Abstract
Paediatric diffuse high grade gliomas are rare, but deadly tumours. The discovery of recurrent mutations in the tail of histone H3, changing lysine 27 to methionine, or glycine 34 to arginine or valine, has illuminated a critical role for epigenetic dysregulation in the aetiology of childhood gliomas and opened new avenues of exploration that have resulted in numerous advances for the field. In this review, we describe the current models of H3K27M mutant cancer that are available to the research community and the insights they have provided on tumour biology and the epigenetic and transcriptional effects of histone mutations. We also review the current understanding of the H3G34R/V mutation and the therapeutic outlook for the treatment of paediatric high grade gliomas.
Keywords: H3K27M, histone H3 mutation, DIPG, epigenetic, high-grade glioma, pediatric, mouse model, therapy
Introduction
While diffuse high grade gliomas (HGG) are predominantly an adult disease, when they occur in children (pHGG) they have a similarly dismal prognosis (1). Worldwide sequencing efforts have unveiled the mutations associated with pHGG and provided researchers with new avenues to explore in the search for therapies to these deadly tumours (2-4). One surprising finding was the observation of recurring mutations in HGG in the tail of histone H3 at lysine 27 (K27M) and glycine 34 (G34R/V) (Figure 1) (5, 6). Histones had previously been thought to be unlikely targets of oncogenic mutations because of their highly redundant nature; for example, there are fifteen genes coding for histone H3 in the human genome. These histone mutations are overwhelmingly a feature of the paediatric versus the adult disease, defining ~50% of pHGG cases compared to less than 1% of adult HGGs (3, 7).
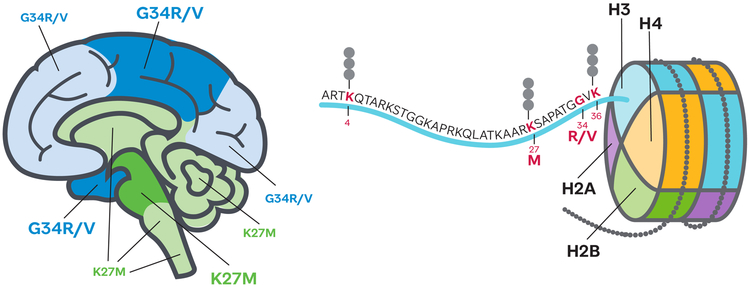
Figure 1. The histone H3 tail mutations found in pHGGs occupy distinct brain locations.
Cartoon brain shows predominant locations of histone H3 K27M (green) and G34R/V (blue) bearing tumours; dark shade indicates regions of highest prevalence. Nucleosome with detail of the histone H3 tail showing the location of residues with post-translational modifications featured in this review.
Histone H3 has three main variants, H3.1 and H3.2, which are deposited in chromatin only during DNA replication and H3.3, which is replication independent and is found primarily in regions of active transcription and heterochromatin (8). G34R/V mutations are found only in H3.3 and heavily favour H3F3A, which is one of only two genes that code for H3.3. K27M also is found most commonly on H3.3 encoded by H3F3A, but can also occur on HIST1H3B and HIST1H3C, two of the ten genes that code for H3.1 and rarely on HIST2H3C one of the three genes coding for H3.2 (3). H3G34R/V mutations occur primarily in cortical tumours and are found in ~16% of pHGG in the cortex (3, 5, 6, 9). By contrast, the H3K27M mutation is largely restricted to pHGG in midline structures including thalamic diffuse glioma and diffuse intrinsic pontine glioma (DIPG) where it is present in ~80% of tumours (6, 9-14) (Figure 1). This molecular hallmark has now been incorporated into the revised classification of gliomas from the World Health Organization to identify diffuse midline glioma- H3K27M mutant as a distinct entity (15). H3G34R/V tumours are found in older patients (median 15 years) than H3K27M tumours and there is a longer average survival time (median 18 months), while K27M mutations on H3.1 tend to be found in slightly younger patients than H3.3 (5 versus 8 years), have a better response to radiation and longer average survival (15 versus 11 months) (3, 16). There is also a difference in the frequency of co-occurring mutations, with ACVR1 mutations often found in H3.1K27M DIPGs and TP53 alterations and PDGFRA amplifications more common in H3.3K27M (3, 10, 11, 13, 14). Despite the fact that a single mutant allele produces only a small fraction of the total histone H3 pool, the H3K27M mutation causes a global loss of H3K27me3 (9, 17-19). This histone modification is deposited by the Polycomb Repressive Complex 2 (PRC2) and is associated with repression of gene expression. By contrast, G34R/V mutations, which consistently co-occur with mutations in ATRX (3, 5, 14), cause disruption of H3K36me2/3 in cis (18, 19); however, no dominant mechanism of action for H3G34R/V has so far been reported.
Analogous to its strong association with midline structures within gliomas, the H3K27M mutation has only been found outside of gliomas at low frequency in a few other cancers such as AML and melanoma (20). Quantitative measurement using mass spectroscopy has shown that a large excess of H3K27M to PRC2 molecules per cell is required to produce the global loss of H3K27me3 associated with H3K27M mutant DIPGs, meaning that cells with a low amount of PRC2 are the most vulnerable to the effects of the H3K27M mutation (21). This prerequisite would be predicted to greatly restrict the types of cells that would be an effective “cell of origin” for an H3K27M driven cancer and also provides a potential explanation for previous reports that the effect of H3K27M overexpression on proliferation was cell type dependent (22). PRC2 mediated repression has been shown to be critical to maintaining stemness and preventing differentiation in embryonic stem (ES) cells (23) and H3K27M-dependent loss of H3K27me3 may act on the cell of origin to repress differentiation and/or reset it to an earlier stem-like state. Evidence from most experimental studies shows an H3K27M-dependent increase in expression of stemness genes in neural precursor (NPC) or gliomas, while the bulk of DIPG tumor cells show expression signatures similar to oligodendrocyte precursor (OPC) cells (19, 22, 24-29).
Intensive efforts by many different laboratories and institutions have begun to elucidate how H3 mutations impact the progression and molecular underpinnings of these unrelenting paediatric tumours and are giving hope for effective therapies. Here we review the current state of knowledge for the H3K27M and H3G34R/V mutations including insights from mouse models, xenografts, cell culture and in vitro studies.
Mouse models of DIPG and patient-derived DIPG xenografts provide critical insights into H3K27M tumourigenesis
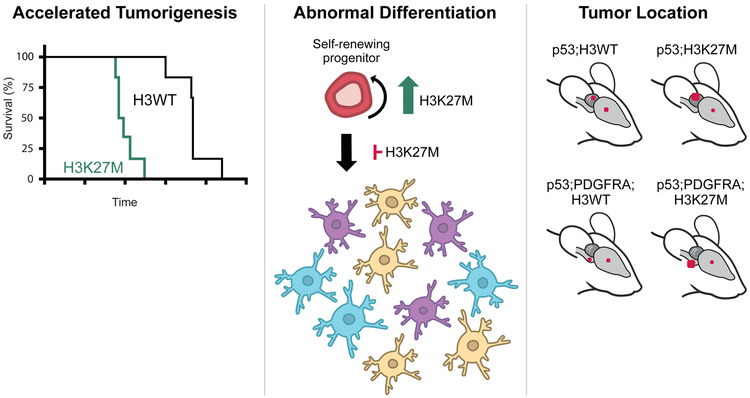
Sequencing data from numerous studies are consistent with a heterozygous H3K27M mutation in 100% of tumour cells, pointing to an initiating event or strong selection pressure (3, 5, 6, 10-14, 30, 31). Recent studies using H3K27M mouse models, xenografts and cell lines have allowed greater insight into the role of the H3K27M mutation in self renewal, tumour initiation and also the requirement of H3K27M for continued tumour growth (Figure 2).
Figure 2. The H3K27M mutation impacts multiple aspects of tumourigenesis in pHGG models, including decreased animal survival time, increased progenitor self-renewal capacity and altered differentiation, and propensity for hindbrain tumour location.
Red dots on mouse brains indicate the approximate location and frequency (size of dot) of tumours with the different genetic driver combinations shown.
Mouse models created by multiple labs using various methodologies have thus far failed to generate tumours using H3K27M alone as a driver (25, 28, 32); however, multiple relevant tumour models have now been generated using combinations of oncogenic drivers. PDGFRA is the most frequently altered receptor tyrosine kinase in DIPG, targeted by focal amplification (30%) and/or activating mutation (5%) (33, 34), and p53 loss of function is seen in 40-50% of DIPGs making these genes the most commonly chosen co-drivers for these models (3, 35). The details of these H3K27M mouse tumour models are laid out below.
Because H3K27M mutant histones only need to represent a fraction of the histone pool in order to exert their influence, several models were generated using an exogenous expression approach. In the first engineered tumour model with H3K27M, human ES cell-derived NPCs were used as the host cell for lentiviral introduction of constitutively active, ligand independent PDGFRA D842V, shRNA targeting TP53 and HA-tagged K27M or WT histone H3.3 (22). These cells were implanted into the brainstem of postnatal day 6 (P6) NOD-SCID mice and resulted in low grade tumours from cells containing H3K27M, but not H3WT. Using a similar strategy, a second mouse model employing exogenous expression of PDGFB and H3K27M or H3WT in mouse NPCs injected into the pons of 6-10 week old SCID mice resulted in significantly faster tumour formation with H3K27M than H3WT (36). In both of these NPC-based systems, the addition of H3K27M markedly improved colony forming potential in soft agar, indicating that the H3K27M mutation enhances self-renewal.
Using a different approach, a third mouse model took advantage of the RCAS/tv-a system to allow targeted exogenous expression of PDGFB, p53 shRNA (or Cre in p53 conditional knockout mice), and H3.3WT or H3.3K27M in Nestin expressing cells in the brainstem of P3-4 immunocompetent mice (32). PDGFB expression with H3K27M produced a small subset of mice with grade 3 or 4 tumours, while PDGFB; H3WT mice had no high-grade lesions. When p53 loss was added, all mice developed HGGs and H3K27M significantly worsened survival. This same model system has been used to establish H3K27M tumours in both cortical and brainstem locations to investigate observations that brainstem pHGGs have poorer blood brain barrier (BBB) permeability (37). This controlled study showed that the brainstem location, rather than the H3K27M mutation is responsible for the difference in the BBB.
A fourth mouse tumour model used in utero electroporation of hindbrain (E12.5) or cortex (E13.5) to investigate the cooperative effects of mutations during embryogenesis. PiggyBAC transposon-based vectors containing combinations of p53 CRISPR/Cas, ATRX shRNA, WT or constitutively active, ligand independent D842V PDGFRA and H3.3K27M or H3.3WT were introduced into developing brain (28). Due to technical challenges, a small number of hindbrain electroporations were carried out only for p53 CRISPR/Cas and H3.3K27M or H3WT, producing only H3K27M; p53 knockout, not H3WT; p53 knockout tumours of unspecified grade by 4-6 months post introduction. In the cortical setting, adding ATRX knockdown to p53 knockout also produced tumours only with H3K27M, not H3WT, while p53 knockout; ATRX knockdown; WT PDGFRA; H3K27M tumours came up faster and with higher penetrance than H3WT. Exchanging WT PDGFRA for PDGFRA D842V produced tumours with 100% penetrance by 4-6 months with or without H3K27M.
The previously described models directed the location of tumors by specifically introducing mutations or cells into brainstem or forebrain. The use of a genetic approach in the fifth mouse model allowed for expression of H3K27M across brain regions and spontaneous tumour formation, making it the first model in which the relative susceptibility of different brain regions for H3K27M-driven tumors was directly tested (25). Here, a conditional knock-in allele of C-terminal FLAG/HA-tagged H3F3a with or without a K27M mutation was introduced into the endogenous mouse locus. H3K27M NSCs from Nestin-Cre embryos (E15.5) grew faster and showed increased self-renewal compared to H3WT. To induce mutations in neonatal neural stem/progenitor cells, Nestin-CreER was induced at P0 and P1, driving p53 conditional knockout, with or without conditional expression of a constitutively active, ligand independent PDGFRA transgene with the V544ins mutation. p53 knockout with H3WT produced a mixture of supratentorial HGGs and cerebellar medulloblastomas and the H3K27M mutation decreased survival time and significantly shifted the proportion towards more medulloblastomas. The addition of PDGFRA V544ins to p53 knockout shifted the tumour spectrum almost exclusively to HGG. 59% of H3WT HGGs were found in the brainstem, while the H3K27M mutation produced 95% brainstem HGGs and again decreased survival time. While the co-occurring mutations had a major influence on the type of tumours formed, the H3K27M mutation consistently drove the tumours to a more hindbrain location and significantly decreased mouse survival time.
Taken together, the diversity of models demonstrated that H3K27M can contribute to gliomagenesis when introduced into embryonic or postnatal neural stem/progenitor cells. Variations in the cell state and developmental context where H3K27M mutation is acquired and where cells become fully transformed may contribute in part to the heterogeneity of the human disease.
Two recent studies have used orthotopic xenografts into mouse to look at the effect of H3K27M loss on tumour growth against an isogenic DIPG background. One study used lentiviral delivery of H3F3A or non-silencing shRNAs to three different H3.3K27M DIPG patient derived xenografts (PDX) and assessed the ability of the cells to form tumours in vivo (29). While loss of the H3K27M mutation increased survival for all three DIPG PDXs, the H3F3A knockdown tumours that grew out still had strong reduction of H3K27M and high H3K27me3, suggesting DIPG tumours can still grow with significantly depleted H3K27M. The same H3F3A knockdown did not alter survival time of an H3WT paediatric midline HGG. A second study utilized the CRISPR/Cas-9 methodology in H3.3K27M DIPG xenografts to generate clones completely lacking the H3K27M mutation and compared tumourigenicity of these clones to both an unedited clone and the parental PDX (38, 39). Here, clones lacking the H3K27M mutation were not able to generate tumours, while both the parental and unedited clones were. Technical limitations in both studies (incomplete H3F3A knockdown by shRNA in the first and the complicating effect of inherent variation between clones in the second) make it difficult to conclusively determine whether tumours are completely dependent upon H3K27M for their growth. However, both studies show clearly that H3K27M has an important ongoing role in tumour growth after the initiating event.
H3K27M impairs differentiation
In addition to effects on tumour growth and self-renewal, H3K27M alters differentiation programs. Gene ontology analysis of genes differentially expressed in response to H3K27M frequently show enrichment for lists associated with neural lineage differentiation (25, 29, 32, 38, 40). In cultured cells, expression of H3K27M blocked astrocytic and impaired oligodendrocytic differentiation of hNPCs expressing constitutively active PDGFRA with p53 loss (22). A recent study by Filbin et al. assessed primary H3K27M DIPGs by single cell RNA-seq and compared them to IDH mutant HGGs (24). Expression programs point to four main cellular subsets common between multiple H3K27M DIPGs: cycling cells, oligodendrocyte precursor (OPC)-like, astrocyte (AC)-like and oligodendrocyte (OC)-like. While the tumour composition varied between individuals, the OPC-like subset was consistently the largest in DIPGs, and when compared to IDH mutant HGGs, DIPGs were found to contain more cycling and undifferentiated cells. H3K27M knockdown in xenografts from three different DIPGs consistently decreased expression of the AC-like and increased expression of the OC-like programs described by Filbin et al., and decreased expression of neural tube progenitor signatures from normal midline brain development (29). Interestingly, H3K27M mutations also occur at much lower frequency in acute myeloid leukaemia, and were shown to increase the frequency of functional haematopoietic stem cells and alter differentiation (41).
H3K27M exerts a dominant negative effect on H3K27me3 through its interaction with PRC2
Since the initial observations that the presence of the K27M mutation on only 5-17% of H3 histones causes global reduction of H3K27me3 (9, 18, 19), several studies have elucidated the mechanism through which this dominant negative effect occurs. The solution of the PRC2 complex interaction with H3K27M showed that the H3K27M peptide binds to the active site of the SET domain of EZH2 with the methionine placed in the pocket that normally receives lysine 27 and prevents hydrolysis of S-adensylmethionine (SAM), the methyl donor and disengagement from EZH2 (42). The PRC2 complex binds to an H3K27M peptide in vitro with a sixteen-fold higher affinity than an H3WT peptide, which is consistent with the dominant effect the mutation exerts (42). Interestingly, recent studies of posterior fossa type A (PFA) ependymomas, which also exhibit very low levels of H3K27me3, showed overexpression and mutations in CXORF67, which binds to PRC2 and reduces levels of H3K27me3 (43). CXORF67, now termed EZH1/2 Inhibitory Protein (EZHIP), is normally expressed predominantly in gonads, where it limits PRC2 enzymatic activity (44). EZHIP contains an amino acid sequence with similarities to the K27M mutant histone H3 tail and also places a methionine in the lysine binding pocket of EZH2, resulting in even more potent inhibition of PRC2 (45, 46). In agreement with this proposed overlap of mechanism, H3K27M mutations are found with low frequency in PFA ependymomas and are always mutually exclusive with high expression of EZHIP (43).
There have been conflicting ideas about how the interaction between H3K27M and PRC2 plays out in a biological setting to produce widespread loss of H3K27me3. One proposed model that is consistent with the in vitro observation of impaired release of EZH2 from the H3K27M peptide suggested that H3K27M might bind to PRC2 and sequester it, thereby preventing appropriate H3K27me3 deposition (9, 19). Analysis of ChIP-seq for EZH2 and H3.3K27M has shown that EZH2 does cooccur on chromatin with H3.3K27M; however, these H3.3K27M-associated EZH2 peaks are relatively weak and it remains to be seen if they represent a biologically relevant sequestration of PRC2 (47). Live cell imaging has shown that while H3K27M lengthens the time EZH2 spends searching for a target and its residence time once it finds it, the fraction of EZH2 bound to chromatin is unchanged (48). Other experimental observations using ChIP-seq have shown that histones bearing the H3.3K27M mutation co-occur with H3K27ac, PolII and bromodomain proteins, such as BRD2 and BRD4 much more frequently than with H3K27me3 and PRC2 components SUZ12 and EZH2 (26, 40) and quantitative MudPIT mass spectroscopy of immunoprecipitated H3.3K27M mononucleosomes did not show enrichment for PRC2 components (49). Interestingly, reports of the localization of H3.1K27M have been inconsistent, with one study reporting that the recruitment pattern mirrors H3.3K27M (and thus H3.3WT) deposition (40) and another showing that H3.1K27M is deposited broadly across the genome, in locations consistent with H3.1WT (26).
Recent studies have provided data that help to resolve the apparent contradictions between experimental observations and the PRC2 sequestration model, and instead propose a model in which interaction with H3K27M causes lasting changes to PRC2 that impair its function (21, 50) (Figure 3A). Utilizing inducible expression of H3K27M and ChIP-seq across multiple timepoints, Stafford . showed that while H3K27M colocalized with PRC2 components on chromatin during an initial phase, the frequency of this cooccurrence decreased with time. While H3K27M was not detected along with PRC2 complex purified from H3K27M mutant cells, the PRC2 complex was found to be less active than that purified from H3K27WT cells. This finding was confirmed using a combination of in vitro and tissue culture-based in vivo experiments that provide evidence to support a new model in which the interaction with H3K27M causes persistent alterations to PRC2 that impair its enzymatic activity, perhaps in part via a stable conformational change. Additionally, work by Lee et al. has shown that H3K27M reduces the level of EZH2 automethylation, which decreases PRC2 methyltransferase activity in a manner that most dramatically affects the conversion of H3K27me2 to me3 (50). This is consistent with the relative effects of H3K27M on H3K27me2 vs H3K27me3 quantity and deposition (38).
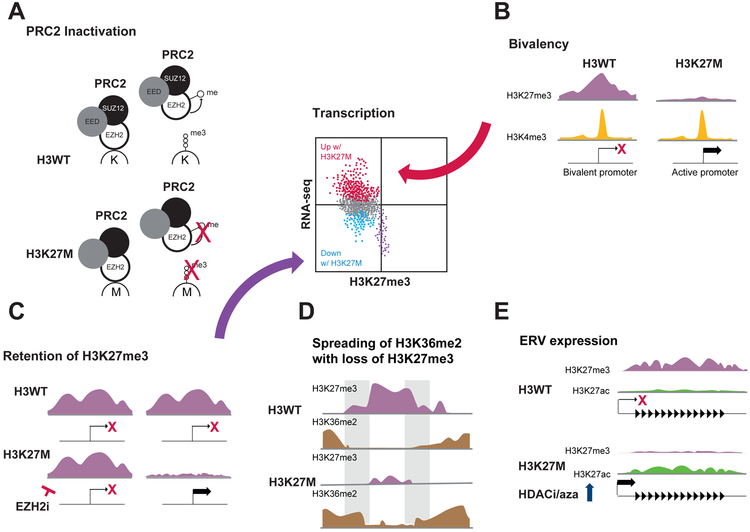
Figure 3.
Proposed mechanisms by which the H3K27M mutation changes the epigenetic landscape and impacts transcription, including (A) inactivation of PRC2 leading to H3K27me3 loss, (B) conversion of bivalent to active promoters, (C) selective retention of H3K27me3 (purple dots in scatter plot), (D) altered spreading of histone marks and (E) increased propensity for ERV expression.
H3K27M-dependent effects on transcription and epigenetic landscape
An enormous amount of work has been done on the epigenetic and transcriptional effects of the H3K27M mutation (Figure 3). Multiple sets of primary patient tumours and/or patient derived cell lines and PDX have been compiled and mined for H3K27M-dependent differences in gene expression (5, 9, 18, 51, 52). Not surprisingly, a common theme between these studies has been the relative upregulation of PRC2 target genes in H3K27M tumours compared to H3WT tumours; however, the set of genes found to be differentially regulated has varied between studies. A difficulty of transcriptional analyses carried out on cohorts of primary patient tumours is the paucity of H3WT tumours that are matched in location with H3K27M tumours. As a result, even beyond the expected heterogeneity between different human subjects, patient age, tumour location and cell of origin have been additional complicating factors that have obscured the effect of the H3K27M mutation on transcription and epigenetics. Now, with the development of the mouse models and isogenic xenograft comparisons described above, a flood of new data has allowed a more nuanced exploration of this topic than ever before.
Histone environment of H3K27M mutation impacts both epigenetic patterns and gene expression
Direct comparisons between H3.1K27M and H3.3K27M tumours have begun to show that the isoform of histone H3 bearing the K27M mutation delivers its own transcriptional and epigenetic signature (16, 26, 53). H3.1K27M could be discriminated from H3.3K27M tumours based on both DNA methylation profile and pattern of H3K27me3 deposition, as well as by gene expression profile, with H3.1K27M tumours having a more mesenchymal and astroglial signature, while H3.3K27M were more proneural and oligodendrocytic (16, 53). A recent study using conditional expression of H3.1K27M or H3.3K27M in isogenic early OPCs (eOPCs) differentiated from human iPSCs found that within one cell division following induction, the pattern of deposition between H3.1K27M and H3.3K27M was different, while the total loss of H3K27me3 was the same (26). In H3.3K27M, but not H3.1K27M, loss of H3K27me3 at CpG islands correlated with strength of H3.3 signal, suggesting that direct chromosomal recruitment of PRC2 by K27M mutant histone H3 helps set up these pattern differences. The pattern of H3K27ac recruitment also differed between the H3.1K27M and H3.3K27M eOPCs and reflected the differences in H3.1K27M versus H3.3K27M DIPG cell lines as well.
Role of bivalency in H3K27M-dependent gene expression changes
Gene promoters with bivalent cooccurrence of repression-associated H3K27me3 and activation-associated H3K4me3 on the same or neighbouring nucleosomes are poised for expression and this promoter state is often found at genes important in development (54). Does the widespread loss of H3K27me3 seen in H3K27M DIPGs have implications for bivalent genes? A recent study used a genetic mouse model of H3WT and H3K27M DIPG to ask this question (25). While only ~10% of all gene promoters were bivalent in H3WT tumours, more than 50% of promoters of genes that are upregulated in the H3K27M tumours are bivalent in H3WT tumours. Similarly, H3K27M-upregulated genes in three different DIPG PDX models were substantially enriched for bivalent promoters in tumours with H3K27M knockdown (29). These results suggest that release of poised promoters is a general mechanism of H3K27M-dependent gene upregulation (Figure 3B).
Regions of H3K27me3 retention against global loss in H3K27M cells
While the loss of H3K27me3 is a hallmark of the H3K27M mutation, regions exist that are able to retain or even gain H3K27me3 in H3K27M cells. These loci represent strong PRC2 targets (36, 38) and may be of critical importance in H3K27M tumourigenicity (Figure 3C). To target these loci, several small molecule inhibitors of EZH2 (EZH2i) have been used to deplete the remaining H3K27me3 in H3K27M cells, but the impact on cell growth has been somewhat inconsistent. Mohammad et al., Piunti et al. and Harutyunyan et al. have all found that EZH2i slows the growth of H3K27M mutant cells; however, whether this represents an H3K27M-specific dependency is unclear (36, 38, 40). Mohammad et al. saw similar effects on both H3WT and H3K27M NSCs, but Harutyunyan et al. found that H3K27M tumour cells were more sensitive than H3WT cells. By contrast, other studies have shown that while EZH2i treatment dramatically reduced H3K27me3, it had no impact on the growth of mouse or human tumour cells regardless of histone mutation status (32, 55).
One locus that shows retention of H3K27me3 in multiple H3K27M models is CDKN2A, and regulation of one isoform, the cell cycle regulator p16/INK4A has been implicated in several studies as an important piece in the H3K27M story. CDKN2A is a target of both PRC1 and PRC2 regulation and multiple groups have reported that H3K27me3 recruitment is often spared or increased at this locus in H3K27M cells, although the magnitude of H3K27me3 occupancy and the extent of Cdkn2a inhibition varies significantly in different studies (25, 29, 32, 36, 40). Piunti et al. found that while p16 expression was upregulated in response to SUZ12 or EED knockdown in SF8628 (H3.3K27M) cells, in SU-DIPG-VI cells (H3.1K27M) it was unchanged (40). Cordero et al. found t hat EZH2i depleted H3K27me3 occupancy at Cdkn2a, but did not alleviate H3K27M-dependent repression of p16, indicating that H3K27me3 did not directly regulate repression of Cdkn2a. However, treatment with the DNA methyltransferase inhibitors decitabine or azacytidine increased p16 mRNA in H3K27M cells to near H3WT levels. H3K27M cells also showed increased sensitivity to palbociclib, a CDK4/6 inhibitor that acts downstream of p16, and knockout of Cdkn2a (encoding both p16/INK4A and p19/ARF) decreased survival of mice with H3WT and H3K27M tumours and eliminated the difference in survival between the H3WT and H3K27M tumours. In addition to the CDKN2A locus, other tumour suppressor genes have been noted to be in regions of retained H3K27me3 and may represent important dependencies for DIPG, including NKX2.2 and WT1 (22, 47).
Other epigenetic changes influenced by H3K27M
In addition to the canonical loss of H3K27me3, other changes in the epigenetic landscape have been noted. DNA methylation can easily separate H3K27M and H3G34R/V histone mutant HGGs from each other and from other HGG subtypes, and the DNA methylation classifier (www.molecularneuropathology.org) is widely used by the scientific and clinical communities to classify brain tumours (4, 56). An H3K27M-driven increase in H3K27ac has been reported by multiple groups (19, 21, 25, 29, 39, 40, 49); however, antibody specificity and ChIP-seq normalization issues often complicate assessment of changes in levels of histone modifications. Mass spectroscopy revealed that H3K27M leads to increased H4 tail acetylation, loss of H3K27me1 and me2 in addition to H3K27me3 and gain of H3K36me2; however, not all of these epigenetic changes have been reported in all mass spec studies, which could reflect differences in models (21, 38, 40). ChIP-seq has shown that in addition to the well-established contraction of H3K27me3 peaks in H3K27M expressing cells, H3K27me2 appears to spread beyond its normal borders (38). Likewise, increased spreading of H3K36me2 into regions that have lost H3K27me3 in H3K27M 293T cells compared to H3WT reflects an interplay between these two histone modifications that has been noted in other studies and may hold additional keys to DIPG biology (Figure 3D) (20, 21, 57).
The H3K27M-dependent increase in H3K27ac has been shown to reflect a genome wide increase, including both regions that show H3K27ac deposition in H3WT cells as well as interstitial gain (25, 39). While these gains are modest at most loci, they may still have biological relevance. Increased H3K27ac over regions containing endogenous retroviral elements (ERVs) have been shown to correlate with an increase in their transcription (39). Since expression of ERVs can induce an innate immune response, it was hypothesized that this increase could represent an H3K27M-dependent vulnerability. Indeed, in H3.3K27M DIPG cells and matched isogenic controls where the mutant allele of H3F3A was knocked out, the H3K27M cells showed greater sensitivity to two agents that further increase ERV expression, 5’-azacytidine, a DNA methyltransferase inhibitor when administered alone or in combination with the HDACi, panobinostat (Figure 3E).
The potential impact of the H3K27M-dependent gain of H3K27ac on the enhancer landscape in DIPGs has also been a topic of inquiry. While deletion of H3K27M in DIPGs has indicated that the size or number of super enhancers (SEs) in DIPGs is not H3K27M-dependent (39), conditional expression of H3.1K27M versus H3.3K27M in eOPCs produced different patterns of H3K27ac deposition (26). This may suggest that the enhancer landscape is H3K27M-dependent during its initial establishment, but that once it is set up, H3K27M is no longer required for its maintenance. Regardless of whether the SE landscape in H3K27M DIPGs is K27M-dependent, the unique combination of factors that make up the core regulatory circuitry of DIPGs provide both clues to their cells of origin and also represent potential dependencies that can be exploited therapeutically (58). Nagaraja et al. defined SEs in four DIPG cultures and determined that many of the SE-driven genes are expressed by the oligodendrocyte lineage, providing further evidence that OPCs are the DIPG cell of origin (27). Genes coding for EPH receptors were also noted to be enriched for being driven by SEs and treatment with LDN-211904, an inhibitor of forward EPH receptor signaling inhibited H3K27M DIPGs in both invasion and migration assays. Krug et al. took the approach of looking for both shared and distinct SE-driven TFs across multiple paediatric brain tumours including H3K27M DIPGs and found many commonalities, but also identified IRX2 and PAX3 as possible DIPG-specific dependencies (39).
Primary dominant mechanisms of H3G34R/V oncogenesis remain to be uncovered
As of yet, H3G34R/V pHGGs have revealed few of their secrets. H3G34R/V tumours are primarily cortical and they segregate clearly from other pHGGs based on their DNA methylation profile, which shows that they are hypomethylated compared to other pHGGs (3, 4). H3G34R tumours frequently co-occur with ATRX and TP53 mutations (3). H3G34R/V mutations affect trimethylation of H3K36 on the same histone H3 tail, but unlike H3K27M, no dominant activity affecting HK36me3 or another histone modification has so far been found (19). Intriguingly, mutations in SETD2, an H3K36 methylase have been found in pHGG and they are mutually exclusive with H3G34R/V mutations, suggesting they may act through a common mechanism (59); however, the low overall frequency of H3G34R/V and SETD2 mutations could also account for this observation. H3G34R/V mutations have been shown to block di- and trimethylation of H3K36 because of steric hindrance (60) and mutation of the H3G34 residue on the same peptide as H3K36M relieves the dominant effect of H3K36M on H3K36me3 by decreasing the stable interaction of the H3 tail with SETD2 (61). Surprisingly, H3G34R has been reported to result in a local increase of H3K36me3 at specific loci, including MYCN (51, 62). The loci exhibiting an increase in H3K36me3 are also enriched for H3.3 and the K9/K36 demethylase KDM4 and show increased H3K9me3 which is consistent with a local and transient inhibition of KDM4 (62).
Fission yeast engineered to express only H3G34R exhibit genome instability and are defective for DNA damage repair by homologous recombination (63) and mammalian cells overexpressing H3G34R/V display a hypermutator phenotype (60). However, unlike H3K27M, H3G34R/V gave no growth advantage to NPCs in culture (22), nor was introduction of H3G34R and p53 loss into NPCs in the embryonic forebrain able to produce tumours, unlike H3K27M in the same setting (28).
Therapeutic outlook
The flurry of interest in histone mutations since their discovery has led to a welcomed variety of preclinical studies as well as the opening of several new clinical trials for DIPG patients. In particular, the availability of mouse and PDX models of DIPG have elevated the preclinical trials that can be carried out for this deadly disease. A recent survey of attempts to generate DIPG PDX and cell lines found that an encouraging 40-85% were successful across implant and culture models for both autopsy and biopsy material (64). Besides the EZH2 inhibitors discussed previously, small molecules targeting multiple other pathways have been tested using these models and proposed as potential therapeutics or tool compounds.
The HDAC inhibitor, panobinostat has been shown to impair growth of DIPGs in multiple labs (27, 28, 39, 52, 65), and multiple mechanisms through which the increase in H3K27ac slows growth have been proposed, including effects on superenhancers and ERVs. However, other studies suggest that the response to panobinostat is not H3K27M-dependent despite increased H3K27ac in H3K27M mutant cells (65). Similarly, bromodomain inhibitors (BETi) have showed efficacy in DIPGs (27, 39, 40, 66), but not always in an H3K27M-dependent manner (39). Interestingly, while cells that have developed panobinostat resistance are not sensitive to a BETi, likely because they impact expression of related genes, they retain sensitivity to the CDK7 inhibitor THZ1, offering potential for combination therapy (27). The histone demethylase JMJD3 inhibitor GSK-J4 has also been shown to decrease the viability of H3K27M DIPG cell lines and can act synergistically with panobinostat (52, 67). The menin inhibitor, MI-2 was identified as a top hit in a drug screen for growth inhibition of tumour-competent H3K27M, PDGFRA D842V expressing p53 knockdown hNPCs compared to normal NPCs, but the lack of mutations in menin or its binding partner MLL1 in gliomas was puzzling (22). A recent study determined that in glioma cells, MI-2 acts via a menin-independent mechanism and instead disrupts cholesterol homeostasis by targeting lanosterol synthase (68).
Additional promising avenues for therapeutic intervention address other features of H3K27M mutant tumours not directly related to the epigenetic machinery. CAR T cells showed dramatic efficacy against H3K27M mutant gliomas xenografted into pons, spinal cord and thalamus in mice. This encouraging outcome was accompanied by important notes of caution due to significant toxicity caused by tumour-associated inflammation in the thalamic tumours that would need to be carefully managed clinically (69). The H3.3K27M mutant is displayed as a neoantigen in HLA-A2+ hosts, providing the rationale for an ongoing clinical trial using a synthetic peptide as a vaccine to stimulate an anti-tumour immune response in patients who are positive for HLA-A2, and have H3.3K27M mutant glioma (70). Other exciting studies have elucidated how neuronal activity regulates glioma growth via the proliferative effects of the secreted factor NLGN3 and depolarization of glioma cells themselves (71-73). Agents that interfere with either of these mechanisms have been shown to decrease glioma proliferation and extend survival of mice bearing H3K27M DIPG xenografts.
In less than a decade, the field has advanced from limited molecular information to an unprecedented view of the genome, transcriptome and epigenome of paediatric high-grade glioma and an increasing understanding of the unique features and potential vulnerabilities of these deadly tumours. Despite many exciting new findings, the outcome for patients with histone mutant HGG remains dismal. The ongoing intense focus on these mutations and the critical contribution of developmental context to tumour pathogenesis will drive accelerated progress to address critical unanswered questions including mechanisms underlying tumourigenesis and therapeutic resistance.
Acknowledgment:
This work was partially funded by ALSAC and National Cancer Institute grant P01-CA096832 to SJB.
References
- 1.Hoffman LM, Veldhuijzen van Zanten SEM, Colditz N, Baugh J, Chaney B, Hoffmann M, et al. Clinical, Radiologic, Pathologic, and Molecular Characteristics of Long-Term Survivors of Diffuse Intrinsic Pontine Glioma (DIPG): A Collaborative Report From the International and European Society for Pediatric Oncology DIPG Registries. J Clin Oncol. 2018;36(19):1963–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones C, Baker SJ. Unique genetic and epigenetic mechanisms driving paediatric diffuse high-grade glioma. Nat Rev Cancer. 2014;14(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mackay A, Burford A, Carvalho D, Izquierdo E, Fazal-Salom J, Taylor KR, et al. Integrated Molecular Meta-Analysis of 1,000 Pediatric High-Grade and Diffuse Intrinsic Pontine Glioma. Cancer Cell. 2017;32(4):520–37 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sturm D, Bender S, Jones DT, Lichter P, Grill J, Becher O, et al. Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nat Rev Cancer. 2014;14(2):92–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–31. [DOI] [PubMed] [Google Scholar]
- 6.Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44(3):251–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell. 2016;164(3):550–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140(5):678–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bender S, Tang Y, Lindroth AM, Hovestadt V, Jones DT, Kool M, et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24(5):660–72. [DOI] [PubMed] [Google Scholar]
- 10.Buczkowicz P, Hoeman C, Rakopoulos P, Pajovic S, Letourneau L, Dzamba M, et al. Genomic analysis of diffuse intrinsic pontine gliomas identifies three molecular subgroups and recurrent activating ACVR1 mutations. Nat Genet. 2014;46(5):451–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fontebasso AM, Papillon-Cavanagh S, Schwartzentruber J, Nikbakht H, Gerges N, Fiset PO, et al. Recurrent somatic mutations in ACVR1 in pediatric midline high-grade astrocytoma. Nat Genet. 2014;46(5):462–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khuong-Quang DA, Buczkowicz P, Rakopoulos P, Liu XY, Fontebasso AM, Bouffet E, et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol. 2012;124(3):439–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor KR, Mackay A, Truffaux N, Butterfield Y, Morozova O, Philippe C, et al. Recurrent activating ACVR1 mutations in diffuse intrinsic pontine glioma. Nat Genet. 2014;46(5):457–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu G, Diaz AK, Paugh BS, Rankin SL, Ju B, Li Y, et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet. 2014;46(5):444–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–20. [DOI] [PubMed] [Google Scholar]
- 16.Castel D, Philippe C, Calmon R, Le Dret L, Truffaux N, Boddaert N, et al. Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol. 2015;130(6):815–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Venneti S, Garimella MT, Sullivan LM, Martinez D, Huse JT, Heguy A, et al. Evaluation of histone 3 lysine 27 trimethylation (H3K27me3) and enhancer of Zest 2 (EZH2) in pediatric glial and glioneuronal tumors shows decreased H3K27me3 in H3F3A K27M mutant glioblastomas. Brain Pathol. 2013;23(5):558–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan KM, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 2013;27(9):985–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340(6134):857–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nacev BA, Feng L, Bagert JD, Lemiesz AE, Gao J, Soshnev AA, et al. The expanding landscape of ‘oncohistone’ mutations in human cancers. Nature. 2019;567(7749):473–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stafford JM, Lee CH, Voigt P, Descostes N, Saldana-Meyer R, Yu JR, et al. Multiple modes of PRC2 inhibition elicit global chromatin alterations in H3K27M pediatric glioma. Sci Adv. 2018;4(10):eaau5935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Funato K, Major T, Lewis PW, Allis CD, Tabar V. Use of human embryonic stem cells to model pediatric gliomas with H3.3K27M histone mutation. Science. 2014;346(6216):1529–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Filbin MG, Tirosh I, Hovestadt V, Shaw ML, Escalante LE, Mathewson ND, et al. Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science. 2018;360(6386):331–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larson JD, Kasper LH, Paugh BS, Jin H, Wu G, Kwon CH, et al. Histone H3.3 K27M Accelerates Spontaneous Brainstem Glioma and Drives Restricted Changes in Bivalent Gene Expression. Cancer Cell. 2019;35(1):140–55 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagaraja S, Quezada MA, Gillespie SM, Arzt M, Lennon JJ, Woo PJ, et al. Histone Variant and Cell Context Determine H3K27M Reprogramming of the Enhancer Landscape and Oncogenic State. Mol Cell. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagaraja S, Vitanza NA, Woo PJ, Taylor KR, Liu F, Zhang L, et al. Transcriptional Dependencies in Diffuse Intrinsic Pontine Glioma. Cancer Cell. 2017;31(5):635–52 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pathania M, De Jay N, Maestro N, Harutyunyan AS, Nitarska J, Pahlavan P, et al. H3.3(K27M) Cooperates with Trp53 Loss and PDGFRA Gain in Mouse Embryonic Neural Progenitor Cells to Induce Invasive High-Grade Gliomas. Cancer Cell. 2017;32(5):684–700 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silveira AB, Kasper LH, Fan Y, Jin H, Wu G, Shaw TI, et al. H3.3 K27M depletion increases differentiation and extends latency of diffuse intrinsic pontine glioma growth in vivo. Acta Neuropathol. 2019;137(4):637–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikbakht H, Panditharatna E, Mikael LG, Li R, Gayden T, Osmond M, et al. Spatial and temporal homogeneity of driver mutations in diffuse intrinsic pontine glioma. Nat Commun. 2016;7:11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vinci M, Burford A, Molinari V, Kessler K, Popov S, Clarke M, et al. Functional diversity and cooperativity between subclonal populations of pediatric glioblastoma and diffuse intrinsic pontine glioma cells. Nat Med. 2018;24(8):1204–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cordero FJ, Huang Z, Grenier C, He X, Hu G, McLendon RE, et al. Histone H3.3K27M Represses p16 to Accelerate Gliomagenesis in a Murine Model of DIPG. Mol Cancer Res. 2017;15(9):1243–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paugh BS, Broniscer A, Qu C, Miller CP, Zhang J, Tatevossian RG, et al. Genome-wide analyses identify recurrent amplifications of receptor tyrosine kinases and cell-cycle regulatory genes in diffuse intrinsic pontine glioma. J Clin Oncol. 2011;29(30):3999–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paugh BS, Zhu X, Qu C, Endersby R, Diaz AK, Zhang J, et al. Novel oncogenic PDGFRA mutations in pediatric high-grade gliomas. Cancer Res. 2013;73(20):6219–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diaz AK, Baker SJ. The genetic signatures of pediatric high-grade glioma: no longer a one-act play. Semin Radiat Oncol. 2014;24(4):240–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohammad F, Weissmann S, Leblanc B, Pandey DP, Hojfeldt JW, Comet I, et al. EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat Med. 2017;23(4):483–92. [DOI] [PubMed] [Google Scholar]
- 37.Subashi E, Cordero FJ, Halvorson KG, Qi Y, Nouls JC, Becher OJ, et al. Tumor location, but not H3.3K27M, significantly influences the blood-brain-barrier permeability in a genetic mouse model of pediatric high-grade glioma. J Neurooncol. 2016;126(2):243–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harutyunyan AS, Krug B, Chen H, Papillon-Cavanagh S, Zeinieh M, De Jay N, et al. H3K27M induces defective chromatin spread of PRC2-mediated repressive H3K27me2/me3 and is essential for glioma tumorigenesis. Nat Commun. 2019;10(1):1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krug B, De Jay N, Harutyunyan AS, Deshmukh S, Marchione DM, Guilhamon P, et al. Pervasive H3K27 Acetylation Leads to ERV Expression and a Therapeutic Vulnerability in H3K27M Gliomas. Cancer Cell. 2019;35(5):782–97 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piunti A, Hashizume R, Morgan MA, Bartom ET, Horbinski CM, Marshall SA, et al. Therapeutic targeting of polycomb and BET bromodomain proteins in diffuse intrinsic pontine gliomas. Nat Med. 2017;23(4):493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boileau M, Shirinian M, Gayden T, Harutyunyan AS, Chen CCL, Mikael LG, et al. Mutant H3 histones drive human pre-leukemic hematopoietic stem cell expansion and promote leukemic aggressiveness. Nat Commun. 2019;10(1):2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Justin N, Zhang Y, Tarricone C, Martin SR, Chen S, Underwood E, et al. Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat Commun. 2016;7:11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pajtler KW, Wen J, Sill M, Lin T, Orisme W, Tang B, et al. Molecular heterogeneity and CXorf67 alterations in posterior fossa group A (PFA) ependymomas. Acta Neuropathol. 2018;136(2):211–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ragazzini R, Perez-Palacios R, Baymaz IH, Diop S, Ancelin K, Zielinski D, et al. EZHIP constrains Polycomb Repressive Complex 2 activity in germ cells. Nat Commun. 2019;10(1):3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hubner JM, Muller T, Papageorgiou DN, Mauermann M, Krijgsveld J, Russell RB, et al. EZHIP / CXorf67 mimics K27M mutated oncohistones and functions as an intrinsic inhibitor of PRC2 function in aggressive posterior fossa ependymoma. Neuro Oncol. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jain SU, Do TJ, Lund PJ, Rashoff AQ, Diehl KL, Cieslik M, et al. PFA ependymoma-associated protein EZHIP inhibits PRC2 activity through a H3 K27M-like mechanism. Nat Commun. 2019;10(1):2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fang D, Gan H, Cheng L, Lee JH, Zhou H, Sarkaria JN, et al. H3.3K27M mutant proteins reprogram epigenome by sequestering the PRC2 complex to poised enhancers. Elife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tatavosian R, Duc HN, Huynh TN, Fang D, Schmitt B, Shi X, et al. Live-cell single-molecule dynamics of PcG proteins imposed by the DIPG H3.3K27M mutation. Nat Commun. 2018;9(1):2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Herz HM, Morgan M, Gao X, Jackson J, Rickels R, Swanson SK, et al. Histone H3 lysine-to-methionine mutants as a paradigm to study chromatin signaling. Science. 2014;345(6200):1065–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee CH, Yu JR, Granat J, Saldana-Meyer R, Andrade J, LeRoy G, et al. Automethylation of PRC2 promotes H3K27 methylation and is impaired in H3K27M pediatric glioma. Genes Dev. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bjerke L, Mackay A, Nandhabalan M, Burford A, Jury A, Popov S, et al. Histone H3.3. mutations drive pediatric glioblastoma through upregulation of MYCN. Cancer Discov. 2013;3(5):512–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grasso CS, Tang Y, Truffaux N, Berlow NE, Liu L, Debily MA, et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat Med. 2015;21(7):827. [DOI] [PubMed] [Google Scholar]
- 53.Castel D, Philippe C, Kergrohen T, Sill M, Merlevede J, Barret E, et al. Transcriptomic and epigenetic profiling of ‘diffuse midline gliomas, H3 K27M-mutant’ discriminate two subgroups based on the type of histone H3 mutated and not supratentorial or infratentorial location. Acta Neuropathol Commun. 2018;6(1):117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12(1):7–18. [DOI] [PubMed] [Google Scholar]
- 55.Wiese M, Schill F, Sturm D, Pfister S, Hulleman E, Johnsen SA, et al. No Significant Cytotoxic Effect of the EZH2 Inhibitor Tazemetostat (EPZ-6438) on Pediatric Glioma Cells with Wildtype Histone 3 or Mutated Histone 3.3. Klin Padiatr. 2016;228(3):113–7. [DOI] [PubMed] [Google Scholar]
- 56.Capper D, Stichel D, Sahm F, Jones DTW, Schrimpf D, Sill M, et al. Practical implementation of DNA methylation and copy-number-based CNS tumor diagnostics: the Heidelberg experience. Acta Neuropathol. 2018;136(2):181–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jani KS, Jain SU, Ge EJ, Diehl KL, Lundgren SM, Muller MM, et al. Histone H3 tail binds a unique sensing pocket in EZH2 to activate the PRC2 methyltransferase. Proc Natl Acad Sci U S A. 2019;116(17):8295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fontebasso AM, Schwartzentruber J, Khuong-Quang DA, Liu XY, Sturm D, Korshunov A, et al. Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta Neuropathol. 2013;125(5):659–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fang J, Huang Y, Mao G, Yang S, Rennert G, Gu L, et al. Cancer-driving H3G34V/R/D mutations block H3K36 methylation and H3K36me3-MutSalpha interaction. Proc Natl Acad Sci U S A. 2018;115(38):9598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y, Shan CM, Wang J, Bao K, Tong L, Jia S. Molecular basis for the role of oncogenic histone mutations in modulating H3K36 methylation. Sci Rep. 2017;7:43906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Voon HPJ, Udugama M, Lin W, Hii L, Law RHP, Steer DL, et al. Inhibition of a K9/K36 demethylase by an H3.3 point mutation found in paediatric glioblastoma. Nat Commun. 2018;9(1):3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yadav RK, Jablonowski CM, Fernandez AG, Lowe BR, Henry RA, Finkelstein D, et al. Histone H3G34R mutation causes replication stress, homologous recombination defects and genomic instability in S. pombe. Elife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsoli M, Shen H, Mayoh C, Franshaw L, Ehteda A, Upton D, et al. International experience in the development of patient-derived xenograft models of diffuse intrinsic pontine glioma. J Neurooncol. 2019;141(2):253–63. [DOI] [PubMed] [Google Scholar]
- 65.Hennika T, Hu G, Olaciregui NG, Barton KL, Ehteda A, Chitranjan A, et al. Pre-Clinical Study of Panobinostat in Xenograft and Genetically Engineered Murine Diffuse Intrinsic Pontine Glioma Models. PLoS One. 2017;12(1):e0169485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taylor IC, Hutt-Cabezas M, Brandt WD, Kambhampati M, Nazarian J, Chang HT, et al. Disrupting NOTCH Slows Diffuse Intrinsic Pontine Glioma Growth, Enhances Radiation Sensitivity, and Shows Combinatorial Efficacy With Bromodomain Inhibition. J Neuropathol Exp Neurol. 2015;74(8):778–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hashizume R, Andor N, Ihara Y, Lerner R, Gan H, Chen X, et al. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat Med. 2014;20(12):1394–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Phillips RE, Yang Y, Smith RC, Thompson BM, Yamasaki T, Soto-Feliciano YM, et al. Target identification reveals lanosterol synthase as a vulnerability in glioma. Proc Natl Acad Sci U S A. 2019;116(16):7957–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mount CW, Majzner RG, Sundaresh S, Arnold EP, Kadapakkam M, Haile S, et al. Potent antitumor efficacy of anti-GD2 CAR T cells in H3-K27M(+) diffuse midline gliomas. Nat Med. 2018;24(5):572–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chheda ZS, Kohanbash G, Okada K, Jahan N, Sidney J, Pecoraro M, et al. Novel and shared neoantigen derived from histone 3 variant H3.3K27M mutation for glioma T cell therapy. J Exp Med. 2018;215(1):141–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Venkatesh HS, Johung TB, Caretti V, Noll A, Tang Y, Nagaraja S, et al. Neuronal Activity Promotes Glioma Growth through Neuroligin-3 Secretion. Cell. 2015;161(4):803–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Venkatesh HS, Morishita W, Geraghty AC, Silverbush D, Gillespie SM, Arzt M, et al. Electrical and synaptic integration of glioma into neural circuits. Nature. 2019;573(7775):539–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Venkatesh HS, Tam LT, Woo PJ, Lennon J, Nagaraja S, Gillespie SM, et al. Targeting neuronal activity-regulated neuroligin-3 dependency in high-grade glioma. Nature. 2017;549(7673):533–7. [DOI] [PMC free article] [PubMed] [Google Scholar]