bmp10 mutant phenotype is reminiscent of the HHT patient phenotype, with variable age of onset and expressivity of vascular lesions concentrated in anterior skin (face), gill vessels (developmental origin of pulmonary arteries), pectoral fins (analogous to hands), and liver. Moreover, lesion development closely correlates with an enlarged heart and HOHF without maladaptive remodeling, similar to that reported in acvrl1 conditional knockout mice [98]. HOHF is increasingly recognized as a complication of HHT in aging patients and correlates with severe liver involvement [99]. There is little information regarding the progression of HOHF in humans and no therapy for this disease short of elimination of the underlying vascular problem [100]. In the case of HHT patients, this means liver transplantation. However, even liver transplantation is not a cure, as vascular malformations reappear in transplanted HHT patients with a cumulative risk of 48% over 15 years [101]. As such, bmp10 mutants afford us an opportunity to study the progression of HOHF and to gain mechanistic insight into the factors that tip the trajectory from compensation to failure.
Supplementary Material
Acknowledgments
We thank J. Keith Joung (Massachusetts General Hospital) for generating TALENs (NIH R01GM088040); the Sanger Institute Zebrafish Mutation Project, Sarah Childs (University of Calgary), Ching-Ling Lien (University of Southern California), Stephan Schulte-Merker (University of Münster), and Deborah Yelon (University of California-San Diego) for providing fish lines; Su Diler for technical assistance; Brenda Diergaarde (University of Pittsburgh) for statistical help; and Rachael Gerheart and Pitt aquatics staff for fish care.
Funding: This work was funded by National Institutes of Health (NIH) R01HL133009, NIH R01HL136566, and the Institute for Transfusion Medicine/Vitalant (BLR), and by NIH UM1HL098160 (HJY). The Vevo2100 small animal imaging system was funded by NIH 1S10RR027383-01 (KK).
Footnotes
Conflict of interest: The authors declare that they have no conflict of interest.
Ethical approval: All procedures performed in studies involving animals were in accordance with the ethical standards of the University of Pittsburgh (PHS approval A3187–01).
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- 1.Roman BL, Hinck AP (2017) ALK1 signaling in development and disease: new paradigms. Cell Mol Life Sci 74:4539–4560 doi: 10.1007/s00018-017-2636-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Droege F, Thangavelu K, Stuck BA, Stang A, Lang S, Geisthoff U (2018) Life expectancy and comorbidities in patients with hereditary hemorrhagic telangiectasia. Vasc Med:1358863X18767761 doi: 10.1177/1358863X18767761 [DOI] [PubMed] [Google Scholar]
- 3.Zarrabeitia R, Farinas-Alvarez C, Santibanez M, Senaris B, Fontalba A, Botella LM, Parra JA (2017) Quality of life in patients with hereditary haemorrhagic telangiectasia (HHT). Health Qual Life Outcomes 15:19 doi: 10.1186/s12955-017-0586-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Gussem EM, Edwards CP, Hosman AE, Westermann CJ, Snijder RJ, Faughnan ME, Mager JJ (2016) Life expextancy of parents with Hereditary Haemorrhagic Telangiectasia. Orphanet journal of rare diseases 11:46 doi: 10.1186/s13023-016-0427-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faughnan ME, Palda VA, Garcia-Tsao G, Geisthoff UW, McDonald J, Proctor DD, Spears J, Brown DH, Buscarini E, Chesnutt MS, Cottin V, Ganguly A, Gossage JR, Guttmacher AE, Hyland RH, Kennedy SJ, Korzenik J, Mager JJ, Ozanne AP, Piccirillo JF, Picus D, Plauchu H, Porteous ME, Pyeritz RE, Ross DA, Sabba C, Swanson K, Terry P, Wallace MC, Westermann CJ, White RI, Young LH, Zarrabeitia R, Group HHTFI-GW (2011) International guidelines for the diagnosis and management of hereditary haemorrhagic telangiectasia. J Med Genet 48:73–87 doi: 10.1136/jmg.2009.069013 [DOI] [PubMed] [Google Scholar]
- 6.Halderman AA, Ryan MW, Marple BF, Sindwani R, Reh DD, Poetker DM (2018) Bevacizumab for Epistaxis in Hereditary Hemorrhagic Telangiectasia: An Evidence-based Review. Am J Rhinol Allergy:1945892418768588 doi: 10.1177/1945892418768588 [DOI] [PubMed] [Google Scholar]
- 7.Mitchell A, Adams LA, MacQuillan G, Tibballs J, vanden Driesen R, Delriviere L (2008) Bevacizumab reverses need for liver transplantation in hereditary hemorrhagic telangiectasia. Liver transplantation : official publication of the American Association for the Study of Liver Diseases and the International Liver Transplantation Society 14:210–213 doi: 10.1002/lt.21417 [DOI] [PubMed] [Google Scholar]
- 8.Dupuis-Girod S, Ginon I, Saurin JC, Marion D, Guillot E, Decullier E, Roux A, Carette MF, Gilbert-Dussardier B, Hatron PY, Lacombe P, Lorcerie B, Riviere S, Corre R, Giraud S, Bailly S, Paintaud G, Ternant D, Valette PJ, Plauchu H, Faure F (2012) Bevacizumab in patients with hereditary hemorrhagic telangiectasia and severe hepatic vascular malformations and high cardiac output. JAMA 307:948–955 doi: 10.1001/jama.2012.250 [DOI] [PubMed] [Google Scholar]
- 9.Massague J (2012) TGFbeta signalling in context. Nature reviews. Molecular cell biology 13:616–630 doi: 10.1038/nrm3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallione CJ, Repetto GM, Legius E, Rustgi AK, Schelley SL, Tejpar S, Mitchell G, Drouin E, Westermann CJ, Marchuk DA (2004) A combined syndrome of juvenile polyposis and hereditary haemorrhagic telangiectasia associated with mutations in MADH4 (SMAD4). Lancet 363:852–859 [DOI] [PubMed] [Google Scholar]
- 11.Johnson DW, Berg JN, Baldwin MA, Gallione CJ, Marondel I, Yoon SJ, Stenzel TT, Speer M, Pericak-Vance MA, Diamond A, Guttmacher AE, Jackson CE, Attisano L, Kucherlapati R, Porteous ME, Marchuk DA (1996) Mutations in the activin receptor-like kinase 1 gene in hereditary haemorrhagic telangiectasia type 2. Nature Genet. 13:189–195 [DOI] [PubMed] [Google Scholar]
- 12.McAllister KA, Grogg KM, Johnson DW, Gallione CJ, Baldwin MA, Jackson CE, Helmbold EA, Markel DS, McKinnon WC, Murrell J, McCormick MK, Pericak-Vance MA, Heutnik P, Oostra BA, Haitjema T, Westerman CJJ, Porteous ME, Guttmacher AE, Letarte M, Marchuk DA (1994) Endoglin, a TGF-b binding protein of endothelial cells, is the gene for hereditary haemorrhagic telangiectasia type 1. Nature Genet. 8:345–351 [DOI] [PubMed] [Google Scholar]
- 13.McDonald J, Wooderchak-Donahue W, VanSant Webb C, Whitehead K, Stevenson DA, Bayrak-Toydemir P (2015) Hereditary hemorrhagic telangiectasia: genetics and molecular diagnostics in a new era. Frontiers in genetics 6:1 doi: 10.3389/fgene.2015.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castonguay R, Werner ED, Matthews RG, Presman E, Mulivor AW, Solban N, Sako D, Pearsall RS, Underwood KW, Seehra J, Kumar R, Grinberg AV (2011) Soluble Endoglin Specifically Binds Bone Morphogenetic Proteins 9 and 10 via Its Orphan Domain, Inhibits Blood Vessel Formation, and Suppresses Tumor Growth. J Biol Chem 286:30034–30046 doi:M111.260133 [pii] 10.1074/jbc.M111.260133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown MA, Zhao Q, Baker KA, Naik C, Chen C, Pukac L, Singh M, Tsareva T, Parice Y, Mahoney A, Roschke V, Sanyal I, Choe S (2005) Crystal structure of BMP-9 and functional interactions with pro-region and receptors. J Biol Chem 280:25111–25118 [DOI] [PubMed] [Google Scholar]
- 16.Mitchell D, Pobre EG, Mulivor AW, Grinberg AV, Castonguay R, Monnell TE, Solban N, Ucran JA, Pearsall RS, Underwood KW, Seehra J, Kumar R (2010) ALK1-Fc inhibits multiple mediators of angiogenesis and suppresses tumor growth. Mol Cancer Ther 9:379–388 doi:1535–7163.MCT-09–0650 [pii] 10.1158/1535-7163.MCT-09-0650 [DOI] [PubMed] [Google Scholar]
- 17.Townson SA, Martinez-Hackert E, Greppi C, Lowden P, Sako D, Liu J, Ucran JA, Liharska K, Underwood KW, Seehra J, Kumar R, Grinberg AV (2012) Specificity and structure of a high affinity activin receptor-like kinase 1 (ALK1) signaling complex. J Biol Chem 287:27313–27325 doi: 10.1074/jbc.M112.377960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.David L, Mallet C, Mazerbourg S, Feige JJ, Bailly S (2007) Identification of BMP9 and BMP10 as functional activators of the orphan activin receptor-like kinase 1 (ALK1) in endothelial cells. Blood 109:1953–1961 [DOI] [PubMed] [Google Scholar]
- 19.Scharpfenecker M, van Dinther M, Liu Z, van Bezooijen RL, Zhao Q, Pukac L, Lowik CW, ten Dijke P (2007) BMP-9 signals via ALK1 and inhibits bFGF-induced endothelial cell proliferation and VEGF-stimulated angiogenesis. J Cell Sci 120:964–972 [DOI] [PubMed] [Google Scholar]
- 20.Miller AF, Harvey SA, Thies RS, Olson MS (2000) Bone morphogenetic protein-9. An autocrine/paracrine cytokine in the liver. J Biol Chem 275:17937–17945 doi:275/24/17937 [pii] [DOI] [PubMed] [Google Scholar]
- 21.Bidart M, Ricard N, Levet S, Samson M, Mallet C, David L, Subileau M, Tillet E, Feige JJ, Bailly S (2012) BMP9 is produced by hepatocytes and circulates mainly in an active mature form complexed to its prodomain. Cell Mol Life Sci 69:313–324 doi: 10.1007/s00018-011-0751-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tillet E, Ouarne M, Desroches-Castan A, Mallet C, Subileau M, Didier R, Lioutsko A, Belthier G, Feige JJ, Bailly S (2018) A heterodimer formed by bone morphogenetic protein 9 (BMP9) and BMP10 provides most BMP biological activity in plasma. J Biol Chem 293:10963–10974 doi: 10.1074/jbc.RA118.002968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen H, Brady Ridgway J, Sai T, Lai J, Warming S, Chen H, Roose-Girma M, Zhang G, Shou W, Yan M (2013) Context-dependent signaling defines roles of BMP9 and BMP10 in embryonic and postnatal development. Proc Natl Acad Sci U S A 110:11887–11892 doi: 10.1073/pnas.1306074110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Breitkopf-Heinlein K, Meyer C, Konig C, Gaitantzi H, Addante A, Thomas M, Wiercinska E, Cai C, Li Q, Wan F, Hellerbrand C, Valous NA, Hahnel M, Ehlting C, Bode JG, Muller-Bohl S, Klingmuller U, Altenoder J, Ilkavets I, Goumans MJ, Hawinkels LJ, Lee SJ, Wieland M, Mogler C, Ebert MP, Herrera B, Augustin H, Sanchez A, Dooley S, Ten Dijke P (2017) BMP-9 interferes with liver regeneration and promotes liver fibrosis. Gut 66:939–954 doi: 10.1136/gutjnl-2016-313314 [DOI] [PubMed] [Google Scholar]
- 25.Chen H, Shi S, Acosta L, Li W, Lu J, Bao S, Chen Z, Yang Z, Schneider MD, Chien KR, Conway SJ, Yoder MC, Haneline LS, Franco D, Shou W (2004) BMP10 is essential for maintaining cardiac growth during murine cardiogenesis. Development 131:2219–2231 doi: 10.1242/dev.01094 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neuhaus H, Rosen V, Thies RS (1999) Heart specific expression of mouse BMP-10 a novel member of the TGF-beta superfamily. Mech Dev 80:181–184 doi:S0925477398002214 [pii] [DOI] [PubMed] [Google Scholar]
- 27.Ricard N, Ciais D, Levet S, Subileau M, Mallet C, Zimmers TA, Lee SJ, Bidart M, Feige JJ, Bailly S (2012) BMP9 and BMP10 are critical for postnatal retinal vascular remodeling. Blood 119:6162–6171 doi:blood-2012–01-407593 [pii] 10.1182/blood-2012-01-407593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kienast Y, Jucknischke U, Scheiblich S, Thier M, de Wouters M, Haas A, Lehmann C, Brand V, Bernicke D, Honold K, Lorenz S (2016) Rapid Activation of Bone Morphogenic Protein 9 by Receptor-mediated Displacement of Pro-domains. J Biol Chem 291:3395–3410 doi: 10.1074/jbc.M115.680009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Baardewijk LJ, van der Ende J, Lissenberg-Thunnissen S, Romijn LM, Hawinkels LJ, Sier CF, Schipper IB (2013) Circulating bone morphogenetic protein levels and delayed fracture healing. Int Orthop 37:523–527 doi: 10.1007/s00264-012-1750-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ola R, Dubrac A, Han J, Zhang F, Fang JS, Larrivee B, Lee M, Urarte AA, Kraehling JR, Genet G, Hirschi KK, Sessa WC, Canals FV, Graupera M, Yan M, Young LH, Oh PS, Eichmann A (2016) PI3 kinase inhibition improves vascular malformations in mouse models of hereditary haemorrhagic telangiectasia. Nature communications 7:13650 doi: 10.1038/ncomms13650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruiz S, Zhao H, Chandakkar P, Chatterjee PK, Papoin J, Blanc L, Metz CN, Campagne F, Marambaud P (2016) A mouse model of hereditary hemorrhagic telangiectasia generated by transmammary-delivered immunoblocking of BMP9 and BMP10. Scientific reports 5:37366 doi: 10.1038/srep37366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baeyens N, Larrivee B, Ola R, Hayward-Piatkowskyi B, Dubrac A, Huang B, Ross TD, Coon BG, Min E, Tsarfati M, Tong H, Eichmann A, Schwartz MA (2016) Defective fluid shear stress mechanotransduction mediates hereditary hemorrhagic telangiectasia. J Cell Biol 214:807–816 doi: 10.1083/jcb.201603106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roman BL, Pham VN, Lawson ND, Kulik M, Childs S, Lekven AC, Garrity DM, Moon RT, Fishman MC, Lechleider RJ, Weinstein BM (2002) Disruption of acvrl1 increases endothelial cell number in zebrafish cranial vessels. Development 129:3009–3019 [DOI] [PubMed] [Google Scholar]
- 34.Corti P, Young S, Chen CY, Patrick MJ, Rochon ER, Pekkan K, Roman BL (2011) Interaction between alk1 and blood flow in the development of arteriovenous malformations. Development 138:1573–1582 doi:dev.060467 [pii] 10.1242/dev.060467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laux DW, Young S, Donovan JP, Mansfield CJ, Upton PD, Roman BL (2013) Circulating Bmp10 acts through endothelial Alk1 to mediate flow-dependent arterial quiescence. Development 140:3403–3412 doi: 10.1242/dev.095307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Westerfield M (1995) The zebrafish book. University of Oregon Press, Eugene [Google Scholar]
- 37.Choi J, Dong L, Ahn J, Dao D, Hammerschmidt M, Chen JN (2007) FoxH1 negatively modulates flk1 gene expression and vascular formation in zebrafish. Dev Biol 304:735–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitesell TR, Kennedy RM, Carter AD, Rollins EL, Georgijevic S, Santoro MM, Childs SJ (2014) An alpha-smooth muscle actin (acta2/alphasma) zebrafish transgenic line marking vascular mural cells and visceral smooth muscle cells. PLoS One 9:e90590 doi: 10.1371/journal.pone.0090590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bussmann J, Bos FL, Urasaki A, Kawakami K, Duckers HJ, Schulte-Merker S (2010) Arteries provide essential guidance cues for lymphatic endothelial cells in the zebrafish trunk. Development 137:2653–2657 doi:dev.048207 [pii] 10.1242/dev.048207 [DOI] [PubMed] [Google Scholar]
- 40.Gordon K, Schulte D, Brice G, Simpson MA, Roukens MG, van Impel A, Connell F, Kalidas K, Jeffery S, Mortimer PS, Mansour S, Schulte-Merker S, Ostergaard P (2013) Mutation in vascular endothelial growth factor-C, a ligand for vascular endothelial growth factor receptor-3, is associated with autosomal dominant milroy-like primary lymphedema. Circ Res 112:956–960 doi: 10.1161/CIRCRESAHA.113.300350 [DOI] [PubMed] [Google Scholar]
- 41.Lin YF, Swinburne I, Yelon D (2012) Multiple influences of blood flow on cardiomyocyte hypertrophy in the embryonic zebrafish heart. Dev Biol 362:242–253 doi: 10.1016/j.ydbio.2011.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang CJ, Tu CT, Hsiao CD, Hsieh FJ, Tsai HJ (2003) Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev Dyn 228:30–40 doi: 10.1002/dvdy.10356 [DOI] [PubMed] [Google Scholar]
- 43.Rottbauer W, Saurin AJ, Lickert H, Shen X, Burns CG, Wo ZG, Kemler R, Kingston R, Wu C, Fishman M (2002) Reptin and pontin antagonistically regulate heart growth in zebrafish embryos. Cell 111:661–672 [DOI] [PubMed] [Google Scholar]
- 44.Kettleborough RN, Busch-Nentwich EM, Harvey SA, Dooley CM, de Bruijn E, van Eeden F, Sealy I, White RJ, Herd C, Nijman IJ, Fenyes F, Mehroke S, Scahill C, Gibbons R, Wali N, Carruthers S, Hall A, Yen J, Cuppen E, Stemple DL (2013) A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature 496:494–497 doi: 10.1038/nature11992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cade L, Reyon D, Hwang WY, Tsai SQ, Patel S, Khayter C, Joung JK, Sander JD, Peterson RT, Yeh JR (2012) Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res 40:8001–8010 doi:gks518 [pii] 10.1093/nar/gks518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.den Dunnen JT, Dalgleish R, Maglott DR, Hart RK, Greenblatt MS, McGowan-Jordan J, Roux AF, Smith T, Antonarakis SE, Taschner PE (2016) HGVS Recommendations for the Description of Sequence Variants: 2016 Update. Human mutation 37:564–569 doi: 10.1002/humu.22981 [DOI] [PubMed] [Google Scholar]
- 47.Isogai S, Horiguchi M, Weinstein BM (2001) The vascular anatomy of the developing zebrafish: an atlas of embryonic and early larval development. Dev. Biol 230:278–301 [DOI] [PubMed] [Google Scholar]
- 48.Wang LW, Huttner IG, Santiago CF, Kesteven SH, Yu ZY, Feneley MP, Fatkin D (2017) Standardized echocardiographic assessment of cardiac function in normal adult zebrafish and heart disease models. Dis Model Mech 10:63–76 doi: 10.1242/dmm.026989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spanjaard B, Hu B, Mitic N, Olivares-Chauvet P, Janjuha S, Ninov N, Junker JP (2018) Simultaneous lineage tracing and cell-type identification using CRISPR-Cas9-induced genetic scars. Nat Biotechnol 36:469–473 doi: 10.1038/nbt.4124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R (2019) Comprehensive Integration of Single-Cell Data. Cell 177:1888–1902 e1821 doi: 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R (2018) Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36:411–420 doi: 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hafemeister C, Satija R (2019) Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. bioRxiv:576827 doi: 10.1101/576827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F, Newell EW (2018) Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol doi: 10.1038/nbt.4314 [DOI] [PubMed] [Google Scholar]
- 54.Dowle M, Srinivasan A (2019) data.table:Extension of data.frame. R packaged version 1.12.2. In: [Google Scholar]
- 55.Wickham H (2016) ggplot2 Elegant Graphics for Data Analysis. Springer Nature, New York, USA [Google Scholar]
- 56.Susan-Resiga D, Essalmani R, Hamelin J, Asselin MC, Benjannet S, Chamberland A, Day R, Szumska D, Constam D, Bhattacharya S, Prat A, Seidah NG (2011) Furin is the major processing enzyme of the cardiac-specific growth factor bone morphogenetic protein 10. J Biol Chem 286:22785–22794 doi:M111.233577 [pii] 10.1074/jbc.M111.233577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Constam DB (2014) Regulation of TGFbeta and related signals by precursor processing. Seminars in cell & developmental biology 32:85–97 doi: 10.1016/j.semcdb.2014.01.008 [DOI] [PubMed] [Google Scholar]
- 58.Rochon ER, Menon PG, Roman BL (2016) Alk1 controls arterial endothelial cell migration in lumenized vessels. Development 143:2593–2602 doi: 10.1242/dev.135392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.El-Brolosy MA, Kontarakis Z, Rossi A, Kuenne C, Gunther S, Fukuda N, Kikhi K, Boezio GLM, Takacs CM, Lai SL, Fukuda R, Gerri C, Giraldez AJ, Stainier DYR (2019) Genetic compensation triggered by mutant mRNA degradation. Nature 568:193–197 doi: 10.1038/s41586-019-1064-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Corti P, Xue J, Tejero J, Wajih N, Sun M, Stolz DB, Tsang M, Kim-Shapiro DB, Gladwin MT (2016) Globin X is a six-coordinate globin that reduces nitrite to nitric oxide in fish red blood cells. Proc Natl Acad Sci U S A 113:8538–8543 doi: 10.1073/pnas.1522670113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang W, Chen H, Wang Y, Yong W, Zhu W, Liu Y, Wagner GR, Payne RM, Field LJ, Xin H, Cai CL, Shou W (2011) Tbx20 transcription factor is a downstream mediator for bone morphogenetic protein-10 in regulating cardiac ventricular wall development and function. J Biol Chem 286:36820–36829 doi: 10.1074/jbc.M111.279679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang J, Elicker J, Bowens N, Liu X, Cheng L, Cappola TP, Zhu X, Parmacek MS (2012) Myocardin regulates BMP10 expression and is required for heart development. J Clin Invest 122:3678–3691 doi: 10.1172/JCI63635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou Y, Cashman TJ, Nevis KR, Obregon P, Carney SA, Liu Y, Gu A, Mosimann C, Sondalle S, Peterson RE, Heideman W, Burns CE, Burns CG (2011) Latent TGF-beta binding protein 3 identifies a second heart field in zebrafish. Nature 474:645–648 doi:nature10094 [pii] 10.1038/nature10094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang W, Chen H, Qu X, Chang CP, Shou W (2013) Molecular mechanism of ventricular trabeculation/compaction and the pathogenesis of the left ventricular noncompaction cardiomyopathy (LVNC). Am J Med Genet C Semin Med Genet 163C:144–156 doi: 10.1002/ajmg.c.31369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen H, Yong W, Ren S, Shen W, He Y, Cox KA, Zhu W, Li W, Soonpaa M, Payne RM, Franco D, Field LJ, Rosen V, Wang Y, Shou W (2006) Overexpression of bone morphogenetic protein 10 in myocardium disrupts cardiac postnatal hypertrophic growth. J Biol Chem 281:27481–27491 doi:M604818200 [pii] 10.1074/jbc.M604818200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peshkovsky C, Totong R, Yelon D (2011) Dependence of cardiac trabeculation on neuregulin signaling and blood flow in zebrafish. Dev Dyn 240:446–456 doi: 10.1002/dvdy.22526 [DOI] [PubMed] [Google Scholar]
- 67.Liu J, Bressan M, Hassel D, Huisken J, Staudt D, Kikuchi K, Poss KD, Mikawa T, Stainier DY (2010) A dual role for ErbB2 signaling in cardiac trabeculation. Development 137:3867–3875 doi:137/22/3867 [pii] 10.1242/dev.053736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shovlin CL, Guttmacher AE, Buscarini E, Faughnan ME, Hyland RH, Westermann CJ, Kjeldsen AD, Plauchu H (2000) Diagnostic criteria for hereditary hemorrhagic telangiectasia (Rendu-Osler-Weber syndrome). American journal of medical genetics 91:66–67 [DOI] [PubMed] [Google Scholar]
- 69.Snellings DA, Gallione CJ, Clark DS, Vozoris NT, Faughnan ME, Marchuk DA (2019) Somatic Mutations in Vascular Malformations of Hereditary Hemorrhagic Telangiectasia Result in Bi-allelic Loss of ENG or ACVRL1. Am J Hum Genet doi: 10.1016/j.ajhg.2019.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bussmann J, Wolfe SA, Siekmann AF (2011) Arterial-venous network formation during brain vascularization involves hemodynamic regulation of chemokine signaling. Development 138:1717–1726 doi:dev.059881 [pii] 10.1242/dev.059881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hasan SS, Tsaryk R, Lange M, Wisniewski L, Moore JC, Lawson ND, Wojciechowska K, Schnittler H, Siekmann AF (2017) Endothelial Notch signalling limits angiogenesis via control of artery formation. Nat Cell Biol 19:928–940 doi: 10.1038/ncb3574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weijts B, Gutierrez E, Saikin SK, Ablooglu AJ, Traver D, Groisman A, Tkachenko E (2018) Blood flow-induced Notch activation and endothelial migration enable vascular remodeling in zebrafish embryos. Nature communications 9:5314 doi: 10.1038/s41467-018-07732-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Harrison MR, Bussmann J, Huang Y, Zhao L, Osorio A, Burns CG, Burns CE, Sucov HM, Siekmann AF, Lien CL (2015) Chemokine-guided angiogenesis directs coronary vasculature formation in zebrafish. Dev Cell 33:442–454 doi: 10.1016/j.devcel.2015.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu C, Hasan SS, Schmidt I, Rocha SF, Pitulescu ME, Bussmann J, Meyen D, Raz E, Adams RH, Siekmann AF (2014) Arteries are formed by vein-derived endothelial tip cells. Nature communications 5:5758 doi: 10.1038/ncomms6758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rasmussen JP, Vo NT, Sagasti A (2018) Fish Scales Dictate the Pattern of Adult Skin Innervation and Vascularization. Dev Cell 46:344–359 e344 doi: 10.1016/j.devcel.2018.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Korzh S, Pan X, Garcia-Lecea M, Winata CL, Wohland T, Korzh V, Gong Z (2008) Requirement of vasculogenesis and blood circulation in late stages of liver growth in zebrafish. BMC Dev Biol 8:84 doi:1471–213X-8–84 [pii] 10.1186/1471-213X-8-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yao Y, Lin J, Yang P, Chen Q, Chu X, Gao C, Hu J (2012) Fine structure, enzyme histochemistry, and immunohistochemistry of liver in zebrafish. Anat Rec (Hoboken) 295:567–576 doi: 10.1002/ar.22416 [DOI] [PubMed] [Google Scholar]
- 78.Daly JJ, Schiller AL (1976) The liver in hereditary hemorrhagic telangiectasia (Osler-Weber-Rendu disease). Am J Med 60:723–726 doi: 10.1016/0002-9343(76)90510-6 [DOI] [PubMed] [Google Scholar]
- 79.Sawabe M, Arai T, Esaki Y, Tsuru M, Fukazawa T, Takubo K (2001) Three-dimensional organization of the hepatic microvasculature in hereditary hemorrhagic telangiectasia. Arch Pathol Lab Med 125:1219–1223 doi: [DOI] [PubMed] [Google Scholar]
- 80.Sugden WW, Meissner R, Aegerter-Wilmsen T, Tsaryk R, Leonard EV, Bussmann J, Hamm MJ, Herzog W, Jin Y, Jakobsson L, Denz C, Siekmann AF (2017) Endoglin controls blood vessel diameter through endothelial cell shape changes in response to haemodynamic cues. Nat Cell Biol 19:653–665 doi: 10.1038/ncb3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Garcia-Tsao G (2007) Liver involvement in hereditary hemorrhagic telangiectasia (HHT). J Hepatol 46:499–507 doi: 10.1016/j.jhep.2006.12.008 [DOI] [PubMed] [Google Scholar]
- 82.Niessen K, Zhang G, Ridgway JB, Chen H, Yan M (2010) ALK1 signaling regulates early postnatal lymphatic vessel development. Blood 115:1654–1661 doi: 10.1182/blood-2009-07-235655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Subileau M, Merdzhanova G, Ciais D, Collin-Faure V, Feige JJ, Bailly S, Vittet D (2019) Bone Morphogenetic Protein 9 Regulates Early Lymphatic-Specified Endothelial Cell Expansion during Mouse Embryonic Stem Cell Differentiation. Stem Cell Reports 12:98–111 doi: 10.1016/j.stemcr.2018.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Levet S, Ciais D, Merdzhanova G, Mallet C, Zimmers TA, Lee SJ, Navarro FP, Texier I, Feige JJ, Bailly S, Vittet D (2013) Bone morphogenetic protein 9 (BMP9) controls lymphatic vessel maturation and valve formation. Blood 122:598–607 doi: 10.1182/blood-2012-12-472142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ginon I, Decullier E, Finet G, Cordier JF, Marion D, Saurin JC, Dupuis-Girod S (2013) Hereditary hemorrhagic telangiectasia, liver vascular malformations and cardiac consequences. European journal of internal medicine 24:e35–39 doi: 10.1016/j.ejim.2012.12.013 [DOI] [PubMed] [Google Scholar]
- 86.Sergeeva IA, Christoffels VM (2013) Regulation of expression of atrial and brain natriuretic peptide, biomarkers for heart development and disease. Biochim Biophys Acta 1832:2403–2413 doi: 10.1016/j.bbadis.2013.07.003 [DOI] [PubMed] [Google Scholar]
- 87.Reddy YNV, Melenovsky V, Redfield MM, Nishimura RA, Borlaug BA (2016) High-Output Heart Failure: A 15-Year Experience. J Am Coll Cardiol 68:473–482 doi: 10.1016/j.jacc.2016.05.043 [DOI] [PubMed] [Google Scholar]
- 88.Wooderchak-Donahue WL, McDonald J, O’Fallon B, Upton PD, Li W, Roman BL, Young S, Plant P, Fulop GT, Langa C, Morrell NW, Botella LM, Bernabeu C, Stevenson DA, Runo JR, Bayrak-Toydemir P (2013) BMP9 Mutations Cause a Vascular-Anomaly Syndrome with Phenotypic Overlap with Hereditary Hemorrhagic Telangiectasia. Am J Hum Genet 93:530–537 doi:S0002–9297(13)00295–4 [pii] 10.1016/j.ajhg.2013.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG, Exome Aggregation C (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536:285–291 doi: 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Graf S, Haimel M, Bleda M, Hadinnapola C, Southgate L, Li W, Hodgson J, Liu B, Salmon RM, Southwood M, Machado RD, Martin JM, Treacy CM, Yates K, Daugherty LC, Shamardina O, Whitehorn D, Holden S, Aldred M, Bogaard HJ, Church C, Coghlan G, Condliffe R, Corris PA, Danesino C, Eyries M, Gall H, Ghio S, Ghofrani HA, Gibbs JSR, Girerd B, Houweling AC, Howard L, Humbert M, Kiely DG, Kovacs G, MacKenzie Ross RV, Moledina S, Montani D, Newnham M, Olschewski A, Olschewski H, Peacock AJ, Pepke-Zaba J, Prokopenko I, Rhodes CJ, Scelsi L, Seeger W, Soubrier F, Stein DF, Suntharalingam J, Swietlik EM, Toshner MR, van Heel DA, Vonk Noordegraaf A, Waisfisz Q, Wharton J, Wort SJ, Ouwehand WH, Soranzo N, Lawrie A, Upton PD, Wilkins MR, Trembath RC, Morrell NW (2018) Identification of rare sequence variation underlying heritable pulmonary arterial hypertension. Nature communications 9:1416 doi: 10.1038/s41467-018-03672-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Long L, Ormiston ML, Yang X, Southwood M, Graf S, Machado RD, Mueller M, Kinzel B, Yung LM, Wilkinson JM, Moore SD, Drake KM, Aldred MA, Yu PB, Upton PD, Morrell NW (2015) Selective enhancement of endothelial BMPR-II with BMP9 reverses pulmonary arterial hypertension. Nat Med 21:777–785 doi: 10.1038/nm.3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kang Q, Sun MH, Cheng H, Peng Y, Montag AG, Deyrup AT, Jiang W, Luu HH, Luo J, Szatkowski JP, Vanichakarn P, Park JY, Li Y, Haydon RC, He TC (2004) Characterization of the distinct orthotopic bone-forming activity of 14 BMPs using recombinant adenovirus-mediated gene delivery. Gene Ther 11:1312–1320 doi: 10.1038/sj.gt.3302298 [DOI] [PubMed] [Google Scholar]
- 93.Little SC, Mullins MC (2009) Bone morphogenetic protein heterodimers assemble heteromeric type I receptor complexes to pattern the dorsoventral axis. Nat Cell Biol 11:637–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Montague TG, Schier AF (2017) Vg1-Nodal heterodimers are the endogenous inducers of mesendoderm. eLife 6 doi: 10.7554/eLife.28183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pashmforoush M, Lu JT, Chen H, Amand TS, Kondo R, Pradervand S, Evans SM, Clark B, Feramisco JR, Giles W, Ho SY, Benson DW, Silberbach M, Shou W, Chien KR (2004) Nkx2–5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell 117:373–386 [DOI] [PubMed] [Google Scholar]
- 96.Grego-Bessa J, Luna-Zurita L, del Monte G, Bolos V, Melgar P, Arandilla A, Garratt AN, Zang H, Mukouyama YS, Chen H, Shou W, Ballestar E, Esteller M, Rojas A, Perez-Pomares JM, de la Pompa JL (2007) Notch signaling is essential for ventricular chamber development. Dev Cell 12:415–429 doi: 10.1016/j.devcel.2006.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee J, Fei P, Packard RR, Kang H, Xu H, Baek KI, Jen N, Chen J, Yen H, Kuo CC, Chi NC, Ho CM, Li R, Hsiai TK (2016) 4-Dimensional light-sheet microscopy to elucidate shear stress modulation of cardiac trabeculation. J Clin Invest 126:1679–1690 doi: 10.1172/JCI83496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Morine KJ, Qiao X, Paruchuri V, Aronovitz MJ, Mackey EE, Buiten L, Levine J, Ughreja K, Nepali P, Blanton RM, Karas RH, Oh SP, Kapur NK (2017) Conditional knockout of activin like kinase-1 (ALK-1) leads to heart failure without maladaptive remodeling. Heart Vessels 32:628–636 doi: 10.1007/s00380-017-0955-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Iyer VN, Saberi B, Heimbach JK, Larson JJ, Raghavaiah S, Ditah I, Swanson K, Kamath PS, Watt KD, Taner T, Krowka MJ, Leise MD (2018) Liver Transplantation Trends and Outcomes for Hereditary Hemorrhagic Telangiectasia In the United States. Transplantation doi: 10.1097/TP.0000000000002491 [DOI] [PubMed] [Google Scholar]
- 100.Mehta PA, Dubrey SW (2009) High output heart failure. QJM : monthly journal of the Association of Physicians 102:235–241 doi: 10.1093/qjmed/hcn147 [DOI] [PubMed] [Google Scholar]
- 101.Dumortier J, Dupuis-Girod S, Valette PJ, Valent A, Guillaud O, Saurin JC, Hervieu V, Robinson P, Plauchu H, Paliard P, Boillot O, Scoazec JY (2019) Recurrence of Hereditary Hemorrhagic Telangiectasia After Liver Transplantation: Clinical Implications and Physiopathological Insights. Hepatology 69:2232–2240 doi: 10.1002/hep.30424 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.