Abstract
As microbiome research has moved from associative to mechanistic studies, the activities of specific microbes and their products have been investigated in development of inflammatory bowel diseases, cancer, metabolic syndrome, and neuropsychiatric disorders. Findings from microbiome research have already been applied to the clinic, such as in fecal microbiota transplantation (FMT) for treatment of recurrent Clostridium difficile infection. We review the evidence for associations between alterations in the intestinal microbiome and gastrointestinal diseases and findings from clinical trials of FMT. We discuss opportunities for treatment of other diseases with FMT, based on findings from small clinical and preclinical studies.
Following the completion of human genome project in 2003, scientists set their eyes on the next big genomic challenge—mapping microbial communities throughout the human body. For example, the HMP consortium analyzed bacterial communities of up to 18 body sites of 242 adults (129 men and 113 women, 18–40 years old) using 16S rRNA and whole-genome metagenome sequencing technologies (1,2). Similar microbiome cataloging efforts were pursued throughout the world, with for example a European consortium (Meta-Hit) focusing on intestinal microbiome composition in health and disease (3). Humans are colonized from birth with microorganisms that rapidly assembled into a complex community comprising archaea, bacteria, viruses, and fungi. It was thought that the combination of microbiome and genomic data would provide a powerful holistic view of the human superorganism that narrowed the gap between basic and clinical research. For example, studies of the human microbiome, combined with genetic information, could increase our understanding of susceptibility to immune disorders (4–6). These efforts have produced a large amount of data and contributed significant resources to the scientific community. However, findings from large microbiome analyses are only slowly being applied to the clinic.
The mammalian microbiome is dominated by bacteria, of 6 phyla: Actinobacteria, Bacteroidetes, Cyanobacteria, Firmicutes, Proteobacteria, and Fusobacteria. The relative abundance of these phyla varies not only among body sites (such as intestine vs skin), but also among individuals, depending on age, diet, health status, and location. The intestinal microbiome has a symbiotic relationship with its host, contributing to energy and nutrient extraction from the diet, development of the immune response, intestinal mucosal barrier integrity, and metabolism of xenobiotics (7–10).
Alterations in the microbiome, determined from analyses of fecal and intestinal samples, have been associated with gastrointestinal disorders such as ulcerative colitis (UC), irritable bowel syndrome (IBS), and constipation (11–13) as well as extraintestinal disorders such as cancer, metabolic syndrome, and neuropsychiatric disorders (14–16). Not surprisingly, efforts directed at modulating intestinal microbiota for therapeutic purposes have attracted wide attention from the scientific community.
This interest in fecal microbiota transplantation (FMT) is not recent—it was described by Ge Hong in 4th century China, who described the oral ingestion of fecal material for treatment of severe diarrhea (17). In 1958, the American surgeon Ben Eiseman reported that 4 patients with antibiotic-induced diarrhea rapidly improved after administration of fecal enemas (18). However, this field of research was put on hold for more than 50 years—the first controlled randomized trial of FMT for recurrent Clostridium difficile-induced colitis was performed by investigators in Amsterdam (19). FMT is now used worldwide to successfully treat patients with recurrent C difficile infection (CDI) that does not respond to antibiotics.
We review the potential for microbiota-based therapies for gastrointestinal disorders, focusing on FMT. For reviews of the clinical effects of probiotics, prebiotics, antibiotics, or bacteriophage on the intestinal microbiota (see ref 20). Research into the human intestinal microbiome and its dysregulation during disease pathogenesis is progressing rapidly. Due to length and topic restriction, we have not addressed all microbiota-associated diseases and excluded interventions in which the microbiota is manipulated with other methods than FMT.
Gastrointestinal Disorders
UC
The most evidence for the benefits of FMT, beyond treatment of CDI, comes studies of patients with inflammatory bowel diseases (IBD). The composition of the microbiota in the feces of patients with UC differs from that of healthy individuals (controls) in diversity is usually lower, with reduced relative abundance of Firmicutes (Clostridium clusters XIVa and IV) and Bacteroidetes. In particular, lower abundance of the butyrate producer Fecalibacterium prausnitzii was associated with UC, as well as an overrepresentation of Proteobacteria and Actinobacteria (21). This microbe imbalance leads reductions in short-chain fatty acids (SCFAs), predominantly butyrate, which are essential nutrients for colonocytes and important immune regulators. Diversion of the fecal stream by stoma can cause a type of colitis that has been treated successfully with topical butyrate preparations (22).
Four controlled trials of the effects of FMT in patients with active UC were published—3 reported positive outcomes (Table 1). Moayyedi et al randomly assigned 70 patients with active UC to groups that received allogeneic FMT or water (placebo), administered in 6 weekly enemas (23). The primary endpoint, a total Mayo score <3 and endoscopic healing (endoscopic Mayo score 0), was achieved by 24% of the patients who received FMT vs 5% of patients who received placebo. Interestingly, most patients who responded to the therapy received FMT from a single super-donor (39% of recipients met the endpoint vs 10% of the other donors), suggesting a donor effect, which has not been observed for CDI treatment. Patients who underwent FMT had greater microbial diversity, based on analyses of fecal samples, than patients who received the placebo.
TABLE 1:
controlled clinical trials with FMT for common gastrointestinal disorders
| Study | Indication | Intervention | Control Group | Study size | Effect size |
|---|---|---|---|---|---|
| Moayyedi et al. (22) | Ulcerative colitis | allegeneic FMT weekly enemas 6 week | Water enemas | 70 | 19% |
| Rossen et al. (23) | Ulcerative colitis | duodenal infusion of donor stool | Autologous feces | 50 | 10% |
| Paramsothy et al. (24) |
Ulcerative colitis | colonoscope injection+ 5 enemas (pooled) | Saline | 81 | 19% |
| Costello et al. (25) | Ulcerative colitis | colonoscope injection+enemas (pooled) | Autologous feces | 73 | 23% |
| Misra et al. (27) | Ulcerative colitis | Vanco+SER-287 | 58 | 40% | |
| Johnsen et al. (36) | Irritable Bowel Syndrome | colonoscope injection (2 mixed donors) | Autologous feces | 90 | 22% |
| Holvoet et al. (37) | Irritable Bowel Syndrome | colonoscope injection (2 mixed donors) | Autologous feces | 64 | 30% |
| Halkjær et al. (38) | Irritable Bowel Syndrome | FMT capsules | PLC Capsules | 52 | NONE |
| Tian et al. (39) | Slow transit constipation | duodenal infusion of donor stool | PLC | 60 | 33% |
A single-center study in Amsterdam studied a different approach for treatment of UC. Based on the reported effects of duodenal infusion of fecal material into patients with Clostridium-induced colitis (19), Rossen et al gave 25 patients with active UC (simple clinical colitis index scores of 4–11) 2 duodenal infusions of 500 ml fresh allogeneic fecal material (or autologous fecal material as control), 3 weeks apart (24). Twenty-five patients received their own feces via the same route as controls. The primary endpoint, clinical response (simple clinical colitis index scores ≤ 2 and mucosal improvement ≥ 1 point), was attained by 30% of patients who received allogeneic fecal material vs 20% of patients who received autologous fecal material, although the difference was not significant. Interestingly, an increase in microbial diversity (Shannon index) was observed in responders to allogeneic or autologous fecal material, but no super donor was identified in this study.
Paramsothy et al administered feces through a colonoscope followed by repeated enemas to patients with mild to moderate UC (25). The patients used colonic lavage prior to FMT and received feces from 3 to 7 pooled donors, rather than from a single donor. Steroid-free clinical remission with endoscopic response or remission was achieved in 1¼1 patients (27%) who received active fecal material and ¾0 (8%) who received the placebo (isotonic saline). Microbial diversity increased in responders and lack of remission was associated with increased relative abundance of Fusobacterium. Costello et al used the same mode of administration with fewer enemas but preserved the donor feces from pooled donors in anaerobic conditions. The outcome was comparable to that of the study by Paramsothy et al (remission in 32% of patients who received fecal material vs 9% who received placebo) (26).
FMT therefore appears to benefit certain patients with UC. However, the net benefit is limited and there is clear need for a better understanding of the kinetics of this treatment. Is duodenal infusion equal or superior to colonic administration? Is pre-treatment with colonic lavage or even antibiotics useful? Are anaerobic conditions necessary to preserve the feces? What types of microbes are included in samples from the super donor, compared with other less effective donors? To what extent is the change in colon flora permanent and are repeated treatments necessary to maintain remission? Which microbes mediate the therapeutic effects of FMT?
Transfer of fecal filtrate, by single nasojejunal administration, to 5 patients with chronic-relapsing CDI restored normal stool habits and eliminate symptoms (27). The fecal filtrate contains bacterial products and other microbes, but not intact bacteria. This finding indicates that the effects of FMT might not require intact bacteria, but instead involved other microorganism, such as bacteriophages, spores, or viruses, or other molecules present in fecal material.
The value of antibiotic pre-treatment was addressed in a randomized controlled trial by Bharat et al (at Seres Therapeutics, in Boston, MA). SER-287 is an investigational product that contains bacterial spores of Firmicutes (28). Patients with ulcerative colitis given vancomycin had higher engraftment of microbes from SER-287 and better outcomes following administration of the product. Antibiotic pretreatment might therefore increase the effects of FMT or other microbe-based therapies. However, further studies of this effect are needed.
Crohn’s disease (CD)
Given the promising effects of FMT in patients with UC, its effects were studied in patients with CD. However, the available evidence of efficacy of FMT for patients CD is weaker than for patients with UC patients and there have been no controlled studies. Only uncontrolled case series studies of 6–30 patients with CD have been published, with poor documentation of outcomes and a great variety of reported effects (29,30). Intriguingly, patients in some of the studies reported high fevers a few hours following FMT. One patient in a trial in Belgium developed aspiration pneumonia as a serious complication. No further trials of the effects of FMT are known to be underway in patients with CD.
Pouchitis
Development of pouchitis following restorative proctocolectomy in patients with refractory IBD is believed to be mediated by bacteria— pouchitis develops only after restoration of the intestinal continuity and usually resolves following administration of antibiotics such as metronidazole and ciprofloxacin (31). Moreover, probiotics have been reported to be effective in treatment of pouchitis, indicating that it responds to changes in the microbiome. (32,33).
In a case series of 8 patients with chronic pouchitis, FMT provided no clear benefit administered via the nasogastric route, although 2 patients regained sensitivity to ciprofloxacin therapy (34). Changes in the composition of the pouch microbiome were observed. Although optimization of the microbiome of the pouch is an attractive approach to reduce morbidities of patients with chronic or recurrent pouchitis, further studies are needed in this difficult population.
IBS
The prevalence of IBS is as high as 20% in areas of the United States (35). IBS is characterized by a variety of symptoms and there is great need for effective treatments. Patients with IBS patients have reduced diversity of the intestinal microbiome, with increased abundance of enterobacteriaceae and relatively lower levels of bifidobacteria and lactobacilli. Lack of butyrate production and increased amounts of acetic and propionic acids have been associated with bloating (36).
IBS is the only disease, beside UC, in which several prospective and controlled clinical trials of FMT have been performed (Table 1). In Norway, 90 patients with IBS with predominant diarrhea or diarrhea and constipation were randomly assigned (2:1) to groups that received FMT or placebo. The response rates at 3 months were 43% in patients who received placebo (quite high) but 65% in patients who received FMT (mixed feces from 2 donors) (a significant increase) (37). Holvoet et al studied 64 patients with IBS with predominant bloating without constipation. Patients were randomly assigned (2:1) to receive FMT (from 2 donors) via colonoscopy or their own feces (controls). The patients who received FMT from the donors had significant reductions in discomfort, abdominal pain, and flatulence, but not patients who received their own fecal samples (38). The microbiome analysis from this study has not been published, but Holvoet et al previously reported that patients with IBS with a response to FMT had higher baseline concentrations of Streptococcus and higher enrichment of the microbiome than non-responders (39).
Halkjær et al studied the effects of fecal microbiota capsules, vs placebo, in patients with IBS. Patients who received the fecal material capsules had an increase in microbial biodiversity, based on analyses of their feces, but, interestingly, symptom improvement was greater in patients who received the placebo (40). In conclusion, although there is some evidence from randomized controlled trials that administration of fecal material is effective for patients with IBS, detailed analyses of microbiome profile are needed before and after administration. Researchers should study changes in composition of microbiomes most associated with symptom improvement.
Other benign GI diseases
Constipation could be associated with dysbiosis of the intestinal microbiome, although there is weaker evidence for this than for other diseases. The effects of conventional constipation treatment were compared with those of 6 sessions of FMT (naso-enteric administration, along with conventional treatment) in 60 adults with slow-transit constipation (Table 1) (41). Patients who received FMT had significant reductions in symptoms and increases in stool consistency and colonic transit time compared to patients given conventional treatment. Further research is warranted in this field. Trials of lactobacilli and bifidobacteria are underway or completed (42).
Doki et al investigated the association between changes in the microbiome and development of graft vs host disease (GVHD) following allogeneic hematopoietic stem cell transplantation. Although the diversity of the intestinal microbiome did not differ among patients who did and did not develop GVHD, patients who did develop GVHD had a significantly higher abundance of Firmicutes and a lower abundance of Bacteroidetes (43). Although this study did not adequately account for diet, the authors concluded that maintenance treatment with Bacteroidetes throughout hematopoietic stem cell transplantation might prevent GVHD. Based on those observations, a number of uncontrolled, small experiments were performed and indicated that FMT is safe and effective for patients undergoing hematopoietic stem cell transplantation, increasing microbial diversity. Higher abundance of Eubacterium limosum reduced risk relapse or progression of GVHD in 541 patients who underwent allogeneic stem cell transplantation and were followed for 2 years, which validated these uncontrolled findings uncontrolled findings (44–46).
Use broad-spectrum antibiotics is often associated with changes in bowel habits, most commonly watery diarrhea, called post-antibiotic colitis. Some researchers have suggested that probiotic preparations might prevent this complication (47). Restitution of the mucosal intestinal microbiome was studied in mice and humans given antibiotics, followed by autologous transplantation of feces (collected prior to the use of antibiotics) or administration of multi-strain probiotics. Whereas autologous feces induced rapid normalization of the microbiome (within days, measured in fecal samples), probiotics delayed normalization (48). FMT might therefore benefit patients with post-antibiotic diarrhea, but larger controlled trials are needed.
Hepatic encephalopathy
End-stage liver cirrhosis often leads to portal hypertension and recurrent hepatic encephalopathy (HE), which often requires hospitalization and has been associated with dysbiosis of the microbiome. In a small open-label randomized trial, 20 male outpatients with cirrhosis received 5 days of broad-spectrum antibiotics followed by FMT, from a single donor and administered via a 1 enema, or the standard of care. Interestingly, the donor was selected based on machine learning data aiming at the highest relative abundances of Lachnospiraceae and Ruminococcaceae among a universal stool donor bank. Encephalopathy recurred in 5/10 patients given the standard of care but in none of 10 patients who underwent FMT; FMT was also reported to improve cognitive function (49). No change in fecal microbiome diversity indices was observed in patients given the standard of care, but changes were observed in patients given FMT (a relative increase in Lactobacillaceae and Bifidobacteriaceae). Further studies of these interesting effects are needed.
Inflammatory Disorders Outside the Gastrointestinal Tract
Psoriasis
Psoriasis is a common inflammatory condition of the skin with a prevalence of approximately 3% worldwide and pathophysiologic similarities with IBD. There is little evidence for the efficacy of FMT in patients with psoriasis. There is evidence that the microbiota of the skin affects the development and severity of psoriasis and possibly also response to therapy (50). The relative abundance of Akkermansia mucinophila appears to be reduced in intestinal microbiota of patients with psoriasis (51) and the ratio of Firmicutes:Bacteroidetes was 3-fold higher in patients with psoriasis compared to controls (52); this disturbance correlated with the psoriasis severity score PASI.
Successful therapies for psoriasis, such as balneotherapy and narrow-band ultraviolet B radiation, have been associated with alterations in the skin microbiota (53). Studies of strategies to alter gut microbiomes of patients with psoriasis are underway. These include a randomized, placebo-controlled trial of FMT into the small intestine of patients with psoriatic arthritis or active peripheral disease that has not responded to methotrexate (54).
Central Nervous System Diseases
Multiple sclerosis
Multiple sclerosis is a chronic autoimmune disease characterized by demyelination and serious neurological disability—few effective treatments available. Many patients with multiple sclerosis have gastrointestinal symptoms and alterations in the intestinal microbiome, compared to healthy individuals (controls) (55). Studies in animal models have established a role of the gut microbiome in disease progression. In mice with autoimmune encephalomyelitis (EAE), inflammatory responses were attenuated under germ-free conditions (56) and reduced by strains butyrate-producing or other bacteria. Butyrate can induce epigenetic modifications such as acetylation of the Foxp3 locus to produce anti-inflammatory effects (57). Multiple sclerosis might be treated with microbes that produce these SCFAs, as in patients with UC. Two prospective trials with FMT are underway.
Parkinson disease
Parkinson disease is an intractable neurodegenerative disorder that has been associated with gastrointestinal conditions such as constipation and IBD. Patients with IBS have an increased risk for developing Parkinson disease (58). The intestinal microbiome of patients with Parkinson disease is characterized by an overabundance of Bacteroidetes, F prausnitzii, Enterococci, Prevotella, and Clostridium species—these alterations are associated with a poor course of Parkinson disease (59). Microbiota transplanted from patients with Parkinson disease into a mouse model of the disease led to worsening of neurological manifestations whereas depletion of gut microbiota in the same model reduced neurologic symptoms (60). Brain derived neurotrophic factor (BDNF) mediates interactions between intestinal cells and the nervous system. BDNF produced by the gut microbiome and is reduced in patients with Parkinson disease, leading to neurodegeneration (61). Changes in the microbiome that increase synthesis of BDNF might reduce symptoms and slow progression of Parkinson disease, but there are few data from controlled studies. Studies to alter the microbiome of Parkinson’s patients are underway.
Autism
Autism is often associated with constipation, bloating, diarrhea, and alterations in the intestinal microbiome. Children with autism had a reduced ratio of Bacteroidetes:Firmicutes (62). Changes in the microbiome might interfere with the tryptophan metabolism to alter behavior. In a pilot trial of FMT for children with autism, preceded by 2 weeks of antibiotics, behavioral symptoms improved in parallel with intestinal symptoms (bloating, constipation, diarrhea); these improvements were maintained for more than 8 weeks after FMT. Overall bacterial diversity increased, with increased abundances of Bifidobacterium, Prevotella, and Desulfovibrio observed for more than 8 weeks after FMT (63). This observation indicates a link between the microbiome and behavior, but results are preliminary. However, in a cohort of simplex families with autism spectrum disorder (ASD) and neurotypical siblings, there was no significant difference detected in diversity or composition of fecal microbiomes of children with ASD vs their siblings without ASD (64). Further studies are needed before FMT can be recommended treatment of autism.
Cancer
Although FMT has not been tested in the patients with cancer, there is great opportunity for ecosystem manipulation for this pathology. A link between carcinogenesis and microorganisms was established decades ago, with for example development of various form of cancers including lymphoma, leukemia, gastric cancer, and hepatocellular carcinoma, following infection with class 1 carcinogenic microorganisms such as Helicobacter pylori, hepatitis B or C viruses, Epstein-Barr virus, or Kaposi sarcoma herpes virus (65). However, over the past decade there has been tremendous progress in our understanding of the role of the entire microbiome in carcinogenesis, as opposed to single microorganisms.
Due to the size and diversity of the intestinal microbiome, it is not surprising that its relationship with colorectal cancer (CRC) has been widely studied (66). Experiments involving transfer of microbial communities from one host to another have demonstrate diseased transmissibility and protection. Fecal microbiota transfer experiments can be performed either passively, through coprophagy (co-housing), or actively, through oral-gastric feeding. For example, mice with disruption of the nucleotide-binding oligomerization domain-containing protein 2 gene (Nod2) gene, which encodes a protein that recognizes bacterial molecules and stimulates the inflammasome pathway, are more susceptible to colitis-associated colorectal cancer than wild-type mice (67). Interestingly, the risk of colitis is increased in wild-type mice passively exposed (co-housed) to fecal microbiota from NOD2-deficient mice. Wild-type mice passively exposed to fecal biota from NLRP6-deficient mice have increased susceptibility to colitis-associated cancer (68). NLRP6 is also part of the inflammasome pathway.
The ability of fecal microbiota from Nod2−/− mice to induce colitis in wild-type mice was associated with changes in Bacteroides, Butyrivibrio, and Lachnobacterium communities. Since bacteria do not have effects on NLRP6 signaling (69), it is not clear which microorganisms determine susceptibility to colitis. Importantly, transplantation of fecal microbiota from healthy wild-type mice reduced development of colitis in Nod2−/− mice. The DNA-sensing molecule AIM2 protects against colorectal carcinogenesis (70,71). Passive exposure of Aim2−/− mice to fecal microbiota from wild-type mice reduced tumor development (70), supporting the concept that a component of the microbiota could prevent colorectal carcinogenesis in animal models.
The importance of the microbial ecosystem in CRC development was demonstrated in an elegant study in which a pool of fecal materials obtained from healthy subjects or patients with CRC was transferred to germ-free wild-type mice or wild-type mice exposed to the carcinogenic compound azoxymethane (72). The proportion of mice with polyps and numbers of colon polyps were significantly higher in mice that received fecal materials from patients with CRC than in mice that received fecal microbiota from healthy subjects. Interestingly, germ-free, wild-type mice that received fecal material from patients with CRC or healthy individuals did not develop polyps, indicating that genetic factors affect the ability of microbes to promote carcinogenesis.
It is important to note that the influence of intestinal microbiota on carcinogenesis extends beyond the intestine. Researchers demonstrated a functional link between intestinal microbiota and the development of pancreatic cancer in mice (73,74). One study showed that development of pancreatic ductal adenocarcinoma (PDAC) was prevented when Pdx1-Cre; LSL-Kras mice were bred under germ-free conditions (74). Moreover, antibiotics reduced development of PDAC in Pdx1Cre;LSL KrasG12D;Trp53R172H (KPC) mice, whereas fecal transferred of KPC-derived feces, but not feces from wild-type mice accelerated tumorigenesis (74). A study of KrasG12D; PTENlox/+ mice showed that PDAC progression was attenuated when the intestinal bacterial community was depleted with antibiotics, compared to microbiota-intact mice (73).
Changes in the intestinal microbiota have also been associated with liver cancer progression, affecting metabolism of bile acid from primary to secondary structures. Secondary bile acids inhibit recruitment of natural killer T cells, which have anti-tumor effects, to the liver (75). Mice with disruption of the tet methylcytosine dioxygenase 2 gene (Tet2−/− mice) have preleukemic myeloproliferation (PMP). These mice have impaired intestinal barrier function, which causes bacterial translocation in the spleen and mesenteric lymph nodes, resulting in increased plasma levels of IL6 (76). This microbiota-dependent increase in IL6 promotes expansion of IL6Rα+ granulocyte-macrophage progenitors—a step in the development of PMP. Importantly, microbiota manipulation through germ-free husbandry conditions or introduction of antibiotics prevented and reversed PMP development inTet2−/− mice. Overall, these findings indicate that alterations to the intestinal microbiota can promote carcinogenesis, revealing therapeutic opportunities.
Researchers have compared intestinal bacteria of healthy individuals with those of patients with CRC, to identify microbial biomarkers of cancer stage and progression (77–81). Species such as Fusobacterium nucleatum, Bacteroides clarus, Roseburia intestinalis, Clostridium hathewayi, and an undefined species named m7 were detected by quantitative PCR and associated with CRC in 2 Asian cohorts (81). Studies are needed to determine whether these markers can be used to identify patients with CRC in different populations. Furthermore, it is not clear whether any preventive action could be taken after identification of individuals at risk for cancer, based on microbe markers—we don’t know if these markers identify patients with early-stage, treatable neoplasia.
Based on evidence showing differences in the intestinal microbiomes of individuals with vs without CRC, and findings from mice that different microbiomes affect risk of colitis-associated cancers, FMT might be used to prevent CRC or slow its progression (Fig.1). FMT might be included with, or performed before or after, cancer surgery or chemotherapy. Prospective studies are needed to test the effects of FMT in patients with CRC.
Figure 1: Potential Application of FMT to Cancer Therapy.
Intrinsic and extrinsic factors can disrupt healthy intestinal microbiota and increase susceptibility to cancer. Replacement of the intestinal microbiota with FMT might be used to prevent or treat different forms of cancer including colorectal, liver, and pancreatic cancer.
Studies have associated the intestinal microbiome with response to cancer therapy (81–84), expanding this field of microbiome research from promoting to treating cancer. For example, in mice with xenograft tumors grown from P815 mastocytoma or MCA205 sarcoma cells, the anti-tumor effects of cyclophosphamide were reduced if mice were germ-free or given antibiotics (86). Furthermore, orally administered Lactobacillus johnsonii and Enterococcus hirae increased the anti-tumor effects of cyclophosphamide in mice (86). In mice with xenograft tumors grown from EL4 lymphoma or MC38 colon carcinoma cells, antibiotics reduced the cytotoxic effects of oxaliplatin and cisplatin (87). Microbes promoted the antitumor effects of CpG-oligonucleotide immunotherapy in mice with xenograft tumors grown from EL4 lymphoma, MC38 colon carcinoma, or B16 melanoma cells. These findings indicate that specific microbes, or their products, can increase the effects of cancer therapies, and that FMT might have effects in patients with cancer undergoing treatment.
Researchers have also studied the effects of the microbiota on immune checkpoint inhibitor therapy (88). Antibodies against CTLA4 did not inhibit growth of xenograft tumors derived from MCA205 sarcoma, MC38 colon carcinoma, or Ret melanoma cells in germ-free mice or mice given antibiotics compared with mice carrying a complete microbiota (89). Alterations in the intestinal microbiota modified the efficacy of antibodies against PD-L1 in mice with xenograft tumors grown from B16.SIY melanoma cells (90). Interestingly, the modulatory effect of the microbiota on therapeutic efficacy was associated with increased myeloid cell-mediated, T-helper (Th) cell-mediated (Th1 and Th17), and CD8+ T-cell responses. Although these findings were made in studies of mice, they indicate that microbes and their products might synergize with anti-cancer agents to slow tumor development.
How relevant is this research to human cancer? Studies of large cohorts of patients with advanced renal cell carcinoma (RCC) or non-small-cell lung cancer (NSCLC), from 2 different cancer centers, showed that administration of antibiotics within 30 days of anti-PD1, anti-PDL1, or anti-CTLA4 agents (alone or in combination) reduced times of progression-free and overall survival, compared to patients who did not receive antibiotics (91–92). These findings indicate that intestinal bacteria might affect patient responses to immune checkpoint inhibitors. A number of observations support this hypothesis; the microbiomes of patients with advanced RCC, NSCLC, or melanoma who respond to anti-PD1 therapy differs from than those of non-responders (92–94). Remarkably, when fecal samples from patients who responded or did not respond to anti-PD1 therapy were administered orally to germ-free mice with tumors, the mice had the same response (or lack of response) to anti-PD1 treatment (91, 93, 94). Although all 3 studies concluded that the composition of the microbiome is an important determinant of response to immune checkpoint inhibitors, the composition of the microbiomes associated with response varied among the studies. For examples, PD1 responsiveness in patients with advanced melanoma was associated with increased relative abundance of Feacalibacterium species (94) or Bifidobacterium (93), whereas Akkermansia muciniphila was associated with treatment efficacy in patients with RCC or NSCLC (91). Importantly, introduction of A muciniphila was able to reverse unresponsiveness in mice given fecal samples from non-responders (91), so specific microbes might determine the effects of certain immunotherapeutic agents. Interestingly, using datasets from these studies (91–93), researchers found microbe composition to have poor predictive power in defining PD1 responsiveness, whereas microbial gene content had better predictive performance (95).
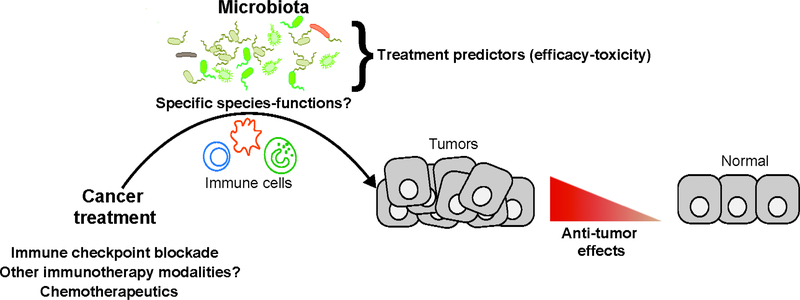
Side effects are an important concern for strategies to manipulate the microbiomes of patients receiving immune checkpoint inhibitor therapy. For example, an adverse effect of the CTLA4 inhibitor ipilimumab is development of a Crohn’s-like colitis, observed in 8%–30% of patients (96). Dubin et al studied the intestinal microbiota of 34 patients with metastatic melanoma treated with ipilimumab and observed increased proportions of the Bacteriodetes phylum in patients who did not develop colitis (97). Whole-genome metagenome analysis of patients given CTLA4 inhibitors revealed that microbial modules for polyamine transport system and the biosynthesis of thiamine, riboflavin, and pantothenate were associated with risk of colitis (97). In a subsequent study of 26 patients with metastatic melanoma treated with ipilimumab, Chaput et al observed that patients with a high abundance of Bacteroidetes before treatment were resistant to colitis whereas patients who developed colitis had an increased abundance Firmicutes, especially of the Faecalibacterium genus (98). Interestingly, patients with a high abundance of Faecalibacterium had longer progression-free survival, revealing a double-edge sword of microbiota manipulation (efficacy vs toxicity). Not Not surprisingly, trials are underway to investigate the effects of FMT combined with cancer immunotherapy or chemotherapy (99). Interactions among the microbiota, immune response, and cancer treatments should be considered in management of patients with cancer (Fig.2).
Figure 2: Synergy of the Intestinal Microbiota With the Immune System in Cancer.
Treatment Specific microbes or their products could increase the activities of chemotherapeutic or immune checkpoint inhibitor therapy, perhaps through interactions with immune cells.
Metabolic syndrome
Metabolic syndrome comprises obesity, type 2 diabetes, hypertension, and cardiovascular diseases; pathogenesis involves a combination of genetic and environmental factors. Changes in the intestinal microbiome have been associated with development of metabolic syndrome (100). Whole-genome metagenome and 16S rDNA analyses revealed differences in microbiome composition and gene richness between feces of obese subjects and healthy lean subjects (101–103), but these differences were not large enough to distinguish between the groups (104). Higher levels of energy from diet were measured in mice colonized with intestinal microbes from obese vs lean individuals (105). In a twin study, transfer of fecal microbiota from only the obese sibling (not the non-obese sibling) to germ-free mice increased body mass and adiposity (106). A trial of 18 patients who received allogenic (n=9) FMT from lean human donors or obese patients who received (control) autologous (n=9) FMT reported improved insulin sensitivity in the group that underwent allogenic FMT, after 6 weeks (107). A subsequent larger trial (n=38) from the same research group showed no benefit of allogenic FMT (n =26) on insulin sensitivity or weight compared to autologous FMT (n =12) at 18 weeks, which correlated with no changes in the composition of the intestinal microbiome (108). Interestingly, in the same cohort, a modest beneficial effect on insulin sensitivity was observed after 6 weeks in the group given allogenic FMT, due to a subgroup of responders who had low microbial diversity at baseline compared to non-responders. This suggests that patients’ responses to microbiome manipulation might be influenced by their original microbiome
The relationship between the composition of the microbiome and metabolic function is complex and unclear. When severely obese patients underwent bariatric surgery, either with adjustable gastric bands or Roux-en-Y-gastric bypass, most patients who underwent Roux-en-Y-gastric bypass had low microbial gene richness 1 year after surgery, yet these patients had more pronounced improvements in metabolic function than patients who received the gastric bands (109), So clinical and metabolic effects do not always correlate with the composition of the intestinal microbiome. This may be related to the capacity of intestinal microbiota to produce a drastically different set of metabolites, depending on nutrient exposure, without altering their phylogeny. This was also observed in a study that compared in vegan individuals with omnivores (110). This concept is important because changes observed in the microbiomes of individuals with obesity, metabolic syndrome, or hypertension are not consistent, and may be unique to each condition.
Hypertension (when blood pressure exceeds the normal range of 120/80 mm Hg) is risk factor for cardiovascular disease. Although genome-wide association studies identified variants at as many as 120 loci that could affect blood pressure, fewer than 4% of cases of hypertension can be accounted for by all these loci, so environmental factors are likely to be involved (111–113). 16S rDNA sequence analyses revealed lower microbial diversity and richness in 10 patients with high blood pressure compared with subjects with normal blood pressure, with clear differences in principal coordinate analysis (114). In a subsequent metagenomic and metabolomic study of 41 healthy individuals (controls), 56 subjects with pre-hypertension, and 99 individuals with primary hypertension, Li et al observed decreased microbiota diversity and richness in pre-hypertensive and hypertensive patients compared to controls (115). At the genus level, principal coordinate analysis showed a distinctive clustering of microbiota between hypertensive patients and controls, mediated by the presence of Prevotella and Bacteroides, respectively. It is important to note that the microbiomes of prehypertensive vs hypertensive patients did not differ, so changes in microbiome composition might precede disease development.
The functional effects of the intestinal microbiota have been studied in animals. Germ-free mice colonized with feces from hypertensive patients had increases in blood pressure (115). Similarly, transferring feces from hypertensive susceptible SRH rats to normotensive WKR rats increased their blood pressure (114). Interestingly, the antibiotic minocycline was able to decrease blood pressure in rats with angiotensin II-induced hypertension, associated with changes in microbiota composition. It is not clear if reverse alterations in the composition of the microbiota, following therapeutic intervention, associate with functions of specific bacteria; FMT studies might investigate this.
Diet has a large effect on the composition of the intestinal microbiome, its homeostasis (116), and development of metabolic syndrome. Metabolites generated from fiber-rich diets include SCFA (butyrate, propionate, and acetate). Interestingly, a meta-analysis of clinical trials investigating the effect of fiber intake in patients with hypertension reported reduced blood pressures of subjects with high-fiber diets (117). In mice, SCFA receptors such as GPR41 and OLFR78 maintain normal systemic blood pressure (118,119). A high-fiber diet or acetate supplementation decreased high blood pressure in mice with deoxycorticosterone acetate salt-induced hypertension (120) However, not all effects of SCFAs are beneficial. For example, butyrate promotes development of CRC in mice with disruption the DNA repair gene encoding MSH2 (121), whereas acetate promotes insulin resistance and metabolic syndrome in Tlr5−/− mice (122). Studies are needed to determine how microbe-derived metabolites affect the metabolism and development of metabolic syndrome.
Regardless, trials are underway to evaluate the effects of altering the intestinal microbiota in patients with metabolic syndrome. A phase 3 trial of 44 participants is underway to evaluate whether FMT from lean healthy donors can reduce insulin-resistance more than lifestyle changes alone in patients with metabolic syndrome (NCT02050607). A phase 2 trial (NCT02970877) of 48 participants will test when stool from healthy lean people transplanted into morbidly obese patients will improve insulin resistance and other obesity-related parameters. Manipulation of the intestinal microbiome, with FMT or by administration of specific microbes or groups of microbes, has been tested in patients with an array of medical conditions (Table 2). Analyses of data from these studies will provide important insights into disease pathogenesis and potential treatment strategies.
TABLE 2:
DISEASES FOR WHICH FMT HAS BEEN/IS BEING TESTED
| INDICATION | REFERENCE | TYPE OF STUDY | EFFECT |
|---|---|---|---|
| GASTROINTESTINAL DISEASES | |||
| Ulcerative Colitis | 23-26 | RCT | overall positive |
| Crohn’s disease | 29, 30 | case series | no effect |
| Pouchitis | 34 | case series | no effect |
| Irritable Bowel Syndrome | 37, 38 | RCT | suggestive |
| Graft versus Host Disease | 43-46 | case series | suggestive |
| Post-antibiotic diarrhea | 47 | case series | suggestive |
| Constipation | 41 | RCT | suggestive |
| Hepatic encephalopathy | 49 | RCT | suggestive |
| Psoriasis | RCT ongoing | unknown | |
| Multiple sclerosis | 2 RCTs ongoing | unknown | |
| Autism | 63 | uncontrolled pilot trial | suggestive |
| Metabolic syndrome | 104.105 | controlled trials | suggestive |
Future Directions
Despite advances in studies of the intestinal microbiome, there have been few controlled trials of therapeutic interventions that alter the microbiome. Strategies to alter the intestinal microbiome could be used to treat a variety of gastrointestinal and other diseases, but there are many important questions to answer first. Are alterations in the microbiome associated with certain conditions the cause or consequence? How long can the effect of a microbiome intervention last, since microbiome profiles seems to be highly specific to individuals and are tolerated by the mucosal immune system? To which extent does the residing dysbiotic microbiome need to be destroyed before externally administered microbiota can successfully engraft? And, importantly, are living bacteria (or other microbes) needed for a therapeutic effect or are there other components of the microbiome, such as microbial products, that mediate the effects?
In modern medicine, FMT was first used to treat patients with C difficile-associated colitis following treatment with antibiotics. CDI is an acute/subacute condition in which the natural intestinal microbiome has been completely wiped out, making the mucosa receptive to colonization by externally administered microbiota. In the controlled prospective trials of patients with UC, it has not been clear whether pretreatment with antibiotics, such as vancomycin, increase the effects of FMT. It is important to answer this question soon. UC and IBS are chronic diseases, so we need to determine whether changes in the microbiome following FMT are permanent, or how long they last—repeated treatments are likely to be necessary. In addition, could FMT be used to maintain remission?
Findings from studies of patients with metabolic syndrome or type 2 diabetes are encouraging in light of the expanding obesity pandemic and require further exploration. Further studies are also needed to determine the effects of microbiome alterations on tumor growth and cancer therapies, not only for intestinal but also other types of cancer. Patients with graft vs host disease are often heavily treated with antibiotics, and evidence is mounting that manipulating the intestinal microbiome could improve outcomes.
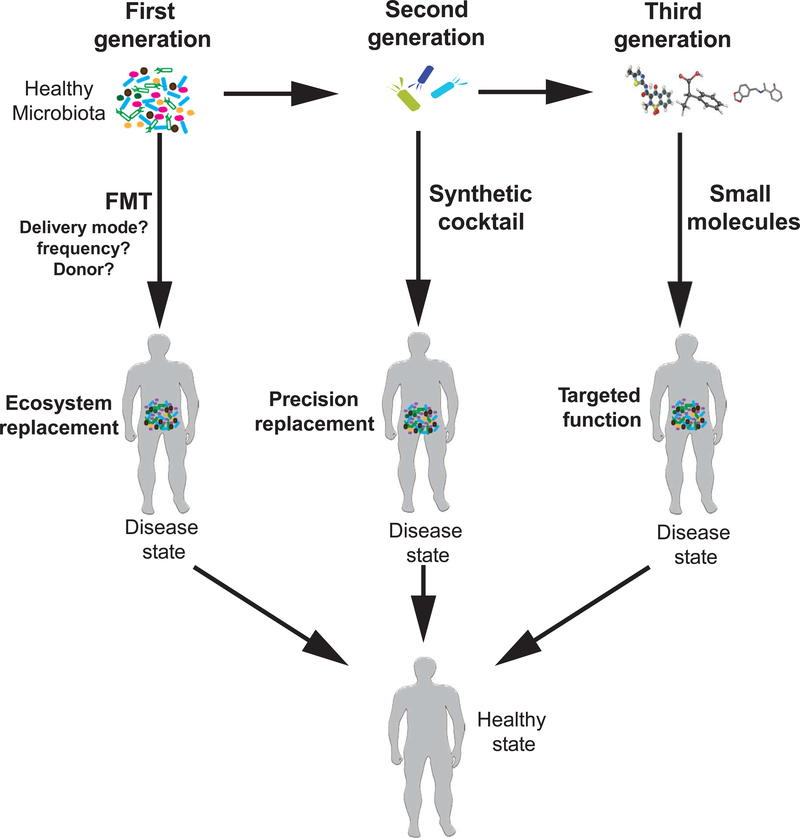
A series of methodology questions must be answered for FMT. Beside the requirement for antibiotic pre-treatment, route of administration (colonic/nasoenteric), frequency of administration, and volume of fecal material required have varied among trials. The features of superdonors should be determined and will vary among diseases (Fig.3). We must also define the factors in fecal material that mediate its therapeutic effects. Answering these questions will help refine and enhance microbe-based therapies.
Figure 3: Searching for Microbe-based Therapeutic Targets.
Microbe-based therapies could replace the entire intestinal microbiota, such as in FMT (first generation), involve specific combinations of microbes (second generation), or microbe-derived compounds (third generation). Studies are underway identify the microbes or products that are altered during disease development and therefore might be therapeutically targeted, or microbes or molecules with therapeutic effects. Once identified, these require validation and prospective clinical studies.
Microbe-based therapies are likely to eventually involve small molecule compounds derived from microbes identified in mechanistic studies, or complex combinations of microbiota or synthetic cocktails (Fig.3). FMT is not a 1 size fits all strategy, and studies are required to identify components of the microbiota that have specific effects in patients with different diseases. The march toward microbe-based precision medicine is underway. We encourage funding of research in this rapidly expanding field of research
ACKNOWLEDGEMENTS
C. Jobin acknowledges support from NIH (R01DK073338, R01AT08623 and R21 CA195226) and University of Florida Department of Medicine Gatorade Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Geert R. D’Haens, Dept of Gastroenterology, Amsterdam University Medical Center, Meibergdreef 9, 1100 DZ Amsterdam, The Netherlands
Christian Jobin, Dept of Medicine, Dept of Anatomy and Cell Biology, Dept of Infectious Diseases and Immunology, University of Florida, 2033 Mowry Rd, PO Box 103633, Gainesville, FL, USA.
REFERENCES
- 1.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012;486: 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Human Microbiome Project Consortium. A framework for human microbiome research. Nature 2012;486:215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arumugam M, Raes J, Pelletier E, et al. , the Metahit Consortium. Enterotypes of the human gut microbiome. Nature. 2011;473:174–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schirmer M, Smeekens SP, Vlamakis H, et al. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell 2016:167:1897. [DOI] [PubMed] [Google Scholar]
- 5.ter Horst R, Jaeger M, Smeekens SP, et al. Host and Environmental Factors Influencing Individual Human Cytokine Responses. Cell 2016:167:1111–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Oosting M, Smeekens SP, et al. A Functional Genomics Approach to Understand Variation in Cytokine Production in Humans. Cell 2016:167:1099–1110. [DOI] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009;587:4153–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sonnenburg JL, Backhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535:56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmes E, Li JV, Marchesi JR, et al. Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell Metab. 2012;16:559–64. [DOI] [PubMed] [Google Scholar]
- 11.Machiels K, Joossens M, Sabino J, et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014;63:1275–83. [DOI] [PubMed] [Google Scholar]
- 12.Codling C, O’Mahony L, Shanahan F, et al. A molecular analysis of fecal and mucosal bacterial communities in irritable bowel syndrome. Dig Dis Sci 2010;55:392–397. [DOI] [PubMed] [Google Scholar]
- 13.Mancabelli L, Milani C, Lugli GA, et al. Unveiling the gut microbiota composition and functionality associated with constipation through metagenomic analyses. Sci Rep. 2017;7:9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zou S, Fang L, Lee MH. Dysbiosis of gut microbiota in promoting the development of colorectal cancer. Gastroenterol Rep (Oxf). 2018;6:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006; 444: 1027–1031. [DOI] [PubMed] [Google Scholar]
- 16.Miyake S, Yamamura T. Gut environmental factors and multiple sclerosis. J Neuroimmunol. 2018; S0165–5728(17)30414–9. [DOI] [PubMed] [Google Scholar]
- 17.Zhang F, Luo W, Shi Y, et al. Should we standardize the 1,700 year-old fecal microbiota transplantation? Am J Gastroenterol 2012; 107: 1755–56. [DOI] [PubMed] [Google Scholar]
- 18.Eiseman B, Silen W, Bascom GS, et al. Fecal enema as an adjunct in the treatment of pseudomembranous enterocolitis. Surgery 1958; 854–859. [PubMed] [Google Scholar]
- 19.Van Nood E, Vrieze A, Nieuwdorp M, et al. Duodendal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 2013; 368: 407–415. [DOI] [PubMed] [Google Scholar]
- 20.Gagliardi A, Totino V, Cacciotti F, et al. Rebuilding the Gut Microbiota Ecosystem. Int J Environ Res Public Health. 2018;15 (8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frank DN, St Amand AL, Feldman RA, et al. Molecular –phylogenic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Nat Acad Sci USA 2007;104:13780–13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guillemot F, Colombel JF, Neut C, et al. Treatment of diversion colitis by short-chain fatty acids. Prospective and double-blind study. Dis Colon Rectum. 1991;34:861–4. [DOI] [PubMed] [Google Scholar]
- 23.Mooayyedi P, Surette MG, Kim PT, et al. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology 2015;149:102–109. [DOI] [PubMed] [Google Scholar]
- 24.Rossen NG, Fuentes S, van der Spek MJ, et al. Findings from a randomized controlled trial of fecal transplantation for patients with active ulcerative colitis. Gastroenterology 2015;149:110–118. [DOI] [PubMed] [Google Scholar]
- 25.Paramsothy S, Kamm MA, Kaakoush NO, et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet 2017;389:1218–28. [DOI] [PubMed] [Google Scholar]
- 26.Costello S, Waters O, Bryant R, et al. Short duration, low intensity pooled faecal microbiota transplantation induces remission in patients with mild-moderately active ulcerative colitis: a randomized controlled trial. Journal of Crohn’s & colitis 2017;11(suppl 1):S23. [Google Scholar]
- 27.Ott SJ, Waetzig GH, Rehman A, et al. Efficacy of Sterile Fecal Filtrate Transfer for Treating Patients With Clostridium difficile Infection. Gastroenterology 2017;152:799–811. [DOI] [PubMed] [Google Scholar]
- 28.Misra Bharat K.. A Multiple Dose Phase 1b Study to Evaluate the Efficacy, Safety, and Microbiome Dynamics of SER-287 in Subjects with Mild-to-Moderate Ulcerative Colitis (UC). Gastroenterology 2018. (abstract). [Google Scholar]
- 29.Vermeire S, Joossens M, Verbeke K, et al. Donor species richness determines faecal microbiota transplantation success in inflammatory bowel disease. J Crohns Colitis. 2016;10:387–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cui B, Feng Q, Wang H, et al. Fecal microbiota transplantation through mid-gut for refractory Crohn’s disease: safety, feasibility, and efficacy trial results. J Gastroenterol Hepatol. 2015;30:51–8. [DOI] [PubMed] [Google Scholar]
- 31.Gionchetti P, Calafiore A, Riso D, et al. The role of antibiotics and probiotics in pouchitis. Ann Gastroenterol. 2012;25:100–105. [PMC free article] [PubMed] [Google Scholar]
- 32.Gionchetti P, Rizzello F, Helwig U, et al. Prophylaxis of pouchitis onset with probiotic therapy: a double-blind, placebo-controlled trial. Gastroenterology 2003;124:1202–9. [DOI] [PubMed] [Google Scholar]
- 33.Mimura T, Rizzello F, Helwig U, et al. Once daily high dose probiotic therapy (VSL#3) for maintaining remission in recurrent or refractory pouchitis. Gut 2004;53:108–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landy J, Walker AW, Li JV, et al. Variable alterations of the microbiota, without metabolic or immunological change, following faecal microbiota transplantation in patients with chronic pouchitis. Sci Rep. 2015; 5: 12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Defrees DN, Bailey J. Irritable Bowel Syndrome: Epidemiology, Pathophysiology, Diagnosis, and Treatment. Prim Care. 2017;44:655–671. [DOI] [PubMed] [Google Scholar]
- 36.Ralilic-Stojanovic M, Biagi E, Heilig HG, et al. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology 2011; 141:1792–1801. [DOI] [PubMed] [Google Scholar]
- 37.Johnsen PH, Hilpüsch F, Cavanagh JP, et al. Faecal microbiota transplantation versus placebo for moderate-to-severe irritable bowel syndrome: a double-blind, randomised, placebo-controlled, parallel-group, single-centre trial. Lancet Gastroenterol Hepatol. 2018;3:17–24. [DOI] [PubMed] [Google Scholar]
- 38.Holvoet T, Joossens M, Boelens J, et al. Fecal microbiota transplantation in irritable bowel syndrome with predominant abdominal bloating: results from a double blind, placebo-controlled clinical trial. Gastroenterology 2018. (Abstract) [DOI] [PubMed] [Google Scholar]
- 39.Holvoet T, Joossens M, Wang J, et al. Assessment of faecal microbial transfer in irritable bowel syndrome with severe bloating. Gut 2017;66:980–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halkjær SI, Christensen AH, Lo BZS, et al. Faecal microbiota transplantation alters gut microbiota in patients with irritable bowel syndrome: results from a randomised, double-blind placebo-controlled study. Gut. 2018;67:2107–2115. [DOI] [PubMed] [Google Scholar]
- 41.Tian H, Ge X, Nie Y, et al. Fecal microbiota transplantation in patients with slow-transit constipation: A randomized, clinical trial. PLoS One. 2017. February 3;12(2):e0171308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. https://clinicaltrials.gov/ct2/results?cond=Constipation+-+Functional.
- 43.Doki N, Suyama M, Sasajima S, et al. Clinical impact of pre-transplant gut microbial diversity on outcomes of allogeneic hematopoietic stem cell transplantation. Ann Hematol. 2017;96:1517–1523. [DOI] [PubMed] [Google Scholar]
- 44.Kakihana K, Fujioka Y, Suda W, et al. Fecal microbiota transplantation for patients with steroid-resistant acute graft-versus-host disease of the gut. Blood. 2016;128:2083–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeFilipp Z, Peled JU, Li S, et al. Third-party fecal microbiota transplantation following allo-HCT reconstitutes microbiome diversity. Blood Adv. 2018;2:745–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peled JU, Devlin SM, Staffas A, et al. Intestinal Microbiota and Relapse After Hematopoietic-Cell Transplantation. J Clin Oncol. 2017;35:1650–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goldenberg JZ, Lytvyn L, Steurich J, et al. Probiotics for the prevention of pediatric antibiotic-associated diarrhea. Cochrane Database Syst Rev 2015; CD004827. [DOI] [PubMed] [Google Scholar]
- 48.Suez J, Zmora N, Zilberman-Schapira G, et al. Post-Antibiotic Gut Mucosal Microbiome Reconstitution Is Impaired by Probiotics and Improved by Autologous FMT. Cell 2018;174:1406–1423. [DOI] [PubMed] [Google Scholar]
- 49.Bajaj JS, Kassam Z, Fagan A, et al. Fecal microbiota transplant from a rational stool donor improves hepatic encephalopathy: A randomized clinical trial. Hepatology 2017; 66:17271738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benhadou F, Minthoff D, Schnebert B, Thio HB. Psoriasis and microbiota: a systematic review. Diseases 2018; 6: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tan L, Zhao S, Zhu W, et al. The Akkermansia muciniphila is a gut microbiota signature in psoriasis. Exp Dermatol. 2018;27:144–149. [DOI] [PubMed] [Google Scholar]
- 52.Masallat D, Moemen D. Afr J Microbiol Res 2016; 10:1337–1343. [Google Scholar]
- 53.Assarsson M et al. , Acta Dermatol Venereol 2018;98:428. [DOI] [PubMed] [Google Scholar]
- 54.Kragsnaes MS, Kjeldsen J, Horn HC, et al. Efficacy and safety of faecal microbiota transplantation in patients with psoriatic arthritis: protocol for a 6-month, double-blind, randomised, placebo-controlled trial. BMJ Open. 2018;8:e019231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen J, Chia R, Kalari KR, et al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci Rep, 2016; 6: 28484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee YK, Menezes JS, Umesaki Y, et al. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci 2011;108: 4615–4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Furusawa Y, Obata Y, Hase K. Commensal microbiota regulates T cell fate in the gut. Semin Immunopathol 2015; 37: 17–25. [DOI] [PubMed] [Google Scholar]
- 58.Lai SW, Liao KF, Lin CL, et al. Irritable bowel syndrome correlates with increased risk of Parkinson’s disease in Taiwan. Eur J Epidemiol, 29: 57–62. [DOI] [PubMed] [Google Scholar]
- 59.Keshavarzian A, Green SJ, Engen PA, et al. Colonic bacterial composition in Parkinson’s disease. Mov Disord, 2015;30:1351–1360. [DOI] [PubMed] [Google Scholar]
- 60.Sampson TR, Debelius JW, Thron T, et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell, 2016;167:1469–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maqsood R, Stone TW. The gut–brain axis, BDNF, NMDA and CNS disorders Neurochem Res. 2016;2016:2819–283. [DOI] [PubMed] [Google Scholar]
- 62.Tomova A, Husarova V, Lakatosova S, et al. Gastrointestinal microbiota in children with autism in Slovakia. Physiol Behav. 2015;138:179–187. [DOI] [PubMed] [Google Scholar]
- 63.Kang DW, Adams JB, Gregory AC, et al. Microbiota transfer therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open-label study. Microbiome 2017;5:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Son JS, Zheng LJ, Rowehl LM, et al. Comparison of fecal microbiota in children with autism spectrum disorders and neurotypical siblings in the Simons Simplex Collection. PLoS One 2015; 10:eD137725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oh J-K, Weiderpass E. Infection and cancer: global distribution and burden of diseases. Ann Glob Health 2014;80:384–392. [DOI] [PubMed] [Google Scholar]
- 66.Jobin C. Colorectal Cancer: Looking for Answers in the Microbiota. Cancer Discov 2013;3:384–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Couturier-Maillard A, Secher T, Rehman A, et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. The Journal of clinical investigation 2013;123:700–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu B, Elinav E, Huber S, et al. Microbiota-induced activation of epithelial IL-6 signaling links inflammasome-driven inflammation with transmissible cancer. Proceedings of the National Academy of Sciences of the United States of America 2013;110:9862–9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mamantopoulos M, Ronchi F, Van Hauwermeiren F, et al. Nlrp6- and ASC-Dependent Inflammasomes Do Not Shape the Commensal Gut Microbiota Composition. IMMUNI 2017;47:339–348.e4. [DOI] [PubMed] [Google Scholar]
- 70.Man SM, Zhu Q, Zhu L, et al. Critical Role for the DNA Sensor AIM2 in Stem Cell Proliferation and Cancer. Cell 2015;162:45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilson JE, Petrucelli AS, Chen L, et al. Inflammasome-independent role of AIM2 in suppressing colon tumorigenesis via DNA-PK and Akt. Nature medicine 2015;21:906–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wong SH, Zhao L, Zhang X, et al. Gavage of Fecal Samples From Patients With Colorectal Cancer Promotes Intestinal Carcinogenesis in Germ-Free and Conventional Mice. Gastroenterology 2017;153:1621–1633. [DOI] [PubMed] [Google Scholar]
- 73.Thomas RM, Gharaibeh RZ, Gauthier J, et al. Intestinal microbiota enhances pancreatic carcinogenesis in preclinical models. Carcinogenesis 2018;6:2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pushalkar S, Hundeyin M, Daley D, et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov 2018;8:403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ma C, Han M, Heinrich B, et al. Gut microbiome–mediated bile acid metabolism regulates liver cancer via NKT cells. Science 2018;360:5931–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meisel M, Hinterleitner R, Pacis A, et al. Microbial signals drive pre-leukaemic myeloproliferation in a Tet2-deficient host. Nature 2018;557:580–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zackular JP, Rogers MAM, Ruffin MT, et al. The Human Gut Microbiome as a Screening Tool for Colorectal Cancer. Cancer Prevention Research 2014;7:1112–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Narayanan V, Peppelenbosch MP, Konstantinov SR. Human Fecal Microbiome-Based Biomarkers for Colorectal Cancer. Cancer Prevention Research 2014;7:1108–1111. [DOI] [PubMed] [Google Scholar]
- 79.Zeller G, Tap J, Voigt AY, et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 2014;10:766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yu J, Feng Q, Wong SH, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut 2017;66:70–78. [DOI] [PubMed] [Google Scholar]
- 81.Liang Q, Chiu J, Chen Y, et al. Fecal Bacteria Act as Novel Biomarkers for Noninvasive Diagnosis of Colorectal Cancer. Clin. Cancer Res. 2017;23:2061–2070. [DOI] [PubMed] [Google Scholar]
- 82.Perez-Chanona E, Jobin C. From promotion to management: The wide impact of bacteria on cancer and its treatment. Bioessays 2014;36:658–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zitvogel L, Ayyoub M, Routy B, et al. Perspective. Cell 2016;165:276–287. [DOI] [PubMed] [Google Scholar]
- 84.Gopalakrishnan V, Helmink BA, Spencer CN, et al. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer cell 2018;33:570–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Roy S, Trinchieri G. Microbiota: a key orchestrator of cancer therapy. Nature reviews 2017;17:271–285. [DOI] [PubMed] [Google Scholar]
- 86.Viaud S, Saccheri F, Mignot G, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013;342:971–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Iida N, Dzutsev A, Stewart CA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013;342:967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Miller JFAP, Sadelain M. The journey from discoveries in fundamental immunology to cancer immunotherapy. Cancer cell 2015;27:439–449. [DOI] [PubMed] [Google Scholar]
- 89.Vétizou M, Pitt JM, Daillère R, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015;350:1079–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015;350:1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1based immunotherapy against epithelial tumors. Science 2018;359:91–97. [DOI] [PubMed] [Google Scholar]
- 92.Derosa L, Hellmann MD, Spaziano M, et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Annals of Oncology 2018;29:1437–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Matson V, Fessler J, Bao R, et al. The commensal microbiome is associated with antiPD-1 efficacy in metastatic melanoma patients. Science 2018;359:104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018;359:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gharaibeh RZ, Jobin C. Microbiota and cancer immunotherapy: in search of microbial signals. (2018). Gut. 2018. December 8 pii: gutjnl-2018–317220. doi: 10.1136/gutjnl-2018-317220. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gupta A, De Felice KM, Loftus EV, et al. Systematic review: colitis associated with antiCTLA-4 therapy. Aliment Pharmacol Ther. 2015;42:406–417. [DOI] [PubMed] [Google Scholar]
- 97.Dubin K, Callahan MK, Ren B, et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun 2016;7:10391–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chaput N, Lepage P, Coutzac C, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol. 2017;28:1368–1379. [DOI] [PubMed] [Google Scholar]
- 99.McQuade JL, Daniel CR, Helmink BA, Wargo JA. Modulating the microbiome to improve therapeutic response in cancer. Lancet Oncol. 2019. February; 20(2) e77–e91. [DOI] [PubMed] [Google Scholar]
- 100.Karlsson FH, Tremaroli V, Nookaew I, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 2013;498:99–103. [DOI] [PubMed] [Google Scholar]
- 101.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature 2009;457:480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Le Chatelier E, Nielsen T, Qin J, et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2014;500:541–546. [DOI] [PubMed] [Google Scholar]
- 103.Cotillard A, Kennedy SP, Kong LC, et al. Dietary intervention impact on gut microbial gene richness. Nature 2014;500:585–588. [DOI] [PubMed] [Google Scholar]
- 104.Pasolli E, Truong DT, Malik F, et al. Machine Learning Meta-analysis of Large Metagenomic Datasets: Tools and Biological Insights. PLoS Comput Biol 2016;12:e1004977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006;444:1027–1031. [DOI] [PubMed] [Google Scholar]
- 106.Ridaura VK, Faith JJ, Rey FE, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013;341:1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vrieze A, van Nood E, Holleman F, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 2012;143:913–916. [DOI] [PubMed] [Google Scholar]
- 108.Kootte RS, Levin E, Salojärvi J, et al. Improvement of Insulin Sensitivity after Lean Donor Feces in Metabolic Syndrome Is Driven by Baseline Intestinal Microbiota Composition. Cell Metab. 2017;26:611–619. [DOI] [PubMed] [Google Scholar]
- 109.Aron-Wisnewsky J, Prifti E, Belda E, et al. Major microbiota dysbiosis in severe obesity: fate after bariatric surgery. Gut 2019;68:70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wu GD, Compher C, Chen EZ, et al. Comparative metabolomics in vegans and omnivores reveal constraints on diet-dependent gut microbiota metabolite production. Gut 2016;65:63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Surendran P, Drenos F, Young R, et al. Trans-ancestry meta-analyses identify rare and common variants associated with blood pressure and hypertension. Nature genetics 2016;48:1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ehret GB, Ferreira T, Chasman DI, et al. The genetics of blood pressure regulation and its target organs from association studies in 342,415 individuals. Nature genetics 2016;48:1171–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu C, Kraja AT, Smith JA, et al. Meta-analysis identifies common and rare variants influencing blood pressure and overlapping with metabolic trait loci. Nature genetics 2016;48:1162–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang T, Santisteban MM, Rodriguez V, et al. Gut dysbiosis is linked to hypertension. Hypertension 2015;65:1331–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li J, Zhao F, Wang Y, et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 2017;5:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ohland CL, Jobin C. Microbial activities and intestinal homeostasis: A delicate balance between health and disease. Cell Mol Gastroenterol Hepatol 2015;1:28–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Effect of dietary fiber intake on blood pressure: a meta-analysis of randomized, controlled clinical trials. Clinical Governance: An International Journal 2007;12:cgij.2007.24812aae.008. [DOI] [PubMed] [Google Scholar]
- 118.Natarajan N, Hori D, Flavahan S, et al. Microbial short chain fatty acid metabolites lower blood pressure via endothelial G protein-coupled receptor 41. Physiol Genomics 2016;48:826–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pluznick JL, Protzko RJ, Gevorgyan H, et al. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proceedings of the National Academy of Sciences USA 2013;110:4410–4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Marques FZ, Nelson E, Chu P-Y, et al. High-Fiber Diet and Acetate Supplementation Change the Gut Microbiota and Prevent the Development of Hypertension and Heart Failure in Hypertensive Mice. Circulation 2017;135:964–977. [DOI] [PubMed] [Google Scholar]
- 121.Belcheva A, Irrazabal T, Robertson SJ, et al. Gut microbial metabolism drives transformation of msh2-deficient colon epithelial cells. Cell 2014;158:288–299. [DOI] [PubMed] [Google Scholar]
- 122.Singh V, Chassaing B, Zhang L, et al. Microbiota-Dependent Hepatic Lipogenesis Mediated by Stearoyl CoA Desaturase 1 (SCD1) Promotes Metabolic Syndrome in TLR5Deficient Mice. Cell Metabolism 2015;22:983–96. [DOI] [PMC free article] [PubMed] [Google Scholar]