Abstract
Eukaroytic RNA-binding proteins (RBPs) recognize and process RNAs through recognition of their sequence motifs via RNA-binding domains (RBDs). RBPs usually consist of one or more RBDs and can include additional functional domains that modify or cleave RNA. Engineered RBPs have been used to answer basic biology questions, control gene expression, locate viral RNA in vivo, as well as many other tasks. Given the growing number of diseases associated with RNA and RBPs, engineered RBPs also have the potential to serve as therapeutics. This review provides an in depth description of recent advances in engineered RBPs and discusses opportunities and challenges in the field.
Introduction
Scientists have worked for many years to understand the complex processing that RNA undergoes within a eukaryotic cell. RNA-binding proteins (RBPs) are key players throughout the life cycle of all RNAs. RBPs are a diverse group of proteins with functions and regulatory roles that include RNA capping, RNA editing, alternative splicing, translation, localization, and degradation of RNA. The data from the growing rise of whole transcriptomic sequencing demonstrates that the number and types of RNAs are immense. This vast number of RNAs serve in a wide variety of functions, that include transcription, splicing, RNA modifications, and translation and do so almost always in the company of RBPs. It is estimated that there are up to 1,500 RBPs in humans revealing that the post-transcriptional regulatory network is complex (Gerstberger, Hafner et al. 2014). These RBPs recognize their respective RNA binding sites via modular RNA-binding domains (RBDs) such as an RNA recognition motif (RRM), double-stranded RNA binding domain (dsRBD), zinc fingers, and many others. These domains recognize RNA through specific RNA sequences and structural motifs or through both modes of recognition (Auweter, Oberstrass et al. 2006, Masliah, Barraud et al. 2013). In general, individual RBDs have modest binding affinity and specificity, such that multiple copies of the same RBD or a combination of different RBDs are often combined to increase affinity and specificity (Figure 1). The modular nature of RBPs and the ability to mix and match RBDs provides a mechanism for RBPs to recognize multiple different sequences and structures to regulate numerous cellular processes. Through a combination of biochemical and structural approaches, an understanding of how many RBDs and RBPs recognize their RNA substrates has emerged (Figure 2). For detailed reviews on this work, see (Auweter, Oberstrass et al. 2006, Masliah, Barraud et al. 2013). Building on the understanding of the natural function of RBPs, researchers are now focusing on designing engineered RBPs with novel specificity and activities.
Figure 1.
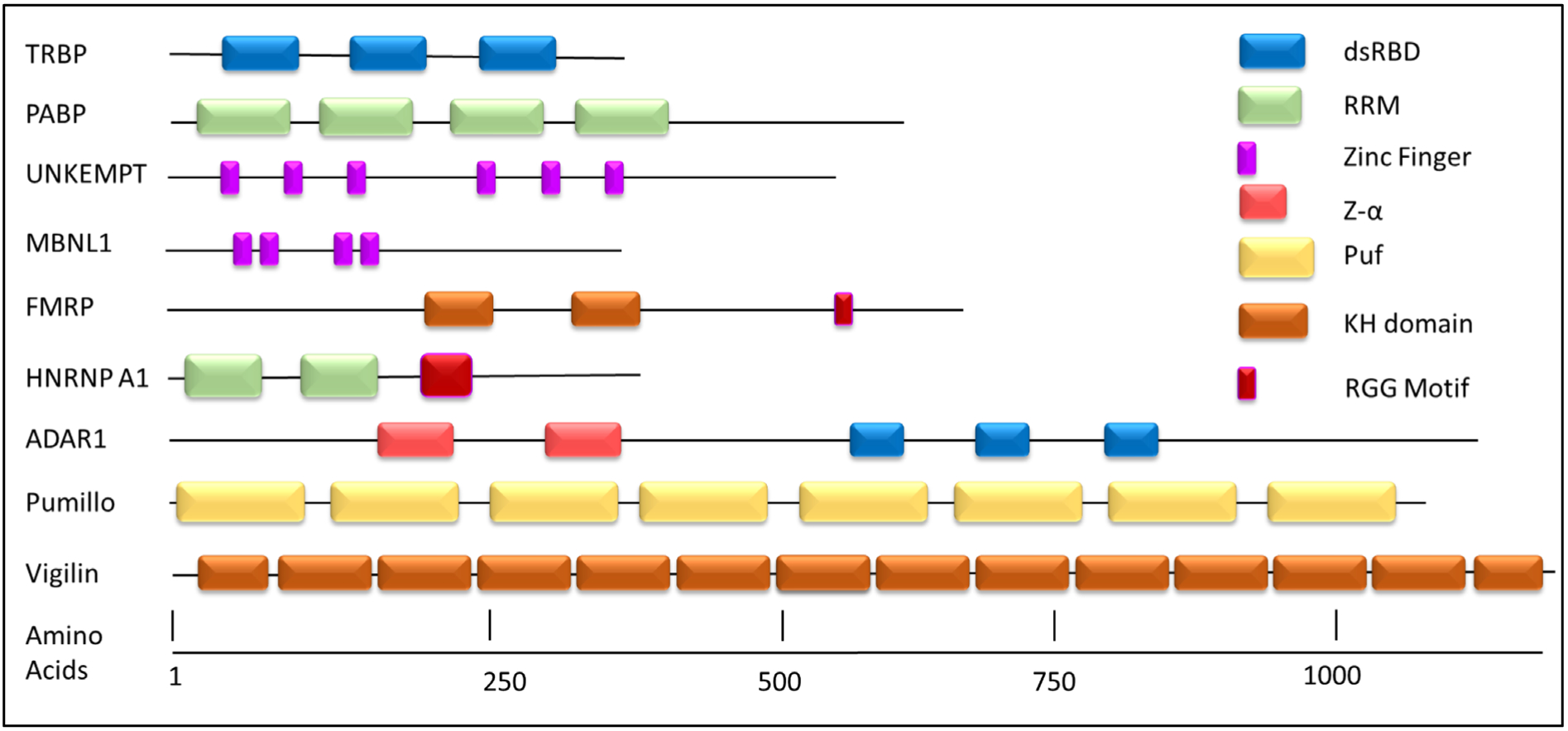
Modular Nature of RNA-Binding Proteins. RNA-binding domains can act in an independent manner and when found in multiple copies can act cooperatively. Proteins are sized according to their amino acid lengths. Domains are represented in block structure.
Figure 2.
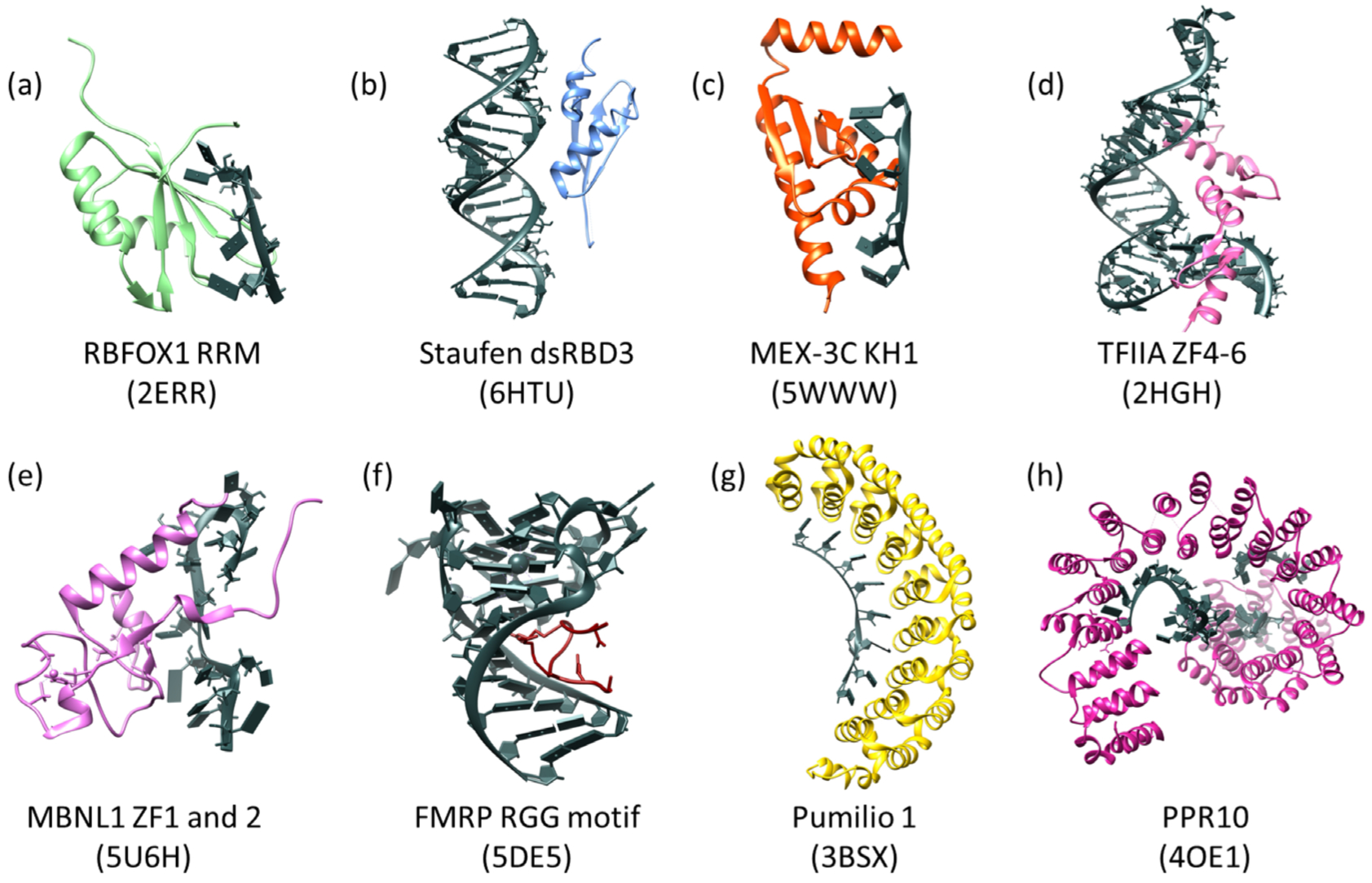
Example RBDs with RNA substrates (shown in dark gray with block nucleotides). (a) The RRM of human RBFOX1 in complex with a 7-mer oligo (UGCAUGU) which interacts through base stacking of aromatic residues and through ionic interactions. (b) DsRBD3 of human Staufen in complex with ARF1 RNA. Recognition through the specific shape of A-form dsRNA and the 2′OH present on the RNA. (c) The KH1 domain of human MEX-3C in complex with a 10-mer RNA oligo. Recognition through hydrogen bonding and shape complementarity. (d) Transcription Factor IIIA zinc fingers 4–6 from Xenopus laevis bound to 5S rRNA (55-mer). Recognition through base stacking of aromatic residues with RNA bases. (e) ZF1 and 2 of human MBNL1 in complex with RNA from Cardiac Troponin T, which interacts with the RNA via base stacking of aromatic residues and hydrogen bonding. (f) The human FMRP RGG motif in complex with G-quadruplex RNA of sc1, which interacts with the RNA via the arginines present in these domains. (g) Human Pumilio 1 in complex with Puf5 RNA. This domain interacts with RNA via base stacking and hydrogen bonds. (h) Zea mays PPR10 in complex with an 18-nt PSAJ RNA element. This domain interacts with RNA similar to PUF domains in that it utilizes hydrogen bonding and base stacking. The PDB entrys are below the names in parentheses for all domains. All panels were made using Chimera (Pettersen EF 2004).
Recent success in engineered DNA-binding proteins (DBPs) have shown the feasibility of designing synthetic proteins to control different aspects of gene expression. Two notable examples are the Zinc finger Transcription Activator-like Effector (TALE) repeat proteins and the CRISPR-CAS system, which have been used to target gene expression and to cleave DNA to add or remove nucleotides (Nomura 2018). The engineered DBP field has progressed to the point that researchers can enter a specific DNA target sequence into an online tool and the program will design a zinc finger protein that specifically recognizes this sequence (Mandell and Barbas 2006). The RBP field has made significant strides in engineering RBPs as summarized in Table 1, but several challenges and opportunities remain as discussed throughout the review. Considerable work has been done with the CRISPR-CAS system to specifically target RNA to address important biological questions and as potential therapeutic strategies (O’Connell 2019, Wang, Wang et al. 2019). Another commonly used technique is to tether proteins of interest to a small viral or bacterieophage RBD to reporter RNAs to study the role of RBPs in RNA metabolism (Coller and Wickens 2002). The MS2 hairpin structure is often placed in the 3’ untranslated region of a reporter RNA to study a protein or region of a protein of interest. See the following review for more information on tethering assays (Bos, Nussbacher et al. 2016). With chose to focus on the successes and challenges in the development of engineered eukaryotic RBPs that target specific RNA sequences and the design of new functions for RBPs. With excellent reviews on this topic available as recently as 2015 (Mackay, Font et al. 2011, Wei and Wang 2015), we have chosen to focus on more recent results in the field of RBP design.
Table 1.
RNA-binding domains and protein engineering attempts.
| RBD | Engineered? | Size (Amino Acids) | RNA Target | Representative References |
|---|---|---|---|---|
| RRM | Yes | 85 | ssRNA, Stem-loop RNA | (Laird-Offringa and Belasco 1995) |
| dsRBD | Yes | 68 | dsRNA (A-form), Stem-loop | (Lee, Cho et al. 2010) |
| PUF | Yes | 36 | ssRNA | (Wang, Wang et al. 2013) |
| Zinc Fingers | Yes | 30 | ssRNA, dsRNA, Stem Loop RNA, Tertiary folded RNA | (Hale, Richardson et al. 2018, De Franco, Vandenameele et al. 2019) |
| KH | No | 70 | ssRNA | (Garrey, Cass et al. 2008, Hollingworth, Candel et al. 2012) |
| RGG | Yes | 8–17 | RNA G-quartet | (Takahama, Miyawaki et al. 2015) |
| PPR | Yes | 35 | ssRNA | (Barkan, Rojas et al. 2012, Okuda, Shoki et al. 2014, Kindgren, Yap et al. 2015, Miranda, Rojas et al. 2017) |
Traditional RNA-Binding Domains
In the following section we discuss some of the traditional RBDs and how they interact with RNA.
RNA Recognition Motif (RRM)
RRMs are the most abundant RBDs in higher vertebrates and are found in over 50% of human RBPs (Maris, Dominguez et al. 2005). Proteins containing these domains function in most posttranscriptional RNA regulatory pathways. The average RRM domain is 85 amino acids long and binds to its target in a sequence-specific manner (Muto and Yokoyama 2012). The canonical structure of an RRM is a βαββαβ topology with the four antiparallel β sheets packing against the two α helices (Figure 2A). The central β sheet of this structure mediates RNA-binding in the majority of cases. There is usually a conserved Arginine or Lysine residue that forms a salt bridge with the backbone of the RNA as well as two conserved aromatic residues that form base stacking interactions with the RNA bases (Muto and Yokoyama 2012). To increase specificity and affinity, multiple RRMs are frequently found in RBPs and the N- and C-terminal regions of RRMs can extend the RRM’s binding site and/or specificity. For example, the C-terminal helix of U1A has been shown to form interactions in the U1A-RNA complex (Oubridge, Ito et al. 1994). Through its canonical structural domain, RRMs in conjunction with other functional domains can bind a complex array of RNAs.
The complexity of RRM-RNA interaction has hampered the design of RRM-containing RBPs. This complexity coupled with the wide range of affinities and specificities of RRMs is likely part of the reason why there is no defined set of RRM amino acids specifically assigned to binding one or more RNA nucleotides (ie a recognition site “code”). However, mutational analyses have revealed how RRMs recognize different RNA sites (Laird-Offringa and Belasco 1995, Melamed, Young et al. 2013). One early study used phage display to identify U1A proteins that had increased affinity for a stem-loop compared to the wild type version of the protein in which it was found that Leucine 49 plays a critical role in RNA binding and a variant of the protein was identified that bound more tightly to the stem loop (Laird-Offringa and Belasco 1995). This type of approach on a larger set of RRMs could prove useful in identifying key amino acid sequences or domain structures that alter the RRMs’ affinity and/or specificity. Interestingly, the linker regions between RRMs form interdomain interactions that affect RNA recognition, presenting an additional challenge to engineering RRM-containing RBPs. The design of synthetic RBPs containing RRMs has proven to be a more complex problem. However, there has been significant mutational analysis and structural studies with the single RRM-containing proteins RBFOX1 and U1A (Auweter, Fasan et al. 2006) (Allain, Howe et al. 1997), providing a potential foundation for tackling the challenge of engineering RRMs.
Double-Stranded RNA-Binding Domain (dsRBD)
Double stranded RNA (dsRNA) is found in many RNAs, including viruses, ribosomal RNAs, pre-miRNAs and throughout many coding and non-coding RNAs. Thus, dsRNA recognition plays a critical role in many cellular processes and one mechanism to accomplish this recognition is through the use of a dsRBD domain. The average dsRBD is 68 amino acids in length and typically adopts an αβββα conformation. The two α helices interact to fold into a “y” shape that packs against the three antiparallel β sheets (Figure 2B). There are two subclasses of dsRBD’s known as “type A” and “type B.” Type A dsRBDs contain the canonical binding domain (St Johnston, Brown et al. 1992) whereas Type B is highly conserved at the C-terminus but not at the N-terminus. Type-B dsRBDs are also referred to as Hal domains and have poor dsRNA binding compared to type-A . The Type B dsRBDs are present in several proteins, cooperate with Type A dsRBDs to weakly bind dsRNA and have roles in protein-protein interactions (St Johnston, Brown et al. 1992, Krovat and Jantsch 1996). In general, dsRBDs recognize the shape of dsRNA and do not bind in a sequence-specific manner. The dsRBD recognizes A-form dsRNA through the recognition of the 2’OH present on the ribose sugar and the shape of the minor and major grooves (Ryter and Schultz 1998). There are three regions of the dsRBDs involved in RNA interaction: the α1 helix, the N-terminal region of α2, and the loop that connects β1 and β2. An A-form dsRNA minor groove 2’ OH is recognized by the α1 helix loop 2 and α1 regions through direct and water mediated hydrogen bonds (Ryter and Schultz 1998). The N-terminal α2 region is rich in arginine and lysine residues that specifically recognize the width of the major groove of the dsRNA (Ramos, Grünert et al. 2000). Finally loop 2 inserts near the minor groove of the RNA and the conserved Histidine 31 forms a direct hydrogen bond with the 2’OH present on the ribose (Masliah, Barraud et al. 2013). Despite the general lack of sequence-specific binding, there are cases of base-specific recognition by dsRBDs such as those present in ADAR2 that specifically edit RNA (Stefl, Oberstrass et al. 2010). Furthermore when found in tandem, dsRBDs may also have independent and new functions (Nanduri, Carpick et al. 1998, Stefl, Oberstrass et al. 2010). For example, in combination TRBP’s dsRBD1 and dsRBD2 slide along dsRNA in an ATP-independent manner, and removal of one domain results in a loss of this function (Koh, Kidwell et al. 2013). Overall dsRBDs play a key role in RNA regulation and consistent with this role, dsRBDs have been shown to be present in prokaryotes (Masliah, Barraud et al. 2013) as well as in the last common ancestor of metazoans (Kerner, Degnan et al. 2011). DsRBD-containing proteins have a wide range of specific activities on RNA including degradation, transport, editing, and localization (Masliah, Barraud et al. 2013). DsRBDs have been shown to have global impacts, such as the binding of viral RNA by TRBP which activates a stress pathway resulting in global translational arrest (Gatignol, Lainé et al. 2005).
Limited engineering studies have been published on dsRBDs, likely due to the lack of sequence specificity. However, there was one study that used the rare, nontraditional PAZ domain RBD found in the Argonaut and Dicer proteins that bind the 3’ end of siRNA and miRNA (Hutvagner and Simard 2008). In this work, the authors fused the PAZ domain with a dsRBD to detect hybridized microRNAs on array surfaces (Lee, Cho et al. 2010). Even though this work has not been followed up on, dsRBDs have the potential to be powerful tools in engineered RBPs. Future studies could include using dsRBDs in combination with other RBDs that are sequence-specific, meaning the RBP could recognize both dsRNA and ssRNA regions in a highly structured RNA and possibly modulate the structure of the RNA so both domains could bind.
K Homology (KH) Domain
KH domains have been shown to bind ssRNA and ssDNA substrates and are found in proteins with numerous cellular functions, including transcriptional and translational regulation. These domains are on average 70 amino acids (Figure 2C) and are found in archaea, bacteria, and eukaryotes. The loss of function in KH domains is associated with several diseases, such as fragile X mental retardation syndrome (Zhang, O’Connor et al. 1995) and paraneoplastic disease (Buckanovich, Yang et al. 1996). As with other RBDs, multiple KH domains are frequently found in proteins such as the 14 domains found in Vigilin (Figure 1). There are two distinct types of KH domains that share the same secondary structure but fold differently (Grishin 2001): Type I KH which have a βααββα topology and type II KH domains which have an αββααβ topology. The RNA-binding surface of both types is formed by a GXXG loop, two consecutive α helices, the terminal β strand, and the variable loop (Lewis, Musunuru et al. 2000). The binding site on KH domains is unique in that they do not use aromatic residues to interact with ssRNA like other RBDs but instead exclusively use hydrogen bonding and shape complementarity. For example, MEX-3C, which regulates the degradation of mRNAs through the 3’ UTR, has two KH domains that were shown to lack base stacking interactions when crystallized with RNA (Figure 2.) (Yang, Wang et al. 2017). The KH domain-RNA interaction can be quite complex and affect the overall structure of complex RNAs (Nicastro, Taylor et al. 2015).
The specificity of the KH domain’s RNA recognition has presented challenges to engineering attempts with this domain. While significant mutational analyses have been done on KH domain-RNA interactions (Siomi, Choi et al. 1994), no general recognition code for how these domains recognize RNA has been revealed. However an engineering study using chimeric KH domains revealed important amino acids for specific RNA recognition (Garrey, Cass et al. 2008). Additional studies demonstrated that mutating the important GxxG loop to GDDG resulted in a KH domain that was unable to bind RNA but maintained the typical fold of the domain (Hollingworth, Candel et al. 2012). A similar approach applied to other KH domains will provide further information on how this RBD recognize RNA and will aid in future engineering attempts. Given that multiple KH domain amino acids are important for sequence-specific RNA-binding and domain folding as well as the fact that amino acids outside of the defined KH domain are also important for RNA-binding, engineering sequence specificity will likely be a complex task for KH domains.
Zinc finger (ZF) Domains
Zing fingers (ZF) are a small protein motif characterized by the presence of one or more zinc (Zn2+) ions. There are a number of different types of zinc finger domains, each with a specific architecture and specific engineering advantages and challenges. Several relevant RNA-binding ZF domain sub-categories are described below:
CCHH ZF domains
The classical Zinc finger (ZF) domain has approximately 30 residues and contains two conserved histidine residues and two conserved cysteine residues (ie CCHH) that coordinate a zinc ion. This configuration allows the domain to fold around the zinc ion into a small β sheet and an α helix (Pavletich and Pabo 1991). Examples of this class of zinc finger are the nine fingers present in TFIIIA, a transcription activator the binds both 5S rDNA and 5S rRNA and increases transcription of the 5S rRNA gene (Figure 2D). The TFIIA ZF domains are versatile in that they can interact with RNA by recognizing both structural and sequence elements of the RNA substrates (Lu, Alexandra Searles et al. 2003). The fifth ZF of TFIIIA interacts with the phosphate backbone of a double helical region, recognizing a unique helical structure. In contrast, ZFs four and six recognize specific nucleotides that are exposed and presented in the folded RNA structure (Friesen and Darby 1997). No CCHH RBPs have been engineered to date, possibly due to the fact that many can bind DNA as well as RNA.
CCCC (Ran-BP2) domains
The CCCC domain ZFs are found in organisms from fungi to humans and like the other ZFs are on average 30 amino acids in length. In humans, there are approximately 30 proteins that contain this domain compared to 56 proteins that contain a CCCH domain (Nguyen, Mansfield et al. 2011). The CCCC domains fold into two distorted β-hairpins on either side of a centralized tryptophan and are stabilized by a single zinc ion. They were first discovered in ZRANB2, a human splicing factor that is proposed to recognize 5’ splice sites (Plambeck, Kwan et al. 2003). The ZFs in this protein bind to ssRNA with micromolar affinity and specifically recognize “GGU” (Plambeck, Kwan et al. 2003). This interaction occurs by hydrogen bonds between the protein residues and the nucleotide bases that form a guanine-tryptophan-guanine ladder (Loughlin, Mansfield et al. 2009). Interestingly there are no interactions observed between the ZF and the backbone of RNA. This lack of backbone interaction shows that unlike other ZFs, CCCC zinc fingers do not appear to depend on a specific conformation of the RNA (Nguyen, Mansfield et al. 2011).
This ZF is well suited for protein engineering because it does not require a specific RNA conformation for recognition. However this ZF domain is limited to a short sequence recognition site, meaning larger RNAs cannot be targeted by one domain and multiple domains must be linked together to resolve this issue. Recently, Ran-BP2’s multiple copies of this ZF domain were used as a starting point to examine the sequence specificity of the domain and their suitability for RBP engineering (De Franco, Vandenameele et al. 2019). In this study the authors linked together several different Ran-BP2 ZFs as well as ZFs from different families and showed that the engineered proteins could target a long RNA sequence in a sequence-specific manner with 25 nM affinity. As this work was done in vitro and in bacteria, it will be interesting to determine if these engineered proteins will respond similarly in vivo in more complex systems.
CCCH Zinc Finger (ZF) domains
So far 56 proteins that encode CCCH ZFs have been discovered in humans, and like the other ZF domains, have an average size of 30 amino acids (Liang, Song et al. 2008). These proteins, which are generally involved in either RNA metabolism or immune response, often contain one or more ZFs as well as other functional domains (Fu and Blackshear 2016). An example of a CCCH-ZF domain containing protein is MBNL1 (Figure 2E), a master regulator of RNA processing. MBNL1 contains four CCCH ZFs that folds into two domains, with on average 60 amino acids in both domains and two zinc ions in each domain (Teplova and Patel 2008, Park, Phukan et al. 2017). The ZFs of MBNL binds to YGCY (Y= C or U) motifs through base stacking with aromatic and non-aromatic residues (Phenylalanine, Tyrosine, Tryptophan, Leucine and Isoleucine) and hydrogen bonding through multiple backbone amides and side chains of residues in the zinc fingers (Park, Phukan et al. 2017). MBNL1 has been well studied because of its role in the RNA splicing defects associated with diseases such as Myotonic Dystrophy type 1 (DM1) and type 2 (DM2), Spinocerebellar Ataxia type 8, and Fuchs Endothelial Corneal Dystrophy (Du, Cline et al. 2010, Fernandez-Costa, Llamusi et al. 2011, Du, Aleff et al. 2015). This connection with disease pathogenesis provides an impetus for designing engineered CCCH zinc fingers with altered or modified functions.
Several engineered RBPs have been developed using these types of ZFs including multiple based on the MBNL1 protein (Hale, Richardson et al. 2018). Previous work by the same group has shown that the first two zinc fingers (ZF1–2) were more responsible for the protein’s RNA-binding and alternative splicing regulation functions than the last two ZFs (ZF3–4) (Purcell, Oddo et al. 2012). Building upon this concept, the 2018 study used a rational design method to replace ZF3–4 with a second ZF1–2 domain in one protein and in another protein replaced ZF1–2 with a ZF3–4 domain. This design strategy allowed the authors to determine the activity of the respective domains (discussed further in functional domain section) (Hale, Richardson et al. 2018). That ZFs of this class can be added or deleted from RBPs to modulate activity has implications for the engineering of other CCCH-ZF containing proteins.
RGG Domain
The arginine/glycine rich (RGG) domains are made up of repeats of the RGG motif with linkers of variable length. The arginines of these domains have been shown to mediate hydrogen bonding and base stacking with both RNA and DNA (Figure 2F). The RGG domain of hnRNP U was one of the first RGG domains shown to bind RNA (Kiledjian and Dreyfuss 1992) and has subsequently been shown to bind G-quadruplexes and increases their stability (Hanakahi et al.; 1999, Schaeffer et al., 2001). In general, this domain binds both primary and secondary nucleic acid structures. Although the primary function of RGG domains has been nucleic acid binding, these domains have also been shown to mediate protein-protein interactions, such as the RGG domain of FMRP, which interacts with Ran binding proteins (Menon et al., 2004). Proteins with RGG domains have been shown to have roles in protein localization, alternative splicing, translational repression, regulation of apoptosis, transcriptional regulation, and DNA damage signaling (reviewed in Thandapani et al 2013). Misregulation or loss of expression of proteins with RGG domains have been shown to be important in a number of human diseases, including Fragile X Mental Retardation Syndrome (Verkerk et al., 1991), Amyotrophic lateral sclerosis (Hoell et al., 2011; Kwiatkowski et al., 2009), Spinal muscular atrophy (Côté and Richard, 2005), macrocephaly (Field et al., 2007), autism spectrum disorders (Sato et al., 2012), Ewing Sarcoma (Araya et al., 2005), and multiple types of cancer (Destouches et al., 2008; Watanabe et al., 2010; Krust et al., 2011).
A major challenge for using the RGG domain in engineering is that it recognizes both RNA and DNA g-quadruplexes. Only one engineered RGG protein that binds RNA, based on the RGG domain of FUS (Takahama et al., 2015), has been developed. FUS is a protein that binds to RNA and interacts with other proteins to regulate transcription, alternative splicing and other aspect of RNA processing as well as DNA damage regulation (Bertolotti, Lutz et al. 1996, Wang, Arai et al. 2008, Schwartz, Ebmeier et al. 2012, Tan, Riley et al. 2012). FUS has been shown to interact with the G-quadruplex of TERRA, or telomeric repeat-containing RNA, which is a non-coding RNA transcribed from telomeres and regulates histone modifications of telomeres (Takahama, Takada et al. 2013). In the engineering study, the RGG domain was mutated to RGGY to specifically bind and stabilize the G-quadruplex of telomeric repeat RNA. This mutation allowed the RGG domain to specifically recognize the RNA and was used to probe how TERRA regulates histone modifications (Takahama, Miyawaki et al. 2015). Another more recent study used RGG domains fused to elastin-like polypeptides to develop RBPs with tuneable phase behaviour in protocells in an effort to study RBPs and RNA granules and showed that they granules can inhibit translation through either reversible or irreversible sequestration of mRNA (Simon, Eghtesadi et al. 2019). Taken together these studies demonstrated that RGG boxes can be engineered for a variety of functions.
Pumillo Family (PUF) of RNA-binding Proteins
The Pumillo family (PUF) of proteins are a unique group of proteins that tend to bind to the 3’UTR’s of their target RNAs and have important roles in stem cell maintenance and memory (Schweers et al. 2002; Wickens et al. 2002). The canonical PUF protein is made up of eight domains or repeats, and each domain contains three imperfect alpha helices that fold together, as illustrated by Pumillo 1 (Figure 2G). Each domain is made up of 36 amino acids that recognize one nucleotide and the domain is repeated multiple times to recognize different sequences with the length of the RNA site generally correlating with the number of PUF domains. RNA binds on the concave face of this domain and interacts via hydrogen bonds and base stacking with the helices present on the face in an antiparallel fashion (Wang et al. 2002). Establishing an RNA recognition code for the PUF domain has been the work of many labs (Wang, McLachlan et al. 2002, Cheong and Hall 2006, Dong, Wang et al. 2011, Koh, Wang et al. 2011, Campbell, Valley et al. 2014). At first glance this code can be considered relatively straightforward: In the PUF domain, the 12th and 16th positions determine which RNA bases are recognized: (1) If glutamate and serine are present in the 12th and 16th position guanine is recognized, (2) if glutamine and cysteine or serine are present adenine is recognized, (Mandell and Barbas) and if glutamine and asparagine are present uracil is recognized (Wang, McLachlan et al. 2002, Cheong and Hall 2006) (Figure 3). Given the relative ease of using this established RNA recognition code, PUF domains have been widely used for protein engineering. The application of PUF domains for protein engineering was greatly enhanced by the development of a synthetic PUF domain that recognizes cytosine (Dong, Wang et al. 2011, Filipovska, Razif et al. 2011). Placing serine at position 12 and an arginine at position 16 was shown to specifically bind cytosine (Dong, Wang et al. 2011, Filipovska, Razif et al. 2011), although it is important to note that this code is not universal to all PUF scaffolds (Campbell, Valley et al. 2014).
Figure 3.
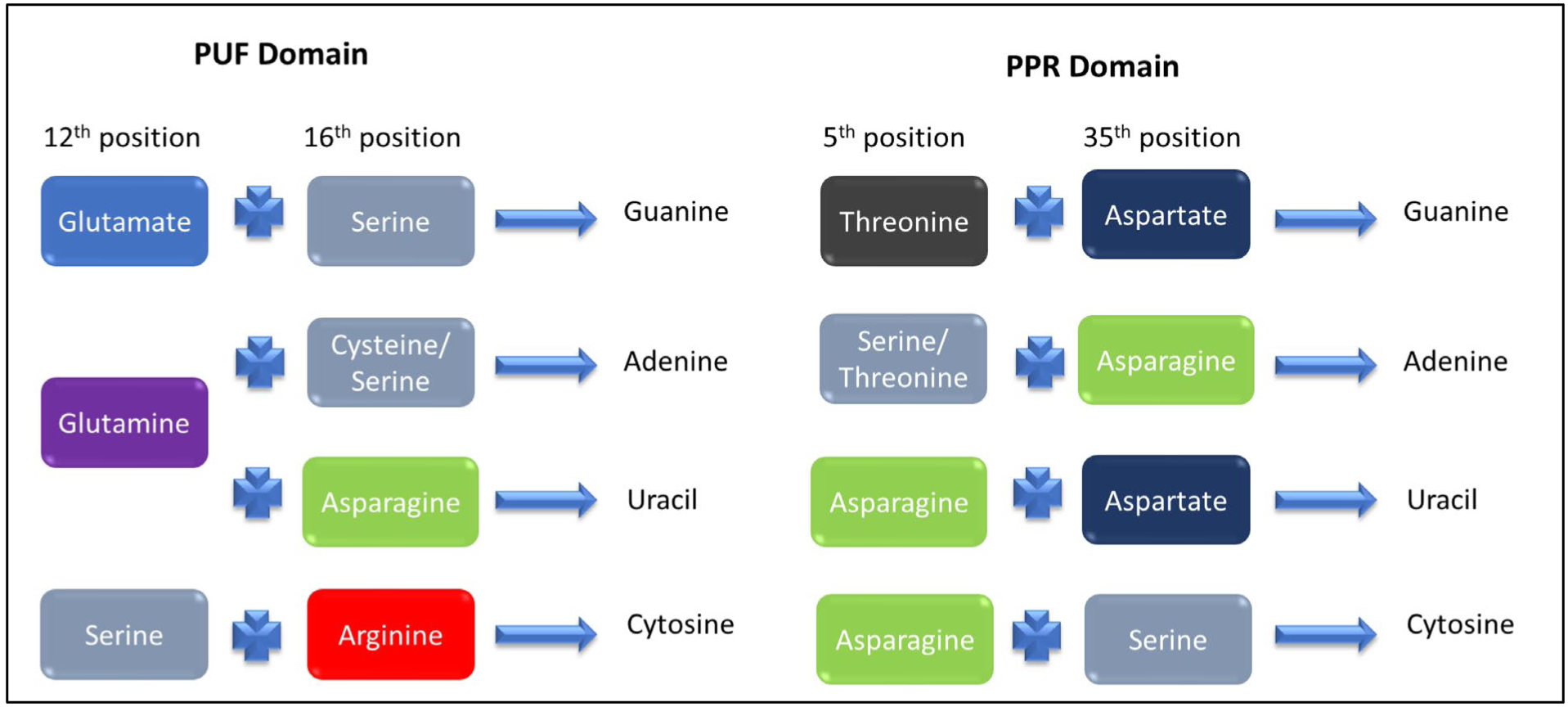
The recognition code for the PUF and PPR repeat domains. The specific amino acids at these positions in each repeat of the domain specify which RNA nucleotide is recognized. Domains can be built from this code to recognize specific sequences of RNA with caveats as discussed in the text.
Despite the ease of use and established recognition code, there are several complicating factors. PUF proteins can flip out undesirable nucleotides, meaning that some engineered PUF proteins do not always bind the predicted linear sequence (Gupta, Nair et al. 2008). Campbell et al. went into rigorous detail on the mechanisms used by domains in fungal PUF proteins and demonstrated that subtle differences in the packaging of the repeats and the backbone of the protein can have large affects on the specificity of these proteins (Campbell, Valley et al. 2014). Despite these complications, engineered PUF proteins have been developed to regulate the translation, localization, stability, and processing of RNA (Wang, Wang et al. 2013) and will be further discussed later in the review.
Pentatricopeptide (PPR) Proteins
Pentatricopeptide (PPR) containing proteins are a family of α-helical proteins that are primarily found in plants and function in the expression and regulation of chloroplast or mitochondrial genes. PPR-repeat proteins have been shown to have anywhere from 2–30 PPR repeats or domains, with an average domain size of 35 amino acids. PPR10, for example, has 19 PPR domains (Figure 2H). PPR domains, like PUF domains, are made of a scaffold of α-helices where each domain recognizes a specific nucleotide (Wang, McLachlan et al. 2002). PPR10 uses PPR domains 3–19 to recognize a sequence of 17–18 nucleotides (Barkan, Rojas et al. 2012). Using the sequence-specific binding of these domains, PPR protein can recognize and bind a wide variety of sequences. Each PPR domain folds into a helix-turn-helix that when repeated, fold together to form a righthanded super helix (Figure 2H) (Small and Peeters 2000, Cheng, Gutmann et al. 2016). A PPR recognition code (Figure 3) has also been developed after it was discovered that amino acids in two positions in the PPR motif determine which nucleotide is recognized (Figure 3) (Barkan, Rojas et al. 2012, Yagi, Hayashi et al. 2013, Shen, Zhang et al. 2016).
The established “PPR code” has led to the extensive use of this domain in engineering RBPs both to reprogram native proteins (Barkan, Rojas et al. 2012, Okuda, Shoki et al. 2014, Kindgren, Yap et al. 2015, Miranda, Rojas et al. 2017) and to generate proteins with customized specificity (Coquille, Filipovska et al. 2014, Shen, Wang et al. 2015, Shen, Zhang et al. 2016). Recent work has shown that ten contiguous repeats are sufficient to reach maximal binding affinity and that purine-PPR domain interactions appear to be more important than pyrimidine-PPR domain interactions. Additionally, experiments have suggested that the recognition of the 5’ end of the RNA may be more important for maximizing affinity (Miranda, McDermott et al. 2018). Refinements to the PPR code were recently made through the study of additional PPR proteins (Yan, Yao et al. 2019), yielding a more accurate code and the web-based server platform PPRCODE to facilitate domain design. As with the PUF domains and most RBDs there are limitations in engineering PPR domains. It has been shown that a degree of mismatching can be acceptable between certain native PPR domains and their RNAs, meaning that the code is not always definitive (Barkan, Rojas et al. 2012, Kindgren, Yap et al. 2015, Miranda, Rojas et al. 2017). Yin et al added another layer of complication when they showed that PPR10 can use both the canonical amino acid code to recognize RNAs and an alternative recognition mechanism (Yin, Li et al. 2013). Furthermore, Miranda et al. later demonstrated that PPR10’s amino acid code was not sufficient to predict where it binds in the chloroplast transcriptome (Miranda, Rojas et al. 2017). This work reveals that when engineering PPR or PUF proteins, the recognition codes cannot be completely relied upon for predicting their RNA-binding sites in vivo.
Intrinsically Disordered Regions
Intrinsically Disordered Domains, defined as regions lacking stable three-dimensional structures under physiological conditions, have been shown recently to have increasing importance in RBPs (Habchi, Tompa et al. 2014, Järvelin, Noerenberg et al. 2016). Several studies have found that there are dozens of nontraditional RBPs with intrinsically disordered domains, some of which may be proteins that moonlight with multiple functions (Baltz, Munschauer et al. 2012, Castello, Fischer et al. 2012). In vivo work showed that of the ~170 RBPs discovered, 20% of those proteins were primarily disordered proteins. These disordered regions are enriched in hydrophobic residues and charged residues and tyrosines (Castello, Fischer et al. 2012, Kwon, Yi et al. 2013). These residues are typically found in the interacting surfaces of traditional RBDs supporting the model that these regions interact with RNA. Disordered regions are emerging as multifunctional RNA-binding modules that can have non-specific to highly selective binding targets. The disorder of these domains may also endow special properties to the parent RBP. For example, the C-terminal region of RBFOX1 contains a disordered domain that when tethered to MS2 was sufficient to promote alternative exon inclusion of RBFOX1 regulated events (Sun, Zhang et al. 2012). While these domains have interesting functions, currently they are difficult to work with because of their disorder and inability to predict their function. As more of these regions and their functions are studied, they can be utilized in the development of engineered RBPs to provide new functions.
Engineering RNA-Binding Proteins
Traditional and engineered RBDs and their ability to interact with RNA form the basis for the design and engineering of RNA-binding proteins. There are a number of important considerations in designing an RBP including selection of binding domain, target function, linker region(s), and limiting off-target effects. While these factors need to be taken into account, the intended function of the RBP is a primary driver of RBP design. Ideally, the protein needs to be relatively small and have a well-defined activity in order to limit the off-target effects and to facilitate delivery. Other design factors to be considered are discussed below and can include: (1) which RBD(s) to use, (2) interdomain linker, (Mandell and Barbas) domain orientation, (4) cellular location and cell delivery, and (5) adding other domains with specific activities (Figure 4).
Figure 4.
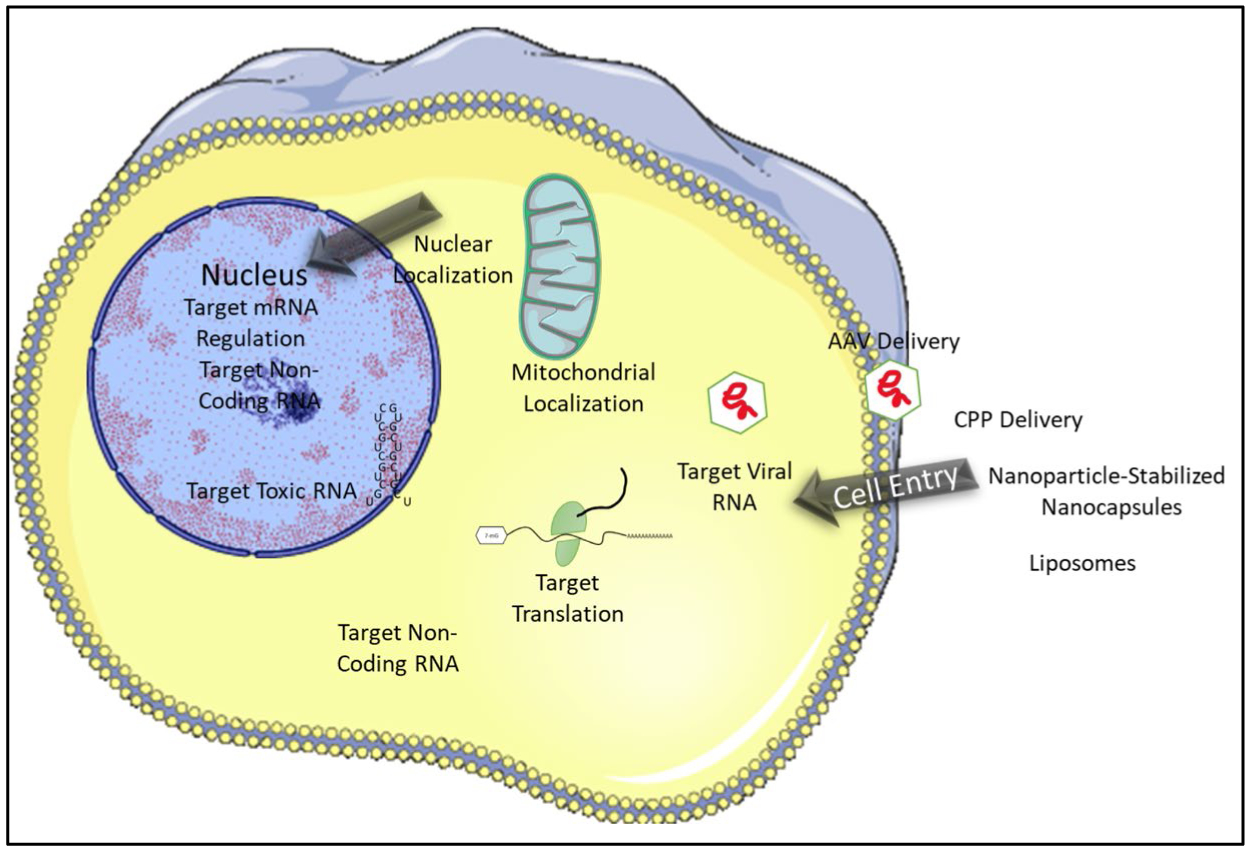
Building Blocks of Engineered RNA Binding Proteins. When designing RBPs as tools and therapeutics the tissue target, cell entry methods, and the localization of the RNA target also need to be considered. Stock images adapted from Servier Medical Art
1. Choosing a Binding Domain
There are two main strategies for binding domain selection when developing an engineered RBP: (1) combining different RBDs that recognize specific RNA sequences and (2) re-engineering existing RBDs to bind specific RNA sequences. The first method is an excellent way to target longer RNAs and/or RNAs with complex secondary/tertiary structure as it builds upon existing knowledge of RBD structure and function. This approach mimics nature, where many endogenous proteins have evolved to bind complex RNA sequences and structures by employ a combination of RRM, RGG, KH, dsRBDs, and ZFs in a modular fashion. The lack of sequence-specificity or our ability to engineer specificity for many RBD domains does hamper this type of approach. This challenge can potentially be overcome with high throughput screens or in vitro evolution methods to select the desired specificity. For the latter, phage display has been used on the U1A RRM to tailor RNA-binding to the TAR RNA (Crawford, Blakeley et al. 2016). The second method for RBD design is to use PUF and PPR domains and their established recognition codes to engineer novel RBPs to bind specific sequences of interest. As previously described, significant advancements have been made with PUF and PPR domains to allow the design of RBPs with the desired sequence specificity. Primary challenges that remain with this approach are that it takes 35 or 36 amino acids to recognize one nucleotide meaning that a PUF/PPR engineered protein that recognizes 8 nucleotides will be 30 kD, which can limit delivery or packaging of the engineered protein. Additionally PUF/PPR domains recognize only single-stranded RNAs and not structured RNAs further limiting their target. Recent work has indicated that PUF domains reach maximum binding affinity at 9 or 10 repeats (Wang and Ye 2017, Zhao, Mao et al. 2018) and decline thereafter. These limitations mean that it will be challenging to engineer PUF/PPR domains to recognize RNA sequences greater than 10 nucleotides and/or for structured RNAs. The choice of binding domain selection strategy is most often driven by the specific question being asked and what is known about the target RNA.
Interdomain Linkers
When engineering proteins with multiple domains, the design of the interdomain linker(s) is important. A naturally occurring linker region is typically 3–25 amino acids (George and Heringa 2002, Chen, Zaro et al. 2013) Apart from simply acting as a spacer to maintain separation between functional domains, Linker regions can have many other functions including: modulating the activity of an RBD; moderating protein-protein interactions; and/or controlling the movement of domains by acting as hinge elements (Gokhale and Khosla 2000, Wriggers, Chakravarty et al. 2005, Reddy Chichili, Kumar et al. 2013). Given that the linker region can function beyond serving as a simple linker joining RBDs, testing how the linker affects the desired activity is important. Another important factor that needs to be considered is the flexibility or rigidity of the linker. An overly flexible linker (usually Gly rich) could lead to the development of an RNA-independent function of any attached functional domain, while an overly rigid linker can prevent the RBDs from adopting favorable conformations for binding RNA (Chen, Zaro et al. 2013). The linker can also affect the solubility of the engineered RBP so it is important to consider solubility when choosing between natural or engineered linkers. Given the importance of linker design and selection, there are a number of linker databases (George and Heringa 2002) and tools for linker prediction or modeling (Crasto and Feng 2000, Samant, Hulgeri et al. 2012) to assist in the overall design of the engineered RBP. Ultimately as adding any region to a protein can have unpredicted functions, testing the function of the linker region within an engineered RBP in vivo will likely be required for successful RBP engineering.
2. Domain Organization
How the RBDs are arranged relatively to each other and other domains is another important factor to consider when engineering RBPs. The order and arrangement of domains can influence individual domain function and/or overall protein function. For example, in one study, investigators tested two versions of engineered RBPs; one with an N-terminal PUF domain and C-terminal PIN endonuclease and another with the order of the two domains reversed (N-PIN-PUF-C) (Choudhury, Tsai et al. 2012). This simple switch changed the engineered RBP from a site-specific cleaver of RNA for the former to a nonspecific cleaver of RNA for the latter (Choudhury, Tsai et al. 2012). Thus, when designing RBPs for desired specific activities it is important to test different arrangements of domains.
3. Cellular location and cell delivery
The delivery of an engineered protein to cells has proved difficult especially in the early days of the field. When delivering an engineered RBP in cell culture (Figure 5), there are many different delivery techniques available such as lentiviral expression, electroporation, and cell-penetrating peptides (Guidotti, Brambilla et al. 2017, Jauset and Beaulieu 2019). There is an excellent review that goes into the details of intracellular protein delivery for therapeutics (Bruce and McNaughton 2017). When delivering proteins as therapeutics, cell type specific and cell-penetrating peptides (Bruce and McNaughton 2017, Guidotti, Brambilla et al. 2017) or adeno associated viruses (AAVs) are good candidates. AAV is the leading method for gene therapy delivery in human disease. Recent advancements in recombinant AAVs have been able to evade the host immune system and help to further improve the tissue specificity of AAV serotypes (Barnes, Scheideler et al. 2019, Wang, Tai et al. 2019). An interesting possibility is that if an engineered protein has broad activity, the off-target effects could be limited by targeted delivery of the protein. One drawback is the size that can be packaged (~5kb total); however, many engineered proteins can easily fit this size range. Other methods that could be utilized are liposomes, Nanoparticle-stabilized nanocapsules, and Fusogenic liposomes (Ray, Lee et al. 2017). This is a rapidly developing field and continued improvements for the delivery of proteins is expected in the future and will be useful for the delivery of engineered RBPs.
Figure 5.
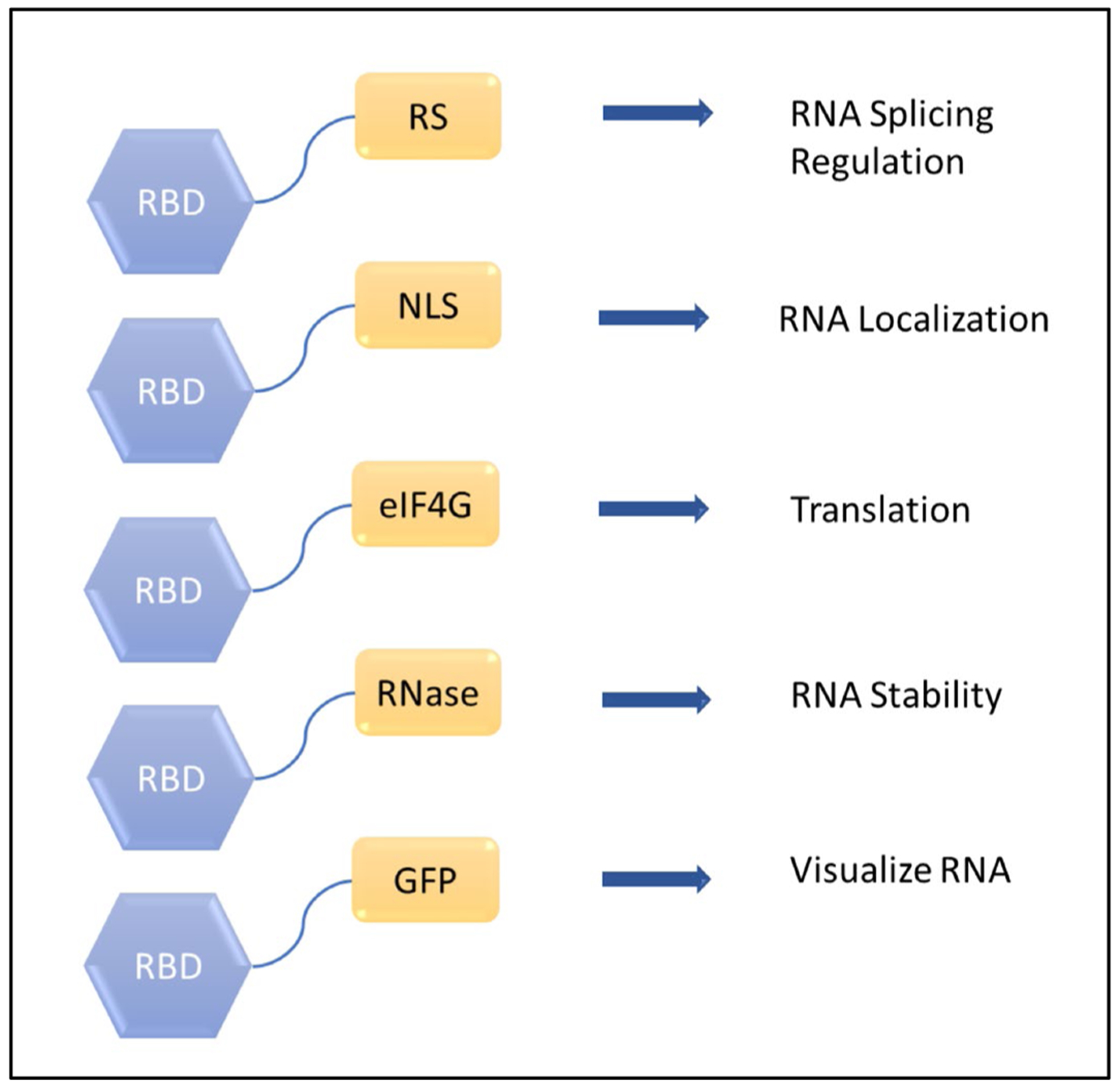
Adding Functional Domains to the RBD. After the specific RBD for the engineered protein has been chosen, a functional domain can be added for functionality.
4. Additional functional domains and example of engineered proteins
Different functional domains can be added to the RBD protein to create specific and new functions beyond simple RNA binding to include modulate of almost any aspect of RNA function (Figure 6). Examples of these functions and associated engineered proteins are included below:
RNA Splicing Factors
Most human transcripts undergo alternative splicing to produce multiple isoforms of a gene with distinct activities. RBPs tightly regulate this activity and serious diseases can occur when the process is mis-regulated (Douglas and Wood 2011). These RBPs recognize and bind short sequences in pre-mRNAs that function to enhance or silence alternative exons. Many natural splicing regulators contains one of the RBDs discussed above with an additional function domain such as an arginine/serine-rich (RS) domain or a glycine (Gly)-rich domain that promote exon inclusion or exclusion (Graveley and Maniatis 1998, Del Gatto-Konczak, Olive et al. 1999). The importance of splicing regulation has motivated researchers to engineer RBPs that can be used to control alternative splicing.
An example of this approach is the fusion of a PUF domain to the RS domain of SRSF1 (PUF-RS) or the Gly domain of hnRNPA1 (PUF-Gly) to study splicing regulation (Wang, Cheong et al. 2009). The engineered PUF-RS domain promotes exon inclusion when bound within an alternatively regulated exon but promotes exon exclusion when it is bound downstream. In contrast, the engineered PUF-Gly protein promotes exon inclusion when binding both within the alternatively regulated exon and downstream of the exon. By altering the isoforms of specific transcripts with this approach, several types of cancer cells were able to be sensitized to anticancer drugs (Wang, Cheong et al. 2009). A similar approach has been used to control splicing through engineered proteins with RS domains and other types of RBDs.
Another example of an engineered RNA splicing factor can be found in the recent study that used a rational design method to replace ZF3–4 of MBNL1 with another copy of ZF1–2 or ZF1–2 with a ZF3–4 domain (Hale, Richardson et al. 2018). The former protein with two ZF1–2’s showed a 5-fold increase in activity compared to wild type MBNL1, and the latter protein with two ZF3–4 domains had a 4-fold decrease in activity (Hale, Richardson et al. 2018). The double ZF1–2 protein also showed rescue of MBNL1 regulated alternative splicing events in a Myotonic dystrophy type 1 disease model (Hale, Richardson et al. 2018). While this approach to engineering an RNA splicing factor produced some promising results, it requires an intimate knowledge of a protein’s domain structure and functions, limiting its usefulness in the design of other splicing factors.
RNA Endonuclease
Artificial site-specific RNA endonucleases (ASREs) were developed by combining a PUF domain with the PilT N-terminus (PIN) nuclease domain of the nonspecific SMG6 endonuclease (Choudhury, Tsai et al. 2012). Because the PUF domain can be engineered to bind any sequence, ASREs are an attractive choice when targeting a specific RNA for research or therapeutics. An example of this approach is the engineered ASRE designed to specifically bind the DM1 toxic expanded RNA (Zhang, Wang et al. 2014). This study was able to significantly reduce the amount of toxic RNA in cell culture by using a custom-designed RNA endonuclease to specifically bind and cleave (CUG)n repeats (Zhang, Wang et al. 2014). Engineered ASREs have also been used to study mitochondrial RNA processing in trypanosomes, the protozoa that cause human African sleeping sickness. This approach specifically targeted an essential transcript involved in ATP synthesis in the trypanosome such that expression of the ASRE was shown to be lethal to the trypanosomes but not the host (Szempruch, Choudhury et al. 2015). Some potential challenges with RNA endonuclease design are the possibility of cleaving off-target RNAs, especially when the PUF/PPR domain targets shorter nucleotide sequences (8–11 nucleotides) which may not be large enough for specific binding. Combining the PUF domains with a different type of RBD might overcome the length limitation.
Translation Regulators
Engineered RBPs have been used to regulate translation through the addition of domains present in known translational regulation proteins. In an example of this approach, researchers combined a PUF domain with a GLD2 translation activation domain of a CAF1 translation repression protein (Cooke, Prigge et al. 2011). The subsequent engineered protein binds to the 3’ UTR of a specific RNA via the PUF domain and elicits poly(A) addition or removal via the GLD2 translation activation domain. A similar approach has been used by tethering eIF4e to a PUF domain to activate translation through increased translation initiation (Cridge, Castelli et al. 2010, Blewett and Goldstrohm 2012). An interesting variant on this approach is the blue light-inducible system for the assembly of an NLS-deficient truncated version of the CIB1 protein (CIBN) fused to a PUF domain with an N-terminal photolyase homology region of CRY2 fused to eIF4e protein. The presence of blue-light induces heterodimerization of CRY2PHR and CIBN, thereby translocating eIF4E to the target mRNA and initiating translation. This system was able to increase the expression of the luciferase reporter gene by over 17-fold expression (Cao, Arha et al. 2014). Additionally, a PUF domain fused to a segment of yeast poly-A binding protein was used to upregulate cyclin B1 translation (~400%) in cancer cells to increase sensitivity to chemotherapeutic drugs (Campbell, Valley et al. 2014). Interestingly fusion with other functional domains is not absolutely necessary to control translation as a PUF protein alone has been used to bind to the 5’ untranslated region of a transcript to block translation machinery (Cao, Arha et al. 2015). Taken together these examples highlight the potential of additional function domains to engineer RBPs for various purposes from targeting control of translation in signalling pathway regulation to the design of cancer therapeutics.
RNA Localization
In recent years it has become increasingly evident that mRNA localization to subcellular compartments is crucial in many different biological processes, and mislocalization is linked to several human diseases (Cody, Iampietro et al. 2013). The addition of subcellular localization tags to an engineered RBP targeted to a specific RNA can change where that RNA is localized in the cell, such as shifting the location of a mutated or toxic ncRNA to prevent downstream functions. A similar approach was recently used with an inducible system that included a PUF domain to control mRNA transport (Abil, Gumy et al. 2017). In a eukaryotic cell’s transport system, molecular motors such as dyneins and kinesins can carry cargo along microtubules towards the positive (+) or negative (−) ends. The PUF domains were fused with one or more FKBP domains, which can be induced to dimerize with partner FRB domains in the presence of the drug rapalog. The FRB domain in turn was fused to either retrograde (N-terminal portion of Bicaudal D2 protein) or anterograde (truncated kinesin-1 heavy chain KIF5B protein without cargo binding tail) molecular motors. This system was used to specifically transport firefly luciferase mRNAs containing PUF binding sites in the 3’ UTR toward the axonal growth cones of primary neurons (Abil, Gumy et al. 2017). This provides an elegant tool for studying how localization of certain RNAs can affect cellular function.
RNA Probes
As a counterpart to controlling the localization of RNA, an engineered RBP approach can be used to localize or identify the presence of the RNA. In 2007 Ozawa et al. designed a split GFP system tethered to artificial PUF proteins to visualize the presence of a target RNA (Ozawa, Natori et al. 2007). In this system, enhanced GFP was split into two parts and fused to different PUF domains, such that when these PUF domains bind to adjacent sequence sites on the same RNA, the two GFP fragments are close enough to reassemble thereby visualizing the target. The authors utilized this system to visualize mitochondrial RNA in live cells (Ozawa, Natori et al. 2007). Using techniques like this, engineered RBPs have widely been used to detect the presence of plant viruses in vivo (Tilsner, Linnik et al. 2009, Wei, Huang et al. 2010, Tilsner, Linnik et al. 2012, Tilsner, Linnik et al. 2013) and retroviral RNA in mammalian cells (Yu, Lujan et al. 2011). A similar approach was utilized more recently with the zinc fingers of TIS11d, a tumor suppressor protein that also plays a critical role in mRNA regulation by interacting with inflammatory cytokine mRNAs. The zinc fingers were reengineered to bind a novel target sequence and fused to either lanthanide and antenna which luminescence when brought into close proximity by the ZF binding their target sequence (Raibaut, Vasseur et al. 2017). This novel approach can serve as an essential tool to probe the mechanisms of inflammation and cancer signaling pathways and serve as a templet for engineered RBPs as probes for different diseases.
In summary, while function is the primary driver of RBP design, these additional design factors must also be considered in the overall engineering approach. While it is possible to learn a tremendous amount from previous engineered RBPs and their design approach, there are some additional challenges that even the best designed RBP will face.
The challenge of off-target effects
When engineering an RBP to be used in vivo, the control of off-target effects is a concern that needs to be taken into careful consideration. Off-target effects could have unforeseen consequences such as the mis-regulation of mRNAs that have similar sequences to the target. Some off-target effects can be minimized not in the design of the RBP itself but in the selection of the target. Selection of a target sequence from the mRNA of interest with the least amount of homology to other RNAs in that transcriptome will limit potential off-target binding and function. The increasing use of transcriptomics has made it easier to identify potential off-targets sites based on the chosen target sequence. While predictive methods of reducing off-target effects are becoming increasingly powerful, experimental testing of the engineered RBP using RNAseq and CLIPseq will provide valuable in vivo assessment of unpredicted off-target activity. Aside from target selection, off-target effects can be minimized through the selection of multiple RBDs for long target RNA sequences. This approach also places a greater emphasis on the choice of linker sequence. When designing RBPs with nonrepetitive domains as well as PUF/PPR proteins, it may be useful to combine RBDs that recognize RNA through different modes of recognition. For example, an RNA stem-loop could be targeted by having one RBD bind the single-stranded sequence in the loop and the dsRNA stem region bound by a dsRBD to engineer an RBP with novel affinity and specificity for an RNA stem-loop. The more specific the target sequence, either through site selection or protein design, the less likely off-target effects will be observed.
Another method to reduce off-target effects is to control the cellular location, cell type, or tissue where the engineered RBP is to be expressed. This approach seeks to minimize off-target effects not through target selection but by target availability. This targeting can be accomplished through the addition of structure-, cell- or tissue-specific tags and can be especially useful in higher-order eukaryotes where a large number of specialized cells mean that cell-specific transcriptomes can vary greatly. This approach is extremely useful if the target sequence is highly specific but the function needs to be limited to a particular tissue or cell type. For example, the target sequence may be highly expressed in two cell-types (brain & muscle) but only one type will produce the desired effect (brain). Thus, a brain-specific delivery (such as with an AAV specific to the brain) could be advantageous (Barnes, Scheideler et al. 2019) by limiting effects outside the target tissue. Controlling the localization of the protein with a tag such as a nuclear (SV40 NLS) or mitochondrial tag (COX4 mitochondrial matrix tag) can direct the engineered RBP to specific cellular locations. This approach can be useful if the engineered protein domains have similar functions but different targets in different cellular locations. This approach can also reduce unwanted and unanticipated effects from interactions with proteins from a particular sub-cellular location.
Overall the off-target effects of the engineered protein can be limited by target selection combined with proper protein design and delivery. However ultimately the complexity of the transcriptome of many high-order eukaryotes is such that it is impossible to eliminate off-target effects entirely by selection and design. In these situations proper in vivo testing and cataloging of all off-target effects is one of the only ways to minimize the deleterious effects of the engineered RBP.
Future Applications
Engineered RBPs have been used successfully to modulate the regulation of alternative splicing, regulate translation, localize RNAs, and as RNA probes. The design, function and performance of these proteins provide an excellent roadmap for pursing additional novel applications for engineered RBPs. Some of these additional novel functions could include:
RNA Modifications
RNA modifications can affect the post-transcriptional regulation of mRNAs and ncRNAs (Roundtree, Evans et al. 2017). There are over a hundred different modifications identified to date and these modifications may impact ~16,000 genes (Roundtree, Evans et al. 2017). RNA modifications function at the level of gene regulation and can dramatically increase the range of activities for many classes of RNAs (Saletore, Meyer et al. 2012). Many of the RNA modifications have been tied to disease mechanisms including mitochondrial disease, Parkinson’s disease, and aging (Sazanov 2015). While there is still a gap in understanding precisely how these modifications affect cellular RNAs, numerous recent discoveries have been made as the technology to study RNA modification improves. Engineered RBPs could potentially play a role in helping to dissect the function of RNA modifications while also providing a platform for modulating this function. Identifying non-traditional RBDs that specifically interact with a particular modification would be an interesting basis for engineered proteins that can be used to subsequently study those modifications. RBPs could also be designed to induce specific modifications to an RNA by combining a sequence specific RBD domain with a domain such as a deaminase domain. Given the growing importance of RNA modifications in the epitranscriptome, it is more than likely that engineered RBPs will play an important role in future discoveries.
Targeting Noncoding RNAs
Non-coding RNAs (ncRNAs) regulate many biological activities and have been linked to numerous diseases, including cancer (Esteller 2011). However, there have been very few efforts to design proteins to target them apart from attempts to engineer proteins for the detection of microRNAs (Lee, Cho et al. 2010) due to the functions of miRNAs being the most understood. The previously mentioned work with FUS RGG and TERRA ncRNA (Takahama, Miyawaki et al. 2015) has also opened the field to study how RBDs recognize noncoding RNA and provides potential to engineer proteins to target other ncRNAs. For example, microRNAs, for which up or down-regulation has been shown to occur in many types of cancers (Hayes, Peruzzi et al. 2014), could be targeted with engineered RBPs to control their expression. Given the increasing evidence for complex RBP–ncRNA interactions that function across multiple biological processes including transcription and epigenetics, engineering RBP are likely to play an increasing role in the ncRNA field.
Engineered RBPs as Therapeutics
There is a vast network of cellular functions that depend on mRNA, non-coding RNA, and the RBPs that bind them. Changes in pre-mRNA splicing (Cooper, Wan et al. 2009), the production of toxic RNAs (Zhang and Ashizawa 2017), mis-regulation of long non coding RNAs (Bhan and Mandal 2014), single point mutations, and many more RNA-related mechanisms have been found to cause disease, many of which currently have no effective treatments. From simply detecting the presence of viral RNA to targeting and degrading coding and noncoding RNAs, engineered RBPs have the potential to provide a therapeutic framework for treating many of these diseases. While challenges such as cell delivery and off targets affects are still a major hurdle, engineered RBPs may have advantages over genetic engineering which can produce permanent and often undesirable genome modifications (Wang, Wang et al. 2019). The growing understanding of the importance and complexity of RNA roles within the cell places a greater emphasis on designing and engineering suitable RNA-binding proteins to study and modulate the vast cellular network of RNAs.
Conclusion
The modular nature of RBP domains provides an excellent template for the design and creation of engineered RBPs. These domains can bind to RNA in a variety of ways, including through both sequence- and structure-specific mechanisms. There exists a strong set of traditional RNA-binding domains, including RRM, dsRBD, KH, ZF and many more, from which a researcher can select and design new RBPs. Each individual RBD has specific advantages and disadvantages that must be considered in the design process. However the lack of a defined RNA recognition code is common for many of these RBDs, which hampers the ability to effectively design a de novo RBP. In contrast, a few RBDs, such as the pentatricopeptide domain, have been studied in sufficient detail that a web-based interface is available to assist in the engineering process. While considerable advances have been made in the field of RBP engineering, careful consideration and testing of the new RBP is still an essential aspect of design.
Function remains the primary driver for the design and engineering of RBPs, although there are several additional important factors to be considered. Selecting and organizing the appropriate RBD along with the linker region can have considerable consequences on the downstream function of the engineered protein. Furthermore adding elements to target the RBP to the appropriate cellular compartment, cell- and/or tissue-type can also be critical for reducing off-target effects and ultimately the effectiveness of the RBP. If the RBP has a function beyond RNA binding, the selection and addition of functional domains must also be taken into account. There are multiple successful examples of engineered RBPs with functions in splicing regulation, RNA degradation, translation control and RNA localization. Ultimately the growing understanding of the importance of RNAs in cellular function and the vast network of transcriptomics and epitranscriptomics data provide a rich environment for the design, engineering and testing of novel RBPs. These new proteins will have an impact on the understanding of RNA mechanisms and the design of the next generation of therapeutics to improve human health.
Nucleotide Expansion Disorders.
To date over 40 diseases have been discovered to be caused by unstable microsatellite expansions (Rohilla and Gagnon 2017). Most of these diseases are neurodegenerative in nature and affect multiple tissue types. The repeat expansion can occur in both the coding and noncoding regions of a gene. After transcription, the expansions can result in a toxic RNA gain–of-function, where the expanded RNA aggregate in the nucleus and sequester RBPs preventing function. Two of the most well-known of these diseases, myotonic dystrophy type 1 and type 2 (DM1 and DM2), are caused by an expansion of CTG and CCTG expansions respectively that, when transcribed, sequester the MBNL family of proteins which are master regulators of RNA metabolism (Meola and Cardani 2015). When these RPBs are sequestered, their normal cellular functions such as RNA splicing regulation, RNA localization/transport, miRNA processing, DNA repair, transcription regulation, protein quality control, and apoptosis can be disrupted. The sequestration of MBNL proteins has been directly linked to many of the disease symptoms in DM1 and DM2 (Savkur, Philips et al. 2001). Thus, targeting these RNAs via engineered RBPs could have great potential as therapeutics.
Acknowledgments
Thank you to all the members of the Berglund Lab especially to Tammy Reid for their helpful comments on the contents of this review.
Funding Information - Myotonic Dystrophy and Wyck Foundation Fellowship, NIH R01 GM121862
Footnotes
No conflicts of interests.
Contributor Information
Carl R. Shotwell, University of Florida Department of Biochemistry and Molecular Biology,.
John D. Cleary, RNA Institute, University at Albany
J. Andrew Berglund, University at Albany, Department of Biological Sciences and RNA Institute,.
References
- Abil Z, Gumy LF, Zhao H and Hoogenraad CC (2017). “Inducible Control of mRNA Transport Using Reprogrammable RNA-Binding Proteins.” ACS Synthetic Biology 6(6): 950–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allain FH, Howe PW, Neuhaus D and Varani G (1997). “Structural basis of the RNA-binding specificity of human U1A protein.” The EMBO journal 16(18): 5764–5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auweter SD, Fasan R, Reymond L, Underwood JG, Black DL, Pitsch S and Allain FHT (2006). “Molecular basis of RNA recognition by the human alternative splicing factor Fox-1.” The EMBO journal 25(1): 163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auweter SD, Oberstrass FC and Allain FHT (2006). “Sequence-specific binding of single-stranded RNA: is there a code for recognition?” Nucleic acids research 34(17): 4943–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz Alexander G., Munschauer M, Schwanhäusser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, Wyler E, Bonneau R, Selbach M, Dieterich C and Landthaler M (2012). “The mRNA-Bound Proteome and Its Global Occupancy Profile on Protein-Coding Transcripts.” Molecular Cell 46(5): 674–690. [DOI] [PubMed] [Google Scholar]
- Barkan A, Rojas M, Fujii S, Yap A, Chong YS, Bond CS and Small I (2012). “A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins.” PLoS genetics 8(8): e1002910–e1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C, Scheideler O and Schaffer D (2019). “Engineering the AAV capsid to evade immune responses.” Current Opinion in Biotechnology 60: 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti A, Lutz Y, Heard DJ, Chambon P and Tora L (1996). “hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II.” The EMBO journal 15(18): 5022–5031. [PMC free article] [PubMed] [Google Scholar]
- Bhan A and Mandal SS (2014). “Long Noncoding RNAs: Emerging Stars in Gene Regulation, Epigenetics and Human Disease.” ChemMedChem 9(9): 1932–1956. [DOI] [PubMed] [Google Scholar]
- Blewett NH and Goldstrohm AC (2012). “A eukaryotic translation initiation factor 4E-binding protein promotes mRNA decapping and is required for PUF repression.” Molecular and cellular biology 32(20): 4181–4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos TJ, Nussbacher JK, Aigner S and Yeo GW (2016). “Tethered Function Assays as Tools to Elucidate the Molecular Roles of RNA-Binding Proteins.” Advances in experimental medicine and biology 907: 61–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce VJ and McNaughton BR (2017). “Inside Job: Methods for Delivering Proteins to the Interior of Mammalian Cells.” Cell Chemical Biology 24(8): 924–934. [DOI] [PubMed] [Google Scholar]
- Buckanovich R, Yang Y and Darnell R (1996). “The onconeural antigen Nova-1 is a neuron-specific RNA-binding protein, the activity of which is inhibited by paraneoplastic antibodies.” The Journal of Neuroscience 16(3): 1114–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell ZT, Valley CT and Wickens M (2014). “A protein-RNA specificity code enables targeted activation of an endogenous human transcript.” Nature structural & molecular biology 21(8): 732–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Arha M, Sudrik C, Mukherjee A, Wu X and Kane RS (2015). “A universal strategy for regulating mRNA translation in prokaryotic and eukaryotic cells.” Nucleic acids research 43(8): 4353–4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Arha M, Sudrik C, Schaffer DV and Kane RS (2014). “Bidirectional Regulation of mRNA Translation in Mammalian Cells by Using PUF Domains.” Angewandte Chemie International Edition 53(19): 4900–4904. [DOI] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Benedikt M. Beckmann, Strein C, Davey Norman E., Humphreys David T., Preiss T, Steinmetz Lars M., Krijgsveld J and Hentze Matthias W. (2012). “Insights into RNA Biology from an Atlas of Mammalian mRNA-Binding Proteins.” Cell 149(6): 1393–1406. [DOI] [PubMed] [Google Scholar]
- Chen X, Zaro J and Shen WC (2013). “Fusion Protein Linkers: Property, Design and Functionality.” Adv Drug Deliv Rev 65(10): 1357–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Gutmann B, Zhong X, Ye Y, Fisher MF, Bai F, Castleden I, Song Y, Song B, Huang J, Liu X, Xu X, Lim BL, Bond CS, Yiu S-M and Small I (2016). “Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants.” The Plant Journal 85(4): 532–547. [DOI] [PubMed] [Google Scholar]
- Cheong C-G and Hall TMT (2006). “Engineering RNA sequence specificity of Pumilio repeats.” Proceedings of the National Academy of Sciences of the United States of America 103(37): 13635–13639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury R, Tsai YS, Dominguez D, Wang Y and Wang Z (2012). “Engineering RNA endonucleases with customized sequence specificities.” Nature communications 3: 1147–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cody NAL, Iampietro C and Lécuyer E (2013). “The many functions of mRNA localization during normal development and disease: from pillar to post.” Wiley Interdisciplinary Reviews: Developmental Biology 2(6): 781–796. [DOI] [PubMed] [Google Scholar]
- Coller J and Wickens M (2002). “Tethered function assays using 3′ untranslated regions.” Methods 26(2): 142–150. [DOI] [PubMed] [Google Scholar]
- Cooke A, Prigge A, Opperman L and Wickens M (2011). “Targeted translational regulation using the PUF protein family scaffold.” Proceedings of the National Academy of Sciences of the United States of America 108(38): 15870–15875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TA, Wan L and Dreyfuss G (2009). “RNA and disease.” Cell 136(4): 777–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquille S, Filipovska A, Chia T, Rajappa L, Lingford JP, Razif MFM, Thore S and Rackham O (2014). “An artificial PPR scaffold for programmable RNA recognition.” Nature Communications 5: 5729. [DOI] [PubMed] [Google Scholar]
- Crasto CJ and Feng J.-a. (2000). “LINKER: a program to generate linker sequences for fusion proteins.” Protein Engineering, Design and Selection 13(5): 309–312. [DOI] [PubMed] [Google Scholar]
- Crawford DW, Blakeley BD, Chen P-H, Sherpa C, Le Grice SFJ, Laird-Offringa IA and McNaughton BR (2016). “An Evolved RNA Recognition Motif That Suppresses HIV-1 Tat/TAR-Dependent Transcription.” ACS chemical biology 11(8): 2206–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cridge AG, Castelli LM, Smirnova JB, Selley JN, Rowe W, Hubbard SJ, McCarthy JEG, Ashe MP, Grant CM and Pavitt GD (2010). “Identifying eIF4E-binding protein translationally-controlled transcripts reveals links to mRNAs bound by specific PUF proteins.” Nucleic acids research 38(22): 8039–8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Franco S, Vandenameele J, Brans A, Verlaine O, Bendak K, Damblon C, Matagne A, Segal DJ, Galleni M, Mackay JP and Vandevenne M (2019). “Exploring the suitability of RanBP2-type Zinc Fingers for RNA-binding protein design.” Scientific reports 9(1): 2484–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Gatto-Konczak F, Olive M, Gesnel MC and Breathnach R (1999). “hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer.” Molecular and cellular biology 19(1): 251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S, Wang Y, Cassidy-Amstutz C, Lu G, Bigler R, Jezyk MR, Li C, Hall TMT and Wang Z (2011). “Specific and modular binding code for cytosine recognition in Pumilio/FBF (PUF) RNA-binding domains.” The Journal of biological chemistry 286(30): 26732–26742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas AGL and Wood MJA (2011). “RNA splicing: disease and therapy.” Briefings in Functional Genomics 10(3): 151–164. [DOI] [PubMed] [Google Scholar]
- Du H, Cline MS, Osborne RJ, Tuttle DL, Clark TA, Donohue JP, Hall MP, Shiue L, Swanson MS, Thornton CA and Ares M (2010). “Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy.” Nature structural & molecular biology 17(2): 187–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Aleff RA, Soragni E, Kalari K, Nie J, Tang X, Davila J, Kocher J-P, Patel SV, Gottesfeld JM, Baratz KH and Wieben ED (2015). “RNA toxicity and missplicing in the common eye disease fuchs endothelial corneal dystrophy.” The Journal of biological chemistry 290(10): 5979–5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M (2011). “Non-coding RNAs in human disease.” Nature Reviews Genetics 12: 861. [DOI] [PubMed] [Google Scholar]
- Fernandez-Costa JM, Llamusi MB, Garcia-Lopez A and Artero R (2011). “Alternative splicing regulation by Muscleblind proteins: from development to disease.” Biological Reviews 86(4): 947–958. [DOI] [PubMed] [Google Scholar]
- Filipovska A, Razif MFM, Nygård KKA and Rackham O (2011). “A universal code for RNA recognition by PUF proteins.” Nature Chemical Biology 7: 425. [DOI] [PubMed] [Google Scholar]
- Friesen WJ and Darby MK (1997). “Phage Display of RNA Binding Zinc Fingers from Transcription Factor IIIA.” Journal of Biological Chemistry 272(17): 10994–10997. [DOI] [PubMed] [Google Scholar]
- Fu M and Blackshear PJ (2016). “RNA-binding proteins in immune regulation: a focus on CCCH zinc finger proteins.” Nature Reviews Immunology 17: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrey SM, Cass DM, Wandler AM, Scanlan MS and Berglund JA (2008). “Transposition of two amino acids changes a promiscuous RNA binding protein into a sequence-specific RNA binding protein.” RNA (New York, N.Y.) 14(1): 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatignol A, Lainé S and Clerzius G (2005). “Dual role of TRBP in HIV replication and RNA interference: viral diversion of a cellular pathway or evasion from antiviral immunity?” Retrovirology 2: 65–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George RA and Heringa J (2002). “An analysis of protein domain linkers: their classification and role in protein folding.” Protein Engineering, Design and Selection 15(11): 871–879. [DOI] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M and Tuschl T (2014). “A census of human RNA-binding proteins.” Nature Reviews Genetics 15: 829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale RS and Khosla C (2000). “Role of linkers in communication between protein modules.” Current Opinion in Chemical Biology 4(1): 22–27. [DOI] [PubMed] [Google Scholar]
- Graveley BR and Maniatis T (1998). “Arginine/Serine-Rich Domains of SR Proteins Can Function as Activators of Pre-mRNA Splicing.” Molecular Cell 1(5): 765–771. [DOI] [PubMed] [Google Scholar]
- Grishin NV (2001). “KH domain: one motif, two folds.” Nucleic acids research 29(3): 638–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti G, Brambilla L and Rossi D (2017). “Cell-Penetrating Peptides: From Basic Research to Clinics.” Trends in Pharmacological Sciences 38(4): 406–424. [DOI] [PubMed] [Google Scholar]
- Gupta YK, Nair DT, Wharton RP and Aggarwal AK (2008). “Structures of Human Pumilio with Noncognate RNAs Reveal Molecular Mechanisms for Binding Promiscuity.” Structure 16(4): 549–557. [DOI] [PubMed] [Google Scholar]
- Habchi J, Tompa P, Longhi S and Uversky VN (2014). “Introducing Protein Intrinsic Disorder.” Chemical Reviews 114(13): 6561–6588. [DOI] [PubMed] [Google Scholar]
- Hale MA, Richardson JI, Day RC, McConnell OL, Arboleda J, Wang ET and Berglund JA (2018). “An engineered RNA binding protein with improved splicing regulation.” Nucleic Acids Research 46(6): 3152–3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J, Peruzzi PP and Lawler S (2014). “MicroRNAs in cancer: biomarkers, functions and therapy.” Trends in Molecular Medicine 20(8): 460–469. [DOI] [PubMed] [Google Scholar]
- Hollingworth D, Candel AM, Nicastro G, Martin SR, Briata P, Gherzi R and Ramos A (2012). “KH domains with impaired nucleic acid binding as a tool for functional analysis.” Nucleic acids research 40(14): 6873–6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G and Simard MJ (2008). “Argonaute proteins: key players in RNA silencing.” Nature Reviews Molecular Cell Biology 9: 22. [DOI] [PubMed] [Google Scholar]
- Järvelin AI, Noerenberg M, Davis I and Castello A (2016). “The new (dis)order in RNA regulation.” Cell Communication and Signaling 14(1): 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauset T and Beaulieu M-E (2019). “Bioactive cell penetrating peptides and proteins in cancer: a bright future ahead.” Current Opinion in Pharmacology 47: 133–140. [DOI] [PubMed] [Google Scholar]
- Kerner P, Degnan SM, Marchand L, Degnan BM and Vervoort M (2011). “Evolution of RNA-Binding Proteins in Animals: Insights from Genome-Wide Analysis in the Sponge Amphimedon queenslandica.” Molecular Biology and Evolution 28(8): 2289–2303. [DOI] [PubMed] [Google Scholar]
- Kiledjian M and Dreyfuss G (1992). “Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box.” The EMBO journal 11(7): 2655–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindgren P, Yap A, Bond CS and Small I (2015). “Predictable alteration of sequence recognition by RNA editing factors from Arabidopsis.” The Plant cell 27(2): 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh HR, Kidwell MA, Ragunathan K, Doudna JA and Myong S (2013). “ATP-independent diffusion of double-stranded RNA binding proteins.” Proceedings of the National Academy of Sciences of the United States of America 110(1): 151–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh YY, Wang Y, Qiu C, Opperman L, Gross L, Tanaka Hall TM and Wickens M (2011). “Stacking interactions in PUF-RNA complexes.” RNA (New York, N.Y.) 17(4): 718–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krovat BC and Jantsch MF (1996). “Comparative Mutational Analysis of the Double-stranded RNA Binding Domains of Xenopus laevis RNA-binding Protein A.” Journal of Biological Chemistry 271(45): 28112–28119. [DOI] [PubMed] [Google Scholar]
- Kwon SC, Yi H, Eichelbaum K, Föhr S, Fischer B, You KT, Castello A, Krijgsveld J, Hentze MW and Kim VN (2013). “The RNA-binding protein repertoire of embryonic stem cells.” Nature Structural & Molecular Biology 20: 1122. [DOI] [PubMed] [Google Scholar]
- Laird-Offringa IA and Belasco JG (1995). “Analysis of RNA-binding proteins by in vitro genetic selection: identification of an amino acid residue important for locking U1A onto its RNA target.” Proceedings of the National Academy of Sciences of the United States of America 92(25): 11859–11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Cho H and Jung Y (2010). “Fabrication of a Structure-Specific RNA Binder for Array Detection of Label-Free MicroRNA.” Angewandte Chemie International Edition 49(46): 8662–8665. [DOI] [PubMed] [Google Scholar]
- Lewis HA, Musunuru K, Jensen KB, Edo C, Chen H, Darnell RB and Burley SK (2000). “Sequence-Specific RNA Binding by a Nova KH Domain: Implications for Paraneoplastic Disease and the Fragile X Syndrome.” Cell 100(3): 323–332. [DOI] [PubMed] [Google Scholar]
- Liang J, Song W, Tromp G, Kolattukudy PE and Fu M (2008). “Genome-wide survey and expression profiling of CCCH-zinc finger family reveals a functional module in macrophage activation.” PloS one 3(8): e2880–e2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughlin FE, Mansfield RE, Vaz PM, McGrath AP, Setiyaputra S, Gamsjaeger R, Chen ES, Morris BJ, Guss JM and Mackay JP (2009). “The zinc fingers of the SR-like protein ZRANB2 are single-stranded RNA-binding domains that recognize 5’ splice site-like sequences.” Proceedings of the National Academy of Sciences of the United States of America 106(14): 5581–5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Alexandra Searles M and Klug A (2003). “Crystal structure of a zinc-finger–RNA complex reveals two modes of molecular recognition.” Nature 426(6962): 96–100. [DOI] [PubMed] [Google Scholar]
- Mackay JP, Font J and Segal DJ (2011). “The prospects for designer single-stranded RNA-binding proteins.” Nature Structural & Molecular Biology 18: 256. [DOI] [PubMed] [Google Scholar]
- Mandell JG and Barbas CF 3rd (2006). “Zinc Finger Tools: custom DNA-binding domains for transcription factors and nucleases.” Nucleic acids research 34(Web Server issue): W516–W523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris C, Dominguez C and Allain FH-T (2005). “The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression.” The FEBS Journal 272(9): 2118–2131. [DOI] [PubMed] [Google Scholar]
- Masliah G, Barraud P and Allain FHT (2013). “RNA recognition by double-stranded RNA binding domains: a matter of shape and sequence.” Cellular and molecular life sciences : CMLS 70(11): 1875–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed D, Young DL, Gamble CE, Miller CR and Fields S (2013). “Deep mutational scanning of an RRM domain of the Saccharomyces cerevisiae poly(A)-binding protein.” RNA 19(11): 1537–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meola G and Cardani R (2015). “Myotonic dystrophies: An update on clinical aspects, genetic, pathology, and molecular pathomechanisms.” Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 1852(4): 594–606. [DOI] [PubMed] [Google Scholar]
- Miranda RG, McDermott JJ and Barkan A (2018). “RNA-binding specificity landscapes of designer pentatricopeptide repeat proteins elucidate principles of PPR-RNA interactions.” Nucleic acids research 46(5): 2613–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda RG, Rojas M, Montgomery MP, Gribbin KP and Barkan A (2017). “RNA-binding specificity landscape of the pentatricopeptide repeat protein PPR10.” RNA (New York, N.Y.) 23(4): 586–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto Y and Yokoyama S (2012). “Structural insight into RNA recognition motifs: versatile molecular Lego building blocks for biological systems.” Wiley Interdisciplinary Reviews: RNA 3(2): 229–246. [DOI] [PubMed] [Google Scholar]
- Nanduri S, Carpick BW, Yang Y, Williams BR and Qin J (1998). “Structure of the double-stranded RNA-binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation.” The EMBO journal 17(18): 5458–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen CD, Mansfield RE, Leung W, Vaz PM, Loughlin FE, Grant RP and Mackay JP (2011). “Characterization of a Family of RanBP2-Type Zinc Fingers that Can Recognize Single-Stranded RNA.” Journal of Molecular Biology 407(2): 273–283. [DOI] [PubMed] [Google Scholar]
- Nicastro G, Taylor IA and Ramos A (2015). “KH–RNA interactions: back in the groove.” Current Opinion in Structural Biology 30: 63–70. [DOI] [PubMed] [Google Scholar]
- Nomura W (2018). “Development of Toolboxes for Precision Genome/Epigenome Editing and Imaging of Epigenetics.” The Chemical Record 18(12): 1717–1726. [DOI] [PubMed] [Google Scholar]
- O’Connell MR (2019). “Molecular Mechanisms of RNA Targeting by Cas13-containing Type VI CRISPR–Cas Systems.” Journal of Molecular Biology 431(1): 66–87. [DOI] [PubMed] [Google Scholar]
- Okuda K, Shoki H, Arai M, Shikanai T, Small I and Nakamura T (2014). “Quantitative analysis of motifs contributing to the interaction between PLS-subfamily members and their target RNA sequences in plastid RNA editing.” The Plant Journal 80(5): 870–882. [DOI] [PubMed] [Google Scholar]
- Oubridge C, Ito N, Evans PR, Teo CH and Nagai K (1994). “Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin.” Nature 372(6505): 432–438. [DOI] [PubMed] [Google Scholar]
- Ozawa T, Natori Y, Sato M and Umezawa Y (2007). “Imaging dynamics of endogenous mitochondrial RNA in single living cells.” Nature Methods 4: 413. [DOI] [PubMed] [Google Scholar]
- Park S, Phukan PD, Zeeb M, Martinez-Yamout MA, Dyson HJ and Wright PE (2017). “Structural Basis for Interaction of the Tandem Zinc Finger Domains of Human Muscleblind with Cognate RNA from Human Cardiac Troponin T.” Biochemistry 56(32): 4154–4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavletich N and Pabo C (1991). “Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A.” Science 252(5007): 809–817. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, G. T, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004). “UCSF Chimera-- a visualization system for exploratory research and analysis.” J Comput Chem. 25(13): 1605–1612. [DOI] [PubMed] [Google Scholar]
- Plambeck CA, Kwan AHY, Adams DJ, Westman BJ, van der Weyden L, Medcalf RL, Morris BJ and Mackay JP (2003). “The Structure of the Zinc Finger Domain from Human Splicing Factor ZNF265 Fold.” Journal of Biological Chemistry 278(25): 22805–22811. [DOI] [PubMed] [Google Scholar]
- Purcell J, Oddo JC, Wang ET and Berglund JA (2012). “Combinatorial Mutagenesis of MBNL1 Zinc Fingers Elucidates Distinct Classes of Regulatory Events.” Molecular and Cellular Biology 32(20): 4155–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raibaut L, Vasseur W, Shimberg GD, Saint-Pierre C, Ravanat J-L, Michel SLJ and Sénèque O (2017). “Design of a synthetic luminescent probe from a biomolecule binding domain: selective detection of AU-rich mRNA sequences.” Chemical Science 8(2): 1658–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A, Grünert S, Adams J, Micklem DR, Proctor MR, Freund S, Bycroft M, St Johnston D and Varani G (2000). “RNA recognition by a Staufen double-stranded RNA-binding domain.” The EMBO journal 19(5): 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray M, Lee Y-W, Scaletti F, Yu R and Rotello VM (2017). “Intracellular delivery of proteins by nanocarriers.” Nanomedicine (London, England) 12(8): 941–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy Chichili VP, Kumar V and Sivaraman J (2013). “Linkers in the structural biology of protein-protein interactions.” Protein science : a publication of the Protein Society 22(2): 153–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohilla KJ and Gagnon KT (2017). “RNA biology of disease-associated microsatellite repeat expansions.” Acta neuropathologica communications 5(1): 63–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree IA, Evans ME, Pan T and He C (2017). “Dynamic RNA Modifications in Gene Expression Regulation.” Cell 169(7): 1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryter JM and Schultz SC (1998). “Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA.” The EMBO journal 17(24): 7505–7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saletore Y, Meyer K, Korlach J, Vilfan ID, Jaffrey S and Mason CE (2012). “The birth of the Epitranscriptome: deciphering the function of RNA modifications.” Genome Biology 13(10): 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samant V, Hulgeri A, Valencia A and Tendulkar A (2012). Accurate Demarcation of Protein Domain Linkers based on Structural Analysis of Linker Probable Region.
- Savkur RS, Philips AV and Cooper TA (2001). “Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy.” Nature Genetics 29: 40. [DOI] [PubMed] [Google Scholar]
- Sazanov LA (2015). “A giant molecular proton pump: structure and mechanism of respiratory complex I.” Nature Reviews Molecular Cell Biology 16: 375. [DOI] [PubMed] [Google Scholar]
- Schwartz JC, Ebmeier CC, Podell ER, Heimiller J, Taatjes DJ and Cech TR (2012). “FUS binds the CTD of RNA polymerase II and regulates its phosphorylation at Ser2.” Genes & development 26(24): 2690–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Wang X, Liu Y, Li Q, Yang Z, Yan N, Zou T and Yin P (2015). “Specific RNA Recognition by Designer Pentatricopeptide Repeat Protein.” Molecular Plant 8(4): 667–670. [DOI] [PubMed] [Google Scholar]
- Shen C, Zhang D, Guan Z, Liu Y, Yang Z, Yang Y, Wang X, Wang Q, Zhang Q, Fan S, Zou T and Yin P (2016). “Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.” Nature communications 7: 11285–11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JR, Eghtesadi SA, Dzuricky M, You L and Chilkoti A (2019). “Engineered Ribonucleoprotein Granules Inhibit Translation in Protocells.” Molecular Cell 75(1): 66–75.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Choi M, Siomi MC, Nussbaum RL and Dreyfuss G (1994). “Essential role for KH domains in RNA binding: Impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome.” Cell 77(1): 33–39. [DOI] [PubMed] [Google Scholar]
- Small ID and Peeters N (2000). “The PPR motif – a TPR-related motif prevalent in plant organellar proteins.” Trends in Biochemical Sciences 25(2): 45–47. [DOI] [PubMed] [Google Scholar]
- St Johnston D, Brown NH, Gall JG and Jantsch M (1992). “A conserved double-stranded RNA-binding domain.” Proceedings of the National Academy of Sciences of the United States of America 89(22): 10979–10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefl R, Oberstrass FC, Hood JL, Jourdan M, Zimmermann M, Skrisovska L, Maris C, Peng L, Hofr C, Emeson RB and Allain FHT (2010). “The solution structure of the ADAR2 dsRBM-RNA complex reveals a sequence-specific readout of the minor groove.” Cell 143(2): 225–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Zhang Z, Fregoso O and Krainer AR (2012). “Mechanisms of activation and repression by the alternative splicing factors RBFOX1/2.” RNA (New York, N.Y.) 18(2): 274–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szempruch AJ, Choudhury R, Wang Z and Hajduk SL (2015). “In vivo analysis of trypanosome mitochondrial RNA function by artificial site-specific RNA endonuclease-mediated knockdown.” RNA (New York, N.Y.) 21(10): 1781–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahama K, Miyawaki A, Shitara T, Mitsuya K, Morikawa M, Hagihara M, Kino K, Yamamoto A and Oyoshi T (2015). “G-Quadruplex DNA- and RNA-Specific-Binding Proteins Engineered from the RGG Domain of TLS/FUS.” ACS Chemical Biology 10(11): 2564–2569. [DOI] [PubMed] [Google Scholar]
- Takahama K, Takada A, Tada S, Shimizu M, Sayama K, Kurokawa R and Oyoshi T (2013). “Regulation of Telomere Length by G-Quadruplex Telomere DNA- and TERRA-Binding Protein TLS/FUS.” Chemistry & Biology 20(3): 341–350. [DOI] [PubMed] [Google Scholar]
- Tan AY, Riley TR, Coady T, Bussemaker HJ and Manley JL (2012). “TLS/FUS (translocated in liposarcoma/fused in sarcoma) regulates target gene transcription via single-stranded DNA response elements.” Proceedings of the National Academy of Sciences of the United States of America 109(16): 6030–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplova M and Patel DJ (2008). “Structural insights into RNA recognition by the alternative-splicing regulator muscleblind-like MBNL1.” Nature structural & molecular biology 15(12): 1343–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilsner J, Linnik O, Christensen NM, Bell K, Roberts IM, Lacomme C and Oparka KJ (2009). “Live-cell imaging of viral RNA genomes using a Pumilio-based reporter.” The Plant Journal 57(4): 758–770. [DOI] [PubMed] [Google Scholar]
- Tilsner J, Linnik O, Louveaux M, Roberts IM, Chapman SN and Oparka KJ (2013). “Replication and trafficking of a plant virus are coupled at the entrances of plasmodesmata.” The Journal of cell biology 201(7): 981–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilsner J, Linnik O, Wright KM, Bell K, Roberts AG, Lacomme C, Santa Cruz S and Oparka KJ (2012). “The TGB1 movement protein of Potato virus X reorganizes actin and endomembranes into the X-body, a viral replication factory.” Plant physiology 158(3): 1359–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B and Ye K (2017). “Nop9 binds the central pseudoknot region of 18S rRNA.” Nucleic acids research 45(6): 3559–3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Tai PWL and Gao G (2019). “Adeno-associated virus vector as a platform for gene therapy delivery.” Nature Reviews Drug Discovery 18(5): 358–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Wang L, Zou X, Duan S, Li Z, Deng Z, Luo J, Lee SY and Chen S (2019). “Advances in CRISPR-Cas systems for RNA targeting, tracking and editing.” Biotechnology Advances. [DOI] [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK and Kurokawa R (2008). “Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription.” Nature 454(7200): 126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, McLachlan J, Zamore PD and Hall TMT (2002). “Modular Recognition of RNA by a Human Pumilio-Homology Domain.” Cell 110(4): 501–512. [DOI] [PubMed] [Google Scholar]
- Wang Y, Cheong C-G, Hall TMT and Wang Z (2009). “Engineering splicing factors with designed specificities.” Nature methods 6(11): 825–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang Z and Tanaka Hall TM (2013). “Engineered proteins with Pumilio/fem-3 mRNA binding factor scaffold to manipulate RNA metabolism.” The FEBS journal 280(16): 3755–3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H and Wang Z (2015). “Engineering RNA-binding proteins with diverse activities.” Wiley Interdisciplinary Reviews: RNA 6(6): 597–613. [DOI] [PubMed] [Google Scholar]
- Wei T, Huang T-S, McNeil J, Laliberté J-F, Hong J, Nelson RS and Wang A (2010). “Sequential recruitment of the endoplasmic reticulum and chloroplasts for plant potyvirus replication.” Journal of virology 84(2): 799–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wriggers W, Chakravarty S and Jennings PA (2005). “Control of protein functional dynamics by peptide linkers.” Peptide Science 80(6): 736–746. [DOI] [PubMed] [Google Scholar]
- Yagi Y, Hayashi S, Kobayashi K, Hirayama T and Nakamura T (2013). “Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants.” PloS one 8(3): e57286–e57286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Yao Y, Hong S, Yang Y, Shen C, Zhang Q, Zhang D, Zou T and Yin P (2019). “Delineation of pentatricopeptide repeat codes for target RNA prediction.” Nucleic acids research 47(7): 3728–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Wang C, Li F, Zhang J, Nayab A, Wu J, Shi Y and Gong Q (2017). “The human RNA-binding protein and E3 ligase MEX-3C binds the MEX-3–recognition element (MRE) motif with high affinity.” Journal of Biological Chemistry 292(39): 16221–16234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P, Li Q, Yan C, Liu Y, Liu J, Yu F, Wang Z, Long J, He J, Wang H-W, Wang J, Zhu J-K, Shi Y and Yan N (2013). “Structural basis for the modular recognition of single-stranded RNA by PPR proteins.” Nature 504: 168. [DOI] [PubMed] [Google Scholar]
- Yu SF, Lujan P, Jackson DL, Emerman M and Linial ML (2011). “The DEAD-box RNA helicase DDX6 is required for efficient encapsidation of a retroviral genome.” PLoS pathogens 7(10): e1002303–e1002303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N and Ashizawa T (2017). “RNA toxicity and foci formation in microsatellite expansion diseases.” Current opinion in genetics & development 44: 17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Wang Y, Dong S, Choudhury R, Jin Y and Wang Z (2014). “Treatment of type 1 myotonic dystrophy by engineering site-specific RNA endonucleases that target (CUG)(n) repeats.” Molecular therapy : the journal of the American Society of Gene Therapy 22(2): 312–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, O’Connor JP, Siomi MC, Srinivasan S, Dutra A, Nussbaum RL and Dreyfuss G (1995). “The fragile X mental retardation syndrome protein interacts with novel homologs FXR1 and FXR2.” The EMBO journal 14(21): 5358–5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y-Y, Mao M-W, Zhang W-J, Wang J, Li H-T, Yang Y, Wang Z and Wu J-W (2018). “Expanding RNA binding specificity and affinity of engineered PUF domains.” Nucleic acids research 46(9): 4771–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]


