Abstract
We analyzed the genomic architecture of neuroanatomical diversity using magnetic resonance imaging and single nucleotide polymorphism (SNP) data from >26 000 individuals from the UK Biobank project and 5 other projects that had previously participated in the ENIGMA (Enhancing NeuroImaging Genetics through Meta-Analysis) consortium. Our results confirm the polygenic architecture of neuroanatomical diversity, with SNPs capturing from 40% to 54% of regional brain volume variance. Chromosomal length correlated with the amount of phenotypic variance captured, r ~ 0.64 on average, suggesting that at a global scale causal variants are homogeneously distributed across the genome. At a local scale, SNPs within genes (~51%) captured ~1.5 times more genetic variance than the rest, and SNPs with low minor allele frequency (MAF) captured less variance than the rest: the 40% of SNPs with MAF <5% captured <one fourth of the genetic variance. We also observed extensive pleiotropy across regions, with an average genetic correlation of rG ~ 0.45. Genetic correlations were similar to phenotypic and environmental correlations; however, genetic correlations were often larger than phenotypic correlations for the left/right volumes of the same region. The heritability of differences in left/right volumes was generally not statistically significant, suggesting an important influence of environmental causes in the variability of brain asymmetry. Our code is available athttps://github.com/neuroanatomy/genomic-architecture.
Keywords: genetics, heritability, neuroimaging, polygenic architecture, subcortical structures
Introduction
Imaging genetics studies have traditionally emphasized the role of candidate genes and specific loci. The underlying hypothesis is that phenotypic diversity results from the action of a reduced number of genes, close to the Mendelian paradigm where a phenotype is determined by a single locus. However, the amount of neuroanatomical variance captured by candidate genes or genome-wide significant loci has remained extremely small. Genome-wide association studies (GWAS) aiming at identifying associated variants through international collaborative efforts such as Enhancing NeuroImaging Genetics through Meta-Analysis (ENIGMA) and Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) have revealed only few statistically significant associated loci (Stein et al. 2012; Hibar et al. 2015; Satizabal et al. 2017), capturing <1% of phenotypic variance. This is in contrast with the large heritability of neuroanatomical diversity estimated by twin and extended pedigree studies (Blokland et al. 2012; Wen et al. 2016), which show that about 80% of neuroanatomical variability is captured by genetic factors.
The genetic architecture of neuroanatomical diversity could result, alternatively, from the aggregated effect of thousands of different loci spread across the genome, a paradigm closer to the infinitesimal model proposed by Fisher (1918). Thanks to the development of methods to estimate heritability from whole-genome genotyping data (reviewed by Yang et al. 2017), several groups have estimated that genotyped variants, taken together, were able to capture up to 55% of the phenotypic variance, retrieving about two thirds of the heritability estimated by family studies (Toro et al. 2015; Ge et al. 2015, 2016; Elliott et al. 2018; Zhao et al. 2018). These results are compatible with the hypothesis of a highly polygenic architecture, where phenotypes are influenced by large numbers of loci of effect sizes too small to reach genome-wide significance (usually P < 5 × 10−8). While information about the function of a few candidate genes can have a strong explanatory power in the case of a few causal loci, a polygenic approach calls for alternative, system-wide, sources of biological insight.
Our aim was to better understand the role of polygenic causes in the determination of neuroanatomical diversity. In a previous work, we used a sample of N = 2011 subjects with whole-genome genotyping data from the (IMAGEN) imaging genetics consortium combined with structural magnetic resonance imaging (MRI) data analyzed according to the pipelines of the ENIGMA consortium and showed that genome-wide complex trait analysis (GCTA) was able to capture a substantial proportion of the variability in regional brain volume (BV)—up to 55% (Toro et al. 2015). Due to sample size limitations, however, standard errors (SEs) were high (about ±25%), and statistical power for post hoc analyses was limited. The advent of collaborative efforts such as the ENIGMA consortium, and large-scale projects such as UK Biobank, in particular, has allowed researchers to greatly increase the number of subjects used for heritability estimation (see e.g., Elliott et al. 2018; Zhao et al. 2018). Here, we replicate and follow up on our original results using a sample 10 times larger, N = 26 818, which combines data from the UK Biobank project as well as 5 other projects that had previously participated in the ENIGMA consortium (IMAGEN, Study of Health in Pomerania [SHIP], Trauma Related Neuronal Dysfunction (TREND), Lothian, Alzheimer’s Disease Neuroimaging Initiative).
We used the same regional BVs estimates than those used in the ENIGMA consortium studies by Stein et al. (2012) and Hibar et al. (2015): several subcortical structures, total BV, and intracranial volume (ICV). In addition, we also studied height and intelligence scores (ISs). BV correlates with height and ISs (Taki et al. 2012), which are both known to be heritable (Yang et al. 2010; Plomin and Deary 2015). We aimed thus at determining to which extent the heritability of regional BVs was given by their relationship with height (i.e., affected by the same genetic factors that determine body size) or if different genetic factors affected them specifically. Similarly for ISs, we aimed at better understanding its relationship with BV.
For all phenotypes, we estimated to what extent genome-wide single nucleotide polymorphism (SNPs) were able to capture the interindividual variability in regional BVs, that is, we estimated the proportion of phenotypic variance captured by SNPs across the genome (also called SNP heritability). Additionally, we used GWAS data to compute genome-wide polygenic scores (GPSs), which provide a phenotypic prediction at the individual level. The analyses of the influence of the complete genome on our phenotypes were complemented with analyses on a series of genomic partitions: genic versus nongenic; preferential expression in the central nervous system (CNS) or by cell type; low, medium, or high minor allele frequency (MAF). This type of analysis can reveal whether specific genomic regions are enriched in the amount of variance they capture. Finally, we looked at the pleiotropy across phenotypes. For this, we computed genetic correlations and phenotypic correlations for all pairs of phenotypes. For brain regions, we also compared the genetic and phenotypic correlations between the left and right parts of the same structure as a means to estimate the role of genetics and environment in brain asymmetry.
Our results confirm the observation that polygenic factors play an important role in the determination of neuroanatomical variability and show different ways in which biological information can be obtained to better understand polygenic effects.
Material and Methods
Data Sharing
We obtained whole-genome genotyping from N = 26 818 subjects from six different projects: UK Biobank, IMAGEN, ADNI, Lothian Birth Cohort 1936, SHIP, and TREND. Extensive efforts have been made to homogenize the neuroanatomical measurements across sites, which were described in the ENIGMA 1 and 2 reports (Stein et al. 2012; Hibar et al. 2015) and the UK Biobank neuroimaging analysis group (Alfaro-Almagro et al. 2018). The UK Biobank project (https://imaging.ukbiobank.ac.uk) is a large, long-term biobank study in the United Kingdom aiming at investigating the contributions of genetic predisposition and environmental exposure to the development of disease. The study is following about 500 000 volunteers enrolled at ages from 40 to 69 years old, 54% females. IMAGEN (https://imagen-europe.com) is a project to identify and characterize specific genetically influenced alterations in reinforcer sensitivity and executive control which manifest in adolescence and carry the risk for overt psychopathology later in life. It includes general population 13–17-year-old adolescents (49% of females) from Germany, France, Ireland, and the United Kingdom. ADNI (http://adni.loni.usc.edu) is a longitudinal multicenter study designed to develop clinical, imaging, genetic, and biochemical biomarkers for the early detection and tracking of Alzheimer’s disease in the United States of America. The dataset combines data from the initial 5-year study (ADNI-1) and the follow-ups ADNI-GO, ADNI-2, and ADNI-3. It includes subjects 54–90 years old, 42% female. The Lothian Birth Cohort 1936 (https://www.lothianbirthcohort.ed.ac.uk) is a follow-up of the Scottish Mental Surveys of 1947, which tested the intelligence of almost every child born in 1936 and attending school in Scotland in the month of June 1947. It includes subjects 71–73 years old, 47% female. The SHIP and TREND cohorts contain data from the SHIP (http://www2.medizin.uni-greifswald.de/cm/fv/ship.html), a population-based epidemiological study consisting of 2 independent cohorts SHIP and SHIP-TREND. These projects investigate common risk factors, subclinical disorders, and diseases in a population of northeast Germany. The dataset included data from subjects 21–90 years old, 44% female for TREND, 48% female for SHIP. All data sharing was approved by our local ethical board as well as by those of the participating projects wherever required. The list of projects and their respective number of subjects are described in Table 1.
Table 1.
Sample sizes and number of variants per project
| Project | Total sample size | Percentage of females | Mean age (standard deviation) | Sample size included | Total number of variants | Number of variants included |
|---|---|---|---|---|---|---|
| IMAGEN | 2011 | 49% | 14.6 (0.4) | 1736 | 573 299 | 267 151 |
| Lothian Birth Cohort 1936 | 1005 | 47% | 72.7 (0.7) | 544 | 529 015 | 256 417 |
| TREND | 858 | 44% | 50.0 (13.5) | 813 | 2 389 858 | 597 902 |
| SHIP | 963 | 48% | 56.5 (12.6) | 941 | 863 230 | 271 635 |
| ADNI | 1189 | 42% | 74.2 (7.1) | 986 | 331 088 | 227 005 |
| UK Biobanka | 20 792 | 54% | 62.6 (7.5) | 19 270 | 734 447 | 490 061 |
aThe UK Biobank dataset was split in 2 parts: N = 14 144 subjects were used for heritability analyses (13 086 included), and N = 6678 subjects were used for validation of GPSs (6184 included)
Regional BVs
The measurements of regional BV coming from projects that had previously participated in ENIGMA (IMAGEN, Lothian Birth Cohort 1936, TREND, SHIP, and ADNI) were the same that had been used in Stein et al. (2012) and Hibar et al. (2015). For the UK Biobank subjects, the estimation of the volumes was performed using FreeSurfer 6.0 (https://surfer.nmr.mgh.harvard.edu). For comparison, we also included the estimates obtained using FSL FIRST (https://fsl.fmrib.ox.ac.uk/fsl) that were made available by UK Biobank (the processing pipeline is described in https://biobank.ctsu.ox.ac.uk/crystal/docs/brain_mri.pdf). In addition to the subjects excluded in the quality control made by the UK Biobank, we excluded 52 additional subjects showing an extreme relationship between total BV and ICV. For this, we used a kernel density estimator to fit a probability density function to the intracranial versus BV data and tagged as outliers all subjects with a local density inferior to 1% of the maximum density for UK Biobank and 2% for other datasets.
The regions included in our analyses were as follows: nucleus accumbens (labelled as Acc), amygdala (Amy), putamen (Pu), pallidum (Pa), caudate nucleus (Ca), hippocampus (Hip), and thalamus (Th), along with BV and ICV. In addition to these regions, we investigated height and IS, available from the UK Biobank and IMAGEN projects. It is important to note that the fluid IS in UK Biobank (a 2 min test aiming at evaluating the capacity to solve problems that require logic and reasoning ability, independent of acquired knowledge; see https://biobank.ctsu.ox.ac.uk/crystal/label.cgi?id=100027) is not the same as the IS used by the IMAGEN project, which was obtained using the Wechsler Intelligence Scale for Children (WISC) test.
Genotype Filtering
All genetic analyses were performed for each project independently. Genotyping data was converted to the hg19 reference wherever required using UCSC LiftOver (http://genome.ucsc.edu/cgi-bin/hgLiftOver). We used genotyped autosomal SNPs which passed UK Biobank quality control for all batches (http://www.ukbiobank.ac.uk/wp-content/uploads/2014/04/UKBiobank_genotyping_QC_documentation-web.pdf). Additionally, we removed SNPs in 24 regions with long range linkage disequilibrium (LD, see Price et al. 2008). SNPs were then filtered to exclude those with MAF <0.1%, missing rate >1%, or Hardy–Weinberg disequilibrium with a P < 10−6. Individuals were removed when >10% of their SNPs were missing. We finally pruned SNPs which were in LD with a variance inflation factor >10, which corresponds to a multiple 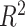 for the regression over linked SNPs <0.9. The filtering was made using PLINK v1.90b3.46 (Purcell et al. 2007).
for the regression over linked SNPs <0.9. The filtering was made using PLINK v1.90b3.46 (Purcell et al. 2007).
Genetic Relationship Matrices
Genetic relationship matrices (GRMs) were computed based on autosomal chromosomes using GCTA v1.91.3 (Yang et al. 2011a). We included only one of each pair of subjects with an estimated relatedness >0.025 (approximately corresponding to cousins 2–3 times removed). GRMs were computed per chromosome and then merged for the whole genome.
Population Structure
Genetic variance estimates based on genomic estimates of relatedness are sensitive to cryptic relatedness and population structure. These factors can influence the phenotypic similarity beyond the estimated degree of genetic relatedness (Browning and Browning 2011; Yang et al. 2011b). In addition to the exclusion of subjects with a degree of genetic relatedness greater than 0.025, we used the first 10 principal components of the GRM as covariates in our statistical analyses.
Genetic Variance
We estimated the amount of phenotypic variance captured by SNPs using a linear mixed model with age, sex, imaging centre, and the first 10 principal components of the GRM as fixed effect covariates, and a random effect with a covariance matrix corresponding to the GRM (Genome-wide complex trait analysis (GCTA) Genome-based restricted maximum likelihood (GREML) method, Yang et al. 2011a, 2011b). We estimated SNP heritability as the ratio of the genetic variance to the phenotypic variance, with genetic variance being the variance of the random component and the phenotypic variance being the sum of random component and residual component with fixed effects removed. We used GCTA v1.91.3 (Yang et al. 2011a) for those computations and did not constrain genetic variance estimates to lie in the range 0–100%, in order to obtain unbiased estimates (option—reml-no-constrain).
Genetic Correlation
Genetic correlation was estimated using GCTA (REML) Restricted maximum likelihood estimation bivariate analysis (Lee et al. 2012b) in constrained mode (option—reml-bivar). Both phenotypic and genetic correlations were adjusted for age, sex, imaging centre, and the first 10 principal components of the GRM. We compared genetic and phenotypic correlations using the delta method to estimate SEs (R package msm https://cran.r-project.org/web/packages/msm/). We report estimates with their SEs.
Genetic Variance Partitioning
In its simplest form, GCTA allows to estimate the amount of variance captured by the matrix of genetic relationships, assuming that each SNP captures the same amount of variance. Through genomic partitions, we can create different GRMs based on nonoverlapping regions of the genome. The SNPs on each of these partitions can capture then a different amount of variance (although, as before, SNPs within a given partition are supposed to capture all the same amount of variance). We grouped SNPs in the following partitions:
Partition based on genic status. Using 66 632 gene boundaries from the UCSC Genome Browser hg19 assembly, we made a first set with all SNPs within these boundaries, 2 further sets that included also SNPs 0–20 and 20–50 kbp upstream and downstream of each gene, and a last set including the SNPs not located in regions less than 50 kbp upstream or downstream of genes. Both exonic and intronic regions were included in the genic regions. These partitions do not correspond exactly to those used by Toro et al. (2015) which were as follows: one with strict genic/nongenic boundaries (0 kbp), another with genic ±20 kbp versus the rest, and finally genic ±50 kbp versus the rest.
Partition based on preferential CNS expression (Raychaudhuri et al. 2010; Lee et al. 2012a) using ±50 kbp as gene boundaries and based on markers of brain cell types as defined by 2 recent scRNA-Seq studies (Li et al. 2018; Skene et al. 2018) using ±20 kbp as gene boundaries.
Partition based on allele frequency. A partition based on MAF with 4 groups: from 0.1% to 5%, from 5% to 20%, from 20% to 35%, and from 35% to 50%. In Toro et al. (2015), only the last 3 partitions were included, covering the range from 5% to 50% of MAF.
Genetic Variance Partition Enrichment
Once the variance captured by each partition was computed, we were able to estimate the significance of the difference in variance captured by an individual partition against a model where each SNP captures exactly the same amount of variance (a null hypothesis of no enrichment). In this latter case, the amount of variance captured by each partition should be directly proportional to the number of SNPs it contains.
We tested whether any of the partitions captured more variance than what could be expected given its number of SNPs. A Z-score was computed by comparing the SNP-set genetic estimated variance 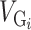 of partition
of partition 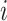 to the SNP-set genetic variance
to the SNP-set genetic variance 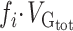 expected under no enrichment:
expected under no enrichment:
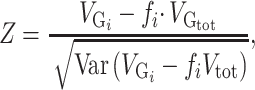 |
(1) |
where 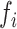 is the fraction of the SNPs included in partition
is the fraction of the SNPs included in partition 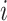 . We estimated the variance of the observed enrichment as:
. We estimated the variance of the observed enrichment as:
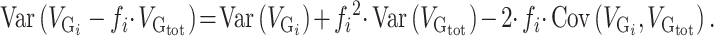 |
(2) |
Here, 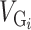 represents the genetic variance of partition
represents the genetic variance of partition 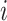 and
and 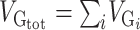 . We preferred the analytic estimation over the estimation by permutations because the permutation methods presented some limitations due to the difficulty of preserving the SNP structure of the partitions and due to the computational resources needed for the permutations (Supplementary Figs 1 and 2).
. We preferred the analytic estimation over the estimation by permutations because the permutation methods presented some limitations due to the difficulty of preserving the SNP structure of the partitions and due to the computational resources needed for the permutations (Supplementary Figs 1 and 2).
Meta-analytical Combination of the Estimatesof Each Project
The independent estimates obtained from each of the projects were combined into a single one using an inverse variance weighting method. We validated the meta-analytical approach by comparing the distribution of genetic variance estimates from simulated heritable phenotypes with their theoretical normal distribution in sub-samples of ADNI and UK Biobank datasets.
Accounting for Measurement Error
The estimation of regional brain volumes, height, and ISs is submitted to measurement errors. In particular, small, poorly defined brain regions are more difficult to measure accurately than larger ones. Measurement errors enter as environmental variance in the decomposition of phenotypic variance into genetic and environmental variance and hence decrease the heritability estimates (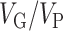 ). Heritability estimates unbiased for measurement errors can be obtained with repeated measurements (Ponzi et al. 2018); however, the datasets we used to measure brain volumes provide only 1 MRI per subject. We used datasets from the Consortium for Reliability and Reproducibility (CoRR) (Zuo et al. 2014), which includes multiple MRIs per individual to estimate intraclass correlation coefficients (ICCs) (Fisher 1970) for each regional volume. We segmented the volumes as we did in the UK Biobank dataset, using FreeSurfer 6.0. ICCs were computed on the volumes of 836 subjects with 2 MRI sessions after covarying for age, sex, and scanning site. For intelligence, we estimated ICC from 1301 subjects in UK Biobank who took the fluid intelligence (FI) test 3 times. The ICC for height was considered to be equal to 1 (no measurement error). Adjusted phenotypic variance and covariance estimates were obtained by multiplying the raw estimates by their corresponding ICC (see Supplementary Materials).
). Heritability estimates unbiased for measurement errors can be obtained with repeated measurements (Ponzi et al. 2018); however, the datasets we used to measure brain volumes provide only 1 MRI per subject. We used datasets from the Consortium for Reliability and Reproducibility (CoRR) (Zuo et al. 2014), which includes multiple MRIs per individual to estimate intraclass correlation coefficients (ICCs) (Fisher 1970) for each regional volume. We segmented the volumes as we did in the UK Biobank dataset, using FreeSurfer 6.0. ICCs were computed on the volumes of 836 subjects with 2 MRI sessions after covarying for age, sex, and scanning site. For intelligence, we estimated ICC from 1301 subjects in UK Biobank who took the fluid intelligence (FI) test 3 times. The ICC for height was considered to be equal to 1 (no measurement error). Adjusted phenotypic variance and covariance estimates were obtained by multiplying the raw estimates by their corresponding ICC (see Supplementary Materials).
GPSs
We used the SNP effects estimated in the association analysis of 13 086 UK Biobank subjects to estimate the phenotypes of 6184 additional unrelated subjects with MRI data from the latest release of the UK Biobank project. The scores were estimated from the filtered SNPs (not LD pruned). SNPs under various association P-value thresholds were selected, and the ones in LD with a more significantly associated SNP were clumped. The P-value threshold that produced the best fit with the target dataset was selected. We used the software PRSice associated with PLINK for the computation of GPSs (Euesden et al. 2015). Each phenotype was regressed on age, sex, scanning centre, and the 10 first principal components of the GRM. The analyses were performed on the residuals of this linear regression. We then estimated the proportion of variance captured by GPSs in each phenotype using the coefficient of determination 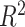 . For comparison, we also computed the predicted height of the 6184 subjects using SNP effects from an independent group of 277 756 unrelated (with a pairwise estimated relatedness <0.025 in the GRM) UK Biobank subjects.
. For comparison, we also computed the predicted height of the 6184 subjects using SNP effects from an independent group of 277 756 unrelated (with a pairwise estimated relatedness <0.025 in the GRM) UK Biobank subjects.
Results
Genome-wide Variants Capture a Large Proportion of the Diversity of Regional Brain Volumes, Height, and IS
The heritability estimates for intracranial volume, total brain volume, as well as the volume of subcortical structures, were substantial and statistically significant (Fig. 1, Supplementary Table 1.1). Estimates for intracranial volume and total brain volume were large: VG/VP (ICV) = 53 ± 4.5% (all our variance estimates are reported as estimation ± SE), VG/VP (BV) = 52 ± 4.5%. Similarly, the genetic variance estimates for subcortical structures were all above 40%: Acc (VG/VP = 40 ± 4.5%), Amy (VG/VP = 45 ± 4.5%), Pu (VG/VP = 48 ± 4.5%), Pa (VG/VP = 40 ± 4.5%), Ca (VG/VP = 52 ± 4.5%), Th (VG/VP = 54 ± 4.5%), Hip (VG/VP = 53 ± 4.5%). All estimates were highly statistically significant with P < 10−11 in all cases (log-likelihood ratio statistics from 49 to 242 in UK Biobank alone) (Fig. 1, Supplementary Table 1.2). The VG/VP estimate for Ca (VG/VP = 52 ± 4.4%) was statistically significant and very different from what was observed in the IMAGEN cohort (VG/VP = 4 ± 24%) which, as mentioned in Toro et al. (2015), may reflect an age bias specific to IMAGEN (where individuals were on average 14 years old).
Figure 1.
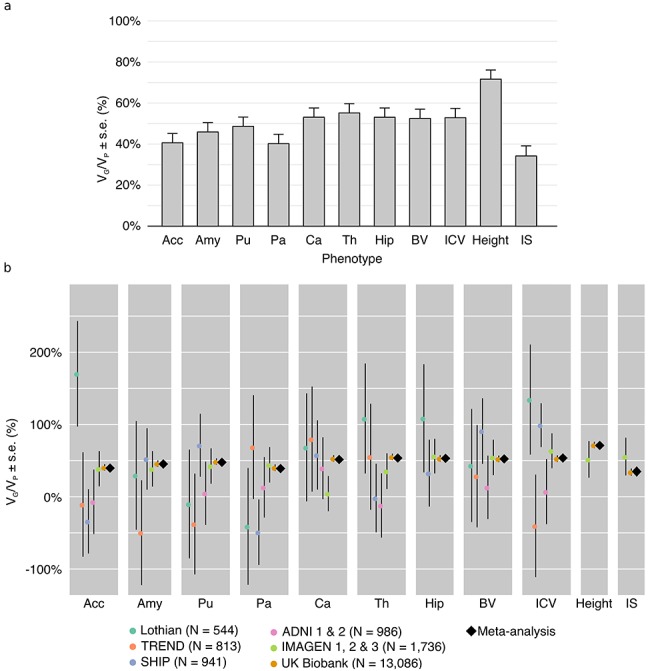
(a) Proportion of variance captured by common genotyped variants (VG/VP) for brain regions, height and IS. Meta-analytic estimates were obtained using inverse variance weighting of the estimates of the different projects studied. (b) VG/VP estimates for each project. The estimates were obtained using GCTA, without constraining the results to lie in the 0–100% range. The diamond shows the meta-analytic estimation. Age, sex, center, and the first 10 principal components were included as covariates. The error bars show the SEs of the VG/VP estimates.
The heritability estimate (VG/VP) for height (which combines only UK Biobank and IMAGEN) was large: VG/VP = 71 ± 3.1% and close to those obtained from twin studies (Polderman et al. 2015) and from previous SNP heritability estimates (VG/VP = 68.5 ± 0.4% in Ge et al. 2017). The fluid IS in UK Biobank had the lowest heritability among all 11 studied phenotypes, VG/VP = 35 ± 4.8%. This estimate is similar to the one obtained by Davies et al. (2011) on 30 801 UK Biobank subjects (VG/VP = 31 ± 1.8%). The verbal intelligence quotient (VIQ) and performance intelligence quotient (PIQ) in IMAGEN, although not based on the same test as FI in UK Biobank, aim at capturing a similar phenotype. The estimate for FI in UK Biobank seems smaller than those for VIQ and PIQ in IMAGEN (~56 ± 26%); however, they were not statistically significantly different.
Our estimates were obtained using a meta-analytic approach to combine the results of the different projects using inverse variance weighting. In all cases, the meta-analytic estimates closely corresponded with the values obtained for the UK Biobank project (Supplementary Table 1.2) because of its large sample size which accounted for ~94% of the weighted estimates. SEs agreed well with the theoretical values proposed by Visscher et al. (2014) and implemented in the GREML statistical power calculator (http://cnsgenomics.com/shiny/gctaPower): they were from 1.01 to 1.05 times higher than the theoretical values when taking into account the variance of the genetic relationships, except for ADNI where they were 1.21 times higher. As expected, the SEs decreased with sample size, from about 76% for ~550 individuals, 43% for ~1000 individuals, 24% for ~1750 individuals (IMAGEN), down to 4% for UK Biobank with more than 13 000 individuals. Our result from simulated phenotypes showed that the estimates of VG/VP obtained with sub-samples of N = 800, N = 400, N = 200, and even N = 100 subjects from the UK Biobank project were unbiased relative to the expected normal distribution. The simulations based on the ADNI project, however, showed a significant bias towards positive values when the sub-samples included N = 100 to N = 400 subjects, probably due to the heterogeneity of the population (Supplementary Figs 12, 13 and Supplementary Tables 6, 7).
Similarly to what we had observed previously (Toro et al. 2015), the genetic variance estimates were not significantly affected by population structure: the noninclusion of the 10 first PCs did not impact the estimates of variance, which changed on average by less than 4% (Supplementary Fig. 3).
Finally, our heritability estimates for the UK Biobank and ADNI projects were comparable with those of previous studies of these projects by Elliott et al. (2018) and Zhao et al. (2018) (see Supplementary Fig. 6 and Supplementary Materials).
Genetic Partitions Show Significant Enrichment of the Proportion of Variance Captured by Specific Gene Sets
Partition per Chromosome (Autosomes)
We observed a strong correlation between VG/VP estimates and chromosome size, which was significant for all phenotypes except Pa (Fig. 2). The correlation coefficients ranged from 0.30 ± 0.21 to 0.80 ± 0.13, r = 0.63 on average, capturing 41% of the variance (estimated as 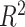 ).
).
Figure 2.
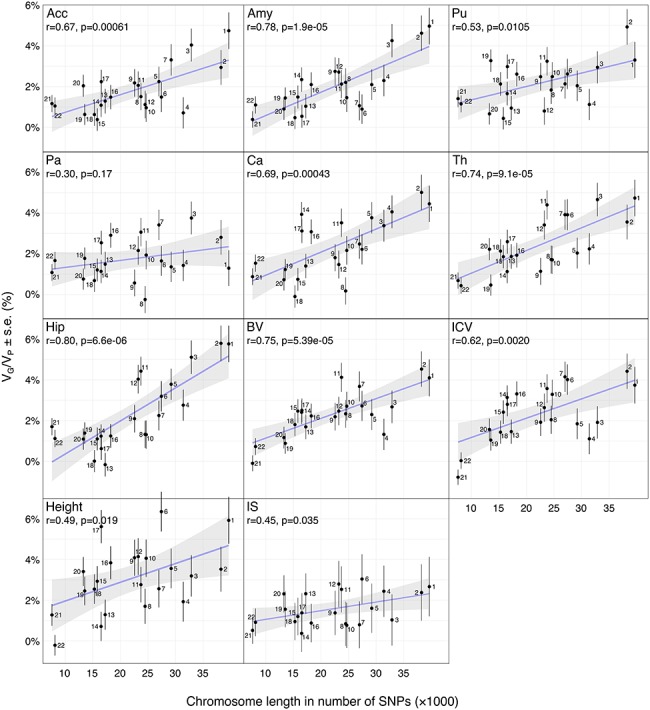
Scatter plots of the number of SNPs per chromosome versus VG/VP estimates computed for each chromosome. VG/VP estimates were obtained by partitioning SNPs across chromosomes and computed using the GCTA REML unconstrained method for total subcortical volumes. Age, sex, center, and the top 10 principal components were included as covariates. The error bars show the SEs of the VG/VP estimates.
Partition between Genic and Non-genic Regions
The genic SNP set (± 0 kbp) contained 51% of all genotyped SNPs and captured on average 69% of the variance attributable to SNPs of most of the studied phenotypes, significantly more than what we would expect from its length (FDR < 5%). The only exceptions were FI and Acc, for which it explained respectively 59 ± 9% and 55 ± 6% of the total genetic variance (Fig. 3, Supplementary Table 4). Height was the only phenotype for which we observed an enrichment of VG/VP captured by one of the nongenic SNP sets: the set of SNPs within 0 ± 20 kbp of the genic set. This set contained 15% of all SNPs but explained 27% of the variance of the height phenotype attributable to SNPs (FDR corrected P < 0.01). In total, the variants located between 0 and 50 kbp away from genes captured 36 ± 4% of the genetic variance of height (FDR corrected P < 0.05).
Figure 3.
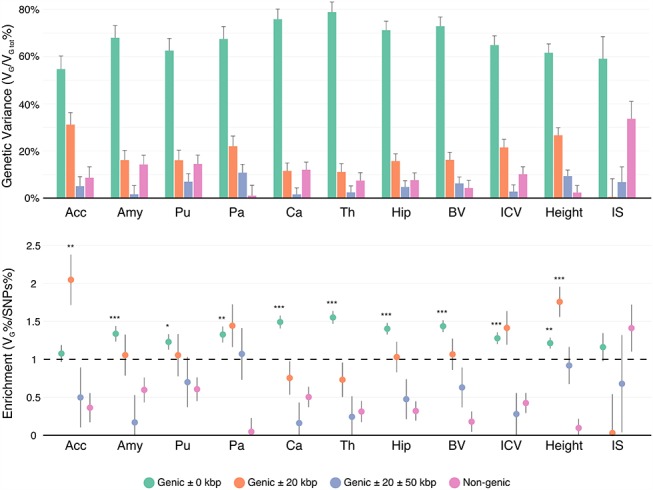
Variance enrichment for partitions based on closeness to genic regions. Meta-analytic estimates were obtained using inverse variance weighting of the estimates of the different projects studied. Top: VG/ estimates computed for 4 sets of SNPs based on their distance to gene boundaries: all SNPs within the boundaries of the 66 632 gene boundaries from the UCSC Genome Browser hg19 assembly, 2 further sets that included also SNPs 0–20 and 20–50 kbp upstream and downstream of each gene, and a remaining set containing SNPs located farther than 50 kb from one of the gene boundaries. VG/
estimates computed for 4 sets of SNPs based on their distance to gene boundaries: all SNPs within the boundaries of the 66 632 gene boundaries from the UCSC Genome Browser hg19 assembly, 2 further sets that included also SNPs 0–20 and 20–50 kbp upstream and downstream of each gene, and a remaining set containing SNPs located farther than 50 kb from one of the gene boundaries. VG/ estimates were computed using the GCTA REML unconstrained method for height, intelligence, and brain, intracranial and total subcortical volumes. The error bars represent the SEs. Bottom: enrichment of variance captured by each partition. The y-axis shows the ratio of the fraction of genetic variance explained by each partition divided by the fraction of SNPs contained in each partition. If all SNPs explained a similar amount of variance, this ratio should be close to 1 (dashed line). A Z-test was used to compare the ratios to 1 and P-values were FDR adjusted (*P < 0.05, **P < 0.01, ***P < 0.001).
estimates were computed using the GCTA REML unconstrained method for height, intelligence, and brain, intracranial and total subcortical volumes. The error bars represent the SEs. Bottom: enrichment of variance captured by each partition. The y-axis shows the ratio of the fraction of genetic variance explained by each partition divided by the fraction of SNPs contained in each partition. If all SNPs explained a similar amount of variance, this ratio should be close to 1 (dashed line). A Z-test was used to compare the ratios to 1 and P-values were FDR adjusted (*P < 0.05, **P < 0.01, ***P < 0.001).
Partition by Involvement in Preferential CNS Expression and in Brain Cell Types
No statistically significant enrichment was observed for CNS expression nor brain cell type partitions.
Partition by Allele Frequency
SNPs within the low MAF partition (MAF < 5%) captured less genetic variance per SNP than those with medium and high MAF (the 3 partitions with MAF > 5%) (Fig. 3), as previously described by Speed et al. (2017). Our 4 MAF-based partitions included the following average proportions of total SNPs: (1) MAF from 0.1% to 5%: comprising 40% of SNPs, (2) 5% to 20%: with 30% of SNPs, (3) 20% to 35%: with 12% of SNPs, and (4) 35% to 50%: with 9% of SNPs. Although the partition of SNPs with low MAF contained 40% of the SNPs, it captured on average only about 16% of the total genetic variance. This is less than expected in the GCTA model where each SNP captures the same amount of phenotypic variance but slightly more than expected in a neutral theory of evolution, where the captured variance is proportional to the size of the MAF bin (Yang et al. 2017; Visscher et al. 2012).
Genetic Factors Influencing Volume Are Shared Among Brain Regions
We observed extensive pleiotropy across brain regions, with an average genetic correlation of rG ~ 0.45 (Fig. 4). Genetic correlations, which represent the correlation between genetic effects of 2 phenotypes, were computed for each pair of brain regions, ICV, height, and IS, using data from the UK Biobank project. Figure 4 shows the genetic correlation matrix together with the phenotypic correlation matrix (see also Supplementary Table 5).
Figure 4.
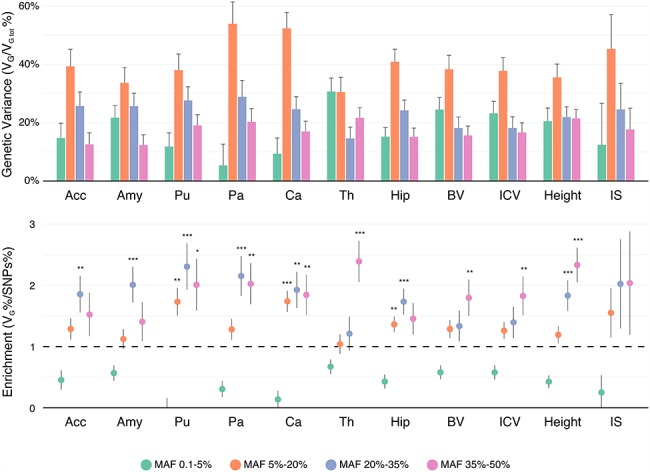
Variance enrichment for partitions based on MAF. Meta-analytic estimates were obtained using inverse variance weighting of the estimates of the different projects studied. Top: VG/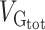 estimates computed for 4 sets of SNPs based on their MAF: from 0.1% to 5%, from 5% to 20%, from 20% to 35% and from 35% to 50%. VG/
estimates computed for 4 sets of SNPs based on their MAF: from 0.1% to 5%, from 5% to 20%, from 20% to 35% and from 35% to 50%. VG/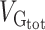 estimates were computed using the GCTA REML unconstrained method for height, intelligence, and brain, intracranial and total subcortical volumes. The error bars represent the SEs. Bottom: enrichment of variance captured by each partition. The y-axis shows the ratio of the fraction of genetic variance explained by each partition divided by the fraction of SNPs contained in each partition. If all SNPs explained a similar amount of variance, this ratio should be close to 1 (dashed line). A Z-test was used to compare the ratios to 1 and P-values were FDR adjusted (*P < 0.05, **P < 0.01, ***P < 0.001).
estimates were computed using the GCTA REML unconstrained method for height, intelligence, and brain, intracranial and total subcortical volumes. The error bars represent the SEs. Bottom: enrichment of variance captured by each partition. The y-axis shows the ratio of the fraction of genetic variance explained by each partition divided by the fraction of SNPs contained in each partition. If all SNPs explained a similar amount of variance, this ratio should be close to 1 (dashed line). A Z-test was used to compare the ratios to 1 and P-values were FDR adjusted (*P < 0.05, **P < 0.01, ***P < 0.001).
Genetic correlations were in general similar to phenotypic and environmental correlations; however, genetic correlations were often larger than phenotypic correlations for the left/right volumes of the same region. The concordance between phenotypic and genetic correlation was high (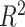 = 0.80, Fig. 5c), consistent with the report by Sodini et al. (2018) for other traits, and the correlation matrices were similar (Supplementary Fig. 7). Phenotypic correlations can be decomposed as a sum of genetic and environmental correlations. The concordance between genetic and environmental correlations was also strong (
= 0.80, Fig. 5c), consistent with the report by Sodini et al. (2018) for other traits, and the correlation matrices were similar (Supplementary Fig. 7). Phenotypic correlations can be decomposed as a sum of genetic and environmental correlations. The concordance between genetic and environmental correlations was also strong (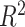 = 0.47, Supplementary Fig. 8).
= 0.47, Supplementary Fig. 8).
Figure 5.
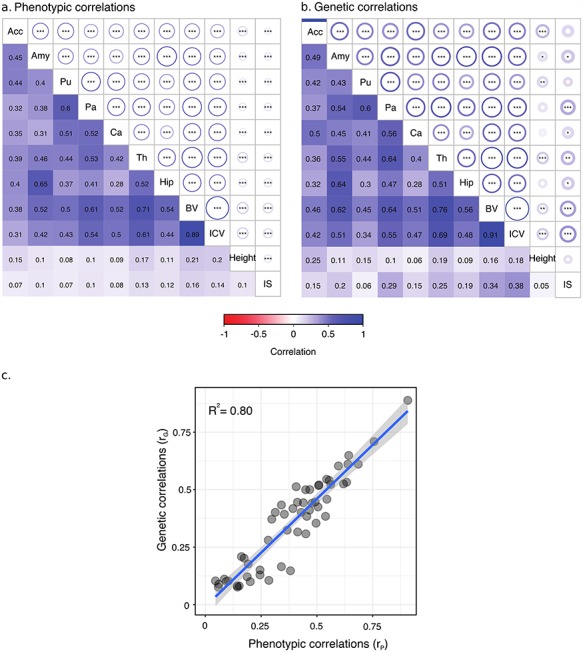
Phenotypic and genetic correlations. Significant phenotypic (a) and genetic (b) correlations were observed for most phenotypes. Correlation estimates are shown in the lower triangular part of the matrices, statistical significance in the upper triangular part. Circle radius represents correlation strength, stars indicate statistical significance of the correlation being non null (*P < 0.05, **P < 0.01, ***P < 0.001). The scatter plot (c) of phenotypic versus genetic correlations.
Environmental Factors Are Important in Shaping Brain Asymmetry
For all brain regions, with the exception of Acc, the differences between their left and right volumes appeared to be of environmental origin. When considering total volumes (left plus right), the differences between genetic and phenotypic correlations were not different from zero (Z-test FDR > 50%, Supplementary Table 5). However, the situation was different when considering the regional volume asymmetries (left minus right). In that case, the genetic correlations of the asymmetries were not statistically different from rG = 1 and were in all cases statistically significantly larger than the phenotypic correlations (Z-test FDR < 5%, Supplementary Fig. 9 and Supplementary Table 5), which suggest an important role of the environment in shaping hemispheric asymmetry. To further test this hypothesis, we measured the VG/VP of the differences between left and right volumes of each structure. Only the heritability of the volume asymmetry of Acc was significantly different from zero (VG/VP = 16 ± 4.5%). Except for this structure, the differences in volumes between right and left hemispheres of all the other brain regions seemed to be only of environmental origin.
Genetic Factors Influencing Brain Volume Are Shared with Height and IS, But Less between Height and IS
The genetic correlation between height and brain volume (rG:BV.height = 0.164 ± 0.053) was smaller than the genetic correlation between brain volume and intelligence (rG:BV.IS = 0.343 ± 0.080). However, the difference was not statistically significant: rG:BV.IS—rG:BV.height = 0.179 ± 0.095, 95% CI: −0.008–0.365 (computed using the formula xxxvii in Pearson and Filon 1898). The phenotypic and genetic correlations between ISs and height were, however, the smallest we observed across all phenotypes: rP = 0.101 ± 0.009 and rG = 0.048 ± 0.073. This is different to what we had previously observed in the IMAGEN cohort (Toro et al. 2015), where the correlation between brain volume and ISs was higher than the correlation between brain volume and height.
Heritability and Genetic Correlation Estimates Remain Statistically Significant after Accounting for Measurement Errors
The estimates of heritability, phenotypic and environmental correlation can be biased by noise in the volume measurements, which could in particular explain the differences between genetic and phenotypic correlations of left and right volumes. We investigated this hypothesis by removing the effects of measurement errors estimated from CoRR (Zuo et al. 2014) for FreeSurfer segmentations and from the repeated measures of IS in UK Biobank. ICCs for ISs were estimated at 63%, whereas the lowest ICCs for subcortical structures were found for Acc (83%), Pa (87%), and Amy (89%). After correction for measurement error, height remained the most heritable phenotype although it was the only one that we did not correct. Disattenuated estimates of VG/VP for intelligence became 54 ± 7.8%, which is similar to the disattenuated heritabilities of brain volumes (between 46 ± 5.3% for Pa and 61 ± 5.0% for Th) (Supplementary Fig. 10). Genetic correlations between left and right hemispheres of subcortical structures remained statistically significantly greater than the adjusted phenotypic correlations except for Pa and Acc (Supplementary Table 5 and Supplementary Fig. 11). Estimates of VG/VP for the differences between left and right volumes remained low and not significantly different from 0 for all subcortical structures (between 5.4 ± 14% for Pa and 16 ± 11% for Th) except for Acc (34 ± 9.8%).
GPSs Captured a Statistically Significant But Small Proportion of Phenotypic Variance
GPSs based on a GWAS of 13 086 subjects captured a statistically significant although very small proportion of brain region volume variance. The GPSs were computed for ~6000 additional participants from the UK Biobank who were not used in the GWAS. The predictions captured an amount of phenotypic variance ranging from 0.5% (P < 10−22) for Amy to 2.2% (P < 10−31) for brain volume (Supplementary Figs 15, 16 and Supplementary Table 8). For height, a GPS was obtained based on GWAS summary statistics from the 13 086 UK Biobank subjects with MRI captured ~3% of the variance. To evaluate the impact of the number of samples used in the GWAS on the amount of variance captured by the GPSs, we also computed the GPSs for height using the GWAS summary statistics from ~277 k unrelated UK Biobank subjects not included in the validation dataset. This allowed us to capture ~27% of the variance of height (an >8 times increase) (Supplementary Fig. 17).
Discussion
Our results suggest that neuroanatomical diversity is the product of a highly polygenic architecture, with SNPs capturing from 40% to 54% of regional brain volume variance, confirming our original findings (Toro et al. 2015) as well as those of others (Elliott et al. 2018; Zhao et al. 2018). At a global scale, causal variants were distributed across the genome: for different brain regions, chromosomes containing a larger number of SNPs captured a proportionally larger amount of variance than smaller chromosomes, with a correlation of r ~ 0.64 on average. At a local scale, however, SNPs within genes (~51%) captured ~1.5 times more genetic variance than the rest, and SNPs with low MAF captured significantly less variance than those with higher MAF.
Influence of rare variants on heritability estimates
When partitioning genetic variance into MAF bins, the lowest MAF partition going from 0.1% to 5% (10% of the total MAF range) contained ~40% of all SNPs but captured only ~16% of the total genetic variance: SNPs with low MAF captured significantly less variance than those with higher MAF. However, they captured more variance than expected under a neutral evolution model, where a MAF bin is expected to capture an amount of variance proportional to its size (10% size but 16% of the variance). This result suggests a negative selection model, where loci of large effect are being removed from the population. This apparent contradiction between high effect size and low captured variance for low MAF variants can be explained by the fact that the relationship between captured heritability 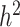 and the allele effect
and the allele effect 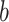 for a given SNP is dependent of its allele frequency
for a given SNP is dependent of its allele frequency 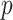 :
: 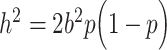 (Schoech et al. 2017; Zeng et al. 2018). Given this heterogeneity in effect sizes, using more flexible models that do not make strong assumptions on the relationships between effect size, LD, and MAF, such as GREML-LDMS (Yang et al. 2015), should improve the accuracy of the heritability estimates. Our study of rare variants is limited because of the use of datasets based on SNP arrays. The availability of whole-genome sequencing data for large cohorts is starting to allow the study of more refined partitions of rare variants and is showing that rare causal variants might be a main source of the variance remaining to be explained (Wainschtein et al. 2019).
(Schoech et al. 2017; Zeng et al. 2018). Given this heterogeneity in effect sizes, using more flexible models that do not make strong assumptions on the relationships between effect size, LD, and MAF, such as GREML-LDMS (Yang et al. 2015), should improve the accuracy of the heritability estimates. Our study of rare variants is limited because of the use of datasets based on SNP arrays. The availability of whole-genome sequencing data for large cohorts is starting to allow the study of more refined partitions of rare variants and is showing that rare causal variants might be a main source of the variance remaining to be explained (Wainschtein et al. 2019).
Environment may play a role in phenotypic asymmetries of regional brain volumes
In addition to showing the heritability of regional volume diversity, our analyses also show an extensive pleiotropy across brain regions. The computation of genetic covariance for pairs of brain regions allowed us to estimate their genetic correlation. We observed an average genetic correlation of rG ~ 0.45. Interestingly, we observed that although genetic correlations were similar to phenotypic correlations across brain regions, if we compared the left and right aspects of the same brain region, their genetic correlations were close to 1 and systematically larger than phenotypic correlations. This could be an indication that the observed phenotypic asymmetries in regional brain volume are of environmental origin. To confirm this result, we used an alternative way of analyzing the genetic/environmental nature of regional asymmetry, by looking at the heritability of the differences in left/right volumes. These heritabilities were generally not statistically significantly >0, which again supports the idea of the environmental nature of regional brain asymmetries.
Genetic correlation between brain, height and intelligence
The analysis of genetic correlations also allows us to explore the link between neuroanatomical diversity and other anatomical parameters or even cognitive functions. Brain volume is correlated with body size, and in the recent years, the polygenic architecture of height has been well described (Yengo et al. 2018). It could be argued that neuroanatomical diversity is simply determined by the same genetic factors that produce body size variability in general. The genetic correlation between brain volume and height was, however, relatively small (rG ~ 0.16), suggesting little overlap between their genetic causes (in Toro et al. 2015, we did not find a significant genetic correlation, most likely due to lack of statistical power). By contrast, the genetic correlation between brain volume and ISs was of medium strength (rG ~ 0.34), suggesting a larger overlap. The difference between these 2 genetic correlations was not significant, and the question deserves further study. The genetic correlation between height and ISs was the smallest of all those we studied, rG < 0.05, suggesting that their relationship with brain volume may be due to different genetic factors.
Measurement errors and centers influence the heritability estimates
One important source of bias in our estimates of heritability and genetic correlation could be related to measurement error. In MRI data, for example, some brain regions are more clearly delimited than others, which make them easier to segment accurately. Furthermore, there is an important variability in volume across brain regions. Segmentation errors would then be comparatively more important for small, poorly delimited regions such as Amy, than for large regions like Th. We sought to take into account errors in automatic segmentation of brain regions by analyzing data from the CoRR project (Zuo et al. 2014), where the same subjects were scanned several times. For ISs, we used the subset of UK Biobank where subjects passed the FI test on multiple occasions. Our results remained for the most part unchanged after adjusting the estimates for phenotypic measurement errors. A limitation of this approach, however, is that we used MRI scans from a different project (although the processing pipeline was the same). Ideally, one would have preferred to have repeated scans for a subset of UK Biobank subjects because segmentation quality depends on MRI quality which varies between datasets. The availability of repeated measurements in the same datasets from which genetic variance is estimated may allow to more precisely distinguish between the different sources of phenotypic variations.
Another possible source of bias could be the multicentric nature of our data. The results presented here come from 6 different projects. Rather than combining our genetic raw data into a single dataset, we chose to estimate the part of phenotypic variance captured by SNPs independently in each dataset and then combine estimates in a meta-analysis. While our chosen solution helps to handle heterogeneity, it trades on statistical power. The reason for this is that the SE of the GCTA GREML heritability estimate is approximately inversely proportional to the number of subjects (Visscher et al. 2014) (Supplementary Fig. 14). In the present analysis, the UK Biobank project accounted for ~94% of the estimates, largely driving the results. Indeed, the inclusion of the other projects reduced the SE of the estimates based on the UK Biobank project only by a factor of ~1.03. If the raw genotyping data had been combined in a single mega-analysis instead of in a meta-analysis, the decrease in SE would have been ~1.36 smaller than what we reported. The meta-analytical approach may however prove interesting in the future when large imaging genetics datasets other than UK Biobank will be available or in cases where raw genotyping data cannot be easily shared.
The advent of very large cohorts should decrease the gap between heritability and GPS analyses
Our analyses are also limited in their ability to provide information at the individual level. With the advent of large GWAS studies, GPS have become increasingly used to predict phenotypes from whole-genome genotyping data. GPSs are based on effect sizes estimated through GWAS in a large population. These estimates can then be used to predict the phenotype in an independent sample. We aimed at evaluating to which extent GWAS of the N ~ 13 k UK Biobank subjects used for our heritability analyses allowed us to predict the phenotypes of additional N ~ 6 k subjects. GPSs captured a very small, although statistically significant, proportion of the variance. For brain volume, for example, GPS captured ~2.5% of the variance. This is the expected result in the presence of a strongly polygenic phenotype (Wray et al. 2013). It is also expected that prediction accuracy will improve as the number of subjects used for effect size estimation increases (the UK Biobank project alone should provide data for N ~ 100 k in the years to come). We aimed at testing the potential increase in predictive power by computing a GPS for height. When effect sizes were estimated from N ~ 13 k subjects, GPS captured ~3% of height variance; however, when effect sizes were estimated from N ~ 277 k subjects, the amount of variance explained increased to ~27%. We expect that effect sizes for regional brain volume estimated from N ~ 100 k will allow us to compute GPSs capturing ~12% of the variance (based on Wray et al. 2013 and Daetwyler et al. 2008).
The study of more complex neuroimaging phenotypes will increase knowledge about cognitive and mental health traits
The detection of candidate genes of large effect is an appealing tool for gaining mechanistic insight on normal and pathological phenotypes. However, this approach is ill adapted to strongly polygenic architectures, where not only a few large-effect alleles are involved but potentially hundreds of thousands of alleles of almost infinitesimal effect (Wray et al. 2018). Neuroimaging endophenotypes such as those obtained using structural and functional MRI could provide an alternative source of mechanistic insight. The brain imaging literature is rich in examples of associations between different brain regions and networks with normal and pathological cognitive phenotypes. The automatic mining of these associations could provide a layer of annotation for brain regions and networks similar to those available today for genome annotation. Further investigation of the genomic architecture of neuroimaging endophenotypes should prove an important tool to better understand the biological basis of brain diversity and evolution in humans, as well as the biological basis of the susceptibility to psychiatric disorders.
Supplementary Material
Funding
Institut Pasteur; Center for Research and Interdisciplinarity, Centre National de la Recherche Scientifique; the University Paris Diderot; the Fondation pour la Recherche Médicale [DBI20141231310]; the European Commission Horizon 2020 [COSYN]; The Human Brain Project; the European Commission Innovative Medicines Initiative [AIMS2-TRIALS; No. 777394]; the Cognacq-Jay foundation; the Bettencourt-Schueller foundation; the Orange foundation; the FondaMental foundation; the Conny-Maeva foundation; and the Agence Nationale de la Recherche (ANR) [SynPathy]; the Laboratory of Excellence GENMED (Medical Genomics) (grant no. ANR-10-LABX-0013, Bio-Psy; by the INCEPTION program ANR-16-CONV-0005, managed by the ANR part of the Investment for the Future program).
NIH-NIBIB P41 EB019936 (ReproNim) NIH-NIMH R01 MH083320 (CANDIShare) and NIH 5 U24 DA039832 (NIF), as well as the Canada First Research Excellence Fund, awarded to McGill University for the Healthy Brains for Healthy Lives initiative (to J.-B.P.).
The Study of Health in Pomerania (SHIP) is part of the Community Medicine Research net (http://www.medizin.uni-greifswald.de/icm) of the University Medicine Greifswald, which is supported by the German Federal State of Mecklenburg-West Pomerania. MRI scans in SHIP and SHIP-TREND have been supported by a joint grant from Siemens Healthineers, Erlangen, Germany, and the Federal State of Mecklenburg-West Pomerania. This study was further supported by the EU-JPND Funding for BRIDGET (FKZ:01ED1615).
References
- Alfaro-Almagro F, Jenkinson M, Bangerter NK, Andersson JLR, Griffanti L, Douaud G, Sotiropoulos SN, Jbabdi S, Hernandez-Fernandez M, Vallee E et al. . 2018. Image processing and quality control for the first 10,000 brain imaging datasets from UK biobank. Neuroimage. 166(Feb):400–424. doi: 10.1016/j.neuroimage.2017.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokland GAM, de ZGI, McMahon KL, Wright MJ. 2012. Genetic and environmental influences on neuroimaging phenotypes: a meta-analytical perspective on twin imaging studies. Twin Research and Human Genetics. 15:351–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL. 2011. Population structure can inflate SNP-based heritability estimates. Am J Human Genet. 89(1):191–193. doi: 10.1016/j.ajhg.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler HD, Villanueva B, Woolliams JA. 2008. Accuracy of predicting the genetic risk of disease using a genome-wide approach. PLOS One. 3(10):e3395. doi: 10.1371/journal.pone.0003395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G, Tenesa A, Payton A, Yang J, Harris SE, Liewald D, Ke X, Le Hellard S, Christoforou A, Luciano M et al. . 2011. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Molecular psychiatry. 16:996–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott LT, Sharp K, Alfaro-Almagro F, Shi S, Miller KL, Douaud G, Marchini J, Smith SM. 2018. Genome-wide association studies of brain imaging phenotypes in UK biobank. Nature. 562(7726):210. doi: 10.1038/s41586-018-0571-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euesden J, Lewis CM, O’Reilly PF. 2015. PRSice: polygenic risk score software. Bioinformatics. 31(9):1466–1468. doi: 10.1093/bioinformatics/btu848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. 1918. The correlation between relatives on the supposition of Mendelian inheritance. Transactions of the Royal Society of Edinburgh. 52:399–433. [Google Scholar]
- Fisher RA. 1970. Statistical Methods for Research Workers. 14th ed, (revised and enlarged) Edinburgh: Oliver and Boyd. [Google Scholar]
- Ge T, Chen C-Y, Neale BM, Sabuncu MR, Smoller JW. 2017. Phenome-wide heritability analysis of the UK biobank. PLOS Genetics. 13:e1006711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge T, Nichols TE, Lee PH, Holmes AJ, Roffman JL, Buckner RL, Sabuncu MR, Smoller JW. 2015. Massively expedited genome-wide heritability analysis (MEGHA). PNAS. 112:2479–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge T, Reuter M, Winkler AM, Holmes AJ, Lee PH, Tirrell LS, Roffman JL, Buckner RL, Smoller JW, Sabuncu MR. 2016. Multidimensional heritability analysis of neuroanatomical shape. Nature Communications. 7:13291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar DP, Stein JL, Renteria ME, Arias-Vasquez A, Desrivières S, Jahanshad N, Toro R, Wittfeld K, Abramovic L, Andersson M et al. . 2015. Common genetic variants influence human subcortical brain structures. Nature. 520(7546):224–229. doi: 10.1038/nature14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, DeCandia TR, Ripke S, Yang J, Sullivan PF, Goddard ME, Keller MC, Visscher PM, Wray NR. 2012a. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 44(3):247–250. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Yang J, Goddard ME, Visscher PM, Wray NR. 2012b. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics(Oxford,England). 28(19):2540–2542. doi: 10.1093/bioinformatics/bts474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Santpere G, Kawasawa YI, Evgrafov OV, Gulden FO, Pochareddy S, Sunkin SM, Li Z, Shin Y, Zhu Y et al. . 2018. Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science. 362(6420):eaat7615. doi: 10.1126/science.aat7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson K, Filon LNG. 1898. Mathematical contributions to the theory of evolution. IV. On the probable errors of frequency constants and on the influence of random selection on variation and correlation. Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences. 191:229–311. [Google Scholar]
- Plomin R, Deary IJ. 2015. Genetics and intelligence differences: five special findings. Mol Psychiatry. 20(1):98–108. doi: 10.1038/mp.2014.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polderman TJC, Benyamin B, de LCA, Sullivan PF, van BA, Visscher PM, Posthuma D. 2015. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nature Genetics. [DOI] [PubMed] [Google Scholar]
- Ponzi E, Keller LF, Bonnet T, Muff S. 2018. Heritability, selection, and the response to selection in the presence of phenotypic measurement error: effects, cures, and the role of repeated measurements. Evolution. 72(10):1992–2004. doi: 10.1111/evo.13573. [DOI] [PubMed] [Google Scholar]
- Price AL, Weale ME, Patterson N, Myers SR, Need AC, Shianna KV, Ge D, Rotter JI, Torres E, Taylor KD et al. . 2008. Long-range LD can confound genome scans in admixed populations. Am J Hum Genet. 83(1):132–135. doi: 10.1016/j.ajhg.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Todd-Brown K, Thomas L, Ferreira MA et al. . 2007. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81(3):559–575. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Korn JM, McCarroll SA, The International Schizophrenia Consortium, Altshuler D, Sklar P, Purcell S, Daly MJ. 2010. Accurately assessing the risk of schizophrenia conferred by rare copy-number variation affecting genes with brain function. PLoS Genet. 6(9):e1001097. doi: 10.1371/journal.pgen.1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoech AP, Jordan DM, Loh P-R, Gazal S, O’Connor LJ, Balick DJ, Palamara PF, Finucane HK, Sunyaev SR, Price AL. 2019. Quantification of frequency-dependent genetic architectures in 25 UK biobank traits reveals action of negative selection. Nat Commun. 10:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satizabal CL, Adams HHH, Hibar DP, White CC, Knol MJ, Stein JL, Scholz M, Sargurupremraj M, Jahanshad N, Roshchupkin GV et al. . 2019. Genetic architecture of subcortical brain structures in 38,851 individuals. Nat Genet. 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene NG, Bryois J, Bakken TE, Breen G, Crowley JJ, Gaspar HA, Giusti-Rodriguez P, Hodge RD, Miller JA, Muñoz-Manchado AB et al. . 2018. Genetic identification of brain cell types underlying schizophrenia. Nat Genet. 50(6):825–833. doi: 10.1038/s41588-018-0129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodini SM, Kemper KE, Wray NR, Trzaskowski M. 2018. Comparison of genotypic and phenotypic correlations: Cheverud’s conjecture in humans. Genetics. genetics. 300630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speed D, Cai N. 2017. the UCLEB Consortium In: Johnson MR, Nejentsev S, Balding DJ, editors. Reevaluation of SNP heritability in complex human traits. Nat Genet. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, Toro R, Appel K, Bartecek R, Bergmann Ø et al. . 2012. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet. 44(5):552–561. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki Y, Hashizume H, Sassa Y, Takeuchi H, Asano M, Asano K, Kotozaki Y, Nouchi R, Wu K, Fukuda H et al. . 2012. Correlation among body height, intelligence, and brain Gray matter volume in healthy children. Neuroimage. 59(2):1023–1027. doi: 10.1016/j.neuroimage.2011.08.092 [DOI] [PubMed] [Google Scholar]
- Toro R, Poline J-B, Huguet G, Loth E, Frouin V, Banaschewski T, Barker GJ, Bokde A, Büchel C, Carvalho FM et al. . 2015. Genomic architecture of human neuroanatomical diversity. Mol Psychiatry. 20(8):1011–1016. doi: 10.1038/mp.2014.99 [DOI] [PubMed] [Google Scholar]
- Visscher PM, Goddard ME, Derks EM, Wray NR. 2012. Evidence-based psychiatric genetics, AKA the false dichotomy between common and rare variant hypotheses. Molecular Psychiatry. 17:474–485. [DOI] [PubMed] [Google Scholar]
- Visscher PM, Hemani G, Vinkhuyzen AAE, Chen G-B, Lee SH, Wray NR, Goddard ME, Yang J, Barsh GS. 2014. Statistical power to detect genetic (co)variance of complex traits using SNP data in unrelated samples. PLoS Genet. 10(4):e1004269. doi: 10.1371/journal.pgen.1004269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Thalamuthu A, Mather KA, Zhu W, Jiang J, de Micheaux PL, Wright MJ, Ames D, Sachdev PS. 2016. Distinct genetic influences on cortical and subcortical brain structures. Scientific Reports. 6:32760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainschtein P, Jain DP, Yengo L, Zheng Z, Trans-Omics for Precision Medicine Consortium TOPMed Anthropometry Working Group, Cupples LA, Shadyab AH, McKnight B, Shoemaker BM, Mitchell BD et al. . 2019. Recovery of trait heritability from whole genome sequence data. (Mar):588020. doi: 10.1101/588020. [DOI] [Google Scholar]
- Wray NR, Wijmenga C, Sullivan PF, Yang J, Visscher PM. 2018. Common disease is more complex than implied by the Core gene Omnigenic model. Cell. 173(7):1573–1580. doi: 10.1016/j.cell.2018.05.051. [DOI] [PubMed] [Google Scholar]
- Wray NR, Yang J, Hayes BJ, Price AL, Goddard ME, Visscher PM. 2013. Pitfalls of predicting complex traits from SNPs. Nat Rev Genet. 14(7):507–515. doi: 10.1038/nrg3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Bakshi A, Zhu Z, Hemani G, Vinkhuyzen AAE, Nolte IM, Vliet-Ostaptchouk V, Snieder H, Lifelines Cohort Study, Esko T et al. . 2015. Genome-wide genetic homogeneity between sexes and populations for human height and body mass index. Hum Mol Genet. 24(25):7445–7449. doi: 10.1093/hmg/ddv443. [DOI] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW et al. . 2010. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 42(7):565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Hong Lee S, Goddard ME, Visscher PM. 2011a. GCTA: a tool for genome-wide complex trait analysis. Am J Human Genet. 88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, Cunningham JM, Andrade M, Feenstra B, Feingold E, Hayes MG et al. . 2011b. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 43(6):519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zeng J, Goddard ME, Wray NR, Visscher PM. 2017. Concepts, estimation and interpretation of SNP-based heritability. Nature Genetics. 49:1304–1310. [DOI] [PubMed] [Google Scholar]
- Yengo L, Sidorenko J, Kemper KE, Zheng Z, Wood AR, Weedon MN, Frayling TM, Hirschhorn J, Yang J, Visscher PM. 2018. Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum Mol Genet. 27(20):3641–3649. doi: 10.1093/hmg/ddy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Vlaming R, Wu Y, Robinson MR, Lloyd-Jones LR, Yengo L, Yap CX, Xue A, Sidorenko J, McRae AF et al. . 2018. Signatures of negative selection in the genetic architecture of human complex traits. Nat Genet. 50(5):746. doi: 10.1038/s41588-018-0101-4. [DOI] [PubMed] [Google Scholar]
- Zhao B, Ibrahim JG, Li Y, Li T, Wang Y, Shan Y, Zhu Z, Zhou F, Zhang J, Huang C et al. . 2019. Heritability of regional brain volumes in large-scale neuroimaging and genetic studies. Cereb Cortex. 29:2904–2914. doi: 10.1093/cercor/bhy157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X-N, Anderson JS, Bellec P, Birn RM, Biswal BB, Blautzik J, Breitner JCS, Buckner RL, Calhoun VD, Castellanos FX et al. . 2014. An Open Science resource for establishing reliability and reproducibility in functional Connectomics. Sci Data. 1(Dec):140049. doi: 10.1038/sdata.2014.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


