Abstract
Proline dehydrogenase (PRODH) catalyzes the first step of proline catabolism, the FAD-dependent 2-electron oxidation of l-proline to Δ1-pyrroline-5-carboxylate. PRODH has emerged as a possible cancer therapy target, and thus the inhibition of PRODH is of interest. Here we show that the proline analogue thiazolidine-2-carboxylate (T2C) is a mechanism-based inactivator of PRODH. Structures of the bifunctional proline catabolic enzyme proline utilization A (PutA) determined from crystals grown in the presence of T2C feature strong electron density for a 5-membered ring species resembling l-T2C covalently bound to the N5 of the FAD in the PRODH domain. The modified FAD exhibits a large butterfly bend angle, indicating that the FAD is locked into the 2-electron reduced state. Reduction of the FAD is consistent with the crystals lacking the distinctive yellow color of the oxidized enzyme and stopped-flow kinetic data showing that T2C is a substrate for the PRODH domain of PutA. A mechanism is proposed in which PRODH catalyzes the oxidation of T2C at the C atom adjacent to the S atom of the thiazolidine ring (C5). Then, the N5 atom of the reduced FAD attacks the C5 of the oxidized T2C species, resulting in the covalent adduct observed in the crystal structure. To our knowledge, this is the first report of T2C inactivating (or inhibiting) PRODH or any other flavoenzyme. These results may inform the design of new mechanism-based inactivators of PRODH for use as chemical probes to study the roles of proline metabolism in cancer.
Graphical Abstract
The proline catabolic pathway catalyzes the 4-electron oxidation of proline to glutamate and consists of the enzymes proline dehydrogenase (PRODH) and L-glutamate-γ- semialdehyde (GSAL) dehydrogenase (GSALDH). PRODH catalyzes the FAD-dependent oxidation of L-proline to Δ1- pyrroline-5-carboxylate (P5C) (Scheme 1).1 The nonenzymatic hydrolysis of P5C produces GSAL, the substrate for GSALDH. The enzyme GSALDH, also known as ALDH4A1 in humans, completes the pathway by catalyzing the NAD+-dependent oxidation of GSAL to L-glutamate. In some bacteria, PRODH and GSALDH are combined into a single bifunctional enzyme, known as proline utilization A (PutA).2
Scheme 1. Reactions of Proline Catabolism.

Proline catabolism has been associated with important metabolic alterations in cancer cells. In some metastatic cancers, upregulation of PRODH,3,4 GSALDH (a.k.a. ALDH4A1), and the proline biosynthetic enzymes PYCR1 and P5C synthase5,6 has been observed. Expression of the catabolic enzymes is p53-dependent7,8 and the expression of the biosynthetic enzymes is MYC-dependent,9 highlighting potential roles in cancer. Reprogramming of proline metabolic pathways allows cancer cells to proliferate faster than normal cells. This is thought to be due to changes in the oxidative state in the cell, which activates downstream cell signaling pathways, as well as providing ATP and redox cofactors to the cell.10
These observations suggest that proline metabolic enzymes are potential targets for cancer therapy. PRODH is of particular interest in this regard. The reversible PRODH inhibitor l-tetrahydro-2-furoic acid (THFA, Scheme 2) has been shown to decrease breast cancer metastases in mouse models,4 and the irreversible, mechanism-based inactivator N-propargylglycine (NPPG) decreases cancer cell growth and viability, especially when used in combination with a glutaminase inhibitor.11
Scheme 2. Chemical Structures of THFA and.

l-T2C
The present study grew out of a larger effort to screen proline analogues for the inhibition of proline metabolic enzymes. Here, we provide evidence that thiazolidine-2- carboxylate (T2C, Scheme 2) is a mechanism-based inactivator that covalently modifies the FAD in PRODH. We report the crystal structure of PutA from Sinorhizobium meliloti (SmPutA), which we are using as a surrogate for human PRODH, featuring an apparent oxidized l-T2C species covalently connected to the N5 of the FAD. The T2C-modified FAD appears to be in the 2-electron reduced state, based on the butterfly bend angle of the isoalloxazine and the lack of yellow color in the crystal. Stopped-flow spectroscopy shows that T2C is rapidly oxidized by PRODH, resulting in the reduction of the FAD. Kinetic measurements show that T2C inactivates PRODH in a time-dependent manner, consistent with mechanism-based covalent inactivation. A possible mechanism for the inactivation reaction is proposed. To our knowledge, this is the first report of mechanism-based inactivation of any flavoenzyme by T2C.
RESULTS AND DISCUSSION
Crystallographic Evidence That T2C Covalently Modifies the FAD of PutA.
Our discovery that T2C covalently modifies the FAD of the PutA PRODH domain was serendipitous. The cocrystallization of SmPutA with T2C was attempted as part of an effort to find noncovalent proline analogue inhibitors of PRODH, which are valued as chemical probes in cancer biology research.10 The addition of T2C to an SmPutA stock solution immediately bleached the yellow color of the enzyme, suggesting that the FAD had been reduced and T2C had been oxidized. Nevertheless, the colorless enzyme solution was used in crystallization trials. Typically, crystals of oxidized SmPutA appear in a few days as bright yellow rectangular solids; however, this did not occur. Instead, after a long time (at least 10 days), colorless crystals were observed. The crystals remained colorless during the process of harvesting, which was done under ordinary aerobic laboratory conditions, without the addition of a reducing agent or extra T2C.
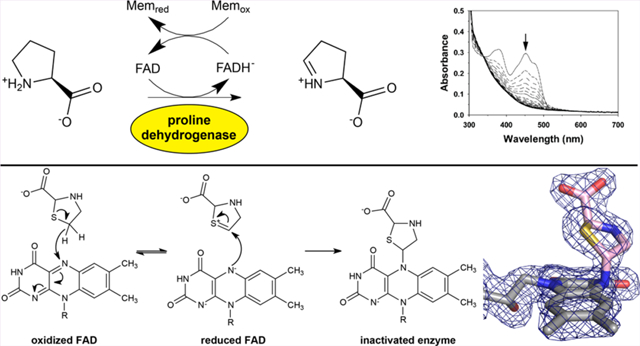
Two structures were determined from crystals harvested on either day 30 or day 14 after setting up the crystallization experiment. The electron density maps from both structures clearly indicated that the FAD was modified at the N5 atom (Figure 1A and Figure S1). The maps were consistent with a proline analogue—presumably a derivative of T2C—bonded to the FAD N5. The electron density provided unambiguous characterization of the stereochemistry and regiochemistry of the modification. For example, the density for the carboxylate moiety was very prominent, which allowed for identification of the C2 atom of the T2C ring. Also, a very strong peak in the Fo−Fc omit map (12σ) was assumed to represent the S atom (Figure 1B). Together, these observations not only confirmed that the attached species was derived from the l-enantiomer of T2C but also implicated that C5 of the l-T2C ring is the atom bonded to N5 of the FAD. We note that a racemic mixture of d,l-T2C was used in cocrystallization.
Figure 1.
Electron density evidence (1.74 Å resolution) for covalent modification of the FAD in PRODH by T2C. (A) Modified FAD of SmPutA covered by a polder omit map (4σ). (B) Polder omit map contoured at 12σ, showing a peak indicating the S atom. The FAD and T2C are colored gray and pink, respectively.
Crystal backsoaking was used to assess the irreversibility of the FAD−T2C covalent adduct. The mother liquor surrounding crystals of T2C-inactivated SmPutA was replaced with a cryobuffer lacking T2C and incubated for 24 h prior to flashcooling and X-ray diffraction data collection. The electron density from the backsoaked crystals clearly showed the covalent adduct, indicating that the covalent modification of the FAD was stable in crystallo for at least 24 h (Figure S2).
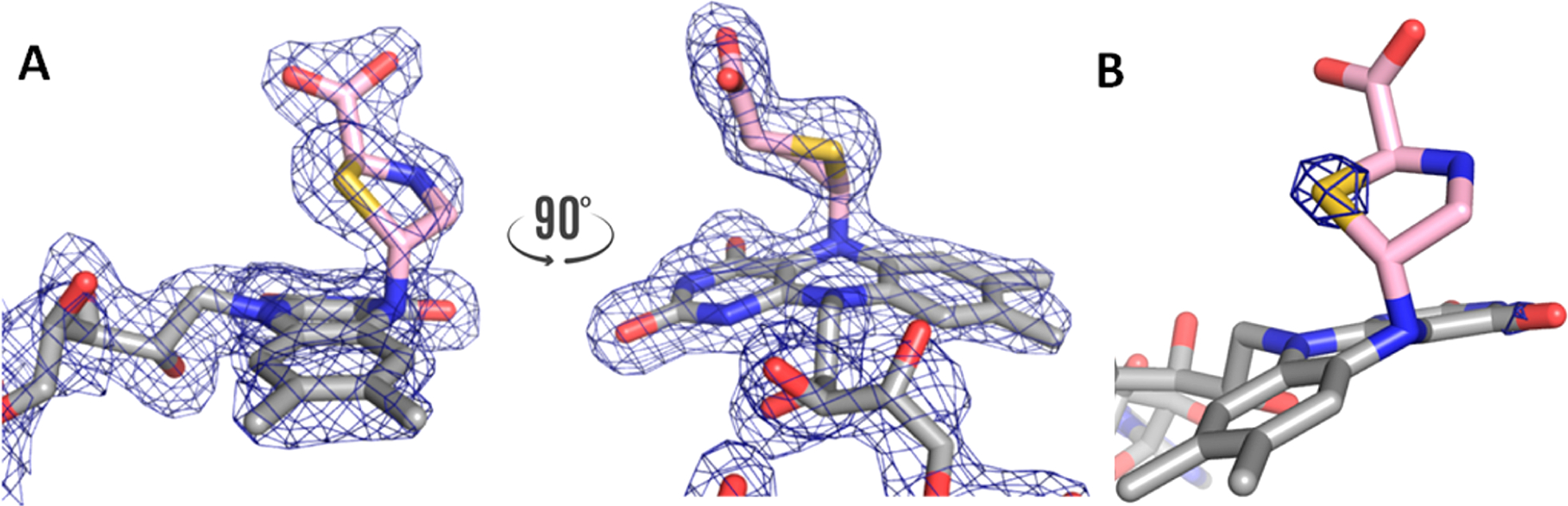
The covalently bound l-T2C occupies the same location as the noncovalent inhibitor THFA. l-T2C is attached to the si face of the FAD. The carboxylate of l-T2C forms ion pairs with Arg488, Arg489, and Lys265 (Figure 2A). These ion pairs are also seen in the THFA complex (Figure 2B). A subtle difference between the l-T2C-modified and THFA-inhibited enzymes is the conformation of Arg489; this is likely due to the different conformations of the ribityl chain of the FAD. The amino group of l-T2C forms a hydrogen bond with a conserved water molecule. Similarly, the heteroatom oxygen of THFA forms an analogous hydrogen bond with a water molecule in the same location.
Figure 2.
Interactions of (A) the T2C covalent adduct with the active site, compared with (B) the noncovalent THFA complex (PDB 5KF6).
The modified FAD appears to be reduced. As noted above, the bleached color of the crystal is consistent with a 2-electron reduction of the FAD. Also, the conformation of the FAD is consistent with reduction. The conformational indicators of the redox state of the FAD in PutA are known from crystal structures of oxidized, reduced, and inactivated PutAs and PRODHs. The conformation of the isoalloxazine is the most reliable structural proxy for the flavin redox state of PRODH. The isoalloxazine of the oxidized FAD is planar, whereas that of the 2-electron reduced FAD exhibits butterfly bend angles of 9°−35° (si face is convex).12–16 The FAD of T2C-modified SmPutA exhibits a convex si face with a butterfly angle of 24°, consistent with the FAD being reduced (Figure 3A).
Figure 3.

Conformation of the modified FAD is consistent with 2-electron reduction. (A) Comparison of PutA structures highlighting the bend of the isoalloxazine. From left to right: oxidized SmPutA complexed with THFA (PDB 5KF6); T2C-modified SmPutA; Geobacter sulfurreducens PutA (GsPutA) reduced by dithionite (PDB 4NMD); NPPG-inactivated GsPutA (PDB 4NME). (B) Comparison of the ribityl chain conformations in PutA structures. From left to right: oxidized SmPutA complexed with THFA (PDB 5KF6); T2C-modified SmPutA; GsPutA reduced by dithionite (PDB 4NMD); NPPG-inactivated GsPutA (PDB 4NME).
The conformation of the ribityl is also suggestive of flavin reduction. In the T2C-modified FAD, the 2′−OH and 3′−OH groups are below the pyrimidine ring of the isoalloxazine and the 4′−OH sits under the dimethylbenzene ring (Figure 3B). This arrangement of hydroxyl groups is associated with the reduced state of PutA13–16 and differs substantially from that of oxidized SmPutA (Figure 3B).
The FAD conformation is similar to that of PutA inactivated by the mechanism-based inactivator NPPG. Inactivation by NPPG locks the FAD into the 2-electron reduced state and results in a 3-carbon covalent link between the FAD N5 atom and a conserved active site lysine residue.13,14,16,17 Both the T2C- and NPPG-modified FADs exhibit large butterfly bending and the ribityl conformation typical of the reduced flavin (Figure 3).
Covalent Modification of Surface Cysteine Residues.
In the structure determined from the crystal harvested on day 30, three cysteine residues of SmPutA (Cys46, Cys470, Cys638) exhibited strong electron density features, indicating covalent modification of their S atoms (Figure 4). The modified residues are on, or near, the surface of SmPutA, with Cys46 being part of the N-terminal arm, Cys470 located on β7 of the PRODH (βα)8 barrel, and Cys638 located in the GSALDH domain (Figure 4A). The tube-like electron density suggested modification by a linear chain of four atoms linked to the cysteine by an S−S bond. A survey of modified cysteines in the PDB uncovered S,S-(2-hydroxyethyl)thiocysteine (PDB ligand ID: CME) as a good fit to the density (Figure 4B). We note that these cysteines were not modified in the absence of T2C, suggesting that the modifying species derives from T2C (see native SmPutA structure, PDB 5KF6). Further, these residues were not modified in the T2C-inactivated SmPutA structure determined from a crystal harvested on day 14, indicating that modification of the flavin occurs before modification of the cysteines (Figure 4C).
Figure 4.

Electron density evidence for covalent modification of cysteines in the crystal harvested on day 30. (A) Location of the modified cysteine residues in SmPutA. SmPutA is colored by domain, and the modified cysteines are shown as spheres. (B) Modified Cys46, Cys470, and Cys638 of SmPutA observed in the structure determined from a crystal harvested on day 30. The cage represents a refined 2Fo−Fc map (1σ). (C) Equivalent cysteine residues are not modified in the structure of T2C-inactivated SmPutA determined from a crystal harvested on day 14 (2Fo−Fc map, 1σ). (D) Depiction of the disorder in helix 7 caused by the modification of Cys470. The native (PDB 5KF6) and T2C-modified enzymes are colored yellow and gray, respectively. THFA in the unmodified structures is colored cyan.
The modification at Cys470 apparently causes helix α7 of the (βα)8 barrel to unfold (Figure 4D). The electron density for the residues of α7 (residues 543−466) was very weak, implying conformational disorder. Consequently, residues 543−466 were omitted from the model deposited in the PDB (6UFP). Superposition of the native and the T2C inactivated structures shows that modified Cys470 overlaps with Tyr457 of α7 (Figure 4D). Thus, the covalent modification of Cys470 necessitates a protein conformational change to avoid steric clash, and apparently, this conformational change involves the unfolding of α7.
Kinetic Data Showing the PutA PRODH Domain Catalyzes the Oxidation of T2C.
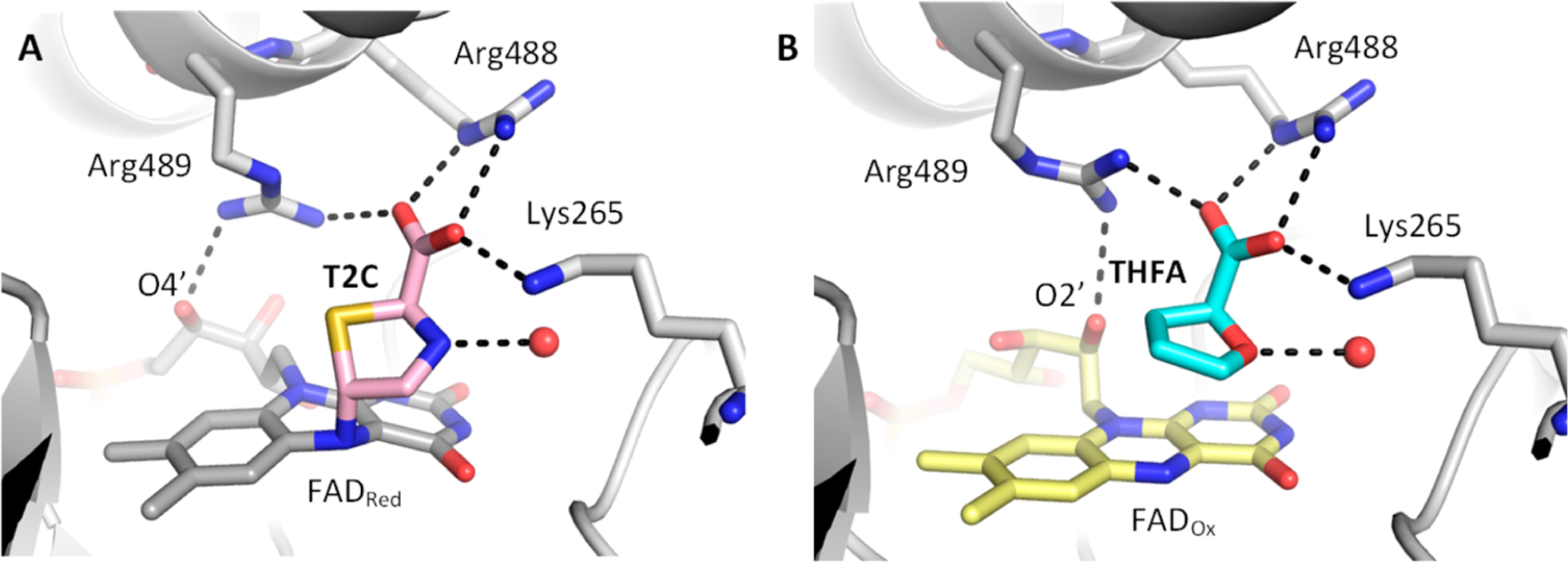
The reaction of T2C with SmPutA was explored by UV−visible stopped-flow kinetics under single turnover conditions in the absence of an electron acceptor. Figure 5 shows the absorbance traces after rapid mixing of SmPutA with l-proline (Figure 5A) and D,L-T2C (Figure 5B). The spectrum of the oxidized enzyme shows the typical features of an oxidized flavoenzyme with peaks at 379 and 452 nm. A rapid decrease in absorbance at 452 nm is observed with proline and T2C (Figure 5C), which indicates the reduction of the FAD. Single-exponential fits of the data revealed observed rate constants of 0.34 and 1.1 s−1 for reduction of the FAD by proline and T2C, respectively. Because NAD+ was included in the SmPutA solution, we were able to determine whether the PRODH reaction generated an aldehyde substrate for the GSALDH active site, which would be indicated by an increase in absorbance at 340 nm. In the reaction with proline, the absorbance at 340 nm increased with an observed rate constant of 0.07 s−1 (Figure 5C), which is consistent with the generation of P5C/GSAL (Scheme 1) and substrate channeling to the GSALDH site, as shown previously for SmPutA.18,19 In contrast, no increase in absorbance at 340 nm was observed with T2C, indicating that the oxidized product of T2C is not a substrate for the GSALDH domain. Incubating SmPutA with T2C for longer times (e.g., 30 min) still did not show evidence of NADH production. The lack of increase at 340 nm suggests that, unlike proline, the oxidation of T2C does not generate an efficient substrate for the GSALDH domain.
Figure 5.
Stopped-flow kinetics of SmPutA with proline and T2C. Oxidized SmPutA (20.3 μM after mixing) was mixed with (A) 20 mM l-proline (after mixing) and (B) 40 mM D,l-T2C (after mixing) and was monitored by stopped-flow multiwavelength absorption. The spectra shown were recorded at 0.005−197 s after mixing. (C) Single wavelength traces with proline at 452 nm (blue) and 340 nm (orange) and with T2C at 452 nm (red) and 340 nm (gray). Observed rate constants (kobs) for the reduction of the FAD by proline and T2C, and formation of NADH in the reaction with proline, were estimated by fitting the traces to a single exponential equation. No formation of NADH was observed in the reaction of SmPutA with T2C.
Kinetic Evidence for Enzyme Inactivation by T2C.
Inactivation in solution was studied by measuring the PRODH activity remaining after preincubating the enzyme with 2 mM T2C for various periods of time and removing the excess T2C by spin filtration. The residual activity decreases with preincubation time, consistent with mechanism-based inactivation (Figure 6). Fitting of the data to an exponential decay function yielded an apparent pseudo-first order rate constant for enzyme inactivation of 0.44 ± 0.08 days−1. The slow kinetics of inactivation are consistent with the long time scale required for crystallization.
Figure 6.

Kinetics of inactivation of SmPutA by T2C. The points represent the PRODH activity measured after preincubating SmPutA with 2 mM T2C for various time periods and removing the excess T2C. The error bars represent the standard deviation of three technical replicates. The curve represents the fit to an exponential decay function, which yields an apparent pseudo-first order rate constant for enzyme inactivation of 0.44 ± 0.08 days−1.
Mechanism of Covalent Modification.
Here, we showed that T2C covalently modifies the FAD of PRODH, locking the enzyme into an inactive, reduced state. The literature on T2C interacting with enzymes is rather limited. Early research on D-amino acid oxidase pursued the idea that T2C might be the physiological substrate of the enzyme.20,21 Fitzpatrick and Massey showed that the FAD in D-amino acid oxidase is reduced very rapidly by D-T2C but only slowly by l-T2C.21 Interestingly, they discovered that the enzymatic activity observed with the l-enantiomer was due to the rapid racemization of l-T2C, which occurred with a half-time of 3.3 min under the conditions used (pH 8.5, 20 °C).
T2C covalently modified our enzyme in two locations:surface cysteines and the FAD. We suggest that these modifications result from two different enzyme-mediated pathways that involve reduction of the FAD: one beginning with oxidation of the C4 atom of T2C leading to cysteine modification (Scheme 3A) and another beginning with oxidation of the C5 atom T2C, resulting in enzyme inactivation (Scheme 3B).
Scheme 3. Proposed Pathways for the Oxidation of T2C by the PRODH Active Site of PutAa.
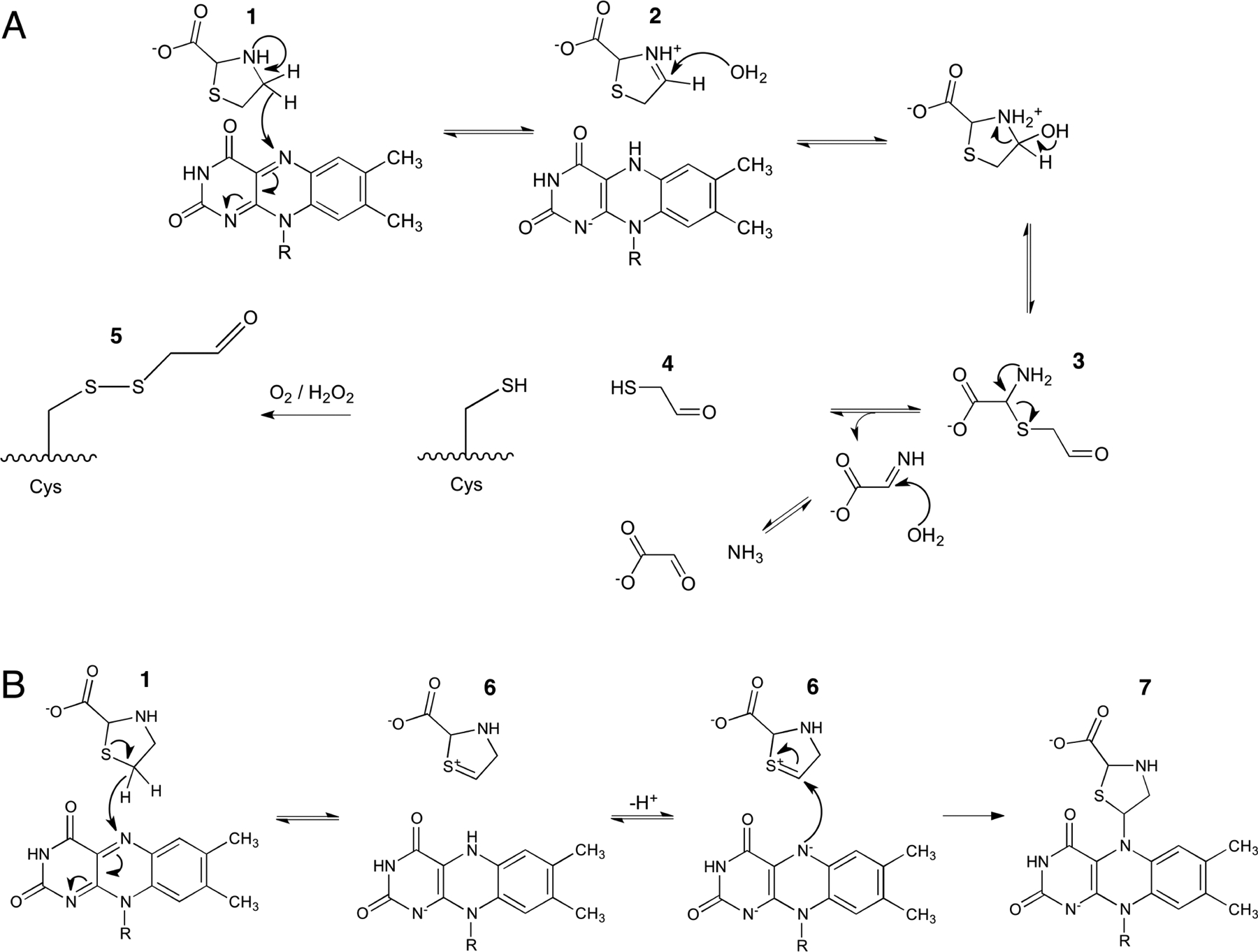
a(A) PRODH-catalyzed oxidation of T2C at the C4 atom, which is analogous to the oxidation of proline to P5C. Species 4 is proposed to modify surface cysteine residues of PutA to produce 5 (see Figure 4). (B) PRODH-catalyzed oxidation of T2C at the C5 atom, resulting in the covalent modification of the FAD and inactivation of the PRODH site (see Figure 1).
We propose that the mechanism of cysteine modification resembles the oxidation of the natural substrate l-proline. The modified cysteines were modeled as S,S-(2-hydroxyethyl)thiocysteine. This modification in protein crystal structures can result from the reaction of 2-mercaptoethanol with cysteine residues (e.g., PDB 1EVF22). Because 2-mercaptoethanol was not present in either the protein stock solution or crystallization reservoir, we entertained the possibility that the modifying compound was derived from T2C. The enzymecatalyzed oxidation of T2C at the C4 atom would produce a cyclic imine analogous to P5C (2 in Scheme 3A) and the reduced FAD. This step is consistent with the rapid reduction of the FAD observed in the stopped-flow experiment with T2C. Hydrolysis of 2 produces an open chain aldehyde analogous to GSAL (3 in Scheme 3A). Finally, the decomposition of 3 is expected to generate a thiol compound (4) that could react with cysteine to produce the electron density features observed in the crystal structure. Since S,S-(2hydroxyethyl)thiocysteine and 5 differ by only a hydrogen atom, they are indistinguishable in electron density maps at 1.74 Å resolution.
A different mechanism can explain the covalent modification of the FAD. This mechanism begins with oxidation of the C5 atom of T2C, producing 6 and the 2-electron reduced FAD (Scheme 3B). The sulfur-stabilized carbocation (6) arising from oxidation at C5 may be more reactive than the iminium ion resulting from oxidation at C4 (2).23 Nucleophilic attack by the N5 of the reduced FAD at the C5 of 6 results in the covalently modified FAD observed in the crystal (7).
We looked for spectroscopic evidence of the T2C-modifed FAD using single-turnover stopped-flow spectroscopy. Previous studies of NPPG-inactivation of PutA showed that modification of the N5 of FAD results in a decrease at 451 nm and an increase at 378 nm.13 The decrease at 451 nm indicates reduction of the FAD, whereas the increase at 378 nm is due to modification of the FAD at the N5 atom. With T2C, we observed a rapid decrease at 451 nm but not a corresponding increase at 378 nm. This result suggests that the proposed mechanism-based inactivation of the enzyme by T2C (Scheme 3B) is very slow compared to reduction of FAD by T2C, and thus, the modified FAD did not accumulate significantly in the time frame of the stopped-flow experiment. Overall, a high ratio of T2C turnover to enzyme inactivation can explain the kinetic and crystallographic data in which the turnover product of T2C, 2-mercaptoethanal (4), modifies surface cysteines and oxidation at C5 generates a sulfur-stabilized carbocation (6) that inactivates the enzyme by covalent modification of the flavin cofactor.
In summary, T2C unexpectedly inactivates the PRODH site of PutA by covalent modification of the FAD at the N5 atom. To our knowledge, this is the first report of T2C covalently modifying a flavoenzyme. Although the kinetics of inactivation appear to be slow, our results suggest a potential new strategy of developing thiazolidine-based compounds as chemical probes to study the role of PRODH in cancer metabolism.
METHODS
Protein Production and Crystallization.
SmPutA was expressed and purified as described previously.18 In brief, the protein was expressed in BL21(DE3) pLysS cells overnight at 18 °C. The protein was purified over a Ni-NTA column and eluted with 500 mM imidazole. The N-terminal His-tag was removed by tobacco etch virus protease. His-cleaved SmPutA was run over the Ni-NTA resin to isolate the tag-free SmPutA. SmPutA was further purified with a Superdex-200 16/600 size exclusion column in a buffer containing 50 mM Tris (pH 8.0), 50 mM NaCl, 5% (w/v) glycerol, and 0.5 mM Tris(2-caboxyethyl)phosphine.
Crystals were grown in sitting drops at 13 °C using a reservoir solution of 20% PEG-3350, 0.2 M ammonium sulfate, 0.1 M magnesium chloride, 0.1 M HEPES (pH 8.0), and 0.1 M sodium formate. We note this recipe is similar to that used previously for native SmPutA.18 Prior to crystallization, the protein (8 mg mL−1) was supplemented with 50 mM D,l-T2C and 10 mM NAD+ (final concentrations). The addition of D,l-T2C immediately bleached the yellow color of the PutA solution, indicating reduction of the FAD. Colorless crystals appeared after 10 days. This result was atypical, because SmPutA crystals grown in the absence of T2C appear in 1−3 days and are yellow in color. Crystals were harvested on day 14 and day 30, cryoprotected in the reservoir solution supplemented with 15% PEG 200, and immediately flash-cooled in liquid nitrogen. No additional T2C was added during cryoprotection. The crystals remained colorless during cryoprotection.
X-ray Diffraction Data Collection and Refinement.
X-ray diffraction data were collected in shutterless mode at ALS beamline 4.2.2 (crystal harvested on day 14) and APS beamline 24-ID-C (crystal harvested on day 30). The data sets were integrated and scaled with XDS.24 Intensities were converted to amplitudes with AIMLESS.25 The space group is P21 with the unit cell dimensions listed in Table 1. The asymmetric unit contains one SmPutA dimer. We note this is the same crystal form that we used previously to determine the structure of oxidized SmPutA complexed with THFA and NAD+.18 Data processing statistics are listed in Table 1.
Table 1.
Data Collection and Refinement Statistics
| harvest timea | 30 days | 14 days |
|---|---|---|
| space group | P21 | P21 |
| unit cell parameters (Å, deg) | a = 101.4, b = 102.1, c = 126.7, = 106.5 | a = 101.0, b = 102.1, c = 126.4, = 106.3 |
| wavelength (Å) | 0.9791 | 1.0000 |
| resolution (Å)b | 121.5–1.74 (1.77–1.74) | 48.4–1.52 (1.55–1.52) |
| observations | 939438 | 1321174 |
| unique refls. | 251763 | 373487 |
| Rmerge (I)b | 0.042 (1.216) | 0.077 (1.377) |
| Rmeas (I)b | 0.049 (1.431) | 0.091 (1.663) |
| Rpim (I)b | 0.025 (0.742) | 0.048 (0.916) |
| mean I/σb | 15.1 (1.0) | 10.9 (0.7) |
| CC1/2b | 0.999 (0.441) | 0.998 (0.354) |
| completeness (%)b | 98.8 (92.3) | 99.1 (91.5) |
| multiplicityb | 3.7 (3.5) | 3.5 (3.1) |
| no. of protein residues | 2394 | 2434 |
| no. of atoms | ||
| protein | 17666 | 18207 |
| FAD | 106 | 106 |
| T2C | 16 | 16 |
| NAD+ | 79 | 88 |
| CME | 30 | N/A |
| water | 1219 | 2366 |
| Rcrystb | 0.168 (0.311) | 0.180 (0.339) |
| Rfreeb,c | 0.200 (0.349) | 0.207 (0.352) |
| rmsd bonds (Å) | 0.006 | 0.005 |
| rmsd angles (deg) | 0.816 | 0.846 |
| Ramachandran plotd | ||
| favored (%) | 98.14 | 98.06 |
| outliers (%) | 0.04 | 0.04 |
| clashscore (PR)d | 2.01 (99) | 2.57 (99) |
| MolProbity score (PR)d | 0.97 (100) | 1.05 (99) |
| average B (Å2) | ||
| protein | 40.3 | 23.5 |
| FDA | 34.7 | 19.5 |
| T2C | 37.2 | 20.5 |
| NAD+ | 56.3 | 20.0 |
| CME | 53.1 | N/A |
| water | 42.0 | 31.0 |
| coord. error (Å)e | 0.21 | 0.21 |
| PDB code | 6UFP | 6VZ9 |
The time between setting up the crystallization experiment and flashcooling the crystal.
Values for the outer resolution shell of data are given in parentheses.
5% test set.
From MolProbity. The percentile ranks (PR) for Clashscore and MolProbity score are given in parentheses.
Maximum likelihood-based coordinate error estimate from PHENIX.
PHENIX26 was used for refinement, and Coot27 was used for model building. The starting model for refinement was derived from the coordinates of oxidized SmPutA (PDB 5KF6). The SMILES string of l-T2C was input to ELBOW28 to generate coordinates for the ligand and a restraint file for refinement. The l-T2C molecule was fit into the density in Coot such that the C5 atom was within covalent bond distance of the FAD N5. The desired covalent bond between the C5 of l-T2C and N5 of FAD was automatically recognized by PHENIX during crystallographic refinement. The structures were validated using MolProbity29 and the wwPDB validation service.30 Refinement statistics are listed in Table 1.
Stopped-Flow Absorbance Spectroscopy.
The reactions of SmPutA with proline and D,L-T2C were performed under anaerobic conditions on a Hi-Tech Scientific SF-61DX2 stopped-flow instrument equipped with a photodiode array detector as previously described.31 Prior to stopped-flow, SmPutA was dialyzed into 50 mM potassium phosphate buffer (pH 7.4, 25 mM NaCl) and degassed with repeated cycles of vacuum and flushing with nitrogen. Solutions of l-proline, D,L-T2C, and NAD+ were made in the same phosphate buffer and similarly degassed. Degassed solutions were then brought into an anaerobic glovebox and transferred to anaerobic syringes under a nitrogen atmosphere. NAD+ was added to the SmPutA solution at a final concentration of 0.4 mM, which, after mixing, was 0.2 mM. As a safeguard to maintain anaerobic conditions, protocatechuate dioxygenase (0.05 U/mL) and protocatechuic acid (100 μM) were added to the SmPutA and substrate solutions just prior to conducting the experiments. The reactions were performed at room temperature (RT) (23 °C). Single wavelength traces of the absorbance decrease at 452 nm (proline and T2C reactions) and absorbance increase at 340 nm (proline reaction) were fit to a single exponential equation using SigmaPlot 12 to obtain an estimate of the observed rate constant (kobs).
Kinetics of Inactivation.
The kinetics of inactivation of the PRODH activity of SmPutA by T2C were studied by measuring the PRODH activity remaining after preincubation with T2C for various periods of time, as follows. Identical samples of SmPutA at 3 mg mL−1 (22.8 μM) in a buffer of 25 mM Tris, 25 mM NaCl, 50 mM magnesium chloride, 50 mM sodium formate, 0.1 M ammonium sulfate, 50 mM HEPES (pH 8.0), and 2.5% (w/v) PEG 3350 were stored at 20 °C for a period of 6 days. Each day, T2C was added to one of the samples to a final concentration of 2 mM. After 6 days, the excess T2C was removed from each sample by spin ultrafiltration using Amicon Ultra-0.5 mL centrifugal filter units in preparation for PRODH assays. Prior to the enzyme assays, the enzyme concentration was measured and used to adjust the kinetic data. The PRODH assays were performed in triplicate in 96-well plates at 20 °C in an Epoch 2 plate reader (BioTek, Winooski, VT). The assays contained the substrate L-proline (20 mM), the electron acceptor menadione (5 μM), and the P5C-trapping agent o-aminobenzaldehyde (4 mM) in a buffer containing 20 mM MOPS (pH 7.5) and 10 mM magnesium chloride. The initial rate was obtained by monitoring production of the P5C−o-aminobenzaldehyde adduct by absorbance at 443 nm. The percent activity remaining as a function of preincubation time was fit to an exponential decay function with Origin 2020.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Schuermann and J. Nix for help with the remote X-ray diffraction data collection at beamlines 24-ID-C and 4.2.2, respectively. This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P30 GM124165). The Pilatus 6M detector on 24-IDC beamline is funded by a NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE- AC02-06CH11357. Beamline 4.2.2 of the Advanced Light Source, a DOE Office of Science User Facility under Contract No. DE-AC02-05CH11231, is supported in part by the ALS- ENABLE program funded by the National Institutes of Health, National Institute of General Medical Sciences, grant P30 GM124169-01.
Funding
This research was supported by NIGMS, National Institutes of Health, Grants R01GM065546 and R01GM061068. A.C.C. was funded in part by a Fulbright Science & Innovation Graduate Award.
ABBREVIATIONS
- ALDH
aldehyde dehydrogenase
- GSAL
l-glutamate-γ-semialdehyde
- GSALDH
l-glutamate-γ-semialdehyde dehydrogenase
- NPPG
N-propargylglycine
- PRODH
proline dehydrogenase
- P5C
Δ1-pyrroline-5-carboxylate
- PutA
proline utilization A
- SmPutA
proline utilization A from Sinorhizobium meliloti
- T2C
thiazolidine-2-carboxylate
- THFA
l-tetrahydro-2-furoic acid
Footnotes
Complete contact information is available at: https://pubs.acs.org/10.1021/acschembio.9b00935
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acschembio.9b00935.
Figures of electron density for modified FAD(PDF)
Accession Codes
Coordinates and structure factor amplitudes have been deposited in the Protein Data Bank under accession codes 6UFP and 6VZ9.
The authors declare no competing financial interest.
Contributor Information
Ashley C. Campbell, Department of Biochemistry, University of Missouri, Columbia, Missouri 65211, United States;.
Donald F. Becker, Department of Biochemistry, Redox Biology Center, University of Nebraska, Lincoln, Nebraska 68588, United States;.
Kent S. Gates, Department of Chemistry and Department of Biochemistry, University of Missouri, Columbia, Missouri 65211, United States;.
John J. Tanner, Department of Biochemistry and Department of Chemistry, University of Missouri, Columbia, Missouri 65211, United States;.
REFERENCES
- (1).Tanner JJ (2019) Structural Biology of Proline Catabolic Enzymes. Antioxid. Redox Signaling 30, 650–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Liu LK, Becker DF, and Tanner JJ (2017) Structure, function, and mechanism of proline utilization A (PutA). Arch. Biochem. Biophys 632, 142–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Polyak K, Xia Y, Zweier JL, Kinzler KW, and Vogelstein B (1997) A model for p53-induced apoptosis. Nature 389, 300–305. [DOI] [PubMed] [Google Scholar]
- (4).Elia I, Broekaert D, Christen S, Boon R, Radaelli E, Orth MF, Verfaillie C, Grunewald TGP, and Fendt SM (2017) Proline metabolism supports metastasis formation and could be inhibited to selectively target metastasizing cancer cells. Nat. Commun 8, 15267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Pors K, and Moreb JS (2014) Aldehyde dehydrogenases in cancer: an opportunity for biomarker and drug development? Drug Discovery Today 19, 1953–1963. [DOI] [PubMed] [Google Scholar]
- (6).Nilsson R, Jain M, Madhusudhan N, Sheppard NG, Strittmatter L, Kampf C, Huang J, Asplund A, and Mootha VK (2014) Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat. Commun 5, 3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Raimondi I, Ciribilli Y, Monti P, Bisio A, Pollegioni L, Fronza G, Inga A, and Campomenosi P (2013) P53 family members modulate the expression of PRODH, but not PRODH2, via intronic p53 response elements. PLoS One 8, No. e69152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Yoon KA, Nakamura Y, and Arakawa H (2004) Identification of ALDH4 as a p53-inducible gene and its protective role in cellular stresses. J. Hum. Genet 49, 134–140. [DOI] [PubMed] [Google Scholar]
- (9).Kardos GR, Wastyk HC, and Robertson GP (2015) Disruption of Proline Synthesis in Melanoma Inhibits Protein Production Mediated by the GCN2 Pathway. Mol. Cancer Res 13, 1408–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Tanner JJ, Fendt SM, and Becker DF (2018) The Proline Cycle As a Potential Cancer Therapy Target. Biochemistry 57, 3433–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Scott GK, Yau C, Becker BC, Khateeb S, Mahoney S, Jensen MB, Hann B, Cowen BJ, Pegan SD, and Benz CC (2019) Targeting Mitochondrial Proline Dehydrogenase with a Suicide Inhibitor to Exploit Synthetic Lethal Interactions with p53 Upregulation and Glutaminase Inhibition. Mol. Cancer Ther 18, 1374–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Zhang W, Zhang M, Zhu W, Zhou Y, Wanduragala S, Rewinkel D, Tanner JJ, and Becker DF (2007) Redox-induced changes in flavin structure and roles of flavin N(5) and the ribityl 2’OH group in regulating PutA-membrane binding. Biochemistry 46, 483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Srivastava D, Zhu W, Johnson WH Jr., Whitman CP, Becker DF, and Tanner JJ (2010) The structure of the proline utilization a proline dehydrogenase domain inactivated by N propargylglycine provides insight into conformational changes induced by substrate binding and flavin reduction. Biochemistry 49, 560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Singh H, Arentson BW, Becker DF, and Tanner JJ (2014) Structures of the PutA peripheral membrane flavoenzyme reveal a dynamic substrate-channeling tunnel and the quinone binding site. Proc. Natl. Acad. Sci. U. S. A 111, 3389–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Korasick DA, Pemberton TA, Arentson BW, Becker DF, and Tanner JJ (2018) Structural Basis for the Substrate Inhibition of Proline Utilization A by Proline. Molecules 23, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Korasick DA, Singh H, Pemberton TA, Luo M, Dhatwalia R, and Tanner JJ (2017) Biophysical investigation of type A PutAs reveals a conserved core oligomeric structure. FEBS J 284, 3029–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).White TA, Johnson WH Jr., Whitman CP, and Tanner JJ (2008) Structural basis for the inactivation of Thermus thermophilus proline dehydrogenase by N-propargylglycine. Biochemistry 47, 5573–5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Luo M, Gamage TT, Arentson BW, Schlasner KN, Becker DF, and Tanner JJ (2016) Structures of Proline Utilization A (PutA) Reveal the Fold and Functions of the Aldehyde Dehydrogenase Superfamily Domain of Unknown Function. J. Biol. Chem 291, 24065–24075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Moxley MA, Sanyal N, Krishnan N, Tanner JJ, and Becker DF (2014) Evidence for Hysteretic Substrate Channeling in the Proline Dehydrogenase and Delta1-Pyrroline-5-carboxylate Dehydrogenase Coupled Reaction of Proline Utilization A (PutA). J. Biol. Chem 289, 3639–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Hamilton GA, Buckthal DJ, Mortensen RM, and Zerby KW (1979) Reactions of cysteamine and other amine metabolites with glyoxylate and oxygen catalyzed by mammalian D-amino acid oxidase. Proc. Natl. Acad. Sci. U. S. A 76, 2625–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Fitzpatrick PF, and Massey V (1982) Thiazolidine-2 carboxylic acid, an adduct of cysteamine and glyoxylate, as a substrate for D-amino acid oxidase. J. Biol. Chem 257, 1166–1171. [PubMed] [Google Scholar]
- (22).Phan J, Mahdavian E, Nivens MC, Minor W, Berger S, Spencer HT, Dunlap RB, and Lebioda L (2000) Catalytic cysteine of thymidylate synthase is activated upon substrate binding. Biochemistry 39, 6969–6978. [DOI] [PubMed] [Google Scholar]
- (23).Madern JM, Hansen T, van Rijssel ER, Kistemaker HAV, van der Vorm S, Overkleeft HS, van der Marel GA, Filippov DV, and Codee JDC (2019) Synthesis, Reactivity, and Stereoselectivity of 4-Thiofuranosides. J. Org. Chem 84, 1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Kabsch W (2010) XDS. Acta Crystallogr., Sect. D: Biol. Crystallogr 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Evans PR, and Murshudov GN (2013) How good are my data and what is the resolution? Acta Crystallogr., Sect. D: Biol. Crystallogr 69, 1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, and Adams PD (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr., Sect. D: Biol. Crystallogr 68, 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010) Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Moriarty NW, Grosse-Kunstleve RW, and Adams PD (2009) electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr., Sect. D: Biol. Crystallogr 65, 1074–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr D66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Gore S, Sanz Garcia E, Hendrickx PMS, Gutmanas A, Westbrook JD, Yang H, Feng Z, Baskaran K, Berrisford JM, Hudson BP, Ikegawa Y, Kobayashi N, Lawson CL, Mading S, Mak L, Mukhopadhyay A, Oldfield TJ, Patwardhan A, Peisach E, Sahni G, Sekharan MR, Sen S, Shao C, Smart OS, Ulrich EL, Yamashita R, Quesada M, Young JY, Nakamura H, Markley JL, Berman HM, Burley SK, Velankar S, and Kleywegt GJ (2017) Validation of Structures in the Protein Data Bank. Structure 25, 1916–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Moxley MA, and Becker DF (2012) Rapid reaction kinetics of proline dehydrogenase in the multifunctional proline utilization A protein. Biochemistry 51, 511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


