Significance
The emergence of influenza viruses with reduced susceptibility to baloxavir marboxil (BXM) would limit the clinical utility of this novel antiviral. To assess the risk of such resistance emerging, we evaluated influenza A and B viruses carrying BXM-reduced susceptibility substitutions and compared their fitness to that of their drug-susceptible wild-type (I38-WT) counterparts. The 38T/F/M substitutions inhibited activity of the virus PA protein, and two of them (38T/F) hindered virus replication in cells. Even so, 38T/F/M viruses could transmit between ferrets, the gold-standard model for human transmission. These findings argue that there is a risk of transmission of BXM-resistant viruses from treated individuals. Whether such viruses could compete with WT viruses in spreading through the wider untreated community is less clear.
Keywords: influenza, endonuclease inhibitor, baloxavir marboxil, PA protein, I38T substitution
Abstract
Baloxavir marboxil (BXM) was approved in 2018 for treating influenza A and B virus infections. It is a first-in-class inhibitor targeting the endonuclease activity of the virus polymerase acidic (PA) protein. Clinical trial data revealed that PA amino acid substitutions at residue 38 (I38T/F/M) reduced BXM potency and caused virus rebound in treated patients, although the fitness characteristics of the mutant viruses were not fully defined. To determine the fitness impact of the I38T/F/M substitutions, we generated recombinant A/California/04/2009 (H1N1)pdm09, A/Texas/71/2017 (H3N2), and B/Brisbane/60/2008 viruses with I38T/F/M and examined drug susceptibility in vitro, enzymatic properties, replication efficiency, and transmissibility in ferrets. Influenza viruses with I38T/F/M substitutions exhibited reduced baloxavir susceptibility, with 38T causing the greatest reduction. The I38T/F/M substitutions impaired PA endonuclease activity as compared to that of wild-type (I38-WT) PA. However, only 38T/F A(H3N2) substitutions had a negative effect on polymerase complex activity. The 38T/F substitutions decreased replication in cells among all viruses, whereas 38M had minimal impact. Despite variable fitness consequences in vitro, all 38T/M viruses disseminated to naive ferrets by contact and airborne transmission, while 38F-containing A(H3N2) and B viruses failed to transmit via the airborne route. Reversion of 38T/F/M to I38-WT was rare among influenza A viruses in this study, suggesting stable retention of 38T/F/M genotypes during these transmission events. BXM reduced susceptibility-associated mutations had variable effects on in vitro fitness of influenza A and B viruses, but the ability of these viruses to transmit in vivo indicates a risk of their spreading from BXM-treated individuals.
Epidemic spread of influenza A and B viruses occurs annually in the Northern and Southern Hemispheres. In the United States alone, the symptomatic influenza infection rates are 3 to 11% for the total population. Epidemiologic parameters vary by season, but it is estimated that influenza-associated infections result in 140,000 to 960,000 hospitalizations and 12,000 to 79,000 deaths each year (1, 2). This disease burden persists despite active vaccination programs, with the vaccine composition requiring annual reevaluation because of the rate of genetic change and associated antigenic drift. Current seasonal influenza virus vaccines are generally less effective in high-risk groups, including the young and the elderly, and they are unlikely to protect against antigenically novel influenza viruses entering humans through zoonotic transmission (3). In such cases, antiinfluenza drugs are critically important for protecting against virus infection and spread. Since the early 2000s, treatment of influenza infection has largely been dependent upon a single class of drugs, the neuraminidase (NA) inhibitors (NAIs). Overreliance on these drugs and natural virus evolution have raised the risk of NAI resistance among circulating viruses. This occurred in prepandemic/former-seasonal influenza A(H1N1) viruses that became resistant to the most commonly prescribed NAI, oseltamivir phosphate (OSE) (4).
Additional influenza inhibitors that target diverse virus proteins are urgently needed. Among these targets are the subunits of the influenza virus replication complex: the polymerase basic 1 (PB1), polymerase basic 2 (PB2), and the polymerase acidic (PA) proteins. Antivirals targeting these polymerase proteins have great potential because of the critical roles these proteins play in virus replication, and because their functional domains remain largely conserved (5). Inhibitors targeting each of these proteins have been developed, leading to drug approval (baloxavir marboxil [BXM], targeting PA) (6, 7), restricted use (favipiravir, targeting PB1) (8), or use in clinical trials (pimodivir, targeting PB2) (9). The most advanced of these antivirals has been BXM, marketed as Xofluza (10, 11) and approved since 2018/2019 in Japan, the United States, Singapore, Thailand, and Taiwan. BXM is a prodrug that is converted to the active metabolite baloxavir acid (BXA). BXA is commonly used for in vitro assays, as in this study. It specifically targets the endonuclease activity of the PA protein, the active site being located within the ∼200 amino acids of the N-terminal domain (PAN). This activity plays a critical role in cleaving away host messenger RNA (mRNA) caps that the virus uses to initiate transcription of its own genes. BXM inhibits both influenza A and B viruses, as their PAN domains are highly conserved (12, 13). BXM is approved for treatment of uncomplicated virus infections and is currently administered as a single age/weight-adjusted oral dose (40 mg to 80 mg). It alleviates symptoms in a time equivalent to that seen with OSE, the standard of care, and it enhances the reduction of patient virus load beginning 1 d posttreatment (14, 15). Thus, BXM shows great promise for treating influenza and bolsters our limited arsenal of antiinfluenza drugs.
The emergence of BXM-reduced susceptibility viruses is a concern given the rapid evolution rates of influenza viruses. In BXM-treated patients, viruses with PA protein substitutions of isoleucine (I) 38 to threonine (T), phenylalanine (F), or methionine (M) were shed at a rate of 2.3% in phase II studies and 9.7% in phase III studies (5, 13–15). These same PA substitutions induce a 7- to 50-fold reduction in drug susceptibility (13). A small pediatric trial reported that >23% of BXM-treated patients (18/77) shed viruses with 38T/M changes (5, 13, 16). In all trials, virus acquisition of 38T/F/M was associated with some rebound of shedding. In the phase III trial, patients shedding viruses with these mutations displayed delayed symptom resolution compared to patients shedding I38-WT virus (14). Routine surveillance for substitutions at residue 38 in the Asia−Pacific region and North America in 2012–2018 revealed a low incidence, with only the I38 to leucine (L) (in two viruses) and I38M (in one virus) being reported (6, 17). In Japan, where antivirals such as BXM are more commonly used (5, 18), at least 20 viruses with reduced susceptibility were isolated in the 2018/2019 influenza season, all from pediatric patients and all with I38T/F/M/N (asparagine)/S (serine) single or mixed substitutions (19–21).
With other antiinfluenza inhibitors, such as the NAIs, the acquisition of reduced susceptibility to a drug is sometimes associated with a loss of virus fitness, and this increases the threshold of resistance (22). Data concerning the fitness of BXM-reduced susceptibility viruses are limited and controversial. Omoto et al. (13) reported that viruses harboring the I38T substitution show severely impaired replicative fitness in cells and correspondingly reduced endonuclease activity in vitro. However, Checkmahomed et al. (23) demonstrated that the I38T substitution did not alter the replication kinetics of A(H1N1)pdm09 and A(H3N2) viruses in vitro or in mice. Further, the I38T/F/M fitness effects in influenza B virus backgrounds have not been well defined.
In the present study, we conducted a comprehensive investigation of I38T/F/M virus fitness in the background of influenza A(H1N1)pdm09, A(H3N2), and two influenza B viruses that represent seasonal circulating subtypes/lineages. We examined the effects of these substitutions on PA protein function, the activity of the polymerase complex, and virus replication in cells. Finally, we examined how these 38T/F/M viruses could transmit among naive ferrets through direct contact (DC) and airborne routes, in an effort to mimic dissemination of these mutant viruses from BXM-treated individuals. Our data indicate that acquisition of 38T/F/M substitutions caused fitness defects at the enzymatic and whole-virus levels in vitro, most notably among 38T/F viruses. However, most of these viruses were still capable of transmission in ferrets.
Results
Effects of I38T/F/M Substitutions on PA Protein Endonuclease Activity.
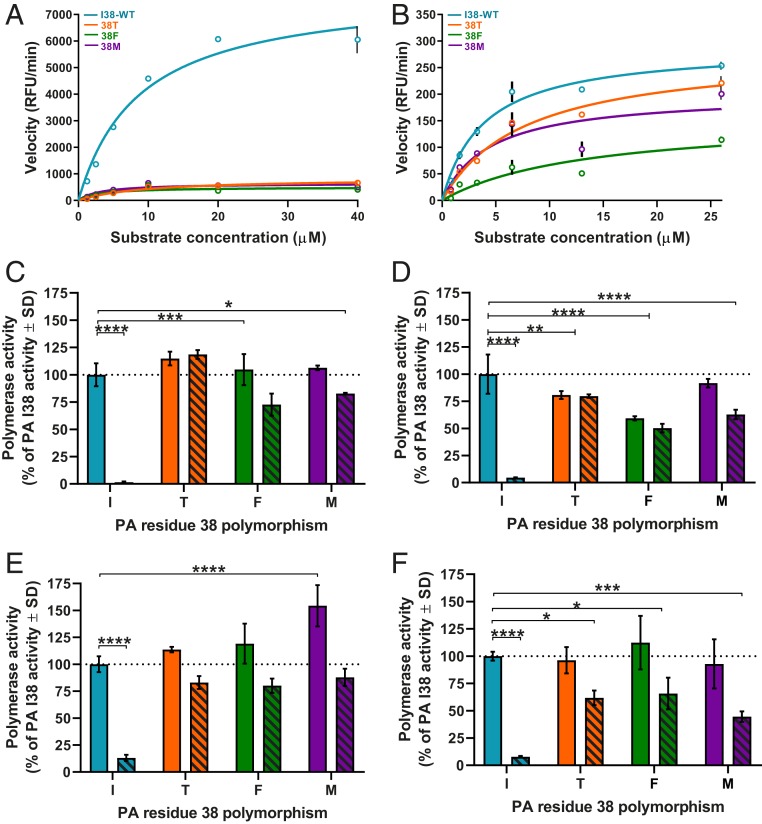
BXM targets the PAN domain of the PA protein. Therefore, we examined the effect of 38T/F/M substitutions on recombinant PAN (rPAN) from influenza A/California/04/2009 (H1N1)pdm09, (A/CA/04), and B/Brisbane/60/2008 (B/BR/60) viruses. PAN was incubated with a single-stranded substrate that fluoresced when cleaved (24). All 38T/F/M substitutions inhibited influenza A and B PAN enzyme progression, with the mutant constructs consistently emitting less relative fluorescence units (RFU) than was seen with I38-WT (Fig. 1 A and B). For influenza A PAN, the maximal velocity (Vmax) was 7,951 RFU/min for I38-WT compared to 835, 497, and 643 RFU/min for 38T, 38F, or 38M, respectively, representing a reduced Vmax of ≥ 9.5-fold compared to I38-WT (Fig. 1A). By comparison, 38T/F/M substitutions’ detrimental impact on enzyme velocity was less severe for influenza B PAN than A PAN. B PAN Vmax was 290 RFU/min for I38-WT compared to 281, 156, and 201 RFU/min for 38T, 38F, or 38M respectively (Fig. 1B), resulting in no reduction of velocity for 38T, and 1.4- to 1.8-fold reduction for 38F/M. Overall, 38T/F/M endonuclease domain substitutions have a negative effect on enzymatic fitness when compared to the I38-WT endonuclease, but this effect is most pronounced in influenza A PA protein.
Fig. 1.
Endonuclease and minireplicon polymerase complex activity of I38-WT and 38T/F/M. The rPAN (5 µg) from (A) A/CA/04 (H1N1)pdm09 or (B) B/BR/60 (Victoria lineage) with a single amino acid substitution at residue 38 of the PA protein (I38-WT and 38T/F/M) was incubated with fluorescently labeled ssDNA substrate. PAN enzymatic activity and reaction rates were determined from measured release of fluorescence upon substrate cleavage over time (RFU/min). Polymerase activity of (C) A/CA/04 (H1N1)pdm09, (D) A/TX/71 (H3N2), (E) B/BR/60 (Victoria lineage), or (F) B/PH/3073 (Yamagata lineage) was determined by minireplicon assay in HEK293T cells transfected with plasmids expressing virus proteins NP, PB1, and PB2, with a luciferase and β-galactosidase reporter. PA plasmids with the indicated PA residue 38 substitution were also transfected. Cells were either mock-treated (vehicle, DMSO, solid bars) or BXA-treated (10 nM, hashed bars) 1 h before transfection and 24 h or 48 h posttransfection. The polymerase activity luciferase output was normalized to β-galactosidase activity for each data point, then compared to the activity of the mock-treated I38-WT (100% activity, dotted line). Endonuclease data (in A and B) are presented as duplicate measures ± SD for each time point and concentration and are representative of at least three independent experiments. Minireplicon data (in C–F) are presented as at least duplicate measures ± SD and are representative of at least three independent assays for each virus polymerase complex. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001. Statistical comparison bar endcaps indicate differences from I38-WT mock-treated activity vs. an individual bar (┐) or both bars of the 38X substitution group (┤). Lack of comparison bars and P values on each graph represents “not significant.”
Effects of I38T/F/M Substitutions on Polymerase Complex Activity in the Minireplicon Assay.
The PAN domain does not function alone during virus replication; it is complexed within a trimer that includes the PB1 and PB2 proteins. Therefore, we studied the effects of 38T/F/M substitutions in the context of the polymerase complex by using a minireplicon reporter assay that produces luciferase as an indicator of polymerase complex activity (25). Accumulated luciferase in cells was measured 24 h to 48 h after transfection. We and others have demonstrated that 38T substitution causes no decrease in polymerase activity in this assay with influenza A(H1N1) polymerase complexes (26, 27). The same effect was observed for A(H1N1)pdm09 complexes with 38F/M substitutions (Fig. 1C). In A/Texas/71/2017 (A/TX/71, H3N2) polymerase complexes, 38M-substituted PA has polymerase activity equivalent to that of I38-WT PA, whereas the 38T/F substitutions decreased the activity by 19% and 41%, respectively (Fig. 1D). The I38M substitution increased polymerase activity by ∼50% in B/BR/60 complexes but had little effect on activity of B/Phuket/3073/2013 (B/PH/3073) complexes, compared to the respective I38-WT. No significant differences were observed with 38T/F with either B virus (Fig. 1 E and F).
PA 38T confers polymerase complex reduced susceptibility to a similar endonuclease inhibitor RO-7 (26). We hypothesized the same may be true for baloxavir. Therefore, minireplicon assays were conducted in the presence of 10 nM BXA. Acquisition of 38T/F/M substitutions allowed the retention of ≥44% polymerase activity of the respective I38-WT complexes, while I38-WT itself was highly susceptible to BXA inhibition (Fig. 1 C–F). Therefore, when PA 38T/F/M substitutions are present in the context of the entire polymerase complex, all provide reduced BXA susceptibility, while 38T/F cause fitness defects that are unique to the A/TX/71 virus.
I38T/F/M Virus Rescue and Confirmation of Reduced Susceptibility.
To understand the effects of PA 38 substitutions on virus replication in vitro, we rescued recombinant viruses containing the I38-WT or 38T/F/M substitution by reverse genetics (rg) (28). To represent seasonally circulating influenza viruses, the I38T/F/M variants were rescued in two influenza A backgrounds (A/CA/04 [H1N1] and A/TX/71 [H3N2]) and both lineages of influenza B (B/BR/60, Victoria, and B/PH/3073, Yamagata). We examined the BXA susceptibility of the 38T/F/M viruses in virus yield reduction assays in Madin−Darby canine kidney (MDCK) cells (Table 1). We observed ≈4-Log10 50% tissue culture infectious dose per ml (TCID50/mL) reduction of virus titers in influenza A I38-WT viruses with 1 nM BXA, and no replication at >1 nM. Influenza B I38-WT viruses had lower BXA susceptibility, replicating in up to 10 nM BXA, but with a 1- to 2-Log10TCID50/mL titer reduction compared to mock-treated wells. In stark contrast, influenza A and B 38T/F/M viruses retained their replication capacity at near−mock-treated levels in the presence of 1 nM or 10 nM BXA. Most 38T/M viruses and the B/PH/3073 38F virus replicated at 100 nM BXA, yielding titers of 0.8- to 5.4-Log10TCID50/mL and, overall, presenting a clear pattern of reduced BXA susceptibility (Table 1). To quantify this effect, we subjected each virus to plaque reduction assays and determined the BXA 50% effective concentrations (EC50s). All I38-WT viruses were highly susceptible to BXA (with EC50s of 0.2 nM to 3.4 nM). EC50s of influenza B viruses were, on average, 12-fold higher than those of influenza A viruses, consistent with previous reports (6, 13). All EC50 values increased upon the acquisition of 38T/F/M substitutions. The largest EC50 fold change relative to I38-WT was observed in 38T viruses (with changes of 12- to 92-fold); this was followed by lower EC50 fold changes among the 38F/M viruses (6- to 21-fold). This phenomenon was observed with both influenza A and B viruses. Therefore, the rescued PA variant viruses displayed reduced BXA susceptibility and corresponded to phenotypes observed after treatment (29), in preclinical studies (12, 13) and antiviral surveillance (6).
Table 1.
BXA susceptibility of recombinant influenza A and B viruses carrying I38T/F/M PA substitution
| Influenza virus | Abbreviation | PA 38 genotype* | Virus titer (Log10TCID50/mL) with BXA treatment (nM)† | Plaque reduction EC50 ± SEM (nM)‡ | Fold increase EC50 over I38-WT | |||
| 0 | 1 | 10 | 100 | |||||
| rg-A/California/04/2009 (H1N1)pdm09 | A/CA/04 | IWT | 5.7 | 1.1 | < | < | 0.3 ± 0.1 | − |
| T | 5.3 | 5.2 | 4.0 | 0.8 | 21.0 ± 7.6 | 70.0 | ||
| F | 5.0 | 4.2 | 2.0 | < | 2.2 ± 0.5 | 7.3 | ||
| M | 4.8 | 3.8 | 3.3 | < | 2.5 ± 1.4 | 8.3 | ||
| rg-A/Texas/71/2017 (H3N2) | A/TX/71 | IWT | 7.0 | 3.2 | < | < | 0.2 ± 0.1 | − |
| T | 6.3 | 6.3 | 5.9 | 4.0 | 18.4 ± 2.1 | 92.0 | ||
| F | 6.9 | 5.6 | 4.9 | < | 3.2 ± 0.6 | 16.0 | ||
| M | 7.6 | 7.0 | 5.1 | 2.3 | 4.1 ± 1.6 | 21.0 | ||
| rg-B/Brisbane/60/2008 (Victoria) | B/BR/60 | IWT | 6.6 | 6.1 | 4.6 | < | 2.9 ± 1.1 | − |
| T | 6.2 | 4.7 | 5.2 | 4.3 | 37.0 ± 9.0 | 15.0 | ||
| F | 3.7 | 3.7 | 3.8 | < | ND | ND | ||
| M | 7.0 | 6.8 | 6.3 | 4.3 | 22.0 ± 5.6 | 8.3 | ||
| rg-B/Phuket/3073/2013 (Yamagata) | B/PH/3073 | IWT | 7.1 | 6.8 | 5.9 | < | 3.4 ± 0.8 | − |
| T | 6.4 | 6.5 | 6.1 | 5.4 | 42.0 ± 3.0 | 12.4 | ||
| F | 5.7 | 5.8 | 5.5 | 3.5 | 27.0 ± 10.5 | 8.0 | ||
| M | 6.8 | 6.8 | 6.3 | 4.6 | 19.0 ± 4.4 | 6.0 | ||
<, mean value was below the assay limit of detection (0.75-Log10TCID50/mL); ND, not determined; −, not applicable.
Amino acid identity at residue 38 of the PA protein, introduced by reverse genetics system.
Virus yield (Log10 TCID50/mL) is from virus-infected MDCK cells (MOI 0.01; n = 3 wells per virus) at 72 or 96 hpi with BXA (0 nM to 100 nM) treatment. Mean values are from one of three experiments. Limit of detection = 0.75-Log10 TCID50/100 mL.
Reduction of plaque formation number from virus-infected MDCK cells (50 PFU to 80 PFU per well; n = 3 wells per drug concentration per virus) at 72 hpi or 120 hpi. Mean values from three independent experiments are presented ± SEM.
I38T/F/M Virus Replication Kinetics in MDCK Cells.
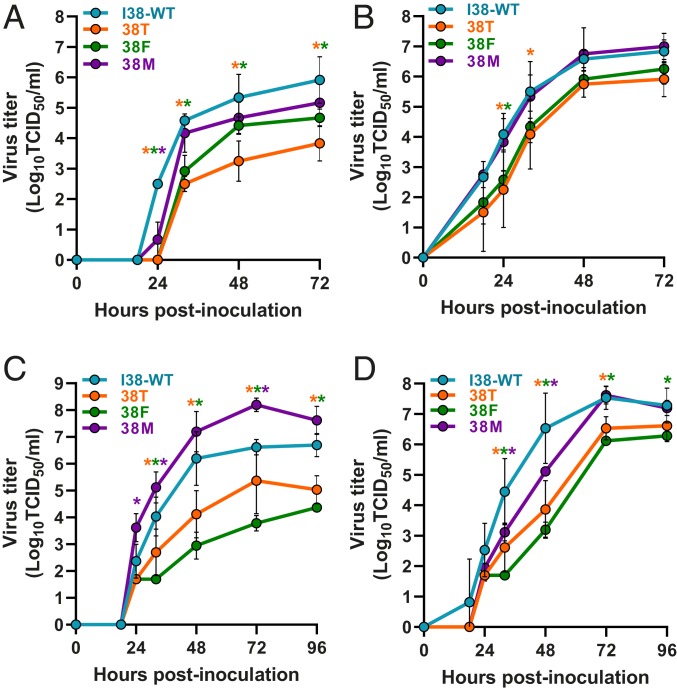
The replication fitness of 38T/F/M viruses was determined in MDCK cells (Fig. 2) which are replication permissive for most influenza viruses. Influenza A viruses with the 38M substitution reached titers comparable to those of I38-WT by 72 h postinfection (hpi), although some moderate delay (24 hpi) was noted with A/CA/04. In contrast, 38T/F viruses replicated to lower titers at all time points (in the case of A/CA/04) or early time points (in the case of A/TX/71) (Fig. 2 A and B). B/PH/3073 viruses with a 38T/M substitution reached titers comparable to those of I38-WT virus by late in the time course, but delayed replication (relative to I38-WT) was noted at earlier time points (32 and 48 hpi) (Fig. 2D). In contrast, the 38M mutant B/BR/60 showed replication rates higher than those of I38-WT virus at 24, 32, 72, and 96 hpi (Fig. 2C). Influenza B viruses with 38T/F substitutions replicated to lower titers than I38-WT virus and have significantly delayed replication at most time points beyond 24 hpi, similar to influenza A viruses with 38T/F substitutions (Fig. 3 C and D). Thus, both types A and B viruses with 38M substitution exhibit replication efficiency nearly equivalent to I38-WT virus, whereas viruses with 38T/F substitutions are impaired to varying degrees depending on the virus background.
Fig. 2.
Replication kinetics of I38-WT and 38T/F/M influenza A and B viruses in MDCK cells. MDCK cells were inoculated at an MOI of 0.001 with (A) A/CA/04, (B) A/TX/71, (C) B/BR/60 (Victoria lineage), or (D) B/PH/3073 (Yamagata lineage) virus carrying a single amino acid substitution at residue 38 of the PA protein (I38-WT and 38T/F/M). Supernatants were collected over time, and virus titers were determined at 18, 24, 32, 48, 72, and 96 hpi by TCID50 assays in MDCK cells. Data are presented as the mean values of three inoculated wells ± the SD for each time point and are representative of three independent experiments. The limit of detection of virus was 0.75-Log10TCID50/mL. P values are color coded according to PA 38 substitution and indicate titer comparisons relative to I38-WT at each time point. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Fig. 3.
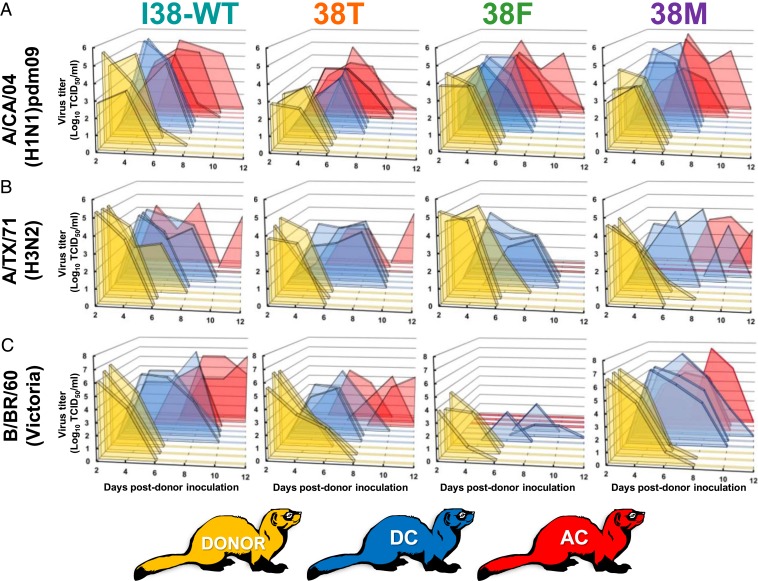
Transmissibility of I38-WT and 38T/F/M influenza A and B viruses in ferrets. Donor ferrets (n = 3, gold) were inoculated with 105 TCID50 units per ferret of recombinant influenza (A) A/CA/04 (H1N1)pdm09, (B), A/TX/71(H3N2), or (C) B/BR/60 (Victoria lineage) virus. Twenty-four hours later, DC cage mates (n = 3, blue) or AC cage mates (separated by a barrier that excluded physical contact, n = 3, red) were introduced. Virus titers (log10TCID50/mL; limit of detection 0.75-Log10TCID50/mL) were assayed in nasal washes collected at the indicated time points after donor inoculation. Each peak represents the shed virus titer of an individual animal. Donor B/BR/60 38F animals received 103.5 TCID50 units per ferret.
I38T/F/M Virus Pathogenesis and Transmission in Ferrets.
To determine the potential for 38T/F/M viruses to transmit among individuals, we modeled such events in ferrets, which are the gold-standard in vivo system for understanding influenza virus spread in humans (30). Naive “donor” ferrets were intranasally inoculated with either I38-WT or 38T/F/M influenza A or B virus. They were then paired with naive animals housed in the same cage (DCs) to understand direct virus transmission. We measured airborne-route transmission by placing naive animals (airborne contacts [ACs]) in an adjacent cage separated by a barrier that prevented donor/DC contact but allowed free airflow.
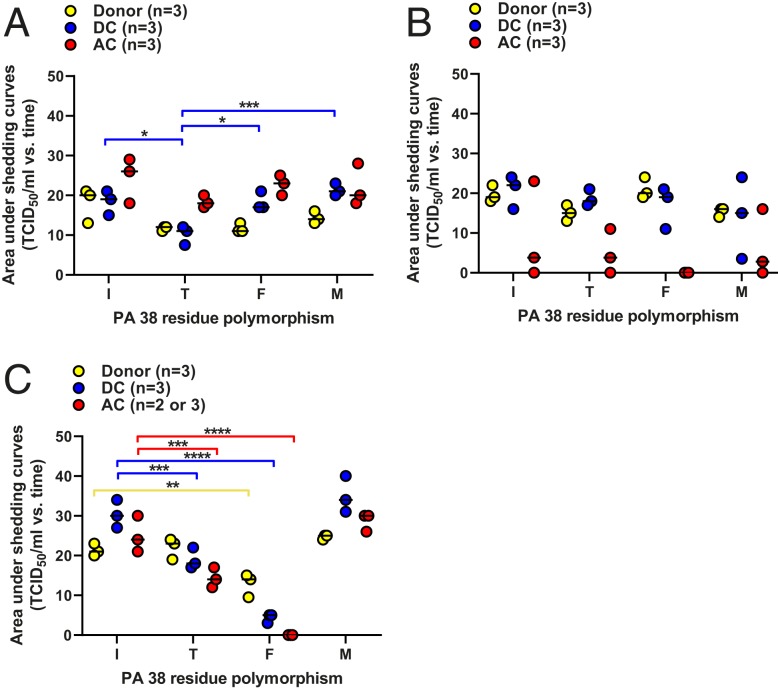
A/CA/04 virus was shed by all donors (three of three), all DCs (three of three), and all ACs (three of three), regardless of PA 38 genotype (Fig. 3 and SI. Appendix, Table S1). We quantified the virus shed from animals under the contact conditions by using area under the curve (AUC) analysis. A/CA/04 virus donors and AC animals had similar shedding curves, whereas 38T virus DCs had statistically lower (45 to 52%) AUC values when compared to all other PA 38 genotypes (Fig. 4A).
Fig. 4.
AUC for virus shedding in ferrets by I38-WT and 38T/F/M influenza A and B viruses. Ferrets were inoculated and housed as described in the legend for Fig. 3. The AUCs for virus shedding from ferrets inoculated with (A) A/CA/04 (H1N1)pdm09, (B), A/TX/71(H3N2), or (C) B/BR/60 (Victoria lineage) viruses for individual donors (gold), DCs (blue), and ACs (red) were calculated in GraphPad Prism 8.0. Data are presented as individual animal shedding curve values ± the SEM. P values and associated bars are color coded to indicate the contact groups(s) being compared. *P < 0.05; **P < 0.01, ***P < 0.001; ****P < 0.0001. Lack of comparison bars and P values on each graph represents “not significant.”
A/TX/71 virus with I38-WT or 38T/M substitutions was shed by all donors (three of three) and DCs (three of three). AC transmission in all these groups was limited to two of three ferrets. The incomplete airborne transmission is consistent with a pilot study conducted with this A/TX/71 I38-WT virus, where only one of two AC animals became infected and shed (SI Appendix, Fig. S1). No 38F virus was shed by AC ferrets at any time point (Fig. 3 and SI Appendix, Table S2). No statistically significant differences in AUC shedding values were observed with any A/TX/71 virus or successful route of transmission (Fig. 4B).
Efforts to propagate B/BR/60 with the 38F substitution to reach the inoculum dose for all other viruses (105 TCID50) were unsuccessful, and donor ferrets were given the maximum allowable dose of 103.5 TCID50. All PA 38 genotype viruses were shed by donors (three of three) and DCs (three of three). All ACs (three of three) shed I38-WT and 38T/M viruses, but no ACs shed 38F virus (Fig. 3 and SI Appendix, Table S3). PA 38F virus-inoculated ferrets had a 40% lower donor AUC, and 89% lower DC AUC when compared to I38-WT. The 38T viruses shed from ferrets displayed 40 to 42% lower AUC in DC/AC animal shedding when compared to I38-WT. PA 38M ferrets displayed AUC values similar to I38-WT (Fig. 4C).
Clinical and respiratory signs, onset of shedding, and peak virus shedding were recorded for all animals; however, no observable patterns were noticed in relation to virus or PA substitution. Seroconversion was detected among all shedding donors and DCs, although it was inconsistent among ACs shedding A/TX/71 and B/BR/60 (SI Appendix, Tables S1–S3).
Overall, 38T/M have minimal or no reduction in contact and airborne transmission of influenza A and B viruses, although lower AUC shedding values were observed with mutants shedding A/CA/04 among DCs, and in all 38T contacts shedding B/BR/60. In contrast, 38F attenuates airborne transmission of A/TX/71 and B/BR/60, decreases AUC shedding levels in B/BR/60, but still allows productive transmission by all routes with A/CA/04.
Stability of the I38T Genotype in Transmission Events.
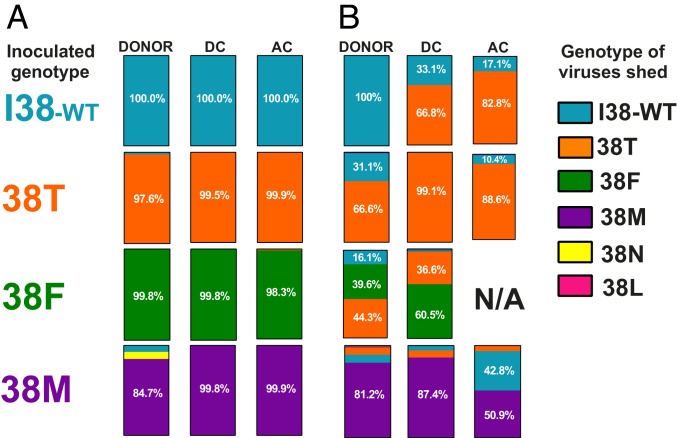
To determine whether the 38T/F/M genotypes reverted to the fitter I38-WT genotype during transmission, we amplified A/CA/04 and A/TX/71 PAN nucleotide segments from ferret nasal washes at the peak shedding time points and subjected samples to deep sequencing and amino acid variance analysis at position 38. No genotype changes were observed in any animals inoculated with A/CA/04 I38-WT virus. In general, few instances of reversion to I38-WT genotype were noted among all A/CA/04 viruses. Reversion of T → I (2.4%), M → I (7.3%), or an M → N substitution (7.9%) was observed in these virus-inoculated donors, but transmission of these altered genotypes onward was minimal (<1%). A minor substitution of F → T (1.2%) was observed in 38F ACs (Fig. 5A).
Fig. 5.
Genomic stability of PA 38 substitution upon ferret transmission of influenza A viruses. The PAN from (A) A/CA/04 (H1N1)pdm09 or (B) A/TX/71 viruses in donor, DC, and AC ferrets was amplified by RT-PCR and subjected to Illumina platform sequencing with variant calling at the nucleotide(s) encoding the amino acid at residue 38 of the PA protein and Qiagen CLC Genomics Workbench software. Data are compiled as a percentage at least 20,000 paired reads. The PA 38 genotypes within a contact group are indicated as parts of a whole. Amino acid polymorphisms ≥10% of the populations from which samples could be amplified have their values indicated on the bar. N/A denotes not applicable, no transmission to this contact group.
While all A/TX/71 I38-WT-inoculated donors shed this genotype, we were surprised to observe two animals that shed 38T in both the DCs and ACs. They were present at I:T ratios of ∼1:3 and ∼1:5, respectively, suggesting DC acquisition of 38T virus was amplified when transmitted to ACs. Some T → I (31.1%), and a minor T → M (1.1%) substitution was noted in 38T-inoculated donors, but both populations remained low or decreased in the transmission to DC and ACs. A similar trend was documented in the 38F-inoculated donors. The donor F → I population (16.1%) dwindled in transmission events, but, interestingly, F → T substitutions predominated in the donors and were retained at a ratio of ∼1:3 during transmission to DCs. Minor populations of less than 19% of M → I/T were also observed in 38M-inoculated donors and DCs. In contrast to other groups, the I38-WT genotype increased in prevalence (42.8%) in transmission to AC animals (Fig. 5B).
Overall, successful transmission of PA 38 mutant influenza A viruses with reduced susceptibility to BXM is likely not due to reversion to I38-WT genotype, as no incidence of dominant (above 50%) reversion to I38-WT was observed in any virus group.
Discussion
The approval of BXM, the first representative of the newest class of influenza PA endonuclease inhibitors, is a critical advance in influenza treatment and therapeutics. BXM is the first influenza drug to be recommended for treating uncomplicated influenza infections since the introduction of the NAIs OSE and zanamivir in 1999. Furthermore, it provides relief of symptoms and resolution of disease similar to OSE but is currently given as a single oral dose, compared to OSE’s 5-d, twice-daily regimen (31). Given the equivalent or superior clinical efficacy of BXM, its route of administration, and its favorable dosing and pharmacodynamics compared to those of some NAIs, it is expected that BXM will play an ever-growing role in influenza therapy for the foreseeable future. It is therefore critical to document any incidences of reduced susceptibility to this drug and report the resulting fitness of such viruses.
Cocrystallization of rPAN and active drug metabolite BXA revealed that the drug’s pyridotriazine motifs interact with PAN residues that coordinate divalent metal cations (Mn2+ or Mg2+) (13), similar to first-generation endonuclease inhibitors like L-742,001 (27, 32). BXA and similar compounds drive substitutions at residue 38 partially as a result of their extensive hydrophobic interaction between the compounds’ dibenzothiazepine, fused-ring motif as it wraps around I38. This phenomenon promotes PA 38 changes, including 38T, that can reduce hydrophobic interactions and severely disrupt drug binding (26). Recent molecular dynamic simulations also allude to mechanisms of decreased drug susceptibility of 38F/M viruses, where the Met38 side chain may be spatially too close to the BXM fused-ring motif to form an interaction, while interaction between BXM’s pyridotriazine motif and one catalytic site Mn2+ metal ion is apparently lost with Phe38 (33). This region is also involved in host pre-mRNA substrate binding (13, 26), so strong selective pressure is probably exerted to decrease drug interaction such that the virus survives patient treatment, while maintaining sufficient RNA affinity to complete transcription and virus replication. The structural mechanism(s) by which 38T/F/M viruses retain replicative fitness may involve the restoration of hydrophobic substrate binding at side-chain functional groups different from those of I38-WT. While structural studies relating to the functional competence of 38T/F/M viruses are underway, our work clearly demonstrates that these viruses are still viable in vitro and in vivo, and we can extrapolate from our in vitro studies which substitution(s) may be most or least favored. In the endonuclease assays, no substitutions provided the same level of enzymatic velocity as I38-WT. However, all of the substitutions allowed some activity (13). The loss of substrate processing velocity was less severe with influenza B PAN, although a limitation to this study is that the preferred substrate for influenza B viruses has not been reported. However, a hierarchy of Vmax values of I > T > M > F was present for PA proteins of both virus types, and this phenomenon can probably be attributed to the interaction between the substrate and the substituted side chains together with minor amino acid differences in PAN that result in type A and type B viruses binding substrate differently (13, 34–36).
PAN−substrate interactions shed light on effects of I38T/F/M substitutions, but during virus infection, a 1:1 interaction between substrate and PA does not occur. Instead, this process is mediated by the entire trimeric polymerase complex, in which multiple contact points on PA and PB2 can anchor the substrate and provide additional “help” to an otherwise impaired PA 38T/F/M. This may explain the fitness discrepancies between our endonuclease assays and the minireplicon assays, and it is consistent with data on mutations conferring reduced susceptibility to the first-generation drugs of this class (27). Although a 38T/F/M substitution causes impairment in the endonuclease assay, it has no effect on A/CA/04 or B viruses in the minireplicon. However, 38T and more significantly, 38F do decrease complex activity with the A/TX/71 virus. Substitutions at residue 38 have been assayed by minireplicon previously (26, 27), with multiple A(H1N1) strains demonstrating that they have no fitness deficiency. A(H1N1)pdm09 and B virus polymerase complexes may be more tolerant overall of minor changes in the catalytic endonuclease framework than the polymerase complex of A/TX/71 or other A(H3N2) viruses. We also considered that the minireplicon assay might accommodate indiscriminate substitutions at PA 38. However, introducing a conformationally restricted proline within the PA of A/CA/04 trimeric complexes completely abolished polymerase activity, whereas the polar, positively charged lysine only halved the activity when compared to I38-WT (SI Appendix, Fig. S2), suggesting that this is not the case. Further, mutagenesis and screening of non-PA 38 residues via this assay have successfully demonstrated discrimination among mutations affecting complex activity (27).
In both A and B viruses, 38T/F substitutions impaired the replication kinetics in comparison to those of I38-WT viruses, although virus titers of 4-Log10TCID50/mL or higher were still obtained in growth curve analyses, demonstrating that these viruses are viable. While our data support the trend toward a cell-replication fitness deficit observed by Omoto et al. (13), others report no change or even intrastudy variation among clinical isolates in cell-based assays of 38T virus fitness (21, 23). Thus, the conclusion on fitness of 38T viruses in tissue culture remains unclear and may be influenced by genetic composition of the virus origin and cell lines utilized. In contrast, 38M viruses showed replication capacity nearly equal to that of I38-WT and even greater in the B/BR/60 virus background. The explanation of the increased replication of the 38M virus in the B/BR/60 background is not understood. However, this finding is bolstered by the B/BR/60 38M minireplicon data where polymerase activity was statistically higher than I38-WT. Further, equivalent virus titers were shed by B/BR/60 38M ferrets, while lower titers were shed by B/BR/60 I38T/M contact ferrets, compared to I38-WT. The analyses of BXM-reduced susceptibility substitutions 38T/F/M are significantly less defined with influenza B viruses. It is therefore critical to include such viruses in ongoing BXM susceptibility studies, as it may be possible that BXM reduced susceptibility patterns will differ between influenza A and B viruses, similar to observations reported for the NAI class of drugs (37).
A recent study demonstrated that two A(H3N2) viruses (A/Louisiana/49/2017 and A/Bangladesh/3007/2017) with either 38T or 38M substitution were as fit as a WT counterpart, replicating in ferrets to equivalent titers, but this study did not examine transmission efficiency (38). To date, there are no published data demonstrating in vivo ferret transmission of 38F/M viruses, and only a single report demonstrating successful ferret airborne transmission of I38T virus isolated from patients (21). The 38M viruses were most consistently fit in the three in vitro assays, and, similarly, transmitted among DC and AC animals with efficiency equivalent to that of I38-WT. Conversely, the 38F viruses, which demonstrated the most instances of impairment, performed the worst, with only the A(H1N1)pdm09 virus transmitting to AC animals. The 38T viruses transmitted with the same efficiency as I38-WT viruses, but generally lower titers were shed from the animals, possibly reflecting the intermediate fitness phenotypes seen in the endonuclease and cell-based assays. However, even the minor fitness decreases observed in vitro did not always predict a lack of transmission; this argues for a more comprehensive analysis of viruses with these PA-associated substitutions that includes at least some in vivo data.
Our data suggest an emerging pattern of general fitness from PA 38T/F/M substitutions. Qualitatively, the 38M substitution appears to result in the fittest viruses relative to I38-WT viruses, demonstrating the least loss of activity in many in vitro assays and similar phenotypes in vivo. This is followed closely by the 38T substitution, which has a slight competitive advantage over 38M with substrate cleavage, but lags in in vitro replication kinetics and overall quantity of virus shed from infected ferrets. Finally, 38F viruses are the most impaired in nearly all observations, except some instances in the minireplicon. This appears to be especially true for influenza B viruses, in which the 38F substitution significantly impairs endonuclease activity and AC transmission in ferrets. The reason(s) for this hierarchy are unresolved, but neither clinical trials nor routine surveillance have revealed 38F substitution (or any other substitution) occurring naturally in influenza B viruses, whereas T, M, and F substitutions occurred by order of decreasing frequencies in influenza A viruses in these same studies (5, 13, 14, 19, 20, 29, 39, 40). The virus may prefer the replication phenotypes provided by 38M, but it settles for higher reduced susceptibility to BXM provided by 38T at the loss of some fitness. Structural biology data, including cocrystallization of 38T/F/M PA with a nucleotide polymer natural enzyme substrate, along with measuring the strength of those interactions, will provide important clues to explain the observed phenotypes.
Sequence analysis of endonuclease domains among the virus shed from inoculated ferrets showed that successful transmission to DC or AC animals was not correlated with a predominance of I38-WT revertant genotypes. The single exception is the change from M → I in ACs of A(H3N2) 38M-inoculated ferrets, but, even in this case, the revertant genotype did not exceed 50% of the population. This stability of the 38 mutant genotypes in ferrets is consistent with other groups who report a lack of reversion to I38-WT genotype in ferrets inoculated with and shedding A(H3N2) PA 38M or 38T viruses (38) or in airborne route ferret transmission events involving clinical A(H1N1)pdm09 or A(H3N2) isolates containing 38T (21). These studies, along with our datasets, support that 38T/M genotypes, and, to some degree, 38F, are stable in transmission events. This presents a risk of their dissemination, even in the absence of drug pressure. This finding also recalls our previous reports of virus passage with a similar endonuclease inhibitor (RO-7), which induced a stable retention of PA I38T for at least five passages (26) in the absence of drug pressure. Further, clinical reports document a common enrichment of the I38T change in tissue culture expansion of 38T virus mixtures shed from BXM-treated patients in Japan in 2018–2019 (19, 20, 39, 40). Non-38T/F/M substitutions were rare in our sequence analyses, but we did observe PA 38N as a variant substitution that was later reported from a virus mixture (I38T/N/S) isolated from a BXM-treated patient (21). We also observed a small population of viruses with a 38L substitution genotype which has been described or used experimentally at least twice (6, 38). That PA 38 substitutions are genetically stable in various systems appears clear, but the ease with which such viruses would transmit among humans, as in our A(H1N1)pdm09, A(H3N2), and B ferret challenge models, is unknown. Two recent studies involving Japanese pediatric patients strongly suggest that 38T virus was transmitted from a BXM-treated child to an untreated child in a daycare or household setting (20, 21). Additionally, household transmission of 38T virus from a BXM-treated child to an untreated infant sibling was recently documented, and the virus shed was genotypically identical (39). Overall, this evidence suggests that, although variably impaired in vitro, influenza A and B viruses carrying 38T/M and F (A[H1N1]pdm09 only) substitutions are probably transmissible, at least in close household settings.
Transmission of BXM-resistant influenza viruses is a significant concern. This can be partially addressed by reevaluating dosing regimens and administration schedules. A recent clinical report documents decreased efficiency of BXM treatment of PA 38T viruses in adult patients, but these infections were successfully treated with peramivir, an NAI drug (41). So, while BXM-reduced susceptibility viruses retain susceptibility to NAIs, it still may be important to consider BXM in a combination therapy approach. For the first time in many years, we have available two classes of influenza antivirals: PA endonuclease inhibitors like BXM and the NAIs. A third class of drugs, the cap-binding inhibitors characterized by pimodivir, may soon be available as well (5, 9). Combination therapy is a successful approach to limit the emergence of antiviral resistance and has been employed successfully to treat human immunodeficiency virus, hepatitis C virus, and other viruses (42). BXA demonstrates synergy with NAIs in vitro, and combination treatment with BXM and OSE in mice produces synergistic responses against influenza virus infections, suggesting that treating humans with the combination could produce beneficial outcomes (43).
In conclusion, we have demonstrated fitness disparities among influenza A and B viruses that acquired reduced susceptibility to BXM through 38T/F/M substitutions in multiple assays. Decreased in vitro fitness often did not correlate with a lack of transmission in ferrets, and reversion to the WT and most fit genotypes was not a prerequisite for successful transmission. Thus, risk of these viruses spreading from treated patients to naive individuals is significantly elevated. Continuous monitoring of virus isolates displaying BXM reduced susceptibility-associated changes in treated patients, and, more broadly, in circulating influenza viruses, is critical to ensure the utility of this class of drugs in the near future.
Materials and Methods
Cell Culture and Antiviral Compound.
MDCK cells and human embryonic kidney (HEK293T) cells were purchased from the American Type Culture Collection and maintained in modified Eagle’s medium (MEM; CellGro) and Opti-MEM (Fisher), respectively, supplemented with 10% fetal calf serum (HyClone) and 1× penicillin/streptomycin/amphotericin B. The free-acid, active form of the BXM prodrug (BXA) was purchased from MedChem Express.
Influenza Viruses.
Recombinant influenza A/California/04/2009 (H1N1pdm09; A/CA/04), A/Texas/71/2017 (H3N2, A/TX/71), B/Brisbane/60/2008 (Victoria lineage; B/BR/60), and B/Phuket/3073/2013 (Yamagata lineage; B/PH/3073) viruses were generated and rescued by the rg method, using either pHW2000 (influenza A) or pAD3000 (influenza B) plasmids (28). The I38T/F/M PA substitutions were inserted into the respective plasmids by using gene-specific primers and the QuikChange Site-Directed Mutagenesis Kit (Agilent). Viruses were propagated in MDCK cells at 37 °C for 72 h (influenza A viruses) or at 33 °C for 96 h (influenza B viruses) in serum-free MEM containing l-tosylamido 2-phenylethyl chloromethyl ketone (TPCK)-treated trypsin (Worthington) (1 μg/mL to 1.8 μg/mL). Full-length PA gene from each virus stock was amplified and subjected to Sanger platform sequence analysis. The presence of the respective, single PA 38X substitution, and the absence of non-PA 38 substitutions, was confirmed. For all subsequent in vitro experiments, influenza A and B viruses were maintained at 37 °C and 33 °C, respectively.
Protein Expression.
Sequences encoding a loop-deleted version (with residues 51 to 72 replaced with a GGS linker) of the 1- to 200-residue PA endonuclease domain (PAN) of A/CA/04 (35) or a non–loop-deleted version of the same domain of B/BR/60 with I38-WT or 38T/F/M substations were synthesized and inserted into pET-28a(+) expression plasmids (GenScript). The proteins were expressed in BL21(DE3) cells and purified by affinity chromatography and gel filtration.
Fluorescence Resonance Energy Transfer Endonuclease Assay.
Endonuclease activity assays were performed as previously described, with minor modifications (24). The rA/PAN or rB/PAN (5 µg) with I38 or 38T/F/M substitutions was incubated with single-strand DNA (ssDNA) substrate (1.25 μM to 40 µM) with a 5′ fluorophore and a 3′ quencher (56-FAM/AAT CGC AGG CAG CAC TC/3BHQ) in reaction buffer (20 mM Tris⋅HCl, 100 mM NaCl, 10 mM β-mercaptoethanol, 1.0 mM MnCl2, pH 8.2). Fluorescence released after PAN substrate cleavage was measured every 2 min (rA/PAN) or every 3 min (rB/PAN) at 485/535 nm Ex/Em. The measurement for substrate concentration without rPAN was subtracted as background. RFU at time periods 1 min to 26 min or 27 min were fitted in GraphPad Prism v8 with nonlinear regression for generation of Michaelis−Menten plots and derivation of Vmax.
Minireplicon Assay.
Influenza virus genes (encoding PB1, PB2, PA I38T/F/M, and nucleoprotein [NP]) from influenza A or B viruses were cloned into pHW2000 (for influenza A) or pAD3000 (for influenza B), propagated in Escherichia coli Top 10 (Invitrogen), and purified with a Qiagen HiSpeed Plasmid Maxi Kit (Qiagen). HEK293Ts were incubated with mock medium or 10 nM BXA medium for 1 h before transfection. Virus gene plasmids were cotransfected with reporter constructs specific to influenza A (pPolI-Δ358 NP noncoding region driving firefly luciferase) or influenza B (pPolI-HA noncoding region driving firefly luciferase). Both reactions included the pCMV-β-galactosidase for transfection control normalization. At 24 h to 48 h posttransfection, cells were lysed with 100 μL to 200 µL of passive lysis buffer (Promega), and the ratio of luciferase (as measured with a Luciferase Assay System [Promega]) to β-galactosidase was determined from a 15- to 30-µL sample lysate. All values were normalized to the ratio from PA I38-WT–transfected cells (100% polymerase activity). The influenza A reporter was kindly provided by Megan Shaw, Mount Sinai School of Medicine, New York, NY.
Virus Yield Reduction Assay.
MDCK cells (1 × 106) were plated in six-well plates, inoculated with influenza viruses (multiplicity of infection [MOI] of 0.001), cultured in MEM containing 1% bovine serum albumin (BSA), 1 μg/mL TPCK trypsin, and BXA (0, 1, 10, or 100 nM) At 72 hpi to 96 hpi, the supernatants were collected, and the TCID50 values were determined (44).
Plaque Reduction Assay.
MDCK cells were plated as in the virus yield reduction assays and inoculated with a virus dilution previously determined to yield 50 to 80 plaque-forming units (PFUs). At 1 h post virus inoculation, cells were washed, overlaid with MEM containing 0.45% immunodiffusion-grade agarose (MP Biomedical), 1% BSA, 1 μg/mL to 1.8 μg/mL TPCK trypsin, and BXA (0.001 nM to 500 nM). At 72 hpi to 120 hpi, the overlays were removed, and the cell monolayers were stained with 1% crystal violet–10% formaldehyde. The number of PFUs per well was calculated, and EC50s were determined by using the log (inhibitor) versus response logistic nonlinear regression equation in GraphPad Prism 8.0 software.
Virus Growth Curves.
Multistep growth curve analyses for influenza A and B viruses were performed in MDCK cells (1 × 106) plated in six-well plates. The cells were inoculated and maintained in culture as in the virus yield reduction experiments. Supernatants were sampled between 18 hpi and 96 hpi and titered in MDCK cells, and TCID50 values were determined (44).
Ferret Transmission Experiments.
Animal experiments were approved by St. Jude Children’s Research Hospital Institutional Animal Care and Use Committee in accordance with the Animal Welfare Act (US Department of Agriculture) and the Guide for the Care and Use of Laboratory Animals (45). Animals were housed in individual isolators, with individual negative airflow/high-efficiency particulate air filtering. Extensive change of personal protective equipment and floor and surface decontamination (5% Micro-Chem Plus, National Chemical Laboratories, followed by 70% ethanol) was performed each time when moving between isolators. Young male ferrets (4 mo to 5 mo of age) were obtained from Triple F Farms and verified as seronegative for influenza A(H1N1)pdm09 and A(H3N2) and both lineages of influenza B virus. Donor ferrets (n = 3 per test virus) were housed individually in a multilevel cage system and inoculated intranasally with 105 TCID50 units of PA I38-WT or 38T/F/M virus in each influenza A or B in 1 mL of sterile phosphate-buffered saline (PBS). Donor B/BR/60 38F animals received 103.5 TCID50 units because of the lower resulting titers of the MDCK-propagated virus stocks. Twenty-four hours later, naive DC ferrets (n = 3, one paired with each donor) were introduced into the cage with the inoculated donor. At the same time, additional naive AC ferrets (n = 3, one per donor/DC pair) were placed in an adjacent cage isolated by a double set of bar dividers (7.6 cm apart) that completely prevented physical contact but allowed free passage of respiratory droplets. The animals were observed for 20 min to 30 min on a daily basis and scored on an activity scale to determine general lethargy, weighed, and temperature-checked and compared to baseline to assess fever/weight loss/anorexia. Respiratory events were also recorded during this period. Beginning at 2 d postinjection (dpi), and every 48 h thereafter, ferrets were anesthetized with ketamine (25 mg/kg) and induced to sneeze by intranasal instillation of 1 mL of sterile PBS. The virus titer (TCID50/mL) of nasal washes was determined in MDCK cells as described previously. At 14 dpi, sera were collected from all animals, and seroconversion to homologous challenge virus was determined by hemagglutination inhibition assay with 0.5% turkey red blood cells (46). The AUC values were calculated for each animal in each virus group in GraphPad Prism 8.0.
Sequencing.
Using an RNeasy Kit (Qiagen), total RNA was isolated from ferret nasal washes on the peak shedding day (as described in Fig. 5 and SI Appendix, Tables S1–S3). The first 600 to 800 bases of the PA gene (encompassing the endonuclease domain) was amplified by a one-step RT-PCR kit (Invitrogen) with gene-specific primers and subjected to PCR purification (Qiagen). Next-generation sequencing of purified products was conducted, and libraries were prepared using a Nextera XT DNA Library Preparation Kit (Illumina) at 0.5× the recommended volumes. The libraries were then run on the MiSeq platform (Illumina), using the 2 × 250 kit for a total of ∼500 cycles.
Statistical Analysis.
Data were analyzed using GraphPad Prism 8.0, with individual significance being determined by unpaired t tests and/or two-way ANOVA as indicated.
Data Availability Statement.
All data needed to support the findings of this manuscript are included in the main text and SI Appendix.
Supplementary Material
Acknowledgments
We are grateful for the excellent scientific editing of the manuscript by Keith Laycock. We thank Chelsi Stultz, John Franks, Daniel Darnell, Heather Smallwood, and Jonah Sacha for experimental assistance and helpful discussion. We also thank the staff of the Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children’s Research Hospital for their help in the next-generation sequencing, and the staff of the St. Jude Protein Production Facility for their help in the production of rPAN. This study was supported by the National Institute of Allergy and Infectious Diseases, NIH, under Contract HHSN272201400006C.
Footnotes
Competing interest statement: E.A.G. reports receiving consultant fees and travel support from Genentech/Roche for serving on an advisory board. The other authors declare no conflicts of interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1916825117/-/DCSupplemental.
References
- 1.Centers for Disease Control and Prevention ,“Burden of influenza in seasonal influenza: About flu” (Centers for Disease Control and Prevention, 2019).
- 2.Tokars J. I., Olsen S. J., Reed C., Seasonal incidence of symptomatic influenza in the United States. Clin. Infect. Dis. 66, 1511–1518 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soema P. C., Kompier R., Amorij J. P., Kersten G. F., Current and next generation influenza vaccines: Formulation and production strategies. Eur. J. Pharm. Biopharm. 94, 251–263 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Boivin G., Detection and management of antiviral resistance for influenza viruses. Influenza Other Respir. Viruses 7 (suppl. 3), 18–23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mifsud E. J., Hayden F. G., Hurt A. C., Antivirals targeting the polymerase complex of influenza viruses. Antiviral Res. 169, 104545 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Gubareva L. V., et al. , Assessing baloxavir susceptibility of influenza viruses circulating in the United States during the 2016/17 and 2017/18 seasons. Euro Surveill. 24, 1800666 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heo Y. A., Baloxavir: First global approval. Drugs 78, 693–697 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Furuta Y., et al. , Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antiviral Res. 100, 446–454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayden F. G., Shindo N., Influenza virus polymerase inhibitors in clinical development. Curr. Opin. Infect. Dis. 32, 176–186 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Genentech , XOFLUZA Brochure. https://www.xofluza.com. Accessed 24 October 2019.
- 11.O’Hanlon R., Shaw M. L., Baloxavir marboxil: The new influenza drug on the market. Curr. Opin. Virol. 35, 14–18 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Noshi T., et al. , In vitro characterization of baloxavir acid, a first-in-class cap-dependent endonuclease inhibitor of the influenza virus polymerase PA subunit. Antiviral Res. 160, 109–117 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Omoto S., et al. , Characterization of influenza virus variants induced by treatment with the endonuclease inhibitor baloxavir marboxil. Sci. Rep. 8, 9633 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayden F. G. et al.; Baloxavir Marboxil Investigators Group , Baloxavir marboxil for uncomplicated influenza in adults and adolescents. N. Engl. J. Med. 379, 913–923 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Watanabe A., et al. , Baloxavir marboxil in Japanese patients with seasonal influenza: Dose response and virus type/subtype outcomes from a randomized phase 2 study. Antiviral Res. 163, 75–81 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Hirotsu N., et al. , Baloxavir marboxil in Japanese pediatric patients with influenza: Safety and clinical and virologic outcomes. Clin. Infect. Dis., 10.1093/cid/ciz908 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koszalka P., Tilmanis D., Roe M., Vijaykrishna D., Hurt A. C., Baloxavir marboxil susceptibility of influenza viruses from the Asia-Pacific, 2012-2018. Antiviral Res. 164, 91–96 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Sugaya N., Widespread use of neuraminidase inhibitors in Japan. J. Infect. Chemother. 17, 595–601 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Takashita E. et al.; On Behalf of the Influenza Virus Surveillance Group of Japan , Detection of influenza A(H3N2) viruses exhibiting reduced susceptibility to the novel cap-dependent endonuclease inhibitor baloxavir in Japan, December 2018. Euro Surveill. 24, 1800698 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takashita E., et al. , Influenza A(H3N2) virus exhibiting reduced susceptibility to baloxavir due to a polymerase acidic subunit I38T substitution detected from a hospitalised child without prior baloxavir treatment, Japan, January 2019. Euro Surveill. 24, 1900170 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imai M., et al. , Influenza A variants with reduced susceptibility to baloxavir isolated from Japanese patients are fit and transmit through respiratory droplets. Nat. Microbiol. 5, 27–33 (2020). [DOI] [PubMed] [Google Scholar]
- 22.McKimm-Breschkin J. L., Influenza neuraminidase inhibitors: Antiviral action and mechanisms of resistance. Influenza Other Respir. Viruses 7 (suppl. 1), 25–36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Checkmahomed L., et al. , Impact of the baloxavir-resistant polymerase acid I38T substitution on the fitness of contemporary influenza A(H1N1)pdm09 and A(H3N2) strains. J. Infect. Dis. 221, 63–70 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fudo S., et al. , Structural and computational study on inhibitory compounds for endonuclease activity of influenza virus polymerase. Bioorg. Med. Chem. 23, 5466–5475 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Song M. S., et al. , The polymerase acidic protein gene of influenza A virus contributes to pathogenicity in a mouse model. J. Virol. 83, 12325–12335 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones J. C., et al. , Identification of the I38T PA substitution as a resistance marker for next-generation influenza virus endonuclease inhibitors. MBio 9, e00430-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stevaert A., et al. , Mutational analysis of the binding pockets of the diketo acid inhibitor L-742,001 in the influenza virus PA endonuclease. J. Virol. 87, 10524–10538 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoffmann E., Krauss S., Perez D., Webby R., Webster R. G., Eight-plasmid system for rapid generation of influenza virus vaccines. Vaccine 20, 3165–3170 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Uehara T., et al. , Treatment-emergent influenza variant viruses with reduced baloxavir susceptibility: Impact on clinical and virologic outcomes in uncomplicated influenza. J. Infect. Dis. 221, 346–355 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Bouvier N. M., Animal models for influenza virus transmission studies: A historical perspective. Curr. Opin. Virol. 13, 101–108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Principi N., et al. , Drugs for influenza treatment: Is there significant news? Front. Med. (Lausanne) 6, 109 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomassini J., et al. , Inhibition of cap (m7GpppXm)-dependent endonuclease of influenza virus by 4-substituted 2,4-dioxobutanoic acid compounds. Antimicrob. Agents Chemother. 38, 2827–2837 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshino R., Yasuo N., Sekijima M., Molecular dynamics simulation reveals the mechanism by which the influenza cap-dependent endonuclease acquires resistance against baloxavir marboxil. Sci. Rep. 9, 17464 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dias A., et al. , The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458, 914–918 (2009). [DOI] [PubMed] [Google Scholar]
- 35.DuBois R. M., et al. , Structural and biochemical basis for development of influenza virus inhibitors targeting the PA endonuclease. PLoS Pathog. 8, e1002830 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kowalinski E., et al. , Structural analysis of specific metal chelating inhibitor binding to the endonuclease domain of influenza pH1N1 (2009) polymerase. PLoS Pathog. 8, e1002831 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burnham A. J., Baranovich T., Govorkova E. A., Neuraminidase inhibitors for influenza B virus infection: Efficacy and resistance. Antiviral Res. 100, 520–534 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chesnokov A., et al. , Replicative fitness of seasonal influenza A viruses with decreased susceptibility to baloxavir. J. Infect. Dis. 221, 367–371 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takashita E., et al. , Human-to-human transmission of influenza A(H3N2) virus with reduced susceptibility to baloxavir, Japan, February 2019. Emerg. Infect. Dis. 25, 2108–2111 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takashita E., et al. , Susceptibility of influenza viruses to the novel cap-dependent endonuclease inhibitor baloxavir marboxil. Front. Microbiol. 9, 3026 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seki M., Sakai-Tagawa Y., Yasuhara A., Watanabe Y., Adult influenza A (H3N2) with reduced susceptibility to baloxavir or peramivir cured after switching anti-influenza agents. IDCases 18, e00650 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hofmann W. P., Soriano V., Zeuzem S., Antiviral combination therapy for treatment of chronic hepatitis B, hepatitis C, and human immunodeficiency virus infection. Handb. Exp. Pharmacol. 189, 321–346 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Fukao K., et al. , Combination treatment with the cap-dependent endonuclease inhibitor baloxavir marboxil and a neuraminidase inhibitor in a mouse model of influenza A virus infection. J. Antimicrob. Chemother. 74, 654–662 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muench H., Reed L. J., A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 27, 493–497 (1938). [Google Scholar]
- 45.National Research Council , Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
- 46.Kim J. K., et al. , Puzzling inefficiency of H5N1 influenza vaccines in Egyptian poultry. Proc. Natl. Acad. Sci. U.S.A. 107, 11044–11049 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to support the findings of this manuscript are included in the main text and SI Appendix.