Abstract
The N6-methyladenosine (m6A) modification regulates mRNA stability and translation. Here, we show that transcriptomic m6A modification can be dynamic and the m6A reader protein YTH N6-methyladenosine RNA binding protein 2 (YTHDF2) promotes mRNA decay during cell cycle. Depletion of YTHDF2 in HeLa cells leads to the delay of mitotic entry due to overaccumulation of negative regulators of cell cycle such as Wee1-like protein kinase (WEE1). We demonstrate that WEE1 transcripts contain m6A modification, which promotes their decay through YTHDF2. Moreover, we found that YTHDF2 protein stability is dependent on cyclin-dependent kinase 1 (CDK1) activity. Thus, CDK1, YTHDF2, and WEE1 form a feedforward regulatory loop to promote mitotic entry. We further identified Cullin 1 (CUL1), Cullin 4A (CUL4A), damaged DNA-binding protein 1 (DDB1), and S-phase kinase-associated protein 2 (SKP2) as components of E3 ubiquitin ligase complexes that mediate YTHDF2 proteolysis. Our study provides insights into how cell cycle mediators modulate transcriptomic m6A modification, which in turn regulates the cell cycle.
Introduction
Methyltransferase Like 3 (METTL3) and METTL14 [1]—together with several key components, such as Wilms tumor 1-associated protein (WTAP) [2], Vir Like m6A Methyltransferase Associated (VIRMA) [3], and Zinc Finger CCCH-Type Containing 13 (ZC3H13) [4]—form a methyltransferase complex to mediate N6-methyladenosine (m6A) modification on mRNAs in a co-transcriptional manner [5,6]. m6A, the most abundant modification on mRNAs, plays multifaceted roles in regulating pre-mRNA processing [7,8], nuclear export [9], stability [10], translation [11], and other biochemical properties [12,13] of mRNA in eukaryotes. The myriad roles of the m6A modification mostly rely on downstream RNA-binding proteins, known as m6A “readers,” that preferentially recognize m6A-modified RNAs [9–11,14,15].
YTH N6-methyladenosine RNA binding protein 2 (YTHDF2) was the first functionally verified m6A reader to promote the degradation of m6A-modified mRNAs in humans [10]. It accelerates the decay of m6A-marked transcripts by directly recruiting the Carbon Catabolite Repressor 4-Negative on TATA (CCR4-NOT) deadenylase complex [16] or the endoribonucleolytic RNase P/Mitochondrial RNA Processing (MRP) complex [17]. In zebrafish, Ythdf2 promotes the clearance of the maternal transcripts during maternal to zygotic transition (MZT) [18]. Loss of Ythdf2 in zebrafish delays MZT initiation and impedes zygotic genome activation [18]. Likewise, Ythdf2 was confirmed maternally essential for early zygotic development in mice [19], and Ythdf2 depletion also negatively impacts the proliferation and differentiation of neural progenitor cells in mouse [20], suggesting broad impact of Ythdf2 on mRNA degradation in different tissues and cell types in vertebrates. The significant role of YTHDF2 in cell differentiation was further shown with depletion of YTHDF2-blocking differentiation and driving self-renewal of both mouse and human hematopoietic stem cells [21,22]. This protein is also critical to acute myeloid leukemia (AML) [23].
The m6A pathway has been implicated in regulating genes related to cell cycle. It was reported that transcripts involved in cell cycle regulation contain m6A modification in mouse embryonic stem cells (mESC) [24]. Down-regulation of METTL3 in AML causes cell cycle arrest because of down-regulation of genes in the cell cycle pathway [25]. AlkB Homolog 5 (ALKBH5), an m6A demethylase, facilitates glioblastoma proliferation by demethylating the transcripts of forkhead box protein M1 (FOXM1), an important cell cycle regulator [26]. METTL14 was shown to be essential for mammalian cortical neurogenesis by facilitating the decay of a few categories of transcripts, including those regulating the cell cycle [27]. These studies suggest that the m6A pathway potentially plays a role in cell cycle regulation; however, mechanistic studies are still limited, and the impact of m6A readers on cell cycle remains unexplored. Here, we investigated changes in transcriptomic m6A during cell cycle and demonstrated a role of YTHDF2 in regulating phase-specific transcript degradation. Furthermore, we identified the proteolysis pathway of YTHDF2, which establishes a mechanistic interconnection between the m6A pathway and cell cycle.
Results
Dynamic changes of transcriptomic m6A during cell cycle
Cells go through dynamic transcriptome changes during cell cycle, especially during G1 and S phases [28]. We hypothesized that m6A modification may also change during cell cycle because m6A on mRNAs are deposited co-transcriptionally [5,6] and are known to affect cell cycle. To understand how transcriptomic m6A changes during cell cycle, we synchronized HeLa cells to the stage of G1/S transition by double thymidine block and collected cells for m6A methylated RNA immunoprecipitation sequencing (MeRIP-seq) from 0, 4, and 8 hours post release, which correspond to G1/S, S, and G2/M phases, respectively (S1A Fig) (S1 Table). Motif search of m6A peaks all identified motifs containing the “GGAC” core motif (Fig 1A). m6A peaks predominantly reside on the 3′ untranslated region (UTR) of transcripts at all phases (Fig 1B), consistent with previous reports [29,30]. Gene ontology (GO) enrichment analysis of the m6A-marked common genes at 3 phases identified a number of GO terms, including “cell cycle” (S1B Fig), suggesting that the m6A pathway regulates genes involved in cell cycle control, consistent with a previous study in mESC [24].
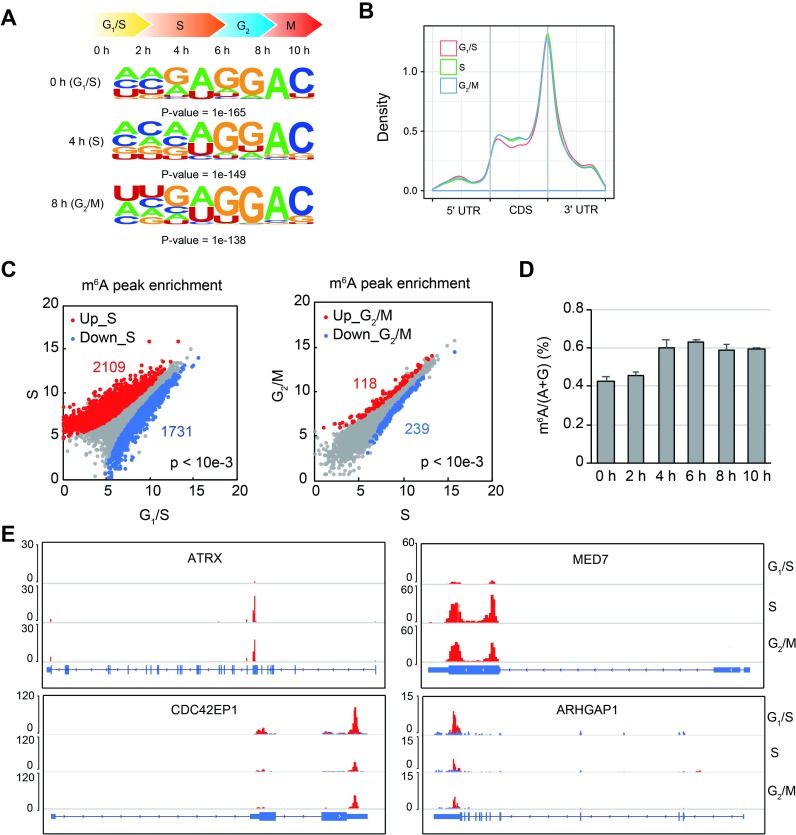
Fig 1. Dynamic transcriptomic m6A changes during cell cycle.
(A) Enriched RNA motifs from the m6A MeRIP-seq data from cells at different phases of cell cycle. (B) Metagene analysis of m6A peak distribution on mRNAs. (C) Differential m6A enrichment analysis across G1/S, S, and G2/M phases of cell cycle. (D) The m6A levels of mRNA quantified by LC-MS/MS at different time points post G1/S synchronization. y-Axis indicates the ratio of m6A to the sum of A and G. (E) Examples of genes with dynamic m6A modification on mRNAs. ATRX and MED7 m6A peak enrichment increases at S phase compared with G1/S and are related to DNA duplex unwinding and Pol II transcription, respectively. m6A peak enrichment of CDC42EP1 and ARHGAP1 reduces at S phase, which are related to Rho protein signaling or GTPase activity. Blue and red bars indicate the input and IP read coverages, respectively. Underlying data for this figure can be found in S1 Data. ARHGAP1, Rho GTPase Activating Protein 1; ATRX, alpha thalassemia/mental retardation syndrome X-linked; CDC42EP1, CDC42 Effector Protein 1; CDS, coding sequence; IP, immunoprecipitation; LC-MS/MS, liquid chromatography–tandem mass spectrometry; MED7, Mediator Complex Subunit 7; m6A, N6-methyladenosine; MeRIP-seq, methylated RNA immunoprecipitation sequencing; Pol II, RNA polymerase II; UTR, untranslated region.
Differential enrichment analysis of m6A peaks was performed to assess the changes in m6A levels with progression across the 3 cell phases. We found that 2,109 peaks from 1,489 genes showed significantly higher enrichment from G1/S to late S phase, while 1,731 peaks from 1,416 genes exhibited reduced enrichment (Fig 1C). In contrast, only 118 and 239 peaks, corresponding to 112 and 210 genes, displayed differential changes from late S to G2/M phase (Fig 1C) (S2 Table). Correlation analysis of these samples showed a higher correlation between late S and G2/M (S1C Fig). These results suggest that transcriptomic m6A is dynamic through G1 and S phase while remaining relatively steady from late S to G2/M phase.
We collected cells at more time points after synchronization and measured m6A levels of mRNA by mass spectrometry. We found that m6A level increased from G1/S (0 hours) to S (4 hours) phase and remained steady thereafter (Fig 1D). This observation is consistent with the transcriptomic m6A changes during cell cycle (Fig 1C). To test whether the observed m6A changes were caused by potential changes of methyltransferase or demethylases, we measured the protein levels of METTL3 and METTL14, members of the core m6A methyltransferase complex, and fat mass and obesity-associated protein (FTO), an m6A demethylase by western blot. We found that METTL3 showed lower protein level at early G1/S (0 h) phase and M (10 h) phase, while METTL14 only decreased at M phase (S1D Fig). FTO remained relatively steady during cell cycle (S1D Fig), indicating that the methyltransferase activity—instead of the demethylase activity—might affect m6A levels during cell cycle. In contrast, RNA sequencing (RNA-seq) data (see subsequently) revealed that the transcript levels of METTL3, METTL14, and FTO all remained relatively steady (S1E Fig), suggesting possible translational or post-translational regulation for METTL3 and METTL14 during cell cycle.
Correlation analysis of gene expression level and m6A enrichment revealed that the overall gene expression showed a weak negative correlation with m6A peak enrichment (S2A Fig), consistent with previous studies [31,32]. To investigate whether gene expression affects m6A modification, we next specifically analyzed genes with differential m6A enrichment during cell cycle phase transitions. We found that genes with higher m6A enrichment in the next phase tend to have significantly higher expression levels; in contrast, genes with reduced m6A modifications concurrently showed lower expression (S2B Fig). These results reveal tight correlation between transcriptional regulation and m6A modifications during phase transitions, when dynamic transcriptomic changes occur during cell cycle.
To understand the biological pathways undergoing differential m6A changes, we performed GO enrichment analysis for the genes exhibiting m6A dynamics. Genes such as ATRX and MED7 with increased m6A from G1/S to late S phase (Fig 1E) were mostly classified as GO terms related to “DNA duplex unwinding” and “regulation of transcription” (S2C Fig), which are typical S phase events [33]. In contrast, terms such as “Rho protein signal” and “GTPase activity” (S2D Fig), which are essential for G1 phase [34], showed enrichment for genes with decreased m6A enrichment from G1/S to late S phase, such as CDC42EP1 and ARHGAP1 (Fig 1E). At the G2/M phase, “microtubule-based process” showed higher m6A enrichment, which is a hallmark of mitosis (S2E Fig) [35]. In addition, genes related to transcription and response to DNA damage, which are active at S phase [33], exhibited a reduction of m6A on transcripts (S2F Fig). Collectively, m6A profiling at different phases revealed that transcriptomic m6A is actively regulated during cell cycle, indicating that m6A modification of the phase-specific transcripts might be important for cell cycle progression in HeLa cells that we examined.
YTHDF2 depletion elevates phase-specific transcripts
During cell cycle, the expression of hundreds of genes dynamically fluctuates along with phase transitions in both human and yeast [36–38]. Like transcription, mRNA decay is also dynamic during cell cycle [39]. However, the mechanism underlying mRNA decay remains elusive. YTHDF2 plays important roles in transcript turnover during animal development [18–20]. We thus hypothesized that YTHDF2 facilitates the timely turnover of phase-specific transcripts to ensure cell cycle progression. To test this, we generated YTHDF2 knockout HeLa cell lines by using CRISPR-Cas9 (Fig 2A and S3A Fig). We found that all the YTHDF2 knockout lines showed slower proliferation compared with the wild type (Fig 2B) and that the proliferation defect can be restored by YTHDF2 transfection (S3B Fig). YTHDF2 knockdown by small interfering RNA (siRNA) displayed consistent slower proliferation (S3C Fig), indicating that YTHDF2 promotes cell proliferation possibly by facilitating mRNA degradation during cell cycle.
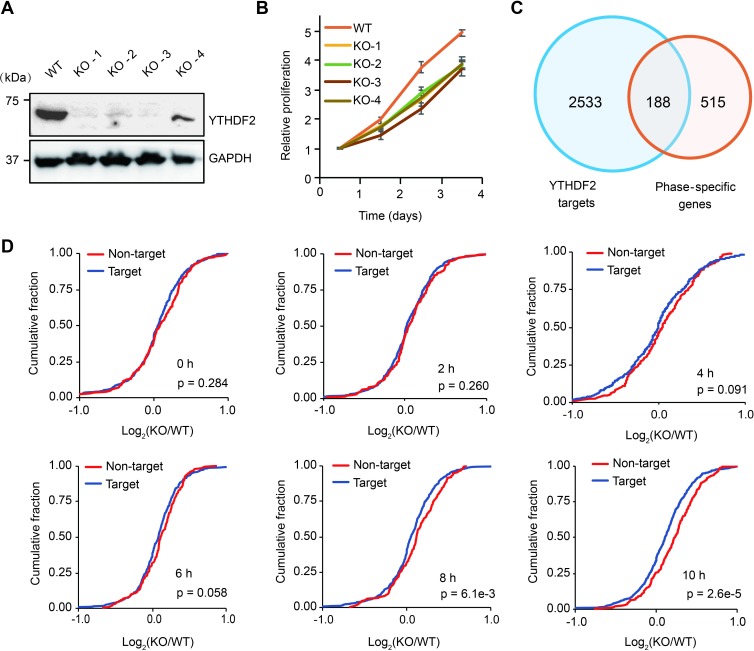
Fig 2. YTHDF2 depletion delays cell proliferation and elevates target transcripts.
(A) Western blot of YTHDF2 in CRISPR-Cas9 KO cell lines. (B) Cell proliferation assays of WT and YTHDF2 KO cell lines. (C) Intersection of genes that are confident YTHDF2 targets and the ones that are phase specific. (D) Cumulative distribution of phase-specific transcripts that are YTHDF2 bound (188 genes, in red) and unbound (515 genes, in blue) by comparing WT and KO cell lines. The x-axes indicate the log2 fold change of gene expression in KO versus WT. P values were calculated using the Mann-Whitney test. Underlying data for this figure can be found in S1 Raw Images and S1 Data. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; KO, knockout; WT, wild type; YTHDF2, YTH N6-methyladenosine RNA binding protein 2.
To further test this hypothesis, we attempted to obtain a more confident set of YTHDF2-bound transcripts by performing YTHDF2 RNA immunoprecipitation (RIP) using unsynchronized cells followed by sequencing. Based on the YTHDF2 RIP sequencing (RIP-seq) data and the published YTHDF2 photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) data [10], we identified a total of 2,701 common genes as confident YTHDF2 targets (S4A Fig) (S3 Table), 1,975 (73.1%) of which were detected with significant m6A enrichment. To compare the expression changes of YTHDF2 target and nontarget genes, wild-type and YTHDF2 knockout cells were collected for RNA-seq at 0, 2, 4, 6, 8, and 10 hours post release from G1/S synchronization. Cumulative fractions of gene expression changes between knockout and wild-type cells showed that the 2,701 YTHDF2 targets displayed significantly higher accumulation from 2 to 10 hours post release in the absence of YTHDF2, compared with the 4,668 nontargets (S4B Fig). These results confirm that YTHDF2 generally plays a role in facilitating mRNA degradation during cell cycle.
A previous study reported a group of genes that express in phase-specific patterns in HeLa cells [36]. We thus examined how these genes are impacted upon YTHDF2 depletion. Among the phase-specific transcripts, 188 are confident YTHDF2 targets (Fig 2C), the majority (80.3%) of which were detected with significant m6A enrichment. We found that the 188 phase-specific YTHDF2 targets displayed significantly higher accumulation at 8 and 10 hours post release in the absence of YTHDF2, compared with the 515 nontargets (Fig 2D) (S4 Table). These results suggest that YTHDF2 binds to a subset of phase-specific transcripts to promote their degradation throughout the late G2 phase and mitosis.
YTHDF2 promotes mitotic entry by negatively regulating Wee1-like protein kinase
We further investigated whether YTHDF2 generally promotes all the phases of the cell cycle or a specific phase by synchronizing both wild-type and YTHDF2 knockout lines to G1/S phase, followed by cytometry analysis at different time points post release. YTHDF2 knockout cells show marked delay at 8 hours post release, corresponding to G2/M transition (Fig 3A), confirming that YTHDF2 plays a role in promoting mitotic entry.
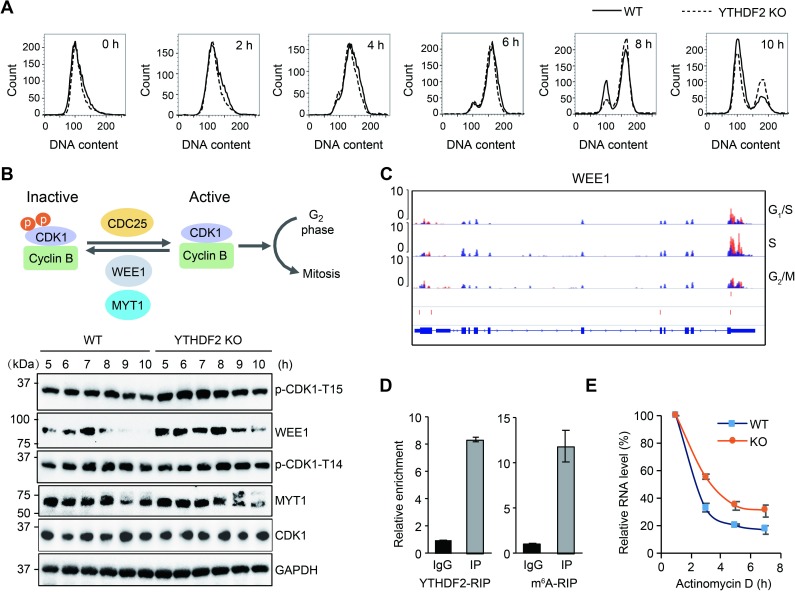
Fig 3. WEE1 is negatively regulated by YTHDF2.
(A) Flow cytometry analysis of wild-type and YTHDF2 knockout cells that were synchronized and released from G1/S phase using double thymidine block. (B) Western blot analysis of key proteins that determine mitosis entry. The upper panel illustrates the key regulators of mitotic entry. The lower panel indicates western blot results from wild-type and YTHDF2 knockout samples that were synchronized to G1/S phase using double thymidine block. Samples were collected at times as indicated in the figure post release. (C) Genome browser view of WEE1 m6A peaks at different phases of the cell cycle. The red bars in lower 2 tracks represent the m6A sites identified by 2 replicates of YTHDF2 PAR-CLIP data [10]. (D) YTHDF2 and m6A RIP-qPCR for WEE1. Rabbit IgG was used as control. (E) WEE1 mRNA decay assay using actinomycin D treatment. The relative levels of WEE1 transcripts were normalized to the housekeeping gene HPRT1. Underlying data for this figure can be found in S1 Data. CDC25, cell division cycle 25; CDK1, cyclin-dependent kinase 1; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IgG, immunoglobulin G; IP, immunoprecipitation; KO, knockout; m6A, N6-methyladenosine; MYT1, membrane associated tyrosine/threonine 1; p-CDK1-T15, phosphorylating CDK1 at threonine 14; PAR-CLIP, photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation; qPCR, quantitative PCR; RIP, RNA immunoprecipitation; WEE1, Wee1-like protein kinase; WT, wild type; YTHDF2, YTH N6-methyladenosine RNA binding protein 2.
We next explored the underlying mechanism of mitotic entry regulation mediated by YTHDF2 by assessing protein levels of genes that are important for G2/M transition or mitosis from 5 to 10 hours after G1/S release by western blots. Mitotic entry is predominantly determined by the phosphorylation status of cyclin-dependent kinase 1 (CDK1), which is controlled by phosphorylation activities of Wee1-like protein kinase (WEE1) and membrane associated tyrosine/threonine 1 (MYT1), and dephosphorylation by cell division cycle 25 (CDC25) [40]. We found that the protein level of CDK1 remained steady across the period in both wild-type and knockout cells (Fig 3B). Interestingly, WEE1—but not MYT1—showed dramatic overaccumulation, suggesting that WEE1 is negatively regulated by YTHDF2 (Fig 3B and S5A Fig). WEE1 and MYT1 are negative regulators of CDK1 by phosphorylating CDK1 at tyrosine 15 (p-CDK1-Y15) and threonine 14 (p-CDK1-T14), respectively [41,42]. Consistent with the higher levels of WEE1 in YTHDF2 knockout cells, p-CDK1-Y15—but not p-CDK1-T14—is accordingly excessive in the knockout cells (Fig 3B and S5A Fig). RNA-seq data also confirmed higher expression of WEE1 after 4 hours post synchronization, corresponding to G2 to M phase (S5B Fig), consistent with the protein level change (S5C Fig). Moreover, overexpression of WEE1 resulted in slower cell proliferation and higher ratio of cells at G2/M phase, suggesting delay of mitotic entry (S5D Fig). These results confirm that YTHDF2 represses the key negative regulator WEE1 from G2 to M phase to maintain CDK1 activity.
From the m6A MeRIP-seq and PAR-CLIP data, we found that WEE1 transcripts indeed contain m6A modification mainly at the 3′ UTR region (Fig 3C), suggesting that WEE1 is regulated at the epitranscriptomic level through YTHDF2. RIP using either m6A or YTHDF2 antibody followed by reverse transcription quantitative PCR (RT-qPCR) confirmed that WEE1 transcript is indeed methylated and bound by YTHDF2 (Fig 3D). RNA decay assay using actinomycin D–mediated transcription inhibition showed that WEE1 transcripts degrade slower in the absence of YTHDF2, demonstrating that YTHDF2 shortens the lifetime of WEE1 transcripts (Fig 3E). Moreover, knockdown of YTHDF2 or METTL3 using siRNAs consistently increased both WEE1 mRNA and protein levels, which led to an overaccumulation of p-CDK1-Y15 (S5E Fig), suggesting that m6A modification of the WEE1 transcript predominantly promotes its degradation through YTHDF2. Knockdown of YTHDF2 also resulted in higher METTL3 transcript accumulation, but not the protein level (S5E Fig), implicating other mechanisms as regulators of METTL3. Moreover, regardless of the effect on WEE1, knockdown of METTL3 resulted in marked delay from G1 to S phase (S5F Fig), suggesting more complex regulatory pathways downstream of METTL3 that regulate cell cycle. Collectively, these results show that YTHDF2 mediates the decay of WEE1 transcripts to ensure timely mitotic entry.
YTHDF2 stability is maintained by CDK1
We assessed YTHDF2 levels in synchronized cells and found substantially higher accumulation of YTHDF2 protein from late S to G2/M phases (Fig 4A), but not the YTHDF2 transcript expression (S6A Fig), indicating that cell cycle controls YTHDF2 at the protein level. To further verify this, we tested a few small molecule inhibitors for cyclin-dependent kinases in cell culture. We found that YTHDF2 is quickly degraded when HeLa cells are incubated with the selective CDK1 inhibitor RO-3306 [43], but not CDK2 Inhibitor III, a selective CDK2 inhibitor [44], or the general kinase inhibitor dinaciclib [45] (Fig 4B). The same effect of RO-3306 was also observed for human embryonic kidney 293T (HEK 293T) cells (S6B Fig), indicating a general effect of CDK1 inhibition on YTHDF2 degradation. To rule out the off-target effect of RO-3306, we tested a different selective CDK1 inhibitor, purvalanol A [46], and found the same effect on YTHDF2 in HeLa cells (S6C Fig). In contrast, any other kinase inhibitors—such as rapamycin and Torin1 for mammalian target of rapamycin (mTOR) [47] and GDC-0994 and SCH772984 for extracellular signal-regulated kinase (ERK) [48]—did not cause YTHDF2 degradation (S6D Fig), suggesting that YTHDF2 degradation is specifically activated by CDK1 inhibition.
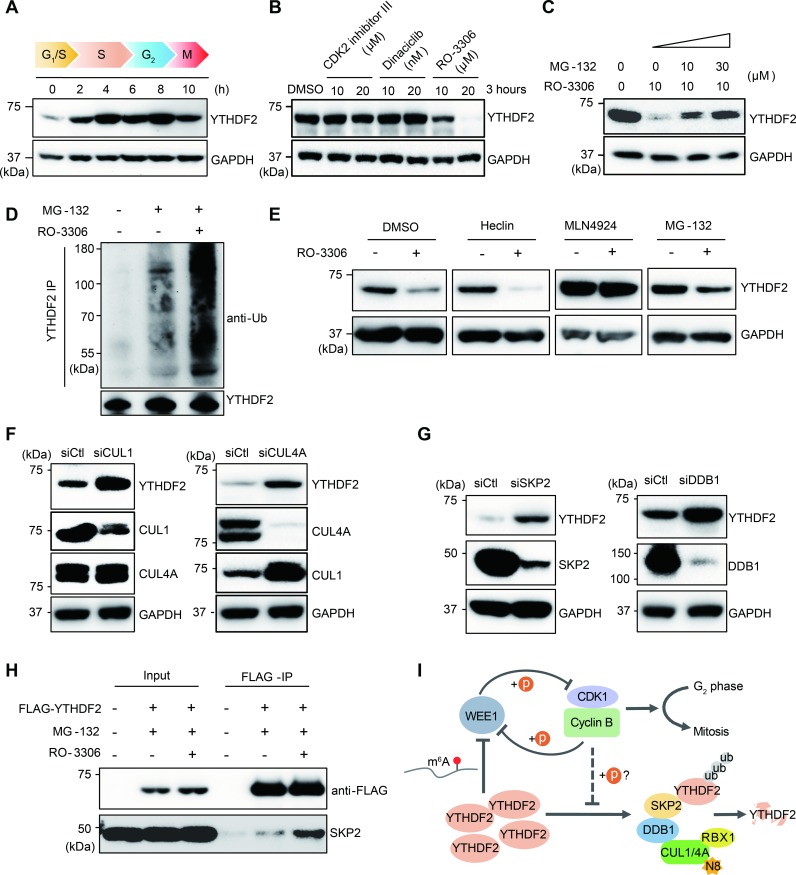
Fig 4. YTHDF2 stability is dependent on CDK1 activity.
(A) Western blot of YTHDF2 post G1/S synchronization. Samples were collected at the indicated hours post release. (B) Western blot of YTHDF2 using cells treated with small molecule inhibitors. Cells were treated with CDK2 inhibitor III, dinaciclib, and the CDK1 inhibitor RO-3306 in different concentrations as indicated for 3 hours. (C) Western blot of YTHDF2 using cells pretreated with the proteasome inhibitor MG-132 before the CDK1 inhibitor RO-3306 treatment. Cells were incubated with MG-132 with indicated concentrations for 3 hours before RO-3306 treatment. (D) YTHDF2 IP followed by western blot with ubiquitin antibody and YTHDF2 antibody. Cells were pretreated with MG-132 3 hours before being incubated for 3 hours with RO-3306. (E) Small molecular inhibitor assays to identify the proteolysis pathway for YTHDF2. Cells were pretreated with indicated inhibitors 3 hours before treating with RO-3306 for 3 hours for western blots. (F–G) siRNA knockdown of CUL1, CUL4A, SKP2, and DDB1, followed by western blot for YTHDF2, CUL1, CUL4A, SKP2, and DDB1. Control siRNA knockdown is indicated as “siCtl.” (H) FLAG-IP using cells sequentially treated with MG-132 and RO-3306 as indicated. Levels of FLAG-YTHDF2 and SKP2 were detected by western blots with anti-FLAG and anti-SKP2 antibodies, respectively. (I) A proposed model of feedforward loop regulating mitosis entry involving CDK1, YTHDF2, and WEE1. WEE1 and CDK1 reciprocally repress each other by phosphorylation to regulate mitotic entry. CDK1 stabilizes YTHDF2 by preventing YTHDF2 from associating with the E3 ligase complexes for degradation. Whether this relies on direct phosphorylation by CDK1 remains to be explored (labeled with question mark). YTHDF2 represses WEE1 by promoting the degradation of WEE1 transcripts via m6A. YTHDF2 is degraded after polyubiquitination by the E3 ligase complexes. Underlying data for this figure can be found in S1 Raw Images. anti-Ub, ubiquitin antibody; CDK1, cyclin-dependent kinase 1; CUL1, Cullin 1; DDB1, damaged DNA-binding protein 1; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IP, immunoprecipitation; m6A, N6-methyladenosine; RBX1, RING-box protein 1; siCtl, control siRNA; siRNA, small interfering RNA; SKP2, S-phase kinase-associated protein 2; WEE1, Wee1-like protein kinase; YTHDF2, YTH N6-methyladenosine RNA binding protein 2.
Recent studies have shown that loss of YTHDF2 results in enhancement of hematopoietic stem cell expansion in both mouse and human [21,22]. In contrast, depletion of YTHDF2 in AML resulted in apoptosis of AML cells [23]. Thus, YTHDF2 represents a potential selective target for AML treatment. We tested whether CDK1 inhibition also causes YTHDF2 degradation in AMLs to trigger apoptosis. Interestingly, YTHDF2 showed dramatic reduction upon RO-3306 and purvalanol A treatment in both NB4 and MonoMac-6 cells (S6E Fig). Consistently, both AML cells underwent apoptosis along with YTHDF2 degradation, as revealed by 2 apoptosis markers poly (ADP-ribose) polymerase (PARP) and cleaved Caspase-3 (S6E Fig). These results suggest that CDK1 inhibitors promote YTHDF2 degradation in AMLs and might serve as a potential therapeutic method in AML treatment.
The ubiquitin-proteasome pathway mediates the proteolysis of important cell cycle regulators [49]. We hypothesized that YTHDF2 might undergo the same degradation pathway. By pretreating cells with the proteasome inhibitor MG-132, we found that the effect of RO-3306 was antagonized (Fig 4C), suggesting that YTHDF2 proteolysis occurs on proteasomes. Immunoprecipitation of YTHDF2 from cells with sequential MG-132 and RO-3306 treatment revealed a dramatic increase of polyubiquitinated YTHDF2 fraction (Fig 4D), confirming that CDK1 inhibition results in the polyubiquitination of YTHDF2 and subsequent degradation by proteasomes. Taken together, these results show that CDK1, YTHDF2, and WEE1 form a feedforward regulatory loop regulating the onset of mitosis.
Proteolysis pathway of YTHDF2
We next sought to identify the ubiquitination pathway that is responsible for YTHDF2 proteolysis. Homologous to the E6-AP Carboxyl Terminus (HECT) and Really Interesting New Gene (RING) E3 ligases are among the major classes of ubiquitin ligases [50]. The NEDD8-activating enzyme (NAE) is an E1 enzyme critical for the neddylation pathway that controls the activity of multi-subunit cullin-RING-type E3 ligases (CRLs) [51]. By comparing the effects of the HECT E3 inhibitor Heclin [52] and NAE inhibitor MLN4924 [53], we found that MLN4924—but not Heclin—profoundly antagonized the effect of RO-3306 (Fig 4E). Moreover, treatment of HeLa cells with different concentrations of MLN4924 for 24 hours caused high levels of YTHDF2 accumulation (S7A Fig), similar to p27, which is known to be controlled by NAE [53]. These results suggest that the NAE-mediated neddylation is an essential step for YTHDF2 degradation.
To rule out that RO-3306 treatment possibly activates NAE activity leading to downstream YTHDF2 degradation, we assessed the level of neddylated form of UBC12 (UBC12-NEDD8) catalyzed by NAE. No increase in UBC12-NEDD8 was observed after RO-3306 treatment (S7B Fig). Consistently, although MLN4924 antagonizes the effect of RO-3306 (Fig 4E), RO-3306 does not counteract the effect of MLN4924 in decreasing the conjugated form of UBC12 and NEDD8 (S7C Fig). These results suggest that CDK1 inhibition activates YTHDF2 degradation downstream of E1 and E2 ligases in the CRL pathway. In addition, the reduction of UBC12-NEDD8 conjugation induced by MLN4924 indicates that RING-box protein 1 (RBX1)—but not RBX2—participates in the YTHDF2 degradation pathway because UBC12 and RBX1 are specific pairs in the CRL pathway [54].
In order to identify the CRL components, we performed siRNA-mediated knockdown of a few cullins, including CUL1, CUL4A, CUL4B, and CUL7. Both CUL1 and CUL4A knockdown, but not CUL4B and CUL7, led to the overaccumulation of YTHDF2 (Fig 4F and S7D Fig), suggesting that YTHDF2 degradation is mediated by distinct E3 ligase complexes. Cullin 1 (CUL1) primarily forms E3 ligase complexes with S-phase kinase-associated protein 1 (SKP1) and SKP2, mediating the degradation of many important cell cycle regulators [55,56]. However, knockdown of SKP1 did not increase YTHDF2 accumulation (S7E Fig), suggesting that CUL1 might form complexes with alternative adaptors rather than SKP1 to mediate YTHDF2 degradation. Unexpectedly, we found that CUL4A knockdown led to the overaccumulation of CUL1 (Fig 4F), implicating a hierarchical cascade of CRLs, in which CUL4A controls the proteolysis of CUL1.
Damaged DNA-binding protein 1 (DDB1) is a known adaptor of CUL4A [57] and might interact with substrate receptor SKP2 [58] or CDT2 [59]. Knockdown of DDB1 and SKP2, but not CDT2, caused dramatic YTHDF2 increase (Fig 4G and S7F Fig), suggesting that CUL4A, DDB1, and SKP2 might form complexes for YTHDF2 degradation. To further verify this, a stable HeLa cell line expressing FLAG-YTHDF2 was sequentially treated with MG-132 and RO-3306. Immunoprecipitation using anti-FLAG M2 beads revealed that a higher fraction of SKP2 was co-immunoprecipitated with FLAG-YTHDF2 when CDK1 was inhibited by RO-3306 (Fig 4H). These results confirm that SKP2 is the substrate receptor of YTHDF2 and that CDK1 inhibition increases the association between YTHDF2 and SKP2.
Discussion
Transcriptomic m6A participates in a variety of biological processes, such as cell differentiation, development, memory, and immunity. Although it has been implicated in cell cycle regulation, systematic and mechanistic studies are still lacking. In this study, we investigated whether transcriptomic m6A is impacted by cell cycle regulation and found that m6A-modified transcripts, especially the ones with phase-specific expression patterns, display dynamic changes during cell cycle. It has been reported that mRNA synthesis and decay show dynamic changes during yeast cell cycle [39]; however, factors that contribute to the fast turnover of mRNAs remain elusive. This study revealed that YTHDF2 regulates a subset of phase-specific genes, confirming the regulatory role of transcriptomic m6A modification in promoting RNA degradation during cell cycle.
Further investigation found that YTHDF2 depletion leads to delayed mitotic entry. By meticulously comparing a number of key regulators gating mitotic entry, we found that WEE1, a suppressor of CDK1 by phosphorylation, is regulated by YTHDF2 by promoting WEE1 transcript decay. Therefore, overaccumulation of WEE1 protein upon YTHDF2 depletion leads to the delay of G2 to M phase transition by increasing the ratio of the inactive CDK1 form—CDK1-Y15. Phosphorylation and dephosphorylation of regulators, such as CDK1 and WEE1, are important mechanisms that regulate mitosis [40,60,61]. Here, we reveal that, in addition to the regulation at the protein level, the m6A modification of cell cycle regulator transcripts represents another pathway to fine-tune cell cycle progression. Future epitranscriptomic studies of important cell cycle regulators in different cell types and biological processes could provide more insights.
By testing a number of small molecule inhibitors, we found that the YTHDF2 protein stability relies on CDK1 activity. YTHDF2 undergoes dramatic polyubiquitination and subsequent degradation upon CDK1 inhibition. This result indicates that CDK1 maintains YTHDF2 stability during cell cycle, which represses WEE1 at the RNA level via m6A. However, whether CDK1 maintains YTHDF2 stability by directly phosphorylating YTHDF2 or through an indirect mechanism remains to be investigated in the future. Considering that CDK1 represses WEE1 by phosphorylation at serine 123 [62], CDK1, YTHDF2, and WEE1 thus form a feedforward regulatory loop that regulates mitotic entry (Fig 4I).
Adopting a similar chemical biology approach, we interrogated the degradation pathway of YTHDF2. We found that YTHDF2 proteolysis occurs on proteasomes mediated by the CRL-NEDD8 pathway, which is known for regulating many cellular processes, including cell cycle progression [49]. Screening for individual components of the potential E3 ligases using siRNAs identified CUL1, CUL4A, DDB1, and SKP2 as responsible for YTHDF2 proteolysis. We found that CDK1 maintains YTHDF2 stability likely by preventing the association between YTHDF2 and the SKP2-involved E3 ligase complexes. Moreover, our results suggested that CUL1 proteolysis is dependent on CUL4A. However, overaccumulation of CUL1 is not sufficient to compensate for YTHDF2 degradation in the absence of CUL4A, suggesting that CUL4A and CUL1 may function in a cascading yet distinct manner in mediating protein degradation.
In summary, we revealed a novel regulatory mechanism involving m6A that serves as an additional level of regulation in cell cycle control, in addition to the well-known regulations through protein phosphorylation and degradation. Moreover, we identified a feedforward regulatory loop that consists of CDK1, YTHDF2, and WEE1 regulating mitotic entry. The recent study revealed that YTHDF2 is essential for AML cells and depletion of YTHDF2 leads to the apoptosis of AML cells [23], rendering YTHDF2 a potential target for AML treatment. Our results highlight the potential of CDK1 inhibitors in inducing YTHDF2 degradation and apoptosis in AMLs. Moreover, the discovery of the YTHDF2 proteolysis pathway could lay a foundation for developing additional methods to manipulate YTHDF2 levels by techniques such as proteolysis targeting chimera (PROTAC) [63]. Therefore, elucidation of how YTHDF2 is regulated and its proteolysis pathway would potentially benefit the development of new therapeutic strategies to manipulate YTHDF2 levels in cancers—such as AML [23]—or for ex vivo amplification of hematopoietic stem cells [21,22].
Materials and methods
Cell culture
Human HeLa and HEK 293T cell lines were purchased from ATCC and maintained in DMEM (Gibco) with 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin/streptomycin (Gibco) in a humidified 37°C incubator with 5% CO2. NB4 cells were grown in RPMI medium 1640 (Gibco) with 10% FBS, 1% HEPES (Gibco), and 1% penicillin/streptomycin. MonoMac-6 cells were grown in RPMI 1640 supplemented with 10% FBS, 1% HEPES, 2 mM L-Glutamine (Gibco), 1% nonessential amino acids solution (Gibco), 1 mM sodium pyruvate (Gibco), 9 μg/ml insulin (MilliporeSigma), and 1% penicillin/streptomycin.
Plasmid construction
Human full-length YTHDF2 cDNA was cloned and inserted into the PiggyBac vector with a FLAG tag at the N-terminus. High-purity plasmids used for mammalian cell transfection and stable cell line preparation were prepared using the HiSpeed Plasmid Midi Kit (Qiagen). The WEE1 construct (Category Number RC209760) and empty vector control (Category Number PS100001) were ordered from OriGene.
siRNA knockdown and plasmid transfection
Cells were resuspended 16 hours prior to both siRNA and plasmid transfections. siRNAs were transfected to cells at approximately 50% confluency using the transfection reagent Lipofectamine RNAiMAX (Invitrogen). Cells reaching approximately 70% confluency were transfected with plasmids using Lipofectamine 2000 (Invitrogen). Both siRNA and plasmid transfections were conducted following the manufacturer’s protocols. Transfected cells were harvested 48 hours post siRNA or plasmid transfections for the following experiments. For stable FLAG-YTHDF2 HeLa cell line generation, DMEM medium supplemented with 1 μg/mL puromycin was used for selection 48 hours post plasmid transfection. Cells were maintained in medium with 1 μg/mL puromycin for at least a week before experiments.
CRISPR-Cas9–mediated knockout
YTHDF2 knockout was performed using the Alt-R CRISPR-Cas9 System from Integrated DNA Technologies (IDT) in HeLa cells. Guide RNA sequences were designed using CHOPCHOP [64] and synthesized as crRNAs from IDT (S5 Table). Both Alt-R CRISPR-Cas9 tracrRNA and synthesized crRNAs were dissolved to 100 μM in nuclease-free IDTE buffer (IDT), while Alt-R S.p. Cas9 Nuclease V3 (IDT) was diluted to 1 μM in Cas9 working buffer (20 mM HEPES [pH 7.5], 150 mM KCI). Equimolar tracrRNA and crRNA were mixed to reach a final concentration of 1 μM of each oligo in nuclease-free duplex buffer (IDT) and annealed by heating at 95°C for 5 minutes and cooling down to room temperature. Annealed oligos (1.5 μL), diluted Cas9 enzyme (1.5 μL), and 22 μL Opti-MEM (Gibco) were mixed and incubated at room temperature for 5 minutes to assemble the RNP complexes. After the incubation, 1.2 μL of Lipofectamine RNAiMAX and 23.8 μL Opti-MEM were added to the RNP complex solution and incubated for 20 minutes at room temperature. The transfection complexes were then added to a 96-well plate containing 40,000 cells/100 μL suspensions per well, followed by incubation at 37°C for 48 hours. Single-cell colonies were obtained by serial dilution of cells harvested from each well. Primers flanking the guide RNA sites were synthesized to amplify the DNA from the knockout cell lines derived from single colonies. PCR products were cloned to plasmids using the TOPO TA Cloning Kit (Invitrogen) for Sanger sequencing, which confirmed that CRISPR mostly caused open reading frame shifts, with at least one copy of the YTHDF2 gene in line KO-4 having 39-bp deletion, resulting in the deletion of 13 amino acid residues.
Cell proliferation assay
Cells were suspended in medium and counted using the Countess Automated Cell Counter (Invitrogen). About 3,000 cells were seeded to each well of the 96-well plates. After the cells adhered to the plate, 10 μL of the Cell Counting Kit-8 (MilliporeSigma) solution was added to each well and incubated for 2 hours in the 37°C CO2 incubator, followed by measuring of the absorbance at 450 nm using the Synergy HTX Multi-Mode Microplate Reader (BioTek). Absorbance was measured for 3 additional days to calculate the relative cell proliferation. To measure the proliferation of cells with siRNA knockdown, cells were transfected with siRNAs using Lipofectamine RNAiMax (Invitrogen) at least 6 hours before trypsinization for cell counting and seeding into 96-well plates. At least 5 wells were measured to calculate the average absorbance for each cell line at each time point.
Cell synchronization
HeLa cells were synchronized to early S phase using the double thymidine block based on a previous protocol [65]. Briefly, HeLa cells were grown in 10 cm plates with 10 mL medium to reach approximately 40% confluency, and then 200 uL of 100 mM thymidine stock solution was added to the cell culture to a final concentration of 2 mM. Cells were incubated in the 37°C CO2 incubator for 14 hours and then washed with 10 mL PBS twice. Growth medium supplemented with 24 μM deoxycytidine was added to the cells and incubated for 9 hours. The amount of 200 μL of 100 mM thymidine stock solution was added to the cell culture and incubated for 14 hours. To release the block, cells were washed with 10 mL PBS twice, and 10 mL medium was added for cell cycle progression. Cells were collected at different time points for up to 10 hours for experiments.
Flow cytometry
Single-cell suspensions were acquired by treating cells with trypsin solution at different time points post double thymidine block and mixed with equal volume of freshly prepared 4% paraformaldehyde (PFA) solution in PBS to reach a final concentration of 2% PFA. Cells were fixed at room temperature for 15 minutes, followed by washing twice with PBS. Cells were stained in 5 μg/mL Hoechst 33342 (MilliporeSigma) in PBS for 15 minutes at 37°C before flow cytometry. Ultraviolet light with 355-nm wavelength and 450/50-nm bandpass filters were used to quantify the DNA content of the cells for flow cytometry.
Small molecule inhibitor treatment
Small molecule inhibitors were dissolved in DMSO as stock solutions. Inhibitors were added to cell cultures with final concentrations and incubation times as indicated in each figure legend. For proteasome or E3 ligase inhibition assays, cells were incubated with 10 μM MG-132 (Selleckchem) [66], 100 μM Heclin (MilliporeSigma) [67], or 5 μM MLN4924 (MilliporeSigma) [53] for 3 hours, followed by 10 μM RO-3306 (MilliporeSigma) [66] before collecting the cells for western blots or protein co-immunoprecipitations. The concentrations of CDK2 inhibitor III, dinaciclib, purvalanol A, mTOR inhibitors, and ERK inhibitors were determined by the manufacturer’s instructions or previous reports [44,45,68–70]. For transcription inhibition assays, a final concentration of 5 μg/mL actinomycin D (MilliporeSigma) [10] was added to cell cultures at 6 hours, 3 hours, and 0 hours before cell collection for RNA extraction.
Protein co-immunoprecipitation and western blot
A stable line of HeLa cells expressing FLAG-YTHDF2 was collected from 15 cm plates and pelleted by centrifuge at 400g for 5 minutes. Cell pellets were resuspended with 3 volumes of immunoprecipitation lysis buffer (25 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% NP-40, and 5% glycerol, 1:50 protease inhibitor cocktail) and incubated on ice for 20 minutes. Cell lysates were then centrifuged at 16,000g for 15 minutes at 4°C. We saved 50 μl of cell lysate as input and incubated the rest with pre-equilibrated 50 μl anti-FLAG M2 magnetic beads (MilliporeSigma) by rotating at 4°C for 2 hours. After incubation, beads were subjected to washing 6 times with lysis buffer and boiled in NuPAGE LDS Sample Buffer (Invitrogen) with 5% 2-mercaptoethanol (2-ME). After magnetic separation, the supernatant was transferred as immunoprecipitation sample for western blotting, together with 1% input.
For samples collected for western blot, cells were lysed using RIPA buffer (Invitrogen) containing 1:100 protease inhibitor cocktail (Invitrogen) and phosphatase inhibitor cocktail (Invitrogen) on ice for 20 minutes. After centrifuging at 16,000g for 15 minutes at 4°C, the supernatant was transferred and boiled in NuPAGE LDS Sample Buffer (Invitrogen) with 5% 2-ME (MilliporeSigma). Samples were loaded to a NuPAGE 4% to 12% Bis-Tris Protein Gel (Invitrogen) for electrophoresis at a constant voltage of 160 V until the front dye reached the bottom of the gel and were transferred to 0.2 or 0.45 μM nitrocellulose membranes (BioRad) using the Trans-Blot SD semi-dry transfer cell (BioRad), followed by blocking with 5% nonfat milk (BioRad) in TBST. Antibody concentrations used for western blots were adjusted based on the manufacturer’s instructions.
RNA purification
Total RNA was purified from cells with the TRIzol reagent (Thermo Scientific). Polyadenylated RNA was purified from total RNA using the Dynabeads mRNA DIRECT kit (Thermo Scientific) according to the manufacturer’s protocol. mRNA concentrations were measured by Qubit fluorometer (Invitrogen) with the Qubit RNA HS Assay Kit (Invitrogen).
LC-MS/MS
The amount of 300 ng of isolated polyadenylated RNA was further processed using RiboMinus Transcriptome Isolation Kit (Invitrogen). The eluted RNA was then digested by nuclease P1 (MilliporeSigma) followed by dephosphorylation by alkaline phosphatase (Invitrogen). The samples were then used for LC-MS/MS. For detailed procedures for sample process and mass spectrometry, refer to the previous report [10].
RNA-seq
Total RNA was extracted from HeLa cells at different time points post release from double thymidine block and was subjected to polyadenylated RNA isolation using the Dynabeads mRNA DIRECT kit (Thermo Scientific). The amount of 100 ng of mRNA samples was used for RNA-seq library construction using the TruSeq stranded mRNA sample preparation kit (Illumina) by following the manufacturer’s protocol. High-throughput libraries for 2 biological replicates were sequenced using Illumina HiSeq 4000 with single-end 50-bp read length at The University of Chicago.
RIP-seq
Unsynchronized HeLa cells with 90% confluency in 15 cm plates were collected by cell lifters, pelleted by centrifuge for 5 minutes at 400g, and washed once with cold PBS. The cell pellets were lysed with 3 volumes of lysis buffer (150 mM KCl, 10 mM HEPES [pH 7.6], 2 mM EDTA, 0.5% NP-40, 0.5 mM DTT, 1:50 protease inhibitor cocktail, 1 U/μL SUPERase•In RNase Inhibitor) on ice for 20 minutes. The lysates were centrifuged at 16,000g for 15 minutes; 50 μL cell lysate was saved as input and mixed with 1 mL TRIzol for RNA extraction; and 20 μg of YTHDF2 antibody was added to the rest of the lysate and incubated by rotating for 2 hours at 4°C. Afterward, 40 μL of Pierce Protein A/G Magnetic Beads was washed twice with lysis buffer and added to the lysate and was incubated by rotating for 1 hour at 4°C. After separation using a magnetic rack, beads were washed with ice-code lysis 6 times. Beads were then mixed with 1 mL TRIzol and recovered for RNA, which was saved as immunoprecipitation. RNA extracted from input was further subjected to mRNA purification using the Dynabeads mRNA DIRECT kit (Thermo Scientific). Input mRNA of 100 ng and immunoprecipitation RNA samples with 2 biological replicates were used for RNA-seq library preparation.
RT-qPCR
For reverse transcription, 100 ng of total RNA or RIP extracted using TRIzol was subjected to DNA depletion using the TURBO DNA-free kit (Invitrogen) according to the manufacturer’'s protocol. The resultant DNA-free RNA was reverse-transcribed using the PrimeScript RT Reagent Kit (Takara, Category Number RR037A). The cDNA products were then used for real-time qPCR on the LightCycler 96 System (Roche) using FastStart Essential DNA Green Master (Roche). Gene-specific and housekeeping gene HPRT1 primers were designed and synthesized from IDT (S5 Table). qPCR analysis was performed using the ΔΔCt method. Average values from 3 replicates were calculated for each sample.
m6A MeRIP-seq
The m6A MeRIP-seq library preparation was adapted from the previous published protocol [71]. Briefly, total RNA extracted from cells synchronized at different phases of the cell cycle was subjected to polyadenylated RNA isolation, as described earlier; 5 μg of isolated poly(A)+ RNA was fragmented to approximately 100 nt by sonication using Bioruptor (Diagenode) with 4°C water bath. A portion (100 ng) of the fragmented RNA was then saved as input. The rest of the fragmented RNA was then subjected to m6A immunoprecipitation using 15 μg anti-m6A antibody (Synaptic Systems) in immunoprecipitation buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.1% NP-40, 1 U/μL SUPERase•In RNase Inhibitor) for 2 hours at 4°C. The amount of 50 μl pre-washed protein A/G beads was added and incubate by rotating for 2 hours, followed by washing 3 times using immunoprecipitation buffer. The m6A-containing RNA fragments from beads were eluted twice using 100 μl elution buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 6.7 mM m6A, 0.1% NP-40, 1 U/μL SUPERase•In RNase Inhibitor) for 1 hour while rotating at 4°C. Eluted RNA was then precipitated in 0.3 M sodium acetate (pH 5.2) and 75% ethanol with GlycoBlue (Invitrogen) at −80°C overnight. RNA was recovered from the pellet by centrifuge at 16,000g for 20 minutes at 4°C. Both input and immunoprecipitation RNA samples were subjected to RNA-seq library construction.
Sequencing data analysis
The quality control of sequencing reads of RNA-seq, RIP-seq, and m6A-seq libraries was performed using FastQC version 0.11.5. After trimming adapter using Cutadapt version 1.1.5 [72], we then mapped to the human genome GRCh37/hg19 using TopHat version 2.1.0, allowing for at most 2 mismatches [73]. For RNA-seq data, cuffnorm from the Cufflinks (version 2.2.1) package was used for geometric normalization and FPKM calculation [74]. For RIP-seq data analysis, Cufflinks—combined with Cuffdiff—was employed to quantify gene expression and differential analysis. Genes with q < 0.05 were considered YTHDF2 targets. More confident YTHDF2 targets were obtained by intersecting targets from RIP-seq of this study and the published YTHDF2 PAR-CLIP data [10].
For m6A-seq data analysis, the m6A-enriched regions in each m6A-immunoprecipitation sample were identified by MACS2 version 2.1.0 with the option of “—nomodel” [75], using corresponding input library as control. Peaks identified with q < 0.05 were used for downstream analysis. Peak annotation and motif search were both accomplished using the software HOMER [76]. Metagene analysis of m6A distribution on transcripts was performed using the MetaPlotR pipeline [77]. IGVtools was used to convert bam files to tdf files, which were loaded into the Integrative Genomics Viewer (IGV) for m6A peak visualization [78]. The R package “DiffBind” was used for m6A peak differential analysis with a cutoff of P < 1 × 10−3. Genes with differential m6A enrichment at different phases of the cell cycle were subjected to GO analysis using DAVID [79]. GO terms were further summarized and visualized using REVIGO [80].
Supporting information
(A) Workflow of cell synchronization by double thymidine block and time points for cell collection after release. (B) GO terms of common genes with m6A modifications across 3 phases of the cell cycle. Color key represents the −log(P value) of enriched GO terms. (C) Correlation heatmap representing pairwise comparison of m6A enrichment for each replicate at 3 phases. (D) Western blot of METTL3, METTL14, and FTO at different time points post synchronization. (E) Expression levels of METTL3, METTL14, and FTO at different time points post synchronization from the RNA-seq data. Underlying data for this figure can be found in S1 Raw Images and S1 Data.
(TIF)
(A) Correlation analysis of gene expression levels and m6A peak enrichment. (B) Cumulative distribution of gene expression changes with differential m6A enrichment at different phases. Left panel shows cumulative distribution by comparing the expression level of transcripts with up- or down-regulated m6A from G1/S to S phase. Right panel shows that from S to G2/M phase. The x-axes indicate the log2 fold change of gene expression level in the next phase compared with the previous phase. P values were calculated using the Mann-Whitney test. (C) GO terms for increased m6A peaks at S phase compared with G1/S. (D) GO terms for decreased m6A peaks at S phase compared with G1/S. (E) GO terms for increased m6A peaks at G2/M phase compared with S. TUBB4B is an example that is related to “microtubule-based process” with higher m6A at G2/M phase. (F) GO terms for decreased m6A peaks at G2/M phase compared with S. SMAD3 is an example that is related to regulation of transcription with reduced m6A from S phase to G2/M phase. Underlying data for this figure can be found in S1 Data.
(TIF)
(A) Design of crRNAs for CRISPR-Cas9 for YTHDF2 knockout. (B) Rescue of YTHDF2 knockout cell lines by FLAG-YTHDF2 transfection. Two knockout cell lines KO-1 and KO-2 were randomly selected for transfection and proliferation assay. (C) Cell proliferation assays for HeLa cells with YTHDF2 siRNA knockdown compared with the siRNA control. Underlying data for this figure can be found in S1 Data. crRNA, CRISPR RNA.
(TIF)
(A) Intersection for a confident YTHDF2 targets in HeLa cells between YTHDF2 RIP-seq and PAR-CLIP data. PAR-CLIP results are from Wang and colleagues [10]. The 4,668 nontarget genes were obtained after filtering out the genes in either RIP-seq or PAR-CLIP list and the ones with FPKM < 1 in the input sample of the RIP-seq data. (B) Cumulative distribution of 2,701 YTHDF2 targets and 4,668 nontargets by comparing WT and knockout cell lines. Genes with FPKM < 1 at each time point were further removed from the analysis. x-Axes indicate the log2 fold change of gene expression in knockout versus wild type. P values were calculated using the Mann-Whitney test. Underlying data for this figure can be found in S1 Data. FPKM, Fragments Per Kilobase of transcript per Million mapped reads.
(TIF)
(A) Quantification of WEE1 and p-CDK1-Y15 by ImageJ from Fig 3B. The protein levels were normalized to the loading control GAPDH. (B) Expression level of WEE1 revealed by RNA-seq in wild-type and knockout cells at different time points post release from G1/S phase. (C) Western blot of WEE1 at different time points post synchronization in wild-type and YTHDF2 knockout HeLa cells. The right panel shows the normalized values of WEE1 quantified by ImageJ. (D) Effect of WEE1 overexpression in HeLa cells. Left panel shows cell proliferation of HeLa cells transfected with Myc-WEE1 compared with the empty vector control. The right panel shows flow cytometry analysis results of each phase during cell cycle. The percentages of each phase were quantified using FlowJo. (E) siRNA knockdown of YTHDF2 and METTL3 in HeLa cells. The left panel shows RT-qPCR results with two-sided Student t test (*P < 0.05; **P < 0.01; ***P < 0.001). The right panel shows western blot results of each protein. (F) Flow cytometry results of each phase in the cell cycle upon YTHDF2 or METTL3 knockdown. Underlying data for this figure can be found in S1 Data and S1 Raw Images.
(TIF)
(A) The level of YTHDF2 at different time points post synchronization. The black line indicates protein level changes of YTHDF2 quantified by ImageJ. The red dots indicate transcript levels of YTHDF2 at each time point, which were normalized to the value at 0 hours. (B) RO-3306 induces YTHDF2 degradation in HEK 393T cells within 3 hours. The concentrations of RO-3306 are indicated. (C) Purvalanol A, a different CDK1 inhibitor, induces YTHDF2 degradation. Concentrations and incubation times are as indicated. (D) Detection of YTHDF2 levels after the treatment of relevant inhibitors, including mTOR and ERK inhibitors. (E) Western blot of YTHDF2 and apoptosis markers PARP and Caspase-3 (cleaved form) after CDK1 inhibitor treatment in NB4 and MonoMac-6 cells. Underlying data for this figure can be found in S1 Data and S1 Raw Images.
(TIF)
(A) Western blot of YTHDF2 after cells incubated with different concentrations of MLN4924 for 24 hours. p27 is a positive control. (B) Detection of the conjugated form of UBC12-NEDD8 after sequential treatment of RO-3306 and MLN4924. Cells were pretreated with RO-3306 for 3 hours before MLN4924 incubation for 3 hours, with concentrations indicated in the figure. (C) Detection of the conjugated form of UBC12-NEDD8 after treating cells with 10 μM RO-3306 for indicated hours. (D–F) Western blot of YTHDF2 after siRNA knockdown of CUL4B, CUL7, SKP1, and CDT2. Underlying data for this figure can be found in S1 Raw Images.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(PDF)
Abbreviations
- 2-ME
2-mercaptoethanol
- ALKBH5
AlkB Homolog 5
- AML
acute myeloid leukemia
- CCR4-NOT
Carbon Catabolite Repressor 4-Negative on TATA
- CDC25
cell division cycle 25
- CDK1
cyclin-dependent kinase 1
- CRL
cullin-RING-type E3 ligase
- CUL1
Cullin 1
- CUL4A
Cullin 4A
- DDB1
damaged DNA-binding protein 1
- ERK
extracellular signal-regulated kinase
- FBS
fetal bovine serum
- FOXM1
forkhead box protein M1
- FTO
fat mass and obesity-associated protein
- GO
gene ontology
- HECT
Homologous to the E6-AP Carboxyl Terminus
- HEK 293T
human embryonic kidney 293T
- IDT
Integrated DNA Technologies
- IGV
Integrative Genomics Viewer
- m6A
N6-methyladenosine
- MeRIP-seq
methylated RNA immunoprecipitation sequencing
- mESC
mouse embryonic stem cell
- METTL3
Methyltransferase Like 3
- MRP
Mitochondrial RNA Processing
- mTOR
mammalian target of rapamycin
- MYT1
membrane associated tyrosine/threonine 1
- MZT
maternal to zygotic transition
- NAE
NEDD8-activating enzyme
- PAR-CLIP
photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation
- PARP
poly (ADP-ribose) polymerase
- p-CDK1-T14
phosphorylating CDK1 at threonine 14
- p-CDK1-Y15
phosphorylating CDK1 at tyrosine 15
- PFA
paraformaldehyde
- PROTAC
proteolysis targeting chimera
- RBX1
RING-box protein 1
- RING
Really Interesting New Gene
- RIP
RNA immunoprecipitation
- RIP-seq
RIP sequencing
- RNA-seq
RNA sequencing
- RT-qPCR
reverse transcription quantitative PCR
- siRNA
small interfering RNA
- SKP2
S-phase kinase-associated protein 2
- UTR
untranslated region
- VIRMA
Vir Like m6A Methyltransferase Associated
- WEE1
Wee1-like protein kinase
- WTAP
Wilms tumor 1-associated protein
- YTHDF2
YTH N6-methyladenosine RNA binding protein 2
- ZC3H13
Zinc Finger CCCH-Type Containing 13
Data Availability
All raw data are available at the Gene Expression Omnibus (GEO) under the accession number GSE134700.
Funding Statement
This work is supported by the U.S. National Institutes of Health (https://www.nih.gov) (GM113194 and HG008935 to C.H.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10(2):93–5. 10.1038/nchembio.1432 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24(2):177–89. 10.1038/cr.2014.3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, et al. VIRMA mediates preferential m(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10 10.1038/s41421-018-0019-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wen J, Lv R, Ma H, Shen H, He C, Wang J, et al. Zc3h13 Regulates Nuclear RNA m(6)A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol Cell. 2018;69(6):1028–38 e6. 10.1016/j.molcel.2018.02.015 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vagbo CB, Geula S, et al. m(6)A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 2017;31(10):990–1006. 10.1101/gad.301036.117 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang H, Weng H, Zhou K, Wu T, Zhao BS, Sun M, et al. Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature. 2019;567(7748):414–9. 10.1038/s41586-019-1016-7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell. 2016;61(4):507–19. 10.1016/j.molcel.2016.01.012 . [DOI] [PubMed] [Google Scholar]
- 8.Louloupi A, Ntini E, Conrad T, Orom UAV. Transient N-6-Methyladenosine Transcriptome Sequencing Reveals a Regulatory Role of m6A in Splicing Efficiency. Cell Rep. 2018;23(12):3429–37. 10.1016/j.celrep.2018.05.077 . [DOI] [PubMed] [Google Scholar]
- 9.Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife. 2017;6 10.7554/eLife.31311 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–20. 10.1038/nature12730 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161(6):1388–99. 10.1016/j.cell.2015.05.014 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518(7540):560–4. 10.1038/nature14234 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roost C, Lynch SR, Batista PJ, Qu K, Chang HY, Kool ET. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J Am Chem Soc. 2015;137(5):2107–15. 10.1021/ja513080v . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27(3):315–28. 10.1038/cr.2017.15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, et al. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27(9):1115–27. 10.1038/cr.2017.99 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626 10.1038/ncomms12626 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, et al. Endoribonucleolytic Cleavage of m(6)A-Containing RNAs by RNase P/MRP Complex. Mol Cell. 2019;74(3):494–507 e8. 10.1016/j.molcel.2019.02.034 . [DOI] [PubMed] [Google Scholar]
- 18.Zhao BS, Wang X, Beadell AV, Lu Z, Shi H, Kuuspalu A, et al. m(6)A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature. 2017;542(7642):475–8. 10.1038/nature21355 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivanova I, Much C, Di Giacomo M, Azzi C, Morgan M, Moreira PN, et al. The RNA m(6)A Reader YTHDF2 Is Essential for the Post-transcriptional Regulation of the Maternal Transcriptome and Oocyte Competence. Mol Cell. 2017;67(6):1059–67 e4. 10.1016/j.molcel.2017.08.003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li M, Zhao X, Wang W, Shi H, Pan Q, Lu Z, et al. Ythdf2-mediated m(6)A mRNA clearance modulates neural development in mice. Genome Biol. 2018;19(1):69 10.1186/s13059-018-1436-y . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z, Qian P, Shao W, Shi H, He XC, Gogol M, et al. Suppression of m(6)A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res. 2018;28(9):904–17. 10.1038/s41422-018-0072-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Zuo H, Liu J, Wen F, Gao Y, Zhu X, et al. Loss of YTHDF2-mediated m(6)A-dependent mRNA clearance facilitates hematopoietic stem cell regeneration. Cell Res. 2018;28(10):1035–8. 10.1038/s41422-018-0082-y . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paris J, Morgan M, Campos J, Spencer GJ, Shmakova A, Ivanova I, et al. Targeting the RNA m(6)A Reader YTHDF2 Selectively Compromises Cancer Stem Cells in Acute Myeloid Leukemia. Cell Stem Cell. 2019. 10.1016/j.stem.2019.03.021 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15(6):707–19. 10.1016/j.stem.2014.09.019 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millan-Zambrano G, Robson SC, et al. Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature. 2017;552(7683):126–31. 10.1038/nature24678 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. m(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell. 2017;31(4):591–606 e6. 10.1016/j.ccell.2017.02.013 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoon KJ, Ringeling FR, Vissers C, Jacob F, Pokrass M, Jimenez-Cyrus D, et al. Temporal Control of Mammalian Cortical Neurogenesis by m(6)A Methylation. Cell. 2017;171(4):877–89 e17. 10.1016/j.cell.2017.09.003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bertoli C, Skotheim JM, de Bruin RA. Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol. 2013;14(8):518–28. 10.1038/nrm3629 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–6. 10.1038/nature11112 . [DOI] [PubMed] [Google Scholar]
- 30.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149(7):1635–46. 10.1016/j.cell.2012.05.003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma C, Chang M, Lv H, Zhang ZW, Zhang W, He X, et al. RNA m(6)A methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 2018;19(1):68 10.1186/s13059-018-1435-z . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He S, Wang H, Liu R, He M, Che T, Jin L, et al. mRNA N6-methyladenosine methylation of postnatal liver development in pig. PLoS ONE. 2017;12(3):e0173421 10.1371/journal.pone.0173421 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sclafani RA, Holzen TM. Cell cycle regulation of DNA replication. Annu Rev Genet. 2007;41:237–80. 10.1146/annurev.genet.41.110306.130308 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coleman ML, Marshall CJ, Olson MF. RAS and RHO GTPases in G1-phase cell-cycle regulation. Nat Rev Mol Cell Biol. 2004;5(5):355–66. 10.1038/nrm1365 . [DOI] [PubMed] [Google Scholar]
- 35.Gadde S, Heald R. Mechanisms and molecules of the mitotic spindle. Curr Biol. 2004;14(18):R797–805. 10.1016/j.cub.2004.09.021 . [DOI] [PubMed] [Google Scholar]
- 36.Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13(6):1977–2000. 10.1091/mbc.02-02-0030. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rustici G, Mata J, Kivinen K, Lio P, Penkett CJ, Burns G, et al. Periodic gene expression program of the fission yeast cell cycle. Nat Genet. 2004;36(8):809–17. 10.1038/ng1377 . [DOI] [PubMed] [Google Scholar]
- 38.Simon I, Barnett J, Hannett N, Harbison CT, Rinaldi NJ, Volkert TL, et al. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell. 2001;106(6):697–708. 10.1016/s0092-8674(01)00494-9 . [DOI] [PubMed] [Google Scholar]
- 39.Eser P, Demel C, Maier KC, Schwalb B, Pirkl N, Martin DE, et al. Periodic mRNA synthesis and degradation co-operate during cell cycle gene expression. Mol Syst Biol. 2014;10:717 10.1002/msb.134886 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol. 2001;2(1):21–32. 10.1038/35048096 . [DOI] [PubMed] [Google Scholar]
- 41.Parker LL, Piwnica-Worms H. Inactivation of the p34cdc2-cyclin B complex by the human WEE1 tyrosine kinase. Science. 1992;257(5078):1955–7. 10.1126/science.1384126 . [DOI] [PubMed] [Google Scholar]
- 42.Liu F, Stanton JJ, Wu Z, Piwnica-Worms H. The human Myt1 kinase preferentially phosphorylates Cdc2 on threonine 14 and localizes to the endoplasmic reticulum and Golgi complex. Mol Cell Biol. 1997;17(2):571–83. 10.1128/mcb.17.2.571 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, et al. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci U S A. 2006;103(28):10660–5. 10.1073/pnas.0600447103 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brooks EE, Gray NS, Joly A, Kerwar SS, Lum R, Mackman RL, et al. CVT-313, a specific and potent inhibitor of CDK2 that prevents neointimal proliferation. J Biol Chem. 1997;272(46):29207–11. 10.1074/jbc.272.46.29207 . [DOI] [PubMed] [Google Scholar]
- 45.Parry D, Guzi T, Shanahan F, Davis N, Prabhavalkar D, Wiswell D, et al. Dinaciclib (SCH 727965), a novel and potent cyclin-dependent kinase inhibitor. Mol Cancer Ther. 2010;9(8):2344–53. 10.1158/1535-7163.MCT-10-0324 . [DOI] [PubMed] [Google Scholar]
- 46.Gray NS, Wodicka L, Thunnissen AM, Norman TC, Kwon S, Espinoza FH, et al. Exploiting chemical libraries, structure, and genomics in the search for kinase inhibitors. Science. 1998;281(5376):533–8. 10.1126/science.281.5376.533 . [DOI] [PubMed] [Google Scholar]
- 47.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284(12):8023–32. 10.1074/jbc.M900301200 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ryan MB, Der CJ, Wang-Gillam A, Cox AD. Targeting RAS-mutant cancers: is ERK the key? Trends Cancer. 2015;1(3):183–98. 10.1016/j.trecan.2015.10.001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bassermann F, Eichner R, Pagano M. The ubiquitin proteasome system—implications for cell cycle control and the targeted treatment of cancer. Biochim Biophys Acta. 2014;1843(1):150–62. 10.1016/j.bbamcr.2013.02.028 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Metzger MB, Hristova VA, Weissman AM. HECT and RING finger families of E3 ubiquitin ligases at a glance. J Cell Sci. 2012;125(Pt 3):531–7. 10.1242/jcs.091777 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene. 2004;23(11):1985–97. 10.1038/sj.onc.1207414 . [DOI] [PubMed] [Google Scholar]
- 52.Mund T, Lewis MJ, Maslen S, Pelham HR. Peptide and small molecule inhibitors of HECT-type ubiquitin ligases. Proc Natl Acad Sci U S A. 2014;111(47):16736–41. 10.1073/pnas.1412152111 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458(7239):732–6. 10.1038/nature07884 . [DOI] [PubMed] [Google Scholar]
- 54.Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, Scott DC, et al. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell. 2009;33(4):483–95. 10.1016/j.molcel.2009.01.011 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakayama KI, Nakayama K. Regulation of the cell cycle by SCF-type ubiquitin ligases. Semin Cell Dev Biol. 2005;16(3):323–33. 10.1016/j.semcdb.2005.02.010 . [DOI] [PubMed] [Google Scholar]
- 56.Vodermaier HC. APC/C and SCF: controlling each other and the cell cycle. Curr Biol. 2004;14(18):R787–96. 10.1016/j.cub.2004.09.020 . [DOI] [PubMed] [Google Scholar]
- 57.Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443(7111):590–3. 10.1038/nature05175 . [DOI] [PubMed] [Google Scholar]
- 58.Bondar T, Kalinina A, Khair L, Kopanja D, Nag A, Bagchi S, et al. Cul4A and DDB1 associate with Skp2 to target p27Kip1 for proteolysis involving the COP9 signalosome. Mol Cell Biol. 2006;26(7):2531–9. 10.1128/MCB.26.7.2531-2539.2006 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell. 2006;23(5):709–21. 10.1016/j.molcel.2006.08.010 . [DOI] [PubMed] [Google Scholar]
- 60.Barr FA, Elliott PR, Gruneberg U. Protein phosphatases and the regulation of mitosis. J Cell Sci. 2011;124(Pt 14):2323–34. 10.1242/jcs.087106 . [DOI] [PubMed] [Google Scholar]
- 61.Barr FA, Sillje HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol. 2004;5(6):429–40. 10.1038/nrm1401 . [DOI] [PubMed] [Google Scholar]
- 62.Watanabe N, Arai H, Iwasaki J, Shiina M, Ogata K, Hunter T, et al. Cyclin-dependent kinase (CDK) phosphorylation destabilizes somatic Wee1 via multiple pathways. Proc Natl Acad Sci U S A. 2005;102(33):11663–8. 10.1073/pnas.0500410102 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gu S, Cui D, Chen X, Xiong X, Zhao Y. PROTACs: An Emerging Targeting Technique for Protein Degradation in Drug Discovery. Bioessays. 2018;40(4):e1700247 10.1002/bies.201700247 . [DOI] [PubMed] [Google Scholar]
- 64.Labun K, Montague TG, Krause M, Torres Cleuren YN, Tjeldnes H, Valen E. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019. 10.1093/nar/gkz365 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ma HT, Poon RY. Synchronization of HeLa cells. Methods Mol Biol. 2011;761:151–61. 10.1007/978-1-61779-182-6_10 . [DOI] [PubMed] [Google Scholar]
- 66.Chow JP, Poon RY, Ma HT. Inhibitory phosphorylation of cyclin-dependent kinase 1 as a compensatory mechanism for mitosis exit. Mol Cell Biol. 2011;31(7):1478–91. 10.1128/MCB.00891-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mund T, Graeb M, Mieszczanek J, Gammons M, Pelham HR, Bienz M. Disinhibition of the HECT E3 ubiquitin ligase WWP2 by polymerized Dishevelled. Open Biol. 2015;5(12):150185 10.1098/rsob.150185 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Villerbu N, Gaben AM, Redeuilh G, Mester J. Cellular effects of purvalanol A: a specific inhibitor of cyclin-dependent kinase activities. Int J Cancer. 2002;97(6):761–9. 10.1002/ijc.10125 . [DOI] [PubMed] [Google Scholar]
- 69.Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332(6035):1317–22. 10.1126/science.1199498 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhan T, Ambrosi G, Wandmacher AM, Rauscher B, Betge J, Rindtorff N, et al. MEK inhibitors activate Wnt signalling and induce stem cell plasticity in colorectal cancer. Nat Commun. 2019;10(1):2197 10.1038/s41467-019-09898-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, Amariglio N, Rechavi G. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc. 2013;8(1):176–89. 10.1038/nprot.2012.148 . [DOI] [PubMed] [Google Scholar]
- 72.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17(1):10–2. [Google Scholar]
- 73.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–11. 10.1093/bioinformatics/btp120 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–5. 10.1038/nbt.1621 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9(9):R137 10.1186/gb-2008-9-9-r137 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–89. 10.1016/j.molcel.2010.05.004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Olarerin-George AO, Jaffrey SR. MetaPlotR: a Perl/R pipeline for plotting metagenes of nucleotide modifications and other transcriptomic sites. Bioinformatics. 2017;33(10):1563–4. 10.1093/bioinformatics/btx002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–6. 10.1038/nbt.1754 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. 10.1038/nprot.2008.211 . [DOI] [PubMed] [Google Scholar]
- 80.Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011;6(7):e21800 10.1371/journal.pone.0021800 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Workflow of cell synchronization by double thymidine block and time points for cell collection after release. (B) GO terms of common genes with m6A modifications across 3 phases of the cell cycle. Color key represents the −log(P value) of enriched GO terms. (C) Correlation heatmap representing pairwise comparison of m6A enrichment for each replicate at 3 phases. (D) Western blot of METTL3, METTL14, and FTO at different time points post synchronization. (E) Expression levels of METTL3, METTL14, and FTO at different time points post synchronization from the RNA-seq data. Underlying data for this figure can be found in S1 Raw Images and S1 Data.
(TIF)
(A) Correlation analysis of gene expression levels and m6A peak enrichment. (B) Cumulative distribution of gene expression changes with differential m6A enrichment at different phases. Left panel shows cumulative distribution by comparing the expression level of transcripts with up- or down-regulated m6A from G1/S to S phase. Right panel shows that from S to G2/M phase. The x-axes indicate the log2 fold change of gene expression level in the next phase compared with the previous phase. P values were calculated using the Mann-Whitney test. (C) GO terms for increased m6A peaks at S phase compared with G1/S. (D) GO terms for decreased m6A peaks at S phase compared with G1/S. (E) GO terms for increased m6A peaks at G2/M phase compared with S. TUBB4B is an example that is related to “microtubule-based process” with higher m6A at G2/M phase. (F) GO terms for decreased m6A peaks at G2/M phase compared with S. SMAD3 is an example that is related to regulation of transcription with reduced m6A from S phase to G2/M phase. Underlying data for this figure can be found in S1 Data.
(TIF)
(A) Design of crRNAs for CRISPR-Cas9 for YTHDF2 knockout. (B) Rescue of YTHDF2 knockout cell lines by FLAG-YTHDF2 transfection. Two knockout cell lines KO-1 and KO-2 were randomly selected for transfection and proliferation assay. (C) Cell proliferation assays for HeLa cells with YTHDF2 siRNA knockdown compared with the siRNA control. Underlying data for this figure can be found in S1 Data. crRNA, CRISPR RNA.
(TIF)
(A) Intersection for a confident YTHDF2 targets in HeLa cells between YTHDF2 RIP-seq and PAR-CLIP data. PAR-CLIP results are from Wang and colleagues [10]. The 4,668 nontarget genes were obtained after filtering out the genes in either RIP-seq or PAR-CLIP list and the ones with FPKM < 1 in the input sample of the RIP-seq data. (B) Cumulative distribution of 2,701 YTHDF2 targets and 4,668 nontargets by comparing WT and knockout cell lines. Genes with FPKM < 1 at each time point were further removed from the analysis. x-Axes indicate the log2 fold change of gene expression in knockout versus wild type. P values were calculated using the Mann-Whitney test. Underlying data for this figure can be found in S1 Data. FPKM, Fragments Per Kilobase of transcript per Million mapped reads.
(TIF)
(A) Quantification of WEE1 and p-CDK1-Y15 by ImageJ from Fig 3B. The protein levels were normalized to the loading control GAPDH. (B) Expression level of WEE1 revealed by RNA-seq in wild-type and knockout cells at different time points post release from G1/S phase. (C) Western blot of WEE1 at different time points post synchronization in wild-type and YTHDF2 knockout HeLa cells. The right panel shows the normalized values of WEE1 quantified by ImageJ. (D) Effect of WEE1 overexpression in HeLa cells. Left panel shows cell proliferation of HeLa cells transfected with Myc-WEE1 compared with the empty vector control. The right panel shows flow cytometry analysis results of each phase during cell cycle. The percentages of each phase were quantified using FlowJo. (E) siRNA knockdown of YTHDF2 and METTL3 in HeLa cells. The left panel shows RT-qPCR results with two-sided Student t test (*P < 0.05; **P < 0.01; ***P < 0.001). The right panel shows western blot results of each protein. (F) Flow cytometry results of each phase in the cell cycle upon YTHDF2 or METTL3 knockdown. Underlying data for this figure can be found in S1 Data and S1 Raw Images.
(TIF)
(A) The level of YTHDF2 at different time points post synchronization. The black line indicates protein level changes of YTHDF2 quantified by ImageJ. The red dots indicate transcript levels of YTHDF2 at each time point, which were normalized to the value at 0 hours. (B) RO-3306 induces YTHDF2 degradation in HEK 393T cells within 3 hours. The concentrations of RO-3306 are indicated. (C) Purvalanol A, a different CDK1 inhibitor, induces YTHDF2 degradation. Concentrations and incubation times are as indicated. (D) Detection of YTHDF2 levels after the treatment of relevant inhibitors, including mTOR and ERK inhibitors. (E) Western blot of YTHDF2 and apoptosis markers PARP and Caspase-3 (cleaved form) after CDK1 inhibitor treatment in NB4 and MonoMac-6 cells. Underlying data for this figure can be found in S1 Data and S1 Raw Images.
(TIF)
(A) Western blot of YTHDF2 after cells incubated with different concentrations of MLN4924 for 24 hours. p27 is a positive control. (B) Detection of the conjugated form of UBC12-NEDD8 after sequential treatment of RO-3306 and MLN4924. Cells were pretreated with RO-3306 for 3 hours before MLN4924 incubation for 3 hours, with concentrations indicated in the figure. (C) Detection of the conjugated form of UBC12-NEDD8 after treating cells with 10 μM RO-3306 for indicated hours. (D–F) Western blot of YTHDF2 after siRNA knockdown of CUL4B, CUL7, SKP1, and CDT2. Underlying data for this figure can be found in S1 Raw Images.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(PDF)
Data Availability Statement
All raw data are available at the Gene Expression Omnibus (GEO) under the accession number GSE134700.