Abstract
Polyglutamine (polyQ) tract polymorphism within the human androgen receptor (AR) shows population heterogeneity. African American men possess short polyQ tracts significantly more frequently than Caucasian American men. The length of polyQ tracts is inversely correlated with the risk of prostate cancer, age of onset, and aggressiveness at diagnosis. Aberrant activation of Wnt signaling also reveals frequently in advanced prostate cancer, and an enrichment of androgen and Wnt signaling activation has been observed in African American patients. Here, we assessed aberrant expression of AR bearing different polyQ tracts and stabilized β-catenin in prostate tumorigenesis using newly generated mouse models. We observed an early onset oncogenic transformation, accelerated tumor cell growth, and aggressive tumor phenotypes in the compound mice bearing short polyQ tract AR and stabilized β-catenin. RNA sequencing analysis showed a robust enrichment of Myc-regulated downstream genes in tumor samples bearing short polyQ AR versus those with longer polyQ tract AR. Upstream regulator analysis further identified Myc as the top candidate of transcriptional regulators in tumor cells from the above mouse samples with short polyQ tract AR and β-catenin. Chromatin immunoprecipitation analyses revealed increased recruitment of β-catenin and AR on the c-Myc gene regulatory locus in the tumor tissues expressing stabilized β-catenin and shorter polyQ tract AR. These data demonstrate a promotional role of aberrant activation of Wnt/β-catenin in combination with short polyQ AR expression in prostate tumorigenesis and suggest a potential mechanism underlying aggressive prostatic tumor development, which has been frequently observed in African American patients.
Keywords: Prostate Cancer, The androgen receptor, β-catenin, Wnt signaling, MYC
INTRODUCTION
Prostate cancer is the most commonly diagnosed malignancy and the second leading cause of cancer-related death among men in the United States 1. Importantly, African American men have been shown to have the highest risk in both prostate cancer incidence and mortality 2. Although socio-economic status and other environmental and cultural factors may contribute to this racial disparity, emerging evidence has shown an important role of genetic and epigenetic factors in the pathogenesis of African American prostate cancer 2.
Androgen signaling, mediated through the androgen receptor (AR) and its ligands, testosterone and 5a-dihydrotestosterone (DHT), plays a central role in prostate tumorigenesis 3, 4. The AR is a ligand-induced transcriptional factor 5, and contains an N-terminal transactivation domain (NTD), a central DNA-binding domain (DBD), a C-terminal ligand-binding domain (LBD), and a hinge region between the DBD and LBD 6. The NTD of human AR is encoded by exon 1, containing a variable region of different length CAG (cytosine, adenine, guanine) triplet repeats that codes the polyglutamine (PolyQ) tract. The length of CAG repeats has been shown to inversely correlate with the transcriptional activity of the AR 7, 8. Expansion of the CAG repeats length within the AR directly associates with spinal and bulbar muscular atrophy and Kennedy’s disease 9, 10. Intriguingly, it has been shown that African American men bear short CAG repeats significantly more frequently than Caucasian American men 11, 12. Moreover, the length of CAG repeats in the AR gene are inversely correlated with the risk of developing prostate cancer, age of onset, and risk of advanced disease at diagnosis 7, 8, suggesting a potential link between the length of polyQ tracts and the pathogenesis and aggressiveness of prostate cancer in African American men.
Aberrant activation of Wnt signaling pathways was revealed as one of the most frequent abnormalities in advanced human prostate cancer and directly contributes to prostate tumorigenesis 13, 14. An increase in nuclear β-catenin expression has been shown to promote prostate cancer cell proliferation 15. Conditional expression of stabilized β-catenin in prostate epithelium induces the development of squamous metaplasia and prostate intraepithelial neoplasia (PIN) 16, 17. An interaction between the AR and β-catenin proteins has been identified in prostate cancer cells 18–20. Co-expression of AR and stabilized β-catenin in the mouse prostate results in accelerated prostate tumor development with aggressive tumor invasion and a decreased survival rate 21. A significant enrichment of the androgen and Wnt signaling pathways was observed in patients with fast growing and early onset prostate cancer 22. Intriguingly, this enrichment was also identified in prostate cancer samples isolated from African American men 23.
To recapitulate aberrant Wnt and androgen activation in human prostate cancer tumorigenesis, particularly in those aggressive diseases observed in African American men, we generated a series of mouse strains, in which humanized AR bearing different lengths of CAG repeats and stabilized β-catenin are co-expressed in mouse prostatic epithelia. An early onset oncogenic transformation, accelerated tumor development, and more aggressive tumor invasion phenotypes were observed in the prostates of the mice with the expression of short polyQ AR and stabilized β-catenin. Castration of these mice bearing prostatic tumors showed significant tumor regression. RNA sequence analysis showed a robust enrichment of Myc regulated downstream target genes in the differentially expressed genes (DEGs) of tumor samples of short polyQ AR versus those with longer polyQ tracts. Upstream Regulator Analysis identified Myc as the top candidate of transcriptional regulators in tumor cells isolated from mice with short polyQ tract AR proteins and stabilized β-catenin expression. Using chromatin immunoprecipitation (ChIP) analyses, we further identified increased recruitment of β-catenin and AR on the c-Myc gene regulatory locus in tumor tissues from the compound mice expressing stabilized β-catenin and short polyQ AR. These data demonstrate a promotional role of short polyQ AR expression in enhancing aberrant activation of Wnt/β-catenin induced prostate tumorigenesis, suggesting a potential mechanism underlying aggressive prostatic tumor development that has been frequently observed in African American patients.
RESULTS
Conditional expression of stabilized β-catenin in the prostate of humanized AR knock-in mice
The dysregulation of Wnt and androgen signaling pathways co-occurs in human prostate cancer 14 (Supplemental Fig. 1 and Supplemental Table 1). African American men bearing short polyQ tract AR proteins appeared more frequently and revealed higher incidence and greater lethality of prostate cancer than Caucasian American men 11, 12. To assess aberrant Wnt and androgen activation in prostate tumorigenesis, we developed a series of compound mice, in which conditional expression of stabilized β-catenin is activated by probasin promoter-driven Cre activity in prostatic epithelia of humanized AR knock-in mice bearing different length polyQ tracts 24–26 (Fig. 1A). Using genomic PCR approaches, we confirmed humanized AR bearing different length of CAG repeats, Ctnnb1 exon 3 floxed, and PB-Cre4 alleles (Fig. 1B). Western blotting analyses further demonstrated the expression of humanized AR with different lengths of polyQ tracts and stabilized β-catenin proteins in mouse prostatic tissues (Fig. 1C). The expression of both humanized AR and stabilized β-catenin was further assessed using immunohistochemistry (IHC) analyses in adjacent tissue sections. Uniform nuclear staining of humanized AR proteins with the antibody against the amino terminal region of the human AR protein was revealed in prostate tissues isolated from each of the three compound mouse lines (Fig. 1D2–4). Cytoplasmic and nuclear β-catenin expression was also confirmed in adjacent tissue sections (Fig. 1E2–4). In contrast, cellular membrane staining of endogenous β-catenin, but no staining for human AR antibody was revealed in prostate tissues of wild type controls (Fig. 1D1 and 1E1). These results demonstrated the co-expression of humanized AR and stabilized β-catenin in the compound mouse prostatic tissues.
Figure 1. Generation of mouse models with stabilized β-catenin in the prostates of mice with humanized AR alleles bearing different length of CAG repeats.
A. Schematic of human AR (hAR) knock-in alleles and Ctnnb1 exon3-floxed allele, as well as the recombined allele. B. Genomic PCR was used to confirm the hAR knock-in (upper panel), Ctnnb1 exon3-floxed and wild type alleles (middle panel), as well as PB-Cre4 allele (bottom panel) in the indicated mouse models. C. Immunoblotting showing protein expression of humanized AR, endogenous β-catenin, and stabilized β-catenin (Ex3 deleted protein) (n=3). D. Representative IHC images of adjacent prostate tissue sections from different mouse tissues as labeled. IHC staining were performed with the indicated antibodies: h-AR (D) and β-catenin (E). The scale bar represents 10 μm.
Expression of humanized AR bearing shorter polyQ tracts enhances β-catenin-mediated oncogenic transformation and tumor development in the mouse prostate
All experimental mice, including ArhAR12Q:Ctnnb1L(ex3)/+:PB-Cre4, ArhAR21Q:Ctnnb1L(ex3)/+:PB-Cre4, and ArhAR48Q:Ctnnb1L(ex3)/+:PB-Cre4 mice were born at the expected Mendelian ratios and appeared normal with no obvious difference from their wild type littermates at birth. We systematically examined male mice at 2, 4, 6, 9, and 12 months of age adhering to recommendations of the Mouse Models of Human Cancers Consortium Prostate Pathology Committee 27. We identified pathologic changes featuring low grade prostatic intraepithelial neoplasia (PIN) in all prostate tissues isolated from the three different compound mouse strains at 2 months of age (Supplemental Fig.2A1–3‘, 2B1–3‘ and 2C1–3‘). Over time, these low-grade mPINs progressed towards more severe lesions. Gross examination showed abnormal growth appearance in the prostate of all 4-month-old mice (Fig. 2A1, 2B1, and 2C1). Histologically, ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4, and ArhAR21Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice showed similar prostatic adenocarcinoma development at four-months of age (Supplemental Fig.3 A1–2 and 3B1–2), while ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 only revealed less severe lesions, similar to high grade PIN (Supplemental Fig. 3C1–2). Nuclear staining for both stabilized β-catenin and humanized AR proteins was revealed in tumor or atypical cells within prostatic adenocarcinoma or PIN lesions in all of the compound mouse samples, respectively (Supplemental Fig. 3A3–4, 3B3–4 and 3C3–4). In contrast, untransformed luminal epithelial cells showed only membrane staining of endogenous β-catenin (red arrows, Supplemental Fig. 3C3). The cellular properties of the above prostatic adenocarcinomas and PIN lesions were further assessed using IHC approaches. Positive staining for E-cadherin, CK8, and mouse AR antibody appeared in most tumor and atypical cells (Supplemental Fig. 3A5–7, 3B5–7 and 3C5–7). Only very few scattered cells showed positive staining for CK5 or p63, but no cells showed staining for synaptophysin in the above sections (Supplemental Fig. 3A8–10, 3B8–10 and 3C8–10). These data demonstrate the luminal cell origin of prostatic adenocarcinomas and PIN from the humanized AR and stabilized β-catenin knock-in mice.
Figure 2. Humanized AR bearing shorter polyQ tracts enhances β-catenin-mediated oncogenic transformation and prostatic tumor development.
A-C. Gross images of prostates with seminal vesicles and urinary bladders from each different genotype mice at indicated ages. A1–4: Ar hAR12Q:Ctnnb1L(ex3)/+:PB-Cre4; B1–4: Ar hAR21Q:Ctnnb1L(ex3)/+:PB-Cre4; C1–4:Ar hAR48Q:Ctnnb1L(ex3)/+:PB-Cre4. The scale bar represents 5 mm. D. Graphical representation of the ratio of prostate wet weight/body weight. The prostate tissues were isolated from mice with indicated genotype at 12M of age. *P<0.05; **P<0.01. (n=6). E-G. Histological and immunohistochemistry analysis of prostate tissues isolated from mice with indicated genotype at 12M of age. E1–3, F1–3 and G1–3: Low and high magnification of representative H&E images for the prostatic tissues isolated from Ar hAR12Q:Ctnnb1L(ex3)/+:PB-Cre4, Ar hAR21Q:Ctnnb1L(ex3)/+:PB-Cre4 and Ar hAR48Q:Ctnnb1L(ex3)/+:PB-Cre mice, respectively. The scale bars represent 200 μm or 50 μm. E4–5, F4–5 and G4–5: Low and high magnification of representative IHC images were shown for Ki67 expression in prostate tissues isolated from Ar hAR12Q:Ctnnb1L(ex3)/+:PB-Cre4, Ar hAR21Q:Ctnnb1L(ex3)/+:PB-Cre4 and Ar hAR48Q:Ctnnb1L(ex3)/+:PB-Cre mice, respectively. The scale bar represents 50 μm or 12.5 μm. H. Summary of pathological abnormalities in the prostates of mice with indicated genotype. I. Quantification of Ki67 positive cells per 1000 epithelial cells from prostate tissues of each genotype, as indicated (n=3). **P<0.01.
The above mice were monitored continuously as their age progressed. Gross examination of aged mice revealed ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice showed faster tumor growth and more severe phenotypes than other genotype mice (Fig. 2A2–4, 2B2–4, 2C2–4, and 2H). The average weight of prostate tumor masses isolated from 12 month-old ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice was significantly higher than those from age-matched ArhAR21Q:Ctnnb1L(Ex3)/wt:PB-Cre4, or ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice (Fig. 2D). Histologically, large and aggressive adenocarcinoma lesions appeared in prostate tissues of both ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 (Fig. 2E1), and ArhAR21Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice (Fig. 2F1). They were predominantly intracystic, typically filling and markedly distending the lumen (Fig. 2E1–2 and 2F1–2). In some areas, neoplastic glandular structures invaded the prostatic stroma (arrows, Fig. 2E3). Irregular outpouchings of the intracystic carcinoma suggested incipient stromal invasion (Arrows, Fig. 2F3). In contrast, ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice showed variably sized cystically dilated glands with prominent squamous differentiation in the prostates (red arrows, Fig. 2G2 and Supplemental Fig. 4A). There was an abrupt transition from glandular epithelium to lamellar keratin (Fig. 2G1–3), and from columnar cyst lining to squamous metaplastic epithelium with prominent keratinization as well as calcification (Supplemental Fig. 4A1–2, respectively). The association between keratin and metaplastic squamous epithelium also appeared (Arrows, Fig. 2G2). Overall, tumor lesions in both Ar hAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 and ArhAR21Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice appeared much more aggressive than those in ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice. A significant increase in Ki67 positive tumor cells was revealed in tumor lesions of both ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 and ArhAR21Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice in comparison to those of ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice (Fig. 2E4–5 and 2F4–5 vs 2G4–5 and Fig. 2I). However, there is no significant difference of Ki67 positive cells between tumor samples of ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 and ArhAR21Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice. Taken together, the above data demonstrate a robust effect of humanized AR proteins bearing 12 and 21 polyQ tracts in cooperatively enhancing stabilized β-catenin mediated mouse prostatic tumor development, growth, and progression.
AR proteins bearing short PolyQ tracts enhance prostatic epithelia growth, oncogenic transformation, and PIN and prostatic tumor development and progression
In our compound mouse model, humanized AR proteins were expressed systematically 24. To directly assess the biological role of these AR proteins in prostatic epithelial cells, we used organoid culture assays to specifically evaluate the role of polyQ tract length in the fate and growth of prostatic epithelial cells. Since both humanized AR proteins bearing 12 and 21 polyQ tracts showed a significant role in enhancing stabilized β-catenin mediated prostatic tumor development, growth, and progression in comparison to AR48Q (Fig. 2), we analyzed prostatic epithelial cells isolated from 6 month old ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 and ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice in organoid culture experiments. Prostatic organoids were formed from prostatic epithelial cells sorted by CD24 antibody and cultured in Matrigel for 7 days (Fig. 3A). The number and size of organoids from prostatic epithelial cells of ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice were significantly greater than those from ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 cells (Fig. 3B1–2 versus Fig.3C1–2, and Fig. 3D). Histologically, organoids from prostatic epithelial cells of ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice showed much severer pathological lesions featuring multilayer atypical cells grown into lumen (Supplemental Fig. 5A1) or grown as cribriform structure (Supplemental Fig.5A5), whereas those from prostatic epithelial cells of ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice only revealed a few bridging structures with one or two layers of atypical cells (Supplemental Fig. 5B1 and 5B5). Expression of stabilized β-catenin (Supplemental Fig. 5A2, 5A6, 5B2 and 5B6) and AR (Supplemental Fig. 5A3 and 5B3) appeared in the above prostatic organoids from both genotypes. The majority of cells in organoids showed positive staining for CK8 but negative staining for CK5, suggesting their glandular cell origin (Supplemental Fig. 5A7–8 and 5B7–8), although a few cells were reactive to both CK5 and CK8 antibodies (Supplemental Fig. 5B7–8). There was no cell staining positively with synaptophysin (Supplemental Fig. 5A4 and 5B4). Additionally, organoids from prostatic epithelial cells of ArhAR12Q and ArhAR48Q mice were also examined (Supplemental Fig. 5C1 and 5D1). The organoids showed single layer epithelial cells and no pathological lesions, such as cribriform (Supplemental Fig. 5C1 and 5D1), and the expression of AR (Supplemental Fig. 5C2 and 5D2) and β-catenin (Supplemental Fig. 5C4 and 5D4) appeared in the above prostatic organoids. The organoids also showed positive staining for CK8, CK5 or both (Supplemental Fig. 5C3, 5C5, 5D3, and 5D5), but showed negative staining to synaptophysin (Supplemental Fig. 5C6 and 5D6). The above data recapitulate oncogenic transformation and abnormal cell growth mediated through the expression of stabilized β-catenin, and further demonstrate a strong effect of humanized AR proteins with the short length of polyQ tracts in enhancing stabilized β-catenin mediated prostatic cell oncogenic transformation and growth in the organoid cultures.
Figure 3. Humanized AR bearing shorter polyQ tracts enhances prostatic organoid formation and accelerates tumor development and progression.
A. Schematic representation of experimental design. Prostatic epithelial cells were dissociated from prostates of either Ar hAR12Q:Ctnnb1L(ex3)/+:PB-Cre4 or Ar hAR48Q:Ctnnb1L(ex3)/+:PB-Cre mice at 6M of age, and sorted based on CD24 staining. The same number of CD24+ cell from each genotype was used for organoid culture. 7 days after the culturing, organoids were collect and subjected to further analysis. B-C. Gross images of organoids derived from prostatic epithelial cells of each genotype (n=3). The scale bar represents 400μm. D. Quantification of organoids after culture for 7days (n=3). E. Schematic representation of experimental design. The anterior prostate lobes of either Ar hAR12Q:Ctnnb1L(ex3)/+:PB-Cre4 or Ar hAR48Q:Ctnnb1L(ex3)/+:PB-Cre mice at 2W of age were isolated and implanted under the kidney capsule of SCID mice. 12W after implantation, the xenografts were collected and subjected for further analysis. F. Gross image of xenografts derived from prostatic tissues of each genotype. (n=6) The scale bar represents 5 mm. G. Weight of xenografts (left panel) (n=6) and quantification of Ki67+ cells in graft tissues (right panel) (n=3). H and I. Histological and immunohistochemistry analysis of graft tissues with indicated genotype. H1-H3 and I1-I3. Representative H&E images were shown for the xenograft tissues isolated from different genotype mice as labeled. H4–7 and I4–7. Representative IHC images of adjacent xenograft tissue sections with indicated genotypes were shown. The prostate tissue sections were stained with different antibodies as labeled. The scale bar represents 200 μm, 50 μm or 12.5 μm.
To further assess the biological role of humanized AR proteins bearing different polyQ tracts, we performed tissue implantation experiments using prostate tissues of ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 or ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice. Similar sizes of prostatic anterior lobe tissues that were isolated from the different genotype mice at postnatal day 14 were grafted under the renal capsule of the same NOD/SCID mice, and harvested and analyzed 12 weeks post-implantation (Fig. 3E). The grafted implants from ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 were grossly bigger and heavier than those isolated from ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 (Fig. 3F–2 and Fig.3G left panel). Histological analysis showed typical adenocarcinoma lesions in graft tissues of ArhAR12Q:Ctnnb1L(Ex3)/wt :PB-Cre4 mice (Fig.3H1–3). The large tumor glands were composed of markedly atypical cells with enlarged, hyperchromatic nuclei and a high nucleus to cytoplasm ratio (Fig. 3H1–3). In contrast, grafts of Ar hAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice implanted in the same SCID mice showed smaller glandular structures with a preserved basal cell layer featuring HGPIN lesions (Fig. 3I1–3 and Supplemental Fig. 6B7). Strong nuclear and cytoplasmic staining for stabilized β-catenin and nuclear staining of humanized AR proteins were revealed in both atypical and tumor cells with the above lesions (Fig. 3H4–7, and 3I4–7), providing a direct link between the expression of humanized AR and stabilized β-catenin and the development of prostatic adenocarcinoma and PIN lesions. More Ki67 positive cells appeared in graft tissues of ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 than those of ArhAR48Q:Ctnnb1L(Ex3)/wt :PB-Cre4 mice (Fig. 3G, right panel; Supplemental Fig. 6A3–4 vs 6B3–4). The majority of tumor or atypical cells were stained with CK8, only scattered cells were positive staining for CK5, but no cell positively staining for synaptophysin within prostatic adenocarcinoma or PIN lesions from the above mice, respectively (Supplemental Fig. 6A5–10 and B5–10). These data provide an additional line of evidence to demonstrate the faster growing and more aggressive tumor phenotypes in prostatic implants of ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice.
The promotional role of short polyQ containing AR in inducing PIN and prostatic tumor growth and progression is androgen dependent
The activation of AR through androgens plays a central role in prostate tumorigenesis 3, 4. We observed a promotional role of the AR bearing short lengths of polyQ tracts in enhancing stabilized β-catenin mediated prostate cancer growth and progression, which is consistent with the previous observations that the length of polyQ tracts of AR is inversely correlated with AR transcriptional activity. High levels of androgens have also been linked to increased risk of prostate cancer development 28. To determine a direct role of androgens in the AR bearing short polyQ tracts in prostate cancer development and progression, we castrated 4-month-old mice, following prostatic tumor development, and analyzed them at 9 months of age (Fig. 4A). Significant tumor regression was revealed in prostatic tissues isolated from the above mice (Fig. 4B1–2, 4C1–2, and 4D1–2), but atypical and tumor cells still remained in regressed prostatic glands with strong cytoplasmic and nuclear β-catenin staining (Fig. 4B3–3’, 4C3–3’ and 4D3–3’). Interestingly, these atypical and tumor cells showed weak AR staining (Fig. 4B4–4’, 4C4–4’ and 4D4–4’). These observations implicate the growth of these prostatic tumors in the above compound mice is dependent on androgens, recapitulating abnormal activation of androgen and Wnt signaling pathways in human prostate cancer pathogenesis.
Figure 4. The promotional role of short polyQ containing AR in inducing PIN and prostatic tumor growth and progression is androgen dependent.
A. Schematic representation of experimental design. The mice with indicated genotypes were castrated at 4 months of age and analyzed at 9 months of age (n=3). B-D. Histological and immunohistochemistry analysis of prostate tissues with indicated genotype. B-B2, C-C2, and D-D2: Representative H&E images of prostatic tissues from Ar hAR12Q:Ctnnb1L(ex3)/+:PB-Cre4, Ar hAR21Q:Ctnnb1L(ex3)/+:PB-Cre4, or Ar hAR48Q:Ctnnb1L(ex3)/+:PB-Cre mice, respectively. The scale bar represents 200 μm or 40 μm, respectively. B3–4, C3–4, and D3–4: Immunohistochemistry analysis of prostate tissues with indicated genotype. Representative IHC images of adjacent prostatic tissue sections with indicated antibodies. The prostate tissues from different genotypes of mice were stained with β-catenin antibody. The scale bar represents 40 μm or 20 μm.
The expression of stabilized β-catenin was controlled by ARR2PB, a modified probasin promoter-driven Cre activity in the above compound mice 25. An increase in endogenous probasin mRNA levels was observed in prostate tissues of ArhAR12Q knock-in mice in comparison to those of ArhAR48Q mice 24. To exclude the possibility that the aggressive tumor phenotypes was due to increased expression of stabilized β-catenin by elevating ARR2PB promoter activity through short polyQ tract containing AR proteins, we performed a set of proof-in-principle experiments using newly generated ArhAR12Q:Ctnnb1L(Ex3)/wt:K8-CreERT2 and ArhAR48Q:Ctnnb1L(Ex3)/wt:K8CreERT2 mice (Fig. 5A), in which conditional expression of stabilized β-catenin is temporally controlled through Krt 8 promoter-driven Cre-ER activation in prostatic luminal epithelial cells 29. Tamoxifen (TM) was administrated at 8 weeks, prostate tissues were collected at 12 weeks and implanted under the kidney capsule of SCID mice, and the grafts were then harvested 8 weeks after implantation (Fig. 5A). Gross analysis showed greater size and increased weight of grafts of ArhAR12Q:Ctnnb1L(Ex3)/wt:K8CreERT2 than those of ArhAR48Q:Ctnnb1L(Ex3)/wt:K8CreERT2 mice (Fig. 5B and 5C). In addition, HGPIN and prostatic tumor lesions were present in the graft tissues from ArhAR12Q:Ctnnb1L(Ex3)/wt:K8CreERT2, featuring atypical cells with high nuclear cytoplasmic ratio (Fig. 5E1–2 and 5F1), whereas only LGPIN lesions were present in the graft tissues from ArhAR48Q:Ctnnb1L(Ex3)/wt:K8CreERT2 mice (Fig. 5G1–2 and 5H1). Both atypical and tumor cells showed strong nuclear and cytoplasmic staining for stabilized β-catenin (Fig. 5F3 and 5H3). IHC analyses showed the cellular properties of prostatic luminal cells within both PIN lesions of ArhAR12Q:Ctnnb1L(Ex3)/wt:K8CreERT2 and ArhAR48Q:Ctnnb1L(Ex3)/wt:K8CreERT2 grafts. Atypical cells showed positive staining for AR and CK8 (Fig. 5E3, 5F2, 5G3 and 5H2), but very few cells appeared to be positive for CK5 and no cells were positive for synaptophysin (Fig. 5E4–5 and 5G4–5). Increased Ki67 staining was revealed in HGPIN lesions in grafts of ArhAR12Q:Ctnnb1fl(Ex3)/wt:K8CreERT2 in comparison to those of ArhAR48Q:Ctnnb1L(Ex3)/wt:K8CreERT2 mice (Fig. 5F4 vs 5H4 and Fig. 5D). The expression of CyclinD1, a downstream target for β-catenin, was also elevated in ArhAR12Q:Ctnnb1L(Ex3)/wt:K8CreERT2 grafts in comparison with the counterparts bearing Q48 AR, implicating a link between higher proliferation status and stabilization of β-catenin in ArhAR12Q:Ctnnb1L(Ex3)/wt:K8CreERT2 grafts (Fig 5F5 vs 5H5). Using RT-qPCR approaches, we further assessed the activation of stabilized β-catenin in grafted prostate tumor tissues. Increased expression of β-catenin target genes, including Axin2, Cd44, Myc and Egr1, were revealed in grafted tumor tissues of ArhAR12Q:Ctnnb1L(Ex3)/wt:K8CreERT2 in comparison with those of ArhAR48Q:Ctnnb1L(Ex3)/wt:K8CreERT2 mice (Fig. 5I). These data demonstrate a direct role of AR proteins bearing short lengths of polyQ tracts in enhancing stabilized β-catenin mediated transcriptional activity.
Figure 5. Humanized AR bearing shorter polyQ tracts enhances β-catenin-mediated oncogenic transformation in CK8 expressing prostatic cells.
A. Schematic representation of experimental design. The male ArhAR12Q :Ctnnb1L(Ex3)/wt :K8CreERT2 and ArhAR48Q:Ctnnb1L(Ex3)/wt:K8CreERT2 mice were generated. Tamoxifen (TM) was administrated at 8 weeks and prostate tissues were collected at 12 weeks and implanted under the kidney capsule of SCID mice. The prostatic xenografts were harvested 8 weeks after implantation (n=5). B. Representative gross images of xenografts derived from prostatic tissues of each genotype, as labeled. The scale bar represents 2 mm. C. Weight of xenografts derived from prostatic tissues of each genotype (n=5). D. Quantification of Ki67+ cells in graft tissues derived from prostatic tissues of each genotype (n=3). E-H. Histological and immunohistochemistry analysis of xenograft tissues with indicated genotype. E1–2 and F1: Representative H&E images of xenograft tissues derived from prostate of ArhAR12Q :Ctnnb1L(Ex3)/wt :K8CreERT2 mice. The scale bar represents 50 μm and 12.5 μm, respectively. E3–5: Representative IHC images of adjacent xenograft tissue sections (ArhAR12Q :Ctnnb1L(Ex3)/wt :K8CreERT2) with CK8, CK5 and Synaptophysin antibodies, respectively. F2–5: Representative IHC images of adjacent xenograft tissue sections (ArhAR12Q :Ctnnb1L(Ex3)/wt :K8CreERT2) with hAR, β-catenin, Ki67 and CyclinD1, respectively. The scale bar represents 12.5 μm. G1–2 and H1: Representative H&E images of xenograft tissues derived from prostate of ArhAR48Q :Ctnnb1L(Ex3)/wt :K8CreERT2 mice. The scale bar represents 50 μm and 12.5 μm, respectively. G3–5: Representative IHC images of adjacent xenograft tissue sections (ArhAR48Q :Ctnnb1L(Ex3)/wt :K8CreERT2) with CK8, CK5 and Synaptophysin antibodies, respectively. H2–5: Representative IHC images of adjacent xenograft tissue sections (ArhAR48Q :Ctnnb1L(Ex3)/wt :K8CreERT2) with hAR, β-catenin, Ki67 and CyclinD1, respectively. The scale bar represents 12.5 μm. I. RT-qPCR analysis shows fold changes of the genes as labeled. Significance was determined by Students’ T test and data were represented as + SD (n=3 replicated per data point); * p<0.05; ** p<0.01.
Enrichment of cell signaling pathways related to aggressive tumor phenotypes and disease progression in prostate tumors of AR bearing shorter length of polyQ tracts
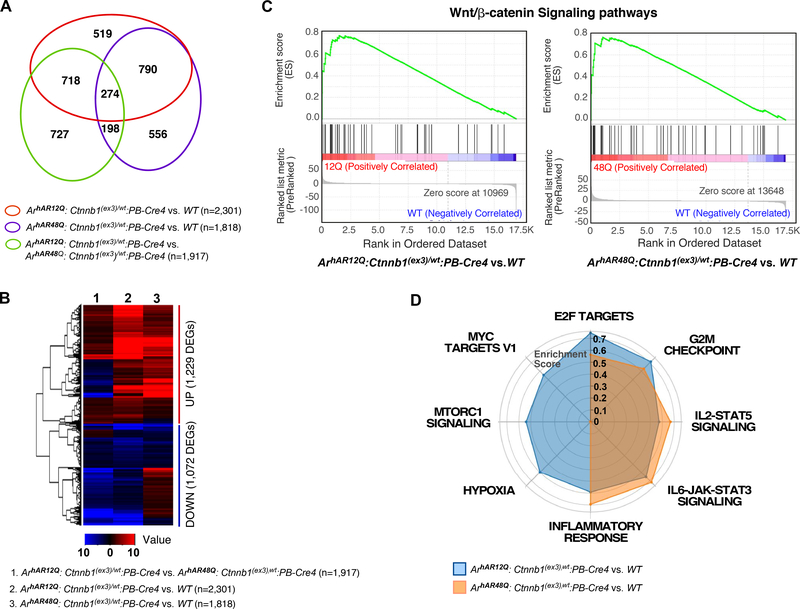
In search of the molecular mechanism by which AR bearing shorter polyQ tracts promotes tumorigenesis mediated by stabilization of β-catenin, we performed RNA sequencing (RNAseq) to examine the global transcriptome profiles using prostatic tumor tissues of ArhAR12Q:Ctnnb1L(Ex3)/wt :PB-Cre4 and ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice, representatively. We identified 1917, 2301, and 1818 differentially expressed genes (DEGs), with FDR<0.01 and fold change >1.5 by comparing RNAseq samples between ArhAR12Q:Ctnnb1fl(Ex3)/wt :PB-Cre4 versus ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4, and ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 or ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 versus wildtype (WT), respectively (Fig. 6A–B and Supplemental Table 2). To assess β-catenin mediated transcription, we compared gene sets from the samples of ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 or ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 versus WT mice and identified a group of overlapping DEGs (n=1064) between these two groups (Fig. 6A). GSEA analysis showed a significant enrichment of Wnt/β-catenin target genes in pre-ranked gene lists of ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 and ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 versus WT mice (Fig. 6C, Supplemental Table 3), implicating a dominant role of stabilized β-catenin in prostatic tumor development in these mice. Significant enrichment of other signaling pathways was also observed in hallmark gene sets using pre-ranked gene lists of ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 or ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 versus WT mice (Fig. 6D, Supplemental Table 4). Among them, E2F mediated signaling, G2/M checkpoint, IL2-Stat5, and IL6-Jak-Stat3 signaling and inflammatory response signaling were enriched in both the gene sets of ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 or ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 versus WT mice (Fig. 6D). While E2F mediated signaling and G2/M checkpoint showed higher enrichment score in the gene list of ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 versus WT mice, IL2-Stat5, and IL6-Jak-Stat3 signaling and inflammatory response signaling showed higher enrichment score in the gene list of ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 versus WT mice (Fig. 6D). Interestingly, the signaling pathways related to tumor progression, including Myc targets V1, Hypoxia and MTORC1 signaling pathways were only significantly enriched in the gene list of ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 versus WT mice, providing evidence of molecular mechanisms for the development of aggressive prostatic tumor phenotypes in ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice, with humanized AR bearing the short length of polyQ tract.
Figure 6. Determining the signaling pathways in mouse prostate tissues using RNA-seq analysis.
A. Venn diagram depicts the number of differentially expressed genes by different genotype mice as labeled above. B. Heatmap shows differential expression patterns of DEGs from the three comparisons as indicated in the figure. Red and blue indicates up- and downregulation, respectively. C. GSEA results show a significant enrichment of Wnt/β-catenin target genes in pre-ranked gene lists of ArhAR12Q:Ctnnb1fl(Ex3)/wt:PB-Cre4 or ArhAR48Q:Ctnnb1fl(Ex3)/wt:PB-Cre4 versus WT mice, respectively. D. Radar chart displays differential enrichment of ArhAR12Q:Ctnnb1fl(Ex3)/wt:PB-Cre4 and ArhAR48Q:Ctnnb1fl(Ex3)/wt:PB-Cre4 in comparison with wildtype. Blue and yellow lines indicate enriched hallmark gene sets in ArhAR12Q:Ctnnb1fl(Ex3)/wt:PB-Cre4 and ArhAR48Q:Ctnnb1fl(Ex3)/wt:PB-Cre4 models, respectively.
Elevated Myc expression and activation in prostate tumors mediated by stabilization of β-catenin and AR bearing shorter length of polyQ tracts
To assess the molecular mechanism for the promotional role of AR bearing shorter length of polyQ tracts on β-catenin mediated tumorigenesis, we searched for the potential regulators in the DEGs of ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 versus ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice using Ingenuity Upstream Regulator analysis in IPA combined with the ChIPseq database from Enrichr (Fig. 7A). Five transcription factors were identified, and, among them, MYC was shown to regulate 185 of the DEGs when comparing ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 versus ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice (Fig. 7B Supplemental Tables 5 and 6). Using RT-qPCR approaches, we validated an increase in the expression of Myc, E2f1, and Foxm1 in prostatic tumor samples of ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 in comparison with those of ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice (Fig. 7C). IHC analyses also showed increased expression of MYC proteins in prostatic tumor cells from samples of ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice in comparison to those of ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice (Fig. 7D–D’ vs Fig.7E–E’). GSEA analyses with hallmark gene sets, using a pre-ranked gene list comparing ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 versus ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice, revealed significant enrichment in Myc downstream target genes (Fig. 7F). A significant increase in the expression of Myc downstream targets, including Cd44, Mcm5, Birc5, Cnna2, Tpi-1 and Plk1 was also shown in the tumor samples of ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 in comparison with those of ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 (Fig. 7G). These data demonstrate a direct role of Myc in promoting fast growing and aggressive tumor phenotypes in ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice, which also implicates that stabilized β-catenin and short polyQ AR proteins contribute to elevating Myc expression in the tumor cells. Intriguingly, co-occurrence of aberrant genetic alterations between the Myc and CTNNB1 or APC genes was also detected in human prostate cancer samples (Supplemental Fig. 7). To further search for the regulatory mechanism for elevated Myc expression in ArhAR12Q:Ctnnb1fl(Ex3)/wt:PB-Cre4 mice, we performed chromatin immunoprecipitaion (ChIP) analyses with the immunoprecipitated genomic DNA samples isolated from prostatic tumor cells to examine the mouse c-Myc locus (Fig. 7H) 30. Increased recruitment of β-catenin was observed in the both β-catenin /TCF4-binding sites in tumor samples isolated from ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice in comparison to those from ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice (Fig. 7I). We further assessed the occupancy of the AR in the above loci to determine the involvement of AR in β-catenin mediated Myc expression. Interestingly, although both 12Q and 48Q AR proteins were recruited on the two TCF/β-catenin binding sites in c-Myc loci, 12Q AR proteins showed increased occupancies compare to their 48Q counterparts (Fig. 7J). Taken together, these results demonstrate a molecular mechanism underlying AR bearing shorter length of polyQ tracts in enhancing stabilized β-catenin regulated Myc expression in prostatic tumor cells, which directly contributes to fast growing and aggressive tumor phenotypes in the mouse prostate.
Figure 7. Investigating the molecular mechanisms for prostate tumors mediated by stabilization of β-catenin and AR bearing shorter length of PolyQ tracts.
A. Heatmap shows differential expression level of indicated transcriptional regulators. B. Bar plot displays the number of DEGs being regulated by the indicated transcriptional regulator (ArhAR12Q:Ctnnb1fl(Ex3)/wt:PB-Cre4 vs ArhAR48Q:Ctnnb1fl(Ex3)/wt:PB-Cre4). C. RT-qPCR analysis shows fold changes of the genes as labeled. Significance was determined by Students’ T test and data were represented as + SD (n=3 replicated per data point); * p<0.05; ** p<0.01. D-E. Representative IHC images with MYC antibody in prostatic tumor samples of different genotype mouse models as labeled in the Figure. The scale bar represents 50 μm or 12.5 μm. F. GSEA results show significant enrichment of hallmark gene sets, MYC target V1 and MYC target V2, in pre-ranked gene lists of samples from ArhAR12Q:Ctnnb1fl(Ex3)/wt:PB-Cre4 vs. that from ArhAR48Q:Ctnnb1fl(Ex3)/wt:PB-Cre4 mice. G. RT-qPCR analysis of alterations in genes comparing from ArhAR12Q:Ctnnb1fl(Ex3)/wt:PB-Cre4 vs ArhAR48Q:Ctnnb1fl(Ex3)/wt:PB-Cre4. * p<0.05, ** p<0.01. H. Schematic diagram showing the mouse c-Myc locus containing two β-catenin/TCF4 binding sites. I. β-catenin ChIP-qPCR (left graph) and AR ChIP-qPCR (right graph) shown as percent input of c-Myc gene on its binding sites A and B. Significance was determined by Students’ T test and data were represented as ± SD (n=3 replicated per data point); * p<0.05, ** p<0.01.
DISCUSSION
Although recent studies have shown significant improvements in overall survival in African American prostate cancer patients 31–33, African American men still possess the highest risk in both prostate cancer incidence and mortality 2. Therefore, there is an urgent need for identifying the molecular mechanisms underlying aggressive prostate tumor phenotypes and developing more effective and specific treatment for African American patients. Emerging evidence has shown a central role of the AR in prostate development and tumorigenesis 34. The polyQ tracts are located within exon 1 of human AR proteins, and their length is inversely correlated to AR transcriptional activity and associated directly with the risk of developing prostate cancer and aggressive phenotype of prostate cancer 8, 35. Intriguingly, African American men carry shorter length of polyQ tracts more frequently and show higher risk of prostate cancer development in comparison with Caucasian American men 35. Dysregulation of Wnt signaling pathways has been shown to contribute to prostate tumorigenesis 13. Aberrant activation of both androgen and Wnt signaling pathways were revealed in prostate cancer samples isolated from African American patients 23. Given the significance and clinical relevance of aberrant androgen and Wnt signaling pathways in prostate tumorigenesis as elucidated above, we directly assessed a collaborative role of aberrant AR and β-catenin signaling pathways using a series of newly generated mouse models in this study. The mouse models used in this study possess the conditional expression of stabilized β-catenin in prostatic epithelia of humanized AR knock-in mice bearing different length polyQ tracts, mimicking what happens in prostate cancer patients 24. The compound mice, which bear humanized AR proteins with short polyQ tracts, displayed an earlier onset of oncogenic transformation, an accelerated tumor development, and aggressive tumor phenotypes in the prostate in comparison to their counterparts with the longer polyQ tract AR proteins. These results provide the scientific evidence to recapitulate the collaborative role of β-catenin and AR proteins bearing short lengths of polyQ tracts in prostate tumorigenesis, and implicate the new mechanistic insight for fast growing and aggressive tumor phenotypes of prostate cancer that have been observed frequently in African American patients.
Emerging evidence has explored an important role of the Wnt/b-catenin signaling pathway in prostate tumorigenesis 13. However, since mutations in β-catenin and the destruction complex are rare in prostate cancer cells, other regulatory mechanisms have been implicated in regulating β-catenin mediated prostate tumorigenesis. The interaction between b-catenin and AR has been identified in prostate cancer cells 18–20. Through the interaction, β-catenin enhances AR mediated transcription as a transcriptional co-activator, and promotes prostate cell growth 19, 21. Since aberrant activations of androgen and Wnt signaling pathways frequently occur in prostate cancer, we assessed the effect of humanized AR proteins bearing 12, 21, and 48 polyQ tracts in enhancing aberrant β-catenin activation in prostate tumorigenesis. Although these humanized AR knock-in mice showed no pathologic abnormality 24, the compound mice with the expression of both humanized AR and stabilized β-catenin developed PIN and prostatic tumors. Interestingly, the compound mice bearing 12Q and 21Q humanized AR proteins showed fast growing and aggressive prostatic tumor phenotypes as well as severer pathologic lesions in comparison to their counterparts with humanized 48Q AR proteins. It should be noted that the humanized AR 48Q mice failed to develop muscle weakness and other abnormalities as observed in human Kennedy disease 24, 36. Our observations in the above compound mice suggest a distinct role of AR protein with a longer polyQ tract in β-catenin mediated prostate tumorigenesis. In addition, significant regression of prostatic tumors was observed in the above castrated compound mice with different AR polyQ proteins and stabilized β-catenin, which further implicates that stabilized β-catenin induced prostate tumorigenesis relies on AR and androgen medicated signaling pathways in the current mouse models.
In this study, we further examined the activity of AR bearing 12Q and 48Q tracts in prostate tumor development using the mouse models in which stabilized β-catenin expression is regulated by Krt8 promoter 29, a non-androgen regulated promoter. We observed an accelerated prostatic tumor development and aggressive tumor phenotypes in ArhAR12Q:Ctnnb1L(Ex3)/wt:K8-CreERT2 mice in comparison to the counterparts with AR 48Q proteins, which is very similar to what we saw in the compound mice with probasin promoter driven Cre. These lines of evidence further indicate a direct role of AR proteins with 12 polyQ tracts in enhancing stabilized β-catenin initiated prostate tumor growth and progression. The co-occurrence of short polyQ AR expression and aberrant activation of Wnt signaling pathways has been observed in prostate cancer samples of African American patients 11, 12, 23. Thus, our current compound mouse models provide the clinically relevant tools for further assessing the molecular mechanisms underlying aberrant AR polyQ proteins and stabilized β-catenin expression in prostate cancer development and progression.
Using RNAseq analysis, we searched for the molecular mechanisms underlying short polyQ tract-containing AR and stabilized β-catenin -induced prostate tumorigenesis. GSEA analyses with pre-ranked gene lists showed significant enrichment in the signaling pathways related to E2F targets, G2/M checkpoint, IL2-Stat5, and IL6-Jak-Stat3 in samples from ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 or ArhAR48Q :Ctnnb1L(Ex3)/wt :PB-Cre4 mice in comparison to WT mice. Interestingly, significant enrichment in Myc-mediated gene sets was observed in the DEGs of ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 versus ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 or wildtype control mice, suggesting a collaborative role of stabilized β-catenin and short polyQ tract AR proteins in regulating Myc activity. Using IPA upstream regulator analyses, we further identified five master transcriptional regulators based on the above DEGs, which have been shown to play a direct role in prostate tumorigenesis 37–41. Among them, the MYC oncoprotein was on the top of the list and reveled to regulate more DEGs than other regulators. Using IHC analyses, we further validated increased MYC expression in prostatic tumor cells in ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice. Elevated expression of MYC downstream target genes, such as Mcm5, Ccna2 and Birc5, was also observed in the above tumor tissues. These data demonstrate increasing expression and activity of MYC in prostate tumors of ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice. Myc is one of β-catenin downstream target genes and its expression is regulated by activation of Wnt/β-catenin signaling 42. The amplification of the MYC gene has been frequently observed in human prostate cancer samples harboring genomic alterations and mutations in CTNNB1 and/or APC, resulting the upregulation of Myc expression in prostatic tumor cells 43. Intriguingly, aberrant expression and activation of Myc also appeared to be enriched in African American prostate cancers 44. Using ChIP analyses, we further demonstrate a collaborative role of stabilized β-catenin and short polyQ tract containing AR on binding to two β-catenin/TCF4-binding regions on the mouse c-Myc loci, which leads to increasing Myc expression in prostate tumor cells. These results implicate a novel molecular insight into the AR proteins bearing shorter length of polyQ tracts in enhancing β-catenin-induced prostate tumorigenesis, which also provide scientific evidence for MYC inhibition in treating aggressive prostate cancer that frequently occurs in African American patients.
MATERIALS AND METHODS
Mouse mating, genotyping, and castration
All animal experiments performed in this study were approved by the Institutional Animal Care and Use Committee at Beckman Research Institute/City of Hope. All mice used in this study were from a C57BL/6 background. The Ctnnb1(Ex3)fl (Ctnnb1(Ex3)L)mice 26 were first crossed with the humanized AR mice 24 to generate Ctnnb1(Ex3)L/L:ArhAR12Q/hAR12Q, Ctnnb1(Ex3)L/L:ArhAR21Q/hAR21Q and Ctnnb1(Ex3)L/L:ArhAR48Q/hAR48Q mice. The above homozygous females were then crossed with PB-Cre4 males 25 to generate the following male mice: ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4, ArhAR21Q:Ctnnb1L(Ex3)/wt:PB-Cre4, and ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice. The male K8-CreERT2 mice 29 were crossed with either Ctnnb1(Ex3)L/L:ArhAR12Q/hAR12Q or Ctnnb1(Ex3)L/L:ArhAR48Q/hAR48Q female mice to generate ArhAR12Q:Ctnnb1L(Ex3)/wt:K8-CreERT2 and ArhAR48Q:Ctnnb1L(Ex3)/wt:K8-CreERT2 male mice. Mice were genotyped by PCR using specific primers (Supplemental Table 7). Three or more mice of each genotype were analyzed at different time points, as indicated in each experiment. Castration of adult male mice was performed 45. Briefly, the mice were anesthetized by intraperitoneal injection of ketamine and xylazine. Both testicles and epididymis were removed through a scrotal approach. The distal end of the spermatic cord was ligated with silk thread.
Pathological analyses
The guidelines recommended by The Mouse Models of Human Cancers Consortium Prostate Pathology Committee in 2013 were used for the pathological analyses in this study 27. Mouse tissues were processed and Hematoxylin-eosin staining (H&E) was performed as described previously 46.
Immunohistochemistry (IHC) and microscope image acquisition
IHC was performed as previously described 46. Tissue sections were treated by boiling in 0.01M citrate buffer (pH 6.0) for antigen retrieval, blocked in 5% normal goat serum, and incubated with primary antibodies (see Supplemental Table 8) diluted in 1% normal goat serum at 4°C overnight. Slides were incubated with biotinylated secondary antibodies for 1 hour then with horseradish peroxidase streptavidin (SA-5004, Vector Laboratories, Burlingame, CA, USA) for 30 min, visualized by DAB kit (SK-4100, Vector Laboratories), then counterstained with 5% (w/v) Harris Hematoxylin, and subsequently mounted with Permount Mounting Medium (SP15–500, Thermo Fisher Scientific, Waltham, MA, USA). Images of H&E and IHC were acquired on an Axio Lab A1 microscope using 10x and 40x Zeiss A-Plan objectives with a Canon EOS 1000D camera and using Axiovision software (Carl Zeiss, Oberkochen, Germany).
Prostate regeneration and organoid culture assays
Mouse prostatic tissues were collected from male mice with desired genotypes and ages and portions of dissected prostatic lobes were implanted under the renal capsule of SCID mice 2. Twelve weeks after the implantation, the SCID mice were euthanized and the grafts were collected for further analyses. Organoid culture with prostatic epithelial cells was performed as previously described 47. The prostatic tissue were collected, minced and subjected to single cell dissociation through collagenase and TrypLE treatment (Gibco). Dissociated cells were stained with anti-mouse CD24 antibody and sorted with an AriaIII. The single cells were then mixed with Matrigel, and cultured for 7 days.
Immunoblotting
The mouse prostate tissues were isolated and the whole cell lysates were prepared and applied for SDS-PAGE gels 46. The proteins were then transferred to nitrocellulose (Schleicher & Schüll, Keene, NH, USA) and blocked in TBS-T (50 mM Tris-HCl, 150 mM NaCl and 0.08% Tween 20) with 5% dry skim milk. Membranes were probed with desired primary antibodies, followed by anti-rabbit or mouse immunoglobulin G conjugated to horseradish peroxidase secondary antibodies (Promega, Madison, WI, USA). Detection was performed with enhanced chemiluminescence reagents (Amersham Biosciences, Pittsburgh, PA, USA).
RNA isolation, reverse transcription-Quantitative PCR (RT-qPCR), and RNA sequencing
RNA samples were isolated from fresh mouse tissues using RNA-Bee (TEL-TEST, Inc., Friendswood, TX, USA) for preparing RNA sequencing libraries with Kapa RNA HyperPrep Kit (Kapa Biosystems, Cat KK8581). Sequencing runs were performed on Illumina Hiseq 2500 in the single read mode of 51 cycles of read 1 and 7 cycles of index with V4 Kits. Real-time analysis (RTA) 2.2.38 software was used to process the image analysis and base calling. Reverse transcription was performed as described previously 21, and RT-qPCR assays were carried out using SYBR GreenER qPCR Super Mix Universal (11762, Invitrogen) with specific primers (Supplemental Table 9) on the 7500 Real-Time PCR system (Thermo Fisher Scientific).
Chromatin immunoprecipitation (ChIP) Assays
ChIP assays were performed as described previously 48. Briefly, mouse tissues were minced and incubated with 1% formaldehyde for 15 min and quenched with 0.150 M glycine for 10 min. Samples were washed sequentially with cold PBS, and resuspended in cell lysis buffer (50 mM Tris-HCl (pH 8.0), 140 mM NaCl, 1 mM EDTA, 10% Glycerol, 0.5% NP-40, and 0.25% Triton X-100), and then homogenized. The chromatin was sheared in nuclear lysis buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 0.5 mM EGTA, and 0.2% SDS) to an average size of 200–500 bp by sonication, and then diluted 3-fold in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.1, and 167 mM NaCl), and was subjected to immunoprecipitation by magnetic protein G beads (Invitrogen) conjugated with AR (ab74272, abcam) or β-catenin antibody (610154, BD Biosciences). Cross-links were reversed and chromatin DNA fragments were analyzed by real-time qPCR with specific primers (Supplemental Table 9).
Data preprocessing, normalization, and analyses
Reads were aligned against the mouse genome (mm10) using TopHat2 (v.2.0.14) 49. Read counts were tabulated using HTSeq-count (v0.6.0) 50, with UCSC known gene annotations (TxDb. Mmusculus. UCSC. Mm10. knownGene) 51. Fold-change values were calculated from Fragments Per Kilobase per Million reads (FPKM) 52 normalized expression values, which were also used for visualization (following a log2 transformation). Aligned reads were counted using GenomicRanges 53. False discovery rate (FDR) and P values were calculated using the method of Benjamini and Hochberg 54 and from raw counts using DEseq2 (v1.14.1) 52. Prior to p-value calculation, genes were filtered to only include transcripts with an FPKM expression level of 0.1 (after a rounded log2-transformation) in at least 50% of samples as well as genes that are greater than 150 bp. Genes were defined as differentially expressed if they had a | fold-change|>1.5 and FDR<0.01. Using pre-ranked gene lists, enriched hallmark gene sets were identified as having nominal P<0.05 from gene set enrichment analysis (GSEA). GSEA analysis with Wnt/β-catenin signaling pathways was performed using a Wnt target gene list (http://web.stanford.edu/group/nusselab/cgi-bin/wnt/). IPA upstream analysis and Erichr ChEA2016 were used to identify the upstream regulators based on the DEGs of ArhAR12Q:Ctnnb1L(Ex3)/wt:PB-Cre4 versus ArhAR48Q:Ctnnb1L(Ex3)/wt:PB-Cre4 mice.
Statistical Analysis
Data are shown as the mean ± SD. Differences between groups were examined by 2-tailed Student’s t test or 2-way ANOVA for comparisons among multiple groups. For all analyses, P < 0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by Public Health Service grants, R01CA070297, R01CA166894, R21CA190021, and R01DK104941.
Footnotes
COMPETING INTERESTS: The authors declare no potential conflicts of interest.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019; 69: 7–34. [DOI] [PubMed] [Google Scholar]
- 2.DeSantis CE, Miller KD, Goding Sauer A, Jemal A, Siegel RL. Cancer statistics for African Americans, 2019. CA Cancer J Clin 2019; 69: 211–233. [DOI] [PubMed] [Google Scholar]
- 3.Culig Z Role of the androgen receptor axis in prostate cancer. Urology 2003; 62: 21–26. [DOI] [PubMed] [Google Scholar]
- 4.Kyprianou N, Isaacs JT. Activation of programmed cell death in the rat ventral prostate after castration. Endocrinology 1988; 122: 552–562. [DOI] [PubMed] [Google Scholar]
- 5.Jenster G, van der Korput HA, van Vroonhoven C, van der Kwast TH, Trapman J, Brinkmann AO. Domains of the human androgen receptor involved in steroid binding, transcriptional activation, and subcellular localization. Mol Endocrinol 1991; 5: 1396–1404. [DOI] [PubMed] [Google Scholar]
- 6.Zhou ZX, Wong CI, Sar M, Wilson EM. The androgen receptor: an overview. Recent Progress Hormone Research 1994; 49: 249–274. [DOI] [PubMed] [Google Scholar]
- 7.Clark PE, Irvine RA, Coetzee GA. The androgen receptor CAG repeat and prostate cancer risk. Methods Mol Med 2003; 81: 255–266. [DOI] [PubMed] [Google Scholar]
- 8.Stanford JL, Just JJ, Gibbs M, Wicklund KG, Neal CL, Blumenstein BA et al. Polymorphic repeats in the androgen receptor gene: molecular markers of prostate cancer risk. Cancer Res 1997; 57: 1194–1198. [PubMed] [Google Scholar]
- 9.Choong CS, Wilson EM. Trinucleotide repeats in the human androgen receptor: a molecular basis for disease. J Mol Endocrinol 1998; 21: 235–257. [DOI] [PubMed] [Google Scholar]
- 10.Mhatre AN, Trifiro MA, Kaufman M, Kazemi-Esfarjani P, Figlewicz D, Rouleau G et al. Reduced transcriptional regulatory competence of the androgen receptor in X-linked spinal and bulbar muscular atrophy. Nat Genet 1993; 5: 184–188. [DOI] [PubMed] [Google Scholar]
- 11.Bennett CL, Price DK, Kim S, Liu D, Jovanovic BD, Nathan D et al. Racial variation in CAG repeat lengths within the androgen receptor gene among prostate cancer patients of lower socioeconomic status. J Clin Oncol 2002; 20: 3599–3604. [DOI] [PubMed] [Google Scholar]
- 12.Sartor O, Zheng Q, Eastham JA. Androgen receptor gene CAG repeat length varies in a race-specific fashion in men without prostate cancer. Urology 1999; 53: 378–380. [DOI] [PubMed] [Google Scholar]
- 13.Verras M, Sun Z. Roles and regulation of Wnt signaling and beta-catenin in prostate cancer. Cancer Lett 2006; 237: 22–32. [DOI] [PubMed] [Google Scholar]
- 14.Robinson D, Van Allen EM, Wu Y-M, Schultz N, Lonigro RJ, Mosquera J-M et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015; 161: 1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chesire DR, Isaacs WB. Ligand-dependent inhibition of beta-catenin/TCF signaling by androgen receptor. Oncogene 2002; 21: 8453–8469. [DOI] [PubMed] [Google Scholar]
- 16.Bierie B, Nozawa M, Renou JP, Shillingford JM, Morgan F, Oka T et al. Activation of beta-catenin in prostate epithelium induces hyperplasias and squamous transdifferentiation. Oncogene 2003; 22: 3875–3887. [DOI] [PubMed] [Google Scholar]
- 17.Gounari F, Signoretti S, Bronson R, Klein L, Sellers WR, Kum J et al. Stabilization of beta-catenin induces lesions reminiscent of prostatic intraepithelial neoplasia, but terminal squamous transdifferentiation of other secretory epithelia. Oncogene 2002; 21: 4099–4107. [DOI] [PubMed] [Google Scholar]
- 18.Mulholland DJ, Cheng H, Reid K, Rennie PS, Nelson CC. The androgen receptor can promote beta-catenin nuclear translocation independently of adenomatous polyposis coli. J Biol Chem 2002; 277: 17933–17943. [DOI] [PubMed] [Google Scholar]
- 19.Truica CI, Byers S, Gelmann EP. Beta-catenin affects androgen receptor transcriptional activity and ligand specificity. Cancer Res 2000; 60: 4709–4713. [PubMed] [Google Scholar]
- 20.Yang F, Li X, Sharma M, Sasaki CY, Longo DL, Lim B et al. Linking beta-catenin to androgen-signaling pathway. J Biol Chem 2002; 277: 11336–11344. [DOI] [PubMed] [Google Scholar]
- 21.Lee SH, Luong R, Johnson DT, Cunha GR, Rivina L, Gonzalgo ML et al. Androgen signaling is a confounding factor for beta-catenin-mediated prostate tumorigenesis. Oncogene 2016; 35: 702–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weischenfeldt J, Simon R, Feuerbach L, Schlangen K, Weichenhan D, Minner S et al. Integrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset prostate cancer. Cancer Cell 2013; 23: 159–170. [DOI] [PubMed] [Google Scholar]
- 23.Wang BD, Yang Q, Ceniccola K, Bianco F, Andrawis R, Jarrett T et al. Androgen receptor-target genes in african american prostate cancer disparities. Prostate Cancer 2013; 2013: 763569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albertelli MA, Scheller A, Brogley M, Robins DM. Replacing the mouse androgen receptor with human alleles demonstrates glutamine tract length-dependent effects on physiology and tumorigenesis in mice. Mol Endocrinol 2006; 20: 1248–1260. [DOI] [PubMed] [Google Scholar]
- 25.Wu X, Wu J, Huang J, Powell WC, Zhang J, Matusik RJ et al. Generation of a prostate epithelial cell-specific Cre transgenic mouse model for tissue-specific gene ablation. Mech Dev 2001; 101: 61–69. [DOI] [PubMed] [Google Scholar]
- 26.Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, Oshima M et al. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. Embo J 1999; 18: 5931–5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ittmann M, Huang J, Radaelli E, Martin P, Signoretti S, Sullivan R et al. Animal models of human prostate cancer: the consensus report of the New York meeting of the Mouse Models of Human Cancers Consortium Prostate Pathology Committee. Cancer Res 2013; 73: 2718–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gann PH, Hennekens CH, Ma J, Longcope C, Stampfer MJ. Prospective study of sex hormone levels and risk of prostate cancer. J Natl Cancer Inst 1996; 88: 1118–1126. [DOI] [PubMed] [Google Scholar]
- 29.Zhang L, Zhang B, Han SJ, Shore AN, Rosen JM, Demayo FJ et al. Targeting CreER(T2) expression to keratin 8-expressing murine simple epithelia using bacterial artificial chromosome transgenesis. Transgenic Res 2012; 21: 1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahmoudi T, Boj SF, Hatzis P, Li VS, Taouatas N, Vries RG et al. The leukemia-associated Mllt10/Af10-Dot1l are Tcf4/beta-catenin coactivators essential for intestinal homeostasis. PLoS Biol 2010; 8: e1000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dess RT, Hartman HE, Mahal BA, Soni PD, Jackson WC, Cooperberg MR et al. Association of Black Race With Prostate Cancer-Specific and Other-Cause Mortality. JAMA Oncol 2019; 5: 975–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McNamara MA, Oyekunle T, Chin BB, Oldan J, Anand A, Ritz M et al. Patterns of response and progression in bone and soft tissue during and after treatment with radium-223 for metastatic castrate-resistant prostate cancer. Prostate 2019; 79: 1106–1116. [DOI] [PubMed] [Google Scholar]
- 33.Spratt DE, Chen YW, Mahal BA, Osborne JR, Zhao SG, Morgan TM et al. Individual Patient Data Analysis of Randomized Clinical Trials: Impact of Black Race on Castration-resistant Prostate Cancer Outcomes. Eur Urol Focus 2016; 2: 532–539. [DOI] [PubMed] [Google Scholar]
- 34.Jenster G The role of the androgen receptor in the development and progression of prostate cancer. Semin Oncol 1999; 26: 407–421. [PubMed] [Google Scholar]
- 35.Irvine RA, Yu MC, Ross RK, Coetzee GA. The CAG and GGC microsatellites of the androgen receptor gene are in linkage disequilibrium in men with prostate cancer. Cancer Res 1995; 55: 1937–1940. [PubMed] [Google Scholar]
- 36.Yu Z, Dadgar N, Albertelli M, Scheller A, Albin RL, Robins DM et al. Abnormalities of germ cell maturation and sertoli cell cytoskeleton in androgen receptor 113 CAG knock-in mice reveal toxic effects of the mutant protein. Am J Pathol 2006; 168: 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koh CM, Bieberich CJ, Dang CV, Nelson WG, Yegnasubramanian S, De Marzo AM. MYC and Prostate Cancer. Genes Cancer 2010; 1: 617–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan H, Zhu Y, Wei W, Shao S, Rui X. Transcription factor FoxM1 is the downstream target of c-Myc and contributes to the development of prostate cancer. World J Surg Oncol 2018; 16: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shukla S, Bhaskaran N, Maclennan GT, Gupta S. Deregulation of FoxO3a accelerates prostate cancer progression in TRAMP mice. Prostate 2013; 73: 1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, Place RF, Huang V, Wang X, Noonan EJ, Magyar CE et al. Prognostic value and function of KLF4 in prostate cancer: RNAa and vector-mediated overexpression identify KLF4 as an inhibitor of tumor cell growth and migration. Cancer Res 2010; 70: 10182–10191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng C, Ren Z, Wang H, Zhang W, Kalvakolanu DV, Tian Z et al. E2F1 Induces tumor cell survival via nuclear factor-kappaB-dependent induction of EGR1 transcription in prostate cancer cells. Cancer Res 2009; 69: 2324–2331. [DOI] [PubMed] [Google Scholar]
- 42.Rennoll S, Yochum G. Regulation of MYC gene expression by aberrant Wnt/beta-catenin signaling in colorectal cancer. World J Biol Chem 2015; 6: 290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fleming WH, Hamel A, MacDonald R, Ramsey E, Pettigrew NM, Johnston B et al. Expression of the c-myc protooncogene in human prostatic carcinoma and benign prostatic hyperplasia. Cancer Res 1986; 46: 1535–1538. [PubMed] [Google Scholar]
- 44.Kumar R, Bhat TA, Walsh EM, Chaudhary AK, O’Malley J, Rhim JS et al. Cytochrome c Deficiency Confers Apoptosome and Mitochondrial Dysfunction in African-American Men with Prostate Cancer. Cancer Res 2019; 79: 1353–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sugimura Y, Cunha GR, Donjacour AA. Morphological and histological study of castration-induced degeneration and androgen-induced regeneration in the mouse prostate. Biol Reprod 1986; 34: 973–983. [DOI] [PubMed] [Google Scholar]
- 46.Zhu C, Luong R, Zhuo M, Johnson DT, McKenney JK, Cunha GR et al. Conditional expression of the androgen receptor induces oncogenic transformation of the mouse prostate. J Biol Chem 2011; 286: 33478–33488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Drost J, Karthaus WR, Gao D, Driehuis E, Sawyers CL, Chen Y et al. Organoid culture systems for prostate epithelial and cancer tissue. Nat Protoc 2016; 11: 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee J, Beliakoff J, Sun Z. The novel PIAS-like protein hZimp10 is a transcriptional co-activator of the p53 tumor suppressor. Nucleic Acids Res 2007; 35: 4523–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013; 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015; 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC Known Genes. Bioinformatics 2006; 22: 1036–1046. [DOI] [PubMed] [Google Scholar]
- 52.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 2008; 5: 621–628. [DOI] [PubMed] [Google Scholar]
- 53.Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R et al. Software for computing and annotating genomic ranges. PLoS Comput Biol 2013; 9: e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J R Stat Soc B 1995; 57: 289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.