Highlights
-
•
There were shifts in the predominance of influenza types or subtypes.
-
•
Drifted A(H3N2) strains circulated during the 2014–2015 season and a higher hospitalisation rate was reported.
-
•
Circulation of B/Vic-like viruses not included in the trivalent vaccine during the 2015–2016 season.
-
•
Virulence and resistance mutations were detected due to an enhanced surveillance.
-
•
Virological surveillance should be strengthened in hospital settings.
Keywords: Influenza viruses, Surveillance, Respiratory tract infection, Genetic diversity, Molecular epidemiology
Abstract
Background
Influenza viruses (FLUV) are continuously evolving, which explain the occurrence of seasonal influenza epidemics and the need to review the vaccine strain composition annually. The aim is to describe the genetic diversity and clinical outcomes of FLUV detected at a tertiary university hospital in Barcelona (Spain) during the 2012–2016 seasons.
Methods
The detection of FLUV from patients attended at the Emergency Department or admitted to the hospital was performed by either immunofluorescence or PCR-based assays. A specific real-time one-step multiplex RT-PCR was performed for influenza A (FLUAV) subtyping. The complete coding haemagglutinin domain 1 (HA1) and neuraminidase (NA) (2015–2016) protein sequences from a representative sampling were molecular characterised.
Results
A total 1774 (66.1%) FLUAV and 910 (33.9%) influenza B (FLUBV) cases were laboratory-confirmed. The hospitalisation rate was different between seasons, being the highest (81.4%) during the 2014–2015 season. FLUV were genetically close to vaccine strains except to the 2014–2015, in which most characterised A(H3N2) viruses belonged to a genetic group different from the vaccine strain. During the 2015–2016 season, B/Victoria-like viruses were the most predominant, but this component was not included in the trivalent vaccine used. Mutations D222G or D222N in HA1-domain were found in 3 A(H1N1)pdm09 strains from ICU-admitted cases. Three A(H1N1)pdm09 strains carried the NA H275Y (2) and S247N (1) mutations, respectively related to resistance or decreased susceptibility to oseltamivir.
Conclusions
The circulation of drifted A(H3N2) strains during the 2014–2015 season was related to the high hospitalisation rate due to the mismatch with the vaccine strains. The predominance of a FLUBV lineage not included in the trivalent influenza vaccine during the 2015–2016 season highlights the need to use a tetravalent influenza vaccine. Virological surveillance of viral variants carrying protein changes that alter tropism and susceptibility to antivirals features should be strengthened in hospital settings.
1. Background
Influenza viruses (FLUV), one of the main causative agents of respiratory infections, are related to high morbidity and mortality in the community, mainly in high-risk patients such as the elderly, those with underlying comorbidities or pregnant women [1], [2].
FLUV are single-stranded, negative-sense, segmented RNA viruses belonging to the Orthomyxoviridae family [3], [4]. Based on genetic and antigenic features, three different types of human FLUV (A, B and C) are currently described. According to antigenic properties of the surface proteins, haemagglutinin (HA) and neuraminidase (NA), influenza A viruses (FLUAV) are divided into different subtypes (H1N1pdm09 and H3N2) [3], [5], as well as influenza B viruses (FLUBV) into two major lineages, B/Victoria/2/87-like (FLUBV/VIC) and B/Yamagata/16/88-like (FLUBV/YAM) [6].
FLUV are continuously evolving through amino acid substitutions altering antigenic properties (antigenic drift) or, less frequently, by segment reassortment events (antigenic shift) [7]. Some amino acid substitutions in the HA receptor binding site (RBS) can alter the cellular host tropism, conditioning the species range and tissue tropism, which may be also related to viral virulence [8]. The high mutation rate of the NA protein can also affect the antiviral susceptibility to currently available neuraminidase inhibitors (NAIs) drugs [9]. In addition, changes in the antigenic features of FLUAV or FLUBV, mainly in HA, explain the occurrence of seasonal influenza epidemics and are the reason for the annual vaccine composition update needed for a more effective prevention of disease and the related complications in high-risk patients [10]. Vaccination is the most effective public health action for preventing influenza. A higher vaccination enhances the achievement of community immunisation [11]. In Catalonia, the influenza vaccination is recommended to individuals over 60 years of age, especially those living in nursing homes, children over 6 months of age and adults with high risk comorbidities for clinical complications due to influenza infection, such as pregnancy or chronic diseases. It is also recommended to healthcare professionals and people working for public services (police, firefighters or teachers). The influenza vaccines available in Catalonia are subunit, adjuvanted and cell-based vaccines. The tetravalent vaccines have only been available since the 2016–2017 season for the vaccination of high-risk patients at hospital settings [12]. The vaccination coverage in our region is variable and goes from 10 to 70% depending on the age group, although the mean coverage is around 35% each season [13].
The aim of this study is to describe the genetic features of detected viruses and the clinical outcome of influenza cases attended at Vall d’Hebron University Hospital (Barcelona, Spain) from the 2012–2013 to the 2015–2016 seasons.
2. Material and methods
2.1. Sampling
Vall d’Hebron University Hospital is the largest tertiary hospital in Catalonia and the second one in Spain, covering more than 450.000 inhabitants. From October 2012 (epidemiological week 40/2012) to May 2016 (epidemiological week 20/2016), upper (nasopharyngeal aspirates or swabs) and lower (bronchoalveolar lavages, bronchoaspirates or tracheal aspirates) respiratory tract specimens were collected for influenza and other respiratory viruses laboratory-confirmation from patients with suspicion of acute respiratory tract infection who were attended at the Emergency Department that accomplished the criteria to be admitted to the hospital; from patients that could benefit from antiviral treatment without needing hospitalisation; or from hospitalised patients that acquired influenza nosocomial infection. Based on medical records, some demographic features (age and sex), hospitalisation, Intensive Care Unit (ICU) admission, outcome and the administration of antiviral treatment were retrospectively collected from influenza laboratory-confirmed cases.
2.2. Definition of SARI cases
According to the Catalan Public Health Agency, a severe hospitalised patient is described as that case with pneumonia due to influenza virus or to bacterial infection, septic shock, multiorganic failure, acute respiratory distress or admission to Intensive Care Unit (ICU). Moreover, antiviral (NAIs) treatment was administered when influenza infection was laboratory-confirmed for those patients who are hospitalised due to the severity of the disease or belonging to high-risk groups [14] to prevent further disease complications.
2.3. Detection of respiratory viruses
The detection of FLUV and other respiratory viruses (respiratory syncytial virus, metapneumovirus, adenovirus, rhinoviruses, enteroviruses, coronaviruses, parainfluenza viruses and bocavirus) was carried out by either immunofluorescence antigen detection (SOFIA Influenza A + B, Quidel, California, USA; and D3 Ultra 8TM DFA Respiratory Virus Screening & Identification Kit Diagnostic HYBRIDS, USA) or PCR-based assays (Anyplex II RV16 Detection Kit, Seegene, Korea; and, GeneXpert Flu, Cepheid, USA). Prior to PCR-based assays, total nucleic acids were extracted using NucliSens easyMAG (bioMérieux, Marcy l’Etoile, France) according to the manufacturer’s instructions, and kept frozen (−20 °C) until use. An in-house real-time one-step multiplex RT-PCR [15] was performed for HA subtyping of seasonal influenza A viruses (H1pdm09 and H3).
2.4. Amplification and sequencing of influenza haemagglutinin and neuraminidase genes
The complete coding region of HA1-domain of a representative number of influenza laboratory-confirmed specimens from hospitalised patients was sequenced for phylogenetic analysis and molecular characterisation. In addition, during the 2015–2016 season, the complete NA coding region was also sequenced for the screening of amino acid substitutions associated with a reduced antiviral susceptibility [9], [16]. HA and NA amplifications were carried out by a one-step RT-PCR assay using the one-step RT-PCR Kit (Qiagen, Hilden, Germany) with the primers and PCR protocols shown in Supplementary Table 1. PCR products purification was subsequently performed using Exo-SAP-IT (USB, Affymetrix Inc. Cleveland, Ohio, USA) and sequenced by the ABI Prism Big Dye Terminator cycle sequencing kit v3.1 (Thermo Fisher Scientific, Waltham, MA, USA) on the ABI PRISM 3130XL Genetic Analyzer (Thermo Fisher Scientific, Waltham, MA, USA) with the sequencing primers (Supplementary Table 1). Nucleotide sequences were edited and assembled using SeqScape v2.6 software (Thermo Fisher Scientific, Waltham, MA, USA). Sequences of the present study were submitted to the Global Initiative on Sharing Avian Influenza Data (GISAID available at www.platform.gisaid.org).
2.5. Detection of polymorphisms and phylogenetic analysis
Phylogenetic analyses of complete HA1-domain coding region sequences were performed using the reference sequences by subtype (FLUAV) or by lineage (FLUBV) by season in the Northern Hemisphere recommended by the European Centre for Disease Prevention and Control (ECDC) (Supplementary Table 2). Both multiple nucleotide sequence alignment using the MUSCLE algorithm [17] and the molecular evolutionary model analysis were conducted in MEGA v5.2 [18]. The phylogenetic trees were constructed using a neighbor-joining (NJ) distance method as implemented in MEGA v5.2 with the nucleotide substitution model with the lowest Bayesian information criterion (BIC) score [18]. The topological accuracy of the internal branch was evaluated by the bootstrap method (1000 replicates).
In order to detect amino acid substitutions in the complete HA1-domain and NA coding regions, the deduced amino acid sequences from aligned nucleotide sequences were compared to those of the vaccine strains recommended by the World Health Organisation (WHO) (A(H1N1)pdm09: A/California/07/2009 (2012–2016); A(H3N2): A/Victoria/361/2011 (2012–2013); A/Texas/50/2012 (2013–2015); and A/Switzerland/9715293/2014 (2015–2016); B/Yamagata: B/Wisconsin/01/2010 (2012–2013); B/Massachusetts/02/2012 (2013–2015); and B/Phuket/3073/2013 (2015–2016); B/Victoria: B/Brisbane/60/2008 (2012–2016)). Primers and protocols are available under request.
2.6. Statistical analysis
Statistical analysis was performed using STATA v14 (StataCorp. 2015 Stata Statistical Software: Release 14. College Station, TX: StataCorp LP). A descriptive analysis was performed, calculating the median as a measure of central tendency as well as the interquartile range as a measure of its dispersion. Categorical variables were described through frequencies and proportions. Chi-squared test was calculated to assess associations between categorical variables. P values < 0.05 were considered to be statistically significant.
3. Results
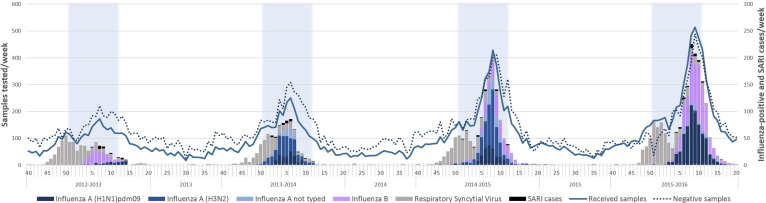
A total of 18,405 specimens from 12,680 cases (median age: 14.5 years; female: 46%) were received for respiratory viruses’ laboratory-confirmation, of which 2866 (16%) specimens from 2684 (21%) patients (median age: 36 years; female: 49%) were laboratory-confirmed for 1774 (66.1%) FLUAV (female: 48.4%) and 910 (33.9%) FLUBV (female: 50.2%). FLUV detection was usually reported in winter months, and only a few sporadic cases were detected during the interseason periods (Fig. 1 ). In addition, detection of FLUV steadily increased during the study period (p < 0.001). A variable co-circulation of the different influenza types was shown, since FLUAV circulated during all seasons, while FLUBV did not circulate during the 2013–2014 season. Moreover, a particular influenza type or subtype was also predominant in each studied season: FLUBV during the 2012–2013 season; A(H3N2) during the 2013–2014 and 2014–2015 seasons; and A(H1N1)pdm09 and FLUBV during the 2015–2016 season, as shown in Fig. 1.
Fig. 1.
Distribution of Severe Acute Respiratory Infections (SARI) cases and the positive cases for Influenza and other respiratory viruses detected from the epidemiological weeks 40/2012 to 20/2016. The period of winter in the North Hemisphere is highlighted in blue. Detections were performed by either immunofluorescence assay or real time RT-PCR. The period of influenza vaccine campaign begins in October (epidemiological week 42) in Catalonia (Spain). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Data regarding demographic features (age and sex), hospitalisation rate, ICU-admission, antiviral treatment administration and fatal outcomes of influenza-confirmed cases by season are summarised in Table 1 . Overall, the median age of FLUAV-confirmed cases (44 years; IQR 5–70 years) was significantly higher (p < 0.001) than that of FLUBV-confirmed cases (23 years; IQR 4–62 years). However, no significant differences (p = 0.489) between A(H1N1)pdm09 (42 years; IQR 3–67 years) and A(H3N2) (46 years; IQR 7–74 years) were found. Though, differences in the distribution by age groups were found between influenza types (p < 0.001) or subtypes (p < 0.001).
Table 1.
Detection, age (median), age categories/groups, female, hospitalisation, ICU-admission, antiviral treatment and mortality rates (N; %) by influenza types or subtypes and season (% in columns, depending on the influenza type/subtype).
| 2012–2013 | 2013–2014 | 2014–2015 | 2015–2016 | TOTAL | p-value | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Received samples | 3156 | 3571 | 5189 | 6489 | 18,405 | |||||||||||
| Influenza type | 181 (5.7) | 397 (11.1) | 883 (17.0) | 12235 (18.8) | 2684 (14.6) | |||||||||||
| FLUAV 49 (27.1) | FLUBV 132 (72.9) | FLUAV 397 (100) | FLUBV 0 (0.0) | FLUAV 667 (75.5) | FLUBV 215 (24.3) | FLUAV 661 (54.0) | FLUBV 563 (46.0) | FLUAV 1774 (66.1) | FLUBV 910 (33.9) | |||||||
| Influenza subtype1 | H1pdm09 | H3 | H1pdm09 | H3 | H1pdm09 | H3 | H1pdm09 | H3 | H1pdm09 | H3 | ||||||
| 38 (77.6) | 6 (12.2) | 113 (28.0) | 236 (59.0) | 172 (25.8) | 394 (59.1) | 593 (89.7) | 13 (2.0) | 916 (51.6) | 649 (36.6) | |||||||
| Age (median) | 9 | 38.5 | 8 | 29 | 30 | 48 | 58 | 56 | 47 | 69 | 13 | 42 | 46 | 23 | ||
| <2 years | 7 (18.4) | 0 | 21 (15.9) | 20 (17.7) | 51 (21.6) | 28 (16.3) | 39 (9.9) | 15 (7.0) | 101 (17.0) | 2 (15.4) | 62 (11.0) | 156 (17.0) | 92 (14.2) | 98 (10.8) | p < 0.0001 | |
| 2–4 years | 9 (23.7) | 1 (16.7) | 19 (14.4) | 18 (15.9) | 21 (8.9) | 19 (11.0) | 25 (6.3) | 9 (4.2) | 53 (8.9) | 0 (0.0) | 112 (19.9) | 99 (10.8) | 47 (7.2) | 140 (15.4) | ||
| 5–14 years | 4 (10.5) | 1 (16.7) | 40 (30.3) | 7 (6.2) | 25 (10.6) | 9 (5.2) | 47 (11.9) | 17 (7.9) | 32 (5.4) | 0 (0.0) | 118 (21.0) | 52 (5.7) | 73 (11.2) | 175 (19) | ||
| 15–64 years | 15 (39.5) | 3 (50) | 40 (30.3) | 50 (44.2) | 87 (36.9) | 70 (40.7) | 105 (26.6) | 88 (40.9) | 217 (36.6) | 3 (23.1) | 157 (27.9) | 352 (38.4) | 198 (30.5) | 285 (31.3) | ||
| >64 years | 3 (7.9) | 1 (16.7) | 12 (9.1) | 18 (15.9) | 52 (22.0) | 46 (26.7) | 179 (45.4) | 86 (40.0) | 190 (32.0) | 8 (61.5) | 114 (20.2) | 257 (28.1) | 240 (37.0) | 212 (23.3) | ||
| Female | 86 (47.5) | 210 (52.9) | 429 (49) | 589 (48.2) | 1314 (49.0) | p = 0.398 | ||||||||||
| 8 (21.1) | 3 (50.0) | 72 (54.5) | 60 (53.1) | 125 (53.0) | 79 (45.9) | 188 (47.7) | 108 (50.2) | 273 (46.0) | 9 (69.2) | 277 (49.2) | 420 (45.9) | 325 (50.1) | 457 (50.2) | |||
| Inpatients2 | 97 (53.6) | 185 (46.6) | 719 (81.4) | 493 (40.3) | 1494 (55.7) | p < 0.001 | ||||||||||
| 23 (60.5) | 4 (66.7) | 68 (70.1) | 52 (46.0) | 102 (43.2) | 141 (82.0) | 307 (77.9) | 181 (84.2) | 296 (49.9) | 6 (46.2) | 166 (29.5) | 512 (55.9) | 419 (64.6) | 415 (45.6) | p < 0.001 | ||
| ICU-admitted3 | 18 (9.9) | 23 (5.8) | 18 (2.0) | 28 (2.3) | 87 (3.2) | p trend < 0.001 | ||||||||||
| 5 (13.0) | 0 (0.0) | 12 (9.1) | 7 (6.2) | 10 (4.2) | 7 (5.0) | 4 (1.0) | 3 (1.4) | 17 (2.3) | 0 (0.0) | 9 (1.6) | 36 (3.9) | 14 (2.2) | 24 (2.6) | p = 0.037 | ||
| Exitus4 | 8 (4.4) | 8 (2.0) | 17 (1.9) | 13 (1.1) | 46 (1.7) | p trend = 0.003 | ||||||||||
| 3 (8.0) | 0 (0.0) | 5 (3.8) | 1 (0.9) | 3 (1.3) | 2 (1.2) | 12 (3.0) | 2 (0.9) | 9 (1.5) | 1 (7.7) | 2 (0.4) | 15 (1.6) | 16 (2.5) | 14 (1.5) | p = 0.116 | ||
Numbers may not add due to the existence of Influenza A viruses not subtyped. Season 2012–2013: 5 (10,2%); season 2013–2014: 48 (12,1%); season 2014–2015: 101 (1,4%); season 2015–2016: 55 (8,3%); global: 209 (11,8%).
Numbers may not add due to the existence of Influenza A viruses not subtyped. Season 2012–2013: 2 (40,0%); season 2013–2014: 31 (64,5%); season 2014–2015: 89 (88,1%); season 2015–2016: 25 (45,5%); global: 147 (70,3%).
Numbers may not add due to the existence of Influenza A viruses not subtyped. Season 2012–2013: 1 (20,0%); season 2013–2014: 6 (12,5%); season 2014–2015: 4 (3,9%); season 2015–2016: 2 (3,6%); global: 13 (6,2%).
Numbers may not add due to the existence of Influenza A viruses not subtyped. Season 2012–2013: 0 (0,0%); season 2013–2014: 4 (8,3%); season 2014–2015: 1 (1,0%); season 2015–2016: 1 (1,8%); global: 6 (2,9%).
During the season 2015–2016 one patient presented a co-infection between FLUAV and FLUBV.
The hospitalisation rate for influenza laboratory-confirmed cases was overall 55.7% (1494/2684 cases), with a balanced male:female proportion (female: 49.0%; p = 0.398). Furthermore, the hospitalisation rate was different between seasons (p < 0.001), being the highest (81.4%) during the 2014–2015 season. The hospitalisation rates were also significantly different between influenza types (FLUAV: 60.8% vs. FLUBV: 45.6%; p < 0.001) or FLUAV subtypes (H1N1pdm09: 55.9% vs. H3N2: 64.6%; p < 0.001). Among hospitalised patients, ICU-admissions were also different by season throughout the study period and showed a decreasing trend (p < 0.001). While no differences on ICU admissions between influenza types (FLUAV: 3.5% vs. FLUBV: 2.6%; p = 0.444) were observed, these were different between FLUAV subtypes (A(H1N1)pdm09: 3.9% vs. A(H3N2): 2.2%; p = 0.037). Through the study period, there was also a decreasing trend in mortality (p = 0.003), but no differences were observed between influenza types (p = 0.116) or FLUAV subtypes (p = 0.572). Nevertheless, the high number of fatal cases attributed to A(H1N1)pdm09 during the 2012–2013 season was remarkable (7.8%) in comparison with later seasons.
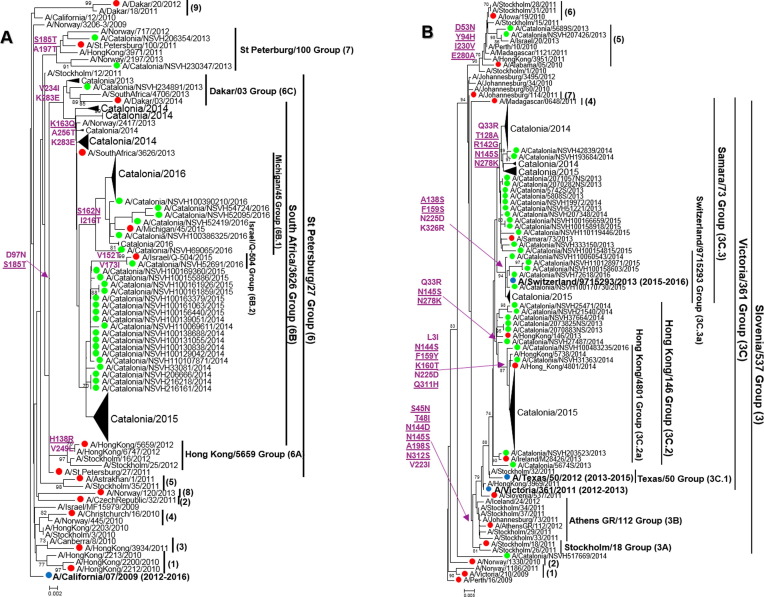
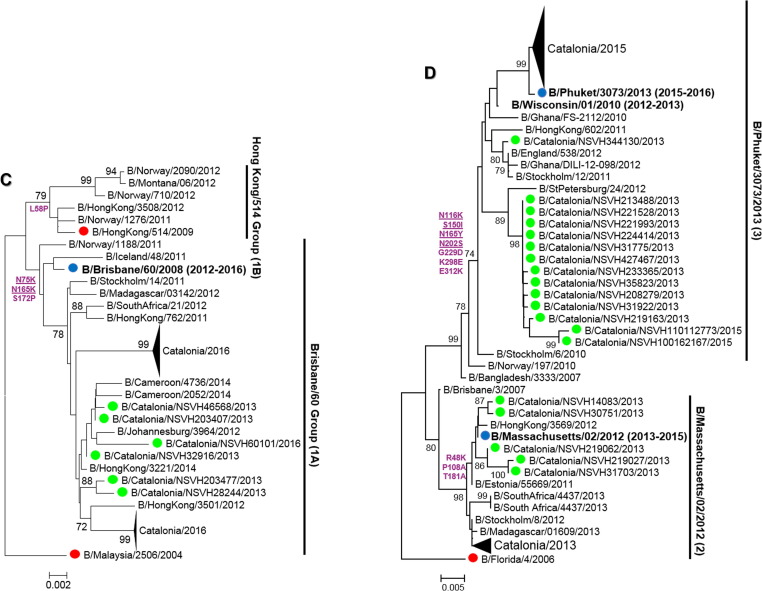
Phylogenetic analyses of HA1 sequences revealed the high genetic diversity of FLUV, distinguishing different genetic clades and subclades, represented in Fig. 2 (A–D) and Supplementary Table 3. Regarding A(H1N1)pdm09 viruses, all sequences (2 6 8) clustered into clades 6 and 7 during the 2012–2013 season. During the following seasons, sequences fell into two subgroups (6B.1 and 6B.2) within clade 6B (Fig. 2A), in which the strains (6B.1) gained a new glycosylation site (S162N) [19]. In addition, all A(H3N2) sequences (386) but one clustered within genetic clade 3 (A/Slovenia/537/2011-like), in different genetic subgroups (3C.2a, A/HongKong/4801/2014-like; 3C.3, A/Samara/73/2013-like; and 3C.3a, A/Switzerland/9715293/2013-like) (Fig. 2B). However, during three seasons the detected strains were closely related to the included vaccine strains, while during the 2014–2015 season A(H3N2) viruses belonged to the 3C.2a genetic subset (A/HongKong/4801/2014-like) that showed different antigenic features from vaccine strain included that season according to WHO/ECDC data [20]. Furthermore, during the 2015–2016 season there was a shift in the predominance of FLUBV lineage to FLUBV/VIC (Fig. 2C) (57 sequences), which was not included in the trivalent vaccine, since during the previous seasons, all FLUBV viruses belonged to FLUBV/YAM lineage (Fig. 2D). FLUBV/YAM viruses (134) detected during the 2012–2013 season belonged mostly to clade 2 (B/Massachusetts/02/2012-like) and only a few to clade 3 (B/Phuket/3073/2013-like), while all FLUBV/YAM detected during 2014–2015 corresponded to clade 3 (B/Phuket/3073/2013-like).
Fig. 2.
(A–D): Phylogenetic trees of HA1-domain sequences from influenza A(H1N1)pdm09 (A), A(H3N2) (B), B/VIC (C) and B/YAM (D) strains. The Tamura-Nei (TN93) method with gamma-distribution was used to carry out the analysis. Almost all sequences corresponding to influenza viruses detected during the 2012–2016 season are compressed, although some of them are labelled in green. Sequences used as reference are labelled in red and seasonal vaccine strains, in blue and bold. Only those bootstrap values over 70% are shown. Amino acid substitutions that define every phylogenetic cluster are marked in purple at the nodes, underlined those related to antigenic sites, and derived from each reference sequence (A(H1N1)pdm09: A/California/07/2009, A(H3N2): A/Perth/16/2009, B/Yamagata: B/Florida/04/2006, B/Victoria: B/Malaysia/2506/2004). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The characterisation of amino acid sequences of coding HA1-domain sequences also revealed the detection of some viral variants carrying the following amino acid substitutions in the RBS [21], [22]: S137F (1 case), I192S (1 case) and I192T (2 cases) among A(H3N2) viruses; and P241S (1 case) among FLUBV, all found in samples from the upper respiratory tract. In addition, D222G (1 case) and D222N (2 cases) mutations of A(H1N1)pdm09 viruses during the 2012–2013 season but not later, were detected in upper (2 cases) and lower (1 case) respiratory specimens from three ICU-admitted patients, one of which with fatal outcome (D222N). These mutations in the RBS were associated to severe disease in previous studies [23], [24], [25], [26].
Regarding amino acid substitutions associated with reduced NAI susceptibility [9], [16], two mutations were found in three 2015–2016 A(H1N1)pdm09 viruses (H275Y: 2 cases; S247N: 1 case) in respiratory samples collected from immunosuppressed patients, who favourably responded to a standard five-day oseltamivir treatment. Samples from two cases (H275Y and S246N) were collected before treatment, and the other one, one day after the onset of start of treatment.
4. Discussion
In the present study, the prevalence, the genetic diversity and the clinical outcomes of detected FLUV in patients attended or hospitalised at a tertiary hospital in Catalonia are reported, which might be considered a continuation of the influenza surveillance report from 2006 to 2012 in Catalonia previously published [15].
As shown in Fig. 1, the number of FLUV detected cases per season increased over the season, which might be related to the improvement of the detection methods by a broader use of PCR-based methods rather than antigen-detection based assays [27] in our laboratory, as well as the strengthened surveillance of respiratory viruses in our hospital.
The co-circulation of FLUAV and FLUBV during these four consecutive seasons was highly variable, with shifts in the predominance of influenza types and subtypes. Overall, the circulating FLUV showed similar proportions to those reported in other Spanish regions [28], but showing some differences in comparison to other European countries [29]. Previous studies already reported some differences between Catalonia and other Spanish regions [15]. FLUAV was predominant in three out of the four studied seasons, with the only exception of the 2012–2013 season when FLUBV was the most frequently detected, similarly to other Spanish regions but different from other European countries [29]. At subtype level, during the 2013–2014 season, A(H3N2) was predominantly detected in Catalonia based on these results and also on data provided by the Public Health Agency [30], in contrast to other Spanish communities and other European countries [28], [29]. This heterogeneity and variability at temporal and geographical level of FLUV circulation reinforce the idea to maintain a local and continuous influenza surveillance.
The clinical impact of influenza epidemics in the community is always considered a valuable data to be reviewed, taking into account the FLUV genetic diversity. Therefore, some differences were found between influenza types (A vs B) or subtypes (H1pdm09 vs H3), or vice versa regarding the age and the clinical outcomes of influenza-confirmed cases, such as hospitalisation, ICU-admission, and mortality rates. In the present study, the population susceptible to FLUAV infection seemed to be older than that susceptible for FLUBV, although FLUAV was predominant in almost all age groups, except to 5–14-year-old group in whom FLUBV prevailed as previously reported by other authors [31], [32]. On the other hand, while no differences were found between A(H1N1)pdm09 and A(H3N2), the distribution of cases by age groups was different. The hospitalisation rate was variable throughout the study reporting differences between seasons, which might be related to the predominant influenza types or subtypes. Overall, FLUAV and A(H3N2) were related to higher hospitalisation rates than FLUBV and A(H1N1)pdm09, respectively. However, the association of A(H3N2) with a higher hospitalisation and lower ICU-admission rates than A(H1N1)pdm09 might be explained due to the high number of hospitalised cases during the 2014–2015 season when a drifted A(H3N2) virus was circulating [20]. Mortality rate was declining along the study period, not showing differences between FLUV types or subtypes, but highlighting the high mortality rate for A(H1N1)pdm09 during the 2012–2013 season when a low number of FLUAV cases was reported. This decreasing mortality rate over the four seasons could be related to the increase frequency of sampling due to the introduction of PCR-based methods to speed diagnostic and treatment and reducing complications.
Based on phylogenetic analysis of HA1 sequences, A(H3N2) showed a higher heterogenicity than A(H1N1)pdm09. Despite the emergence, the spread and the genetic divergence of A(H1N1)pdm09 strains into different genetic subgroups (6B.1 and 6B.2) within A(H1N1)pdm09 6B clade, as reported worldwide [33], the current A(H1N1)pdm09 viruses remained antigenically similar to the vaccine strain since the beginning of the pandemic [20], [34]. However, despite antigenic stability, A(H1N1)pdm09 is still causing severe hospitalisations.
On the other hand, A(H3N2) virus has shown a great capability to evolve quickly due to maybe the immune pressure from the human population infected because it has been globally circulating for more than 50 years. During the last seasons, A(H3N2) viruses acquired a great number of amino acid substitutions, diverging in different genetic clades and subclades as shown in the present study, and continuously changing the potential glycosylation features of HA. In fact, as previously commented, the high activity of A(H3N2) viruses during the 2014–2015 season was related to the circulation of A(H3N2) viruses antigenically different from the vaccine strain used [20]. However, the unpredictable evolution of FLUV together with the current challenges for the correct antigenic characterisation of A(H3N2) viruses, make difficult to forecast the vaccine composition for upcoming seasons [20].
Regarding FLUBV, there was co-circulation of both lineages, but with the predominance of FLUBV/YAM in two out of three seasons with FLUBV detection reported as well as the no correspondence with the strain lineage included in the trivalent vaccine composition. The shift on the predominance of both lineages is well-known, and sometimes the circulating lineage is not included in the trivalent vaccine, which highlights the need of using the tetravalent influenza vaccine to give a better and wider protection to the high-risk population. Therefore, the vaccination with tetravalent vaccines in primary care would both improve the attitude towards influenza vaccination and have a beneficial effect on population coverage.
In addition to the circulation of both FLUBV lineages, FLUBV also showed a continuous evolution based on HA sequences. Therefore, FLUBV/YAM viruses belonged to two genetically and antigenically divergent genetic clades (clade 2 and 3) [29], one of which (clade 3) was not represented by the corresponding vaccine strain during the 2012–2013 season.
In regard to the detection of virulence and resistance markers, A(H1N1)pdm09 has the capability to acquire mutations that can alter the tropism or the antiviral susceptibility during the infection [24], [35]. On one hand, A(H1N1)pdm09 strains from ICU-admitted cases carrying mutations D222G or D222N in HA1, which are still the unique genetic markers related to disease severity [23], [24], [25], [26], were detected during the 2012–2013 season but not in later seasons, when a higher A(H1N1)pdm09-related mortality rate was reported. Surely, the prevalence of variants carrying these mutations might be underestimated due to a suboptimal characterisation of severe influenza cases, since the virological surveillance of influenza viruses is overall carried out from samples collected from community-based surveillance networks. On the other hand, the emergence of genetically resistant variants to antiviral drugs is one of the main concerns of influenza surveillance and Public Health worldwide [9], [16]. In the present study, three viruses carrying the mutations H275Y and S247N, associated with resistance or reduced susceptibility to NAIs, were found in respiratory specimens from hospitalised patients under standard oseltamivir treatment during the 2015–2016 season. The fact that immunosuppression status and antiviral treatment administration are considered high-risk factors for the selection and emergence of resistant strains [35], conditions that are accomplished by many of the patients hospitalised in tertiary centres, highlights the importance to strengthen the virological surveillance of influenza viruses in hospital settings. While surveillance tasks in community-based networks is highly valuable, the information reported in hospital settings, where cases of severe disease or antiviral resistance are attended, should be always considered to monitor circulating viruses, in particular those with novel phenotypic features.
In summary, our results highlight the prevalence of FLUV in patients admitted to a tertiary hospital and its variable and unpredictable circulation. The capability of FLUV to acquire genetic diversity that can affect viral features related to antigenicity, tropism, and susceptibility to antivirals makes virological surveillance, where these variants can be monitored, highly recommended in hospital settings.
Ethical approval
Institutional Review Board approval (PR(AG)329/2014) was obtained from the Vall d’Hebron University Hospital (HUVH) Clinical Research Ethics Committee.
Conflict of interest
The authors have no conflicts of interest to declare.
Acknowledgements
This work has been partially supported by Fondo de Investigación Sanitaria (grants FIS PI08/0118, FIS PI11/01864, FIS PI14/01838) from the Spanish Ministry Science, Innovation and Universities; Agència d’Avaluació de Tecnologia i Recerca Mèdiques (AATRM) (grant 402/02/2008); Catalan Association of Public Universities (ACUP) (grants 2010ACUP_00437); the Spanish Network for the Research in Infectious Diseases (REIPI RD16/0016/0003); and the European Regional Development Fund (ERDF). Cristina Andrés has been a recipient of a predoctoral fellowship from the Vall d’Hebron Research Institute (PRED-VHIR-2013). We would like to acknowledge GISAID as we used their reference sequences.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.vaccine.2019.03.046.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References:
- 1.Lin T., Wang G., Li A., Zhang Q., Wu C., Zhang R. The hemagglutinin structure of an avian H1N1 influenza A virus. Virology. 2009;392:73–81. doi: 10.1016/j.virol.2009.06.028. [DOI] [PubMed] [Google Scholar]
- 2.Medina R.A., García-Sastre A. Influenza A viruses: new research developments. Nat Rev Microbiol. 2011;9:590–603. doi: 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schrauwen E.J., de Graaf M., Herfst S., Rimmelzwaan G.F., Osterhaus A.D., Fouchier R.A. Determinants of virulence of influenza A virus. Eur J Clin Microbiol Infect Dis. 2014;33:479–490. doi: 10.1007/s10096-013-1984-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouvier N.M., Palese P. The biology of influenza viruses. Vaccine. 2008;26(Suppl. 4):D49–D53. doi: 10.1016/j.vaccine.2008.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamb R.A., Choppin P.W. The gene structure and replication of influenza virus. Annu Rev Biochem. 1983;52:467–506. doi: 10.1146/annurev.bi.52.070183.002343. [DOI] [PubMed] [Google Scholar]
- 6.Tewawong N., Suwannakarn K., Prachayangprecha S., Korkong S., Vichiwattana P., Vongpunsawad S. Molecular epidemiology and phylogenetic analyses of influenza B virus in Thailand during 2010 to 2014. PLoS ONE. 2015;10:e0116302. doi: 10.1371/journal.pone.0116302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steinhauer D.A. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology. 1999;258:1–20. doi: 10.1006/viro.1999.9716. [DOI] [PubMed] [Google Scholar]
- 8.Glaser L., Stevens J., Zamarin D., Wilson I.A., García-Sastre A., Tumpey T.M. A single amino acid substitution in 1918 influenza virus hemagglutinin changes receptor binding specificity. J Virol. 2005;79:11533–11536. doi: 10.1128/JVI.79.17.11533-11536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pozo Francisco, Lina Bruno, de Andrade Helena Rebelo, Enouf Vincent, Kossyvakis Athanasios, Broberg Eeva, Daniels Rob, Lackenby Angie, Meijer Adam. Guidance for clinical and public healt laboratories testing influenza virus antiviral drug susceptibility in Europe. J Clincal Virol Sci. 2013:7. doi: 10.1016/j.jcv.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Smith D.J., Lapedes A.S., de Jong J.C., Bestebroer T.M., Rimmelzwaan G.F., Osterhaus A.D. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 11.Muñoz M.P., Soldevila N., Martínez A., Carmona G., Batalla J., Acosta L.M. Influenza vaccine coverage, influenza-associated morbidity and all-cause mortality in Catalonia (Spain) Vaccine. 2011;29:5047–5052. doi: 10.1016/j.vaccine.2011.04.067. [DOI] [PubMed] [Google Scholar]
- 12.Manual de vacunacions de Catalunya. Agència de Salut Pública de Catalunya (ASPCAT). Generalitat de Catalunya. p. 166-74. Available at: <http://salutpublica.gencat.cat/web/.content/minisite/aspcat/promocio_salut/vacunacions/00manual_de_vacunacions/Manual-de-vacunacions.pdf>.
- 13.Sistemes d'Informació dels Serveis d'Atenció Primària (SISAP). Institut Català de la Salut (ICS). Available at: <https://www.ics.gencat.cat/sisap/grip/principal>.
- 14.Pla d’actuació a Catalunya enfront d’una infecció per virus de la grip en fase post-pandèmica. Sub-direcció General de Vigilància i Resposta a Emergències de Salut Pública. Generalitat de Catalunya, Departament de Salut.; Gener 2018. Available at: <http://canalsalut.gencat.cat/web/.content/home_canal_salut/professionals/temes_de_salut/grip/documents/grippostpan2010.pdf>.
- 15.Antón A., Marcos M.A., Torner N., Isanta R., Camps M., Martínez A. Virological surveillance of influenza and other respiratory viruses during six consecutive seasons from 2006 to 2012 in Catalonia, Spain. Clin Microbiol Infect. 2016;22(564):e1–e9. doi: 10.1016/j.cmi.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen H.T., Fry A.M., Gubareva L.V. Neuraminidase inhibitor resistance in influenza viruses and laboratory testing methods. Antivir Ther. 2012;17:159–173. doi: 10.3851/IMP2067. [DOI] [PubMed] [Google Scholar]
- 17.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.World Health Organisation (WHO). Recommended composition of influenza virus vaccines for use in the 2016-2017 northern hemisphere influenza season; 2016. Available at: <www.who.int/influenza/vaccines/virus/recommendations/201602_recommendation.pdf2016>.
- 20.World Health Organisation (WHO). WHO recommended candidate viruses and potency testing reagents for the development and production of vaccines for use in specific influenza seasons. Available at: <http://www.who.int/influenza/vaccines/virus/candidates_reagents/home/en/>.
- 21.Brownlee G.G., Fodor E. The predicted antigenicity of the haemagglutinin of the 1918 Spanish influenza pandemic suggests an avian origin. Philos Trans R Soc Lond B Biol Sci. 2001;356:1871–1876. doi: 10.1098/rstb.2001.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu R., Ekiert D.C., Krause J.C., Hai R., Crowe J.E., Wilson I.A. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328:357–360. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu L., Bao L., Lv Q., Deng W., Ma Y., Li F. A single-amino-acid substitution in the HA protein changes the replication and pathogenicity of the 2009 pandemic A (H1N1) influenza viruses in vitro and in vivo. Virol J. 2010;7:325. doi: 10.1186/1743-422X-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vazquez-Perez J.A., Isa P., Kobasa D., Ormsby C.E., Ramírez-Gonzalez J.E., Romero-Rodríguez D.P. A (H1N1) pdm09 HA D222 variants associated with severity and mortality in patients during a second wave in Mexico. Virol J. 2013;10:41. doi: 10.1186/1743-422X-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corcioli F., Arvia R., Pierucci F., Clausi V., Bonizzoli M., Peris A. HA222 polymorphism in Influenza A(H1N1) 2009 isolates from Intensive Care Units and ambulatory patients during three influenza seasons. Virus Res. 2014;180:39–42. doi: 10.1016/j.virusres.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 26.Voltersvik P., Aqrawi L.A., Dudman S., Hungnes O., Bostad L., Brokstad K.A. Pulmonary changes in Norwegian fatal cases of pandemic influenza H1N1 (2009) infection: a morphologic and molecular genetic study. Influenza Other Respir Viruses. 2016;10:525–531. doi: 10.1111/irv.12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pérez-Ruiz M., Pedrosa-Corral I., Sanbonmatsu-Gámez S., Navarro-Marí M. Laboratory detection of respiratory viruses by automated techniques. Open Virol J. 2012;6:151–159. doi: 10.2174/1874357901206010151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Instituto de Salud Carlos III (ISCIII). Sistema de Vigilancia de la Gripe en España. Available at: <http://vgripe.isciii.es/inicio.do>.
- 29.Medical Research Council (MRC) CRU, the Wellcome Trust, UCL (University College London), Imperial College London and King's College London. The Francis Crick Institute. Available at: <https://www.crick.ac.uk/partnerships/worldwide-influenza-centre/annual-and-interim-reports>.
- 30.Pla d'Informació de les Infeccions Respiratories Agudes a Catalunya (PIDIRAC). Generalitat de Catlunya; 1999. Available at: <http://canalsalut.gencat.cat/ca/professionals/vigilancia-epidemiologica/pla-dinformacio-de-les-infeccions-respiratories-agudes-a-catalunya-pidirac/>.
- 31.Wie S.H., So B.H., Song J.Y., Cheong H.J., Seo Y.B., Choi S.H. A comparison of the clinical and epidemiological characteristics of adult patients with laboratory-confirmed influenza A or B during the 2011–2012 influenza season in Korea: a multi-center study. PLoS ONE. 2013;8:e62685. doi: 10.1371/journal.pone.0062685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beauté J., Zucs P., Korsun N., Bragstad K., Enouf V., Kossyvakis A. Age-specific differences in influenza virus type and subtype distribution in the 2012/2013 season in 12 European countries. Epidemiol Infect. 2015;143:2950–2958. doi: 10.1017/S0950268814003422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Broberg E.M.A., Prosenc K., Bragstad K., Hungnes O., on behalf of the WHO European Region and the European Influenza Surveillance Network members of, countries tr Predominance of influenza A(H1N1)pdm09 virus genetic subclade 6B.1 and influenza B/Victoria lineage viruses at the start of the 2015/16 influenza season in Europe. Euro Surveill. 2016:30184. doi: 10.2807/1560-7917.ES.2016.21.13.30184. [DOI] [PubMed] [Google Scholar]
- 34.Nelson M., Spiro D., Wentworth D., Beck E., Fan J., Ghedin E. The early diversification of influenza A/H1N1pdm. PLoS Curr. 2009;1:RRN1126. doi: 10.1371/currents.RRN1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hurt A.C., Chotpitayasunondh T., Cox N.J., Daniels R., Fry A.M., Gubareva L.V. Antiviral resistance during the 2009 influenza A H1N1 pandemic: public health, laboratory, and clinical perspectives. Lancet Infect Dis. 2012;12:240–248. doi: 10.1016/S1473-3099(11)70318-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.