A 4-year surveillance of carbapenem-resistant Acinetobacter spp. isolates in Argentina identified 40 strains carrying blaNDM-1. Genome sequencing revealed that most were Acinetobacter baumannii, whereas seven represented other Acinetobacter spp. The A. baumannii genomes were closely related, suggesting recent spread. blaNDM-1 was located in the chromosome of A. baumannii strains and on a plasmid in non-A. baumannii strains.
KEYWORDS: Acinetobacter baumannii, NDM-1, carbapenem resistance, Acinetobacter, carbapenems
ABSTRACT
A 4-year surveillance of carbapenem-resistant Acinetobacter spp. isolates in Argentina identified 40 strains carrying blaNDM-1. Genome sequencing revealed that most were Acinetobacter baumannii, whereas seven represented other Acinetobacter spp. The A. baumannii genomes were closely related, suggesting recent spread. blaNDM-1 was located in the chromosome of A. baumannii strains and on a plasmid in non-A. baumannii strains. A resistance gene island carrying blaPER-7 and other resistance determinants was found on a plasmid in some A. baumannii strains.
TEXT
The predominant species expressing the NDM-1 carbapenemase are Klebsiella pneumoniae and Escherichia coli. However, Acinetobacter spp. isolates are recognized as intermediate reservoirs for the blaNDM-1 resistance determinant (1, 2). blaNDM is a metallo-β-lactamase (MBL) generally found on a plasmid or other mobile element that carries resistance determinants for other antibiotic classes, rendering many NDM-positive isolates extensively drug resistant. Infections caused by carbapenem-resistant Acinetobacter baumannii isolates are associated with mortality rates as high as 60% (3, 4).
The Argentina National Reference Laboratory (NRL) identified an increase in the prevalence of NDM-containing Acinetobacter spp. isolates beginning in 2015. Of the 20,028 clinical isolates screened since 2010, 15,621 were carbapenem resistant and 144 had an MBL phenotype (5–7). PCR assays for common MBLs confirmed that 68 (47%), i.e., 40 blaNDM and 28 blaIMP, of 144 isolates were producers; whereas in the remaining strains, the MBL-like phenotype observed in the institution of origin was due to the presence of blaOXA-23 or blaOXA-58.
NDM-producing strains were recovered from 19 hospitals in nine cities and seven provinces in Argentina (see Fig. S1 in the supplemental material). A. baumannii isolates had more extensive antimicrobial resistance profiles than the non-baumannii isolates (see Table S1 in the supplemental material). Genome sequences were obtained on an Illumina NextSeq 500 and assembled using Velvet (8). A BLASTN search at NCBI classified seven genomes as representing five different non-baumannii Acinetobacter species. (see Table S1). MLST analysis showed that all 33 A. baumannii isolates belonged to sequence type ST25 (9). The ST25 genomes were most closely related to isolates found throughout the world, including HEU3 (Honduras), HWBA8 (South Korea), NM3 (United Arab Emirates), and two genomes with no geographic origin provided, i.e., AR_0088 and AB5256. The AR_0088 genome has been completely sequenced (GenBank accession no. CP027530.1) and was used as the reference genome for single nucleotide polymorphisms (SNPs) and insertion element (IS) annotation.
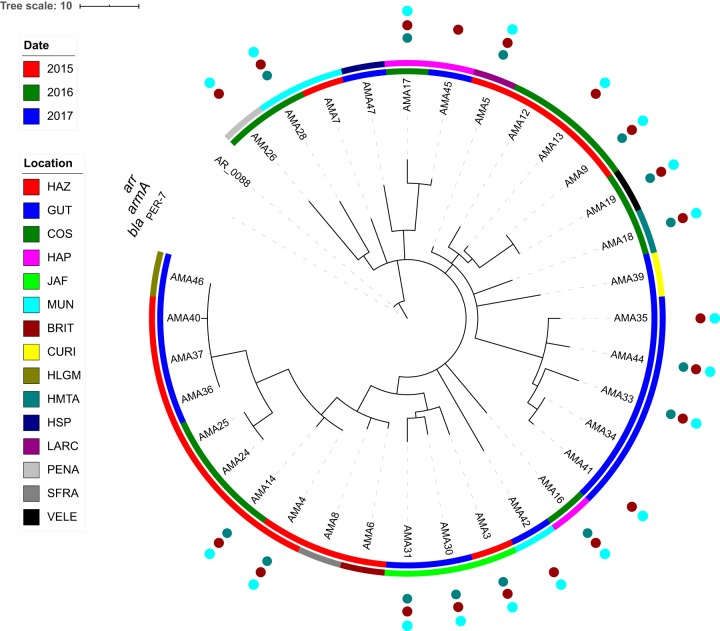
The A. baumannii genomes differed by only 14 to 36 sequence variants, suggesting recent divergence. Patterns of shared SNPs (10), IS locations (11), and epidemiological data were highly concordant (Fig. 1). Genomes were predominantly clustered by isolation location, with a few exceptions. For example, AMA19 from Hospital General de Agudos Vélez Sarsfield (VELE) was essentially identical to AMA9 from Hospital Dr. Cosme Argerich (COS) and on the same branch as other COS isolates. These hospitals belong to the public care sector in the Buenos Aires capital district, where patient exchange is frequent, suggesting a possible transmission event.
FIG 1.
Phylogenetic tree of A. baumannii genomes. A neighbor-joining tree was constructed using shared SNP and IS insertion sites. Inner circle shows year of isolation. Outer circle shows hospital in which the isolate was recovered. See Table S1 for further information on the location of each hospital. Isolates that are positive for components of RI-PER-7 are denoted by colored circles outside of the tree. The scale bar represents the combined number of SNPs and IS insertion events. The AR_0088 genome was used as the out group, but the branch length is not shown to highlight the relationships of the AMA strains. The figure was created using iTOL (24).
The AR_0088 reference genome contained two plasmids, i.e., pAR_0088_1 (GenBank accession no. CP027531.1) and pAR_0088_2 (GenBank accession no. CP027532.1). pAR_0088_2 carries the blaNDM-1 gene in Tn125, and this plasmid is the likely location of the blaNDM-1 gene in the non-baumannii Acinetobacter genomes (1, 12, 13). pAR_0088_2 sequences were not present in the A. baumannii ST25 genomes. To ascertain the location of Tn125 in the A. baumannii genomes, we identified the locations of ISAba125 insertions, which flank the transposon, in the draft genome assembly of a representative strain (AMA16). Six ISAba125 insertion sites were inferred, all in the chromosome, based on alignment of flanking sequences to the AR_0088 reference sequence. PCR and Sanger sequencing were used to demonstrate that Tn125 was inserted at base 3,921,386 of the AR_0088 genome in AMA16 and all other A. baumannii isolates, interrupting a gene encoding a hypothetical protein, AM467_RS18915 (see Table S2 in the supplemental material).
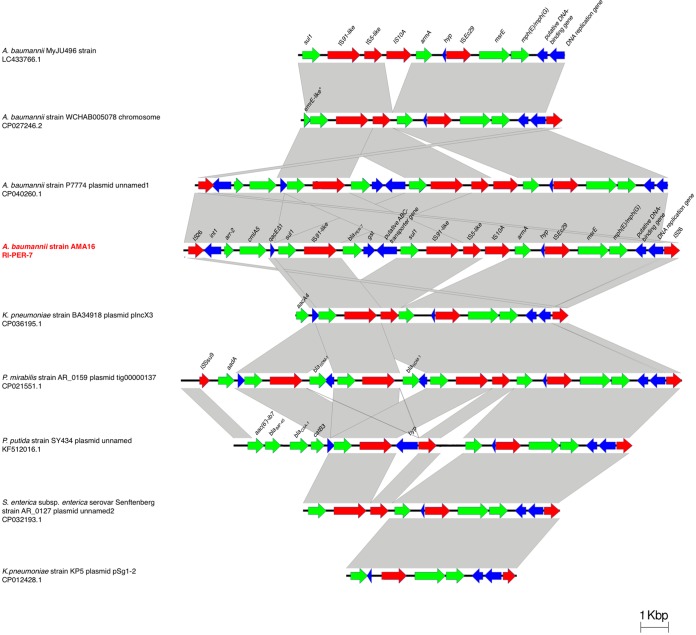
A large resistance gene island (RI) was identified in 14 of the ST25 strains (RI-PER-7). This ∼23.8-kb sequence is bound by a pair of IS26 elements in direct-repeat orientation (Fig. 2). The island carries genes encoding resistance to aminoglycosides (armA), rifampin (arr), cephalosporins (blaPER-7), and fosfomycin (GST) (Fig. 2). This RI also carries two copies of ISCR1 (IS91-like) and three other IS elements that are found predominantly in Enterobacteriaceae isolates but are rare in Acinetobacter isolates, i.e., IS10A, ISEc28, and ISEc29. The 7.5 kb at the 3′ end of the island is found in several non-Acinetobacter genomes. The complete structure was identified in seven accessions in GenBank, mostly plasmid sequences. Five of the ST25 genomes harbored a shorter version of the RI, lacking the blaPER-7 gene. Genetic structures similar to RI-PER-7 were identified in other species, including K. pneumoniae, Proteus mirabilis, and E. coli. In these structures, blaPER-7 was not present, but other β-lactamases were found (Fig. 2).
FIG 2.
Comparison of genetic structure of RI-PER-7 genomic island. Gray bars, regions shared between isolates; red arrows, IS elements; green arrows, antibiotic resistances genes. The figure was created using EasyFig, version 2.2.2.
RI-PER-7 is not present in AR_0088, but it was found on one of the plasmids in HWBA8 (14). pHWBA8_1 (GenBank accession no. CP020596.1) is 195,838 bases, and a large portion of this sequence is present in the ST25 A. baumannii genomes, including the complete RI-PER-7. Interestingly, pHWBA8_1 is also similar to pAR_0088_1, but the latter plasmid lacks the RI and another ∼17.5-kb segment and has extensive rearrangements covering another ∼20 kb of the sequence. The location of RI-PER-7 in the plasmid was confirmed by PCR amplification and Sanger sequencing of the junction regions (see Table S2). This large plasmid carries five additional AR genes located outside RI-PER-7.
The comM gene, a common location for insertion of RIs in A. baumannii isolates, is intact in ST25 strains. IS elements were not found upstream of the chromosomal blaADC or blaOXA genes in this study. The ST25 strains carry S84L and S81L substitutions in the parC and gyrA genes, consistent with their nonsusceptibility to ciprofloxacin.
The NRL also confirmed the presence of blaNDM-producing Enterobacteriaceae in 12 of 19 hospitals (data not shown). In 8 of the 12 hospitals, this emergence occurred 1 to 4 months after the first detection of NDM-producing Acinetobacter isolates. Additional work is needed to determine whether Acinetobacter spp. could have played a role in the interspecies dissemination of NDM in these institutions.
Although reports of blaNDM in Latin America are limited, sentinel investigations have described the presence of Tn125 in Acinetobacter spp. (6, 15–17). An ST25 A. baumannii strain from a patient with an abdominal infection in Honduras was determined to have Tn125 on a plasmid (18), but this plasmid was not present in the ST25 strains analyzed here. Tn125 has been reported in the chromosome of Acinetobacter spp. as well (19, 20).
ST25 strains in Argentina typically carry blaOXA-23 β-lactamase (17). According to available data, the cooccurrence of blaNDM-1 and blaOXA-23/58 seems to be an uncommon event (12, 21–23), a scenario that may change due to the increasing emergence of NDM-1 in A. baumannii isolates.
In summary, the genetic context of blaNDM-1 differs between A. baumannii and non-baumannii isolates in Argentina, with non-baumannii strains mostly retaining susceptibility to some antibiotics. We also describe an RI in a subset of A. baumannii genomes that likely contributes substantially to the MDR phenotype of these strains. The escalating number of reports of NDM-1 among A. baumannii isolates suggests a switch regarding the genetic basis of carbapenem resistance in this species, and intensive tracking of patient contacts is warranted for ST25.
Data availability.
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under BioProject accession no. PRJNA562922. Contig sequences for each genome are available under GenBank accession nos. VYSH00000000 to VYTU00000000.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to the institutions and professionals of the antimicrobial surveillance systems who referred the clinical isolates for characterization to NRL: Ana Maria Zalof Dakoff (Hospital Dr. Avelino Castelan), Marta Giovanakis (Hospital Britanico de Buenos Aires), Nora Gomez (Hospital Cosme Argerich), Maria Laura Chaves (Hospital Oncologica Marie Curie), Ana Maria Togneri (Hospital Evita Lanus), Claudia Hernandez (Hospital de Pediatria Garrahan), Estefania Biondi (Hospital de Pediatria Ricardo Gutierrez), Flavia Amalfa (Hospital Piñero), Gabriela Rivollier (Hospital Artimides Zatti), Mariana Carol Rey (Hospital Dr. Julio Perrando), A. Lopez (Hospital Lagomaggiore), Gallo Romina (Hospital Lucio Molas), Johanna Perez (Hospital de Trauma Malvinas Argentinas), Norma Cech (Hospital Pablo Soria), Laura Errecalde and Sandra Cogut (Hospital Dr. Juan A Fernandez), Sabrina Laura Rech (Hospital Dr. Raúl F. Larcade), Myriam Mortarini (Hospital de Infecciosas F. Muñiz), Monica Millara (Hospital Jose M. Penna), Claudia Etcheves (Sanatorio Franchin-UOCRA), and Silvana Manganello (Hospital Dr. Velez Sarsfield).
We have no conflicts of interest to declare.
The work was supported by R01AI100560 to R.A.B. and A.J.V.; R01AI063517 and R01AI072219 to R.A.B.; Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT) to A.J.V.; and NIH SC3GM125556 to M.S.R. The study was supported in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs (award no. 1I01BX001974) from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development and the Geriatric Research Education and Clinical Center VISN 10 to R.A.B.
The content is solely the responsibility of the authors and does not necessarily represent the official view of the National Institutes of Health or the Department of Veterans Affairs.
C.L. is the recipient of a doctoral fellowship from CONICET. L.J.G. and A.J.V. are staff members from CONICET.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Bontron S, Nordmann P, Poirel L. 2016. Transposition of Tn125 encoding the NDM-1 carbapenemase in Acinetobacter baumannii. Antimicrob Agents Chemother 60:7245–7251. doi: 10.1128/AAC.01755-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toleman MA, Spencer J, Jones L, Walsh TR. 2012. blaNDM-1 is a chimera likely constructed in Acinetobacter baumannii. Antimicrob Agents Chemother 56:2773–2776. doi: 10.1128/AAC.06297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jamulitrat S, Arunpan P, Phainuphong P. 2009. Attributable mortality of imipenem-resistant nosocomial Acinetobacter baumannii bloodstream infection. J Med Assoc Thai 92:413–419. [PubMed] [Google Scholar]
- 4.Kumar SH, De AS, Baveja SM, Gore MA. 2012. Prevalence and risk factors of metallo beta-lactamase producing Pseudomonas aeruginosa and Acinetobacter species in burns and surgical wards in a tertiary care hospital. J Lab Physicians 4:39–42. doi: 10.4103/0974-2727.98670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fiorilli G, Faccone D, Lopardo H, Callejo R, Rapoport M, Prieto M, Galas M, Pasteran F. 2010. Emergence of metallo-beta-lactamases in Acinetobacter spp clinical isolates from Argentina. Rev Esp Quimioter 23:100–102. [PubMed] [Google Scholar]
- 6.Faccone D, Martino F, Pasteran F, Albornoz E, Biondi E, Vazquez M, Rapoport M, Rodrigo V, De Belder D, Gomez S, Corso A. 2019. Multiple clones of metallo-beta-lactamase-producing Acinetobacter ursingii in a children hospital from Argentina. Infect Genet Evol 67:145–149. doi: 10.1016/j.meegid.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 7.Pasteran F, Mora MM, Albornoz E, Faccone D, Franco R, Ortellado J, Melgarejo N, Gomez S, Riquelme I, Matheu J, Ramon-Pardo P, Corso A. 2014. Emergence of genetically unrelated NDM-1-producing Acinetobacter pittii strains in Paraguay. J Antimicrob Chemother 69:2575–2578. doi: 10.1093/jac/dku139. [DOI] [PubMed] [Google Scholar]
- 8.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. 2010. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. doi: 10.1371/journal.pone.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poplin R, Ruano-Rubio V, DePristo MA, Fennell TJ, Carneiro MO, Van der Auwera GA, Kling DE, Gauthier LD, Levy-Moonshine A, Roazen D, Shakir K, Thibault J, Chandran S, Whelan C, Lek M, Gabriel S, Daly MJ, Neale B, MacArthur DG, Banks E. 2018. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv doi: 10.1101/201178:201178. [DOI]
- 11.Adams MD, Bishop B, Wright MS. 2016. Quantitative assessment of insertion sequence impact on bacterial genome architecture. Microb Genom 2:e000062. doi: 10.1099/mgen.0.000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krizova L, Bonnin RA, Nordmann P, Nemec A, Poirel L. 2012. Characterization of a multidrug-resistant Acinetobacter baumannii strain carrying the blaNDM-1 and blaOXA-23 carbapenemase genes from the Czech Republic. J Antimicrob Chemother 67:1550–1552. doi: 10.1093/jac/dks064. [DOI] [PubMed] [Google Scholar]
- 13.Poirel L, Bonnin RA, Boulanger A, Schrenzel J, Kaase M, Nordmann P. 2012. Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob Agents Chemother 56:1087–1089. doi: 10.1128/AAC.05620-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon EJ, Kim JO, Yang JW, Kim HS, Lee KJ, Jeong SH, Lee H, Lee K. 2017. The blaOXA-23-associated transposons in the genome of Acinetobacter spp. represent an epidemiological situation of the species encountering carbapenems. J Antimicrob Chemother 72:2708–2714. doi: 10.1093/jac/dkx205. [DOI] [PubMed] [Google Scholar]
- 15.Brasiliense D, Cayo R, Streling AP, Nodari CS, Barata RR, Lemos PS, Massafra JM, Correa Y, Magalhaes I, Gales AC, Sodre R. 2019. Diversity of metallo-beta-lactamase-encoding genes found in distinct species of Acinetobacter isolated from the Brazilian Amazon region. Mem Inst Oswaldo Cruz 114:e190020. doi: 10.1590/0074-02760190020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pillonetto M, Arend L, Vespero EC, Pelisson M, Chagas TP, Carvalho-Assef AP, Asensi MD. 2014. First report of NDM-1-producing Acinetobacter baumannii sequence type 25 in Brazil. Antimicrob Agents Chemother 58:7592–7594. doi: 10.1128/AAC.03444-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez CH, Nastro M, Famiglietti A. 2018. Carbapenemases in Acinetobacter baumannii. Review of their dissemination in Latin America. Rev Argent Microbiol 50:327–333. doi: 10.1016/j.ram.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 18.Waterman PE, McGann P, Snesrud E, Clifford RJ, Kwak YI, Munoz-Urbizo IP, Tabora-Castellanos J, Milillo M, Preston L, Aviles R, Sutter DE, Lesho EP. 2013. Bacterial peritonitis due to Acinetobacter baumannii sequence type 25 with plasmid-borne New Delhi metallo-β-lactamase in Honduras. Antimicrob Agents Chemother 57:4584–4586. doi: 10.1128/AAC.00275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang L, Yu Y, Zeng W, Guo J, Lv F, Wang X, Liu X, Zhao Z. 2019. Whole-genome analysis of New Delhi metallo-β-lactamase-1-producing Acinetobacter haemolyticus from China. J Glob Antimicrob Resist 20:204–208. doi: 10.1016/j.jgar.2019.05.012. [DOI] [PubMed] [Google Scholar]
- 20.Krahn T, Wibberg D, Maus I, Winkler A, Bontron S, Sczyrba A, Nordmann P, Pühler A, Poirel L, Schlüter A. 2016. Intraspecies transfer of the chromosomal Acinetobacter baumannii blaNDM-1 carbapenemase gene. Antimicrob Agents Chemother 60:3032–3040. doi: 10.1128/AAC.00124-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Z, Qlu S, Wang Y, Wang Y, Liu S, Wang Z, Du X, Wang L, Guo J, Wang Z, Liu N, Yuan J, Song H, Huang L. 2011. Coexistence of blaNDM-1 with the prevalent blaOXA23 and blaIMP in pan-drug resistant Acinetobacter baumannii isolates in China. Clin Infect Dis 52:692–693. doi: 10.1093/cid/ciq231. [DOI] [PubMed] [Google Scholar]
- 22.El-Mahdy TS, Al-Agamy MH, Al-Qahtani AA, Shibl AM. 2017. Detection of blaOXA-23-like and blaNDM-1 in Acinetobacter baumannii from the eastern region, Saudi Arabia. Microb Drug Resist 23:115–121. doi: 10.1089/mdr.2015.0304. [DOI] [PubMed] [Google Scholar]
- 23.Karthikeyan K, Thirunarayan MA, Krishnan P. 2010. Coexistence of blaOXA-23 with blaNDM-1 and armA in clinical isolates of Acinetobacter baumannii from India. J Antimicrob Chemother 65:2253–2254. doi: 10.1093/jac/dkq273. [DOI] [PubMed] [Google Scholar]
- 24.Letunic I, Bork P. 2019. Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under BioProject accession no. PRJNA562922. Contig sequences for each genome are available under GenBank accession nos. VYSH00000000 to VYTU00000000.