Abstract
The mTORC1 (mechanistic target of rapamycin complex 1) protein kinase regulates growth in response to nutrients and growth factors. Nutrients promote its translocation to the lysosomal surface, where its Raptor subunit interacts with the Rag guanosine triphosphatase (GTPase)–Ragulator complex. Nutrients switch the heterodimeric Rag GTPases among four different nucleotide-binding states, only one of which (RagA/B•GTP–RagC/D•GDP) permits mTORC1 association. We used cryo–electron microscopy to determine the structure of the supercomplex of Raptor with Rag-Ragulator at a resolution of 3.2 angstroms. Our findings indicate that the Raptor α-solenoid directly detects the nucleotide state of RagA while the Raptor “claw” threads between the GTPase domains to detect that of RagC. Mutations that disrupted Rag-Raptor binding inhibited mTORC1 lysosomal localization and signaling. By comparison with a structure of mTORC1 bound to its activator Rheb, we developed a model of active mTORC1 docked on the lysosome.
INTRODUCTION
The mTORC1 protein kinase controls growth in response to diverse environmental cues, such as nutrients and growth factors. Deregulated mTORC1 signaling is associated with many diseases, including some cancers and neurological disorders (1–5). Amino acids promote the translocation of mTORC1 to the surface of the lysosome, where it can interact with and be activated by the Rheb GTPase (6–10). mTORC1, composed of the core mTOR, Raptor, and mLST8 subunits, docks on the lysosome through the direct interaction of Raptor with the lysosome-associated Rag GTPase–Ragulator complex (11, 12). Through their C-terminal roadblock domains (CRDs), the Rag GTPases form heterodimers consisting of RagA or RagB bound to RagC or RagD (13, 14). The obligate heterodimeric nature of the Rags allows cross-talk between their GTPase domains, which is necessary for mTORC1 signaling to respond quickly to changes in nutrient levels (15). Ragulator consists of five subunits and is necessary for targeting the Rag GTPases to the lysosomal surface (11).
As in other GTPases, the GTPase domains of the Rags consist of a network of secondary structural elements, known as switches, that undergo conformational changes upon the exchange or hydrolysis of bound guanosine di- or triphosphate (GDP or GTP), respectively (16). Under the control of several nutrient-regulated GTPase-activating proteins (GAPs) and guanine nucleotide exchange factors (GEFs), such as GATOR1 (17, 18), FLCN-FNIP (19, 20), and SLC38A9 (21), the Rag heterodimer can oscillate among four nucleotide configurations, only one of which (RagA/B•GTP–RagC/D•GDP) represents nutrient sufficiency and interacts with mTORC1 (8, 19). To understand how mTORC1 discriminates between the Rag nucleotide states, we used cryo–electron microscopy (cryo-EM) to determine the structure of the Raptor subunit of mTORC1 bound to the Rag-Ragulator complex. This structure not only sheds light on the conformations of the Rag GTPases that underlie nutrient sensing by mTORC1, but also allows us to develop a structural model of mTORC1 docked on the lysosome.
RESULTS
Reconstitution of the Raptor-Rag-Ragulator supercomplex
Because the interactions that promote the association of mTORC1 with the lysosomal surface underlie signaling events and are thus transient, we built the Raptor-Rag-Ragulator complex using a bottom-up approach instead of trying to isolate an intact complex from human cells. We generated the RagA-RagC heterodimer and the pentameric Ragulator complex in bacteria, and produced Raptor in a human embryonic kidney (HEK)–293 cell expression system (see supplementary materials). To stabilize the interaction of Raptor with the Rag GTPases, we introduced two point mutations into RagC (S75N, T90N) that are found in patients with follicular lymphoma (22). Each of these mutations independently stabilizes the GDP-bound state of RagC and promotes the interaction of the heterodimer with Raptor (22), and we found that in combination they have additive effects. We used the wild-type version of RagA because we assumed that its slow GTPase rate (15) would keep it bound to GTP. Indeed, analysis of the RagA-RagC (S75N, T90N) heterodimer confirmed that it contained close to stoichiometric amounts of GTP and GDP (fig. S1A). By mixing together the Rag GTPase heterodimer, Ragulator, and Raptor, we produced a fairly homogeneous supercomplex that was suitable for structural studies (Fig. 1A).
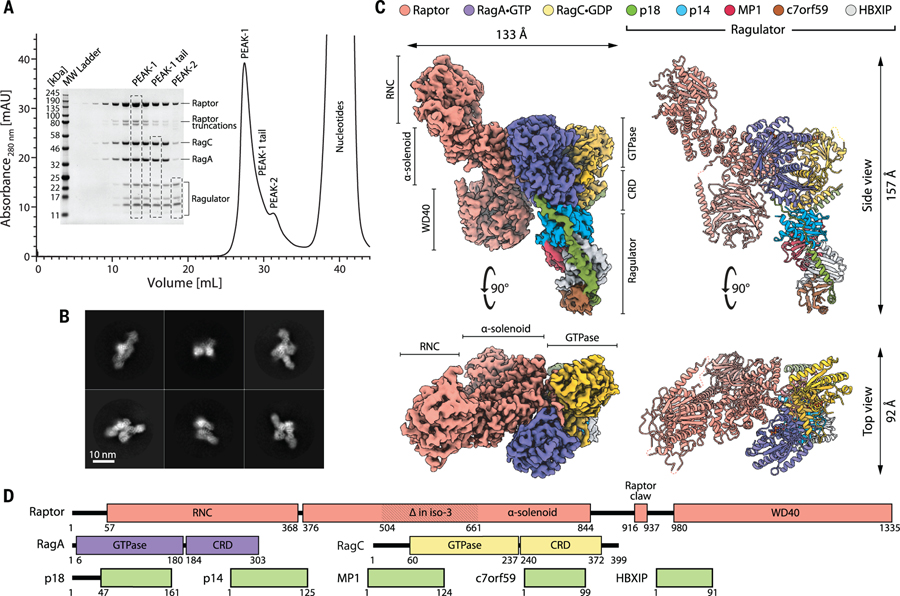
Figure 1. Purification, assembly, and structure determination of the Raptor-Rag-Ragulator supercomplex.
(A) Gel filtration profile and corresponding SDS–polyacrylamide gel electrophoresis of the reconstituted Raptor-Rag-Ragulator supercomplex as visualized with Coomassie Blue staining. The fully assembled complex (peak 1) partially overlaps with two subcomplexes: Rag-Ragulator (peak 1 tail) and Ragulator (peak 2). (B) Representative two-dimensional class averages of the Raptor-Rag-Ragulator supercomplex. (C) Cryo-EM structure of the supercomplex, determined to 3.2 Å resolution. Two orthogonal views of the experimental electron density (left) are shown next to corresponding views of the molecular model (right). (D) Domain organization of all components that make up the supercomplex.
Cryo-EM structure of the Raptor-Rag-Ragulator supercomplex
In initial applications of the supercomplex to holey carbon microscopy grids, the particles strongly adsorbed to carbon, but after extensive optimization of blotting and freezing conditions, we obtained grids with a gradient of particle distributions. The edges of holes attracted many overlapping particles, whereas the centers had well-distributed individual complexes. The uneven particle distribution proved to be a major obstacle to many particle-picking algorithms, and only the Topaz and crYOLO pickers (23, 24) discriminated well enough between particles on carbon and in holes to provide a uniform range of orientations and yield good two-dimensional classes (Fig. 1B).
After a series of refinements and particle sorting, we selected 112,037 particles to reconstruct the cryo-EM structure of the Raptor-Rag-Ragulator complex at 3.18 Å resolution (Fig. 1C and movie S1). The structure reveals the binding mode between Raptor and the Rag-Ragulator complex. The central region of Raptor (amino acids 376 to 844), which constitutes an α-solenoid (Fig. 1D), engages the Rag GTPases, whereas its N-terminal RNC (Raptor N-terminal conserved) domain and C-terminal WD40 domain do not. Although Ragulator makes extensive contacts with the CRDs of the Rags (fig. S1B), it does not directly interact with Raptor. Overall, Raptor and Rag-Ragulator form two elongated halves of the structure, positioned off-parallel to one another. The local resolution of the core of the supercomplex, formed by Rag-Raptor(α-solenoid), extends to ~2.3 Å resolution; the periphery of the structure was resolved to ~4.2 Å (fig. S9). Variability analysis revealed that such nonuniform resolution stems from the dynamics of the two halves of the complex, which swing with respect to one another. The highest amplitudes are observed at the tip of Ragulator and in the RNC and WD40 domains of Raptor (movie S2). The interface between Rag GTPase and CRD domains serves as a hinge region for the swinging movement of the Rag(CRD)-Ragulator substructure, which explains why capturing the full-length Rag-Ragulator with x-ray crystallography was likely an impossible task (25, 26).
As in the structure of the Rag GTPases bound to GATOR1 (27), the relative positions of the GTPase domains in our current structure are altered with respect to those in previously reported structures of the Rag homologs in budding yeast, the Gtr proteins [PDB IDs 3R7W and 4ARZ (28, 29)], (fig. S1C). In fact, the Gtr1•GTP-Gtr2•GDP active state would clash with Kog1 (the yeast homolog of Raptor) in a binding mode modeled on human Raptor-Rag. This suggests either that the structure of Kog1 adapts to accommodate the different Gtr structure or that the Gtr GTPases shift their conformation upon Kog1 binding. A structure of the yeast Gtr1-Gtr2-Kog1 complex will be needed to resolve the true situation among these possibilities.
The interaction of Raptor with RagA
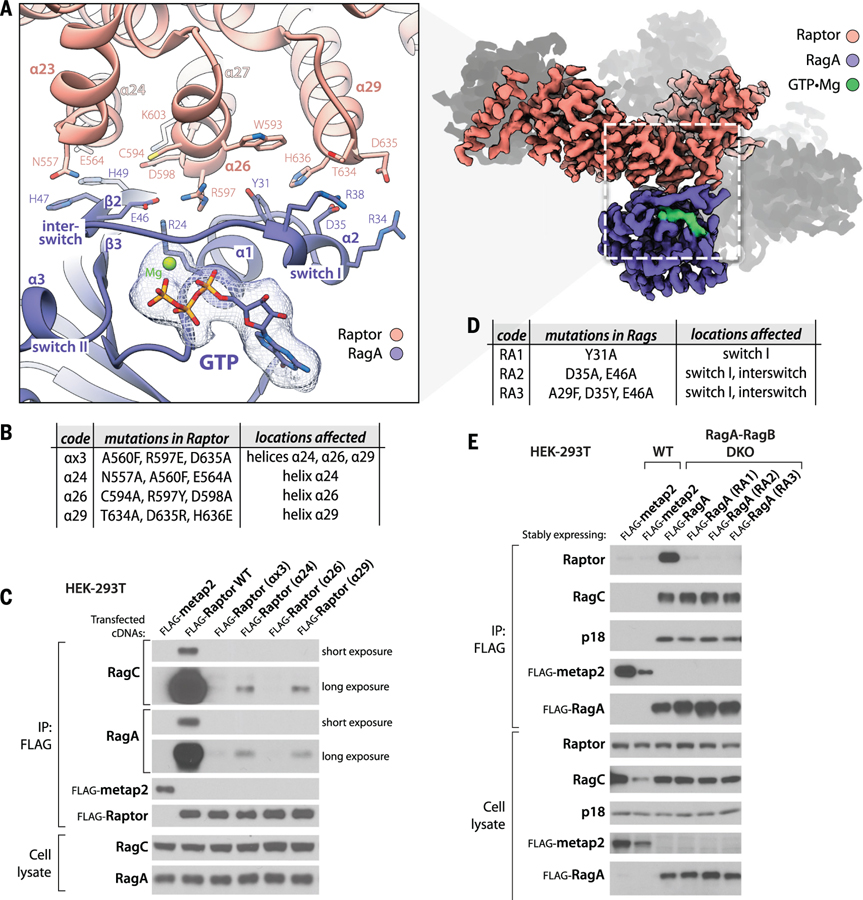
Raptor and RagA interact extensively with each other within the Raptor-Rag-Ragulator supercomplex. Three helices (α24, α26, α29) within a ~100–amino acid region (amino acids 546 to 650) of the Raptor α-solenoid (amino acids 376 to 844) directly engage the switch I–containing face of RagA and form a network of hydrogen bonds and salt bridges (Fig. 2A). The E564 and N557 residues of helix α24 of Raptor contact the RagA interswitch residues H49 and H47. R597 of helix α26 forms extensive interactions by tightly fitting the pocket formed by E46, R24, and Y31 of RagA. These interactions are supported by hydrogen bonds between the backbone of H49 of RagA and D598 and K603 of Raptor. The third helix α29 of Raptor engages helix α2 of RagA, with a number of salt bridges between T634, D635, and H636 of Raptor and R38, R34, and D35 of RagA (Fig. 2A).
Figure 2. Raptor-RagA interaction.
(A) Three helices (α24, α26, α29) in the HEAT-repeat domain of Raptor directly engage the switch-I face of GTP-loaded RagA. (B) Description of mutations introduced in the RagA-binding helices of Raptor. (C) Mutations in these helices render Raptor expressed via cDNA transient transfection unable to co-immunoprecipitate the endogenous RagA-RagC heterodimer. (D) Description of mutations introduced in the Raptor-binding region of RagA. (E) In RagA-RagB DKO HEK-293T cells, the transiently expressed RagA mutants in (D) cannot bind endogenous Raptor but have an intact capacity to bind to RagC and Ragulator, as assessed by its p18 subunit. Flag-metap2 was used as a control protein.
To test the importance of these contacts in mediating the Raptor-Rag interaction, we mutated interface residues on either Raptor or RagA. In Raptor, we disrupted either all three interacting helices involved in the interaction with one mutation per helix, or each of the three helices individually by introducing three mutations in each helix (Fig. 2B). All three helices, α24, α26, and α29, appear necessary for the binding of Raptor to the Rag heterodimer, with either mutations in all helices or mutations in helix α26 showing the strongest effect; this finding suggests that helix α26 plays a primary role in complex formation (Fig. 2C). None of the mutations affected the binding of Raptor to mTOR (fig. S2A).
Because the Raptor-RagA interface involves the critical switch I and interswitch regions of RagA, we only targeted, singly or in combination, residues on RagA that point away from the core of the protein (Fig. 2D) and validated that these mutations do not affect its capacity to bind GTP (fig. S2B). Consistent with the structural data, when expressed in RagA-RagB double-knockout (DKO) HEK-293T cells lacking RagA and its redundant homolog RagB (fig. S2C), none of the RagA mutants interacted with Raptor, whereas all still heterodimerized with RagC and bound to Ragulator, as assessed by coimmunoprecipitation of its p18/LAMTOR1 subunit (Fig. 2E).
On the basis of precedence with many small GTPases to which RagA is related (29, 30), we predict that in the GDP-bound state, both switch I and strand β2 of the interswitch of RagA would rearrange and disrupt the binding sites for the three key Raptor helices (α24, α26, and α29), explaining why Raptor only interacts with the Rag heterodimer when RagA (or RagB) is GTP-bound (8).
The Raptor claw
The heterodimeric Rag GTPases exhibit rotational two-fold symmetry, such that their switch machineries are on opposite ends of the heterodimer and their nucleotide-binding pockets face the dimeric center (Fig. 3, A and B). Thus, if Raptor directly engages the switch I–containing face of RagA, how does it also detect the nucleotide state of RagC when its switch region is on the other side of the heterodimer?
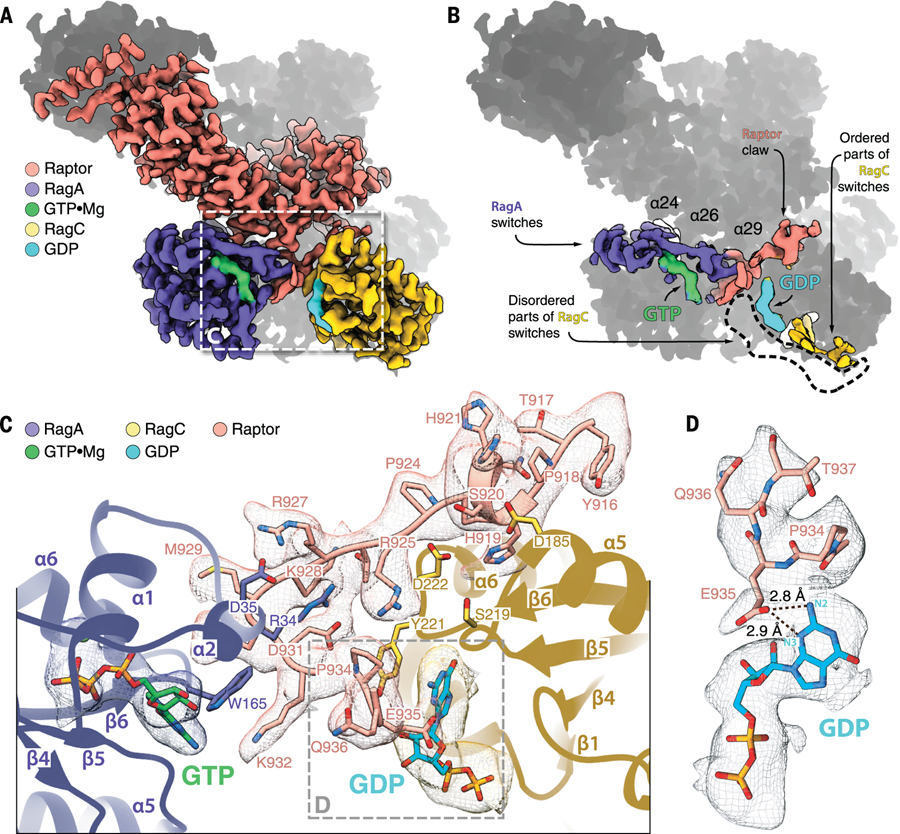
Figure 3. Raptor-RagC interaction.
(A) The binding interface between the Raptor α-solenoid and Rag GTPases. (B) The GTPase switch machineries of Rags are positioned at the opposite ends of the heterodimer. Note that in our structure, the switch machinery of RagC is largely disordered, and the circled region marks the space it would have occupied if it was fully resolved. (C) The Raptor claw is a 22–amino acid loop that enters the inter-Rag space depending on the nucleotide state of RagC. The claw forms interactions with both RagA and RagC, and its residue E935 engages RagC-loaded GDP directly. (D) The E935-GDP interaction is driven by hydrogen bonding between the E935 side chain and N2 and N3 of the guanine of GDP.
Surprisingly, the refined map of the Raptor-Rag-Ragulator complex revealed a large stretch of electron density in the space between the two GTPase domains of the Rags. This density is acutely angled; it enters the space between the Rags from the Raptor side, loops back, and exits near the bound GDP on RagC. At first, it was not clear which protein contributed the density: RagA, RagC, Raptor, or a Ragulator component. By mining our particle images for rare Rag-Ragulator and Raptor-Rag subcomplexes and determining their structures to low resolution (see supplementary materials), we used a process of elimination to determine that the electron density belonged to Raptor (fig. S3A). We traced an amino acid backbone through the density, and the distinct positions of prolines assigned the side chains to amino acids 916 to 937 of Raptor (Fig. 3C). The electron density of this region is unaccounted for in the previous structure of Raptor (31), and considering its triangular shape, we named it the “Raptor claw.” The primary sequence of the claw appears to be conserved in vertebrates and their closest invertebrate kin (fig. S3B).
The N-terminal end of the claw contains a very short helical fragment α33a (amino acids 919 to 922) that anchors it in a groove between the GTPase (D185, D186, K188) and CRD (M279) domains of RagC (fig. S3C). It enters the inter-GTPase domain space with a coil (R925, R927, M929) that first extends towards the CRD (Q237) and switch I (A33, R34, D35) of RagA, and then loops back 130° and exits (P934, E935) near the nucleotide-binding site of RagC (Y221) (Fig. 3C and fig. S3C). Interestingly, E935 of the claw is positioned close enough to the RagC-bound GDP to form hydrogen bonds with N2 and N3 of the guanine base (Fig. 3D). The switch machinery of RagC in the GDP-bound form in our cryo-EM structure is largely disordered, allowing us to model only parts of switch II and the interswitch. The remaining modules of the switch machinery appear dynamic.
Detection by Raptor of the RagC nucleotide state
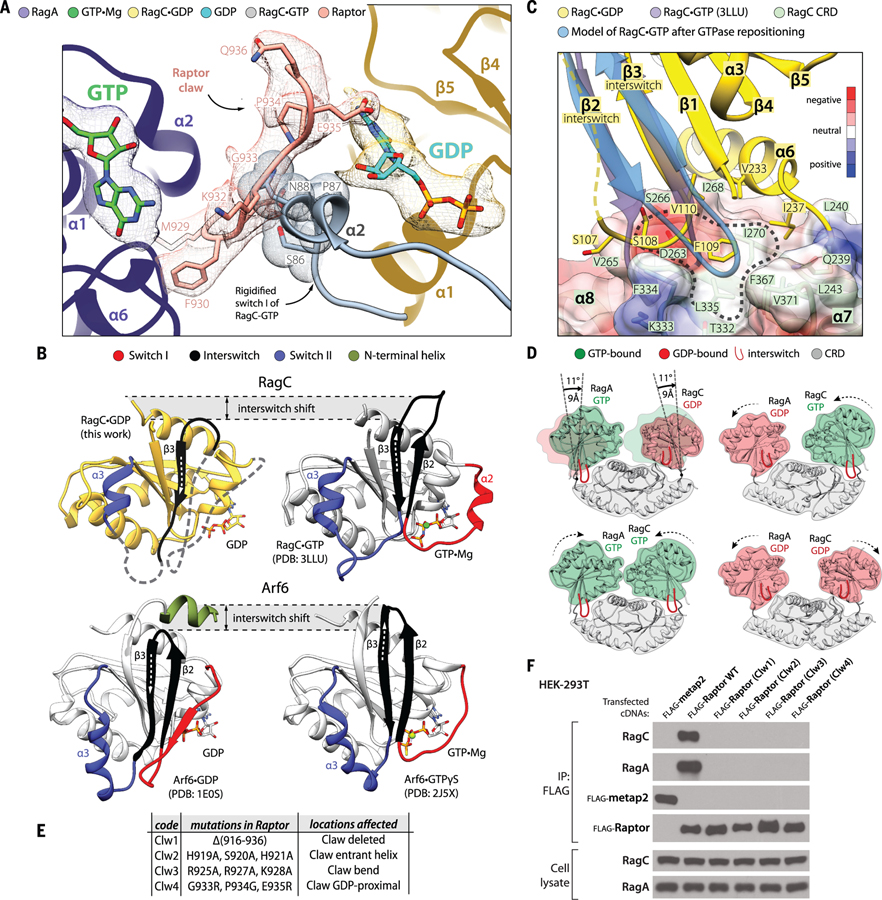
Because the Raptor claw can reach through the inter-Rag space to contact the switch machinery of RagC, we reasoned that it could serve as a nucleotide-state detector and explain why Raptor interacts with the Rag heterodimer when RagC (or RagD) is GDP-bound (19). To examine this possibility, we first superimposed a publicly available structure of the GTP-bound GTPase domain of RagC (PDB ID 3LLU) onto that of GDP-bound RagC from our cryo-EM structure. This analysis revealed that by causing switch I to rigidify, the loading of RagC with GTP would lead residues S86 and N88 of switch I to clash with the tip of the claw (residues: K932 and G933; see Fig. 4A).
Figure 4. The dynamics of the Rag-Raptor interaction.
(A) Loading of GTP by RagC would trigger its switch I to rigidify and clash with the Raptor claw. The structure of the GTP-loaded GTPase domain of RagC (PDB ID 3LLU) was superimposed with our cryo-EM structure of RagC•GDP. (B) The organization of the switches in RagC (top) and Arf6 (bottom) [PDB ID: 1E0S, 2J5X (37, 38)]. Note that although the switches change their positions in GTP- versus GDP-loaded states, the core of the structure does not move. Even though a large proportion of the switches in our GDP-loaded RagC structure are disordered, we observed that the interswitch changes its register by two residues, in a manner similar to Arf GTPases. (C) The movement of the interswitch during the GDP-to-GTP transition would cause its loop to clash with the CRD pocket (circled area). Instead, the interswitch repositions itself such that it engages with the more central part of the CRD. The disordered interswitch strand β2 of RagC•GDP is drawn with a dashed line. The surface of the CRD is colored according to electrostatic potential (see the color key). (D) The shifting of the GTPase domains in the Rag heterodimer during GTP-GDP binding exchanges. GTPase domains loaded with GDP are positioned away from the central axis of the Rag heterodimer, and their interswitches are retracted. Loading of GTP causes the interswitch to extend and press the CRD pocket such that the entire GTPase domain becomes repositioned closer to the Rag central axis. The models were created by superimposing RagA•GTP with RagC•GDP through their CRDs (specifically by matching their β7 and α9). (E) Description of the Raptor claw mutants used in (F). (F) Elimination of the Raptor claw or mutations in its critical Rag-interacting regions prevent Raptor from coimmunoprecipitating the RagA-RagC heterodimer. Longer exposures of the RagA and RagC immunoblots did not reveal any signal for Raptor. Flag-metap2 was used as a negative control protein.
Moreover, upon the GDP-to-GTP transition, the interswitch strand β3 of RagC shifts its register by two residues (Cα shift of 5.8 Å) in a manner similar to what has been observed in Arf GTPases (32) (Fig. 4B and movie S3). In GDP-bound RagC, S108 and F109 from the β2-β3 loop (107–110) of the interswitch rest on the side of a CRD pocket formed by four strands of the antiparallel β sheet and two bottom-face α helices (Fig. 4C). The two-residue register shift caused by GTP binding makes the interswitch stick outward, such that its β2-β3 loop would clash with the CRD pocket. To avoid such a clash, the interswitch loop would have to shift its position and engage with a larger area of the CRD pocket, leading the entire GTPase domain to reposition itself closer to the central axis of the Rag heterodimer (Fig. 4D). We modeled such domain repositionings in RagA and RagC in the context of the intact heterodimer because we observed no major domain rearrangements in the Rags upon Raptor binding, as evidenced by fitting the Rag heterodimer from the supercomplex into an 8.9 Å resolution map of the Rag-Ragulator subcomplex (fig. S3, A and D). This analysis showed that depending on the nature of the nucleotide, the space between the two GTPase domains becomes closed (RagA•GTP-RagC•GTP state), shifted toward the RagA (GDP-GTP) or RagC (GTP-GDP) side, or widened (GDP-GDP) (Fig. 4D).
This altered accessibility of the inter-Rag space likely is the basis of the ability of the claw to detect the nucleotide state of RagC. In the RagA•GTP-RagC•GDP state we captured, the α-solenoid of Raptor engages switch I of RagA, and the claw penetrates the inter-Rag space because switch I of RagC is disordered (movie S4). In the RagA•GTP-RagC•GTP configuration, not only would switch I of RagC rigidify and push the claw away from the bound nucleotide, but also the entire GTPase-domain of RagC would shift to close the intersubunit space. The opposite state, RagA•GDP-RagC•GTP, appears completely incompatible with interacting with Raptor because RagA•GDP cannot support α-solenoid binding and RagC•GTP expels the claw. The primary sequences of the RagA and RagC switches matter. The α-solenoid of Raptor is able to associate with the switch machinery of RagA•GTP, but it would clash with the equivalent nucleotide state of RagC (fig. S3E). Technically, the open RagA•GDP-RagC•GDP state should allow the claw to make interactions within the inter-Rag space. However, threading of the claw involves not only RagC residues but also those from RagA. In the GDP-loaded state, the switch I residues of RagA, R34, and D35 that contact the middle part of the claw (Fig. 3C) would become disordered. Ultimately, the entire RagA GTPase domain would shift away from the claw, leaving very little surface for any potential interactions. Beyond affecting the intersubunit space, the shifts in the GTPase domains we have modeled here are also likely involved in the cross-talk phenomenon that we previously observed in kinetic studies (15).
Given the extensive binding surface between RagA•GTP and the α-solenoid of Raptor, we considered that the claw might simply be a nucleotide detection mechanism for ensuring that RagC is GDP-bound and perhaps does not contribute to the strength of the Raptor-Rag interaction. Surprisingly, however, Raptor variants lacking the claw or with mutations in three critical regions—the entrant helix, the bend, and the part proximal to GDP (Fig. 4E)—could not bind the Rags (Fig. 4F) but still interacted with mTOR (fig. S3F).
In addition to the claw, the Raptor-Rag-Ragulator structure revealed a contact between the CRD of RagC and a loop (amino acids 795 to 806) extending from the α-solenoid of Raptor (fig. S4, A and B). This interface is largely driven by side chain–backbone hydrogen bonding (e.g. RagC R342 and Raptor L799) and some backbone-to-backbone interactions (Raptor G801 and RagC S281; fig. S4, C and D). Like the claw, this loop is also disordered in the previously reported structure of Raptor (31), and presumably its proximity to RagC rigidified it into a defined shape in our complexes. By mutating interfacing residues on Raptor, we found that deletion of residues 799 to 805 or a combination of three mutations (L799A, G801Y, F804D) weakened, but did not eliminate, the interaction of Raptor with the Rag GTPases without impacting that with mTOR (fig. S4E).
Collectively, our analyses of the role of interfacing residues between Raptor and the Rag heterodimer suggest a model in which Raptor first transiently binds to GTP-loaded RagA, forming interactions with its switch I region, and also weakly touches the CRD of RagC. Threading of the Raptor claw in between the GTPase domains of the Rags then generates a more stable interaction. The claw can fully engage only when RagC is loaded with GDP and its switch I is disordered. This would explain why RagC must be GDP-bound for the Rag heterodimer to interact with Raptor (19).
Model of mTORC1 docked on the lysosomal membrane
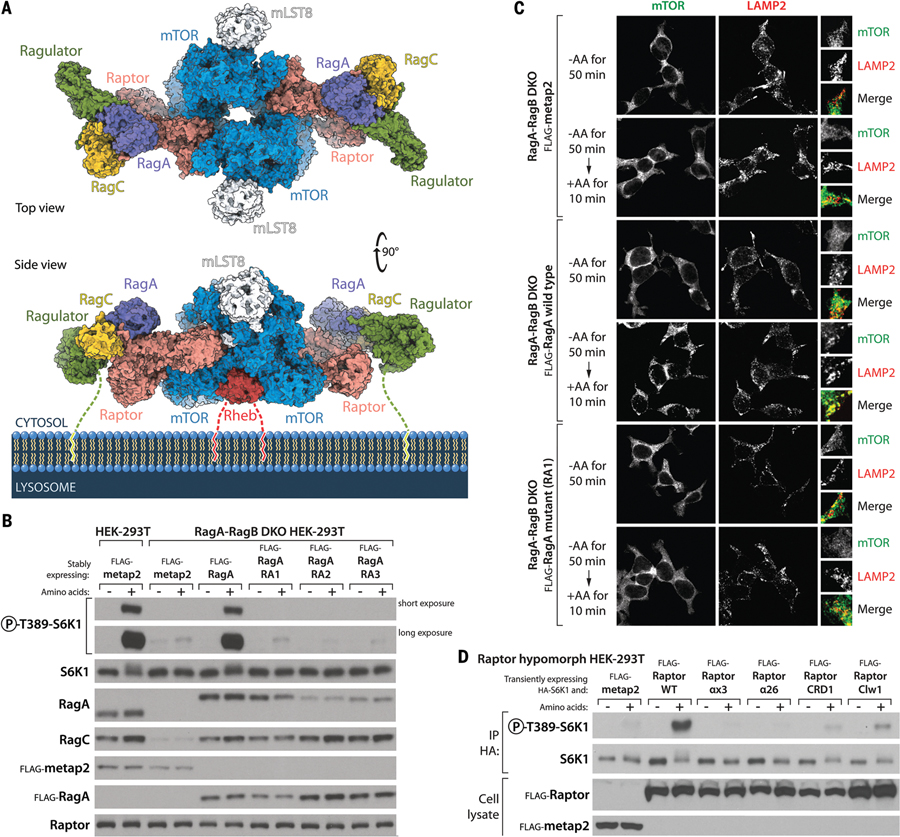
Translocation to the lysosomal surface is critical for the activation of mTORC1 because there it can interact with its kinase activator, the Rheb GTPase. A recent cryo-EM structure of mTORC1 bound to Rheb [PDB ID 6BCU (31)] provided a likely orientation of mTORC1 toward the lysosomal membrane because, like most small GTPases, Rheb associates with membranes through a C-terminal farnesyl group (33). Because both the Rheb-bound mTORC1 and Raptor-Rag-Ragulator structures contain Raptor, we superimposed them to generate a structural model of the Rheb-activated mTORC1 dimer tethered to the lysosome via Rag-Ragulator (Fig. 5A and movie S5).
Figure 5. mTORC1 on the lysosomal surface.
(A) Model of a Rheb-activated mTORC1 dimer bound to two lysosomal-targeting Rag-Ragulator complexes. The Rag-Ragulator complex clamps mTORC1 to the surface of the lysosome. Note that both Ragulator and Rheb carry lipid-modified terminal residues that tether the supercomplex to the membrane. Our cryo-EM structure of the Raptor-Rag-Ragulator complex was superimposed with that of Rheb-bound mTORC1 [PDB ID 6BCU (31)]. (B) Stable expression in RagA-RagB DKO HEK-293T cells of RagA mutants that cannot bind Raptor (Fig. 2, D and E) does not restore mTORC1 activity, as assayed by the phosphorylation of S6K1 in amino acid–replete cells. Note that although the levels of the stably expressed wild-type and mutant Flag-RagA are relatively even as assessed with the anti-Flag antibody, the recognition of the RA2 and RA3 RagA mutants by the RagA antibody (monoclonal; CST D8B5) is reduced, which suggests that the mutated residues affect the epitope recognized by this antibody, which has not been disclosed. (C) Unlike expression of wild-type RagA, expression in RagA-RagB DKO HEK-293T cells of the Y31A RagA (RA1) mutant that cannot bind Raptor does not rescue the amino acid–induced colocalization of mTOR with lysosomes, as marked by the lysosomal protein LAMP2. (D) In cells hypomorphic for Raptor, transient expression of wild-type Raptor promotes S6K1 phosphorylation, whereas that of Raptor mutants defective in the RagA-Raptor interaction (αx3 and α26) does not. Mutants that cannot bind the CRD of RagC (CRD1) or lacking the claw (Clw1) activate S6K1 phosphorylation to a small degree. See supplementary materials and fig. S6B for a description of the Raptor hypomorph cell line.
In our original cartoons of mTORC1 on the lysosomal surface, we envisioned Rag-Ragulator as a pillar protruding from the lysosome with mTORC1 sitting atop it (11). This appears not to be the case. Instead, the Rag-Ragulator complex binds to the top and side of mTORC1, serving as a clamp that pushes mTORC1 down onto the lysosome (Fig. 5A). The p18/LAMTOR1 subunit of Ragulator has a 45–amino acid N-terminal tail with myristoyl and palmitoyl modifications on its end that is necessary for localizing Ragulator to the lysosome (34). Although we cannot resolve this tail in our structure, we predict that if extended, its length of ~14 nm would provide enough room to properly orient the assembled supercomplex of mTORC1-Rag-Ragulator on the lysosomal surface, as depicted in Fig. 5A. In fact, given the broad diameter range of lysosomes (0.1 to 1 μm) (35), the p18 tail should allow mTORC1 to make expansive searches of the lysosomal surface for the activated Rheb GTPase (fig. S5, A and B). Once two Raptor subunits of mTORC1 are clamped down by the Rag-Ragulator complexes, the search radius will be restricted. However, the resulting avidity effect of such dimeric binding will further improve the overall strength of the interaction with the lysosome and will facilitate the lasting activation of the mTOR kinase.
To study the consequences of eliminating the Raptor-Rag interaction on mTORC1 signaling, we took advantage of the HEK-293T RagA-RagB DKO cells lacking RagA and RagB (fig. S2C). These cells do not activate mTORC1 upon amino acid stimulation, as judged by the phosphorylation of S6K1, a canonical mTORC1 substrate. Although stable expression of wild-type RagA readily restored mTORC1 activity in amino acid–stimulated cells, that of the RagA mutants that cannot bind Raptor did not (Fig. 2, D and E, and Fig. 5B). Consistent with this finding, in the cells expressing RagA mutants deficient in Raptor binding, mTOR no longer localized with lysosomes upon stimulation with amino acids (Fig. 5C and fig. S6A).
It is not trivial to study the effects of Raptor mutations on mTORC1 signaling because Raptor is a cell-essential gene, and therefore it is not possible to generate cells lacking it. However, using CRISPR-Cas9, we were able to generate a HEK-293T cell line that has defective mTORC1 signaling because its Raptor alleles have partial loss-of-function mutations that also reduce their expression (fig. S6B) (see supplementary materials). Transient expression of wild-type Raptor in these cells activated mTORC1 signaling, whereas that of the Raptor mutants affecting the RagA binding site did not (Fig. 5D). The claw- and CRD-binding mutants were slightly less defective, as they provided a small fraction of the signal that wild-type Raptor did.
Intriguingly, a fairly abundant isoform of Raptor (isoform 3) that lacks the Rag-interacting α-solenoid (Δ504–661 compared to the canonical isoform 1) has been described (36), raising the possibility that dimeric mTORC1 complexes may exist containing two different Raptor isoforms, only one of which can bind to the Rag GTPases. Such mTORC1 heterodimers would likely associate more weakly with lysosomes, which may be useful in certain signaling contexts. mTORC1 dimers may also exist having only the non–Rag-binding Raptor isoform, which might play roles in cells (e.g., erythrocytes) that lack lysosomes. We attempted experiments with this isoform of Raptor but found that it expresses poorly in HEK-293T cells.
Our cryo-EM structure of Raptor, the defining subunit of mTORC1, in complex with Rag-Ragulator, allows us to model how mTORC1 would dock on the lysosomal surface, which is a key step in its activation. We have described the regulated interface between Raptor and Rag GTPases, the details of which are critical for understanding nutrient sensing at the molecular level and also for designing small molecules that are more specific to mTORC1 signaling than existing drugs such as rapamycin. These drugs also target the related protein complex mTORC2, which has other roles in the cell.
Supplementary Material
ACKNOWLEDGMENTS
We thank all members of the Sabatini Lab for helpful insights and suggestions, particularly Kuang Shen for help with the GTP-binding assays; the labs of Drs. Thomas Schwartz, Catherine Drennan, Iain Cheeseman, Jing-Ke Weng, Tania Baker, Eric Fischer, and Roger Williams, as well as the Broad Institute Center for the Development of Therapeutics and MIT Biology Structural Biology Core Facility for advice, reagents, or access to instruments; the cryo-EM facilities of UMass Medical School, Brandeis University and the Whitehead Institute, particularly Kangkang Song (UMass), Michael Rigney (Brandeis), and Nicki Watson (Whitehead), for training or access to electron microscopes; the high-performance computing team at Whitehead, Craig Andrew, Paul McCabe, Robert Taylor, Peter Macfarlane, for the installation and maintenance of data-processing servers.
Funding: Supported by NIH grants R01 CA103866, R01 CA129105, and R37 AI47389, Department of Defense grant W81XWH-07–0448, and the Lustgarten Foundation (D.M.S.) and fellowships from the Tuberous Sclerosis Association (K.B.R.), the Koch Institute (X.G.), NIH F30 CA236179 (J.F.K.), the Charles A. King Trust (M.A.-R.), and a Saudi Aramco Ibn Khaldun Fellowship for Saudi Women (N.N.L.). D.M.S. is an investigator of the Howard Hughes Medical Institute and an American Cancer Society Research Professor.
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: Cryo-EM maps were deposited in the Electron Microscopy Data Bank under accession numbers EMD-20660 (Raptor-Rag-Ragulator) and EMD-20661 (Rag-Ragulator). The atomic model of Raptor-Rag-Ragulator was deposited in the Protein Data Bank under accession number 6U62. pFloat and pDarmo cloning plasmids were deposited on Addgene under accession numbers specified in table S1.
REFERENCES AND NOTES
- 1.Saxton RA, Sabatini DM, mTOR Signaling in Growth, Metabolism, and Disease. Cell. 168 (2017), pp. 960–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sabatini DM, Twenty-five years of mTOR: Uncovering the link from nutrients to growth. Proceedings of the National Academy of Sciences. 114, 11818–11825 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim J, Guan KL, mTOR as a central hub of nutrient signalling and cell growth. Nature Cell Biology. 21 (2019), pp. 63–71. [DOI] [PubMed] [Google Scholar]
- 4.Mossmann D, Park S, Hall MN, mTOR signalling and cellular metabolism are mutual determinants in cancer. Nature Reviews Cancer. 18 (2018), pp. 744–757. [DOI] [PubMed] [Google Scholar]
- 5.Valvezan AJ, Manning BD, Molecular logic of mTORC1 signalling as a metabolic rheostat. Nature Metabolism. 1, 321–333 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buerger C, DeVries B, Stambolic V, Localization of Rheb to the endomembrane is critical for its signaling function. Biochemical and Biophysical Research Communications. 344, 869–880 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Huang J, Manning BD, The TSC1–TSC2 complex: a molecular switchboard controlling cell growth. Biochemical Journal. 412, 179–190 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sancak Y et al. , The Rag GTPases bind Raptor and mediate amino acid signaling to mTORC1. Science. 320, 1496–1501 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL, Regulation of TORC1 by Rag GTPases in nutrient response. Nature Cell Biology. 10, 935–945 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Efeyan A et al. , Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature. 493, 679–683 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sancak Y et al. , Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 141, 290–303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM, Ragulator is a GEF for the Rag GTPases that signal amino acid levels to mTORC1. Cell. 150, 1196–1208 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakashima N, Noguchi E, Nishimoto T, Saccharomyces cerevisiae putative G protein, Gtr1p, which forms complexes with itself and a novel protein designated as Gtr2p, negatively regulates the Ran/Gsp1p G protein cycle through Gtr2p. Genetics. 152, 853–867 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sekiguchi T, Hirose E, Nakashima N, Ii M, Nishimoto T, Novel G Proteins, Rag C and Rag D, Interact with GTP-binding Proteins, Rag A and Rag B. Journal of Biological Chemistry. 276, 7246–7257 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Shen K, Choe A, Sabatini DM, Intersubunit Crosstalk in the Rag GTPase Heterodimer Enables mTORC1 to Respond Rapidly to Amino Acid Availability. Molecular Cell. 68, 552–565.e8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wittinghofer A, Vetter IR, Structure-Function Relationships of the G Domain, a Canonical Switch Motif. Annual Review of Biochemistry. 80, 943–971 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Bar-Peled L et al. , A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 340, 1100–1106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panchaud N, Péli-Gulli MP, De Virgilio C, Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Science Signaling. 6 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Tsun ZY et al. , The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Molecular Cell. 52, 495–505 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petit CS, Roczniak-Ferguson A, Ferguson SM, Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. Journal of Cell Biology. 202, 1107–1122 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen K, Sabatini DM, Ragulator and SLC38A9 activate the Rag GTPases through noncanonical GEF mechanisms. Proceedings of the National Academy of Sciences. 115, 9545–9550 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okosun J et al. , Recurrent mTORC1-activating RRAGC mutations in follicular lymphoma. Nature Genetics. 48, 183–188 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bepler T et al. , Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. ArXiv (2018). https://arxiv.org/abs/1803.08207 [PMC free article] [PubMed]
- 24.Wagner T et al. , SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Communications Biology. 2 (2019), doi: 10.1038/s42003-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yonehara R et al. , Structural basis for the assembly of the Ragulator-Rag GTPase complex. Nature Communications. 8 (2017), doi: 10.1038/s41467-017-01762-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Araujo MEG et al. , Crystal structure of the human lysosomal mTORC1 scaffold complex and its impact on signaling. Science. 358, 377–381 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Shen K et al. , Architecture of the human GATOR1 and GATOR1-Rag GTPases complexes. Nature. 556, 64–69 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gong R et al. , Crystal structure of the Gtr1p-Gtr2p complex reveals new insights into the amino acid-induced TORC1 activation. Genes and Development. 25, 1668–1673 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeong JH et al. , Crystal structure of the Gtr1pGTP-Gtr2pGDP protein complex reveals large structural rearrangements triggered by GTP-to-GDP conversion. Journal of Biological Chemistry. 287, 29648–29653 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicastro R, Sardu A, Panchaud N, De Virgilio C, The architecture of the Rag GTPase signaling network. Biomolecules. 7 (2017), doi: 10.3390/biom7030048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang H et al. , Mechanisms of mTORC1 activation by Rheb and inhibition by PRAS40. Nature. 552, 368–373 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pasqualato S, Renault L, Cherfils J, Arf, Arl, Arp and Sar proteins: A family of GTP-binding proteins with a structural device for “front-back” communication. EMBO Reports. 3, 1035–1041 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clark GJ et al. , The Ras-related protein Rheb is farnesylated and antagonizes Ras signaling and transformation. Journal of Biological Chemistry. 272, 10608–10615 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Nada et al. , The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK-ERK pathway to late endosomes. EMBO Journal. 28, 477–489 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novikoff AB, Beaufay H, De Duve C, Electron microscopy of lysosome-rich fractions from rat liver. Journal of Cell Biology. 2, 179–184 (1956). [PMC free article] [PubMed] [Google Scholar]
- 36.Sun C, Southard C, Di Rienzo A, Characterization of a novel splicing variant in the RAPTOR gene. Mutation Research - Fundamental and Molecular Mechanisms of Mutagenesis. 662, 88–92 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ménétrey J, Macia E, Pasqualato S, Franco M, Cherfils J, Structure of Arf6-GDP suggests a basis for guanine nucleotide exchange factors specificity. Nature Structural Biology. 7, 466–469 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Pasqualato S, Ménétrey J, Franco M, Cherfils J, The structural GDP/GTP cycle of human Arf6. EMBO Reports. 2, 234–238 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rogala KB et al. , The Caenorhabditis elegans protein SAS-5 forms large oligomeric assemblies critical for centriole formation. eLife. 4, 1–51 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rogala KB, Molecular architecture of SAS-5 enables construction of a daughter centriole (PhD thesis). University of Oxford (2016). [Google Scholar]
- 41.Bolivar F et al. , Construction and characterization of new cloning vehicle. II. A multipurpose cloning system. Gene. 2, 95–113 (1977). [PubMed] [Google Scholar]
- 42.Qing G et al. , Cold-shock induced high-yield protein production in Escherichia coli. Nature Biotechnology. 22, 877–882 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Casini A et al. , One-pot DNA construction for synthetic biology: The Modular Overlap-Directed Assembly with Linkers (MODAL) strategy. Nucleic Acids Research. 42 (2013), doi: 10.1093/nar/gkt915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mariati S. C. L. Ho, Yap MGS, Yang Y, Evaluating post-transcriptional regulatory elements for enhancing transient gene expression levels in CHO K1 and HEK293 cells. Protein Expression and Purification. 69, 9–15 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Kost TA, Condreay JP, Jarvis DL, Baculovirus as versatile vectors for protein expression in insect and mammalian cells. Nature Biotechnology. 23 (2005), pp. 567–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bernard P, Couturier M, Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. Journal of Molecular Biology. 226, 735–745 (1992). [DOI] [PubMed] [Google Scholar]
- 47.Gibson DG et al. , Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods. 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Andersen KR, Leksa NC, Schwartz TU, Optimized E. coli expression strain LOBSTR eliminates common contaminants from His-tag purification. Proteins: Structure, Function and Bioinformatics. 81, 1857–1861 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mansouri M et al. , Highly efficient baculovirus-mediated multigene delivery in primary cells. Nature Communications. 7 (2016), doi: 10.1038/ncomms11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luckow VA, Lee SC, Barry GF, Olins PO, Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. Journal of Virology. 67, 4566–79 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mastronarde D, SerialEM: A Program for Automated Tilt Series Acquisition on Tecnai Microscopes Using Prediction of Specimen Position. Microscopy and Microanalysis, 9(S02), 1182–1183 (2003). [Google Scholar]
- 52.Zheng SQ et al. , MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nature Methods. 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zivanov J, Nakane T, Scheres SHW, A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ. 6, 5–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang K, Gctf: Real-time CTF determination and correction. Journal of Structural Biology. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wagner T (2019, August 27). MPI-Dortmund/sphire-janni: JANNI 0.0.5 (Version v0.0.5). Zenodo. 10.5281/zenodo.3378300 [DOI] [Google Scholar]
- 56.Zivanov J et al. , New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 7 (2018), doi: 10.7554/elife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scheres SHW, Beam-induced motion correction for sub-megadalton cryo-EM particles. eLife. 3, e03665 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA, CryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nature Methods. 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Zi Tan Y et al. , Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nature Methods. 14, 793–796 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pettersen EF et al. , UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 25, 1605–12 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallographica Section D Biological Crystallography. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Afonine PV et al. , Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallographica Section D: Structural Biology. 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen VB et al. , MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallographica Section D: Biological Crystallography. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barad BA et al. , EMRinger: Side chain-directed model and map validation for 3D cryo-electron microscopy. Nature Methods. 12, 943–946 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goddard TD et al. , UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Science. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith SJM, Rittinger K Preparation of GTPases for Structural and Biophysical Analysis In GTPase Protocols (Humana Press, 2003), pp. 013–024. [DOI] [PubMed] [Google Scholar]
- 67.Wang T et al. , Identification and characterization of essential genes in the human genome. Science. 350, 1096–1101 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gu X et al. , SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science. 358, 813–818 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abraham MJ et al. , Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 1–2, 19–25 (2015). [Google Scholar]
- 70.Huang J et al. , CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nature Methods. 14, 71–73 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Parrinello M, Rahman A, Polymorphic transitions in single crystals: A new molecular dynamics method. Journal of Applied Physics. 52, 7182–7190 (1981). [Google Scholar]
- 72.Berendsen HJC, Postma JPM, Van Gunsteren WF, Dinola A, Haak JR, Molecular dynamics with coupling to an external bath. The Journal of Chemical Physics. 81, 3684–3690 (1984) [Google Scholar]
- 73.Darden T, York D, Pedersen L, Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. The Journal of Chemical Physics. 98, 10089–10092 (1993). [Google Scholar]
- 74.Zhang T et al. , Structural basis for Ragulator functioning as a scaffold in membrane-anchoring of Rag GTPases and mTORC1. Nature Communications. 8 (2017), doi: 10.1038/s41467-017-01567-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mu Z, Wang L, Deng W, Wang J, Wu G, Structural insight into the Ragulator complex which anchors mTORC1 to the lysosomal membrane. Cell Discovery. 3 (2017), doi: 10.1038/celldisc.2017.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Su MY et al. , Hybrid Structure of the RagA/C-Ragulator mTORC1 Activation Complex. Molecular Cell. 68, 835–846.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Katoh K, Rozewicki J, Yamada KD, MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics (2017), doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Robert X, Gouet P, Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Research. 42 (2014), doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Afonine PV et al. , New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallographica Section D: Structural Biology. 74, 814–840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.