ABSTRACT
Despite the importance of Wnt signaling for adult intestinal stem cell homeostasis and colorectal cancer, relatively little is known about its role in colon formation during embryogenesis. The development of the colon starts with the formation and extension of the hindgut. We show that Wnt3a is expressed in the caudal embryo in a dorsal-ventral (DV) gradient across all three germ layers, including the hindgut. Using genetic and lineage-tracing approaches, we describe novel dorsal and ventral hindgut domains, and show that ventrolateral hindgut cells populate the majority of the colonic epithelium. A Wnt3a-β-catenin-Sp5/8 pathway, which is active in the dorsal hindgut endoderm, is required for hindgut extension and colon formation. Interestingly, the absence of Wnt activity in the ventral hindgut is crucial for proper hindgut morphogenesis, as ectopic stabilization of β-catenin in the ventral hindgut via gain- or loss-of-function mutations in Ctnnb1 or Apc, respectively, leads to severe colonic hyperplasia. Thus, the DV Wnt gradient is required to coordinate growth between dorsal and ventral hindgut domains to regulate the extension of the hindgut that leads to colon formation.
KEY WORDS: Wnt3a, Hindgut, Endoderm, Intestine, β-Catenin, Colon, Mouse
Highlighted Article: A dorsal-ventral gradient of Wnt3a distinguishes embryonic dorsal and ventral hindgut domains, and regulates the growth necessary for proper colon formation.
INTRODUCTION
The colon is the caudal-most part of the gastrointestinal (GI) tract and functions to absorb water, process and store waste before elimination, and is home to hundreds of species of gut flora that impact multiple aspects of host physiology (Sommer and Bäckhed, 2013). The formation of the GI tract begins with the generation and anterior-posterior patterning of the definitive endoderm during gastrulation. This two-dimensional sheet of cells is converted into a three-dimensional tube through complex morphogenetic processes that occur at the anterior and posterior ends of the embryo to form the foregut and hindgut, respectively (McGrath and Wells, 2015; Nerurkar et al., 2019; Spence et al., 2011; Wells and Spence, 2014; Zorn and Wells, 2009). The hindgut arises ventral to the primitive streak (PS), starting at ∼E8.25 with the formation of the caudal intestinal portal and the outpocketing of the hindgut endoderm. The posterior extension of the hindgut occurs over several subsequent days (∼E8.5-13.5) to generate a tube that will eventually form the colon; however, the mechanisms underlying tubulogenesis and extension are not well understood.
Studies performed in vertebrate model organisms and in human pluripotent stem cells (PSCs) collectively demonstrate that the Wnt, Fgf, Nodal, retinoic acid and Bmp signaling pathways play important roles in the early development of the GI tract (Chin et al., 2017; McGrath and Wells, 2015; Wells and Spence, 2014; Zorn and Wells, 2009). For example, the Wnt, Fgf and Bmp pathways are active during gastrulation to promote hindgut fates from posterior definitive endoderm through the regulation of hindgut determinants such as Cdx2 (Engert et al., 2013; Gregorieff et al., 2004; Munera et al., 2017; Rankin et al., 2018; Sherwood et al., 2011; Spence et al., 2011; Stevens et al., 2017). Recent studies in the chick demonstrate that Fgfs also regulate collective cell movements of the endoderm to direct the formation of the hindgut (Nerurkar et al., 2019). It is unclear whether similar mechanisms function during mouse hindgut formation, as visceral endoderm cells are dispersed by the intercalation of definitive endoderm and are not displaced by mass unidirectional movements of endoderm (Kwon et al., 2008).
Following the formation of the nascent hindgut, the subsequent extension of the epithelial hindgut tube occurs simultaneously with the posteriorly directed growth of the embryonic body axis, which is largely thought to be driven by the proliferation of axial progenitors in the caudal epiblast, the generation and movement of mesenchymal cells from the primitive streak (PS), and the morphogenesis of the presomitic mesoderm (PSM), trunk somites and spinal cord (Garriock et al., 2015; Wilson et al., 2009; Yamaguchi et al., 1999b). How these processes are coordinated remains unclear; however, caudal gradients of Wnt, Fgf and Bmp proteins are likely candidates, given their demonstrated roles in hindgut specification (see above) and body axis extension (Bénazéraf et al., 2010; Dubrulle and Pourquié, 2004; Edri et al., 2019; Garriock et al., 2015; Stevens et al., 2017; Wymeersch et al., 2016; Yamaguchi et al., 1999a). Interestingly, tetraploid complementation experiments show that the hindgut has organizer activity and can communicate with the overlying epiblast and PS to regulate axial extension (Engert et al., 2013) suggesting that signaling between germ layers is reciprocal. Recent single cell studies have begun to catalog the many signaling pathways that are active in the developing gut endoderm (Nowotschin et al., 2019).
The Wnt/β-catenin signaling pathway regulates the transcription of target genes (Nusse and Clevers, 2017; Perochon et al., 2018). The function of this pathway in the adult intestine is particularly well-studied as it controls intestinal stem cell fate (Gehart and Clevers, 2019; Kretzschmar and Clevers, 2017), and because activating mutations are causative for colon cancer (Cancer Genome Atlas, 2012; Clevers and Batlle, 2013; van Es et al., 2012). In the absence of Wnt ligand, nuclear β-catenin levels are kept low through the activity of the β-catenin destruction complex involving the tumor suppressor adenomatous polyposis coli (Apc). Upon Wnt ligand binding to cell-surface receptors, the β-catenin destruction complex is attenuated, leading to elevated nuclear β-catenin levels, the formation of β-catenin/Tcf1-Lef1 complexes, and the transcriptional activation of target genes (Cadigan and Waterman, 2012; Clevers and Nusse, 2012). We have shown in ESCs and in vivo, that the Sp1/Klf family transcription factors Sp5 and Sp8 bind directly to DNA, Tcf1 and Lef1 to participate in the transcriptional activation of a select subset of Wnt target genes (Dunty et al., 2014; Kennedy et al., 2016). We show here that Wnt3a is expressed in a DV gradient in the elongating hindgut, where it signals via a β-catenin-Sp5/8 pathway to regulate Bmp signaling and direct the hindgut expansion that is an essential prerequisite for colon formation.
RESULTS
Hindgut cell proliferation during axial extension occurs primarily at the caudal terminus
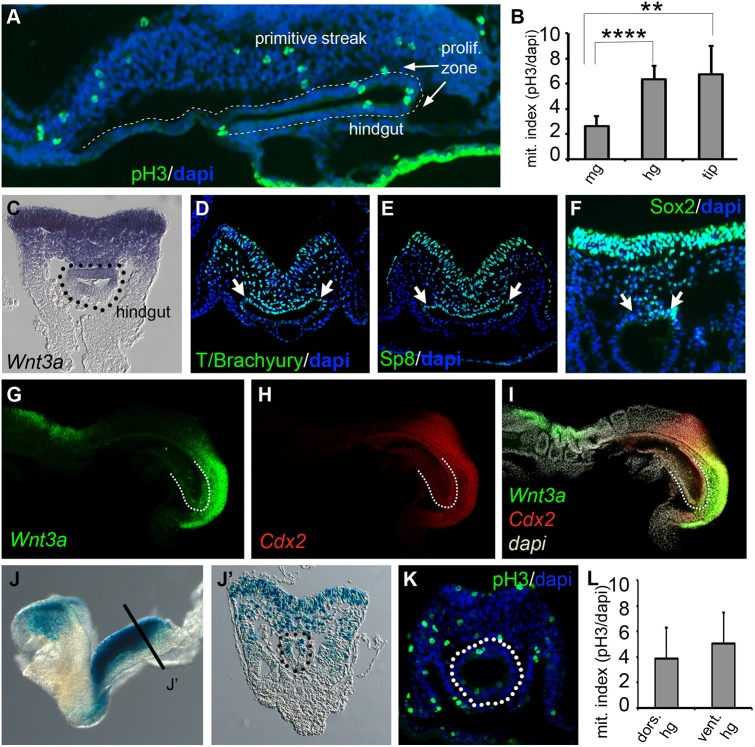
To investigate the mechanisms underlying colon formation, we first examined cell proliferation along the developing hindgut using phospho-histone H3 (pH3) antibody staining (Hans and Dimitrov, 2001). Mitotic nuclei were detected in hindgut endoderm at E8.5, particularly in caudal regions of the hindgut that underlie the PS ventrally (Fig. 1A). Quantification showed 2.5 times more pH3+ cells in the caudal hindgut endoderm than in midgut located rostral to the PS region (6.4% versus 2.6%, respectively) but no dorsal-ventral bias was observed (Fig. 1B,L). This suggested that the hindgut endoderm might be exposed to more mitogenic signals than the midgut.
Fig. 1.
Hindgut proliferation and analysis of Wnt signaling in the E8.5 hindgut. (A) Sagittal section through the E8.5 primitive streak and hindgut showing anti-phospho-HistoneH3 labeling (green) and nuclei counterstained with DAPI (blue). The endodermal DAPI staining is outlined (dashed line) to highlight the hindgut endoderm. (B) Graph of cell proliferation in the midgut (mg), hindgut (hg) and posterior tip of the hindgut (n=5 E8.5 embryos). The hindgut epithelium showed a significantly greater mitotic index than the midgut epithelium (data are mean±s.e.m. ****P<1.2×10−4). Additionally, the posterior tip of the hindgut also showed a significantly higher number of mitotic cells than the midgut epithelium (data are mean±s.e.m. **P<7.9×10−3). (C) Transverse section through the PS region of an E8.5 embryo processed for whole-mount in situ hybridization and displaying Wnt3a mRNA gradient along the DV axis. (D-F) Antibody detection of T/Bra, Sp8 and Sox2 protein, respectively, in transverse sections through the PS. There is strong expression in the dorsolateral hindgut (arrows). (G-I) In situ HCR to detect the expression of Wnt3a (G,I) and Cdx2 (H,I) at E8.5. (J,J′) In situ Wnt/β-catenin reporter (BATlacZ) detected by β-galactosidase staining viewed laterally in whole-mount (J) and in transverse (J′) sections through PS and hindgut regions. (K) Transverse section through the PS and hindgut (dotted circle) stained for phospho-Histone H3 using antibody (green) with DAPI nuclear counterstain (blue). (L) Graph of mitotic indexes in dorsal and ventral hindgut (n=5, E8.5 embryos). No significant difference was observed.
We considered if the hindgut endoderm was responding to Wnt3a signals from the PS, as Wnt/β-catenin signals are known drivers of cell proliferation (Clevers and Nusse, 2012) and Wnt3a is a known regulator of posterior development (Takada et al., 1994). As expected, Wnt3a was readily detected in the PS region at E8.5 (Fig. 1C,G,I; Fig. S1A,B). Sections revealed that Wnt3a was expressed in all three germ layers, in a DV gradient that included the hindgut, with highest expression observed dorsally in the streak epiblast, lower levels in the streak mesoderm and dorsal hindgut, and little to no expression in the ventral hindgut. Sagittal sections additionally showed that Wnt3a is largely restricted to the dorsoposterior hindgut terminus (Fig. 1G,I; Fig. S1). The Wnt3a targets T/brachyury, Sp8 and Cdx2 (Dunty et al., 2014; Sherwood et al., 2011; Yamaguchi et al., 1999b) were co-expressed with Wnt3a, suggesting that the dorsal hindgut is a site of active Wnt/β-catenin signaling (arrows, Fig. 1D,E,H; Fig. S1B). Consistent with this proposal, examination of BATlacZ transgenic embryos for the expression of a β-catenin/Tcf lacZ reporter (Nakaya et al., 2005) also showed that β-galactosidase (βgal) activity was restricted to the dorsal hindgut (Fig. 1J,J′). Interestingly, the stem cell marker and Wnt target Sox2, commonly referred to as a foregut and stomach marker (McGrath and Wells, 2015; Sherwood et al., 2011), was also detected in a subset of dorsal hindgut cells (Fig. 1F). The overlapping detection of Wnt3a, Cdx2, Sox2, T/brachyury and Sp8 suggested that the dorsal hindgut might harbor a unique endodermal progenitor population as the same markers are co-expressed in the PS where they define Wnt-regulated multipotent neuromesodermal progenitors (NMPs) (Dunty et al., 2014; Garriock et al., 2015; Gouti et al., 2014; Martin and Kimelman, 2012; Tsakiridis et al., 2014). Unlike NMPs, the dorsal hindgut does not express the NMP and PSM markers Nkx1.2 and Tbx6 (Chapman et al., 2003; Rodrigo Albors et al., 2018), demonstrating that these cells have unique but overlapping gene expression profiles (Fig. S2). Taken together, these data raise the intriguing possibility that the dorsal hindgut is a Wnt3a-responsive colon progenitor population.
A Wnt3a/β-catenin/Sp5/8 signaling axis is required for hindgut development and colon formation
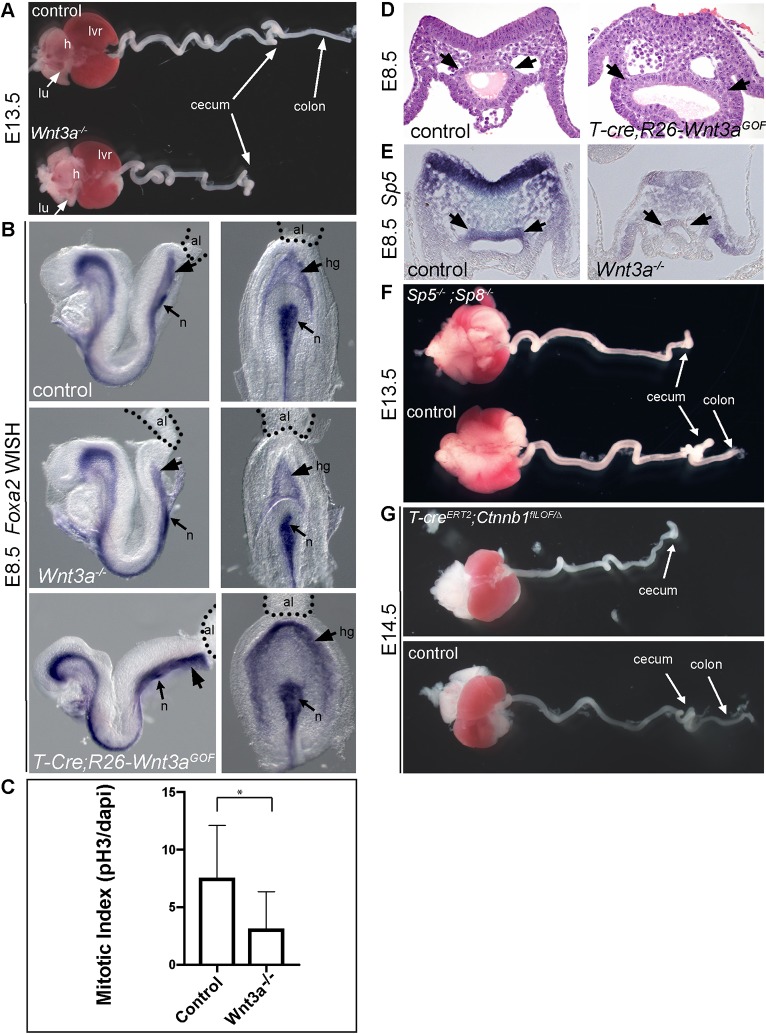
GI malformations were not previously reported in the Wnt3a−/− null fetus (Takada et al., 1994). Examination of the GI tract of wild-type embryos at E13.5 showed an elongated intestinal tract with distinct small and large intestine/colon domains separated by the cecum (Fig. 2A). Remarkably, while anterior regions of the GI tract appear to be normally sized in Wnt3a−/− mutants, the colon was completely absent (Fig. 2A). To determine whether colon agenesis was caused by an early defect in hindgut formation, we examined control and Wnt3a−/− mutants at E8.5 when the embryonic hindgut first forms. Examination of Foxa2 expression in the hindgut of Wnt3a−/− mutants showed a narrow and underdeveloped hindgut pocket that did not appear to fully extend to the posterior terminus and allantois, as occurred in control embryos (Fig. 2B). Quantification of mitotic cells in the E8.5 hindgut epithelium revealed a significant decrease in the number of proliferating cells in Wnt3a−/− mutants compared with wild-type littermates (Fig. 2C). By E9.5, it was clear that hindgut extension had failed in Wnt3a−/− mutants (Fig. S3A). The Foxa2-expressing hindgut had a narrow balloon-like shape in wild-type embryos that was absent, or severely underdeveloped, in the mutants. In contrast, constitutive overexpression of Wnt3a in the PS and hindgut using the T-Cre mouse to express Cre in T (brachyury)-expressing mesendoderm progenitors (T-Cre;R26-Wnt3aGOF) (Chalamalasetty et al., 2016; Perantoni et al., 2005) resulted in a broad posterior and lateral expansion of the Foxa2-expressing hindgut (Fig. 2B,D). These data demonstrate that Wnt3a is not required for hindgut initiation or for the formation of the caudal intestinal portal, but instead regulates hindgut extension.
Fig. 2.
The Wnt3a/β-catenin signaling pathway is required for colon formation and hindgut development. (A) Isolated E13.5 GI tract and organs from control littermate (top) and Wnt3a−/− mutant (bottom) fetus. The colon region is missing in Wnt3a−/− mutants. (B) WISH detection of Foxa2 in E8.5 control, Wnt3a−/− and T-Cre;R26-Wnt3aGOF mutants. (C) Graph of mitotic indexes in control (n=15) and Wnt3a−/− (n=8) hindgut. Data are mean±s.d. (*P=0.02; unpaired two-tailed t-test). (D) Hematoxylin and Eosin-stained transverse sections through the PS and hindgut (black arrows) of E8.5 control and T-cre;R26-Wnt3aGOF embryos. (E) Transverse sections through the PS and hindgut showing WISH detection of the Wnt-responsive gene Sp5 in E8.5 control littermate and Wnt3a−/− mutants. (F) E13.5 isolated GI tract and organs from control (bottom) and Sp5−/−;Sp8−/− mutant (top) fetus. (G) E14.5 isolated GI tract and organs from control (bottom) and T-CreERT2;Ctnnb1flLOF/Δ mutant (top) fetus. TAM was administered at E7.5. lu, lung; lvr, liver; h, heart; al, allantois; hg, hindgut; n, node.
To further support the premise that the hindgut responds directly to Wnt3a, we examined sections of E8.5 control and Wnt3a−/− embryos processed for Sp5 expression by whole-mount in situ hybridization. Sp5 is a direct Wnt/β-catenin target gene in mice and fish, and is highly regulated by Wnt3a in the PS (Dunty et al., 2014; Takahashi et al., 2005; Thorpe et al., 2005; Weidinger et al., 2005). In control E8.5 embryos, Sp5 was expressed in the PS ectoderm, mesoderm and dorsal hindgut (arrows) (Fig. 2E), as anticipated based on the expression of Wnt3a and other Wnt target genes (Fig. 1). Sp5 mRNA was not detected in the smaller mis-shaped hindgut of Wnt3a−/− mutants (Fig. 2E), demonstrating that the expression of Wnt target genes in the hindgut depend upon Wnt3a. We have previously shown that Sp5 and the closely related Sp8 transcription factors are co-expressed in the primitive streak region (Dunty et al., 2014) and function in the Wnt/β-catenin signaling pathway as Tcf/Lef co-factors (Kennedy et al., 2016). The reduced expression of Sp5 raises the question of whether Sp5 plays an essential role in hindgut extension. Although Sp5 mutants have no demonstrable gut phenotypes, the Sp5−/−; Sp8−/− GI tracts were truncated at the cecum, similar to the Wnt3a−/− gut, and did not form colons (Fig. 2F), further corroborating the role of the Wnt/β-catenin pathway in hindgut extension.
As both the Wnt3a and Sp5/8 mutants display severe defects in caudal development that result in the truncation of the axial skeleton beginning at the level of the forelimbs (Dunty et al., 2014), it is possible that colon agenesis is a secondary consequence of this axial truncation. We therefore examined colon formation in T-CreERT2;Ctnnb1flLOF/Δ embryos in which β-catenin (encoded by Ctnnb1) is conditionally inactivated in the PS and hindgut with the Tamoxifen (TAM)-inducible T-CreERT2 mouse (Anderson et al., 2013) and which display more extensive caudal development, as evidenced by the presence of the posterior trunk and hindlimbs (Fig. S3B). Administration of TAM to T-CreERT2;R26R embryos at E7.5 efficiently targeted both dorsal and ventral hindgut populations by E8.5, in addition to the PS epiblast and mesoderm (Garriock et al., 2015). Examination of the GI tracts of E14.5 T-CreERT2;Ctnnb1flLOF/Δ fetuses that had received TAM at E7.5 revealed that they were also truncated at the cecum (Fig. 2G), similar to the Wnt3a−/− and Sp5−/−;Sp8−/− gut, despite considerably more extensive trunk development. These results suggest that the colon agenesis phenotype does not simply result from a failure of trunk caudal extension.
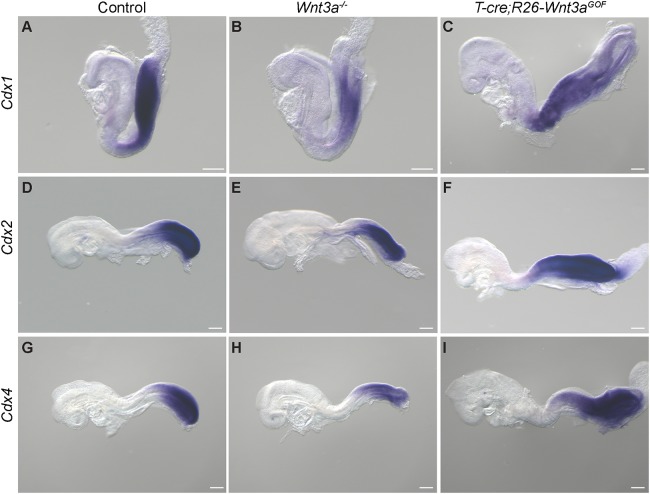
To determine whether the hindgut phenotypes could arise from an inability to maintain posterior identity, we examined Wnt3a−/− embryos for the expression of the Caudal-related Cdx genes, which are well characterized Wnt target genes and posterior determinants (Gao et al., 2009; Gaunt et al., 2003; Ikeya and Takada, 2001; Prinos et al., 2001; Sherwood et al., 2011; van de Ven et al., 2011). Cdx1, Cdx2 and Cdx4 were co-expressed in posterior domains in control embryos at hindgut extension stages (E8.5-9) (Figs 1H,I and 3A,D,G, Fig. S4A). Cdx1 and Cdx4 were both downregulated but remained properly posteriorly localized in Wnt3a−/− embryos, while Cdx2 continued to be highly expressed in the posterior terminus, including the hindgut epithelium, as in wild type (Fig. 3B,E,H, Fig. S4A). Similarly, the posteriorly restricted expression of Hoxd13 was downregulated but remained detectable in a caudal domain of Wnt3a−/− embryos (Fig. S4B). These results demonstrate that posterior identity is maintained in Wnt3a null mutants at E8.5-9. In contrast, overexpression of Wnt3a in posterior progenitors led to the posterior extension of the Cdx spatial domains (Fig. 3C,F,I), consistent with the proposed role for Wnt3a in axial and hindgut extension.
Fig. 3.
Analysis of Cdx gene expression in Wnt3a loss- and gain-of-function embryos at hindgut extension stages. Cdx1 (A-C), Cdx2 (D-F) and Cdx4 (G-I) expression in control, Wnt3a−/− and T-cre; R26-Wnt3aGOF embryos at E8.5. Scale bars: 200 µm.
Bmp signaling plays important roles in the development of the posterior gut (Munera et al., 2017; Wills et al., 2008). To examine whether Wnt3a is regulating hindgut development by modulating Bmp signaling, we assessed the expression of phosphorylated Smad1/5 (pSmad1/5), which reports active Bmp signaling (Kitisin et al., 2007). Anti-pSmad1/5 antibodies strongly labeled the PSM and lateral plate mesoderm (LPM), as well as the ventrolateral hindgut endoderm of E8.5 control embryos (Fig. 4A,C). Notably, pSmad1/5 was not detected in the dorsal hindgut or in the PS epiblast. pSmad1/5 was maintained in the LPM of Wnt3a−/− embryos but was downregulated in the PSM region, concomitant with the transformation of PSM-to-neural fates (Yoshikawa et al., 1997), and was nearly absent from the hindgut (Fig. 4B,D). These results suggest that Wnt3a is required for the expression of a Bmp ligand or, alternatively, for the hindgut to be competent to respond to Bmp signals.
Fig. 4.
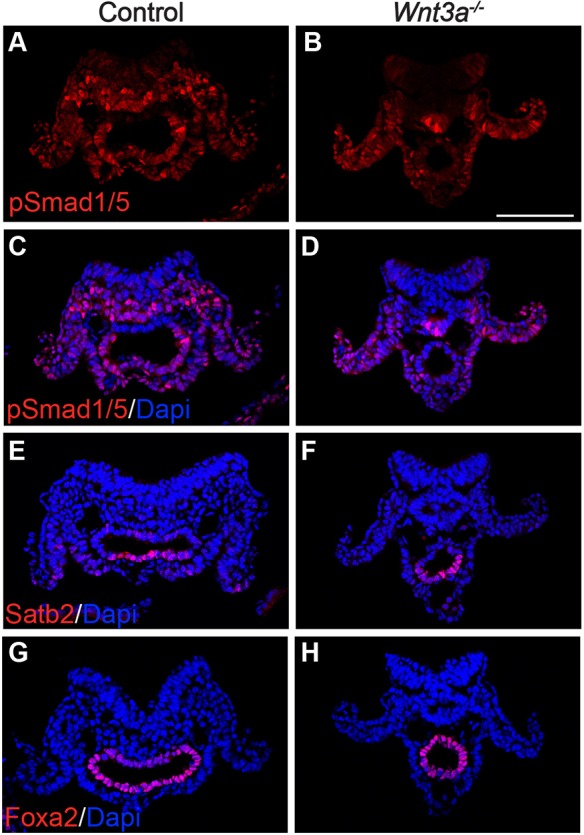
Analysis of Bmp signaling in Wnt3a−/− embryos at hindgut extension stages. Sections of E8.5 control (Wnt3a+/−) or Wnt3a−/− embryos stained using antibodies to phosphorylated Smad1/5 (pSmad1/5) (A,B), pSmad1/5/DAPI (C,D), Satb2/DAPI (E,F) or Foxa2/DAPI (G,H). Scale bar: 100 µm.
The homeodomain protein Satb2 is expressed in the hindgut endoderm and is a target of the Bmp pathway (Munera et al., 2017). Analysis of Satb2 expression in sections of the hindgut at E8.5 revealed that Satb2 is restricted to the ventral hindgut (Fig. 4E). Satb2 remained expressed in the Wnt3a−/− ventral hindgut, suggesting that Satb2 expression does not depend upon Wnt3a or on Bmp signaling (Fig. 4F). Although the Satb2+ population appeared similar between control and mutant embryos, the dorsal Satb2− population was reduced in the mutants. Analysis of the gut endoderm transcription factor Foxa2 showed that Foxa2 is expressed throughout the gut endoderm, but also displays a dorsal-ventral bias with high expression in ventrolateral nuclei and lower expression in the dorsal hindgut (Fig. 4G). Notably, this dorsal population appeared absent from the smaller Wnt3a−/− mutant hindgut, with predominantly high Foxa2-expressing cells populating the mutant hindgut (Fig. 4H). Together, these results highlight stark differences between the dorsal and ventral hindgut, and suggest that dorsal hindgut cells are maintained by Wnt3a.
Dorsal and ventral hindgut progenitors differentially contribute to the developing colon
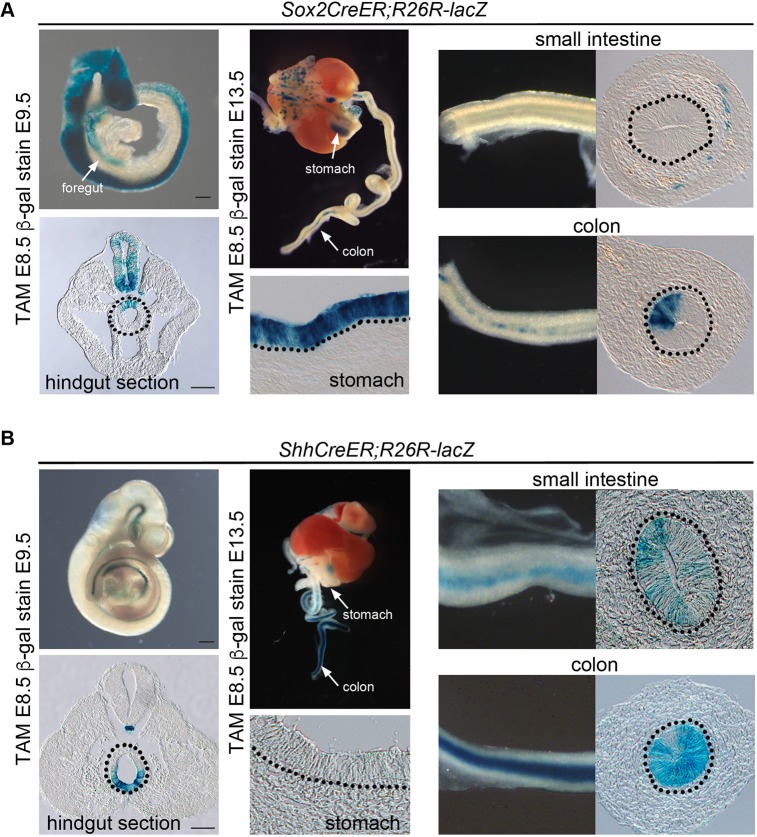
The dorsally restricted expression of Wnt3a, and the Wnt3a target genes and stem cell markers Sp5, Sp8, Cdx2, T/brachyury and Sox2 suggests that dorsal hindgut cells are a unique Wnt-responsive progenitor population. To understand their potential contribution to colon development, we attempted to perform a genetic lineage tracing analysis using a TAM-inducible Sox2-CreER driver, which has CreER knocked-in to the Sox2 locus (Arnold et al., 2011), crossed to the R26RlacZ reporter to label dorsal hindgut progenitors. TAM was administered with a single injection at E8.5 and embryos were examined 24 h later, revealing that dorsal hindgut progenitors (and neural tube) were labeled as expected (Fig. 5A, left). Given that a single pulse of TAM labels cells between 6 and 24 h post-injection in TAM-inducible CreER transgenics (Anderson et al., 2013; Nakamura et al., 2006), the βgal+ cell populations at stages beyond E9.5 should consist of descendants of cells recombined to express lacZ between E8.5 and 9.5. Assessment of Sox2-CreER;R26R-lacZ embryos at E13.5, following TAM injections at E8.5, detected patches of βgal+ cells in the epithelial layer of the colon (Fig. 5A, right). Counting of these βgal+ cells in sections of Sox2-CreER;R26R-lacZ embryos (n=3 embryos sectioned) revealed that 19% of the colon epithelium was derived from the dorsal hindgut. We detected many traced cells throughout the stomach epithelium that originated from the labeled foregut, demonstrating that our cell tracing was effective in these embryos (Fig. 5A). Thus, the dorsal hindgut contains a progenitor population that contributes to a minor component of the colon epithelium at E13.5.
Fig. 5.
Genetic lineage tracing of dorsal and ventrolateral hindgut populations at E8.5. TAM-inducible transgenics (Sox2-CreER and Shh-CreER) and the R26R-lacZ reporter were employed to trace the fate of hindgut progenitors. (A) TAM-induced labeling of Sox2-CreER;R26R-lacZ embryos at E8.5 marked the CNS, foregut and dorsal hindgut (dotted circle) progenitors at E9.5 (left column). Isolated GI tract dissected at E13.5 showed high contribution of β-gal+ cells to the stomach (middle column) but relatively few β-gal+ cells in the small and large intestine/colon (right column). Scale bar: 200 µm (top left), 100 µm (bottom left). (B) TAM-induced labeling of Shh-CreER;R26R-lacZ embryos at E8.5 marked the notochord and ventrolateral hindgut at E9.5 (left column) and the intestines at E13.5 with particularly high contribution to the large intestine/colon (middle and right columns). Scale bar: 200 µm (top left), 100 µm (bottom left).
We then traced the ventrolateral part of the hindgut using the Shh-CreER transgene in conjunction with R26R-lacZ (Fig. 5B, left). Labeling a cohort of cells with an injection of TAM at E8.5 and examination of their distribution at E9.5 revealed βgal+ cells restricted to the ventrolateral hindgut that were distributed in a pattern complementary to that observed with Sox2-CreER (Fig. 5A,B). Examination of Shh-CreER;R26R-lacZ tracings at E13.5 showed that the colon, cecum and distal small intestine were highly labeled, whereas the proximal small intestine showed a mosaic distribution of labeled cells (Fig. 5B, right). Quantitative analysis of three sectioned embryos showed that 84% of the colon epithelium was derived from Shh-CreER;R26R-lacZ traced cells. As the majority of the colon epithelium was populated by Shh-CreER-expressing ventrolateral hindgut cells, it suggests that the ventrolateral hindgut is the predominant source of colon intestinal epithelial progenitors.
The DV Wnt gradient is necessary for proper hindgut morphogenesis
Our demonstration of a DV gradient of Wnt/β-catenin signaling in the early hindgut, with little to no Wnt signaling ventrally, makes two predictions: (1) Wnt/β-catenin signaling is required in the dorsal, but not the ventral, hindgut; and (2) Wnt/β-catenin signaling must be prohibited in the ventral hindgut for proper hindgut development to proceed. To investigate the function of β-catenin specifically in the dorsal hindgut compartment, we attempted to remove β-catenin activity with Sox2-CreER at E8.5 (Fig. S5). Although brain phenotypes were evident at E9.5 and E13.5, as expected given the broad activity of Sox2CreER throughout the CNS (Fig. 5A), GI phenotypes were not observed. This is likely due to the mosaic expression of Sox2CreER-expressing cells in the dorsal hindgut (Fig. 5A). Notably, endogenous Sox2 is also expressed only within a subset of the dorsal hindgut domain (Fig. 1F). To test whether Wnt/β-catenin signaling is required in ventral hindgut progenitors, Shh-Cre was used to remove β-catenin activity. Examination of Shh-Cre;Ctnnb1flLOF/Δ embryos at E13.5 showed that the colon was present despite the abnormal posterior development (Fig. S6C,D). Lung agenesis was evident at this stage, as previously reported (Goss et al., 2009), indicating that the Shh-Cre driver was functional. Thus, β-catenin is not required in the ventral hindgut epithelium for hindgut extension, as predicted.
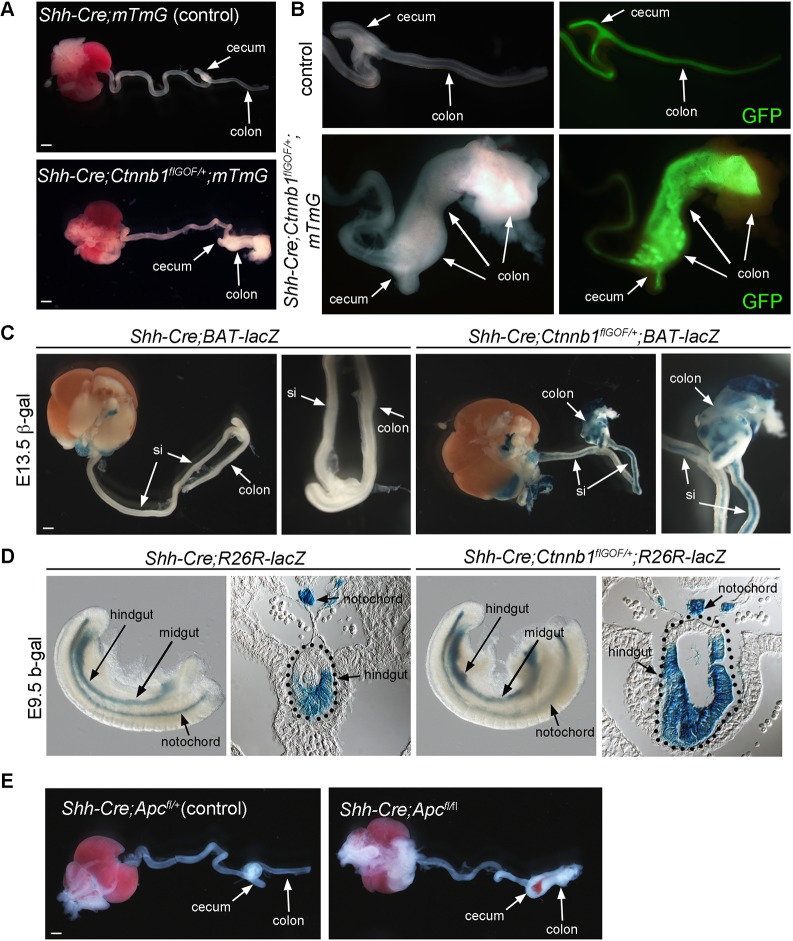
To test whether Wnt signaling must be suppressed in the ventral hindgut, we ectopically activated β-catenin in the ventral hindgut using Shh-Cre and Ctnnb1lox(ex3), a floxed β-catenin gain-of-function allele referred to here as Ctnnb1flGOF (Harada et al., 1999). Examination of the GI tracts dissected from E13.5 Shh-Cre;Ctnnb1flGOF/+ fetuses revealed a remarkable expansion of the colon epithelium and the appearance of large polyp-like nodules (Fig. 6A,B). Although this phenotype was only observed in the colon, Shh-Cre was functional throughout the anterior-posterior extent of the intestinal epithelium, as indicated by the GFP expression of the R26mT/mG Cre reporter strain at E13.5 (Fig. 6B) as well as βgal expression from the R26R-lacZ Cre reporter line at E8.5 and 9.5 (Fig. S6A,B; Garriock et al., 2015). To confirm that β-catenin activity was enhanced, we examined the expression of the β-catenin/Tcf reporter (BATlacZ) in the Shh-Cre;Ctnnb1flGOF/+ fetus (Fig. 6C). In E13.5 control fetuses, we observed little to no BATlacZ activity in the intestine, although activity was observed in the stomach and kidneys (Fig. 6C). Shh-Cre;Ctnnb1flGOF/+;BATlacZ fetuses showed abundant β-catenin/Tcf reporter activity throughout the intestinal tract, including the small intestine that did not undergo substantial enlargement (Fig. 6C), demonstrating that the β-catenin/Tcf pathway was indeed ectopically activated in anterior domains. We speculate that the selective growth of the colon was due to the involvement of additional posteriorly localized co-factors.
Fig. 6.
Effects of elevated Wnt signaling on colon development. (A) GI tracts dissected from E13.5 control and Shh-Cre;Ctnnb1flGOF;mTmG mutants. (B) Magnified view of cecum and colon region of the intestine imaged fluorescently to detect GFP, which reports the Shh-Cre;mT/mG-expressing intestinal epithelium. The large intestinal epithelium in Shh-Cre;Ctnnb1flGOF;mT/mG mutants is expanded when compared with the Shh-Cre;mT/mG control. (C) Isolated GI tract from E13.5 transgenic Wnt reporter line (BATlacZ) showing no β-gal staining in Shh-Cre;BATlacZ control intestine but staining throughout the intestinal tract in Shh-Cre;Ctnnb1flGOF;BATlacZ mutants. (D) E9.5 β-gal staining of Shh-Cre;R26R-lacZ embryos marking the endodermal tissues (hindgut and midgut) and notochord. The hindgut region is expanded in Shh-Cre;Ctnnb1flGOF;R26R-lacZ mutants. Sections through the hindgut region show that the expanded hindgut tissue occurred in the β-gal stained ventrolateral Shh-Cre;R26R-lacZ region. (E) GI tracts removed from E13.5 control Shh-Cre;Apcfl/+ embryos (left) and Shh-Cre;Apcfl/fl mutants (right). Scale bar: 1 mm. si, small intestine.
Our T-Cre;R26-Wnt3aGOF experiments demonstrating an early growth response in the hindgut to elevated Wnt3a signals (Fig. 2B) suggest that the enlarged colon observed in Shh-Cre;Ctnnb1flGOF fetuses may occur early in embryonic development. We therefore examined Shh-Cre;Ctnnb1flGOF;R26R-lacZ embryos at E9.5 and found an early expansion of the βgal labeled ventral hindgut, but not midgut, in these mutants compared to controls although overall embryonic morphology of the trunk tissue was similar between controls and mutants (Fig. 6D). Analysis of pH3+ nuclei in Shh-Cre;Ctnnb1flGOF mutants showed that cell proliferation was similar in the ventral midgut of Shh-Cre;Ctnnb1flGOF mutants compared to controls, but 2.6 times higher in the ventral hindgut (Fig. S7A-D).
To further demonstrate that the ventral hindgut is typically maintained in a state of low Wnt signaling, we conditionally deleted the tumor suppressor Apc (adenomatous polyposis coli) there. Apc functions in Wnt signaling to limit the levels of nuclear β-catenin (Clevers and Nusse, 2012) and is essential to terminate the Wnt signaling response after ligand-induced activation. Consequently, loss-of-function mutations in Apc lead to unregulated Wnt signaling and cancer (Clevers and Nusse, 2012; Nelson and Nathke, 2013). Conditionally deleting the Apc loss-of-function allele Apctm1Tyj (referred to here as Apcfl) in ventrolateral hindgut progenitors (Shh-Cre;Apcfl/fl) resulted in a large intestine hyperplasia phenotype similar to that observed in the Shh-Cre;Ctnnb1flGOF fetus (n=9) (Fig. 6E), suggesting that a precise level of Wnt signaling is crucial for proper development of the large intestine.
DISCUSSION
We have characterized the role of the Wnt3a/β-catenin signaling pathway in the development of the GI tract and have shown that it is essential for colon formation. Mutations that lead to the loss of several components in the pathway, including Wnt3a, β-catenin or Sp5 and Sp8, lead to colon agenesis. This colon phenotype appears to be similar to that described in Tcf1−/−;Tcf4−/− mutants, although fetal GI tracts were not analyzed directly (Gregorieff et al., 2004). Together, these results strongly support a role for a Wnt3a-β-catenin-Sp5/8-Tcf1/4 pathway in the development of the colon.
As the fetal colon develops from the embryonic hindgut, we analyzed Wnt3a−/− mutants at early hindgut stages (E8.5) and found that the hindgut forms but is reduced in size and fails to elongate by E9.5. In contrast, overexpression of Wnt3a in the posterior embryo leads to an expansion and posterior extension of the E8.5-9.5 hindgut (Chalamalasetty et al., 2016), suggesting that Wnt3a regulates colon formation by promoting hindgut extension. Unfortunately, the T-Cre;R26-Wnt3aGOF embryos do not turn and display highly abnormal morphology by E9.5, precluding an analysis of the consequences of Wnt3a overexpression on the fetal colon. Interestingly, a conditional, endoderm-specific knockout of β-catenin has demonstrated an early, cell-autonomous requirement for β-catenin signaling in the definitive endoderm leading to the complete absence of the hindgut (Engert et al., 2013). This suggests that two early stages of colon formation depend upon Wnt/β-catenin signaling, an early stage that presumably relies on Wnt3 secreted from posterior visceral endoderm to specify the definitive endoderm necessary for the formation of the hindgut pocket (Engert et al., 2013), followed by the caudal extension of the hindgut mediated by Wnt3a secreted by the dorsoposterior hindgut.
We have presented evidence for a novel dorsal-ventral gradient of Wnt signaling in the caudal embryo that crosses all three germ layers, extending into the dorsal hindgut during hindgut extension. A recent transcriptional profiling study of single gut endoderm cells confirmed that Wnt3a and associated Wnt/β-catenin pathway components are highly expressed in the hindgut but did not detect the dorsal expression bias (Nowotschin et al., 2019). Several lines of evidence argue that Wnt3a functions as an autocrine factor to regulate dorsal, but not ventral, hindgut progenitors. The expression of Wnt3a, Wnt3a/β-catenin target genes and proteins, as well as a transgenic reporter of β-catenin/Tcf activity are all restricted to the dorsal hindgut, and the expression of target genes there depends upon functional Wnt3a. Our demonstration that the conditional removal of Ctnnb1 from the dorsal and ventral hindgut using T-CreERT2 leads to colon agenesis, while the deletion of Ctnnb1 in the ventral gut with Shh-Cre does not, strongly suggests that Wnt3a/β-catenin signaling is specifically required in the dorsal hindgut for hindgut extension. Although this conclusion does not initially appear to be supported by our observation of normal colons in Sox2-CreER;Ctnnb1flLOF/Δ embryos, the absence of a phenotype may be readily explained by the relatively few dorsal hindgut cells that are targeted by Sox2-CreER. We suggest that β-catenin activity is removed from only a subset of dorsal hindgut cells. As T-CreERT2 is active in the PS mesoderm, in addition to the hindgut endoderm (Anderson et al., 2013; Garriock et al., 2015), we cannot rule out the possibility that the colon agenesis phenotype arises from a requirement for Ctnnb1 in the mesoderm. Future studies employing more efficient dorsal endoderm and mesoderm-specific Cre expressors will be required to address this issue.
The lineage-tracing studies showed that the ventral hindgut contributes the majority of the colon progenitors. Thus, the extending hindgut likely derives primarily from ventrally located cells, as illustrated by the ShhCreER;R26R-lacZ labeling that initially marks the ventral hindgut. As Wnt signaling appears to be required in the dorsal hindgut for hindgut extension, the data suggest that the dorsal hindgut is necessary for the growth of the ventral hindgut. We propose that the dorsal hindgut acts as a signaling center to maintain the ventral hindgut, and that Wnt3a may function dorsally to control the expression of another signaling molecule that maintains the ventral hindgut. The absence of pSmad1/5 expression in the Wnt3a−/− hindgut is consistent with the Bmp signaling pathway functioning downstream of Wnt3a.
The absence of Wnt target gene expression and β-catenin/Tcf reporter activity in the ventral hindgut suggests that Wnt signaling is suppressed ventrally. This is crucial for appropriate hindgut morphogenesis, as ectopic activation of the Wnt/β-catenin pathway in the ventral hindgut using Shh-Cre to activate β-catenin or inactivate Apc results in a massive hyperplasia of the fetal colon. The mechanisms underlying Wnt inhibition in the ventral gut are currently unknown; however, the Wnt antagonists Dkk1, Dkk2 and Dkk3 are good candidates, as they are expressed in the hindgut (Fig. S1B; Nowotschin et al., 2019). Parallels can be drawn between the responsiveness of the ventral hindgut to pathological growth stimuli, such as mis-regulated β-catenin signaling, and adenoma formation in the adult colon through activating mutations in β-catenin or inactivating mutations in Apc (Cancer Genome Atlas, 2012; Clevers and Nusse, 2012; Nelson and Nathke, 2013). Although we currently do not understand why the hindgut is particularly responsive to stabilized β-catenin, despite overexpression throughout the intestinal epithelium, the answer may lie in the observation that Tcf7l1/Tcf3, which functions as a transcriptional repressor of Wnt target genes, is expressed throughout the E8.75 gut, except in the hindgut (Fig. S1B).
It has become increasingly clear from live imaging and single cell transcriptome studies that the gut endoderm is composed of intermingled descendants from the epiblast-derived definitive endoderm and the visceral endoderm. These studies have shown that visceral endoderm cells are not evenly distributed along the body axes; they are enriched in the hindgut, particularly so in the dorsal hindgut, and are excluded from lateral regions that are largely populated by definitive endoderm (Kwon et al., 2008; Nowotschin et al., 2019). Although visceral endoderm cells do appear to contribute to ventral hindgut, we speculate that these different lineal origins for dorsal and lateral hindgut may contribute to their differential response to growth stimuli during intestine formation.
Embryos lacking the transcription factor Cdx2 display a malformed cecum and do not form colons (Gao et al., 2009). Cdx2 has also been proposed to be a Wnt target (Sherwood et al., 2011), suggesting that it could be an important effector of Wnt3a in the hindgut; however, high levels of Cdx2 RNA were maintained in Wnt3a−/− mutants, suggesting that the colon agenesis phenotype is not caused by loss of Cdx2 expression. Our results showing that Cdx1 and Cdx4 were downregulated confirm that they are Wnt3a target genes. The Cdx genes are known to function redundantly to regulate axial extension, including the extension of the hindgut (Savory et al., 2009; van Rooijen et al., 2012; Young et al., 2009), raising the possibility that Cdx1 and Cdx4 are transducing Wnt3a signals. However, the grafting of Cdx mutant cells to wild-type recipients has shown that there is no cell-autonomous requirement for Cdx genes in axial progenitors (Bialecka et al., 2010). Instead, Cdx genes are regulating these cells non-cell autonomously by functioning in a positive-feedback loop to maintain Wnt and Fgf pathway gene expression and thereby establish a signaling niche (Amin et al., 2016). Thus, Wnt3a is effectively functioning ‘downstream’ of Cdx genes. The continued posteriorly restricted expression of the Cdx genes in Wnt3a−/− embryos is consistent with this argument and further suggests that Wnt3a is not required to maintain posterior identity in the gut.
The results of our loss- and gain-of-function approaches show that Wnt3a controls colon formation by regulating embryonic hindgut extension; however, the precise mechanisms remain unclear. Analysis of mitotic cells suggests that the hindgut terminus is a proliferative zone. Despite the demonstration of a dorsal bias in caudal Wnt3a gene expression in the hindgut, no differences in the mitotic indices were detected between the dorsal and ventral hindgut. Nevertheless, fewer mitotic cells were present in the Wnt3a−/− hindgut, and ectopic activation of β-catenin in the ventrolateral hindgut elevated the mitotic index and grossly expanded the hindgut and colon, illustrating the importance of restricting Wnt signaling to the dorsal region, and suggesting that Wnt3a functions, at least in part, as a mitogen to promote hindgut expansion. Considering the role that Wnt3a plays in the maintenance of neuromesodermal progenitors, which supply cells to the caudally extending neural tube and presomitic mesoderm of the trunk and tail (Garriock et al., 2015; Gouti et al., 2017; Koch et al., 2017; Wymeersch et al., 2019), it is tempting to speculate that Wnt3a is also maintaining hindgut progenitors in the hindgut terminus. Future studies will address the origins of the hindgut progenitors and explore the mechanisms through which Wnt3a coordinates the posterior extension of multiple tissue types.
MATERIALS AND METHODS
Embryos and transgenic strains
Wild-type embryos for whole-mount in situ hybridization were dissected from NIH-Swiss female mice in PBS prior to fixation. The BATlacZ transgenic mouse was used to assess Wnt/β-catenin activity in vivo (Nakaya et al., 2005). To modulate Wnt/β-catenin signaling, we used the following strains: a non-conditional Wnt3atm1Amc null allele, Ctnnb1tm2Kem (a conditional loss-of-function allele of β-catenin), Ctnnb1lox(ex3) (a conditional gain-of-function allele that results in the stabilization of β-catenin), Apctm1Tyj (a conditional Apc loss-of-function allele obtained from the Jackson Laboratory) and a Cre-activatable Wnt3aGOF strain targeted into the Rosa26 locus (Brault et al., 2001; Chalamalasetty et al., 2016; Harada et al., 1999; Takada et al., 1994). To control the excision of conditional alleles, we used T-Cre, T-CreERT2 (Perantoni et al., 2005; Anderson et al., 2013), Sox2-CreER (Jackson Laboratory; Arnold et al., 2011), Shh-CreER and Shh-Cre (Arnold et al., 2011; Harfe et al., 2004; Perantoni et al., 2005). Crosses were generally established as follows: ♂CreER/+;Ctnnb1Δ/+×♀Ctnnb1fl/fl. Control embryos have the following genotypes: CreER/+;Ctnnb1fl/+, +/+;Ctnnb1fl/+, +/+;Ctnnb1fl/Δ. For lineage-tracing experiments, we used the mouse strains Gt(ROSA)26Sortm1SorR26R−LacZ (R26R-lacZ) for color detection (Soriano, 1999) and Gt(ROSA)26Sortm4(ACTB−tdTomato,−EGFP)Luo (mT/mG) for fluorescent detection (Muzumdar et al., 2007), both acquired from the Jackson Laboratory. This study was carried out in compliance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (Frederick National Lab Animal Care and Use Committee Proposal #18-407). Rodents were euthanized by CO2 inhalation in accordance with the most recent AVMA Guidelines on Euthanasia.
Histology and immunohistochemistry
Embryos or dissected tissues were fixed in a 4% paraformaldehyde solution in PBS for 1 h at 4°C and processed for paraffin or frozen sections. For paraffin embedding and sectioning, embryos were dehydrated in ethanol, cleared in citrisolv (Fisher) twice for 20 min, embedded in three 20 min changes of molten paraffin wax (Fisher) and then sectioned to 8 µm on charged glass slides (Superfrost). For detection of phospho-Histone H3 in paraffin wax sections, retrieval of antigenicity was achieved with a 20 min boil in 10 mM sodium citrate buffer (pH 6.0). Sections were permeabilized in 0.2% Triton-X100 in PBS for 20 min then washed three times in PBS plus 0.1% Tween20 (PBT) prior to incubating with the phospho-Histone H3 antibody (Upstate, 06-570, 1:100) in 0.2% Roche blocking buffer in PBT overnight. Sections were washed three times in PBT and the primary antibody was detected using Alexa Fluor fluorescent secondary antibodies (Invitrogen, 1:400). Slides were counterstained with 1 nM DAPI (Molecular Probes) and mounted using aqua poly/mount (Polysciences). Embryos and tissues for frozen section were incubated in 20% sucrose in PBS overnight at 4°C, embedded in OCT and sectioned at 10 µm. Antibodies used were pSmad1/5 Ser463/465 (41D10) (Cell Signaling, rabbit mAb, 9516, 1:200), Satb2 [Cell Marque (Millipore Sigma), EP281, 1:100], Cdx2 (Biogenex, AM392, 1:2), FoxA2 (D56D6) (Cell Signaling, rabbit mAb 8186, 1:100), Tbx6 (R&D Systems, AF4744, 1:100), Sox2 (R&D Systems, mab2018, 1:100), T/brachyury (R&D Systems, AF2085, 1:100) and Sp8 (Santa Cruz, #sc-104661, 1:100). Sectioned material was imaged on either a Leica Sp8 confocal microscope or Zeiss Axioplan II microscope. For generation of the mitotic index in Fig. 2C, images were imported into Imaris software (Oxford Instruments) to count the number of DAPI- and pH3-positive nuclei in control (n=15) and Wnt3a−/− (n=8) hindguts.
Lineage tracings and analysis
For lineage tracing, the R26R-lacZ was used to permanently trace Sox2-CreER and Shh-CreER descendants. Tamoxifen (TAM) was administered intraperitoneally as previously described (Anderson et al., 2013; Soriano, 1999). Total TAM dose per pregnant female mouse were 6 mg for Sox2-CreER and 2 mg for Shh-CreER matings. For determination of lineage contribution to the colon, whole-mount β-gal stained isolated colon tissue was paraffin sectioned as described above except that tissues were rehydrated and stained with DAPI to mark cell nuclei. From a minimum of three serial sections the number of DAPI stained nuclei was compared with β-gal stained cells to determine the average percentage of cells traced over three separate experiments.
Detection of gene products
Whole-mount β-gal staining was performed on whole embryos or isolated gut organs as previously described (Whiting et al., 1991). For determination of cell proliferation in Sox2CreER or Shh-CreER lineage-traced populations, phospho-Histone H3 antibody detection was performed on paraffin sections of whole-mount β-gal staining as described above. Colorimetric whole-mount in situ hybridization (WISH) was performed as previously described (Biris et al., 2007) with digoxigenin-UTP antisense RNA probes. After staining, embryos or isolated tissues were re-fixed in 4% PFA in PBS and photographed and/or prepared for paraffin sectioning. Whole embryos and/or tissues were photographed in PBT using a Leica MZFllII microscope, AxioCam digital camera and Axiovision software (Zeiss). For fluorescence in situ hybridization chain reaction (HCR), v3.0 HCR probes were ordered from Molecular Instruments (Choi et al., 2018). Embryos were fixed in 4% PFA, digested in Proteinase K and processed as described for colorimetric WISH (Biris et al., 2007). Embryos were incubated in 1 ml probe hybridization buffer and processed according to manufacturer's instructions. For spinning disk confocal microscopy, embryos were embedded in 1% agarose on glass bottomed culture dishes (MatTEK) and cleared overnight with Ce3D++ (V. Magidson, unpublished), a modified Ce3D solution (Li et al., 2017) designed to match the refractive index of microscopy oil. To make Ce3D++, we added 20.83 g of Nycodenz (Alere Technologies ) to a 50 ml falcon tube, pipetted 10.5 g of 40% v/v N-methylacetamide solution and 22.5 mg of Triton-x-100 (0.1% v/v final concentration), and gently mixed on a rotator overnight at room temperature. Embryos were imaged using a 20× multi-immersion objective on a Leica Sp8 confocal microscopy equipped with a Yokogawa CSU-W1 spinning disk system and a Borealis illumination system (Andor). Optical sections (2 µm) of whole embryos were acquired and images processed using Metamorph and Fiji software.
Supplementary Material
Acknowledgements
We thank Dr Alan Perantoni for providing materials and support, Dr Nikolaos Mandalos for assistance with cryo sections, Ms Audrey Badjouen-Dzouabet for assistance with cell counts and Ms Ruth Wolfe for excellent assistance with mouse colony management. We thank Brian Harfe for providing the Shh-Cre and Shh-CreER mouse lines and Dr Valentin Magidson (NCI Optical Microscopy Laboratory) for assistance with spinning disk confocal microscopy.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: R.J.G., T.P.Y.; Methodology: R.J.G., T.P.Y.; Validation: R.B.C.; Formal analysis: R.J.G.; Investigation: R.J.G., R.B.C., M.W.K., A.K.; Resources: J.Z., M.W.K., A.K., S.M.; Data curation: R.J.G., R.B.C.; Writing - original draft: R.J.G., T.P.Y.; Writing - review & editing: S.M., T.P.Y.; Visualization: R.B.C., T.P.Y.; Supervision: T.P.Y.; Project administration: T.P.Y.; Funding acquisition: T.P.Y.
Funding
This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.185108.supplemental
Peer review history
The peer review history is available online at https://dev.biologists.org/lookup/doi/10.1242/dev.185108.reviewer-comments.pdf
References
- Amin S., Neijts R., Simminifig.s S., van Rooijen C., Tan S. C., Kester L., van Oudenaarden A., Creyghton M. P. and Deschamps J. (2016). Cdx and T brachyury co-activate growth signaling in the embryonic axial progenitor niche. Cell Rep. 17, 3165-3177. 10.1016/j.celrep.2016.11.069 [DOI] [PubMed] [Google Scholar]
- Anderson M. J., Naiche L. A., Wilson C. P., Elder C., Swing D. A. and Lewandoski M. (2013). TCreERT2, a transgenic mouse line for temporal control of Cre-mediated recombination in lineages emerging from the primitive streak or tail bud. PLoS ONE 8, e62479 10.1371/journal.pone.0062479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K., Sarkar A., Yram M. A., Polo J. M., Bronson R., Sengupta S., Seandel M., Geijsen N. and Hochedlinger K. (2011). Sox2+ adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell 9, 317-329. 10.1016/j.stem.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bénazéraf B., Francois P., Baker R. E., Denans N., Little C. D. and Pourquié O. (2010). A random cell motility gradient downstream of FGF controls elongation of an amniote embryo. Nature 466, 248-252. 10.1038/nature09151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialecka M., Wilson V. and Deschamps J. (2010). Cdx mutant axial progenitor cells are rescued by grafting to a wild type environment. Dev. Biol. 347, 228-234. 10.1016/j.ydbio.2010.08.032 [DOI] [PubMed] [Google Scholar]
- Biris K. K., Dunty W. C. Jr and Yamaguchi T. P. (2007). Mouse Ripply2 is downstream of Wnt3a and is dynamically expressed during somitogenesis. Dev. Dyn. 236, 3167-3172. 10.1002/dvdy.21342 [DOI] [PubMed] [Google Scholar]
- Brault V., Moore R., Kutsch S., Ishibashi M., Rowitch D. H., McMahon A. P., Sommer L., Boussadia O. and Kemler R. (2001). Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development 128, 1253-1264. [DOI] [PubMed] [Google Scholar]
- Cadigan K. M. and Waterman M. L. (2012). TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb. Perspect Biol. 4, a007906 10.1101/cshperspect.a007906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. (2012). Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330-337. 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalamalasetty R. B., Ajima R., Garriock R. J., Kennedy M. W., Tessarollo L. and Yamaguchi T. P. (2016). A new gain-of-function mouse line to study the role of Wnt3a in development and disease. Genesis 54, 497-502. 10.1002/dvg.22959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman D. L., Cooper-Morgan A., Harrelson Z. and Papaioannou V. E. (2003). Critical role for Tbx6 in mesoderm specification in the mouse embryo. Mech. Dev. 120, 837-847. 10.1016/S0925-4773(03)00066-2 [DOI] [PubMed] [Google Scholar]
- Chin A. M., Hill D. R., Aurora M. and Spence J. R. (2017). Morphogenesis and maturation of the embryonic and postnatal intestine. Semin. Cell Dev. Biol. 66, 81-93. 10.1016/j.semcdb.2017.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H. M. T., Schwarzkopf M., Fornace M. E., Acharya A., Artavanis G., Stegmaier J., Cunha A. and Pierce N. A. (2018). Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development 145, dev165753 10.1101/285213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. and Batlle E. (2013). SnapShot: the intestinal crypt. Cell 152, 1198-1198.e1192. 10.1016/j.cell.2013.02.030 [DOI] [PubMed] [Google Scholar]
- Clevers H. and Nusse R. (2012). Wnt/beta-catenin signaling and disease. Cell 149, 1192-1205. 10.1016/j.cell.2012.05.012 [DOI] [PubMed] [Google Scholar]
- Dubrulle J. and Pourquié O. (2004). fgf8 mRNA decay establishes a gradient that couples axial elongation to patterning in the vertebrate embryo. Nature 427, 419-422. 10.1038/nature02216 [DOI] [PubMed] [Google Scholar]
- Dunty W. C. Jr, Kennedy M. W., Chalamalasetty R. B., Campbell K. and Yamaguchi T. P. (2014). Transcriptional profiling of Wnt3a mutants identifies Sp transcription factors as essential effectors of the Wnt/β-catenin pathway in neuromesodermal stem cells. PLoS ONE 9, e87018 10.1371/journal.pone.0087018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edri S., Hayward P., Baillie-Johnson P., Steventon B. J. and Martinez Arias A. (2019). An epiblast stem cell-derived multipotent progenitor population for axial extension. Development 146, dev168187 10.1242/dev.168187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engert S., Burtscher I., Liao W. P., Dulev S., Schotta G. and Lickert H. (2013). Wnt/beta-catenin signalling regulates Sox17 expression and is essential for organizer and endoderm formation in the mouse. Development 140, 3128-3138. 10.1242/dev.088765 [DOI] [PubMed] [Google Scholar]
- Gao N., White P. and Kaestner K. H. (2009). Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev. Cell 16, 588-599. 10.1016/j.devcel.2009.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriock R. J., Chalamalasetty R. B., Kennedy M. W., Canizales L. C., Lewandoski M. and Yamaguchi T. P. (2015). Lineage tracing of neuromesodermal progenitors reveals novel Wnt-dependent roles in trunk progenitor cell maintenance and differentiation. Development 142, 1628-1638. 10.1242/dev.111922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt S. J., Drage D. and Cockley A. (2003). Vertebrate caudal gene expression gradients investigated by use of chick cdx-A/lacZ and mouse cdx-1/lacZ reporters in transgenic mouse embryos: evidence for an intron enhancer. Mech. Dev. 120, 573-586. 10.1016/S0925-4773(03)00023-6 [DOI] [PubMed] [Google Scholar]
- Gehart H. and Clevers H. (2019). Tales from the crypt: new insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol. 16, 19-34. 10.1038/s41575-018-0081-y [DOI] [PubMed] [Google Scholar]
- Goss A. M., Tian Y., Tsukiyama T., Cohen E. D., Zhou D., Lu M. M., Yamaguchi T. P. and Morrisey E. E. (2009). Wnt2/2b and β-catenin signaling are necessary and sufficient to specify lung progenitors in the foregut. Dev. Cell 17, 290-298. 10.1016/j.devcel.2009.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouti M., Tsakiridis A., Wymeersch F. J., Huang Y., Kleinjung J., Wilson V. and Briscoe J. (2014). In vitro generation of neuromesodermal progenitors reveals distinct roles for wnt signalling in the specification of spinal cord and paraxial mesoderm identity. PLoS Biol. 12, e1001937 10.1371/journal.pbio.1001937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouti M., Delile J., Stamataki D., Wymeersch F. J., Huang Y., Kleinjung J., Wilson V. and Briscoe J. (2017). A gene regulatory network balances neural and mesoderm specification during vertebrate trunk development. Dev. Cell 41, 243-261.e247. 10.1016/j.devcel.2017.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorieff A., Grosschedl R. and Clevers H. (2004). Hindgut defects and transformation of the gastro-intestinal tract in Tcf4(−/−)/Tcf1(−/−) embryos. EMBO J. 23, 1825-1833. 10.1038/sj.emboj.7600191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans F. and Dimitrov S. (2001). Histone H3 phosphorylation and cell division. Oncogene 20, 3021-3027. 10.1038/sj.onc.1204326 [DOI] [PubMed] [Google Scholar]
- Harada N., Tamai Y., Ishikawa T., Sauer B., Takaku K., Oshima M. and Taketo M. M. (1999). Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. EMBO J. 18, 5931-5942. 10.1093/emboj/18.21.5931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe B. D., Scherz P. J., Nissim S., Tian H., McMahon A. P. and Tabin C. J. (2004). Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell 118, 517-528. 10.1016/j.cell.2004.07.024 [DOI] [PubMed] [Google Scholar]
- Ikeya M. and Takada S. (2001). Wnt-3a is required for somite specification along the anteroposterior axis of the mouse embryo and for regulation of cdx-1 expression. Mech. Dev. 103, 27-33. 10.1016/S0925-4773(01)00338-0 [DOI] [PubMed] [Google Scholar]
- Kennedy M. W., Chalamalasetty R. B., Thomas S., Garriock R. J., Jailwala P. and Yamaguchi T. P. (2016). Sp5 and Sp8 recruit β-catenin and Tcf1-Lef1 to select enhancers to activate Wnt target gene transcription. Proc. Natl. Acad. Sci. USA 113, 3545-3550. 10.1073/pnas.1519994113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitisin K., Saha T., Blake T., Golestaneh N., Deng M., Kim C., Tang Y., Shetty K., Mishra B. and Mishra L. (2007). Tgf-Beta signaling in development. Sci. STKE 2007, cm1 10.1126/stke.3992007cm1 [DOI] [PubMed] [Google Scholar]
- Koch F., Scholze M., Wittler L., Schifferl D., Sudheer S., Grote P., Timmermann B., Macura K. and Herrmann B. G. (2017). Antagonistic activities of Sox2 and brachyury control the fate choice of neuro-mesodermal progenitors. Dev. Cell 42, 514-526.e517. 10.1016/j.devcel.2017.07.021 [DOI] [PubMed] [Google Scholar]
- Kretzschmar K. and Clevers H. (2017). Wnt/β-catenin signaling in adult mammalian epithelial stem cells. Dev. Biol. 428, 273-282. 10.1016/j.ydbio.2017.05.015 [DOI] [PubMed] [Google Scholar]
- Kwon G. S., Viotti M. and Hadjantonakis A. K. (2008). The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extraembryonic lineages. Dev. Cell 15, 509-520. 10.1016/j.devcel.2008.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Germain R. N. and Gerner M. Y. (2017). Multiplex, quantitative cellular analysis in large tissue volumes with clearing-enhanced 3D microscopy (Ce3D). Proc. Natl. Acad. Sci. USA 114, E7321-E7330. 10.1073/pnas.1708981114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin B. L. and Kimelman D. (2012). Canonical Wnt signaling dynamically controls multiple stem cell fate decisions during vertebrate body formation. Dev. Cell 22, 223-232. 10.1016/j.devcel.2011.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath P. S. and Wells J. M. (2015). SnapShot: GI tract development. Cell 161, 176-176.e171. 10.1016/j.cell.2015.03.014 [DOI] [PubMed] [Google Scholar]
- Munera J. O., Sundaram N., Rankin S. A., Hill D., Watson C., Mahe M., Vallance J. E., Shroyer N. F., Sinagoga K. L., Zarzoso-Lacoste A. et al. (2017). Differentiation of human pluripotent stem cells into colonic organoids via transient activation of BMP signaling. Cell Stem Cell 21, 51-64.e56. 10.1016/j.stem.2017.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar M. D., Tasic B., Miyamichi K., Li L. and Luo L. (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593-605. 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Nakamura E., Nguyen M. T. and Mackem S. (2006). Kinetics of tamoxifen-regulated Cre activity in mice using a cartilage-specific CreER(T) to assay temporal activity windows along the proximodistal limb skeleton. Dev. Dyn. 235, 2603-2612. 10.1002/dvdy.20892 [DOI] [PubMed] [Google Scholar]
- Nakaya M.-A., Biris K., Tsukiyama T., Jaime S., Rawls J. A. and Yamaguchi T. P. (2005). Wnt3a links left-right determination with segmentation and anteroposterior axis elongation. Development 132, 5425-5436. 10.1242/dev.02149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson S. and Nathke I. S. (2013). Interactions and functions of the adenomatous polyposis coli (APC) protein at a glance. J. Cell Sci. 126, 873-877. 10.1242/jcs.100479 [DOI] [PubMed] [Google Scholar]
- Nerurkar N. L., Lee C., Mahadevan L. and Tabin C. J. (2019). Molecular control of macroscopic forces drives formation of the vertebrate hindgut. Nature 565, 480-484. 10.1038/s41586-018-0865-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotschin S., Setty M., Kuo Y.-Y., Liu V., Garg V., Sharma R., Simon C. S., Saiz N., Gardner R., Boutet S. C. et al. (2019). The emergent landscape of the mouse gut endoderm at single-cell resolution. Nature 569, 361-367. 10.1038/s41586-019-1127-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse R. and Clevers H. (2017). Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985-999. 10.1016/j.cell.2017.05.016 [DOI] [PubMed] [Google Scholar]
- Perantoni A. O., Timofeeva O., Naillat F., Richman C., Pajni-Underwood S., Wilson C., Vainio S., Dove L. F. and Lewandoski M. (2005). Inactivation of FGF8 in early mesoderm reveals an essential role in kidney development. Development 132, 3859-3871. 10.1242/dev.01945 [DOI] [PubMed] [Google Scholar]
- Perochon J., Carroll L. R. and Cordero J. B. (2018). Wnt signalling in intestinal stem cells: lessons from mice and flies. Genes (Basel) 9, E138 10.3390/genes9030138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinos P., Joseph S., Oh K., Meyer B. I., Gruss P. and Lohnes D. (2001). Multiple pathways governing Cdx1 expression during murine development. Dev. Biol. 239, 257-269. 10.1006/dbio.2001.0446 [DOI] [PubMed] [Google Scholar]
- Rankin S. A., McCracken K. W., Luedeke D. M., Han L., Wells J. M., Shannon J. M. and Zorn A. M. (2018). Timing is everything: reiterative Wnt, BMP and RA signaling regulate developmental competence during endoderm organogenesis. Dev. Biol. 434, 121-132. 10.1016/j.ydbio.2017.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo Albors A., Halley P. A. and Storey K. G. (2018). Lineage tracing of axial progenitors using Nkx1-2CreER(T2) mice defines their trunk and tail contributions. Development 145 10.1242/dev.164319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savory J. G., Pilon N., Grainger S., Sylvestre J.-R., Béland M., Houle M., Oh K. and Lohnes D. (2009). Cdx1 and Cdx2 are functionally equivalent in vertebral patterning. Dev. Biol. 330, 114-122. 10.1016/j.ydbio.2009.03.016 [DOI] [PubMed] [Google Scholar]
- Sherwood R. I., Maehr R., Mazzoni E. O. and Melton D. A. (2011). Wnt signaling specifies and patterns intestinal endoderm. Mech. Dev. 128, 387-400. 10.1016/j.mod.2011.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F. and Bäckhed F. (2013). The gut microbiota--masters of host development and physiology. Nat. Rev. Microbiol. 11, 227-238. 10.1038/nrmicro2974 [DOI] [PubMed] [Google Scholar]
- Soriano P. (1999). Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70-71. 10.1038/5007 [DOI] [PubMed] [Google Scholar]
- Spence J. R., Lauf R. and Shroyer N. F. (2011). Vertebrate intestinal endoderm development. Dev. Dyn. 240, 501-520. 10.1002/dvdy.22540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens M. L., Chaturvedi P., Rankin S. A., Macdonald M., Jagannathan S., Yukawa M., Barski A. and Zorn A. M. (2017). Genomic integration of Wnt/β-catenin and BMP/Smad1 signaling coordinates foregut and hindgut transcriptional programs. Development 144, 1283-1295. 10.1242/dev.145789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S., Stark K. L., Shea M. J., Vassileva G., McMahon J. A. and McMahon A. P. (1994). Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes Dev. 8, 174-189. 10.1101/gad.8.2.174 [DOI] [PubMed] [Google Scholar]
- Takahashi M., Nakamura Y., Obama K. and Furukawa Y. (2005). Identification of SP5 as a downstream gene of the beta-catenin/Tcf pathway and its enhanced expression in human colon cancer. Int. J. Oncol. 27, 1483-1487. [PubMed] [Google Scholar]
- Thorpe C. J., Weidinger G. and Moon R. T. (2005). Wnt/beta-catenin regulation of the Sp1-related transcription factor sp5l promotes tail development in zebrafish. Development 132, 1763-1772. 10.1242/dev.01733 [DOI] [PubMed] [Google Scholar]
- Tsakiridis A., Huang Y., Blin G., Skylaki S., Wymeersch F., Osorno R., Economou C., Karagianni E., Zhao S., Lowell S. et al. (2014). Distinct Wnt-driven primitive streak-like populations reflect in vivo lineage precursors. Development 141, 1209-1221. 10.1242/dev.101014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Ven C., Bialecka M., Neijts R., Young T., Rowland J. E., Stringer E. J., Van Rooijen C., Meijlink F., Novoa A., Freund J. N. et al. (2011). Concerted involvement of Cdx/Hox genes and Wnt signaling in morphogenesis of the caudal neural tube and cloacal derivatives from the posterior growth zone. Development 138, 3451-3462. 10.1242/dev.066118 [DOI] [PubMed] [Google Scholar]
- van Es J. H., Haegebarth A., Kujala P., Itzkovitz S., Koo B.-K., Boj S. F., Korving J., van den Born M., van Oudenaarden A., Robine S. et al. (2012). A critical role for the Wnt effector Tcf4 in adult intestinal homeostatic self-renewal. Mol. Cell. Biol. 32, 1918-1927. 10.1128/MCB.06288-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooijen C., Simmini S., Bialecka M., Neijts R., van de Ven C., Beck F. and Deschamps J. (2012). Evolutionarily conserved requirement of Cdx for post-occipital tissue emergence. Development 139, 2576-2583. 10.1242/dev.079848 [DOI] [PubMed] [Google Scholar]
- Weidinger G., Thorpe C. J., Wuennenberg-Stapleton K., Ngai J. and Moon R. T. (2005). The Sp1-related transcription factors sp5 and sp5-like act downstream of Wnt/β-catenin signaling in mesoderm and neuroectoderm patterning. Curr. Biol. 15, 489-500. 10.1016/j.cub.2005.01.041 [DOI] [PubMed] [Google Scholar]
- Wells J. M. and Spence J. R. (2014). How to make an intestine. Development 141, 752-760. 10.1242/dev.097386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiting J., Marshall H., Cook M., Krumlauf R., Rigby P. W., Stott D. and Allemann R. K. (1991). Multiple spatially specific enhancers are required to reconstruct the pattern of Hox-2.6 gene expression. Genes Dev. 5, 2048-2059. 10.1101/gad.5.11.2048 [DOI] [PubMed] [Google Scholar]
- Wills A., Dickinson K., Khokha M. and Baker J. C. (2008). Bmp signaling is necessary and sufficient for ventrolateral endoderm specification in Xenopus. Dev. Dyn. 237, 2177-2186. 10.1002/dvdy.21631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson V., Olivera-Martinez I. and Storey K. G. (2009). Stem cells, signals and vertebrate body axis extension. Development 136, 1591-1604. 10.1242/dev.021246 [DOI] [PubMed] [Google Scholar]
- Wymeersch F. J., Huang Y., Blin G., Cambray N., Wilkie R., Wong F. C. and Wilson V. (2016). Position-dependent plasticity of distinct progenitor types in the primitive streak. Elife 5, e10042 10.7554/eLife.10042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wymeersch F. J., Skylaki S., Huang Y., Watson J. A., Economou C., Marek-Johnston C., Tomlinson S. R. and Wilson V. (2019). Transcriptionally dynamic progenitor populations organised around a stable niche drive axial patterning. Development 146, dev168161 10.1242/dev.168161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T. P., Bradley A., McMahon A. P. and Jones S. (1999a). A Wnt5a pathway underlies outgrowth of multiple structures in the vertebrate embryo. Development 126, 1211-1223. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T. P., Takada S., Yoshikawa Y., Wu N. and McMahon A. P. (1999b). T (Brachyury) is a direct target of Wnt3a during paraxial mesoderm specification. Genes Dev. 13, 3185-3190. 10.1101/gad.13.24.3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa Y., Fujimori T., McMahon A. P. and Takada S. (1997). Evidence that absence of Wnt-3a signaling promotes neuralization instead of paraxial mesoderm development in the mouse. Dev. Biol. 183, 234-242. 10.1006/dbio.1997.8502 [DOI] [PubMed] [Google Scholar]
- Young T., Rowland J. E., van de Ven C., Bialecka M., Novoa A., Carapuco M., van Nes J., de Graaff W., Duluc I., Freund J.-N. et al. (2009). Cdx and Hox genes differentially regulate posterior axial growth in mammalian embryos. Dev. Cell 17, 516-526. 10.1016/j.devcel.2009.08.010 [DOI] [PubMed] [Google Scholar]
- Zorn A. M. and Wells J. M. (2009). Vertebrate endoderm development and organ formation. Annu. Rev. Cell Dev. Biol. 25, 221-251. 10.1146/annurev.cellbio.042308.113344 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.