Abstract
Heavy reliance on plants is rare in Carnivora and mostly limited to relatively small species in subtropical settings. The feeding behaviors of extinct cave bears living during Pleistocene cold periods at middle latitudes have been intensely studied using various approaches including isotopic analyses of fossil collagen. In contrast to cave bears from all other regions in Europe, some individuals from Romania show exceptionally high δ15N values that might be indicative of meat consumption. Herbivory on plants with high δ15N values cannot be ruled out based on this method, however. Here we apply an approach using the δ15N values of individual amino acids from collagen that offsets the baseline δ15N variation among environments. The analysis yielded strong signals of reliance on plants for Romanian cave bears based on the δ15N values of glutamate and phenylalanine. These results could suggest that the high variability in bulk collagen δ15N values observed among cave bears in Romania reflects niche partitioning but in a general trophic context of herbivory.
Subject terms: Palaeontology, Biogeochemistry, Palaeoecology
Introduction
Bears represent the largest terrestrial members within the Carnivora alive today and the vast majority of them have carnivorous or omnivorous feeding habits. Until around 25,000 years ago, the coldest period in the Pleistocene, additional, now extinct bear species were living1–4, among which the so-called cave bears, a very large type of bear that formed the sister lineage of extant brown bears and polar bears (e.g., ref. 5). The paradox of the cave bear is that their diet has been said to be herbivorous despite their large body sizes while extant herbivorous Carnivora species are smaller6,7. After their divergence from the brown bear lineage 1.2–1.6 million years ago, cave bear populations showed substantial morphological and genetic variability and multiple forms have been recognized8, although their taxonomic status and the relationships among them continue to be debated9. The possible causes of the extinction of these bears are also intensively debated, involving climate change, human impacts, and (lack of) flexibility in feeding behavior10–15. Understanding cave bear feeding behavior is therefore important as it might give insights into the extinction of this species, and also it could be relevant for the conservation of extant, herbivorous carnivoran species that are under threat of extinction (e.g., binturong, red panda, giant panda16,17).
More recent studies have shown mixed results based on different lines of evidence including anatomical properties like craniodental morphologies, tooth wear analyses, mortality patterns (e.g., sex ratio), etc. wherein the conclusions were highly context dependent and differed by sample-sets18–23. This is also the case for stable carbon and nitrogen isotope analyses (δ13C and δ15N) on collagen extracted from bone/teeth24–31. Relatively low δ15N values of most of these bears so far indicate their highly-plant-dependent feeding habits with possible exceptions for some groups in today’s Romania that exhibited relatively high δ15N values30,32,33. The δ15N value of bulk bone/teeth collagen is known to increase from prey to its consumer in a food web, thus being an important indicator to evaluate the trophic position of organisms34,35. Other factors also influence δ15N values of collagen, such as variability in the consumed plants and environmental factors including climate36–38, leading to possible spatiotemporal shifts of the isotopic baseline due to heterogeneities among local environments39–42. Hence, some herbivores may exhibit similar δ15N values as predatory species in the same context30,43–45.
To solve this uncertainty, a new isotopic approach has been developed. Indeed, it has been demonstrated that δ15N values of individual amino acids (AA) can be classified into several categories, “trophic” and “source” AAs that exhibit more 15N enrichment and less/little 15N enrichment, respectively, in each trophic step of food webs46–53. It is worth mentioning that the comparison of the “source” AAs like phenylalanine (Phe) with the “trophic” AAs like glutamate (Glx) enables the evaluation of animal trophic position regardless of baseline δ15N fluctuations in ecosystems54. We aim at testing the two hypotheses that high collagen δ15N values observed for cave bears in the Romanian region were attributed to (i) omnivorous/carnivorous feeding behavior associated with a trophic level effect as a corollary or (ii) consumption of plants with high bulk δ15N values. To this end, δ15N values of individual AAs in collagen were measured for adult cave bears from several sites in this region (Fig. 1). One cave lion and one horse have been analyzed together with the adult cave bear collagen to represent end-members for trophic positions for a terrestrial carnivore and a terrestrial herbivore in the Late Pleistocene.
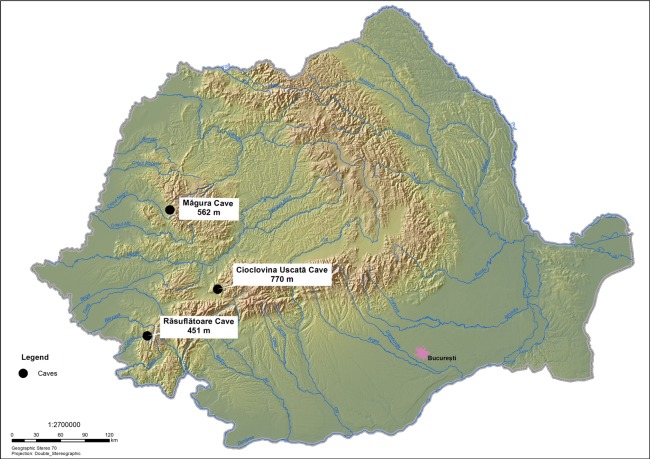
Figure 1.
Map showing locations of the cave sites in Romania mentioned in the text.
Results
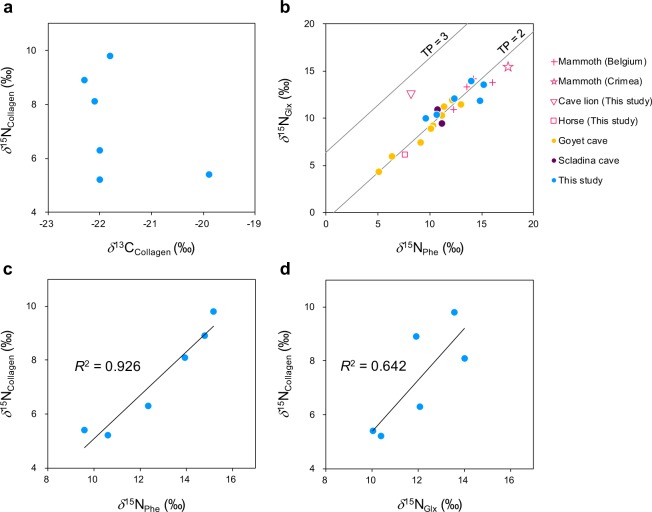
Judging from the C content (26.0–46.1%), N content (9.1–16.2%) and C/N atomic ratio (3.3–3.5), all the cave bear as well as the cave lion and horse bone collagen are well preserved (Table 1)55,56. Large variations in δ13C and δ15N values of bulk collagen for the cave bears were observed: −19.9 to −22.3‰ for δ13C and 5.2 to 9.8‰ for δ15N values, respectively (Fig. 2). The bears sampled from each cave corresponded to different time horizons. Out of six cave bear samples, two have been molecular dated (USR10 from Măgura cave and USR67 from Răsuflătoarei cave), and the other four have been radiocarbon dated (Table 1). Based on calibrated 14C age and the mean of the posterior samples for molecular dated samples, ages of the samples from Măgura fell within the interval 28–31 ka BP, while the oldest bears were from Cioclovina Uscată with 14C ages of c. 43 and 48 cal ka BP. The samples from Răsuflătoarei cover a wider time-span (20–49 ka BP), due to the wide confidence interval depicted based on molecular tip dating, given that USR67 is the only representative within its clade. In each cave and related timespan, both high (above 8‰) and lower δ15N values of bulk collagen have been recorded (see more details in Table 1).
Table 1.
Collagen samples for cave bears from the three Romanian caves and reference fauna analyzed in this study.
| Lab. ID | Common name/Species name | Site | Uncalibrated 14C date/Molecular tip date* (yrs BP) | Calibrated dates (95.4%) (cal BC)/95% credibility interval for molecular estimates* | Lab. number | d13C (‰) | d15N (‰) | %C | %N | C/N atom |
|---|---|---|---|---|---|---|---|---|---|---|
| USR10 | Cave bear/− | Măgura | 30,866* | 28,885-32,518* | — | −22.0 | 6.3 | 26.0 | 9.1 | 3.3 |
| USR11 | Cave bear/− | Măgura | 24,615 ± 101 | 28,392-28,891 | ETH-87394 | −22.1 | 8.1 | 36.8 | 12.3 | 3.5 |
| USR22 | Cave bear/− | Cioclovina Uscată | 40,567 ± 594 | 43,125-45,230 | ETH-87395 | −22.0 | 5.2 | 33.9 | 11.8 | 3.3 |
| USR25 | Cave bear/− | Cioclovina Uscată | 43,119 ± 807 | 44,990-48,401 | ETH-87396 | −22.3 | 8.9 | 36.7 | 12.9 | 3.3 |
| USR65 | Cave bear/− | Răsuflătoarei | 31,005 ± 183 | 34,541-35,333 | ETH-95736 | −21.8 | 9.8 | 38.3 | 13.5 | 3.3 |
| USR67 | Cave bear/− | Răsuflătoarei | 38,313* | 19,932-49,692* | — | −19.9 | 5.4 | 46.1 | 16.2 | 3.3 |
| Reference fauna | ||||||||||
| LOM 16 | Horse /Equus ferus | Lommersum | −20.9 | 4.2 | 38.8 | 13.7 | 3.3 | |||
| Leo 01 | Cave lion /Panthera spelaea | North Sea | 37110 ± 1070 | −19.5 | 8.3 | 36.8 | 13.1 | 3.3 | ||
Figure 2.
Isotope results for the cave bears investigated from the three cave sites in Romania. Scatterplots of: (a) δ13C and δ15N values of bulk collagen; (b) δ15N values of glutamate and phenylalanine. The solid lines indicate the theoretical lines for TP = 2 and TP = 3, respectively, in terrestrial C3-vascular-plant ecosystems. The spacing between these lines indicate the inter trophic enrichment (see Methods). Values for mammoth and cave bears from Belgium were taken from the literature57,58,60; (c) δ15N values of bulk bone collagen and phenylalanine; (d) δ15N values of bulk collagen and glutamate.
Strong correlations between the δ15N values of several AAs and those of bulk collagen were observed for the investigated cave bears, among which phenylalanine showed the strongest one (R2 = 0.926, P < 0.01). Other AA such as hydroxyproline (R2 = 0.704, P < 0.05) also showed strong correlations with bulk collagen δ15N values. The scatterplot presenting δ15NPhe vs. δ15NGlx values shows that some cave bear individuals from the Romanian caves exhibited isotope values similar to those analyzed in previous studies for cave bears from Late Pleistocene Belgian sites, though three individuals with bulk collagen δ15N values above +8‰ analyzed in our study exhibited higher δ15NPhe values than the others without any overlaps. Most relevant for this study are the canonical trophic position (TP; 1 = primary producers, 2 = primary consumers, 3 = secondary consumers, etc…) estimates based on eqn. 1 (see Methods section) indicating that all cave bears, including those with high δ15N values for their bulk collagen, had a TP around 2 (TP = 1.8–2.2), similar to published TP-values for cave bears from other sites (Supplementary Table 1).
Discussion
Feeding behavior of the cave bears based on AA δ15N values
The bulk collagen δ13C and δ15N values are within the range of published data for cave bears from Romanian caves in general (Fig. 3)24,26. Thus, the isotopic results that were obtained on single AAs in the present study are likely to apply also to most other cave bears from Romania, including the ones claimed to have been omnivores, carnivores or piscivores, based on the high δ15N values of their bulk collagen. Our results rule out at least the following possibilities; (i) significant contribution of aquatic resources such as fish to their diets; (ii) carnivory as the main feeding behaviour54. The TP estimates for all individuals based on δ15N of glutamate and phenylalanine were around 2, which rather indicates a highly plant-dependent feeding behavior for all the analyzed cave bears, including those with a high δ15N value of their bulk collagen.
Figure 3.
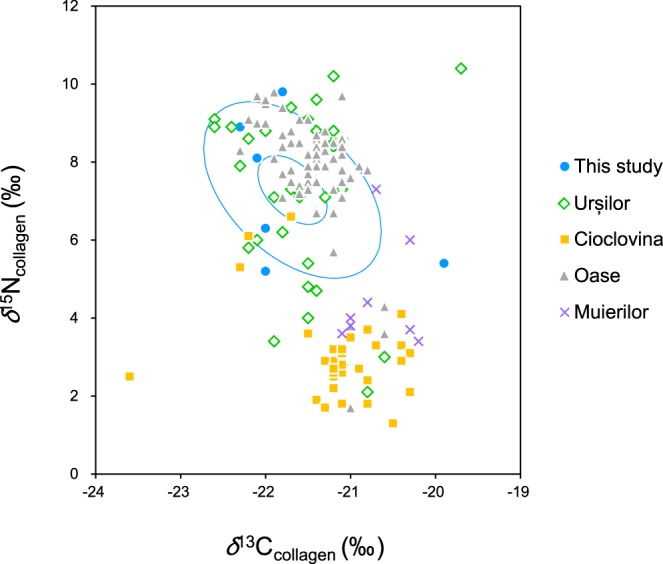
Carbon and nitrogen isotopic composition of bulk collagen for cave bears from the Romanian region. Ellipses indicate 90% and 50% confidence intervals for isotopic compositions of the cave bears analyzed in this study; except for a few individuals like the one with values plotted at the right-upper corner, many cave bears with δ15N value of collagen around 7–8‰ that might be interpreted as an omnivorous/carnivorous signal can be categorized to highly-plant-dependent feeders.
The strongest correlation was observed between the δ15N value of bulk bone collagen and the δ15N value of phenylalanine that represents δ15N of the nitrogen source in a food web. This indicates that the bulk collagen δ15N values of the Romanian cave bears reported here can be explained by the following two possibilities, that are not mutually exclusive: (i) a δ15N shift of the baseline in this local ecosystem, and/or (ii) the consumption of some specific plants with high δ15N values by these cave bear individuals. This means that the reported high δ15N values in the Romanian cave bears’ bulk collagen can be explained by the exclusive consumption of plants rather than by the trophic level effect caused by consumption of animal meat. As shown in Fig. 2, δ15NGlx and δ15NPhe values for some individuals, especially the ones with high bulk δ15N values, overlap with those of strictly herbivorous species, such as late Pleistocene mammoths from other regions, suggesting that a purely herbivorous species could exhibit such carbon and nitrogen isotopic values of bulk collagen in this palaeoenvironmental context57,58.
It was shown that there is currently a potential error of ~0.3 units for the TP estimation calculated by the propagation of errors for each parameter in eqn.159. Considering this error together with a general offset of 0.1–0.2 (accuracy) between calculated TP based on eqn.1 and known TP that were estimated based on well-controlled field observations or laboratory feeding experiments for extant animals, our estimates for all individuals are well within the range of acceptable values for herbivorous mammals. In addition to predatory carnivores, fossils that so far yielded TP-values above 2.3 originate from brown bears, wild boars, foxes and badgers54,60, in contrast to the cave bears with TPs less than 2.2 analyzed in the present study.
The question remains which kind of plants could have been consumed by the cave bears with bulk collagen high δ15N values. Mammoth and some Pleistocene fallow deer have been found to exhibit higher δ15N value and lower δ13C values relative to the other coeval herbivorous species43,44. Some horses from the Late Pleistocene of western Germany also exhibited similar high δ15N and low δ13C values and are interpreted as having the same ecological niche as mammoths61,62. It is conceivable that the cave bears with high bone collagen δ15N values fed on plants, since the high values were also found in species with indisputable herbivorous dietary behavior. Possible candidates of such plants with high bulk δ15N value consumed by cave bears were those consumed by some fallow deer in the Pleistocene44, while those consumed by mammoth such as dry, mature grasses and sedges were less likely based on the lower resistance of cave bear teeth against abrasion. Mushrooms, another plant food with high δ15N values, can be also excluded as an explanation for the high δ15N values of some cave bears for the two following reasons: (1) mushrooms typically have high δ15N and high δ13C values (not low δ13C values), (2) mushrooms are consumers, like animals, and they display a trophic position value higher than 252, therefore their consumption would lead to a TP significantly higher than 2.
One additional factor specific to bears that has to be considered is hibernation, since unlike the other mammalian taxa discussed above, including Homo sapiens, bears hibernate. Indeed, it has been demonstrated that a longer hibernation period caused by climate cooling could result in higher δ15N of cave bear bulk collagen63,64. Although we did not see any statistically significant chronological trend in δ15N values of AAs, only a limited number of individuals have been analyzed so far with this approach. Further investigations on the possible influence of this factor on the δ15N values of individual AAs of Ursids should be performed on more extinct cave bears as well as extant bears in order to clarify how nitrogen metabolism such as urea recycling during hibernation possibly affects the δ15N of AAs in bone collagen65,66. Yet, our data currently does not support a major physiological control of the high bulk collagen δ15N value in bone that remodels intensively during the warm season30.
Evolution of feeding behavior and niche partitioning among cave bear populations
Despite a predominantly herbivorous diet, Romanian cave bears exhibit a very large variation of their isotopic values that probably reflects ecological differences. Such isotopic differences in bone collagen reflect long-term dietary differences, since the turnover of collagen is slow in bone, averaging diet isotopic composition over many years67. It is possible that, in contrast to other regions of Europe, the absence of mammoths played a role in allowing cave bears to have access to a more diverse feeding niche and that competition was stronger among cave bears in Late Pleistocene Romania than in other regions, although an exact contemporaneity between the populations with diverse bulk collagen δ15N values must be examined. If the observed variability in δ15N values of bulk collagen among the Romanian cave bears was the result of high competition among cave bears in a context of a feeding niche restricted to herbivory, it is possible that some individuals specialized on other plants rather than turned to omnivory including animal food resources. Such a dietary strategy is a very unusual case among bears. Assuming that the herbivory for individual cave bears observed in our study was a general trait across all Late Pleistocene cave bear taxa, the most parsimonious interpretation would be the loss of omnivorous feeding behavior after the divergence of cave bears from the brown/polar bear lineage and before the basal divergence of cave bear lineages (e.g., U. ingressus and U. spelaeus; see also low bulk δ15N values for U. kudarensis68). However, it still remains an open question why this extinct animal that could live over a wide geographical range reaching Transbaikalia in Eastern Siberia with diverse ecological conditions within the mammoth steppe ecosystem finally became extinct69,70. It is possible that a dietary adaptation similar to that seen in extant giant panda that consumes large quantities of bamboos but has a macronutrient profile, namely the percent of energy derived from protein, similar to that of carnivores, might have facilitated the full shift for the cave bear lineage towards highly plant-dependent feeding behavior while keeping some ecological plasticity in the mammoth steppe71.
Our findings have interesting implications regarding the evolution of herbivory among carnivorans. The foraging strategy of an animal is largely determined by body mass via energetic needs72. Cave bear populations were likely adapted to their local habitats, with different altitudes being associated with different features, such as body size, tooth morphology, and tooth wear patterns (e.g., ref. 21,33,73). Although the tooth microwear and the isotopic composition of bone collagen reflect the diet of the analyzed individuals at different pre-mortem time periods (e.g., several weeks vs several years or more before the death74), the observed variation in the bulk collagen δ15N values for cave bears in the Romanian region possibly reflected a difference in foraging strategy on plants associated with different body sizes24. Extant Carnivora species with herbivorous feeding behavior (giant panda, red panda and binturong) have smaller body size and live only in temperate and tropical/subtropical Asia, highlighting the paradox of cave bears that had much larger body sizes and lived in a colder and drier environment. Besides, these three extant species are all on very long evolutionary branches, namely they diverged more than 10 million years ago from their closest relatives. Cave bears, in contrast, share a much more recent ancestor within an omnivorous/carnivorous clade, which is another ecological paradox of this extinct taxon. It is possible that the modern herbivorous carnivorans are relicts of a dietary trend that was much more widespread in the past, and that their rarity is rather due to the recent extinction of the large mammalian species rather than to the impossibility for Carnivora to have a diet restricted to plant food. Our results emphasize the need to investigate more in detail the diversity in food sources, especially plants, consumed by cave bears, as a unique case among Carnivora to further understand the benefits, costs and limitations of herbivory.
Conclusions
A highly plant-dependent feeding behavior for cave bears was demonstrated by the TP estimates based on δ15NGlx and δ15NPhe values. The high δ15N of bulk bone collagen of some cave bears can be explained by a higher δ15N baseline of the exploited food chain that is reflected in the δ15NPhe values, namely plants with high δ15N values. Therefore, the high δ15N value of bulk collagen of some cave bears in the Romanian region, a unique feature compared to cave bears from the other regions of Europe analyzed so far, can be explained by the consumption of plants. Further research is needed to clarify which plant species were eaten by those cave bears by using other approaches such as δ13C analysis of individual AAs to identify protein sources75,76. It is also of importance to see if this trend towards herbivory was common for other cave bear populations from broader regions including Asia.
Methods
Samples
The Romanian cave sites of Măgura, Cioclovina and Răsuflătoarei, among others, yielded skeletal remains of cave bears77. Well-preserved bone specimens of cave bears from these three sites allowed us to examine the feeding behavior of the individuals from the Romanian region. Măgura (46.53169, 22.595969) is a 1,500m-long cave located in the eastern part of the Sighiștel valley part of the Apuseni Natural Park; Cioclovina Uscată (45.576628, 23.134228) is a 2,002m-long cave formed in the Early Cretaceous reef limestones of the Luncani karst platform and situated on the foot of the Southern Carpathians, in the Grădiștea-Muncelului Cioclovina Natural Park; Răsuflătoare cave (45.187800, 21.928000) is located in Caraş-Severin County, Banat Mountains and is a part of the Semenic-Cheile Carașului National Park.
Bone collagen from 6 adult cave bears from the three caves was subjected to δ13C and δ15N analysis as well as AA-specific δ15N analysis. One cave lion collagen sample from the North Sea, an area that was terrestrial during the Late Pleistocene and one horse sample from the site of Lommersum (Germany) from the late Pleistocene, have been analyzed together with the cave bear collagen using the same analytical protocol.
Collagen extraction and bulk isotopic analysis
The samples were washed in an ultrasonic bath in acetone, rinsed several times with demineralized water, dried at 35 °C for 72 h and crushed to powder of 0.7 mm grain size. Collagen extraction was performed after ref. 78 with a minor modification as described in ref. 79. In summary, this acid-base-acid method includes a first step of demineralization with 1 M HCl, a second step in 0.125 M NaOH to remove humic acids and lipids, and a final step of gelatinization at pH 2 during 17 h at 100 °C. Isotopic measurements were done using an NC2500 elemental analyzer connected to a Thermo Quest Delta þ XL mass spectrometer. Using the ‘δ’ (delta) value, the isotopic compositions are expressed as follows: δ (‰) ≡ 103 [Rsample/Rstandard − 1], where the R denotes the 13C/12C ratio for carbon and the 15N/14N ratio for nitrogen, with the international reference (standards) being V-PDB for δ13C values and atmospheric nitrogen (AIR) for δ15N values. Isotopic measurements were normalized to δ13C values of USGS24 (δ13C = −16.00‰) and to δ15N values of IAEA 305 A (δ15N = +39.80‰). The reproducibility was ±0.1‰ for δ13C measurements and ±0.2‰ for δ15N measurements, based on one standard-deviation of the mean of multiple analyses of purified collagen from modern bones and international standards.
Analytical conditions for δ15N values of AAs
Bone collagen extracted from the bone samples was hydrolysed with 12 N HCl at 110 °C for 12 h, and derivatized with thionyl chloride/2-propanol (1:4, v/v) at 110 °C for 2 h and subsequently pivaloyl chloride/dichloromethane (1:4, v/v) at 110 °C for 2 h. The δ15N values of the individual AA derivatives were measured at University of Tübingen using the following systems; A gas chromatography/IRMS (GC/IRMS) using an Agilent Technology 7890B GC coupled to an Elementar Isoprime100 IRMS (Elementar, Germany) via combustion and reduction furnaces. AA derivatives were injected into the GC column (Ultra-2, 50 m × 0.32 mm-i.d. 0.52-µm film thickness; Agilent Technology) in splitless mode at 275 °C. The GC oven temperature was programmed as follows: isothermal hold at 40 °C for 3 min; temperature ramp to 110 °C at 15 °C min−1; ramp to 150 °C at 3 °C min−1; ramp to 240 °C at 2.5 °C min−1; ramp to 270 °C at 6 °C min−1; and subsequent holding isothermally at 270 °C for 4 min. Carrier gas (He) flow rate through the GC column was 1.4 ml min−1. The CO2 generated in the combustion furnace was eliminated by a liquid nitrogen trap. Standard mixtures of ten AAs with known δ15N values were injected into the GC/IRMS every two to five runs to confirm reproducibility of the isotope measurements. The precision of the reference mixtures was 0.65–0.90‰. Nitrogen isotopic composition of maximally the following eight AAs was determined as their N2 gas peaks from derivatives showed fine resolution on the GC/IRMS chromatogram: valine (Val), leucine (Leu), isoleucine (Ile), proline (Pro), serine (Ser), glutamate (Glx), phenylalanine (Phe) and hydroxyproline (Hyp). All reported δ15N values for glutamate included the contributions from the α-amino group of glutamic acid and glutamine, as glutamine is converted to glutamic acid during acid hydrolysis. Data for alanine and glycine were not obtained for all samples due to relatively large peak sizes in this study.
Using the equation below based on δ15N of glutamate and phenylalanine, we estimated the trophic position of cave bears in terrestrial C3-plant-based ecosystem: TP = [(δ15NGlx − δ15NPhe − β)/TDF] + 1 (eqn. 1), where δ15NGlx, δ15NPhe, β and TDF indicate δ15N of glutamate, δ15N of phenylalanine and the isotopic offset between glutamate and phenylalanine (δ15NGlx − δ15NPhe value) in primary producers, the trophic discrimination factor, respectively60. The values in the equation are based on the fact that some “source” AAs show little isotopic change through each trophic step (e.g. 0.4 ± 0.4‰ for phenylalanine) whereas other “trophic” AAs fractionate with trophic steps (e.g. 8.0 ± 1.1‰ for glutamate)47–49. We adopted 8.4‰ for the β value for terrestrial C3 vascular plants49,80, and 7.2‰ for the TDF value81, in this study.
Radiocarbon dating
Cave bear collagen was dated directly using AMS radiocarbon dating. Radiocarbon dating was performed by the Labor für Ionenstrahlphysik at Eidgenössische Technische Hochschule—ETH in Zurich (Switzerland). The obtained radiocarbon ages were calibrated using the OxCal v4.2.4 software using the IntCal13 atmospheric curve82,83. All dates were calibrated to BP dates with 2σ (95.4%) probability.
Inferring molecular ages
Ancient DNA extraction and library building
For two samples lacking radiocarbon dates (USR10 and USR67), molecular tip-dating was performed. DNA was extracted from 50 mg of cave bear petrous bones following the protocol of ref. 84, with reduced centrifugation speeds as described in ref. 85. Prior to library building, each DNA extract has been quantified with the Qubit 2.0 fluorometer with high sensitivity reagents (Thermo Fisher Scientific) by using one microlitre out of 25 μL extract. To support the validity of the measurements, positive and negative controls were also measured. Illumina single-stranded libraries were then prepared following the protocol of ref. 86, after the removal of uracil residues and abasic sites. The optimal number of library amplification PCR cycles has been assessed with qPCR as described in ref. 85. A volume of 80 μL including 20 μL template library, Accuprime Pfx DNA polymerase and eight base-pair tailed-primers was used for Indexing PCR to generate dual-indexed library molecules. Amplified libraries were purified using commercial silica spin-columns (Qiagen MinElute). Prior to single-end sequencing on Illumina platforms, library concentration and length distribution were quantified using Qubit 2.0 and 2200 TapeStation (Agilent Technologies), respectively.
Sequencing and bioinformatic analyses
Shotgun sequencing was performed on an Illumina NextSeq 500 sequencing platform. We obtained 75 bp single-end reads using the NextSeq 500/550 High Output Kit v2 (75 cycles) sequencing kit, following the procedures described in ref. 87. Raw reads were trimmed with minimum overlap of one nucleotide and reads shorter than 30 bp were discarded using cutadapt v1.1288. The processed reads were mapped to the reference mitogenome sequence of Ursus spelaeus (Genbank Acc. No. EU327344, ref. 89) with default parameters using the bwa v0.7.15 “aln” algorithm90. The alignment was then filtered for mapping quality (Q > 30), sorted by read position and potential PCR duplicates were removed using SAMtools v0.1.1990.
Mitochondrial genome reconstruction and alignment
ANGSDv0.920 was used to generate consensus sequences for both individuals91. Using the option -doFasta 3 that takes into account bases with highest effective depth92, the mitochondrial consensus has been constructed only considering read mapping and Phred base quality scores >30. The two mitogenomes were aligned to a published alignment of cave bear mitochondrial sequences15. The repetitive section of the d-loop was removed from the alignment because it could not be aligned unambiguously. The final alignment comprised 19 sequences of 16,468 nucleotides, publicly available (Dryad database and GenBank). The GenBank accession numbers of the mitogenomes generated in this study are: MN311249 for USR10 and MN311250 for USR67.
Molecular tip-dating
Ages of the two samples were estimated using Bayesian phylogenetic tip dating93, a method shown to provide accurate age estimates for Late Pleistocene European cave bears15. PartitionFinder v.2.1.1 has been used for data partitioning and substitution model selection94. All substitution models available in BEAST v.1.8.2 were considered and the best partitioning scheme has been computed under the Bayesian Information Criterion using linked branch lengths and the greedy search algorithm (Supplementary Table 2)95.
Tip-dating was performed separately for each sample by analyzing its mitochondrial sequence in BEAST v1.8.2, using median 14C dates as calibration from the dataset in ref. 15 (Supplementary Tables 3). For each tip date its posterior distribution has been sampled using a uniform, uninformative prior of zero to one million yBP. To accommodate changes in effective population size during the time-span of the tree, a piecewise-constant coalescent Bayesian Skyline population model was selected. Unlinked strict molecular clocks were set for each data partition with the posterior distribution of the substitution rate of each sample with a uniform, uninformative prior of zero to 20% mutations per million years. The software Tracer v1.6 was used to assess if the MCMC chain had run for a sufficient length to achieve burn-in and adequate sampling of all parameters (ESS > 200)96. The Maximum Clade Credibility Tree was selected from the posterior sample using TreeAnnotator and visualised in FigTree (Supplementary Fig. 1).
Supplementary information
Acknowledgements
The authors thank Peter Tung and Thomas Wendel of University of Tübingen for their contributions during experiments. We would also like to thank Viorel Traian Lascu, Bogdan Bădescu and Iosif Morac for help during fieldwork. Lastly, we would like to thank the administrations of the Apuseni and Grădiștea Muncelului Cioclovina Natural Parks, and Semenic-Cheile Carașului National Park for access to the caves studied. This study was financially supported by the Alexander von Humboldt Foundation and the Romanian Academy (INM), by the Grant-in-Aid for Scientific Research on Innovative Areas (Grant No. 1802 for FY2016–2020) from MEXT, and the JSPS (Japan Society for the Promotion of Science) Grant-in-Aid for Young Scientists (A) (17H05018) to YIN. Lastly, we acknowledge support by Open Access Publishing Fund of University of Tübingen.
Author contributions
Y.I.N., C.W., D.G.D., M.R., M.V. and I.N.M. performed the experiments; Y.I.N., H.B., I.N.M., M.H. and A.B. analyzed the data; the manuscript was written by Y.I.N. and I.N.M., with input from all authors.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yuichi I. Naito, Email: ynaito@num.nagoya-u.ac.jp
Ioana N. Meleg, Email: ioana.meleg@gmail.com
Supplementary information
is available for this paper at 10.1038/s41598-020-62990-0.
References
- 1.Bocherens H, et al. The last of its kind? Radiocarbon, ancient DNA and stable isotope evidence from a late cave bear (Ursus spelaeus ROSENMÜLLER, 1794) from Rochedane (France) Quat. Int. 2014;339–340:179–188. doi: 10.1016/j.quaint.2013.05.021. [DOI] [Google Scholar]
- 2.Baca, M. et al. Retreat and extinction of the Late Pleistocene cave bear (Ursus spelaeus sensu lato). Sci. Nat. 103 (2016). [DOI] [PMC free article] [PubMed]
- 3.Pacher M, Stuart AJ. Extinction chronology and palaeobiology of the cave bear (Ursus spelaeus) Boreas. 2009;38:189–206. doi: 10.1111/j.1502-3885.2008.00071.x. [DOI] [Google Scholar]
- 4.Terlato G, et al. Chronological and Isotopic data support a revision for the timing of cave bear extinction in Mediterranean. Europe. Hist. Biol. 2018;2963:1–11. [Google Scholar]
- 5.Krause J, et al. Mitochondrial genomes reveal an explosive radiation of extinct and extant bears near the Miocene-Pliocene boundary. BMC Evol. Biol. 2008;8:1–12. doi: 10.1186/1471-2148-8-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christiansen P. What size were Arctodus simus and Ursus spelaeus (Carnivora: Ursidae)? Annales Zoologici Fennici. 1999;36:93–102. [Google Scholar]
- 7.Kurtén, B. The Cave Bears Story: Life and Death of a Vanished Animal. (Columbia University Press, 1976).
- 8.Stiller M, et al. Mitochondrial DNA diversity and evolution of the Pleistocene cave bear complex. Quat. Int. 2014;339–340:224–231. doi: 10.1016/j.quaint.2013.09.023. [DOI] [Google Scholar]
- 9.Barlow A, et al. Partial genomic survival of cave bears in living brown bears. Nat. Ecol. Evol. 2018;2:1563–1570. doi: 10.1038/s41559-018-0654-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mondanaro A, et al. Additive effects of climate change and human hunting explain population decline and extinction in cave bears. Boreas. 2019;48:605–615. doi: 10.1111/bor.12380. [DOI] [Google Scholar]
- 11.Cooper A, et al. Abrupt warming events drove Late Pleistocene Holarctic megafaunal turnover. Science. 2015;349:602–606. doi: 10.1126/science.aac4315. [DOI] [PubMed] [Google Scholar]
- 12.Barnosky AD, Koch PL, Feranec RS, Wing SL, Shabel AB. Assessing the causes of late Pleistocene extinctions on the continents. Science. 2004;306:70–75. doi: 10.1126/science.1101476. [DOI] [PubMed] [Google Scholar]
- 13.Beilman DW, et al. Pattern of extinction of the woolly mammoth in Beringia. Nat. Commun. 2012;3:893. doi: 10.1038/ncomms1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kosintsev P, et al. Evolution and extinction of the giant rhinoceros Elasmotherium sibiricum sheds light on late Quaternary megafaunal extinctions. Nat. Ecol. Evol. 2019;3:31–38. doi: 10.1038/s41559-018-0722-0. [DOI] [PubMed] [Google Scholar]
- 15.Fortes GG, et al. Ancient DNA reveals differences in behaviour and sociality between brown bears and extinct cave bears. Molec. Ecol. 2016;25:4907–4918. doi: 10.1111/mec.13800. [DOI] [PubMed] [Google Scholar]
- 16.Wei F, et al. Progress in the ecology and conservation of giant pandas. Conserv. Biol. 2015;29:1497–1507. doi: 10.1111/cobi.12582. [DOI] [PubMed] [Google Scholar]
- 17.Dietl GP, Flessa KW. Conservation paleobiology: putting the dead to work. Trends Ecol. Evol. 2011;26:30–37. doi: 10.1016/j.tree.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 18.Figueirido B, Palmqvist P, Pérez-Claros JA. Ecomorphological correlates of craniodental variation in bears and paleobiological implications for extinct taxa: an approach based on geometric morphometrics. J. Zool. 2009;277:70–80. doi: 10.1111/j.1469-7998.2008.00511.x. [DOI] [Google Scholar]
- 19.Grandal-d’Anglade A. Bite force of the extinct Pleistocene Cave bear Ursus spelaeus Rosenmüller from Europe. Force de la morsure de l’ours des cavernes Ursus spelaeus Rosenmüller du Pléistocène d’Europe. Comptes Rendus Palevol. 2010;9:31–37. doi: 10.1016/j.crpv.2009.10.003. [DOI] [Google Scholar]
- 20.Peigné S, et al. Predormancy omnivory in European cave bears evidenced by a dental microwear analysis of Ursus spelaeus from Goyet, Belgium. Proc. Natl. Acad. Sci. USA. 2009;106:15390–15393. doi: 10.1073/pnas.0907373106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peigné S, Merceron G. Palaeoecology of cave bears as evidenced by dental wear analysis: a review of methods and recent findings. Hist. Biol. 2019;31:448–460. doi: 10.1080/08912963.2017.1351441. [DOI] [Google Scholar]
- 22.Münzel SC, et al. Behavioural ecology of Late Pleistocene bears (Ursus spelaeus, Ursus ingressus): Insight from stable isotopes (C, N, O) and tooth microwear. Quat. Int. 2014;339–340:148–163. doi: 10.1016/j.quaint.2013.10.020. [DOI] [Google Scholar]
- 23.Stiner MC. Mortality analysis of Pleistocene bears and its paleoanthropological relevance. J. Hum. Evol. 1998;34:303–326. doi: 10.1006/jhev.1997.0198. [DOI] [PubMed] [Google Scholar]
- 24.Robu M, et al. Isotopic evidence for dietary flexibility among European Late Pleistocene cave bears (Ursus spelaeus) Can. J. Zool. 2013;91:227–234. doi: 10.1139/cjz-2012-0222. [DOI] [Google Scholar]
- 25.Robu M, et al. The diverse dietary profiles of MIS 3 cave bears from the Romanian Carpathians: insights from stable isotope (δ13C and δ15N) analysis. Palaeontology. 2018;61:209–219. doi: 10.1111/pala.12338. [DOI] [Google Scholar]
- 26.Richards MP, et al. Isotopic evidence for omnivory among European cave bears: Late Pleistocene Ursus spelaeus from the Peştera cu Oase, Romania. Proc. Natl. Acad. Sci. USA. 2008;105:600–604. doi: 10.1073/pnas.0711063105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Münzel SC, Stiller M, Hofreiter M, Mittnik A, Conard NJ. Pleistocene bears in the Swabian Jura (Germany): Genetic replacement, ecological displacement, extinctions and survival. Quat. Int. 2011;245:225–237. doi: 10.1016/j.quaint.2011.03.060. [DOI] [Google Scholar]
- 28.Bocherens H, et al. Niche partitioning between two sympatric genetically distinct cave bears (Ursus spelaeus and Ursus ingressus) and brown bear (Ursus arctos) from Austria: Isotopic evidence from fossil bones. Quat. Int. 2011;245:238–248. doi: 10.1016/j.quaint.2010.12.020. [DOI] [Google Scholar]
- 29.Bocherens H, Fizet M, Mariotti A. Diet, physiology and ecology of fossil mammals as inferred from stable carbon and nitrogen isotope biogeochemistry: implications for Pleistocene bears. Palaeogeogr. Palaeoclimatol. Palaeoecol. 1994;107:213–225. doi: 10.1016/0031-0182(94)90095-7. [DOI] [Google Scholar]
- 30.Bocherens H. Isotopic tracking of large carnivore palaeoecology in the mammoth steppe. Quat. Sci. Rev. 2015;117:42–71. doi: 10.1016/j.quascirev.2015.03.018. [DOI] [Google Scholar]
- 31.Bocherens H, Grandal-d’Anglade A, Hobson KA. Pitfalls in comparing modern hair and fossil bone collagen C and N isotopic data to reconstruct ancient diets: a case study with cave bears (Ursus spelaeus) Isotopes Environ. Health Stud. 2014;50:291–299. doi: 10.1080/10256016.2014.890193. [DOI] [PubMed] [Google Scholar]
- 32.Bocherens H. Isotopic insights on cave bear palaeodiet. Hist. Biol. 2019;31:410–421. doi: 10.1080/08912963.2018.1465419. [DOI] [Google Scholar]
- 33.Krajcarz M, et al. Isotopic variability of cave bears (δ15N, δ13C) across Europe during MIS 3. Quat. Sci. Rev. 2016;131:51–72. doi: 10.1016/j.quascirev.2015.10.028. [DOI] [Google Scholar]
- 34.Bocherens H, Drucker D. Trophic level isotopic enrichment of carbon and nitrogen in bone collagen: case studies from recent and ancient terrestrial ecosystems. Int. J. Osteoarchaeol. 2003;13:46–53. doi: 10.1002/oa.662. [DOI] [Google Scholar]
- 35.Minagawa M, Wada E. Stepwise enrichment of 15N along food chains: Further evidence and the relation between δ15N and animal age. Geochim. Cosmochim. Acta. 1984;48:1135–1140. doi: 10.1016/0016-7037(84)90204-7. [DOI] [Google Scholar]
- 36.Amundson R, et al. Global patterns of the isotopic composition of soil and plant nitrogen. Global Biogeochem. Cycles. 2003;17:31/1–31/10. doi: 10.1029/2002GB001903. [DOI] [Google Scholar]
- 37.Bowen GJ. Isoscapes: spatial pattern in isotopic biogeochemistry. Annu. Rev. Earth Planet. Sci. 2010;38:161–187. doi: 10.1146/annurev-earth-040809-152429. [DOI] [Google Scholar]
- 38.Stevens RE, Hedges RE. Carbon and nitrogen stable isotope analysis of northwest European horse bone and tooth collagen, 40,000BP–present: Palaeoclimatic interpretations. Quat. Sci. Rev. 2004;23:977–991. doi: 10.1016/j.quascirev.2003.06.024. [DOI] [Google Scholar]
- 39.Bocherens H, Drucker DG, Madelaine S. Evidence for a 15N positive excursion in terrestrial foodwebs at the Middle to Upper Palaeolithic transition in south-western France: Implications for early modern human palaeodiet and palaeoenvironment. J. Hum. Evol. 2014;69:31–43. doi: 10.1016/j.jhevol.2013.12.015. [DOI] [PubMed] [Google Scholar]
- 40.Drucker DG, Bocherens H, Billiou D. Evidence for shifting environmental conditions in Southwestern France from 33000 to 15000 years ago derived from carbon-13 and nitrogen-15 natural abundances in collagen of large herbivores. Earth Planet. Sci. Lett. 2003;216:163–173. doi: 10.1016/S0012-821X(03)00514-4. [DOI] [Google Scholar]
- 41.Drucker D, Bocherens H. Carbon and nitrogen stable isotopes as tracers of change in diet breadth during Middle and Upper Palaeolithic in. Europe. Int. J. Osteoarchaeol. 2004;14:162–177. doi: 10.1002/oa.753. [DOI] [Google Scholar]
- 42.Wißing C, Matzerath S, Turner E, Bocherens H. Paleoecological and climatic implications of stable isotope results from late Pleistocene bone collagen, Ziegeleigrube Coenen, Germany. Quat. Res. 2015;84:96–105. doi: 10.1016/j.yqres.2015.05.005. [DOI] [Google Scholar]
- 43.Bocherens H, et al. Palaeoenvironmental and palaeodietary implications of isotopic biogeochemistry of last interglacial Neanderthal and mammal bones in Scladina Cave (Belgium) J. Archaeol. Sci. 1999;26:599–607. doi: 10.1006/jasc.1998.0377. [DOI] [Google Scholar]
- 44.Bocherens H. Isotopic biogeochemistry and the palaeoecology of the mammoth steppe fauna. Deinsea. 2003;9:57–76. [Google Scholar]
- 45.Kuitems M, et al. Carbon and nitrogen stable isotopes of well-preserved Middle Pleistocene bone collagen from Schöningen (Germany) and their paleoecological implications. J. Hum. Evol. 2015;89:105–113. doi: 10.1016/j.jhevol.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 46.Mccarthy MD, Benner R, Lee C, Fogel ML. Amino acid nitrogen isotopic fractionation patterns as indicators of heterotrophy in plankton, particulate, and dissolved organic matter. Geochim. Cosmochim. Acta. 2007;71:4727–4744. doi: 10.1016/j.gca.2007.06.061. [DOI] [Google Scholar]
- 47.McClelland JW, Montoya JP. Trophic relationships and the nitrogen isotopic composition of amino acids in plankton. Ecology. 2002;83:2173–2180. doi: 10.1890/0012-9658(2002)083[2173:TRATNI]2.0.CO;2. [DOI] [Google Scholar]
- 48.O’Connell TC. ‘Trophic’ and ‘source’ amino acids in trophic estimation: a likely metabolic explanation. Oecologia. 2017;184:317–326. doi: 10.1007/s00442-017-3881-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chikaraishi Y, Ogawa NO, Doi H, Ohkouchi N. 15N/14N ratios of amino acids as a tool for studying terrestrial food webs: a case study of terrestrial insects (bees, wasps, and hornets) Ecol. Res. 2011;26:835–844. doi: 10.1007/s11284-011-0844-1. [DOI] [Google Scholar]
- 50.Chikaraishi Y, et al. Determination of aquatic food-web structure based on compound-specific nitrogen isotopic composition of amino acids. Limnol. Oceanogr. Methods. 2009;7:740–750. doi: 10.4319/lom.2009.7.740. [DOI] [Google Scholar]
- 51.Naito YI, Honch NV, Chikaraishi Y, Ohkouchi N, Yoneda M. Quantitative evaluation of marine protein contribution in ancient diets based on nitrogen isotope ratios of individual amino acids in bone collagen: an investigation at the Kitakogane Jomon site. Am. J. Phys. Anthropol. 2010;143:31–40. doi: 10.1002/ajpa.21287. [DOI] [PubMed] [Google Scholar]
- 52.Steffan SA, et al. Microbes are trophic analogs of animals. Proc. Natl. Acad. Sci. USA. 2015;112:15119–15124. doi: 10.1073/pnas.1508782112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohkouchi N, et al. Advances in the application of amino acid nitrogen isotopic analysis in ecological and biogeochemical studies. Org. Geochem. 2017;113:150–174. doi: 10.1016/j.orggeochem.2017.07.009. [DOI] [Google Scholar]
- 54.Naito YI, et al. An overview of methods used for the detection of aquatic resource consumption by humans: compound-specific delta N-15 analysis of amino acids in archaeological materials. J. Archaeol. Sci. Reports. 2016;6:720–732. doi: 10.1016/j.jasrep.2015.11.025. [DOI] [Google Scholar]
- 55.Ambrose SH. Preparation and characterization of bone and tooth collagen for isotopic analysis. J. Archaeol. Sci. 1990;17:431–451. doi: 10.1016/0305-4403(90)90007-R. [DOI] [Google Scholar]
- 56.DeNiro MJ. Postmortem preservation and alteration of in vivo bone collagen isotope ratios in relation to palaeodietary reconstruction. Nature. 1985;317:806–809. doi: 10.1038/317806a0. [DOI] [Google Scholar]
- 57.Drucker DG, et al. Isotopic analyses suggest mammoth and plant in the diet of the oldest anatomically modern humans from far southeast Europe. Sci. Rep. 2017;7:6833. doi: 10.1038/s41598-017-07065-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Naito YI, et al. Ecological niche of Neanderthals from Spy Cave revealed by nitrogen isotopes of individual amino acids in collagen. J. Hum. Evol. 2016;93:82–90. doi: 10.1016/j.jhevol.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 59.Naito YI, et al. Reply to “Comment on ‘Ecological niche of Neanderthals from Spy Cave revealed by nitrogen isotopes of individual amino acids in collagen.’ [J. Hum. Evol. 93 (2016) 82–90]” [J. Hum. Evol. 117 (2018) 53–55] J. Hum. Evol. 2018;117:56–60. doi: 10.1016/j.jhevol.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 60.Naito YI, et al. Evidence for herbivorous cave bears (Ursus spelaeus) in Goyet Cave, Belgium: implications for palaeodietary reconstruction of fossil bears using amino acid δ15N approaches. J. Quat. Sci. 2016;31:598–606. doi: 10.1002/jqs.2883. [DOI] [Google Scholar]
- 61.Drucker DG, et al. Tracking possible decline of woolly mammoth during the Gravettian in Dordogne (France) and the Ach Valley (Germany) using multi-isotope tracking (13C, 14C, 15N, 34S, 18O) Quat. Int. 2015;359–360:304–317. doi: 10.1016/j.quaint.2014.11.028. [DOI] [Google Scholar]
- 62.Wißing C, et al. Stable isotopes reveal patterns of diet and mobility in the last Neandertals and first modern humans in Europe. Sci. Rep. 2019;9:4433. doi: 10.1038/s41598-019-41033-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grandal d’Anglade, A. & Mosquera, D. F. Hibernation can also cause high δ15N values in cave bears: a response to Richards et al. Proc. Natl. Acad. Sci. USA105, E14; author reply E15 (2008). [DOI] [PMC free article] [PubMed]
- 64.Grandal-d’Anglade A, Pérez-Rama M, García-Vázquez A, González-Fortes GM. The cave bear’s hibernation: reconstructing the physiology and behaviour of an extinct animal. Hist. Biol. 2019;31:429–441. doi: 10.1080/08912963.2018.1468441. [DOI] [Google Scholar]
- 65.Guppy M. The hibernating bear: why is it so hot, and why does it cycle urea through the gut? Trends Biochem. Sci. 1986;11:274–276. doi: 10.1016/0968-0004(86)90022-8. [DOI] [Google Scholar]
- 66.Nakashita R, et al. Ecological application of compound-specific stable nitrogen isotope analysis of amino acids - A case study of captive and wild bears. Reseaches Org. Geochemistry. 2011;27:73–79. [Google Scholar]
- 67.Matsubayashi J, Tayasu I. Collagen turnover and isotopic records in cortical bone. J. Archaeol. Sci. 2019;106:37–44. doi: 10.1016/j.jas.2019.03.010. [DOI] [Google Scholar]
- 68.Bocherens H, Baryshnikov G, Van Neer W. Were bears or lions involved in salmon accumulation in the Middle Palaeolithic of the Caucasus? An isotopic investigation in Kudaro 3 cave. Quat. Int. 2014;339–340:112–118. doi: 10.1016/j.quaint.2013.06.026. [DOI] [Google Scholar]
- 69.Knapp M, et al. First DNA sequences from Asian cave bear fossils reveal deep divergences and complex phylogeographic patterns. Mol. Ecol. 2009;18:1225–1238. doi: 10.1111/j.1365-294X.2009.04088.x. [DOI] [PubMed] [Google Scholar]
- 70.Szpak P, et al. Regional differences in bone collagen δ13C and δ15N of Pleistocene mammoths: Implications for paleoecology of the mammoth steppe. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2010;286:88–96. doi: 10.1016/j.palaeo.2009.12.009. [DOI] [Google Scholar]
- 71.Nie Y, et al. Giant pandas are macronutritional carnivores. Curr. Biol. 2019;29:1677–1682.e1-e2. doi: 10.1016/j.cub.2019.03.067. [DOI] [PubMed] [Google Scholar]
- 72.Nagy KA. Field metabolic rate and food requirement scaling in mammals and birds. Ecol. Monogr. 1987;57:111–128. doi: 10.2307/1942620. [DOI] [Google Scholar]
- 73.Rabeder G, Debeljak I, Hofreiter M, Withalm G. Morphological responses of cave bears (Ursus spelaeus group) to high-alpine habitats. Die Höhle. 2008;59:59–72. [Google Scholar]
- 74.Lidén K, Angerbjörn A. Dietary change and stable isotopes: a model of growth and dormancy in cave bears. Proc. R. Soc. B. 1999;266:1779–1783. doi: 10.1098/rspb.1999.0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smith CI, Fuller BT, Choy K, Richards MP. A three-phase liquid chromatographic method for δ13C analysis of amino acids from biological protein hydrolysates using liquid chromatography-isotope ratio mass spectrometry. Anal. Biochem. 2009;390:165–172. doi: 10.1016/j.ab.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 76.Larsen T, et al. Tracing carbon sources through aquatic and terrestrial food webs using amino acid stable isotope fingerprinting. PLoS One. 2013;8:e73441. doi: 10.1371/journal.pone.0073441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ponta, G. M. L. & Onac, B. P. Cave and Karst Systems of Romania. (Springer, 2019).
- 78.Bocherens H, Billiou D, Bonjean D, Otte M, Mariotti A. Paleobiological implications of the isotopic signatures (13C, 15N) of fossil mammal collagen in Scladina Cave (Sclayn, Belgium) Quat. Res. 1997;48:370–380. doi: 10.1006/qres.1997.1927. [DOI] [Google Scholar]
- 79.Longin R. New method of collagen extraction for radiocarbon dating. Nature. 1971;230:241–242. doi: 10.1038/230241a0. [DOI] [PubMed] [Google Scholar]
- 80.Chikaraishi, Y., Ogawa, N. O. & Ohkouchi, N. Further evaluation of the trophic level estimation based on nitrogen isotopic composition of amino acids. In Earth, Life, and Isotopes (eds. Ohkouchi, N., Tayasu, I. & Koba, K.) 37–51 (Kyoto University Press, 2010).
- 81.Blanke CM, et al. Comparing compound-specific and bulk stable nitrogen isotope trophic discrimination factors across multiple freshwater fish species and diets. Can. J. Fish. Aquat. Sci. 2017;74:1291–1297. doi: 10.1139/cjfas-2016-0420. [DOI] [Google Scholar]
- 82.Ramsey CB, Scott EM, van der Plicht J. Calibration for archaeological and environmental terrestrial samples in the time range 26–50 ka cal BP. Radiocarbon. 2013;55:2021–2027. doi: 10.2458/azu_js_rc.55.16935. [DOI] [Google Scholar]
- 83.Reimer PJ, et al. IntCal13 and Marine13 radiocarbon age calibration curves 0–50,000 years cal BP. Radiocarbon. 2013;55:1869–1887. doi: 10.2458/azu_js_rc.55.16947. [DOI] [Google Scholar]
- 84.Dabney J, et al. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl. Acad. Sci. USA. 2013;110:15758–15763. doi: 10.1073/pnas.1314445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Basler N, et al. Reduction of the contaminant fraction of DNA obtained from an ancient giant panda bone. BMC Res. Notes. 2017;10:754. doi: 10.1186/s13104-017-3061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gansauge M-T, Matthias M. Single-stranded DNA library preparation for the se-quencing of ancient or damaged DNA. Nat. Protoc. 2013;8:737–748. doi: 10.1038/nprot.2013.038. [DOI] [PubMed] [Google Scholar]
- 87.Paijmans, J. L. A. et al. Sequencing single-stranded libraries on the Illumina NextSeq 500 platform. arXiv [q-bio.OT] arXiv. http://arxiv.org/abs/1711.11004 (2017).
- 88.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 89.C. Bon N, et al. Deciphering the complete mitochondrial genome and phylogeny of the extinct cave bear in the Paleolithic painted cave of Chauvet. Proc. Natl. Acad. Sci. USA. 2008;105:17447–17452. doi: 10.1073/pnas.0806143105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler trans-form. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Korneliussen TS, Albrechtsen A, Nielsen R. ANGSD: Analysis of next generation sequencing data. BMC Bioinformatics. 2014;15:356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang Y, Lu J, Yu J, Gibbs RA, Yu F. An integrative variant analysis pipeline for accurate genotype/haplotype inference in population NGS data. Genome Res. 2013;23:833–842. doi: 10.1101/gr.146084.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shapiro B, et al. A Bayesian phylogenetic method to estimate unknown sequence ages. Mol. Biol. Evol. 2011;28:879–887. doi: 10.1093/molbev/msq262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lanfear R, Calcott B, Ho SY, Guindon S. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012;29:1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- 95.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rambaut, A., Suchard, M. A., Xie, D. & Drummond, A. J. Tracer v1.6. (2014).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.