Abstract
Influenza D virus (IDV) utilizes bovines as a primary reservoir with periodical spillover to other mammalian hosts. By using traditional hemagglutination assay coupled with sialoglycan microarray (SGM) platform and functional assays, we demonstrated that IDV is more efficient in recognizing both 9-O-acetylated N-acetylneuraminic acid (Neu5,9Ac2) and 9-O-acetylated N-glycolylneuraminic acid (Neu5Gc9Ac) than influenza C virus (ICV), a ubiquitous human pathogen. ICV seems to strongly prefer Neu5,9Ac2 over Neu5Gc9Ac. Since Neu5Gc9Ac is different from Neu5,9Ac2 only by an additional oxygen in the group at the C5 position, our results reveal that the hydroxyl group in Neu5Gc9Ac plays a critical role in determining receptor binding specificity, which as a result may discriminate IDV from ICV in communicating with 9-O-acetylated SAs. These findings shall provide a framework for further investigation towards better understanding of how newly discovered multiple-species-infecting IDV exploits natural 9-O-acetylated SA variations to expand its host range.
Keywords: influenza D virus, influenza C virus, glycan receptors
1. Introduction
The Orthomyxoviridae family has four influenza genera, A, B, C, and D, which are classified according to their antigenic differences residing in nucleoprotein (NP) and matrix (M) proteins. Influenza D virus (IDV) was first isolated in 2011 (Hause et al., 2013) and officially named in 2016 (https://www.cdc.gov/flu/about/viruses/types.htm). IDV represents a novel type of virus, which is more closely related to influenza C (ICV) than influenza A (IAV) or influenza B (IBV) (Hause et al., 2014; Hause et al., 2013). IDV is unique among four influenza types in that it utilizes bovines as a primary reservoir and amplification host with periodical spillover to other mammalian hosts (Ducatez et al., 2015; Ferguson et al., 2015; Ferguson et al., 2018; Foni et al., 2017; Hause et al., 2014; Hause et al., 2013; Murakami et al., 2016; Nedland et al., 2018; Salem et al., 2017; White et al., 2016; Zhai et al., 2017). Since its first isolation from a pig with influenza-like symptoms in Oklahoma of the United States in 2011, IDVs have been found in cattle and swine populations in North America and Eurasia (Collin et al., 2015; Ducatez et al., 2015; Ferguson et al., 2015; Flynn et al., 2018; Foni et al., 2017; Hause et al., 2014; Hause et al., 2013; Jiang et al., 2014; Murakami et al., 2016; Salem et al., 2017; Snoeck et al., 2018; Zhai et al., 2017). Susceptibility to infection by this novel virus has also been demonstrated in sheep, goats, horses, camelids, guinea pigs, mice, wild boars, and ferrets (Collin et al., 2015; Ducatez et al., 2015; Ferguson et al., 2015; Flynn et al., 2018; Foni et al., 2017; Gorin et al., 2019; Hause et al., 2014; Hause et al., 2013; Jiang et al., 2014; Murakami et al., 2016; Nedland et al., 2018; Oliva et al., 2020; Quast et al., 2015; Salem et al., 2017; Skelton et al., 2019; Snoeck et al., 2018; Sreenivasan et al., 2015; Zhai et al., 2017). Of public health importance, serological evidence of IDV infections in humans has been demonstrated (Hause et al., 2013; Trombetta et al., 2019; White et al., 2016), and increasing IDV outbreaks in pigs have been recently observed in China and Italy (Foni et al., 2017; Zhai et al., 2017).
Viral attachment to terminal sialic acids (SAs) of sialoglycans exposed on the cell surface is a determinant of tissue tropism and host range. To date, little is known about the receptor usage of IDV and its impact on virus replication. It is also unclear about the similarity and difference between IDV and its closely related ICV in functional engagement of sialic acid (SA) receptor on the cell surface. As such, further investigation into the receptor biology of IDV and biological relevance of SA receptor in IDV infection is needed because it will offer novel insights into how influenza viruses such as IDV originating from a novel bovine reservoir frequently transmits to spillover hosts such as pigs and humans. Comparative studies involving both IDV and ICV may provide some clues as to how two closely related seven-segment genome-containing influenza viruses read the glycan receptors differentially, which as a result influences their tissue and species tropism.
2. Materials and methods
2.1. Viruses and cells
Madin-Darby Canine Kidney (MDCK) cells (ATCC) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) at 37 °C with 5 % CO2. IAV (H1N1 A/WSN/1933 virus), IBV (B/Brisbane/60/2008), ICV (C/Johannesburg/1/66), IDVs (D/swine/Oklahoma/1334/2011 and D/bovine/Oklahoma/660/2013) used for the hemagglutination or the cell-based inhibition assay were propagated in MDCK cells with DMEM containing 1 μg/ml Tosyl phenylalanyl chloromethyl ketone (TPCK)-trypsin (Sigma-Aldrich). ICV (C/Johannesburg/1/66) and bIDV (D/bovine/Oklahoma/660/2013) used for the glycan array were propagated in 9 day-old embryonated specific-pathogen-free (SPF) chicken eggs.
The passage three virus stocks of swine and bovine IDVs (prepared in MDCK cells) were used for further propagation in eggs or MDCK cells towards glycan array experiments. For C/Johannesburg/1/66 virus provided by Peter Palese (Mt. Sinai Medical School, New York) (Gao et al., 2008), two more passages were made in MDCK cells from the initial gift stock prior to further propagation in eggs to generate labeled ICV virions for glycan array experiments. Utilization of eggs over MDCK cells to generate virus particles for determining and comparing the receptor binding preference between ICV and IDV was because ICV replicates very poorly in MDCK cells, while both ICV and IDV can undergo a productive replication in eggs so sufficient amounts of virus particles can be readily produced for glycan array experiments. Note that MDCK cell culture system was used to generate virus particles for comparative studies of two IDVs (swine D/OK and bovine D/660) in terms of receptor binding preference in glycan array experiments.
2.2. Sialic acids removal assay
1 ml 5% Turkey red blood cells (RBCs) (Lampire) were treated with 200 milliunits (mU) of neuraminidase from Clostridium perfringens (Sigma-Aldrich) at 37 °C for 1 hour (h). After 1 h treatment, RBCs were diluted to a final 1% concentration in PBS for the hemagglutination (HA) assay. 4 HA/25 μl of each virus were mixed with the same amount of neuraminidase-treated RBCs, followed by 1 h incubation at 4 °C before observing results. Non-treated RBCs were used as controls.
2.3. Hemagglutination (HA) assay-based competitive inhibition assay
Bovine submaxillary mucin (BSM) (Sigma-Aldrich), synthetic 9-O-acetyl-N-acetyl-neuraminic acid (Neu5,9Ac2) and 4-O-acetyl-N-acetylneuraminic acid (Neu4,5Ac2) (Applied Biotech, Austria) were added to different types of viruses containing four HA units each. Mixtures were incubated for 30 min at room temperature. 25 μl of 0.5% Turkey RBCs were added to the mixtures followed by reading results after 30 min incubation at room temperature.
2.4. 9-O-Acetyl group removal assay
20 μg pure, synthetic Neu5,9Ac2 (Applied Biotech) was treated with 5 mU of sialate-9-O-acetylesterase (9-O-SE) (Applied Biotech) at 37 °C for 3 h. Four HA units of different types of viruses were incubated with 9-O-SE-treated or non-treated Neu5,9Ac2 for 30 min at room temperature. Then 25 μl of 0.5% Turkey RBCs were added and results were observed after 30 min incubation at room temperature. All HA assays were performed in three independent assays with each in triplicate.
2.5. Cell-based inhibition assay by receptor analogs (Digital Droplet PCR)
Aliquotes of IDV and ICV (25 μl each) containing 100 TCID50 were respectively pretreated with an equal volume of Neu5,9Ac2, Neu5Gc9Ac, and Neu5Ac receptor analogs at 20, 80 and 320 ng/μl concentrations for 30 min at 4 °C. MDCK cells in 96-well plates were infected with virus-receptor analog mixtures for 1 h followed by additional 5 h incubation in DMEM containing 1 μg/ml of TPCK-trypsin. Supernatants were then removed and cells in each well were lysed with 200 μl of Trizol (Life Technologies). Total RNAs were extracted according to manufacturer’s instructions, which were then followed by reverse transcription reactions with oligo (dT) primer and High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). NP segment-derived mRNA molecules of ICV and IDV were selected as our target for determining the effects of various receptor analogs on viral replication, while the mRNA of canine TATA-Box binding protein (TBP) was used as the reference gene for PCR data normalization. The detailed information for primers and probes is provided in Table 1. The ddPCR reaction consisted of 10 μl of 2x Supermix for Probes (Bio-Rad), 900 nM primers, 25 nM probes and 8 μl undiluted cDNA into a final volume of 20 μl. Droplets were generated and PCR amplified according to manufacturer’s recommendation and were analyzed on a QX200 Bio-Rad Droplet Reader. All samples were tested in three independent experiments with each assayed in triplicate. No template controls (NTC) were included in every run. Viral mRNA copies were normalized with TBP references and results were reported as NP mRNA copies per million TBP.
Table 1.
Primers and probes used for digital droplet PCR.
| Target genes | Primers/probes name | Primers/probes sequence |
|---|---|---|
| IDV NP | qD660NP-For | 5’-TGCCGATTGGTGGAGTCAA-3’ |
| qD660NP-Rev | 5’-TTTCAGTGCCATTCCCAATCT-3’ | |
| qD660NP-Probe | 5’−6FAM-AGCTGGGAAATGTAGTGC-MGBNFQ-3’ | |
| ICV NP | qJHBNP-For | 5’-TGAAGCCTACATTGCCATTTGT-3’ |
| qJHBNP-Rev | 5’-GCCATTTTCCAGGATCAACATT-3’ | |
| qJHBNP-Probe | 5’−6FAM-AGGAAGTGGGCCTTAA-MGBNFQ-3’ | |
| Canine TBP | Canine-TBP-For | 5’-AGGATGATCAAACCCAGAATTGTT-3’ |
| Canine-TBP-Rev | 5’-GCCCTTTAGAATAGGGTAGATGTTTTC-3’ | |
| Canine-TBP-Probe | 5’-VIC-TTGTACTAACAGGTGCTAAAG-MGBNFQ-3’ |
2.6. Virus labeling and sialylated glycan microarray (SGM)
IDV and ICV were propagated in 9d-old embryonated SPF chicken eggs. Allantoic fluid was harvested at 72 hours. The clarified supernatant was layered over a 20% sucrose cushion in PBS buffer and then ultracentrifuged at 100,000 × g for 2 h at 4 °C in a SW28 rotor (Beckman Coulter). Pellets were resuspended in PBS and further purified over a 30–60% cold sucrose gradient at 28,000 rpm for 4 h at 4 °C in an SW 28 rotor. Fractions (1.0 ml) were collected from the top and all fractions showing greater or equal to 6 log2 HA units were pooled together, followed by a final ultracentrifugation through a 20 % sucrose cushion (w/v) in PBS at 100, 000 g for 2 h at 4 °C. The pellets were resuspended in CMS buffer (0.15 M NaCl, 0.25 mM CaCl2, 0.8 mM MgCl2, pH 7.4) and diluted to a solution containing 1.0 × 105 HAU (HA unit)/ml.
We employed the standard protocol to generate dye-labeled IDV and ICV virions as described previously (Song et al., 2011). In brief, 10 μl of 1.0 M sodium bicarbonate (pH 9.0) was added into 100 μl purified viruses containing ~1.0 × 104 HA units followed by addition of Alexa Fluor-488 succinimidyl ester (Molecular Probes) in a ratio of 0.005 μg Alexa per HAU. This ratio was determined by the HA titration experiment to give maximal labeling without loss of binding activity. After stirring for 1 h at room temperature in the dark, the labeling reaction mixtures were dialyzed (Slide-A-Lyzer Mini Dialysis Units 7000 MWCO, Pierce) at 4 °C for overnight. After functional evaluation (HA activity), labeled viruses were ready for glycan receptor binding experiment.
To determine the glycan-binding specificities of labeled IDV and ICV, a glycan screen array was performed at the Consortium for Functional Glycomics. The printed glycan microarray consists of 77 sialoglycans incorporating 16 sialic acid forms in α2–3 and α2–6-linkages to different underlying structures (Song et al., 2011). Briefly, fluorescent labeled virus was incubated on a glycan microarray slide under a coverslip at 4 °C for 1 h, washed to remove unbound virus, and scanned using a ProScan Array (Perkin-Elmer Life sciences) equipped with multiple lasers. The data were processed using the manufacturer’s software as described previously, which provided raw values in relative fluorescence units (RFU) from each spot in an Excel spreadsheet. The intensity of binding to each of the 77 glycans on the array was graphed and shown as values representing means ± S.D.s of four replicate samples.
3. Results
3.1. 9-O-Acetyl group is a critical sialic acid receptor determinant of bovine IDV.
We first employed the traditional hemagglutination (HA) assay to determine the glycan receptor of IDV. To determine whether sialic acid serves as a component of the receptor for bovine IDV (bIDV), we treated turkey RBCs with neuraminidase from Clostridium perfringens. As summarized in Table 2, pretreatment with neuraminidase resulted in a complete loss of RBC agglutination mediated by bIDV as well as by IAV A/WSN/1933 (H1N1), IBV (B/Brisbane/60/2008), or ICV (C/Johannesburg/1/66). This result indicated that sialic acid was involved in bIDV-mediated RBC agglutination. Agglutination of RBCs by bIDV was also inhibited in the presence of bovine submaxillary mucin (BSM) used at concentrations ranging from 6.25 to 400 ng/μl, which is rich in Neu5,9Ac2 and Neu5Gc9Ac (Table 3) (Langereis et al., 2015). Intriguingly, the level and pattern of inhibition of bIDV-mediated RBC agglutination by BSM were similar to that observed in ICV, which is known to utilize Neu5,9Ac2-containing glycans as receptors for viral entry (Rosenthal et al., 1998), thereby suggesting that bIDV likely utilizes Neu5,9Ac2-glycans as receptors for attachment. To further confirm these results, we performed the HA-based competitive inhibition assay involving a synthetic Neu5,9Ac2. Neu4,5Ac2 was used as a control, which has O-acetyl group at the C4, not the C9, position. RBC agglutination by bIDV and ICV was not inhibited by Neu4,5Ac2 (up to 1280 ng/μl, the maximum concentration used). In contrast, agglutination was completely lost for bIDV at the presence of 20 ng/μl Neu5,9Ac2 and for ICV at 80 ng/μl Neu5,9Ac2 (Table 4). This result further demonstrated that Neu5,9Ac2 is a receptor sialic acid form of bIDV. It is interesting to note that the minimal concentration to abolish ICV-mediated RBC agglutination was 4 times higher than that needed for bIDV. Finally, pretreatment of the synthetic Neu5,9Ac2-glycan with recombinant sialate-9-O-acetylesterase (9-O-SE) (e.g., removes 9-O-acetylated group) abolished its inhibitory effect of bIDV- or ICV-mediated RBC agglutination, whereas binding and agglutination of RBCs by IAV were not affected (Table 5). In summary, the results of our qualitative HA-based experiments suggest that bIDV uses Neu5,9Ac2 as a receptor sialic acid form for infection.
Table 2.
The effects of neuraminidase (NA) treatment on agglutination of Turkey red blood cells by viruses.
| Virus | Untreated RBCs | NA-treated RBCs |
|---|---|---|
| A/WSN/1933 (H1N1) | −a | +b |
| B/Brisbane/60/2008 | − | + |
| C/Johannesburg/1/66 | − | + |
| D/bovine/Oklahoma/660/2013 | − | + |
the minus (−) sign indicates evident hemagglutination in the wells.
the plus (+) sign denotes non-hemagglutination in the wells.
Table 3.
The effects of bovine submaxillary mucin (BSM) treatment on agglutination of Turkey red blood cells by viruses.
| Virus | BSM (ng/μl) | ||||
|---|---|---|---|---|---|
| PBS | 6.25 | 25 | 100 | 400 | |
| A/WSN/1933 (H1N1) | −a | − | − | − | − |
| B/Brisbane/60/2008 | − | − | − | − | − |
| C/Johannesburg/1/66 | − | − | +b | + | + |
| D/bovine/Oklahoma/660/2013 | − | − | + | + | + |
| DMEMc | + | + | + | + | + |
the minus (−) sign indicates evident hemagglutination (no inhibitory effect) in the wells.
the plus (+) sign denotes non-hemagglutination (inhibitory effect) in the wells.
DMEM serves as a negative control to replace the virus solution in the HA assay to determine any non-specific inhibitory effect on hemagglutination caused by BSM. Non-hemagglutination indicated by (+) sign is anticipated.
Table 4.
The effects of synthetic Neu5,9Ac2 and Neu4,5Ac2 receptor analogs on agglutination of Turkey red blood cells by viruses.
| Virus | Neu5,9Ac2 (ng/μl) | |||||
|---|---|---|---|---|---|---|
| PBS | 5 | 20 | 80 | 320 | 1280 | |
| A/WSN/1933 (H1N1) | −a | − | − | − | − | − |
| B/Brisbane/60/2008 | − | − | − | − | − | − |
| C/Johannesburg/1/66 | − | − | − | +b | + | + |
| D/bovine/0klahoma/660/2013 | − | − | + | + | + | + |
| DMEMc | + | + | + | + | + | + |
| Virus | Neu4,5Ac2 (ng/μl) | |||||
| PBS | 5 | 20 | 80 | 320 | 1280 | |
| A/WSN/1933 (H1N1) | − | − | − | − | − | − |
| B/Brisbane/60/2008 | − | − | − | − | − | − |
| C/Johannesburg/1/66 | − | − | − | − | − | − |
| D/bovine/0klahoma/660/2013 | − | − | − | − | − | − |
| DMEM | + | + | + | + | + | + |
the minus (−) sign indicates evident hemagglutination (no inhibitory effect) in the wells.
the plus (+) sign denotes non-hemagglutination (inhibitory effect) in the wells.
DMEM serves as a negative control to replace the virus solution in the HA assay to determine any non-specific effect on hemagglutination caused by indicated receptor analogs. Non-hemagglutination indicated by (+) sign is anticipated.
Table 5.
The effects of synthetic Neu5,9Ac2 receptor analog and recombinant sialate-9-O-acetylesterase (9-O-SE) pretreated Neu5,9Ac2 on agglutination of Turkey red blood cells by viruses.
| Virus | PBS | Neu5,9Ac2 | Neu5,9Ac2 with 9-O-SE | 9-O-SE with PBS |
|---|---|---|---|---|
| A/WSN/1933 (H1N1) | −a | − | − | − |
| C/Johannesburg/1/66 | − | +b | − | − |
| D/bovine/0klahoma/660/2013 | − | + | − | − |
the minus (−) sign indicates evident hemagglutination (no inhibitory effect) in the wells.
the plus (+) sign denotes non-hemagglutination (inhibitory effect) in the wells.
3.2. Receptor binding characteristics of labeled bIDV and ICV on a sialoglycan microarray (SGM).
In parallel, we utilized sialoglycan microarray (SGM) approach to examine the specific glycans that serve as potential receptors for bIDV and ICV. The SGM used consists of 77 sialoglycans incorporating 16 sialic acid forms in α2–3 and α2–6-linkages to different underlying structures (Song et al., 2011). Under an identical condition, we found that bIDV preferred to bind glycans terminated in either Neu5,9Ac2 (IDs#38 and 52) or Neu5Gc9Ac (IDs#39 and 53) (Fig. 1A). Other forms of glycans failed to achieve significant interactions with bIDV, indicating a specific binding directed largely by the 9-O-acetyl group. Binding of bIDV to 9-O-acetylated glycans was not dependent on the specific linkage (α2–3 or α2–6) (IDs#39 Vs. 53 and 38 Vs. 52) (Fig. 1C). ICV array did not give clear data (low signal/noise ratio). Nevertheless, only #38 glycan terminated with Neu5,9Ac2 (α2–6-linkage) showed consistent interactions with ICV (Fig. 1B) in our repeated SGM experiments. Neu5,9Ac2 has been shown previously as a receptor determinant of ICV infection (Rogers et al., 1986). In summary, the glycan array-based data are consistent with the observations of the HA-based experiments, thereby further demonstrating that bIDV utilizes 9-O-acetylated SAs-containing glycans as receptors for viral entry and infection.
Figure 1. Identification of 9-O-acetylated SA receptors of IDV.
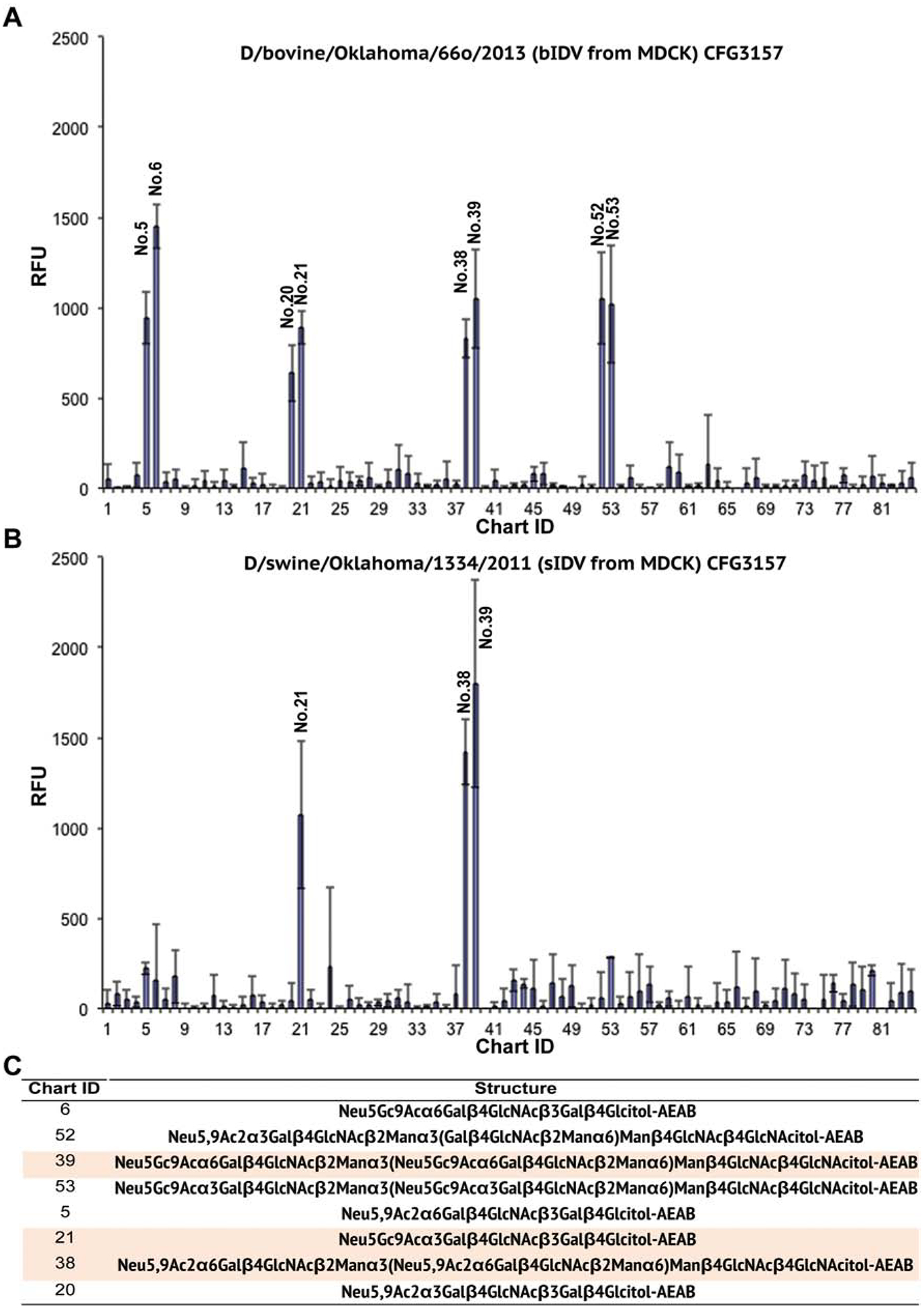
Glycan binding analyses as measured by relative fluorescence intensity (RFU) were presented for labeled bIDV (D/bovine/Oklahoma/660/2013) (A) and ICV (C/Johannesburg/1/66) (B) respectively. The structures of the sialoglycans showing a significant interaction with bovine IDV are listed in (C). Note a “*” sign indicates glycans that bind to both IDV and ICV with gray yellow shade (C). Chemical structures of Neu5,9Ac2 and Neu5Gc9Ac are showed in (D). For virus labeling and sialylated glycan microarray (SGM) experiment, in brief, Alexa488 fluorescence-labeled bovine IDV or ICV was incubated at 4°C for 1 h on the printed glycan array that consists of 77 siaoglycans incorporating 16 SA forms in α2–3 and α2–6 linkages to different underlying structures. Following washing 6 times to remove unbound viruses, the slides were scanned using a ProScan Array (Perkin-Elmer Life sciences) equipped with four lasers covering an excitation range from 488 to 637 nm. For Alexa488 fluorescence, 495 nm excitation and 510 nm emission were used. The data were processed using the manufacturer’s software, which provided raw values in relative fluorescence units (RFU) from each spot in an Excel spreadsheet. The intensity of binding to each of the 77 glycans on the array was graphed and shown as values representing mean±S.D. of 4 replicates. Note that an equal amount of purified IDV or ICV (500 HA units) grown in embryonated eggs was used for the glycan array analysis. The data presented were representative of 2 independent glycan microarray experiments performed in 4 replicates.
Binding to both Neu5,9Ac2- and Neu5Gc9Ac- containing glycans by bIDV was also confirmed in additional microarray experiment involving another IDV strain, swine D/OK (D/swine/Oklahoma/1334/2011) (sIDV used here). sIDV is a representative strain of IDV D/OK lineage, while bIDV represents IDV D/660 lineage (Collin et al., 2015). As summarized in Fig. 2, despite the observed fine receptor binding differences between sIDV and bIDV, two IDV strains overall behaved similarly in interacting with glycans terminated with Neu5,9Ac2 or Neu5Gc9Ac. For example, both viruses displayed good binding to Glycan ID#: 21, 38, and 39 (Figs. 2A, 2B, and 2C). This result indicates that sIDV is similar to bIDV in engagement of glycan receptors. It should be noted that the receptor binding specificity of bIDV appeared to be reproduced nicely between two independent experiments presented in Figs. 1 and 2, respectively. In addition to Glycan IDs: 6, 38, 39, 52, and 53 (Fig. 1A), in this comparative glycan array experiment involving two IDV strains, bIDV also picked up three more glycans (IDs#5, 20, and 21) terminated with Neu5,9Ac2 or Neu5Gc9Ac (Figs. 2A and 2C). Note that IDs# 20 and 21 were relative weak binding partners for bIDV. The slightly different glycan-binding patterns between two separate experiments may be caused by the propagation system that derived virus particles (eggs used for Fig. 1 and MDCK cells for Fig. 2).
Figure 2. Receptor binding profiles of bovine IDV D/660 (bIDV) and swine IDV D/OK (sIDV).

Glycan binding analyses as measured by relative fluorescence intensity (RFU) were presented for labeled bIDV (A) and sIDV (B) respectively. The structures of the sialoglycans showing a significant interaction with bIDV are listed in (C). Note a “*” sign indicates glycans that bind to both bIDV and bICV shaded in gray yellow (C). Experimental procedures were similar to those described in Figure 1 except that MDCK cells instead of eggs were used to grow two viruses. The data presented were representative of 2 independent glycan microarray experiments performed in 4 replicates.
Further analysis of glycan array data related to two influenza D viruses (Fig. 2) revealed that bIDV and sIDV possessed distinct receptor fine specificities, despite having overlapping binding partners. For example, bIDV displayed a broader receptor specificity and its glycan-binding was not affected substantially by sialyl linkages (α2–3/6) and internal glycans bearing 9-O-acetyl group. In contrast, sIDV had a narrower receptor specificity and preferred to bind to sialosides with specific linkages and internal glycans, including α2–6-sialylated biantennary complex N-glycans with Neu5,9Ac2 (ID#38) or Neu5Gc9Ac (ID#39) and α2–3- sialylated LNnT with Neu5Gc9Ac form only (ID#21). Because sIDV and bIDV belong to two distinctive antigenic clades represented by D/OK and D/660 plus these two viruses differing in 6 surface-exposed positions around the receptor-binding pocket of the hemagglutinin-esterase-fusion protein (data not shown), we speculate that these critical mutations confer broad receptor specificity to bIDV, which certainly warrants further investigation.
3.3. Functional studies of the roles of Neu5,9Ac2- or Neu5Gc9Ac-containing glycans in IDV and ICV infection.
To determine the functional relevance of these receptor candidates, we first tested four synthetic glycans and examined their ability to disrupt the agglutination of turkey RBCs by IDV and ICV in hemagglutination inhibition format where we replaced antibody with receptor analogs with four HA unit-working concentration used for each virus per reaction. These four glycans were Neu5Acα2–3LNnTβProN3, Neu5Gcα2–3LNnTβProN3, Neu5Ac9Acα2–3LNnTβProN3, and Neu5Gc9Acα2–3LNnTβProN3. As shown in Fig. 3, the presence of both glycans containing Neu5,9Ac2 (Fig. 3C) or Neu5Gc9Ac (Fig. 3D) at a concentration of 20 ng/μl and above resulted in a complete loss of RBC agglutination by IDV. In contrast, neither glycan containing Neu5Ac (Fig. 3A) nor that containing Neu5Gc (Fig. 3B) affected IDV-mediated RBC agglutination, thereby confirming our glycan array data. Interestingly, we noticed that ICV needed 4 and 16 times higher concentrations of Neu5,9Ac2 and Neu5Gc9Ac-glycans, respectively, than IDV for completely losing RBC agglutination ability. This result suggests that IDV binds both glycans terminated with 9-O-acetylated SAs with higher affinity than ICV, and ICV prefers Neu5,9Ac2 over Neu5Gc9Ac.
Figure 3. Inhibition of viral hemagglutination by receptor analogs.
The receptor analogs Neu5Ac (A), Neu5Gc (B), Neu5,9Ac2 (C), and Neu5Gc9Ac (D) at indicated concentrations were added to IDV (D/bovine/Oklahoma/660/2013) or ICV (C/Johannesburg/1/66) containing 4 HA units each. Mixtures were incubated for 30 min at room temperature. Aliquotes of turkey RBCs were then added to the mixtures and results were read after 30 min. The data shown represents 4 independent experiments performed in duplicate. Note that “+” sign indicates no hemagglutination, while “−” denotes evident hemagglutination. DMEM serves as a mock negative control to replace the virus solution in the HA assay to determine any non-specific inhibitory effect on hemagglutination caused by indicated receptor analogs.
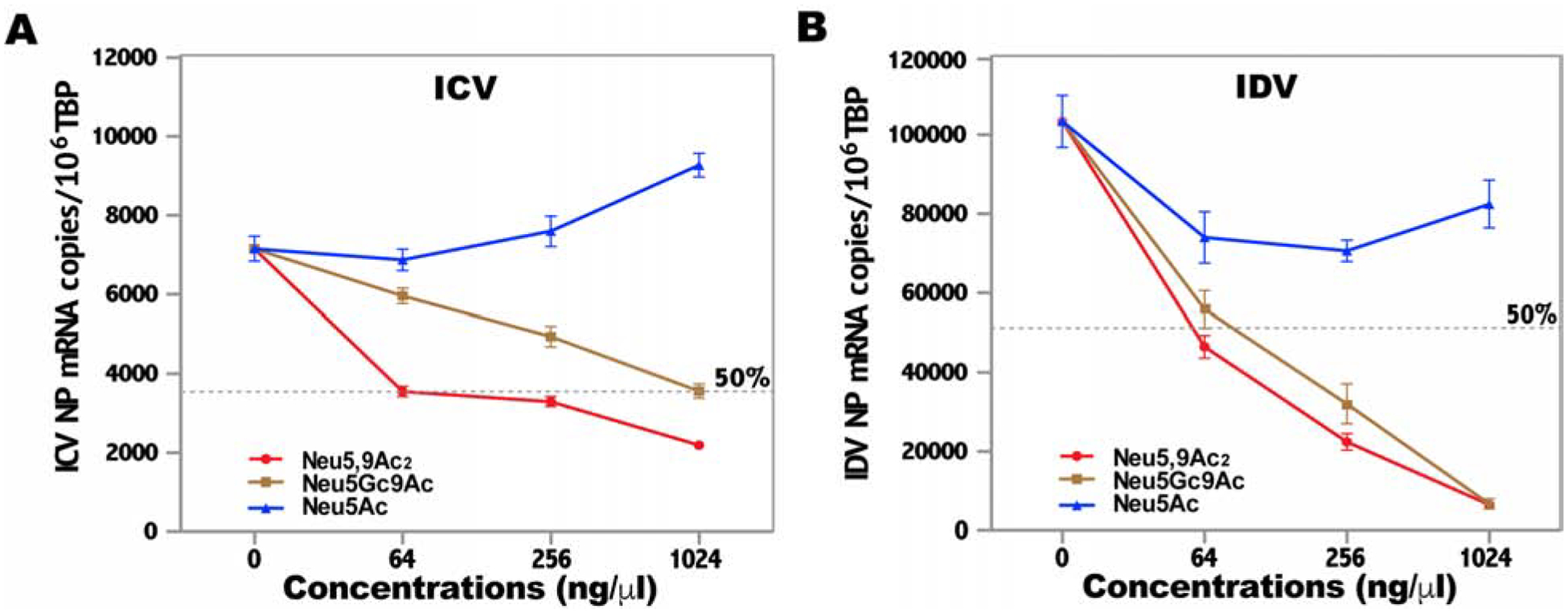
In parallel, four synthetic glycans were examined for their ability to inhibit the replication of ICV (Fig. 4A) and IDV (Fig. 4B) in a MDCK cell-based replication assay. After pretreatment for 30 min at 4 °C, MDCK cells were infected with 100 TCID50 of IDV or ICV, followed by an additional 6 h-incubation. Extracted cellular RNAs were subjected to reverse transcription with oligo(dT) followed by Bio-Rad QX200 Droplet Digital PCR (ddPCR) with primers and probe targeting viral NP gene. These ddPCR reactions included another set of primers and probe targeting canine TATA box binding protein (TBP) gene as an internal reference to derive the relative copy number of viral NP mRNA. The detailed information for primers and probes is provided in Table 1. As shown in Fig. 4B, both Neu5,9Ac2 and Neu5Gc9Ac-glycans exhibited a dose-dependent inhibition of IDV replication. For example, IDV infectivity was reduced approximately by 50% with Neu5,9Ac2-glycan and by 40% with Neu5Gc9Ac-glycan at 64 ng/μl, respectively, whereas the infectivity was reduced by 90% with either treatment at 1024 ng/μl. Similarly, a dose-dependent inhibition of ICV replication was observed by the above receptor analogs but the inhibition levels were less pronounced (Fig. 4A). Interestingly, ICV replication was inhibited more significantly by glycans containing Neu5,9Ac2 than the one containing Neu5Gc9Ac (Fig. 4A). The concordance between the cell-based and HA-based inhibition assays further validates our findings from glycan array experiments. In summary, our data suggest that IDV binds both glycans terminated with Neu5,9Ac2 and Neu5Gc9Ac equally well, while ICV prefers Neu5,9Ac2 over Neu5Gc9Ac-glycan. Our results also indicate that IDV is more efficient in recognizing Neu5,9Ac2 and Neu5Gc9Ac receptors than ICV.
Figure 4. Inhibition of viral replication in MDCK cells by receptor analogs.

MDCK cell-based multiple-round replication assay was used to determine the inhibition of ICV (A) and IDV (B) replication by receptor analogs. Aliquotes of IDV (D/bovine/Oklahoma/660/2013) and ICV (C/Johannesburg/1/66) containing 100 TCID50 were respectively pretreated with Neu5,9Ac2, Neu5Gc9Ac, or Neu5Ac receptor analogs at indicated concentrations for 30 min at 4 °C. MDCK cells were washed once with PBS and then infected with the virus-receptor mixtures for 1 h. Cells were washed 3 times and further incubated in DMEM containing 1 μg/ml of TPCK-trypsin for 6 h, and then lysed for RNA purification. The RNA samples were reverse transcribed using oligo(dT) primer. IDV or ICV NP gene and canine TBP housekeeping gene primers and probes were used in ddPCRs. Non-template controls (NTC) were included in every ddPCR run. The normalized results are shown as NP mRNA copies per million TBP, which represent mean±S.D. of 3 independent experiments performed in triplicate. Note that the different scale (10-fold difference) in Y-axis is used for panel 4A and 4B towards better visualization of influenza C virus replication and the inhibitory effects mediated by receptor analogs.
4. Discussion
Since its identification in 2011, to date IDVs have evolved into two genetic and antigenic lineages, represented by well-characterized swine IDV (sIDV) isolate D/swine/Oklahoma/1334/2011 (D/OK) and bovine IDV (bIDV) isolate D/bovine/Oklahoma/660/2013 (D/660) in North America (Collin et al., 2015). Recently, new genetic lineage of IDV has been reported in cattle herds in Japan (Murakami et al., 2020). Despite the seroprevalence of IDV in humans and pigs and other mammals, it is generally believed that IDV utilizes bovines as a primary reservoir and host with frequently spillover to other animal hosts (Hause et al., 2014; Sreenivasan et al., 2015).
The recognition of terminal sialic acids (SAs) of sialoglycans exposed on the cell surface is the first and critical step of the influenza virus life cycle. Our study has presented multiple lines of evidence supporting that IDV binds to Neu5,9Ac2 and/or Neu5Gc9Ac and utilizes them for viral entry, while ICV prefers Neu5,9Ac2 over Neu5Gc9Ac. The present work reported here has extended and further supports a previous structure-based study of IDV receptor binding (Song et al., 2016). Using a fluorescent dye-labeled recombinant Hemagglutinin-Esterase-Fusion (HEF) protein of IDV swine D/OK (sIDV), Song and colleagues found that sIDV HEF protein interacted with both α2–3 and α2–6-linked 9-O-acetylated SAs (Song et al., 2016), suggesting that sIDV likely utilizes them as its receptors for virus entry. Furthermore, this study resolved the crystal structure of sIDV HEF, showing that sIDV differed from its related ICV in that its HEF protein possessed an open receptor-binding cavity, which may allow IDV to accommodate more diverse SA 9-O-acetyl moieties that could broaden its tissue and species tropism. Taken together, these studies demonstrate that IDV is more efficient in recognizing both Neu5,9Ac2- and Neu5Gc9Ac-containing glycans than ICV, and ICV seems to preferentially bind to glycans with Neu5,9Ac2 over Neu5Gc9Ac as a terminal sialic acid.
Neu5Gc and Neu5Gc9Ac are generally abundant in many agricultural animals such as cattle and swine (Samraj et al., 2015), but are absent in humans and ferrets due to frame-shift mutations of cytidine 5’-monophospho-N-acetylenuraminic acid hydroxylase (CMAH) that converts CMP-Neu5Ac to CMP-Neu5Gc (Irie et al., 1998; Ng et al., 2014), the precursor of the Neu5Gc9Ac-glycans. The efficient usage of either Neu5,9Ac2 or Neu5Gc9Ac receptor likely gives IDV an ecological niche to infect multiple agricultural animals with abundant expression of Neu5Gc9Ac as well as humans only expressing Neu5,9Ac2. The zoonotic risk to humans by IDV is further supported by our previous study showing that ferrets are susceptible to IDV infection (Hause et al., 2013). It should be noted that despite the genetic inability to synthesize Neu5Gc and its derivative Neu5Gc9Ac, humans still possess the cellular 9-O-acetylation machinery for the synthesis of 9-O-acetylated sialic acids. In this regard, humans can take Neu5Gc from dietary sources, convert it into Neu5Gc9Ac, and express it on the cell surfaces of various cell types (Bardor et al., 2005). It has been shown previously that Neu5Gc and possible its 9-O-acetylated form (Neu5Gc9Ac) can be accumulated in human cancer cells through an unknown mechanism (Inoue et al., 2010). In light of the fact that exogenous expression of nonhuman Neu5Gc can make humans vulnerable to pathogens utilizing Neu5Gc as a receptor for infection (Byres et al., 2008), it will be interesting to address in the near future whether humans or ferrets expressing exogenous Neu5Gc9Ac under certain conditions are more susceptible to IDV infection.
Humans are thought to be the primary host and reservoir of ICV, although this virus has been identified in other hosts probably after reverse zoonotic transmission from humans (Matsuzaki et al., 2006). Our observation that ICV preferentially uses Neu5,9Ac2 over Neu5Gc9Ac receptors appears to support this theory. First, as discussed above, the evolutionary loss of CMAH gene for conversion of Neu5Ac to Neu5Gc may make humans exclusively and abundantly express Neu5Ac and Neu5,9Ac2 (Irie et al., 1998). As such humans become a perfect host for ICV that selectively prefers Neu5,9Ac2 for infection. Second, agricultural animals such as cattle and pigs that are rich in Neu5Gc can express high levels of Neu5Gc9Ac (Samraj et al., 2015). Because a significant portion of Neu5Ac has been converted to Neu5Gc in these animals, the number of Neu5Ac molecules available for synthesizing Neu5,9Ac2 is substantially reduced. Reduced expression of the Neu5,9Ac2 receptor may render agricultural animals less susceptible to ICV infection when compared to humans. In case these animals would be infected by ICV after reverse zoonotic transmission from humans, it is questionable that ICV transmission can be sustained. On the other hand, IDV differs from ICV in that it effectively engages with no clear preference Neu5,9Ac2 and/or Neu5Gc9Ac. One can envision that the differential expression levels of these two 9-O-acetylayed sialic acids in animals and humans may not substantially affect IDV replication and spread. Therefore, this unique receptor binding property should enable IDV to effectively infect and transmit among different mammalian hosts including humans.
9-O-Acetylated derivatives (Neu5,9Ac2 and Neu5Gc9Ac) and their precursors (Neu5Ac and Neu5Gc) are the most common sialic acids in nature (Song et al., 2011). Neu5Gc9Ac is different from Neu5,9Ac2 only by an additional oxygen atom at the C5 position (Song et al., 2011) (Fig. 1D). The differential usage of these two nearly identical 9-O-acetylated SAs between IDV and ICV implies that these two seven-segmented influenza viruses diverge in communicating with both O-acetyl group at the C9 position and acetyl/glycolyl groups at the C5 position in terminal 9-carbon sialic acids (Fig. 1D). We interpret our experimental data presented here that IDV in general may have high binding affinity and broad specificity, while ICV may have lower binding affinity and narrow selectivity for 9-O-acetylated SAs. Such qualitative and quantitative differences in the virus-glycan receptor interaction that may ultimately discriminate IDV from its related ICV in infection landscape and ecology, which clearly warrants further investigation.
ACKNOWLEDGMENTS
We thank all the members of the Li and Wang laboratories for their input into this work. We thank Dr. Reinhard Vlasak at University of Salzburg, Austria, for many valuable suggestions about our experiments. We also thank Peter Palese (Mt. Sinai Medical School, New York) for providing the C/Johannesburg/1/66 virus. We thank the Consortium for Functional Glycomics (Core H) that conducted glycan microarray assays. which was supported by NIH Grant GM62116.
Funding information
This work was partially supported by NIH grants R01AI141889 and R01AI130684 and NIH Grant GM62116, by SDSU AES 3AH-477, by National Science Foundation/EPSCoR (http://www.nsf.gov/od/iia/programs/epscor/index.jsp) award IIA-1335423, and by the state of South Dakota’s Governor’s Office of Economic Development as a South Dakota Research Innovation Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest
The authors have read the journal’s policy and have no conflicts of interest.
REFERENCES
- Bardor M, Nguyen DH, Diaz S, Varki A, 2005. Mechanism of uptake and incorporation of the non-human sialic acid N-glycolylneuraminic acid into human cells. The Journal of biological chemistry 280, 4228–4237. [DOI] [PubMed] [Google Scholar]
- Byres E, Paton AW, Paton JC, Lofling JC, Smith DF, Wilce MC, Talbot UM, Chong DC, Yu H, Huang S, Chen X, Varki NM, Varki A, Rossjohn J, Beddoe T, 2008. Incorporation of a non-human glycan mediates human susceptibility to a bacterial toxin. Nature 456, 648–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin EA, Sheng Z, Lang Y, Ma W, Hause BM, Li F, 2015. Cocirculation of two distinct genetic and antigenic lineages of proposed influenza D virus in cattle. Journal of virology 89, 1036–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducatez MF, Pelletier C, Meyer G, 2015. Influenza D Virus in Cattle, France, 2011–2014. Emerging Infectious Diseases 21, 368–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson L, Eckard L, Epperson WB, Long LP, Smith D, Huston C, Genova S, Webby R, Wan XF, 2015. Influenza D virus infection in Mississippi beef cattle. Virology 486, 28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson L, Luo K, Olivier AK, Cunningham FL, Blackmon S, Hanson-Dorr K, Sun H, Baroch J, Lutman MW, Quade B, Epperson W, Webby R, DeLiberto TJ, Wan XF, 2018. Influenza D Virus Infection in Feral Swine Populations, United States. Emerg Infect Dis 24, 1020–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn O, Gallagher C, Mooney J, Irvine C, Ducatez M, Hause B, McGrath G, Ryan E, 2018. Influenza D Virus in Cattle, Ireland. Emerg Infect Dis 24, 389–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foni E, Chiapponi C, Baioni L, Zanni I, Merenda M, Rosignoli C, Kyriakis CS, Luini MV, Mandola ML, Bolzoni L, Nigrelli AD, Faccini S, 2017. Influenza D in Italy: towards a better understanding of an emerging viral infection in swine. Sci Rep 7, 11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Brydon EW, Palese P, 2008. A seven-segmented influenza A virus expressing the influenza C virus glycoprotein HEF. Journal of virology 82, 6419–6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorin S, Fablet C, Queguiner S, Barbier N, Paboeuf F, Herve S, Rose N, Simon G, 2019. Assessment of Influenza D Virus in Domestic Pigs and Wild Boars in France: Apparent Limited Spread within Swine Populations Despite Serological Evidence of Breeding Sow Exposure. Viruses 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause BM, Collin EA, Liu R, Huang B, Sheng Z, Lu W, Wang D, Nelson EA, Li F, 2014. Characterization of a novel influenza virus in cattle and Swine: proposal for a new genus in the Orthomyxoviridae family. mBio 5, e00031–00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause BM, Ducatez M, Collin EA, Ran Z, Liu R, Sheng Z, Armien A, Kaplan B, Chakravarty S, Hoppe AD, Webby RJ, Simonson RR, Li F, 2013. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS pathogens 9, e1003176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Sato C, Kitajima K, 2010. Extensive enrichment of N-glycolylneuraminic acid in extracellular sialoglycoproteins abundantly synthesized and secreted by human cancer cells. Glycobiology 20, 752–762. [DOI] [PubMed] [Google Scholar]
- Irie A, Koyama S, Kozutsumi Y, Kawasaki T, Suzuki A, 1998. The molecular basis for the absence of N-glycolylneuraminic acid in humans. The Journal of biological chemistry 273, 15866–15871. [DOI] [PubMed] [Google Scholar]
- Jiang WM, Wang SC, Peng C, Yu JM, Zhuang QY, Hou GY, Liu S, Li JP, Chen JM, 2014. Identification of a potential novel type of influenza virus in Bovine in China. Virus Genes 49, 493–496. [DOI] [PubMed] [Google Scholar]
- Langereis MA, Bakkers MJ, Deng L, Padler-Karavani V, Vervoort SJ, Hulswit RJ, van Vliet AL, Gerwig GJ, de Poot SA, Boot W, van Ederen AM, Heesters BA, van der Loos CM, van Kuppeveld FJ, Yu H, Huizinga EG, Chen X, Varki A, Kamerling JP, de Groot RJ, 2015. Complexity and Diversity of the Mammalian Sialome Revealed by Nidovirus Virolectins. Cell reports 11, 1966–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Y, Katsushima N, Nagai Y, Shoji M, Itagaki T, Sakamoto M, Kitaoka S, Mizuta K, Nishimura H, 2006. Clinical features of influenza C virus infection in children. J Infect Dis 193, 1229–1235. [DOI] [PubMed] [Google Scholar]
- Murakami S, Endoh M, Kobayashi T, Takenaka-Uema A, Chambers JK, Uchida K, Nishihara M, Hause B, Horimoto T, 2016. Influenza D Virus Infection in Herd of Cattle, Japan. Emerg Infect Dis 22, 1517–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S, Sato R, Ishida H, Katayama M, Takenaka-Uema A, Horimoto T, 2020. Influenza D Virus of New Phylogenetic Lineage, Japan. Emerg Infect Dis 26, 168–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedland H, Wollman J, Sreenivasan C, Quast M, Singrey A, Fawcett L, Christopher-Hennings J, Nelson E, Kaushik RS, Wang D, Li F, 2018. Serological evidence for the co-circulation of two lineages of influenza D viruses in equine populations of the Midwest United States. Zoonoses Public Health 65, e148–e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PS, Bohm R, Hartley-Tassell LE, Steen JA, Wang H, Lukowski SW, Hawthorne PL, Trezise AE, Coloe PJ, Grimmond SM, Haselhorst T, von Itzstein M, Paton AW, Paton JC, Jennings MP, 2014. Ferrets exclusively synthesize Neu5Ac and express naturally humanized influenza A virus receptors. Nat Commun 5, 5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva J, Mettier J, Sedano L, Delverdier M, Bourges-Abella N, Hause B, Loupias J, Pardo I, Bleuart C, Bordignon PJ, Meunier E, Le Goffic R, Meyer G, Ducatez MF, 2020. Murine Model for the Study of Influenza D Virus. Journal of virology 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast M, Sreenivasan C, Sexton G, Nedland H, Singrey A, Fawcett L, Miller G, Lauer D, Voss S, Pollock S, Cunha CW, Christopher-Hennings J, Nelson E, Li F, 2015. Serological evidence for the presence of influenza D virus in small ruminants. Veterinary microbiology 180, 281–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GN, Herrler G, Paulson JC, Klenk HD, 1986. Influenza C virus uses 9-O-acetyl-N-acetylneuraminic acid as a high affinity receptor determinant for attachment to cells. The Journal of biological chemistry 261, 5947–5951. [PubMed] [Google Scholar]
- Rosenthal PB, Zhang X, Formanowski F, Fitz W, Wong CH, Meier-Ewert H, Skehel JJ, Wiley DC, 1998. Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature 396, 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem E, Cook EAJ, Lbacha HA, Oliva J, Awoume F, Aplogan GL, Hymann EC, Muloi D, Deem SL, Alali S, Zouagui Z, Fevre EM, Meyer G, Ducatez MF, 2017. Serologic Evidence for Influenza C and D Virus among Ruminants and Camelids, Africa, 1991–2015. Emerg Infect Dis 23, 1556–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samraj AN, Pearce OM, Laubli H, Crittenden AN, Bergfeld AK, Banda K, Gregg CJ, Bingman AE, Secrest P, Diaz SL, Varki NM, Varki A, 2015. A red meat-derived glycan promotes inflammation and cancer progression. Proc Natl Acad Sci U S A 112, 542–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelton RM, Shepardson KM, Hatton A, Wilson PT, Sreenivasan C, Yu J, Wang D, Huber VC, Rynda-Apple A, 2019. Contribution of Host Immune Responses Against Influenza D Virus Infection Toward Secondary Bacterial Infection in a Mouse Model. Viruses 11. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Snoeck CJ, Oliva J, Pauly M, Losch S, Wildschutz F, Muller CP, Hubschen JM, Ducatez MF, 2018. Influenza D Virus Circulation in Cattle and Swine, Luxembourg, 2012–2016. Emerg Infect Dis 24, 1388–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Qi J, Khedri Z, Diaz S, Yu H, Chen X, Varki A, Shi Y, Gao GF, 2016. An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism. PLoS pathogens 12, e1005411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Yu H, Chen X, Lasanajak Y, Tappert MM, Air GM, Tiwari VK, Cao H, Chokhawala HA, Zheng H, Cummings RD, Smith DF, 2011. A sialylated glycan microarray reveals novel interactions of modified sialic acids with proteins and viruses. The Journal of biological chemistry 286, 31610–31622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasan C, Thomas M, Sheng Z, Hause BM, Collin EA, Knudsen DE, Pillatzki A, Nelson E, Wang D, Kaushik RS, Li F, 2015. Replication and Transmission of the Novel Bovine Influenza D Virus in a Guinea Pig Model. Journal of virology 89, 11990–12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta CM, Marchi S, Manini I, Kistner O, Li F, Piu P, Manenti A, Biuso F, Sreenivasan C, Druce J, Montomoli E, 2019. Influenza D Virus: Serological Evidence in the Italian Population from 2005 to 2017. Viruses 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SK, Ma W, McDaniel CJ, Gray GC, Lednicky JA, 2016. Serologic evidence of exposure to influenza D virus among persons with occupational contact with cattle. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology 81, 31–33. [DOI] [PubMed] [Google Scholar]
- Zhai SL, Zhang H, Chen SN, Zhou X, Lin T, Liu R, Lv DH, Wen XH, Wei WK, Wang D, Li F, 2017. Influenza D Virus in Animal Species in Guangdong Province, Southern China. Emerg Infect Dis 23, 1392–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]


