Abstract
The family Picornaviridae comprises of small, non-enveloped, positive-strand RNA viruses and contains many human and animal pathogens including enteroviruses (e.g. poliovirus, coxsackievirus, enterovirus 71 and rhinovirus), cardioviruses (e.g. encephalomyocarditis virus), hepatitis A virus and foot-and-mouth disease virus. Picornavirus infections activate a cytosolic RNA sensor, MDA5, which in turn, induces a type I interferon response, a crucial component of antiviral immunity. Moreover, picornaviruses activate the formation of stress granules (SGs), large aggregates of preassembled mRNPs (messenger ribonucleoprotein particles) to temporarily store these molecules upon cellular stress. Meanwhile, picornaviruses actively suppress these antiviral responses to ensure efficient replication. In this review we provide an overview of the induction and suppression of the MDA5-mediated IFN-α/β response and the cellular stress pathway by picornaviruses.
Keywords: Picornaviruses, Interferon, MDA5, Stress granules, Viral evasion
1. Introduction
On the front line of antiviral immunity is the production of type I interferons (IFN-α/β) and other antiviral cytokines at the site of infection. IFN-α/β can be produced by virtually all nucleated cell types and act in autocrine and paracrine manners (i.e. on both the infected cells and neighboring non-infected cells). IFN-α/β-receptor engagement induces the expression of large numbers of interferon-stimulated genes (ISGs), which together establish a so-called antiviral state in the recipient cells. To recognize the invading pathogens timely and correctly, cells employ specialized pattern recognition receptors (PRRs) to detect specific pathogen-associated molecular patterns (PAMPs). RIG-I-like receptors (RLRs) are a family of ubiquitously expressed PRRs that detect “non-self” viral RNAs in the cytoplasm. Two RLRs, RIG-I and MDA5, mediate IFN-α/β production upon infection of various RNA viruses [1], [2]. While RIG-I is required for sensing, among others, paramyxoviruses, influenza virus, Japanese encephalitis virus and hepatitis C virus, MDA5 recognizes picornaviruses, mouse norovirus, mouse hepatitis virus and defective interfering particles of paramyxoviruses [1], [2]. Another RLR, LGP2, lacks functional domains required for downstream signaling, and therefore has been long suspected to play regulatory roles on RIG-I and MDA5. Besides the IFN-α/β and inflammatory responses, cells also engage various other mechanisms to cope with undesirable conditions. One of such mechanisms is the formation of stress granules (SGs) in the presence of stress such as oxidative, heat, or nutrient stress, UV radiation and viral infections. Although the stress pathway was initially thought to function independently of classical innate antiviral responses such as IFN-α/β, it was recently suggested that the stress response may also directly or indirectly play an antiviral role [3].
During the long co-evolution with their hosts, viruses have acquired strategies to actively counteract host antiviral responses. Both the RLR-mediated IFN-α/β induction pathway as well as the stress pathway are targeted by various viruses [3], [4]. This review summarizes our current knowledge on the recognition and suppression of host antiviral pathways by picornaviruses, a large family of human and animal pathogens. We focus on two important and well-studied genera of picornaviruses, namely Enterovirus and Cardiovirus, and discuss how their RNAs are recognized by RLRs, and how they antagonize the IFN-α/β induction pathway and SG formation in infected cells.
2. Picornaviruses
2.1. Classification and genetics of picornaviruses
Picornaviridae is a large and diverse virus family currently containing 26 genera. The Enterovirus genus contains hundreds of (sero)types of important human pathogens [4], [5], [6]. Poliovirus (PV) is the causative agent of poliomyelitis. Various types of coxsackieviruses (CVs), echoviruses, and other enteroviruses (EVs) can cause viral meningitis, encephalitis, myocarditis and pancreatitis, and have also been implicated in the development of type I diabetes [5], [7]. Enterovirus 71 (EV71) is an emerging virus that can cause severe neurological symptoms in young children, and have caused many outbreaks in the past decade, mostly in Southeast Asia [8]. Human rhinoviruses (HRVs) are responsible for approximately one third of common colds in adults, and are also associated with asthma exacerbations and chronic obstructive pulmonary disease (COPD) [9]. The Cardiovirus genus contains mostly animal pathogens such as encephalomyocarditis virus (EMCV) and Theiler's murine encephalomyelitis virus (TMEV). Both EMCV and TMEV are primarily rodent pathogens, but EMCV also causes severe, sometimes lethal, infections in other animals such as pigs, elephants, lions and primates, posing problems in zoos and national parks [10], [11]. Recently, a new cardiovirus that infects humans has been discovered, namely the Saffold virus, which has been associated with gastroenteritis, and respiratory and neurological infections [12]. Other well-known genera are Hepatovirus (e.g. hepatitis A virus [HAV]), Aphthovirus (e.g. the foot-and-mouth disease virus [FMDV]), and Parechovirus (e.g. human parechovirus [HPeV]).
2.2. Life cycle of picornaviruses
Members of the Picornaviridae family are small, non-enveloped, positive-strand RNA viruses. The viral genome, a single-stranded (ss) RNA molecule of 7.5–8.5 kb, encodes a single open reading frame (ORF), an untranslated region (UTR) at either terminus, and a poly(A) tail at the extreme 3′ end. The 5′ terminus of the viral RNA is coupled to a small viral peptide VPg (also known as 3B) via a phosphodiester bond as a result of VPg-primed viral RNA replication. The genomic RNA also contains several structured RNA elements that are crucial for virus replication. The internal ribosomal entry site (IRES) in the 5′ UTR contains several stem–loop structures and drives viral cap-independent translation. Stem–loop structures in the UTRs and a cis-acting RNA element (CRE) – the position of which varies across different genera – are crucial for viral RNA replication [13].
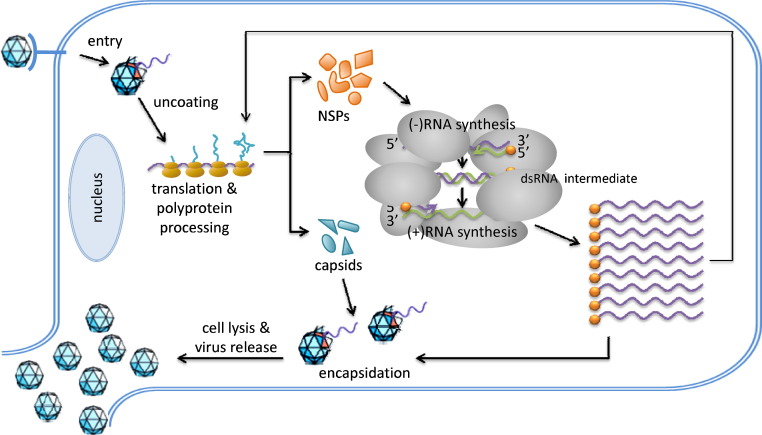
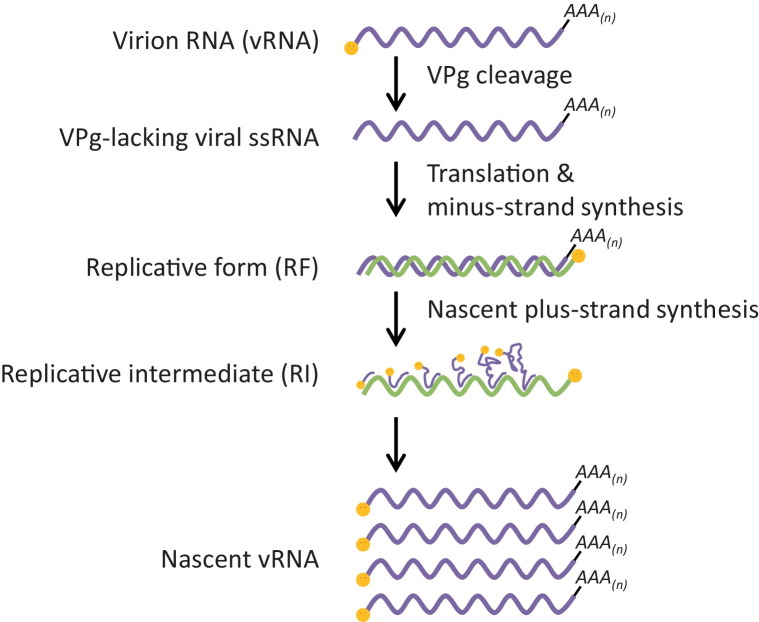
Picornaviruses share a similar life cycle (Fig. 1 ), with some details varying across genera [13]. Infection is initiated via receptor-mediated endocytosis, followed by uncoating to release the genomic RNA in the cytoplasm. A cellular enzyme then releases the VPg peptide from the genomic RNA [14], for reasons not yet understood, generating a single-stranded viral RNA carrying a 5′ monophosphate group. The viral genome is then immediately translated by the host translation machinery to generate a large polyprotein, which undergoes proteolytic processing by the virally encoded proteinases. All picornaviruses carry a 3Cpro, which mediates most of the proteolytic processing, and members of some genera carry an additional proteinase that also participates (e.g. 2Apro for enteroviruses and Lpro for aphthoviruses). Additionally, these viral proteinases also cleave host factors to aid virus RNA replication and/or to evade host antiviral responses. Next, several viral non-structural proteins hijack regulatory mechanisms of host membrane metabolism to induce extensive remodeling of the intracellular membranous structures to form the so-called replication organelles (ROs) where viral RNA replication takes place. The process of RNA replication is carried out by the virally encoded RNA-dependent RNA polymerase 3Dpol (Fig. 2 ). First, 3Dpol uridylylyates VPg, and uses the resulting VPg-pU-pU as a primer to transcribe the positive-strand RNA into a complementary, negative-strand RNA molecule. During this process, a long dsRNA intermediate product is produced, which is referred to as the replicative form (RF). Next, 3Dpol uses the negative-strand RNA as a template, and again VPg-pU-pU as a primer, to produce a large number of nascent positive strands. This step leads to the production of another intermediate product, namely the replicative intermediate (RI), which comprises of a single negative-strand RNA and multiple incomplete positive-strand RNAs that are undergoing active transcription. The completed nascent positive-strand RNAs either enter a new round of translation and RNA replication, or are encapsidated to form new virions. At the end of the replication cycle, progeny virus particles are released by cell lysis [13].
Fig. 1.
Life cycle of picornaviruses. NSPs, non-structural proteins. Purple line, viral positive-strand (+)RNAs. Green line, viral negative-strand (−)RNAs. Orange circle, VPg/3B.
Fig. 2.
Steps of picornavirus RNA replication. Purple line, viral positive-strand RNAs. Green line, viral negative-strand RNAs. Orange circle, VPg/3B. Modified from [24].
2A and L are the two most divergent picornaviral proteins. Both can act as proteinases in some genera but not in others [13]. Because of their activities in inducing host shutoff, modulating cell death and counter-acting immune responses, these proteins have been classified as viral security proteins [15]. In fact, the L proteins of both cardioviruses and aphthoviruses have been found to actively suppress IFN-α/β response ([16], [17], see also Sections 4.2, 4.3).
3. Recognition of picornavirus RNAs by RIG-I-like receptors
3.1. RIG-I-like receptors in antiviral responses
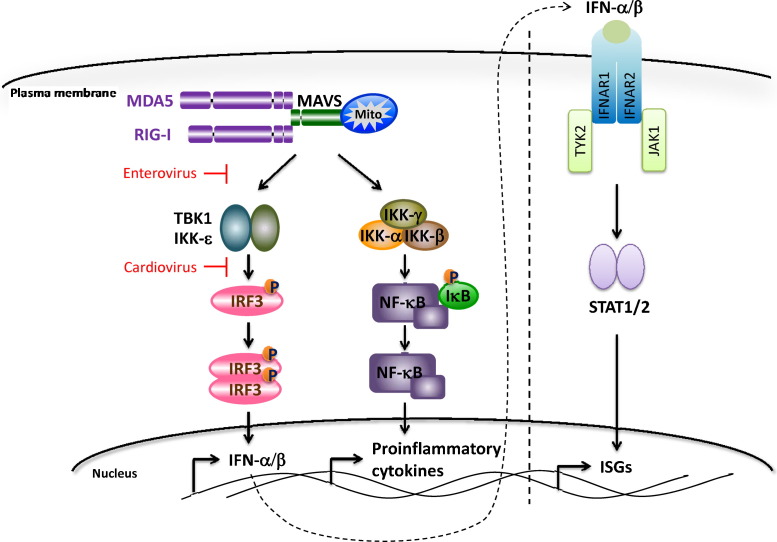
RIG-I-like receptors (RLRs) are cytoplasmic RNA sensors that initiate a type I interferon response upon RNA virus infections. Currently, the RLR family contains three DExH/D box helicases, namely RIG-I, MDA5 and LGP2. Upon ligand recognition, RIG-I and MDA5 interact with a mitochondrial adaptor molecule MAVS (also known as VISA, Cardif and IPS-I), which in turn, activates the TBK1/IKK-ɛ and IKK-α/β/γ pathways (Fig. 3 ). TBK1 then phosphorylates and activates IRF3, leading to transcription activation of IFN-α/β genes, whereas the IKK-α/β/γ complex leads to NF-κB activation and the transcription of many proinflammatory cytokine genes. LGP2 also binds RNAs but lacks the signaling domains, thereby the ability to initiate downstream signaling cascades. Instead, it has been shown to play regulatory roles on RIG-I and MDA5 [1], [2].
Fig. 3.
RLR-mediated IFN-α/β induction pathway and the IFN-α/β signaling pathway. ISGs, interferon-stimulated genes. Blockade by enteroviruses and cardioviruses are indicated in red.
Lots of effort have been invested into characterizing RIG-I and MDA5 ligands [1], [2]. In vitro studies using synthetic ligands showed that RIG-I requires relatively short dsRNAs, or 5′ triphosphate (5′ppp)-containing ssRNAs with base-paired regions for activation. In agreement with these findings, the 5′ppp-containing pan-handle RNA structures of many negative-strand RNA viruses (e.g. influenza virus and Sendai virus) have been shown to potently activate RIG-I. MDA5 is activated by long dsRNAs, as evidenced by transfection studies using poly(I:C). To date, there is no evidence that specific terminal groups are required on MDA5 ligands.
3.2. What is the viral PAMP(s) that activates MDA5 in picornavirus-infected cells?
It is well established that picornavirus infections induce MDA5-mediated IFN-α/β response in both cultured cells as well as mice [18], [19], [20], [21], [22], [23]. MDA5-stimulatory activity was observed in total RNA extracts from EMCV- or TMEV-infected cells [23]. Further analysis in this study suggested that the MDA5-stimulating RNA was a high molecular weight RNA complex that consisted of both ssRNAs and dsRNAs, hinting that cardiovirus infections may lead to production of various PAMPs that can activate MDA5. As mentioned above, several viral RNA species are produced during picornavirus RNA replication, all bearing “non-self” features that could potentially be recognized by cellular sensors including MDA5. Picornavirus ssRNAs lack a 5′ cap structure but, instead, contain either a covalently linked VPg peptide or a monophosphate at the 5′ terminus. The RF is a dsRNA molecule of 7.5–8.5 kbp, and the RI is a complex, partially double-stranded RNA. Recently, Triantafilou et al. and our group both examined the abilities of purified picornaviral RNA species in stimulating MDA5. Together, the results clearly show that the two viral ssRNA species are in fact poor IFN-α/β inducers upon transfection, despite of the highly structured regions and the “non-self” features they carry [24], [25]. In contrast, both viral RNA replication intermediate products, namely the RF and the RI, induced potent IFN-α/β response upon transfection, which was completely dependent on MDA5 [24], [25]. Additionally, the RF was shown to activate the ATPase activity of recombinant MDA5 in the absence of any additional proteins [24], demonstrating that this RNA can serve as a direct ligand of MDA5.
While these recent data from transfection and in vitro experiments provided significant insights into picornavirus RNA recognition by MDA5, they cannot be taken for granted to reflect MDA5 activation in infected cells. During infection, the viral RNAs may be less, if at all, accessible because they are bound by various viral and host factors that participate in viral RNA replication, and may also be (partially) shielded by the virus-induced membranous ROs. Recently, this question was tackled by using compounds that inhibited viral RNA replication at different steps in infected cells. Inhibition of negative-strand RNA synthesis, and therefore RF formation, caused a complete abrogation of IFN-α/β response in infected cells. In contrast, a significant IFN-α/β response was observed when RF formation was allowed to proceed but the subsequent step, nascent positive-strand RNA synthesis, thereby the formation of RI, was inhibited [24]. These results strongly suggest that picornavirus RF is a physiological MDA5 ligand in virus-infected cells. The role of RI in stimulating MDA5 under physiological conditions is less clear. In contrast to RF, the RI is a primarily ssRNA molecule containing double-stranded regions and many protruding single-stranded ends [26]. MDA5 activation, however, requires long dsRNAs [1], [2], [27]. The average length of the double-stranded regions in RI molecules is unknown. Yet, RI induced high levels of MDA5-mediated IFN-α/β response upon transfection [25], suggesting that there is sufficient length of base-paired regions in purified RI to induce MDA5 activation. Whether this is also the case for the dynamic structure of RI during active viral RNA replication remains to be established.
3.3. Role of LGP2 in picornavirus-induced MDA5 activation
Besides the classically recognized viral RNA species of picornaviruses, Deddouche and colleagues suggested in a recent study that an additional MDA5-stimulating viral PAMP might be produced in EMCV-infected cells. LGP2-associated RNAs from EMCV-infected cells were found to exert MDA5-stimulatory activity upon transfection of naive cells [28]. Subsequent deep-sequencing analysis of the RNA pool that co-immunoprecipitated with LGP2 revealed a clear enrichment of a 170-nucleotide fragment that is complementary to the coding region of the viral protein L. While this is a remarkable and unexpected finding, it also raises a number of questions.
Firstly, it is not yet clear whether this L antisense RNA is produced as a specific entity during EMCV infection since northern blotting analysis failed to detect this RNA species in total RNA preparation from EMCV-infected cells [28]. Therefore this 170-nucleotide fragment might represent the most stable fragment of a larger trunk of, or even the full-length, negative-strand RNA during the isolation procedure. Alternatively, small RLR-stimulating RNAs can be generated by RNase L from host and viral RNAs during virus infections [29], [30], [31], and perhaps the L antisense RNA fragment is such a product. However, there was no evidence of the involvement of RNase L, which is also in line with a previous observation that EMCV-induced IFN-α/β response was comparable in wt and RNase L knockout cells [24]. It remains to be clarified if and how this EMCV L antisense RNA is produced during infection.
Secondly, it is unclear how such a small ssRNA could activate MDA5 in an LGP2-dependent manner. Involvement of LGP2 in EMCV-induced IFN-α/β response has been previously reported [32]. The newly discovered L antisense RNA was recovered from LGP2 but not MDA5 immunoprecipitations [28], suggesting that LGP2 directly binds to this small RNA and then facilitate MDA5 activation. Both MDA5 and LGP2 have been shown to bind short ssRNAs [33], [34], [35], [36], although MDA5 can only be functionally activated by long dsRNAs [27], [36]. LGP2 has also been shown to interact with MDA5, though this strictly depended on the presence of dsRNAs [37]. It remains to be demonstrated whether the base-paired regions in EMCV L antisense RNA are sufficient to facilitate such LGP2/MDA5 complex formation, or that LGP2 induces MDA5 activation via a yet unknown mechanism. In any case, the L antisense RNA seems to contain some immune-stimulatory signature since in vitro transcripts (without 5′ppp to exclude RIG-I-mediated recognition) of this fragment, but not that of the complementary sequence (L sense), induced an MDA5-mediated IFN-α/β response upon transfection [28].
Lastly, it is important to realize that the L antisense RNA is not the sole MDA5 ligand produced during EMCV infection (see Section 3.2). A recombinant EMCV carrying deletions in the L-encoding region (ΔL), which thereby could not produce the L antisense RNA (or the L protein, the known IFN-α/β antagonist), induced a potent IFN-α/β response. Yet, this response was less strong as compared to another recombinant EMCV that lacked a functional L protein due to mutations (Lmut) but could still produce the L antisense RNA [28]. Whether picornaviruses from other genera also produce small MDA5-stimulating ssRNAs remains to be further investigated.
4. Strategies of picornaviruses to evade the RLR-mediated type I interferon response
Many viruses have evolved to actively suppress the IFN-α/β system (often at multiple steps) [1], [2], [4], [5], and picornaviruses are no exception. It has long been suspected that picornavirus-induced host gene expression shutoff and secretory pathway blockade (the latter of which has only been reported for some genera such as Enterovirus) contribute to IFN-α/β suppression [5]. However, we and others have shown that little IRF3 activation can be detected during enterovirus and cardiovirus infections [38], [39], [40], implying that the primary blockade of this pathway lies upstream of IFN production.
4.1. Enteroviral strategies to restrict IFN-α/β transcription
4.1.1. Antagonization of the RLR signaling pathway
Enterovirus infections trigger little IRF3 phosphorylation [38], [39], [40]. A recent report showed that TBK1 phosphorylation, a prerequisite to IRF3 phosphorylation, is inhibited in CVB3-infected cells, suggesting that inhibition of the RLR-mediated IFN-α/β induction pathway lies upstream of TBK1 [41]. In line with this observation, several enteroviruses have been reported to directly interfere with upstream factors. MDA5 was shown to be degraded during infections of PV and EV71 in a caspase-dependent manner, and in the case of PV, also via proteasome activities [39], [42]. Also the downstream adaptor molecule MAVS has been shown to be targeted by several enteroviruses including HRV1A [40], CVB3 [38] and EV71 [43], via various proposed mechanisms. 2Apro of EV71 and 3Cpro of CVB3 were suggested to be responsible for MAVS cleavage during infection of these viruses, whereas 2Apro, 3Cpro as well as caspase 3 were all implicated in HRV1A-induced MAVS cleavage. It was somewhat surprising that enteroviruses seemed to employ a variety of means to shut down the RLR pathway, since these viruses often utilize the same strategies to target a particular host factor or pathway. Recently, a systematic examination of signaling components of the RLR pathway revealed that both MDA5 and MAVS are cleaved during CVB3 infection, and could be reproduced by recombinant viral proteinase 2Apro. 2Apro's of three enteroviruses belonging to different species, namely EV71 (Enterovirus A), CVB3 (Enterovirus B) and PV (Enterovirus C), also targeted MDA5 and MAVS for cleavage when inserted into the genome of a EMCV (which does not induce these cleavages by itself) [41]. These data suggest that enteroviruses very likely employ a unified mechanism to interfere with the RLR-mediated IFN-α/β induction pathway. Recently, we also observed that insertion of enterovirus 2Apro in EMCV Lmut not only resulted in cleavage of MDA5 and MAVS, but also almost a complete inhibition of IFN-α/β induction (data not shown). These data show that 2Apro is indeed the enterovirus IFN-α/β antagonist.
That MDA5 is specifically targeted by the viral 2Apro [41] seems to contradict earlier reports that MDA5 is degraded by caspases and the proteasome during enterovirus infections [39], [42]. However, it is important to point out that the caspase- and proteasome-mediated MDA5 cleavage events were observed under conditions where MDA5 expression level was artificially upregulated prior to infection (either by poly(I:C) or viral RNA transfections) [39], [42], which may sensitize cells for virus-induced apoptosis, thereby promote caspase- and proteasome-mediated protein degradations. In contrast, the 2Apro-mediated MDA5 cleavage was observed without manipulations of its expression levels [41]. The importance of studying cleavage of endogenous proteins is also illustrated by the observation that 3Cpro can cleave overexpressed MAVS [38], [41] but not endogenous MAVS. Cleavage products observed in infected cells are only observed upon expression of 2Apro [41], [43], suggesting that 2Apro-mediated MAVS cleavage is likely a more prominent event under physiological conditions.
Interestingly, RIG-I is also cleaved by various enteroviruses via their 3Cpro activity [41], [44]. However, it remains to be elucidated why these viruses target a RNA sensor that does not participate in their recognition [1], [2]. It has been proposed that RIG-I may directly turn on ISG transcription via STAT1 activation in an IFN-α/β-independent fashion [45]. Whether enteroviruses cleave RIG-I to prevent an augmentation of ISG expression via the STAT1 pathway remains to be investigated.
4.1.2. Effect of nucleocytoplasmic trafficking disorder?
As mentioned above, enterovirus 2Apro is an effective IFN-α/β antagonist. While it is tempting to attribute this IFN-α/β-suppressing effect to the cleavage of MDA5 and MAVS, it is known that 2Apro also targets many other host proteins, thereby affecting many cellular processes, which may possibly influence IFN-α/β response against these viruses. One group of cellular substrates of 2Apro are the nucleoporins (Nups), proteins that form the nuclear pore complex and regulate protein and mRNA trafficking through the nuclear pore. Nup cleavage during enterovirus infection results in a bidirectional loss of selective macromolecule trafficking across the pore [46], [47], [48], [49], a phenomenon often referred to as the nucleocytoplasmic trafficking (NCT) disorder. This virus-induced NCT disorder may interfere with IFN-α/β transcription activation by affecting shuttling and/or activation of IRF3, which must translocate to the nucleus to activate target gene transcription [50]. Alternatively localization of other (regulatory) factors necessary for IFN-α/β transcription may be deregulated by the NCT disorder.
The potential blockade at the level of IRF3 activation may seem unnecessary since upstream TBK1 activation is already inhibited in infected cells [41]. However, it is common for viruses to employ redundant strategies to interfere with the IFN system to ensure an effective inhibition. Furthermore, these two mechanisms may also be important at different stages of infection. The NCT disorder can be observed extremely rapidly upon enterovirus infections – hours before the cleavages of MDA5 and MAVS could be detected. Thus, it is possible that enteroviruses rely on the NCT disorder to keep IFN-α/β under control before MDA5 and MAVS could be inactivated. Of note, it cannot be excluded that MDA5 and MAVS are already cleaved earlier during infection, locally at the site of viral protein synthesis and RNA replication, but are undetectable when analyzing these factors in whole cytoplasmic lysates [41]. The relative contributions of the NCT disorder and MDA5/MAVS cleavage to IFN-α/β antagonization remain to be demonstrated.
4.2. Cardiovirus evasion strategies
4.2.1. L protein-induced nucleocytoplasmic trafficking disorder
Cardioviruses do not target MDA5 or MAVS, however, still effectively prevent IRF3 phosphorylation [41]. It was also shown that TBK1 is phosphorylated – believed to reflect activation – during EMCV infection [41], pinpointing the viral antagonization to a step between TBK1 activation and IRF3 phosphorylation. The cardiovirus IFN-α/β antagonist has long been established, namely the L protein, a small peptide (67 amino acids in the case of EMCV) produced at the extreme 5′ end of the viral polyprotein. The mature L protein contains an N-terminal CHCC Zn-finger domain, a hinge region and a C-terminal highly acidic domain [51], [52]. This protein is non-essential for viral RNA replication, but very important for counteracting host antiviral responses. Lmut viruses replicate to similar levels as wt viruses in IFN-α/β-deficient cells/mice, but are attenuated in IFN-α/β-competent systems [16], [53], [54], suggesting that it is an important player in IFN-α/β antagonization. L has neither enzymatic activity, nor known homologs [51], [52].
It is thus far unknown how L inhibits IRF3 activation and subsequently IFN-α/β production. This effect of L may be linked to its role in the NCT disorder that is induced by cardioviruses, which like enteroviruses also disrupt macromolecule trafficking across nuclear pores [55], [56]. L has been shown to form a tight complex with the small GTPase Ran, which is currently the only reported interaction partner of L [51], [57]. Hereby, L disrupts the RanGDP/GTP gradient across the nuclear pore, a crucial regulatory mechanism for nuclear import and export [51], [57]. L also induces hyperphosphorylation of several Nups (e.g. Nup 62, 98, 153 & 214) within the domains that form the physical barriers of the nuclear pore and provide important docking sites for transport receptors [58], [59], [60]. Hereby L may physically interfere with interactions of cargo transporters with Nups [58]. Mutant L proteins harboring substitutions in the Zn-finger domain or acidic domain that fail to induce the NCT disorder [51], [55], [56], [59] – due to impaired Ran binding and Nup hyperphosphorylation [51], [58], [59] – are also deficient in suppressing IFN-α/β [16], [17], [41], [54], [61], [62]. These results suggest that IFN-α/β suppression by L may be in one way or another caused by its activity to deregulate NCT. As mentioned in Section 4.1.2, NCT disorder may interfere with IRF3 activation by preventing its shuttling and/or nuclear translocation upon activation by TBK1. Future research is necessary to demonstrate a clear cause–effect relationship between the NCT disorder and IFN-α/β suppression, as well as the exact underlying mechanism.
4.2.2. RIG-I antagonization?
It has been reported that EMCV also proteolytically targets RIG-I in infected cells, either via the viral 3Cpro [44], or caspases [63]. However, RIG-I cleavage was not observed throughout infection of either wt or Lmut EMCV in another study [41]. It is currently unclear what causes this disagreement in literature. Nonetheless, assuming RIG-I is targeted by EMCV, how could this benefit virus replication? As discussed above (see Section 4.1.2), RIG-I does not participate in detecting picornaviruses [1], [2]. It has been suggested that RIG-I may directly activate STAT1, thereby activating ISG expression independently of IFN-α/β production [45]. Thus, it is possible that cardioviruses inactivate RIG-I to prevent ISG transcription activation, though experimental proof remains to be provided.
4.3. Evasion strategies of other picornaviruses
Picornaviruses from other genera than Enterovirus and Cardiovirus are also known to actively interfere with IFN-α/β induction. HAV, a member of the Hepatovirus genus, proteolytically targets MAVS. In this case, a precursor of the viral 3Cpro proteinase, namely the 3ABC, is responsible, and both the proteinase activity of 3Cpro and a transmembrane domain in 3A, which targets 3ABC to mitochondria where MAVS is localized, are required [64]. Another picornavirus, FMDV from the Aphthovirus genus, also targets the RLR pathway. The FMDV viral proteinase Lpro (which is unrelated to the cardiovirus L) exerts deubiquitinating activity on K48 as well as K63 ubiquitin chains, and has been shown to reduce ubiquitination of, among others, RIG-I and TBK1 [65], which is known to modulate the activity of these factors. In addition, the 3Cpro of FMDV has been reported to cleave NF-κB essential modulator (NEMO, also known as IKK-γ), a factor required for NF-κB activation [66]. These results clearly demonstrate that picornaviruses employ a diverse range of mechanisms to suppress host antiviral responses, and knowledge obtained from one genus cannot be directly applied to other genera. Of note, another important genus of picornaviruses, Parechovirus, which can cause several neonatal infections in humans [6], do not encode for an L protein, and neither do their 2A proteins exert any protease activity [13], [15]. How these viruses interfere with host antiviral responses remains to be elucidated.
5. Interaction between picornaviruses and the stress response
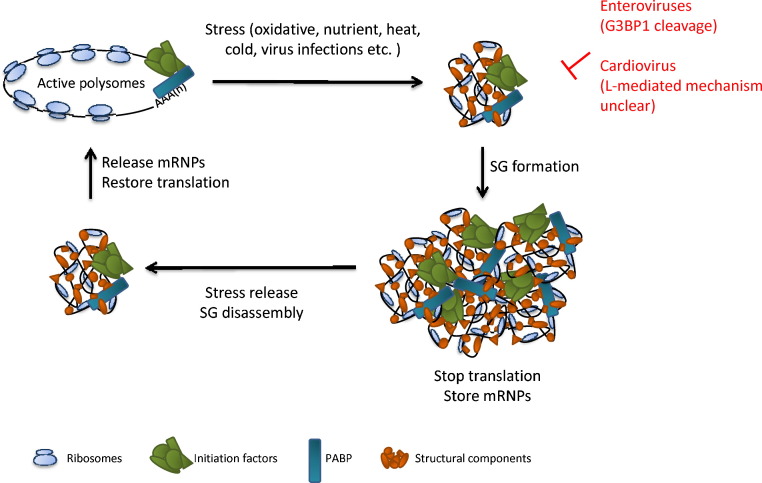
Stress granule (SG) formation is part of the cellular stress response and is important for cell survival during stress conditions. These granules are dynamic and serve to temporarily store pre-initiation mRNA complexes (Fig. 4 ) [3], [67]. They contain Ras GTPase-activating protein-binding proteins (G3BP1/2), T-cell intracellular antigen 1 (TIA-1), TIA-related protein (TIAR), many translation initiation factors, 40S ribosomes, mRNAs, and numerous other RNA-binding proteins. Many more proteins have recently been reported to associate with SGs formed under specific types of stress [3], indicating that there is some level of specificity of the cellular stress response. In most cases, SG formation is triggered by a halt in cap-dependent translation, which is often caused by phosphorylation of eukaryotic translation initiation factor 2α (eIF2α) by one of the four kinases – protein kinase R (PKR), PKR-like endoplasmic reticulum kinase (PERK), heme-regulated kinase (HRI) and general control non-depressible 2 kinase (GCN2). However, eIF2α phosphorylation-independent SG formation after oxidative stress or direct inhibition of eIF4G or eIF4A has also been reported [68].
Fig. 4.
The formation and disassembly of stress granules (SGs). PABP, poly(A)-binding protein. mRNPs, messenger ribonucleoproteins. Blockade by enteroviruses and cardioviruses are indicated in red.
5.1. The antiviral role of the stress pathway
Growing evidence suggests that SGs exert an antiviral function, though the exact mechanism is largely unknown. Since the primary function of the stress pathway is to halt cap-dependent translation and temporarily store preformed translation initiation complexes in SGs [3], [67], it has been suggested that also viral translation is inhibited. Indeed, in the case of influenza virus, which relies solely on cap-dependent translation for viral protein synthesis, SG formation was associated with reduced viral protein accumulation levels and thus virus replication [69]. Also for picornaviruses, which use IRES-dependent translation, SG formation appears to be disadvantageous for virus replication, albeit only to moderate levels [70], [71], [72]. How SG formation acts on picornavirus replication is incompletely understood. Hypothetically, viral mRNAs can get trapped in the SGs, thereby precluding them from translation and/or replication. However, it is still under debate whether picornavirus RNAs localize to SGs (see Section 5.2.1). Alternatively, it has been suggested that host and/or viral proteins, which play a role in virus replication, are scavenged in SGs, thereby limiting virus replication [71], but this has not been investigated. Recent observations suggest that there is a link between the stress pathway and the IFN-α/β pathway – which is discussed here below – providing novel insights into the possible antiviral role of SGs.
5.2. Link between the stress response and the IFN-α/β pathway
5.2.1. SG as platform for viral RNA recognition
Recently, SGs were suggested to function as sites for viral RNA recognition by RLRs. The group of Fujita showed that both RIG-I and influenza viral ssRNA (ligand of RIG-I) translocate to SGs during influenza virus ΔNS1 infection. Inhibition of SG formation, either by G3BP1 knockdown or PKR knockout, resulted in a 3- to 10-fold reduction in IFN-β response [73]. These results led the authors to propose that SGs may serve as a platform for viral RNA recognition by RIG-I. Also MDA5, the sensor for picornavirus RNA [24], localizes to SGs [61]. This observation suggests that MDA5 may also use SGs as sites for viral RNA recognition. However, it is thus far unclear whether picornaviral dsRNA (ligand of MDA5) also localizes to SGs. On the basis of propidium iodide staining it was concluded that dsRNA localized to SGs in EMCV-infected cells [72]. However, detection of EMCV or TMEV viral RNAs using either a dsRNA-specific antibody or in situ hybridization, respectively, failed to show any viral RNA in virus-induced SGs [61], [74]. These observations suggest that it is unlikely that MDA5 utilizes SGs as (primary) site for viral RNA recognition during picornavirus infections.
5.2.2. Cross-talk between stress and IFN-α/β pathways
Accumulating evidence suggest that there is cross-talk between the stress pathway and RLR-mediated IFN-α/β response. It has been reported that PKR and G3BP2 enhance activation of the NF-κB pathway [75], [76] and consequently also IFN-α/β production [77]. Vice versa, DHX36, a novel activator of RIG-I [78], as well as MAVS [79] were recently shown to be involved in PKR activation. Clearly, activation of the one pathway does not necessarily lead to full-blown activation of the other. Langereis et al. showed that activation of the stress pathway and subsequent SG formation did not induce IFN-α/β production, and conversely, activation of the RLR signaling pathway or IFN-α/β treatment of cells did not lead to SG formation [61]. Notwithstanding this, IFN-α/β treatment did sensitize cells for stress pathway activation [80], [81], possibly due to IFN-induced increase in PKR expression. It remains to be established to what extent, and under what circumstances, the cross-talk between these two pathways occurs.
5.3. Strategies of picornaviruses to evade the stress pathway
Since stress pathway activation acts as an antiviral response, viruses have found ways to inhibit this pathway. Enteroviruses (e.g. PV and CVB3) induce SG formation early during infection, which may partly result from virus-induced cap-dependent translation shutoff via eIF4G cleavage [3]. These granules disappear as infection proceeds due to cleavage of G3BP1 by the viral proteinase 3Cpro [61], [70], [71]. Overexpression of a G3BP1 mutant that can no longer be cleaved by the viral 3Cpro (G3BP1-Q325E) led to stable SG formation and stronger repression of PV replication than wt G3BP1 overexpression. This observation underscores the important role of 3Cpro-mediated cleavage of G3BP1 on enhancing enterovirus replication [71]. Besides enteroviruses, also alphaviruses like Semliki Forest virus [82] and Chickungunya virus [83] prevent SG formation by interacting with G3BP1, suggesting that targeting this protein is an effective way to inhibit the antiviral role of SGs.
Cardioviruses also suppress SG formation. As influenza A virus, which utilizes its NS1 protein to suppress both IFN-α/β activation and SG formation [69], [73], cardioviruses also employ a single viral protein, namely L, to antagonize these two antiviral pathways [61], [74]. While no SGs are observed during wt cardiovirus infections, Lmut EMCV and TMEV induced PKR-dependent activation of the stress pathway [61] and persistent SGs throughout infection [61], [74]. Moreover, overexpression of L alone was sufficient to repress de novo SG formation induced via HRI and PERK activation [74], suggesting that L expression inhibits SG formation at a common step downstream of PKR, HRI and PERK, possibly at the level of eIF2α phosphorylation or a yet unknown downstream step. As mentioned in Section 4.2.1, cardiovirus L also induces NCT disorder. Although there is no proof that coordinated trafficking of proteins between the nucleus and the cytoplasm is essential for stress pathway activation and SG formation, TIA-1 is known to migrate from the nucleus to the cytoplasm upon stress pathway activation. Since TIA-1 is an essential structural component of SGs, it is possible that the L-induced NCT disorder affects TIA-1 migration and thus SG formation. Of course, it cannot be excluded that L acts on a central host factor or process that controls NCT, IFN-α/β response as well as SG formation, though no plausible candidate has been suggested, to date.
Recently, it was reported that EMCV induced SG formation early in infection, and the SGs disappeared later in infection as a result of G3BP1 cleavage via viral 3Cpro, similar to what is described for enteroviruses [72]. While this is a remarkable observation, it provides no explanation as to how Lmut cardioviruses, which all express active 3Cpro, potently induce SG formation [61], [74].
In summary, both enteroviruses and cardioviruses are able to inactivate the stress pathway. However, the mechanism how this is achieved, especially for cardioviruses, remains to be investigated in more detail. Studying how picornaviruses target the stress pathway and how SG formation acts as an antiviral response may also help reveal novel insights into the interplay between stress and innate immune responses.
6. Future perspectives and concluding remarks
The interaction between pathogens and the innate antiviral responses is extremely intricate. Recent studies provided a first glimpse on MDA5 activation during the course of a normal picornavirus infection, however, much is still to be learned about MDA5 activation during virus infections in general. What RNA(s) are directly bound by MDA5? What is the minimum length of MDA5 filaments that can activate MAVS in cells? Is LGP2 a positive regulator of MDA5 in general, or is it only needed for specific (types of) RNA ligands? In addition, the immune evasion strategies of many important picornaviruses are still incompletely understood. Furthermore, research on the antiviral activities of the stress response, and the underlying mechanisms, are also still in its infancy.
While addressing these questions in future research, it is important to realize that viruses replicate at specific subcellular microenvironments that can largely differ from in vitro experimental conditions. Efforts should be made to study events during natural infections. In addition, the spatial and temporal aspects of viral RNA recognition and viral evasion strategies are important issues that need to be clarified. At what stage during a picornavirus infection does MDA5 become activated? Where does MDA5 gain access to viral RNA PAMPs that are produced in restricted subcellular microenvironments? Where and when do viral proteinases encounter and target crucial factors of immune pathways? These important questions await future investigation and warrant an exciting research area of the interaction between picornaviruses and innate antiviral responses.
Acknowledgements
We would like to thank the Netherlands Organisation for Scientific Research (NWO) for the following funding: Q.F. is supported by a Mosaic grant (NWO-017.006.043), M.A.L. by a Veni grant (NWO-863.13.008), and F.J.M.K. by an ECHO grant (NWO-CW-700.59.007).
Biographies

Qian Feng, Ph.D., just earned her Ph.D. on the induction and suppression of innate antiviral responses by picornaviruses at the Utrecht University, in the Department of Infectious Diseases and Immunology from the Faculty of Veterinary Medicine. She is a laureate of a personal Ph.D. grant from the Netherlands Organisation for Scientific Research (NWO). Qian earned her M.Sc. degree at the Radboud University Nijmegen. Her areas of expertise include molecular virology and innate antiviral responses.

Martijn A. Langereis, Ph.D., is a post-doctoral fellow at the Utrecht University, in the Department of Infectious Diseases and Immunology from the Faculty of Veterinary Medicine. He earned his Ph.D. in the same department studying viral attachment of coronaviruses. He is a laureate of a personal Dutch Rubicon and Veni grant from the Netherlands Organisation for Scientific Research (NWO). His current area of expertise includes molecular virology, innate antiviral responses like type I interferon and the stress pathway, and viral evasion strategies.

Frank J.M. van Kuppeveld, Ph.D., is an established (picorna)virologist and is a Full Professor in Molecular Virology at the Utrecht University. Dr. van Kuppeveld is also a laureate of a prestigious Dutch Vici research grant, and the coordinator of a large European consortium EUVIRNA, which focuses on positive-strand RNA virus replication and the development of antivirals. Dr. van Kuppeveld earned his doctorate in Virology at the Radboud University Nijmegen, where he also completed his post-doctoral training. Dr. van Kuppeveld's research has focused on the replication of picornaviruses, especially the formation of the virus-induced membranous replication organelles, the interaction between picornaviruses and innate antiviral responses, and the development of antivirals against picornaviruses.
References
- 1.Kato H., Takahasi K., Fujita T. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol Rev. 2011;243:91–98. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- 2.Goubau D., Deddouche S., Reis E., Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reineke L.C., Lloyd R.E. Diversion of stress granules and P-bodies during viral infection. Virology. 2013;436:255–267. doi: 10.1016/j.virol.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris K.G., Coyne C.B. Enter at your own risk: how enteroviruses navigate the dangerous world of pattern recognition receptor signaling. Cytokine. 2013;63:230–236. doi: 10.1016/j.cyto.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chase A.J., Semler B.L. Viral subversion of host functions for picornavirus translation and RNA replication. Future Virol. 2012;7:179–191. doi: 10.2217/fvl.12.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tapparel C., Siegrist F., Petty T.J., Kaiser L. Picornavirus and enterovirus diversity with associated human diseases. Infect Genet Evol. 2013;14:282–293. doi: 10.1016/j.meegid.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 7.Yeung W.-C.G., Rawlinson W.D., Craig M.E. Enterovirus infection and type 1 diabetes mellitus: systematic review and meta-analysis of observational molecular studies. BMJ. 2011;342:d35. doi: 10.1136/bmj.d35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ooi M.H., Wong S.C., Lewthwaite P., Cardosa M.J., Solomon T. Clinical features, diagnosis, and management of enterovirus 71. Lancet Neurol. 2010;9:1097–1105. doi: 10.1016/S1474-4422(10)70209-X. [DOI] [PubMed] [Google Scholar]
- 9.Kurai D., Saraya T., Ishii H., Takizawa H. Virus-induced exacerbations in asthma and COPD. Front Microbiol. 2013;4:293. doi: 10.3389/fmicb.2013.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canelli E., Luppi A., Lavazza A., Lelli D., Sozzi E., Martin A.M.M. Encephalomyocarditis virus infection in an Italian zoo. Virol J. 2010;7:64. doi: 10.1186/1743-422X-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reddacliff L.A., Kirkland P.D., Hartley W.J., Reece R.L. Encephalomyocarditis virus infections in an Australian zoo. J Zoo Wildl Med. 1997;28:153–157. [PubMed] [Google Scholar]
- 12.Himeda T., Ohara Y. Saffold virus, a novel human Cardiovirus with unknown pathogenicity. J Virol. 2012;86:1292–1296. doi: 10.1128/JVI.06087-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehrenfeld E., Domingo E., Roos R.P. ASM Press; Washington, DC: 2010. The picornaviruses. [Google Scholar]
- 14.Virgen-Slane R., Rozovics J.M., Fitzgerald K.D., Ngo T., Chou W., van der Heden van Noort G.J. An RNA virus hijacks an incognito function of a DNA repair enzyme. Proc Natl Acad Sci USA. 2012;109:14634–14639. doi: 10.1073/pnas.1208096109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agol V.I., Gmyl A.P. Viral security proteins: counteracting host defences. Nat Rev Microbiol. 2010;8:867–878. doi: 10.1038/nrmicro2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hato S.V., Ricour C., Schulte B.M., Lanke K.H., de Bruijni M., Zoll J. The mengovirus leader protein blocks interferon-alpha/beta gene transcription and inhibits activation of interferon regulatory factor 3. Cell Microbiol. 2007;9:2921–2930. doi: 10.1111/j.1462-5822.2007.01006.x. [DOI] [PubMed] [Google Scholar]
- 17.Van Pesch V., van Eyll O., Michiels T. The leader protein of Theiler's virus inhibits immediate-early alpha/beta interferon production. J Virol. 2001;75:7811–7817. doi: 10.1128/JVI.75.17.7811-7817.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 19.Abe Y., Fujii K., Nagata N., Takeuchi O., Akira S., Oshiumi H. The toll-like receptor 3-mediated antiviral response is important for protection against poliovirus infection in poliovirus receptor transgenic mice. J Virol. 2012;86:185–194. doi: 10.1128/JVI.05245-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gitlin L., Barchet W., Gilfillan S., Cella M., Beutler B., Flavell R.A. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci USA. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin Y.-H., Kim S.J., So E.Y., Meng L., Colonna M., Kim B.S. Melanoma differentiation-associated gene 5 is critical for protection against Theiler's virus-induced demyelinating disease. J Virol. 2012;86:1531–1543. doi: 10.1128/JVI.06457-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J.P., Cerny A., Asher D.R., Kurt-Jones E.A., Bronson R.T., Finberg R.W. MDA5 and MAVS mediate type I interferon responses to coxsackie B virus. J Virol. 2010;84:254–260. doi: 10.1128/JVI.00631-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pichlmair A., Schulz O., Tan C.P., Rehwinkel J., Kato H., Takeuchi O. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J Virol. 2009;83:10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng Q., Hato S.V., Langereis M.A., Zoll J., Virgen-Slane R., Peisley A. MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells. Cell Rep. 2012;2:1187–1196. doi: 10.1016/j.celrep.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Triantafilou K., Vakakis E., Kar S., Richer E., Evans G.L., Triantafilou M. Visualisation of direct interaction of MDA5 and the dsRNA replicative intermediate form of positive strand RNA viruses. J Cell Sci. 2012;125:4761–4769. doi: 10.1242/jcs.103887. [DOI] [PubMed] [Google Scholar]
- 26.Richards O.C., Martin S.C., Jense H.G., Ehrenfeld E. Structure of poliovirus replicative intermediate RNA electron microscope analysis of RNA cross-linked in vivo with psoralen derivative. J Mol Biol. 1984;173:325–340. doi: 10.1016/0022-2836(84)90124-4. [DOI] [PubMed] [Google Scholar]
- 27.Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deddouche S., Goubau D., Rehwinkel J., Chakravarty P., Begum S., Maillard P.V. Identification of an LGP2-associated MDA5 agonist in picornavirus-infected cells. Elife. 2014;3:e01535. doi: 10.7554/eLife.01535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luthra P., Sun D., Silverman R.H., He B. Activation of IFN-beta; expression by a viral mRNA through RNase L and MDA5. Proc Natl Acad Sci USA. 2011;108:2118–2123. doi: 10.1073/pnas.1012409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malathi K., Saito T., Crochet N., Barton D.J., Gale M., Jr., Silverman R.H. RNase L releases a small RNA from HCV RNA that refolds into a potent PAMP. RNA. 2010;16:2108–2119. doi: 10.1261/rna.2244210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malathi K., Dong B., Gale M., Jr., Silverman R.H. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Satoh T., Kato H., Kumagai Y., Yoneyama M., Sato S., Matsushita K. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci USA. 2010;107:1512–1517. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X., Ranjith-Kumar C.T., Brooks M.T., Dharmaiah S., Herr A.B., Kao C. The RIG-I-like receptor LGP2 recognizes the termini of double-stranded RNA. J Biol Chem. 2009;284:13881–13891. doi: 10.1074/jbc.M900818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saito T., Hirai R., Loo Y.-M., Owen D., Johnson C.L., Sinha S.C. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci USA. 2007;104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahasi K., Kumeta H., Tsuduki N., Narita R., Shigemoto T., Hirai R. Solution structures of cytosolic RNA sensor MDA5 and LGP2 C-terminal domains: identification of the RNA recognition loop in RIG-I-like receptors. J Biol Chem. 2009;284:17465–17474. doi: 10.1074/jbc.M109.007179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peisley A., Lin C., Wu B., Orme-Johnson M., Liu M., Walz T. Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. Proc Natl Acad Sci USA. 2011;108:21010–21015. doi: 10.1073/pnas.1113651108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Childs K.S., Randall R.E., Goodbourn S. LGP2 plays a critical role in sensitizing mda-5 to activation by double-stranded RNA. PLOS ONE. 2013;8:e64202. doi: 10.1371/journal.pone.0064202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mukherjee A., Morosky S., Delorme-Axford A.E., Dybdahl-Sissoko N., Oberste M.S., Wang T. The coxsackievirus B 3C protease cleaves MAVS and TRIF to attenuate host type I interferon and apoptotic signaling. PLOS Pathog. 2011;7:e1001311. doi: 10.1371/journal.ppat.1001311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuo R.-L., Kao L.-T., Lin S.-J., Wang R.Y.-L., Shih S.-R. MDA5 plays a crucial role in enterovirus 71 RNA-mediated IRF3 activation. PLOS ONE. 2013;8:e63431. doi: 10.1371/journal.pone.0063431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drahos J., Racaniello V.R. Cleavage of IPS-1 in cells infected with human rhinovirus. J Virol. 2009;83:11581–11587. doi: 10.1128/JVI.01490-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng Q., Langereis M.A., Lork M., Nguyen M., Hato S.V., Lanke K. Enterovirus 2Apro targets MDA5 and MAVS in infected cells. J Virol. 2014;88:3369–3378. doi: 10.1128/JVI.02712-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barral P.M., Morrison J.M., Drahos J., Gupta P., Sarkar D., Fisher P.B. MDA-5 is cleaved in poliovirus-infected cells. J Virol. 2007;81:3677–3684. doi: 10.1128/JVI.01360-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang B., Xi X., Lei X., Zhang X., Cui S., Wang J. Enterovirus 71 protease 2Apro targets MAVS to inhibit anti-viral type I interferon responses. PLOS Pathog. 2013;9:e1003231. doi: 10.1371/journal.ppat.1003231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barral P.M., Sarkar D., Fisher P.B., Racaniello V.R. RIG-I is cleaved during picornavirus infection. Virology. 2009;391:171–176. doi: 10.1016/j.virol.2009.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang L.-J., Zhang N.-N., Ding F., Li X.-Y., Chen L., Zhang H.-X. RA-inducible gene-I induction augments STAT1 activation to inhibit leukemia cell proliferation. Proc Natl Acad Sci USA. 2011;108:1897–1902. doi: 10.1073/pnas.1019059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Belov G.A., Lidsky P.V., Mikitas O.V., Egger D., Lukyanov K.A., Bienz K. Bidirectional increase in permeability of nuclear envelope upon poliovirus infection and accompanying alterations of nuclear pores. J Virol. 2004;78:10166–10177. doi: 10.1128/JVI.78.18.10166-10177.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park N., Katikaneni P., Skern T., Gustin K.E. Differential targeting of nuclear pore complex proteins in poliovirus-infected cells. J Virol. 2008;82:1647–1655. doi: 10.1128/JVI.01670-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watters K., Palmenberg A.C. Differential processing of nuclear pore complex proteins by rhinovirus 2A proteases from different species and serotypes. J Virol. 2011;85:10874–10883. doi: 10.1128/JVI.00718-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Castelló A., Izquierdo J.M., Welnowska E., Carrasco L. RNA nuclear export is blocked by poliovirus 2A protease and is concomitant with nucleoporin cleavage. J Cell Sci. 2009;122:3799–3809. doi: 10.1242/jcs.055988. [DOI] [PubMed] [Google Scholar]
- 50.Reich N.C. Nuclear/cytoplasmic localization of IRFs in response to viral infection or interferon stimulation. J Interferon Cytokine Res. 2002;22:103–109. doi: 10.1089/107999002753452719. [DOI] [PubMed] [Google Scholar]
- 51.Porter F.W., Bochkov Y.A., Albee A.J., Wiese C., Palmenberg A.C. A picornavirus protein interacts with Ran-GTPase and disrupts nucleocytoplasmic transport. Proc Natl Acad Sci USA. 2006;103:12417–12422. doi: 10.1073/pnas.0605375103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Basta H.A., Bacot-Davis V.R., Ciomperlik J.J., Palmenberg A.C. Encephalomyocarditis virus leader is phosphorylated by CK2 and Syk as a requirement for subsequent phosphorylation of cellular nucleoporins. J Virol. 2014;88(4):2219–2226. doi: 10.1128/JVI.03150-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zoll J., Galama J.M., van Kuppeveld F.J., Melchers W.J. Mengovirus leader is involved in the inhibition of host cell protein synthesis. J Virol. 1996;70:4948–4952. doi: 10.1128/jvi.70.8.4948-4952.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ricour C., Delhaye S., Hato S.V., Olenyik T.D., Michel B., van Kuppeveld F.J.M. Inhibition of mRNA export and dimerization of interferon regulatory factor 3 by Theiler's virus leader protein. J Gen Virol. 2009;90:177–186. doi: 10.1099/vir.0.005678-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lidsky P.V., Hato S., Bardina M.V., Aminev A.G., Palmenberg A.C., Sheval E.V. Nucleocytoplasmic traffic disorder induced by cardioviruses. J Virol. 2006;80:2705–2717. doi: 10.1128/JVI.80.6.2705-2717.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Delhaye S., van Pesch V., Michiels T. The leader protein of Theiler's virus interferes with nucleocytoplasmic trafficking of cellular proteins. J Virol. 2004;78:4357–4362. doi: 10.1128/JVI.78.8.4357-4362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bacot-Davis V.R., Palmenberg A.C. Encephalomyocarditis virus leader protein hinge domain is responsible for interactions with Ran GTPase. Virology. 2013;443:177–185. doi: 10.1016/j.virol.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Porter F.W., Brown B., Palmenberg A.C. Nucleoporin phosphorylation triggered by the encephalomyocarditis virus leader protein is mediated by mitogen-activated protein kinases. J Virol. 2010;84:12538–12548. doi: 10.1128/JVI.01484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bardina M.V., Lidsky P.V., Sheval E.V., Fominykh K.V., van Kuppeveld F.J.M., Polyakov V.Y. Mengovirus-induced rearrangement of the nuclear pore complex: hijacking cellular phosphorylation machinery. J Virol. 2009;83:3150–3161. doi: 10.1128/JVI.01456-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tu L.-C., Fu G., Zilman A., Musser S.M. Large cargo transport by nuclear pores: implications for the spatial organization of FG-nucleoporins. EMBO J. 2013;32:3220–3230. doi: 10.1038/emboj.2013.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Langereis M.A., Feng Q., van Kuppeveld F.J. MDA5 localizes to stress granules but this localization is not required for the induction of type I interferon. J Virol. 2013;87(11):6314–6325. doi: 10.1128/JVI.03213-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zoll J., Melchers W.J.G., Galama J.M.D., van Kuppeveld F.J.M. The mengovirus leader protein suppresses alpha/beta interferon production by inhibition of the iron/ferritin-mediated activation of NF-kappa B. J Virol. 2002;76:9664–9672. doi: 10.1128/JVI.76.19.9664-9672.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Papon L., Oteiza A., Imaizumi T., Kato H., Brocchi E., Lawson T.G. The viral RNA recognition sensor RIG-I is degraded during encephalomyocarditis virus (EMCV) infection. Virology. 2009;393:311–318. doi: 10.1016/j.virol.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 64.Yang Y., Liang Y., Qu L., Chen Z., Yi M., Li K. Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc Natl Acad Sci USA. 2007;104:7253–7258. doi: 10.1073/pnas.0611506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang D., Fang L., Li P., Sun L., Fan J., Zhang Q. The leader proteinase of foot-and-mouth disease virus negatively regulates the type I interferon pathway by acting as a viral deubiquitinase. J Virol. 2011;85:3758–3766. doi: 10.1128/JVI.02589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang D., Fang L., Li K., Zhong H., Fan J., Ouyang C. Foot-and-mouth disease virus 3C protease cleaves NEMO to impair innate immune signaling. J Virol. 2012;86:9311–9322. doi: 10.1128/JVI.00722-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.White J.P., Lloyd R.E. Regulation of stress granules in virus systems. Trends Microbiol. 2012;20:175–183. doi: 10.1016/j.tim.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Buchan J.R., Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khaperskyy D.A., Hatchette T.F., McCormick C. Influenza A virus inhibits cytoplasmic stress granule formation. FASEB J. 2012;26:1629–1639. doi: 10.1096/fj.11-196915. [DOI] [PubMed] [Google Scholar]
- 70.Fung G., Ng C.S., Zhang J., Shi J., Wong J., Piesik P. Production of a dominant-negative fragment due to G3BP1 cleavage contributes to the disruption of mitochondria-associated protective stress granules during CVB3 infection. PLOS ONE. 2013;8:e79546. doi: 10.1371/journal.pone.0079546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.White J.P., Cardenas A.M., Marissen W.E., Lloyd R.E. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe. 2007;2:295–305. doi: 10.1016/j.chom.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 72.Ng C.S., Jogi M., Yoo J.-S., Onomoto K., Koike S., Iwasaki T. Encephalomyocarditis virus disrupts stress granules, the critical platform for triggering antiviral innate immune responses. J Virol. 2013;87:9511–9522. doi: 10.1128/JVI.03248-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Onomoto K., Jogi M., Yoo J.-S., Narita R., Morimoto S., Takemura A. Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PLOS ONE. 2012;7:e43031. doi: 10.1371/journal.pone.0043031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Borghese F., Michiels T. The leader protein of cardioviruses inhibits stress granule assembly. J Virol. 2011;85:9614–9622. doi: 10.1128/JVI.00480-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bonnet M.C., Daurat C., Ottone C., Meurs E.F. The N-terminus of PKR is responsible for the activation of the NF-kappaB signaling pathway by interacting with the IKK complex. Cell Signal. 2006;18:1865–1875. doi: 10.1016/j.cellsig.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 76.Prigent M., Barlat I., Langen H., Dargemont C. IkappaBalpha and IkappaBalpha/NF-kappa B complexes are retained in the cytoplasm through interaction with a novel partner, RasGAP SH3-binding protein 2. J Biol Chem. 2000;275:36441–36449. doi: 10.1074/jbc.M004751200. [DOI] [PubMed] [Google Scholar]
- 77.Chu W.M., Ostertag D., Li Z.W., Chang L., Chen Y., Hu Y. JNK2 and IKKbeta are required for activating the innate response to viral infection. Immunity. 1999;11:721–731. doi: 10.1016/s1074-7613(00)80146-6. [DOI] [PubMed] [Google Scholar]
- 78.Yoo J.-S., Takahasi K., Ng C.S., Ouda R., Onomoto K., Yoneyama M. DHX36 enhances RIG-I signaling by facilitating PKR-mediated antiviral stress granule formation. PLOS Pathog. 2014;10:e1004012. doi: 10.1371/journal.ppat.1004012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang P., Li Y., Xia J., He J., Pu J., Xie J. IPS-1 plays an essential role in stress granule formation induced by dsRNA through interacting with PKR and mediating its activation. J Cell Sci. 2014;127:2471–2482. doi: 10.1242/jcs.139626. [DOI] [PubMed] [Google Scholar]
- 80.Ruggieri A., Dazert E., Metz P., Hofmann S., Bergeest J.-P., Mazur J. Dynamic oscillation of translation and stress granule formation mark the cellular response to virus infection. Cell Host Microbe. 2012;12:71–85. doi: 10.1016/j.chom.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.John L., Samuel C.E. Induction of stress granules by interferon and down-regulation by the cellular RNA adenosine deaminase ADAR1. Virology. 2014;454–455:299–310. doi: 10.1016/j.virol.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Panas M.D., Varjak M., Lulla A., Eng K.E., Merits A., Karlsson Hedestam G.B. Sequestration of G3BP coupled with efficient translation inhibits stress granules in Semliki Forest virus infection. Mol Biol Cell. 2012;23:4701–4712. doi: 10.1091/mbc.E12-08-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fros J.J., Domeradzka N.E., Baggen J., Geertsema C., Flipse J., Vlak J.M. Chikungunya virus nsP3 blocks stress granule assembly by recruitment of G3BP into cytoplasmic foci. J Virol. 2012;86:10873–10879. doi: 10.1128/JVI.01506-12. [DOI] [PMC free article] [PubMed] [Google Scholar]