Abstract
The crucial participation of cellular RNA‐binding proteins (RBPs) in virtually all steps of virus infection has been known for decades. However, most of the studies characterizing this phenomenon have focused on well‐established RBPs harboring classical RNA‐binding domains (RBDs). Recent proteome‐wide approaches have greatly expanded the census of RBPs, discovering hundreds of proteins that interact with RNA through unconventional RBDs. These domains include protein–protein interaction platforms, enzymatic cores, and intrinsically disordered regions. Here, we compared the experimentally determined census of RBPs to gene ontology terms and literature, finding that 472 proteins have previous links with viruses. We discuss what these proteins are and what their roles in infection might be. We also review some of the pioneering examples of unorthodox RBPs whose RNA‐binding activity has been shown to be critical for virus infection. Finally, we highlight the potential of these proteins for host‐based therapies against viruses.
This article is categorized under:
RNA Interactions with Proteins and Other Molecules > Protein–RNA Interactions: Functional Implications
RNA in Disease and Development > RNA in Disease
RNA Interactions with Proteins and Other Molecules > RNA–Protein Complexes
Keywords: infection, RBPome, RNA, RNA‐binding domain, RNA‐binding protein, RNA interactome capture, RNA metabolism, translation, virus
The compendium of RNA‐binding proteins related to virus infection reveals an unexplored world of unconventional virus–host interactions.
1. INTRODUCTION
As obligate intracellular pathogens with small‐sized genomes, viruses rely on host cell resources for the completion of their biological cycle and have developed sophisticated mechanisms to hijack key host machineries (Davey, Trave, & Gibson, 2011). On the other hand, the host cell activates a complex response to generate an “antiviral state”, transitioning from a virus permissive to a hostile environment (Barbalat, Ewald, Mouchess, & Barton, 2011; Rehwinkel & Reis e Sousa, 2013). The infected cell thus becomes a battlefield where the virus and the host cell deploy their molecular “arsenals” to take control of the processes required for infection. RNA is a central molecule in virus biology, because it functions as a genome and template for replication and transcription in RNA viruses, and also as a messenger (m)RNA for the synthesis of viral proteins in both DNA and RNA viruses (Baltimore, 1971). While viral RNA‐binding proteins (RBPs) guide the viral (v)RNA through the series of RNA metabolic steps, including synthesis, splicing, localization, translation, and packaging, cellular RBPs are also necessary for these processes to occur. Work accumulated over the last two decades has shown that cellular RBPs are indeed involved in virtually every stage of vRNA metabolism in the cycle of both RNA and DNA viruses. Moreover, RBPs are also essential for antiviral defense, because many sensors of the innate immunity recognize unusual RNA products of viral replication, such as double‐stranded (ds)RNA and triphosphate ends (Barbalat et al., 2011; Vladimer, Gorna, & Superti‐Furga, 2014).
Many of the present‐day pandemics involve viruses, including human immunodeficiency virus (HIV), hepatitis C virus (HCV), and hepatitis B virus (HBV), leading to millions of deaths every year. However, most virus infections still lack effective treatments or vaccines. So far, antiviral treatments have mainly focused on the use of compounds acting directly on viral enzymes. However, drug resistance can arise due to the high mutation rate of viral polymerases and the co‐existence of genetically diverse viral populations called quasispecies (Irwin, Renzette, Kowalik, & Jensen, 2016; Sanjuan & Domingo‐Calap, 2016). The selective pressure driven by the drug promotes the expansion of resistant viral strains, shifting the viral population from drug sensitive to resistant (Clutter, Jordan, Bertagnolio, & Shafer, 2016; Garbelli, Riva, Crespan, & Maga, 2017). To prevent the emergence of resistant strains, novel antiviral treatments combine multiple drugs targeting different viral enzymes (Broder, 2010; Emery & Feld, 2017; Valuev‐Elliston & Kochetkov, 2017). However, a broad catalogue of antivirals is required to make this strategy feasible, and this is only available for a small number of viruses such as HIV. Although some drugs can inhibit multiple viruses (e.g., nucleoside analogues with activity against both HIV and HBV [Debing, Neyts, & Delang, 2015; Mendes‐Correa & Nunez, 2010]), viral enzyme‐targeting drugs usually fail at protecting even against closely related virus species. By contrast, inhibition of cellular factors required for virus infection is expected to provide a higher barrier to the emergence of resistance. This is due to two key factors: (a) the higher stability of human genomes and (b) the difficulty for the virus to develop new strategies to utilize alternative host factors or pathways (Law, Korth, Benecke, & Katze, 2013). One of the best examples of targeting host factors is alisporivir (also known as Debio 025), an inhibitor of cyclophilin A (PPIA) that strongly reduces HCV infection (Flisiak et al., 2008; Paeshuyse et al., 2006). Because different viruses often exploit common cellular pathways, host proteins are promising targets for the development of broad‐spectrum antiviral therapies (Martinez, Sasse, Bronstrup, Diez, & Meyerhans, 2015; Zhu, Meng, Wang, & Wang, 2015). Cellular kinases (Schor & Einav, 2018), cyclophilins (Dawar, Tu, Khattak, Mei, & Lin, 2017; Debing et al., 2015), and heat‐shock proteins (Geller, Taguwa, & Frydman, 2012; Y. Wang et al., 2017) represent potential candidates for such antiviral intervention.
A substantial proportion of the host proteins that benefit or hinder viral replication are RBPs, and many efforts have been undertaken in the last years to characterize the roles of protein–RNA interactions in infected cells. Many cellular RBPs containing well‐established RNA‐binding domains (RBDs) are known to be critical for the infection of specific viruses. Next generation, proteome‐wide methods have discovered hundreds of unconventional RBPs in the past years (Baltz et al., 2012; Castello et al., 2012; Hentze, Castello, Schwarzl, & Preiss, 2018), raising the possibility of their potential involvement in virus infection as well. Some of these novel RBPs have already been linked to diverse virus‐related processes, but in most cases, their ability to interact with RNA was unknown. Here, we present a compendium of RBPs involved in virus infection and review several pioneering studies about the roles of unconventional RBPs in infection.
2. THE ERA OF THE RNA INTERACTOMES: NEW INSIGHTS INTO VIRUS INFECTION
2.1. Identifying “virus‐related” RBPs
In the past, proteins were classified as RBPs largely based on the presence of well‐established, classical RBDs such as the RNA recognition motif (RRM), K‐homology (KH) domain, DEAD box helicase domain, dsRNA motif (DSRM), zinc‐fingers (ZF), and others, as reviewed in detail by Anantharaman, Koonin, and Aravind (2002) and Lunde, Moore, and Varani (2007). Most of these classical RBDs have been well characterized biochemically and structurally, and they fold into well‐established stable secondary and tertiary structures (Lunde et al., 2007). RBPs are usually modular, combining multiple RBDs that cooperate to recognize different sequence and/or structural features in target RNAs (Gronland & Ramos, 2017; Lunde et al., 2007). This modularity enables interactions that can range from very selective to promiscuous, supporting a wide diversity of functions (Hentze et al., 2018).
A recently developed proteome‐wide method called “RNA interactome capture” (RNA‐IC) has greatly expanded the census of cellular RBPs (Baltz et al., 2012; Castello et al., 2012; Hentze et al., 2018). RNA‐IC employs ultraviolet (UV) crosslinking of living cells to induce covalent bonds between proteins and RNA placed at zero distance. Cells are then lysed under denaturing conditions, and polyadenylated [poly(A)] RNA and proteins covalently linked to it are captured under very stringent conditions with oligo(dT) magnetic beads. After elution, RBPs are released by RNase treatment and analyzed by quantitative proteomics. This method has been applied to numerous cell lines and organisms, and has revealed RBPs acting in living systems in a comprehensive manner (Hentze et al., 2018). Proteins identified by this method are highly enriched in classical RBDs, as expected. However, hundreds of unorthodox RBPs were also reported to bind RNA. A recent review compiled the scope of RBPs identified by all the RNA‐IC studies so far, classifying 1,392 proteins as RBPs in human cells (Hentze et al., 2018). Among them, 278 RBPs harbor at least one classical RBD, and 1,114 can be classified as unorthodox RBPs based on lack of such domains (Figure 1a and File S1, Supporting Information).
Figure 1.
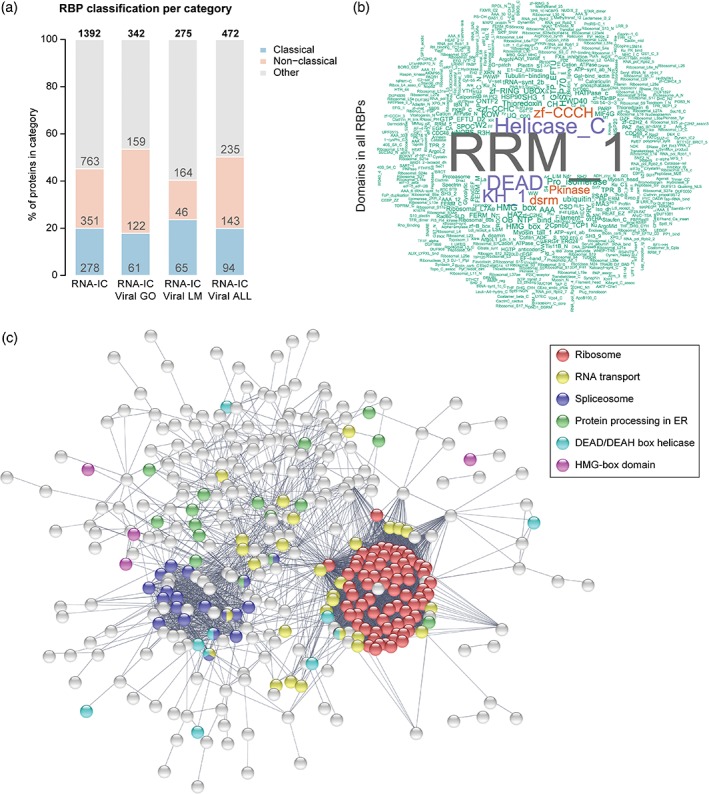
RNA‐binding proteome (RBPome) implicated in virus infection and immunity. (a) Classification by RNA‐binding domain (i.e., classical, nonclassical, and other) of RNA‐binding proteins (RBPs) in different datasets: human RNA interactome capture (RNA‐IC); RNA‐IC linked to virus by gene ontology (GO) terms; RNA‐IC linked to virus by literature mining (LM); RNA‐IC linked to virus by GO and LM. (b) Word cloud of protein domains present in all virus‐linked RBPs. (c) STRING protein network showing connections between all virus‐linked RBPs. ER, endoplasmic reticulum
The newly established RNA‐binding proteome (RBPome) represents a unique resource to explore the interconnections between host RBPs and viruses. In order to do so, we first analyzed how many of these experimentally identified RBPs have been annotated by gene ontology (GO) terms related to viruses, infection, and immunity (File S1), leading to the identification of 342 “virus‐linked” RBPs. Furthermore, complementary literature mining analysis revealed 130 additional RBPs with more than five publications in PubMed linking them to viruses. Taking together, GO and literature mining lead to a superset of 472 virus‐linked RBPs (Table S1 and Figure 1a).
Among the compendium of virus‐linked RBPs, the most prevalent domains are classical RBDs including RRM, DEAD box helicase, KH, DSRM, and ZF (Figure 1b), as expected due to the frequent occurrence of these domains in the human RBPome. Virus‐linked RBPs are involved in every step of posttranscriptional control of gene expression, including splicing and RNA processing, RNA transport, and translation (Figure 1c). Strikingly, 378 virus‐linked RBPs lack canonical RBDs, suggesting that many newly discovered RBPs are involved in virus infection. We further grouped these unorthodox RBPs into “noncanonical” and “other” based on the presence or absence of nonclassical domains previously reported to bind RNA, respectively [Castello et al., 2012; Castello et al., 2016]. We discuss these unconventional RBPs in detail below.
2.2. Well‐known RBPs meet viruses
Well‐established RBPs, in particular classical RBPs, have been linked to virtually every step of virus infection (Table 1). Even though most RNA viruses encode their own RNA polymerases to synthesize vRNAs, host RBPs participate in: (a) selective recognition of the viral template RNA; (b) recruitment of vRNA to the membranous compartments where viral replication takes place and activation of viral replicase complexes; (c) switch from translation to replication; and (d) synthesis of negative, positive, and subgenomic vRNAs (Z. Li & Nagy, 2011; Nagy & Pogany, 2011; R. Y. Wang & Li, 2012). Other viruses, such as HIV, rely on the host RNA transcription machinery, mainly RNA polymerase II (POLR2). RBPs such as non‐POU domain containing octamer binding (NONO) or splicing factor proline and glutamine rich (SFPQ) can be recruited co‐transcriptionally to regulate vRNA synthesis, as described for the DNA virus Epstein–Barr (Harwig et al., 2017). Furthermore, many viruses fine‐tune or simply shut off host transcription to evade antiviral defenses. To do so, viruses often target cellular RBPs such as cleavage and polyadenylation specific factor 4 (CPSF4), poly(A) binding protein nuclear 1 (PABPN1), transcription factors, or, directly, POLR2 (Herbert & Nag, 2016).
Table 1.
Examples of well‐established cellular RBPs involved in virus infection
| RNA‐related process | Examples of RBPs | References |
|---|---|---|
| Recognition of vRNA template | EEF1A1, PCBP1, PTBP1, NCL | (Z. Li & Nagy, 2011; Nagy & Pogany, 2011; R. Y. Wang & Li, 2012) |
| Recruitment of vRNA to replication factory | HNRNPA1, HNRNPC, EEF1A1 | (Z. Li & Nagy, 2011; Nagy & Pogany, 2011; R. Y. Wang & Li, 2012) |
| Switch from translation to replication | PTBP1, PCBP2, PABPC1, SSB, ILF3 | (Z. Li & Nagy, 2011; Nagy & Pogany, 2011; R. Y. Wang & Li, 2012) |
| vRNA synthesis | HNRNPC, TIA1, PTBP1, HNRNPK, POLR2, NONO, SFPQ, RBM14, EEF1A1 | (Harwig, Landick, & Berkhout, 2017; Z. Li & Nagy, 2011; Nagy & Pogany, 2011; R. Y. Wang & Li, 2012) |
| Host mRNA transcription shutoff | CPSF4, PABPN1, POLR2 | (Herbert & Nag, 2016) |
| Splicing | HNRNPA1, HNRNPA2B1, HNRNPA3, HNRNPH, HNRNPC, HNRNPM, U2AF1, U2AF2, SR proteins | (Meyer, 2016; Stoltzfus & Madsen, 2006; Tazi et al., 2010) |
| RNA editing/reading of edited RNA | ADAR, ADARB1, ADARB2, RBM15, ALKBH5, YTHDF1, YTHDF3 | (Gonzales‐van Horn & Sarnow, 2017; Samuel, 2011) |
| Nucleus–cytoplasm shuttling | PTBP1, PCBP2, SSB, CSDE1, HNRNPC, HNRNPA1, HNRNPK, HNRNPM, DHX9, SYNCRIP, TIA1, TIAL1, G3BP1 | (Lloyd, 2015) |
| vRNA export from nucleus | NXF1, ALYREF, RAN, SR proteins, DDX1, DDX3X, DDX5, HNRNPA2B1, SFPQ, MATR3 | (Kuss, Mata, Zhang, & Fontoura, 2013; Stake, Bann, Kaddis, & Parent, 2013) |
| vRNA trafficking and packaging | HNRNPA2B1, EEF1A1, STAU1 | (Cochrane, McNally, & Mouland, 2006; Kaddis Maldonado & Parent, 2016; Stake et al., 2013) |
| Extracellular trafficking | HNRNPA2B1, AGO2, SYNCRIP, YBX1 | (Kouwaki, Okamoto, Tsukamoto, Fukushima, & Oshiumi, 2017) |
| vRNA stability | ELAVL1, PCBP2, HNRNPD, YBX1, ILF3 | (Dickson & Wilusz, 2011; Moon & Wilusz, 2013) |
| 5′→3′ RNA degradation | XRN1, PATL1, XRN2 | (Molleston & Cherry, 2017; Moon, Barnhart, & Wilusz, 2012; Narayanan & Makino, 2013; Oshiumi, Mifsud, et al., 2016; Rigby & Rehwinkel, 2015) |
| 3′→5′ RNA degradation | DDX17, ZC3HAV1, UPF1, UPF3, RRP6 | (Molleston & Cherry, 2017; Moon, Barnhart, & Wilusz, 2012; Narayanan & Makino, 2013; Oshiumi, Mifsud, et al., 2016; Rigby & Rehwinkel, 2015) |
| Translation | PABPC1, EIF4E, EIF4G, EIF4A, EIF2, EIF3, ribosomal proteins, EIF5B, EEF1A1, EEF2, ETF1 | (McCormick & Khaperskyy, 2017; Smith & Gray, 2010; Walsh & Mohr, 2011) |
| RNP granules | G3BP1, G3BP2, EIF2AK2, LSM14A, XRN1, TIA1, TIAL1, STAU1, DDX3X, DDX6 | (Beckham & Parker, 2008; Lloyd, 2013; McCormick & Khaperskyy, 2017; Onomoto, Yoneyama, Fung, Kato, & Fujita, 2014; Reineke & Lloyd, 2013; Yoneyama, Jogi, & Onomoto, 2016) |
| Antiviral sensors and cofactors | TLR3, DDX58, IFIH1, ADAR, ADARB1, EIF2AK2, OAS, DHX9, DDX1, DDX3X, DHX29, DHX36 | (Beachboard & Horner, 2016; Gelinas, Clerzius, Shaw, & Gatignol, 2011; Masliah, Barraud, & Allain, 2013; Oshiumi, Kouwaki, et al., 2016; Peisley & Hur, 2013; Ranji & Boris‐Lawrie, 2010; Steimer & Klostermeier, 2012) |
Note. mRNA = messenger RNA; RBP = RNA‐binding protein; vRNA = viral RNA.
Viruses also hijack RBPs involved in co/post‐transcriptional RNA processing. For example, several RNA and DNA viruses usurp the cellular capping and polyadenylation machineries to add a 5′ cap structure and 3′ poly(A) tail to their mRNAs through a broad variety of molecular mechanisms (Decroly, Ferron, Lescar, & Canard, 2011; Schrom, Moschall, Schuch, & Bodem, 2013). The spliceosome and regulators of splicing are hijacked by viruses that need to splice their RNAs (Meyer, 2016; Stoltzfus & Madsen, 2006; Tazi et al., 2010). Several RBPs involved in RNA editing (e.g., 5′‐methylcytosine, N6‐methyladenosine, or adenosine‐to‐inosine modifications) or recognition of edited RNA can positively or negatively modulate virus infection, affecting, for example, vRNA stability, translation, and antiviral sensing (Gonzales‐van Horn & Sarnow, 2017; Samuel, 2011). Interestingly, many of the RBPs involved in the aforementioned processes have a nuclear distribution, but cytoplasmic RNA viruses have the capacity to alter their subcellular localization and co‐opt them for new functions (Lloyd, 2015). Heterogeneous nuclear ribonucleoproteins (HNRNPs) are typically identified as interactors of vRNAs with an altered localization in infected cells. While these interactions are likely to be functionally relevant for different steps of infection (Table 1), HNRNPs are very abundant proteins and thus easy to detect in proteomic‐based analyses, probably representing the tip of the iceberg.
Host RBPs involved in RNA trafficking are also hijacked by viruses and contribute to vRNA export from the nucleus, transport throughout the cytoplasm, and packaging into viral capsids (Cochrane et al., 2006; Kaddis Maldonado & Parent, 2016; Stake et al., 2013). On the other hand, viruses can impair host RNA trafficking by targeting RBPs, including the nuclear RNA export factor 1 (NXF1) and GTP‐binding nuclear protein Ran (RAN). By doing so, viruses prevent nuclear export of cellular mRNAs encoding antiviral factors and cytokines and block import into the nucleus of relevant transcription factors (Castello, Alvarez, & Carrasco, 2011; Kuss et al., 2013).
Cellular RBPs that modulate RNA stability, such as ELAV‐like RBP 1 (ELAVL1 or HuR), can be utilized by viruses to preserve the integrity of their vRNAs (Dickson & Wilusz, 2011; Moon & Wilusz, 2013). On the other hand, viruses can promote sequestering of cellular RBPs that enhance RNA stability, leading to destabilization of host transcripts. Cellular exonucleases are also key players in infection. Viruses have developed multiple ways of evading the cellular 5′→3′ and 3′→5′ RNA degradation machineries that would otherwise target vRNAs for decay (Molleston & Cherry, 2017; Moon et al., 2012; Narayanan & Makino, 2013; Oshiumi, Mifsud, & Daito, 2016; Rigby & Rehwinkel, 2015). Viruses have also evolved strategies to use the 5′→3’ mRNA decay machinery, including decapping activators (Jungfleisch, Blasco‐Moreno, & Diez, 2016) and the nonsense‐mediated decay machinery (Balistreri, Bognanni, & Muhlemann, 2017), to the benefit of vRNA translation and replication.
Viruses strongly rely on the host translation machinery to synthesize viral proteins (McCormick & Khaperskyy, 2017; Walsh & Mohr, 2011). Nevertheless, many viruses disrupt specific host RBPs critical for translation initiation, such as poly(A) binding protein cytoplasmic 1 (PABPC1) (Smith & Gray, 2010), eukaryotic translation initiation factor (EIF)4E, EIF4G, EIF2, or EIF3; translation elongation (EIF5B, EEF1A1, or EEF2) and termination (e.g., ETF1) as well as ribosomal proteins. vRNA can bypass host translation shutoff through noncanonical translation mechanisms (Au & Jan, 2014; Firth & Brierley, 2012; Mailliot & Martin, 2018), possibly by recruiting specific RBPs and specialized ribosomes. For example, picornaviruses and other viruses employ cellular trans‐acting factors (ITAFs) to facilitate internal ribosome entry site (IRES)‐mediated, cap‐independent translation (Mailliot & Martin, 2018).
RNA helicases are a family of proteins often implicated in virus infection. They can promote different steps of viral replication or mediate the activation of the antiviral response (Ranji & Boris‐Lawrie, 2010; Steimer & Klostermeier, 2012). Several antiviral sensors and effectors, such as the retinoic acid‐inducible gene I protein (RIG‐I, also known as DDX58), harbor classical helicase cores to recognize replication intermediates (Barbalat et al., 2011; Rehwinkel & Reis e Sousa, 2013). These antiviral sensors cooperate with different RNA helicases, including DExH‐box helicase (DHX)9, DEAD‐box helicase 3 X‐linked (DDX3X), or DHX36, that act as accessory factors in antiviral pathways (Oshiumi, Kouwaki, & Seya, 2016).
3. UNORTHODOX RBPs: NEW PLAYERS IN THE VIRUS–HOST BATTLEFIELD
3.1. The discovery of unorthodox RBPs
Classical RBDs do not account for all RNA‐binding activities observed in the cell (Hentze et al., 2018). Initially, dozens of unorthodox RBPs were discovered in a somewhat sporadic manner. Further mechanistic studies demonstrated that these proteins have pivotal functions in RNA metabolism and gene expression (Arif et al., 2018; Castello, Hentze, & Preiss, 2015; Chang et al., 2013; Muckenthaler, Galy, & Hentze, 2008; Mukhopadhyay, Jia, Arif, Ray, & Fox, 2009). The scope of unconventional RBPs has been greatly expanded by RNA‐IC, and other recently developed techniques have uncovered in a proteome‐wide scale their modes of RNA binding (Box 1) (Castello et al., 2016; He et al., 2016; Kramer et al., 2014). A number of conclusions arose from these studies: (a) half of the RNA‐binding regions within RBPs map to intrinsically disordered regions (IDRs) (Box 2); (b) globular domains with diverse known functions (e.g., protein–protein interaction domains, DNA‐binding domains, and enzymatic cores) and protein folds with unknown function can also interact with RNA; (c) RBDs are hotspots for posttranslational modifications (PTMs), including phosphorylation, acetylation, and methylation, which are expected to regulate RBP activity; (d) novel RNA‐binding regions are more evolutionary conserved than adjacent regions that do not interact with RNA; and (e) Mendelian mutations linked to disease and mapping to RBPs occur predominantly at RNA‐binding disordered regions and PTM sites, highlighting their regulatory relevance (Castello et al., 2016; Castello, Fischer, Hentze, & Preiss, 2013; Moore, Jarvelin, Davis, Bond, & Castello, 2017).
BOX 1. IDENTIFICATION OF RNA‐BINDING SITES BY RBDmap .
RBDmap is a novel system‐wide method built on RNA‐IC for the comprehensive in vivo identification of RNA‐interacting sites within RBPs (Castello et al., 2016; Castello et al., 2017). Living cells are irradiated with UV light at 254 nm to induce covalent bonds between nucleotides and amino acids interacting directly (zero distance) in physiological conditions. After cell lysis, poly(A) RNAs are captured by hybridization with oligo(dT) magnetic beads under very stringent denaturing conditions. Because protein–RNA interactions are covalently preserved, noncovalent binders (i.e., protein–protein interactors) are removed and only direct RNA binders are purified together with the RNA. RBPs are digested with a protease that cleaves every 17 amino acids on average and poly(A) RNAs are purified via a second oligo(dT) capture. In this way, the protein regions still bound to RNA (termed “RBDpeps”) are isolated from the released peptides, which represent the protein regions that do not interact with RNA. RBDpeps are crosslinked to (at least) one nucleotide, making their identification by mass spectrometry challenging. To circumvent this problem, RBDpeps are digested with trypsin that cleaves every seven to nine amino acids on average, thus releasing a peptide still crosslinked to RNA and another peptide with native mass that is referred to as “neighboring peptide”. These latter peptides are readily identified by proteomics and the original RBDpep is reconstructed in silico. RNA‐binding sites are identified by comparing the peptides released upon the protease treatment (released fraction) against those remaining bound to the RNA (RNA‐bound fraction) (Castello et al., 2017). RBDmap revealed 1,174 high‐confidence RNA‐binding sites within 529 proteins in human cells (Castello et al., 2016) and 568 sites within 368 RBPs in murine cardiomyocytes (Liao et al., 2016). Forty‐five percent of the identified RNA‐binding sites mapped to well‐established RBDs, while 55% mapped to previously unknown or poorly characterized RBDs and IDRs.
BOX 2. INTRINSICALLY DISORDERED REGIONS IN RNA BINDING.
Approximately 25% of the experimentally identified cellular RBPs have classical RBDs in human and murine cell lines, and other 25% contain nonclassical but known RBDs (Castello et al., 2012; Castello et al., 2016). However, about half of the human and mouse RBPomes lack known RBDs (Castello et al., 2012; Castello et al., 2016; Liao et al., 2016). In many cases, the regions responsible for RNA binding are intrinsically disordered, lacking stable 3D structure and containing multiple interaction surfaces (Castello et al., 2016; Jarvelin, Noerenberg, Davis, & Castello, 2016). These RNA‐binding IDRs are rich in serine (S), glycine (G), proline (P), lysine (K), arginine (R), and tyrosine (Y) amino acids, and their contribution to protein–RNA interactions has recently been reviewed (Jarvelin et al., 2016). Interestingly, IDRs show a modular organization in RBPs as is typical for classical RBPs: sequences of different length are repeated in a nonrandom mode, including RS dipeptides, RGG boxes, YGG boxes (also called [G/S]Y[G/S]), and poly(K) motifs, among others. In many cases, they can mediate both specific and nonspecific interactions with RNA in cooperation with globular RBDs. It is still unclear how specificity is generated by disordered RNA‐binding regions.
3.2. Unorthodox globular RBDs in “virus‐linked” RBPs
To analyze the scope of newly discovered RBPs linked to infection, we first removed all RBPs harboring a classical RBD. The resulting set of unconventional RBPs was then divided into two groups: (a) “nonclassical” RBPs if the protein lacks classical RBDs but harbors other domains described experimentally to bind RNA (Castello et al., 2012; Castello et al., 2016; File S1) and (b) “other” RBPs when lacking both classical and nonclassical RBDs. Ribosomal proteins were prevalent among nonclassical virus‐linked RBPs (Figure 2a,b), which is expected as viral genomes cannot encode the large amount of protein and RNA components required to build this complex machinery. Another group of nonclassical RBPs linked to virus infection comprises protein chaperones from the heat shock protein (HSP)90, HSP70, and HSP20 families, known to harbor discrete RNA‐binding regions conserved across homologous proteins (Castello et al., 2016; Moore et al., 2017). These proteins are important for the co‐translational folding of proteins and the assembly and remodeling of cellular ribonucleoproteins (RNPs) (Iwasaki et al., 2010; Willmund et al., 2013). The involvement of protein chaperones in virus infection has been known for a long time; however, the relevance of their newly described RNA‐binding activity for their recruitment and function remains unknown (Geller et al., 2012; Y. Wang et al., 2017). Peptidyl‐prolyl cis–trans isomerases from the FKBP and PPI families are represented in the noncanonical RBP group (Figure 2a,b). This finding expands the well‐documented impact of PPIA on different viruses (D. Zhou, Mei, Li, & He, 2012) to other members of the PPI and FKBP families. Proteins containing thioredoxin enzymatic domain, 14‐3‐3 protein–protein interaction domain, high mobility (HGMB) protein family, cytoskeleton‐related proteins, and metabolic enzymes are also present in the group of nonclassical, virus‐linked RBPs.
Figure 2.
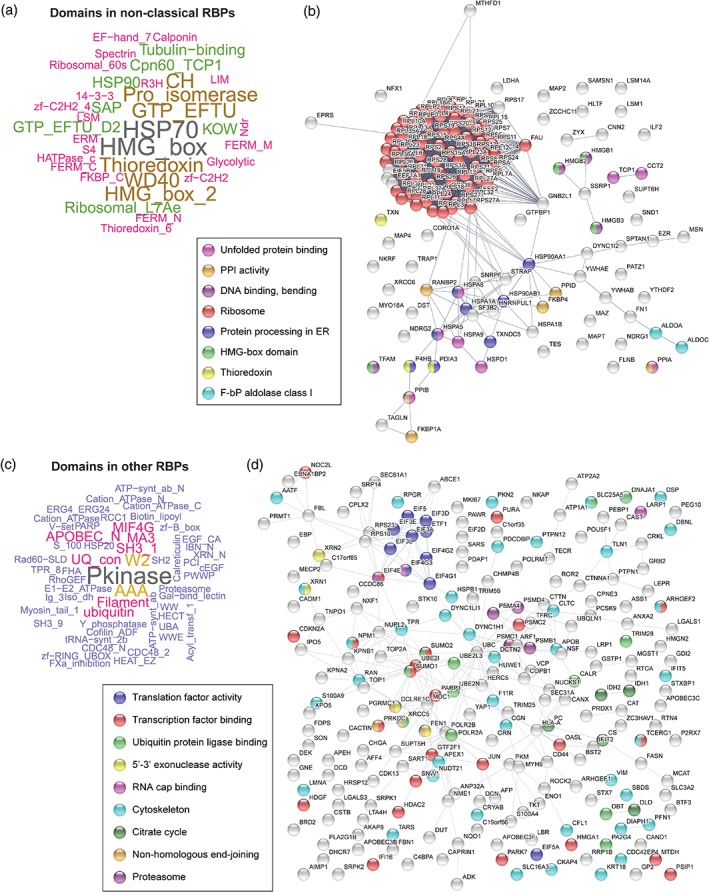
Virus‐linked RNA‐binding proteins (RBPs) with nonclassical or other RNA‐binding domains (RBDs). (a and c) Word cloud of protein domains present in nonclassical (a) and other (c) virus‐linked RBPs. (b and d) STRING protein network showing connections between nonclassical (b) and other (d) virus‐linked RBPs. ER, endoplasmic reticulum; F‐bP, fructose‐biphosphate; PPI, peptidyl‐prolyl cis–trans isomerase
In the “other” group of virus‐linked RBPs, we find proteins with clear roles in RNA biology such as translation initiation factors as well as proteins involved in splicing and RNA transport, but also kinases, metabolic enzymes, ubiquitin‐binding proteins (including E3 ubiquitin ligases), proteasome components, and others (Figure 2c,d). Future work should focus on elucidating the poorly understood connections between virus infection and the RNA‐binding capacity of these proteins.
3.3. Disordered regions in RBPs: Roles in virus infection?
Cellular RNP granules, such as stress granules and processing (P)‐bodies, are complex aggregates of RNA and proteins that regulate RNA translation, turnover, and storage. Cytoplasmic granules have primarily an antiviral nature and many viruses inhibit their formation by targeting mainly G3BP stress granule assembly factor 1 (G3BP1), G3BP2, and EIF2AK2 (PKR) (Lloyd, 2013; McCormick & Khaperskyy, 2017; Onomoto et al., 2014; Yoneyama et al., 2016). On the other hand, many of the host proteins found in stress granules and/or P‐bodies can be co‐opted by viruses to enhance replication (Beckham & Parker, 2008; Reineke & Lloyd, 2013).
Transition from a soluble phase (dispersed proteins and RNA) to a condensed state (concentrated components) is responsible for RNP granule formation. Different lines of evidence have shown that IDRs within RBPs are important for this process to occur (Jarvelin et al., 2016). IDRs with specific sequence features (Box 2) have been shown to bind RNA (Basu & Bahadur, 2016; Castello et al., 2016; Jarvelin et al., 2016). In 170 out of 529 RBPs identified by RBDmap (Box 1) the only detectable RNA‐binding sites were located at IDRs, suggesting that they are sufficient to mediate interactions with RNA (Castello et al., 2016). IDRs involved in RNA binding are present in both classical and unorthodox RBPs (Table S1), and many of these proteins display different roles in virus infection. For example, interleukin enhancer binding factor 3 (ILF3, 76% disordered) harboring a GQSY‐rich region at the C‐terminus (Figure 3a), G3BP1 (76% disordered), G3BP2 (80% disordered), HNRNPA1 (80% disordered), and cell cycle associated protein 1 (CAPRIN1, 86% disordered, Figure S1a) contain RNA‐binding IDRs. The importance in virus infection of IDRs and their properties to bind RNA and promote phase transitions deserves further attention.
Figure 3.
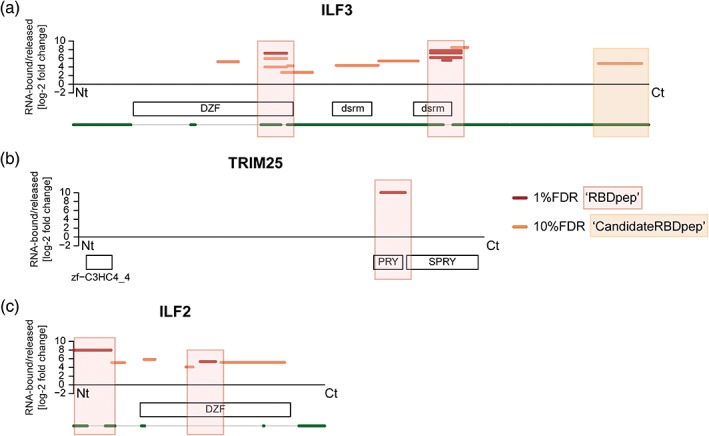
RNA‐binding domains (RBDs) of virus‐related RNA‐binding proteins. RBDmap profiles of ILF3 (a), TRIM25 (b), and ILF2 (c). Boxes represent high confidence (red, 1% FDR) or relevant candidate (orange, 10% FDR) RBDs. White boxes symbolize Pfam‐annotated domains. Green lines indicate predicted disordered regions (IUPred score > 0.4). FDR, false discovery rate
Strikingly, viruses mimic short disordered motifs to regulate key processes for virus infection (Davey et al., 2011). For example, dengue virus (DENV) and HCV IDRs are involved in protein–protein binding as well as in interaction with nucleic acids, metals, and other small ligands (Fan et al., 2014; Meng et al., 2015). These viral IDRs are phosphorylation hotspots with regulatory roles. The control of protein–RNA interactions and viral replication by host kinases phosphorylating viral proteins has been reviewed by Jakubiec and Jupin (2007) and Keck, Ataey, Amaya, Bailey, and Narayanan (2015). Several viral RBPs interact with RNA through disordered regions, including HIV Tat and Rev that harbor an arginine‐rich motif with nucleic acid‐binding capacity (Jarvelin et al., 2016; Xue, Mizianty, Kurgan, & Uversky, 2012). These short, disordered peptides co‐fold with the target RNA forming stable structures (Daugherty, Liu, & Frankel, 2010; Puglisi, Tan, Calnan, Frankel, & Williamson, 1992). Other arginine‐rich RNA‐binding motifs have been described in Ebola virus (EBOV) VP30 (John et al., 2007), Flaviviridae capsid (Ivanyi‐Nagy, Lavergne, Gabus, Ficheux, & Darlix, 2008), and Nodamura virus RNA‐dependent RNA polymerase (Gitlin, Hagai, LaBarbera, Solovey, & Andino, 2014). Short motifs confer viral proteins the ability to interact with many different viral and host components despite the limited coding capacity of viral genomes (Xue et al., 2014). This is a cost‐effective RNA (and protein)‐binding solution that requires only a dozen amino acids (Jarvelin et al., 2016; Phan et al., 2011), while the most common classical RBDs (e.g., RRM) contain 75–85 amino acids (Lunde et al., 2007).
4. FUNCTIONS OF NOVEL RBPs IN VIRUS INFECTION
4.1. Ubiquitination routes in antiviral responses mediated by tripartite motif proteins
Virus infection activates diverse immune signaling pathways involving many RBPs that are highly regulated through PTMs such as phosphorylation or ubiquitination (Chiang & Gack, 2017; Y. Zhou, He, Wang, & Ge, 2017). Ubiquitination of antiviral sensors and effectors can modulate protein function in many ways and requires three different classes of enzymes: ubiquitin‐activating (E1), ‐conjugating (E2), and ‐ligating (E3). Tripartite motif (TRIM) is a large family of proteins that harbor E3 ligase activity and restrict virus infection through direct and indirect mechanisms (van Tol, Hage, Giraldo, Bharaj, & Rajsbaum, 2017; Versteeg et al., 2013). TRIM proteins have broad roles in the cell, but these were not linked to RNA biology until recently. Among the upward of 80 TRIM family members, four (TRIM25, TRIM28, TRIM56, and TRIM71) were found to interact directly with RNA in living cells by RNA‐IC experiments (Hentze et al., 2018). The RNA‐binding activity of TRIM25 and TRIM71 was further validated by orthogonal approaches in mouse embryonic stem cells (Kwon et al., 2013). TRIM25 carries the critical function of ubiquitinating RIG‐I, a cellular receptor that recognizes triphosphate ends in vRNAs and triggers the type I interferon response (Gack et al., 2007). The high‐precision proteome‐wide method to identify RBDs, RBDmap (Box 1), revealed that TRIM25 interacts with RNA via its PRY/SPRY domain (Figure 3b) (Castello et al., 2016), which had been previously implicated in protein–protein interactions. This finding was confirmed and further characterized by orthogonal in vitro and in vivo methods (Choudhury et al., 2017). Importantly, this study showed that the E3‐ubiquitin ligase activity of TRIM25 is regulated by RNA binding. For example, in vitro experiments revealed that auto‐ubiquitination of TRIM25 and ubiquitination of other targets such as zinc finger CCCH‐type containing antiviral 1 (ZC3HAV1 or ZAP) is impaired in mutants lacking the RNA‐binding region identified by RBDmap, or in the presence of RNases. DENV2 was recently shown to prevent TRIM25 antiviral potential through an ingenious mechanism (Manokaran et al., 2015). The exonuclease XRN1 degrades the DENV2 genomic RNA up to a pseudoknot that stalls it. This produces an abundant noncoding subgenomic flavivirus RNA (sfRNA) with 5′ monophosphate end, and thereby it is not recognized by intracellular sensors. This sfRNA is bound (directly or indirectly) by TRIM25 and sequesters it, thus reducing RIG‐I ubiquitination and the subsequent stimulation of type I interferon synthesis (Manokaran et al., 2015). ZC3HAV1 is a broad but not universal antiviral RBP that promotes translation inhibition and degradation of target RNAs (Todorova, Bock, & Chang, 2015). TRIM25 can ubiquitinate ZC3HAV1, but this is not directly responsible for the increase in ZC3HAV1 antiviral activity against Sindbis virus (SINV) (M. M. Li et al., 2017). The interaction of TRIM25 with other host factors (mainly involved in RNA metabolism [Choudhury et al., 2017]) and/or their ubiquitination may contribute to ZC3HAV1 antiviral effects. Interestingly, the RNA‐binding SPRY domain of TRIM25 is critical for the interaction with ZC3HAV1 (M. M. Li et al., 2017) possibly through the formation of an RNA bridge. Many other RBPs (including ZC3HAV1) have been proven to interact with TRIM25 in an RNA‐dependent manner (Choudhury et al., 2017). Therefore, RNA may not only modulate the E3 ligase activity of TRIM25 but could also serve as a scaffold to concentrate substrates in the proximity of TRIM25‐binding sites. Other viral strategies to usurp the host ubiquitylation and SUMOylation machineries have been reviewed by Wimmer and Schreiner (2015).
TRIM56 was initially linked to the host response against DNA viruses, contributing to the cGAS‐STING pathway in a ubiquitin‐ligase‐dependent manner (Tsuchida et al., 2010). TRIM56 also regulates the TLR3/TRIF signaling pathway for dsRNA sensing but, in this case, its ubiquitin ligase activity is not required (Shen et al., 2012). Furthermore, TRIM56 can specifically restrict several RNA viruses through diverse antiviral mechanisms (Kane et al., 2016; Liu et al., 2016; Liu et al., 2014; J. Wang et al., 2011). Distinct regions of TRIM56 have been linked to different viruses and stages of the viral cycle: (a) both the N‐terminal E3 ubiquitin ligase and the integrity of the C‐terminus are required for the suppression of bovine viral diarrhea virus (BVDV), yellow fever virus, and DENV2 RNA replication; (b) the E3 ubiquitin ligase activity is sufficient to inhibit late steps (i.e., packaging and/or egress) of human coronavirus infection; and (c) the C‐terminus suffices to impede influenza A virus (IAV) and influenza B virus (IBV) RNA synthesis. The C‐terminus of TRIM56 has no known domain architecture, but it is known to interact with different cellular (STING and TRIF) and viral (BVDV N‐protease) proteins. TRIM56 C‐terminal part is also believed to function as a scaffold for the formation of multiprotein complexes that are essential for TLR3 signaling (Shen et al., 2012). The recent discovery that TRIM56 also interacts with RNA in living cells suggests that its E3 ubiquitin ligase activity may be regulated by RNA (Hentze et al., 2018), in analogy to TRIM25 (Choudhury et al., 2017). However, TRIM56 lacks the PRY/SPRY domain present in TRIM25 and thus must employ a different yet unknown RBD. TRIM56 antiviral activities in BVDV, IAV, and IBV infections rely on the last 130 amino acids of the C‐terminal part. This sequence has been proposed to interact with vRNA through conserved residues at the NHL repeats (Liu et al., 2016; J. Wang et al., 2011). However, the proposed ability of the C‐terminus of TRIM56 to interact with RNA should be experimentally assessed.
4.2. Recognition of vRNA 5′ ends by interferon‐induced proteins
The minimal cellular cap structure, named “cap0”, consists of a N7‐methylated guanosine (m7G) linked to the 5′ end of the mRNA by a 5′ triphosphate bond. This structure controls mRNA export, maturation, stability, and translation. The first and second nucleotides of cellular mRNAs are further methylated at the 2′‐O positions, leading to the formation of “cap1” and “cap2”. Several antiviral sensors interact with the 5′ end of vRNAs, which usually bear unusual cap structures (Decroly et al., 2011) or triphosphate ends, in contrast to uncapped host RNAs that have 5′ monophosphate ends. Incubation of cellular extracts with immobilized RNA probes harboring 5′ triphosphate ends coupled to proteomics led to the identification of a family of proteins with high affinity for this unusual molecular pattern, called IFIT1‐5 or interferon‐induced protein with tetratricopeptide repeats (TPRs) (Pichlmair et al., 2011). The expression of these proteins is triggered rapidly after activation of the host antiviral defenses. IFIT proteins target nonself, single‐stranded (ss)RNAs in a 5′‐end‐dependent manner (sensing 5′ triphosphate and cap0 RNAs) to inhibit infection by different mechanisms, mainly by affecting viral translation (Habjan et al., 2013; Kumar et al., 2014). The unconventional RNA‐binding activity of IFIT1, 2, and 5 has been recently unraveled and resides in the TPRs, which usually mediate protein–protein interactions. These repeats generate an α‐helix‐rich fold with a positively charged, small pocket that, together with the participation of acidic residues that coordinate divalent cations, embraces the negatively charged triphosphate ends (Abbas, Pichlmair, Gorna, Superti‐Furga, & Nagar, 2013). The different proteins of the IFIT family appear to have developed different roles and modes of action. Both IFIT1 and IFIT5 interact only with ssRNA. IFIT5 binds RNA with cap0, 5′ monophosphate, or 5′ triphosphate ends with similar affinities and do not interact with cap1 (Katibah et al., 2014). IFIT1 forms part of a multiprotein complex together with IFIT2 and IFIT3 (Pichlmair et al., 2011) and can bind cap0 with very high affinity in addition to 5′ triphosphate RNAs (with lower affinity than IFIT5) (Kumar et al., 2014). Functionally, IFIT1 can outcompete the binding of EIF4F complex to the cap0, thus inhibiting viral translation initiation, while IFIT5 can inhibit 48S translation initiation complex formation on mRNAs with 5′ triphosphate ends (Fleith et al., 2018; Kumar et al., 2014). IFIT1 and IFIT2 can also block viral protein synthesis by interacting with the EIF3 complex (Diamond & Farzan, 2013; Vladimer et al., 2014), which has a central role in translation initiation of cellular and viral transcripts (Cate, 2017). IFIT3 has no known RNA‐binding capacity, but it has been recently described to allosterically modulate the IFIT1 RNA‐binding pocket thus enhancing its affinity for cap0 (Fleith et al., 2018; Johnson et al., 2018). IFIT2 is an RBP that can selectively bind AU‐rich RNAs and might recognize specific sequences in vRNAs (Yang et al., 2012). Some viruses such as flavivirus, poxvirus, and coronavirus can escape from IFIT1 and IFIT2 by methylating their vRNA 5′ cap (Daffis et al., 2010). Other vRNAs lack a 2′‐O methylated cap but can still evade IFIT1 restriction. For example, a stable stem‐loop structure located in the 5′ untranslated region (UTR) of the genomic RNA of alphaviruses antagonizes IFIT1 antiviral activity (Hyde et al., 2014).
4.3. The multiple pro‐ and antiviral roles of interleukin enhancer factors
Interleukin enhancer‐binding factor 3 (ILF3) is a multifunctional protein with broad roles in cellular and viral RNA metabolism, including regulation of transcription and translation, mRNA stabilization and localization, and noncoding RNA biogenesis (Castella, Bernard, Corno, Fradin, & Larcher, 2015; Patino, Haenni, & Urcuqui‐Inchima, 2015). ILF3 is a well‐established RBP that harbors three RBDs: two DSRM in positions 417–478 and 540–601, and an intrinsically disordered RGG box in position 653–669 that binds single‐stranded RNA and DNA (Figure 3a). These RBDs cooperate in an intricate dynamic mechanism to recognize diverse RNA substrates (Schmidt et al., 2016). ILF3 can act alone as homodimer or form an RNA regulatory complex with ILF2 and other factors, promoting or restricting virus infection depending on the context. ILF3/ILF2 heterodimerization is mediated by the domain associated with ZF (DZF), which is a nucleotidyltransferase domain that lacks enzymatic activity (Wolkowicz & Cook, 2012). RBDmap identified a novel RNA‐binding activity within this DZF domain, which is present in both ILF3 and ILF2 (Figure 3a,c) and was further validated by orthogonal assays (Castello et al., 2016). However, the target RNAs and the biological importance of this interaction are still unknown. PKR is a dsRNA‐dependent protein kinase that phosphorylates the threonine (Thr)188 and Thr315 of ILF3, inducing ILF3/ILF2 complex disassembly and shuttling of ILF3 to the cytoplasm. Once there, ILF3 binds to viral transcripts and blocks viral translation (Harashima, Guettouche, & Barber, 2010). Thr315 resides in the novel RNA‐binding site, but this is also a protein‐binding interface; therefore, it is still uncertain which of these two activities is affected by Thr315 phosphorylation. One possibility is that disruption of the ILF3/ILF2 complex (or the interactions with other proteins) affects vRNA recognition. In this regard, ILF2 can modulate in vitro the RNA‐binding specificity and efficiency of ILF3 (Schmidt, Friedrich, Golbik, & Behrens, 2017; Schmidt, Knick, et al., 2017), and this may also occur in cells. ILF3 and ILF2 may have additional roles in RNA metabolism as homodimers. Accordingly, ILF2 binds to AU‐rich IRES and enhances translation of cellular and viral RNAs (Faye et al., 2013; Graber, Baird, Kao, Mathews, & Holcik, 2010). The IRES trans‐acting factor (ITAF) activity of ILF2 does not involve ILF3 and, in fact, it is even possible that ILF3 reduces ILF2 ITAF activity when forming the heterodimer (Graber et al., 2010). ILF3 associates preferentially with 25–30 nt AU‐rich motifs present at the 3′ UTRs of cellular RNAs, blocking translation (Kuwano et al., 2010). Strikingly, ILF2 and ILF3, together with the helicase DHX9, bind to AU‐rich sequences present in the 5′ and 3’ UTRs of HCV and BVDV RNAs triggering their circularization in vitro (Isken et al., 2007). These proteins are recruited to HCV replication complexes in infected cells. Deletion of their binding sites in HCV UTRs completely blocks viral replication, suggesting that the circularization favors infection. In a reconstituted in vitro system, ILF2 interaction with ILF3 enhances ILF3 RNA chaperone activity and stimulates the first step of HCV RNA synthesis mediated by the viral polymerase NS5B (Schmidt, Friedrich, et al., 2017). In the related BVDV, ILF2/ILF3‐mediated circularization of the vRNA may also be important for IRES translation (Isken et al., 2003; Isken, Grassmann, Yu, & Behrens, 2004). Finally, ILF3 and ILF2 promote HIV infection by interacting directly with vRNA. Deletion of the DSRMs of ILF3 or the N‐terminal RGG domain of ILF2 disrupts binding to HIV RNA (T. Li et al., 2016), thus confirming their importance for RNA binding (Figure 3a,c). It is believed that ILF3 stabilizes vRNA while ILF2 upregulates HIV gene expression without affecting RNA stability, but the exact mechanism is still not well documented.
There are two main isoforms of ILF3, NF90 and NF110. NF110 contains an extended C‐terminus that is intrinsically disordered and is enriched in GQSY motifs that mediate protein–protein interactions and increase the RNA‐binding activity of the DSRMs (Reichman & Mathews, 2003). RBDmap assigns RNA‐binding activity to the GQSY region (Figure 3a), which suggests that NF110 may have different RNA‐binding properties than the NF90 isoform. Recent reports showed that both NF90 and NF110 promote RNA circularization in the nucleus and associate with mature circular RNAs in the cytoplasm (X. Li et al., 2017). Circular RNAs have regulatory roles in the antiviral immune response and are generated by back‐splicing from exons with flanking complementary Alu repeats. Both NF90 and NF110 bind dsRNA formed by inverted Alu pairing; however, NF110 also interacts with single‐stranded Alu RNA (X. Li et al., 2017). NF90 binds preferentially to circularized RNA compared to linear RNA with the same sequence, while NF110 strongly associates with both. NF90/NF110 binds vesicular stomatitis virus RNAs in the cytoplasm to inhibit viral replication, and simultaneously causes a reduction in host circular RNAs (X. Li et al., 2017). The specificity (if any) of the GQSY‐rich region of NF110 and the biological importance of this region in infected cells still requires further study.
ILF3 and ILF2 have also been implicated in the regulation of stress granules. The GQSY‐rich region of the NF110 isoform of ILF3 interacts with CAPRIN1 (Figure 3a and Figure S1a) and promotes stress granule formation. This region is similar in sequence and functionally exchangeable with the IDR present in RBP FUS that can trigger RNP aggregation (Shiina & Nakayama, 2014). This region might be sufficient but not essential to promote the formation of RNA granules, because the short ILF3 isoform (i.e., NF90) that lacks it can also induce the assembly of stress granules in response to dsRNA (Wen et al., 2014). Furthermore, ILF proteins directly influence the fate of viral proteins. For example, the nucleoprotein of IAV is sequestered in stress granules by a mechanism that involves NF90, and this harms viral replication. This process is antagonized by the multifunctional IAV NS1 protein (Khaperskyy, Hatchette, & McCormick, 2012; Mok et al., 2012).
4.4. Hijacking the ribosome: The EIF3 complex
The cap structure and the poly(A) tail, present in most cellular mRNAs, are recognized by EIF4E and PABPC1, respectively. These two proteins interact with EIF4G to form a closed loop between the 5′ and 3′ ends of the mRNA. To recruit ribosomes, EIF4G interacts with a complex formed by 13 subunits, called EIF3, which serves as a molecular bridge between the ribosomes and the RNA (Hinnebusch, 2014). Interestingly, virtually all EIF3 subunits have been linked to virus infection.
While EIF3B and EIF3G harbor RRMs, other subunits employ unconventional modes of RNA binding (Table S1 and Figure S1b–d). For example, RBDmap revealed a region between the amino acids 112–131 of EIF3D that interact with RNA (Figure S1b), in agreement with previous in vitro experiments (Asano et al., 1997). Strikingly, HIV protease cleaves EIF3D in positions 114–115, and the knockdown of this EIF3 subunit, but not others, increases HIV infectivity (Jager et al., 2011). The latter study proposed that EIF3D binds the vRNA and inhibits reverse transcription, and that this antiviral mechanism is counteracted by cleaving EIF3D (Jager et al., 2011). Notably, it has been recently described that EIF3D is a novel cap‐binding protein that interacts with a specific pool of cellular mRNAs that do not employ the EIF4F complex to initiate translation (Lee, Kranzusch, Doudna, & Cate, 2016). The translation mechanism proposed implicates both a specific RNA structure that opens the EIF3D cap‐binding pocket and another RNA element that prevents EIF4F recruitment. The possibility that similar structures are present in vRNAs has been proposed, but requires experimental confirmation (Carrasco, Sanz, & Gonzalez‐Almela, 2018). SINV subgenomic mRNA follows a noncanonical translation initiation mechanism that employs a cap‐dependent “scanning” but lacks the participation of several EIFs, including the EIF4E, EIF4A, and EIF2α, and intact EIF4G (Castello, Sanz, Molina, & Carrasco, 2006; Garcia‐Moreno, Sanz, & Carrasco, 2015; Garcia‐Moreno, Sanz, Pelletier, & Carrasco, 2013). A stem‐loop structure present at the 5′ end of this mRNA is required for efficient translation (McInerney, Kedersha, Kaufman, Anderson, & Liljestrom, 2005; Ventoso et al., 2006). Thus, novel cap‐binding proteins such as EIF3D may replace the EIF4F complex and promote viral translation after activation by the hairpin structure (Carrasco et al., 2018).
Other vRNAs lack a 5′ cap structure, but instead a viral protein (VPg) is covalently linked to the 5′ end of the RNA. The calicivirus VPg interacts in vitro and in vivo with EIF3D as well as with other EIFs and ribosomal proteins (Chung et al., 2014; Daughenbaugh, Fraser, Hershey, & Hardy, 2003; Daughenbaugh, Wobus, & Hardy, 2006). The role of EIF3D in calicivirus translation is not yet well understood, but this interaction points toward its importance in cap‐independent translation initiation by binding viral proteins covalently linked to the 5′ end of vRNAs.
4.5. Utilizing the cellular transport network: Viruses and dynein
4.5.1. RNA transport mediated by the dynein molecular motor
RNA localization over long distances often involves the active bidirectional transport along microtubules by molecular motors, mainly dynein and kinesin proteins and adapter proteins (Marchand, Gaspar, & Ephrussi, 2012; Tekotte & Davis, 2002). Recently, RNA‐IC experiments in different cells and organisms revealed that numerous dynein and kinesin proteins are endowed with RNA‐binding activity (Table S1) (Hentze et al., 2018), and RBDmap in human cells identified potential RNA‐binding sites in some members of the dynein family (Figure S2a–e). Interestingly, Drosophila dynein light chain 1 (highly conserved in humans) can strongly bind in vitro to a particular region in the 3′ UTR of gurken RNA, and mediates its correct localization during oogenesis in vivo (Rom, Faicevici, Almog, & Neuman‐Silberberg, 2007). Active transport has also been linked to P‐body and stress granule formation in mammalian and Drosophila cells because silencing of dynein cytoplasmic 1 heavy chain 1 (DYNC1H1) leads to a dramatic impairment of their formation (Loschi, Leishman, Berardone, & Boccaccio, 2009). Most viruses can subvert cellular transport motors. For example, dyneins and kinesins facilitate uncoating, movement of preintegration complexes, and transport of retrovirus proteins (e.g., Gag and Env) as well as the movement of the vRNA to the perinuclear region for virion assembly (Arriagada, 2017).
Movement of newly synthesized retroviral RNA throughout the cytoplasm may require the dynein motor complex, although results are conflicting. Some studies suggest that, after nuclear export, the HIV unspliced RNA moves toward the microtubule organizing center (MTOC) where Gag interacts with the psi packaging signal. Gag also associates with endosomes that are transported to the virion assembly sites. The dynein complex, including members such as DYNC1H1 and dynactin subunit 2 (DCTN2), would mediate the transport of both the vRNA to the MTOC and the vesicles containing viral Gag/RNA RNP (Lehmann et al., 2009; Molle et al., 2009). However, another study suggests that HIV unspliced RNA traffics throughout the cytoplasm by diffusion after leaving the nucleus en masse through a Rev‐dependent mechanism (Pocock, Becker, Swanson, Ahlquist, & Sherer, 2016). Further studies are required to clarify the vRNA transport mechanisms followed by retroviruses and the implications of the newly discovered RNA‐binding activity of dyneins in this process.
4.5.2. The role of dyneins in HIV uncoating and reverse transcription
HIV uncoating is an incompletely understood multistep process during which the capsid shell enclosing the genome is disassembled. Recent studies suggest that dyneins promote HIV uncoating and this process is linked to the intracellular movement toward the nucleus (Lukic, Dharan, Fricke, Diaz‐Griffero, & Campbell, 2014; Pawlica & Berthoux, 2014). Accordingly, DYNC1H1 knockdown or disruption of the dynein complex by overexpressing DCTN2 interferes with uncoating (Pawlica & Berthoux, 2014). Whether dynein‐mediated uncoating affects reverse transcription is unclear, because microtubule network disruption with nocodazole does not inhibit HIV reverse transcription (Lukic et al., 2014), and perturbation of the dynein complex only causes a transient defect in reverse transcription (Pawlica & Berthoux, 2014). A different study showed that cytoplasmic dynein light chain 1 (DYNLL1), acting outside the microtubule‐associated dynein complex, is required for both HIV uncoating and reverse transcription (Jayappa et al., 2015).
The novel RNA‐binding activities of several dynein and adapter proteins such as DYNC1H1, dynein cytoplasmic 1 intermediate chain 2 (DYNC1I2), dynein cytoplasmic 1 light intermediate chain 1 (DYNC1LI1), and DCTN2 may contribute to both early and late events of HIV infection by binding directly to vRNA. In agreement, DYNC1H1 was identified in a recent approach aimed at deciphering the in vivo HIV RNA interactome (Knoener, Becker, Scalf, Sherer, & Smith, 2017).
4.5.3. Involvement of dyneins in viral replication factory assembly
Most cytoplasmic viruses induce the remodeling of intracellular membranes to assemble stable compartments (referred to as “viral factories”) for vRNA replication, translation, and packaging protected from host antiviral defenses. In HCV‐infected cells, the viral core protein directs a dynein‐mediated redistribution of lipid droplets around the MTOC (Boulant et al., 2008). HCV nonstructural protein 5A (NS5A) is essential for the regulation of vRNA synthesis and the formation of endoplasmic reticulum (ER)‐derived viral factories. Newly synthesized NS5A associates with core‐coated lipid droplets and facilitates viral genome transfer, thus linking HCV replication with virion assembly (Eyre et al., 2014). Interestingly, microtubule polymerization inhibitors and DYNC1H1 knockdown impair NS5A movement and HCV RNA replication (Eyre et al., 2014). The dynein complex may also be involved in the movement of DENV2 proteins (and possibly vRNA) to the ER and from the ER to the Golgi apparatus for viral replication and virion assembly (Shrivastava, Sripada, Kaur, Shah, & Cecilia, 2011). Moreover, dynein complex disruption by DCTN2 overexpression and depolymerization of microtubules with specific drugs affect the distribution of viral nucleocapsid and inhibit vRNA synthesis in Hantavirus‐infected cells (Ramanathan et al., 2007). The roles of dyneins and their RNA‐binding activities in viral factory biogenesis should be assessed in the future.
4.6. RBPs resident in the ER affect virus infection
The protein transport protein Sec31A (SEC31A) is a component of the coat protein complex II, which mediates the budding of transport vesicles from the ER, and is endowed with RNA‐binding activity (Figure 4a) as identified by RNA‐IC and RBDmap (Castello et al., 2016; Hentze et al., 2018). SEC31A redistributes from the perinuclear region to the entire cytoplasm and is degraded as poliovirus infection progresses, suggesting that it may have an important antiviral role (Richards, Soares‐Martins, Riddell, & Jackson, 2014). Future investigations will shed light on the role of SEC31A and its RNA‐binding activity in virus infection.
Figure 4.
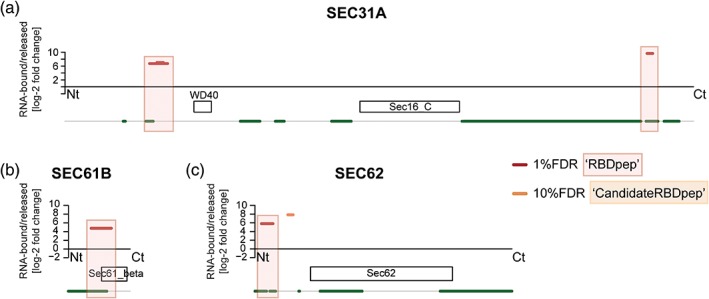
RNA‐binding domains (RBDs) of virus‐related RNA‐binding proteins. RBDmap profiles of SEC31A (a), SEC61B (b), and SEC62 (c). Boxes represent high confidence (red, 1% FDR) or relevant candidate (orange, 10% FDR) RBDs. White boxes symbolize Pfam‐annotated domains. Green lines indicate predicted disordered regions (IUPred score > 0.4). FDR, false discovery rate
Several proteins of the ER translocation machinery such as Sec61 translocon alpha 1 subunit (SEC61A1), Sec61 translocon beta subunit (SEC61B, Figure 4b), SEC62 homolog preprotein translocation factor (SEC62, Figure 4c), or SEC63 homolog protein translocation regulator (SEC63) were described to bind RNA in different RNA‐IC experiments (Hentze et al., 2018). Orthogonal proteomic studies aimed at identifying the native ER‐associated RNA interactome confirmed that several ER integral membrane proteins have noncanonical RNA‐binding activity, including SEC61A1 and SEC61B (SEC61 complex) (Jagannathan et al., 2014). They are involved in a novel mechanism that selectively anchors mRNAs encoding endomembrane proteins to the ER (Jagannathan et al., 2014). The SEC61 complex may play multiple roles in virus infection. Zika virus promotes the formation of cytoplasmic vacuoles, and viral RNA and E protein (that contains a transmembrane domain) accumulate in their borders (Monel et al., 2017). Inhibition of SEC61A1 with specific drugs impairs both vacuole formation and virus production. The SEC61 complex is also essential for glycoprotein proteostasis of IAV hemagglutinin and neuraminidase proteins and HIV gp120 protein, as well as for DENV RNA replication (Heaton et al., 2016). SEC61A1 also interacts and co‐localizes with the membrane‐associated VP24 protein of EBOV, and is proposed to regulate transcription and replication of the viral genome (Iwasa et al., 2011). The SEC61B subunit, endowed with noncanonical RNA‐binding capacity (Figure 4b), was also identified as an important factor for flavivirus replication in siRNA (Savidis et al., 2016) and CRISPR screens (Zhang et al., 2016). Whether the SEC61 complex anchors vRNA directly to ER‐derived membranous compartments, as described for specific cellular mRNAs (Jagannathan et al., 2014), remains unknown.
The dimeric SEC62/SEC63 complex interacts with the SEC61 complex and acts in posttranslational translocation into the ER. SEC62 novel RNA‐binding site maps to the disordered cytosolic N‐terminal part (Figure 4c), while SEC63 RBD is still unknown. SEC62 abundance increases after FMDV infection and its depletion reduces FMDV titre (Guo et al., 2015). Moreover, SEC63 knock out decreases virus yield in both mammalian and insect cells infected with different flaviviruses (Zhang et al., 2016). Although SEC62 and SEC63 are important for viral fitness, their exact roles in infection and the potential of their RNA‐binding activity to promote or restrict viral replication remain unknown.
5. OUTLOOK
RBPs are key regulators of cellular pathways that respond to infection or are hijacked by viruses. Functions of many classical RBPs have been characterized in the context of infection. However, recent RNA‐IC experiments have expanded the census of RBPs and identified unanticipated modes of RNA binding (Hentze et al., 2018). About 200 unorthodox RBPs have been linked to virus infection or immunity but, in many cases, their RNA‐binding properties had remained unnoticed. In this review, we have discussed several examples of unconventional RBPs that either support viral replication or mediate the host antiviral response. Most virus‐linked, unorthodox RBPs remain poorly understood. Importantly, many of these newly discovered RBPs have enzymatic activities such as chaperone, kinase, isomerase, E3 ubiquitin ligase, and others, and are therefore susceptible to inhibition by small compounds in host‐based antiviral therapies. Further research should aim at characterizing their roles in infection to better understand the interplay between unorthodox RBPs and viruses.
System‐wide approaches to discover the vRNA–host RBP landscape have contributed to our understanding of the role of host RBPs in infected cells (Knoener et al., 2017; LaPointe, Gebhart, Meller, Hardy, & Sokoloski, 2018; Lenarcic, Landry, Greco, Cristea, & Thompson, 2013; Phillips, Soderblom, Bradrick, & Garcia‐Blanco, 2016; Viktorovskaya, Greco, Cristea, & Thompson, 2016). Although the currently available methods have technical limitations, we are confident that global analyses of protein–RNA interactions can determine the arsenal of RBPs deployed in the host–virus battlefield. Such approaches must be complemented with techniques such as crosslinking immunoprecipitation followed by sequencing (CLIP‐seq) and its variants (Wheeler, Van Nostrand, & Yeo, 2018) to determine in a global scale the footprints of RBPs on viral and host RNAs. We believe that there is a whole universe of host–virus interactions to be discovered that will not only further our understanding of virus and cell biology, but also lead to the development of novel antiviral approaches.
CONFLICT OF INTEREST
The authors have declared no conflicts of interest for this article.
RELATED WIREs ARTICLE
Supporting information
Figure S1 RNA‐binding domains (RBDs) of virus‐related RNA‐binding proteins. RBDmap profiles of CAPRIN1 (a), EIF3D (b), EIF3I (c), and EIF3G (d). Boxes represent high confidence (red, 1% FDR) or relevant candidate (orange, 10% FDR) RBDs. White boxes symbolize Pfam‐annotated domains. Green lines indicate predicted disordered regions (IUPred score > 0.4). FDR, false discovery rate.
Figure S2 RNA‐binding domains (RBDs) of virus‐related RNA‐binding proteins. RBDmap profiles of DYNC1H1 C‐terminal (a), DYNC1I2 (c), DYNC1LI1 (d), and DCTN2 (e). DYNC1H1 Pfam domain architecture (b). Boxes represent high confidence (red, 1% FDR) or relevant candidate (orange, 10% FDR) RBDs. White boxes symbolize Pfam‐annotated domains. Green lines indicate predicted disordered regions (IUPred score > 0.4). FDR, false discovery rate.
File S1 Description of how the virus‐linked RNA interactome was constructed.
Table S1 Virus‐linked RNA interactome, GO terms used in its construction, and domains used in classification of RNA‐binding proteins (RBPs). A particular virus‐linked RBP would be missing if (a) it has not been identified by RNA‐IC due to its low abundance, low crosslink ability, incompatibility with mass spectrometry or because RNA‐IC studies have only been performed in steady‐state conditions; (b) it is not (yet) annotated by GO terms related to viruses, infection, or immunity; or/and (c) it appears in fewer than six publication in PubMed linked to the term “virus.”
ACKNOWLEDGMENTS
A.C. is funded by MRC Career Development Award (number MR/L019434/1) and John Fell funds from the University of Oxford. M.G.M. is funded by the European Union's Horizon 2020 Research and Innovation programme under the Marie‐Sklodowska‐Curie Grant agreement number 700184.
Garcia‐Moreno M, Järvelin AI, Castello A. Unconventional RNA‐binding proteins step into the virus–host battlefront. WIREs RNA. 2018;9:e1498. 10.1002/wrna.1498
Funding information Marie‐Sklodowska‐Curie Grant, Grant/Award Number: 700184; European Commission; University of Oxford through the John Fell Fund programme; MRC: Medical Research Council (UK), Grant/Award Number: MR/L019434/1
REFERENCES
- Abbas, Y. M. , Pichlmair, A. , Gorna, M. W. , Superti‐Furga, G. , & Nagar, B. (2013). Structural basis for viral 5'‐PPP‐RNA recognition by human IFIT proteins. Nature, 494(7435), 60–64. 10.1038/nature11783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman, V. , Koonin, E. V. , & Aravind, L. (2002). Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Research, 30(7), 1427–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arif, A. , Yao, P. , Terenzi, F. , Jia, J. , Ray, P. S. , & Fox, P. L. (2018). The GAIT translational control system. WIREs RNA, 9(2), e1441 10.1002/wrna.1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arriagada, G. (2017). Retroviruses and microtubule‐associated motor proteins. Cellular Microbiology, 19(9), e12759. 10.1111/cmi.12759 [DOI] [PubMed] [Google Scholar]
- Asano, K. , Vornlocher, H. P. , Richter‐Cook, N. J. , Merrick, W. C. , Hinnebusch, A. G. , & Hershey, J. W. (1997). Structure of cDNAs encoding human eukaryotic initiation factor 3 subunits. Possible roles in RNA binding and macromolecular assembly. The Journal of Biological Chemistry, 272(43), 27042–27052. [DOI] [PubMed] [Google Scholar]
- Au, H. H. , & Jan, E. (2014). Novel viral translation strategies. WIREs RNA, 5(6), 779–801. 10.1002/wrna.1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balistreri, G. , Bognanni, C. , & Muhlemann, O. (2017). Virus escape and manipulation of cellular nonsense‐mediated mRNA decay. Viruses, 9(1), 24. 10.3390/v9010024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltimore, D. (1971). Expression of animal virus genomes. Bacteriological Reviews, 35(3), 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz, A. G. , Munschauer, M. , Schwanhausser, B. , Vasile, A. , Murakawa, Y. , Schueler, M. , … Landthaler, M. (2012). The mRNA‐bound proteome and its global occupancy profile on protein‐coding transcripts. Molecular Cell, 46(5), 674–690. [DOI] [PubMed] [Google Scholar]
- Barbalat, R. , Ewald, S. E. , Mouchess, M. L. , & Barton, G. M. (2011). Nucleic acid recognition by the innate immune system. Annual Review of Immunology, 29, 185–214. 10.1146/annurev-immunol-031210-101340 [DOI] [PubMed] [Google Scholar]
- Basu, S. , & Bahadur, R. P. (2016). A structural perspective of RNA recognition by intrinsically disordered proteins. Cellular and Molecular Life Sciences, 73(21), 4075–4084. 10.1007/s00018-016-2283-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beachboard, D. C. , & Horner, S. M. (2016). Innate immune evasion strategies of DNA and RNA viruses. Current Opinion in Microbiology, 32, 113–119. 10.1016/j.mib.2016.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham, C. J. , & Parker, R. (2008). P bodies, stress granules, and viral life cycles. Cell Host & Microbe, 3(4), 206–212. 10.1016/j.chom.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulant, S. , Douglas, M. W. , Moody, L. , Budkowska, A. , Targett‐Adams, P. , & McLauchlan, J. (2008). Hepatitis C virus core protein induces lipid droplet redistribution in a microtubule‐ and dynein‐dependent manner. Traffic, 9(8), 1268–1282. 10.1111/j.1600-0854.2008.00767.x [DOI] [PubMed] [Google Scholar]
- Broder, S. (2010). The development of antiretroviral therapy and its impact on the HIV‐1/AIDS pandemic. Antiviral Research, 85(1), 1–18. 10.1016/j.antiviral.2009.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco, L. , Sanz, M. A. , & Gonzalez‐Almela, E. (2018). The regulation of translation in alphavirus‐infected cells. Viruses, 10(2), 70. 10.3390/v10020070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castella, S. , Bernard, R. , Corno, M. , Fradin, A. , & Larcher, J. C. (2015). Ilf3 and NF90 functions in RNA biology. WIREs RNA, 6(2), 243–256. 10.1002/wrna.1270 [DOI] [PubMed] [Google Scholar]
- Castello, A. , Alvarez, E. , & Carrasco, L. (2011). The multifaceted poliovirus 2A protease: Regulation of gene expression by picornavirus proteases. Journal of Biomedicine & Biotechnology, 2011, 369648 10.1155/2011/369648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello, A. , Fischer, B. , Eichelbaum, K. , Horos, R. , Beckmann, B. M. , Strein, C. , … Hentze, M. W. (2012). Insights into RNA biology from an atlas of mammalian mRNA‐binding proteins. Cell, 149(6), 1393–1406. [DOI] [PubMed] [Google Scholar]
- Castello, A. , Fischer, B. , Frese, C. K. , Horos, R. , Alleaume, A. M. , Foehr, S. , … Hentze, M. W. (2016). Comprehensive identification of RNA‐binding domains in human cells. Molecular Cell, 63(4), 696–710. 10.1016/j.molcel.2016.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello, A. , Fischer, B. , Hentze, M. W. , & Preiss, T. (2013). RNA‐binding proteins in Mendelian disease. Trends in Genetics, 29(5), 318–327. [DOI] [PubMed] [Google Scholar]
- Castello, A. , Frese, C. K. , Fischer, B. , Jarvelin, A. I. , Horos, R. , Alleaume, A. M. , … Hentze, M. W. (2017). Identification of RNA‐binding domains of RNA‐binding proteins in cultured cells on a system‐wide scale with RBDmap. Nature Protocols, 12(12), 2447–2464. 10.1038/nprot.2017.106 [DOI] [PubMed] [Google Scholar]
- Castello, A. , Hentze, M. W. , & Preiss, T. (2015). Metabolic enzymes enjoying new partnerships as RNA‐binding proteins. Trends in Endocrinology and Metabolism, 26(12), 746–757. 10.1016/j.tem.2015.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello, A. , Sanz, M. A. , Molina, S. , & Carrasco, L. (2006). Translation of Sindbis virus 26S mRNA does not require intact eukariotic initiation factor 4G. Journal of Molecular Biology, 355(5), 942–956. 10.1016/j.jmb.2005.11.024 [DOI] [PubMed] [Google Scholar]
- Cate, J. H. (2017). Human eIF3: From 'blobology' to biological insight. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 372(1716), 20160176 10.1098/rstb.2016.0176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutter, D. S. , Jordan, M. R. , Bertagnolio, S. , & Shafer, R. W. (2016). HIV‐1 drug resistance and resistance testing. Infection, Genetics and Evolution, 46, 292–307. 10.1016/j.meegid.2016.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane, A. W. , McNally, M. T. , & Mouland, A. J. (2006). The retrovirus RNA trafficking granule: From birth to maturity. Retrovirology, 3, 18 10.1186/1742-4690-3-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C. H. , Curtis, J. D. , Maggi, L. B., Jr. , Faubert, B. , Villarino, A. V. , O'Sullivan, D. , … Pearce, E. L. (2013). Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell, 153(6), 1239–1251. 10.1016/j.cell.2013.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, C. , & Gack, M. U. (2017). Post‐translational control of intracellular pathogen sensing pathways. Trends in Immunology, 38(1), 39–52. 10.1016/j.it.2016.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury, N. R. , Heikel, G. , Trubitsyna, M. , Kubik, P. , Nowak, J. S. , Webb, S. , … Michlewski, G. (2017). RNA‐binding activity of TRIM25 is mediated by its PRY/SPRY domain and is required for ubiquitination. BMC Biology, 15(1), 105 10.1186/s12915-017-0444-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, L. , Bailey, D. , Leen, E. N. , Emmott, E. P. , Chaudhry, Y. , Roberts, L. O. , … Goodfellow, I. G. (2014). Norovirus translation requires an interaction between the C terminus of the genome‐linked viral protein VPg and eukaryotic translation initiation factor 4G. The Journal of Biological Chemistry, 289(31), 21738–21750. 10.1074/jbc.M114.550657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffis, S. , Szretter, K. J. , Schriewer, J. , Li, J. , Youn, S. , Errett, J. , … Diamond, M. S. (2010). 2'‐O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature, 468(7322), 452–456. 10.1038/nature09489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughenbaugh, K. F. , Fraser, C. S. , Hershey, J. W. , & Hardy, M. E. (2003). The genome‐linked protein VPg of the Norwalk virus binds eIF3, suggesting its role in translation initiation complex recruitment. The EMBO Journal, 22(11), 2852–2859. 10.1093/emboj/cdg251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughenbaugh, K. F. , Wobus, C. E. , & Hardy, M. E. (2006). VPg of murine norovirus binds translation initiation factors in infected cells. Virology Journal, 3, 33 10.1186/1743-422X-3-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty, M. D. , Liu, B. , & Frankel, A. D. (2010). Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nature Structural & Molecular Biology, 17(11), 1337–1342. 10.1038/nsmb.1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey, N. E. , Trave, G. , & Gibson, T. J. (2011). How viruses hijack cell regulation. Trends in Biochemical Sciences, 36(3), 159–169. 10.1016/j.tibs.2010.10.002 [DOI] [PubMed] [Google Scholar]
- Dawar, F. U. , Tu, J. , Khattak, M. N. , Mei, J. , & Lin, L. (2017). Cyclophilin A: A key factor in virus replication and potential target for anti‐viral therapy. Current Issues in Molecular Biology, 21, 1–20. 10.21775/cimb.021.001 [DOI] [PubMed] [Google Scholar]
- Debing, Y. , Neyts, J. , & Delang, L. (2015). The future of antivirals: Broad‐spectrum inhibitors. Current Opinion in Infectious Diseases, 28(6), 596–602. 10.1097/QCO.0000000000000212 [DOI] [PubMed] [Google Scholar]
- Decroly, E. , Ferron, F. , Lescar, J. , & Canard, B. (2011). Conventional and unconventional mechanisms for capping viral mRNA. Nature Reviews. Microbiology, 10(1), 51–65. 10.1038/nrmicro2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond, M. S. , & Farzan, M. (2013). The broad‐spectrum antiviral functions of IFIT and IFITM proteins. Nature Reviews. Immunology, 13(1), 46–57. 10.1038/nri3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson, A. M. , & Wilusz, J. (2011). Strategies for viral RNA stability: Live long and prosper. Trends in Genetics, 27(7), 286–293. 10.1016/j.tig.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery, J. S. , & Feld, J. J. (2017). Treatment of hepatitis B virus with combination therapy now and in the future. Best Practice & Research. Clinical Gastroenterology, 31(3), 347–355. 10.1016/j.bpg.2017.04.007 [DOI] [PubMed] [Google Scholar]
- Eyre, N. S. , Fiches, G. N. , Aloia, A. L. , Helbig, K. J. , McCartney, E. M. , McErlean, C. S. , … Beard, M. R. (2014). Dynamic imaging of the hepatitis C virus NS5A protein during a productive infection. Journal of Virology, 88(7), 3636–3652. 10.1128/JVI.02490-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, X. , Xue, B. , Dolan, P. T. , LaCount, D. J. , Kurgan, L. , & Uversky, V. N. (2014). The intrinsic disorder status of the human hepatitis C virus proteome. Molecular BioSystems, 10(6), 1345–1363. 10.1039/c4mb00027g [DOI] [PubMed] [Google Scholar]
- Faye, M. D. , Graber, T. E. , Liu, P. , Thakor, N. , Baird, S. D. , Durie, D. , & Holcik, M. (2013). Nucleotide composition of cellular internal ribosome entry sites defines dependence on NF45 and predicts a posttranscriptional mitotic regulon. Molecular and Cellular Biology, 33(2), 307–318. 10.1128/MCB.00546-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth, A. E. , & Brierley, I. (2012). Non‐canonical translation in RNA viruses. The Journal of General Virology, 93(Pt 7), 1385–1409. 10.1099/vir.0.042499-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleith, R. C. , Mears, H. V. , Leong, X. Y. , Sanford, T. J. , Emmott, E. , Graham, S. C. , … Sweeney, T. R. (2018). IFIT3 and IFIT2/3 promote IFIT1‐mediated translation inhibition by enhancing binding to non‐self RNA. Nucleic Acids Research, 46(10), 5269–5285. 10.1093/nar/gky191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flisiak, R. , Horban, A. , Gallay, P. , Bobardt, M. , Selvarajah, S. , Wiercinska‐Drapalo, A. , … Scalfaro, P. (2008). The cyclophilin inhibitor Debio‐025 shows potent anti‐hepatitis C effect in patients coinfected with hepatitis C and human immunodeficiency virus. Hepatology, 47(3), 817–826. 10.1002/hep.22131 [DOI] [PubMed] [Google Scholar]
- Gack, M. U. , Shin, Y. C. , Joo, C. H. , Urano, T. , Liang, C. , Sun, L. , … Jung, J. U. (2007). TRIM25 RING‐finger E3 ubiquitin ligase is essential for RIG‐I‐mediated antiviral activity. Nature, 446(7138), 916–920. 10.1038/nature05732 [DOI] [PubMed] [Google Scholar]
- Garbelli, A. , Riva, V. , Crespan, E. , & Maga, G. (2017). How to win the HIV‐1 drug resistance hurdle race: Running faster or jumping higher? The Biochemical Journal, 474(10), 1559–1577. 10.1042/BCJ20160772 [DOI] [PubMed] [Google Scholar]
- Garcia‐Moreno, M. , Sanz, M. A. , & Carrasco, L. (2015). Initiation codon selection is accomplished by a scanning mechanism without crucial initiation factors in Sindbis virus subgenomic mRNA. RNA, 21(1), 93–112. 10.1261/rna.047084.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Moreno, M. , Sanz, M. A. , Pelletier, J. , & Carrasco, L. (2013). Requirements for eIF4A and eIF2 during translation of Sindbis virus subgenomic mRNA in vertebrate and invertebrate host cells. Cellular Microbiology, 15(5), 823–840. 10.1111/cmi.12079 [DOI] [PubMed] [Google Scholar]
- Gelinas, J. F. , Clerzius, G. , Shaw, E. , & Gatignol, A. (2011). Enhancement of replication of RNA viruses by ADAR1 via RNA editing and inhibition of RNA‐activated protein kinase. Journal of Virology, 85(17), 8460–8466. 10.1128/JVI.00240-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller, R. , Taguwa, S. , & Frydman, J. (2012). Broad action of Hsp90 as a host chaperone required for viral replication. Biochimica et Biophysica Acta‐Molecular Cell Research, 1823(3), 698–706. 10.1016/j.bbamcr.2011.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin, L. , Hagai, T. , LaBarbera, A. , Solovey, M. , & Andino, R. (2014). Rapid evolution of virus sequences in intrinsically disordered protein regions. PLoS Pathogens, 10(12), e1004529 10.1371/journal.ppat.1004529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales‐van Horn, S. R. , & Sarnow, P. (2017). Making the mark: The role of adenosine modifications in the life cycle of RNA viruses. Cell Host & Microbe, 21(6), 661–669. 10.1016/j.chom.2017.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber, T. E. , Baird, S. D. , Kao, P. N. , Mathews, M. B. , & Holcik, M. (2010). NF45 functions as an IRES trans‐acting factor that is required for translation of cIAP1 during the unfolded protein response. Cell Death and Differentiation, 17(4), 719–729. 10.1038/cdd.2009.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronland, G. R. , & Ramos, A. (2017). The devil is in the domain: Understanding protein recognition of multiple RNA targets. Biochemical Society Transactions, 45(6), 1305–1311. 10.1042/BST20160362 [DOI] [PubMed] [Google Scholar]
- Guo, H. C. , Jin, Y. , Han, S. C. , Sun, S. Q. , Wei, Y. Q. , Liu, X. J. , … Liu, X. T. (2015). Quantitative proteomic analysis of BHK‐21 cells infected with foot‐and‐mouth disease virus serotype Asia 1. PLoS One, 10(7), e0132384. 10.1371/journal.pone.0132384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habjan, M. , Hubel, P. , Lacerda, L. , Benda, C. , Holze, C. , Eberl, C. H. , … Pichlmair, A. (2013). Sequestration by IFIT1 impairs translation of 2'O‐unmethylated capped RNA. PLoS Pathogens, 9(10), e1003663. 10.1371/journal.ppat.1003663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima, A. , Guettouche, T. , & Barber, G. N. (2010). Phosphorylation of the NFAR proteins by the dsRNA‐dependent protein kinase PKR constitutes a novel mechanism of translational regulation and cellular defense. Genes & Development, 24(23), 2640–2653. 10.1101/gad.1965010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwig, A. , Landick, R. , & Berkhout, B. (2017). The battle of RNA synthesis: Virus versus host. Viruses, 9(10), 309. 10.3390/v9100309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, C. , Sidoli, S. , Warneford‐Thomson, R. , Tatomer, D. C. , Wilusz, J. E. , Garcia, B. A. , & Bonasio, R. (2016). High‐resolution mapping of RNA‐binding regions in the nuclear proteome of embryonic stem cells. Molecular Cell, 64(2), 416–430. 10.1016/j.molcel.2016.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton, N. S. , Moshkina, N. , Fenouil, R. , Gardner, T. J. , Aguirre, S. , Shah, P. S. , … Marazzi, I. (2016). Targeting viral proteostasis limits influenza virus, HIV, and dengue virus infection. Immunity, 44(1), 46–58. 10.1016/j.immuni.2015.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze, M. W. , Castello, A. , Schwarzl, T. , & Preiss, T. (2018). A brave new world of RNA‐binding proteins. Nature Reviews. Molecular Cell Biology, 19, 327–341. 10.1038/nrm.2017.130 [DOI] [PubMed] [Google Scholar]
- Herbert, K. M. , & Nag, A. (2016). A tale of two RNAs during viral infection: How viruses antagonize mRNAs and small non‐coding RNAs in the host cell. Viruses, 8(6), 154. 10.3390/v8060154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch, A. G. (2014). The scanning mechanism of eukaryotic translation initiation. Annual Review of Biochemistry, 83, 779–812. 10.1146/annurev-biochem-060713-035802 [DOI] [PubMed] [Google Scholar]
- Hyde, J. L. , Gardner, C. L. , Kimura, T. , White, J. P. , Liu, G. , Trobaugh, D. W. , … Diamond, M. S. (2014). A viral RNA structural element alters host recognition of nonself RNA. Science, 343(6172), 783–787. 10.1126/science.1248465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin, K. K. , Renzette, N. , Kowalik, T. F. , & Jensen, J. D. (2016). Antiviral drug resistance as an adaptive process. Virus Evolution, 2(1), vew014. 10.1093/ve/vew014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken, O. , Baroth, M. , Grassmann, C. W. , Weinlich, S. , Ostareck, D. H. , Ostareck‐Lederer, A. , & Behrens, S. E. (2007). Nuclear factors are involved in hepatitis C virus RNA replication. RNA, 13(10), 1675–1692. 10.1261/rna.594207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken, O. , Grassmann, C. W. , Sarisky, R. T. , Kann, M. , Zhang, S. , Grosse, F. , … Behrens, S. E. (2003). Members of the NF90/NFAR protein group are involved in the life cycle of a positive‐strand RNA virus. The EMBO Journal, 22(21), 5655–5665. 10.1093/emboj/cdg562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken, O. , Grassmann, C. W. , Yu, H. , & Behrens, S. E. (2004). Complex signals in the genomic 3′ nontranslated region of bovine viral diarrhea virus coordinate translation and replication of the viral RNA. RNA, 10(10), 1637–1652. 10.1261/rna.7290904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanyi‐Nagy, R. , Lavergne, J. P. , Gabus, C. , Ficheux, D. , & Darlix, J. L. (2008). RNA chaperoning and intrinsic disorder in the core proteins of Flaviviridae. Nucleic Acids Research, 36(3), 712–725. 10.1093/nar/gkm1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasa, A. , Halfmann, P. , Noda, T. , Oyama, M. , Kozuka‐Hata, H. , Watanabe, S. , … Kawaoka, Y. (2011). Contribution of Sec61alpha to the life cycle of Ebola virus. The Journal of Infectious Diseases, 204(Suppl 3), S919–S926. 10.1093/infdis/jir324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki, S. , Kobayashi, M. , Yoda, M. , Sakaguchi, Y. , Katsuma, S. , Suzuki, T. , & Tomari, Y. (2010). Hsc70/Hsp90 chaperone machinery mediates ATP‐dependent RISC loading of small RNA duplexes. Molecular Cell, 39(2), 292–299. 10.1016/j.molcel.2010.05.015 [DOI] [PubMed] [Google Scholar]
- Jagannathan, S. , Hsu, J. C. , Reid, D. W. , Chen, Q. , Thompson, W. J. , Moseley, A. M. , & Nicchitta, C. V. (2014). Multifunctional roles for the protein translocation machinery in RNA anchoring to the endoplasmic reticulum. The Journal of Biological Chemistry, 289(37), 25907–25924. 10.1074/jbc.M114.580688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager, S. , Cimermancic, P. , Gulbahce, N. , Johnson, J. R. , McGovern, K. E. , Clarke, S. C. , … Krogan, N. J. (2011). Global landscape of HIV‐human protein complexes. Nature, 481(7381), 365–370. 10.1038/nature10719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubiec, A. , & Jupin, I. (2007). Regulation of positive‐strand RNA virus replication: The emerging role of phosphorylation. Virus Research, 129(2), 73–79. 10.1016/j.virusres.2007.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvelin, A. I. , Noerenberg, M. , Davis, I. , & Castello, A. (2016). The new (dis)order in RNA regulation. Cell Communication and Signaling: CCS, 14, 9 10.1186/s12964-016-0132-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayappa, K. D. , Ao, Z. , Wang, X. , Mouland, A. J. , Shekhar, S. , Yang, X. , & Yao, X. (2015). Human immunodeficiency virus type 1 employs the cellular dynein light chain 1 protein for reverse transcription through interaction with its integrase protein. Journal of Virology, 89(7), 3497–3511. 10.1128/JVI.03347-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John, S. P. , Wang, T. , Steffen, S. , Longhi, S. , Schmaljohn, C. S. , & Jonsson, C. B. (2007). Ebola virus VP30 is an RNA binding protein. Journal of Virology, 81(17), 8967–8976. 10.1128/JVI.02523-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, B. , VanBlargan, L. A. , Xu, W. , White, J. P. , Shan, C. , Shi, P. Y. , … Amarasinghe, G. K. (2018). Human IFIT3 modulates IFIT1 RNA binding specificity and protein stability. Immunity, 48(3), 487–499.e5. 10.1016/j.immuni.2018.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungfleisch, J. , Blasco‐Moreno, B. , & Diez, J. (2016). Use of cellular decapping activators by positive‐strand RNA viruses. Viruses, 8(12), 340. 10.3390/v8120340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaddis Maldonado, R. J. , & Parent, L. J. (2016). Orchestrating the selection and packaging of genomic RNA by retroviruses: An ensemble of viral and host factors. Viruses, 8(9), 257. 10.3390/v8090257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane, M. , Zang, T. M. , Rihn, S. J. , Zhang, F. , Kueck, T. , Alim, M. , … Bieniasz, P. D. (2016). Identification of interferon‐stimulated genes with antiretroviral activity. Cell Host & Microbe, 20(3), 392–405. 10.1016/j.chom.2016.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katibah, G. E. , Qin, Y. , Sidote, D. J. , Yao, J. , Lambowitz, A. M. , & Collins, K. (2014). Broad and adaptable RNA structure recognition by the human interferon‐induced tetratricopeptide repeat protein IFIT5. Proceedings of the National Academy of Sciences of the United States of America, 111(33), 12025–12030. 10.1073/pnas.1412842111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck, F. , Ataey, P. , Amaya, M. , Bailey, C. , & Narayanan, A. (2015). Phosphorylation of single stranded RNA virus proteins and potential for novel therapeutic strategies. Viruses, 7(10), 5257–5273. 10.3390/v7102872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaperskyy, D. A. , Hatchette, T. F. , & McCormick, C. (2012). Influenza A virus inhibits cytoplasmic stress granule formation. The FASEB Journal, 26(4), 1629–1639. 10.1096/fj.11-196915 [DOI] [PubMed] [Google Scholar]
- Knoener, R. A. , Becker, J. T. , Scalf, M. , Sherer, N. M. , & Smith, L. M. (2017). Elucidating the in vivo interactome of HIV‐1 RNA by hybridization capture and mass spectrometry. Scientific Reports, 7(1), 16965 10.1038/s41598-017-16793-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouwaki, T. , Okamoto, M. , Tsukamoto, H. , Fukushima, Y. , & Oshiumi, H. (2017). Extracellular vesicles deliver host and virus RNA and regulate innate immune response. International Journal of Molecular Sciences, 18(3), 666. 10.3390/ijms18030666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer, K. , Sachsenberg, T. , Beckmann, B. M. , Qamar, S. , Boon, K. L. , Hentze, M. W. , … Urlaub, H. (2014). Photo‐cross‐linking and high‐resolution mass spectrometry for assignment of RNA‐binding sites in RNA‐binding proteins. Nature Methods, 11(10), 1064–1070. 10.1038/nmeth.3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, P. , Sweeney, T. R. , Skabkin, M. A. , Skabkina, O. V. , Hellen, C. U. , & Pestova, T. V. (2014). Inhibition of translation by IFIT family members is determined by their ability to interact selectively with the 5′‐terminal regions of cap0‐, cap1‐ and 5'ppp‐ mRNAs. Nucleic Acids Research, 42(5), 3228–3245. 10.1093/nar/gkt1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuss, S. K. , Mata, M. A. , Zhang, L. , & Fontoura, B. M. (2013). Nuclear imprisonment: Viral strategies to arrest host mRNA nuclear export. Viruses, 5(7), 1824–1849. 10.3390/v5071824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwano, Y. , Pullmann, R., Jr. , Marasa, B. S. , Abdelmohsen, K. , Lee, E. K. , Yang, X. , … Gorospe, M. (2010). NF90 selectively represses the translation of target mRNAs bearing an AU‐rich signature motif. Nucleic Acids Research, 38(1), 225–238. 10.1093/nar/gkp861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, S. C. , Yi, H. , Eichelbaum, K. , Fohr, S. , Fischer, B. , You, K. T. , … Kim, V. N. (2013). The RNA‐binding protein repertoire of embryonic stem cells. Nature Structural & Molecular Biology, 20(9), 1122–1130. 10.1038/nsmb.2638 [DOI] [PubMed] [Google Scholar]
- LaPointe, A. T. , Gebhart, N. N. , Meller, M. E. , Hardy, R. W. , & Sokoloski, K. J. (2018). The identification and characterization of Sindbis virus RNA:Host protein interactions. Journal of Virology, 92, e02171‐17 10.1128/JVI.02171-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law, G. L. , Korth, M. J. , Benecke, A. G. , & Katze, M. G. (2013). Systems virology: Host‐directed approaches to viral pathogenesis and drug targeting. Nature Reviews. Microbiology, 11(7), 455–466. 10.1038/nrmicro3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, A. S. , Kranzusch, P. J. , Doudna, J. A. , & Cate, J. H. (2016). eIF3d is an mRNA cap‐binding protein that is required for specialized translation initiation. Nature, 536(7614), 96–99. 10.1038/nature18954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann, M. , Milev, M. P. , Abrahamyan, L. , Yao, X. J. , Pante, N. , & Mouland, A. J. (2009). Intracellular transport of human immunodeficiency virus type 1 genomic RNA and viral production are dependent on dynein motor function and late endosome positioning. The Journal of Biological Chemistry, 284(21), 14572–14585. 10.1074/jbc.M808531200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenarcic, E. M. , Landry, D. M. , Greco, T. M. , Cristea, I. M. , & Thompson, S. R. (2013). Thiouracil cross‐linking mass spectrometry: A cell‐based method to identify host factors involved in viral amplification. Journal of Virology, 87(15), 8697–8712. 10.1128/JVI.00950-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M. M. , Lau, Z. , Cheung, P. , Aguilar, E. G. , Schneider, W. M. , Bozzacco, L. , … MacDonald, M. R. (2017). TRIM25 enhances the antiviral action of zinc‐finger antiviral protein (ZAP). PLoS Pathogens, 13(1), e1006145. 10.1371/journal.ppat.1006145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T. , Li, X. , Zhu, W. , Wang, H. , Mei, L. , Wu, S. , … Han, X. (2016). NF90 is a novel influenza A virus NS1‐interacting protein that antagonizes the inhibitory role of NS1 on PKR phosphorylation. FEBS Letters, 590(16), 2797–2810. 10.1002/1873-3468.12311 [DOI] [PubMed] [Google Scholar]
- Li, X. , Liu, C. X. , Xue, W. , Zhang, Y. , Jiang, S. , Yin, Q. F. , … Chen, L. L. (2017). Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Molecular Cell, 67(2), 214–227.e7. 10.1016/j.molcel.2017.05.023 [DOI] [PubMed] [Google Scholar]
- Li, Z. , & Nagy, P. D. (2011). Diverse roles of host RNA binding proteins in RNA virus replication. RNA Biology, 8(2), 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y. , Castello, A. , Fischer, B. , Leicht, S. , Foehr, S. , Frese, C. K. , … Preiss, T. (2016). The cardiomyocyte RNA‐binding proteome: Links to intermediary metabolism and heart disease. Cell Reports, 16(5), 1456–1469. 10.1016/j.celrep.2016.06.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B. , Li, N. L. , Shen, Y. , Bao, X. , Fabrizio, T. , Elbahesh, H. , … Li, K. (2016). The C‐terminal tail of TRIM56 dictates antiviral restriction of influenza A and B viruses by impeding viral RNA synthesis. Journal of Virology, 90(9), 4369–4382. 10.1128/JVI.03172-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B. , Li, N. L. , Wang, J. , Shi, P. Y. , Wang, T. , Miller, M. A. , & Li, K. (2014). Overlapping and distinct molecular determinants dictating the antiviral activities of TRIM56 against flaviviruses and coronavirus. Journal of Virology, 88(23), 13821–13835. 10.1128/JVI.02505-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschi, M. , Leishman, C. C. , Berardone, N. , & Boccaccio, G. L. (2009). Dynein and kinesin regulate stress‐granule and P‐body dynamics. Journal of Cell Science, 122(Pt 21), 3973–3982. 10.1242/jcs.051383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukic, Z. , Dharan, A. , Fricke, T. , Diaz‐Griffero, F. , & Campbell, E. M. (2014). HIV‐1 uncoating is facilitated by dynein and kinesin 1. Journal of Virology, 88(23), 13613–13625. 10.1128/JVI.02219-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde, B. M. , Moore, C. , & Varani, G. (2007). RNA‐binding proteins: Modular design for efficient function. Nature Reviews. Molecular Cell Biology, 8(6), 479–490. 10.1038/nrm2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, R. E. (2013). Regulation of stress granules and P‐bodies during RNA virus infection. WIREs RNA, 4(3), 317–331. 10.1002/wrna.1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, R. E. (2015). Nuclear proteins hijacked by mammalian cytoplasmic plus strand RNA viruses. Virology, 479–480, 457–474. 10.1016/j.virol.2015.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailliot, J. , & Martin, F. (2018). Viral internal ribosomal entry sites: Four classes for one goal. WIREs RNA, 9(2), e1458. 10.1002/wrna.1458 [DOI] [PubMed] [Google Scholar]
- Manokaran, G. , Finol, E. , Wang, C. , Gunaratne, J. , Bahl, J. , Ong, E. Z. , … Ooi, E. E. (2015). Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science, 350(6257), 217–221. 10.1126/science.aab3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand, V. , Gaspar, I. , & Ephrussi, A. (2012). An intracellular transmission control protocol: Assembly and transport of ribonucleoprotein complexes. Current Opinion in Cell Biology, 24(2), 202–210. 10.1016/j.ceb.2011.12.014 [DOI] [PubMed] [Google Scholar]
- Martinez, J. P. , Sasse, F. , Bronstrup, M. , Diez, J. , & Meyerhans, A. (2015). Antiviral drug discovery: Broad‐spectrum drugs from nature. Natural Product Reports, 32(1), 29–48. 10.1039/c4np00085d [DOI] [PubMed] [Google Scholar]
- Masliah, G. , Barraud, P. , & Allain, F. H. (2013). RNA recognition by double‐stranded RNA binding domains: A matter of shape and sequence. Cellular and Molecular Life Sciences, 70(11), 1875–1895. 10.1007/s00018-012-1119-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick, C. , & Khaperskyy, D. A. (2017). Translation inhibition and stress granules in the antiviral immune response. Nature Reviews. Immunology, 17(10), 647–660. 10.1038/nri.2017.63 [DOI] [PubMed] [Google Scholar]
- McInerney, G. M. , Kedersha, N. L. , Kaufman, R. J. , Anderson, P. , & Liljestrom, P. (2005). Importance of eIF2alpha phosphorylation and stress granule assembly in alphavirus translation regulation. Molecular Biology of the Cell, 16(8), 3753–3763. 10.1091/mbc.E05-02-0124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes‐Correa, M. , & Nunez, M. (2010). Management of HIV and hepatitis virus coinfection. Expert Opinion on Pharmacotherapy, 11(15), 2497–2516. 10.1517/14656566.2010.500615 [DOI] [PubMed] [Google Scholar]
- Meng, F. , Badierah, R. A. , Almehdar, H. A. , Redwan, E. M. , Kurgan, L. , & Uversky, V. N. (2015). Unstructural biology of the dengue virus proteins. The FEBS Journal, 282(17), 3368–3394. 10.1111/febs.13349 [DOI] [PubMed] [Google Scholar]
- Meyer, F. (2016). Viral interactions with components of the splicing machinery. Progress in Molecular Biology and Translational Science, 142, 241–268. 10.1016/bs.pmbts.2016.05.008 [DOI] [PubMed] [Google Scholar]
- Mok, B. W. , Song, W. , Wang, P. , Tai, H. , Chen, Y. , Zheng, M. , … Chen, H. (2012). The NS1 protein of influenza A virus interacts with cellular processing bodies and stress granules through RNA‐associated protein 55 (RAP55) during virus infection. Journal of Virology, 86(23), 12695–12707. 10.1128/JVI.00647-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molle, D. , Segura‐Morales, C. , Camus, G. , Berlioz‐Torrent, C. , Kjems, J. , Basyuk, E. , & Bertrand, E. (2009). Endosomal trafficking of HIV‐1 gag and genomic RNAs regulates viral egress. The Journal of Biological Chemistry, 284(29), 19727–19743. 10.1074/jbc.M109.019844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molleston, J. M. , & Cherry, S. (2017). Attacked from all sides: RNA decay in antiviral defense. Viruses, 9(1), 2. 10.3390/v9010002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monel, B. , Compton, A. A. , Bruel, T. , Amraoui, S. , Burlaud‐Gaillard, J. , Roy, N. , … Schwartz, O. (2017). Zika virus induces massive cytoplasmic vacuolization and paraptosis‐like death in infected cells. The EMBO Journal, 36(12), 1653–1668. 10.15252/embj.201695597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, S. L. , Barnhart, M. D. , & Wilusz, J. (2012). Inhibition and avoidance of mRNA degradation by RNA viruses. Current Opinion in Microbiology, 15(4), 500–505. 10.1016/j.mib.2012.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, S. L. , & Wilusz, J. (2013). Cytoplasmic viruses: Rage against the (cellular RNA decay) machine. PLoS Pathogens, 9(12), e1003762. 10.1371/journal.ppat.1003762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, S. , Jarvelin, A. I. , Davis, I. , Bond, G. L. , & Castello, A. (2017). Expanding horizons: New roles for non‐canonical RNA‐binding proteins in cancer. Current Opinion in Genetics & Development, 48, 112–120. 10.1016/j.gde.2017.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckenthaler, M. U. , Galy, B. , & Hentze, M. W. (2008). Systemic iron homeostasis and the iron‐responsive element/iron‐regulatory protein (IRE/IRP) regulatory network. Annual Review of Nutrition, 28, 197–213. 10.1146/annurev.nutr.28.061807.155521 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay, R. , Jia, J. , Arif, A. , Ray, P. S. , & Fox, P. L. (2009). The GAIT system: A gatekeeper of inflammatory gene expression. Trends in Biochemical Sciences, 34(7), 324–331. 10.1016/j.tibs.2009.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, P. D. , & Pogany, J. (2011). The dependence of viral RNA replication on co‐opted host factors. Nature Reviews. Microbiology, 10(2), 137–149. 10.1038/nrmicro2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan, K. , & Makino, S. (2013). Interplay between viruses and host mRNA degradation. Biochimica et Biophysica Acta, 1829(6–7), 732–741. 10.1016/j.bbagrm.2012.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onomoto, K. , Yoneyama, M. , Fung, G. , Kato, H. , & Fujita, T. (2014). Antiviral innate immunity and stress granule responses. Trends in Immunology, 35(9), 420–428. 10.1016/j.it.2014.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiumi, H. , Kouwaki, T. , & Seya, T. (2016). Accessory factors of cytoplasmic viral RNA sensors required for antiviral innate immune response. Frontiers in Immunology, 7, 200. 10.3389/fimmu.2016.00200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiumi, H. , Mifsud, E. J. , & Daito, T. (2016). Links between recognition and degradation of cytoplasmic viral RNA in innate immune response. Reviews in Medical Virology, 26(2), 90–101. 10.1002/rmv.1865 [DOI] [PubMed] [Google Scholar]
- Paeshuyse, J. , Kaul, A. , De Clercq, E. , Rosenwirth, B. , Dumont, J. M. , Scalfaro, P. , … Neyts, J. (2006). The non‐immunosuppressive cyclosporin DEBIO‐025 is a potent inhibitor of hepatitis C virus replication in vitro. Hepatology, 43(4), 761–770. 10.1002/hep.21102 [DOI] [PubMed] [Google Scholar]
- Patino, C. , Haenni, A. L. , & Urcuqui‐Inchima, S. (2015). NF90 isoforms, a new family of cellular proteins involved in viral replication? Biochimie, 108, 20–24. 10.1016/j.biochi.2014.10.022 [DOI] [PubMed] [Google Scholar]
- Pawlica, P. , & Berthoux, L. (2014). Cytoplasmic dynein promotes HIV‐1 uncoating. Viruses, 6(11), 4195–4211. 10.3390/v6114195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley, A. , & Hur, S. (2013). Multi‐level regulation of cellular recognition of viral dsRNA. Cellular and Molecular Life Sciences, 70(11), 1949–1963. 10.1007/s00018-012-1149-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan, A. T. , Kuryavyi, V. , Darnell, J. C. , Serganov, A. , Majumdar, A. , Ilin, S. , … Patel, D. J. (2011). Structure‐function studies of FMRP RGG peptide recognition of an RNA duplex‐quadruplex junction. Nature Structural & Molecular Biology, 18(7), 796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, S. L. , Soderblom, E. J. , Bradrick, S. S. , & Garcia‐Blanco, M. A. (2016). Identification of proteins bound to dengue viral RNA in vivo reveals new host proteins important for virus replication. MBio, 7(1), e01865‐15. 10.1128/mBio.01865-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair, A. , Lassnig, C. , Eberle, C. A. , Gorna, M. W. , Baumann, C. L. , Burkard, T. R. , … Superti‐Furga, G. (2011). IFIT1 is an antiviral protein that recognizes 5′‐triphosphate RNA. Nature Immunology, 12(7), 624–630. 10.1038/ni.2048 [DOI] [PubMed] [Google Scholar]
- Pocock, G. M. , Becker, J. T. , Swanson, C. M. , Ahlquist, P. , & Sherer, N. M. (2016). HIV‐1 and M‐PMV RNA nuclear export elements program viral genomes for distinct cytoplasmic trafficking behaviors. PLoS Pathogens, 12(4), e1005565. 10.1371/journal.ppat.1005565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi, J. D. , Tan, R. , Calnan, B. J. , Frankel, A. D. , & Williamson, J. R. (1992). Conformation of the TAR RNA‐arginine complex by NMR spectroscopy. Science, 257(5066), 76–80. [DOI] [PubMed] [Google Scholar]
- Ramanathan, H. N. , Chung, D. H. , Plane, S. J. , Sztul, E. , Chu, Y. K. , Guttieri, M. C. , … Jonsson, C. B. (2007). Dynein‐dependent transport of the hantaan virus nucleocapsid protein to the endoplasmic reticulum‐Golgi intermediate compartment. Journal of Virology, 81(16), 8634–8647. 10.1128/JVI.00418-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranji, A. , & Boris‐Lawrie, K. (2010). RNA helicases: Emerging roles in viral replication and the host innate response. RNA Biology, 7(6), 775–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel, J. , & Reis e Sousa, C. (2013). Targeting the viral Achilles' heel: Recognition of 5′‐triphosphate RNA in innate anti‐viral defence. Current Opinion in Microbiology, 16(4), 485–492. 10.1016/j.mib.2013.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichman, T. W. , & Mathews, M. B. (2003). RNA binding and intramolecular interactions modulate the regulation of gene expression by nuclear factor 110. RNA, 9(5), 543–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke, L. C. , & Lloyd, R. E. (2013). Diversion of stress granules and P‐bodies during viral infection. Virology, 436(2), 255–267. 10.1016/j.virol.2012.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, A. L. , Soares‐Martins, J. A. , Riddell, G. T. , & Jackson, W. T. (2014). Generation of unique poliovirus RNA replication organelles. MBio, 5(2), e00833‐13 10.1128/mBio.00833-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby, R. E. , & Rehwinkel, J. (2015). RNA degradation in antiviral immunity and autoimmunity. Trends in Immunology, 36(3), 179–188. 10.1016/j.it.2015.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rom, I. , Faicevici, A. , Almog, O. , & Neuman‐Silberberg, F. S. (2007). Drosophila dynein light chain (DDLC1) binds to gurken mRNA and is required for its localization. Biochimica et Biophysica Acta, 1773(10), 1526–1533. 10.1016/j.bbamcr.2007.05.005 [DOI] [PubMed] [Google Scholar]
- Samuel, C. E. (2011). Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral. Virology, 411(2), 180–193. 10.1016/j.virol.2010.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan, R. , & Domingo‐Calap, P. (2016). Mechanisms of viral mutation. Cellular and Molecular Life Sciences, 73(23), 4433–4448. 10.1007/s00018-016-2299-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savidis, G. , McDougall, W. M. , Meraner, P. , Perreira, J. M. , Portmann, J. M. , Trincucci, G. , … Brass, A. L. (2016). Identification of Zika virus and dengue virus dependency factors using functional genomics. Cell Reports, 16(1), 232–246. 10.1016/j.celrep.2016.06.028 [DOI] [PubMed] [Google Scholar]
- Schmidt, T. , Friedrich, S. , Golbik, R. P. , & Behrens, S. E. (2017). NF90‐NF45 is a selective RNA chaperone that rearranges viral and cellular riboswitches: Biochemical analysis of a virus host factor activity. Nucleic Acids Research, 45(21), 12441–12454. 10.1093/nar/gkx931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, T. , Knick, P. , Lilie, H. , Friedrich, S. , Golbik, R. P. , & Behrens, S. E. (2016). Coordinated action of two double‐stranded RNA binding motifs and an RGG motif enables nuclear factor 90 to flexibly target different RNA substrates. Biochemistry, 55(6), 948–959. 10.1021/acs.biochem.5b01072 [DOI] [PubMed] [Google Scholar]
- Schmidt, T. , Knick, P. , Lilie, H. , Friedrich, S. , Golbik, R. P. , & Behrens, S. E. (2017). The properties of the RNA‐binding protein NF90 are considerably modulated by complex formation with NF45. The Biochemical Journal, 474(2), 259–280. 10.1042/BCJ20160790 [DOI] [PubMed] [Google Scholar]
- Schor, S. , & Einav, S. (2018). Repurposing of kinase inhibitors as broad‐spectrum antiviral drugs. DNA and Cell Biology, 37(2), 63–69. 10.1089/dna.2017.4033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrom, E. M. , Moschall, R. , Schuch, A. , & Bodem, J. (2013). Regulation of retroviral polyadenylation. Advances in Virus Research, 85, 1–24. 10.1016/B978-0-12-408116-1.00001-X [DOI] [PubMed] [Google Scholar]
- Shen, Y. , Li, N. L. , Wang, J. , Liu, B. , Lester, S. , & Li, K. (2012). TRIM56 is an essential component of the TLR3 antiviral signaling pathway. The Journal of Biological Chemistry, 287(43), 36404–36413. 10.1074/jbc.M112.397075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina, N. , & Nakayama, K. (2014). RNA granule assembly and disassembly modulated by nuclear factor associated with double‐stranded RNA 2 and nuclear factor 45. The Journal of Biological Chemistry, 289(30), 21163–21180. 10.1074/jbc.M114.556365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava, N. , Sripada, S. , Kaur, J. , Shah, P. S. , & Cecilia, D. (2011). Insights into the internalization and retrograde trafficking of dengue 2 virus in BHK‐21 cells. PLoS One, 6(10), e25229. 10.1371/journal.pone.0025229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, R. W. , & Gray, N. K. (2010). Poly(A)‐binding protein (PABP): A common viral target. The Biochemical Journal, 426(1), 1–12. 10.1042/BJ20091571 [DOI] [PubMed] [Google Scholar]
- Stake, M. S. , Bann, D. V. , Kaddis, R. J. , & Parent, L. J. (2013). Nuclear trafficking of retroviral RNAs and Gag proteins during late steps of replication. Viruses, 5(11), 2767–2795. 10.3390/v5112767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steimer, L. , & Klostermeier, D. (2012). RNA helicases in infection and disease. RNA Biology, 9(6), 751–771. 10.4161/rna.20090 [DOI] [PubMed] [Google Scholar]
- Stoltzfus, C. M. , & Madsen, J. M. (2006). Role of viral splicing elements and cellular RNA binding proteins in regulation of HIV‐1 alternative RNA splicing. Current HIV Research, 4(1), 43–55. [DOI] [PubMed] [Google Scholar]
- Tazi, J. , Bakkour, N. , Marchand, V. , Ayadi, L. , Aboufirassi, A. , & Branlant, C. (2010). Alternative splicing: Regulation of HIV‐1 multiplication as a target for therapeutic action. The FEBS Journal, 277(4), 867–876. 10.1111/j.1742-4658.2009.07522.x [DOI] [PubMed] [Google Scholar]
- Tekotte, H. , & Davis, I. (2002). Intracellular mRNA localization: Motors move messages. Trends in Genetics, 18(12), 636–642. [DOI] [PubMed] [Google Scholar]
- Todorova, T. , Bock, F. J. , & Chang, P. (2015). Poly(ADP‐ribose) polymerase‐13 and RNA regulation in immunity and cancer. Trends in Molecular Medicine, 21(6), 373–384. 10.1016/j.molmed.2015.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida, T. , Zou, J. , Saitoh, T. , Kumar, H. , Abe, T. , Matsuura, Y. , … Akira, S. (2010). The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double‐stranded DNA. Immunity, 33(5), 765–776. 10.1016/j.immuni.2010.10.013 [DOI] [PubMed] [Google Scholar]
- Valuev‐Elliston, V. T. , & Kochetkov, S. N. (2017). Novel HIV‐1 non‐nucleoside reverse transcriptase inhibitors: A combinatorial approach. Biochemistry (Moscow), 82(13), 1716–1743. 10.1134/S0006297917130107 [DOI] [PubMed] [Google Scholar]
- van Tol, S. , Hage, A. , Giraldo, M. I. , Bharaj, P. , & Rajsbaum, R. (2017). The TRIMendous role of TRIMs in virus‐host interactions. Vaccines (Basel), 5(3), 23. 10.3390/vaccines5030023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventoso, I. , Sanz, M. A. , Molina, S. , Berlanga, J. J. , Carrasco, L. , & Esteban, M. (2006). Translational resistance of late alphavirus mRNA to eIF2alpha phosphorylation: A strategy to overcome the antiviral effect of protein kinase PKR. Genes & Development, 20(1), 87–100. 10.1101/gad.357006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg, G. A. , Rajsbaum, R. , Sanchez‐Aparicio, M. T. , Maestre, A. M. , Valdiviezo, J. , Shi, M. , … Garcia‐Sastre, A. (2013). The E3‐ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern‐recognition receptors. Immunity, 38(2), 384–398. 10.1016/j.immuni.2012.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viktorovskaya, O. V. , Greco, T. M. , Cristea, I. M. , & Thompson, S. R. (2016). Identification of RNA binding proteins associated with dengue virus RNA in infected cells reveals temporally distinct host factor requirements. PLoS Neglected Tropical Diseases, 10(8), e0004921. 10.1371/journal.pntd.0004921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladimer, G. I. , Gorna, M. W. , & Superti‐Furga, G. (2014). IFITs: Emerging roles as key anti‐viral proteins. Frontiers in Immunology, 5, 94. 10.3389/fimmu.2014.00094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, D. , & Mohr, I. (2011). Viral subversion of the host protein synthesis machinery. Nature Reviews. Microbiology, 9(12), 860–875. 10.1038/nrmicro2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Liu, B. , Wang, N. , Lee, Y. M. , Liu, C. , & Li, K. (2011). TRIM56 is a virus‐ and interferon‐inducible E3 ubiquitin ligase that restricts pestivirus infection. Journal of Virology, 85(8), 3733–3745. 10.1128/JVI.02546-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R. Y. , & Li, K. (2012). Host factors in the replication of positive‐strand RNA viruses. Chang Gung Medical Journal, 35(2), 111–124. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Jin, F. , Wang, R. , Li, F. , Wu, Y. , Kitazato, K. , & Wang, Y. (2017). HSP90: A promising broad‐spectrum antiviral drug target. Archives of Virology, 162(11), 3269–3282. 10.1007/s00705-017-3511-1 [DOI] [PubMed] [Google Scholar]
- Wen, X. , Huang, X. , Mok, B. W. , Chen, Y. , Zheng, M. , Lau, S. Y. , … Chen, H. (2014). NF90 exerts antiviral activity through regulation of PKR phosphorylation and stress granules in infected cells. Journal of Immunology, 192(8), 3753–3764. 10.4049/jimmunol.1302813 [DOI] [PubMed] [Google Scholar]
- Wheeler, E. C. , Van Nostrand, E. L. , & Yeo, G. W. (2018). Advances and challenges in the detection of transcriptome‐wide protein‐RNA interactions. WIREs RNA, 9(1). 10.1002/wrna.1436, e1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmund, F. , del Alamo, M. , Pechmann, S. , Chen, T. , Albanese, V. , Dammer, E. B. , … Frydman, J. (2013). The cotranslational function of ribosome‐associated Hsp70 in eukaryotic protein homeostasis. Cell, 152(1–2), 196–209. 10.1016/j.cell.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer, P. , & Schreiner, S. (2015). Viral mimicry to usurp ubiquitin and SUMO host pathways. Viruses, 7(9), 4854–4872. 10.3390/v7092849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkowicz, U. M. , & Cook, A. G. (2012). NF45 dimerizes with NF90, Zfr and SPNR via a conserved domain that has a nucleotidyltransferase fold. Nucleic Acids Research, 40(18), 9356–9368. 10.1093/nar/gks696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, B. , Blocquel, D. , Habchi, J. , Uversky, A. V. , Kurgan, L. , Uversky, V. N. , & Longhi, S. (2014). Structural disorder in viral proteins. Chemical Reviews, 114(13), 6880–6911. 10.1021/cr4005692 [DOI] [PubMed] [Google Scholar]
- Xue, B. , Mizianty, M. J. , Kurgan, L. , & Uversky, V. N. (2012). Protein intrinsic disorder as a flexible armor and a weapon of HIV‐1. Cellular and Molecular Life Sciences, 69(8), 1211–1259. 10.1007/s00018-011-0859-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. , Liang, H. , Zhou, Q. , Li, Y. , Chen, H. , Ye, W. , … Liu, Y. (2012). Crystal structure of ISG54 reveals a novel RNA binding structure and potential functional mechanisms. Cell Research, 22(9), 1328–1338. 10.1038/cr.2012.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama, M. , Jogi, M. , & Onomoto, K. (2016). Regulation of antiviral innate immune signaling by stress‐induced RNA granules. Journal of Biochemistry, 159(3), 279–286. 10.1093/jb/mvv122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, R. , Miner, J. J. , Gorman, M. J. , Rausch, K. , Ramage, H. , White, J. P. , … Diamond, M. S. (2016). A CRISPR screen defines a signal peptide processing pathway required by flaviviruses. Nature, 535(7610), 164–168. 10.1038/nature18625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, D. , Mei, Q. , Li, J. , & He, H. (2012). Cyclophilin A and viral infections. Biochemical and Biophysical Research Communications, 424(4), 647–650. 10.1016/j.bbrc.2012.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y. , He, C. , Wang, L. , & Ge, B. (2017). Post‐translational regulation of antiviral innate signaling. European Journal of Immunology, 47(9), 1414–1426. 10.1002/eji.201746959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J. D. , Meng, W. , Wang, X. J. , & Wang, H. C. (2015). Broad‐spectrum antiviral agents. Frontiers in Microbiology, 6, 517. 10.3389/fmicb.2015.00517 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 RNA‐binding domains (RBDs) of virus‐related RNA‐binding proteins. RBDmap profiles of CAPRIN1 (a), EIF3D (b), EIF3I (c), and EIF3G (d). Boxes represent high confidence (red, 1% FDR) or relevant candidate (orange, 10% FDR) RBDs. White boxes symbolize Pfam‐annotated domains. Green lines indicate predicted disordered regions (IUPred score > 0.4). FDR, false discovery rate.
Figure S2 RNA‐binding domains (RBDs) of virus‐related RNA‐binding proteins. RBDmap profiles of DYNC1H1 C‐terminal (a), DYNC1I2 (c), DYNC1LI1 (d), and DCTN2 (e). DYNC1H1 Pfam domain architecture (b). Boxes represent high confidence (red, 1% FDR) or relevant candidate (orange, 10% FDR) RBDs. White boxes symbolize Pfam‐annotated domains. Green lines indicate predicted disordered regions (IUPred score > 0.4). FDR, false discovery rate.
File S1 Description of how the virus‐linked RNA interactome was constructed.
Table S1 Virus‐linked RNA interactome, GO terms used in its construction, and domains used in classification of RNA‐binding proteins (RBPs). A particular virus‐linked RBP would be missing if (a) it has not been identified by RNA‐IC due to its low abundance, low crosslink ability, incompatibility with mass spectrometry or because RNA‐IC studies have only been performed in steady‐state conditions; (b) it is not (yet) annotated by GO terms related to viruses, infection, or immunity; or/and (c) it appears in fewer than six publication in PubMed linked to the term “virus.”


