SUMMARY
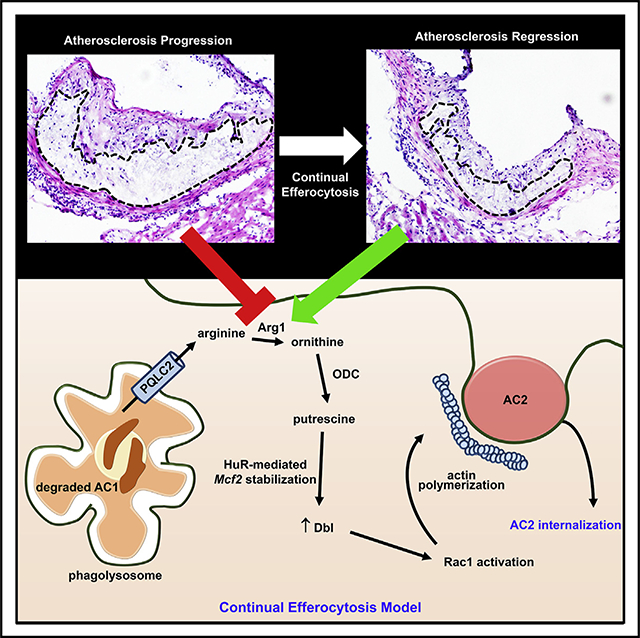
Continual efferocytic clearance of apoptotic cells (ACs) by macrophages prevents necrosis and promotes injury resolution. How continual efferocytosis is promoted is not clear. Here, we show that the process is optimized by linking the metabolism of engulfed cargo from initial efferocytic events to subsequent rounds. We found that continual efferocytosis is enhanced by the metabolism of AC-derived arginine and ornithine to putrescine by macrophage arginase 1 (Arg1) and ornithine decarboxylase (ODC). Putrescine augments HuR-mediated stabilization of the mRNA encoding the GTP-exchange factor Dbl, which activates actin-regulating Rac1 to facilitate subsequent rounds of AC internalization. Inhibition of any step along this pathway after first-AC uptake suppresses second-AC internalization, whereas putrescine addition rescues this defect. Mice lacking myeloid Arg1 or ODC have defects in efferocytosis in vivo and in atherosclerosis regression, while treatment with putrescine promotes atherosclerosis resolution. Thus, macrophage metabolism of AC-derived metabolites allows for optimal continual efferocytosis and resolution of injury.
In Brief
Macrophages engulf apoptotic cells (ACs) by continual efferocytosis to prevent inflammation and necrosis. Here, Yurdagul et. al. show that this process is linked to the metabolism of AC-derived arginine or ornithine to putrescine, which is required for the optimization of subsequent rounds of efferocytosis and proper resolution of atherosclerosis.
Graphical Abstract
INTRODUCTION
Impaired clearance, or “efferocytosis,” of apoptotic cells (ACs) by macrophages has dire consequences for the proper homeostasis of tissues and organisms, as uncleared ACs become secondarily necrotic and promote inflammation and tissue damage (Arandjelovic and Ravichandran, 2015; Vandivier et al., 2006; Yurdagul et al., 2018). This scenario has been implicated in chronic lung disease, neurodegenerative diseases, and atherosclerosis (Arandjelovic and Ravichandran, 2015; Khanna et al., 2010; Thorp and Tabas, 2009; Vandivier et al., 2006). Moreover, resolution and repair processes can also be compromised owing to loss of activation of efferocytosis receptor signaling pathways (Bäck et al., 2019; Cai et al., 2018). In advanced atherosclerosis, for example, impaired efferocytosis promotes plaque necrosis, inflammation, and impaired resolution, leading to plaque disruption, acute thrombosis, and tissue ischemia or infarction (Fredman and Tabas, 2017; Tabas and Lichtman, 2017; Yurdagul et al., 2018). Impaired efferocytosis in disease can have multiple causes (Cai et al., 2017; Kojima et al., 2016; Morioka et al., 2019), but defects in the ability of individual macrophages to efficiently internalize multiple ACs over consecutive rounds of engulfment, termed “continual efferocytosis,” may be particularly important in chronic inflammatory diseases, where ACs often far outnumber macrophages (Park et al., 2011; Wang et al., 2017).
Macrophages with a pro-resolving phenotype execute efferocytosis more efficiently than inflammatory macrophages (Xu et al., 2006). Furthermore, macrophage phenotypes are linked to their metabolic state (Saha et al., 2017). In general, proinflammatory macrophages have higher rates of glycolysis, whereas pro-resolving macrophages acquire more of their energy through oxidative phosphorylation (Saha et al., 2017). However, much less is known about the role of amino acid metabolism in macrophage biology, and there have been no links to efferocytosis. In inflammatory macrophages, arginine is catalyzed by inducible nitric oxide synthase (iNOS) to generate nitric oxide (NO), while in pro-resolving mouse macrophages, arginine is converted by arginase 1 (Arg1) into the polyamine precursor, ornithine. In human macrophages, Arg1 expression is low in certain settings, and thus, less is known about arginine metabolism in these cells.
We provide here evidence in vitro and in vivo that macrophage metabolism of arginine or ornithine derived from the engulfment of a first-encountered AC is necessary for optimal internalization of a subsequent AC in subsequent rounds of efferocytosis. Mechanistically, macrophages metabolize ornithine, either synthesized from AC-derived arginine or taken up directly from ACs, into the polyamine putrescine to drive increased expression of the Rac1 guanosine triphosphate (GTP)-exchange factor (GEF), Dbl. The increase in Dbl promotes Rac1 activation and thereby enhances actin-mediated internalization of a subsequent AC. Thus, pro-resolving macrophages link metabolism of AC-derived arginine and ornithine to their ability to optimally execute continual efferocytosis.
RESULTS
Arg1-Mediated Arginase Activity in Resolving-Type Macrophages Enhances Continual Efferocytosis in Mouse Macrophages
As an unbiased approach to initially understand how macrophages might metabolize AC-derived cargo in efferocytosis, liquid chromatography/tandem mass spectroscopy (LC-MS/MS) was used to examine two classes of metabolites—amino acids and acylcarnitines—in macrophages that had or had not ingested an AC. Bone-marrow-derived macrophages were incubated with ACs labeled with the fluorescent dye PKH67 dye for 45 min followed by AC removal. After 2 h of further incubation, the macrophages were detached and sorted by flow cytometry to isolate AC-positive (AC+) and AC-negative (AC−) macrophages, which were then subjected to LC-MS/MS analysis. Of the metabolites surveyed, arginine and ornithine showed the highest increases in AC+ versus AC− macrophages (Figure 1A; Table S1).
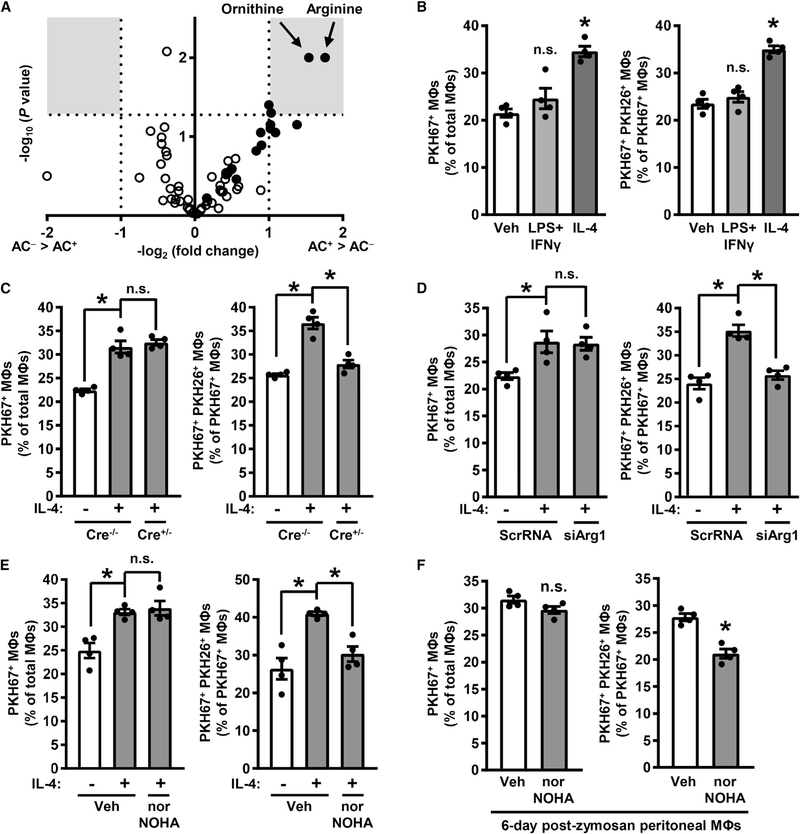
Figure 1. Arg1 Drives Continual Efferocytosis in Resolving-Type Mouse Macrophages.
(A) A volcano plot of AC− and AC+ mouse bone-marrow-derived macrophages from an efferocytosis assay followed by mass spectrometry analysis for acylcarnitines (open circles) and amino acids (closed circles). Gray boxes show areas of the plot with changes ≥2-fold at p < 0.05. Raw values are shown in Table S1 (n = 3 biological replicates).
(B) The percentages of PKH67+ macrophages versus total macrophages (left panel) and PKH67+ PKH26+ macrophages versus PKH67+ macrophages (right panel) as determined by epifluorescence microscopy of macrophages treated with either vehicle (Veh), LPS + IFNγ, or IL-4 in a two-step efferocytosis assay utilizing PKH67-labeled ACs in the first round of efferocytosis and PKH26-labeled ACs in the second round. Illustrative images are shown in Figure S1A.
(C) Single- and double-AC efferocytosis (as in B) by macrophages isolated from Arg1fl/fl (Cre−/−) and Arg1fl/fl Lysz2-Cre+/− (Cre+/−) mice and then treated for 24 h with vehicle or IL-4.
(D) Single- and double-AC efferocytosis in macrophages transfected with scrambled RNA (ScrRNA) or siArg1 and then treated 24 h with vehicle or IL-4.
(E) Single- and double-AC efferocytosis in macrophages that were treated with vehicle or IL-4 and then with vehicle or 500 μM nor-NOHA for 1 h before the addition of the first AC.
(F) Single- and double-AC efferocytosis in peritoneal macrophages harvested from mice 6 days after i.p. injection with 0.1 mg of zymosan A and then incubated for 1 h with vehicle or 500 μM nor-NOHA.
All values are means ± SEM; *p < 0.05; n.s., not significant; n = 3–4 biological replicates.
Arginine can be metabolized to ornithine by Arg1 in pro-resolving macrophages, whereas arginine is predominantly metabolized to NO by iNOS in inflammatory macrophages (Rath et al., 2014). Thus, to explore the role of arginine metabolism, we first verified that macrophages treated with interleukin-4 (IL-4), as a model of resolving-type macrophages, expressed high levels of Arg1, while those treated with LPS and interferon γ (IFNγ), as a model of inflammatory-type macrophages, expressed high levels of iNOS (Figure S1A). We then assessed the ability of macrophages treated with vehicle, lipopolysaccharide (LPS) + IFNγ, or IL-4 to ingest one AC or, as a model of continual efferocytosis, two sequential ACs. For this purpose, macrophages were first incubated for 45 min with ACs labeled with PKH67 (green) and then, after AC removal and a 120-min interval, incubated with a second round of ACs labeled with PKH26 (red). Macrophages that had ingested only a PKH67-labeled AC (single efferocytosis) were distinguished from those ingesting both PKH67- and PKH26-labeled ACs (continual efferocytosis) by epifluorescence microscopy. As expected for resolving-type macrophages (Chinetti-Gbaguidi et al., 2015; Dalli and Serhan, 2012; Korns et al., 2011; Nepal et al., 2019; Ramon et al., 2016; Sun et al., 2015; Zhong et al., 2018), IL-4-treated macrophages displayed a higher level of single and continual efferocytosis than other types of macrophages (Figures 1B and S1B). Next, focusing on IL-4-treated resolving-type macrophages, we tested if arginine metabolism by Arg1 was linked to single-AC or double-AC efferocytosis. Macrophages were isolated from Arg1fl/fl (Arg1-WT [wild-type]) and Arg1fl/fl Lyz2-Cre+/− (myeloid-Arg1-KO [knockout]) mice, treated with IL-4, and then subjected to the sequential AC uptake assay. Arg1 deletion (documented in Figure S1C) had no effect on single-AC efferocytosis but inhibited double-AC efferocytosis to levels seen in Arg1-WT macrophages not treated with IL-4 (Figure 1C). Similar results were obtained when macrophages were transfected with siArg1 (Figures 1D and S1D) or treated with the Arg1 inhibitor nor-NOHA (Figure 1E); when the first AC was labeled with a pH-sensitive dye to document delivery of the first AC to lysosomes (Figures S1E and S1F), or when the macrophages were initially treated with the other major Th2 cytokine, IL-13, instead of IL-4 (Figure S1G).
To examine macrophages isolated after a setting of physiological resolution, we harvested peritoneal macrophages from mice 6 days after injecting them intraperitoneally (i.p.) with 0.1 mg of the yeast cell wall component zymosan A (Newson et al., 2014; Proto et al., 2018). Exudate macrophages from the zymosan-A-injected mice had higher Arg1 expression (Figure S1H) and arginase activity compared with resident macrophages from control PBS-injected mice, with no change in NO content (Figure S1I). As with IL-4-treated bone-marrow-derived macrophages, Arg1 inhibition in exudate macrophages suppressed double-AC efferocytosis but not single-AC efferocytosis (Figure 1F). Thus, Arg1 activity in resolving-type macrophages optimizes uptake of a second AC in a sequential two-AC efferocytosis assay.
Arg1-Mediated Arginase Activity in Macrophages Enables Optimal Efferocytosis In Vivo
After mice are injected with dexamethasone, there is robust thymocyte apoptosis, followed by AC clearance by thymic macrophages. Importantly, there is a high AC:macrophage ratio, a scenario in which continual efferocytosis is important (Park et al., 2011). Consistent with this point, Mac2+ macrophages from thymi of dexamethasone-treated mice had 3-fold greater Arg1 expression than thymic macrophages of vehicle-treated mice (Figure 2A). We then compared efferocytosis by thymic macrophages in Arg1-WT (Cre−/−) and myeloid-Arg1-KO (Cre+/−) mice 18 h after i.p. injection with PBS or dexamethasone. As expected, dexamethasone-treated control Cre−/− mice had marked reductions in thymus weight and cellularity owing to the coupled processes of thymocyte apoptosis and efferocytosis (Figures S2A and S2B; Cre−/− groups). In contrast, thymus weight and cellularity were not decreased in dexamethasone-treated Cre+/− mice (Figures S2A and S2B; Cre+/− groups). Consistent with impaired efferocytosis in Cre+/− thymi, the number of annexin V+ ACs was greater in Cre+/− versus Cre-−/− thymi (Figure 2B). More importantly, the thymi of the Cre+/− cohort had a lower ratio of macrophage-associated TUNEL+ ACs to free ACs, with no difference in macrophage number, indicating impaired efferocytosis (Figures 2C and S2C) Moreover, the number of annexin V+ ACs was similar in Cre+/− versus Cre-−/− 4 h after dexamethasone injection (Figure S2D), indicating that time is needed for continual efferocytosis to show its effect in vivo and that myeloid Arg1 deficiency does not affect thymocyte apoptosis per se. Indeed, thymocytes isolated from Cre−/− and Cre+/− mice were equally susceptible to dexamethasone-induced apoptosis in vitro (Figure S2E). Thus, macrophage Arg1 is necessary for optimal continual efferocytosis in a setting where the AC burden is high.
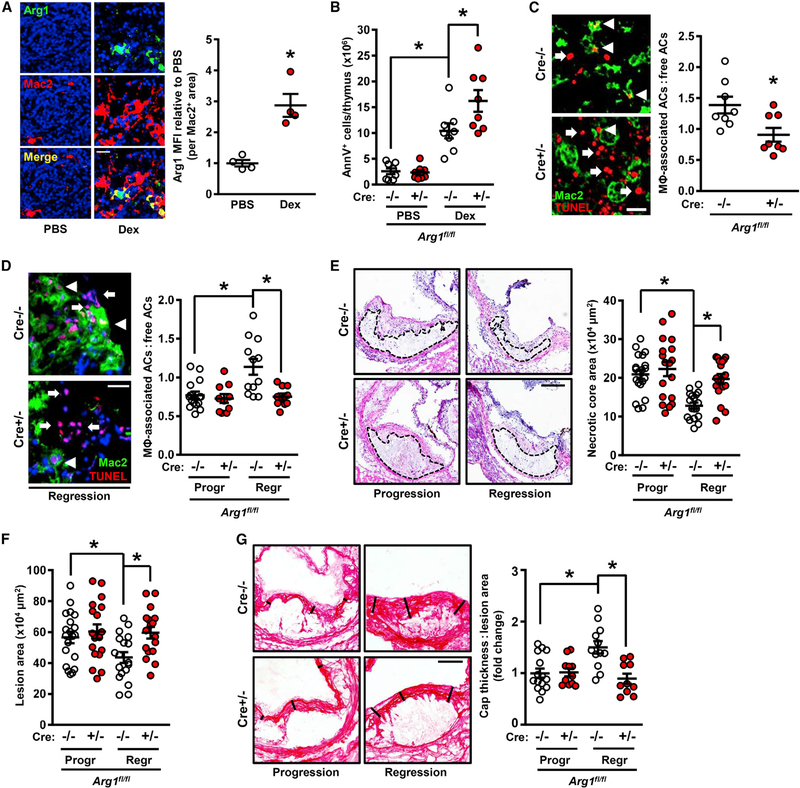
Figure 2. Myeloid-Arg1 Deletion Impairs Efferocytosis In Vivo.
(A) Arg1 immunostaining with quantification of thymic sections from mice 18 h after i.p. injection with PBS or dexamethasone (Dex) (n = 4 mice per group). Scale bar, 50 μm.
(B) Annexin V+ cells in thymi from myeloid-Arg1 WT (Cre−/−) and KO (+/−) mice harvested 18 h after injection with PBS or dexamethasone (n = 8 mice per group).
(C) Quantification of efferocytosis (macrophage-associated ACs:free ACs ratio) in thymic sections from the mice in (B) (n = 8 per group). Arrowheads and arrows depict macrophage-associated and free TUNEL+ cells, respectively.
(D–G) Arg1fl/fl Ldlr−/− (Cre−/−) and Arg1fl/fl Lysz2-Cre+/− Ldlr−/− (Cre+/−) mice placed on atherosclerosis progression (Progr) or regression (Regr) protocols were assayed for the following parameters: (D) efferocytosis (scale bar, 40 μm), (E and F) aortic root (E) necrotic core and (F) lesion areas (n = 17–20 mice per group; scale bar, 200 μm), and (G) collagen cap thickness (n = 10–15 mice per group; scale bar, 100 μm).
All values are means ± SEM; *p < 0.05; n.s., not significant.
Efferocytosis in general and continual efferocytosis in particular are important processes during atherosclerosis, where efficient clearance of the large number of ACs in atherosclerotic lesions is needed to prevent the accumulation of secondarily necrotic cells (Yurdagul et al., 2018). As plaques progress, however, efferocytosis becomes impaired due to changes in macrophage processes required for efferocytosis as well as recognition signals on ACs, leading to the clinically dangerous feature of plaque necrosis (Fredman and Tabas, 2017; Linton et al., 2016). Interestingly, plaque macrophages in progressing atherosclerotic lesions have been reported to express low levels of Arg1 (Teupser et al., 2006; Thomas et al., 2007). We verified this finding by showing that immunoreactive macrophage Arg1 was reduced in advanced versus early atherosclerotic lesions in both humans and mice (Figures S2F and S2G). In contrast, lesional macrophages in regressing atherosclerotic lesions have much higher Arg1 expression (Feig et al., 2012). We therefore hypothesized that Arg1 would play a more important role in efferocytosis in regressing lesions than in progressing lesions. In support of this notion, hematopoietic-Arg1 deficiency was shown not to affect lesion area in progressing atherosclerotic lesions in Western diet (WD)-fed Ldlr−/− mice (Ren et al., 2017).
To test the role of myeloid Arg1 in efferocytosis in progressing versus regressing lesions, lethally irradiated Ldlr−/− mice were transplanted with Arg1fl/fl Lyz2-Cre−/− or Arg1fl/fl Lyz2-Cre+/− bone marrow cells and then placed on a WD for 16 weeks (progression model). To induce regression, some of the 16-week WD-fed mice were switched to a normal chow diet for an additional 5 weeks while simultaneously receiving a helper-dependent adenovirus containing the human LDLR gene (HDAd-LDLR) to further reduce plasma LDL-cholesterol (regression model) (Willecke et al., 2015). This virus is non-integrating and primarily targets the liver, restoring expression of the LDL receptor in hepatocytes to levels that are sufficient to reduce plasma cholesterol (Table S2). Blood glucose; body weight; and blood leukocytes, neutrophils, monocytes, lymphocytes, eosinophils, basophils, Ly6Chigh, and Ly6Clow cells were similar in the Cre−/− and Cre+/− groups in both the progression and regression models (Table S2). Moreover, in accordance with the Arg1 expression data cited above, macrophage Arg1 immunostaining was much higher in the lesions of the Cre−/− regression versus progression cohort, and as validation of the Cre-lox model, lesional Arg1 was not detectable in Mac2+ regions in the Cre+/− regression cohort (Figure S3A). Further, as expected from the inflammatory nature of progressing atherosclerosis, macrophage iNOS expression and lesional NO were higher in the progression cohort. Although it was theoretically possible that Arg1 deletion in myeloid cells would increase iNOS and/or lesional NO, no changes were observed (Figures S3A and S3B).
In control mice, 5 weeks of lowering plasma cholesterol promoted increased lesional efferocytosis (Figure 2D; Cre−/− data). Most importantly, the increase in efferocytosis in regressing lesions was abrogated by myeloid-Arg1 deletion (Figure 2D). Acritical consequence of defective efferocytosis in atherosclerotic lesions is plaque necrosis, and plaque necrosis was reduced in the regressing lesions of Cre−/− mice, but not Cre+/− mice (Figure 2E). Overall lesion area was also greater in the regressing lesions of Cre+/− mice (Figure 2F). Fibrous cap thickness, which is a sign of resolution and plaque stability, paralleled the changes in lesional efferocytosis (Figure 2G), consistent with the concept that efferocytosis and resolution are coupled processes (Fredman et al., 2016; Fredman and Tabas, 2017). Finally, IL-1β and tumor necrosis factor α (TNFα) were reduced in regressing versus progressing lesions, and consistent with the idea that reduced necrosis should dampen inflammation, the decrease in these two cytokines in regressing lesions was partially abrogated by myeloid-Arg1 deletion (Figures S3C and S3D). Collectively, these data indicate that targeting macrophage Arg1 in regressing atherosclerotic lesions causes defects in efferocytosis, which is associated with impaired lesion regression. Thus, in two in vivo models where AC burden is high and Arg1-expressing macrophages are present, macrophage Arg1 plays a key role in efferocytosis.
Apoptotic Cell-Derived Arginine Drives ODC-Mediated Putrescine Production to Promote Continual Efferocytosis
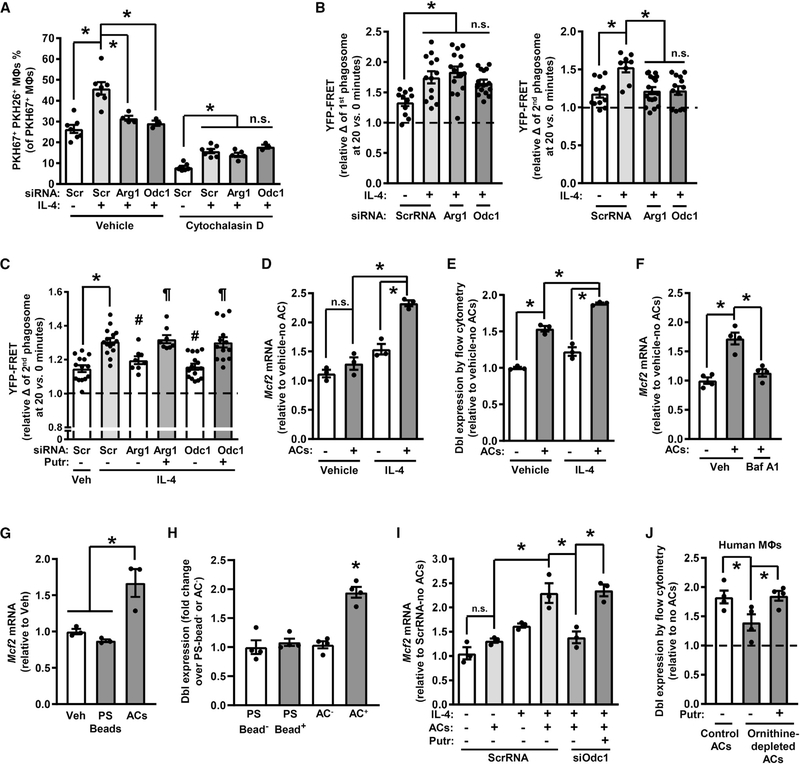
Arg1+ macrophages can metabolize arginine into ornithine, and ornithine, like arginine, was highly elevated in AC+ versus AC− macrophages (Figure 1A; Table S1). Ornithine can be metabolized into citrulline via ornithine transcarbamoylase (OTC) as part of the urea cycle or into the polyamine putrescine via ornithine decarboxylase (ODC; gene name Odc1) (Kierszenbaum et al., 1987; Tarasenko et al., 2015). To determine if one or both of these pathways promotes continual efferocytosis, single- and double-AC uptake assays were conducted using control macrophages versus those with silenced Otc or Odc1. While there were no effects of Otc or Odc1 silencing on the uptake of a single AC, Odc1 silencing, but not Otc silencing, suppressed the efferocytosis of two ACs (Figures 3A and S4A). These data suggested that the polyamine pathway rather than the urea cycle was involved in continual efferocytosis.
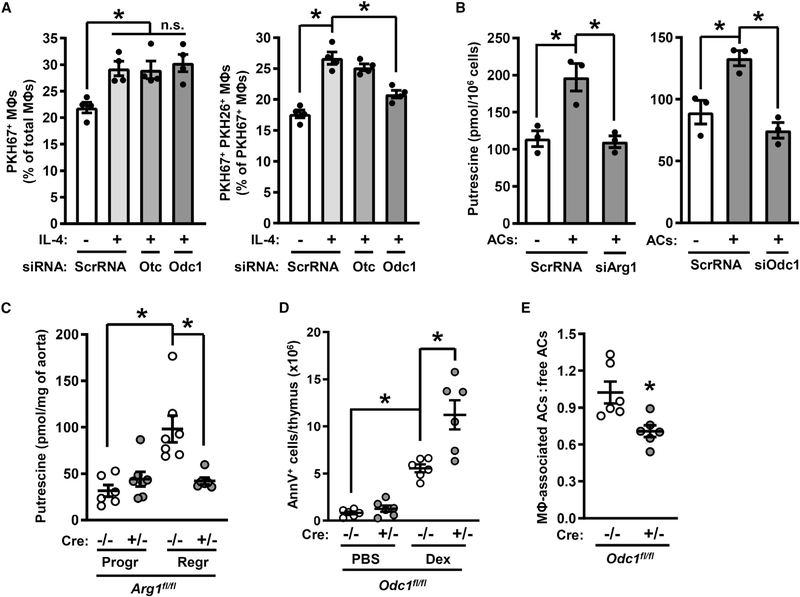
Figure 3. Deletion of Odc1 Causes Defective Continual Efferocytosis.
(A) Efferocytosis by ScrRNA-, siOtc-, or siOdc1-transfected macrophages treated with vehicle or IL-4 (n = 4 biological replicates).
(B) Putrescine content of IL-4-treated macrophages that were incubated with PKH67-labeled ACs for 45 min and then after 2 h sorted into AC− and AC+ populations (n = 3 biological replicates).
(C) Putrescine content of aortas from Arg1fl/fl Ldlr−/− (Cre−/−) and Arg1fl/fl Lysz2-Cre+/− Ldlr−/− (Cre+/−) mice that had been placed on atherosclerosis progression or regression protocols (n = 6–7 mice per group).
(D and E) Annexin V+ cells and efferocytosis in thymi harvested 18 h after injection of PBS or dexamethasone into C57BL/6J mice reconstituted with bone marrow from Odc1fl/fl (Cre−/−) or Odc1fl/fl Lysz2-Cre+/− (Cre+/−) bone marrow (n = 6 mice per group).
All values are means ± SEM; *p < 0.05; n.s., not significant.
Accordingly, we examined if cellular polyamines levels rose in macrophages after AC ingestion. Vehicle-treated or IL-4-treated macrophages were sorted into AC− and AC+ populations as described above and analyzed for putrescine, spermidine, and spermine content. While IL-4 alone and ACs alone cause a modest increase in putrescine, the combination of these treatments caused a marked increase in putrescine (Figure S4B, left graph). Spermidine and spermine levels were also higher in AC+ versus AC− macrophages, but these polyamines were not elevated by IL-4 treatment (Figure S4B, middle and right graphs). Most importantly, silencing Arg1 or Odc1 prevented the increase in putrescine in AC+ macrophages (Figure 3B) but had no significant effect on spermidine or spermine in these cells (Figures S4C and S4D). The contribution of AC-derived putrescine was minimal, as putrescine levels in ACs were ~7 times lower than that in IL-4-treated macrophages (Figure S4E). Further, treatment of IL-4 macrophages with N1,N11-diethylnorspermine (DENSPM), which induces a polyamine catabolizing enzyme and thereby reduces all three polyamines in cells (Alhonen et al., 1998), did not affect the uptake of a first AC (Figures S4F and S4G). We then re-examined the lesions of the progression-regression atherosclerosis experiment and found that putrescine content was higher in regressing versus progressing lesions and that myeloid Arg1 deficiency abolished the increase in putrescine in regressing lesions (Figure 3C). These combined data support the presence of an Arg1-ODC-putrescine pathway in resolving-type macrophages.
To test the role of myeloid ODC in efferocytosis in vivo, C57BL/6J mice were irradiated and then transplanted with the bone marrow of Odc1fl/fl (Cre−/−) or Odc1fl/fl Lysz2-Cre+/− (Cre+/−) mice. 5 weeks later, after chimerism was established, the mice were injected i.p. with PBS or dexamethasone, and the thymi were examined 18 h later. ODC expression was evident in the macrophages in the thymi of Cre−/− mice, but not in thymi of Cre+/− mice (Figure S4H), with similar results in bone-marrow-derived macrophages from these mice (Figure S4I). Similar to the findings with Arg1-KO mice, deletion of Odc1 in myeloid cells impaired dexamethasone-induced decreases in both thymus weight and cellularity (Figures S4J and S4K) and caused an increase in the accumulation of annexin V+ cells (Figure 3D). Most importantly, myeloid-specific deletion of Odc1 decreased efferocytosis by thymic macrophages (Figure 3E) despite similar macrophage numbers in the thymi of the two cohorts of mice (Figure S4L). Thus, myeloid ODC, like myeloid Arg1, is required for optimal efferocytosis in vivo.
As further evidence of an ODC-putrescine-continual efferocytosis pathway, the addition of exogenous putrescine, but not spermidine or spermine, restored continual efferocytosis to Odc1-deficient macrophages (Figure 4A). Moreover, silencing the spermidine-synthesizing enzyme spermidine synthase (gene name Srm) showed no effect on continual efferocytosis in IL-4-treated macrophages (Figures 4B and S5A). We next examined the pathway in zymosan-induced peritonitis, which, as noted above, is a model of inflammation resolution at 6 days post zymosan injection. Zymosan-injected mice were treated with nor-NOHA to inhibit arginase activity, or with vehicle control, at 24 h and 2 h before sacrifice. The two nor-NOHA injections were sufficient to reduce arginase activity by ~60% without affecting NO production (Figure S5B). Some mice were also dosed with putrescine 3 h prior to sacrifice. 1 h before sacrifice, mice were injected i.p. with a 1:1 ratio of PKH67 (green)- and CellVue Claret (far-red)-labeled apoptotic neutrophils (Figure S5C). At sacrifice, peritoneal exudate cells were collected and analyzed by flow cytometry for green and far-red fluorescence. Consistent with the in vitro data, nor-NOHA treatment reduced the percentage of macrophages labeled with both fluorophores, and putrescine rescued this defect (Figures 4C and S5D). Interestingly, the defect and rescue of continual efferocytosis in this in vivo model was substantially greater than what was observed in vitro, suggesting that relatively small changes observed in cultured cells may reflect larger effects in relevant settings in vivo. Changes observed in vivo reflect cumulative effects over time and may also be influenced by closer AC-macrophage contact and other niche effects that are not present in cell culture.
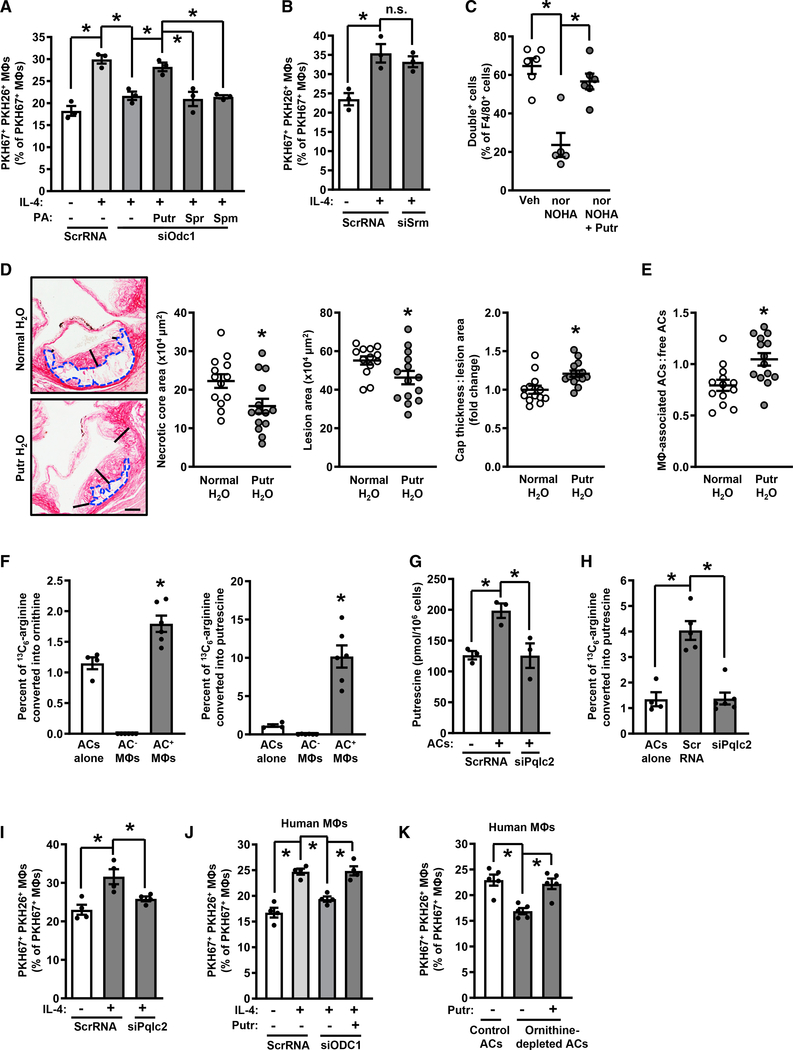
Figure 4. Metabolism of AC-Derived Arginine into Putrescine Enables Continual Efferocytosis.
(A) Efferocytosis by ScrRNA- and siOdc1-transfected macrophages treated with vehicle or IL-4 and with the indicated polyamines (100 μM) added with the first AC (n = 3 biological replicates).
(B) Efferocytosis by ScrRNA- and siSrm-transfected macrophages treated with vehicle or IL-4 (n = 3 biological replicates).
(C) Double-cell efferocytosis by peritoneal lavage macrophages from mice injected i.p. with 0.1 mg of zymosan A for 6 days and then with 2 × 106 PKH67-labeled and 2 × 106 CellVue Claret-labeled apoptotic neutrophils 1 h before sacrifice (n = 5–6 mice per group).
(D and E) Necrotic core area (blue dashed outline), total lesion area, fibrous cap thickness (indicated by black lines), and efferocytosis in aortic root lesions of Ldlr−/− mice fed a Western diet for 16 weeks ± supplementation of drinking water with 3 mM putrescine starting at 8 weeks (n = 13–14 mice per group). Scale bar, 200 μm.
(F) Percent of 13C5-ornithine and 13C4-putrescine of total 13C label in sorted PKH67+ macrophages that had been incubated with 13C6-arginine-loaded, PKH67-labeled apoptotic Jurkat cells (n = 4–6 biological replicates).
(G) Putrescine content of ScrRNA- and siPqlc2-transfected macrophages (n = 3 biological replicates).
(H) Percent of 3C4-putrescine of total 13C label in sorted PKH67+ macrophages that had been transfected with ScrRNA or siPqlc2 (n = 4–6 biological replicates).
(I) Double-AC efferocytosis by ScrRNA- and siPqlc2-transfected macrophages that were treated with vehicle or IL-4 (n = 4 biological replicates).
(J) Double-AC efferocytosis by ScrRNA- and siODC1-transfected HMDMs that were treated with vehicle or IL-4, with 100 μM putrescine added with the first AC to one group of siODC1-treated cells (n = 4 biological replicates).
(K) Double-AC efferocytosis by IL-4-treated HMDMs that were incubated first with PKH67-labeled and then with PKH26-labeled control or ornithine-depleted ACs, with putrescine added with the first AC to one group as indicated (n = 5 biological replicates).
All values are means ± SEM; *p < 0.05; n.s., not significant.
As Arg1 expression and putrescine content are both low in progressing versus regressing lesions, we sought to determine if putrescine supplementation in mice with pre-established atherosclerosis could enhance efferocytosis in lesional macrophages and reduce atherosclerosis progression. Accordingly, Ldlr−/− mice were fed a WD for 8 weeks with ad libitum access to normal drinking water. After 8 weeks of WD-feeding, mice were continued on WD with normal drinking water or water supplemented with 3 mM putrescine for 2 or 8 more weeks. Aortic putrescine content after 2 weeks of treatment was significantly elevated in the aorta of mice given the putrescine-supplemented water, with no change in spermidine or spermine content (Figure S5E). After 8 weeks of treatment, the atherosclerotic lesions of the mice given putrescine-supplemented water showed reduced necrotic core area and lesion size and thicker fibrous caps (Figure 4D). Most importantly, putrescine supplementation enhanced efferocytosis in lesional macrophages (Figure 4E). Whereas body weight; total cholesterol; blood glucose; and blood leukocytes, neutrophils, lymphocytes, eosinophils, and basophils were not affected by putrescine, blood monocytes were modestly reduced (Figures S5F–S5N). However, when a large subgroup of putrescine-supplemented and control mice with overlapping monocyte counts were compared, the relevant atherosclerotic endpoints remained significantly different, and lesional efferocytosis was still elevated (Figures S5O–S5S). These data provide further in vivo data for the role of putrescine in efferocytosis.
To determine the source of arginine used to fuel putrescine synthesis in AC+ macrophages, Jurkat cells were cultured for 3 days in the presence of heavy 13C6-arginine using the stable isotope labeling with amino acids in cell culture (SILAC) method. The Jurkat cells were then induced to undergo apoptosis by UV-irradiation, labeled with PKH67, and then added to IL-4-treated macrophages. After 45 min, unbound ACs were removed, and the macrophages were cultured for an additional 120 min. 13C5-ornithine and 13C4-putrescine in sorted AC− and AC+ macrophages, as well as in the ACs themselves, were then quantified by reversed phase ultra-high-performance LC-MS. The data show enrichment of label in both ornithine and putrescine in the AC+ macrophages compared with the original ACs (Figure 4F), indicating conversion of AC-derived arginine into macrophage ornithine and putrescine. Because AC-derived arginine would have to be transported from the phagolysosome into the cytosol for metabolism by Arg1 and Odc1, we tested whether silencing the lysosomal arginine transporter Pqlc2 (Liu et al., 2012) would suppress the efferocytosis-induced increases in macrophage putrescine, conversion of heavy 13C6-arginine into 13C4-putrescine, and continual efferocytosis. Pqlc2-silenced macrophages showed significant defects in putrescine content after AC ingestion, conversion of AC-derived 13C6-arginine into putrescine, and continual efferocytosis (Figures 4G–4I and S6A). Similar findings were evident when the lysosomal vacuolar ATPase inhibitor bafilomycin A1 was used to block phagolysosomal AC degradation (Figures S6B and S6C). Thus, AC-protein degradation generates arginine to drive an ornithine-putrescine pathway in mouse macrophages that enhances continual efferocytosis.
IL-4 treatment does not increase Arg1 in human monocyte-derived macrophages (HMDMs) in cell culture (Raes et al., 2005), and consistent with this feature, the arginase inhibitor nor-NOHA failed to block continual efferocytosis in these cells (Figure S6D). However, as ACs contain ornithine, we wondered whether HMDMs might utilize AC-derived ornithine for the ODC-putrescine-continual efferocytosis pathway. Consistent with this idea, siODC1 suppressed continual efferocytosis in IL-4-treated HMDMs, and efferocytosis could be restored in these cells by adding back putrescine (Figures 4J and S6E). Next, to test the role of AC ornithine, we incubated HMDMs with ornithine-depleted ACs. Ornithine depletion was achieved by incubating Jurkat cells in arginine-free media containing dialyzed fetal bovine serum (FBS) and nor-NOHA prior to induction of apoptosis (Figure S6F). Whereas putrescine content was increased in IL-4-treated macrophages that had engulfed control ACs, this was not the case with macrophages that had engulfed ornithine-depleted ACs (Figure S6G). Most importantly, continual efferocytosis of ornithine-depleted ACs was reduced compared to that of control ACs, and efferocytosis could be restored by adding back exogenous putrescine to macrophages exposed to ornithine-depleted ACs (Figure 4K). Thus, HMDMs utilize an alternative means to link AC metabolism to continual efferocytosis through the putrescine pathway. However, there may be settings in vivo where elevated Arg1 expression in human macrophages, e.g., in early atherosclerosis in humans (Thomas and Mattila, 2014), enables functionality of the arginine-Arg1-ornithine-putrescine pathway.
The Arg1-ODC-Putrescine Pathway Promotes Continual Efferocytosis by Increasing Dbl-Rac1-Mediated Second-AC Internalization
Efferocytosis is a multi-step process in which macrophages first bind ACs via specific efferocytosis receptors and then internalize the ACs (Erwig and Henson, 2008). We tested if the defect in second AC uptake in Arg1- or Odc1-silenced macrophages was due to a decrease in AC binding or AC internalization. For this purpose, the actin polymerization inhibitor, cytochalasin D, was added after the first round of efferocytosis to inhibit AC internalization. This protocol allows one to determine whether macrophage binding of the second AC, which is an actin-independent process, was intact or impaired. In the cytochalasin-D-treated cells, macrophage-AC association was similar among scrambled RNA, siArg1, and siOdc1 groups (Figure 5A), indicating that Arg1 and ODC promote second AC internalization, not binding.
Figure 5. The Arg1-ODC-Putrescine Pathway Drives the Expression of the Rac-GEF Dbl.
(A) Double-cell efferocytosis by ScrRNA-, siArg1-, or siOdc1-transfected macrophages that were treated with vehicle or IL-4, with some of the macrophages receiving 5 μM cytochalasin D 15 min before the addition of the second AC (n = 3–7 biological replicates).
(B) First AC and second AC phagosomal Rac1 activity (phagosome YFP:CFP fluorescence ratio [YFP-FRET]—20 min relative 0 min) in macrophages from Raichu-Rac FRET transgenic mice that were transfected with ScrRNA, siArg1, or siOdc1; treated with vehicle or IL-4; incubated with a first set of ACs for 45 min; and then, 2 h later, incubated with a second set of ACs (n = 11–17 cells per group, 2 plates of macrophages per condition).
(C) Second AC phagosomal Rac1 activity as in (B), except that some of the cells were treated with 100 μM putrescine with the first AC (n = 8–15 macrophages per group, 2 plates of macrophage per condition).
(D and E) (D) Mcf2 mRNA and (E) Dbl expression in control or IL-4-treated macrophages that were incubated ± ACs for 45 min and then 2 h later harvested (n = 3 biological replicates).
(F) Mcf2 mRNA in IL-4-treated macrophages that were pre-treated with bafilomycin A1 and incubated ± ACs for 45 min and then 2 h later harvested (n = 4 biological replicates).
(G and H) (G) Mcf2 mRNA and (H) Dbl expression in IL-4-treated macrophages that were incubated ± 6-μm phosphatidylserine-coated latex beads or ACs for 45 min and then 2 h later harvested (n = 3 biological replicates).
(I) Mcf2 mRNA in ScrRNA- or siOdc1-transfected macrophages that were treated with vehicle or IL-4, incubated ± ACs for 45 min with or without 100 μM putrescine, and then harvested 2 h later (n = 3 biological replicates).
(J) Dbl expression in IL-4-treated HMDMs that were incubated with control or ornithine-depleted ACs, with 100 μM putrescine added to one group (n = 4 biological replicates).
All values are means ± SEM. For (A), (B), and (D)–(H), *p < 0.05; n.s., not significant. For (C), *p < 0.05 for group 2 versus 1; #p < 0.05 for groups 3 and 5 versus group 2; and ¶p < 0.05 for groups 4 and 6 versus groups 2, 3, and 5.
The key cell biological process in AC internalization is actin remodeling around the forming phagosome, and previous studies have shown that this process requires the small GTPase Rac1 (Leverrier and Ridley, 2001). To determine if Arg1 and Odc1 were necessary for full Rac1 activation during continual efferocytosis, we used bone-marrow-derived macrophages isolated from transgenic mice expressing the Raichu Rac-fluorescence resonance energy transfer (FRET) biosensor (Johnsson et al., 2014). As expected, these macrophages showed a progressively increasing phagosomal YFP:CFP fluorescence ratio during the 20-min interval after AC addition, indicating increasing phagosomal Rac activity during this period (Figure S6H). Most importantly, this increase in YFP:CFP fluorescence ratio in the first AC phagosome, i.e., 20 min after first AC addition, was not affected by silencing Arg1 or Odc1, but the YFP:CFP fluorescence ratio in the second-AC phagosome, i.e., 20 min after second AC addition, was decreased by Arg1 and Odc1 silencing in a manner that could be rescued by adding exogenous putrescine (Figures 5B and 5C). These data suggest that Rac activation during second AC internalization, but not first AC internalization, is dependent on the Arg1-ODC-putrescine pathway.
Rac1 activation requires a GEF, and the specific GEF used depends on the cellular context (Marei and Malliri, 2017). Studies examining intestinal epithelial cell migration have shown a link between polyamines and the Rac GEF, Dbl (gene name Mcf2) (Ray et al., 2003, 2011). We therefore assayed Mcf2 mRNA expression by qRT-PCR and Dbl expression by flow cytometry in mouse macrophages treated with or without IL-4 and in the absence or presence of ACs. The combination of IL-4 and ACs led to a robust increase in both Mcf2 mRNA and Dbl protein (Figures 5D, 5E, and S6I). Increased Mcf2 expression under these conditions was blocked by inhibiting AC degradation with bafilomycin (Figure 5F) and could not be induced by cargo-free phosphatidylserine-coated beads (Figures 5G and 5H). Furthermore, silencing ODC with siOdc1 also blocked Mcf2 expression, which was restored by putrescine (Figure 5I). Alternative activation of cultured macrophages can also be achieved with IL-13 (Martinez et al., 2009). Accordingly, we found that the AC-Arg1-Mcf2 pathway was evident in macrophages treated with IL-13, as Mcf2 was increased when IL-13-treated macrophages were exposed to ACs, and this increase was blocked by inhibiting Arg1 with nor-NOHA (Figure S6J). We then tested relevance to the putrescine pathway in human macrophages, which as shown in the previous section, is activated by AC-derived ornithine. Dbl expression was impaired in human macrophages incubated with ornithine-depleted ACs compared with cells incubated with control ACs, and Dbl expression was completely restored by adding back putrescine (Figure 5J).
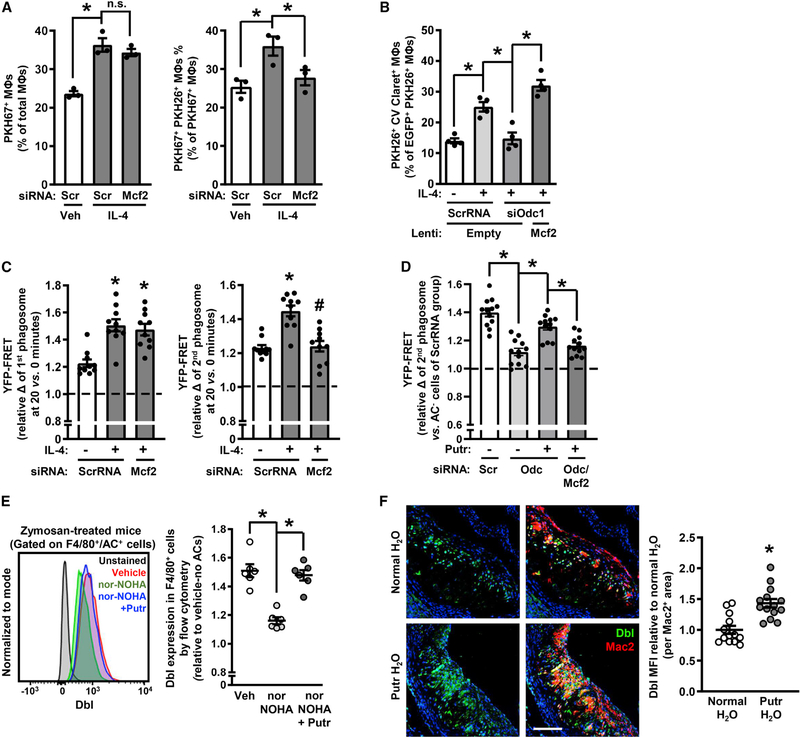
To determine a causative role for Dbl, we treated macrophages with siMcf2 and assayed continual efferocytosis. Dbl silencing in both IL-4- and IL-13-treated macrophages decreased second AC internalization, but not first AC internalization (Figures 6A, S6K, and S6L). Furthermore, the reduction in continued efferocytosis that occurs with ODC silencing was prevented by transducing the macrophages with Mcf2-expressing lentivirus (Figure 6B). Further, silencing Dbl in macrophages decreased Rac activation in the second AC phagosome, but not in the first AC phagosome (Figure 6C), and putrescine was unable to rescue Rac activity in Odc1-silenced macrophages when Dbl was also silenced, demonstrating that the effect of putrescine on Rac activation is Dbl-dependent (Figure 6D).
Figure 6. Putrescine Promotes Continual Efferocytosis by Increasing Dbl Expression and Rac1 Activity.
(A) Single- and double-cell efferocytosis by ScrRNA- and siMcf2-transfected macrophages treated with vehicle or IL-4 (n = 3 biological replicates).
(B) Double-cell efferocytosis by ScrRNA- and siOdc1-transfected macrophages, with one group of the siOdc1-treated cells transduced with Mcf2 and all groups receiving IL-4 except for one group of the ScrRNA-treated cells (n = 4 biological replicates).
(C) First AC and second AC phagosomal Rac1 activity (20 min relative 0 min) of ScrRNA- and siMcf2-transfected macrophages treated with vehicle or IL-4 (n = 10 cells per group, 2 plates macrophages per condition).
(D) Second AC phagosomal Rac1 activity relative to AC− cells of ScrRNA-, siOdc1-, and Mcf2-transduced macrophages, with or without 100 μM putrescine added with the first AC (n = 12 cells per group, 2 plates macrophages per condition).
(E) Dbl expression in F4/80+ peritoneal macrophages 6 days after zymosan injection (n = 6 mice per group).
(F) Dbl immunostaining with quantification in aortic root cross-sections from the mice described in Figure 4D. Scale bar, 250 μm.
All values for are means ± SEM; *p < 0.05; n.s., not significant.
To assess the Arg1-putrescine-Dbl pathway in vivo, we turned to the 6-day zymosan peritonitis model. The data show that i.p. nor-NOHA treatment suppressed Dbl expression in exudate macrophages and that Dbl expression could be restored by i.p. putrescine treatment (Figure 6E). Furthermore, in line with the atherosclerosis data in Figure 2 and the above findings, Dbl expression in atherosclerotic lesional macrophages was higher in regressing lesions versus progressing lesions in WT-Arg1 Ldlr−/− mice, but this increase was not seen in myeloid-Arg1-KO Ldlr−/− mice (Figure S6M). Plaque macrophages from mice drinking putrescine-supplemented water also showed elevations in Dbl expression (Figure 6F). Collectively, these findings support the idea that the AC-arginine-ornithine-putrescine pathway activates Dbl-Rac1 to maximize continual efferocytosis.
Putrescine Augments Dbl Expression by Promoting HuR-Mediated Stability of Mcf2 mRNA
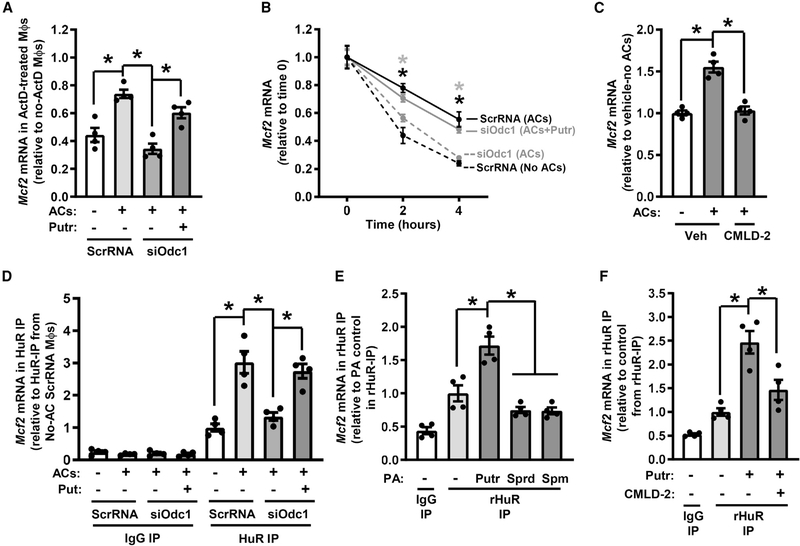
To determine the mechanism by which putrescine increases Mcf2, we first tested if the regulation of Mcf2 in IL-4-treated macrophages by ACs, siOdc1, and putrescine was due to increased Mcf2 transcription or increased Mcf2 mRNA stability. Although basal Mcf2 mRNA was decreased in actinomycin-D-treated IL-4 macrophages by approximately 60%, Mcf2 transcripts were still increased by ACs, decreased by siOdc1, and increased by putrescine treatment of Odc1-silenced cells (Figure 7A). These data suggested that Mcf2 regulation by the AC-ODC-putrescine pathway occurs primarily by affecting Mcf2 mRNA turnover. To directly test this idea, we conducted a 5-ethynyl uridine (EU)-labeled pulse-chase experiment. EU is an alkyne-modified nucleoside that is incorporated into nascent mRNA such that after removal of EU from the media, the decay of EU-labeled mRNAs can be quantified using a biotin-streptavidin pull-down protocol followed by qRT-PCR (Jao and Salic, 2008). The data show that the decay of Mcf2 was decreased by ACs, increased by siOdc1, and decreased by putrescine treatment of Odc1-silenced cells (Figure 7B). Thus, the AC-ODC-putrescine pathway augments the stability of Mcf2 mRNA.
Figure 7. Putrescine Promotes Mcf2 mRNA Stability.
(A) Relative Mcf2 mRNA in ScrRNA- and siOdc1-transfected macrophages treated with IL-4 and then incubated ± ACs, 100 μM putrescine, and/or 2 μg/mL actinomycin D (ActD) (n = 4 biological replicates).
(B) Mcf2 mRNA in biotinylated RNA pelleted from 5-EU-labeled macrophages that were transfected with ScrRNA or siOdc1, treated with IL-4, incubated for 45 min with or without ACs ± putrescine, and then harvested 2 h later (n = 4 biological replicates).
(C) Mcf2 mRNA in IL-4- and ActD-treated macrophages that were incubated with vehicle or 30 μM CMLD-2 for 30 min, incubated with or without ACs, and then harvest 2 h later (n = 4 biological replicates).
(D) Relative Mcf2 mRNA in anti-HuR or control precipitates from macrophages that were transfected with ScrRNA or siOdc1, treated with IL-4, incubated for 45 min with or without ACs ± putrescine, and then harvested 2 h later (n = 4 biological replicates).
(E) Relative Mcf2 mRNA in anti-HuR or control precipitates from a reaction mix in which recombinant HuR (rHuR, 10 nM), polyamines (100 nM for putrescine and spermine, 1 μM for spermidine), and poly(A)-RNA (8 μg/mL) from IL-4-treated macrophages were incubated for 24 h (n = 4 technical replicates).
(F) As in (E) but with the addition of a group treated with 350 nM CMLD-2 (n = 4 technical replicates).
All values are means ± SEM; *p < 0.05.
To identify one or more RNA-binding proteins that might be involved in Mcf2 transcript stabilization by the AC-ODC1-putrescine pathway, we conducted an in silico analysis of the 3′ UTR sequence of Mcf2 using RBPmap, a web-based server for mapping binding sites of RNA-binding proteins (Paz et al., 2014). We identified nine conserved, high-stringency HuR-binding sites (UURKUUU). HuR binds adenine-uridine rich (AUR) elements in the 3′ UTR of mRNA transcripts and promotes mRNA stability, leading to increased protein translation (Brennan and Steitz, 2001). To test for a possible role for HuR-mRNA interaction in the regulation of Mcf2 by ACs, we pre-treated IL-4 macrophages with CMLD-2, a small-molecule compound that prevents HuR binding to AUR elements (Wu et al., 2015). Notably, CMLD-2 treatment abrogated AC-mediated increase in Mcf2 mRNA (Figure 7C). To assess if HuR directly bound to Mcf2 mRNA, a parallel set of cells was subjected to immunoprecipitation using anti-HuR, followed by qRT-PCR for Mcf2. Mcf2 in the HuR immunoprecipitate was increased by ACs, decreased by siOdc1, and restored by putrescine treatment of siOdc1-silenced macrophages (Figure 7D).
We next tested whether AC-induced putrescine increased HuR expression or HuR localization to the cytoplasm, where stabilization of mRNA transcripts occurs (Brennan and Steitz, 2001). Neither AC addition to macrophages nor ODC silencing affected HuR expression (Figure S7A), but addition of ACs to IL-4-treated macrophages enhanced localization of HuR to the cytosol (Figure S7B). HuR cytoplasmic localization was not a direct effect of putrescine itself, as cytoplasmic localization of HuR was not affected by adding exogenous putrescine to ODC-silenced macrophages (Figure S7C). To test if putrescine could directly increase the association of HuR with Mcf2 mRNA, we co-incubated recombinant HuR (rHuR), polyamines, and poly-A mRNA isolated from IL-4-treated macrophages, followed by HuR immunoprecipitation and then Mcf2 PCR. The data show that putrescine, but not spermidine or spermine, increased Mcf2 in rHuR immunoprecipitates (Figure 7E), and this effect of putrescine was blocked by treatment CMLD-2 (Figure 7F). In the absence of an AC, when HuR localization to the cytosol is low, putrescine was unable to increase Mcf2 mRNA when added back to ODC-silence macrophages (Figure S7D), and putrescine did not increase single-AC efferocytosis (Figure S7E). Collectively, these data support a model in which AC uptake by resolving-type macrophages leads to an increase in cytoplasmic HuR, which then stabilizes Mcf2 when putrescine is generated from the metabolism of that AC. The stabilization of Mcf2 then results in increased Dbl expression, which increases Rac1 activity to enhance second AC phagosome internalization (Figure S7F).
DISCUSSION
Efferocytosis maintains normal tissue function, and impaired efferocytosis promotes a number of prevalent non-resolving inflammatory diseases (Arandjelovic and Ravichandran, 2015). We and others have demonstrated that macrophages must ingest multiple ACs to adequately clear dead cells and prevent necrosis in vivo (Park et al., 2011; Wang et al., 2017). We show here that Arg1+ resolving-type macrophages are uniquely programmed to optimize continued AC uptake in a manner that requires proper metabolism of metabolites derived from engulfment of the preceding AC. This process, which increases a Rac1 GEF, may be necessary to counter a decrease in cell-surface Rac1 during first AC internalization (Schille et al., 2018). We showed previously that phagolysosome-to-plasma membrane vesicular trafficking is also needed for continual efferocytosis, as it maintains cell-surface membrane area for second AC phagosome formation (Wang et al., 2017). That process and the one described here likely represent sequential steps in continual efferocytosis, i.e., phagosome formation followed by phagosome internalization.
Arg1/ODC-mediated polyamine synthesis may contribute to other resolution programs in macrophages. For example, induction of Arg1 in macrophages by IL-4 or IL-13 contributes to the rapid resolution of inflammation after helminth infection (Grencis, 2015; Wynn and Vannella, 2016). Accordingly, infection of myeloid-Arg1 knockout mice with Schistosoma mansoni leads to granuloma necrosis and decreased survival (Pesce et al., 2009). Further, an Arg1 pathway in brain microglia and macrophages was shown to play a role in clearing dead neurons and promoting inflammation resolution after stroke, though the mechanism linking Arg1 to efferocytosis in this setting remains to be elucidated (Cai et al., 2019). As another example, deletion of myeloid-specific ODC drives M1-like macrophage activation in gastrointestinal infections and worsens colitis, which can be reversed by putrescine (Hardbower et al., 2017). In contrast, it is possible that the efferocytosis-putrescine pathway described here is usurped by Trypanosoma cruzi, the parasite that causes Chagas disease, as macrophages infected with T. cruzi that also happen to take up ACs produce putrescine, which fuels parasite growth (Freire-de-Lima et al., 2000). Finally, spermidine and spermine may also play important roles in macrophage resolution pathways. For example, spermidine promotes eIF5A hypusination, a process that causes eIF5A-dependent translation elongation (Dever et al., 2014), and inhibiting this pathway blocks oxidative phosphorylation-dependent alternative activation of macrophages (Puleston et al., 2019).
The metabolism of other AC components by efferocytosing macrophages may also contribute to resolution. For example, deletion of the DNase that fragments DNA in phagolysosomes causes the accumulation of unfragmented AC-derived DNA, elevates production of the proinflammatory cytokine TNFα, and exacerbates polyarthritis (Kawane et al., 2006). As another example, efferocytosing macrophages rapidly upregulate the cholesterol transporters ABCA1 and ABCG1 to enable efflux of AC-derived cholesterol (Fond et al., 2015; Kiss et al., 2006; N et al., 2009), and deleting Abca1 and Abcg1 renders macrophages susceptible to cell death after efferocytosis (Yvan-Charvet et al., 2010). Further, macrophage metabolism of fatty acids during efferocytosis increases the expression of Il10 (Zhang et al., 2019). There may also be metabolic changes in efferocytes that precede AC engulfment. For example, exposure of phagocytes to ACs increases cell-surface expression of the glucose transporter SLC2A1, which increases glucose uptake, promotes aerobic glycolysis and enhances subsequent AC engulfment (Morioka et al., 2018).
Impairment of the pathway described here is predicted to occur in chronic inflammatory settings owing to a paucity of Arg1+ macrophages and possibility other defects in arginine/ornithine metabolism, leading to defective efferocytosis, further inflammation, impaired resolution, and tissue necrosis. We illustrate this point in progressing atherosclerosis. Conversely, therapeutic measures that promote the resolving macrophage phenotype or enable inflammatory macrophages to metabolize AC-derived arginine and ornithine may be able to break this pathological cycle by enhancing continual efferocytosis.
Limitation of Study
While human macrophages can express Arg1 in certain settings in vivo, cultured HMDMs do not express appreciable levels of Arg1, and this may reflect other settings in vivo. Accordingly, we showed that treating HMDMs with nor-NOHA did not impair continual efferocytosis. However, AC-derived ornithine is also available for metabolism by ODC into putrescine, and we show that macrophages can use ornithine to form putrescine and boost continual efferocytosis by the putrescine-Dbl pathway. A key goal for the future will be to determine the relevance of this pathway in human atherosclerosis, including (1) whether the absence of the pathway in progressing lesions, owing to the prevalence of Arg1− macrophages, contributes to clinically significant plaque progression, and (2) whether the presence of Arg1+ macrophages in regressing lesions, which can be achieved in humans by marked lowering of plasma cholesterol, contributes to plaque stabilization by the pathway described here.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ira Tabas (iat1@columbia.edu). This study did not generate new unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Lines
Jurkat (human T lymphocytes) and L-929 (mouse fibroblasts) cells were obtained from ATCC and cultured in DMEM (GIBCO) supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (HI-FBS; GIBCO), and 10 U/mL penicillin and 100 mg/mL streptomycin (Corning). Cells were cultured in a humidified CO2 incubator at 37°C. The Jurkat cell line was established from the peripheral blood of a 14-year-old male diagnosed with T cell leukemia. L-929 cells were originally isolated from adipose tissue of a 100-day-old male C3H/An mouse.
Primary Cell Cultures
For bone marrow-derived macrophages (BMDMs), bone marrow cells from 8–12 week old mice (either male or female) were cultured for 7–10 days in DMEM supplemented with 10% (vol/vol) heat-inactivated (HI) FBS, 10 U/mL penicillin,100 mg/mL streptomycin, and 20% (vol/vol) L-929 fibroblast-conditioned media. University Institutional Review Board and Health Insurance Portability and Accountability Act guidelines were followed for isolating peripheral human blood leukocytes. For human macrophages, peripheral human blood leukocytes were isolated from buffy coats of anonymous, de-identified healthy adult volunteers, with informed consent (New York Blood Center) and subsequently purified in a discontinuous gradient of Histopaque solution. After 4 h of adhesion on 24-well plates, cells were rinsed, and medium was changed to RPMI-1640 (GIBCO) containing 10% HI-FBS, 10 U/mL penicillin and 100 mg/mL streptomycin, and 10 ng/mL of M-CSF (Peprotech). These cells were then used for experiments after 7–10 days when they were more than 75% confluent. To deplete ornithine, Jurkat cells were cultured for 4 days in arginine-free media in 10% (vol/vol) dialyzed FBS in the presence of 500 μM nor-NOHA.
Experimental Animals
Animal protocols were approved by Columbia University’s institutional animal care and use committee. All mice were cared for according to the NIH guidelines for the care and use of laboratory animals, and all were in good general health based on appearance and activity. The mice were socially housed in standard cages at 22°C under a 12–12 h light-dark cycle in a barrier facility with ad libitum access to water and food. Arg1fl/fl mice were purchased from The Jackson Laboratory (Stock No. 008817), Odc1fl/fl mice were generated as described (Hardbower et al., 2017), and Lyz2-Cre mice were a gift from Irmgard Förster (Clausen et al., 1999). Littermate control mice were randomly assigned to experimental groups by investigators. Investigators were blinded for the atherosclerosis studies but were not blinded for the zymosan-induced sterile peritonitis or the dexamethasone-induced thymus injury experiments.
METHOD DETAILS
Induction of Apoptosis and Fluorescent Labeling of Jurkat Cells
Jurkat cells were irradiated under a 254-nm UV lamp for 15 min, followed by incubation under normal cell culture conditions for 2–3 h. This method routinely yields more than 85% Annexin V+ cells. The apoptotic cells (ACs) were rinsed once with serum-free DMEM, resuspended at a concentration of 2 × 107 cells/mL in Diluent C (Sigma-Aldrich), and incubated for 3 min with four mL of Diluent C containing concentrated PKH or CellVue Claret membrane-intercalating dyes (Sigma-Aldrich). The cells were then rinsed twice with DMEM containing 10% heat-inactivated FBS and used for experiments. Alternatively, ACs were rinsed with PBS twice, resuspended at a concentration of 4 × 106 and incubated for 30 min with 20 ng/mL pHrodo green or pHrodo red (Life Technologies), rinsed twice with DMEM containing 10% heat-inactivated FBS, and then used for experiments.
In Vitro Continual Efferocytosis Assay
Bone marrow-derived macrophages or peritoneal macrophages isolated six days after a single injection of zymosan A (0.1 mg/mouse, i.p.; Sigma-Aldrich) were plated in 24-well dishes at a density of 0.18 × 106 cells per well. PKH67- or PKH26-labeled ACs were incubated with the macrophages for 45 min at a 5:1 AC:macrophage ratio followed by washing three times with PBS. Some groups of macrophages were then incubated for another 2 h in normal cell culture media, followed by the addition of PKH27- or CellVue Claret-labeled ACs. After 45 min, macrophages were washed three times with PBS to remove unbound ACs, and then the macrophages were fixed with 4% formaldehyde for 20 min, rinsed three times with PBS, and imaged on a Leica epifluorescence microscope (DMI6000B).
Amino Acid Measurements in AC− and AC+ Sorted Macrophages
Bone marrow-derived macrophages were incubated with PKH67-labeled apoptotic Jurkat cells for 45 min. Macrophages were then vigorously rinsed with PBS three times to remove unbound ACs. After a 2 h incubation period, macrophages were detached and sorted into PKH67− and PKH67+ populations. The cells were then pelleted by centrifugation and resuspended in a solution containing 50% acetonitrile and 0.3% formic acid to extract metabolites. Quantification of amino acids was performed via stable isotope dilution techniques as described previously using a Waters Acquity UPLC system equipped with a TQD and MassLynx 4.1 operating system (An et al., 2004; Wu et al., 2004).
Immunoblotting and Immunofluorescence Microscopy of Macrophages
Cells were lysed in 2X Laemmli lysis buffer (Bio-Rad) containing 50 mM DTT. Cell lysates were heated at 95°C for 10–15 min and then separated on 4%–20% SDS-PAGE gradient gels (Invitrogen) at 120V for 1.5–2 h, and electro-transferred to 0.45-μm PVDF membranes at 100V for 2–3 h. The membranes were incubated overnight at 4°C with primary antibodies in PBS containing 1% BSA and detected using HRP-conjugated secondary antibodies (Pierce). Densitometry was performed using ImageJ software. For immunofluorescence microscopy, macrophages were fixed in 4% formaldehyde, permeabilized with 0.1% Triton X-100, blocked with 1% denatured bovine serum albumin (BSA) for one h, and incubated with 1:200 anti-HuR antibody for at least 18 h. Cells were washed in TBS with 0.1% Tween 20 (TBST) and incubated for 2 h with 1:1000 AlexaFluor-546-conjugated secondary antibody. Unbound secondary antibody was removed by washing three times with TBST, and then the cells were visualized and image-captured using a Leica epifluorescence microscope (DMI6000B).
siRNA-Mediated Gene Silencing
Scrambled siRNA control and oligo-targeting siRNAs were transfected into macrophages using Lipofectamine RNAiMAX (Life Technologies) at 50 nM of siRNA in 24-well plates following the manufacturer’s instructions. Briefly, the cells were incubated for 18 h with 1.5 mL of Opti-MEM (GIBCO) containing Lipofectamine RNAiMAX per 0.18 × 106 cells and 50 nM siRNA. Experiments and mRNA analyses were conducted three days later.
Quantitative Real Time PCR
RNA was extracted from samples using the DirectZol miniprep kits (Zymo Research). The purity of the RNA was assessed by absorbance at 260 and 280 nm using a NanoDrop spectrophotometer (Thermo Scientific). Complementary DNA was synthesized from 250 ng of RNA, which had a 260/280 ratio of > 1.8, using oligo (dT) and Superscript II (Applied Biosystems). Quantitative RT-PCR was performed using a 7500 Real-Time PCR system (Applied Biosystems) and SYBR Green Master Mix reagents (Applied Biosystems).
Zymosan-Induced Peritonitis and Peritoneal Uptake of Labeled Apoptotic Cells
Sterile peritonitis was induced by a single 500 μL i.p. injection of 0.1 mg zymosan A (Sigma-Aldrich) (day 0). Mice were then injected i.p. with 200 μg of nor-NOHA in a 500 μL volume of PBS or vehicle control on day 5 and again on day 6 two h before sacrifice. For experiments where putrescine was used, 100 μg of putrescine in a 500 μL volume of PBS or vehicle control was i.p. injected three h before sacrifice. To prepare ACs for i.p. injection into these recipient mice, neutrophils were harvested by peritoneal lavage from donor mice 6 h after a single i.p. injection with 1 mg of zymosan A. The collected lavage fluid was passed through a 40 μm strainer and neutrophils were isolated with the EasySep Mouse Neutrophil Enrichment Kit according to the manufacturer’s instructions (STEMCELL Technologies). Isolated neutrophils were then cultured overnight in medium containing 1% HI-FBS and 10 U/mL penicillin and 100 mg/mL streptomycin in DMEM, after which they became apoptotic. The apoptotic neutrophils were then labeled with PKH67, PKH26, or CellVue Claret. The recipient mice were injected i.p. with 2 × 106 of either labeled apoptotic cell population, and 45 min later, the mice were sacrificed and the peritoneum was lavaged with three mL of PBS. Efferocytosis was quantified as the percentage of F4/80+ macrophages that had taken up one AC (sum of the percent of either population) or two ACs (percent positive for both of populations).
Arginase Activity and Nitric Oxide Measurements
To assess arginase activity, cells were lysed for 10 min in 10 mM Tris-HCl, pH 7.4, containing 1 μM pepstatin A, 1 μM leupeptin, and 0.4% (w/v) Triton X-100, followed by centrifugation at 13,000 × g for 10 min. Supernatant fractions were then assayed for arginase activity, according to the manufacturer’s protocol (Sigma-Aldrich). For nitric oxide detection, reductive acidic tri-iodide (KI/I3)–based chemiluminescence was used on a Sievers NO analyzer to quantify NOx concentrations in samples. Samples were lysed in a nitrite-preserving buffer (100 mM NEM, 0.8 M K3Fe(CN6), and 10% NP-40) and injected into a purge vessel with a solution containing potassium iodide (66.8 mM), iodine (28.5 mM), and acetic acid (78% [vol/vol]). Chemiluminescence values were obtained and compared with sodium nitrite standard curves to calculate concentrations.
Flow Cytometry
Cells were suspended in FACS staining buffer (PBS containing 2% FBS and 1 mM EDTA) at a density of 1 × 106 cells/100 μL and incubated with Fc block (anti-mouse CD16/32; Biolegend) for 30 min on ice. Cell surface and cytoplasmic immunostaining, which was facilitated by permeabilization with 0.1% saponin, was carried out with fluorescent antibodies for 60 min on ice. Cells were washed in FACS buffer twice and then resuspended for analysis on a BD FACS Canto II flow cytometer. For apoptosis detection, cells were washed twice with cold FACS buffer, resuspended in annexin V-binding buffer at a concentration of 1 × 106 cells/1 ml, and incubated with FITC-conjugated Annexin V for 15 min at room temperature. Samples were then analyzed on a BD FACS Canto II flow cytometer. Data analysis was carried out using FlowJo software.
In Vivo Thymus Efferocytosis Assay
Eight-week-old male mice were injected i.p. with 250 μL PBS containing 250 μg dexamethasone (Sigma-Aldrich) dissolved in DMSO. Eighteen h after injection, the mice were euthanized, and thymi were harvested and weighed. One lobe of the thymus was mechanically disaggregated, and cells were enumerated. Flow cytometry was then performed to determine the number of Annexin V+ and F4/80+ cells (Biolegend). The other thymus lobe was formalin-fixed, paraffin-embedded, and sectioned, followed by staining of the sections (5 μm) with TUNEL reagents (Roche) and an antibody to Mac2 (Cedarlane). In situ efferocytosis was quantified by counting TUNEL+ nuclei that were associated with Mac2+ macrophages (“associated”), indicative of efferocytosis, or not associated with macrophages (“free”). Macrophage-associated apoptotic cells followed the criteria of TUNEL+ nuclei surrounded by or in contact with neighboring Mac2+ macrophages. Free apoptotic cells exhibited nuclear condensation, loss of antibody Mac2 reactivity, and were not in contact with neighboring macrophages.
Tissue Collection and Lesion Analysis
All experiments using human tissue were deemed exempt human subjects research by the LSUHSC-Shreveport IRB due to the exclusive use of postmortem samples. Human samples were excised postmortem during routine autopsies at LSUHSC-Shreveport. For atherosclerosis studies, 10-week-old male Ldlr−/− recipient mice were given acidic water containing neomycin and polymyxin B (10 mL of 10 mg/mL neomycin in saline and 400 mL of 25 mg/mL polymyxin B sulfate in water added to 1 L of acidic water) one week before irradiation. The mice were then irradiated using 1,000 rads from a 137 Cesium Gammacell source. Later in the same day, the mice were injected via tail vein with 2.5 × 106 bone marrow cells from eight to ten-week-old male Arg1fl/fl (Arg1-WT) and Arg1fl/fl LysM+/− (Arg1-KO) mice. Arg1fl/fl mice were purchased from The Jackson Laboratory (stock no. 008817) The bone marrow cells were prepared as follows: femurs from donor mice were isolated and flushed with RPMI-1640, supplemented with 10 units/mL of heparin, and a cocktail of 10 U/mL penicillin and 100 mg/mL streptomycin. The cells were filtered through a 40 μm cell strainer and centrifuged at 500 × g for 10 min at 4°C. Bone marrow cells were rinsed and pelleted twice more. Six weeks after injection of bone marrow cells, the mice were placed on a Western diet (Envigo, TD 88137) for 16 weeks. To induce plaque regression, Arg1-WT and Arg1-KO mice were then switched back to chow diet for an additional five weeks while simultaneously receiving a helper-dependent adenovirus containing the human Ldlr gene (HDAd-LDLR). This virus is a non-integrating vector that primarily targets the liver and restores expression of the Ldlr in hepatocytes to levels sufficient to significantly reduce circulating cholesterol in Ldlr−/− mice. To test the effect of putrescine in atherosclerosis, 8-week-old male Ldlr−/− mice were placed on a Western diet for a total of 16 weeks. Between 8–16 weeks of Western diet feeding, drinking water of the experimental group was supplemented with 3 mM putrescine, whereas control mice remained on regular drinking water. Control and putrescine-supplemented water was exchanged every two or three days. Upon sacrifice, mice were perfused with PBS through left ventricular cardiac puncture, and aortic roots were collected and processed for histology. For some mice, aortas were collected for polyamine analysis (see the section on Polyamine Detection). Sections were stained with hematoxylin and eosin for morphometric lesion analysis. Atherosclerotic lesion area, defined as the space from the internal elastic lamina to the lumen, was quantified by taking the average of 6 sections spaced 30 μm apart, beginning at the base of the aortic root. Boundary lines were drawn around these regions, and the area measurements were obtained by image analysis software. A 3,000-μm2 threshold cut-off was applied to acellular regions to calculate areas of necrosis. Collagen staining was performed using picrosirius red (Polysciences, catalog 24901A) per the manufacturer’s instructions. Collagen cap thickness was quantified at the lesional midpoint and both shoulder regions and then averaged and quantified as the ratio of collagen cap thickness to lesion area. Fasting blood glucose levels were measured using ONETOUCH Ultra after food was withdrawn for 18 h. Total plasma cholesterol was measured using a kit from WAKO Diagnostics. Complete blood cell counts, including leukocyte differential, were obtained using a FORCYTE Hematology Analyzer (Oxford Science).
Tissue Immunohistochemistry and Immunofluorescence Microscopy
Frozen specimens were immersed in OCT and cryosectioned, and 6-μm sections were placed on glass slides and then exposed to ice-cold acetone for 20 min to fix and permeabilize. Paraffin-embedded specimens were sectioned, de-paraffinized with xylene, and rehydrated in decreasing concentrations of ethanol. Sections were incubated with TUNEL staining reagents at 37°C for 60 min and then washed three times with PBS. Sections were then blocked for 60 min, incubated overnight at 4°C with the following antibodies: anti-Arg1 (1:200), anti-iNOS (1:200), anti-TNFα (1:200), anti-IL-1β (1:200), anti-Dbl (1:50), anti-CD68 (1:400), anti-ODC (1:200), or anti-Mac2 (1:10,000) antibodies, incubated with fluorescently-labeled secondary antibodies, and counterstained with DAPI. Images were captured using a Leica epifluorescence microscope (DMI6000B).
Polyamine Detection
Quantitation of putrescine, spermidine, and spermine was performed using ultra performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS). Standards and deuterated internal standards were purchased from TRC Chemicals (Ontario, Canada). Bone marrow-derived macrophages were incubated with PKH67-labeled apoptotic Jurkat cells for 45 min. Macrophages were then vigorously rinsed with PBS three times to remove unbound ACs. After a 2 h incubation period, macrophages were detached and sorted into PKH67− and PKH67+ populations. Cells were then pelleted by centrifugation and resuspended in H2O. For in vivo polyamine analysis, aortic segments from the ascending to thoracic aorta, where plaque burden is highest, were harvested on ice and placed in cold PBS. The tissue was minced with dissecting scissors, placed in 1.7 mL microcentrifuge tubes, and further sonicated on ice. Samples were then spun at 15,000 × g for 15 min to pellet cell debris, and extracellular matrix, and supernatants were collected for polyamine analysis. A 50 μL aliquot of sample was spiked with the internal standard, and the protein was precipitated with 0.4M perchloric acid and centrifuged at 12,000 × g for 5min. The supernatant was mixed with 1M sodium bicarbonate and 0.1% dansyl chloride, vortexed and heated at 60°C for 10 min. After chilled on ice for 2 min, samples were then centrifuged at 12,000 × g for 5 min and the upper phase was mixed with 200 μL of ethyl acetate. After being centrifuged at 12,000 × g for 5 min again, the supernatant was evaporated to dryness under a nitrogen stream and suspended in 50 μL of acetonitrile. UPLC-MS/MS was carried out on a Waters Xevo TQ MS integrated with an ACQUITY UPLC system (Waters). The system was controlled by MassLynx Software 4. 1. Five microliters of the sample was loaded onto a Waters ACQUITY UPLC BEH Phenyl column (3 mm ID × 100 mm length, 1.7 μm particle size), maintained at 40°C and at a flow rate of 300 μL/min. The initial conditions were 50% mobile phase A (water containing 0.1% formic acid) and 50% mobile phase B (acetonitrile containing 0.1% formic acid). Solvent B was increased linearly to 99% over 5 min and maintained until 7.5 min with a total run time of 8 min. Positive ESI-MS/MS with multiple reaction monitoring (MRM) mode was performed using the following parameters: capillary voltage 4 kV, desolvation temperature at 500°C, desolvation gas flow at 1,000 L/h. Concentrations of polyamines were quantitated by comparing integrated peak areas for those of each species against those of known amounts of purified standards.
13C6-Arginine Tracer Experiments
Metabolite Extraction for Targeted Isotope Tracer Analysis
Jurkat cells were cultured for four days in arginine-depleted DMEM/10% FBS containing 84 μg/mL 13C6-arginine and 500 μM nor-NOHA. Jurkat cells were then made apoptotic, as stated above, and labeled with PKH67. These ACs were then added to IL-4-treated macrophages for 45 min. Macrophages were then vigorously rinsed with PBS three times to remove unbound ACs. After a 2 h incubation period, macrophages were detached and sorted into PKH67− and PKH67+ populations. Metabolites from cell culture samples were then extracted with 0.5 mL of 80% methanol (−80°C) followed by vortexing and incubation on dry ice for 20 min. Insoluble material was pelleted by centrifugation at 14,000 × g for 5 min at 4°C, and the supernatant containing extracted metabolites was transferred to a clean tube. There were two additional extractions of the pellet were performed using the same procedure. The 1.5 mL total metabolite extract from the pooled supernatants was dried in a vacuum centrifuge (Savant SC210A SpeedVac Concentrator, Thermo Scientific) and resuspended in 20 μL of HPLC grade water (Burdick & Jackson, Honeywell, Muskegon, MI).
Liquid Chromatography-Selected Ion Monitoring Mass Spectrometry
For targeted isotope tracer analysis, an aliquot (5 μL) of each sample was analyzed using an ultra-high performance liquid chromatograph (UPLC model U3000, Dionex) interfaced with a QExactive Focus mass spectrometer (ThermoFisher Scientific) using full MS as well as selected ion monitoring (SIM) for three labeled and unlabeled endogenous metabolites; arginine, ornithine, and putrescine. Samples were separated using an Accucore reversed phase C18 column (27826–153030, 2.1 mm ID × 100 mm length, 2.6 μm particle size, ThermoFisher Scientific) at 350 μL/min. Solvent A was 100% HPLC grade water (LC365–4, Burdick & Jackson) containing 0.05% heptafluorobutyric acid (LC6206, HFBA, Proteomics Grade, ProteoChem), and solvent B was 100% HPLC grade methanol (LC230–4, Burdick & Jackson) containing 0.05% heptafluorobutyric acid (LC6206, HFBA, Proteomics Grade, ProteoChem); HFBA was used as counter-ion to enable analysis of the polyamines. The gradient starting at 20% solvent B was held for 2 min, then ramped from 20% B to 50% B from 2 to 4 min and from 50% B to 80% B from 4 to 6 min; followed by re-equilibration over 3 min, for a total of 9 min for each experiment. Samples were analyzed in positive ion mode using an ESI voltage of 3.5 kV. Mass spectrometry instrument parameters for SIM included the following: resolution 70,000; isolation window width 1.0 m/z; AGC target 2e5; maximum IT 200 ms; an inclusion list containing the m/z for each labeled and unlabeled targeted metabolite and a scheduled time window for metabolite elution. Peak heights for unlabeled metabolites and 13C-labeled metabolites were extracted from the chromatograms using Xcalibur Quan Browser (version 3.0.63, ThermoFisher Scientific). Conversion of 13C6-arginine into 13C5-ornithine was quantified as 13C5-ornithine peak height values relative to the sum of 13C6-arginine, 13C5-ornithine, and 13C4-putrescine peak height values; and conversion of 13C6-arginine into 13C4-putrescine was quantified as 13C4-putrescine peak height values relative to the sum of 13C6-arginine, 13C5-ornithine, and 13C4-putrescine peak height values.
Ornithine Detection
Metabolite Extraction
Cell pellets (2 million apoptotic Jurkat cells per replicate, n = 5) were spiked with 5 μL of internal standard solution containing 1 μg/mL of 13C5-ornithine (CLM-4724), purchased from Cambridge Isotope Labs (Tewksbury, MA). An aliquot (200 μL) of HPLC grade methanol (AH365–4, Burdick & Jackson, Honeywell, Muskegon, MI) heated to 80°C was added to each sample, vortexed and incubated for 5 min at 80°C. Samples were centrifuged at 16,200 × g for 15 min at 4°C and the supernatant collected. The extraction process was repeated using 100 μL of hot methanol at 80°C. The supernatants containing metabolites from both extractions were pooled and dried in a vacuum centrifuge (Savant SC210A SpeedVac Concentrator, ThermoFisher Scientific). Samples were resuspended in 50 μL of HPLC grade water (Burdick & Jackson, Honeywell, Muskegon, MI).
Liquid Chromatography-Selected Ion Monitoring
Metabolites were analyzed using an ultra-high performance liquid chromatograph (Vanquish, ThermoFisher Scientific, San Jose, CA) interfaced with an electrospray Q Exactive Focus mass spectrometer (ThermoFisher Scientific, San Jose, CA) using selected ion monitoring (LC-SIM) for quantification of ornithine. The following solvent system was used: solvent A was 100% HPLC grade water (Burdick & Jackson, Honeywell, Muskegon, MI) containing 0.05% Heptafluorobutyric acid (LC6206, HFBA, Proteomics Grade, ProteoChem, Hurricane, UT), and solvent B was aqueous 100% methanol (LC230–4, Burdick & Jackson, Honeywell, Muskegon, MI) with 0.05% HFBA. For each sample, a 5 μl aliquot of the metabolite mixture was loaded onto an Accucore reverse phase C18 column (27826–153030, 2.1 mm × 100 mm, 2.6 μm particle size, ThermoFisher Scientific). A gradient of 20% B to 80% B was applied over 6 min with a flow rate of 0.350 mL/min followed by re-equilibration over 3 min, for a total of 9 min for the LC experiment. Mass spectrometry instrument parameters for SIM on the Q Exactive Focus included the following: resolution 35,000; isolation window width 1.0 m/z; AGC target 2e5; maximum IT 200 ms; an inclusion list containing the m/z for ornithine, its respective stable isotope standard (SIS) and a scheduled time window for ornithine elution.
Data Analysis for Metabolite Quantification
Xcalibur Qual Browser (version 4.0.27.19, ThermoFisher Scientific, San Jose, CA) was used for data analysis. Ornithine amounts (pmol) were calculated using the peak height ratio of ornithine to its respective SIS.
Rac-FRET Detection and Quantification
Rac activity was assessed in bone marrow-derived macrophages isolated from Rac-FRET transgenic mice, which express the validated intramolecular Raichu-Rac FRET biosensor (Johnsson et al., 2014). Alternatively, macrophages were transfected with Raichu Rac-FRET plasmid via electroporation. 1.5 μg of Raichu-Rac FRET plasmid DNA was added directly to 5×105 pelleted macrophages. Cells and DNA were resuspended in 80 μL of R-buffer (Invitrogen) and subjected to electroporation at 1550 mV with 10 ms intervals for 2 pulses. The macrophages were then plated on glass chambers, and fluorescence microscopy experiments were conducted two days later. Confocal fluorescence images were collected using an A1RMP scanning confocal microscope (Nikon Instruments, Melville NY) and a 40×/1.3 Plan-Fluor objective lens (Nikon) using excitation at 405 nm for CFP, and a spectral detector with emission bandpasses set at 450–490 nm for CFP and either 525–540 nm for YFP. To prevent bleed-through, PKH26 and CellVue Claret-labeled AC images were captured using emission bandpasses set at 561–620 nm and 650–750, respectively. Ratiometric images of YFP:CFP were then captured every 5 min from time 0, defined as the time when an AC fell into the frame of the image, to 20 min unless otherwise noted. The CFP and YFP fluorescence intensity images were background-subtracted from an area devoid of cells.
Viral Transduction
Myc-DDK-tagged Mcf2 was cloned into the Lenti-P2A-tGFP vector and packaged into lentiviral particles by Origene. Mouse bone marrow-derived macrophages were incubated for 18 h with empty or Myc-DDK-Mcf2-containing Lenti-P2A-tGFP at an MOI of 10 (day 1). Cells were then transfected with ScrRNA or Odc1-targeting siRNA on day 2 and treated with IL-4 on day 4. The double AC-efferocytosis assay was then conducted using PKH26- and CellVue Claret-labeled ACs and analyzed on day 5.
Click-iT Detection of mRNA Turnover
Turnover of mRNA was assessed by the Click-iT Nascent RNA Capture kit (Life Technologies). Briefly, macrophages were cultured in the presence of 0.25 mM 5-ethynyl uridine (EU, an alkyne-modified nucleoside) for 2 days. After transfections and treatments, cell culture media was removed, and cells were rinsed five times to remove the uridine analog before the addition of ACs. After the indicated times, cells were lysed, and mRNA was isolated as previously described. Click-iT reactions were performed to attach biotinylated azide to EU-containing mRNA using 0.5 mM of biotinylated azide for 1 μg of mRNA. EU-containing mRNAs were captured and precipitated with Streptavidin T1 magnetic beads, which were then extensively washed and processed for cDNA synthesis and analyzed by qRT-PCR.
RNA-Binding Protein Immunoprecipitation (RIP)
RIP on mouse bone marrow-derived macrophages was performed using the Magna RIP kit (Millipore Sigma), according to the manufacturer’s instructions. Briefly, cells were treated as described and then washed twice with ice-cold PBS. Cells were then lysed in RIP lysis buffer for 5 min and freeze-thawed once to completely lyse the cells. Samples were precleared using prewashed magnetic beads for one h. Subsequently, magnetic beads conjugated to either normal rabbit IgG or a Millipore RIPAb+ validated mouse anti-HuR antibody (5 μg of antibody/sample) were added to immunoprecipitate HuR-bound mRNA. The RNA was isolated, and qRT-PCR was performed. For some experiments, recombinant HuR (Novus Biologicals, 10 nM), polyamines (100 nM for putrescine, 1 μM for spermidine, and 100 nM for spermine), and RNA containing poly(A)-mRNA (8 μg/mL) from IL-4-treated macrophages were incubated for 24 h in RIP wash buffer, and then HuR pulldown and Mcf2 mRNA analysis by qRT-PCR were conducted as described above.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data were tested for normality using either the D’Agostino-Pearson or Shapiro-Wilk test, and statistical significance was determined using GraphPad Prism software. P values for normally distributed data were calculated using either the Student’s t test for two groups or 1-way ANOVA with Fisher’s LSD post hoc analysis when three or more groups were tested. Data that were not normally distributed were calculated using the non-parametric Mann-Whitney U test or the uncorrected Dunn’s test. Data are shown as mean values ± SEM. Differences were considered statistically significant at p ≤ 0.05. Based on our previous mouse atherosclerosis studies and power calculations, the numbers of mice chosen for each cohort was sufficient to enable the testing of our hypotheses based on an expected 20%–30% coefficient of variations and an 80% chance of detecting a 33% difference in key plaque parameters (lesion size and necrotic core area). Exclusion criteria prior to the start of any of the in vivo studies were death, injury requiring euthanasia, or weight loss > 15%.
DATA AND CODE AVAILABILITY
This study did not generate any unique datasets or code.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-Arg1 | Cell Signaling Technology | Cat# 93668; RRID:AB_2800207 (1:1000 dilution) |
| Rabbit anti-Arg1 | Sigma Aldrich | Cat# HPA024006; RRID:AB_1844987 (1:200 dilution) |
| Rabbit anti-iNOS | Abcam | Cat# ab213987 (1:200 dilution) |
| Rat anti-Mac2 | Cederlane | Cat# CL8942AP; RRID:AB_10060357 (1:10,000 dilution) |
| Rabbit anti-Dbl | Cell Signaling Technology | Cat# 2089; RRID:AB_2143816 (1:50 dilution) |
| Mouse anti-β-actin | Santa Cruz Biotechnology | Cat# SC-47778; RRID:AB_626632 (1:5000 dilution) |
| Rabbit anti-IL-1β | Abcam | Cat# ab9722; RRID:AB_308765 (1:200 dilution) |
| Rabbit anti-TNFα | Abcam | Cat# ab9739; RRID:AB_308774 (1:200 dilution) |
| Rabbit anti-HuR | Sigma-Aldrich | Cat# 03-102; RRID:AB_11211202 (1:1000 dilution) |
| Rabbit anti-Otc | Abcam | Cat# ab203859 (1:500 dilution) |
| Rabbit anti-Odc | Abcam | Cat# ab97395 RRID: AB_10679334 (1:200 dilution) |
| Rabbit anti-Srm | Abcam | Cat# ab241496 (1:250 dilution) |
| Rabbit anti-Pqlc2 | Thermo Scientific | Cat# PA5-48512; RRID:AB_2633969 (1:1000 dilution) |
| Mouse anti-CD68 | DAKO | Cat# M0814; RRID:AB_2314148 (1:400 dilution) |
| Rabbit IgG | Sigma-Aldrich | Cat# PP64B; RRID:AB_97852 |
| Goat anti-mouse (HRP) | Invitrogen | Cat# 32230 (1:2000 dilution) |
| Goat anti-rabbit (HRP) | Invitrogen | Cat# 32260 (1:2000 dilution) |
| Goat anti-mouse (AF488) | Invitrogen | Cat# A11001; RRID:AB_2534069 (1:200 dilution) |
| Goat anti-mouse (AF555) | Invitrogen | Cat# A28180; RRID:AB_2536164 (1:200 dilution) |
| Goat anti-rat (AF488) | Invitrogen | Cat# A11006; RRID:AB_141373 (1:200 dilution) |
| Goat anti-rabbit (AF488) | Invitrogen | Cat# A11034 (1:200 dilution) |
| Goat anti-rabbit (AF555) | Invitrogen | Cat# A21428; RRID:AB_141784 (1:200 dilution) |
| Goat anti-Rat (AF647) | Invitrogen | Cat# A21247; RRID:AB_141778 (1:200 dilution) |
| Rat anti-CD16/CD32 | BD Biosciences | Cat# 553142; RRID:AB_394657 (1:100 dilution) |
| FITC anti-Arginase 1 | Thermo Scientific | Cat# 53-3697-82; RRID:AB_2734831 (1:50 dilution) |
| FITC anti-DBL | LSBio | Cat# LS-C262830-200 (1:50 dilution) |
| Pacific Blue anti-F4/80 | Biolegend | Cat# 123123; RRID:AB_893487 (1:100 dilution) |
| Alexa Fluor 488 anti-CD115 | Biolegend | Cat# 135511; RRID:AB_11218605 (1:100 dilution) |
| PE anti-Ly6C | Biolegend | Cat# 128007; RRID_AB1186133 (1:100 dilution) |
| Bacterial and Virus Strains | ||
| pLenti-P2A-tGFP Lentiviral Particles | Origene | Cat# PS100108 |
| Mcf2-pLenti-P2A-tGFP Lentiviral Particles | Origene | N/A, Custom Request |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Dulbecco’s Modified Eagle Media (DMEM) | Corning | Cat# 10-013-CV |
| Roswell Park Memorial Institute (RPMI) 1640 Media | Corning | Cat# 10-040-CV |
| Opti-MEM | GIBCO | Cat# 31985-070 |
| 1X HBSS | Corning | Cat# 21-022-CV |
| 1X PBS | Corning | Cat# 21-040-CV |
| CellStripper | Corning | Cat# 25-056-CI |
| Heat-Inactivated Fetal Bovine Serum | GIBCO | Cat# 10438-026 |
| Penicillin/Streptomycin | Corning | Cat# 30-002-CI |
| Human Macrophage Colony Stimulating Factor (M-CSF) | Peprotech | Cat# 300-25 |
| HISTOPAQUE-1077 | Sigma-Aldrich | Cat# 10771-100ML |
| 2-Well Glass Slides | Thermo Scientific | Cat# 155380 |
| 4-Well Glass Slides | Thermo Scientific | Cat# 155383 |
| 8-Well Glass Slides | Thermo Scientific | Cat# 155411 |
| Novex 4–20% Tris-Glycine Mini Gels, 15-well | Invitrogen | Cat# XP04205BOX |
| 4X Laemmli Buffer | Bio-Rad | Cat# 1610747 |
| PKH67 Fluorescent Cell Linker | Sigma-Aldrich | Cat# PKH67GL-1KT |
| PKH26 Fluorescent Cell Linker | Sigma-Aldrich | Cat# PKH26GL-1KT |
| Cell-Vue Claret Fluorescent Cell Linker | Sigma-Aldrich | Cat# MINCLARET-1KT |
| Diluent C | Sigma-Aldrich | Cat# CGLDIL |
| Interleukin-4 | Sigma-Aldrich | Cat# SRP3211 |
| Interleukin-13 | Sigma-Aldrich | Cat# I1896 |
| Cytochalasin D | Sigma-Aldrich | Cat# C8273 |
| nor-NOHA | Sigma-Aldrich | Cat# 399275 |
| pHrodo Green | Thermo Scientific | Cat# P35373 |
| pHrodo Red | Thermo Scientific | Cat# P35372 |
| Hoechst 33342 | Thermo Scientific | Cat# 62249 |
| CMLD-2 | Sigma-Aldrich | Cat# 538339 |
| Recombinant HuR | Novus Biologicals | Cat# H00001994-P01 |
| Bafilomycin A1 | Sigma-Aldrich | Cat# B1793 |
| Biotinylated-Phosphatidylserine | Echelon Biosciences | Cat# L-31B16 |
| Streptavidin-Coated Polystyrene Beads | Spherotech | Cat# SVP-60-5 |
| Dexamethasone | Calbiochem | Cat# 265005 |
| Actinomycin D | Sigma-Aldrich | Cat# A1410 |
| Picrosirius Red Solution | Polysciences | Cat# 24901B-250 |
| Putrescine dihydrochloride | Sigma-Aldrich | Cat# P7505 |
| Spermidine trihydrochloride | Sigma-Aldrich | Cat# S2501 |
| Spermine tetrahydrochloride | Sigma-Aldrich | Cat# 85610 |
| N1, N11-Diethylnorspermine tetrahydrochloride | Tocris | Cat# 0468 |
| 13C6-Arginine | Cambridge Isotope Laboratories | Cat# CLM-2265 |
| 13C5-Ornithine | Cambridge Isotope Laboratories | Cat# CLM-4724-H-PK |
| 13C4-Putrescine (1,4-Butanediamine) | Cambridge Isotope Laboratories | Cat# CLM-6574-PK |
| Annexin V (FITC) | BD Biosciences | Cat# 560931 |
| Zymosan A | Sigma-Aldrich | Cat# Z4250 |
| Lipofectamine RNAiMax | Life Technologies | Cat# 13778-150 |
| Neomycin | Sigma-Aldrich | Cat# N1142 |
| Polymyxin B | Sigma-Aldrich | Cat# P4932 |
| 40 μm Nylon Cell Strainers | BD Falcon | Cat# 352340 |
| Power SYBR Green PCR Master Mix | Applied Biosystems | Cat# 4367659 |
| Savant SpeedVac High Capacity Concentrators | Thermo Scientific | Cat# SC210A |
| Accucore C18 HPLC Columns (2.1 mm ID × 100 mm, 2.6 μm) | Thermo Scientific | Cat# 17126-102130 |
| Water Burdick & Jackson Brand LC-MS | Honeywell Research Chemicals | Cat# LC365-4 |
| Methanol Burdick & Jackson Brand LC-MS | Honeywell Research Chemicals | Cat# LC230-4 |
| Heptafluorobutyric Acid (HFBA) | ProteoChem | Cat# LC6206 |
| Acetyl Carnitine Hydrochloride | Sigma | Cat# A6706 |
| Propionyl Carnitine Hydrochloride | Larodan | Cat# 17-0300-9 |
| Butyryl Carnitine Hydrochloride | Larodan | Cat# 17-0400-9 |
| Isovaleryl Carnitine | Supelco | Cat# 91403 |
| Hexanoyl Carnitine Hydrochloride | Larodan | Cat# 17-0060-9 |
| Octanoyl Carnitine Hydrochloride | Larodan | Cat# 17-0800-9 |
| Decanoyl Carnitine Hydrochloride | Larodan | Cat# 17-1000-9 |
| Lauroyl Carnitine Hydrochloride | Sigma | Cat# L3131 |
| Myristoyl Carnitine Hydrochloride | Sigma | Cat# M0135 |
| Palmitoyl Carnitine Hydrochloride | Sigma | Cat# P4509 |
| Stearoyl Carnitine Hydrochloride | Sigma | Cat# S2381 |
| D3-Acetylcarnitine | Cambridge Isotopes Laboratory | Cat# DLM-1871 |
| D3-Propionylcarnitine | Cambridge Isotopes Laboratory | Cat# DLM-3973 |
| D3-Butyrylcarnitine | Cambridge Isotopes Laboratory | Cat# DLM-3861 |
| D9-Isovalerylcarnitine | Cambridge Isotopes Laboratory | Cat# DLM-3974 |
| D3-Octanoylcarnitine | Cambridge Isotopes Laboratory | Cat# DLM-755 |
| D3-Palmitoylcarnitine | Cambridge Isotopes Laboratory | Cat# DLM-1263 |
| Glycine | Sigma | Cat# G7126 |
| L-Alanine | Sigma | Cat# A7627 |
| L-Valine | Sigma | Cat# V0500 |
| L-Proline | Sigma | Cat# P0380 |
| L-Serine | Sigma | Cat# S4500 |
| L-Methionine | Sigma | Cat# M9625 |
| L-Phenylalanine | Sigma | Cat# P2126 |
| L-Tyrosine | Sigma | Cat# T3754 |
| L-Aspartic Acid | Sigma | Cat# A9256 |
| L-Glutamic Acid | Sigma | Cat# G1251 |
| L-Ornithine Monohydrochloride | Sigma | Cat# O4386 |
| L-Citrulline | Sigma | Cat# C7629 |
| L-Arginine | Sigma | Cat# A5131 |
| L-Histidine Monohydrochloride Monohydrate | Sigma | Cat# H8125 |
| Glycine, 2,2-D2, | CDN Isotopes | Cat# D-0064 |
| L-Alanine, –2,3,3,3-D4, | CDN Isotopes | Cat# D-1488 |
| L-Valine-D8, | CDN Isotopes | Cat# D-1076 |
| L-Proline-D7, | Cambridge Isotopes Laboratory | Cat# DLM-487 |
| L-Serine,-2,3,3-D3 | Cambridge Isotopes Laboratory | Cat# DLM-582 |
| L-Leucine-D3 | CDN Isotopes | Cat# D-1973 |
| L-Methionine,-D3, | Cambridge Isotopes Laboratory | Cat# DLM-431 |
| DL-Phenylalanine-D5 | CDN Isotopes | Cat# D-1587 |
| L-Tyrosine-D4 | Isotec | Cat# 489808 |
| L-Aspartic Acid, 2,3,3-D3 | CDN Isotopes | Cat# D-0898 |
| L-Glutamic Acid 2,4,4-D3 | Cambridge Isotopes Laboratory | Cat# DLM-335 |
| L-Ornithine 5,5-D2 | Cambridge Isotopes Laboratory | Cat# DLM-4261 |
| L-Citrulline 5,5-D2 | Cambridge Isotopes Laboratory | Cat# DLM-3860 |
| L-Arginine 5 13C, 4,4,5,5-D4 | Cambridge Isotopes Laboratory | Cat# DLM-3789 |
| L-Histidine-15N3 | Cambridge Isotopes Laboratory | Cat# NLM-1513 |
| N-ethylamaleimide | Sigma-Aldrich | Cat# 04259 |
| Sodium Iodide | Sigma-Aldrich | Cat# 383112 |
| Potassium hexacyanoferrate(III) | EMD-Millipore | Cas# 13746-66-22 |
| Nonidet P40 | Biochemika | Cat# 74385 |
| Glacial acetic acid | VWR | Cat# 0714 |
| Critical Commercial Assays | ||
| Direct-Zol RNA Miniprep Plus | Zymo Research | Cat# R2072 |
| Click-iT Nascent RNA Capture Kit | Thermo Scientific | Cat# C10365 |
| TUNEL Kit | Roche | Cat# 12156792910 |
| Total Cholesterol Kit | Wako Diagnostics | Cat# 999-02601 |
| Arginase Activity Assay Kit | Sigma-Aldrich | Cat# MAK112 |
| Nitric Oxide Assay Kit | Abcam | Cat# ab65328 |
| Experimental Models: Cell Lines | ||
| Human: Jurkat cells | ATCC | ATCC TIB-152 |
| Mouse: L-929 Fibroblast cells | ATCC | ATCC CCL-1 |
| Mouse: Bone Marrow-Derived Macrophage cells | This paper | N/A |
| Mouse: Elicited Peritoneal Macrophage cells | This paper | N/A |
| Mouse: Raichu Rac-FRET Transgenic Macrophage cells | Johnsson et al., 2014 | N/A |
| Human: Peripheral Blood Monocyte-Derived Macrophage cells | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | JAX: 000664 |
| Mouse: Ldlr−/−: C57BL/6J | The Jackson Laboratory | JAX: 002207 |
| Mouse: Arg1/fl: C57BL/6J | The Jackson Laboratory | JAX: 008817 |
| Mouse: LysMcre: C57BL/6J | Clausen et al., 1999 | N/A |
| Mouse: Odc1/fl: C57BL/6J | Hardbower et al., 2017 | N/A |
| Oligonucleotides | ||
| ON-TARGETplus non-targeting pool | GE Healthcare Dharmacon | D-001810-10-05 |
| Mouse: ON-TARGETplus Arg1 siRNA | GE Healthcare Dharmacon | L-40721-00-0005 |
| Mouse: ON-TARGETplus Otc siRNA | GE Healthcare Dharmacon | L-060377-01-0005 |
| Mouse: ON-TARGETplus Odc siRNA | GE Healthcare Dharmacon | L-041731-01-0005 |
| Mouse: ON-TARGETplus Srm siRNA | GE Healthcare Dharmacon | L-062372-01-0005 |
| Mouse: ON-TARGETplus Pqlc2 siRNA | GE Healthcare Dharmacon | L-051498-01-0005 |
| Mouse: ON-TARGETplus Mcf2 siRNA | GE Healthcare Dharmacon | L-046870-01-0005 |
| Human: ON-TARGETplus ODC siRNA | GE Healthcare Dharmacon | L-006668-00-0005 |
| Mouse: Mcf2 Forward GGGAGCTTACACTTAGTCCTGG |
PrimerBank | www.pga.mgh.harvard.edu/primerbank |
| Mouse: Mcf2 Reverse TCGAAATCCAATGTCCGTGAAAG |
PrimerBank | www.pga.mgh.harvard.edu/primerbank |
| Mouse: Hprt Forward TCAGTCAACGGGGGACATAAA |
PrimerBank | www.pga.mgh.harvard.edu/primerbank |
| Mouse: Hprt Reverse GGGGCTGTACTGCTTAACCAG |
PrimerBank | www.pga.mgh.harvard.edu/primerbank |
| Human: ODC Forward TTTACTGCCAAGGACATTCTGG |
PrimerBank | www.pga.mgh.harvard.edu/primerbank |
| Human: ODC Reverse GGAGAGCTTTTAACCACCTCAG |
PrimerBank | www.pga.mgh.harvard.edu/primerbank |
| Human: HPRT Forward CCTGGCGTCGTGATTAGTGAT |
PrimerBank | www.pga.mgh.harvard.edu/primerbank |
| Human: HPRT Reverse AGACGTTCAGTCCTGTCCATAA |
PrimerBank | www.pga.mgh.harvard.edu/primerbank |
| Recombinant DNA | ||
| HDAd-LDLR | Baylor College of Medicine | Willecke et.al., 2015 |
| Mouse: Mcf2 ORF Clone | Origene | Cat# MR215353 |
| Software and Algorithms | ||
| NIS-Elements | Nikon | Advanced Research |
| Leica Application Suite | Leica | Advanced Fluorescence |
| ImageJ | NIH | www.imagej.nih.gov/ij |
| FIJI | Schindelin et al., 2012 | www.fiji.sc |
| PRISM | GraphPad Software | Version 6 |
| FlowJo | FlowJo | Version 10 |
| RBPmap | Paz et al., 2014 | http://rbpmap.technion.ac.il/ |
| Xcalibur Software | Thermo Scientific | Version 3.0.63 |
| Other | ||
| Mouse Diet: High-Fat Western Diet | Envigo | Cat# TD.88137 |
| Genotyping Service | Genetyper | www.genetyper.com |
| Lentivirus Cloning and Packaging Services | Origene | www.origene.com |
| Helper-Dependent Adenovirus Cloning and Packaging Services | Baylor College of Medicine | N/A |
Highlights.
Macrophages take up arginine and ornithine from apoptotic cells during efferocytosis
Arg1 and ODC convert apoptotic cell-derived arginine and ornithine into putrescine
Putrescine augments subsequent rounds of efferocytosis by increasing Rac1 activation
Lack of Arg1 or ODC blunts continual efferocytosis and resolution of atherosclerosis
Context and Significance.
Macrophages engulf dead cells in a process called efferocytosis to prevent inflammation and promote tissue repair. Such clearance occurs over multiple rounds, and failure of this process promotes chronic inflammatory diseases, such as atherosclerosis. Here, Ira Tabas and colleagues show that continual efferocytosis is dependent on the metabolites macrophages take up from dead cells in previous rounds of efferocytosis. They found that engulfment of arginine and ornithine and their metabolization to putrescine allows for optimization of subsequent rounds of efferocytosis. Mice lacking an enzyme required for such metabolization have defects in efferocytosis and the clearance of plaques during the resolution phase of atherosclerosis. Treatment with exogenous putrescine overcomes this defect and suppresses atherosclerosis progression, suggesting a possible future therapy for this chronic condition.
ACKNOWLEDGMENTS
We thank Dr. Heidi Welch (Babraham Institute) for bone marrow cells from Raichu-Rac FRET transgenic mice and Dr. Michiyuki Matsuda (Kyoto University) for the Raichu-Rac FRET plasmid, Theresa Swayne and Laura Munteanu for assistance with confocal imaging and analysis, Drs. Wayne Orr (LSUHSC-Shreveport) and James Traylor (LSUHSC-Shreveport) for de-identified human atherosclerosis specimens, and Carrolyn Hennecken for assistance with efferocytosis assays. Confocal microscopy was performed in the Confocal and Specialized Microscopy Shared Resource of the Herbert Irving Comprehensive Cancer Center at Columbia University, supported by NIH grants P30 CA013696 and S10 RR025686. Flow cytometry was conducted in the Columbia Center for Translational Immunology Core Facility, funded by NIH grants P30 DK063608 and S10 OD020056. Isotope-labeled tracer experiments were supported by the Proteomics and Metabolomics Core at Moffitt Cancer Center, funded by NIH grant P30 CA076292 and the Moffitt Foundation. Polyamine detection was carried out by the Translational Biomarker Analytical Core facility, supported by NIH grant UL1TR001873. Co-authors were supported by the following NIH grants: T32 HL007343-28 (A.Y. and Z.Z.); K99 HL145131 (A.Y.); F32 HL137398 (S.B.C.); P20 GM121307 (C.G.K.); R01 HL128349 and DK089312 (D.M.M.); and R01 HL075662, HL127464, and HL132412 (I.T.). X.W. was supported by a Liver Scholar Award, and J.L.C. is the Cortner-Couch Endowed Chair for Cancer Research, University of South Florida School of Medicine.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.cmet.2020.01.001.
REFERENCES
- A-Gonzalez N, Bensinger SJ, Hong C, Beceiro S, Bradley MN, Zelcer N, Deniz J, Ramirez C, Díaz M, Gallardo G, et al. (2009). Apoptotic cells promote their own clearance and immune tolerance through activation of the nuclear receptor LXR. Immunity 31, 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhonen L, Karppinen A, Uusi-Oukari M, Vujcic S, Korhonen VP, Halmekytö M, Kramer DL, Hines R, Jänne J, and Porter CW (1998). Correlation of polyamine and growth responses to N1,N11-diethylnorspermine in primary fetal fibroblasts derived from transgenic mice overexpressing spermidine/spermine N1-acetyltransferase. J. Biol. Chem 273, 1964–1969. [DOI] [PubMed] [Google Scholar]
- An J, Muoio DM, Shiota M, Fujimoto Y, Cline GW, Shulman GI, Koves TR, Stevens R, Millington D, and Newgard CB (2004). Hepatic expression of malonyl-CoA decarboxylase reverses muscle, liver and whole-animal insulin resistance. Nat. Med 10, 268–274. [DOI] [PubMed] [Google Scholar]
- Arandjelovic S, and Ravichandran KS (2015). Phagocytosis of apoptotic cells in homeostasis. Nat. Immunol 16, 907–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäck M, Yurdagul A Jr., Tabas I, Öörni K, and Kovanen PT (2019). Inflammation and its resolution in atherosclerosis: mediators and therapeutic opportunities. Nat. Rev. Cardiol 16, 389–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CM, and Steitz JA (2001). HuR and mRNA stability. Cell. Mol. Life Sci 58, 266–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai B, Thorp EB, Doran AC, Sansbury BE, Daemen MJ, Dorweiler B, Spite M, Fredman G, and Tabas I (2017). MerTK receptor cleavage promotes plaque necrosis and defective resolution in atherosclerosis. J. Clin. Invest 127, 564–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai B, Kasikara C, Doran AC, Ramakrishnan R, Birge RB, and Tabas I (2018). MerTK signaling in macrophages promotes the synthesis of inflammation resolution mediators by suppressing CaMKII activity. Sci. Signal 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W, Dai X, Chen J, Zhao J, Xu M, Zhang L, Yang B, Zhang W, Rocha M, Nakao T, et al. (2019). STAT6/Arg1 promotes microglia/macrophage efferocytosis and inflammation resolution in stroke mice. JCI Insight 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinetti-Gbaguidi G, Colin S, and Staels B (2015). Macrophage subsets in atherosclerosis. Nat. Rev. Cardiol 12, 10–17. [DOI] [PubMed] [Google Scholar]
- Clausen BE, Burkhardt C, Reith W, Renkawitz R, and Förster I (1999). Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 8, 265–277. [DOI] [PubMed] [Google Scholar]
- Dalli J, and Serhan CN (2012). Specific lipid mediator signatures of human phagocytes: microparticles stimulate macrophage efferocytosis and pro-resolving mediators. Blood 120, e60–e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Gutierrez E, and Shin BS (2014). The hypusine-containing translation factor eIF5A. Crit. Rev. Biochem. Mol. Biol. 49, 413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwig LP, and Henson PM (2008). Clearance of apoptotic cells by phagocytes. Cell Death Differ. 15, 243–250. [DOI] [PubMed] [Google Scholar]
- Feig JE, Vengrenyuk Y, Reiser V, Wu C, Statnikov A, Aliferis CF, Garabedian MJ, Fisher EA, and Puig O (2012). Regression of atherosclerosis is characterized by broad changes in the plaque macrophage transcriptome. PLoS ONE 7, e39790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fond AM, Lee CS, Schulman IG, Kiss RS, and Ravichandran KS (2015). Apoptotic cells trigger a membrane-initiated pathway to increase ABCA1. J. Clin. Invest 125, 2748–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredman G, and Tabas I (2017). Boosting Inflammation Resolution in Atherosclerosis: The Next Frontier for Therapy. Am. J. Pathol 187, 1211–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredman G, Hellmann J, Proto JD, Kuriakose G, Colas RA, Dorweiler B, Connolly ES, Solomon R, Jones DM, Heyer EJ, et al. (2016). An imbalance between specialized pro-resolving lipid mediators and proinflammatory leukotrienes promotes instability of atherosclerotic plaques. Nat. Commun 7, 12859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire-de-Lima CG, Nascimento DO, Soares MB, Bozza PT, Castro-Faria-Neto HC, de Mello FG, DosReis GA, and Lopes MF (2000). Uptake of apoptotic cells drives the growth of a pathogenic trypanosome in macrophages. Nature 403, 199–203. [DOI] [PubMed] [Google Scholar]
- Grencis RK (2015). Immunity to helminths: resistance, regulation, and susceptibility to gastrointestinal nematodes. Annu. Rev. Immunol 33, 201–225. [DOI] [PubMed] [Google Scholar]
- Hardbower DM, Asim M, Luis PB, Singh K, Barry DP, Yang C, Steeves MA, Cleveland JL, Schneider C, Piazuelo MB, et al. (2017). Ornithine decarboxylase regulates M1 macrophage activation and mucosal inflammation via histone modifications. Proc. Natl. Acad. Sci. USA 114, E751–E760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao CY, and Salic A (2008). Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. USA 105, 15779–15784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson AE, Dai Y, Nobis M, Baker MJ, McGhee EJ, Walker S, Schwarz JP, Kadir S, Morton JP, Myant KB, et al. (2014). The Rac-FRET mouse reveals tight spatiotemporal control of Rac activity in primary cells and tissues. Cell Rep. 6, 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawane K, Ohtani M, Miwa K, Kizawa T, Kanbara Y, Yoshioka Y, Yoshikawa H, and Nagata S (2006). Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature 443, 998–1002. [DOI] [PubMed] [Google Scholar]
- Khanna S, Biswas S, Shang Y, Collard E, Azad A, Kauh C, Bhasker V, Gordillo GM, Sen CK, and Roy S (2010). Macrophage dysfunction impairs resolution of inflammation in the wounds of diabetic mice. PLoS ONE 5, e9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierszenbaum F, Wirth JJ, McCann PP, and Sjoerdsma A (1987). Impairment of macrophage function by inhibitors of ornithine decarboxylase activity. Infect. Immun 55, 2461–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss RS, Elliott MR, Ma Z, Marcel YL, and Ravichandran KS (2006). Apoptotic cells induce a phosphatidylserine-dependent homeostatic response from phagocytes. Curr. Biol 16, 2252–2258. [DOI] [PubMed] [Google Scholar]
- Kojima Y, Volkmer JP, McKenna K, Civelek M, Lusis AJ, Miller CL, Direnzo D, Nanda V, Ye J, Connolly AJ, et al. (2016). CD47-blocking antibodies restore phagocytosis and prevent atherosclerosis. Nature 536, 86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korns D, Frasch SC, Fernandez-Boyanapalli R, Henson PM, and Bratton DL (2011). Modulation of macrophage efferocytosis in inflammation. Front. Immunol 2, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverrier Y, and Ridley AJ (2001). Requirement for Rho GTPases and PI 3-kinases during apoptotic cell phagocytosis by macrophages. Curr. Biol 11, 195–199. [DOI] [PubMed] [Google Scholar]
- Linton MF, Babaev VR, Huang J, Linton EF, Tao H, and Yancey PG (2016). Macrophage Apoptosis and Efferocytosis in the Pathogenesis of Atherosclerosis. Circ. J 80, 2259–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Du H, Rutkowski R, Gartner A, and Wang X (2012). LAAT-1 is the lysosomal lysine/arginine transporter that maintains amino acid homeostasis. Science 337, 351–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marei H, and Malliri A (2017). GEFs: Dual regulation of Rac1 signaling. Small GTPases 8, 90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez FO, Helming L, and Gordon S (2009). Alternative activation of macrophages: an immunologic functional perspective. Annu. Rev. Immunol 27, 451–483. [DOI] [PubMed] [Google Scholar]
- Morioka S, Perry JSA, Raymond MH, Medina CB, Zhu Y, Zhao L, Serbulea V, Onengut-Gumuscu S, Leitinger N, Kucenas S, et al. (2018). Efferocytosis induces a novel SLC program to promote glucose uptake and lactate release. Nature 563, 714–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morioka S, Maueröder C, and Ravichandran KS (2019). Living on the Edge: Efferocytosis at the Interface of Homeostasis and Pathology. Immunity 50, 1149–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepal S, Tiruppathi C, Tsukasaki Y, Farahany J, Mittal M, Rehman J, Prockop DJ, and Malik AB (2019). STAT6 induces expression of Gas6 in macrophages to clear apoptotic neutrophils and resolve inflammation. Proc. Natl. Acad. Sci. USA 116, 16513–16518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newson J, Stables M, Karra E, Arce-Vargas F, Quezada S, Motwani M, Mack M, Yona S, Audzevich T, and Gilroy DW (2014). Resolution of acute inflammation bridges the gap between innate and adaptive immunity. Blood 124, 1748–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Han CZ, Elliott MR, Kinchen JM, Trampont PC, Das S, Collins S, Lysiak JJ, Hoehn KL, and Ravichandran KS (2011). Continued clearance of apoptotic cells critically depends on the phagocyte Ucp2 protein. Nature 477, 220–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz I, Kosti I, Ares M Jr., Cline M, and Mandel-Gutfreund Y (2014). RBPmap: a web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res. 42, W361–W367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesce JT, Ramalingam TR, Mentink-Kane MM, Wilson MS, El Kasmi KC, Smith AM, Thompson RW, Cheever AW, Murray PJ, and Wynn TA (2009). Arginase-1-expressing macrophages suppress Th2 cytokine-driven inflammation and fibrosis. PLoS Pathog. 5, e1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proto JD, Doran AC, Gusarova G, Yurdagul A Jr., Sozen E, Subramanian M, Islam MN, Rymond CC, Du J, Hook J, et al. (2018). Regulatory T Cells Promote Macrophage Efferocytosis during Inflammation Resolution. Immunity 49, 666–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puleston DJ, Buck MD, Klein Geltink RI, Kyle RL, Caputa G, O’Sullivan D, Cameron AM, Castoldi A, Musa Y, Kabat AM, et al. (2019). Polyamines and eIF5A Hypusination Modulate Mitochondrial Respiration and Macrophage Activation. Cell Metab. 30, 352–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes G, Van den Bergh R, De Baetselier P, Ghassabeh GH, Scotton C, Locati M, Mantovani A, and Sozzani S (2005). Arginase-1 and Ym1 are markers for murine, but not human, alternatively activated myeloid cells. J. Immunol 174, 6561. [DOI] [PubMed] [Google Scholar]
- Ramon S, Dalli J, Sanger JM, Winkler JW, Aursnes M, Tungen JE, Hansen TV, and Serhan CN (2016). The Protectin PCTR1 Is Produced by Human M2 Macrophages and Enhances Resolution of Infectious Inflammation. Am. J. Pathol 186, 962–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath M, Müller I, Kropf P, Closs EI, and Munder M (2014). Metabolism via Arginase or Nitric Oxide Synthase: Two Competing Arginine Pathways in Macrophages. Front. Immunol 5, 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray RM, McCormack SA, Covington C, Viar MJ, Zheng Y, and Johnson LR (2003). The requirement for polyamines for intestinal epithelial cell migration is mediated through Rac1. J. Biol. Chem 278, 13039–13046. [DOI] [PubMed] [Google Scholar]
- Ray RM, Bavaria MN, Bhattacharya S, and Johnson LR (2011). Activation of Dbl restores migration in polyamine-depleted intestinal epithelial cells via Rho-GTPases. Am. J. Physiol. Gastrointest. Liver Physiol 300, G988–G997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Van Kampen E, Van Berkel TJ, Cruickshank SM, and Van Eck M (2017). Hematopoietic arginase 1 deficiency results in decreased leukocytosis and increased foam cell formation but does not affect atherosclerosis. Atherosclerosis 256, 35–46. [DOI] [PubMed] [Google Scholar]
- Saha S, Shalova IN, and Biswas SK (2017). Metabolic regulation of macrophage phenotype and function. Immunol. Rev 280, 102–111. [DOI] [PubMed] [Google Scholar]
- Schille S, Crauwels P, Bohn R, Bagola K, Walther P, and van Zandbergen G (2018). LC3-associated phagocytosis in microbial pathogenesis. Int. J. Med. Microbiol 308, 228–236. [DOI] [PubMed] [Google Scholar]
- Sun L, Zhou H, Zhu Z, Yan Q, Wang L, Liang Q, and Ye RD (2015). Ex vivo and in vitro effect of serum amyloid a in the induction of macrophage M2 markers and efferocytosis of apoptotic neutrophils. J. Immunol 194, 4891–4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas I, and Lichtman AH (2017). Monocyte-Macrophages and T Cells in Atherosclerosis. Immunity 47, 621–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasenko TN, Rosas OR, Singh LN, Kristaponis K, Vernon H, and McGuire PJ (2015). A new mouse model of mild ornithine transcarbamylase deficiency (spf-j) displays cerebral amino acid perturbations at baseline and upon systemic immune activation. PLoS ONE 10, e0116594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teupser D, Burkhardt R, Wilfert W, Haffner I, Nebendahl K, and Thiery J (2006). Identification of macrophage arginase I as a new candidate gene of atherosclerosis resistance. Arterioscler. Thromb. Vasc. Biol 26, 365–371. [DOI] [PubMed] [Google Scholar]
- Thomas AC, and Mattila JT (2014). “Of mice and men”: arginine metabolism in macrophages. Front. Immunol 5, 479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas AC, Sala-Newby GB, Ismail Y, Johnson JL, Pasterkamp G, and Newby AC (2007). Genomics of foam cells and nonfoamy macrophages from rabbits identifies arginase-I as a differential regulator of nitric oxide production. Arterioscler. Thromb. Vasc. Biol 27, 571–577. [DOI] [PubMed] [Google Scholar]
- Thorp E, and Tabas I (2009). Mechanisms and consequences of efferocytosis in advanced atherosclerosis. J. Leukoc. Biol 86, 1089–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandivier RW, Henson PM, and Douglas IS (2006). Burying the dead: the impact of failed apoptotic cell removal (efferocytosis) on chronic inflammatory lung disease. Chest 129, 1673–1682. [DOI] [PubMed] [Google Scholar]
- Wang Y, Subramanian M, Yurdagul A Jr., Barbosa-Lorenzi VC, Cai B, de Juan-Sanz J, Ryan TA, Nomura M, Maxfield FR, and Tabas I (2017). Mitochondrial Fission Promotes the Continued Clearance of Apoptotic Cells by Macrophages. Cell 171, 331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willecke F, Yuan C, Oka K, Chan L, Hu Y, Barnhart S, Bornfeldt KE, Goldberg IJ, and Fisher EA (2015). Effects of High Fat Feeding and Diabetes on Regression of Atherosclerosis Induced by Low-Density Lipoprotein Receptor Gene Therapy in LDL Receptor-Deficient Mice. PLoS ONE 10, e0128996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JY, Kao HJ, Li SC, Stevens R, Hillman S, Millington D, and Chen YT (2004). ENU mutagenesis identifies mice with mitochondrial branched-chain aminotransferase deficiency resembling human maple syrup urine disease. J. Clin. Invest 113, 434–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Lan L, Wilson DM, Marquez RT, Tsao WC, Gao P, Roy A, Turner BA, McDonald P, Tunge JA, et al. (2015). Identification and validation of novel small molecule disruptors of HuR-mRNA interaction. ACS Chem. Biol 10, 1476–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn TA, and Vannella KM (2016). Macrophages in Tissue Repair, Regeneration, and Fibrosis. Immunity 44, 450–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Roos A, Schlagwein N, Woltman AM, Daha MR, and van Kooten C (2006). IL-10-producing macrophages preferentially clear early apoptotic cells. Blood 107, 4930–4937. [DOI] [PubMed] [Google Scholar]
- Yurdagul A Jr., Doran AC, Cai B, Fredman G, and Tabas IA (2018). Mechanisms and consequences of defective efferocytosis in atherosclerosis. Front. Cardiovasc. Med 4, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yvan-Charvet L, Pagler TA, Seimon TA, Thorp E, Welch CL, Witztum JL, Tabas I, and Tall AR (2010). ABCA1 and ABCG1 protect against oxidative stress-induced macrophage apoptosis during efferocytosis. Circ. Res 106, 1861–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Weinberg S, DeBerge M, Gainullina A, Schipma M, Kinchen JM, Ben-Sahra I, Gius DR, Yvan-Charvet L, Chandel NS, et al. (2019). Efferocytosis Fuels Requirements of Fatty Acid Oxidation and the Electron Transport Chain to Polarize Macrophages for Tissue Repair. Cell Metab. 29, 443–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Lee HN, Kim SH, Park SA, Kim W, Cha YN, and Surh YJ (2018). Myc-nick promotes efferocytosis through M2 macrophage polarization during resolution of inflammation. FASEB J. 32, 5312–5325. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code.