Abstract
Global demand for recombinant proteins has steadily accelerated for the last 20 years. These recombinant proteins have a wide range of important applications, including vaccines and therapeutics for human and animal health, industrial enzymes, new materials and components of novel nano‐particles for various applications. The majority of recombinant proteins are produced by traditional biological “factories,” that is, predominantly mammalian and microbial cell cultures along with yeast and insect cells. However, these traditional technologies cannot satisfy the increasing market demand due to prohibitive capital investment requirements. During the last two decades, plants have been under intensive investigation to provide an alternative system for cost‐effective, highly scalable, and safe production of recombinant proteins. Although the genetic engineering of plant viral vectors for heterologous gene expression can be dated back to the early 1980s, recent understanding of plant virology and technical progress in molecular biology have allowed for significant improvements and fine tuning of these vectors. These breakthroughs enable the flourishing of a variety of new viral‐based expression systems and their wide application by academic and industry groups. In this review, we describe the principal plant viral‐based production strategies and the latest plant viral expression systems, with a particular focus on the variety of proteins produced and their applications. We will summarize the recent progress in the downstream processing of plant materials for efficient extraction and purification of recombinant proteins. J. Cell. Physiol. 216: 366–377, 2008. © 2008 Wiley‐Liss, Inc.
Recombinant DNA technology was initially used to express proteins that were difficult to produce in their native organisms. Increasing efforts, however, have been focused on designing new molecules with more desirable characteristics and/or functionality. Pharmaceuticals and industrial enzymes were the first recombinant biotech products on the world market and biopharmaceuticals were the majority of commercialized recombinant proteins (Pavlou and Reichert, 2004). Many protein‐based drugs, similar to traditional small molecule pharmaceuticals, function as antagonists by binding to and thereby inhibiting the activity of their target, such as an enzyme or a receptor. Classical protein antagonists include full monoclonal antibodies (mAbs), their single‐chain derivatives (ScFv) and mAb‐fusion proteins. Recent research programs have also focused on non‐antibody antagonists that consist of a scaffold protein displaying the inserted affinity peptide (Walsh, 2006). Recombinant DNA technology also provided an excellent alternative for developing safer vaccines. Subunit vaccines are based on immunodominant protein components of a pathogen, but do not contain its genetic material. Consequently they cannot replicate, cause disease, or introduce pathogens into non‐endemic regions. Viral coat proteins are exceptional subunit vaccine candidates and in some cases are able to form virus‐like particles (VLPs) when expressed in heterologous systems. In fact, the only recombinant subunit vaccines presently available are based on VLPs. They are highly immunogenic and able to induce both humoral and cellular responses (Chackerian, 2007).
In addition to the pharmaceutical industry, many other fields are also relying intensely on recombinant proteins. Areas as diverse as agro‐food technology, chemistry, detergent production, bioremediation, biosensoring, petroleum, and paper industries all receive significant contribution from applications of recombinant proteins. For example, increasing needs for a diversity of food processing enzymes, for example, amylase, lipase, xylanase, pullulanase and pectin modifying enzymes, demand a substantial involvement of recombinant protein technology (Olempska‐Beer et al., 2006).
In the coming years, there will be a significant increase in demand for high quality recombinant proteins. In response, biological systems used for the production of proteins must be scalable, cost‐effective, safe and flexible enough to meet market requirements. Current systems rely on “bio‐factories,” that is, mammalian, insect, yeast, and microbial cell cultures. The majority of the recombinant proteins are currently produced in Escherichia coli or mammalian cells with a few exceptions of yeast or insect cells (Yin et al., 2007). All of these bio‐factories are based on fermentation technology of suspension cells in bioreactors, which requires an enormous upfront capital investment and, thereby, severely constrains their scalability.
The use of plants as production systems for recombinant proteins has been actively investigated over the last two decades. Plants are attractive as protein factories because they can produce large volumes of products efficiently and sustainably and, under certain conditions, can have significant advantages in decreasing manufacturing costs (Hood et al., 1999; Giddings, 2001). Plant systems are far less likely to harbor microbes pathogenic to humans than mammalian cells or whole transgenic animal systems. In addition, one of the major advantages of plants is that they possess an endomembrane system and secretory pathway that are similar to mammalian cells (Vitale and Pedrazzini, 2005). Thus, proteins are generally efficiently assembled with appropriate post‐translational modifications. These cost, scale, and safety advantages make plant‐made pharmaceuticals very promising for both commercial pharmaceutical production and for manufacturing products destined for the developing world.
Three approaches are commonly used to express heterologous proteins in plants: (1) stable transformation of the nuclear genome, (2) stable transformation of the chloroplastic genome, and (3) viral transient transformation.
In stable transformation technology, an expression cassette harboring the exogenous gene of interest is integrated into the nuclear or plastid genome of plant cells, which results in the acquired character to be stably inherited over generations. These lines can be propagated vegetatively or by seed and thus, readily scaled up for protein production. Stable nuclear transformation is often achieved using Agrobacterium tumefaciens, which delivers fragments of DNA into the plant cell nucleus at random positions. Alternatively, the “biolistic” method (microprojectile bombardment) can be used for plant hosts that are difficult to transform by Agrobacterium (Hansen and Wright, 1999).
Due to their double membrane structure, chloroplast transformation can be achieved only by bombardment with tungsten or gold particles coated with fragments of DNA. Stable transformation of the chloroplast offers several distinct advantages in areas of transgene targeting, product yield, and regulatory compliance. Since the chloroplast genome allows for homologous recombination, transgenes can be precisely targeted to a specific locus of the genome to avoiding positional effect or accidental gene knock‐outs. Each plant cell has several chloroplasts and each of these contains many circular genomes. Therefore, the shear copy number of the transgene enables high‐level expression of the target recombinant protein. Since the chloroplast is inherited maternally, this technology reduces the risk of potential transgene escape by pollen dissemination. Similar to bacterial cells, however, the chloroplast is unable to perform typical eukaryotic posttranslational modifications, such as glycosylation. As a result, this technology cannot be used to produce proteins when such modifications are essential for their function (Bock, 2007).
The third strategy relies on replicating plant viruses. These viruses are small, can be easily manipulated, and their infection process is relatively simple. The above features make viral vectors an attractive alternative to stable transgenic systems for the expression of foreign proteins in plants. In this strategy, the gene of interest is inserted among viral replicating elements, episomically amplified, and subsequently translated in the plant cell cytosol. Most of the transient expression systems are based on non‐food, non‐feed plants like tobacco, therefore, requiring further purification prior to application.
Production of recombinant proteins with stably or transiently transformed plants has been performed both in green‐houses and in field. Neither practice requires large investments in hardware or culture media, thus making scale‐up more economical than fermentation cultures. The possibility of producing recombinant protein agents on an agricultural scale by “molecular farming” is extremely attractive.
Although the use of plants as expression systems for recombinant proteins was first conceived for oral delivery of antigenic proteins (Walmsley and Arntzen, 2000), plant‐made recombinant proteins have also been purified and used in many different applications. An enormous number of proteins different in size, structure, origin and biological function have been successfully expressed in stably transformed monocot and dicot plants such as maize (Chikwamba et al., 2003), rice (Nochi et al., 2007), wheat (Brereton et al., 2007), potato (Youm et al., 2007), tomato (Huang et al., 2005), tobacco (Watson et al., 2004), lettuce (Sun et al., 2006), alfalfa (Huang et al., 2006), lupin (Smart et al., 2003), carrots (Marquet‐Blouin et al., 2003), barley (Joensuu et al., 2006), soybean (Moravec et al., 2007), and thale cress (Carrillo et al., 1998).
Many antigenic monomeric proteins from viruses (Webster et al., 2006) and prokaryotes (Alvarez et al., 2006) have been produced and in a few cases, their immunogenic properties have been successfully evaluated in human volunteers during phase I clinical studies (Tacket et al., 1998, 2000; Thanavala et al., 2005).
Complex oligomeric mAbs were first expressed in stable transgenic plants by Hiatt et al. (1989). Since then, the number and type of plant‐expressed antibodies, such as secretory antibodies, antibody fragments and, more recently, immune complexes, have increased steadily. Several commercial candidates of mAbs have been developed, with five mAbs under clinical evaluation, and two having reached Phase II clinical studies (Fischer et al., 2003).
In addition to pharmaceuticals, recombinant proteins for a variety of applications have also been expressed in transgenic plants. Examples include alpha‐amylase enzyme (Pen et al., 1992), the protein brazzein for commercial sweeteners (Lamphear et al., 2005), and the fungal enzyme manganese peroxidase (Clough et al., 2006). Moreover the chicken avidin diagnostic protein (Hood et al., 1999) and the bovine trypsin enzyme (Woodard et al., 2003) from transgenic maize, and the human anti‐microbic proteins lysozyme (Huang et al., 2002) and lactoferrin (Humphrey et al., 2002) from transgenic rice have been commercially produced and are currently marketed by Sigma Chemical Company (St. Louis, MO).
The major challenges facing transgenic plant technology lay in increasing the quantity of protein, the optimization of expression systems, and improvement of downstream processing. In this review, we will focus on transient production strategies using plant viral expression systems, with a particular focus on the variety of proteins produced, and their applications. We will also discuss the latest developments in the downstream processing of plant materials for efficient extraction and purification of recombinant proteins.
Plant Viral Vectors
The history of genetic engineering and applied virology are intimately connected; the first recombinant molecule assembled was a chimeric SV40 containing genes from the bacteriophage λ (Jackson et al., 1972). The unique properties of viruses such as ease of manipulation, high level amplification, site specific recombination, strong infectivity, enhanced translation and compact and repetitive morphological structure have enabled their broad application, from basic research to product development, including the generation of robust expression systems. Viruses with a variety of host ranges such as bacteriophages, mammalian retroviruses, invertebrate infecting baculoviruses, and plant viruses have been genetically modified to express heterologous proteins.
From the discovery of viruses in 1898 (tobacco mosaic virus, TMV) (Bos, 1999), to the first demonstration of RNAs role in virus replication by turnip yellow mosaic virus (TYMV) (Matthews, 1989), to the very recent discovery of gene silencing and its implication in host response to infection, gene regulation and transgene expression (Baulcombe, 1999; Lu et al., 2003; Waterhouse and Helliwell, 2003), plant virology has played a crucial role in the understanding of the most fundamental concepts of modern biology. In addition, plant viral elements such as promoters, terminators, translational enhancers and various cis‐regulatory sequences have been extensively used in plant biotechnology.
Plant viruses have been used to introduce foreign genes in plants since the early 1980s and technical advances in molecular biology and plant virology have allowed the generation of many improved expression systems. These recent systems provide many advantages including rapid, high‐level transgene expression, and, in the case of movement defective systems, better transgene containment due to the lack of vertical and horizontal gene transfer.
The earlier plant virus expression systems were based on the cauliflower mosaic virus (CaMV) of the Caulimoviridae family, the only plant viruses with a double stranded DNA genome. However, limited packaging capacity and the restricted amount of viral DNA that can be removed without affecting essential functions hindered the application of these initial expression systems. Fortunately, technical advances in molecular biology such as generating cDNA from an RNA template have enabled us to expand our search for expression systems into single stranded RNA viruses, the most represented types of plant viruses in nature. Another technical breakthrough was the use of Agrobacterium tumefaciens to promote viral infection (Grimsley et al., 1986; Turpen et al., 1993). Generally, viral infection can be initiated by mechanical inoculation of infectious viral particles on leaves or alternatively by transfection of nucleic acids. The use of Agrobacterium tumefaciens, known as “agroinfection,” allows the direct and efficient targeting of cDNA to the plant cell nucleus (Fig. 1). The cDNA constructs in the plant expression cassette would result in an infectious, autonomously replicating nucleic acid after host cell nuclear transcription and processing. Moreover agroinfection allowed the use of viruses that in nature are not mechanically transmissible, but instead need a special insect vector to initiate the infection process. These viral vectors are commonly divided into gene substitution vectors, gene insertion vectors, modular or deconstructed vector systems and peptide display vectors.
Figure 1.
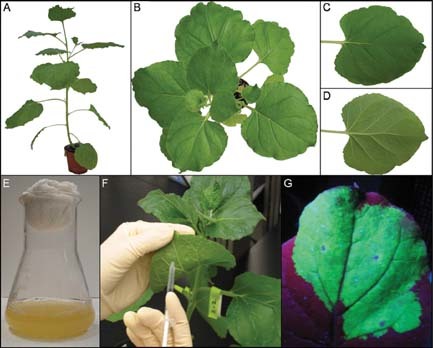
Agroinfection method. Nicotiana benthamiana plant (A,B), leaves adaxial side (C) and abaxial side (D), Agrobacteriun tumefaciens liquid culture (E), small scale infiltration procedure (F), GFP expression under U.V. light (G). [Color figure can be viewed in the online issue, which is available at http://www.interscience.wiley.com/.]
Gene substitution or replacement vectors are based on the exchange of an endogenous viral sequence with a heterologous gene of interest. The first successful expression of a foreign gene ever achieved in plants was obtained using a substitution vector based on the CaMV. Usually, the coat protein (CP) gene is the viral coding sequence of choice to be replaced. However, with a few exceptions, the CP is essential for cell to cell and/or systemic movement and infection. However, caution has to be taken if leaves of the whole plants are intended to be infected. For example, a CP replacement vector was developed using the tomato bushy stunt virus (TBSV). In this case the CP is not essential for systemic movement of the virus in certain species of Nicotiana (Scholthof et al., 1993) and it was indeed demonstrated that the insertion of a heterologous gene supports a systemic infection (Scholthof et al., 1993). In TMV‐based substitution vectors, where the CP is necessary for systemic infection, a chloramphenicol acetyl transferase (CAT)–CP gene replacement vector generated only local lesions and CAT activity was confined on the inoculated leaf (Takamatsu et al., 1987). In another recently developed TMV‐based gene substitution vector (Musiychuk et al., 2007) the CP gene was exchanged with the gene of interest fused to a thermostable carrier molecule, the beta‐1,3–1,4‐glucanase enzyme (lichenase) of Clostridium thermocellum, to facilitate target expression, stability and purification. The heat tolerant property of the lichenase (65°C) allows easy target proteins purification by heat treatment which precipitates up to 50% of contaminating plant proteins. The protein of interest, fused at the N‐ or C‐terminus of lichenase, or inserted in the central loop of an engineered version of the enzyme, is protected from the degradation and thus purified.
Gene insertion vectors consist of complete functional viruses with the addition of an extra open reading frame (ORF) for the target protein. They are capable of cell to cell and systemic movement and infection. Viruses with both spherical and rod shaped virions have been investigated. Viruses with rod‐shaped particles have a better potential due to fewer constraints on the amount of nucleic acid inserted. However, there is still a limit on the genome size of the chimeric rod‐shaped virion based vectors. It has been proven that chimeric vectors with genome size beyond certain limits resulted in unsustained virion assembly. Two of the most famous gene insertion vectors have been derived from TMV and potato virus X (PVX). Both viruses have a single stranded positive RNA genome. They use subgenomic promoters and consequently subgenomic RNAs to express some of their ORFs and have a helical virion symmetry which results in rod‐shaped particles. A chimeric TMV was constructed with the CAT gene inserted between the movement protein (MP) gene and the CP gene; the expression was regulated by an additional copy of the subgenomic promoter of the CP gene that, as a result, was duplicated in the hybrid vector. The vector was able to replicate, to form subgenomic RNAs, to assemble correctly, and to produce reporter gene activity. Nevertheless the duplication of the subgenomic promoter sustained homologous recombination causing instability and the consequent loss of the exogenous gene (Dawson et al., 1989). This problem was solved by using a subgenomic CP promoter derived from a different virus belonging to the Tobamovirus genus to prevent homologous recombination. Similar problems and solutions of recombination between duplicated subgenomic promoters were reported in a recent insertion vector system based on the grapevine virus A (GVA) for N. benthamiana (Haviv et al., 2006). Analogous to TMV, a PVX‐based vector was generated using the duplicated promoter of the CP gene. However in this case the vector was stable and able to retain the additional coding sequence (Chapman et al., 1992). Nevertheless, recently it has been demonstrated that the size of the inserted gene of interest can play a crucial role in the stability of the modified PVX expression vector, with a positive correlation between the elimination rate of the gene of interest and its length (Avesani et al., 2007). Recombination can involve homologous CP promoter sequences but also other mechanisms mediated by both the host plant and the virus (Avesani et al., 2007).
Another well established system is based on Cowpea Mosaic Virus (CPMV) the type member of Comoviridae whose bipartite genome is constituted by single stranded positive RNA molecules. RNA‐1 encodes for proteins involved in replication while RNA‐2 carries the genetic information for the movement and structural proteins. The system is very versatile; it can be utilized in a totally transient fashion, co‐infiltrating wild‐type plants with two separate c‐DNA constructs representing RNA‐1 to sustain amplification and RNA‐2 harboring the gene of interest, or it can be utilized combining transgenic plants with virus‐mediated transient expression. In this case the RNA‐2 is used for stable transformation and the RNA‐1, necessary to prime the active replication of RNA‐2 and of the heterologous gene, is supplemented by agro‐infiltration. It is worth noticing that a deleted version of RNA‐2, unable to produce infectious clones, it has also been utilized to prevent possible environmental containment concerns (Liu et al., 2005; Sainsbury et al., 2008).
In conclusion it is possible to obtain an inducible expression system if RNA‐1 is supplemented by agroinfiltration and a constitutive stable expression system if it is supplemented by crossing with an RNA‐1 stable transgenic line (Canizares et al., 2006).
Modular or deconstructed vectors are a new generation of viral expression systems. Their development was driven by the understanding that: (1) not all viral components are essential or beneficial for an expression vector and (2) viruses can be broken down into different genomic elements that would still operate together in the infection process as wild‐type multipartite genome viruses. Moreover, agroinfection provided the technical possibility to co‐deliver multiple different components. Hence in modular systems, viral components are separated into distinct portions and inserted into binary vectors contained in an Agrobacterium strain. The strains are mixed together and co‐infiltrated into plant leaves. This strategy allows the replicative portion, the so called replicon, to be reduced in size, down to minimal to accommodate the insertion of transgene, while other viral components necessary for the vector function can be provided in trans during the infection process. At present, one of the most famous and broadly used vector is the deconstructed system based on TMV and developed by Icon Genetics (Halle, Germany), recently acquired by Bayer Innovation GmbH. The vector has been engineered to divide the TMV genome into two major cDNA modules: a 5′ module which contains the viral RNA dependent RNA polymerase and the MP, and a 3′ module that carries the gene of interest and the 3′ untranslated region (UTR) of the virus essential for the efficient replication and amplification of the vector. In this particular case, viral functions are not complemented in trans but the two modules actually assemble together in vivo by a site specific recombinase delivered by a third Agrobacterium cell line. Furthermore, different 5′ modules carry different organelle targeting signals which, fuse in frame with the gene of interest after recombination and nuclear processing, allow a single construct to be combined pair wise with various targeting elements in separate constructs. This feature greatly facilitates the experimental procedure to test optimal expression in terms of accumulation in different sub‐cellular compartments. An additional attribute of the vector, that permitted a dramatic enhancement of expression levels, was the introduction of several introns in the coding sequences of the 5′ module (Marillonnet et al., 2004, 2005). Levels of expression are impressive and can reach up to 5 mg of recombinant protein per gram of fresh weight (FW). The system was extensively tested with different genes, coding for antibodies and antibody‐derivatives, interferons, growth hormones, bacterial and viral antigens, adjuvants and enzymes (Gleba et al., 2005), confirming each time its versatility and robustness.
Another interesting modular system was developed using the bean yellow dwarf virus (BeYDV) of the Geminiviridae family. Geminivirus are single stranded circular DNA viruses known to replicate through the rolling circle process, promoting high level of genome amplification in the nucleus of the infected cell. The BeYDV based system is depleted of the MP and CP gene and consequently the expression of the protein of interest is confined to the inoculation area. The basic version involves two modules, one containing the gene of interest inserted between the BeYDV long and short intragenic regions, LIR and SIR respectively. Both intragenic regions are necessary to sustain rolling circle replication. The gene of interest is driven by a strong plant specific constitutive promoter. The second module is responsible for the expression of the Rep protein which catalyzes various aspects of the rolling circle replication. Initially, the system was used as an expression vector in tobacco plant cell culture (Mor et al., 2003) but its use has recently been extended to whole leaves with agroinfection. The modular nature of this system also permits the co‐expression of a variety of sequences of different viral or non‐viral origin that are beneficial for replicon amplification and transgene expression. For example, genes of post‐transcriptional gene silencing (PTGS) inhibiting proteins can be co‐expressed to prevent PTGS and hence enhance target protein accumulation.
Display viral vectors have been extensively used to provide a molecular scaffolding for different kinds of peptides to be fused with the viral CP and, therefore, to be exposed and displayed on the surface of chimeric virus particles (CVPs). Coat protein genes of several plant viruses were genetically modified to support fusions with coding sequences of heterologous peptides in specific regions known to be well exposed on the virion surface (Johnson et al., 1997; Porta and Lomonossoff, 1998). Therefore, the virus and the plant host would provide the expression system to produce large quantities of the chimeric fusion protein. Clearly this is only feasible for well characterized viruses in terms of assembly, movement strategies and structure. Fusions must be compatible with the normal assembly and viral fitness, and must avoid any possible steric hindrance or interference with virus movement. Defining peptide features for their correct display on CVP is of particular interest among studies in this area (Bendahmane et al., 1999; Porta et al., 2003; Lico et al., 2006). Two strategies have been developed to overcome assembly problems for long or difficult peptides. One strategy is to modify the 3′ terminus of the CP gene so that the CP stop codon would function in a leaky fashion. This would cause most of the CP produced to be unfused, while a small portion would be fused to the peptide displayed on the surface (Hamamoto et al., 1993; Sugiyama et al., 1995; Borovsky et al., 2006). The other strategy employs the 2A peptide of the small foot and mouth disease virus (FMDV). This sequence, inserted between the epitope coding sequence and the 5′ terminus of the CP gene, produces a ribosomal skip during the translational procedure (Donnelly et al., 2001; de Felipe et al., 2003), with the result that only a small portion of the translational products will consist in a fusion between the heterologous sequence and the CP (Fig. 2). The use of these strategies, however, opens the question of the precise dose of the antigen in the vaccine formulation. Many plant viruses have been used to display different kinds of peptides. The most widely used viruses for this purpose are the CPMV, the TMV and the PVX. Extensive research regarding the symmetry, shape and structure at atomic level for these viruses have allowed the precise definition of specific sites for peptide insertion to ensure the successful display of fusion peptides (Klug and Caspar, 1960; Lomonossoff and Johnson, 1991; Baratova et al., 1992a,b; Johnson et al., 1997; Parker et al., 2002).
Figure 2.
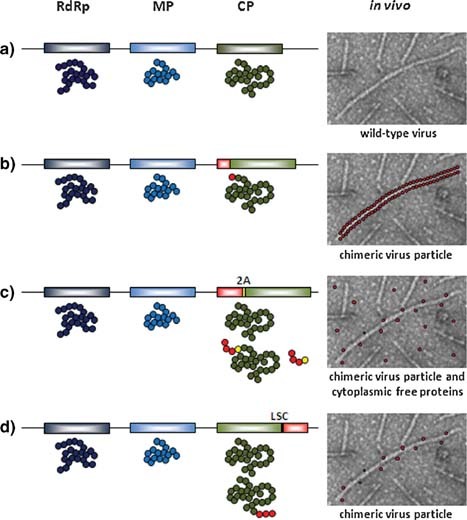
Display strategies. Wild‐type viral genome organization and encoded proteins (a), standard CP fusion strategy (b), Foot and Mouth Disease Virus 2A peptide CP fusion strategy (c), leaky stop codon CP fusion strategy (d). RdRp, RNA dependent RNA polymerase; MP, movement protein; CP, coat protein; 2A, Foot and Mouth Disease Virus 2A peptide; LSC, amber leaky stop codon. [Color figure can be viewed in the online issue, which is available at http://www.interscience.wiley.com/.]
Major efforts are now directed to the development of new viral vector typologies in terms of both improvement of the expression strategies and virus choice. Host plants have also been extended to a wider variety of species including herbaceous plants and cucurbitaceous family. Besides the viruses mentioned above, other viruses have also been investigated as useful vectors, such as tomato mosaic virus (ToMV) (Dohi et al., 2006); maize streak virus (MSV) (Palmer and Rybicki, 2001); bean pod mottle virus (BPMV) (Zhang and Ghabrial, 2006); beet necrotic yellow vein virus (BNYVV) (Schmidlin et al., 2005); tomato golden mosaic virus (TGMV) (Hayes et al., 1989); potato virus A (PVA) (Kelloniemi et al., 2006); zucchini yellow mosaic virus (ZYMV) (Hsu et al., 2004); cucumber mosaic virus (CMV) (Nuzzaci et al., 2007); and cucumber green mottle mosaic virus (CGMMV) (Ooi et al., 2006).
Monoclonal antibodies
Antibodies are key molecules of the vertebrates' immune system. They are responsible for the recognition and binding of target antigens with high affinity and specificity. Monoclonal antibodies (mAbs) are used in a wide range of applications, including diagnosis, prevention and treatment of many diseases. For example, they are being successfully utilized as “targeting vectors” to direct drugs, radioisotopes and other therapeutic molecules to their target cells or tissues, as immunomodulators in pathology‐related functions, as well as diagnostic tools to identify and localize molecular alterations. In addition to their pharmaceutical applications, mAbs can also be used to modulate plant traits, generate disease or pest resistant crops (Tavladoraki et al., 1993; Nolke et al., 2004; Peschen et al., 2004; Villani et al., 2005), and to study plant metabolic pathways (Nolke et al., 2005). Antibodies are complex glycoproteins consisting of four polypeptides, linked by disulfide bridges and non‐covalent bonds. Despite their complexity, mAbs and their derivatives can be easily expressed in heterologous systems including plants (Fischer et al., 2003). However, mAbs produced by heterologous systems may not share the precise structural and functional properties of the native molecules. For example, mAbs produced by a bacterial expression system will lack post‐translational modifications including glycosylation. Non‐mammalian eukaryotic expression systems could fail to use the appropriate glycans groups resulting in different glycosylation patterns. Yeasts, for example, tend to hyperglycosylate (Harashima, 1994; Malissard et al., 1996); while plants preferentially introduce xylose and fucose residues and fail to use galactose and syalic acid (Gomord and Faye, 2004). Although proper glycosylation is required for functions of many mAbs, it is to be noted that different glycosylation patterns do not always lead to loss of function in vitro or in vivo, or cause side‐effects in humans (Chargelegue et al., 2000). Different strategies have been adopted to circumvent these limitations such as targeting mAbs to the endoplasmic reticulum, or abolishing glycosylation sites by mutagenesis to avoid inappropriate glycosylated products. In fact, it has been reported that the absence of glycans in some cases did not interfere with correct assembly and activity (Rodriguez et al., 2005; Nuttall et al., 2005). In addition a fascinating approach that requires sophisticated genetic and metabolic engineering resides in the inactivation of endogenous glycosyltransferases and/or expression of heterologous mammalian specific enzymes in the host plant system (Saint‐Jore‐Dupas et al., 2007).
Complete antibodies and their derivatives have been produced with stable plant transformation technologies (Ma et al., 2005). Thanks to the recent improvements of viral‐based vectors, mAbs have been produced with transient expression systems to quickly achieve much higher production levels along with other complex proteins. Single chain fragments, that preserve antigen‐recognition elements of the full length immunoglobulin, are often more effective as drugs or diagnostic tools than mAbs due to their increased ability to penetrate target tissues, reduced immunogenicity, and more rapid clearance. A chimeric gene insertion TMV based vector was used for the expression of a human scFv for treatment of non‐Hodgkin's lymphoma (NHL) (McCormick et al., 1999, 2003). The plant‐derived scFv generated an antibody response in vaccinated animals, and was an effective vaccine in a murine NHL tumor challenge model. The TMV strategy provides the speed and versatility required for the development of a patient specific treatment and this anti‐idiotype vaccine is currently undergoing clinical trials for safety and immunogenicity evaluation in humans.
Another example is a scFv derivative (small immune protein, SIP) to treat transmissible gastroenteritis virus (TGEV), a porcine coronavirus. The SIP was expressed by a PVX gene insertion vector and by a CPMV based display vector using the 2A expression strategy. The aim was to provide passive protection against enteric infections upon oral delivery in crude plant extracts (Monger et al., 2006). The SIP folded correctly and preserved the ability to bind and neutralize TGEV in tissue cultures. Moreover, it provided in vivo protection against challenge with TGEV in piglets vaccinated orally with crude extracts of scFv‐expressing tobacco plants. The same researchers also expressed a full length TGEV‐specific IgA by co‐infecting plants with two separate PVX insertion vectors for the light chain (LC) and the heavy chain (HC) respectively (Alamillo et al., 2006). The IgA was correctly assembled in plant tissue and provided in vivo protection against TGEV in piglets after oral administration. A previous work (Verch et al., 1998) also indicated the possibility to produce a full size mAb in plants through the co‐infection of two modified TMV based viral vectors. These results clearly indicated the possibility to obtain functional mAbs through plant expression systems.
Nonetheless, expression of heterooligomeric proteins such as mAbs might be inefficient due to the co‐delivery of viral vectors built on the same virus backbone. In fact, this feature may result in early segregation and subsequent preferential amplification of one of the vectors in one cell. This problem has been recently overcome by utilizing two sets of vectors derived from non‐competing TMV and PVX to express the two different mAb chains (Giritch et al., 2006). TMV and PVX may interact with different host proteins to sustain their replication and movement without interfering with each other. As a result, neither of the two vectors gains a replicative advantage over the other, allowing efficient co‐expression of HC and LC in the same cells. The level of expression by each vector is independent within the same cell and can reach up to 0.5 mg/g FW of full assembled mAb in few weeks. This strategy represents a useful platform for rapid large‐scale manufacturing of mAbs and other hetero‐oligomeric proteins especially in situations requiring rapid responses such as pandemic threats and bioterrorism events.
Antigens
A vaccine is an antigenic preparation used to establish immunity to a disease. Vaccination has been one of the greatest revolutions in medical science and has dramatically improved the quality and length of life expectancy. Although vaccines are the most cost effective form of disease protection in healthcare, their cost is still too excessive for many people in the world, especially in developing countries. Vaccines can be therapeutic, to overcome an infection already established, or prophylactic, to prevent a future infection. The aim of vaccination strategies is to obtain efficient and safe vaccine formulations to induce a long lasting immunity against various pathogens. To achieve this goal, the vaccine component should generate not only a neutralizing antibody response, a long‐term memory B cells stimulation, but also a T cell mediated immunity to eliminate infected cells which represent the pathogen reservoirs. Recombinant subunit vaccines, based on a single protein (antigen) or a single peptide (epitope) derived from the pathogen, are particularly attractive strategies when compared to classic inactivated, attenuated or live recombinant vaccines. Recombinant subunit vaccines offer potentially equivalent efficacy but are much safer and in some cases easier to produce. Plants offer unique advantages for the production of subunit vaccines in terms of scale, speed, costs, yield, and safety. The first work that describes the expression of the hepatitis B surface antigen in stable transgenic tobacco in 1992 marked the beginning for developing low cost vaccine formulations in plants (Mason et al., 1992). Researchers focused on expressing their antigens in edible plant tissues as oral vaccines, opening a new prospect for vaccine delivery (Mor et al., 1998). Since then, many research groups have adopted this system and carried out studies in the areas of improving target protein expression levels, analyzing the administration route and schedule, choosing the appropriate animal models, and exploring the use of possible adjuvants.
The first report in this area described the expression of the FMDV structural protein VP1 with a TMV‐based gene insertion vector (Wigdorovitz et al., 1999). A yield of approximately 0.15 mg/g FW was achieved and subsequently the antigen was administered to mice without further purification. All mice immunized intraperitoneally developed a protective immune response against experimental challenge with virulent FMDV. A great number of antigens have been produced in plants with different immunologic results (Streatfield and Howard, 2003). Altogether, these works demonstrated that plant‐derived antigens retained their antigenicity and were able to induce active protective humoral and cell‐mediated immune responses. The development of plant derived vaccines for human and veterinary uses against viral pathologies, tumors, allergies, bacterial infections, and diabetes‐associated autoantigens has been extensively investigated (Table 1).
Table 1.
Representative antigens expressed through viral vectors
| Pathogen | Antigen | Viral vector | References |
|---|---|---|---|
| Human immunodeficiency virus‐1 | Capsid epitopes | CPMV (D) |
Porta et al. (1994) |
| Plasmodium falciparum | Several epitopes | TMV (D) |
Turpen et al. (1995) |
| Influenza virus | HA epitope | TMV (D) |
Sugiyama et al. (1995) |
| Human immunodeficiency virus‐1 | Capsid epitopes | AMV (D) |
Yusibov et al. (1997) |
| TBSV (D) |
Joelson et al. (1997) |
||
| Mink enteritis virus | VP2 protein peptide | CPMV (D) |
Dalsgaard et al. (1997) |
| Canine parvovirus | VP2 protein peptide | PPV (D) |
Fernandez‐Fernandez et al. (1998) |
| Staphylococcus aureus | FnBP epitope | CPMV (D) |
Brennan et al. (1999) |
| Foot and mouth disease virus | VP1 protein | TMV (I) |
Wigdorovitz et al. (1999) |
| Hepatitis C virus | Region 1 of E2 | TMV (I) |
Nemchinov et al. (2000) |
| Rotavirus | VP6 protein | PVX (D) |
O'Brien et al. (2000) |
| Human immunodeficiency virus‐1 | P24 protein | TBSV (I) |
Zhang et al. (2000) |
| Pseudomonas aeruginosa | F protein peptides | TMV (D) |
Staczek et al. (2000) |
| Human immunodeficiency virus‐1 | Capsid epitopes | PVX (D) |
Marusic et al. (2001) |
| Rabbit haemorrhagic disease virus | VP60 protein | PPV (I) |
Fernandez‐Fernandez et al. (2001) |
| Rabies virus | Chimeric peptide | AMV (D) |
Yusibov et al. (2002) |
| Human papilloma virus | E7 oncoprotein | PVX (I) |
Franconi et al. (2002) |
| Bovine herpes virus | Glycoprotein D | TMV (I) |
Perez Filgueira et al. (2003) |
| Hepatitis C virus | Mimotope | CMV (D) |
Natilla et al. (2004) |
| Colorectal antigen | GA733‐2 antigen | TMV (I) |
Verch et al. (2004) |
| Human immunodeficiency virus‐1 | Tat protein | TMV (I) |
Karasev et al. (2005) |
| Respiratory syncytial virus | G protein epitope | AMV (D) |
Yusibov et al. (2005) |
| Classical swine fever | E2 glycoprotein peptides | PVX (D) |
Marconi et al. (2006) |
| Canine oral papillomavirus | L2 protein | TMV (D) |
Smith et al. (2006) |
| Melanoma | p15e‐Trp2 epitopes | TMV (D) |
McCormick et al. (2006b) |
| Yersinia pestis | F1 and V proteins | TMV (M) |
Santi et al. (2006) |
| Influenza A virus | M2 protein ectodomain | PVX (I) |
Nemchinov and Natilla (2007) |
| Dengue virus | Domain III of E protein | TMV (I) |
Saejung et al. (2007) |
| Mycobacterium tuberculosis | ESAT6—Ag85B antigens | TMV (I, D) |
Dorokhov et al. (2007) |
| Human papilloma virus | E7 protein | TMV (S) |
Massa et al. (2007) |
| Smallpox | pB5 antigenic domain | TMV (M) |
Golovkin et al. (2007) |
| Yersinia pestis | F1 and V proteins | TMV (S) |
Mett et al. (2007) |
| Bacillus anthracis | PA peptide | CPMV (D) |
Phelps et al. (2007) |
| Bacillus anthracis | PA and LF domains | TMV (S) |
Chichester et al. (2007) |
CPMV, cowpea mosaic virus; TMV, tobacco mosaic virus; AMV, alfalfa mosaic virus; TBSV, tomato bushy stunt virus; PPV, plum pox virus; PVX, potato virus X; CMV, cucumber mosaic virus; I, insertion vector; S, substitution vector; M, modular/deconstructed vector; D, viral display.
Yusibov et al., expressed the E7 oncoprotein from human papilloma virus (HPV) in N. benthamiana plants with a gene replacement vector. The antigen was subsequently purified from infected leaves using the thermotolerance properties of lichenase (Massa et al., 2007). Mice, immunized subcutaneously with the plant‐produced E7, showed a specific antibody and cytotoxic T‐cell response. The dose also protected the animals against challenge with E7 tumor cells. Moreover, the recombinant antigen was able to prevent tumor development when administered after virus challenge, supporting the ultimate goal to produce an anti‐tumor vaccine with both therapeutic and prophylactic uses.
A fundamental revolution within the last few years came from the use of modular deconstructed systems. A recent work using the magnICON TMV based system is worth noting. Santi et al. (2006) focused on the production of antigens for agents of biological warfare, for example, the Yersinia pestis bacterium, the causative agent of plague. In this work the protective efficacy of the fraction 1 capsular antigen (F1) and the low calcium response virulent antigen (V) expressed at levels up to 2 mg/g FW in N. benthamiana leaves was evaluated. Guinea pigs, immunized with purified antigens, showed antigen‐specific serum IgG titers and, more importantly, 75% of animals were protected from an aerosolized challenge of virulent Y. pestis. In a subsequent work Mett et al. (2007) expressed the same Y. pestis F1 and V antigens in N. benthamiana plants with an average yield of 0.38 mg/g FW with their lichenase optimized vector. The purified proteins were administered subcutaneously to non‐human primates, Cynomolgus Macaques, producing antigen specific IgG and IgA responses which resulted in complete protection against lethal challenge with Y. pestis.
Recent work show a particular interest on emerging and re‐emerging diseases, as well as agents of biological warfare, and have extended plant‐derived vaccine development to smallpox (Golovkin et al., 2007), anthrax (Chichester et al., 2007), dengue virus (Saejung et al., 2007), and avian influenza A virus (Nemchinov and Natilla, 2007). Altogether, these data collectively show the establishment of a portfolio of well characterized and novel, improved strategies for the expression of effective vaccines in plants.
Viral display for vaccine formulations
“Peptide” vaccines are a particular subclass of subunit vaccines based on the ability of immunodominant epitopes to induce specific immune responses. Epitopes are the most important regions of an antigen. They interact directly with surface receptors on B and T cells and induce an immune response. A single protein can have several epitopes with different immunological features and structures. Epitopes can be linear, determined by the amino acid sequence, or conformational, determined by neighboring amino acids only in the tridimensional tertiary structure. An epitope alone could be poorly immunogenic and often possess a short half‐life in the serum (Lien and Lowman, 2003). As a result, extensive efforts are being directed to the development of new delivery strategies to increase the immunogenicity and half‐life of epitope vaccines (Purcell et al., 2007). Plant viral display vectors have the potential to play an important role in such strategic development. Plant virus particles can structurally function as a scaffold to support, stabilize and display epitopic peptides. For example, a target epitope can be genetically fused to the CP protein. The chimeric virion will be formed by the CP‐epitope fusion, representing the vaccine by itself. As a result, plant viruses can be used as a carrier to stabilize epitopes and to present them correctly to the immune system.
Strategies such as the use of the amber leaky stop codon and 2A peptide (described earlier) have been adopted successfully to display peptides (Turpen et al., 1995; Marconi et al., 2006), as well as for full‐length protein fusions (O'Brien et al., 2000). In addition, new strategies involving biotinylation of the capsid and the subsequent binding of streptavidin‐conjugated target protein have been developed for displaying long sequences. The canine oral papillomavirus L2 protein, displayed on the TMV surface with this strategy, was significantly more immunogenic in animal models compared to the uncoupled antigen (Smith et al., 2006). With this strategy, it is also possible to combine helper epitopes and peptides known to facilitate cellular uptake or other additional T‐cell targets to avoid the use of adjuvants. Coexpression of melanoma‐associated cytotoxic T lymphocytes (CTL) epitopes p15e and Trp2 on a TMV scaffold stimulated effective tumor protection from challenge and showed a significant survival improvement over the single peptides alone (McCormick et al., 2006a).
The capability of CVPs in inducing an antibody response specifically to the displayed epitope upon intranasal, intraperitoneal or oral administration, have been extensively demonstrated in different animal models (Table 1). Other data show CVPs ability to elicit a strong specific neutralizing immune response in the absence of adjuvants (Yusibov et al., 1997; Brennan et al., 1999; Marusic et al., 2001). Oral delivery of spinach leaves infected with CVPs elicited a strong antibody response to the displayed rabies virus peptide in mice and human volunteers (Yusibov et al., 2002), indicating a potential use of this system as a supplementary oral booster for rabies vaccinations. Alfalfa mosaic virus (AMV)‐derived CVPs were able to elicit T‐ and B‐cell responses, in non‐human primates (Yusibov et al., 2005). Chimeric TMV particles was shown to be able to directly interact with and stimulate mammalian antigen presenting cells to induce a strong cellular anti‐tumor immune response (McCormick et al., 2006b). Studies of body distribution for orally administered CVPs also suggested their application as nanocapsules in novel drug delivery systems (Rae et al., 2005; Smith et al., 2007).
Applications of peptide‐displaying CVPs extend far beyond the area of vaccine development. Of particular interest are a killer peptide to confer broad‐spectrum resistance to phytopathogens (Donini et al., 2005), a metal‐binding peptide converting CVPs into an artificial metal‐adsorbing sink for metal tolerance and phytoremediation (Shingu et al., 2006), and a mosquito hormone‐derived decapeptide as a larvicide to protect plants against agricultural insect pests and to control vector mosquitoes (Borovsky et al., 2006). Finally Werner et al. (2006) fused a fragment of protein A (133aa) to the TMV CP C‐terminus via a 15‐aa linker. This chimeric nanoparticle allowed a simple purification of mAbs with 50% recovery yield and product purity of greater than 90%. This technology provides an inexpensive self‐assembling matrix that could be used as industrial immuno‐adsorbent for antibody purification.
Nanotechnology applications
Due to their simple macromolecular organization, assembly capabilities, structure stability, easy scalability and facile purification, plant viruses can offer a cheap source of biopolymers and nanoparticles. Well characterized viruses with stable, highly ordered and repetitive structures, such as TMV and CPMV, are of particular interests. For example, CPMV, cowpea chlorotic mottle virus (CCMV) and other icosahedral viruses have been used as nucleation cages for the mineralization of materials (Douglas and Young, 1998). Another promising application is in noninvasive in vivo vascular imaging techniques (Manchester and Singh, 2006). CPMV for example has been fluorescently labeled to visualize blood flow for periods of at least 72 h to identify vessels and to monitor tumor neovascularization (Lewis et al., 2006). Moreover, CPMV can be modified to generate new surface properties for developing novel biomaterials, electrochemical biosensors, or nanoelectronic devices (Wang et al., 2002; Chatterji et al., 2004; Steinmetz et al., 2006). Rod‐shaped viruses, like TMV, have been extensively used in the synthesis of a variety of metals nanowires, magnetic materials and semiconductors. TMV can be used as organic templates for the controlled deposition of gold, silver and platinum to prepare 1‐D arrays (Dujardin et al., 2003). Self‐assembled, modified CP monomers of TMV were also employed for the construction of photovoltaic components in a novel light‐harvesting system (Miller et al., 2007). CVPs can be also incorporated in liquid crystal systems or used to generate thin films and fibers (Flynn et al., 2003). These examples clearly demonstrate the broad application and emerging potential of plant viruses in nanotechnology fields.
Downstream Processing of Plant‐Derived Pharmaceuticals
Over the last decade, plant based production of pharmaceuticals has made remarkable progress as the expression of a diverse set of proteins has been demonstrated, several proteins have moved into clinical testing, and a plant‐derived veterinary vaccine has been approved. Recent developments in transient expression systems and production in a controlled green house environment are directly addressing the issues of low expression levels and under developed regulatory standards for plant made pharmaceuticals (PMPs).
In spite of this progress, barriers remain that prevent the broad adoption of this technology platform. One of these is the lack of translational research to bridge the gap between bench discoveries and their corresponding clinical products. In fact, many product failures during development are ultimately caused by problems of transition from laboratory prototype to industrial product, as stated by the FDA Critical Path Report (http://www.fda.gov/oc/initiatives/criticalpath/). Product industrialization programs are routinely delayed or derailed by inadequate efforts in downstream manufacturing, scale‐up development, and quality control. Therefore, as tremendous progress has been realized in recombinant protein expression, research focus has gradually been shifted to improvements in downstream processes including extraction, purification and recovery of the final products. Downstream processing is fundamentally important to the commercial viability of the specific PMPs and the PMP technology in general. An optimized downstream process will not only provide the additional cost‐saving measures for the overall product cost, it will also enable the large‐scale production of the products to meet the market demands. In addition, downstream process development is an essential step in compliance with FDA's current Good Manufacture Practice (cGMP) regulations.
The general downstream processing steps for extraction and purification of pharmaceutical proteins are similar to those of other systems. These steps include harvest and fractionation of the tissue containing the targeted protein, extraction of target protein into designed buffer, clarification of cell debris and particles from the extract, purification of the target protein, and vialing of the purified products into the proper container in the desired formulation buffer. The unique structure and biochemistry of plant cells and tissues present different challenges and opportunities for each of the above processing steps. In addition, the choice of host plant species, tissue and subcellular organelle targeting of the product will have profound impact on the strategy and outcome of downstream processing efforts. Since this review concerns primarily viral vector based transient production of PMPs, our discussion will focus on the downstream bioprocessing of proteins targeted to the cytoplasm of leafy plants. PMP production with seeds has been extensively reviewed by others (Menkhaus et al., 2004; Nikolov and Woodard, 2004).
Biomaterial harvest and fractionation
The first step of biomaterial processing is extraction and purification from tissue with a high concentration of the target recombinant protein. The selective harvest of the target tissue will reduce the volume of initial biomaterial, and in turn, reduce the unnecessary need to eliminate the extra host proteins introduced by the inclusion of non‐target tissue, and the total operational cost. The selectivity will also allow the avoidance of tissues which are hard to process, prime to introducing environmental contaminants (e.g., roots), or rich in other undesirable molecules such as proteases, alkaloids, and phenolics.
For transient expression with viral vectors, leaves are primarily the target tissue for protein accumulation. Currently most of the biomaterial production with these vectors is accomplished in greens houses with a controlled environment. For example, 4–18 days after inoculation, N. benthamiana leaves will be selectively harvested away from roots and stems (Werner et al., 2006). For small scale preparation, some researchers perform further fractionation of leaves by manually separating the central vein from the rest of leaves. For large‐scale production, this kind of operation becomes impractical and whole leaves are processed thoroughly in the next step. If the biomaterial is produced in open fields, extra steps have to be performed to remove the diverse array of environmental contaminants. Rinsing and light washing of plants, before harvesting the target tissue, is a common practice for this purpose. Regardless of where the biomaterial is generated, it is important to lower the bioburden and other contaminants as much as possible at this step. This measure will prevent microorganisms and other contaminants from entering purification feed streams and thus, greatly simplify the subsequent purification process and ensure the regulatory compliance of the final product.
One of critical factors in choosing plant expression platforms is the protein stability in the targeted tissue. With all the advantages of the viral based transient expression system, recombinant proteins generally do have less long‐term stability in leaves due to higher water content in contrast to the seed‐based stable transformed expression systems (Doran, 2006). However, some proteins did show relative long‐term stability in leaves of up to 7 days at room temperature (Fiedler et al., 1997) or even longer (12 weeks) in dry alfalfa tissue (Khoudi et al., 1999). Depending on the nature of the target protein, some of them need to be further processed as fresh leaf tissue, while others can be stored up to months as frozen tissue without losing their recovery and/or biological activity (Chen, unpublished work). The possibility of storing tissue at a cold temperature for certain proteins offers and expands the flexibility of this production platform.
Protein extraction
The major goal of this processing step is to release the target protein into a liquid buffer solution from the plant tissue. To achieve this goal in a leaf‐based transient expression system, leaf tissue is first ground in the presence of a desired extraction buffer to break the tissue and cells. The tissue homogenate will then be further pressed to release the protein into the aqueous buffer. The crude extract will be clarified to separate the protein‐containing buffer from plant debris, insoluble proteins (see below) and other particulate matters. In comparison to other production hosts, plants produce more solid debris (up to 30% of totally weight of the biomaterial (Menkhaus et al., 2004)). The larger and denser solids make it impossible to achieve effective clarification by filtration methods. As a result, continuous centrifugation remains the most effective and scalable technique for plant extract clarification. It should be noted that it is possible to extract small recombinant proteins (<50 kDa) that are targeted through the endomembrane system to the apoplast without breaking the plant cells. For example, some of the smaller proteins can be released directly into the extraction buffer by simply rinsing the tissue or by a specialized centrifugation technique (Lohaus et al., 2001). The purification process for this class of proteins can be greatly simplified with the result of a reduction in host protein contaminants.
The choice of extraction buffer has to be carefully determined based on the properties (e.g., pI, size, hydrophobicity, and stability) of the target protein and the major contaminating host molecules. For example, the major contaminating protein in plant leaves is ribulose 1,5‐bisphosphate carboxylase‐oxygenase (RuBisCo). Correspondingly, the choice of low pH buffers (∼pH 5.3) should be encouraged to keep RuBisCo insoluble and prevent it being extracted into the aqueous phase. At laboratory bench scale, protease inhibitors and antioxidants are routinely added to the extraction buffer to counter the denaturation, degradation and structural modification by proteases and phenolic compounds. Due to regulatory restraints and low‐cost requirement, these molecules should only be included when deemed absolutely necessary for large‐scale production.
Purification
Both chromatographic and non‐chromatographic methods have been employed to purify plant‐derived pharmaceutical proteins. Like proteins from other production hosts, purification strategies are formulated for each individual protein based on its solubility, size, pI, charge, hydrophobicity, and affinity to specific ligands and the parallel characteristics of host proteins. For mAb‐based PMPs, while protein A or G‐based chromatography provides a superb and convenient step (Langone, 1982), much effort is needed to eliminate plant host molecules which cause resin fouling or/and interfere with the binding of the target protein to protein A resin, and thus, reduce its capacity. Non‐chromatographic scalable processes such as aqueous two‐phase partitioning systems (ATPS) are being developed to address this key issue (Platis and Labrou, 2006).
For other non‐mAb‐based vaccines and therapeutics, a purification scheme has to be developed individually which is based on multiple steps of conventional chromatographic methods (Menkhaus et al., 2004). This time‐consuming and challenging process calls for the need to develop more “universal” or versatile purification methods. The employment of affinity tags, particularly the tandem affinity tag purification (TAP) strategy, provides possibilities for such versatile solution (Lichty et al., 2005; Arnau et al., 2006; Tagwerker et al., 2006). However, the search for both efficient and precise proteases for tag removal still presents serious challenges. In addition, the inclusion of proteases in the manufacturing process further complicates product purification, as well as raises cost and regulatory concerns (Feeney et al., 2006; Kenig et al., 2006). Several non‐enzymatic affinity tag removal techniques have been explored (Rais‐Beghdadi et al., 1998; Wood et al., 1999), but still require further optimization before becoming practical for product manufacturing.
In addition to native proteins, carbohydrates and other host molecules, unique plant endogenous molecules such as phenolics needed to be removed during purification. Phenolics tend to modify proteins by forming complexes, thus impeding purification if not removed earlier during purification (Kusnadi et al., 1998; Menkhaus et al., 2004). Some plants, such as tobacco, also produce a high‐level of toxic alkaloids that have to be eliminated from final product (Twyman et al., 2003). Regarding agroinfection, the potential elevated‐level of endotoxin (lipopolysaccharide, LPS) from a gram‐negative bacterium is another concern. Specific purification strategies to lower the LPS to acceptable levels must be incorporated into the overall scheme to ensure the safety of PMPs produced using this platform (Magalhaes et al., 2007).
The overall purification design has to be robust, scalable, cost‐effective and compliant to cGMP regulations. Ideally, no more than three chromatographic steps should be included to obtain a highly purified product. Otherwise, recovery will drop to an impractical level. Whenever possible, resins widely accepted by standard pharmaceutical industry should be considered first during process‐development to minimize future regulatory expenditure. The requirement for an independent Quality Management System (QMS) to govern the manufacture of pharmaceuticals also applies to the PMPs. In addition to cGMP compliance during downstream processing, analytical tests for releasing the final purified products have to address plant specific contaminants in addition to the standard required assays. Even though plants do not contain animal viruses and other infectious agents, it must be validated that LPS, phenolics, and alkaloids, as well as herbicides and insecticides have been adequately removed from the final product.
Conclusion
A significant increase in demand for high quality recombinant proteins is already on the horizon. New biological systems for the production of these proteins must be developed to meet market demands. Plant expression systems based on viral vectors have the greatest potential to provide such technology. Once optimized and implemented at a commercial scale, these expression systems should create a technology platform to produce recombinant proteins with scalability, speed, efficiency, cost‐effectiveness and safety.
Literature Cited
- Alamillo JM, Monger W, Sola I, Garcia B, Perrin Y, Bestagno M, Burrone OR, Sabella P, Plana‐Duran J, Enjuanes L, Lomonossoff GP, Garcia JA. 2006. Use of virus vectors for the expression in plants of active full‐length and single chain anti‐coronavirus antibodies. Biotechnol J 1: 1103–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez ML, Pinyerd HL, Crisantes JD, Rigano MM, Pinkhasov J, Walmsley AM, Mason HS, Cardineau GA. 2006. Plant‐made subunit vaccine against pneumonic and bubonic plague is orally immunogenic in mice. Vaccine 24: 2477–2490. [DOI] [PubMed] [Google Scholar]
- Arnau J, Lauritzen C, Petersen GE, Pedersen J. 2006. Current strategies for the use of affinity tags and tag removal for the purification of recombinant proteins. Protein Expr Purif 48: 1–13. [DOI] [PubMed] [Google Scholar]
- Avesani L, Marconi G, Morandini F, Albertini E, Bruschetta M, Bortesi L, Pezzotti M, Porceddu A. 2007. Stability of Potato virus X expression vectors is related to insert size: Implications for replication models and risk assessment. Transgenic Res 16: 587–597. [DOI] [PubMed] [Google Scholar]
- Baratova LA, Grebenshchikov NI, Dobrov EN, Gedrovich AV, Kashirin IA, Shishkov AV, Efimov AV, Jarvekulg L, Radavsky YL, Saarma M. 1992a. The organization of potato virus X coat proteins in virus particles studied by tritium planigraphy and model building. Virology 188: 175–180. [DOI] [PubMed] [Google Scholar]
- Baratova LA, Grebenshchikov NI, Shishkov AV, Kashirin IA, Radavsky JL, Jarvekulg L, Saarma M. 1992b. The topography of the surface of potato virus X: Tritium planigraphy and immunological analysis. J Gen Virol 73: 229–235. [DOI] [PubMed] [Google Scholar]
- Baulcombe DC. 1999. Fast forward genetics based on virus‐induced gene silencing. Curr Opin Plant Biol 2: 109–113. [DOI] [PubMed] [Google Scholar]
- Bendahmane M, Koo M, Karrer E, Beachy RN. 1999. Display of epitopes on the surface of tobacco mosaic virus: Impact of charge and isoelectric point of the epitope on virus‐host interactions. J Mol Biol 290: 9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R. 2007. Plastid biotechnology: Prospects for herbicide and insect resistance. Metabolic engineering and molecular farming. Curr Opin Biotechnol 18: 100–106. [DOI] [PubMed] [Google Scholar]
- Borovsky D, Rabindran S, Dawson WO, Powell CA, Iannotti DA, Morris TJ, Shabanowitz J, Hunt DF, DeBondt HL, DeLoof A. 2006. Expression of Aedes trypsin‐modulating oostatic factor on the virion of TMV: A potential larvicide. Proc Natl Acad Sci USA 103: 18963–18968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos L. 1999. Beijerinck's work on tobacco mosaic virus: Historical context and legacy. Philos Trans R Soc Lond B Biol Sci 354: 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan FR, Bellaby T, Helliwell SM, Jones TD, Kamstrup S, Dalsgaard K, Flock JI, Hamilton WD. 1999. Chimeric plant virus particles administered nasally or orally induce systemic and mucosal immune responses in mice. J Virol 73: 930–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brereton HM, Chamberlain D, Yang R, Tea M, McNeil S, Coster DJ, Williams KA. 2007. Single chain antibody fragments for ocular use produced at high levels in a commercial wheat variety. J Biotechnol 129: 539–546. [DOI] [PubMed] [Google Scholar]
- Canizares MC, Liu L, Perrin Y, Tsakiris E, Lomonossoff GP. 2006. A bipartite system for the constitutive and inducible expression of high levels of foreign proteins in plants. Plant Biotechnol J 4: 183–193. [DOI] [PubMed] [Google Scholar]
- Carrillo C, Wigdorovitz A, Oliveros JC, Zamorano PI, Sadir AM, Gomez N, Salinas J, Escribano JM, Borca MV. 1998. Protective immune response to foot‐and‐mouth disease virus with VP1 expressed in transgenic plants. J Virol 72: 1688–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chackerian B. 2007. Virus‐like particles: Flexible platforms for vaccine development. Expert Rev Vaccines 6: 381–390. [DOI] [PubMed] [Google Scholar]
- Chapman S, Kavanagh T, Baulcombe D. 1992. Potato virus X as a vector for gene expression in plants. Plant J 2: 549–557. [DOI] [PubMed] [Google Scholar]
- Chargelegue D, Vine ND, van Dolleweerd CJ, Drake PM, Ma JK. 2000. A murine monoclonal antibody produced in transgenic plants with plant‐specific glycans is not immunogenic in mice. Transgenic Res 9: 187–194. [DOI] [PubMed] [Google Scholar]
- Chatterji A, Ochoa WF, Paine M, Ratna BR, Johnson JE, Lin T. 2004. New addresses on an addressable virus nanoblock; uniquely reactive Lys residues on cowpea mosaic virus. Chem Biol 11: 855–863. [DOI] [PubMed] [Google Scholar]
- Chichester JA, Musiychuk K, de la Rosa P, Horsey A, Stevenson N, Ugulava N, Rabindran S, Palmer GA, Mett V, Yusibov V. 2007. Immunogenicity of a subunit vaccine against Bacillus anthracis. Vaccine 25: 3111–3114. [DOI] [PubMed] [Google Scholar]
- Chikwamba RK, Scott MP, Mejia LB, Mason HS, Wang K. 2003. Localization of a bacterial protein in starch granules of transgenic maize kernels. Proc Natl Acad Sci USA 100: 11127–11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough RC, Pappu K, Thompson K, Beifuss K, Lane J, Delaney DE, Harkey R, Drees C, Howard JA, Hood EE. 2006. Manganese peroxidase from the white‐rot fungus Phanerochaete chrysosporium is enzymatically active and accumulates to high levels in transgenic maize seed. Plant Biotechnol J 4: 53–62. [DOI] [PubMed] [Google Scholar]
- Dalsgaard K, Uttenthal A, Jones TD, Xu F, Merryweather A, Hamilton WD, Langeveld JP, Boshuizen RS, Kamstrup S, Lomonossoff GP, Porta C, Vela C, Casal JI, Meloen RH, Rodgers PB. 1997. Plant‐derived vaccine protects target animals against a viral disease. Nat Biotechnol 15: 248–252. [DOI] [PubMed] [Google Scholar]
- Dawson WO, Lewandowski DJ, Hilf ME, Bubrick P, Raffo AJ, Shaw JJ, Grantham GL, Desjardins PR. 1989. A tobacco mosaic virus‐hybrid expresses and loses an added gene. Virology 172: 285–292. [DOI] [PubMed] [Google Scholar]
- de Felipe P, Hughes LE, Ryan MD, Brown JD. 2003. Co‐translational, intraribosomal cleavage of polypeptides by the foot‐and‐mouth disease virus 2A peptide. J Biol Chem 278: 11441–11448. [DOI] [PubMed] [Google Scholar]
- Dohi K, Nishikiori M, Tamai A, Ishikawa M, Meshi T, Mori M. 2006. Inducible virus‐mediated expression of a foreign protein in suspension‐cultured plant cells. Arch Virol 151: 1075–1084. [DOI] [PubMed] [Google Scholar]
- Donini M, Lico C, Baschieri S, Conti S, Magliani W, Polonelli L, Benvenuto E. 2005. Production of an engineered killer peptide in Nicotiana benthamiana by using a potato virus X expression system. Appl Environ Microbiol 71: 6360–6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly ML, Luke G, Mehrotra A, Li X, Hughes LE, Gani D, Ryan MD. 2001. Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: A putative ribosomal ‘skip’. J Gen Virol 82: 1013–1025. [DOI] [PubMed] [Google Scholar]
- Doran PM. 2006. Foreign protein degradation and instability in plants and plant tissue cultures. Trends Biotechnol 24: 426–432. [DOI] [PubMed] [Google Scholar]
- Dorokhov YL, Sheveleva AA, Frolova OY, Komarova TV, Zvereva AS, Ivanov PA, Atabekov JG. 2007. Superexpression of tuberculosis antigens in plant leaves. Tuberculosis (Edinb) 87: 218–224. [DOI] [PubMed] [Google Scholar]
- Douglas T, Young M. 1998. Host‐guest encapsulation of materials by assembled virus protein cages. 393: 152–155. [Google Scholar]
- Dujardin E, Peet C, Stubbs G, Culver JN, Mann S. 2003. Organization of metallic nanoparticles using tobacco mosaic virus templates. Nano Lett 3: 413–417. [Google Scholar]
- Feeney B, Soderblom EJ, Goshe MB, Clark AC. 2006. Novel protein purification system utilizing an N‐terminal fusion protein and a caspase‐3 cleavable linker. Protein Expr Purif 47: 311–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez‐Fernandez MR, Martinez‐Torrecuadrada JL, Casal JI, Garcia JA. 1998. Development of an antigen presentation system based on plum pox potyvirus. FEBS Lett 427: 229–235. [DOI] [PubMed] [Google Scholar]
- Fernandez‐Fernandez MR, Mourino M, Rivera J, Rodriguez F, Plana‐Duran J, Garcia JA. 2001. Protection of rabbits against rabbit hemorrhagic disease virus by immunization with the VP60 protein expressed in plants with a potyvirus‐based vector. Virology 280: 283–291. [DOI] [PubMed] [Google Scholar]
- Fiedler U, Phillips J, Artsaenko O, Conrad U. 1997. Optimization of scFv antibody production in transgenic plants. Immunotechnology 3: 205–216. [DOI] [PubMed] [Google Scholar]
- Fischer R, Twyman RM, Schillberg S. 2003. Production of antibodies in plants and their use for global health. Vaccine 21: 820–825. [DOI] [PubMed] [Google Scholar]
- Flynn CE, Lee S‐W, Peelle BR, Belcher AM. 2003. Viruses as vehicles for growth, organization and assembly of materials. Acta Materialia 51: 5867–5880. [Google Scholar]
- Franconi R, Di Bonito P, Dibello F, Accardi L, Muller A, Cirilli A, Simeone P, Dona MG, Venuti A, Giorgi C. 2002. Plant‐derived human papillomavirus 16 E7 oncoprotein induces immune response and specific tumor protection. Cancer Res 62: 3654–3658. [PubMed] [Google Scholar]
- Giddings G. 2001. Transgenic plants as protein factories. Curr Opin Biotechnol 12: 450–454. [DOI] [PubMed] [Google Scholar]
- Giritch A, Marillonnet S, Engler C, van Eldik G, Botterman J, Klimyuk V, Gleba Y. 2006. Rapid high‐yield expression of full‐size IgG antibodies in plants coinfected with noncompeting viral vectors. Proc Natl Acad Sci USA 103: 14701–14706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleba Y, Klimyuk V, Marillonnet S. 2005. Magnifection—A new platform for expressing recombinant vaccines in plants. Vaccine 23: 2042–2048. [DOI] [PubMed] [Google Scholar]
- Golovkin M, Spitsin S, Andrianov V, Smirnov Y, Xiao Y, Pogrebnyak N, Markley K, Brodzik R, Gleba Y, Isaacs SN, Koprowski H. 2007. Smallpox subunit vaccine produced in Planta confers protection in mice. Proc Natl Acad Sci USA 104: 6864–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomord V, Faye L. 2004. Posttranslational modification of therapeutic proteins in plants. Curr Opin Plant Biol 7: 171–181. [DOI] [PubMed] [Google Scholar]
- Grimsley N, Hohn B, Hohn T, Walden R. 1986. “Agroinfection,” an alternative route for viral infection of plants by using the Ti plasmid. Proc Natl Acad Sci 83: 3282–3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto H, Sugiyama Y, Nakagawa N, Hashida E, Matsunaga Y, Takemoto S, Watanabe Y, Okada Y. 1993. A new tobacco mosaic virus vector and its use for the systemic production of angiotensin‐I‐converting enzyme inhibitor in transgenic tobacco and tomato. Biotechnology (NY) 11: 930–932. [DOI] [PubMed] [Google Scholar]
- Hansen G, Wright MS. 1999. Recent advances in the transformation of plants. Trends Plant Sci 4: 226–231. [DOI] [PubMed] [Google Scholar]
- Harashima S. 1994. Heterologous protein production by yeast host‐vector systems. Bioprocess Technol 19: 137–158. [PubMed] [Google Scholar]
- Haviv S, Galiakparov N, Goszczynski DE, Batuman O, Czosnek H, Mawassi M. 2006. Engineering the genome of Grapevine virus A into a vector for expression of proteins in herbaceous plants. J Virol Methods 132: 227–231. [DOI] [PubMed] [Google Scholar]
- Hayes RJ, Coutts RH, Buck KW. 1989. Stability and expression of bacterial genes in replicating geminivirus vectors in plants. Nucleic Acids Res 17: 2391–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiatt A, Cafferkey R, Bowdish K. 1989. Production of antibodies in transgenic plants. Nature 342: 76–78. [DOI] [PubMed] [Google Scholar]
- Hood EE, Kusnadi A, Nikolov Z, Howard JA. 1999. Molecular farming of industrial proteins from transgenic maize. Adv Exp Med Biol 464: 127–147. [DOI] [PubMed] [Google Scholar]
- Hsu C‐H, Lin S‐S, Liu F‐L, Su W‐C, Yeh S‐D. 2004. Oral administration of a mite allergen expressed by zucchini yellow mosaic virus in cucurbit species downregulates allergen‐induced airway inflammation and IgE synthesis. J Allergy Clin Immunol 113: 1079–1085. [DOI] [PubMed] [Google Scholar]
- Huang J, Wu L, Yalda D, Adkins Y, Kelleher SL, Crane M, Lonnerdal B, Rodriguez RL, Huang N. 2002. Expression of functional recombinant human lysozyme in transgenic rice cell culture. Transgenic Res 11: 229–239. [DOI] [PubMed] [Google Scholar]
- Huang Z, Elkin G, Maloney BJ, Beuhner N, Arntzen CJ, Thanavala Y, Mason HS. 2005. Virus‐like particle expression and assembly in plants: Hepatitis B and Norwalk viruses. Vaccine 23: 1851–1858. [DOI] [PubMed] [Google Scholar]
- Huang LK, Liao SC, Chang CC, Liu HJ. 2006. Expression of avian reovirus sigma C protein in transgenic plants. J Virol Methods 134: 217–222. [DOI] [PubMed] [Google Scholar]
- Humphrey BD, Huang N, Klasing KC. 2002. Rice expressing lactoferrin and lysozyme has antibiotic‐like properties when fed to chicks. J Nutr 132: 1214–1218. [DOI] [PubMed] [Google Scholar]
- Jackson DA, Symons RH, Berg P. 1972. Biochemical method for inserting new genetic information into DNA of Simian Virus 40: Circular SV40 DNA molecules containing lambda phage genes and the galactose operon of Escherichia coli . Proc Natl Acad Sci USA 69: 2904–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joelson T, Akerblom L, Oxelfelt P, Strandberg B, Tomenius K, Morris TJ. 1997. Presentation of a foreign peptide on the surface of tomato bushy stunt virus. J Gen Virol 78: 1213–1217. [DOI] [PubMed] [Google Scholar]
- Joensuu JJ, Kotiaho M, Teeri TH, Valmu L, Nuutila AM, Oksman‐Caldentey KM, Niklander‐Teeri V. 2006. Glycosylated F4 (K88) fimbrial adhesin FaeG expressed in barley endosperm induces ETEC‐neutralizing antibodies in mice. Transgenic Res 15: 359–373. [DOI] [PubMed] [Google Scholar]
- Johnson J, Lin T, Lomonossoff G. 1997. Presentation of heterologous peptides on plant viruses: Genetics, structure, and function. Annu Rev Phytopathol 35: 67–86. [DOI] [PubMed] [Google Scholar]
- Karasev AV, Foulke S, Wellens C, Rich A, Shon KJ, Zwierzynski I, Hone D, Koprowski H, Reitz M. 2005. Plant based HIV‐1 vaccine candidate: Tat protein produced in spinach. Vaccine 23: 1875–1880. [DOI] [PubMed] [Google Scholar]
- Kelloniemi J, Makinen K, Valkonen JP. 2006. A potyvirus‐based gene vector allows producing active human S‐COMT and animal GFP, but not human sorcin, in vector‐infected plants. Biochimie 88: 505–513. [DOI] [PubMed] [Google Scholar]
- Kenig M, Peternel S, Gaberc‐Porekar V, Menart V. 2006. Influence of the protein oligomericity on final yield after affinity tag removal in purification of recombinant proteins. J Chromatogr A 1101: 293–306. [DOI] [PubMed] [Google Scholar]
- Khoudi H, Laberge S, Ferullo JM, Bazin R, Darveau A, Castonguay Y, Allard G, Lemieux R, Vezina LP. 1999. Production of a diagnostic monoclonal antibody in perennial alfalfa plants. Biotechnol Bioeng 64: 135–143. [DOI] [PubMed] [Google Scholar]
- Klug A, Caspar DL. 1960. The structure of small viruses. Adv Virus Res 7: 225–325. [DOI] [PubMed] [Google Scholar]
- Kusnadi AR, Evangelista RL, Hood EE, Howard JA, Nikolov ZL. 1998. Processing of transgenic corn seed and its effect on the recovery of recombinant beta‐glucuronidase. Biotechnol Bioeng 60: 44–52. [DOI] [PubMed] [Google Scholar]
- Lamphear BJ, Barker DK, Brooks CA, Delaney DE, Lane JR, Beifuss K, Love R, Thompson K, Mayor J, Clough R, Harkey R, Poage M, Drees C, Horn ME, Streatfield SJ, Nikolov Z, Woodard SL, Hood EE, Jilka JM, Howard JA. 2005. Expression of the sweet protein brazzein in maize for production of a new commercial sweetener. Plant Biotechnol J 3: 103–114. [DOI] [PubMed] [Google Scholar]
- Langone JJ. 1982. Protein A of Staphylococcus aureus and related immunoglobulin receptors produced by streptococci and pneumonococci. Adv Immunol 32: 157–252. [PubMed] [Google Scholar]
- Lewis JD, Destito G, Zijlstra A, Gonzalez MJ, Quigley JP, Manchester M, Stuhlmann H. 2006. Viral nanoparticles as tools for intravital vascular imaging. Nat Med 12: 354–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichty JJ, Malecki JL, Agnew HD, Michelson‐Horowitz DJ, Tan S. 2005. Comparison of affinity tags for protein purification. Protein Expr Purif 41: 98–105. [DOI] [PubMed] [Google Scholar]
- Lico C, Capuano F, Renzone G, Donini M, Marusic C, Scaloni A, Benvenuto E, Baschieri S. 2006. Peptide display on Potato virus X: Molecular features of the coat protein‐fused peptide affecting cell‐to‐cell and phloem movement of chimeric virus particles. J Gen Virol 87: 3103–3112. [DOI] [PubMed] [Google Scholar]
- Lien S, Lowman HB. 2003. Therapeutic peptides. Trends Biotechnol 21: 556–562. [DOI] [PubMed] [Google Scholar]
- Liu L, Canizares MC, Monger W, Perrin Y, Tsakiris E, Porta C, Shariat N, Nicholson L, Lomonossoff GP. 2005. Cowpea mosaic virus‐based systems for the production of antigens and antibodies in plants. Vaccine 23: 1788–1792. [DOI] [PubMed] [Google Scholar]
- Lohaus G, Pennewiss K, Sattelmacher B, Hussmann M, Hermann Muehling K. 2001. Is the infiltration‐centrifugation technique appropriate for the isolation of apoplastic fluid? A critical evaluation with different plant species. Physiol Plant 111: 457–465. [DOI] [PubMed] [Google Scholar]
- Lomonossoff GP, Johnson JE. 1991. The synthesis and structure of comovirus capsids. Prog Biophys Mol Biol 55: 107–137. [DOI] [PubMed] [Google Scholar]
- Lu R, Martin‐Hernandez AM, Peart JR, Malcuit I, Baulcombe DC. 2003. Virus‐induced gene silencing in plants. Methods 30: 296–303. [DOI] [PubMed] [Google Scholar]
- Ma JK, Drake PM, Chargelegue D, Obregon P, Prada A. 2005. Antibody processing and engineering in plants, and new strategies for vaccine production. Vaccine 23: 1814–1818. [DOI] [PubMed] [Google Scholar]
- Magalhaes PO, Lopes AM, Mazzola PG, Rangel‐Yagui C, Penna TC, Pessoa A Jr. 2007. Methods of endotoxin removal from biological preparations: A review. J Pharm Pharm Sci 10: 388–404. [PubMed] [Google Scholar]
- Malissard M, Borsig L, Di Marco S, Grutter MG, Kragl U, Wandrey C, Berger EG. 1996. Recombinant soluble beta‐1,4‐galactosyltransferases expressed in Saccharomyces cerevisiae. Purification, characterization and comparison with human enzyme. Eur J Biochem 239: 340–348. [DOI] [PubMed] [Google Scholar]
- Manchester M, Singh P. 2006. Virus‐based nanoparticles (VNPs): Platform technologies for diagnostic imaging. Adv Drug Deliv Rev 58: 1505–1522. [DOI] [PubMed] [Google Scholar]
- Marconi G, Albertini E, Barone P, De Marchis F, Lico C, Marusic C, Rutili D, Veronesi F, Porceddu A. 2006. In planta production of two peptides of the Classical Swine Fever Virus (CSFV) E2 glycoprotein fused to the coat protein of potato virus X. BMC Biotechnol 6: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marillonnet S, Giritch A, Gils M, Kandzia R, Klimyuk V, Gleba Y. 2004. In planta engineering of viral RNA replicons: Efficient assembly by recombination of DNA modules delivered by Agrobacterium. Proc Natl Acad Sci USA 101: 6852–6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marillonnet S, Thoeringer C, Kandzia R, Klimyuk V, Gleba Y. 2005. Systemic Agrobacterium tumefaciens‐mediated transfection of viral replicons for efficient transient expression in plants. Nat Biotechnol 23: 718–723. [DOI] [PubMed] [Google Scholar]
- Marquet‐Blouin E, Bouche FB, Steinmetz A, Muller CP. 2003. Neutralizing immunogenicity of transgenic carrot (Daucus carota L.)‐derived measles virus hemagglutinin. Plant Mol Biol 51: 459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusic C, Rizza P, Lattanzi L, Mancini C, Spada M, Belardelli F, Benvenuto E, Capone I. 2001. Chimeric plant virus particles as immunogens for inducing murine and human immune responses against human immunodeficiency virus type 1. J Virol 75: 8434–8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason HS, Lam DM, Arntzen CJ. 1992. Expression of hepatitis B surface antigen in transgenic plants. Proc Natl Acad Sci USA 89: 11745–11749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massa S, Franconi R, Brandi R, Muller A, Mett V, Yusibov V, Venuti A. 2007. Anti‐cancer activity of plant‐produced HPV16 E7 vaccine. Vaccine 25: 3018–3021. [DOI] [PubMed] [Google Scholar]
- Matthews REF. 1989. Roy Markham: Pioneer in plant pathology. Ann Rev Phytopathology 27: 13–23. [DOI] [PubMed] [Google Scholar]
- McCormick AA, Kumagai MH, Hanley K, Turpen TH, Hakim I, Grill LK, Tuse D, Levy S, Levy R. 1999. Rapid production of specific vaccines for lymphoma by expression of the tumor‐derived single‐chain Fv epitopes in tobacco plants. Proc Natl Acad Sci USA 96: 703–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick AA, Reinl SJ, Cameron TI, Vojdani F, Fronefield M, Levy R, Tuse D. 2003. Individualized human scFv vaccines produced in plants: Humoral anti‐idiotype responses in vaccinated mice confirm relevance to the tumor Ig. J Immunol Methods 278: 95–104. [DOI] [PubMed] [Google Scholar]
- McCormick AA, Corbo TA, Wykoff‐Clary S, Nguyen LV, Smith ML, Palmer KE, Pogue GP. 2006a. TMV‐peptide fusion vaccines induce cell‐mediated immune responses and tumor protection in two murine models. Vaccine 24: 6414–6423. [DOI] [PubMed] [Google Scholar]
- McCormick AA, Corbo TA, Wykoff‐Clary S, Palmer KE, Pogue GP. 2006b. Chemical conjugate TMV‐peptide bivalent fusion vaccines improve cellular immunity and tumor protection. Bioconjug Chem 17: 1330–1338. [DOI] [PubMed] [Google Scholar]
- Menkhaus TJ, Bai Y, Zhang C, Nikolov ZL, Glatz CE. 2004. Considerations for the recovery of recombinant proteins from plants. Biotechnol Prog 20: 1001–1014. [DOI] [PubMed] [Google Scholar]
- Mett V, Lyons J, Musiychuk K, Chichester JA, Brasil T, Couch R, Sherwood R, Palmer GA, Streatfield SJ, Yusibov V. 2007. A plant‐produced plague vaccine candidate confers protection to monkeys. Vaccine 25: 3014–3017. [DOI] [PubMed] [Google Scholar]
- Miller RA, Presley AD, Francis MB. 2007. Self‐assembling light‐harvesting systems from synthetically modified tobacco mosaic virus coat proteins. J Am Chem Soc 129: 3104–3109. [DOI] [PubMed] [Google Scholar]
- Monger W, Alamillo JM, Sola I, Perrin Y, Bestagno M, Burrone OR, Sabella P, Plana‐Duran J, Enjuanes L, Garcia JA, Lomonossoff GP. 2006. An antibody derivative expressed from viral vectors passively immunizes pigs against transmissible gastroenteritis virus infection when supplied orally in crude plant extracts. Plant Biotechnol J 4: 623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor TS, Gomez‐Lim MA, Palmer KE. 1998. Perspective: Edible vaccines—A concept coming of age. Trends Microbiol 6: 449–453. [DOI] [PubMed] [Google Scholar]
- Mor TS, Moon Y‐S, Palmer KE, Mason HS. 2003. Geminivirus vectors for high‐level expression of foreign proteins in plant cells. Biotechnol Bioeng 81: 430–437. [DOI] [PubMed] [Google Scholar]
- Moravec T, Schmidt MA, Herman EM, Woodford‐Thomas T. 2007. Production of Escherichia coli heat labile toxin (LT) B subunit in soybean seed and analysis of its immunogenicity as an oral vaccine. Vaccine 25: 1647–1657. [DOI] [PubMed] [Google Scholar]
- Musiychuk K, Stevenson N, Bi H, Farrance CE, Orozovic C, Brodelius M, Brodelius P, Horsey A, Ugulava N, Shamloul A‐M, Mett V, Rabindran S, Streatfield SJ, Yusibov V. 2007. A launch vector for the production of vaccine antigens in plants. Influenza 1: 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natilla A, Piazzolla G, Nuzzaci M, Saldarelli P, Tortorella C, Antonaci S, Piazzolla P. 2004. Cucumber mosaic virus as carrier of a hepatitis C virus‐derived epitope. Arch Virol 149: 137–154. [DOI] [PubMed] [Google Scholar]
- Nemchinov LG, Natilla A. 2007. Transient expression of the ectodomain of matrix protein 2 (M2e) of avian influenza A virus in plants. Protein Expr Purif 56: 153–159. [DOI] [PubMed] [Google Scholar]
- Nemchinov LG, Liang TJ, Rifaat MM, Mazyad HM, Hadidi A, Keith JM. 2000. Development of a plant‐derived subunit vaccine candidate against hepatitis C virus. Arch Virol 145: 2557–2573. [DOI] [PubMed] [Google Scholar]
- Nikolov ZL, Woodard SL. 2004. Downstream processing of recombinant proteins from transgenic feedstock. Curr Opin Biotechnol 15: 479–486. [DOI] [PubMed] [Google Scholar]
- Nochi T, Takagi H, Yuki Y, Yang L, Masumura T, Mejima M, Nakanishi U, Matsumura A, Uozumi A, Hiroi T, Morita S, Tanaka K, Takaiwa F, Kiyono H. 2007. From the cover: Rice‐based mucosal vaccine as a global strategy for cold‐chain‐ and needle‐free vaccination. Proc Natl Acad Sci 104: 10986–10991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolke G, Fischer R, Schillberg S. 2004. Antibody‐based pathogen resistance in plants. J Plant Pathol 86: 5–17. [Google Scholar]
- Nolke G, Schneider B, Fischer R, Schillberg S. 2005. Immunomodulation of polyamine biosynthesis in tobacco plants has a significant impact on polyamine levels and generates a dwarf phenotype. Plant Biotechnol J 3: 237–247. [DOI] [PubMed] [Google Scholar]
- Nuttall J, Ma JK, Frigerio L. 2005. A functional antibody lacking N‐linked glycans is efficiently folded, assembled and secreted by tobacco mesophyll protoplasts. Plant Biotechnol J 3: 497–504. [DOI] [PubMed] [Google Scholar]
- Nuzzaci M, Piazzolla G, Vitti A, Lapelosa M, Tortorella C, Stella I, Natilla A, Antonaci S, Piazzolla P. 2007. Cucumber mosaic virus as a presentation system for a double hepatitis C virus‐derived epitope. Arch Virol 152: 915–928. [DOI] [PubMed] [Google Scholar]
- O'Brien GJ, Bryant CJ, Voogd C, Greenberg HB, Gardner RC, Bellamy AR. 2000. Rotavirus VP6 expressed by PVX vectors in Nicotiana benthamiana coats PVX rods and also assembles into viruslike particles. Virology 270: 444–453. [DOI] [PubMed] [Google Scholar]
- Olempska‐Beer ZS, Merker RI, Ditto MD, DiNovi MJ. 2006. Food‐processing enzymes from recombinant microorganisms—A review. Regul Toxicol Pharmacol 45: 144–158. [DOI] [PubMed] [Google Scholar]
- Ooi A, Tan S, Mohamed R, Rahman NA, Othman RY. 2006. The full‐length clone of cucumber green mottle mosaic virus and its application as an expression system for Hepatitis B surface antigen. J Biotechnol 121: 471–481. [DOI] [PubMed] [Google Scholar]
- Palmer KE, Rybicki EP. 2001. Investigation of the potential of maize streak virus to act as an infectious gene vector in maize plants. Arch Virol 146: 1089–1104. [DOI] [PubMed] [Google Scholar]
- Parker L, Kendall A, Stubbs G. 2002. Surface features of potato virus X from fiber diffraction. Virology 300: 291–295. [DOI] [PubMed] [Google Scholar]
- Pavlou AK, Reichert JM. 2004. Recombinant protein therapeutics—Success rates, market trends and values to 2010. 22: 1513–1519. [DOI] [PubMed] [Google Scholar]
- Pen J, van Ooyen AJ, van den Elzen PJ, Rietveld K, Hoekema A. 1992. Direct screening for high‐level expression of an introduced alpha‐amylase gene in plants. Plant Mol Biol 18: 1133–1139. [DOI] [PubMed] [Google Scholar]
- Perez Filgueira DM, Zamorano PI, Dominguez MG, Taboga O, Del Medico Zajac MP, Puntel M, Romera SA, Morris TJ, Borca MV, Sadir AM. 2003. Bovine herpes virus gD protein produced in plants using a recombinant tobacco mosaic virus (TMV) vector possesses authentic antigenicity. Vaccine 21: 4201–4209. [DOI] [PubMed] [Google Scholar]
- Peschen D, Li HP, Fischer R, Kreuzaler F, Liao YC. 2004. Fusion proteins comprising a Fusarium‐specific antibody linked to antifungal peptides protect plants against a fungal pathogen. Nat Biotechnol 22: 732–738. [DOI] [PubMed] [Google Scholar]
- Phelps JP, Dang N, Rasochova L. 2007. Inactivation and purification of cowpea mosaic virus‐like particles displaying peptide antigens from Bacillus anthracis. J Virol Methods 141: 146–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platis D, Labrou NE. 2006. Development of an aqueous two‐phase partitioning system for fractionating therapeutic proteins from tobacco extract. J Chromatogr A 1128: 114–124. [DOI] [PubMed] [Google Scholar]
- Porta C, Lomonossoff GP. 1998. Scope for using plant viruses to present epitopes from animal pathogens. Rev Med Virol 8: 25–41. [DOI] [PubMed] [Google Scholar]
- Porta C, Spall VE, Loveland J, Johnson JE, Barker PJ, Lomonossoff GP. 1994. Development of cowpea mosaic virus as a high‐yielding system for the presentation of foreign peptides. Virology 202: 949–955. [DOI] [PubMed] [Google Scholar]
- Porta C, Spall VE, Findlay KC, Gergerich RC, Farrance CE, Lomonossoff GP. 2003. Cowpea mosaic virus‐based chimaeras. Effects of inserted peptides on the phenotype, host range, and transmissibility of the modified viruses. Virology 310: 50–63. [DOI] [PubMed] [Google Scholar]
- Purcell AW, McCluskey J, Rossjohn J. 2007. More than one reason to rethink the use of peptides in vaccine design. Nat Drug Rev Discov 6: 404–414. [DOI] [PubMed] [Google Scholar]
- Rae CS, Khor IW, Wang Q, Destito G, Gonzalez MJ, Singh P, Thomas DM, Estrada MN, Powell E, Finn MG, Manchester M. 2005. Systemic trafficking of plant virus nanoparticles in mice via the oral route. Virology 343: 224–235. [DOI] [PubMed] [Google Scholar]
- Rais‐Beghdadi C, Roggero MA, Fasel N, Reymond CD. 1998. Purification of recombinant proteins by chemical removal of the affinity tag. Appl Biochem Biotechnol 74: 95–103. [DOI] [PubMed] [Google Scholar]
- Rodriguez M, Ramirez NI, Ayala M, Freyre F, Perez L, Triguero A, Mateo C, Selman‐Housein G, Gavilondo JV, Pujol M. 2005. Transient expression in tobacco leaves of an aglycosylated recombinant antibody against the epidermal growth factor receptor. Biotechnol Bioeng 89: 188–194. [DOI] [PubMed] [Google Scholar]
- Saejung W, Fujiyama K, Takasaki T, Ito M, Hori K, Malasit P, Watanabe Y, Kurane I, Seki T. 2007. Production of dengue 2 envelope domain III in plant using TMV‐based vector system. Vaccine 25: 6646–6654. [DOI] [PubMed] [Google Scholar]
- Sainsbury F, Lavoie PO, D'Aoust MA, Vezina LP, Lomonossoff GP. 2008. Expression of multiple proteins using full‐length and deleted versions of cowpea mosaic virus RNA‐2. Plant Biotechnol J 6: 82–92. [DOI] [PubMed] [Google Scholar]
- Saint‐Jore‐Dupas C, Faye L, Gomord V. 2007. From planta to pharma with glycosylation in the toolbox. Trends Biotechnol 25: 317–323. [DOI] [PubMed] [Google Scholar]
- Santi L, Giritch A, Roy CJ, Marillonnet S, Klimyuk V, Gleba Y, Webb R, Arntzen CJ, Mason HS. 2006. Protection conferred by recombinant Yersinia pestis antigens produced by a rapid and highly scalable plant expression system. Proc Natl Acad Sci USA 103: 861–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidlin L, Link D, Mutterer J, Guilley H, Gilmer D. 2005. Use of a beet necrotic yellow vein virus RNA‐5‐derived replicon as a new tool for gene expression. J Gen Virol 86: 463–467. [DOI] [PubMed] [Google Scholar]
- Scholthof HB, Morris TJ, Jackson AO. 1993. The capsid protein gene of Tomato bushy stunt virus is dispensable for systemic movement and can be replaced for localized expression of foreign genes. Mol Plant Microbe Interact 6: 309–322. [Google Scholar]
- Shingu Y, Yokomizo S, Kimura M, Ono Y, Yamaguchi I, Hamamoto H. 2006. Conferring cadmium resistance to mature tobacco plants through metal‐adsorbing particles of tomato mosaic virus vector. Plant Biotechnol J 4: 281–288. [DOI] [PubMed] [Google Scholar]
- Smart V, Foster PS, Rothenberg ME, Higgins TJ, Hogan SP. 2003. A plant‐based allergy vaccine suppresses experimental asthma via an IFN‐gamma and CD4+CD45RBlow T cell‐dependent mechanism. J Immunol 171: 2116–2126. [DOI] [PubMed] [Google Scholar]
- Smith ML, Lindbo JA, Dillard‐Telm S, Brosio PM, Lasnik AB, McCormick AA, Nguyen LV, Palmer KE. 2006. Modified tobacco mosaic virus particles as scaffolds for display of protein antigens for vaccine applications. Virology 348: 475–488. [DOI] [PubMed] [Google Scholar]
- Smith ML, Corbo T, Bernales J, Lindbo JA, Pogue GP, Palmer KE, McCormick AA. 2007. Assembly of trans‐encapsidated recombinant viral vectors engineered from Tobacco mosaic virus and Semliki Forest virus and their evaluation as immunogens. Virology 358: 321–333. [DOI] [PubMed] [Google Scholar]
- Staczek J, Bendahmane M, Gilleland LB, Beachy RN, Gilleland HE Jr. 2000. Immunization with a chimeric tobacco mosaic virus containing an epitope of outer membrane protein F of Pseudomonas aeruginosa provides protection against challenge with P. aeruginosa. Vaccine 18: 2266–2274. [DOI] [PubMed] [Google Scholar]
- Steinmetz NF, Calder G, Lomonossoff GP, Evans DJ. 2006. Plant viral capsids as nanobuilding blocks: Construction of arrays on solid supports. Langmuir 22: 10032–10037. [DOI] [PubMed] [Google Scholar]
- Streatfield SJ, Howard JA. 2003. Plant‐based vaccines. Int J Parasitol 33: 479–493. [DOI] [PubMed] [Google Scholar]
- Sugiyama Y, Hamamoto H, Takemoto S, Watanabe Y, Okada Y. 1995. Systemic production of foreign peptides on the particle surface of tobacco mosaic virus. FEBS Lett 359: 247–250. [DOI] [PubMed] [Google Scholar]
- Sun HJ, Cui ML, Ma B, Ezura H. 2006. Functional expression of the taste‐modifying protein, miraculin, in transgenic lettuce. FEBS Lett 580: 620–626. [DOI] [PubMed] [Google Scholar]
- Tacket CO, Mason HS, Losonsky G, Clements JD, Levine MM, Arntzen CJ. 1998. Immunogenicity in humans of a recombinant bacterial antigen delivered in a transgenic potato. Nat Med 4: 607–609. [DOI] [PubMed] [Google Scholar]
- Tacket CO, Mason HS, Losonsky G, Estes MK, Levine MM, Arntzen CJ. 2000. Human immune responses to a novel norwalk virus vaccine delivered in transgenic potatoes. J Infect Dis 182: 302–305. [DOI] [PubMed] [Google Scholar]
- Tagwerker C, Flick K, Cui M, Guerrero C, Dou Y, Auer B, Baldi P, Huang L, Kaiser P. 2006. A tandem affinity tag for two‐step purification under fully denaturing conditions: Application in ubiquitin profiling and protein complex identification combined with in vivo cross‐linking. Mol Cell Proteomics 5: 737–748. [DOI] [PubMed] [Google Scholar]
- Takamatsu N, Ishikawa M, Meshi T, Okada Y. 1987. Expression of bacterial chloramphenicol acetyltransferase gene in tobacco plants mediated by TMV‐RNA. EMBO J 6: 307–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavladoraki P, Benvenuto E, Trinca S, De Martinis D, Cattaneo A, Galeffi P. 1993. Transgenic plants expressing a functional single‐chain Fv antibody are specifically protected from virus attack. Nature 366: 469–472. [DOI] [PubMed] [Google Scholar]
- Thanavala Y, Mahoney M, Pal S, Scott A, Richter L, Natarajan N, Goodwin P, Arntzen CJ, Mason HS. 2005. Immunogenicity in humans of an edible vaccine for hepatitis B. Proc Natl Acad Sci USA 102: 3378–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turpen TH, Turpen AM, Weinzettl N, Kumagai MH, Dawson WO. 1993. Transfection of whole plants from wounds inoculated with Agrobacterium tumefaciens containing cDNA of tobacco mosaic virus. J Virol Methods 42: 227–239. [DOI] [PubMed] [Google Scholar]
- Turpen TH, Reinl SJ, Charoenvit Y, Hoffman SL, Fallarme V, Grill LK. 1995. Malarial epitopes expressed on the surface of recombinant tobacco mosaic virus. Biotechnology (NY) 13: 53–57. [DOI] [PubMed] [Google Scholar]
- Twyman RM, Stoger E, Schillberg S, Christou P, Fischer R. 2003. Molecular farming in plants: Host systems and expression technology. Trends Biotechnol 21: 570–578. [DOI] [PubMed] [Google Scholar]
- Verch T, Yusibov V, Koprowski H. 1998. Expression and assembly of a full‐length monoclonal antibody in plants using a plant virus vector. J Immunol Methods 220: 69–75. [DOI] [PubMed] [Google Scholar]
- Verch T, Hooper DC, Kiyatkin A, Steplewski Z, Koprowski H. 2004. Immunization with a plant‐produced colorectal cancer antigen. Cancer Immunol Immunother 53: 92–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villani ME, Roggero P, Bitti O, Benvenuto E, Franconi R. 2005. Immunomodulation of cucumber mosaic virus infection by intrabodies selected in vitro from a stable single‐framework phage display library. Plant Mol Biol 58: 305–316. [DOI] [PubMed] [Google Scholar]
- Vitale A, Pedrazzini E. 2005. Recombinant pharmaceuticals from plants: The Plant endomembrane system as bioreactor. Mol Interv 5: 216–225. [DOI] [PubMed] [Google Scholar]
- Walmsley AM, Arntzen CJ. 2000. Plants for delivery of edible vaccines. Curr Opin Biotechnol 11: 126–129. [DOI] [PubMed] [Google Scholar]
- Walsh G. 2006. Biopharmaceutical benchmarks 2006. Nat Biotechnol 24: 769–776. [DOI] [PubMed] [Google Scholar]
- Wang Q, Kaltgrad E, Lin T, Johnson JE, Finn MG. 2002. Natural supramolecular building blocks. Wild‐type cowpea mosaic virus. Chem Biol 9: 805–811. [DOI] [PubMed] [Google Scholar]
- Waterhouse PM, Helliwell CA. 2003. Exploring plant genomes by RNA‐induced gene silencing. Nat Rev Genet 4: 29–38. [DOI] [PubMed] [Google Scholar]
- Watson J, Koya V, Leppla SH, Daniell H. 2004. Expression of Bacillus anthracis protective antigen in transgenic chloroplasts of tobacco, a non‐food/feed crop. Vaccine 22: 4374–4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster DE, Smith SD, Pickering RJ, Strugnell RA, Dry IB, Wesselingh SL. 2006. Measles virus hemagglutinin protein expressed in transgenic lettuce induces neutralising antibodies in mice following mucosal vaccination. Vaccine 24: 3538–3544. [DOI] [PubMed] [Google Scholar]
- Werner S, Marillonnet S, Hause G, Klimyuk V, Gleba Y. 2006. Immunoabsorbent nanoparticles based on a tobamovirus displaying protein A. Proc Natl Acad Sci USA 103: 17678–17683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigdorovitz A, Perez Filgueira DM, Robertson N, Carrillo C, Sadir AM, Morris TJ, Borca MV. 1999. Protection of mice against challenge with foot and mouth disease virus (FMDV) by immunization with foliar extracts from plants infected with recombinant tobacco mosaic virus expressing the FMDV structural protein VP1. Virology 264: 85–91. [DOI] [PubMed] [Google Scholar]
- Wood DW, Wu W, Belfort G, Derbyshire V, Belfort M. 1999. A genetic system yields self‐cleaving inteins for bioseparations. Nat Biotechnol 17: 889–892. [DOI] [PubMed] [Google Scholar]
- Woodard SL, Mayor JM, Bailey MR, Barker DK, Love RT, Lane JR, Delaney DE, McComas‐Wagner JM, Mallubhotla HD, Hood EE, Dangott LJ, Tichy SE, Howard JA. 2003. Maize (Zea mays)‐derived bovine trypsin: Characterization of the first large‐scale, commercial protein product from transgenic plants. Biotechnol Appl Biochem 38: 123–130. [DOI] [PubMed] [Google Scholar]
- Yin J, Li G, Ren X, Herrler G. 2007. Select what you need: A comparative evaluation of the advantages and limitations of frequently used expression systems for foreign genes. J Biotechnol 127: 335–347. [DOI] [PubMed] [Google Scholar]
- Youm JW, Won YS, Jeon JH, Ryu CJ, Choi YK, Kim HC, Kim BD, Joung H, Kim HS. 2007. Oral immunogenicity of potato‐derived HBsAg middle protein in BALB/c mice. Vaccine 25: 577–584. [DOI] [PubMed] [Google Scholar]
- Yusibov V, Modelska A, Steplewski K, Agadjanyan M, Weiner D, Hooper DC, Koprowski H. 1997. Antigens produced in plants by infection with chimeric plant viruses immunize against rabies virus and HIV‐1. Proc Natl Acad Sci USA 94: 5784–5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusibov V, Hooper DC, Spitsin SV, Fleysh N, Kean RB, Mikheeva T, Deka D, Karasev A, Cox S, Randall J, Koprowski H. 2002. Expression in plants and immunogenicity of plant virus‐based experimental rabies vaccine. Vaccine 20: 3155–3164. [DOI] [PubMed] [Google Scholar]
- Yusibov V, Mett V, Davidson C, Musiychuk K, Gilliam S, Farese A, Macvittie T, Mann D. 2005. Peptide‐based candidate vaccine against respiratory syncytial virus. Vaccine 23: 2261–2265. [DOI] [PubMed] [Google Scholar]
- Zhang C, Ghabrial SA. 2006. Development of Bean pod mottle virus‐based vectors for stable protein expression and sequence‐specific virus‐induced gene silencing in soybean. Virology 344: 401–411. [DOI] [PubMed] [Google Scholar]
- Zhang G, Leung C, Murdin L, Rovinski B, White KA. 2000. In planta expression of HIV‐1 p24 protein using an RNA plant virus‐based expression vector. Mol Biotechnol 14: 99–107. [DOI] [PubMed] [Google Scholar]


