Abstract
Porcine reproductive and respiratory syndrome (PRRS), an economically-important disease caused by PRRS virus (PRRSV), has become endemic to most pig-producing countries. Point mutation and recombination are responsible for genetic heterogeneity, resulting in circulation of genetically-diverse strains. However, no natural recombinant PRRSV has yet been identified in Korea. Here, we successfully isolated natural recombinant PRRSV-2 (KU-N1202) using cell culture, investigated its genomic characteristics, and further evaluated its pathogenicity. KU-N1202 is a recombinant strain between Korean MN184-like and VR-2332-like strains. Specifically, ORF5 to partial ORF7 of the VR-2332-like strain was inserted into the backbone of a CP07-626-2-like strain. KU-N1202 induced mild-to-moderate clinical signs and mild histopathological changes with low viral loads in challenged pigs. Contact pigs showed minimal clinical signs and lower viral loads than those in the challenge group. This study demonstrates the genomic characteristics and pathogenicity of natural recombinant PRRSV-2, illustrating the potential importance of recombination in the field.
Keywords: Pathogenicity, Pig, Porcine reproductive and respiratory syndrome virus, Recombination
Highlights
-
•
A natural recombinant PRRSV-2 virus (KU-N1202) was isolated using cell culture.
-
•
The virus harbored the genes from field strain and VR-2332-like strain.
-
•
KU-N1202 induced mild-to-moderate clinical signs with low viral loads in challenged pig.
-
•
Contact pigs showed minimal clinical signs with relatively low viral loads.
1. Introduction
Porcine reproductive and respiratory syndrome virus (PRRSV) causes a variety of problems including reproductive failure in sows and respiratory distress in growing pigs, which are collectively named porcine reproductive and respiratory syndrome (PRRS). PRRS was almost simultaneously recognized on two different continents (US in the late 1980s and Europe in 1990) (Collins et al., 1992, Wensvoort et al., 1991). Since its emergence, PRRSV has spread widely throughout most pig-producing countries and continues to cause enormous economic losses in the swine industry due to the complexity of the disease. Despite the similar clinical manifestations and nearly simultaneous emergence, the two viruses from different continents exhibit remarkable genetic heterogeneity with approximately 56% nucleotide identity (Zimmerman et al., 2012). Now, two viruses have been considered different species; PRRSV-1 (formerly known as European and type 1) and PRRSV-2 (formerly known as North American and type 2) (Adams et al., 2016, Kuhn et al., 2016). Specifically, PRRSV-1 consists of three subtypes, whereas PRRSV-2 can be classified into nine lineages (Shi et al., 2010a, Shi et al., 2010b). The genome of PRRSV is greater than 15 kb in length and consists of two large nonstructural open reading frames (ORFs) and eight structural ORFs.
In Korea, PRRSV-2 was the only species of PRRSV until 2005, when PRRSV-1 was first identified in a commercial pig herd (Cha et al., 2006, Kim et al., 2006). Currently, both PRRSV-1 and PRRSV-2 co-circulate in Korean pig populations and cause huge economic losses to the swine industry (Kwon et al., 2019, Kwon et al., 2018, Lee et al., 2017). Over the past two decades, Korean PRRSV-2 has expanded its genetic diversity through the continuous evolution and emergence of novel strains, which results in the independent evolution of the virus in Korea (Kwon et al., 2018). Specifically, some of Korean PRRSV-2 strains are phylogenetically related to MN184 strain as well as NADC30 strain, both of which were identified in United States in 2001 and 2008, respectively (Brockmeier et al., 2012, Han et al., 2006). Since 2005, MN184-like strains have emerged and become widespread in Korean pig population (Choi et al., 2013). Korean MN184-like strains are closely related to the MN184 strain and they share identical discontinuous nucleotide deletions within nsp2 (Choi et al., 2014). Korean MN184-like viruses account for a large proportion of PRRSV-2 in Korea and form a unique clade distinct from other MN184-like viruses identified in other countries (Kwon et al., 2018). Since the first commercialization of a PRRSV-2 vaccine in 1997, the use of a modified live vaccine (MLV) has become an important strategy to control PRRSV infection. Especially, routine vaccination using MLV has been carried out in most PRRSV-endemic farms. However, this environment provides favorable condition for two different viruses to infect a single pig, and even a single cell.
Recombination plays a critical role in viral epidemiology and results in the expansion of genetic variation of PRRSV. Evidence regarding the possible involvement of recombination in PRRSV evolution was first documented in 1996 (Kapur et al., 1996). Subsequently, a recombination event was identified following co-infection of two strains of PRRSV-1 (van Vugt et al., 2001) and PRRSV-2 (Yuan et al., 1999) in vitro. The importance of recombination in PRRSV evolution has been highlighted by the emergence of recombinant highly pathogenic PRRSV (HP-PRRSV) in China. After the first emergence of HP-PRRSV, secondary outbreaks of novel HP-PRRSV strains in 2009–2010 were associated with recombination events between two Chinese PRRSV strains (Shi et al., 2013). Interestingly, tertiary outbreaks of HP-PRRSV in 2013 were also related to recombination in the field. After the introduction of a novel strain, NADC30, into China, it became localized and continuously recombined with Chinese strains at a high rate, which resulted in at least five different patterns of recombination (Tian, 2017, Zhao et al., 2015). In addition, possible recombination event between vaccine strains was described (Renson et al., 2017).
We isolated a PRRSV-2 strain from a conventional pig farm reporting signs of respiratory distress and reproductive problems in its animals. The isolate contained large nucleotide deletions in nsp2 regions, which were identical to those of a Korean MN184-like strain but showed a high level of nucleotide identity to VR-2332 in ORF5–ORF7 regions. Despite the increasing amount of information regarding Korean PRRSV, there is no evidence of natural recombinant strains. Therefore, the purpose of this study was to analyze the complete genome sequence and further investigate the pathogenicity of a novel recombinant Korean PRRSV-2 isolate.
2. Materials and methods
2.1. Sample collection
In 2012, blood samples were submitted for laboratory diagnostics from a commercial farrow-to-finisher farm with 220 sows in Gyeonggi-do, Korea. Approximately 25% of weaned pigs in the herd experienced respiratory distress as well as increased mortality. Abortion and early farrowing were observed in 20–30% of late-gestation sows, and the stillbirth rate increased to 30%. Despite the presence of suspicious PRRS clinical signs, PRRSV vaccine had not been administered to the herd.
2.2. Virus isolation
For virus isolation, a filtered serum sample was inoculated into confluent layers of MARC-145 cells. MARC-145 cells were cultured in Dulbecco's Modified Eagle Medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (GenDEPOT, Barker, TX, USA) and antibiotic-antimycotic solutions (Gibco, Grand Island, NY, USA). The cells were maintained at 37 °C in a humidified 5% CO2 incubator. After incubation for 4 days, samples with visible cytopathic effects (CPEs) in greater than 70% of cells were passaged for further analysis. Three rounds of plaque purification were performed in MARC-145 cells to harvest progeny viruses generated from a single infectious viral particle.
2.3. RT-PCR and sequencing analysis
To determine the full-length genome sequence of the novel strain, KU-N1202, viral RNA was extracted from selected plaque-purified clones (passage 10; P10) using Qiazol Lysis Reagent (Qiagen, MD, USA) according to the manufacturer's instructions. Reverse transcription was then performed with gene-specific primers using M-MLV reverse transcriptase (Promega, Madison, WI, USA). PCR was conducted to amplify eight overlapping fragments using Takara Ex Taq (TaKaRa Bio, Shiga, Japan). Primers used in this study were based on a previous study with some modifications (Deng et al., 2015). The 5′ and 3′ ends of genome of the virus were determined using classical rapid amplification of cDNA ends (RACE) (Scotto-Lavino et al., 2006a, Scotto-Lavino et al., 2006b). PCR products from RT-PCR and classical RACE were gel-purified and then directly sequenced in both directions. The complete genome sequence of KU-N1202 in this study was deposited in GenBank under accession number MK057529. Furthermore, ORF2–7 of original serum sample and other five plaque-purified clones were amplified and sequenced to verify the recombination event in the field as well as to exclude any contaminations during cell culture.
2.4. Recombination analysis
The complete genome sequence of KU-N1202 was aligned to those of six Korean strains and two US strains (VR-2332 and MN184C) using MUSCLE. Recombination was detected based on three different methods (Edgar, 2004). First, the complete genome was scanned for possible recombination events using Simplot software v 3.5.1. with a window size of 200 bp and a step size of 20 bp (Lole et al., 1999). Second, the recombination detection program 4 (RDP4) was used to search for statistically significant recombination breakpoints; a set of six recombination detection methods were implemented with the default settings (Martin et al., 2015). Last, putative recombination events were supported by phylogenies of parental regions using the neighbor-joining method with 1000 bootstrapping values in MEGA 6 (Tamura et al., 2013).
2.5. Animal study
The experimental protocol was approved by the Konkuk University Institutional Animal Care and Use Committee. Twelve 3-week-old conventional pigs were obtained from a PRRSV-negative herd with high health status. Pigs were randomly assigned to three groups as follows: six pigs for the challenge group, three for the contact group, and three for the control group. Virus challenge was performed after 1 week of acclimation in experimental facility. The pigs in the challenge group were intranasally administered 4 ml of 104.903 TCID50 ml−1 of P10 virus. Two hours after virus challenge, the contact group was housed with the challenged pigs in the same pen until the end of the study. Pigs from the control group received 4 ml of PBS intranasally. The rectal temperature was recorded daily. All pigs were examined daily for clinical respiratory disease, and were assigned a score that ranged from 0 to 6; other relevant clinical observations were noted separately (Halbur et al., 1996). Blood and nasal swabs were collected individually from all pigs daily, and the swab samples were diluted in 1 ml of sterile phosphate buffered saline (PBS). Pen-based oral fluid samples were collected by pig group, and challenge and contact groups were separated during the collection of oral fluid samples. The samples were centrifuged at 2000×g for 10 min and stored at − 80 °C until further processing. Four pigs (two from the challenge group, one from the contact group, and one from the control group) were euthanized at 3, 5, and 7 days post-infection (dpi). In addition, lung, lymph nodes (submandibular, mesenteric, inguinal, and tracheobronchial), tonsil, spleen, kidney, heart, liver, small intestine, and brain were collected to evaluate the quantity of viral RNA and histopathology.
2.6. Quantitative RT-PCR (qRT-PCR) and histopathology
For all tissues, 0.1 g of sample was diluted 1:10 in sterile PBS, homogenized, and centrifuged at 2000×g for 10 min. Viral RNA was extracted from serum samples, nasal swabs, oral fluid samples, and tissue homogenates using a commercial viral RNA extraction kit. TaqMan probe-based qRT-PCR was performed with primer and probe sets using iTaq™ Universal Probes One-Step Kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The sequences of the primers and probe were as follows: forward primer, 5′-TGCAGGGCTTGTTTGATCTTCC-3′; reverse primer, 5′-CTGAATTTATCCCCGTTGCCTAGAG-3′; probe, 5′-HEX-AGGCACACCCGTCAACCTCGCAGT-BHQ1–3′. Thermal cycling conditions were 10 min at 50 °C, 3 min 95 °C, and 40 cycles of 15 s at 95 °C and 1 min at 60 °C.
For the histopathological examination, lung and tracheobronchial lymph node were fixed in 10% neutral-buffered formalin. After the fixation, tissue samples were embedded in paraffin and stained with hematoxylin and eosin (H&E) stain.
3. Results
3.1. PRRSV diagnosis and isolation
PRRSV-specific RNA was detected from two of 26 sows and 12 of 45 piglets. In addition, the seroprevalence of anti-PRRSV antibodies in sows and piglets on the farm was 100% and 77%, respectively. Preliminary diagnostic sequencing of three selected samples in piglets showed that ORF2–ORF7 shared greater than 99% nucleotide identity among the isolates. KU-N1202 was isolated from one of the three serum samples. The virus continuously induced CPE in MARC-145 cells and was plaque-purified three times.
3.2. Complete genomic characterization of recombinant PRRSV-2
Nucleotide sequences of ORF2–7 of original sample and P10 virus shared 99.9% identity with a nucleotide difference, which was a synonymous mutation at position 48 of ORF7. In addition, other five plaque-purified clone contained one to three nucleotide mutations in ORF2–7 regions, when compared to P10 virus ( Table 1).
Table 1.
Comparison of ORF2–7 sequences of the recombinant virus from original sample, KU-N1202 (passage 10) and other five plaque-purified clones (passage 10).
| Sample | GP2 |
GP5 | M | N | |
|---|---|---|---|---|---|
| 555a | 593a | 101a | 186a | 48a | |
| Serum | C | G | G | C | C |
| KU-N1202 | C | G | G | C | Tb |
| Plaque-purified clone 1 | C | G | G | Tb | C |
| Plaque-purified clone 2 | C | A (G198D)c | G | C | C |
| Plaque-purified clone 3 | C | G | G | C | C |
| Plaque-purified clone 4 | C | G | G | C | C |
| Plaque-purified clone 5 | Tb | G | A (G34D)c | C | C |
Nucleotide position in each gene.
Synonymous mutations.
Non-synonymous mutations.
To determine the genomic characteristics of KU-N1202, its full-length genome was sequenced. The results indicated that the complete genome comprised 15,016 nucleotides (nt), excluding the poly (A) tail. When compared to that of the PRRSV-2 prototype VR-2332, the viral genome was 395 nt shorter. The 5′ UTR of KU-N1202 was 1 nt longer than that of VR-2332, and remarkable nucleotide deletions were found in the nsp2 region of the recombinant virus. The complete nucleotide sequence of the virus was compared to that of other PRRSV-2 strains including VR-2332, MN184C, and Korean isolates. While the recombinant virus shared 85.0% nucleotide identity with VR-2332, the virus was the most similar to the CP07-626-2 strain at the nucleotide level (92.5% identity). KU-N1202 had higher nucleotide identity to recently identified Korean strains than MN184C strain. Nucleotide and amino acid identities between KU-N1202 and reference strains are summarized in Table 2.
Table 2.
Comparisons of genomic sequence identities between porcine reproductive and respiratory syndrome virus KU-N1202 and VR-2332, five Korean PRRSV-2 isolates, and MN184C.
| VR2332 | CP07–626–2 | CA-2 | KNU-12-KJ4 | e417-2 | LMY | MN184C | |
|---|---|---|---|---|---|---|---|
| Nucleotides | |||||||
| Complete | 85.0 | 92.1 | 89.4 | 90.4 | 89.9 | 83.2 | 86.8 |
| 5′ UTR | 92.1 | N.D.a | 90.5 | 95.2 | N.D.a | 91.5 | 90.5 |
| ORF1a | 77.3 | 91.3 | 88.7 | 88.8 | 89.4 | 75.8 | 86.0 |
| ORF1b | 90.7 | 95.1 | 90.1 | 92.5 | 92.0 | 89.0 | 86.3 |
| ORF2 | 93.5 | 90.5 | 88.4 | 90.1 | 92.3 | 94.0 | 90.0 |
| ORF2b | 96.3 | 91.8 | 91.8 | 93.2 | 94.1 | 95.9 | 93.2 |
| ORF3 | 90.0 | 94.1 | 88.4 | 92.5 | 85.0 | 87.0 | 85.2 |
| ORF4 | 88.4 | 95.1 | 94.5 | 94.0 | 86.0 | 87.8 | 86.7 |
| ORF5 | 99.1 | 84.9 | 85.4 | 86.0 | 88.0 | 90.8 | 86.4 |
| ORF5a | 99.3 | 88.4 | 89.1 | 87.8 | 88.4 | 90.3 | 86.5 |
| ORF6 | 99.0 | 92.1 | 92.9 | 91.8 | 95.0 | 96.7 | 92.7 |
| ORF7 | 96.5 | 93.2 | 94.0 | 92.4 | 89.5 | 93.5 | 92.2 |
| 3′ UTR | 93.3 | N.D.a | 97.3 | 99.3 | N.D.a | 92.7 | 96.0 |
| Amino acids | |||||||
| nsp1a | 96.1 | 94.4 | 92.7 | 93.3 | 93.3 | 96.1 | 93.3 |
| nsp1b | 76.3 | 90.1 | 86.6 | 87.1 | 86.6 | 78.3 | 86.6 |
| nsp2 | 67.7 | 88.5 | 83.9 | 83.0 | 84.3 | 65.6 | 82.9 |
| nsp3 | 95.6 | 94.3 | 96.9 | 95.6 | 96.5 | 93.9 | 96.9 |
| nsp4 | 93.1 | 93.6 | 97.5 | 96.5 | 98.0 | 92.1 | 94.6 |
| nsp5 | 85.2 | 91.7 | 91.7 | 92.3 | 91.7 | 86.4 | 84.7 |
| nsp6 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| nsp7a | 93.2 | 95.3 | 94.6 | 96.6 | 95.3 | 92.6 | 89.9 |
| nsp7b | 76.3 | 88.1 | 87.2 | 86.3 | 85.4 | 74.5 | 76.3 |
| nsp8 | 86.6 | 86.6 | 86.6 | 88.8 | 80.0 | 86.6 | 84.4 |
| nsp9 | 95.9 | 98.1 | 96.6 | 97.9 | 96.6 | 95.3 | 95.4 |
| nsp10 | 96.8 | 97.9 | 96.1 | 97.5 | 95.4 | 97.2 | 94.1 |
| nsp11 | 95.5 | 96.4 | 92.8 | 94.1 | 95.0 | 94.6 | 93.2 |
| nsp12 | 93.4 | 96.7 | 92.8 | 96.0 | 94.1 | 90.8 | 93.4 |
| GP2 | 93.7 | 90.2 | 88.6 | 90.2 | 91.7 | 92.5 | 88.2 |
| E | 95.8 | 91.7 | 90.4 | 94.5 | 93.1 | 97.2 | 93.1 |
| GP3 | 88.9 | 92.1 | 87.7 | 91.3 | 84.6 | 87.4 | 84.2 |
| GP4 | 86.5 | 94.3 | 94.9 | 92.6 | 87.0 | 89.3 | 89.8 |
| GP5 | 99.5 | 86.0 | 86.0 | 87.5 | 85.5 | 88.5 | 84.5 |
| ORF5a protein | 100.0 | 78.4 | 80.3 | 76.4 | 80.3 | 92.1 | 78.4 |
| M | 99.4 | 95.9 | 95.9 | 93.6 | 97.7 | 98.8 | 95.9 |
| N | 100.0 | 97.5 | 96.7 | 95.1 | 95.9 | 99.1 | 95.1 |
Not determined in this study because there is no information regarding each 5′ or 3′ end of the reference strains.
KU-N1202 shared the nsp2 deletion signature of MN184C and harbored three additional nucleotide deletions at positions 3013–3015 of VR-2332. These nucleotide deletions resulted in 111, 1, 19, and 1 amino acid deletion patterns, when compared to the corresponding sequences of VR-2332. In contrast to hypervariable regions, residues that play an important role in nsp2/3 proteolytic cleavage in a papain-like protease domain (PLP2), were conserved (Han et al., 2009). The genome of KU-N1202 also encoded a conserved catalytic motif, Cys54 (Gly) and His123 (Trp), and cysteine residues (Cys110, Cys141, and Cys146). The viral residues crucial for trans-cleavage activities, specifically Asp84, Trp85, and Asp88, were identical to those of VR-2332 (Han et al., 2009).
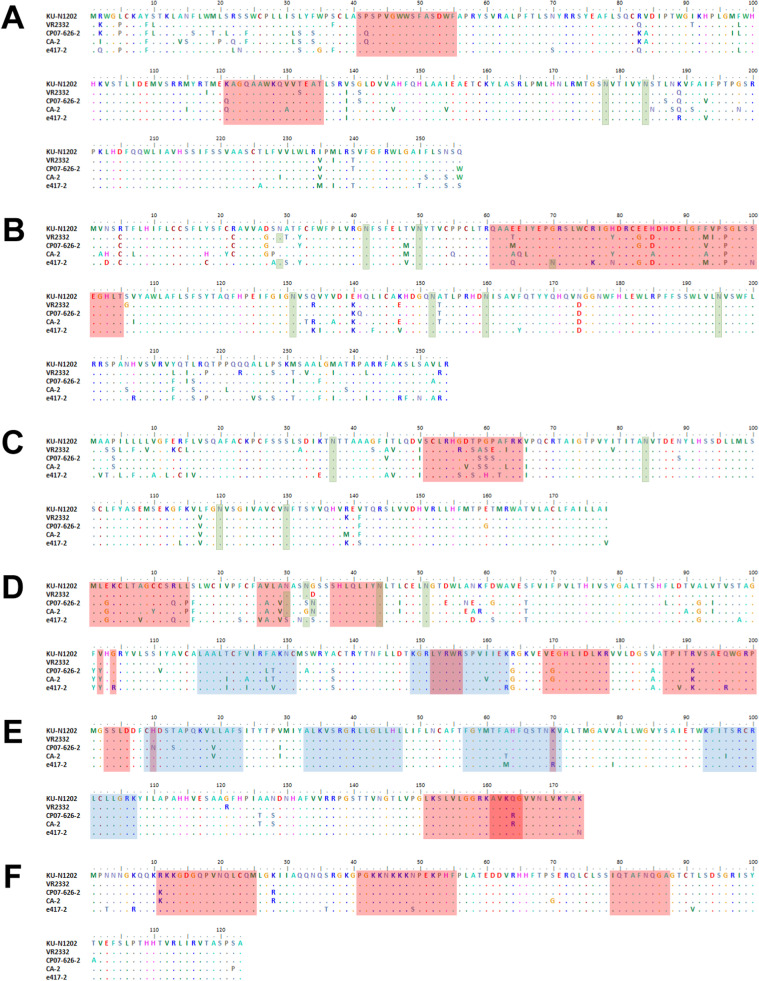
The main distinguishing feature of KU-N1202 was the genetic organization of the structural protein. While minor structural proteins of KU-N1202 were similar to those of Korean field strains, major structural regions of the virus shared high levels of nucleotide and amino acid identity to those of VR-2332. For GP2, the aa 41–55 epitope was identical to that of VR-2332, but the aa 131–145 epitope had an amino acid substitution at position 152. GP3 consists of four consecutive B-cell epitopes at 61–105, including a neutralizing epitope at 61–75 (de Lima et al., 2006). KU-N1202 harbored one, three, four, and three substitutions within 61–75, 71–85, 81–95, and 91–105 epitopes, respectively. The GP4 epitope, aa 51–65, of the virus was also highly variable, when compared to that of VR-2332. For major structural proteins, amino acid sequences of all epitopes were identical to those of VR-2332, including a neutralizing epitope and key residues that participate in neutralizing activity (Fan et al., 2015, Ostrowski et al., 2002). Putative glycosylation analysis indicated that the glycosylation pattern of KU-N1202 was similar to that of VR-2332, but N29 of GP3 was deleted. Amino acid sequences of the structural protein of KU-N1202 are shown in Fig. 1A–F.
Fig. 1.
Alignment of amino acid sequences of porcine reproductive and respiratory syndrome virus KU-N1202 structural proteins. (A) GP2a, (B) GP3, (C) GP4, (D) GP5, (E) M, and (F) N. Red shade, previously identified B-cell epitope; blue shade, previously identified T-cell epitope; green shade, potential glycosylation site.
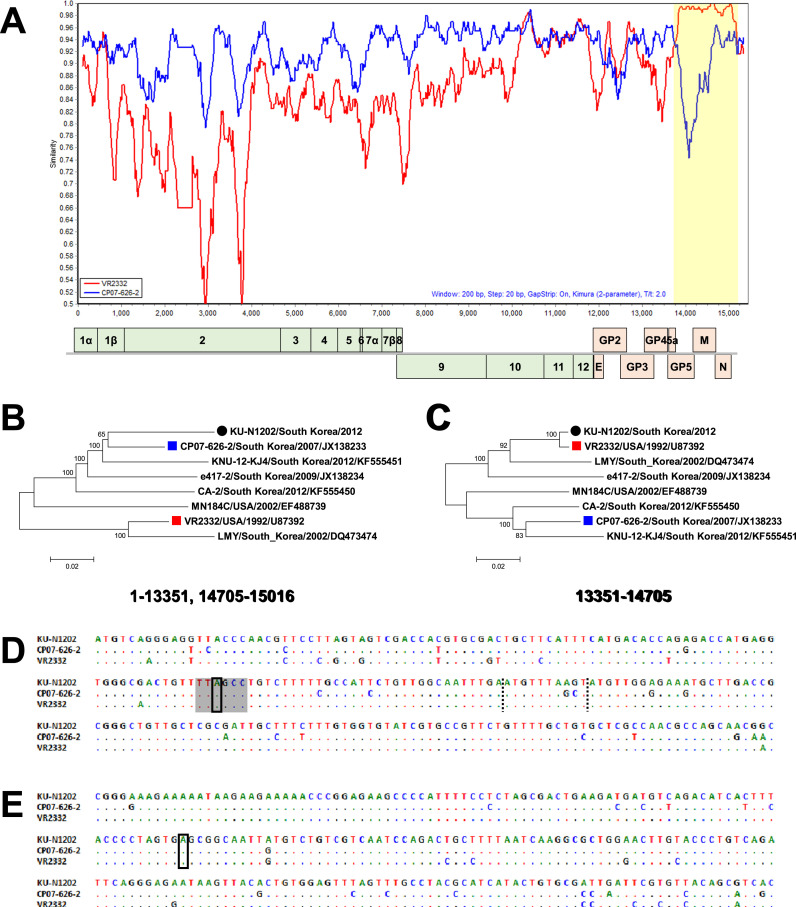
3.3. Recombination analysis
Recombination events within the complete genome of KU-N1202 were detected by three different analytical methods. First, Simplot analysis revealed that KU-N1202 is the result of recombination between CP07–626–2-like and VR-2332-like strains circulating in Korea. In addition, six different methods in the RDP4 program identified KU-N1202 as a recombinant virus (data not shown). Based on these analyses, we identified two recombination breakpoints at positions 13,351 and 14,705 ( Fig. 2A). Results indicated that ORF5 to partial ORF7 of the minor parental strain VR-2332 was inserted into the major parental strain, a CP07–626–2-like strain. Finally, KU-N1202 was differentially positioned based on phylogenies of its parental regions (Fig. 2B and C). Whereas the minor parental region of KU-N1202 shared a close relationship with VR-2332, the virus clustered with a recently isolated Korean PRRSV-2 strain in the phylogenetic tree based on the major parental region.
Fig. 2.
Recombination analyses of porcine reproductive and respiratory syndrome virus KU-N1202. (A) Similarity plot analysis of KU-N1202 (query) compared to CP07–626–2 (blue) and VR-2332 (red). Yellow indicates recombination region between 13,351 and 14,705, which covers ORF5, ORF6, and partial ORF7. Phylogenetic trees of major (B) and minor (C) parental regions. The major parental region of KU-N1202 was found to be related to Korean MN184-like strains, but the minor parental region of KU-N1202 was closely related to VR-2332. The beginning (D) and ending (E) of recombination breakpoints were elucidated. The beginning breaking point (black box) was found to be located in the transcription-regulating sequence (gray shade box) at the 5′ end of sub-genomic RNA 5. Left and right dashes indicate the stop codon of ORF5 and the start codon of ORF6, respectively. The ending breakpoint (black box) was found to be located in the middle region of ORF7.
Furthermore, we analyzed the possible mechanism of recombination that occurred in regions encoding structural genes. Surprisingly, the beginning of the breakpoint was located in a transcription-regulating sequence (TRS) at the 5′ end of sub-genomic RNA 5 (sgRNA5), which mainly encodes GP5 (Fig. 2D). However, the end of the breakpoint was situated in the middle region of ORF7 independent of the TRS region (Fig. 2E).
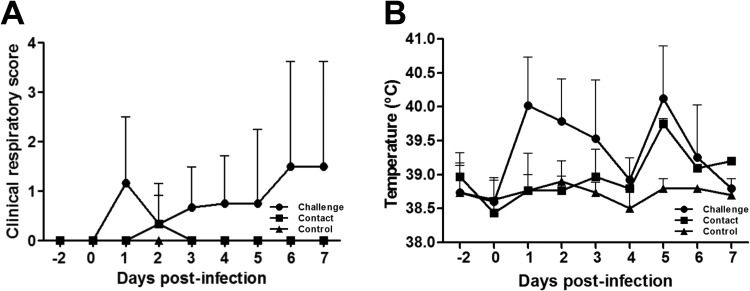
3.4. Clinical signs
The rectal temperature of challenged pigs increased up to 40.0 °C at 1 dpi ( Fig. 3A). Three KU-N1202-challenged pigs exhibited signs of high fever, greater than 40.5 °C. However, the average rectal temperature was not greater than 40.5 °C, defined as a high fever, throughout the study. Three pigs in the challenge group showed mild to moderate respiratory distress after 1 dpi and two additional pigs exhibited mild clinical respiratory signs after 3 dpi (Fig. 3B). Regarding the contact group, one pig showed mild respiratory distress signs only at 2 dpi; however, the other two pigs exhibited no clinical respiratory signs until the end of study. No clinical respiratory distress signs were observed in control pigs, and their rectal temperature remained normal throughout the study.
Fig. 3.
Characteristics of pigs infected with porcine reproductive and respiratory syndrome virus KU-N1202. Mean clinical respiratory score (A) and rectal temperature (B) of challenged (•), contact (■), and control groups (▲).
3.5. Viral loads in serum, nasal swabs, and different tissues
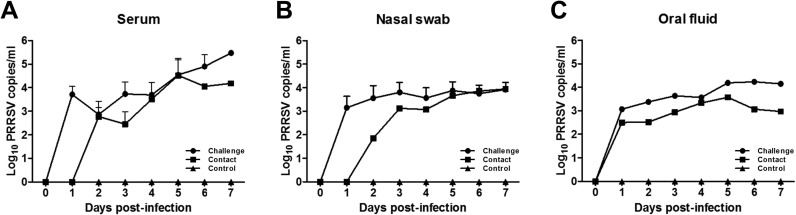
All challenged pigs were viremic throughout the study ( Fig. 4A). PRRSV loads in the challenge group continuously increased and reached the highest levels at the end of the study. In contrast, pig serum in the contact group was negative for PRRSV RNA at 1 dpi, and viral loads were lower than those in the serum of challenged pigs throughout the study. Nasal virus shedding was observed in challenged pigs (Fig. 4B). For the contact group, viral shedding was not detected at 1 dpi, but the load reached similar levels to those in the challenge group by 6 dpi. PRRSV in the pen-based oral fluid of the challenge group exhibited a similar shedding pattern to that observed based on nasal swabs (Fig. 4C). PRRSV RNA was detected in the oral fluid of the contact group from 1 to 7 dpi, but the load was lower than that in the challenge group.
Fig. 4.
Viral loads of porcine reproductive and respiratory syndrome virus KU-N1202 towards pigs. Mean 50% tissue culture infectious doses/ml (log10) of KU-N1202 in serum (A), nasal swab (B), and pen-based oral fluid samples (C).
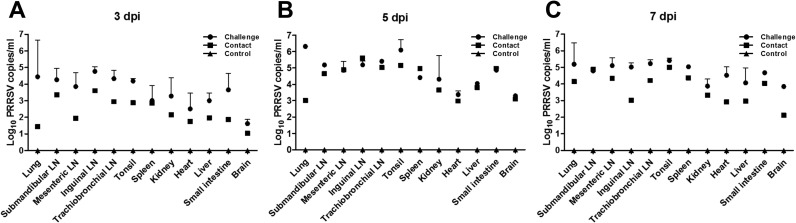
qRT-PCR confirmed PRRSV RNA in all tissues of challenged and contact pigs at the time of the euthanasia, on 3, 5 and 7 dpi. PRRSV loads in different tissues are shown in Fig. 5A–C. For the challenge group, PRRSV loads in the lung, lymph nodes, and tonsil were higher than those in other organs at 3 dpi. At 5 dpi, the viral loads in all organs of the challenged pigs increased and loads in the lung and tonsil significantly increased, up to 103 TCID50/ml. Whereas viral loads in the heart and brain increased, loads in the lung and tonsil of challenged pigs decreased at 7 dpi. Viral loads in various tissues of the contact group were generally lower than those in the challenge group. Peak viral loads in most tissues were detected at 5 dpi, but PRRSV loads in the lung and submandibular lymph node increased until 7 dpi. All tissues in control pigs were negative for PRRSV.
Fig. 5.
Distribution of porcine reproductive and respiratory syndrome virus KU-N1202 in tissues of infected pigs. Mean 50% tissue culture infectious doses/ml (log10) of KU-N1202 in different tissues from pigs that were euthanized at 3 days post-infection (dpi) (A), 5 dpi (B), and 7 dpi (C).
3.6. Histopathology
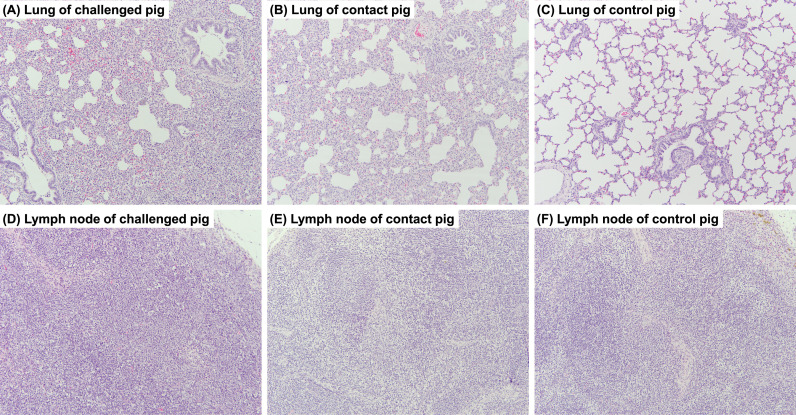
Infection of the recombinant virus induced mild interstitial pneumonia in four challenged pigs, specifically, such as one pig at 3 dpi, one pig at 5 dpi, and two pigs at 7 dpi, respectively. The major histopathological features were alveolar wall thickening, perivascular cuffing, and apoptosis of inflammatory cells in the alveoli ( Fig. 6A–C). In contrast, interstitial pneumonia was not observed in contact and control pigs, and only minimal to mild alveolar wall thickening was found in contact pigs. In addition, mild to moderate lymphoid hyperplasia was found in the lymph nodes of pigs challenged with the recombinant virus (Fig. 6D–F). Specially, lymphoid necrosis in the germinal centers was found in one pig at 5 dpi and two pigs at 7 dpi in the challenge group.
Fig. 6.
Histopathology of porcine reproductive and respiratory syndrome virus KU-N1202-infected pigs that were euthanized at 7 days post-infection. Lung and tracheobronchial lymph node in the challenged pig (A and D), contact pig (B and E), and control pig (C and F), respectively. x100, H&E staining.
4. Discussion
It has been three decades since PRRS was first recognized in the USA and Europe, and the disease has become one of the most economically important diseases worldwide. Beyond its first emergence on two different continents, PRRS spread has become endemic to most pig-producing countries. The continuous evolution of PRRSV has resulted in the recent emergence of novel highly pathogenic variants of both PRRSV-1 in Eastern Europe (Karniychuk et al., 2010), PRRSV-2 in North America (Han et al., 2006) and Asia (Tian et al., 2007, Tong et al., 2007). Rapid evolution is a feature of PRRSV and results from two evolutionary mechanisms, specifically, high mutation rates and recombination, which lead to an expansion of genetic diversity and a gain of viral fitness (Domingo and Holland, 1997, Hanada et al., 2005, Shi et al., 2010a). Vast genetic heterogeneity was initially regarded as the accumulation of point mutations based on the high error rate of viral polymerase. However, the recent emergence of various forms of recombinant HP-PRRSV in China led to an emphasis on importance of recombination during PRRSV evolution. In this study, we successfully isolated a natural recombinant virus that was circulating in a commercial pig herd using a stable cell line and evaluated its pathogenicity in pigs.
First, we determined the complete genome sequence of KU-N1202 to investigate its genetic and recombination characteristics. Three rounds of plaque purification in a stable cell line excluded the possibility of co-infection of two different PRRSV-2 strains and other contamination. Amino acid sequence analysis showed that the virus was closely related to Korean nsp2-deletion isolates, sharing three discontinuous deletions in nsp2 regions (Choi et al., 2014). KU-N1202 also harbored an additional 1-amino acid deletion at position 559 based on nsp2 of VR-2332. All deletions within nsp2 of KU-N1201 occurred in a region that is not essential for virus viability (Han et al., 2007). The most distinct feature of KU-N1202 was the chimeric architecture of structural proteins. Major structural proteins were found to be highly similar to those of VR-2332, but minor structural proteins were closely related to those of Korean strains. Many previous studies have highlighted the important role of structural proteins in PRRSV pathogenesis. A heterodimer of GP5 and M interacts with heparin-like receptor and minor structural proteins attach to the CD163 receptor on porcine alveolar macrophages (PAMs) for viral entry (Das et al., 2010, Delputte et al., 2002). In addition, neutralizing epitopes were found in GP2, GP3, and GP4 of PRRSV-1, and antibodies against these neutralizing epitopes were previously found to effectively reduce the growth of PRRSV in PAMs (Vanhee et al., 2011). GP5 has been considered a major determinant for immunogenicity and the protective properties of PRRSV, as immunization with GP5 in experimental conditions elicits the production of serum-neutralizing antibodies and further, a linear neutralizing epitope was previously identified (Ostrowski et al., 2002, Pirzadeh and Dea, 1998). Recent studies showed that key residues of GP5 and M participate in neutralizing activity against swine polyclonal sera (Fan et al., 2015, Fan et al., 2016, Trible et al., 2015). Given the importance of structural proteins in viral pathogenesis, KU-N1202 successfully utilizes structural proteins from different origins for viral attachment. In addition, it is plausible that the virus might display broad immunogenicity against Korean PRRSV-2 as well as VR-2332-like strains. Further study is needed to investigate the effect of the chimeric composition of structural proteins on viral pathogenesis and immunogenicity.
The order Nidovirales consists of two families, Arterviridae and Coronaviridae, and all viruses within this order produce a 3′ co-terminal nested set of subgenomic mRNAs during infection. The current model regarding the production of this set of sgRNAs proposes that RdRp pauses at the body TRS near the 5′ end of each structural ORF and then jumps to the leader TRS at the 3′ end of the 5′ UTR, guided by the complementarity of the two TRSs (Fehr and Perlman, 2015). It is generally known that the TRS forms a stem-loop structure to generate a nested set of 5′ and 3′ co-terminal sgRNAs. This discontinuous sgRNA transcription strategy resembles a copy-choice mechanism that is a widely accepted mechanism of viral recombination (Kappes and Faaberg, 2015). For this reason, the discontinuous sgRNA transcription strategy is the most plausible reason for the high frequency of recombination in coronaviruses (van Marle et al., 1999). However, recombination breakpoints appear to be random throughout the genome of coronaviruses with little evidence of a role for TRS in recombination (Su et al., 2016). For PRRSV, homologous recombination events were continuously identified, especially in China, and their breakpoints were found to be scattered throughout the PRRSV genome, regardless of any strong tendency for recombination breakpoints. These results implied the occurrence of PRRSV recombination at random breakpoints in the field. However, KU-N1202 had unique recombinant characteristics; specifically, the beginning breakpoint was located at the body TRS of ORF5. This might indicate that the stem-loop structure of TRS acts as a recombination hotspot, at which point RNA-dependent RNA polymerase disjoined and switched the genome of PRRSV.
KU-N1202 is a recombinant virus between a Korean field strain and VR-2332-like strain. The major parental strain of the virus was found to be Korean MN184-like strain, which resulted from independent evolution. The fact that the minor parent strain of KU-N1202 shared 99% nucleotide identity to VR-2332 is remarkable because VR-2332 is a parental strain of the commercial MLV that has been widely used in Korean pig herds. Under these circumstances, the possible mechanisms underlying this recombination event in the field are suggested as follows: 1) vaccination using a commercial MLV in a Korean PRRSV-2-infected pig or 2) co-infection with Korean PRRSV-2 and a reverted vaccine-like virus. Inappropriate vaccination strategies and the use of genetically distant vaccines might contribute to the emergence of recombinant viruses in the field. Furthermore, most diagnostic laboratories use ORF5 sequencing as a common approach to identify PRRSV strains in Korea. If the sample had been submitted to other laboratories, the virus might have been diagnosed as a vaccine-like strain. However, the virus was actually a Korean field strain that recombined with a vaccine-like strain. This point highlights the importance of an advanced method to identify PRRSV strains using broader regions, for example entire structural genes (Kwon et al., 2018).
In this study, mild-to-moderate respiratory clinical signs and an increase in rectal temperature were observed in challenged pigs. A low level of viremia, virus shedding, and viral loads in tissues were also identified in the challenge group. Histopathological findings confirmed the extent of respiratory and lymphoid lesions as mild-to-moderate grade in challenged pigs. Taken together, KU-N1202, a Korean recombinant virus, could be considered a mildly-to-moderately virulent strain, although severe PRRS at a high prevalence was observed in the infected farm. There are various possible explanations for differences in clinical manifestations between experimental and field conditions. One is that PRRSV is one of the most important pathogens responsible for the porcine respiratory disease complex in the field, in which co-infection with other respiratory pathogens exacerbates the severity of clinical signs and pathology in pigs (Opriessnig et al., 2011). In addition, poor housing conditions and poor management might also contribute to respiratory disease in the field. In contrast, our experiment condition minimized the effect of other factors by using PRRSV-negative pigs with high health status and housing them in research facilities which maintained appropriate temperature, uniform light and good air flow with high efficiency particulate air (HEPA) filter system. Another possibility is that the virus used in this study adapted to MARC-145 during plaque-purification, which might contribute to differences in pathogenicity in pigs. Furthermore, it is interesting that pigs in the contact group were healthy throughout the study, although low viremia, low virus shedding, and low virus distribution in tissues were identified. This implied that the recombinant virus could be transmitted between pigs, possibly via nasal secretions, but displays limited capability to induce clinical manifestations in contact pigs. However, in this study, we could not determine the effect of recombination on pathogenicity and transmissibility in pigs because we were not able to isolate some of Korean strains that shared over 98% nucleotide identities. In animal study, other Korean field strains were not included because the involvement of genetically distant virus, not parental strain, could unintentionally mislead about role of recombination in pathogenicity and transmissibility. In this respect, a reverse genetic system could provide important clues regarding the possible relationship between recombination and pathogenicity.
For the first time, this study provided evidence of a PRRSV recombination event in Korean PRRSV and investigated the genomic characteristics and pathogenicity of the recombinant virus. The virus was a natural product of recombination between a Korean field strain and a VR-2332-like strain and its genome consisted of ORF5 to partial ORF7 of the VR-2332-like strain, with the remainder from the Korean MN184-like strain. Animal studies indicated mild-to-moderate pathogenicity in challenged pigs, whereas contact pigs remained healthy with lower viral loads compared to those in challenged pigs. This study contributes to a comprehensive understanding of recombination and associated PRRSV pathogenicity in the field.
References
- Adams M.J., Lefkowitz E.J., King A.M., Harrach B., Harrison R.L., Knowles N.J., Kropinski A.M., Krupovic M., Kuhn J.H., Mushegian A.R., Nibert M., Sabanadzovic S., Sanfacon H., Siddell S.G., Simmonds P., Varsani A., Zerbini F.M., Gorbalenya A.E., Davison A.J. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2016) Arch. Virol. 2016;161:2921–2949. doi: 10.1007/s00705-016-2977-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmeier S.L., Loving C.L., Vorwald A.C., Kehrli M.E., Jr., Baker R.B., Nicholson T.L., Lager K.M., Miller L.C., Faaberg K.S. Genomic sequence and virulence comparison of four Type 2 porcine reproductive and respiratory syndrome virus strains. Virus Res. 2012;169:212–221. doi: 10.1016/j.virusres.2012.07.030. [DOI] [PubMed] [Google Scholar]
- Cha S.H., Choi E.J., Park J.H., Yoon S.R., Song J.Y., Kwon J.H., Song H.J., Yoon K.J. Molecular characterization of recent Korean porcine reproductive and respiratory syndrome (PRRS) viruses and comparison to other Asian PRRS viruses. Vet. Microbiol. 2006;117:248–257. doi: 10.1016/j.vetmic.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Choi E.-J., Lee C.-H., Song J.-Y., Song H.-J., Park C.-K., Kim B., Shin Y.-K. Genetic diversity of porcine reproductive and respiratory syndrome virus in Korea. J. Vet. Sci. 2013;14:115. doi: 10.4142/jvs.2013.14.2.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H.W., Nam E., Lee Y.J., Noh Y.H., Lee S.C., Yoon I.J., Kim H.S., Kang S.Y., Choi Y.K., Lee C. Genomic analysis and pathogenic characteristics of Type 2 porcine reproductive and respiratory syndrome virus nsp2 deletion strains isolated in Korea. Vet. Microbiol. 2014;170:232–245. doi: 10.1016/j.vetmic.2014.02.027. [DOI] [PubMed] [Google Scholar]
- Collins J.E., Benfield D.A., Christianson W.T., Harris L., Hennings J.C., Shaw D.P., Goyal S.M., McCullough S., Morrison R.B., Joo H.S. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J. Vet. Diagn. Investig. 1992;4:117–126. doi: 10.1177/104063879200400201. [DOI] [PubMed] [Google Scholar]
- Das P.B., Dinh P.X., Ansari I.H., de Lima M., Osorio F.A., Pattnaik A.K. The minor envelope glycoproteins GP2a and GP4 of porcine reproductive and respiratory syndrome virus interact with the receptor CD163. J. Virol. 2010;84:1731–1740. doi: 10.1128/JVI.01774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lima M., Pattnaik A.K., Flores E.F., Osorio F.A. Serologic marker candidates identified among B-cell linear epitopes of Nsp2 and structural proteins of a North American strain of porcine reproductive and respiratory syndrome virus. Virology. 2006;353:410–421. doi: 10.1016/j.virol.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Delputte P.L., Vanderheijden N., Nauwynck H.J., Pensaert M.B. Involvement of the matrix protein in attachment of porcine reproductive and respiratory syndrome virus to a heparinlike receptor on porcine alveolar macrophages. J. Virol. 2002;76:4312–4320. doi: 10.1128/JVI.76.9.4312-4320.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M.C., Chang C.Y., Huang T.S., Tsai H.J., Chang C., Wang F.I., Huang Y.L. Molecular epidemiology of porcine reproductive and respiratory syndrome viruses isolated from 1991 to 2013 in Taiwan. Arch. Virol. 2015;160:2709–2718. doi: 10.1007/s00705-015-2554-4. [DOI] [PubMed] [Google Scholar]
- Domingo E., Holland J.J. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan B., Liu X., Bai J., Zhang T., Zhang Q., Jiang P. The amino acid residues at 102 and 104 in GP5 of porcine reproductive and respiratory syndrome virus regulate viral neutralization susceptibility to the porcine serum neutralizing antibody. Virus Res. 2015;204:21–30. doi: 10.1016/j.virusres.2015.04.015. [DOI] [PubMed] [Google Scholar]
- Fan B., Liu X., Bai J., Zhang T., Zhang Q., Jiang P. Influence of the amino acid residues at 70 in M protein of porcine reproductive and respiratory syndrome virus on viral neutralization susceptibility to the serum antibody. Virol. J. 2016;13:51. doi: 10.1186/s12985-016-0505-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbur P.G., Paul P.S., Meng X.J., Lum M.A., Andrews J.J., Rathje J.A. Comparative pathogenicity of nine US porcine reproductive and respiratory syndrome virus (PRRSV) isolates in a five-week-old cesarean-derived, colostrum-deprived pig model. J. Vet. Diagn. Investig. 1996;8:11–20. doi: 10.1177/104063879600800103. [DOI] [PubMed] [Google Scholar]
- Han J., Liu G., Wang Y., Faaberg K.S. Identification of nonessential regions of the nsp2 replicase protein of porcine reproductive and respiratory syndrome virus strain VR-2332 for replication in cell culture. J. Virol. 2007;81:9878–9890. doi: 10.1128/JVI.00562-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Rutherford M.S., Faaberg K.S. The porcine reproductive and respiratory syndrome virus nsp2 cysteine protease domain possesses both trans- and cis-cleavage activities. J. Virol. 2009;83:9449–9463. doi: 10.1128/JVI.00834-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Wang Y., Faaberg K.S. Complete genome analysis of RFLP 184 isolates of porcine reproductive and respiratory syndrome virus. Virus Res. 2006;122:175–182. doi: 10.1016/j.virusres.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Hanada K., Suzuki Y., Nakane T., Hirose O., Gojobori T. The origin and evolution of porcine reproductive and respiratory syndrome viruses. Mol. Biol. Evol. 2005;22:1024–1031. doi: 10.1093/molbev/msi089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappes M.A., Faaberg K.S. PRRSV structure, replication and recombination: origin of phenotype and genotype diversity. Virology. 2015 doi: 10.1016/j.virol.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur V., Elam M.R., Pawlovich T.M., Murtaugh M.P. Genetic variation in porcine reproductive and respiratory syndrome virus isolates in the midwestern United States. J. Gen. Virol. 1996;77(Pt 6):1271–1276. doi: 10.1099/0022-1317-77-6-1271. [DOI] [PubMed] [Google Scholar]
- Karniychuk U.U., Geldhof M., Vanhee M., Van Doorsselaere J., Saveleva T.A., Nauwynck H.J. Pathogenesis and antigenic characterization of a new East European subtype 3 porcine reproductive and respiratory syndrome virus isolate. BMC Vet. Res. 2010;6:30. doi: 10.1186/1746-6148-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-Y., Lee S.-Y., Sur J.H., Lyoo Y.S. Serological and genetic characterization of the European strain of the porcine reproductive and respiratory syndrome virus isolated in Korea. Korean. J. Vet. Res. 2006;46:363–370. [Google Scholar]
- Kuhn J.H., Lauck M., Bailey A.L., Shchetinin A.M., Vishnevskaya T.V., Bao Y., Ng T.F., LeBreton M., Schneider B.S., Gillis A., Tamoufe U., Diffo Jle D., Takuo J.M., Kondov N.O., Coffey L.L., Wolfe N.D., Delwart E., Clawson A.N., Postnikova E., Bollinger L., Lackemeyer M.G., Radoshitzky S.R., Palacios G., Wada J., Shevtsova Z.V., Jahrling P.B., Lapin B.A., Deriabin P.G., Dunowska M., Alkhovsky S.V., Rogers J., Friedrich T.C., O'Connor D.H., Goldberg T.L. Reorganization and expansion of the nidoviral family Arteriviridae. Arch. Virol. 2016;161:755–768. doi: 10.1007/s00705-015-2672-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon T., Yoo S.J., Lee D.U., Sunwoo S.Y., Je S.H., Park J.W., Kim M.H., Park C.K., Lyoo Y.S. Differential evolution of antigenic regions of porcine reproductive and respiratory syndrome virus 1 before and after vaccine introduction. Virus Res. 2019;260:12–19. doi: 10.1016/j.virusres.2018.11.004. [DOI] [PubMed] [Google Scholar]
- Kwon T., Yoo S.J., Sunwoo S.Y., Lee D.U., Je S.H., Park J.W., Park C.K., Lyoo Y.S. Independent evolution of porcine reproductive and respiratory syndrome virus 2 with genetic heterogeneity in antigenic regions of structural proteins in Korea. Arch. Virol. 2018 doi: 10.1007/s00705-018-4048-7. [DOI] [PubMed] [Google Scholar]
- Lee D.U., Yoo S.J., Kwon T., Je S.H., Shin J.Y., Byun J.J., Kim M.H., Lyoo Y.S. Genetic diversity of ORF 4-6 of type 1 porcine reproductive and respiratory syndrome virus in naturally infected pigs. Vet. Microbiol. 2017;199:54–61. doi: 10.1016/j.vetmic.2016.12.026. [DOI] [PubMed] [Google Scholar]
- Lole K.S., Bollinger R.C., Paranjape R.S., Gadkari D., Kulkarni S.S., Novak N.G., Ingersoll R., Sheppard H.W., Ray S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D.P., Murrell B., Golden M., Khoosal A., Muhire B. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1:vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opriessnig T., Gimenez-Lirola L.G., Halbur P.G. Polymicrobial respiratory disease in pigs. Anim. Health Res. Rev. 2011;12:133–148. doi: 10.1017/S1466252311000120. [DOI] [PubMed] [Google Scholar]
- Ostrowski M., Galeota J.A., Jar A.M., Platt K.B., Osorio F.A., Lopez O.J. Identification of neutralizing and nonneutralizing epitopes in the porcine reproductive and respiratory syndrome virus GP5 ectodomain. J. Virol. 2002;76:4241–4250. doi: 10.1128/JVI.76.9.4241-4250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirzadeh B., Dea S. Immune response in pigs vaccinated with plasmid DNA encoding ORF5 of porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 1998;79(Pt 5):989–999. doi: 10.1099/0022-1317-79-5-989. [DOI] [PubMed] [Google Scholar]
- Renson P., Touzain F., Lebret A., Le Dimna M., Quenault H., Normand V., Claude J.B., Pez F., Rose N., Blanchard Y., Bourry O. Complete genome sequence of a recombinant porcine reproductive and respiratory syndrome virus strain from two genotype 1 modified live virus vaccine strains. Genome Announc. 2017;5 doi: 10.1128/genomeA.00454-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotto-Lavino E., Du G.W., Frohman M.A. 3′ end cDNA amplification using classic race. Nat. Protoc. 2006;1:2742–2745. doi: 10.1038/nprot.2006.481. [DOI] [PubMed] [Google Scholar]
- Scotto-Lavino E., Du G.W., Frohman M.A. 5′ end cDNA amplification using classic race. Nat. Protoc. 2006;1:2555–2562. doi: 10.1038/nprot.2006.480. [DOI] [PubMed] [Google Scholar]
- Shi M., Holmes E.C., Brar M.S., Leung F.C.C. Recombination is associated with an outbreak of novel highly pathogenic porcine reproductive and respiratory syndrome viruses in China. J. Virol. 2013;87:10904–10907. doi: 10.1128/JVI.01270-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., Lam T.T., Hon C.C., Hui R.K., Faaberg K.S., Wennblom T., Murtaugh M.P., Stadejek T., Leung F.C. Molecular epidemiology of PRRSV: a phylogenetic perspective. Virus Res. 2010;154:7–17. doi: 10.1016/j.virusres.2010.08.014. [DOI] [PubMed] [Google Scholar]
- Shi M., Lam T.T., Hon C.C., Murtaugh M.P., Davies P.R., Hui R.K., Li J., Wong L.T., Yip C.W., Jiang J.W., Leung F.C. Phylogeny-based evolutionary, demographical, and geographical dissection of North American type 2 porcine reproductive and respiratory syndrome viruses. J. Virol. 2010;84:8700–8711. doi: 10.1128/JVI.02551-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian K. NADC30-Like porcine reproductive and respiratory syndrome in China. Open Virol. J. 2017;11:59–65. doi: 10.2174/1874357901711010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian K., Yu X., Zhao T., Feng Y., Cao Z., Wang C., Hu Y., Chen X., Hu D., Tian X., Liu D., Zhang S., Deng X., Ding Y., Yang L., Zhang Y., Xiao H., Qiao M., Wang B., Hou L., Wang X., Yang X., Kang L., Sun M., Jin P., Wang S., Kitamura Y., Yan J., Gao G.F. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One. 2007;2:e526. doi: 10.1371/journal.pone.0000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong G.Z., Zhou Y.J., Hao X.F., Tian Z.J., An T.Q., Qiu H.J. Highly pathogenic porcine reproductive and respiratory syndrome, China. Emerg. Infect. Dis. 2007;13:1434–1436. doi: 10.3201/eid1309.070399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trible B.R., Popescu L.N., Monday N., Calvert J.G., Rowland R.R. A single amino acid deletion in the matrix protein of porcine reproductive and respiratory syndrome virus confers resistance to a polyclonal swine antibody with broadly neutralizing activity. J. Virol. 2015;89:6515–6520. doi: 10.1128/JVI.03287-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marle G., Dobbe J.C., Gultyaev A.P., Luytjes W., Spaan W.J.M., Snijder E.J. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc. Natl. Acad. Sci. 1999;96:12056–12061. doi: 10.1073/pnas.96.21.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vugt J.J., Storgaard T., Oleksiewicz M.B., Botner A. High frequency RNA recombination in porcine reproductive and respiratory syndrome virus occurs preferentially between parental sequences with high similarity. J. Gen. Virol. 2001;82:2615–2620. doi: 10.1099/0022-1317-82-11-2615. [DOI] [PubMed] [Google Scholar]
- Vanhee M., Van Breedam W., Costers S., Geldhof M., Noppe Y., Nauwynck H. Characterization of antigenic regions in the porcine reproductive and respiratory syndrome virus by the use of peptide-specific serum antibodies. Vaccine. 2011;29:4794–4804. doi: 10.1016/j.vaccine.2011.04.071. [DOI] [PubMed] [Google Scholar]
- Wensvoort G., Terpstra C., Pol J.M., ter Laak E.A., Bloemraad M., de Kluyver E.P., Kragten C., van Buiten L., den Besten A., Wagenaar F. Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet. Q. 1991;13:121–130. doi: 10.1080/01652176.1991.9694296. [DOI] [PubMed] [Google Scholar]
- Yuan S., Nelsen C.J., Murtaugh M.P., Schmitt B.J., Faaberg K.S. Recombination between North American strains of porcine reproductive and respiratory syndrome virus. Virus Res. 1999;61:87–98. doi: 10.1016/S0168-1702(99)00029-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Ye C., Chang X.B., Jiang C.G., Wang S.J., Cai X.H., Tong G.Z., Tian Z.J., Shi M., An T.Q. Importation and recombination are responsible for the latest emergence of highly pathogenic porcine reproductive and respiratory syndrome virus in China. J. Virol. 2015;89:10712–10716. doi: 10.1128/JVI.01446-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman J.J., Benfield D.A., Scott A.D., Murtaugh M.P., Stadejek T., Stevenson G.W., Torremorell M. Porcine reproductive and respiratory syndrome virus (Porcine arterivirus) In: Zimmerman M.P., Karriker L.A., Ramirez A., Schwartz K.J., Stevenson G.W., editors. Diseases of Swine. 10th ed. Wiley-Blackwell; 2012. pp. 387–418. [Google Scholar]