Summary
Autophagy signaling pathway is involved in cellular homeostasis, developmental processes, cellular stress responses, and immune pathways. The aim of this review is to summarize the relationship between autophagy and viruses. It is not possible to be fully comprehensive, or to provide a complete “overview of all viruses”. In this review, we will focus on the interaction of autophagy and viruses and survey how human viruses exploit multiple steps in the autophagy pathway to help viral propagation and escape immune response. We discuss the role that macroautophagy plays in cells infected with hepatitis C virus, hepatitis B virus, rotavirus gastroenteritis, immune cells infected with human immunodeficiency virus, and viral respiratory tract infections both influenza virus and coronavirus.
Keywords: autophagy, host cell, interaction, virus
1. INTRODUCTION
The presence of numerous membrane‐enclosed structures is the most remarkable morphological feature of eukaryotic cells.1 Macroautophagy is one of the best examples of membrane mobilization which occurs during the process of self‐degradation of cellular component pathway in the cytoplasm. This process aids cell survival in response to multiple stress situations such as nutrient or growth factor deprivation, reactive oxygen species (ROS), hypoxia, and the presence of intracellular pathogens.2
Autophagy (from the Greek, “auto” meaning oneself, “phagy” meaning to eat) is a conserved intracellular homeostatic process where waste cellular components and infectious agents are surrounded by a double layer membrane to create a compartment called the phagosome.3 The phagosome then is delivered and fuses with the lysosome compartments to degrade its contents.4 The double layer membrane (phagophore) is most likely originated from a lipid bilayer donated by the endoplasmic reticulum (ER) and/or the trans‐Golgi.5 Autophagy is as a cellular recycling mechanism that generates ATP and controls damage by removing non‐functional proteins and organelles.6 In addition to its homeostatic role, autophagy also functions in caspase‐independent autophagic cell death, a subset of type II programmed cell death. Autophagic cell death is characterized by the extensive sequestration of portions of the cytoplasm into autophagosomes, giving the cell a characteristic vacuolated appearance.7, 8
1.1. History of autophagy
In 1962, Ashford and Porter detected membrane‐bound vesicles encompassing semi‐digested mitochondria and ER in the hepatocytes of rats that had been treated with glucagon.9 One year later, in 1963, at the Ciba Foundation symposium on lysosomes, Christian de Duve coined the term “autophagy” to define the presence of single‐membrane or double‐membrane vesicles that contain parts of the cytoplasm and organelles.10 Studies into the molecular control of autophagy, starting in the early 1990s by pioneering work from Ohsumi's group reported that the morphology of autophagy in yeast was similar to that recognized in mammals.11 In addition, several selective modes of autophagy such as lipophagy (the autophagy‐mediated degradation of lipid droplets) and mitophagy (selective degradation of mitochondria by autophagy) were first proposed based on the experiments with cultured hepatocytes and whole livers.12, 13, 14, 15
1.2. Autophagy steps
Three forms of autophagy that play different physiological functions have been identified: chaperone‐mediated autophagy, microautophagy, and macroautophagy.6 This review focuses on the mutual interaction between viruses and macroautophagy (hereafter autophagy). Currently, 40 autophagy‐related genes (ATG) have been discovered and characterized.16, 17 There are 5 key steps in autophagy: (1) phagophore formation and nucleation; (2) conjugation of Atg5‐Atg12 complex, interaction with Atg16L and polymerization of the complex Atg12‐Atg5/Atg16L at the immature phagophore; (3) extension of the phagophore membrane by microtubule‐associated protein 1 light chain 3 alpha (LC3) processing and insertion; (4) engulfing of random or selective targets for degradation; and (5) fusion with the lysosome to form autolysosome, followed by proteolytic degradation of captured contents by lysosomal proteases of captured contents.18
1.3. Autophagy induction
Autophagy induction is regulated through 3 main pathways. The first pathway takes place by mammalian target of rapamycin (mTOR), a serine/threonine‐specific protein kinase, and negatively modulates autophagy.19 The mTOR inhibits serine/threonine‐protein kinase ULK1 from recruiting its partners to form the autophagosome.20, 21 TOR, a vital regulator of cell growth, plays an important role in cell growth and autophagy formation in response to nutritional status. During lack of nutrients, AMP protein‐activated kinase (AMPK) negatively regulates mTOR and results in autophagy induction.22 However, in response to insulin or growth factors, class I phosphatidylinositol 3‐kinase (PI3K)‐triggered phosphorylation of protein kinase B (also known as Akt) activates mTOR, thus inhibiting autophagy.21
Atg6/Beclin‐1 is involved in the second pathway of autophagy induction. The interaction of Beclin‐1 with vesicular protein sorting 34 (Vps34) stimulates the catalytic function of Vps34 and increases levels of phosphatidylinositol triphosphate.23 Vps34 is specifically involved in autophagy when it interacts with Beclin‐1.24 Vps34 only phosphorylates phosphatidylinositol (PI) as substrate to generate phosphatidylinositol triphosphate, which is indispensable for phagophore elongation and recruitment of auxiliary Atg proteins to the phagophore.23 Furthermore, Beclin‐1 is a binding partner of the anti‐apoptotic protein B‐cell chronic lymphocytic leukemia/ lymphoma 2 (Bcl‐2) protein, and BCL2‐bound Beclin‐1 is inaccessible for autophagosome formation.25 Antagonistic interactions between viral proteins and Beclin‐1 subvert autophagosome formation and maturation by blocking fusion with lysosomes and exacerbating virulence.26 Other regulatory proteins, such as Rubicon, UV radiation resistance‐associated gene (UVRAG), BIF‐1, Atg14L, and Ambra, form complexes with Vps34 and Beclin‐1 at the ER and nucleated phagophore to both stimulate and suppress autophagy.27, 28, 29
The third pathway that contributes to autophagosome formation and elongation involves ubiquitin‐like conjugation processes that generate membrane‐bound protein complexes.30 Atg7 and Atg10 mediate the conjugation of Atg12 to Atg5, which subsequently interact with Atg16. The Atg12‐Atg5/Atg16L complex binds to the outer membrane of autophagosome and then dissociates upon completion of the autophagosome.31 The second conjugation reaction involves LC3, a mammalian homolog of yeast Atg8. The precursor proLC3 is constitutively cleaved by Atg4 to produce LC3‐I. Upon receiving the signal to induce autophagy, Atg7 and Atg3 mediate the conjugation of LC3‐I to the membrane lipid phosphatidylethanolamine to form LC3‐II. The LC3‐II integrates the outer and inner membranes of the autophagosome and helps complete membrane elongation and closure.31
LC3 is the only reliable marker of autophagosome and autolysosome formation and is routinely detected in several cell lines.32 The LC3 exists in 2 forms: Cytosolic LC3‐I and its lipidated derivative LC3‐II. During autophagy processing, the level of membrane‐associated 16 kDa LC3‐II increases, while the level of soluble 18 kDa LC3‐I decreases. Thus, the relative amount of LC3‐II is linked to the dynamic turnover of LC3‐II via the lysosomal activity.33, 34 In this process, LC3‐I is covalently conjugated to phosphatidylethanolamine through LC3 lipid coupling mechanism, resulting in LC3‐II. LC3‐II then moves from the cytosol to the autophagosome membranes34, 35 where it remains associated with the expanding limiting membrane, sealed autophagosomes, and mature autophagosomes/autolysosomes.36, 37, 38 Thus, LC3‐II levels usually correlate with autophagosome numbers.34 To evaluate if substrate degradation is autophagy dependent, it is useful to perform assays measuring LC3‐II levels assays in the presence and absence of a lysosomal vacuolar‐type H(+)‐ATPase inhibitor such as Bafilomycin A1 (Baf‐A1).38
2. VIRUSES AND AUTOPHAGY
Autophagy acts as a double‐edged sword in pathogenesis of viral disease.39 Xenophagy describes autophagy that serves as an innate immune response to eliminate intracellular infectious pathogens.40 Hosts deficiency in autophagy are prone to microbial infections.41 Autophagy occurs at basal levels to maintain cellular homeostasis, but the stress of viral infection can trigger autophagy induction. During infection, autophagy can play either a pro‐viral or antiviral role, depending on the virus, the cell type, and the cellular environment.42, 43 Autophagy can function differently in response to positive‐strand and negative‐strand RNA viruses, DNA viruses, viral genotypes, step of viral life cycle, and stage of pathogenesis.44 During host cell infection, viruses and viral proteins can be targeted for degradation by autophagy to stop viral replication. Conversely, some viruses have evolved strategies to avoid autophagy‐induced lysosomal degradation by subverting autophagy into autophagosomal shelters harboring viral replication. In fact, these viruses exploit the autophagy pathway by replicating in the autophagosome‐like structures themselves.45, 46, 47, 48, 49, 50, 51, 52, 53 Manipulation of autophagy can be a promising strategy to eliminate intracellular pathogens.38
2.1. Antiviral function of autophagy and the role of autophagy in suppression of anti‐viral immune response
Autophagy is an integral part of immune response to viral infections and plays a key role both in innate and adaptive immunity responses. Autophagy is essential for delivering cytoplasmic viral RNA to the endosomal pathway through unique ability to deliver cytosolic pathogen‐associated molecular patterns into the closeness of endosomal pattern recognition receptors (PRRs) and MHC loading compartments. For example, influenza viruses are sensed in the lysosomes of plasmacytoid dendritic cells (pDCs) through TLR7 after endocytosis of virions.54 Human immunodeficiency virus (HIV) 1 can escape immune system through a new mechanism in DCs. HIV‐1 was revealed to block macroautophagy initiation via inducing the mTOR pathway. This suppression resulted in a deficiency of TLR4 and TLR8 activation through viral replication intermediates and of antigen presentation to CD4+ T cells. Macroautophagy was presented to be essential for MHC class II antigen processing of a HIV gag‐derived CD4+ T cell epitope. HIV gag‐specific CD4+ T cell clone significantly decreased TNF‐α secretion upon induction with macroautophagy‐deficient DCs, pulsed with inactivated HIV virus. While CD8+ T cell activation was not compromised when macroautophagy was suppressed. This study indicated that efficient HIV antigen processing for MHC class II presentation needs macroautophagy.55, 56 HCV abuses autophagy to inhibit IFN signaling. It also temporally regulates the autophagic flux to sequester and deplete TRAF6 to promote viral RNA replication and control of host innate immune responses.57 It is interesting to note that a paradigm has been appeared in which Th1 cytokines induce autophagy, while, Th2 cytokines suppress autophagy activation.58, 59 In HIV infection, inhibition of autophagy in bystander macrophage/monocytic was dependent on IL‐10 and Src‐Akt and STAT3 triggered by HIV‐1 Tat.60 Autophagy has been involved in both the delivery of cytosolic antigens to the major histocompatibility complex classes I and II. The HIV envelope proteins are recognized to subvert MHC class II presentation via enhancing mTOR signaling.61 Importantly, immunity‐associated GTPase family M(IRGM) IRGM‐interacting viral proteins, such as HCV‐NS3 and HIV‐1‐Nef, induce autophagy in an IRGM‐dependent pathway in order to improve viral infectivity.62 The result of recent study suggests that efficient cross‐presentation of viral antigens needed macroautophagy in antigen donor cells. To assess cross‐presentation via macroautophagy wild‐type mouse embryonic fibroblasts (MEFs) and Bax/Bak−/− MEFs cells were applied. While wild‐type MEFs can complete caspase‐dependent apoptosis, Bax/Bak−/− MEFs cannot undergo this type of cell death but can upregulate macroautophagy under the treatment conditions. Both cells were infected with influenza A virus, then treated with pro‐apoptotic compounds and subsequently transferred in vivo to prime experiment. Mice immunized with Bax/Bak−/− MEFs expressed a significantly higher CD8+ T cell response specific to both immunodominant of influenza haemagglutinin and neucleoprotein.63 Last but not least, autophagy hijacked by nonstructural protein 4 viroporin‐activated calcium/calmodulin‐dependent kinase kinase‐β signaling is indispensable for rotavirus replication.64
2.2. Interplay between HCV and autophagy
Autophagy is involved in different liver physiology and pathophysiology processes, including the removal of mis‐folded proteins, nutrient and lipid metabolism, regulation of selective organelle degradation, and response to hepatitis virus infection.12 Hepatitis C virus (HCV) and hepatitis B virus (HBV) are the main causes of chronic liver infection and lead to liver fibrosis, cirrhosis, and ultimately malignant transformation of the hepatocytes.65 HCV is a hepatotropic membrane‐enveloped, positive‐sense, single‐stranded RNA.66
Infections of the Flaviviridae family such as HCV, Dengue virus, and Japanese encephalitis virus are reported to activate autophagy in cell culture experiments.67 Upon binding to the host cell, HCV is internalized through clathrin‐mediated endocytosis.68 To release the viral nucleocapsid into the cytoplasm, the viral and endosomal membranes fuse in a pH‐dependent manner.69 HCV RNA is transported to the ER, where HCV non‐structural proteins NS4B and NS5A induce ER membrane alteration (known as “membranous web” formation). This alteration provides a scaffold for the replication of the HCV and protects the virus from host immune defenses.70 HCV NS4B, a membranous web formation inducer, may elicit a stress response that triggers autophagy as shown in Figure 1.71 Autophagy that is basal, rather than stress‐induced, may provide an initial membranous platform for incoming RNA translation. Once replication is established, this platform becomes unnecessary for translation of HCV RNA progeny.72
Figure 1.
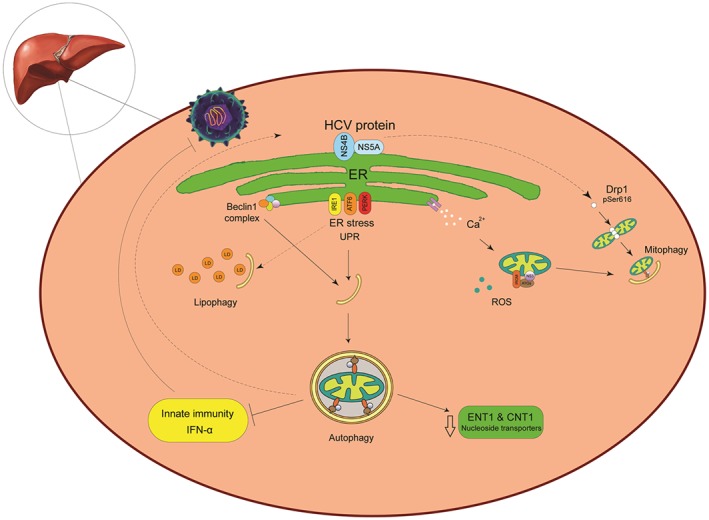
The cross‐talk between autophagy and HCV. The hepatitis C virus (HCV) induces autophagy to enhance virus replication rather than to block it. After HCV decapsidation, the virus genome is transferred to endoplasmic reticulum (ER) and initiates replication in the ER‐derived membranous web as the HCV replication platform. HCV‐induced ER stress disrupts ER function and results in accumulation of unfolded proteins in the ER. Cells adapt to ER stress by triggering an unfolded protein response (UPR) to restore ER homeostasis. The UPR signaling pathway includes 3 central elements, PKR‐like ER kinase (PERK), ATF6, and IRE1. The UPR could stimulate an autophagy response. Under these conditions, calcium released from the ER stimulates impaired mitochondrial activity connect to excessive production of ROS. Dysfunctional mitochondria are engulfed for degradation via mitophagy. In addition, HCV promoted dynamin‐related protein 1‐mediated mitochondrial fission protein 1 (Drp1) leads to mitophagy. HCV replication takes place in close proximity to lipid droplets (LDs), the neutral lipid storage organelles, and this closeness enables the downstream viral morphogenetic events. A selective autophagy for lipids (lipophagy) protects cells from an excessive lipid accumulation elicited by HCV. Upon HCV infection, the host stimulates the antiviral interferon‐mediated frontline. HCV employs autophagy to escape host innate immune surveillance by inhibiting IFNα production. Furthermore, HCV replication reduced ribavirin uptake, due to reduced expression of the nucleoside transporters ENT1 and CNT1
Growing evidence indicates that HCV infection‐triggered cellular stress indirectly induces autophagy. The accumulation of mis‐folded or unfolded protein in the ER causes ER stress which in turn results in the activation of 3 distinct pathways: the inositol‐requiring enzyme 1 (IRE1), activating transcription factor 6 (ATF6), and double‐stranded (ds) RNA‐activated protein kinase, including ER kinase (PERK). These proteins trigger downstream signaling pathways to cope with mis‐folded proteins.73 This process, called the unfolded protein response (UPR), alleviates ER stress by reducing protein synthesis, upregulating ER chaperone protein expression to facilitate protein folding, and augmenting protein degradation via autophagy and the ER‐associated degradation pathway. If the UPR fails to alleviate ER stress, it will induce apoptosis.73, 74 In addition, ER stress releases calcium from the ER resulting in an increase of mitochondrial metabolic activity. This activity leads to an overproduction of ROS, which leads to mitochondrial oxidative. Damaged mitochondria are then selectively targeted for degradation by mitophagy via activation of the mitochondrial fission protein dynamin‐related protein‐1; this process is reported to occur during HCV infection.75 HCV upregulates expression levels of serine/threonine kinase PTEN‐induced putative kinase 1 and Parkin (an E3 ubiquitin ligase), both of which then translocate to the outer mitochondrial membrane. The new Parkin on the mitochondrial surface recruits its substrate to induce mitophagy.76 HCV‐activated mitophagy could decrease HCV‐induced apoptosis in order to promote viral persistence.77 The HCV core protein has been revealed to stimulate both the ATF6 and PERK pathways that, in turn, enhance the expression of the ATG12 and LC3 genes.78 Induction of autophagy and mitophagy is essential to sustain the survival of infected cells and establish a chronic HCV infection.77
Interaction between autophagy and HCV is time dependent. The co‐localization of ATG5 and HCV RNA polymerase NS5B is detected 48 hours following infection. In contrast after 5 days post‐infection, the co‐localization has disappeared.79 In the normal autophagic pathway, the UVRAG enables the fusion between autophagosomes and lysosomes. Induced by HCV in the early stages of infection, Rubicon, RUN domain and cysteine‐rich domain containing Beclin 1‐interacting protein, blocks UVRAG function thus prevents fusion of autophagosomes and lysosomes.80 By contrast, the induction of UVRAG in the late stages of HCV infection disables the inhibitory effect of Rubicon, leading to the maturation of autophagosomes.80, 81, 82 Another study demonstrated that knockdown of ATG7 and Beclin‐1 reduced viral shedding without any significant effect on the intracellular viral proteins and RNA levels.83
Lipophagy is a selective type of autophagy responsible for the degradation of intracellular lipid stores during the HCV life cycle.84 This indicates that HCV possibly develops lipoprotein degradation using autophagy, thereby, bypassing virus‐produced lipid droplet accumulation in host cells. Additionally, pharmacological inhibition of autophagy leads to a substantial accumulation of intracellular cholesterol deposits; inhibiting cholesterol synthesis by statins considerably drops autophagy levels, as well as viral replication.85 Taken together, these data indicate that inhibition of lipid‐selective autophagy might be connected to the beginning of steatosis in chronically HCV‐infected patients.
Induced autophagy response in persistent HCV infection selectively downregulates the functional type I IFN‐α receptor, but not the type II (IFN‐γ) or type III (IFN‐λ) IFN receptors. In the same context, the expression of the nucleoside transporters ENT1 and CNT1 is also reduced, suggesting a possible contribution of autophagy in the IFN‐α combined with ribavirin resistance mechanisms against HCV.86 By contrast, by studying transgenic mice that expressed HCV proteins, it was clear that IFN‐β could arouse the autophagic degradation of HCV core and NS3/4A proteins, negatively regulating HCV replication.86, 87 Pharmacological autophagy inhibitors, such as chloroquine (CQ) and bafilomycin A1, inhibit HCV infection specifically by stimulation of type I IFN antiviral immunity.71
2.3. HBV interaction with autophagy
Hepatitis B virus (HBV), a member of the Hepadnaviridae family, with a DNA virus that infects hepatocytes, enhances and recruits autophagy in favor of its DNA replication.88 HBV is responsible for one of the most common chronic infections of the liver in humans, and chronic hepatitis caused by HBV is the main etiologic agent of hepatocellular carcinoma.89 HBV small envelope proteins (SHBs) mediate ER stress and UPR induction, and the effect of SHBs on autophagy was eliminated when the 3 sensors of ER stress (ie, IRE1, PERK, and ATR6) were silenced with siRNAs.90 Recently, it has been verified that HBV is able to activate the ER‐associated degradation pathway and trigger the expression of ER degradation‐enhancing mannosidase‐like proteins, which then facilitate the degradation of HBV surface proteins through the autophagic pathway.91
Autophagy dysfunction has been related to several diseases including cancer. However, a recent report showed that in HBV‐associated hepatocellular carcinoma (HCC), autophagy was downregulated, and there was a reverse relation between the autophagic activity and the level of microRNA‐224 (miR‐224) in these tumor cells.92 Autophagosomes can sequester miR‐224 and subsequently degrade it. Increased miR‐224 results in downregulation of its target gene Smad4, a transcription factor that activates the TGF‐β signaling pathway.92 Smad4 inhibition can shift TGF‐β's role from a tumor suppressor to a tumor enhancer factor.93 The induction of autophagy is mediated by HBV X‐Protein (Trans‐Activator X Gene) through interaction and activation of phosphatidylinositol‐3‐kinase class 3 (PI3KC3).88
2.4. Rotaviruses and autophagy
Rotavirus consists of 11 segments of double‐stranded RNA94 and causes age‐dependent and life‐threatening dehydrating viral gastroenteritis in children worldwide. The full mechanism by which rotavirus induces diarrhea remains unclear, but it is triggered to some extent by virus‐encoded non‐structural protein 4 (NSP4), which acts as a viral enterotoxin through viroporin activity.95, 96 During rotavirus infection, NSP4 co‐localizes with the LC3 protein and surrounds viroplasms, the viral factories in the cell.97 Rotavirus hijacks the autophagy membrane trafficking pathway through NSP4 to transfer ER‐associated viral proteins NSP4 and VP7 to virus producing viroplasms.64 The interaction of NSP4 with immature virus particles at the boundary between viroplasms and NSP4/LC3‐containing membranes mediates the assembly of the outer capsid proteins onto the double‐layered particle, finally forming the infectious virion which is a triple‐layered particle.98 NSP4 stimulates the release of ER lumenal calcium into the cytoplasm of the host cell, thereby stimulating calcium/calmodulin‐dependent kinase kinase‐β (CaMKK‐β) that phosphorylates 5′ adenosine monophosphate‐activated protein kinase (AMPK) to initiate autophagy.64 Phosphorylated AMPK is able to inhibit the mTOR complex 1 (mTORC1) or directly phosphorylate ULK1, both of which result in autophagy induction.99
Rotavirus‐induced autophagosomes are unable to fuse with lysosomes, suggesting that rotavirus infection triggers autophagosome formation but blocks maturation. Inhibiting autophagosome maturation may help the rotavirus evade the antiviral function of autophagy, although it is unclear whether rotavirus inhibits autophagosome maturation directly or indirectly. Rotaviruses that use 2 non‐structural proteins, NSP4 and NSP2, can de‐polymerize the microtubule network100, 101 and render the cell incompetent for autophagy processing, because microtubules are essential for autophagosome membrane trafficking.102 Recently, it was reported that rotavirus induces autophagic signals to promote virus replication while also suppressing protein degradation. Rotavirus infection enhances expression of several stress‐related genes, which suggests that NSP4 may be involved in the lipolysis of viroplasms via lipophagy.103 In addition, it was reported that lipidated LC3 plays an important role exclusively in the late stages of the viral life cycle to enhance virus replication.104, 105
2.5. Autophagy involved in HIV infection in a cell type dependent manner
Acquired immunodeficiency syndrome (AIDS) is caused by HIV. HIV targets and replicates in vital cells of the human immune system, mainly CD4+ T cells, macrophages, and dendritic cells (DCs).106, 107 HIV infection causes acquired immunodeficiency, primarily due to the depletion of CD4 lymphocytes (< 200 cells μL−1 blood volume). When CD4+ T cell numbers drop below this critical level, the immune system loses its ability to fight opportunistic diseases, which are the principal causes of mortality in HIV‐infected patients. The mechanism by which the virus depletes these cells has not been fully elucidated but appears to result from viral strategies employed during multiple parts of its life cycle to evade the innate and adaptive immune responses and to ensure viral persistence.
A major challenge for developing a cure for HIV is the existence of a quiescent pool of infected cells. In the latent phase, HIV pro‐viruses persist in long‐lived cells, such as hematopoietic stem cells, central memory CD4+ T cells, DCs, and cells from the monocyte‐macrophage lineage including microglial cells, which are the main HIV reservoirs in the central nervous system (CNS).107 The potential mechanisms of HIV persistence have been discussed recently in a review by Hong and Mellors108 . Viral latency is commonly a reversible state defined by non‐productive infection of individual cells. This state provides an important mechanism for viral persistence and escape from immune recognition.109 However, the term latency is quite complex for HIV‐1 because the virus is still able to infect the resting CD4+ T cells at moderate levels.110
Autophagy plays multiple roles in immunity. Besides its degradative function, the autophagy pathway plays a role in innate immunity; the pathway delivers cytosolic microbial and viral products to PRRs, known as Toll‐like receptors (TLRs), in antigen‐presenting cells such as macrophages and DCs.111, 112 Autophagy also controls inflammation. For example, autophagy supports the secretion of the pro‐inflammatory cytokine interleukin (IL)‐1β from macrophages,113 curbs inflammation by downregulating type I IFN signaling,86 captures and removes endogenous inflammasome agonists,114 and causes immune mediator secretion.115 In T cells, autophagy contributes to antigen presentation by MHC class II from macrophages and DCs and affects T cell repertoires and polarization.115
In recent years, several studies uncovered that autophagy, used by the cell as an antiviral immune defense, is usurped by HIV‐1 to facilitate viral protein processing and virion assembly.116 Moreover, autophagy is an intracellular degradation process responsible for the clearance of aggregate‐prone cytoplasmic proteins that cause neurodegenerative diseases such as Parkinson's and Alzheimer's diseases. Aberrant activation and dysregulation of autophagy in HIV infection contribute to HIV‐associated neurocognitive disorders (HAND). Here, we discuss the dysregulation of autophagy by HIV‐1 proteins in the immune system and the CNS.
Findings in this field can help elucidate the mechanisms involved in HIV‐1 infection and viral replication, and contributing to HIV therapeutic development. HIV‐1 infects a variety of immune cells, but its largest cytopathic effect is the shrinkage of CD4+ T cell population117, 118 through mechanisms including pyroptosis of abortively infected T cells,119 apoptosis of uninfected bystander cells,120 direct viral killing of infected cells, and killing of infected CD4+ T cells by CD8 cytotoxic lymphocytes.121
In the context of autophagy, depletion of the CD4+ T cell population during HIV‐1 infection is proposed to occur by Env‐mediated cell death in uninfected CD4+ T cells by induction of autophagy as depicted in Figure 2.122, 123 While autophagy is induced in uninfected CD4+ T cells, it is inhibited in infected cells through the transcriptional level downregulation of Beclin‐1.123, 124 In 2002, Castedo described the precise mechanism by which uninfected T cells die upon co‐receptor engagement with HIV Env.125 This study demonstrates that CXCR4 engagement leads to the activation of mTOR and subsequent phosphorylation and activation of p53. Activated p53 increases the expression of Bax and activates the mitochondrial death pathway. These data suggest that HIV‐1 manipulates autophagy during infection to circumvent the immune response. Accordingly, a significant increase in the autophagic activity has been detected in the peripheral blood mononuclear cells of elite controllers; individuals may remain asymptomatic for more than 10 years due to their ability to maintain a viral load below the limits of detection in the absence of antiretroviral therapy.126 A study by Sagnier et al found that autophagy restricts HIV‐1 infection by selectively degrading the viral trans‐activator Tat, highlighting the anti‐HIV effect of authophagy.127
Figure 2.
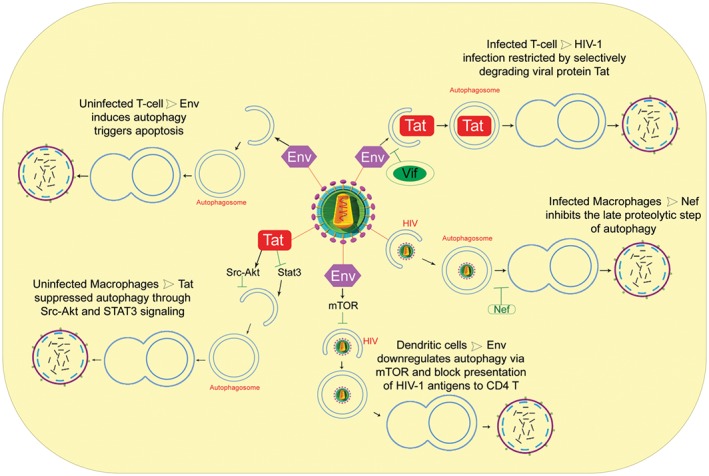
Schematic overview of the autophagy and HIV‐1 in target cells. Autophagy, as a cell‐autonomous innate defense, plays an important role against HIV‐1 infection, it restricts hiv‐1 infection by Tat degradation and limits virion production. However, HIV‐1 proteins are able to modulate autophagy pathway by different strategy at the initiation and maturation step (Tat in uninfected macrophages; Env in dendritic cells; Vif in infected T cells; Nef in infected macrophages). In particular, in infected macrophages, Nef acts as an “antiautophagic maturation factor” and blocks the autophagosome maturation step. In uninfected CD4 T cells, Env‐mediated autophagy leads to apoptotic cell death
Interestingly, HIV‐1 has evolved strategies to counteract the antiviral effect of autophagy. For example, the viral protein Vif can inhibit the early steps of autophagy, allowing for the productive infection of CD4+ T cells.128 HIV‐1 negative factor protein (Nef) inhibits autophagosome maturation in infected macrophages through its association with Beclin 1, thereby avoiding degradation.22 In DCs, HIV‐1 Env has been proposed to increase cell‐associated HIV‐1 by activating mTOR and S6‐kinase (S6K), thus inhibiting autophagy and transferring infection into CD4+ T cells (trans‐infection).55
Cells belonging to the monocyte‐macrophage lineage are more resistant to cytopathic effects and are able to harbor viruses for longer time periods. A study by Espert et al demonstrated the resistance of macrophages to HIV Env‐mediated bystander cell death, in contrast to CD4+ T cells.123 The secreted form of HIV‐Tat blocks autophagy by activating Src‐Akt and STAT3 signaling pathways, previously known to inhibit authophagy.60 Moreover, HIV‐Tat induces production of IL‐10.129 Interestingly, IL‐10 inhibits autophagy initiation in uninfected bystander macrophages, independently of the presence of Env.60 The Tat protein also dysregulates IFN‐γ signaling and suppresses autophagy in macrophages. The protein inhibits STAT1 phosphorylation and reduces upregulation of LC3B and autophagosome formation. Additionally, HIV‐Tat restricts mycobacteria capture by autophagosomes, thereby providing a favorable environment for opportunistic microbes in HIV‐infected individuals.130 Furthermore, the HIV‐1 precursor Gag co‐localized and interacted with the autophagy factor LC3, suggesting that autophagy promoted productive Gag processing and viral particle production in macrophages.116
Taken together, these studies demonstrate that HIV evades degradation by autophagy to enhance the success of its infection. The role of autophagy in HIV infection is different in macrophages than it is in DCs or CD4+ T cells because, HIV‐1 infected cells from the monocyte‐macrophage lineage are more resistant to apoptosis, a major obstacle to eradication of the virus. Therefore, autophagy could be a possible key player in macrophage reservoir promotion for the virus.
Dendritic cells (DCs), which are located in the mucosa (including oral and vaginal mucosal surfaces) and lymphoid tissues, are crucial in the generation and regulation of immune responses to HIV infection. DCs play a pivotal role in the early dissemination of HIV, likely among the first cells that encounter HIV during sexual transmission.131
However, the role autophagy plays in DCs during HIV infection has been understudied. Blanchet et al showed HIV Env‐mediated downregulation of autophagy by activating mTOR.55 This study found that downregulation of autophagy leads to a decrease in TLR‐mediated innate immune response and blocks presentation of HIV‐1 antigens to CD4+ T cells in the context of MHC class II (adaptive immunity).55 Thus, disruption of autophagy in DCs could significantly contribute to HIV‐1 disease progression, allowing for opportunistic infections and cancers to evade innate and adaptive immune responses.
Conversely, pDCs secrete IFN‐α in response to infectious or non‐infectious HIV‐1 through induction of autophagy following TLR7 signaling.132, 133 Natural killer (NK) cells are activated by pDCs responding to HIV‐1.134 The ability of NK cells to lyse infected cells is increased by IFN‐α.135 Thus, autophagy is required for suppression of HIV‐1 replication by cytolytic function of the innate immune system.
2.6. Autophagy in HIV‐associated neurological disorders
The CNS is considered to be a viral reservoir, because antiretroviral therapy drugs have limited access to this area.136, 137, 138 There are several lines of evidence that suggest that brain cells harbor genome‐integrated HIV.139 HIV‐associated neurological disorders occur almost immediately after systemic infection.140 Autophagy as an intracellular clearance pathway for mis‐folded and aggregate proteins plays a protective role against various neurodegenerative disorders, such as Huntington's and Parkinson's diseases.141, 142 Defects of autophagy in the CNS have emerged as a feature of neuroAIDS.143 Autophagy markers are moderately increased in postmortem brains of individuals with HIV when compared with brains of uninfected individuals or HIV‐infected individuals without encephalitis. This suggests that dysregulation of autophagy may be important in the pathogenesis of neuroAIDS.143 Previous reports demonstrated that inflammatory molecules released by simian immunodeficiency virus‐infected microglia significantly reduced the autophagy processing in neurons resulting in decreased neuronal survival. This reduction of autophagy was correlated with an accumulation of sequestosome‐1/p62 in neurons from cell cultures. Similar phenotype of p62 aggregations have been discovered in the post‐mortem brains of HIV demented patients when compared with subjects without neurological disease.144
Moreover, dysregulated autophagy also contributes to aging‐related neuropathology in HIV patients who develop HAND.145 These results are similar to the bystander effect of HIV‐1 Env that increases autophagy in CD4+ cell, described previously.
In addition to gp120, the HIV‐1 transactivator protein Tat causes neuronal injury. One study suggests that Tat contributes to HIV‐1 neuropathogenesis by altering the autophagy pathway.146 The authors show that Tat induces autophagic degradation by promoting fusion of autophagosomes and lysosomes, leading to abnormal neuronal autophagy and dysregulated degradation of critical intracellular components.146
In conclusion, autophagy plays key roles in immune defenses against invading bacterial and viral pathogens. These data support the role autophagy plays in HIV‐1 infection and pathogenesis. HIV‐1 counteracts autophagy‐mediated degradation and is able to manipulate autophagy for its own benefit, favoring viral replication in the targeted cell types. Meanwhile, HIV proteins induce autophagy and promote autophagic T cell death in uninfected cells, thereby contributing to HIV‐1 pathogenesis. Although challenges still remain, particularly the dual role autophagy plays in HIV‐1 infection, these observations provide novel insight into the pathogenesis of HIV‐1 and allow us to consider new strategies toward a cure for HIV.116, 142, 147, 148, 149
2.7. Cross‐talk between influenza virus and autophagy
Influenza A virus (IAV) (family Orthomyxoviridae, genus Influenza virus A) has caused severe respiratory diseases for hundreds of years in humans and different animal species, resulting in considerable morbidity and mortality.150 IAVs are multi‐step infectious agents that recruit host cell machinery to support the replication and transportation of their own viral components.151, 152 Understanding this viral strategy requires characterization of the intracellular pathways by which the influenza virus replicate.151 Here, we focus on autophagy as one of the most prominent processes involved in the efficient replication of IAV inside the host cell.
Autophagy was originally characterized as a critical process for influenza virus replication. This infection triggers autophagy in different types of cells and enhances autophagosome formation and autophagic flux.153 Baf‐A1 is a highly specific inhibitor for vacuolar‐type proton (V‐H+) pumps, which are responsible for the acidification of endosomes and lysosomes, and are necessary for influenza virus replication.154 Administration of Baf‐A1 at low concentrations effectively inhibits IAV replication without impacting host cell viability.155 This dose of Baf‐A1 inhibits the formation of autophagosomes, resulting from either an inhibition of autophagosome formation or an increase in degradative flux. However, the most common scenario with an inhibitor of autophagy like Baf‐A1 is an increase in levels of LC3‐II, and is indicator of increased autophagosome formation.38
In addition, autophagy may act as an innate immune mechanism to restrict virus replication.156 Autophagy inhibition by IAV can be considered a novel immune escape mechanism or a by‐product of altered autophagosome physiology. If the viral escape mechanism is broken, virus replication and virulence are impaired37, 157, 158 suggesting that induction of autophagosome degradation during viral infections might restrict virus replication within infected cells.159 Viruses can upregulate autophagosome formation, or, alternatively, autophagosome degradation in lysosomes could be blocked. Several pieces of evidence support the latter scenario, whereby after influenza infection, autophagosomes do not fuse with acidic compartments.160 Blocking autophagosome degradation would lead to accumulation of autophagosomes and LC3‐II48, 160 to evade presentation of viral antigens.48, 160
It has been reported that autophagy is involved in the replication of IAV, and inhibiting autophagy will inhibit the replication of this virus.161 In 2007, Dorothee Schmid found that autophagy in IAV‐infected lung epithelial cells is inhibited. Autophagosomes accumulate because they do not fuse with lysosomes, leading to increased virus survival.36 A study on the antiviral activity of statins found that the expression level of LC3‐II was independent of the presence of Baf‐A1, confirming that influenza virus increases autophagosome formation by LC3‐II accumulation and inhibits autophagosome maturation.162
Two viral proteins, NS1 and M2, are associated with autophagy signaling. NS1 binds to p85β, the regulatory subunit of PI3K,163 while M2 keeps autophagy at moderate levels by limiting the degree of lysosome fusion with autophagosomes.160 M2 protein, a virus‐specific membrane protein,164 is expressed at the surface of infected cells. It has a short (24 residues long), highly conserved extracellular N‐terminal domain, an internal hydrophobic anchorage domain (approximately 19 residues long), and a C‐terminal amphipathic cytoplasmic domain (54 residues long) facing inside the viral particle.165 The M2 protein makes a proton channel involved in 2 virus replication functions. First, M2 serves as a proton selective ion channel, causing un‐coating of the virus and exposure of its contents to the host cytoplasm. Second, it initiates viral replication and maturation of the haemagglutinin glycoprotein.166, 167 In addition, the M2 protein is necessary and sufficient to block and inhibit autophagy (autophagosome maturation) and to trigger autophagy initiation.48, 168 IAV takes advantage of autophagy inhibition. Autophagy inhibition hides IAV from immune attack despite not being useful for its replication.48 The first 60 amino acids of M2 seem sufficient for inhibition of autophagosome maturation after binding to Beclin‐1. The specific inhibitor of the M2 H+ channel, amantadine, could not abrogate the inhibition of autophagy flux. This suggests that a block of autophagosome maturation does not involve M2 ion channel activity. Thus, the main functional part of the M2 protein is believed to be located within the internal domain that is responsible for proton conductive activity.48
The mTOR signaling pathway negatively regulates autophagy.169, 170 The phosphorylated form of the effector protein kinase mTOR is known to inhibit autophagy.171 It has been proposed that the highly pathogenic avian influenza virus H5N1 induces autophagy by suppressing phosphorylated mTOR signaling. Thus, inhibition of autophagy could reduce H5N1‐mediated cellular damage.172 In terminal autophagy, mTORC2 upregulates p70S6K activity (the original natural substrate of mTORC1) required for LC3‐II formation. Interestingly, when the complex mTORC2/p70S6K blocks lethal autophagy in the absence of apoptosis, drug inhibitors of lethal autophagy limit viral production in cells.173 Increased autophagy shows altered autophagy signaling with increased mTORC1 and PI3K/mTORC2 activity and p70S6K phosphorylation. Blocking PI3K, mTORC2, or p70S6K activity prevents lethal autophagy and decreases infectious virus production.173
Pan H et al (2914) reported that the activation of NF‐kB signaling promoted H5N1 pseudotyped particles‐induced autophagosome formation both in human lung epithelial cell lines and mouse lung tissues. The positive feedback between autophagy and NF‐kB and p38 MAPK signaling cascades could be an important mechanism contributing to H5N1 pseudotyped particles‐induced lung inflammation.174
Autophagy also performs as a key mechanism contributing to the inflammatory responses induced by H1N1 and H9N2.175 Ectopic P‐granules autophagy protein 5 homolog (EPG5), which is essential for basal autophagy and functions of ATG genes complex in the formation of degradative autolysosomes,176 regulates basal expression of multiple cytokines in the lung. Epg5 deficiency in myeloid cells in the lung causes an increase in IL‐1b, IL‐6, and IL‐13 cytokines in lung macrophages. Optimal cytokine levels exert protective effects against viral replication, thus enhancing the innate immune response to influenza.176, 177
One study demonstrated that the activation of the PI3K/AKT pathway, which is closely related to the autophagic process, has a biphasic effect on influenza virus replication.178 Deficiency in autophagy causes impaired survival of memory CD8+ T cells during infection with influenza virus.179 Delayed apoptosis and highly stimulated autophagy were observed in IAV‐infected cells.180 In a report that studied H3N2 infection, it was shown that lipidated LC3‐II began to accumulate a few hours post‐infection in A549 cells and very early post‐infection in Ana‐1 cells. This also confirmed that H3N2 induced autophagy in both A549 and Ana‐1 cells.181
Influenza A virus induces the nucleotide‐binding domain and leucine‐rich repeat (NLR) family, including the pyrin domain containing 3 (NLRP3) inflammasome, causing mitochondrial damage and leading to the release of ROS. NLRP3 forms an inflammasome complex with ASC (essential adaptor of inflammasomes) and caspase‐1, thus inducing the production of IL‐1α and IL‐18 in response to mitochondrial ROS. Deficiency in autophagy results in the accumulation of dysfunctional mitochondria. This event produces excessive ROS upon stimulation, which potently activates the NLRP3 inflammasome.182 Upon infection with IAV, the cytosolic PRR Nod2 and its downstream regulator RIPK2 induce mitophagy by inducing phosphorylation of ULK1 to prevent excessive activation of the NLRP3 inflammasome.183
As illustrated in Figure 3, influenza A virus can be involved in IAV interaction with the host autophagy pathway in several ways, including the following:
ROS production, which prevents the conversion of LC3‐II to LC3‐I by degrading Atg4 and leads to increased levels of LC3‐II;
Binding between viral M2 protein and Beclin 1;
Upregulation of the expression of several ATG, which increase autophagic flux; and
IAV subversion of autophagy through the LC3‐interacting region (LIR) on M2, which leads to LC3 redistribution to the plasma membrane in infected cells.184
Figure 3.
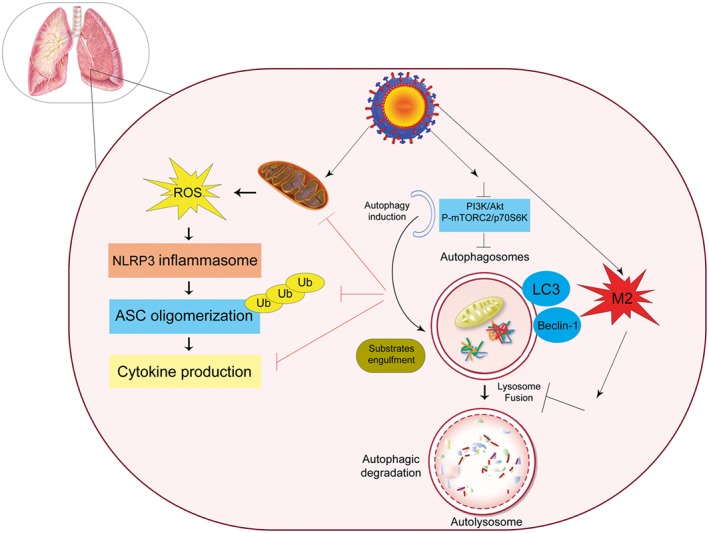
Regulatory signaling of autophagy during influenza A virus infection. Influenza A virus as inducer of the NLRP3 inflammasome causes mitochondrial damage, which leads to the release of reactive oxygen species (ROS). NLRP3 forms an inflammasome complex with ASC and induces the production of inflammatory cytokines. Influenza A virus also binds to Beclin1 by viral M2 protein. It upregulates the expression of several autophagy related genes, which can increase the autophagic flux, and M2 also contains an LC3‐interacting region (LIR), which is required for influenza virus subversion of autophagy which leads to LC3 redistribution to the plasma membrane in infected cells
Influenza virus induces the autophagosome accumulation by blocking autophagosome degradation. Previously, it was hypothesized that M2 protein causes this accumulation by preventing autophagosome fusion with lysosomes, and that the proton channel activity of M2 is involved in this blocking activity. In fact, Gannage et al showed that M2 proton channel activity is not involved in blocking autophagosome fusion with lysosomes. They verified this result by using amantadine hydrochloride, an inhibitor of M2 ion channel activity, which was unable to prevent autophagosome accumulation in IAV‐infected cells. Finally, they showed that the N‐terminal domain of M2 blocks autolysosome formation and acts independently of its proton channel function.160, 185, 186
Targeting cellular proteins could be effective in fighting influenza infections. To harness the full potential of this strategy, we need a greater understanding of the host intracellular pathways that influenza viruses use to replicate.151 The mechanism that blocks autophagosome‐lysosome fusion by the virus requires particular focus on its molecular details.
2.8. Other respiratory viruses and autophagy
Coronaviruses, which infect a variety of hosts from human to birds, mainly cause mild respiratory infections, with the exception of severe acute respiratory syndrome coronavirus (SARS‐CoV).187 The main symptoms of SARS‐CoV infection are fever, dry cough, and shortness of breath. It has been shown that membrane‐associated papain‐like protease PLP2 (PLP2‐TM) of SARS‐CoV is an autophagy‐inducing protein. PLP2‐TM induces the accumulation of autophagosomes and blocks the fusion of autophagosomes with lysosomes. Furthermore, PLP2‐TM interacts with the key autophagy regulators, LC3 and Beclin 1, and promotes Beclin 1 interaction with STING, the key regulator for antiviral IFN signaling.188 Coronavirus NSP6 proteins limit autophagosome expansion to remove host proteins that would inhibit replication. This may favor coronavirus infection by compromising the ability of autophagosomes to deliver viral components to lysosomes for degradation.189 As evidenced by the large quantity of information gathered here, the relationship between viruses and autophagy is complex,190 and fundamental mechanisms of autophagy involvement in viral infection are still in being elucidated.
3. PERSPECTIVE
The autophagy pathway interacts with viruses. As a result of co‐evolution, viruses developed numerous approaches to usurp the host autophagy system. The effect of autophagy on virus fate is time dependent and may play both pro‐viral and antiviral roles during the multiplication and virus pathogenesis stages.
Autophagy is a determining factor when considering negative‐sense RNA viruses such as influenza viruses. A study by our research team showed that when autophagy induction is promoted before virus infection, virus titer enhanced significantly 24 hours post‐infection, but it was not significant 48 hours post‐infection. In contrast when autophagy formation is induced after virus infection, the virus replication was inhibited 24 and 48 hours post‐infection.191 Additionally, we showed that inhibition of autophagy using 3‐MA significantly reduced viral replication.191
In chronic HCV infection, selectively autophagy induction downregulates the IFN‐α receptor‐1 chain and the expression of the nucleoside transporters ENT1 and CNT1, thus leading to drug resistance.86 Therefore, in new treatments of HCV chronic infection, it is necessary to include an autophagy inhibitor drug. On the other hand, telomerase activation is crucial for immortalization and establishment of malignant tumor cells.192 It is well documented that telomerase activation by viral oncoproteins such as HBV X protein (HBx)193 and HCV core194 is connected to carcinoma. The results of our recent study suggest that the induction of autophagy reduces telomerase activity in tumor cells.195 Thus, pharmacological induction of autophagy may reduce telomerase activity and tumor progression.
Growing evidence suggests that autophagy contributes to MHC class I and II antigen presentation and processing of certain endogenously produced peptides. Hence, precisely targeting antigens to autophagy pathways via fusion with the LC3 protein may be a promising strategy for eliciting CD4+ and CD8+ T cell responses. Autophagy‐targeted candidate vaccine composed of 19‐kDa Mycobacterium tuberculosis lipoprotein (LpqH) DNA and microtubule‐associated protein light chain‐3 (LC3) gene, which transfers LpqH to autophagosomes able to enhance protective efficiency against Mycobacterium tuberculosis. Mice immunized with the aforementioned candidate vaccine had lower mycobacterial loads in the lung and spleen with increased IgG2a and IFN‐γ and IL‐2 levels.196 Our team formulated Beclin‐1, an autophagy initiator, as an adjuvant with an HEV candidate DNA vaccine in a mouse model. The immunized mice induced robust immune response197 but have not significant effect on HEV protein candidate vaccine.198 The role of autophagy in apoptosis of uninfected bystander cells, adaptive immunity by viral processing and MHC‐antigen presentation, and in innate immunity by affecting pathogen sensing opens novel approaches for an HIV‐1 vaccine and combination treatment strategies.199 Autophagy controls immunogenic signaling throughout cancer therapy which can be exploited to design therapeutic combinations with methods that either activate or inhibit autophagy to promote the therapeutic efficacy of oncolytic virus immunotherapy.200
Many viruses use the host autophagy pathway, but relatively little is known regarding the specific mechanisms by which viruses manipulate autophagy for their gain. We hypothesize that autophagy may be important for the maturation of viral particles and proteins that need to be processed and cleaved with cellular protease under acidic condition. Upon virus escape to cytoplasm, positive strand RNA viruses initiate polyprotein synthesis. The viral precursor protein may be engulfed by autophagy and delivered to the lysosome for processing by cellular protease. For example, the HIV‐1 precursor Gag interacts with the autophagy factor LC3, suggesting that autophagy promoted productive Gag processing and that viral particles were produced in macrophages.116
4. CONCLUSION
In this review, we summarize the multifaceted roles of autophagy in normal physiology and viral infection and pathogenesis (Table 1). We highlight recent advances in understanding the relationship between autophagy and viral infection, and how autophagy plays dual roles in the disease progression. These data provide an overview of the molecular mechanisms that underlie autophagy, the role of this pathway in the pathogenesis of viruses, and strategies for therapeutic modulation. Manipulation of autophagy heralds the potential for highly effective treatments for a wide range of clinical diseases, including both bacterial and viral infections as well as autoimmune and inflammatory disease states which deserve attention. Although our knowledge of the autophagy pathway is quickly progressing, there is still much to be learned regarding the specific molecules that regulate this pathway and the mechanisms by which viruses target these molecules to facilitate their replication. Further investigations are required to be examined in preclinical and clinical trials.
Table 1.
Cross‐talking between autophagy and viral infections
| Viruses | Autophagy Induction Mechanism | Autophagy Inhibition Mechanism |
|---|---|---|
| HCV |
‐HCV NS4B triggers a stress response that induces autophagy. Also, it leads to the lipidation of LC3 and forms complexes with Rab5, Vps34, and Beclin‐171 ‐HCV triggers lipoprotein degradation through lipophagy.84 ‐HCV causes mitochondrial damage and oxidative stress, and impaired mitochondria are selectively eliminated mitophagy.75 ‐NS5B/ATG5 interaction could be necessary for the establishment of HCV replication.79
HCV‐NS3 induces autophagy in an IRGM‐dependent pathway.62
HCV activates a selective autophagy for lipids (lipophagy) protects cells from an excessive lipid accumulation.13 |
‐‐‐ |
| HBV |
‐the X protein of HBV interacts to, and induces, both phosphatidylinositol‐3‐kinase class 3 (PI3KC3) and death‐associated protein kinase (DAPK), activating autophagic signaling.193
‐autophagy inhibits tumorigenesis of HBV‐associated Hepato cellular carcinoma (HCC) by degradation of microRNA‐224.92
‐HBV small surface protein (SHB) activates autophagy.201 |
‐HBV X protein blocks autophagic degradation via inhibition of lysosomes.202 |
| Rotavirus |
‐rotavirus NSP4 cause to release of calcium from the ER lumen, triggering autophagic signaling.64 |
‐‐‐ |
| HIV |
‐Tat: Tat enhances autophagic degradation through increasing fusion process in neurons.127
‐Gag: Gag macrophages Gag‐derived proteins co‐localized and interacted with the autophagy factor LC3, and autophagy promoted productive Gag processing.116
‐Env: Env attaches to CXCR4 and leads to autophagy triggers cell death in neurons and uninfected T cells.122 ‐Env induces autophagy and promotes autophagic T cell death, in bystander T cell.123
‐Nef: Nef induces autophagy in an IRGM‐dependent pathway.62 |
‐Nef serves as an “antiautophagic maturation factor” and blocks the late proteolytic stage of autophagy.116 ‐Nef mimics Baf A1 and blocks the formation of autophagolysosome in human astrocytes.203 ‐Tat suppressed IFN‐γ‐induced autophagy in human macrophages.130 ‐Tat blocks autophagy through Src‐Akt and STAT3 signaling in uninfected macrophages and monocytes.60 |
| Influenza virus |
‐NS1 is able to upregulate autophagy.163
‐proteolytic cleavage of viral HA activates autophagy. |
‐M2 blocks autophagosome–lysosome fusion by means of its viroporin activity.48 |
CONFLICTS OF INTEREST
None declared.
ACKNOWLEDGEMENTS
We greatly appreciate Mehrdad Araiinejad for thoughtful comments that helped improve this manuscript.
Abdoli A, Alirezaei M, Mehrbod P, Forouzanfar F. Autophagy: The multi‐purpose bridge in viral infections and host cells. Rev Med Virol. 2018;28:e1973 10.1002/rmv.1973
REFERENCES
- 1. Cao Y, Klionsky DJ. Physiological functions of Atg6/Beclin 1: a unique autophagy‐related protein. Cell Res. 2007;17(10):839‐849. [DOI] [PubMed] [Google Scholar]
- 2. Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12(9):814‐822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Levine B, Klionsky DJ. Development by self‐digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6(4):463‐477. [DOI] [PubMed] [Google Scholar]
- 4. Eskelinen E‐L. Maturation of autophagic vacuoles in mammalian cells. Autophagy. 2005;1(1):1‐10. [DOI] [PubMed] [Google Scholar]
- 5. Axe EL, Walker SA, Manifava M, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3‐phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182(4):685‐701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221(1):3‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9(12):1004‐1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kroemer G, Galluzzi L, Vandenabeele P, et al. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16(1):3‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ashford TP, Porter KR. Cytoplasmic components in hepatic cell lysosomes. J Cell Biol. 1962;12(1):198‐202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Reuck AVS, Cameron MP. Ciba Foundation Symposium on Lysosomes. 1963. London: JA Churchill; 2008. [Google Scholar]
- 11. Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase‐deficient mutants and conditions for its induction. J Cell Biol. 1992;119(2):301‐311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee YJ, Jang BK. The role of autophagy in hepatocellular carcinoma. Int J Mol Sci. 2015;16(11):26629‐26643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Singh R, Cuervo AM. Lipophagy: connecting autophagy and lipid metabolism. Int J Cell Biol. 2012;2012:1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim I, Rodriguez‐Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462(2):245‐253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rodriguez‐Enriquez S, Kim I, Currin RT, Lemasters JJ. Tracker dyes to probe mitochondrial autophagy (mitophagy) in rat hepatocytes. Autophagy. 2006;2(1):39‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reggiori F, Klionsky DJ. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics. 2013;194(2):341‐361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10(7):458‐467. [DOI] [PubMed] [Google Scholar]
- 18. Mizushima N. Autophagy: process and function. Genes Dev. 2007;21(22):2861‐2873. [DOI] [PubMed] [Google Scholar]
- 19. Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18(16):1926‐1945. [DOI] [PubMed] [Google Scholar]
- 20. Shrivastava S, Bhanja Chowdhury J, Steele R, Ray R, Ray RB. Hepatitis C virus upregulates Beclin1 for induction of autophagy and activates mTOR signaling. J Virol. 2012;86(16):8705‐8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584(7):1287‐1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132‐141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9(10):1102‐1109. [DOI] [PubMed] [Google Scholar]
- 24. Backer JM. The regulation and function of class III PI3Ks: novel roles for Vps34. Biochem J. 2008;410(1):1‐17. [DOI] [PubMed] [Google Scholar]
- 25. Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy, and tumor suppressor function. Autophagy. 2005;1(1):46‐52. [DOI] [PubMed] [Google Scholar]
- 26. Kuballa P, Nolte WM, Castoreno AB, Xavier RJ. Autophagy and the immune system. Annu Rev Immunol. 2012;30(1):611‐646. [DOI] [PubMed] [Google Scholar]
- 27. Matsunaga K, Saitoh T, Tabata K, et al. Two Beclin 1‐binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11(4):385‐396. [DOI] [PubMed] [Google Scholar]
- 28. Liang C, Feng P, Ku B, et al. Autophagic and tumour suppressor activity of a novel Beclin1‐binding protein UVRAG. Nat Cell Biol. 2006;8(7):688‐698. [DOI] [PubMed] [Google Scholar]
- 29. Fimia GM, Stoykova A, Romagnoli A, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447(7148):1121‐1125. [DOI] [PubMed] [Google Scholar]
- 30. Hanada T, Noda NN, Satomi Y, et al. The Atg12‐Atg5 conjugate has a novel E3‐like activity for protein lipidation in autophagy. J Biol Chem. 2007;282(52):37298‐37302. [DOI] [PubMed] [Google Scholar]
- 31. Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19(5):2092‐2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mann SS, Hammarback JA. Molecular characterization of light chain 3. A microtubule binding subunit of MAP1A and MAP1B. J Biol Chem. 1994;269(15):11492‐11497. [PubMed] [Google Scholar]
- 33. Kabeya Y, Mizushima N, Yamamoto A, Oshitani‐Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form‐II formation. J Cell Sci. 2004;117(13):2805‐2812. [DOI] [PubMed] [Google Scholar]
- 34. Kabeya Y, Mizushima N, Ueno T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. The EMBO J. 2000;19(21):5720‐5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rubinsztein DC, Cuervo AM, Ravikumar B, et al. In search of an “autophagomometer”. 2009;5:585‐589. Taylor & Francis [DOI] [PubMed] [Google Scholar]
- 36. Schmid, D. , Autophagy delivers viral antigens for MHC class II presentation and is regulated by viral infection, 2007, The Rockefeller University. p. 205.
- 37. Cheng J, Ohsaki Y, Tauchi‐Sato K, Fujita A, Fujimoto T. Cholesterol depletion induces autophagy. Biochem Biophys Res Commun. 2006;351(1):246‐252. [DOI] [PubMed] [Google Scholar]
- 38. Rubinsztein DC, Cuervo AM, Ravikumar B, et al. In search of an autophagomometer. Autophagy. 2009;5(5):585‐589. [DOI] [PubMed] [Google Scholar]
- 39. Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Alexander D, Leib DA. Xenophagy in herpes simplex virus replication and pathogenesis. Autophagy. 2008;4(1):101‐103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Orvedahl A, MacPherson S, Sumpter R Jr, Tallóczy Z, Zou Z, Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe. 2010;7(2):115‐127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jackson WT. Viruses and the autophagy pathway. Virology. 2015;479–480:450‐456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lennemann NJ, Coyne CB. Catch me if you can: the link between autophagy and viruses. PLoS Pathog. 2015;11(3):e1004685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shi J, Luo H. Interplay between the cellular autophagy machinery and positive‐stranded RNA viruses. Acta Biochim Biophys Sin. 2012;44(5):375‐384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rubinsztein DC, DiFiglia M, Heintz N, et al. Autophagy and its possible roles in nervous system diseases, damage and repair. Autophagy. 2005;1(1):11‐22. [DOI] [PubMed] [Google Scholar]
- 46. Munz C. Autophagy and antigen presentation. Cell Microbiol. 2006;8(6):891‐898. [DOI] [PubMed] [Google Scholar]
- 47. Deretic V, Singh S, Master S, et al. Mycobacterium tuberculosis inhibition of phagolysosome biogenesis and autophagy as a host defence mechanism. Cell Microbiol. 2006;8(5):719‐727. [DOI] [PubMed] [Google Scholar]
- 48. Rossman JS, Lamb RA. Autophagy, apoptosis, and the influenza virus M2 protein. Cell Host Microbe. 2009;6(4):367‐380. [DOI] [PubMed] [Google Scholar]
- 49. Wileman T. Aggresomes and autophagy generate sites for virus replication. Science. 2006;312(5775):875‐878. [DOI] [PubMed] [Google Scholar]
- 50. Matarrese P, Nencioni L, Checconi P, et al. Pepstatin A alters host cell autophagic machinery and leads to a decrease in influenza A virus production. J Cell Physiol. 2011;226(12):3368‐3377. [DOI] [PubMed] [Google Scholar]
- 51. Shintani T, Klionsky DJ. Autophagy in health and disease: a double‐edged sword. Science. 2004;306(5698):990‐995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kemball CC, Alirezaei M, Flynn CT, et al. Coxsackievirus infection induces autophagy‐like vesicles and megaphagosomes in pancreatic acinar cells in vivo. J Virol. 2010;84(23):12110‐12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Alirezaei M, Flynn CT, Wood MR, Harkins S, Whitton JL. Coxsackievirus can exploit LC3 in both autophagy‐dependent and‐independent manners in vivo. Autophagy. 2015;11(8):1389‐1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Iwasaki A. Role of autophagy in innate viral recognition. Autophagy. 2007;3(4):354‐356. [DOI] [PubMed] [Google Scholar]
- 55. Blanchet FP, Moris A, Nikolic DS, et al. Human immunodeficiency Virus‐1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity. 2010;32(5):654‐669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gannage M, Münz C. MHC presentation via autophagy and how viruses escape from it. Semin Immunopathol. 2010;32(4):373‐381. [DOI] [PubMed] [Google Scholar]
- 57. Chan ST, Jing‐hsiung JO. Hepatitis C virus‐induced autophagy and host innate immune response. Virus. 2017;9(8):224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Desai M, Fang R, Sun J. The role of autophagy in microbial infection and immunity. Immunotargets and Therapy. 2015;4:13‐26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lee HK, Iwasaki A. Autophagy and antiviral immunity. Curr Opin Immunol. 2008;20(1):23‐29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Grol JV, Subauste C, Andrade RM, Fujinaga K, Nelson J, Subauste CS. HIV‐1 inhibits autophagy in bystander macrophage/monocytic cells through Src‐Akt and STAT3. PLoS One. 2010;5(7):e11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Oh JE, Lee HK. Autophagy in innate recognition of pathogens and adaptive immunity. Yonsei Med J. 2012;53(2):241‐247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Grégoire IP, Richetta C, Meyniel‐Schicklin L, et al. IRGM is a common target of RNA viruses that subvert the autophagy network. PLoS Pathog. 2011;7(12):e1002422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Uhl M, Kepp O, Jusforgues‐Saklani H, Vicencio JM, Kroemer G, Albert ML. Autophagy within the antigen donor cell facilitates efficient antigen cross‐priming of virus‐specific CD8+ T cells. Cell Death Differ. 2009;6(7):991‐1005. [DOI] [PubMed] [Google Scholar]
- 64. Crawford SE, Hyser JM, Utama B, Estes MK. Autophagy hijacked through viroporin‐activated calcium/calmodulin‐dependent kinase kinase‐β signaling is required for rotavirus replication. Proc Natl Acad Sci U S A. 2012;109(50):E3405‐E3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Alavian S, Ande SR, Coombs KM, et al. Virus‐triggered autophagy in viral hepatitis—possible novel strategies for drug development. J Viral Hepat. 2011;18(12):821‐830. [DOI] [PubMed] [Google Scholar]
- 66. Choo Q, Weiner AJ, Overby LR, Kuo G, Houghton M, Bradley DW. Hepatitis C virus: the major causative agent of viral non‐A, non‐B hepatitis. Br Med Bull. 1990;46(2):423‐441. [DOI] [PubMed] [Google Scholar]
- 67. Ke P‐Y, Chen SS. Autophagy in hepatitis C virus‐host interactions: potential roles and therapeutic targets for liver‐associated diseases. World J Gastroenterol. 2014;20(19):5773‐5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Blanchard E, Belouzard S, Goueslain L, et al. Hepatitis C virus entry depends on clathrin‐mediated endocytosis. J Virol. 2006;80(14):6964‐6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tscherne DM, Jones CT, Evans MJ, Lindenbach BD, McKeating JA, Rice CM. Time‐and temperature‐dependent activation of hepatitis C virus for low‐pH‐triggered entry. J Virol. 2006;80(4):1734‐1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Romero‐Brey I, Merz A, Chiramel A, et al. Three‐dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 2012;8(12):e1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ke P‐Y, Chen SS‐L. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest. 2011;121(1):37‐56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106(33):14046‐14051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012;13(2):89‐102. [DOI] [PubMed] [Google Scholar]
- 74. Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334(6059):1081‐1086. [DOI] [PubMed] [Google Scholar]
- 75. Vescovo T, Refolo G, Romagnoli A, et al. Autophagy in HCV infection: keeping fat and inflammation at bay. Biomed Res Int. 2014;2014:1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kim S‐J, Syed GH, Siddiqui A. Hepatitis C virus induces the mitochondrial translocation of Parkin and subsequent mitophagy. PLoS Pathog. 2013;9(3):e1003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kim S‐J, Syed GH, Khan M, et al. Hepatitis C virus triggers mitochondrial fission and attenuates apoptosis to promote viral persistence. Proc Natl Acad Sci U S A. 2014;111(17):6413‐6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wang J, Kang R, Huang H, et al. Hepatitis C virus core protein activates autophagy through EIF2AK3 and ATF6 UPR pathway‐mediated MAP1LC3B and ATG12 expression. Autophagy. 2014;10(5):766‐784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Guévin C, Manna D, Bélanger C, Konan KV, Mak P, Labonté P. Autophagy protein ATG5 interacts transiently with the hepatitis C virus RNA polymerase (NS5B) early during infection. Virology. 2010;405(1):1‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wang L, Tian Y, Ou J‐hJ. HCV induces the expression of Rubicon and UVRAG to temporally regulate the maturation of autophagosomes and viral replication. PLoS Pathog. 2015;11(3):e1004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wang L, Ou J‐h J. Hepatitis C virus and autophagy. Biol Chem. 2015;396(11):1215‐1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Huang H, Kang R, Wang J, Luo G, Yang W, Zhao Z. Hepatitis C virus inhibits AKT‐tuberous sclerosis complex (TSC), the mechanistic target of rapamycin (MTOR) pathway, through endoplasmic reticulum stress to induce autophagy. Autophagy. 2013;9(2):175‐195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tanida I, Fukasawa M, Ueno T, Kominami E, Wakita T, Hanada K. Knockdown of autophagy‐related gene decreases the production of infectious hepatitis C virus particles. Autophagy. 2009;5(7):937‐945. [DOI] [PubMed] [Google Scholar]
- 84. Liu K, Czaja M. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 2013;20(1):3‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Vescovo T, Romagnoli A, Perdomo AB, et al. Autophagy protects cells from HCV‐induced defects in lipid metabolism. Gastroenterology. 2012;142(3):644, e3‐653. [DOI] [PubMed] [Google Scholar]
- 86. Chandra PK, Bao L, Song K, et al. HCV infection selectively impairs type I but not type III IFN signaling. Am J Pathol. 2014;184(1):214‐229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Desai MM, Gong B, Chan T, et al. Differential, type I interferon‐mediated autophagic trafficking of hepatitis C virus proteins in mouse liver. Gastroenterology. 2011;141(2):674, e6‐685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sir D, Tian Y, Chen WL, Ann DK, Yen TSB, Ou JHJ. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc Natl Acad Sci U S A. 2010;107(9):4383‐4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lavanchy D. Worldwide epidemiology of HBV infection, disease burden, and vaccine prevention. J Clin Virol. 2005;34:S1‐S3. [DOI] [PubMed] [Google Scholar]
- 90. Li J, Liu Y, Wang Z, et al. Subversion of cellular autophagy machinery by hepatitis B virus for viral envelopment. J Virol. 2011;85(13):6319‐6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lazar C, Macovei A, Petrescu S, Branza‐Nichita N. Activation of ERAD pathway by human hepatitis B virus modulates viral and subviral particle production. PLoS One. 2012;7(3):e34169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lan SH, Wu SY, Zuchini R, et al. Autophagy suppresses tumorigenesis of hepatitis B virus‐associated hepatocellular carcinoma through degradation of microRNA‐224. Hepatology. 2014;59(2):505‐517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Zhang B, Halder SK, Kashikar ND, et al. Antimetastatic role of Smad4 signaling in colorectal cancer. Gastroenterology. 2010;138(3):969, e3‐980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wickner R. Double‐stranded RNA Virus replication and packaging. J Biol Chem. 1993;268:3797‐3797. [PubMed] [Google Scholar]
- 95. Ball JM, Tian P, Zeng CQY, Morris AP, Estes MK. Age‐dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science. 1996;272(5258):101‐104. [DOI] [PubMed] [Google Scholar]
- 96. Estes MK, Kang G, Zeng CQ‐Y, Crawford SE, Ciarlet M. Pathogenesis of Rotavirus Gastroenteritis. in Novartis Foundation Symposium. 1999 Chichester; New York: John Wiley; 2001. [DOI] [PubMed] [Google Scholar]
- 97. Berkova Z, Crawford SE, Trugnan G, Yoshimori T, Morris AP, Estes MK. Rotavirus NSP4 induces a novel vesicular compartment regulated by calcium and associated with viroplasms. J Virol. 2006;80(12):6061‐6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Crawford SE, Estes MK. Viroporin‐mediated calcium‐activated autophagy. Autophagy. 2013;9(5):797‐798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22(2):132‐139. [DOI] [PubMed] [Google Scholar]
- 100. Yang W, McCrae MA. The rotavirus enterotoxin (NSP4) promotes re‐modeling of the intracellular microtubule network. Virus Res. 2012;163(1):269‐274. [DOI] [PubMed] [Google Scholar]
- 101. Martin D, Duarte M, Lepault J, Poncet D. Sequestration of free tubulin molecules by the viral protein NSP2 induces microtubule depolymerization during rotavirus infection. J Virol. 2010;84(5):2522‐2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Jahreiss L, Menzies FM, Rubinsztein DC. The itinerary of autophagosomes: from peripheral formation to kiss‐and‐run fusion with lysosomes. Traffic. 2008;9(4):574‐587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Cuadras MA, Feigelstock DA, An S, Greenberg HB. Gene expression pattern in Caco‐2 cells following rotavirus infection. J Virol. 2002;76(9):4467‐4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Campagna M, Eichwald C, Vascotto F, Burrone OR. RNA interference of rotavirus segment 11 mRNA reveals the essential role of NSP5 in the virus replicative cycle. J Gen Virol. 2005;86(5):1481‐1487. [DOI] [PubMed] [Google Scholar]
- 105. Arnoldi F, de Lorenzo G, Mano M, et al. Rotavirus increases levels of lipidated LC3 supporting accumulation of infectious progeny virus without inducing autophagosome formation. PLoS One. 2014;9(4):e95197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Alexaki A, Liu Y, Wigdahl B. Cellular reservoirs of HIV‐1 and their role in viral persistence. Curr HIV Res. 2008;6(5):388‐400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Svicher V, Ceccherini‐Silberstein F, Antinori A, Aquaro S, Perno CF. Understanding HIV compartments and reservoirs. Curr HIV/AIDS Rep. 2014;11(2):186‐194. [DOI] [PubMed] [Google Scholar]
- 108. Hong FF, Mellors JW. Changes in HIV reservoirs during long‐term antiretroviral therapy. Curr Opin HIV AIDS. 2015;10(1):43‐48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Perng G‐C, Jones C. Towards an understanding of the herpes simplex virus type 1 latency‐reactivation cycle. Interdiscip Perspect Infect Dis. 2010;2010:1‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Siliciano RF, Greene WC. HIV latency. Cold Spring Harb Perspect Med. 2011;1(1):a007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Delgado M, Singh S, de Haro S, et al. Autophagy and pattern recognition receptors in innate immunity. Immunol Rev. 2009;227(1):189‐202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Shi C‐S, Kehrl JH. TRAF6 and A20 regulate lysine 63–linked ubiquitination of beclin‐1 to control TLR4‐induced autophagy. Sci Signal. 2010;3(123): p. ra42‐ra42):ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V. Autophagy‐based unconventional secretory pathway for extracellular delivery of IL‐1β. The EMBO J. 2011;30(23):4701‐4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Shi C‐S, Shenderov K, Huang NN, et al. Activation of autophagy by inflammatory signals limits IL‐1 [beta] production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. 2012;13(3):255‐263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13(10):722‐737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Kyei GB, Dinkins C, Davis AS, et al. Autophagy pathway intersects with HIV‐1 biosynthesis and regulates viral yields in macrophages. J Cell Biol. 2009;186(2):255‐268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Ameisen JC, Capron A. Cell dysfunction and depletion in AIDS: the programmed cell death hypothesis. Immunol Today. 1991;12(4):102‐105. [DOI] [PubMed] [Google Scholar]
- 118. Terai C, Kornbluth RS, Pauza CD, Richman DD, Carson DA. Apoptosis as a mechanism of cell death in cultured T lymphoblasts acutely infected with HIV‐1. J Clin Invest. 1991;87(5):1710‐1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Doitsh G, Galloway NLK, Geng X, et al. Pyroptosis drives CD4 T‐cell depletion in HIV‐1 infection. Nature. 2014;505(7484):509‐514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Garg H, Mohl J, Joshi A. HIV‐1 induced bystander apoptosis. Virus. 2012;4(11):3020‐3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Kumar V. Robbins Basic Pathology. 9th Edition | Vinay Kumar, Abul Abbas, Jon Aster | ISBN 9781437717815. 2012 ed.; 2012. 2016/12/15/17:47:37; Available from: http://store.elsevier.com/Robbins-Basic-Pathology/Vinay-Kumar/isbn-9781437717815. [Google Scholar]
- 122. Denizot M, Varbanov M, Espert L, et al. HIV‐1 gp41 fusogenic function triggers autophagy in uninfected cells. Autophagy. 2008;4(8):998‐1008. [DOI] [PubMed] [Google Scholar]
- 123. Espert L, Varbanov M, Robert‐Hebmann V, et al. Differential role of autophagy in CD4 T cells and macrophages during X4 and R5 HIV‐1 infection. PLoS One. 2009;4(6):e5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Zhou D, Spector SA. Human immunodeficiency virus type‐1 infection inhibits autophagy. AIDS. 2008;22(6):695‐699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Castedo M, Roumier T, Blanco J, et al. Sequential involvement of Cdk1, mTOR and p53 in apoptosis induced by the HIV‐1 envelope. The EMBO J. 2002;21(15):4070‐4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Nardacci R, Amendola A, Ciccosanti F, et al. Autophagy plays an important role in the containment of HIV‐1 in nonprogressor‐infected patients. Autophagy. 2014;10(7):1167‐1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Sagnier S, Daussy CF, Borel S, et al. Autophagy restricts HIV‐1 infection by selectively degrading Tat in CD4+ T lymphocytes. J Virol. 2015;89(1):615‐625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Borel S, Robert‐Hebmann V, Alfaisal J, et al. HIV‐1 viral infectivity factor interacts with microtubule‐associated protein light chain 3 and inhibits autophagy. AIDS. 2015;29(3):275‐286. [DOI] [PubMed] [Google Scholar]
- 129. Badou A, Bennasser Y, Moreau M, Leclerc C, Benkirane M, Bahraoui E. Tat protein of human immunodeficiency virus type 1 induces interleukin‐10 in human peripheral blood monocytes: implication of protein kinase C‐dependent pathway. J Virol. 2000;74(22):10551‐10562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Li JCB, Au KY, Fang JW, et al. HIV‐1 trans‐activator protein dysregulates IFN‐γ signaling and contributes to the suppression of autophagy induction. AIDS. 2011;25(1):15‐25. [DOI] [PubMed] [Google Scholar]
- 131. Piguet V, Blauvelt A. Essential roles for dendritic cells in the pathogenesis and potential treatment of HIV disease. J Invest Dermatol. 2002;119(2):365‐369. [DOI] [PubMed] [Google Scholar]
- 132. Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy‐dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315(5817):1398‐1401. [DOI] [PubMed] [Google Scholar]
- 133. Zhou D, Kang KH, Spector SA. Production of interferon α by human immunodeficiency virus type 1 in human plasmacytoid dendritic cells is dependent on induction of autophagy. J Infect Dis. 2012;205(8):1258‐1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Alter G, Suscovich TJ, Teigen N, et al. Single‐stranded RNA derived from HIV‐1 serves as a potent activator of NK cells. J Immunol. 2007;178(12):7658‐7666. [DOI] [PubMed] [Google Scholar]
- 135. Hervas‐Stubbs S, Perez‐Gracia JL, Rouzaut A, Sanmamed MF, le Bon A, Melero I. Direct effects of type I interferons on cells of the immune system. Clin Cancer Res. 2011;17(9):2619‐2627. [DOI] [PubMed] [Google Scholar]
- 136. Cory TJ, Schacker TW, Stevenson M, Fletcher CV. Overcoming pharmacologic sanctuaries. Curr Opin HIV AIDS. 2013;8(3):190‐195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Eisfeld C, Reichelt D, Evers S, Husstedt I. CSF penetration by antiretroviral drugs. CNS Drugs. 2013;27(1):31‐55. [DOI] [PubMed] [Google Scholar]
- 138. Letendre S, Marquie‐Beck J, Capparelli E, et al. Validation of the CNS penetration‐effectiveness rank for quantifying antiretroviral penetration into the central nervous system. Arch Neurol. 2008;65(1):65‐70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Kramer‐Hämmerle S, Rothenaigner I, Wolff H, Bell JE, Brack‐Werner R. Cells of the central nervous system as targets and reservoirs of the human immunodeficiency virus. Virus Res. 2005;111(2):194‐213. [DOI] [PubMed] [Google Scholar]
- 140. Valcour V, Chalermchai T, Sailasuta N, et al. Central nervous system viral invasion and inflammation during acute HIV infection. J Infect Dis. 2012;206(2):275‐282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. García‐Arencibia M, Hochfeld WE, Toh PPC, Rubinsztein DC. Autophagy, a guardian against neurodegeneration. Semin Cell Dev Biol. 2010;21(7):691‐698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov. 2012;11(9):709‐730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Zhou D, Masliah E, Spector SA. Autophagy is increased in postmortem brains of persons with HIV‐1‐associated encephalitis. J Infect Dis. 2011;203(11):1647‐1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Alirezaei M, Kiosses WB, Fox HS. Decreased neuronal autophagy in HIV dementia: a mechanism of indirect neurotoxicity. Autophagy. 2008;4(7):963‐966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Fields J, Dumaop W, Rockenstein E, et al. Age‐dependent molecular alterations in the autophagy pathway in HIVE patients and in a gp120 tg mouse model: reversal with beclin‐1 gene transfer. J Neurovirol. 2013;19(1):89‐101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Fields J, Dumaop W, Elueteri S, et al. HIV‐1 Tat alters neuronal autophagy by modulating autophagosome fusion to the lysosome: implications for HIV‐associated neurocognitive disorders. J Neurosci. 2015;35(5):1921‐1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Espert L, Denizot M, Grimaldi M, et al. Autophagy is involved in T cell death after binding of HIV‐1 envelope proteins to CXCR4. J Clin Invest. 2006;116(8):2161‐2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Levine B, Packer M, Codogno P. Development of autophagy inducers in clinical medicine. J Clin Invest. 2015;125(1):14‐24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Mostowy S. Autophagy and bacterial clearance: a not so clear picture. Cell Microbiol. 2013;15(3):395‐402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Webster R, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56(1):152‐179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Haidari M, Zhang W, Ganjehei L, Ali M, Chen Z. Inhibition of MLC phosphorylation restricts replication of influenza virus—a mechanism of action for anti‐influenza agents. PLoS One. 2011;6(6):e21444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Whittaker GR. Intracellular trafficking of influenza virus: clinical implications for molecular medicine. Expert Rev Mol Med. 2001;3:1‐13. [DOI] [PubMed] [Google Scholar]
- 153. Zhou Z, Jiang X, Liu D, et al. Autophagy is involved in influenza A virus replication. Autophagy. 2009;5(3):321‐328. [DOI] [PubMed] [Google Scholar]
- 154. Ochiai H, Sakai S, Hirabayashi T, Shimizu Y, Terasawa K. Inhibitory effect of bafilomycin A1, a specific inhibitor of vacuolar‐type proton pump, on the growth of influenza A and B viruses in MDCK cells. Antiviral Res. 1995;27(4):425‐430. [DOI] [PubMed] [Google Scholar]
- 155. Yeganeh B, Ghavami S, Kroeker AL, et al. Suppression of influenza a virus replication in human lung epithelial cells by noncytotoxic concentrations bafilomycin A1. Am J Physiol Gastrointest Liver Physiol. 2015;308(3):L270‐L286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Sumpter R, Levine B. Autophagy and innate immunity: triggering, targeting and tuning In: Semin Cell Dev Biol. Elsevier; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Ghavami S, Mutawe MM, Hauff K, et al. Statin‐triggered cell death in primary human lung mesenchymal cells involves p53‐PUMA and release of Smac and Omi but not cytochrome c. Biochim Biophys Acta. 2010;1803(4):452‐467. [DOI] [PubMed] [Google Scholar]
- 158. Codogno P, Meijer AJ. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 2005;12(2):1509‐1518. [DOI] [PubMed] [Google Scholar]
- 159. Pattingre S, Tassa A, Qu X, et al. Bcl‐2 antiapoptotic proteins inhibit Beclin 1‐dependent autophagy. Cell. 2005;122(6):927‐939. [DOI] [PubMed] [Google Scholar]
- 160. Gannagé M, Dormann D, Albrecht R, et al. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe. 2009;6(4):367‐380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Dai JP, Li WZ, Zhao XF, et al. A drug screening method based on the autophagy pathway and studies of the mechanism of evodiamine against influenza A virus. PLoS One. 2012;7(8):e42706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Mehrbod P, Hair‐Bejo M, Tengku Ibrahim TA, et al. Simvastatin modulates cellular components in influenza A virus‐infected cells. Int J Mol Med. 2014;34(1):61‐73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Hale BG, Jackson D, Chen YH, Lamb RA, Randall RE. Influenza A virus NS1 protein binds p85beta and activates phosphatidylinositol‐3‐kinase signaling. Proc Natl Acad Sci U S A. 2006;103(38):14194‐14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Esghaei M, Monavari SHR, Tavassoti‐Kheiri M, et al. Expression of the influenza M2 protein in three different eukaryotic cell lines. J Virol Methods. 2012;179(1):161‐165. [DOI] [PubMed] [Google Scholar]
- 165. Betakova T, Hay JA. Stability and function of the influenza A virus M2 ion channel protein is determined by both extracellular and cytoplasmic domains. Arch Virol. 2009;154(1):147‐151. [DOI] [PubMed] [Google Scholar]
- 166. Zebedee SL, Lamb RA. Influenza A virus M2 protein: monoclonal antibody restriction of virus growth and detection of M2 in virions. J Virol. 1988;62(8):2762‐2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Lamb RA, Zebedee SL, Richardson CD. Influenza virus M2 protein is an integral membrane protein expressed on the infected‐cell surface. Cell. 1985;40:627‐633. [DOI] [PubMed] [Google Scholar]
- 168. Ciampor F, Thompson CA, Grambas S, Hay AJ. Regulation of pH by the M2 protein of influenza A viruses. Virus Res. 1992;22(3):247‐258. [DOI] [PubMed] [Google Scholar]
- 169. Egan DF, Kim J, Shaw RJ, Guan KL. The autophagy initiating kinase ULK1 is regulated via opposing phosphorylation by AMPK and mTOR. Autophagy. 2011;7(6):645‐646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Liu G, Zhong M, Guo C, et al. Autophagy is involved in regulating influenza A virus RNA and protein synthesis associated with both modulation of Hsp90 induction and mTOR/p70S6K signaling pathway. Int J Biochem Cell Biol. 2016;72:100‐108. [DOI] [PubMed] [Google Scholar]
- 171. Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717‐1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Ma J, Sun Q, Mi R, Zhang H. Avian influenza A virus H5N1 causes autophagy‐mediated cell death through suppression of mTOR signaling. J Genet Genomics. 2011;38(11):533‐537. [DOI] [PubMed] [Google Scholar]
- 173. Datan E, Shirazian A, Benjamin S, et al. mTOR/p70S6K signaling distinguishes routine, maintenance‐level autophagy from autophagic cell death during influenza a infection. Virology. 2014;452–453:175‐190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Pan H, Zhang Y, Luo Z, et al. Autophagy mediates avian influenza H5N1 pseudotyped particle‐induced lung inflammation through NF‐κB and p38 MAPK signaling pathways. Am J Physiol Lung Cell Mol Physiol. 2014;306(2):L183‐L195. [DOI] [PubMed] [Google Scholar]
- 175. Law AH‐Y, Lee DCW, Yuen KY, Peiris M, Lau ASY. Cellular response to influenza virus infection: a potential role for autophagy in CXCL10 and interferon‐alpha induction. Cell Mol Immunol. 2010;7(4):263‐270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Zhao H, Zhao YG, Wang X, et al. Mice deficient in EPG5 exhibit selective neuronal vulnerability to degeneration. J Cell Biol. 2013;200(6):731‐741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Lu Q, Yokoyama CC, Williams JW, et al. Homeostatic control of innate lung inflammation by Vici Syndrome Gene Epg5 and additional autophagy genes promotes influenza pathogenesis. Cell Host Microbe. 2016;19(1):102‐113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Ehrhardt C, Marjuki H, Wolff T, et al. Bivalent role of the phosphatidylinositol‐3‐kinase (PI3K) during influenza virus infection and host cell defence. Cell Microbiol. 2006;8(8):1336‐1348. [DOI] [PubMed] [Google Scholar]
- 179. Puleston DJ, Zhang H, Powell TJ, et al. Autophagy is a critical regulator of memory CD8(+) T cell formation. Elife. 2014;3:e03706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Zhirnov OP, Klenk HD. Influenza a virus proteins NS1 and hemagglutinin along with M2 are involved in stimulation of autophagy in infected cells. J Virol. 2013;87:13107‐13114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Zhu H‐y, Han L, Shi XL, et al. Baicalin inhibits autophagy induced by influenza A virus H3N2. Antiviral Res. 2015;113:62‐70. [DOI] [PubMed] [Google Scholar]
- 182. Nakahira K, Haspel JA, Rathinam VAK, et al. Autophagy proteins regulate innate immune response by inhibiting NALP3 inflammasome‐mediated mitochondrial DNA release. Nat Immunol. 2011;12(3):222‐230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Lupfer C, Thomas PG, Anand PK, et al. Receptor interacting protein kinase 2‐mediated mitophagy regulates inflammasome activation during virus infection. Nat Immunol. 2013;14(5):480‐488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Beale R, Wise H, Stuart A, Ravenhill BJ, Digard P, Randow F. A LC3‐interacting motif in the influenza A virus M2 protein is required to subvert autophagy and maintain virion stability. Cell Host Microbe. 2014;15(2):239‐247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Bouvier PP. The biology of influenza viruses. Vaccine. 2008;26(4):D49‐D53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Haque KV, Axelsen PH, Lentz BR. Properties and structures of the influenza and HIV fusion peptides on lipid membranes: implications for a role in fusion. Biophys J. 2005;89:3183‐3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Maier H, Britton P. Involvement of autophagy in coronavirus replication. Virus. 2012;4(12):3440‐3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Chen X, Wang K, Xing Y, et al. Coronavirus membrane‐associated papain‐like proteases induce autophagy through interacting with Beclin1 to negatively regulate antiviral innate immunity. Protein Cell. 2014;5(12):912‐927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Cottam EM, Whelband MC, Wileman T. Coronavirus NSP6 restricts autophagosome expansion. Autophagy. 2014;10(8):1426‐1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Klein KA, Jackson WT. Human rhinovirus 2 induces the autophagic pathway and replicates more efficiently in autophagic cells. J Virol. 2011;85(18):9651‐9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Feizi N, Mehrbod P, Romani B, et al. Autophagy induction regulates influenza virus replication in a time‐dependent manner. J Med Microbiol. 2017;66(4):536‐541. [DOI] [PubMed] [Google Scholar]
- 192. Park YM, Choi JY, Byun BH, Cho CH, Kim HS, Kim BS. Telomerase is strongly activated in hepatocellular carcinoma but not in chronic hepatitis and cirrhosis. Exp Mol Med. 1998;30(1):35‐40. [DOI] [PubMed] [Google Scholar]
- 193. Liu H, Shi W, Luan F, et al. Hepatitis B virus X protein upregulates transcriptional activation of human telomerase reverse transcriptase. Virus Genes. 2010;40(2):174‐182. [DOI] [PubMed] [Google Scholar]
- 194. Zhu Z, Wilson AT, Gopalakrishna K, Brown KE, Luxon BA, Schmidt WN. Hepatitis C virus core protein enhances telomerase activity in Huh7 cells. J Med Virol. 2010;82(2):239‐248. [DOI] [PubMed] [Google Scholar]
- 195. Taji F, Kouchesfahani HM, Sheikholeslami F, et al. Autophagy induction reduces telomerase activity in HeLa cells. Mech Ageing Dev. 2016. [DOI] [PubMed] [Google Scholar]
- 196. Hu D, Wu J, Zhang R, et al. Autophagy‐targeted vaccine of LC3–LpqH DNA and its protective immunity in a murine model of tuberculosis. Vaccine. 2014;32(20):2308‐2314. [DOI] [PubMed] [Google Scholar]
- 197. Naziri H, Naziri H, Abdoli A, et al. Enhancement of hepatitis E virus DNA vaccine immunity by beclin‐1‐mediated autophagy. Jundishapur J Microbiol. 2018. in press [Google Scholar]
- 198. Khateri M, Abdoli A, Motevalli F, et al. Evaluation of autophagy induction on HEV 239 vaccine immune response in a mouse model. IUBMB Life. 2018. In Press [DOI] [PubMed] [Google Scholar]
- 199. Jagannath C, Lindsey DR, Dhandayuthapani S, Xu Y, Hunter RL, Eissa NT. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat Med. 2009;15(3):267‐276. [DOI] [PubMed] [Google Scholar]
- 200. Hu L, Jiang K, Ding C, Meng S. Targeting autophagy for oncolytic immunotherapy. Biomedicine. 2017;5(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Wu S‐Y, Lan S‐H, Liu H‐S. Autophagy and microRNA in hepatitis B virus‐related hepatocellular carcinoma. World J Gastroenterol. 2016;22(1):176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Liu B, Fang M, Hu Y, et al. Hepatitis B virus X protein inhibits autophagic degradation by impairing lysosomal maturation. Autophagy. 2014;10(3):416‐430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Saribas AS, Khalili K, Sariyer IK. Dysregulation of autophagy by HIV‐1 Nef in human astrocytes. Cell Cycle. 2015;14(18):2899‐2904. [DOI] [PMC free article] [PubMed] [Google Scholar]


