Abstract
Limited sampling means that relatively little is known about the diversity and evolutionary history of mammalian members of the Hepadnaviridae (genus Orthohepadnavirus). An important case in point are shrews, the fourth largest group of mammals, but for which there is limited knowledge on the role they play in viral evolution and emergence. Here, we report the discovery of a novel shrew hepadnavirus. The newly discovered virus, denoted shrew hepatitis B virus (SHBV), is divergent to be considered a new species of Orthohepadnavirus. Phylogenetic analysis revealed that these viruses were usually most closely related to TBHBV (tent-making bat hepatitis B virus), known to be able to infect human hepatocytes, and had a similar genome structure, although SHBV fell in a more basal position in the surface protein phylogeny. In sum, these data suggest that shrews are natural hosts for hepadnaviruses and may have played an important role in their long-term evolution.
Keywords: Hepadnaviruses, Shrews, Phylogeny, Evolution, Cross-species transmission
Highlights
-
•
A highly divergent hepadnavirus was identified in shrews in China.
-
•
The shrew virus represents a novel species of mammalian orthohepadnaviruses.
-
•
The shrew virus grouped with TBHBV in some genes, previously shown to be able to infect human hepatocytes.
-
•
Cross-species virus transmission occurred among the three shrew species.
1. Introduction
Hepadnaviruses (family Hepadnaviridae) are characterized by partially circular, double-stranded DNA genomes (~3.2 kb in length). Interestingly, the replication of these viruses requires an RNA intermediate produced by reverse transcription (Urban et al., 2010). At present, the Hepadnaviridae are classified into three genera: Avihepadnavirus, Orthohepdnavirus and one unassigned genus (https://talk.ictvonline.org/taxonomy/). As their names imply, avihepadnaviruses have been identified in birds (e.g. ducks and parrots) while orthohepadnaviruses are associated with mammals, including human hepatitis B virus (HBV). The unassigned genus contains two hepadnavirus species found in fish - white sucker hepatitis B virus and bluegill hepatitis B virus - and a single virus associated with the Tibetan frog (Tibetan frog hepatitis B virus). Recently, highly divergent non-enveloped viruses that are the sister-group to the hepadnaviruses, denoted the nackednaviruses, were discovered in fish (Lauber et al., 2017). Despite their diversity, all members of the mammalian genus Orthohepadnavirus contain four overlapped open reading frames (ORFs): polymerase (P), core protein (C), surface protein (S) and X protein. The large S mRNA is thought to encode three proteins - the preS1, preS2 and S. The preS1 is a domain involved in virus-host interaction via cell receptors (Neurath et al., 1986).
HBV is an important and globally distributed viral pathogen affecting approximately 257 million people worldwide and causing around 887,000 deaths annually, mostly as a consequence of cirrhosis and hepatocellular carcinoma (http://www.who.int/en/news-room/fact-sheets/detail/hepatitis-b). While the majority of human pathogens causing emerging infectious diseases have an animal origin (Wolfe et al., 2007), the host range of the known orthohepadnavirus is limited and the evolutionary history of orthohepadnavirus is therefore unclear. Although rodents comprise approximately 42% of living mammalian species (Wilson and Reeder, 2005), to date only ground squirrels (genus Marmota) appear to be natural rodent hosts of the virus (Summers et al., 1978, Marion et al., 1980, Testut et al., 1996). Similarly, bats comprise around 20% of living mammalian species and are also natural hepadnavirus reservoirs (He et al., 2013, He et al., 2015, Drexler et al., 2013, Wang et al., 2017a, Nie et al., 2018). Although a divergent orthohepadnavirus was recently identified in domestic cats (cat hepadnavirus, CHV) (Aghazadeh et al., 2018), the only other mammalian species that appear to be natural hosts of orthohepadnavirus are primates, including divergent viruses present in the woolly monkey (WMHBV) (Lanford et al., 1998, Rasche et al., 2016) and capuchin monkey (CMHBV) (de Carvalho Dominguez Souza et al., 2018). Importantly, a phylogenetically distinct virus harboured by tent-making bats (TBHBV) has the ability to infect human hepatocytes (Drexler et al., 2013). Interestingly, those viruses harboured by rodents, woolly monkey, capuchin monkey and tent-making bat were all identified in New World species (Rasche et al., 2016).
Shrews are small, mouse/mole-like mammals that belong to the family Soricidae (Mammalia: Eulipotyphla) that have a worldwide distribution, and are also one of the largest and most abundant mammalian groups (Wilson and Reeder, 2005). While rodents and bats are recognised as important hosts of zoonotic pathogens, including lassavirus (Smith and Wang, 2013), filoviruses (Smith and Wang, 2013), hantaviruses (Guo et al., 2013) and coronaviruses (Wang et al., 2015, Lin et al., 2017, Woo et al., 2012), relatively little is known about the role played by shrews in shaping virus diversity, evolution and transmission. Recently, several viruses - arenaviruses (Li et al., 2015), coronaviruses (Wang et al., 2017b), hantaviruses (Lin et al., 2014, Radosa et al., 2013, Zhang, 2014) and rotaviruses (Li et al., 2016) - have been discovered in shrews. The shrew arenavirus is particularly important as it is considered a zoonotic pathogen that could infect humans causing fever and respiratory symptoms (Blasdell et al., 2016). Hence, shrews may be key hosts for a diversity of RNA viruses of which only a tiny proportion have been sampled to date (Wang et al., 2017b), but have been largely neglected as reservoirs for infectious agents.
In this study we screened shrew samples collected in Zhejiang and Hubei provinces, China, for the presence of hepadnaviruses. From the data obtained, we further explored the evolutionary history of orthohepadnaviruses based on the discovery of a phylogenetically distinct shrew virus.
2. Results
2.1. Novel hepadnavirus identified in shrews
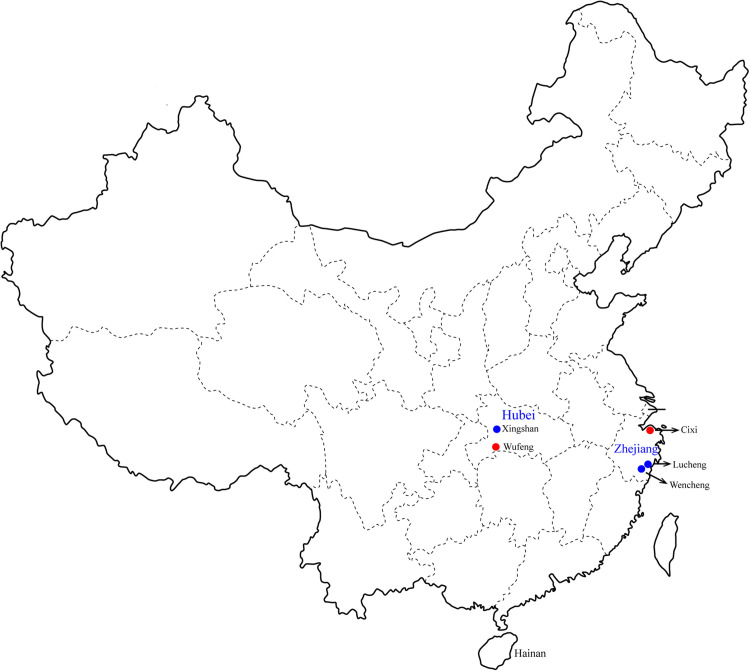
During 2013–2018, 449 shrews, comprising four species from three genera, were collected from Wufeng county of Hubei province, China, as well as two localities from Zhejiang province - Cixi county, Longwan district Lin et al. (2014) and Wencheng county ( Fig. 1). A total of 200 Anourosorex squamipes and 39 Crocidura attenuata were captured from Wufeng, eight Crocidura attenuata from Xingshan, 32 Crocidura lasiura from Cixi, and 60 and 110 Suncus murinus from Longwan and Wencheng, respectively ( Table 1). Shrews were identified to the species level by sequencing of the mitochondrial b (mt-cyt b) gene following morphological identification. Liver tissues were collected from all sampled animals to screen for hepadnaviruses by amplifying a 360-bp fragment of the hepadnavirus polymerase (P) gene. Of 449 shrews screened, eight were found to be positive for hepadnavirus, with a total detection rate of 1.8%. Separated by species, the viral positive rate was 2.5% in Anourosorex squamipes (Wufeng, Hubei province), 4.3% in Crocidura attenuate (Wufeng, Hubei province), and 3.1% in Crocidura lasiura (Cixi, Zhejiang province) (Table 1). Interestingly, the virus was only identified at two sampling sites, Wufeng and Cixi, with positive rates of 2.9% and 3.1%, respectively. Five complete genomes were successfully recovered from the hepadnavirus positive samples collected in Cixi (Cixi-Cl-88) and Wufeng (Wufeng-Ca-70, Wufeng-Ca-111, Wufeng-As-178, Wufeng-As-205) based on host species and sampling sites (Table 1).
Fig. 1.
A map of China illustrating the location of trap sites (circles) in which shrews were captured. Virus positive sites are marked in red and negative sites in blue.
Table 1.
Prevalence of hepadnaviruses in shrews by species and geographic location in China.
| Species |
Location |
|||||
|---|---|---|---|---|---|---|
|
Hubei province |
Zhejiang province |
Total (%) | ||||
| Wufeng | Xingshan | Cixi | Longwan | Wencheng | ||
| Anourosorex squamipes | 5/200 | – | – | – | – | 5/200 (2.5) |
| Crocidura attenuata | 2/39 | 0/8 | – | – | – | 2/47 (4.3) |
| Crocidura lasiura | – | – | 1/32 | – | – | 1/32 (3.1%) |
| Suncus murinus | 0/60 | 0/110 | 0/170 (0) | |||
| Total (%) | 7/239 (2.9) | 0/8 (0) | 1/32 (3.1%) | 0/60 (0) | 0/110 (0) | 8/449 (1.8) |
Note: ‘‘-’’ indicates that no animals were captured.
Notably, no hepadnavirus was detected in Suncus murinus (Asian house shrews), although 170 of these animals were captured and screened (Table 1). Moreover, other rodent species from the same district captured during the same years and from the same geographic were also screened for hepadnaviruses, but none were positive (data not shown). Clearly, however, larger-scale sampling is needed to accurately document the prevalence of hepadnaviruses in shrews and rodents.
2.2. Phylogenetic relationships between SHBV, known hepadnaviruses and their hosts
Although these viruses were discovered in three species of shrews at two distant sample sites (~1200 km), they were relatively closely to each other, with 91.3–99.8% nucleotide sequence similarity at the genome level, and 90.8–99.8%, 93.2–100%, 92.7–99.9% and 92.4–99.8% sequence similarity in the polymerase, core, surface and X genes, respectively. In marked contrast, these shrew viruses exhibited more than 50% genome divergence compared to all known hepadnaviruses (Table 3). Together with the phylogenetic analyses (see below), these results showed that the shrew viruses discovered in this study were distinct from the known orthhepadnaviruses, such that they represent a new member of the genus Orthohepadnavirus. Following the naming tradition of a new orthohepadnaviruses we named these as representatives of shrew hepatitis B virus (SHBV) based on its host association.
Table 3.
Genomic similarity (%; upper triangle) and divergence (%; lower triangle) of SHBV (bold) and other orthohepadnaviruses.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HBV_ayw | 78.1 | 61.6 | 61.0 | 63.2 | 62.3 | 63.4 | 61.9 | 57.9 | 57.8 | 57.8 | 57.8 | 57.9 | |
| 2 | WMHBV | 26.0 | 62.3 | 61.2 | 63.0 | 63.3 | 64 | 62.1 | 57.8 | 57.8 | 57.7 | 57.9 | 58.5 | |
| 3 | WHV | 53.9 | 52.5 | 58.8 | 62.3 | 62.0 | 63.0 | 60.5 | 57.3 | 57.2 | 57.3 | 57.3 | 56.9 | |
| 4 | TBHBV | 55.2 | 54.8 | 59.7 | 60.1 | 60.6 | 60.8 | 58.5 | 59.1 | 59.1 | 59.1 | 59.1 | 59.1 | |
| 5 | RBHBV | 50.7 | 51.4 | 52.7 | 57.1 | 73.5 | 70.4 | 63.7 | 56.6 | 56.6 | 56.7 | 56.7 | 56.9 | |
| 6 | PBHBV | 52.6 | 50.5 | 53.3 | 56.0 | 32.8 | 71.3 | 63.8 | 56.1 | 56.1 | 56.1 | 56.1 | 56.5 | |
| 7 | LBHBV | 50.3 | 49.3 | 51.2 | 55.6 | 37.8 | 36.3 | 65.1 | 57.5 | 57.4 | 57.5 | 57.5 | 58.0 | |
| 8 | CHV | 53.7 | 53.2 | 56.4 | 60.6 | 50.1 | 49.6 | 47.0 | 55.0 | 54.9 | 55.0 | 55.0 | 54.9 | |
| 9 | Wufeng-As-178 | 62.2 | 62.2 | 63.4 | 59.2 | 65.4 | 66.3 | 63.2 | 69.3 | 99.8 | 99.5 | 99.7 | 91.3 | |
| 10 | Wufeng-As-205 | 62.4 | 62.3 | 63.6 | 59.2 | 65.4 | 66.4 | 63.3 | 69.4 | 0.2 | 99.6 | 99.7 | 91.3 | |
| 11 | Wufeng-Ca-70 | 62.4 | 62.5 | 63.3 | 59.2 | 65.2 | 66.4 | 63.1 | 69.2 | 0.5 | 0.4 | 99.7 | 91.5 | |
| 12 | Wufeng-Ca-111 | 62.4 | 62.1 | 63.3 | 59.2 | 65.2 | 66.4 | 63.0 | 69.2 | 0.3 | 0.3 | 0.3 | 91.5 | |
| 13 | Cixi-Cl-88 | 62.2 | 60.6 | 64.5 | 59.1 | 64.7 | 65.3 | 61.9 | 69.2 | 9.3 | 9.3 | 9.1 | 9.1 |
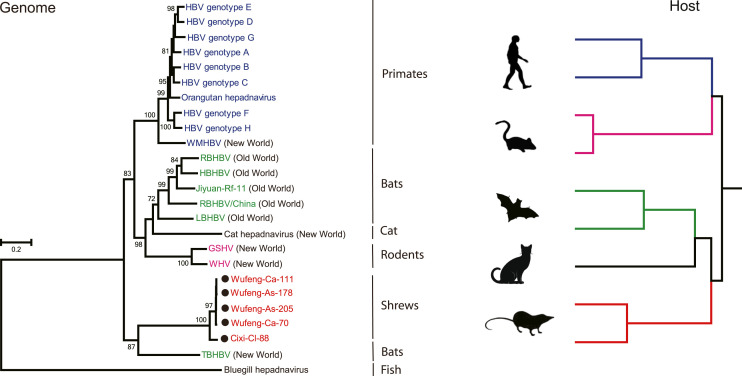
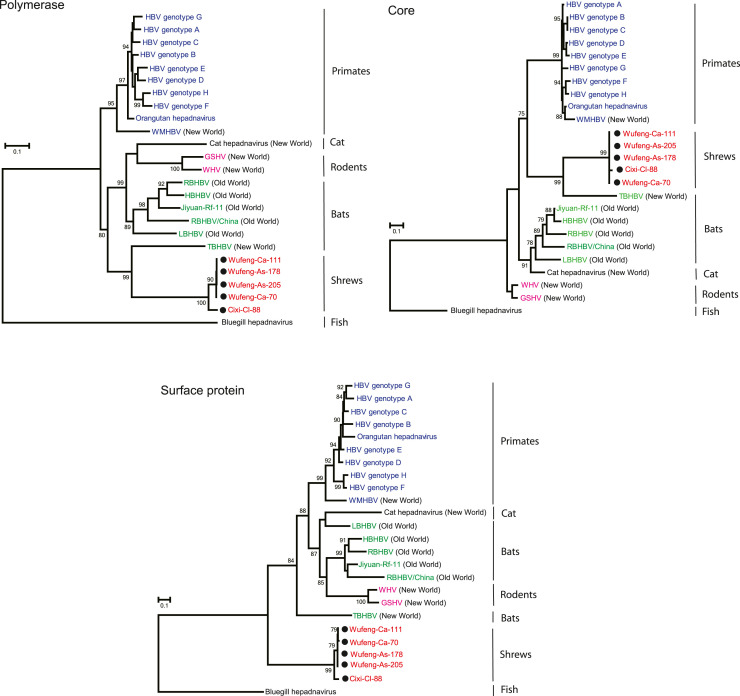
To better understand the evolutionary relationship of SHBV with other orthohepadnaviruses, we estimated phylogenetic trees of full length of genome, the polymerase, surface protein and core protein using a Maximum Likelihood (ML) method ( Fig. 2 and Fig. 3) and utilizing the hepadnavirus from bluegill (Dill et al., 2016) as an outgroup as it is the virus most closely related to the orthohepadnaviruses. In the full genome tree (Fig. 2), SHBVs and other orthohepadnaviruses largely clustered according to their host order (with the exception of bats) and could be grouped into six lineages: (i) primate, (ii) bat (Old World), (iii) cat, rodent, (iv) shrew, and (v) Tent-making bat (New World). Notably, the novel shrew hepadnaviruses formed a distinct clade within the orthohepadnaviruses, and grouped with TBHBV sampled from New World bats (Uroderma bilobatum) in Panama (Drexler et al., 2013) with strong bootstrap support, although separated by a long branch. Additionally, the phylogenies based on both polymerase and core genes had the same grouping as seen in the whole genome, with SHBV forming a clade with TBHBV (Fig. 3). The only tree in which SHBV exhibited a different phylogenetic position was that for the surface protein, in which SHBV and TBHBV did not group together, with the shrew viruses falling in a well-supported basal phylogenetic position (Fig. 3). Although this topological movement is suggestive of inter-genic recombination, no statistically significant evidence for recombination was found using the RDP and Simplot recombination analyses (Fig. S4). The conflicting signals among gene trees may in part reflect the short alignment lengths and the highly divergent sequences involved. Finally, it is notable that the newly discovered shrew hepadnaviruses formed two lineages according to their geographic origins rather than their host species (Fig. 3), indicative of a local cross-species transmission event.
Fig. 2.
Phylogenetic history of the virus genome sequences and of their vertebrate hosts. On the virus phylogeny, the sequences obtained in this study were marked with solid circles. Bootstrap support values (>70%) are shown at relevant nodes. The scale bar depicts the number of amino acid substitutions per site.
Fig. 3.
Phylogenetic history of the amino acid sequences of the P, C and S genes of SHBV inferred using an ML method. The sequences obtained in this study were marked with solid circles. Bootstrap support values (>70%) are shown at relevant nodes. The scale bar depicts the number of amino acid substitutions per site. Note the divergent position of SHBV in the S gene tree.
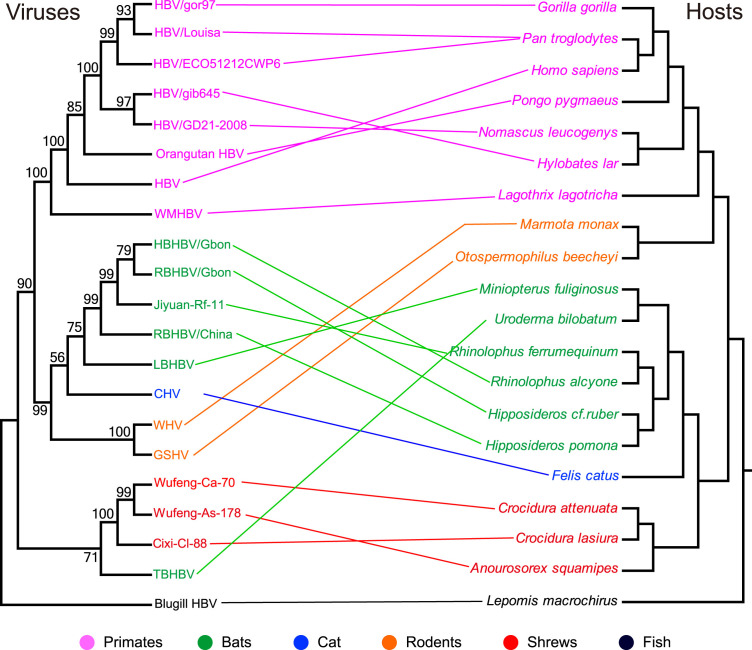
The orthohepadnavirus phylogenies also provided evidence for a combination of virus-host co-divergence and cross-species transmission. As an example of the latter, closely related SHBV were identified in three species of shrews that belong to two genera in two shrew subfamilies (Anourosorex squamipes, genus Anourosorex, subfamily Soricinae; Crocidura attenuate and Crocidura lasiura, genus Crocidura, subfamily Crocidurinae), indicative of cross-species virus transmission at the level of family or genus. However, at the level of host order, the topology of these viruses was in part similar to that of their hosts (Fig. 2). To better understand the association of the viruses and their hosts, we inferred a virus-host tanglegram ( Fig. 4). This reveals relatively frequent cross-species virus transmission both the family and order levels. Similarly, a co-phylogenetic analysis of all orthohepadnaviruses and their hosts using two methods (a distance-based method and a heuristic event-based method) revealed that co-divergence was evident overall (p < 0.02, copycat), but that cross-species events occurred frequently on this background of long-term virus-host associations (p < 0.01) (Fig. S1).
Fig. 4.
Tanglegram presenting the evolution associations between orthohepadnaviruses (with bluegill hepadnavirus as an outgroup) and their hosts. The virus tree was estimated using the genome sequences (left) and the host tree was based on topology implied in the web of Time tree of life (http://www.timetree.org/).
2.3. Genome features of the newly identified and known orthohepadnaviruses
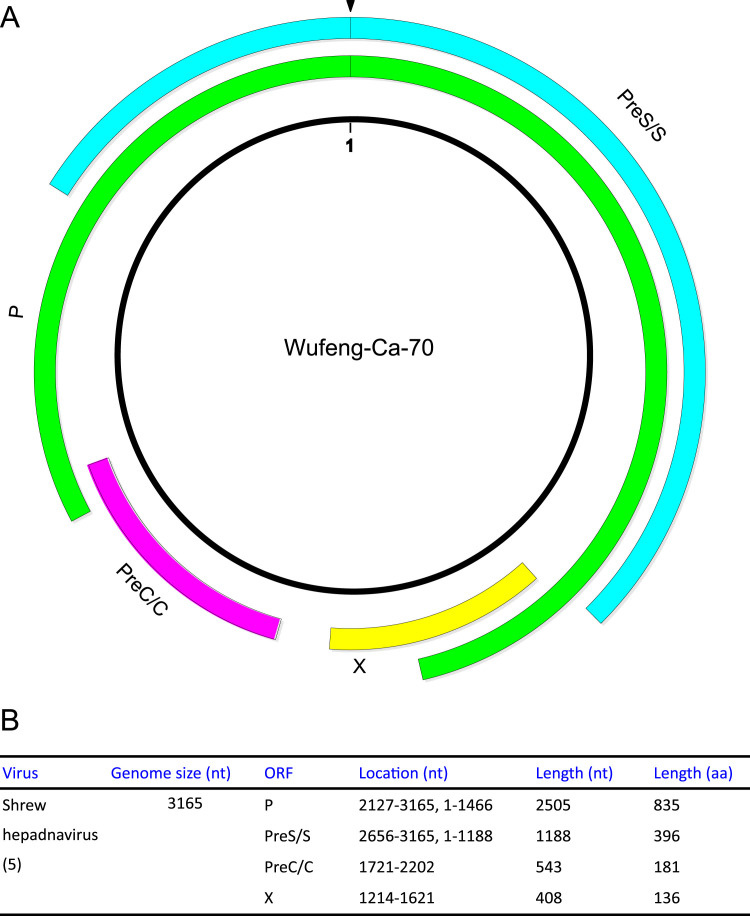
The key genomic features of the newly identified shrew hepadnaviruses are described in Table 2 and Fig. 5. The genomic structure of the shrew hepadnaviruses were similar to that of other known orthohepadnaviruses with four open reading frames (ORFs) (Polymerase, Surface gene, Core gene and X gene) and a low G+C content of 45.9–46.1%. In addition, direct repeats (DRs) (Table 2) and the conservative YMDD were also found in the genomes.
Table 2.
Key genome features of the novel shrew hepadnaviruses described here.
| Host (#) | Genome size (nt) | Pol (nt) | PreS/S (nt) | preC/C (nt) | X (nt) | G+C (%) |
|---|---|---|---|---|---|---|
| Primate (10) | 3179–3248 | 2499–2538 | 1170–1203 | 636–675 | 459–465 | 48.25–49.52 |
| Rodent (3) | 3302–3323 | 2634–2655 | 1284–1296 | 654–675 | 417–426 | 43.43–44.48 |
| Bat (5) | 3230–3368 | 2562–2709 | 1200–1338 | 654 | 426–435 | 48.02–53.48 |
| Shrew (5) | 3165 | 2505 | 1188 | 543 | 408 | 45.88–46.10 |
| Tent-making bat (4) | 3149 | 2484 | 1146 | 663 | 408 | 48.46–48.62 |
| Cat (1) | 3187 | 2517 | 1149 | 657 | 438 | 50.96 |
Note: ”#” indicates the number of sequences used.
Fig. 5.
Genome structure characteristics of SHBV. A: ORFs are indicated by colours with polymerase (green), surface protein (cyan), core protein (purple) and X protein (yellow). Numbering starts at the junction of preS1 and preS2 domains in the surface protein. B: Genome features of SHBV. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Variations in genome and gene length are apparent within the orthohepadnaviruses. The genome length, polymerase gene length and surface gene length of SHBV fell between those of TBHBV and primate HBV (Table 2). It was notable that the SHBV had the shortest core gene (543) and X gene (408) compared to all other known orthohepadnaviruses. Compared with RBHBV and HBHBV, SHBV exhibited similar amino acid deletions with primate HBV and TBHBV, with three gaps (positions 228–242, 287–300 and 310–330) in the polymerase, two (position 29–49 and 106–146) in surface protein, and one (position 181–189) in the core protein (Fig. S2).
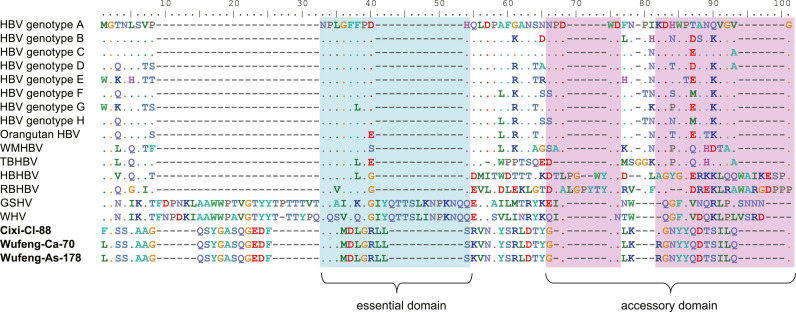
Given that TBHBV has the ability to infect human hepatocytes (Drexler et al., 2013), and that SHBV often grouped with TBHBV in the phylogeny, we further analysed the potential ability of SHBV to infect humans by comparing the sequence of pre-S1 (in the N terminal of large surface protein) which is documented to be the essential domain to bind to its receptor (Neurath et al., 1986, Drexler et al., 2013). However, SHBV showed fewer conserved amino acid residues with primate HBV than TBHBV in main receptor binding region ( Fig. 6). Specifically, the shrew viruses had only three (NPLXXXXXX) and four amino acid residues (XXDWD and XXXXXXXXXXXXXG) that were conserved with primate viruses in the essential domain (NPLGFFPDH) and adjacent accessory domains (NPDWD and NKDHWPEANKVGVG). Additionally, TBHBV and orangutan HBV exhibited more conserved amino acid residues than SHBV, with two and one mutant residues in essential domain, respectively. Notably, although there were some differences between SHBV and HBV in the "a" determinant region (aa 99–172), SHBV had no mutation at key cysteine residues (aa 121, 124, 137, 138, 139, 147 and 149) which play a crucial role associated with the entry of HBV into human hepatocytes (Abou-Jaoude and Sureau, 2007). Clearly, however, additional work is needed to determine whether SHBV is able to infect human hepatocytes.
Fig. 6.
Comparison of the essential and accessory domain in preS1. Essential (green) and accessory (reddish) domains are compared among the hepadnaviruses from primates, rodents, bats and shrews. The first amino acid “M” for genotypes A to H is at amino acid position 12 based on preS1 protein. The exception of genotype D, for which PreS1 is 11 amino acids shorter. The viruses documented in this study are shown in bold.
2.4. Discussion
We identified a novel hepadnavirus, denoted shrew hepatitis B virus (SHBV), in three species of shrews collected in Zhejiang and Hubei provinces, China. SHBV had the same genome structure as other orthohepadnaviruses, but was highly divergent compared to any known members of this genus, with around 50% nucleotide differences. According to the proposals for the delineation of recent bat hepadnaviruses species, viruses with greater than 20% genome divergence and a new host species may represent new viral species. Hence, the SHBV identified in this study is sufficiently genetically divergent to be considered a new species of the genus Orthohepadnavirus.
Orthohepadnaviruses clearly have a complex evolutionary history. Although co-divergence and cross-species transmission have both been reported in primate hepadnaviruses (Geoghegan et al., 2017, Rasche et al., 2016, Locarnini et al., 2013, Littlejohn et al., 2016), both in this and our previous study of bat hepadnaviruses (Nie et al., 2018) revealed that cross-species transmission was commonplace. The SHBVs newly identified here were found in three shrew species from two subfamilies (Fig. 2). Notably, they clustered according to their geographic origins rather than their host species, again indicative of the cross-species hepadnavirus transmission in shrews. Indeed, our additional analysis of all known hepadnaviruses revealed cross-species transmission at both the family and order levels. Thus, like RNA viruses (Shi et al., 2016, Shi et al., 2018), the evolutionary history of hepadnaviruses is likely characterized by the occurrence of cross-species virus transmission on a background of virus-host co-divergence.
As WMHBV, WHV, ASHV, GSHV and TBHBV were all identified from New World animal species, with WMHBV located basally to other primate HBVs, it has been argued that orthohepadnaviruses may have originated in the New World (Rasche et al., 2016). However, the SHBV identified in this study were from shrews captured in China (Old World) and fell as a sister group to TBHBV in the genome phylogeny. This argues against a New World origin, as does the very deep phylogenetic position of the newly identified cat hepadnavirus identified in Australia (Aghazadeh et al., 2018). Moreover, SHBV was discovered in multiple shrew species and at more than one sampling site (two distant sampling sites, >1200 km), indicating that SHBV has likely been present in these animals for an extended time period. Similarly, the hepadnaviruses discovered in Rhinolophus ferrumequinum bats sampled from Henan province, China clustered with the RBHBV and HBHBV identified in bats sampled from West Africa (Nie et al., 2018), and the ancestor of Noctilionoidea (U. bilobatum belongs) has been proposed to have an Old Word origin (Teeling et al., 2005), again indicative of a long evolutionary history. The virus positive shrew species identified here, Anourosorex squamipes, Crocidura attenuate and Crocidura lasiura, have only been identified in the Old World (http://oldredlist.iucnredlist.org/), and although 170 Asian house shrews were captured (37.9% of all shrew samples) in this study, no virus positive samples were identified in this globally distributed shrew species. In sum, more data is clearly needed to resolve the geographic origins of the hepadnaviruses.
Shrews are the fourth most diverse species of mammals (385 species in 26 genera) rivalled only by the families Muridae, Cricetidae and Vespertilionidae, and distributed almost worldwide (Wilson and Reeder, 2011). However, these mammals have attracted far less attention in studies of virus emergence, transmission and evolution. In recent years, however, the discovery of several viruses in shrews have highlighted their potential as virus hosts (Wang et al., 2017b, Tsoleridis et al., 2016, Li et al., 2015, Li et al., 2016, Zhang, 2014). We identified a novel and divergent orthohepadnavirus, SHBV, in three species of shrews sampled from two counties in China, but not in mice and rats captured from the same location. We therefore propose that shrews are likely to comprise natural host of hepadnaviruses and that other shrew species should be investigated to understand their potential role in virus evolution and transmission.
3. Materials and methods
3.1. Trapping of small animals and sample collection
Most of shrews used in this study were collected during 2016–2017 in Wufeng county, Hubei province, and Longwan district and Wencheng county of Zhejiang province, China. The exception were 32 liver sample nucleic acids from Crocidura lasiura taken from previous research (Lin et al., 2014). Following morphological examination, sequence analysis of the mitochondrial cyt b gene was used to further identify the shrew species. Before necropsy, euthanasia was performed and every effort was made to minimize animal suffering. Liver samples were collected for hepadnavirus detection and immediately frozen at −80 °C after collection.
This research was approved by the ethics committee of the National Institute for Communicable Disease Control and Prevention of the Chinese Center for Disease control and Prevention (CDC). All shrews were treated according to the guidelines for Laboratory Animal Use and Care of the Chinese CDC and the Rules for the Implementation of Laboratory Animal Medicine (1998) from the ministry of Health, China.
3.2. DNA extraction, hepadnavirus detection and genome amplification
An approximately 30 mg liver sample was used for DNA and RNA extraction. The DNA/RNA extraction kit (Omega bio-tek, USA) was used for total DNA extraction and the procedure was performed exactly following the manufacture's instruction. The DNA was eluted in 100 μl ddH2O and RNA in 50 μl diethyl pyrocarbonate (DEPC) water. The same primers used previously for screening bat hepadnaviruses (Nie et al., 2018) were used here to detect hepadnaviruses in shrews. Complete viral genomes were amplified using nested PCR. Based on the ~360 bp fragments recovered and the circular viral genome, primers were designed to bridge the gap of the genome by PCR. The two segments were stitched together using the SeqMan program (DNASTAR, Madison, WI). Finally, the viral genomes were manually edited by removing the overlap sequences for a cyclised genome.
The pMD18-T vector (TaKaRa, Dalian, China) was used to clone PCR amplicons (> 700 bp) after the amplicons purified by QIAquick Gel Extraction kit (Qiagen, Valencia, USA) according to the manufacturer's recommendations. JM109–143 competent cells were used for plasmid transformation.
3.3. Genome construction and similarity analysis
Virus strain Wufeng-Ca-70 was used as a representative for construction of the SHBV genome, which was undertaken using the SeqBuilder program implemented in Lasergene (v7). Manual adjustments were made using Adobe Illustrator CS6. The MegAlign program implemented in Lasergene (v7) was used to estimate both the nucleic acid and amino acid genetic distances between the sequences of the P, S, C and X genes.
3.4. Recombination analysis
Before phylogenetic analysis, we screened the full-length genomes of the five newly identified viruses for recombination using multiple methods with the RDP package (Version 4; 3SEQ, BOOTSCAN, CHIMAERA, GENECONV, MAXCHI, RDP, SISCAN) (Martin et al., 2010). Any putative recombination event had to identified by at least two methods at a P < 0.05 significance level. Finally, we also performed a Simplot analysis (Version 3.5.1) (Lole et al., 1999) to reveal putative recombination events.
3.5. Phylogenetic analysis
All the sequences generated here were initially aligned using the Mafft program (Katoh and Standley, 2013), with ambiguously aligned regions and gaps then removed using TrimAL (version 1.2) (Capella-Gutierrez et al., 2009). Phylogenetic trees were then estimated on these data using the maximum likelihood (ML) method implemented in PhyML v3.0 (Guindon et al., 2010) based on the amino acid sequences of the polymerase (595 aa), surface (322 aa) and core (180 aa) proteins of orthohepadnaviruses. Bootstrap support values were calculated from 1000 replicate trees using the best-fit model of amino acid substitution and a Subtree Prunning and Regrafting (SPR) branch-swapping algorithm. The amino acid alignment of the X protein was too short to establish a reliable phylogenetic tree.
3.6. Co-phylogenetic analysis of orthohepadnaviruses and their hosts
To better understand the evolutionary relationship between the orthohepadnaviruses and their mammalian hosts we performed a co-phylogenetic analysis using both the Jane (Conow et al., 2010) and CopyCat (Meier-Kolthoff et al., 2007) packages and building on our previous work in this area (Nie et al., 2018). As in previous studies (Geoghegan et al., 2017, Lin et al., 2017, Nie et al., 2018), weights of 1 were used for duplication, lineage loss, host switching and failure to diverge, and 0 for co-divergence, employing 100 generations and a population size of 100 were used as parameters for genetic algorithm. The random tip mapping method was used to test the probability of observing the events by chance, with the following parameters: sample size = 50, generation number = 100 and population size = 100.
Acknowledgments
This study was supported by National Natural Science Foundation of China (Grants 81861138003 and 81672057) and the Special National Project on Research and Development of Key Biosafety Technologies (2016YFC1201900 and 2016YFC1200101). ECH is supported by an ARC Australian Laureate Fellowship (FL170100022).
Acknowledgments
GenBank accession numbers
All the SHBV sequences recovered in this study have been submitted to GenBank and assigned accession numbers MH484438 to MH484442.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2019.03.007.
Appendix A. Supplementary material
Supplementary material Fig. S1. Co-phylogenetic analyses of mammal hosts and their associated hepadnaviruses. The host tree was based on topology implied in the web of Time tree of life (http://www.timetree.org/), and the orthohepadnavirus tree was based on the polymerase, core and surface genes (ie. rooted with the bluegill virus). A, summary of phylogenetic test of virus-host association and co-divergence; B, estimation of co-phylogenetic events across the evolution history of mammalian hepadnaviruses: co-divergence (red), duplication (blue), host switching (green) and extinction (purple).
Supplementary material Fig. S2. Genome features of orthohepadnaviruses. The main deletion/expansion regions associated with genome size among the representative viruses in genus orthohepadnavirus are shown with alignment numbering. The viruses identified in this study are shown in bold.
Supplementary material Fig. S3. Comparison of the “a” determinant region of SHBV with human HBV and TBHBV. Numbering was according to amino acid position of genotype A. The viruses documented in this study are shown in bold.
Supplementary material Fig. S4. Recombination analyses of SHBV (Wufeng-Ca-70) and other known orthohepadnaviruses.
Supplementary material.
References
- Abou-Jaoude G., Sureau C. Entry of hepatitis delta virus requires the conserved cysteine residues of the hepatitis B virus envelope protein antigenic loop and is blocked by inhibitors of thiol-disulfide exchange. J. Virol. 2007;81:13057–13066. doi: 10.1128/JVI.01495-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghazadeh M., Shi M., Barrs V.R., McLuckie A.J., Lindsay S.A., Jameson B., Hampson B., Holmes E.C., Beatty J.A. A novel hepadnavirus identified in an immunocompromised domestic cat in Australia. Viruses. 2018;10 doi: 10.3390/v10050269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasdell K.R., Duong V., Eloit M., Chretien F., Ly S., Hul V., Deubel V., Morand S., Buchy P. Evidence of human infection by a new mammarenavirus endemic to Southeastern Asia. eLife. 2016;5 doi: 10.7554/eLife.13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S., Silla-Martinez J.M., Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conow C., Fielder D., Ovadia Y., Libeskind-Hadas R. Jane: a new tool for the cophylogeny reconstruction problem. Algorithms Mol. Biol. 2010;5:16. doi: 10.1186/1748-7188-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho Dominguez Souza B.F., Konig A., Rasche A., de Oliveira Carneiro I., Stephan N., Corman V.M., Roppert P.L., Goldmann N., Kepper R., Muller S.F., Volker C., de Souza A.J.S., Gomes-Gouvea M.S., Moreira-Soto A., Stocker A., Nassal M., Franke C.R., Rebello Pinho J.R., Soares M., Geyer J., Lemey P., Drosten C., Netto E.M., Glebe D., Drexler J.F. A novel hepatitis B virus species discovered in capuchin monkeys sheds new light on the evolution of primate hepadnaviruses. J. Hepatol. 2018;68:1114–1122. doi: 10.1016/j.jhep.2018.01.029. [DOI] [PubMed] [Google Scholar]
- Dill J.A., Camus A.C., Leary J.H., Di Giallonardo F., Holmes E.C., Ng T.F. Distinct viral lineages from fish and amphibians reveal the complex evolutionary history of hepadnaviruses. J. Virol. 2016;90:7920–7933. doi: 10.1128/JVI.00832-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J.F., Geipel A., Konig A., Corman V.M., van Riel D., Leijten L.M., Bremer C.M., Rasche A., Cottontail V.M., Maganga G.D., Schlegel M., Muller M.A., Adam A., Klose S.M., Carneiro A.J., Stocker A., Franke C.R., Gloza-Rausch F., Geyer J., Annan A., Adu-Sarkodie Y., Oppong S., Binger T., Vallo P., Tschapka M., Ulrich R.G., Gerlich W.H., Leroy E., Kuiken T., Glebe D., Drosten C. Bats carry pathogenic hepadnaviruses antigenically related to hepatitis B virus and capable of infecting human hepatocytes. Proc. Natl. Acad. Sci. USA. 2013;110:16151–16156. doi: 10.1073/pnas.1308049110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan J.L., Duchene S., Holmes E.C. Comparative analysis estimates the relative frequencies of co-divergence and cross-species transmission within viral families. PLoS Pathog. 2017;13:e1006215. doi: 10.1371/journal.ppat.1006215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Guo W.P., Lin X.D., Wang W., Tian J.H., Cong M.L., Zhang H.L., Wang M.R., Zhou R.H., Wang J.B., Li M.H., Xu J., Holmes E.C., Zhang Y.Z. Phylogeny and origins of hantaviruses harbored by bats, insectivores, and rodents. PLoS Pathog. 2013;9:e1003159. doi: 10.1371/journal.ppat.1003159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Fan Q., Yang F., Hu T., Qiu W., Feng Y., Li Z., Li Y., Zhang F., Guo H., Zou X., Tu C. Hepatitis virus in long-fingered bats, Myanmar. Emerg. Infect. Dis. 2013;19:638–640. doi: 10.3201/eid1904.121655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Zhang F., Xia L., Hu T., Chen G., Qiu W., Fan Q., Feng Y., Guo H., Tu C. Identification of a novel Orthohepadnavirus in pomona roundleaf bats in China. Arch. Virol. 2015;160:335–337. doi: 10.1007/s00705-014-2222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford R.E., Chavez D., Brasky K.M., Burns R.B., 3rd, Rico-Hesse R. Isolation of a hepadnavirus from the woolly monkey, a New World primate. Proc. Natl. Acad. Sci. USA. 1998;95:5757–5761. doi: 10.1073/pnas.95.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber C., Seitz S., Mattei S., Suh A., Beck J., Herstein J., Borold J., Salzburger W., Kaderali L., Briggs J.A.G., Bartenschlager R. Deciphering the origin and evolution of hepatitis b viruses by means of a family of non-enveloped fish viruses. Cell Host Microbe. 2017;22(387–399):e386. doi: 10.1016/j.chom.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Lin X.D., Huang K.Y., Zhang B., Shi M., Guo W.P., Wang M.R., Wang W., Xing J.G., Li M.H., Hong W.S., Holmes E.C., Zhang Y.Z. Identification of novel and diverse rotaviruses in rodents and insectivores, and evidence of cross-species transmission into humans. Virology. 2016;494:168–177. doi: 10.1016/j.virol.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Lin X.D., Wang W., Shi M., Guo W.P., Zhang X.H., Xing J.G., He J.R., Wang K., Li M.H., Cao J.H., Jiang M.L., Holmes E.C., Zhang Y.Z. Isolation and characterization of a novel arenavirus harbored by rodents and shrews in Zhejiang province, China. Virology. 2015;476:37–42. doi: 10.1016/j.virol.2014.11.026. [DOI] [PubMed] [Google Scholar]
- Lin X.D., Zhou R.H., Fan F.N., Ying X.H., Sun X.Y., Wang W., Holmes E.C., Zhang Y.Z. Biodiversity and evolution of Imjin virus and Thottapalayam virus in Crocidurinae shrews in Zhejiang Province, China. Virus Res. 2014;189:114–120. doi: 10.1016/j.virusres.2014.05.014. [DOI] [PubMed] [Google Scholar]
- Lin X.D., Wang W., Hao Z.Y., Wang Z.X., Guo W.P., Guan X.Q., Wang M.R., Wang H.W., Zhou R.H., Li M.H., Tang G.P., Wu J., Holmes E.C., Zhang Y.Z. Extensive diversity of coronaviruses in bats from China. Virology. 2017;507:1–10. doi: 10.1016/j.virol.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlejohn M., Locarnini S., Yuen L. Origins and evolution of hepatitis B virus and hepatitis D virus. Cold Spring Harb. Perspect. Med. 2016;6:a021360. doi: 10.1101/cshperspect.a021360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locarnini S., Littlejohn M., Aziz M.N., Yuen L. Possible origins and evolution of the hepatitis B virus (HBV) Semin. Cancer Biol. 2013;23:561–575. doi: 10.1016/j.semcancer.2013.08.006. [DOI] [PubMed] [Google Scholar]
- Lole K.S., Bollinger R.C., Paranjape R.S., Gadkari D., Kulkarni S.S., Novak N.G., Ingersoll R., Sheppard H.W., Ray S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion P.L., Oshiro L.S., Regnery D.C., Scullard G.H., Robinson W.S. A virus in Beechey ground squirrels that is related to hepatitis B virus of humans. Proc. Natl. Acad. Sci. USA. 1980;77:2941–2945. doi: 10.1073/pnas.77.5.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D.P., Lemey P., Lott M., Moulton V., Posada D., Lefeuvre P. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff J.P., Auch A.F., Huson D.H., Goker M. COPYCAT: cophylogenetic analysis tool. Bioinformatics. 2007;23:898–900. doi: 10.1093/bioinformatics/btm027. [DOI] [PubMed] [Google Scholar]
- Neurath A.R., Kent S.B., Strick N., Parker K. Identification and chemical synthesis of a host cell receptor binding site on hepatitis B virus. Cell. 1986;46:429–436. doi: 10.1016/0092-8674(86)90663-x. [DOI] [PubMed] [Google Scholar]
- Nie F.Y., Lin X.D., Hao Z.Y., Chen X.N., Wang Z.X., Wang M.R., Wu J., Wang H.W., Zhao G., Ma R.Z., Holmes E.C., Zhang Y.Z. Extensive diversity and evolution of hepadnaviruses in bats in China. Virology. 2018;514:88–97. doi: 10.1016/j.virol.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasche A., Souza B., Drexler J.F. Bat hepadnaviruses and the origins of primate hepatitis B viruses. Curr. Opin. Virol. 2016;16:86–94. doi: 10.1016/j.coviro.2016.01.015. [DOI] [PubMed] [Google Scholar]
- Radosa L., Schlegel M., Gebauer P., Ansorge H., Heroldova M., Janova E., Stanko M., Mosansky L., Fricova J., Pejcoch M., Suchomel J., Purchart L., Groschup M.H., Kruger D.H., Ulrich R.G., Klempa B. Detection of shrew-borne hantavirus in Eurasian pygmy shrew (Sorex minutus) in Central Europe. Infect. Genet. Evol. 2013;19:403–410. doi: 10.1016/j.meegid.2013.04.008. [DOI] [PubMed] [Google Scholar]
- Shi M., Lin X.D., Chen X., Tian J.H., Chen L.J., Li K., Wang W., Eden J.S., Shen J.J., Liu L., Holmes E.C., Zhang Y.Z. The evolutionary history of vertebrate RNA viruses. Nature. 2018;556:197–202. doi: 10.1038/s41586-018-0012-7. [DOI] [PubMed] [Google Scholar]
- Shi M., Lin X.D., Tian J.H., Chen L.J., Chen X., Li C.X., Qin X.C., Li J., Cao J.P., Eden J.S., Buchmann J., Wang W., Xu J., Holmes E.C., Zhang Y.Z. Redefining the invertebrate RNA virosphere. Nature. 2016 doi: 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- Smith I., Wang L.F. Bats and their virome: an important source of emerging viruses capable of infecting humans. Curr. Opin. Virol. 2013;3:84–91. doi: 10.1016/j.coviro.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers J., Smolec J.M., Snyder R. A virus similar to human hepatitis B virus associated with hepatitis and hepatoma in woodchucks. Proc. Natl. Acad. Sci. USA. 1978;75:4533–4537. doi: 10.1073/pnas.75.9.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling E.C., Springer M.S., Madsen O., Bates P., O'Brien S., Murphy, W.J J. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307:580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- Testut P., Renard C.A., Terradillos O., Vitvitski-Trepo L., Tekaia F., Degott C., Blake J., Boyer B., Buendia M.A. A new hepadnavirus endemic in arctic ground squirrels in Alaska. J. Virol. 1996;70:4210–4219. doi: 10.1128/jvi.70.7.4210-4219.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoleridis T., Onianwa O., Horncastle E., Dayman E., Zhu M., Danjittrong T., Wachtl M., Behnke J.M., Chapman S., Strong V., Dobbs P., Ball J.K., Tarlinton R.E., McClure C.P. Discovery of novel alphacorona viruses in European rodents and shrews. Viruses. 2016;8:84. doi: 10.3390/v8030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban S., Schulze A., Dandri M., Petersen J. The replication cycle of hepatitis B virus. J. Hepatol. 2010;52:282–284. doi: 10.1016/j.jhep.2009.10.031. [DOI] [PubMed] [Google Scholar]
- Wang B., Yang X.L., Li W., Zhu Y., Ge X.Y., Zhang L.B., Zhang Y.Z., Bock C.T., Shi Z.L. Detection and genome characterization of four novel bat hepadnaviruses and a hepevirus in China. Virol. J. 2017;14:40. doi: 10.1186/s12985-017-0706-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Lin X.D., Liao Y., Guan X.Q., Guo W.P., Xing J.G., Holmes E.C., Zhang Y.Z. Discovery of a highly divergent coronavirus in the Asian house shrew from China illuminates the origin of the alphacoronaviruses. J. Virol. 2017;98:764. doi: 10.1128/JVI.00764-17. (-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Lin X.D., Guo W.P., Zhou R.H., Wang M.R., Wang C.Q., Ge S., Mei S.H., Li M.H., Shi M., Holmes E.C., Zhang Y.Z. Discovery, diversity and evolution of novel coronaviruses sampled from rodents in China. Virology. 2015;474:19–27. doi: 10.1016/j.virol.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D.E., Reeder D.M. 3rd ed. Johns Hopkins University Press; Bsltimore, MD: 2005. Mammal Species of the World. A Taxonomic and Geographic Reference. [Google Scholar]
- Wilson D.E., Reeder D.M. Class Mammalia Linnaeus, 1758. In: Zhang Z.-Q., editor. Animal biodiversity: An outline of higher-level classification and survey of taxonomic richness. Vol. 3148. Zootaxa; 2011. pp. 56–60. [DOI] [PubMed] [Google Scholar]
- Wolfe N.D., Dunavan C.P., Diamond J. Origins of major human infectious diseases. Nature. 2007;447:279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.Z. Discovery of hantaviruses in bats and insectivores and the evolution of the genus Hantavirus. Virus Res. 2014;187:15–21. doi: 10.1016/j.virusres.2013.12.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material Fig. S1. Co-phylogenetic analyses of mammal hosts and their associated hepadnaviruses. The host tree was based on topology implied in the web of Time tree of life (http://www.timetree.org/), and the orthohepadnavirus tree was based on the polymerase, core and surface genes (ie. rooted with the bluegill virus). A, summary of phylogenetic test of virus-host association and co-divergence; B, estimation of co-phylogenetic events across the evolution history of mammalian hepadnaviruses: co-divergence (red), duplication (blue), host switching (green) and extinction (purple).
Supplementary material Fig. S2. Genome features of orthohepadnaviruses. The main deletion/expansion regions associated with genome size among the representative viruses in genus orthohepadnavirus are shown with alignment numbering. The viruses identified in this study are shown in bold.
Supplementary material Fig. S3. Comparison of the “a” determinant region of SHBV with human HBV and TBHBV. Numbering was according to amino acid position of genotype A. The viruses documented in this study are shown in bold.
Supplementary material Fig. S4. Recombination analyses of SHBV (Wufeng-Ca-70) and other known orthohepadnaviruses.
Supplementary material.