The transfer of reducing power in the form of electrons, generated in the catabolism of nutrients, from a bacterium to an extracellular acceptor appears to be common in nature. The electron acceptor can be another cell or abiotic material. Such extracellular electron transfer contributes to syntrophic metabolism and is of wide environmental, industrial, and medical importance. Electron transfer between microorganisms and electrodes is fundamental in microbial fuel cells for energy production and for electricity-driven synthesis of chemical compounds in cells. In contrast to the much-studied extracellular electron transfer mediated by cell surface exposed cytochromes, little is known about components and mechanisms for such electron transfer in organisms without these cytochromes and in Gram-positive bacteria such as E. faecalis, which is a commensal gut lactic acid bacterium and opportunistic pathogen.
KEYWORDS: extracellular electron transfer, ferric reductase, EetA, PplA, Enterococcus faecalis, type 2 NADH dehydrogenase
ABSTRACT
Enterococcus faecalis cells are known to have ferric reductase activity and the ability to transfer electrons generated in metabolism to the external environment. We have isolated mutants defective in ferric reductase activity and studied their electron transfer properties to electrodes mediated by ferric ions and an osmium complex-modified redox polymer (OsRP). Electron transfer mediated with ferric ions and ferric reductase activity were both found to be dependent on the membrane-associated Ndh3 and EetA proteins, consistent with findings in Listeria monocytogenes. In contrast, electron transfer mediated with OsRP was independent of these two proteins. Quinone in the cell membrane was required for the electron transfer with both mediators. The combined results demonstrate that extracellular electron transfer from reduced quinone to ferric ions and to OsRP occurs via different routes in the cell envelope of E. faecalis.
IMPORTANCE The transfer of reducing power in the form of electrons, generated in the catabolism of nutrients, from a bacterium to an extracellular acceptor appears to be common in nature. The electron acceptor can be another cell or abiotic material. Such extracellular electron transfer contributes to syntrophic metabolism and is of wide environmental, industrial, and medical importance. Electron transfer between microorganisms and electrodes is fundamental in microbial fuel cells for energy production and for electricity-driven synthesis of chemical compounds in cells. In contrast to the much-studied extracellular electron transfer mediated by cell surface exposed cytochromes, little is known about components and mechanisms for such electron transfer in organisms without these cytochromes and in Gram-positive bacteria such as E. faecalis, which is a commensal gut lactic acid bacterium and opportunistic pathogen.
INTRODUCTION
Enterococci are Gram-positive bacteria present as commensals in the intestines of animals, and some species are opportunistic pathogens, for example, Enterococcus faecalis and Enterococcus faecium. The enterococci are lactic acid bacteria that are phylogenetically closely related to common human pathogens, for example, streptococci and pneumococci, and to bacteria that are important for the food industry, for example, lactococci. Enterococci are also found in other environments, such as in consortia of microbes in microbial fuel cells, where energy in the form of electricity is harvested by wiring metabolism of electroactive microbes to electrodes (1, 2). With E. faecalis cells, we have demonstrated and characterized extracellular electron transfer (EET) mediated by various mediators on graphite and gold electrodes (3, 4). Electricigenicity seems to be a physiological property of many microorganisms (5). Recent studies have revealed EET in the mammalian gut and identified other electroactive gut bacteria (6–8). Knowledge about the mechanisms behind EET in bacteria not only is of clinical relevance but is very important for biotechnical applications such as the design of microbial fuel cells. The nature of electron transfer pathways in electrode-bacterium interactions remains the primary issue in microbial electrochemical techniques (2, 9, 10).
Being a lactic acid bacterium with a mainly fermentative metabolism, E. faecalis grows well also under anoxic conditions. If supplied with heme, the bacterium can respire with molecular oxygen due to the assembly of a membrane-bound cytochrome bd, which is a terminal quinol oxidase that reduces molecular oxygen to water (11, 12). E. faecalis contains demethylmenaquinone (DMK) as the only quinone in the membrane. Electrons are generated in glycolysis in the cytoplasm via the activity of membrane-associated dehydrogenases, for example, NADH dehydrogenase, which reduces DMK, which is oxidized by cytochrome bd or by EET processes. We have demonstrated that DMK is essential for EET and that cytochrome bd activity attenuates EET efficiency in E. faecalis (4). Effective electron transfer from the bacterial cell to an electrode requires a mediator—either a highly flexible redox polymer that forms a hydrogel together with the bacterial cells and precipitates onto the electrode surface, i.e., osmium-containing redox polymer {[Os(2,2′-bipyridine)2-poly(N-vinylimidazole) 10Cl]2+/+ (OsRP)}, or an added diffusible compound such as ferricyanide (1, 3, 4).
E. faecalis and several other Enterococcus species can reduce exogenous ferric ions (13–16). This dissimilatory activity apparently occurs as an electron sink to oxidize accumulating intracellular reducing equivalents, e.g., NADH, and the activity influences the yield and composition of carbon-containing end products obtained from catabolism of, e.g., glucose (14). Research with Listeria monocytogenes has revealed a single gene cluster on the chromosome that is important for both ferric reductase activity and EET (13). Genes in the cluster encode a type II NADH dehydrogenase, a flavoprotein (PplA), two small proteins (EetA and EetB), and quinone synthesis enzymes. A similar gene cluster is present in many other Gram-positive bacteria, including E. faecalis (13) (Fig. 1). Studies with E. faecalis have shown that ferric ion metabolism is associated with biofilm formation and identified genes important for this property, but these genes are not orthologous to those found to be important for ferric reductase activity in L. monocytogenes (16). These recent findings together prompted us to investigate the extent to which EET to electrodes and ferric reductase activity are connected processes in E. faecalis. While our research work was in progress it, was reported that EET to ferric iron in E. faecalis depends on genes corresponding to those in the gene cluster originally found in L. monocytogenes and on a gene encoding the tip adhesion protein EbpA (17). To identify proteins involved in EET to ferric ions and determine if these also are crucial for EET, we have here screened a collection of mutants for ferric reductase deficiency, identified the responsible mutations, and analyzed the bioelectrochemical properties of the mutants.
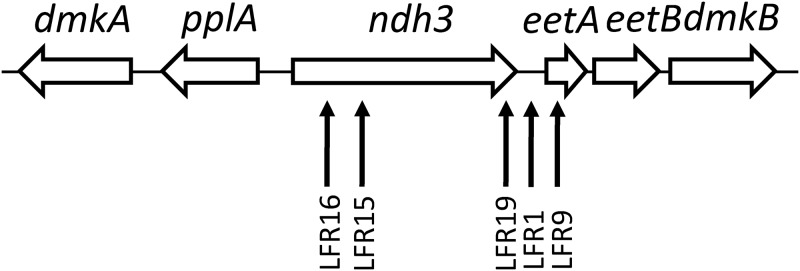
FIG 1.
Map of the ndh3 gene region in the chromosome of E. faecalis. Transposon insertion positions in the five ferric reductase mutants isolated in this work are indicated with vertical arrows. For more information see Table 1.
RESULTS
Requirements for ferric reductase activity of E. faecalis cells.
Ferric reductase activity of washed E. faecalis cells in growth medium supplemented with glucose was determined with ferric sulfate and ferricyanide as alternative substrates as described in Materials and Methods. E. faecalis cells cannot synthesize heme but encode two heme proteins, cytochrome bd and catalase (12). To determine the effect of heme and cytochrome bd on the ferric reductase activity, the laboratory model strain OG1RF (Table 1) was grown in tryptic soy broth without dextrose (TSB) with 1% (wt/vol) glucose added (TSBG) with and without hemin added. Heme in the growth medium caused attenuation of ferric reductase activity (Table 2), most likely because cytochrome bd oxidase activity decreases the reduction level of the DMK pool in the cytoplasmic membrane (4). Ferric reductase activity was found to be dependent on DMK in the cell membrane; i.e., strain WY84, which lacks DMK, showed low (∼15%) activity compared to the parental strain grown under the same conditions (Table 2 and Fig. 2). These results indicated that the reduction of ferric ions on the outside of E. faecalis cells is mediated by reduced DMK.
TABLE 1.
E. faecalis strains used in this worka
| Strain | EfaMarTn insertion position | Locus tag | Gene | Source of strain or reference |
|---|---|---|---|---|
| OG1RF | NAb | NA | NA | 24 |
| LFR1 | 2659891 | NA | Intergenic (ndh3-eetA) | This work |
| LFR9 | 2660001 | OG1RF_12511 | eetA | This work |
| LFR15 | 2658400 | OG1RF_12510 | ndh3 | This work |
| LFR16 | 2658088 | OG1RF_12510 | ndh3 | This work |
| LFR19 | 2659648 | OG1RF_12510 | ndh3 | This work |
| ndh3::Tn | 2658965 | OG1RF_12510 | ndh3 | 25 (strain RS12800) |
| pplA::Tn | 2657173 | OG1RF_12509 | pplA | 25 (strain RS12795) |
| WY84 | NAc | OG1RF_10330 | menB | 26 |
NA, not applicable.
OG1RF is the parental strain with no transposon insertion.
This strain has the menB gene deleted and contains no transposon insertion.
TABLE 2.
Ferric reductase activity of E. faecalis cells grown in TSBG with and without hemin supplementation
| Strain | Relevant genotype | Hemin (8 μM) added to the growth medium | Ammonium ferric sulfate reductase activity (%) | Ferricyanide reductase activity (%) |
|---|---|---|---|---|
| OG1RF | Wild type | No | 100 ± 15 | 100 ± 7 |
| Yes | 10 ± 3 | 13 ± 7 | ||
| WY84 | menB | No | 15 ± 4 | 13 ± 8 |
| Yes | 11 ± 7 | 18 ± 6 |
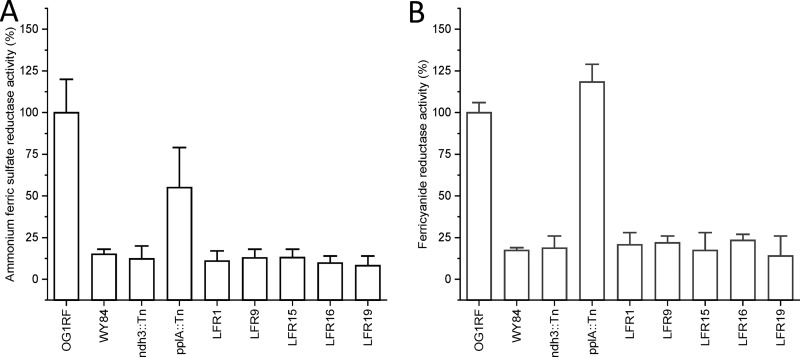
FIG 2.
Ferric reductase activity of E. faecalis parental (OG1RF) and mutant strains grown without heme. The strains are described in Table 1. (A) Activity with ammonium ferric sulfate. (B) Activity with potassium ferricyanide. Error bars show the standard error of the mean (SEM) based on more than four measurements with at least two biological replicates.
Identification of genes important for ferric reductase activity.
The genes ndh2 and pplA, which encode the type II NADH dehydrogenase Ndh2 and the flavoprotein PplA, respectively, have in L. monocytogenes been shown to be important for ferric reductase activity. We therefore analyzed ferric reductase activity of E. faecalis strains with the corresponding genes, ndh3 and pplA, respectively, inactivated (Table 1). The ndh3 mutant (ndh3::Tn) showed low activity with both ammonium ferric sulfate and ferricyanide (Fig. 2). That with pplA inactivated (pplA::Tn) showed slightly reduced activity with ammonium ferric sulfate and increased activity with ferricyanide (Fig. 2).
To find additional genes important for ferric reductase activity in E. faecalis, we subsequently screened a library containing clones with a random transposon insertion in the chromosome for defective ferric reductase activity. The library was previously constructed in our laboratory using E. faecalis OG1RF and the mini-mariner transposon (EfaMarTn) (18). Clones of the library in a 96-well format were spotted on agar plates containing growth medium supplemented with 0.2 mM ammonium ferric sulfate and screened for ferric reductase activity using an agar plate colony overlay staining procedure developed for L. monocytogenes (19) and adjusted for E. faecalis (see Materials and Methods).
Five ferric reductase-defective mutants (LFR1, -9, -15, -16, and -19) were identified among ∼2,000 screened clones (Table 1). The mutant cells grown in TSBG showed low ferric reductase activity compared to the parental strain (Fig. 2). This low activity was similar to that of parental cells grown in the presence of hemin (Table 2) and to that of strain WY84 lacking DMK (Fig. 2). Thus, a residual ferric reductase activity remained in both the isolated mutants and in strain WY84.
The insertion site for EfaMarTn in the chromosome of each mutant was determined using inverse PCR with transposon-specific primers, PCR with region-specific primers (Table S2), and PCR combined with DNA sequence analysis. Three mutants (LFR15, -16, and -19) were found to have the ndh3 gene disrupted by insertion of the transposon, one (LFR9) had the insertion in the eetA gene, and one (LFR1) had it in the ndh3-eetA intergenic region (Fig. 1 and Table 1).
Properties of mutants defective in ndh3 or eetA.
Isolated membranes of the ferric reductase-defective mutants grown in the presence of heme, to generate an active cytochrome bd in the cells, showed ≥43% NADH oxidase activity compared to the parental strain OG1RF (Table 3). This rather high membrane-bound NADH oxidase activity remaining in Ndh3-defective mutants can be explained by the presence of two other type II NADH dehydrogenases in E. faecalis, Ndh and Ndh2. As expected, the DMK-deficient strain WY84 and all strains grown in the absence of hemin lacked NADH oxidase activity because of the absence of cytochrome bd activity (Table 3). The results show that Ndh3 is required for ferric reductase activity but contributes only marginally to respiration with molecular oxygen. The colony size on agar plates and the growth of ferric reductase-defective mutants in liquid medium (TSBG with and without hemin) were similar to those of the parental strain OG1RF, indicating no gross defect in metabolism in the mutants.
TABLE 3.
NADH oxidase activity of isolated membranes from E. faecalis strains grown in TSBG with or without hemin supplementation
| Strain | Gene inactivated | Hemin (2 μM) added to growth medium | NADH oxidase activity (%)a |
|---|---|---|---|
| OG1RF | Yes | 100 ± 6 | |
| No | <8 | ||
| ndh3::Tn | ndh3 | Yes | 75 ± 11 |
| No | <8 | ||
| pplA::Tn | pplA | Yes | 126 ± 6 |
| No | <8 | ||
| WY84 | menB | Yes | <8 |
| No | <8 | ||
| LFR1 | Intergenic ndh3-eetA | Yes | 46 ± 1 |
| No | <8 | ||
| LFR9 | eetA | Yes | 68 ± 2 |
| No | <8 | ||
| LFR16 | ndh3 | Yes | 43 ± 2 |
| No | <8 |
One hundred percent activity was 0.53 nmol NADH oxidized per minute and mg of protein.
Electrochemical characterization of mutants.
It has been reported that ferric reductase activity in L. monocytogenes is important for EET (13) and that E. faecalis biofilm production depends on the environmental iron ion level and on EET (16). We therefore analyzed the electrochemical properties of E. faecalis cells defective in ferric reductase activity to perform EET to a graphite electrode in response to added glucose. Cells were grown without heme, and the analysis was done with washed cells that do not multiply during the experiment but are metabolically active. As we demonstrated before (3, 4), effective electron transfer from E. faecalis cells to an electrode requires an artificial redox mediator—either a polymeric or a monomeric shuttle. In the current work, we used redox mediators with similar redox potentials (see Fig. S1 in the supplemental material) as follows: OsRP (420 ± 4 mV) coimmobilized on the electrode and freely diffusible ferricyanide (426 ± 3 mV), respectively.
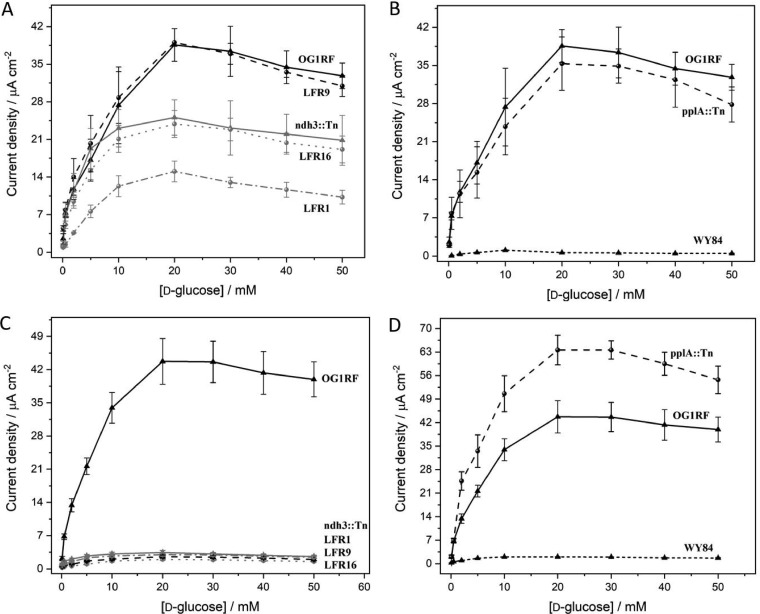
Mutants defective in pplA, ndh3, and eetA, respectively, and mediated by OsRP showed electron transfer ability to the electrodes in response to glucose (Fig. 3A). Compared to the parental strain OG1RF, the ndh3 mutants were slightly affected in EET (Fig. 3A). The pplA mutant showed a high activity (Fig. 3B). With ferricyanide as the mediator, in contrast, the ndh3- and eetA-defective mutants were found to essentially lack EET activity (<5 μA cm−1) (Fig. 3C and Fig. S2), while the parental strain and the pplA-defective mutant showed current densities of >35 μA cm−1 (Fig. 3D), consistent with high ferricyanide reductase activity (Fig. 2). EET with both OsRP and ferricyanide as the mediator was strictly dependent on the presence of DMK in the cell as demonstrated using strain WY84, which is blocked in quinone synthesis (Fig. 3B and D).
FIG 3.
Electrochemical communication between heme-free E. faecalis parental (OG1RF) and mutant strains and graphite electrodes in the presence of various d-glucose concentrations. The strains are described in Table 1. (A and B) Current density responses with cells immobilized on OsRP-coated graphite electrodes. (C and D) Results for the cells immobilized on graphite electrodes and with ferricyanide (Fe3+) as the redox mediator. The electrochemical behavior of the mutant strains presented in panel C is shown in Fig. S2 with an expanded current density scale.
The results from the electrochemical experiments show that Ndh3- and EetA-dependent ferric reductase activities of E. faecalis cells are important for EET with ferric ions as the redox mediator. However, with OsRP as the mediator, Ndh3 and EetA are not involved in the electron transfer from reduced DMK in the cell to the electrode.
DISCUSSION
In this work, we screened a collection of E. faecalis clones with random transposon insertions in the chromosome for mutants defective in ferric reductase activity. The isolated mutants were investigated for their ability to transfer electrons generated in glucose catabolism to a graphite electrode depending on the type of provided redox mediator. In parallel to our work, Lam et al. (17) studied the properties of E. faecalis strains with transposon insertion in genes corresponding to those known to be important for ferric reductase activity in L. monocytogenes (13) and for DMK synthesis in E. faecalis. Although the approaches of our research groups were different and EET was not measured using the same type of experimental setup, ndh3 (OG1RF_12510), eetA (OG1RF_12511), and menB (OG1RF_10330) were identified as important for EET in both studies. Lam et al. also showed that eetB (OG1RF_12512), dmkA (OG1RF_12507), and menE (OG1RF_10331) are of major importance for EET (i.e., defective mutants showed <50% cumulative charge production) (17).
MenB, MenE, and DmkA catalyze steps in DMK synthesis from chorismate. EetA (15.8 kDa) is a predicted membrane protein anchored to the outer side of the cytoplasmic membrane by a noncleaved N-terminal signal sequence. EetB (20.5 kDa) is an integral membrane protein with 4 predicted transmembrane segments and a large periplasmic loop connecting transmembrane segments 3 and 4. EetA and EetB might form a complex in the membrane. Ndh3 (71.8 kDa) and the paralogs Ndh (49.7 kDa) and Ndh2 (44.0 kDa) are from the predicted amino acid sequences suggested to be type II NADH:quinone oxidoreductases. Ndh3 is atypical for this class of enzymes in that it has a C-terminal extension of the polypeptide containing two predicted transmembrane segments. Ndh3 is not important for aerobic respiration and seems to be specialized for ferric reductase activity in E. faecalis (this work) and in L. monocytogenes (where the orthologue is Ndh2) (13). Streptococcus agalactiae (a lactic acid bacterium closely related to E. faecalis) contains only one type II NADH dehydrogenase. The gene for this protein is in the same operon as the cydABCD genes encoding cytochrome bd polypeptides. In E. faecalis, the ndh2 gene is on the chromosome colocalized with cydABCD, and the sequence of Ndh2 is very similar to that of S. agalactiae NADH dehydrogenase. Based on these findings, Ndh2 is probably the major respiratory NADH dehydrogenase in E. faecalis.
The collected data for E. faecalis (17; this work) and L. monocytogenes (13) indicate that an atypical NADH dehydrogenase (Ndh3 in E. faecalis) with the flavin adenine dinucleotide (FAD)-containing domain presumably on the cytoplasmic side of the membrane, EetA that is mainly exposed to the periplasm, and EetB in the membrane are required for ferric reductase activity dependent on DMK. The basic role of Ndh3 is probably to couple the oxidation of NADH in the cytoplasm to reduction of a quinone species that might be special and synthesized with the help of DmkAB, as suggested by Light et al. (13). The function of Ndh3 seems specifically connected to ferric ion reduction involving EetA and EetB. In the presence of active cytochrome bd oxidase and under oxic conditions, the reduction level of the quinone pool in the membrane of the E. faecalis cell is low, and this attenuates both ferric reductase activity (Table 2) and EET (4). Thus, cytochrome bd is apparently more efficient than the ferric reductase system in oxidizing quinol. Alternatively, the Ndh3 enzyme might show low NADH dehydrogenase activity when the NADH/NAD+ ratio is low in the cytoplasm. Since Ndh3 only marginally contributes to NADH oxidase activity (Table 3), it seems likely that the quinol generated by the activity of Ndh3 is reserved for the reduction of ferric ions via EetAB. The phenotype of ndh3-defective mutants could alternatively be explained by the fact that the Ndh3 protein via its transmembrane C-terminal extension binds EetA and EetB in a, for enzyme function, obligatory protein complex. The role(s) of EetA and EetB in electron transfer from reduced quinone to ferric ion and the actual site of ferric ion reduction in the cell envelope remain enigmatic. It has, however, been shown that the EbpA pilin of the Ebp fiber, which is covalently bound to the peptidoglycan of the cell wall, can bind iron, and it has been suggested that the Ebp fiber is involved in iron acquisition in E. faecalis and might play some role in ferric ion reduction (17).
Electron-conducting redox polymers, such as OsRP, have significant potential in bioelectrochemical applications within environmental monitoring and energy transformation. As demonstrated in this work, Ndh3 and EetA are not important for electron transfer from the quinol pool to OsRP (Fig. 3). OsRP apparently accepts electrons directly from reduced DMK. Variants of redox hydrogel containing covalently bound quinone can make up for the lack of DMK in E. faecalis cells and mediate glucose-dependent EET (20, 21). In conclusion, the function of OsRP as a mediator in EET does not rely on components involved in EET mediated by ferric ions; i.e., electron transfer mediated to an electrode by OsRP occurs via a route distinct from that mediated with ferric ions. This aspect of the material-cell interface is directly applicable in the development and optimization of microbial electrochemical systems to achieve efficient energy conversion designs.
MATERIALS AND METHODS
Growth of bacteria.
The bacterial strains used in this work are listed in Table 1. The E. faecalis strains were grown in tryptic soy broth without dextrose (Difco) (TSB) with 1% (wt/vol) glucose added (TSBG) and with or without hemin added, as previously described (18), or on 1.5% (wt/vol) agar plates containing TSBG, Todd-Hewitt agar (Difco), or brain heart infusion (Difco) medium.
Isolation of ferric reductase mutants.
An available collection of E. faecalis OG1RF EfaMarTn insertion mutants (18), stored frozen as individual clones in 20% (wt/vol) glycerol in 96-well microtiter plates, was replicated to Todd-Hewitt agar plates supplemented with 0.2 mM ammonium ferric sulfate dodecahydrate. The plates were incubated at 37°C for 1 or 2 days to obtain colonies. Then the agar plates were overlaid with soft agar (0.8% wt/vol agar) containing 0.5% glucose and 2 mM FerroZine [3-(2-pyridyl)-5,6-diphenyl-1,2,4-triazine-4′,4ʺ-disulfonic acid monosodium salt hydrate]. Ferric reductase activity was observed as pink coloration of colonies. Poorly staining colonies were immediately picked and streaked on the same type of plates, and the phenotype was confirmed first by repetition of the ferric reductase activity staining of colonies and subsequently by ferric reductase activity determination with washed cells grown in liquid medium.
Identification of EfaMarTn insertion sites in chromosomal DNA.
The insertion site of the 2.1-kb transposon EfaMarTn (22) in the chromosome of E. faecalis strains LFR9 and LFR16 was determined using inverse PCR after cleavage of DNA with HindIII and exploiting primers invCATR2 and invGFPR1 for both DNA amplification and sequence analysis, as described before (18). The transposon insertion sites in strains LFR1, LFR15, and LFR19 were determined by sequence analysis of PCR products obtained using primers ndh3up and ndh3dw. The genotypes of strains ndh3::Tn and pplA::Tn were confirmed using primers ndh3up, ndh3dwn, pplAup, pplAmid, invCATR2, and invGFPR1. Phusion DNA polymerase (Thermo Scientific) was used for PCR. The sequences of the primers are presented in Table S1.
Ferric reductase activity assay of cells.
Cells were grown in 25 ml TSBG in a 250-ml Erlenmeyer flask at 200 rpm and 37°C to the mid-exponential growth phase at an optical density at 600 nm (OD600) of 0.6 to 0.8 (90 to 180 min). After the cells were harvested by centrifugation (2,700 × g for 10 min at room temperature) and washed once in 5 ml of TSB, they were suspended in TSB to an OD600 of 1.2 and stored on ice and analyzed the same day. Ferric reductase activity assayed using FerroZine was determined at 37°C in 0.92 ml in a 1-ml cuvette containing TSB, 0.4% (wt/vol) glucose, 1 mM ammonium ferric sulfate, 2 mM FerroZine, and cells of OD600 of 0.4. The assay was started by the addition of 500 μl 2 mM ammonium ferric sulfate in TSB to the cuvette containing the other ingredients. Formation of ferrous-FerroZine complex was monitored at 562 nm. Reductase activity was calculated based on the initial rate of increase in absorbance after subtraction of the background value obtained in the absence of FerroZine and using the extinction coefficient 27.9 mM−1 cm−1 (19).
Activity with potassium ferricyanide as the substrate was determined at 37°C in 1 ml in a 1-ml cuvette containing 35 mM Tris-HCl (pH 7.5), 0.4% (wt/vol) glucose, 1 mM K3Fe(CN)6, and cells of OD600 of 0.4. Two cuvettes and a double beam spectrophotometer were used. Ferricyanide (40 μl, 25 mM) was added only to the sample cuvette. This started the reaction, which was monitored as a reduction in absorption at 420 nm over 2 min. Activity was calculated based on the extinction coefficient 1.04 mM−1 cm−1.
Ferric reductase activities were determined using at least duplicate cell samples grown independently, and two or more assays were done per sample.
Electrochemical measurements.
E. faecalis cells were grown in 10 ml of TSBG at 37°C overnight. The cells were harvested by centrifugation and washed in 50 mM phosphate buffer, pH 7.4, containing 100 mM KCl. The washed cell suspension was adjusted with the buffer to a cell density of 1 g of wet weight per ml and used in electrochemical experiments.
Electrochemical measurements were done at room temperature in phosphate buffer (50 mM sodium phosphate buffer, pH 7.4, containing 100 mM KCl) using a three-electrode system. When indicated, 0.5 mM potassium ferricyanide added to the buffer or osmium complex modified redox polymer (OsRP) {[Os(2,2′-bipyridine)2-poly(N-vinylimidazole)10Cl]2+/+} (23) with a formal potential of 420 mV versus the standard hydrogen electrode (SHE), 10 mg ml−1 in water, was applied as a mediator. Spectrographic graphite rods (Alfa Aesar GmbH & Co.; diameter, 3.05 mm) were prepared and modified with E. faecalis cells and OsRP as described in reference 4 and used as working electrodes.
Amperometric measurements were performed at a constant potential of 588 mV versus the standard hydrogen electrode (SHE) controlled by a potentiostat (Zäta Electronik, Höör, Sweden) and using a wall jet electrochemical flow cell with a connected working graphite electrode, platinum counter electrode, and Ag-AgCl (0.1 M KCl) reference electrode. All potentials in this work are given in reference to SHE. The electrolyte solution was pumped through the system at a flow rate of 0.5 ml min−1.
Cyclic voltammetry was realized using a stationary solution three-electrode electrochemical cell including a working graphite electrode, a platinum counter electrode, and an Ag-AgCl (saturated solution KCl) reference electrode controlled by a potentiostat (Autolab PGSTAT 30; Eco Chemie, Utrecht, The Netherlands).
Miscellaneous methods.
Isolation of membranes from E. faecalis strains and protein determination were done as described before (4). NADH oxidase activity of isolated membranes at 30°C in 50 mM aerated potassium phosphate buffer (pH 7.4) with an initial NADH concentration of 0.16 mM was determined using an upgraded Aminco DW-2 spectrophotometer (OLIS Instruments) (absorption at 340 nm versus 400 nm) and a 3-ml cuvette with magnetic bar stirrer.
Supplementary Material
ACKNOWLEDGMENTS
The work was supported by the Swedish Research Council (grant 2015-02547) to L.H.
We thank Dónal Leech for providing the OsRP, Garry Dunny for strains, Samuel Light for discussions and strains and for providing unpublished information, and undergraduate students Raymilda Ekua Abaka, Carl-Johan Hörberg, Sebastian Kaiser, Elena Lopez, and Ariana Seira Calderon Moreno of the Molecular Microbiology course BIOR63 at Lund University for their assistance in screening for mutants.
For a commentary on this article, see https://doi.org/10.1128/JB.00029-20.
Supplemental material is available online only.
REFERENCES
- 1.Pankratova G, Hederstedt L, Gorton L. 2019. Extracellular electron transfer features of Gram-positive bacteria. Anal Chim Acta 1076:32–47. doi: 10.1016/j.aca.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 2.Logan BE. 2009. Exoelectrogenic bacteria that power microbial fuel cells. Nat Rev Microbiol 7:375–381. doi: 10.1038/nrmicro2113. [DOI] [PubMed] [Google Scholar]
- 3.Pankratova G, Hasan K, Leech D, Hederstedt L, Gorton L. 2017. Electrochemical wiring of the Gram-positive bacterium Enterococcus faecalis with osmium redox polymer modified electrodes. Electrochem Commun 75:56–59. doi: 10.1016/j.elecom.2016.12.010. [DOI] [Google Scholar]
- 4.Pankratova G, Leech D, Gorton L, Hederstedt L. 2018. Extracellular electron transfer by the Gram-positive bacterium Enterococcus faecalis. Biochemistry 57:4597–4603. doi: 10.1021/acs.biochem.8b00600. [DOI] [PubMed] [Google Scholar]
- 5.Doyle LE, Marsili E. 2018. Weak electricigens: a new avenue for bioelectrochemical research. Bioresour Technol 258:354–364. doi: 10.1016/j.biortech.2018.02.073. [DOI] [PubMed] [Google Scholar]
- 6.Wang W, Du Y, Yang S, Du X, Li M, Lin B, Zhou J, Lin L, Song Y, Li J, Zuo X, Yang C. 2019. Bacterial extracellular electron transfer occurs in mammalian gut. Anal Chem 91:12138–12141. doi: 10.1021/acs.analchem.9b03176. [DOI] [PubMed] [Google Scholar]
- 7.Schwab L, Rago L, Koch C, Harnisch F. 2019. Identification of Clostridium cochleariumas as an electroactive microorganism from the mouse gut microbiome. Bioelectrochemistry 130:107334. doi: 10.1016/j.bioelechem.2019.107334. [DOI] [PubMed] [Google Scholar]
- 8.Naradasu D, Miran W, Sakamoto M, Okamoto A. 2019. Isolation and characterization of human gut bacteria capable of extracellular electron transport by electrochemical techniques. Front Microbiol 9:3267. doi: 10.3389/fmicb.2018.03267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar A, Hsu LH, Kavanagh P, Barriére F, Lens PNL, Lapinsonniére L, Lienhard JH V, Schröder U, Jiang X, Leech D. 2017. The ins and outs of microorganism-electrode electron transfer reactions. Nature Rev Chem 1:1–13. [Google Scholar]
- 10.Lusk BG. 2019. Thermophiles; or, modern Prometheus: the importance of extreme microorganisms for understanding and applying extracellular electron transfer. Front Microbiol 10:818. doi: 10.3389/fmicb.2019.00818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winstedt L, Frankenberg L, Hederstedt L, von Wachenfeldt C. 2000. Enterococcus faecalis V583 contains a cytochrome bd-type respiratory oxidase. J Bacteriol 182:3863–3866. doi: 10.1128/jb.182.13.3863-3866.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baureder M, Hederstedt L. 2013. Heme proteins in lactic acid bacteria. Adv Microb Physiol 62:1–43. doi: 10.1016/B978-0-12-410515-7.00001-9. [DOI] [PubMed] [Google Scholar]
- 13.Light SH, Su L, Rivera-Lugo R, Cornejo JA, Louie A, Iavarone AT, Ajo-Franklin CM, Portnoy DA. 2018. A flavin-based extracellular electron transfer mechanism in diverse Gram-positive bacteria. Nature 562:140–144. doi: 10.1038/s41586-018-0498-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim GT, Hyun MS, Chang IS, Kim HJ, Park HS, Kim BH, Kim SD, Wimpenny JWT, Weightman AJ. 2005. Dissimilatory Fe(III) reduction by an electrochemically active lactic acid bacterium phylogentically related to Enterococcus gallinarium isolated from submerged soil. J Appl Microbiol 99:978–987. doi: 10.1111/j.1365-2672.2004.02514.x. [DOI] [PubMed] [Google Scholar]
- 15.Deneer HG, Healey V, Boychuk I. 1995. Reduction of exogenous ferric iron by a surface-associated ferric reductase of Listeria spp. Microbiology 141:1985–1992. doi: 10.1099/13500872-141-8-1985. [DOI] [PubMed] [Google Scholar]
- 16.Keogh D, Lam LN, Doyle LE, Matysik A, Pavagadhi S, Umashankar S, Low PM, Dale JL, Song Y, Ng SP, Boothroyd CB, Dunny GM, Swarup S, Williams RBH, Marsili E, Kline KA. 2018. Extracellular electron transfer powers Enterococcus faecalis biofilm metabolism. mBio 9:e00626-17. doi: 10.1128/mBio.00626-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lam LN, Wong JJ, Matysik A, Paxman JJ, Chong LKK, Low PM, Chua ZS, Heras B, Marsili E, Kline KA. 2019. Sortase-assembled pili promote extracellular electron transfer and iron acquisition in Enterococcus faecalis biofilm. bioRxiv doi: 10.1101/601666. [DOI]
- 18.Baureder M, Hederstedt L. 2012. Genes important for catalase activity in Enterococcus faecalis. PLoS One 7:e36725. doi: 10.1371/journal.pone.0036725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deneer H, I B. 1993. Reduction of ferric iron by Listeria monocytogenes and other species of Listeria. Can J Microbiol 39:480–485. doi: 10.1139/m93-068. [DOI] [PubMed] [Google Scholar]
- 20.Pankratova G, Szypulska E, Pankratov D, Leech D, Gorton L. 2019. Electron transfer between the Gram-positive Enterococcus faecalis bacterium and electrode surface via osmium redox polymers. ChemElectroChem 6:110–113. doi: 10.1002/celc.201800683. [DOI] [Google Scholar]
- 21.Pankratova G, Pankratov D, Milton R, Minteer S, Gorton L. 2019. Following nature: bioinspired mediation strategy for Gram-positive bacterial cells. Adv Energy Mater 9:1900215. doi: 10.1002/aenm.201900215. [DOI] [Google Scholar]
- 22.Kristich CJ, Nguyen VT, Le T, Barnes AM, Grindle S, Dunny GM. 2008. Development and use of an efficient system for random mariner transposon mutagenesis to identify novel genetic determinants of biofilm formation in the core Enterococcus faecalis genome. Appl Environ Microbiol 74:3377–3386. doi: 10.1128/AEM.02665-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrière F, Ferry Y, Rochefort D, Leech D. 2004. Targetting redox polymers as mediators for laccase oxygen reduction in a membrane-less biofuel cell. Electrochem Commun 6:237–241. doi: 10.1016/j.elecom.2003.12.006. [DOI] [Google Scholar]
- 24.Dunny G, Brown B, Clewell D. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc Natl Acad Sci U S A 75:3479–3483. doi: 10.1073/pnas.75.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dale JL, Beckman KB, Willett JLE, Nilson JL, Palani NP, Baller JA, Hauge A, Gohl DM, Erickson R, Manias DA, Sadowsky MJ, Dunny GM. 2018. Comprehensive functional analysis of the Enterococcus faecalis core genome using an ordered, sequence-defined collection of insertional mutations in strain OG1RF. mSystems 3:e00062-18. doi: 10.1128/mSystems.00062-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Yang Y, Moore D, Nimmo S, Lightfoot S, Huycke M. 2012. 4-Hydroxy-2-nonenal mediates genotoxicity and bystander effects caused by Enterococcus faecalis infected macrophages. Gastroenterology 142:543–551. doi: 10.1053/j.gastro.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.