Highlights
► WNV is a member of the flavivirus family. ► Alexander the Great died of WNV infection in Babylon, around 325 years BC. ► WNV core protein mediates genomic RNA circularization by virtue of its RNA chaperoning activity.
Abbreviations: CS, conserved sequence; DAR, downstream AUG region; DB, dumbbell-like structure; DENV, dengue virus; JEV, Japanese encephalitis virus; ORF, open reading frame; RdRp, RNA-dependent RNA polymerase; sfRNA, subgenomic flavivirus RNA; TBEV, tick-borne encephalitis virus; UAR, upstream AUG region; UTR, untranslated region; WNV, West Nile virus; YFV, yellow fever virus
Keywords: West Nile virus, Core protein, Flaviviruses, Viral replication, Genome cyclization, RNA chaperoning
Abstract
Genome cyclization through conserved RNA sequences located in the 5′ and 3′ terminal regions of flavivirus genomic RNA is essential for virus replication. Although the role of various cis-acting RNA elements in panhandle formation is well characterized, almost nothing is known about the potential contribution of protein cofactors to viral RNA cyclization. Proteins with nucleic acid chaperone activities are encoded by many viruses (e.g., retroviruses, coronaviruses) to facilitate RNA structural rearrangements and RNA–RNA interactions during the viral replicative cycle. Since the core protein of flaviviruses is also endowed with potent RNA chaperone activities, we decided to examine the effect of West Nile virus (WNV) core on 5′–3′ genomic RNA annealing in vitro. Core protein binding resulted in a dramatic, dose-dependent increase in 5′–3′ complex formation. Mutations introduced in either the UAR (upstream AUG region) or CS (conserved sequence) elements of the viral RNA diminished core protein-dependent annealing, while compensatory mutations restored the 5′–3′ RNA interaction. The activity responsible for stimulating RNA annealing was mapped to the C-terminal RNA-binding region of WNV core protein. These results indicate that core protein – besides its function in viral particle formation – might be involved in the regulation of flavivirus genomic RNA cyclization, and thus virus replication.
1. Introduction
Flaviviruses constitute a large and diverse group within the Flaviviridae family of positive-strand enveloped RNA viruses (Lindenbach et al., 2007). Medically important flaviviruses include the mosquito-borne West Nile virus (WNV), yellow fever virus (YFV), Japanese encephalitis virus (JEV), and dengue virus (DENV serotypes 1–4), as well as tick-borne encephalitis viruses (TBEV). Infection by arthropod-borne flaviviruses is associated with significant morbidity and mortality worldwide (Gould and Solomon, 2008, Griffin, 2011, Guzman et al., 2010). Increased urbanization, intercontinental travel, failure of vector mosquito control and increasing global temperatures have collectively resulted in the recent emergence or re-emergence of mosquito-borne flaviviruses in previously disease-free areas (Gould and Higgs, 2009, Kilpatrick, 2011). West Nile virus first appeared in the New World in 1999, causing a viral encephalitis outbreak in New York City (Petersen, 2009). Since then, the virus has rapidly spread throughout the US, Central and South America, reaching Argentine by 2006 (Petersen, 2009). Due to the increased geographic dispersal of the Asian tiger mosquito Aëdes albopictus, dengue virus is currently re-emerging in Southern Europe (Gjenero-Margan et al., 2010, La Ruche et al., 2010), after almost a hundred-year hiatus (Rosen, 1986).
Flaviviruses are small, enveloped viruses with a single-stranded, positive sense RNA genome of ∼11 kb. A single open reading frame (ORF) encodes a large polyprotein precursor which is co- and post-translationally processed to yield the viral structural proteins (C-prM-E), and the non-structural proteins (NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5) directing genome replication (Lindenbach et al., 2007). The ORF is flanked by a short (∼100 nt) 5′ untranslated region (UTR) and a longer (∼400–800 nt) 3′ UTR, both of which contain highly conserved RNA secondary structures and RNA sequences involved in the regulation of viral translation and RNA replication [reviewed in (Brinton, 2002, Markoff, 2003); see Fig. 1 ]. The 5′ UTR begins with a Y-shaped stem-loop (SLA) (Brinton and Dispoto, 1988) necessary for viral replication (Filomatori et al., 2006). The initiator AUG codon for polyprotein synthesis is situated in a second stem-loop structure (SLB), which is followed by the capsid-coding region hairpin (cHP) regulating start codon selection (Clyde et al., 2008, Clyde and Harris, 2006). The 3′ UTR consists of a variable region, followed by tandem dumbbell-like structures (DB1 and DB2), and a highly stable and conserved terminal stem-loop (3′ SL) (Brinton et al., 1986). In addition, a complex network of pseudoknot interactions is thought to influence the topology and function of the 3′ UTR (Funk et al., 2010, Olsthoorn and Bol, 2001, Pijlman et al., 2008, Shi et al., 1996, Silva et al., 2010).
Fig. 1.
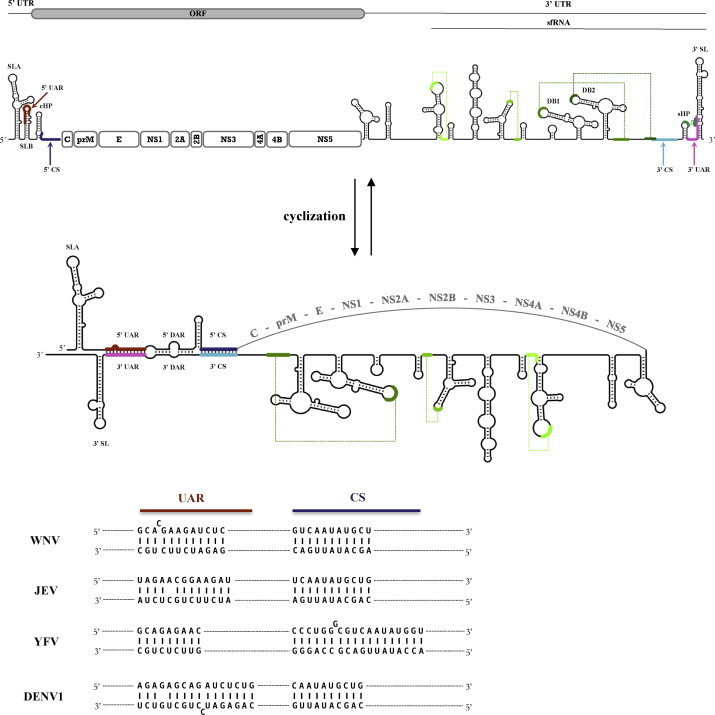
Schematic representation of the linear and circular forms of WNV genomic RNA. Flavivirus infected cells accumulate two major RNA species, the viral genomic RNA and the small sfRNA (subgenomic flavivirus RNA). RNA secondary structures were predicted by Mfold (Zuker, 2003), using constraints on experimentally characterized RNA structures (see references in text). The conserved RNA structures and sequences include: SLA—stem-loop A; UAR—upstream AUG region; DAR—downstream AUG region; CS—conserved sequence; cHP—capsid region hairpin; sHP—small hairpin; DB1 and DB2—dumbbell structures; 3′ SL—3′ stem-loop. 5′ and 3′ UAR sequences are highlighted in dark red and magenta, respectively. 5′ and 3′ CS elements are highlighted in dark and light blue. Conserved pseudoknot structures are illustrated in green. The core protein-coding sequence partly overlaps with RNA secondary structures in the 5′ region of the genome. Note that the lower stem of 3′ SL is opened in the circular RNA form, facilitating binding of the viral replication complex. Cyclization sequences of representative mosquito-borne flaviviruses are shown below the circular RNA structure (GenBank accession for West Nile virus—AF260968; Japanese encephalitis virus—NC_001437.1; yellow fever virus—NC_002031.1; dengue virus type 1—NC_001477). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of the article.)
The positive-sense genomic RNA of flaviviruses is first copied by the viral RNA-dependent RNA polymerase (RdRp) to generate a complementary minus-strand RNA that, in turn, serves as a template for the amplification of plus-strand viral genomes. Interestingly, in vitro polymerase assays demonstrated that minus-strand RNA synthesis requires the presence of both the 5′ and 3′ UTRs (You et al., 2001, You and Padmanabhan, 1999). The promoter element for minus-strand RNA synthesis was later identified as stem-loop A (SLA, Fig. 1) in the 5′ UTR, which binds the viral RdRp with high affinity (Filomatori et al., 2011, Filomatori et al., 2006, Lodeiro et al., 2009). According to current models, based mostly on work in dengue viruses, panhandle formation through complementary sequences in the 5′-core and 3′ UTR regions would reposition the 3′ end of the genome in the vicinity of the promoter–RdRp complex, coupling transcription initiation to genome cyclization (reviewed in Gebhard et al., 2011). At the same time, the long-distance RNA interaction is believed to be accompanied by structural rearrangements in the highly conserved 3′ stem-loop (3′ SL, Fig. 1) (Dong et al., 2008), allowing RdRp binding to the terminal nucleotides of the genome (Gebhard et al., 2011). Genomic RNA cyclization in mosquito-borne flaviviruses is mediated by three pairs of long distance RNA interactions: 5′–3′ UAR (upstream AUG region) (Alvarez et al., 2005b), 5′–3′ DAR (downstream AUG region) (Dong et al., 2008, Friebe and Harris, 2010), and 5′–3′ CS (conserved sequence) (Hahn et al., 1987) (Fig. 1). Mutations disrupting complementarity in any of these elements abrogate viral replication in subgenomic replicon model systems (Alvarez et al., 2008, Alvarez et al., 2005b, Bredenbeek et al., 2003, Corver et al., 2003, Friebe and Harris, 2010, Friebe et al., 2011, Khromykh et al., 2001, Lo et al., 2003, Men et al., 1996, Villordo et al., 2010, Zhang et al., 2008), while compensatory mutations rescue viability (Alvarez et al., 2008, Alvarez et al., 2005b, Friebe and Harris, 2010, Friebe et al., 2011, Khromykh et al., 2001, Lo et al., 2003, Zhang et al., 2008).
Flavivirus genomic RNA is encapsidated by the small, highly basic core protein. Core corresponds to the N-terminus of the polyprotein, from where it is released in its mature form by the action of the NS2B-NS3 serine protease complex (Amberg et al., 1994, Lobigs, 1993, Yamshchikov and Compans, 1994). Despite relatively little sequence conservation, all flavivirus core proteins share a common functional and structural domain organization. The N- and C-terminal extremities contain a high concentration of basic amino acids, and bind to the viral genomic RNA independently (Khromykh and Westaway, 1996). Flanked by the RNA-binding regions, an internal hydrophobic domain is responsible for the dimerization/oligomerization (Bhuvanakantham and Ng, 2005, Wang et al., 2004), as well as for the membrane/lipid association of core protein (Markoff et al., 1997, Samsa et al., 2009). Structural studies by circular dichroism, X-ray crystallography and nuclear magnetic resonance (NMR) on DENV, WNV, and YFV core proteins (Dokland et al., 2004, Ivanyi-Nagy et al., 2008, Jones et al., 2003, Ma et al., 2004) also support a conserved structure, with a highly flexible N-terminal RNA-binding region followed by three or four alpha-helices. Interestingly, deletion analyses demonstrated a remarkable functional flexibility of core protein, suggesting that precisely folded three-dimensional structures are not required for RNA binding and membrane association, and these functions are rather determined by the overall physico-chemical nature of the domains. Large deletions are well tolerated in the internal hydrophobic region of both mosquito-borne and tick-borne flaviviruses (Kofler et al., 2002, Kofler et al., 2003, Schlick et al., 2009, Zhu et al., 2007). Similarly, the N- and C-terminal extremities can function (at least partly) redundantly in RNA packaging (Patkar et al., 2007).
Core proteins of WNV (Ivanyi-Nagy et al., 2008), dengue virus (Pong et al., 2011), as well as cores in the related hepaciviruses (Cristofari et al., 2004, Ivanyi-Nagy et al., 2006) and pestiviruses (Ivanyi-Nagy et al., 2008), were shown to facilitate nucleic acid rearrangements without ATP consumption, acting as efficient RNA chaperones in vitro. The RNA chaperone activity of WNV core was mapped to the C-terminal RNA-binding region of the protein (Ivanyi-Nagy et al., 2008). In this study, we examined the effect of WNV core protein chaperoning on viral 5′–3′ UTR annealing, using an in vitro model system with separate 5′ and 3′ RNAs. We found that core protein binding greatly increases the rate of 5′–3′ complex formation, and is required for the interaction when full-length 3′ UTR RNAs are used (Fig. 2 ). Mutations abolishing either the UAR or CS interaction diminished, but did not completely abrogate core-protein induced annealing, while compensatory mutations restored the interaction (Fig. 3 ). In agreement with the results of in vitro chaperone assays (Ivanyi-Nagy et al., 2008), stimulation of RNA annealing was mapped to the C-terminal RNA-binding region of core protein (Fig. 4 ).
Fig. 2.
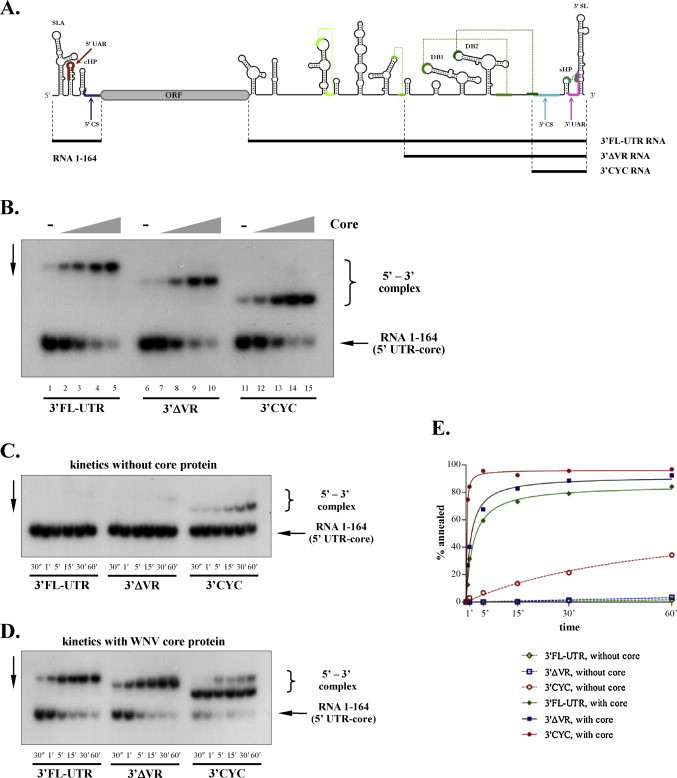
Core protein facilitates the annealing of WNV 5′ UTR-core and 3′ UTR RNAs. (A) Position and secondary structure of RNA molecules used in this study. (B) Core protein enhances 5′–3′ WNV RNA interactions. Radioactively labelled RNA 1–164 was incubated with the full-length 3′FL-UTR RNA (lanes 1–5), 3′ΔVR RNA (lanes 6–10), or 3′CYC RNA (lanes 11–15) in the presence of increasing concentrations of full-length WNV core protein, as described in Section 2. Protein-to-nucleotide molar ratios were 1/40 (lanes 2, 7, 12), 1/20 (lanes 3, 8, 13), 1/10 (lanes 4, 9, 14), or 1/5 (lanes 5, 10, 15). Lanes 1, 6, and 11 show RNA annealing in the absence of core protein. (C) Kinetics of 5′–3′ RNA annealing without core protein. Radioactively labelled RNA 1–164 was incubated with 3′FL-UTR RNA, 3′ΔVR RNA, or 3′CYC RNA at 37 °C for different length of time, as indicated below the lanes. (D) Kinetics of 5′–3′ RNA annealing in the presence of core protein. The experiment was carried out the same way as in (C), but a constant 1 protein/5 nucleotide molar ratio of full-length core protein was added to the RNA molecules. (E) Quantification of the annealing reactions shown in (C) and (D). Annealing curves were fitted using Graphpad Prism©, as described in Doetsch et al. (2011).
Fig. 3.
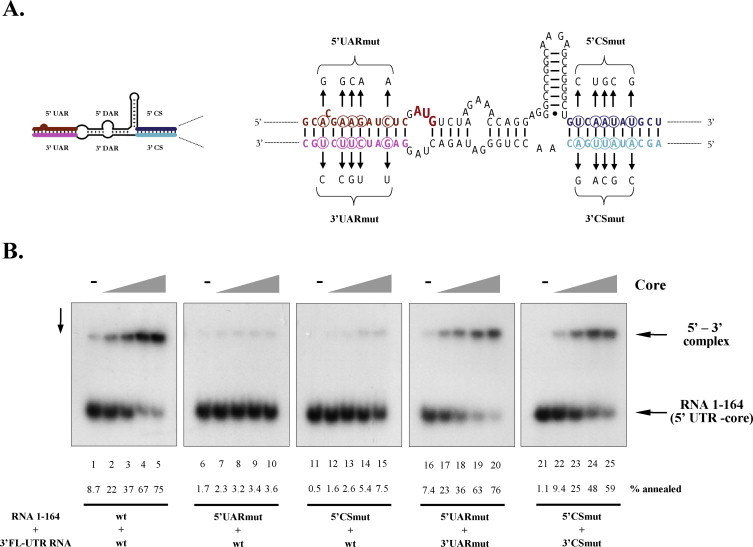
Core protein-induced annealing is mediated by the UAR and CS elements. (A) Mutations introduced in the UAR and CS elements of WNV. The initiator AUG is indicated in red. (B) Wild-type (wt) or mutant radioactively labelled RNA 1–164 was incubated with wild-type 3′FL-UTR RNA or with 3′ UTR RNAs containing mutations restoring the 5′–3′ UAR or 5′–3′ CS interactions. Protein-to-nucleotide molar ratios were 1/40 (lanes 2, 7, 12, 17, 21), 1/20 (lanes 3, 8, 13, 18, 23), 1/10 (lanes 4, 9, 14, 19, 24), and 1/5 (lanes 5, 10, 15, 20, 25). Lanes 1, 6, 11, 16, and 21 show RNA annealing in the absence of core protein. The percentage of 5′–3′ complex formation is indicated below the lanes. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of the article.)
Fig. 4.
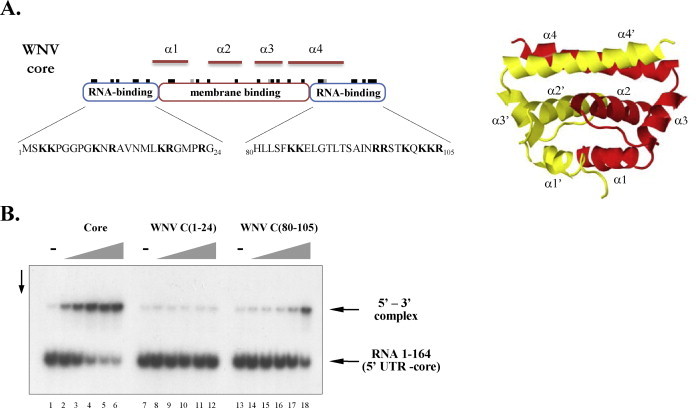
The C-terminal RNA-binding region of core directs RNA annealing. (A) Domain organization and structure of WNV core protein. The position of basic and acidic amino acids is illustrated by black and grey notches, respectively, on top of the domain outlines. The sequence of WNV C(1–24) and WNV C(80–105) peptides is indicated below the RNA-binding domains. The 3D structure of the WNV core dimer, determined by X-ray crystallography, is shown to the right (based on PDB entry 1SFK (Dokland et al., 2004), rendered by Jmol). (B) Radioactively labelled RNA 1–164 was incubated with 3′FL-UTR RNA in the presence of increasing concentrations of full-length WNV core (lanes 2–6), WNV C(1–24) (lanes 8–12), or WNV C(80–105) (lanes 14–18). Protein-to-nucleotide molar ratios were 1/40 (lanes 2, 8, 14), 1/20 (lanes 3, 9, 15), 1/10 (lanes 4, 10, 16), 1/5 (lanes 5, 11, 17), and 1/2.5 (lanes 6, 12, 18). Lanes 1, 7, and 13 show RNA annealing in the absence of core protein.
2. Materials and methods
2.1. Plasmid construction
For cloning the 3′ UTR of West Nile virus (Eg101 strain, GenBank accession AF260968), total RNA was extracted from virus infected Vero cells and reverse transcribed using the ThermoScript RT-PCR system (Invitrogen). Reverse transcription was carried out at 60 °C, using ODN Eg101-3′UTR-ss as a primer (Table 1 ). cDNA was amplified by Eg101-3′UTR-ss and Eg101-3′UTR-as and cloned between the SalI and HindIII sites of pSP64 (Promega), resulting in pSP64-3′UTR vector. Deletions in the 3′ UTR were introduced by amplifying the desired regions in pSP64-3′UTR by PCR, and re-cloning the fragments between the SalI and HindIII sites of pSP64.
Table 1.
Oligonucleotides used in this study.
| Name | Sense | Sequence (5′–3′) | Description | Use |
|---|---|---|---|---|
| Eg101-3′UTR-ss | + | GGGCTCGAGTAATACGACTCACTATAGGATACTTTATTAATTGTAAATAGA | XhoI site, followed by T7 promoter and nt 10,399–10,421 of the WNV genome | Cloning of the 3′ UTR of the WNV Eg101 genome |
| Eg101-3′UTR-as | − | GGGGAAGCTTAGATCCTGTGTTCTCGCACCACCA | HindIII site, followed by nt 11,029–11,006 of the WNV genome | |
| Eg101-5′UTR-ss01 | + | AATTCTAATACGACTCACTATAGGGAGTAGTTCGCCTGTGTGAGCTGACAAACTTAGTAGT | EcoRI site, followed by T7 promoter and nt 1–36 of the WNV genome | Overlapping ODNs to clone the 5′ UTR-core region of the WNV Eg101 genome |
| Eg101-5′UTR-01 | − | ATCCTCACAAACACTACTAAGTTTGTCAGCTCACACAGGCGAACTACTCCCTATAGTGAGTCGTATTAG | nt 48–1 of the WNV genome, followed by T7 promoter | |
| Eg101-5′UTR-02 | + | GTTTGTGAGGATTAACAACAATTAACACGGTGCGAGCTGTTTCTTAGCACGAAGATCTCGAT | nt 37–98 of the WNV genome | |
| Eg101-5′UTR-03 | − | GGTTTCTTAGACATCGAGATCTTCGTGCTAAGAAACAGCTCGCACCGTGTTAATTGTTGTTA | nt 110–49 of the WNV genome | |
| Eg101-5′UTR-04 | + | GTCTAAGAAACCAGGAGGGCCCGGCAAGAGCCGGGCTGTCAATATGCTAAAACGCGGAATGCC | nt 99–161 of the WNV genome | |
| Eg101-5′UTR-05 | − | CCGGGGCATTCCGCGTTTTAGCATATTGACAGCCCGGCTCTTGCCGGGCCCTCCT | XmaI site, followed by nt 160–111 of the WNV genome | |
| 5′UARmut | − | TCTTAGACATCGAtATtgcCGcGCTAAGAAACAGC | Introduced mutations are indicated in lower case | Introducing mutations in the cyclization sequences |
| 5′CSmut | − | CCGCGTTTTAGCcTgcaGgCAGCCCGGCTC | ||
| 3′UARmut | − | GTTGTGCAGAGCgGgcaATaTCCTAGTCATTCC | ||
| 3′CSmut | − | CTATCCCAGGTGcCtgcAgGCTGTTTTGTTGTG | ||
| pSP64 | + | TGGAATTGTGAGCGGATAAC | Common oligos annealing to the vector backbone | |
| pSP64-T7 | + | AGGTTATGCGTAATACGACTCACTATAGG | ||
| SP6 | − | TATTTAGGTGACACTATAG | ||
| ΔVR | + | GGGCTCGAGTAATACGACTCACTATAGGGATCAACGCCCCACGCGGC | XhoI site, followed by T7 promoter and nt 10,748–10,765 of the WNV genome | Cloning of 5′ deleted 3′ UTR regions |
| CYC | + | GGGCTCGAGTAATACGACTCACTATAGGGAAACAGCATATTGACACCTGG | XhoI site, followed by T7 promoter and nt 10,919–10,939 of the WNV genome |
The 5′ UTR-core region (nt 1–164 of the viral genome) was cloned by annealing of overlapping ODNs (Eg101-5′UTR-01 to -05), followed by ligation between the XmaI and EcoRI sites of pSP64 vector (Promega).
Mutations were introduced in the UAR or CS region of pSP64-3′UTR or pSP64-5′UTR-core by using a PCR-based mutagenesis protocol (Mikaelian and Sergeant, 1992), with three common ODNs (pSP64, pSP64-T7, and SP6) and one mutation-specific ODN (mut-UAR or mut-CS) for each mutant.
All plasmid constructs were verified by sequencing.
2.2. Proteins and peptides
Full length WNV core protein (amino acids 2–105; GenBank accession number AF481864) was expressed in Escherichia coli and purified as previously described (Ivanyi-Nagy et al., 2008). Core peptides WNV C(1–24) and WNV C(80–105), corresponding to the N- and C-terminal RNA-binding regions of the core protein, were synthesized by Fmoc-OH/DCC/Hobt chemistry and purified as previously described (Ivanyi-Nagy et al., 2008).
2.3. In vitro RNA synthesis
Plasmids containing the 5′ UTR-core region or fragments of the 3′ untranslated region of WNV strain Eg101 were linearized by digestion with XmaI or HindIII restriction enzymes, respectively. In vitro transcription was carried out using T7 RNA polymerase, according to the manufacturer's instructions (Promega). 5′ UTR-core RNAs were labelled by incorporation of α32-P UMP during in vitro transcription. RNAs were purified on 6% denaturing polyacrylamide gels containing 7 M urea in 50 mM Tris-borate (pH 8.3)–1 mM EDTA (0.5× TBE), and recovered by elution in 0.3 M sodium acetate–0.1% SDS overnight at 37 °C, followed by ethanol precipitation.
2.4. RNA annealing assays
In vitro synthesized RNAs were heat denatured for 2 min at 95 °C and chilled on ice. 0.1 pmol of each 5′ UTR-core and 3′ UTR RNA were mixed with annealing buffer to a final concentration of 20 mM Tris–Cl, pH 7.0, 30 mM NaCl, 0.1 mM MgCl2, 10 μM ZnCl2, 10 U RNasin and 5 mM DTT in 10 μl final volume. WNV core protein or core peptides were added to final protein to RNA nucleotide molar ratios as indicated in the figure legends (typically between 1/40 and 1/5 protein/nucleotide ratios). Reactions were incubated at 37 °C for 10–15 min and quenched by adding stop solution (0.5% SDS–25 mM EDTA). Proteins were removed by proteinase K digestion and phenol–chloroform extraction. The purified RNA samples were resolved by 8% native polyacrylamide gel electrophoresis in 0.5× TBE and analysed by autoradiography and Phosphorimager quantification.
3. Results
3.1. Chaperoning 5′–3′ RNA interactions by the core protein
Cyclization of the genomic RNA is essential for viral replication in all mosquito-borne flaviviruses (Alvarez et al., 2008, Alvarez et al., 2005b, Corver et al., 2003, Khromykh et al., 2001, Lo et al., 2003, Zhang et al., 2008), but the determinants and regulation of panhandle formation are still poorly understood. Although complex formation between 5′ and 3′ UTR RNAs can be readily detected in vitro in the absence of protein cofactors (Alvarez et al., 2008, Alvarez et al., 2005b, Villordo et al., 2010, Zhang et al., 2008, Zhang et al., 2010), these interactions usually require high magnesium and RNA concentrations and involve short RNA molecules, thus minimizing the possibility for the RNA to become kinetically trapped in non-functional conformation(s). But in the cellular milieu, RNA folding and RNA–RNA interactions are perhaps universally facilitated by RNA chaperones and/or specific RNA-binding proteins (Cristofari and Darlix, 2002), either encoded by viruses or hijacked from the host for chaperoning viral translation, replication, and packaging.
We have previously shown that the core protein of WNV possesses potent RNA chaperone activities in vitro (Ivanyi-Nagy et al., 2008), facilitating nucleic acid annealing and RNA structural rearrangements. In order to analyse the possible effect of core protein on panhandle formation in the WNV genome, RNA molecules corresponding to nucleotides 1–164 of the genomic RNA (RNA 1–164; Fig. 2A) were in vitro synthesized, radioactively labelled and incubated with equal amounts of non-labelled 3′FL-UTR RNA (Fig. 2A) in the presence of varying amounts of full-length WNV core protein. Following proteinase K digestion of proteins and phenol–chloroform purification of RNAs, 5′–3′ RNA complex formation was assessed by electrophoretic mobility shift assays (Fig. 2B, lanes 1–5). WNV core induced a dose-dependent increase in RNA–RNA interactions, resulting in almost complete annealing at 1 protein to 5 nt molar ratio (Fig. 2B, lane 5; compared to lane 1 in the absence of core protein). In order to examine the potential effect of intramolecular interactions on annealing in the 3′ UTR, 5′ deleted 3′ UTR RNAs, lacking either the variable region (3′ΔVR RNA), or most of the UTR except for the cyclization sequences and the 3′ stem-loop (3′CYC RNA) were examined (Fig. 2B, lanes 6–10 and 11–15, respectively). For all RNA molecules, incubation with core protein induced efficient annealing in a dose-dependent manner (Fig. 2B, lanes 7–10 and 12–15). Interestingly, core protein-independent annealing was more pronounced for the shortest 3′CYC RNA (lane 11 vs lanes 6 and 1), suggesting that RNA sequences 5′ to the cyclization signals might change the topology of the 3′ UTR and interfere with the annealing reaction. To further characterize these differences, a time-course analysis of annealing, either without core protein (Fig. 2C), or in the presence of full-length core (Fig. 2D) was carried out. Without core protein, annealing for the 3′FL-UTR RNA and 3′ΔVR RNA was hardly detectable during the 1 h incubation period, while for the shortest 3′CYC RNA, around one third of the molecules formed 5′–3′ RNA interaction (Fig. 2C and E). In contrast, core protein dramatically increased complex formation, resulting in high levels of annealing as early as 30 s (Fig. 2D and E). Indeed, based on second-order kinetics, the initial annealing rate was estimated to increase by ∼500-fold in the presence of core (data not shown). Nevertheless, annealing of the longer RNAs was still significantly delayed compared to 3′CYC RNA, suggesting that a kinetic barrier must be overcome for the reaction to proceed.
Phylogenetic analyses suggest that the current discontinuous cyclization sequence of mosquito-borne flaviviruses originated – possibly by template switching – as a perfectly complementary continuous region, conserved to this day in the tick-borne members of the flavivirus genus (Gritsun and Gould, 2007). Thus, the UAR-DAR-CS region probably acts as a single regulatory sequence, where replication is determined by the overall stability of the long distance interaction between the 5′ and 3′ regions. In support of this, replication of WNV with a complete deletion of the 3′ CS can be rescued by second-site mutations stabilizing the UAR and DAR interactions (Zhang et al., 2010). In order to examine whether the core protein-dependent annealing of the 5′ and 3′ UTR RNAs depends on the UAR and/or CS interaction, mutations were introduced separately in these elements of RNA 1–164 (Fig. 3A), and cyclization with wild-type 3′FL-UTR RNA was assessed by electrophoretic mobility shift assays. Mutations in either the 5′ UAR or 5′ CS led to a significant decrease in annealing (Fig. 3B, lanes 6–10 and 11–15), while cyclization was rescued by compensatory mutations introduced in the 3′ UAR or 3′ CS of the 3′FL-UTR RNA (lanes 16–20 and 21–25, respectively). These results suggest that the core protein-dependent in vitro annealing recapitulates the features of the cyclization sequence analysed in subgenomic or genome-length cellular models. Interestingly in vitro RNA annealing was readily detectable even with 5 mutations at high core protein levels, while even single point mutations result in a lethal phenotype in subgenomic replicons. This suggests that chaperoning by core protein may stabilize the weak interaction present in the mutants, thus partially relaxing the complementarity requirements of the long distance interaction (Basu and Brinton, 2011, Zhang et al., 2010; see Section 4).
3.2. The C-terminal domain of WNV core protein chaperones 5′–3′ RNA interactions
Flavivirus core proteins have two independent RNA-binding regions at the N- and C-terminal extremities of the protein (Khromykh and Westaway, 1996; Fig. 4A). We have previously shown that the two RNA-binding domains do not act in synergy and that only the C-terminal basic region was active in in vitro nucleic acid chaperone assays (Ivanyi-Nagy et al., 2008). In order to delineate the requirements of core protein for facilitating 5′–3′ UTR interaction, synthetic peptides corresponding to the N- and C-terminal RNA-binding regions (WNV C(1–24) and WNV C(80–105), respectively; Fig. 4A) were used in the annealing reactions with wild-type RNAs. While WNV C(1–24) did not influence 5′–3′ annealing (Fig. 4B, lanes 7–12), WNV C(80–105) induced an increase in 5′–3′ complex formation (Fig. 4B, lanes 13–18). Nevertheless, at equal molar ratios, incubation with full-length core protein resulted in higher levels of complex formation than with WNV C(80–105), indicating that sequences or structural features outside the C-terminal region contribute to full chaperoning activity.
4. Discussion
Complementarity between the genome-terminal cyclization sequences, presumably resulting in panhandle formation in vivo, is absolutely required for flavivirus RNA replication (Khromykh et al., 2001). Although formation of the long-distance interaction is believed to be thermodynamically favoured, the annealing reaction is prohibitively slow (or may not even reach completion) under physiologically relevant conditions (Fig. 2). RNA chaperones are able to disrupt transient or non-functional RNA interactions, thereby decreasing the kinetic barrier hindering the formation of the most stable RNA conformation (reviewed in Cristofari and Darlix, 2002, Schroeder et al., 2004). Indeed, RNA chaperoning might increase the speed of intramolecular RNA rearrangements or intermolecular RNA–RNA interactions several thousand-fold without ATP consumption. We have previously shown that core proteins in the Flaviviridae family, including that of WNV, hepatitis C virus and bovine viral diarrhoea virus, possess potent RNA chaperone activities (Cristofari et al., 2004, Ivanyi-Nagy et al., 2006, Ivanyi-Nagy et al., 2008), facilitating nucleic acid annealing reactions with various DNA and RNA substrates. The molecular mechanism of chaperone action is still poorly understood, and it probably involves a combination of charge neutralization, molecular crowding, and local melting of nucleic acid structures by entropy transfer (Tompa and Csermely, 2004). High affinity binding of a protein to the substrate DNA/RNA is not sufficient, in itself, to trigger the conformational changes observed upon core protein chaperoning (Cristofari et al., 2004, Ivanyi-Nagy et al., 2008).
In this study, we analysed the RNA chaperone activity of WNV core using its cognate target molecules, corresponding to the highly structured terminal regions of the viral genomic RNA. WNV core protein was found to induce a dramatic acceleration in the 5′–3′ UTR annealing reaction (Fig. 2, Fig. 3, Fig. 4), suggesting a possible link between genome cyclization, replication, and packaging.
Paradoxically, with the exception of a short N-terminal sequence which contains the 5′ CS RNA element, the structural protein-coding region is not essential for WNV genomic RNA replication (Khromykh and Westaway, 1997). The discrepancy between the in vitro activity of core protein and its in vivo dispensability suggests that flaviviruses might hijack cellular RNA-binding proteins that may compensate for the loss of core chaperoning. Indeed, an RNA–protein–RNA co-immunoprecipitation assay identified seven distinct proteins (with molecular weights of 35, 37, 40, 45, 52, 76, and 97 kDa) interacting with both the 5′ UTR and 3′ UTR RNAs of dengue virus (Garcia-Montalvo et al., 2004). Although most of these cellular proteins remain to be identified and their interaction with the genomic RNA verified, the 52 kDa band was confirmed as being La protein, an abundant cellular RNA chaperone (Belisova et al., 2005, Chakshusmathi et al., 2003). La protein was shown to interact with the UTRs of dengue virus and Japanese encephalitis virus (De Nova-Ocampo et al., 2002, Garcia-Montalvo et al., 2004, Vashist et al., 2009, Vashist et al., 2011, Yocupicio-Monroy et al., 2007), and to bind to the viral replication proteins NS5 and NS3 (Garcia-Montalvo et al., 2004). At present, the effect of La binding on genomic RNA circularization is still controversial. Recombinant La protein led to decreased RNA synthesis in an in vitro viral replicase assay, leading to the suggestion that its binding inhibited RNA cyclization (Yocupicio-Monroy et al., 2007). However, co-precipitation of the 5′ and 3′ genomic RNA regions was stimulated by the presence of increasing amounts of La protein (Vashist et al., 2011). A possible explanation for these seemingly contradictory findings might be provided by the results of Villordo and co-workers, who have shown that a balance between the linear and circular conformations of the genomic RNA, rather than cyclization per se, is important for efficient viral replication (Villordo et al., 2010). Thus, overly efficient RNA cyclization, as well as the lack of it, might both be deleterious for RNA synthesis. Besides La protein, a number of other cellular RNA chaperones, including heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1), hnRNP A2, hnRNP Q, Y-box binding protein (YB-1), and polypyrimidine tract binding protein (PTB), have been described to interact with flaviviral genomic RNA (Agis-Juarez et al., 2009, De Nova-Ocampo et al., 2002, Katoh et al., 2011, Paranjape and Harris, 2007). Although the effect of these chaperones on genome cyclization is currently unknown, they might participate in the regulation of panhandle formation in the absence of (or in addition to) core protein, thus masking the in vivo effect of core on genome replication.
Nevertheless, recent mutagenesis studies have provided indirect evidence for the involvement of the core protein region in genome circularization and RNA replication (Basu and Brinton, 2011, Zhang et al., 2010). Genome-length viral RNAs with multiple adjacent mutations in CS or with a complete deletion of the 3′ CS element were replication competent and generated revertants or second-site mutations upon passaging, restoring efficient panhandle formation (Basu and Brinton, 2011, Zhang et al., 2010). In contrast, disruption of the 5′–3′ CS interaction invariably resulted in a lethal phenotype when analysed in subgenomic replicons lacking core protein (Corver et al., 2003, Khromykh et al., 2001, Lo et al., 2003). Similarly, a deletion in the core coding region of a genome-length replicon also precluded the rescue of the CS mutant (Zhang et al., 2010), although it is still debated whether this was due to the lack of core protein expression or to the absence of RNA secondary structures in the core-coding region (Friebe et al., 2012). These results, together with our findings (Fig. 3B), suggest that cellular proteins can only partially substitute for core protein chaperoning in genome cyclization, and the requirements for panhandle formation might be more relaxed in the presence of core. This is especially important since mutations in the cyclization sequences are considered for the design of live-attenuated flavivirus vaccines, with the aim to reduce the risk of recombination with naturally circulating viruses (Suzuki et al., 2008).
The annealing efficiency of 3′ UTR RNAs was found to be highly dependent on the length of the RNA used (Fig. 2). While 3′CYC RNA, containing the cyclization sequences and the 3′ SL structure, could interact with the 5′ region even in the absence of core protein, the longer 3′ΔVR and 3′FL-UTR RNAs required core for 5′–3′ complex formation, suggesting that the presence of the dumbbell structures (DB1 and DB2) might interfere with annealing. Pseudoknot interactions, conserved in mosquito-borne flaviviruses, have been suggested to form between the loop of the dumbbells and a single-stranded region, including the CS element, in the linear form of the viral RNA (Olsthoorn and Bol, 2001; Fig. 1, Fig. 2). Total or partial deletion of the individual dumbbell structures was found to seriously compromise RNA replication (Alvarez et al., 2005a, Manzano et al., 2011, Men et al., 1996), resulting in attenuated viruses that are actively pursued as a vaccine candidate for dengue viruses (Durbin et al., 2001, Troyer et al., 2001, Whitehead et al., 2003). Our results suggest that core protein chaperoning might be required to resolve the pseudoknots, yielding an RNA conformation competent for 5′–3′ annealing. Thus, the pseudoknot structures may constitute an additional layer of regulation in genome cyclization, contributing to the delicate balance between the linear and circular RNA forms.
In addition to the genomic RNA, a small subgenomic RNA (sfRNA), corresponding to the last 300–500 nts of the positive-strand viral RNA, also accumulates in cells infected by flaviviruses (Lin et al., 2004, Pijlman et al., 2008, Urosevic et al., 1997). sfRNA is generated by the incomplete degradation of the viral genome by the host exonuclease XRN1, where the 3′ RNA region is protected by a conserved pseudoknot structure (Funk et al., 2010, Pijlman et al., 2008, Silva et al., 2010). Kunjin virus sfRNA was not required for viral replication but was found to be essential for cytopathicity in cell culture and for pathogenicity in infected mice (Pijlman et al., 2008). As sfRNA contains all the RNA elements required for interaction with 5′ cyclization sequences, its accumulation might regulate (−)-strand RNA synthesis by inhibiting panhandle formation or by sequestering host or viral proteins (including core protein) interacting with the 3′ UTR (Fan et al., 2011). The interaction of core protein with sfRNA and the possible consequences remain to be experimentally verified.
Acknowledgements
Work was supported by ANRS, INSERM and FINOVI. Thanks are due to Hervé Zeller for providing WNV infected cells used to clone the 5′ and 3′ regions of the WNV genome.
References
- Agis-Juarez R.A., Galvan I., Medina F., Daikoku T., Padmanabhan R., Ludert J.E., del Angel R.M. Polypyrimidine tract-binding protein is relocated to the cytoplasm and is required during dengue virus infection in Vero cells. Journal of General Virology. 2009;90(Pt 12):2893–2901. doi: 10.1099/vir.0.013433-0. [DOI] [PubMed] [Google Scholar]
- Alvarez D.E., De Lella Ezcurra A.L., Fucito S., Gamarnik A.V. Role of RNA structures present at the 3′UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology. 2005;339(2):200–212. doi: 10.1016/j.virol.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Alvarez D.E., Filomatori C.V., Gamarnik A.V. Functional analysis of dengue virus cyclization sequences located at the 5′ and 3′UTRs. Virology. 2008;375(1):223–235. doi: 10.1016/j.virol.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Alvarez D.E., Lodeiro M.F., Luduena S.J., Pietrasanta L.I., Gamarnik A.V. Long-range RNA–RNA interactions circularize the dengue virus genome. Journal of Virology. 2005;79(11):6631–6643. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg S.M., Nestorowicz A., McCourt D.W., Rice C.M. NS2B-3 proteinase-mediated processing in the yellow fever virus structural region: in vitro and in vivo studies. Journal of Virology. 1994;68(6):3794–3802. doi: 10.1128/jvi.68.6.3794-3802.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu M., Brinton M.A. West Nile virus (WNV) genome RNAs with up to three adjacent mutations that disrupt long distance 5′–3′ cyclization sequence basepairs are viable. Virology. 2011;412(1):220–232. doi: 10.1016/j.virol.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belisova A., Semrad K., Mayer O., Kocian G., Waigmann E., Schroeder R., Steiner G. RNA chaperone activity of protein components of human Ro RNPs. RNA. 2005;11(7):1084–1094. doi: 10.1261/rna.7263905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuvanakantham R., Ng M.L. Analysis of self-association of West Nile virus capsid protein and the crucial role played by Trp 69 in homodimerization. Biochemical and Biophysical Research Communications. 2005;329(1):246–255. doi: 10.1016/j.bbrc.2005.01.121. [DOI] [PubMed] [Google Scholar]
- Bredenbeek P.J., Kooi E.A., Lindenbach B., Huijkman N., Rice C.M., Spaan W.J. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. Journal of General Virology. 2003;84(Pt 5):1261–1268. doi: 10.1099/vir.0.18860-0. [DOI] [PubMed] [Google Scholar]
- Brinton M.A. The molecular biology of West Nile Virus: a new invader of the western hemisphere. Annual Review of Microbiology. 2002;56:371–402. doi: 10.1146/annurev.micro.56.012302.160654. [DOI] [PubMed] [Google Scholar]
- Brinton M.A., Dispoto J.H. Sequence and secondary structure analysis of the 5′-terminal region of flavivirus genome RNA. Virology. 1988;162(2):290–299. doi: 10.1016/0042-6822(88)90468-0. [DOI] [PubMed] [Google Scholar]
- Brinton M.A., Fernandez A.V., Dispoto J.H. The 3′-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology. 1986;153(1):113–121. doi: 10.1016/0042-6822(86)90012-7. [DOI] [PubMed] [Google Scholar]
- Chakshusmathi G., Kim S.D., Rubinson D.A., Wolin S.L. A La protein requirement for efficient pre-tRNA folding. EMBO Journal. 2003;22(24):6562–6572. doi: 10.1093/emboj/cdg625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyde K., Barrera J., Harris E. The capsid-coding region hairpin element (cHP) is a critical determinant of dengue virus and West Nile virus RNA synthesis. Virology. 2008;379(2):314–323. doi: 10.1016/j.virol.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyde K., Harris E. RNA secondary structure in the coding region of dengue virus type 2 directs translation start codon selection and is required for viral replication. Journal of Virology. 2006;80(5):2170–2182. doi: 10.1128/JVI.80.5.2170-2182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corver J., Lenches E., Smith K., Robison R.A., Sando T., Strauss E.G., Strauss J.H. Fine mapping of a cis-acting sequence element in yellow fever virus RNA that is required for RNA replication and cyclization. Journal of Virology. 2003;77(3):2265–2270. doi: 10.1128/JVI.77.3.2265-2270.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofari G., Darlix J.L. The ubiquitous nature of RNA chaperone proteins. Progress in Nucleic Acid Research and Molecular Biology. 2002;72:223–268. doi: 10.1016/s0079-6603(02)72071-0. [DOI] [PubMed] [Google Scholar]
- Cristofari G., Ivanyi-Nagy R., Gabus C., Boulant S., Lavergne J.P., Penin F., Darlix J.L. The hepatitis C virus Core protein is a potent nucleic acid chaperone that directs dimerization of the viral (+) strand RNA in vitro. Nucleic Acids Research. 2004;32(8):2623–2631. doi: 10.1093/nar/gkh579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nova-Ocampo M., Villegas-Sepulveda N., del Angel R.M. Translation elongation factor-1alpha, La, and PTB interact with the 3′ untranslated region of dengue 4 virus RNA. Virology. 2002;295(2):337–347. doi: 10.1006/viro.2002.1407. [DOI] [PubMed] [Google Scholar]
- Doetsch M., Furtig B., Gstrein T., Stampfl S., Schroeder R. The RNA annealing mechanism of the HIV-1 Tat peptide: conversion of the RNA into an annealing-competent conformation. Nucleic Acids Research. 2011;39(10):4405–4418. doi: 10.1093/nar/gkq1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokland T., Walsh M., Mackenzie J.M., Khromykh A.A., Ee K.H., Wang S. West Nile virus core protein; tetramer structure and ribbon formation. Structure. 2004;12(7):1157–1163. doi: 10.1016/j.str.2004.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H., Zhang B., Shi P.Y. Terminal structures of West Nile virus genomic RNA and their interactions with viral NS5 protein. Virology. 2008;381(1):123–135. doi: 10.1016/j.virol.2008.07.040. [DOI] [PubMed] [Google Scholar]
- Durbin A.P., Karron R.A., Sun W., Vaughn D.W., Reynolds M.J., Perreault J.R., Thumar B., Men R., Lai C.J., Elkins W.R., Chanock R.M., Murphy B.R., Whitehead S.S. Attenuation and immunogenicity in humans of a live dengue virus type-4 vaccine candidate with a 30 nucleotide deletion in its 3′-untranslated region. American Journal of Tropical Medicine and Hygiene. 2001;65(5):405–413. doi: 10.4269/ajtmh.2001.65.405. [DOI] [PubMed] [Google Scholar]
- Fan Y.H., Nadar M., Chen C.C., Weng C.C., Lin Y.T., Chang R.Y. Small noncoding RNA modulates Japanese encephalitis virus replication and translation in trans. Virology Journal. 2011;8:492. doi: 10.1186/1743-422X-8-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filomatori C.V., Iglesias N.G., Villordo S.M., Alvarez D.E., Gamarnik A.V. RNA sequences and structures required for the recruitment and activity of the dengue virus polymerase. Journal of Biological Chemistry. 2011;286(9):6929–6939. doi: 10.1074/jbc.M110.162289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filomatori C.V., Lodeiro M.F., Alvarez D.E., Samsa M.M., Pietrasanta L., Gamarnik A.V. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes and Development. 2006;20(16):2238–2249. doi: 10.1101/gad.1444206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebe P., Harris E. Interplay of RNA elements in the dengue virus 5′ and 3′ ends required for viral RNA replication. Journal of Virology. 2010;84(12):6103–6118. doi: 10.1128/JVI.02042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebe P., Pena J., Pohl M.O., Harris E. Composition of the sequence downstream of the dengue virus 5′ cyclization sequence (dCS) affects viral RNA replication. Virology. 2012;422(2):346–356. doi: 10.1016/j.virol.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebe P., Shi P.Y., Harris E. The 5′ and 3′ downstream AUG region elements are required for mosquito-borne flavivirus RNA replication. Journal of Virology. 2011;85(4):1900–1905. doi: 10.1128/JVI.02037-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk A., Truong K., Nagasaki T., Torres S., Floden N., Balmori Melian E., Edmonds J., Dong H., Shi P.Y., Khromykh A.A. RNA structures required for production of subgenomic flavivirus RNA. Journal of Virology. 2010;84(21):11407–11417. doi: 10.1128/JVI.01159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Montalvo B.M., Medina F., del Angel R.M. La protein binds to NS5 and NS3 and to the 5′ and 3′ ends of Dengue 4 virus RNA. Virus Research. 2004;102(2):141–150. doi: 10.1016/j.virusres.2004.01.024. [DOI] [PubMed] [Google Scholar]
- Gebhard L.G., Filomatori C.V., Gamarnik A.V. Functional RNA elements in the dengue virus genome. Viruses. 2011;3(9):1739–1756. doi: 10.3390/v3091739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjenero-Margan I., Aleraj B., Krajcar D., Lesnikar V., Klobucar A., Pem-Novosel I., Kurecic-Filipovic S., Komparak S., Martic R., Duricic S., Betica-Radic L., Okmadzic J., Vilibic-Cavlek T., Babic-Erceg A., Turkovic B., Avsic-Zupanc T., Radic I., Ljubic M., Sarac K., Benic N., Mlinaric-Galinovic G. Autochthonous dengue fever in Croatia, August–September 2010. Euro Surveillance. 2010;16(9) [PubMed] [Google Scholar]
- Gould E.A., Higgs S. Impact of climate change and other factors on emerging arbovirus diseases. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2009;103(2):109–121. doi: 10.1016/j.trstmh.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould E.A., Solomon T. Pathogenic flaviviruses. Lancet. 2008;371(9611):500–509. doi: 10.1016/S0140-6736(08)60238-X. [DOI] [PubMed] [Google Scholar]
- Griffin D.E. Viral encephalomyelitis. PLoS Pathogens. 2011;7(3):e1002004. doi: 10.1371/journal.ppat.1002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritsun T.S., Gould E.A. Origin and evolution of flavivirus 5′UTRs and panhandles: trans-terminal duplications? Virology. 2007;366(1):8–15. doi: 10.1016/j.virol.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Guzman M.G., Halstead S.B., Artsob H., Buchy P., Farrar J., Gubler D.J., Hunsperger E., Kroeger A., Margolis H.S., Martinez E., Nathan M.B., Pelegrino J.L., Simmons C., Yoksan S., Peeling R.W. Dengue: a continuing global threat. Nature Reviews Microbiology. 2010;8(12 Suppl.):S7–S16. doi: 10.1038/nrmicro2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn C.S., Hahn Y.S., Rice C.M., Lee E., Dalgarno L., Strauss E.G., Strauss J.H. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. Journal of Molecular Biology. 1987;198(1):33–41. doi: 10.1016/0022-2836(87)90455-4. [DOI] [PubMed] [Google Scholar]
- Ivanyi-Nagy R., Kanevsky I., Gabus C., Lavergne J.P., Ficheux D., Penin F., Fosse P., Darlix J.L. Analysis of hepatitis C virus RNA dimerization and core-RNA interactions. Nucleic Acids Research. 2006;34(9):2618–2633. doi: 10.1093/nar/gkl240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanyi-Nagy R., Lavergne J.P., Gabus C., Ficheux D., Darlix J.L. RNA chaperoning and intrinsic disorder in the core proteins of Flaviviridae. Nucleic Acids Research. 2008;36(3):712–725. doi: 10.1093/nar/gkm1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C.T., Ma L., Burgner J.W., Groesch T.D., Post C.B., Kuhn R.J. Flavivirus capsid is a dimeric alpha-helical protein. Journal of Virology. 2003;77(12):7143–7149. doi: 10.1128/JVI.77.12.7143-7149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh H., Mori Y., Kambara H., Abe T., Fukuhara T., Morita E., Moriishi K., Kamitani W., Matsuura Y. Heterogeneous nuclear ribonucleoprotein A2 participates in the replication of Japanese encephalitis virus through an interaction with viral proteins and RNA. Journal of Virology. 2011;85(21):10976–10988. doi: 10.1128/JVI.00846-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh A.A., Meka H., Guyatt K.J., Westaway E.G. Essential role of cyclization sequences in flavivirus RNA replication. Journal of Virology. 2001;75(14):6719–6728. doi: 10.1128/JVI.75.14.6719-6728.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh A.A., Westaway E.G. RNA binding properties of core protein of the flavivirus Kunjin. Archives of Virology. 1996;141(3–4):685–699. doi: 10.1007/BF01718326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh A.A., Westaway E.G. Subgenomic replicons of the flavivirus Kunjin: construction and applications. Journal of Virology. 1997;71(2):1497–1505. doi: 10.1128/jvi.71.2.1497-1505.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick A.M. Globalization, land use, and the invasion of West Nile virus. Science. 2011;334(6054):323–327. doi: 10.1126/science.1201010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R.M., Heinz F.X., Mandl C.W. Capsid protein C of tick-borne encephalitis virus tolerates large internal deletions and is a favorable target for attenuation of virulence. Journal of Virology. 2002;76(7):3534–3543. doi: 10.1128/JVI.76.7.3534-3543.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R.M., Leitner A., O’Riordain G., Heinz F.X., Mandl C.W. Spontaneous mutations restore the viability of tick-borne encephalitis virus mutants with large deletions in protein C. Journal of Virology. 2003;77(1):443–451. doi: 10.1128/JVI.77.1.443-451.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Ruche G., Souares Y., Armengaud A., Peloux-Petiot F., Delaunay P., Despres P., Lenglet A., Jourdain F., Leparc-Goffart I., Charlet F., Ollier L., Mantey K., Mollet T., Fournier J.P., Torrents R., Leitmeyer K., Hilairet P., Zeller H., Van Bortel W., Dejour-Salamanca D., Grandadam M., Gastellu-Etchegorry M. First two autochthonous dengue virus infections in metropolitan France, September 2010. Euro Surveillance. 2010;15(39):19676. [PubMed] [Google Scholar]
- Lin K.C., Chang H.L., Chang R.Y. Accumulation of a 3′-terminal genome fragment in Japanese encephalitis virus-infected mammalian and mosquito cells. Journal of Virology. 2004;78(10):5133–5138. doi: 10.1128/JVI.78.10.5133-5138.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach B., Thiel H.J., Rice C.M. Lippincott-Raven Publishers; Philadelphia: 2007. Flaviviridae: The Viruses and their Replication, Fields Virology. pp. 1101–1152. [Google Scholar]
- Lo M.K., Tilgner M., Bernard K.A., Shi P.Y. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. Journal of Virology. 2003;77(18):10004–10014. doi: 10.1128/JVI.77.18.10004-10014.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobigs M. Flavivirus premembrane protein cleavage and spike heterodimer secretion require the function of the viral proteinase NS3. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(13):6218–6222. doi: 10.1073/pnas.90.13.6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodeiro M.F., Filomatori C.V., Gamarnik A.V. Structural and functional studies of the promoter element for dengue virus RNA replication. Journal of Virology. 2009;83(2):993–1008. doi: 10.1128/JVI.01647-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Jones C.T., Groesch T.D., Kuhn R.J., Post C.B. Solution structure of dengue virus capsid protein reveals another fold. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(10):3414–3419. doi: 10.1073/pnas.0305892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano M., Reichert E.D., Polo S., Falgout B., Kasprzak W., Shapiro B.A., Padmanabhan R. Identification of cis-acting elements in the 3′-untranslated region of the dengue virus type 2 RNA that modulate translation and replication. Journal of Biological Chemistry. 2011;286(25):22521–22534. doi: 10.1074/jbc.M111.234302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markoff L. 5′- and 3′-noncoding regions in flavivirus RNA. Advances in Virus Research. 2003;59:177–228. doi: 10.1016/S0065-3527(03)59006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markoff L., Falgout B., Chang A. A conserved internal hydrophobic domain mediates the stable membrane integration of the dengue virus capsid protein. Virology. 1997;233(1):105–117. doi: 10.1006/viro.1997.8608. [DOI] [PubMed] [Google Scholar]
- Men R., Bray M., Clark D., Chanock R.M., Lai C.J. Dengue type 4 virus mutants containing deletions in the 3′ noncoding region of the RNA genome: analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. Journal of Virology. 1996;70(6):3930–3937. doi: 10.1128/jvi.70.6.3930-3937.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikaelian I., Sergeant A. A general and fast method to generate multiple site directed mutations. Nucleic Acids Research. 1992;20(2):376. doi: 10.1093/nar/20.2.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsthoorn R.C., Bol J.F. Sequence comparison and secondary structure analysis of the 3′ noncoding region of flavivirus genomes reveals multiple pseudoknots. RNA. 2001;7(10):1370–1377. [PMC free article] [PubMed] [Google Scholar]
- Paranjape S.M., Harris E. Y box-binding protein-1 binds to the dengue virus 3′-untranslated region and mediates antiviral effects. Journal of Biological Chemistry. 2007;282(42):30497–30508. doi: 10.1074/jbc.M705755200. [DOI] [PubMed] [Google Scholar]
- Patkar C.G., Jones C.T., Chang Y.H., Warrier R., Kuhn R.J. Functional requirements of the yellow fever virus capsid protein. Journal of Virology. 2007;81(12):6471–6481. doi: 10.1128/JVI.02120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen L.P. vol. 1937. 2009. Global epidemiology of West Nile Virus, emerging infectious diseases of the 21st century. (West Nile Encephalitis Virus Infection). pp. 1–23. [Google Scholar]
- Pijlman G.P., Funk A., Kondratieva N., Leung J., Torres S., van der Aa L., Liu W.J., Palmenberg A.C., Shi P.Y., Hall R.A., Khromykh A.A. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host and Microbe. 2008;4(6):579–591. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Pong W.L., Huang Z.S., Teoh P.G., Wang C.C., Wu H.N. RNA binding property and RNA chaperone activity of dengue virus core protein and other viral RNA-interacting proteins. FEBS Letters. 2011;585(16):2575–2581. doi: 10.1016/j.febslet.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen L. Dengue in Greece in 1927 and 1928 and the pathogenesis of dengue hemorrhagic fever: new data and a different conclusion. American Journal of Tropical Medicine and Hygiene. 1986;35(3):642–653. doi: 10.4269/ajtmh.1986.35.642. [DOI] [PubMed] [Google Scholar]
- Samsa M.M., Mondotte J.A., Iglesias N.G., Assuncao-Miranda I., Barbosa-Lima G., Da Poian A.T., Bozza P.T., Gamarnik A.V. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathogens. 2009;5(10):e1000632. doi: 10.1371/journal.ppat.1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlick P., Taucher C., Schittl B., Tran J.L., Kofler R.M., Schueler W., von Gabain A., Meinke A., Mandl C.W. Helices alpha2 and alpha3 of West Nile virus capsid protein are dispensable for assembly of infectious virions. Journal of Virology. 2009;83(11):5581–5591. doi: 10.1128/JVI.02653-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder R., Barta A., Semrad K. Strategies for RNA folding and assembly. Nature Reviews Molecular Cell Biology. 2004;5(11):908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- Shi P.Y., Brinton M.A., Veal J.M., Zhong Y.Y., Wilson W.D. Evidence for the existence of a pseudoknot structure at the 3′ terminus of the flavivirus genomic RNA. Biochemistry. 1996;35(13):4222–4230. doi: 10.1021/bi952398v. [DOI] [PubMed] [Google Scholar]
- Silva P.A., Pereira C.F., Dalebout T.J., Spaan W.J., Bredenbeek P.J. An RNA pseudoknot is required for production of yellow fever virus subgenomic RNA by the host nuclease XRN1. Journal of Virology. 2010;84(21):11395–11406. doi: 10.1128/JVI.01047-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R., Fayzulin R., Frolov I., Mason P.W. Identification of mutated cyclization sequences that permit efficient replication of West Nile virus genomes: use in safer propagation of a novel vaccine candidate. Journal of Virology. 2008;82(14):6942–6951. doi: 10.1128/JVI.00662-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P., Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB Journal. 2004;18(11):1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- Troyer J.M., Hanley K.A., Whitehead S.S., Strickman D., Karron R.A., Durbin A.P., Murphy B.R. A live attenuated recombinant dengue-4 virus vaccine candidate with restricted capacity for dissemination in mosquitoes and lack of transmission from vaccinees to mosquitoes. American Journal of Tropical Medicine and Hygiene. 2001;65(5):414–419. doi: 10.4269/ajtmh.2001.65.414. [DOI] [PubMed] [Google Scholar]
- Urosevic N., van Maanen M., Mansfield J.P., Mackenzie J.S., Shellam G.R. Molecular characterization of virus-specific RNA produced in the brains of flavivirus-susceptible and -resistant mice after challenge with Murray Valley encephalitis virus. Journal of General Virology. 1997;78(Pt 1):23–29. doi: 10.1099/0022-1317-78-1-23. [DOI] [PubMed] [Google Scholar]
- Vashist S., Anantpadma M., Sharma H., Vrati S. La protein binds the predicted loop structures in the 3′ non-coding region of Japanese encephalitis virus genome: role in virus replication. Journal of General Virology. 2009;90(Pt 6):1343–1352. doi: 10.1099/vir.0.010850-0. [DOI] [PubMed] [Google Scholar]
- Vashist S., Bhullar D., Vrati S. La protein can simultaneously bind to both 3′- and 5′-noncoding regions of Japanese encephalitis virus genome. DNA and Cell Biology. 2011;30(6):339–346. doi: 10.1089/dna.2010.1114. [DOI] [PubMed] [Google Scholar]
- Villordo S.M., Alvarez D.E., Gamarnik A.V. A balance between circular and linear forms of the dengue virus genome is crucial for viral replication. RNA. 2010;16(12):2325–2335. doi: 10.1261/rna.2120410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.H., Syu W.J., Hu S.T. Identification of the homotypic interaction domain of the core protein of dengue virus type 2. Journal of General Virology. 2004;85(Pt 8):2307–2314. doi: 10.1099/vir.0.80067-0. [DOI] [PubMed] [Google Scholar]
- Whitehead S.S., Falgout B., Hanley K.A., Blaney J.E., Jr.JJr., Markoff L., Murphy B.R. A live, attenuated dengue virus type 1 vaccine candidate with a 30-nucleotide deletion in the 3′ untranslated region is highly attenuated and immunogenic in monkeys. Journal of Virology. 2003;77(2):1653–1657. doi: 10.1128/JVI.77.2.1653-1657.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamshchikov V.F., Compans R.W. Processing of the intracellular form of the west Nile virus capsid protein by the viral NS2B-NS3 protease: an in vitro study. Journal of Virology. 1994;68(9):5765–5771. doi: 10.1128/jvi.68.9.5765-5771.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yocupicio-Monroy M., Padmanabhan R., Medina F., del Angel R.M. Mosquito La protein binds to the 3′ untranslated region of the positive and negative polarity dengue virus RNAs and relocates to the cytoplasm of infected cells. Virology. 2007;357(1):29–40. doi: 10.1016/j.virol.2006.07.042. [DOI] [PubMed] [Google Scholar]
- You S., Falgout B., Markoff L., Padmanabhan R. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. Journal of Biological Chemistry. 2001;276(19):15581–15591. doi: 10.1074/jbc.M010923200. [DOI] [PubMed] [Google Scholar]
- You S., Padmanabhan R. A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. Journal of Biological Chemistry. 1999;274(47):33714–33722. doi: 10.1074/jbc.274.47.33714. [DOI] [PubMed] [Google Scholar]
- Zhang B., Dong H., Stein D.A., Iversen P.L., Shi P.Y. West Nile virus genome cyclization and RNA replication require two pairs of long-distance RNA interactions. Virology. 2008;373(1):1–13. doi: 10.1016/j.virol.2008.01.016. [DOI] [PubMed] [Google Scholar]
- Zhang B., Dong H., Ye H., Tilgner M., Shi P.Y. Genetic analysis of West Nile virus containing a complete 3′CSI RNA deletion. Virology. 2010;408(2):138–145. doi: 10.1016/j.virol.2010.09.033. [DOI] [PubMed] [Google Scholar]
- Zhu W., Qin C., Chen S., Jiang T., Yu M., Yu X., Qin E. Attenuated dengue 2 viruses with deletions in capsid protein derived from an infectious full-length cDNA clone. Virus Research. 2007;126(1–2):226–232. doi: 10.1016/j.virusres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]


