Abstract
Objective
There are no validated approaches to predict benefit from adjuvant chemotherapy for resected patients with non-small-cell lung cancer (NSCLC). The aim of this study was to translate a 15-gene mRNA expression profile published by Zhu et al, shown to be prognostic and predictive of benefit, into a readily applicable immunohistochemistry (IHC) panel.
Methods
For seven of the genes in the gene expression profile (GEP) for which suitable commercial antibodies were available, we semiquantitatively assessed the IHC expression and prognostic significance for 173 patients treated at the Saint John Regional Hospital (SJRH). Cut-offs for high and low expression were defined for each marker and applied to IHC scores from 291 of the 482 patients in JBR.10, including patients on both the adjuvant chemotherapy and observation arms. The prognostic and predictive value of these markers on overall survival (OS) or recurrence-free survival (RFS) was assessed by Cox regression models.
Results
In the SJRH cohort, in 62 patients with resected stage II–III NSCLC, the prognostic significance of IHC assays for four proteins were concordant with Zhu’s GEP results. Low FOSL2 (OS, HR=0.15; p=0.0001; RFS, HR=0.14; p<0.0001) and high STMN2 (RFS, HR=2.501; p=0.0197) were adverse prognostic factors. Low ATP1B1 and low TRIM14 expression trended toward worse OS and RFS. Validation of these markers with JBR.10 patients failed to show prognostic significance either individually or in combined risk classifications. Additionally, the interaction between these markers and chemotherapy treatment in predicting OS (FOSL2, p=0.52; STMN2 p=0.14; ATP1B1, p=0.33; TRIM14, p=0.81) or RFS (FOSL2, p=0.63; STMN2, p=0.12; ATP1B1, p=0.66; TRIM14, p=0.57) did not reach significance, individually or in combination panels.
Conclusions
Zhu’s GEP could not be translated into an IHC panel predictive of benefit from adjuvant chemotherapy. Future predictive biomarker analysis in the adjuvant NSCLC setting may need to focus on novel therapies.
Keywords: non-small-cell lung cancer, immunohistochemistry, gene expression profile, biomarker, predictive, adjuvant chemotherapy
Key questions.
What is already known about this subject?
Adjuvant chemotherapy for non-small-cell lung cancer (NSCLC) improves overall survival in approximately 5%–15% of patients.
There are no validated clinical tools to identify those patients likely to benefit from adjuvant chemotherapy.
What does this study add?
Using published predictive gene expression profiles, we developed an immunohistochemistry-based prognostic biomarker panel.
Analysis of this biomarker panel with the JBR.10 clinical dataset was unable to validate any predictive benefit.
How might this impact on clinical practice?
The opportunity to identify predictive biomarkers for adjuvant chemotherapy in NSCLC is closing.
Future predictive biomarker analysis in the adjuvant NSCLC setting may need to focus on novel therapies.
Introduction
Based on several randomised clinical trials including JBR.10,1 Adjuvant Navelbine International Trialist Association (ANITA),2 CALGB3 and International Adjuvant Lung Trial (IALT),4 platinum-based adjuvant chemotherapy is standard of care for patients with resected stage II–IIIA non-small-cell lung cancer (NSCLC). The most significant result came from the JBR.10 study which combined cisplatin with vinorelbine to achieve a 15% improvement in overall survival (OS).1 A meta-analysis of these studies showed an absolute benefit of 5.4% in 5-year OS.5 Although this result is clinically significant, it is important to note that for the remaining 85%–95% of patients, the addition of chemotherapy to surgery may not provide any long-term benefit and may, in some cases, cause more harm. Therefore, identifying patients who will not benefit from adjuvant chemotherapy by developing predictive biomarkers will prevent unnecessary toxicity and decrease resource utilisation needed to administer this treatment.
The LACE-Bio group has been active in the development of such biomarkers. Using immunohistochemistry (IHC), low protein expression of ERCC1 was shown to be predictive of benefit from platinum-based chemotherapy using the large IALT.6 Similarly, high expression of TUBB3 was shown to be predictive using the JBR.10 clinical trial.7 However, attempts to validate ERCC1 or TUBB3 as predictive biomarkers fell short in large cross-validation studies.8 9 Similar attempts to validate other single biomarkers such as p53, RRM1 and p2710–12 also failed to show any clinical utility.
Panels that consist of several biomarkers may prove more informative and reflect the complex heterogeneity of tumours. Recently, several gene expression panels have been published that are both prognostic and predictive of benefit from adjuvant chemotherapy in patients with NSCLC.13–19 Of these panels, only four13 15 16 18 were derived from genome-wide mRNA expression profiling (GEP). The 15-gene signature of Zhu et al was derived from GEP of the JBR.10 study, the only such dataset from a randomised trial with a no-treatment control arm. The Moon et al signature was computationally derived from the GEP of the Zhu’s study18 but lacked any validation on additional datasets. The remaining two GEP datasets were validated on this same JBR.10 dataset but were not derived from it a priori.
The 15-gene mRNA signature published by Zhu et al13 was derived from 172 genes that were independently prognostic and further validated as predictive as a biomarker panel. Despite its significance, this 15-gene mRNA signature has limitations that have prevented it from being routinely applied. The primary objective of the study presented here was to translate the 15-gene GEP published by Zhu et al into a readily applicable IHC protein-based biomarker panel that is not only prognostic, but more importantly, is predictive of benefit from adjuvant chemotherapy. For this, we assessed protein expression and the prognostic significance of these biomarkers using tissues from patients treated locally. These biomarkers were then validated on tissues collected as part of the JBR.10 study.
Patients and methods
Patients and tumour tissue
The Saint John Regional Hospital (SJRH) cohort comprised 173 patients with stage I–III NSCLC (American Joint Committee on Cancer, Seventh Edition) under approval of the Horizon Health Network Research Ethics Board. JBR.10 was a North American Intergroup trial led by the Canadian Clinical Trials Group that evaluated benefit from adjuvant chemotherapy in patients with stage IB–II NSCLC.1 Of the 482 patients enrolled in the trial, formalin-fixed paraffin-embedded (FFPE) tissue from 291 patients was available to us in the form of tissue microarrays (TMAs). This subset of patients on the TMA is representative of the overall trial population.7 For both the SJRH and JBR.10 cohorts, TMAs were constructed from 0.6 mm cores from three separate tumour areas. Serial 4 µm sections from each tissue block were mounted on glass slides for immunostaining.
Immunohistochemistry
TMA slides from the SJRH cohort or the JBR.10 cohort were stained with antibodies following optimised protocols. Briefly, following deparaffinisation, rehydration and antigen retrieval, antibodies were applied and left overnight. Next day, chromogenic detection involved the Elite ABC HRP biotin–avidin peroxidase system (Vectastain) followed by 3,3′-diaminobenzidine (Sigma). Antibodies were as follows: TRIM14, STMN2, UMPS from Proteintech; ATP1B1 and FOSL2 from Sigma; HEXIM1 from Abcam and FAM64A from Novus Biologicals. Antibody lots remained consistent between the SJRH and JBR.10 cohorts. Expression of each protein was scored by two independent observers using the H-score method, generating a semiquantitative measure between 0 and 200 based on the percentage and intensity of staining in malignant cells. Individual scores were averaged and scores that differed by more than 20% were resolved by consensus. Due to limited material, TRIM14 staining only included four of eight SJRH TMAs.
Statistical analysis
Cut-offs for each marker were determined in the SJRH cohort using Cutoff Finder20 (http://molpath.charite.de/cutoff/) by optimising cut-offs based on survival outcomes. These cut-offs were then applied to the JBR.10 cohort. Associations between biomarker expression groups and covariates were assessed using the χ2 test. Differences in OS and recurrence-free survival (RFS) between high-expressing and low-expressing groups were compared using the log-rank test. The Cox proportional hazards model was used to assess the independent value of each marker adjusted for other identified covariates including age, sex, histology, disease stage and type of surgery. This model also included the interaction between treatment assignment and biomarker expression to determine the predictive value of these biomarkers. For the subset of JBR.10 patients who had GEP performed on frozen tissues,13 mRNA levels were correlated with H-scores derived from IHC for each of the four assayed proteins. All p-values are two sided.
Results
Immunohistochemical data and SJRH baseline characteristics
Of the 15 genes in the Zhu GEP, we were able to validate antibodies for IHC for seven proteins: FOSL2, ATP1B1, STMN2, TRIM14, HEXIM1, UMPS and FAM64A (figure 1 and online supplementary figure S1). Applying these antibodies to the SJRH TMAs, there was significant correlation of immunohistochemical H-scores between biomarkers (online supplementary table S1). Specifically, FOSL2 expression negatively correlated with ATP1B1 expression and positively correlated with STMN2 and HEXIM1. STMN2 associated with TRIM14, UMPS and FAM64A; TRIM14 correlated with UMPS expression and FAM64A correlated with both ATP1B1 and UMPS. A comparison of the baseline patient characteristics in the SRJH cohort between high and low expression for each biomarker is shown in table 1 and online supplementary table S2. Significant imbalances were seen in FOSL2, where patients who received chemotherapy after surgery were more likely to have high FOSL2 expression (p=0.02), and in STMN2 where patients with high expression were more likely to be older than 65 years (p=0.01).
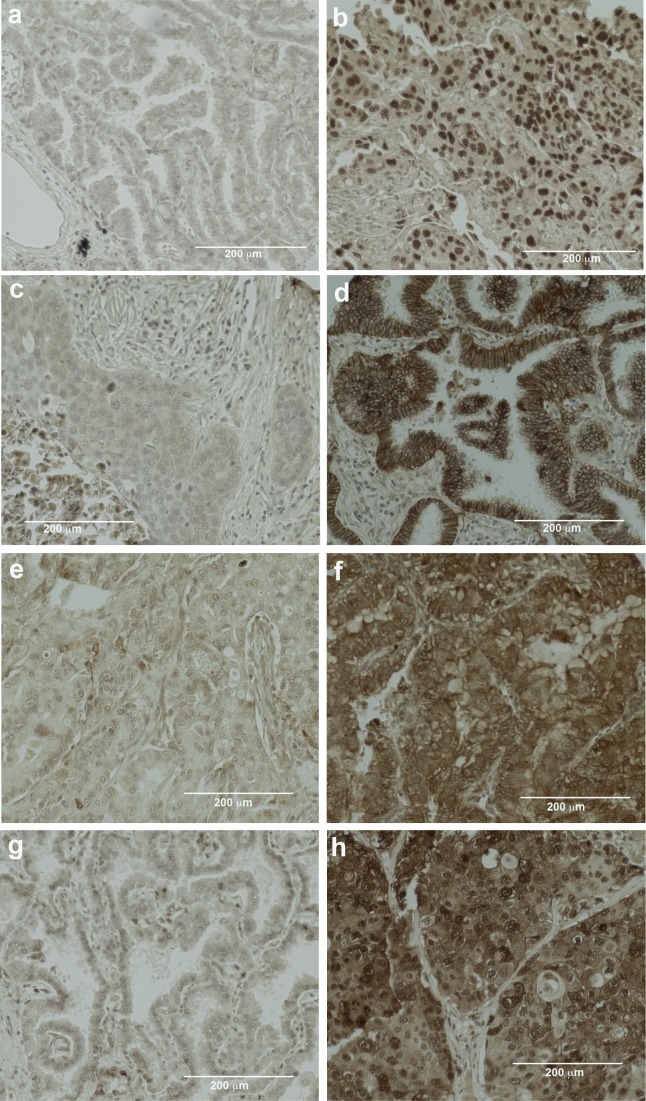
Figure 1.
Immunohistochemistry staining of biomarkers on the Saint John Regional Hospital tissue microarrays. Representative images demonstrating low and high nuclear staining of FOSL2 (A, B), membrane and cytoplasmic staining of ATP1B1 (C, D), cytoplasmic staining of STMN2 (E, F) and cytoplasmic staining of TRIM14 (G, H), respectively.
Table 1.
Association of biomarkers with clinicopathological parameters in the SJRH cohort
| FOSL2 | P value | ATP1B1 | P value | STMN2 | P value | TRIM14 | P value | |||||
| Characteristics | High (n=125) | Low (n=43) | High (n=32) | Low (n=142) | High (n=59) | Low (n=112) | High (n=50) | Low (n=41) | ||||
| Age, years | 0.85 | 0.68 | 0.01 | 0.66 | ||||||||
| <65 | 39 (31.2%) | 14 (32.6%) | 9 (28.1%) | 46 (32.4%) | 11 (18.6%) | 42 (37.5%) | 19 (38.0%) | 13 (31.7%) | ||||
| ≥65 | 86 (68.8) | 29 (67.4%) | 23 (78.9%) | 96 (67.6%) | 48 (81.4%) | 70 (62.5%) | 31 (62.0%) | 28 (68.3%) | ||||
| Sex | 0.22 | 0.12 | 0.34 | 0.29 | ||||||||
| Male | 61 (48.8%) | 26 (60.5%) | 12 (37.5%) | 77 (54.2%) | 27 (45.8%) | 60 (53.6%) | 26 (52.0%) | 16 (39.1%) | ||||
| Female | 64 (51.2%) | 17 (39.5%) | 20 (62.5%) | 66 (46.4%) | 32 (54.2%) | 52 (46.4%) | 24 (48.0%) | 25 (60.9%) | ||||
| TNM stage | 0.38 | 0.29 | 0.1 | 0.79 | ||||||||
| I | 74 (59.2%) | 30 (69.8%) | 18 (56.2%) | 90 (63.4%) | 43 (72.9%) | 63 (56.3%) | 34 (68.0%) | 26 (63.4%) | ||||
| II | 33 (26.4%) | 7 (16.3%) | 11 (34.4%) | 31 (21.8%) | 10 (16.9%) | 31 (27.7%) | 10 (20.0%) | 8 (19.5%) | ||||
| III | 18 (14.4%) | 6 (13.9%) | 3 (9.4%) | 21 (14.8%) | 6 (10.2%) | 18 (16.0%) | 6 (12.0%) | 7 (17.1%) | ||||
| Histology | 0.5 | 0.64 | 0.29 | 0.2 | ||||||||
| Adeno | 64 (51.2%) | 21 (48.8%) | 14 (43.8%) | 72 (50.7%) | 29 (49.2%) | 56 (50.0%) | 29 (58.0%) | 16 (39.1%) | ||||
| SCC | 44 (25.2%) | 13 (30.2%) | 11 (34.3%) | 47 (33.1%) | 17 (28.8%) | 41 (36.6%) | 16 (32.%) | 19 (46.3%) | ||||
| Other | 17 (13.6%) | 9 (20.9%) | 7 (21.9%) | 22 (15.5%) | 13 (22.0%) | 15 (13.4%) | 5 (10.0%) | 6 (14.6%) | ||||
| Type of surgery | 1 | 0.7 | 0.75 | 1 | ||||||||
| Pneumonectomy | 9 (7.2%) | 3 (7.0%) | 1 (3.1%) | 11 (7.7%) | 5 (8.5%) | 7 (6.3%) | 3 (6.0%) | 3 (7.3%) | ||||
| Lesser surgery | 116 (92.8%) | 40 (93.0%) | 31 (96.9%) | 131 (92.3%) | 54 (91.5%) | 105 (93.7%) | 47 (94.0%) | 38 (92.7%) | ||||
| Chemotherapy | 0.02 | 0.47 | 0.63 | 0.68 | ||||||||
| Post | 44 (35.2%) | 9 (20.9%) | 8 (25.0% | 47 (33.1%) | 16 (27.1%) | 39 (34.8%) | 15 (30.0%) | 9 (22.0%) | ||||
| None | 77 (61.6%) | 28 (65.1%) | 21 (65.6%) | 88 (62.0%) | 39 (66.1%) | 67 (59.8%) | 32 (64.0)% | 29 (70.7%) | ||||
| Other | 4 (3.2%) | 6 (14.5%) | 3 (9.4%) | 7 (4.9%) | 4 (6.8%) | 6 (5.4%) | 3 (6.0%) | 3 (7.3%) | ||||
| KRAS mutation | 0.24 | 0.51 | 0.37 | 0.63 | ||||||||
| Present | 33 (26.4%) | 16 (37.2%) | 7 (21.9%) | 41 (28.9%) | 14 (23.7%) | 35 (31.2%) | 14 (28.0%) | 9 (22.0%) | ||||
| Absent | 92 (73.6%) | 27 (62.8%) | 25 (78.1%) | 101 (71.1%) | 45 (76.3%) | 77 (68.8%) | 36 (72.0%) | 32 (78.0%) | ||||
*Bold values represent statistical significance
adeno, adenocarcinoma; SCC, squamous cell carcinoma; SJRH, Saint John Regional Hospital.
esmoopen-2020-000679supp001.pdf (1.6MB, pdf)
Prognostic significance of SJRH cohort
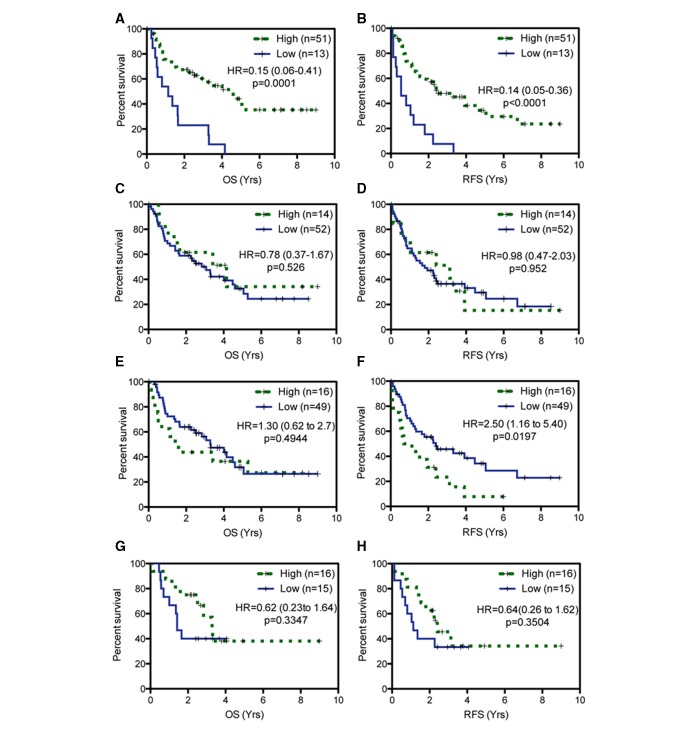
In all stage I–III patients in the SJRH cohort, significant differences in OS or recurrence-free survival (RFS) between high and low expressions were found for FOSL2 (OS, HR=0.5, p=0.0089; RFS, HR=0.56, p=0.0184) and UMPS (OS, HR=0.52, p=0.0109; RFS, HR=0.62, p=0.0439) (online supplementary table S3). To better reflect the population at high risk of recurrence, correlations between survival and biomarker expression were restricted to patients with stage II–III disease (figure 2, online supplementary figure S2). In 62 patients with resected stage II–III NSCLC, in univariate analysis, the prognostic significance for four proteins were concordant with the Zhu GEP results.13 Low FOSL2 (OS, HR=0.15, p=0.0001; RFS, HR=0.14, p<0.0001) and high STMN2 (RFS, HR=2.501, p=0.0197) were adverse prognostic factors. Low ATP1B1 (HR=0.78, p=0.526) and low TRIM14 expression (HR=0.64, p=0.3504) trended toward worse OS and RFS, respectively. The remaining three proteins, HEXIM1, UMPS and FAM64A, (online supplementary table S3) were not concordant with Zhu’s GEP and therefore excluded from subsequent analysis.
Figure 2.
Prognostic evaluation of individual biomarkers in Saint John Regional Hospital cohort. The overall survival (OS) and relapse-free survival (RFS) curves according to FOSL2 (A, B), ATP1B1 (C, D), STMN2 (E, F) and TRIM14 (G, H) expression, respectively.
Prognostic significance in the JBR.10 cohort
TMAs from 291 participants of the JBR.10 trials were stained, scored and cut-offs applied for FOSL2, ATP1B1, TRIM14 and STMN2 following protocols established for the SJRH cohort. As shown in table 2, patients with high expression of TRIM14 were less likely to be of squamous cell histology (p=0.01). No other significant correlations were found between the other biomarkers and any of the defined covariates. Validation of these biomarkers in univariate or multivariate analysis with JBR.10 patients failed to show prognostic significance for either OS or RFS (table 3). High STMN2 (OS, HR=1.25, p=0.3936; RFS, HR=1.33, p=0.2567) and low TRIM14 (OS, HR=0.78, p=0.3142; RFS, HR=0.80, p=0.337) trended toward being predictors of worse OS or RFS, as they did in the SJRH cohort. For the subset of patients who had matching mRNA and protein expression scores, we evaluated their correlation. As shown in online supplementary table S4, only ATP1B1 protein and mRNA expression values were significantly associated (p=0.0226).
Table 2.
Association of biomarkers with clinicopathological parameters in the JBR.10 cohort
| FOSL2 | P value | ATP1B1 | P value | STMN2 | P value | TRIM14 | P value | |||||
| Characteristics | High (n=227) | Low (n=64) | High (n=13) | Low (n=264) | High (n=67) | Low (n=219) | High (n=121) | Low (n=164) | ||||
| Age, years | 0.77 | 0.77 | 0.96 | 0.51 | ||||||||
| <65 | 148 (65.2%) | 43 (67.2%) | 8 (61.5%) | 173 (65.5%) | 44 (65.7%) | 143 (65.3%) | 82 (67.8%) | 105 (64.0%) | ||||
| ≥65 | 79 (34.8) | 21 (38.2%) | 5 (38.5%) | 91 (34.5%) | 23 (34.3%) | 76 (34.7%) | 39 (32.2%) | 59 (36.0%) | ||||
| Sex | 0.69 | 0.37 | 0.66 | 0.52 | ||||||||
| Male | 150 (66.1%) | 44 (68.8%) | 7 (53.8%) | 177 (67.0%) | 43 (64.2%) | 147 (67.1%) | 79 (65.30%) | 113 (68.9%) | ||||
| Female | 77 (33.9%) | 20 (31.2%) | 6 (46.2%) | 87 (33.0%) | 24 (35.8%) | 72 (32.9%) | 42 (34.8%) | 51 (31.1%) | ||||
| Node | 0.59 | 0.45 | 0.3 | 0.81 | ||||||||
| N1 | 115 (50.7%) | 30 (46.9%) | 8 (61.5%) | 134 (50.8%) | 30 (44.8%) | 114 (52.1%) | 63 (52.1%) | 83 (50.6%) | ||||
| N0 | 112 (49.3%) | 34 (53.1%) | 5 (38.5%) | 130 (49.2%) | 37 (55.2%) | 105 (47.9%) | 58 (47.9%) | 81 (49.4%) | ||||
| Histology | 0.29 | 0.44 | 0.96 | 0.01 | ||||||||
| Adeno | 118 (52.0%) | 40 (62.5%) | 8 (61.5%) | 141 (53.4%) | 36 (53.7%) | 118 (53.9%) | 67 (55.4%) | 83 (50.6%) | ||||
| SCC | 87 (38.3%) | 18 (28.1%) | 3 (23.1%) | 98 (37.1%) | 25 (37.3%) | 79 (36.1%) | 35 (28.9%) | 70 (42.7%) | ||||
| Other | 22 (9.7%) | 6 (9.4%) | 2 (15.4%) | 25 (9.5%) | 6 (9.0%) | 22 (10.0%) | 19 (15.7%) | 11 (6.7%) | ||||
| Type of surgery | 0.64 | 1 | 0.65 | 0.93 | ||||||||
| Pneumonectomy | 56 (24.7%) | 14 (21.9%) | 3 (23.1%) | 64 (24.2%) | 15 (22.4%) | 55 (25.1%) | 29 (24.0%) | 40 (24.4%) | ||||
| Lesser surgery | 171 (75.3%) | 50 (78.1%) | 10 (76.9%) | 200 (75.8%) | 52 (77.6%) | 164 (74.9%) | 92 (76.0%) | 124 (75.6%) | ||||
| KRAS mutation | 0.85 | 0.45 | 0.42 | 0.07 | ||||||||
| Present | 52 (22.9%) | 14 (21.9%) | 0 (0%) | 61 (23.1%) | 17 (25.4%) | 46 (21.0%) | 33 (27.3%) | 30 (18.3%) | ||||
| Absent | 174 (76.7%) | 50 (78.1%) | 13 (100%) | 202 (76.5%) | 49 (73.1) | 173 (79.0%) | 87 (71.9%) | 134 (81.7%) | ||||
| Missing | 1 (4.4%) | 0 (0%) | 0 (0%) | 1 (0.4%) | 1 (1.5%) | 0 (0%) | 1 (0.8%) | 0 (0%) | ||||
*Bold values represent statistical significance
adeno, adenocarcinoma; SCC, squamous cell carcinoma.
Table 3.
Prognostic significance of biomarkers in JBR.10 cohort
| Overall survival | ||||
| Univariate | Multivariate | |||
| Biomarker | HR (CI) | P value | HR (CI) | P value |
| FOSL2 | 1.27 (0.78 to 2.26) | 0.4077 | 1.22 (0.68 to 2.17) | 0.51 |
| ATP1B1 | 1.06 (0.33 to 3.39) | 0.9237 | 0.83 (0.25 to 2.77) | 0.76 |
| STMN2 | 1.25 (0.75 to 2.10) | 0.3936 | 1.22 (0.72 to 2.06) | 0.46 |
| TRIM14 | 0.78 (0.48 to 1.27) | 0.3142 | 0.66 (0.40 to 1.09) | 0.1 |
| Recurrence-free survival | ||||
| Univariate | Multivariate | |||
| HR (CI) | P value | HR (CI) | P value | |
| FOSL2 | 1.13 (0.68 to 1.89) | 0.6408 | 1.08 (0.64 to 1.82) | 0.77 |
| ATP1B1 | 1.33 (0.49 to 3.66) | 0.5768 | 1.65 (0.59 to 4.58) | 0.34 |
| STMN2 | 1.33 (0.81 to 2.16) | 0.2567 | 1.33 (0.81 to 2.17) | 0.26 |
| TRIM14 | 0.80 (0.50 to 1.27) | 0.337 | 0.69 (0.43 to 1.11) | 0.12 |
CI, 95% Confidence Interval; HR, Hazard ratio.
Interaction between treatment assignment and biomarker status in predicting OS and RFS in the JBR.10 cohort
Importantly, JBR.10 includes patients randomised to observation and adjuvant chemotherapy treatment arms, allowing for the investigation of the predictive aspect of these biomarkers. Following defined gene expression classifications by Zhu et al,13 low FOSL2, ATP1B1, TRIM14 and high STMN2 are considered high-risk factors and likely to predict benefit from chemotherapy. The interaction between the four biomarkers and chemotherapy treatment in predicting OS (FOSL2, p=0.52; STMN2, p=0.14; ATP1B1, p=0.33; TRIM14, p=0.81) or RFS (FOSL2, p=0.63; STMN2, p=0.12; ATP1B1, p=0.66; TRIM14, p=0.57) did not reach significance individually (table 4). Only high STMN2 and low TRIM14 trended toward being high-risk biomarkers. However, a combined risk classification system that incorporated high STMN2 and low TRIM14 also failed to reach statistical significance (table 4).
Table 4.
Predictive significance of biomarkers in JBR.10 cohort
| Overall survival | |||||
| High risk | Low risk | Interaction | |||
| Biomarker | HR (CI) | P value | HR (CI) | P value | P value |
| FOSL2 | 0.99 (0.46 to 2.11) | 0.97 | 0.76 (0.51 to 1.11) | 0.15 | 0.52 |
| ATP1B1 | 0.88 (0.61 to 1.25) | 0.47 | 0.30 (0.03 to 2.87) | 0.27 | 0.33 |
| STMN2 | 0.47 (0.22 to 0.99) | 0.04 | 0.94 (0.63 to 1.40) | 0.75 | 0.14 |
| TRIM14 | 0.76 (0.49 to 1.18) | 0.21 | 0.80 (0.46 to 1.40) | 0.44 | 0.81 |
| STMN2/TRIM14 | 0.77 (0.44–1.75) | 0.72 | 0.88 (0.51–1.13) | 0.17 | 0.62 |
Bolded values represent statistical significance
*Bold values indicate statistical signficance
CI, 95% confidence interval; HR, Hazard ratio.
| Recurrence-free survival | |||||
| High risk | Low risk | Interaction | |||
| Biomarker | HR (CI) | P value | HR (CI) | P value | P value |
| FOSL2 | 0.81 (0.41 to 1.61) | 0.54 | 0.68 (0.47to 0.98) | 0.04 | 0.63 |
| ATP1B1 | 0.76 (0.54 to 1.06) | 0.11 | 0.44 (0.09 to 2.12) | 0.3 | 0.66 |
| STMN2 | 0.44 (0.22 to 0.89) | 0.02 | 0.82 (0.56 to 1.88) | 0.29 | 0.12 |
| TRIM14 | 0.64 (0.42 to 0.98) | 0.04 | 0.78 (0.47 to 1.31) | 0.34 | 0.57 |
| STMN2/TRIM14 | 0.65 (0.45 to 0.96) | 0.03 | 0.86 (0.461.63) | 0.64 | 0.43 |
Discussion
We successfully translated four genes from the GEP published by Zhu et al13 into prognostically significant biomarkers evaluable by IHC (figure 1). However, despite the use of appropriate methodology, we were unable to validate either the prognostic or the predictive aspect of FOSL2, ATP1B1, STMN2 or TRIM14 individually or in combined risk stratifications using the JBR.10 clinical trial dataset (tables 3 and 4). Additionally, mRNA and protein expression in the JBR.10 dataset only weakly correlated or were not associated at all (online supplementary table S4). These results demonstrate that translating mRNA-based predictive biomarkers into IHC-based protein biomarkers is difficult.
A recent study by the LACE-Bio group showed that validation of individual predictive markers initially identified in the original studies was also not possible.12 Many of these original biomarkers were chosen because they were hypothesised to be biologically relevant for mediating a response to adjuvant chemotherapy. For example, expression or mutation of the tumour suppressor p53 that regulates cell cycle progression and apoptosis was found to be both prognostic and predictive in the JBR.10 dataset,21 but this result could not be replicated with either the CALGB22 or the IALT23 studies. Similarly, expression of β-tubulin, which was previously shown to be important in mediating resistance to antitubulin agents, was identified as a prognostic and predictive factor in the JBR.10 dataset7 but could not be replicated in LACE-Bio cross-validation studies.8
Biological impact can strengthen the adoption of biomarkers in clinical practice.24 Importantly, the four proteins that we successfully validated as prognostic markers in the SJRH cohort have some biological significance in the context of mediating chemotherapeutic resistance. FOSL2 functions as a transcription factor that regulates numerous genes25; TRIM14 was recently shown to act as a tumour suppressor in NSCLC by inhibiting cancer cell growth and promoting apoptosis.26 ATP1B1 encodes the beta subunit of the Na+K+ ATPase pump and has been implicated as a tumour suppressor in renal cell carcinoma.27 28 Additionally, higher expression of Na+K+ ATPase has also been shown to increase platinum-based chemotherapy sensitivity.29 30 STMN2 is primarily a neuronal protein involved in differentiation and tubulin destabilisation but has been implicated in cisplatin resistance in testicular embryonic carcinoma cells31 and shown to be upregulated in NSCLC in response to deregulation of a long non-coding RNA.32
Our study focused on translating a GEP that was derived from an unbiased statistical approach. From the JBR.10 dataset, 172 genes were identified as prognostic using a median cut-off, and from here, a minimum set of 15 genes were identified that could classify patients into high-risk and low-risk groups.13 Several key differences exist between the SJRH and JBR.10 cohorts. First, the patient populations between these two cohorts were not the same, as JBR.10 did not include stage III patients. Second, in the Zhu13 study, a median cut-off was applied to define univariate prognostic significance. In the SJRH dataset, median cut-offs were not significant for any of the markers and there was no linear relationship between protein expression and prognosis. Therefore, we used a cut-off-finding algorithm that determines the best separation between high-expressing and low-expressing groups.20 These cut-offs were then applied to the JBR.10 cohort for validation. Despite these efforts, we were unable to validate the prognostic significance of the biomarkers identified in the SJRH cohort.
The inability to validate these biomarkers may stem from some of the inherent limitations to our study. Many biomarkers fail validation studies due to low sample size.33 Although we cannot rule out that a larger sample size would have identified more significant biomarkers, despite the relatively low sample size in the SJRH (n=173) and further narrowed by only those with stage II–III disease (n=62), this cohort was still large enough to uncover statistically significant prognostic biomarkers. Second, issues with immunohistochemical biomarker standardisation, validation and reproducibility are well established.24 Antibody validation is also paramount for effective IHC analysis. Different antibodies against the well-studied ERCC1 biomarker showed different outcomes34 and the most widely used antibody was recently shown to be non-specific.35 Although antibody specificity was carefully assessed and validation of our results from SJRH cohort was prospectively planned for the JBR.10 cohort, maintaining assay and antibody conditions, differences in tissue fixation, collection and age, as well as scoring differences and antibody specificity cannot be ruled out.
The central dogma of molecular biology states that DNA is translated into mRNA, which is subsequently transcribed to protein. Success of our study relied on the assumption that this occurs in a linear manner such that mRNA and protein levels would correlate. However, discordance between mRNA and protein level has been observed in lung cancer tissues, among other tissue types.36 A lack of correlation between quantitative reverse transcription PCR and IHC has been reported for several NSCLC biomarkers including ERCC1, TUBB3, RRM1and BRCA1.37 This lack of association in our (online supplementary table S4) and these studies may reflect the dynamic transition from mRNA to protein and the available intracellular resources for that function.38 Specifically, this dynamic transition involves factors that not only regulate translation efficiency and protein stability39 but also the many factors that affect mRNA such as microRNAs, long non-coding RNAs, RNA-binding proteins40 and epigenetic control of RNA expression.41 The dynamic molecular interactions involved in these processes limit the assumption that mRNA and protein levels should correlate in biomarker studies.
The tumour tissue collected as part of the LACE-Bio trials like JBR.10 provided a unique opportunity to identify and validate biomarkers predictive of benefit from adjuvant chemotherapy. These trials were all randomised phase III studies from the early-mid 1990s that included watchful waiting as the standard-of-care control arms. Because these trials will never be repeated, the ability to define predictive biomarkers relies on the tissues available from these trials. Only the JBR.10 trial collected frozen tissue on a subset (133 of 482) of patients, from which the GEP developed by Zhu was derived.13 JBR.10 and the other pivotal trials did include the collection of FFPE tissue on ~1400 patients in total,7 8 but attempts to validate the signature in FFPE tissue using nanostring technology proved difficult (personal communication). IHC is the most widely used application for FFPE tissue, which is why our study focused on translating a predictive GEP into an IHC-based protein biomarker panel.
With the failure to translate a GEP into an immunohistochemical protein-based biomarker panel, as shown in our study, the opportunity to define predictive biomarkers for adjuvant chemotherapy for early-stage NSCLC is closing. Instead, the focus should shift toward identifying novel therapeutic approaches to better treat this patient population, with integral identification of predictive biomarkers.
Footnotes
Contributors: Conception and design of work: SG, CA and TR. Acquisition of data: SG, JM, MF, AR, ML, FH, AM, JA and CA. Data analysis and interpretation: SG, KD and TR. Writing the manuscript and intellectual input: SG, JT, FAS and TR.
Funding: This work was supported by a Canadian Cancer Society Research Chair and Canadian Institutes of Health (CIHR) Strategy for Patient-Oriented Research (SPOR) Mentorship Chair to TR and a fellowship award from the Beatrice Hunter Cancer Research Institute, New Brunswick Health Research Foundation and Lung Cancer Canada to SG.
Competing interests: None declared.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available on reasonable request.
References
- 1.Winton T, Livingston R, Johnson D, et al. Vinorelbine plus cisplatin vs. observation in resected non-small-cell lung cancer. N Engl J Med 2005;352:2589–97. 10.1056/NEJMoa043623 [DOI] [PubMed] [Google Scholar]
- 2.Douillard J-Y, Rosell R, De Lena M, et al. Adjuvant vinorelbine plus cisplatin versus observation in patients with completely resected stage IB-IIIA non-small-cell lung cancer (Adjuvant Navelbine International Trialist Association [ANITA]): a randomised controlled trial. Lancet Oncol 2006;7:719–27. 10.1016/S1470-2045(06)70804-X [DOI] [PubMed] [Google Scholar]
- 3.Strauss GM, Herndon JE, Maddaus MA, et al. Adjuvant paclitaxel plus carboplatin compared with observation in stage Ib non–small-cell lung cancer: CALGB 9633 with the cancer and leukemia group B, radiation therapy Oncology group, and North central cancer treatment group study groups. JCO 2008;26:5043–51. 10.1200/JCO.2008.16.4855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arriagada R, Bergman B, Dunant A, et al. Cisplatin-Based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N Engl J Med 2004;350:351–60. 10.1056/NEJMoa031644 [DOI] [PubMed] [Google Scholar]
- 5.Pignon J-P, Tribodet H, Scagliotti GV, et al. Lung adjuvant cisplatin evaluation: a pooled analysis by the lacE Collaborative group. J Clin Oncol 2008;26:3552–9. 10.1200/JCO.2007.13.9030 [DOI] [PubMed] [Google Scholar]
- 6.Olaussen KA, Dunant A, Fouret P, et al. Dna repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med 2006;355:983–91. 10.1056/NEJMoa060570 [DOI] [PubMed] [Google Scholar]
- 7.Sève P, Lai R, Ding K, et al. Class III beta-tubulin expression and benefit from adjuvant cisplatin/vinorelbine chemotherapy in operable non-small cell lung cancer: analysis of NCIC JBR.10. Clin Cancer Res 2007;13:994–9. 10.1158/1078-0432.CCR-06-1503 [DOI] [PubMed] [Google Scholar]
- 8.Reiman T, Lai R, Veillard AS, et al. Cross-Validation study of class III beta-tubulin as a predictive marker for benefit from adjuvant chemotherapy in resected non-small-cell lung cancer: analysis of four randomized trials. Ann Oncol 2012;23:86–93. 10.1093/annonc/mdr033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friboulet L, Olaussen KA, Pignon J-P, et al. Ercc1 isoform expression and DNA repair in non-small-cell lung cancer. N Engl J Med 2013;368:1101–10. 10.1056/NEJMoa1214271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olaussen KA, Postel-Vinay S. Predictors of chemotherapy efficacy in non-small-cell lung cancer: a challenging landscape. Ann Oncol 2016;27:2004–16. 10.1093/annonc/mdw321 [DOI] [PubMed] [Google Scholar]
- 11.Wallerek S, Sørensen JB. Biomarkers for efficacy of adjuvant chemotherapy following complete resection in NSCLC stages I-IIIA. Eur Respir Rev 2015;24:340–55. 10.1183/16000617.00005814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seymour L, Le Teuff G, Brambilla E, et al. LACE-Bio: validation of predictive and/or prognostic Immunohistochemistry/Histochemistry-based biomarkers in resected non-small-cell lung cancer. Clin Lung Cancer 2019;20:66–73. 10.1016/j.cllc.2018.10.001 [DOI] [PubMed] [Google Scholar]
- 13.Zhu C-Q, Ding K, Strumpf D, et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J Clin Oncol 2010;28:4417–24. 10.1200/JCO.2009.26.4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen D-T, Hsu Y-L, Fulp WJ, et al. Prognostic and predictive value of a malignancy-risk gene signature in early-stage non-small cell lung cancer. J Natl Cancer Inst 2011;103:1859–70. 10.1093/jnci/djr420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang H, Xiao G, Behrens C, et al. A 12-gene set predicts survival benefits from adjuvant chemotherapy in non-small cell lung cancer patients. Clin Cancer Res 2013;19:1577–86. 10.1158/1078-0432.CCR-12-2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Laar RK. Genomic signatures for predicting survival and adjuvant chemotherapy benefit in patients with non-small-cell lung cancer. BMC Med Genomics 2012;5:30 10.1186/1755-8794-5-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wistuba II, Behrens C, Lombardi F, et al. Validation of a proliferation-based expression signature as prognostic marker in early stage lung adenocarcinoma. Clin Cancer Res 2013;19:6261–71. 10.1158/1078-0432.CCR-13-0596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moon H, Zhao Y, Pluta D, et al. Subgroup analysis based on prognostic and predictive gene signatures for adjuvant chemotherapy in early-stage non-small-cell lung cancer patients. J Biopharm Stat 2018;28:750–62. 10.1080/10543406.2017.1397006 [DOI] [PubMed] [Google Scholar]
- 19.Buhl IK, Santoni-Rugiu E, Ravn J, et al. Molecular prediction of adjuvant cisplatin efficacy in Non-Small Cell Lung Cancer (NSCLC)-validation in two independent cohorts. PLoS One 2018;13:e0194609 10.1371/journal.pone.0194609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Budczies J, Klauschen F, Sinn BV, et al. Cutoff Finder: a comprehensive and straightforward web application enabling rapid biomarker cutoff optimization. PLoS One 2012;7:e51862 10.1371/journal.pone.0051862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsao M-S, Aviel-Ronen S, Ding K, et al. Prognostic and predictive importance of p53 and Ras for adjuvant chemotherapy in non small-cell lung cancer. J Clin Oncol 2007;25:5240–7. 10.1200/JCO.2007.12.6953 [DOI] [PubMed] [Google Scholar]
- 22.Graziano SL, Gu L, Wang X, et al. Prognostic significance of mucin and p53 expression in stage Ib non-small cell lung cancer: a laboratory companion study to CALGB 9633. J Thorac Oncol 2010;5:810–7. 10.1097/JTO.0b013e3181d89f95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma X, Le Teuff G, Lacas B, et al. Prognostic and Predictive Effect of TP53 Mutations in Patients with Non-Small Cell Lung Cancer from Adjuvant Cisplatin-Based Therapy Randomized Trials: A LACE-Bio Pooled Analysis. J Thorac Oncol 2016;11:850–61. 10.1016/j.jtho.2016.02.002 [DOI] [PubMed] [Google Scholar]
- 24.Buyse M, Sargent DJ, Grothey A, et al. Biomarkers and surrogate end points--the challenge of statistical validation. Nat Rev Clin Oncol 2010;7:309–17. 10.1038/nrclinonc.2010.43 [DOI] [PubMed] [Google Scholar]
- 25.Milde-Langosch K. The Fos family of transcription factors and their role in tumourigenesis. Eur J Cancer 2005;41:2449–61. 10.1016/j.ejca.2005.08.008 [DOI] [PubMed] [Google Scholar]
- 26.Hai J, Zhu C-Q, Wang T, et al. TRIM14 is a putative tumor suppressor and regulator of innate immune response in non-small cell lung cancer. Sci Rep 2017;7:39692 10.1038/srep39692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selvakumar P, Owens TA, David JM, et al. Epigenetic silencing of Na, K-ATPase β 1 subunit gene ATP1B1 by methylation in clear cell renal cell carcinoma. Epigenetics 2014;9:579–86. 10.4161/epi.27795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inge LJ, Rajasekaran SA, Yoshimoto K, et al. Evidence for a potential tumor suppressor role for the Na, K-ATPase beta1-subunit. Histol Histopathol 2008;23:459–67. 10.14670/HH-23.459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tummala R, Wolle D, Barwe SP, et al. Expression of Na, K-ATPase-beta(1) subunit increases uptake and sensitizes carcinoma cells to oxaliplatin. Cancer Chemother Pharmacol 2009;64:1187–94. 10.1007/s00280-009-0985-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahmed Z, Deyama Y, Yoshimura Y, et al. Cisplatin sensitivity of oral squamous carcinoma cells is regulated by Na+, K+-ATPase activity rather than copper-transporting P-type ATPases, ATP7A and ATP7B. Cancer Chemother Pharmacol 2009;63:643–50. 10.1007/s00280-008-0781-z [DOI] [PubMed] [Google Scholar]
- 31.Abada PB, Howell SB. Cisplatin induces resistance by triggering differentiation of testicular embryonal carcinoma cells. PLoS One 2014;9:e87444 10.1371/journal.pone.0087444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, He R-Q, Dang Y-W, et al. Comprehensive analysis of the long noncoding RNA HOXA11-AS gene interaction regulatory network in NSCLC cells. Cancer Cell Int 2016;16:89 10.1186/s12935-016-0366-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McShane LM, Altman DG, Sauerbrei W, et al. Reporting recommendations for tumour marker prognostic studies (REMARK). Br J Cancer 2005;93:387–91. 10.1038/sj.bjc.6602678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arbogast S, Behnke S, Opitz I, et al. Automated ERCC1 immunohistochemistry in non-small cell lung cancer. Appl Immunohistochem Mol Morphol 2011;19:99–105. 10.1097/PAI.0b013e3181f1feeb [DOI] [PubMed] [Google Scholar]
- 35.Ma D, Baruch D, Shu Y, et al. Using protein microarray technology to screen anti-ERCC1 monoclonal antibodies for specificity and applications in pathology. BMC Biotechnol 2012;12:88 10.1186/1472-6750-12-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen G, Gharib TG, Huang C-C, et al. Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteomics 2002;1:304–13. 10.1074/mcp.M200008-MCP200 [DOI] [PubMed] [Google Scholar]
- 37.Vilmar A, Garcia-Foncillas J, Huarriz M, et al. RT-PCR versus immunohistochemistry for correlation and quantification of ERCC1, BRCA1, Tubb3 and RRM1 in NSCLC. Lung Cancer 2012;75:306–12. 10.1016/j.lungcan.2011.08.016 [DOI] [PubMed] [Google Scholar]
- 38.Liu Y, Beyer A, Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell 2016;165:535–50. 10.1016/j.cell.2016.03.014 [DOI] [PubMed] [Google Scholar]
- 39.de Sousa Abreu R, Penalva LO, Marcotte EM, et al. Global signatures of protein and mRNA expression levels. Mol Biosyst 2009;5:1512–26. 10.1039/b908315d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barrett LW, Fletcher S, Wilton SD. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell Mol Life Sci 2012;69:3613–34. 10.1007/s00018-012-0990-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haruehanroengra P, Zheng YY, Zhou Y, et al. Rna modifications and cancer. RNA Biol 2020:1–16. 10.1080/15476286.2020.1722449 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
esmoopen-2020-000679supp001.pdf (1.6MB, pdf)