Significance
Because genetic diversity in the Mycobacterium tuberculosis complex (MTBC) is less pronounced than in other pathogens, the variable outcome of infection has been attributed mainly to host and environmental factors. Here, we reveal widely different metabolic phenotypes among MTBC members, suggesting that strain diversity may play an important role during infection. To unravel the genetic basis for metabolic diversity, we developed a approach that integrates metabolomic and genomic data for 18 MTBC clinical strains. Our approach allowed us to investigate the metabolic effect of rare genetic variants and to predict mutations that associate with strain-specific metabolic vulnerabilities and inherent baseline susceptibility to antibiotics. Our model-based approach is broadly applicable to many organisms, opening possibilities for identifying more selective treatment strategies.
Keywords: metabolomics, tuberculosis, constraint-based model
Abstract
Human tuberculosis is caused by members of the Mycobacterium tuberculosis complex (MTBC) that vary in virulence and transmissibility. While genome-wide association studies have uncovered several mutations conferring drug resistance, much less is known about the factors underlying other bacterial phenotypes. Variation in the outcome of tuberculosis infection and diseases has been attributed primarily to patient and environmental factors, but recent evidence indicates an additional role for the genetic diversity among MTBC clinical strains. Here, we used metabolomics to unravel the effect of genetic variation on the strain-specific metabolic adaptive capacity and vulnerability. To define the functionality of single-nucleotide polymorphisms (SNPs) systematically, we developed a constraint-based approach that integrates metabolomic and genomic data. Our model-based predictions correctly classify SNP effects in pyruvate kinase and suggest a genetic basis for strain-specific inherent baseline susceptibility to the antibiotic para-aminosalicylic acid. Our method is broadly applicable across microbial life, opening possibilities for the development of more selective treatment strategies.
The Mycobacterium tuberculosis complex (MTBC) consists of closely related bacteria that are etiological agents of tuberculosis (TB) in humans and animals. Whole-genome sequencing distinguishes seven phylogenetic lineages within the MTBC that are adapted to humans and feature limited genetic diversity (1–4). Given the low mutation rate, strictly clonal reproduction, and limited genetic diversity of the MTBC (5, 6), the variable outcomes of TB infection and disease have been attributed mainly to host and environmental factors rather than to bacterial variation (7). However, a growing body of evidence links genetic diversity among clinical MTBC strains to distinct geographic distributions (8), variation in the natural course of infection (9–11), and key selective advantages such as evolvability and transmissibility (12–15). Strain diversity also appears to influence the development of antibiotic resistance (16) and the inherent baseline susceptibility to drugs (i.e., minimal inhibitory concentrations) of clinical isolates generally considered drug susceptible (17, 18). However, in most of the examples above, there is currently limited knowledge of the biological mechanisms that underpin these differences.
A key element of MTBC’s pathogenic success is its ability to evade the host immune response (19) and to survive long periods of hypoxia, nutrient limitation, and oxidative or nitro-oxidative stresses (20–22), which requires plasticity for different metabolic responses. It is an open question how much of MTBC’s genetic diversity confers strain-specific metabolic characteristics, ultimately affecting transmissibility, disease progression, and baseline susceptibility, as opposed to resistance, to antibiotics. Indeed, evolution of metabolism has been demonstrated to play a key role in successful adaptation to complex and dynamic in vivo environments (23–25), in bacterial drug tolerance (26–28), and in survival during infection of macrophages (29–32). Beyond analyses of individual laboratory strains, however, no systematic characterization and comparative analysis of intrinsic metabolic differences across human-adapted MTBC clinical strains has been performed.
If the metabolic and other phenotypic diversity between MTBC strains contributes to and modulates pathogenicity, an obvious question is: Which elements of the limited genetic diversity in the MTBC are responsible for phenotypic strain diversification? In many species, associations between observable traits and single-nucleotide polymorphisms (SNPs) have been detected through genome-wide association studies (GWASs) (33, 34). GWASs typically estimate correlations between individual SNPs and phenotypes, in MTBC antibiotic resistance, but large sample sizes are necessary to achieve adequate statistical power and to identify genes responsible for complex phenotypes. This is particularly challenging for the MTBC, given the sporadic nature of genetic differences and the practical difficulties in systematically characterizing strain-specific phenotypes beyond drug resistance. Alternative computational solutions to establish causal links between genotype and phenotype, such as homology-based methods like SIFT (Sorting Intolerant from Tolerant) (35) or VIPUR (Variant Interpretation and Prediction Using Rosetta) (36), often exhibit low specificity (i.e., high risk of false positives) (37) and do not consider correlations between genetic variants. Moreover, we do not know of any validation of these methods in microbes. As a result, only a small proportion of SNPs in the MTBC have been mechanistically linked to phenotypes such as virulence or acquired drug resistance (38, 39).
To overcome the limitations of GWASs and homology-based predictions, we developed a computational approach that integrates strain-specific exometabolomes, genomes, and genome-scale metabolic networks into a single model. Application to 18 representative human-adapted MTBC clinical strains allowed us to predict the effects of SNPs in enzyme-encoding genes on metabolic phenotypes and to identify functional SNPs that associate with strain-specific metabolic vulnerabilities and an intrinsically reduced susceptibility to antibiotics.
Results
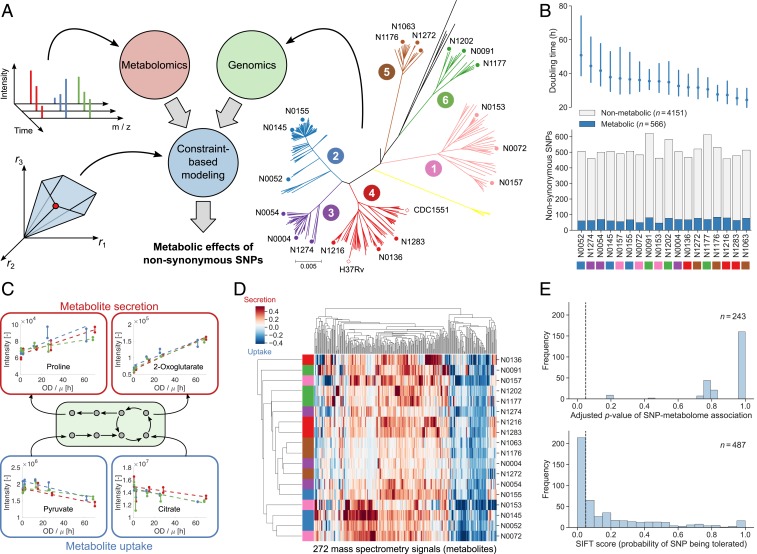
To investigate metabolic diversity in the human-adapted MTBC, we selected three strains each from six of the seven known MTBC lineages (8), covering much of the intralineage diversity (40) (Fig. 1A). Lineage 7 has only recently been characterized (41) and was not included here because strains were not readily available. All 18 MTBC strains were grown in batch cultures with a modified 7H9 medium containing pyruvate as the main carbon source and without common additives such as Tween 80, oleic acid, and glycerol. This medium supports growth of all strains, including Mycobacterium africanum (MTBC lineages 5 and 6), while simplifying the composition to facilitate downstream analysis. We collected supernatant samples from exponentially growing cultures over a period of 5 d and monitored cell growth by optical density. The supernatants were analyzed with a nontargeted metabolomics method using a time-of-flight mass spectrometer (42).
Fig. 1.
Connecting genetic and phenotypic diversity in the MTBC. (A) We integrated the genomes and dynamic exometabolomes of 18 clinical MTBC strains (3 strains each from six lineages, as indicated in the phylogenetic tree) into a single constraint-based model that predicts the effects of nonsynonymous SNPs on metabolic fluxes. (B) Mean doubling times with 95% confidence intervals estimated from three pooled replicate experiments for each strain (Top) and number of metabolic and nonmetabolic nonsynonymous SNPs (Bottom) for all strains (colored by lineage). (C) Relative metabolite uptake and secretion rates were inferred by fitting linear models (dashed lines) to dynamic exometabolomes. To compare metabolite exchange rates between strains with different growth rates, µ (h−1), metabolite intensities were plotted against OD divided by the respective growth rate. Here, uptake of pyruvate and citrate and secretion of proline and 2-oxoglutarate are shown for strain N1202. Biological replicates (three per strain) are indicated by colors, and solid lines connect technical replicates (two per time point). (D) Relative uptake (blue) and secretion (red) rates for all strains (rows, colored by lineage as in A) and metabolites in the exometabolomes (columns). Strains and metabolites are clustered by Euclidean distance. Each cell shows the mean relative rate across biological replicates for one SNP-metabolite pair. (E) Corrected P values of associations between SNPs and relative uptake and secretion rates inferred from exometabolomes for all SNPs affecting at least two strains (Top) and SIFT scores for all SNPs for which SIFT predictions were available (Bottom). In both cases, SNPs to the left of the dashed vertical line would be classified as functional with a significance level of 0.05.
There were 4,717 nonsynonymous SNPs in our 18 MTBC strains (40), 566 of which could be mapped to enzyme-encoding genes in a genome-scale metabolic model (43) (Datasets S1 and S2). The range of doubling times, from 25 to 50 h, of our MTBC strains (Fig. 1B and Dataset S3) suggests different metabolic strategies for nutrient uptake and utilization. To link growth and metabolic phenotypes, we used nontargeted mass spectrometry to profile dynamic exometabolome changes of 272 putative metabolites and derived relative nutrient uptake and by-product secretion rates (Fig. 1 C and D and Dataset S4). These rates can be directly compared between strains for each metabolite but not between different metabolites. About 50% of the metabolites were differentially secreted or consumed in at least one of the 18 strains (maximum pairwise log2 fold change larger than 2 between any two strains). Overall, the relative uptake and secretion rates of metabolites varied across strains by up to two orders of magnitude (SI Appendix, Fig. S1A). The most significant (ANOVA, q ≤ 0.01) exometabolome differences across MTBC strains pertained to amino acids and intermediates of glycolysis and the tricarboxylic acid (TCA) cycle, suggesting divergent evolutionary strategies in central carbon and nitrogen metabolism (SI Appendix, Fig. S1B and Datasets S5 and S6). By correlating growth rates and individual metabolite uptake/secretion rates across strains, we found only 14 metabolites that exhibited an absolute Pearson correlation larger than or equal to 0.5. Hence, metabolite exchange rates can explain at most 25% of the variance of growth rates across MTBC strains (SI Appendix, Fig. S1C). This result suggests that the metabolic diversity in MTBC cannot be explained by indirect, growth-related effects but rather is caused by genetic differences. Consistent with this hypothesis, we found pairwise genetic and metabolic distances between strains to significantly correlate (Pearson’s r = 0.34, P = 2.0 × 10−10), showing that strains from the same lineage had more similar exometabolome profiles than strains from different lineages (Fig. 1D and SI Appendix, Fig. S2).
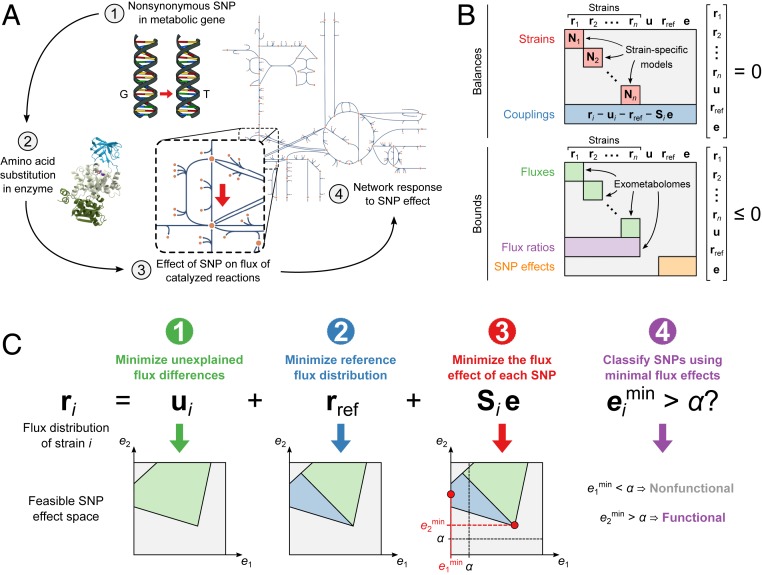
Because of the small sample size, we were not able to find any significant statistical associations between SNPs and differences in exometabolomes, and predictions from SIFT reduced the set of potentially functional SNPs only by half (Fig. 1E). Therefore, to identify genetic variations responsible for the observed metabolic differences among MTBC strains, we built on flux balance analysis (FBA) (44), a constraint-based approach that predicts metabolic fluxes at steady state in a stoichiometric reaction model by optimizing a cellular objective function. Here, we developed an FBA-based differential analysis to predict the effects of strain-specific SNPs via perturbation of the associated enzymes and propagation of this perturbation through the metabolic network (Fig. 2A). Specifically, we considered only nonsynonymous (missense) SNPs in enzyme-encoding genes that potentially reduce enzymatic activity. We represented all 18 MTBC strains in a single constraint-based model (Fig. 2B) that contained genome-scale metabolic networks specific for each strain. These networks incorporated experimentally determined bounds on growth and carbon source uptake rates (Dataset S7) and strain-specific genotypes. We derived the latter by identifying deletions and SNPs that represented a deviation from a phylogenetically derived reconstruction of the ancestral state of the MTBC genome (45). The model coupled strains to each other through mass balances, SNP effects (we assumed that a SNP had the same effect in all affected strains), and measured ratios of exchange fluxes. Only metabolites with relative uptake or secretion rates significantly different from zero were allowed to be exchanged with the medium (Dataset S4). In total, we added exometabolome-derived constraints on uptake and secretion rates to the model for 140 of the 272 metabolites in the exometabolome (Dataset S8). For metabolites with measured uptake or secretion but no exchange reaction in the model we added new exchange reactions. A strain’s steady-state flux distribution, ri (Fig. 2C), is given by the reference strain flux distribution, rref, and deviations from this reference that are either caused by SNP effects, e, and propagated through the network by a strain-specific structural sensitivity matrix (46), Si, or due to effects that cannot be explained by metabolic SNPs, ui. These unexplained metabolic effects allow feasible flux solutions even though the model with experimentally derived constraints was overconstrained. To infer the unknowns ri, rref, e, and ui, we sequentially minimized the following: 1) the L1 norm (sum of absolute values) of unexplained flux differences between strains (ui), 2) the L1 norm of fluxes in the reference strain (rref), and 3) each SNP effect (e). This approach aimed at 1) finding flux solutions that best described the experimental data, 2) identifying biologically justifiable flux distributions without infeasible loops (47), and 3) explaining as much of the observed metabolic variation across the strains as possible by as few SNPs as possible. A common problem of constraint-based models is that predicted flux distributions or, in our case, combinations of SNP effects, are not unique (48). We overcome this issue by finding the minimum value for each SNP effect across all possible optimal solutions. We classified a SNP as functional if its smallest possible effect in all optimal solutions was above a flux threshold, α, and nonfunctional otherwise (Fig. 2C). We chose the threshold value α = 10−4 mmol⋅gDW−1⋅h−1 by systematically testing all possible values and selecting the one at which the number of SNPs classified as functional started decreasing more slowly with increasing (stricter) α (SI Appendix, Fig. S3).
Fig. 2.
Predicting the metabolic effects of nonsynonymous SNPs. (A) Our constraint-based model integrates the metabolic effects of all nonsynonymous SNPs in enzyme-encoding genes. Each SNP effect is represented in the following way: 1) a nonsynonymous SNP causes 2) an amino acid substitution in an enzyme, possibly leading to reduced enzyme activity and 3) reduced flux through catalyzed reactions, and 4) the metabolic network responds by adjusting fluxes to a new steady state. The last two steps, as defined by structural sensitivity analysis, are represented in the model. (B) The model consists of balances and bounds, which are equality and inequality constraints, respectively. The constrained variables are the strain-specific flux distributions (r1, r2, …, rn), fluxes in a reference strain shared by all strains (rref), unexplained flux differences between strains (u), and flux differences between strains explained by SNP effects (e). The balance constraints are the steady-state mass balances of strain-specific models (red) and couplings that ensure that differences between strain-specific fluxes and reference fluxes are accounted for by unexplained effects and SNP effects (blue). Bounds are applied to all fluxes in the strain-specific models, including bounds on exchange fluxes obtained from exometabolomes (green), the ratio of exchange fluxes between pairs of strains, also from exometabolomes (purple), and the size of SNP effects relative to their affected reference fluxes (orange). (C) SNP effects are predicted by sequentially minimizing 1) the unexplained flux differences between strains 2), the reference fluxes, and 3) the flux effect of each SNP. The effect of each minimization step on a hypothetical feasible space defined by two SNP effects (e1 and e2) is shown below. Finally, 4) SNPs with a minimal effect larger than a chosen threshold α are classified as functional.
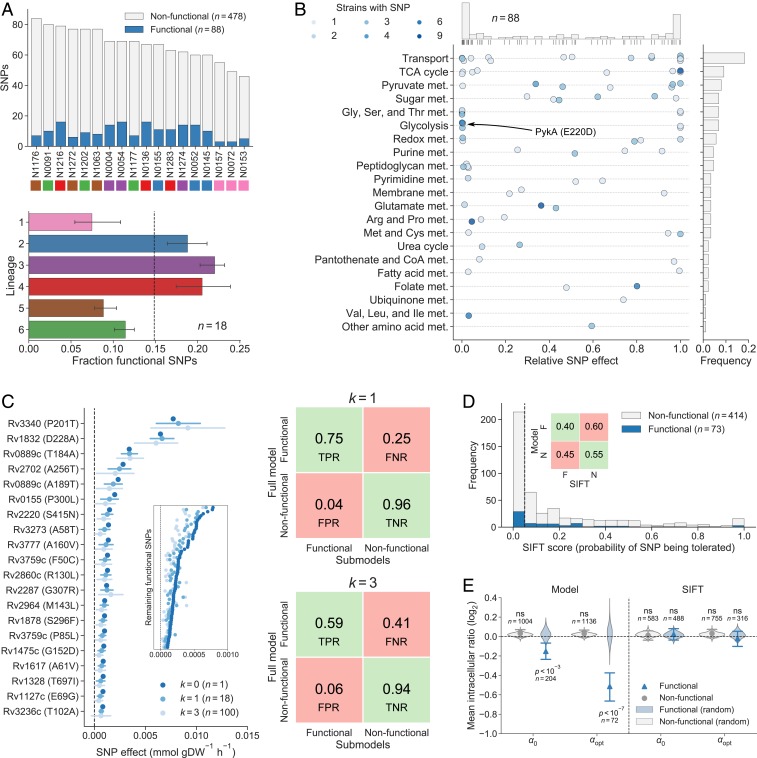
Of the 566 nonsynonymous SNPs in enzyme-encoding genes represented in the model, 88 (16%) were classified as functional. Overall, strains from lineages 2, 3, and 4 had a slightly larger fraction of functional SNPs than strains from lineages 1, 5, and 6 (Fig. 3A). The functional SNPs affected 67 unique enzymes distributed across most metabolic pathways, and SNPs were most often predicted to either abolish the flux of a reaction (i.e., complete loss-of-function) or have a relatively minor effect under the tested condition (Fig. 3B and Dataset S2). Metabolic pathways with many functional SNPs and large relative SNP effects included the TCA cycle, pyruvate metabolism, and glycolysis as well as glycine, serine, and threonine metabolism, consistent with our previous observation of significant differences in secretion rates of central metabolism intermediates and amino acids. Results were robust to small changes in the threshold value and to random sampling of 15 of the 18 TB strains (Fig. 3C and SI Appendix, Figs. S4 and S5, and Dataset S9). Notably, of the 216 functional SNPs predicted by SIFT, 29 SNPs were also predicted to be functional by our constraint-based approach, illustrating the difference in predicted functional SNPs between the two methods (P > 0.5, hypergeometric test) (Fig. 3D and Dataset S2).
Fig. 3.
Predicted functional SNPs in the MTBC and their metabolic effects. (A) Number of nonsynonymous SNPs classified as functional and nonfunctional in our 18 MTBC strains (above) and the fraction of SNPs classified as functional by lineage (below). Lineages are indicated by color. Error bars indicate the range of the three data points for each lineage. (B) Metabolic pathway distribution of functional SNPs and their relative effects on affected enzymes. Each dot represents a functional SNP and indicates its minimal effect divided by the mean predicted reference flux of reactions catalyzed by affected enzymes (darker color means that more strains have the SNP). The distribution of SNP effects is shown on top. The bars on the right indicate the frequency of functional SNPs in each pathway. (C) Sensitivity analysis of model-based functional SNP predictions. On the left, the minimal effect of the 20 functional SNPs with the largest minimal effect in the full model are shown for the full model as well as in sets of submodels in which k = 1 or k = 3 strains were removed. The mean minimal effect is indicated by a dot, and error bars indicate one SD of the observations. (Inset) The mean minimal effects of the remaining 68 functional SNPs without error bars. The true positive rate (TPR), false positive rate (FPR), true negative rate (TNR), and false negative rate (FNR) of functional SNP classification using submodels are shown on the right for k = 1 and k = 3. (D) SIFT scores of functional and nonfunctional SNPs classified by our model. SIFT classifies SNPs to the left of the vertical dashed line (significance level 0.05) as functional. The TPR, FPR, TNR, and FNR of the SIFT classification are shown in the Inset. (E) Relative intracellular levels of metabolites participating in reactions catalyzed by enzymes affected by SNPs (ratios of affected metabolite levels between strains with and without SNPs). Mean values are shown with 95% confidence intervals from bootstrapping (1,000 samples) and distributions of mean levels obtained from assigning SNPs randomly to strains while preserving the number of strains affected by each SNP. Results are shown for our model and SIFT and for the threshold used for SNP classification (α0) as well as a stricter threshold that maximizes the difference between functional and nonfunctional SNPs (αopt). The mean relative levels of functional SNPs predicted by our model were significantly different from random distributions for α0 (Z-test, P = 2.0 × 10−4, n = 1,000) as well as for αopt (Z-test, P = 9.4 × 10−8, n = 1,000), but we did not find any significant difference using predictions from SIFT. Significance is indicated by P value and nonsignificance is indicated by “ns.”
To systematically benchmark our predictions, we tested whether the levels of intracellular metabolites directly linked to enzymes with predicted functional SNPs exhibited larger variation across strains than metabolites proximal to enzymes with nonfunctional SNPs. Our premise was that functional mutations affect enzyme kinetics (e.g., KM, kcat), resulting in local adaptive changes of metabolite levels (49). Such local metabolic changes are typically found in response to genetic (49–51) or chemical perturbations of enzymes (52, 53). We therefore determined intracellular levels of 294 putative metabolites in the 18 MTBC strains and identified substrates and products of the reactions catalyzed by each enzyme with an annotated SNP. For each metabolite, we then calculated the ratio between average levels in strains with and without the SNP (Dataset S10). Finally, we compared the ratios of functional and nonfunctional SNPs to the distributions of ratios obtained from randomizing the strain assignment of SNPs. On average, these ratios were significantly different between functional and nonfunctional SNPs (t test, P = 2.8 × 10−4, n = 1,208). Moreover, the effects of functional SNPs were significantly different from distributions obtained by randomly assigning SNPs to strains (Z-test, P = 2.0 × 10−4, n = 1,000), whereas the ratios of nonfunctional SNPs were not (Z-test, P = 0.91, n = 1,000). By making the functional SNP classification more stringent (α = 4.1 × 10−4 mmol⋅gDW−1⋅h−1), the separation between functional and nonfunctional SNPs became more pronounced, indicating that SNPs with larger predicted effects had a greater impact on intracellular metabolite levels (Fig. 3E and SI Appendix, Fig. S6). Prediction of functional SNPs with SIFT (35) did not achieve a significant separation for any classification threshold (SI Appendix, Fig. S6), possibly because this homology-based method is invariant to conditions while our functionality predictions are condition-specific. Thus, the independent genome-scale analysis of intracellular metabolome data across the 18 MTBC strains indirectly supported our model-based classification of SNP functionality over homology-based predictions.
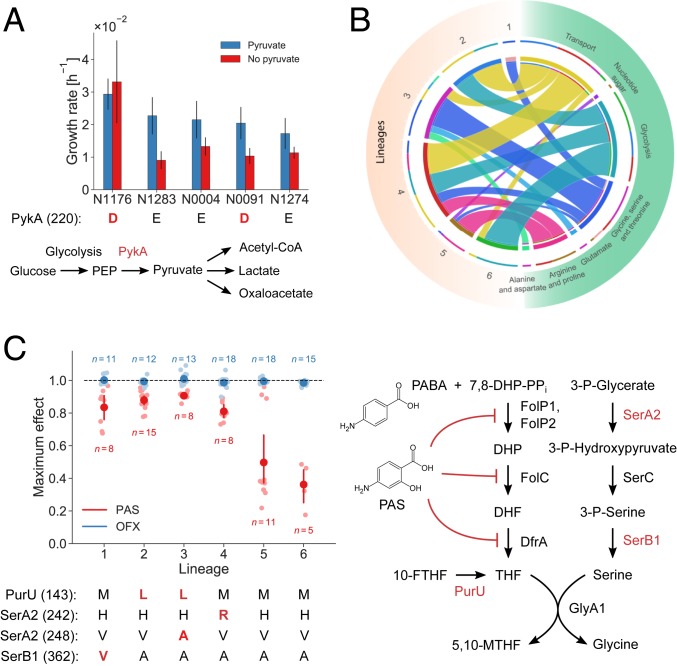
For a direct validation of our SNP functionality predictions, we focused on the known functional E220D substitution in the glycolytic enzyme pyruvate kinase (PykA) (54, 55), a mutation reported to abolish enzyme activity in all animal-adapted MTBC strains and the human-adapted lineages 5 and 6, also known as M. africanum (56). The SNP induces a pyruvate requirement, and these strains cannot grow on glucose as the sole carbon source (54, 55). Deleting pykA in the laboratory strain H37Rv also leads to glucose toxicity, presumably due to accumulation of methylglyoxal (57). Unexpectedly, the effect size of PykA E220D was ranked 78 out of the 88 predicted functional SNPs. Hence, differently from what has been reported in animal strains, our analysis, while classifying the PykA E220D substitution as functional, predicted only a relatively small flux effect under the tested conditions in PykA E220D-carrying strains (Fig. 4A). To test this prediction, we grew five MTBC strains on 7H9 glucose medium with and without pyruvate. Clinical isolates with the E220D substitution were able to grow on glucose both with and without pyruvate, in agreement with our prediction (Fig. 4A; SI Appendix, Figs. S8 and S9; and Dataset S11). Consistently, the intracellular levels of methylglyoxal were comparable in strains with and without the E220D SNP (SI Appendix, Fig. S7). Thus, our physiological data support the model prediction of PykA E220D functionality and indicate that, under the tested conditions, clinical isolates behave differently from the animal-adapted strains.
Fig. 4.
Phenotypic consequences of predicted functional SNPs in the MTBC. (A) Estimated growth rates of five strains grown in glucose medium with (blue) and without (red) supplemented pyruvate. Strains affected by the PykA E220D substitution are indicated in red. One experiment was performed for each strain and condition, and error bars indicate two SEs of the growth rate estimate. The pathway diagram shows PykA in its metabolic context. (B) Metabolic subsystems of predicted synthetic lethal interactions involving enzymes affected by functional SNPs. Each ribbon connects an MTBC lineage to a metabolic subsystem, and its size indicates the number of functional SNPs in the lineage that epistatically interact with an enzyme in the metabolic pathway. (C) Sensitivity of strains to the antibiotics PAS and OFX. The plot shows the maximum observed drug effect by lineage with dark dots indicating means and error bars indicating 95% confidence intervals from bootstrapping (1,000 samples). Lineages 1 to 4 were significantly less sensitive to PAS than lineages 5 and 6 (t test, P = 4.5 × 10−13, n = 55) while all strains were similarly sensitive to OFX. The pathway diagram illustrates the mode of action of PAS, which is an analog of para-aminobenzoic acid (PABA), and its connection to enzymes affected by four predicted functional SNPs in folate metabolism that span lineages 1 to 4 (indicated in red below the plot).
Next, we systematically investigated whether the predicted functional SNPs could reveal strain-specific metabolic vulnerabilities or carbon-source dependencies. We used FBA to find all synthetic lethal pairs of gene deletions in which one of the genes contained at least one of our predicted functional SNPs (modeled as deletions in this analysis). The predicted epistatic interactions of SNP-affected enzymes differed between strains and were mostly with enzymes involved in glycolysis, amino acid biosynthesis, and nutrient transport (Fig. 4B and Dataset S12). For example, predicted epistatic interactions in lineages 1 and 3 were significantly enriched in glycine, serine, and threonine metabolism, while strains from lineages 2, 4, and 6 were predicted to be significantly more vulnerable to disruptions in glycolysis (Dataset S13). Moreover, by analyzing the total predicted effects of SNPs on exchange fluxes we could connect SNPs to strain-specific differences in metabolite uptake and secretion. Specifically, we multiplied our predicted SNP effects by the strain-specific sensitivity matrices (network responses) and found significant differences in uptake and secretion fluxes caused by SNPs (ANOVA, P = 3.43 × 10−12). The largest total SNP effects on uptake and secretion across strains were predicted for proline, phosphoenolpyruvate, and citrate, and the metabolic subsystems with the largest total effects were amino acid metabolism and the TCA cycle, reflecting the exometabolome data (SI Appendix, Fig. S10, and Dataset S14). These analyses suggest that MTBC genetic diversity can confer strain-specific sensitivity to the inhibition of metabolic functions and strain-specific nutrient dependencies.
Several functional SNPs conferring strain-specific vulnerabilities were found in pathways dependent on folate cofactors. Specifically, we found six functional SNPs in glycine and serine biosynthesis, four in purine metabolism, three in methionine and cysteine metabolism, and two in pantothenate metabolism (Fig. 3B). Since folate biosynthesis is a validated metabolic target for antibiotics (58), we tested the possibility that metabolomic differences manifest as differential baseline susceptibilities of MTBC strains to inhibitors of folate metabolism. To this end, we determined the inherent baseline sensitivity of 15 of the 18 MTBC strains to para-aminosalicylic acid (PAS), an inhibitor of tetrahydrofolate (THF) biosynthesis and one of the antimycobacterial drugs currently used for multidrug-resistant TB (52, 59). Although no strain carried the genetic determinants of PAS resistance (60), strains from lineages 1 to 4 were significantly more susceptible to PAS than strains from lineages 5 and 6 (t test, P = 4.5 × 10−13, n = 55) (Fig. 4C; SI Appendix, Figs. S11 and S12; and Dataset S15). Notably, because of the small sample size, classical GWAS analysis yielded no significant (corrected P < 0.05) associations between any of the 566 SNPs and PAS sensitivity (SI Appendix, Fig. S13 and Dataset S2). Thus, a key advantage of our approach is the ability to reduce the space of functional enzyme SNPs that could interfere with folate metabolism, even with small sample size, and potentially explain differential drug susceptibility. Eight of the 88 predicted functional SNPs were in genes encoding enzymes proximal to THF, and 4 of these SNPs, affecting three different enzymes, were found in strains with increased inherent baseline susceptibility to PAS (Fig. 4C). These substitutions are present in drug-sensitive strains; they affect SerA2 and SerB1 (needed for serine biosynthesis) and PurU (which converts 10-formyl-THF to THF). In contrast, all of the strains were similarly susceptible to the DNA gyrase inhibitor ofloxacin (OFX) used as a control (Fig. 4C). We speculate that, similar to previous observations in Escherichia coli (61), recycling of 10-formyl-THF and 5,10-methylene-THF via PurU, SerA2, and SerB1 plays a key role in replenishing the THF pool to compensate for the effect of PAS and that the SNPs impair this role.
Discussion
Current GWAS methods for relating MTBC genetic variation to phenotype were successful in identifying clinically prevalent mutations that confer strong phenotypes such as drug resistance (38). However, because the genetic diversity in MTBC strains is small compared to other pathogens (6, 62), such standard methods struggle to relate genetic variation to more subtle phenotypes such as host adaptation and transmissibility. Moreover, it is unclear what the relevant phenotypes for host adaptation are. Because metabolic adaptability is crucial for MTBC strains to scavenge nutrients in the context of infection (63), we hypothesized that metabolic phenotypes could discriminate between MTBC strains and their clinical phenotypes. Our data for 18 MTBC strains from six lineages demonstrate substantial lineage-specific phenotypic differences between MTBC strains, reflected in different growth rates and preferences for nutrient uptake and by-product secretion, potentially revealing divergent metabolic specialization in MTBC lineages (64, 65). This suggests that integrating genome and metabolome profiling can help find strain-specific factors for host adaptation as well as metabolic vulnerabilities that are strain-specific.
To link metabolic phenotypes to genomic information, we developed a constraint-based method to identify condition-dependent functional SNPs affecting enzymes, that is, nonsynonymous SNPs that are required to explain the experimental observations such as growth, uptake, and secretion rates. The central concept of the method is to represent all strains and experimental information in a single constraint-based model to consider all possible dependencies between SNPs and their effects on metabolic phenotypes. This systematic data integration allowed us to predict the functionality of enzyme SNPs despite a relatively small sample size and restricted genetic variability, making it a valid alternative to GWAS. Our computational strategy is broadly applicable to other organisms, in particular because of the large and rapidly increasing number of microbes for which sequencing information and genome-scale metabolic models are available. Notably, it is likely that genetic variation in nonmetabolic genes contributes to the metabolic variability observed among strains. While the current approach is restricted to enzymatic SNPs and does not account for potential genetic interactions with mutations in nonenzymatic genes, extensions could include control sequences of metabolic genes (e.g., by representing SNP-modulated enzyme concentrations) as well as other forms of genetic variability. Despite this limitation, our model-based prediction of functional SNPs has a key advantage over homology-based methods such as SIFT (35) in that it predicts condition-specific functionality of genetic variations and investigates SNP functionality in the context of potential compensatory mechanisms such as alternative pathways and other genetic variations. This was crucial to correctly predict the E220D SNP functionality in PykA and to anticipate its mild impact under the tested conditions.
While nonmetabolic SNPs are dominant in MTBC strains, our analysis shed light on the underlying genetic basis for metabolic diversity and enabled a systematic characterization of divergent evolution in MTBC lineages. In addition, we showed that the approach could open the door to strain-specific drug treatments. As a proof of principle, we demonstrated significant differences in the inherent baseline susceptibility to PAS among MTBC strains. These experiments were motivated by our model-based results, and the strains’ differential baseline sensitivities could be rationally linked to predicted functional SNPs in enzymes involved in the utilization and recycling of folate cofactors. Such predictions can guide the identification of natural genetic variants that facilitate emergence of resistance, while not conferring resistance directly. This is especially relevant in the context of the recently described relationship between minor variations in the inherent baseline susceptibility to drugs and treatment outcomes (18). More generally, we envisage our model-based approach to enable a more systematic understanding of the key metabolic dependencies in pathogenic bacteria and to aid the development of selective treatments that exploit metabolic vulnerabilities during infection.
Materials and Methods
Cultivation and Sampling.
Three biological replicates of the 18 employed MTBC strains (40) were cultured in 50-mL conical tubes containing 15 mL of medium incubated at 37 °C and shaken continuously on an orbital rotator. We used a modified 7H9 medium (BD) supplemented with 0.5% (wt/vol) pyruvate, 0.05% (vol/vol) tyloxapol, 0.2% (wt/vol) glucose, 0.5% (wt/vol) bovine serum albumin (Fraction V), and 14.5 mM NaCl. Compared to the usual composition of 7H9 we omitted glycerol, Tween 80, oleic acid, and catalase from the medium. The choice of the medium used in this study was primarily driven by the established practice of growing M. africanum (MTBC lineages 5 and 6) on media containing pyruvate. We monitored growth by determining optical density measured at 600 nm (OD600). To test pykA SNP functionality, we inoculated 10 mL of appropriate medium (with or without pyruvate) with ∼107 bacterial cells of N0004, N0091, N1176, N1274, or N1283 (starting at OD600 < 0.01). We incubated the cultures on a rotatory shaker at 37 °C and quantified their growth by optical density after 10 d. For analysis of the exometabolome, we periodically withdrew 1-mL aliquots, pelleted the cells by centrifugation (10,000 × g, 5 min), filtered the supernatant through 0.22-µm syringe filters twice to remove viable bacteria, and kept the resulting supernatant at −80 °C until mass spectrometric analysis. For the analysis of the intracellular metabolome, we harvested three OD600 unit equivalents of cells during midexponential growth phase (OD600 = 0.50 ± 0.10) using rapid filtration through a 0.45-μm filter. We washed the pellets with 2 mL of 75 mM ammonium carbonate buffer (pH 6.6, prewarmed to 37 °C). Metabolites were extracted by transferring the filter with cell pellets into a 15-mL conical tube containing 7.5 mL of ice-cold acetonitrile:methanol:water (2:2:1) solution and incubated at −20 °C for 1 to 4 h. After incubation we filtered the extraction solution through 0.22-µm syringe filters twice to remove viable bacteria and kept the resulting supernatant at −80 °C until mass spectrometric analysis.
Genome Sequencing.
Details on MTBC genome sequencing and bioinformatics analysis for sequence read alignment and variant determination are described in detail in ref. 40.
Metabolomics Profiling.
Intracellular extracts and supernatant aliquots were directly injected into an Agilent 6550 time-of-flight mass spectrometer (ESI-iFunnel Q-TOF, Agilent Technologies) (42). The platform consists of an Agilent Series 1100 LC pump coupled to a Gerstel MPS2 autosampler and an Agilent 6550 Series Quadrupole Time of Flight mass spectrometer (Agilent, Santa Clara, CA). Mass spectra were recorded from m/z 50 to 1,000 using the highest resolving power (4 GHz HiRes). Detected ions were matched to a list of metabolites based on the corresponding molar mass. We compiled a comprehensive list of metabolites from the following genome-scale metabolic models: an automatically constructed M. tuberculosis H37Rv model from the Model SEED resource (Seed83332.1) (66), a manually curated M. tuberculosis H37Rv model (sMtb) (67), an E. coli K12 model (iJO1366) (68), and a Mycobacterium smegmatis mc2155 model from the BioModels database (BMID000000141548, https://www.ebi.ac.uk/biomodels-main) (69). The chemical formula of each metabolite was used to calculate the deprotonated monoisotopic molecular mass. Detected ions within a mass tolerance difference of less than 0.003 Da were associated with the nearest reference metabolites. This method is not able to separate compounds with similar m/z and relies on direct ionization without liquid chromatography (LC) separation. Spectral data processing allowed annotation of 294 intracellular and 272 extracellular metabolites.
Statistical Analysis of Metabolome Data.
A one-way analysis of variance (ANOVA) was used to find metabolites exhibiting significant secretion/uptake difference across lineages. Eighteen strains and 6 lineages were analyzed, yielding a between-groups degree of freedom of 12. Estimated P values were corrected for multiple tests using the procedure described in ref. 70.
SNP-Phenotype Associations.
For each SNP affecting at least two strains, we performed a t test comparing the slopes inferred from exometabolomes (mean across biological replicates) between strains with and without the SNP. We assigned the smallest P value across all metabolites as the P value of SNP-exometabolome association for each SNP. We also performed a GWAS with the online tool easyGWAS (https://easygwas.ethz.ch) (71) using the mean PAS sensitivity of each strain as phenotype and the 566 SNPs in our model as genotype. In both cases, we used the Holm-Šidák method to correct for multiple testing.
SIFT Scores.
We obtained SIFT scores for SNPs relative to the laboratory strain H37Rv from SIFT4G (https://sift.bii.a-star.edu.sg/sift4g/) (35) and filtered out predictions with median sequence information above 3.25 to avoid prediction based on closely related sequences. SNPs with a SIFT score (probability of amino acid change being tolerated) below the recommended threshold of 0.05 were classified as functional according to SIFT.
Constraint-Based Modeling.
All details on the implementation of the model, from inference of constraints on growth, uptake, and secretion rates to functional SNP prediction, are given in SI Appendix, Supplementary Methods.
Intracellular Metabolome Analysis.
For each nonsynonymous SNP and each metabolite in the intracellular metabolome participating in a reaction catalyzed by the affected enzyme, the mean intracellular level in strains with the SNP was divided by the mean intracellular level in strains without the SNP. The resulting relative intracellular levels were used to compute the mean relative intracellular levels of functional and nonfunctional SNPs (as classified by our model and SIFT), and bootstrapping with 1,000 samples was used to compute 95% confidence intervals. The data were resampled 1,000 times with replacement, the mean of each sample was calculated, and the 95% confidence interval was defined from the 2.5th and 97.5th percentiles of these means after sorting. Random distributions of mean relative intracellular intensities were obtained by randomizing the strain assignment of SNPs 1,000 times while preserving the number of strains affected by each SNP.
Antibiotic Sensitivity Analysis.
The sensitivity of strains to PAS and OFX was determined as described elsewhere (72). Briefly, we prepared 96-well plates such that they contained a twofold serial dilution of a drug of interest (e.g., PAS, starting from a concentration of 4 μg/mL) in 90 μL of our modified 7H9 medium. We added 10 μL of bacterial culture adjusted to a starting OD600 of 0.02. Plates were incubated for 10 d at 37 °C after which time we added 10 μL of 0.02% alamar blue solution and incubated for a further 24 h. After 24 h we added 100 μL of 3.7% formalin solution to kill the bacteria and scored the plates by measuring the fluorescence (λex = 545 nm, λem = 590 nm) using a Molecular Devices spectraMAX microplate reader. For each drug and each strain, fluorescence signals were normalized by 1) subtracting the signal of the drug without any cells and 2) dividing by the signal of the cells without drug. For each biological replicate, the maximum drug effect was quantified as one minus the ratio of fluorescences at the highest and lowest drug dose, respectively. The mean maximum effect was calculated for each lineage with 95% confidence intervals obtained from bootstrapping with 1,000 samples, as for intracellular metabolome analysis.
Software.
Models were built in Python using COBRApy (73), and optimization problems were solved with the Gurobi Optimizer 7.5.2 (Gurobi Optimization, Beaverton, OR). Inference of metabolite uptake and secretion and all steps of mass spectrometry data processing and analysis were performed in MATLAB R2018 (The MathWorks, Natick, MA). Remaining data analysis was performed in Python.
Data and Code Availability.
All data presented in this study are available in SI Appendix. The model and the code needed to solve it and reproduce our results are available at https://gitlab.com/csb.ethz/MtbSNP.
Supplementary Material
Acknowledgments
We thank Dr. Hans-Michael Kaltenbach and Charlotte Ramon for helpful contributions to the analysis. This work was supported by the Swiss National Science Foundation (Grants 310030_166687, IZRJZ3_164171, IZLSZ3_170834, and CRSII5_177163); the European Research Council (Project 309540-EVODRTB); and SystemsX.ch (TbX).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915551117/-/DCSupplemental.
References
- 1.Achtman M., Insights from genomic comparisons of genetically monomorphic bacterial pathogens. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 860–867 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gagneux S., Ecology and evolution of Mycobacterium tuberculosis. Nat. Rev. Microbiol. 16, 202–213 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Köser C. U., Feuerriegel S., Summers D. K., Archer J. A. C., Niemann S., Importance of the genetic diversity within the Mycobacterium tuberculosis complex for the development of novel antibiotics and diagnostic tests of drug resistance. Antimicrob. Agents Chemother. 56, 6080–6087 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Namouchi A., Didelot X., Schöck U., Gicquel B., Rocha E. P. C., After the bottleneck: Genome-wide diversification of the Mycobacterium tuberculosis complex by mutation, recombination, and natural selection. Genome Res. 22, 721–734 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coscolla M., Gagneux S., Consequences of genomic diversity in Mycobacterium tuberculosis. Semin. Immunol. 26, 431–444 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eldholm V., Balloux F., Antimicrobial resistance in Mycobacterium tuberculosis: The Odd One Out. Trends Microbiol. 24, 637–648 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Gagneux S., Small P. M., Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect. Dis. 7, 328–337 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Stucki D., et al. , Mycobacterium tuberculosis lineage 4 comprises globally distributed and geographically restricted sublineages. Nat. Genet. 48, 1535–1543 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holt K. E., et al. , Frequent transmission of the Mycobacterium tuberculosis Beijing lineage and positive selection for the EsxW Beijing variant in Vietnam. Nat. Genet. 50, 849–856 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Jong B. C., et al. , Progression to active tuberculosis, but not transmission, varies by Mycobacterium tuberculosis lineage in The Gambia. J. Infect. Dis. 198, 1037–1043 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guerra-Assunção J. A., et al. , Large-scale whole genome sequencing of M. tuberculosis provides insights into transmission in a high prevalence area. eLife 4, e05166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manson A. L. et al.; TBResist Global Genome Consortium , Genomic analysis of globally diverse Mycobacterium tuberculosis strains provides insights into the emergence and spread of multidrug resistance. Nat. Genet. 49, 395–402 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Neill M. B., Mortimer T. D., Pepperell C. S., Diversity of Mycobacterium tuberculosis across Evolutionary Scales. PLoS Pathog. 11, e1005257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coscolla M., Gagneux S., Does M. tuberculosis genomic diversity explain disease diversity? Drug Discov. Today Dis. Mech. 7, e43–e59 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gagneux S., Ed., Strain Variation in the Mycobacterium Tuberculosis Complex: Its Role in Biology, Epidemiology and Control (Springer International Publishing, 2017). [Google Scholar]
- 16.Castro R. A. D., et al. , The genetic background modulates the evolution of fluoroquinolone-resistance in Mycobacterium tuberculosis. Mol. Biol. Evol. 37, 195–207 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ängeby K., Juréen P., Kahlmeter G., Hoffner S. E., Schön T., Challenging a dogma: Antimicrobial susceptibility testing breakpoints for Mycobacterium tuberculosis. Bull. World Health Organ. 90, 693–698 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colangeli R. et al.; DMID 01-009/Tuberculosis Trials Consortium Study 22 Teams , Bacterial factors that predict relapse after tuberculosis therapy. N. Engl. J. Med. 379, 823–833 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Behar S. M., Divangahi M., Remold H. G., Evasion of innate immunity by Mycobacterium tuberculosis: Is death an exit strategy? Nat. Rev. Microbiol. 8, 668–674 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nathan C., Shiloh M. U., Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. U.S.A. 97, 8841–8848 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berney M., Berney-Meyer L., Mycobacterium tuberculosis in the face of host-imposed nutrient limitation. Microbiol. Spectr. 5, 1–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voskuil M. I., et al. , Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198, 705–713 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eoh H., et al. , Metabolic anticipation in Mycobacterium tuberculosis. Nat. Microbiol. 2, 17084 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zimmermann M., et al. , Integration of metabolomics and transcriptomics reveals a complex diet of Mycobacterium tuberculosis during early macrophage infection. mSystems 2, e00057–e17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beste D. J. V., et al. , 13C metabolic flux analysis identifies an unusual route for pyruvate dissimilation in mycobacteria which requires isocitrate lyase and carbon dioxide fixation. PLoS Pathog. 7, e1002091 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vilchèze C., et al. , Enhanced respiration prevents drug tolerance and drug resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 114, 4495–4500 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koul A., et al. , Delayed bactericidal response of Mycobacterium tuberculosis to bedaquiline involves remodelling of bacterial metabolism. Nat. Commun. 5, 3369 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manina G., Dhar N., McKinney J. D., Stress and host immunity amplify Mycobacterium tuberculosis phenotypic heterogeneity and induce nongrowing metabolically active forms. Cell Host Microbe 17, 32–46 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Mehrotra P., et al. , Pathogenicity of Mycobacterium tuberculosis is expressed by regulating metabolic thresholds of the host macrophage. PLoS Pathog. 10, e1004265 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKinney J. D., et al. , Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406, 735–738 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Eoh H., Rhee K. Y., Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 110, 6554–6559 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wakamoto Y., et al. , Dynamic persistence of antibiotic-stressed mycobacteria. Science 339, 91–95 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Power R. A., Parkhill J., de Oliveira T., Microbial genome-wide association studies: Lessons from human GWAS. Nat. Rev. Genet. 18, 41–50 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Desjardins C. A., et al. , Genomic and functional analyses of Mycobacterium tuberculosis strains implicate ald in D-cycloserine resistance. Nat. Genet. 48, 544–551 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaser R., Adusumalli S., Leng S. N., Sikic M., Ng P. C., SIFT missense predictions for genomes. Nat. Protoc. 11, 1–9 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Baugh E. H., et al. , Robust classification of protein variation using structural modelling and large-scale data integration. Nucleic Acids Res. 44, 2501–2513 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grimm D. G., et al. , The evaluation of tools used to predict the impact of missense variants is hindered by two types of circularity. Hum. Mutat. 36, 513–523 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hicks N. D., et al. , Clinically prevalent mutations in Mycobacterium tuberculosis alter propionate metabolism and mediate multidrug tolerance. Nat. Microbiol. 3, 1032–1042 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carey A. F., et al. , TnSeq of Mycobacterium tuberculosis clinical isolates reveals strain-specific antibiotic liabilities. PLoS Pathog. 14, e1006939 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borrell S., et al. , Reference set of Mycobacterium tuberculosis clinical strains: A tool for research and product development. PLoS One 14, e0214088 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yimer S. A., et al. , Genomic characterization of Mycobacterium tuberculosis lineage 7 and a proposed name: ‘Aethiops vetus.’ Microb. Genomics 2, e000063 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fuhrer T., Heer D., Begemann B., Zamboni N., High-throughput, accurate mass metabolome profiling of cellular extracts by flow injection-time-of-flight mass spectrometry. Anal. Chem. 83, 7074–7080 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Jamshidi N., Palsson B. Ø., Investigating the metabolic capabilities of Mycobacterium tuberculosis H37Rv using the in silico strain iNJ661 and proposing alternative drug targets. BMC Syst. Biol. 1, 26 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orth J. D., Thiele I., Palsson B. Ø., What is flux balance analysis? Nat. Biotechnol. 28, 245–248 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Comas I., et al. , Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat. Genet. 42, 498–503 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uhr M., Stelling J., Structural sensitivity analysis of metabolic networks. IFAC Proc. Vol. 41, 15879–15884 (2008). [Google Scholar]
- 47.Lewis N. E., et al. , Omic data from evolved E. coli are consistent with computed optimal growth from genome-scale models. Mol. Syst. Biol. 6, 390 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahadevan R., Schilling C. H., The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab. Eng. 5, 264–276 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Fendt S.-M., et al. , Tradeoff between enzyme and metabolite efficiency maintains metabolic homeostasis upon perturbations in enzyme capacity. Mol. Syst. Biol. 6, 356 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fuhrer T., Zampieri M., Sévin D. C., Sauer U., Zamboni N., Genomewide landscape of gene-metabolome associations in Escherichia coli. Mol. Syst. Biol. 13, 907 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ishii N., et al. , Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science 316, 593–597 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Zampieri M., et al. , High-throughput metabolomic analysis predicts mode of action of uncharacterized antimicrobial compounds. Sci. Transl. Med. 10, eaal3973 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Campos A. I., Zampieri M., Metabolomics-driven exploration of the chemical drug space to predict combination antimicrobial therapies. Mol. Cell 74, 1291–1303.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Keating L. A., et al. , The pyruvate requirement of some members of the Mycobacterium tuberculosis complex is due to an inactive pyruvate kinase: Implications for in vivo growth. Mol. Microbiol. 56, 163–174 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Chavadi S., et al. , Global effects of inactivation of the pyruvate kinase gene in the Mycobacterium tuberculosis complex. J. Bacteriol. 191, 7545–7553 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Jong B. C., Antonio M., Gagneux S., Mycobacterium africanum: Review of an important cause of human tuberculosis in West Africa. PLoS Negl. Trop. Dis. 4, e744 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Noy T., et al. , Central role of pyruvate kinase in carbon co-catabolism of Mycobacterium tuberculosis. J. Biol. Chem. 291, 7060–7069 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hawser S., Lociuro S., Islam K., Dihydrofolate reductase inhibitors as antibacterial agents. Biochem. Pharmacol. 71, 941–948 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Chakraborty S., Gruber T., Barry C. E. III, Boshoff H. I., Rhee K. Y., Para-aminosalicylic acid acts as an alternative substrate of folate metabolism in Mycobacterium tuberculosis. Science 339, 88–91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coll F., et al. , Genome-wide analysis of multi- and extensively drug-resistant Mycobacterium tuberculosis. Nat. Genet. 50, 307–316 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Zampieri M., Zimmermann M., Claassen M., Sauer U., Nontargeted metabolomics reveals the multilevel response to antibiotic perturbations. Cell Rep. 19, 1214–1228 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Achtman M., Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu. Rev. Microbiol. 62, 53–70 (2008). [DOI] [PubMed] [Google Scholar]
- 63.Berney M., et al. , Essential roles of methionine and S-adenosylmethionine in the autarkic lifestyle of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 112, 10008–10013 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Asante-Poku A., et al. , Mycobacterium africanum is associated with patient ethnicity in Ghana. PLoS Negl. Trop. Dis. 9, e3370 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Otchere I. D., et al. , Comparative genomics of Mycobacterium africanum Lineage 5 and Lineage 6 from Ghana suggests distinct ecological niches. Sci. Rep. 8, 11269 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Henry C. S., et al. , High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat. Biotechnol. 28, 977–982 (2010). [DOI] [PubMed] [Google Scholar]
- 67.Rienksma R. A., Suarez-Diez M., Spina L., Schaap P. J., Martins dos Santos V. A. P., Systems-level modeling of mycobacterial metabolism for the identification of new (multi-)drug targets. Semin. Immunol. 26, 610–622 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Orth J. D., et al. , A comprehensive genome-scale reconstruction of Escherichia coli metabolism: 2011. Mol. Syst. Biol. 7, 535 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Büchel F., et al. , Path2Models: Large-scale generation of computational models from biochemical pathway maps. BMC Syst. Biol. 7, 116 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Storey J. D., A direct approach to false discovery rates. J. R. Stat. Soc. B 64, 479–498 (2002). [Google Scholar]
- 71.Grimm D. G., et al. , easyGWAS: A cloud-based platform for comparing the results of genome-wide association studies. Plant Cell 29, 5–19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Palomino J.-C., et al. , Resazurin microtiter assay plate: Simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 46, 2720–2722 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ebrahim A., Lerman J. A., Palsson B. O., Hyduke D. R., COBRApy: COnstraints-based reconstruction and analysis for Python. BMC Syst. Biol. 7, 74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data presented in this study are available in SI Appendix. The model and the code needed to solve it and reproduce our results are available at https://gitlab.com/csb.ethz/MtbSNP.