Summary
Long noncoding RNAs (lncRNAs) represent a key class of cellular regulators, involved in the modulation and control of multiple biological processes. Distinct classes of lncRNAs are now known to be induced by host cytokines following viral infections. Current evidence demonstrates that lncRNAs play essential roles at the host‐pathogen interface regulating viral infections by either innate immune responses at various levels including activation of pathogen recognition receptors or by epigenetic, transcriptional, and posttranscriptional effects. We review the newly described mechanisms underlying the interactions between lncRNAs, cytokines, and metabolites differentially expressed following viral infections; we highlight the regulatory networks of host antiviral responses and emphasize the need for interdisciplinary research between lncRNA biology and immunology to deepen understanding of viral pathogenesis.
Keywords: cell metabolism, epigenetic regulation, innate immune response, long noncoding RNA, pathogen recognition receptor
Abbreviations
- CARDs
caspase activation and recruitment domains
- CTD
C‐terminal domain
- EVA71
enterovirus A71
- GOT2
glutamic‐oxaloacetic transaminase 2
- HBV
hepatitis B virus
- HCMV
human cytomegalovirus
- IFN‐α
interferon alpha
- IKK
IkB kinase
- IRF3
interferon regulatory factor 3
- IRGs
immune response genes
- ISGs
interferon‐stimulated genes
- KSHV
Kaposi's sarcoma‐associated herpesvirus
- L‐ASP
l‐aspartic acid
- lncRNA
long noncoding RNA
- MDA5
melanoma differentiation‐associated gene 5
- miRNA
microRNA
- NADPH
nicotinamide adenine dinucleotide phosphate
- ncRNAs
noncoding RNAs
- NEMO
NF‐κB essential modulator
- P
phosphorylation
- piRNAs
piwi‐interacting RNA
- PRRs
pathogen recognition receptors
- RIG‐I
retinoic acid‐inducible gene‐I
- SARS‐CoV
severe acute respiratory syndrome‐associated coronavirus
- TLRs
Toll‐like receptors
- TMEV
Theiler's murine encephalomyelitis virus
- TNF‐α
tumor necrosis factor‐α
- VSV
vesicular stomatitis virus
- α‐KG
α‐ketoglutarate
1. INTRODUCTION
In recent years, with the advent of microRNA (miRNA) research and the development of high‐throughput sequencing technologies, numerous novel long noncoding RNAs (lncRNAs) have been discovered and functionally annotated.1 The discovery of lncRNAs dates back to 1990 when the first lncRNA, H19, was identified in mammals and the first functionally characterized lncRNA, Xist, was implicated in the X chromosome inactivation in the female embryos of eutherian mammals in 1991.2, 3 In 2002, a large‐scale discovery of lncRNAs was achieved by sequence analyses of 60 770 murine full‐length complementary DNAs (cDNAs).4 As of November 2018, approximately 16 066 human lncRNAs and nearly 29 566 transcripts have been released by GenCode (Version 29).5 However, we still possess an extremely limited understanding of lncRNAs, which is exemplified by the fact that less than 200 lncRNAs have been investigated and functionally annotated thus far.6 New genetic and cell regulatory mechanisms have been revealed by the discoveries and functional annotation of noncoding RNAs, which have made a major impact on the development of life sciences. Studies have found that lncRNAs participate in the regulation of various biological processes by interactions with both nucleic acids and proteins and are also intimately involved in the development of malignancy and in host response to infectious diseases.7, 8 In addition, research has also revealed that viral infections can cause abnormal lncRNA expression in host cells and that some lncRNAs are also encoded by viruses.9, 10 These host and virally encoded lncRNAs regulate infections through various regulatory mechanisms, such as participating in innate immune responses and altering cellular metabolic pathways.11, 12 Therefore, increased investigation and the functional characterization of the lncRNA repertoire elicited during the innate immune response and alterations in cellular metabolism following viral infection will help to elucidate the host defense mechanisms against viruses and provide potential targets for therapeutic intervention and a deeper understanding of viral pathogenesis.
1.1. Origin and types of lncRNAs
lncRNAs are arbitrarily defined as a class of RNA molecules greater than 200 nucleotides (nts) in length, transcribed by RNA polymerase II or III, without a protein‐coding open reading frame. Similar to protein‐coding mRNAs, most lncRNAs are 5′ capped, spliced,13 and can be either polyadenylated or nonpolyadenylated (ie, are bimorphic).14, 15 lncRNAs are distributed in both the nucleus and cytoplasm, typically have low expression levels, and exhibit poor conservation between species, although their structure is well conserved.16, 17 The abundance of lncRNA transcripts is typically at least an order of magnitude lower than that of mRNAs, with an estimated 80% of lncRNAs tissue‐specific.18, 19, 20 According to the positional relationship between lncRNAs and their target genes on the chromosome, lncRNAs can be classified into five groups21, 22: (1) sense lncRNAs: located on the positive DNA strand and partially overlap with protein‐coding genes (see Figure 1A); (2) intronic lncRNAs: located entirely within an intron sequence of a target gene (Figure 1B); (3) long intergenic noncoding RNAs (lincRNAs): coexist on the same strand and are located upstream of the target gene with a spacing of less than 10 kb (Figure 1C); (4) antisense lncRNAs: located on the opposite strand of their target gene, and partially, or completely, overlap with the coding region (Figure 1D); and (5) bidirectional lncRNAs: located on the negative strand of the target gene with a spacing of less than 1 kb whereby transcriptional orientation is opposing and the transcripts do not overlap (Figure 1E).
Figure 1.
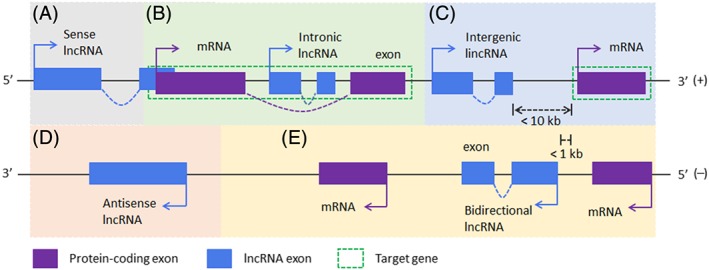
Current classification of long noncoding RNAs (lncRNAs) based on the positional relationship between lncRNAs and adjacent protein‐encoding genes. A, Sense lncRNAs partially overlap with protein‐coding genes. B, Intronic lncRNAs are located on an intron of the target gene. C, Intergenic lncRNAs are located between two protein‐coding genes (<10 kb apart from the target gene). D, Antisense IncRNAs are located on the negative strand and partially or completely overlap with the protein‐coding gene. E, Bidirectional lncRNAs are located on the negative strand, which are transcribed in the opposite direction, but does not overlap with the target gene. Arrows indicate the direction of gene transcription. Genomic DNA plus (+) and minus (−) strands are labeled accordingly. The target gene of sense lncRNA, intronic lncRNA, and antisense lncRNA is shown in panel B and that of intergenic lncRNA and bidirectional lncRNA is shown in panel C
1.2. The currently known functional properties of lncRNAs
It has been estimated that about 70% of the human genome can be transcribed, among which, only approximately 1% to 2% of the transcripts encode proteins, and the remaining 98% are noncoding RNAs (ncRNAs).5, 23 Among the ncRNAs, tRNAs and rRNAs are well known for their structural and scaffolding roles, while the vast majority of the remaining ncRNA fraction have been regarded as “transcriptional noise,” and, until recently, their biology and roles in transcriptional processes are largely ignored. Gradually, studies have confirmed the functional importance of ncRNAs and that they play pivotal roles in cellular and metabolic activities,8 with small ncRNAs and lncRNAs now known to account for an estimated 20% to 42% of ncRNAs.24 Small ncRNAs, such as miRNAs, are involved in the regulation of a variety of cellular activities leading to gene silencing by base pairing with specific complementary nucleotide sequences of the mRNAs of their target genes.25 In contrast, lncRNAs greater than 200 nts in length exert their functions by a greater variety of known mechanisms through interactions with either target proteins or nucleic acids via their specific and complex secondary structures.26, 27, 28, 29, 30 These known mechanisms can be classified into seven distinct groups: (1) lncRNA binds to transcription factors to form a transcription complex, which affects downstream gene expression by inhibiting RNA polymerase II or by mediating chromatin remodeling and histone modifications31, 32, 33, 34; (2) lncRNA forms a complementary double‐stranded region with the mRNA transcript of its target gene, thereby interfering with the cleavage, transport, and/or translation of the targeted mRNA35; (3) lncRNA competes with a target gene for transcription factors, thereby silencing the target gene expression; (4) lncRNA acts as a scaffold to bring multiple proteins together to regulate gene transcription36; (5) lncRNA binds to a specific protein, altering the activity or cytoplasmic localization of the protein37; (6) a lncRNA gene transcribes an additional ncRNA, such as pseudogenes and circular RNAs, and because of the similarity to the mRNA sequence of the target gene, the ncRNA can operate as a molecular sponge that attracts miRNAs, thereby insulating the transcription and translation of the functional genes from miRNA effects and enabling cross talk between different ncRNA classes38; and finally (7) lncRNAs can be transcribed as the precursors of small molecule RNAs, such as miRNAs and piwi‐interacting RNAs (piRNAs).39 In summary, lncRNAs can regulate gene expression through DNA and RNA modifications by a variety of modes, such as by serving as signal molecules, decoy molecules, guide molecules, molecular sponges, and scaffolds.27, 40 lncRNAs participate in multiple biological processes such as cell differentiation, carcinogenesis, and immune responses and play important roles in the development and, thereby, potentially in the prevention of human diseases.28, 29, 41, 42 In the past decade, lncRNA biology has become a new frontier in life sciences and attracted many researchers to the field. In this review, we will focus on the present state of understanding of lncRNAs in the context of innate antiviral immunity.
2. DIFFERENTIAL EXPRESSION OF LNCRNAS
2.1. Viral infection and differential expression of lncRNAs
Virus infection elicits changes in the expression of cellular lncRNAs. By means of gene chip analyses, Winterling and colleagues revealed 17 differentially expressed lncRNAs in human alveolar epithelial cells (A549) infected with influenza virus A/WSN/33 (H1N1).43 Subsequently, using high‐throughput sequencing, numerous research groups identified thousands of differentially expressed lncRNAs following viral infection. In order to systematically study the regulatory effects of the neurotropic enterovirus A71 (EVA71) infection on lncRNAs, Yin et al performed a comprehensive analysis of EVA71‐infected rhabdomyosarcoma (RD) cells and observed differential expression of greater than 4800 lncRNAs.44 Similarly, Meng et al identified 8541 differentially expressed lncRNAs in EVA71‐infected human peripheral blood mononuclear cells.45 Also, using the RNA sequencing (RNA‐Seq) technique, Shi et al identified a total of 760 and 1210 lncRNAs that were upregulated and downregulated, respectively, in RD cells infected with Coxsackievirus A16 (CVA16). Interestingly, there was an apparent bias in the lncRNA class differentially regulated upon CVA16 infection as approximately 43.6% and 22.3% were intergenic and sense lncRNAs, respectively, demonstrating a significant enrichment upon viral infection of the former lncRNA class.46 Peng and colleagues have analyzed the lung transcriptome in severe acute respiratory syndrome‐associated coronavirus (SARS‐CoV)‐infected mice and reported that the expression of 504 annotated and 1406 unannotated lncRNAs had significant changes.47 In addition, other viral families, such as Zika virus,48 hepatitis B virus (HBV),49, 50, 51 and hepatitis C virus,52 have been shown upon host infection to lead to differential expression of host lncRNAs compared with uninfected controls. Therefore, the differential expression of lncRNAs in various cell types following viral infection appears to be a widespread phenomenon and not a property attributable to one viral family or host target cell.
Surprisingly, some viruses also encode their own lncRNAs to modulate the host immune response. For example, human cytomegalovirus (HCMV) encodes a 2.7 kb lncRNA, β2.753; β2.7 inhibits the induction of stress responses and apoptosis in infected cells, which facilitates HCMV replication.9 Gammaherpesviruses transcribe their own structurally and functionally intricate lncRNAs, which modulate cellular and viral gene expression to maintain viral latency or to induce lytic reactivation.54 In addition, a 3.91 kb lncRNA M3‐04 generated by gammaherpesviruses regulates viral replication in mice by interacting with antisense miRNAs and the latency gene M2.55 Other herpesviruses also encode multiple functional lncRNAs, such as the Epstein‐Barr virus encoding ncRNA, BamH I‐A rightward transcripts,56 Kaposi's sarcoma‐associated herpesvirus (KSHV) encoding UCA1,57 and herpesvirus saimiri encoding U‐rich RNAs.58 Additionally, the 3′‐untranslated region (UTR) of the flavivirus RNA genome is capable of transcribing an active lncRNA, termed the subgenomic flavivirus RNA (sfRNA), which protects viral RNAs from degradation by the host nuclease Xrn1 and suppresses antiviral RNA interference (RNAi) in infected human cells in culture and, also, interestingly, in mosquitoes by direct interaction with the RNAi machinery.10, 59 Therefore, viral infection can induce differential expression of a series of lncRNAs in cells, suggesting that lncRNAs may play important functional roles in viral infection.
2.2. Cytokines and differential expression of lncRNAs
Cytokines play an essential role in regulating the development, maturation, and differentiation of nonspecific innate immune cells. lncRNAs are found to be deregulated upon viral infection or following interferon (IFN) treatment, and some lncRNAs can modulate viral infection in an IFN‐dependent manner.60 To investigate whether IFN regulates the transcription of lncRNAs, Carnero et al stimulated human hepatoma (Huh7) cells with high doses of type I interferon alpha (IFN‐α) and found that IFN‐α significantly upregulated the expression of lncRNAs ISR2, ISR8, and ISR12.61 Similarly, treatment of Huh7 cells by IFN‐α2 or type III IFN‐λ induced an upregulation of the expression of lncRNAs BST2 and ISG15. Inhibition experiments showed that lncRNA BST2 is a positive regulator of the host antiviral factor BST2.62 Tumor necrosis factor‐α (TNF‐α) also induces the differential expression of the lncRNA Lethe, which responds to NF‐κB stimulation and reduces inflammation.63 The lncRNA NKILA which is induced by proinflammatory cytokines binds to NF‐κB/IκB and directly masks the phosphorylation motifs of IκB, thereby inhibiting IKK‐induced IκB phosphorylation and downstream NF‐κB activation, and thereby preventing excessive activation of the NF‐κB pathway in epithelial cells.64 Therefore, the inductive effect of cytokines is closely related to and modulated by the differential expression of host lncRNAs.
2.3. Metabolites and differential expression of lncRNAs
Metabolites not only function within cellular metabolic pathways but have also been implicated in the regulation and differential expression of lncRNAs. The specific β3‐adrenergic receptor agonist, CL‐316,243, induces the differentiation of brown adipocytes, and a total of 21 differentially expressed lncRNAs have been detected in both cellular and adipose tissues.65 This has led to the identification of the lncRNA Blnc1 as a key regulator of brown cell differentiation and function. Blnc1 forms a nuclear ribonucleoprotein complex with the transcription factor EBF2, to stimulate and activate the thermogenic adipose program.65 In addition, lncRNA Blnc1 itself is a target of EBF2, and through a feedforward regulatory loop, the cells and tissues differentiate into a pyrogenic phenotype that favors adipogenesis.
The prostate‐specific lncRNA PCGEM1 can be induced by androgens, which promotes glucose uptake. Coupling with the pentose phosphate pathway, PCGEM1 promotes the synthesis of nucleic acids and lipids and balances the redox reaction by generating NADPH.66 In addition, lncRNA PCGEM1 affects glutamine metabolism at the transcriptional level. Taken together, lncRNA PCGEM1 is a key transcriptional regulator of cellular metabolic pathways.
In the process of viral infection, lncRNAs can act as mediators to link viral infection to innate immunity and cellular metabolism. lncRNAs are involved in not only cytokine‐mediated innate antiviral immune responses but also the regulation of cellular metabolic pathways, altering the efficiency of viral replication in cells. The perturbation of the transcription of ncRNAs elicited by virus infection often leads to disturbance of homeostasis, resulting in disease.67 lncRNAs regulate viral infections by modifying innate immune responses and cellular metabolic pathways at various levels including the activation of pathogen recognition receptors (PRRs), epigenetic modulation, and transcriptional and posttranscriptional modification.29
3. REGULATION OF VIRAL REPLICATION BY LNCRNA‐MEDIATED INNATE IMMUNITY
3.1. Activation of pathogen recognition receptor‐related signals by lncRNAs
Innate immunity is the first line of defense against viral infections and plays a key role in identification of viral RNAs, induction of interferon‐stimulated genes (ISGs), and proinflammatory responses in the early stages of the infection process.68, 69 Whether viral infection activates innate immunity is dependent on the activation of PRRs and PRR‐dependent signaling pathways. Virus infection can also activate retinoic acid‐inducible gene‐I (RIG‐I), Toll‐like receptors (TLRs), melanoma differentiation‐associated gene 5 (MDA5), and Nod‐like receptor (NLR) pathways,70 which in turn activate interferon regulatory factors, such as IRF3, IRF7, and the major proinflammatory transcription factor NF‐κB.71, 72 The lncRNAs Lethe and lnc‐Lsm3b have been shown to bind directly to immunosensors and block downstream signaling of the innate immune pathway (Table 1). The lncRNA Lethe competitively binds with and blocks NF‐κB from binding to specific promoter regions of its target genes, thereby preventing a cascade of signal transduction events.63 The IFN‐inducible, host‐derived lncRNA, lnc‐Lsm3b, directly competes with vesicular stomatitis virus (VSV) RNA for the binding to the RIG‐I monomer, limiting the conformational transition of the RIG‐I PRR. Feedback in the later stages of the innate immune response inactivates RIG‐I (Figure 2A),73 thereby inhibiting the development of virus infection or activating type I interferon‐mediated immune cells, such as dendritic, NK, and T cells, either directly or indirectly eliminating viral infections.87 In addition, lnc‐Lsm3b interferes with the interaction between caspase activation and recruitment domain (CARD) proteins, present in numerous innate immune effectors and the mitochondrial antiviral signaling (MAVS) protein by stabilizing the interaction between the N‐terminal CARD of RIG‐I and the helicase domain. It has been demonstrated that TRIM25‐mediated ubiquitination of RIG‐I K63 is also inhibited by lnc‐Lsm3b upon RNA virus stimulation. Taken together, these findings implicate lnc‐Lsm3b as a potent negative regulator of innate RIG‐I signaling pathway upon RNA virus infections.73
Table 1.
Host lncRNAs implicated in the innate antiviral immune response
| lncRNAs | Name | Stimuli | Functions/Mechanisms | Refs. |
|---|---|---|---|---|
| PRR‐related signal transduction | Lethe | TNF‐α | lncRNA Lethe competitively binds with NF‐κB and inhibits the interaction between NF‐κB and the promoter, thereby preventing downstream signal transduction. | Rapicavoli et al63 |
| Lsm3b | VSV | Binding of lnc‐Lsm3b restricts the conformational change of the RIG‐I protein, which inactivates the function of RIG‐I, thereby limiting the production of type I interferon IFN‐β. | Jiang et al73 | |
| Pretranscriptional level | lincRNA‐Cox2 | Pam3CKS4, LPS | lincRNA‐Cox2 interacts with hnRNPA/B or the SWI/SNF complex to regulate the expression of the innate immune‐associated cytokines, chemokines, and ISGs that mediate inflammatory responses. | Hu et al74 and Carpenter et al75 |
| NeST | Theiler's virus | NeST binds to WDR5, which is implicated in nucleosomal core histone methylation, thereby negatively regulating the transcription of type II interferons in CD8+ T cells. | Gomez et al76 | |
| NRAV | IAV | NRAV inhibits the transcriptional initiation of antiviral genes by affecting the histone modifications of MxA and IFITM3 (H3K4me3 and H3K27me3). | Ouyang et al77 | |
| Aspro5 | HIV | Aspro5 recruits a variety of histone modification enzymes to form a heterochromatic structure inhibiting the transcriptional activity of the viral promoter. | Saayman et al78 | |
| Transcriptional level | NRON | HIV | A complex of the lincRNA NRON, a scaffold protein, and three NFAT kinases forms a large cytoplasmic RNA‐protein scaffold to inhibit the function of the transcription factor NFAT. | Sharma et al36 |
| NEAT1 | HIV | NEAT1 interacts with the NONO protein to promote the formation of paraspeckles in the nucleus and to deactivate the splicing factor SFPQ in the transcription of IL‐8. | Imamura et al37 | |
| lincRNA‐EPS | LPS | lincRNA‐EPS controls nucleosome localization and inhibits IRG transcription by interacting with heterogeneous nuclear ribonucleoprotein L. | Atianand et al79 | |
| sfRNA | Flaviviruses | sfRNA prevents the activation of the IFN‐β promoter and the initiation of transcription by inhibiting the phosphorylation of IRF3. | Pijlman et al80 and Chang et al81 | |
| sfRNA | Dengue virus | sfRNA binds to the host RNA binding protein and CAPRIN1 and inhibits the transcription of IFITM2 and PKR; sfRNA also binds to the ubiquitin ligase TRIM25 and negatively regulates RIG‐I expression. | Manokaran et al82 and Bidet et al83 | |
| Posttranscriptional level | NEAT1 | HIV | NEAT1 inhibits nuclear export of HIV‐I mRNAs and reduces replication efficiency in viral cells. | Zhang et al84 |
| ISG20 | Sendai virus; IAV; poly I:C | Acts as a competitive endogenous RNA (ceRNA) of ISG20 mRNA, lncRNA ISG20 binds to miRNA‐326, releases its transcriptional repression of ISG20 mRNA, and activates the innate immune response. | Chai et al85 | |
| Metabolic level | ACOD1 | VSV | ACOD1 binds to GOT2, an important aminotransferase during metabolism in the cytoplasm, and elevates the activity of GOT2, which in turn regulates the metabolism of L‐aspartate and α‐ketoglutarate, thereby promoting viral replication. | Wang et al86 |
Figure 2.
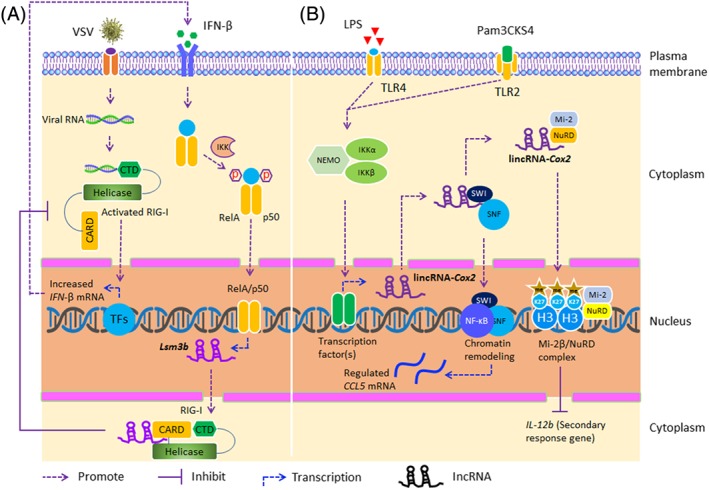
Long noncoding RNAs (lncRNAs) regulate innate immune responses through activation of RIG‐I and via deregulation of epigenetic mechanisms. A, Under normal circumstances in the resting state of macrophages, RIG‐I exists in the autoinhibited state with the CARD2 domain of RIG‐I binding to the insertion region of the helicase. With the invasion of the 5′‐triphosphate double‐stranded viral RNA, the CTD senses and binds the viral RNA, which leads to the structural transformation of RIG‐I into an open conformation and the release of CARDs, thereby mediating downstream signaling and induction of IFN‐β expression. After sensing the extracellular IFN‐β signal, TLR2 upregulates the expression of lnc‐Lsm3b, which inhibits RIG‐I‐mediated immune responses by binding to RIG‐I and suppresses the production of IFN‐β and other anti‐inflammatory factors. B, TLR2 and TLR4 induce the expression of lincRNA‐Cox2 through the NF‐κB signal pathway upon LPS or Pam3CKS4 stimulation. On the one hand, lincRNA‐Cox2 promotes the binding of the Mi‐2/NuRD complex in the promoter region of IL‐12b, decreasing histone H3K27 acetylation and increasing H3K27 dimethylation, which leads to lincRNA‐Cox2‐mediated transinhibition of the secondary response gene IL‐12b. Alternatively, upon LPS stimulation, lincRNA‐Cox2 is assembled into the SWI/SNF complex and acts to recruit NF‐κB to the complex, which plays an important role in the regulation of SWI/SNF‐associated chromatin remodeling and the transactivation of advanced inflammatory response genes
3.2. lncRNAs regulate innate immunity through epigenetic regulation of gene transcription
Epigenetic regulation is a reversible modification of genetic functions by modifying expression without alteration of the underlying nucleotide sequence during the pretranscriptional regulation of eukaryotic genes.88 lncRNAs can regulate gene expression at the epigenetic level through chromatin remodeling pathways.29, 89 For example, lipopolysaccharide (LPS) (or the synthetic triacylated lipopeptide Pam3CKS4) can induce formation of a lincRNA‐Cox2 and SWI/SNF complex which modulates the assembly of NF‐κB subunits to the SWI/SNF complex in macrophages, triggering chromatin remodeling and transactivation of the late‐primary inflammatory response genes in response to microbial challenge (Figure 2B and Table 1).74, 75 lincRNA‐Cox2 regulates the expression of numerous ISGs, such as OAS1, OAS2, OASL, IFI204, and ISG15 through the binding of hnRNP‐A/B or A2/B1.75, 90 Surprisingly, the expression of the adjacent gene Cox2, which is also an essential immunoregulatory factor, is not affected by lincRNA‐Cox2 raising the possibility that another layer of transcriptional regulation is potentially affected by ncRNAs and other protein effectors remains undiscovered.
It is well established that many lncRNA‐mediated epigenetic regulations occur through modification of the nucleosomal core histone H3. Trimethylation of the N‐terminal lysines of H3 at residues 4 and 27 (H3K4me3 and H3K27me3, respectively) can determine the structural state of the chromatin (ie, “loose” euchromatin or “tight” heterochromatin), and correspondingly, the expression of the cis‐linked genes can be switched to “on” or “off.”76, 91 In A549 cells, by regulating the abundance of the lncRNA negative regulator of antiviral response (NRAV), located on chromosome 12q24.31, H3K27me3 is significantly enriched, thereby promoting the expression of ISGs, such as the immune effector proteins MxA and IFITM3. In turn, innate antiviral immunity is activated, and viral genome replication is inhibited.77 In addition, the lincRNA‐Cox2 promotes the assembly of the reconstituted Mi‐2 nucleosome and the deacetylase (Mi‐2/NuRD) repressor complex in the promoter region of the secondary response gene IL‐12b, resulting in a decrease of histone H3K27 acetylation and an elevation of H3K27 dimethylation, respectively. This epigenetic mechanism contributes to the lincRNA‐Cox2‐mediated transinhibition of the IL‐12b gene.92
The lncRNA NeST can promote the formation of H3K4me3 by forming a complex with the WD (Trp‐Asp) repeat‐containing protein WDR5, thereby repressing the IFN‐γ promoter. This ultimately results in an increase in type II interferon transcript levels in the activated CD8+ T cells.76 Interestingly, this enhancer‐like lncRNA NeST is located adjacent to the IFN‐γ‐encoding locus in both the mouse (Ifng) and human (IFNG) genomes. NeST was initially discovered as a candidate susceptibility locus for infection by Theiler's murine encephalomyelitis virus (TMEV), a single‐stranded, positive sense RNA cardiovirus, family Picornaviridae. The acronym NeST abbreviates in French as nettoie Salmonella pas Theiler's (“cleanup Salmonella not Theiler's”). In both the murine and human genomes, not only is synteny conserved but the NeST lncRNA is also encoded on the opposite DNA strand to that coding for IFN‐γ in both species. Thereby, the survival of TMEV is prolonged in mice, but the pathogenicity of Salmonella enterica is attenuated.76 Whether such host‐derived lncRNAs, such as NeST, represent lncRNA targets with pharmaceutical potential where bacterial infection could be attenuated without predisposing to viral infections in humans is clearly worthy of further study.
Apart from lncRNAs encoded by the human host, lncRNAs derived from the virus itself are also emerging as important elements for epigenetic regulation during infection.93 The HIV‐encoded lncRNA aspro5 recruits the host de novo DNA (cytosine‐5)‐methyltransferase (DNMT3a), the histone deacetylase HDAC1, the histone‐lysine N‐methyltransferase, and the polycomb protein enhancer of zeste homolog 2 (EZH2) to the 5′ long terminal repeat of the integrated proviral DNA. This leads to the formation of H3K9me2, H3K27me3, histone deacetylation, and associated transcriptional inert heterochromatic structures. These changes in the chromatin structure inhibit transcriptional activity of the promoter, thereby silencing the expression of the viral gene and likely contributing to latency and immune evasion of the integrated retroviral genome.78
3.3. lncRNAs regulate innate immunity at the transcriptional level
Transcriptional regulation is one of the most important mechanisms in controlling eukaryotic gene expression. lncRNAs can saturate the binding sites of transcription factors and thus inhibit the transcription and expression of IFNs and ISGs.94, 95 On the other hand, lncRNAs can also recruit transcription factors to promoters and enhancers to initiate transcription of innate immune genes.82, 96, 97 For example, lncRNA‐DC has been shown to regulate the activity of a number of transcription factors. When cells are stimulated by pathogens, lncRNA‐DC, which has been found to be specifically expressed in dendritic cells, binds to the transcription factor STAT3 and blocks the binding site for SHP1. The sustained activation of STAT3 induced by the phosphorylation of tyrosine residue 705 promotes the expression of genes involved in dendritic cell differentiation.98 lncRNA‐DC is also implicated in the immune response to HBV and acts by reducing the concentration of secreted TNF‐α, IL‐6, IL‐12, and IFN‐γ, as well as increasing the IL‐1β concentration in dendritic cells.99
lncRNAs can form a complex with heterogeneous nuclear ribonucleoproteins (hnRNPs) to regulate the expression of ISGs. lncRNA #32 is one such antiviral factor; knockdown of lncRNA #32 significantly reduces the mRNA level of the interferon‐inducible proteins OASL, IP‐10, RSAD2, and APOBEC3A. lncRNA #32 forms a stable complex with hnRNPU, which activates transcription factor 2 (ATF2) to promote the transcription of ISG effectors.100 Similarly, the complex of lncRNA NRON with GTPase‐activating proteins and three NFAT kinases in the cytoplasm of T cells serves as a molecular scaffold for the activation of the nuclear factor NFAT. Upon stimulation, activated NFAT is released from the complex and promotes nuclear transport, thereby initiating gene transcription (Table 1).36 In addition, by reducing or increasing the abundance of lncRNA NRON (an inhibitor of NFAT) through the early viral protein Nef and the late protein Vpu, respectively, the function of NFAT is controlled to achieve a subtle balance between HIV proliferation and cell death.101
In addition, some lncRNAs, such as the lncRNA THRIL (linc‐1992), NEAT1, and lincRNA‐EPS, can indirectly regulate the transcription of immune‐related genes. TNF‐α is a critical cytokine that activates NF‐κB‐mediated inflammatory responses. THRIL forms a complex with hnRNPL, which in turn activates the TNF‐α transcription by binding to the TNF‐α promoter.90 TNF‐α regulates THRIL expression through a negative feedback. The expression of additional cytokines and chemokines, such as IL‐8, CCL1, CSF1, and CXCL10, is also regulated by the lncRNA THRIL.
NEAT1 is an important regulator of antiviral responses, whose expression is elevated by viral infections including HIV‐1, Japanese encephalitis virus, rabies virus, influenza A virus (IAV), herpes simplex virus, and Hantavirus.37, 102, 103 In the cytoplasm, NEAT1 interacts with the NONO protein to promote the formation of paraspeckles in the nucleus. Paraspeckles are subnuclear structures, termed ribonucleoprotein bodies, found in the interchromatin spaces in nuclei and are thought to regulate gene expression by inhibiting nuclear RNA export. The targeting of paraspeckles by retroviruses may serve to impede host responses to viral infection and help establish latency. NEAT1 also functions in the relocalization of the splicing factor SFPQ from the promoter region of the antiviral cytokine IL‐8 to the paraspeckle. In this way, the inhibition of transcription by SFPQ is released, leading to the induction of IL‐8 expression and inhibition of HIV‐1 replication (Figure 3A).37
Figure 3.
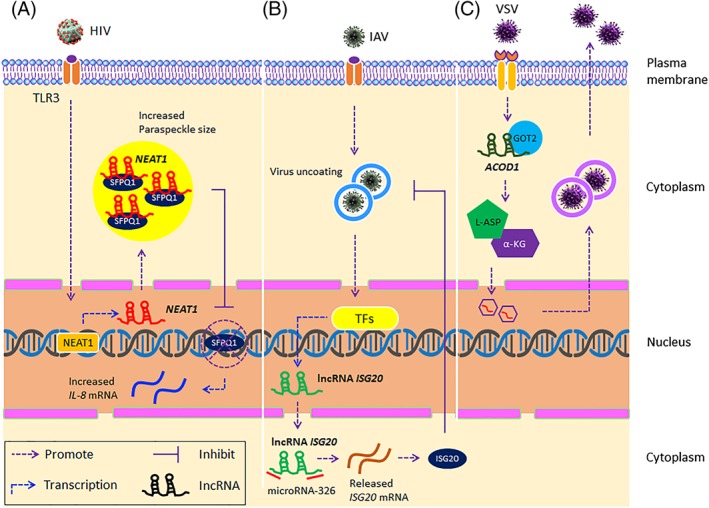
Additional roles of long noncoding RNAs (lncRNAs) in innate antiviral immunity. A, In A549 cells, NEAT1 relocates the negative regulatory factor SFPQ from the IL‐8 promoter to subnuclear structures known as paraspeckles, thereby reducing the inhibitory effect of SFPQ on the IL‐8 promoter and promoting the transcription of IL‐8. B, miRNA‐326 regulates the expression of ISG20 by binding to the 3′‐untranslated region (UTR) of the ISG20 mRNA. lncRNA ISG20 competitively interacts with miRNA‐326, thereby releasing free ISG20 mRNA and facilitating the translation of the ISG20 mRNA. C, lncRNA‐ACOD1 causes a conformational change of GOT2, an essential aminotransferase during cell metabolism in the cytoplasm, at a position close to the substrate binding site, which enhances the catalytic activity of GOT2, thereby increasing the production of L‐ASP and α‐KG, increasing metabolic efficiency, and promoting VSV replication and infection
Without interference from pathogens, lincRNA‐EPS, which possesses immunoregulatory properties in macrophages, accumulates in the regulatory region of immune response genes (IRGs). By interacting with hnRNPL via a CANACA motif located at its 3′‐end, lincRNA‐EPS regulates nucleosome localization and IRG transcription, thereby reducing proinflammatory responses.79
Interestingly, certain virus‐encoded lncRNAs with transcriptional regulatory functions play important roles in host‐virus interactions.78, 104 The flavivirus‐encoded lncRNA, sfRNA, is highly conserved in the 3′‐UTR of the flavivirus genome.80 Studies have shown that sfRNA inhibits the activation of interferon regulatory factor 3 (IRF3) by blocking IRF3 phosphorylation, thereby preventing the activation of the IFN‐β promoter and inhibiting the transcription of IFN‐β.81, 105 Dengue virus serotype 2 (DENV2)‐induced sfRNA inhibits the transcription of IFITM2 and the double‐stranded RNA‐dependent protein kinase R (PKR) by binding to the host RNA‐binding proteins G3BP1, G3BP2, and CAPRIN1, which in turn negatively regulates the expression of IFITM2 and PKR.83 It has also been confirmed that DENV2‐induced sfRNA prevents deubiquitination of the ubiquitin ligase TRIM25 by direct binding to TRIM25, thereby negatively regulating RIG‐I expression and inhibiting innate immune responses.82
KSHV‐infected cells express a lncRNA, termed polyadenylated nuclear RNA (PAN RNA), which modulates the cellular immune response by interacting with both viral and cellular DNA and proteins.106 The PAN RNA plays an important role in controlling viral gene transcription and subversion of the host immune response.107, 108 The PAN lncRNA exerts these effects by interacting with specific histone demethylases (UTX and JMJD3) and physically binds to the KSHV genome to mediate activation of viral gene expression by removing suppressive trimethylated H3K23 marks.109 In addition to interacting with histone demethylases, PAN RNA also interacts with SUZ12 and EZH2 which are components of the Polycomb repression complex 2 (PRC2), a chromatin‐modifying (histone methylation) complex, which represses gene expression at numerous loci.104
Despite these advances, limited information has been revealed to date about virus‐encoded lncRNAs with regulatory functions at the transcriptional level. In the future, the discovery and functional characterization of this lncRNA class of virally encoded transcripts will help in the delineation of virus‐host interactions and may afford the opportunity to develop therapeutics that inhibit virus‐specific lncRNAs.
3.4. Posttranscriptional regulation of innate immunity by lncRNAs
lncRNAs also regulate posttranscriptional modification of mRNAs by involvement in splicing and processing, enhancing mRNA stability, and improving nuclear export efficiency. Upon exposure to amyloid β peptide (Aβ peptide), the expression level of the antisense lncRNA BACE1‐AS, transcribed by the beta‐secretase 1 gene (BACE1), is elevated. This increases the stability of the BACE1 mRNA and generates additional Aβ peptide through a posttranscriptional, feedforward mechanism.110 Some lncRNAs can reduce the nuclear export efficiency of viral mRNAs and inhibit viral replication. HIV‐1 infection can induce a differential expression of the lncRNA NEAT1 in T cells, which serves as a binding scaffold maintaining the integrity of paraspeckles and prevents the transport of spliced HIV‐1 pre‐mRNA to the cytoplasm for translation (Table 1).84 lincRNA‐p21 acts as an inhibitor and reduces the translation efficiency of its target gene by pairing with the target mRNA.111
lncRNA can also minimize the inhibitory effect of miRNAs through additional mechanisms, such as competitive binding or via so‐called sponge effects where miRNAs are titrated from other targets, thereby promoting the translation of the miRNA‐targeted mRNAs.112 In A549 cells treated with poly(I:C) and Sendai virus, lncRNA ISG20, acting as a competitive endogenous RNA (ceRNA), competes with miRNA‐326, resulting in a decreased abundance of miRNA‐326 at the 3′‐UTR of the ISG20 mRNA. In addition, the reduced binding efficiency of microRNA‐326 to ISG20 mRNA also promotes ISG20 translation and inhibits IAV replication (Figure 3B).85 Similarly, lncRNA‐BGL3, acting as a ceRNA, regulates the translation of phosphatase and tensin homolog (PTEN) genes by competitively binding with a series of miRNAs, including miR‐17, miR‐93, miR‐20a, miR‐20b, miR‐106a, and miR‐106b.113
3.5. lncRNAs affect viral replication by regulating cell metabolism
Viruses as obligate intracellular parasites lack the basic metabolic mechanism necessary for replication in host cells. To solve this problem, they hijack host metabolic pathways to derive the resources for viral replication. Hence, viruses act to regulate the metabolism of the host to improve their own replication efficiency. HNF4α is a nuclear receptor that serves as a master regulator of hepatocyte differentiation by functioning in the activation of glycolysis in hepatocytes, inhibition of apoptosis of host cells, and promotion of flavivirus hepatitis C replication.114 It has also been revealed by lipidomics that fatty acid and lipid mediators are critical in the replication of IAV in A549 cells.115, 116 The involvement of lncRNAs in cellular metabolism plays a crucial role in innate immunity induced by viral infection. Wang et al found that deletion of lncRNA ACOD1 greatly attenuates VSV infection mediated by the IFN‐I/IRF3‐independent pathway. In addition, the lncRNA ACOD1 elevates the catalytic activity of glutamic‐oxaloacetic transaminase 2 (GOT2) by directly binding to GOT2 in the cytoplasm, and through which the metabolism of l‐aspartic acid (L‐ASP) and α‐ketoglutarate (α‐KG) is promoted, leading to the immune escape for VSV (Figure 3C).86 Taken together, these studies reveal a new regulatory mechanism of lncRNAs through which host metabolism, rather than classical innate immune pathways, is regulated to promote viral genomic replication and virus proliferation.
4. SUMMARY AND PROSPECTS
lncRNAs interact with proteins and nucleic acids through specific binding to tertiary conformational structures or complementary nucleotide base pairing within genes, respectively, thereby playing essential roles in the regulatory network of innate immune responses and viral replication. Firstly, virus‐induced lncRNAs promote immune responses following a self‐recognition mode. Acting as a potent molecular decoy, the inducible “self” lncRNA blocks the RIG‐I binding site to limit viral RNA‐induced innate immune responses and maintain immune homeostasis. Secondly, ncRNAs can regulate innate immunity at the transcriptional level by activating transcription initiation complexes or competing for transcription factor binding sites. In addition, host lncRNAs can release more free mRNAs and enhance the expression of innate immune‐related genes by increasing mRNA stability and relieving the restrictions imposed by miRNAs. Thirdly, lncRNAs can also promote the assembly of posttranslational histone modification complexes by recruiting histone methyltransferases to regulate the expression of innate immune factors, such as IFNs and ISGs at the epigenetic level. Taken together, these findings provide strong evidence for the role of the ubiquitous and versatile classes of lncRNAs in antiviral regulation. Although many breakthroughs have been achieved towards the understanding of lncRNA‐mediated innate immune responses, the study of lncRNAs is only in its infancy when compared with other ncRNAs, such as microRNAs. IFN‐I‐independent lncRNAs can also promote viral replication by regulating cellular metabolism, and this serves as a new paradigm for the regulation of viral infection other than via effects on innate immunity.86 This mechanism adds a further layer of complexity to the regulatory networks previously established in viral infection. The connection between lncRNAs, metabolism, and virus infection has set a new direction for the study of immune regulatory mechanisms; however, this is far from fully understood, and the discovery and functional annotation of numerous lncRNAs remain to be further explored. With a deeper understanding of host and virally encoded lncRNAs and the regulatory mechanisms involved in the innate antiviral immune response they impact upon, this will likely provide new druggable targets and therapeutic strategies for the treatment of infectious disease.
CONFLICT OF INTEREST
The authors have no competing interest.
FUNDING INFORMATION
The study was supported by the National Natural Science Foundation of China (81802009 and 81601773), the Natural Science Foundation of Shandong Province, China (ZR2018PC012), and the Science and Technology Development Plan of Traditional Chinese Medicine of Shandong Province, China (2015‐259), and W.S. was supported by the Taishan Scholars Program of Shandong Province (ts201511056).
Liu S, Liu X, Li J, et al. Long noncoding RNAs: Novel regulators of virus‐host interactions. Rev Med Virol. 2019;29:e2046 10.1002/rmv.2046
Contributor Information
Zhenjie Zhang, Email: zjzhang@tsmc.edu.cn.
Weifeng Shi, Email: shiwf@ioz.ac.cn.
REFERENCES
- 1. Sun M, Kraus WL. From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocr Rev. 2015;36(1):25‐64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10(1):28‐36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown CJ, Ballabio A, Rupert JL, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349(6304):38‐44. [DOI] [PubMed] [Google Scholar]
- 4. Okazaki Y, Furuno M, Kasukawa T, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full‐length cDNAs. Nature. 2002;420(6915):563‐573. [DOI] [PubMed] [Google Scholar]
- 5. Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775‐1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res. 2011;39(suppl_1):D146‐D151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mercer TRDM, Mattick JS. Long non‐coding RNAs: insights into functions. Genetics. 2009;10:155‐159. [DOI] [PubMed] [Google Scholar]
- 8. Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193(3):651‐669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reeves MB, Davies AA, McSharry BP, Wilkinson GW, Sinclair JH. Complex I binding by a virally encoded RNA regulates mitochondria‐induced cell death. Science. 2007;316(5829):1345‐1348. [DOI] [PubMed] [Google Scholar]
- 10. Funk A, Truong K, Nagasaki T, et al. RNA structures required for production of subgenomic flavivirus RNA. J Virol. 2010;84(21):11407‐11417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ouyang J, Hu J, Chen JL. lncRNAs regulate the innate immune response to viral infection. Wiley Interdiscip Rev RNA. 2016;7(1):129‐143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ma L, Bajic VB, Zhang Z. On the classification of long non‐coding RNAs. RNA Biol. 2013;10:925‐933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Quinn JJ, Chang HY. Unique features of long non‐coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47‐62. [DOI] [PubMed] [Google Scholar]
- 14. Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013;9(6):e1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dykes IM, Emanueli C. Transcriptional and post‐transcriptional gene regulation by long non‐coding RNA. Genomics Proteomics Bioinformatics. 2017;15(3):177‐186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154(1):26‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81(1):145‐166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cabili MN, Trapnell C, Goff L, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915‐1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yan L, Yang M, Guo H, et al. Single‐cell RNA‐Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol. 2013;20(9):1131‐1139. [DOI] [PubMed] [Google Scholar]
- 20. Guttman M, Garber M, Levin JZ, et al. Ab initio reconstruction of cell type‐specific transcriptomes in mouse reveals the conserved multi‐exonic structure of lincRNAs. Nat Biotechnol. 2010;28(5):503‐510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kapusta A, Feschotte C. Volatile evolution of long noncoding RNA repertoires: mechanisms and biological implications. Trends Genet. 2014;30(10):439‐452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Meng XY, Luo Y, Anwar MN, et al. Long non‐coding RNAs: emerging and versatile regulators in host‐virus interactions. Front Immunol. 2017;8:1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. ENCODE Project Consortium , Birney E, Stamatoyannopoulos JA, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799‐816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Atianand MK, Caffrey DR, Fitzgerald KA. Immunobiology of long noncoding RNAs. Annu Rev Immunol. 2017;35(1):177‐198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215‐233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904‐914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guttman M, Rinn JL. Modular regulatory principles of large non‐coding RNAs. Nature. 2012;482(7385):339‐346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Geisler S, Coller J. RNA in unexpected places: long non‐coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14(11):699‐712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298‐1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fitzgerald KA, Caffrey DR. Long noncoding RNAs in innate and adaptive immunity. Curr Opin Immunol. 2014;26:140‐146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nagano T, Mitchell JA, Sanz LA, et al. The air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322(5908):1717‐1720. [DOI] [PubMed] [Google Scholar]
- 32. Gupta RA, Shah N, Wang KC, et al. Long non‐coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071‐1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Arab K, Park YJ, Lindroth AM, et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell. 2014;55(4):604‐614. [DOI] [PubMed] [Google Scholar]
- 34. Zhang XD, Huang GW, Xie YH, et al. The interaction of lncRNA EZR‐AS1 with SMYD3 maintains overexpression of EZR in ESCC cells. Nucleic Acids Res. 2018;46(4):1793‐1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ebralidze AK, Guibal FC, Steidl U, et al. PU.1 expression is modulated by the balance of functional sense and antisense RNAs regulated by a shared cis‐regulatory element. Genes Dev. 2008;22(15):2085‐2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sharma S, Findlay GM, Bandukwala HS, et al. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA‐protein scaffold complex. Proc Natl Acad Sci U S a. 2011;108(28):11381‐11386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Imamura K, Imamachi N, Akizuki G, et al. Long noncoding RNA NEAT1‐dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell. 2014;53(3):393‐406. [DOI] [PubMed] [Google Scholar]
- 38. Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358‐369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494‐1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484‐1488. [DOI] [PubMed] [Google Scholar]
- 41. Satpathy AT, Chang HY. Long noncoding RNA in hematopoiesis and immunity. Immunity. 2015;42(5):792‐804. [DOI] [PubMed] [Google Scholar]
- 42. Zhang Y, Cao X. Long noncoding RNAs in innate immunity. Cell Mol Immunol. 2016;13(2):138‐147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Winterling C, Koch M, Koeppel M, Garcia‐Alcalde F, Karlas A, Meyer TF. Evidence for a crucial role of a host non‐coding RNA in influenza A virus replication. RNA Biol. 2014;11(1):66‐75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yin Z, Guan D, Fan Q, et al. lncRNA expression signatures in response to enterovirus 71 infection. Biochem Biophys Res Commun. 2013;430(2):629‐633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Meng J, Yao Z, He Y, et al. Long non‐coding RNA expression profiles in different severity EV71‐infected hand foot and mouth disease patients. Biochem Biophys Res Commun. 2017;493(4):1594‐1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shi Y, Tu H, Chen X, et al. The long non‐coding RNA expression profile of Coxsackievirus A16 infected RD cells identified by RNA‐seq. Virol Sin. 2016;31(2):131‐141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Peng X, Gralinski L, Armour CD, et al. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. MBio. 2010;1(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hu B, Huo Y, Yang L, et al. ZIKV infection effects changes in gene splicing, isoform composition and lncRNA expression in human neural progenitor cells. Virol J. 2017;14(1):217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. He B, Peng F, Li W, Jiang Y. Interaction of lncRNA‐MALAT1 and miR‐124 regulates HBx‐induced cancer stem cell properties in HepG2 through PI3K/Akt signaling. J Cell Biochem. 2018;120(3):2908‐2918. [DOI] [PubMed] [Google Scholar]
- 50. Hu Z, Huang P, Yan Y, Zhou Z, Wang J, Wu G. Hepatitis B virus X protein related lncRNA WEE2‐AS1 promotes hepatocellular carcinoma proliferation and invasion. Biochem Biophys Res Commun. 2019;508(1):79‐86. [DOI] [PubMed] [Google Scholar]
- 51. Yilmaz Susluer S, Kayabasi C, Ozmen Yelken B, et al. Analysis of long non‐coding RNA (lncRNA) expression in hepatitis B patients. Bosn J Basic Med Sci. 2018;18(2):150‐161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xie Q, Chen S, Tian R, et al. Long noncoding RNA ITPRIP‐1 positively regulates the innate immune response through promotion of oligomerization and activation of MDA5. J Virol. 2018;92(17). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Greenaway PJWG. Nucleotide sequence of the most abundantly transcribed early gene of human cytomegalovirus strain AD169. Virus Res. 1987;7(1):17‐31. [DOI] [PubMed] [Google Scholar]
- 54. Chavez‐Calvillo G, Martin S, Hamm C, Sztuba‐Solinska J. The structure‐to‐function relationships of gammaherpesvirus‐encoded long non‐coding RNAs and their contributions to viral pathogenesis. Noncoding RNA. 2018;4(4):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kara M, O'Grady T, Feldman ER, et al. Gammaherpesvirus readthrough transcription generates a long non‐coding RNA that is regulated by antisense miRNAs and correlates with enhanced lytic replication in vivo. Noncoding RNA. 2019;5(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Marquitz AR, Mathur A, Edwards RH, Raab‐Traub N. Host gene expression is regulated by two types of noncoding RNAs transcribed from the Epstein‐Barr virus BamHI A rightward transcript region. J Virol. 2015;89(22):11256‐11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sethuraman S, Gay LA, Jain V, Haecker I, Renne R. microRNA dependent and independent deregulation of long non‐coding RNAs by an oncogenic herpesvirus. PLoS Pathog. 2017;13(7):e1006508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Guo YE, Oei T, Steitz JA. Herpesvirus saimiri microRNAs preferentially target host cell cycle regulators. J Virol. 2015;89(21):10901‐10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Moon SL, Dodd BJ, Brackney DE, Wilusz CJ, Ebel GD, Wilusz J. Flavivirus sfRNA suppresses antiviral RNA interference in cultured cells and mosquitoes and directly interacts with the RNAi machinery. Virology. 2015;485:322‐329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Qiu L, Wang T, Tang Q, Li G, Wu P, Chen K. Long non‐coding RNAs: regulators of viral infection and the interferon antiviral response. Front Microbiol. 2018;9:1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Carnero E, Barriocanal M, Segura V, et al. Type I interferon regulates the expression of long non‐coding RNAs. Front Immunol. 2014;5:548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Barriocanal M, Carnero E, Segura V, Fortes P. Long non‐coding RNA BST2/BISPR is induced by IFN and regulates the expression of the antiviral factor Tetherin. Front Immunol. 2014;5:655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge RM, Chang HY. A mammalian pseudogene lncRNA at the interface of inflammation and anti‐inflammatory therapeutics. Elife. 2013;2:e00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Liu B, Sun L, Liu Q, et al. A cytoplasmic NF‐kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27(3):370‐381. [DOI] [PubMed] [Google Scholar]
- 65. Zhao XY, Li S, Wang GX, Yu Q, Lin JD. A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol Cell. 2014;55(3):372‐382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hung CL, Wang LY, Yu YL, et al. A long noncoding RNA connects c‐Myc to tumor metabolism. Proc Natl Acad Sci U S a. 2014;111(52):18697‐18702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Esteller M. Non‐coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861‐874. [DOI] [PubMed] [Google Scholar]
- 68. Medzhitov R, Janeway CA Jr. Decoding the patterns of self and nonself by the innate immune system. Science. 2002;296(5566):298‐300. [DOI] [PubMed] [Google Scholar]
- 69. Hajishengallis G, Lambris JD. Microbial manipulation of receptor crosstalk in innate immunity. Nat Rev Immunol. 2011;11(3):187‐200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schneider WM, Chevillotte MD, Rice CM. Interferon‐stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32(1):513‐545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Iwasaki A, Pillai PS. Innate immunity to influenza virus infection. Nat Rev Immunol. 2014;14(5):315‐328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Goubau D, Schlee M, Deddouche S, et al. Antiviral immunity via RIG‐I‐mediated recognition of RNA bearing 5'‐diphosphates. Nature. 2014;514(7522):372‐375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jiang M, Zhang S, Yang Z, et al. Self‐recognition of an inducible host lncRNA by RIG‐I feedback restricts innate immune response. Cell. 2018;173(4):906‐919 e913. [DOI] [PubMed] [Google Scholar]
- 74. Hu G, Gong AY, Wang Y, et al. LincRNA‐Cox2 promotes late inflammatory gene transcription in macrophages through modulating SWI/SNF‐mediated chromatin remodeling. J Immunol. 2016;196(6):2799‐2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Carpenter S, Aiello D, Atianand MK, et al. A long noncoding RNA mediates both activation and repression of immuneresponse genes. Science. 2013;341(6147):789‐792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gomez JA, Wapinski OL, Yang YW, et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon‐gamma locus. Cell. 2013;152(4):743‐754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ouyang J, Zhu X, Chen Y, et al. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon‐stimulated gene transcription. Cell Host Microbe. 2014;16(5):616‐626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Saayman S, Ackley A, Turner AW, et al. An HIV‐encoded antisense long noncoding RNA epigenetically regulates viral transcription. Mol Ther. 2014;22(6):1164‐1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Atianand MK, Hu W, Satpathy AT, et al. A long noncoding RNA lincRNA‐EPS acts as a transcriptional brake to restrain inflammation. Cell. 2016;165(7):1672‐1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Pijlman GP, Funk A, Kondratieva N, et al. A highly structured, nuclease‐resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe. 2008;4(6):579‐591. [DOI] [PubMed] [Google Scholar]
- 81. Chang RY, Hsu TW, Chen YL, et al. Japanese encephalitis virus non‐coding RNA inhibits activation of interferon by blocking nuclear translocation of interferon regulatory factor 3. Vet Microbiol. 2013;166(1‐2):11‐21. [DOI] [PubMed] [Google Scholar]
- 82. Manokaran G, Finol E, Wang C, et al. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science. 2015;350(6257):217‐221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bidet K, Dadlani D, Garcia‐Blanco MA. G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non‐coding RNA. PLoS Pathog. 2014;10(7):e1004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhang Q, Chen CY, Yedavalli VS, Jeang KT. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV‐1 posttranscriptional expression. MBio. 2013;4:e00596‐e00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Chai W, Li J, Shangguan Q, et al. Lnc‐ISG20 inhibits influenza a virus replication by enhancing ISG20 expression. J Virol. 2018;92(16). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wang P, Xu J, Wang Y, Cao X. An interferon‐independent lncRNA promotes viral replication by modulating cellular metabolism. Science. 2017;358(6366):1051‐1055. [DOI] [PubMed] [Google Scholar]
- 87. Crouse J, Kalinke U, Oxenius A. Regulation of antiviral T cell responses by type I interferons. Nat Rev Immunol. 2015;15(4):231‐242. [DOI] [PubMed] [Google Scholar]
- 88. Bird A. Perceptions of epigenetics. Nature. 2007;447(7143):396‐398. [DOI] [PubMed] [Google Scholar]
- 89. Betancur JG. Pervasive lncRNA binding by epigenetic modifying complexes—the challenges ahead. Biochim Biophys Acta. 2016;1859(1):93‐101. [DOI] [PubMed] [Google Scholar]
- 90. Li Z, Chao TC, Chang KY, et al. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc Natl Acad Sci U S a. 2014;111(3):1002‐1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ramirez‐Carrozzi VR, Braas D, Bhatt DM, et al. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138(1):114‐128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Tong Q, Gong AY, Zhang XT, et al. LincRNA‐Cox2 modulates TNF‐alpha‐induced transcription of Il12b gene in intestinal epithelial cells through regulation of Mi‐2/NuRD‐mediated epigenetic histone modifications. FASEB J. 2016;30(3):1187‐1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lazar DC, Morris KV, Saayman SM. The emerging role of long non‐coding RNAs in HIV infection. Virus Res. 2016;212:114‐126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Blume SW, Meng Z, Shrestha K, Snyder RC, Emanuel PD. The 5'‐untranslated RNA of the human dhfr minor transcript alters transcription pre‐initiation complex assembly at the major (core) promoter. J Cell Biochem. 2003;88(1):165‐180. [DOI] [PubMed] [Google Scholar]
- 95. Pruneski JA, Hainer SJ, Petrov KO, Martens JA. The Paf1 complex represses SER3 transcription in Saccharomyces cerevisiae by facilitating intergenic transcription‐dependent nucleosome occupancy of the SER3 promoter. Eukaryot Cell. 2011;10(10):1283‐1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Carpenter S. Long noncoding RNA: novel links between gene expression and innate immunity. Virus Res. 2016;212:137‐145. [DOI] [PubMed] [Google Scholar]
- 97. Kambara H, Niazi F, Kostadinova L, et al. Negative regulation of the interferon response by an interferon‐induced long non‐coding RNA. Nucleic Acids Res. 2014;42(16):10668‐10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Wang P, Xue Y, Han Y, et al. The STAT3‐binding long noncoding RNA lnc‐DC controls human dendritic cell differentiation. Science. 2014;344(6181):310‐313. [DOI] [PubMed] [Google Scholar]
- 99. Zhuang L, Tian J, Zhang X, Wang H, Huang C. Lnc‐DC regulates cellular turnover and the HBV‐induced immune response by TLR9/STAT3 signaling in dendritic cells. Cell Mol Biol Lett. 2018;23(1):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Nishitsuji H, Ujino S, Yoshio S, et al. Long noncoding RNA #32 contributes to antiviral responses by controlling interferon‐stimulated gene expression. Proc Natl Acad Sci U S a. 2016;113(37):10388‐10393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Imam H, Bano AS, Patel P, Holla P, Jameel S. The lncRNA NRON modulates HIV‐1 replication in a NFAT‐dependent manner and is differentially regulated by early and late viral proteins. Sci Rep. 2015;5(1):8639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Saha S, Murthy S, Rangarajan PN. Identification and characterization of a virus‐inducible non‐coding RNA in mouse brain. J Gen Virol. 2006;87(7):1991‐1995. [DOI] [PubMed] [Google Scholar]
- 103. Ma H, Han P, Ye W, et al. The long noncoding RNA NEAT1 exerts antihantaviral effects by acting as positive feedback for RIG‐I signaling. J Virol. 2017;91(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Rossetto CC, Tarrant‐Elorza M, Verma S, Purushothaman P, Pari GS. Regulation of viral and cellular gene expression by Kaposi's sarcoma‐associated herpesvirus polyadenylated nuclear RNA. J Virol. 2013;87(10):5540‐5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Donald CL, Brennan B, Cumberworth SL, et al. Full genome sequence and sfRNA interferon antagonist activity of Zika virus from Recife, Brazil. PLoS Negl Trop Dis. 2016;10(10):e0005048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Sztuba‐Solinska J, Rausch JW, Smith R, Miller JT, Whitby D, Le Grice SFJ. Kaposi's sarcoma‐associated herpesvirus polyadenylated nuclear RNA: a structural scaffold for nuclear, cytoplasmic and viral proteins. Nucleic Acids Res. 2017;45(11):6805‐6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Rossetto CC, Pari GS. Kaposi's sarcoma‐associated herpesvirus noncoding polyadenylated nuclear RNA interacts with virus‐ and host cell‐encoded proteins and suppresses expression of genes involved in immune modulation. J Virol. 2011;85(24):13290‐13297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Conrad NK. New insights into the expression and functions of the Kaposi's sarcoma‐associated herpesvirus long noncoding PAN RNA. Virus Res. 2016;212:53‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Rossetto CC, Pari G. KSHV PAN RNA associates with demethylases UTX and JMJD3 to activate lytic replication through a physical interaction with the virus genome. PLoS Pathog. 2012;8(5):e1002680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Faghihi MA, Modarresi F, Khalil AM, et al. Expression of a noncoding RNA is elevated in Alzheimer's disease and drivesrapid feed‐forward regulation of beta‐secretase. Nat Med. 2008;14(7):723‐730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Yoon JH, Abdelmohsen K, Srikantan S, et al. LincRNA‐p21 suppresses target mRNA translation. Mol Cell. 2012;47(4):648‐655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Du Z, Sun T, Hacisuleyman E, et al. Integrative analyses reveal a long noncoding RNA‐mediated sponge regulatory network in prostate cancer. Nat Commun. 2016;7(1):10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Guo G, Kang Q, Zhu X, et al. A long noncoding RNA critically regulates Bcr‐Abl‐mediated cellular transformation by acting as a competitive endogenous RNA. Oncogene. 2015;34(14):1768‐1779. [DOI] [PubMed] [Google Scholar]
- 114. Levy G, Habib N, Guzzardi MA, et al. Nuclear receptors control pro‐viral and antiviral metabolic responses to hepatitis C virus infection. Nat Chem Biol. 2016;12(12):1037‐1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Tam VC, Quehenberger O, Oshansky CM, et al. Lipidomic profiling of influenza infection identifies mediators that induce and resolve inflammation. Cell. 2013;154(1):213‐227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Morita M, Kuba K, Ichikawa A, et al. The lipid mediator protectin D1 inhibits influenza virus replication and improves severe influenza. Cell. 2013;153(1):112‐125. [DOI] [PubMed] [Google Scholar]


