Ceftobiprole is an advanced-generation broad-spectrum cephalosporin antibiotic with potent and rapid bactericidal activity against Gram-positive pathogens, including methicillin-resistant Staphylococcus aureus, as well as susceptible Gram-negative pathogens, including Pseudomonas sp. pathogens. In the case of Pseudomonas aeruginosa, ceftobiprole acts by inhibiting P. aeruginosa penicillin-binding protein 3 (PBP3). Structural studies were pursued to elucidate the molecular details of this PBP inhibition.
KEYWORDS: beta-lactam antibiotic, penicillin-binding protein, protein crystallography
ABSTRACT
Ceftobiprole is an advanced-generation broad-spectrum cephalosporin antibiotic with potent and rapid bactericidal activity against Gram-positive pathogens, including methicillin-resistant Staphylococcus aureus, as well as susceptible Gram-negative pathogens, including Pseudomonas sp. pathogens. In the case of Pseudomonas aeruginosa, ceftobiprole acts by inhibiting P. aeruginosa penicillin-binding protein 3 (PBP3). Structural studies were pursued to elucidate the molecular details of this PBP inhibition. The crystal structure of the His-tagged PBP3-ceftobiprole complex revealed a covalent bond between the ligand and the catalytic residue S294. Ceftobiprole binding leads to large active site changes near binding sites for the pyrrolidinone and pyrrolidine rings. The S528 to L536 region adopts a conformation previously not observed in PBP3, including partial unwinding of the α11 helix. These molecular insights can lead to a deeper understanding of β-lactam-PBP interactions that result in major changes in protein structure, as well as suggesting how to fine-tune current inhibitors and to develop novel inhibitors of this PBP.
INTRODUCTION
Pseudomonas aeruginosa infections pose a serious health threat and are difficult to treat due to antibiotic resistance. This resistance is often multifactorial and can involve antibiotic-inactivating enzymes or changes in expression and/or mutations in porins and efflux pumps; it can also lead to a carbapenem-resistant phenotype (1, 2).
The major class of antibiotics used throughout the the world for the treatment of infection is the β-lactams. They target penicillin-binding proteins (PBPs), a family of proteins responsible for generating cross-links in the peptidoglycan layer that strengthen the cell wall. For P. aeruginosa, P. aeruginosa PBP3 (PaPBP3) is the most essential PBP for cell growth (3). The enzyme belongs to the high-molecular-weight class B of PBPs, with the C-terminal transpeptidase cross-linking domain harboring the conserved catalytic S294 residue (4).
Ceftobiprole (Fig. 1A), which is marketed in several countries within and outside Europe, is very active against both Gram-positive and Gram-negative bacteria, including Staphylococcus aureus (including methicillin-resistant S. aureus), Enterococcus faecalis, and non-extended-spectrum β-lactamase-phenotype Enterobacteriaceae strains. Ceftobiprole activity against P. aeruginosa was reported (with 77% of P. aeruginosa species being susceptible at 4 mg/ml) (5–7). Davies et al. showed that the cephalosporin bound to PaPBP3 with a 50% inhibitory concentration of 0.1 μg/ml, which was confirmed by the production of filaments observed by microscopy in the presence of the substance (8).
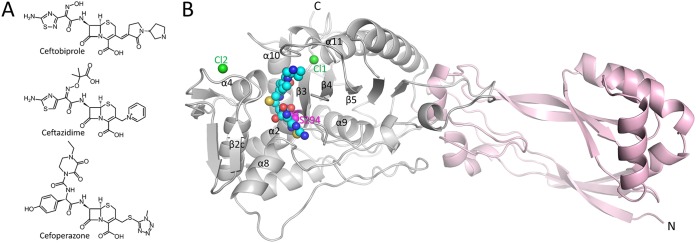
FIG 1.
Structures of ceftobiprole and PBP3. (A) Chemical structures of ceftobiprole, ceftazidime, and cefoperazone. (B) Overall structure of PaPBP3 in complex with ceftobiprole. The C-terminal catalytic transpeptidase domain (gray) and the N-terminal domain (pink) are drawn as a ribbon diagram. Atoms in ceftobiprole and the chloride ions are depicted as spheres (cyan carbons, blue nitrogens, red oxygens, yellow sulfur, and green chlorides). The catalytic S294 is shown as pink spheres and is labeled. Key secondary structural elements are labeled as in reference 4.
PaPBP3 complexes with β-lactams have been described previously (4, 9–12). The inhibitors form a covalent complex with S294, thereby blocking the active site. To explore the mechanism of inhibition of ceftobiprole against PaPBP3 and to examine the interaction with the unique C3 side chain, we determined the atomic structure of ceftobiprole complexed to PaPBP3, using protein crystallography. This complexed structure allowed further comparisons to other inhibitor-PaPBP3 structures, as well as to other previously determined ceftobiprole-PBP complexes. In addition to the structural analyses, we present complementary thermal shift assay and mass spectrometry data characterizing ceftobiprole binding to PaPBP3.
RESULTS AND DISCUSSION
Crystal structure.
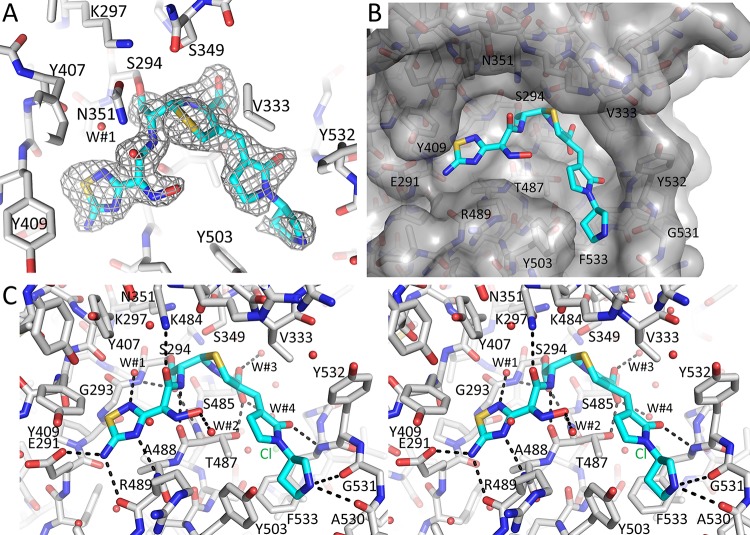
The crystal structure of the His-tagged ceftobiprole-PaPBP3 complex reveals ceftobiprole binding to the active site in the C-terminal catalytic domain (Fig. 1B). Most of the ceftobiprole ligand is well ordered in the unbiased omit |Fo|-|Fc| electron density map except for the terminal pyrrolidine ring, for which the corresponding density is somewhat weaker (Fig. 2A). The adjacent pyrrolidinone and connected 6-membered dihydrothiazine rings of ceftobiprole are also well ordered in the electron density map. These two ceftobiprole moieties are partially covered by a small bridge composed of two juxtaposed residues, V333 and Y532 (Fig. 2B). Therefore, ceftobiprole fits the active site groove well, making numerous hydrogen-bonding and van der Waals interactions, as well as the covalent bond to the catalytic S294. This covalent bond is formed with the β-lactam carbonyl carbon atom, resulting in the 4-membered β-lactam ring being opened during this acylation step. The carboxyl moiety attached to the 6-membered dihydrothiazine ring of ceftobiprole donates a hydrogen bond with the side chain of T487 and two water-mediated interactions (W#3 and W#4 in Fig. 2C). The hydrophobic face of the dihydrothiazine ring makes a hydrophobic interaction with V333. The thiadiazole moiety of ceftobiprole also forms hydrogen bonds with the side chain of E291 and with main chain atoms of R489 (Fig. 2C). This thiadiazole ring further creates a water-mediated interaction with a deeply buried water (W#1 in Fig. 2C). This water also forms a hydrogen bond with the backbone oxygen of Y407 and is situated near the carbonyl bond attached to S294 (Fig. 2C). The hydrophobic faces of the thiadiazole ring are wedged by PBP3 residues A488, Y409, and G293. The amide moiety of ceftobiprole interacts with both walls of the active site; the amide oxygen hydrogen bonds with the nitrogen of the side chain of N351, whereas the amide nitrogen hydrogen bonds with the backbone oxygen of T487 (these atoms in N351 and T487 are 7.3 Å from each other across the active site cleft, with the ceftobiprole amide moiety between them). The hydroxyimino moiety makes only water-mediated interactions (W#2 in Fig. 2C). On the other end of ceftobiprole, the oxygen of the pyrrolidinone moiety interacts with the backbone nitrogen of F533. Although somewhat less well ordered, the terminal pyrrolidine ring interacts, via its nitrogen, with the backbone oxygens of residues A530 and G531 (Fig. 2C) (note that this secondary amine is likely positively charged).
FIG 2.
Ceftobiprole binding to PBP3. (A) Unbiased omit |Fo|-|Fc| difference density contoured at 3σ, showing the presence of the covalently bound ceftobiprole in the active site. Ceftobiprole is shown in stick representation, with cyan carbon atoms. (B) Surface representation of the PBP3 active site, with ceftobiprole bound. (C) Side-by-side stereo figure showing the interactions between ceftobiprole and the active site residues. Hydrogen bonds involving ceftobiprole atoms are depicted as dashed lines. Ceftobiprole and bound water molecules are shown in stick and spherical representation, respectively. The chloride ion (Cl-1) and water molecules (W#1 to W#4) are shown.
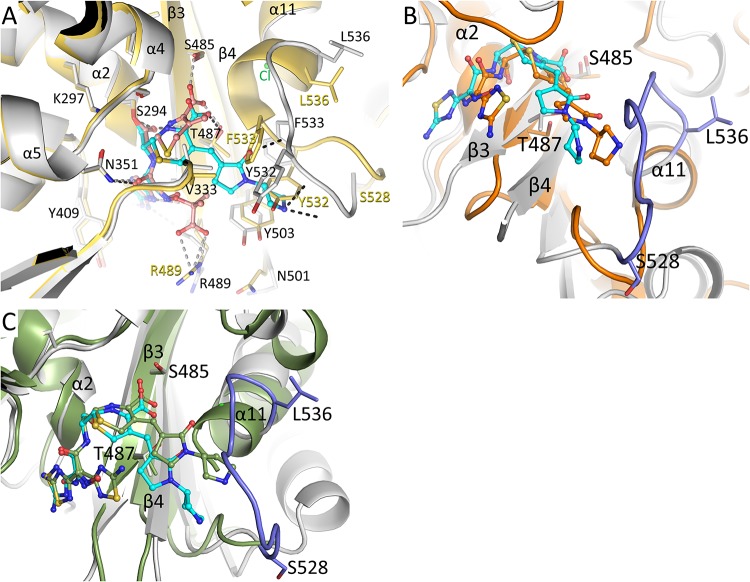
Before comparing PBP3 conformational changes upon ceftobiprole binding to, for example, ceftazidime binding (Fig. 3A), it is helpful to first discuss previously observed conformational changes in unbound and liganded complexes of PaPBP3 with cephalosporins, carbapenems, and monobactams. Previous studies identified a number of regions of significant conformational changes in the active site upon ligand binding (4, 9–12). The largest conformational changes were observed in residues S528 to L536, which represent the loop before the beginning of the α11 helix (Fig. 3A) (4, 9–12). In addition, the end of the β3 strand (T487-A488-R489-K490) and the beginning of the β4 strand (N501 to R504) yielded conformational variability between these different structures (the imipenem and meropenem complexes have a different conformation for these regions, compared to the other complexes) (Fig. 3A). These β3 strand residues are near the ceftobiprole thiadiazole binding site; imipenem and meropenem both lack the presence of such a functional group. Also nearby, residues Y409 to L411 can have various conformations (Fig. 3A) (4, 9–12). Finally, residues V333 to S337, forming one wall of the active site cleft, were also observed to shift upon ligand binding, albeit more modestly than the aforementioned regions of conformational changes (Fig. 3A) (4, 9–12).
FIG 3.
Structural comparison of the ceftobiprole-PBP3 structure. (A) Superposition of the ceftobiprole- and ceftazidime-bound PBP3 structures. The root mean square deviation (RMSD) of the 466 aligned C-α atoms is 1.00 Å (the ceftazidime-PaPBP3 structure has PDB accession number 3PBO [4]). The PBP3 protein residues from the ceftobiprole and ceftazidime complexes are colored gray and yellow, respectively. The cephalosporin ligands are both shown in ball-and-stick representation, ceftobiprole with cyan carbon atoms and ceftazidime with orange carbon atoms. Hydrogen bonds of ceftobiprole and ceftazidime with the protein are shown as black and gray dashed lines, respectively. Residues that differ significantly between the two structures have the two different conformations labeled in black or yellow; other residues have a single black label. Key secondary structural elements are also labeled. (B) Superposition of the ceftobiprole-bound PaPBP3 and ceftobiprole-bound Staphylococcus aureus PBP4 (PDB accession number 5TXI [14]) structures. The PBP4 protein is colored orange, as are the carbon atoms of its bound ceftobiprole; the PBP3-ceftobiprole atoms are colored as in panel A except for residues 528 to 536, which are colored light blue. The following residues were used for superpositioning of the active sites: PBP3 residues 290 to 307, 345 to 360, 406 to 409, 484 to 488, and 505 to 511 were superimposed on PBP4 residues 71 to 88, 135 to 150, 179 to 182, 259 to 263, and 268 to 274, respectively; the RMSD for 50 C-α atoms was 1.34 Å. (C) Superposition of the ceftobiprole-bound PaPBP3 and ceftobiprole-bound Staphylococcus aureus PBP2a (PDB accession number 4DKI [15]) structures. The PBP2a protein is colored green, as are the carbon atoms of its bound ceftobiprole; the PBP3-ceftobiprole atoms are colored as in panel B. The following residues were used for superpositioning of the active sites: PBP3 residues 290 to 307, 340 to 360, 406 to 409, 482 to 488, and 503 to 510 were superimposed on PBP2a residues 399 to 416, 453 to 473, 518 to 521, 595 to 601, and 613 to 620, respectively; the RMSD for 58 C-α atoms was 1.06 Å.
With these previously observed conformational changes in mind, we compared the ceftobiprole-PaPBP3 complex against the previously published complexes. Ceftobiprole binding to PBP yields the largest conformational change in the S528 to L536 region, with the unwinding of the first turn of the α11 helix (Fig. 3A). The backbone nitrogen atom of α11 A539 forms a hydrogen bond either with the backbone oxygen of L536 in the unbound PBP3 structure (in a short 310 helix region of α11) or with the backbone oxygen of G534 in the ceftazidime-bound structure (4). In the ceftobiprole complex, this region is unwound, allowing a chloride ion (Cl-1) to interact with the N terminus of the shortened α11 helix (Fig. 2C and 3A). The shifts in this region (in particular, movements of Y532 and F533) open up a new active site groove to accommodate the connected pyrrolidinone and pyrrolidine rings (Fig. 3A). Both Y532 and F533 side chains would otherwise sterically clash with these two ceftobiprole rings. The normally solvent-exposed F533 reorients itself inward to form the bottom of this new active site groove section, whereas Y532 and its backbone atoms form the outside wall of this groove in which the pyrrolidinone and pyrrolidine rings reside. The observation of a chloride ion at the N terminus of the α11 helix could be due to favorable helix dipole interactions, as observed previously in an unrelated protein (13). The aforementioned conformational changes are in the PBP3 region observed to be more flexible in the apoprotein structure (PDB accession number 3PBN) (4); the S528 to L536 region in the apoprotein structure contains higher temperature factors ranging from ∼45 to 55 Å2, compared to the core active site regions (the S294-containing α2 helix, which has temperature factors of ∼25 Å2, and the β3 strand, which has temperature factors ranging from ∼25 to 55 Å2). Even upon ceftobiprole binding, the temperature factors for the S528 to L536 main chain atoms do not decrease, as they range from ∼55 to 67 Å2, except for G534, which peaks at 87 Å2. The core active site regions, the α2 helix and β3 strand, have temperature factors in the range of ∼20 to 25 Å2 for the ceftobiprole complex.
Comparing the different cephalosporin-PaPBP3 interactions involving the ceftazidime and cefoperazone ligands reveals several conserved features in both of these ligands, including the amide moiety, the dihydrothiazine ring, the carbonyl moiety attached to S294, and the carboxyl moiety (4, 9). The latter functional group makes two hydrogen bonds in the PBP3 active site, with the side chains of S485 and T487 (in both the ceftazidime and cefoperazone complexes). In contrast, we find that the carboxyl moiety in the complex with the cephalosporin ceftobiprole does not go as deeply into the active site and makes only the hydrogen bond with T487 (Fig. 2C and 3A). A possible explanation is that the pyrrolidinone and pyrrolidine rings of ceftobiprole, which now bind in the newly opened active site groove, sterically prevent the connected 6-membered dihydrothiazine ring and its carboxyl moiety from becoming more deeply embedded in the active site. An ∼1-Å shift of this carboxyl moiety is needed to make the hydrogen bond with S485. Ceftazidime and cefoperazone do not have such a large substituent. Compared to cefoperazone, ceftazidime is more similar to ceftobiprole, with its amino-thiazole ring being very similar to the amino-thiadiazole moiety of ceftazidime. When bound to PBP3, these moieties have nearly identical binding modes and interactions (Fig. 3A). One difference is that the extra nitrogen in the thiadiazole ring of ceftobiprole allows for the interaction with the deeply buried water (W#1 in Fig. 2C); this nitrogen is absent in ceftazidime. Furthermore, the additional carboxyl moiety of ceftazidime forms a salt bridge interaction with R489, and ceftobiprole lacks this moiety.
Previously, two other structures of ceftobiprole bound to different PBPs, i.e., Staphylococcus aureus PBP2a and S. aureus PBP4, were determined (14, 15). Comparison of the ceftobiprole-PBP3 complex with the PBP2a and PBP4 complexes could yield insights into conserved conformational similarities and differences of ceftobiprole interacting in these different but related PBP active sites. First, when the conformation and position of the pyrrolidinone and pyrrolidine rings of ceftobiprole in the three different PBP complex structures are compared, this section of ceftobiprole adopts similar conformations and points in similar directions in the three structures (Fig. 3B and C). However, PBP2a and PBP4 do not have an active site region, corresponding to PBP3 S528 to L536, that could sterically interfere with the pyrrolidinone and pyrrolidine rings of ceftobiprole (Fig. 3B and C). Therefore, unlike in PaPBP3, the carboxyl moiety of ceftobiprole in the PBP2a and PBP4 structures forms hydrogen bonds with both respective S/T residues in the carboxyl binding pocket (PBP3 S485 corresponds to S598 in PBP2a and T260 in PBP4; PBP3 T487 corresponds to T600 in PBP2a and S262 in PBP4).
Second, regarding the thiadiazole hydroxyimino portion of ceftobiprole, this moiety is flipped 180° in the PBP4 complex, compared to its binding mode when bound to PBP3, such that the hydroxyimino moiety points toward the protein (Fig. 3B). Remarkably, both conformations of this thiadiazole hydroxyimino section are observed in the PBP2a complex (15) (Fig. 3C). These observations suggest that this ceftobiprole moiety has conformational plasticity and can alter its position to accommodate binding to different PBPs.
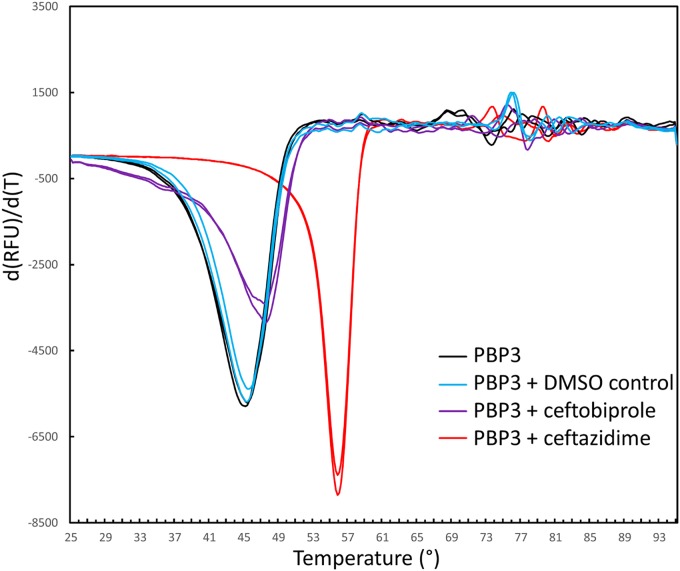
Thermal shift binding assay.
To probe the stabilizing effects of ceftobiprole binding to PBP3, we carried out a differential scanning fluorimetry (DSF) thermal shift assay in the presence and absence of the ligand (Fig. 4). The experiments were performed in duplicate. In the presence of 200 μM ceftobiprole, the two melting temperature measurements for PBP3 were 47.4°C and 47.4°C. For the control sample with the protein without the ligand but with the 4% dimethyl sulfoxide (DMSO) ligand solvent, the melting temperature measurements were observed to be 45.4°C and 45.4°C; the two melting temperature measurements for the PBP3 protein without either ligand or DMSO were 45.2°C and 45.4°C. These data indicate that ceftobiprole binding to PBP3 leads to a 2.0°C increase in the thermal stability of the protein.
FIG 4.
DSF thermal shift assay of ceftobiprole and ceftazidime binding to PaPBP3. The derivative of the change in fluorescence is plotted versus temperature. Experiments were performed in duplicate.
To complement our structural comparison of ceftobiprole and the structurally similar cephalosporin ceftazidime (Fig. 3A), we also carried out the thermal shift assay with PBP3 in the presence of 200 μM ceftazidime (also with 4% DMSO); the observed melting temperature was measured in duplicate as 55.8°C and 55.8°C, which is a 10.4°C increase in thermal stability (Fig. 4). The latter significant increase is more consistent with that observed for two other ligands binding to PaPBP3, namely, cefoperazone and azlocillin, which yielded increases in thermal stability of 15°C and 12°C, respectively (9). To explain the lower increase in thermal stability upon ceftobiprole binding, compared to ceftazidime binding, we reasoned that it is likely due to the presence of the pyrrolidinone and pyrrolidine rings, which are not present in the similar ceftazidime. These moieties appear to have both direct and indirect effects on the interactions that ceftobiprole makes in the PBP3 active site. The direct effect is that, to accommodate these large moieties, the S528 to L536 region undergoes a significant conformational change, with partial unwinding of the N-terminal part of the α11 helix (Fig. 2C and 3A); these conformational changes open a new active site groove region for the pyrrolidinone and pyrrolidine rings. The indirect effect is that, despite these accommodating active site changes, the bulky pyrrolidinone and pyrrolidine rings prevent the carboxyl moiety from being more deeply embedded in the active site to make hydrogen-bonding interactions with both T487 and S485 in the conserved carboxyl binding pocket (Fig. 3A). The sum of these described conformational changes and inability to form key carboxylate interactions is the likely cause of the relatively modest increase in PBP3 thermal stability upon the binding of ceftobiprole, compared to the other ligands. Note that the thermal stability differences cannot be explained by the change in accessible protein surface upon binding of either ligand; the buried protein surfaces upon binding of ceftobiprole and ceftazidime are not very different from each other, being 383 Å and 353 Å, respectively, as calculated using CCP4 Areaimol (16). A clear correlation between the change in protein stability and the affinity of a ligand due to combined enthalpic and entropic effects when affecting the thermal stability of a protein is not evident (17); microbiologically and biochemically, ceftobiprole was quite effective against P. aeruginosa PBP3, as noted above. Note that ceftobiprole binding to S. aureus PBP2a also resulted in an only ∼2°C increase in thermal stability (15).
Mass spectrometry of ceftobiprole-PBP3 and ceftazidime-PBP3 complexes.
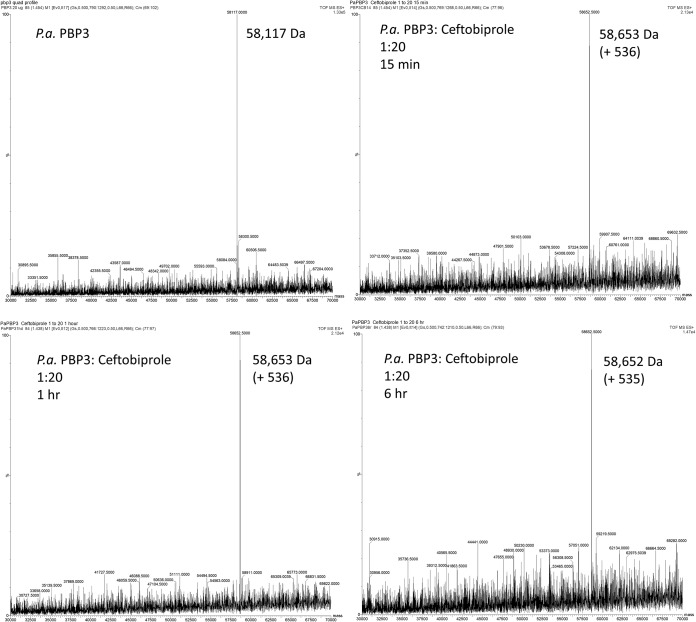
Some β-lactam cephalosporins can have the R2 group in the C3 position eliminated when bound to a β-lactamase (18) or a PBP (4). To investigate whether the ceftobiprole remains intact when bound as an acyl complex, as suggested by the crystal structure, we carried out timed mass spectrometry analyses of the PBP3 complex with ceftobiprole. PBP3 by itself has a mass of 58,117 Da (Fig. 5 and Table 1); the calculated mass of the PBP3 construct, including the His tag, is 58,253 Da (the 136-Da difference is likely due to loss of the N-terminal methionine). Adding ceftobiprole to PBP3 increased the mass to 58,652 Da (+535 Da) or 58,653 Da (+536 Da) at the 15-min, 1-h, and 6-h time points (Table 1). The increased molecular mass of PBP3 is due the covalent addition of an intact ceftobiprole molecule, which has a molecular mass of 535 Da; this indicates that no R groups of ceftobiprole have been eliminated upon the covalent inhibition of PBP3. For comparison, we also carried out a timed mass spectrometry experiment with ceftazidime binding to PBP3 (Table 1; also see Fig. S1 in the supplemental material). Adding ceftazidime to PBP3 increased the mass to 58,587 Da (+470 Da), 58,589 Da (+472 Da), or 58,585 Da (+468 Da) at the 15-min, 1-h, and 6-h time points, respectively (Table 1). The average of these mass additions is 470 Da, which is smaller than the mass of ceftazidime, i.e., 547 Da. This finding suggests that ceftazidime has fragmented, because a 77-Da fragment is missing. This fragment is likely the pyridine R2 group, because pyridine has a calculated molecular mass of 79 Da. This finding is in agreement with the crystal structure of ceftazidime bound to PaPBP3, in which the acylated ceftazidime also lacks its pyridine moiety (4). Ceftobiprole does not show this R2 group elimination, because ceftobiprole has a noncleavable electron-withdrawing substituent at this position, the pyrrolidinone and adjacent pyrrolidine moiety (19). In agreement, ceftobiprole bound to Staphylococcus aureus PBP2a and S. aureus PBP4 also does not show cleavage of the R2 group at the C3 position (14, 15).
FIG 5.
Mass spectra for PaPBP3 alone and after 15 min, 1 h, and 6 h of preincubation with ceftobiprole. The PaPBP3/ceftobiprole molar ratio was 1:20. Mass accuracy is ±5 Da.
TABLE 1.
Summary of mass spectrometry analysis of ceftobiprole and ceftazidime binding to PaPBP3a
| Protein | Ligand | Incubation time | Molecular mass (Da) |
|---|---|---|---|
| PaPBP3 | 58,117 | ||
| PaPBP3 | Ceftobiprole | 15 min | 58,653 (+536) |
| PaPBP3 | Ceftobiprole | 1 h | 58,653 (+536) |
| PaPBP3 | Ceftobiprole | 6 h | 58,652 (+535) |
| PaPBP3 | Ceftazidime | 15 min | 58,587 (+470) |
| PaPBP3 | Ceftazidime | 1 h | 58,589 (+472) |
| PaPBP3 | Ceftazidime | 6 h | 58,585 (+468) |
The protein/ligand concentration ratio was 1:20 for each ligand-binding experiment.
In summary, the 1.76-Å-resolution crystal structure of PaPBP3 in complex with the advanced-generation broad-spectrum cephalosporin ceftobiprole reveals a covalent complex of the ligand with the catalytic S294 and a key conformational role for the S528 to L536 region. Our studies have revealed a new conformational state and mode of inhibition in P. aeruginosa PBP3, the key PBP for growth of this pathogen. These molecular insights can lead to fine-tuning of current inhibitors or to development of novel inhibitors of this PBP.
MATERIALS AND METHODS
N-terminally His6-tagged PaPBP3 was expressed using a pET28a plasmid and was purified as described previously (4). The plasmid was transformed into Rosetta (DE3)pLysS competent cells (Novagen) for expression. After cell lysis and Ni-nitrilotriacetic acid purification (4), the protein was purified using a Superdex 200 Increase size exclusion column (GE Life Sciences). Crystals of PaPBP3 were grown using conditions published previously (4), i.e., 30% polyethylene glycol 4000, 0.2 M MgCl2, 0.1 M Tris (pH 8.5). The unbound PBP3 crystals were soaked for 45 h with 0.5 mM ceftobiprole before the crystal was frozen in liquid nitrogen. Data were collected at SSRL beamline 14-1 and processed to 1.76-Å resolution using AutoXDS (20) (Table 2). The structure was solved by molecular replacement using the PBP3 protein coordinates from its imipenem complex structure (PDB accession number 3PBQ) (4) and was refined using REFMAC (21). The program Coot (22) was used for model building. Final refined R and Rfree were 0.204 and 0.252 with the model composed of residues 50 to 491, residues 501 to 560, 1 covalently bound ceftobiprole ligand, 2 chloride ions, and 242 water molecules (Table 2). Figures were generated using PyMOL (https://pymol.org/2).
TABLE 2.
Data collection and refinement statistics for the PaPBP3-ceftobiprole complex
| Parameter | Valuea |
|---|---|
| Wavelength (Å) | 1.194990 |
| Data range (Å) | 1.76–39.61 (1.76–1.79) |
| Space group | P212121 |
| Cell dimensions | |
| Distances (Å) | 68.19, 83.74, 89.92 |
| Angles (°) | 90, 90, 90 |
| Completeness (%) | 99.8 (96.6) |
| No. of unique reflections | 51,816 (2,811) |
| Total no. of observations | 692,945 |
| Average multiplicity | 13.4 (11.8) |
| Mean I/σ(I) | 25.2 (2.3) |
| Rmerge | 0.047 (0.984) |
| Rwork | 0.204 |
| Rfree | 0.252 |
| Resolution (Å) | 1.76–39.61 |
| Ligands | Ceftobiprole, 2 chloride ions |
| No. of water molecules | 242 |
| RMSD | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.278 |
Values in parentheses are for the highest-resolution shell.
For the DSF thermal shift assay, we used SYPRO orange dye (Fisher Scientific) at 10× concentration. DSF was performed on a CFX96 Touch thermocycler (Bio-Rad). Reactions were run in duplicate, in a sample volume of 30 μl, with 5 μM PBP3 either with or without 200 μM inhibitor (ceftobiprole or ceftazidime, with the latter similar β-lactam being used as a positive control), in 14 mM Tris (pH 8.0), 280 mM NaCl, 7% glycerol. Since both ligands were dissolved in DMSO, we also included a control with protein plus 4% DMSO (the same concentration as in the ligand experiments). Temperatures were ramped from 25°C to 95°C during the DSF experiment.
For mass spectrometry measurements, 20 μg of PaPBP3 was incubated with ceftobiprole or ceftazidime at a molar ratio of 1:20 in 10 mM phosphate-buffered saline (pH 7.4), in a total reaction volume of 20 μl, for 15 min, 1 h, and 6 h. Reactions were quenched with 10 μl acetonitrile and added to 1 ml 0.1% formic acid in water. Samples were analyzed using a Waters SYNAPT G2-Si mass spectrometer and a Waters Acquity ultrapressure liquid chromatography BEH C18 column (1.7-μm pore size; 2.1 by 50 mm). MassLynx v4.1 was used to deconvolute protein peaks. The tune settings for each data run were as follows: capillary voltage, 3.25 kV; sampling cone, 35 V; source offset, 35 V; source temperature, 100°C; desolvation temperature, 500°C; cone gas flow, 100 liters/h; desolvation gas flow, 800 liters/h; nebulizer pressure, 6.0 × 105 Pa. Quad profile mass settings were 900, 1,200, and 1,700. Mobile phase A was 0.1% formic acid in water, and mobile phase B was 0.1% formic acid in acetonitrile. The mass accuracy for this system is 5 Da.
Data availability.
Coordinates and structure factors have been deposited in the PDB with accession number 6VJE.
Supplementary Material
ACKNOWLEDGMENTS
We thank Seungil Han for the PBP3 expression plasmid and Basilea Pharmaceutica International Ltd. for providing ceftobiprole and funds to support this project. We also thank Stanford Synchrotron Radiation Lightsource (SSRL) for help with data collection.
Use of the SSRL, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the U.S. Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including grant P41GM103393). This research was also supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (NIH) to R.A.B. under award numbers R01AI100560, R01AI063517, and R01AI072219 and by funds and/or facilities provided by the Louis Stokes Cleveland Veterans Affairs Medical Center to R.A.B. and Veterans Affairs Merit Review Program Awards 1I01BX001974 to R.A.B. and 1I01BX002872 to K.M.P.-W. from the Biomedical Laboratory Research and Development Service of the Veterans Health Administration Office of Research and Development and the Geriatric Research Education and Clinical Center, VISN 10. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the U.S. Department of Veterans Affairs, or the U.S. government.
V.K. carried out the crystallization and higher-resolution structure determination of the complex. C.T. carried out the initial protein expression, purification, crystallization, and initial (lower-resolution) structure determination of the complex. C.T. also carried out the DSF experiments. C.R.B. carried out the mass spectrometry experiments, guided by K.M.P.-W. J.W. carried out initial DSF experiments. E.D. initiated the project idea and edited the manuscript. R.A.B. directed the mass spectrometry aspects and edited the manuscript. F.V.D.A. directed the overall project, finished the crystallographic refinement, prepared the figures, wrote the first draft of the manuscript, and edited the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Tacconelli E, Magrini N, Carmeli Y, Harbath S, Kahlmeter G, Kluytmans J, Mendelson M, Pulcini C, Singh N, Theuretzbacher U. 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. World Health Organization, Geneva, Switzerland: https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf. [Google Scholar]
- 2.Pang Z, Raudonis R, Glick BR, Lin TJ, Cheng Z. 2019. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol Adv 37:177–192. doi: 10.1016/j.biotechadv.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zhang YM, Davies C. 2017. Penicillin-binding protein 3 is essential for growth of Pseudomonas aeruginosa. Antimicrob Agents Chemother 61:e01651-16. doi: 10.1128/AAC.01651-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han S, Zaniewski RP, Marr ES, Lacey BM, Tomaras AP, Evdokimov A, Miller JR, Shanmugasundaram V. 2010. Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 107:22002–22007. doi: 10.1073/pnas.1013092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfaller MA, Flamm RK, Mendes RE, Streit JM, Smart JI, Hamed KA, Duncan LR, Sader HS. 2018. Ceftobiprole activity against Gram-positive and -negative pathogens collected from the United States in 2006 and 2016. Antimicrob Agents Chemother 63:e01566-18. doi: 10.1128/AAC.01566-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfaller MA, Flamm RK, Duncan LR, Shortridge D, Smart JI, Hamed KA, Mendes RE, Sader HS. 2019. Ceftobiprole activity when tested against contemporary bacteria causing bloodstream infections in the United States (2016–2017). Diagn Microbiol Infect Dis 94:304–313. doi: 10.1016/j.diagmicrobio.2019.01.015. [DOI] [PubMed] [Google Scholar]
- 7.Hebeisen P, Heinze-Krauss I, Angehrn P, Hohl P, Page MG, Then RL. 2001. In vitro and in vivo properties of Ro 63-9141, a novel broad-spectrum cephalosporin with activity against methicillin-resistant staphylococci. Antimicrob Agents Chemother 45:825–836. doi: 10.1128/AAC.45.3.825-836.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies TA, Page MG, Shang W, Andrew T, Kania M, Bush K. 2007. Binding of ceftobiprole and comparators to the penicillin-binding proteins of Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus, and Streptococcus pneumoniae. Antimicrob Agents Chemother 51:2621–2624. doi: 10.1128/AAC.00029-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ren J, Nettleship JE, Males A, Stuart DI, Owens RJ. 2016. Crystal structures of penicillin-binding protein 3 in complexes with azlocillin and cefoperazone in both acylated and deacylated forms. FEBS Lett 590:288–297. doi: 10.1002/1873-3468.12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Starr J, Brown MF, Aschenbrenner L, Caspers N, Che Y, Gerstenberger BS, Huband M, Knafels JD, Lemmon MM, Li C, McCurdy SP, McElroy E, Rauckhorst MR, Tomaras AP, Young JA, Zaniewski RP, Shanmugasundaram V, Han S. 2014. Siderophore receptor-mediated uptake of lactivicin analogues in Gram-negative bacteria. J Med Chem 57:3845–3855. doi: 10.1021/jm500219c. [DOI] [PubMed] [Google Scholar]
- 11.Mitton-Fry MJ, Arcari JT, Brown MF, Casavant JM, Finegan SM, Flanagan ME, Gao H, George DM, Gerstenberger BS, Han S, Hardink JR, Harris TM, Hoang T, Huband MD, Irvine R, Lall MS, Lemmon MM, Li C, Lin J, McCurdy SP, Mueller JP, Mullins L, Niosi M, Noe MC, Pattavina D, Penzien J, Plummer MS, Risley H, Schuff BP, Shanmugasundaram V, Starr JT, Sun J, Winton J, Young JA. 2012. Novel monobactams utilizing a siderophore uptake mechanism for the treatment of Gram-negative infections. Bioorg Med Chem Lett 22:5989–5994. doi: 10.1016/j.bmcl.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 12.Murphy-Benenato KE, Dangel B, Davis HE, Durand-Reville TF, Ferguson AD, Gao N, Jahic H, Mueller JP, Manyak EL, Quiroga O, Rooney M, Sha L, Sylvester M, Wu F, Zambrowski M, Zhao SX. 2015. SAR and structural analysis of siderophore-conjugated monocarbam inhibitors of Pseudomonas aeruginosa PBP3. ACS Med Chem Lett 6:537–542. doi: 10.1021/acsmedchemlett.5b00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fiedler TJ, Davey CA, Fenna RE. 2000. X-ray crystal structure and characterization of halide-binding sites of human myeloperoxidase at 1.8 Å resolution. J Biol Chem 275:11964–11971. doi: 10.1074/jbc.275.16.11964. [DOI] [PubMed] [Google Scholar]
- 14.Alexander JAN, Chatterjee SS, Hamilton SM, Eltis LD, Chambers HF, Strynadka N. 2018. Structural and kinetic analyses of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance in Staphylococcus aureus. J Biol Chem 293:19854–19865. doi: 10.1074/jbc.RA118.004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lovering AL, Gretes MC, Safadi SS, Danel F, de Castro L, Page MG, Strynadka NC. 2012. Structural insights into the anti-methicillin-resistant Staphylococcus aureus (MRSA) activity of ceftobiprole. J Biol Chem 287:32096–32102. doi: 10.1074/jbc.M112.355644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee B, Richards FM. 1971. The interpretation of protein structures: estimation of static accessibility. J Mol Biol 55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 17.Bergsdorf C, Wright SK. 2018. A guide to run affinity screens using differential scanning fluorimetry and surface plasmon resonance assays. Methods Enzymol 610:135–165. doi: 10.1016/bs.mie.2018.09.015. [DOI] [PubMed] [Google Scholar]
- 18.Faraci WS, Pratt RF. 1985. Mechanism of inhibition of the PC1 β-lactamase of Staphylococcus aureus by cephalosporins: importance of the 3′-leaving group. Biochemistry 24:903–910. doi: 10.1021/bi00325a014. [DOI] [PubMed] [Google Scholar]
- 19.Hubschwerlen C. 2007. β-Lactam antibiotics, p 479–518. In Taylor JB, Triggle DJ (ed), Comprehensive medicinal chemistry II, vol 7 Elsevier, London, England. [Google Scholar]
- 20.Gonzalez A, Tsay Y. 2010. A quick XDS tutorial for SSRL. http://smb.slac.stanford.edu/facilities/software/xds/#autoxds_script.
- 21.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors have been deposited in the PDB with accession number 6VJE.