Abstract
Motivation
Accurate estimation of transcript isoform abundance is critical for downstream transcriptome analyses and can lead to precise molecular mechanisms for understanding complex human diseases, like cancer. Simplex mRNA Sequencing (RNA-Seq) based isoform quantification approaches are facing the challenges of inherent sampling bias and unidentifiable read origins. A large-scale experiment shows that the consistency between RNA-Seq and other mRNA quantification platforms is relatively low at the isoform level compared to the gene level. In this project, we developed a platform-integrated model for transcript quantification (IntMTQ) to improve the performance of RNA-Seq on isoform expression estimation. IntMTQ, which benefits from the mRNA expressions reported by the other platforms, provides more precise RNA-Seq-based isoform quantification and leads to more accurate molecular signatures for disease phenotype prediction.
Results
In the experiments to assess the quality of isoform expression estimated by IntMTQ, we designed three tasks for clustering and classification of 46 cancer cell lines with four different mRNA quantification platforms, including newly developed NanoString’s nCounter technology. The results demonstrate that the isoform expressions learned by IntMTQ consistently provide more and better molecular features for downstream analyses compared with five baseline algorithms which consider RNA-Seq data only. An independent RT-qPCR experiment on seven genes in twelve cancer cell lines showed that the IntMTQ improved overall transcript quantification. The platform-integrated algorithms could be applied to large-scale cancer studies, such as The Cancer Genome Atlas (TCGA), with both RNA-Seq and array-based platforms available.
Availability and implementation
Source code is available at: https://github.com/CompbioLabUcf/IntMTQ.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Recent studies have shown that the majority of human genes produce multiple isoforms with diverse functions through alternative transcription and splicing (Hu et al., 2013; Shen et al., 2014; Wang et al., 2015). It provides cells with the opportunity to create protein isoforms from the same gene to participate in different functional pathways (David and Manley, 2010). Therefore, elucidation of gene expressions at the isoform resolution enables the detection of better molecular signatures for phenotype prediction, and the identified biomarkers may provide insights into the functional sequences of disease. High-throughput mRNA sequencing (RNA-Seq) is the most commonly used platform for quantifying isoform expressions across transcriptome and identification of novel isoforms (Conesa et al., 2016). Accurate RNA-Seq-based transcript quantification plays an important role in downstream transcriptome analyses, such as biomarker detection, isoform function prediction and differential transcript expression analysis (Zhang et al., 2015). However, transcript quantification remains a challenging problem due to sampling biases in the library preparation process and read mapping uncertainty as a result of alternative splicing events in most eukaryotic genes (Dapas et al., 2016; Huang et al., 2012).
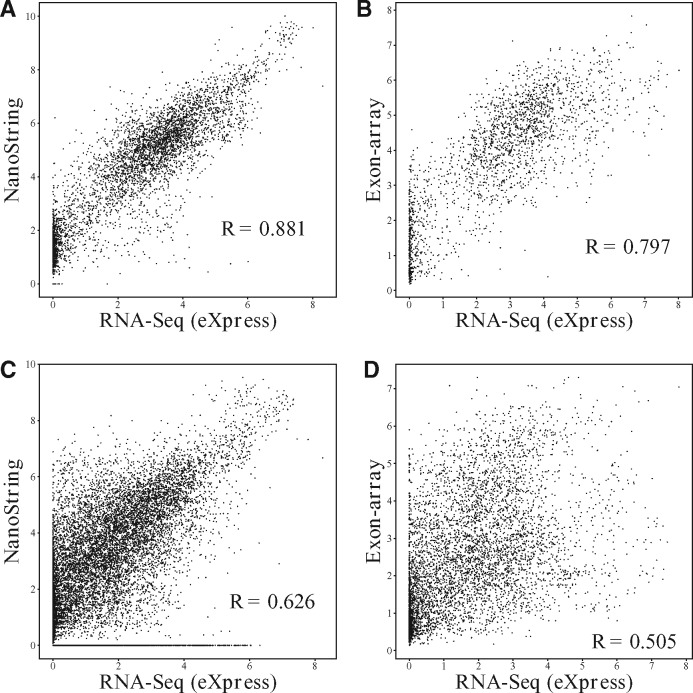
During the last decades, several technology platforms have been developed besides RNA-Seq to quantify gene/isoform expressions, including array-based technologies (e.g. Microarray, Exon-array), reverse transcriptome-quantitative polymerase chain reaction (RT-qPCR) and single-molecule imaging-based NanoString nCounter technology. It is commonly admitted that RNA-Seq is particularly useful for measuring mRNA expressions across transcriptome which performs high sensitivity and accuracy, and broad dynamic range of expression levels (Dapas et al., 2016; Wang et al., 2009; Zhao et al., 2014). Compared to RNA-Seq technology, array-based platforms encounter the limitations of cross-hybridization and the low dynamic range of expression levels. Exon-array platform provides the ability of evaluating many samples for alternative splicing with time and cost advantages which is attractive for large-scale post-transcriptome analyses. NanoString nCounter technology employs target-specific, color-coded probes to directly detect mRNA molecules and measure mRNA expressions without enzymatic reactions or bias (Geiss et al., 2008). RT-qPCR serves as a gold standard method that determines the exact amount of amplified DNA in samples. It has been used as a third platform to validate the findings and evaluate the performance of RNA-Seq and array-based expression data most of the time. Few studies assessed the strengths and weaknesses of each platform and simulated the consistency among different platforms so as to deliver better strategy for transcriptome expression profiling (Dapas et al., 2016). Gene expression analyses are widely applied for biomarker identification in cancer studies (Zhang et al., 2013, 2017), while expressions on isoform level are strongly related to protein functionality and thus provide molecular signatures at higher resolution for cancer outcome prediction. Accurate quantification of expression at the isoform level is limited with current technology although it is a common practice in cancer research (Safikhani et al., 2017; Vitting-Seerup and Sandelin, 2017), and the abundances estimated from RNA-Seq, NanoString and Exon-array platforms are wildly inconsistent on isoform level expression compared to gene level expression as shown in Figure 1 and Supplementary Figure S1–S10. In this article, we propose a novel integrative method IntMTQ to combine isoform abundances from other platforms to improve the resolution of RNA-Seq-based isoform quantification. The article is organized as follows. In Section 2, we describe the probabilistic model for transcript quantification with RNA-Seq data, the proposed IntMTQ model and the evaluation methods to assess the quantification methods. In Section 3, we first describe the data preparation of RNA-Seq, NanoString, Exon-Array and RT-qPCR on the same cancer cell lines. We then compared the predictive power of isoform expressions estimated by IntMTQ and the methods using RNA-Seq data only. The isoform expression was applied to stratify and classify cell lines into different cancer domains. An RT-qPCR experiment was performed to evaluate the quantification accuracy of IntMTQ.
Fig. 1.
Scatter plots of gene expression and isoform expression estimated by RNA-Seq and other two platforms. (A and B) show the correlation of gene expressions between RNA-Seq and NanoString/Exon-array. (C and D) show the correlation of isoform expressions between RNA-Seq and NanoString/Exon-array. eXpress (Roberts and Pachter, 2013) with sequence-specific bias correction was applied for isoform/gene expression quantification with RNA-Seq data
Algorithm 1 IntMTQ
1: Input: T, r, , E, λ.
2: Output: P
3: Initialize
4: Fordo
5: /*update the scaling factor*/
6: E-step:
7:
8: M-step:
9:
10: Ifthen
11: Break
12: End If
13: End For
14: Return P
2 Materials and methods
In this section, we first review a generative probabilistic model, BaseEM (Base Expectation-Maximization), for transcript quantification on RNA-Seq data to handle read mapping uncertainty. We then introduce the platform-integrated isoform quantification model (IntMTQ) to improve the quantification performance of RNA-Seq data by integrating the mRNA expression generated from the other platforms. The notations used in the equations are summarized in Table 1. At last, several methods to evaluate the quality of the estimated isoform expressions are described.
Table 1.
Notations
| Notation | Description |
|---|---|
| T | set of transcripts in a gene; Ti denotes the ith transcript of the gene |
| l | lengths of transcripts; li denotes the length of transcript Ti |
| r | set of reads aligned to a gene; rj is the jth read aligned to the gene |
| P | the probability of a read generated by the transcripts in the gene, |
| specifically, P= | |
| the effective lengths of the transcripts, where , | |
| li and lr are the length of Ti and the length of the read respectively | |
| qij | read sampling probability, if read rj is aligned to Ti, |
| otherwise qij = 0 | |
| E | the transcripts’ expressions in NanoString or Exon-array platform |
| aij | a soft assignment of read rj to transcript Ti in the EM algorithm |
| α | a scaling factor between the transcript expressions in different platforms |
| λ | hyper-parameter |
2.1 Base model for transcript quantification
We first consider the method proposed in Li et al., (2010), Pachter (2011), Xing et al. (2006) and Zhang et al. (2015) as the base model (BaseEM) for estimation of the isoforms in a single gene. In this model, T is denoted as the set of transcripts in a gene and Ti be the ith transcript in T, and be the total number of transcripts in the gene. The set of RNA-Seq reads aligned to the gene is represented as r. The probability of a read originated from the ith transcript is modeled by a categorical distribution with parameter pi, where and . For simplicity, .
The goal is to estimate the parameter P such that the likelihood of the observation r is maximized. Assuming that each read is sampled independently from one transcript, the likelihood function from the observed RNA-Seq read alignments is shown in Equation (1), where is denoted as qij, and the second equality in Equation (1) follows the assumption that the reads are sampled independently.
| (1) |
Specifically, for read rj aligned to transcript Ti, the probability of generating rj from Ti, denoted as is , where li and lr are the length of transcript Ti and the length of the read respectively. If the read rj was not aligned to transcript Ti, then qij = 0. Note that in transcript Ti, the number of positions in which a read can start is . The adjusted length is called the effective length of Ti (Pachter, 2011).
The log-likelihood function of Equation (1), , is concave with respect to P, and EM algorithm is adopted to learn the optimal P. The EM algorithm estimates the expectation of read assignments to transcripts in the E-step and maximizes the likelihood function given the expected assignments in the M-step as follows:
E-step: A soft assignment of read rj to transcript Ti, namely aij, is estimated in the expectation step:
where t is the tth iteration in the EM algorithm.
M-step: Given qij and the distribution of read assignments estimated in E-step, the probability of originating r from T is maximized when
After pi is estimated, the abundance of transcript Ti can be derived as .
2.2 Integrative model for transcript quantification
In our proposed platform integrative model IntMTQ, transcript expressions of NanoString/Exon-array data are introduced as penalization term(s) to encourage the consistency between transcript expressions learned from RNA-Seq platform and NanoString/Exon-array platforms. The model assumes that the transcript expression learned from different platforms for the same sample should be similar to each other. Based on the assumption, IntMTQ appends a constraint to the log-likelihood function of the BaseEM model as follows:
| (2) |
The penalized log-likelihood function consists of two terms. The first term represents the estimated log-likelihood of the observed read alignments from RNA-Seq data. The second term learns the consistency between transcript expressions of two platforms, i.e. RNA-Seq and NanoString or RNA-Seq and Exon-array, where = ,…, represent the transcript expression values from RNA-Seq, E denotes the transcript expressions provided by NanoString/Exon-array data, and is a scaling factor between transcript expressions learned from RNA-Seq data and NanoString/Exon-array data. The k-norm () is applied to minimize the differences between two platforms in the second term. The hyper-parameter λ modulates the consistency between RNA-Seq data and NanoString/Exon-array data. A larger λ shows more confidence on NanoString/Exon-array data. Therefore, a reasonable choice of λ is important, and it can be learned based on the log-likelihood function described in Section 2.1, where is the optimal P learned by IntMTQ in Equation (2) with parameter λ. The penalized log-likelihood function is concave as well, thus EM algorithm is applied to estimate the optimal P for each gene by maximizing with cvxpy package (Diamond and Boyd, 2016). The detailed IntMTQ algorithm is outlined in the Algorithm 1.
In the algorithm, P is initialized as . The For loop between line 4–13 updates P for all the transcripts in one gene by applying EM algorithm. In the M-step (line 9), two constraints are applied to maximizing the equation: (1) and (2) . The scaling factor α is updated with current P in each iteration in the EM algorithm. The expression of transcript Ti is then obtained with .
The IntMTQ framework can be generalized to multiple platforms. For example, one more constraint can be added to Equation (2) when data of the three platforms (RNA-Seq, NanoString and Exon-array) are all available for the genes in the samples of our interest. Then, the objective function is as follows, which combining both expression values from NanoString and Exon-array with the read density information from RNA-Seq.
| (3) |
2.3 Evaluation methods
To evaluate the quality of transcript quantification proposed by IntMTQ, we designed cancer cell line clustering and classification tasks with the assumption that better isoform quantification will lead to molecular signatures with higher resolution for cancer outcome prediction and patient stratification.
Hierarchical clustering was applied to distinguish groups of cancer cell lines with different cancer types by constructing a hierarchy of clusters step-by-step. A bottom-up approach which is also known as hierarchical agglomerative clustering merges pairs of clusters with the least dissimilarity and finally creates a dendrogram. The Euclidean distance served as the measure of dissimilarity between two cell lines, and the linkage method which minimizes the variance was applied to determine the distance between clusters being merged. Adjusted Rand Index (ARI) (Hubert and Arabie, 1985) and Normalized Mutual Information (NMI) (Studholme et al., 1999) were considered as performance evaluation strategies for hierarchical clustering tasks. NMI or ARI = 1 means the clustering result is the same as the ground truth, and NMI or ARI = 0 means the samples are randomly grouped.
SVM and Random Forest were applied for binary cancer type classification. Canonical machine learning techniques Support Vector Machine (SVM) and Random Forest were considered as classifiers for evaluation of isoform quantification quality reported by IntMTQ and baseline methods. Hierarchical clustering, Random Forest and SVM were implemented via scikit-learn (Pedregosa et al., 2011).
3 Results
In this section, we first describe the data preparation of the same cancer cell lines generated from RNA-Seq, NanoString and Exon-array platforms which were applied in this study, and the RT-qPCR experiment which was employed to evaluate the performance of IntMTQ. In the experiment section, IntMTQ was compared with BaseEM (Equation 1), Kallisto (Bray et al., 2016), Salmon (Patro et al., 2017), eXpress (Roberts and Pachter, 2013) and RSEM (Li and Dewey, 2011) to further investigate the quality and predictive power of the transcript expressions derived by different models. The results of cancer outcome prediction and cancer stratification on 46 cancer cell lines in more than four cancer types are illustrated in this section. In addition, the RT-qPCR experiment was applied to evaluate the quantification accuracy of IntMTQ. Running time of IntMTQ is reported at the end of this section.
3.1 Data preparation
This study was dedicated to improving the RNA-Seq-based isoform quantification by taking advantage of estimated expressions across platforms. 46 cancer cell lines were applied to evaluate the performance of IntMTQ, including 12 ovarian cancer cell lines, 7 colon cancer cell lines, 8 breast cancer cell lines, 9 lung cancer cell lines, 4 pancreas cancer cell lines, 2 prostate cancer cell lines and 4 other cell lines. Both RNA-Seq and NanoString platforms of all the 46 cancer cell lines were involved in this study. 28 out of the 46 cell lines were identical to the Exon-array data in Gene Expression Omnibus (GEO). We also designed RT-qPCR experiments on 12 of them to evaluate the performance of IntMTQ. The complete lists of the cancer cell lines for each platform in this project is available in Table 2 and Supplementary Table S1.
Table 2.
Summary of cell lines in each platform
| Number of cell lines | Number of genes/isoforms | Resource | Platform | |
|---|---|---|---|---|
| NanoString | 46 | 99/304 | nCounter | |
| RNA-Seq | 46 | 19278/50470 | CCLE | Illumina HiSeq 2000 |
| Exon-array | 28 | 3317/9400 | GEO | Affy HuEx.1.0.st.v2 |
| RT-qPCR | 12 | 7/14 |
3.1.1 RNA-Seq data preprocessing
The raw Bam files of the 46 CCLE (Cancer Cell Line Encyclopedia) cancer cell lines (Barretina et al., 2012) were downloaded from NCI GDC Legacy Archive (Grossman et al., 2016). The Bam file of each cell line was first converted back to paired-end fastq files by SAMTools (Li et al., 2009). Then the paired-end short reads were aligned to the hg19 RefSeq reference to match to the annotations to the other platforms by TopHat2 (Kim et al., 2013). According to the aligned bam file, reads aligned to each individual gene were extracted and utilized to build up qij, the read sampling probability, introduced in Equation (1). With the hg19 RefSeq annotation downloaded from UCSC Genome Browser, there were 19 278 genes and 50 470 transcripts in total after filtering out uncertain and duplicate isoforms.
3.1.2 NanoString probe design and transcript abundance estimation
We designed a NanoString experiment with 404 probes to estimate the 304 isoform expressions of 99 multi-isoform genes for the same 46 cancer cell lines based on hg19 RefSeq annotation. The 99 genes were all cancer genes from the literature (Futreal et al., 2004) for better reliability in annotations and better signatures for cancer studies. The NanoString signal in each cell line was normalized by the geometric mean of the ten house keeping genes. The expression of isoforms was estimated by minimizing the differences between the predicted and observed intensities for probes as , s.t. , where A is a m-by-n indicator matrix with values 0 and 1. m and n denote the total number of designed probes and total number of isoforms in the gene. If the probe j covers the coding region of isoform i in the gene, , otherwise . Vector y represents the probe intensities from the NanoString experiment, and vector x represents the isoform expressions we want to learn. Then the learned isoform expressions were normalized by in all the experiments in this study. The NanoString data is deposited in GEO database (Series GSE133226).
3.1.3 Exon-array data and transcript abundance estimation
The raw Exon-array data (.CEL file) of the 28 cancer cell lines were downloaded from GEO based upon availability. The isoform expressions were quantified using Multi-Mapping Bayesian Gene eXpression (MMBGX) method (Turro et al., 2010) with hg19 Ensembl annotation. This method disaggregates the signal of the probes matched to multiple isoforms in the same gene to estimate the expression of each individual isoform. To match the annotations between RefSeq and Ensembl for the data integration in our IntMTQ model, we only considered the multi-isoform genes in the two annotations containing the exact same isoforms for further analysis. In the end, 9400 isoforms in 3317 genes from Exon-array data were applied in this study.
3.1.4 RT-qPCR experiment design
In most cancer studies, RT-qPCR experiments were considered as gold standard to evaluate the identified biomarker from high-throughput sequencing or array-based data. Therefore, an independent RT-qPCR experiments was designed to measure the isoform proportions of seven multi-isoform genes (LRIG3, CD79A, ARID1A, TPM4, BCL2, BCR, NOTCH2) in twelve cell lines (Supplementary Table S1) to evaluate the accuracy of isoform expressions quantified by IntMTQ compared to other baseline methods. The seven genes were selected based on the availability of the primers to distinguish the isoforms in the genes. The twelve human cell lines were selected based on the availability of cell culture in our labs. The cancer cell lines were obtained from the American Type Culture Collection (ATCC) and cultured according to standard mammalian tissue culture protocols and sterile technique.
In the experiments, the total RNAs from cells were isolated by Trizol method, and reverse transcription reaction using Oligo-d(T) priming and superscript III (Invitrogen) was carried out according to the manufacturer’s protocol. SYBR Green was used to detect and quantify the PCR products in real-time reactions. When comparative Ct method was used, we normalized the Ct values to total RNAs. Primer sequences to measure the expression for each transcript are available in the Supplementary Material. The RT-qPCR data is also deposited in GEO database (Series GSE133226).
3.2 Experiments
To verify that the isoform expression estimated by IntMTQ produces more highly discriminative molecular signatures which are strongly related to disease phenotype and can be applied for accurate cancer outcome prediction, we developed three transcript abundance estimation experiments (E1, E2 and E3) on RNA-Seq, NanoString, Exon-array and RT-qPCR platforms as illustrated in Table 3. In all three experiments, IntMTQ was compared with five baseline methods, Kallisto, Salmon, eXpress, RSEM and BaseEM. Kallisto, Salmon, eXpress and RSEM (with and without sequence-specific bias correction) are involved in comparison. The command lines of running baseline methods are reported in the Supplementary Material. As mentioned in data preparation section, we expected sufficient utilization of data across different platforms to simulate the remarkable variety in transcriptome study. In experiment one (E1), IntMTQ integrated RNA-Seq and NanoString platforms on 46 cancer cell lines based on Equation (2) to quantify the expression of 304 transcripts in 99 multi-isoform genes overlapped between the two platforms. Similarly, in experiment two (E2), IntMTQ integrated RNA-Seq and Exon-array platforms on 28 cancer cell lines to quantify the expression of 9400 transcripts in 3317 multi-isoform genes overlapped between the two platforms. The 28 cancer cell lines in E2 is a subset of the 46 cell lines in E1. In the last experiment (E3), IntMTQ considered transcript expressions from both NanoString and Exon-array platforms in the constrained RNA-Seq-based log-likelihood function in Equation (3). The same 28 cancer cell lines and 215 isoforms in 75 genes involved in experiment E3 were the overlapped ones in all three platforms (RNA-Seq, NanoString and Exon-array). The raw Kallisto/Salmon/eXpress/RSEM abundance that reported as Transcripts Per Million (TPM) was normalized by (expression + 1). BaseEM and IntMTQ isoform expression values were normalized by total number of pair-end reads of each cancer cell line and then being transferred to log scale expression values.
Table 3.
Summary of experiments
| Experiments | Number of cell lines | Number of genes/isoforms | Platform integrated |
|---|---|---|---|
| E1 | 46 | 99/304 | NanoString |
| E2 | 28 | 3317/9400 | Exon-array |
| E3 | 28 | 75/215 | NanoString and Exon-array |
We evaluated the proposed integrative model IntMTQ based on the estimated transcript expressions collected from the three experiments. IntMTQ was compared with the five baseline methods on cancer cell line clustering and cancer type predictions to investigate the potential of using transcript expressions as predictive biomarkers in disease diagnosis and treatment. Statistical assessment was also performed directly on the data to demonstrate the informative significance of the isoform expression results. In addition, an independent RT-qPCR experiment on seven genes in twelve cancer cell lines was performed to directly evaluate the quantification accuracy of IntMTQ.
3.3 Parameter tuning
In all the experiments, the optimal parameter in the IntMTQ was estimated based on the log-likelihood function , where is the optimal P learned by IntMTQ in Equation (2) with parameter λ. In particular, λ was chosen from {1e-3, 1e-2, 1e-1, 1, 10, 100, 1000, 10 000, 100 000, 1 000 000}. Then the λ with the estimated P that maximizes the log-likelihood function (the first term in Equation 2) was chosen as the optimal parameter. The plot of the log-likelihood function with different λ values on experiment one (E1) is shown in Supplementary Figure S12. Similarly, the in Equation (3) were selected in a same strategy. λ in both experiments E1 and E2 were 10 000, and λ1 and λ2 in experiment E3 were 1000.
The Equation (2) with l1, l2 and infinity norms (k = 1, 2 and ) were tested on experiment one (E1) with a fixed λ. The estimated isoform proportion correlation are shown in Supplementary Figure S13. From the result we can see that the correlation coefficients between different norms are close to one and the l2-norm (k = 2) was applied to all the experiments in this study.
3.4 IntMTQ improved cancer cell line clustering
In this section, we evaluated the quality of transcript expression estimated by each method based on correctly clustering the cancer cell lines from the same cancer type into the same group. We applied hierarchical clustering to measure the performance of the isoform expressions quantified by IntMTQ compared with five baselines, Kallisto, Salmon, eXpress, RSEM and BaseEM. For each experiment setting, the same cancer cell lines and isoforms were used based upon the availability in Table 3. The clusters being grouped were cell lines in ovarian cancer, colon cancer, lung cancer and breast cancer. The clustering performance of all three methods were evaluated with ARI and NMI to illustrate how the clustering outcomes from each method agree with the ground truth which represented by cancer types. In comparisons corresponding to the three experiments, the transcript expressions estimated by IntMTQ achieved best clustering results in three out of six cases as shown in Table 4. In addition, IntMTQ consistently performs better compared to Kallisto without bias correction, eXpress with and without bias correction, RSEM with and without bias correction and BaseEM in all the experiments. The incorporating information from NanoString/Exon-array platforms could carry out isoform expressions with more discriminative power which lead to better performance on cancer cell line clustering by comparing with RNA-Seq platform alone.
Table 4.
Results of hierarchical clustering on four cancer types
| Experiment | Kallisto | Kallisto (bias) | Salmon | Salmon (bias) | eXpress | eXpress (bias) | RSEM | RSEM (bias) | BaseEM | IntMTQ | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| E1 | ARI | 0.032 | 0.135 | 0.209 | 0.150 | 0.142 | 0.171 | 0.092 | 0.067 | 0.152 | 0.251 |
| NMI | 0.187 | 0.295 | 0.417 | 0.351 | 0.329 | 0.355 | 0.293 | 0.273 | 0.342 | 0.390 | |
| E2 | ARI | 0.267 | 0.267 | 0.267 | 0.267 | 0.267 | 0.267 | 0.267 | 0.267 | 0.267 | 0.310 |
| NMI | 0.543 | 0.543 | 0.543 | 0.543 | 0.543 | 0.543 | 0.543 | 0.543 | 0.543 | 0.569 | |
| E3 | ARI | 0.094 | 0.272 | 0.252 | 0.171 | 0.137 | 0.035 | 0.137 | 0.095 | 0.137 | 0.158 |
| NMI | 0.300 | 0.464 | 0.482 | 0.406 | 0.369 | 0.261 | 0.369 | 0.296 | 0.369 | 0.419 |
Note: (bias) means the baseline methods were performed with sequence-specific bias correction. The best results across the ten methods are bold.
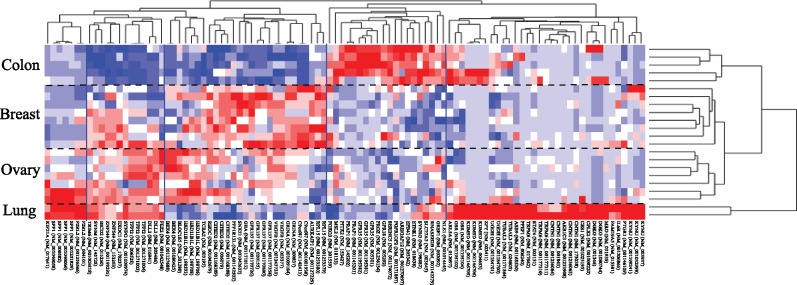
Complex diseases such as cancer are highly heterogeneous with different types or subtypes that lead to varying clinical outcomes including prognosis, response to treatment and changes of recurrence and metastasis. By selecting appropriate number of molecular signatures with non-redundant predictive signals, we could best capture the dissimilarity information that truly stratify the cancer patients into correct sub-groups. To confirm that the isoform expression estimated by IntMTQ is able to provide better signals for cancer patient stratification, we applied one-way ANOVA with a P-value cut-off 0.05 on the isoform expression data estimated by different methods in each experiment. The numbers of selected isoforms are shown in Table 5. Based on the results, IntMTQ is proved to be capable of identifying more marker isoforms relevant to cancer patient stratification. In addition, we selected the top 100 isoforms in E2 with the expression quantified by IntMTQ and run hierarchical clustering on both isoform and cell line dimensions of the data. The clustering result with a heat map of the isoform expression is displayed in Figure 2. Based on the expression of the top 100 isoforms quantified by IntMTQ, we can perfectly cluster the cell lines into correct groups. The four clusters from top to bottom are colon, breast, ovarian and lung cancer cell lines. Since breast and ovarian cancers are two related cancer types, they shared some similar bi-cluster patterns in the heat map.
Table 5.
Number of significant molecular features identified by ANOVA
| Experiment | Kallisto | Kallisto (bias) | Salmon | Salmon (bias) | eXpress | eXpress (bias) | RSEM | RSEM (bias) | BaseEM | IntMTQ |
|---|---|---|---|---|---|---|---|---|---|---|
| E1 | 54 | 51 | 52 | 53 | 56 | 51 | 49 | 51 | 58 | 67 |
| E2 | 895 | 893 | 869 | 885 | 903 | 910 | 857 | 881 | 1029 | 1190 |
| E3 | 24 | 22 | 24 | 28 | 25 | 24 | 26 | 26 | 27 | 33 |
Note: A P-value cutoff 0.05 is applied for the molecular feature selection. The best results across the ten methods are bold.
Fig. 2.
Cancer cell line clustering by 100 marker isoforms estimated by IntMTQ. The black dashed horizontal lines separate the clusters of cancer cell lines. The four clusters from top to bottom are colon, breast, ovarian and lung cancer cell lines. The solid vertical blue lines indicate the isoform clusters derived by hierarchical clustering. The official gene symbols with the RefSeq isoform names in the parentheses are listed at the bottom
3.5 IntMTQ improved cancer type classification
To provide an additional evaluation of the quality of transcript quantification, we designed two cancer outcome prediction tasks by the assumption that higher quality of transcript quantification can recommend better molecular signatures for cancer outcome prediction. IntMTQ was compared with BaseEM, Kallisto, Salmon, eXpress, and RSEM by classification with the quantification of isoforms in two different tasks. The first task was to classify ovarian cancer cell lines versus all the other cell lines in the three experiments since ovarian cancer has the largest cohort of cell lines compared to all the other cancer types in this study. The second task was to classify ovarian cancer and breast cancer cell lines versus all the other ones since ovarian and breast cancers are two related cancer types, and the bi-cluster patterns of the two cancer types in Figure 2 are close to each other. The dataset was randomly split into 70% as training set, and 30% as independent test set. The classification performance was measured on the test set. SVM with linear kernel and random forest were chosen as the classifiers to evaluate the classification performance. Due to the limited sample size (46 samples for E1 experiment, 28 samples for E2 and E3 experiments), no separated validation set was used to tune the parameters in the classification model. Instead, the default parameters of each classifier was applied to train the model. We repeated the random splitting 1000 times for each classifier in each experiment. To make the classification results comparable among different quantification methods, the same setup of training and test sets were used for all the methods in each splitting. For SVM, only the isoforms with t-test P-value < 0.1 were selected on training data to build up the classifier with default parameter (i.e. C = 1). Since the random forest classifier consists of an ensemble of decision trees with the advantages of dealing with the datasets with high-dimensional feature space but low sample size, the feature selection step was skipped and all the isoforms were used to build up the classifier on the training data with a minimum number of samples per leaf node equal to two.
The average area under the curve (AUC) of receiver operating characteristic of the 1000 repeats for SVM and random forest are reported in Table 6. The standard derivation of AUC scores are shown in the parentheses. The results show that the isoform expression estimated by IntMTQ performs significantly better than those by baseline methods. Specifically, in all the twelve classification tasks, the isoform expression quantified by IntMTQ outperforms all the baseline methods in eight tasks and IntMTQ is reported to be second best in two tasks. Overall the isoform expression estimated by IntMTQ has very competitive performance compared to the expressions quantified by baseline methods. The classification results suggest that this integrative strategy produce better RNA-Seq-based quantification on isoform level for disease phenotype prediction and human cancer studies.
Table 6.
Classification performance of estimated isoform expressions on three experiments
| Experiment | Task | Kallisto | Kallisto (bias) | Salmon | Salmon (bias) | eXpress | eXpress (bias) | RSEM | RSEM (bias) | BaseEM | IntMTQ |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SVM | |||||||||||
| E1 | OV | 0.626(0.101) | 0.606(0.099) | 0.631(0.101) | 0.662(0.100) | 0.624(0.091) | 0.605(0.096) | 0.631(0.097) | 0.648(0.098) | 0.628(0.100) | 0.691(0.090) |
| OV+BRCA | 0.607(0.086) | 0.598(0.085) | 0.572(0.085) | 0.573(0.087) | 0.634(0.082) | 0.604(0.083) | 0.595(0.087) | 0.596(0.084) | 0.551(0.083) | 0.562(0.082) | |
| E2 | OV | 0.643(0.153) | 0.637(0.151) | 0.611(0.152) | 0.648(0.150) | 0.621(0.146) | 0.612(0.149) | 0.595(0.149) | 0.613(0.145) | 0.594(0.143) | 0.660(0.145) |
| OV+BRCA | 0.852(0.072) | 0.851(0.070) | 0.856(0.071) | 0.893(0.062) | 0.852(0.073) | 0.863(0.072) | 0.862(0.068) | 0.869(0.068) | 0.855(0.072) | 0.881(0.055) | |
| E3 | OV | 0.533(0.135) | 0.543(0.134) | 0.540(0.126) | 0.596(0.139) | 0.534(0.130) | 0.558(0.137) | 0.524(0.128) | 0.546(0.133) | 0.532(0.140) | 0.608(0.140) |
| Random Forest | |||||||||||
| OV+BRCA | 0.643(0.120) | 0.721(0.113) | 0.644(0.123) | 0.695(0.110) | 0.655(0.116) | 0.672(0.118) | 0.671(0.122) | 0.672(0.112) | 0.767(0.103) | 0.779(0.095) | |
| E1 | OV | 0.701(0.092) | 0.706(0.089) | 0.717(0.090) | 0.724(0.087) | 0.734(0.091) | 0.744(0.089) | 0.712(0.091) | 0.707(0.094) | 0.719(0.092) | 0.763(0.080) |
| OV+BRCA | 0.628(0.088) | 0.648(0.090) | 0.632(0.091) | 0.664(0.090) | 0.660(0.090) | 0.663(0.092) | 0.622(0.090) | 0.636(0.091) | 0.650(0.087) | 0.660(0.086) | |
| E2 | OV | 0.686(0.134) | 0.685(0.135) | 0.624(0.143) | 0.627(0.144) | 0.616(0.146) | 0.633(0.140) | 0.690(0.135) | 0.693(0.132) | 0.698(0.131) | 0.717(0.130) |
| OV+BRCA | 0.804(0.090) | 0.798(0.090) | 0.721(0.111) | 0.742(0.106) | 0.728(0.109) | 0.729(0.105) | 0.803(0.094) | 0.807(0.095) | 0.799(0.088) | 0.851(0.079) | |
| E3 | OV | 0.540(0.152) | 0.542(0.149) | 0.591(0.147) | 0.632(0.145) | 0.582(0.152) | 0.592(0.147) | 0.569(0.150) | 0.569(0.153) | 0.576(0.148) | 0.628(0.144) |
| OV+BRCA | 0.639(0.117) | 0.638(0.111) | 0.664(0.112) | 0.671(0.112) | 0.686(0.115) | 0.689(0.111) | 0.641(0.115) | 0.648(0.114) | 0.680(0.114) | 0.694(0.111) | |
Note: The mean AUC scores (standard deviation in parentheses) of classifying cell lines by estimated transcript expression in 1000 repeats for each experiment are reported. The best AUCs across the ten methods are bold.
3.6 RT-qPCR confirmed improved isoform quantification
To evaluate the quality of the isoform expression estimated by IntMTQ, an independent RT-qPCR experiment on seven genes in twelve cancer cell lines was performed to compare with the isoform proportion estimated by IntMTQ and baseline methods. Root Mean Square Errors (RMSE) were calculated between the isoform proportions estimated by each RNA-Seq quantification method and RT-qPCR experiment. The results are reported in Table 7 for each cell line. The smaller the RMSE, the closer the isoform proportion to the RT-qPCR data. From the results, we can see that IntMTQ achieved the lowest RMSE in six out of twelve cell lines. Both of the baseline methods Salmon and eXpress perform better than all the other methods in two cell lines. In summary, IntMTQ is much closer to the RT-qPCR results and improve the overall isoform quantification significantly.
Table 7.
RT-qPCR experimental results to evaluate the isoform quantification accuracy
| Cell Line | Kallisto | Kallisto (bias) | Salmon | Salmon (bias) | eXpress | eXpress (bias) | RSEM | RSEM (bias) | BaseEM | IntMTQ |
|---|---|---|---|---|---|---|---|---|---|---|
| HT-29 | 3.431 | 3.084 | 3.509 | 3.166 | 2.799 | 2.563 | 3.339 | 2.968 | 3.068 | 2.379 |
| A549 | 2.146 | 2.126 | 2.034 | 2.092 | 1.633 | 1.705 | 2.051 | 1.888 | 2.355 | 2.090 |
| PC-3 | 1.354 | 1.557 | 1.406 | 1.523 | 1.196 | 0.959 | 1.408 | 1.330 | 2.230 | 1.910 |
| T-47D | 1.596 | 1.438 | 1.624 | 1.533 | 1.855 | 1.911 | 1.530 | 1.366 | 2.162 | 1.847 |
| DU_145 | 2.433 | 2.475 | 2.483 | 2.576 | 2.272 | 2.289 | 2.651 | 2.599 | 2.490 | 2.054 |
| MCF7 | 2.563 | 2.642 | 2.421 | 2.514 | 2.006 | 2.070 | 2.372 | 2.560 | 2.882 | 2.428 |
| HT-1080 | 1.606 | 1.806 | 1.579 | 1.900 | 1.988 | 2.315 | 1.634 | 1.781 | 2.181 | 1.673 |
| BT-549 | 2.482 | 2.736 | 2.790 | 2.930 | 3.189 | 3.216 | 3.130 | 2.976 | 2.912 | 2.108 |
| HCT_116 | 1.622 | 1.657 | 1.593 | 1.608 | 1.815 | 1.860 | 1.830 | 1.834 | 2.707 | 1.987 |
| AGS | 2.456 | 2.511 | 2.476 | 2.543 | 2.212 | 2.206 | 2.702 | 2.643 | 2.174 | 1.798 |
| MDA-MB-231 | 1.863 | 2.162 | 1.950 | 2.149 | 1.902 | 1.971 | 2.193 | 2.241 | 2.242 | 1.842 |
| HCT-15 | 2.313 | 2.296 | 2.417 | 2.405 | 2.241 | 2.341 | 2.422 | 2.212 | 2.747 | 1.888 |
Note: Root Mean Square Errors (RMSE) of the isoform proportion predictions by each method compared to the RT-qPCR results in each cell line. The smallest RMSEs across the ten methods are bold.
3.7 Running time
To measure the scalability of IntMTQ, we tested the algorithm on the cancer cell lines in three experiments. In the experiments with small gene lists, IntMTQ took 338 CPU seconds and 270 CPU seconds to run one sample in experiments E1 and E3 respectively. In the experiment with large gene list, IntMTQ took 9798 CPU seconds to run one sample in experiment E2. The CPU time was measured on Intel Core i7-7700 CPU with 3.60 GHz.
4 Discussion and conclusion
In this article, we explore the possibility of improving RNA-Seq-based transcript quantification by integrating isoform expression from the other mRNA quantification and identification platforms. RNA-Seq has been the most used platform for the whole transcriptome study compared to all the other platforms. The estimated isoform expressions are demonstrated to be inconsistent across platforms and in the absence of a gold standard evaluation method. The proposed model IntMTQ formulates one unified machine learning framework to integrate the quantification information from RNA-Seq and NanoString/Exon-array platforms to estimate the isoform expressions based on a global optimization strategy.
The idea of integrating two existing datasets from different platforms will shift the paradigm in transcriptome analysis. RNA-Seq and Microarray gene expression data have been integrated together to improve cancer outcome profiling (Castillo et al., 2017) and biomarker identification (Ma et al., 2017). PolyA-Seq has been incorporated with RNA-Seq to improve alternative polyadenylation detection (Chang et al., 2018). These studies provide evidence that multiple datasets produced by different profiling platforms can be combined to provide a higher resolution in data analysis, though the challenges still remain: (i) high cost by requiring the same sample source for the profiling; (ii) more difficult optimization problem compared to single platform data analysis; (iii) low scalability due to the different platforms may generate different number of transcriptome features (e.g. RNA-Seq and NanoString platforms). Therefore, a solution for a low cost, highly comprehensive transcriptome analysis tool with a flexible application to existing data generated from different platforms can be our future research direction.
The experimental results in this study consistently demonstrate the better performance of isoform expression learned by IntMTQ compared with the ones estimated by baseline methods in quantification accuracy, cancer cell line clustering and cancer type classification. Statistical assessment also shows that IntMTQ can provide more isoform features as molecular signatures for disease phenotype predictions. An independent experiment described in the Supplementary Material shows that IntMTQ is more robust to the factors in isoform quantification (e.g. number of isoforms in the gene, abundance of gene expression level and sequencing depth). The experimental results suggested a great potential of integrating high-throughput multi-omics data to overcome the limitations of using the data in a single platform and improve the quality of the genomic features for better phenotype prediction. Overall, the work in this article simulates the possibility of integrating different platforms for transcriptome studies to effectively enhance the contribution of isoform expression analysis in downstream applications.
Funding
This research work was supported by grant from the National Science Foundation (IIS 1755761) and National Institutes of Health (1R01GM113952-01A1).
Conflict of Interest: none declared.
Supplementary Material
References
- Barretina J. et al. (2012) The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature, 483, 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N.L. et al. (2016) Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol., 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Castillo D. et al. (2017) Integration of RNA-seq data with heterogeneous microarray data for breast cancer profiling. BMC Bioinf., 18, 506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J.-W. et al. (2018) An integrative model for alternative polyadenylation, IntMAP, delineates mTOR-modulated endoplasmic reticulum stress response. Nucleic Acids Res., 46, 5996–6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A. et al. (2016) A survey of best practices for RNA-seq data analysis. Genome Biol., 17, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dapas M. et al. (2016) Comparative evaluation of isoform-level gene expression estimation algorithms for RNA-seq and exon-array platforms. Brief. Bioinf., 18, 260–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David C.J., Manley J.L. (2010) Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev., 24, 2343–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond S., Boyd S. (2016) CVXPY: a Python-embedded modeling language for convex optimization. J. Mach. Learn. Res., 17, 1–5. [PMC free article] [PubMed] [Google Scholar]
- Futreal P.A. et al. (2004) A census of human cancer genes. Nat. Rev. Cancer, 4, 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss G.K. et al. (2008) Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol., 26, 317–325. [DOI] [PubMed] [Google Scholar]
- Grossman R.L. et al. (2016) Toward a shared vision for cancer genomic data. N. Engl. J. Med., 375, 1109–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y. et al. (2013) DiffSplice: the genome-wide detection of differential splicing events with RNA-seq. Nucleic Acids Res., 41, e39–e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. et al. (2012) A robust method for transcript quantification with RNA-Seq data. In: Annual International Conference on Research in Computational Molecular Biology, pp. 127–147. Springer. [DOI] [PMC free article] [PubMed]
- Hubert L., Arabie P. (1985) Comparing partitions. J. Class., 2, 193–218. [Google Scholar]
- Kim D. et al. (2013) Tophat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol., 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C.N. (2011) RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinf., 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. (2009) The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B. et al. (2010) RNA-seq gene expression estimation with read mapping uncertainty. Bioinformatics, 26, 493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T. et al. (2017) A joint Bayesian model for integrating microarray and RNA sequencing transcriptomic data. J. Comput. Biol., 24, 647–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachter L. (2011) Models for transcript quantification from RNA-Seq. arXiv Preprint arXiv: 1104.3889.
- Patro R. et al. (2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods, 14, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F. et al. (2011) Scikit-Learn: machine learning in Python. J. Mach. Learn. Res., 12, 2825–2830. [Google Scholar]
- Roberts A., Pachter L. (2013) Streaming fragment assignment for real-time analysis of sequencing experiments. Nat. Methods, 10, 71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safikhani Z. et al. (2017) Gene isoforms as expression-based biomarkers predictive of drug response in vitro. Nat. Commun., 8, 1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S. et al. (2014) rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-seq data. Proc. Natl. Acad. Sci. USA, 111, E5593–E5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studholme C. et al. (1999) An overlap invariant entropy measure of 3D medical image alignment. Pattern Recogn., 32, 71–86. [Google Scholar]
- Turro E. et al. (2010) MMBGX: a method for estimating expression at the isoform level and detecting differential splicing using whole-transcript Affymetrix arrays. Nucleic Acids Res., 38, e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitting-Seerup K., Sandelin A. (2017) The landscape of isoform switches in human cancers. Mol. Cancer Res., 15, 1206–1220. [DOI] [PubMed] [Google Scholar]
- Wang Y. et al. (2015) Mechanism of alternative splicing and its regulation. Biomed. Rep., 3, 152–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. et al. (2009) RNA-seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet., 10, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y. et al. (2006) An expectation-maximization algorithm for probabilistic reconstructions of full-length isoforms from splice graphs. Nucleic Acids Res., 34, 3150–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. et al. (2013) Network-based survival analysis reveals subnetwork signatures for predicting outcomes of ovarian cancer treatment. PLoS Comput. Biol., 9, e1002975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. et al. (2015) Network-based isoform quantification with RNA-seq data for cancer transcriptome analysis. PLoS Comput. Biol., 11, e1004465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. et al. (2017) Network-based machine learning and graph theory algorithms for precision oncology. NPJ Precision Oncol., 1, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S. et al. (2014) Comparison of RNA-seq and microarray in transcriptome profiling of activated T cells. PLoS One, 9, e78644. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.