Highlights
-
•
Autophagy is beneficial to PEDV replication.
-
•
PEDV nonstructural protein 6 (nsp6) is a key protein involved in PEDV-induced autophagy.
-
•
Nsp6 of PEDV induced autophagy via the PI3K/Akt/mTOR signaling pathway.
Keywords: Porcine epidemic diarrhea virus, Nonstructural protein 6, Autophagy, Replication
Abstract
Porcine epidemic diarrhea virus (PEDV) has caused, and continues to cause, severe economic losses to the swine industry worldwide. The pathogenic mechanism and immune regulatory interactions between PEDV and the host remain largely unknown. In this study, the interaction between autophagy and PEDV replication in intestinal porcine epithelial (IPEC-J2) cells was investigated. The effects of the structural and nonstructural proteins of PEDV on the autophagy process and the autophagy-related signaling pathways were also examined. The results shown that PEDV replication increased the autophagy flux in IPEC-J2 cells, and that autophagy was beneficial to PEDV replication, which may be one of the reasons for the rapid damage to intestinal epithelial cells and the enhanced virulence of PEDV in both newborn piglets and finishing pigs. When autophagy was pharmacologically induced by rapamycin, PEDV replication increased from 8.5 × 105 TCID50/mL to 8.8 × 106 TCID50/mL in IPEC-J2 cells. When autophagy was pharmacologically suppressed by hydroxychloroquine, PEDV replication decreased from 8.5 × 105 TCID50/mL to 7.9 × 104 TCID50/mL. To identify which PEDV proteins were the key inducers of autophagy, all 4 structural proteins and 17 nonstructural proteins of PEDV were eukaryotic expressed. It was found that the nonstructural protein 6 (nsp6) and ORF3 of PEDV were able to induce significant autophagy in IPEC-J2 cells, but the other proteins were unable to induce autophagy. It was indicated that nsp6-induced autophagy mainly occurred via the PI3K/Akt/mTOR signaling pathway. The results accelerate the understanding of the biology and pathogenesis of PEDV infection and provide new insights into the development of effective therapeutic strategies.
1. Introduction
Porcine epidemic diarrhea virus (PEDV) is a reemerging swine enteropathogenic coronavirus belonging to the genus Alphacoronavirus of the family Coronaviridae. PEDV is the causative agent of porcine epidemic diarrhea, a condition that presents as severe diarrhea, vomiting, and dehydration. As PEDV is highly contagious, the resulting economic burden to the swine industry worldwide is substantial (Bertasio et al., 2016; Gerber et al., 2016; Wang et al., 2014a). Since December 2010, swine farms in China have seen large-scale outbreaks of PEDV infection with typical mortality rates of 50–90% in suckling piglets (Bi et al., 2012; Chen et al., 2012; Wei et al., 2012). In the United States, PEDV pandemic strains have caused more than 80 % mortality in suckling piglets and have been rapidly transmitted between farms since 2013. There were over 8 million piglets lost, which represents nearly 10 % of all U.S. farm piglets in 2014 (Stevenson et al., 2013). There were some new features of this PEDV epidemic, including the high mortality rates in suckling piglets. Another feature was the rapid occurrence of diarrhea, dehydration and death, not only in suckling piglets but also in finishing pigs. Numerous studies have focused on viral isolation and molecular epidemiology surveys (Park et al., 2012), which indicated that PEDV pandemic strains could rapidly replicate in vivo, cause damage to intestinal epithelial cells, and ultimately induce diarrhea, dehydration and death in both newborn piglets and finishing pigs.
PEDV is an enveloped, single-stranded, positive-sense RNA virus that is composed primarily of four structural proteins, the spike protein (S), membrane protein (M), envelope protein (E), and nucleocapsid protein (N), and 17 nonstructural proteins (nsp), nsp1-nsp16 and ORF3. The S protein is located on the surface of the virus particle and is a type I membrane fusion protein. S protein binds to receptors on the target cell, and then, the virus enters the cell through fusion with the plasma membrane or via endocytosis (Chang et al., 2002; Hou et al., 2019; Park et al., 2014; Supekar et al., 2004). Several viral proteins have a suppressive effect on innate immunity. The N protein directly interacts with TBK1, resulting in the inhibition of IFN regulatory factor 3 (IRF3) activation and type I IFN production (Ding et al., 2014). Nsp1 blocks the nuclear translocation of IRF1, resulting in a reduction in peroxisomes and thereby suppressing IRF1-mediated type III IFNs (Zhang et al., 2017). Nsp5, a 3C-like protease, disrupts type I IFN signaling by cleaving NF-κB essential modulator (NEMO), which is an essential adaptor that bridges interferon-regulatory factor and NF-κB activation (Wang et al., 2015). Nsp6 of coronaviruses is integral membrane protein believed to be involved in double membrane vesicle (DMV) formation (Angelini et al., 2013; Oostra et al., 2008). Nsp6 of the infectious bronchitis virus (IBV) generates autophagosomes from the endoplasmic reticulum (Cottam et al., 2014).
Autophagy is a lysosome-dependent, ubiquitous physiological mechanism that serves as a type of quality control in eukaryotic cells (Alexander et al., 2007; Lussignol et al., 2013; Singh et al., 2017; Surviladze et al., 2013). Autophagy is not only a physiological cellular pathway for self-degradation but also a defense mechanism that protects host cells against pathogen infection (Chiramel and Best, 2017; Maier et al., 2013; O’Connell and Liang, 2016; Rozieres et al., 2017). Autophagy plays important roles in modulating viral replication and antiviral immune response (McFarlane et al., 2011; Mohl et al., 2012; Monastyrska et al., 2013; Paul and Munz, 2016; Radtke et al., 2013). Increasing evidence indicates that autophagy plays both anti-viral and pro-viral roles in the life cycles and pathogenesis of a wide range of viruses (Chiramel et al., 2013; Jackson, 2015; Zang et al., 2016). Previous studies have shown that coronavirus infection is associated with the autophagic process, and that the nonstructural proteins of coronavirus induce the formation of autophagosomes from the endoplasmic reticulum via an omegasome intermediate (Cottam et al., 2011).
PEDV pandemic strains can proliferate rapidly in vivo and cause damage to the intestinal epithelial cells of both newborn piglets and finishing pigs (Jung et al., 2015; Lin et al., 2016). Whether these biological effects are associated with autophagy is unclear. In our previous research, it was found that the PEDV pandemic strain could activate more significantly autophagy-related signaling pathway compared with the PEDV classical strain in intestinal porcine epithelial (IPEC-J2) cells, and the proliferation rate of the PEDV pandemic strain was significantly higher than that of the PEDV classical strain, which indicated that there may be a positive correlation between autophagy and PEDV replication (Lin et al., 2017). It was reported that autophagy was beneficial to PEDV replication in African green monkey kidney (Vero) cells, and the PEDV-induced autophagy exhibited a positive feedback loop with the NF-κB signaling pathway (Guo et al., 2017). As an intestinal proliferated virus, the interactions between PEDV and the intestinal porcine epithelial cells are still unknown, which is extremely important for determining the pathogenic mechanism and immune regulatory interactions between PEDV and the host. Moreover, which proteins (structural or nonstructural proteins) are the key inducers of autophagy has yet to be explored. In this study, it was investigated the mechanisms by which PEDV infection affected IPEC-J2 cells autophagy, and by which autophagy affected PEDV replication in IPEC-J2 cells. The effects of structural and nonstructural proteins of PEDV on the process of PEDV-induced autophagy and the autophagy-related signaling pathway activated by PEDV replication were also examined.
2. Materials and methods
2.1. Cells, virus, and reagents
The intestinal porcine epithelial cell line (IPEC-J2) was originally obtained from Creative Bioarray (New York, USA). Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Life Technologies, USA) supplemented with 10 % heat-inactivated fetal bovine serum (FBS, Invitrogen, USA) at 37 °C in a humidified 5% CO2 atmosphere. The PEDV strain used in this study, YC2014 (GenBank: KU252649.1), was isolated in our laboratory from a sucking piglet with acute diarrhea, and was propagated in IPEC-J2 cells. PEDV was made replication-incompetent by irradiating 10 mL of suspended virus, dispensed to form a layer of fluid in an open cell culture dish, with UV light for 5 h with gentle shaking at intervals. The absence of virus replication after UV treatment was confirmed by indirect immunofluorescence assay (IFA). The monoclonal antibody (mAb) 3A6 anti-PEDV N protein was prepared in our laboratory. Rapamycin, hydroxychloroquine, and anti-LC3 antibody were obtained from Sigma-Aldrich (Shanghai, China). The Akt-specific inhibitor GSK690693 was obtained from Selleck (Shanghai, China). The lentivirus LV-mRFP-GFP-LC3 was obtained from Hanbio (HB-LP210 0001, Shanghai, China) (Chen et al., 2017; Wang et al., 2014b; Xu et al., 2014). The LV-mRFP-GFP-LC3 double fluorescence labeling indicating system produces mRFP to label LC3, and the decrease of GFP indicates the fusion of lysosomes and autophagosomes to form autolysosomes, which is due to the sensitivity of GFP to acid. Thus, when autophagosomes fuses with lysosomes, GFP fluorescence is quenched and only RFP fluorescence can be detected. The yellow spots shown after red and green fluorescence merge together in microscope imaging indicate autophagosomes. The red spots indicate autolysosomes. The intensity of the autophagy flux can be clearly observed by counting the spots in different colors. The primary antibodies against mTOR, phospho-mTOR, p70S6K, phospho-p70S6K, p53, phospho-p53, and β-actin were purchased from Santa Cruz Biotechnology (CA, USA), or Cell Signaling Technology (MA, USA).
2.2. The eukaryotic expression of the PEDV nsps
RNA was extracted from the PEDV YC2014 strain, and the genes encoding the nsps were amplified using different primers (Table 1 ). The genes of nsp were then cloned into the EcoR I/Xho I site of pCAGGS-HA (BioVector NTCC Inc., Beijing, China) and transfected into IPEC-J2 cells using Lipofectamine 2000 (Invitrogen). The expression of the nsps was analyzed by western blot and IFA.
Table 1.
Primers used in this research.
| Primer | Sequence (5’–3’) |
|---|---|
| PEDV-F | TATTCCCGTTGATGAGG |
| PEDV-R | AGAGGCCAAAGTATCC |
| SF | CGCGGATCCGAAGAATGGTAAGTTGCTAGTGCGTAA |
| SR | CCCAAGCTTTGGGCAATAAAGAACAATGACAGC |
| MF | CGCGGATCCATGTCCGCACAAAGGGA |
| MR | CCGCTCGAGTGACTGGATTACATGTTACCTCTATA |
| EF | CGCGGATCCATGACACGGGTCGCAAT |
| ER | ATGGTCGACGCATATACTTATACAGGCGAGC |
| NF | CGCGGATCCATGAATAATCGTAAGTATTCAA |
| NR | CCGCTCGAGAATAAAAACATCATCATCG |
| nsp1F | GAATTCATGGCTAGCAACCATGTTAC |
| nsp1R | CTCGAGACCACCACGACGACCAA |
| nsp2 F | GAATTCAACATCGTGCCAGTTGACCAA |
| nsp2R | CTCGAGACCATTGAGTGCTGGTGGTTTAA |
| nsp3 F | GAATTCAGTATTGCTATTGTTGATGGCTTTG |
| nsp3R | CTCGAGACCCTTCTTATTTGCAATGCAAA |
| nsp4 F | GAATTCGCAGGTCTTCCTAGTTTTTC |
| nsp4R | CTCGAGCTGTAGAGTTGAATTGTAACTCA |
| nsp5F | GAATTCGCTGGCTTGCGTAAGATG |
| nsp5R | CTCGAGCTGAAGATTAACGCCATACATT |
| nsp6F | GAATTCAGTGGTTATGTTTCACGCGCC |
| nsp6R | CTCGAGCTGAACGGAAGAAATCTTAATATTC |
| nsp7F | GAATTCTCTAAACTGACTGATATTAAGTGTAGTAACGT |
| nsp7R | CTCGAGACTCTGCAACATACTATTGTCATTAAAATAG |
| nsp8F | GAATTCGTTGCATCTACTTATGTCGGTTTGC |
| nsp8R | CTCGAGCTGGAGCTTGACAATACGCTCA |
| nsp9F | GAATTCAATAATGAAATTATTCCTGGTAAGCTGA |
| nsp9R | CTCGAGCTGCAAGCGTACAGTGGCAC |
| nsp10F | GAATTCGCTGGTAAACAAACAGAACAGGC |
| nsp10R | CTCGAGTTGCATAATGGATCTGTCACAAGTG |
| nsp11F | GAATTCTTCAATAATGAAATTATTCCTGGTAAGCT |
| nsp11R | CTCGAGAGCTGCAAGCGTACAGTGGCA |
| nsp12 F | GAATTCAATTCTCTGCAGGGCTTTGTGT |
| nsp12R | CTCGAGTCGAGCTGCAAATCAGACAATTTAAG |
| nsp13 F | GAATTCTCTGCAGGGCTTTGTGTTGTT |
| nsp13R | CTCGAGCTGCAAATCAGACAATTTAAGCTCA |
| nsp14 F | GAATTCGCTAATGAGGGTTGTGGTCTTTTT |
| nsp14R | CTCGAGTTGCAAATTGTTACTAAATGTCTGCC |
| nsp15F | GAATTCGGTCTTGAGAACATTGCTTTCAATG |
| nsp15R | CTCGAGCTGAAGTTGCGGATAAAATGTCTG |
| nsp16F | GAATTCGCCAGTGAATGGAAGTGTGGTT |
| nsp16R | CTCGAGTTTGTTTACGTTGACCAAATGATTAG |
| ORF3 F | GAATTCATGTTTCTTGGACTTTTTCA |
| ORF3R | CTCGAGTTCACTAATTGTAGCATACTCG |
2.3. Viral infection and biochemical intervention
The IPEC-J2 cells were grown to 80 % confluence in 6-well plates. PEDV, multiplicity of infection (MOI) of 10 with 8 μg/mL trypsin, was allowed to adsorb to cells for 2 h at 37 °C. After being washed three times with D-Hanks, the cells were cultured in fresh medium in the presence of 8 μg/mL trypsin. The replication of PEDV in the IPEC-J2 cells was measured by IFA with the monoclonal antibody (mAb) 3A6 anti-PEDV N protein (prepared in our laboratory) as the primary antibody, and the FITC-conjugated goat anti-mouse IgG (Abcam, China) as the secondary antibody.
Rapamycin and its analogs are strong inducers of autophagy by inhibiting mTOR activity (Ravikumar et al., 2004; Russo et al., 2018), and hydroxychloroquine can inhibit autophagy by blocking the fusion of autophagosomes with lysosomes (Yang et al., 2013). For the autophagy induction and inhibition experiments, cells grown to 60 % confluence in 6-well plates were treated with 100 nM rapamycin for 2 h, or with 50 μM hydroxychloroquine for 4 h prior to viral infection. Viral adsorption was performed at 37 °C for 2 h. Then, the inoculum was removed, and the cells were washed three times with PBS. Then, the cells were incubated in fresh medium containing rapamycin (100 nM) or hydroxychloroquine (50 μM) until the cells or the culture medium were harvested. At 0, 12, 24, and 36 h post infection, the cells were collected for subsequent analysis. The same volume of DMSO was added to separate samples as the carrier control.
2.4. Establishment of ATG5 knockout IPEC-J2 cell lines
It was reported that coronavirus nsp6 generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate, and the autophagosomes formation was dependent on ATG5 and class III PI3 kinase (Cottam et al., 2011). To establish ATG5 knockout cell lines, the optimized sgRNA construct, targeting ATG5 (Gene ID: 100739102), and the Cas9 expression construct were obtained from Hanbio (HB-LP210 0001, Shanghai, China). The ATG5 knockout IPEC-J2 cell lines were constructed as previously described (Yasunaga et al., 2018).
2.5. Confocal immunofluorescence microscopy
For the detection of autophagosomes, the IPEC-J2 cells were grown to 70–80 % confluence on glass coverslips in 6-well plates, then infected with lentivirus LV-mRFP-GFP-LC3 at a MOI of 10 for 2 h. After three washes with PBS, the cells were transfected with pCAGGS-nsp6 using Lipofectamine 2000. After 24 h the cells were examined under a confocal laser scanning microscope (LSM 510 Meta, Carl Zeiss, Munich, Germany).
2.6. MTT assay
The MTT method was used to determine the cytotoxic effects of the reagents on the IPEC-J2 cells as previously described (Irvine et al., 2006). Briefly, the IPEC-J2 cells were seeded in 96-well plates and incubated in I/R medium for 12 h. Then, 20 μl of MTT 3-(4,5-dmethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) dissolved in Hank’s balanced salt solution was added to each well to a final concentration of 5 μg/mL. The plates were incubated in 5% CO2 at 37 °C. After 2−4 h, the medium was aspirated from each well, and 200 μl of DMSO was added to dissolve the formazan crystals. The absorbance of each well was measured at 570 nm and 630 nm using a plate reader. Each experiment was repeated at least three times.
2.7. Transmission electron microscopy
IPEC-J2 cells were mock infected or infected with PEDV at a MOI of 10 and collected 12 h post infection (hpi) for ultrastructural analysis. Ultra-thin sections were viewed on a JEM-1400 transmission electron microscope (JEOL Ltd., Tokyo, Japan). Autophagosome-like vesicles were defined as double- or single-membrane vesicles measuring 0.3–2.0 μm in diameter that contained clearly recognizable cytoplasmic elements. Quantitative electron microscopy is one of the most sensitive methods to detect the presence of Autophagosome-like vesicles (Yla-Anttila et al., 2009). For cytological studies, five cells per sample were examined under electron microscopy using uniform random sampling. The number of autophagosome-like vesicles per cell was estimated by point counting.
2.8. Reverse transcription and quantitative real-time PCR (qRT-PCR) analysis
Total RNA was extracted from the PEDV-infected cells using TRIzol® Reagent (Invitrogen) and treated with RNase-free DNase I according to the manufacturer’s instructions. The RNA was reverse-transcribed into cDNA using the oligo (dT) primers listed in Table 1. qRT-PCR, using SYBR premix EX Taq (TaKaRa) and SYBR Green I, was performed to compare the relative expression levels of the gene messenger RNAs. β-actin was used as an internal control.
2.9. Western blot analysis
IPEC-J2 cell monolayers were washed twice with cold PBS and were incubated on ice with RIPA lysis buffer containing 1 mM PMSF (Beyotime) for 10 min. The lysates were clarified by centrifugation at 13,000 rpm for 20 min at 4 °C, and the protein concentration was quantified using a BCA protein assay kit (Beyotime). Equal amounts of protein samples were boiled for 5 min in 5 × SDS-PAGE loading buffer. The proteins (20 μg) were separated by 12 % SDS-PAGE and then electro-transferred onto polyvinylidene fluoride (PVDF) membranes. Membranes were blocked at 25 °C with 5% (w/v) skim milk in PBST. After 2 h the membranes were incubated with primary antibody (mTOR, p70S6K, or p53) at 37 °C for 1–2 h and then with the appropriate HRP-conjugated secondary antibody at 37 °C for 1 h. The protein bands were detected using an ECL Plus kit (Beyotime) and measured with Image-Pro Plus 6.0 software (Media Cybernetics). The images were captured with a CanoScan LiDE 100 scanner (Canon).
2.10. Statistical analysis
All the experiments were performed independently three times. Variables are expressed as the mean ± standard deviation (SD) and were analyzed by one-way ANOVA using SPSS software (version 22.0). A P value of < 0.05 was considered statistically significant, a P value of < 0.01 was considered highly significant, and a P value of < 0.001 was considered extremely significant.
3. Results
3.1. PEDV infection increases autophagy in IPEC-J2 cells
The replication of PEDV in IPEC-J2 cells was measured by IFA with the monoclonal antibody (mAb) 3A6 anti-PEDV N protein as the primary antibody, and the FITC-conjugated goat anti-mouse IgG as the secondary antibody. The results of IFA shown that the PEDV YC2014 strain replicated efficiently in IPEC-J2 cells (Fig. S1).
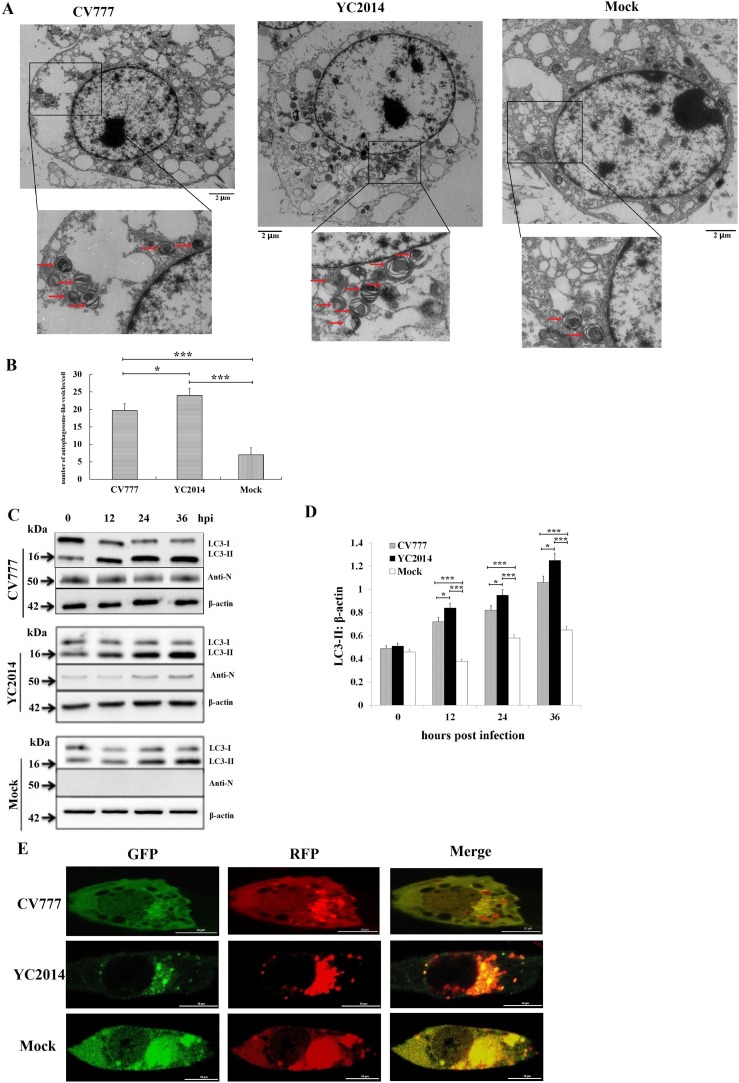
By transmission electron microscopy (TEM), it was revealed that the number of double- and single-membrane vesicles containing cytosolic components or sequestered organelles were abundant in the cytoplasm of PEDV-infected IPEC-J2 cells (both of the classical strain CV777 and the pandemic strain YC2014), while these autophagosome-like vesicles were rarely observed in mock-infected cells (Fig. 1 A and B). The number of autophagosome-like vesicles in the pandemic strain YC2014 infected IPEC-J2 cells was significantly higher than the classical strain CV777 infected cells (P < 0.05, Fig. 1B).
Fig. 1.
PEDV infection increases autophagy in IPEC-J2 cells. (A, B) Photos of transmission electron microscopy (TEM) shown that the number of autophagosome-like vesicles (double- and single-membrane vesicles containing cytosolic components or sequestered organelles) were significantly higher in the cytoplasm of both the PEDV pandemic strain YC2014 and the classical strain CV777 infected cells than in mock-infected cells. The number of autophagosome-like vesicles in the pandemic strain YC2014 infected cells was significantly higher than the classical strain CV777 infected cells. IPEC-J2 cells were infected at a MOI of 10 for 12 h. *P < 0.05, ***P < 0.001. (C, D) PEDV infection increases the conversion of LC3-I to LC3-II. C, Western blot of PEDV-infected (MOI of 10) or mock-infected IPEC-J2 cells. D, The ratio of LC3-II to β-actin was quantitated by densitometry (n = 3). The ratios of LC3-II to β-actin were both significantly higher in the classical strain CV777 and the pandemic strain YC2014 infected cells than in uninfected cells 12 hpi. The ratios of LC3-II to β-actin in the pandemic strain YC2014 infected cells were significantly higher than the classical strain CV777 infected cells. Data were expressed as the mean ± SD of three independent experiments and were analyzed by one-way ANOVA. **P < 0.01, *P < 0.05, ***P < 0.001. (E) Both GFP-LC3 and RFP-LC3-labeled puncta were detectable in IPEC-J2 cells after PEDV YC2014 strain and CV777 strain infection.
LC3 is a specific marker protein for monitoring autophagic vesicle formation, due to its role in vesicle formation and lipidation reactions. The ratio of LC3-II to β-actin is commonly used to assess the activity of autophagy. To further analyze the autophagy activity triggered by PEDV infection, IPEC-J2 cells were infected with the PEDV classical strain CV777 and the pandemic strain YC2014, respectively, and at the indicated time points, the cells were harvested and subjected to SDS-PAGE and electroblotting. The proteins were detected using an anti-LC3 antibody that recognizes both LC3-II and LC3-I, and an anti-PEDV N-protein monoclonal antibody. The western blot analyses shown that the level of LC3-II increased with increasing incubation time (Fig. 1C). The ratios of LC3-II to β-actin were both significantly higher in the classical strain CV777 and the pandemic strain YC2014 infected cells than in uninfected cells after 12 h of infection (P < 0.001, Fig. 1D). The ratios of LC3-II to β-actin in the pandemic strain YC2014 infected IPEC-J2 cells were significantly higher than the classical strain CV777 infected cells (P < 0.05, Fig. 1D). The lentivirus LV-mRFP-GFP-LC3 was used to measure the autophagic flux. The green fluorescence of this tandem autophagosome reporter is attenuated in the acidic pH of the lysosome by lysosomal hydrolysis, whereas the red fluorescence is not. Therefore, autophagosomes have both GFP and RFP signals, whereas autolysosomes have only RFP signals. As shown in Fig. 1E, both GFP-LC3 and RFP-LC3 labeled puncta were observed in both of the PEDV classical strain CV777 and the pandemic strain YC2014 infected cells. Taken together, these data indicate that autophagosomes accumulate in IPEC-J2 cells infected with PEDV.
3.2. Autophagy promotes PEDV replication
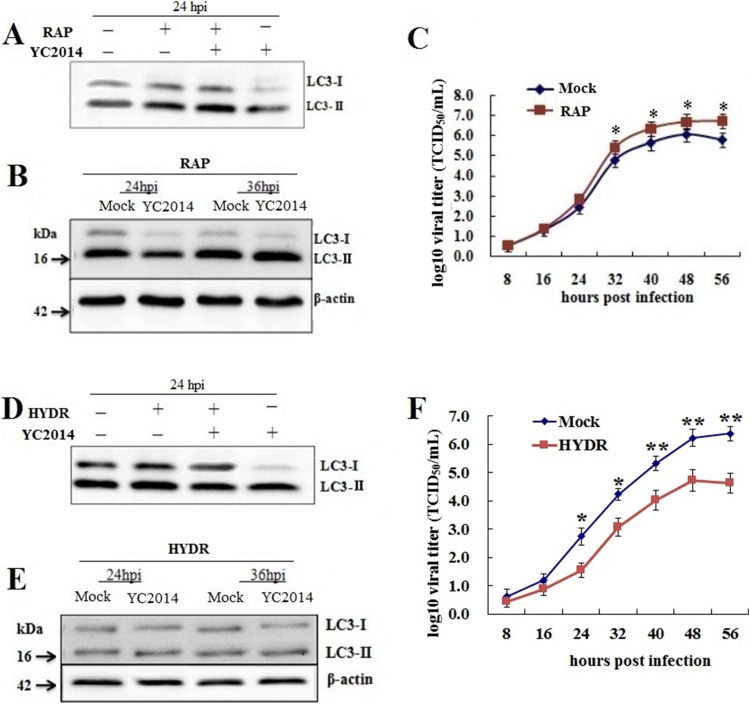
To determine whether autophagy enhances PEDV replication, PEDV-infected IPEC-J2 cells were treated with 100 nM rapamycin, which induces autophagy by inhibiting the mTOR signaling pathway. As expected rapamycin treatment resulted in increased levels of LC3-II in infected and uninfected cells (Fig. 2 A and B). The PEDV titers (TCID50/mL) were significantly higher (P < 0.05) in the rapamycin-treated cells than in the mock-treated cells after 32 h of infection (Fig. 2C). At 48 h after infection, the PEDV titer in the rapamycin-treated cells reached 8.8 × 106 TCID50/mL, which was ten times more than that in the mock group (8.5 × 105 TCID50/mL). These results suggest that autophagy promotes PEDV replication.
Fig. 2.
Autophagy affects PEDV replication. (A, B) Induction of autophagy with rapamycin in IPEC-J2 cells. IPEC-J2 cells were treated with 100 nM rapamycin for 2 h prior to PEDV infection (MOI of 10). The cells were lysed and analyzed by western blotting with antibodies against LC3 and β-actin. (C) The viral titers of the rapamycin-treated cells were significantly higher than those of the mock-treated cells (n = 3). Data were expressed as the mean ± SD of three independent experiments and were analyzed by one-way ANOVA. *P < 0.05. (D, E) Inhibition of autophagy with hydroxychloroquine. IPEC-J2 cells were treated with 50 μM hydroxychloroquine for 4 h prior to PEDV infection (MOI of 10). The cells were lysed and analyzed by western blotting with antibodies against LC3 and β-actin. (F) The viral titers of the hydroxychloroquine-treated cells were significantly lower than those of the mock-treated cells (n = 3). Data were expressed as the mean ± SD of three independent experiments and were analyzed by one-way ANOVA. *P < 0.05, **P < 0.01.
The effect of autophagy on PEDV replication was further investigated by treating IPEC-J2 cells with hydroxychloroquine. As shown in Fig. 2D and E, hydroxychloroquine treatment resulted in decreased ratios of LC3-II in PEDV-infected cells, although not in mock-infected cells, demonstrating that hydroxychloroquine effectively inhibits PEDV-induced autophagy. The PEDV titers (TCID50/mL) in the hydroxychloroquine-treated cells were significantly lower (P < 0.05) than those in the mock-treated cells after 24 h of infection (Fig. 2F). At 48 h after infection, the PEDV titer in the hydroxychloroquine-treated cells reached 7.9 × 104 TCID50/mL, which was ten times lower than the mock group (8.5 × 105 TCID50/mL). These results demonstrate that early-stage inhibition of autophagy inhibits PEDV replication.
3.3. PEDV replication but not PEDV particles enhances the autophagy process
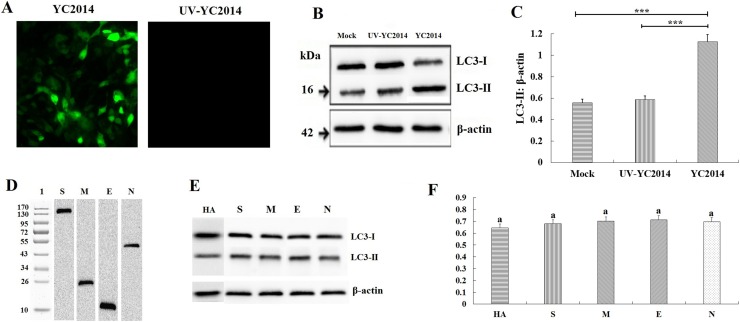
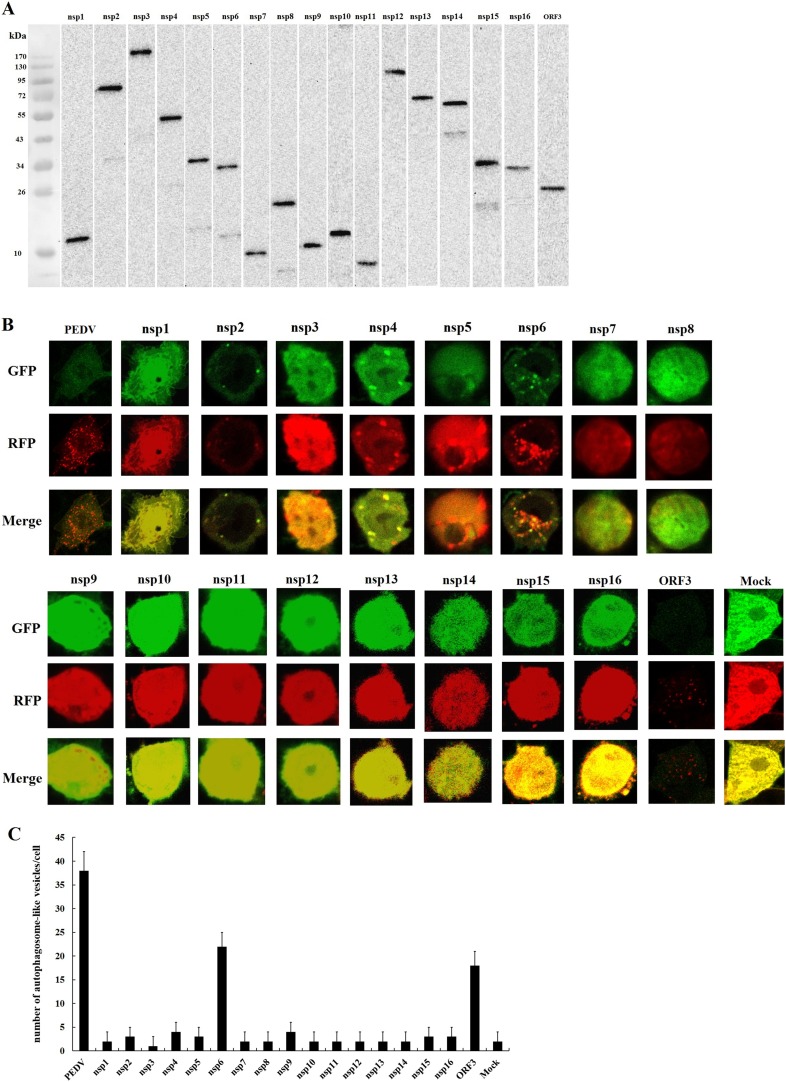
To determine whether PEDV-induced autophagy is the result of viral entry or if viral replication is necessary, we used UV-treated PEDV. The polyclonal antibody against nucleocapsid protein (N) of PEDV was used as the primary antibody. IFA was used to demonstrate that UV-treated PEDV was replication defective (Fig. 3 A). The western blot results shown that there was no detectable conversion of LC3-I to LC3-II in cells infected with UV-treated PEDV after 24 h, while autophagy was triggered normally by the nonirradiated, replication-competent virus (Fig. 3B and C). To determine whether the structural proteins could induce autophagy during PEDV replication, all four structural proteins (S, M, E, N) were eukaryotic expressed in IPEC-J2 cells after cloning the coding genes into the pCAGGS-HA plasmid. The expression of the four structural proteins was determined by western blot (Fig. 3D). The levels of LC3-II were not elevated significantly in cells expressing the structural proteins (Fig. 3E and F). These results indicate that viral replication is required for PEDV-induced autophagy, and the nonstructural proteins (nsps) rather than the structural proteins are the key inducers in PEDV-induced autophagy.
Fig. 3.
PEDV replication is required for PEDV-induced autophagy. (A) IFA of replication-competent PEDV or replication-incompetent PEDV (UV-treated) in IPEC-J2 cells. (B) Western blot showing the levels of LC3-I and LC3-II in IPEC-J2 cells 24 hpi with PEDV and UV-treated PEDV. (C) The ratio of LC3-II to β-actin was quantitated by densitometry (n = 3). Data were expressed as the mean ± SD of three independent experiments and were analyzed by one-way ANOVA. ***P < 0.001. (D) Western blot analysis of the eukaryotic expressed the structural proteins. (E) Western blot analysis of LC3-I and LC3-II in IPEC-J2 cells expressing structural proteins. (F) The ratio of LC3-II to β-actin was quantitated by densitometry (n = 3). Data were expressed as the mean ± SD of three independent experiments and were analyzed by one-way ANOVA. Same letter (a) indicates no significant difference between the groups.
3.4. PEDV replication was inhibited in ATG5 knockout IPEC-J2 cell lines
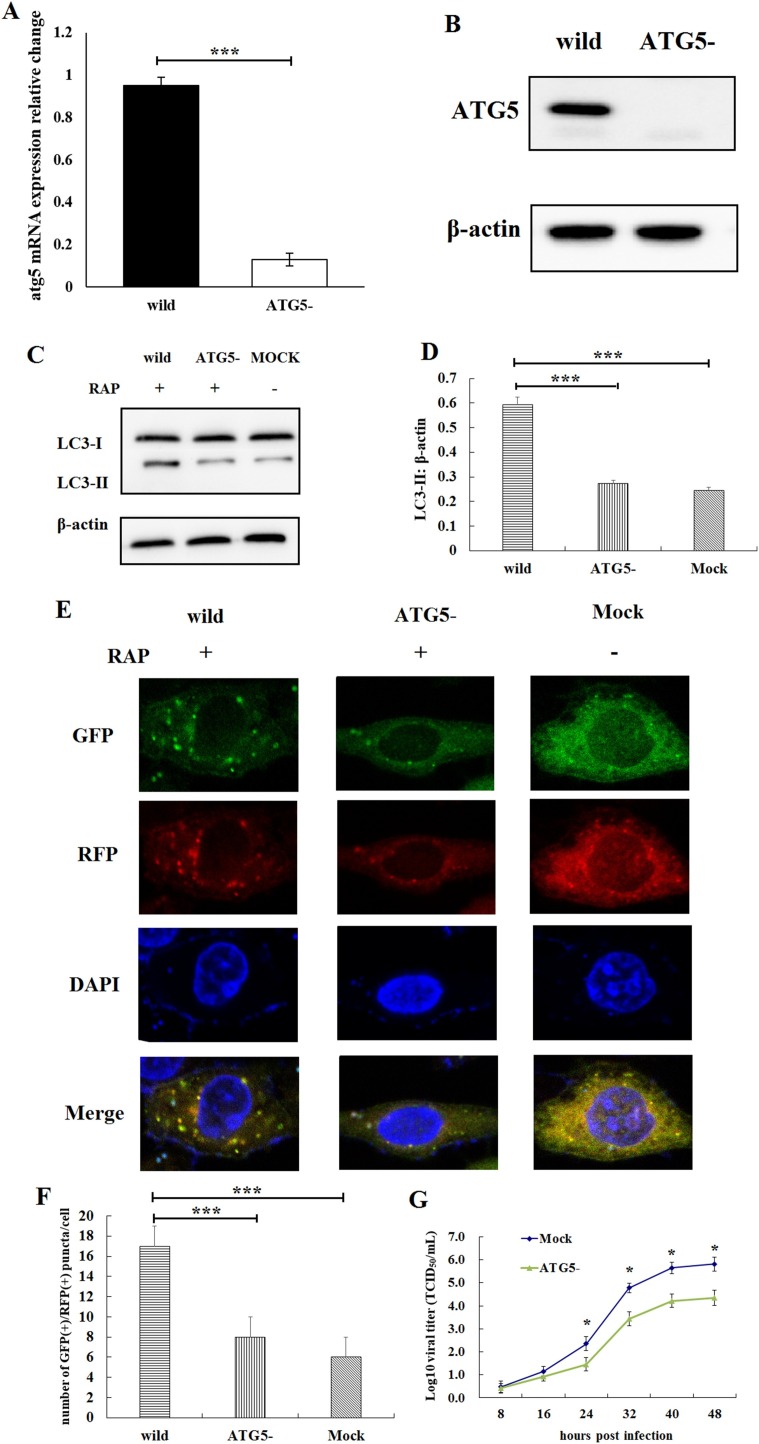
It was reported that coronavirus generate autophagosomes dependent on ATG5 and class III PI3 kinase (Cottam et al., 2011). To evaluate the mechanism by which PEDV replicates in autophagy-defective cells, ATG5 knockout IPEC-J2 cell lines were constructed by CRISPR/Cas9 genome editing. The results of qRT-PCR (Fig. 4 A) and western blot (Fig. 4B) shown that the ATG5 gene was knockout from IPEC-J2 cells. The atg5 mRNA in ATG5 knockout IPEC-J2 cells was significantly lower than the wild-type IPEC-J2 cells (P < 0.001, Fig. 4A). The expression of ATG5 was completely suppressed in ATG5 knockout IPEC-J2 cells (Fig. 4B). The lack of autophagy in ATG5-knockout IPEC-J2 cells was further confirmed by analyzing the conversion of LC3-I to LC3-II in response to 100 nM rapamycin (Fig. 4C and D), and confocal immunofluorescence microscopy analysis (Fig. 4E and F). The ratio of LC3-II to β-actin in ATG5-knockout IPEC-J2 cells was significantly lower than that in the wild-type IPEC-J2 cells after the cells been treated by rapamycin (P < 0.001, Fig. 4C and D). The number of autophagosome-like vesicles per cell in ATG5-knockout IPEC-J2 cells was significantly lower than that in the wild-type IPEC-J2 cells after PEDV infection (P < 0.001, Fig. 4E and F). The results of qRT-PCR shown that the proliferation rate of PEDV in ATG5-knockout (autophagy-defective) IPEC-J2 cells was significantly lower than that in normal IPEC-J2 cells (P < 0.05, Fig. 4G).
Fig. 4.
Examination of PEDV replication in ATG5 knockout IPEC-J2 cell lines. (A) qRT-PCR detection of the atg5 mRNA in ATG5 knockout IPEC-J2 cell lines. The atg5 mRNA in ATG5 knockout IPEC-J2 cells was significantly lower than the wild-type IPEC-J2 cells. Data were expressed as the mean ± SD of three independent experiments and were analyzed by one-way ANOVA, ***P < 0.001. (B) Western blot identification of the ATG5 knockout IPEC-J2 cell lines. The expression of ATG5 was completely suppressed in ATG5 knockout IPEC-J2 cells. (C) Western blot analysis of LC3-I and LC3-II. (D) The ratio of LC3-II to β-actin was quantitated by densitometry (n = 3). The ratio of LC3-II to β-actin in ATG5-knockout IPEC-J2 cells was significantly lower than that in the wild-type IPEC-J2 cells after the cells been treated by rapamycin. ***P < 0.001. (E, F) Confocal immunofluorescence microscopy analysis of the autophagosome-like vesicles. The number of autophagosome-like vesicles per cell in ATG5-knockout IPEC-J2 cells was significantly lower than that in the wild-type IPEC-J2 cells after PEDV infection. ***P < 0.001. (G) The results shown that the proliferation rate of PEDV in autophagy-defective IPEC-J2 cells was significantly lower than that in normal IPEC-J2 cells (n = 3). Data are presented as the mean ± SD of three independent experiments. *P < 0.05.
3.5. Nsp6 and ORF3 are two of the key inducers of PEDV-induced autophagy
To identify which nsp could induce autophagy during PEDV replication, all 17 nsps (nsp1-nsp16 and ORF3) were eukaryotic expressed in IPEC-J2 cells after cloning the nsps into the pCAGGS-HA plasmid, respectively. The expression of the nsps was determined by western blot (Fig. 5 A), with a polyclonal antibody against the prokaryotically expressed nsps as the primary antibody, respectively. The lentivirus LV-mRFP-GFP-LC3 was used to measure the autophagic flux. Both GFP-LC3 and RFP-LC3-labeled puncta were present in nsp6-expressing cells and ORF3-expressing cells but not in the other nsp-expressing cells (Fig. 5B and C). The results demonstrate that nsp6 or ORF3 expression induces autophagy in IPEC-J2 cells, and indicate that nsp6 and ORF3 are two of the key inducers of PEDV-induced autophagy.
Fig. 5.
Examination of nsps-induced autophagy in IPEC-J2 cells. (A) Western blot analysis of the eukaryotic expression of the nsps. (B, C) Confocal immunofluorescence microscopy analysis of autophagosomes-like vesicles in IPEC-J2 cells expressing nsps. Nsp6 or ORF3 expression causes an increase of autophagosome-like vesicles, but the expression of the other nsps does not.
3.6. Nsp6 induces autophagy through the PI3K/Akt/mTOR signaling pathway
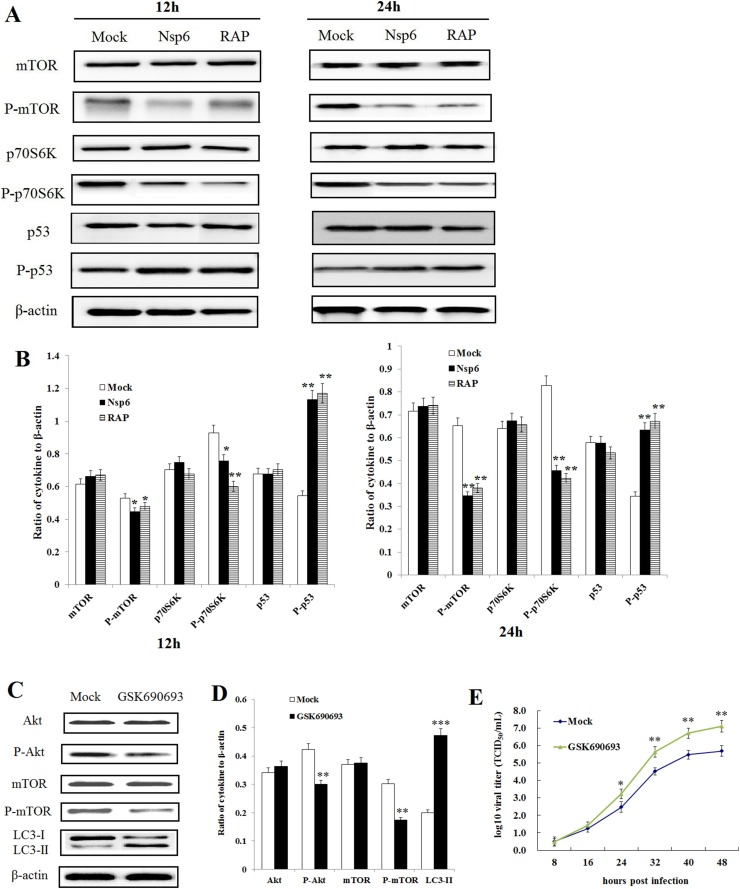
It was reported that, ORF3-induced autophagy is dependent on the ER stress response (Zou et al., 2019), however, the mechanism of nsp6-induced autophagy has yet to be determined. It is well established that the PI3K/Akt/mTOR signaling pathway plays a pivotal role in autophagy (Saiki et al., 2011; Surviladze et al., 2013). We investigated the effect of nsp6 expression on the PI3K/Akt/mTOR pathway by quantitating the expression of phosphorylated and unphosphorylated mTOR, p70S6K, and p53 in nsp6 expressing IPEC-J2 cells. As shown in Fig. 6 A and B, the levels of phosphorylation of mTOR and p70S6K were decreased in nsp6-expressing cells, while the phosphorylation levels of p53 were increased.
Fig. 6.
Nsp6-induced autophagy occurs via the PI3K/Akt/mTOR signaling pathway. (A) Western blot detection of cytokines and their phosphorylation levels in nsp6-expressing IPEC-J2 cells, rapamycin-treated IPEC-J2 cells or mock-treated IPEC-J2 cells. (B) The ratio of cytokines to β-actin as determined by densitometry (n = 3). Data were expressed as the mean ± SD of three independent experiments and were analyzed by one-way ANOVA. *P < 0.05, **P < 0.01. (C, D) The inhibition efficiency of Akt-specific inhibitor GSK690693 in IPEC-J2 cells as determined by western blot 24 hpi (n = 3). Data were expressed as the mean ± SD of three independent experiments and were analyzed by one-way ANOVA. **P < 0.01, ***P < 0.001. (E) PEDV titers were increased in cells with Akt-specific inhibitor GSK690693 compared to the titers in mock cells (n=3). Data are presented as the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01.
To further investigate the role of the PI3K/Akt/mTOR signaling pathway in PEDV replication, the Akt-specific inhibitor GSK690693 (dissolved with DMSO) was used at a final concentration of 25 μM. The IPEC-J2 cells were pretreated with GSK690693 for 2 h before PEDV infection. Viral adsorption was performed at 37 °C for 2 h. Then, the inoculum was removed, and the cells were washed three times with PBS. The cells were subsequently incubated in fresh medium containing GSK690693 (10 μM) until the cells were harvested. The results of western blot (Fig. 6C and D) shown that the levels of phosphorylated Akt and phosphorylated mTOR were significantly decreased in cells exposed to GSK690693 (P < 0.01), while the levels of LC3-II were increased (P < 0.001). The results of qRT-PCR (Fig. 6E) shown that inhibiting phosphorylated Akt expression could increase PEDV viral titers significantly. These results indicate that inhibition of the PI3K/Akt/mTOR signaling pathway could increase PEDV replication by increasing IPEC-J2 autophagy.
4. Discussion
The role of autophagy in the viral infection process has been widely investigated (Lv et al., 2015; Paul and Munz, 2016). While autophagy may eliminate viruses from the host cell by delivering them to the lysosomal compartment for degradation (Hernaez et al., 2013), many viruses, such as human cytomegalovirus and herpes simplex virus type 1, have developed mechanisms to block autophagy, or to utilize the process for their benefit, (Yordy and Iwasaki, 2013). Viruses such as classical swine fever virus, porcine reproductive and respiratory syndrome virus, and rotavirus utilize the membranes of autophagosome-like vesicles during replication (Crawford et al., 2012). New PED outbreaks caused by PEDV pandemic strains have been documented in China since late 2010 and are now distributed all over the world. It was characterized by acute enteric infection and high mortality in suckling piglets and finishing pigs. In this study, we demonstrated that PEDV replication triggers autophagy in IPEC-J2 cells and that autophagy enhances replication of PEDV. It was also demonstrated that PEDV structural proteins (S, M, E, N) are not sufficient to trigger autophagy, and the nonstructural proteins (nsp1-nsp16 and ORF3) of PEDV are required in autophagy programs. These results were consistent with previous research on PEDV-induced autophagy in Vero cells by Guo X. et al. (Guo et al., 2017). However, Ko S. et al. reported that PEDV infection is suppressed in IPEC-J2 cells by an increase in autophagy induced by rapamycin (Ko et al., 2017). These different results might occur because of the different degrees of virulence of different PEDV strains. By expressing all 4 structural proteins and 17 nsps of PEDV in eukaryotic cells, we found that autophagy was triggered by the expression of nsp6 or ORF3, but not by the expression of the other proteins, in IPEC-J2 cells.
Productive PEDV infection promoted the formation of double-membrane vesicles (DMVs), which are a hallmark of coronavirus replication, and potentially a platform for viral replication (Zhou et al., 2017). During coronavirus infection, the formation of DMVs is believed to be associated with autophagy (Angelini et al., 2013; Blanchard and Roingeard, 2015). This observation was supported by the increased expression of autophagic marker proteins during PEDV replication. Using PEDV that was rendered replication-incompetent (but still infectious) by UV irradiation, it was found that viral replication induces autophagy. The LV-mRFP-GFP-LC3 lentivirus was used to measure the autophagic flux. The green fluorescence of this tandem autophagosome reporter is attenuated in the acidic pH of the lysosome by lysosomal hydrolysis, whereas the red fluorescence is not. In PEDV-infected cells, the autophagosomes exhibit both GFP and mRFP signals, whereas autolysosomes exhibit only mRFP signals.
Replication mechanisms vary among members of the Coronaviridae family. Infectious bronchitis virus is able to induce autophagy but does not require autophagy for its replication (Maier et al., 2013). Transmissible gastroenteritis virus infection induces autophagy, which negatively regulates its replication (Guo et al., 2016). In this study, the impact of autophagy on PEDV replication was examined by using a pharmacological enhancer and a pharmacological suppressor of autophagy. The PEDV yield was suppressed when autophagy was inhibited by hydroxychloroquine at an early stage, while the viral titers were increased when autophagy was induced by rapamycin. These results suggest that autophagy enhances PEDV replication in IPEC-J2 cells.
The severe inflammatory response induced by viral infection is one of the causes of rapid host death (Wang et al., 2018b). It was reported that transmissible gastroenteritis virus (TGEV) infection activates the NF-κB pathway in porcine intestinal epithelial cells and results in severe inflammation (Wang et al., 2018a). The differentially expressed mRNAs and ncRNAs in mock- and TGEV-infected IPEC-J2 cells were primarily enriched in inflammation-related pathways, and ssc_circ_009380 promoted the activation of the NF-κB pathway by binding to miR-22 during TGEV-induced inflammation (Ma et al., 2018). SARS-coronavirus can mediate myocardial inflammation and damage that are associated with the downregulation of the myocardial ACE2 system, which may be responsible for the myocardial dysfunction and adverse cardiac outcomes in patients with SARS (Oudit et al., 2009). In our previous studies, PEDV pandemic strains induced both autophagy-related and inflammation-related signaling pathways more significantly than the PEDV classical strains in IPEC-J2 cells (Lin et al., 2017). This may be associated with the rapid proliferation of PEDV pandemic strains in vivo, which induces inflammatory cytokine storms in intestinal epithelial cells and ultimately results in rapid damage to intestinal epithelial cells and the occurrence of diarrhea, dehydration and death. The mutual relationships between the autophagy and inflammation induced by PEDV pandemic strains will be studied in our following research.
The PI3K/Akt/mTOR signaling pathway plays a pivotal role in autophagy (Saiki et al., 2011; Surviladze et al., 2013). By analyzing the expression of unphosphorylated and phosphorylated mTOR, p70S6K, and p53 in nsp6-expressing IPEC-J2 cells, we shown that nsp6 induces autophagy through the PI3K/Akt/mTOR signaling pathway. The relationship between autophagy and PEDV replication was further confirmed through RNAi experiments, in which the endogenous p53 gene was specifically silenced. RNAi-mediated silencing of p53 expression decreased the viral titer compared to the mock-treated group. These data also demonstrate that the autophagy triggered by PEDV is via the PI3K/Akt/mTOR signaling pathway.
In summary, the induction of autophagy by PEDV in IPEC-J2 cells enhances viral replication, which induces rapid damage to intestinal epithelial cells and enhances the virulence of PEDV in both newborn piglets and finishing pigs. Nsp6 is one of the key inducers of PEDV-induced autophagy, and it acts through the PI3K/Akt/mTOR signaling pathway. These results improve the understanding of the biology and pathogenesis of PEDV infection and provide novel insights into the development of effective therapeutic strategies.
Declaration of Competing Interest
The authors declare they have no competing interests.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31702278), the Fundamental Research Funds for the Central Universities (KYDS201801, KJQN201826), the Jiangsu Agriculture Science and Technology Innovation Fund (CX(19)2020), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.vetmic.2020.108684.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Alexander D.E., Ward S.L., Mizushima N., Levine B., Leib D.A. Analysis of the role of autophagy in replication of herpes simplex virus in cell culture. J. Virol. 2007;81:12128–12134. doi: 10.1128/JVI.01356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini M.M., Akhlaghpour M., Neuman B.W., Buchmeier M.J. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio. 2013:4. doi: 10.1128/mBio.00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertasio C., Giacomini E., Lazzaro M., Perulli S., Papetti A., Lavazza A., Lelli D., Alborali G., Boniotti M.B. Porcine epidemic diarrhea virus shedding and antibody response in swine farms: a longitudinal study. Front. Microbiol. 2016;7:2009. doi: 10.3389/fmicb.2016.02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi J., Zeng S., Xiao S., Chen H., Fang L. Complete genome sequence of porcine epidemic diarrhea virus strain AJ1102 isolated from a suckling piglet with acute diarrhea in China. J. Virol. 2012;86:10910–10911. doi: 10.1128/JVI.01919-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard E., Roingeard P. Virus-induced double-membrane vesicles. Cell. Microbiol. 2015;17:45–50. doi: 10.1111/cmi.12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S.H., Bae J.L., Kang T.J., Kim J., Chung G.H., Lim C.W., Laude H., Yang M.S., Jang Y.S. Identification of the epitope region capable of inducing neutralizing antibodies against the porcine epidemic diarrhea virus. Mol. Cells. 2002;14:295–299. [PubMed] [Google Scholar]
- Chen F., Pan Y., Zhang X., Tian X., Wang D., Zhou Q., Song Y., Bi Y. Complete genome sequence of a variant porcine epidemic diarrhea virus strain isolated in China. J. Virol. 2012;86:12448. doi: 10.1128/JVI.02228-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.H., Wang W.T., Huang W., Fang K., Sun Y.M., Liu S.R., Luo X.Q., Chen Y.Q. The lncRNA HOTAIRM1 regulates the degradation of PML-RARA oncoprotein and myeloid cell differentiation by enhancing the autophagy pathway. Cell Death Differ. 2017;24:212–224. doi: 10.1038/cdd.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiramel A.I., Best S.M. 2017. Role of Autophagy in Zika Virus Infection and Pathogenesis. Virus Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiramel A.I., Brady N.R., Bartenschlager R. Divergent roles of autophagy in virus infection. Cells. 2013;2:83–104. doi: 10.3390/cells2010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam E.M., Maier H.J., Manifava M., Vaux L.C., Chandra-Schoenfelder P., Gerner W., Britton P., Ktistakis N.T., Wileman T. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy. 2011;7:1335–1347. doi: 10.4161/auto.7.11.16642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam E.M., Whelband M.C., Wileman T. Coronavirus NSP6 restricts autophagosome expansion. Autophagy. 2014;10:1426–1441. doi: 10.4161/auto.29309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford S.E., Hyser J.M., Utama B., Estes M.K. Autophagy hijacked through viroporin-activated calcium/calmodulin-dependent kinase kinase-beta signaling is required for rotavirus replication. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E3405–3413. doi: 10.1073/pnas.1216539109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Fang L., Jing H., Zeng S., Wang D., Liu L., Zhang H., Luo R., Chen H., Xiao S. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J. Virol. 2014;88:8936–8945. doi: 10.1128/JVI.00700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber P.F., Xiao C.T., Lager K., Crawford K., Kulshreshtha V., Cao D., Meng X.J., Opriessnig T. Increased frequency of porcine epidemic diarrhea virus shedding and lesions in suckling pigs compared to nursery pigs and protective immunity in nursery pigs after homologous re-challenge. Vet. Res. 2016;47:118. doi: 10.1186/s13567-016-0402-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Yu H., Gu W., Luo X., Li R., Zhang J., Xu Y., Yang L., Shen N., Feng L., Wang Y. Autophagy negatively regulates transmissible gastroenteritis virus replication. Sci. Rep. 2016;6:23864. doi: 10.1038/srep23864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Zhang M., Zhang X., Tan X., Guo H., Zeng W., Yan G., Memon A.M., Li Z., Zhu Y., Zhang B., Ku X., Wu M., Fan S., He Q. Porcine epidemic diarrhea virus induces autophagy to benefit its replication. Viruses. 2017:9. doi: 10.3390/v9030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernaez B., Cabezas M., Munoz-Moreno R., Galindo I., Cuesta-Geijo M.A., Alonso C. A179L, a new viral Bcl2 homolog targeting Beclin 1 autophagy related protein. Curr. Mol. Med. 2013;13:305–316. [PubMed] [Google Scholar]
- Hou Y., Meulia T., Gao X., Saif L.J., Wang Q. Deletion of both the tyrosine-based endocytosis signal and the endoplasmic reticulum retrieval signal in the cytoplasmic tail of spike protein attenuates porcine epidemic diarrhea virus in pigs. J. Virol. 2019:93. doi: 10.1128/JVI.01758-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine K.M., Burns C.J., Wilks A.F., Su S., Hume D.A., Sweet M.J. A CSF-1 receptor kinase inhibitor targets effector functions and inhibits pro-inflammatory cytokine production from murine macrophage populations. FASEB J. 2006;20:1921–1923. doi: 10.1096/fj.06-5848fje. [DOI] [PubMed] [Google Scholar]
- Jackson W.T. Viruses and the autophagy pathway. Virology. 2015;479-480:450–456. doi: 10.1016/j.virol.2015.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Annamalai T., Lu Z., Saif L.J. Comparative pathogenesis of US porcine epidemic diarrhea virus (PEDV) strain PC21A in conventional 9-day-old nursing piglets vs. 26-day-old weaned pigs. Vet. Microbiol. 2015;178:31–40. doi: 10.1016/j.vetmic.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko S., Gu M.J., Kim C.G., Kye Y.C., Lim Y., Lee J.E., Park B.C., Chu H., Han S.H., Yun C.H. Rapamycin-induced autophagy restricts porcine epidemic diarrhea virus infectivity in porcine intestinal epithelial cells. Antiviral Res. 2017;146:86–95. doi: 10.1016/j.antiviral.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Chen L., Gao L., Yuan X., Ma Z., Fan H. Epidemic strain YC2014 of porcine epidemic diarrhea virus could provide piglets against homologous challenge. Virol. J. 2016;13:68. doi: 10.1186/s12985-016-0529-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Li B., Chen L., Ma Z., He K., Fan H. Differential protein analysis of IPEC-J2 cells infected with porcine epidemic diarrhea virus pandemic and classical strains elucidates the pathogenesis of infection. J. Proteome Res. 2017;16:2113–2120. doi: 10.1021/acs.jproteome.6b00957. [DOI] [PubMed] [Google Scholar]
- Lussignol M., Queval C., Bernet-Camard M.F., Cotte-Laffitte J., Beau I., Codogno P., Esclatine A. The herpes simplex virus 1 Us11 protein inhibits autophagy through its interaction with the protein kinase PKR. J. Virol. 2013;87:859–871. doi: 10.1128/JVI.01158-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv S., Xu Q., Sun E., Yang T., Li J., Feng Y., Zhang Q., Wang H., Zhang J., Wu D. Autophagy activated by bluetongue virus infection plays a positive role in its replication. Viruses. 2015;7:4657–4675. doi: 10.3390/v7082838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Zhao X., Zhang Z., Guo J., Guan L., Li J., Mi M., Huang Y., Tong D. Differentially expressed non-coding RNAs induced by transmissible gastroenteritis virus potentially regulate inflammation and NF-kappaB pathway in porcine intestinal epithelial cell line. BMC Genomics. 2018;19:747. doi: 10.1186/s12864-018-5128-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier H.J., Cottam E.M., Stevenson-Leggett P., Wilkinson J.A., Harte C.J., Wileman T., Britton P. Visualizing the autophagy pathway in avian cells and its application to studying infectious bronchitis virus. Autophagy. 2013;9:496–509. doi: 10.4161/auto.23465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane S., Aitken J., Sutherland J.S., Nicholl M.J., Preston V.G., Preston C.M. Early induction of autophagy in human fibroblasts after infection with human cytomegalovirus or herpes simplex virus 1. J. Virol. 2011;85:4212–4221. doi: 10.1128/JVI.02435-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohl B.P., Tedbury P.R., Griffin S., Harris M. Hepatitis C virus-induced autophagy is independent of the unfolded protein response. J. Virol. 2012;86:10724–10732. doi: 10.1128/JVI.01667-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monastyrska I., Ulasli M., Rottier P.J., Guan J.L., Reggiori F., de Haan C.A. An autophagy-independent role for LC3 in equine arteritis virus replication. Autophagy. 2013;9:164–174. doi: 10.4161/auto.22743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell D., Liang C. Autophagy interaction with herpes simplex virus type-1 infection. Autophagy. 2016;12:451–459. doi: 10.1080/15548627.2016.1139262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostra M., Hagemeijer M.C., van Gent M., Bekker C.P., te Lintelo E.G., Rottier P.J., de Haan C.A. Topology and membrane anchoring of the coronavirus replication complex: not all hydrophobic domains of nsp3 and nsp6 are membrane spanning. J. Virol. 2008;82:12392–12405. doi: 10.1128/JVI.01219-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudit G.Y., Kassiri Z., Jiang C., Liu P.P., Poutanen S.M., Penninger J.M., Butany J. SARS-coronavirus modulation of myocardial ACE2 expression and inflammation in patients with SARS. Eur. J. Clin. Invest. 2009;39:618–625. doi: 10.1111/j.1365-2362.2009.02153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Kim H.K., Song D.S., An D.J., Park B.K. Complete genome sequences of a Korean virulent porcine epidemic diarrhea virus and its attenuated counterpart. J. Virol. 2012;86:5964. doi: 10.1128/JVI.00557-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.E., Cruz D.J., Shin H.J. Clathrin- and serine proteases-dependent uptake of porcine epidemic diarrhea virus into Vero cells. Virus Res. 2014;191:21–29. doi: 10.1016/j.virusres.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul P., Munz C. Autophagy and mammalian viruses: roles in immune response, viral replication, and beyond. Adv. Virus Res. 2016;95:149–195. doi: 10.1016/bs.aivir.2016.02.002. [DOI] [PubMed] [Google Scholar]
- Radtke K., English L., Rondeau C., Leib D., Lippe R., Desjardins M. Inhibition of the host translation shutoff response by herpes simplex virus 1 triggers nuclear envelope-derived autophagy. J. Virol. 2013;87:3990–3997. doi: 10.1128/JVI.02974-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B., Vacher C., Berger Z., Davies J.E., Luo S., Oroz L.G., Scaravilli F., Easton D.F., Duden R., O’Kane C.J., Rubinsztein D.C. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- Rozieres A., Viret C., Faure M. Autophagy in measles virus infection. Viruses. 2017:9. doi: 10.3390/v9120359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo R., Varano G.P., Adornetto A., Nazio F., Tettamanti G., Girardello R., Cianfanelli V., Cavaliere F., Morrone L.A., Corasaniti M.T., Cecconi F., Bagetta G., Nucci C. Rapamycin and fasting sustain autophagy response activated by ischemia/reperfusion injury and promote retinal ganglion cell survival. Cell Death Dis. 2018;9:981. doi: 10.1038/s41419-018-1044-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki S., Sasazawa Y., Imamichi Y., Kawajiri S., Fujimaki T., Tanida I., Kobayashi H., Sato F., Sato S., Ishikawa K., Imoto M., Hattori N. Caffeine induces apoptosis by enhancement of autophagy via PI3K/Akt/mTOR/p70S6K inhibition. Autophagy. 2011;7:176–187. doi: 10.4161/auto.7.2.14074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S.S., Vats S., Chia A.Y., Tan T.Z., Deng S., Ong M.S., Arfuso F., Yap C.T., Goh B.C., Sethi G., Huang R.Y., Shen H.M., Manjithaya R., Kumar A.P. Dual role of autophagy in hallmarks of cancer. Oncogene. 2017 doi: 10.1038/s41388-017-0046-6. [DOI] [PubMed] [Google Scholar]
- Stevenson G.W., Hoang H., Schwartz K.J., Burrough E.R., Sun D., Madson D., Cooper V.L., Pillatzki A., Gauger P., Schmitt B.J., Koster L.G., Killian M.L., Yoon K.J. Emergence of Porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Invest. 2013;25:649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- Supekar V.M., Bruckmann C., Ingallinella P., Bianchi E., Pessi A., Carfi A. Structure of a proteolytically resistant core from the severe acute respiratory syndrome coronavirus S2 fusion protein. Proc. Natl. Acad. Sci. U. S. A. 2004;101:17958–17963. doi: 10.1073/pnas.0406128102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surviladze Z., Sterk R.T., DeHaro S.A., Ozbun M.A. Cellular entry of human papillomavirus type 16 involves activation of the phosphatidylinositol 3-kinase/Akt/mTOR pathway and inhibition of autophagy. J. Virol. 2013;87:2508–2517. doi: 10.1128/JVI.02319-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. New variant of porcine epidemic diarrhea virus, United States, 2014. Emerging Infect. Dis. 2014;20:917–919. doi: 10.3201/eid2005.140195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Liu J., Zhen J., Zhang C., Wan Q., Liu G., Wei X., Zhang Y., Wang Z., Han H., Xu H., Bao C., Song Z., Zhang X., Li N., Yi F. Histone deacetylase 4 selectively contributes to podocyte injury in diabetic nephropathy. Kidney Int. 2014;86:712–725. doi: 10.1038/ki.2014.111. [DOI] [PubMed] [Google Scholar]
- Wang D., Fang L., Shi Y., Zhang H., Gao L., Peng G., Chen H., Li K., Xiao S. Porcine epidemic diarrhea virus 3C-LiK.e protease regulates its interferon antagonism by cleaving NEMO. J. Virol. 2015;90:2090–2101. doi: 10.1128/JVI.02514-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Qiao X., Zhang S., Qin Y., Guo T., Hao Z., Sun L., Wang X., Wang Y., Jiang Y., Tang L., Xu Y., Li Y. Porcine transmissible gastroenteritis virus nonstructural protein 2 contributes to inflammation via NF-kappaB activation. Virulence. 2018;9:1685–1698. doi: 10.1080/21505594.2018.1536632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Li G., De W., Luo Z., Pan P., Tian M., Wang Y., Xiao F., Li A., Wu K., Liu X., Rao L., Liu F., Liu Y., Wu J. Zika virus infection induces host inflammatory responses by facilitating NLRP3 inflammasome assembly and interleukin-1beta secretion. Nat. Commun. 2018;9:106. doi: 10.1038/s41467-017-02645-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z.Y., Lu W.H., Li Z.L., Mo J.Y., Zeng X.D., Zeng Z.L., Sun B.L., Chen F., Xie Q.M., Bee Y.Z., Ma J.Y. Complete genome sequence of novel porcine epidemic diarrhea virus strain GD-1 in China. J. Virol. 2012;86:13824–13825. doi: 10.1128/JVI.02615-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Fu Y., Tong J., Fan S., Xu K., Sun H., Liang Y., Yan C., Yuan Z., Ge Y. MicroRNA-216b/Beclin 1 axis regulates autophagy and apoptosis in human Tenon’s capsule fibroblasts upon hydroxycamptothecin exposure. Exp. Eye Res. 2014;123:43–55. doi: 10.1016/j.exer.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Yang Y.P., Hu L.F., Zheng H.F., Mao C.J., Hu W.D., Xiong K.P., Wang F., Liu C.F. Application and interpretation of current autophagy inhibitors and activators. Acta Pharmacol. Sin. 2013;34:625–635. doi: 10.1038/aps.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasunaga K., Ito T., Miki M., Ueda K., Fujiyama T., Tachibana Y., Fujimori N., Kawabe K., Ogawa Y. Using CRISPR/Cas9 to knock out amylase in acinar cells decreases pancreatitis-induced autophagy. Biomed Res. Int. 2018;2018 doi: 10.1155/2018/8719397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yla-Anttila P., Vihinen H., Jokitalo E., Eskelinen E.L. Monitoring autophagy by electron microscopy in Mammalian cells. Meth. Enzymol. 2009;452:143–164. doi: 10.1016/S0076-6879(08)03610-0. [DOI] [PubMed] [Google Scholar]
- Yordy B., Iwasaki A. Cell type-dependent requirement of autophagy in HSV-1 antiviral defense. Autophagy. 2013;9:236–238. doi: 10.4161/auto.22506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang F., Chen Y., Lin Z., Cai Z., Yu L., Xu F., Wang J., Zhu W., Lu H. Autophagy is involved in regulating the immune response of dendritic cells to influenza A (H1N1) pdm09 infection. Immunology. 2016;148:56–69. doi: 10.1111/imm.12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Ke H., Blikslager A., Fujita T., Yoo D. Type III interferon restriction by porcine epidemic diarrhea virus and the role of viral protein nsp1 in IRF1 signaling. J. Virol. 2017 doi: 10.1128/JVI.01677-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Cong Y., Veenendaal T., Klumperman J., Shi D., Mari M., Reggiori F. Ultrastructural characterization of membrane rearrangements induced by porcine epidemic diarrhea virus infection. Viruses. 2017:9. doi: 10.3390/v9090251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou D., Xu J., Duan X., Xu X., Li P., Cheng L., Zheng L., Li X., Zhang Y., Wang X., Wu X., Shen Y., Yao X., Wei J., Yao L., Li L., Song B., Ma J., Liu X., Wu Z., Zhang H., Cao H. Porcine epidemic diarrhea virus ORF3 protein causes endoplasmic reticulum stress to facilitate autophagy. Vet. Microbiol. 2019;235:209–219. doi: 10.1016/j.vetmic.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.