Abstract
Although immune checkpoint blockade (ICB) improves clinical outcome in several types of malignancies, pancreatic ductal adenocarcinoma (PDA) remains refractory to this therapy. Preclinical studies have demonstrated that the relative abundance of suppressive myeloid cells vs. cytotoxic T cells determines the efficacy of combination immunotherapies, which include ICB. Here, we evaluated the role of the ubiquitin-specific protease 22 (USP22) as a regulator of the immune tumor microenvironment (TME) in PDA. We report that deletion of USP22 in pancreatic tumor cells reduced the infiltration of myeloid cells and promoted the infiltration of T cells and NK cells, leading to an improved response to combination immunotherapy. We also showed that ablation of tumor cell–intrinsic USP22 suppressed metastasis of pancreatic tumor cells in a T cell–dependent manner. Finally, we provide evidence that USP22 exerted its effects on the immune TME by reshaping the cancer cell transcriptome through its association with the deubiquitylase module of the SAGA/STAGA transcriptional co-activator complex. These results indicated that USP22 regulates immune infiltration and immunotherapy sensitivity in preclinical models of pancreatic cancer.
Keywords: Pancreatic cancer, tumor microenvironment, immunotherapy, USP22, deubiquitinase
INTRODUCTION
Immune checkpoint blockade (ICB) has led to improved clinical outcomes in several types of cancer (1,2). However, the majority of patients treated with ICB fail to respond, leading to overall response rates of 20-40% (2). The abundance of tumor-infiltrating T cells is a major factor predicting response to immunotherapy, as T cell–inflamed tumors are more sensitive to ICB than non-T cell–inflamed tumors (3). Thus, there is an urgent need to understand the factors regulating intratumoral T-cell infiltration. Studies have shown that the tumor mutational burden does not fully explain the abundance of tumor-infiltrating T cells (4), and there is an increasing agreement that various tumor cell intrinsic factors, including signaling molecules, secreted factors, and transcriptional regulators, influence the level of T-cell infiltration into tumors and the resulting response to immunotherapy (5).
Pancreatic ductal adenocarcinoma (PDA) is predicted to become the second leading cause of cancer-related death in the United States within the next five years (38). Pancreatic tumors are characterized by T-cell exclusion and an immunosuppressive tumor microenvironment (TME). As a result, PDA is largely resistant to immunotherapy (6). We and others have reported that tumor cell–intrinsic factors – secreted molecules, signaling pathways, and epigenetic status – play a central role in shaping the immune TME in PDA and other types of cancer (5,7-9). Importantly, heterogeneous mechanisms may contribute to the suppression of antitumor immunity (8). Several studies have demonstrated that a drug combination including gemcitabine, nab-paclitaxel, CD40 agonistic antibody, PD-1 blocking antibody, and CTLA-4 blocking antibody promotes clinical outcomes in preclinical mouse models of PDA (8,10,11). Importantly, preclinical studies indicate that the abundance of tumor-infiltrating activated CD8+ T cells predicts sensitivity to this therapy (8). In this study, we evaluated the behavior of ubiquitin-specific protease 22 (USP22) as a tumor cell–intrinsic factor shaping the immune TME in PDA.
MATERIALS AND METHODS
Animals
All animal procedures were conducted following National Institutes of Health guidelines. All mouse protocols were in accordance with, and with the approval of the Institutional Animal Care and Use committee (IACUC) of the University of Pennsylvania. All wild-type C57BL/6 were purchased from The Jackson Laboratory and/or bred at the University of Pennsylvania. Kras-LSL-G12D/+;Trp53-LSL-R172H/+, Pdx1-Cre, and Rosa-LSL-YFP (KPCY) mice were bred in-house, backcrossed for over 10 generations with C57BL/6J mice (Jackson Laboratories) and assessed at the DartMouse Speed Congenic Core facility at the Geisel School of Medicine at Dartmouth college.
Tumor cells
All mouse pancreatic tumor cell clones were tested by the Research Animal Diagnostic Laboratory (RADIL) at the University of Missouri, using the Infectious Microbe PCR Amplification Test (IMPACT) II. Tumor cells were cultured in DMEM (high glucose without sodium pyruvate) with 10% FBS (Gibco) and glutamine (2 mM). The clones were regularly tested using the MycoAlert Mycoplasma Detection Kit (Lonza). Mouse pancreatic tumor cell clones 6419c5, 6694c2, and 6422c1 were generated in our lab (8) and were used for less than 18 passages. These tumor cell clones were derived from the KPCY mice (as described above) and, thus, expressed YFP at the time of generation. We used 293T cells (Clontech, 632180) for lentivirus packaging.
Implantation of tumor cells
Pancreatic tumor cells (as described above in the “Tumor Cells” section) were dissociated into single cells with 0.25% trypsin (Gibco), washed with serum-free Dulbecco’s Modified Eagle’s medium (DMEM) twice, and counted in preparation for subcutaneous or orthotopic implantations. 2.0×105 tumor cells were implanted subcutaneously and 5.0×104 tumor cells were implanted orthotopically into the pancreas of 6-8-week old female C57BL/6 mice via laparotomy as previously described (8). Tumors were harvested 18-24 days following implantation. Endpoint criteria included tumor volume exceeding 500 mm3, severe cachexia, or weakness and inactivity.
Subcutaneous tumor growth and regression assessments
For tumor growth kinetics, tumors were measured every 3 days. Tumor length and width were measured with calipers, and tumor volumes were then calculated as length*width2/2. Tumor volumes of 500 mm3 were used as an endpoint for survival analysis. Tumor regressions and were calculated using the initial tumor size at the start of treatment to tumor size 15 days later.
Lung metastatic colonization assay
1.0×105 Usp22-WT and Usp22-KO tumor cells were injected via tail vein into C57BL/6 mice. After 14 days, lungs were harvested, weighed, and subjected to flow cytometry for immune profiling as described below in the flow analysis section. The tail vein injection experiment was performed in one experiment for each using two CRISPR knockout clones, with 5 mice per group for analysis.
Treatments and T-cell depletions in C57BL/6J mice
Gemcitabine (Hospira) and Nab-paclitaxel (Abraxane, Celgene) were purchased from the Hospital of the University of Pennsylvania Pharmacy. Gemcitabine (G) was procured as pharmaceutical grade suspension at 38 mg/mL, and further diluted to 12 mg/mL in PBS and administered at 120 mg/kg via intraperitoneal (i.p.) injection. Abraxane (A) was purchased as a pharmaceutical grade powder resuspended at 12 mg/mL in PBS for i.p. injection at dose of 120 mg/kg. Vehicle control mice received the equivalent to nab-paclitaxel dose of human albumin (huAlb; Sigma). Chemotherapy was injected when tumor was 3-5 mm. For anti-CD40 agonist treatment, mice were injected i.p. with 100 μg of either agonistic CD40 rat anti-mouse IgG2a (clone FGK45, endotoxin-free) or the isotype control IgG2a (clone 2A3, BioXcell) 48hrs after chemotherapy. For checkpoint blockade treatment, mice were injected i.p. with 200 μg of anti–PD-1 (clone RMP 1-14, BioXcell) and 200 μg anti–CTLA-4 (clone 9H10, BioXcell), starting day 10 (therapy start timepoint), with 6 and 3 doses, respectively. Control mice received the isotype control IgG2a (clone 2A3, BioXcell) on treatment days. CD4+ and CD8+ T cells were depleted using i.p. injections of 200 mg anti-CD4 (clone GK1.5, BioXcell) and anti-CD8 (clone 2.43, BioXcell), three days prior tumor implantation, and three times a week for the duration of the experiment. Control groups received IgG2b isotype control (BioXcell). The treatment experiment was performed in one experiment for each using two CRISPR knockout clones, with 7-8 mice per group for analysis.
Analysis of RNA-seq, differential gene expression, GSEA analysis, and EnrichR analysis
RNA-seq data is deposited at GEO (GSE140088). RNA samples were extracted from sorted YFP+ tumor cells from subcutaneous tumors 21 days post-implantation using the Qiagen RNeasy Micro Kit following manufacturer’s instructions. RNA was sent to Novogene (California, USA), for library preparation and high-throughput sequencing using Illumina sequencers (Hiseq 2500) to generated paired-end 150 bp data. Fastq files were checked for quality using FastQC (Babraham). Raw counts of gene transcripts were obtained using alignment-independent tool, Salmon (https://combine-lab.github.io/salmon/), using standard settings. The raw count matrix was subsequently imported into R-studio (R version 3.3.3) and used as input for DESeq2 following the vignette of the package for normalization and differential gene expression analysis. Salmon was also used to normalize and quantitate gene expression in transcripts-per-million (tpm) through quasi-alignment. Differentially expressed genes from the DESeq2 analysis were used as input for GSEA MSigDB geneset enrichment analysis. Differentially expressed genes (padj value < 0.01 and absolute fold change >2) were used as input for EnrichR analysis (ENCODE and ChEA Consensus TFs from ChIP-X & ChEA datasets).
Lentiviral transduction of tumor cells for shRNA-mediated knockdown and CRISPR-mediated ablation
The shRNA knockdown vector, pLKO.1.puro, was a gift from Robert Weinberg (Addgene plasmid #8453). The CRISPR vector, lentiCRISPR v2, was a gift from Feng Zhang (Addgene plasmid # 52961). The vector and pVSVg and psPAX2 lentiviral packaging plasmids (Addgene) were co-transfected into 293T cells (Clontech, 632180) using PEI reagent (Polysciences, 23966-2). Lentiviral particles were collected 48 or 72 hours after transfection and filtered for usage. Tumor cells transduced either with Cas9-Puro (control, ctrl) or Cas9-guide-Puro (knockout, KO)(ctrl is from Addgene, #52961, and KOs were cloned following the instruction from Addgene) were selected with 8 μg/mL puromycin (Invitrogen, A1113803). Single-cell clones were picked from bulk knockout cell line using single-cell sorting using a BD Jazz FACS sorter into 96-well plates. Knockout efficiencies were assessed by gene-specific qPCR analysis and immunofluorescence to detect target proteins (staining methods described below). shRNA sequences used were: Atxn7l3-sh1-F-CCGG-AGGCGAACCGTACGGATTTAT-CTCGAG-ATAAATCCGTACGGTTCGCCT-TTTTTG; Atxn7l3-sh1-R-AATT-CAAAAA-AGGCGAACCGTACGGATTTAT-CTCGAG-ATAAATCCGTACGGTTCGCCT; Atxn7l3-sh2-F-CCGG-TCGAAGATCCAAGTCTCTAAA-CTCGAG-TTTAGAGACTTGGATCTTCGATTTTTG; Atxn7l3-sh2-R-AATT-CAAAAA-TCGAAGATCCAAGTCTCTAAA-CTCGAGTTTAGAGACTTGGATCTTCGA. CRISPR sgRNA sequences used were: Usp22-A-TCT-GCG-TGG-ACT-GAT-CAA-CC; Usp22-B-AGT-TCC-AGC-TCC-CGT-TTA-GT.
Quantitative PCR (qPCR) analysis for gene expression
RNA was prepared from cultured tumor cells using RNeasy Mini Kit or RNeasy Micro Kit (Qiagen). cDNA was generated using High-capacity cDNA Reverse Transcription Kit from 1 μg RNA in 20 μl reaction volume and diluted 1:10 for qPCR analysis (Life Technologies). qPCR analysis was performed using 2 μl diluted cDNA with biological (2-3) and technical replicates (2-3) using SsoAdvanced SYBR reagent (Bio-Rad) and Bio-Rad qPCR platform, and results were normalized to the expression of Tbp using the Bio-Rad software. Primer sequences utilized for qPCR were: Usp22-F-CTC-CCC-ACA-CAT-TCC-ATA-CAA-G; Usp22-R-TGG-AGC-CCA-CCC-GTA-AAG-A; Atxn7l3-F-TTG-TCT-GGC-CTG-GAT-AAC-AGC; Atxn7l3-R-CCG-GTG-TAC-TTC-AAA-GCA-GAA-TC; Tbp-F-AGA-ACA-ATC-CAG-ACT-AGC-AGC-A; Tbp-R-GGG-AAC-TTC-ACA-TCA-CAG-CTC.
Flow cytometry of implanted tumors and lung
For the flow cytometric analyses, subcutaneous or orthotopic tumors following 18-24 days of implantation were chopped into small pieces and digested in collagenase (1 mg/mL in DMEM; Sigma-Aldrich) at 37°C for 45 minutes and filtered through a 70-μM cell strainer. Single-cell suspensions were stained with antibodies on ice for 30 minutes and washed twice with PBS with 5% FBS for flow cytometric analysis. No intracellular staining is needed for this analysis. Cells were then analyzed by flow cytometry using BD FACS (BD Biosciences) and FlowJo software (Treestar).
Antibodies used for the analysis: CD279 (PD-1) FITC (29F.1A12; Biolegend 135214), CD335 (NKp46) PE (29A1.4; Biolegend 137604), CD103 PE/Dazzle 594 (2E7; Biolegend 121430), CD3e PE/Cy5 (145-2C11; Biolegend 100310), CD45 AF700 (30-F11; Biolegend 103128), CD8a PE/Cy7 (53-6.7; Biolegend 100722), I-A/I-E (MHCII) PE/Cy7 (M5/114.15.2; Biolegend 107630), Ly-6G V450 (1A8; BD 560603), H-2Kb/H-2Db (MHCI) AF647 (28-8-6; Biolegend 114612), F4/80 APC/Cy7 (BM8; Biolegend 123118), CD11b PerCP-Cy5.5 (M1/70; BD 550993), CD11c BV605 (N418; Biolegend 117334), Ly-6C BV570 (HK1.4; Biolegend 128030), CD4 BV650 (RM4-5; Biolegend 100546).
Gating Strategies for immune cells: myeloid cells - Live CD45+CD11b+; granulocytic (g)MDSCs/neutrophils - Live CD45+CD11b+Gr1+; macrophages - Live CD45+F4/80+CD11b+; CD11c+ dendritic cells - Live CD45+F4/80− CD11c+; CD103+ dendritic cells - Live CD45+CD11b−F4/80−CD11c+ CD103+; T cells - Live CD45+CD11b−F4/80−NKp46−CD3+; CD4+ T cells - Live CD45+CD11b−F4/80−NKp46−CD3+CD4+; CD8+ T cells - Live CD45+CD11b−F4/80−NKp46−CD3+CD8+. The flow analysis of immune infiltration of USP22-WT vs. KO tumors were performed in two experiments using six CRISPR knockout clones, with 4-5 mice per tumor cell clone.
Flow cytometry of in vitro tumor cells for EdU
Tumor cells were incubated with EdU for 3 hours in DMEM with 10% FBS and Glutamax (Thermo 35050061) and then fixed with fixation buffer following the instruction of the reagent (eBioscience 00-5123-43 and 00-5223-56). Samples were further processed for EdU staining as per protocol (Thermo Fisher, C10086). Samples were stained for EdU and then analyzed by flow cytometry using BD FACS LSR machine (BD Biosciences) and FlowJo software (Treestar).
In vitro treatment with IFNγ for MHC I expression analysis
Usp22-WT and Usp22-KO tumor cells were treated with 100ng/mL of IFNγ (Peprotech) in DMEM with 10% FBS and Glutamax (Thermo 35050061) for 24 hours. Tumor cells were trypsinized from culture plates and re-suspended in PBS with 5% FBS for staining of antibodies. Single-cell suspensions were stained with antibodies on ice for 30 minutes and washed twice with PBS with 5% FBS for flow cytometric analysis. Samples were stained for MHCI (Biolegend 114612) and then analyzed by flow cytometry using BD FACS LSR machine (BD Biosciences) and FlowJo software (Treestar).
Immunofluorescent and immunohistochemistry staining
For CD3, USP22, and Gr1 staining, collected implanted tumor tissues were fixed in Zn-formalin for 24 hours and embedded in paraffin. Sections were deparaffinized, rehydrated, and prepared by antigen retrieval for 6 minutes each, and then blocked in 5% donkey serum (Sigma, D9663) for 1 hour at room temperature, incubated with primary antibodies overnight at 4°C, washed with 0.1% PBST (PBS with Tween-20), incubated with secondary antibodies for 1 hour at room temperature, and then washed and mounted. Slides were visualized and imaged using an Olympus IX71 inverted multicolor fluorescent microscope and a DP71 camera. For CD3 and Gr1 staining quantification, stained cells were counted for CD3+ T cells manually in 5-8 fields per sample.
Primary antibodies: CD3 (Abcam ab5690, 1:100 dilution), USP22 (Abcam ab195289, 1:100 dilution), Gr-1 (eBioscience 14-5931-85, 1:50 dilution), YFP (Abcam, ab6673). Secondary antibodies were purchased from Invitrogen (A-11055, A-21207, A-21209) and were used as 1:250 dilution for all staining.
Cancer Dependency Map Portal (depmap) data analysis
The depmap portal (https://depmap.org/portal/) and the CRISPR (Avana) Public 19Q3 dataset were used for this analysis. No samples were excluded from the dataset, and 625 cell lines in total were included in this analysis. Following the depmap instruction, the dependency scores of genes were downloaded and plotted as dot plots. Pearson correlation was calculated for all the plots using Prism.
Statistical analysis
Statistical comparisons between two groups were performed using Student’s unpaired t test. For comparisons between multiple groups, one-way ANOVA with Tukey’s HSD post-test was used. For survival comparison between two groups, log-rank p-values of Kaplan-Meier curves were determined in GraphPad Prism 8 (GraphPad). On graphs, error bars represent either range or standard error of mean (SEM), as indicated in legends. For all figures, p<0.05 was considered statistically significant, * indicates p<0.05, **p<0.01, and ***p<0.001.
Software
PRISM software and R were used for the data processing, statistical analysis, and result visualization (http://www.graphpad.com). The R language and environment for statistical computing and graphics (https://www.r-project.org) was utilized in this study for the statistical and bioinformatics analysis of RNA-seq data. The R packages used for all analysis described in this manuscript were obtained from the Bioconductor and CRAN.
RESULTS
Tumor cell intrinsic USP22 regulates immune cell infiltration in implanted PDA tumors
We previously established an experimental system that recapitulates the heterogeneity of immune cell infiltration in pancreatic cancer (8). This platform consists of a group of primary murine PDA tumors stratified into two main subsets: a T cell–low group, enriched for myeloid-derived suppressor cells (MDSCs), and a T cell–high group, enriched for T cells and dendritic cells (DCs), with reduced MDSCs. We and others have previously demonstrated that several tumor cell intrinsic factors, including CSF2, CXCL1, CSF3, EPHA2, and PTGS2, can control the abundance of tumor-infiltrating myeloid cells and T cells, which, in turn, affects the sensitivity of PDA tumors to combination immunotherapy (7). In these studies, we found that mechanisms underlying the establishment of the non-T cell–inflamed TME varied from tumor to tumor. Here, we aimed to identify alternative tumor cell–intrinsic regulators of the immune TME that might lead to novel therapeutic targets for precision immunotherapy.
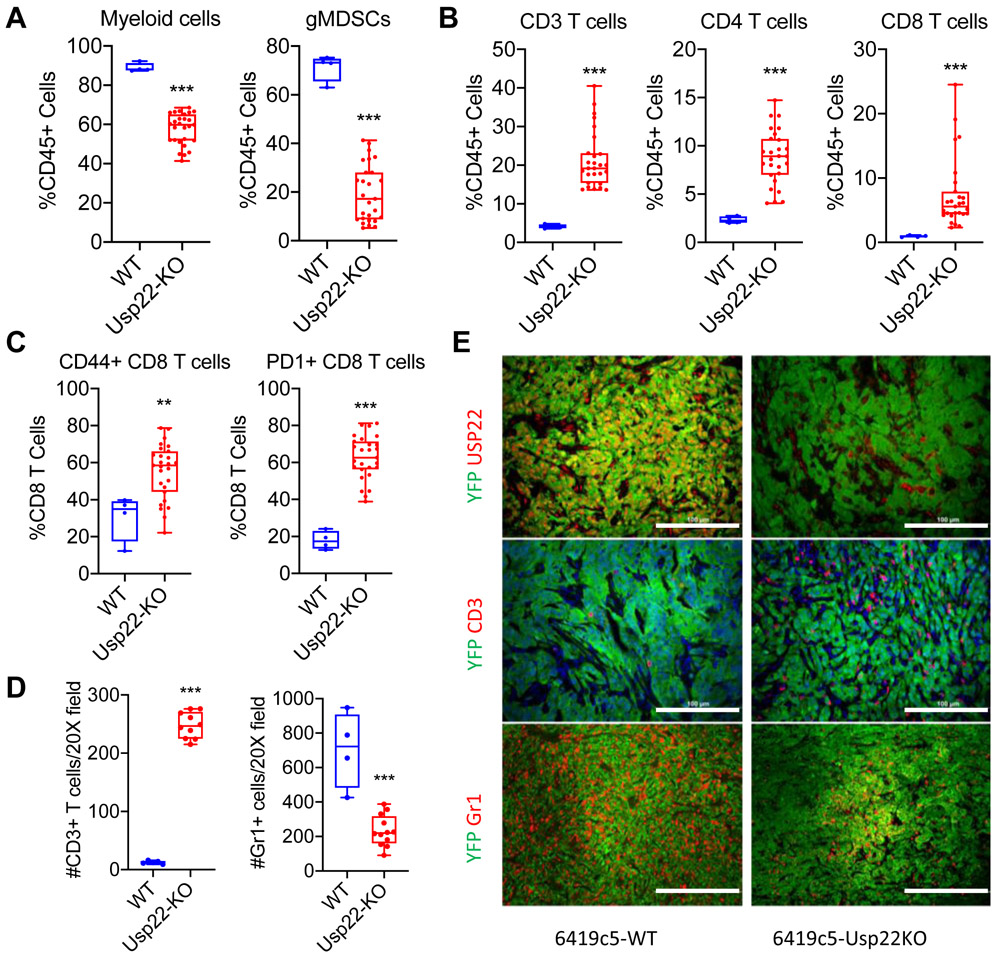
Ablation of USP22 in liver tumor cells has been shown to increase tumor immunogenicity and promote infiltration of T cells into the resulting liver tumors (12). An examination of our PDA clone library revealed that T cell–low tumors expressed higher USP22 at the RNA and protein level compared to T cell-high tumors (Supplementary Figure S1A-B). These findings are consistent with the idea that USP22 promotes the non-T cell–inflamed TME and suppresses antitumor immunity in PDA. To functionally examine the role of USP22 in regulating immune TME, we utilized CRISPR/Cas9 strategy to generated six independent Usp22 knockout (Usp22-KO) cell clones (Supplementary Figure S1C) from a well-established T cell–low PDA tumor cell clone, 6419c5. We then performed flow cytometric analysis to assess the phenotype of immune cells that infiltrated into subcutaneously implanted Usp22-WT and Usp22-KO tumors. USP22 ablation resulted in decrease in total myeloid cells and granulocytic MDSCs (gMDSCs) and an increase in CD3+, CD4+, and CD8+ T cells in the TME (Figure 1A-B). CD8+ T cells in Usp22-KO tumors were more frequently positive for CD44 and PD-1 (Figure 1C). These changes in myeloid and T-cell populations within the TME were confirmed by immunofluorescence staining of CD3 and Gr1 (Figure 1D-E). Usp22-KO tumors also exhibited increased NK cell infiltration but no significant change in the number of macrophages or DCs (Supplementary Figure S1D).We also observed a minor decrease of tumor growth in vivo and 10% decrease of EdU incorporation in vitro following ablation of USP22 (Supplementary Figure S1E-F), consistent with previous reports suggesting that USP22 regulates cell cycle progression and tumor growth (13-15). Likewise, T-cell depletion had no effect on the growth of either Usp22-WT and Usp22-KO tumors (Supplementary Figure S1G-H), suggesting that the growth effects of Usp22 ablation are not T cell–dependent.
Figure 1. Loss of tumor cell–intrinsic USP22 increases T-cell infiltration and decreases myeloid cell infiltration in subcutaneously implanted PDA tumors.
(A-C) Flow cytometric analysis of tumor-infiltrating immune cells in subcutaneously implanted Usp22-WT and Usp22-KO (6 independent knockout tumor cell clones, n=4-27 tumors analyzed per group in 3 experiments). (D) Quantification and (E) representative immunofluorescent staining images (E, middle & bottom) of CD3+ T cells and Gr1+ myeloid cells in Usp22-WT and Usp22-KO tumors (2 independent knockout clones, n=4-12 tumors analyzed per group) stained for CD3 or Gr1 (red), YFP (green), and DAPI (blue). (E, top) Representative image stained for USP22 (red) and YFP (green) in Usp22-WT or Usp22-KO tumors. In (A-D), data are presented as boxplots with horizontal lines and error bars indicating mean and range, respectively. Statistical differences between groups were calculated using Student’s unpaired t-test. *p<0.05, **p<0.01, ***p< 0.001. Scale bars, 100 μm for CD3 and USP22; 200 μm for Gr1.
To expand these observations to other tumors, we generated Usp22 knockouts from two other T cell–low tumor cell clones, 6422c1 and 6694c2. Flow analysis of the implanted tumors showed that loss of tumor cell–intrinsic USP22 decreased the abundance of gMDSCs and increased the number of CD3+ and CD4+ T cells in the TME of both 6422c1 and 6694c2 tumors (Supplementary Figure S1I-L). A non-significant trend towards increased CD8+ T cells was seen in Usp22-KO 6422c1 tumors (Supplementary Figure S1H, right-most panel) and no difference seen in Usp22-KO 6694c2 tumors (Supplementary Figure 1J, right-most panel). These results suggested that different T cell–low tumors depended on USP22 to regulate CD8+ T-cell infiltration to different degrees, further highlighting the importance of identifying patient-specific regulators of the immune TME.
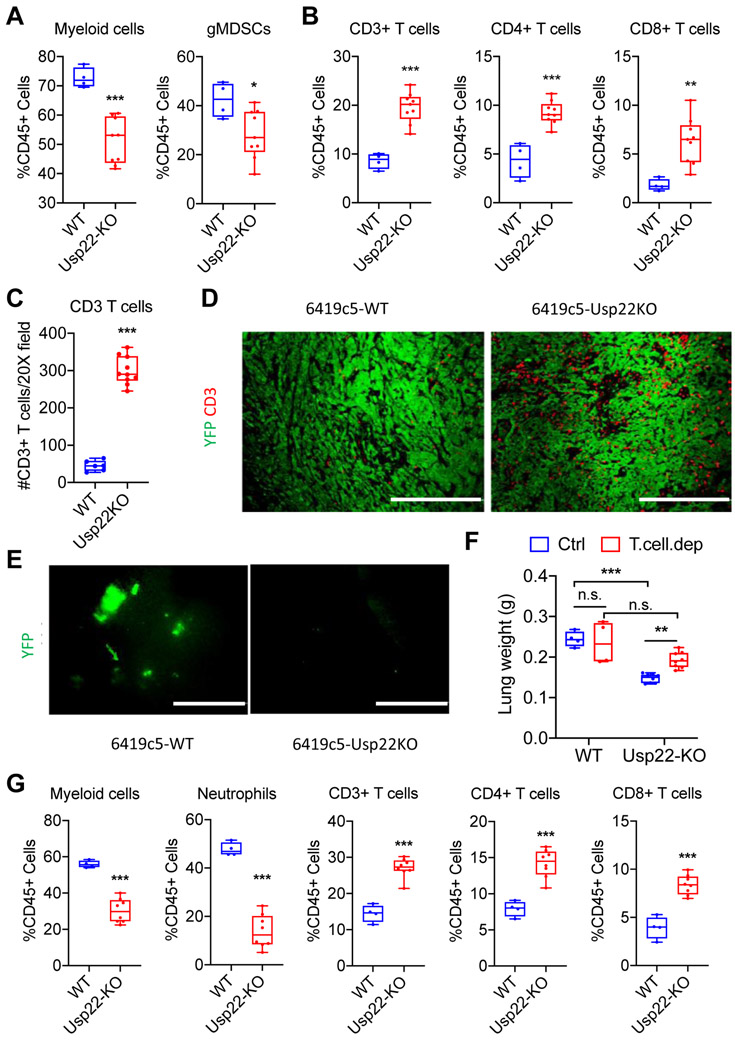
Next, we assessed the infiltration of immune cells in tumors arising from 6419c5 Usp22-WT and Usp22-KO tumors that were orthotopically injected into the pancreas. Flow cytometry and immunofluorescence staining revealed that Usp22 deletion resulted in a significant decrease of total myeloid cells and gMDSCs and an increase of CD3+, CD4+, and CD8+ T cells in the TME, reproducing the findings from subcutaneous tumors (Figure 2A-D, Supplementary Figure S2A). Collectively, these results indicated that tumor cell–intrinsic USP22 establishes a non-T cell–inflamed immune TME dominated by myeloid cells and a paucity of tumor-infiltrating T cells.
Figure 2. Loss of tumor cell–intrinsic USP22 promotes antitumor immunity in orthotopic PDA tumors and suppresses lung metastasis.
(A-B) Flow cytometric analysis of tumor-infiltrating immune cells in orthotopically implanted Usp22-WT and Usp22-KO tumors (2 independent knockout clones, n=4-9 tumors analyzed per group in 2 experiments). (C) Quantification and (D) representative immunofluorescent staining of CD3+ T cells in Usp22-WT and Usp22-KO orthotopic tumors (2 independent knockout clones, n=6-10 tumors analyzed per group) stained for CD3 (red) and YFP (green). (E) Representative images of lungs from animals in (D) with YFP+ tumor cells shown in green. (F) Quantification of lung weight 14 days after tail vein injection of Usp22-WT or Usp22-KO tumor cells (2 independent knockout clones) with or without T cell depletion (n=5 mice/group). (G) Flow cytometric analysis of lungs following tail veil injection of Usp22-WT or Usp22-KO tumor cells (2 independent KO clones, n=5-10 mice/group in 1 experiment). In (A-C & F-G), data are presented as boxplots with horizontal lines and error bars indicating mean and range, respectively. In (A-C & G), statistical differences between groups were calculated using Student’s unpaired t-test. In (F), statistical differences between groups were calculated using 2-way ANOVA analysis with multiple comparison. *p<0.05, **p<0.01, *** p< 0.001. Scale bars, 200 μm in (D), 1 mm in (E).
USP22 regulates T cell–dependent lung metastasis
Studies have demonstrated the significant role of both adaptive and innate immune cells in regulating the metastasis of tumor cells, including pancreatic tumor cells (8,16). Using the orthotopic model, we observed that mice injected with 6419c5 Usp22-KO tumors harbored fewer lung metastases compared to mice injected with 6419c5 Usp22-WT tumors (Figure 2E). To functionally examine the effect of tumor cell–intrinsic USP22 on lung metastatic colonization, we performed tail vein injection of Usp22-WT and Usp22-KO tumor cells into immunocompetent mice and observed that Usp22-KO tumor cells resulted in decreased metastatic burden compared to the Usp22-WT tumor cells (Figure 2F). T–cell depletion had no effect on metastatic burden in mice bearing Usp22-WT tumors but led to an increased metastatic burden in mice bearing Usp22-KO tumor cells (Figure 2F, Supplementary Figure S2B). Metastatic lung lesions from Usp22-KO tumors exhibited fewer infiltrating myeloid cells and more T cells (Figure 2G, Supplementary Figure S2C). Similar results on metastatic burden and immune infiltration were seen when another T cell–low tumor cell clone, 6694c2, was introduced by tail vein injection (Supplementary Figure S2D-E). These results suggest that Usp22-KO tumor cells have a reduced ability to form metastases secondary to T-cell surveillance.
Tumor cell–intrinsic USP22 affects the sensitivity of PDA tumors to immunotherapy
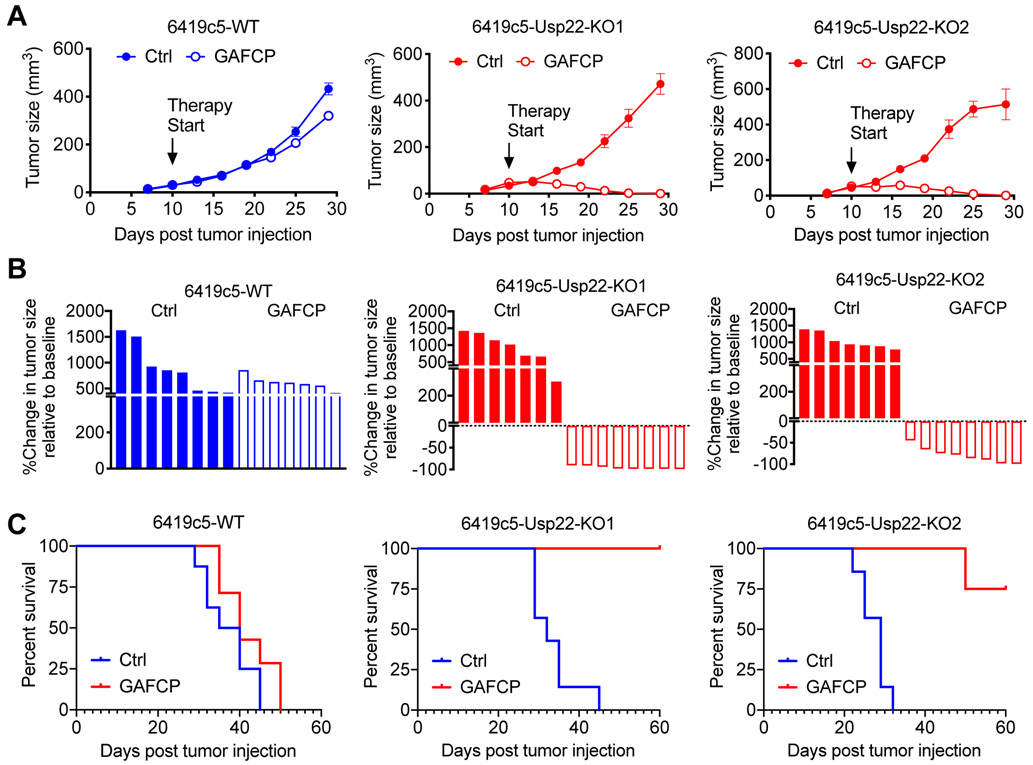
We previously reported that relative to T cell–high tumors, T cell–low tumors have decreased sensitivity to combination immunotherapy with GAFCP (gemcitabine, G; nab-paclitaxel, A; anti-CD40 agonist, F; anti–CTLA-4, C; and anti–PD-1, P). A similar combination therapy, including chemotherapy, checkpoint blockade, and CD40 agonist is currently tested in two clinical trials for PDA patients ( NCT02588443, NCT03214250). Given that loss of tumor cell–intrinsic USP22 resulted in increased T-cell infiltration, we assessed the sensitivity of 6419c5 Usp22-WT and Usp22-KO tumors to this combination. USP22-deficient PDA tumor cells exhibited a significantly better response to GAFCP therapy, resulting in tumor regressions and prolonged survival (Figure 3A-C). Collectively, these experiments suggest that USP22 expression in PDA tumors cells suppresses antitumor immunity and confers resistance to immunotherapy.
Figure 3. USP22 loss increases the sensitivity of implanted PDA tumors to a combined immunotherapy.
(A) Growth curves (error bars represent SEM) of Usp22-WT or Usp22-KO tumors with or without GAFCP treatment. Therapy was started on day 10 after implantation, when tumors were 3-5 mm in diameter. (B) Waterfall plots from the cohort in (A) showing change in size of Usp22-WT and Usp22-KO tumors relative to the baseline (day 10) measured 15 days after treatment with or without GAFCP. (C) Survival of animals from the cohort with Usp22-WT or Usp22-KO tumors with or without GAFCP treatment. Tumor cells implanted subcutaneously into C57BL/6 mice (n=7-8/group). *p<0.05, **p<0.01, ***p<0.001.
Ablation of USP22 leads to transcriptional reprogramming of tumor cells
USP22 has been reported to regulate the immune TME by controlling the protein stability of PD-L1 in liver tumor cells (12). Immunofluorescent staining revealed that USP22 protein is principally located in the nucleus of PDA tumor cells (Figure 1E, Supplementary Figure S1B, Supplementary Figure S3A), consistent with data from the human protein atlas (17,18). We, therefore, hypothesized that USP22 might influence tumor immunity through nuclear functions independent of its effects on PD-L1 protein stability.
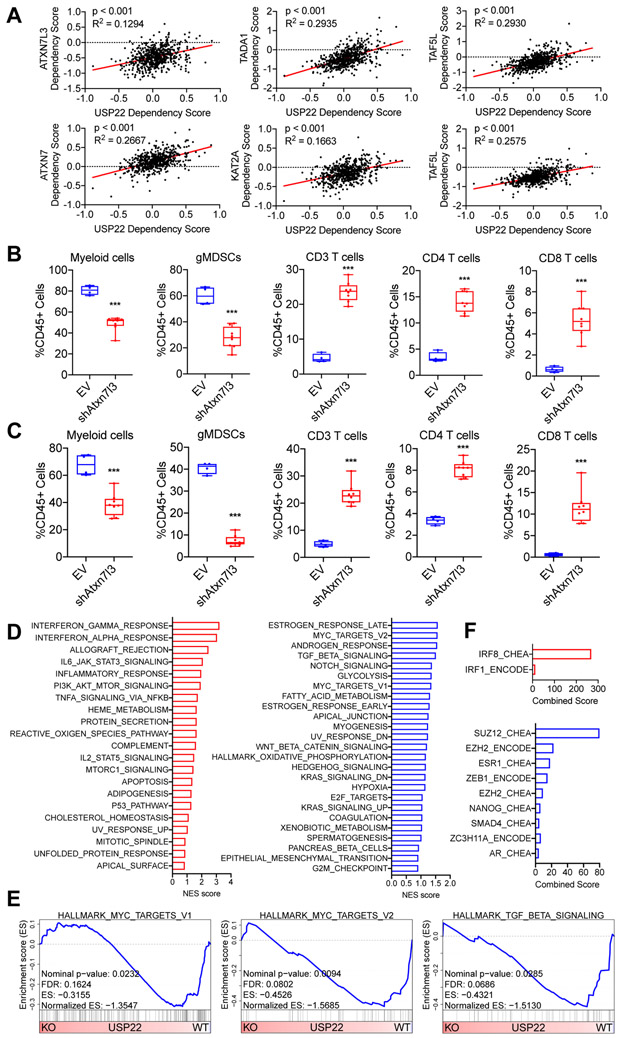
USP22 is a major component of the SAGA/STAGA complex, a multiprotein complex involved in transcriptional regulation (13). As the major deubiquitylase in the deubiquitylation module of this complex, USP22 is thought to regulate transcription by controlling ubiquitin levels of two histones – H2A and H2B – as well as other transcription factors including c-MYC, thereby regulating their stabilization and/or activity (13,19-24). Analyzing the data from the Cancer Dependency Map Portal (DepMap), we found USP22 shared dependencies with other factors in the deubiquitylation module of the SAGA/STAGA complex, including ATXN7, ATXN7L3, KAT2A, TADA1, TAF5L, and TAF6L (Figure 4A). To further explore whether these other components of the SAGA/STAGA complex regulated the immune TME, we used short-hairpin RNA (shRNA) to generate 6419c5 Atxn7l3-knockdown cell lines and performed flow analysis on the resulting tumors. Knockdown of Atxn7l3 resulted in a decrease in total myeloid cells and gMDSCs, as well as an increase in CD3+, CD4+, CD8+ T cells, NK cells, and DCs (Figure 4B, Supplementary Figure S3B-C). Likewise, Atxn7l3-knockdown tumors had a higher frequency of PD-1+ CD8+ T cells (Supplementary Figure S3C). These results were confirmed by knocking down Atxn7l3 in a second T cell–low clone, 6694c2 (Figure 4C, Supplementary Figure S3D-E). These data suggested that at least one other component of the deubiquitylation module of the nuclear SAGA/STAGA complex - ATXN7L3 – may regulate the immune TME of PDA tumors.
Figure 4. USP22 loss reprograms tumor cells towards a T cell–high transcriptional program.
(A) Dot plots showing the dependency scores for the indicated members of the SAGA/STAGA complex relative to USP22 across all tumor cell line samples in the Cancer Dependency Map. (B) Flow cytometric analysis of tumor-infiltrating immune cells in subcutaneously implanted 6419c5-EV– and 6419c5-Atxn7l3–knockdown tumors (2 independent knockdown cell lines, n=4-8 tumors analyzed per group in 1 experiment). (C) Flow cytometric analysis of tumor-infiltrating immune cells in subcutaneously implanted 6694c2-EV– and 6694c2-Atxn7l3–knockdown tumors (2 independent knockdown cell lines, n=4-8 tumors analyzed per group in 1 experiment). (D) Hallmark Gene Set Enrichment Analysis comparing differentially expressed genes in sorted Usp22-WT or Usp22-KO tumor cells (n=3-6 tumors analyzed per group). Gene sets enriched in Usp22-KO tumor cells are labeled in red (left); gene sets enriched in Usp22-WT tumor cells are labeled in blue (right). (E) Leading-edge plots from the GSEA analysis highlighting three Usp22-WT tumor cell-enriched gene sets: Hallmark_MYC_targets_V1, Hallmark_MYC_targets_V2, and Hallmark_TGF_beta_signaling. (F) Bar graphs showing predicted transcriptional regulators. Differentially expressed genes (p-adjusted<0.01 and absolute fold change >2) were used as input for EnrichR analysis (ENCODE and ChEA Consensus TFs from ChIP-X dataset). In (A), data are presented as dot plots with the red line showing the linear regression result. Correlation metrics are shown. In (B-C), data are presented as boxplots with horizontal lines and error bars indicating mean and range, respectively. Statistical differences between groups were calculated using Student’s unpaired t-test. *p<0.05, **p<0.01, ***p<0.001.
Based on these findings, we hypothesized that ablation of tumor cell–intrinsic USP22 would result in significant transcriptional changes in tumor cells, thereby contributing to USP22’s regulation of the immune TME. We used a YFP-lineage marker to sort the cancer cells from implanted tumors and then performed RNA-sequencing. Usp22-WT and Usp22-KO tumor cells exhibited global differences in transcription (Supplementary Figure S4A). Gene Set Enrichment Analysis (GSEA) of genes whose expression changed with Usp22 loss revealed an enrichment of hallmark gene sets associated with naturally arising T cell–low and T cell–high tumor cells (8). Specifically, Usp22-KO tumor cells were enriched for interferon response signatures, whereas Usp22-WT tumor cells were enriched for MYC, TGFβ, and cell cycle signatures (Figure 4D-E, Supplementary Figure S4B-D). We checked the expression of a group of factors regulating function and trafficking of myeloid cells and regulatory T cells and found that most of them were not significantly differentially expressed between Usp22-WT and Usp22-KO tumor cells. We found Csf3 was significantly decreased in Usp22-KO tumor cells. In our previous publication (8), we identified CSF3 as one tumor cell–intrinsic regulator promoting the establishment of a non-T cell–inflamed TME. One paper (12) has demonstrated that USP22 can regulate PD-L1 at the post-translational level to control tumor immunity in liver cancer. Here, we assessed the expression of both PD-L1 and MHCI on tumor cell surface with or without treatment of IFNγ and found that PD-L1 was not differentially expressed between Usp22-WT and Usp22-KO tumor cells (Supplementary Figure S4E). However, consistent with our RNA-seq result, Usp22-KO tumor cells had increased expression of MHCI on cell surface compared to Usp22-WT tumor cells (Supplementary Figure S4E). We then used the list of genes that were differentially expressed between Usp22-WT and Usp22-KO tumor cells as input for EnrichR (25,26). EnrichR is an analytical tool that uses information from the encyclopedia of DNA elements (ENCODE) and ChIP-X enrichment analysis (ChEA) datasets to identify potential transcriptional regulators. This analysis revealed an enrichment of an IRF8 signature in Usp22-KO tumor cells (Figure 4F, Supplementary Figure S4F). This analysis nominated SUZ12 and EZH2, two key components of the polycomb repressive complex 2 (PRC2 complex), as transcriptional regulators active in Usp22-WT tumor cells whose activity is lost following Usp22 deletion (Supplementary Figure S4F). Together, these evidences suggest potential mechanisms underlying USP22-mediated regulation of the immune TME of pancreatic cancer. Collectively, these data demonstrated that loss of tumor cell–intrinsic USP22 reprogramed the tumor cell transcriptome, resulting in a T cell–high phenotype at molecular and cellular levels.
DISCUSSION
Understanding the determinants of T-cell infiltration into tumors has important therapeutic implications. We and others have shown that regulatory factors operating in tumor cells play a central role in determining the makeup of their surrounding microenvironments (5,7,27). Here, we reported that USP22, a highly conserved component of the nuclear multiprotein SAGA/STAGA complex, is one such factor. These findings expand our understanding of tumor-intrinsic epigenetic factors that shape the TME (28,29) and are in line with a report describing a role for USP22 in regulating the immune microenvironment in liver cancer (12).
The SAGA/STAGA complex promotes transcription, an activity mediated in part through a deubiquitylation module that contains USP22 and its binding partners ATXN7L3, ATXN7, and ENY2 (23,30-33). We found that Atxn7l3 knockdown phenocopied Usp22 loss, supporting the notion that USP22-dependent effects on the immune microenvironment are mediated at least in part by the SAGA/STAGA complex. Deletion of Usp22 associated with the decreased expression of genes enriched for consensus target gene signatures of SUZ12 and EZH2, two components of the repressive PRC2 complex. These findings raise the possibility that the SAGA/STAGA and PRC2 complexes act coordinately to regulate the activation and repression of genes which control the immune makeup of PDA tumors.
Usp22 is overexpressed in multiple tumor types and has been shown to regulate cell cycle activity in association with cancer progression (13,14,34-37) and has been reported to play a role in antitumor immunity through the stabilization of PD-L1 in tumor cells (12). The present study suggests that transcriptional regulation may be another mechanism by which Usp22 controls the immune TME, thereby converting cells that are fully resistant to immunotherapy to a sensitive state. To date, no small molecule inhibitors of USP22 have been reported, but given mounting evidence in multiple contexts that USP22 promotes tumor progression, the development of such inhibitors would be desirable. Such compounds may be useful in the context of novel immunotherapy combinations, such as those described in this study and currently under clinical investigation for PDA ( NCT03214250).
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Stanger laboratories for technical help and scientific discussions. This work was supported by grants from the NIH (R01-CA229803), and the Blavatnik Family Foundation (JL).
Financial support: This work was supported by grants from the NIH (R01-CA229803 to BZS), and the Blavatnik Family Foundation Fellowship (JL).
Footnotes
Conflict of interest: Dr. Stanger has received research funding from Boehringer-Ingelheim.
REFERENCES
- 1.Ribas A, and Wolchok JD 2018. Cancer immunotherapy using checkpoint blockade. Science 359:1350–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanmamed MF, and Chen L 2018. A Paradigm Shift in Cancer Immunotherapy: From Enhancement to Normalization. Cell 175:313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, et al. 2014. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515:568–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spranger S, Luke JJ, Bao R, Zha Y, Hernandez KM, Li Y, Gajewski AP, Andrade J, and Gajewski TF 2016. Density of immunogenic antigens does not explain the presence or absence of the T-cell-inflamed tumor microenvironment in melanoma. Proc Natl Acad Sci U S A 113:E7759–E7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spranger S, and Gajewski TF 2018. Impact of oncogenic pathways on evasion of antitumour immune responses. Nat Rev Cancer 18:139–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morrison AH, Byrne KT, and Vonderheide RH 2018. Immunotherapy and Prevention of Pancreatic Cancer. Trends Cancer 4:418–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Markosyan N, Li J, Sun YH, Richman LP, Lin JH, Yan F, Quinones L, Sela Y, Yamazoe T, Gordon N, et al. 2019. Tumor cell-intrinsic EPHA2 suppresses anti-tumor immunity by regulating PTGS2 (COX-2). J Clin Invest 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Byrne KT, Yan F, Yamazoe T, Chen Z, Baslan T, Richman LP, Lin JH, Sun YH, Rech AJ, et al. 2018. Tumor Cell-Intrinsic Factors Underlie Heterogeneity of Immune Cell Infiltration and Response to Immunotherapy. Immunity 49:178–193 e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bayne LJ, Beatty GL, Jhala N, Clark CE, Rhim AD, Stanger BZ, and Vonderheide RH 2012. Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer. Cancer Cell 21:822–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byrne KT, and Vonderheide RH 2016. CD40 Stimulation Obviates Innate Sensors and Drives T Cell Immunity in Cancer. Cell Rep 15:2719–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winograd R, Byrne KT, Evans RA, Odorizzi PM, Meyer AR, Bajor DL, Clendenin C, Stanger BZ, Furth EE, Wherry EJ, et al. 2015. Induction of T-cell Immunity Overcomes Complete Resistance to PD-1 and CTLA-4 Blockade and Improves Survival in Pancreatic Carcinoma. Cancer Immunol Res 3:399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang X, Zhang Q, Lou Y, Wang J, Zhao X, Wang L, Zhang X, Li S, Zhao Y, Chen Q, et al. 2019. USP22 deubiquitinates CD274 to suppress anti-cancer immunity. Cancer Immunol Res. [DOI] [PubMed] [Google Scholar]
- 13.Melo-Cardenas J, Zhang Y, Zhang DD, and Fang D 2016. Ubiquitin-specific peptidase 22 functions and its involvement in disease. Oncotarget 7:44848–44856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, and McMahon SB 2008. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell 29:102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lv L, Xiao XY, Gu ZH, Zeng FQ, Huang LQ, and Jiang GS 2011. Silencing USP22 by asymmetric structure of interfering RNA inhibits proliferation and induces cell cycle arrest in bladder cancer cells. Mol Cell Biochem 346:11–21. [DOI] [PubMed] [Google Scholar]
- 16.Wculek SK, and Malanchi I 2015. Neutrophils support lung colonization of metastasis-initiating breast cancer cells. Nature 528:413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, et al. 2015. Proteomics. Tissue-based map of the human proteome. Science 347:1260419. [DOI] [PubMed] [Google Scholar]
- 18.Uhlen M, Zhang C, Lee S, Sjostedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, et al. 2017. A pathology atlas of the human cancer transcriptome. Science 357. [DOI] [PubMed] [Google Scholar]
- 19.Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, and Berger SL 2003. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev 17:2648–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim D, Hong A, Park HI, Shin WH, Yoo L, Jeon SJ, and Chung KC 2017. Deubiquitinating enzyme USP22 positively regulates c-Myc stability and tumorigenic activity in mammalian and breast cancer cells. J Cell Physiol 232:3664–3676. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Osdal T, Ho Y, Chun S, McDonald T, Agarwal P, Lin A, Chu S, Qi J, Hsieh YT, et al. 2014. SIRT1 activation by a c-MYC oncogenic network promotes the maintenance and drug resistance of human FLT3-ITD acute myeloid leukemia stem cells. Cell Stem Cell 15:431–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonnet J, Wang CY, Baptista T, Vincent SD, Hsiao WC, Stierle M, Kao CF, Tora L, and Devys D 2014. The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes Dev 28:1999–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lang G, Bonnet J, Umlauf D, Karmodiya K, Koffler J, Stierle M, Devys D, and Tora L 2011. The tightly controlled deubiquitination activity of the human SAGA complex differentially modifies distinct gene regulatory elements. Mol Cell Biol 31:3734–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang XY, Pfeiffer HK, Thorne AW, and McMahon SB 2008. USP22, an hSAGA subunit and potential cancer stem cell marker, reverses the polycomb-catalyzed ubiquitylation of histone H2A. Cell Cycle 7:1522–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44:W90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, and Ma’ayan A 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, and Stanger BZ 2019. The tumor as organizer model. Science 363:1038–1039. [DOI] [PubMed] [Google Scholar]
- 28.Sheng W, LaFleur MW, Nguyen TH, Chen S, Chakravarthy A, Conway JR, Li Y, Chen H, Yang H, Hsu PH, et al. 2018. LSD1 Ablation Stimulates Anti-tumor Immunity and Enables Checkpoint Blockade. Cell 174:549–563 e519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng D, Kryczek I, Nagarsheth N, Zhao L, Wei S, Wang W, Sun Y, Zhao E, Vatan L, Szeliga W, et al. 2015. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature 527:249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sowa ME, Bennett EJ, Gygi SP, and Harper JW 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell 138:389–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohler A, Zimmerman E, Schneider M, Hurt E, and Zheng N 2010. Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell 141:606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samara NL, Datta AB, Berndsen CE, Zhang X, Yao T, Cohen RE, and Wolberger C 2010. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science 328:1025–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, et al. 2008. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell 29:92–101. [DOI] [PubMed] [Google Scholar]
- 34.Liang JX, Ning Z, Gao W, Ling J, Wang AM, Luo HF, Liang Y, Yan Q, and Wang ZY 2014. Ubiquitinspecific protease 22induced autophagy is correlated with poor prognosis of pancreatic cancer. Oncol Rep 32:2726–2734. [DOI] [PubMed] [Google Scholar]
- 35.Ning Z, Wang A, Liang J, Xie Y, Liu J, Feng L, Yan Q, and Wang Z 2014. USP22 promotes the G1/S phase transition by upregulating FoxM1 expression via beta-catenin nuclear localization and is associated with poor prognosis in stage II pancreatic ductal adenocarcinoma. Int J Oncol 45:1594–1608. [DOI] [PubMed] [Google Scholar]
- 36.Schrecengost RS, Dean JL, Goodwin JF, Schiewer MJ, Urban MW, Stanek TJ, Sussman RT, Hicks JL, Birbe RC, Draganova-Tacheva RA, et al. 2014. USP22 regulates oncogenic signaling pathways to drive lethal cancer progression. Cancer Res 74:272–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu YL, Yang YM, Xu H, and Dong XS 2010. Increased expression of ubiquitin-specific protease 22 can promote cancer progression and predict therapy failure in human colorectal cancer. J Gastroenterol Hepatol 25:1800–1805. [DOI] [PubMed] [Google Scholar]
- 38.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, and Matrisian LM 2014. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 74: 2913–2921. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.