Abstract
The genus Termitomyces (Lyophyllaceae, Basidiomycota) is often associated with fungus-feeding termites (Macrotermitinae) due to their strong symbiotic relationships. The genus is widely found exclusively in certain regions of Africa and Asia. They are recognized as edible mushroom within Southeast Asia as well. But it is often misidentified based on morphology by the local communities especially in Malaysia for Chlorophyllum molybdites which is a highly poisonous mushroom. Thus, it is necessary to study the genus for Malaysia with the synergy of using both morphological and molecular identification. In this study, we aim to describe another new species as an addition to the genus Termitomyces found within Sabah, Malaysia. We generated two new sequences (nrLSU and mtSSU) for the new species and a total of 28 nrLSU and mtSSU sequences were retrieved from GenBank for the phylogenetic analysis using maximum likelihood and Bayesian inferences. We identified that the new collection from Sabah province is a new species and named as Termitomyces gilvus based on the termites found in the mound. A phylogeny tree made from the concatenated genes of LSU and mtSSU suggests that T. gilvus is closely related to T. bulborhizus from China. According to our results, the combination of molecular and morphology proved to be a robust approach to re-evaluate the taxonomic status of Termitomyces species in Malaysia. Additional surveys are needed to verify the species diversity and clarify their geographic distribution.
Keywords: Malaysian Borno, molecular phylogeny, mtSSU rDNA, nrLSU, Termitomyces
1. Introduction
The genus Termitomyces R. Heim, is a paleotropical and edible mushroom classified under the family Lyophyllaceae (Basidiomycota). The genus forms an obligate symbiotic or mutualistic association with the fungus-feeding termites [1,2]. The fruiting bodies of the Termitomyces are the main food source of the fungus-growing termites, under the family Macrotermitinae (Isoptera). The Macrotermitinae termites are exclusively found in Africa and Southeast Asia [1–3]. Termitomyces species are able to decompose and degrade lignin in order to utilize the cellulose more efficiently by termites [4]. Termitomyces mushroom also provide digestive enzymes and vitamins to their hosts [5,6].
Globally, about 30 species of Termitomyces have been estimated so far [7]. Based on the Index Fungorum database, there are currently 92 legitimate names available within this group. In addition, Mossebo [8] had revised the current distribution of Termitomyces taxa which they described two more additional new taxa and one new species combination. Termitomyces species have been widely documented from equatorial and throughout southern Africa and Southeast Asia [7,8]. All species within Termitomyces are delicious in flavor, edible, nutritious and consumed locally in many South-East Asian [9–11]. Termitomyces contain biologically active compounds that are potential uses as antioxidants, immunomodulators, antitumors, antimicrobials and treating neurodegenerative disorders [11–14].
Termitomyces and their host termite species relationships have been widely studied [1–3,15]. The fungi live symbiotically with different genera of termites including Odontotermes, Microtermes, Macrotermes, Hypotermes, Protermes and Canthotermes [16]. Different species of termites cultivate different species of Termitomyces [17]. Morphological characteristics alone are insufficient for Termitomyces species identification [15]. Thus, molecular identification is useful and widely has been accepted for species-level identification within the genus. For fungal identification, molecular markers such as the internal transcribed spacer (ITS) region, nuclear large subunit ribosomal DNA (nrLSU) region and mitochondrial small subunit ribosomal DNA (mtSSU) region have been mainly used in Termitomyces studies [18,19]. Some studies used ITS barcode for Termitomyces but all of them were unidentified strains of Termitomyces [8]. Recently, Moseebo et al. [8] have generated new sequences with a large number of species collections, sequences and an update information on Termitomyces from Africa and Asia using both nLSU and mtSSU.
In Malaysia, Termitomyces species is locally known as “Cendawan busut”, “Cendawan melukut”, “Cendawan susu pelanduk” (mousedeer hoof mushroom), “Cendawan anai-anai (termite mushroom),” “Cendawan guruh” (thunder mushroom), “Kulat tahun” (annual mushroom), “Cendawan Tali” or “Kulat Taun” [20–22]. The most common species such as the T. eurhizus, T. heimii and T. clypeatus are mainly found in the oil palm plantation areas and have been delicious for many local people of Malay Peninsula [20,23] and Malaysian Borneo [24,25]. Eight Termitomyces species have been reported in Malaysia: Termitomyces clypeatus, T. entolomoides, T. heimii, T. eurhizus, T. microcarpus, T. aurantiacus, T. radicatus, and T. striatus. There were mainly reported from Peninsular Malaysia [9,10,26].
Sabah (Northern Borneo), the eastern part of Malaysia (Borneo Island), is well known for its high biodiversity flora and fauna. Mycological studies in this region started in the early 1930–1990s [27–34]. However, there were no documentation of Termitomyces species from this region. As part of the Borneensis-Agaricomycetes DNA Barcoding project, we have started exploring the diversity of Bornean mushrooms and during the study; we encountered an interesting collection of Termitomyces from the surroundings of University Malaysia Sabah campus area (mainly secondary forest). The morphological features of the collected Termitomyces species are similar to those of Termitomyces bulborhizus, but they showed a clear difference by phylogenetic analyses based on nrLSU and mtSSU sequences. Therefore, we examined its taxonomic status using morphology and phylogenetic analysis of concatenated nrLSU-mtSSU sequences and provide detailed description of Termitomyces as a new species.
2. Materials and methods
2.1. Sample collections
The Termitomyces specimens were collected from Universiti Malaysia Sabah campus area, Kota Kinabalu, Sabah, Malaysia Borneo (6˚2′ N, 116˚6′ E) between May and June 2018. The fruit bodies of Termitomyces were collected and labeled with the field number. Information of the specimen habitat and location were recorded using the Global Positioning System (GPS). Fresh morphological characteristics were recorded and photographs were taken using Olympus Digital camera (Model TG-4 16MP). All color names and alphanumeric codes follow the Methuen Handbook of Color [35]. All samples were collected from the field and were brought to the laboratory for further identification and analysis. Specimens were dried using the food dehydrator around 40 °C for 1–2 days. The dried specimens were placed in a paper bag with silica gel and stored at BORNEENSIS (BORH) herbaria, Institute of Tropical Biology and Conservation (ITBC), Universiti Malaysia Sabah. As for termite species identification, the soldier and worker termites were collected. The sample was identified at the species level based on the identification key following Ahmad [36] and Eggleton [37].
2.2. Morphological studies
The macro-morphological characteristics of the fresh specimens were observed. The microscopic features were observed under microscope by using fresh specimens and dried specimen. Sections of pileus, lamellae, and context were prepared with a razor blade. They were rehydrated in 3% KOH and stained with 1% (w/v) Phloxine and Melzer’s reagent [38] and then observed using an 80i compound light microscope (Nikon, Tokyo, Japan) at either 400× or 1000× magnification. A total of 20 basidiospores and basidia were measured. Q value denotes the length/width ratio of the basidiospores. Basidiospore statistics include: Xm, the arithmetic mean of the basidiospore length by basidiospore width (±standard deviation) for n basidiospores measured in a single specimen. Morphological identification was assisted using the literatures [8,10,11].
2.3. DNA extraction, PCR amplification and sequencing
Genomic DNA was extracted from fresh or dried fruit bodies of Termitomyces using CTAB method [39] with slight modifications. The fungal primer pair LR0R/LR5 [40] and SSUFW/SSUREV [1] were used to amplify the nrLSU region and the mtSSU, respectively. PCR reactions were performed in a C 1000 thermal cycler (Bio-Rad, Hercules, CA) using AccuPower PCR premix (Bioneer Co., Daejon, Korea) in a final volume of 20 µl containing 10 pmol of each primer and 1 µl of genomic DNA. PCR amplification was performed as described by Mossebo et al. [8]. The PCR products were electrophoresed through a 1% agarose gel stained with EcoDye DNA staining solution (SolGent Co., Daejeon, Korea) and purified with the Expin PCR Purification Kit (GeneAll Biotechnology, Seoul, Korea) according to manufacturer’s instructions. DNA sequencing was performed in both directions using the PCR primers at Macrogen (Seoul, Korea) in an ABI3700 automated DNA sequencer.
2.4. Phylogenetic analysis
New sequences were generated for nrLSU and mtSSU. Sequences were manually corrected by reviewing the chromatograms of the DNA sequences. All sequences were assembled and edited using MEGA 6 software [41] and were deposited in GenBank (www.ncbi.nlm.nih.gov) with the accession number (GenBank accession numbers: MK472701 for nrLSU and MK478904 for mtSSU). A total of 28 nrLSU and 29 mtSSU sequences were retrieved from GenBank (Table 1). Lyophyllum semitale and L. decastes were selected as outgroup. All sequences were aligned using MAFFT v7 [42] with the default settings.
Table 1.
List of species, voucher number, geographic origin and GenBank accession numbers of nrLSU and mtSSU sequences used in the molecular analysis.
| Species | Herbarium number | Geographic origin | GenBank accession numbers |
|
|---|---|---|---|---|
| nrLSU | mtSSU | |||
| T. aurantiacus | HUY1-DM 152E | Cameroon | KY809234 | KY809186 |
| T. brunneopileatus | K(M) 144300 | Cameroon | KY809273 | KY809225 |
| T. bulborhizus | K(M) 128338 | China | KY809261 | KY809213 |
| T. cartilagineus | K(M) 109565 | South Africa | KY809259 | KY809211 |
| T. clypeatus | K(M) 128340 | China | KY809262 | KY809214 |
| T. entolomoides | tgf103 | Africa | AY232693 | AY232680 |
| T. eurrhizus | K(M) 142 419 | Zambia | KY809266 | KY809218 |
| T. globulus | HUY1-DM 770 | Cameroon | KY809252 | KY809204 |
| T. heimii | K(M) 16 528 | Malaysia | KY809253 | KY809205 |
| T. heimii | K(M) 109 538 | Pakistan | KY809257 | KY809209 |
| T. letestui | HUY1-DM 666A | Cameroon | KY809248 | KY809200 |
| T. gilvus | BORH/FUMS-A03 | Malaysia (Borneo) | MK472701 | MK478904 |
| T. mammiformis | HUY1-DM 25G | Cameroon | KY809230 | KY809183 |
| T. mboudaeina | HUY1-DM 223E | Cameroon | KY809237 | KY809189 |
| T. medius | K(M) 16 685 | Nigeria | KY809254 | KY809206 |
| T. medius | HUY1-DM 372G | Cameroon | KY809243 | KY809195 |
| T. microcarpus | HUY1-DM 268E | Cameroon | / | KY809191 |
| PRU3900 | / | AF042587 | AF357092 | |
| T. robustus | HUY1-DM 436 | Tanzania | KY809265 | KY809217 |
| T. sagittaeformis | K(M) 109566 | South Africa | KY809260 | KY809212 |
| T. schimperi | HUY1-DM 24E | Cameroon | KY809228 | KY809181 |
| T. singidensis | tgf74 | Tanzania | AY232713 | AY232687 |
| T. striatus | K(M) 142436 | Malawi | KY809267 | KY809219 |
| T. striatus f. bibasidiatus | HUY1-DM 280 | Cameroon | KY809240 | KY809192 |
| HUY1-DM 280B | Cameroon | KY809241 | KY809193 | |
| T. subumkowaan | HUY1-DM 260B | Cameroon | KY809275 | KY809227 |
| HUY1-DM 260F | Cameroon | KY809239 | KY809190 | |
| T. titanicus | K(M) 142 416 | Zambia | KY809264 | KY809216 |
| Lyophyllum semitale | CBS 369.47 | – | AF223207 | AF357124 |
| L. decastes | JM 87/16 | – | AF042583 | AF357136 |
Accession numbers of the newly generated sequences are indicated in bold. BORH-BORNEENSIS Herbarium, Universiti Malaysia Sabah.
Bold sequences indicate new sequences produced in this study.
Maximum likelihood (ML) and Bayesian Analysis (BA) were performed with the following parameters (i) ML: the analysis was run in the RAxML v. 3.3 in CIPRES web portal (http://www.phylo.org/portal2/; [43]) under a GTR model with 1000 bootstrap replicates; (ii) BA: the analysis was run using MrBayes 3.2.2 [44] for 10 million generations, under a HKY + G + I and HKY + G model for nrLSU and mtSSU, respectively, with four chains, and trees sampled every 100 generations; after examining the graphic representation of the likelihood scores, using Tracer (http://tree.bio.ed.ac.uk/software/tracer/), the burn-in period was set to 1.5 million generations for all datasets. Bootstrap values (BS) ≥70% and posterior probability (PP) ≥90% values were considered significant. The edited alignment sequence dataset was deposited in TreeBase (http://purl.org/phylo/treebase/phylows/study/TB2:S23814).
3. Results
The sequencing of nrLSU and mtSSU from the specimen was successful. Additional sequences of the two loci were retrieved from GenBank and using in constructing the alignment datasets. The concatenated alignment contained 30 specimens, of which sequences from the two loci were present. The alignments of nrLSU and mtSSU consisted of 566 and 393 characters, respectively.
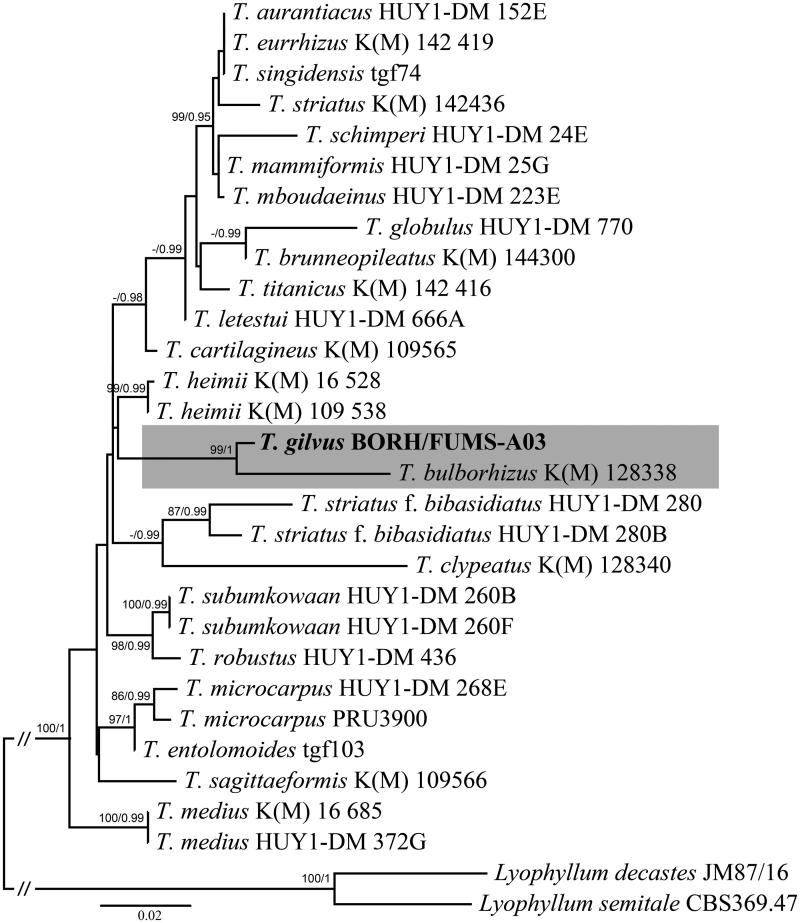
A phylogeny tree made from the concatenated genes of LSU and mtSSU suggests that Termitomyces specimen is closely related to T. bulborhizus from China [45,46]. ML analysis revealed that Termitomyces specimen is monophyletic with strong support (BS = 100%; PP = 1.0) and distinctive compared to T. bulborhizus from China. The same topology was observed for single gene analysis using nrLSU and mtSSU (not shown here).
4. Taxonomy
Termitomyces gilvus C.S. Yee, and J.S. Sathiya Seelan, sp. nov. (Figures 1 and 2)
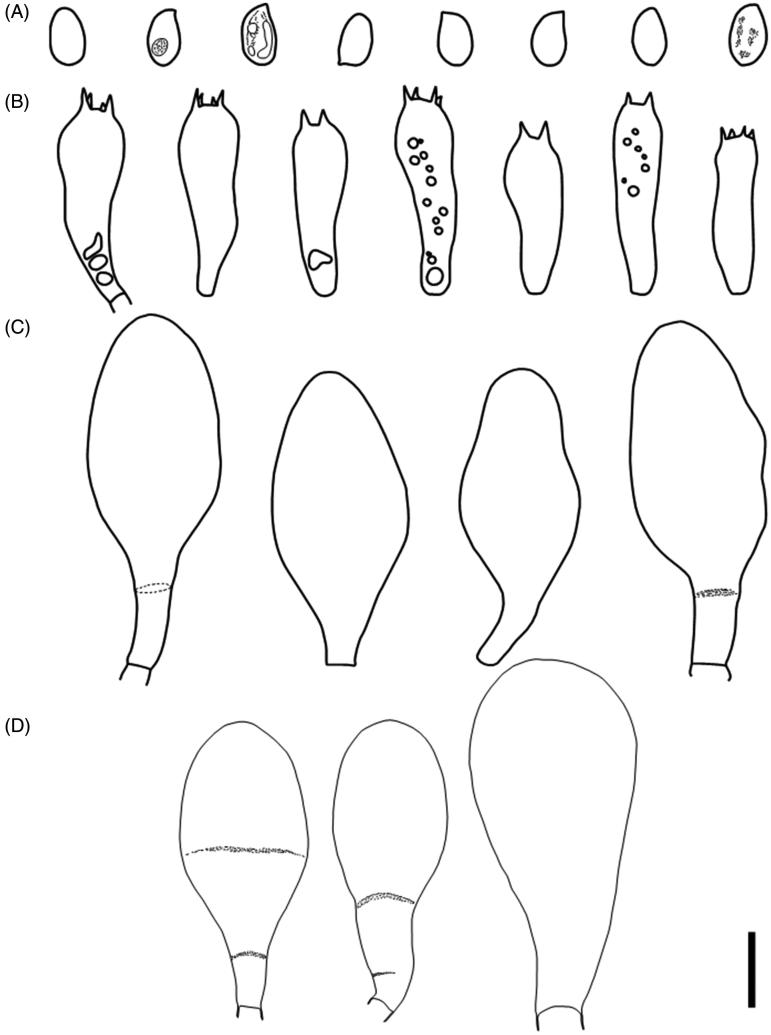
Figure 1.
Fruit body of Termitomyces gilvus sp. nov. (BORH/FUMS-A03, holotype). (A–B, E) surface of pileus; (C) lamellae; (D, F) stipe.
Figure 2.
Microscopic features of Termitomyces gilvus sp. nov (BORH/FUMS-A03, holotype). (A) basidiospores; (B) basidia; (C) cheilocystidia; (D) pleurocystidia. Scale bar = 10 µm.
MycoBank MB829475.
Diagnosis: Termitomyces gilvus differs morphologically from the closely related species of T. bulborhizus by pileus size and color, length of bulbous stipe, short pseudorrhiza, and larger basidia and pleurocystidia.
Type: Malaysia. Sabah, Kota Kinabalu, Universiti Malaysia Sabah campus (UMS), 50 m elev., N6˚2′ 116˚6′ E, 10 May 2018, leg. J.S Sathiya Seelan, collected by J.S. Sathiya Seelan, BORH/FUMS-A03 (Holotype, BORH!).
rDNA and mtDNA sequence ex holotype: MK472701 (nLSU); MK478904 (mtDNA)
Pileus 80–130 mm in diam, fleshy, at first convex becoming convexo-applanate with strongly blunt pointed perforatorium and irregularly lobed margin; Surface brownish orange (5C6) to dark brown at the center, brownish yellow (5C7) to orange white (5A2) toward the margin, rough to slightly smooth; perforatorium usually dark brown (6F5) velar squamules; margin enrolled when young and expanding toward the age, never uplifting, often crenulate. Lamellae free, white to pinkish (11A2) to 11 mm wide, densely crowded, with two series of lamellulae between lamellae. Context up to 40 mm thick, white, comprising repent, thin-walled hyphae, 2.7–7.7 µm diam, inflating to 28 µm. Spore print pink. Odor pleasant. Stipe 9–13 cm long above ground, 5–11 cm thick, enlarged to 1–6 cm diam. at ground level and usually abruptly forming a prominent globose bulb below ground, solid, robust, fibrous, surface above and pale brown on the bulb; with concolorous, sparsely distributed floccules. Partial veil ephemeral and often absent after the initial stage. Pseudorrhiza to 3 cm long, narrowing to 30–60 mm immediately below the bulb or tapering toward the base, surface white to pale brown, with longitudinal grooves and cracks.
Basidiospores 6–8.5 × 3.7–5.4 µm (Q = 1.5–1.6(1.8), n = 20), ovoid to ellipsoid, subhyaline, thin-walled. Basidia 21.8–29.2 × 6.1–8.3 µm, clavate, bearing four sterigmata. Hymenophoral trama regular, 8.0–20 µm wide, of hyaline hyphae, 2.5–22.0 µm diam. Subhymenial layer 12.0–30.0 µm diam. wide, repent hyphae, 2.4–2.9 µm diam. Lamella-edge heteromorphous, with crowded cheilocystidia, dispersed with basidia. Cheilocystidia clavate to pyriform, 36.8–51.1 × 12.7–24.8 µm, thin-walled. Pleurocystidia abundant, clavate to pyriform, occasionally turbinate, 47.0–66.3 × 21.2–31.5 µm, thin-walled. Pileipellis a repent epicutis of narrow, radial hyphae, 14.2–14.9 µm. Clamp connection present.
Habitat: Scattered to gregarious around termite (Macrotermes gilvus) mounds or on soil within campus area.
Etymology: The species epithet is named after the host termite species that was found.
Comments: Termitomyces gilvus mainly found scattered to gregarious near termite mound (colonies of Macrotermes gilvus). The species is morphologically and phylogenetically similar to T. bulborhizus. The pileal surface color of T. gilvus is brownish orange compared to T. bulborhizus (KM128338, China) which has a darker reddish brown in color. Termitomyces gilvus has smaller bulbous stipe and shorter pseudorrhiza compared to T. bulborhizus. Microscopically, T. gilvus has slightly larger basidia and pleurocystidia compared to T. bulborhizus.
5. Discussion
Morphological characters and concatenated analyses of nrLSU-mtSSU of the novel edible T. gilvus are produced herein (Figure 3). The type specimen BORH/F-UMSA03 T. gilvus is different from T. bulborhizus specimens from China and Thailand based on their pileus size and color, lamellae, stipe, pseudorrhiza, basidia, pleurocystidia and the termite host (Table 2). Termitomyces gilvus is mainly characterized by brownish orange to dark brown colored dry pileus, thick stipe, wide lamellae, shorter pseudorrhiza with floccose, and larger basidia and pleurocystidia. Meanwhile, T. bulborhizus is mainly distinguished by the large basidioma, bulbous stipe base and floccose stipe surface [45,46].
Figure 3.
Phylogenetic position of Termitomyces species inferred from concatenated nrLSU-mtSSU sequences using maximum likelihood analysis. Bootstrap and posterior probability values (ML/PP: 100%/1.0) are indicated above/below branches. The new species in in bold.
Table 2.
Macroscopic, microscopic features, termite hosts and distribution of Termitomyces gilvus and closely related species of T. bulborhizus from China and Thailand.
| Characters/Host |
T. gilvus (Malaysia) (This study) |
T. bulborhizus (China) [46,47] |
T. bulborhizus (Thailand) [19] |
|---|---|---|---|
| Pileus size | 8−13 cm | 10−22 cm | 9.2−21 cm |
| Pileus color | Surface brownish orange (5C6) to dark brown at the center, brownish yellow (5C7) to orange white (5A2) toward the margin | Surface reddish brown to dark brown at center; pale brown to brown toward the margin | Surface dark brown at the center, elsewhere pale brown to brown, paling toward the margin |
| Pileus shape | Convex to convexo-applanate | Convex to convexo-applanate | Convex then expanding to convexo-applanate |
| Lamellae | White to pink; 11 mm wide; densely crowded; lamellulae 2 series | Free; white to pink; 8 mm wide; densely crowded; lamellulae | White, to 9 mm wide, crowded, with lamellulae |
| Stipe | 9–13 cm long; 5−11 cm thick, enlarged to 1−6 cm diam. | 3–13 cm long; 0.8−6 cm thick; enlarged 3−9 cm diam. | 5−9 cm long; 6−8 cm thick; enlarged to 8.7−14.5 cm diam. |
| Pseudorrhiza | 3 cm long | 80 cm long | 1−4 cm long |
| Spore size | 6−8.5 × 3.7−5.4 µm | 6−9 × 4−6 µm | 5−8.5 × 3.5−6 µm |
| Basidia | 21.8−29.2 × 6.1−8.3 µm | 17.5−27 × 5.5−9 µm | 16−25 × 4−9 µm |
| Cheilocystidia | 36.8−51.1 × 12.7−24.8 µm | 19−60 × 12−34 µm | NA |
| Pleurocystidia | 47.0−66.3 × 21.2−31.5 µm | 19−78 × 10−32 µm | NA |
| Termite Host | Macrotermes gilvus | Odontotermes formosanus; Macrotermes barneyi | Hypotermes makhamensis |
| Distribution | Sabah (Malaysia) | South and southwest of (China) | Sai Yok district (Thailand) |
Characters with bold font are distinguishing features between the species.
NA: not available.
The most prominent morphological characters for T. gilvus in comparison to T. bulborhizus are presented in Table 2. Morphological features of T. gilvus mainly distinguished by the golden orange pilei, shorter pseudorrhiza, larger basidia and pleurocystidia. Termitomyces bulborhizus possess sub-globulose base at the end of the stipe similar to T. bulborhizus [45,46]. However, the bulbous stipe of type specimen BORH/FUMS-A03 tend to be smaller in size than the T. bulborhizus (Table 2). Pileus size were smaller (80–130 mm) and lamellae with 2-series were observed in the Bornean mushroom (BORH/FUMS-A03). However, T. bulborhizus from China tend to have larger pileus size (100–220 mm) and without lamellulae. Although similar morphology were observed as in T. bulborhizus, somehow, the bulbous stipe of T. gilvus is smaller and the presence of floccose stipe surface when it was freshly collected. Pseudorrhiza of T. bulborhizus were longer (80 cm long) for Chinese specimen however the Bornean specimen tend to be smaller in size (<3 cm long) which is similar to the pseudorrhiza of T. bulbohizus found in Thailand (1–4 cm long) [19].
Termitomyces bulborhizus were first recorded and described in Sichuan and Yunnan province in China. Later, it was recorded in the central region of Thailand [19,47]. Based on the ITS sequences, Sawhassan et al. [19] have reported that the Thai specimen was T. bulborhizus and similar to the Chinese specimen. In terms of host, T. clypeatus and T. bulborhizus are usually associated with Macrotermes gilvus Sawhassan et al. [19]. They recorded T. bulborhizus was associated with Hypotermes makhamensis in the central region of Thailand. This species is a fairly rare species in the area, and only one fruiting body was found. Besides, it occurs only nearby the termite mound of Macrotermes gilvus colonies below the Acacia mangium tree.
Phylogenetic analysis showed that the Bornean collection clusters within Termitomyces, and is closely related to T. bulborhizus. Sawhassan et al. [19] produced ITS gene of T. bulborhizus from Thailand specimen. Later, Mossebo et al. [8] reported that ITS collections for Termitomyces specimens were mostly limited and unidentified strains. The African and Asian specimens have been revised by Mossebo et al. [8] using the nrLSU and mtSSU for Termitomyces taxa which includes the T. bulborhizus specimen (KM 128338) from China in their study. The most representation of Termitomyces sequences (nrLSU and mtSSU) from Mossebo et al. [8] were used in this study for the phylogenetic position of the Bornean specimen, T. gilvus. Thus, T. gilvus is the first report for Sabah, using both morphology and with molecular data. Previously, T. clypeatus and T. eurrhizus were reported in Sabah [25,34] without any molecular work. So far, there are eight recognized Termitomyces species in Malaysia, but the results of this study suggest that there are more novel species awaiting for discovery. Further studies of Termitomyces using both morphology and phylogenetic analysis will be revised on this genus for Malaysia.
In this study, we propose that Termitomyces gilvus is a new species based on the differences in morphology and their phylogenetic placement, which is closely related to T. bulborhizus from China. Termitomyces gilvus is easily recognized due to their short bulbous stipe, golden orange pileus, large sized basidia and pleurocystidia. To our knowledge, this is the first report for Sabah (Northern Borneo) on this genus with adequate morphological description and molecular data. The erection of the new species has prompted the interest on how many species of Termitomyces are actually distributed within Borneo. The new species discovery within this genus suggests that more studies and sampling should be conducted to revise the Malaysian Termitomyces since it is regarded as one of the seasonal delicacy in Malaysia. The cultivation of Termitomyces could sustain the economic development among the local people in Sabah. In the long run through Borneensis-Agaricomycetes 2020–2025 project, many novel species discoveries are possible and this will be a major contribution to tropical mushrooms study. Thus, this study could serve as a baseline information to gather more information on this genus in Malaysia especially in Borneo.
Funding Statement
The authors would like to thank Universiti Malaysia Sabah for providing the financial support under early career grant SLB0119-STWN-2016.
Acknowledgements
J.S.S.S. was supported under the Korean Foundation for Advanced Studies (KFAS) to conduct this research project at Seoul National University (SNU) as part of the BORNEENSIS-AGARICOMYCETES Project 2019-2024. J.S.S.S., F.S.F., M.M.D. and C.S.Y. thanking to Sabah Parks and Sabah Biodiversity Council (SaBC) for providing the access license, TTS/IP/100-6/2 Jld. 7 (111) and export permit (JKM/MBS.1000-2/3 JLD.3 (89)). Fieldwork and laboratory experiments undertaken in this study comply with the current laws of Malaysia. We are grateful for the help of two anonymous reviewers for their comments on the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1.Aanen DK, Eggleton P, Rouland-Lefèvre C, et al. The evolution of fungus-growing termites and their mutualistic fungal symbionts. Proc Natl Acad Sci USA. 2002;99(23):14887–14892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aanen DK, Eggleton P. Fungus-growing termites originated in African rain forest. Curr Biol. 2005;15(9):851–855. [DOI] [PubMed] [Google Scholar]
- 3.Aanen DK. As you reap, so shall you sow: coupling of harvesting and inoculating stabilizes the mutualism between termites and fungi. Biol Lett. 2006;2(2):209–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hyodo F, Tayasu I, Inoue T, et al. Differential role of symbiotic fungi in lignin degradation and food provision for fungus-growing termites (Macrotermitinae: Isoptera). Funct Ecol. 2003;17(2):186–193. [Google Scholar]
- 5.Batra LR, Batra S. The fungus gardens of insects. Sci Am. 1967;217(5):112–120. [Google Scholar]
- 6.Batra LR, Batra S. Termite-fungus mutualism In: Batra LR, editor. Insect-fungus symbiosis. Nutrition, mutualism and commensalism. New York: Allanheld, Osmun and Co; 1979. p. 117–163. [Google Scholar]
- 7.Kirk PM, Cannon PF, Minter DW, et al. Dictionary of the fungi. 10th ed Wallingford, UK: CABI; 2008. p. 771. [Google Scholar]
- 8.Mossebo DC, Essouman EPF, Machouart MC, et al. Phylogenetic relationships, taxonomic revision and new taxa of Termitomyces (Lyophyllaceae, Basidiomycota) inferred from combined nLSU- and mtSSU-rDNA sequences. Phytotaxa. 2017;321(1):71–102. [Google Scholar]
- 9.Pegler DN, Vanhaecke M. Termitomyces of Southeast Asia. Kew Bull. 1994;49(4):717–736. [Google Scholar]
- 10.Turnbull E, Watling R. Some records of Termitomyces from old world rainforests. Kew Bull. 1999;54(3):731–738. [Google Scholar]
- 11.Sangvichien E, Taylor-Hawksworth PA. Termitomyces mushrooms: a tropical delicacy. Mycologist. 2001;15(1):31–33. [Google Scholar]
- 12.Venkatachalapathi A, Paulsamy S. Exploration of wild medicinal mushroom species in Walayar valley, the Southern Western Ghats of Coimbatore District Tamil Nadu. Mycosphere. 2016;7(2):118–130. [Google Scholar]
- 13.Teke NA, Kinge TR, Bechem E, et al. Ethnomycological study in the Kilum-Ijim mountain forest, Northwest Region, Cameroon. J Ethnobiol Ethnomed. 2018;14(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh HM, Ju YM. Medicinal components in Termitomyces mushrooms. Appl Microbiol Biotechnol. 2018;102(12):4987–4994. [DOI] [PubMed] [Google Scholar]
- 15.Frøslev TG, Aanen DK, Rosendahl S, et al. Phylogenetic relationship of Termitomyces and related taxa. Mycol Res. 2003;107(11):1277–1286. [DOI] [PubMed] [Google Scholar]
- 16.Chang ST, Quimio TH. Tropical mushrooms biological nature and cultivation methods. Hong Kong: The Chinese University Press; 1982. p. 493. [Google Scholar]
- 17.Rouland-Lefevre C, Diouf MN, Brauman A, et al. Phylogenetic relationships in Termitomyces (family Agaricaceae) based on the nucleotide sequence of ITS: a first approach to elucidate the evolutionary history of the symbiosis between fungus-growing termites and their fungi. Mol Phylogenetics Evol. 2002;22(3):423–429. [DOI] [PubMed] [Google Scholar]
- 18.Taprab Y, Ohkuma M, Johjima T, et al. Molecular phylogeny of symbiotic Basidiomycetes of fungus-growing termites in Thailand and their relationship with the host. Biosci Biotechnol Biochem. 2002;66(5):1159–1163. [DOI] [PubMed] [Google Scholar]
- 19.Sawhassan P, Worapong J, Vinijsanun T. Morphological and molecular studies of selected Termitomyces species collected from 8 districts of Kanchanaburi province, Thailand. Thai J Agric Sci. 2011;44:183–196. [Google Scholar]
- 20.Chang YS, Lee SS. Utilisation of macrofungi in Malaysia. Fungal Divers. 2004;15:15–22. [Google Scholar]
- 21.Azliza MA, Ong HC, Vikineswary S, et al. Ethno-medicinal resources used by the Temuan in Ulu Kuang Village. Stud Ethno-Med. 2012;6(1):17–22. [Google Scholar]
- 22.Azliza MA. A comparison of ethnobiological knowledge between the Mah Meri and Temuan tribes in Selangor [master’s thesis]. Kuala Lumpur: University of Malaya; 2013. [Google Scholar]
- 23.Faridah A, Kanapathipillay V, Whalley M, et al. Termitomyces mushrooms: a tropical delicacy. Mycologist. 2002;16:9. [Google Scholar]
- 24.Abdullah F, Rusea G. Documentation of inherited knowledge on wild edible fungi from Malaysia. Blumea – J Plant Tax Plant Geog. 2009;54(1):35–38. [Google Scholar]
- 25.Foo FS, Saikim FH, Kulip J, et al. Distribution and ethnomycological knowledge of wild edible mushrooms in Sabah (Northern Borneo, Malaysia. J Trop Biol Conserv. 2018;15:203–222. [Google Scholar]
- 26.Siddiquee S, Rovina K, Naher L, et al. Phylogenetic relationships of Termitomyces aurantiacus inferred from internal transcribed spacers DNA sequences. Adv Biosci Biotechnol. 2015;06(05):358–367. [Google Scholar]
- 27.Corner E. The seasonal fruiting of Agarics in Malaya. Gardens Bull Straits Settlement. 1935;9:79–88. [Google Scholar]
- 28.Corner EJH. A discussion of the results of the Royal Society Expedition to North Borneo organized by E.J.H. Corner. J Proc Linn Soc Bot Series B. 1964;161:3–6 and 90–91. [Google Scholar]
- 29.Corner E. 1981. The agaric genera Lentinus, Panus, and Pleurotus with particular reference to Malaysian species. Beih Nova Hedwigia. 1981;69:1–169. [Google Scholar]
- 30.Corner EJH. I am part of all that I have met (Tennyson’s Ulysses) In: Isaac S, Frankland JC, Watling R, Whalley AJS, editors. Aspects of tropical mycology. Cambridge: Cambridge University Press; 1993. p. 1–13. [Google Scholar]
- 31.Corner E. Agarics in Malesia I. Tricholomatoid. II. Mycenoid. Beih Nova Hedwigia. 1994;109:1–271. [Google Scholar]
- 32.Corner E. The Agaric Genera Marasmius, Chaetocalathus, Crinipellis, Heimiomyces, Resupinatus, Xerula and Xerulina in Malesia. Beih Nova Hedwigia. 1996;111:1–175. [Google Scholar]
- 33.Hjortstam K, Spooner BM, Oldridge SG. Some Aphyllophorales and Heterobasidiomycetes from Sabah, Malaysia. Kew Bull. 1990;45(2):303–322. [Google Scholar]
- 34.Pegler DN. The larger fungi of Borneo. Kota Kinabalu: Natural History Publications; 1997. p. 95. [Google Scholar]
- 35.Kornerup A, Wanscher JH. Methuen handbook of colour. 3rd ed London: Eyre Methuen; 1978. p. 252. [Google Scholar]
- 36.Ahmad M. Termites (Isoptera) of Thailand. B Am Mus Nat Hist. 1965;131:1–111. [Google Scholar]
- 37.Eggleton P, Homathevi R, Jeeva D, et al. The species richness and composition of termites (Isoptera) in primary and regenerating lowland dipterocarp forest in Sabah, East Malaysia. Ecotropica. 1997;3:119–128. [Google Scholar]
- 38.Largent DL, Johnson D, Watling R. How to identify mushrooms to genus, III Microscopic features. Eureka: Mad River Press; 1977. p. 25–26. [Google Scholar]
- 39.Bruns TD, Fogel R, Taylor JW. Amplification and sequencing of DNA from fungal herbarium specimens. Mycologia. 1990;82(2):175–184. [Google Scholar]
- 40.White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: a guide to methods and applications. New York: Academic Press; 1990. p. 315–322. [Google Scholar]
- 41.Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES science gateway for inference of large phylogenetic trees SC10 workshop on gateway computing environments (GCE10); 2010 Nov 13–19; New Orleans (LA): IEEE Computer Society; 2010. p. 1–8. [Google Scholar]
- 44.Ronquist F, Teslenko M, Van Der Mark P, et al. MrBAYES 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei TZ, Tang BH, Yao YJ. Revision of Termitomyces in China. Mycotaxon. 2009;108(1):257–285. [Google Scholar]
- 46.Wei T, Yao Y, Wang B, et al. Termitomyces bulborhizus sp. nov. from China, with a key to allied species. Mycol Res. 2004;108(12):1458–1462. [DOI] [PubMed] [Google Scholar]
- 47.Kosakul TA, Boasri A, Chalermpongse , Kuhiran M. Genetic diversity of Termitomyces in central Thailand using isozyme markers. J Sci Res Chula Univ. 2007;32:63–72. [Google Scholar]