Abstract
The human liver reacts to hepatitis C virus (HCV) with a balanced response consisting of host anti‐ and proviral activities. To explore these subtle host responses, we used oligonucleotide microarrays to investigate the differential gene expression between two groups of liver samples with high and low HCV loads (>100‐fold difference). We identified and validated 26 genes that were up‐regulated in livers with high HCV loads, including transmembrane protease serine 2 (TMPRSS2). Trypsin inhibitors inhibited the infection of Huh7‐25‐CD81 cells with cell‐culture–derived HCV (HCVcc) of Japanese fulminant hepatitis 1 isolate at the postbinding and entry step, and trypsin enhanced HCVcc infection at an early stage of infection. Several major transmembrane serine proteases, in particular, furin and hepsin, were detected in Huh7‐25‐CD81 cells, but TMPRSS2 was not. Huh7‐25‐CD81 cell clones stably expressing TMPRSS2‐ WT (wild type) and inactive TMPRSS2‐mutant genes showed positive and negative enhancement of their susceptibility to HCVcc infection, respectively. The enhanced susceptibility of TMPRSS2‐WT Huh7‐25‐CD81 cells was confirmed by knockdown of TMPRSS2 using small interfering RNA. The cell‐surface protease activity of TMPRSS2‐WT cells was markedly active in the cleavage of QAR and QGR, corresponding to amino acid residues at P3 to P1. Conclusion: The cell‐surface activity of a trypsin‐like serine protease, such as TMPRSS2, activates HCV infection at the postbinding and entry stage. Host transmembrane serine proteases may be involved in the sensitivity, persistence, and pathogenesis of HCV infection and be possible targets for antiviral therapy. (Hepatology 2015;61:438‐447)
Abbreviations
- Ab
antibody
- AEBSF
4‐(2‐aminoethyl)‐benzenesulfonyl fluoride hydrochloride
- APO
apolipoprotein
- Boc
t‐butyloxycarbony
- cDNA
complementary DNA
- CLDN1
claudin‐1
- cRNA
complementary RNA
- DAB
3,3′‐diaminobenzidine
- DAPI
4′,6‐diamidino‐2‐phenylindole
- DMEM
Dulbecco's modified Eagle's medium
- EGFP
enhanced green fluorescent protein
- EMT
epithelial‐mesenchymal transition
- FBS
fetal bovine serum
- HA
hemagglutinin
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- HCVcc
cell‐culture–derived HCV
- HPN
hepsin
- IF
immunofluorescence
- IFN
interferon
- IHC
immunohistochemistry
- ISG
IFN‐stimulating gene
- JFH‐1
Japanese fulminant hepatitis 1 isolate
- MCA
4‐methyl‐coumaryl‐7‐amide
- mRNA
messenger RNA
- MTT
3‐(4,5‐Dimethyl‐2‐thiazolyl)‐2,5‐diphenyl‐2H‐tetrazolium bromide
- OASL
2′‐5′‐oligoadenylate synthetase‐like
- OCLN
occludin
- PBS
phosphate‐buffered saline
- qPCR
quantitative real‐time polymerase chain reaction
- SBTI
soybean trypsin inhibitor
- siRNA
small interfering RNA
- TMPRSS2
transmembrane protease, serine 2
- WT
wild type
Hepatitis C virus (HCV) is a causative agent of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (HCC). Therapy for chronic hepatitis C has markedly progressed following the recent approval of direct‐acting antiviral agents, which can be administered in addition to standard care with pegylated interferon (IFN)‐α plus ribavirin.1, 2 However, the pathogenesis of chronic liver diseases caused by HCV infection remains to be well elucidated. For instance, what type of host responses induced by HCV infection contribute to HCV pathogenesis, and what is their mechanism of action? An analysis of HCV‐induced changes in the host transcriptome may help resolve these issues. The responses of Huh7 cells infected with HCV have been examined in vitro,3, 4, 5 while the in vivo liver responses of chimpanzees and patients with acute resolving and chronic HCV infection have been examined.6, 7, 8, 9 HCV infection elevates IFN‐stimulating genes (ISGs), much more notably in vivo than in vitro, but not in Huh7.5 cells that are defective in their response to IFN because of a mutation in retinoic acid‐inducible gene I.3, 5 The analysis of the gene expression profiles of these cells revealed not only the antiviral responses of proinflammatory and proapoptotic signaling, but also proviral responses required for HCV replication and secretion at the cellular level. This complex virus‐host interaction may be relevant to the pathogenesis of HCV infection, including steatosis, fibrosis, and persistence. The analysis of the tissue‐level responses to HCV infection in vivo identified genes responsive to HCV infection, including those promoting inflammatory cell infiltrations and the IFN response.6, 7, 8, 9 Using the differential hybridization of complementary DNA (cDNA) library, we also isolated two ISG genes that were markedly up‐regulated in chimpanzee livers during the acute phase of HCV infection: major histocompatibility complex, class I, C, and IFN‐α‐inducible protein 6.10 Thus, it is clear that the IFN response is actually activated by HCV infection in vivo, and various downstream responses contribute to HCV‐host interaction. Currently, ISGs (including those mentioned above) have been investigated in detail, and their roles as critical molecules in intra‐ and extracellular innate immunity against HCV infection are becoming clear.11, 12 However, there remain differentially expressed genes whose functions are unknown.
The gene expression profiles of human livers are more useful than those of cultured cells for elucidating natural HCV‐host interactions and studying the control of host antiviral responses at the level of the entire organism. To identify the critical factors responsible for HCV control in the human liver, it is useful to determine the subtle differences in the host responses to different HCV loads in the liver, rather than the marked differences between human livers with and without HCV infection. To achieve this, we investigated the differences between human livers with chronic hepatitis containing high or low loads of HCV. By comparing gene expression profiles between livers containing quite different HCV loads, we focused on the limited, more critical factors that govern the HCV‐host interaction in vivo. After verifying the differential gene expression between the two groups of liver samples, the functions of genes with potential anti‐ or proviral effects were analyzed during HCV infection. In this study, we identified 26 genes that were up‐regulated in livers with high HCV loads. Among several genes with unknown functions in HCV infection, we characterized transmembrane protease serine 2 (TMPRSS2) as a new host factor activating HCV infection.
Materials and Methods
Liver Specimens and Cells
HCV‐positive liver samples were obtained by surgical resection of nontumorous liver tissues from 59 patients with HCC. All patients had a solitary HCC that was less than 5 cm in diameter and showed no metastasis to lymph nodes or other organs. After quantifying the HCV‐RNA concentrations in the livers, followed by normalizing with 18S ribosomal RNA, we classified the 59 samples into three groups according to the HCV load (measured in arbitrary units of HCV RNA): high (>30,000); middle (>299 and <30,001); and low (<300). Of the 59 patients with chronic hepatitis, but no liver cirrhosis, were subjects belonging to the HCV‐high (9 cases) and HCV‐low groups (8 cases; http://onlinelibrary.wiley.com/doi/10.1002/hep.27426/suppinfo). HCV‐negative liver samples were obtained from the nontumorous liver tissues of 5 patients with hepatic metastases of colon cancer. Written informed consent was obtained from each patient. Our study protocol was previously approved by the ethics committee of our school in accord with the 1975 Declaration of Helsinki. Huh7‐25‐CD81 cells13 and Japanese fulminant hepatitis 1 isolate (JFH‐1) HCV subgenomic replicon cells14 were cultured in Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% fetal bovine serum (FBS), containing 400 µg/mL and 1 mg/mL of G418 (Invitrogen, Carlsbad, CA), respectively.
Reagents
α1‐antitrypsin from human plasma, phosphoramidon, bestatin, and N‐acetylated‐trypsin were purchased from Sigma‐Aldrich (St. Louis, MO). Trypsin from porcine pancreas (1,000‐2,000 BAEE units/mg), soybean trypsin inhibitor (SBTI; ∼10,000 BAEE units/mg), 4‐(2‐aminoethyl)‐benzenesulfonyl fluoride hydrochloride (AEBSF), calpain inhibitor I, pepstatin, X‐tremeGENE 9, and HP were from Roche Diagnostics GmbH (Mannheim, Germany). Small interfering RNA (siRNA) and DharmaFECT4 were from Dharmacon, Thermo Fisher Scientific (Waltham, MA). Rabbit anti‐TMPRSS2 monoclonal antibody (Ab; ab92323) was from Abcam (Cambridge, UK). The secondary Ab conjugated with Alexa Fluor 555 was from Invitrogen. 3‐(4,5‐Dimethyl‐2‐thiazolyl)−2,5‐diphenyl‐2H‐tetrazolium bromide (MTT) was purchased from Dojindo (Kumamoto, Japan).
Comprehensive Analysis of Messenger RNA Expression
A summary of messenger RNA (mRNA) expression is shown in http://onlinelibrary.wiley.com/doi/10.1002/hep.27426/suppinfo. Five random samples each from the HCV‐high and HCV‐low groups of nontumorous liver tissues without liver cirrhosis (http://onlinelibrary.wiley.com/doi/10.1002/hep.27426/suppinfo) were analyzed for their gene expression profiles. We also analyzed the gene expression in seven and three samples each from the HCV‐high and HCV‐low groups of nontumorous liver tissues with liver cirrhosis (http://onlinelibrary.wiley.com/doi/10.1002/hep.27426/suppinfo). Biotin‐labeled complementary RNA (cRNA) was synthesized from total RNA using the MEGAscript T7 kit (Ambion, Austin, TX). Then, fragmented cRNA was hybridized to a Human Genome U133 Plus 2.0 array (Affymetrix, Santa Clara, CA), which comprises 54,675 probes. After washing and staining with Fluidics Station 450 (Affymetrix), the array was scanned with a Scanner 3000 (Affymetrix), according to the protocol described in the Affymetrix GeneChip Expression Analysis Manual. The signal of each gene was analyzed using GeneSpring version 7 (Silicon Genetics, Redwood, CA). http://onlinelibrary.wiley.com/doi/10.1002/hep.27426/suppinfo shows 130 candidate differentially expressed genes isolated from chronic hepatitis liver samples with and without liver cirrhosis.
Quantification of mRNA
Total RNA was isolated from liver tissues and subjected to DNase I treatment, followed by cDNA synthesis, as described previously.15 Synthesis of cDNA from cultured cells was performed using 1 µg of total RNA and ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo, Osaka, Japan). Expression levels of specific mRNA were determined by quantitative real‐time polymerase chain reaction (qPCR) using SYBR green or TaqMan gene expression assays. The primer sequences for SYBR green qPCR are shown together with TaqMan gene expression assay ID in http://onlinelibrary.wiley.com/doi/10.1002/hep.27426/suppinfo.
Immunohistochemical Staining
Formalin‐fixed, paraffin‐embedded thin sections, 4 µm in thickness, were treated with 10 mM of citrate buffer (pH 6.0 at 120°C) for 15 minutes for antigen retrieval. After blocking with 1% hydrogen peroxide followed by 5% skim milk in phosphate‐buffered saline (PBS) for 30 minutes at 37°C, sections were incubated with 2.5 µg/mL of anti‐TMPRSS2 Ab at 37°C for 60 minutes and then stained with a Histofine Simple Stain MAX PO (MULTI; Nichirei, Tokyo, Japan) for 30 minutes at room temperature. The sections were reacted with 3,3′‐diaminobenzidine (DAB) chromogen (Wako Pure Chemical Industries, Ltd., Tokyo, Japan) and counterstained with hematoxylin.
Infection With Cell‐Culture–Derived HCV
Cell‐culture–derived HCV (HCVcc) derived from JFH‐1 (genotype 2a) were prepared as previously described.16 Huh7‐25‐CD81 cells (2 × 104) were seeded in 24‐well plates containing coverslips and incubated overnight at 37°C. Cells were inoculated with JFH‐1 HCVcc (4 × 105 copies of HCV RNA, 200 focus‐forming units) at 37°C for 2 hours in 200 µL of PBS. Then, after removing the inoculum, the cells were incubated in complete medium for 1 or 2 days. After culture, coverslips were rinsed in PBS and stained for HCV proteins with Alexa Fluor 555, as described previously.15 Nuclei were stained with 4′,6‐diamidino‐2‐phenylindole (DAPI; Invitrogen), and the stained cells were visualized with a BIOREVO BZ‐9000 microscope (Keyence, Osaka, Japan) and a IX71 fluorescent microscope (Olympus, Tokyo, Japan). HCV infectivity was determined by the number of foci containing HCV‐positive cells. When the cell number was occasionally decreased after staining, the number of foci was normalized against the total number of cells using the area of DAPI‐positive nuclei instead of cell number.
Gene Knockdown by siRNA
After 24‐hour cultivation of 1.5 × 104 cells in 24‐well plates, cells were transfected with 5 nM of siRNAs using 0.15 µL of DharmaFECT 4. Cells were cultured in complete medium and then harvested or infected with JFH‐1 HCVcc at 48 hours post‐transfection.
Stable Huh7‐25‐CD81 Cell Clones Producing TMPRSS2–Wild Type and TMPRSS2‐Mutant
The pcDNA3‐TMPRSS2 containing cDNA encoding human wild‐type (WT) TMPRSS2 and pCA7‐TMPRSS2 (S441A) encoding inactive mutant TMPRSS2 (S441A)17 were digested with EcoRI and NotI. The isolated cDNA fragments were inserted into the EcoRI/NotI sites of the pEF6/V5‐His vector containing a blasticidin‐resistance gene (Invitrogen). The pEF6/V5‐His vector was used as a negative control. Huh7‐25‐CD81 cells were transfected with the complex of the plasmid and X‐tremeGENE 9, and stable clones of transfected cells were established by culture with 7 µg/mL of blasticidin. The TMPRSS2‐expressing clones were screened by immunofluorescence (IF) staining with anti‐TMPRSS2 Ab and a secondary Ab conjugated with Alexa Fluor 555, after fixation with 4% paraformaldehyde at room temperature for 10 minutes.
Assessment of Protease Activity
Cell‐surface serine protease activity was assessed using the following synthetic peptide analogs, t‐butyloxycarbony (Boc)‐peptideanalog‐4‐methyl‐coumaryl‐7‐amide (MCA): Boc‐RVRR‐MCA; Boc‐FSR‐MCA; Boc‐QGR‐MCA; Boc‐QAR‐MCA; Boc‐QRR‐MCA (Peptide Institute, Inc., Osaka, Japan); and Boc‐QSR‐MCA (Genenet, Fukuoka, Japan). After preculturing Huh7‐25‐CD81 cells (2 × 104) for 24 hours in 96‐well black plates followed by a PBS wash, 200 µL of 100 µM of peptide substrate in Opti‐MEM were added to cells and the fluorescence of released aminomethyl coumarin was measured immediately using an Infinite F200 (Tecan Schweiz AG, Männedorf, Switzerland) at 360(35)/ 465(35) nm of excitation/emission wavelength every 15 minutes for 3 hours at 28°C. Fluorescence was normalized against the cell number determined by MTT assay. Enzyme activity assessed was kinetically specific and was inhibited by SBTI in a dose‐dependent manner (http://onlinelibrary.wiley.com/doi/10.1002/hep.27426/suppinfo).
Cell Fusion Induced by the Cleavage of Influenza A Virus Hemagglutinin
Cell‐to‐cell fusion was assessed as described previously.18, 19 Stable cell clones of Huh7‐25‐CD81 were grown in 12‐well plates and transfected with a mixture of 1 µg of the hemagglutinin (HA; H3 subtype)‐expressing plasmid, pCAGGS‐HA,20 and 0.5 µg of enhanced green fluorescent protein (EGFP)‐expressing plasmid, pCAGGS‐EGFP, using X‐tremeGENE HP DNA Transfection Reagent. At 1 day post‐transfection, cells were incubated with DMEM in the absence of exogenous trypsin (−) or supplemented with 3 µg/mL of acetylated‐trypsin (+; Sigma‐Aldrich). At 2 days post‐transfection, cells were washed with PBS and treated with prewarmed low‐pH buffer (145 mM of NaCl and 20 mM of sodium citrate; pH 5.4) for 2 minutes. The low‐pH buffer was then replaced with DMEM containing 10% FBS, and cells were incubated at 37°C for 5 hours. Cells were observed using a fluorescence microscope. Percentage of fluorescent areas was measured by using imageJ software (version 1.36b, National Institutes of Health, Bethesda, MD).
Statistical Analysis
Mann‐Whitney's U test was used to analyze the two‐group comparisons. The Student t test was used in the experiments in vitro. P values less than 0.05 were considered statistically significant.
Results
TMPRSS2 Expression Was Increased in the HCV‐High Group Livers
To investigate anti‐ and proviral host responses to HCV infection of human livers, we used oligonucleotide microarrays to examine the differential gene expression between two groups of liver samples, containing high and low HCV loads. To exclude the involvement of other variables, we divided nontumorous liver tissues into two groups, one with and one without liver cirrhosis, which were obtained from patients with chronic hepatitis in the early stages of HCC. HCV load differed markedly (>100‐fold) between the two groups, but the clinicopathological features were not significantly different according to the statistical analyses (http://onlinelibrary.wiley.com/doi/10.1002/hep.27426/suppinfo). We identified candidate genes from livers with chronic hepatitis in the presence or absence of liver cirrhosis and verified 26 genes that were up‐regulated in livers with high HCV loads using chronic hepatitis livers without liver cirrhosis (http://onlinelibrary.wiley.com/doi/10.1002/hep.27426/suppinfo). Of the 26 genes, seven were ISGs; OASL (2′‐5′‐oligoadenylate synthetase‐like), an ISG of unknown function, inhibited HCV replication in vitro , as described previously.15 Thus, most genes that were up‐regulated in the HCV‐high group were related to an antiviral response. However, it is not known whether several genes are associated with anti‐ or proviral responses or how they function during infection. Among the genes up‐regulated in the HCV‐high group (http://onlinelibrary.wiley.com/doi/10.1002/hep.27426/suppinfo), we focused on TMPRSS2, which is a member of the hepsin (HPN) subfamily of type II transmembrane serine proteases.21 TMPRSS2 was one of the top three most differentially expressed genes in addition to OASL and HLA‐DQA1; these genes had the lowest P value (<0.003 by Mann‐Whitney's U test) and showed a more than 3‐fold change. Because the current understanding of the physiological functions of type II transmembrane serine proteases has highlighted their contribution to human disease, it seemed important to determine whether TMPRSS2 is involved in the life cycle of HCV or its pathogenesis. Up‐regulation of TMPRSS2 in HCV‐high livers, compared with HCV‐low and HCV‐negative livers, was confirmed by qPCR (Fig. 1A). The TMPRSS2 protein was also highly expressed in the HCV‐high liver, as shown by immunohistochemistry (IHC); it was localized heterogeneously in liver tissue and strongly positive at the apical membrane of hepatocytes (Fig. 1B).
Figure 1.
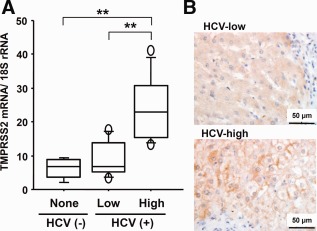
TMPRSS2 expression in human liver tissues. (A) Quantification of TMPRSS2 mRNA in livers of the HCV‐negative (None), HCV‐low (Low), and HCV‐high (High) groups. mRNA levels were quantified by qPCR using a standard curve from serial dilution of standard cDNA. The quantity of TMPRSS2 was normalized against that of 18S rRNA and is expressed in arbitrary units. HCV (−), nontumorous livers from 5 patients with hepatic metastases of colon cancer; HCV (+), nontumorous livers from 8 and 9 patients containing low and high viral loads, respectively, as shown in http://onlinelibrary.wiley.com/doi/10.1002/hep.27426/suppinfo. Two asterisks indicate a significant difference (P < 0.01) by Mann‐Whitney's U test. (B) IHC for TMPRSS2 protein in nontumorous liver with the low and high HCV loads. Brown, immunostain for TMPRSS2 with DAB; blue, counterstain for nuclei with hematoxylin; bar, 50 µm.
Inhibitors of Trypsin‐Like Serine Protease Inhibited an Early Stage of JFH‐1 HCVcc Infection In Vitro
To investigate the involvement of the cell‐surface trypsin‐like serine protease in HCV infection in vitro, we analyzed HCV infectivity by inoculating JFH‐1 HCVcc for 2 hours in the presence of several different protease inhibitors, followed by removing the inoculum and culturing for a further 2 days. Among the inhibitors to five classes of proteases, only serine protease inhibitors, including α1‐antitrypsin, SBTI, and AEBSF, inhibited the infectivity in a dose‐dependent manner (Fig. 2). To confirm the role of trypsin‐like serine proteases on the cell surface in an early stage of HCV infection, HCVcc were inoculated in the presence of trypsin. HCV infectivity was enhanced up to 2‐fold by 0.2 mg/mL of trypsin (Fig. 2), which had no effect on cell morphology. To determine which step during the early stage of infection (attachment or entry of HCVcc to the cells) was inhibited by SBTI, HCVcc were incubated with cells at 4°C for the first hour for attachment, and then, after removing the HCVcc inoculum, cells were incubated at 37°C for a further hour to allow entry (Fig. 3A). Then, cells were incubated for 1 day to determine HCV infectivity by IF staining of HCV‐positive cells. Even when SBTI was present only during the second 1‐hour step of entry, most of the HCV infectivity was inhibited (Fig. 3Ac). HCV replication postentry was not inhibited by SBTI (Fig. 3Ae,3B). These results suggest that the trypsin‐like serine protease is involved at an early stage of infection, particularly during entry of HCVcc into cells. SBTI also inhibited HCV infection, to some extent, with chimeric viruses containing other genotypes 3a and 1b of structural genes, which was not statistically significant (http://onlinelibrary.wiley.com/doi/10.1002/hep.27426/suppinfo).
Figure 2.
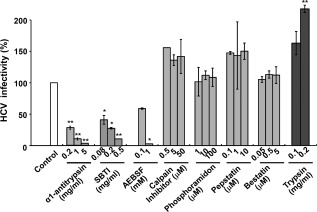
HCV infectivity in the presence of protease inhibitors and trypsin. Huh7‐25‐CD81 cells were inoculated with JFH‐1 HCVcc in the presence of various protease inhibitors (against serine protease, cysteine protease, metal protease, aspartate protease, and aminopeptidase) and trypsin at the concentrations indicated. After 2 hours, the inoculum was removed and the cells were cultured in fresh complete medium for 2 days. HCV infectivity was determined by the number of foci containing HCV‐positive cells. The control experiment was conducted in the absence of inhibitors and trypsin, and infectivity is expressed as 100%. Each experiment was performed in duplicate, and two independent experiments gave the same result. The results are expressed as the mean ± standard error of a representative experiment. *P < 0.05 and **P < 0.01 by the Student t test, respectively.
Figure 3.
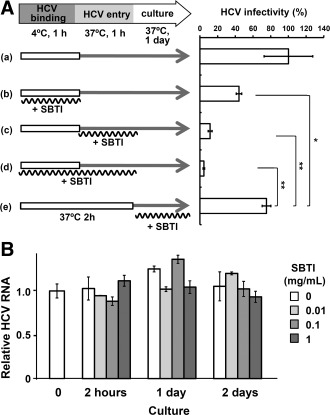
SBTI inhibited an early stage of HCV infection. (A) Two steps of HCV binding and entry were separated by incubation at 4°C and 37°C, respectively. Huh7‐25‐CD81 cells were precultured for 1 day, and JFH‐1 HCVcc were inoculated in the presence or absence of 1 mg/mL of SBTI at 4°C for 1 hour. After removing the inoculum, the cells were incubated with fresh complete medium in the presence or absence of 1 mg/mL of SBTI at 37°C for 1 hour. Then, after removing the SBTI, the cells were cultured in fresh complete medium for 24 hours and HCV infectivity was determined as described in Fig. 2. (a) Control experiment without SBTI. (b) SBTI at the binding step. (c) SBTI at entry. (d) SBTI at both steps. (e) SBTI after inoculation. Two independent experiments gave the same result, and a representative is shown. Data are shown as the mean ± standard error in duplicate experiments. *P < 0.05 and **P < 0.01 by the Student t test, respectively. (B) HCV RNA replication in HCV replicon cells after treatment with various concentrations of SBTI. HCV replicon cells were treated with SBTI (0, 0.01, 0.1, and 1 mg/mL) at 37°C for 2 hours. After removing SBTI, cells were incubated with fresh complete medium for 2 days. HCV RNA was determined at four time points and normalized to glyceraldehyde‐3‐phosphate dehydrogenase expression by a relative quantitation method. Relative HCV RNA was expressed as a ratio to that of replicon cells before treatment with SBTI.
Transmembrane Serine Proteases Expressed in Huh7‐25‐CD81 Cells Are Involved in JFH‐1 HCVcc Infection
We asked what type of cell‐surface serine proteases are expressed in Huh7‐25‐CD81 cells and whether the expression of these proteases are increased or decreased by HCVcc infection. We determined the mRNA levels of 10 transmembrane serine proteases pre‐ and postinfection with JFH‐1 HCVcc. FURIN and HPN were expressed in Huh7‐25‐CD81 cells, but TMPRSS2 was absent (Fig. 4A). FURIN mRNA was up‐regulated 2‐fold after infection with HCVcc (Fig. 4A). On the other hand, normal human livers predominantly expressed four proteases. HPN was the most abundant, followed by FURIN, TMPRSS6, and TMPRSS2 in descending order (http://onlinelibrary.wiley.com/doi/10.1002/hep.27426/suppinfo). Only TMPRSS2 was up‐regulated in HCV‐high livers (http://onlinelibrary.wiley.com/doi/10.1002/hep.27426/suppinfo and Fig. 1). Cell‐surface serine proteases in Huh7‐25‐CD81 cells were measured using synthetic fluorogenic substrates containing various amino acid sequences of P3‐P2‐P1 in the cleavage site (Fig. 4B). The highest protease activity was against QSR. The activities against QAR, QRR, FSR, and RVRR were of similar levels and less than half that of QSR. The substrate specificity profile of the Huh7‐25‐CD81 cells was quite different from that of pancreatic trypsin (Fig. 4B). Knockdown of FURIN and HPN by their respective siRNAs had no effect on the focus formation of infected cells (http://onlinelibrary.wiley.com/doi/10.1002/hep.27426/suppinfo). These results suggest that a serine protease other than FURIN and HPN is involved in the susceptibility of Huh7‐25‐CD81 cells to HCV infection.
Figure 4.
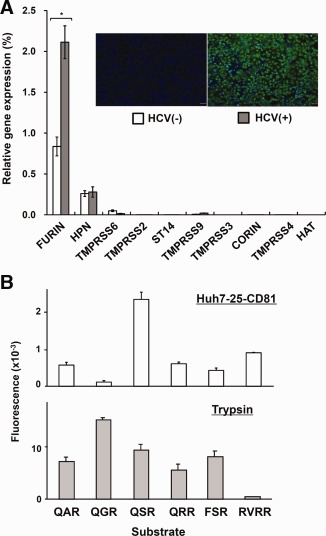
mRNA expression of transmembrane serine proteases and cell‐surface serine protease activity in Huh7‐25‐CD81 cells. (A) mRNA levels of the following 10 serine proteases were quantified by qPCR: type I serine protease (FURIN); HPN/TMPRSS2 subfamily, type II (HPN, TMPRSS2, TMPRSS3, and TMPRSS4); matriptase subfamily, type II (TMPRSS6, TMPRSS9, and suppression of tumorigenicity 14 [ST14]); CORIN subfamily, type II (CORIN); HAT subfamily, type II (human airway trypsin‐like protease; HAT). HCV (+) cells were prepared by inoculating JFH‐1 HCVcc (4 × 106 HCV RNA copies) to Huh7‐25‐CD81 cells (2 × 104, precultured for 1 day) and culturing in fresh complete medium for 6 days with one passage. HCV (−) cells were not infected and cultured in the same way. Relative gene expression is expressed as a percentage ratio of the expression of the target gene to that of glyceraldehyde‐3‐phosphate dehydrogenase. Insert panel, fluorescence staining of HCV proteins (green) with Alexa Fluor 555 and nuclei (blue) with DAPI. *P < 0.05 by the Student t test. (B) Cell‐surface serine protease activity of Huh7‐25‐CD81 cells was measured using the various peptide analog‐MCA substrates. The fluorescence released was determined for 3 hours and was normalized against the cell number determined by MTT assay. The protease activity of 20 ng per mL of pancreatic trypsin was also measured against the same peptides for 30 minutes. Data are shown as the mean ± standard error in duplicate experiments.
TMPRSS2 Activates HCVcc Infection
To investigate whether TMPRSS2 is involved in HCV infection, we established stable Huh7‐25‐CD81 cell clones producing WT TMPRSS2 and inactive mutant TMPRSS2 (S441A) (Fig. 5A,B). The cell‐surface proteases of the TMPRSS2‐WT clones were specifically activated against the substrates, QAR and QGR, putative substrates of TMPRSS2, but not the proteases of the TMPRSS2‐mutant and ‐negative control clones (Fig. 5C). When influenza A virus HA was introduced by transfection of the expression plasmid into the cell clones, HA induced cell‐to‐cell fusion only in the TMPRSS2‐WT cell clones 18 and 25 (Fig. 5D,E). The fusion depended on TMPRSS2 activity and was not induced in the TMPRSS2‐mutant and ‐negative cell clones. However, trypsin treatment induced cell fusion in the TMPRSS2‐mutant and ‐negative cell clones. Immunoblotting of TMPRSS2 showed WT‐dependent autocleavage of TMPRSS2; a 24‐kDa peptide was generated from the C‐terminal active domain in TMPRSS2‐WT cell clones, but not in the TMPRSS2‐mutant cell clones (http://onlinelibrary.wiley.com/doi/10.1002/hep.27426/suppinfo). Thus, TMPRSS2‐WT cell clones showed functional protease activity against synthetic peptides in vitro and native substrates in vivo. HCV infectivity was also enhanced in a WT‐dependent manner by the TMPRSS2‐positive cell clones. TMPRSS2‐WT cell clones were more sensitive to HCV infection than the TMPRSS2‐mutant and ‐negative cell clones (Fig. 5F).
Figure 5.
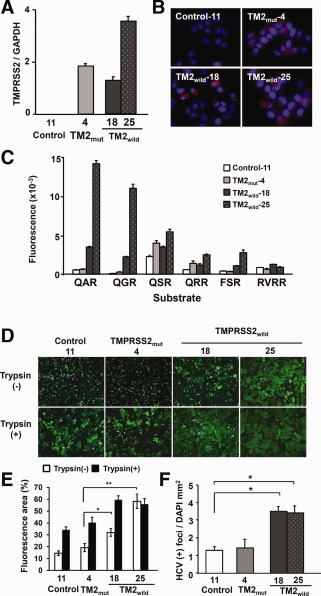
Stable Huh7‐25‐CD81 cell clones producing TMPRSS2. (A) Expression of TMPRSS2 mRNA. mRNA level was quantified by qPCR in stable Huh7‐25‐CD81 cell clones producing TMPRSS2‐WT (TM2wild−18, −25) and TMPRSS2‐mutant (TM2mut−4). A stable clone transfected with pEF6/V5‐His vector was used as a control (Control‐11). The quantity of TMPRSS2 mRNA was normalized against that of GAPDH mRNA by a relative quantitation method. (B) IF staining of TMPRSS2. Stable cell clones, as shown in (A), were stained for TMPRSS2 (red) by IF and for nuclei (blue) with DAPI. The TMPRSS2‐positive cell ratios were 0%, 80%, 20%, and 100% in Control‐11, TMmut−4, TM2wild−18, and −25, respectively. (C) Cell‐surface protease activity against various peptide analog‐MCA substrates. Four stable cell clones, as shown in (A), were examined for surface protease activity, as described in Fig. 4B. (D) Cell fusion induced by influenza A virus HA. Four stable cell clones, as shown in (A), were transfected with an HA‐expressing plasmid and EGFP‐expressing plasmid, and then treated with (+) or without (−) trypsin for 2 days. After treatment with low‐pH buffer, cells were observed using a fluorescence microscope. (E) Percentage of fluorescent areas in Fig. 5D. Data shown are the mean ± standard deviation of three different microscopic field areas. *P < 0.05 and **P < 0.01 by the Student t test, respectively. (F) HCV infectivity. Four stable cell clones were infected with JFH‐1 HCVcc and the number of HCV‐positive cell foci was measured and normalized to the area of DAPI. *P < 0.05 by the Student t test. Abbreviation: GAPDH, glyceraldehyde‐3‐phosphate dehydrogenase.
When TMPRSS2 mRNA was knocked down by siRNA, protease activity against QAR and QGR was markedly reduced in the TMPRSS2‐WT cell clones (Fig. 6A,B). Protease activity against QSR and FSR also responded to knockdown of WT TMPRSS2 (Fig. 6B). Enhanced HCV infectivity was also reduced to the level in the negative control by the knockdown of TMPRSS2 (Fig. 6C). The ability of HCV to infect TMPRSS2‐mutant cells was not changed, even though the mRNA was markedly knocked down (Fig. 6A,C). Thus, the TMPRSS2 protease activity activates HCVcc infectivity in vitro.
Figure 6.
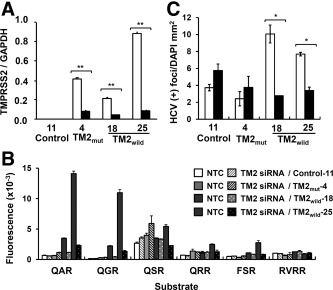
Knockdown of TMPRSS2 in four stable cell clones, Control‐11, TMmut−4, TM2wild−18, and −25. (A) Expression of TMPRSS2 mRNA. Level of TMPRSS2 mRNA was quantified by qPCR 2 days after transfection with a nontargeting control siRNA (open bar) or TMPRSS2 siRNA (closed bar). Expression is expressed as described in Fig. 5A. (B) Cell‐surface protease activity. Cells, 2 days post‐transfection with the siRNA (nontargeting control and TMPRSS2), as shown in (A), were incubated with six peptide analog‐MCA substrates, and the fluorescence released was measured as described in Fig. 5C. (C) HCV infectivity. Cells, 2 days post‐transfection with the siRNA (nontargeting control and TMPRSS2), as shown in (A), were inoculated with JFH‐1 HCVcc for 2 hours. Infectivity was determined as described in Fig. 5E. *P < 0.05 and **P < 0.01 by the Student t test, respectively. Abbreviation: GAPDH, glyceraldehyde‐3‐phosphate dehydrogenase.
Discussion
We found that 26 genes were significantly elevated in nontumorous, noncirrhotic livers with chronic hepatitis containing high HCV loads, compared with those containing low HCV loads (http://onlinelibrary.wiley.com/doi/10.1002/hep.27426/suppinfo and http://onlinelibrary.wiley.com/doi/10.1002/hep.27426/suppinfo). Unexpectedly, well‐known antiviral genes were rather highly expressed in livers with HCV‐high loads, including ISGs, inflammation/immune response‐related genes, and anti‐viral genes. For example, a markedly up‐regulated and typical ISG, OASL (http://onlinelibrary.wiley.com/doi/10.1002/hep.27426/suppinfo), the function of which was initially unknown, inhibited replication of JFH‐1 HCVcc, as described previously.15 Interestingly, epithelial‐mesenchymal transition (EMT)‐related genes22, 23 are notably up‐regulated (http://onlinelibrary.wiley.com/doi/10.1002/hep.27426/suppinfo), because an active EMT state was inhibitory to tight junction‐mediated viral infection, such as measles virus infection,24 and, most likely, for HCV infection. Thus, our comparative study showed that among two possible host responses to HCV infection and anti‐ and proviral responses, the antiviral response was the chief response in livers with high HCV loads. Antiviral responses are considered to result from high HCV loads, whereas proviral responses are a cause of high HCV loads. The persistence of high HCV loads may be caused by an unknown proviral mechanism by which the response is not sufficient to reduce HCV loads. Only six genes, including 4 ISGs, overlapped with previous studies that compared HCV‐negative and ‐positive livers (http://onlinelibrary.wiley.com/doi/10.1002/hep.27426/suppinfo).6, 8, 9, 25, 26 The genes first identified in this study are useful in elucidating the mechanisms of HCV control and persistence in the human liver. Genes such as TMPRSS2, whose roles are unknown in HCV control or persistence, remained to be described. We asked how TMPRSS2, which was quite significantly up‐regulated in HCV‐high livers, was associated with HCV infectivity in vitro.
We demonstrate here that trypsin‐like serine proteases are involved in the early stage of HCV infection, and that TMPRSS2 activates HCVcc infection in vitro. TMPRSS2 is a member of the type II transmembrane serine protease family21 and an activating protease in the airway epithelium for influenza virus infection. TMPRSS2 cleaves the surface glycoprotein, HA, to produce a fusion peptide and, so, facilitates the spread of the virus.27, 28, 29, 30 Thus, tissue expression of TMPRSS2 affects viral pneumotropism and pathogenicity. Recently, TMPRSS2 has also been shown to contribute to the spread of other respiratory viruses, such as severe acute respiratory syndrome coronavirus and parainfluenza viruses, by cleaving the viral spike protein and F protein, respectively, leading to cell‐to‐virus and cell‐to‐cell fusion.31, 32, 33, 34, 35 Current reports using TMPRSS2‐knockout mice indicate that TMPRSS2 is essential for influenza virus pathogenesis in mice.36, 37, 38 It is not known yet what the target protein of TMPRSS2 is in HCV infection or whether fusion activity is induced. TMPRSS2 is preferentially localized at the apical membrane of hepatocytes in human livers with high HCV loads (Fig. 1B), and TMPRSS2‐induced protease activity was detected at the surface of Huh7 cells (Fig. 5C). Therefore, TMPRSS2 might also be involved in HCV entry or spread at or close to the tight junctions of hepatocytes in the human liver, where the components, claudin‐1 (CLDN1) and occludin (OCLN), are key molecules for HCV entry and spread.39, 40 On the other hand, TMPRSS2 cleaves newly synthesized HA in the secretory pathway within the influenza virus‐infected cell30 and also plays a role in the release of porcine epidemic diarrhea virus.41 To determine whether activation by TMPRSS2 occurs at the early stage of HCV entry and/or spread or at the late stage of HCV release, it is crucial to identify a protein cleaved and activated by TMPRSS2 during HCV infection.
Our study suggests possible cleavage‐site sequences targeted by TMPRSS2; QAR and QGR at P3 to P1 are the most sensitive of the sequences examined (Figs. 5C and 6B). QSR and QTR are also sensitive sequences, shown by native proteins as targets. An active form of TMPRSS2 generated by autocleavage at QSR was produced in stably transfected cells in an enzyme‐dependent manner (http://onlinelibrary.wiley.com/doi/10.1002/hep.27426/suppinfo). TMPRSS2‐activated, HA‐mediated fusion occurred using the H3 subtype of HA (Udorn/72), in which the cleavage site is QTR (Fig. 5D,E). Thus, TMPRSS2 is broadly active on several cleavage‐site sequences; QXR, QAR, and QGR are the most sensitive, but RVRR is clearly resistant (Figs. 5C and 6B). Recently, the substrate for TMPRSS2 was characterized using various HA subtypes; most of the 18 subtypes of HA examined were cleaved by TMPRSS2, but with varying efficiency.42 Surveying the cleavage‐site sequences, such as (Q/E)XR, in the surface proteins of lipoviroparticles and host entry factors in HCV infection identifies several possible proteins as candidate targets of TMPRSS2: E2; apolipoprotein (APO)B; APOE; low‐density lipoprotein receptor; scavenger receptor class B, type I; CLDN1; and OCLN. The HCV fusion process depends on pH, the lipid composition of viral and target membranes, HCV E2, CD81, and CLDN1, though it remains unclear whether cellular proteases are required for this step.40 To identify the target of TMPRSS2 will be helpful in highlighting a novel activation mechanism for HCV infection.
Major endogenous transmembrane serine proteases, such as FURIN and HPN, are not involved in the infection of Huh7 cells by JFH‐1 HCVcc (http://onlinelibrary.wiley.com/doi/10.1002/hep.27426/suppinfo). We demonstrated that the cell‐surface serine proteases of Huh7 cells were mainly active against QSR, and secondly to RVRR and QAR cleavage‐site sequences, but nearly negative to QGR (Fig. 4B). TMPRSS2 and exogenous trypsin were both unable to cleave RVRR (Figs. 5C and 4B) and also enhanced HCV infection. These findings suggest that an endogenous transmembrane serine protease other than FURIN and HPN, which does not preferentially cleave QGR and RVRR, is most likely helpful to the HCVcc infectivity similar to TMPRSS2.
In conclusion, we demonstrated that TMPRSS2 was up‐regulated in human livers containing HCV‐high loads, and that TMPRSS2 activated JFH‐1 HCVcc infection in vitro. The cell‐surface, trypsin‐like serine protease was required for an early event in HCV infection/HCV entry. These results suggest that host transmembrane serine proteases, such as TMPRSS2, support HCV entry at the cell surface in human liver cells. The target of TMPRSS2 is a key molecule for proviral infection and its discovery will provide a new insight into antiviral strategy.
Author names in bold designate shared co‐first authorship.
Supporting information
Additional Supporting Information may be found at http://onlinelibrary.wiley.com/doi/10.1002/hep.27426/suppinfo.
Supplementary Informationpublishers website.
Acknowledgment
The authors thank Dr. Masamichi Oshima, National Institute of Infectious Diseases, Japan, for providing us with the Huh7‐it cells that were used for HCVcc production in this study. The authors also thank Drs. Sulyi Kim and Makoto Hijikata, Institute for Virus Research, Kyoto University, for helpfully conducting trials of the experiments. The authors also thank Drs. Keiko Takagi, Takuichi Oikawa, and Takao Mamiya, Nihon University Itabashi Hospital, Japan, for providing data on the patients and Prof. Shin Aizawa, Nihon University School of Medicine, for a helpful discussion.
Potential conflict of interest: Nothing to report.
This work was supported by a grant from the “Open Research Center” Project for Private Universities: matching fund subsidy from MEXT (2005), Nihon University Joint Research Grant for 2006, Nihon University Multidisciplinary Research Grant for 2009, Grants‐in‐Aid for Scientific Research (C) 22590350 and 25430142 from MEXT (2010), and “Strategic Research Base Development” Program for Private Universities subsidized by MEXT (2010).
References
- 1. Liang TJ, Ghany MG. Current and future therapies for hepatitis C virus infection. N Engl J Med 2013;368:1907‐1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Scheel TK, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med 2013;19:837‐849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Walters KA, Syder AJ, Lederer SL, Diamond DL, Paeper B, Rice CM, et al. Genomic analysis reveals a potential role for cell cycle perturbation in HCV‐mediated apoptosis of cultured hepatocytes. PLoS Pathog 2009;5:e1000269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blackham S, Baillie A, Al‐Hababi F, Remlinger K, You S, Hamatake R, et al. Gene expression profiling indicates the roles of host oxidative stress, apoptosis, lipid metabolism, and intracellular transport genes in the replication of hepatitis C virus. J Virol 2010;84:5404‐5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Woodhouse SD, Narayan R, Latham S, Lee S, Antrobus R, Gangadharan B, et al. Transcriptome sequencing, microarray, and proteomic analyses reveal cellular and metabolic impact of hepatitis C virus infection in vitro. Hepatology 2010;52:443‐453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bigger CB, Brasky KM, Lanford RE. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. J Virol 2001;75:7059‐7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Smith MW, Yue ZN, Korth MJ, Do HA, Boix L, Fausto N, et al. Hepatitis C virus and liver disease: global transcriptional profiling and identification of potential markers. Hepatology 2003;38:1458‐1467. [DOI] [PubMed] [Google Scholar]
- 8. Bigger CB, Guerra B, Brasky KM, Hubbard G, Beard MR, Luxon BA, et al. Intrahepatic gene expression during chronic hepatitis C virus infection in chimpanzees. J Virol 2004;78:13779‐13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Helbig KJ, Lau DT, Semendric L, Harley HA, Beard MR. Analysis of ISG expression in chronic hepatitis C identifies viperin as a potential antiviral effector. Hepatology 2005;42:702‐710. [DOI] [PubMed] [Google Scholar]
- 10. Kato T, Esumi M, Yamashita S, Abe K, Shikata T. Interferon‐inducible gene expression in chimpanzee liver infected with hepatitis C virus. Virology 1992;190:856‐860. [DOI] [PubMed] [Google Scholar]
- 11. Horner SM, Gale M, Jr. Regulation of hepatic innate immunity by hepatitis C virus. Nat Med 2013;19:879‐888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Metz P, Reuter A, Bender S, Bartenschlager R. Interferon‐stimulated genes and their role in controlling hepatitis C virus. J Hepatol 2013;59:1331‐1341. [DOI] [PubMed] [Google Scholar]
- 13. Akazawa D, Date T, Morikawa K, Murayama A, Miyamoto M, Kaga M, et al. CD81 expression is important for the permissiveness of Huh7 cell clones for heterogeneous hepatitis C virus infection. J Virol 2007;81:5036‐5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kato T, Date T, Miyamoto M, Furusaka A, Tokushige K, Mizokami M, et al. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 2003;125:1808‐1817. [DOI] [PubMed] [Google Scholar]
- 15. Ishibashi M, Wakita T, Esumi M. 2′,5′‐Oligoadenylate synthetase‐like gene highly induced by hepatitis C virus infection in human liver is inhibitory to viral replication in vitro. Biochem Biophys Res Commun 2010;392:397‐402. [DOI] [PubMed] [Google Scholar]
- 16. Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 2005;11:791‐796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shirogane Y, Takeda M, Iwasaki M, Ishiguro N, Takeuchi H, Nakatsu Y, et al. Efficient multiplication of human metapneumovirus in Vero cells expressing the transmembrane serine protease TMPRSS2. J Virol 2008;82:8942‐8946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sakai K, Nagata N, Ami Y, Seki F, Suzaki Y, Iwata‐Yoshikawa N, et al. Lethal canine distemper virus outbreak in cynomolgus monkeys in Japan in 2008. J Virol 2013;87:1105‐1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sakai K, Yoshikawa T, Seki F, Fukushi S, Tahara M, Nagata N, et al. Canine distemper virus associated with a lethal outbreak in monkeys can readily adapt to use human receptors. J Virol 2013;87:7170‐7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takeda M, Leser GP, Russell CJ, Lamb RA. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc Natl Acad Sci U S A 2003;100:14610‐14617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Choi SY, Bertram S, Glowacka I, Park YW, Pohlmann S. Type II transmembrane serine proteases in cancer and viral infections. Trends Mol Med 2009;15:303‐312. [DOI] [PubMed] [Google Scholar]
- 22. Thiery JP, Sleeman JP. Complex networks orchestrate epithelial‐mesenchymal transitions. Nat Rev Mol Cell Biol 2006;7:131‐142. [DOI] [PubMed] [Google Scholar]
- 23. Tikhmyanova N, Golemis EA. NEDD9 and BCAR1 negatively regulate E‐cadherin membrane localization, and promote E‐cadherin degradation. PLoS One 2011;6:e22102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shirogane Y, Takeda M, Tahara M, Ikegame S, Nakamura T, Yanagi Y. Epithelial‐mesenchymal transition abolishes the susceptibility of polarized epithelial cell lines to measles virus. J Biol Chem 2010;285:20882‐20890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bieche I, Asselah T, Laurendeau I, Vidaud D, Degot C, Paradis V, et al. Molecular profiling of early stage liver fibrosis in patients with chronic hepatitis C virus infection. Virology 2005;332:130‐144. [DOI] [PubMed] [Google Scholar]
- 26. De Giorgi V, Monaco A, Worchech A, Tornesello M, Izzo F, Buonaguro L, et al. Gene profiling, biomarkers and pathways characterizing HCV‐related hepatocellular carcinoma. J Transl Med 2009;7:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bottcher E, Matrosovich T, Beyerle M, Klenk HD, Garten W, Matrosovich M. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J Virol 2006;80:9896‐9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chaipan C, Kobasa D, Bertram S, Glowacka I, Steffen I, Tsegaye TS, et al. Proteolytic activation of the 1918 influenza virus hemagglutinin. J Virol 2009;83:3200‐3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bertram S, Glowacka I, Blazejewska P, Soilleux E, Allen P, Danisch S, et al. TMPRSS2 and TMPRSS4 facilitate trypsin‐independent spread of influenza virus in Caco‐2 cells. J Virol 2010;84:10016‐10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bottcher‐Friebertshauser E, Freuer C, Sielaff F, Schmidt S, Eickmann M, Uhlendorff J, et al. Cleavage of influenza virus hemagglutinin by airway proteases TMPRSS2 and HAT differs in subcellular localization and susceptibility to protease inhibitors. J Virol 2010;84:5605‐5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Matsuyama S, Nagata N, Shirato K, Kawase M, Takeda M, Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol 2010;84:12658‐12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bertram S, Glowacka I, Muller MA, Lavender H, Gnirss K, Nehlmeier I, et al. Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin‐like protease. J Virol 2011;85:13363‐13372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abe M, Tahara M, Sakai K, Yamaguchi H, Kanou K, Shirato K, et al. TMPRSS2 is an activating protease for respiratory parainfluenza viruses. J Virol 2013;87:11930‐11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bertram S, Dijkman R, Habjan M, Heurich A, Gierer S, Glowacka I, et al. TMPRSS2 activates the human coronavirus 229E for cathepsin‐independent host cell entry and is expressed in viral target cells in the respiratory epithelium. J Virol 2013;87:6150‐6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Glowacka I, Bertram S, Muller MA, Allen P, Soilleux E, Pfefferle S, et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol 2011;85:4122‐4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hatesuer B, Bertram S, Mehnert N, Bahgat MM, Nelson PS, Pohlman S, et al. Tmprss2 is essential for influenza H1N1 virus pathogenesis in mice. PLoS Pathog 2013;9:e1003774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tarnow C, Engels G, Arendt A, Schwalm F, Sediri H, Preuss A, et al. TMPRSS2 is a host factor that is essential for pneumotropism and pathogenicity of H7N9 influenza A virus in mice. J Virol 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sakai K, Ami Y, Tahara M, Kubota T, Anraku M, Abe M, et al. The host protease TMPRSS2 plays a major role for in vivo replication of emerging H7N9 and seasonal influenza viruses. J Virol 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brimacombe CL, Grove J, Meredith LW, Hu K, Syder AJ, Flores MV, et al. Neutralizing antibody‐resistant hepatitis C virus cell‐to‐cell transmission. J Virol 2011;85:596‐605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zeisel MB, Fofana I, Fafi‐Kremer S, Baumert TF. Hepatitis C virus entry into hepatocytes: molecular mechanisms and targets for antiviral therapies. J Hepatol 2011;54:566‐576. [DOI] [PubMed] [Google Scholar]
- 41. Shirato K, Matsuyama S, Ujike M, Taguchi F. Role of proteases in the release of porcine epidemic diarrhea virus from infected cells. J Virol 2011;85:7872‐7880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Galloway SE, Reed ML, Russell CJ, Steinhauer DA. Influenza HA subtypes demonstrate divergent phenotypes for cleavage activation and pH of fusion: implications for host range and adaptation. PLoS Pathog 2013;9:e1003151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found at http://onlinelibrary.wiley.com/doi/10.1002/hep.27426/suppinfo.
Supplementary Informationpublishers website.


