Abstract
Oaks are dominant forest tree species widely distributed across the Northern Hemisphere, where they constitute natural resources of economic, ecological, social and historical value. Hybridization and adaptive introgression have long been thought to be major drivers of their ecological success. Thus, the maintenance of species barriers remains a key question, given the extent of interspecific gene flow.
In this study, we made use of the tremendous genetic variation among four European white oak species (31 million SNPs) to infer the evolutionary history of these species, study patterns of genetic differentiation and identify reproductive barriers.
We first analyzed the ecological and historical relationships among these species and inferred a long-term strict isolation followed by a recent and extensive postglacial contact using Approximate Bayesian Computation. Assuming this demographic scenario, we then performed backward simulations to generate the expected distributions of differentiation under neutrality to scan their genomes for reproductive barriers. We finally identified important intrinsic and ecological functions driving the reproductive isolation.
We discussed the importance of identifying the genetic basis for the ecological preferences between these oak species and its implications for the renewal of European forests under global warming.
Keywords: Demographic inferences, reproductive isolation, speciation, intrinsic and ecological barriers, Genome scan, approximate Bayesian computation
Introduction
Oaks are a diverse group of about 350 to 500 species widely distributed throughout the Northern Hemisphere (Hubert et al., 2014; Denk et al., 2017). The variability in the number of recorded oak species highlights the challenge of delineating species limits within a genus displaying a high degree of morphological diversity, sometimes described as a “botanic horror” by taxonomists (Darwin, 1859; Palmer, 1948; Rieseberg et al., 2006; Leroy et al., 2019a). Genetic markers have corroborated these taxonomic concerns, particularly in European white oaks, which have been the subject of a large number of genetic surveys. Studies based on nuclear DNA markers have reported unambiguously high levels of admixture between European white oak species, confirming the reported taxonomic issues for oaks (Lepais et al., 2009). Several detailed empirical studies based on chloroplast DNA markers have revealed an absence of private chlorotypes between European white oak species, but congruent associations between chlorotypes and expansion routes during the last postglacial recolonization, suggesting cytoplasmic capture via recurrent hybridization and backcrossing (Petit et al., 1997; 2002). Recent advances in oak genomics (Plomion et al., 2016; 2018) have made it possible to investigate interspecific gene flow at the whole-genome scale. Thus, Leroy et al. (2017) have provided evidence suggesting that extensive secondary contacts have occurred between four European white oak species, probably at the start of the current interglacial period. These results reconcile earlier findings of contrasting species differentiation at the nuclear and organelle levels. Indeed, secondary contacts explain present-day patterns of species differentiation, including complete sharing of haplotypes in mixed oak stands (Petit et al., 2002) and the partial maintenance of nuclear genetic divergence at some loci (e.g. Scotti-Saintagne et al., 2004). The inferences drawn are also consistent with the persistence of genomic regions impermeable to gene flow due to reproductive barriers, corresponding to a typical case of semi-isolated species (Leroy et al., 2017). However, the genetic basis of these barriers remains unknown.
Controlled pollination trials have provided empirical evidence for the existence of strong reproductive barriers in these four European white oak species (Abadie et al., 2012; Lepais et al., 2013). Ecological preferences in situ have also been previously reported, with tolerance to dry (Q. petraea) or wet (Q. robur) sites (Eaton et al., 2016), or acidic (Q. pyrenaica) or limy (Q. pubescens) soils (Timbal & Aussenal, 1996) but fine-grained ecological surveys do not yet exist for all these four species. The four species occupy different geographic ranges (Fig. 1A): extending up to Scandinavia for Q. petraea and Q. robur, whereas the other two species are present mostly in Mediterranean and sub-Mediterranean regions. However, the distribution ranges of these species overlap in some areas, mostly in South-West France, but the four species are rarely found together in the same stand (but see Lepais et al., 2009). The overlapping species ranges in South-West France thus provide an ideal “natural laboratory” (Hewitt, 1988) for investigating reproductive barriers between these European white oak species.
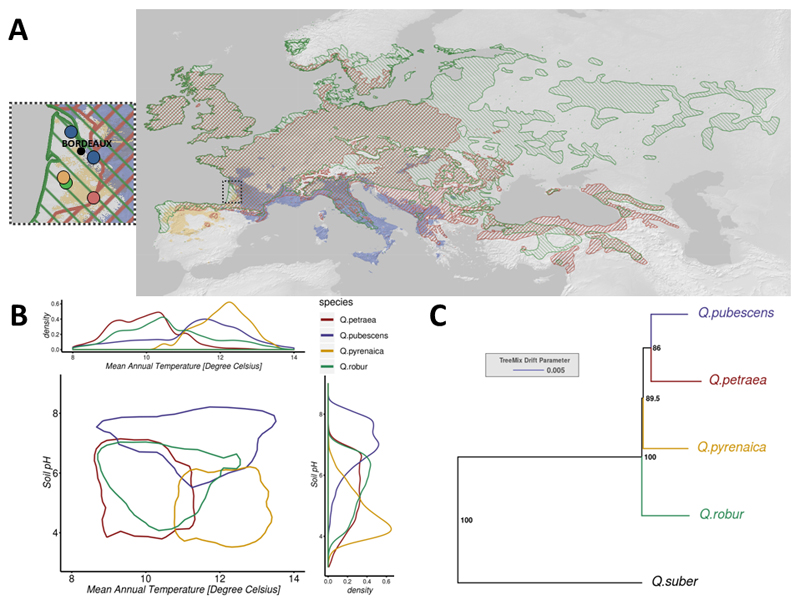
Fig. 1. Continental-scale species distributions and origin of the study material (A) and, ecological (B) and phylogenic relationships of the four European white oak species under investigation using TreeMix (C).
B) The central plot shows the contour of the two-dimensional density between the soil pH and the mean annual temperature enclosing 95% of the ecological data values, based on the French data only. The above and right charts show the one-dimensional density curves for mean annual temperature and soil pH, respectively.
Here we combined state-of-the-art methods in population genomics to explore the genomic patterns of species differentiation (Fig. 2): (i) we used approximate Bayesian computation (ABC) to perform ascertainment bias-free demographic inferences in order to refine estimates of the timing of secondary contacts, and (ii) scan genomes for reproductive barriers. Our findings identified important intrinsic and ecological functions driving the reproductive isolation of these four oak species including tolerance to biotic and abiotic constraints, and intrinsic mating barriers.
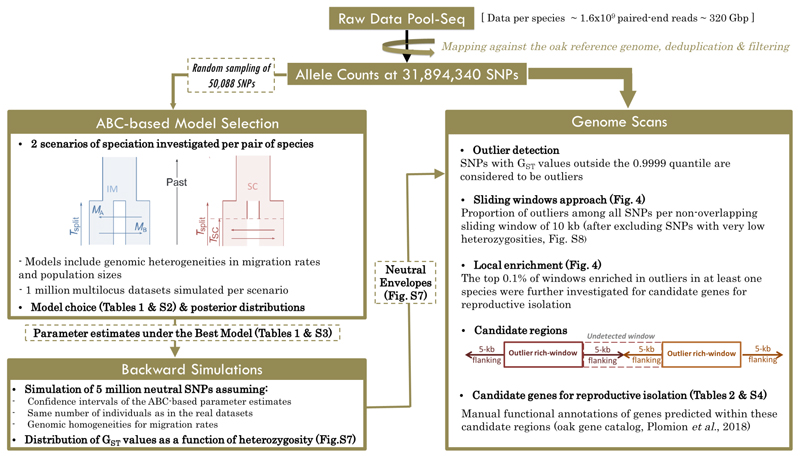
Fig. 2. Workflow used to identify genes contributing to reproductive isolation between four European white oak species.
A subset of 50 thousand of the called SNPs was selected at random and used for model selection under an ABC framework and the generation of parameter estimates under the best model. Large neutral datasets were then simulated to create null envelopes for the identification of SNPs displaying significant departure from expectations under neutrality. We searched for candidate genes in regions enriched in outliers.
Materials & Methods
Ecological niche of the four species
French data
We delineated the extant ecological niche of the four oak species in France (Figs. 1B, S1 and S2) by using their distribution maps based on the National Forest Inventory (Fig. S3) and climatic data extracted from the Chelsa data base (Karger et al., 2017). In addition to the climatic data we added pH values of the soil. Proxies of pH values were derived from National Forest inventory floristic plots installed since 2005. Floristic composition of these inventory plots was compared to existing database (comprising floristic and physical data) to calculate proxies of pH values (Gégout et al., 2005). We intersected the distribution maps with the climatic rasters (30” resolution) and using the R package “ggplot2” v. 2.2.1 (Wickham, 2009) calculated a 2D density plot of species presence in the ecological space as defined by climate (mean temperature) and soil pH.
European data
European distributions maps were constructed based on presence data of species made available by the European Forest Genetic Resources Programme (EUFORGEN, for Q. robur and Q. petraea, de Vries et al., 2015) and the European atlas of forest tree species (JCR, San-Miguel-Ayanz et al., 2016, for Q. pyrenaica & Q. pubescens). Climatic data are based on the Chelsa database (Karger et al., 2017) and soil pH were derived from the European atlas of forest tree species data (JCR, San-Miguel-Ayanz et al., 2016). Since these data were collected from different sites, we computed univariate (rather than bivariate) density distributions using ggplot2, using a procedure similar to that used for the French data.
Sampling and sequencing
We sampled populations of the four Quercus species in stands of natural origin located in South-West France. We sampled 13 Q. petraea trees from Laveyron (Landes, France), and 20 Q. robur and 20 Q. pyrenaica trees from the Landes EVOLTREE “Intensive Study Site” (ISS), 18 Q. pubescens trees from two sites in Gironde: 12 in Branne and 6 in Blaignan (Gironde, France) (see Table S1 for details). Samples of reference species populations came from the same geographic region. Such sampling strategy does not bias species comparison, as shown by an earlier methodological study showing that species differentiation is only moderately impacted by the geographical origin of populations (Bodénès et al. 1997).
DNA was extracted from individual trees with the Invisorb Spin Plant Mini kit, according to the manufacturer’s specification (Startec Molecular, GmbH, Berlin, Germany). DNA yields were evaluated with a NanoDrop 1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) and DNA samples were mixed in equimolar amounts to obtain a single pool for each species.
In this study, we sequenced libraries of pooled DNA samples. Such a pool-seq approach is indeed a cost-and time-effective alternative to individual- sequencing. The main advantages include the reduction in the total sequencing effort and the costs for the preparation of the genomic libraries (e.g. Gautier et al., 2013; Schlötterer et al., 2014). If pool-seq data is expected to generate efficient allele frequency estimates under various experimental designs (Gautier et al., 2013), a pool-seq strategy has also some limitations, especially the difficulty to reconstruct haplotype blocks and estimate linkage disequilibrium, at least for short reads, as used in this study. To meet the requirement of independence in some analyses (see the ABC section below), we randomly selected a low proportion of SNPs (one SNP every 15 Kb, on average), a far larger physical distance than the level of linkage disequilibrium generally assumed for oaks (e.g. <500 bp for the Q. mongolica var. crispula oak, Kremer et al., 2012). For each pool, a paired-end DNA genomic library was generated with the Paired-End DNA Sample Preparation Kit (Illumina, San Diego, CA, USA). The library was then sequenced on a HiSeq2000 sequencer (Illumina, San Diego, CA, USA) with 2x100 paired-end reads. For each pool, we used nine to ten sequencing lanes.
Raw reads were trimmed to remove low-quality bases (<20) from the ends, and sequences between the second unknown nucleotide and the end of the reads were removed. Reads less than 30 nucleotides long after trimming were discarded.
Overall, between 1,617,465,418 and 1,813,403,677 reads per pool were retained for analysis, corresponding to 313 Gb (425X) to 356 Gb (483X) of raw data. Raw data have been deposited in the Sequence Read Archive (SRA): PRJEB23847.
Mapping and calling
All reads were then mapped against the v2.3 oak haplome assembly (Plomion et al., 2018), with bowtie2 v. 2.1.0 (Langmead & Salzberg, 2012), using standard parameters for the “sensitive end-to-end” mode. PCR duplicates were removed with Picard v. 1.106 (http://broadinstitute.github.io/picard/). Samtools v.1.1 (Li et al., 2009) and Popoolation2 v. 1.201 (Kofler et al., 2011) were then used to call biallelic SNPs with at least 10 copies of the alternate allele and a depth between 50 and 2000X at each position. To ensure a reasonably low rate of false positives due to Illumina sequencing errors, all SNPs with a MAF lower than 0.02 were discarded. We obtained allele counts for a total of 31,894,340 SNPs. FST in non-overlapping sliding-window was calculated from allele frequencies with the popoolation2 bioinformatics software suite (Kofler et al., 2011).
Approximate Bayesian computation (ABC) analysis
Observed dataset
For all subsequent ABC analyses, we randomly selected 50,088 of the 31 million SNPs. For each of these SNPs, we multiplied allele frequencies by the number of set of chromosomes in each pool (26 for Q. petraea, 36 for Q. pubescens and 40 for both Q. pyrenaica and Q. robur) to generate the corresponding number of a specific allele in each pool. At this stage, we assumed that any departure from equimolarity due to bias in the mixing of DNA samples would have a negligible effect on our summary statistics calculated for a set of 50 thousand SNPs. Seventeen summary statistics were computed by mscalc: 1) the number of polymorphic sites specific to each gene pool, 2) the number of polymorphic sites existing in both gene pools, 3) nucleotide diversity (π) for each gene pool and, between gene pools, the mean value and standard variation for 4) gross divergence (Dxy), 5) net divergence (Da), 6) FST, and 7) Pearson's R2 correlation coefficient in pi (see also Leroy et al., 2017; Roux et al., 2013; 2016). Demographic inferences based on summary statistics of the site frequency spectrum (SFS) are known to be robust to many sources of variation including the number of individuals and loci (Fraïsse et al. 2018). Besides, SFS-based inferences from pool-seq data have been shown to be robust (Christe et al. 2017).
Demographic scenarios
We used an ABC procedure similar to that described by Leroy and coworkers (2017). Briefly, we compared two different scenarios: isolation-with-migration (IM) and secondary contact (SC). Both scenarios assumed the subdivision of an ancestral panmictic population (PopAnc) into two daughter populations (Pop1 & Pop2) at time TSPLIT, with population sizes remaining constant over time (NPopAnc, NPop1, NPop2). The IM model assumed uninterrupted gene flow since TSPLIT. The SC model assumed that populations initially evolved in strict isolation, with secondary gene flow beginning at some time before the present (at time TSC, Leroy et al., 2017).
Coalescent simulations
For coalescent simulations, we adapted the pipeline described by Leroy et al. (2017) for the calculation of summary statistics to large datasets (up to 100,000 SNPs). This pipeline includes a modified version of the random prior generator priorgen (Leroy et al., 2017; Roux et al., 2013; 2016), and the msnsam (Hudson, 2002; Ross-Ibarra et al., 2008) and mscalc (Ross-Ibarra et al., 2008; Roux et al., 2016) programs. For each of the two scenarios (IM and SC), 1,000,000 random multilocus simulations were performed. Both models made use of genomic heterogeneity in effective migration rates (M) and population sizes (Ne) to take into account the occurrence of genomic barriers to gene flow and the confounding effect of linked selection. For both sources of heterogeneity, we used the strategy described by Leroy et al. (2017).
Model selection
The 500 best replicate simulations closest to the observed values were selected, and the posterior probabilities for each of the two scenarios (IM or SC) were estimated with a feed-forward neural network, by nonlinear multilocus regression (Leroy et al., 2017 for details). ABC computations were performed with 20 feed-forward trained neural and 8 hidden networks.
Parameter estimation
Posterior distributions of the parameters were estimated with a two-step hierarchical procedure. We first evaluated the parameters under the best model, to check the consistency of our estimates, particularly concerning our previous support for very recent secondary contacts (Leroy et al., 2017). We then ran an additional set of 1,000,000 coalescent simulations under an SC model, assuming that TSC occurred in the last 5% of the divergence time, using a modified version of priorgen. This strategy was used to obtain more precise parameter estimates. For both rounds of estimation, we used a logit transformation of the parameters on the 500 best simulations providing the smallest Euclidean distance. The posterior probability of parameters was then estimated by the neural network procedure, from the means of weighted nonlinear multivariate regressions of the parameters on the summary statistics for 25 feed-forward trained neural and 10 hidden networks.
Genome scans
Coalescent simulations for outlier detection
For each pair of species, we ran 5,000,000 coalescent simulations, using parameter values sampled from the 95% confidence interval (CI) of the posterior distribution of all parameters for the pair considered. The simulations assumed genomic homogeneity for effective migration rates (M1 & M2) but heterogeneity for population size. Random values were generated with a modified version of priorgen. For each simulated locus, we then calculated He and GST with a custom-developed script. All scripts and datasets are publicly available for download from a GitHub repository (https://github.com/ThibaultLeroyFr/GenomeScansByABC). Extreme quantiles of the distribution of GST (99.99% of simulated values) relative to the expected heterozygosity of the locus were then calculated, with a strategy similar to that used for Fdist (Beaumont & Nichols, 1996). More specifically, a null envelope was computed from GST quantiles, with heterozygosity intervals of 2%. This strategy has the advantage of providing a null distribution of GST under much more complex scenarios (i.e. the best inferred demographic scenario) than other state-of-the art genome-scan methods.
For each of the 31 million SNPs, we computed the same summary statistics as for simulated neutral markers. The observed GST values conditioned by heterozygosity at the locus were compared to the previously generated null envelope. Markers with a GST value above this envelope were considered to be outliers. We then used a non-overlapping sliding-windows approach to estimate the proportion of outliers per 10-kb window. Windows containing fewer than 10 SNPs were discarded.
We generated parameter estimates under the best-fitting secondary contact model for each pair of species. Taking into account the 95% confidence intervals for each TSC/TSPLIT ratio (Tables 1 & S2) and the divergence time between these species (1-10 million years (Hubert et al., 2014; Hipp et al., 2018), the analysis yielded quite large estimates with secondary contact occurring between 100 and 62,400 years ago, corresponding to up to 1,225 generations, assuming a generation time of 50 years (Gregorius et al., 2007).
Table 1. Posterior probabilities of the SC scenario and timing of secondary contacts.
Mean (bold) relative posterior probability of the secondary contact scenario and standard deviation (round brackets). Median (bold) and 95% confidence intervals (square brackets) for both the inferred ratio between divergence time (TSPLIT) and time of the secondary contact (TSC) and the secondary contact ‘expressed in number of years) after setting TSPLIT to 10 million years (the upper bound for the divergence of this species complex, Hipp et al. 2018). More details are given in Tables S2 & S3.
| Pair | Post. Probability SC | TSC/TSPLIT estimates | TSC (years ago) |
|---|---|---|---|
| Q. robur – Q. petraea | 0.98883 (±0.01089) | 1.487 x 10-3 [0.43-4.05] x 10-3 | 14,870 [4,300-40,500] |
| Q. robur – Q. pyrenaica | 0.99249 (±0.01152) | 1.197 x 10-3 [0.31-4.95} x 10-3 | 11,970 [3,100-40,500] |
| Q. robur – Q. pubescens | 0.98719 (±0.01342) | 0.245 x 10-3 [0.09-0.72} x 10-3 | 2,450 [900-7,200] |
| Q. pubescens – Q. petraea | 0.98549 (±0.02231) | 2.176 x 10-3 [0.77-6.24] x 10-3 | 21,760 [7,700-62,400] |
| Q. pubescens – Q. pyrenaica | 0.99087 (±0.01152) | 0.865 x 10-3 [0.32-2.16] x 10-3 | 8,650 [3,200-21,600] |
| Q. pyrenaica – Q. petraea | 0.99378 (±0.00697) | 0.383 x 10-3 [0.10-1.14] x 10-3 | 3,830 [1,000-11,400] |
Functional annotations
For the 227 genes found within regions enriched in outliers or in close vicinity to these regions (5 kb on both sides to exclude border effects, see “candidate regions” in Fig. 2), we conducted BlastP searches in both the SwissProt and nr protein databases. Only BlastP results with e-values lower than 10e-5 were considered for protein function annotation. After identification of the protein function by BlastP analyses, functional annotations were performed using extensive manual literature searches rather than using automatic approaches based on gene ontology (GO)-oriented methods to ensure high quality gene annotations. We also reported information from a previous identification of orthologous and paralogous genes in 16 plant species, including Q. robur, performed with OrthoMCL (Plomion et al., 2018 for details).
Results
Ecological preferences of the four species
We intersected the distribution maps of the four species (Fig. S3) with climatic and soil data derived from a large-scale floristic survey in France. Bivariate density distributions (Fig. 1B) show clear patterns of ecological preferences among the four white oak species. As expected, the two so called temperate white oaks (Q. petraea and Q. robur) are more frequently observed under cooler climates than Mediterranean and sub-Mediterranean species (Q. pubescens and Q. pyrenaica). Mean annual temperatures of the areas occupied by the two latter species extend up to 20°c, while the two former species occupy much cooler climates (mean temperature below 15°C). pH of the soils segregates particularly Q. pyrenaica from Q. pubescens. The modal value of the former species is close to 5, while the mode of the latter is around 7. Although we could not combine climatic and soil data over the whole species’ ranges, univariate density distributions for both climate (Fig. S1) and soil pH (Fig. S2) based on continental-scale data showed similar trends.
Divergence and post-glacial secondary contact between European white oaks
A total of 31,894,340 SNPs were identified after the filtering of variants with a low minor allele frequency (MAF<0.02) in population samples for the four species (Q. petraea, Q. robur, Q. pubescens, Q. pyrenaica), corresponding to one SNP every 23.2 bp, on average. We also used genome-wide data for a Q. suber accession described by Leroy et al. (2017) to root a phylogenetic tree and investigate relationships between species for 9,084,835 of the 31.9 million SNPs. The best maximum-likelihood tree suggested that Q. robur initially diverged from the ancestor of the other three species (Fig. 1C).
We then randomly selected 50,088 SNPs from the entire set of 31.9 million SNPs for ascertainment bias-free demographic ABC inference. We compared two models of divergence with gene flow (Fig. 2) for each of the six possible species pairs: an isolation-with-migration model assuming constant gene flow since the divergence time (TSPLIT), and a model assuming secondary contact with gene flow starting at TSC, a time point after divergence (TSC < TSPLIT). For all pairs, we obtained strong statistical support for the secondary contact model (>98% posterior probability, Tables 1 & S3 & Fig. S4), consistent with our previous findings based on individual data for 3,524 SNPs (Leroy et al., 2017).
Interspecific genetic differentiation
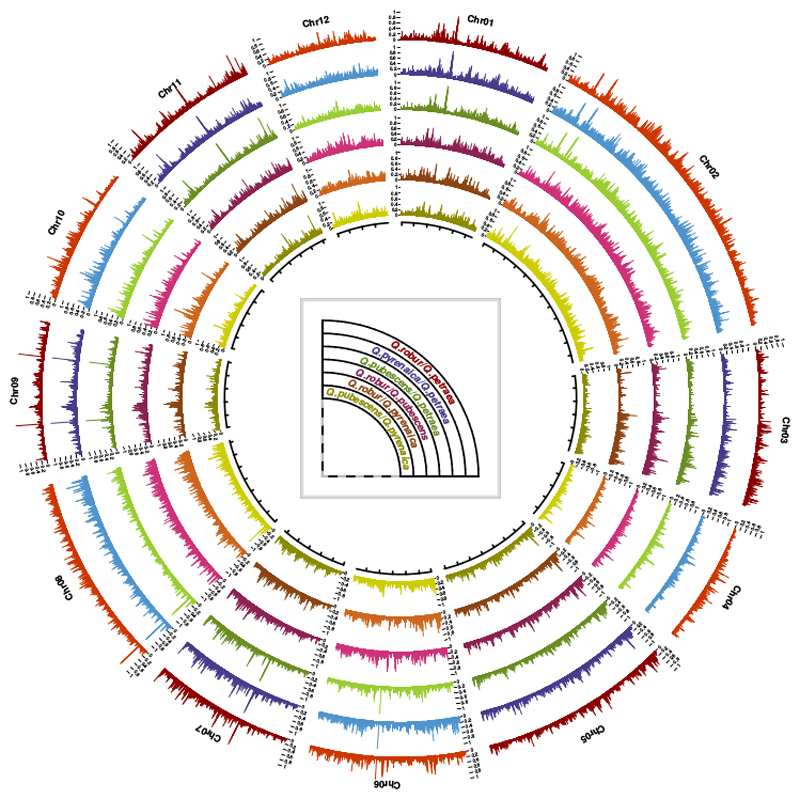
Based on FST values calculated over 10-kb genome segments, the genetic differentiation between species pairs differed considerably between chromosomes (Fig. S5), and, more interestingly, between segments within chromosomes (Fig. 3)
Fig. 3. Circular representation of FST values along the 12 oak chromosomes.
FST was calculated from allele frequencies, using non-overlapping 10 kb sliding windows (detailed patterns are accessible from: https://github.com/ThibaultLeroyFr/GenomeScansByABC/).
First, with rare exceptions, the three most strongly differentiated species pairs, for all oak chromosomes, were Q. robur/Q. pubescens, Q. robur/Q. petraea and Q. pyrenaica/Q. petraea. Lower levels of differentiation were observed for Q. pubescens/Q. pyrenaica and Q.pubescens/Q.petraea (Fig. S5A). These results suggest that the phylogenetic history of these species shaped the genomic landscape of differentiation such that the ranking of pairs of species according to mean FST values is conserved over chromosomes. Conversely, regardless of the pair of species considered, the interchromosomal variation of mean FST values was considerable (Fig. S5B), with significantly higher mean FST values for chromosomes 2 and 6 and significantly lower values for chromosome 4. For all pairs of species, the relationship between mean FST and the rate of recombination was significant at the chromosome scale (P < 0.05; Fig. S6), for calculations based on the comparison of the genetic length of each of the 12 linkage groups (Bodénès et al., 2016) and the physical size of the corresponding pseudo-chromosomes (Plomion et al., 2018), thus suggesting that the chromosomal recombination rate is a good predictor of the mean chromosomal FST.
Second, genome-wide patterns of FST variation plotted over the entire genome with 10-kb sliding windows (Fig. 3) revealed a highly heterogeneous differentiation landscape, even for the pair of species displaying the lowest range of differentiation according to FST values (Q. pubescens/Q. robur). Indeed 10-kb FST estimates between Q. pubescens and Q. robur ranged from 0 to 0.765. For the pairs of species including Q. petraea, the values for the window corresponding to the highest level of differentiation were 0.998 for Q. petraea/Q. pyrenaica, 0.999 for Q. petraea/Q. robur and 1 for Q. petraea/Q. pubescens. On closer inspection, very narrow regions of very high FST were identified on several chromosomes, for most pairs of species (Fig. 3, chromosomes 1, 7, 9, and 11, for example).
Narrow regions of non-neutral evolution
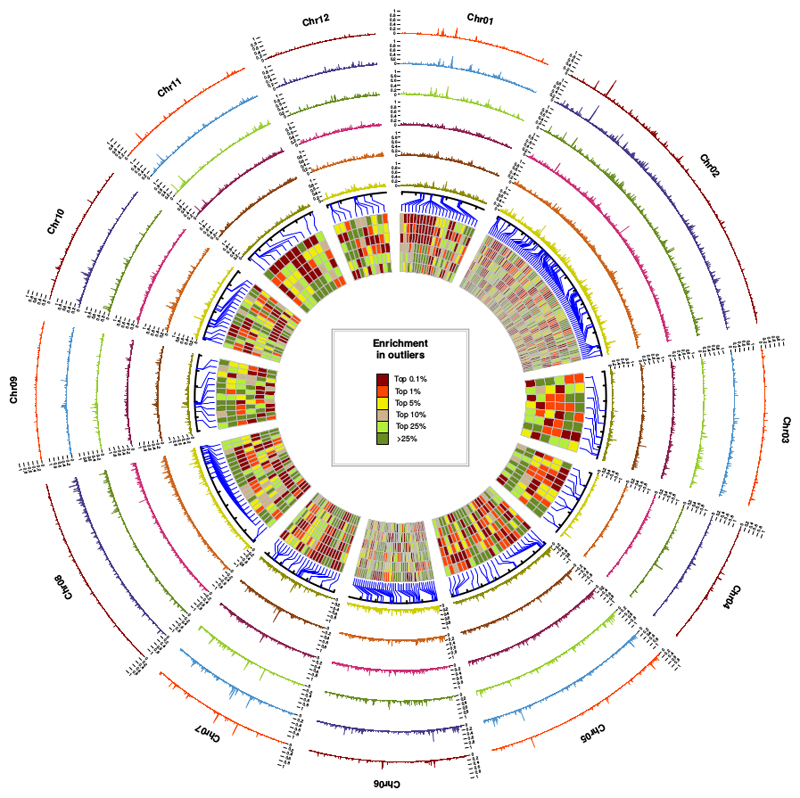
We then took advantage of these demographic inferences to perform differentiation outlier tests. We performed extensive backward simulations (5,000,000 independent SNPs) under the best inferred scenario to generate null distributions for each pairwise comparison (Fig. S7, see also Notes S1). The most outlier-enriched windows were retained for the identification of candidate genes underlying species barriers (after excluding SNPs with very low heterozygosities, Figs. 2 & S8). We identified 281 windows containing the highest proportion of outliers (top 0.1% of window enriched in outliers for at least one pair of species). We then analyzed the clustering of these outlier-enriched windows. We defined a candidate genomic region by merging close windows, e.g. two contiguous sequences of two outlier-enriched windows, with possible interruption by a single undetected window (Fig. 2). The 281 windows were distributed over 215 candidate genomic regions, distributed over all chromosomes (blue lines, Fig. 4).
Fig. 4. Local density in outliers per non-overlapping 10 kb sliding window.
From outside to inside, the species pairs are Q. robur/Q. petraea, Q. pyrenaica/Q. petraea, Q. pubescens/Q. petraea, Q. robur/Q. pubescens, Q. robur/Q. pyrenaica and Q. pubescens/Q. pyrenaica. An outlier corresponds to a SNP with a level of differentiation exceeding the expectations assuming the inferred evolutionary history (0.9999 quantile, see Fig.2 for details). Detailed patterns are accessible from: https://github.com/ThibaultLeroyFr/GenomeScansByABC/. Each rectangle in the inner circle represents the level of enrichment in outliers with He ≤ 0.2 for each pair of species at a given position in the genome, assuming the same order of pairs. These rectangles correspond to the 281 most outlier-enriched windows found in at least one of the six pairs (top 0.1%).
We listed all the Quercus genes located within or flanking these 215 genomic regions. We identified 227 genes distributed over 133 of the 215 regions, with very few candidates per region (mean: 1.71±1.76 genes per region, 1.49 ±0.82 genes after excluding 5 regions with chloroplast-like DNA signatures, see also Notes S2). On all these genes, we performed extensive literature searches to generate manually curated gene annotations. Albeit non-exhaustive by definition, manual literature searches represent more than ever a relevant alternative to methods based on automatically extracted information from literature. Improving the accuracy and traceability of the gene annotations is especially important in oaks since genetic-engineering methods, such as forward and reverse genetic approaches, are not yet available for oaks and gene functions cannot be fully validated based on their phenotypic impacts.
In the following sections, we discuss three major functional categories on the basis of their known implications for ecological and intrinsic reproductive isolation (Dataset S1 for all information). These 3 functional categories contain at least 32 candidate genes (Table 2) among the 227 detected genes (Table S4 and Dataset S1 for details). The first category comprises genes underlying the ecological preferences of the four species: tolerance of water deficit, cold tolerance, adaptation to alkaline soils. The second includes genes involved in biotic interactions, such as immune responses, resistance to biotic stresses, and mycorrhization. The third gathers genes probably involved in intrinsic barriers, and includes genes with functions related to flowering time, pollen recognition, pollen growth and embryo development.
Table 2. Genomic positions, gene names, annotations for 32 selected candidate genes for intrinsic and ecological functions driving reproductive isolation.
All other annotations are available in Table S3 (see Dataset S1 for details, including references). Species patterns were determined from the analysis of the most outlier-enriched windows between pairs, as detailed in Table S5.
| chr. | positions (regions) | Candidate gene | Gene name | Gene functions | #Paralogs | Pattern |
|---|---|---|---|---|---|---|
| Intrinsic barriers | ||||||
| Flowering | ||||||
| Chr08 | 62493250-62513250 | Qrob_P0412860.2 | Two-component response regulator-like PRR73 | photoperiodic flowering response, circadian clock | 1 | Complex |
| Chr08 | 62673250-62693250 | Qrob_P0749650.2 | Floral homeotic protein APETALA 2 | Transcr. factor RAP2-7 | Delay transition to flowering time, biotic/abiotic stresses | 0 | Complex |
| Chr07 | 44592072-44632072 | Qrob_P0088630.2 | Transcr. activator DEMETER (DME) / protein ROS1-like | Transcriptional activator required for floral development | 1 | Complex |
| Pollen development (Cycloartenol synthases) | ||||||
| Chr06 | 28660694-28680694 | Qrob_P0684330.2 | Cycloartenol synthase 2 | Sterol or triterpenoid synthesis, pollen development | 13 | Complex |
| Chr06 | 28690694-28710694 | Qrob_P0684400.2 | Cycloartenol synthase 2 | Sterol or triterpenoid synthesis, pollen development | 13 | Shared |
| Chr06 | 28770694-28810694 | Qrob_P0684360.2 | Cycloartenol synthase 2 | Sterol or triterpenoid synthesis, pollen development | 13 | Shared |
| Pollen recognition & seed germination (G-type lectin S-receptor-like Serine/threonine kinase genes) | ||||||
| Chr03 | 26118565-26138565 | Qrob_P0538230.2 | G-type lectin S-receptor-like STK At2g19130 | Putatively involved in recognition of pollen | 0 | Complex |
| Chr05 | 4001785-4021785 | Qrob_P0641650.2 | G-type lectin S-receptor-like STK At1g11300 | Putatively involved in recognition of pollen | 190 | Complex |
| Chr06 | 1453464-1473464 | Qrob_P0430150.2 | G-type lectin S-receptor-like STK LECRK | Regulates expression of immunity genes & seed germination | 36 | Q. pyrenaica-specific |
| Chr06 | 1513464-1563464 | Qrob_P0430190.2 | G-type lectin S-receptor-like STK LECRK | Regulates expression of immunity genes & seed germination | 36 | Q. pyrenaica-specific |
| Chr08 | 1749735-1769735 | Qrob_P0138480.2 | G-type lectin S-receptor-like STK LECRK | Regulates expression of immunity genes & seed germination | 36 | Q. pyrenaica-specific |
| Embryo development & organogenesis | ||||||
| Chr12 | 30514806-30534806 | Qrob_P0436820.2 | Transcriptional corepressor LEUNIG-like isoform | Leaf, flower (gynoecium) and embryo development | 5 | All except. Q. pub/Q.pyr |
| Chr02 | 31611918-31651918 | Qrob_P0297580.2 | CHD3-type chromatin-remodeling factor PICKLE | Repressor of LEC1, activator of embryo development | 0 | Complex |
| Chr06 | 43006752-43026752 | Qrob_P0309300.2 | Probable N-acetyltransferase HLS1-like | Auxin-responsive gene expression, shoot organogenesis | 1 | All except. Q. rob/Q.pet |
| Chr08 | 49805801-49825801 | Qrob_P0248780.2 | Receptor-like protein 12 (RLP12) | Meristem maintenance control, organogenesis | 0 | Complex |
| Photoreceptor & UV-B tolerance | ||||||
| Chr02 | 37788250-37808250 | Qrob_P0338870.2 | Ultraviolet-B receptor UVR8 | Photoreceptor/response to UV, circadian clock, stomata | 2 | Shared |
| Chr03 | 34667252-34687252 | Qrob_P0500290.2 | DNA mismatch repair protein MSH2 | UV-B-induced DNA damage response pathway | 0 | Shared |
| Chr06 | 11400887-11420887 | Qrob_P0577750.2 | Transcription factor MYB12 | Positive regulator of flavonoid biosynthesis, UV-B tolerance | 0 | All except. Q. rob/Q.pet |
| Ecological barriers - abiotic stresses | ||||||
| Nramp metal transporters | ||||||
| Chr09 | 7739510-7759510 | Qrob_P0191830.2 | Metal transporter Nramp5 | Manganese and cadmium uptake | 8 | Q. pubescens-specific |
| Chr06 | 38764748-38784748 | Qrob_P0097150.2 | Metal transporter Nramp6 | Involved in iron ion homeostasis | 1 | All except. Q. rob/Q.pet |
| Dehydration/lateral root growth | ||||||
| Chr08 | 68418235-68443235 | Qrob_P0457680.2 | Protein DEHYDRATION-INDUCED 19-like | Stress-induced sensor, interacting with CPK11 & S-Rnase | 0 | All except. Q. rob/Q.pet |
| Chr02 | 27061274-27081274 | Qrob_P0304800.2 | Transcription factor WER | Controls cell fate specification, e.g. hairy roots or stomata | 0 | All except. Q. rob/Q.pet |
| Chr02 | 28240136-28260136 | Qrob_P0299670.2 | Root Primordium Defective 1 (RPD1) | Lateral root morphogenesis; active cell proliferation | 0 | Complex |
| Chr02 | 36392234-36442234 | Qrob_P0422470.2 | Alkaline/neutral invertase CINV2 | Regulator of root growth, sucrose catabolism | 7 | Shared |
| Chr09 | 19506093-19526093 | Qrob_P0418880.2 | 1-aminocyclopropane-1-carboxylate synthase | Ethylene biosynthetic process, lateral root formation | 4 | All except. Q. rob/Q.pet |
| Chr04 | 19182244-19222244 | Qrob_P0652510.2 | Phosphatidylinositol-3-phosphatase myotubularin-1 (or 2) | Role in soil-water-deficit stress | 2 | Shared |
| Chr02 | 96008743-96028743 | Qrob_P0387640.2 | Protein WVD2-like 6 | Organ stockiness (periph. root cap, trichomes & leafs) | 0 | All except. Q. rob/Q.pet |
| Freezing/cold adaptation | ||||||
| Chr09 | 18536093-18556093 | Qrob_P0768740.2 | Dehydration-responsive element-binding protein 1 | Key role in freezing tolerance and cold acclimation | 6 | Complex |
| Chr02 | 112153196-112173196 | Qrob_P0339800.2 | B3 domain-containing transcription factor VRN1 | Vernalization responsiveness, repressor of FLC | 3 | All except. Q. pub/Q.pyr |
| Ecological barriers - biotic stresses | ||||||
| Pathogen resistance/mycorrhization | ||||||
| Chr10 | 12310682-12340682 | Qrob_P0070130.2 | Transportin MOS14 | Plant immunity (splicing of resistant genes) | 0 | Q. pyrenaica-specific |
| Chr01 | 26763631-26783631 | Qrob_P0648170.2 | Ubiquitin carboxyl-terminal hydrolase 12-like | Protein deubiquitination, regulator of disease resistance | 3 | Complex |
| Chr05 | 3053435-3073435 | Qrob_P0622110.2 | Nodulation-signaling pathway 1 (NSP1) | Nodulation & mycorrhization | 0 | Complex |
Species-specific ecological and non-ecological reproductive barriers
Unlike studies aiming at interpreting every region enriched in outliers, our objective was rather to focus on genes displaying distinct patterns among pairs of species. This is especially important since these patterns are unexpected to arise via background selection (see Notes S3 for details). After excluding genes with a “shared” pattern (see Table 2), several different cases were observed (Table S5): (i) 9 regions enriched in outliers for all but one pair of species (including 7 regions for all pairs except Q. robur – Q. petraea and 2 regions for all pairs except Q. pubescens – Q. pyrenaica pairs), (ii) 5 regions specific to all pairs sharing the same species (4 for Q. pyrenaica and 1 for Q. pubescens) and (iii) 11 regions with more complex patterns.
Among the nine regions with an “all-versus-one-pair” relationship, seven excluded the Q. robur/Q. petraea pair and the other two excluded the Q. pubescens /Q. pyrenaica pair. Four of the seven candidate genes for which a pattern “all except Q. robur/Q. petraea pair” was observed are known to be involved in drought tolerance or in lateral root growth (Table 2). This pattern is consistent with the higher drought tolerance of Q. pyrenaica and Q. pubescens compared to Q. petraea or Q. robur (Fonti et al., 2013). Reciprocally, we observed an “all vs. one” pattern (undetected for the Q. robur-/Q. petraea pair) for a VRN1 gene involved in responsiveness to vernalization and known to play a key role in cold acclimation in many plant species (Levy et al., 2002). Overall, the genomic variation of these nine genes parallels the Northern-Southern distribution of the studied species, suggesting that the underlying barriers are driven by climate preferences (Fig. 1 A, B).
Among the five candidate genes from genomic regions with branch-specific patterns, i.e. deviating from neutrality in all pairs containing a given species, four have Q. pyrenaica-specific patterns, including three encoding G-type lectin S-receptor-like serine/threonine kinases (LECRKs) and one encoding a transportin (MOS14). The former is known to be essential for proper splicing of several resistance genes in Arabidopsis (Xu et al., 2011), and therefore suggests substantial interspecific differences in plant immunity. Along with the four regions containing genes with Q. pyrenaica specific alleles, we identified a fifth-species-specific pattern for Q. pubescens. The gene encodes a metal transporter (Nramp5) involved in the assimilation of manganese and cadmium in rice and barley (Wu et al., 2016).
Among genes with complex patterns, we identified many candidate genes for intrinsic premating and postmating barriers. We identified several genes involved in the timing of flowering, including APETALA2 and PRR73. APETALA2 is a key transcription factor for the establishment of the floral meristem (Irish & Sussex, 1990). Similarly, PRR73 contributes to flowering time variation in barley and wheat (Higgins et al., 2010 and references therein).
Discussion
The increasing availability of genomic resources for phylogenetically related species has the potential to greatly improve our understanding of their evolutionary trajectories and the molecular basis of their reproductive isolation as shown here for European temperate oaks. Our demographic reconstruction supports long periods of isolation between these oak species for most of their history leading to the gradual loss of shared alleles and the accumulation of reproductive barriers.
Systematic shift in the evolutionary trajectories
We found evidence of a systematic shift in the evolutionary trajectories that occurred at the transition between the last glacial maximum and subsequent postglacial period. More precisely, this shift took place while the oak species were migrating northwards as the climate became warmer, and resulted in their encountering in central Europe. Even if the results reported here are in line with our previous findings (Leroy et al., 2017), this study is based on ascertainment bias-free analyses and use of 10 times as many SNPs. The genome-wide investigation performed here provided stronger support for the occurrence of secondary contact. Indeed, the previous inferences were drawn from a dataset with a strong ascertainment bias. Such a deviation from the true site frequency spectrum is known to have a negative impact on the performance of likelihood-free analyses (Albrechtsen et al., 2010). As in our previous report, our ABC inferences were found to be highly robust to alternative models based on 1000 pseudo-observed datasets, indicating that the probability of an incorrect inference of secondary contact was very low (Fig. S4).
In addition, our use of 10 times as many SNPs and a two-round procedure to generate parameter estimates made it possible to narrow down the time frame over which these contacts took place. After confirming that recent secondary contact had occurred between these four species, we ran additional backward simulations explicitly assuming recent secondary contact (Tsc < 5% Tsplit), to ensure high accuracy for parameter estimates. Assuming that TSPLIT occurred between 1 and 5 million years ago (MYA) (Hubert et al., 2014) or between 7 and 10 MYA (Denk et al., 2017), and taking into account the 95% confidence intervals for each TSC/TSPLIT ratio (Table S2), we estimated that secondary contacts occurred between 100 and 62,400 years ago, corresponding to up to 1,225 generations, assuming a generation time of 50 years (Gregorius et al. 2007, but see also in this issue Leroy et al., 2019a, for a discussion about the uncertainties with regard to the oak generation times), whereas the upper bound for our previous estimate was 11,200 generations (Leroy et al., 2017). Overall, the median estimates of the timing of secondary contact ranged from 2,450 to 21,760 years depending on the species pair considered (49 to 435 generations), consistent with the general hypothesis of a resumption of secondary gene flow at the start of the current interglacial period.
In line with our previous conclusions (Leroy et al., 2017), our inferences cannot however exclude that a few secondary contact periods had already taken place earlier. Since we did not allow for the possibility of multiple periods of contact and isolation in our model, it remains impossible to know if some (few) other periods of contacts had already taken place earlier during the divergence of these species. Indeed, we used summary statistics that are not expected to capture this information well enough in order to be conclusive. However, it should be noted that a scenario with lot of secondary contact periods (e.g. once every past postglacial period) remains unlikely, since it would have generated a much higher posterior probability for IM scenarios. In addition, historical variations in effective population sizes are not taken into account. Further work to infer evolutionary history of these species based on summary statistics at the gene scale (ie based on summary statistics accounting for linkage disequilibrium) rather than those at the base scale will likely have more power to capture these more complex scenarios. Lastly, our analyses are only based on pairwise comparisons. Joint inferences of the evolutionary history of the four (or more) European white oak species should provide additional information about the evolutionary history of these species, especially regarding the direction of interspecific gene flow.
Highly heterogeneous genetic differentiation landscape
The mixture of different species and populations in central Europe occurring during the Holocene was so massive that private (or near private alleles) were redistributed among these interfertile species. Indeed, current levels of interspecific differentiation are extremely low along almost all the genome (mean interchromosomal 10-kb estimates of FST below 0.08 for all pairs) confirming earlier reports by Scotti-Saintagne et al. (2004). Recently, Lang et al. (2018) reported a slightly higher mean FST value for the Q. robur /Q. petraea pair (mean FST=0.13 over ~12,500 SNPs) based on a restricted representation strategy, representing a total sequence data of ~530 Kb (0.072% of the oak genome, Plomion et al., 2018). The differences need to be treated with caution because both estimates are potentially slightly biased. Indeed, we do not rule out the possibility that our estimate is negatively impacted by some DNA quantification and pipetting errors or some unfiltered sequencing errors (Gautier et al., 2013; Hivert et al., 2018). Reciprocally, Lang et al. (2018) targeted some genes potentially involved in species ecological preferences that may have resulted in an overestimation of the mean FST value. Notwithstanding the uncertainty in both estimates, these reported values are consistent with an overall low current level of differentiation and are at a level compatible with many reports of within-species population structure in the literature (Roux et al., 2016).
Interestingly, chromosomal FST estimates differed considerably between pairs of species and chromosomes. For all pairs, inter-chromosomal FST variation was significantly correlated with variations of chromosomal recombination rate (Fig. S6). This is consistent with the increasing number of reports for a correlation between the recombination rate and the rates of introgression, e.g. in the house mouse (Janoušek et al., 2015), in Mimulus monkey flowers (Aeschbacher et al., 2017), in Heliconius butterflies (Martin et al., 2019) or in humans (Juric et al., 2016) and expected to be due to the variation of linkage to introgressed deleterious alleles (background selection, Charlesworth et al., 1993). The recent report of a remarkably high rate of deleterious mutations relative to 28 other plant species (Plomion et al., 2018) highlights the putative role of deleterious mutations in shaping the genomic landscape of species differentiation in oaks. The contribution of linkage to the genomic width of reproductive barriers appears to be limited, as no long stretches of extensive differentiation were observed. We identified very restricted, widely distributed “islands” of high differentiation. The high-resolution genetic landscape of differentiation observed with scattered microislands of high FST on otherwise poorly differentiated chromosomes contrasts sharply with many other FST scan studies reporting either continents of differentiation (e.g. Tine et al., 2014) or islands of high differentiation in highly structured populations (e.g. Renaut et al., 2013).
To summarize thus far, extensive secondary gene flow over the last 20,000 years, together with very high levels of prezygotic and postzygotic selection (Abadie et al., 2012; Lepais et al., 2013), probably eroded interspecific genetic structures other than those at barrier loci, thereby generating a highly heterogeneous differentiation landscape. At some narrow regions distributed throughout the genome, interspecific differentiation however reaches extremely high levels (10-kb estimates of FST above 0.8). These peaks most likely correspond to narrow regions where selection counteracted the homogenizing effect of gene flow, thus leading to the present-day highly heterogeneous landscape of differentiation.
Candidate genes for drought tolerance and insights for the renewal of oak forests
The highest differentiated SNPs contributing to reproductive barriers mostly set apart Southern (Q. pyrenaica & Q. pubescens) from Northern species (Q. robur & Q. petraea). While this observation is inconsistent with the inferred phylogeny (Fig. 1C and Leroy et al., 2017), it however coincides with the climatic preferences of the four species (Fig. 1 A,B). Indeed, all genes with an “all-versus-one-pair” relationship parallel the Northern-Southern distribution of the studied species, suggesting that the underlying barriers are driven by ecological preferences. Given that dehydration-associated genes currently act as strong barriers between these species in South-West France, we question whether drought tolerance alleles from Q. pubescens and Q. pyrenaica would introgress easily into the Q. petraea or Q. robur genomes. In a companion paper (Leroy et al. 2019b), we found evidence for adaptive introgression from Q. robur to Q. petraea populations located at the northern and higher elevational margins. Such introgressed genes were enriched in alleles exhibiting higher Q. petraea/Q.robur differentiation. These results suggest that introgression may override species barriers between these four white oaks under peculiar ecological contexts, and potentially contribute to adaptation (e.g. Bontrager & Angert, 2019; Suarez-Gonzalez et al., 2018; Taylor & Larson, 2019). As Mediterranean oak species (Q. pyrenaica & Q. pubescens) are likely to migrate northwards due to climate change, we anticipate that opportunities for hybridization with temperate species (Q. petraea - Q. robur) may increase, leading potentially to introgression enhancing adaptation of the temperate species to warmer climates.
In addition, we found genetic support for Q. pubescens preference for alkaline soils and found evidence for one key gene, Nramp5. Indeed Nramp5 is involved in the assimilation of manganese and cadmium in rice and barley (Wu et al., 2016). Manganese assimilation is known to be essential for many plant functions, but manganese availability in the soil tends to decrease with increasing pH, and becomes limiting beyond a soil pH of 6.5. This metal transporter gene probably indicates a greater ecological preference of Q. pubescens for dolomitic soils (Fig. 1B) in comparison to the other three species.
We also found differences between Q. pyrenaica and the three other species at genes involved in plant immunity, in line with previous reports for higher mortality rates in this species due to pathogens (Desprez-Loustau et al., 2011 and references therein). Indeed, Q. pyrenaica is known to be extremely sensitive to oak powdery mildew, a pathogen that was introduced into Europe at the start of the 20th century. Soon after the first detection of the fungus in Europe, high mortality rates were reported for Q. pyrenaica in the humid warm-temperate forests of Southwestern and Western France (Desprez-Loustau et al., 2011). Q. pyrenaica-specific alleles at these genes may be the signature of the high susceptibility of Q. pyrenaica to biotic stresses in moist environments. It however remains to be studied why these genes act as reproductive barriers.
In addition, we identified several candidate genes for intrinsic barriers likely involved in pollen or embryo development, suggesting that both premating and postmating intrinsic barriers operate in oaks as suggested by Bodénès et al. (2016). Three of these genes encode cycloartenol synthases known to be essential for pollen development in Arabidopsis (Badiychuk et al., 2008). Interestingly, some genes can have pleiotropic effects, that may act at both ecological and mating levels.VRN1 is a case of point. In addition to its primary role in vernalization, VRN1 is involved in the repression of FLC (itself a known repressor of flowering) in Arabidopsis, through a vernalization-independent floral pathway (Levy et al., 2002).
Overall, our results suggest that key selective abiotic and biotic factors triggered by post glacial environmental changes have molded the extant landscape of species reproductive barriers in European temperate oak species. We anticipate that these drivers will operate during ongoing climate changes as Mediterranean oak species (Q. pyrenaica & Q. pubescens) are migrating northwards getting in contact in more Northern latitudes with local temperate species (Q. petraea and Q. robur).
Supplementary Material
Acknowledgments
This research was funded by the French ANR (GENOAK project, 11-BSV6-009-021) and by the European Research Council under the European Union’s Seventh Framework Programme (TREEPEACE project, FP/2014-2019; ERC Grand Agreement no. 339728). We thank the Genotoul Bioinformatics Platform Toulouse Midi-Pyrenees (Bioinfo Genotoul) and the Biogenouest BiRD core facility (Université de Nantes) for providing computing and storage resources. We also thank Jorge A. P. Paiva for providing access to Q. suber data and Camille Roux for fruitful discussions concerning ABC. We would like to thank fellow members of the pedunculate oak genome consortium for helpful advice and suggestions.
Footnotes
Author Contribution
T.L designed and performed the research, analyzed the data and drafted the manuscript; Q.R, and C.Bo. contributed to data analysis and interpretation. J-L.D. performed the analysis of the climatic and soil data. C.L. performed the DNA extractions and equimolarly pooled DNA. C. Be., K.L. and J-M.A generated the sequencing data. GLP and CP were involved in the sampling and selection of genotypes. C.P and A.K contributed to the design of the research, interpretation and drafted the manuscript.
References
- Abadie P, Roussel G, Dencausse B, Bonnet C, Bertocchi E, Louvet J-M, Kremer A, Garnier-Géré P. Strength, diversity and plasticity of postmating reproductive barriers between two hybridizing oak species (Quercus robur L. and Quercus petraea (Matt) Liebl.) Journal of Evolutionary Biology. 2012;25:157–173. doi: 10.1111/j.1420-9101.2011.02414.x. [DOI] [PubMed] [Google Scholar]
- Aeschbacher S, Selby JP, Willis JH, Coop G. Population-genomic inference of the strength and timing of selection against gene flow. Proceedings of the National Academy of Sciences. 2017;114:7061. doi: 10.1073/pnas.1616755114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrechtsen A, Nielsen FC, Nielsen R. Ascertainment Biases in SNP Chips Affect Measures of Population Divergence. Molecular Biology and Evolution. 2010;27:2534–2547. doi: 10.1093/molbev/msq148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiychuk E, Bouvier-Navé P, Compagnon V, Suzuki M, Muranaka T, Van Montagu M, Kushnir S, Schaller H. Allelic mutant series reveal distinct functions for Arabidopsis cycloartenol synthase 1 in cell viability and plastid biogenesis. Proceedings of the National Academy of Sciences. 2008;105:3163. doi: 10.1073/pnas.0712190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont MA, Nichols RA. Evaluating loci for use in the genetic analysis of population structure. Proceedings of the Royal Society of London Series B: Biological Sciences. 1996;263:1619–1626. [Google Scholar]
- Bodénès C, Chancerel E, Ehrenmann F, Kremer A, Plomion C. High-density linkage mapping and distribution of segregation distortion regions in the oak genome. DNA Res. 2016;23:115–124. doi: 10.1093/dnares/dsw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodénès C, Joandet S, Laigret F, Kremer A. Detection of genomic regions differentiating two closely related oak species Quercus petraea (Matt.) Liebl. and Quercus robur L. Heredity. 1997;78:433–444. [Google Scholar]
- Bontrager M, Angert AL. Gene flow improves fitness at a range edge under climate change. Evolution letters. 2018;3:55–68. doi: 10.1002/evl3.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Morgan MT, Charlesworth D. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993;134:1289. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christe C, Stölting KN, Paris M, Fraïsse C, Bierne N, Lexer C. Adaptive evolution and segregating load contribute to the genomic landscape of divergence in two tree species connected by episodic gene flow. Molecular Ecology. 2017;26:59–76. doi: 10.1111/mec.13765. [DOI] [PubMed] [Google Scholar]
- Darwin C. On the Origin of Species by Means of Natural Selection: Or the Preservation of Favoured Races in the Struggle for Life. (5th edition) London, UK: John Murray; 1869. [PMC free article] [PubMed] [Google Scholar]
- Denk T, Grimm GW, Manos PS, Deng M, Hipp AL. An updated infrageneric classification of the oaks: review of previous taxonomic schemes and synthesis of evolutionary patterns. bioRxiv. 2017 doi: 10.1101/168146. 168146. [DOI] [Google Scholar]
- Desprez-Loustau M-L, Feau N, Mougou-Hamdane A, Dutech C. Interspecific and intraspecific diversity in oak powdery mildews in Europe: coevolution history and adaptation to their hosts. Mycoscience. 2011;52:165–173. [Google Scholar]
- Eaton E, Caudullo G, Oliveira S, de Rigo D. Quercus robur and Quercus petraea in Europe: distribution, habitat, usage and threats. In: San-Miguel-Ayanz J, de Rigo D, Caudullo G, Houston Durrant T, Mauri A, editors. European Atlas of Forest Tree Species. Luxembourg City, Luxembourg: The publications office of the European Union; 2016. European Atlas of Forest Tree Species. pp. e01c6df+ [Google Scholar]
- Fonti P, Heller O, Cherubini P, Rigling A, Arend M. Wood anatomical responses of oak saplings exposed to air warming and soil drought. Plant Biology. 2013;15:210–219. doi: 10.1111/j.1438-8677.2012.00599.x. [DOI] [PubMed] [Google Scholar]
- Fraïsse C, Roux C, Gagnaire P-A, Romiguier J, Faivre N, Welch JJ, Bierne N. The divergence history of European blue mussel species reconstructed from Approximate Bayesian Computation: the effects of sequencing techniques and sampling strategies. PeerJ. 2018;6:e5198. doi: 10.7717/peerj.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier M, Foucaud J, Gharbi K, Cézard T, Galan M, Loiseau A, Thomson M, Pudlo P, Kerdelhué C, Estoup A. Estimation of population allele frequencies from next-generation sequencing data: pool-versus individual-based genotyping. Molecular Ecology. 2013;22:3766–3779. doi: 10.1111/mec.12360. [DOI] [PubMed] [Google Scholar]
- Gégout JC, Coudun C, Bailly G, Jabiol B. EcoPlant: A forest site database linking floristic data with soil and climate variables. Journal of Vegetation Science. 2005;16:257–260. [Google Scholar]
- Gregorius HR, Degen B, König A. Problems in the Analysis of Genetic Differentiation Among Populations – a Case Study in Quercus robur. Silvae Genetica. 2007;56:190–199. [Google Scholar]
- Hewitt GM. Hybrid zones-natural laboratories for evolutionary studies. Trends in Ecology & Evolution. 1988;3:158–167. doi: 10.1016/0169-5347(88)90033-X. [DOI] [PubMed] [Google Scholar]
- Higgins JA, Bailey PC, Laurie DA. Comparative Genomics of Flowering Time Pathways Using Brachypodium distachyon as a Model for the Temperate Grasses. PLOS ONE. 2010;5:e10065. doi: 10.1371/journal.pone.0010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp AL, Manos PS, González-Rodríguez A, Hahn M, Kaproth M, McVay JD, Avalos SV, Cavender-Bares J. Sympatric parallel diversification of major oak clades in the Americas and the origins of Mexican species diversity. New Phytologist. 2018;217:439–452. doi: 10.1111/nph.14773. [DOI] [PubMed] [Google Scholar]
- Hivert V, Leblois R, Petit EJ, Gautier M, Vitalis R. Measuring Genetic Differentiation from Pool-seq Data. Genetics. 2018;210:315. doi: 10.1534/genetics.118.300900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorius HR, Degen B, König A. Problems in the Analysis of Genetic Differentiation Among Populations – a Case Study in Quercus robur. Silvae Genetica. 2007;56:190–199. [Google Scholar]
- Hubert F, Grimm GW, Jousselin E, Berry V, Franc A, Kremer A. Multiple nuclear genes stabilize the phylogenetic backbone of the genus Quercus. Systematics and Biodiversity. 2014;12:405–423. [Google Scholar]
- Hudson RR. Generating samples under a Wright–Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- Irish VF, Sussex IM. Function of the apetala-1 gene during Arabidopsis floral development. The Plant Cell. 1990;2:741. doi: 10.1105/tpc.2.8.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoušek V, Munclinger P, Wang L, Teeter KC, Tucker PK. Functional organization of the genome may shape the species boundary in the house mouse. Molecular biology and evolution. 2015;32:1208–1220. doi: 10.1093/molbev/msv011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gégout JC, Coudun C, Bailly G, Jabiol B. EcoPlant: A forest site database linking floristic data with soil and climate variables. Journal of Vegetation Science. 2005;16:257–260. [Google Scholar]
- Juric I, Aeschbacher S, Coop G. The Strength of Selection against Neanderthal Introgression. PLOS Genetics. 2016;12:e1006340. doi: 10.1371/journal.pgen.1006340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karger DN, Conrad O, Böhner J, Kawohl T, Kreft H, Soria-Auza RW, Zimmermann NE, Linder HP, Kessler M. Climatologies at high resolution for the earth’s land surface areas. Scientific Data. 2017;4 doi: 10.1038/sdata.2017.122. 170122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer A, Abbott AG, Carlson JE, Manos PS, Plomion C, Sisco P, Staton ME, Ueno S, Vendramin GG. Genomics of Fagaceae. Tree Genetics & Genomes. 2012;8:583–610. [Google Scholar]
- Kofler R, Pandey RV, Schlötterer C. PoPoolation2: identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq) Bioinformatics. 2011;27:3435–3436. doi: 10.1093/bioinformatics/btr589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang T, Abadie P, Léger V, Decourcelle T, Frigerio J-M, Burban C, Bodénès C, Guichoux E, Le Provost G, Robin C, et al. High-quality SNPs from genic regions highlight introgression patterns among European white oaks (Quercus petraea and Q. robur) bioRxiv. 2018 doi: 10.1101/388447. 388447. [DOI] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepais O, Petit RJ, Guichoux E, Lavabre JE, Alberto F, Kremer A, Gerber S. Species relative abundance and direction of introgression in oaks. Molecular Ecology. 2009;18:2228–2242. doi: 10.1111/j.1365-294X.2009.04137.x. [DOI] [PubMed] [Google Scholar]
- Lepais O, Roussel G, Hubert F, Kremer A, Gerber S. Strength and variability of postmating reproductive isolating barriers between four European white oak species. Tree Genetics & Genomes. 2013;9:841–853. [Google Scholar]
- Leroy T, Kremer A, Plomion C. Oak symbolism in the light of genomics. New Phytologist. 2019a doi: 10.1111/nph.15987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy T, Louvet J-M, Lalanne C, Le Provost G, Labadie K, Aury J-M, Delzon S, Plomion C, Kremer A. Adaptive introgression as driver of local adaptation to climate in European white oaks. bioRxiv. 2019b doi: 10.1101/584847. 584847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy T, Roux C, Villate L, Bodénès C, Romiguier J, Paiva JAP, Dossat C, Aury J-M, Plomion C, Kremer A. Extensive recent secondary contacts between four European white oak species. New Phytologist. 2017;214:865–878. doi: 10.1111/nph.14413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy YY, Mesnage S, Mylne JS, Gendall AR, Dean C. Multiple Roles of Arabidopsis VRN1 in Vernalization and Flowering Time Control. Science. 2002;297:243. doi: 10.1126/science.1072147. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SH, Davey JW, Salazar C, Jiggins CD. Recombination rate variation shapes barriers to introgression across butterfly genomes. PLOS Biology. 2019;17:e2006288. doi: 10.1371/journal.pbio.2006288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer EJ. HYBRID OAKS OF NORTH AMERICA. Journal of the Arnold Arboretum. 1948;29:1–48. [Google Scholar]
- Petit RJ, Csaikl UM, Bordács S, Burg K, Coart E, Cottrell J, van Dam B, Deans JD, Dumolin-Lapègue S, Fineschi S, et al. Chloroplast DNA variation in European white oaks: Phylogeography and patterns of diversity based on data from over 2600 populations. Range wide distribution of chloroplast DNA diversity and pollen deposits in European white oaks: inferences about colonisation routes and management of oak genetic resources. Forest Ecology and Management. 2002;156:5–26. [Google Scholar]
- Petit RJ, Pineau E, Demesure B, Bacilieri R, Ducousso A, Kremer A. Chloroplast DNA footprints of postglacial recolonization by oaks. Proceedings of the National Academy of Sciences. 1997;94:9996–10001. doi: 10.1073/pnas.94.18.9996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomion C, Aury J-M, Amselem J, Alaeitabar T, Barbe V, Belser C, Bergès H, Bodénès C, Boudet N, Boury C, et al. Decoding the oak genome: public release of sequence data, assembly, annotation and publication strategies. Molecular Ecology Resources. 2016;16:254–265. doi: 10.1111/1755-0998.12425. [DOI] [PubMed] [Google Scholar]
- Plomion C, Aury J-M, Amselem J, Leroy T, Murat F, Duplessis S, Faye S, Francillonne N, Labadie K, Le Provost G, et al. Oak genome reveals facets of long lifespan. Nature Plants. 2018;4:440–452. doi: 10.1038/s41477-018-0172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaut S, Grassa CJ, Yeaman S, Moyers BT, Lai Z, Kane NC, Bowers JE, Burke JM, Rieseberg LH. Genomic islands of divergence are not affected by geography of speciation in sunflowers. Nature Communications. 2013;4:1827. doi: 10.1038/ncomms2833. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Wood TE, Baack EJ. The nature of plant species. Nature. 2006;440:524–527. doi: 10.1038/nature04402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Ibarra J, Wright SI, Foxe JP, Kawabe A, DeRose-Wilson L, Gos G, Charlesworth D, Gaut BS. Patterns of Polymorphism and Demographic History in Natural Populations of Arabidopsis lyrata. PLoS ONE. 2008;3:e2411. doi: 10.1371/journal.pone.0002411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux C, Fraïsse C, Romiguier J, Anciaux Y, Galtier N, Bierne N. Shedding Light on the Grey Zone of Speciation along a Continuum of Genomic Divergence. PLOS Biology. 2016;14:e2000234. doi: 10.1371/journal.pbio.2000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux C, Tsagkogeorga G, Bierne N, Galtier N. Crossing the species barrier: genomic hotspots of introgression between two highly divergent Ciona intestinalis species. Molecular Biology and Evolution. 2013;30:1574–1587. doi: 10.1093/molbev/mst066. [DOI] [PubMed] [Google Scholar]
- San-Miguel-Ayanz J, de Rigo D, Caudullo G, Houston Durrant T, Mauri A, Tinner W, Ballian D, Beck P, Birks H, Eaton E, et al. European atlas of forest tree species. Luxembourg City, Luxembourg: The publications office of the European Union; 2016. [Google Scholar]
- Schlötterer C, Tobler R, Kofler R, Nolte V. Sequencing pools of individuals — mining genome-wide polymorphism data without big funding. Nature Reviews Genetics. 2014;15:749. doi: 10.1038/nrg3803. [DOI] [PubMed] [Google Scholar]
- Scotti-Saintagne C, Mariette S, Porth I, Goicoechea PG, Barreneche T, Bodénès C, Burg K, Kremer A. Genome Scanning for Interspecific Differentiation Between Two Closely Related Oak Species [Quercus robur L. and Q. petraea (Matt.) Liebl.] Genetics. 2004;168:1615. doi: 10.1534/genetics.104.026849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Gonzalez A, Lexer C, Cronk QCB. Adaptive introgression: a plant perspective. Biology letters. 2018;14 doi: 10.1098/rsbl.2017.0688. 20170688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SA, Larson EL. Insights from genomes into the evolutionary importance and prevalence of hybridization in nature. Nature Ecology & Evolution. 2019;3:170–177. doi: 10.1038/s41559-018-0777-y. [DOI] [PubMed] [Google Scholar]
- Timbal J, Aussenac G. An overview of ecology and silviculture of indigenous oaks in France. Annals of Forest Science. 1996;53:649–661. [Google Scholar]
- Tine M, Kuhl H, Gagnaire P-A, Louro B, Desmarais E, Martins RST, Hecht J, Knaust F, Belkhir K, Klages S, et al. European sea bass genome and its variation provide insights into adaptation to euryhalinity and speciation. Nature Communications. 2014;5:5770. doi: 10.1038/ncomms6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries S, Murat A, Bozzano M, Burianek V, Collin E, Cottrell J, Ivankovic M, Kelleher C, Koskela J, Rotach P, et al. Pan-European strategy for genetic conservation of forest trees and establishment of a core network of dynamic conservation units. Rome, Italy: Bioversity International; 2015. p. 40. [Google Scholar]
- Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag; New York: 2016. [Google Scholar]
- Wu D, Yamaji N, Yamane M, Kashino-Fujii M, Sato K, Feng Ma J. The HvNramp5 Transporter Mediates Uptake of Cadmium and Manganese, But Not Iron. Plant Physiology. 2016;172:1899. doi: 10.1104/pp.16.01189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Zhang Z, Jing B, Gannon P, Ding J, Xu F, Li X, Zhang Y. Transportin-SR Is Required for Proper Splicing of Resistance Genes and Plant Immunity. PLOS Genetics. 2011;7:e1002159. doi: 10.1371/journal.pgen.1002159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.