Abstract
Infectious bronchitis coronavirus (IBV), Newcastle disease virus (NDV), and avian influenza virus (AIV) H9 subtype are major pathogens of chickens causing serious respiratory tract disease and heavy economic losses. To better understand the replication features of these viruses in their target organs and molecular pathogenesis of these different viruses, comparative proteomic analysis was performed to investigate the proteome changes of primary target organ during IBV, NDV, and AIV H9 infections, using 2D‐DIGE followed MALDI‐TOF/TOF‐MS. In total, 44, 39, 41, 48, and 38 proteins were identified in the tracheal tissues of the chickens inoculated with IBV (ck/CH/LDL/97I, H120), NDV (La Sota), and AIV H9, and between ck/CH/LDL/97I and H120, respectively. Bioinformatics analysis showed that IBV, NDV, and AIV H9 induced similar core host responses involved in biosynthetic, catabolic, metabolic, signal transduction, transport, cytoskeleton organization, macromolecular complex assembly, cell death, response to stress, and immune system process. Comparative analysis of host response induced by different viruses indicated differences in protein expression changes induced by IBV, NDV, and AIV H9 may be responsible for the specific pathogenesis of these different viruses. Our result reveals specific host response to IBV, NDV, and AIVH9 infections and provides insights into the distinct pathogenic mechanisms of these avian respiratory viruses.
Keywords: Animal proteomics, Avian influenza virus H9 subtype, Infectious bronchitis coronavirus, Newcastle disease virus, Pathogenesis
Abbreviations
- AIV
avian influenza virus
- ANXA1
annexin A1
- ANXA2
annexin A2
- dpi
days postinoculation
- EID50
egg infective dose 50%
- EIF5A
eukaryotic translation initiation factor 5A
- HI
hemagglutination inhibition
- HSPB1
HSP 27
- IBV
infectious bronchitis coronavirus
- LGALS1
beta‐galactoside‐binding lectin
- LMNA
lamin‐A
- NDV
Newcastle disease virus
- NUMA
nuclear mitotic apparatus protein
- PPIA
peptidyl‐prolyl cis‐trans isomerase A
- PRDX
peroxiredoxin
- SPF
specific pathogen‐free
- UPP
ubiquitin‐proteasome pathway
1. Introduction
Avian respiratory viruses, including infectious bronchitis coronavirus (IBV), Newcastle disease virus (NDV), and avian influenza virus (AIV), are the primary cause of morbidity and mortality worldwide in the poultry industry and have been identified as the most economically important etiological agents of acute, highly contagious poultry diseases. Among these viruses, NDV is a member of the Avulavirus genus within the Paramyxoviridae family and is prevalent worldwide and infects almost all avian species. In fact, highly contagious respiratory, enteric, and neurological diseases in chickens are often caused by a variety of infectious NDV strains 1, 2. AIV is an enveloped virus with a segmented, single‐stranded, negative‐sense RNA genome belonging to the Orthomyxoviridae family and can cause clinical diseases in a wide range of mammalian and avian species. Low pathogenic avian influenza virus subtype H9 circulates worldwide in multiple avian species thereby posing a potential threat to not only the poultry industry, but also to public health 3, 4. IBV is a gammacoronavirus of the subfamily Coronavirinae, family Coronaviridae, and order Nidovirales 5, and primarily replicates in epithelial cells of the respiratory tract and causes lower respiratory tract infections in chickens (Gallus domesticus). In addition, emerging IBV isolates that vary in their replicative abilities in nonrespiratory tissues have been associated with proventriculus and kidney lesions 6, 7.
Proteomic analyses of pathogen‐infected organisms can offer significant insight to the “battle” between pathogens and host during infection, hence proteomic analyses of host cellular responses against viral infections could aid in identification of potential host factors involved in the viral life cycle and host defense mechanisms. To date, various proteomic strategies have been applied to elucidate protein dynamics during influenza infections, such as differential expression of brain proteins in response to the neurovirulent AIV H5N1 infection 8, host responses of murine lung tissues infected with AIV H5N1 9, and cellular protein changes in H1N1‐infected human lung A549 cells 10; while cellular responses to AIV H9N2 subtype infections has only been reported by Liu et al. in human cells 11. For IBV, proteomic changes have been reported in the cytoplasm, nuclei, and nucleoli in mammalian Vero cells and nucleolar proteomes in avian cells (DF‐1) infected with IBV (Beaudette strain) 12, 13. We previously reported alterations in the proteome of tracheal and kidney cells during IBV strain H120 infection in vitro, and IBV strain ck/CH/LDL/97I and its attenuated strain ck/CH/LDL/97I P115 infection in vivo 14, 15. However, alterations of cellular protein expression profiles, whether in NDV‐infected cell lines or tissues, and the differences in induced protein expression changes between different avian respiratory viruses (IBV, NDV, and AIV H9) have not yet been reported.
The tracheal epithelium of chicken is not only the primary target for replication and infection of avian respiratory viruses, but also the first line of defense in the innate immune response. The avian respiratory viruses, including IBV, NDV and AIV H9, can infect tracheal epithelial cells and induce damage to tracheal epithelium resulting in respiratory disease. However, the severity and outcome of the clinical illness caused by these different viruses are different 16, 17, 18. To better understand the molecular basis of pathogenesis and specific virus–host interactions, herein, we described the alterations of protein expression profiles in chicken tracheal tissues caused by IBV ck/CH/LDL/97I and H120, NDV La Sota, and AIV H9 infection by using 2D‐DIGE analyses. A series of differentially expressed proteins associated with viral life cycle and host response to IBV, NDV, and AIV H9 infections were identified. The present proteome results using the same virus of different strains (IBV virulent ck/CH/LDL/97I and H120 vaccine) and different viruses (IBV, NDV and AIV H9) might provide more significant information on specific aspects of molecular pathogenesis and virus–host interactions of these different viruses. Furthermore, these findings might also provide useful clues for the development of novel prevention or therapeutics strategies against these viruses.
2. Materials and methods
2.1. Viral strains
A commercial vaccine against IBV strain H120 was obtained in freeze‐dried 1000‐dose vials that each contained at least 103 egg infective dose 50% (EID50) per dose (isbi® BIO H120; batch no. F48355). IBV strain ck/CH/LDL/97I 19, NDV strain La Sota vaccine, and AIV subtype H9 20 were used in this study. The virus stock for this study was produced by inoculating the virus into embryonated specific pathogen‐free (SPF) chicken eggs via the allantoic cavity and collecting the infectious allantoic fluid 72 h postinoculation. The allantoic fluid was clarified by centrifugation at 3000 × g for 10 min and filtered with a Teflon membrane and the presence of IBV was confirmed by negative contrast electron microscopy (JEM‐1200, EX; JEOL Ltd., Tokyo, Japan) and RT‐PCR as previously described 21. NDV and subtype H9 AIV viral particles in the allantoic fluids of inoculated eggs were confirmed using hemagglutination inhibition (HI) analysis with specific antibodies, respectively 22. The titers of the viruses were determined by inoculation at tenfold dilutions into groups of five 10‐day‐old embryonated chicken eggs. The EID50 was calculated by the method of Reed and Muench 23.
2.2. Experimental design
One hundred twenty 1‐day‐old SPF White Leghorn chicks were divided into five groups of 24 birds each and housed in different isolators with negative pressure. Chickens assigned to groups 1–4 were inoculated with IBV ck/CH/LDL/97I, H120, NDV La Sota, and AIV H9, respectively, by oculonasal application at 3 days of age with a dose of log105 EID50/100 μL per chick. Birds in group 5 were mock‐inoculated with sterile allantoic fluid as a negative control. This study was approved by the Animal Welfare Committee of Heilongjiang Province, China. All experiments were conducted at appropriate biosafety levels.
Three birds from each group were killed humanely at 3, 7, 12, and 21 days postinoculation (dpi) and fresh tissue of trachea samples were quickly harvested and washed with ice‐cold Tris‐sucrose buffer (10 mM Tris, 250 mM sucrose, pH 7.5), snap‐frozen in liquid nitrogen, and stored at −80°C for subsequent 2D‐DIGE analysis. Sera samples from ten chickens in each group were collected at 3, 7, 12, and 21 dpi for ELISA and HI analyses. Sera samples from groups 1, 2, and 5 were assayed using a commercial IBV antibody test kit (IDEXX Corporation, Westbrook, ME, USA) according to the manufacturer's instructions. Serum samples from groups 3, 4, and 5 were detected for HI antibody titer (positive when HI ≥ 3) against NDV and AIV, respectively, using an HI assay as reported previously 24. Viral load of IBV, NDV, and AIV in tracheal tissues were analyzed using methods described previously. Briefly, viral RNA was extracted from the tracheal tissues of sacrificial birds from each group using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA). For detection of the viruses, specific primers and TaqMan probes against IBV 25, NDV 26, and AIV subtype H9 4 viral RNA were selected. Real‐time RT‐PCR was performed on a LightCycler® 480 real‐time PCR system with One Step PrimeScript™ RT‐PCR Kit (TaKaRa, Dalian, China) 27. All of the samples were tested in triplicate in each reaction. Standard templates with known concentration and no template negative control were contained for quantitative analysis.
2.3. 2D‐DIGE analysis of tracheal proteins
The tracheal tissues were ground into powder in liquid nitrogen. Fifty milligrams of powder were extracted with 500 μL of DIGE‐compatible lysis buffer containing 7 M urea, 2 M thiourea, 4% CHAPS, 1% nuclease mix, and 1% Protease Inhibitor Mix (GE Healthcare, Waukesha, WI, USA), vortexed for 3 h in an ice bath, and then centrifuged at 15 000 × g for 1 h at 4°C. The supernatant was collected and purified using the Ettan 2‐D Clean‐Up Kit (GE Healthcare). The pH of the protein sample was adjusted to 8.5 and the protein concentration was determined using the Ettan 2‐D Quant Kit (GE Healthcare). The purified protein lysates (50 μg) were labeled with 400 pmol of Cy2, Cy3, and Cy5 dyes (GE Healthcare) following the manufacturer's instructions. Briefly, the Cy2‐labeled internal standard was created by pooling an aliquot from all involved biological samples, while Cy3 and Cy5 were used randomly to label samples from the IBV ck/CH/LDL/97I‐, H120‐, NDV La Sota‐, AIV H9‐, and mock‐infected groups. The experimental design of the different fluorescent dye labeling protocols is listed in Supporting Information Table 1. The labeling reaction was performed as described in our previous study 15 and labeled samples were subsequently loaded onto 24‐cm, linear pH 3–10 IPG strips (GE Healthcare) for integrated IEF that was performed using an Ettan™ IPGphor 3 fully integrated IEF system (GE Healthcare). After IEF, the IPG strips were treated in an equilibration buffer (6 M urea, 50 mM Tris‐HCl, pH 8.8, 30% glycerol, 2% SDS, and 0.002% bromophenol blue) containing 1% DTT and 2.5% iodoacetamide. A second dimension SDS‐PAGE was performed using an Ettan DALT six system electrophoresis system (GE Healthcare).
After SDS‐PAGE, the CyDye‐labeled gels were scanned using the Typhoon 9400 scanner (GE Healthcare) at a resolution of 100 μm. The intensity was adjusted to ensure that the maximum volume of each image was within 60 000–80 000. DIGE analysis was performed using DeCyder software version V6.5 (GE Healthcare). Protein spots were pairwise compared among control and different infection groups and differentially expressed spots with statistically significant results via the Student's t‐test (p < 0.05) and average ratio ≥1.2 or ≤−1.2 were selected for MS identification. Spot picking was performed using preparative gels under the conditions as described above for 2D‐DIGE except that the IPG strips were loaded with 1000 μg of protein and the gels were stained with Coomassie Blue R‐350, then the protein spots were excised manually from the gels and subjected to MALDI‐TOF/TOF MS identification.
2.4. MALDI‐TOF/TOF MS and database search
The gel spots were excised, trypsin‐digested, and subjected to MALDI‐TOF/TOF MS analysis as described in our previous study 15. The PMF combined MS/MS data were submitted to MASCOT version 3.0 (Matrix Science, Inc., Boston, MA, USA) for identification according to the National Center for Biotechnology Information nr database (release 16/01/2010, 10,343,571 sequences, and 3,528,215,794 residues). The search parameters were set as follows: Gallus, trypsin cleavage (one missed cleavage allowed), carbamidomethylation of cysteine as a fixed modification, oxidation of methionine as a variable modification, peptide mass tolerance set at 100 ppm, and fragment tolerance set at 0.8 Da. The criterion for successful identification of a protein was a protein score confidence interval (CI) ≥95%.
2.5. GO annotation and classification
To investigate the potential roles played by the differentially expressed proteins during infection, GI numbers of identified proteins were submitted to GORetriever (http://www.agbase.msstate.edu/) to obtain the GO annotations. If no annotation was acquired from GORetriever, GOanna was used to retrieve GO annotations assigned on the basis of searches using the Basic Local Alignment Search Tool. Then, the resulting annotations were summarized using GOSlimViewer.
2.6. Analysis of mRNA abundance by real‐time RT‐PCR
Ten target proteins that extensively changed during IBV, NDV, and AIV H9 infections shown by 2D‐DIGE and potentially involved in the viral infection were selected for transcriptional analysis. The expression of selected proteins were not all altered at four time points (Supporting Information Table 2), therefore, the mRNA levels of selected proteins at their corresponding time point were analyzed, respectively. Specific primers for the target genes of MS/MS‐identified proteins were designed according to the published gene sequences using Beacon Designer software 7.5 (Premier Biosoft International, Palo Alto, CA, USA) and information on the primers is listed in Supporting Information Table 3. Total tracheal tissue RNA was extracted from sacrificed birds of each group at each time point using TRIzol Reagent (Invitrogen) and then reverse‐transcribed to cDNA. The real‐time PCR was performed using a LightCycler® 480 real‐time PCR system (Roche Diagnostics Deutschland GmbH, Mannheim, Germany). Melting curve and quantitative analyses of the data were determined using the LightCycler® 480 real‐time PCR system with the relative quantification 2−ΔΔCt model, Cp values of target gene in control group at each time point were used as the calibrator (relative expression = 1), and 28S ribosomal RNA gene was used as an internal reference gene.
2.7. Western blot analysis
To verify the 2D‐DIGE results, several representative proteins were selected for Western blot analysis based on the commercial availability of the corresponding antibodies. Tracheal proteins from sacrificed birds of each group at all four time points were extracted and equivalent lysate amounts were subjected to 12% SDS‐PAGE and then transferred to NC membranes. After blocking overnight at 4°C, the membranes were incubated with goat polyclonal antibody to annexin A2 (ANXA2; sc‐1924; Santa Cruz Biotechnology), rabbit polyclonal antibody to annexin A1 (ANXA1; sc‐11387; Santa Cruz Biotechnology), goat polyclonal antibody to peroxiredoxin I (PRDX1; sc‐23970; Santa Cruz Biotechnology), mouse mAb to HSP 27 (HSPB1; sc‐51956, Santa Cruz Biotechnology), rabbit polyclonal antibody to phospho‐HSPB1 (pSer82; SAB4504434, Sigma‐Aldrich), and mouse mAb to β‐tubulin (sc‐55529; Santa Cruz Biotechnology) for 1 h at 37°C, respectively. Then, the membranes were incubated with IRDye800DX‐conjugated anti‐mouse, anti‐goat, and anti‐rabbit secondary antibodies (610–130–121, 605–430–002, and 611–730–127; Rockland Immunochemicals, Gilbertsville, PA, USA), respectively, for 1 h at 37°C. After washing, the membranes were scanned using a LI‐COR infrared imaging system with Odyssey software (Li‐Cor Biosciences, Lincoln, NE, USA).
3. Results
3.1. Clinical observations, serology, and virus detection in trachea
All chickens infected with the IBV ck/CH/LDL/97I strain exhibited clinical respiratory signs at approximately 3–12 dpi and one of the diseased birds died at 12 dpi. The clinical signs included tracheal rales, watery eyes, nasal mucus, and sneezing. Gross lesions within the chickens were confined mainly to the kidneys 15, whereas the chickens in the control group and those inoculated with the IBV H120 and NDV La Sota strains showed no apparent respiratory clinical signs and no gross lesions. AIV H9‐infected chickens appeared lethargy, depression, coughing, sneezing, and dyspnea during the experimental period.
The serological responses induced by infections with IBV ck/CH/LDL/97I, H120, NDV La Sota, and AIV H9 are summarized in Supporting Information Table 4. No seroconversion was observed in any group at 3 dpi. Antibodies appeared at 7 dpi and all chickens showed seroconversion after 21 dpi in the IBV, NDV, and AIV infection groups. The chickens in the control group showed negative serum antibody responses against IBV, NDV, and AIV.
IBV, NDV, and AIV infections were verified using real‐time RT‐PCR. As shown in Fig. 1, negative results were obtained following viral detection in the tracheal tissues from chickens in the control group. In the IBV ck/CH/LDL/97I‐infected group, the IBV genome was detected at 3, 7, 12, and 21 dpi. The viral load gradually decreased during the experimental period after the viral copy number peaked at 3 dpi (Fig. 1A). Similar results were obtained from group H120, except no viral genome was detected at 21 dpi (Fig. 1B). In the NDV La Sota‐ and AIV H9‐infected groups, viruses were detected at 3 dpi and viral copy number peaked at 7 dpi, whereas the results at 12 and 21 dpi were negative (Fig. 1C and D). The differences in viral kinetics among IBV ck/CH/LDL/97I, H120, NDV La Sota, and AIV H9 were demonstrated in Fig. 1E. Taken together, these results confirmed the successful virus infections to SPF chicken.
Figure 1.
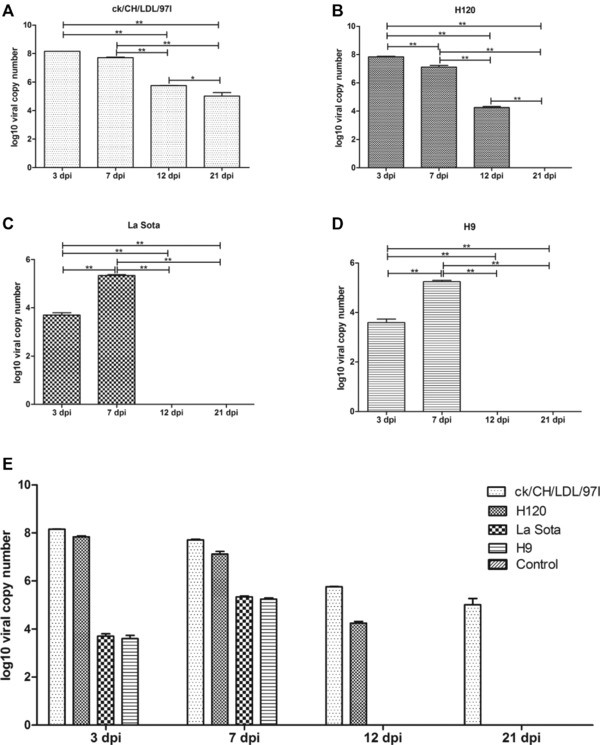
The viral load was quantified using real‐time RT‐PCR. The viral loads in tracheal samples of three sacrificial birds from each group at each time point were determined. The average viral copy number per 1 μg RNA of each group was calculated through standard curve based on tenfold dilution series of standard template with known concentration. The LOD of the real‐time RT‐PCR reaction was 102, 103 and 103 copies of template for IBV, NDV and AIV, respectively. (A) Kinetics of viral loads of IBV ck/CH/LDL/97I. (B) Kinetics of viral loads of IBV H120. (C) Kinetics of viral loads of NDV La Sota. (D) Kinetics of viral loads of and AIV H9. (E) The differences in viral kinetics among IBV ck/CH/LDL/97I, H120, NDV La Sota, and AIV H9 infected groups. Data represent means of three biological replicates per group. Error bars indicate SEM, and dpi represents days postinoculation. The symbol * indicate significant differences (*p < 0.05 and **p < 0.01) between different groups.
3.2. 2D‐DIGE profiles of tracheal tissues following infections with avian respiratory viruses (IBV, NDV, and AIV H9)
To obtain a detailed comparison of the differential protein expression profiles, the tracheal proteins from avian respiratory virus‐infected (IBV ck/CH/LDL/97I, H120, NDV La Sota, and AIV H9) and mock‐inoculated chickens were extracted for quantitative and comparative analysis via 2D‐DIGE at 3, 7, 12, and 21 dpi. Comparative analysis revealed that total 53, 44, 70, and 33 spots were significantly differentially expressed at 3, 7, 12, and 21 dpi, respectively, which are indicated by arrows in the corresponding representative 2D‐DIGE map (Fig. 2A, B, C, and D). In the IBV ck/CH/LDL/97I‐, H120‐, NDV La Sota‐, and AIV H9‐infected groups, and between ck/CH/LDL/97I‐ and H120‐infected groups, 72, 65, 63, 86 and 63 differentially expressed protein spots were identified, respectively (Table 1 and Supporting Information Fig. 1). In general, these data showed that the expression of more host protein spots were changed in the chicken tracheal tissues after virulent IBV ck/CH/LDL/97I and AIV H9 infections, comparing with those of the vaccine strains IBV H120 and NDV La Sota strains.
Figure 2.
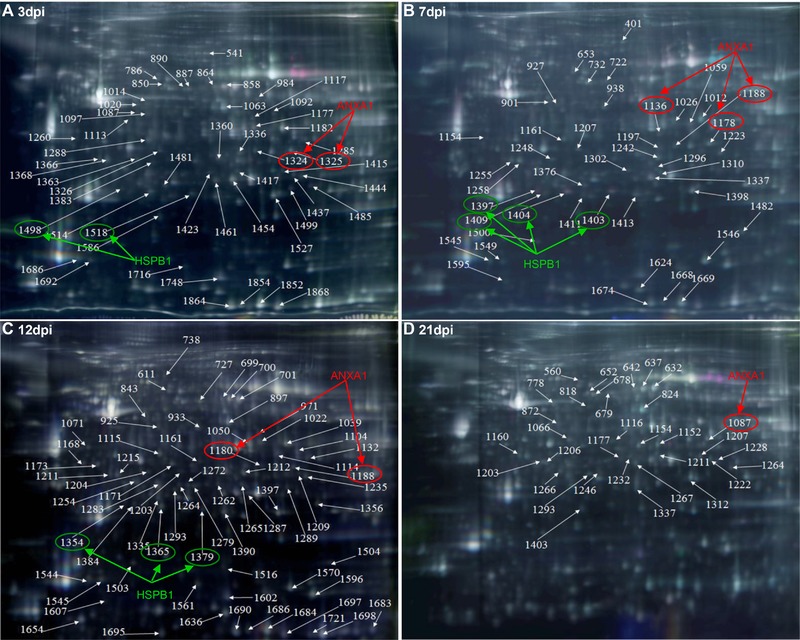
The representative gel images showing 2D‐DIGE maps labeled the relative locations of spots that displayed significant quantitative differential expression (p ≤ 0.05 and average ratio ≥1.2 or ≤−1.2) during different sampling times. (A) 3 dpi; (B) 7 dpi; (C) 12 dpi; and (D) 21 dpi. Representative 2D‐DIGE images of IBV ck/CH/LDL/97I, H120, NDV La Sota, and AIV H9 infected groups with their respective sampling points were represented as Supporting Information Fig. 1. Fluor‐labeled protein samples were separated on 24‐cm pH 3–10 linear IPG strips, subjected to SDS‐PAGE, and then gels were scanned on a Typhoon 9400 scanner, and image analysis was performed with Ettan™ DeCyder Software version v6.5. The identified protein spots were marked with arrows and labeled with the respective Match ID listed in Supporting Information Table 2. The green and red arrows and ellipses indicating HSPB1 and ANXA1 identified from separated spots.
Table 1.
Summary of differentially expressed spots and proteins in IBV ck/CH/LDL/97I, H120, NDV La Sota, and AIV H9 infected groups and between IBV ck/CH/LDL/97I and H120‐infected groups
| IBV ck/CH/LDL/97I/control | IBV H120/control | NDV La Sota/control | AIV H9/control | IBV ck/CH/LDL/97I/ H120 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Spots | Proteins | Spots | Proteins | Spots | Proteins | Spots | Proteins | Spots | Proteins | |
| Totala | 72 | 44 | 65 | 39 | 63 | 41 | 86 | 48 | 63 | 38 |
| Upregulatedb | 26 | 19 | 20 | 15 | 38 | 27 | 39 | 30 | 39 | 28 |
| Downregulatedc | 46 | 29 | 45 | 28 | 25 | 21 | 47 | 25 | 24 | 17 |
Total number of differentially expressed spots and protein.
Number of upregulated spots and proteins.
Number of downregulated spots and proteins.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
3.3. Identification and classification of differentially expressed proteins
To identify the differentially expressed proteins in viral infected tracheal tissues, 200 spots with thresholds >1.2‐fold (Supporting Information Table 2) were excised from preparative gels and subjected to MALDI‐TOF/TOF MS identification. Detailed information of the identified proteins is provided in Supporting Information Table 5 and Supporting Information Fig. 2. Totally, 200 differentially expressed protein spots corresponding to 75 altered cellular proteins were successfully identified; the biological functions of the differentially expressed proteins were summarized in Supporting Information Table 6. Number of differentially expressed spots and proteins induced by IBV ck/CH/LDL/97I, H120, NDV La Sota, and AIV H9 infections were summarized in Table 1. Among these identified proteins, some were identified from two or more separated spots, indicating that they may be differently modified (e.g., phosphorylation of ANXA1 and HSPB1) in the process of host response to IBV, NDV, and AIV H9 infections (Fig. 2). In order to verify the expression of phosphorylated HSPB1, tracheal proteins from sacrificed birds of each group were examined by Western blot analysis with specific antibodies to phospho‐HSPB1 (pSer82). The upregulation of phosphorylated HSPB1 in IBV‐, NDV‐ and AIV H9‐infected groups at 7 and 12 dpi were indentified (Fig. 3A and B); these results were consistent with the expression pattern of HSPB1 spots showed by 2D‐DIGE, confirming the expression of phosphorylated HSPB1 during IBV, NDV, and AIV H9 infections.
Figure 3.
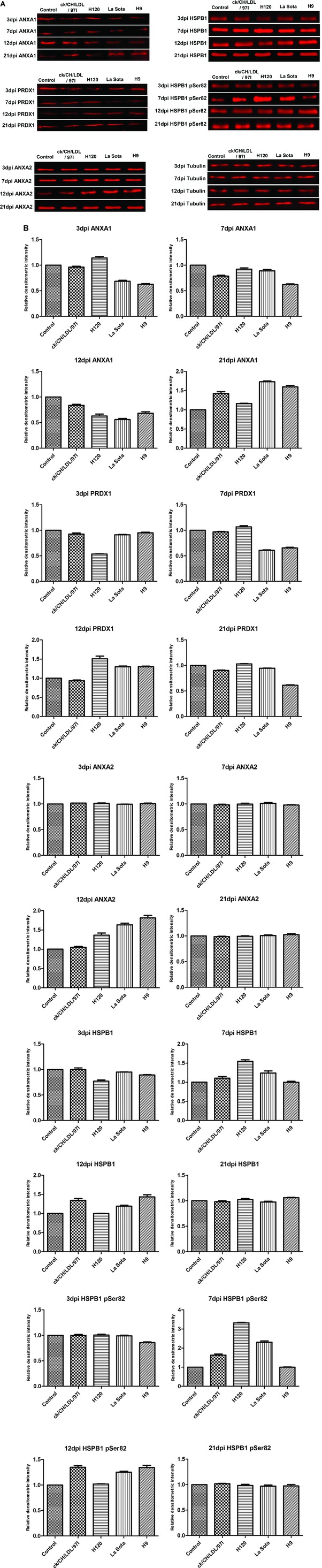
Western blot confirmation of representative proteins in IBV ck/CH/LDL/97I, H120, NDV La Sota, and AIV H9 infected chickens. (A) The immunoblot analysis of ANXA1, ANXA2, PRDX1, HSPB1, and phosphorylated HSPB1. (B) The relative densitometric intensity of ANXA1, ANXA2, PRDX1, HSPB1, and phosphorylated HSPB1 in immunoblot analysis. β‐Tubulin was used as an internal control to normalize the quantitative data.
To better understand the potential role of host factors involved in viral infection, differentially expressed proteins were classified based on GO annotations derived from the AgBase database (http://www.agbase.msstate.edu/). As illustrated in Fig. 4A, the biological process of proteins differentially expressed in IBV ck/CH/LDL/97I, H120, NDV La Sota, and AIV H9 infected groups and between IBV ck/CH/LDL/97I and H120‐infected groups (Fig. 4B), all mainly involved in anatomical structure development, biosynthetic process, transport, signal transduction, protein complex assembly, cell death, cell differentiation, translation, cellular protein modification process, response to stress, cytoskeleton organization, growth, macromolecular complex assembly, immune system process, vesicle‐mediated transport, embryo development, cell proliferation, transmembrane transport, and various metabolic‐associated process. These analyses indicated that all of the four viruses affect host global response similarly.
Figure 4.
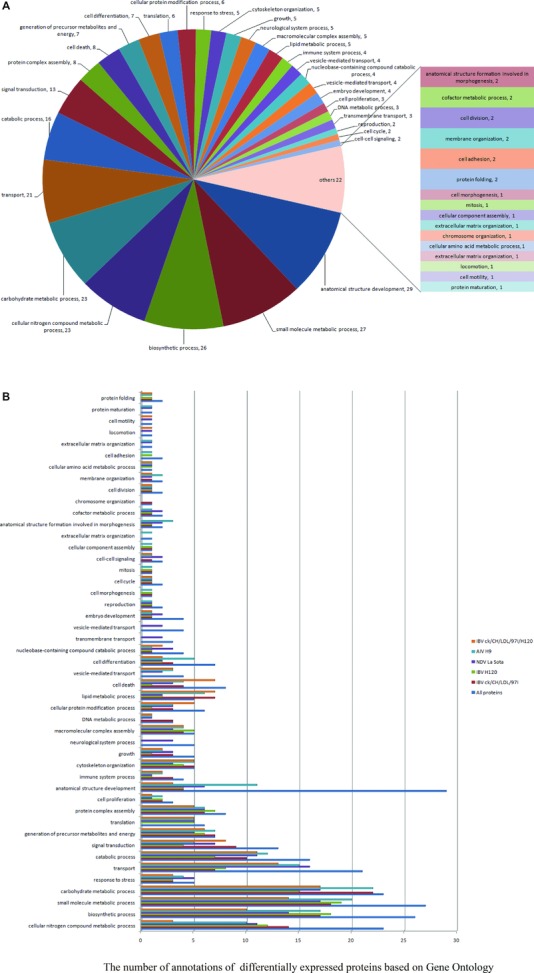
GO annotation analyses of differentially expressed proteins according to their biological process categories. This classification was produced by using the GOSlimViewer tool of the Agbase database (http://www.agbase.msstate.edu/). (A) Biological process classification of all identified proteins. (B) Comparison of biological process classification of differentially expressed proteins in IBV ck/CH/LDL/97I, H120, NDV La Sota, and AIV H9 infected groups and between IBV ck/CH/LDL/97I and H120‐infected groups.
In order to highlight the differences of proteome changes in trachea induced by different viruses, a side‐by‐side comparison of the differentially expressed proteins in trachea between different groups was performed (Table 2 and Supporting Information Table 7). The comparisons revealed that alteration of certain proteins and corresponding biological process appeared to be general or specific to host response in the process of different virus infections. For example, ENO1, ENO3, PGAM1, ANXA1, creatine kinase (CKB), GST, HSPB1, hemoglobin subunit alpha‐A (HBAA), hemoglobin subunit alpha‐D (HBAD), lamin‐A (LMNA), aldo‐keto reductase (akr), and PRDX1 were commonly altered in all viruses incubated groups, likely suggesting that these proteins were involved in and had similar functions in response to the infection of the three avian respiratory viruses. Glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH), L‐lactate dehydrogenase A chain (LDHA), peptidyl‐prolyl cis–trans isomerase A (PPIA), PRDX6, and hydroxyacyl‐coenzyme A dehydrogenase (HADH) were altered in pathogenic IBV ck/CH/LDL/97I and AIV H9 infected groups, while proteasomal subunit beta type 2 was more abundant in vaccine strains IBV H120 and NDV La Sota incubated groups. And we also found that some proteins were specifically affected by IBV ck/CH/LDL/97I (S100A11, ANXA8, and PHB), H120 (UQCRFS1), NDV La Sota (rbf and VDAC1), and AIV H9 (RPSA, CTHRC1, proteasomel subunit alpha type 6, and TAGLN) infections, respectively (Table 2).
Table 2.
List of differentially expressed proteins commonly induced by IBV ck/CH/LDL/97I, H120, NDV La Sota, and AIV H9 infections
| Groups containing commonly expressed proteinsa | Number of commonly expressed proteins | Gene name of commonly expressed proteins | Biological processb |
|---|---|---|---|
|
12 |
|
|
|
2 |
|
|
|
1 | C21 | – |
|
6 |
|
|
|
1 | ARPC5 |
|
|
1 | EIF5A1 |
|
|
4 |
|
|
|
2 |
|
|
|
1 | NDUFV2 | – |
|
5 |
|
|
|
3 |
|
|
|
1 | PSMB2 | catabolic process 2 |
|
1 | COL2A1 |
|
|
1 | THBS1 | – |
|
3 |
|
– |
|
3 |
|
|
|
1 | ECI1 | – |
|
1 | GNB2L1 |
|
|
3 |
|
|
| IBV ck/CH/LDL/97I/Control | 3 |
|
|
| IBV H120/Control | 1 | UQCRFS1 | Generation of precursor metabolites and energy 1 |
| NDV La Sota/Control | 2 |
|
|
| AIV H9/Control | 4 |
|
– |
Groups incubated different viruses (IBV ck/CH/LDL/97I, H120, NDV La Sota, and AIV H9) containing commonly expressed proteins.
Biological process of commonly expressed proteins in different viruses infected groups. This annotation was obtained using GOSlimViewer tool at Agbase database (http://www.agbase.msstate.edu/).
–: Indicated no GO annotation was acquired; PSMA6: proteasomel subunit alpha type 6; SOD1: Cu/Zn superoxide dismutase.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Expectedly, some proteins identified in this study have also been reported in proteomic analyses of cells or tissues infected with IBV or AIV (Table 3). For example, upregulation of beta‐galactoside‐binding lectin 1 (LGALS1), PPIA, PRDX4, and HSPB1 has been found in AIV H9 infected human cells 11. ANXA2, LGALS1, eukaryotic translation initiation factor 5A 2 (EIF5A2), and PPIA were also upregulated in IBV‐infected avian DF‐1 cells 12. And downregulation of ANXA1 was reported in IBV‐infected chicken trachea in our previous works 14, 15. The common alteration of these proteins in different models of infection with the same virus of different strains suggests that these proteins certainly involves in the course of corresponding virus infection.
Table 3.
Proteins identified both in our study and previous reports in proteomes of the infected tissues or cells induced by AIV and IBV infectionsa
| Tissues/cells | AIV H9b | H5N1c | H5N1c | H1N1c | H9c | IBV (ck/CH/LDL/97I)d | IBV (H120)d | IBV (Beaudette)d | IBV (Beaudette)e | IBV (H120)e | IBV (ck/CH/LDL/97I P5 and P115)e | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chicken trachea | Chicken trachea | Chicken trachea | DF‐1 cells [12] | Vero cells [13] | Chicken trachea [15] | |||||||||
| Protein name | RPSA | EIF5A2 | RPSA | RPSA | ARPC5 | CKB | ARPC5 | TPI1 | ARPC5 | |||||
| (abbreviation) | LGALS1 | EIF5A1 | LGALS1 | akr | HBAA | akr | TTR | akr | ||||||
| CKB | GST | CKB | ENO1 | HBAD | ENO1 | CKB | ENOI | ENOI | ENOI | |||||
| HSPB1 | GAPDH | HSPB1 | LGALS1 | NDUFV2 | UQCRFS1 | EPYC | LGALS1 | LGALS1 | ||||||
| LMNA | HBAA | LMNA/C | ANXA1 | NUMA | ANXA1 | HBAA | ANXA1 | ANXA1 | ANXA1 | |||||
| PPIA | HBAD | PPIA | ERP29 | OGN | THBS1 | HBAD | ERP29 | ERP29 | ||||||
| PRDX4 | LDHA | PRDX4 | GST | PPIA | GST | NUMA | GST | GST | ||||||
| SEPT2 | NUMA | SEPT2 | HSPB1 | PRDX6 | HSPB1 | NME1 | HSPB1 | HSPB1 | ||||||
| ARPC5 | NME1 | LMNA | PGAM1 | LMNA | PGAM1 | LMNA | LMNA | |||||||
| akr | PRDX6 | GAPDH | ANXA8 | ADA | ECI1 | GAPDH | ||||||||
| ENO1 | PGAM1 | ADA | HADH | EIF5A1 | HEBP1 | EIF5A1 | EIF5A1 | |||||||
| ANXA1 | HEBP1 | PHB | NDUFB10 | SEPT2 | MYOZ1 | PHB | ||||||||
| ANXA2 | HADH | SEPT2 | PPIC | ANXA2 | NDUFB10 | ANXA2 | ANXA2 | |||||||
| ENO3 | MYOZ1 | LDHA | PRDX4 | ENO3 | PPIC | LDHA | ||||||||
| HAPLN1 | PRDX1 | PRDX1 | APOBEC1 | PRDX1 | PSMB2 | PRDX1 | ||||||||
| TPI1 | PSMA6 | PRDX3 | THBS1 | HAPLN1 | NDUFV2 | PRDX3 | ||||||||
| TTR | APOBEC1 | C21 | TPI1 | C21 | C21 | |||||||||
| COPE | CTHRC1 | ACTA2 | S100A11 | ACTA2 | ACTA2 | |||||||||
| COL2A1 | PRDX3 | EIF5A2 | TTR | EIF5A2 | ||||||||||
| SOD1 | SRL | ALDOC | C4 | |||||||||||
| ERP29 | TAGLN | ENO3 | ||||||||||||
| EPYC | TPI1 | |||||||||||||
NDV was not included in this table because the proteome changes of cells or tissues induced by NDV have not yet been reported.
Proteins identified in this study that induced by AIV H9 infection.
Proteins identified both in our study and previous reports induced by AIV infection.
Proteins identified in this study that induced by IBV ck/CH/LDL/97I and H120 infections.
Proteins identified both in our study and previous reports induced by IBV ck/CH/LDL/97I and H120 infections.
PSMA6: proteasomel subunit alpha type 6; SOD1: Cu/Zn superoxide dismutase.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
3.4. Transcriptional profiles of differentially expressed proteins
Transcriptional alteration analysis of ten selected genes at 3, 7, 12, and 21 dpi were achieved by quantifying the mRNA transcripts, in which the housekeeping gene 28S rRNA was used as an internal control (Fig. 5A). In this study, primers designed for transcriptional alteration analysis were located at positions across intron–exon boundaries to avoid the interference of genomic DNA, so quantitative comparison of total amount of mRNA transcripts of each gene was conducted. In general, the trends in the change in mRNA abundance of selected genes at four time points (Fig. 5A) were similar to the change patterns of their corresponding proteins showed by 2D‐DIGE (Fig. 5B and Supporting Information Table 2). And degrees of change in mRNA abundance of ANXA1, HSPB1, Cu/Zn superoxide dismutase 1, EIF5A2, LGALS1, and PRDX1 were more obvious than the abundance change of corresponding proteins showed by 2D‐DIGE. This is not unexpected because posttranscriptional mechanisms, including protein translation, PTM, and degradation may also contribute to the abundance change of these proteins during virus infection 28. These data provided transcriptional information complementary to the differentially expressed proteins revealed by proteomics analysis.
Figure 5.
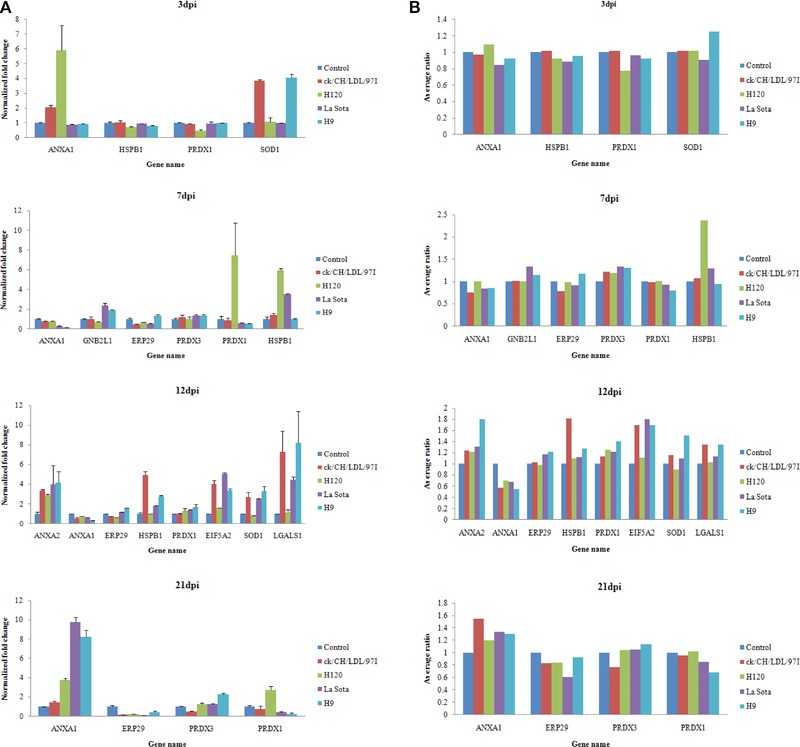
Transcript alteration of selected differentially expressed proteins in IBV ck/CH/LDL/97I, H120, NDV La Sota, and AIV H9 infected chicken. (A) Comparison of mRNA levels of selected proteins differentially expressed at 3, 7, 12, and 21 dpi. Total RNA extracts were prepared from the tracheal tissues of sacrificial birds in control and all virus‐infected groups at four time points (3, 7, 12, and 21 dpi), measured by real‐time RT‐PCR. Data represent means of three biological replicates per group. Error bars indicate standard error. Gene names refer to Supporting Information Table 6. Samples were normalized with the expression of the 28S ribosomal RNA gene in the control group. (B) Average ratios of selected proteins determined by 2D‐DIGE using DeCyder Software.
3.5. Validation of the differentially expressed proteins by Western blot analysis
To confirm the dynamic alterations of protein abundance during ck/CH/LDL/97I, H120, La Sota, and H9 infections, four protein candidates ANXA1, PRDX1, ANXA2, and HSPB1 were chosen for Western blot analysis with specific antibodies (Fig. 3A and B). The abundance changes of ANXA1 and PRDX1 (both altered at all four time points), ANXA2 (altered only at 12dpi) and HSPB1 (altered at 3, 7, and 12 dpi) were consistent with the expression changes showed by 2D‐DIGE (Fig. 5B and Supporting Information Table 2). Altogether, Western blot validated the results obtained from proteomics analyses during IBV, NDV, and AIV H9 infections.
4. Discussion
In this study, host responses against different avian respiratory viruses that result in distinct disease phenotypes were analyzed using comparative proteomics. IBV ck/CH/LDL/97I and AIV H9 are two kinds of pathogenic viruses for chicken causing significantly clinical signs, while, IBV H120 and NDV La Sota are avirulent vaccine strains inducing effective immune response. Our proteomic analyses of the alterations in protein expression induced by different viruses under the same physiological conditions provide more specific information of molecular pathogenesis and virus–host interactions of these different viruses. Although GO functional annotation and classification showed that all four viruses affect host global cellular processes similarly, the trends and degrees of alteration of protein expression were not identical. What apparent from the results is that pathogenic IBV ck/CH/LDL/97I and AIV H9 induced slightly more changes in protein expression than the vaccine strains IBV H120 and NDV La Sota in chicken trachea. In addition, the differences in protein expression changes induced by IBV ck/CH/LDL/97I, H120, NDV La Sota, and AIV H9 indicated that different interactions likely occurred in the host response to infection by these viruses and resulted in the different pathogenicity. This is the first report of comparative proteomics analysis performed to study the host response to infections of different avian respiratory viruses; the information obtained herein might provide critical clues for future studies.
4.1. Inflammatory responses in IBV, NDV, and AIV H9 infections
IBV, NDV, and AIV H9 subtype viruses can initially infect the chicken tracheal epithelium and elicit inflammatory responses associated with the disease 16, 29, 30. In this study, several proteins involved in inflammatory responses were identified, including ANXA1, PPIA, LGALS1, and PRDX4.
Our previous works have reported downregulation of the ANXA1 in IBV‐infected trachea 14, 15. In this study, the endogenous anti‐inflammatory protein ANXA1 31 was found downregulated in both pathogenic IBV ck/CH/LDL/97I and AIV H9 infected groups at 7dpi and in all virus‐infected groups at 12 dpi, suggesting that the decreased expression of ANXA1 may be associated with the inflammatory phenotype during the course of avian respiratory viruses infections. More notably, ANXA1 was identified in more than one spots in our study, suggesting that ANXA1 phosphorylation during IBV, NDV, and AIV H9 infections may associate with their pathogenesis 32. The PPIA has been shown to elicit inflammatory responses and inhibit CD4+ T‐cell activation through its interaction with Itk 33. We noticed that proinflammatory PPIA was only upregulated in pathogenic IBV CK/CH/LDL/97I and AIV H9 infected groups; there was no alteration of PPIA expression in trachea of chickens infected with IBV H120 and NDV La Sota. This difference may indicate that pathogenic IBV ck/CH/LDL/97I and AIV H9 trigger more severe inflammatory responses and immune suppression than IBV H120 and NDV La Sota vaccine strains that associate with their different pathogenetic processes. In addition, PPIA can facilitate replication of human immunodeficiency virus 1 (HIV‐1) 34 and coronaviruses including IBV 35. However, PPIA can inhibit the influenza A viral replication through enhancing the degradation of M1 protein 36. Chicken PPIA also showed anti‐influenza virus activity 37. Upregulated PPIA has been detected in human cells infected with AIV H9N2 11. Hence, we speculate that the upregulation of PPIA may contribute to the replication and pathogenesis of IBV ck/CH/LDL/97I and participate in host defense against AIV H9.
PRDX4 can protect pancreatic beta‐cells against injury by suppressing oxidative stress and inflammatory signaling activation 38. Galectin‐1 (LGALS1) is an important contributor to immune homeostasis and exhibits anti‐inflammatory activity during infection 39. We found that anti‐inflammatory PRDX4 and LGALS1 were generally upregulated in IBV ck/CH/LDL/97I, NDV La Sota, and AIV H9 infected groups at 12 dpi. In the case of avian respiratory viral infections, the inflammatory process diminished at the recovery stage 16. Hence, the upregulation of anti‐inflammatory PRDX4 and LGALS1 may be involved in tracheal epithelium recovery from inflammation damage induced by IBV ck/CH/LDL/97I, NDV La Sota, and AIV H9. Besides as an immunomodulator, galectin‐1 can function as pattern recognition receptor to facilitate pathogen recognition 39. The interaction of galectin‐1 with viral envelope glycoproteins can inhibit infectivity of AIV and Nipah virus 40, 41. Galectin‐1 promotes HIV‐1 infection through stabilization of viral adsorption 42. Moreover, upregulation of galectin‐1 was also found in human cells infected with AIV H9N2 11, suggesting that galectin‐1 may participate in innate immune response to AIV H9 infection. Our results also indicate that galectin‐1 potentially involved in IBV ck/CH/LDL/97I and NDV La Sota infections, whether galectin‐1 binds to the surface of IBV and NDV, and functions as effective pattern recognition receptor or promotes infection by facilitating viral adsorption need to be further elucidated.
4.2. IBV, NDV, and AIV H9 may induce alterations of nuclear structure and hijack host translational machinery
Nucleus‐associated proteins, including nuclear mitotic apparatus protein (NUMA), LMNA, and EIF5A1, 2, were identified in the present study. Though the primary site of replication of IBV, NDV, and AIV is the cytoplasm, IBV nucleocapsid protein, NDV matrix protein, and AIV nucleoprotein can localize to the nucleolus 43, 44, 45. In this study, LMNA was universally downregulated in all virus‐infected groups and NUMA was downregulated in IBV ck/CH/LDL/97I, H120‐infected and AIV H9 infected groups. NUMA and LMNA are components of nuclear matrix and nuclear lamina, downregulations of NUMA and LMNA might implicate that IBV, NDV, and AIV H9 infections commonly result in disruption of nuclear architecture due to nuclear entry and accumulation of viral proteins in the nucleolus. Downregulation of LMNA has also been detected in influenza virus‐infected cells 10 and IBV‐infected trachea 15.
As well known that virus may hijack host translational machinery by regulating distinct eIFs 46, 47, 48, 49. Previous reports showed EIF5A can regulate translation and replication of HIV‐1 48, and EIF5A mutants inhibit HIV‐1 replication 49. In this study, EIF5A1 was downregulated in IBV H120, NDV La Sota and AIV H9 infected groups, while EIF5A2 was upregulated in IBV ck/CH/LDL/97I, NDV La Sota, and AIV H9 infected groups. We speculate that IBV, NDV, and AIV H9 may interfere with the host protein synthesis by downregulating constitutively expressed EIF5A1. While the upregulation of EIF5A2 facilitates viral mRNAs translation in infected cells; the expression of EIF5A2 is extremely low in normal conditions. Furthermore, the differences in expression of EIF5A1 and EIF5A2 indicate that the two IBV strains differentially hijack host translational apparatus for their own benefits. Our results provide additional evidences that RNA viruses can interact with the nucleolus to hijack cellular functions and recruit nucleolus proteins to facilitate virus replication.
4.3. IBV, NDV, and AIV H9 manipulate host UPP through different strategies
The ubiquitin‐proteasome pathway (UPP), a cellular ubiquitin‐dependent protein degradation system, has been implicated in viral infection cycle and virus–host interactions 50. Inhibition of the UPP has been reported to affect the replication of several viruses, including influenza virus 51 and coronavirus 52. Moreover, the interaction of HIV‐1 Tat protein with proteasomal β subunits contributes to the immune escape of HIV‐1 53. However, the potential roles of host UPP during IBV and NDV infections and precise interaction mechanisms between UPP and AIV are not well elucidated. Previous reports showed that proteasome subunit alpha and beta types were upregulated in IBV‐infected DF‐1 cells 12. In this study, proteasomel subunit alpha type 6 was upregulated only in AIV H9 infected group, whereas proteasomal subunit beta type 2 was commonly upregulated in IBV H120 and NDV La Sota infected groups and more abundant in H120‐infected group than ck/CH/LDL/97I‐infected group, indicating that different viruses (IBV, NDV, and AIV) and different strains of the same virus (ck/CH/LDL/97I and H120) may evolve different strategies to utilize UPP to facilitate replication and infection by specific interaction with distinct proteasomal subunits.
4.4. Different roles of ANXA2 in IBV, NDV, and AIV H9 infections
ANXA2 has been shown involved in exocytosis, cell proliferation, membrane trafficking, and apoptosis 54, the organized ANXA2 membrane domains are necessary for virion budding from infected cells 55. Notably, many viruses utilize ANXA2 for effective infection and replication, such as respiratory syncytial virus (RSV) 56, HIV 57, and hepatitis C virus (HCV) 58. In this study, ANXA2 was commonly upregulated during IBV H120, NDV La Sota, and AIV H9 infections. We speculate that the upregulated ANXA2 may support replication of AIV H9 by incorporating influenza virus particles to enhance influenza virus replication via mediated activation of plasminogen 59. However, upregulated ANXA2 might act as an anti‐viral protein to inhibit replication of IBV H120 as it was reported that ANXA2 reduces viral frameshifting efficiency and successful replication of IBV as a cellular RNA‐binding protein 54. Upregulation of ANXA2 has also been found in avirulent IBV‐infected DF‐1 cells 12. In our previous work, the abundance of ANXA2 was only increased following attenuated IBV ck/CH/LDL/97I P115 infection, no significantly abundance changes of ANXA2 were observed during virulent IBV ck/CH/LDL/97I P5 infection, suggesting that the ANXA2 may be specific host response to avirulent IBV strains (ck/CH/LDL/97I P115 and H120). The function of ANXA2 in life cycle of NDV needs further investigation.
4.5. IBV, NDV, and AIV H9 infections and stress response proteins
HSPB1 is a multifunctional protein involved in various cellular processes and viral infection 60. Phosphorylated HSPB1 has been shown to participate in the replication of adenovirus 60, HCV 61, and herpes simplex virus type 1 62. In this study, the upregulation of phosphorylated HSPB1 during IBV, NDV, and AIV H9 infections was confirmed by Western bolt (Fig. 3). It was reported that RSV‐induced phosphorylation of HSPB1 in human bronchial epithelial cells ultimately results in epithelial membrane permeability 63. Avian respiratory viruses infections can result in tracheal epithelium lesion, but the severity and persistence of the lesions differed among different viruses 16, 17, 18. Therefore, phosphorylated HSPB1 that mainly upregulated in IBV H120 and NDV La Sota infected groups at 7 dpi and in IBV ck/CH/LDL/97I and AIV H9 infected groups at 12 dpi may be responsible for the tracheal epithelium lesions at different stages of avian respiratory viruses infections. Further studies should focus on the exact roles of phosphorylated HSPB1 in the replication and pathogenesis of IBV, NDV, and AIV H9.
Viral infections are capable of induce oxidative stress associated with the activation of phagocytosis and release of ROS that positively modulate immune activation, eliminate viral infection, and immune‐induced cellular injury 64. In this study, antioxidant stress proteins including PRDX1, 3, 4, 6; GST; and Cu/Zn superoxide dismutase 1 (Table 2 and Supporting Information Table 2), were universally altered in all viruses‐infected groups. Most of altered proteins were upregulated, which may be related to relief of cellular oxidative stress induced by IBV, NDV, and AIV H9 infections. Moreover, more extensive changes were observed in AIV H9 infected group, suggesting that AIV H9 may activate more intense oxidative stress. Among these antioxidant stress proteins, PRDX1 and PRDX4 have been reported to protect RSV‐induced oxidative damage to the nuclear cytoskeletal proteins in epithelial cells 65, while downregulation of antioxidant enzymes (SOD, GST, and PRDX) induced by RSV contributes to RSV pathogenesis 66. In addition, PRDX1 can activate secretion of proinflammatory cytokines by interacting with TLR4 67. PRDX4 plays critical protective roles in inflammatory diseases 38. Similarly, we considered that the upregulation of PRDX1 and PRDX4 during IBV, NDV, and AIV H9 infections may activate host antiviral immunity to eliminate viral infection and provide protective cellular mechanisms to reduce oxidative stress and inflammatory damage. However, the potential roles of downregulated oxidative stress‐associated proteins in the pathogenesis of IBV, NDV, and AIV needed to be investigated further.
In this study, trachea was chosen for global analysis of the host response because it is the primary target organ of avian respiratory viruses. The comparative analysis of changes in tracheal protein profile induced by IBV, NDV, and AIV H9 in vivo provides insight into mechanisms of global host response to different avian respiratory virus infections. However, the sites of IBV, NDV, and AIV infection are actually not completely consistent, IBV replicates in the epithelium of lower respiratory tract of chicken 68, NDV and AIV H9 subtypes induce lesions in the upper respiratory tract 18, 69. The roles of different sites of respiratory tract in this study had not been effectively measured. Moreover, infections of IBV, NDV, and AIV H9 subtype viruses can elicit inflammatory responses characterized by recruitment of myeloid or lymphoid cells. Consequently, the cellular types of tracheal tissues harvested from mock and virus‐infected chickens for proteomics analysis may differ due to immune cell infiltration. As a result, the changes revealed by proteomic analysis of some proteins, such as LGALS1, PPIA, and PRDX1, that could also be expressed in immune cells in the course of viral infection may be caused or partly caused by cellular heterogeneity occurred during the inflammatory response to viral infection (including infiltration by innate and adaptive immune cells). This needed to be further investigated. In addition, though a number of differentially expressed proteins that have potential roles in revealing the common or specific aspects of viral pathogenesis and virus–host interactions were identified, the precise roles of these proteins during viral infection had not proved in this study. Nevertheless, the similar and different protein expression profiles induced by different avian respiratory viruses after infection might be indicators of the essential response of infected target cells, provide insights into viral pathogenesis and host antiviral defense, and supply useful clues for the development of novel prevention or therapeutics strategies against these viruses.
The authors have declared no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Table S1. Experimental design for fluorescent dye labeling.
Table S2. Summary of average ratio of differentially expressed proteins in IBV (ck/CH/LDL/97I, H120)‐, NDV (La Sota)‐ and AIV (H9)‐infected groups and between IBV ck/CH/LDL/97I‐ and H120‐infected groups.
Table S3. The primers for Real‐time RT‐PCR.
Table S4. Serological results post‐inoculation with IBV (ck/CH/LDL/97I, H120), NDV (La Sota), and AIV (H9).
Table S5. Identification information of differentially expressed proteins in tracheal tissues of chickens infected with IBV (ck/CH/LDL/97I, H120), NDV (La Sota), and AIV (H9) identified by MALDI‐TOF/TOF MS.
Table S6. Biological function annotation of differentially expressed proteins.
Table S7. Summary of up and down‐regulated proteins during IBV (ck/CH/LDL/97I, H120), NDV (La Sota) and AIV (H9) infection, and proteins differentially expressed between IBV ck/CH/LDL/97I‐ and H120‐infected groups.
Figure S1. Representative 2D‐DIGE images of IBV (ck/CH/LDL/97I, H120)‐, NDV (La Sota)‐, and AIV (H9)‐infected groups with their respective sampling points.
Figure S2. This file includes the PMF spectrum and confirmed MALDI‐TOF/TOF MS spectrum of differentially expressed proteins.
Acknowledgments
This work was supported by grants from the China Agriculture Research Systerm (no. CARS‐41‐K12) and Special Fund for Agro‐scientific Research in the Public Interest (no. 201203056).
Colour Online: See the article online to view Figs. 2–5 in colour.
5 References
- 1. Rout, S. N. , Samal, S. K. , The large polymerase protein is associated with the virulence of Newcastle disease virus. J. Virol. 2008, 82, 7828–7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Peeters, B. , Gruijthuijsen, Y. , De Leeuw, O. , Gielkens, A. , Genome replication of Newcastle disease virus: involvement of the rule‐of‐six. Arch. Virol. 2000, 145, 1829–1845. [DOI] [PubMed] [Google Scholar]
- 3. Sun, Y. , Qin, K. , Wang, J. , Pu, J. et al., High genetic compatibility and increased pathogenicity of reassortants derived from avian H9N2 and pandemic H1N1/2009 influenza viruses. Proc. Natl. Acad. Sci. USA 2011, 108, 4164–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ben Shabat, M. , Meir, R. , Haddas, R. , Lapin, E. et al., Development of a real‐time TaqMan RT‐PCR assay for the detection of H9N2 avian influenza viruses. J. Virol. Methods 2010, 168, 72–77. [DOI] [PubMed] [Google Scholar]
- 5. Carstens, E. , Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2009). Arch. Virol. 2010, 155, 133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Han, Z. , Sun, C. , Yan, B. , Zhang, X. et al., A 15‐year analysis of molecular epidemiology of avian infectious bronchitis coronavirus in China. Infect. Genet. Evol. 2011, 11, 190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cook, J. K. A. , Jackwood, M. , Jones, R. , The long view: 40 years of infectious bronchitis research. Avian Pathol. 2012, 41, 239–250. [DOI] [PubMed] [Google Scholar]
- 8. Zou, W. , Ke, J. , Zhang, A. , Zhou, M. et al., Proteomics analysis of differential expression of chicken brain tissue proteins in response to the neurovirulent H5N1 avian influenza virus infection. J. Proteome Res. 2010, 9, 3789–3798. [DOI] [PubMed] [Google Scholar]
- 9. Zhao, D. , Liang, L. , Li, Y. , Liu, L. et al., Proteomic analysis of the lungs of mice infected with different pathotypes of H5N1 avian influenza viruses. Proteomics 2012, 12, 1970–1982. [DOI] [PubMed] [Google Scholar]
- 10. Coombs, K. M. , Berard, A. , Xu, W. , Krokhin, O. et al., Quantitative proteomic analyses of influenza virus‐infected cultured human lung cells. J. Virol. 2010, 84, 10888–10906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu, N. , Song, W. , Wang, P. , Lee, K. et al., Proteomics analysis of differential expression of cellular proteins in response to avian H9N2 virus infection in human cells. Proteomics 2008, 8, 1851–1858. [DOI] [PubMed] [Google Scholar]
- 12. Emmott, E. , Smith, C. , Emmett, S. R. , Dove, B. K. , Hiscox, J. A. , Elucidation of the avian nucleolar proteome by quantitative proteomics using SILAC and changes in cells infected with the coronavirus infectious bronchitis virus. Proteomics 2010, 10, 3558–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Emmott, E. , Rodgers, M. A. , Macdonald, A. , McCrory, S. et al., Quantitative proteomics using stable isotope labeling with amino acids in cell culture reveals changes in the cytoplasmic, nuclear, and nucleolar proteomes in vero cells infected with the coronavirus infectious bronchitis virus. Mol. Cell. Proteomics 2010, 9, 1920–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cao, Z. , Han, Z. , Shao, Y. , Geng, H. et al., Proteomic analysis of chicken embryonic trachea and kidney tissues after infection in ovo by avian infectious bronchitis coronavirus. Proteome Sci. 2011, 9, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cao, Z. , Han, Z. , Shao, Y. , Liu, X. et al., Proteomics analysis of differentially expressed proteins in chicken trachea and kidney after infection with the highly virulent and attenuated coronavirus infectious bronchitis virus in vivo. Proteome Sci. 2012, 10, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Benyeda, Z. , Szeredi, L. , Mato, T. , Süveges, T. et al., Comparative histopathology and immunohistochemistry of QX‐like, Massachusetts and 793/B serotypes of infectious bronchitis virus infection in chickens. J. Comp. Pathol. 2010, 143, 276–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brown, C. C. , Sullivan, L. , Dufour‐Zavala, L. , Kulkarni, A. et al., Comparing presence of avian paramyxovirus‐1 through immunohistochemistry in tracheas of experimentally and naturally infected chickens. Avian Dis. 2012, 57, 36–40. [DOI] [PubMed] [Google Scholar]
- 18. Nili, H. , Asasi, K. , Natural cases and an experimental study of H9N2 avian influenza in commercial broiler chickens of Iran. Avian Pathol. 2002, 31, 247–252. [DOI] [PubMed] [Google Scholar]
- 19. Liu, S. , Han, Z. , Chen, J. , Liu, X. et al., S1 gene sequence heterogeneity of a pathogenic infectious bronchitis virus strain and its embryo‐passaged, attenuated derivatives. Avian Pathol. 2007, 36, 231–234. [DOI] [PubMed] [Google Scholar]
- 20. Han, Z. , Zhao, F. , Shao, Y. , Liu, X. et al., Fine level epitope mapping and conservation analysis of two novel linear B‐cell epitopes of the avian infectious bronchitis coronavirus nucleocapsid protein. Virus Res. 2013, 171, 54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu, S. , Kong, X. , A new genotype of nephropathogenic infectious bronchitis virus circulating in vaccinated and non‐vaccinated flocks in China. Avian Pathol. 2004, 33, 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Majiyagbe, K. , Hitchner, S. , Antibody response to strain combinations of Newcastle disease virus as measured by hemagglutination‐inhibition. Avian Dis. 1977, 576–584. [PubMed] [Google Scholar]
- 23. Reed, L. J. , Muench, H. , A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar]
- 24. Dortmans, J. , Koch, G. , Rottier, P. , Peeters, B. , A comparative infection study of pigeon and avian paramyxovirus type 1 viruses in pigeons: evaluation of clinical signs, virus shedding and seroconversion. Avian Pathol. 2011, 40, 125–130. [DOI] [PubMed] [Google Scholar]
- 25. Jones, R. , Ellis, R. , Cox, W. , Errington, J. et al., Development and validation of RT‐PCR tests for the detection and S1 genotyping of infectious bronchitis virus and other closely related gammacoronaviruses within clinical samples. Transbound. Emerg. Dis. 2011, 58, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wise, M. G. , Suarez, D. L. , Seal, B. S. , Pedersen, J. C. et al., Development of a real‐time reverse‐transcription PCR for detection of newcastle disease virus RNA in clinical samples. J. Clin. Microbiol. 2004, 42, 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu, X. , Ma, H. , Xu, Q. , Sun, N. et al., Characterization of a recombinant coronavirus infectious bronchitis virus with distinct S1 subunits of spike and nucleocapsid genes and a 3'untranslated region. Vet. Microbiol. 2013, 162, 429–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen, G. , Gharib, T. G. , Huang, C.‐C. , Taylor, J. M. et al., Discordant protein and mRNA expression in lung adenocarcinomas. Mol. Cell. Proteomics 2002, 1, 304–313. [DOI] [PubMed] [Google Scholar]
- 29. Seal, B. , King, D. , Sellers, H. , The avian response to Newcastle disease virus. Dev. Comp. Immunol. 2000, 24, 257–268. [DOI] [PubMed] [Google Scholar]
- 30. Julkunen, I. , Melén, K. , Nyqvist, M. , Pirhonen, J. et al., Inflammatory responses in influenza A virus infection. Vaccine 2000, 19, S32–S37. [DOI] [PubMed] [Google Scholar]
- 31. McArthur, S. , Cristante, E. , Paterno, M. , Christian, H. et al., Annexin A1: a central player in the anti‐inflammatory and neuroprotective role of microglia. J. Immunol. 2010, 185, 6317–6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. D'Acunto, C. W. , Gbelcova, H. , Festa, M. , Rum, T. , The complex understanding of Annexin A1 phosphorylation. Cell. Signal. 2013, 26, 173–178. [DOI] [PubMed] [Google Scholar]
- 33. Brazin, K. N. , Mallis, R. J. , Fulton, D. B. , Andreotti, A. H. , Regulation of the tyrosine kinase Itk by the peptidyl‐prolyl isomerase cyclophilin A. Proc. Natl. Acad. Sci. USA 2002, 99, 1899–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sokolskaja, E. , Sayah, D. M. , Luban, J. , Target cell cyclophilin A modulates human immunodeficiency virus type 1 infectivity. J. Virol. 2004, 78, 12800–12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pfefferle, S. , Schöpf, J. , Kögl, M. , Friedel, C. C. et al., The SARS‐coronavirus‐host interactome: identification of cyclophilins as target for pan‐coronavirus inhibitors. PLoS Pathog. 2011, 7, e1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu, X. , Zhao, Z. , Xu, C. , Sun, L. et al., Cyclophilin A restricts influenza A virus replication through degradation of the M1 protein. PLoS ONE 2012, 7, e31063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xu, C. , Meng, S. , Liu, X. , Sun, L. , Liu, W. , Chicken cyclophilin A is an inhibitory factor to influenza virus replication. Virol. J. 2010, 7, 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yamada, S. , Ding, Y. , Sasaguri, Y. , Peroxiredoxin 4: critical roles in inflammatory diseases. J. Uoeh. 2012, 34, 27–39. [DOI] [PubMed] [Google Scholar]
- 39. Vasta, G. R. , Roles of galectins in infection. Nat. Rev. Microbiol. 2009, 7, 424–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Garner, O. B. , Aguilar, H. C. , Fulcher, J. A. , Levroney, E. L. et al., Endothelial galectin‐1 binds to specific glycans on Nipah virus fusion protein and inhibits maturation, mobility, and function to block syncytia formation. PLoS Pathog. 2010, 6, e1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang, M. L. , Chen, Y. H. , Wang, S. W. , Huang, Y. J. et al., Galectin‐1 binds to influenza virus and ameliorates influenza virus pathogenesis. J. Virol. 2011, 85, 10010–10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. St‐Pierre, C. , Manya, H. , Ouellet, M. , Clark, G. F. et al., Host‐soluble galectin‐1 promotes HIV‐1 replication through a direct interaction with glycans of viral gp120 and host CD4. J. Virol. 2011, 85, 11742–11751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hiscox, J. A. , Wurm, T. , Wilson, L. , Britton, P. et al., The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J. Virol. 2001, 75, 506–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Peeples, M. E. , Wang, C. , Gupta, K. C. , Coleman, N. , Nuclear entry and nucleolar localization of the Newcastle disease virus (NDV) matrix protein occur early in infection and do not require other NDV proteins. J. Virol. 1992, 66, 3263–3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Neumann, G. , Castrucci, M. R. , Kawaoka, Y. , Nuclear import and export of influenza virus nucleoprotein. J. Virol. 1997, 71, 9690–9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xiao, H. , Xu, L. , Yamada, Y. , Liu, D. , Coronavirus spike protein inhibits host cell translation by interaction with eIF3f. PLoS ONE 2008, 3, e1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang, X. , Liao, Y. , Yap, P. L. , Png, K. J. et al., Inhibition of protein kinase R activation and upregulation of GADD34 expression play a synergistic role in facilitating coronavirus replication by maintaining de novo protein synthesis in virus‐infected cells. J. Virol. 2009, 83, 12462–12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu, J. , Henao‐Mejia, J. , Liu, H. , Zhao, Y. , He, J. J. , Translational regulation of HIV‐1 replication by HIV‐1 Rev cellular cofactors Sam68, eIF5A, hRIP, and DDX3. J. Neuroimmune Pharmacol. 2011, 6, 308–321. [DOI] [PubMed] [Google Scholar]
- 49. Bevec, D. , Jaksche, H. , Oft, M. , Wöhl, T. et al., Inhibition of HIV‐1 replication in lymphocytes by mutants of the Rev cofactor eIF‐5A. Science 1996, 271, 1858–1860. [DOI] [PubMed] [Google Scholar]
- 50. Gao, G. G. G. , Luo, H. L. H. , The ubiquitin‐proteasome pathway in viral infections. Can. J. Physiol. Pharmacol. 2006, 84, 5–14. [DOI] [PubMed] [Google Scholar]
- 51. Widjaja, I. , de Vries, E. , Tscherne, D. M. , García‐Sastre, A. et al., Inhibition of the ubiquitin‐proteasome system affects influenza A virus infection at a postfusion step. J. Virol. 2010, 84, 9625–9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Raaben, M. , Posthuma, C. C. , Verheije, M. H. , te Lintelo, E. G. et al., The ubiquitin‐proteasome system plays an important role during various stages of the coronavirus infection cycle. J. Virol. 2010, 84, 7869–7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Apcher, G. S. , Heink, S. , Zantopf, D. , Kloetzel, P. M. et al., Human immunodeficiency virus‐1 Tat protein interacts with distinct proteasomal alpha and beta subunits. FEBS Lett. 2003, 553, 200–204. [DOI] [PubMed] [Google Scholar]
- 54. Kwak, H. , Park, M. W. , Jeong, S. , Annexin A2 binds RNA and reduces the frameshifting efficiency of infectious bronchitis virus. PLoS ONE 2011, 6, e24067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gerke, V. , Moss, S. E. , Annexins: from structure to function. Physiol. Rev. 2002, 82, 331–371. [DOI] [PubMed] [Google Scholar]
- 56. Malhotra, R. , Ward, M. , Bright, H. , Priest, R. et al., Isolation and characterisation of potential respiratory syncytial virus receptor (s) on epithelial cells. Microbes. Infect. 2003, 5, 123–133. [DOI] [PubMed] [Google Scholar]
- 57. Ryzhova, E. V. , Vos, R. M. , Albright, A. V. , Harrist, A. V. et al., Annexin 2: a novel human immunodeficiency virus type 1 Gag binding protein involved in replication in monocyte‐derived macrophages. J. Virol. 2006, 80, 2694–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Backes, P. , Quinkert, D. , Reiss, S. , Binder, M. et al., Role of annexin A2 in the production of infectious hepatitis C virus particles. J. Virol. 2010, 84, 5775–5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. LeBouder, F. , Morello, E. , Rimmelzwaan, G. F. , Bosse, F. et al., Annexin II incorporated into influenza virus particles supports virus replication by converting plasminogen into plasmin. J. Virol. 2008, 82, 6820–6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kostenko, S. , Moens, U. , Heat shock protein 27 phosphorylation: kinases, phosphatases, functions and pathology. Cell. Mol. Life Sci. 2009, 66, 3289–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Choi, Y. W. , Tan, Y. J. , Lim, S. G. , Hong, W. , Goh, P. Y. , Proteomic approach identifies HSP27 as an interacting partner of the hepatitis C virus NS5A protein. Biochem. Bioph. Res. Co. 2004, 318, 514–519. [DOI] [PubMed] [Google Scholar]
- 62. Mathew, S. S. , Della Selva, M. P. , Burch, A. D. , Modification and reorganization of the cytoprotective cellular chaperone Hsp27 during herpes simplex virus type 1 infection. J. Virol. 2009, 83, 9304–9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Singh, D. , McCann, K. L. , Imani, F. , MAPK and heat shock protein 27 activation are associated with respiratory syncytial virus induction of human bronchial epithelial monolayer disruption. Am. J. Physiol. Lung. Cell. Mol. Physiol. 2007, 293, L436–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schwarz, K. B. , Oxidative stress during viral infection: a review. Free Radic. Bio. Med. 1996, 21, 641–649. [DOI] [PubMed] [Google Scholar]
- 65. Jamaluddin, M. , Wiktorowicz, J. E. , Soman, K. V. , Boldogh, I. et al., Role of peroxiredoxin 1 and peroxiredoxin 4 in protection of respiratory syncytial virus‐induced cysteinyl oxidation of nuclear cytoskeletal proteins. J. Virol. 2010, 84, 9533–9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hosakote, Y. M. , Jantzi, P. D. , Esham, D. L. , Spratt, H. et al., Viral‐mediated inhibition of antioxidant enzymes contributes to the pathogenesis of severe respiratory syncytial virus bronchiolitis. Am. J. Resp. Crit. Care. 2011, 183, 1550–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Riddell, J. R. , Wang, X. Y. , Minderman, H. , Gollnick, S. O. , Peroxiredoxin 1 stimulates secretion of proinflammatory cytokines by binding to TLR4. J. Immunol. 2010, 184, 1022–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cavanagh, D. , Coronavirus avian infectious bronchitis virus. Vet. Res. 2007, 38, 281–297. [DOI] [PubMed] [Google Scholar]
- 69. Bang, F. , Foard, M. , Bang, B. , Acute Newcastle viral infection of the upper respiratory tract of the chicken: I. A model for the study of environmental factors on upper respiratory tract infection. Am. J. Pathol. 1974, 76, 333–348. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Table S1. Experimental design for fluorescent dye labeling.
Table S2. Summary of average ratio of differentially expressed proteins in IBV (ck/CH/LDL/97I, H120)‐, NDV (La Sota)‐ and AIV (H9)‐infected groups and between IBV ck/CH/LDL/97I‐ and H120‐infected groups.
Table S3. The primers for Real‐time RT‐PCR.
Table S4. Serological results post‐inoculation with IBV (ck/CH/LDL/97I, H120), NDV (La Sota), and AIV (H9).
Table S5. Identification information of differentially expressed proteins in tracheal tissues of chickens infected with IBV (ck/CH/LDL/97I, H120), NDV (La Sota), and AIV (H9) identified by MALDI‐TOF/TOF MS.
Table S6. Biological function annotation of differentially expressed proteins.
Table S7. Summary of up and down‐regulated proteins during IBV (ck/CH/LDL/97I, H120), NDV (La Sota) and AIV (H9) infection, and proteins differentially expressed between IBV ck/CH/LDL/97I‐ and H120‐infected groups.
Figure S1. Representative 2D‐DIGE images of IBV (ck/CH/LDL/97I, H120)‐, NDV (La Sota)‐, and AIV (H9)‐infected groups with their respective sampling points.
Figure S2. This file includes the PMF spectrum and confirmed MALDI‐TOF/TOF MS spectrum of differentially expressed proteins.


