Significance
New approaches to prevention and treatment of drug-resistant bacterial infections are needed. Here we show how synthetic molecular evolution, screening of iterative libraries, can identify membrane permeabilizing antimicrobial peptides (AMPs) that do not have the usual impediments to activity in vivo. Screening in the presence of concentrated human erythrocytes to mimic a generic cell-rich environment enabled identification of peptides with potent, broad-spectrum bactericidal activity under conditions where most known AMPs have little or no activity. One optimized peptide was tested in the cell- and protein-rich environment of an infected, purulent wound in mice. Infection was significantly reduced, and biofilm formation in the wound was prevented, demonstrating the power of synthetic molecular evolution of AMPs.
Keywords: drug resistant bacteria, antimicrobial peptide, Pseudomonas, MRSA, wound care
Abstract
Novel classes of antibiotics and new strategies to prevent and treat infections are urgently needed because the rapid rise in drug-resistant bacterial infections in recent decades has been accompanied by a parallel decline in development of new antibiotics. Membrane permeabilizing antimicrobial peptides (AMPs) have long been considered a potentially promising, novel class of antibiotic, especially for wound protection and treatment to prevent the development of serious infections. Yet, despite thousands of known examples, AMPs have only infrequently proceeded as far as clinical trials, especially the chemically simple, linear examples. In part, this is due to impediments that often limit their applications in vivo. These can include low solubility, residual toxicity, susceptibility to proteolysis, and loss of activity due to host cell, tissue, and protein binding. Here we show how synthetic molecular evolution can be used to evolve potentially advantageous antimicrobial peptides that lack these impediments from parent peptides that have at least some of them. As an example of how the antibiotic discovery pipeline can be populated with more promising candidates, we evolved and optimized one family of linear AMPs into a new generation with high solubility, low cytotoxicity, potent broad-spectrum sterilizing activity against a panel of gram-positive and gram-negative ESKAPE pathogens, and antibiofilm activity against gram-positive and gram-negative biofilms. The evolved peptides have these activities in vitro even in the presence of concentrated host cells and also in vivo in the complex, cell- and protein-rich environment of a purulent animal wound model infected with drug-resistant bacteria.
Multidrug-resistant bacterial pathogens are a significant, rapidly expanding, and costly threat to human health (1–8). Mathematical models predict that if current rates of increase continue, by 2050 more than 10 million people will die annually from drug-resistant bacterial infections (DRBIs), surpassing all other causes of death, including cancer (9). Nosocomial DRBIs are now commonplace in healthcare settings and community-acquired DRBIs are increasing (3, 4, 10). DRBIs frequently begin by first contaminating wounds, burns, surgical sites, chronic skin ulcers, and medical devices such as catheters. For example, at least 40% of war trauma wounds (11) and >75% of diabetic foot ulcers (12, 13) are contaminated with bacteria, with the majority of cultures containing drug-resistant bacteria. Improved infection control procedures in healthcare settings have had a positive impact, yet up to 6% of US hospitals have ongoing outbreaks of drug-resistant bacteria at any point in time (14) causing substantial morbidity and mortality and creating a very high financial burden (1, 6, 7, 15). Exacerbating the problem, the development of new classes of antibiotics has dramatically decreased such that only two new classes of antibiotic have been approved by the Food and Drug Administration (FDA) or European Medicines Agency since 2000 (16). There have also been very few new antibiotics specifically designed to protect wounds from DRBIs. New therapeutic targets and novel chemotypes are needed to protect and treat wounds against DRBIs, just as new targets and chemotypes are needed for systemic antibiotics. A key element of solving this problem will be the revitalization of the dwindling antibiotic development pipeline.
Antimicrobial peptides (AMPs), which act on microbial membranes, have long been considered a unique and potentially useful class of antibiotic (17, 18). There are many known examples, natural and synthetic, that have excellent, broad-spectrum bactericidal activity in vitro (19), and a few have undergone clinical trials (20), mostly for treatment of infections in chronic wounds (21). Despite their promise, most known AMPs have impediments to therapeutic utility. For example, AMPs can have limited solubility, residual host cell toxicity, susceptibility to proteolytic degradation, and binding to host cells, tissue, and serum proteins, all of which are expected to reduce or eliminate the activity of an AMP in vivo (22, 23).
Membrane disrupting AMPs act by interfacial activity (24) through heterogeneous, dynamic interactions with membranes and other structures (lipopolysaccharides [LPS], cell wall components, DNA, and intracellular proteins). Very few AMPs have sequence-specific interactions with any individual biochemical entity. However, we note that the two membrane permeabilizing AMPs which have been approved for limited use in humans, colistins (polymyxin B and E) and daptomycin, are complex, natural, cyclic peptides that specifically interact with bacterial lipid precursors (25), suggesting that better selectivity for bacteria over host cells will be important for development of useful AMPs.
The AMP field has continued to advance toward better activity in vivo. For example, database-driven design of AMPs has been used to identify active peptides that are unusual in their low cationicity (26). Low cationicity may be one way to reduce host cell inhibition. Others have demonstrated some utility for in silico optimization of AMPs (27, 28). Despite these examples, it remains difficult to intentionally populate the antibiotic pipeline with AMPs designed to be active in vivo because there are few reliable quantitative structure activity relationship (QSAR) rules that are useful in rational design, engineering, and optimization of AMPs to avoid these impediments. It is also difficult to rationally design an AMP to act more selectively on microbes over host cells. With a few exceptions, discovery of new AMPs has been achieved by the trial-and-error modification of known sequences. Even then, the major impediments to therapeutic applications discussed here, one or more of which likely apply to most known AMPs (22, 23), are not usually assessed until after the peptide design/discovery phase. This approach reduces opportunities for feedback and down-selection of AMPs that have impediments, ultimately limiting the flow of promising new AMP candidates into the drug development pipeline.
Here, we use iterative library design and orthogonal screening (29–33), which we call synthetic molecular evolution (SME), to identify AMPs that are specifically designed to have selectivity for microbes over host cells and specifically designed to lack other impediments to activity in vivo. We achieve this gain of function by applying selective pressures during initial AMP screening for bactericidal activity with down-selection based on solubility, hemolysis, and host cell inhibition. Antibacterial screens are done in the presence of concentrated human red blood cells, a consistent and experimentally reliable generic surrogate for all host cells and tissue. We do this to identify “host cell compatible” AMPs that retain activity against microbes in the presence of concentrated host cells. The resulting host cell compatible AMPs we have evolved are highly effective in vitro against many bacterial pathogens even in the presence of concentrated host cells, and they have antibiofilm activity and gram-negative and gram-positive biofilms. In a clinically relevant wound model in mice (34), the evolved peptides are active against both gram-negative and gram-positive drug-resistant bacteria and have antibiofilm activity in infected wounds.
Results
Library Design and Screening.
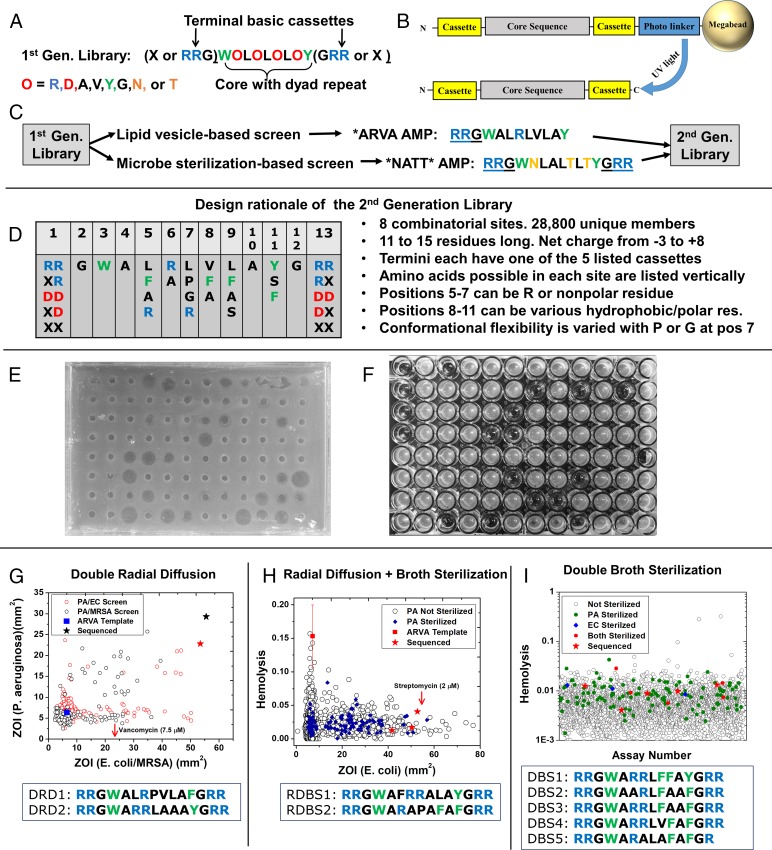
To identify host cell compatible AMPs, we screened an iterative, second generation library that was built on the results of screening and characterization of a de novo designed first-generation library of β-sheet-rich peptides (35–37). The first-generation library of 9 to 15 residue peptides is shown in Fig. 1A. Libraries are synthesized as one-bead one-compound libraries with the architecture shown in Fig. 1B. The first-generation library was previously screened for synthetic lipid vesicle permeabilization (36, 37) and for potent, broad-spectrum microbial sterilization (35). Each of these first-generation screens yielded ∼10 optimal sequences, exemplified by the peptides ARVA (RRGWALRLVLAY-NH2) and NATT (RRGWNLALTLTYGRR-NH2) (Fig. 1C) from the vesicle-based and antimicrobial screens, respectively. These two sequences, which belong to distinct families (35), both have potent, sterilizing antimicrobial activity in standard laboratory assays (38) against a range of gram-negative and gram-positive bacteria, including drug-resistant strains (35). However, like many known AMPs, ARVA and NATT are strongly inhibited by host cells (23), are susceptible to proteolysis (22), have poor solubility, and are somewhat cytotoxic (35).
Fig. 1.
Design and screening of the second-generation AMP library. (A) The first-generation, de novo designed library that was previously screened for membrane permeabilization (37) and bacterial sterilization (35). RRG−/−GRR cassettes were present or absent as tripeptides. (B) One-bead, one-sequence library architecture. Library members are attached to nominal 0.3-mm diameter polystyrene/PEG Tentagel Megabeads by a photocleavable linker. Each bead releases about 0.5 nmol of a single peptide sequence. (C) Schematic of prior work leading to the second-generation library, see text. (D) Second-generation library design. The 2 to 5 residues listed vertically under each position, and the native residue, are all equally probable. Acidic or basic terminal groups are cassettes of 0, 1, or 2 residues. A list of design principles are shown to the Right. See text. (E) Example radial diffusion assay plate for E. coli. Small holes in the bacteria-seeded agar layer were filled with 10 μL of an 8-μM solution of a single-library member extracted from a single bead. Bacteria grow at the interface after addition of nutrient agar, except where inhibited. Radii of zones of inhibition were measured after overnight incubation. (F) Example plate from broth sterilization screening assay P. aeruginosa. Library members at 8 μM plus 1 × 109 human RBC/mL were added to 105 P. aeruginosa/mL in growth media. After overnight incubation, a second assay plate containing media was inoculated and grown overnight. Clear wells in the second plate indicate that the bacteria were sterilized in the presence of concentrated RBCs. (G) Results of small-scale screen by DRD of 7.5 μM of peptide in the presence of 1 × 109 human RBC/mL against P. aeruginosa and either E. coli or MRSA. The sequences of two exceptional peptides, marked by stars, are shown. The activity of 7.5 μM vancomycin against S. aureus is shown for comparison. (H) Results of small-scale hybrid screen using radial diffusion with ∼7.5 μM of each peptide in the presence of 1 × 109 human RBC/mL against E. coli, and broth sterilization with RBCs against P. aeruginosa. Hemolysis was also measured. Sequences of two exceptional peptides, with activity against both microbes and low hemolysis, marked by red stars, are shown. The activity of 2 μM streptomycin against E. coli is shown for comparison. (I) Results from ∼3,800 library members that were screened by a DBS assay in which sterilization of E. coli and P. aeruginosa in the presence of 1 × 109 human RBC/mL were assessed in addition to hemolysis. The sequences of five exceptional peptides with low hemolysis are shown.
In this work we sought to evolve optimized AMPs that do not have these impediments. We used ARVA and NATT and their extensive characterization (22, 23, 35–37) to guide the design of the second-generation library shown in Fig. 1D. See also SI Appendix, Fig. S1. Our approach to SME uses libraries that are constrained in diversity and that test specific hypotheses through limited variation of rationally identified positions in the sequence. Since the RRG- and -GRR terminal cassettes were found in many of the first-generation hits, we included these terminal motifs, and variants of them, in the second-generation library (positions 1 and 13). We also included anionic cassettes to test for anionic AMPs, but we ultimately did not select any. The terminal cassettes are varied independently (Fig. 1D), giving 25 possible combinations of termini. The library has a nine-residue core that is similar to ARVA. This core has a potentially more polar N-terminal region, positions 3 to 7, and a more hydrophobic C-terminal region, positions 8 to 11. A single, critical R is present in ARVA at position 6 so we permitted library positions 5, 6, and 7 to have either a cationic R, or a nonpolar residue. To test if conformational flexibility affects activity, we allowed the central position 7 to have either G (more flexible) or P (less flexible) in addition to the native L and cationic R. To test if the hydrophobicity of the C-terminal region of the core sequence is critical for activity, hydrophobicity was varied in the library by allowing for some of the set A, F, Y, or S in positions 8, 9, and 11. The library contains 28,800 unique, 9 to 15 residue sequences with net charges from −3 to +8. The library was synthesized as a one-bead one-sequence library (Fig. 1B) and was validated as described in detail previously (29–32). Upon irradiation of the UV-cleavable linker, each bead released about 0.5 nmol of a single peptide sequence, sufficient to perform the multiple, parallel assays used here for selection.
To select host cell-compatible AMPs, we performed antimicrobial screens in the presence of 1 × 109 human red blood cells (RBC)/mL, a dense cell suspension, 20% of cell density in blood. Compared to nucleated cells or homogenized tissues from experimental animals, concentrated human RBCs are a more readily available, highly consistent, stable, and sterile reagent to mimic host cells and tissue, including the environment one would find in a wound. In the presence of 1 × 109 RBC/mL the activity of many AMPs is reduced or eliminated by RBC binding and RBC-dependent proteolysis (22, 23).
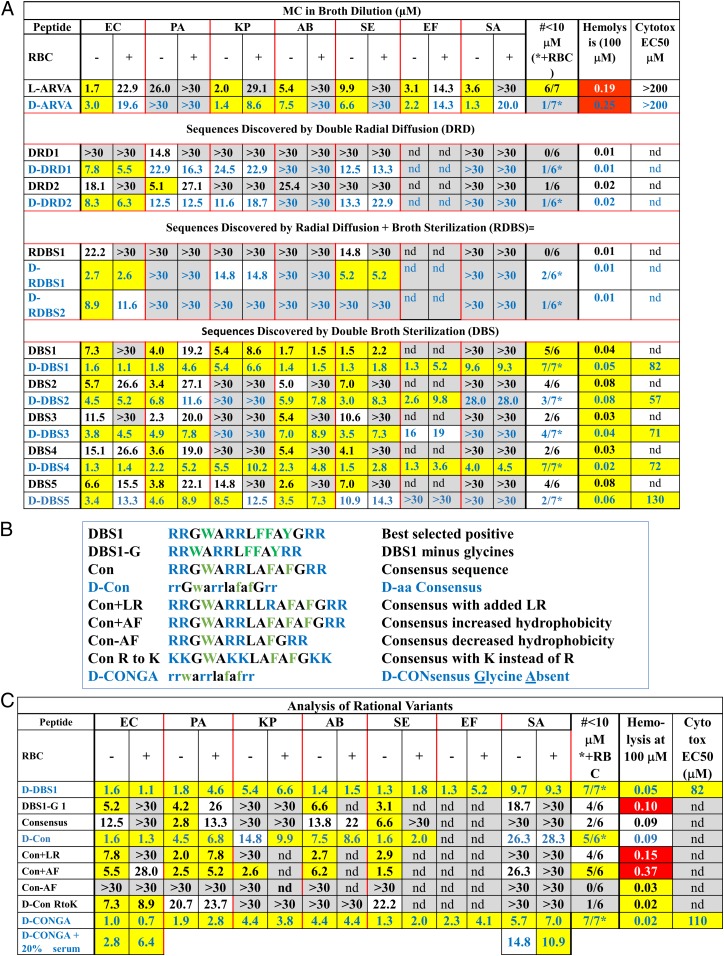
We tested three screening strategies, utilizing either radial diffusion (Fig. 1E) or broth sterilization (Fig. 1F), performed in the presence of concentrated human RBCs (23). Results of the three screening strategies are shown in Fig. 1 G–I. With one exception we tested activity in parallel against two gram-negative pathogens, Escherichia coli and Pseudomonas aeruginosa, and also measured hemolysis. An apparent advantage of radial diffusion as a screening tool is that it provides the zone of inhibition (ZOI), a quantitative measure of potency, from a single point, enabling hits to be rank ordered. In the double radial diffusion (DRD) screen, we assayed a small sample of the library in parallel against E. coli and P. aeruginosa in one test, and against P. aeruginosa and methicillin-resistant Staphylococcus aureus (MRSA) in another test. Compared to the template ARVA, which is inactive in this screen due to host cell inhibition, the library contained some clearly superior sequences (Fig. 1G). Two of these peptides that had low hemolysis were sequenced. These peptides were then synthesized tested against a panel of ESKAPE organisms (Fig. 2A) in broth sterilization assays. The ESKAPE bacteria include the common human pathogens: Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and the Enterobacteriaceae, which includes Enterobacter, Escherichia, and Salmonella species. Assays were performed with and without 1 × 109 human RBC/mL using both protease susceptible l-amino acid peptides and protease resistant D-amino acid peptides. The selected peptides, even in D-form, generally had poor sterilizing activity against the panel, including the bacteria they inhibit in the screen. The DRD screen did not yield the sterilizing antimicrobial activity we sought.
Fig. 2.
Assessment of antibacterial activity and toxicity of selected peptides. (A) MIC values are reported in micromolar peptide against a panel of ESKAPE bacterial pathogens: E. coli (EC), P. aeruginosa (PA), K. pneumoniae (KP), A. baumannii (AB), S. enterica (SE), E. faecium (EF), and S. aureus (SA). The two columns under each organism are for assays performed in the absence (−) and presence (+) of 1 × 109 human RBC/mL yellow: MIC ≤ 10 μM; gray: MIC ≥ 25 μM. “>30” means that sterilization was not observed at 30 μM, the highest concentration tested. Values in blue text are for D-amino acid peptides. The column marked “#≤10 μM” is a count of the number of organisms, out of seven, with MIC ≤ 10 μM. Counts are in the absence of RBC for l-amino acid peptides and in the presence of RBCs for D-amino acid peptides. The column marked “hemolysis” is the fractional hemolysis of 1 × 108 human RBC/mL at 100 μM peptide. Yellow indicates ≤5% hemolysis. The column marked “Cytotox EC50” contains the concentration of peptide that kills 50% of WI-38 human fibroblast cells assayed by entry of SYTOX Green DNA binding dye. Yellow signifies EC50 ≥ 50 μM. SDs of MIC are typically about 50% of the MIC value. SDs in hemolysis are ±0.02 and SDs in cytotoxicity EC50 are ±8 μM. nd, not determined. (B) Rational variants of DBS peptides that were tested and the rationale for each variant. (C) MIC values for rational variants tested, compared to D-DBS1. Definitions and color codes are identical to those in A.
Next, we developed a hybrid radial diffusion-broth sterilization (RDBS) screen. We measured ZOI by radial diffusion against E. coli in the presence of concentrated RBCs, as above. But in parallel, we performed a broth dilution sterilization assay against P. aeruginosa in the presence of RBCs (Fig. 1H). Two of the best sequences in this small-scale screen, which have large ZOIs against E. coli, sterilize P. aeruginosa, and have low hemolysis, were sequenced. These RDBS peptides appear to be very similar to the DRD positives. They were also tested against the panel of ESKAPE pathogens with and without RBCs (Fig. 2A). Again, the selected peptides did not have potent sterilizing activity, and, in fact, did not sterilize P. aeruginosa in the slightly more stringent postscreen assay, despite their ability to do so under the screening conditions. Radial diffusion-based screens do not identify peptides with sterilizing activity against multiple pathogens when tested in broth dilution.
Ultimately, we screened the library with a double broth sterilization (DBS) assay (Fig. 1I) in which library members were assayed in parallel for 1) sterilization of E. coli in the presence of concentrated RBCs, 2) sterilization of P. aeruginosa in the presence of concentrated RBCs, and 3) lack of hemolysis of human RBCs. The screen was performed in 96-well format, where each well had about 8 μM of one library member, 1 × 109 human RBC/mL, and either E. coli or P. aeruginosa. A parallel assay for hemolysis was performed with 8 μM peptide and 1 × 109 RBC/mL. After a preliminary test showed that DBS positives had the sterilizing activity, we assayed 3,800 library members under high stringency conditions. A total of 13 (0.3%) of the library members sterilized only E. coli and 127 (3.3%) sterilized only P. aeruginosa. Nine library members (0.2%) sterilized both organisms in the presence of 1 × 109 RBC/mL and caused less than 2% hemolysis. We sequenced five of these positives, giving the DBS sequences shown in Fig. 1I.
The five DBS-positive peptides were very similar in sequence, suggesting convergence to a small part of the sequence space of the library. Thus, we terminated the screening to focus on characterization of the activity of the DBS hits against seven ESKAPE pathogens (Fig. 2A). To verify that we had selected peptides without host cell inhibition, we synthesized all DBS peptides in both l- and D-amino acid forms, and tested them in the absence and presence of 1 × 109 human RBC/mL. This approach is useful because we have previously shown that human RBCs have very high cytosolic exo- and endopeptidase activity (22) which is similar to the proteolytic activity in human serum. Thus, postscreening assays done in the presence of RBCs test for host cell inhibition due to binding and conveniently mimic serum proteolysis for l-amino acid peptides. In the postscreening assays, the proteolytic activity released from concentrated RBCs by a small amount of background autolysis and/or a small amount of peptide-induced hemolysis is enough to digest l-amino acid peptides (SI Appendix, Fig. S2).
The DBS screening strategy was highly successful. The screen positives had significant sterilizing activity against the panel of ESKAPE pathogens and low hemolysis (Fig. 2A). In the absence of RBCs, the activities of the l- and D-peptides were indistinguishable. The presence of human RBCs did not significantly decrease the activity of D-amino acid DBS peptides, but reduced the activity of the l-DBS peptides by proteolysis, as expected (22, 23) (SI Appendix, Fig. S2). We compared l- and D-amino acid variants of the five selected DBS peptides against all seven organisms. In the absence of host cells, 25 comparisons are possible; 18 showed that the D-peptide was more active (P = 0.05, not significant for abundance). The mean ratio of measured minimum inhibitory concentration (MIC) (D/L) was 0.85 ± 0.60 (SD, n = 20, P > 0.05) again indicating no difference in activity in the absence of RBCs. In the presence of RBCs, however, 26/26 available MIC comparisons showed the D-peptide to have MIC equal to or lower than l-peptides (P < 1 × 10−5) for abundance. The mean MIC ratio (D/L) is 0.31 ± 0.28 (SD, n = 20, P < 0.001). D-peptides had much better activity in the presence of RBCs, due to their protease resistance. For the DBS and related peptides, very similar MICs are also obtained using radial diffusion (SI Appendix, Fig. S3). Two of the peptides, DBS1 and DBS4, have sterilizing activity at ≤10 μM against all seven organisms, including the gram-positive S. aureus and E. faecium, in the presence of concentrated RBCs. The other DBS peptides have sterilizing activity against most of the gram-negative organisms. K. pneumoniae, a gram-negative organism, was susceptible to three of the five DBS peptides in D-form. These results show that our strategy of screening by broth sterilization in parallel against E. coli and P. aeruginosa enables selection of broad-spectrum antibiotics. In SI Appendix, Fig. S4, we show that the five l-DBS peptides also have potent sterilizing activity against multidrug-resistant clinical isolates of K. pneumoniae and A. baumannii.
We note that the postscreen assays shown in Fig. 2 are much more stringent than typical MIC assays for AMPs reported in the literature. This is not only because peptides have been preincubated with concentrated RBCs, but also because we do not use the typical 1- to 3-h preincubation of peptide with bacteria in minimal media prior to the addition of growth media. Instead, we add bacteria in full growth media directly to peptides with RBCs. In this case, bacteria are energized and replicating from time 0 and can readily overcome any antibiotic that does not rapidly sterilize 100% of cells. Under these conditions, many well-known AMPs, natural and synthetic, are inactive (22, 23).
Sequence Analysis.
The sequences selected in the three screens are very similar, representing a motif that may itself have utility in future AMP evolution and optimization. There were 25 possible combinations of N and C termini in the library (Fig. 1A). Yet, in the nine selected peptides we found essentially only one; all positives have RR- on the N terminus and -RR on the C terminus, except for DBS5, which has RR- and -R. A similarly specific selection for terminal basic cassettes was observed in the biological screen of the first-generation library (35). The more polar N-terminal core begins with the invariant WA sequence followed by three positions that could contain either R or one of several nonpolar amino acids. The selected positive peptides all have one or two R residues in that region, more often in positions 5 and 6. The sequence WARRL in positions 3 to 7 was observed in four of the nine total positives, including the highly active DBS1, DBS3, and DBS4. In the central position 7 we allowed the library to have G and P in addition to L to select for variants with altered conformational flexibility. However, G was never selected, and P was selected only in two sequences from the inferior DBS and RDBS screens. In the five superior DBS-positive peptides, position 7 was always L. The C-terminal region of the library core is, by design, hydrophobic, but the selected sequences are more hydrophobic than the average library member, always containing two or three aromatic residues in the three varied positions. Based on this analysis, we recognize a DBS consensus sequence of RRGWARRLAFAFGRR.
Rational Variants.
Next, we tested hypotheses that were not directly addressed in the library design by characterizing the variants described in Fig. 2B and comparing them to D-DBS1 (Fig. 2C). We modified the N core of the consensus sequence by adding LR, and added or removed an AF unit from the C core. Core changes all had consequential, negative effects on activity. The addition of a second LR to the N-terminal core increased hemolysis (Fig. 2B). Removal of one AF unit eliminated all bactericidal activity. Addition of an extra AF unit dramatically increased hemolysis. The balance between core charge and hydrophobicity is an important determinant of selectivity and bactericidal activity. Replacement of all six R residues in the consensus sequence with K eliminated almost all activity, highlighting the critical role of R. Removal of the two invariant G residues from DBS1 (positions 3 and 13) caused small loss of bactericidal activity and an increase in hemolysis. The consensus sequence (CON) in l- and D-form had slightly lower bactericidal activity than DBS1. Finally, removal of the invariant glycines from the consensus sequence to make D-CONGA (D-amino acid CONsensus with Glycine Absent) resulted in a peptide with superior bactericidal activity against all seven ESKAPE pathogens, including the gram-positive species S. aureus and E. faecium. We also tested the effect of 20% human serum and 20% serum + 20% human RBCs on the activity of D-CONGA and found that the activity was not strongly affected (Fig. 2C).
The mechanism of bacterial cell killing by these evolved peptides includes membrane disruption which is typical of cationic/hydrophobic AMPs. In SI Appendix, Fig. S5 A and B, we show that D-DBS4 and D-CONGA enable the entry of SYTOX Green, a membrane impermeant DNA binding dye, into bacterial cells, and that SYTOX Green entry is maximum at around the MIC of the peptides (1 to 2 μM). These peptides may also interact strongly with DNA as shown in SI Appendix, Fig. S5C where D-DBS4 and D-CONGA are shown to displace SYTOX Green from bacterial DNA at low micromolar concentrations. Further, like most cationic AMPs, D-CONGA binds strongly to synthetic bacteria-like bilayers (SI Appendix, Fig. S6A) and inserts hydrophobic residues into the bilayer (SI Appendix, Fig. S6B). D-CONGA causes leakage of entrapped dyes from anionic vesicles (SI Appendix, Fig. S6C) at peptide-to-lipid ratios that are typical of synthetic AMPs (39).
Ultimately, the data in Fig. 2 A and C show that D-DBS1, D-DBS4, and D-CONGA, which differ by two deleted glycines and by one or two conservative variations in the C-terminal core, are the most optimal peptides identified in this work. They are active against all ESKAPE pathogens tested and are not inhibited by the presence of concentrated host cells. These peptides have solubility in saline solution ≥1 mM and are resistant to proteolysis. Their therapeutic index appears to be high as they cause little or no hemolysis, even at high peptide concentration, and have low toxicity against human fibroblasts (Fig. 2 and SI Appendix, Fig. S7 A and B).
Absence of Resistance.
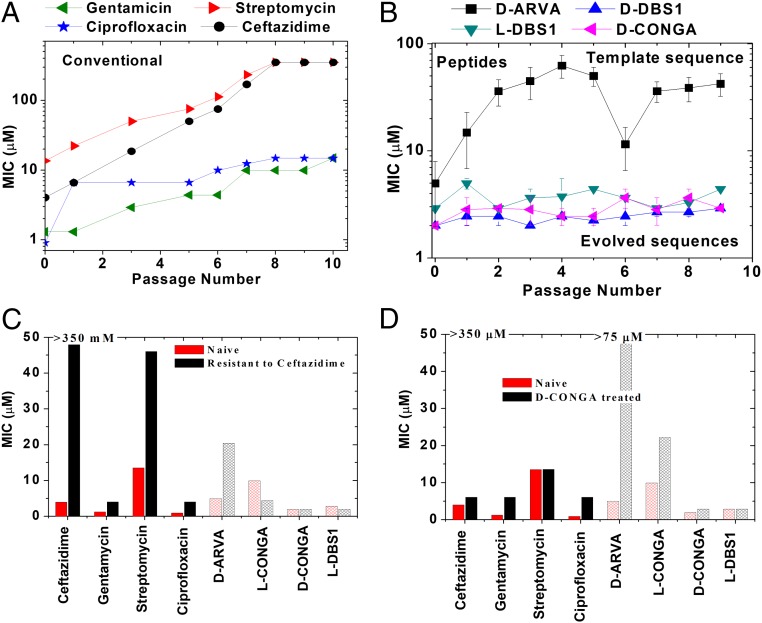
We tested the lead peptides in a P. aeruginosa model of stable antibiotic resistance. P. aeruginosa is inherently resistant, or can rapidly become resistant, to many conventional antibiotics through multiple mechanisms (40). It can also become resistant to some AMPs through alteration of the composition of its outer membrane lipopolysaccharide (40). We tested whether P. aeruginosa could develop stable resistance to four conventional small-molecule antibiotics, to the template peptide D-ARVA, or to positive sequences, l- and D-DBS1 and D-CONGA. We grew bacteria in serial dilutions of antibiotic and selected the bacteria that grew at the highest concentration of antibiotic to be recultured overnight in the absence of antibiotic. The latter step enables only the selection of stable resistance without a decrease in fitness. Each passage was again treated with a concentration series of antibiotic and the process was repeated for 10 passages. Complete resistance (MIC ≥ 350 μM) to streptomycin and ceftazidime were observed after 8 passages (Fig. 3A), while resistance to ciprofloxacin and gentamycin steadily increased at each passage. When treated with the library template sequence, D-ARVA, P. aeruginosa became resistant (MIC ≥ 50 μM) over the first 4 passages (Fig. 3B). Against l-ARVA, P. aeruginosa became resistant in a single passage. However, against D-DBS1, l-DBS1, and D-CONGA, no measurable increase in resistance was observed over 10 passages.
Fig. 3.
Selection for resistance. (A and B) A culture of P. aeruginosa in log phase growth in TSB media was treated with serial dilutions of four conventional antibiotics (A) and four antimicrobial peptides (B). The bacteria that grew in the highest concentration of antibiotic were cultured overnight in the absence of antibiotic and then treated similarly the next day. This was continued for 10 passages to select for stable resistance mutants. MICs for each passage are shown in A and B. The highest concentrations tested were 350 μM for conventional antibiotics and 67.5 μM for AMPs. (C and D) Cross-resistance was measured by testing the zeroth passage (unselected) and the 10th passages of P. aeruginosa selected in the presence of either ceftazidime (C) or D-CONGA (D) against the whole panel of four conventional and four peptide antibiotics.
Cross-resistance profiles were determined by measuring changes in the susceptibility profile after 10 passages with either ceftazidime or D-CONGA. Despite very different biochemical mechanisms, ceftazidime treatment leads to significant streptomycin resistance and some resistance to the other small-molecule antibiotics (Fig. 3 C and D), possibly through multidrug efflux mechanisms (40). Complete resistance to ceftazidime and streptomycin does not lead to resistance to DBS1 or D-CONGA. Treatment with D-CONGA for 10 passages did not induce any change in susceptibility to DBS1 in l- or D-form; however, it did induce resistance to D-ARVA.
Antibiofilm Activity.
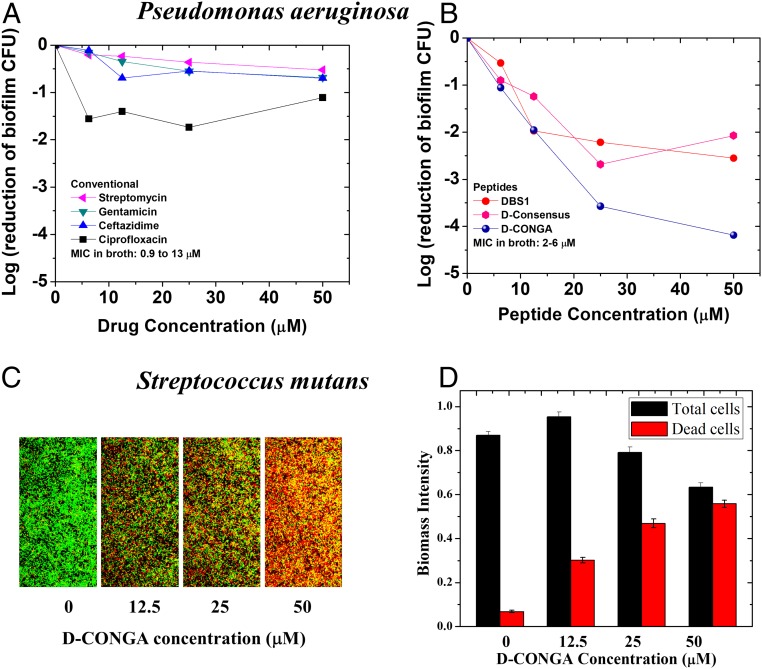
We also tested the activity of the lead peptides against bacterial biofilms, notoriously drug-resistant communal states of microbes (41–43). In a stringent assay for total viable P. aeruginosa biofilm colony-forming units (CFUs), small-molecule antibiotics had little effect, even at concentrations much higher than their MIC in broth dilution assays (Fig. 4A). On the other hand, the lead peptides D-DBS1 and D-CONGA reduced viable CFUs throughout the biofilm. Killing of 99% of biofilm cells was achieved with ∼12.5 μM of D-CONGA (Fig. 4B). We also tested D-CONGA against robust biofilms formed by the gram-positive bacterium Streptococcus mutans which forms oral biofilms that contribute to tooth decay in humans (44). D-CONGA treatment of stable S. mutans biofilms on glass slides resulted in rapid and almost complete killing of biofilm organisms (Fig. 4 C and D). Below, we show that D-CONGA is also efficacious at preventing P. aeruginosa and MRSA biofilm formation in an in vivo model of wound infection.
Fig. 4.
Activity against bacterial biofilms. (A and B) Bacterial biofilms of P. aeruginosa were grown as described elsewhere (59), washed extensively to remove loosely adhered bacteria, and treated with conventional antibiotics or antimicrobial peptides. After treatment, the biofilms were scraped and homogenized. Counting of viable colony forming units was done by serial dilution and plating on nutrient agar. (C) Robust biofilms formed by S. mutans were treated with increasing concentrations of D-CONGA and imaged by confocal microscopy in the presence of Syto9 (green) which stains all cells and propidium iodide (red) which stains only dead cells. (D) Quantitation of total biomass (total cell count) and dead cell counts was done as described elsewhere (58).
Treatment of Deep Tissue Wound Infection.
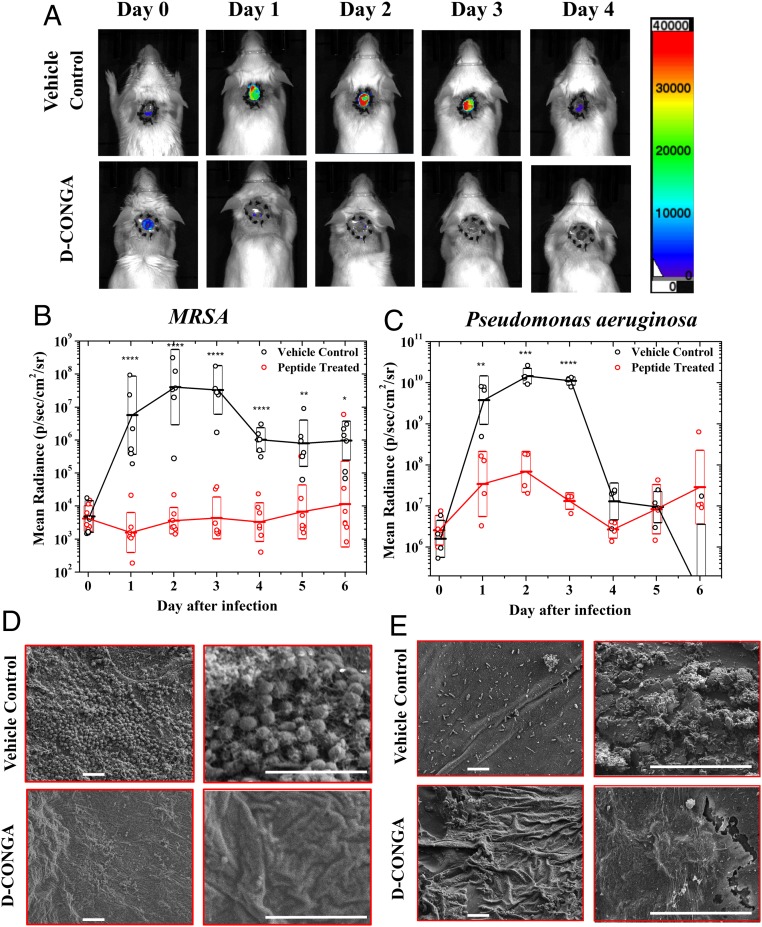
To begin assessing the efficacy of our lead candidate, D-CONGA, in a complex wound environment we used an established mouse model of deep surgical wound infection (34, 45). Dorsal, full thickness surgical punch wounds 5 mm in diameter and 3 mm deep that penetrated the epidermis, dermis, and panniculus carnosis were made in healthy adult CD1 mice. To roughly mimic human wound healing, skin contraction was prevented with a silicone ring sutured around the wound opening. A Tegaderm dressing was used to prevent wound drying. Wounds were infected either with 104 CFUs of luminescent MRSA (46) or with 104 CFUs of luminescent P. aeruginosa, strain PA01, which has inherent resistance to many antibiotics. Bacterial burdens in the wound bed were monitored with whole animal luminescence imaging (Fig. 5A). Within the first 2 h of infection, high bacterial burdens are established and wound beds become filled with purulence within 12–24 h. This wound environment presents a useful in vivo model for our host cell-compatible AMPs because it is rich with immune cells and concentrated serum proteins.
Fig. 5.
Animal model of deep surgery wound infection. (A) Circular, dorsal puncture wounds were surgically created in healthy, adult CD1 mice, stabilized with a sutured silicon ring, and covered with Tegaderm dressing to better mimic infection and wound healing in humans. Wounds were infected with luminescent MRSA or P. aeruginosa and were treated with D-CONGA peptide or vehicle control every 8 h until day 4. An IVIS whole animal imager was used to measure luminescence in all animals once per day for 13 d after infection. Examples of daily images of two mice infected with P. aeruginosa are shown. (B and C) Total integrated radiance from the wound bed was measured daily in MRSA (B) and P. aeruginosa (C) infected mice. Antilogs of average of log (radiance) values are shown with boxes indicating the upper and lower bounds of the 68% confidence interval. Raw values from individual animals are shown as open circles. Statistical significance of the pairwise differences are shown, determined with a t test on log values after applying the Bonferroni correction for multiple comparisons. *P < 0.05, **P < 1 × 10−2 , ***P < 1 × 10−3, ****P < 1 × 10−4(D and E) Examples of scanning electron microscopy images of glutaraldehyde-fixed Tegaderm dressing, which was removed from experimental animals on day 3. Two different magnifications are shown of different areas. (Scale bars, 5 μm in all images.)
Using this model, we tested D-CONGA against P. aeruginosa and MRSA infections, comparing it to vehicle only as a negative control or gentamycin treatment as a positive control. Peptide antibiotics tested in clinical trials for wound protection and treatment are often used in viscous ointment formulations containing 0.8 or 1% active compound (21). Here, we tested 0.38% D-CONGA in 0.025% acetic acid, applied every 8 h to avoid confounding effects of formulation. When wounds were infected with luminescent MRSA on day 0, and treated with vehicle only, they remained infected with high bacterial burdens for 3 subsequent days before spontaneous resolution, aided by the removal of the Tegaderm dressing on day 3 and the silicon ring on days 5 to 8. Treatment every 8 h for the first 5 d with 75 μg of D-CONGA in 20 μL of 0.025% acetic acid resulted in a 4-log decrease in luminescence to close to background levels, at the peak of infection, days 2 and 3 (Fig. 5B). When wounds were infected with P. aeruginosa, and treated with vehicle only, bacterial burdens similarly increased dramatically for 3 d before beginning to resolve on day 4. Treatment every 8 h with 75 μg of D-CONGA in 20 μL of 0.025% acetic acid resulted in 2 to 3 logs of reduction in luminescence at the peak of infection (Fig. 5C). In both cases, D-CONGA was efficacious at treating the infection in the wound bed and preventing significant bacterial colonization. Animal health-related scores as well as auxiliary wound scores indicate that peptide-treated animals remain healthy and have gross wound pathology that is similar to, or better than, the control wound healing response (SI Appendix, Fig. S8).
The Tegaderm wound dressings, which were removed and fixed with glutaraldehyde 3 d after infection, were examined by electron microscopy for bacteria and bacterial biofilms. In vehicle control samples, Tegaderm from infected animals show unmistakable evidence of individual MRSA-like cocci or Pseudomonas-like rods as well as some complex three-dimensional (3D) matrices containing bacteria, consistent with organized bacterial biofilms (Fig. 5 D and E). Tegaderm films from animals treated with D-CONGA or gentamycin showed essentially no bacteria-like objects and no evidence of organized biofilms. In images of randomly selected areas, large numbers of MRSA-like bacteria were observed in 4/10 vehicle-treated controls. In peptide-treated samples no bacteria were observed in any of 11 samples (P = 0.035 for proportion of positive samples, P < 1 × 10−5 for cell count). For P. aeruginosa-infected animals, Pseudomonas-like bacteria were observed in 12/15 vehicle control samples, but no bacteria were observed in any of the 15 peptide-treated samples (P < 1 × 10−5).
We also characterized the systemic effects of D-DBS4, DBS2, and D-CONGA in CD1 mice using 5 d of intraperitoneal (i.p.) injection of peptide in phosphate buffered saline (PBS), or in a few cases in PBS with liposomes (47). We monitored behavior and weight, and measured liver alanine aminotransferase (ALT)enzyme for evidence of liver toxicity at the end of the study. D-DBS4 in PBS was well tolerated in mice at 100, 200, and 400 μg per dose. Higher doses were not tested. No weight loss or liver toxicity was observed (SI Appendix, Fig. S9). Some temporary effects on animal behavior (slight hunching and lethargy) were observed immediately after administration of peptide. Similarly, DBS2 was well tolerated at 50 and 100 μg per dose, per day. Higher doses were not tested. D-CONGA in PBS was tolerated up to 25 μg per dose for 5 d, showing no behavioral effects, weight loss, or liver toxicity at these doses. At 50 or 100 μg dose per day, D-CONGA in PBS caused significant hunching and moribund behavior after one to three treatments. D-CONGA in PBS with liposomes, to slow systemic dispersal, was well tolerated with minor behavioral effects at doses of up to 200 μg per day, without weight loss or liver toxicity (SI Appendix, Fig. S9).
Discussion
The only membrane permeabilizing AMPs approved by the FDA for use in humans are two complex, cyclic lipopeptides, polymyxin B and E, which are active against gram-negative bacteria, and daptomycin, which is active against gram-positive bacteria. It is probably significant that these AMPs have specific interactions with bacterial lipid precursors. While such interactions improve selectivity in vivo, they also provide opportunities for resistance (25). At the level of basic research, more than a thousand examples of membrane-disrupting AMPs with activity in vitro have been described (48, 49). Despite the longstanding promise, decades of research have yielded no other approved AMP drugs, although a few clinical trials of AMPs have taken place, or are underway (18, 20, 21, 50–52). Notably, few AMP trials have been for systemic applications. Instead trials have been for indications such as chronic diabetic foot ulcers, oral candidiasis, or pneumonia.
The failure of a few AMP trials is not, by itself, a concern for the field because this is a common outcome of late-stage clinical trials for all classes of drugs. Instead, the most significant obstacle may be that so few AMPs have advanced beyond the laboratory. The work described here is inspired by the idea that there are no insurmountable impediments to AMP drugs, especially for applications such as wound protection and treatment against drug-resistant bacteria. Instead, there are impediments to populating the preclinical pipeline with peptides that have a reasonable chance of success.
Molecular optimization of new AMPs has previously been done by trial-and-error variation of known examples, sometimes guided by machine learning algorithms, followed by testing under standard laboratory conditions. But optimization under standard conditions does not include early down-selection for solubility, toxicity, host cell inhibition, and proteolytic degradation. Instead, these factors are tested at a later stage, when feedback is less effective, if they are tested at all. The limitations imposed by this classical approach are compounded by the fact that there are few, if any, reliable QSAR rules for engineering or optimizing the activity of AMPs, especially for host cell inhibition, toxicity, and degradation.
The inexplicable differences in activities of the closely related peptides in Fig. 2 emphasize the difficulty of rationally optimizing antimicrobial peptides. For example, the DRD peptides and the DBS peptides are very similar (Fig. 1 G and I) yet have very different activities in broth dilution (Fig. 2A). DBS1 and the consensus sequence are very similar, with only two conservative substitutions in the C core. Yet the consensus sequence is consistently a bit less active against all microbes, compared to DBS1. Further, removal of the invariant glycines from DBS1 decreased activity against most organisms. Yet, removal of the glycines from the consensus sequence created D-CONGA, the most effective peptide overall, which improved the activity toward every organism. The fact that these differences would not have been predicted by any known QSAR rules strengthens the case for using SME as an efficient and powerful means to perform large-scale optimization of AMPs.
Our goal in this work was to show that one can use synthetic molecular evolution and rational variation to identify host cell- and tissue-compatible AMPs. To demonstrate success, we tested the evolved peptides in vitro in the presence of concentrated host cells, and in vivo using a relevant animal model of wounds infected with drug-resistant bacteria. We successfully identified short linear peptides that do not have the impediments listed above. The peptides we discovered are host cell compatible in the sense that they are much more selective for microbes over host cells than most AMPs (23). They retain activity in the presence of concentrated RBCs and serum, do not cause hemolysis, and have little cytotoxicity. The best peptides, D-DBS4 and D-CONGA, have excellent activity against all ESKAPE pathogens under all conditions studied. Finally, evolved peptides were tested in an animal wound model (34), with peptide added to infected wounds that are initially filled with opaque and viscous purulence (pus) due to leukocytes and secreted proteins. The results show that the selection conditions identified peptides that are active in the very challenging environment of purulent wounds infected with drug-resistant bacteria and can treat infections and prevent biofilm formation under these conditions.
Although resistance to AMPs is known and can be selected (53), one of the largest potential advantages of AMPs is that, sometimes, resistance is slower or more difficult to induce compared to conventional antibiotics (54, 55). Importantly, resistance to conventional antibiotics is also associated with “widespread collateral susceptibility” to antimicrobial peptides (56), suggesting that simultaneous resistance to both may be unlikely. The evolved peptides we studied here do not induce resistance in P. aeruginosa under conditions that readily enable selection of resistance to conventional antibiotics and to other AMPs, such as the template sequence D-ARVA. This would appear to be a fortuitous activity as it was not intentionally part of the selection criteria. These peptides may make it possible to understand the mechanism of resistance avoidance in gram-negative bacteria so that it can be selected intentionally. Among the resistance mechanisms of P. aeruginosa, addition of cationic groups to the outer membrane LPS, via amino rabinosylation of lipid A, is the most likely explanation for resistance to cationic AMPs such as D-ARVA (25, 40). Like many AMPs, D-ARVA accumulates massively on LPS as part of its mechanism (23). The lack of inducible resistance against the D-DBS peptides and against D-CONGA suggests that they do not have to accumulate via electrostatic interactions on LPS and permeabilize the outer membrane, but perhaps can pass through the outer membrane, by translocation or by passing through porins, to access the inner membrane more directly. The fact that P. aeruginosa passaged against D-CONGA gains resistance to D-ARVA, but not to D-CONGA itself, further supports this mechanistic hypothesis.
Here we demonstrate that synthetic molecular evolution can be used to identify host cell-compatible antimicrobial peptides with excellent activities in vitro and in vivo. The peptides discovered here, their next-generation derivatives, or other peptides discovered with this approach, may have a greater probability of success in the clinic in protecting wounds against drug-resistant bacterial infections or treating wounds infected by drug-resistant bacteria, compared to other AMPs described in the primary literature. Most importantly we anticipate that the approach used here, screening iterative rational libraries under physiologically relevant conditions that allow for orthogonal selection and down-selection based on known impediments, could change the paradigm for AMP development and could increase the population of potentially viable AMP therapeutics that make it through the preclinical pipeline. This, in turn, could increase the number of AMPs that might proceed into clinical trials, which ultimately would increase the probability that one, or some, will succeed.
Methods
DRD Screen.
To a rectangular, one-well plate from Nunc, 20 mL of underlay agarose, inoculated with 8 × 106 CFUs of bacteria, was added. A sterile, 96-well plate replicator from Sigma-Aldrich was set in the molten agarose and removed once the agarose solidified. Peptide plus type O+ human RBCs at 2 × 108 cells/mL was added to an equal volume of 15 μM peptide. For each assay, 10 µL of the peptide/RBC mixture was added to a set of two radial diffusion plates, each harboring a different bacterial strain. The plates were incubated upside down for 3 h at 37 °C. A nutrient-rich overlay agar was added and plates were incubated upside down overnight at 37 °C and zones of inhibition were assessed using ImageJ.
Radial Diffusion-Broth Dilution Screen.
RBCs at 6 × 109 cells/mL were added and the plates were incubated with peptide for 1 h with shaking. Following incubation, the plates were centrifuged to measure hemolysis, then resuspended in a radial diffusion plate harboring E. coli. These plates were processed as described above. To the remaining peptide/RBC solution, 20 µL of P. aeruginosa at 5 × 105 CFU/mL was added. Following incubation, tryptic soy broth (TSB) was added and the plate was incubated at 37 °C overnight. Bacterial growth was detected by RBC coloration and sterilized wells were verified by CFU counts.
DBS.
Extracted peptide in PBS was added to type O+ human RBCs at 3.7 × 109 cells/mL, incubated for 30 min, and centrifuged at 1,000 × g to measure hemolysis. The remaining solution was split and added to 96-well plates with either E. coli, 2 × 104 CFU/mL, or P. aeruginosa, 2 × 106 CFU/mL in PBS. The plates were incubated at 37 °C for 3 h, and 2× TSB was added for overnight incubation. Inhibition of bacterial growth was detected by a lack of deoxygenation in the wells (as evidenced by RBC coloration) and sterilization was verified by CFU counting on TSB agar.
Postscreen Broth Dilution.
Peptides were serially diluted by a factor of 2:3 horizontally across a 96-well, conical-bottomed plate. Type O+ human RBCs at 0 or 2.5 × 109 cells/mL were added. Following a 30-min incubation, TSB, inoculated with 5 × 105 CFU/mL, was added to all wells for overnight incubation at 37 °C. A second inoculation was performed with 10 μL of solution from the original plate added to 100 μL of sterile TSB for overnight incubation. Optical density (OD600) was measured. Values of less than 0.1 were considered sterilized.
E. coli (ATCC 25922), S. aureus (ATCC 25923), P. aeruginosa (PA01), E. faecium (ATCC 19434), K. pneumoniae subsp. pneumoniae (ATCC 13883), A. baumannii (ATCC 19606), and S. enterica subsp. enterica (ATCC 14028) were used in this study.
Postscreen Hemolysis.
Peptide was serially diluted in PBS starting at a concentration of 100 μM. An equal volume of RBCs in PBS at 2 × 108 cells/mL was added. Controls were PBS only and 1% Triton. The mixtures were incubated at 37 °C for 1 h and centrifuged at 1,000 × g for 5 min. The absorbance of released hemoglobin at 410 nm was recorded.
SYTOX Green Cytotoxicity Assay.
WI-38 fibroblasts were grown overnight from a seeded density of 10,000 cells/well in a 96-well tissue-culture plate. In a separate plate, peptides were serially diluted in Dulbecco's Modified Eagle Medium (DMEM) containing 5 μM SYTOX Green. After addition of peptide to cells, SYTOX Green fluorescence was measured every 5 min, for 1 h. MelP5, a highly lytic peptide, was used as a positive.
Selection for Stable Resistance.
Antibiotics were prepared at five times the final concentration and serially diluted by a factor of 2:3 horizontally across a 96-well plate. TSB, inoculated with 5 × 105 CFU/mL P. aeruginosa (PA01) was added to all wells, followed by overnight incubation at 37 °C. The well that contained the highest concentration of antibiotic that allowed for bacterial growth was chosen for overnight passaging without antibiotic. Repeat treatments by the same antibiotics was carried out the next day. Glycerol stocks of all generated resistance strains of PA01 were snap frozen in liquid nitrogen and stored for later testing. These selected strains from the tenth passage were then cross-treated against antibiotics other than the one they were selected against.
Biofilm Assay Using CFU Reduction Method.
An overnight culture of P. aeruginosa (PA01) was diluted 10 times in Luria broth (LB) and plated 125 μL per well onto a 96-well plate. The plate was incubated for 48 h at room temperature to drive biofilm formation. On the third day, wells were washed with distilled water twice and then treated with 1:2 dilution of the antibiotics. Wells with PBS were used as controls. After a 2-h incubation, wells were washed twice with PBS and scraped and homogenized using a pipette tip with aspiration. The cell suspension was serially diluted 1:100 five times and plated on LB agar for overnight incubation at 37 °C, followed by colony counting.
Biofilm Assay Using Confocal Microscopy.
S. mutans UA159 was grown in a semidefined biofilm medium (57) with glucose (18 mM) and sucrose (2 mM) added as supplemental carbohydrate sources. Aerobic S. mutans biofilms were cultured on an eight-well chambered coverglass at 37 °C and 5% CO2 for 3 d with medium changed daily. After washing, biofilms were then treated with 12.5, 25, 50 µM AMP, or PBS, respectively, for 4 h. Biofilms after the treatments were stained with LIVE/DEAD BacLight fluorescent dye and imagined using an inverted 40× oil objective. Confocal microscopy was performed with a Zeiss LSM 700 microscope. Confocal z-stacks and simulated xyz 3D images were acquired and generated using Zeiss 10.0 software. Images were further analyzed using Comstat2 software (58).
Murine Wound Infection Model.
All animal studies strictly adhered to protocol 131, which was approved by Tulane School of Medicine’s Institutional Animal Care and Use Committee. As described (34), female CD1 mice at 8 to 12 wk of age were anesthetized via i.p. ketamine and xylazine. A full thickness biopsy wound was generated using a 5-mm biopsy punch (Integra) on depilated and chlorhexidine-scrubbed dorsal surfaces. A silicon (Invitrogen) ring 0.5 mm thick with an outer diameter of 10 mm and a hole with a 5-mm diameter was placed over the wound and held to the skin with a surgical adhesive. The silicon ring was covered with Tegaderm (3M), and further adhered using 4–0 braided silk interrupted sutures (Ethicon). Mice were given 0.05 mg/kg buprenorphine immediately following surgery as well as daily for the next 2 d to alleviate pain from the procedure. Wound beds were infected by penetrating the Tegaderm with an insulin syringe and injecting 1 × 104 CFUs of bioluminescent MRSA (MW2) or P. aeruginosa (PA01) suspended in 10 µL sterile PBS directly onto the wound bed. The MRSA strain used was generated and described by Plaut et al. (46) and consists of MW2 strain MRSA with the plasmid pRP1195 to confer luminescence. Four hours after infection, mice were topically treated with 75 μg D-CONGA (2 mM), 1.2 mM gentamicin (Gibco), or 0.025% acetic acid (vehicle control) in a 20 µL volume injecting directly into the wound bed. Treatment was every 8 h for the first 5 d of infection. Mice were imaged daily for 2 wk using the in vivo imaging system (IVIS)-XMRS (PerkinElmer), and bioluminescence generated from the bacteria was quantified in values of radiance (photons/sec/centimeter2/steradian).
Scanning Electron Microscopy.
Tegaderm dressings removed from wounds were washed with PBS and fixed in 2.5% glutaraldehyde overnight at 4 °C. Tegaderm was attached to hydroxyapatite discs placed horizontally in 24-well microtiter plates. The fixed samples were dehydrated using increasing concentrations of ethanol and then desiccated with CO2 critical point drying. The samples were coated with carbon and subjected to scanning electron microscopy with a Hitachi S-4800 high-resolution microscope.
Systemic Toxicity.
Healthy adult CD1 mice, 25 to 32 g were treated once per day with i.p. administration of 500 μL of peptide solution or vehicle. Vehicle was PBS in most experiments. For a few samples, we used PBS containing 5 mM of 0.1 μm unilamellar lipid vesicles composed of 1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphoglycerol (POPG) to slow dispersal of peptide as demonstrated previously (56). Mice were monitored for 1 h after treatment with subsequent checks performed three times daily. Weight was measured each day prior to treatment. At the study endpoint, after 5 d of treatment, mice were killed by CO2 asphyxiation and livers were collected. Liver ALT enzyme was measured using a fluorometric kit from Abcam. Liver samples were homogenized in buffer and the supernatant was used to measure liver ALT enzyme by fluorescence using a coupled enzyme reaction. Fluorescence from pyruvate standards of known concentrations were used to convert fluorescence to milliunits (mU) of ALT activity.
Data Availability.
All data described herein are available and can be requested from the corresponding author.
Supplementary Material
Acknowledgments
This work was funded by NIH 1R21AI119104. We thank Michael Schurr for luminescent P. aeruginosa and Roger Plaut for luminescent MRSA. We thank Alan Grossfield for naming D-CONGA.
Footnotes
Competing interest statement: Authors C.G.S., J.G., S.G., and W.C.W. are inventors on a US Patent application filed by Tulane University.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918427117/-/DCSupplemental.
References
- 1.Arias C. A., Murray B. E., Antibiotic-resistant bugs in the 21st century–A clinical super-challenge. N. Engl. J. Med. 360, 439–443 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Uhlemann A. C., Otto M., Lowy F. D., Deleo F. R., Evolution of community- and healthcare-associated methicillin-resistant Staphylococcus aureus. Infect. Genet. Evol. 21, 563–574 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Otto M., MRSA virulence and spread. Cell. Microbiol. 14, 1513–1521 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otto M., Basis of virulence in community-associated methicillin-resistant Staphylococcus aureus. Annu. Rev. Microbiol. 64, 143–162 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Hospenthal D. R., et al. ; Prevention of Combat-Related Infections Guidelines Panel , Infection prevention and control in deployed military medical treatment facilities. J. Trauma 71 (suppl. 2), S290–S298 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Calhoun J. H., Murray C. K., Manring M. M., Multidrug-resistant organisms in military wounds from Iraq and Afghanistan. Clin. Orthop. Relat. Res. 466, 1356–1362 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott P., et al. , An outbreak of multidrug-resistant Acinetobacter baumannii-calcoaceticus complex infection in the US military health care system associated with military operations in Iraq. Clin. Infect. Dis. 44, 1577–1584 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Thorpe K. E., Joski P., Johnston K. J., Antibiotic-resistant infection treatment costs have doubled since 2002, now exceeding $2 billion annually. Health Aff. (Millwood) 37, 662–669 (2018). [DOI] [PubMed] [Google Scholar]
- 9.O’Neill J., Tackling Drug resisrtant infections globally: Final report and recommendations (2017). https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf. Accessed 2 February 2020.
- 10.Goto M., et al. , Antimicrobial nonsusceptibility of gram-negative bloodstream isolates, veterans health administration system, United States, 2003-20131. Emerg. Infect. Dis. 23, 1815–1825 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petersen K., et al. , Trauma-related infections in battlefield casualties from Iraq. Ann. Surg. 245, 803–811 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saltoglu N., et al. ; Turkish Society of Clinical Microbiology and Infectious Diseases ; Diabetic Foot Infections Study Group , Influence of multidrug resistant organisms on the outcome of diabetic foot infection. Int. J. Infect. Dis. 70, 10–14 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Sánchez-Sánchez M., et al. , Bacterial prevalence and antibiotic resistance in clinical isolates of diabetic foot ulcers in the Northeast of Tamaulipas, Mexico. Int. J. Low. Extrem. Wounds 16, 129–134 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Snitkin E. S., et al. ; NISC Comparative Sequencing Program Group , Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci. Transl. Med. 4, 148ra116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D’Agata E. M., Rapidly rising prevalence of nosocomial multidrug-resistant, gram-negative bacilli: A 9-year surveillance study. Infect. Control Hosp. Epidemiol. 25, 842–846 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Luepke K. H., et al. , Past, present, and future of antibacterial economics: Increasing bacterial resistance, limited antibiotic pipeline, and societal implications. Pharmacotherapy 37, 71–84 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Zasloff M., Antimicrobial peptides of multicellular organisms. Nature 415, 389–395 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Jenssen H., Hamill P., Hancock R. E., Peptide antimicrobial agents. Clin. Microbiol. Rev. 19, 491–511 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Easton D. M., Nijnik A., Mayer M. L., Hancock R. E., Potential of immunomodulatory host defense peptides as novel anti-infectives. Trends Biotechnol. 27, 582–590 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costa F., Teixeira C., Gomes P., Martins M. C. L., Clinical application of AMPs. Adv. Exp. Med. Biol. 1117, 281–298 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Lipsky B. A., Holroyd K. J., Zasloff M., Topical versus systemic antimicrobial therapy for treating mildly infected diabetic foot ulcers: A randomized, controlled, double-blinded, multicenter trial of pexiganan cream. Clin. Infect. Dis. 47, 1537–1545 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Starr C. G., Wimley W. C., Antimicrobial peptides are degraded by the cytosolic proteases of human erythrocytes. Biochim. Biophys. Acta Biomembr. 1859, 2319–2326 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Starr C. G., He J., Wimley W. C., Host cell interactions are a significant barrier to the clinical utility of peptide antibiotics. ACS Chem. Biol. 11, 3391–3399 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wimley W. C., Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem. Biol. 5, 905–917 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H., Srinivas S., Xu Y., Wei W., Feng Y., Genetic and biochemical mechanisms for bacterial lipid A modifiers associated with polymyxin resistance. Trends Biochem. Sci. 44, 973–988 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Mishra B., Lakshmaiah Narayana J., Lushnikova T., Wang X., Wang G., Low cationicity is important for systemic in vivo efficacy of database-derived peptides against drug-resistant Gram-positive pathogens. Proc. Natl. Acad. Sci. U.S.A. 116, 13517–13522 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haney E. F., et al. , Computer-aided discovery of peptides that specifically attack bacterial biofilms. Sci. Rep. 8, 1871 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Porto W. F., et al. , In silico optimization of a guava antimicrobial peptide enables combinatorial exploration for peptide design. Nat. Commun. 9, 1490 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kauffman W. B., Guha S., Wimley W. C., Synthetic molecular evolution of hybrid cell penetrating peptides. Nat. Commun. 9, 2568 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li S., et al. , Potent macromolecule-sized poration of lipid bilayers by the macrolittins, A synthetically evolved family of pore-forming peptides. J. Am. Chem. Soc. 140, 6441–6447 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Krauson A. J., He J., Wimley A. W., Hoffmann A. R., Wimley W. C., Synthetic molecular evolution of pore-forming peptides by iterative combinatorial library screening. ACS Chem. Biol. 8, 823–831 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiedman G., Kim S. Y., Zapata-Mercado E., Wimley W. C., Hristova K., pH-triggered, macromolecule-sized poration of lipid bilayers by synthetically evolved peptides. J. Am. Chem. Soc. 139, 937–945 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiedman G., et al. , Highly efficient macromolecule-sized poration of lipid bilayers by a synthetically evolved peptide. J. Am. Chem. Soc. 136, 4724–4731 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffmann J. P., et al. , In situ treatment with novel microbicide inhibits methicillin resistant Staphylococcus aureus in a murine wound infection model. Front. Microbiol. 10, 3106 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rathinakumar R., Wimley W. C., High-throughput discovery of broad-spectrum peptide antibiotics. FASEB J. 24, 3232–3238 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rathinakumar R., Walkenhorst W. F., Wimley W. C., Broad-spectrum antimicrobial peptides by rational combinatorial design and high-throughput screening: The importance of interfacial activity. J. Am. Chem. Soc. 131, 7609–7617 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rathinakumar R., Wimley W. C., Biomolecular engineering by combinatorial design and high-throughput screening: Small, soluble peptides that permeabilize membranes. J. Am. Chem. Soc. 130, 9849–9858 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiegand I., Hilpert K., Hancock R. E., Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3, 163–175 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Guha S., Ghimire J., Wu E., Wimley W. C., Mechanistic landscape of membrane-permeabilizing peptides. Chem. Rev. 119, 6040–6085 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joo H. S., Fu C. I., Otto M., Bacterial strategies of resistance to antimicrobial peptides. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371, 20150292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolfmeier H., Pletzer D., Mansour S. C., Hancock R. E. W., New perspectives in biofilm eradication. ACS Infect. Dis. 4, 93–106 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Davies D., Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2, 114–122 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Stewart P. S., Costerton J. W., Antibiotic resistance of bacteria in biofilms. Lancet 358, 135–138 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Pitts N. B., et al. , Dental caries. Nat. Rev. Dis. Primers 3, 17030 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Dunn L., et al. , Murine model of wound healing. J. Vis. Exp., e50265 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Plaut R. D., Mocca C. P., Prabhakara R., Merkel T. J., Stibitz S., Stably luminescent Staphylococcus aureus clinical strains for use in bioluminescent imaging. PLoS One 8, e59232 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahmad I., Perkins W. R., Lupan D. M., Selsted M. E., Janoff A. S., Liposomal entrapment of the neutrophil-derived peptide indolicidin endows it with in vivo antifungal activity. Biochim. Biophys. Acta 1237, 109–114 (1995). [DOI] [PubMed] [Google Scholar]
- 48.Fjell C. D., Hancock R. E., Cherkasov A., AMPer: A database and an automated discovery tool for antimicrobial peptides. Bioinformatics 23, 1148–1155 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Wang G., Li X., Wang Z., APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 44, D1087–D1093 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naafs M., The antimicrobial peptides: Ready for clinical trials? Biomed. J. Sci. Tech. Res. 7, 001536 (2018). [Google Scholar]
- 51.Mahlapuu M., Håkansson J., Ringstad L., Björn C., Antimicrobial peptides: An emerging category of therapeutic agents. Front. Cell. Infect. Microbiol. 6, 194 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trotti A., et al. , A multinational, randomized phase III trial of iseganan HCl oral solution for reducing the severity of oral mucositis in patients receiving radiotherapy for head-and-neck malignancy. Int. J. Radiat. Oncol. Biol. Phys. 58, 674–681 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Perron G. G., Zasloff M., Bell G., Experimental evolution of resistance to an antimicrobial peptide. Proc. Biol. Sci. 273, 251–256 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pollard J. E., et al. , In vitro evaluation of the potential for resistance development to ceragenin CSA-13. J. Antimicrob. Chemother. 67, 2665–2672 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Breij A., et al. , The antimicrobial peptide SAAP-148 combats drug-resistant bacteria and biofilms. Sci. Transl. Med. 10, eaan4044 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Lázár V., et al. , Antibiotic-resistant bacteria show widespread collateral sensitivity to antimicrobial peptides. Nat. Microbiol. 3, 718–731 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lobos O., Padilla A., Padilla C., In vitro antimicrobial effect of bacteriocin PsVP-10 in combination with chlorhexidine and triclosan against Streptococcus mutans and Streptococcus sobrinus strains. Arch. Oral Biol. 54, 230–234 (2009). [DOI] [PubMed] [Google Scholar]
- 58.Vorregaard M., Comstat2 - A Modern 3D Image Analysis Environment for Biofilms, Masters Thesis, Informatics and Mathematical Modelling (Technical University of Denmark, 2008). [Google Scholar]
- 59.O’Toole G. A., Microtiter dish biofilm formation assay. J. Vis. Exp., 2437 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data described herein are available and can be requested from the corresponding author.