Abstract
The success of RNA viruses as pathogens of plants, animals, and humans depends on their ability to reprogram the host cell metabolism to support the viral infection cycle and to suppress host defense mechanisms. Plus-strand (+)RNA viruses have limited coding potential necessitating that they co-opt an unknown number of host factors to facilitate their replication in host cells. Global genomics and proteomics approaches performed with Tomato bushy stunt virus (TBSV) and yeast (Saccharomyces cerevisiae) as a model host have led to the identification of 250 host factors affecting TBSV RNA replication and recombination or bound to the viral replicase, replication proteins, or the viral RNA. The roles of a dozen host factors involved in various steps of the replication process have been validated in yeast as well as a plant host. Altogether, the large number of host factors identified and the great variety of cellular functions performed by these factors indicate the existence of a truly complex interaction between TBSV and the host cell. This review summarizes the advantages of using a simple plant virus and yeast as a model host to advance our understanding of virus–host interactions at the molecular and cellular levels. The knowledge of host factors gained can potentially be used to inhibit virus replication via gene silencing, expression of dominant negative mutants, or design of specific chemical inhibitors leading to novel specific or broad-range resistance and antiviral tools against (+)RNA plant viruses.
Keywords: Virus replication, Viral replicase complex, Host factors, Genome-wide screens, RNA–protein interaction, Yeast as a host, Protein–protein interaction
I. Introduction
The success of plus-strand (+)RNA viruses as pathogens of plants, animals, and humans depends on the ability of these viruses to reprogram the host cell metabolism to support the infection process and to avoid/suppress host defense mechanisms. (+)RNA viruses have limited coding potential with usually 4–10 genes, yet they can replicate efficiently in the infected host cells. They accomplish this feat by recruiting an unknown number of host factors, such as host proteins, membranes, and ribonucleotides for their replication that can produce thousands to millions of progeny viral RNAs per cell in 24 h. The virus can trick the recruited host factors to perform novel functions that are frequently targeted against the host cells. (+)RNA viruses can also induce strong responses of the infected host cells leading to the activation of the innate immune responses. Altogether, the (+)RNA virus infected cells go through major changes during the infection process. Many of the original cellular processes/pathways are getting reprogrammed by the infecting virus and these changes make the cells dramatically different from the uninfected cells. The outcome of the virus infection often resembles to the chaotic situation and destruction caused by having two opposing armies fighting a well-planned out, but expensive war for gaining full control over the same country.
In spite of the significance of virus–host interaction for human, animal, and plant health, our current understanding of the host factors involved in (+)RNA virus infections is still incomplete. Therefore, one of the major frontiers of ongoing research is to identify all changes in the infected cells that can potentially result in better, more efficient antiviral strategies and/or reduce the damage in the host cells caused by viral infections.
A. (+)RNA virus replication is a multistep process in the infected cells
After entry of virus into the cell, translation of the viral RNA leads to production of the viral replication proteins. These proteins then facilitate the rescue of the viral RNA from translation and the viral RNA is selected/recruited for replication. This is followed by the assembly of the viral replicase on subcellular membrane surfaces. The assembled replicase complex produces complementary minus-strand (−)RNA using the original (+)RNA as a template. Then, the minus-stranded RNA intermediate is used by the viral replicase to synthesize excess amount of new (+)RNA progeny, which is released from the site of replication to the cytosol and/or become encapsidated to form new viral particles (Ahlquist et al., 2003, Panavas et al., 2005a). One of the amazing things about (+)RNA viruses is that they complete an infection cycle within 6–24 h in the primary infected cells. Thus, (+)RNA virus infection is often a fast race between the parasite and the host to gain control over the resources of the host cell.
B. Selection of the viral RNA template for replication and the recruitment of the replication proteins to the subcellular sites of replication
Viral (+)RNA replication takes place on the cytosolic surfaces of intracellular membranes, where the viral RNA and replication proteins, together with co-opted host factors, are sequestered and reach high local concentrations that facilitate robust replication. Many RNA viruses are known to form spherules (small membrane invaginations) with small openings towards to the cytosol (Fig. 1 ) (Kopek et al., 2007, Schwartz et al., 2002). These spherules are the sites of RNA replication and frequently connected with the sites of virus assembly (Ahlquist, 2006). But how are these spherules formed and what is the role of the co-opted host factors? Although we do not yet know the answers to these fascinating questions, it is likely that efficient recruitment of all the viral and host factors are critical for replication.
Figure 1.
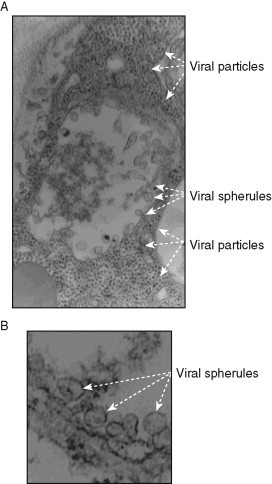
Representative electron micrographs of portions of N. benthamiana cells infected with the tombusvirus Cucumber necrosis virus (CNV). (A) The CNV-induced spherules in the center of the image and the assembled large number of virions in plant cells are depicted with arrows. Note that the entire cytosol of the portion of the cell shown is completely filled by CNV virions, demonstrating robust CNV replication. Magnification is 49,000×. (B) Several characteristic CNV-induced spherules are marked with arrowheads on the EM images. These 50–80 nm spherules are formed via membrane invagination into peroxisomal or ER-derived membranes. Narrow openings (necks) are visible likely connecting the spherules to the cytosol. Control samples lacking CNV do not show similar structures (not shown). Magnification is 98,000×. The images were taken by Dr. Barajas.
The selection of the viral (+)RNA for replication is thought to be mediated by a viral-coded protein binding selectively to a specific sequence/structure in the viral (+)RNA. For example, the Tomato bushy stunt virus (TBSV) p33 auxiliary replication protein recognizes a C⋅C mismatch within an extended hairpin in the viral RNA (Monkewich et al., 2005, Pogany et al., 2005). The Brome mosaic virus (BMV) 1a helicase-like protein binds to the subgenomic promoter region in RNA3 to facilitate the selection of the RNA for replication (Wang et al., 2005). The flock house virus (FHV) protein A replication protein specifically recognizes the 5′ sequence in the RNA facilitating its recruitment to the mitochondrial outer membrane (Van Wynsberghe and Ahlquist, 2009). In spite of these advances in our understanding of the template selection process, we do not yet know how the translating ribosomes are removed from the (+)RNA templates selected for replication. Models have been proposed that viral replication proteins or host factors, such as Lsm proteins, promote the switch of the RNA from translation to replication (Beckham et al., 2007).
Another important step is the recruitment of the viral (+)RNA and the viral replication proteins together with co-opted host proteins to the sites of replication. The viral (+)RNA likely travels together with the viral proteins as an RNP complex. The targeting of key viral proteins in the cells is guided by signal sequences present in the replication proteins. Other viral proteins, such as the viral RdRp protein, and co-opted host factors might be “piggy-backing” on the targeted viral protein to reach the destination of the subcellular membranes.
C. The assembly of the replicase complexes of (+)RNA viruses is a complex process
A key step in viral replication is the assembly of the viral replicase on the cytosolic surfaces of intracellular membranes, which is a poorly understood process (Salonen et al., 2005). The viral replicase consists of viral-coded RNA-dependent RNA polymerase (RdRp), viral auxiliary replication proteins, the subcellular membrane, and co-opted host proteins. Among these factors, the contribution of host factors to the viral replication process is the least understood. The host factors likely complement the functions of the viral replication proteins to regulate RNA replication. Moreover, the host components in the replicase complex likely provide protection from host cellular ribonucleases, including the powerful gene silencing machinery, as well as they might be involved in delaying the recognition of viral components by the host surveillance system. The host factors are also likely responsible for activation of RdRps of several (+)RNA viruses, such as p92pol of TBSV, 2apol of BMV, P2 of Alfalfa mosaic virus (AMV), 180K of Tomato mosaic virus (ToMV), and the hepatitis C virus (HCV) NS5B. These viral RdRps become activated only after the assembly of the viral replicase in membranous spherules or vesicles (Panaviene et al., 2004, Panaviene et al., 2005, Quadt et al., 1995, Vlot et al., 2001). Although the role of host factors during the assembly of the replicase and the activation of RdRp is currently unknown, they likely work together with the viral RNA as well, which also plays a key role in these processes.
D. RNA synthesis by the viral replicase is a two-step process
(+)RNA virus replication occurs within membrane-bound structures and is performed by the viral replicase complex using the recruited (+)RNA as the template (Ahlquist et al., 2003). The viral RNA contains specific cis-acting elements, including promoters, silencers, and enhancers, which regulate de novo initiation of RNA synthesis and the efficiency of replication (Dreher, 1999, Kao et al., 2001, Nagy & Pogany, 2006, White & Nagy, 2004, Wu et al., 2009). Interestingly, the (−)RNA produced on the (+)RNA template is the more efficient template, resulting in 20–100-fold more (+)RNA progeny than the amount of (−)RNA intermediate. It is currently thought that the (−)RNA is always present in the replicase complex and never gets released from the site of replication, whereas the majority of (+)RNA progeny is released to the cytosol or assembled into viral particles (Panavas et al., 2005a). Host factors are certainly involved in these processes, although detailed mechanistic studies on their roles are not yet available.
E. Tombusviruses are simple model (+)RNA viruses of plants
TBSV and other tombusviruses are model plant RNA viruses with a single 4.8-kb genomic (g)RNA component. The gRNA codes for two replication proteins, termed p33 and p92pol, and produces two subgenomic RNAs for the expression of three viral proteins involved in cell-to-cell movement, encapsidation, and suppression of gene silencing (Nagy & Pogany, 2008, White & Nagy, 2004). Recent advances with tombusviruses have been accelerated by the development of yeast (Saccharomyces cerevisiae) as a model host to study TBSV replication and recombination (Nagy, 2008, Panavas & Nagy, 2003). Yeast expressing p33 and p92pol replication proteins can efficiently replicate a short TBSV-derived replicon (rep)RNA (Panavas & Nagy, 2003, Panaviene et al., 2004). Importantly, replication of the TBSV repRNA in yeast depends on the same cis-acting RNA elements and trans-acting p33/p92 replication proteins as in plants (Nagy, 2008, Panavas & Nagy, 2003). The DI-72 repRNA, which is derived naturally from TBSV infections of plants, does not encode proteins and it can replicate efficiently in yeast cells without maintaining an artificial selection pressure. Moreover, the tombusvirus repRNA plays several functions, including serving as a template for replication and as a platform for the assembly of the viral replicase complex (Nagy & Pogany, 2008, Panaviene et al., 2005, Pogany et al., 2005). The viral RNA also participates in RNA recombination (Serviene et al., 2005, White & Morris, 1994, White & Nagy, 2004), which likely plays a major role in virus evolution. Altogether, the development of yeast as a model host for TBSV facilitated the application of the available genomics and proteomics tools to identify host components required or affecting TBSV replication and virus–host interactions (Jiang et al., 2006, Li et al., 2008, Li et al., 2009, Nagy, 2008, Nagy & Pogany, 2006, Panavas et al., 2005b, Serva & Nagy, 2006, Serviene et al., 2005, Serviene et al., 2006). The results generated with these genomics and proteomics tools will be described below.
II. Genome-Wide Screens for Systematic Identification of Host Factors Affecting TBSV Replication
An advantage to perform genome-wide screens in an organism is the availability of a collection of knockout mutants, such as the yeast single-gene-knockout (YKO) library, or a library containing gene sets with regulatable expression, such as yTHC library for the essential genes in yeast (Fig. 2 ). RNAi-based genome-wide screens can also be performed with large siRNA libraries, as shown for fruit fly and mammalian cells (Cherry et al., 2005, Hao et al., 2008, Kok et al., 2009, Krishnan et al., 2008). This chapter focuses on the results obtained with TBSV and the yeast libraries.
Figure 2.
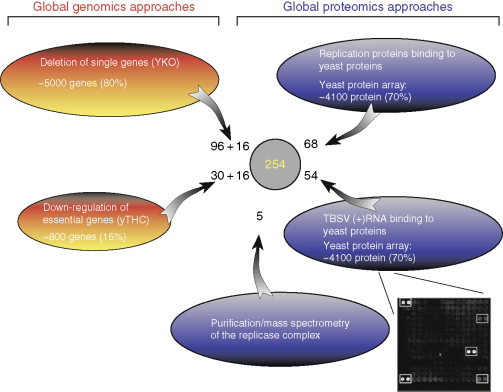
Summary of global genomics and proteomics screens performed with TBSV RNA/replication proteins. The number of host factors identified in the screens are shown, while the number behind “+” shows the number of host factors identified that affected TBSV recombination in yeast. Note that the sum of all factors from the individual screens and the total number of identified host factors (total of 254) are not the same due to identification of several common host factors in different screens. A representative subarray from the protein array is also shown. The two circled dots on the top right side of the subarray indicate a strong and a weak yeast protein interactor, while the other circled dots represent standards.
A. Single-gene-knockout YKO library
This library contains ∼5000 yeast strains representing ∼80% of yeast genes, and ∼66% of those genes are characterized based on cellular function (Fig. 2). The advantage of the YKO library is that the strains are well defined and they lack the expression of the given gene, unlike the siRNA-based screens, where the expression of the particular gene is only knocked down. The repeatability of siRNA screens also depends on the level of knockdown, which could be influenced by several factors/parameters of the experiments.
The disadvantage of the use of the YKO library or other mutant libraries is the significant gene redundancy in biological systems. Namely, many genes have two or more homologous copies, like the heat-shock protein 70 (Hsp70), which can efficiently complement each other, leading to false negatives during the screens. Also, many proteins have pleiotropic effects in cells, affecting the functions of several other proteins and pathways. These host proteins might contribute indirectly to virus replication. Therefore, usually it is not yet known at the end of the genome-wide screens if the identified host genes affect virus replication directly or indirectly.
The systematic screen of the YKO library has revealed that TBSV repRNA replication is affected by 96 different host genes (Fig. 2) (Panavas et al., 2005b). Single deletion of 90 genes reduced, while 6 increased TBSV repRNA accumulation. Grouping of the identified host genes based on their known cellular functions revealed that TBSV replication depends on a wide variety of gene functions belonging to 11 different groups. These include five genes involved in protein biosynthesis by either being part of the ribosome or acting as a translation elongation factor. The other groups are involved in protein metabolism, such as ubiquitination pathway and posttranslational modification (eight genes), RNA metabolism (five genes), and in lipid metabolism (five genes). A large group includes 20 genes implicated in vesicle-mediated transport, affecting endoplasmic reticulum (ER), Golgi, vacuole transport, or membrane fusion. Additional groups include membrane-associated proteins (seven genes) or stress responsive genes (five genes) or have variable functions in general metabolism (11 genes). The remaining groups contain genes involved in transcription and DNA remodeling or include genes with unknown function and hypothetical ORFs.
An additional screen with the YKO library and the TBSV repRNA led to the identification of 16 host genes affecting TBSV RNA recombination (Cheng et al., 2006, Serviene et al., 2005, Serviene et al., 2006). The identified host genes code for proteins involved in various cellular processes, such as translation, RNA metabolism, protein modifications and intracellular transport, or membrane modifications. Since viral RNA recombination is a major mechanism for viruses in their rapid evolution, the involvement of host factors in RNA recombination suggests that the evolution of (+)RNA viruses might not depend on totally random events, but it is affected by the host.
Subsequent, more detailed studies have led to the characterization of the roles of the following host factors: Xrn1p in TBSV replication and recombination (Cheng et al., 2006, Cheng et al., 2007, Jaag & Nagy, 2009, Serviene et al., 2005); Nsr1p for inhibition of TBSV replication (Jiang et al., 2010); seven ESCRT (endosomal sorting complexes required for transport) proteins for affecting the quality of the assembly process for the tombusvirus replicase complex (Barajas et al., 2009a); Erg4p in TBSV replication due to its affect on sterol biosynthesis (Sharma et al., 2009), and Pmr1p affecting TBSV recombination and replication (Jaag et al., 2010). These examples covering ∼10% of the identified genes from the YKO library vindicate genome-wide screens as powerful tools for identification of host factors in TBSV replication.
Altogether, the identification of over 100 genes with rather diverse functions that affected TBSV replication/recombination via the genome-wide screens suggests that the interaction between a host and an (+)RNA virus is likely very complex and the replication of the virus is affected by many factors and pathways inside the cells. This unexpectedly high complexity in virus–host interaction validates the use of high-throughput approaches to identify all the players from the host participating in the interaction.
B. yTHC library/essential genes
The Tet-promoter-based Hughes collection (yTHC) contains ∼800 out of ∼1100 essential yeast genes (Mnaimneh et al., 2004). In the yTHC collection, the expression of a given essential yeast gene is under the control of a Tet-titratable promoter in the genome. The expression of the essential gene can be turned off by the addition of doxycycline to the yeast growth medium (Mnaimneh et al., 2004).
Using the yTHC collection, a total of 30 essential host genes have been identified that affected TBSV replication (Fig. 2) (Jiang et al., 2006). The identified genes have different molecular functions in various cellular processes including RNA binding/processing (nine genes), RNA helicase/unwinding/RNA metabolism (four genes), or RNA polymerase/RNA transcription (five genes). Others are involved in protein synthesis/modification (four genes), protein transport (one gene), or lipid biosynthesis (one gene). Other genes are involved in general metabolism, in chromatin remodeling, or function as putative GTPases, while two genes have currently unknown functions.
Among the yTHC collection, 16 strains showed altered recombination frequency (Serviene et al., 2006). The identified genes included five affecting RNA binding/processing/unwinding, three genes are known to code for proteins with RNA polymerase/RNA transcription function. Others are involved in protein modification/catabolism, or protein transport.
Follow-up studies characterized the roles of the following host factors: Erg25p in TBSV replication due to its affect on sterol biosynthesis (Sharma et al., 2010), and the indirect role of Rpb11p transcription factor, which affected TBSV recombination via changing the ratio of p33 and p92 proteins produced in yeast (Jaag et al., 2007). Overall, the host genes identified as being essential are represented almost at twice the ratio (3.75%) when compared with the nonessential genes (2%), suggesting that tombusviruses might have adapted to use and/or dependent on essential genes to higher extent than on nonessential genes. However, additional follow-up studies with the essential genes are more difficult than with the nonessential genes, due to the shared requirement of the identified essential factors in cell growth and TBSV replication/recombination.
III. Proteomics-Based Screens for Systematic Identification of Host Factors Affecting TBSV Replication
The accumulation levels and molecular functions/activities of proteins in cells are affected not only by the level of mRNA transcription, but also by many other processes, such as the efficiency of translation of a given mRNA controlled by cis-acting RNA sequences, stability of the protein, subcellular localization of the protein and posttranslational modifications (phosphorylation, ubiquitination, acetylation, etc.), as well as the availability of interacting protein/RNA/DNA partners and substrates. Therefore, proteome-wide screens based on proteomics approaches are needed to identify all the host factors interacting with selected viral proteins or to determine what molecular networks and cellular pathways are affected by the viral proteins.
One major advantage of proteomics approaches in general is that they are not limited by gene redundancy. Therefore, proteomics can efficiently complement the above genomics approaches to identify host factors affecting (+)RNA replication. Another advantage is that direct protein interaction networks can be established. The disadvantages of many proteomics approaches are that the abundance of a particular protein in the cell could be a critical factor, since low abundance proteins are more difficult to identify in biological samples than high abundance proteins. On the contrary, low or high expression level for a given protein is a lesser concern in the above genomics screens with gene deletions or downregulation of mRNA transcripts/protein levels. Also, weak molecular interactions important in regulatory networks are notoriously difficult to detect by many proteomics approaches. In spite of these disadvantages, the ever-improving proteomics approaches are gaining in popularity to identify host factors affecting viral replication.
Based on these considerations, we have introduced proteomics approaches to study the interaction of tombusviruses with their hosts. The two major approaches discussed in this chapter are based on protein copurification/mass spectrometry and a protein array approach.
A. Replicase purification/mass spectrometry
Identification of cellular factors recruited into the viral replicase complex for helping viral replication is important to determine the players and their functions in the replicase complex. What makes this a really challenging task is the membrane-association of the replicase complex for all eukaryotic (+)RNA viruses. Therefore, the replicase complex should be solubilized from the membranes prior to further analysis. Both the solubilization step and the following purification step likely remove proteins that are weakly/loosely associated with the replicase complex. Thus, it is highly possible that the copurified proteins in the replicase complex represent only the most abundant and strongly bound cellular proteins.
To identify the host factors present in the viral replicase complex, recent proteomics approaches revealed that 4–10 host proteins were part of the highly purified functional tombusvirus replicase (Fig. 3 ) (Serva and Nagy, 2006). Additional studies have determined at least seven proteins in the replicase complex, including the viral p33 and p92pol, the heat-shock protein 70 chaperones (Hsp70, Ssa1/2p in yeast), glyceraldehyde-3-phosphate dehydrogenase (GAPDH, encoded by TDH2 and TDH3 in yeast), pyruvate decarboxylase (Pdc1p), Cdc34p ubiquitin ligase (Li et al., 2008, Serva & Nagy, 2006, Wang & Nagy, 2008), and eukaryotic translation elongation factor 1A (eEF1A) (Li et al., 2009). The functions of GAPDH and Hsp70 have been studied in some details (Pogany et al., 2008, Wang & Nagy, 2008, Wang et al., 2009a, Wang et al., 2009b), but the roles of the other host proteins in the replicase complex are currently undefined. Also, the number of the identified host proteins within the tombusvirus replicase complex is likely an underestimation of the actual number of host proteins being permanent or temporally residents in the replicase complex.
Figure 3.
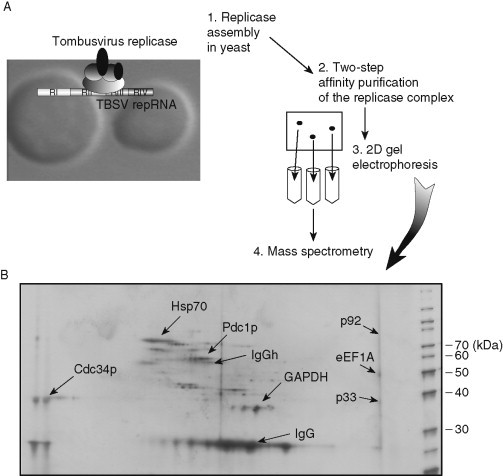
Proteomics analysis of the tombusvirus replicase complex. (A) The proteomics approach. (B) A representative silver-stained 2D gel image of the two-step affinity-purified tombusvirus replicase preparation. The identified proteins on the 2D gel are indicated. IgG and IgGh represent proteins derived from affinity purification.
B. A yeast protein microarray approach to identify host proteins interacting with the viral replication proteins
To reprogram and exploit cellular processes, tombusvirus-coded p33 and p92pol replication proteins likely interact with a currently unknown number of host proteins. The recruited host proteins could be part of the replicase complex to aid viral replication. Moreover, the interacting host proteins might facilitate the transport of viral proteins in the cells or affect the assembly of the viral replicase as well as provide regulatory functions during viral replication. To catalogue the list of host proteins interacting with the viral replication proteins, we have taken a proteome-wide approach with the yeast protein array carrying ∼4100 purified proteins that covers ∼70% of yeast proteins (Fig. 2). This has led to the identification of 57 proteins binding to p33, whereas an additional 11 host proteins bound only to the unique portion of p92, but not to p33.
Among the identified host proteins interacting with p33, there are three protein chaperones (Gim3p, Jjj1p, and Jjj3p), five proteins involved in protein ubiquitination (Cdc34p, Rsp5p, Uba1p, Ubp10p, and Ubp15p), six translation factors involved in mRNA translation (Bfr1p, Efb1p, Hbs1p, Rpl8Ap, Tif1p, and Tif11p), and 10 proteins involved in RNA processing and metabolism (Ala1p, Bud21p, Erb1, Rib2p, Sas10p, Stm1p, Trm1p, Trz1p, Tsr2p, and Urn1p). The remaining list of identified host proteins are involved in various cellular processes and the functions of nine proteins are not yet defined. The 11 host proteins bound only to p92 includes an RNA helicase (Dpb3p), a methylase (Dot1p), an aminopeptidase (Map1p), an RNA-binding protein (Npl3p), and a translation factor (eEF1A/Tef2p).
Similar to other genome-wide approaches, the use of protein arrays might lead to false positives and false negatives as well. The false negatives could be due to many factors, including (i) the use of general binding conditions, which are not optimized for individual protein–protein interactions; (ii) the absence of cofactors or membrane surfaces under the in vitro conditions; and (iii) inactive or denatured proteins on the chip. Indeed, we did not detect significant binding between the purified p33 and Ssa1p, an Hsp70, which has been shown to be part of the replicase complex (Serva and Nagy, 2006). This suggests that multiple complementary approaches are needed to identify all host proteins interacting with the replication proteins.
We used additional approaches to confirm the data from the protein array, including protein pull-down experiments with purified recombinant p33 and yeast proteins as well as the split-ubiquitin yeast two-hybrid assay (Li et al., 2008). The split-ubiquitin assay, unlike the original yeast two-hybrid system, allows the analysis of protein interactions on the cytosolic surfaces of membranes, which is the natural subcellular location of the membrane-bound p33 protein (McCartney et al., 2005, Panavas et al., 2005a).
C. A yeast protein microarray approach to identify host proteins binding to the viral RNA
Many RNA-binding host proteins likely play multiple roles during tombusvirus replication. A proteome-wide approach using the yeast protein array identified 57 host proteins bound to either TBSV or BMV RNAs. Among these host proteins, 11 proteins bound selectively to TBSV RNA, including two known helicases (DBP2 and YFR038W), a translation initiation factor (GCD2), and two RNA modifying proteins (DEG1 and UTP7). An additional 43 host proteins identified with both TBSV and BMV RNA probes are involved in a variety of cellular processes, such as translation, transcription activation, ribosomal RNA processing/binding, mRNA transport, and protein-membrane targeting with various biochemical activities—such as helicase, tRNA ligase, tRNA methyltransferase, rRNA dimethylase, ribonuclease, cochaperone, and protein kinase.
More detailed experiments with translation elongation factor eEF1A have shown that this host protein is part of the tombusvirus replicase and binds to p33/p92 replication proteins and the 3′-UTR of the TBSV (+)RNA as well. Interestingly, eEF1A has been shown to bind to the BMV RNA (Bastin and Hall, 1976). In addition, the identified pseudouridine synthase Pus4p might be involved in pseudouridinylation-based modification of TBSV RNA and BMV RNA, which has been shown to occur for the BMV RNA in vivo (Baumstark and Ahlquist, 2001). Interestingly, Pus4p has also been identified in a similar screen with a unique yeast protein array using a 3′-end cis-acting element from BMV RNA (Zhu et al., 2007), although its actual function in BMV replication is currently unknown.
To validate the above proteome-wide approach for identification of host proteins binding to the viral RNA, several recombinant yeast proteins have been shown to bind to the TBSV (+)repRNA in a gel mobility shift assay and via protein/RNA copurification approach from yeast cells (Li et al., 2009). Moreover, several of the identified host RNA-binding proteins affected TBSV repRNA replication, supporting the idea that a number of RNA-binding proteins play a role in the tombusvirus replication process.
IV. Grouping of Host Factors and Identification of Networks Involved in TBSV Replication
The above genome-wide genomics and proteomics approaches have led to the identification of 254 host proteins that either affected TBSV repRNA accumulation or bound to the viral replicase, replication proteins, or the viral RNA (Fig. 2). The large number of factors identified and the great variety of cellular function for these factors (Fig. 4 ) indicate the existence of a truly complex interaction between a simple (+)RNA virus and the yeast model host. This complexity might be due to several levels of interaction taking place between the virus and the host. For example, it is possible that many recruited host factors play a direct, well-defined function for promoting virus replication; others could inhibit TBSV accumulation, while many more host proteins might have only indirect roles in TBSV replication by affecting the general metabolisms/pathways in the cells that also influence virus replication. Many of the identified host factors could be part of protein networks and pathways that are recruited in an orchestrated way for virus replication, while other factors might be recruited and function individually. Another complication is that virus replication might utilize either the known function or an unknown function of a given host factor or even multiple functions of the same protein. Moreover, some host factors might perform completely novel functions (not performed during regular cellular processes) during virus replication. Detailed further analysis of the identified host factors based on bioinformatics in combination with biochemical, genetic, and cellular analyses will be needed to dissect the functions of the host proteins during TBSV replication. These are enormous challenges waiting for virology research to uncover the mechanism of (+)RNA virus replication in order to understand virus–host interaction.
Figure 4.
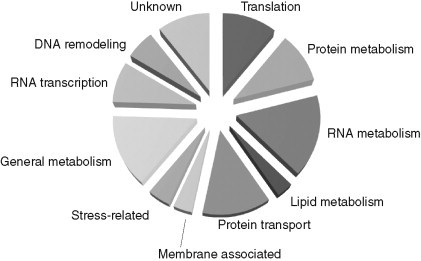
The frequency of identification of host factors representing one of the shown functional groups as described in Table I. The total number of host factors identified in the global screens is 254 (see Fig. 2).
Below, we will summarize our current grouping of host factors, view those host factors that have been characterized in more detail, and propose protein networks that could be involved in TBSV replication.
A. Translation factors and cellular proteins involved in protein biosynthesis
Translation of the viral RNA precedes viral (+)RNA replication, which uses the same RNA. Therefore, translation and replication must be coordinated to regulate temporally and spatially these processes and prevent the collision between the ribosomes and the viral replicase. Although the mechanism of the switch of the viral (+)RNA from translation to replication is currently unknown, it is likely that host factors in combination with the newly translated viral replication proteins play a role in this process. Translation factors are especially good candidates to be involved in the switch, since they are already present during translation and several of them have been identified to bind to either the replication proteins or the viral RNA (Li et al., 2008, Li et al., 2009). The large number of the identified host factors in this group, however, makes it difficult to narrow down the actual candidates. Also, these factors could play direct roles in subsequent steps in replication.
Additional experiment was performed on eEF1A to dissect its role in TBSV replication. eEF1A is a highly abundant cellular protein and its best-known function is to deliver aminoacyl-tRNA to the elongating ribosome in a GTP-dependent manner. eEF1A has many additional functions, such as quality control of newly produced proteins, ubiquitin-dependent protein degradation, and organization of the actin cytoskeleton (Chuang et al., 2005, Gross & Kinzy, 2005).
The experiments with eEF1A revealed that it is a permanent resident of the tombusvirus replicase complex (Li et al., 2009). Mutational analysis of eEF1A suggests that it might be involved in promoting (−)RNA synthesis by the replicase complex (Z. Li and P.D. Nagy, unpublished data). Another function of eEF1A is to stabilize the p33 replication protein, while it did not affect the half-life of the less abundant p92pol replication protein in yeast (Li et al., 2009). Interestingly, a prokaryotic homolog of eEF1A, called Tu translation elongation factor, plays a role in replication of bacteriophage Qβ (Blumenthal et al., 1976). In addition, eEF1A was found to bind to many viral (+)RNAs, including the 3′-UTR of Turnip yellow mosaic virus (TYMV) (Dreher, 1999), of West Nile virus (WNV), Dengue virus, Tobacco mosaic virus (TMV) and Turnip mosaic virus (De Nova-Ocampo et al., 2002, Nishikiori et al., 2006, Thivierge et al., 2008, Zeenko et al., 2002). In addition, eEF1A has also been shown to interact with the NS5A replication protein of Bovine viral diarrhea virus (BVDV) (Johnson et al., 2001), NS4A of HCV (Kou et al., 2006), the TMV replicase (Yamaji et al., 2006), and the Gag polyprotein of HIV-1 (Cimarelli and Luban, 1999). The biochemical functions provided by eEF1A for (+)RNA virus replication are currently poorly understood. It has been proposed to affect minus-strand synthesis for WNV (Davis et al., 2007), albeit it repressed minus-strand synthesis of TYMV in vitro (Dreher, 1999, Dreher et al., 1999, Matsuda et al., 2004). Overall, the interaction of eEF1A with viral RNAs and viral replication proteins and its high abundance in cells might facilitate recruitment of eEF1A into virus replication.
The identification of 28 host factors (11% of all host factors) (Fig. 4) suggests that translation factors and/or the ribosome itself might be involved in correct folding of the viral RNA or the newly made viral replication proteins. Several proteins might also be involved in modification of the viral (+)RNA, similar to tRNAs and rRNAs. These processes could affect the stability and/or subsequent localization of the viral proteins/RNA. Albeit additional translation factors/ribosomal proteins, similar to eEF1A, might be recruited for replication, it is unlikely that all 28 proteins in this group would be directly targeted for assisting replication.
B. Protein modification enzymes
Posttranslational modification serves as an important means switching protein molecules between active and inactive forms, in regulating their stability, their subcellular localization, and their interactions with other proteins, nucleic acids, or membranes. The TBSV p33 replication protein can be phosphorylated (Shapka et al., 2005, Stork et al., 2005) and ubiquitinated (Barajas et al., 2009a, Li et al., 2008) with both modifications likely playing roles in TBSV replication. Accordingly, Cka1p, Mob1p, Mps1p, Sln1p kinases and Siw14p phosphatase have been identified in the genome-wide screens (Table I ). Albeit it is yet unknown what host kinases phosphorylate p33 in cells, protein kinase C (PKC) was shown to phosphorylate purified p33 in vitro. Interestingly, the sites of p33 phosphorylation are located proximal to an essential RNA-binding domain (Shapka et al., 2005, Stork et al., 2005). Since the phosphorylated form of p33 lost its ability to bind to (+)rep RNA and in vitro phosphorylation of the p33:repRNA complex led to the release of the RNA from the complex, it has been proposed that phosphorylation of p33 and possibly p92, which carries the same p33 sequence at its N-terminus due to its overlapping ORF, might lead to the release of the viral RNA from the replicase complex (Stork et al., 2005).
Table I.
Functional grouping and roles of the identified host genes affecting TBSV RNA replication and recombinationa
| Gene | Cellular function | Viral replicationb | Viral recombination | Interaction |
|---|---|---|---|---|
| 1. Translation/protein biosynthesis | ||||
| BFR1 | mRNP complexes/polyribosomes | p33/RNA | ||
| DED1 | DEAD-box RNA helicase, translation | Required | ||
| EFB1 | Translation elongation factor 1β | p33 | ||
| ERB1 | Maturation of ribosomal RNAs | Stimulatoryc | p33, p92 | |
| GCD2 | δ-subunit of eIF2B | Stimulatoryc | RNA | |
| HBS1 | GTP binding, similarity to EF-1α | Stimulatoryc | p33 | |
| IPI3 | Rix1 complex, pre-rRNA processing | p33 | ||
| MRPL32 | Protein biosynthesis | Inhibitory | ||
| NOG1 | Putative GTPase, ribosome biogenesis | Inhibitory | ||
| NOG2 | Putative GTPase, ribosome biogenesis | Inhibitory | ||
| NOP53 | Processing of 27S pre-rRNA | Inhibitoryc | RNA | |
| RPL1B | Protein biosynthesis | Inhibitory | ||
| RPL4A | Component of the large ribosomal subunit | RNA | ||
| RPL7A | Protein biosynthesis | Inhibitory | ||
| RPL8A | Ribosomal protein L4 | p33/p92/RNA | ||
| RPL17A | Structural constituent of ribosome | Inhibitory | ||
| RPL26B | Component of the large ribosomal subunit | RNA | ||
| RPS21B | Protein biosynthesis | Required | ||
| SAS10 | Ribosomal processome | p33/p92/RNA | ||
| SSF2 | rRNA binding | RNA | ||
| STM1 | Required for optimal translation | Stimulatoryc | p33/p92/RNA | |
| TEF1d | Translational elongation factor eEF1A | Required | p33/p92/RNA/replicase | |
| TEF2d | Translational elongation factor eEF1A | Required | p33/p92/RNA/replicase | |
| TEF4 | Translation elongation factor | Required | ||
| TIF1 | Translation initiation factor eIF4A | p33 | ||
| TIF11 | Translation initiation factor eIF1A | p33/p92 | ||
| TSR2 | pre-rRNA processing | p33 | ||
| YCR016W | Ribosome biogenesis (predicted) | p33/p92/RNA | ||
| 2. Protein metabolism, posttranslation modification | ||||
| ARO1 | Aromatic amino acid synthesis | Required | ||
| BRE1 | Ubiquitin-protein ligase | Required | ||
| CDC34d | Ubiquitin-conjugating enzyme or E2 | Required, stimulatoryc | p33/replicase | |
| CKA1 | α-subunit of protein kinase CK2 | RNA | ||
| DOA4d | Protein deubiquitination | Required | ||
| EPL1 | Histone acetyltransferase activity | Required | ||
| LGE1 | Protein monoubiquitination | Required | ||
| MAK3 | Protein amino acid acetylation | Required | ||
| MAP1 | Methionine aminopeptidase | p92/RNA | ||
| MET1 | Uroporphyrin methyltransferase | Required | ||
| MOB1 | Protein amino acid phosphorylation | Required | ||
| MPS1 | Protein threonine/tyrosine kinase | Accelerator | ||
| NOB1 | Protein involved in proteasome maturation | p92/RNA | ||
| OTU2 | Predicted cysteine proteases | Inhibitoryc | p33/p92/RNA | |
| RAD6 | Ubiquitin-conjugating enzyme | Required | ||
| RPT4 | Endopeptidase | Accelerator | ||
| RSP5d | Ubiquitin-protein ligase | Inhibitory | p33/p92 | |
| SLN1 | Protein histidine kinase activity | Required | ||
| SIW14 | Protein tyrosine phosphatase | Required | ||
| UBA1 | Ubiquitin-activating enzyme | p33 | ||
| UBP3 | Ubiquitin-specific protease | Suppressor | ||
| UBP10 | Ubiquitin-specific protease | p33/p92/RNA | ||
| UBP15 | Ubiquitin-specific protease | p33 | ||
| YDR161W | ER-associated protein degradation | p33 | ||
| 3. RNA-binding proteins/RNA metabolism | ||||
| BUD21 | snoRNA binding | Inhibitory, inhibitoryc | p33/RNA | |
| CCR4 | 3′–5′ exoribonuclease | Required | ||
| CTL1 | Polynucleotide 5′-phosphatase | Suppressor | ||
| CWC25 | pre-mRNA splicing | RNA | ||
| DBP2 | RNA helicase of the DEAD-box protein family | Stimulatoryc | RNA | |
| DBP3 | Putative RNA helicase/DEAD-box family | p92 | ||
| DIM1 | Essential 18S rRNA dimethylase | RNA | ||
| DEG1 | Nonessential tRNA: pseudouridine synthase | Inhibitoryc | RNA | |
| GLO3 | GTPase activation, ER-Golgi transport | RNA | ||
| GRC3 | Possibly involved in rRNA processing | Inhibitory | ||
| HAS1 | Putative ATP-dependent RNA helicase | Inhibitoryc | RNA | |
| IRC5 | DEAD-box helicase | RNA | ||
| LHP1 | RNA-binding protein/maturation of tRNA | RNA | ||
| LRP1 | Nuclear cofactor for exosome activity | RNA | ||
| MET22/HAL2 | 3′(2′),5′-bisphosphate nucleotidase | Suppressor | ||
| MEX67 | Poly(A)RNA-binding protein | Required | ||
| MSE1 | Glutamate-tRNA ligase activity | RNA | ||
| NAB2 | Polyadenylated RNA binding; hnRNPs | Required | ||
| NOP4 | RNA binding, ribosomal RNA processing | Inhibitory | ||
| NOP10 | RNA binding, pseudouridylation, 18S rRNA | Modifier | ||
| NPL3 | mRNA binding | Required, inhibitoryc | p92/RNA | |
| NSR1d | RNA binding/rRNA processing | Required | ||
| PRP5 | RNA helicase in the DEAD-box family | Inhibitory | ||
| PRP39 | RNA binding, nuclear mRNA splicing | Required | ||
| PUS4 | Pseudouridine synthase | Inhibitoryc | RNA | |
| RIB2 | Cytoplasmic tRNA pseudouridine synthase | p33 | ||
| RNA14 | RNA binding/mRNA cleavage | Required | ||
| RNY1 | RNAse; endoribonucleases | Inhibitoryc | RNA | |
| RPL15A | Binds to 5.8S rRNA | Inhibitory | ||
| RPM2 | Ribonuclease P activity | Modifier | ||
| RRP9 | RNA binding, pre-rRNA processing | Required | Accelerator | |
| RRP42 | 3′–5′ exoribonuclease activity | Required | ||
| SEN1 | RNA helicase, processing of tRNA, rRNA | Required | Accelerator | |
| TRM1 | tRNA methyltransferase | p33/p92/RNA | ||
| TRZ1 | tRNase Z, involved in RNA processing | Stimulatoryc | p33 | |
| URN1 | Pre-mRNA splicing factor | p33/p92 | ||
| UTP7 | Small subunit (SSU) processome | Inhibitoryc | p92/RNA | |
| UTP9 | snoRNA binding, interacts with UTP15 | Required | ||
| UTP15 | snoRNA binding, interacts with UTP9 | Required | ||
| XRN1/KEM1d | 5′–3′ exoribonuclease | Required | Suppressor | |
| YBL055C | 3′–5′ exoribonuclease, endoribonuclease | RNA | ||
| YKL023W | mRNA degradation | p92/RNA | ||
| 4. Lipid metabolism | ||||
| ERG4d | δ24(24-1)-sterol reductase | Required/sterol level | ||
| ERG25d | Ergosterol biosynthesis | Required/sterol level | ||
| FAS2 | α-subunit of fatty acid synthetase | Modifier | ||
| FOX2 | Peroxisomal fatty acid β-oxidation pathway | p33/RNA | ||
| INO2 | Phospholipid biosynthesis | Required | ||
| MCT1 | S-malonyltransferase/fatty acid metabolism | Required | ||
| POX1 | Acyl-CoA oxidase/fatty acid β-oxidation | Required | ||
| TGL2 | Triacylglycerol lipase/lipid metabolism | Required | ||
| 5. Protein and vesicle-mediated transport | ||||
| APM2 | Vesicle-mediated transport | Inhibitoryc | RNA | |
| ARL3 | Small monomeric GTPase | Required | ||
| BRE5 | Vesicle-mediated transport | Required | ||
| COP1 | Protein transporter, COPI vesicle | Required | Suppressor | |
| DID2d | ESCRT/protein–vacuolar targeting | Required | ||
| GOS1 | v-SNARE activity/intra-Golgi transport | Required | ||
| MCH5 | Transporter/membrane associated | Required | ||
| MON1 | Protein–vacuolar targeting | Required | ||
| NUP53 | Subunit of the nuclear pore complex | RNA | ||
| PEP3 | Transporter/vacuolar membrane | Required | ||
| PEP7/VPS19 | Unknown/Golgi to vacuole transport | Accelerator | ||
| PEX19d | Chaperone/import to peroxisome | p33 | ||
| PTH1/VAM3 | Golgi to vacuole transport | Accelerator | ||
| RIC1 | Guanyl-nucleotide exchange factor | Required | ||
| SEC62 | SRP-dependent/protein-membrane targeting | RNA | ||
| SNF7d | ESCRT/late endosome | Required | ||
| SNL1 | Nuclear pore organization and biogenesis | RNA | ||
| SRP40 | Nucleocytoplasmic transport/chaperone | RNA | ||
| TLG2 | t-SNARE, v-SNARE/vesicle fusion | Required | ||
| TOM71 | Component of the TOM translocase | p33/p92/RNA | ||
| VPS4d | ESCRT/ATPase/late endosome | Required | ||
| VPS23/STP22d | ESCRT/protein–vacuolar targeting | Required | ||
| VPS24d | ESCRT/late endosome | Required | ||
| VPS28d | Protein–vacuolar targeting | Required | ||
| VPS29 | Retrograde/endosome to Golgi/transport | Required | Accelerator | |
| VPS35 | Endosome to Golgi transport | Accelerator | ||
| VPS41 | Rab guanyl-nucleotide exchange factor | Required | ||
| VPS43/VAM7 | Golgi to vacuole transport | Accelerator | ||
| VPS51 | Protein–vacuolar targeting | Required | ||
| VPS61 | Protein–vacuolar targeting | Required | ||
| VPS66 | Cytoplasmic protein/vacuolar protein sorting | p92/RNA | ||
| VPS69 | Protein–vacuolar targeting | Required | ||
| YOS9 | Protein transporter/ER to Golgi transport | Required | ||
| 6. Membrane associated | ||||
| KEG1 | Integral membrane protein of the ER | Inhibitoryc | RNA | |
| MSP1 | ATPase/mitochondrial translocation | Required | ||
| OPT1 | Oligopeptide transporter | Required | ||
| PMR1(HUR1)d | Ca2+/Mn2+ ion pump | Suppressor | ||
| SAC1 | Inositol/phosphatidylinositol phosphatase | Required | ||
| SNF4 | Protein kinase activator | Required | ||
| STE14 | Isoprenylcysteine methyltransferase | Required | ||
| STV1 | Hydrogen-transporting ATPase | Required | ||
| TOK1 | Potassium channel | Required | ||
| 7. Stress-related/chaperone | ||||
| DDR48 | DNA damage-response, heat-shock stress | Inhibitoryc | p33 | |
| JJJ1 | Cochaperone of Ssa1p | Stimulatoryc | p33/p92/RNA | |
| JJJ3 | Contains J-domain | p33 | ||
| GIM3 | Heterohexameric cochaperone prefoldin complex | p33 | ||
| GRE3 | Aldehyde reductase | Required | ||
| GTT1 | Glutathione transferase | Required | ||
| IRA2 | Ras GTPase activator | Required | ||
| SSA1d | HSP70 chaperone | Required | p33/p92/replicase | |
| SSA2d | HSP70 chaperone | Required | p33/p92/replicase | |
| UGA2 | Glutamate catabolism | Required | ||
| WHI2 | Phosphatase activator | Required | ||
| 8. General metabolism | ||||
| ALA1 | Cytoplasmic alanyl-tRNA synthetase | p33 | ||
| BEM4 | Rho protein signal transduction | Required | ||
| COX12 | Cytochrome c oxidase | Required | ||
| CHO2/PEM1 | Phosphatidylethanolamine N-methyltransferase | Accelerator | ||
| DCI1 | Dodecenoyl-CoA δ-isomerase | Accelerator | ||
| DSE1 | Cell wall organization and biogenesis | Required | ||
| ERR2 | Phosphopyruvate hydratase | p33 | ||
| GLO2 | Hydroxyacylglutathione hydrolase | Required | ||
| GPH1 | Glycogen phosphorylase | Required | p33 | |
| GSY2 | Glycogen synthase | p33/p92 | ||
| HAP3 | Regulation of carbohydrate metabolism | Required | ||
| HOR2 | dl-glycerol-3-phosphatase | p33 | ||
| IPK1 | Inositol/phosphatidylinositol kinase | Accelerator | ||
| ISN1 | Inosine 5′-monophosphate 5′-nucleotidase | p33 | ||
| LPD1 | Pyruvate dehydrogenase | Required | ||
| MAM33 | Mitochondrial matrix/oxidative phosphorylation | p33 | ||
| MDH3 | Cytoplasmic malate dehydrogenase | RNA | ||
| MDM38 | Mitochondrial inner membrane protein | Stimulatoryc | RNA | |
| MSB1 | Establishment of cell polarity | Required | ||
| NAP1 | Regulation of microtubule dynamics | p33 | ||
| PCS60 | Peroxisomal AMP-binding protein | RNA | ||
| PDC1 | Pyruvate decarboxylase | Replicase | ||
| PDI1 | Protein disulfide isomerase, ER lumen | p33 | ||
| PHD1 | Pseudohyphal growth | Required | ||
| PLP2 | Actin binding/similarity to phosducins | p33 | ||
| PYC1 | Pyruvate carboxylase isoform | p33 | ||
| QCR6 | Ubiquinol–cytochrome c reductase complex | p33/p92 | ||
| RIB7 | Deaminase, riboflavin biosynthesis | Modifier | ||
| RMD7 | Cell wall organization and biogenesis | Required | ||
| SHO1 | Transmembrane osmosensor | p33 | ||
| SPE3 | Spermidine synthase | Modifier | ||
| TDH2d | Glyceraldehyde-3-phosphate dehydrogenase | Required | RNA/replicase | |
| TDH3d | Glyceraldehyde-3-phosphate dehydrogenase | Required | RNA/replicase | |
| THI3 | Carboxy-lyase/thiamin biosynthesis | Required | ||
| TUM1 | Mitochondrial, similar to rhodanase | p33 | ||
| YJL218W | Acetyltransferase activity | RNA | ||
| YIL064W | S-adenosylmethionine methyltransferase | Required | ||
| 9. RNA transcription | ||||
| ARP9 | RNA polymerase, actin-related protein | Inhibitory | Accelerator | |
| CDC50 | Transcription regulator | Required | ||
| HAA1 | Transcriptional activator | Inhibitoryc | RNA | |
| MED6 | RNA polymerase II transcription mediator | Required | ||
| ELF1 | A zinc finger transcription elongation factor | p33 | ||
| IWR1 | Affects transcription by pol II | p33/p92 | ||
| NGG1 | Transcription cofactor | Accelerator | ||
| POL1 | α-DNA polymerase, synthesis of RNA primer | Suppressor | ||
| RDS2 | Zinc cluster transcription activator | p92 | ||
| RGR1 | Transcription mediator | Suppressor | ||
| RPB11d | RNA polymerase II subunit B12.5 | Required | Accelerator | |
| RPO21 | RNA polymerase | Required | ||
| ROX3 | RNA polymerase II transcription mediator | Required | ||
| SUB1 | Transcriptional coactivator | Inhibitoryc | RNA | |
| SPT3 | Transcription cofactor | Modifier | ||
| SPT16 | Pol II transcription elongation factor | p33/p92 | ||
| SRB8 | RNA polymerase II transcription mediator | Required | ||
| SWI3 | General RNA polymerase II transcription factor | Required | ||
| TEA1 | Transcription regulator | Required | ||
| TFA2 | General RNA polymerase II transcription factor | Required | ||
| UME6 | Transcription regulator | Required | ||
| 10. DNA remodeling, metabolism | ||||
| ADA2d | Chromatin modification, histone acetylation | Required | ||
| ARP8 | Nuclear actin-related, chromatin remodeling | Stimulatoryc | p33 | |
| DOT1 | Nucleosomal histone methylase | p92 | ||
| DPB4 | ε-DNA polymerase | Required | ||
| HEX3 | DNA recombination | Required | ||
| NGG1 | Chromatin modification, histone acetylation | Required | ||
| ORC6 | DNA replication | Modifier | ||
| POL30 | Proliferating cell nuclear antigen (PCNA) | Stimulatoryc | p33 | |
| RSC8 | Chromatin remodeling | Required | ||
| RTT106 | Histone chaperone/Ty transposition | p33 | ||
| SAS3 | Acetyltransferase/chromatin silencing | Required | ||
| SIN3 | Histone deacetylase | Required | ||
| SLX8 | DNA metabolism | Required | ||
| SLX9 | DNA metabolism | Inhibitory | ||
| SNF6 | Chromatin modeling/SWI/SNF complex | Required | ||
| 11. Function unknown | ||||
| BSC2 | Unknown | Required | ||
| EMI2 | Protein of unknown function | p33 | ||
| FMP40 | Protein of unknown function | p33 | ||
| LDB7 | Unknown | Required | ||
| YBR007C | Unknown | Required | ||
| YBR032W | Unknown | Required | ||
| YCR099C | Unknown | Required | ||
| YDR327W | Unknown | Inhibitory | Modifier | |
| YFL043C | Unknown | Required | ||
| YGL140C | Unknown | Required | ||
| YGL242C | Unknown | p33/p92 | ||
| YGR017W | Unknown | p33/p92 | ||
| YGR026W | Unknown | Inhibitoryc | RNA | |
| YGR027W | Retrotransposon TYA gag gene | p33 | ||
| YGR064W | Unknown | Required | ||
| YHR009C | Unknown | p33 | ||
| YHR029C | Unknown | Required | ||
| YIL090W | Unknown | Required | ||
| YJL175W | Unknown | Required | ||
| YKL033W | Cytoplasmic protein with unknown function | Modifier | ||
| YLR125W | Unknown/Ty3 transposition | p33 | ||
| YLR358C | Unknown | Required | ||
| YNL196C | Unknown/leucine zipper protein | RNA | ||
| YNL321W | Unknown | Required | ||
| YOR309C | Hypothetical protein | p92/RNA | ||
| YPR050C | Unknown | Required | ||
| YPR174C | Unknown | RNA | ||
The shown data are from Jiang et al., 2006, Li et al., 2008, Li et al., 2009, Panavas et al., 2005b, Serva & Nagy, 2006, Serviene et al., 2005, Serviene et al., 2006.
“Required” is based on more than twofold drop in TBSV replication when the host gene is deleted or its expression is downregulated.
Based on protein overexpression in yeast.
Host genes, whose roles/functions have been characterized in details in TBSV replication.
Ubiquitination also plays a role in TBSV replication. Ubiquitination of host proteins by the highly conserved 76 aa ubiquitin (Ub) regulates many cellular processes, such as protein degradation, protein trafficking, transcription, immune response, signal transduction, and autophagy. Protein ubiquitination/deubiquitination requires four types of enzymes. E1 proteins activate ubiquitin, E2s (Ub-conjugating enzymes) function in transferring Ub to the client proteins, whereas E3s are involved in substrate selection, while DUBs remove the Ub from the proteins.
The genome-wide screens with TBSV identified 11 host proteins involved in the ubiquitin-dependent pathway of protein modification/degradation. These proteins include E2 ubiquitin-conjugating enzymes (CDC34 and RAD6), ubiquitin-protein ligases (RSP5 and BRE1), an ubiquitin-activating enzyme (UBA1), and four ubiquitin-specific proteases (DOA4, UBP3, UBP10, and UBP15), while LGE1 is involved in protein monoubiquitination, and BRE5 is a ubiquitin protease cofactor (Table I) (Jiang et al., 2006, Li et al., 2008, Panavas et al., 2005b, Serviene et al., 2005, Serviene et al., 2006). Binding of p33 with ubiquitin-specific proteins suggests that p33 could be modified posttranslationally by ubiquitination as shown in vivo and in vitro (Barajas et al., 2009b, Li et al., 2008).
Among the identified proteins in the ubiquitin pathway, Cdc34p, Rsp5p, and Doa4p (described in Section E) have been characterized in more details (Table I). Cdc34p (also called Ubc3p) was found to bind to p33 and is a permanent resident of the tombusvirus replicase complex (Li et al., 2008). A purified preparation of Cdc34p ubiquitinated p33 in vitro, indicating that Cdc34p is active on the p33 substrate in the absence of an E3 enzyme. Downregulation of Cdc34p level decreased TBSV repRNA accumulation and the activity of the tombusvirus replicase by three- to fivefold. Interestingly, a Cdc34p mutant inactive in ubiquitin conjugation could not complement the reduced amount of wt Cdc34p based on the activity of the isolated tombusvirus replicase (Li et al., 2008), suggesting that the ubiquitination activity of Cdc34p is critical for TBSV replication. However, the actual function of Cdc34p within the replicase complex is not yet known.
Rsp5p E3 ubiquitin ligase has been shown to bind to p33 and p92pol replication proteins and it can ubiquitinate p33 in the presence of E1 and E2 proteins in vitro (Barajas et al., 2009b). However, unlike Cdc34p, Rsp5p inhibits TBSV replication by binding via its three WW repeats to p92pol and destabilizing p92pol and reducing the replicase activity (Fig. 5 ). Surprisingly, the HECT domain involved in protein ubiquitination is not required for the inhibitory activity of Rsp5p (Barajas et al., 2009b). Future experiments should address what is the role of Rsp5p in regulation of TBSV replication.
Figure 5.
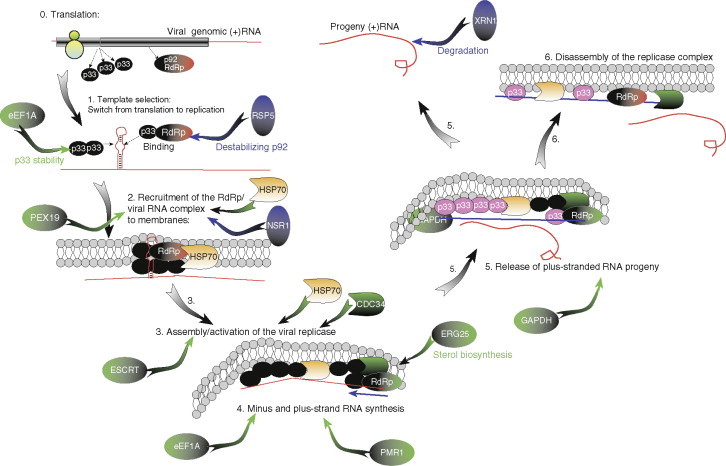
A summary model on the defined and proposed functions of all the characterized host factors during TBSV replication. TBSV replication is divided into six sequential steps and preceded by translation of the TBSV replication proteins. Host factors shown in green circles and green arrows are required, while factors shown in blue circles and blue arrows are inhibitory for TBSV replication. Note that the ESCRT protein circle represents seven of the identified ESCRT proteins. Hsp70 is colored uniquely due to the essential role of Hsp70 in several steps in TBSV replication. The active p33 proteins are represented by black circles, while the inactive (phosphorylated form) is shown with pink circles. Also, the inactive p92 protein is shown as a red circle, while the activated p92 is represented by a green circle.
Other viruses are also known to take advantage of the ubiquitination pathway (Barry & Fruh, 2006, Shackelford & Pagano, 2004, Shackelford & Pagano, 2005, Taylor & Barry, 2006), by using it to regulate protein stability via ubiquitination and deubiquitination of viral proteins (Geoffroy et al., 2006, Mechali et al., 2004, Miller et al., 2004, Nerenberg et al., 2005, Ott et al., 2000, Poon et al., 2006, Wang et al., 2006, Woo & Berk, 2007). For example, ubiquitination has been documented for replication proteins of Turnip mosaic virus, HCV, coxsackievirus, and coronaviruses (Barretto et al., 2005, Hericourt et al., 2000, Ratia et al., 2006, Sulea et al., 2005, Wong et al., 2007). Moreover, a host ubiquitin gene sequence was found inserted in the bovine viral diarrhea virus genomic RNA via RNA recombination (Baroth et al., 2000, Tautz & Thiel, 2003). In spite of intensive efforts, the current knowledge on the roles of ubiquitination in (+)RNA replication and infections is incomplete.
Altogether, the identification of many host proteins involved in the ubiquitin pathway that affected TBSV replication/recombination suggests that ubiquitination plays a critical role in TBSV replication. This was indeed demonstrated in studies with p33 and the so-called ESCRT proteins described in Section E.
C. RNA-binding proteins, RNA modification enzymes, and proteins involved in RNA metabolism
This group of proteins constitutes the largest group among the identified host proteins from our genome-wide studies. The 42 host proteins in this group represent ∼17% of all the identified proteins (Fig. 4), suggesting that RNA-binding host proteins likely play important as well as diverse roles during TBSV replication. For example, the RNA-binding host proteins might affect (i) translation of the viral (+)RNA, (ii) selection and recruitment of the viral (+)RNA template for replication, (iii) the quality or efficiency of RNA synthesis, and/or (iv) stability of the viral RNA (Ahlquist et al., 2003, Brinton, 2001, Cristea et al., 2006, Nagy, 2008, Nagy & Pogany, 2006, Shi & Lai, 2005).
Since the known cellular functions of this group of proteins are amazingly diverse as well as some functions are possibly redundant, it seems that dissecting of the functions of these proteins during TBSV replication will be of a great challenge. Nevertheless, follow-up experiments with this group of proteins provided some insight into the possible functions of two proteins in TBSV replication, namely Nsr1p and Xrn1p (Fig. 5) as summarized below.
Nsr1p, also known as nucleolin, binds to the TBSV RNA and exerts its effect on viral replication directly (Jiang et al., 2010). Nucleolin/Nsr1p is found in various cell compartments and it is especially abundant in the nucleolus. Nucleolin is a ubiquitous and abundant protein involved in multiple processes, such as ribosome biogenesis, transcription of rDNA, processing and modification of rRNA and nuclear to cytosolic transport of ribosomal protein, and ribosomal subunits by shuttling between the nucleus and the cytoplasm (Tuteja and Tuteja, 1998).
Nsr1p was discovered during screening of the yeast YKO library for TBSV replication (Panavas et al., 2005b). Nsr1p seems to be an inhibitor of TBSV replication since virus replication was boosted threefolds in the absence of NSR1. Nsr1p binds to the upstream portion of the 3′-UTR, known as RIII in DI-72 (+)RNA (Jiang et al., 2010). The binding of Nsr1p to RIII(+) is relevant since the inhibitory effect of Nsr1p on DI-72 repRNA accumulation in vivo was lost when DI-72 repRNA lacked RIII sequence. Regulated overexpression of Nsr1p revealed that Nsr1p must be present at the beginning of viral replication for efficient inhibition. Moreover, the purified recombinant Nsr1p inhibited the in vitro replication of the viral RNA in a yeast cell-free assay only when preincubated with the viral RNA before the in vitro replication assay (Jiang et al., 2010). These data suggest that Nsr1p likely inhibits an early step, such as RNA recruitment, in the replication process. We propose that Nsr1p may inhibit TBSV replication via specific binding to the viral RNA and, thus, resulting in inefficient viral RNA recruitment for replication.
Nucleolin is also involved in replication/pathogenesis of various RNA and DNA viruses. Similar to its inhibitory role in tombusvirus replication, nucleolin also inhibits replication of simian virus 40 (SV40) DNA virus by interfering with the unwinding of SV40 origin (Daniely and Borowiec, 2000). In several other cases, nucleolin stimulates viral infections by, for example, interacting with the 3′-UTR of poliovirus and stimulating an early step of virus replication in vitro (Waggoner and Sarnow, 1998). The NS1 protein of influenza A virus binds to nucleolin and colocalizes with nucleolin in the nucleolus, possibly affecting cellular events, such as shut down of host protein synthesis (Murayama et al., 2007).
Another RNA-binding host protein identified during the YKO screens is Xrn1p/Kem1p 5′–3′ exoribonuclease (Xrn4p in plants/mammals) (Panavas et al., 2005b, Serviene et al., 2005). Xrn1p inhibits tombusvirus replication and might be a component of the host innate immunity. Xrn1p is a major enzyme in the RNA degradation pathway in yeast (Johnson, 1997, Sheth & Parker, 2003). Xrn1p is involved in degradation of tombusvirus RNA, including partially degraded viral RNAs generated by endoribonucleases (Cheng et al., 2006, Cheng et al., 2007, Jaag & Nagy, 2009). In the absence of Xrn1p/Xrn4p, accumulation of tombusvirus RNA increased several fold as well as novel viral recombinant RNAs emerged rapidly in yeast and in plants. Moreover, based on a yeast cell-free TBSV replication assay, which supports authentic replication and recombination of TBSV, it has been shown that the purified recombinant Xrn1p efficiently inhibited the accumulation of recombinants and partly degraded viral RNAs. Altogether, the data from yeast and plant hosts and a cell-free assay confirmed a central role for the cytosolic 5′–3′ exoribonuclease in TBSV replication, recombination and viral RNA degradation (Cheng et al., 2006, Cheng et al., 2007, Jaag & Nagy, 2009).
D. Proteins involved in lipid/membrane biosynthesis and metabolism
Many tombusviruses replicate on the cytosolic surface of peroxisomes, where the replicase complexes form (McCartney et al., 2005, Pathak et al., 2008). Electron microscopic images of cells replicating tombusviruses have revealed extensive remodeling of membranes and indicated active lipid biosynthesis (Fig. 1) (Barajas et al., 2009a, McCartney et al., 2005, Navarro et al., 2006). Therefore, it was expected that the genome-wide screens would identify lipid biosynthesis genes affecting TBSV replication. Indeed, the systematic genome-wide screens in yeast identified a list of 14 host genes involved in lipid biosynthesis/metabolism, which affected tombusvirus replication and recombination (Fig. 4) (Jiang et al., 2006, Panavas et al., 2005b, Serviene et al., 2005, Serviene et al., 2006). The 14 identified host genes involved in lipid biosynthesis/metabolism included eight genes affecting phospholipid biosynthesis, four genes affecting fatty acid biosynthesis/metabolism, and two genes affecting ergosterol synthesis (Table I).
Further studies have been performed with Erg25p, a critical enzyme in the sterol biosynthesis pathway. Sterols are ubiquitous and essential membrane components in all eukaryotes, affecting membrane rigidity, fluidity and permeability by interacting with other lipids and proteins within the membranes (Bloch, 1983, Bloch, 1992). Sterols are important for the organization of detergent-resistant lipid rafts (Roche et al., 2008). Erg25p in yeast and the orthologous SMO1 (sterol-4α-methyl-oxidase) and SMO2 in plants perform the removal of two methyl groups at C4 position, which is critical and rate limiting during sterol synthesis (Darnet and Rahier, 2004). Indeed, sterol molecules become functional structural components of membranes only after the removal of the two methyl groups at C4. Downregulation or pharmacological inhibition of ERG25 in yeast led to four- to fivefold decreased TBSV RNA accumulation (Sharma et al., 2010). Among the functions provided by sterols during tombusvirus replication, two roles for sterols have been identified. The first function is to facilitate the assembly of the viral replicase complex based on the reduced in vitro activity of the tombusvirus replicase when isolated from yeast cells with reduced level of sterols. The second function is related to the stability of p92pol viral replication protein, which showed approximately threefold reduced half-life when expressed in yeast treated with a chemical inhibitor of ERG25. The bulky p92pol replicase protein might be exposed to cytosolic proteases in sterol-poor microenvironment. Alternatively, the structure of p92pol is different under sterol-depleted condition, leading to premature degradation of p92pol. Moreover, the subcellular localization of p92pol could be different if less than normal level of sterols was available in cells.
Replication of other viruses, such as Dengue virus, Norwalk virus, and HCV, also depends on sterols (Chang, 2009, Kapadia & Chisari, 2005, Rothwell et al., 2009, Sagan et al., 2006). For example, infection with WNV has been shown to result in redistribution of cholesterol to the sites of virus replication, possibly from the plasma membrane, and reduce antiviral responses (Mackenzie et al., 2007). The HCV replicase complex has been shown to be associated with cholesterol-rich lipid rafts (Aizaki et al., 2004). These findings are expected to promote further studies on dissecting the functional/structural roles of sterols during virus replication.
E. Cellular proteins involved in vesicle-mediated transport/intracellular protein targeting
After translation of the replication proteins from the TBSV RNA by the host ribosome, the replication proteins together with the viral RNA must be localized to the peroxisomal membranes. The replication proteins might have additional functions in the infected cells that could require targeting and transportation. Therefore, it is interesting that 33 host genes have been identified, which are involved in intracellular protein targeting and vesicle-mediated transport (Fig. 4). However, the functions of the identified host proteins during TBSV replication are currently not understood.
Follow-up experiments have been conducted with the so-called ESCRT proteins (Table I), since the YKO screen revealed the involvement of seven ESCRT proteins in TBSV replication (Panavas et al., 2005b). The large number of ESCRT proteins identified during the screens suggests that TBSV might hijack the ESCRT proteins to assist replication. For example, recruitment of ESCRT proteins for TBSV replication could facilitate the assembly of the replicase complex, including the formation of TBSV-induced spherules and vesicles in infected cells (Fig. 1) (McCartney et al., 2005). Indeed, induction of membranous spherule-like replication structures in infected cells might be common for many plus-stranded RNA viruses (Kopek et al., 2007).
ESCRT proteins are known to be involved in the endosome pathway, which is a major protein-sorting pathway in eukaryotic cells. The endosome pathway is used to downregulate plasma membrane proteins; and sort newly synthesized membrane proteins from trans-Golgi vesicles to the lysosome or the plasma membrane (Hurley & Emr, 2006, Katzmann et al., 2002, Slagsvold et al., 2006). The ESCRT proteins have a major role in sorting of cargo proteins from the endosomal limiting membrane to multivesicular bodies (MVBs) via membrane invagination and vesicle formation. Defects in the endosome/MVB pathway can cause serious diseases, including early embryonic lethality, defect in growth control, and cancer (Bowers & Stevens, 2005, Hurley & Emr, 2006, Katzmann et al., 2002, Slagsvold et al., 2006).
TBSV replication was inhibited in the absence of the following ESCRT proteins: Vps23p and Vps28p (ESCRT-I complex), Snf7p and Vps24p (ESCRT-III complex); Doa4p ubiquitin isopeptidase, Did2p having Doa4p-related function; and Vps4p AAA-type ATPase (Table I) (Panavas et al., 2005b). Intriguingly, the ubiquitinated TBSV p33 replication protein was found to interact with Vps23p ESCRT-I and Bro1p accessory ESCRT factors (Barajas et al., 2009a). The interaction has been shown to lead to the recruitment of Vps23p and possibly Bro1p to the peroxisomes, the sites of TBSV replication. This is followed by the recruitment of ESCRT-III proteins, Snf7p and Vps24p, which could help the optimal assembly of the replicase complex, facilitate the grouping of p33/p92 molecules together in the membrane and/or promote the formation of viral spherules by deforming the membrane (membrane invagination). Then, Doa4p deubiquitination enzyme is predicted to remove ubiquitin from the ubiquitinated p33, while Vps4p ATPase likely recycles the ESCRT proteins from the replicase complex at the end of the assembly (Barajas et al., 2009a).
The above model is strongly supported by experimental data, such as the reduced activity of the tombusviral replicase when derived from vps23Δ or vps24Δ yeast (Barajas et al., 2009a). Moreover, the minus-stranded viral RNA in the replicase from vps23Δ or vps24Δ yeast became more accessible to ribonuclease, suggesting that the protection of the viral RNA is compromised within the replicase complex assembled in the absence of ESCRT proteins. Thus, the role of ESCRT proteins seems to control the quality of the replicase complex assembly, making the viral RNAs within replicase complex well protected from ribonucleases. Based on these observations, we propose that ESCRT proteins help tombusviruses hide from host defense recognition and avoid the attack by the host defense machinery during viral replication. Similar role for ESCRT proteins or other host factors might help other (+)RNA viruses, which are also known to deform membranes and form spherules during replication.
The ESCRT proteins are also recruited by various viruses, such as enveloped retro-, filo-, arena-, rhabdo-, and paramyxoviruses to the plasma membrane, leading to budding and fission of the viral particles from infected cells (Morita & Sundquist, 2004, Perlman & Resh, 2006).
F. Membrane-associated cellular proteins
Since tombusvirus replication takes place on the peroxisomal and alternatively on the ER membranes, it is possible that some membrane-bound host proteins could affect TBSV replication directly or indirectly. Among the identified nine host proteins in this group that affected TBSV replication, only the role of Pmr1p has been characterized in detail. Inactivation of PMR1, which codes for the highly conserved Ca2+/Mn2+ pump in yeast, led to greatly increased level of TBSV RNA recombination as well as higher viral RNA accumulation (Jaag et al., 2010).
Pmr1p (for “plasma membrane ATPase related”) is an ATPase-driven Ca2+/Mn2+ exporter/pump in yeast (Ton and Rao, 2004). Pmr1p controls Ca2+ and Mn2+ influx to the Golgi from the cytosol, which is important for signal transduction and protein sorting in yeast.
Inactivation of PMR1 has been shown to lead to an ∼160-fold increase in TBSV RNA recombination (Jaag et al., 2010). Expression of separation-of-function mutants of Pmr1p revealed that the ability of Pmr1p to control the Mn2+ concentration in the cytosol is a key factor in viral RNA recombination. Based on the known cellular function of Pmr1p and in vitro and in vivo TBSV recombination assays, it has been proposed that the Pmr1p Ca2+/Mn2+ ion pump regulates TBSV RNA recombination by keeping the Mn2+ concentration low in the cytosol (Jaag et al., 2010). When Mn2+ concentration is low in the cell, then the RdRp within the viral replicase utilizes the far more abundant Mg2+ over Mn2+, leading to low-frequency RNA recombination. On the other hand, deletion/inhibition of the Pmr1p Ca2+/Mn2+ pump leads to an increased level of cytosolic Mn2+ (Mandal et al., 2000), promoting the more efficient use of Mn2+ by the viral RdRp, which leads to high-frequency RNA recombination. Thus, Pmr1p activity in the cell affects TBSV RNA replication and recombination through regulating the cytosolic Mn2+ level (Jaag et al., 2010). Overall, the emerging picture from the genome-wide studies and the more detailed studies on PMR1 is that complex interactions between TBSV and its host affect not only TBSV replication, but viral adaptation and evolution as well.
Interestingly, high Mn2+ also affects the activity of the reverse transcriptase (Bolton et al., 2002, Vartanian et al., 1999) and the template activity of several RNA virus RdRps, making the polymerase action less specific for templates, and stimulating nucleotide misincorporation (Alaoui-Lsmaili et al., 2000, Arnold et al., 2004, Hardy et al., 2003, Poranen et al., 2008, Yi et al., 2003). Thus, the roles of Ca2+/Mn2+ ion pumps could be general and widespread among viruses.
G. Proteins with stress-related functions
Viruses are known to induce cellular stress during the infection process. The stress stimuli then lead to the activation and high-level expression of stress-related proteins, such as chaperones, including heat-shock proteins (Brodsky & Chiosis, 2006, Mayer, 2005). The genome-wide screens identified 11 proteins in this group that affected TBSV replication (Fig. 4). Among the stress-related proteins, further studies were conducted with Ssa1/2p Hsp70.
The tombusvirus replicase complex contains Hsp70, an abundant cytosolic chaperone, which is required for TBSV replication (Pogany et al., 2008, Serva & Nagy, 2006). The interaction between Hsp70 and the tombusvirus replication proteins occurs in the functional replicase (Fig. 3), since affinity purification of Ssa1p from the solubilized membrane fraction of yeast resulted in copurification of tombusvirus replicase activity (Wang et al., 2009b). Hsp70 chaperone seems to play multiple and essential roles during TBSV replication (Fig. 5). For example, using a temperature-sensitive mutant of Hsp70 at nonpermissive temperature has led to cytosolic localization of p33 replication protein (Wang et al., 2009a). Shifting down from nonpermissive to permissive temperature resulted in relocalization of p33 to the peroxisome membrane surface in yeast. Subcellular fractionation experiments have shown that the viral replication proteins are mostly cytosolic in ssa1ssa2 yeast at the early time point (Wang et al., 2009b). Interestingly, the viral proteins become partly membrane-bound at a latter time point likely due to partial complementation by the cytosolic, stress-inducible Ssa3p and Ssa4p, which operate at higher levels in ssa1ssa2 cells (Becker et al., 1996, Werner-Washburne et al., 1987). This suggests that Hsp70 is involved in localization/transportation of the viral replication proteins. It has been proposed that the binding of p33 and p92pol replication proteins to Hsp70 results in shielding the hydrophobic transmembrane domains in the replication protein that could prevent their aggregation and promote binding to Pex19p transport protein (see below). The latter interaction is needed for peroxisomal targeting of the replication proteins (Pathak et al., 2008).
The second demonstrated function of Hsp70 is the insertion of the replication proteins into intracellular membranes (Wang et al., 2009b). Integration of p33 replication protein into subcellular membranes, such as peroxisomal and ER (Jonczyk et al., 2007), is thought to be critical for tombusvirus replication. This is because p33 mutants localized in the cytosol do not support TBSV replication in yeast or in plant cells (McCartney et al., 2005, Panavas et al., 2005a). The insertion of the replication proteins into the membrane might require additional cellular factors as shown for cellular membrane-associated proteins (Brodsky & Chiosis, 2006, Young et al., 2004).
The third function of Hsp70 is to assist the assembly of the TBSV replicase. This was demonstrated by Pogany et al. (2008) using a yeast extract depleted in Hsp70. The addition of purified recombinant Hsp70 to the cell-free assay was necessary for the assembly of the viral replicase complex and active replication of the repRNA in vitro (Pogany et al., 2008).
Taken together, the available data support a functional role for the cytosolic Hsp70 in several steps of tombusvirus replication, including subcellular localization, membrane insertion, and assembly of the viral replicase complex (Fig. 5) (Pogany et al., 2008).
The above findings with TBSV and Hsp70 might be common among other viruses as well. Accordingly, host-coded heat-shock proteins, such as Hsp70 chaperone family, the J-domain chaperones, and Hsp90, are implicated in replication of HCV, FHV, influenza, vesicular stomatitis virus, retroviruses (HIV), hepatitis B virus, and other RNA viruses (Brown et al., 2005, Castorena et al., 2007, Connor et al., 2007, Kumar & Mitra, 2005, Momose et al., 2002, Naito et al., 2007, Nakagawa et al., 2007, Okamoto et al., 2006, Qanungo et al., 2004, Sohn et al., 2006, Tomita et al., 2003, Weeks & Miller, 2008). The host chaperones have been proposed to stimulate polymerase (RdRp) activity (Momose et al., 2002), and activate the reverse transcriptase for hepadnaviruses (Hu et al., 2004, Tavis et al., 1998). The cytosolic Hsp70 proteins might also affect stability and function of viral proteins during infections since a subset of HSP70 genes are expressed at enhanced levels in plants infected by various plant viruses (Aparicio et al., 2005, Aranda et al., 1996, Whitham et al., 2003, Whitham et al., 2006).
H. Proteins involved in general metabolism of the cell
Virus replication absolutely depends on the resources provided by the host cells. Thus, many proteins involved in general cellular metabolism could indirectly affect virus replication. However, most host proteins have multiple functions and viruses might exploit alternative, less characterized functions of cellular proteins as explained below for a host metabolic enzyme co-opted for TBSV replication. Overall, surprisingly large number of host proteins has been identified (15% of all identified proteins; Fig. 4) in this group that affected TBSV replication. Future studies will be needed to dissect the direct or indirect roles of these proteins for tombusvirus replication.
The metabolic enzyme studied in more detail is called glyceraldehyde-3-phosphate dehydrogenase (GAPDH, coded by TDH1 and TDH2 genes in yeast), which was discovered as a component of the tombusvirus replicase complex via a proteomics analysis of a purified viral replicase preparation (Fig. 3) (Serva and Nagy, 2006). The cellular distribution of GAPDH changed dramatically due to relocalization from the cytosol to the site of TBSV repRNA replication (peroxisome) in yeast cells (Wang and Nagy, 2008). Downregulation of GAPDH levels in yeast correlated with reduced level of (+)-strand repRNA, suggesting an important role for this protein in TBSV replication.
GAPDH is a ubiquitous, highly conserved, very abundant protein (Sirover, 1999) with glyceraldehyde-3-phosphate dehydrogenase function, which is a key component of cytosolic energy production. However, GAPDH displays many additional activities that are unrelated to its glycolytic function. These cellular activities include roles in modulation of the cytoskeleton, vesicular secretary transport, endocytosis, nuclear membrane fusion, nuclear tRNA transport, apoptosis, DNA replication and repair, maintenance of telomere structure, and transcriptional control of histone gene expression (Sirover, 1999, Sirover, 2005). An emerging new function of GAPDH is to bind to various RNAs, such as AU-rich sequences at the 3′-terminus of mRNAs, which can lead to stabilization of the RNA in the cell (Bonafe et al., 2005).
In addition to relocalization from the cytosol to the peroxisomal membrane surface during TBSV replication in yeast, and being part of the replicase complex, GAPDH has been shown to affect viral RNA synthesis. For example, downregulation of GAPDH inhibited TBSV replication, resulting in an ∼1:1 ratio of (+) and (−)RNAs, instead of the hallmark asymmetric RNA synthesis leading to excess (+)RNA. Since GAPDH binds to an AU pentamer sequence in the TBSV (−)RNA, GAPDH has been proposed to play a role in asymmetric viral RNA synthesis by selectively retaining the TBSV (−)RNA template in the replicase complex (Fig. 5) (Wang and Nagy, 2008).
The novel functional role of GAPDH in TBSV RNA replication expands the battery of activities for this multifunctional enzyme. GAPDH is likely co-opted by RNA viruses due to its ability to bind to AU-rich sequences (Nagy & Rigby, 1995, Nagy et al., 2000). Accordingly, GAPDH has been shown to bind to AU-rich sequences present in various RNA viruses, including hepatitis A virus (HAV), HCV, and human parainfluenza virus type 3 (De et al., 1996, Dollenmaier & Weitz, 2003, Randall et al., 2007). The functional role of binding of GAPDH to the above viruses is not yet clear. It has been proposed that binding of GAPDH to the internal entry site (IRES) element in HAV could suppress cap-independent translation of HAV RNA (Yi et al., 2000). GAPDH may also be involved in the posttranscriptional regulation of hepatitis B virus gene expression (Zang et al., 1998).
I. Cellular transcription factors
It is currently not yet known if the identified 21 host factors in this group would have direct or indirect roles in TBSV replication (Fig. 4). It is possible that a given transcription factor affects mRNA levels for a set of host proteins that are involved in TBSV replication. It is also feasible that the TBSV replication proteins and/or RNA could interact with cellular transcription factors to reprogram host transcription, for example, in order to increase lipid biosynthesis that could be beneficial for TBSV. The interaction with a cellular transcription factor could also inhibit the production of the components of the innate antiviral pathways, thus reducing host responses to TBSV infections. Future experiments will be needed to answer these questions.
Experiments with Rpb11p, which is part of the pol II complex, revealed that this transcription factor affected the levels of p33 and p92pol in yeast (Jaag et al., 2007). As predicted, downregulation of Rpb11p inhibited TBSV repRNA replication and altered RNA recombination. An in vitro tombusvirus replicase assay supported that Rpb11p affects TBSV replication and recombination only indirectly, via regulating p33 and p92pol levels. A model has been proposed that the local concentration of replication proteins, described as molecular crowdedness, within the viral replicase is a factor affecting viral replication and recombination (Jaag et al., 2007).
J. Cellular proteins involved in DNA remodeling/metabolism
The functions of any of these 15 host proteins (Fig. 4) in TBSV replication are not yet known. It is possible that they might affect TBSV replication by regulating transcription of cellular genes important for TBSV replication, similar to the above transcription factors.
K. Cellular and hypothetical proteins with unknown functions
The functions of any of these 27 host proteins in this group (10% of all proteins identified during the screens; Fig. 4) in TBSV replication are not yet known. However, the number of genes in this group keeps decreasing due to advances in yeast research in general and TBSV host factors in particular. For example, the original YKO screens on TBSV replication and recombination led to the identification of HUR1 with unknown function (Panavas et al., 2005b, Serviene et al., 2005). Subsequent research on HUR1, however, revealed that its effect on TBSV replication and recombination was due to the PMR1 gene (see Section F), which overlaps with HUR1 on the yeast chromosome (Jaag et al., 2010). Overall, additional research on the cellular functions of this group of genes and bioinformatics analysis to predict their putative functions/activities will likely help understanding if these factors affect TBSV RNA replication directly or indirectly.
L. Host factors missed during the global genomics and proteomics screens
In spite of the systematic and multiple genome-wide screens to identify factors affecting TBSV replication, it is possible that there are still additional host factors not yet identified. Indeed, it has recently been shown that the host shuttle protein Pex19p, which is involved in peroxisomal membrane protein transport, plays a role in TBSV protein transportation to the site of replication (Fig. 5) (Pathak et al., 2008). Pex19p not only binds to the peroxisomal targeting signals in p33 in vitro, but also it is temporarily associated with the replicase complex. When Pex19p was mistargeted to mitochondrial membranes, the wt p33 was also colocalized to the same mitochondrial membranes (Pathak et al., 2008). However, the role of Pex19p is not essential for TBSV replication since TBSV repRNA replicated as efficiently in pex19Δ yeast defective in peroxisome biogenesis as in the wt yeast (Jonczyk et al., 2007, Panavas et al., 2005b). Indeed, confocal microscopy-based approach revealed that the wt tombusvirus p33 replication protein accumulated in the ER in pex3Δ or pex19Δ yeast lacking peroxisomes, suggesting that tombusvirus replication could occur on the surface of the ER membrane. Moreover, the activity of the isolated tombusvirus replicase from wt, pex3Δ, or pex19Δ yeasts was comparable, indicating that the assembly of the replicase was as efficient in the ER as in the authentic subcellular environment (Jonczyk et al., 2007). Overall, these data demonstrated that TBSV, relying on the wt replication proteins, could efficiently replicate on an alternative intracellular membrane. Thus, RNA viruses might have remarkable flexibility for using various host membranes for their replication.
It is currently not known how many other host proteins, similar to Pex19p, have been “missed” in the previous screens. Performing additional genome-wide screens will tell us if we are getting closer to identification of all host factors affecting TBSV replication and recombination at the cellular level.
V. Validation of Host Factors in a Plant Host and Induction of Resistance Against TBSV
Viruses are cellular pathogens that utilize abundant cellular resources for their replication. A small and simple eukaryotic cell, such as yeast, likely can provide most host factors needed for virus replication as demonstrated in case of TBSV (Nagy, 2008). Yet, it is important to demonstrate the relevance of the host factors identified in yeast in a natural plant host as well. The validation of the identified yeast factors has now been done for a growing number of host genes as discussed below. Moreover, the gained knowledge on these host factors can be used to interfere with their functions during TBSV replication, resulting in induction of resistance or development of antiviral treatments.
Knocking down the expression level of the plant NbGAPDH, the ortholog of yeast TDH1 and TDH2 genes, in Nicotiana benthamiana host led to 85–90% reduction in TBSV and Cucumber necrosis virus (CNV, another tombusvirus) genomic RNA accumulation (Wang and Nagy, 2008). Importantly, the GAPDH-silenced plants showed resistance against tombusviruses, indicating that this approach could lead to a new antiviral approach. The same strategy did not interfere with the accumulation of an unrelated plant RNA virus (i.e., TMV), which could be due to the lack of function for GAPDH in TMV replication. This is not surprising since TMV is more similar to BMV than to TBSV and it has been shown previously that TBSV is affected by a vastly different set of host factors as BMV (Kushner et al., 2003, Panavas et al., 2005b).
The second example that TBSV replication requires similar host factors in yeast and in plants is based on the cytosolic Hsp70. Knockdown and chemical inhibition experiments showed that Hsp70 is required for TBSV genomic RNA replication in plant cells and whole N. benthamiana host (Wang et al., 2009b). Interestingly, the pharmacologic inhibition of Hsp70 also inhibited the accumulation of other plant RNA viruses, suggesting that several plant viruses also depend on Hsp70 during their infection cycles (Wang et al., 2009b). Accordingly, Hsp70 was shown to be part of the CNV and TMV replicase complexes (Nishikiori et al., 2006, Serva & Nagy, 2006). Altogether, the chemical inhibition of Hsp70 functions in plant hosts could be a broad antiviral approach.
The third example is that inhibition of sterol biosynthesis in plant protoplasts or in plant leaves with chemical inhibitors or silencing of SMO1/SMO2 genes (involved in phytosterol synthesis) in N. benthamiana also resulted in reduced TBSV RNA accumulation. These data strongly support the role of sterols and host membranes in tombusvirus replication in plants as well (Sharma et al., 2010). Moreover, using chlorpromazine (CPZ) to alter membrane properties of the host cells led to strong inhibition of TBSV accumulation in plants (Sasvari et al., 2009). Interestingly, CPZ was also an effective inhibitor of other plant viruses, including TMV and Turnip crinkle virus, suggesting that CPZ has a broad range of antiviral activity.
Another example in plants is the successful inhibition of tombusvirus replication in N. benthamiana via overexpression of dominant negative mutants of ESCRT-III and Vps4p (Barajas et al., 2010). This inhibition by the dominant negative ESCRT mutants seems to be specific for tombusviruses, since the distantly related Tobacco rattle virus (TRV) RNA accumulation was not inhibited in these plants. The inhibitory effect on tombusvirus replication by the overexpressed dominant negative ESCRT mutants seems to be direct, since the activity of the tombusvirus replicase was also reduced when isolated from these plants (Barajas et al., 2010).
It seems that similar to replication factors, plant recombination factors, such as the 5′–3′ exoribonuclease and the Ca2+/Mn2+ pump, affect TBSV RNA recombination. For example, knockdown of XRN4 (Jaag and Nagy, 2009), the ortholog of the yeast XRN1, or cosilencing of LCA1 and ECA3 Ca2+/Mn2+ pumps (Jaag et al., 2010), orthologs of the yeast PMR1, in N. benthamiana plants resulted in enhanced TBSV recombination and replication, similar to the picture obtained in yeast with knockout mutants.
In conclusion, the above examples provide strong evidence that host factors identified and characterized in the yeast model host are also relevant for TBSV replication in a native plant host infected with the full-length wt TBSV RNA. The discussed examples also showed convincingly that inhibition of host factors could lead to the development of specific or broad-range resistance or other antiviral strategies against TBSV and possibly other plant viruses as well.
VI. Summary and Outlook
Multiple genome-wide screens with TBSV and intensive research on individual host genes using yeast as a model host led to the identification of over 250 host genes. The roles of several of these host genes in TBSV replication and recombination have been validated. However, one of the surprising observations is that the genome-wide screens led to the identification of mostly unique host factors that were not identified in other genome-wide screens. It is possible that many genes involved in TBSV repRNA replication and recombination are functionally redundant. Indeed, the host proteins that were identified in the highly purified functional tombusvirus replicase complex (such as Hsp70, GAPDH, and eEF1A) are coded by two or more genes in the yeast genome. This functional redundancy in host genes justifies the need for additional genome- or proteome-wide approaches in the identification of host genes affecting TBSV replication and recombination. Characterization of the functions of all host factors in the infected plant cells will likely result in better, more efficient antiviral strategies and/or reduce the damage to the plants caused by viral infections.
Acknowledgments
We are grateful to Dr. Daniel Barajas for the unpublished EM images in Fig. 1. This work was supported by NSF (MCB0078152), NIH, and by the University of Kentucky to PDN.
References
- Ahlquist P. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat. Rev. Microbiol. 2006;4:371–382. doi: 10.1038/nrmicro1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlquist P., Noueiry A.O., Lee W.M., Kushner D.B., Dye B.T. Host factors in positive-strand RNA virus genome replication. J. Virol. 2003;77:8181–8186. doi: 10.1128/JVI.77.15.8181-8186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizaki H., Lee K.J., Sung V.M., Ishiko H., Lai M.M. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology. 2004;324:450–461. doi: 10.1016/j.virol.2004.03.034. [DOI] [PubMed] [Google Scholar]
- Alaoui-Lsmaili M.H., Hamel M., L'Heureux L., Nicolas O., Bilimoria D., Labonte P., Mounir S., Rando R.F. The hepatitis C virus NS5B RNA-dependent RNA polymerase activity and susceptibility to inhibitors is modulated by metal cations. J. Hum. Virol. 2000;3:306–316. [PubMed] [Google Scholar]
- Aparicio F., Thomas C.L., Lederer C., Niu Y., Wang D., Maule A.J. Virus induction of heat shock protein 70 reflects a general response to protein accumulation in the plant cytosol. Plant Physiol. 2005;138:529–536. doi: 10.1104/pp.104.058958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda M.A., Escaler M., Wang D., Maule A.J. Induction of HSP70 and polyubiquitin expression associated with plant virus replication. Proc. Natl. Acad. Sci. USA. 1996;93:15289–15293. doi: 10.1073/pnas.93.26.15289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold J.J., Gohara D.W., Cameron C.E. Poliovirus RNA-dependent RNA polymerase (3Dpol): Pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mn2+ Biochemistry. 2004;43:5138–5148. doi: 10.1021/bi035213q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barajas D., Jiang Y., Nagy P.D. A unique role for the host ESCRT proteins in replication of Tomato bushy stunt virus. PLoS Pathog. 2009;5:e1000705. doi: 10.1371/journal.ppat.1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barajas D., Li Z., Nagy P.D. The Nedd4-type Rsp5p ubiquitin ligase inhibits tombusvirus replication by regulating degradation of the p92 replication protein and decreasing the activity of the tombusvirus replicase. J. Virol. 2009;83:11751–11764. doi: 10.1128/JVI.00789-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroth M., Orlich M., Thiel H.J., Becher P. Insertion of cellular NEDD8 coding sequences in a pestivirus. Virology. 2000;278:456–466. doi: 10.1006/viro.2000.0644. [DOI] [PubMed] [Google Scholar]
- Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry M., Fruh K. Viral modulators of cullin RING ubiquitin ligases: Culling the host defense. Sci. STKE. 2006;2006:pe21. doi: 10.1126/stke.3352006pe21. [DOI] [PubMed] [Google Scholar]
- Bastin M., Hall T.C. Interaction of elongation factor 1 with aminoacylated Brome mosaic virus and tRNA's. J. Virol. 1976;20:117–122. doi: 10.1128/jvi.20.1.117-122.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumstark T., Ahlquist P. The Brome mosaic virus RNA3 intergenic replication enhancer folds to mimic a tRNA TpsiC-stem loop and is modified in vivo. RNA. 2001;7:1652–1670. [PMC free article] [PubMed] [Google Scholar]
- Becker J., Walter W., Yan W., Craig E.A. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol. Cell. Biol. 1996;16:4378–4386. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham C.J., Light H.R., Nissan T.A., Ahlquist P., Parker R., Noueiry A. Interactions between Brome mosaic virus RNAs and cytoplasmic processing bodies. J. Virol. 2007;81:9759–9768. doi: 10.1128/JVI.00844-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch K.E. Sterol structure and membrane function. CRC Crit. Rev. Biochem. 1983;14:47–92. doi: 10.3109/10409238309102790. [DOI] [PubMed] [Google Scholar]
- Bloch K. Sterol molecule: Structure, biosynthesis, and function. Steroids. 1992;57:378–383. doi: 10.1016/0039-128x(92)90081-j. [DOI] [PubMed] [Google Scholar]
- Blumenthal T., Young R.A., Brown S. Function and structure in phage Qbeta RNA replicase. Association of EF-Tu-Ts with the other enzyme subunits. J. Biol. Chem. 1976;251:2740–2743. [PubMed] [Google Scholar]
- Bolton E.C., Mildvan A.S., Boeke J.D. Inhibition of reverse transcription in vivo by elevated manganese ion concentration. Mol. Cell. 2002;9:879–889. doi: 10.1016/s1097-2765(02)00495-1. [DOI] [PubMed] [Google Scholar]
- Bonafe N., Gilmore-Hebert M., Folk N.L., Azodi M., Zhou Y., Chambers S.K. Glyceraldehyde-3-phosphate dehydrogenase binds to the AU-rich 3′ untranslated region of colony-stimulating factor-1 (CSF-1) messenger RNA in human ovarian cancer cells: Possible role in CSF-1 posttranscriptional regulation and tumor phenotype. Cancer Res. 2005;65:3762–3771. doi: 10.1158/0008-5472.CAN-04-3954. [DOI] [PubMed] [Google Scholar]
- Bowers K., Stevens T.H. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2005;1744:438–454. doi: 10.1016/j.bbamcr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Brinton M.A. Host factors involved in West Nile virus replication. Ann. NY Acad. Sci. 2001;951:207–219. doi: 10.1111/j.1749-6632.2001.tb02698.x. [DOI] [PubMed] [Google Scholar]
- Brodsky J.L., Chiosis G. Hsp70 molecular chaperones: Emerging roles in human disease and identification of small molecule modulators. Curr. Top. Med. Chem. 2006;6:1215–1225. doi: 10.2174/156802606777811997. [DOI] [PubMed] [Google Scholar]
- Brown G., Rixon H.W., Steel J., McDonald T.P., Pitt A.R., Graham S., Sugrue R.J. Evidence for an association between heat shock protein 70 and the respiratory syncytial virus polymerase complex within lipid-raft membranes during virus infection. Virology. 2005;338:69–80. doi: 10.1016/j.virol.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Castorena K.M., Weeks S.A., Stapleford K.A., Cadwallader A.M., Miller D.J. A functional heat shock protein 90 chaperone is essential for efficient flock house virus RNA polymerase synthesis in Drosophila cells. J. Virol. 2007;81:8412–8420. doi: 10.1128/JVI.00189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K.O. Role of cholesterol pathways in norovirus replication. J. Virol. 2009;83:8587–8595. doi: 10.1128/JVI.00005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.P., Serviene E., Nagy P.D. Suppression of viral RNA recombination by a host exoribonuclease. J. Virol. 2006;80:2631–2640. doi: 10.1128/JVI.80.6.2631-2640.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.P., Jaag H.M., Jonczyk M., Serviene E., Nagy P.D. Expression of the Arabidopsis Xrn4p 5′–3′ exoribonuclease facilitates degradation of tombusvirus RNA and promotes rapid emergence of viral variants in plants. Virology. 2007;368:238–248. doi: 10.1016/j.virol.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Cherry S., Doukas T., Armknecht S., Whelan S., Wang H., Sarnow P., Perrimon N. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev. 2005;19:445–452. doi: 10.1101/gad.1267905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang S.M., Chen L., Lambertson D., Anand M., Kinzy T.G., Madura K. Proteasome-mediated degradation of cotranslationally damaged proteins involves translation elongation factor 1A. Mol. Cell. Biol. 2005;25:403–413. doi: 10.1128/MCB.25.1.403-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimarelli A., Luban J. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 1999;73:5388–5401. doi: 10.1128/jvi.73.7.5388-5401.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor J.H., McKenzie M.O., Parks G.D., Lyles D.S. Antiviral activity and RNA polymerase degradation following Hsp90 inhibition in a range of negative strand viruses. Virology. 2007;362:109–119. doi: 10.1016/j.virol.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristea I.M., Carroll J.W., Rout M.P., Rice C.M., Chait B.T., MacDonald M.R. Tracking and elucidating alphavirus–host protein interactions. J. Biol. Chem. 2006;281:30269–30278. doi: 10.1074/jbc.M603980200. [DOI] [PubMed] [Google Scholar]
- Daniely Y., Borowiec J.A. Formation of a complex between nucleolin and replication protein A after cell stress prevents initiation of DNA replication. J. Cell Biol. 2000;149:799–810. doi: 10.1083/jcb.149.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnet S., Rahier A. Plant sterol biosynthesis: Identification of two distinct families of sterol 4alpha-methyl oxidases. Biochem. J. 2004;378(Pt. 3):889–898. doi: 10.1042/BJ20031572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis W.G., Blackwell J.L., Shi P.Y., Brinton M.A. Interaction between the cellular protein eEF1A and the 3′-terminal stem-loop of West Nile virus genomic RNA facilitates viral minus-strand RNA synthesis. J. Virol. 2007;81:10172–10187. doi: 10.1128/JVI.00531-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De B.P., Gupta S., Zhao H., Drazba J.A., Banerjee A.K. Specific interaction in vitro and in vivo of glyceraldehyde-3-phosphate dehydrogenase and LA protein with cis-acting RNAs of human parainfluenza virus type 3. J. Biol. Chem. 1996;271:24728–24735. doi: 10.1074/jbc.271.40.24728. [DOI] [PubMed] [Google Scholar]
- De Nova-Ocampo M., Villegas-Sepulveda N., del Angel R.M. Translation elongation factor-1alpha, La, and PTB interact with the 3′ untranslated region of dengue 4 virus RNA. Virology. 2002;295:337–347. doi: 10.1006/viro.2002.1407. [DOI] [PubMed] [Google Scholar]
- Dollenmaier G., Weitz M. Interaction of glyceraldehyde-3-phosphate dehydrogenase with secondary and tertiary RNA structural elements of the hepatitis A virus 3′ translated and non-translated regions. J. Gen. Virol. 2003;84(Pt. 2):403–414. doi: 10.1099/vir.0.18501-0. [DOI] [PubMed] [Google Scholar]
- Dreher T.W. Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol. 1999;37:151–174. doi: 10.1146/annurev.phyto.37.1.151. [DOI] [PubMed] [Google Scholar]
- Dreher T.W., Uhlenbeck O.C., Browning K.S. Quantitative assessment of EF-1alpha.GTP binding to aminoacyl-tRNAs, aminoacyl-viral RNA, and tRNA shows close correspondence to the RNA binding properties of EF-Tu. J. Biol. Chem. 1999;274:666–672. doi: 10.1074/jbc.274.2.666. [DOI] [PubMed] [Google Scholar]
- Geoffroy M.C., Chadeuf G., Orr A., Salvetti A., Everett R.D. Impact of the interaction between herpes simplex virus type 1 regulatory protein ICP0 and ubiquitin-specific protease USP7 on activation of adeno-associated virus type 2 rep gene expression. J. Virol. 2006;80:3650–3654. doi: 10.1128/JVI.80.7.3650-3654.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S.R., Kinzy T.G. Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nat. Struct. Mol. Biol. 2005;12:772–778. doi: 10.1038/nsmb979. [DOI] [PubMed] [Google Scholar]
- Hao L., Sakurai A., Watanabe T., Sorensen E., Nidom C.A., Newton M.A., Ahlquist P., Kawaoka Y. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature. 2008;454:890–893. doi: 10.1038/nature07151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy R.W., Marcotrigiano J., Blight K.J., Majors J.E., Rice C.M. Hepatitis C virus RNA synthesis in a cell-free system isolated from replicon-containing hepatoma cells. J. Virol. 2003;77:2029–2037. doi: 10.1128/JVI.77.3.2029-2037.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hericourt F., Blanc S., Redeker V., Jupin I. Evidence for phosphorylation and ubiquitinylation of the Turnip yellow mosaic virus RNA-dependent RNA polymerase domain expressed in a baculovirus-insect cell system. Biochem. J. 2000;349(Pt. 2):417–425. doi: 10.1042/0264-6021:3490417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Flores D., Toft D., Wang X., Nguyen D. Requirement of heat shock protein 90 for human hepatitis B virus reverse transcriptase function. J. Virol. 2004;78:13122–13131. doi: 10.1128/JVI.78.23.13122-13131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J.H., Emr S.D. The ESCRT complexes: Structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaag H.M., Nagy P.D. Silencing of Nicotiana benthamiana Xrn4p exoribonuclease promotes tombusvirus RNA accumulation and recombination. Virology. 2009;386:344–352. doi: 10.1016/j.virol.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Jaag H.H.M., Pogany J., Nahy P.D. A host Ca2+/Mn2+ ion pump is a factor in the emergence of viral RNA recombinants. Cell Host Microbe. 2010;7:74–81. doi: 10.1016/j.chom.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Jaag H.M., Stork J., Nagy P.D. Host transcription factor Rpb11p affects tombusvirus replication and recombination via regulating the accumulation of viral replication proteins. Virology. 2007;368:388–404. doi: 10.1016/j.virol.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Serviene E., Gal J., Panavas T., Nagy P.D. Identification of essential host factors affecting tombusvirus RNA replication based on the yeast Tet promoters Hughes Collection. J. Virol. 2006;80:7394–7404. doi: 10.1128/JVI.02686-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Li Z., Nagy P.D. Nucleolin/Nsr1p binds to the 3′ noncoding region of the tombusvirus RNA and inhibits replication. Virology. 2010;396:10–20. doi: 10.1016/j.virol.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A.W. Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 1997;17:6122–6130. doi: 10.1128/mcb.17.10.6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C.M., Perez D.R., French R., Merrick W.C., Donis R.O. The NS5A protein of bovine viral diarrhoea virus interacts with the alpha subunit of translation elongation factor-1. J. Gen. Virol. 2001;82(Pt. 12):2935–2943. doi: 10.1099/0022-1317-82-12-2935. [DOI] [PubMed] [Google Scholar]
- Jonczyk M., Pathak K.B., Sharma M., Nagy P.D. Exploiting alternative subcellular location for replication: Tombusvirus replication switches to the endoplasmic reticulum in the absence of peroxisomes. Virology. 2007;362:320–330. doi: 10.1016/j.virol.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Kao C.C., Singh P., Ecker D.J. De novo initiation of viral RNA-dependent RNA synthesis. Virology. 2001;287:251–260. doi: 10.1006/viro.2001.1039. [DOI] [PubMed] [Google Scholar]
- Kapadia S.B., Chisari F.V. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc. Natl. Acad. Sci. USA. 2005;102:2561–2566. doi: 10.1073/pnas.0409834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann D.J., Odorizzi G., Emr S.D. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- Kok K.H., Lei T., Jin D.Y. siRNA and shRNA screens advance key understanding of host factors required for HIV-1 replication. Retrovirology. 2009;6:78. doi: 10.1186/1742-4690-6-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopek B.G., Perkins G., Miller D.J., Ellisman M.H., Ahlquist P. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 2007;5:e220. doi: 10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou Y.H., Chou S.M., Wang Y.M., Chang Y.T., Huang S.Y., Jung M.Y., Huang Y.H., Chen M.R., Chang M.F., Chang S.C. Hepatitis C virus NS4A inhibits cap-dependent and the viral IRES-mediated translation through interacting with eukaryotic elongation factor 1A. J. Biomed. Sci. 2006;13:861–874. doi: 10.1007/s11373-006-9104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan M.N., Ng A., Sukumaran B., Gilfoy F.D., Uchil P.D., Sultana H., Brass A.L., Adametz R., Tsui M., Qian F., Montgomery R.R., Lev S. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Mitra D. Heat shock protein 40 is necessary for human immunodeficiency virus-1 Nef-mediated enhancement of viral gene expression and replication. J. Biol. Chem. 2005;280:40041–40050. doi: 10.1074/jbc.M508904200. [DOI] [PubMed] [Google Scholar]
- Kushner D.B., Lindenbach B.D., Grdzelishvili V.Z., Noueiry A.O., Paul S.M., Ahlquist P. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc. Natl. Acad. Sci. USA. 2003;100:15764–15769. doi: 10.1073/pnas.2536857100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Barajas D., Panavas T., Herbst D.A., Nagy P.D. Cdc34p ubiquitin-conjugating enzyme is a component of the tombusvirus replicase complex and ubiquitinates p33 replication protein. J. Virol. 2008;82:6911–6926. doi: 10.1128/JVI.00702-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Pogany J., Panavas T., Xu K., Esposito A.M., Kinzy T.G., Nagy P.D. Translation elongation factor 1A is a component of the tombusvirus replicase complex and affects the stability of the p33 replication co-factor. Virology. 2009;385:245–260. doi: 10.1016/j.virol.2008.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie J.M., Khromykh A.A., Parton R.G. Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host Microbe. 2007;2:229–239. doi: 10.1016/j.chom.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Mandal D., Woolf T.B., Rao R. Manganese selectivity of pmr1, the yeast secretory pathway ion pump, is defined by residue gln783 in transmembrane segment 6. Residue Asp778 is essential for cation transport. J. Biol. Chem. 2000;275:23933–23938. doi: 10.1074/jbc.M002619200. [DOI] [PubMed] [Google Scholar]
- Matsuda D., Yoshinari S., Dreher T.W. eEF1A binding to aminoacylated viral RNA represses minus strand synthesis by TYMV RNA-dependent RNA polymerase. Virology. 2004;321:47–56. doi: 10.1016/j.virol.2003.10.028. [DOI] [PubMed] [Google Scholar]
- Mayer M.P. Recruitment of Hsp70 chaperones: A crucial part of viral survival strategies. Rev. Physiol. Biochem. Pharmacol. 2005;153:1–46. doi: 10.1007/s10254-004-0025-5. [DOI] [PubMed] [Google Scholar]
- McCartney A.W., Greenwood J.S., Fabian M.R., White K.A., Mullen R.T. Localization of the Tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell. 2005;17:3513–3531. doi: 10.1105/tpc.105.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechali F., Hsu C.Y., Castro A., Lorca T., Bonne-Andrea C. Bovine papillomavirus replicative helicase E1 is a target of the ubiquitin ligase APC. J. Virol. 2004;78:2615–2619. doi: 10.1128/JVI.78.5.2615-2619.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C.L., Parker J.S., Dinoso J.B., Piggott C.D., Perron M.J., Nibert M.L. Increased ubiquitination and other covariant phenotypes attributed to a strain- and temperature-dependent defect of reovirus core protein mu2. J. Virol. 2004;78:10291–10302. doi: 10.1128/JVI.78.19.10291-10302.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mnaimneh S., Davierwala A.P., Haynes J., Moffat J., Peng W.T., Zhang W., Yang X., Pootoolal J., Chua G., Lopez A., Trochesset M., Morse D. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Momose F., Naito T., Yano K., Sugimoto S., Morikawa Y., Nagata K. Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. J. Biol. Chem. 2002;277:45306–45314. doi: 10.1074/jbc.M206822200. [DOI] [PubMed] [Google Scholar]
- Monkewich S., Lin H.X., Fabian M.R., Xu W., Na H., Ray D., Chernysheva O.A., Nagy P.D., White K.A. The p92 polymerase coding region contains an internal RNA element required at an early step in tombusvirus genome replication. J. Virol. 2005;79:4848–4858. doi: 10.1128/JVI.79.8.4848-4858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E., Sundquist W.I. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- Murayama R., Harada Y., Shibata T., Kuroda K., Hayakawa S., Shimizu K., Tanaka T. Influenza A virus non-structural protein 1 (NS1) interacts with cellular multifunctional protein nucleolin during infection. Biochem. Biophys. Res. Commun. 2007;362:880–885. doi: 10.1016/j.bbrc.2007.08.091. [DOI] [PubMed] [Google Scholar]
- Nagy P.D. Yeast as a model host to explore plant virus–host interactions. Annu. Rev. Phytopathol. 2008;46:217–242. doi: 10.1146/annurev.phyto.121407.093958. [DOI] [PubMed] [Google Scholar]
- Nagy P.D., Pogany J. Yeast as a model host to dissect functions of viral and host factors in tombusvirus replication. Virology. 2006;344:211–220. doi: 10.1016/j.virol.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Nagy P.D., Pogany J. Multiple roles of viral replication proteins in plant RNA virus replication. Methods Mol. Biol. 2008;451:55–68. doi: 10.1007/978-1-59745-102-4_4. [DOI] [PubMed] [Google Scholar]
- Nagy E., Rigby W.F. Glyceraldehyde-3-phosphate dehydrogenase selectively binds AU-rich RNA in the NAD(+)-binding region (Rossmann fold) J. Biol. Chem. 1995;270:2755–2763. doi: 10.1074/jbc.270.6.2755. [DOI] [PubMed] [Google Scholar]
- Nagy E., Henics T., Eckert M., Miseta A., Lightowlers R.N., Kellermayer M. Identification of the NAD(+)-binding fold of glyceraldehyde-3-phosphate dehydrogenase as a novel RNA-binding domain. Biochem. Biophys. Res. Commun. 2000;275:253–260. doi: 10.1006/bbrc.2000.3246. [DOI] [PubMed] [Google Scholar]
- Naito T., Momose F., Kawaguchi A., Nagata K. Involvement of Hsp90 in assembly and nuclear import of influenza virus RNA polymerase subunits. J. Virol. 2007;81:1339–1349. doi: 10.1128/JVI.01917-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S., Umehara T., Matsuda C., Kuge S., Sudoh M., Kohara M. Hsp90 inhibitors suppress HCV replication in replicon cells and humanized liver mice. Biochem. Biophys. Res. Commun. 2007;353:882–888. doi: 10.1016/j.bbrc.2006.12.117. [DOI] [PubMed] [Google Scholar]
- Navarro B., Russo M., Pantaleo V., Rubino L. Cytological analysis of Saccharomyces cerevisiae cells supporting cymbidium ringspot virus defective interfering RNA replication. J. Gen. Virol. 2006;87(Pt. 3):705–714. doi: 10.1099/vir.0.81325-0. [DOI] [PubMed] [Google Scholar]
- Nerenberg B.T., Taylor J., Bartee E., Gouveia K., Barry M., Fruh K. The poxviral RING protein p28 is a ubiquitin ligase that targets ubiquitin to viral replication factories. J. Virol. 2005;79:597–601. doi: 10.1128/JVI.79.1.597-601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikiori M., Dohi K., Mori M., Meshi T., Naito S., Ishikawa M. Membrane-bound Tomato mosaic virus replication proteins participate in RNA synthesis and are associated with host proteins in a pattern distinct from those that are not membrane bound. J. Virol. 2006;80:8459–8468. doi: 10.1128/JVI.00545-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto T., Nishimura Y., Ichimura T., Suzuki K., Miyamura T., Suzuki T., Moriishi K., Matsuura Y. Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J. 2006;25:5015–5025. doi: 10.1038/sj.emboj.7601367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott D.E., Coren L.V., Chertova E.N., Gagliardi T.D., Schubert U. Ubiquitination of HIV-1 and MuLV Gag. Virology. 2000;278:111–121. doi: 10.1006/viro.2000.0648. [DOI] [PubMed] [Google Scholar]
- Panavas T., Nagy P.D. Yeast as a model host to study replication and recombination of defective interfering RNA of Tomato bushy stunt virus. Virology. 2003;314:315–325. doi: 10.1016/s0042-6822(03)00436-7. [DOI] [PubMed] [Google Scholar]
- Panavas T., Hawkins C.M., Panaviene Z., Nagy P.D. The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of Cucumber necrosis tombusvirus. Virology. 2005;338:81–95. doi: 10.1016/j.virol.2005.04.025. [DOI] [PubMed] [Google Scholar]
- Panavas T., Serviene E., Brasher J., Nagy P.D. Yeast genome-wide screen reveals dissimilar sets of host genes affecting replication of RNA viruses. Proc. Natl. Acad. Sci. USA. 2005;102:7326–7331. doi: 10.1073/pnas.0502604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaviene Z., Panavas T., Serva S., Nagy P.D. Purification of the Cucumber necrosis virus replicase from yeast cells: Role of coexpressed viral RNA in stimulation of replicase activity. J. Virol. 2004;78:8254–8263. doi: 10.1128/JVI.78.15.8254-8263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaviene Z., Panavas T., Nagy P.D. Role of an internal and two 3′-terminal RNA elements in assembly of tombusvirus replicase. J. Virol. 2005;79:10608–10618. doi: 10.1128/JVI.79.16.10608-10618.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak K.B., Sasvari Z., Nagy P.D. The host Pex19p plays a role in peroxisomal localization of tombusvirus replication proteins. Virology. 2008;379:294–305. doi: 10.1016/j.virol.2008.06.044. [DOI] [PubMed] [Google Scholar]
- Perlman M., Resh M.D. Identification of an intracellular trafficking and assembly pathway for HIV-1 gag. Traffic. 2006;7:731–745. doi: 10.1111/j.1398-9219.2006.00428.x. [DOI] [PubMed] [Google Scholar]
- Pogany J., White K.A., Nagy P.D. Specific binding of tombusvirus replication protein p33 to an internal replication element in the viral RNA is essential for replication. J. Virol. 2005;79:4859–4869. doi: 10.1128/JVI.79.8.4859-4869.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogany J., Stork J., Li Z., Nagy P.D. In vitro assembly of the Tomato bushy stunt virus replicase requires the host heat shock protein 70. Proc. Natl. Acad. Sci. USA. 2008;105:19956–19961. doi: 10.1073/pnas.0810851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon A.P., Gu H., Roizman B. ICP0 and the US3 protein kinase of herpes simplex virus 1 independently block histone deacetylation to enable gene expression. Proc. Natl. Acad. Sci. USA. 2006;103:9993–9998. doi: 10.1073/pnas.0604142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poranen M.M., Salgado P.S., Koivunen M.R., Wright S., Bamford D.H., Stuart D.I., Grimes J.M. Structural explanation for the role of Mn2+ in the activity of phi6 RNA-dependent RNA polymerase. Nucleic Acids Res. 2008;36:6633–6644. doi: 10.1093/nar/gkn632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qanungo K.R., Shaji D., Mathur M., Banerjee A.K. Two RNA polymerase complexes from vesicular stomatitis virus-infected cells that carry out transcription and replication of genome RNA. Proc. Natl. Acad. Sci. USA. 2004;101:5952–5957. doi: 10.1073/pnas.0401449101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadt R., Ishikawa M., Janda M., Ahlquist P. Formation of Brome mosaic virus RNA-dependent RNA polymerase in yeast requires coexpression of viral proteins and viral RNA. Proc. Natl. Acad. Sci. USA. 1995;92:4892–4896. doi: 10.1073/pnas.92.11.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall G., Panis M., Cooper J.D., Tellinghuisen T.L., Sukhodolets K.E., Pfeffer S., Landthaler M., Landgraf P., Kan S., Lindenbach B.D., Chien M., Weir D.B. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl. Acad. Sci. USA. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratia K., Saikatendu K.S., Santarsiero B.D., Barretto N., Baker S.C., Stevens R.C., Mesecar A.D. Severe acute respiratory syndrome coronavirus papain-like protease: Structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. USA. 2006;103:5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche Y., Gerbeau-Pissot P., Buhot B., Thomas D., Bonneau L., Gresti J., Mongrand S., Perrier-Cornet J.M., Simon-Plas F. Depletion of phytosterols from the plant plasma membrane provides evidence for disruption of lipid rafts. FASEB J. 2008;22:3980–3991. doi: 10.1096/fj.08-111070. [DOI] [PubMed] [Google Scholar]
- Rothwell C., Lebreton A., Young Ng C., Lim J.Y., Liu W., Vasudevan S., Labow M., Gu F., Gaither L.A. Cholesterol biosynthesis modulation regulates dengue viral replication. Virology. 2009;389(1–2):8–19. doi: 10.1016/j.virol.2009.03.025. [DOI] [PubMed] [Google Scholar]
- Sagan S.M., Rouleau Y., Leggiadro C., Supekova L., Schultz P.G., Su A.I., Pezacki J.P. The influence of cholesterol and lipid metabolism on host cell structure and hepatitis C virus replication. Biochem. Cell Biol. 2006;84:67–79. doi: 10.1139/o05-149. [DOI] [PubMed] [Google Scholar]
- Salonen A., Ahola T., Kaariainen L. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 2005;285:139–173. doi: 10.1007/3-540-26764-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasvari Z., Bach S., Blondel M., Nagy P.D. Inhibition of RNA recruitment and replication of an RNA virus by acridine derivatives with known anti-prion activities. PLoS One. 2009;4:e7376. doi: 10.1371/journal.pone.0007376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M., Chen J., Janda M., Sullivan M., den Boon J., Ahlquist P. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell. 2002;9:505–514. doi: 10.1016/s1097-2765(02)00474-4. [DOI] [PubMed] [Google Scholar]
- Serva S., Nagy P.D. Proteomics analysis of the tombusvirus replicase: Hsp70 molecular chaperone is associated with the replicase and enhances viral RNA replication. J. Virol. 2006;80:2162–2169. doi: 10.1128/JVI.80.5.2162-2169.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serviene E., Shapka N., Cheng C.P., Panavas T., Phuangrat B., Baker J., Nagy P.D. Genome-wide screen identifies host genes affecting viral RNA recombination. Proc. Natl. Acad. Sci. USA. 2005;102:10545–10550. doi: 10.1073/pnas.0504844102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serviene E., Jiang Y., Cheng C.P., Baker J., Nagy P.D. Screening of the yeast yTHC collection identifies essential host factors affecting tombusvirus RNA recombination. J. Virol. 2006;80:1231–1241. doi: 10.1128/JVI.80.3.1231-1241.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford J., Pagano J.S. Tumor viruses and cell signaling pathways: Deubiquitination versus ubiquitination. Mol. Cell. Biol. 2004;24:5089–5093. doi: 10.1128/MCB.24.12.5089-5093.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford J., Pagano J.S. Targeting of host-cell ubiquitin pathways by viruses. Essays Biochem. 2005;41:139–156. doi: 10.1042/EB0410139. [DOI] [PubMed] [Google Scholar]
- Shapka N., Stork J., Nagy P.D. Phosphorylation of the p33 replication protein of Cucumber necrosis tombusvirus adjacent to the RNA binding site affects viral RNA replication. Virology. 2005;343:65–78. doi: 10.1016/j.virol.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Sharma M., Sasvari Z., Nagy P.D. Inhibition of sterol biosynthesis reduces tombusvirus replication in yeast and plants. J Virol. 2009 doi: 10.1128/JVI.02003-09. (published on line) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M., Sasvari Z., Nagy P.D. Inhibition of sterol biosynthesis reduces tombusvirus replication in yeast and plants. J Virol. 2010;84:2270–2281. doi: 10.1128/JVI.02003-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U., Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S.T., Lai M.M. Viral and cellular proteins involved in coronavirus replication. Curr. Top. Microbiol. Immunol. 2005;287:95–131. doi: 10.1007/3-540-26765-4_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirover M.A. New insights into an old protein: The functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim. Biophys. Acta. 1999;1432:159–184. doi: 10.1016/s0167-4838(99)00119-3. [DOI] [PubMed] [Google Scholar]
- Sirover M.A. New nuclear functions of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in mammalian cells. J. Cell. Biochem. 2005;95:45–52. doi: 10.1002/jcb.20399. [DOI] [PubMed] [Google Scholar]
- Slagsvold T., Pattni K., Malerod L., Stenmark H. Endosomal and non-endosomal functions of ESCRT proteins. Trends Cell Biol. 2006;16:317–326. doi: 10.1016/j.tcb.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Sohn S.Y., Kim S.B., Kim J., Ahn B.Y. Negative regulation of hepatitis B virus replication by cellular Hsp40/DnaJ proteins through destabilization of viral core and X proteins. J. Gen. Virol. 2006;87(Pt. 7):1883–1891. doi: 10.1099/vir.0.81684-0. [DOI] [PubMed] [Google Scholar]
- Stork J., Panaviene Z., Nagy P.D. Inhibition of in vitro RNA binding and replicase activity by phosphorylation of the p33 replication protein of Cucumber necrosis tombusvirus. Virology. 2005;343:79–92. doi: 10.1016/j.virol.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Sulea T., Lindner H.A., Purisima E.O., Menard R. Deubiquitination, a new function of the severe acute respiratory syndrome coronavirus papain-like protease? J. Virol. 2005;79:4550–4551. doi: 10.1128/JVI.79.7.4550-4551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz N., Thiel H.J. Cytopathogenicity of pestiviruses: Cleavage of bovine viral diarrhea virus NS2–3 has to occur at a defined position to allow viral replication. Arch. Virol. 2003;148:1405–1412. doi: 10.1007/s00705-003-0106-9. [DOI] [PubMed] [Google Scholar]
- Tavis J.E., Massey B., Gong Y. The duck hepatitis B virus polymerase is activated by its RNA packaging signal, epsilon. J. Virol. 1998;72:5789–5796. doi: 10.1128/jvi.72.7.5789-5796.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J.M., Barry M. Near death experiences: Poxvirus regulation of apoptotic death. Virology. 2006;344:139–150. doi: 10.1016/j.virol.2005.09.032. [DOI] [PubMed] [Google Scholar]
- Thivierge K., Cotton S., Dufresne P.J., Mathieu I., Beauchemin C., Ide C., Fortin M.G., Laliberte J.F. Eukaryotic elongation factor 1A interacts with Turnip mosaic virus RNA-dependent RNA polymerase and VPg-Pro in virus-induced vesicles. Virology. 2008;377:216–225. doi: 10.1016/j.virol.2008.04.015. [DOI] [PubMed] [Google Scholar]
- Tomita Y., Mizuno T., Diez J., Naito S., Ahlquist P., Ishikawa M. Mutation of host DnaJ homolog inhibits Brome mosaic virus negative-strand RNA synthesis. J. Virol. 2003;77:2990–2997. doi: 10.1128/JVI.77.5.2990-2997.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton V.K., Rao R. Functional expression of heterologous proteins in yeast: Insights into Ca2+ signaling and Ca2+-transporting ATPases. Am. J. Physiol. Cell Physiol. 2004;287:C580–C589. doi: 10.1152/ajpcell.00135.2004. [DOI] [PubMed] [Google Scholar]
- Tuteja R., Tuteja N. Nucleolin: A multifunctional major nucleolar phosphoprotein. Crit. Rev. Biochem. Mol. Biol. 1998;33:407–436. doi: 10.1080/10409239891204260. [DOI] [PubMed] [Google Scholar]
- Van Wynsberghe P.M., Ahlquist P. 5′cis elements direct nodavirus RNA1 recruitment to mitochondrial sites of replication complex formation. J. Virol. 2009;83:2976–2988. doi: 10.1128/JVI.02040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian J.P., Sala M., Henry M., Wain-Hobson S., Meyerhans A. Manganese cations increase the mutation rate of human immunodeficiency virus type 1 ex vivo. J. Gen. Virol. 1999;80(Pt. 8):1983–1986. doi: 10.1099/0022-1317-80-8-1983. [DOI] [PubMed] [Google Scholar]
- Vlot A.C., Neeleman L., Linthorst H.J., Bol J.F. Role of the 3′-untranslated regions of Alfalfa mosaic virus RNAs in the formation of a transiently expressed replicase in plants and in the assembly of virions. J. Virol. 2001;75:6440–6449. doi: 10.1128/JVI.75.14.6440-6449.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner S., Sarnow P. Viral ribonucleoprotein complex formation and nucleolar–cytoplasmic relocalization of nucleolin in poliovirus-infected cells. J. Virol. 1998;72:6699–6709. doi: 10.1128/jvi.72.8.6699-6709.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R.Y., Nagy P.D. Tomato bushy stunt virus co-opts the RNA-binding function of a host metabolic enzyme for viral genomic RNA synthesis. Cell Host Microbe. 2008;3:178–187. doi: 10.1016/j.chom.2008.02.005. [DOI] [PubMed] [Google Scholar]
- Wang X., Lee W.M., Watanabe T., Schwartz M., Janda M., Ahlquist P. Brome mosaic virus 1a nucleoside triphosphatase/helicase domain plays crucial roles in recruiting RNA replication templates. J. Virol. 2005;79:13747–13758. doi: 10.1128/JVI.79.21.13747-13758.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Loveland A.N., Kattenhorn L.M., Ploegh H.L., Gibson W. High-molecular-weight protein (pUL48) of human cytomegalovirus is a competent deubiquitinating protease: Mutant viruses altered in its active-site cysteine or histidine are viable. J. Virol. 2006;80:6003–6012. doi: 10.1128/JVI.00401-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R.Y., Stork J., Pogany J., Nagy P.D. A temperature sensitive mutant of heat shock protein 70 reveals an essential role during the early steps of tombusvirus replication. Virology. 2009;394:28–38. doi: 10.1016/j.virol.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R.Y., Stork J., Nagy P.D. A key role for heat shock protein 70 in the localization and insertion of tombusvirus replication proteins to intracellular membranes. J. Virol. 2009;83:3276–3287. doi: 10.1128/JVI.02313-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks S.A., Miller D.J. The heat shock protein 70 cochaperone YDJ1 is required for efficient membrane-specific flock house virus RNA replication complex assembly and function in Saccharomyces cerevisiae. J. Virol. 2008;82:2004–2012. doi: 10.1128/JVI.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Washburne M., Stone D.E., Craig E.A. Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 1987;7:2568–2577. doi: 10.1128/mcb.7.7.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K.A., Morris T.J. Recombination between defective tombusvirus RNAs generates functional hybrid genomes. Proc. Natl. Acad. Sci. USA. 1994;91:3642–3646. doi: 10.1073/pnas.91.9.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K.A., Nagy P.D. Advances in the molecular biology of tombusviruses: Gene expression, genome replication, and recombination. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:187–226. doi: 10.1016/S0079-6603(04)78005-8. [DOI] [PubMed] [Google Scholar]
- Whitham S.A., Quan S., Chang H.S., Cooper B., Estes B., Zhu T., Wang X., Hou Y.M. Diverse RNA viruses elicit the expression of common sets of genes in susceptible Arabidopsis thaliana plants. Plant J. 2003;33:271–283. doi: 10.1046/j.1365-313x.2003.01625.x. [DOI] [PubMed] [Google Scholar]
- Whitham S.A., Yang C., Goodin M.M. Global impact: Elucidating plant responses to viral infection. Mol. Plant Microbe Interact. 2006;19:1207–1215. doi: 10.1094/MPMI-19-1207. [DOI] [PubMed] [Google Scholar]
- Wong J., Zhang J., Si X., Gao G., Luo H. Inhibition of the extracellular signal-regulated kinase signaling pathway is correlated with proteasome inhibitor suppression of coxsackievirus replication. Biochem. Biophys. Res. Commun. 2007;358:903–907. doi: 10.1016/j.bbrc.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Woo J.L., Berk A.J. Adenovirus ubiquitin-protein ligase stimulates viral late mRNA nuclear export. J. Virol. 2007;81:575–587. doi: 10.1128/JVI.01725-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B., Pogany J., Na H., Nicholson B.L., Nagy P.D., White K.A. A discontinuous RNA platform mediates RNA virus replication: Building an integrated model for RNA-based regulation of viral processes. PLoS Pathog. 2009;5:e1000323. doi: 10.1371/journal.ppat.1000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji Y., Kobayashi T., Hamada K., Sakurai K., Yoshii A., Suzuki M., Namba S., Hibi T. In vivo interaction between Tobacco mosaic virus RNA-dependent RNA polymerase and host translation elongation factor 1A. Virology. 2006;347:100–108. doi: 10.1016/j.virol.2005.11.031. [DOI] [PubMed] [Google Scholar]
- Yi M., Schultz D.E., Lemon S.M. Functional significance of the interaction of hepatitis A virus RNA with glyceraldehyde 3-phosphate dehydrogenase (GAPDH): Opposing effects of GAPDH and polypyrimidine tract binding protein on internal ribosome entry site function. J. Virol. 2000;74:6459–6468. doi: 10.1128/jvi.74.14.6459-6468.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi G.H., Zhang C.Y., Cao S., Wu H.X., Wang Y. De novo RNA synthesis by a recombinant classical swine fever virus RNA-dependent RNA polymerase. Eur. J. Biochem. 2003;270:4952–4961. doi: 10.1046/j.1432-1033.2003.03897.x. [DOI] [PubMed] [Google Scholar]
- Young J.C., Agashe V.R., Siegers K., Hartl F.U. Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 2004;5:781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- Zang W.Q., Fieno A.M., Grant R.A., Yen T.S. Identification of glyceraldehyde-3-phosphate dehydrogenase as a cellular protein that binds to the hepatitis B virus posttranscriptional regulatory element. Virology. 1998;248:46–52. doi: 10.1006/viro.1998.9255. [DOI] [PubMed] [Google Scholar]
- Zeenko V.V., Ryabova L.A., Spirin A.S., Rothnie H.M., Hess D., Browning K.S., Hohn T. Eukaryotic elongation factor 1A interacts with the upstream pseudoknot domain in the 3′ untranslated region of Tobacco mosaic virus RNA. J. Virol. 2002;76:5678–5691. doi: 10.1128/JVI.76.11.5678-5691.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Gopinath K., Murali A., Yi G., Hayward S.D., Zhu H., Kao C. RNA-binding proteins that inhibit RNA virus infection. Proc. Natl. Acad. Sci. USA. 2007;104:3129–3134. doi: 10.1073/pnas.0611617104. [DOI] [PMC free article] [PubMed] [Google Scholar]


