Summary
Digit loss/reductions are evolutionary adaptations in cursorial mammals such as pigs. To gain mechanistic insight into these processes, we performed a comparative molecular analysis of limb development in mouse and pig embryos, which revealed a loss of anterior-posterior polarity during distal progression of pig limb bud development. These alterations in pig limb buds are paralleled by changes in the mesenchymal response to Sonic hedgehog (SHH) signaling, which is altered upstream of the reduction and loss of Fgf8 expression in the ectoderm that overlaps the reduced and vestigial digit rudiments of the pig handplate, respectively. Furthermore, genome-wide open chromatin profiling using equivalent developmental stages of mouse and pig limb buds reveals the functional divergence of about one-third of the regulatory genome. This study uncovers widespread alterations in the regulatory landscapes of genes essential for limb development that likely contributed to the morphological diversion of artiodactyl limbs from the pentadactyl archetype of tetrapods.
Keywords: morphological evolution, limb development, cis-regulation, SHH signaling, artiodactyl, pig, mouse, ATAC-seq
Graphical Abstract
Highlights
-
•
AP polarity is lost during distal progression of pig limb bud development
-
•
The mesenchymal response to SHH is anteriorly expanded in pig limb buds
-
•
AER-Fgf8 expression dynamics correlates with the loss and reduction of digits in pigs
-
•
Widespread divergence of the mouse and pig limb bud regulatory genomes
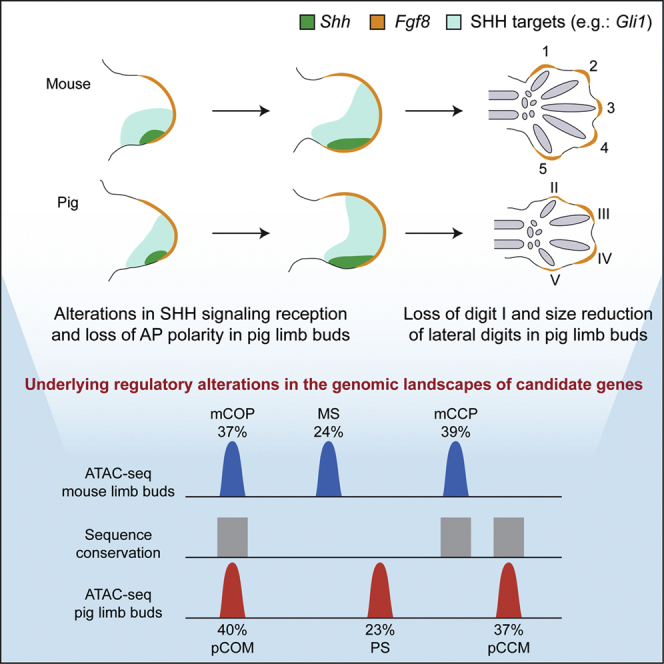
Tissières et al. show that, in comparison to the pentadactyl mouse limb, the expression and regulation of key developmental pathways are altered during pig limb bud development. These changes likely contributed to the morphological evolution of the highly specialized pig limb skeleton, with two weight-bearing and two reduced digits.
Introduction
The tetrapod limb is a paradigm for studying the mechanisms controlling how animal morphology is encoded in the genome (Lopez-Rios, 2016, Petit et al., 2017, Zuniga, 2015). Extensive molecular and genetic analysis of human congenital malformations and of animal models has identified most of the genes with critical functions during limb skeletal development. However, the understanding of the topology and dynamics of the gene regulatory networks (GRNs) that coordinate limb patterning, growth, and differentiation, such as those controlled by HH, WNT, FGF, and BMP signaling, is still sketchy. Genome-wide functional genomics approaches allow for systematic cataloging of cis-regulatory elements (CREs) that interact and are regulated by transcriptional regulators that are ultimately responsible for wiring these GRNs (Osterwalder et al., 2014, Shen et al., 2012, Yue et al., 2014). These advances in our understanding of the complexity of gene regulation are also highly relevant to studying the mechanisms underlying the diversification of animal morphology, as cis-regulatory variations impacting the expression of developmental genes are thought to be a major driver of morphological evolution (Acemel et al., 2017, Carroll, 2008, Long et al., 2016, Stern and Frankel, 2013). Hence, genome-wide analyses of gene regulation have advanced our understanding of the regulatory changes underlying limb development in humans (Cotney et al., 2013), bats (Eckalbar et al., 2016), birds (Seki et al., 2017), and reptiles (Roscito et al., 2018).
Here, we use gene expression analysis and profiling of accessible chromatin in mouse and pig (Sus scrofa) limb buds to map the extent of regulatory divergence between the pentadactyl (mouse) and the streamlined artiodactyl (pig) limb skeletons (Polly, 2007). Previous analysis established that Sonic hedgehog (SHH) signal transduction is changed and Fgf8 expression is reduced in the apical ectodermal ridge (AER) of bovine and pig limb buds in comparison to pentadactyl mouse limb buds (Cooper et al., 2014, Lopez-Rios et al., 2014). Our study uncovers that in contrast to mouse, pig limb development is characterized by a progressive loss of anterior-posterior (AP) polarity in the distal limb bud mesenchyme, which correlates with changes in SHH signal transduction. These events are followed by a reduction and loss of Fgf8 signaling in the AER overlaying the digit primordia in pig limb buds, providing a likely molecular explanation for both digit reductions and loss. In addition, the open chromatin profiling in developing limb buds of both species uncovers the extent of the variation in conserved regulatory regions located in the genomic landscapes of morpho-regulatory genes, which provides an entry point to investigate the molecular mechanisms underlying the evolutionary diversification of the artiodactyl limb skeleton.
Results
Comparative Gene Expression Analysis during the Progression of Mouse and Pig Limb Bud Outgrowth
The order Artiodactyla appears in the fossil record in the early Eocene, around 55 million years (Ma) ago, and soon diversified into several groups, including the suborder Suina (pigs and peccaries; Figure 1A) (Prothero and Foss, 2007). In contrast to mice or humans, which have retained the ancestral pentadactylous blueprint of tetrapods, the limb skeleton of pigs has diverged significantly. As in all extant terrestrial artiodactyls, pig limbs are long and slender, with elongated metacarpal/metatarsal bones and a symmetric handplate (indicative of loss of AP digit polarity; Cooper et al., 2014, Lopez-Rios et al., 2014, Polly, 2007). In pigs, the anterior-most digit (digit I) is lost, digits II and V are very much reduced, and all the weight of the animal rests onto the terminal phalanges of digits III and IV (unguligrade posture), which are hoofed and of equal length (Figures 1B, 1C, and S1; Clifford, 2010, Prothero and Foss, 2007).
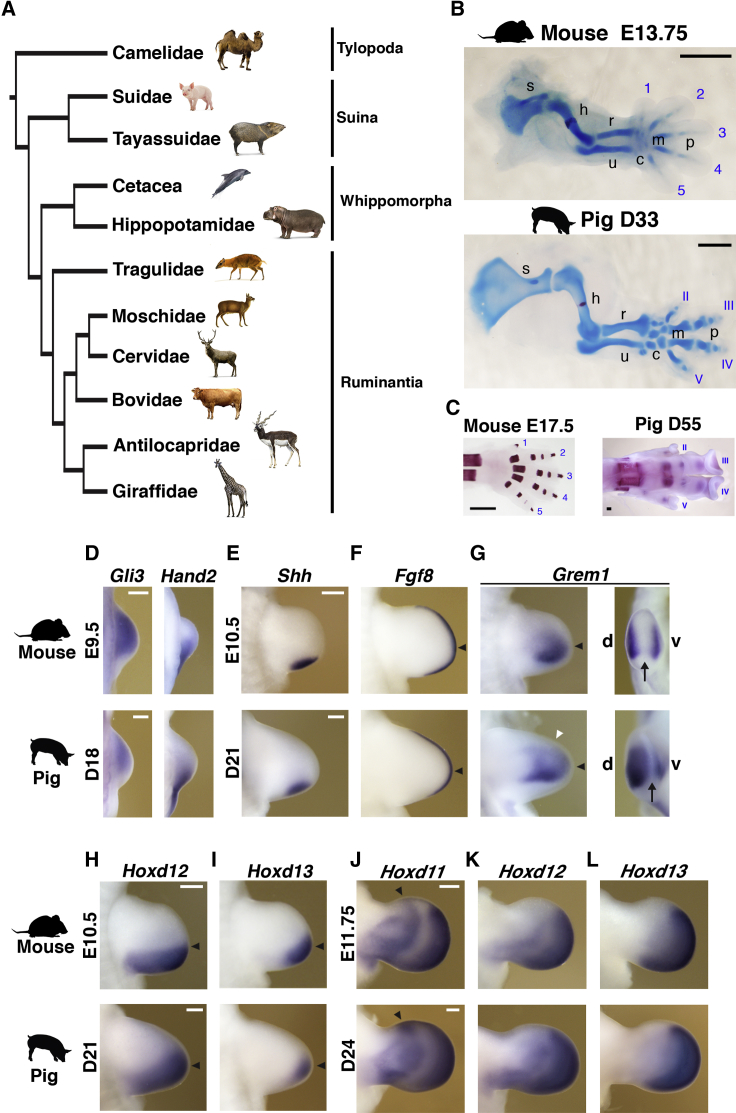
Figure 1.
Comparative Analysis of Mouse and Pig Limb Bud Development
(A) Phylogenetic tree of Artiodactyla.
(B) Developing mouse and pig (Sus scrofa) forelimb skeletons at equivalent embryonic stages (mouse: embryonic day [E] 13.75; pig: gestational day [D] 33). Cartilage is stained with Alcian blue and bone with alizarin red.
(C) Mineralization pattern of the mouse (E17.5) and pig (D55) handplate skeleton. Digits are numbered from 1 to 5 (mouse) and II to V (pig) from anterior to posterior. s, scapula; h, humerus; r, radius; u, ulna; c, carpals; m, metacarpals; p, phalanges.
(D–L) Whole-mount RNA in situ hybridization (ISH) analysis of mouse and pig forelimb buds at equivalent developmental stages.
(D) Expression of Gli3 and Hand2 in mouse (E9.5) and pig limb buds (D18).
(E–I) Spatial distribution of Shh (E), Fgf8 (F), Grem1 (G), Hoxd12 (H), and Hoxd13 (I) transcripts in mouse (E10.5) and pig forelimb buds (D21). Black arrowheads indicate the limb bud apex. A white arrowhead points to the anterior Grem1 expression in pig limb buds (left panels in G). Black arrows point to the gap between the dorsal (d) and ventral (v) Grem1 domains in limb bud apices (right panels in G).
(J–L) Expression of Hoxd11 (J), Hoxd12 (K), and Hoxd13 (L) in mouse (E11.75) and pig (D24) handplates. Black arrowheads indicate the anterior Hoxd11 expression domain in pig handplates.
Pig gene names are not capitalized for simplicity. n = 2 per stage for all pig ISH probes and skeletal stainings; n = 3 per stage for all mouse ISH probes and skeletal stainings. All limbs shown are oriented with anterior to the top, except right panels in (G). Scale bars, 1 mm (B and C) and 0.25 mm (D–L).
See also Figures S1 and S2.
To gain insight into the underlying potentially evolutionarily relevant molecular changes during limb bud and skeletal development, we initially performed a comparative RNA in situ hybridization (ISH) analysis using equivalent stages of mouse and pig limb buds (Figures 1D–1L, 2, 3, and S2). Initiation and early limb bud development were analyzed by studying the expression of key transcriptional regulators such as Gli3 and Hand2, which function in the AP polarization upstream of activating SHH signaling (Figure 1D; Osterwalder et al., 2014, te Welscher et al., 2002), and Tbx4/Tbx5 and Pitx1, which are required for initiating limb bud outgrowth and establishing hindlimb identity (Figure S2; Agarwal et al., 2003, Logan and Tabin, 1999, Minguillon et al., 2005, Naiche and Papaioannou, 2003, Rallis et al., 2003). This analysis showed that the expression of these genes is very similar in developmentally equivalent early limb bud stages (mouse embryonic day [E] 9.5 and pig gestational day [D] 18; Figures 1D, S1, and S2).
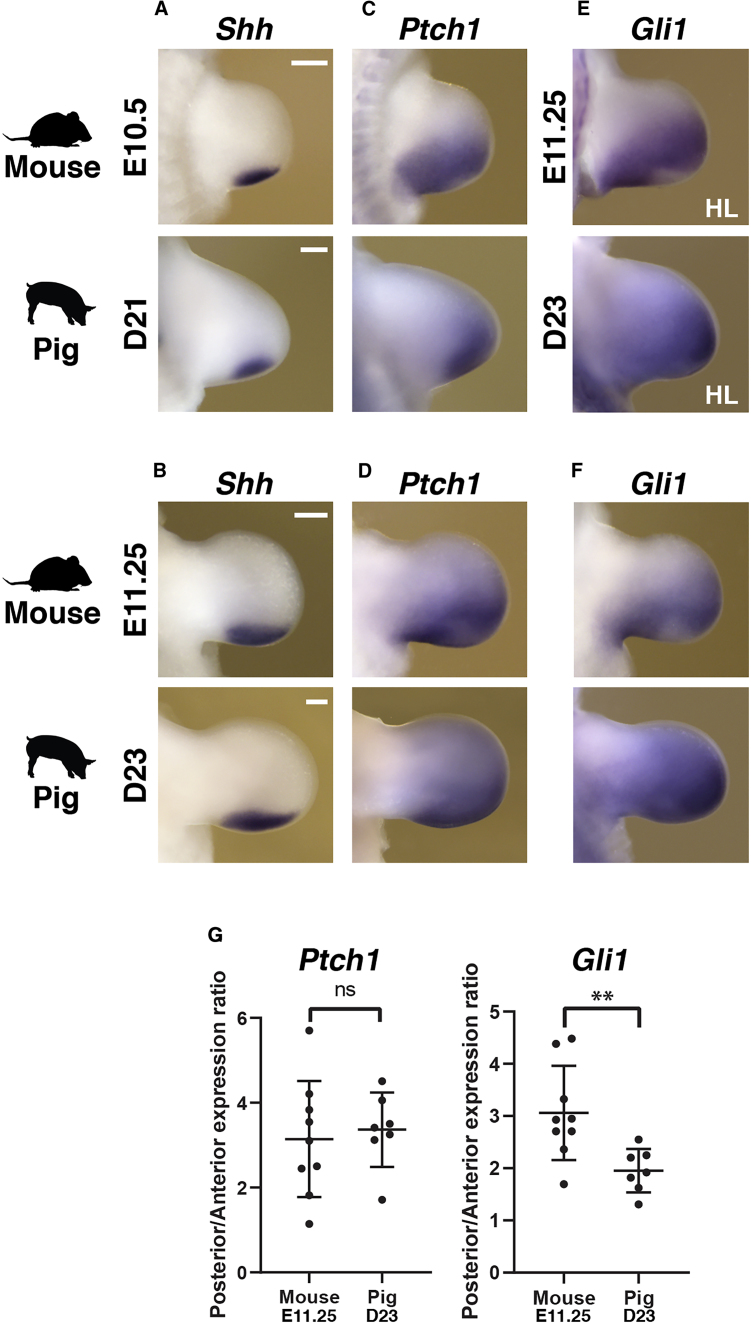
Figure 2.
Divergent SHH Signal Transduction in Mouse and Pig Limb Buds
(A–F) Expression of Shh (A and B), Ptch1 (C and D), and Gli1 (E and F) in mouse and pig limb buds at equivalent stages of limb development.
(G) Expression ratio of Ptch1 and Gli1 in mouse (E11.25) and pig (D23) limb buds dissected into anterior and posterior halves using qRT-PCR analysis (data are represented as mean ± SD; n = 9 for mouse and n = 7 for pig limb buds). HL, hindlimb bud. Scale bars, 0.25 mm. ns, non-significant (p = 0.68); ∗∗p = 0.0073. n = 2 per stage for all pig ISH probes; n = 3 per stage for all mouse ISH probes.
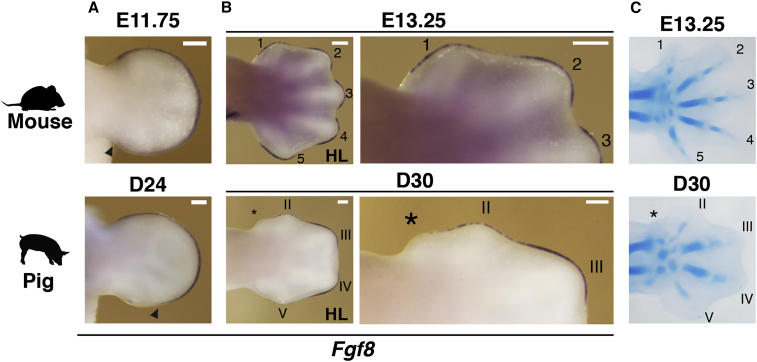
Figure 3.
Progressive Reduction of AER-Fgf8 Expression Correlates with the Loss of Digit I and the Reduced Length of Digits II and V in Pig Limb Buds
(A and B) Comparative analysis of Fgf8 expression during handplate (A) and digit ray development (B) in mouse and pig limb buds. Black arrowheads in (A) point to the posterior-proximal limit of Fgf8 expression. Right panels in (B) are close-ups of the anterior margins of the limb buds shown in the left panels.
(C) Mouse (E13.25) and pig forelimbs (D30) stained with Alcian blue to reveal the cartilage condensation patterns that prefigure the definitive digit rays.
Asterisks indicate vestigial digit I condensation. Scale bars, 0.25 mm. n = 2 per stage for all pig ISH probes and skeletal stainings; n = 3 per stage for all mouse ISH probes and skeletal stainings.
Outgrowth and patterning of the vertebrate limb bud are coordinately controlled by an epithelial-mesenchymal feedback signaling system operating between the SHH-producing polarizing region in the posterior mesenchyme and the AER, which expresses Fgf8 throughout development (Crossley and Martin, 1995, Riddle et al., 1993). This self-regulatory system is established by bone morphogenetic protein (BMP)-mediated induction and SHH-dependent upregulation of the BMP antagonist Grem1 (Bénazet et al., 2009, Michos et al., 2004, Zúñiga et al., 1999). Whereas Shh is expressed in a comparable manner in limb buds of both species (mouse E10.5 and pig D21; Figure 1E; Cooper et al., 2014), the AER-Fgf8 expression domain is more distally restricted in the posterior margin of pig limb buds (Figure 1F). Concurrently, the mesenchymal Grem1 expression extends more anteriorly (white arrowhead in Figure 1G) and the characteristic gap in the distal limb bud apex between the dorsal and ventral Grem1 domains is reduced in pig limb buds (arrows in right panels in Figure 1G). In addition, the Hoxd12 and Hoxd13 expression domains are more distally restricted in pig limb buds at early stages in comparison to their mouse orthologs (Figures 1H and 1I). Later on, the 5′Hoxd expression domains appear slightly expanded anteriorly, which is most evident for the rather symmetric (pig) than asymmetric (mouse) Hoxd11 expression during autopod development (black arrowheads in Figure 1J; see also Figures 1K and 1L).
Alterations of SHH Signal Transduction in Pig Limb Buds Upstream of Losing AP Digit Asymmetry
Morphogenetic SHH signaling is key for AP patterning and outgrowth of vertebrate limb buds (Ahn and Joyner, 2004, Chiang et al., 2001, Harfe et al., 2004, Lopez-Rios et al., 2012, Riddle et al., 1993, Scherz et al., 2007, Towers et al., 2008, Zhu et al., 2008). Previous analysis showed that, in contrast to mouse embryos, the transcriptional upregulation of Ptch1 is disrupted in bovine and pig limb buds (Cooper et al., 2014, Lopez-Rios et al., 2014). Given the morphological differences in digits and molecular changes during handplate formation between mouse and pig limb buds (Figure 1), we next assessed the dynamics of Shh expression and of its direct transcriptional targets Gli1 and Ptch1 (Marigo et al., 1996a, Marigo et al., 1996b, Vokes et al., 2008). Whereas Shh expression is similar in both early and developmentally advanced mouse and pig limb buds (Figures 2A and 2B), Ptch1 expression appears more distally restricted in the posterior mesenchyme of pig in comparison to mouse limb buds at early stages (Figure 2C). However, during handplate development, the posterior Ptch1 expression domains in both species appear rather similar (Figure 2D; see also Cooper et al., 2014), which is corroborated by qRT-PCR analysis (Figure 2G). qRT-PCR analysis shows that the ratio of Ptch1 transcripts in anterior-to-posterior handplates is rather similar in pig and mouse limb buds (Figure 2G). In contrast, the expression domain of Gli1, a transcriptional sensor of SHH signal transduction (Marigo et al., 1996a), is expanded in pig limb buds, in particular at handplate stages (Figures 2E and 2F). This was confirmed by qRT-PCR analysis, as the ratio of posterior to anterior Gli1 transcripts is lower in pig than mouse autopods (Figure 2G). These changes in SHH signal transduction could be linked to the progressive alteration of AP polarity as observed by the differences in the expression of other SHH target genes such as Grem1 and 5′Hoxd (Figure 1; Probst et al., 2011, Zuniga et al., 2012).
Progressive Reduction of AER-Fgf8 Activity Correlates with Digit Length in Pig Limb Buds
AER-FGF signaling is required for the elongation of digit primordia (Lewandoski et al., 2000, Mariani et al., 2008, Sanz-Ezquerro and Tickle, 2003). As in other tetrapods (de Bakker et al., 2013), the only digit lost in pig limbs is the most anterior digit I, whereas the reduced digits II and V retain three phalanges and their metacarpal/metatarsal bones (Figures 1C and S1). Previous studies showed that digit reductions (II and V) and loss (I) in pig limbs are not caused by increased apoptosis (Sears et al., 2011), but might be a consequence of restricted AER-Fgf8 expression during digit development (Cooper et al., 2014). Therefore, we assessed the spatiotemporal AER-Fgf8 expression from early to late pig limb bud development in comparison to mouse limb buds (Figures 3A and 3B). Whereas Fgf8 is expressed by the entire AER until regression in mouse limb buds (Figures 3A and 3B; Crossley and Martin, 1995), Fgf8 is not expressed in the proximal part of the posterior AER from early pig limb bud stages onward (Figures 1F and 3A). During mouse digit development (E13.25), the digit ray primordia are clearly apparent and Fgf8 remains expressed in the AER overlaying all five growing digits (upper panels in Figures 3B and 3C). In contrast, the strongest Fgf8 expression in pig handplates (D30) is detected in the AER overlaying the two central digit primordia (III and IV), whereas it is decreased over the prospective digits II and V (reduced) and lacking over the vestigial condensation for digit I (asterisks in lower panels in Figures 3B and 3C). This differential Fgf8 expression correlates well with the observed definitive digit pattern in pig embryos and provides a straightforward molecular explanation for the reduction of digits II and V and loss of the anterior-most digit I from the pig limb skeleton (Figures 1B, 1C, and S1).
Shared and Differential Chromatin Accessibility during Pig and Mouse Limb Bud Development
We next aimed to identify regulatory changes that might have contributed to the morphological diversification of the pig limb skeleton. To do so, we used the assay for transposase-accessible chromatin using sequencing (ATAC-seq) (Buenrostro et al., 2013) to generate a genome-wide map of the accessible chromatin regions, which characterizes promoters, CREs, and insulators (i.e., all genomic regions with cis-regulatory activity). ATAC-seq of dissected forelimb buds from mouse and pig embryos at early (E10.5/D21) and handplate stages (E11.5/D24) in combination with bioinformatic analysis revealed a very high correlation of the two biological replicates per stage and a broad overlap between the two datasets for both mouse and pig (Figures 4A and 4B). Statistical analysis of the combined datasets provided a global view of the limb bud regulome for both species, detecting 97,528 regions of accessible chromatin in mouse and 87,378 in pig limb buds (Figures 4A and 4B; Tables S1 and S2; see STAR Methods). These open chromatin regions are distributed in a similar manner in both genomes with respect to their positions in relation to promoters, exons, introns, or intergenic regions and their distances to the closest transcriptional start site (TSS; Figures 4A and 4B).
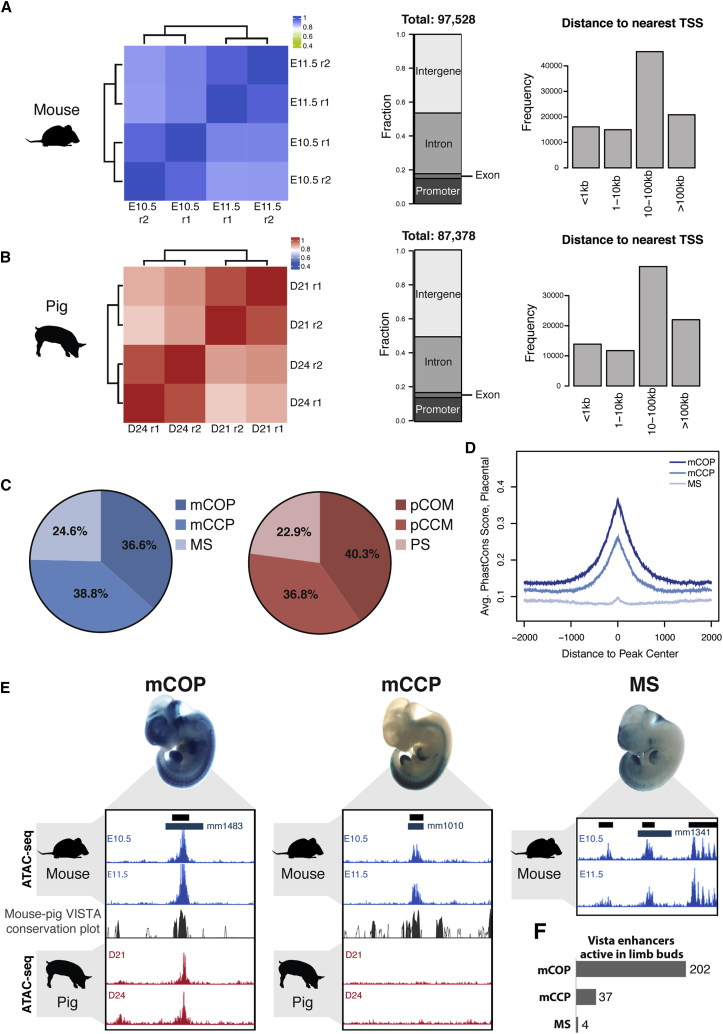
Figure 4.
Comparative ATAC-Seq and Regulome Analysis of Two Equivalent Stages of Mouse and Pig Limb Buds
(A and B) Pearson correlation heatmaps comparing the different ATAC-seq samples (left panels) and cataloging of regions of open chromatin (central and right panels) in mouse (A) and pig (B) (r, biological replicate; n = 2 per stage).
(C) Pie chart graphs indicating the proportion of evolutionarily conserved and/or open chromatin regions in mouse (blue; mCOP, mCCP, and MS) and pig (red; pCOM; pCCM; PS).
(D) Average PhastCons conservation score for placental mammals for mCOP, mCCP, and MS open chromatin regions.
(E) UCSC genome browser windows corresponding to 10 kb around representative mCOP (mm1483; n = 8/8), mCCP (mm1010; n = 9/9), and MS (mm1341; n = 6/6) regulatory regions that have been functionally validated as VISTA enhancers (Visel et al., 2007). Blue bars: VISTA enhancer; black bars: called ATAC-seq peak region.
(F) Number of mCOP, mCCP, and MS regions identified in the VISTA database as enhancers active in mouse limb buds.
See also Tables S1 and S2 and Figures S3–S6.
Next, we performed an inter-species comparison of the ATAC-seq datasets taking into account evolutionary sequence conservation and accessibility profiles. This strategy allowed us to categorize the open chromatin regions in mouse limb buds as follows (Figure 4C, left panel): (1) mCOP (36.6%): conserved orthologous regions of open chromatin in both mouse and pig limb buds; (2) mCCP (38.8%): conserved and open chromatin in mouse but closed in pig limb buds; and (3) MS (mouse specific; 24.6%): open chromatin regions in the mouse that are not scored as conserved in the pig genome using the liftOver tool (Table S1; see STAR Methods).
Plotting these mouse subsets according to their level of sequence conservation in placental mammals indicates that the mCOP regions are evolutionarily more conserved than mCCP regions (Figure 4D). This analysis also reveals that the sequences of MS regions are not well conserved among placental mammals. Gene ontology analysis of the three categories using GREAT (McLean et al., 2010) revealed that both the mCOP and mCCP regions are enriched in the genomic landscapes of genes functioning in skeletal and bone morphogenesis (Figure S3). In contrast, the MS sequences were not significantly enriched in genes that function in these two biological processes (Figure S3).
The pig limb bud regulome was categorized using the same approach, which resulted in strikingly similar overall distributions (Figure 4C, right panel; Table S2): (1) pCOM (40.3%): conserved open chromatin regions in both pig and mouse limb buds; (2) pCCM (36.8%): conserved and open regions in pig that are not accessible (i.e., closed) in mouse limb buds; and (3) PS (pig specific): 22.9% of the open chromatin regions in pig limb buds are not present in the mouse genome as determined by the liftOver tool (Table S2; see STAR Methods).
We next validated our mouse dataset by overlapping the different subgroups of open chromatin regions identified in mouse limb buds with the VISTA-expressed enhancer database (Visel et al., 2007; representative examples for all three subgroups are shown in Figure 4E). This analysis reveals that most of the VISTA enhancers expressed in mouse limb buds overlap with regions of open chromatin detected in mouse limb bud ATAC-seq datasets (243/336; Table S3) and are evolutionarily conserved, i.e., belong to the mCOP and mCCP subgroups (Figure 4F; Tables S1 and S2). In particular, our ATAC-seq analysis of limb buds (mouse E10.5/11.5 and pig D21/D24) detected many, but not all, of the previously identified limb cis-regulatory regions and enhancer clusters in the HoxD genomic landscape (Andrey et al., 2013, Gonzalez et al., 2007, Montavon et al., 2011), and two of them displayed differential accessibility between limb buds of the two species (Figure S4). In contrast, neither of the two known Tbx5 enhancers (Int2 [Minguillon et al., 2012] and CNS12sh [Adachi et al., 2016]) are part of the open chromatin domains in pig and mouse limb buds at the stages analyzed (Figure S5). However, our studies identified additional mCOP/pCOM regions, some of which could encode additional Tbx5 cis-regulatory regions active in mouse and pig forelimb buds at E10.5/E11.5 (Figure S5), which may explain why Tbx5 remains expressed in mouse embryos lacking both the Int2 and CNS12sh enhancers (Cunningham et al., 2018). We also annotated all differentially accessible chromatin regions in the genomic landscape of Gli1, as this transcriptional sensor of SHH signal transduction is differentially expressed in mouse and pig limb buds (Figures 2E–2G). This analysis identified seven pCCM/PS and two mCCP regions that could encode CREs contributing to the anteriorly expanded Gli1 expression domain in pig limb buds (Figure S6). Alternatively, the enhanced SHH signaling input in the anterior pig limb bud mesenchyme could also be integrated at the level of the Gli1 promoter, which is an mCOP/pCOM region that has been previously shown to be enriched in GLI-chromatin complexes in mouse limb buds (Vokes et al., 2008).
Regulatory Divergence between Mouse and Pig Limb Buds Affects the GRNs Controlling Limb Bud and Skeletal Morphogenesis
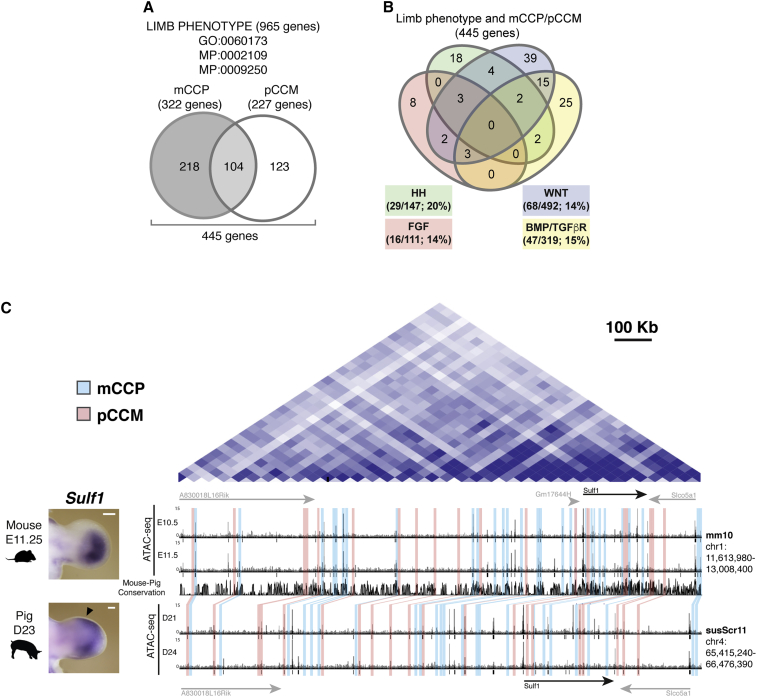
The analysis of the mouse and pig limb bud ATAC-seq datasets uncovered a broad range of evolutionarily conserved regions with limb regulatory activities in only one of the two species. From the perspective of the pig genome, these changes represent either loss (mCCP; 37,797 regions; Table S1) or gain (pCCM; 32,175 regions; Table S2) of chromatin accessibility. Each of these mCCP/pCCM regions was assigned to the closest TSS within their corresponding mouse and pig topologically associating domains (TADs), which delimit the genomic domains in which chromatin interactions are favored (Dixon et al., 2012, Foissac et al., 2019; Tables S1 and S2). Next, we sought to determine which of the genes with known important functions in limb buds were preferentially affected by this divergence in chromatin accessibility in evolutionarily conserved regions. We used gene and mammalian phenotype ontology tools to compile a list of 965 genes with essential functions during limb bud development and skeletal morphogenesis, which are referred to as “limb phenotype” genes (LP genes; Table S4). The overlap of this LP dataset and the list of genes assigned to the mCCP and pCCM peaks (Tables S1 and S2) reveals that 218 LP genes have at least one associated mCCP region, whereas 123 LP genes are associated with one or more pCCM regions. Furthermore, 104 LP genes associate with both mCCP and pCCM regions (Figure 5A; Table S5). These 445 genes encompass components of all major signaling pathways required for limb bud development (Figure 5B; Table S6).
Figure 5.
Evolutionarily Conserved Regions with Divergent Chromatin Accessibility Associated with Genes Essential for Limb Morphogenesis
(A) Limb regulatory genes (see Table S4) with associated mCCP and/or pCCM regions (assigned to the closest TSS).
(B) The list of 445 limb phenotype (LP) genes associated with regions of differential chromatin accessibility in the two species (see Table S5). This list encompasses genes functioning in the SHH, WNT, BMP/TGFβ, and FGF signaling pathways (see Table S6).
(C) Sulf1 expression in mouse (n = 3) and pig (n = 2) handplates. Scale bars, 0.25 mm. Right: HiC contact profile (Dixon et al., 2012) of the mouse Sulf1 TAD (top panel). Additional genes located in the TAD are indicated in gray. ATAC-seq profiles from mouse (E10.5/E11.5) and pig forelimb buds (D21/D24) (bottom panel).
To exemplify this regulatory variation in its chromosomal context, we selected Sulf1, a gene that modulates SHH, WNT, FGF, and BMP signaling during limb skeletal development by modifying extracellular matrix heparan sulfate proteoglycans (HSPGs; Danesin et al., 2006, Dhoot et al., 2001, Fellgett et al., 2015, Ratzka et al., 2008, Viviano et al., 2004, Wang et al., 2004). The systematic annotation of all mCCP (in blue) and pCCM (in pink) regions present in the Sulf1 TAD detected widespread alterations of the open chromatin accessibility between the mouse and pig syntenic regions (Figure 5C). The potential functional relevance of these changes is corroborated by the distinct spatial differences in Sulf1 expression in mouse and pig limb buds (arrowhead in Figure 5C). It is likely that not all these chromatin regions encode essential cis-regulatory regions due to functional redundancy among different CREs (Cannavò et al., 2016, Osterwalder et al., 2018) or because they correspond to CTCF-binding regions that mark chromatin domain boundaries (Rao et al., 2014). However, this approach allows pinpointing differences in the chromatin state that are candidates for the functionally relevant genomic and cis-regulatory changes that underlie the evolutionary diversification from pentadactyly of the artiodactyl limb skeleton.
Discussion
Our comparative molecular analysis of mouse and pig limb buds reveals a progressive loss of AP polarity during distal progression of pig limb bud development. These changes are overall very similar to the ones previously reported for bovine limb bud development (Lopez-Rios et al., 2014), suggesting that they were present in the last common ancestor of pigs and cows that lived ∼50 Ma ago (Benton and Donoghue, 2007, Cooper et al., 2014, Prothero and Foss, 2007). This loss of asymmetry in the handplate is paralleled by the distal restriction of Ptch1 expression in early pig limb buds (this study) and the failure to upregulate its expression in bovine limb buds (Lopez-Rios et al., 2014). The spatiotemporal upregulation of Ptch1 is key to regulating SHH signal transduction, as PTCH1 controls the spread of the SHH ligand by sequestering it close to the source (Briscoe et al., 2001, Chen and Struhl, 1996). Hence, the observed spatiotemporal difference of Ptch1 expression in both these artiodactyl limb buds affects the range of SHH signal transduction, which is evidenced by the anteriorly expanded expression domains of the SHH targets Gli1, Grem1, and 5′Hoxd genes in pig and bovine limb buds (this study and Lopez-Rios et al., 2014). In the present study, we show that the expression of Sulf1 is also anteriorly expanded in pig limb buds, which could further potentiate changes in morphogenetic and SHH signaling (Danesin et al., 2006).
Previous observations correlated digit loss in bovines and pigs with the precocious downregulation of AER activity as monitored by Fgf8 expression (Cooper et al., 2014, Lopez-Rios et al., 2014) but did not provide insight into the alterations underlying the differences in reduction to two (bovine) or four (pig) toes. In contrast to cattle, the cartilage rudiments of the future digits II and V continue to be exposed to AER-FGF8 signaling in pig limb buds, which supports their development into short digits (this study). Hence, the observed spatial changes in AER-Fgf8 expression in pig limb buds appear to support the formation of two elongated central digits and two smaller digits forming the dewclaws. Dewclaws with tri-phalangeal reduced digits have been retained in different artiodactyl species such as pigs, moose, and reindeers, and provide additional support for walking on soft terrains such as mud or snow (Prothero and Foss, 2007). Interestingly, it has been proposed that the hyperphalangy in whale and dolphin flippers could be a consequence of the extended exposure of the central digit primordia to AER-FGF8 signaling (Holder, 1983, Richardson and Oelschläger, 2002, Sanz-Ezquerro and Tickle, 2003). It is therefore plausible that evolutionary tampering with the regulation of AER-FGF signaling during digit ray morphogenesis is a general phenomenon that contributed to the loss, reduction, or elongation of digits during artiodactyl diversification, which in turn enabled their adaptation to different habitats.
In this study, we also used ATAC-seq to map and compare all regions of open chromatin in developing mouse and pig limb buds. Although this analysis is challenging due to their evolutionary distance of >95 Ma (Benton and Donoghue, 2007) and the fact that limb buds of only two tetrapod species could be analyzed, we reasoned that at least some of the regulatory differences between mouse and pig limb buds could be indicative of the morphological divergence of the pig limb skeleton from the prototypic pentadactyly of tetrapods as represented by the mouse model. The classification of regions according to sequence conservation and chromatin accessibility allowed us to directly compare regulatory variation between mouse and pig limb buds. Although our conservative analysis will not detect regulatory divergence in conserved regions that are still accessible (mCOP/pCOM), we used the pCCM and mCCP categories as proxies for regulatory variation. It is important to note that the alterations in chromatin accessibility in the two categories can be due to changes affecting the CRE in cis—without significantly decreasing overall conservation—and/or the transcriptional/chromatin complexes in trans. Interestingly, the conserved but differentially accessible regions in the two species represent about one-third of the mouse (mCCP) and pig (pCCM) limb bud regulome, suggesting that widespread rewiring of the respective limb cis-regulatory networks occurred since their last common ancestor. By taking advantage of the wealth of knowledge of the genetic mechanisms that regulate mouse limb morphogenesis, we focused our analysis on regions located in proximity to the TSS of genes with known functions during limb bud and skeletal development. This analysis identified genes that may be good candidates for future gene-centric studies of the regulatory variation underlying limb diversification. In particular, it is likely that causal regulatory alterations are linked to changes in levels and/or spatiotemporal expression of genes that function in essential morphogenetic signaling systems (Davidson and Erwin, 2006). The present study therefore provides a general strategy to uncover regulatory changes affecting the expression of developmental genes that might be functionally relevant for the evolutionary diversification of homologous structures.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Sheep Anti-Digoxigenin Fab fragments Antibody, AP Conjugated | Roche | Cat# 11093274910; RRID:AB_514497 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DIG RNA Labeling Mix | Roche | Cat# 11277073910 |
| T3 RNA Polymerase | Roche | Cat# 11031163001 |
| SP6 RNA Polymerase | Roche | Cat# 10810274001 |
| T7 RNA Polymerase, HC | ThermoFisher Scientific | Cat# EP0113 |
| BM Purple | Roche | Cat# 11442074001 |
| Alcian blue 8GX | Sigma | Cat# A-3157 |
| Alizarin red S | Sigma | Cat# A-5533 |
| SuperScript III reverse transcriptase | Invitrogen | Cat# 18080093 |
| 2x SYBR Green qPCR Master Mix | Bimake | Cat# B21203 |
| Critical Commercial Assays | ||
| Nextera DNA Library Prep Kit | Illumina | FC-121-1030 |
| Deposited Data | ||
| ATAC-seq data | this manuscript | GEO: GSE126293 |
| Experimental Models: Organisms/Strains | ||
| Mouse:NMRI | Janvier Labs | https://www.janvier-labs.com/ RjHan:NMRI |
| Pig: Sus scrofa domesticus | Own facility | N/A |
| Oligonucleotides | ||
| Anchored oligo dT (20) | Integrated DNA Technologies | Cat# 51-01-15-08 |
| Primers for amplification of ISH probes, see Table S7 | this manuscript | N/A |
| Primers for RT-qPCR, see Table S8 | this manuscript | N/A |
| Recombinant DNA | ||
| Plasmid encoding the mouse Shh ISH probe | Echelard et al., 1993 | N/A; available from the authors |
| Plasmid encoding the mouse Gli1 ISH probe | Hui and Joyner, 1993 | N/A; available from the authors |
| Plasmid encoding the mouse Ptch1 ISH probe | Büscher et al., 1997 | N/A; available from the authors |
| Plasmid encoding the mouse Hand2 ISH probe | Srivastava et al., 1995 | N/A; available from the authors |
| Plasmid encoding the mouse Gli3 ISH probe | Büscher et al., 1997 | N/A; available from the authors |
| Plasmid encoding the mouse Tbx4 ISH probe | Koshiba-Takeuchi et al., 2006 | N/A; available from the authors |
| Plasmid encoding the mouse Tbx5 ISH probe | Bruneau et al., 1999 | N/A; available from the authors |
| Plasmid encoding the mouse Pitx1 ISH probe | Logan et al., 1998 | N/A; available from the authors |
| Plasmid encoding the mouse Grem1 ISH probe | Zúñiga et al., 1999 | N/A; available from the authors |
| Plasmid encoding the mouse Fgf8 ISH probe | Crossley and Martin, 1995 | N/A; available from the authors |
| Plasmid encoding the mouse Hoxd11 ISH probe | Dollé et al., 1989 | N/A; available from the authors |
| Plasmid encoding the mouse Hoxd12 ISH probe | Dollé et al., 1989 | N/A; available from the authors |
| Plasmid encoding the mouse Hoxd13 ISH probe | Dollé et al., 1989 | N/A; available from the authors |
| ISH probes generated in this study, see Table S7 | this manuscript | N/A |
| Software and Algorithms | ||
| R/Bioconductor |
https://www.r-project.org/ https://www.bioconductor.org/ |
|
| Prism 8 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Javier Lopez-Rios (jloprio@upo.es)
Materials Availability
All plasmids generated in this study are available from the Lead Contact without restriction.
Data and Code Availability
ATAC-seq raw data and analysis files have been deposited in the GEO database under accession number GEO: GSE126293.
Experimental Model and Subject Details
Mouse (Mus musculus)
Mouse embryos were collected according to the regulations and permits defined under Spanish, Swiss and EU regulations. All animal experiments were designed according to the 3R principles and the Basel Declaration. Wild-type mouse embryos between 9.5-17.5 days postcoitum (NMRI background) were produced by natural mating. Embryos of both sexes were used in the analysis.
Pig (Sus scrofa)
Pig embryos were collected according to the regulations and permits defined under German and EU regulations. All animal experiments were designed according to the 3R principles and the Basel Declaration. Domestic pig (Sus scrofa domesticus) embryos between 18-33 days postcoitum were produced by hormonal synchronization of recipient sows followed by artificial insemination. Date of insemination was considered gestational day zero (D0) and pregnancy was assessed via ultrasound scans. Pig fetuses at the second month of gestation (D55) were purchased from Nebraska Scientific (Omaha, NE, USA). As in mice, staging of pig embryos refers to gestational day. The equivalency between mouse and pig limb developmental stages was defined during the study according to morphological criteria to generate the comparative table shown in Figure S1 (Kaufman, 1992, Marrable, 1971, Patten, 1948, Sears et al., 2011). Embryos of both sexes were used in the analysis.
Method Details
Skeletal preparations
Mouse and pig embryonic and fetal limbs were stained with Alcian blue, which labels cartilage, and/or Alizarin red to stain ossified tissue. All steps were carried at room temperature, unless stated otherwise. Mouse and pig embryos at the appropriate age were collected in ice-cold phosphate-buffered saline (PBS), euthanized by decapitation and stored overnight in tap water. The following morning, embryos were boiled in water for 15-20 s, deskinned and eviscerated under the stereomicroscope and placed in 95% ethanol for storage until further processing. Cartilage staining was performed by incubating overnight the samples in Alcian Blue stain (150 mg/l Alcian Blue 8GX; 20% glacial acetic acid in ethanol; filtered) with gentle rotation. Next, embryos were rinsed twice with 95% ethanol and extensively washed for > 16 hours with several changes of 95% ethanol. Next, samples were cleared by placing the embryos into 1% KOH for 10-15 minutes, followed by bone staining in Alizarin Red solution (50 mg/l Alizarin Red in 1% KOH; filtered) for 1 hour. Embryos were further cleared by incubation in 1% KOH for 30 min, followed by a 80:20, 60:40, 40:60 and 20:80 1%KOH/glycerol series. For long-term storage, cleared samples were placed in 20% glycerol.
Whole mount in situ hybridization analysis
Endogenous transcripts were detected by whole mount in situ hybridization (ISH) in pig and mouse limb buds. Unless indicated, all incubations were performed at room temperature and each wash corresponds to a five-minute incubation with gentle agitation. Mouse or pig embryos were collected in ice-cold PBS and fixed overnight in 4% paraformaldehyde (PFA) in PBS at 4°C. Samples were subsequently washed twice in PBT (0.1% Tween-20 in PBS), dehydrated through a 25%/50%/75% methanol/PBT series (5 minutes each) and stored in 100% methanol at −20°C until further processing. For whole-mount in situ hybridization, embryos were rehydrated in a reverse methanol/PBT series, washed 3x in PBT and bleached in 6% H2O2/PBT for 15 minutes. Subsequently, embryos were washed 3x in PBT and treated with 10 μg/mL proteinase K (PK) in PBT as follows: 15 minutes (E9.5 mouse and D18 pig embryos); 20 minutes (E10.5 mouse embryos); 25 min (E11.25-E11.75 mouse embryos); 35 minutes (D21-D24 pig embryos; extended digestion time required to allow sufficient probe penetration in the larger pig limb buds). All mouse and pig embryos hybridized with Fgf8 riboprobes were incubated for 5 minutes in 5 μg/mL proteinase K in PBT. After PK permeabilization, samples were treated with freshly prepared 2mg/ml glycine in PBT for 5 minutes, washed twice in PBT and post-fixed in 0.2% glutaraldehyde/4%PFA in PBT for 20 minutes. After three washes in PBT, embryos were placed in 2ml of pre-warmed pre-hybridization buffer (50% deionized formamide; 5x SSC pH 4.5; 2% Roche Blocking Reagent; 0.1% Tween-20; 0.5% CHAPS; 50 μg/mL yeast RNA; 5 mM EDTA; 50 μg/ml heparin). After 1 hour of incubation at 70°C, the solution was replaced by 1ml of hybridization buffer containing 1 μg/ml DIG-labeled riboprobe and samples were incubated overnight at 70°C with gentle rotation. The following morning, probe solution was removed and post-hybridization washes (70°C, 5 minutes each) performed in a waterbath with increasing concentrations of 2xSSC pH 4.5, as follows: 100% pre-hybridization buffer; 75% pre-hybridization buffer/25% 2xSSC; 50% pre-hybridization buffer/50% 2xSSC; 25% pre-hybridization buffer/75% 2xSSC, followed by 2xSCC, 0.1% CHAPS, twice for 30 minutes at 70°C with gentle rotation. Subsequently, samples were treated with 20 μg/ml RNase A in 2x SSC, 0.1% CHAPS for 45 minutes at 37°C and washed twice in maleic acid buffer (100 mM Maleic acid disodium salt hydrate; 150mM NaCl; pH 7.5) for 10 minutes at room temperature, followed by two additional 30 minute washes in the same buffer at 70°C. Embryos where then washed three times in TBST (140mM NaCl; 2.7mM KCl; 25mM Tris-HCl; 1% Tween 20; pH 7.5), blocked in 10% lamb serum/TBST for one hour and finally incubated overnight at 4°C in a 1% lamb serum containing Anti-Dig-AP antibody (Roche, 1:5000). Unbound antibody was washed away by incubating the samples in TBST with gentle agitation (3x5 minutes), followed by 5 one-hour washes in TBST and a final overnight incubation in TBST at 4°C with gentle agitation. The following morning, embryos were equilibrated in NTMT (100mM NaCl, 100mM Tris-HCl; 50mM MgCl2; 1% Tween-20; pH 9.5) and alkaline phosphatase activity was detected by incubating the samples in the dark in BM purple reagent (Roche) at room temperature and with gentle agitation. The staining reaction was stopped by washing the samples 5x10 minutes in PBT. ISH-processed samples were kept in 4%PFA/PBS for long-term storage.
Digoxigenin-labeled antisense riboprobes were in vitro synthesized from linearized plasmids using RNA Labeling Mix (Roche) and T3, T7 or SP6 RNA polymerases. All mouse probes have been previously described (see Key Resources Table), except for Sulf1, which was amplified using specific oligos (Table S7) and cloned into pBluescript KS(+). Pig probes for ISH were generated by PCR from limb bud cDNA (Grem1, Gli1, Gli3, Hoxd11, Hoxd12, Hoxd13, Sulf1) or synthesized as dsDNA fragments (Hand2, Fgf8, Ptch1, Shh, Tbx4, Tbx5; gBlocks, Integrated DNA Technologies) based on the equivalent region covered by the mouse probes and RNA-seq data available (Sears et al., 2015) and cloned into pBluescript KS(+) or pGEM-T Easy. Pig gene names are not capitalized for simplicity. All pig riboprobes were first validated in mouse embryos. The sequence of all probes specifically produced for this publication is provided in Table S7. All embryos were split so that left and right fore- and hindlimbs were processed separately for whole mount ISH. As pig embryonic samples are difficult to obtain and given that the handplate skeleton is very similar between fore and hindlimbs, both fore- and hindlimb buds were used in this study taking into account the developmental delay between anterior and posterior primordia in both mouse and pig (Figure S1). This yielded equivalent results in all cases, being the obvious exception Tbx5 and Tbx4/Pitx1, which are specific of fore- and hindlimbs, respectively. Mostly forelimbs are shown, except when noted in the figures (HL: hindlimb).
Microscopy
Dissected limb buds after ISH or skeletal staining were photographed using a Leica MZ16FA stereo-microscope with a DFC300FX camera or an Olympus SZX16 stereo-microscope equipped with Nikon DS-Fi3 camera. The images were processed in Adobe Photoshop CS6 and brightness and contrast were adjusted uniformly.
RT-qPCR
The handplates of E11.25 mouse and D23 pig forelimbs were dissected in ice-cold PBS into posterior and anterior halves and stored in RNAlater at −20°C until processed. Total RNA was extracted using the Quick-RNA MicroPrep Kit (Zymo Research) following the manufacturer’s instructions. cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen) and Anchored oligo dT20 primers (Integrated DNA Technologies). RT-qPCR analysis was performed using a Bio-Rad CFX96 Real Time PCR system and 2x SYBR Green qPCR Master Mix (Bimake). Ptch1 and Gli1 transcript levels were normalized using the expression of the housekeeping genes Hmbs and Hprt1. Primers used for mouse and pig genes amplify equivalent amplicons. For primer sequences see Table S8.
ATAC-seq
Two independent biological replicates consisting each of the entire left and right forelimbs from a single individual were dissected from mouse (E10.5 and E11.5) and pig (D21 and D24) embryos. After dissociation, 75,000 cells were processed for ATAC-seq as described (Buenrostro et al., 2013), using the Nextera DNA Library Prep Kit (Illumina). Library construction for both biological replicates by species and stage was performed in parallel and multiplexed for sequencing in an Illumina NextSeq500 system (PE41; sequencing performed by the Genomics Facility of the University of Basel). ATAC-seq raw data and analysis files have been deposited in the GEO database with Accession number GSE126293. Reads were aligned with bowtie2 to mouse (UCSC genome version mm10) and pig (UCSC genome version susScr11), respectively. Duplicated reads were marked with picard to asses overall duplication levels. For each sample, regions of accessible chromatin where called with macs2 using the option ‘--nomodel --shift 100 --extsize 200 --broad--keep-dup all --qvalue 0.01’. The resulting peak lists were depleted from peaks called in mitochondrial DNA (∼20%) and ENCODE blacklist regions (for mouse). Additionally, only peaks with a log-fold-change bigger than 1 and a FDR smaller than 0.05 were included in the further analysis. The same threshold was used for both species, as there is a similar signal-to-noise ratio in both datasets. Called peaks were merged across biological replicates and stages and joined below a peak distance of 250bp to arrive at a final peak set per species.
All subsequent analysis was performed within the R/Bioconductor framework. Initially, a genome-wide comparison of the ATAC-seq replicates from mouse forelimb buds at E10.5 and E11.5 (two replicates each) with an available ENCODE DNase hypersensitivity sites (DNase-seq) dataset from mouse forelimb buds at E11.5 (GEO: GSM1014174; Yue et al., 2014) was performed. This analysis validated the mouse forelimb ATAC-seq datasets, as there is an overall high correlation with the ENCODE DNase-seq dataset, with R values between 0.76 and 0.79. For the comparison of open chromatin patterns between mouse and pig, ATAC-seq peaks were lifted between species using the liftOver function of the R package rtracklayer based on the UCSC provided lift-over chain files (mm10ToSusScr11.over.chain for mouse; susScr11ToMm10.over.chain for pig) and subsequently joining regions below 10bp. The proportion of candidate regulatory regions distributed in the different categories (mCOP, mCCP, MS, pCOM, pCCM and PS, as defined by our nomenclature) is similar to that of previous cross-species comparisons of chromatin accessibility (Yue et al., 2014). ATAC peak lists were further annotated using the detailRanges function of the R package csaw. Peak annotation in the mouse is based on the RefGene transcript annotation (downloaded from UCSC 2015-12-18) and for pig on Ensemble version 90. Additionally, overlap or proximity to specific genomic features (TADs, VISTA enhancers, TSS, etc.) was evaluated using functions from the R GRanges package. TAD coordinates had been previously reported in mouse ES cells (Dixon et al., 2012) and pig liver (Foissac et al., 2019). Comparison of evolutionary conservation in mouse is based on the PhastCons conservation score, which is not available for the pig. Specifically, the UCSC-provided 60-way placental bigWig track (downloaded from UCSC 2017-05-02) was imported with the R package rtracklayer and the average score of regions ± 2kb around the center of all peaks was visualized. Visualization of genomic landscapes was based on HiC (Dixon et al., 2012) and SMC1A ChIA-PET (Dowen et al., 2014) data.
Gene and mammalian phenotype ontology
All gene lists were retrieved from Gene Ontology (GO; http://amigo.geneontology.org/amigo) and Mammalian Phenotype (MP; http://www.informatics.jax.org/vocab/mp_ontology) browsers. The “limb phenotype” LP gene list (Table S4) was compiled by merging genes associated to the GO term “limb development” (GO:0060173) and the MP terms “abnormal limb morphology” (MP:0002109) and “abnormal appendicular skeleton morphology” (MP:0009250). This gene set was selected due to its biological relevance, but no claims are made with respect to a differential representation of these genes among the list of relevant conserved and diverging accessible regions. GO codes used to retrieve signaling pathway components in mouse and human were as follows: HH/SMO (GO:0007224), WNT (GO:0016055), BMP/TGFβ (GO:0030509 and GO:0007179) or FGF (GO:0008543)(Table S6). Gene ontology analyses of the mouse regulome shown in Figure S3 was performed using GREAT (McLean et al., 2010) with the following settings: mm10 mouse genome assembly, using the whole genome as “background” regions, single nearest gene peak association and statistically significance of the association determined using the binomial test.
Quantification and Statistical Analysis
Number of biological replicates and statistical significance are indicated in figures and figure legends. Representative results are reported for ISH and skeletal staining data. For the pig ISH, two independent biological replicates were used per gene probe and stage (three independent biological replicates for the mouse). For ATAC-seq data generation, two independent biological replicates were used per species and stage (Pearson correlation shown in Figures 4A and 4B). Transgenic embryo images were obtained from the VISTA enhancer database (https://enhancer.lbl.gov/; transgenic embryos showing expression in the limb bud: mm1483 n = 8/8; mm1010 n = 9/9; mm1341 = 6/6). The significance of the differences between posterior to anterior expression ratios in mouse and pig handplates as defined by RT-qPCR analysis was determined using the unpaired, nonparametric Mann-Whitney test. Sample size was n = 7 in pig and n = 9 in mouse. Results are represented as mean ± SD. Statistical analysis and plotting of results was performed using Prism 8 (GraphPad Software).
Acknowledgments
We thank G. Nusspaumer for critical input on the manuscript and the rest of the members of the group for scientific discussions and technical help. We are also grateful to J. Tena for bioinformatic advice. We wish to thank as well A. Franco, C. Mateos, A. López, E. Terszowska, A. Offinger, A. Brülhart, and A. Siegrist for mouse colony maintenance and H. Paul and J. Bichler for pig care. We are also grateful to L. Selleri and I. Welsh for providing D30 pig embryonic samples and C. Beisel, I. Nisen, and K. Eschbach for next-generation sequencing. Calculations were performed at the sciCORE scientific computing center at the University of Basel (http://scicore.unibas.ch/). This research has been funded by the Spanish Ministerio de Ciencia, Innovación y Universidades (BFU2017-82974-P to J.L.-R.); the “Unidad de Excelencia María de Maeztu 2017–2021” (MDM-2016-0687); an SNF Doc.Mobility fellowship (to V.T.); SNF grant 310030B_166685 and ERC advanced grant INTEGRAL ERC-2015-AdG, Project ID 695032 (to R.Z.); and the University of Basel.
Author Contributions
V.T., R.Z., and J.L.-R. conceived the project and generated the ATAC-seq data. V.T. performed the majority of the experiments and analysis. F.G. performed all the bioinformatic analysis of the ATAC-seq data. B.K. and E.W. provided all the pig embryos used in the ISH analysis and ATAC-seq. J.L.-R. designed the experiments together with V.T., supervised the study, and wrote the manuscript together with V.T. and R.Z. and input from all co-authors.
Declaration of Interests
The authors declare no competing interests.
Published: April 7, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.03.054.
Supplemental Information
Each region of open chromatin received a unique identifier (m1, m2, etc.). In addition to its chromosomal coordinates, the TAD where the peak is located (Dixon et al., 2012) and the closest TSS are indicated. Regulatory regions can also be sorted using specify criteria such as their association with LP (limb phenotype) genes or VISTA enhancers. The column “liftOverAble.susScr11” provides the coordinates of the pig genome for peaks conserved in both species. The column “open.region.susScr11” indicates if the equivalent chromatin region is also accessible in pig limb buds. All peaks can be sorted according to their categories (mCOP, mCCP or MS; see Figure 4).
Each region of open chromatin received a unique identifier (p1, p2, etc). In addition to its chromosomal coordinates, the TAD where the peak is located (Foissac et al., 2019) and the closest TSS are indicated. Regulatory regions can also be sorted using specify criteria such as their association with LP (limb phenotype) genes or VISTA enhancers. The column “liftOverAble.mm10” provides the coordinates of the mouse genome for peaks conserved in both species The column “open.region.mm10” indicates if the equivalent chromatin region is accessible in mouse limb buds. All peaks can be sorted according to their categories (pCOM, pCCM or PS; see Figure 4).
The “limb phenotype” (LP) list consists of 965 genes associated either to the Gene Ontology term “limb development” (GO:0060173) or the Mammalian Phenotype terms “abnormal limb morphology” (MP:0002109) or “abnormal appendicular skeleton morphology” (MP:0009250).
References
- Acemel R.D., Maeso I., Gómez-Skarmeta J.L. Topologically associated domains: a successful scaffold for the evolution of gene regulation in animals. Wiley Interdiscip. Rev. Dev. Biol. 2017;6 doi: 10.1002/wdev.265. [DOI] [PubMed] [Google Scholar]
- Adachi N., Robinson M., Goolsbee A., Shubin N.H. Regulatory evolution of Tbx5 and the origin of paired appendages. Proc. Natl. Acad. Sci. USA. 2016;113:10115–10120. doi: 10.1073/pnas.1609997113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal P., Wylie J.N., Galceran J., Arkhitko O., Li C., Deng C., Grosschedl R., Bruneau B.G. Tbx5 is essential for forelimb bud initiation following patterning of the limb field in the mouse embryo. Development. 2003;130:623–633. doi: 10.1242/dev.00191. [DOI] [PubMed] [Google Scholar]
- Ahn S., Joyner A.L. Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell. 2004;118:505–516. doi: 10.1016/j.cell.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Andrey G., Montavon T., Mascrez B., Gonzalez F., Noordermeer D., Leleu M., Trono D., Spitz F., Duboule D. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science. 2013;340:1234167. doi: 10.1126/science.1234167. [DOI] [PubMed] [Google Scholar]
- Bénazet J.D., Bischofberger M., Tiecke E., Gonçalves A., Martin J.F., Zuniga A., Naef F., Zeller R. A self-regulatory system of interlinked signaling feedback loops controls mouse limb patterning. Science. 2009;323:1050–1053. doi: 10.1126/science.1168755. [DOI] [PubMed] [Google Scholar]
- Benton M.J., Donoghue P.C. Paleontological evidence to date the tree of life. Mol. Biol. Evol. 2007;24:26–53. doi: 10.1093/molbev/msl150. [DOI] [PubMed] [Google Scholar]
- Briscoe J., Chen Y., Jessell T.M., Struhl G. A Hedgehog-insensitive form of Patched provides evidence for direct long-range morphogen activity of Sonic hedgehog in the neural tube. Mol. Cell. 2001;7:1279–1291. doi: 10.1016/s1097-2765(01)00271-4. [DOI] [PubMed] [Google Scholar]
- Bruneau B.G., Logan M., Davis N., Levi T., Tabin C.J., Seidman J.G., Seidman C.E. Chamber-specific cardiac expression of Tbx5 and heart defects in Holt-Oram syndrome. Dev. Biol. 1999;211:100–108. doi: 10.1006/dbio.1999.9298. [DOI] [PubMed] [Google Scholar]
- Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büscher D., Bosse B., Heymer J., Rüther U. Evidence for genetic control of Sonic hedgehog by Gli3 in mouse limb development. Mech. Dev. 1997;62:175–182. doi: 10.1016/s0925-4773(97)00656-4. [DOI] [PubMed] [Google Scholar]
- Cannavò E., Khoueiry P., Garfield D.A., Geeleher P., Zichner T., Gustafson E.H., Ciglar L., Korbel J.O., Furlong E.E. Shadow enhancers are pervasive features of developmental regulatory networks. Curr. Biol. 2016;26:38–51. doi: 10.1016/j.cub.2015.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S.B. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Chen Y., Struhl G. Dual roles for Patched in sequestering and transducing Hedgehog. Cell. 1996;87:553–563. doi: 10.1016/s0092-8674(00)81374-4. [DOI] [PubMed] [Google Scholar]
- Chiang C., Litingtung Y., Harris M.P., Simandl B.K., Li Y., Beachy P.A., Fallon J.F. Manifestation of the limb prepattern: limb development in the absence of Sonic hedgehog function. Dev. Biol. 2001;236:421–435. doi: 10.1006/dbio.2001.0346. [DOI] [PubMed] [Google Scholar]
- Clifford A.B. The evolution of the unguligrade manus in artiodactyls. J. Vertebr. Paleontol. 2010;30:1827–1839. [Google Scholar]
- Cooper K.L., Sears K.E., Uygur A., Maier J., Baczkowski K.S., Brosnahan M., Antczak D., Skidmore J.A., Tabin C.J. Patterning and post-patterning modes of evolutionary digit loss in mammals. Nature. 2014;511:41–45. doi: 10.1038/nature13496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotney J., Leng J., Yin J., Reilly S.K., DeMare L.E., Emera D., Ayoub A.E., Rakic P., Noonan J.P. The evolution of lineage-specific regulatory activities in the human embryonic limb. Cell. 2013;154:185–196. doi: 10.1016/j.cell.2013.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley P.H., Martin G.R. The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development. 1995;121:439–451. doi: 10.1242/dev.121.2.439. [DOI] [PubMed] [Google Scholar]
- Cunningham T.J., Lancman J.J., Berenguer M., Dong P.D.S., Duester G. Genomic knockout of two presumed forelimb Tbx5 enhancers reveals they are nonessential for limb development. Cell Rep. 2018;23:3146–3151. doi: 10.1016/j.celrep.2018.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danesin C., Agius E., Escalas N., Ai X., Emerson C., Cochard P., Soula C. Ventral neural progenitors switch toward an oligodendroglial fate in response to increased Sonic hedgehog (Shh) activity: involvement of Sulfatase 1 in modulating Shh signaling in the ventral spinal cord. J. Neurosci. 2006;26:5037–5048. doi: 10.1523/JNEUROSCI.0715-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E.H., Erwin D.H. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- de Bakker M.A., Fowler D.A., den Oude K., Dondorp E.M., Navas M.C., Horbanczuk J.O., Sire J.Y., Szczerbińska D., Richardson M.K. Digit loss in archosaur evolution and the interplay between selection and constraints. Nature. 2013;500:445–448. doi: 10.1038/nature12336. [DOI] [PubMed] [Google Scholar]
- Dhoot G.K., Gustafsson M.K., Ai X., Sun W., Standiford D.M., Emerson C.P., Jr. Regulation of Wnt signaling and embryo patterning by an extracellular sulfatase. Science. 2001;293:1663–1666. doi: 10.1126/science.293.5535.1663. [DOI] [PubMed] [Google Scholar]
- Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollé P., Izpisúa-Belmonte J.C., Falkenstein H., Renucci A., Duboule D. Coordinate expression of the murine Hox-5 complex homoeobox-containing genes during limb pattern formation. Nature. 1989;342:767–772. doi: 10.1038/342767a0. [DOI] [PubMed] [Google Scholar]
- Dowen J.M., Fan Z.P., Hnisz D., Ren G., Abraham B.J., Zhang L.N., Weintraub A.S., Schujiers J., Lee T.I., Zhao K., Young R.A. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echelard Y., Epstein D.J., St-Jacques B., Shen L., Mohler J., McMahon J.A., McMahon A.P. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993;75:1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- Eckalbar W.L., Schlebusch S.A., Mason M.K., Gill Z., Parker A.V., Booker B.M., Nishizaki S., Muswamba-Nday C., Terhune E., Nevonen K.A. Transcriptomic and epigenomic characterization of the developing bat wing. Nat. Genet. 2016;48:528–536. doi: 10.1038/ng.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellgett S.W., Maguire R.J., Pownall M.E. Sulf1 has ligand-dependent effects on canonical and non-canonical Wnt signalling. J. Cell Sci. 2015;128:1408–1421. doi: 10.1242/jcs.164467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foissac S., Djebali S., Munyard K., Vialaneix N., Rau A., Muret K., Esquerré D., Zytnicki M., Derrien T., Bardou P. Multi-species annotation of transcriptome and chromatin structure in domesticated animals. BMC Biol. 2019;17:108. doi: 10.1186/s12915-019-0726-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F., Duboule D., Spitz F. Transgenic analysis of Hoxd gene regulation during digit development. Dev. Biol. 2007;306:847–859. doi: 10.1016/j.ydbio.2007.03.020. [DOI] [PubMed] [Google Scholar]
- Harfe B.D., Scherz P.J., Nissim S., Tian H., McMahon A.P., Tabin C.J. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Holder N. The vertebrate limb: patterns and constraints in development and evolution. In: Goodwin B.C., Wylie C.C., Holder N., editors. Development and Evolution. Cambridge University Press; 1983. pp. 399–425. [Google Scholar]
- Hui C.C., Joyner A.L. A mouse model of Greig cephalopolysyndactyly syndrome: the extra-toesJ mutation contains an intragenic deletion of the Gli3 gene. Nat. Genet. 1993;3:241–246. doi: 10.1038/ng0393-241. [DOI] [PubMed] [Google Scholar]
- Kaufman M.H. Academic Press; 1992. The Atlas of Mouse Development. [Google Scholar]
- Koshiba-Takeuchi K., Takeuchi J.K., Arruda E.P., Kathiriya I.S., Mo R., Hui C.C., Srivastava D., Bruneau B.G. Cooperative and antagonistic interactions between Sall4 and Tbx5 pattern the mouse limb and heart. Nat. Genet. 2006;38:175–183. doi: 10.1038/ng1707. [DOI] [PubMed] [Google Scholar]
- Lewandoski M., Sun X., Martin G.R. Fgf8 signalling from the AER is essential for normal limb development. Nat. Genet. 2000;26:460–463. doi: 10.1038/82609. [DOI] [PubMed] [Google Scholar]
- Logan M., Tabin C.J. Role of Pitx1 upstream of Tbx4 in specification of hindlimb identity. Science. 1999;283:1736–1739. doi: 10.1126/science.283.5408.1736. [DOI] [PubMed] [Google Scholar]
- Logan M., Simon H.G., Tabin C. Differential regulation of T-box and homeobox transcription factors suggests roles in controlling chick limb-type identity. Development. 1998;125:2825–2835. doi: 10.1242/dev.125.15.2825. [DOI] [PubMed] [Google Scholar]
- Long H.K., Prescott S.L., Wysocka J. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell. 2016;167:1170–1187. doi: 10.1016/j.cell.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rios J. The many lives of SHH in limb development and evolution. Semin. Cell Dev. Biol. 2016;49:116–124. doi: 10.1016/j.semcdb.2015.12.018. [DOI] [PubMed] [Google Scholar]
- Lopez-Rios J., Speziale D., Robay D., Scotti M., Osterwalder M., Nusspaumer G., Galli A., Holländer G.A., Kmita M., Zeller R. GLI3 constrains digit number by controlling both progenitor proliferation and BMP-dependent exit to chondrogenesis. Dev. Cell. 2012;22:837–848. doi: 10.1016/j.devcel.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rios J., Duchesne A., Speziale D., Andrey G., Peterson K.A., Germann P., Unal E., Liu J., Floriot S., Barbey S. Attenuated sensing of SHH by Ptch1 underlies evolution of bovine limbs. Nature. 2014;511:46–51. doi: 10.1038/nature13289. [DOI] [PubMed] [Google Scholar]
- Mariani F.V., Ahn C.P., Martin G.R. Genetic evidence that FGFs have an instructive role in limb proximal-distal patterning. Nature. 2008;453:401–405. doi: 10.1038/nature06876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marigo V., Johnson R.L., Vortkamp A., Tabin C.J. Sonic hedgehog differentially regulates expression of GLI and GLI3 during limb development. Dev. Biol. 1996;180:273–283. doi: 10.1006/dbio.1996.0300. [DOI] [PubMed] [Google Scholar]
- Marigo V., Scott M.P., Johnson R.L., Goodrich L.V., Tabin C.J. Conservation in hedgehog signaling: induction of a chicken Patched homolog by Sonic hedgehog in the developing limb. Development. 1996;122:1225–1233. doi: 10.1242/dev.122.4.1225. [DOI] [PubMed] [Google Scholar]
- Marrable A.W. Pitman; 1971. The Embryonic Pig: A Chronological Account. [Google Scholar]
- McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michos O., Panman L., Vintersten K., Beier K., Zeller R., Zuniga A. Gremlin-mediated BMP antagonism induces the epithelial-mesenchymal feedback signaling controlling metanephric kidney and limb organogenesis. Development. 2004;131:3401–3410. doi: 10.1242/dev.01251. [DOI] [PubMed] [Google Scholar]
- Minguillon C., Del Buono J., Logan M.P. Tbx5 and Tbx4 are not sufficient to determine limb-specific morphologies but have common roles in initiating limb outgrowth. Dev. Cell. 2005;8:75–84. doi: 10.1016/j.devcel.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Minguillon C., Nishimoto S., Wood S., Vendrell E., Gibson-Brown J.J., Logan M.P. Hox genes regulate the onset of Tbx5 expression in the forelimb. Development. 2012;139:3180–3188. doi: 10.1242/dev.084814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montavon T., Soshnikova N., Mascrez B., Joye E., Thevenet L., Splinter E., de Laat W., Spitz F., Duboule D. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147:1132–1145. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- Naiche L.A., Papaioannou V.E. Loss of Tbx4 blocks hindlimb development and affects vascularization and fusion of the allantois. Development. 2003;130:2681–2693. doi: 10.1242/dev.00504. [DOI] [PubMed] [Google Scholar]
- Osterwalder M., Speziale D., Shoukry M., Mohan R., Ivanek R., Kohler M., Beisel C., Wen X., Scales S.J., Christoffels V.M. HAND2 targets define a network of transcriptional regulators that compartmentalize the early limb bud mesenchyme. Dev. Cell. 2014;31:345–357. doi: 10.1016/j.devcel.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder M., Barozzi I., Tissières V., Fukuda-Yuzawa Y., Mannion B.J., Afzal S.Y., Lee E.A., Zhu Y., Plajzer-Frick I., Pickle C.S. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature. 2018;554:239–243. doi: 10.1038/nature25461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten B.M. Third Edition. The Blakiston Company; 1948. Embryology of the Pig. [Google Scholar]
- Petit F., Sears K.E., Ahituv N. Limb development: a paradigm of gene regulation. Nat. Rev. Genet. 2017;18:245–258. doi: 10.1038/nrg.2016.167. [DOI] [PubMed] [Google Scholar]
- Polly P.D. Limbs in mammalian evolution. In: Hall B.K., editor. Fins into Limbs: Evolution, Development, and Transformation. University of Chicago Press; 2007. pp. 245–268. [Google Scholar]
- Probst S., Kraemer C., Demougin P., Sheth R., Martin G.R., Shiratori H., Hamada H., Iber D., Zeller R., Zuniga A. SHH propagates distal limb bud development by enhancing CYP26B1-mediated retinoic acid clearance via AER-FGF signalling. Development. 2011;138:1913–1923. doi: 10.1242/dev.063966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prothero D.R., Foss S.E. The Johns Hopkins University Press; 2007. The Evolution of Artiodactyls. [Google Scholar]
- Rallis C., Bruneau B.G., Del Buono J., Seidman C.E., Seidman J.G., Nissim S., Tabin C.J., Logan M.P. Tbx5 is required for forelimb bud formation and continued outgrowth. Development. 2003;130:2741–2751. doi: 10.1242/dev.00473. [DOI] [PubMed] [Google Scholar]
- Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., Aiden E.L. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratzka A., Kalus I., Moser M., Dierks T., Mundlos S., Vortkamp A. Redundant function of the heparan sulfate 6-O-endosulfatases Sulf1 and Sulf2 during skeletal development. Dev. Dyn. 2008;237:339–353. doi: 10.1002/dvdy.21423. [DOI] [PubMed] [Google Scholar]
- Richardson M.K., Oelschläger H.H. Time, pattern, and heterochrony: a study of hyperphalangy in the dolphin embryo flipper. Evol. Dev. 2002;4:435–444. doi: 10.1046/j.1525-142x.2002.02032.x. [DOI] [PubMed] [Google Scholar]
- Riddle R.D., Johnson R.L., Laufer E., Tabin C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75:1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- Roscito J.G., Sameith K., Parra G., Langer B.E., Petzold A., Moebius C., Bickle M., Rodrigues M.T., Hiller M. Phenotype loss is associated with widespread divergence of the gene regulatory landscape in evolution. Nat. Commun. 2018;9:4737. doi: 10.1038/s41467-018-07122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Ezquerro J.J., Tickle C. Fgf signaling controls the number of phalanges and tip formation in developing digits. Curr. Biol. 2003;13:1830–1836. doi: 10.1016/j.cub.2003.09.040. [DOI] [PubMed] [Google Scholar]
- Scherz P.J., McGlinn E., Nissim S., Tabin C.J. Extended exposure to Sonic hedgehog is required for patterning the posterior digits of the vertebrate limb. Dev. Biol. 2007;308:343–354. doi: 10.1016/j.ydbio.2007.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears K.E., Bormet A.K., Rockwell A., Powers L.E., Noelle Cooper L., Wheeler M.B. Developmental basis of mammalian digit reduction: a case study in pigs. Evol. Dev. 2011;13:533–541. doi: 10.1111/j.1525-142X.2011.00509.x. [DOI] [PubMed] [Google Scholar]
- Sears K.E., Maier J.A., Rivas-Astroza M., Poe R., Zhong S., Kosog K., Marcot J.D., Behringer R.R., Cretekos C.J., Rasweiler J.J., IV, Rapti Z. The relationship between gene network structure and expression variation among individuals and species. PLoS Genet. 2015;11:e1005398. doi: 10.1371/journal.pgen.1005398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki R., Li C., Fang Q., Hayashi S., Egawa S., Hu J., Xu L., Pan H., Kondo M., Sato T. Functional roles of Aves class-specific cis-regulatory elements on macroevolution of bird-specific features. Nat. Commun. 2017;8:14229. doi: 10.1038/ncomms14229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Yue F., McCleary D.F., Ye Z., Edsall L., Kuan S., Wagner U., Dixon J., Lee L., Lobanenkov V.V., Ren B. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D., Cserjesi P., Olson E.N. A subclass of bHLH proteins required for cardiac morphogenesis. Science. 1995;270:1995–1999. doi: 10.1126/science.270.5244.1995. [DOI] [PubMed] [Google Scholar]
- Stern D.L., Frankel N. The structure and evolution of cis-regulatory regions: the shavenbaby story. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013;368:20130028. doi: 10.1098/rstb.2013.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Welscher P., Fernandez-Teran M., Ros M.A., Zeller R. Mutual genetic antagonism involving GLI3 and dHAND prepatterns the vertebrate limb bud mesenchyme prior to SHH signaling. Genes Dev. 2002;16:421–426. doi: 10.1101/gad.219202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers M., Mahood R., Yin Y., Tickle C. Integration of growth and specification in chick wing digit-patterning. Nature. 2008;452:882–886. doi: 10.1038/nature06718. [DOI] [PubMed] [Google Scholar]
- Visel A., Minovitsky S., Dubchak I., Pennacchio L.A. VISTA Enhancer Browser—a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viviano B.L., Paine-Saunders S., Gasiunas N., Gallagher J., Saunders S. Domain-specific modification of heparan sulfate by Qsulf1 modulates the binding of the bone morphogenetic protein antagonist Noggin. J. Biol. Chem. 2004;279:5604–5611. doi: 10.1074/jbc.M310691200. [DOI] [PubMed] [Google Scholar]
- Vokes S.A., Ji H., Wong W.H., McMahon A.P. A genome-scale analysis of the cis-regulatory circuitry underlying Sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev. 2008;22:2651–2663. doi: 10.1101/gad.1693008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Ai X., Freeman S.D., Pownall M.E., Lu Q., Kessler D.S., Emerson C.P., Jr. QSulf1, a heparan sulfate 6-O-endosulfatase, inhibits fibroblast growth factor signaling in mesoderm induction and angiogenesis. Proc. Natl. Acad. Sci. USA. 2004;101:4833–4838. doi: 10.1073/pnas.0401028101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F., Cheng Y., Breschi A., Vierstra J., Wu W., Ryba T., Sandstrom R., Ma Z., Davis C., Pope B.D., Mouse ENCODE Consortium A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Nakamura E., Nguyen M.T., Bao X., Akiyama H., Mackem S. Uncoupling Sonic hedgehog control of pattern and expansion of the developing limb bud. Dev. Cell. 2008;14:624–632. doi: 10.1016/j.devcel.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga A. Next generation limb development and evolution: old questions, new perspectives. Development. 2015;142:3810–3820. doi: 10.1242/dev.125757. [DOI] [PubMed] [Google Scholar]
- Zúñiga A., Haramis A.P., McMahon A.P., Zeller R. Signal relay by BMP antagonism controls the SHH/FGF4 feedback loop in vertebrate limb buds. Nature. 1999;401:598–602. doi: 10.1038/44157. [DOI] [PubMed] [Google Scholar]
- Zuniga A., Laurent F., Lopez-Rios J., Klasen C., Matt N., Zeller R. Conserved cis-regulatory regions in a large genomic landscape control SHH and BMP-regulated Gremlin1 expression in mouse limb buds. BMC Dev. Biol. 2012;12:23. doi: 10.1186/1471-213X-12-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Each region of open chromatin received a unique identifier (m1, m2, etc.). In addition to its chromosomal coordinates, the TAD where the peak is located (Dixon et al., 2012) and the closest TSS are indicated. Regulatory regions can also be sorted using specify criteria such as their association with LP (limb phenotype) genes or VISTA enhancers. The column “liftOverAble.susScr11” provides the coordinates of the pig genome for peaks conserved in both species. The column “open.region.susScr11” indicates if the equivalent chromatin region is also accessible in pig limb buds. All peaks can be sorted according to their categories (mCOP, mCCP or MS; see Figure 4).
Each region of open chromatin received a unique identifier (p1, p2, etc). In addition to its chromosomal coordinates, the TAD where the peak is located (Foissac et al., 2019) and the closest TSS are indicated. Regulatory regions can also be sorted using specify criteria such as their association with LP (limb phenotype) genes or VISTA enhancers. The column “liftOverAble.mm10” provides the coordinates of the mouse genome for peaks conserved in both species The column “open.region.mm10” indicates if the equivalent chromatin region is accessible in mouse limb buds. All peaks can be sorted according to their categories (pCOM, pCCM or PS; see Figure 4).
The “limb phenotype” (LP) list consists of 965 genes associated either to the Gene Ontology term “limb development” (GO:0060173) or the Mammalian Phenotype terms “abnormal limb morphology” (MP:0002109) or “abnormal appendicular skeleton morphology” (MP:0009250).
Data Availability Statement
ATAC-seq raw data and analysis files have been deposited in the GEO database under accession number GEO: GSE126293.