Abstract
The origin and evolution of polyploids have been studied extensively in angiosperms and ferns but very rarely in gymnosperms. With the exception of three species of conifers, all natural polyploid species of gymnosperms belong to Ephedra, in which more than half of the species show polyploid cytotypes. Here, we investigated the origin and evolution of polyploids of Ephedra distributed in the Qinghai–Tibetan Plateau (QTP) and neighbouring areas. Flow cytometry (FCM) was used to measure the ploidy levels of the sampled species that are represented by multiple individuals from different populations, and then, two single‐copy nuclear genes (LFY and DDB2) and two chloroplast DNA fragments were used to unravel the possible origins and maternal donors of the polyploids. The results indicate that the studied polyploid species are allopolyploids, and suggest that allotetraploidy is a dominant mode of speciation in Ephedra. The high percentage of polyploids in the genus could be related to some of its biological attributes such as vegetative propagation, a relatively high rate of unreduced gamete formation, and a small genome size relative to most other gymnosperms. Significant ecological divergences between allotetraploids and their putative progenitors were detected by PCAs and anova and Tukey's tests, with the exception of E. saxatilis. The overlap of geographical distributions and ecological niches of some diploid species could have provided opportunities for interspecific hybridization and allopolyploid speciation.
Keywords: ecological divergence, Ephedra, gymnosperm, hybridization, molecular phylogeny, polyploid speciation
Introduction
Polyploidy or whole‐genome duplication (WGD) has long been recognized as an important process in plant evolution (Otto & Whitton 2000; Soltis et al. 2009). At least one round of WGD occurred before the divergence of seed plants (Jiao et al. 2011), and multiple rounds of WGD have been reported in angiosperms (Vision et al. 2000; Simillion et al. 2002; Bowers et al. 2003; also see review by Leitch & Leitch 2012). Polyploidy may have broad‐scale effects on genomic repatterning, gene expression and genetic networks due to the inheritance of an additional set of chromosomes, and can produce immediate shifts in morphology, breeding system and ecological tolerances (Otto 2007; Soltis et al. 2010; Fawcett et al. 2013; Weiss‐Schneeweiss et al. 2013), although some studies found no correlation between polyploidization rate and species richness (Wood et al. 2009; Mayrose et al. 2011). While it has been recognized that a polyploid species often has multiple origins (see review by Soltis et al. 2014a), it remains challenging to evaluate the direct effect of polyploidy on evolutionary success of the species (Madlung 2013; Soltis et al. 2014b). Moreover, although some studies indicate that ecological divergence is an important driver of polyploid speciation, especially allopolyploid speciation (Fawcett et al. 2009, 2013; Van de Peer et al. 2009; Ramsey 2011), it remains unresolved whether diploid and polyploid plants can share broad‐scale climatic niches (Glennon et al. 2014).
In contrast to the high frequency of polyploids documented in angiosperms, polyploidy is exceedingly rare in gymnosperms excluding Ephedra (Khoshoo 1959; Ahuja 2005; Murray 2013; Wang & Ran 2014). When excluding three species of conifers, that is the two tetraploids Juniperus chinensis ‘Pfitzeriana’ and Fitzroya cupressoides and the hexaploid Sequoia sempervirens (Ahuja 2005), the remaining natural polyploids of gymnosperms all belong to the genus Ephedra, in which 50–65% of species show tetraploid or very rarely octoploid cytotypes (Khoshoo 1959; Huang et al. 2005). It is of interest to investigate why and how so many polyploids have evolved in this genus.
Ephedra comprises about 50 extant species that are mainly shrubs distributed in both temperate and subtropical arid environments in the Northern Hemisphere and South America (Kubitzki 1990; Ickert‐Bond et al. 2009), with the basal‐most lineages distributed in the Mediterranean area (Rydin & Korall 2009; Qin et al. 2013). These species originated by radiative speciation in the Cenozoic with a crown age of about 30 Ma (Ickert‐Bond et al. 2009), although the earliest fossil record of the genus is from the Early Cretaceous (Yang & Wang 2013). As a secondary diversification centre, the Qinghai–Tibetan Plateau (QTP) and adjacent regions harbour approximately 16 species of Ephedra (Fu et al. 1999; Yang 2002; Yang et al. 2003), of which nine were reported to be polyploids or have polyploid cytotypes, including E. distachya, E. equisetina, E. gerardiana, E. glauca, E. intermedia, E. likiangensis, E. monosperma, E. saxatilis and E. sinica. All of the 16 species form a monophyletic clade with several other species from northern and western Asia and Horn of Africa (Huang & Price 2003; Rydin & Korall 2009) and are generally geographically isolated from the other species of the clade, whereas the nine polyploids are respectively located in three well supported subclades, southern QTP, eastern QTP and northern China (Qin et al. 2013). It was inferred that the subclade divergence occurred in the Miocene and was very likely linked to the uplift of the QTP and the Asian aridification (Qin et al. 2013). However, the origin of these polyploid species remains unknown.
The DNA sequence markers, single‐/low‐copy nuclear genes in particular, are increasingly and successfully used to study allopolyploid speciation in plants, such as in Oryza (Ge et al. 1999), Paeonia (Ferguson & Sang 2001), Persicaria (Kim et al. 2008), Solanum (Cai et al. 2012), Nicotiana (Kelly et al. 2013) and Sequoia (Yang et al. 2012a, b). Also, in recent years, the whole‐genome or whole‐transcriptome analysis has greatly advanced our knowledge of origin and evolution of polyploids in angiosperms (e.g. Roulin et al. 2012; Page et al. 2013; Renny‐Byfield et al. 2013). However, genome sequencing of any gymnosperm remains a huge and expensive task, although a draft assembly of the genome has been generated for a couple of conifers, including Norway spruce (Nystedt et al. 2013), white spruce (Birol et al. 2013) and loblolly pine (Neale et al. 2014). Therefore, current phylogenetic analysis using single‐/low‐copy nuclear genes is still the best approach for exploring the evolution of polyploids in Ephedra.
The previous inference of several allotetraploids from karyomorphological data (Mehra 1946) and the presence of hybridogenic speciation in Ephedra (Cutler 1939; Wendt 1993) allow us to hypothesize that allopolyploid speciation might be common in the genus. This study aims to investigate the origin and evolution of polyploids of Ephedra distributed in the QTP and neighbouring areas. First, flow cytometry was used to measure ploidy levels of the sampled species, most of which were represented by multiple individuals from different populations. Then, two single‐copy nuclear genes (LFY and DDB2) and two chloroplast DNA (cpDNA) fragments (trnT‐trnF and trnS‐trnfM) were used to investigate whether these polyploids are allopolyploids or autopolyploids and which species could be the maternal donors of the allopolyploids. Finally, based on a comprehensive analysis of biological attributes, phylogenetic relationships, geographical distributions and ecological factors of the habitats, we discussed the possible correlation between allopolyploid speciation and some biological and ecological features.
Materials and methods
Sampling
A total of 48 populations of twelve Ephedra species (one with two varieties) distributed in the QTP and adjacent regions were sampled for the measurement of ploidy levels, and the cpDNA and nuclear gene analyses (Table 1). Young branchlets were collected from the individuals that were at least 50 m apart from each other. The remaining four species that also occur in the QTP and neighbouring areas, namely E. distachya, E. lomatolepis, E. rituensis and E. fedtschenkoae, were not included in the study due to the controversy of their species status (Fu et al. 1999; Yang 2002; Yang et al. 2003), or a lack of enough population samples. Among them, E. fedtschenkoae seems unique in being monoecious, but Florin (1933) reported that monoecious individuals are common in Ephedra. The other three species are tetraploids based on previous studies (e.g. Leitch et al. 2001) and our preliminary investigation. Therefore, the exclusion of these species should not greatly affect our inference about the origins of other tetraploid species (mostly allotetraploids, see Results).
Table 1.
The chlorotypes, nuclear gene alleles and ploidy levels detected in the sampled populations of the studied Ephedra species
| Species | Pop. | Location | Lat. (N) | Long. (E) | Alt. (m) | Na | Nb | Haplotypes (Individuals) | Nc | LFY | DDB2 | Nd | Ploidy level | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CA | CB | CC | CA | CB | CC | ||||||||||||
| E. equisetina | JJS | Altay, XJ | 47°49′ | 88°10′ | 1291 | 7 | 7 | H11 (14) | 1 | 2 | 2 | 5 | 2x | ||||
| SBG | Erdos, IM | 39°40′ | 106°59′ | 1443 | 0 | 3 | H12 (3) | 1 | 2 | 2 | 3 | 2x | |||||
| SM | Alxa Zuoqi, IM | 38°57′ | 105°52′ | 1924 | 5 | 10 | H1 (1), H12 (14) | 2 | 2 | 1 | 5 | 2x | |||||
| XHD | Zhangjiakou, HB | 40°30′ | 115°20′ | 811 | 0 | 6 | H12 (6) | 1 | 2 | 2 | 5 | 2x | |||||
| XWT | Zhangjiakou, HB | 39°55′ | 114°59′ | 1300 | 0 | 10 | H12 (2), H15 (8) | 1 | 2 | 2 | 5 | 2x | |||||
| E. gerardiana | DZ | Tingri, Tibet | 28°17′ | 86°48′ | 4737 | 16 | 0 | H23 (16) | 1 | 2 | 2 | 5 | 2x | ||||
| RCZ | Rikaze, Tibet | 29°16′ | 88°51′ | 4000 | 24 | 0 | H16 (24) | 1 | 1 | 2 | 2 | 1 | 5 | 4x | |||
| ZXG | Gar, Tibet | 32°24′ | 79°44′ | 4700 | 24 | 0 | H22 (24) | 1 | 1 | 2 | 5 | 2x | |||||
| E. glauca | WBB | Urumqi, XJ | 43°37′ | 87°57′ | 1450 | 5 | 0 | H1 (1), H3 (4) | 1 | 2 | 2 | 1 | 1 | 5 | 4x | ||
| E. intermedia | GT | Minhe, QH | 35°53′ | 102°48′ | 1866 | 5 | 0 | H1 (5) | 2 | 1 | 2 | 2 | 1 | 5 | 4x | ||
| MNS | Manas, XJ | 44°14′ | 86°20′ | 478 | 5 | 0 | H10 (5) | 2 | 2 | 2 | 1 | 1/2 | 5 | 4x | |||
| QHH | Qinghai Lake, QH | 36°33′ | 100°28′ | 3565 | 22 | 0 | H1 (22) | 1 | 1 | 2 | 1 | 1 | 5 | 4x | |||
| E. likiangensis | AN | Jinchuan, SC | 31°17′ | 101°59′ | 2900 | 21 | 0 | H12 (21) | 1 | 1 | 2 | 1 | 1 | 5 | 4x | ||
| GHZ | Lijiang, YN | 27°07′ | 100°15′ | 3040 | 4 | 0 | H13 (4) | 1 | 1 | 1 | 2 | 1 | 4 | 4x | |||
| MRK | Barkam, SC | 31°54′ | 102°13′ | 2800 | 23 | 0 | H12 (23) | 1 | 2 | 1 | 1 | 1 | 5 | 4x | |||
| E. minuta | HZ | Huzhu, QH | 36°53′ | 102°21′ | 2960 | 3 | 0 | H12 (3) | 1 | 1 | 1 | 3 | 2x | ||||
| QT | Ledu, QH | 36°15′ | 102°15′ | 3358 | 20 | 0 | H12 (20) | 1 | 1 | 1 | 5 | 2x | |||||
| E. monosperma | JFS | Baotou, IM | 40°41′ | 110°45′ | 2198 | 0 | 28 | H12 (28) | 2 | 1 | 1 | 5 | 2x | ||||
| KBX | Zhangjiakou, HB | 42°02′ | 114°47′ | 1691 | 0 | 15 | H12 (8), H14 (7) | 1 | 2 | 1 | 10 | 2x | |||||
| MGD | Mongolia | 43°29′ | 104°03′ | 2279 | 0 | 4 | H14 (4) | 1 | 1 | 2 | 4 | 2x | |||||
| MGE | Mongolia | 43°01′ | 106°04′ | 2200 | 0 | 4 | H14 (4) | 1 | 2 | 1 | 4 | 2x | |||||
| NMC | Damxung, Tibet | 30°46′ | 90°52′ | 4800 | 8 | 0 | H12 (8) | 2 | 1 | 1 | 5 | 2x | |||||
| YMT | Delhi, QH | 37°21′ | 98°07′ | 3473 | 17 | 0 | H12 (17) | 2 | 2 | 1 | 5 | 2x | |||||
| YX | Urumqi, XJ | 43°19′ | 87°12′ | 2033 | 6 | 9 | H12 (15) | 2 | 1/2 | 1 | 5 | 2x | |||||
| E. przewalskii | GB | Delhi, QH | 37°16′ | 97°10′ | 2910 | 5 | 12 | H1 (17) | 1 | 1 | 1 | 5 | 2x | ||||
| GSE | Golmud, QH | 36°13′ | 94°48′ | 3053 | 0 | 17 | H1 (17) | 1 | 1 | 1 | 5 | 2x | |||||
| KLMY | Karamay, XJ | 45°33′ | 84°49′ | 360 | 0 | 16 | H4 (1), H8 (15) | 1 | 1 | 1 | 5 | 2x | |||||
| MGF | Mongolia | 44°35′ | 101°42′ | 1597 | 0 | 12 | H1 (6), H3 (6) | 1 | 1 | 2 | 5 | 4x | |||||
| NCT | Golmud, QH | 35°53′ | 94°22′ | 3716 | 0 | 12 | H1 (12) | 1 | 1 | 1 | 5 | 2x | |||||
| WB | Urumqi, XJ | 43°33′ | 87°53′ | 1128 | 5 | 14 | H3 (19) | 1 | 2 | 1 | 5 | 4x | |||||
| E. regeliana | BYG | Urumqi, XJ | 43°25′ | 87°13′ | 1723 | 5 | 0 | H2 (5) | 1 | 2 | 1 | 5 | 2x | ||||
| XAB | Burqin to Altay, XJ | 47°40′ | 86°48′ | 543 | 6 | 2 | H4 (5), H9 (3) | 1 | 3 | 2 | 5 | 4x | |||||
| XJJ | Urumqi, XJ | 43°39′ | 87°39′ | 1130 | 5 | 3 | H2 (8) | 1 | 1 | 1 | 5 | 2x | |||||
| XZN | Shawan, XJ | 43°57′ | 85°46′ | 1159 | 5 | 12 | H2 (17) | 1 | 2 | 1 | 5 | 2x | |||||
| E. rhytidosperma | HLB | Alxa Zuoqi, IM | 38°51′ | 105°50′ | 2150 | 6 | 0 | H12 (6) | 1 | D1 | D1 | 5 | 2x | ||||
| XK | Yinchuan, NX | 38°36′ | 105°56′ | 1500 | 6 | 0 | H12 (6) | 1 | D1 | D1 | 5 | 2x | |||||
| E. saxatilis | JC | Gyaca, Tibet | 29°08′ | 92°35′ | 3249 | 25 | 0 | H16 (5), H17 (20) | 1 | 2 | 2 | 1 | 1 | 5 | 4x | ||
| LX | Nang, Tibet | 29°02′ | 93°04′ | 3100 | 29 | 0 | H17 (29) | 2 | 1 | 1/2 | 1 | 1/2 | 5 | 4x | |||
| PM | Bomi, Tibet | 29°51′ | 95°45′ | 2700 | 20 | 0 | H17 (20) | 1 | 2 | 2 | 1 | 2 | 5 | 4x | |||
| YD | Yadong, Tibet | 27°31′ | 88°56′ | 3600 | 25 | 0 | H17 (25) | 1 | 2 | 1 | 1 | 1 | 5 | 4x | |||
| E. saxatilis var. mairei | DC | Daocheng, SC | 28°26′ | 100°21′ | 3936 | 12 | 0 | H12 (3), H18 (4), H19 (5) | 1 | 1 | 2 | 1 | 2 | 5 | 4x | ||
| LY | Lijiang, YN | 27°03′ | 100°11′ | 3744 | 24 | 0 | H18 (24) | 1 | 1 | 2 | 1 | 2 | 5 | 4x | |||
| YJG | Kangding, SC | 29°55′ | 102°00′ | 4100 | 28 | 0 | H20 (23), H21 (5) | 1 | 1 | 2 | 1 | 1 | 5 | 4x | |||
| E. sinica | KBD | Zhangjiakou, HB | 42°02′ | 114°51′ | 1425 | 0 | 18 | H5 (18) | 1 | 2 | 1 | 1 | 1 | 10 | 4x | ||
| KSKT | Hexigten Qi, IM | 43°34′ | 117°10′ | 1350 | 13 | 0 | H5 (13) | 2 | 1 | 2 | 1 | 1/2 | 5 | 4x | |||
| LWG | Baotou, IM | 40°35′ | 110°39′ | 1028 | 0 | 31 | H5 (31) | 1 | 2 | 2 | 2 | 1 | 5 | 4x | |||
| MGT | Erdos, IM | 38°29′ | 107°30′ | 1350 | 0 | 40 | H1 (32), H5 (6), H6 (1), H7 (1) | 1 | 2 | 2 | 2 | 2 | 5 | 4x | |||
| XHY | Zhangjiakou, HB | 40°30′ | 115°20′ | 787 | 0 | 21 | H5 (21) | 2 | 2 | 1 | 1 | 2 | 10 | 4x | |||
| All species | 48 | 740 | 248 | ||||||||||||||
| E. foeminea | Saudi Arabia | 1 | H24 (1) | 1 | 1 | 1 | |||||||||||
YN, Yunnan; SC, Sichuan; QH, Qinghai; NX, Ningxia; IM, Inner Mongolia; XJ, Xinjiang; HB, Hebei; Pop., population; Lat., latitude; Long., longitude; Alt., altitude. Na, number of individuals sampled for analysis of cytoplasmic DNAs by Qin et al. (2013); Nb, number of individuals sampled for analysis of cytoplasmic DNAs in this paper; Nc, number of individuals sampled for analysis of nuclear genes; Nd, number of individuals sampled for analysis of flow cytometry. Numbers of alleles in CA, CB and CC correspond, respectively, to those in clade A, clade B and clade C in the phylogenies of DDB2 and LFY, and D1 indicates that one allele was placed in clade D. 1/2, one individual has one allele and the other has two alleles.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Information on the geographical variation of cytotypes is critical for studies of origin and evolution of polyploids. To determine the ploidy levels, we initially analysed 10 individuals from each of the three populations KBD, XHY and KBX, and did not find ploidy variation within populations. Consequently, we analysed five individuals from each of the other populations with the exception of four populations (SBG, GHZ, MGD and MGE), from which fewer than five individuals were available. To investigate variation patterns and evolutionary relationships of the maternally inherited cpDNA, sequences of two fragments (trnT‐trnF and trnS‐trnfM) were analysed for a total of 740 samples, including 306 individuals sampled in this study and 434 individuals reported in Qin et al. (2013).
Based on the analyses of ploidy and cpDNA variation, we further chose 58 individuals to explore nuclear gene relationships of the 12 species, one with two varieties. Also, one individual of the Saudi Arabian E. foeminea was sampled as outgroup based on the results of previous phylogenetic analyses (Ickert‐Bond et al. 2009; Rydin & Korall 2009). The samples were consistently used in the three analyses (ploidy, cpDNA and nuclear genes), with the exception that different sample sizes were used.
Chromosome number counts
Chromosome numbers were counted for two species, E. equisetina (cultivated in the Beijing Botanical Garden) and E. intermedia. Fresh root tips were pretreated with 0.01% colchicine solution for 5–6 h and fixed in a mixture of ethanol/acetic acid (3:1) for 12 h at room temperature. After being macerated in 1N HCl at 60 °C for 5–10 min, the materials were stained with 1% carbol‐fuchsin, and then were squashed and observed under a light microscope. The chromosome number of each species was counted based on at least five cells.
Determination of ploidy levels
The flow cytometry (FCM) has made it convenient to detect the variation of DNA ploidy level in large samples from herbaria and silica gel‐dried materials (Suda & Trávníček 2006; Schönswetter et al. 2007; Krejcikova et al. 2013; Vrána et al. 2014). In Ephedra, the leaves are reduced to small membranous sheaths. Therefore, we used the silica gel‐dried young branchlets for the FCM measurement, mainly following the protocol of Suda & Trávníček (2006). Approximately 0.3 g silica gel‐dried branchlets per individual was chopped with a razor blade in a Petri dish containing 1 mL of Otto I buffer (0.1 mol/L citric acid monohydrate, 0.5% (v/v) Tween‐20, pH 2–3). After filtering through a 50‐μm nylon mesh and centrifuging at 100 g for 8 min, the pellet was resuspended in 200 μL buffer of a 1:2 mixture of Otto I and Otto II (0.4 mol/L Na2HPO4 12H2O) and stained with 50 μg/mL PI including 50 μg/mL RNase. The FCM measurements were taken using an Elite flow cytometer (BD FACSCalibur, USA). To guarantee the reliability of the measurements, several samples were reanalysed (up to four times) on different days to assess between‐run fluctuations, and the results showed that the measurements are very consistent. If the coefficient of variation (CV) of the histogram peak exceeded 5%, the sample was discarded or remeasured. The DNA ploidy levels were inferred based on DNA contents measured in plants with known chromosome numbers. That is, based on chromosome number counts (see Results), the two species of Ephedra, E. equisetina (2n = 14, diploid) and E. intermedia (2n = 28, tetraploid), were used as external reference standards, with their DNA contents measured and shown in Fig. S1 (Supporting information).
DNA extraction, PCR amplification, cloning and sequencing
Total DNA was isolated from silica gel‐dried young branchlets by the modified CTAB method (Rogers & Bendich 1985). Two cpDNA regions, trnT‐trnF and trnS‐trnfM, were amplified and sequenced following the protocols of Qin et al. (2013). The LFY gene was amplified with the forward primer LFYE2F2 (5′‐ GACAGTTGGTGCTTTAATAGG ‐3′) located at the second intron and the reverse primer LFYE3R1 (5′‐ CCTCATCTTTGGCTTGTTTAT ‐3′) at the third exon, and the DDB2 gene was amplified with DDB2S2 at the second exon (5′‐ ACAGCCAGGTGATTGTTATGAG ‐3′) and DDB2A1 at the fifth exon (5′‐ TCTAAGGAGGTGACCCGTCTACT ‐3′).
The PCRs were conducted in a volume of 25 mL, containing 50–75 ng total DNA, 6.25 pmol of each primer, 200 mmol/L of each dNTP and 0.75 unit of Taq DNA polymerase (TaKaRa Biotech Co., China). PCR cycles were as follows: 4 min at 94 °C, 36 cycles of 30 s at 94 °C, 30 s at 58 °C and 1–2 min at 72 °C, with a final extension of 10 min at 72 °C. The PCR products were purified using a TIANgel Midi Purification Kit (TIANGEN, China) for the nuclear markers and then cloned with the pGEM‐T Easy Vector System II (Promega). For each individual, 6–20 clones (6–12 for diploids, and 8–20 for tetraploids and the outgroup) were sequenced using primer T7 or LFY E3R1. The sequencing products were separated on an ABI PRISM 3730XL DNA analyzer (Applied Biosystems). The sequences generated in this study are deposited in GenBank under accession numbers KT033384–KT033389 (trnS‐trnfM), KT033390–KT033395 (trnT‐trnF), KT033145–KT033275 (LFY) and KT033276–KT033383 (DDB2).
Data analyses
The DNA sequences were aligned and manually adjusted in bioedit v.7.0.9 (Hall 1999). Haplotype networks were constructed with network 4.6.1.2 (Bandelt et al. 1999), and each indel was treated as a single mutation event. Phylogenetic trees of cpDNA haplotypes and the two nuclear genes were constructed by maximum parsimony (MP), maximum likelihood (ML) and Bayesian inference (BI) methods, using paup*4.0b10 (Swofford 2002), phyml3.0 (Guindon et al. 2010) and mrbayes 3.1.2 (Ronquist & Huelsenbeck 2003), respectively. The gaps were treated as missing data.
The MP analysis used a heuristic search with 1000 random addition sequence replicates, tree‐bisection–reconnection (TBR) and MULTREES on. Branch support was evaluated by a bootstrap analysis (Felsenstein 1985) of 1000 replicates using the same heuristic search settings, and a 50% majority‐rule consensus was used. In the ML analysis, jmodeltest 2 (Guindon & Gascuel 2003; Darriba et al. 2012) was used to determine the best‐fit nucleotide substitution models under the Akaike Information Criterion (AIC), which were GTR+I for cpDNA, TVM+G for LFY and TVM+I for DDB2. As a starting point for the ML search, a BIONJ tree was used (Gascuel 1997). Branch support was estimated by bootstrap analysis with 1000 replicates. The BI analysis used the best‐fit models determined also by jmodeltest 2, including GTR+G for cpDNA [nst = 6, rates = gamma, Prset statefreqpr = dirichlet(1,1,1,1)], GTR+G for LFY [nst = 6, rates = gamma, Prset statefreqpr = dirichlet(1,1,1,1)] and HKY+G for DDB2 [nst = 2, rates = gamma, and Prset statefreqpr = dirichlet(1,1,1,1)]. One cold and three incrementally heated Markov chain Monte Carlo (MCMC) chains were run for 1 000 000 generations each, and trees were sampled every 100 generations with the first 300 samples discarded as burn‐in.
To show explicitly the origin, particularly the putative progenitors, of the allopolyploid species, the program padre (Lott et al. 2009a,b) was used to generate a reticulate phylogenetic network of the studied species based on a collection of LFY, DDB2 and cpDNA trees under default settings. The input topologies were 50% majority‐rule consensus MP trees that were generated from the reduced matrices comprising 13 species (including outgroup) and 19 representative individuals, which included one allele (diploids) or two alleles (polyploids) distributed in different main clades of the two nuclear gene phylogenies.
Ecological niche analysis
To investigate whether there was an association between speciation and climatic factors, ecological niche divergence among the species was also investigated. A total of 557 georeferenced occurrence records (herbarium collections at the Chinese Virtual Herbarium (CVH) http://www.cvh.org.cn/search; records in Freitag & Maier‐Stolte 1994; and extensive field surveys by ourselves) were collected for 13 taxa of Ephedra, including 49 for E. equisetina, 12 for E. gerardiana, 9 for E. glauca, 180 for E. intermedia, 16 for E. likiangensis, 5 for E. minuta, 74 for E. monosperma, 63 for E. przewalskii, 76 for E. regeliana, 3 for E. rhytidosperma, 17 for E. saxatilis, 9 for E. saxatilis var. mairei and 44 for E. sinica (Fig. S2, Supporting information). We retrieved 19 bioclimatic layers (BIO1–BIO19) from Worldclim (http://www.worldclim.org; Hijmans et al. 2005) and the mean annual potential evapotranspiration (PET) from CGIAR‐CSI (http://www.cgiar-csi.org; Trabucco et al. 2008) at a resolution of 30 arc seconds (about 1 km) based on the geocoordinates using arcgis 10.2. The 20 bioclim variables were examined for pairwise Pearson correlations, and 10 variables (BIO2, 3, 4, 8, 12, 14, 15, 18, 19 and PET, shown in Table S2, Supporting information) with correlation coefficients lower than 0.8 were finally selected.
To determine the ecological characteristics of all studied species in the QTP and neighbouring areas and investigate whether ecological divergence had driven polyploid speciation, we used two approaches. First, to identify the divergence of ecological niches, a principal component analysis (PCA) using 10 bioclim variables implemented in ade4‐r package (Dray et al. 2007) was conducted for all taxa, and allotetraploids vs. their putative progenitors, respectively. Then, to identify bioclim variables associated with speciation, a permutational analysis of variance (permanova) was performed to assess the variation of principal components (PC1 and PC2) and each bioclim variable among and within species using the lmperm package (Wheeler 2010). The principal components and bioclim variables with significant differences indicated by the anova were further assessed by the Tukey‘s honestly significant difference (HSD) test for every two taxa using the stats package. The variation of these variables was shown by boxplots. The analyses of PCA, anova and Tukey's HSD were conducted in r version 3.1.2 (R Development Core Team, 2014).
Results
Variation in ploidy levels
To confirm the chromosome numbers of the two samples used as external standards for the FCM analysis, we observed mitotic cell divisions in root tips by light microscopy and found that the chromosome number was 2n = 14 for E. equisetina and 2n = 28 for E. intermedia. Therefore, the FCM fluorescence histograms of the two samples, as shown in Fig. S1 (Supporting information), were used to represent diploid (2x) and tetraploid (4x) nuclei, respectively. The peak ratio of E. intermedia to E. equisetina is 1.98. For all the Ephedra samples (12 species, 48 populations and 248 individuals) analysed by FCM, none of the CV values exceeded 5%. Based on the peak positions that indicate the relative DNA contents, the fluorescence histograms could be clearly divided into two cytotypes, corresponding to diploids and tetraploids, respectively. The peak ratios of diploids to E. equisetina ranged from 0.95 to 1.08, and those of tetraploids to E. intermedia ranged from 0.94 to 1.07. The FCM measurement indicated that three species (E. gerardiana, E. przewalskii and E. regeliana) harboured both diploid and tetraploid cytotypes, four species (E. equisetina, E. minuta, E. monosperma and E. rhytidosperma) showed only the diploid cytotype, and six taxa (E. likiangensis, E. glauca, E. intermedia, E. saxatilis, E. saxatilis var. mairei and E. sinica) exhibited only the tetraploid cytotype (Table 1).
In our survey, two ploidy levels (2n = 14, 28) were found in different populations of E. przewalskii and E. regeliana (Table 1), from which only diploids were previously reported (Kong et al. 2001; Jiang 2006; Wu et al. 2009). Moreover, for some species which were reported to have two or more cytotypes (Table S3, Supporting information), we only found a single cytotype, such as only diploids in E. equisetina and E. monosperma (Table 1).
Distributions and evolutionary relationships of cpDNA haplotypes
The trnT‐trnF and trnS‐trnfM sequences were obtained from all of the 740 samples, including 306 individuals determined in this study and 434 individuals reported in Qin et al. (2013) (Table 1). The new sequences generated are mainly from populations in northern China. The alignment of the combined two cpDNA fragments is 1115 bp in length, including 27 nucleotide substitutions and eight indels that were used to designate 24 haplotypes (H1‐H23 for ingroups and H24 for outgroup, see Table S1, Supporting information), of which five (H6–8, H14, H15) were newly detected. The distributions of the ingroup haplotypes are shown in Fig. 1.
Figure 1.
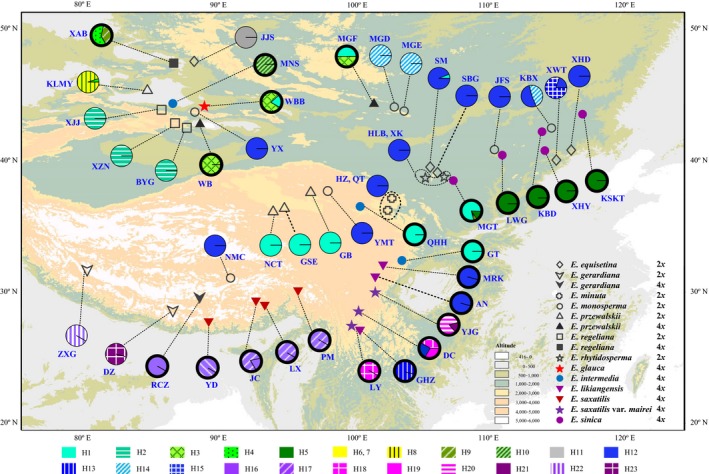
Sampling locations and distribution frequencies of the cpDNA haplotypes detected in the studied 12 Ephedra species. Thick and thin outlines of the pie charts indicate tetraploids and diploids, respectively. Population names correspond to those in Table 1.
When the sequence of E. foeminea (H24) was used as outgroup, three main lineages (I–III) were consistently resolved in the network and phylogenetic tree of cpDNA haplotypes (Figs 2 and S3). Lineage I consisted of 10 haplotypes (H1–H10) that occurred in five species (E. glauca, E. intermedia, E. przewalskii, E. regeliana and E. sinica) mainly distributed in northern China, but only three of them were shared among species, including H1 among E. glauca, E. intermedia, E. przewalskii and E. sinica, H3 between E. przewalskii and E. glauca, and H4 between E. przewalskii and E. regeliana. Lineage II comprised four haplotypes (H12–H15), of which H12 was the most widely distributed, and was shared by six species (E. equisetina, E. likiangensis, E. minuta, E. monosperma, E. rhytidosperma and E. saxatilis var. mairei). Lineage III harboured eight haplotypes (H16–H23) that occurred in two species and a variety (E. gerardiana, E. saxatilis and E. saxatilis var. mairei) from the QTP. The haplotype H11 detected in the population JJS of E. equisetina was not grouped into any of the three lineages. Of the 48 Ephedra populations analysed, 37 (77%) harboured a single haplotype, 9 (19%) exhibited two haplotypes, and only two had more than two haplotypes (Fig. 1; Table 1).
Figure 2.
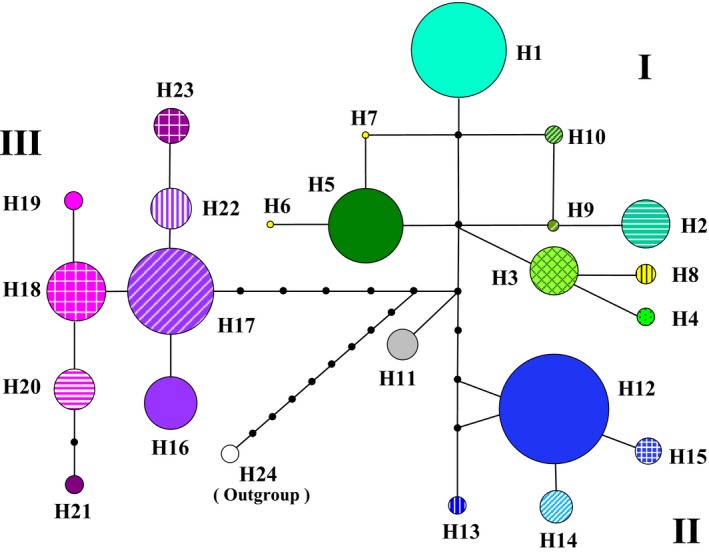
A network of the cpDNA haplotypes constructed by network 4.6.1.2. The sizes of the circles in the network are proportional to the observed frequencies of the haplotypes.
Distributions and evolutionary relationships of the nuclear gene alleles
For each of the nuclear genes LFY and DDB2 that were PCR‐amplified and cloned, 1–2 distinct clones (alleles) were obtained from each diploid individual, and 2–4 alleles were detected in each tetraploid individual, with the exception of two tetraploid individuals of E. przewalskii from populations MGF and WB, each of which contained only 1–2 alleles (Table 1). The distributions of the alleles of LFY and DDB2 are shown in Figs 3 and 4, respectively.
Figure 3.
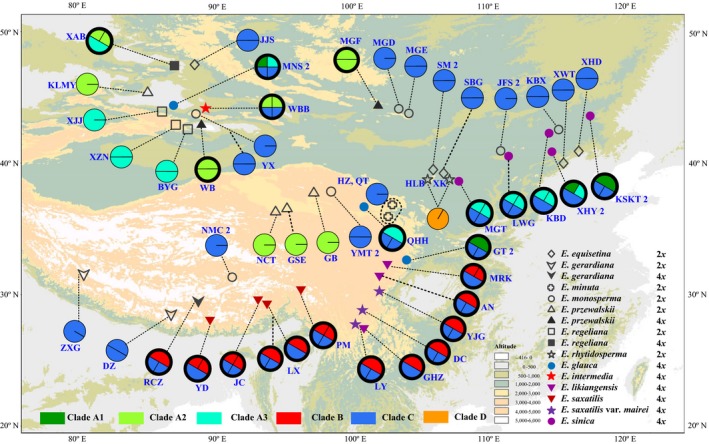
Sampling locations and distributions of the LFY alleles detected in the studied Ephedra species. Pie charts show the proportions of alleles, and thick and thin outlines indicate tetraploids and diploids, respectively. The number 2 following a population name indicates that two individuals were studied. Different colours of the pie charts indicate the positions of the alleles in the gene phylogenies shown in Fig. 5. Population names correspond to those in Table 1.
Figure 4.
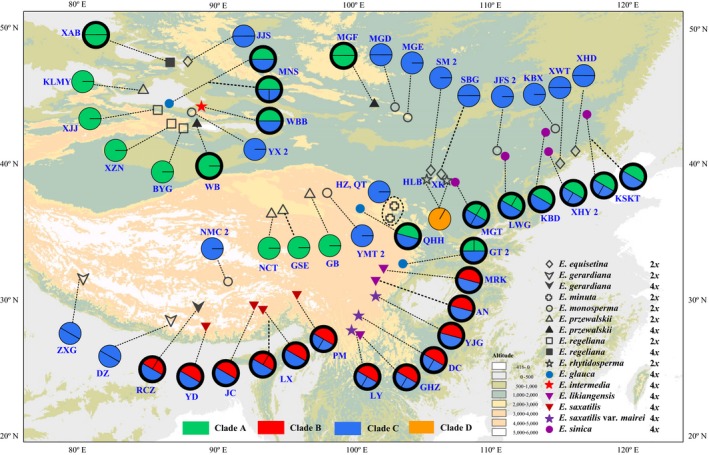
Sampling locations and distributions of the DDB2 alleles detected in the studied Ephedra species. Pie charts show the proportions of alleles, and thick and thin outlines indicate tetraploids and diploids, respectively. The number 2 following a population name indicates that two individuals were studied. Different colours of the pie charts indicate the positions of the alleles in the gene phylogenies shown in Fig. 5. Population names correspond to those in Table 1.
The LFY gene sequences (alleles) were 462–791 bp in length, and the sequence alignment contained 886 sites, of which 244 were variable and 167 were parsimony‐informative. The DDB2 gene sequences ranged from 546 to 551 bp, and the sequence alignment contained 554 sites, of which 87 were variable and 52 were parsimony‐informative.
The generated MP, ML and BI trees of each gene were highly congruent, and the LFY and DDB2 gene trees were also congruent in deep branches (Fig. S4, Supporting information). The simplified strict consensus MP trees of the two genes are shown in Fig. 5, both strongly supporting clades A, B, C and D. Clade A (A‐type) sequences (alleles) were from northern China and northern QTP, clade B (B‐type) alleles occurred in southern and eastern QTP, and clade C (C‐type) alleles had a very wide distribution (Figs 3 and 4). The clade D (type D) sequences were all from E. rhytidosperma, a species narrowly distributed in the Helan Mountain.
Figure 5.
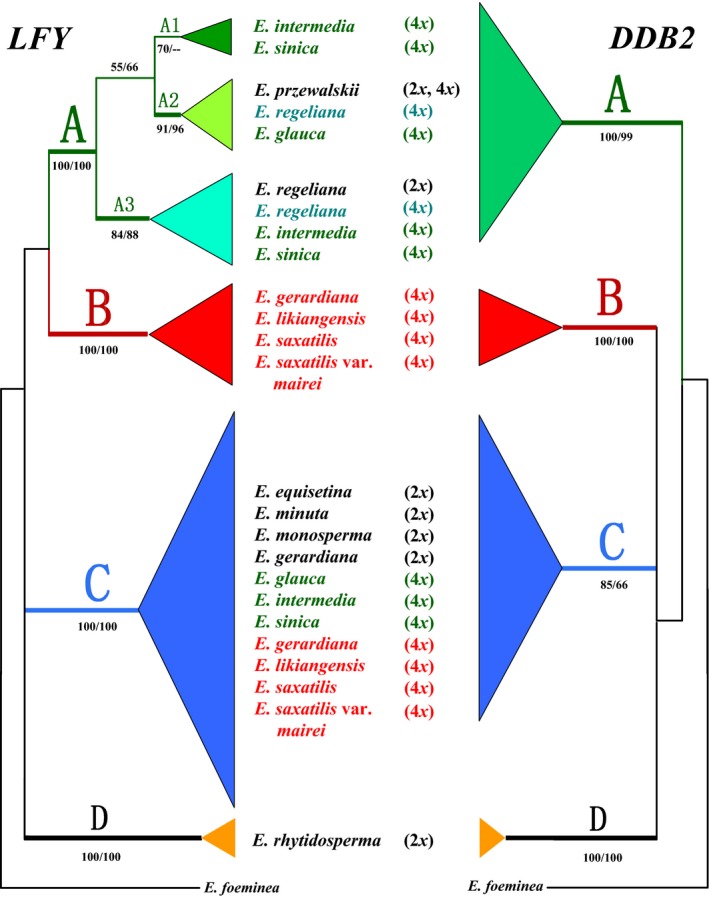
Majority‐rule consensus trees obtained from maximum parsimony analysis of the LFY and DDB2 data sets. Numbers associated with branches are bootstrap percentages of MP and ML greater than 50%, respectively. Bold lines indicate Bayesian posterior probabilities greater than 0.90. Diploids and autotetraploids are in black, and allotetraploids are in colour.
The diploid species or populations contained only one type of sequences, type A (diploid populations of E. przewalskii and E. regeliana), type C (E. equisetina, E. minuta, E. monosperma, and diploid populations of E. gerardiana) or type D (E. rhytidosperma). In contrast, the tetraploid species contained two types of sequences, types A and C (E. glauca, E. intermedia and E. sinica) or types B and C (E. likiangensis, E. saxatilis, E. saxatilis var. mairei, and the tetraploid population of E. gerardiana). However, the tetraploid populations of przewalskii and E. regeliana only had A‐type sequences (Fig. 5).
Reticulate network
A phylogenetic network generated from the integration of all three gene trees (cpDNA, LFY and DDB2) is shown in Fig. 6, from which eight allotetraploid taxa could be inferred, including E. gerardiana, E. glauca, E. intermedia, E. likiangensis, E. regeliana, E. saxatilis, E. saxatilis var. mairei and E. sinica. The diploid progenitors of these allotetraploids were not well resolved, but it seems that all studied diploid ingroup species, with the exception of E. rhytidosperma, could have been involved. For example, the three tetraploids E. glauca, E. intermedia and E. sinica possibly originated from hybridization with diploids most closely related to E. przewalskii in clade A as the maternal parents and diploids of clade C (E. equisetina, E. minuta and E. monosperma) as the paternal parents (see Discussion). The diploid E. regeliana does not share chlorotypes with any of the tetraploids, and therefore is not very likely to have acted as a maternal parent in the allotetraploid speciation.
Figure 6.
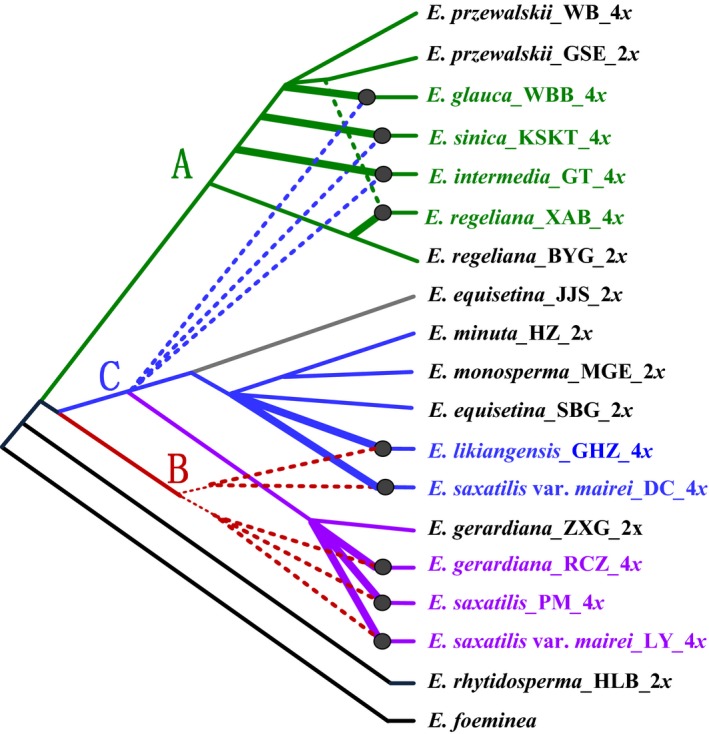
A reticulate network constructed from the reduced 50% majority‐rule consensus MP trees of LFY,DDB2 and cpDNA using the program padre. Different colours of lines indicate the positions of the species in the cpDNA network (lineages I–III in Fig. 2) and the alleles in the two nuclear gene phylogenies (clades A–C in Fig. 5): green, lineage I, clade A; blue, lineage II, clade C; purple, lineage III, clade C; red, clade B; grey, the lineage of cpDNA haplotype H11. Bold solid and dashed lines represent putative maternal and paternal progenitors of the tetraploids, respectively. Diploids and autotetraploids are in black, and allotetraploids are in colour. Letters following species names are population names corresponding to those in Table 1.
Ecological differentiation
More than 10 georeferenced occurrence records were collected for each of the studied species, with the exception of E. glauca, E. minuta and E. rhytidosperma due to their narrow distributions. Results of the PCAs are shown in Figs 7 and 8, with the factor loadings shown in Table S2 (Supporting information).
Figure 7.
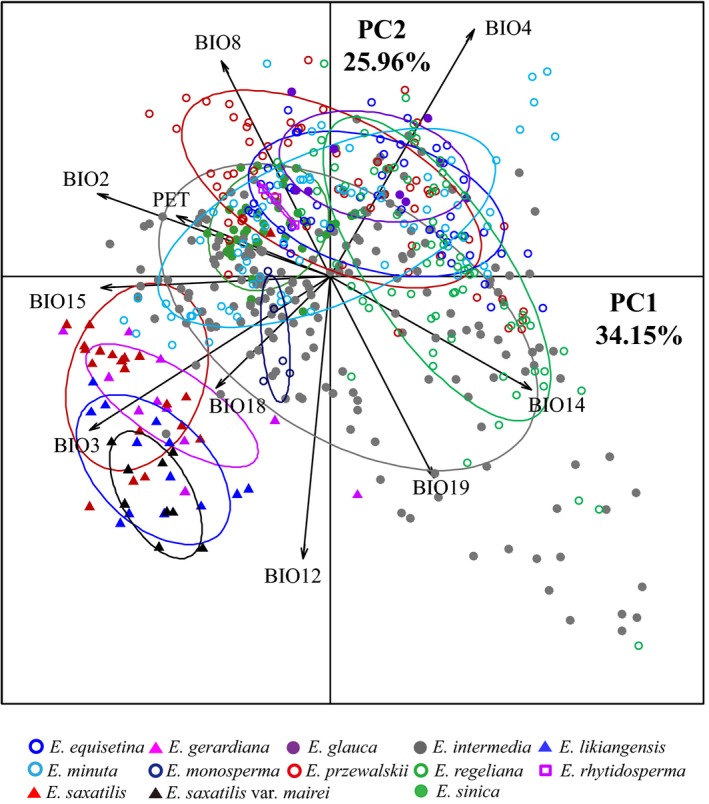
Scatter plots of PC1 and PC2 showing ecological differentiation among the studied Ephedra species based on 10 bioclim variables at sampling locations.
Figure 8.
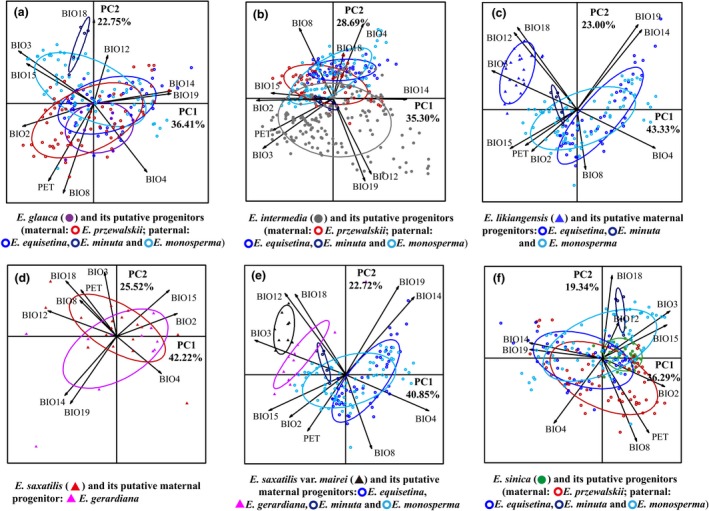
Scatter plots of PC1 and PC2 (a‐f) showing ecological differentiation between allotetraploids and their putative progenitors based on 10 bioclim variables at sampling locations.
For all of the 13 taxa, the PCA revealed two components that cumulatively explained 60.11% of variation, and the scatter plot showed that these taxa were clearly divided into two groups by PC1 and PC2 (Fig. 7). Group 1 include four taxa distributed in the south and east of QTP (E. gerardiana, E. likiangensis, E. saxatilis and E. saxatilis var. mairei), which show a distinct ecological niche with higher isothermality (BIO3) and precipitation in the warmest quarter (BIO18) and a lower temperature seasonality (BIO4), and group 2 comprise the remaining nine taxa. Although the anovas detected significant differences of 10 bioclim variables among the 13 taxa, the Tukey‘s HSD tests indicated that only two bioclim variables (BIO3 and BIO4) were significantly differentiated between group 1 and group 2 species (P < 0.01; Table S4 and Fig. S5A, B, Supporting information), with the exception of a nonsignificant difference between group 1 species and E. minuta in BIO4. In addition, E. likiangensis and E. saxatilis var. mairei show significant differences from group 2 species in BIO12 and BIO18 (P < 0.001 for HSD; Fig. S5C, D, Supporting information). The two taxa occur in a moist climate with higher annual precipitation (BIO12) and precipitation of the warmest quarter (BIO18).
For comparisons between the allotetraploids and their putative progenitors, the PCA revealed two components (PC1 and PC2) that collectively explained 55.63–67.74% of variation (Table S2, Supporting information). All but one (E. saxatilis) of the allotetraploids show ecological divergence from their putative progenitors (Fig. 8). Two divergence patterns were found: (i) the allotetraploids, including E. likiangensis and E. saxatilis var. mairei, occupy separate ecological niches from their putative maternal progenitors; and (ii) the allotetraploids, including E. glauca, E. intermedia and E. sinica, show separate ecological niches from one of their putative paternal progenitors but have partially overlapped ecological niches with their other putative progenitors. For example, the niche of E. glauca is completely different from that of E. minuta, but is slightly overlapped with that of E. monosperma and more overlapped with those of E. equisetina and E. przewalskii (Fig. 8a); E. intermedia has a much wider niche than E. minuta and a separate niche from E. equisetina, and is clearly differentiated from E. equisetina, E. monosperma and E. przewalskii along PC2 (Fig. 8b). The allotetraploid E. saxatilis has a similar niche with its putative maternal progenitor E. gerardiana (Fig. 8d).
Significant ecological divergences between the allotetraploids and their putative progenitors were also detected by the anova and Tukey's tests of the two principal components (PC1, PC2) and the 10 bioclim variables (Tables 2 and S4). Results of the anova indicate that the mean squares of all comparisons among species are higher than those within species with the exception of the comparison between E. saxatilis and E. gerardiana, and almost all interspecific divergences are significant (P < 0.05; Table S4, Supporting information). The Tukey's test detected significant niche divergence between the allotetraploids and their putative progenitors in 2–7 bioclim variables (Table 2). The divergence between E. glauca and E. minuta and between E. glauca and E. monosperma occurred, respectively, in seven and five bioclim variables, which is consistent with the results of the PCA. Compared to E. minuta and E. monosperma, E. glauca is higher in mean temperature of the wettest quarter (BIO8) and annual potential evapotranspiration (PET), intermediate in annual precipitation (BIO12), and lower in precipitation seasonality (BIO15) and precipitation of the warmest quarter (BIO18) (Fig. S5E–I, Supporting information). Relative to their putative progenitors, E. intermedia is lower in temperature seasonality (BIO4) and BIO8 but higher in BIO12 and precipitation of the coldest quarter (BIO19), with the exception of the comparison between it and E. minuta (Fig. S5J–M, Supporting information), and E. likiangensis is higher in isothermality (BIO3), BIO12, BIO18 and PET but lower in BIO4, with the exception of the comparison between it and E. minuta in BIO4 (Fig. S5N–R, Supporting information). Like E. likiangensis, E. saxatilis var. mairei is higher in BIO3, BIO12 and BIO18 except the comparison between it and E. gerardiana in BIO3 (Fig. S5S–U, Supporting information). The allotetraploid E. sinica also shows niche divergence from most of its putative progenitors and tends to occupy a niche with higher BIO12, BIO15 and PET (Fig. S5V–X, Supporting information).
Table 2.
Statistical differences of ecological differentiation between allotetraploids and their putative progenitors based on the Tukey‘s HSD test for the two principal components revealed by the PCA and the 10 bioclim variables
| Allotetraploids | Putative progenitors | PC1 | PC2 | BIO2 | BIO3 | BIO4 | BIO8 | BIO12 | BIO14 | BIO15 | BIO18 | BIO19 | PET |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E. glauca | E. equisetina | ns | ns | — | ns | ns | ns | ** | ns | ns | *** | — | ns |
| E. minuta | ns | *** | — | * | * | ** | *** | ns | * | *** | — | ** | |
| E. monosperma | ns | *** | — | ns | ns | * | * | ns | ** | *** | — | ** | |
| E. przewalskii | ns | ns | — | ns | ns | ns | ns | ns | ns | ns | — | ns | |
| E. intermedia | E. equisetina | — | *** | — | *** | *** | *** | *** | ns | ns | *** | *** | *** |
| E. minuta | — | ns | — | ns | ns | ns | ns | ns | ns | *** | ** | ** | |
| E. monosperma | — | *** | — | ns | *** | *** | *** | ns | *** | *** | *** | *** | |
| E. przewalskii | — | *** | — | ** | *** | *** | *** | ns | ns | ns | *** | ns | |
| E. likiangensis | E. equisetina | *** | *** | — | *** | *** | ** | *** | ns | *** | *** | ns | * |
| E. minuta | * | ns | — | *** | ns | ns | *** | ns | ns | *** | ns | ** | |
| E. monosperma | *** | *** | — | *** | *** | ns | *** | ns | ns | *** | ns | *** | |
| E. saxatilis var. mairei | E. equisetina | *** | *** | ns | *** | *** | ** | *** | ns | * | *** | — | ns |
| E. gerardiana | ns | ns | ns | ns | ns | ns | *** | ns | ns | *** | — | ns | |
| E. minuta | ns | ns | ns | ** | ns | ns | *** | ns | ns | *** | — | ns | |
| E. monosperma | *** | *** | ns | *** | *** | ns | *** | ns | ns | *** | — | ns | |
| E. sinica | E. equisetina | *** | ns | ns | ns | ns | ns | *** | *** | *** | * | *** | ** |
| E. minuta | ns | *** | ns | ns | * | *** | ns | ns | ns | ns | ns | ** | |
| E. monosperma | ** | ns | ns | ns | ns | *** | *** | ns | *** | ns | ns | *** | |
| E. przewalskii | * | *** | ns | ns | ns | ns | *** | ns | *** | *** | * | ns |
ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001; —, P > 0.05 in the anovas.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Discussion
A high frequency of allopolyploid speciation in Ephedra in the QTP and its vicinities
All tetraploid species of Ephedra from the QTP and adjacent regions originated by allopolyploid speciation. Phylogenetic analysis of single‐/low‐copy nuclear genes can be effective in revealing allopolyploid parental lineages (e.g. Ge et al. 1999; Ferguson & Sang 2001; Kim et al. 2008; Cai et al. 2012; Yang et al. 2012a, b; Kelly et al. 2013), when an allopolyploid species, particularly of recent origin, has paralogous sequences inherited from its two or more parental species. These paralogues can place an allotetraploid in different parental clades. In the present study, the FCM measurement of the ploidy levels in a number of individuals from different populations indicates that six (46%) of the studied 13 Ephedra taxa, including E. glauca, E. intermedia, E. likiangensis, E. saxatilis, E. saxatilis var. mairei and E. sinica, are tetraploids (Table 1). In both single‐copy nuclear gene trees (LFY and DDB2), each of the six taxa contains two types of sequences that are distributed in different major clades A and C or B and C (Figs 5 and 6), strongly suggesting an allopolyploid origin involving diploid parents from these clades. Compared to clades A and C, clade B only contains tetraploids. This could be attributed to the extinction or lack of sampling of diploids in this clade.
The three tetraploid species E. glauca, E. intermedia and E. sinica exhibit nuclear gene alleles in both clades A and C (Fig. 5). These species are mainly distributed in northern China, and their chlorotypes belong to lineage I which is also confined to northern China (Figs 1 and 2). In particular, all of them share chlorotypes with diploid E. przewalskii rather than with diploid E. regeliana (Fig. 1). Therefore, the three tetraploid species possibly originated from hybridization with diploids most closely related to E. przewalskii (in clade A of Fig. 5 and lineage I of Fig. 2) from northern China as the maternal parents and diploids most closely related to the widespread E. equisetina–E. minuta–E. monosperma (in clade C of Fig. 5 and lineage II in Fig. 2) as the paternal parents (see the reticulate network in Fig. 6).
In contrast, the three tetraploid taxa E. likiangensis, E. saxatilis and E. saxatilis var. mairei exhibit nuclear gene alleles in both clades B and C (Fig. 5). The chlorotypes of E. saxatilis belong to lineage III (Fig. 2), and are narrowly distributed in southern QTP (Fig. 1), corresponding to the geographical distribution of this species. Hence, this species possibly originated by allopolyploidy with a maternal progenitor from southern QTP, very likely the diploid cytotype of E. gerardiana (in lineage III of Fig. 2 and clade C of Fig. 5), and a paternal progenitor from clade B (Fig. 6). However, as mentioned earlier, the ancient diploids in clade B could be currently extinct. The chlorotypes of E. likiangensis belong to lineage II with a relatively wide distribution (Figs 1 and 2), and thus, its maternal progenitor could be a diploid species in this lineage, such as E. equisetina, E. minuta and E. monosperma (with nuclear gene alleles in clade C), whereas its paternal progenitor should belong to clade B (Figs 5 and 6). The E. saxatilis var. mairei harbours a high frequency of chlorotypes of lineage III and a much lower frequency of chlorotypes of lineage II (Figs 1 and 2). Therefore, the maternal progenitor of this taxon could be diploid species from the two lineages such as E. gerardiana (2x), E. equisetina, E. minuta and E. monosperma, and its paternal progenitor could be from clade B (Figs 5 and 6).
As discussed above, the widespread diploids of E. equisetina–E. minuta–E. monosperma or their progenitors may have played important roles in allopolyploid speciation of Ephedra in the QTP and adjacent regions. Interestingly, most of the above six tetraploid taxa harbour two or more chlorotypes, even chlorotypes from different main lineages such as in E. saxatilis var. mairei (Figs 1 and 2). This may indicate multiple origins of a tetraploid taxon, divergence of chlorotypes subsequent to polyploidy, or interspecific chloroplast introgression.
Both diploid and tetraploid cytotypes are present in E. gerardiana, E. przewalskii and E. regeliana (Table 1). The tetraploids of E. przewalskii only have A2‐type sequences of the nuclear gene LFY (in clade A, Fig. 5) as its diploids (Figs 3 and 4), but do not share all chlorotypes with the diploids (Table 1; Fig. 1), although their chlorotypes all belong to lineage I (Fig. 2). In particular, the chlorotype H3 is shared between the tetraploids of E. przewalskii and E. glauca (Table 1; Fig. 1). Therefore, the tetraploids of E. przewalskii may be autopolyploids or young allopolyploids with very closely related parental species. In the LFY tree (Fig. 5), the tetraploids of E. regeliana harbour two subtypes of sequences in clade A, including A2 with the diploids and tetraploids of E. przewalskii and the tetraploid E. glauca, and A3 with the diploid E. regeliana and two tetraploid species (E. sinica and E. intermedia). Notably, the diploids and tetraploids of E. regeliana do not share chlorotypes (Table 1; Fig. 1). Thus, the tetraploids of E. regeliana could be allopolyploids that might have originated by hybridization between the diploids of E. regeliana and E. przewalskii (Fig. 6), although an origin by hybridization between an autotetraploid of E. regeliana and a tetraploid of other species cannot be ruled out. In addition, there could be incomplete linage sorting or introgression at the tetraploid level. However, it is unknown why this tetraploid population (XAB) only exhibits private chlorotypes. The diploids and tetraploids of E. gerardiana occur in clade C and clade B+ clade C, respectively (Fig. 5), and they do not share chlorotypes (Table 1; Fig. 1). This may also suggest an allopolyploid origin or a complicated origin of the tetraploid cytotype like in the tetraploid E. regeliana (Fig. 6), although the two cytotypes of E. gerardiana do not show clear morphological difference. To understand the origin of the tetraploids of this species, more samples need to be studied in the future.
Polyploid evolution in Ephedra and its correlation with some biological and ecological features
Evolution of polyploids
Of the 36 Ephedra species that have been cytologically studied, 24 are polyploids or contain polyploid cytotypes (66% of species). Although intraspecific polyploidy has been documented in about half of the species, the results of some early cytological studies need to be checked carefully as mentioned earlier, and more population samples are necessary to study the ploidy levels of a species. Notably, all polyploids in the 24 species are tetraploids with the exception of an octoploid cytotype reported in E. funerea and E. gerardiana (Table S3, Supporting information). According to the available information (Table S3, Supporting information), tetraploids are present in about 75% of the Old World species and 62.5% of the New World species. Our present study found that all tetraploid Ephedra species from the QTP and neighbouring areas, excluding the species with both diploid and tetraploid populations, are allotetraploids. Therefore, we may conclude that allotetraploidy is a dominant mode of speciation in Ephedra, although the origin of other polyploids in this genus, especially from Europe and America, needs to be further studied. In fact, all of the remaining three polyploid species from other gymnosperm lineages were also deduced to be allopolyploids by Yang et al. (2012a, b).
The biological features related to polyploid speciation
The high percentage of polyploids in Ephedra could be related to some attributes of the genus such as a shrub habit, vegetative propagation, a relatively high rate of unreduced gamete formation, and a relatively low basic chromosome number and small genome size (at diploid level) for a gymnosperm (Leitch & Leitch 2012). Previous studies in angiosperms indicate that perennial herbaceous plants with clonality are more likely to form polyploids, which can persist for long periods of time until a suitable mate is found (Otto & Whitton 2000; Leitch & Bennett 2007; Otto 2007; Husband et al. 2013; Weiss‐Schneeweiss et al. 2013). Grif (2000) reported that plant taxa with a small DNA value per genome have a high percentage of polyploidy and show higher ploidy levels, and Wood et al. (2009) found that the generic base count (the minimum number of chromosomes reported in a genus) is negatively associated with polyploid incidence in angiosperms. In particular, the union of unreduced gametes has been considered as the most likely way of polyploid formation in plants (Harlan & de Wet 1975; de Wet 1980; Soltis et al. 2010; Brownfield & Köhler 2011), and the different rates of unreduced gamete formation could influence the establishment of polyploids in different lineages (Bretagnolle & Thompson 1996; Ramsey & Schemske 1998, 2002; Ramsey 2006; Younis et al. 2014). According to the karyomorphological study of Ephedra based on pollen germination, three of five tetraploid species exhibited unreduced pollen grains that accounted for 2–5% of the total amount (Mehra 1946). Also, dimorphism of pollen size in the same herbarium specimen has been reported from seven species of Ephedra, including E. alata, E. americana, E. aphylla, E. breana, E. chilensis, E. ochreata, E. torreyana, E. trifurca and E. tweediana (Beug 1956; Kedves 1987; Ickert‐Bond et al. 2003). Hence, a relatively high rate of unreduced gamete formation in Ephedra has very likely contributed to the high frequency of polyploids in the genus.
The ecological differentiation associated with polyploid speciation
Polyploids possess the potential for evolving into new species with evolutionary novelty, and some previous studies suggest that ecological divergence may play a prominent role in polyploid speciation (Brochmann et al. 2004; Hijmans et al. 2007; Dušková et al. 2010; McIntyre 2012; Theodoridis et al. 2013). In this study, we also found that ecological divergence was associated with the speciation or divergence of polyploids in Ephedra.
Significant ecological divergences between the allotetraploids and their putative progenitors were detected by the PCAs and the anova and Tukey's tests, with the exception of E. saxatilis (Figs 8 and S5). The allotetraploids have separate ecological niches from their putative maternal progenitors, or occupy separate ecological niches from one of their putative paternal progenitors but have partially overlapped ecological niches with their other putative progenitors (Figs 8 and S5). For example, E. likiangensis and E. saxatilis var. mairei endemic to the QTP inhabit dense grass swards or dwarf‐shrub communities and prefer a moister climate with higher annual precipitation and precipitation of the warmest quarter than their putative progenitors E. equisetina, E. minuta and E. monosperma (Table S5; Fig. S5P, Q, T, U, Supporting information); E. intermedia favours conditions that have lower temperature seasonality and mean temperature of the wettest quarter and favours higher annual precipitation and precipitation of the coldest quarter, compared to its putative maternal progenitor E. przewalskii and paternal progenitors E. equisetina and E. monosperma (Table 2; Figs 8b and S5J–M).
The allotetraploid E. sinica has a vast distribution from northwestern China northward to Mongolia and Russia and eastward up to the Gulf of Bohai. Similarly, the allotetraploid E. intermedia is widely distributed in Irano‐Turanian and central Asian floristic regions. Both these species widely occur in open areas such as zonal steppe, desert steppe or coarse‐textured‐skeletal‐sandy soils (Our unpublished observations; Freitag & Maier‐Stolte 1994; Fu et al. 1999). However, their putative progenitors E. equisetina, E. minuta and E. monosperma are usually subordinate components of acrophytia with narrow distributions in dry rocky slopes (Table S5; Fig. S2, Supporting information). It is particularly interesting that most of the allotetraploid taxa we found, including E. intermedia, E. likiangensis, E. saxatilis var. mairei and E. sinica, have adapted to a moister climate with a higher annual precipitation than their putative progenitors (Fig. S5L, P, T, V, Supporting information), considering that the genus Ephedra generally occurs in dry habitats.
The ecological divergence associated with allotetraploidy or its divergence could be associated with new habitats triggered by the fast uplift of the QTP and the Asian aridification in the Middle to Late Miocene (An et al. 2001; Guo et al. 2002; Spicer et al. 2003; Dupont‐Nivet et al. 2007; Jiang & Ding 2008; Royden et al. 2008), during which the tetraploid Ephedra species originated (Qin et al. 2013). Such a scenario has been argued for angiosperm polyploids (Wen et al. 2014), for example Aconitum subgenus Lycoctonum (Yuan & Yang 2006), Anaphalis (Meng et al. 2010), Buddleja (Chen et al. 2007), Leontopodium (Meng et al. 2012), Meconopsis (Yang et al. 2012a), Melampodium (Rebernig et al. 2010), Rheum (Liu et al. 2010), Rhodiola (Zhang et al. 2014) and Silene (Luo et al. 2011).
Both geographical distributions and ecological niches of some diploid species, such as E. equisetina, E. monosperma and E. regeliana (2x), mostly or partially overlap (Figs 7 and S2), which could have provided opportunities for the hybridization between these species, giving rise to allotetraploid species by allopolyploid speciation. To investigate whether ecological divergence had driven polyploid speciation of Ephedra in the QTP and neighbouring areas, more molecular markers or phylogenomic approaches should be used to resolve the parental species of these polyploids, and the molecular mechanisms underlying the adaptation to a specific ecological factor could be explored in future studies.
X.Q.W. designed the study. H.W. and Z.M. performed the laboratory work. M.M.W. and A.L.Q. contributed plant materials. H.W., X.Q.W., Z.M. and J.H.R. analysed the data. X.Q.W. and H.W. wrote the article.
Data accessibility
DNA sequences: GenBank accessions KT033145–KT033395.
Sequence alignments, tree files and climate data: Dryad (doi:10.5061/dryad.kb508).
Supporting information
Fig. S1. Histograms of fluorescence intensity in diploid Ephedra equisetina (I) and tetraploid E. intermedia (II).
Fig. S2. Sampling locations for the statistic analysis of ecological differentiation.
Fig. S3. The ML tree of Ephedra constructed by the cpDNA haplotypes.
Fig. S4. Majority‐rule consensus trees obtained from maximum parsimony analyses of the LFY and DDB2 data sets.
Fig. S5. Boxplots showing variation of bioclim variables among the studied Ephedra species.
Table S1. The cpDNA haplotypes detected in the studied Ephedra species and their GenBank accessions.
Table S2. A summary of the principal component analysis.
Table S3. The chromosome numbers in Ephedra L.
Table S4. Results of the anova and Tukey's HSD tests for the two principal components revealed by the PCA and the 10 bioclim variables.
Table S5. Morphological characteristics, habitat preference and distribution of Ephedra species from the QTP and adjacent regions.
Acknowledgements
We thank Drs. Yu‐Zhi Cun and Fu‐Sheng Yang, and Mr. Cai‐Yuan Qiao for their help in sample collection; Ms. Wan‐Qing Jin for her assistance in DNA sequencing. We also thank the Royal Botanic Garden, Edinburgh, for providing the sample of Ephedra foeminea for DNA analysis. We appreciate the Subject Editor and the three anonymous reviewers for their insightful comments and suggestions on the manuscript. This study was supported by the National Natural Science Foundation of China (Grant Nos 31330008, 31170197 and 30730010) and the Chinese Academy of Sciences (KJZD‐EW‐L07 and the CAS/SAFEA International Partnership Program for Creative Research Teams).
References
- Ahuja MR (2005) Polyploidy in gymnosperms: revisited. Silvae Genetica, 54, 59–69. [Google Scholar]
- An Z, Kutzbach JE, Prell WL, Porter SC (2001) Evolution of Asian monsoons and phased uplift of the Himalaya‐Tibetan plateau since Late Miocene times. Nature, 411, 62–66. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Rohl A (1999) Median‐joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16, 37–48. [DOI] [PubMed] [Google Scholar]
- Beug HJ (1956) Pollendimorphismus bei Ephedra . Naturwissenschaften, 43, 332–333. [Google Scholar]
- Birol I, Raymond A, Jackman SD et al (2013) Assembling the 20 Gb white spruce (Picea glauca) genome from whole‐genome shotgun sequencing data. Bioinformatics, 29, 1492–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong JK, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature, 422, 433–438. [DOI] [PubMed] [Google Scholar]
- Bretagnolle F, Thompson JD (1996) An experimental study of ecological differences in winter growth between sympatric diploid and autotetraploid Dactylis glomerata . Journal of Ecology, 84, 343–351. [Google Scholar]
- Brochmann C, Brysting AK, Alsos IG et al (2004) Polyploidy in arctic plants. Biological Journal of the Linnean Society, 82, 521–536. [Google Scholar]
- Brownfield L, Köhler C (2011) Unreduced gamete formation in plants: mechanisms and prospects. Journal of Experimental Botany, 62, 1659–1668. [DOI] [PubMed] [Google Scholar]
- Cai DY, Rodriguez F, Teng YW et al (2012) Single copy nuclear gene analysis of polyploidy in wild potatoes (Solanum section Petota). BMC Evolutionary Biology, 12, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Sun WB, Sun H (2007) Ploidy variation in Buddleja L. (Buddlejaceae) in the Sino‐Himalayan region and its biogeographical implications. Botanical Journal of the Linnean Society, 154, 305–312. [Google Scholar]
- Cutler HC (1939) Monograph of the North American species of the genus Ephedra . Annals of the Missouri Botanical Garden, 26, 373–428. [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Methods, 9, 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dray S, Dufour AB, Chessel D (2007) The ade4 package‐II: two‐table and K‐table methods. R News, 7, 47–52. [Google Scholar]
- Dupont‐Nivet G, Krijgsman W, Langereis CG, Abels HA, Dai S, Fang XF. (2007) Tibetan plateau aridification linked to global cooling at the Eocene‐Oligocene transition. Nature, 445, 635–638. [DOI] [PubMed] [Google Scholar]
- Dušková E, Kolář F, Sklenář P et al (2010) Genome size correlates with growth form, habitat and phylogeny in the Andean genus Lasiocephalus (Asteraceae). Preslia, 82, 127–148. [Google Scholar]
- Fawcett JA, Maere S, Van de Peer Y (2009) Plants with double genomes might have had a better chance to survive the Cretaceous‐Tertiary extinction event. Proceedings of the National Academy of Sciences of the United States of America, 106, 5737–5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett JA, Van de Peer Y, Maere S (2013) Significance and biological consequences of polyploidization in land plant evolution In: Plant Genome Diversity 2: Physical Structure, Behaviour and Evolution of Plant Genomes(eds Leitch IJ, Greilhuber J, Doležel J, Wendel JF.), pp. 277–293. Springer, Wien. [Google Scholar]
- Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution, 39, 783–791. [DOI] [PubMed] [Google Scholar]
- Ferguson D, Sang T (2001) Speciation through homoploid hybridization between allotetraploids in peonies (Paeonia). Proceedings of the National Academy of Sciences of the United States of America, 98, 3915–3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florin CR (1933) Über einige neue oder wenig bekannte asiatische Ephedra‐Arten der Sektion Pseudobaccatae Stapf.‐Kongl. Svenska Vetenskapsakad. Handle, 12, 1–49. [Google Scholar]
- Freitag H, Maier‐Stolte M (1994) Ephedraceae In: Chorology of Trees and Shrubs in Southwest Asia and Adjacent Regions 10(ed. Browicz K.), pp. 5–16, 39–52. Polish Scientific Publishers, Poznan. [Google Scholar]
- Fu L, Yu Y, Riedl H (1999) Ephedraceae In: Flora of China(eds Wu CY, Raven P.), pp. 97–101. Science Press, Beijing, and Missouri Botanical Garden Press, St. Louis. [Google Scholar]
- Gascuel O (1997) BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Molecular Biology and Evolution, 14, 685–695. [DOI] [PubMed] [Google Scholar]
- Ge S, Sang T, Lu BR, Hong DY (1999) Phylogeny of rice genomes with emphasis on origins of allotetraploid species. Proceedings of the National Academy of Sciences of the United States of America, 49, 14400–14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glennon KL, Ritchie ME, Segraves KA (2014) Evidence for shared broad‐scale climatic niches of diploid and polyploid plants. Ecology Letters, 17, 574–582. [DOI] [PubMed] [Google Scholar]
- Grif VG (2000) Some aspects of plant karyology and karyosystematics. International Review of Cytology, 196, 131–175. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O (2003) A simple, fast and accurate method to estimate large phylogenies by maximum‐likelihood. Systematic Biology, 52, 696–704. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard J, Lefort V, Anisimova M, Hordijk W, Gascuel O. (2010) New algorithms and methods to estimate maximum‐likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology, 59, 307–321. [DOI] [PubMed] [Google Scholar]
- Guo ZT, Ruddiman WF, Hao QZ et al (2002) Onset of Asian desertification by 22 Myr ago inferred from loess deposits in China. Nature, 416, 159–163. [DOI] [PubMed] [Google Scholar]
- Hall TA (1999) BioEdit: a user‐friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95–98. [Google Scholar]
- Harlan JR, de Wet JMJ (1975) On Ö. Winge and a prayer: the origins of polyploidy. The Botanical Review, 41, 361–390. [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A (2005) Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology, 25, 1965–1978. [Google Scholar]
- Hijmans RJ, Gavrilenko T, Stephenson S, Bamberg J, Salas A, Spooner DM (2007) Geographical and environmental range expansion through polyploidy in wild potatoes (Solanum section Petota). Global Ecology and Biogeography, 16, 485–495. [Google Scholar]
- Huang J, Price RA (2003) Estimation of the age of extant Ephedra using chloroplast rbcL sequence data. Molecular Biology and Evolution, 20, 435–440. [DOI] [PubMed] [Google Scholar]
- Huang J, Giannasi DE, Price RA (2005) Phylogenetic relationships in Ephedra (Ephedraceae) inferred from chloroplast and nuclear DNA sequences. Molecular Phylogenetics and Evolution, 35, 48–59. [DOI] [PubMed] [Google Scholar]
- Husband BC, Baldwin SJ, Suda J (2013) The incidence of polyploidy in natural plant populations: major patterns and evolutionary processes In: Plant Genome Diversity 2: Physical Structure, Behaviour and Evolution of Plant Genomes(eds Leitch IJ, Greilhuber J, Doležel J, Wendel JF.), pp. 255–276. Springer, Wien. [Google Scholar]
- Ickert‐Bond SM, Skvarla JJ, Chissoe WF (2003) Pollen dimorphism in Ephedra L. (Ephedraceae). Review of Palaeobotany and Palynology, 124, 325–334. [Google Scholar]
- Ickert‐Bond SM, Rydin C, Renner SS (2009) A fossil‐calibrated relaxed clock for Ephedra indicates an Oligocene age for the divergence of Asian and New World clades and Miocene dispersal into South America. Journal of Systematics and Evolution, 47, 444–456. [Google Scholar]
- Jiang HY (2006) Study on the genetic relationship analysis and determination of main medical composition for Ephedra. Bachelor's degree Thesis, Gansu Agricultural University, Lanzhou, China.
- Jiang H, Ding Z (2008) A 20 Ma pollen record of East‐Asian summer monsoon evolution from Guyuan, Ningxia, China. Palaeogeography, Palaeoclimatology, Palaeoecology, 265, 30–38. [Google Scholar]
- Jiao YN, Wickett NJ, Ayyampalayam S et al (2011) Ancestral polyploidy in seed plants and angiosperms. Nature, 473, 97–100. [DOI] [PubMed] [Google Scholar]
- Kedves M (1987) LM and EM studies on pollen grains of recent Welwitschia mirabilis Hook. and Ephedra species. Acta Botanica Hungarica, 33, 81–103. [Google Scholar]
- Kelly LJ, Leitch AR, Clarkson JJ, Knapp S, Chase MW (2013) Reconstructing the complex evolutionary origin of wild allopolyploid tobaccos (Nicotiana section Suaveolentes). Evolution, 67, 80–94. [DOI] [PubMed] [Google Scholar]
- Khoshoo TN (1959) Polyploidy in gymnosperms. Evolution, 13, 24–39. [Google Scholar]
- Kim ST, Sultan SE, Donoghue MJ (2008) Allopolyploid speciation in Persicaria (Polygonaceae): insights from a low‐copy nuclear region. Proceedings of the National Academy of Sciences of the United States of America, 105, 12370–12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong H, Chen Q, Ma J (2001) A study on karyotypes of two species in Ephedra . Journal of Lanzhou University, 37, 101–103. [Google Scholar]
- Krejcikova J, Sudova R, Lucanova M et al (2013) High ploidy diversity and distinct patterns of cytotype distribution in a widespread species of Oxalis in the Greater Cape Floristic Region. Annals of Botany, 111, 641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubitzki K (1990) The Families and Genera of Vascular Plants. Springer, Berlin. [Google Scholar]
- Leitch IJ, Bennett MD (2007) Genome size and its uses: the impact of flow cytometry In: Flow Cytometry with Plant Cells: Analysis of Genes, Chromosomes and Genomes(eds Dolezel J, Greilhuber J, Suda J.), pp. 153–176. Wiley, Weinheim. [Google Scholar]
- Leitch AR, Leitch IJ (2012) Ecological and genetic factors linked to contrasting genome dynamics in seed plants. New Phytologist, 194, 629–646. [DOI] [PubMed] [Google Scholar]
- Leitch IJ, Hanson L, Winfield M, Parker J, Bennett MD (2001) Nuclear DNA C‐values complete familial representation in gymnosperms. Annals of Botany, 88, 843–849. [Google Scholar]
- Liu R, Wang A, Tian X, Wang D, Liu J (2010) Uniformity of karyotypes in Rheum (Polygonaceae), a species‐rich genus in the Qinghai‐Tibetan Plateau and adjacent regions. Caryologia, 63, 82. [Google Scholar]
- Lott M, Spillner A, Huber KT, Moulton V (2009a) PADRE: a package for analyzing and displaying reticulate evolution. Bioinformatics, 25, 1199–1200. [DOI] [PubMed] [Google Scholar]
- Lott M, Spillner A, Huber KT, Petri A, Oxelman B, Moulton V (2009b) Inferring polyploid phylogenies from multiply‐labeled gene trees. BMC Evolutionary Biology, 9, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Liu D, Xu B, Nie ZL, Sun H, Li ZM. (2011) A karyological study of six species of Silene L. (Caryophyllaceae) from the Hengduan Mountains, SW China. Caryologia, 64, 10–13. [Google Scholar]
- Madlung A (2013) Polyploidy and its effect on evolutionary success: old questions revisited with new tools. Heredity, 110, 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayrose I, Zhan SH, Rothfels CJ et al (2011) Recently formed polyploid plants diversify at lower rates. Science, 333, 1257. [DOI] [PubMed] [Google Scholar]
- McIntyre PJ (2012) Polyploidy associated with altered and broader ecological niches in the Claytonia perfoliata (Portulacaceae) species complex. American Journal of Botany, 99, 655–662. [DOI] [PubMed] [Google Scholar]
- Mehra PN (1946) A study of the karyotypes and the occurrence of diploid male gametophytes in some species of the genus Ephedra . Proceedings of the National Academy of Sciences of the United States of America, 16, 259–286. [Google Scholar]
- Meng Y, Sun H, Yang YP, Nie ZL (2010) Polyploidy and new chromosome counts in Anaphalis (Asteraceae: Gnaphalieae) from the Qinghai‐Tibet Plateau of China. Journal of Systematics and Evolution, 48, 58–64. [Google Scholar]
- Meng Y, Nie ZL, Sun H, Yang YP (2012) Chromosome numbers and polyploidy in Leontopodium (Asteraceae: Gnaphalieae) from the Qinghai‐Tibet Plateau of SW China. Caryologia, 65, 87–93. [Google Scholar]
- Murray BG (2013) Karyotype variation and evolution in gymnosperms In: Plant Genome Diversity 2: Physical Structure, Behaviour and Evolution of Plant Genomes(eds Leitch IJ, Greilhuber J, Doležel J, Wendel JF.), pp. 231–244. Springer, Wien. [Google Scholar]
- Neale DB, Wegrzyn JL, Stevens KA et al (2014) Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies. Genome Biology, 15, R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystedt B, Street NR, Wetterbom A et al (2013) The Norway spruce genome sequence and conifer genome evolution. Nature, 497, 579–584. [DOI] [PubMed] [Google Scholar]
- Otto SP (2007) The evolutionary consequences of polyploidy. Cell, 131, 452–462. [DOI] [PubMed] [Google Scholar]
- Otto SP, Whitton J (2000) Polyploid incidence and evolution. Annual Review of Genetics, 34, 401–437. [DOI] [PubMed] [Google Scholar]
- Page JT, Huynh MD, Liechty ZS et al (2013) Insights into the evolution of cotton diploids and polyploids from whole‐genome re‐sequencing. G3: Genes, Genomes, Genetics, 3, 1809–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin AL, Wang MM, Cun YZ et al (2013) Phylogeographic evidence for a link of species divergence of Ephedra in the Qinghai‐Tibetan Plateau and adjacent regions to the Miocene Asian aridification. PLoS One, 8, e56243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2014) A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna: URL http://www.R-project.org/. [Google Scholar]
- Ramsey J (2006) Unreduced gametes and neopolyploids in natural populations of Achillea borealis (Asteraceae). Heredity, 98, 143–150. [DOI] [PubMed] [Google Scholar]
- Ramsey J (2011) Polyploidy and ecological adaptation in wild yarrow. Proceedings of the National Academy of Sciences of the United States of America, 108, 7096–7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey J, Schemske DW (1998) Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annual Review of Ecology and Systematics, 29, 467–501. [Google Scholar]
- Ramsey J, Schemske DW (2002) Neopolyploidy in flowering plants. Annual Review of Ecology and Systematics, 33, 589–639. [Google Scholar]
- Rebernig CA, Weiss‐Schneeweiss H, Schneeweiss GM et al (2010) Quaternary range dynamics and polyploid evolution in an arid brushland plant species (Melampodium cinereum, Asteraceae). Molecular Phylogenetics and Evolution, 54, 594–606. [DOI] [PubMed] [Google Scholar]
- Renny‐Byfield S, Kovarik A, Kelly LJ et al (2013) Diploidization and genome size change in allopolyploids is associated with differential dynamics of low‐ and high‐copy sequences. The Plant Journal, 74, 829–839. [DOI] [PubMed] [Google Scholar]
- Rogers SO, Bendich AJ (1985) Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Molecular Biology, 5, 69–76. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19, 1572–1574. [DOI] [PubMed] [Google Scholar]
- Roulin A, Auer PL, Libault M et al (2012) The fate of duplicated genes in a polyploid plant genome. The Plant Journal, 73, 143–153. [DOI] [PubMed] [Google Scholar]
- Royden LH, Burchfiel BC, van der Hilst RD (2008) The geological evolution of the Tibetan plateau. Science, 321, 1054–1058. [DOI] [PubMed] [Google Scholar]
- Rydin C, Korall P (2009) Evolutionary relationships in Ephedra (Gnetales), with implications for seed plant phylogeny. International Journal of Plant Sciences, 170, 1031–1043. [Google Scholar]
- Schönswetter P, Lachmayer M, Lettner C et al (2007) Sympatric diploid and hexaploid cytotypes of Senecio carniolicus (Asteraceae) in the Eastern Alps are separated along an altitudinal gradient. Journal of Plant Research, 120, 721–725. [DOI] [PubMed] [Google Scholar]
- Simillion C, Vandepoele K, Van Montagu MCE, Zabeau M, Van de Peer Y (2002) The hidden duplication past of Arabidopsis thaliana . Proceedings of the National Academy of Sciences of the United States of America, 99, 13627–13632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Albert VA, Leebens‐Mack J et al (2009) Polyploidy and angiosperm diversification. American Journal of Botany, 96, 336–348. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Buggs RJA, Doyle JJ, Soltis PS (2010) What we still don't know about polyploidy. Taxon, 59, 1387–1403. [Google Scholar]
- Soltis DE, Segovia‐Salcedo MC, Jordon‐Thaden I et al (2014a) Are polyploids really evolutionary dead‐ends (again)? A critical reappraisal of Mayrose et al. (2011). New Phytologist, 202, 1105–1117. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Visger CJ, Soltis PS (2014b) The polyploidy revolution then…and now: stebbins revisited. American Journal of Botany, 101, 1057–1078. [DOI] [PubMed] [Google Scholar]
- Spicer RA, Harris NBW, Widdowson M et al (2003) Constant elevation of southern Tibet over the past 15 million years. Nature, 421, 622–624. [DOI] [PubMed] [Google Scholar]
- Suda J, Trávníček P (2006) Reliable DNA ploidy determination in dehydrated tissues of vascular plants by DAPI flow cytometry—new prospects for plant research. Cytometry Part A, 69, 273–280. [DOI] [PubMed] [Google Scholar]
- Swofford DL (2002) PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods) 4.0 Beta. Sinauer Associates, Sunderland, Maryland. [Google Scholar]
- Theodoridis S, Randin C, Broennimann O, Patsiou T, Conti E (2013) Divergent and narrower climatic niches characterize polyploid species of European primroses in Primula sect. Aleuritia. Journal of Biogeography, 40, 1278–1289. [Google Scholar]
- Trabucco A, Zomer RJ, Bossio DA, Van Straaten O, Verchot LV (2008) Climate change mitigation through afforestation reforestation: a global analysis of hydrologic impacts with four case studies. Agriculture, Ecosystems & Environment, 126, 81–97. [Google Scholar]
- Van de Peer Y, Fawcett JA, Proost S, Sterck L, Vandepoele K (2009) The flowering world: a tale of duplications. Trends in Plant Science, 14, 680–688. [DOI] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD (2000) The origins of genomic duplications in Arabidopsis . Science, 290, 2114–2117. [DOI] [PubMed] [Google Scholar]
- Vrána J, Cápal P, Bednářová M, Doležel J (2014) Flow cytometry in plant research: a success story. Applied Plant Cell Biology, 22, 395–430. [Google Scholar]
- Wang XQ, Ran JH (2014) Evolution and biogeography of gymnosperms. Molecular Phylogenetics and Evolution, 75, 24–40. [DOI] [PubMed] [Google Scholar]
- Weiss‐Schneeweiss H, Emadzade K, Jang TS, Schneeweiss GM (2013) Evolutionary consequences, constraints and potential of polyploidy in plants. Cytogenetic and Genome Research, 140, 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, Zhang JQ, Nie ZL, Zhong Y, Sun H (2014) Evolutionary diversifications of plants on the Qinghai‐Tibetan Plateau. Frontiers in Genetics, 5, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt T (1993) A new variety of Ephedra torreyana (Ephedraceae) from west Texas and Chihuahua, with notes on hybridization in the E. torreyana complex. Phytologia, 74, 141–150. [Google Scholar]
- de Wet JM (1980) Origins of polyploids In: Polyploidy: Biological Relevance(ed. Lewis WH.), pp. 3–15. Plenum Press, New York. [Google Scholar]
- Wheeler B (2010) lmPerm: Permutation tests for linear models. R package version, 1.
- Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH. (2009) The frequency of polyploid speciation in vascular plants. Proceedings of the National Academy of Sciences of the United States of America, 106, 13875–13879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JL, Li S, Jiang HY, Gao YH, Niu JY (2009) Staining and slide‐preparing technique of mitotic chromosomes and application in karyotype determination of Ephedra . China Journal of Chinese Materia Medica, 34, 2725–2729. [PubMed] [Google Scholar]
- Yang Y (2002) Systematics and Evolution of Ephedra L. (Ephedraceae) from China. Chinese Academy of Sciences, Beijing. [Google Scholar]
- Yang Y, Wang Q (2013) The earliest fleshy cone of Ephedra from the early Cretaceous Yixian formation of northeast China. PLoS One, 8, e53652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Fu DZ, Zhu GH (2003) A new species of Ephedra (Ephedraceae) from China. Novon, 13, 153–155. [Google Scholar]
- Yang FS, Qin AL, Li YF, Wang XQ (2012a) Great genetic differentiation among populations of Meconopsis integrifolia and its implication for plant speciation in the Qinghai‐Tibetan Plateau. PLoS One, 7, e37196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZY, Ran JH, Wang XQ (2012b) Three genome‐based phylogeny of Cupressaceae s.l.: further evidence for the evolution of gymnosperms and Southern Hemisphere biogeography. Molecular Phylogenetics and Evolution, 64, 452–470. [DOI] [PubMed] [Google Scholar]
- Younis A, Hwang YJ, Lim KB (2014) Exploitation of induced 2n‐gametes for plant breeding. Plant Cell Reports, 33, 215–223. [DOI] [PubMed] [Google Scholar]
- Yuan Q, Yang QE (2006) Polyploidy in Aconitum subgenus Lycoctonum (Ranunculaceae). Botanical Journal of the Linnean Society, 150, 343–353. [Google Scholar]
- Zhang JQ, Meng SY, Wen J, Rao GY (2014) Phylogenetic relationships and character evolution of Rhodiola (Crassulaceae) based on nuclear ribosomal ITS and plastid trnL‐F and psbA‐trnH sequences. Systematic Botany, 39, 441–451. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Histograms of fluorescence intensity in diploid Ephedra equisetina (I) and tetraploid E. intermedia (II).
Fig. S2. Sampling locations for the statistic analysis of ecological differentiation.
Fig. S3. The ML tree of Ephedra constructed by the cpDNA haplotypes.
Fig. S4. Majority‐rule consensus trees obtained from maximum parsimony analyses of the LFY and DDB2 data sets.
Fig. S5. Boxplots showing variation of bioclim variables among the studied Ephedra species.
Table S1. The cpDNA haplotypes detected in the studied Ephedra species and their GenBank accessions.
Table S2. A summary of the principal component analysis.
Table S3. The chromosome numbers in Ephedra L.
Table S4. Results of the anova and Tukey's HSD tests for the two principal components revealed by the PCA and the 10 bioclim variables.
Table S5. Morphological characteristics, habitat preference and distribution of Ephedra species from the QTP and adjacent regions.
Data Availability Statement
DNA sequences: GenBank accessions KT033145–KT033395.
Sequence alignments, tree files and climate data: Dryad (doi:10.5061/dryad.kb508).


