Summary:
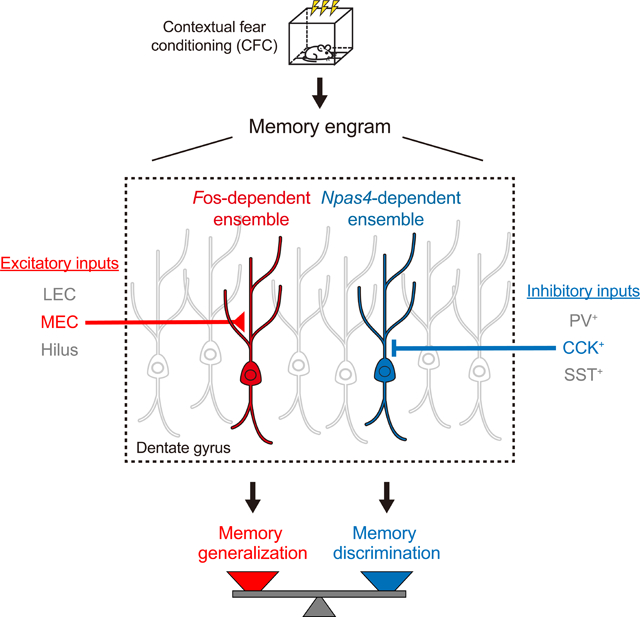
Memories are believed to be encoded by sparse ensembles of neurons in the brain. However, it remains unclear whether there is functional heterogeneity within individual memory engrams, i.e., if separate neuronal subpopulations encode distinct aspects of the memory and drive memory expression differently. Here we show that contextual fear memory engrams in the mouse dentate gyrus contain functionally distinct neuronal ensembles, genetically defined by the Fos- or Npas4-dependent transcriptional pathways. The Fos-dependent ensemble promotes memory generalization and receives enhanced excitatory synaptic inputs from the medial entorhinal cortex, which we find itself also mediates generalization. The Npas4-dependent ensemble promotes memory discrimination and receives enhanced inhibitory drive from local cholecystokinin-expressing interneurons, the activity of which is required for discrimination. Our study provides causal evidence for functional heterogeneity within the memory engram, and reveals synaptic and circuit mechanisms used by each ensemble to regulate the memory discrimination-generalization balance.
Keywords: memory engram, heterogeneity, neuronal ensembles, activity-dependent pathways, dentate gyrus, discrimination, generalization
Graphical Abstract
eTOC blurb
Two functionally distinct neuronal ensembles within a single memory engram undergo different learning-induced synaptic modifications and drive memory-guided behaviors in opposite directions.
Introduction
Engrams of individual memories are the long-lasting biological changes that take place in the brain to encode specific experiences (Josselyn et al., 2015; Tonegawa et al., 2015). Each engram is thought to contain a sparse population of neurons that are activated by the specific learning experience, undergo long-lasting synaptic modifications (Choi et al., 2018; Ryan et al., 2015), and mediate the expression of the encoded memory (Cai et al., 2016; Han et al., 2009; Liu et al., 2012; Yokose et al., 2017). However, a fundamental question remains: whether neurons within a single memory engram are functionally homogeneous, as generally assumed, or heterogeneous, as hypothesized to allow different aspects of a memory to be individually represented and regulated (Ghandour et al., 2019; Mallory and Giocomo, 2018; Richter et al., 2016; Tanaka and McHugh, 2018). The latter would favor flexible memory expression in an ever-changing environment (Eichenbaum, 2004; Xu and Sudhof, 2013). Indeed, heterogeneous population activities have been observed during learning (Grewe et al., 2017; Grosmark and Buzsáki, 2016; Herry et al., 2008; Tanaka et al., 2018) and are theorized to be critical for efficient information coding (Chelaru and Dragoi, 2008; Osborne et al., 2008). Nevertheless, causal evidence for functional heterogeneity within a single engram is still lacking. It is also unknown whether functionally distinct neuronal ensembles can be distinguished within an engram at the molecular and cellular levels, and if they engage different synaptic and circuit mechanisms to modulate memory-guided adaptive behaviors.
Genetically encoded activity reporters based on immediate early genes (IEGs) such as Fos, and Arc (Barth, 2007; Kawashima et al., 2014), commonly considered proxies of neuronal activity (Guzowski et al., 2005), have been used to identify neuronal ensembles in engrams. Until now, most studies have focused on ensembles defined by a single activity-dependent pathway (Barth et al., 2004; Denny et al., 2014; Kawashima et al., 2013; Reijmers et al., 2007). However, activity-dependent pathways are known to be highly diverse: they respond differently to external stimuli and mediate distinct cellular and synaptic processes (Yap and Greenberg, 2018). The relationship between ensembles defined by different pathways has not been investigated. It is conceivable that different activity-dependent pathways may define functionally distinct ensembles within engrams, providing the means to genetically dissect them.
Here we directly examine the functionalities of neuronal ensembles defined by two different activity-dependent pathways, downstream of the IEGs Fos and Npas4, both of which are highly induced by learning but likely trigger distinct synaptic changes in activated neurons (Flavell and Greenberg, 2008; Sun and Lin, 2016). The widely used activity marker Fos mediates long-term potentiation of excitatory synapses (Fleischmann et al., 2003). Npas4, a neuron-specific IEG that is selectively activated by neuronal activity, differs from Fos in that it preferentially recruits inhibitory synapses, or sometimes diminishes excitatory synapses, onto excitatory neurons (Lin et al., 2008; Weng et al., 2018). These different synaptic functions of Fos and Npas4 strongly suggest that the ensembles they define may be functionally distinct.
To investigate the functionalities of the Fos- and Npas4-dependent ensembles, we focus on the dentate gyrus (DG) of the hippocampus and the roles of these ensembles in memory discrimination and generalization, opposed processes that must be balanced appropriately to differentiate between different stimuli (Frankland et al., 1998; Grosso et al., 2018; van Dijk and Fenton, 2018) but also allow recognition of shared features (Dunsmoor and Paz, 2015; Wiltgen and Silva, 2007). The discrimination-generalization balance is critical to adaptive learning and avoiding maladaptive conditions such as post-traumatic stress disorder (PTSD) (Mahan and Ressler, 2012) and panic disorder (Kheirbek et al., 2012). The DG has traditionally been believed to mediate memory discrimination (Leutgeb et al., 2007; McHugh et al., 2007; Treves and Rolls, 1994), but recent studies suggest that this region can also promote memory generalization (Hainmueller and Bartos, 2018; Nakashiba et al., 2012). Very little is known about the involvement of neuronal ensembles in the DG in these processes. Here we show that the Fos-and Npas4-dependent neuronal ensembles within the DG contextual fear memory engram regulate memory generalization and discrimination, respectively, by engaging distinct synaptic and circuit mechanisms.
Results
Fos- and Npas4-dependent RAM (F-RAM and N-RAM) Reporters
To identify Fos- and Npas4-dependent ensembles within the engram, we used our RAM reporter system (Sørensen et al., 2016) to create Fos-dependent (F-RAM) and Npas4-dependent (N-RAM) RAM reporters. FOS and NPAS4 are transcription factors and exert their functions via their downstream transcriptional targets, so we designed reporters that reflect these transcriptional outputs. The promoters in F-RAM and N-RAM (PF-RAM and PN-RAM) consist of the consensus DNA binding sequences for FOS and NPAS4 (Figure S1A). Both promoters drive the expression of reporter genes under the temporal control of a modified doxycycline (Dox)-dependent Tet-Off system developed for the RAM system (Figure 1A).
Figure 1. The F-RAM and N-RAM Reporters Selectively Capture Fos- and Npas4-dependent Neuronal Ensembles.
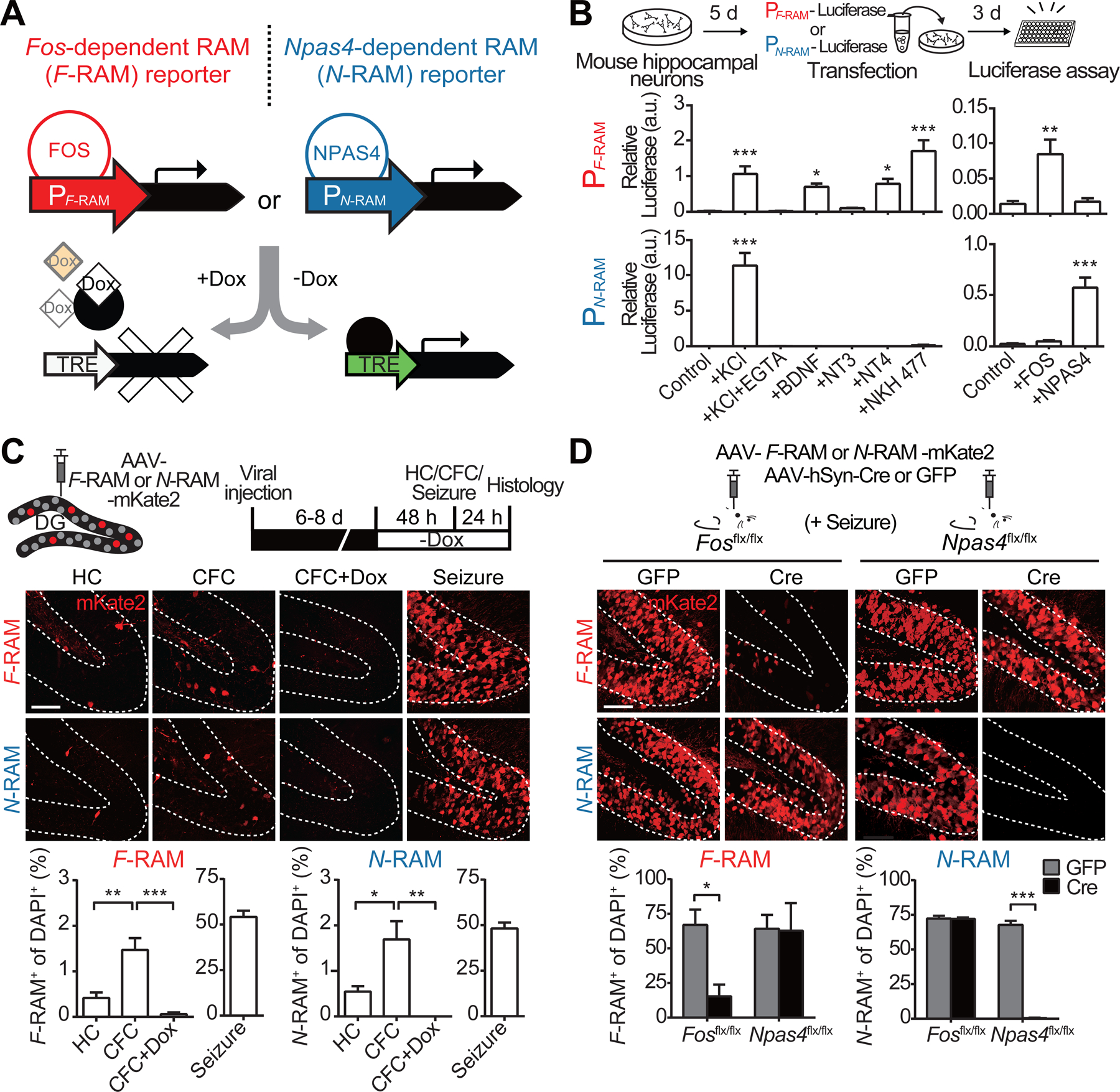
(A) Design of the F-RAM and N-RAM reporters.
(B) Characterization of PF-RAM and PN-RAM. Cultured hippocampal neurons were transfected with luciferase expression plasmids and treated with various extracellular stimuli. The induction of PF-RAM or PN-RAM by various extracellular stimuli was measured by luciferase assay. One-way ANOVA, Dunnett’s test, n = 8–9.
(C) F-RAM and N-RAM label experience-activated neuronal ensembles in vivo. Representative images of the dorsal DG granule cell layers (GCL, dashed lines) and quantifications showing ensembles labeled under the home cage (HC), contextual fear conditioning (CFC), CFC but on Dox (CFC+Dox), and seizure conditions. One-way ANOVA, Tukey’s test, n = 4 per condition.
(D) F-RAM and N-RAM are dependent on endogenous Fos and Npas4, respectively. Representative images and quantifications showing seizure-induced reporter activation in Fos and Npas4 conditional knockout animals. Two-way ANOVA, Sidak’s test, n = 3–4.
Both PF-RAM and PN-RAM were robustly activated by membrane depolarization in cultured mouse hippocampal neurons (Figure 1B; numerical data and statistical information for all figures are included in Table S1), suggesting that they are regulated by neuronal activity. PF-RAM was activated by various extracellular stimuli known to activate Fos, and PN-RAM was selectively activated by neuronal activity, recapitulating this unique property of Npas4 (Lin et al., 2008). Overexpression of Fos, but not Npas4, drives PF-RAM activity, whereas overexpression of Npas4, but not Fos, activates PN-RAM (Figure 1B), indicating that activation of PF-RAM and PN-RAM is selective. Both the F-RAM and N-RAM reporters showed low background activation and were robustly induced by neuronal activity in a Dox-dependent manner (Figure S1D).
We used AAV-F-RAM-mKate2 or AAV-N-RAM-mKate2 to label active neuronal ensembles in vivo, in mouse dorsal DG following contextual fear conditioning (CFC). Labeled cells were mostly located in the granule cell layer (Figure 1C), suggesting that they are mostly dentate granule cells (DGCs). Both reporters showed low activation in the home cage (HC) condition, were robustly activated by kainic acid-induced seizures, and were mostly inhibited in the presence of Dox. Consistent with previous reports (Deng et al., 2013; Lacagnina et al., 2019; Nakayama et al., 2015), on average 1–2% of the DGCs were labeled after CFC (Figure 1C). Both reporters also labeled small populations of cells, mostly mossy cells, in the dentate hilus regions. The number of labeled hilar cells was similar in the HC and CFC conditions, suggesting that contextual fear learning didn’t significantly activate these reporters in the hilus (Figures S1E–S1I).
We next verified that activation of F-RAM and N-RAM in vivo specifically required Fos and Npas4, respectively. In Fos and Npas4 conditional knockout mice (Fosflx/flx and Npas4flx/flx), we deleted the Fos and Npas4 gene, respectively, in DG using AAVs expressing the DNA recombinase Cre (Figures S1J and S1K). Seizure-induced F-RAM activity was largely abolished in the absence of Fos, but not after removal of Npas4 (Figure 1D). The remaining activity of F-RAM may be due to the recruitment of other FOS/JUN family members such as c-JUN and FOSB, which may also bind and activate PF-RAM (Hess et al., 2004). Npas4 deletion, but not Fos deletion, eliminated N-RAM activity. Taken together, these results indicate that the F-RAM and N-RAM reporters specifically label the Fos- and Npas4-dependent active neuronal ensembles.
The F-RAM and N-RAM Reporters Define Distinct Neuronal Ensembles Within the Memory Engram
To determine whether the neuronal ensembles identified by F-RAM and N-RAM are distinct populations within the DG engram, we first examined synaptic properties of DGCs in each ensemble. Whole-cell patch-clamp recordings were carried out on F-RAM+ or N-RAM+ cells 24 hours after CFC (Figure 2A), measuring spontaneous miniature excitatory and inhibitory postsynaptic currents (mEPSCs and mIPSCs). Compared to their unlabeled (F-RAM− or N-RAM−) neighbors, F-RAM+ neurons received stronger excitatory inputs, specifically higher mEPSC frequencies (Figure 2B), while N-RAM+ neurons received stronger inhibitory inputs, specifically increased mIPSC frequencies (Figure 2C). mIPSCs of F-RAM+ neurons and mEPSCs of N-RAM+ neurons were indistinguishable from those of their unlabeled neighbors. F-RAM+ and N-RAM+ neurons were rarely seen in the inner granule cell layer and did not show the high intrinsic excitability typical of adult new-born DGCs (Kuhn et al., 1996; Mongiat et al., 2009) (Figures S2A–S2C), suggesting that they are mature granule cells. Together, these data indicate that the F-RAM and N-RAM ensembles are mostly composed of mature granule cells with distinct synaptic properties.
Figure 2. The F-RAM and N-RAM Ensembles are Distinct Neuronal Populations with Distinct Synaptic Properties.
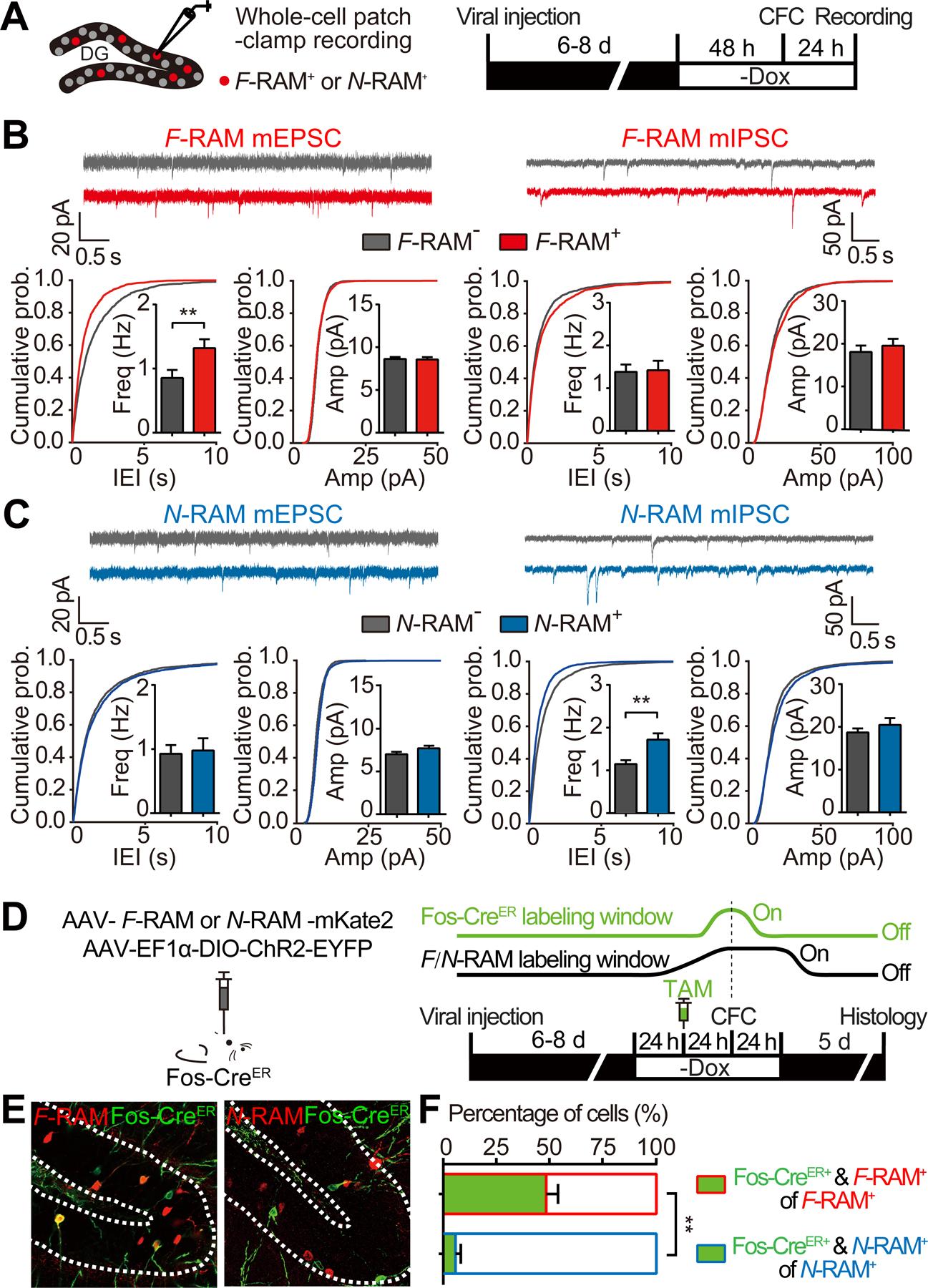
(A) Experimental scheme to measure mEPSCs and mIPSCs on labeled F-RAM+ or N-RAM+ neurons and unlabeled neighboring neurons.
(B) F-RAM+ neurons showed increased mEPSC frequency (Mann-Whitney test; F-RAM−, n = 17; F-RAM+, n = 17) but unchanged mIPSCs (n = 18, 19).
(C) N-RAM+ neurons showed unchanged mEPSCs (N-RAM−, n = 20; N-RAM+, n = 20), but increased mIPSC frequency (n = 25, 26).
(D) Experimental scheme to co-label the F-RAM or N-RAM and Fos-CreER ensembles in Fos-CreER mice. Dox was removed 48 hours before CFC to label the F-RAM or N-RAM ensemble (black line). Tamoxifen (TAM) was injected i.p. 24 hours before CFC to label the Fos-CreER ensemble (green line).
(E and F) Representative images (E) and quantification (F) showing overlap between the F-RAM or N-RAM and Fos-CreER ensembles. Mann-Whitney test, n = 5 per group.
We next measured co-localization of each ensemble and the Fos-expressing neurons that have been postulated to be engram cells (Garner et al., 2012; Tanaka et al., 2014; Tonegawa et al., 2015). We used the FosTRAP mouse (Guenthner et al., 2013), in which the endogenous Fos promoter drives the expression of CreER to label Fos-expressing cells in green via a Cre-dependent EYFP reporter (Figure 2D). The F-RAM or N-RAM ensemble was simultaneously labeled red with mKate2 (Figure 2E). The F-RAM and Fos-CreER ensembles, both of which should reflect Fos expression, showed substantial co-localization (49.902 ± 5.053%; Figure 2F); the incomplete overlap could be due to incomplete viral co-infection, different reporter temporal control (Dox-Off vs. CreER) and different activity promoters (see Discussion). In addition, DGCs labeled by either F-RAM or Fos-CreER alone, or both, all had similar electrophysiological properties: increased mEPSC frequencies compared to unlabeled neurons (Figure S2D). In contrast, the N-RAM and Fos-CreER ensembles showed little overlap (5.756 ± 2.522%; Figure 2F) and were electrophysiologically distinct: DGCs labeled by N-RAM, including the small population labeled by both N-RAM and Fos-CreER, had higher mIPSC frequencies than unlabeled DGCs and those labeled only by Fos-CreER (Figure S2E). These data further indicate that the F-RAM and N-RAM reporters likely capture distinct neuronal populations.
The F-RAM and N-RAM Ensembles Display Distinct Activity Patterns During Memory Recall
The distinct synaptic properties of the F-RAM and N-RAM ensembles suggest different roles in memory expression. We therefore investigated their functions in contextual fear memory discrimination-generalization, which is known to be mediated by the DG (Danielson et al., 2017; Guo et al., 2018; Knierim and Neunuebel, 2016; Senzai and Buzsaki, 2017). In an adopted memory discrimination-generalization assay (Huckleberry et al., 2016; Rozeske et al., 2018), mice were fear conditioned in context A, and exposed 24 hours later to either the conditioned context A, an unconditioned context B very similar to A, or a highly distinct context C (Figures 3A and S3B). Strong fear responses (freezing) were observed in the conditioned context A (Figure 3B). Clear fear responses were also observed in context B, suggesting a degree of memory generalization due to the similarity of the two contexts. Memory discrimination also occurred, however, as freezing levels in context B were significantly lower than in A. In context C mice displayed the basal level of freezing expected for a novel context, indicating strong memory discrimination and minimal generalization.
Figure 3. The F-RAM and N-RAM Ensembles Show Distinct Activity Patterns During the Contextual Fear Memory Discrimination-Generalization Assay.
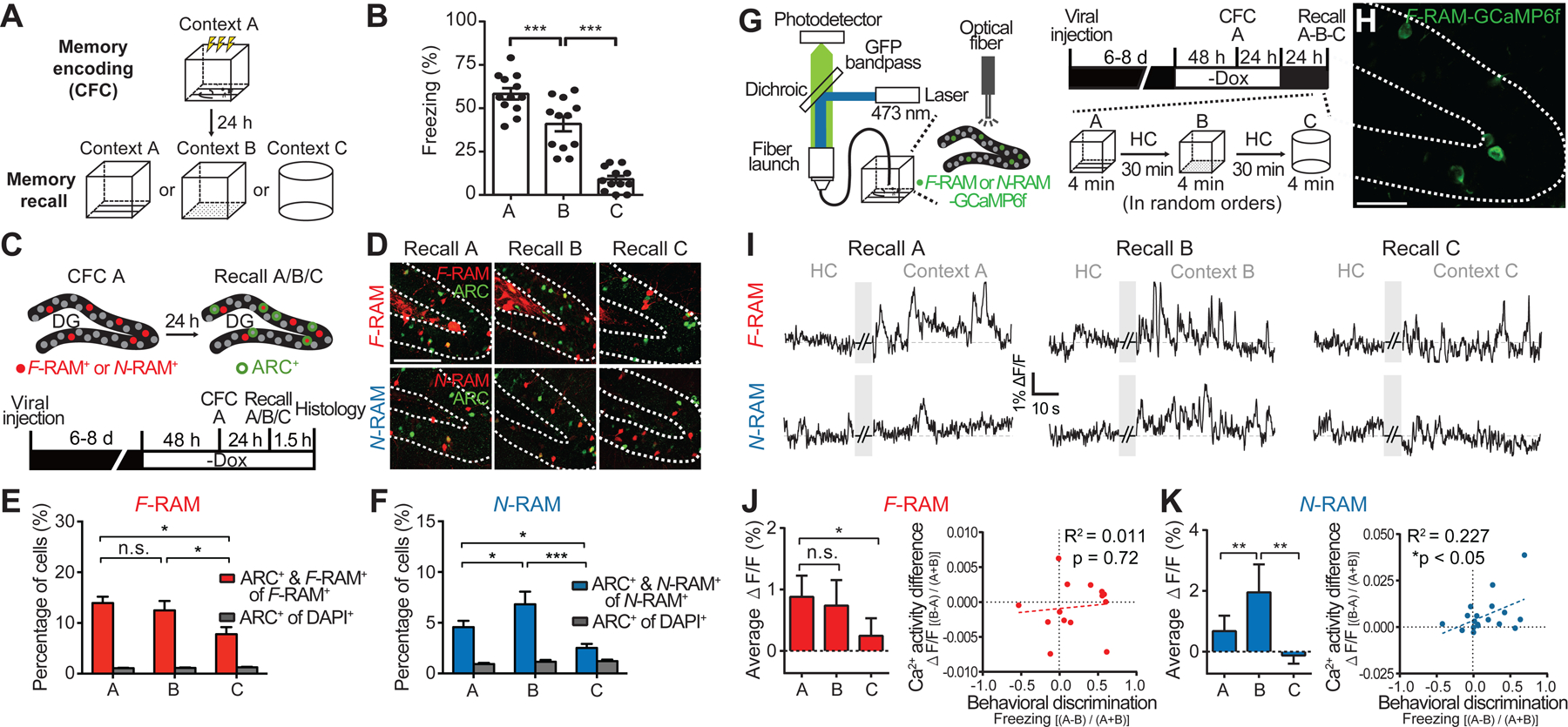
(A) Schematic of the behavioral assay.
(B) Freezing levels during memory recall in the three contexts. One-way ANOVA, Tukey’s test, n = 12 per group.
(C) Experimental scheme to identify recall-activated F-RAM and N-RAM ensemble neurons using Arc expression (ARC+).
(D) Representative images showing overlap (white arrows) of the labeled F-RAM+ or N-RAM+ neurons and ARC+ neurons.
(E and F) Percentages of the F-RAM+ (E) and N-RAM+ neurons (F) reactivated during memory recall. Two-way mixed ANOVA, Tukey’s test, n = 7–9.
(G) Schematic and timeline for examination of Ca2+ activity in ensemble neurons using fiber photometry.
(H) Representative image showing the expression of GCaMP6f in the F-RAM ensemble after CFC.
(I) Representative traces showing Ca2+ activity in F-RAM+ and N-RAM+ neurons.
(J) Left, F-RAM ensemble activity (ΔF/F) during memory recall in different contexts (Friedman’s one-way repeated measures ANOVA, Dunn’s test, n = 14). Right, correlation between animal’s ability to discriminate contexts A and B (Freezing[(A-B)/(A+B)]) and the difference in F-RAM ensemble activities in these two contexts (ΔF/F [(B-A)/(A+B)]; linear regression, n = 14).
(K) The same as (J), except for N-RAM (n = 19).
To understand the roles of the F-RAM and N-RAM ensembles, we first examined their reactivation during memory recall in the different contexts using expression of the IEG ARC (Figures 3C and 3D). The total numbers of ARC+ cells were similar in contexts A, B and C (Table S1), suggesting that all three contexts triggered similar levels of overall activity in the DG. The F-RAM ensemble showed similar rates of reactivation in contexts A and B but less in context C (Figure 3E), suggesting that the activity of F-RAM+ neurons is not sensitive to small differences between contexts and thus favors memory generalization. N-RAM+ neurons, however, were differentially reactivated, significantly less in context A than in B (Figure 3F). The greater reactivation of N-RAM+ neurons in context B was not an effect of novelty, because a lower rate of reactivation was observed in the novel context C. These results suggested that N-RAM+ neurons might be involved in mediating differential memory expression in similar contexts. Similar results were obtained using FOS instead of ARC as the activity marker (Figures S3C).
To confirm that the F-RAM and N-RAM ensembles were reactivated differently in vivo, we measured their calcium (Ca2+) activity during memory recall using fiber photometry (Figure 3G). GCaMP6f, a genetically encoded Ca2+ indicator, was expressed in F-RAM+ or N-RAM+ neurons after CFC (Figure 3H) and GCaMP6f signals were measured when animals recalled memories in the three contexts in random order. Consistent with the IEG staining, the F-RAM ensemble showed similar strong Ca2+ activity during recall in contexts A and B, as quantified by the average Ca2+ signal (ΔF/F) over the entire recall session (Figures 3I and 3J). The N-RAM ensemble showed significantly less Ca2+ activity in context A than in B (Figures 3I and 3K). Remarkably, the difference in Ca2+ activity levels between contexts A and B (ΔF/F[(B-A)/(A+B)]) of the N-RAM (but not the F-RAM) ensemble was significantly correlated with the performance of an animal in discriminating between these two contexts (Freezing[(A-B)/(A+B)], Figure 3K), indicating that the N-RAM ensemble is important for memory discrimination.
The F-RAM and N-RAM Ensembles Differentially Regulate the Memory Discrimination-Generalization Balance
The previous results suggest that the F-RAM ensemble promotes memory generalization and the N-RAM ensemble mediates memory discrimination. To test this hypothesis, we chemogenetically manipulated the activity of F-RAM+ and N-RAM+ neurons during memory recall. By expressing the inhibitory (hM4Di) or excitatory (hM3Dq) DREADDs (Designer Receptors Exclusively Activated by Designer Drugs) (Roth, 2016) in the ensemble neurons (Figures 4A and 4B), we were able to inhibit or activate their activity with the DREADD agonist clozapine N-oxide (CNO), delivered by intraperitoneal (i.p.) injection 30 minutes before memory recall (Figures 4C and S4A). In addition to freezing levels in different contexts, discrimination indices (DIs) for context A vs. B and A vs. C were measured to detect bi-directional shifts in the discrimination-generalization balance.
Figure 4. F-RAM and N-RAM Ensembles Oppositely Regulate the Memory Discrimination-Generalization Balance.
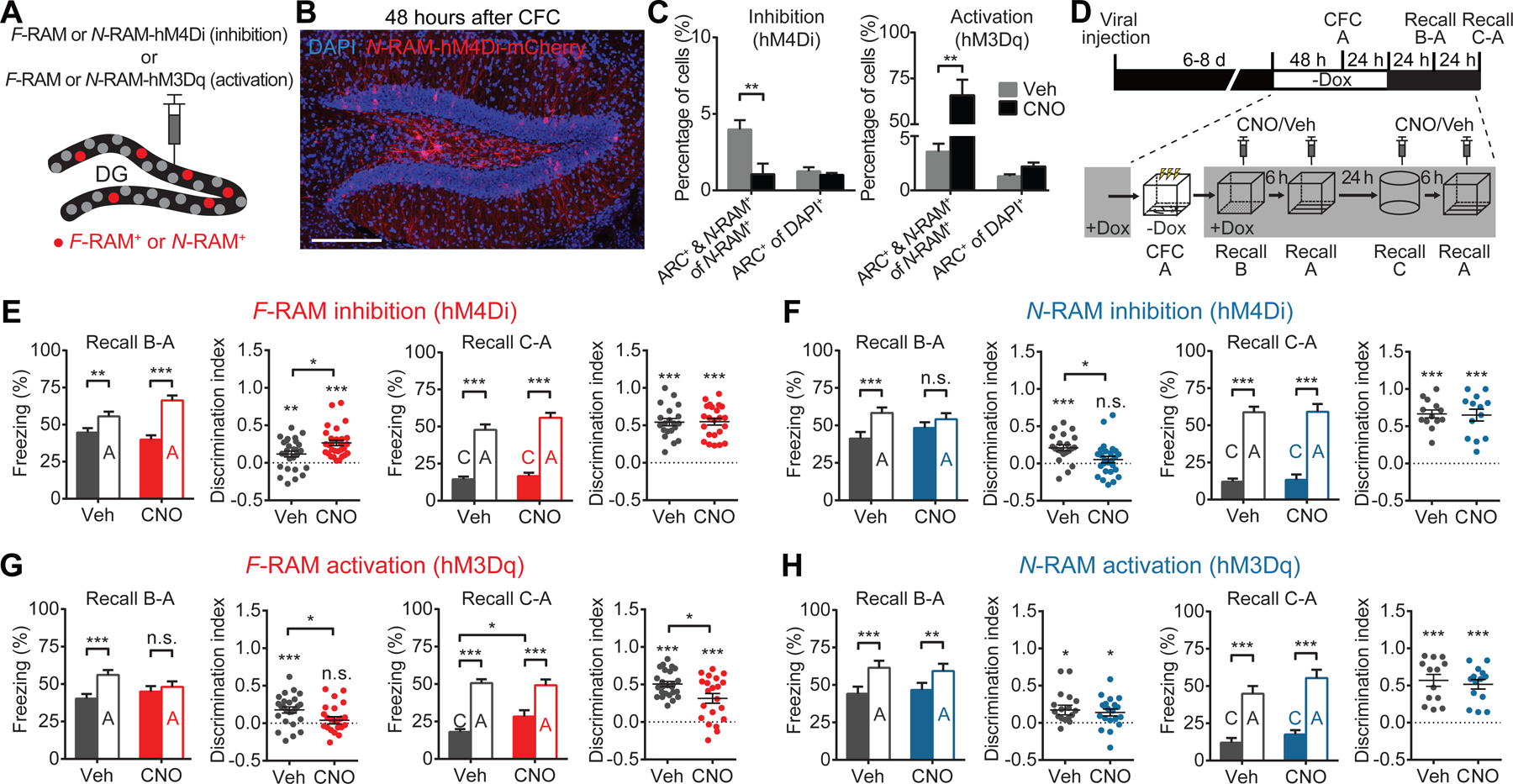
(A) Schematics for expression of hM4Di or hM3Dq in ensembles.
(B) Representative image showing expression of hM4Di-mCherry in the N-RAM ensemble.
(C) Percentages of ensemble neurons reactivated during memory recall, labeled by Arc expression (ARC+), showing that hM4Di inhibited and hM3Dq activated ensemble neurons (two-way mixed ANOVA, Sidak’s test, n = 5 per group).
(D) Experimental scheme to probe the functions of the F-RAM and N-RAM ensembles in memory discrimination-generalization.
(E) Inhibition of F-RAM ensembles enhanced discrimination between contexts A and B (Veh, n = 27; CNO, n = 29), but not A and C (n = 20, 23). Discrimination indices are calculated as Freezing[(A-B)/(A+B)] or Freezing[(A-C)/(A+C)].
(F) Inhibition of N-RAM ensembles reduced discrimination between contexts A and B (n = 21, 26), but not A and C (n = 13, 13).
(G) Activation of F-RAM ensembles reduced discrimination between contexts A and B (n = 25, 23), and between A and C (n = 21, 25).
(H) Activation of N-RAM ensembles did not affect discrimination between contexts A and B (n = 18, 19), or A and C (n = 13, 14).
Two-way mixed ANOVA with Sidak’s test was used for freezing levels, Mann-Whitney test for comparing discrimination indices, and one-sample t-test to compare discrimination indices with zero.
Inhibiting the F-RAM ensemble enhanced discrimination between contexts A and B (Figures 4D and 4E), while inhibiting the N-RAM ensemble reduced memory discrimination (Figure 4F). These results suggest that the F-RAM and N-RAM ensembles shift the discrimination-generalization balance in opposite directions. Notably, these manipulations only affected discrimination between similar contexts (A and B), but not distinct contexts (A and C), consistent with the DG being involved primarily in distinguishing similar patterns (Bernier et al., 2017; Kheirbek et al., 2013; Treves and Rolls, 1994). These behavioral effects were not due to any non-specific effects of CNO (Figure S4C) or altered general locomotion or anxiety levels, as freezing levels in context C were similar across all treatments. Inhibiting the F-RAM or N-RAM ensembles labeled under the home cage condition did not affect memory discrimination-generalization either (Figures S4D–S4F). Activation of the F-RAM ensemble by hM3Dq enhanced memory generalization and diminished discrimination between contexts (Figure 4G), consistent with the idea that this ensemble promotes memory generalization. Activation of the N-RAM ensemble, however, did not affect discrimination (Figure 4H), suggesting that it is required for, but does not actively drive, memory discrimination.
Taken together, our results so far indicate that the contextual fear memory engram consists of functionally distinct neuronal ensembles, and those labeled by F-RAM and N-RAM oppositely modulate the balance between memory discrimination and generalization.
The F-RAM Ensemble Receives Increased Excitatory Inputs from the MEC, while the Ensemble Receives Enhanced Inhibitory Inputs from CCK+ Interneurons
For two ensembles within the same engram to differentially modulate memory expression, we hypothesized that each ensemble would undergo different circuit modifications necessary for their distinct functions. Since F-RAM+ DGCs receive stronger excitatory inputs than their neighbors (Figure 2B), we asked which of the three major afferent pathways of the DG provide this increased excitatory drive: the lateral perforant path (LPP) from the lateral entorhinal cortex (LEC), the medial perforant path (MPP) from the medial entorhinal cortex (MEC) or the mossy cell fibers (MCF) from the dentate hilus (Amaral and Witter, 1989) (Figure 5A). We electrically stimulated the axonal fibers of the LPP, MPP or MCF, which respectively target the outer, middle or inner molecular layers (OML, MML, IML) of the DG (Scharfman and Myers, 2013; Witter, 2007), and simultaneously measured evoked excitatory postsynaptic currents (eEPSCs) of F-RAM+ and unlabeled neighboring neurons in pairs. Selective stimulation of the input pathways was confirmed by observation of their characteristic short-term plasticity: paired-pulse facilitation for the LPP and MCF and paired-pulse depression for the MPP (Hashimotodani et al., 2017; Petersen et al., 2013), and by selective reduction of the MPP response in the presence of the mGluR2/3 agonist DCG-IV (Macek et al., 1996) (Figure 5B). The amplitude of eEPSCs originating from the MPP only was significantly larger on F-RAM+ than F-RAM− cells (Figure 5C). In contrast, eEPSCs from all three pathways were similar in N-RAM+ neurons and their unlabeled neighbors (Figure 5F). Paired-pulse ratios (PPRs) and AMPA/NMDA ratios were comparable in F-RAM+ and F-RAM− neurons (Figures 5D and 5E). These findings were confirmed using optogenetic stimulation (Figure S5C). Therefore, inputs to F-RAM+ neurons from the MEC, but not the LEC or dentate hilus, were selectively strengthened.
Figure 5. The F-RAM Ensemble Receives Increased Excitatory Inputs from the MEC and the N-RAM Ensemble Receives Enhanced Inhibitory Inputs from DG CCK+ Interneurons.
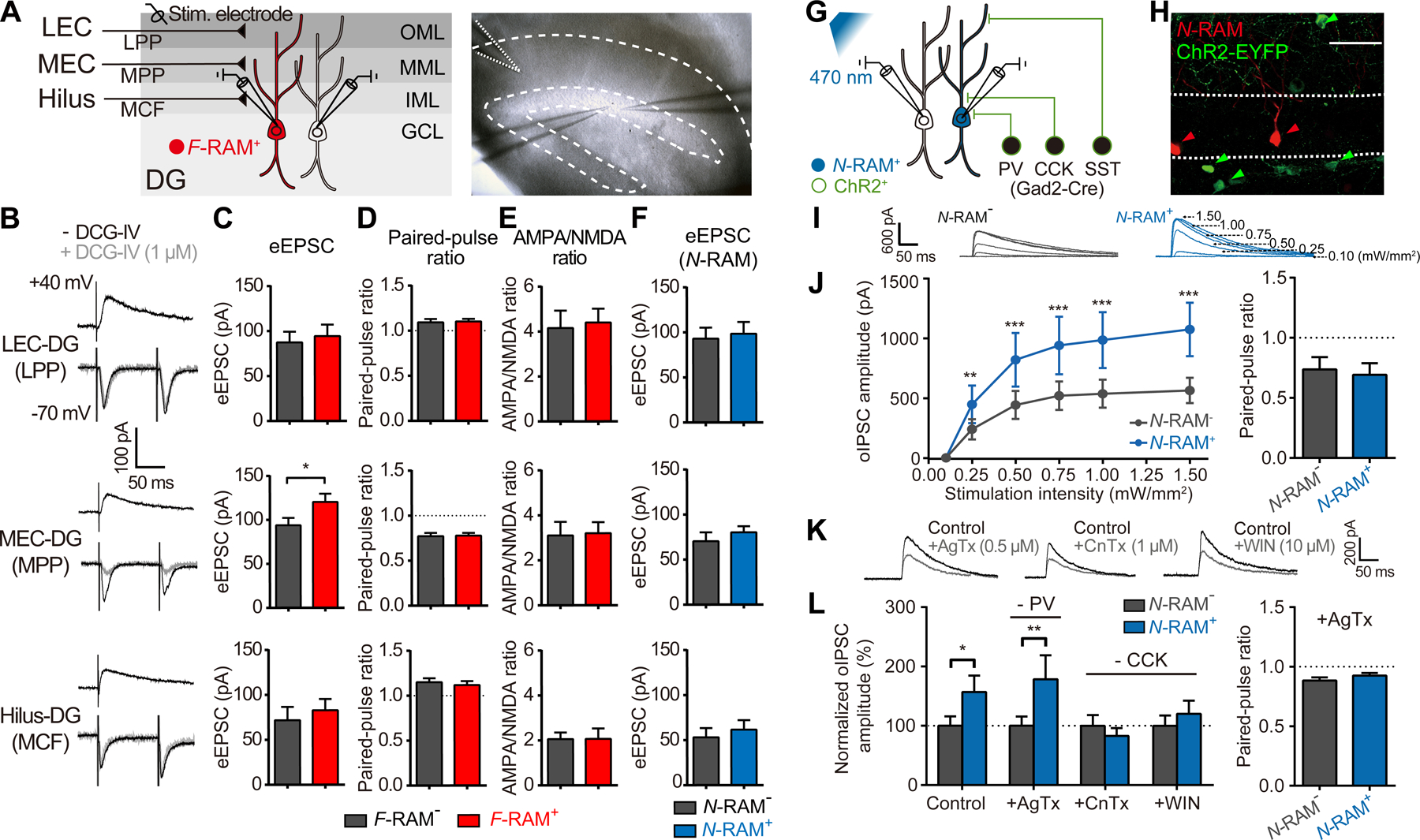
(A) Left, experimental scheme for simultaneous recording of eEPSCs, elicited by stimulating the LPP, MPP or MCF, on pairs of F-RAM+ or N-RAM+ neurons and nearby unlabeled neurons. Right, representative image of MPP stimulation.
(B) Representative traces of eEPSCs during LPP, MPP and MCF stimulation. Grey traces denote eEPSCs in the presence of DCG-IV.
(C-E) Bar graphs comparing eEPSC amplitudes (C, Wilcoxon signed-rank test, n = 15, 18, 21 pairs for LPP, MPP and MCF, respectively), paired-pulse ratios (D, n = 18, 20, 13) and AMPA/NMDA ratios (E, n = 15, 19, 13) between pairs of F-RAM+ and F-RAM− neurons.
(F) Bar graph comparing eEPSC amplitudes between pairs of N-RAM+ and N-RAM− neurons (n = 10, 13, 11).
(G) Experimental scheme for simultaneous recording of oIPSCs on pairs of N-RAM+ or F-RAM+ and nearby unlabeled neurons. oIPSCs were elicited by optogenetically stimulating ChR2-expressing GABAergic interneurons in Gad2-Cre mice.
(H) Representative image showing N-RAM and ChR2 expression.
(I) Representative traces showing oIPSCs recorded in pairs of nearby N-RAM+ and N-RAM− neurons.
(J) Comparison of oIPSC amplitudes (left; two-way repeated measures ANOVA, Sidak’s test, n = 10 pairs) and paired-pulse ratios (right; 200 ms interval, 0.50 mW/mm2, Wilcoxon signed-rank test, n = 10) between N-RAM+ and N-RAM− neurons.
(K) Representative traces showing that oIPSC amplitudes were reduced with bath application of AgTx, CnTx and WIN.
(L) Left, oIPSC amplitudes in N-RAM+ and N-RAM− neurons with and without blockers (two-way mixed ANOVA, Sidak’s test, n = 16, 10, 12, 13 pairs for control, AgTx, CnTx and WIN). Right, paired-pulse ratios in the presence of AgTx (n = 12).
To identify the source of the inhibitory inputs recruited onto N-RAM+ DGCs (Figure 2C), we expressed light-activated channelrhodopsin (ChR2) in DG GABAergic neurons, by delivering AAV-EF1α-DIO-ChR2-EYFP into Gad2-Cre mice (Figures 5G and 5H). Optically evoked inhibitory postsynaptic currents (oIPSCs) were then measured on N-RAM+ or F-RAM+ DGCs and their unlabeled neighbors (Figure 5I). Consistent with our previous finding that N-RAM+ neurons had higher mIPSC frequencies, robustly larger oIPSC amplitudes were observed on N-RAM+ cells, with PPRs being similar in N-RAM+ and N-RAM− cells (Figure 5J). In contrast, oIPSCs on F-RAM+ neurons were similar to those of their unlabeled neighbors (Figure S5E).
The three major subtypes of GABAergic interneuron in DG are parvalbumin- (PV+), somatostatin- (SST+) and cholecystokinin-expressing (CCK+) cells (Freund and Buzsaki, 1996; Pelkey et al., 2017). To determine whether inputs from certain of these interneuron subtypes were recruited to N-RAM+ DGCs, oIPSCs were measured on pairs of N-RAM+ and N-RAM− cells in the presence of the P/Q-type Ca2+ channel blocker ω-agatoxin IVa (AgTx), to selectively inhibits transmission from PV+ interneurons, or the N-type Ca2+ channel antagonist ω-conotoxin GVIa (CnTx), to selectively inhibits transmission from CCK+ interneurons (Hefft and Jonas, 2005) (Figure 5K). The difference in oIPSC amplitudes between N-RAM+ and N-RAM− neurons was abolished in the presence of CnTx, but not AgTx (Figure 5L), suggesting that the increased inhibition on N-RAM+ neurons comes mainly from CCK+ interneurons. Consistently, the increase in oIPSC amplitudes on N-RAM+ neurons was also abolished by the CB1R agonist WIN 55,212–2 (WIN), which selectively inhibits CCK+ interneurons via the endocannabinoid pathway (Tsou et al., 1999). These results suggest that N-RAM+ neurons recruit enhanced inhibitory drive selectively from CCK+ interneurons.
The Distinct Synaptic Properties of Ensemble Neurons are Induced by Learning
Are the distinct synaptic properties of F-RAM+ and N-RAM+ neurons intrinsic features of the cells or acquired through learning? We hypothesized that these features were triggered by contextual fear learning via activation of the Fos- and Npas4-dependent pathways. We first determined whether these synaptic properties can be induced by the expression of Fos or Npas4. We artificially induced Fos and Npas4 expression (FOSind and NPAS4ind) in sparse and randomly selected DGCs (Figures 6A and 6B), and compared mEPSCs and mIPSCs on those neurons to their un-manipulated neighbors. Fos expression led to an increase in mEPSC frequencies (Figure 6C), recapitulating this synaptic feature of F-RAM+ neurons. Npas4 expression resulted in elevated mIPSC frequencies (Figure 6D), mimicking the synaptic changes observed on N-RAM+ cells. Neither mIPSCs of FOSind neurons nor mEPSCs of NPAS4ind neurons were altered (Figures S6A and S6B). These results suggest that activation of Fos- and Npas4-dependent pathways may be sufficient to induce the synaptic properties observed in F-RAM+ and N-RAM+ neurons, respectively.
Figure 6. Synaptic Changes on Ensemble Neurons Are Correlated with the Strength of Learning.
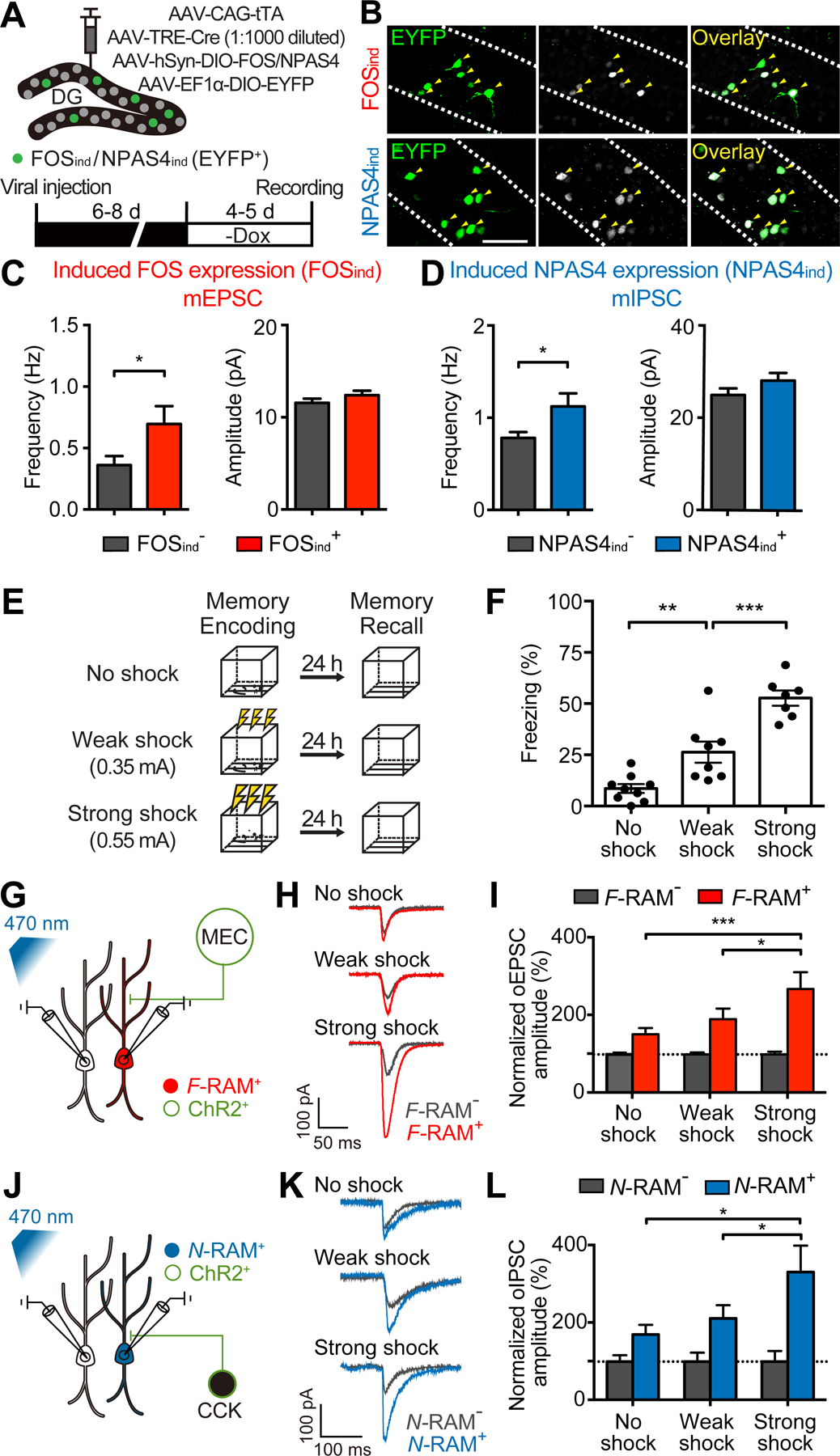
(A) Experimental scheme for inducing FOS or NPAS4 expression in sparse and randomly selected DG neurons.
(B) Representative images showing induced FOS or NPAS4 expression (FOSind or NPAS4ind) in randomly selected neurons (EYFP+, white arrows).
(C) FOSind increased mEPSC frequency (Mann-Whitney test; EYFP−, n = 19; EYFP+, n = 20).
(D) NPAS4ind increased mIPSC frequency (n = 19, 20).
(E and F) Experimental scheme (E) showing that training of animals in context A with no shock, weak (0.35 mA) or strong shock (0.55 mA) resulted in different memory strengths, as indicated by levels of freezing (F; one-way ANOVA, Tukey’s test, n = 7–9).
(G and H) Experimental scheme (G) and representative traces (H) showing simultaneous recording of oEPSCs on F-RAM+ and F-RAM− cells with optogenetic stimulation of the MEC inputs.
(I) F-RAM+ cells showed the largest oEPSC amplitudes in the strong shock group (two-way mixed ANOVA, Tukey’s test, n = 24, 28, 21 pairs for no shock, weak shock and strong shock, respectively).
(J and K) Experimental scheme (J) and representative traces (K) showing simultaneous recording of oIPSCs on N-RAM+ and N-RAM− cells with optogenetic stimulation of the CCK+ interneurons.
(L) N-RAM+ cells showed the largest oIPSC amplitudes in the strong shock group (n = 19, 15, 9).
Since Fos and Npas4 expressions are very low at baseline and highly activated by learning (Ramamoorthi et al., 2011), we predicted that the synaptic properties of the ensemble neurons are not pre-existing, but rather are induced by learning. Like all activity reporters, F-RAM and N-RAM can only identify ensembles after learning, so we cannot compare ensemble neurons before and after CFC. However, if the synaptic properties of interest are indeed induced by learning, we would expect that a stronger learning experience, resulting in a stronger memory, will induce stronger synaptic modifications (Choi et al., 2018). We therefore trained mice in a novel context with no foot shocks, weak foot shocks (0.35 mA) or strong foot shocks (0.55 mA; as used for other experiments) (Figure 6E). These 3 treatments resulted in correspondingly different strengths of contextual fear memory, as the animals displayed freezing levels that were closely correlated with the intensity of shocks they received (Figure 6F). The numbers of labeled F-RAM+ and N-RAM+ neurons were similar in the three groups (Figures S6C and S6D). Measuring oEPSCs from the MEC onto paired F-RAM+ and F-RAM− DGCs and oIPSCs from CCK+ interneurons onto paired N-RAM+ and N-RAM− DGCs, we found that MEC input to F-RAM+ and CCK inhibition on N-RAM+ cells were strongest in mice that received strong foot shocks, followed by animals that received weak foot shocks and then those that received no foot shocks (Figures 6G–6L; CCK+ interneurons were labeled using a genetic intersection strategy described in Figure 7I). These results, together with our previous data, suggest that the distinct synaptic properties of the ensemble neurons are the result of learning-induced synaptic modifications driven by distinct activity-dependent pathways.
Figure 7. The MEC-DG Pathway Mediates Memory Generalization and DG CCK+ Interneurons Mediate Memory Discrimination.
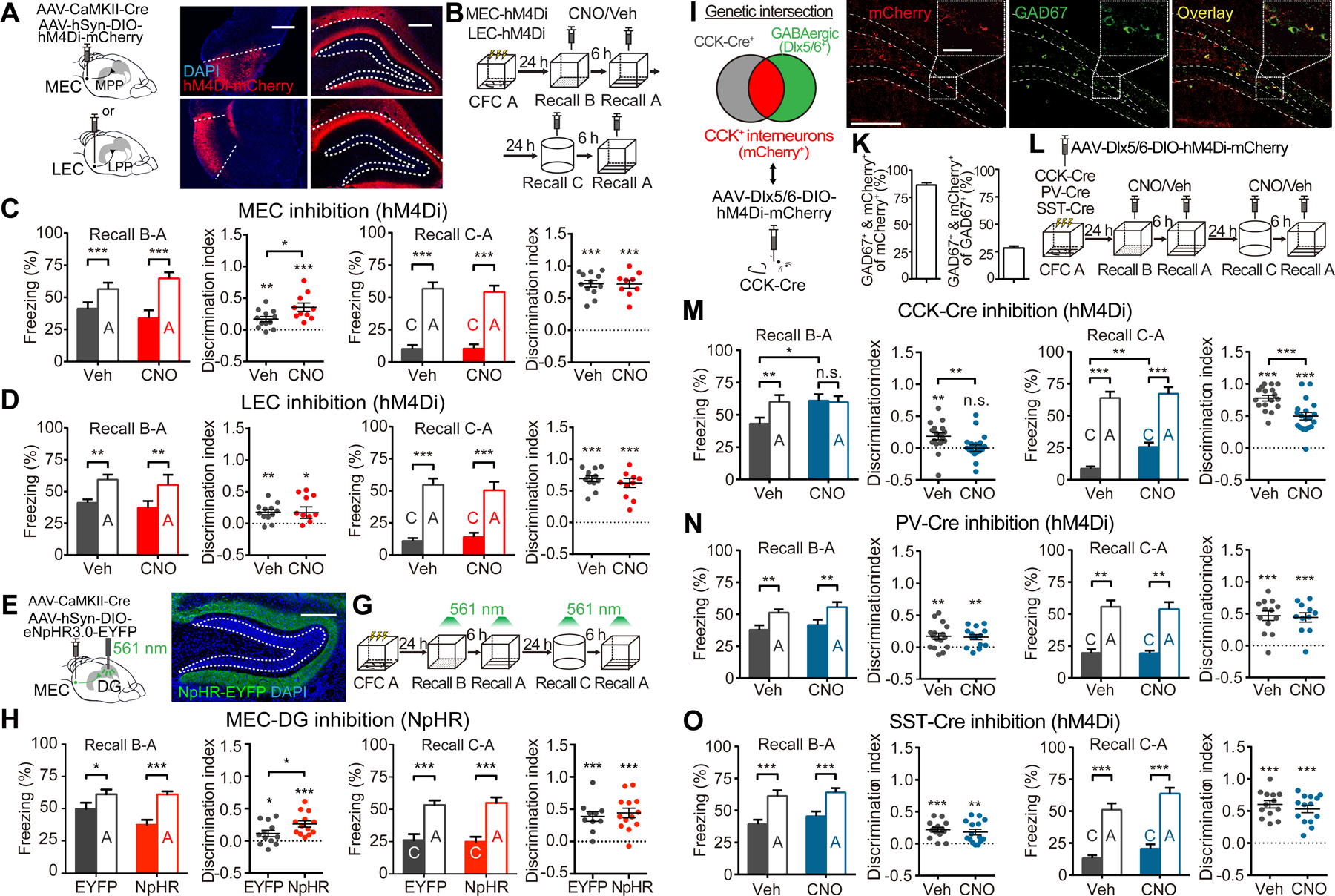
(A) Schematics and representative images showing selective expression of hM4Di in MEC and LEC projection neurons, which send their axonal terminals to the DG MML or OML, respectively.
(B) Experimental scheme for inhibition of MEC or LEC during memory discrimination-generalization.
(C) Inhibiting the MEC enhanced discrimination between contexts A and B (Veh, n = 12; CNO, n = 10), but not A and C (n = 12, 9).
(D) Inhibiting the LEC did not affect generalization between contexts A and B (n = 12, 11) or A and C (n = 12, 10).
(E and F) Schematic (E) and representative image (F) for selective expression of NpHR in MEC projection neurons, which project to the MML of the DG.
(G) Experimental scheme to optogenetically inhibit the MEC-DG pathway during memory recall with 561 nm constant light.
(H) Inhibition of the MEC-DG pathway enhanced discrimination between contexts A and B (EYFP, n = 12; NpHR, n = 13), but not between contexts A and C (n = 10, 13).
(I) Genetic intersection strategy to target CCK+ GABAergic interneurons in DG.
(J) Representative images showing co-localization (white arrows) of labeled cells (mCherry+) with GAD67.
(K) Quantifications showing that labeled neurons were mostly GABAergic (GAD67+) and made up 28% of the GABAergic interneurons in the DG (n = 3).
(L) Experimental scheme to determine which interneuron subtypes are involved in memory discrimination-generalization.
(M) Inhibiting CCK+ interneurons in the DG reduced discrimination between contexts A and B (Veh, n = 17; CNO, n = 20), and between A and C (n = 17, 19).
(N and O) Discrimination was not affected by inhibiting PV+ interneurons (N, A and B: n = 16, 14; A and C: n = 13, 11) or SST+ interneurons (O, A and B: n = 14, 15; A and C: n = 13, 14).
Two-way mixed ANOVA with Sidak’s test was used for freezing levels, Mann-Whitney test for comparing discrimination indices, and one-sample t-test to compare discrimination indices with zero.
The MEC and its Inputs to the DG Mediate Memory Generalization while DG CCK+ Interneurons Mediate Memory Discrimination
These learning-induced circuit modifications in the F-RAM and N-RAM ensembles suggest important roles for the MEC and DG CCK+ interneurons in memory discrimination-generalization. We first determined whether the MEC, like the F-RAM ensemble, promotes memory generalization. MEC activity was inhibited during memory recall by virally delivering hM4Di to MEC projection neurons, precisely and strongly targeting their axonal projections in the MML (Figure 7A). CNO injection then robustly suppressed, although did not completely abolish, the activity of MEC neurons (Figure S7C). In the same memory discrimination-generalization assay described above (Figure 7B), inhibiting the MEC in this way suppressed memory generalization and enhanced discrimination between contexts A and B (Figure 7C), recapitulating the effect of inhibiting the F-RAM ensemble. Inhibiting the LEC in the same way did not affect memory generalization (Figures 7A and 7D).
To confirm that the MEC-DG pathway promotes memory generalization, we optogenetically inhibited MEC terminals within the DG (Figures 7E and 7F). Illuminating the DG with 561 nm light in animals expressing the inhibitory opsin eNpHR3.0 in the MEC significantly reduced the number of FOS+ cells in the DG, but not in the nearby CA1 region that also receives MEC inputs, indicating selective inhibition of the MEC-DG pathway (Figure S7E). Inhibiting the MEC-DG projections in this way enhanced discrimination between contexts A and B (Figures 7G and 7H), similar to inhibition of the F-RAM ensemble. In control experiments without light stimulation mice injected with NpHR were indistinguishable from those injected with EYFP (Figure S7G). These results reveal an important role for the MEC-DG pathway in mediating memory generalization, likely involving the F-RAM+ ensemble neurons.
We next examined the role of CCK+ interneurons in mediating memory discrimination, by chemogenetically inhibiting DG CCK+ interneurons during memory recall. CCK is expressed in both GABAergic and some glutamatergic neurons (Taniguchi et al., 2011), so we adopted a genetic intersection strategy to label only the GABAergic CCK+ cells: using AAV-Dlx5/6-DIO-hM4Di-mCherry to drive Cre-dependent expression of hM4Di from an interneuron-specific promoter, Dlx5/6 (Dimidschstein et al., 2016), in CCK-Cre mice (Figure 7I). The majority of labeled neurons were GAD67+, indicating successful intersectional labeling (Figures 7J and 7K). Labeled neurons displayed electrophysiological properties reported for CCK+ interneurons, namely adaptive firing, depolarization-induced suppression of inhibition (DSI) and asynchronous synaptic transmission (Bartos and Elgueta, 2012) (Figures S7H–S7M). Using the same behavioral assay (Figure 7L), we then found that inhibiting DG CCK+ interneurons abolished discrimination between contexts A and B and also significantly reduced discrimination between A and C (Figure 7M). In contrast, inhibiting PV+ or SST+ interneurons in the DG did not affect memory discrimination (Figures 7N and 7O). These results suggest that DG CCK+ interneurons are involved in memory discrimination.
Discussion
Here we provide direct experimental evidence for functional heterogeneity within memory engrams. We show that two distinct neuronal ensembles (F-RAM and N-RAM) in representations of the same memory exhibit different synaptic properties and promote opposing behavioral outputs: memory generalization and memory discrimination. In addition, we describe synaptic mechanisms underlying this functional heterogeneity: the F-RAM and N-RAM ensembles selectively recruit excitatory and inhibitory inputs, respectively, from different upstream circuits. We show that these circuits regulate the discrimination-generalization balance: the MEC is important for memory generalization and DG CCK+ interneurons for memory discrimination. While the present study focuses on contextual fear conditioning in the DG, heterogeneity of the engram could be a widespread phenomenon (Grewe et al., 2017; Jun et al., 2010; Mallory and Giocomo, 2018): memory engrams may generally be composed of multiple functional subcomponents that differentially modulate experience-dependent behavioral outputs.
How Should We Genetically Define Functionally Distinct Neuronal Ensembles?
Critical questions remain regarding how many distinct ensembles there are and how best to define them genetically. Our findings suggest that the emergence of distinct subpopulations within memory engrams can result from different activity-dependent signaling cascades, triggered within individual ensembles by learning (Figure 6). Neuronal activity-dependent pathways are therefore key molecular mechanisms determining the functions of these ensembles, as well as providing activity markers to define them. Here we have investigated ensembles associated with two pathways, but other activity-dependent pathways such as Arc and CREB may define additional functionally distinct subpopulations (Denny et al., 2014; Park et al., 2016). Many of these pathways have overlapping upstream signaling and may be co-activated in some neurons (Lonergan et al., 2010). Although multiple lines of evidence from our study indicate that the F-RAM and N-RAM ensembles in DG are distinct neuronal populations (Figures 2–4), our data do not exclude the possibility of some overlap. A challenge for the future is to identify sets of orthogonal activity-dependent reporters that define distinct populations and provide a complete description of the memory engram.
Unlike most activity reporters, F-RAM and N-RAM report IEG-dependent downstream transcriptional output rather than expression of IEGs themselves. It is conceivable that the F-RAM and N-RAM ensembles are not identical to those defined by the native Fos and Npas4 promoters, which may help to explain the incomplete overlap of the F-RAM and Fos-CreER ensembles (Figure 2F). Since transcription factors exert their functions through their downstream transcriptional targets, reporters such as F-RAM and N-RAM that monitor transcriptional outputs may better capture functional populations that undergo learning-induced changes.
It is also not known how neurons are recruited from the general population into the kinds of functionally distinct ensembles we describe here. Factors including intrinsic excitability (Yiu et al., 2014; Zhou et al., 2009), connectivity (Ryan et al., 2015) and firing patterns (Tanaka et al., 2018) have been suggested. Future work, for example using time-lapse Ca2+ imaging and single-cell sequencing, will determine whether extrinsic or intrinsic factors or a mixture of both contribute to the formation of functionally distinct neuronal ensembles.
What Information is Encoded in the F-RAM and N-RAM Ensembles?
Intriguingly, neither the F-RAM nor N-RAM ensemble is required for memory retrieval in the conditioned context (Figure 4), and neither is the DG Fos-expressing ensemble (Figures S4G–S4K) (Pignatelli et al., 2018). These results are not unexpected given that lesion or optogenetic inhibition of the entire DG does not impair memory retrieval (Kheirbek et al., 2013; Lee and Kesner, 2004), although inhibiting the DG Arc-expressing ensemble reduced freezing (Denny et al., 2014). The F-RAM and N-RAM ensembles therefore resemble the postulated “higher-order memory trace” (Thompson, 2005). Unlike the “essential memory trace”, the “higher-order memory trace” is not absolutely required for memory retrieval, but is involved in modulating memory-guided adaptive behaviors. Future work that combines in vivo imaging with functional perturbation to decipher the specific information encoded by each ensemble is urgently needed to test and revise current models of memory.
Learning-induced Synaptic Changes in Neuronal Ensembles
The synaptic properties of F-RAM+ and N-RAM+ neurons (Figures 2A–2C) are most likely the result of neuronal activity triggered during contextual fear learning, because they correlate with memory strength and can be induced by the expression of FOS and NPAS4, respectively (Figure 6). However, since the ensembles can only be labeled after they have been activated, we cannot exclude the possibility that other differences between these two groups of neurons already exist prior to the experience.
In addition to the potentiation of excitatory inputs, which has been the primary focus of work on mechanisms of memory encoding, our data suggest that the potentiation of inhibitory inputs is also important. Furthermore, strengthening of excitatory and inhibitory inputs occurs on two different neuronal ensembles within the memory engram, which could provide a mechanism for a neural system to balance the behavioral output (e.g., discrimination vs. generalization) through the “push and pull” of different neuronal ensembles. It remains to be seen whether the F-RAM and N-RAM ensembles interact with each other.
Circuit Components Involved in the Discrimination-Generalization Balance
Mossy cells.
Both F-RAM and N-RAM reporters label small numbers of hilar mossy cells (1–6%), which have recently been implicated in pattern separation (Danielson et al., 2017; GoodSmith et al., 2017; Jinde et al., 2012; Senzai and Buzsaki, 2017). However, the numbers of mossy cells labeled by either reporter were similar between the HC and CFC conditions (Figures S1E and S1F). Although other explanations are possible, given that animals were treated identically throughout the 2–3 day window for reporter labeling (Figure 1C) except for the few minutes during which one group received CFC treatment, it is likely that the reporters captured mossy cells that were active in the home cage and CFC did not activate additional mossy cells. Furthermore, memory discrimination-generalization was not affected when the mossy cells labeled in the HC condition were inhibited (Figures S4D–S4F). These results suggest that the F-RAM and N-RAM reporters do not capture the mossy cell CFC engram. Genetic tools that can specifically label mossy cells activated during contextual learning will be needed to define the role of mossy cells in memory discrimination-generalization.
MEC.
Our data strongly support a role for the MEC-DG pathway in memory generalization, in line with observations that grid cells in the MEC maintain relatively stable spatial firing patterns across similar contexts (Fyhn et al., 2007; Leutgeb et al., 2007). In our study, memory retrieval (in context A) was not impaired by suppressing MEC activity (Figure 7C). This observation is consistent with some published studies (Hales et al., 2014; Kanter et al., 2017) but contradictory to others (Kitamura et al., 2015; Miao et al., 2015; Zhao et al., 2016). The discrepancy may be explained by the different MEC cell populations being targeted and the different manner in which they are being inhibited (lesion, optogenetics, chemogenetics). Future work is needed to pinpoint which MEC subpopulations (Diehl et al., 2017) relay generalization signals to the DG.
CCK+ interneurons.
Of the three major subtypes of interneurons providing inhibitory inputs to DG granule cells, we find that CCK+ and not PV+ or SST+ interneurons are important for memory discrimination. The reason for this is not known. In fact, the roles of CCK+ interneurons in learning and memory have not been extensively studied. CCK+ interneurons are known to have unique synaptic outputs, such as endocannabinoid-dependent short-term and long-term synaptic plasticity (Castillo et al., 2012; Hartzell et al., 2018) and asynchronous synaptic release (Hefft and Jonas, 2005). Our results present an exciting opportunity to identify specific synaptic mechanisms uniquely provided by CCK+ interneurons to regulate memory discrimination.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for reagents may be directed to and will be fulfilled by the lead contact, Dr. Yingxi Lin (yingxi@mit.edu or linyi@upstate.edu).
The F-RAM and N-RAM reporters generated in this study have been deposited to Addgene (ID: 140274 and 140275). All other unique reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Subjects
7–11 week old C57BL/6 male and female mice were used for all experiments. Wildtype mice were purchased from the Charles River Laboratory. Npas4flx/flx (Npas4 conditional knockout) mice were generated previously (Lin et al., 2008) and Fosflx/flx (Fos conditional knockout) mice were generously provided by Dr. Ming Xu at the University of Chicago. Gad2-Cre (Gad2tm2(cre)Zjh/J), CCK-Cre (Ccktm1.1(cre)Zjh/J), PV-Cre (Pvalbtm1(cre)Arbr/J), SST-Cre (Ssttm2.1(cre)Zjh/J) and Fos-CreER (Fostm1.1(cre/ERT2)Luo/J) mice were purchased from the Jackson Laboratory. To produce Cre-expressing animals, heterozygous or homozygous mice carrying the Cre allele were bred with wildtype animals from the Charles River Laboratory. All mice were housed with a 12 hour light-dark cycle. Animal protocols were performed in accordance with NIH guidelines and approved by the Massachusetts Institute of Technology Committees on Animal Care and the Institutional Animal Care and Use Committee at SUNY Upstate Medical University.
METHOD DETAILS
Cell cultures for luciferase assays
Primary hippocampal neurons were prepared from mouse pups at postnatal day 0 as previously described (Ramamoorthi et al., 2011) and plated at 100,000 cells per well on coated 24-well plates. Cultures were kept in dissection medium [Neurobasal A Medium (NBA, Invitrogen), 10% horse serum (Invitrogen) and GlutaMAX (Invitrogen)] for 3 hours before being switched to culture medium [NBA supplemented B27 (Invitrogen) and GlutaMAX]. Cultures were kept at 37 °C in humidified incubators supplemented with 5% CO2 until use.
At 5 days in vitro (DIV), neurons were transfected with plasmids using lipofectamine 2000 (Life Technologies). Plasmid cocktails contained: luciferase reporter plasmids (PF-RAM-Luc or PN-RAM-Luc) expressing firefly luciferase under the control of PF-RAM or PN-RAM, control plasmids (TK-Renilla) expressing renilla luciferase under the control of the constitutively active thymidine kinase promoter (Promega) and plasmids that overexpress Fos or Npas4 under the control of a constitutive promoter (EF1α) in some cases.
At 8 DIV, cultures were blocked with tetrodotoxin (TTX, 1 μM, Tocris) and APV (100 μM, Tocris) for 1 hour prior to stimulation and then stimulated for 6 hours with 34 mM KCl or 34 mM KCl plus 5 mM EGTA (Sigma). Normal culture medium without a high concentration of KCl was used to treat the unstimulated control group. Neurotrophic factors and other drugs were directly added to the cultures in 10 μL culture medium and the neurons were incubated for 6 hours. At the end of the experiment, neurons were rinsed briefly in phosphate buffered saline (PBS, Invitrogen) and lysed in passive lysis buffer (Promega) for 20 minutes at room temperature. Dual-Glo Luciferase Assay System (Promega) reagents were used to measure luciferase levels using the SpectraMax Microplate Reader (Molecular Devices). For each experiment, relative luciferase values (a.u.) were calculated by normalizing the firefly luciferase level to the renilla luciferase level. Data were compiled from 3 separate batches of cultures, each conducted with 2 to 3 replicates.
Neurotrophic factors and other drugs were dissolved in water and used at the following final concentrations: recombinant human BDNF (50 ng/μL, PeproTech), recombinant human NT3 (50 ng/μL, PeproTech), recombinant human NT4 (50 ng/μL, PeproTech), NKH 477 (20 μM, Tocris), Bicuculline (50 μM, Sigma) and 4-aminopyridine (250 μM, Tocris).
Viral vectors
AAV1-hSyn-GFP, AAV1-hSyn-GFP-Cre, AAV1-EF1α-DIO-ChR2-EYFP were obtained from the University of Pennsylvania Vector Core via Addgene. All other AAVs were produced in house. AAVs were generated in HEK293T cells and purified using an adapted h gradient purification protocol as previously described (Sørensen et al., 2016). Viral dilutions were determined for individual experiments using pilot injections.
In experiments to label the F-RAM and N-RAM ensembles in vitro and in vivo, AAV2/AAV8-F-RAM-d2tTA-TRE-mKate2 or AAV2/AAV8-N-RAM-d2tTA-TRE-mKate2 virus was used. AAV2/AAV8s were mixtures of AAV2/2 (rep/cap) and AAV2/8 serotypes at a 1:1 ratio (Sørensen et al., 2016). In most experiments a control virus (AAV2/AAV8-EF1α-EGFP) was co-injected to determine injection accuracy and infection efficiency. To determine whether the F-RAM and N-RAM reporters were dependent on endogenous Fos and Npas4, AAV9-F-RAM-d2tTA-TRE-mKate2 or AAV9-N-RAM-d2tTA-TRE-mKate2 virus was co-injected with AAV1-hSyn-GFP or AAV1-hSyn-GFP-Cre into Npas4 or Fos conditional knockout mice. To examine co-localization of the Fos-CreER ensemble and either the F-RAM or N-RAM ensemble, AAV9-F-RAM-d2tTA-TRE-mKate2 or AAV9-N-RAM-d2tTA-TRE-mKate2 virus was co-injected with a Cre-dependent reporter virus, AAV1-EF1α-DIO-ChR2-EYFP. For the fiber photometry experiments, viral cocktails containing AAV9-TRE-GCaMP6f and either AAV9-F-RAM-d2tTA-TRE-mKate2 or AAV9-N-RAM-d2tTA-TRE-mKate2 were used.
To manipulate ensemble activity with DREADDs, viral cocktails containing AAV9-TRE-hM3Dq-mCherry or AAV9-TRE-hM4Di-mCherry and either AAV9-F-RAM-d2tTA-sEF1α-GFP or AAV9-N-RAM-d2tTA-sEF1α-GFP were used (the sEF1α-GFP component was used to determine injection accuracy and infection efficiency). For control experiments testing the effects of CNO injections alone, AAV9-TRE-mCherry was used instead of the DREADD-expressing viruses and was co-injected with AAV9-N-RAM-d2tTA-sEF1α-GFP. In experiments to manipulate the activity of Fos-expressing ensembles, AAV9-pFos-d2tTA was co-injected with the DREADD viruses. pFos is an engineered Fos promoter similar to that used in previous studies (Liu et al., 2012; Reijmers et al., 2007).
To examine optically evoked IPSCs (oIPSCs) from DG interneurons, AAV2/AAV8-F-RAM-d2tTA-TRE-mKate2 or AAV2/AAV8-N-RAM-d2tTA-TRE-mKate2 virus was co-injected with AAV2/AAV8-EF1α-DIO-ChR2-EYFP virus into Gad2-Cre mice. To examine oIPSCs from DG CCK+ interneurons, AAV9-F-RAM-d2tTA-TRE-mKate2 or AAV9-N-RAM-d2tTA-TRE-mKate2 virus was co-injected with AAV9-Dlx5/6-DIO-ChR2-EYFP virus into CCK-Cre mice. To examine optically evoked EPSCs (oEPSCs) that originate from the medial entorhinal cortex (MEC) and the lateral entorhinal cortex (LEC), AAV9-EF1α-DIO-ChR2-EYFP and AAV1-CaMKII-Cre-GFP were co-injected into these regions and AAV9-F-RAM-d2tTA-TRE-mKate2 was injected into the DG. To examine oEPSCs that originate from the dentate hilus, AAV9-EF1α-DIO-ChR2-EYFP and AAV1-CaMKII-Cre-GFP were co-injected into the contralateral DG and AAV9-F-RAM-d2tTA-TRE-mKate2 was injected into the ipsilateral DG, where whole-cell patch-clamp recordings were carried out.
To over-express FOS or NPAS4 in sparse populations of DG neurons, animals were injected with the following viral cocktail: AAV9-CAG-tTA, AAV1-TRE-Cre (1:1,000 dilution), AAV9-hSyn-DIO-FOS/NPAS4, and AAV1-EF1α-DIO-EYFP.
In experiments to suppress the activity of MEC or LEC projection neurons, AAV1-CaMKII-Cre-GFP and AAV9-hSyn-DIO-hM4Di-mCherry were co-injected into the MEC or LEC. In experiments to optogenetically inhibit the MEC-DG pathway, AAV9-EF1α-DIO-eNpHR3.0-EYFP or AAV9-EF1α-DIO-EYFP was co-injected with AAV1-CaMKII-Cre-GFP into the MEC. To manipulate the activity of DG interneurons with DREADDs, AAV9-Dlx5/6-DIO-hM4Di-mCherry was injected into CCK-Cre, PV-Cre or SST-Cre mice.
Stereotaxic viral injection and fiber implantation
Mice were anesthetized with 1.5–2% isoflurane in O2. To label and manipulate F-RAM and N-RAM ensembles in the DG, stereotaxic injections were performed bilaterally into the dorsal hippocampal DG with the following coordinates (relative to bregma) and volumes: AP −1.90 mm, ML ±1.30 mm, DV −2.00 mm, 150–200 nL per hemisphere. Viruses were infused at a rate of 100 nL per minute and needles were kept at the injection site for 5 minutes. The same coordinates and volumes were used to manipulate interneurons in the DG. To manipulate the activities of MEC and LEC projection neurons, the following coordinates and volumes were used: MEC (AP −4.75 mm, ML ±3.25 mm, DV −3.60 mm, 250–300 nL per hemisphere), LEC (AP −3.40 mm, ML ±4.35 mm, DV −4.10 mm, 250–300 nL per hemisphere). Mice were typically allowed to recover for 6–8 days following surgery. For experiments with Dox-dependent ensemble labeling, animals were put on Dox diet (40 mg/kg, Bio-Serv) after surgery.
For fiber photometry experiments, DG injections were performed unilaterally, followed by fiber implantation in the same procedure. The left or right hemisphere was randomly chosen for unilateral surgeries. Fiber implants were made of multimode fiber (400 μm core, 0.48 NA, Thorlabs) with plastic ferrules (2.5 mm, Thorlabs). Implants were placed unilaterally in the DG 0.20 mm above the injection site (AP −1.90 mm, ML ±1.30 mm, DV −1.80 mm).
For optogenetics experiments, viral injections into the MEC were followed by fiber implantation in the DG. Fiber implants were made of multimode fiber (200 μm core, 0.22 NA, Thorlabs) with plastic ferrules (1.25 mm, Thorlabs). Implants were placed bilaterally in the DG (AP −1.90 mm, ML ±1.30 mm, DV −1.80 mm).
For DG injections, only animals with viral infection of more than 80% of the dorsal DG and less than 20% of CA3 were included in behavioral analyses. For MEC or LEC injections, only animals with selective infection of MEC or LEC axonal terminals in either the middle (MML) or outer (OML) third of the DG molecular layers were included. These histological analyses were performed by investigators blind to the genotypes or conditions.
Drug injections
To induce seizure, kainic acid (KA, Sigma) was dissolved in saline (1 mg/mL, Teleflex Medical) and injected i.p. at 18 mg/kg. Animals were sacrificed 1.5 hours later to check immediate early gene expression, or 24 hours later to check F-RAM and N-RAM expression.
In experiments using Fos-CreER mice, tamoxifen (Sigma) was dissolved in corn oil (Sigma) overnight at 37 °C (15 mg/mL) and injected i.p at 150 mg/kg 24 hours prior to behavioral assays. Tamoxifen solution was stored at 4 °C for no more than 24 hours before use.
Clozapine N-oxide (CNO, Tocris) was dissolved in DMSO (Sigma) at 20 mM and stored at −20 °C. Before experiments, stock CNO was diluted in saline to the desired concentration, 0.4 mg/mL for hM4Di or 0.05 mg/mL for hM3Dq. The same amount of DMSO was diluted in saline as vehicle control. Fresh CNO was injected i.p. at 4 mg/kg for hM4Di or 0.5 mg/kg for hM3Dq 30 minutes before memory recall. CNO doses were determined by pilot experiments and are similar to published studies. For the control experiment testing the effects of CNO injection alone without DREADD expression, a 4 mg/kg dose was used. Diluted CNO solution was kept at 4 °C for up to 24 hours before use.
Contextual fear conditioning (CFC)
Prior to CFC, mice were handled daily in a holding room for 3 days. For experiments with Dox-dependent ensemble labeling, on the third handling day the Dox diet was replaced by regular diet (off Dox) after the last handling session. In control experiments testing reporter expression in the presence of Dox (CFC + Dox), animals were kept on Dox diet throughout. Behavioral assays were typically carried out 48 hours after the last handling session.
On the training day, mice were first transported into the holding room and allowed to acclimatize for at least 30 minutes, then transported into the behavioral room and placed in context A: 24 cm (L) × 19 cm (W) × 17.5 cm (H), with steel grid floors and 1% acetic acid (Sigma). Mice were conditioned as follows: animals were allowed to explore the conditioned chamber freely for 4 minutes and received 2 second 0.55 mA foot shocks at 1 minute intervals 3 times, starting at the 58th second. After each experiment, the chamber was cleaned with 70% ethanol and then with water. In experiments to examine whether synaptic features of ensemble neurons depend on memory strength, no foot shock, weak foot shocks (0.35 mA), or strong foot shocks (0.55 mA, the same as other CFC experiments) were delivered. For experiments with Dox-dependent ensemble labeling, animals were switched back to Dox diet 24 hours after CFC if subsequent behavioral experiments were needed.
To test contextual fear memory discrimination-generalization, 24–48 hours after CFC conditioned mice were returned to the same behavioral room and placed in the conditioned context (A), a similar context (B) or a distinct context (C) for memory recall for 4 minutes. Context B shares all features with context A except that a soft pad insert is used as the floor. Context C uses a different chamber that has white opaque walls, is 32 cm (L) × 25 cm (W) × 32 cm (H), has a soft padded floor, and is scented with 0.25% benzaldehyde (Sigma). In some of the experiments, animals were tested for memory recall in only one context, either A, B or C, and sacrificed 1.5 hours later for immunohistochemistry. In chemogenetics and fiber photometry experiments, the same animals were tested in multiple contexts in order to examine discrimination between contexts. In those cases, fear conditioned animals were tested in contexts B and A on the first day (with at least 6 hours in between), and contexts C and A on the following day.
Contextual fear memory expression was measured by manually scoring freezing behavior over the 4 minute recall period, sampling every 5 seconds, with freezing defined as absence of movement for at least 1 second. Discrimination indices (DIs) are calculated as Freezing[(A-B)/(A+B)] when comparing freezing levels in contexts A and B, or Freezing[(A-C)/(A+C)] when comparing freezing levels in contexts A and C. Scorers were blind to the experimental conditions. For example, for chemogenetics experiments researcher 1 would randomly assign CNO or Veh treatments to the animals and perform the drug injection; researcher 2 would carry out the behavioral testing and quantify the freezing behaviors without knowing the treatments.
Immunohistochemistry and confocal imaging
Animals were either perfused or drop fixed with 4% paraformaldehyde (PFA, Sigma) in PBS. For experiments examining IEG expression, animals were sacrificed 1.5 hours after the last behavioral test. Coronal sections were cut at 50 μm thickness. For typical immunohistochemistry experiments, brain slices were washed in PBS for 10 minutes 3 times and blocked for 2 hours at room temperature with blocking solution: 1% Triton X-100 (Sigma) and 10% goat serum (Invitrogen) in PBS. Slices were then incubated with primary antibody overnight at 4 °C in the antibody solution: 0.1% Triton X-100 and 5% goat serum in PBS. Next day slices were washed in PBS for 10 minutes 3 times and incubated with secondary antibody for 2 hours at room temperature in antibody solution. Finally, slices were washed in PBS for 10 minutes 3 times and mounted with DAPI Fluoromount-G (Southern Biotech). When staining GAD67, a minimum-detergent protocol was adopted from a previous study (Dimidschstein et al., 2016): after initial washing with PBS following PFA fixation, brain slices were permeabilized with 0.1% Triton X-100 for 30 minutes. Slices were then blocked and stained in a detergent-free solution: 3% bovine serum albumin (Sigma) and 5% goat serum in PBS. In this case slices were incubated with anti-GAD67 primary antibody for 48 hours at room temperature.
Antibodies used and dilutions are as follows: rabbit anti-Npas4 (1:10,000, home-made), rabbit anti-Fos (1:1,000, Santa Cruz), rabbit anti-Fos (1:1,000, Synaptic Systems), guinea pig anti-Arc (1:1,000, Synaptic Systems), rabbit anti-mGluR2/3(1:500, Millipore), mouse IgG2a anti-GAD67 (1:500, Millipore) and Alexa 488, 555, 647 secondary antibodies (1:500, Invitrogen).
All images were acquired using an Olympus Fluoview FV1000 confocal microscope. For most cell counting experiments high-resolution z-stack images of the DG were acquired using either a 20× or a 60× oil-immersion objective. Numbers of positive cells were determined manually from 6–12 images per animal as previously described (Sørensen et al., 2016). Analyses were performed by investigators who were blind to the experimental conditions.
Electrophysiology
Mice were subjected to electrophysiology 24–48 hours after CFC. Mice were anesthetized and perfused with carbogenated (95% O2, 5% CO2) ice-cold cutting solution containing (in mM): 210 sucrose, 2.5 KCl, 1.24 NaH2PO4, 8 MgCl2, 1 CaCl2, 26 NaHCO3, 20 Glucose and 1.3 sodium-L-ascorbate, ~340 mOsm osmolarity, pH 7.3. Brains were immediately dissected out. 300 μm transverse slices were cut using a vibratome (VT1200, Leica). Slices were transferred to warm (32 °C) carbogenated recovery solution, containing 50% cutting solution and 50% ACSF. ACSF contains (in mM): 119 NaCl, 2.5 KCl, 1.24 NaH2PO4, 1.3 MgCl2, 2.5 CaCl2, 26 NaHCO3, and 10 glucose, 305 mOsm osmolarity, pH 7.3. Slices were recovered at 32 °C for 15 minutes, then transferred to another chamber filled with room temperature (~23 °C) ACSF. Slices were kept in ACSF for at least 30 minutes before recording.
All recordings were performed in a chamber perfused with carbogenated ACSF at room temperature at a flow rate of 2 mL/min. Whole-cell patch-clamp recordings were performed using borosilicate glass pipettes (3–6 MΩ tip resistance). mEPSCs were pharmacologically isolated with 0.5 μM tetrodotoxin (TTX, Tocris) and 50 μM picrotoxin (Tocris), and recorded from cells voltage clamped at −70 mV using Cs-based internal solution containing (in mM): 130 CsMeSO3, 10 phosphocreatine, 1 MgCl2, 10 HEPES, 0.2 EGTA, 4 Mg-ATP, 0.5 Na-GFP, pH adjusted to 7.25 with CsOH, 295 mOsm osmolarity. mIPSCs were isolated with 0.5 μM TTX, 50 μM APV (Tocris) and 20 μM DNQX (Tocris), and recorded from cells voltage clamped at −70 mV using high-Cl internal solution containing (in mM): 103 CsCl, 12 CsMeSO3, 5 TEA-Cl, 10 HEPES, 4 Mg-ATP, 0.5 Na-GTP, 0.5 EGTA, 1 MgCl2, pH adjusted to 7.25 with CsOH, 295 mOsm osmolarity. Intrinsic excitability and passive membrane properties were recorded in the current clamp mode with the following internal solution (in mM): 130 K+-D-gluconate, 10 KCl, 10 HEPES, 4 Mg-ATP, 0.5 Na-GTP, 0.2 EGTA, 1 MgCl2, pH adjusted to 7.25 with CsOH, 295 mOsm osmolarity. To determine the excitability of cells, membrane potentials were measured in response to intracellular injection of step currents (500 ms duration, magnitudes ranging from −20 to 200 pA in steps of 20 pA). Data were collected using a Multiclamp 700B (Molecular Devices), filtered at 3 kHz and digitized at 10 kHz using a Digidata 1440A and Clampex 10.2 software (Molecular Devices). Recordings with access resistance greater than 30 MΩ or changes exceeding 15% were discarded.
To record evoked EPSCs (eEPSCs) from different excitatory pathways to the DG, stimulating theta glass pipettes (World Precision Instruments) with broken tips (~10–20 μm) were filled with ACSF and placed in the inner molecular layer (< 40 um from the border of the granule cell layer), middle molecular layer (middle third of the molecular layer) or outer molecular layer (outer third of the molecular layer) to stimulate different excitatory inputs onto DG granule cells (GCs). Bipolar square-wave voltage pulses (100–200 μs, 5–25 V) were delivered through a stimulus isolator (ISO-Flex, AMPI). Stimulation intensity was adjusted to obtain comparable amplitudes of AMPA eEPSCs (50–150 pA) across conditions. Paired whole-cell recordings were carried out on labeled (F-RAM+ or N-RAM+) and unlabeled neighboring neurons. Cells were first voltage clamped at −70 mV to record AMPA currents and paired pulses with a 100 ms interval were stimulated. The peak amplitude of the first pulse was used to calculate eEPSC amplitudes. The cells were subsequently voltage clamped at +40 mV and a single pulse was stimulated. NMDA currents were measured at 100 ms after the onset of stimulation. Values were averaged from 10 sweeps repeated every 20–30 seconds. Picrotoxin (50 μM) was added throughout the recording to block GABAA receptors. To confirm the specificity of stimulation, the mGluR2/3 agonist DCG-IV (1 μM) was routinely bath applied at the end of experiments, which reduced the amplitudes of eEPSCs selectively evoked by stimulating the MEC-DG pathway (medial perforant path, MPP).
To record optically evoked IPSCs (oIPSCs) or optically evoked EPSCs (oEPSCs) in the DG, 2 ms 470 nm LED stimulations at 0.1, 0.25, 0.50, 0.75, 1.00, 1.50 mW/mm2, generated from a mounted LED (Thorlabs), were controlled by TTL input and delivered through the 40× objective. Paired whole-cell recordings were carried out on labeled (F-RAM+ or N-RAM+) and unlabeled neighboring neurons. oIPSCs were recorded in the presence of DNQX (20 μM) and APV (50 μM) from cells voltage clamped at 0 mV with Cs-based internal solution. In the experiments where CCK+ interneurons were specifically stimulated, oIPSCs were recorded from cells voltage clamped at −70 mV with high-Cl internal solutions to minimize the effects of depolarization-induced suppression of inhibition (DSI) from CCK+ interneurons. Average peak responses were calculated from 10 sweeps, stimulated every 10–15 seconds. Paired light pulses with an interval of 200 ms were given at 0.50 mW/mm2 to measure paired-pulse ratios. To block transmission from individual interneuron subtypes, ω-agatoxin IVa (AgTx, 0.5 μM, Bachem), ω-conotoxin GVIa (CnTx, 1 μM, Bachem) or WIN 55,212–2 (WIN, 10 μM, Tocris) were bath applied in the ACSF during oIPSC recordings. oEPSCs were recorded in the presence of picrotoxin (50 μM), TTX (0.5 μM) and 4-aminopyridine (100 μM) from cells voltage clamped at −70 mV with Cs-based internal solution.
To characterize the CCK+ interneurons, depolarization-induced suppression of inhibition (DSI) was induced by elevating the holding potential of the postsynaptic GCs from −70 mV to 0 mV for 5 seconds. oIPSCs were measured in GCs voltage clamped at −70 mV and recorded with high-Cl− internal solution in the presence of APV (50 μM) and DNQX (20 μM). oIPSCs recorded before and 200 ms after depolarization, averaged from 10 sweeps, were used to calculate DSI. To examine asynchronous release from CCK+ interneurons, trains of ten action potentials (50 Hz) were evoked in the presynaptic CCK+ interneurons and unitary IPSCs (uIPSCs) recorded in the postsynaptic GCs voltage clamped at −70 mV. uIPSCs were also measured with WIN (10 μM) in the ACSF.
Data were analyzed using MiniAnalysis (Synaptosoft) and Clampfit 10.2 (Molecular Devices) by investigators that were blind to the experimental conditions.
Fiber photometry
A customized fiber photometry system was used. A 473 nm diode laser (OEM Laser Systems) was controlled by a function generator (Agilent, 33599B Series, 400 Hz, square wave). The light was coupled to a 400 μm 0.48 NA optical fiber (Thorlabs) using a 40× 0.65 NA microscope objective (Olympus) and a fiber launch (Thorlabs). Optical fiber with the same diameter was glued to a ceramic ferrule (Thorlabs) and implanted into the mice. Before experiments, implants were connected to the laser with ceramic split mating sleeves (Thorlabs). Laser intensity was set to a level that generates fluorescence signals of about 50% of the maximum detectable values. The GCaMP6f fluorescence signal was collected by the implanted optical fiber and passed through the objective and dichroic mirror. The signal was then transmitted through a longpass filter (Semrock) onto a NewFocus 2151 femtowatt silicon photoreceiver (Newport, DC High Mode). The photoreceiver was connected to a lock-in amplifier (SR830 DSP, Stanford Research Systems, 8 ms time constant) and data were recorded with customized LabVIEW software (National Instruments).
Fear conditioned animals were subjected to fiber photometry experiments 48 hours after training. On the testing day, after connecting their fiber implants, animals were allowed 10–20 minutes to recover in their home cages. Animals were then gently transferred from the home cage to context A, B or C and allowed free exploration for 4 minutes. Animals were then returned to their home cage and allowed to recover for 30 minutes before being transferred to the next context. The order of context exposure was randomized. GCaMP6f signal was collected throughout the experiment.
Fiber photometry data were analyzed with a custom MATLAB (MathWorks) program. Average ΔF/F signals over the entire 4-minute recall session were used to reflect neural activity. Signals were calculated as ΔF/F by normalizing the measured values during recall to the baseline fluorescence values. Baseline values were calculated as the median of the signals during the 60 seconds in the home cage prior to each context exposure. Signals recorded during physical transfer from the home cage to the contexts (30 seconds) were removed from the analysis.
Optogenetics
To optogenetically inhibit the MEC-DG pathway, animals were bilaterally implanted with optical fibers (Thorlabs) in the DG after viral injection into the MEC. After CFC, animals were recalled in different contexts with optogenetic inhibition. Laser stimulation (561 nm, constant, 10 mW) was generated using a 561 nm DPSS laser (Opto Engine LLC) and delivered bilaterally to the DG through 200 μm 0.22 NA optical fibers (Thorlabs). A rotatory joint (Thorlabs) was used to allow animals to move freely. The laser was constantly on during the entire 4-minute recall session.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses were performed using Prism (GraphPad Software). Means of two groups were compared using either a Mann-Whitney test when the samples were not matched, or a Wilcoxon matched-pairs signed-rank test when the samples were matched. One-way ANOVA was used to compare group means between more than two groups, followed by either Dunnett’s multiple comparisons test when all other groups were compared to the control group, or Tukey’s multiple comparisons test when means of each pair of groups were compared. Two-way ANOVA was used to compare group means when there were two or more variables, followed by either Tukey’s or Sidak’s multiple comparisons tests (as noted in the figure legends). Depending on the experimental design, two-way ANOVA, two-way mixed ANOVA or two-way repeated measures ANOVA was used. A one-sample t-test was used to compare group means to hypothetical values. Linear regression was used to examine the correlation between two variables. All comparisons are two-sided. Statistical significance was determined as follows: *p < 0.05, **p < 0.01, ***p < 0.001. Data are shown as mean ± SEM. The statistical details of all experiments, including the number of samples, statistical tests, p values, can be found in Table S1.
DATA AND CODE AVAILABILITY
Data and custom MATLAB code are available upon request from the Lead Contact.
Supplementary Material
Characterization of the F-RAM and N-RAM Reporters, Related to Figure 1.
(A) The promoters of the two reporters, PF-RAM and PN-RAM, were based on the existing RAM system. Both promoters contain 4 tandem repeats of 24-bp enhancer modules and a minimal promoter from the human FOS gene. The enhancer module in PF-RAM contains the Activator Protein 1 (AP1) site (TGANTCA), a consensus binding sequence for FOS/JUN family transcription factors. The enhancer module in PN-RAM contains the NPAS4 Responsive Element (NRE) (TCGTG), a consensus binding motif for NPAS4.
(B) Experimental scheme. Cultured mouse hippocampal neurons were infected with AAVs carrying either the F-RAM or N-RAM reporter and a control virus (AAV-hSyn-GFP) at 7 days in vitro (DIV), stimulated with bicuculline (Bic, 50 μM) and 4-aminopyridine (4-AP, 250 μM) at 14 DIV and harvested for immunocytochemistry and imaging at 15 DIV.
(C) Representative images showing expression of the reporters under three conditions: Stim, stimulated with Bic and 4-AP as described above; Control, without stimulation; Stim + Dox, same as Stim but in the presence of Dox (40 ng/mL).
(D) Quantification of the percentage of mKate2+ neurons among infected neurons (AAV-hSyn-GFP+). Both F-RAM and N-RAM were substantially induced by stimulation and inhibited by Dox. One-way ANOVA, Dunnett’s test, n = 6 per condition.
(E) F-RAM labeled comparable numbers of hilar cells under home cage (HC) and contextual fear conditioning (CFC) conditions (Mann-Whitney test, n = 3 per group). White arrows indicate F-RAM+ cells in the hilus and dashed lines outline the hilus region.
(F) N-RAM labeled comparable numbers of hilar cells under HC and CFC conditions (Mann-Whitney test, n = 4 per group). White arrows indicate N-RAM+ cells in the hilus and dashed lines outline the hilus region.
(G) Representative images showing that F-RAM+ cells co-localize with the mossy cell marker mGluR2/3 (white arrows) but not the GABAergic interneuron marker GAD67.
(H) The majority of CFC-induced F-RAM+ and N-RAM+ cells in the hilus were mossy cells (mGluR2/3+, n = 3 per group).
(I) Percentage of DG cells labeled with F-RAM or N-RAM under the CFC condition that were in the hilus (n = 3–4).
(J) Fosflx/flx animals were injected with AAVs expressing Cre or GFP (control) into the DG. Seizures were induced by kainic acid (18 mg/kg) via i.p. injection and animals sacrificed 1.5 hours later to stain for FOS or NPAS4. Fos expression was abolished in Fosflx/flx animals infected with Cre-expressing virus, indicating effective Fos deletion.
(K) Same as (J), but with Npas4flx/flx animals. Npas4 expression was abolished in Npas4flx/flx animals infected with Cre-expressing virus, indicating effective Npas4 deletion.
Data are shown as mean ± SEM. ***p < 0.001.
Figure S2. Electrophysiological Properties of F-RAM+ and N-RAM+ Neurons, Related to Figure 2.
(A) Example traces showing membrane potentials of F-RAM+ and N-RAM+ neurons compared to their unlabeled neighbors, in response to −20, 0, 20, 100 and 200 pA current injections.
(B) Input-output curves of firing rates versus injected currents for F-RAM+ and F-RAM− neurons (left), or N-RAM+ and N-RAM− neurons (right). F-RAM+ neurons show comparable excitability to F-RAM− neurons (F-RAM−, n = 19; F-RAM+, n = 23). N-RAM+ neurons show decreased excitability compared to N-RAM− neurons (n = 16, 16). Two-way mixed ANOVA, Sidak’s test.
(C) F-RAM+ neurons (F-RAM−, n = 19; F-RAM+, n = 23) and N-RAM+ neurons (n = 16, 16) show similar resting membrane potentials and action potential (AP) thresholds to their neighboring unlabeled cells. Mann-Whitney test.
(D) Whole-cell patch-clamp recordings were carried out on hippocampal slices from animals in which the F-RAM and Fos-CreER ensembles were co-labeled. Neurons labeled by F-RAM, Fos-CreER or both showed increased mEPSC frequencies. One-way ANOVA, Dunnett’s test comparing to the F-RAM− Fos-CreER− group, n = 17, 15, 11, 16 (from left to right). mIPSC frequencies were similar for all groups (n = 13, 12, 12, 10, from left to right).
(E) Whole-cell patch-clamp recordings were carried out on hippocampal slices from animals in which the N-RAM and Fos-CreER ensembles were co-labeled. Neurons labeled by Fos-CreER, but not those labeled by N-RAM, showed increased mEPSC frequencies (n = 12, 11, 13, 17 from left to right). Neurons labeled by N-RAM, including the small population labeled by both N-RAM and Fos-CreER, showed increased mIPSC frequencies (n = 19, 17, 14, 25 from left to right). Data are shown as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
Figure S3. The Memory Discrimination-Generalization Assay and the Activity Patterns of the F-RAM and N-RAM Ensembles Measured by Fos Induction During the Assay, Related to Figure 3.
(A) Images of contexts A, B and C.
(B) Detailed features of the three contexts. Context A is made of transparent plastic walls with a steel grid floor for foot-shock delivery. Acetic acid (1%) is used as the odor cue. Context B shares all features with context A except for a different floor: a soft pad insert instead of the steel grid. Context C is a completely different chamber, with a soft padded floor and distinct odor cues (benzaldehyde, 0.25%) and lighting.
(C) Using FOS rather than ARC as the activity marker also showed that similar percentages of F-RAM+ neurons were reactivated (FOS+) in contexts A and B, while fewer N-RAM+ neurons were reactivated during recall in context A than in B. Two-way mixed ANOVA, Tukey’s test, n = 7–9.
Figure S4. Control Experiments for Chemogenetic Manipulation, Related to Figure 4.
(A) Validation of DREADDs in F-RAM+ cells. hM4Di inhibited and hM3Dq activated tagged F-RAM+ cells. Animals were injected i.p. with CNO or vehicle (Veh) 30 minutes before recall in context A. Percentages of ensemble neurons that were reactivated (ARC+) were quantified (two-way mixed ANOVA, Sidak’s test, n = 4 per group).
(B) Experimental scheme to determine the effect of CNO injection alone on memory discrimination-generalization. AAVs expressing mCherry without DREADD (AAV-N-RAM-mCherry) were injected into the DG and mice were subjected to the contextual fear memory discrimination-generalization assay. CNO (4 mg/kg) and vehicle (Veh) were injected i.p. 30 minutes before memory recall.
(C) CNO and Veh groups showed similar discrimination between contexts A and B (Veh, n = 19; CNO, n = 18), as well as A and C (n = 13, 15). Two-way mixed ANOVA with Sidak’s test was used for the freezing levels, Mann-Whitney test for comparing the discrimination indices, and one-sample t-test to compare discrimination indices with zero.
(D) Experimental scheme to inhibit cells that are labeled under the HC condition during memory discrimination-generalization. Animals remained in their home cages during the off-Dox window to label cells activated by background activity, including hilar cells and other granule cells. Labeled cells (hM4Di+) were inhibited during memory recall.
(E) Inhibiting HC-labeled F-RAM+ cells did not affect discrimination between contexts A and B (Veh, n = 9; CNO, n = 9) or A and C (n = 9, 9).
(F) Inhibiting HC-labeled N-RAM+ cells did not affect discrimination between contexts A and B (n = 10, 9) or A and C (n = 9, 10).
(G) Schematics for expression of the inhibitory DREADD hM4Di or excitatory DREADD hM3Dq in Fos-expressing ensembles after CFC. Fos-expressing ensembles were labeled by AAV-pFos-d2tTA, which uses an engineered Fos promoter to drive d2tTA.
(H) Representative image showing expression of hM4Di-mCherry in the Fos-expressing ensemble after CFC.
(I) Experimental scheme to probe the function of the Fos-expressing ensemble in memory discrimination-generalization.
(J) Inhibition of the Fos-expressing ensemble enhanced discrimination between contexts A and B (Veh, n = 18; CNO, n = 14), but not A and C (n = 17, 14).
(K) Activation of Fos-expressing ensembles reduced discrimination between contexts A and B (n = 14, 15) and between A and C (n = 14, 15).
Two-way mixed ANOVA with Sidak’s test was used for freezing levels, Mann-Whitney test for comparing discrimination indices and one-sample t-test to compare discrimination indices with zero. Data are shown as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
Figure S5. F-RAM+ Neurons Showed Increased oEPSC Amplitudes upon Optogenetic Stimulation of the MPP, but Unchanged oIPSCs upon Optogenetic Stimulation of Local Interneurons, Related to Figure 5.
(A) Experimental scheme. To target the LPP and MPP pathways, animals were co-injected with AAV-CaMKII-Cre and AAV- EF1α-DIO-ChR2-EYFP to express ChR2 in the LEC or MEC, and also injected with AAV-F-RAM-mKate2 in the DG. To target the MCF pathway, AAVCaMKII-Cre and AAV- EF1α-DIO-ChR2-EYFP were co-injected unilaterally into the contralateral DG, which labeled the hilar mossy cells that send axonal projections to the ipsilateral DG through MCFs. AAV-F-RAM-mKate2 was injected unilaterally into the ipsilateral DG, where whole-cell patch-clamp recordings were carried out.
(B) Representative images showing ChR2 and F-RAM expression.
(C) F-RAM+ neurons had increased optically evoked EPSC (oEPSC) amplitudes upon MPP but not LPP or MCF stimulation (Wilcoxon signed-rank test, n = 16, 11, 10 pairs for LPP, MPP and MCF). Tetrodotoxin (TTX, 0.5 μM) and 4-aminopyridine (4-AP, 100 μM) were added to isolate monosynaptic inputs. Picrotoxin (50 μM) was added to block GABAA receptors. Data are shown as mean ± SEM. **p < 0.01.
(D) Example traces showing oIPSCs recorded simultaneously in F-RAM+ and F-RAM− neurons stimulated by 2 ms of 0.10, 0.25, 0.50, 0.75, 1.00, 1.50 mW/mm2 470 nm LED light pulses. oIPSCs were elicited by optogenetically stimulating ChR2-expressing GABAergic interneurons in Gad2-Cre mice.
(E) oIPSC amplitudes were similar in the two groups (two-way repeated measures ANOVA, Sidak’s test, n = 12 pairs).
Figure S6. Induced FOS Expression Does Not Affect mIPSCs and Induced NPAS4 Expression Does Not Affect mEPSCs, Related to Figure 6.
(A) mIPSC frequencies and amplitudes were similar in neurons with (EYFP+) and without (EYFP−) induced FOS expression (FOSind). Mann-Whitney test; EYFP−, n = 18; EYFP+, n = 16.
(B) mEPSC frequencies and amplitudes were similar in neurons with (EYFP+) and without (EYFP−) induced NPAS4 expression (NPAS4ind, n = 17, 19).
(C) There is no significant difference in the number of F-RAM+ cells under no shock, weak shock and strong shock conditions (one-way ANOVA, n = 3–4).
(D) There is no significant difference in the number of N-RAM+ cells under no shock, weak shock and strong shock conditions (n = 4 per condition).
Data are shown as mean ± SEM.
Figure S7. Validation of Manipulation of MEC and CCK+ Interneurons, Related to Figure 7.
(A) Experimental scheme. Mice were co-injected with AAV-CaMKII-Cre and AAV-hSyn-DIO-hM4Di-mCherry to express the inhibitory DREADD hM4Di in MEC excitatory neurons (CaMKII+). Mice were fear conditioned in context A, memory was recalled 24 hours later and animals sacrificed 1.5 hours after recall. CNO (4 mg/kg) or vehicle (Veh) was injected i.p. 30 minutes before recall. Neuronal activity was examined by FOS staining.
(B) Representative images of the MEC (dashed lines) showing hM4Di and FOS expression. White arrows indicate double-positive cells.
(C) Chemogenetic inhibition robustly suppressed, but did not completely abolish, the activity of MEC projection neurons. Percentages of hM4Di-mCherry+ cells that were also FOS+ were quantified (Mann-Whitney test, n = 4 per group).
(D) Representative images showing FOS+ cells (white arrows) in the DG GCL (dashed lines) after recall in context A.
(E) Optogenetic inhibition of the MEC-DG pathway reduced the number of FOS+ cells in DG but not in CA1 after recall (Mann-Whitney test, n = 4 per group).
(F) Experimental scheme. Memory discrimination-generalization was examined in the absence of 561 nm laser stimulation (light OFF). Animals were injected with NpHR or EYFP in the MEC and implanted with optical fibers in the DG.
(G) In the absence of laser stimulation, NpHR and EYFP groups showed comparable discrimination between the similar contexts A and B (two-way mixed ANOVA with Sidak’s test was used for freezing levels, Mann-Whitney test for comparing discrimination indices, and one-sample t-test to compare discrimination indices with zero; EYFP, n = 11; NpHR, n = 13).
(H) Experimental scheme. Putative CCK+, PV+ and SST+ interneurons were identified by injecting a reporter virus (AAV-Dlx5/6-DIO-ChR2-EYFP) into CCK-Cre, PV-Cre and SST-Cre mice. Whole-cell patch-clamp recordings were carried out on labeled cells to examine their firing properties.
(I) CCK+ interneurons (identified by EYFP expression) showed the strongest firing adaptation among the three interneuron subtypes. Example traces showing the membrane potentials of CCK+, PV+ and SST+ interneurons in response to −50, 0, 50, and 500 pA current injections.
(J) Schematic for measurement of depolarization-induced suppression of inhibition (DSI). ChR2 (AAV-Dlx5/6-DIO-ChR2-EYFP) was expressed in CCK+, PV+ or SST+ interneurons to induce optically evoked IPSCs (oIPSCs).
(K) DSI was observed in the CCK+ interneuron (CCK) to granule cell (GC) synapses. oIPSCs were induced by 2 ms 470 nm LED light stimulation and recorded in GCs voltage clamped at −70 mV, with high-Cl− internal solution, in the presence of APV (50 μM) and DNQX (20 μM). DSI was induced by elevating the holding potential of GCs to 0 mV for 5 s. oIPSCs recorded before (oIPSC1) and 200 ms after (oIPSC2) depolarization, averaged from ten sweeps, were compared to calculate DSI.
(L) Quantification of DSI (one-way ANOVA, one-sample t-test for individual columns, n = 6 per group).
(M) Asynchronous releases were observed in the CCK to GC synapses, and were blocked by bath application of WIN 55,212–2 (WIN, 10 uM). Paired whole-cell patch-clamp recordings targeting presynaptic CCK+ interneurons and postsynaptic GCs were carried out. Superimposed traces (black) from 15 sweeps are shown. Trains of ten action potentials (50 Hz) were evoked in the presynaptic CCK+ interneurons, and unitary IPSCs (uIPSCs) were recorded in the postsynaptic GCs voltage clamped at −70 mV. uIPSCs were recorded with high-Cl− internal solution in the presence of APV (50 μM) and DNQX (20 μM). Red traces show superimposed traces of 15 sweeps after bath application of WIN.
Data are shown as mean ± SEM. *p < 0.05, **p < 0.01.
Highlights.
Functionally distinct neuronal ensembles exist within a single memory engram
Fos- and Npas4-dependent ensembles undergo distinct synaptic modifications after CFC
Fos- and Npas4-dependent ensembles drive memory-guided behaviors in opposite directions
Memory generalization and discrimination respectively require MEC and CCK+ interneurons
Acknowledgments:
We thank C. M. Fletcher and J. Radulovic for critical reading of the manuscript; M. Xu for providing Fosflx/flx mice; G. Fishell for sharing Dlx5/6 constructs; L. Paul, Y. Zhang, J. Xue, T.D. Yelhekar and F.-J. Weng for technical assistance; and W. Xu, D.S. Roy, P.E. Castillo, G. Vargish, E.LQ. Lee, J. Guo, and the entire Lin laboratory for helpful suggestions.
Funding: This work was funded by a Henry E. Singleton Fellowship, Leventhal Graduate Student Fellowship and Hugo Shong Fellowship (X.S.), Simons Postdoctoral Fellowship (S.R.), Beijing Municipal Science & Technology Commission (Z181100001518001, X.Z.), MIT Great China Fund for Innovation (Y.L. & X.Z.), NIH BRAIN Initiative 1R01MH111872 (P.O.A.), Solomon Buchsbaum Research Foundation Award and NIH grants MH091220, NS088421 and DC014701 (Y.L.). The authors declare no competing financial interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests: The authors have no competing interests.
References
- Amaral DG, and Witter MP (1989). The three-dimensional organization of the hippocampal formation: A review of anatomical data. Neuroscience 31, 571–591. [DOI] [PubMed] [Google Scholar]
- Barth AL (2007). Visualizing circuits and systems using transgenic reporters of neural activity. Current opinion in neurobiology 17, 567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth AL, Gerkin RC, and Dean KL (2004). Alteration of neuronal firing properties after in vivo experience in a FosGFP transgenic mouse. The Journal of neuroscience : the official journal of the Society for Neuroscience 24, 6466–6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartos M, and Elgueta C (2012). Functional characteristics of parvalbumin- and cholecystokinin-expressing basket cells. J Physiol-London 590, 669–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier BE, Lacagnina AF, Ayoub A, Shue F, Zemelman BV, Krasne FB, and Drew MR (2017). Dentate Gyrus Contributes to Retrieval as well as Encoding: Evidence from Context Fear Conditioning, Recall, and Extinction. Journal of Neuroscience 37, 6359–6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai DJ, Aharoni D, Shuman T, Shobe J, Biane J, Song W, Wei B, Veshkini M, La-Vu M, Lou J, et al. (2016). A shared neural ensemble links distinct contextual memories encoded close in time. Nature 534, 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo PE, Younts TJ, Chavez AE, and Hashimotodani Y (2012). Endocannabinoid Signaling and Synaptic Function. Neuron 76, 70–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelaru MI, and Dragoi V (2008). Efficient coding in heterogeneous neuronal populations. Proceedings of the National Academy of Sciences of the United States of America 105, 16344–16349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Sim SE, Kim JI, Choi DI, Oh J, Ye S, Lee J, Kim T, Ko HG, Lim CS, et al. (2018). Interregional synaptic maps among engram cells underlie memory formation. Science 360, 430–435. [DOI] [PubMed] [Google Scholar]
- Danielson NB, Turi GF, Ladow M, Chavlis S, Petrantonakis PC, Poirazi P, and Losonczy A (2017). In Vivo Imaging of Dentate Gyrus Mossy Cells in Behaving Mice. Neuron 93, 552-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Mayford M, and Gage FH (2013). Selection of distinct populations of dentate granule cells in response to inputs as a mechanism for pattern separation in mice. eLife 2, e00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny CA, Kheirbek MA, Alba EL, Tanaka KF, Brachman RA, Laughman KB, Tomm NK, Turi GF, Losonczy A, and Hen R (2014). Hippocampal memory traces are differentially modulated by experience, time, and adult neurogenesis. Neuron 83, 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl GW, Hon OJ, Leutgeb S, and Leutgeb JK (2017). Grid and Nongrid Cells in Medial Entorhinal Cortex Represent Spatial Location and Environmental Features with Complementary Coding Schemes. Neuron 94, 83–92.e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimidschstein J, Chen Q, Tremblay R, Rogers SL, Saldi GA, Guo L, Xu Q, Liu R, Lu C, Chu J, et al. (2016). A viral strategy for targeting and manipulating interneurons across vertebrate species. Nature neuroscience. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunsmoor JE, and Paz R (2015). Fear Generalization and Anxiety: Behavioral and Neural Mechanisms. Biological psychiatry 78, 336–343. [DOI] [PubMed] [Google Scholar]
- Eichenbaum H (2004). Hippocampus: cognitive processes and neural representations that underlie declarative memory. Neuron 44, 109–120. [DOI] [PubMed] [Google Scholar]
- Flavell SW, and Greenberg ME (2008). Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annual review of neuroscience 31, 563–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann A, Hvalby O, Jensen V, Strekalova T, Zacher C, Layer LE, Kvello A, Reschke M, Spanagel R, Sprengel R, et al. (2003). Impaired long-term memory and NR2A-type NMDA receptor-dependent synaptic plasticity in mice lacking c-Fos in the CNS. The Journal of neuroscience : the official journal of the Society for Neuroscience 23, 9116–9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankland PW, Cestari V, Filipkowski RK, McDonald RJ, and Silva AJ (1998). The dorsal hippocampus is essential for context discrimination but not for contextual conditioning. Behavioral Neuroscience 112, 863–874. [DOI] [PubMed] [Google Scholar]
- Freund TF, and Buzsaki G (1996). Interneurons of the hippocampus. Hippocampus 6, 347–470. [DOI] [PubMed] [Google Scholar]
- Fyhn M, Hafting T, Treves A, Moser MB, and Moser EI (2007). Hippocampal remapping and grid realignment in entorhinal cortex. Nature 446, 190–194. [DOI] [PubMed] [Google Scholar]
- Garner AR, Rowland DC, Hwang SY, Baumgaertel K, Roth BL, Kentros C, and Mayford M (2012). Generation of a synthetic memory trace. Science 335, 1513–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandour K, Ohkawa N, Fung CCA, Asai H, Saitoh Y, Takekawa T, Okubo-Suzuki R, Soya S, Nishizono H, Matsuo M, et al. (2019). Orchestrated ensemble activities constitute a hippocampal memory engram. Nature communications 10, 2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GoodSmith D, Chen XJ, Wang C, Kim SH, Song HJ, Burgalossi A, Christian KM, and Knierim JJ (2017). Spatial Representations of Granule Cells and Mossy Cells of the Dentate Gyrus. Neuron 93, 677-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewe BF, Grundemann J, Kitch LJ, Lecoq JA, Parker JG, Marshall JD, Larkin MC, Jercog PE, Grenier F, Li JZ, et al. (2017). Neural ensemble dynamics underlying a long-term associative memory. Nature 543, 670-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosmark AD, and Buzsáki G (2016). Diversity in neural firing dynamics supports both rigid and learned hippocampal sequences. Science 351, 1440–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosso A, Santoni G, Manassero E, Renna A, and Sacchetti B (2018). A neuronal basis for fear discrimination in the lateral amygdala. Nature communications 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenthner CJ, Miyamichi K, Yang HH, Heller HC, and Luo L (2013). Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron 78, 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo NN, Soden ME, Herber C, Kim MT, Besnard A, Lin PY, Ma X, Cepko CL, Zweifel LS, and Sahay A (2018). Dentate granule cell recruitment of feedforward inhibition governs engram maintenance and remote memory generalization. Nat Med 24, 438-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, Timlin JA, Roysam B, McNaughton BL, Worley PF, and Barnes CA (2005). Mapping behaviorally relevant neural circuits with immediate-early gene expression. Current opinion in neurobiology 15, 599–606. [DOI] [PubMed] [Google Scholar]
- Hainmueller T, and Bartos M (2018). Parallel emergence of stable and dynamic memory engrams in the hippocampus. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales JB, Schlesiger MI, Leutgeb JK, Squire LR, Leutgeb S, and Clark RE (2014). Medial Entorhinal Cortex Lesions Only Partially Disrupt Hippocampal Place Cells and Hippocampus-Dependent Place Memory. Cell reports 9, 893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JH, Kushner SA, Yiu AP, Hsiang HL, Buch T, Waisman A, Bontempi B, Neve RL, Frankland PW, and Josselyn SA (2009). Selective erasure of a fear memory. Science 323, 1492–1496. [DOI] [PubMed] [Google Scholar]
- Hartzell AL, Martyniuk KM, Brigidi GS, Heinz DA, Djaja NA, Payne A, and Bloodgood BL (2018). NPAS4 recruits CCK basket cell synapses and enhances cannabinoid-sensitive inhibition in the mouse hippocampus. eLife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimotodani Y, Nasrallah K, Jensen KR, Chavez AE, Carrera D, and Castillo PE (2017). LTP at Hilar Mossy Cell-Dentate Granule Cell Synapses Modulates Dentate Gyrus Output by Increasing Excitation/Inhibition Balance. Neuron 95, 928-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefft S, and Jonas P (2005). Asynchronous GABA release generates long-lasting inhibition at a hippocampal interneuron-principal neuron synapse. Nature neuroscience 8, 1319–1328. [DOI] [PubMed] [Google Scholar]
- Herry C, Ciocchi S, Senn V, Demmou L, Muller C, and Luthi A (2008). Switching on and off fear by distinct neuronal circuits. Nature 454, 600–606. [DOI] [PubMed] [Google Scholar]
- Hess J, Angel P, and Schorpp-Kistner M (2004). AP-1 subunits: quarrel and harmony among siblings. Journal of cell science 117, 5965–5973. [DOI] [PubMed] [Google Scholar]
- Huckleberry KA, Ferguson LB, and Drew MR (2016). Behavioral mechanisms of context fear generalization in mice. Learning & memory 23, 703–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinde S, Zsiros V, Jiang Z, Nakao K, Pickel J, Kohno K, Belforte JE, and Nakazawa K (2012). Hilar mossy cell degeneration causes transient dentate granule cell hyperexcitability and impaired pattern separation. Neuron 76, 1189–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josselyn SA, Kohler S, and Frankland PW (2015). Finding the engram. Nature reviews Neuroscience 16, 521–534. [DOI] [PubMed] [Google Scholar]
- Jun JK, Miller P, Hernández A, Zainos A, Lemus L, Brody CD, and Romo R (2010). Heterogenous Population Coding of a Short-Term Memory and Decision Task. The Journal of Neuroscience 30, 916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanter BR, Lykken CM, Avesar D, Weible A, Dickinson J, Dunn B, Borgesius NZ, Roudi Y, and Kentros CG (2017). A Novel Mechanism for the Grid-to-Place Cell Transformation Revealed by Transgenic Depolarization of Medial Entorhinal Cortex Layer II. Neuron 93, 1480-+. [DOI] [PubMed] [Google Scholar]
- Kawashima T, Kitamura K, Suzuki K, Nonaka M, Kamijo S, Takemoto-Kimura S, Kano M, Okuno H, Ohki K, and Bito H (2013). Functional labeling of neurons and their projections using the synthetic activity-dependent promoter E-SARE. Nature methods 10, 889–895. [DOI] [PubMed] [Google Scholar]
- Kawashima T, Okuno H, and Bito H (2014). A new era for functional labeling of neurons: activity-dependent promoters have come of age. Frontiers in neural circuits 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheirbek MA, Drew LJ, Burghardt NS, Costantini DO, Tannenholz L, Ahmari SE, Zeng H, Fenton AA, and Hen R (2013). Differential control of learning and anxiety along the dorsoventral axis of the dentate gyrus. Neuron 77, 955–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheirbek MA, Klemenhagen KC, Sahay A, and Hen R (2012). Neurogenesis and generalization: a new approach to stratify and treat anxiety disorders. Nature neuroscience 15, 1613–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T, Sun C, Martin J, Kitch LJ, Schnitzer MJ, and Tonegawa S (2015). Entorhinal Cortical Ocean Cells Encode Specific Contexts and Drive Context-Specific Fear Memory. Neuron 87, 1317–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knierim JJ, and Neunuebel JP (2016). Tracking the flow of hippocampal computation: Pattern separation, pattern completion, and attractor dynamics. Neurobiology of learning and memory 129, 38–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn HG, DickinsonAnson H, and Gage FH (1996). Neurogenesis in the dentate gyrus of the adult rat: Age-related decrease of neuronal progenitor proliferation. Journal of Neuroscience 16, 2027–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacagnina AF, Brockway ET, Crovetti CR, Shue F, McCarty MJ, Sattler KP, Lim SC, Santos SL, Denny CA, and Drew MR (2019). Distinct hippocampal engrams control extinction and relapse of fear memory. Nature neuroscience 22, 753–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, and Kesner RP (2004). Encoding versus retrieval of spatial memory: double dissociation between the dentate gyrus and the perforant path inputs into CA3 in the dorsal hippocampus. Hippocampus 14, 66–76. [DOI] [PubMed] [Google Scholar]
- Leutgeb JK, Leutgeb S, Moser MB, and Moser EI (2007). Pattern separation in the dentate gyrus and CA3 of the hippocampus. Science 315, 961–966. [DOI] [PubMed] [Google Scholar]
- Lin Y, Bloodgood BL, Hauser JL, Lapan AD, Koon AC, Kim TK, Hu LS, Malik AN, and Greenberg ME (2008). Activity-dependent regulation of inhibitory synapse development by Npas4. Nature 455, 1198–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ramirez S, Pang PT, Puryear CB, Govindarajan A, Deisseroth K, and Tonegawa S (2012). Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature 484, 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonergan ME, Gafford GM, Jarome TJ, and Helmstetter FJ (2010). Time-dependent expression of Arc and zif268 after acquisition of fear conditioning. Neural plasticity 2010, 139891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macek TA, Winder DG, Gereau RW, Ladd CO, and Conn PJ (1996). Differential involvement of group II and group III mGluRs as autoreceptors at lateral and medial perforant path synapses. Journal of neurophysiology 76, 3798–3806. [DOI] [PubMed] [Google Scholar]
- Mahan AL, and Ressler KJ (2012). Fear conditioning, synaptic plasticity and the amygdala: implications for posttraumatic stress disorder. Trends in neurosciences 35, 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory CS, and Giocomo LM (2018). Heterogeneity in hippocampal place coding. Current opinion in neurobiology 49, 158–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh TJ, Jones MW, Quinn JJ, Balthasar N, Coppari R, Elmquist JK, Lowell BB, Fanselow MS, Wilson MA, and Tonegawa S (2007). Dentate gyrus NMDA receptors mediate rapid pattern separation in the hippocampal network. Science 317, 94–99. [DOI] [PubMed] [Google Scholar]
- Miao CL, Cao QC, Ito HT, Yamahachi H, Witter MP, Moser MB, and Moser EI (2015). Hippocampal Remapping after Partial Inactivation of the Medial Entorhinal Cortex. Neuron 88, 590–603. [DOI] [PubMed] [Google Scholar]
- Mongiat LA, Esposito MS, Lombardi G, and Schinder AF (2009). Reliable Activation of Immature Neurons in the Adult Hippocampus. PloS one 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashiba T, Cushman JD, Pelkey KA, Renaudineau S, Buhl DL, McHugh TJ, Rodriguez Barrera V, Chittajallu R, Iwamoto KS, McBain CJ, et al. (2012). Young dentate granule cells mediate pattern separation, whereas old granule cells facilitate pattern completion. Cell 149, 188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama D, Iwata H, Teshirogi C, Ikegaya Y, Matsuki N, and Nomura H (2015). Long-delayed expression of the immediate early gene Arc/Arg3.1 refines neuronal circuits to perpetuate fear memory. The Journal of neuroscience : the official journal of the Society for Neuroscience 35, 819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne LC, Palmer SE, Lisberger SG, and Bialek W (2008). The neural basis for combinatorial coding in a cortical population response. The Journal of neuroscience : the official journal of the Society for Neuroscience 28, 13522–13531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Kramer EE, Mercaldo V, Rashid AJ, Insel N, Frankland PW, and Josselyn SA (2016). Neuronal Allocation to a Hippocampal Engram. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkey KA, Chittajallu R, Craig MT, Tricoire L, Wester JC, and McBain CJ (2017). Hippocampal Gabaergic Inhibitory Interneurons. Physiol Rev 97, 1619–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RP, Moradpour F, Eadie BD, Shin JD, Kannangara TS, Delaney KR, and Christie BR (2013). Electrophysiological Identification of Medial and Lateral Perforant Path Inputs to the Dentate Gyrus. Neuroscience 252, 154–168. [DOI] [PubMed] [Google Scholar]
- Pignatelli M, Ryan TJ, Roy DS, Lovett C, Smith LM, Muralidhar S, and Tonegawa S (2018). Engram Cell Excitability State Determines the Efficacy of Memory Retrieval. Neuron. [DOI] [PubMed] [Google Scholar]
- Ramamoorthi K, Fropf R, Belfort GM, Fitzmaurice HL, McKinney RM, Neve RL, Otto T, and Lin Y (2011). Npas4 regulates a transcriptional program in CA3 required for contextual memory formation. Science 334, 1669–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijmers LG, Perkins BL, Matsuo N, and Mayford M (2007). Localization of a stable neural correlate of associative memory. Science 317, 1230–1233. [DOI] [PubMed] [Google Scholar]
- Richter FR, Cooper RA, Bays PM, and Simons JS (2016). Distinct neural mechanisms underlie the success, precision, and vividness of episodic memory. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth BL (2016). DREADDs for Neuroscientists. Neuron 89, 683–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozeske RR, Jercog D, Karalis N, Chaudun F, Khoder S, Girard D, Winke N, and Herry C (2018). Prefrontal-Periaqueductal Gray-Projecting Neurons Mediate Context Fear Discrimination. Neuron 97, 898-+. [DOI] [PubMed] [Google Scholar]
- Ryan TJ, Roy DS, Pignatelli M, Arons A, and Tonegawa S (2015). Engram cells retain memory under retrograde amnesia. Science 348, 1007–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharfman HE, and Myers CE (2013). Hilar mossy cells of the dentate gyrus: a historical perspective. Frontiers in neural circuits 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senzai Y, and Buzsaki G (2017). Physiological Properties and Behavioral Correlates of Hippocampal Granule Cells and Mossy Cells. Neuron 93, 691-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen AT, Cooper YA, Baratta MV, Weng F-J, Zhang Y, Ramamoorthi K, Fropf R, LaVerriere E, Xue J, Young A, et al. (2016). A robust activity marking system for exploring active neuronal ensembles. eLife 5, e13918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, and Lin Y (2016). Npas4: Linking Neuronal Activity to Memory. Trends in neurosciences 39, 264–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka KZ, He HS, Tomar A, Niisato K, Huang AJY, and McHugh TJ (2018). The hippocampal engram maps experience but not place. Science 361, 392–397. [DOI] [PubMed] [Google Scholar]
- Tanaka KZ, and McHugh TJ (2018). The Hippocampal Engram as a Memory Index. Journal of experimental neuroscience 12, 1179069518815942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka KZ, Pevzner A, Hamidi AB, Nakazawa Y, Graham J, and Wiltgen BJ (2014). Cortical representations are reinstated by the hippocampus during memory retrieval. Neuron 84, 347–354. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsani D, Fu Y, Lu J, Lin Y, et al. (2011). A Resource of Cre Driver Lines for Genetic Targeting of GABAergic Neurons in Cerebral Cortex. Neuron 71, 995–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RF (2005). In search of memory traces. Annual review of psychology 56, 1–23. [DOI] [PubMed] [Google Scholar]
- Tonegawa S, Liu X, Ramirez S, and Redondo R (2015). Memory Engram Cells Have Come of Age. Neuron 87, 918–931. [DOI] [PubMed] [Google Scholar]
- Treves A, and Rolls ET (1994). Computational Analysis of the Role of the Hippocampus in Memory. Hippocampus 4, 374–391. [DOI] [PubMed] [Google Scholar]
- Tsou K, Mackie K, Sanudo-Pena MC, and Walker JM (1999). Cannabinoid CB1 receptors are localized primarily on cholecystokinin-containing gabaergic interneurons in the rat hippocampal formation. Neuroscience 93, 969–975. [DOI] [PubMed] [Google Scholar]
- van Dijk MT, and Fenton AA (2018). On How the Dentate Gyrus Contributes to Memory Discrimination. Neuron 98, 832-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng F-J, Garcia RI, Lutzu S, Alviña K, Zhang Y, Dushko M, Ku T, Zemoura K, Rich D, Garcia-Dominguez D, et al. (2018). Npas4 Is a Critical Regulator of Learning-Induced Plasticity at Mossy Fiber-CA3 Synapses during Contextual Memory Formation. Neuron 97, 1137–1152.e1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltgen BJ, and Silva AJ (2007). Memory for context becomes less specific with time. Learning & memory 14, 313–317. [DOI] [PubMed] [Google Scholar]
- Witter MP (2007). The perforant path: projections from the entorhinal cortex to the dentate gyrus. Prog Brain Res 163, 43–61. [DOI] [PubMed] [Google Scholar]
- Xu W, and Sudhof TC (2013). A neural circuit for memory specificity and generalization. Science 339, 1290–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap E-L, and Greenberg ME (2018). Activity-Regulated Transcription: Bridging the Gap between Neural Activity and Behavior. Neuron 100, 330–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiu AP, Mercaldo V, Yan C, Richards B, Rashid AJ, Hsiang HL, Pressey J, Mahadevan V, Tran MM, Kushner SA, et al. (2014). Neurons are recruited to a memory trace based on relative neuronal excitability immediately before training. Neuron 83, 722–735. [DOI] [PubMed] [Google Scholar]
- Yokose J, Okubo-Suzuki R, Nomoto M, Ohkawa N, Nishizono H, Suzuki A, Matsuo M, Tsujimura S, Takahashi Y, Nagase M, et al. (2017). Overlapping memory trace indispensable for linking, but not recalling, individual memories. Science 355, 398–403. [DOI] [PubMed] [Google Scholar]
- Zhao R, Grunke SD, Keralapurath MM, Yetman MJ, Lam A, Lee TC, Sousounis K, Jiang YY, Swing DA, Tessarollo L, et al. (2016). Impaired Recall of Positional Memory following Chemogenetic Disruption of Place Field Stability. Cell reports 16, 793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Won J, Karlsson MG, Zhou M, Rogerson T, Balaji J, Neve R, Poirazi P, and Silva AJ (2009). CREB regulates excitability and the allocation of memory to subsets of neurons in the amygdala. Nature neuroscience 12, 1438–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characterization of the F-RAM and N-RAM Reporters, Related to Figure 1.
(A) The promoters of the two reporters, PF-RAM and PN-RAM, were based on the existing RAM system. Both promoters contain 4 tandem repeats of 24-bp enhancer modules and a minimal promoter from the human FOS gene. The enhancer module in PF-RAM contains the Activator Protein 1 (AP1) site (TGANTCA), a consensus binding sequence for FOS/JUN family transcription factors. The enhancer module in PN-RAM contains the NPAS4 Responsive Element (NRE) (TCGTG), a consensus binding motif for NPAS4.
(B) Experimental scheme. Cultured mouse hippocampal neurons were infected with AAVs carrying either the F-RAM or N-RAM reporter and a control virus (AAV-hSyn-GFP) at 7 days in vitro (DIV), stimulated with bicuculline (Bic, 50 μM) and 4-aminopyridine (4-AP, 250 μM) at 14 DIV and harvested for immunocytochemistry and imaging at 15 DIV.
(C) Representative images showing expression of the reporters under three conditions: Stim, stimulated with Bic and 4-AP as described above; Control, without stimulation; Stim + Dox, same as Stim but in the presence of Dox (40 ng/mL).
(D) Quantification of the percentage of mKate2+ neurons among infected neurons (AAV-hSyn-GFP+). Both F-RAM and N-RAM were substantially induced by stimulation and inhibited by Dox. One-way ANOVA, Dunnett’s test, n = 6 per condition.
(E) F-RAM labeled comparable numbers of hilar cells under home cage (HC) and contextual fear conditioning (CFC) conditions (Mann-Whitney test, n = 3 per group). White arrows indicate F-RAM+ cells in the hilus and dashed lines outline the hilus region.
(F) N-RAM labeled comparable numbers of hilar cells under HC and CFC conditions (Mann-Whitney test, n = 4 per group). White arrows indicate N-RAM+ cells in the hilus and dashed lines outline the hilus region.
(G) Representative images showing that F-RAM+ cells co-localize with the mossy cell marker mGluR2/3 (white arrows) but not the GABAergic interneuron marker GAD67.
(H) The majority of CFC-induced F-RAM+ and N-RAM+ cells in the hilus were mossy cells (mGluR2/3+, n = 3 per group).
(I) Percentage of DG cells labeled with F-RAM or N-RAM under the CFC condition that were in the hilus (n = 3–4).
(J) Fosflx/flx animals were injected with AAVs expressing Cre or GFP (control) into the DG. Seizures were induced by kainic acid (18 mg/kg) via i.p. injection and animals sacrificed 1.5 hours later to stain for FOS or NPAS4. Fos expression was abolished in Fosflx/flx animals infected with Cre-expressing virus, indicating effective Fos deletion.
(K) Same as (J), but with Npas4flx/flx animals. Npas4 expression was abolished in Npas4flx/flx animals infected with Cre-expressing virus, indicating effective Npas4 deletion.
Data are shown as mean ± SEM. ***p < 0.001.
Figure S2. Electrophysiological Properties of F-RAM+ and N-RAM+ Neurons, Related to Figure 2.
(A) Example traces showing membrane potentials of F-RAM+ and N-RAM+ neurons compared to their unlabeled neighbors, in response to −20, 0, 20, 100 and 200 pA current injections.
(B) Input-output curves of firing rates versus injected currents for F-RAM+ and F-RAM− neurons (left), or N-RAM+ and N-RAM− neurons (right). F-RAM+ neurons show comparable excitability to F-RAM− neurons (F-RAM−, n = 19; F-RAM+, n = 23). N-RAM+ neurons show decreased excitability compared to N-RAM− neurons (n = 16, 16). Two-way mixed ANOVA, Sidak’s test.
(C) F-RAM+ neurons (F-RAM−, n = 19; F-RAM+, n = 23) and N-RAM+ neurons (n = 16, 16) show similar resting membrane potentials and action potential (AP) thresholds to their neighboring unlabeled cells. Mann-Whitney test.
(D) Whole-cell patch-clamp recordings were carried out on hippocampal slices from animals in which the F-RAM and Fos-CreER ensembles were co-labeled. Neurons labeled by F-RAM, Fos-CreER or both showed increased mEPSC frequencies. One-way ANOVA, Dunnett’s test comparing to the F-RAM− Fos-CreER− group, n = 17, 15, 11, 16 (from left to right). mIPSC frequencies were similar for all groups (n = 13, 12, 12, 10, from left to right).
(E) Whole-cell patch-clamp recordings were carried out on hippocampal slices from animals in which the N-RAM and Fos-CreER ensembles were co-labeled. Neurons labeled by Fos-CreER, but not those labeled by N-RAM, showed increased mEPSC frequencies (n = 12, 11, 13, 17 from left to right). Neurons labeled by N-RAM, including the small population labeled by both N-RAM and Fos-CreER, showed increased mIPSC frequencies (n = 19, 17, 14, 25 from left to right). Data are shown as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
Figure S3. The Memory Discrimination-Generalization Assay and the Activity Patterns of the F-RAM and N-RAM Ensembles Measured by Fos Induction During the Assay, Related to Figure 3.
(A) Images of contexts A, B and C.
(B) Detailed features of the three contexts. Context A is made of transparent plastic walls with a steel grid floor for foot-shock delivery. Acetic acid (1%) is used as the odor cue. Context B shares all features with context A except for a different floor: a soft pad insert instead of the steel grid. Context C is a completely different chamber, with a soft padded floor and distinct odor cues (benzaldehyde, 0.25%) and lighting.
(C) Using FOS rather than ARC as the activity marker also showed that similar percentages of F-RAM+ neurons were reactivated (FOS+) in contexts A and B, while fewer N-RAM+ neurons were reactivated during recall in context A than in B. Two-way mixed ANOVA, Tukey’s test, n = 7–9.
Figure S4. Control Experiments for Chemogenetic Manipulation, Related to Figure 4.
(A) Validation of DREADDs in F-RAM+ cells. hM4Di inhibited and hM3Dq activated tagged F-RAM+ cells. Animals were injected i.p. with CNO or vehicle (Veh) 30 minutes before recall in context A. Percentages of ensemble neurons that were reactivated (ARC+) were quantified (two-way mixed ANOVA, Sidak’s test, n = 4 per group).
(B) Experimental scheme to determine the effect of CNO injection alone on memory discrimination-generalization. AAVs expressing mCherry without DREADD (AAV-N-RAM-mCherry) were injected into the DG and mice were subjected to the contextual fear memory discrimination-generalization assay. CNO (4 mg/kg) and vehicle (Veh) were injected i.p. 30 minutes before memory recall.
(C) CNO and Veh groups showed similar discrimination between contexts A and B (Veh, n = 19; CNO, n = 18), as well as A and C (n = 13, 15). Two-way mixed ANOVA with Sidak’s test was used for the freezing levels, Mann-Whitney test for comparing the discrimination indices, and one-sample t-test to compare discrimination indices with zero.
(D) Experimental scheme to inhibit cells that are labeled under the HC condition during memory discrimination-generalization. Animals remained in their home cages during the off-Dox window to label cells activated by background activity, including hilar cells and other granule cells. Labeled cells (hM4Di+) were inhibited during memory recall.
(E) Inhibiting HC-labeled F-RAM+ cells did not affect discrimination between contexts A and B (Veh, n = 9; CNO, n = 9) or A and C (n = 9, 9).
(F) Inhibiting HC-labeled N-RAM+ cells did not affect discrimination between contexts A and B (n = 10, 9) or A and C (n = 9, 10).
(G) Schematics for expression of the inhibitory DREADD hM4Di or excitatory DREADD hM3Dq in Fos-expressing ensembles after CFC. Fos-expressing ensembles were labeled by AAV-pFos-d2tTA, which uses an engineered Fos promoter to drive d2tTA.
(H) Representative image showing expression of hM4Di-mCherry in the Fos-expressing ensemble after CFC.
(I) Experimental scheme to probe the function of the Fos-expressing ensemble in memory discrimination-generalization.
(J) Inhibition of the Fos-expressing ensemble enhanced discrimination between contexts A and B (Veh, n = 18; CNO, n = 14), but not A and C (n = 17, 14).
(K) Activation of Fos-expressing ensembles reduced discrimination between contexts A and B (n = 14, 15) and between A and C (n = 14, 15).
Two-way mixed ANOVA with Sidak’s test was used for freezing levels, Mann-Whitney test for comparing discrimination indices and one-sample t-test to compare discrimination indices with zero. Data are shown as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
Figure S5. F-RAM+ Neurons Showed Increased oEPSC Amplitudes upon Optogenetic Stimulation of the MPP, but Unchanged oIPSCs upon Optogenetic Stimulation of Local Interneurons, Related to Figure 5.
(A) Experimental scheme. To target the LPP and MPP pathways, animals were co-injected with AAV-CaMKII-Cre and AAV- EF1α-DIO-ChR2-EYFP to express ChR2 in the LEC or MEC, and also injected with AAV-F-RAM-mKate2 in the DG. To target the MCF pathway, AAVCaMKII-Cre and AAV- EF1α-DIO-ChR2-EYFP were co-injected unilaterally into the contralateral DG, which labeled the hilar mossy cells that send axonal projections to the ipsilateral DG through MCFs. AAV-F-RAM-mKate2 was injected unilaterally into the ipsilateral DG, where whole-cell patch-clamp recordings were carried out.
(B) Representative images showing ChR2 and F-RAM expression.
(C) F-RAM+ neurons had increased optically evoked EPSC (oEPSC) amplitudes upon MPP but not LPP or MCF stimulation (Wilcoxon signed-rank test, n = 16, 11, 10 pairs for LPP, MPP and MCF). Tetrodotoxin (TTX, 0.5 μM) and 4-aminopyridine (4-AP, 100 μM) were added to isolate monosynaptic inputs. Picrotoxin (50 μM) was added to block GABAA receptors. Data are shown as mean ± SEM. **p < 0.01.
(D) Example traces showing oIPSCs recorded simultaneously in F-RAM+ and F-RAM− neurons stimulated by 2 ms of 0.10, 0.25, 0.50, 0.75, 1.00, 1.50 mW/mm2 470 nm LED light pulses. oIPSCs were elicited by optogenetically stimulating ChR2-expressing GABAergic interneurons in Gad2-Cre mice.
(E) oIPSC amplitudes were similar in the two groups (two-way repeated measures ANOVA, Sidak’s test, n = 12 pairs).
Figure S6. Induced FOS Expression Does Not Affect mIPSCs and Induced NPAS4 Expression Does Not Affect mEPSCs, Related to Figure 6.
(A) mIPSC frequencies and amplitudes were similar in neurons with (EYFP+) and without (EYFP−) induced FOS expression (FOSind). Mann-Whitney test; EYFP−, n = 18; EYFP+, n = 16.
(B) mEPSC frequencies and amplitudes were similar in neurons with (EYFP+) and without (EYFP−) induced NPAS4 expression (NPAS4ind, n = 17, 19).
(C) There is no significant difference in the number of F-RAM+ cells under no shock, weak shock and strong shock conditions (one-way ANOVA, n = 3–4).
(D) There is no significant difference in the number of N-RAM+ cells under no shock, weak shock and strong shock conditions (n = 4 per condition).
Data are shown as mean ± SEM.
Figure S7. Validation of Manipulation of MEC and CCK+ Interneurons, Related to Figure 7.
(A) Experimental scheme. Mice were co-injected with AAV-CaMKII-Cre and AAV-hSyn-DIO-hM4Di-mCherry to express the inhibitory DREADD hM4Di in MEC excitatory neurons (CaMKII+). Mice were fear conditioned in context A, memory was recalled 24 hours later and animals sacrificed 1.5 hours after recall. CNO (4 mg/kg) or vehicle (Veh) was injected i.p. 30 minutes before recall. Neuronal activity was examined by FOS staining.
(B) Representative images of the MEC (dashed lines) showing hM4Di and FOS expression. White arrows indicate double-positive cells.
(C) Chemogenetic inhibition robustly suppressed, but did not completely abolish, the activity of MEC projection neurons. Percentages of hM4Di-mCherry+ cells that were also FOS+ were quantified (Mann-Whitney test, n = 4 per group).
(D) Representative images showing FOS+ cells (white arrows) in the DG GCL (dashed lines) after recall in context A.
(E) Optogenetic inhibition of the MEC-DG pathway reduced the number of FOS+ cells in DG but not in CA1 after recall (Mann-Whitney test, n = 4 per group).
(F) Experimental scheme. Memory discrimination-generalization was examined in the absence of 561 nm laser stimulation (light OFF). Animals were injected with NpHR or EYFP in the MEC and implanted with optical fibers in the DG.
(G) In the absence of laser stimulation, NpHR and EYFP groups showed comparable discrimination between the similar contexts A and B (two-way mixed ANOVA with Sidak’s test was used for freezing levels, Mann-Whitney test for comparing discrimination indices, and one-sample t-test to compare discrimination indices with zero; EYFP, n = 11; NpHR, n = 13).
(H) Experimental scheme. Putative CCK+, PV+ and SST+ interneurons were identified by injecting a reporter virus (AAV-Dlx5/6-DIO-ChR2-EYFP) into CCK-Cre, PV-Cre and SST-Cre mice. Whole-cell patch-clamp recordings were carried out on labeled cells to examine their firing properties.
(I) CCK+ interneurons (identified by EYFP expression) showed the strongest firing adaptation among the three interneuron subtypes. Example traces showing the membrane potentials of CCK+, PV+ and SST+ interneurons in response to −50, 0, 50, and 500 pA current injections.
(J) Schematic for measurement of depolarization-induced suppression of inhibition (DSI). ChR2 (AAV-Dlx5/6-DIO-ChR2-EYFP) was expressed in CCK+, PV+ or SST+ interneurons to induce optically evoked IPSCs (oIPSCs).
(K) DSI was observed in the CCK+ interneuron (CCK) to granule cell (GC) synapses. oIPSCs were induced by 2 ms 470 nm LED light stimulation and recorded in GCs voltage clamped at −70 mV, with high-Cl− internal solution, in the presence of APV (50 μM) and DNQX (20 μM). DSI was induced by elevating the holding potential of GCs to 0 mV for 5 s. oIPSCs recorded before (oIPSC1) and 200 ms after (oIPSC2) depolarization, averaged from ten sweeps, were compared to calculate DSI.
(L) Quantification of DSI (one-way ANOVA, one-sample t-test for individual columns, n = 6 per group).
(M) Asynchronous releases were observed in the CCK to GC synapses, and were blocked by bath application of WIN 55,212–2 (WIN, 10 uM). Paired whole-cell patch-clamp recordings targeting presynaptic CCK+ interneurons and postsynaptic GCs were carried out. Superimposed traces (black) from 15 sweeps are shown. Trains of ten action potentials (50 Hz) were evoked in the presynaptic CCK+ interneurons, and unitary IPSCs (uIPSCs) were recorded in the postsynaptic GCs voltage clamped at −70 mV. uIPSCs were recorded with high-Cl− internal solution in the presence of APV (50 μM) and DNQX (20 μM). Red traces show superimposed traces of 15 sweeps after bath application of WIN.
Data are shown as mean ± SEM. *p < 0.05, **p < 0.01.
Data Availability Statement
Data and custom MATLAB code are available upon request from the Lead Contact.


