Abstract
The Zika virus (ZIKV) outbreak, which started in the year 2015, is considered the fastest and most widely spread outbreak reported for this flavivirus. The polymerase domain of the NS5 protein has been targeted in other viral infections and is recognized as a suitable target in ZIKV infection. Different novel modified compounds against ZIKV NS5 have been tested in silico. A few structures have been solved for ZIKV polymerase and deposited in the protein data bank website. Two of these solved structures (with a resolution of less than 1.9 A) are used in this study to test the binding of 74 novel compounds in silico. Molecular docking is used to quantify the binding affinities of ZIKV polymerase and compare it to the hepatitis C virus NS5B. A total of 19 novel compounds revealed results that are either similar to or better than the physiological molecule, guanosine triphosphate. Water molecules are found to facilitate the binding of the compounds to ZIKV RNA‐dependent RNA polymerase (RdRp) structures. The presented 19 novel compounds represent good binders to ZIKV RdRp and could be suitable candidates for developing a new and effective anti‐ZIKV polymerase nucleotide inhibitor.
Keywords: drug‐protein interaction, guanosine derivatives, molecular docking, NS5, RNA‐dependent RNA polymerase, Zika virus
Highlight
ZIKV inhibition using nucleotide inhibitors.
Novel guanosine derivatives against ZIKV polymerase.
Oxidanyl derivatives at position 2' can inhibit the polymerase for both HCV and ZIKV.
1. INTRODUCTION
Seventy years ago, in Uganda, the Zika virus (ZIKV) was reported for the first time.1, 2 About six decades later, newer incidents of emergence recorded in Nigeria, Senegal, and Gabon.3, 4, 5 Despite its spread, the reported ZIKV infections were not like the latest emerging outbreak in the year 2015 in Latin America.6 ZIKV is transmitted through body fluids and sexually. A direct link between newborn microcephaly and pregnant women infection was confirmed in the year 2016.7, 8, 9, 10 Mosquito bites are the main route of spread of ZIKV infections along with sexual intercourse.11, 12 The mild symptomatic ZIKV infection can be easily detected in body fluids like blood urine and saliva.10, 13 Severe neurological diseases and sterility are reported in some patients as the virus targets all cells of the nervous system.14
The ZIKV genome is a single‐stranded RNA that encodes a 3400 amino acid polyprotein, which is processed by viral and host proteases to ten functional proteins.15 The nonstructural 5 (NS5) protein is the most widely conserved protein and has been targeted in previous viral infections like hepatitis C virus (HCV).16, 17, 18 The ZIKV RNA‐dependent RNA polymerase (RdRp) domain of the NS5 protein has been targeted by anti‐HCV drugs (repurposing).19, 20, 21 This protein domain is vital in the virus life cycle as it builds the new RNA strand from the complementary strand (the externalized RNA to the host cell) by utilizing the free nucleotides in the cytoplasm.15, 22 Targeting such a protein with nucleotide inhibitors stops the virus life cycle and eradicates the infection. The active site of NS5 polymerase lies in the palm subdomain (motif C), where two consecutive aspartates (D665 and D666) protrude from a beta‐turn structure and are surface accessible.19, 23 Nucleotide inhibitors mimic the native nucleotides in their ability to selectively target the active site of NS5 to be added into the primer RNA strand. Once bound to the protein, nucleotide inhibitors block the polymerization process and cause protein inhibition.24, 25, 26 Guanosine derivatives were studied against HCV polymerase and a candidate, IDX‐184, was under the clinical trials phase IIb before this was halted due to a side effect in the year 2013.18 This gave better results compared with the uridine derivative (sofosbuvir), adenine derivative (MK‐0608), and the wide‐range antiviral ribavirin against HCV and human coronaviruses in silico.16, 18
Molecular modeling represents a successful method used to predict the drug/target binding potency and mode of interaction. Based on a suitable model, it can differentiate between active and inactive inhibitors and suggest new compounds (in silico screening).27, 28, 29, 30, 31
A few solved structures have been deposited in the protein data bank recently for the ZIKV NS5 protein.32, 33, 34, 35, 36, 37 In this study, the author used two structures with PDB codes 5U04 and 5WZ3 as a drug target due to their appropriate resolution (1.9 and 1.8 Å, respectively) compared with other solved structures of ZIKV NS5. AutoDock Vina was used as the docking calculation method after cross‐docking with HCV NS5B RdRp. Seventy‐four new compounds were tested in this study against ZIKV NS5 RdRp structures. These compounds were derivatives of the guanine nucleotide.
2. MATERIALS AND METHODS
2.1. Ligands preparation for the docking study
Structures of the ligands are prepared using SCIGRESS 3.4 tools.31 The modifications were based on previous work, where three groups of modifications were introduced in the 2′ position in the ribose ring of the guanosine derivative.38 Structural geometry optimization was performed using the following scheme; classical mechanical geometry optimization (MM3 force field) followed by the semi‐empirical parameterization method 6 (PM6) and finally, quantum mechanical density functional theory using the B3LYP functional. All these calculations were performed on the 3.4 GHz intel core‐i7 processor PC (12 GB RAM) using SCIGRESS 3.4 software.39, 40, 41, 42
2.2. Target retrieval, preparation, and docking
The solved structures of ZIKV polymerase (NS5 RdRp domain) deposited in the protein data bank37 were examined, and two were selected, namely (PDB ID: 5U04 and 5WZ3). The structures were solved by x‐ray crystallography over the last 2 years with a resolution of 1.9 and 1.8 A, respectively. The HCV NS5B RdRp solved structure (PDB ID: 2XI3) was retrieved for comparison with ZIKV structures.43 Ions and ligands were removed using SCIGRESS docking preparation tools. The grid box was chosen to be of a 10 Å side length cube for all the structures. The box centers were selected to be at the active site (D665 & D666 and D318 & D319 in ZIKV and HCV NS5 RdRp, respectively). The grid boxes centers for 5U04, 5WZ3, and 2XI3 structures were (21.3 × 70.1 × 96.5), (52.1 × 2.0 × 80.2), and (9.8 × 5.6 × 10.0) Å, respectively. Missing hydrogen atoms were added using the SCIGRESS docking preparation tools, as the targets used in this study were solved by x‐ray crystallography. AutoDock Vina implemented on SCIGRESS 3.4 software was used in this study with the flexible target's active site and the flexible ligand approach.30, 44 The binding affinities were represented for the best complexes with the aid of PyMOL, Maestro, and Microsoft Excel software.45, 46, 47
3. RESULTS
3.1. Ligand preparation
Seventy‐four modified guanosine derivatives (see Table S1) were sketched and optimized using the same procedure of that of Elfiky.38 The modified compounds were based on modifying the 2′‐position of the ribose ring of the guanosine triphosphate (GTP). The addition of a bulky group at this position gave good results against HCV NS5B RdRp in previous studies.38, 48
3.2. Binding affinity calculation
AutoDock Vina implemented on SCIGRESS 3.4 software was used in this study to calculate the binding energies between the ligands and the target polymerase for both ZIKV and HCV. Figure 1A‐C shows the calculated binding energies (docking scores) for the three groups of modifications (groups I, II, and III, respectively) against ZIKV NS5 RdRp (orange line) and HCV NS5B RdRp ((PDB ID: 2XI3) blue line). Average values, with the standard deviations as error bars, are represented in Figure 1 for ZIKV solved structures (PDB ID: 5U04 and 5WZ3).
Figure 1.
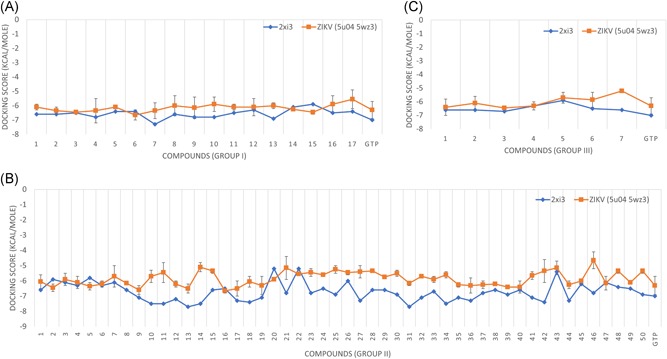
The docking scores calculated by AutoDock Vina software for three groups of modifications to GTP, group I (A), group II (B), and group III (C). The docking study was performed using two ZIKV RdRp solved structures (PDB ID: 5WZ3 and 5U04), and one HCV RdRp solved structure (PDB ID: 2XI3) for comparison. The average values of the docking scores for ZIKV RdRp are represented by the orange line while HCV RdRp docking values in blue lines. GTP, guanosine triphosphate; HCV, hepatitis C virus; RdRp, RNA‐dependent RNA polymerase; ZIKV, Zika virus
Table 1 summarizes the interactions between the top‐ranked ligands and ZIKV NS5 RdRp structures. The selection is based on the docking scores. Values equal to or less than (better) the binding energy of the parent nucleotide (GTP) are listed in Table 1. Group I of the modifications have six compounds that are equal to or better binders compared with GTP. These molecules include the modified GTP at the 2′ position with fluoromethyl, difluoromethyl, trifluoromethyl, 2,2‐difluoroethyl, 2,2,2‐trifluoroethyl, and selenanylmethyl. On the other hand, 10 compounds from group II have better (compared with GTP) docking scores. These molecules include the modified GTP at the 2′ position with ethyloxidanyl, phosphanyl, phenyloxidanyl, 3,5‐dihydroxyphenyl, (2,6‐dihydroxyphenyl)oxidanyl, (2‐hydroxyphenyl)oxidanyl, (3‐hydroxyphenyl)oxidanyl, (2,6‐difluorophenyl)oxidanyl, (3‐fluorophenyl)oxidanyl, and (4‐fluorophenyl)oxidanyl. Finally, group III has only three compounds that have better binding energies than ZIKV RdRp structures compared with GTP. These compounds have two modifications to the 2′ position, which are two methyl groups, ethyl and fluorenyl, and two fluorenyl groups. The interacting amino acids with the ligands are also listed in Table 1 with the number of water molecules that take part in the interactions. Some water molecules mediate the interaction by binding to both the ligand and protein binding pocket.
Table 1.
GTP selected modifications, with the average docking scores less than (better) or equal to GTP, calculated using AutoDock Vina for ZIKV structures (PDB ID: 5U04 and 5WZ3)
| R1 | Substitution name | Average docking scores (kcal/mol) ± SD | H2O and amino acids involved in H‐bond formation | |
|---|---|---|---|---|
| 5U04 | 5WZ3 | |||
| GTP | The parent compound | −6.30 ± 0.60 | H 2 O (5), N612, D665, S798 | H 2 O (13), D540, D665 (2), D666 |
| Group I (R1) | ||||
| 2 | Fluoromethyl | −6.35 ± 0.25 | H 2 O (9), T608, D666, I799, | H 2 O (7), N612 (3), S712 |
| 3 | Difluoromethyl | −6.45 ± 0.05 | H 2 O (7), D665, D666, S712 | H 2 O (7), N612 (2), D665, S712 |
| 4 | Trifluoromethyl | −6.35 ± 0.85 | H 2 O (7), T608, D666, S712 | H 2 O (8), N612 (2), S663(2), D666 |
| 6 | 2,2‐Difluoroethyl | −6.65 ± 0.35 | H 2 O (7), T608, N612, D666, S712 | H 2 O (10), N612 (2), S663 (3), D665, S712 (4) |
| 7 | 2,2,2‐Trifluoroethyl | −6.35 ± 0.55 | H 2 O (7), D666, S712, I799 | H 2 O (9), D540, S663, D665, 2 S712 |
| 15 | Selenanylmethyl | −6.45 ± 0.15 | H 2 O (6), T608, N612, S663, D665, D666 | H 2 O (9), D540 (2), D665 (3), D666, S712, I799 |
| Group II (R2) | ||||
| 2 | Ethyloxidanyl | −6.45 ± 0.25 | H 2 O (7), S663, D665, D666, S712, W797, S798 | H 2 O (8), W539, D540 (4), N612 (2), S663, D665 |
| 5 | Phosphanyl | −6.35 ± 0.25 | H 2 O (5), N612, D665, S712, I799 | H 2 O (10), W539, D540, N612, S663, 2 D665, S798 |
| 9 | Phenyloxidanyl | −6.60 ± 0.30 | H 2 O (9), D540, N612, D665 (3), D666, I779 | H 2 O (9), D540, N612, D665 (3), D666, I779 |
| 13 | 3,5‐Dihydroxyphenyl | −6.50 ± 0.30 | H 2 O (7), T608 (2), S663 (2), D665, D666 (2) | H 2 O (12), D540, S603, S663 (2), D665, D666 |
| 16 | (2,6‐Dihydroxyphenyl)oxidanyl | −6.65 ± 0.15 | H 2 O (8), T608 (2), Y609, D666 (2), S712 | H 2 O (9), D535, N612 (2), D665, S712 |
| 17 | (2‐Hydroxyphenyl)oxidanyl | −6.50 ± 0.50 | H 2 O (8), N612, D665, S712 (2) | H 2 O (9), D535 (2), N612 (2), S663, D665, K691 |
| 19 | (3‐Hydroxyphenyl)oxidanyl | −6.30 ± 0.60 | H 2 O (6), T608, D665, D666 (2) | H 2 O (9), D535, D540, N612, S663 (2), D665 |
| 36 | (2,6‐Difluorophenyl)oxidanyl | −6.30 ± 0.05 | H 2 O (7), T608, D665 (2), D666 (3), S712 | H 2 O (9), D540 (2), D665 (3), S712 (2) |
| 39 | (3‐Fluorophenyl)oxidanyl | −6.40 ± 0.10 | H 2 O (6), T608 (2), D665, D666, S712 (2) | H 2 O (14), D540, S712 |
| 40 | (4‐Fluorophenyl)oxidanyl | −6.40 ± 0.40 | H 2 O (6), D665, D666, S712, W797, I799 | H 2 O (8), W539, D540 (3), N612, S663, D665 (3) |
| Group III (R3 and R4) | ||||
| 1 | Methyl + methyl | −6.40 ± 0.60 | H 2 O (7), D666 (2), S712, S798 | H 2 O (11), N612 (3), D665 |
| 3 | Ethyl + fluoranyl | −6.45 ± 0.15 | H 2 O (5), T608, N612, D666, S712 (2), I799 | H 2 O (7), W539, D540 (3), S663 (2) |
| 4 | Fluoranyl + fluoranyl | −6.30 ± 0.30 | H 2 O (5), T608, D665, D666 (3) | H 2 O (8), D540, N612, S663 (2), D665 (2) |
Note: The H bonds formed between the ligands and the proteins are listed with their number. The amino acids involved in the interaction are listed with the number in brackets for the number of H bonds if greater than 1. It is based on a baseline value below it the values are marked as bold or underlined.
Abbreviations: GTP, guanosine triphosphate; ZIKV, Zika virus.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Figure 2A and 2B shows the docked structures of GTP to ZIKV NS5 RdRp solved structures (PDB ID: 5WZ3 and 5U04, respectively). GTP is represented by atom type in stick color: carbon in green, hydrogen in white, nitrogen in blue, oxygen in red, and sulfur in orange. Water molecules are represented by red spheres. The ZIKV RdRp protein is represented by a rainbow‐colored cartoon. The R groups of the amino acids that take part in the binding to GTP are represented by lines of the same color as the cartoon. Polar interactions are represented by dashed yellow lines.
Figure 2.
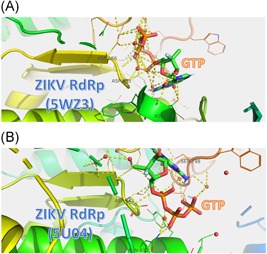
The docking complexes of GTP to ZIKV NS5 RdRp (PDB ID: 5WZ3 (A) and 5U04 (B)). ZIKV RdRp is represented by the colored cartoon while the amino acids involved in the interactions are represented by lines and labeled by their three‐letter code. GTP is represented according to the atom type in stick color ; C, green; H, white; O, red; N, blue; P, orange. Water is represented bye red spheres while yellow dashed lines represente the polar contact. GTP, guanosine triphosphate; NS5, non‐structural 5; RdRp, RNA‐dependent RNA polymerase; ZIKV, Zika virus
4. DISCUSSION
Suggesting a potent inhibitor against ZIKV RdRp is the primary goal of this study, Figure 1 shows that almost all compounds have good binding energies to ZIKV polymerase that are comparable to that for HCV RdRp. For the groups I and III of the modifications, the docking scores of ZIKV are almost in the same range as HCV except for compound 7 in group III that has a bit higher docking score value (−5.2 kcal/mol). On the other hand, group II of the modifications has some compounds that bind more like HCV, and other compounds that have less binding potency compared to HCV. Overall, the docking scores for the modifications lie between −6.65 and −5.1 for ZIKV polymerase while the values for HCV RdRp are between −7.7 and −5.2 (see Table S1). This implies the effectiveness of the modified compounds in competing for the active site of ZIKV polymerase.
Table 1 shows the best compounds based on their binding energies (docking scores) to ZIKV RdRp structures. A total of 19 compounds show better values for binding ZIKV NS5 RdRp compared with the parent compound GTP. These compounds have a better chance to bind to the active site of the polymerase upon its presence in solution with GTP, the parent, and physiological molecule.
The amino acids involved in the binding of the top‐ranked ligands to ZIKV polymerases are listed in Table 1. We can notice that the active site aspartates (D665 and D666) mediate the interaction in almost all binding trials. This supports the conservation of these residues in viral and even human polymerases.
The ZIKV solved structure has water molecules surrounding the binding pocket. The author decided to perform the docking experiment without removing the water molecules as water may affect the binding of the hydrophilic ligands to the hydrophilic active site aspartates. Surprisingly, the number of water molecules that interact with the ligands varies from five (GTP, compound 5 in group II and compounds 3 and 4 of group III) up to 14 (compound 39 in group II). It has been reported that water mediates the dynamics of biomolecules.49 Figure 2 shows how the water molecule interacts in a network facilitating and supporting the binding of GTP into ZIKV NS5 RdRp. Panel A of Figure 2 represents the GTP docked into the ZIKV RdRp structure (PDB ID: 5WZ3) while panel B represents the structure (PDB ID: 5U04). The water molecules network through H‐bonds. Besides this, water connects with both the ligand (GTP) and the protein (ZIKV NS5 RdRp) polar residues through the formation of polar contacts. These polar interactions stabilize the protein‐ligand complexes facilitating the polymerase function. The water network that is formed upon ligand‐protein docking is key for the effectiveness of the ligand as an inhibitor.
5. CONCLUSION
RdRp is suggested to be a suitable target for HCV and other viruses due to its vital role in the replication of the virus. Due to the conservation of the RdRp's active site, it is also targeted in ZIKV and other viral infections. Previous studies show that anti‐HCV NS5B drugs are able to bind to ZIKV RdRp (repurposing trials) but with relatively lower affinity compared with HCV. In this study, the author presents 19 modified guanosine derivatives that show in silico effectiveness against ZIKV NS5 RdRp. Further experimental work is suggested to characterize these modifications further and apply it to the virus in vitro and in vivo assays.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
Supporting information
Supplementary information
Elfiky AA. Novel guanosine derivatives against Zika virus polymerase in silico. J Med Virol 2020;92:11‐16. 10.1002/jmv.25573
References
REFERENCES
- 1. MacNamara FN. Zika virus: a report on three cases of human infection during an epidemic of jaundice in Nigeria. Trans R Soc Trop Med Hyg. 1954;48(2):139‐145. [DOI] [PubMed] [Google Scholar]
- 2. Dick GWA, Kitchen SF, Haddow AJ. Zika virus (I). Isolations and serological specificity. Trans R Soc Trop Med Hyg. 1952;46(5):509‐520. [DOI] [PubMed] [Google Scholar]
- 3. Kuno G, Chang GJJ. Full‐length sequencing and genomic characterization of Bagaza, Kedougou, and Zika viruses. Arch Virol. 2007;152(4):687‐696. [DOI] [PubMed] [Google Scholar]
- 4. Duffy MR, Chen T‐H, Hancock WT, et al. Zika virus outbreak on Yap Island, federated states of micronesia. N Engl J Med. 2009;360(24):2536‐2543. [DOI] [PubMed] [Google Scholar]
- 5. Grard G, Caron M, Mombo IM, et al. Zika virus in Gabon (Central Africa)—2007: a new threat from Aedes albopictus? PLOS Negl Trop Dis. 2014;8(2):e2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Musso D, Nilles EJ, Cao‐Lormeau VM. Rapid spread of emerging Zika virus in the Pacific area. Clin Microbiol Infect. 2014;20(10):O595‐O596. [DOI] [PubMed] [Google Scholar]
- 7. Mlakar J, Korva M, Tul N, et al. Zika virus associated with microcephaly. N Engl J Med. 2016;374(10):951‐958. [DOI] [PubMed] [Google Scholar]
- 8. Althaus CL, Low N. How relevant is sexual transmission of Zika virus? PLOS Med. 2016;13(10):e1002157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Musso D NT, Robin E, Roche C, et al. Potential for Zika virus transmission through blood transfusion demonstrated during an outbreak in French Polynesia, November 2013 to February 2014. Euro Surveill. 2014;19(14):1‐3. [DOI] [PubMed] [Google Scholar]
- 10. Musso D, Rouault E, Teissier A, et al. Molecular detection of Zika virus in blood and RNA load determination during the French Polynesian outbreak. J Med Virol. 2017;89(9):1505‐1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cao‐Lormeau VM, Roche C, Teissier A, et al. Zika virus, French polynesia, South pacific, 2013. Emerg Infect Dis. 2014;20(6):1084‐1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Foy BD, Kobylinski KC, Foy JLC, et al. Probable non–vector‐borne transmission of Zika virus, Colorado, USA. Emerg Infect Dis. 2011;17(5):880‐882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tauro LB, Bandeira AC, Ribeiro GS, et al. Potential use of saliva samples to diagnose Zika virus infection. J Med Virol. 2017;89(1):1‐2. [DOI] [PubMed] [Google Scholar]
- 14. Ojha CR, Rodriguez M, Lapierre J, et al. Complementary mechanisms potentially involved in the pathology of Zika virus. Front Immunol. 2018;9:2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elfiky AA, Elshemey WM. Molecular dynamics simulation revealed binding of nucleotide inhibitors to ZIKV polymerase over 444 nanoseconds. J Med Virol. 2018;90(1):13‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Elfiky AA, Mahdy SM, Elshemey WM. Quantitative structure‐activity relationship and molecular docking revealed a potency of anti‐hepatitis C virus drugs against human corona viruses. J Med Virol. 2017;89(6):1040‐1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Elfiky AA, Ismail AM. Molecular modeling and docking revealed superiority of IDX‐184 as HCV polymerase inhibitor. Future Virol. 2017;12(7):339‐347. [Google Scholar]
- 18. Elfiky AA, Elshemey WM. IDX‐184 is a superior HCV direct‐acting antiviral drug: a QSAR study. Med Chem Res. 2016;25(5):1005‐1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Elfiky AA. Zika viral polymerase inhibition using anti‐HCV drugs both in market and under clinical trials. J Med Virol. 2016;88(12):2044‐2051. [DOI] [PubMed] [Google Scholar]
- 20. Elfiky AA. Zika virus: novel guanosine derivatives revealed strong binding and possible inhibition of the polymerase. Future Virol. 2017;12(12):721‐728. [Google Scholar]
- 21. Bullard‐Feibelman KM, Govero J, Zhu Z, et al. The FDA‐approved drug sofosbuvir inhibits Zika virus infection. Antiviral Res. 2017;137:134‐140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. WHO . Zika strategic response framework and joint operations plan. January‐June 2016.
- 23. Singh A, Jana NK. Discovery of potential Zika virus RNA polymerase inhibitors by docking‐based virtual screening. Comput Biol Chem. 2017;71:144‐151. [DOI] [PubMed] [Google Scholar]
- 24. Ganesan A, Barakat K. Applications of computer‐aided approaches in the development of hepatitis C antiviral agents. Expert Opin Drug Discovery. 2017;12(4):407‐425. [DOI] [PubMed] [Google Scholar]
- 25. González‐Grande R. New approaches in the treatment of hepatitis C. World J Gastroenterol. 2016;22(4):1421‐1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ferrer‐Orta C, Ferrero D, Verdaguer N. RNA‐dependent RNA polymerases of picornaviruses: from the structure to regulatory mechanisms. Viruses. 2015;7(8):4438‐4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Leach A. Molecular Modelling: Principles and Applications. 2nd ed. Upper Saddle River, NJ: Prentice Hall; 2001. [Google Scholar]
- 28. Muegge I. A knowledge‐based scoring function for protein‐ligand interactions: probing the reference state In: Klebe G, ed. Virtual Screening: An Alternative or Complement to High Throughput Screening? Proceedings of the Workshop ‘New Approaches in Drug Design and Discovery,’ special topic ‘Virtual Screening,’ Schloβ Rauischholzhausen, Germany, March 15‐18, 1999. Dordrecht, Netherlands: Springer; 2002:99‐114. [Google Scholar]
- 29. van Dijk ADJ, Bonvin AMJJ. Solvated docking: introducing water into the modelling of biomolecular complexes. Bioinformatics. 2006;22(19):2340‐2347. [DOI] [PubMed] [Google Scholar]
- 30. Morris GM, Huey R, Lindstrom W, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785‐2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Summers KL, Mahrok AK, Dryden MDM, Stillman MJ. Structural properties of metal‐free apometallothioneins. Biochem Biophys Res Commun. 2012;425(2):485‐492. [DOI] [PubMed] [Google Scholar]
- 32. Wang B, Tan X‐F, Thurmond S, et al. The structure of Zika virus NS5 reveals a conserved domain conformation. Nat Commun. 2017;8:14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Upadhyay AK, Cyr M, Longenecker K, Tripathi R, Sun C, Kempf DJ. Crystal structure of full‐length Zika virus NS5 protein reveals a conformation similar to Japanese encephalitis virus NS5. Acta Crystallogr F Struct Biol Commun. 2017;73(3):116‐122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lu G, Gong P. A structural view of the RNA‐dependent RNA polymerases from the Flavivirus genus. Virus Res. 2017;234:34‐43. [DOI] [PubMed] [Google Scholar]
- 35. Godoy AS, Lima GMA, Oliveira KIZ, et al. Crystal structure of Zika virus NS5 RNA‐dependent RNA polymerase. Nat Commun. 2017;8:14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Duan W, Song H, Wang H, et al. The crystal structure of Zika virus NS5 reveals conserved drug targets. EMBO J. 2017;36(7):919‐933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Berman H, Henrick K, Nakamura H. Announcing the worldwide protein data bank. Nat Struct Mol Biol. 2003;10(12):980‐980. [DOI] [PubMed] [Google Scholar]
- 38. Elfiky AA. Novel guanosine derivatives as anti‐HCV NS5b polymerase: a QSAR and Molecular Docking Study. Med Chem. 2019;15(2):130‐137. [DOI] [PubMed] [Google Scholar]
- 39. Becke AD. Density‐functional thermochemistry. III. The role of exact exchange. J Chem Phys. 1993;98(7):5648‐5652. [Google Scholar]
- 40. Lii JH, Allinger NL. Molecular mechanics. The MM3 force field for hydrocarbons. 3. The van der Waals’ potentials and crystal data for aliphatic and aromatic hydrocarbons. J Am Chem Soc. 1989;111(23):8576‐8582. [Google Scholar]
- 41. Stewart JJP. Optimization of parameters for semiempirical methods. III. Extension of PM3 to Be, Mg, Zn, Ga, Ge, As, Se, Cd, In, Sn, Sb, Te, Hg, Tl, Pb, and Bi. J Comput Chem. 1991;12(3):320‐341. [Google Scholar]
- 42. Stewart JJP. Optimization of parameters for semiempirical methods V: Modification of NDDO approximations and application to 70 elements. J Mol Model. 2007;13(12):1173‐1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Harrus D, Ahmed‐El‐Sayed N, Simister PC, et al. Further insights into the roles of GTP and the C terminus of the hepatitis C virus polymerase in the initiation of RNA synthesis. J Biol Chem. 2010;285(43):32906‐32918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455‐461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. The PyMOL Molecular Graphics System , Version 1.7.6, V. Schrödinger, LLC.
- 46. Levine DM, Berenson ML, Stephan D, Lysell D. Statistics for Managers using Microsoft Excel. 660 Upper Saddle River, NJ: Prentice Hall; 1999. [Google Scholar]
- 47. Schrödinger Release 2017‐1: Maestro, Schrödinger, LLC, New York, NY, 2017.
- 48. Mayhoub AS. Hepatitis C RNA‐dependent RNA polymerase inhibitors: a review of structure‐activity and resistance relationships; different scaffolds and mutations. Bioorg Med Chem. 2012;20(10):3150‐3161. [DOI] [PubMed] [Google Scholar]
- 49. Bellissent‐Funel M‐C, Hassanali A, Havenith M, et al. Water determines the structure and dynamics of proteins. Chem Rev. 2016;116(13):7673‐7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information


