Significance
We uncovered a molecular mechanism by which multiple importins regulate nuclear import, phase separation, and stress granule recruitment of an RNA-binding protein. Our work permitted the identification of two types of nuclear localization signal (NLS) in cold-inducible RNA-binding protein (CIRBP). We show that CIRBP, which lacks any classical NLS, binds both the importin Transportin-1 (TNPO1) and Transportin-3 (TNPO3). These interactions and nuclear import of CIRBP involve two distinct CIRBP regions, the arginine-glycine(-glycine) (RG/RGG) region and the discovered arginine-serine-tyrosine (RSY)–rich region. We show that these two regions harbor different specificity toward TNPO1 and TNPO3, with the RG/RGG-NLS being specialized in TNPO1 recognition and the RSY-NLS being specialized in TNPO3 recognition.
Keywords: CIRBP, Transportin-1, Transportin-3, nuclear import, phase separation
Abstract
The specific interaction of importins with nuclear localization signals (NLSs) of cargo proteins not only mediates nuclear import but also, prevents their aberrant phase separation and stress granule recruitment in the cytoplasm. The importin Transportin-1 (TNPO1) plays a key role in the (patho-)physiology of both processes. Here, we report that both TNPO1 and Transportin-3 (TNPO3) recognize two nonclassical NLSs within the cold-inducible RNA-binding protein (CIRBP). Our biophysical investigations show that TNPO1 recognizes an arginine-glycine(-glycine) (RG/RGG)–rich region, whereas TNPO3 recognizes a region rich in arginine-serine-tyrosine (RSY) residues. These interactions regulate nuclear localization, phase separation, and stress granule recruitment of CIRBP in cells. The presence of both RG/RGG and RSY regions in numerous other RNA-binding proteins suggests that the interaction of TNPO1 and TNPO3 with these nonclassical NLSs may regulate the formation of membraneless organelles and subcellular localization of numerous proteins.
Import receptors (importins) mediate active nuclear import of cargo proteins across the nuclear pore complex. Recently, we and others have reported a second function of importins, namely chaperoning and inhibiting liquid–liquid phase separation (LLPS) of aggregation-prone RNA-binding proteins (RBPs): for example, fused in sarcoma (FUS), TAF15, EWSR1, hnRNPA1, and hnRNPA2 by the importin Transportin-1 (TNPO1; also known as Karyopherin β2) and TDP-43 by importin α/β (1–4). Both nuclear import and chaperoning rely on the specific interaction of the importin with a nuclear localization signal (NLS) in its cargo protein (5–7). This interaction and hence, directionality of transport are usually regulated by the small guanosine triphosphate (GTP) hydrolase (GTPase) Ran as binding of Ran to the importin promotes release of the cargo in the nucleus (8).
TNPO1 is known to bind to cargoes containing a so-called proline-tyrosine (PY)-NLS, which has been defined as a sequence with an overall high degree of structural disorder, and a C-terminal R/H/K-X(2-5)-PY consensus sequence preceded by a hydrophobic or basic region (9). However, TNPO1 has also been reported to bind to and mediate nuclear import of cargoes not containing a PY-NLS, such as ribosomal proteins or several viral proteins (10), suggesting that other regions are able to serve as NLS for TNPO1. For the TNPO1 cargo FUS, for example, we have recently demonstrated that TNPO1 can directly interact with several positively charged arginines in the C-terminal RGG3-PY region in the absence of its PY-NLS and that this interaction is regulated by arginine methylation (11, 12). Such arginine-glycine(-glycine) (RG/RGG) regions are highly abundant in RBPs, are involved in both protein and RNA binding (13), and have recently been identified as key drivers in LLPS (14, 15). Importantly, the interaction of TNPO1 with arginines in RG/RGG regions is also crucial for suppression of LLPS of FUS (1). We hypothesized that RG/RGG-rich regions could serve as a type of NLS for TNPO1 and scrutinized published lists of TNPO1 cargoes (16, 17) for the presence of RG/RGG regions. Indeed, we observed that 94 reported TNPO1 cargoes contain RG/RGG regions (Fig. 1A and Dataset S1). Among these proteins, 61 have both a PY-NLS and an RG/RGG region and could thus follow the same recognition mode as FUS. Among the reported TNPO1 cargo proteins, 33 have an RG/RGG region but no PY-NLS and hence, could be proteins in which the RG/RGG region serves as sole NLS for TNPO1. Thus, we set out to test whether RG/RGG regions could serve as a previously unrecognized class of NLS for TNPO1.
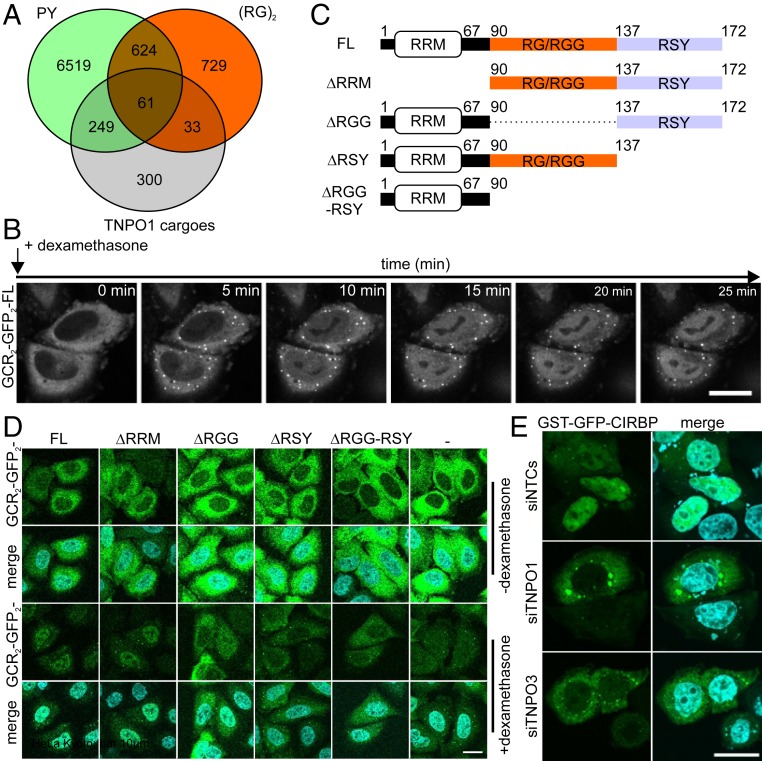
Fig. 1.
The RG/RGG- and RSY-rich regions in CIRBP mediate TNPO1- and 3-dependent nuclear import in HeLa cells. (A) Venn diagram corresponding to the PROSITE analysis (https://prosite.expasy.org/scanprosite/) of two motifs against the nonredundant protein sequences database filtered for human proteins (taxid:9606) harboring either 1) a di-RG motif (orange) each spaced by zero to five amino acids [R-G-x(0-5)-R-G] or 2) a PY motif (green) preceded by a basic amino acid spaced by zero to nine residues [[RKH]-x(0,9)-P-Y]. These two groups of proteins were compared with the published list of TNPO1 cargoes identified by Kimura et al. (16) and Mackmull et al. (17). (B) Time-dependent nuclear import of GCR2-GFP2-CIRBP full length in HeLa Kyoto cells on addition of dexamethasone visualized by live cell imaging. (Scale bar: 20 µm.) (C) Scheme of CIRBP illustrating its domain organization and deletion of individual domains for analysis of their involvement in nuclear import of CIRBP. (D) HeLa Kyoto cells were transiently transfected with constructs coding for GCR2-GFP2-CIRBP full length (FL) or CIRBP mutants lacking either the RRM (DRRM), the RG/RGG (DRGG), the RSY (DRSY), or both RG/RGG and RSY regions (DRGG-RSY). A construct coding for only GCR2-GFP2 (−) was used as control for diffusion. Cells were either fixed before (−dexamethasone) or after (+dexamethasone) the addition of 5 µM dexamethasone for 15 min at 37 °C. Nuclei were stained with DAPI, and cells were analyzed by fluorescence microscopy. (Scale bar: 10 µm.) (E) HeLa Kyoto cells transfected with either control small inhibitory RNA (siRNA) or siRNA against TNPO1 or TNPO3, respectively, were transfected with a construct coding for GST-GFP-CIRBP. Cells were fixed, nuclei were counterstained with DAPI, and cells were analyzed by fluorescence microscopy. (Scale bar: 20 µm.)
Cold-inducible RNA-binding protein (CIRBP) seems to be a promising candidate as deletion of an RG/RGG-rich region was shown to cause cytoplasmic mislocalization of human CIRBP or its Xenopus laevis ortholog (xCIRBP2), demonstrating the importance of this region for proper nuclear localization (18, 19). Nevertheless, the molecular mechanism of CIRBP nuclear import remains elusive. CIRBP belongs to the family of cold shock proteins and is up-regulated in response to a large variety of cellular stresses, including mild cold shock, ultraviolet irradiation, osmotic shock, and hypoxia (20, 21). Under these conditions, CIRBP relocalizes from the nucleus to the cytoplasm and partitions into stress granules (SGs) (19). Cytosolic CIRBP stabilizes a subset of messenger RNAs (mRNAs), facilitating their translation (19, 21, 22). Thus, CIRBP participates in cell adaptation to stress and confers antiapoptotic and antisenescent cytoprotective functions (23–25). In addition to its role in promoting cell survival, CIRBP is associated with various types of cancers, such as liver, breast, brain, and prostate cancers (26). Down-regulation of CIRBP impairs cancer cell survival and enhances response to chemotherapy (27), making CIRBP a potential target for anticancer therapy. Nevertheless, the mechanisms governing CIRBP localization and its redistribution under cellular stress are largely unknown.
Here, we show that both TNPO1 and Transportin-3 (TNPO3) mediate nuclear import and suppress LLPS and SG recruitment of CIRBP. CIRBP lacks a classical NLS or PY-NLS, but we identified two so far uncharacterized NLSs: an RG/RGG region that serves as NLS for TNPO1 and an arginine-serine-tyrosine (RSY)–rich region that serves as NLS for TNPO3. NMR spectroscopy and isothermal titration calorimetry (ITC) revealed multiple interactions between CIRBP-TNPO1 and CIRBP-TNPO3 involving both the RG/RGG and RSY regions. We show that the binding sites for the RG/RGG, RSY, and the classical PY-NLS are overlapping on TNPO1, with a long flexible loop in TNPO1 creating an additional and highly specific binding site for the RG/RGG region. Arginine methylation of the CIRBP RG/RGG region via protein arginine methyltransferase 1 (PRMT1) decreases both TNPO1 and TNPO3 binding and consequently, reduces CIRBP nuclear import. These results suggest that nuclear import, LLPS, and SG recruitment of RBPs containing multiple NLSs can be controlled by an intricate interplay of multiple importins and can be further regulated by posttranslational modifications, such as arginine methylation.
Results
The RG/RGG- and RSY-Rich Regions Mediate Nuclear Import of CIRBP.
To investigate the nuclear import mechanism of CIRBP, we made use of a hormone-inducible nuclear transport assay adapted from Love et al. (28). In this assay, the protein of interest is fused to two hormone-binding domains of the glucocorticoid receptor (GCR) and two green fluorescent protein (GFP) moieties. The GCR domain traps the fusion protein in the cytoplasm until addition of a steroid hormone, such as dexamethasone, triggers nuclear import. Fusion with two GCR and two GFP domains increases the molecular mass of CIRBP by ∼120 kDa and thus, efficiently reduces nuclear import due to passive diffusion across the nuclear pore complex. In the absence of dexamethasone, GCR2-GFP2-CIRBP was predominantly cytoplasmic but rapidly translocated into the nucleus on dexamethasone addition (Fig. 1B and Movie S1). In comparison, a GCR2-GFP2 construct lacking an NLS (GCR2-GFP2-stop-M9) did not become enriched in the nucleus relative to the cytoplasm after dexamethasone addition but at most, equilibrated between nucleus and cytoplasm due to passive diffusion (SI Appendix, Fig. S1A and Movie S2).
In order to identify the regions of CIRBP that mediate nuclear import, we next examined a series of deletion mutants lacking the N-terminal RNA recognition motif (RRM; CIRBPΔRRM), the RG/RGG-rich region (CIRBPΔRGG), the C-terminal RSY-rich region (CIRBPΔRSY), or both the RG/RGG-rich region and the C-terminal RSY-rich region (CIRBPΔRGG-RSY) (schematic diagram in Fig. 1C). Deletion of the RRM did not impair nuclear import, whereas deletion of either the RG/RGG or the RSY region strongly impaired nuclear translocation compared with the GCR2-GFP2 control (Fig. 1D; SI Appendix, Fig. S1B shows the quantification). Combined deletion of both the RG/RGG and the RSY regions did not result in stronger impairment of nuclear import than deletion of the RSY region alone, indicating that the RSY region of CIRBP is the predominant NLS, at least in our cellular system. Together, our data demonstrate that both the RG/RGG region and the RSY region are required for efficient nuclear import of CIRBP.
Nuclear Import of CIRBP Is Mediated by TNPO1 and TNPO3.
We and others have previously demonstrated an interaction of RG/RGG regions in FUS with TNPO1 (2, 11, 12) and therefore, hypothesized that TNPO1 may be responsible for nuclear import of CIRBP via its RG/RGG region. Moreover, since the RSY region is also essential for efficient nuclear import CIRBP (Fig. 1D), we speculated that this arginine- and serine-rich region could be recognized by the import receptor TNPO3 (also known as TNPO-SR), which is known to recognize regions enriched in serine and arginine, such as R-S or R-D/E repeats, termed RS- or RS-like NLS (29–31). Even though the RS(-like) region in CIRBP with only three “RS” dipeptides scattered over a 30-residue region shows only weak similarities with previously reported RS-NLSs, we tested this hypothesis. To this end, we silenced TNPO1 or TNPO3 expression in HeLa cells using small interfering RNA (siRNAs) (SI Appendix, Fig. S1C shows knockdown efficiencies) and then, examined localization of GST-GFP–tagged CIRBP. The GST-GFP tag (∼55 kDa) serves to reduce passive diffusion through nuclear pores, allowing us to address active, receptor-mediated import of CIRBP (32–34). Consistent with our hypothesis, silencing of either TNPO1 or TNPO3 caused a significant accumulation of GST-GFP-CIRBP in the cytoplasm (Fig. 1E; SI Appendix, Fig. S1D shows the quantification), indicating that both TNPO1 and TNPO3 mediate nuclear localization of CIRBP.
TNPO1 and TNPO3 Suppress Phase Separation of CIRBP In Vitro and Its Recruitment to Stress Granules in Cells.
It has been previously shown that import receptors, in addition to mediating nuclear import, also suppress phase separation of RBPs containing prion-like low-complexity domains and prevent their recruitment into SGs (1–4). As CIRBP contains an extended disordered region including the RG/RGG and RSY regions (SI Appendix, Fig. S2A), we tested its ability to phase separate using purified recombinant full-length CIRBP. Indeed, in a turbidity assay, which uses the optical density (OD) of a protein solution as a measure of phase separation, the turbidity of full-length CIRBP increased in a concentration-dependent manner beyond 15 µM (Fig. 2A). Considering that CIRBP is an RBP and that both the RRM domain and the RG/RGG region are involved in RNA recognition (35–37), we next tested the effect of RNA on CIRBP phase separation. Here, titration of (UG)12 RNA to a fixed CIRBP concentration (30 µM) led to a progressive turbidity increase at low RNA concentrations, whereas higher amounts of RNA had a suppressive effect on CIRBP phase separation (Fig. 2B). This is in accordance with a previous study showing that the RNA:protein ratio regulates the phase separation behavior of prion-like RBPs in the above-described manner (38). Importantly, addition of increasing amounts of either TNPO1 or TNPO3 to full-length CIRBP in the presence of RNA led to a concentration-dependent decrease in turbidity, demonstrating that both TNPO1 and TNPO3 have the ability to inhibit phase separation of CIRBP (Fig. 2C). In order to confirm these findings, we monitored CIRBP phase separation by differential interference contrast (DIC) microscopy. In the presence of substoichiometric amounts of RNA, CIRBP immediately formed small condensates that increased in size over time, most likely due to fusion of condensates, indicating their “liquid-like” behavior (Fig. 2D). In line with the turbidity assay, both TNPO1 and TNPO3 were able to suppress CIRBP condensate formation.
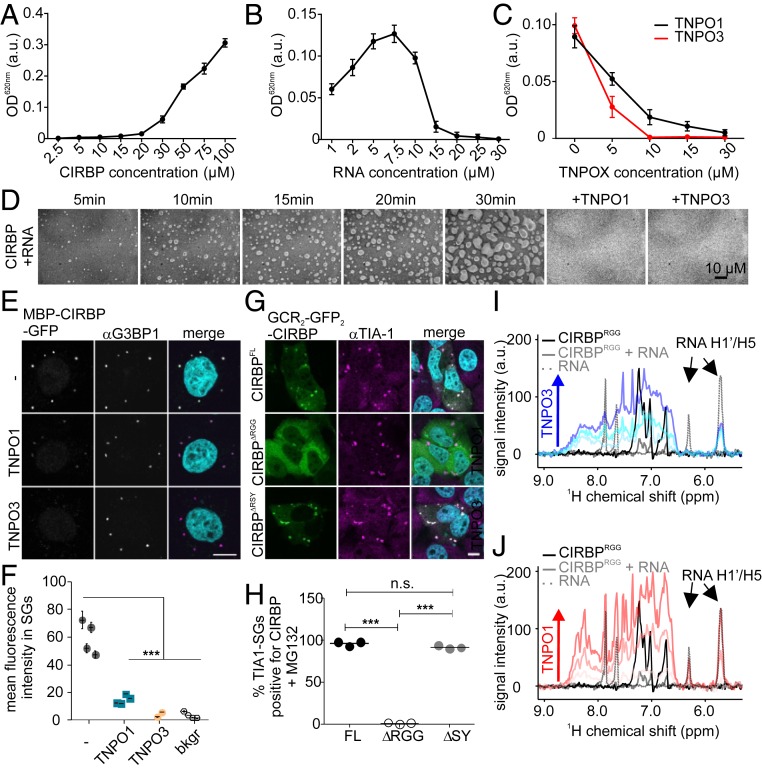
Fig. 2.
TNPO1 and TNPO3 suppress phase separation of CIRBP in vitro and its recruitment to SGs in cells. Turbidity assay to quantify phase separation of full-length CIRBP with increasing CIRBP concentration is in A, with fixed CIRBP concentration (30 µM) and increasing RNA concentration in B and with fixed CIRBP and RNA concentration (30 and 10 µM, respectively) and increasing TNPO1 or TNPO3 concentration in C (black and red lines, respectively). Values represent means ± SEM (n = 3). (D) Images obtained from differential interference contrast microscopy of full-length CIRBP at 30 µM in the presence of 10 µM RNA and/or 30 µM TNPO1 or −3. (Scale bar: 10 µm.) (E) Association of MBP-CIRBP-EGFP with SGs in semipermeabilized cells is suppressed by TNPO1 or TNPO3. SGs in HeLa P4 cells were induced with 10 μM MG132 for 3 h. Afterward, cells were permeabilized using digitonin, cytosol was washed out, and nuclear pores were blocked by WGA. MBP-FUS-EGFP (200 nM) was added in the absence or presence of 2 µM TNPO1 or TNPO3, respectively. SGs were visualized by anti-G3BP1 immunostaining, and recruitment of CIRBP into SGs was monitored directly by enhanced green fluorescent protein (EGFP) fluorescence using confocal microscopy. Nuclei were counterstained with DAPI (displayed in turquoise). Please note that this figure presents a subset of the TNPO1 concentrations tested and is also shown in more detail in SI Appendix, Fig. S2B. (Scale bar: 10 µm.) (F) A quantification of the mean fluorescence intensity of MBP-CIRBP-EGFP in SGs in the absence or presence of increasing concentrations of either TNPO1 or TNPO3 for four or two (2 µM TNPO3) independent replicates with ≥28 SGs ± SEM. ***P < 0.001 by one-way ANOVA Dunett’s multiple comparison test. (G) Recruitment of CIRBP into SGs is dependent on its RG/RGG region. HeLa Kyoto cells were transiently transfected with GCR2-GFP2-CIRBP-full length or constructs lacking either the RG/RGG (CIRBP ΔRGG) or the RSY region (CIRBP ΔRSY). SGs were induced by treatment with 10 µM MG132 for 3 h, and SG recruitment of the GFP reporter was analyzed by colocalization with the SG marker TIA-1. (Scale bar: 10 µm.) (H) Quantification of cells showing GCR2-GFP2-CIRBP-full length (FL), ΔRGG, or ΔRSY localizing to TIA-1–positive SGs. Values represent means ± SEM (n = 3; at least 80 cells per experiment). ***P < 0.001 by one-way ANOVA with Tukey’s multiple comparison test. n.s.: nonsignificant. (I and J) Overlay of 1H NMR spectra of 30 µM 15N-labeled CIRBPRGG (black) in the presence of 10 µM RNA (gray) and with increasing amount of TNPO1 (red; J) or TNPO3 (blue; I). The reference 1H NMR spectra of RNA at the same concentration (10 µM) are shown in gray (dotted lines).
CIRBP has previously been shown to be recruited into SGs in response to various cellular stresses (18, 19). In order to confirm the chaperoning activity of both TNPO1 and TNPO3 for CIRBP in the cellular context, we made use of our previously established SG recruitment assay in semipermeabilized cells (1, 39). Addition of recombinant GFP-tagged maltose-binding protein (MBP)-tagged CIRBP to these semipermeabilized cells resulted in its accumulation in GTPase-activating protein-binding protein (G3BP1)-positive SGs (Fig. 2E). Importantly, concomitant addition of either TNPO1 or TNPO3 suppressed the SG association of MBP-CIRBP (Fig. 2E; Fig. 2F shows the quantification) in a concentration-dependent manner (SI Appendix, Fig. S2 B and C). This result demonstrates that both importins not only suppress phase separation of CIRBP in vitro but also, reduce its association with SGs in cells. Taken together, both TNPO1 and TNPO3 exert all known functions of importins toward CIRBP and hence, are bona fide import receptors of CIRBP.
The RG/RGG Region of CIRBP Contributes to Phase Separation and Is Required for SG Recruitment.
Recently, we and others have shown that RG/RGG regions in FUS are essential for LLPS in vitro (1, 4, 40, 41). Both the RG/RGG and C-terminal RSY regions of CIRBP are strongly predicted to be disordered (SI Appendix, Fig. S2A), suggesting that these regions may be responsible for phase separation and SG recruitment of CIRBP. We first investigated the contribution of these two regions for SG recruitment of CIRBP in intact cells. In order to analyze the tendency of CIRBP deletion mutants to localize to SGs irrespective of their different nuclear import capabilities, we made use of our cytosolically anchored GCR2-GFP2-CIRBP reporter. In the absence of steroid hormones, the reporter protein remained in the cytoplasm and was efficiently recruited into T-cell intracellular antigen 1 (TIA-1)–positive SGs on MG132 treatment (Fig. 2 G and H shows the quantification). In contrast, deletion of the RG/RGG region abolished SG recruitment of the reporter nearly completely, while a CIRBP reporter lacking the RSY region still localized to SGs. In order to investigate whether the region that drives SG recruitment in cells is also the region that drives phase separation in vitro, we examined the ability of individual recombinant CIRBP regions CIRBPRRM, CIRBPRGG, and CIRBPRSY to undergo phase separation in vitro. In contrast to full-length CIRBP, none of the domains alone were able to phase separate in the concentration range tested by turbidity assay or DIC microscopy (up to 100 or 30 µM, respectively) (SI Appendix, Fig. S2 D and E). However, successive addition of RNA strongly promoted phase separation of CIRBPRGG in a concentration-dependent manner in the turbidity assay (SI Appendix, Fig. S2F). In line with this, DIC microscopy showed that CIRBPRGG formed liquid-like condensates in the presence of RNA similar to the full-length protein (SI Appendix, Fig. S2E), suggesting that the interaction of the RG/RGG region with RNA contributes to phase separation of the full-length protein. In both assays, addition of either TNPO1 or TNPO3 was able to suppress the RNA-induced phase separation of CIRBPRGG (SI Appendix, Fig. S2 E and G). When we analyzed the RNA-driven phase separation of CIRBPRGG using NMR spectroscopy, addition of unlabeled (UG)12 RNA to a solution of 15N-labeled CIRBPRGG led to disappearance of 1H-15N cross-peaks (SI Appendix, Fig. S2H) and decreased signal intensity in the corresponding one-dimensional (1D)-1H NMR spectra (Fig. 2 I and J). This is in agreement with the formation of high-molecular mass CIRBPRGG/RNA condensates, which leads to extensive broadening of NMR signals due to the increased rotational tumbling time of CIRBPRGG within the condensates. Nevertheless, we cannot exclude that intermediate timescale chemical exchange processes contribute to the observed line broadening. RNA was bound by CIRBPRGG condensates as NMR signals of unlabeled RNA were absent in the 1D-1H NMR spectra (Fig. 2 I and J). Addition of increasing amounts of TNPO1 or TNPO3 to the CIRBPRGG/RNA sample caused reappearance of the RNA 1H NMR signals and CIRBPRGG 1H-15N cross-peaks (Fig. 2 I and J and SI Appendix, Fig. S2H). This indicates that both TNPO1 binding and TNPO3 binding displace the RNA from the RG/RGG region of CIRBP and hence, lead to dissolution of the condensates.
Taken together, we show that the CIRBP RG/RGG region is required for SG localization in cells. Moreover, it undergoes phase separation in vitro in the presence of RNA, which is suppressed by TNPO1 and TNPO3 via RNA displacement. As both TNPO1 and TNPO3 are able to import CIRBP and suppress its phase separation and SG recruitment, we sought to characterize the binding of these two import receptors to CIRBP on a molecular level in more detail.
Distinct CIRBP Regions Are Directly Recognized by TNPO1 and TNPO3.
In order to obtain residue-resolved information for the interaction of CIRBP with TNPO1 and TNPO3, we performed NMR spectroscopy. The 1H-15N heteronuclear single quantum coherence (HSQC) NMR spectrum of isotope-labeled full-length CIRBP in isolation showed NMR signals characteristic for both folded and intrinsically disordered regions (Fig. 3 A and B). Stepwise addition of unlabeled TNPO1 or TNPO3 resulted in a progressive disappearance of the CIRBP 1H-15N HSQC cross-peaks characteristic for disordered residues (Fig. 3 A and B), which indicates binding of both TNPO1 and TNPO3 to the corresponding residues. The identified complex shows extensive line broadening and disappearance of CIRBP NMR signals at a low stoichiometry of TNPO1 and TNPO3, which are characteristic for a complex with a high nanomolar to micromolar dissociation constant (Kd). In line with this, ITC-derived dissociation constants (Kd) are 741 ± 106 and 289 ± 36 nM for CIRBP binding to TNPO1 and TNPO3, respectively (Table 1 and SI Appendix, Fig. S3A).
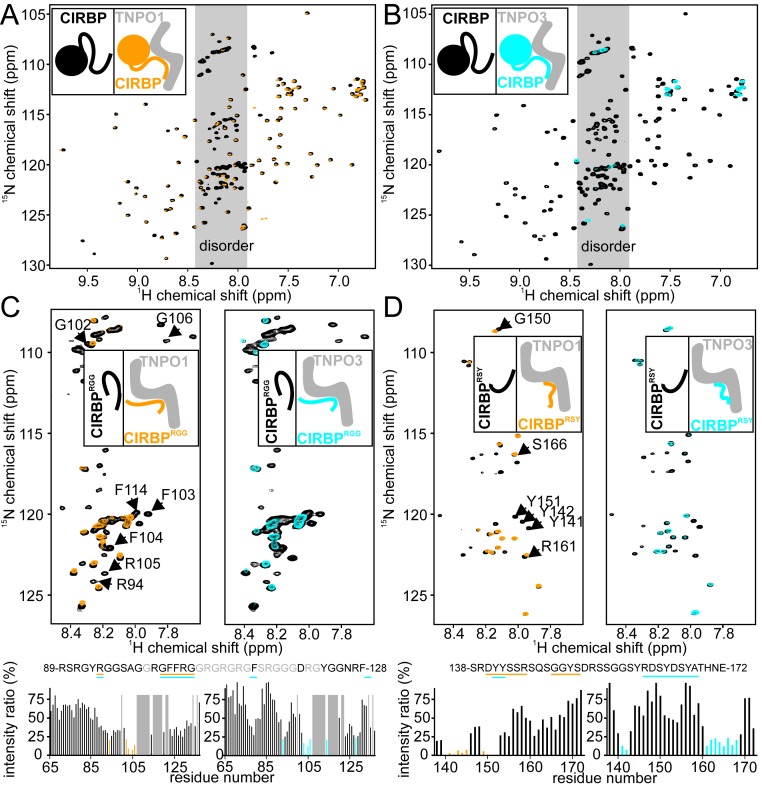
Fig. 3.
Distinct CIRBP regions are directly recognized by TNPO1 and TNPO3. (A and B) 1H-15N HSQC spectrum of full-length 15N-labeled CIRBP at 50 µM without (black) or with addition of one equivalent of TNPO1 (orange in A) or TNPO3 (cyan in B). (C) 1H-15N HSQC spectrum of 15N-labeled CIRBPRGG at 50 µM without (black) or with addition of one equivalent of TNPO1 (orange; Left) or TNPO3 (cyan; Right). The intensity ratio of the 1H-15N HSQC CIRBPRGG cross-peaks between the TNPO1 or TNPO3 free and bound forms are shown in a bar plot, with the CIRBPRGG amino acids strongly affected (ratio < 25%) by TNPO1 or TNPO3 binding colored in orange and cyan, respectively, and highlighted in the CIRBP primary amino acid sequence. The unassigned residues are indicated in gray. (D) Similar to C but using the CIRBPRSY construct in place of the CIRBPRGG construct. A representative cartoon is associated with each NMR spectrum with the unlabeled, NMR-invisible, protein colored gray and the labeled, NMR-visible, partner colored black (unbound), orange (TNPO1-bound), or cyan (TNPO3-bound), respectively.
Table 1.
Thermodynamic parameters of ITC titrations
| Cell | Syringe | Kd (nM) | ΔH (kcal⋅mol−1) | ΔS (cal⋅mol−1⋅K) |
| TNPO1 | CIRBP | 741 ± 106 | −21 ± 0.66 | −42.7 |
| TNPO1 | CIRBPRGG | 75 ± 13 | −11.3 ± 0.16 | −5.3 |
| TNPO1 | meCIRBPRGG | No detectable binding | ||
| TNPO1 | meCIRBP | No detectable binding | ||
| TNPO1 | CIRBPRSY | 7,800 ± 545 | −10 ± 0.77 | −12.2 |
| TNPO1 | FUSPY | 48 ± 6 | −21.9 ± 0.15 | −40 |
| TNPO1Δloop | CIRBPRGG | 364 ± 15 | −22.3 ± 0.18 | −45.3 |
| TNPO1Δloop | CIRBPRSY | 8,075 ± 556 | −19 ± 0.55 | −43.9 |
| TNPO1Δloop | FUSPY | 62 ± 4 | −20.6 ± 0.1 | −36 |
| TNPO1/CIRBPRGG | FUSPY | 413 ± 41 | −13.4 ± 0.21 | −15.8 |
| TNPO1/FUSPY | CIRBPRGG | 391 ± 77 | −12.4 ± 0.36 | −12.4 |
| TNPO3 | CIRBPRGG | 584 ± 77 | −27 ± 0.56 | −62 |
| TNPO3 | CIRBPRSY | 114 ± 5 | −34.9 ± 0.14 | −91.6 |
| TNPO3 | CIRBP | 289 ± 36 | −58.6 ± 0.93 | −167 |
| TNPO3 | meCIRBPRGG | 3,300 ± 320 | −23 ± 1.4 | −48.6 |
| TNPO3 | meCIRBP | 1,002 ± 100 | −63 ± 1.4 | −184 |
The reported errors correspond to the SD of the fit. All of the stoichiometry associated with the complex formation was equal to one.
To further define the CIRBP regions involved in TNPO1 binding, we examined interaction of individual recombinant regions (CIRBPRRM, CIRBPRGG, and CIRBPRSY) with recombinant full-length TNPO1 and TNPO3 in vitro by NMR spectroscopy. In line with the cellular import assay (Fig. 1D), addition of TNPO1 or TNPO3 to a solution of isotope-labeled CIRBPRRM had only a minor effect on the 1H-15N HSQC spectrum, showing that neither TNPO1 nor TNPO3 binds to the RRM alone (SI Appendix, Fig. S3D). In contrast, both TNPO1 and TNPO3 bind to CIRBPRGG and CIRBPRSY as shown by disappearance of CIRBP 1H-15N HSQC cross-peaks on addition of unlabeled TNPO1 or TNPO3 (Fig. 3 C and D). Close inspection of the affected 1H-15N CIRBPRGG cross-peaks revealed that the RG/RGG region of CIRBP (CIRBPRGG) interacts with both TNPO1 and TNPO3 (Fig. 3C). Similarly, close inspection of 1H-15N CIRBPRSY NMR cross-peaks affected by addition of TNPO1 and TNPO3 revealed that the N-terminal part of the CIRBPRSY region interacts with TNPO1, whereas both the N-terminal and C-terminal parts bind to TNPO3 (Fig. 3D). Extensive line broadening is observed across both RG/RGG and RSY CIRBP regions on addition of TNPO1 or TNPO3, suggesting dynamic exchange involving multiple elements across these disordered regions.
Importantly, the interaction of TNPO1 and TNPO3 with CIRBPRGG and CIRBPRSY can be displaced by RanGTP as stepwise addition of RanGTP to a solution of isotope-labeled CIRBPRGG or CIRBPRSY in complex with TNPO1 or TNPO3 leads to progressive signal recovery of the CIRBP 1H-15N HSQC cross-peaks, indicating competition between the CIRBP-NLSs and RanGTP for TNPO1 and TNPO3 binding (SI Appendix, Fig. S3 G and H). Moreover, even though both TNPO1 and TNPO3 can interact with the CIRBP-RG/RGG region as well as the RSY region yet with different affinities, the FUS PY-NLS specifically bound only to TNPO1 but not TNPO3 as shown by NMR (SI Appendix, Fig. S3I).
ITC analysis demonstrated that TNPO1 and TNPO3 have different preferences for the RG/RGG and RSY regions. CIRBPRGG binds TNPO1 and TNPO3 with Kd values of 75 ± 13 and 589 ± 70 nM, respectively (Table 1 and SI Appendix, Fig. S3 B and C), while CIRBPRSY binds TNPO1 and TNPO3 with Kd values of 7,800 ± 545 and 114 ± 5 nM, respectively (Table 1 and SI Appendix, Fig. S3 B and C). The high affinity of CIRBPRSY for TNPO3 was striking given that it contains a much lower number of RS dipeptide sequences than typically observed in classical RS regions. However, CIRBPRSY contains several tyrosine residues that interacted strongly with TNPO3 (Fig. 3D). Mutation of either all tyrosines or all arginines into alanines (CIRBPRSA, CIRBPASY) completely abrogated both TNPO1 and TNPO3 binding by NMR spectroscopy (SI Appendix, Fig. S3 E and F), indicating that both tyrosines and arginines in the RSY-rich region are essential for importin binding.
To summarize, we show that TNPO1 and TNPO3 bind both the RG/RGG region and the RSY region of CIRBP in a RanGTP-competitive manner and that these regions constitute classes of nuclear import signals. Our NMR data suggest that the RG/RGG and RSY NLSs bind to tranportins via extensive multivalent interactions rather than a single short discrete binding motif engaged in a static interaction. Given the relative affinity of these regions for the two different importins, TNPO3 seems to be specialized in RSY-NLS binding, whereas TNPO1 preferentially binds to the RG/RGG-NLS region.
CIRBPRGG and the PY-NLS of FUS Use Overlapping Binding Sites on TNPO1.
Importantly, a number of RBPs, such as FUS, possess both RG/RGG regions and a PY-NLS (Fig. 1A and Dataset S1). We, therefore, sought to investigate whether the binding sites for the RG/RGG-NLS and PY-NLS within TNPO1 are independent of each other or overlapping and hence, might lead to an overall enhanced or decreased affinity for TNPO1 if present within the same RBP. We addressed this question using NMR spectroscopy by employing specific isotope labeling of methyl groups in isoleucine and methionine side chains in a perdeuterated background and methyl transverse relaxation optimized spectroscopy (methyl-TROSY) (42, 43). Using this method, we monitored the interaction of isotope-labeled and NMR-visible TNPO1 with unlabeled and therefore, NMR-invisible CIRBPRGG and/or a construct harboring the PY-NLS of FUS.
Addition of either CIRBPRGG or FUSPY-NLS to a solution of perdeuterated [U-2H, 13C, 15N, isoleucine-methionine (IM)-methyl 1H,13C] TNPO1 induced different chemical shift perturbations of TNPO1 methyl cross-peaks (Fig. 4A). The affected residues can be grouped in two sets: while we detected for some residues chemical shift perturbations specific for either CIRBPRGG or FUSPY-NLS, we also identified residues affected by addition of either CIRBPRGG or FUSPY-NLS. This indicates that the binding site of these NLSs on TNPO1 could partially overlap. However, we cannot exclude that these chemical shift perturbations arise from allosteric changes. To further validate that both CIRBPRGG and FUSPY-NLS compete for binding in TNPO1, we carried out ITC competition experiments. In line with the NMR experiments, binding affinity of CIRBPRGG to TNPO1 was reduced in the presence of FUSPY-NLS with an associated Kd of 391 ± 77 nM compared with 75 ± 13 nM in the absence of FUSPY-NLS (Table 1 and SI Appendix, Figs. S3B and S4A). Vice versa, FUSPY-NLS bound weaker to TNPO1 in the presence of CIRBPRGG with an associated Kd of 413 ± 41 nM compared with 48 ± 6 nM in the absence of CIRBPRGG (Table 1 and SI Appendix, Fig. S4A). In both cases, the ITC-derived stoichiometry was 1:1, demonstrating that both regions can still bind to TNPO1 simultaneously, although with weaker affinity.
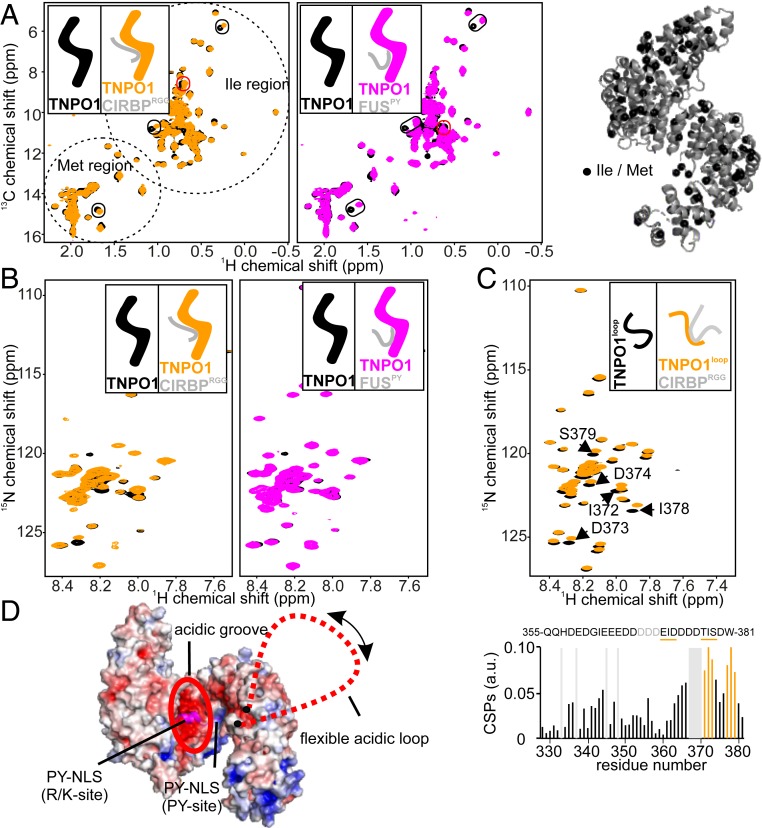
Fig. 4.
CIRBP-RG/RGG region and the PY-NLS of FUS use overlapping binding sites on TNPO1. (A) 1H-13C TROSY-HSQC spectrum of perdeuterated 2H-15N-13C-methyl-IM–labeled TNPO1 at 100 µM without (black) or with addition of one equivalent of CIRBPRGG (orange; Left) or FUSPY (magenta; Right). The peaks specifically affected by CIRBPRGG or FUSPY binding are surrounded in red, and the peaks affected by both CIRBPRGG and FUSPY are surrounded in black. The location of Ile and Met amino acids can be visualized on the 3D X-ray structure of TNPO1 (Protein Data Bank [PDB] ID code 2QMR) (45) with black spheres. (B) 1H-15N TROSY-HSQC spectrum of perdeuterated 2H-15N-13C–labeled TNPO1 at 50 µM without (black) or with addition of one equivalent of CIRBPRGG (orange; Left) or FUSPY (magenta; Right). (C) 1H-15N HSQC spectrum of 15N-labeled TNPO1loop at 50 µM with (orange) or without (black) addition of one equivalent of CIRBPRGG. The chemical shift perturbation (CSP) of the 1H-15N HSQC TNPO1 loop cross-peaks between the CIRBPRGG free and bound forms is shown in a bar plot, with the TNPO1loop amino acids strongly affected (CSP > 0.07) by CIRBPRGG binding colored in orange and highlighted in bold in the TNPO1loop primary amino acid sequence. A representative cartoon is associated with each NMR spectrum with the unlabeled, NMR-invisible, protein colored gray and the labeled, NMR-visible, partner colored black (unbound), orange (CIRBPRGG-bound), or magenta (FUSPY-bound), respectively. (D) Electrostatic surface potential representation of TNPO1 based on its 3D structure solved in complex with FUSPY (PDB ID code 4FQ3) using pymol 2.3.1.
Given that CIRBPRGG and the classical FUSPY-NLS do not share any common sequence motifs, we hypothesized that a distinct region in TNPO1 must be involved in CIRBPRGG recognition. Interestingly, TNPO1 possesses a 30-amino acid-long acidic loop enriched in glutamic acid and aspartic acid residues, which could be involved in binding to basic residues in the CIRBPRGG region via electrostatic interactions. This loop has been removed previously for crystallization of TNPO1 or TNPO1-NLS complexes because of its high degree of disorder but was demonstrated to adopt an ordered conformation on binding of RanGTP (44–47). We recorded 1H-15N HSQC spectra of full-length TNPO1. Here, only flexible regions can be seen due to the high molecular mass of TNPO1. All visible 1H-15N HSQC cross-peaks of TNPO1 correspond to residues located within the TNPO1 loop region as identified by NMR resonance assignment (Fig. 4B). Next, we examined binding of CIRBPRGG and FUSPY-NLS to full-length TNPO1. Addition of CIRBPRGG to a solution of perdeuterated 2H-1H-15N TNPO1 induces chemical shift perturbations of 1H-15N TROSY-HSQC cross-peaks related to the TNPO1 loop region, whereas addition of FUSPY-NLS showed no detectable binding (Fig. 4B). To test whether the TNPO1 acidic loop is sufficient to bind to CIRBP, we carried out NMR chemical shift titrations of isotope-labeled TNPO1loop with CIRBPRGG and FUSPY-NLS. Stepwise addition of CIRBPRGG to a solution of 15N-labeled TNPO1loop induced progressive chemical shift perturbations of the 1H-15N HSQC cross-peaks corresponding to the C-terminal acidic patch of TNPO1loop (Fig. 4C) with a Kd of 57 ± 5 µM (SI Appendix, Fig. S4C). Conversely, neither FUSPY-NLS nor CIRBPRSY binds to TNPO1loop (SI Appendix, Fig. S4B). To determine the contribution of the TNPO1 loop region to the overall affinity of TNPO1 for its cargo, we compared the affinity of CIRBPRGG and FUSPY-NLS for TNPO1 in the absence and presence of the disordered acidic loop. In line with our NMR data, deletion of the acidic loop decreased the affinity of CIRBPRGG for TNPO1 with an associated Kd of 364 ± 15 nM, whereas the deletion had no impact on binding of FUSPY-NLS and CIRBPRSY (Table1 and SI Appendix, Fig. S4D).
When in complex with TNPO1, it is possible that multiple NLSs occupy different or similar binding sites. Three-dimensional (3D) structures of human TNPO1 in complex with PY-NLSs of different RBPs show that the PY-NLSs bind at two sites on TNPO1 involving PY-specific and hydrophobic/basic-rich regions, respectively (Fig. 4D) (46). Using NMR spectroscopy and ITC, we show that the RG/RGG region of CIRBP contacts two key sites on TNPO1, a site within the folded region of TNPO1 and competing with FUSPY-NLS binding and a unique, nonoverlapping site within the disordered negatively charged loop of TNPO1. Based on the structure of TNPO1 bound to the FUSPY-NLS and the electrostatic potential of the importin, we propose that RG/RGG regions bind to the acidic groove of TNPO1, where it competes with the positively charged N-terminal part of PY-NLS (Fig. 4D). In the previously solved RanGTP-bound TNPO1 structure, the TNPO1 disordered loop points toward the acidic groove (47), suggesting that it could stabilize the interaction with the RG/RGG region by creating an extended negatively charged patch.
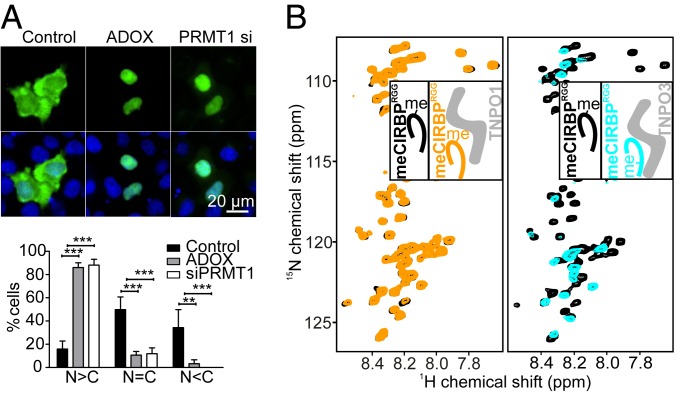
PRMT1-Mediated Arginine Methylation of CIRBPRGG Weakens TNPO1 and TNPO3 Binding.
Recently, we have shown that asymmetric dimethylation of arginines in the RGG3 region of FUS by PRMT1 reduces its affinity for TNPO1 and hereby, regulates FUS nuclear import (11, 48). Interestingly, in Xenopus xCIRBP2 has previously been demonstrated to be methylated by xPRMT1 in vitro, and overexpression of PRMT1 results in increased cytosolic localization of GFP-xCIRBP2 (18), although the molecular mechanisms underlying this relocalization remain elusive. In line with these data, we found that both PRMT1 silencing and inhibition of arginine methylation by adenosine dialdehyde (AdOx) led to enhanced nuclear localization of GST-GFP–tagged CIRBP, which was mostly cytoplasmic in HeLa S3 cells under control conditions (Fig. 5A). One possible mechanism could be that arginine methylation weakens TNPO1 and/or TNPO3 binding and consequently, decreases nuclear import of CIRBP. To test this hypothesis, we carried out in vitro arginine methylation of full-length CIRBP and CIRBPRGG using purified recombinant PRMT1 and tested the effect of arginine methylation on TNPO1 and TNPO3 binding using NMR spectroscopy and ITC. Successful methylation of several arginines, including several previously published residues (49), was confirmed by NMR using 1H-15N-13C, (H)CC(CO)NH experiments (SI Appendix, Fig. S5A). Interestingly, the CIRBP methylation sites are located adjacent to the main TNPO1- and TNPO3-interacting residues of CIRBP. When we tested binding of TNPO1 or TNPO3 with in vitro methylated CIRBPRGG or full-length CIRBP by NMR and ITC, we detected no binding to TNPO1 and reduced binding to TNPO3 (Fig. 5B, Table 1, and SI Appendix, Fig. S5 B and C). In line with this, unmethylated recombinant CIRBP pulled down more TNPO1 and TNPO3 from HeLa cell lysates than methylated CIRBP did in a biochemical pull-down experiment (SI Appendix, Fig. S5D).
Fig. 5.
PRMT1-mediated arginine methylation of the CIRBP RG/RGG region weakens TNPO1 and TNPO3 binding. (A) HeLa cells, either transfected with a control siRNA or siRNA against PRMT1 or treated with AdOx, were transfected with a construct coding for GST-GFP-CIRBP. Nuclei were counterstained with Hoechst 33342, and cells were analyzed by fluorescence microscopy. A quantification of the subcellular localization of the individual reporter proteins (n > C, n = C, and n < C) is shown. Values represent the mean of five independent experiments ± SEM. (Scale bar: 20 µm.) **P < 0.01 by two-way ANOVA + Bonferroni multiple comparison test; ***P < 0.001 by two-way ANOVA + Bonferroni multiple comparison test. (B) 1H-15N HSQC spectrum of 15N-labeled PRMT1-mediated methylated CIRBPRGG at 50 µM without (black) or with addition of one equivalent of TNPO1 (orange; Left) or TNPO3 (cyan; Right). A representative cartoon is associated with each NMR spectrum with the unlabeled, NMR-invisible, protein colored gray and the labeled, NMR-visible, partner colored black (unbound), orange (TNPO1-bound), or cyan (TNPO3-bound), respectively.
Summarizing, we show that PRMT1-mediated arginine methylation of the CIRBP RG/RGG region regulates nuclear import of CIRBP by reducing its binding affinity to both TNPO1 and TNPO3. This implies that arginine methylation of RG/RGG-rich NLSs may generally regulate importin binding and hence, nuclear import of cargoes containing this class of NLS.
Discussion
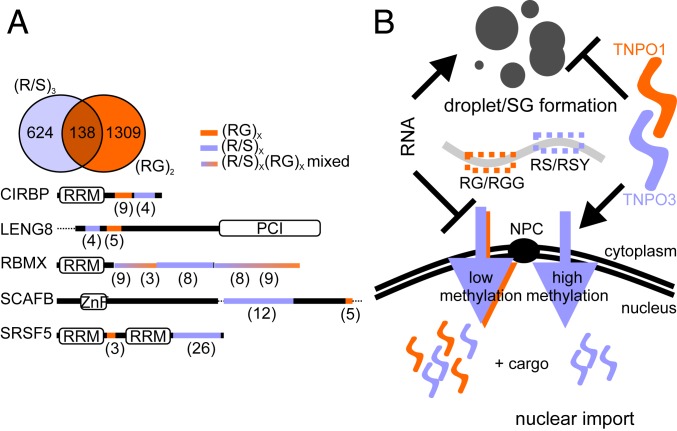
A large number of RBPs contain RG/RGG- or RS-rich regions and combinations thereof (Fig. 6A and Dataset S2). Here, we report two types of NLS and demonstrate that they function in regulating nuclear import, phases separation, and SG recruitment of CIRBP (Fig. 6B). We show that CIRBP, which lacks a PY-NLS or RS-NLS, binds both TNPO1 and TNPO3 in a RanGTP-dependent manner (Fig. 3). This interaction involves two distinct CIRBP regions, the RG/RGG region and the RSY region. We show that these two regions harbor different specificity toward TNPO1 and TNPO3, the RG/RGG NLS being specialized in TNPO1 recognition and the RSY-NLS being specialized in TNPO3 recognition.
Fig. 6.
Analysis of NLSs distribution and their function in nuclear import and phase separation/SG formation. (A, Upper) Venn diagram corresponding to the PROSITE analysis (https://prosite.expasy.org/scanprosite/) of two motifs against the nonredundant protein sequences database filtered for human proteins (taxid:9606) harboring either 1) a di-RG motif (orange) each spaced by zero to five amino acids [R-G-x(0-5)-R-G] and/or 2) a tri-RS or SR motif (violet) each spaced by zero to five amino acids [R/S-R/S-x(0-5)-R/S-R/S-x(0-5)-R/S-R/S]. (A, Lower) Architectural organization of five putative TNPO1 and three cargos containing both RG and RS motifs (displayed in orange and violet, respectively) chosen among the 138 candidates identified. The numbers of RS and RG motifs in the corresponding regions are indicated in parentheses. (B) Model of the regulation of CIRBP nuclear import and phase separation by TNPO1 and TNPO3 depending on its methylation status. RRM: RNA-recognition motif; PCI: proteasome component domain; ZnF: zinc-finger motif; NPC: nuclear pore complex.
TNPO1 Recognizes RG/RGG Regions via Multiple Interactions.
The molecular determinants involved in TNPO1-mediated cargo recognition via the PY-NLS have been studied extensively (2, 50–52). In contrast, little is known about the role of RG/RGG regions in TNPO1 recognition. Among the reported TNPO1 cargoes, we identified 61 proteins harboring a combination of a PY-NLS and an RG/RGG region (Fig. 1A and Dataset S1). Here, we show that a flexible, acidic loop in TNPO1 allows specific binding to the RG/RGG region, whereas the inner acidic groove of TNPO1 is involved in both RG/RGG and PY-NLS recognition (Fig. 4). Several models or a combination thereof could explain the role of the flexible TNPO1 acidic loop in cargo binding: the “fly-casting” effect or an increase in the number of collisions leading to productive binding (53). In the two-step fly-casting binding mechanism (54, 55), the flexible TNPO1 acidic loop screens, with a high capture radius, for binding partners and binds the positively charged RG/RGG regions even at a relatively large distance from the structured core of TNPO1. In the second step, the PY motif gets recruited, binds, and folds inside the inner groove of TNPO1. Through this mechanism, the binding rate can be enhanced over the rate of binding to a fully folded protein. However, the bound state could still undergo conformational exchange both within the PY-NLS site as well as within the RG/RGG regions, with the RG/RGG region being more likely to undergo conformational exchange in the bound state. Although fly casting is difficult to prove or disprove experimentally, it is further supported by the observation that electrostatic attraction, as in this case between the positively charged RG/RGG motif and the negatively charged flexible TNPO1 loop, can enhanced binding rates (55). On the contrary, the slower translational diffusion of TNPO1 with the disordered loop could, in part, offset the kinetic advantage of an increased capture radius (53, 56). Alternatively, the TNPO1 disordered loop could speed up association with RG/RGG and PY-NLS–containing cargo proteins by a reduction of the free energy barrier between initial and final complex due to an increase in the number of collisions leading to productive binding (53, 56). As in the fly-casting process, electrostatic interactions might play an important role in this process by reducing the redissociation rate after initial encounter. Moreover, electrostatic interactions might increase the probability of native-like topologies in the collision complexes (53, 57). In contrast to the fly-casting mechanism, the flexible TNPO1 loop would help avoiding orientational restraints and steric hindrance (53, 58) and “guide” the PY-NLS to the structured core of TNPO1, with the help of the RG/RGG region.
Our study provides a mechanistic explanation for the reported involvement of RG/RGG regions in nuclear import of several RBPs, including hnRNPA2 (59), TAF15 (60), SERBP1 (61), PABPN1 (62), and Sam68 (63). As CIRBP, SERBP1 does not possess a PY-NLS or any other known NLS, suggesting that the RG/RGG region in SERBP1 may be responsible for nuclear import. The interaction between TNPO1 and RG/RGG regions can be of high affinity, in line with our observation that the RG/RGG region of CIRBP binds TNPO1 with high affinity (Table 1 and SI Appendix, Fig. S3B), over 100 times stronger than the C-terminal RG/RGG region of FUS (11). It is tempting to speculate that TNPO1-mediated nuclear import could generally be used for the large class of RG/RGG-containing proteins (13).
TNPO3 Recognizes an RSY-Rich NLS with High Affinity.
The molecular determinants involved in TNPO3-mediated cargo recognition are poorly understood. TNPO3 has been proposed to recognize phosphorylated RS-rich regions in RBPs (classical RS-NLS) (29). Recently, Jang et al. (64) showed that, while an (RS)4 repeat peptide does not interact with TNPO3, its phosphorylated version binds to TNPO3 with micromolar affinity. Here, we present quantitative affinity data of TNPO3 for its native cargo CIRBP and show that its RSY-rich region binds with higher affinity at a low-nanomolar Kd in the absence of phosphorylation. This is comparable with the affinity reported to be necessary for nuclear import of cargo in cells (9, 11, 65). This finding furthermore suggests that unmodified RS-like regions could serve as NLS for TNPO3 in absence of any posttranslational modifications, extending our knowledge of the RS/RS-like NLS consensus sequence. In addition to the arginine residues, the tyrosine residues in this region are essential for TNPO3 binding (SI Appendix, Fig. S3 E and F). Basic Local Alignment Tool for proteins analysis of the CIRBP RSY-NLS against the human protein database identified several similar regions present in RS domain-containing proteins involved in RNA metabolism (SI Appendix, Fig. S6). Notably, such RSY-rich regions are present in SRSF4 and SRFS6 and are located in the flexible linker between the two RRM domains. In addition to the putative RSY-rich NLS, SRSF4 and SRSF6 contain a classical RS-NLS for TNPO3 in their C-terminal tails (SI Appendix, Fig. S6). In contrast, the RSY-rich region of TRA2A is located next to its RS-2 domain (SI Appendix, Fig. S6). RSY-rich regions can be found in several other proteins (SI Appendix, Fig. S6). The lack of a specific and conserved amino acid consensus sequence in the superposition of these regions suggests that the RSY NLS is degenerate and a dynamic binding region. Nevertheless, the detailed relationship of transportin binding/specificity and distribution of R, S, Y and R, G in RSY-rich and RG/RGG-rich regions remains to be determined. Similar to as in CIRBP, the presence of a putative high-affinity TNPO3 binding region (i.e., RSY region) could mediate phosphorylation-independent nuclear import of these RBPs.
Given that we identified two distinct regions (RG/RGG rich and RSY rich) in CIRBP to be involved in its nuclear import, we carried out a bioinformatics analysis focusing on the co-occurrence of RG/RGG and RS/RS-like regions. Strikingly, other than CIRBP, numerous other proteins harbor a combination of RG/RGG and RS/RS-like regions (e.g., LENG8, RBMX, SCAF15, SRSF5) (Fig. 6A and Dataset S2). We propose that nuclear import of these proteins may be mediated by both TNPO1 and TNPO3. We envision that the presence of multiple TNPO1 and/or TNPO3 binding regions together with posttranslational modifications of these regions, such as arginine methylation and phosphorylation, could fine tune nuclear import, phase separation, and SG recruitment of numerous RBPs and in turn, regulate their (patho-)physiological function.
TNPO1 and TNPO3 Suppress CIRBP Phase Separation and Stress Granule Recruitment.
We found that TNPO3 has “chaperone”-like activity toward CIRBP and inhibits its phase separation and SG recruitment (Fig. 2), like the recently discovered chaperone TNPO1 (1–3). As both TNPO1 and TNPO3 suppress phase separation of CIRBP and its localization in SGs, they might be involved in regulating CIRBP function in cell adaption to stress, possibly allowing release of CIRBP-bound RNA targets and making them available for the translation machinery. Other than CIRBP, several proteins that contain an NLS for TNPO3 localize to cellular condensates, including ASF/SF2 (66) and SRSF3 (67, 68) in humans and several serine/arginine-rich proteins in Caenorhabditis elegans (69), suggesting a general role of TNPO3 in chaperoning its cargo RBPs. Numerous studies have underscored a critical role of SGs in neurodegenerative disorders (70–72). Indeed, several proteins that undergo LLPS are found mutated in such disorders, including TDP-43, FUS, hnRNPA1, and Tau, and disease-linked mutations promote aberrant phase transitions and SG recruitment of the mutant proteins (1, 70, 73–77). Therefore, a deeper understanding of the molecular mechanisms regulating phase separation and SG dynamics is a critical step toward the development of therapeutic approaches. Our data revealing regulators of phase separation and SG recruitment will contribute to this end.
CIRBP Nuclear Import Is Regulated by Arginine Methylation of the RG/RGG Region.
RG/RGG regions are often subject to arginine methylation (13), which can regulate transportin–cargo interaction as previously shown for FUS (11, 48). Here, we show that the RG/RGG region of CIRBP is methylated by PRMT1 and that arginine methylation interferes with both TNPO1 and TNPO3 binding (Fig. 5). Depending on the methylation status of the CIRBP RG/RGG region and the relative affinity of each NLS for TNPO1 and TNPO3, we propose two different import scenarios: 1) if the RG/RGG region of CIRBP is unmethylated, active nuclear import of CIRBP is mediated by both the RG/RGG-NLS (TNPO1) and the RSY-NLS (TNPO3), and 2) if the RG/RGG region of CIRBP is methylated, active nuclear import of CIRBP is mediated solely by the RSY-NLS (TNPO3) (Fig. 6B). Although detailed studies are needed in the future, first indications of the roles of arginine methylation in regulating nuclear import and disease pathophysiology have been published by us and others. In the case of the RBPs CIRBP (Fig. 5), FUS (11), and SERBP1 (61), pharmacological inhibition of arginine methylation enhances nuclear localization the corresponding RBP. Alteration in arginine methylation of RG/RGG-containing proteins is associated with a plethora of neurodegenerative diseases and cancers (11, 13, 48, 63, 78–80). In line with this, PRMT1 mRNA levels are increased in several cancer cells and tumors (81–83). Moreover, PRMT1 has been shown to regulate epithelial–mesenchymal transition, cancer cell migration, and invasion, which are hallmarks of cancer progression and metastasis (84). Anomalous PRMT1 expression/distribution might alter cell localization and function of the RG/RGG proteome. Detailed understanding of the molecular mechanisms of RG/RGG-mediated nuclear import is, therefore, essential for the development of therapeutic approaches that may benefit patients with amyotrophic lateral sclerosis or frontotemporal dementia (FUS), brain ischemia (CIRBP), or neurodegenerative disease and cancer in general. Given that the RG/RGG region is highly abundant in the human proteome (13), we anticipate that many more diseases linked to misregulation of RG/RGG regions will appear in the upcoming years.
Materials and Methods
Plasmids.
Constructs for mammalian cell expression of CIRBP or for Escherichia coli expression of human CIRBP, TNPO1, TNPO3, PRMT1, and FUS have been obtained as described in SI Appendix.
Protein Expression and Purification.
For expression of recombinant ZZ-His6 proteins, the different bacterial expression pETM11-ZZ-His6 vectors were transformed into E. coli BL21-DE3 Star strain and purified using Ni-NTA agarose beads (Qiagen), and the ZZ-His6 tag was cleaved with Tobacco etch virus (TEV) protease treatment. A final size exclusion chromatography purification step was performed in the buffer of interest. Detailed information and protocols are in SI Appendix.
cDNA and siRNA Transfection.
Complementary DNA (cDNA) and siRNA transfection for analysis of cellular localization in HeLa Kyoto cells and HeLa S3 cells is described in detail in SI Appendix.
Cell Culture.
HeLa S3 cells were seeded on 1.5-H high-precision glass coverslips (Marienfeld-Superior) and cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Sigma-Aldrich) containing 10% fetal bovine serum (FBS) (Gibco; Thermo Fisher Scientific), penicillin (100 U/mL), streptomycin (100 µg/mL), and amphotericin B (1.25 µg/mL; all Gibco) in a humidified incubator (37 °C, 5% CO2/95% air). HeLa Kyoto cells were grown in DMEM high-glucose GlutaMAX (Invitrogen) supplemented with 10% FBS and 10 µg/mL gentamicin. Note that for GCR reporter assays, cells were maintained using DMEM supplemented with 10% dialyzed FBS (Invitrogen). Cells were maintained in a humidified incubator at 37 °C with 5% CO2. In experiments involving siRNAs and for cDNA transfections, antibiotics were omitted from the cell culture medium. To induce the import of GCR2-GFP2 reporter proteins, cells were grown on poly-l-lysine–coated coverslips and were treated with 5 μM dexamethasone (Sigma) for the times indicated at 37 °C.
Localization Analysis.
For analysis of the localization of GST-GFP-CIRBP, HeLa Kyoto cells were fixed in 3.7% formaldehyde/phosphate-buffered saline, nuclei were stained using 4′,6-diamidino-2-phenylindole (Sigma; 0.5 µg/mL final concentration), and coverslips mounted in Prolong Diamond (Thermo Fisher Sc). Cells were subsequently analyzed by fluorescence microscopy (SI Appendix). For the analysis of import efficiencies, cells were scored on a wide-field fluorescence microscope (Zeiss) (SI Appendix) into the following categories: n > C (more protein in the nucleus than the cytoplasm), n = C (equal distribution of the reporter protein between nucleus and cytoplasm), and n < C (more reporter protein in the cytoplasm than the nucleus).
Semipermeabilized Cell Assay.
HeLa P4 cells were grown on poly-l-lysine–coated 12-mm coverslips No. 1.5. Cells were stressed with 10 µM MG132 for 3 h to induce SGs, permeabilized with 0.003 to 0.005% digitonin in potassium phosphate buffer (KPB) (20 mM potassium phosphate, pH 7.4, 5 mM MgCl2, 200 mM KOAc, 1 mM ethylene glycol tetraacetic acid, 2 mM dithiothreitol (DTT), 1 μg/mL each aprotinin, pepstatin, and leupeptin). After several washes, nuclear pores were blocked by 15 min of incubation with 200 μg/mL wheat germ agglutinin (WGA) on ice. Cells were then incubated for 30 min at room temperature with 200 nM MBP-CIRBP-EGFP in the absence or presence of increasing concentrations of His-TNPO1 or His-TNPO3, respectively, in KPB. After several stringent washes, cells were fixed, and SGs were visualized by immunostaining of G3BP1 and confocal microscopy (SI Appendix).
Image Processing and Statistics.
All microscopy images were processed using Fiji/ImageJ software, applying linear enhancement for brightness and contrast. Statistical analyses were performed in GraphPad Prism 5.
Pull-Down Assay.
Ni-NTA beads (Qiagen) were equilibrated in wash buffer (50 mM Na2HPO4/NaH2PO4, pH 7.5, 150 mM NaCl, 20 mM imidazole, 4 mM β-mercaptoethanol [βME]) and blocked in wash buffer supplemented with 0.3 mg/mL bovine serum albumin (BSA). HeLa cells were lysed in lysis buffer (50 mM Na2HPO4/NaH2PO4, pH 7.5, 150 mM NaCl, 20 mM imidazole, 4 mM βME, 1× protease inhibitor). Supernatant was preincubated with half of previously blocked Ni-NTA beads for 1 h at 4 °C. Precleared cell lysate was equally distributed to 50 µL BSA-blocked Ni-NTA beads and supplemented with 200 µg unmethylated and methylated CIRBP full-length protein. Reactions in a final volume of 1.5 mL were incubated for 2 h at 4 °C on a rotary shaker. Beads were washed three times in wash buffer, resuspended in 2× sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS/PAGE) sample buffer, and boiled for 5 min. Eluted proteins were separated by SDS/PAGE (10 to 20%) and detected by western blot using specific antibodies for TNPO1 (Sigma; clone D45, #T0825), TNPO3 (abcam; clone 3152C2a, #ab54353), and His6 (abcam; ab18184). Note that, due to the small size of recombinant CIRBP, CIRBP could not be detected by standard western blotting techniques. Instead, two layers of 0.22 µm of nitrocellulose membrane were used. TNPO1 and TNPO3 were detected on the first layer, CIRBP was detected on the second layer, and band intensities were analyzed with ImageJ software and normalized to signal intensity of unmethylated CIRBP full length. Statistical analyses were performed in GraphPad Prism 5.
ITC.
All proteins samples were equilibrated in the same buffer containing 50 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, and 2 mM Tris(2-carboxyethyl)phosphine. ITC measurements were taken with a MicroCal VP-ITC instrument (Microcal) with 36 rounds of 8-μL injections at 25 °C. Integration of peaks corresponding to each injection, subtraction of the contribution of protein dilution, and correction for the baseline were performed using the Origin-based 7.0 software provided by the manufacturer. Curve fitting was done with a standard one-site model and gives the equilibrium binding constant (Ka) and enthalpy of the complex formation (ΔH).
NMR.
Binding assay.
All proteins samples were equilibrated in the same buffer containing 50 mM Tris⋅HCl, pH 6.7, 150 mM NaCl, and 2 mM Tris(2-carboxyethyl)phosphine. NMR experiments were performed at 25 °C on Bruker 600- and 900-MHz spectrometers equipped with a triple-resonance cryoprobe. Other than the 1H-15N HSQC spectrum, the following 3D spectra were acquired for assignment of the CIRBP and TNPO1 reference samples (CIRBPRGG, CIRBPRSY, TNPO1, and TNPO1loop): HNCO, HN(CA)CO, HNCACB, CBCA(CO)NH, (H)CC(CO)NH-TOCSY, HN(CA)NNH, and H(NCA)NNH using between 400 and 700 µM 1H-15N-13C–labeled proteins and 10% deuterium oxide. Spectra were processed using NMRpipe (85).
Phase separation assay.
All proteins/RNA (12 × UG repeats) samples were prepared in 20 mM Na2HPO4/NaH2PO4, pH 7.5, 75 mM NaCl, 2.5% glycerol, 1 mM DTT, and 10% 2H2O added for the lock signal. NMR experiments were performed at 25 °C on a Bruker 600-MHz spectrometer. Spectra were processed using Topspin 3.5 and Mnova 11.
Turbidity Assay.
All proteins/RNA (12 × UG repeats) samples were prepared in 20 mM Na2HPO4/NaH2PO4, pH 7.5, 75 mM NaCl, 2.5% glycerol, and 1 mM DTT. Turbidity measurements were conducted at 620 nm in 96-well plates with 90-µL samples using a BioTek Power Wave HT plate reader (BioTek). All experiments were performed in triplicates.
Differential Interference Contrast Microscopy.
All proteins/RNA (12 × UG repeats) samples were prepared in 20 mM Na2HPO4/NaH2PO4, pH 7.5, 75 mM NaCl, 2.5% glycerol, and 1 mM DTT. The 25-µL sample was plated on a 30-mm No. 1 round glass coverslip and mounted on an Observer D1 microscope with 100×/1.45 oil immersion objective (Zeiss). Protein droplets were viewed using HAL 100 halogen lamp, and images were captured with an OrcaD2 camera (Hamamatsu) using VisiView 4.0.0.13 software (Visitron Systems GmbH). For the time course experiment, full-length CIRBP droplet formation was induced by diluting CIRBP from a 10× buffer to a 1× buffer at the concentration of interest in the absence or presence of RNA. For CIRBPRGG, droplet formation was induced after addition of RNA.
In Vitro Methylation Assay.
Untagged CIRBPRGG and full-length CIRBP recombinant proteins and His6-PRMT1 were equilibrated in methylation buffer containing 50 mM Na-phosphate, pH 8, 150 mM NaCl, and 2 mM Tris(2-carboxyethyl)phosphine; 50 µM CIRBPRGG or full-length CIRBP was incubated with 7 µM His6-PRMT1 in the presence of 2 mM S-Adenosylmethionine (New England Biolabs) for 16 h at room temperature. Untagged methylated CIRBPRGG (meCIRBPRGG) and full-length CIRBP (meCIRBP) were then isolated from PRMT1 performing a second affinity purification using Ni-NTA beads, further equilibrated in the buffer of interest, and analyzed using NMR (SI Appendix).
Data Availability Statement.
All data discussed in this study are included in the text and SI Appendix.
Supplementary Material
Acknowledgments
B.G. is a fellow of the Doctoral College Metabolic and Cardiovascular Disease at the Medical University of Graz and was funded by the Austrian Science Foundation (FWF) Grant W 1226-B18 (to W.F.G.) and Nikon Austria. D.D. was supported by Deutsche Forschungsgemeinschaft (DFG; German Research Foundation) Emmy Noether Grants DO1804/1-1, DO1804/3-1, and Schwerpunktprogramm SPP2191 DO1804/4-1 and Germany’s Excellence Strategy within the framework of the Munich Cluster for Systems Neurology Grant EXC 2145 SyNergy ID 390857198. D.D. also acknowledges support from the Fritz Thyssen Foundation and the Paul Ehrlich Foundation. T.M. was supported by Austrian Science Foundation Grants P28854, I3792, and W1226; Austrian Research Promotion Agency (FFG) Grants 864690 and 870454; the Integrative Metabolism Research Center Graz; Austrian Infrastructure Program 2016/2017, the Styrian Government (Zukunftsfonds), and BioTechMed-Graz. Equipment for superresolution microscopy is part of the Nikon Center of Excellence, Graz that is supported by Austrian Infrastructure Program 2013/2014, Nikon Austria Inc., and BioTechMed. We thank the Center for Medical Research, Medical University of Graz, Graz, Austria for laboratory access. We thank Michael Kiebler for providing laboratory infrastructure, reagents, and access to the Zeiss Axio Observer.Z1 microscope and the spinning disc microscope (DFG; Grant INST 86/1581-1 FUGG). We also thank Dr. Vanessa Morris for carefully reading the manuscript. We thank the BioMedical Center core facility Bioimaging.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918944117/-/DCSupplemental.
References
- 1.Hofweber M., et al. , Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell 173, 706–719.e3 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Yoshizawa T., et al. , Nuclear import receptor inhibits phase separation of FUS through binding to multiple sites. Cell 173, 693–705.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo L., et al. , Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with prion-like domains. Cell 173, 677–692.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qamar S., et al. , FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell 173, 720–734.e15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chook Y. M., Blobel G., Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 11, 703–715 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Soniat M., Chook Y. M., Nuclear localization signals for four distinct karyopherin-β nuclear import systems. Biochem. J. 468, 353–362 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Frey S., et al. , Surface properties determining passage rates of proteins through nuclear pores. Cell 174, 202–217.e9 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Cook A., Bono F., Jinek M., Conti E., Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 76, 647–671 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Lee B. J., et al. , Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell 126, 543–558 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Twyffels L., Gueydan C., Kruys V., Transportin-1 and transportin-2: Protein nuclear import and beyond. FEBS Lett. 588, 1857–1868 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Dormann D., et al. , Arginine methylation next to the PY-NLS modulates Transportin binding and nuclear import of FUS. EMBO J. 31, 4258–4275 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Göbl C., et al. , Increasing the chemical-shift dispersion of unstructured proteins with a covalent lanthanide shift reagent. Angew. Chem. Int. Ed. Engl. 55, 14847–14851 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thandapani P., O’Connor T. R., Bailey T. L., Richard S., Defining the RGG/RG motif. Mol. Cell 50, 613–623 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Chong P. A., Vernon R. M., Forman-Kay J. D., RGG/RG motif regions in RNA binding and phase separation. J. Mol. Biol. 430, 4650–4665 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Boeynaems S., et al. , Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol. Cell 65, 1044–1055.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimura M., et al. , Extensive cargo identification reveals distinct biological roles of the 12 importin pathways. eLife 6, e21184 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mackmull M. T., et al. , Landscape of nuclear transport receptor cargo specificity. Mol. Syst. Biol. 13, 962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aoki K., Ishii Y., Matsumoto K., Tsujimoto M., Methylation of Xenopus CIRP2 regulates its arginine- and glycine-rich region-mediated nucleocytoplasmic distribution. Nucleic Acids Res. 30, 5182–5192 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Leeuw F., et al. , The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor. Exp. Cell Res. 313, 4130–4144 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Fornace A. J. Jr, Alamo I. Jr, Hollander M. C., DNA damage-inducible transcripts in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 85, 8800–8804 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan F., Zarate J., Choudhury A., Rupprecht R., Bradley T. M., Osmotic stress of salmon stimulates upregulation of a cold inducible RNA binding protein (CIRP) similar to that of mammals and amphibians. Biochimie 86, 451–461 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Zhou M., Yang W. L., Ji Y., Qiang X., Wang P., Cold-inducible RNA-binding protein mediates neuroinflammation in cerebral ischemia. Biochim. Biophys. Acta 1840, 2253–2261 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakurai T., et al. , Cirp protects against tumor necrosis factor-alpha-induced apoptosis via activation of extracellular signal-regulated kinase. Biochim. Biophys. Acta 1763, 290–295 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Yang C., Carrier F., The UV-inducible RNA-binding protein A18 (A18 hnRNP) plays a protective role in the genotoxic stress response. J. Biol. Chem. 276, 47277–47284 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Lee H. N., Ahn S. M., Jang H. H., Cold-inducible RNA-binding protein, CIRP, inhibits DNA damage-induced apoptosis by regulating p53. Biochem. Biophys. Res. Commun. 464, 916–921 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Lujan D. A., Ochoa J. L., Hartley R. S., Cold-inducible RNA binding protein in cancer and inflammation. Wiley Interdiscip. Rev. RNA 9, e1462 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng Y., Kulkarni P., Inoue T., Getzenberg R. H., Down-regulating cold shock protein genes impairs cancer cell survival and enhances chemosensitivity. J. Cell. Biochem. 107, 179–188 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love D. C., Sweitzer T. D., Hanover J. A., Reconstitution of HIV-1 rev nuclear export: Independent requirements for nuclear import and export. Proc. Natl. Acad. Sci. U.S.A. 95, 10608–10613 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai M. C., Lin R. I., Tarn W. Y., Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc. Natl. Acad. Sci. U.S.A. 98, 10154–10159 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yun C. Y., Velazquez-Dones A. L., Lyman S. K., Fu X. D., Phosphorylation-dependent and -independent nuclear import of RS domain-containing splicing factors and regulators. J. Biol. Chem. 278, 18050–18055 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Maertens G. N., et al. , Structural basis for nuclear import of splicing factors by human Transportin 3. Proc. Natl. Acad. Sci. U.S.A. 111, 2728–2733 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Popken P., Ghavami A., Onck P. R., Poolman B., Veenhoff L. M., Size-dependent leak of soluble and membrane proteins through the yeast nuclear pore complex. Mol. Biol. Cell 26, 1386–1394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Timney B. L., et al. , Simple rules for passive diffusion through the nuclear pore complex. J. Cell Biol. 215, 57–76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dormann D., et al. , ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 29, 2841–2857 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ozdilek B. A., et al. , Intrinsically disordered RGG/RG domains mediate degenerate specificity in RNA binding. Nucleic Acids Res. 45, 7984–7996 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maris C., Dominguez C., Allain F. H., The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 272, 2118–2131 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Loughlin F. E., et al. , The solution structure of FUS bound to RNA reveals a bipartite mode of RNA recognition with both sequence and shape specificity. Mol. Cell 73, 490–504.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Maharana S., et al. , RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ukmar-Godec T., et al. , Lysine/RNA-interactions drive and regulate biomolecular condensation. Nat. Commun. 10, 2909 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bogaert E., et al. , Molecular dissection of FUS points at synergistic effect of low-complexity domains in toxicity. Cell Rep. 24, 529–537.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J., et al. , A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174, 688–699.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu Y., Matthews S., TROSY NMR spectroscopy of large soluble proteins. Top. Curr. Chem. 335, 97–119 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Tugarinov V., Hwang P. M., Kay L. E., Nuclear magnetic resonance spectroscopy of high-molecular-weight proteins. Annu. Rev. Biochem. 73, 107–146 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Imasaki T., et al. , Structural basis for substrate recognition and dissociation by human transportin 1. Mol. Cell 28, 57–67 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Cansizoglu A. E., Chook Y. M., Conformational heterogeneity of karyopherin beta2 is segmental. Structure 15, 1431–1441 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Z. C., Chook Y. M., Structural and energetic basis of ALS-causing mutations in the atypical proline-tyrosine nuclear localization signal of the Fused in Sarcoma protein (FUS). Proc. Natl. Acad. Sci. U.S.A. 109, 12017–12021 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chook Y. M., Blobel G., Structure of the nuclear transport complex karyopherin-beta2-Ran x GppNHp. Nature 399, 230–237 (1999). [DOI] [PubMed] [Google Scholar]
- 48.Suárez-Calvet M., et al. , Monomethylated and unmethylated FUS exhibit increased binding to Transportin and distinguish FTLD-FUS from ALS-FUS. Acta Neuropathol. 131, 587–604 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Guo A., et al. , Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol. Cell. Proteomics 13, 372–387 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huber F. M., Hoelz A., Molecular basis for protection of ribosomal protein L4 from cellular degradation. Nat. Commun. 8, 14354 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cansizoglu A. E., Lee B. J., Zhang Z. C., Fontoura B. M., Chook Y. M., Structure-based design of a pathway-specific nuclear import inhibitor. Nat. Struct. Mol. Biol. 14, 452–454 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soniat M., et al. , Crystal structure of human Karyopherin β2 bound to the PY-NLS of Saccharomyces cerevisiae Nab2. J. Struct. Funct. Genomics 14, 31–35 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mollica L., et al. , Binding mechanisms of intrinsically disordered proteins: Theory, simulation, and experiment. Front. Mol. Biosci. 3, 52 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shoemaker B. A., Portman J. J., Wolynes P. G., Speeding molecular recognition by using the folding funnel: The fly-casting mechanism. Proc. Natl. Acad. Sci. U.S.A. 97, 8868–8873 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou H. X., Bates P. A., Modeling protein association mechanisms and kinetics. Curr. Opin. Struct. Biol. 23, 887–893 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang Y., Liu Z., Kinetic advantage of intrinsically disordered proteins in coupled folding-binding process: A critical assessment of the “fly-casting” mechanism. J. Mol. Biol. 393, 1143–1159 (2009). [DOI] [PubMed] [Google Scholar]
- 57.Ganguly D., Zhang W., Chen J., Electrostatically accelerated encounter and folding for facile recognition of intrinsically disordered proteins. PLoS Comput. Biol. 9, e1003363 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou H. X., Pang X., Lu C., Rate constants and mechanisms of intrinsically disordered proteins binding to structured targets. Phys. Chem. Chem. Phys. 14, 10466–10476 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nichols R. C., et al. , The RGG domain in hnRNP A2 affects subcellular localization. Exp. Cell Res. 256, 522–532 (2000). [DOI] [PubMed] [Google Scholar]
- 60.Marko M., Vlassis A., Guialis A., Leichter M., Domains involved in TAF15 subcellular localisation: Dependence on cell type and ongoing transcription. Gene 506, 331–338 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Lee Y. J., Hsieh W. Y., Chen L. Y., Li C., Protein arginine methylation of SERBP1 by protein arginine methyltransferase 1 affects cytoplasmic/nuclear distribution. J. Cell. Biochem. 113, 2721–2728 (2012). [DOI] [PubMed] [Google Scholar]
- 62.Fronz K., et al. , Arginine methylation of the nuclear poly(a) binding protein weakens the interaction with its nuclear import receptor, transportin. J. Biol. Chem. 286, 32986–32994 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Côté J., Boisvert F. M., Boulanger M. C., Bedford M. T., Richard S., Sam68 RNA binding protein is an in vivo substrate for protein arginine N-methyltransferase 1. Mol. Biol. Cell 14, 274–287 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jang S., et al. , Differential role for phosphorylation in alternative polyadenylation function versus nuclear import of SR-like protein CPSF6. Nucleic Acids Res. 47, 4663–4683 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chook Y. M., Süel K. E., Nuclear import by karyopherin-βs: Recognition and inhibition. Biochim. Biophys. Acta 1813, 1593–1606 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Delestienne N., et al. , The splicing factor ASF/SF2 is associated with TIA-1-related/TIA-1-containing ribonucleoproteic complexes and contributes to post-transcriptional repression of gene expression. FEBS J. 277, 2496–2514 (2010). [DOI] [PubMed] [Google Scholar]
- 67.Kano S., et al. , Oxidative stress-inducible truncated serine/arginine-rich splicing factor 3 regulates interleukin-8 production in human colon cancer cells. Am. J. Physiol. Cell Physiol. 306, C250–C262 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Jayabalan A. K., et al. , NEDDylation promotes stress granule assembly. Nat. Commun. 7, 12125 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J. T., et al. , Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. eLife 3, e04591 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Y. R., King O. D., Shorter J., Gitler A. D., Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 201, 361–372 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bentmann E., Haass C., Dormann D., Stress granules in neurodegeneration–lessons learnt from TAR DNA binding protein of 43 kDa and fused in sarcoma. FEBS J. 280, 4348–4370 (2013). [DOI] [PubMed] [Google Scholar]
- 72.Zhang P., et al. , Chronic optogenetic induction of stress granules is cytotoxic and reveals the evolution of ALS-FTD pathology. eLife 8, e39578 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Conicella A. E., Zerze G. H., Mittal J., Fawzi N. L., ALS mutations disrupt phase separation mediated by α-Helical structure in the TDP-43 low-complexity C-terminal domain. Structure 24, 1537–1549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Walker A. K., et al. , ALS-associated TDP-43 induces endoplasmic reticulum stress, which drives cytoplasmic TDP-43 accumulation and stress granule formation. PLoS One 8, e81170 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Murakami T., et al. , ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron 88, 678–690 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Patel A., et al. , A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 (2015). [DOI] [PubMed] [Google Scholar]
- 77.Wegmann S., et al. , Tau protein liquid-liquid phase separation can initiate tau aggregation. EMBO J. 37, e98049 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tockman M. S., et al. ; LCEWDG Investigators. Lung Cancer Early Detection Group , The early detection of second primary lung cancers by sputum immunostaining. Chest 106 (suppl. 6), 385S–390S (1994). [DOI] [PubMed] [Google Scholar]
- 79.Zhou J., et al. , Purification and characterization of a protein that permits early detection of lung cancer. Identification of heterogeneous nuclear ribonucleoprotein-A2/B1 as the antigen for monoclonal antibody 703D4. J. Biol. Chem. 271, 10760–10766 (1996). [DOI] [PubMed] [Google Scholar]
- 80.Liao W. T., et al. , High expression level and nuclear localization of Sam68 are associated with progression and poor prognosis in colorectal cancer. BMC Gastroenterol. 13, 126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 81.Chuang C. Y., et al. , PRMT1 expression is elevated in head and neck cancer and inhibition of protein arginine methylation by adenosine dialdehyde or PRMT1 knockdown downregulates proliferation and migration of oral cancer cells. Oncol. Rep. 38, 1115–1123 (2017). [DOI] [PubMed] [Google Scholar]
- 82.Goulet I., Gauvin G., Boisvenue S., Côté J., Alternative splicing yields protein arginine methyltransferase 1 isoforms with distinct activity, substrate specificity, and subcellular localization. J. Biol. Chem. 282, 33009–33021 (2007). [DOI] [PubMed] [Google Scholar]
- 83.Yoshimatsu M., et al. , Dysregulation of PRMT1 and PRMT6, Type I arginine methyltransferases, is involved in various types of human cancers. Int. J. Cancer 128, 562–573 (2011). [DOI] [PubMed] [Google Scholar]
- 84.Avasarala S., et al. , PRMT1 is a novel regulator of epithelial-mesenchymal-transition in non-small cell lung cancer. J. Biol. Chem. 290, 13479–13489 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Delaglio F., et al. , NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data discussed in this study are included in the text and SI Appendix.