Abstract
Laboratory diagnosis is important to distinguish influenza from other respiratory virus infections. It will be especially important in detecting the first cases of pandemic influenza.
Good quality respiratory tract sampling is needed to maximise diagnostic yield in influenza infection.
In the appropriate clinical setting, pandemic strain‐specific nucleic acid testing is the initial test of choice for suspected pandemic influenza. It is more sensitive than virus isolation, and more sensitive and specific than serology, immunofluorescence and other antigen detection methods.
Virus isolation is needed to monitor new influenza strains and for vaccine development. Analysis of influenza isolates is undertaken by the World Health Organization Global Influenza Surveillance Network.
Monitoring for antiviral resistance will be needed with widespread use of neuraminidase inhibitors for treatment and prophylaxis during a pandemic.
Keywords: Infectious diseases
Laboratory diagnosis is important in managing influenza virus infection, either in the context of the annual winter outbreaks or in a pandemic. 1 Rapid and accurate influenza diagnosis improves medical management by allowing timely provision of antiviral therapy and prophylaxis, implementation of appropriate infection control strategies for individuals and public health responses to outbreaks, and limitation of unnecessary investigations or antibiotic therapy. 2
In the context of a threatened pandemic (whether due to the highly pathogenic influenza A/H5N1 virus currently causing disease among birds and some human contacts, or another novel virus), rapid laboratory detection of the first cases or clusters in Australia will be crucial. Australia's early response will hinge on containment and production of a vaccine to the pandemic strain. These responses depend on highly sensitive strain‐specific labor‐atory testing that will exclude other respiratory pathogens. Should a pandemic spread around the world with the same impact as the 1918 pandemic, maintaining health care and other essential services will be paramount. To make treatment decisions in this situation, less reliance may be placed on immediate laboratory confirmation and more on the clinical diagnosis. Nevertheless, laboratory detection will still be required for atypical and serious cases, for situations where there are special public health concerns, and to determine whether circulating strains are still sensitive to antiviral drugs or whether they are undergoing antigenic drift.
Influenza virus infection cannot be reliably diagnosed on clinical features alone. 3 The clinical picture may be difficult to distinguish from infections with respiratory syncytial virus, parainfluenza viruses, adenoviruses, coronaviruses and meta‐pneumovirus, among others. Rational application of antiviral chemotherapy, public health responses, and hospital or community resources requires accurate and timely diagnosis. The laboratory is also needed to diagnose some of the complications of influenza, such as secondary bacterial pneumonia due to Streptococcus pneumoniae, Staphylococcus aureus and Haemophilus influenzae.
Influenza virus surveillance is needed to monitor both antigenic shift in the haemagglutinin and neuraminidase glycoproteins leading to pandemic strains, and antigenic drift causing the annual variation in human influenza strains. Worldwide, this laboratory surveillance is undertaken by the World Health Organization Global Influenza Surveillance Network, which consists of four Collaborating Centres and some 110 National Influenza Centres (NICs). Australia is well served by this system, with a WHO Collaborating Centre in Melbourne, and three NICs, in Sydney (Institute of Clinical Pathology and Medical Research), Melbourne (Victorian Infectious Diseases Reference Laboratory) and Perth (PathWest Laboratory Medicine WA). These and other laboratories, including those in the Public Health Laboratory Network of Australasia and in the Asia‐Pacific region, contribute influenza strains to the WHO Collaborating Centre in Melbourne, which definitively characterises circulating influenza virus strains, collates and disseminates data on regional and global influenza epidemiology, evaluates drug resistance, distributes reagents for influenza diagnosis and identification, and develops laboratory techniques. 4 Box 1 shows the winter peaks of influenza virus detection over the past 2.5 years in Australia.
Box 1.
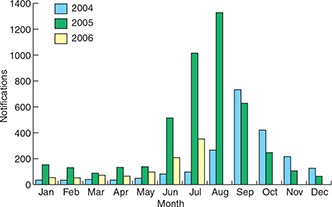
National Notifiable Diseases Surveillance System records of laboratory detection of influenza viruses from 2004 to mid 2006
Laboratory diagnosis
A definitive diagnosis of seasonal human influenza A or B is made by isolating the virus or detecting it by properly validated antigen detection or nucleic acid testing methods. A presumptive diagnosis can be made by a validated rapid antigen or “point‐of‐care” test. 1 Serologically, rising antibody levels between acute and convalescent serum samples confirm recent infection, and a high antibody level in serum during convalescence usually indicates recent infection, provided there was a consistent clinical illness in the context of community influenza activity.
Similar criteria apply to any potential pandemic influenza infection, although pandemic virus‐specific nucleic acid testing methods (and virus isolation) are the tests of choice, at least in the initial stages of a pandemic.
At a minimum, clinical microbiology laboratories should offer a validated rapid diagnostic assay, such as immunofluoresence or nucleic acid testing, during the influenza season. Wherever poss‐ible, conventional cell culture should be performed to obtain influenza virus isolates for characterisation. The relative advantages and disadvantages of diagnostic techniques for influenza are summarised in Box 2. Box 3 lists possible testing approaches for a potential pandemic strain, such as avian H5N1. The pathology guidelines in the Australian health management plan for pandemic influenza discuss various issues related to laboratory diagnosis, including specimen collection and transport (Box 4), and testing methods and strategies.
Box 2.
Comparison of diagnostic techniques for human influenza virus infection
| Test | Sensitivity | Turnaround time | Advantages | Disadvantages |
|---|---|---|---|---|
| Conventional cell culture | About 100% (less than RT‐PCR) | At least 4–5 days |
|
|
| Rapid cell culture (shell vial with IF) |
56%–100% (generally 70%–90%) |
1–4 days |
|
|
| IF for rapid antigen detection |
60%–100% (generally 70%–90%) |
2–4 hours |
|
|
| Nucleic acid testing (RT‐PCR) | About 100% (greater than cell culture) | < 1 day |
|
|
| Rapid antigen (point‐of‐care) tests |
59%–93% (generally about 70%) |
15–30 minutes |
|
|
| Serology (CF, HAI, IF, neutralisation, EIA) | Up to 100% | 1–3 weeks |
|
|
RT‐PCR = reverse transcription polymerase chain reaction. PC3 = Physical Containment level 3. IF = immunofluorescence. CF = complement fixation. HAI = haemagglutination inhibition assay. EIA = enzyme immunoassay.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Box 3 Laboratory methods to diagnose pandemic influenza A infection
A. Pandemic strain‐specific assays
Nucleic acid testing
Pandemic strain‐specific primers (eg, H5)
Quantitation not routinely available
Virus isolation
Limited to laboratories with PC3 facilities and virus culture expertise
Used for vaccine strain determination and genotyping
B. Non‐pandemic strain‐specific assays
Nucleic acid testing
Nucleic acid testing that detects all influenza A/H subtypes (eg, using nucleoprotein or matrix primers) or seasonal human influenza (primers for A/H3N2 and A/H1N1, B)
Nucleic acid testing for other respiratory pathogens
Antigen detection assays
Immunofluorescence that detects all influenza A (or human A/H3 and A/H1) or B
Immunochromatographic (point‐of‐care) tests that detect all influenza A and/or B
Limited experience with A/H5‐specific rapid antigen assays
Serology
Assays to detect recent influenza A or B infection are in use, but A/H5‐specific assays are not routinely available
Antiviral drug resistance testing
Genotypic and phenotypic assays currently limited to the World Health Organization Collaborating Centre and research facilities
PC3 = Physical Containment level 3.
Box 4 Collection and transport of samples
The key to successful patient and community management of influenza is the collection of good quality respiratory tract samples for laboratory testing.
Samples should be collected early in the clinical illness (within the first 96 hours, during maximal viral shedding), transported to the laboratory at 4°C for virus isolation, or room temperature for other assays, and processed as rapidly as possible.
Combined nose (one collected deeply from each nostril) and throat swabs are the most practical samples to collect from adults. Nasopharyngeal aspirates are the sample of choice from children younger than 3 years, provided they can be collected safely.
Collection of respiratory tract specimens
Recovery of virus from nasopharyngeal aspirates, nasal washes, and bronchoalveolar lavages is superior to that from nasopharyngeal and throat swabs and expectorated sputum, 5 ‐ 7 as the latter generally contain less columnar and more squamous epithelial cells. Single‐use swabs containing viral transport media are readily available commercially. Specimen requirements for antigen and nucleic acid testing are similar to those for virus isolation.
Early experience with human H5N1 infections suggests that virus is more readily detectable in lower respiratory tract samples. Upper respiratory tract samples are easier to collect, with the rate and load of detectable H5N1 RNA higher in samples collected from the pharynx (and the trachea) than the nose. Virus has been detected in serum, faeces and cerebrospinal fluid, reflecting the systemic nature of H5N1 infection, with viraemia associated with increased mortality. 8
Serum specimens for serology should be collected during the acute phase within 7–10 days of symptom onset, and the convalescent phase 14–21 days after symptom onset, and tested in parallel.
Virus isolation
The traditional method of influenza virus recovery from clinical specimens is by cell culture. However, the relatively slow turn‐around time (the time from receipt of specimens by a laboratory to a result being available to the clinician) of at least 4–5 days has led to the use of more rapid assays, such as nucleic acid testing or immunofluoresence, and fewer laboratories isolating virus. Nevertheless, virus isolation is usually more sensitive than the rapid culture and antigen detection assays, it recovers novel or highly divergent strains missed by other tests, it provides an isolate for subsequent characterisation and consideration as potential vaccine strains, and it allows the simultaneous recovery of other respiratory viruses if an appropriate range of cell lines is used.
Influenza virus replication within cell culture, often using Madin Darby canine kidney cells, is detected by observing the cytopathic effect, generally manifested by 5 days. 1 Rapid culture techniques that detect influenza virus replication using labelled monoclonal antibodies within 1–3 days (before the development of cytopathic effect) have a sensitivity of 56%–100% compared with conventional cell culture. 9 Virus is readily isolated (PC3 facilities are recommended) from the nose and throat of humans with H5N1 infection, as well as from blood and rectal swabs. 8
Virus isolation in the allantoic and amniotic cavities of 10–11‐day‐old embryonated chicken eggs is labour intensive and not commonly performed by diagnostic laboratories. However, it yields higher viral titres, and remains vital for vaccine production.
Virus typing
Initial typing of influenza virus isolates is most rapidly and conveniently accomplished by immunofluoresence using commercially available type‐specific monoclonal antibodies, and should be performed as soon as possible after isolation. The reference virus subtyping method is the haemagglutination inhibition assay using specific antisera, a technique that characterises both antigenic drift and shift. More rapid subtyping techniques using monoclonal antibodies that differentiate between influenza A/H1N1, A/H3N2, and B virus subtypes have been developed for use on virus isolates, in rapid culture assays, and directly on clinical specimens. 10 , 11 Subtyping using reverse transcription polymerase chain reaction (RT‐PCR) with primers specific for various human and avian influenza strains can be performed on virus isolates or directly on clinical specimens. 12 ‐ 14 DNA microarrays have been used to detect type‐ and subtype‐specific amplification sequences. 15
Sequencing of amplified haemagglutinin and neuraminidase genes is an important subtyping method, as it allows rapid identification of novel or highly divergent strains, the analysis of strain variation, and the determination of the origin of outbreaks. 16 Rapid sequencing of avian and human strains has been used to monitor the evolution of H5N1 viruses around the world, allowing an understanding of its spread, drug resistance and likely candidate vaccine strains. 17
Rapid antigen detection assays
Immunofluoresence or enzyme immunoassay using commercial type‐specific monoclonal antibodies directed against conserved influenza antigens are the most common rapid assays performed directly on clinical specimens (Box 5), although they are starting to be replaced by nucleic acid testing. Indirect or direct immunofluoresence is 60%–100% sensitive compared with cell culture. 1 , 2 As specimens are examined with a fluorescent microscope, an assessment of specimen quality can be made (and reported to the clinician): where few columnar epithelial cells are seen, the sensitivity of immunofluoresence is low and repeat specimen collection should be requested.
Box 5.
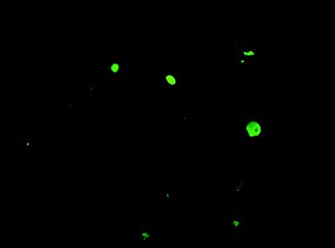
Rapid antigen immunofluorescence assay
Assay performed on cells from a combined nose and throat swab, showing typical nuclear and cytoplasmic “apple‐green” fluorescence after staining with monoclonal antibodies specific for influenza A.
Monoclonal antibodies to other common respiratory pathogens, such as parainfluenza viruses types 1–3, adenovirus and respiratory syncytial virus, can be applied to the same specimen. H5N1‐specific immunofluoresence reagents are not yet routinely available.
The availability of the neuraminidase inhibitors has added impetus to the development of simple, rapid antigen detection assays, sometimes called point‐of‐care tests. 1 , 2 Typically, these tests produce a visual result on an immunochromatographic strip using influenza A or B nucleoprotein‐specific monoclonal antibodies within about 15 minutes of adding an extracted specimen (Box 6). Evaluations of these assays for H5N1 infection are limited, but for A/H3N2, A/H1N1 or B virus infections, they are 56%–100% sensitive and 73%–99% specific compared with cell culture or immunofluoresence, and are 56%–74% sensitive compared with RT‐PCR. 1 , 2 , 18 As with other techniques, specimen quality is a major determinant of their performance. Ideally, when using rapid antigen tests, specimens for cell culture and immunofluoresence or nucleic acid testing should be submitted in parallel to improve sensitivity and to obtain viral isolates for further typing and vaccine analysis, although this strategy increases costs.
Box 6.
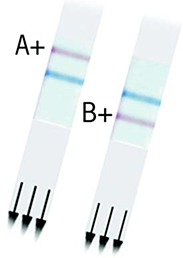
Positive rapid antigen immunochromatographic strips (Quidel Quickvue*)
The strips show positive reactions to influenza A and B. The central blue line is a control reaction to ensure a valid assay.
* Quidel Corporation, San Diego, Calif, USA.
Rapid antigen tests are attractive where virology laboratory support is limited or remote from clinical services, or where laboratories may be overwhelmed (for example, at the height of seasonal outbreaks or during a pandemic). They may also have a role in doctors’ surgeries, emergency departments, or “fever clinics”, to guide the early use of neuraminidase inhibitors, both for individuals or in outbreaks in closed environments, or during a pandemic. 19 They can also be used in community‐based surveillance programs.
Nucleic acid testing
Influenza virus RNA may be detected in clinical specimens by nucleic acid testing techniques such as RT‐PCR. A variety of “in‐house” nucleic acid testing assays using primers specific for a range of influenza virus genes, and various extraction, amplification and product detection methods have been described. 12 ‐ 14 , 18 , 20 Depending on primer selection, these assays may be type‐ or subtype‐specific. Primers that target nucleoprotein or matrix gene sequences are popular, as they detect all influenza subtypes.
Although some studies documented a similar sensitivity of nucleic acid testing to cell culture, others have reported 5%–15% more influenza virus detections using RT‐PCR. 12 , 13 , 18 Specimen quality, timing, and transportation conditions may be less critical for nucleic acid testing than for culture or antigen detection, as viable virus and intact infected cells need not be preserved. Influenza virus RNA is detectable for several days longer into the clinical course than is cultivable virus, potentially allowing a diagnosis to be made in late‐presenting patients. When specimen quality is limited, such as with community‐based surveillance programs involving postal submission of specimens, yields from RT‐PCR are significantly higher than from cell culture. 12 , 13 , 21 Nucleic acid testing using H5N1‐specific primers is the most sensitive assay for the diagnosis of human H5N1 infection. 8
Although the turnaround time for nucleic acid testing is intermediate between cell culture and direct antigen detection, newer techniques can reduce this to 4–5 hours or less. 20 Multiplex RT‐PCR assays have been developed, and these can simultaneously detect influenza and other viral and bacterial respiratory pathogens directly in clinical specimens. 1 , 20 RT‐PCR can quantify the viral load in clinical specimens, as has been done in human H5N1‐infected patients to assess prognosis and measure antiviral efficacy. 8 , 22 Nucleic acid testing can subtype influenza viruses using subtype‐specific primers, and analyse strain variation through genetic sequencing, thus complementing the traditional role of antigenic characterisation of viral isolates.
Serology
Serology can be used where specimens for virus isolation or antigen detection are negative, inadequate, or unavailable. It also provides useful, if delayed, surveillance data. Serological diagnosis of influenza is retrospective, as it requires acute and convalescent serum samples. Furthermore, it generally does not provide information on the antigenic composition of circulating strains.
As influenza virus infection generally represents reinfection, most people have some pre‐existing immunity. Thus, the detection of influenza‐specific IgG or total antibodies on a single serum specimen is not diagnostic of recent infection. The definitive serological diagnosis of acute influenza requires the demonstration of increasing antibody titres on paired acute and convalescent serum samples. Unfortunately, acute specimens are frequently not collected. However, a single high antibody titre in the context of a compatible clinical illness during a period of circulating influenza activity provides presumptive evidence of recent infection.
A variety of influenza‐specific assays are available, including complement fixation, haemagglutination inhibition assay, single radial haemolysis, neutralisation, immunofluorescence and enzyme immunoassay. The traditional “gold standard” techniques for detecting influenza‐specific antibodies are neutralisation and the haemagglutination inhibition assay, as they can differentiate subtype‐specific (including H5N1) and strain‐specific serological responses. Antibody levels correlate accurately with protection from (or susceptibility to) influenza, and with vaccination responses. Complement fixation is more commonly used as it is easier to perform than the haemagglutination inhibition assay and neutralisation, but it does not distinguish between subtypes. Recent influenza infection is suggested by a fourfold rise in complement fixation titre or by a single high titre. 1 , 23
Antiviral susceptibility testing
Two classes of drugs are effective against influenza virus infection: the adamantane M2 channel inhibitors (amantadine and rimantadine), and the neuraminidase inhibitors (zanamivir and oseltamivir). 24 , 25
Amantadine resistance emerges within a few days of treatment onset in 25%–35% of patients, and is mediated by amino acid substitutions in the M2 transmembrane region. Adamantane resistance is increasing around the world. 24 , 25 Neuraminidase inhibitor resistance is most commonly associated with a variety of amino acid substitutions in the conserved residues of the neuraminidase enzyme active site. To date, however, viruses with these mutations have shown impaired replication in cell culture and animal models. Mutations in the haemagglutinin receptor‐binding site can occur, leading to reduced viral dependence on neuraminidase function. 24 Resistance to the neuraminidase inhibitors was rarely seen in immunocompetent humans during clinical trials, although more recent reports of children infected with influenza A in Japan, and some Vietnamese human H5N1 patients treated with neuraminidase inhibitors, showed that resistance may be more common than initially supposed. 22 , 25 Resistant mutations have been detected after treatment of immunocompromised individuals, a situation that requires further study.
Drug resistance is tested using genotyping (sequence analyses of the neuraminidase gene) or phenotyping assays. 24 , 25 These assays are not routinely available, but after therapeutic failure or for surveillance purposes, isolates can be referred to the appropriate reference laboratory. Newer molecular methods that allow rapid resistance determination in a clinically relevant time frame (such as during a pandemic when neuraminidase inhibitors may be used extensively) are under development.
Discussion
Various laboratory techniques are available to detect influenza viruses, whether current human strains or a novel pandemic strain. During a pandemic, the level of laboratory service will vary between centres. Specialist virology laboratories will undertake a full range of influenza virus tests, including nucleic acid testing, rapid antigen detection, culture and typing of isolates for potential use in vaccines, and detection of antiviral drug resistance. Other laboratories may follow a more limited strategy depending on their location (eg, rural areas), their population (eg, public or private hospitals), and access to specialised laboratories.
A challenge for laboratories is ensuring that their assays are accurate for viruses (such as the current H5N1 strains) not yet circulating in Australia. Genetic variation may particularly affect nucleic acid testing, and the lack of clinical samples in Australia from H5N1‐infected individuals makes assessment of the various methods uncertain. Recent urgent funding from the National Health and Medical Research Council (NHMRC) has allowed the assessment and quality assurance of various H5N1‐specific assays.
Another major challenge will be determining diagnostic strat‐egies during the different pandemic phases. These will vary between laboratories depending on their virology expertise, capacity, patient populations, availability of various assays, and the stage of the pandemic. In the very early phases, where containment is paramount, highly sensitive tests such as pandemic strain‐specific nucleic acid testing are required. Testing will have to be rapid to identify the first arrivals and clusters of disease, and to allow early antiviral treatment, prophylaxis and infection control. Other causes of respiratory infection will need to be excluded. Early identification will require cooperation and communication between public health officers, treating clinicians, and public and private laboratories. If the pandemic becomes widespread in Australia, testing strategies may change. One could argue that clinical identification will then suffice, and that laboratory testing should be limited to patients requiring hospital admission or for particular outbreaks. Non‐pandemic strain‐specific influenza assays, such as immunofluorescence or immunochromatographic rapid antigen tests, may be useful in identifying influenza infection during these pandemic stages. Antiviral resistance testing is not yet routinely available in Australia, but will be important if antiviral drugs are used extensively during a pandemic.
Acknowledgements
This project was partially supported by NHMRC grant 382917. The image in Box 5 was provided by David Adams and Ken McPhie at the Centre for Infectious Diseases and Microbiology Laboratory Services, Institute of Clinical Pathology and Medical Research, Westmead Hospital.
Competing Interests
Our laboratories have undertaken clinical research projects funded by laboratory supply companies related to laboratory techniques discussed in this paper. Roche Products Pty Ltd paid travel expenses for Dominic Dwyer and David Smith to attend the Lancet pandemic influenza meeting, Singapore, April 2006.
References
- 1. Playford EG, Dwyer DE. Laboratory diagnosis of influenza virus infection. Pathology 2002; 34: 115–125. . [DOI] [PubMed] [Google Scholar]
- 2. Nicholson KG, Wood JM, Zambon M. Influenza. Lancet 2003; 362: 1733–1744. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nicholson KG. Human influenza In: Nicholson KG, Webster RG, Hays AJ, editors. Textbook of influenza. Oxford: Blackwell Science, 1998. [Google Scholar]
- 4. World Health Organization Collaborating Centre for Reference and Research on Influenza, Melbourne [website]. https://www.influenzacentre.org (accessed Oct 2006).
- 5. Donaldson A, Lewis FA, Kennett ML, et al. The 1976 influenza epidemic in Melbourne. Med J Aust 1978; 2: 45–49. . [DOI] [PubMed] [Google Scholar]
- 6. Cruz JR, Quinonez E, Fernandez A, Devalte F. Isolation of viruses from nasopharyngeal secretions. Comparison of aspiration and swabbing as means of sample collection. J Infect Dis 1987; 156: 415–416. . [DOI] [PubMed] [Google Scholar]
- 7. Schmid ML, Kudesia G, Wake S, Read RC. Prospective comparative study of culture specimens and methods in diagnosing influenza in adults. BMJ 1998; 316: 275. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Jong MD, Simmons CP, Thanh TT, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med 2006; 12: 1203–1207. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Espy MJ, Smith TF, Harmon MW, Kendal AP. Rapid detection of influenza virus by shell vial assay with monoclonal antibodies. J Clin Microbiol 1986; 24: 677–679. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tkácová M, Varecová E, Baker IC, et al. Evaluation of monoclonal antibodies for subtyping of currently circulating human type A influenza viruses. J Clin Microbiol 1997; 35: 1196–1198. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ziegler T, Hall H, Sánchez‐Fauquier A, et al. Type‐ and subtype‐specific detection of influenza viruses in clinical specimens by rapid culture assay. J Clin Microbiol 1995; 33: 318–321. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ellis JS, Fleming DM, Zambon MC. Multiplex reverse transcription‐PCR for surveillance of influenza A and B viruses in England and Wales in 1995 and 1996. J Clin Microbiol 1997; 35: 2076–2082. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schweiger B, Zadow I, Heckler R, et al. Application of a fluorogenic PCR assay for typing and subtyping of influenza viruses in respiratory samples. J Clin Microbiol 2000; 38: 1552–1558. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stockton J, Ellis JS, Saville M, et al. Multiplex PCR for typing and subtyping influenza and respiratory syncytial viruses. J Clin Microbiol 1998; 36: 2990–2995. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li J, Chen S, Evans DH. Typing and subtyping using DNA microarrays and multiplex reverse transcriptase PCR. J Clin Microbiol 2001; 39: 696–704. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Young LC, Dwyer DE, Harris M, et al. Summer outbreak of respiratory disease in an Australian prison due to an influenza A/Fujian/411/2002(H3N2)‐like virus. Epidemiol Infect 2005; 133: 107–112. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. The World Health Organization Global Influenza Program Surveillance Network . Evolution of H5N1 avian influenza viruses in Asia. Emerg Infect Dis 2005; 11: 1515–1521. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Herrmann B, Larsson C, Zweygberg BW. Simultaneous detection and typing of influenza viruses A and B by a nested reverse transcription‐PCR: comparison to virus isolation and antigen detection by immunofluorescence and optical immunoassay (FLU OIA). J Clin Microbiol 2001; 39: 134–138. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sintchenko V, Gilbert GL, Coiera E, Dwyer DE. Treat or test first? Decision analysis of empirical antiviral treatment of influenza virus infection versus treatment based on rapid results. J Clin Virol 2002; 25: 15–21. . [DOI] [PubMed] [Google Scholar]
- 20. Van Elden LJR, Nijhuis PS, Schuurman R, van Loon AM. Simultaneous detection of influenza viruses A and B using real‐time quantitative PCR. J Clin Microbiol 2001; 39: 196–200. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carman WF, Wallace LA, Walker J, et al. Rapid virological surveillance of community influenza infection in general practice. BMJ 2000; 321: 736–737. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Jong MD, Thanh TT, Khanh THJ, et al. Oseltamivir resistance during treatment of influenza A (H5N1) infection. N Engl J Med 2005; 353: 2667–2672. . [DOI] [PubMed] [Google Scholar]
- 23. Rothbarth PH, Groen J, Bohnen AM, et al. Influenza virus serology — a comparative study. J Virol Methods 1999; 78: 163–169. . [DOI] [PubMed] [Google Scholar]
- 24. McKimm‐Breschkin JL. Resistance of influenza viruses to neuraminidase inhibitors — a review. Antivir Res 2000; 47: 1–17. . [DOI] [PubMed] [Google Scholar]
- 25. Hayden FG. Antiviral resistance in influenza viruses — implications for management and pandemic response. N Engl J Med 2006; 354: 785–788. . [DOI] [PubMed] [Google Scholar]


